Abstract
Purpose
Lung cancer (LC) is the leading cause of cancer death in the US. The molecular events preceding the onset of disease are poorly understood and no effective tools exist to identify smokers with premalignant lesions (PMLs) that will progress to invasive cancer. Prior work identified molecular alterations in the smoke-exposed airway field of injury associated with LC. Here we focus on an earlier stage in the disease process leveraging the airway field of injury to study PMLs and its utility in LC chemoprevention.
Experimental Design
Bronchial epithelial cells from normal appearing bronchial mucosa were profiled by mRNA-Seq from subjects with (n=50) and without (n=25) PMLs. Using surrogate variable and gene set enrichment analysis we identified genes, pathways, and LC-related gene sets differentially expressed between subjects with and without PMLs. A computational pipeline was developed to build and test a chemoprevention-relevant biomarker.
Results
We identified 280 genes in the airway field associated with the presence of PMLs. Among the up-regulated genes, oxidative phosphorylation was strongly enriched and immunohistochemistry and bioenergetics studies confirmed pathway findings in PMLs. The relationship to PMLs and squamous cell carcinomas (SCC) was also confirmed using published LC datasets. The biomarker performed well predicting the presence of PMLs (AUC=0.92, n=17), and changes in the biomarker score associated with progression/stability vs. regression of PMLs (AUC=0.75, n=51).
Conclusions
Transcriptomic alterations in the airway field of smokers with PMLs reflect metabolic and early lung SCC alterations and may be leveraged to stratify smokers at high-risk for PML progression and monitor outcome in chemoprevention trials.
Keywords: Bronchial premalignant lesions, Lung cancer, Field of injury, RNA sequencing
Introduction
Exposure to carcinogens such as cigarette smoke induces smoking-related mRNA and microRNA expression alterations in the cytologically normal epithelium that lines the respiratory tract, creating an airway field of injury (1–6). Gene expression alterations in the airway field of injury were leveraged to build a diagnostic test for early lung cancer lung cancer detection (7–10). Examination of gene signatures for p63 (TP63) and the phosphatidylinositol 3-kinase (PIK3C) pathway, revealed increased PIK3C activation in the airway field of smokers with lung cancer or bronchial premalignant lesions (PMLs) (11). These results suggest the airway field of injury reflects processes associated with a precancerous disease state, however, the molecular changes have not been adequately characterized.
This is an important shortcoming because bronchial PMLs are precursors of squamous cell lung carcinoma (SCC), yet we lack effective tools to identify smokers with PMLs at highest risk of progression to invasive cancer. Several studies report loss of heterozygosity, chromosomal aneusomy, and aberrant methylation and protein expression in bronchial PMLs (12–21). These molecular events are associated with histological changes that can be reproducibly graded by a pathologist prior to the development of invasive carcinoma. Autofluorescence bronchoscopy can be used to detect and sample PMLs. In high-risk smokers between 40 and 74 years of age who had smoked at least 20 pack-years, the prevalence of PMLs is approximately 13% for moderate dysplasia and 1.6% for carcinoma in situ (CIS) (22,23). The presence of high grade PMLs (severe dysplasia or CIS) is a marker of increased lung cancer risk in both the central and peripheral airways indicating the presence of changes throughout the airway field (24–26).
Molecular characterization of the airway field of injury in smokers with PMLs may provide novel insights into the earliest stages of lung carcinogenesis and identify relatively accessible biomarkers to guide early lung cancer detection and early intervention.
Methods
Subject population used to derive gene signature and biomarker
Bronchial airway brushings were obtained during autofluorescence bronchoscopy procedures between June 2000 and March 2011 from subjects in the British Columbia Lung Health Study (BC-LHS) at the British Columbia Cancer Agency (BCCA) (Vancouver, BC) (27) and between December 2009 and March 2013 from subjects in the High-Risk Lung Cancer-Screening Program at Roswell Park Cancer Institute (RPCI) (Buffalo, NY) (detailed cohort information in the Supplemental Methods). PMLs were sampled (if present) using endobronchial biopsy, graded by a team of pathologists at BCCA or RPCI, and the worst histology observed was recorded. Bronchial brushes of normal-appearing epithelium from 84 BCCA subjects (1 brush / subject) with and without PMLs were selected to undergo mRNA-Seq while ensuring balanced clinical covariates. Fifty-one bronchial brushes of normal appearing epithelium from 23 RPCI subjects were also profiled by mRNA-Seq (18 subjects had 2 procedures, and 5 subjects had 3 procedures) and utilized as a secondary biomarker validation set. Changes in the biomarker score were calculated between sequential procedures within a subject. Sets of samples were classified as stable/progressive if the worst histological grade at the second time point for a given patient remained the same or worsened, and regressive if the worst histological grade at the second time point improved. The Institutional Review Boards (IRBs) of all participating institutions approved the study and all subjects provided written informed consent.
RNA-Seq library preparation, sequencing and data processing
Total RNA was extracted from bronchial brushings using miRNeasy Mini Kit (Qiagen). Sequencing libraries were prepared from total RNA samples using Illumina® TruSeq® RNA Kit v2 and multiplexed in groups of four using Illumina® TruSeq® Paired-End Cluster Kit. Each sample was sequenced on the Illumina® HiSeq® 2500 to generate paired-end 100 nucleotide reads. Demultiplexing and creation of FASTQ files were performed using Illumina CASAVA (all software versions are reported in Supplemental Materials). For the BCCA samples, reads were aligned to hg19 using TopHat. The insert size mean and standard deviation were determined using the alignments and MISO (28). Reads were realigned using TopHat and the insert size parameters. Alignment and quality metrics were calculated using RSeQC. Gene count estimates were derived using HTSeq-count (29) and the Ensembl v64 GTF file. Gene filtering was conducted on normalized counts per million (cpm) calculated using R and edgeR using a modified version of the mixture model in the SCAN.UPC Bioconductor package (30). A gene was included in downstream analyses if the mixture model classified it as “on” (i.e. “signal”) in at least 15% of the samples. For the RPCI samples, gene counts were computed using RSEM (31) and Bowtie (32) with Ensembl 74 annotation. The data is available from NCBI’s Gene Expression Omnibus (GEO) using the accession ID GSE79315.
Data analysis for the BCCA samples
Sample and gene filtering yielded 13,870 out of 51,979 genes and 82 samples (n=2 excluded due to quality or sex annotation mismatches) for analysis. Data from Beane et al. (1) was used to predict the smoking status of the 82 samples that was utilized in all subsequent analyses (Supplementary: Dataset SD1, Fig. S1, and Methods). Airway brushings were dichotomized into two groups: samples with no evidence of PMLs (samples with no abnormal fluorescing areas or biopsies having normal or hyperplasia histology, n=25); and samples with evidence of PMLs (biopsies having mild, moderate, or severe dysplasia, n=50). Brushes with a worst histology of metaplasia (n=7) were excluded from the dichotomized groups. The limma (33), edgeR (34) and sva packages (35) were used to identify differentially expressed genes associated with presence of PMLs using normalized voom-tranformed (36) data and surrogate variable analysis using the first 7 surrogate variables (Table S1). Gene set enrichment analyses were conducted using ROAST (37) and GSEA (38), and GSVA (39). The Molecular Signatures Database (MSigDb) v4 Entrez ID Gene Sets were converted to Ensembl IDs using BioMart. Additional gene sets were created from CEL files or RNA-Seq counts from The Cancer Cell Line Compendium (CCLE), SCC tumor and adjacent normal tissue from TCGA, GSE19188, GSE18842, and GSE4115 (Supplemental Methods).
Cell culture
Human bronchial epithelial biopsy cell cultures (Supplementary Table S2) were obtained from the Colorado Lung SPORE Tissue Bank and cultured in Bronchial Epithelial Growth Media (BEGM). Human non-small cell lung cancer (NSCLC) cell lines were purchased from ATCC and short tandem repeat (STR) profiles were verified at the time of use by the Promega Gene Print® 10 system at the Dana Faber Cancer Institute. H1299, H2085 and SW900 cells were cultured in RPMI supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin, and H2085 cells were cultured in ALC-4 media. All cells were grown in a 37°C humidified incubator with 5% CO2.
Bioenergetics studies
Oxygen consumption rates (OCR) and extracellular acidification rates (ECAR) were measured using the XF96 Extracellular Flux Analyzer instrument (Seahorse Bioscience Inc). Briefly, ~30,000 cancer cells/well or ~40,000 bronchial epithelial biopsy cells/well (higher numbers due to slow growth rate) were seeded on XF96 cell culture plates and grown overnight. Prior to running the assay, media was replaced with Seahorse base media (2mM (milimole/L) L-glutamine) and placed at 37°C and 0% CO2 for ~30 minutes. The XF Cell Mito Stress Test kit and protocol were utilized to examine mitochondrial function. Measurements were taken every 5 minutes over 80 minutes. To modulate mitochondrial respiration, 5 uM oligomycin, 1 uM FCCP and 5 uM antimycin A were used. Prism software v6 was used to calculate t-statistics for baseline OCR/ECAR comparisons and a 2-way ANOVA was conducted to compare OCR and ECAR measurements.
Mitochondrial Enumeration using Flow Cytometry
Using an established protocol (40), cell cultures (5X105 cells/10cc dish of bronchial biopsy cultures and cancer cell cultures) were grown overnight and exposed to 120 uM MitoTracker Green FM in media free of FBS for 30 min at 37°C humidified incubator with 5% CO2. Cells were subsequently collected, washed in PBS and resuspended in 0.5 mL PBS-EDTA and 1 uL of propidium iodide (PI) was added to distinguish live/dead cells. MitoTracker FM and PI were measured using a BD LSRII flow cytometer and BD FACS Diva software (6.2.1). Data was analyzed using FlowJo (10.2), gating out doublets and dead cells, and normalizing mean fluorescence to the number of cell counts.
Immunohistochemistry
Formalin-fixed, paraffin-embedded (FFPE) sections of human PMLs sampled from high-risk subjects undergoing screening for lung cancer were provided by RPCI as part of an IRB-approved study detailed below (Supplementary Table S3). Dr. Candace Johnson at RPCI provided the FFPE lung sections from the N-nitroso-tris-chloroethylurea (NTCU) mouse model of lung SCC, from SWR/J mice treated with 25ul of 40mmol/L NTCU for 25 weeks in accordance with the Institutional Animal Care and Use Committee approved protocol (41). Briefly, slides were de-paraffinized and rehydrated. For antigen retrieval, slides were heated in citrate buffer. Slides were subsequently incubated in primary antibody (Translocase of the Outer Mitochondrial Membrane 22 (TOMM22): mouse tissue 1:300 and human 1:1,200 (Abcam), and Cytochrome C Oxidase subunit IV (COX4I1) : mouse tissue 1:500 and human 1:5,000 (Abcam)) diluted in 1% Bovine Serum Albumin (BSA). Signal was amplified using an ABC kit (Vector Labs). To reveal endogenous peroxidase activity, slides were incubated in a 3,3′-Diaminobenzidine (DAB) solution. Slides were rinsed, counterstained with hematoxylin, dehydrated in graded alcohol followed by xylene and cover slipped.
Biomarker Development and Validation
A gene expression biomarker discovery pipeline was developed to test thousands of parameter combinations (6,160 predictive models) to identify a biomarker capable of distinguishing between samples from subjects with and without PMLs. Samples were first assigned by batch (sequencing lane) to either a discovery set (n=58) or a validation set (n=17), and the validation set was excluded from biomarker development (Fig. S2 and Supplemental Methods). The biomarker was developed using subsets of the discovery set established by randomly splitting the samples into training (80%, n=46) and test (20%, n=12) sets 500 times. Model performance was assessed using standard metrics for both the training and test sets (Supplemental Methods). The biomarker pipeline was also used to develop biomarkers for sex and smoking status as well as randomized class labels for all phenotypes (serving as positive and negative controls, respectively). A final model (biomarker) was selected (Supplemental Methods) and its ability to distinguish between samples with and without PMLs was tested in a validation set (n=17). In addition, using the bronchial brushings collected longitudinally from subjects at RPCI, we tested whether or not differences in biomarker scores over time were reflective of progression of PMLs (n=28 matched time point pairs) (Supplemental Methods).
Results
Subject population
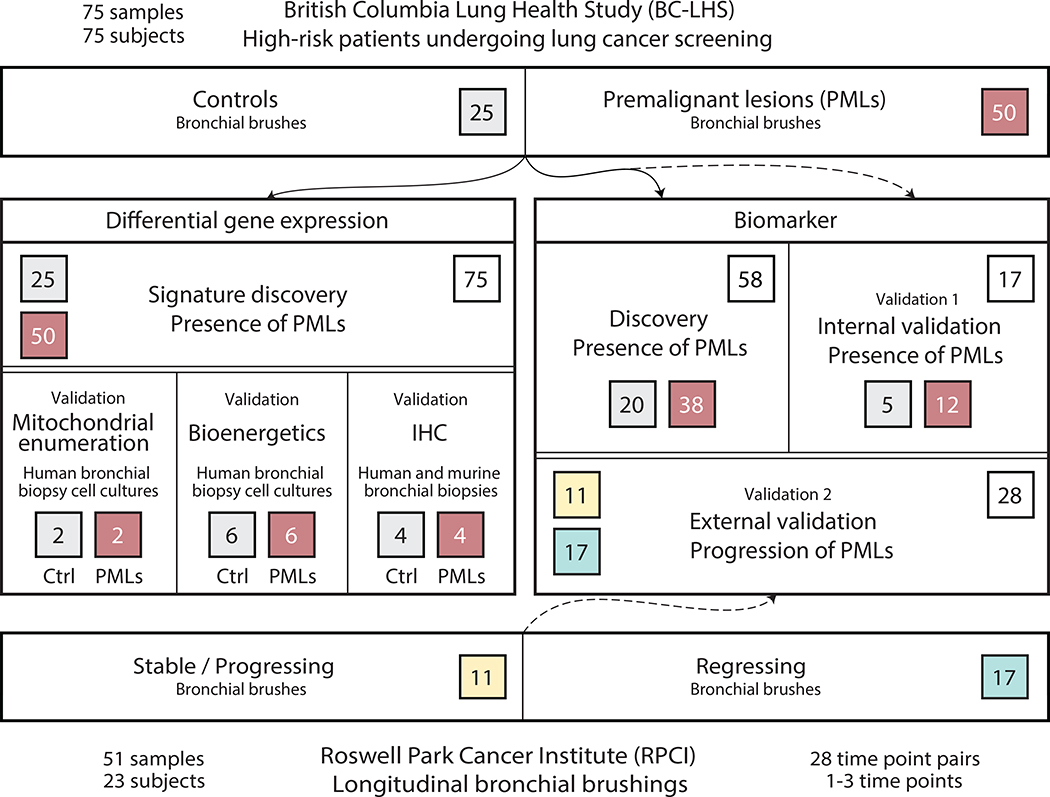
The study design used 126 bronchial brushings obtained via autofluorescence bronchoscopy at the BCCA and RPCI for differential gene expression and pathway analysis, as well as for biomarker development and validation (Fig. 1). A dataset consisting of samples (n=75) collected from BCCA subjects with PMLs (n=50) and without PMLs (n=25) was used to derive a gene expression signature associated with the presence of PMLs. Important clinical covariates such as COPD and reported smoking history as well as alignment statistics from the mRNA-Seq data were not significantly different between the two groups (Table 1 and Table 2). For biomarker development, the 75 BCCA samples were split by batch and used in biomarker discovery (n=58) and validation (n=17) (Supplementary Tables S4 and S5). The change in biomarker score as a predictor of progression of PMLs was then tested in the 51 RPCI samples (Supplementary Tables S5 and S6).
Figure 1. Study Design.
Flow diagram depicting use of bronchial brushings collected from subjects with (red, n=50) and without (gray, n=25) PMLs from the BCCA as part of the BC-LHS for differential gene expression/pathway analysis and for biomarker development. Independent human and mouse bronchial biopsies and biopsy cell cultures were used to validate these findings via mitochondrial enumeration, bioenergetics, and immunohistochemistry (left panel). Biomarker development was conducted by splitting samples from the BC-LHS into a discovery (n=58) and a validation set (Validation 1, n=17) (right panel).. The discovery set was used to create the gene expression-based biomarker to detect the presence of PMLs in the airway field of injury. The biomarker was tested on the BC-LHS validation set and an external validation set (bottom) from RPCI (Validation 2, n=28 matched time point pairs, stable/progressing pairs in yellow and regressing pairs in blue).
Table 1.
Demographic and clinical characteristics stratified by premalignant lesion status.
| Factor | Overall (n=82) | No Lesions (n=25) | Lesions (n=50) | P* |
|---|---|---|---|---|
| Age | 62.9 (7.2) | 64.5 (5.8) | 62.2 (8.0) | 0.16 |
| Male | 54/82 (65.9) | 16/25 (64) | 35/50 (70) | 0.61 |
| Current smoker | 40/82 (48.8) | 11/25 (44) | 25/50 (50) | 0.81 |
| Pack-years | 47.3 (15.7) | 47.6 (17.9) | 47.2 (15.2) | 0.93 |
| FEV1% Predicted | 82.5 (18.6) | 84.5 (17.9) | 81.7 (19.2) | 0.54 |
| FEV1/FVC Ratio | 71.2 (7.9) | 73.4 (7.4) | 69.6 (8.1) | 0.05 |
| COPD (FEV1%<80 & FEV1/FVC<70) | 24/82 (29.3) | 5/25 (20) | 17/50 (34) | 0.28 |
| Histology | <0.001 | |||
| Normal | 12/82 (14.6) | 12/25 (48) | ||
| Hyperplasia | 13/82 (15.9) | 13/25 (52) | ||
| Metaplasia | 7/82 (8.5) | |||
| Mild Dysplasia | 35/82 (42.7) | 35/50 (70) | ||
| Moderate Dysplasia | 12/82 (14.6) | 12/50 (24) | ||
| Severe Dysplasia | 3/82 (3.7) | 3/50 (6) |
Data are means (SD) for continuous variables and proportions with percentages for dichotomous variables. P* values are for the comparison of subjects with and without premalignant lesions. Two sample t-tests were used for continuous variables: Fisher’s exact test was used for categorical variables.
Table 2.
Alignment statistics stratified by premalignant lesion status
| Factor | Overall (n=82) | No Lesions (n=25) | Lesions (n=50) | P* |
|---|---|---|---|---|
| Total Alignments | 90M (17M) | 90M (15M) | 91M (19M) | 0.78 |
| Unique Alignments | 83M (16M) | 83M (14M) | 84M (17M) | 0.76 |
| Properly Paired Alignments | 66M (12M) | 66M (11M) | 67M (14M) | 0.75 |
| Genebody 80/20 Ratio | 1.3 (0.2) | 1.3 (0.1) | 1.3 (0.2) | 0.84 |
| Mean GC Content | 47.8 (3.4) | 47.4 (2.9) | 48.2 (3.7) | 0.34 |
Data are means (SD) for continuous variables and proportions with percentages for dichotomous variables. Reads are expressed in millions denoted by M. P* values are for the comparison of subjects with and without premalignant lesions. Two sample t-tests were used for continuous variables; Fisher’s exact test was used for factors.
Transcriptomic alterations in the airway field of injury associated with the presence of PMLs
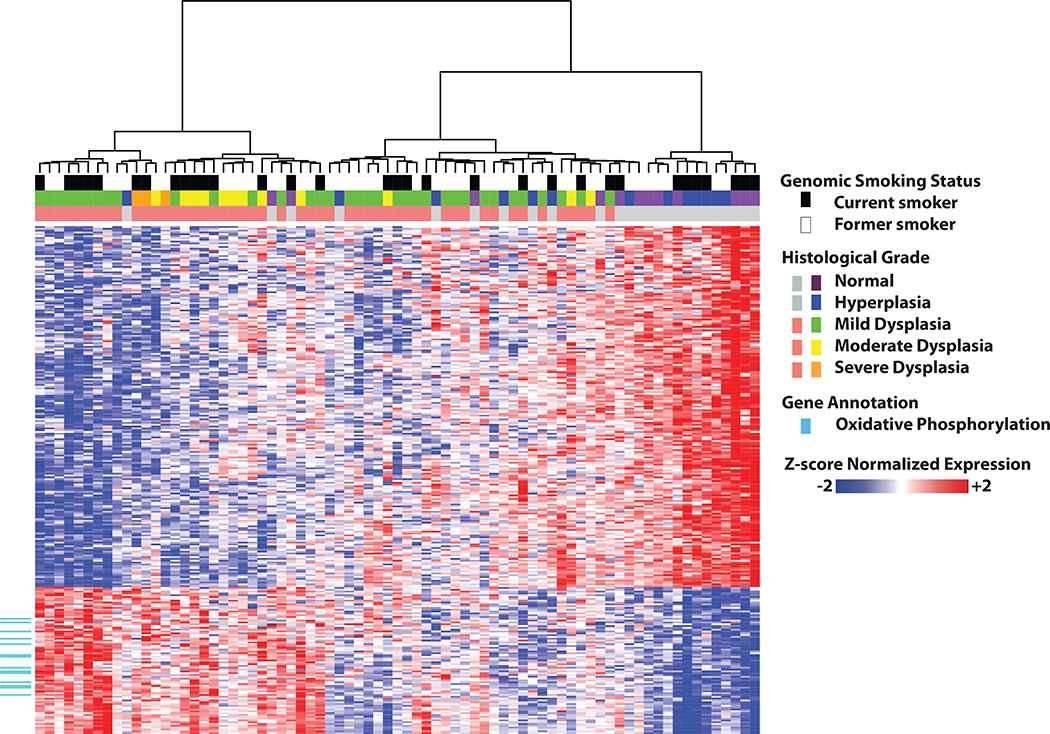
We identified 280 genes significantly differentially expressed between subjects with and without PMLs (FDR<0.002, Fig. 2). Utilizing the MSigDB v4 canonical pathways, we identified 170 pathways significantly enriched in genes up- or down-regulated in the presence of PMLs using ROAST (37) (FDR<0.05, Supplementary Dataset SD2). Pathways involved in oxidative phosphorylation (OXPHOS), the electron transport chain (ETC), and mitochondrial protein transport were strongly enriched among genes up-regulated in the airways of subjects with PMLs. Other up-regulated pathways included DNA repair and the HIF1A pathway. Down-regulated pathways included the STAT3 pathway, the JAK/STAT pathway, IL4 signaling, RAC1 regulatory pathway, NCAM1 interactions, collagen formation, and extracellular matrix organization.
Figure 2. Unsupervised hierarchal clustering of genes associated with the presence of premalignant lesions.
Residual gene expression of the 280 genes differentially expressed between subjects with PMLs (red) and without PMLs (gray). Top color bars represent the worst biopsy histological grade observed during bronchoscopy and genomically-derived smoking status of the subjects. The 14 genes in the KEGG oxidative phosphorylation pathway are indicated in cyan. The residual values after adjusting for the 7 surrogate variables were z-score normalized prior to Ward hierarchal clustering.
OXPHOS is increased in PML cell cultures and biopsies of increasing severity
The ETC and OXPHOS pathways, which involve genes distributed between the complexes I-IV of the ETC and ATP synthase, were highly activated in the airway field in the presence of PMLs. We wanted to determine if the functional activity of these pathways was similarly altered in PMLs compared to normal tissue. We conducted cellular bioenergetics by measuring OCR as a measure of ETC/OXPHOS (aerobic respiration), ECAR as a measure of glycolysis (anerobic respiration) and MitoTraker Green FM as a measure of mitochondrial content in primary cell cultures derived from bronchial biopsies. Additionally, we performed immunohistochemistry of select OXPHOS-related genes in mouse and human dysplastic lesions and normal tissue to measure protein levels.
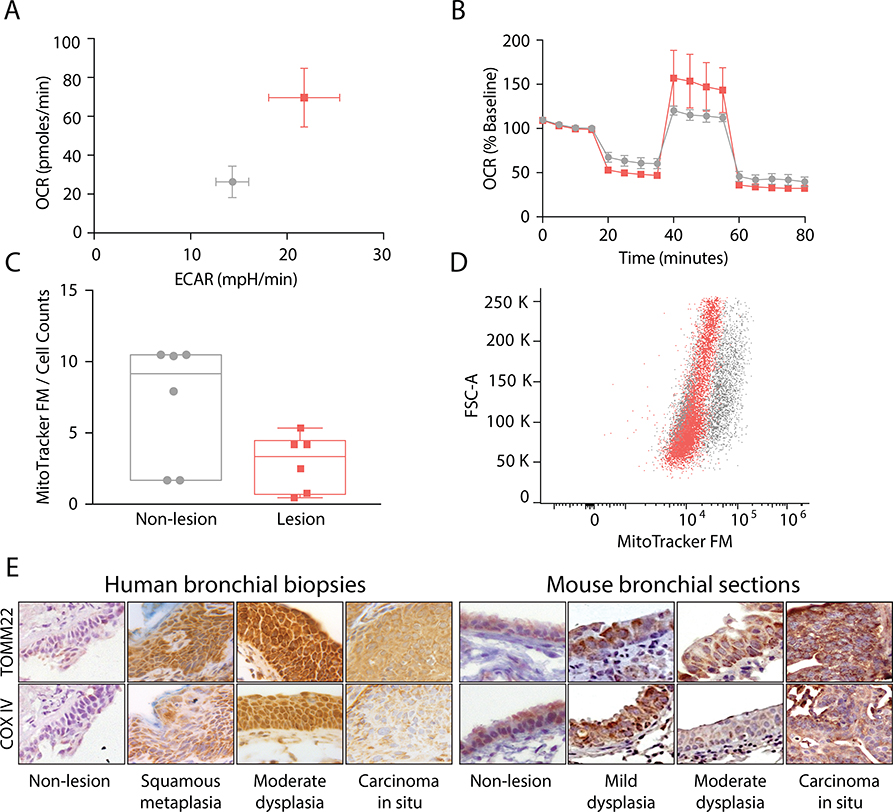
We established a significant concordance between ETC/OXPHOS gene expression and cellular bioenergetics in NSCLC cell lines (Supplementary Fig. S3 A–F). Next, using primary cell cultures derived from normal to severe dysplastic tissue (Table S2), we observed that the mean baseline OCR/ECAR values were 2.5/1.5 fold higher in the cultures from PMLs compared to controls (p<0.035, Fig. 3A), reflecting predictions based on mRNA-Seq field data (Supplementary Fig. S3 G–H). There was a greater reduction in OCR in PMLs immediately following oligomycin treatment (p<0.022) suggesting an increased dependence on OXPHOS for ATP production to meet energetic demands. In addition, the mean spare respiratory capacity following the release of the proton gradient was elevated by ~1.5 fold in the PML cultures compared to controls indicating increased ability to respond to energy demands (42). Lastly, treatment with antimycin A resulted in a greater reduction of OCR in PML cultures (p<0.001, Fig. 3B), suggesting that oxygen consumption in the lesions is dependent on increased ETC components in complex III. No significant changes to ECAR were detected in response to mitochondrial perturbations. Furthermore to examine if the increased OXPHOS was a result of increased mitochondrial biogenesis in PML cultures, cells were incubated with MitoTraker FM to stain for mitochondria content and fluorescence enumerated using flow cytometry revealed no significant difference between PML and controls (p=0.15, Fig. 3C–D).
Figure 3. OXPHOS up-regulation in premalignant lesion biopsies.
(A) The mean baseline OCR/ECAR ratio measured in human bronchial biopsies cultures from PMLs (pink, n=6) was 2.5 fold higher than the biopsies of normal airway epithelium (gray n=6) (p=0.035). (B) Bioenergetic studies testing mitochondrial function demonstrate PMLs (pink) have a significantly (~1.5 fold) higher maximal respiration (p=0.022). Error bars in A and B represent standard error of the mean. (C&D) Mitochonrial enumeration by FACS analysis of MitoTraker GFP suggests increased OCR is not reliant on increase mitochondria as the difference in GFP per cell was not significant (p=0.150). (E) Representative images of TOMM22 and COX IV staining in which expression of both proteins is increased in low and moderate dysplastic lesions in both human and NTCU-mouse PMLs. (Magnification 400X)
Additionally, we found elevated protein levels of Translocase of the Outer Mitochondrial Membrane 22 (TOMM22) and Cytochrome C Oxidase subunit IV (COX4I1) in low/moderate grade dysplastic lesions compared to normal tissue (Fig. 3E-F) using tissues from human bronchial biopsy FFPE sections (Supplementary Table S3) and whole lung sections from the NTCU mouse model of SCC. The results suggest that PMLs are more ETC- and OXPHOS-dependent and express OXPHOS-related proteins at higher levels compared to normal tissue.
PML-associated gene expression alterations in the airway field are involved in lung squamous cell carcinogenesis
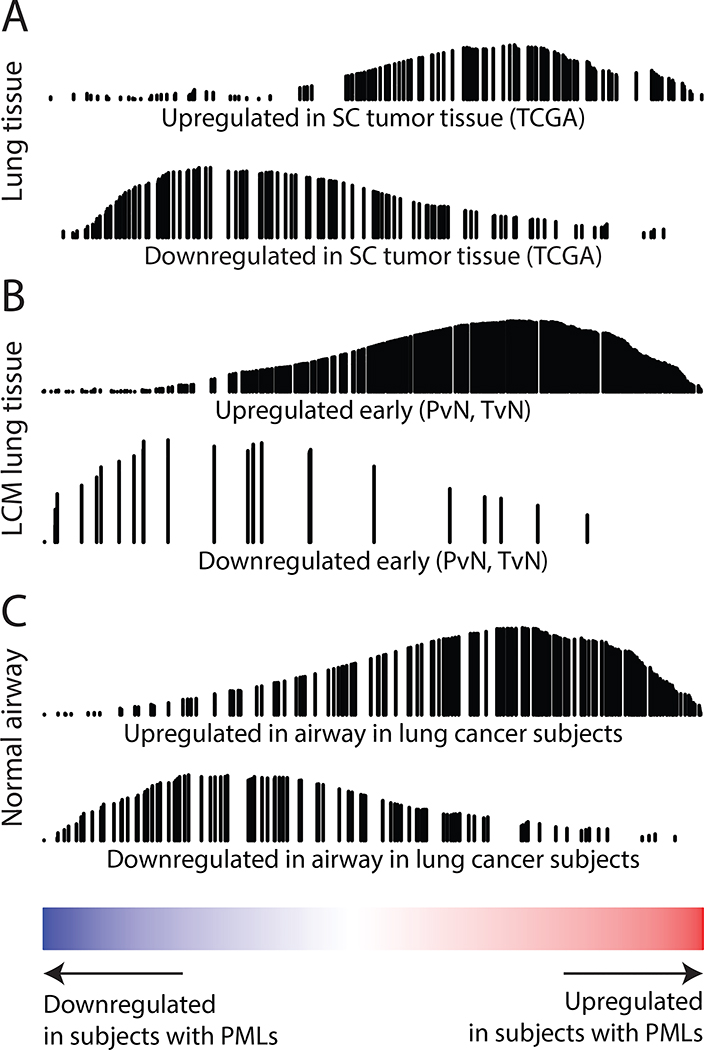
To further extend the connection between the airway field and PMLs, we examined the relationship between PML-associated genes in the airway field and other lung cancer-related datasets. We identified genes differentially expressed between lung tumor tissue (primarily squamous) and normal lung tissue in three different datasets (TCGA, GSE19188, and GSE18842). Genes associated with lung cancer in all datasets were significantly (FDR<0.05) enriched by GSEA, concordantly with gene expression changes associated with the presence of PMLs in the field (Fig. 4A and Supplementary Dataset SD3). Extending beyond the lung tumor, similar enrichment (FDR<0.05) was found using early, stepwise, and late gene expression changes in SCC identified by Ooi et al. (43) (Fig. 4B and Supplementary Dataset SD3) and among genes associated with lung cancer in the airway field of injury (GSE4115, Fig. 4C and Supplementary Dataset SD3). These results support the concept that early events in lung carcinogenesis can be observed throughout the respiratory tract, even in cells that appear normal.
Figure 4. PML-associated gene expression alterations in the field are concordant with SCC-related datasets.
The genes up-regulated in the field of subjects with PMLs are red and genes down regulated in blue (x-axis). GSEA identified the significant enrichment of the lung cancer-related gene expression signatures shown in this ranked list. The black vertical lines represent the position of the genes in the gene set in the ranked list and the height corresponds to the magnitude of the running enrichment score from GSEA (y-axis). (A) Top differentially expressed genes from analysis of TCGA RNA-Seq data comparing lung SCC and matched adjacent normal tumor tissue. (B) Ooi et al. gene sets for early gene expression changes defined by genes altered between premalignant and normal tissue and between tumor and normal tissue (p<0.05) using laser capture microdissected (LCM) epithelium from the margins of resected SCC tumors. (C) Top differentially expressed genes from analysis of cytologically normal bronchial epithelial cells from smokers with and without lung cancer (GSE4115).
Development and validation of a biomarker for PML detection and monitoring
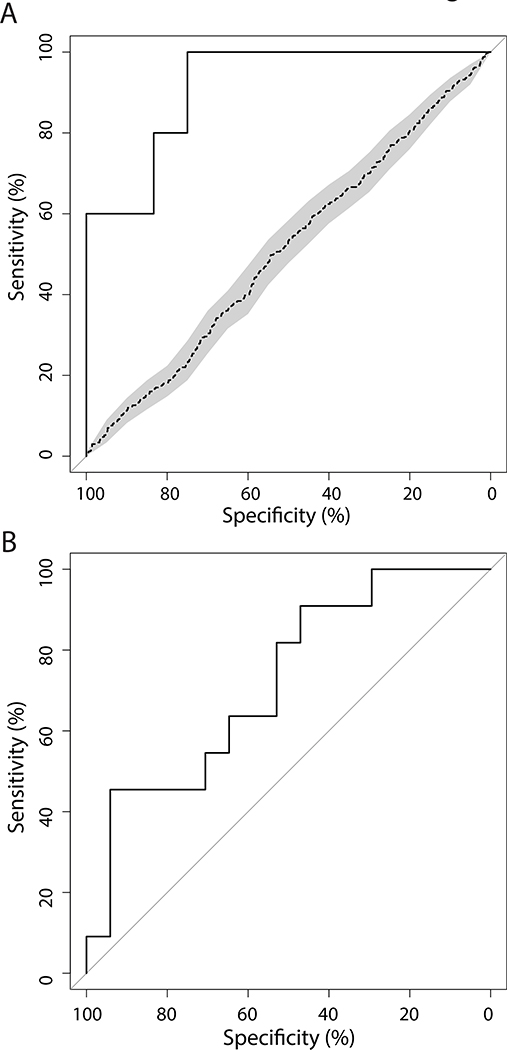
The airway brushings from BCCA subjects with and without PMLs were leveraged to build a biomarker predictive of the presence of PMLs. The biomarker consisted of 200 genes (of which 91 overlapped with the gene signature in Fig. 2) and achieved a ROC-curve AUC of 0.92, sensitivity of 0.75 (9/12 samples with PMLs predicted correctly), and specificity of 1.00 (5/5 samples without PMLs predicted correctly) in independent validation samples (n=17, Fig. 5A). In addition, the biomarker was used to score an independent set of longitudinally collected bronchial brushings from RPCI subjects (Fig. 1). Biomarker scores were calculated for each sample, and the difference in biomarker scores between sequential procedures (n=28 time point pairs, Supplemental Methods) was predictive of whether the worst PML histology observed during the baseline procedure regressed or whether it was stable or progressed with an AUC of 0.75 (Fig. 5B).
Figure 5. Performance of an airway biomarker in detecting the presence and progression of premalignant lesions.
The ROC curves demonstrate the biomarker performance. (A) ROC curve (AUC=0.92) showing biomarker performance based on predictions of the presence of PMLs in the validation samples (n=17), black line. Shuffling of class labels (n=100 permutations) produced an average ROC curve (dotted line) with a significantly lower AUC (p<<0.001). (B) ROC curve (AUC=0.75) showing biomarker performance based on changes in biomarker score over time in detecting PML regression or stable/progression.
Discussion
In this study, we identified a PML-associated gene expression signature in bronchial brushings obtained from normal appearing mucosa and characterized the biological pathways that are dysregulated in the airway field of injury. We established that the PML-associated airway field harbors alterations observed in PMLs and in SCC. This evidence motivated the development of a biomarker that reflects the presence of PMLs and their outcome over time. Our findings provide novel insights into the earliest molecular events associated with lung carcinogenesis and have the potential to impact lung cancer prevention by providing novel targets (e.g., OXPHOS) and potential biomarkers for risk stratification and monitoring the efficacy of chemoprevention agents.
The first major finding of our study was the identification of a PML-associated field of injury. The most significantly enriched pathways among up-regulated genes in subjects with PMLs were OXPHOS, ETC, and mitochondrial protein transport. These pathways efficiently generate energy in the form of ATP by utilizing the ETC in the mitochondria. During cancer development, energy metabolism alterations are described as an increase in glycolysis and suppression of OXPHOS, known as the Warburg effect (44); however, recent studies demonstrate that OXPHOS is maintained in many tumors and can be important for progression (45). We wanted to assay for OXPHOS activation in PMLs as it may support PML progression by generating reactive oxygen species (ROS) that can induce oxidative stress, increase DNA damage, and HIF1A pathway activation (pathways observed in our analysis).
We observed increases in both the basal OCR and the spare respiratory capacity in the PML biopsies, suggesting that PML-derived cell cultures are more ETC and OXPHOS dependent that the non-PML cultures. We also further demonstrated the increase in ETC activity marked by positive COX IV staining associated with increasing PML histological grade. Several members of the mitochondrial protein import machinery (46) were significantly up-regulated (FDR<0.05) in airways with PMLs including members of the TOM complex (TOMM22, TOMM7, and TOMM20) and TIM23 complex (TIMM23, TIMM21, and TIMM17A). We observed positive staining of TOMM22 with increasing PML grade, suggesting that increased import of precursor proteins from the endoplasmic reticulum may be required to meet the energy demands of PMLs. Measurements of mitochondrial content indicated no significant differences between the normal and PML-derived cultures, and transcriptional levels of PPARGC1A, aassociated with mitochondrial biogenesis, were not different between subjects with and without PML indicating that increases in OXPHOS are likely independent of mitochondrial number (47–49). Increases in OXPHOS have also been demonstrated to be associated with PML progression in Barret’s esophagus and esophageal dysplasia (50), cervical dysplasia (51), and the dysplastic lesions that precede oral SCC (52). Collectively, these data suggest that the OXPHOS pathway may be a target for early intervention. Pre-clinical studies in the NTCU mouse model of lung SCC demonstrate the potential for targeting mitochondrial respiration by using the natural product honokiol to inhibit tumor development (53). Further investigations into the role of cellular energy metabolism in the development and progression of PMLs are needed to fully understand how to best target it for intervention in lung cancer.
Additionally, we extended the connection between the PML-associated airway field and PMLs beyond the OXPHOS pathway to processes associated with squamous cell lung carcinogenesis. By examining gene sets from multiple external studies representative of lung cancer-related processes occurring in the tumor, adjacent to the tumor, and in the upper airway, we found significant concordant relationships between the PML-associated field and processes associated with SCC tumors. Genes are similarly altered in these varied cancer-associated contexts and thus tissues in the field both adjacent to and far away from the tumor may reflect basic processes and mechanisms of lung carcinogenesis such as DNA damage as hypothesized earlier.
These observations motivated us to pursue the most translational aspect of this study, a biomarker that can detect PMLs and monitor their progression over time. The 200-gene biomarker, measured in normal appearing bronchial mucosa, achieved high performance detecting the presence of PMLs in a small test set (AUC=0.92). This biomarker may increase the sensitivity of bronchoscopy in detecting the presence of PMLs (which can be difficult to observe under white light), and thus improve identification of high-risk smokers that should be targeted for aggressive lung cancer screening programs. Additionally, the biomarker may offer wider clinical utility in early intervention trials by serving as an intermediate endpoint of efficacy (beyond Ki-67 staining for proliferation, and changes in biopsy histology). Towards this goal, we demonstrated that the change in biomarker scores over time reflects contemporaneous regressive or progressive/stable disease (AUC = 0.75). This result suggests that the airway field of injury in the presence of PMLs is dynamic and that capturing the gene expression longitudinally may allow for further stratification of high-risk subjects. The potential clinical utility of the biomarker is further supported by recent work demonstrating a significant association between the development of incident lung squamous cell carcinoma and the frequency of sites that persist or progress to high-grade dysplasia (24).
Further development and testing in a larger cohort is needed to confirm the biomarker’s performance, utility, and ability to predict future PML progression or regression. Additionally, longitudinal and spatial sampling would provide a greater understanding of the dynamic relationship between the normal epithelium and the PMLs as they regress or progress to SCC. Longitudinal studies would allow for more accurate characterization of the time intervals needed to observe gene expression dynamics both in the PMLs and in the airway field of injury. Spatial sampling throughout the respiratory tract, including the more accessible nasal airway that shares the tobacco-related injury with the bronchial airways (54), would allow for evaluation of the impact of distance between the PMLs and the brushing site, the range of PML histologies, and the multiplicity of PMLs that can be present simultaneously in a patient and influence the PML-associated airway field.
In light of these challenges and opportunities for future work, we have comprehensively profiled gene expression changes in airway epithelial cells in the presence of PMLs that suggest great clinical utility. Moving therapeutics and detection strategies towards an earlier stage in the disease process via molecular characterization of premalignant disease holds great promise (55,56), and this study represents an important step towards a precision medicine approach to lung cancer prevention.
Supplementary Material
Translational Relevance.
Lung cancer prevention could be transformed by novel approaches that enhance the identification of high-risk patients, safe and effective prevention agents, and biomarkers of therapeutic response. We have used mRNA sequencing to identify gene expression alterations in the airway epithelium associated with the presence of bronchial premalignant lesions. Bronchial premalignant lesions are precursors of lung squamous cell carcinoma and are a risk factor for developing lung cancer at the lesion site or elsewhere in the lung. The early molecular changes that we have identified in normal appearing epithelial cells in the premalignant airway may represent possible targets for lung cancer chemoprevention. Additionally, gene expression alterations in airway epithelium can be leveraged to develop a biomarker associated with presence and progression of premalignant lesions that could be used to stratify patients into chemoprevention trials or serve as surrogate markers of efficacy.
Acknowledgements
We thank Daniel Merrick (University of Colorado, Denver) and the Denver SPORE for facilitating the acquisition of premalignant primary cultures used in this study. We also thank Candace Johnson (Roswell Park Cancer Institute) for providing NTCU-mouse tissue for analysis in this study as well as Mary Beth Pine for providing clinical data for the human bronchial brushes from the Roswell Park Cancer Institute.
Financial Support: LUNGevity Foundation Grant #2012-01 (J. Beane) and Janssen Pharmaceutical Research and Development, L.L.C. Sponsorship (A. Spira).
Conflict of Interest: J.B., S.A.M, A.M.T, S.S.D, M.L., M.E.R, and A.S. receive research support from Janssen Pharmaceuticals. A.S. and M.L. are consultants to Veractye, Inc. Boston University owns intellectual property related to the subject matter of this manuscript. S.P. worked for Janssen Pharmaceuticals and owns stock in the company.
References
- 1.Beane J, Sebastiani P, Liu G, Brody JS, Lenburg ME, Spira A. Reversible and permanent effects of tobacco smoke exposure on airway epithelial gene expression. Genome Biol. 2007;8:R201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hackett NR, Heguy A, Harvey B-G, O’Connor TP, Luettich K, Flieder DB, et al. Variability of Antioxidant-Related Gene Expression in the Airway Epithelium of Cigarette Smokers. Am J Respir Cell Mol Biol. 2003;29:331–43. [DOI] [PubMed] [Google Scholar]
- 3.Spira A, Beane J, Shah V, Liu G, Schembri F, Yang X, et al. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci U S A. 2004;101:10143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beane J, Vick J, Schembri F, Anderlind C, Gower A, Campbell J, et al. Characterizing the impact of smoking and lung cancer on the airway transcriptome using RNA-Seq. Cancer Prev Res Phila Pa. 2011;4:803–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sridhar S, Schembri F, Zeskind J, Shah V, Gustafson AM, Steiling K, et al. Smoking-induced gene expression changes in the bronchial airway are reflected in nasal and buccal epithelium. BMC Genomics. 2008;9:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chari R, Lonergan KM, Ng RT, MacAulay C, Lam WL, Lam S. Effect of active smoking on the human bronchial epithelium transcriptome. BMC Genomics. 2007;8:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spira A, Beane JE, Shah V, Steiling K, Liu G, Schembri F, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;13:361–6. [DOI] [PubMed] [Google Scholar]
- 8.Beane J, Sebastiani P, Whitfield TH, Steiling K, Dumas Y-M, Lenburg ME, et al. A prediction model for lung cancer diagnosis that integrates genomic and clinical features. Cancer Prev Res Phila Pa. 2008;1:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitney DH, Elashoff MR, Porta-Smith K, Gower AC, Vachani A, Ferguson JS, et al. Derivation of a bronchial genomic classifier for lung cancer in a prospective study of patients undergoing diagnostic bronchoscopy. BMC Med Genomics. 2015;8:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silvestri GA, Vachani A, Whitney D, Elashoff M, Porta Smith K, Ferguson JS, et al. A Bronchial Genomic Classifier for the Diagnostic Evaluation of Lung Cancer. N Engl J Med. 2015;373:243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gustafson AM, Soldi R, Anderlind C, Scholand MB, Qian J, Zhang X, et al. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci Transl Med. 2010;2:26ra25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wistuba II, Gazdar AF. Lung cancer preneoplasia. Annu Rev Pathol. 2006;1:331–48. [DOI] [PubMed] [Google Scholar]
- 13.Wistuba II, Lam S, Behrens C, Virmani AK, Fong KM, LeRiche J, et al. Molecular damage in the bronchial epithelium of current and former smokers. J Natl Cancer Inst. 1997;89:1366–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wistuba II, Behrens C, Virmani AK, Mele G, Milchgrub S, Girard L, et al. High resolution chromosome 3p allelotyping of human lung cancer and preneoplastic/preinvasive bronchial epithelium reveals multiple, discontinuous sites of 3p allele loss and three regions of frequent breakpoints. Cancer Res. 2000;60:1949–60. [PubMed] [Google Scholar]
- 15.Wistuba II, Behrens C, Milchgrub S, Bryant D, Hung J, Minna JD, et al. Sequential molecular abnormalities are involved in the multistage development of squamous cell lung carcinoma. Oncogene. 1999;18:643–50. [DOI] [PubMed] [Google Scholar]
- 16.Belinsky SA, Palmisano WA, Gilliland FD, Crooks LA, Divine KK, Winters SA, et al. Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res. 2002;62:2370–7. [PubMed] [Google Scholar]
- 17.Lamy A, Sesboüé R, Bourguignon J, Dautréaux B, Métayer J, Frébourg T, et al. Aberrant methylation of the CDKN2a/p16INK4a gene promoter region in preinvasive bronchial lesions: a prospective study in high-risk patients without invasive cancer. Int J Cancer. 2002;100:189–93. [DOI] [PubMed] [Google Scholar]
- 18.Nakachi I, Rice JL, Coldren CD, Edwards MG, Stearman RS, Glidewell SC, et al. Application of SNP microarrays to the genome-wide analysis of chromosomal instability in premalignant airway lesions. Cancer Prev Res Phila Pa. 2014;7:255–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rahman SMJ, Gonzalez AL, Li M, Seeley EH, Zimmerman LJ, Zhang XJ, et al. Lung cancer diagnosis from proteomic analysis of preinvasive lesions. Cancer Res. 2011;71:3009–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massion PP, Zou Y, Uner H, Kiatsimkul P, Wolf HJ, Baron AE, et al. Recurrent genomic gains in preinvasive lesions as a biomarker of risk for lung cancer. PloS One. 2009;4:e5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Boerdonk RAA, Sutedja TG, Snijders PJF, Reinen E, Wilting SM, van de Wiel MA, et al. DNA copy number alterations in endobronchial squamous metaplastic lesions predict lung cancer. Am J Respir Crit Care Med. 2011;184:948–56. [DOI] [PubMed] [Google Scholar]
- 22.Ishizumi T, McWilliams A, MacAulay C, Gazdar A, Lam S. Natural history of bronchial preinvasive lesions. Cancer Metastasis Rev. 2010;29:5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edell E, Lam S, Pass H, Miller YE, Sutedja T, Kennedy T, et al. Detection and localization of intraepithelial neoplasia and invasive carcinoma using fluorescence-reflectance bronchoscopy: an international, multicenter clinical trial. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. 2009;4:49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merrick DT, Gao D, Miller YE, Keith RL, Baron AE, Feser W, et al. Persistence of Bronchial Dysplasia Is Associated with Development of Invasive Squamous Cell Carcinoma. Cancer Prev Res Phila Pa. 2016;9:96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Boerdonk RAA, Smesseim I, Heideman DAM, Coupé VMH, Tio D, Grünberg K, et al. Close Surveillance with Long-Term Follow-up of Subjects with Preinvasive Endobronchial Lesions. Am J Respir Crit Care Med. 2015;192:1483–9. [DOI] [PubMed] [Google Scholar]
- 26.Jeremy George P, Banerjee AK, Read CA, O’Sullivan C, Falzon M, Pezzella F, et al. Surveillance for the detection of early lung cancer in patients with bronchial dysplasia. Thorax. 2007;62:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tammemagi MC, Lam SC, McWilliams AM, Sin DD. Incremental value of pulmonary function and sputum DNA image cytometry in lung cancer risk prediction. Cancer Prev Res Phila Pa. 2011;4:552–61. [DOI] [PubMed] [Google Scholar]
- 28.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinforma Oxf Engl. 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piccolo SR, Sun Y, Campbell JD, Lenburg ME, Bild AH, Johnson WE. A single-sample microarray normalization method to facilitate personalized-medicine workflows. Genomics. 2012;100:337–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma Oxf Engl. 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinforma Oxf Engl. 2012;28:882–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu D, Lim E, Vaillant F, Asselin-Labat M-L, Visvader JE, Smyth GK. ROAST: rotation gene set tests for complex microarray experiments. Bioinforma Oxf Engl. 2010;26:2176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dingley S, Chapman KA, Falk MJ. Fluorescence-activated cell sorting analysis of mitochondrial content, membrane potential, and matrix oxidant burden in human lymphoblastoid cell lines. Methods Mol Biol Clifton NJ. 2012;837:231–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazzilli SA, Hershberger PA, Reid ME, Bogner PN, Atwood K, Trump DL, et al. Vitamin D Repletion Reduces the Progression of Premalignant Squamous Lesions in the NTCU Lung Squamous Cell Carcinoma Mouse Model. Cancer Prev Res Phila Pa. 2015;8:895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chacko BK, Kramer PA, Ravi S, Benavides GA, Mitchell T, Dranka BP, et al. The Bioenergetic Health Index: a new concept in mitochondrial translational research. Clin Sci Lond Engl 1979. 2014;127:367–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ooi AT, Gower AC, Zhang KX, Vick JL, Hong L, Nagao B, et al. Molecular profiling of premalignant lesions in lung squamous cell carcinomas identifies mechanisms involved in stepwise carcinogenesis. Cancer Prev Res Phila Pa. 2014;7:487–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26:877–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen X, Qian Y, Wu S. The Warburg effect: evolving interpretations of an established concept. Free Radic Biol Med. 2015;79:253–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wenz L-S, Opaliński Ł, Wiedemann N, Becker T. Cooperation of protein machineries in mitochondrial protein sorting. Biochim Biophys Acta. 2015;1853:1119–29. [DOI] [PubMed] [Google Scholar]
- 47.Tan Z, Luo X, Xiao L, Tang M, Bode AM, Dong Z, et al. The Role of PGC1α in Cancer Metabolism and its Therapeutic Implications. Mol Cancer Ther. 2016;15:774–82. [DOI] [PubMed] [Google Scholar]
- 48.LeBleu VS, O’Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16:992–1003, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan W, Evans R. PPARs and ERRs: molecular mediators of mitochondrial metabolism. Curr Opin Cell Biol. 2015;33:49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phelan JJ, MacCarthy F, Feighery R, O’Farrell NJ, Lynam-Lennon N, Doyle B, et al. Differential expression of mitochondrial energy metabolism profiles across the metaplasia-dysplasia-adenocarcinoma disease sequence in Barrett’s oesophagus. Cancer Lett. 2014;354:122–31. [DOI] [PubMed] [Google Scholar]
- 51.Xylas J, Varone A, Quinn KP, Pouli D, McLaughlin-Drubin ME, Thieu H-T, et al. Noninvasive assessment of mitochondrial organization in three-dimensional tissues reveals changes associated with cancer development. Int J Cancer. 2015;136:322–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grimm M, Cetindis M, Lehmann M, Biegner T, Munz A, Teriete P, et al. Association of cancer metabolism-related proteins with oral carcinogenesis - indications for chemoprevention and metabolic sensitizing of oral squamous cell carcinoma? J Transl Med. 2014;12:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pan J, Zhang Q, Liu Q, Komas SM, Kalyanaraman B, Lubet RA, et al. Honokiol inhibits lung tumorigenesis through inhibition of mitochondrial function. Cancer Prev Res Phila Pa. 2014;7:1149–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X, Sebastiani P, Liu G, Schembri F, Zhang X, Dumas YM, et al. Similarities and differences between smoking-related gene expression in nasal and bronchial epithelium. Physiol Genomics. 2010;41:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Campbell JD, Mazzilli SA, Reid ME, Dhillon SS, Platero S, Beane J, et al. The Case for a Pre-Cancer Genome Atlas (PCGA). Cancer Prev Res Phila Pa 2016;9:119–24. [DOI] [PubMed] [Google Scholar]
- 56.Kensler TW, Spira A, Garber JE, Szabo E, Lee JJ, Dong Z, et al. Transforming Cancer Prevention through Precision Medicine and Immune-oncology. Cancer Prev Res Phila Pa. 2016;9:2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.