Fecal microorganisms can enter water bodies in diverse ways, including runoff, sewage discharge, and direct fecal deposition. Once in water, the microorganisms experience conditions that are very different from intestinal habitats. The transition from host to aquatic environment may lead to rapid inactivation, some degree of persistence, or growth. Microorganisms may remain planktonic, be deposited in sediment, wash up on beaches, or attach to aquatic vegetation.
KEYWORDS: decay rate, persistence, aquatic, enteric pathogens, fecal organisms, habitat, indicator organisms, survival, water quality, waterborne pathogens
SUMMARY
Fecal microorganisms can enter water bodies in diverse ways, including runoff, sewage discharge, and direct fecal deposition. Once in water, the microorganisms experience conditions that are very different from intestinal habitats. The transition from host to aquatic environment may lead to rapid inactivation, some degree of persistence, or growth. Microorganisms may remain planktonic, be deposited in sediment, wash up on beaches, or attach to aquatic vegetation. Each of these habitats offers a panoply of different stressors or advantages, including UV light exposure, temperature fluctuations, salinity, nutrient availability, and biotic interactions with the indigenous microbiota (e.g., predation and/or competition). The host sources of fecal microorganisms are likewise numerous, including wildlife, pets, livestock, and humans. Most of these microorganisms are unlikely to affect human health, but certain taxa can cause waterborne disease. Others signal increased probability of pathogen presence, e.g., the fecal indicator bacteria Escherichia coli and enterococci and bacteriophages, or act as fecal source identifiers (microbial source tracking markers). The effects of environmental factors on decay are frequently inconsistent across microbial species, fecal sources, and measurement strategies (e.g., culture versus molecular). Therefore, broad generalizations about the fate of fecal microorganisms in aquatic environments are problematic, compromising efforts to predict microbial decay and health risk from contamination events. This review summarizes the recent literature on decay of fecal microorganisms in aquatic environments, recognizes defensible generalizations, and identifies knowledge gaps that may provide particularly fruitful avenues for obtaining a better understanding of the fates of these organisms in aquatic environments.
INTRODUCTION
Many waterborne pathogens originate in the gastrointestinal tracts of humans or other animals (the primary habitat) and enter water bodies (a secondary habitat) via fecal or sewage contamination (Fig. 1). The fate of populations of fecal microorganisms in aquatic environments is generally progression toward nonviability (frequently termed decay); however, some aquatic environments support long-term survival or growth of pathogens and commensal microorganisms shed in feces (1, 2). The trajectory (growth, stasis, or death) and rate of change in fecal-microorganism populations in secondary (extraintestinal) habitats have profound implications for our understanding of the human health risk from fecal contamination. These implications affect practices such as wastewater treatment, ambient water quality assessment, modeling of water quality, risk assessment, and management of environmental waters.
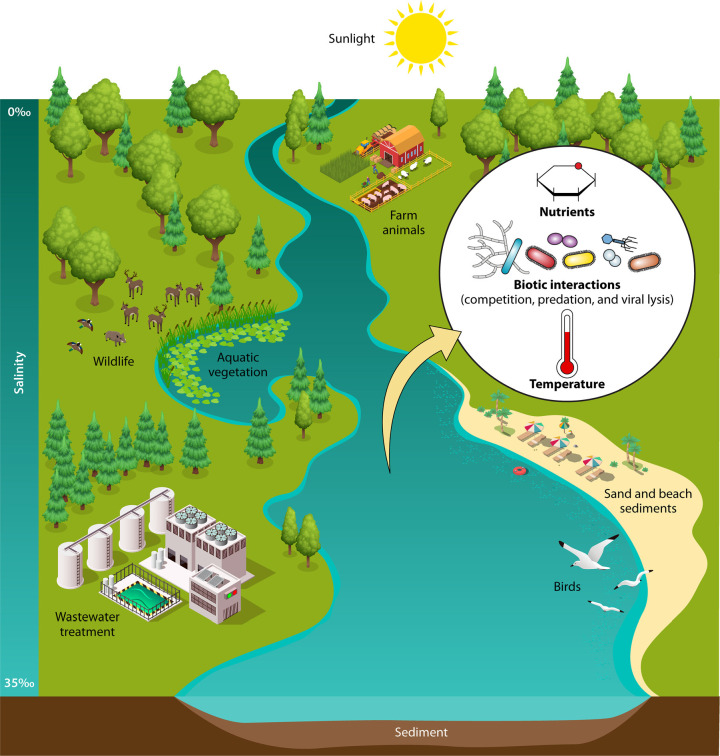
FIG 1.
A variety of habitats for FIB, MST markers, and enteric pathogens are associated with water and watersheds, including primary (e.g., gastrointestinal tracts of humans, farm animals, and wildlife) and secondary (e.g., wastewater, freshwater, and marine water) habitats. Abiotic (e.g., sunlight, nutrients, temperature, salinity, and sediments) and biotic (e.g., competition, predation, and viral lysis) environmental variables that influence microbial decay are depicted.
An important mode of transmission for enteric pathogens in human infections is the fecal-oral route, which frequently entails a transitory period in one or more secondary habitats (e.g., water, sediment, or vegetation). Fecal indicator bacteria (FIB), such as fecal coliforms, enterococci, and Escherichia coli, are used to indicate the presence and level of fecal contamination in water. Microbial source tracking (MST) markers target genes from microorganisms closely associated with human and animal hosts and are used to identify the source of fecal contamination. FIB and MST markers can be used as a general warning system in monitoring programs and as an indication of fecal contamination and the possible presence of enteric pathogens associated with host sources of fecal contamination. These fecal indicators can also help indicate which pathogens to test for based on the contamination source and can serve as surrogates for pathogens in risk assessment or as a direct measurement of human health risk if the marker is a pathogen (e.g., adenovirus). Understanding the extent to which fecal microorganisms can persist or grow in aquatic environments is essential to estimating risk from sewage spills, animal inputs, and other contamination events, yet few general principles about the fate of fecal microorganisms in water bodies have emerged from many decades of research.
The metrics used to express changes in microbial concentrations over time vary according to the subdiscipline of microbiology and author preference, which vastly complicates comparisons across studies. Decay rates can be calculated in many different ways, but all express the change in microbial concentrations over time. The terms T90 and T99 refer to the time required to reduce a microbial population by 90% or 99%, respectively, and are generally expressed in hours or days. A first-order (linear) decay equation is frequently used to describe changes in microbial densities [e.g., k (decay rate constant) = ln(C0/CT)], where C0 is the initial concentration, and CT is the concentration at time T. The more rapid the decay, the higher the decay rate. In this model, also called the Chick-Watson model, a consistent, log-linear rate of decay is assumed. Biphasic and sigmoidal curves are also observed (3). In reality, however, decay may be delayed, producing a shoulder, or the decay rate may decrease as the population decreases, producing a tailing-off effect. More complex models can also be employed to capture these variations in decay curves (3, 4).
Measurement techniques can also affect the observed decay rate. Values obtained by culture, which is a stringent and sometimes selective measure of viability, are almost always higher in a given population than those obtained by direct microscopic counts or molecular methods (e.g., quantitative PCR [qPCR]). Wherever possible, we refer to changes in microbial concentrations over time as decay rates for quantitative measurements or decay for qualitative comparisons. As decay rates increase, the rate of microbial decline increases, and survival (persistence) decreases. The term “decay” works well when a treatment is expected to increase the decay rate (e.g., UV light or the presence of predators) (see Table S1 in the supplemental material). This terminology becomes counterintuitive in a table format when a treatment is expected to decrease decay (increase persistence), such as nutrient addition (Table 1). Both decay and persistence are therefore used in this review and have essentially opposite meanings.
TABLE 1.
Effects of nutrients on bacterial persistence
| Nutrient variable(s) | Organism | Effect on persistencea | Commentb | Source of nutrients | Laboratory or field study | Reference(s) |
|---|---|---|---|---|---|---|
| Organic carbon | E. coli | + | Enhanced survival attributed to organic carbon and influence of sediment properties. | Fecal source, organic sediments | Laboratory | 81, 88 |
| E. coli | + | Artificial nutrients containing organic carbon stimulated greater E. coli growth at higher nutrient concns. | Artificial nutrients | Laboratory | 90 | |
| E. coli | + | Vegetation extract type and extract concn less influential on FIB concn than the interaction between concn and extract type. | Vegetation extracts (turf grass and leaf litter) | Laboratory | 82 | |
| Organic carbon, nitrogen, and phosphorus | E. coli | + | Addition of nutrients stimulated indicator growth in water and sediment. Effect of nutrient addition on FIB growth was minimal at lower nutrient levels than at higher nutrient levels. | Fecal source | Laboratory | 89 |
| E. coli, enterococci | + | Concn-dependent stimulation of growth. | Runoff, wastewater effluent | Laboratory and field | 83 | |
| E. coli | +/0 | Artificial nutrients stimulated growth of culturable heterotrophs, but not E. coli. | Artificial nutrients | Laboratory | 91 | |
| E. coli, enterococci, GenBac3 | + | Conducted in sediments; smaller particle size was also associated with decreased decay rate. | Sediments | Laboratory | 94 | |
| Nitrogen and phosphorus | E. coli | + | DOC, DON, and PO4-P had a major influence on FIB concns. | Sewage effluent | Field | 82 |
| E. coli, enterococci | + | Greater survival of FIB after addition of inorganic fertilizer. | Inorganic fertilizer | Field and laboratory | 84–86 | |
| Fecal coliforms | +/0 | P stimulated growth of fecal coliforms; N did not. | Artificial nutrients | Laboratory | 92 |
+, greater persistence in the presence of nutrients; 0, no effect of nutrients on persistence.
DOC, dissolved organic carbon; DON, dissolved organic nitrogen.
The literature abounds with studies of the fates of fecal viruses, bacteria, and protozoa in aquatic habitats, many of which have seemingly conflicting conclusions. A general understanding exists that intrinsic (microbial type/species and host) and extrinsic (competition and predation) biotic factors play important roles in survival, as do abiotic factors, such as UV light intensity and temperature (Fig. 1). The great variability in experimental design, measurement tools, and data analyses, however, may influence measured decay rates and study conclusions. The ability to generalize from individual studies to general principles and effective monitoring regimens, which are needed to advance efforts to improve or maintain water quality, is then hindered.
The objective of this review is to examine the literature on the inactivation, survival, and growth of fecal microorganisms, and sometimes their associated nucleic acids, in aquatic environments. Constraints to study inclusion were that they must have been conducted in a natural or simulated aquatic habitat, including submerged sediment and aquatic vegetation but excluding engineered or completely simplified habitats, such as wastewater treatment plants and drinking water. All the included studies needed to juxtapose control versus treatment (e.g., sunlight exposure versus dark controls) or two different treatments (e.g., freshwater versus marine water) and report findings in some quantitative manner, such as decay rate or log10 reduction.
EFFECTS OF ABIOTIC FACTORS ON DECAY RATES
The characteristics of secondary habitats encountered by fecal microorganisms released into aquatic ecosystems can contribute to rapid decay or extended microbial survival and growth (5–9). Decay rates in aquatic habitats are influenced by a variety of abiotic (physical and chemical) environmental factors that affect their physiological status. Here, we explore the influence of sunlight, water type, nutrients, temperature, and physical location (e.g., water column, sediment/sand, and aquatic vegetation) on the decay rates of FIB, MST markers, and various pathogens.
Sunlight
Enteric microorganisms that enter aquatic habitats are subjected to numerous environmental pressures that are absent from the gastrointestinal tract, including sunlight and associated electromagnetic radiation (e.g., UV light). UVA (315 to 400 nm) and UVB (280 to 315 nm) radiation are predominantly responsible for germicidal properties associated with ambient sunlight (10, 11). Major mechanisms of microbial inactivation by UV light include (i) UVB-induced formation of pyrimidine dimers, leading to mutations, and (ii) UVA-induced photo-oxidative damage by free radicals and reactive oxygen species (ROS) (10, 11). Photo-oxidative damage can be either exogenous or endogenous, depending on the location of free radicals and ROS. In the exogenous scenario, the source of free radicals and ROS is the natural organic matter present in the water column, whereas in the endogenous mechanism, photons absorbed by internal cellular components create damaging molecules inside the cell.
Bacteria possess many different repair mechanisms that are activated following direct DNA damage caused by UV light (e.g., photolyase-mediated repair, base/nucleotide excisions, and SOS repair) (10). In addition, global regulators (e.g., oxyR and soxRS) become activated following oxidative stress, such as indirect DNA damage caused by free radicals and ROS (12, 13). DNA repair mechanisms have been identified in a variety of proteobacteria, including FIB and many bacterial pathogens, as well as Bacteroides (10), a common target of the MST markers (2). The small genomes of viruses do not usually include genes for nucleic acid repair, although the large megavirales possess a suite of DNA repair genes (14). DNA repair genes have been found in other double-stranded DNA viruses, including fowlpox virus (15, 16) and bacteriophage T4 (10, 17). The susceptibility of Cryptosporidium and Giardia (oo)cysts to UV radiation characteristic of drinking water treatment processes is well documented (reviewed in reference 18); however, little is known about how exposure to ambient sunlight in an aquatic environment affects these protozoa. Some evidence for repair exists (19, 20), and necessary genes have been identified (21, 22).
Here, we focused on studies that investigated the effect of ambient (or simulated) sunlight directly either by including dark (or shaded) controls (4, 23–38) or by conducting experimentation at different depths (39–41). The sources of inocula ranged from laboratory-propagated strains to a variety of human (feces, sewage, septage, and a waste stabilization pond) and/or other animal (cattle manure/feces, dog feces, and meat-processing facility effluent) sources (see Table S1 in the supplemental material), and experimental design varied from indoor, laboratory-based studies (25, 42, 43) to experiments conducted outdoors (ex situ and in situ) (4, 23, 24, 26–36, 38–41, 44). Table S1 and Fig. 2 summarize the effects of sunlight on the decay of various indicator organisms and some enteric pathogens in brackish and marine water and a variety of freshwater (groundwater and lake, river, creek, wetland, lagoon, and pond water).
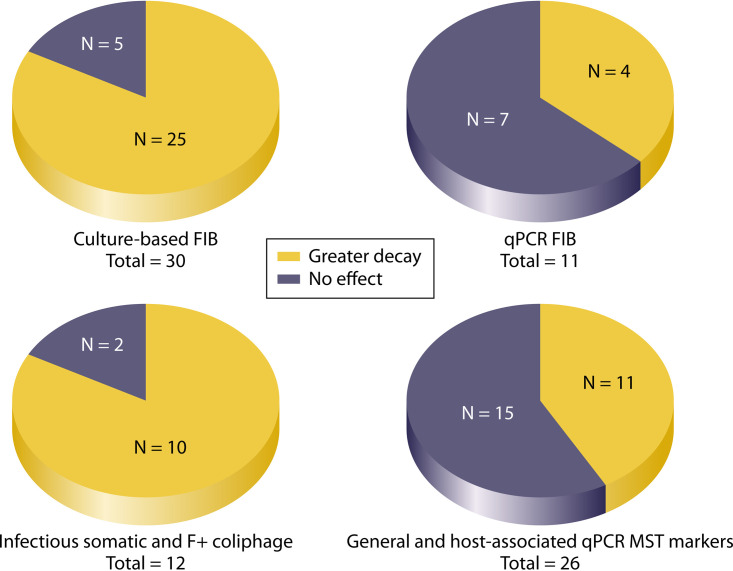
FIG 2.
Effect of sunlight exposure on culture and molecular (i.e., qPCR) measurements of FIB, infectious coliphage, and various MST markers. N represents the number of observations from all studies; in some cases, more than one species or target is included per study.
Twenty-five of 30 (83.3%) FIB analyses by culture methods found that exposure to sunlight increased decay rates (Fig. 2); however, this effect was sometimes either not statistically significant or the statistical significance of the parameter was not reported (see Table S1). Within these studies, other factors contributing to decay included temperature (29, 32, 33, 35, 38, 39), biotic interactions (4, 25, 28, 39, 45), seasonality (31, 33, 34, 41), source of FIB (4, 27), salinity/water type (26, 34), presence of sediments (25), and oxidative stress (30). In contrast, only 4 of 11 (36.4%) observations where FIB were measured by qPCR found a significant effect of sunlight (Fig. 2) (23, 24, 26, 30, 46). The few exceptions were noted for experiments that were conducted in situ (4, 28, 41). The effect of sunlight on cultured (infectious) coliphages was similar to its effect on cultured FIB, as 10 of 12 (83.3%) measurements found greater decay rates for somatic and F+ coliphage in the presence of sunlight (Fig. 2) (4, 29, 33, 34, 42–44) (see Table S1), although statistical analysis was not always reported. Collectively, these studies determined that, while the effect of sunlight is particularly significant for cultured FIB and bacteriophages, the magnitude of the effect likely depends on other parameters, such as UVB intensity and seasonality (41), the source of the fecal material (4), or the duration of exposure to the aquatic environment (28).
The effect of sunlight on decay rates of MST marker genes measured by qPCR followed the trend of decreased impact compared to the strong effects of sunlight noted on cultured microorganisms (23, 24, 26, 35, 36, 41), although several studies found accelerated decay rates for certain markers (4, 25, 28). Overall, in 11 of 26 (42.3%) instances, sunlight increased the decay rates of MST markers (Fig. 2). Sunlight significantly increased the decay rates of general Bacteroidales (GenBac3) and human-associated (HF183 and HumM2) MST markers in three of nine (33.3%) studies (4, 25, 28) (see Table S1). Interestingly, all three studies found that sunlight was a dominant factor in decay rates in the first 2 to 5 days of the study; however, the effect of the indigenous microbiota was the principal factor influencing decay rates in later stages. Decay studies conducted using animal-associated MST markers are much less numerous in the literature. Out of the five studies investigating the effect of sunlight on decay of cattle-associated MST markers (BacCow-UCD, BacR, CF 128/193, CowM2, CowM3, and Rum2Bac), one study conducted in marine water (23) reported a significant effect of sunlight on decay whereas the three conducted in freshwater and groundwater generally did not observe a similar effect (24, 35, 36) (see Table S1). The remaining study, conducted in both marine water and freshwater, reported a significant effect of sunlight on decay of CowM2 and Rum2Bac, but not CowM3 (46) (see Table S1).
Studies investigating pathogen decay are less numerous, but the discrepancy between culture and qPCR holds, with detrimental effects of sunlight on decay noted on cultured Salmonella enterica, Campylobacter jejuni, and E. coli O157:H7 (31, 38, 39), but not for S. enterica or C. jejuni measured by qPCR (24) (see Table S1). A study determining the effect of ambient sunlight on infectious Cryptosporidium parvum also noted faster decay in sunlight-exposed treatments than in the dark controls (37). Sunlight also accelerated the decay rates of cultured adenovirus type 2 (42, 43) and poliovirus type 3 (42, 44) (see Table S1). The exception was the qPCR signal for adenovirus type 2, which was not significantly affected by sunlight (24).
Studies that attempted to assess the impacts of multiple environmental variables on decay rates are more ambiguous about the detrimental effects of sunlight than the simplistic laboratory studies. While differences in study designs are likely contributing factors, it should also be noted that the germicidal effects of sunlight depend on factors such as dissolved oxygen concentrations (34, 47–51), humic acids (48, 52), and temperature and pH (48, 53, 54), as well as water depth and turbidity (55–57). In addition, other variables affecting decay that are not uniform across the studies (e.g., seasonality, salinity/water type, biotic interactions, presence of sediment, oxidative stress, inoculum source, and water composition) may interact with the observed effects of sunlight (see Table S1). The relative sensitivity to sunlight of bacteria and viruses measured by culture methods compared to those measured by qPCR is the most generalizable conclusion gained from comparison of these studies.
Water Type
The salinity and the associated ionic content of marine and estuarine waters presents a hypertonic environment for most enteric microorganisms, which can induce osmotic shock and negatively impact survival. Exposure of bacteria to waters with high salinity content induces osmoregulatory systems, leading to accumulation of compatible solutes (58–61) and differential expression of genes affecting membrane composition (58). As a result, changes in salinity may cause immediate loss of culturability due to sublethal injury, but some enteric microorganisms can adapt to survive in waters with higher salt content (62, 63). Less is known about the effect of osmotic shock on viruses, although earlier research indicated that osmotic shock can lead to rupture of viral capsids and loss of infectivity (64).
Here, we focus on studies that compared decay rates under contrasting salinity conditions. Our criteria for study inclusion required direct comparisons between different water types (9, 26, 27, 34, 45, 65–73) or within the same water type but with varied levels of salinity obtained by addition of artificial salts (74, 75). We found 18 studies conducted over the last 4 decades that investigated the effect of water type on the decay of FIB; bacterial, viral, and protozoan pathogens; coliphage; and a variety of MST markers and also met our selection criteria (Fig. 3). Since a number of these studies measured more than one microorganism and often used more than one analytical technique for the same organisms, this combined data set produced 74 unique observations or data points regarding the effect of the water type (Fig. 3). Specifically, there were 24 data points for culturable FIB (9, 27–29, 34, 45, 65, 67–72), 5 for FIB measured by qPCR (26, 46, 69), 2 for bacterial pathogens measured by qPCR (69), 4 for infectious coliphage (29, 34, 68), 7 for infectious enteric viruses (73, 75), 1 for enteric viruses measured by qPCR (65), 1 for infectious C. parvum (76), and 30 for a variety of MST marker genes (7 for general MST, 12 for human-associated, and 11 for non-human-associated) (26, 46, 65, 66, 68, 69, 71, 72, 74).
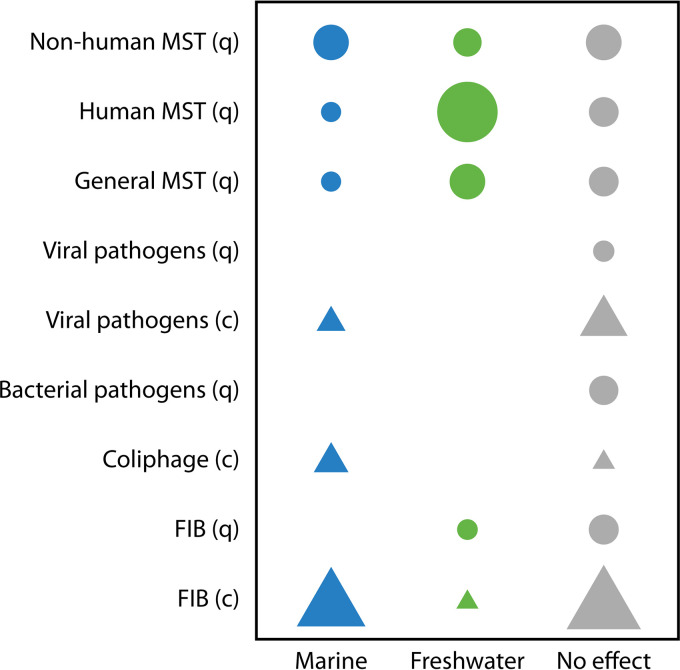
FIG 3.
Effect of water type on decay of fecal microorganisms measured by qPCR (q; circles) or culture (c; triangles). The symbols represent studies in which decay rates were significantly greater in marine water or freshwater or in which the water type had no effect. The symbol size corresponds to the number of studies with results that fall within each category (greater decay in marine water or freshwater or no effect).
The water type affected the decay rate of microorganisms in only 55.4% of all comparisons (46, 65–70, 73, 75) (Table 2). Faster decay in estuarine/marine waters was noted in approximately 32% of all comparisons (9, 27, 34, 45, 46, 66, 68, 72, 73, 76), while ∼23% of observations recorded faster decay in freshwater (Table 2) (26, 27, 46, 65, 68, 71, 74). Faster decay in estuarine/marine waters was most frequently observed for microorganisms measured by culture-dependent methods, including FIB (9, 27, 34, 45, 65, 68), coliphage (34, 68), and viral/protozoan pathogens (73, 76) (Fig. 3 and Table 2). Freshwater rarely increased the decay rates of culturable microorganisms (2.8% of observations) but more frequently increased the decay rates of molecular targets (42.1%), e.g., human MST markers (Fig. 3). The trend of faster decay in freshwater versus estuarine/marine waters was observed for FIB, pathogens, and various MST markers measured by qPCR (26, 46, 65, 68, 71, 74) (Table 2 and Fig. 3). This differential effect of water type on the persistence of culture-based versus molecular targets was statistically significant (Fisher exact test; P < 0.0001).
TABLE 2.
Summary of the influence of water type on decay rates of various microbial targets for culture versus molecular measurement strategies
| Measurement strategy | Water type with more rapid decay [no. of observations (%)] |
||
|---|---|---|---|
| Marine | Freshwater | No effect | |
| Culture based | 17 (47.2) | 1 (2.8) | 18 (50.0) |
| Molecular (qPCR based) | 7 (18.4) | 16 (42.1) | 15 (39.5) |
While the salinity level is the most apparent difference between marine water, estuarine water, and freshwater, several other factors may influence microbial decay. For example, it has been noted that predatory protozoan populations vary within and among the water types (77–80); thus, it is difficult to separate the effects of salinity and biotic interactions on decay rates. Furthermore, differences in clarity among the water types can also affect the magnitude of exposure to ambient sunlight, where more suspended organic matter in the water column can inhibit the penetration of UV radiation (56, 57).
Nutrients
The gastrointestinal tracts of animals and humans are copiotrophic environments characterized by abundant nutrient supplies; therefore, the transition to oligotrophic environments, such as aquatic habitats, is generally detrimental to the survival of enteric bacteria. Alternatively, an abundance of nutrients, such as in eutrophic aquatic systems, can facilitate proliferation or extended survival of enteric bacteria, including pathogens. The influx of nutrients into aquatic environments can originate from various sources, including urban runoff, sewage effluent, fecal material, and resuspension from sediments (81–83).
To simulate the effect of elevated nutrients in eutrophic aquatic systems, studies have employed various nutrient sources, including vegetation extracts (82), inorganic fertilizer (84–86), fecal material (87–89), and synthetic/artificial nutrients (89–91). The majority of these studies have been conducted in the laboratory and can be divided into several categories based on experimental design; some studies utilized both sediments and the overlying water column (81, 87–89), while others focused on the water column only (83, 92). Another distinguishing characteristic was the source of the bacterial inoculum, which ranged from laboratory-propagated strains (81) to organisms in feces (83, 87, 88) to environmental strains from water/sediments (83, 89, 92). Of note, most of the studies utilized nonsterilized water and sediment that contained a full complement of indigenous microbiota, including predators and competitors. Table 1 summarizes how certain nutrients (organic carbon, phosphorus, and nitrogen) influenced the decay of various indicators. It is important to note that most studies investigating the effects of nutrients focused only on changes in culturable-FIB levels, and therefore, there are few data available for other groups of interest (e.g., MST genetic markers and various pathogens).
Organic carbon is a limiting nutrient for bacteria in many environments and generally constrains the survival of enteric bacteria in aquatic environments (93). Several studies reported increased E. coli persistence after addition of organic carbon in the form of fecal material (88, 89) or organic-rich sediment (81) (Table 1). FIB decay rates were often reduced in sediment compared to the overlying water column (81, 88), and culturable E. coli and enterococci, as well as the GenBac3 MST marker, decayed more slowly in sediments with higher organic carbon levels (94). Extended survival in sediments is attributed to factors including greater access to nutrients, such as organic carbon, and protection from UV light and predators. Other factors, such as sediment properties (e.g., particle size and clay content), as well as the presence of biofilms, can also promote extended survival in sediment (81, 88, 94).
In surface waters, FIB concentrations were often correlated with organic carbon concentrations (82, 83). In particular, elevated indicator concentrations were observed when organic carbon levels were at or above 7 mg/liter (83). Other studies conducted using incremental levels of organic carbon also reported enhanced survival of E. coli in the water column (90, 91). In both studies, the positive effect of nutrient addition on E. coli varied depending on nutrient levels and/or the presence of biota (predators and competitors) (90, 91).
Both phosphorus and nitrogen are necessary for bacterial growth and metabolism, and either may be a limiting nutrient in certain aquatic environments (82, 83, 87, 89, 92). Table 1 lists several laboratory studies in which phosphorus and nitrogen were added in the form of artificial nutrients (84–86, 91, 92) or fecal matter (87–89). In addition, other studies that assessed the relationship between FIB and elevated phosphorus and nitrogen in surface waters are listed (82, 83). In general, organic and/or inorganic phosphorus addition increased the survival of fecal coliforms, enterococci, and E. coli (84–86, 89, 92). In one study comparing the effects of the addition of phosphorus in the form of fecal matter (bovine, deer, and goose), only deer feces, which had a comparatively high phosphorus content, decreased the decay rate of FIB (87).
The results of studies conducted in surface waters were similar to those of laboratory studies, finding that FIB concentrations (E. coli and enterococci) were often correlated with phosphorus concentrations (82, 83). At lower nutrient concentrations, studies have shown that the presence of biota can confound the effect of nutrient addition in the water column (89, 92), diminishing the effect of nutrients on FIB persistence. This finding is consistent with those of another laboratory-based study in which the effect of organic carbon addition on FIB persistence was concentration dependent (90).
Temperature
Enteric bacteria (indicators and pathogens alike) are mesophiles, with optimal growth temperatures within the moderate range (20°C to 40°C), consistent with the temperatures characteristic of human and other animals’ gastrointestinal tracts. In many ecosystems, temperature fluctuates on a daily and seasonal basis, presenting adaptational challenges for microbes entering their secondary habitats. While higher temperatures (within the tolerance range of a given organism) typically result in elevated metabolic activities when nutrients are abundant, survival in nutrient-poor environments tends to be prolonged at low temperatures (93).
The extended survival at lower temperatures has been attributed to various stress response mechanisms. For example, exposure of Enterococcus faecalis to lowered temperature triggers a cold shock response (95). In E. coli, a temperature downshift from 37°C to 15°C induces expression of various cold shock proteins (mostly belonging to the CspA family), leading to the adjustment of the membrane lipid composition and the ability to overcome deleterious effects of cold shock on transcription and translation (96). Less is known about temperature adaptation in enteric viruses, but available research on the λ bacteriophage suggests greater stability at lower temperatures (97–99). For example, at low temperatures, the λ bacteriophage follows a lysogenic pathway that leads to a stable prophage, but at higher temperatures, it follows a lytic cycle that results in production of phage progeny (99, 100).
Here, our criteria for study inclusion required direct comparison of decay at a minimum of two temperatures under controlled laboratory settings (25, 29, 66, 71, 74, 75, 81, 88, 101–113) or a seasonal comparison in field studies (32, 33, 35, 101, 111, 114). Temperatures reported in the individual studies ranged from 4°C to 45°C, but since 20°C is the lower bound for the optimal growth of mesophilic organisms, we binned study conditions into temperatures of ≤20°C or ≥20°C. This strategy yielded 63 data points or observations: 6 for culturable fecal coliforms (32, 33, 71, 101, 112, 114); 11 for E. coli measured by culture and qPCR (25, 29, 32, 35, 81, 88, 104, 105, 110, 111); 6 for enterococci measured by culture and qPCR (29, 35, 101, 104, 105); 4 (33, 35, 107, 112) and 3 (33, 109, 113) for infectious somatic and F+ coliphage, respectively; 4 for culturable bacterial pathogens (Salmonella enterica serovar Typhi and Shigella sonnei) (111); 8 for viral pathogens (coxsackievirus B5, echovirus type 6, norovirus, and poliovirus type 1) measured by culture and qPCR (75, 108, 111, 112); 5 for general MST markers (AllBac, BacPre1, and BacUni) (25, 66, 71, 102, 104, 114); 8 for human MST markers (BacH, BacHum, HF183, Human-Bac1, and the alpha-1,6-mannanase gene) (25, 35, 66, 71, 74, 104, 105); 7 for other animal MST markers (BacR, CowBac2, BacCow, BacCan, and PigBac2) (35, 66, 71, 104); and 1 for infectious C. parvum (115).
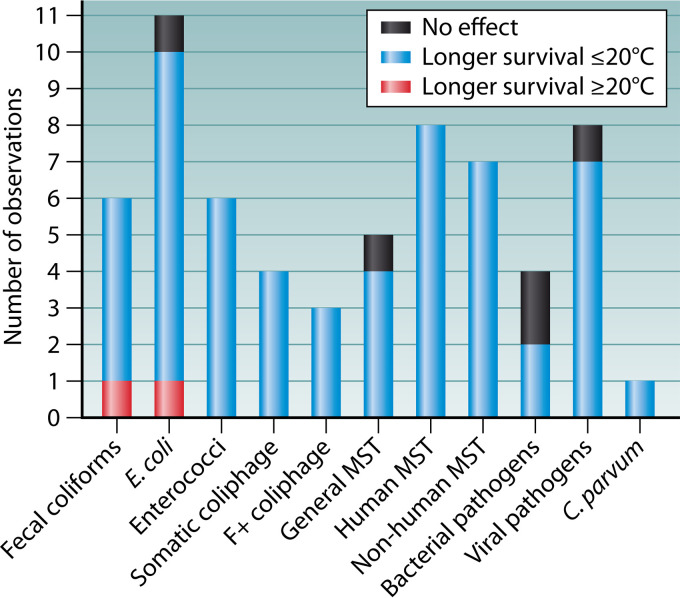
Our findings are summarized in Fig. 4. A strong trend among all the microorganisms indicated extended persistence at temperatures of ≤20°C (55/63; 87%). Only 3% (2/63) of the comparisons noted the opposite trend, while 8% (5/63) showed no effect on the decay rate. The null findings were observed for culturable E. coli, S. Typhi, S. sonnei, infectious poliovirus type 1 (111), and a general MST marker (AllBac) (25). The studies that observed no effect shared dark-only (no-sunlight) conditions, as one was an outdoor in situ study conducted at depths of 3 and 10 m (111) and the other was an indoor study without exposure to UV light (25). Extended persistence at temperatures of ≥20°C was observed in only two instances (for culturable E. coli and fecal coliforms) (101, 104), and it is more difficult to understand. However, it should be noted that in the study reporting on fecal coliforms, (i) the lengths of the experiments for the summer (≥20°C) and winter seasons were uneven (1 week versus 2 weeks, respectively), possibly confounding the findings, and (ii) enterococci persisted longer in the winter than in the summer, as would be expected (101). The study that noted extended persistence of E. coli at 20°C versus 6°C was conducted in anaerobic sediments (104), where nutrients may have been abundant enough to support growth of E. coli at the higher temperature.
FIG 4.
Effect of temperature on survival of FIB (fecal coliforms, E. coli, and enterococci); coliphage (somatic and F+); bacterial (S. Typhi and S. sonnei), viral (coxsackievirus B5, echovirus type 6, norovirus, and poliovirus type 1), and protozoan pathogens (C. parvum); and MST markers.
Alternative Habitats
The many types of microbial habitats offered by aquatic environments differ widely in terms of benefits and stressors, thereby affecting the persistence of various indicators and pathogens. Microorganisms tend to attach to particles, which can mediate relatively rapid transport to sediments (116). They may also form biofilms on aquatic vegetation or other structures (117). Although not strictly an aquatic environment, sand on beaches can provide a habitat in a sand-water continuum (118). The stressors and advantages of these various environments, and their effects on the survival of enteric microorganisms, are explored below.
Sediments.
In a rare consensus, all the studies comparing decay in sediments versus the overlying water column found extended persistence in sediments. This finding applied to culturable FIB (fecal coliforms, E. coli, and enterococci) (9, 45, 69, 85, 88, 104, 119–122), FIB quantified by qPCR (Entero1a and uidA) (69, 104), a variety of MST markers (BacCan, BacCow, BacHum, and LA35) (69, 104), and bacterial pathogens quantified by both culture and qPCR (Campylobacter coli, E. coli O157:H7, Salmonella spp., and Vibrio parahaemolyticus) (69, 120, 122). The trend was also consistent regardless of the water type (i.e., freshwater or marine water), experimental design (outdoor or indoor), or inoculum source (strains, human and other animal fecal material, or organisms indigenous to aquatic habitats). The only notable exception (no difference in decay between sediments and the overlying water column) was reported for Entero1a qPCR in an indoor study utilizing lake-derived water and sediments (104). Of note, this particular study was conducted in the dark, which potentially contributed to extended persistence in the water column (104). Other studies focused on elucidating the effects of sediment characteristics on decay, rather than direct comparisons between the two matrices. In general, smaller grain size was frequently associated with elevated nutrient content and extended survival of culturable FIB (E. coli and enterococci) (94, 121, 123, 124), FIB quantified by qPCR (Entero1a) (94), general (GenBac3) as well human-associated (HF183) MST markers (94), and a variety of bacterial pathogens (Vibrio cholerae, Salmonella enterica serovar Typhimurium, and Shigella dysenteriae) (124).
Vegetation.
Decay studies exploring the effect of aquatic vegetation on microbial decay are comparatively rare, although filamentous green algae, such as Cladophora glomerata, contribute to water quality degradation in the Great Lakes (reviewed in reference 117). Three available reports investigated the effects of (i) freshwater submerged aquatic vegetation (SAV) on the decay of enterococci (125), (ii) marine wrack on the decay of E. coli and enterococci (126), and (iii) senescing seaweeds on the survival of E. coli (127). All the studies noted extended persistence of FIB in vegetated mesocosms as opposed to unvegetated controls (125, 126) and enhanced survival in the presence of aquatic vegetation (127).
Sand.
A recent review (118) provided an exhaustive summary of our understanding of the community structure, ecology, fate, transport, and public health implications of various microbes in beach sand. Our goal here was to focus exclusively on decay studies that examined the survival of FIB (103, 106, 110, 128–135) and bacterial pathogens (130, 134, 136, 137) analyzed by culture and qPCR, coliphage (culture) (134), and various MST markers (128–130, 134, 138) in lake and marine beach sands. However, the effects of additional factors studied (e.g., moisture, grain size, temperature, interactions with the indigenous microbiota, the quantification technique, and the intrinsic properties of the microorganisms studied) varied in direction and magnitude across the studies (Table 3). In summary, the decay rates of various indicators and pathogens tended to be greater in the water column than in sediments/sands, and aquatic vegetation could augment persistence in marine and freshwater environments alike.
TABLE 3.
Summary of factors affecting persistence of various indicators and pathogens in beach sanda
| Reference | Organism type | Organism(s) | Observations | Spike source | Sand source | Analytical method |
|---|---|---|---|---|---|---|
| 106 | FIB | E. coli | Persisted over 40 days at 4, 10, or 20°C. | Environmental isolate | Wisconsin (Lake Winnebago) | Colisure |
| 103 | E. coli | Sand supports growth and provides a shield from UV radiation. | Laboratory-grown strain | Great Lakes (Bradford Beach, Lake Michigan) | Membrane filtration on mTEC | |
| 132 | E. coli, enterococci | Biotic interactions are important determinants of survival. | Environmental isolates | Florida (Hollywood Beach) | Membrane filtration on mTEC (E. coli) and mEI (enterococci) | |
| 133 | E. coli, enterococci | Moisture was a dominant factor for E. coli survival, but not for enterococci. | Laboratory-grown strains | California (Santa Monica Beach) | Membrane filtration on mTEC (E. coli) and mEI (enterococci) | |
| 131 | E. coli, enterococci | Protozoan predation and viral lysis are not important contributors to decay, but competition with indigenous bacterial populations is. | Laboratory-grown strains | Hawaii (Sand Island and Kailua Beach) | Membrane filtration on mTEC (E. coli) and mE-EIA (enterococci) | |
| 136 | C. perfringens | Greater survival in higher moisture content. | Laboratory-grown strain | Florida (Hollywood Beach) | Plating on mCP agar | |
| 134 | E. coli, enterococci, Enterococcus by qPCR | E. coli less persistent than enterococci. qPCR signal more persistent than for culturable enterococci. | Primary treated sewage | California (Lovers Points) | Membrane filtration on mTEC (E. coli) and mEI (enterococci) and by qPCR (Entero1a) | |
| 130 | E. coli, enterococci | Decay rates similar to those of bacterial pathogens. | Raw sewage | Great Lakes (Lake Superior, Duluth Boat Club Beach) | Membrane filtration on mTEC (E. coli) and mEI (enterococci) and by qPCR (Entero1a) | |
| Enterococcus by qPCR | Decay rates dissimilar to those of bacterial pathogens. | |||||
| 128 | Enterococci, Enterococcus by qPCR | Wetting of sand resulted in faster decay than with dry controls. | Seagull feces | California (Cowell Beach, Santa Cruz) | Membrane filtration on mEI (enterococci) and by qPCR (Entero1a) | |
| 135 | E. coli, enterococci, Enterococcus by qPCR, C. perfringens | C. perfringens persisted longer than E. coli and enterococci. | Raw sewage | Hawaii (Kualoa Beach) | Membrane filtration on mTEC (E. coli), mEI (enterococci) and mCP (C. perfringens), enterococci by qPCR (Entero1a). | |
| 110 | E. coli | Survival was greater in finer-grain sand and at lower temperatures. | Environmental isolate | Canada (Burlington and Marie Curtis Beaches) | Membrane filtration on mTEC | |
| 129 | E. coli, E. coli by qPCR | All FIB were detected for the duration of the experiment (∼2 mo). | Seagull feces, raw sewage | Great Lakes (Bradford Beach, Lake Michigan) | Membrane filtration on mTEC and by qPCR (uidA) | |
| Enterococci, Enterococcus by qPCR | Membrane filtration on mEI (enterococci) and by qPCR (Entero1a) | |||||
| 134 | Coliphage | Somatic, F+ | Somatic coliphage more persistent than F+. | Primary treated sewage | California (Lovers Points) | Double agar layer |
| 137 | Bacterial pathogens | E. coli O157:H7 | Grain size did not affect survival. | Environmental isolate | Great Britain (Porth Dafarch, Traeth Llydan, Porth Trecastell) | Plating on CT-SMAC agar |
| 136 | Staphylococcus aureus | Decay pattern similar to that of C. perfringens. | Environmental isolate | Florida (Hollywood Beach) | Plating on S110 agar and confirmation on MSA | |
| 134 | Salmonella spp. | Culturable pathogens more persistent than bacterial and viral indicators, but opposite was the case for the qPCR signal. | Primary treated sewage | California (Lovers Points) | Enrichment on TSB/Rappaport (MSRV) medium followed by streaking on XLD agar; confirmation on LIA/TSIA agar and by PCR (invA); also by qPCR (ttrA-ttrC) | |
| Campylobacter spp. | Enrichment in Bolton broth followed by streaking on modified Karmali agar and confirmation by PCR (16S rRNA); also by qPCR (16S rRNA) | |||||
| 130 | Campylobacter jejuni | Decayed more rapidly than FIB and MST markers. | Laboratory-grown strains | Great Lakes (Lake Superior, Duluth Boat Club Beach) | qPCR (16S rRNA) | |
| Methicillin-resistant S. aureus | Concns increased at 28% moisture. | qPCR (mecA) | ||||
| S. enterica serovar Typhimurium | Greater decay at lower moisture content (14%). | qPCR (ttrRSBCA) | ||||
| Shigella flexneri | qPCR (ipaH) | |||||
| 134 | MST markers | Human associated (HF183) | Persistence similar to that of Entero1a. | Primary treated sewage | California (Lovers Points) | qPCR |
| 130 | General (AllBac) | Concns increased at 28% moisture. | Raw sewage | Great Lakes (Lake Superior, Duluth Boat Club Beach) | qPCR | |
| Human associated (HF183) | Decay rates similar to those of bacterial pathogens. | |||||
| 128 | Seagull associated (Cat) | Greater decay than enterococci in dry sands, but similar decay in wetted sands. | Seagull feces | California (Cowell Beach, Santa Cruz) | qPCR | |
| 129 | Human associated (Lachno2, BacHum), seagull associated (Gull2) | All MST markers decayed more rapidly than FIB (culture or qPCR). | Seagull feces, raw sewage | Great Lakes (Bradford Beach, Lake Michigan) | qPCR | |
| 138 | Seagull associated (Gull2) | Remained detectable for the duration of the study (28 days). | Laboratory-grown strain | Florida (Inner Cabrillo Beach) | qPCR |
TSB, tryptic soy broth; XLD, xylose lysine deoxycholate; LIA, lysine iron agar; TSIA, triple-sugar iron agar; mTEC, membrane thermotolerant E. coli; mEI, membrane Enterococcus indoxyl-β-d-glucoside agar; mE-EIA, membrane Enterococcus esculin iron agar; mCP, membrane Clostridium perfringens; CT-SMAC agar, cefixime-tellurite-sorbitol MacConkey agar; S110 agar, Staphylococcus agar no. 110; MSA, mannitol salt agar.
EFFECTS OF BIOTIC FACTORS ON DECAY RATES
Biological characteristics of microorganisms, relationships among microbial species, and interactions within microbial communities influence FIB, MST genetic marker, and pathogen decay rates in aquatic environments. Below, we categorize these factors as extrinsic or intrinsic, where the former encompasses the effects of other microorganisms on the target microorganism and the latter refers to the physiological and genetic characteristics of the target microorganism itself.
Extrinsic Factors
Ecosystem functions at all scales are influenced by top-down (consumer-based) and bottom-up (production-based) processes. Resource scarcity increases the influence of interspecies competition on reproductive success, while predation acts as a top-down control on populations. Field observations and modeling studies in freshwater and marine environments suggest that top-down processes are more applicable in oligotrophic systems while bottom-up processes are more important in eutrophic systems (139–143). Interactions among members of microbial communities are frequently dominated by competition, which may be characterized by exploitative (use of scarce nutrients) or interference (production of antagonistic substances) mechanisms (144).
Protozoan grazing accounts for up to 90% of bacterial mortality in freshwater and marine systems (70) and is attributed to several key players, including flagellated and ciliated protozoa and amoebas in certain environments, such as soil (77, 145). Small heterotrophic flagellates contribute ∼30% of the total plankton biomass, but they are important grazers of bacteria (141, 146). Similarly, ciliated protists are important bacterial grazers, especially in highly productive environments (e.g., ponds and throughout surface marine waters) (147–149). Although lytic activity by some bacteria, e.g., Bdellovibrio, can be a factor in decay rates (150), none of the studies included here explored this facet of predation.
Unlike protozoan grazing and competition with indigenous bacteria, the contribution of viral lysis to mortality of FIB, MST genetic markers, and pathogens in fresh and marine waters is far less clear and may be system specific, as some authors have suggested that virus-mediated lysis is greater in oxygen-poor or highly productive systems (145). The only viral study included here tested the effect of bacteriophage-mediated lysis on E. coli in beach sand and found little influence (131). This result is not surprising, considering that certain threshold densities of both coliphage and its bacterial host (as well as the appropriate physiological condition of the host) are required for bacteriophage replication, conditions rarely found in ambient waters or other extraintestinal environments (151). Therefore, our focus here is on predation by bacterivorous protozoa and competition with the indigenous microbiota.
Various experimental designs have been employed to assess the effects of predation and/or competition in aquatic systems. Some studies have allowed discrimination between the effects of protozoa and those of indigenous bacteria (roughly, predation versus competition) via the use of inhibitory compounds (e.g., cycloheximide and various antibiotics) (28, 90, 122, 131, 152–156) that affect only one group, while others have excluded all indigenous microbiota by filtration (4, 27, 28, 45, 90, 102, 153, 155–161), autoclaving (25, 101, 131, 156), or “baking” of sediments (45, 122, 156), which does not allow comparison of the effects of different protozoa and bacteria but instead evaluates the effect of the total indigenous microbiota.
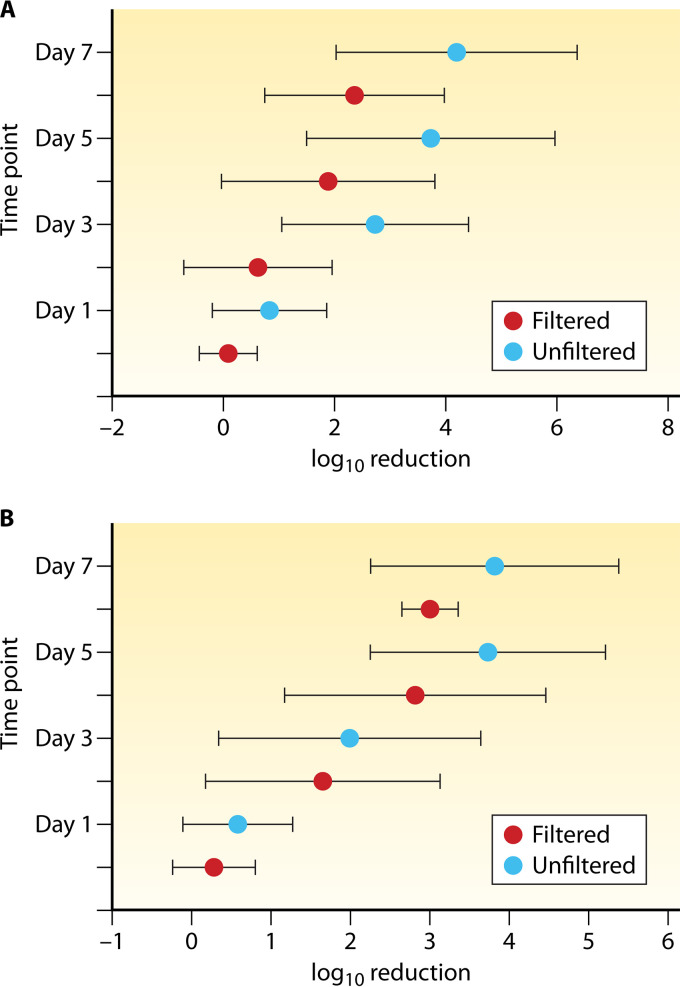
The trend toward greater decay rates in the presence of the indigenous microbiota is consistently seen in the literature and is exemplified by a collection of studies that assessed the decay of culturable E. coli and enterococci in the presence (unfiltered water) and absence (filtered water) of the indigenous biota (27, 28, 45, 90, 156). While decay rates (log10 reduction) were greater for E. coli (Fig. 5A) and enterococci (Fig. 5B) as the length of exposure to water increased, both FIB consistently decayed more rapidly in the presence of the indigenous microbiota than in their absence. Removal of all indigenous microbiota resulted in reduced decay rates of FIB (culture-based and molecular measurements) (4, 25, 27, 28, 39, 45, 90, 122, 131, 152–154, 156–158, 162–164); culturable Bacteroides fragilis and Bacteroides distasonis (101, 155); Bacteroides spp. measured by qPCR (102); various general and human-associated MST markers (GenBac3, HF183, HumBac, and HumM2) (4, 25, 28); and bacterial pathogens, including C. jejuni (162), S. enterica (122), and, in some instances, E. coli O157:H7 (122, 161, 165). Certain studies reported no effect of the indigenous microbiota (39), while others noted preferential grazing of Platophyra spp. and Colpoda spp. on E. coli O157:H7 over autochthonous bacteria (165). Another study observed higher decay rates of E. coli O157:H7 in water from an area impacted by livestock than in less impacted waters (161). Taken together with reports of preferential protozoan grazing on the C+ phenotype of E. coli O157:H7 (compared to the C− phenotype) (166) and the differential effects of motility on grazing effectiveness (122), these studies suggest that decay of this bacterial pathogen (and likely others) in aquatic habitats is influenced by individual characteristics of pathogenic strains.
FIG 5.
Decay (log10 reduction) of culturable E. coli (A) and enterococci (B). Means ± standard deviations for aggregate values from several studies are shown for each time point of exposure to the indigenous freshwater and marine microbiota (blue symbols) or to filtered water with no indigenous microbiota (red symbols) (27, 28, 45, 122, 156).
Some studies conducted in the presence of native microbes noted a temporal trend in which the decay rates of FIB and MST genetic markers increased in the later stages of the experiment (generally after 72 h of exposure) (4, 28). This time frame corresponds well to the time required for protozoan populations to adjust to an influx of prey and start feeding (167–169), and it is substantiated by a next-generation-sequencing decay study of 16S and 18S metagenomes, which indicated an increase in the relative abundance of Bodo sp. flagellates after 72 h (170).
Studies that selectively excluded either predators or bacterial competitors noted that predation is the dominant mechanism governing culturable-FIB decay in many marine waters and freshwater, although the effects of competition are also significant in most systems (45, 90, 153, 155–157, 164). The interplay between nutrient levels, predation, and competition in aquatic habitats creates complex interactions. For example, elevated levels of nutrients mitigated the effects of predation on E. coli (90). E. coli and enterococcus levels were also influenced by dissolved organic carbon and phosphorus concentrations, where decay was observed when nutrients were below a certain threshold (83). Above the threshold, FIB either grew or appeared to be in a steady state, which the authors attributed to predator-prey oscillations (83). Lastly, it is important to point out that rates of predation on FIB, MST genetic markers, and bacterial pathogens in environmental waters are influenced by various factors, including temperature (101, 102, 155, 157), location (45, 122, 156), water type (27, 28), nutrient availability (83, 90), source (4, 27), prey characteristics (39, 122, 154, 158, 163, 166), and predator/prey densities (157, 158) (Table 4).
TABLE 4.
Extrinsic biotic factors that influence decay rates in aquatic environments
| Extrinsic factor | Modifying factor | Organism/indicator | Notes | References |
|---|---|---|---|---|
| Protozoan predation | Temp | Culturable E. coli, enterococci, B. fragilis, B. distasonis, and Bacteroides spp. measured by qPCR | Higher decay rate at warmer temps coinciding with increased numbers of protozoan grazers. | 101, 102, 155, 157 |
| Prey characteristics | Culturable E. coli, E. faecalis, Staphylococcus epidermidis, E. coli O157:H7, Klebsiella pneumoniae, S. enterica | Higher decay rates for larger allochthonous organisms (e.g., E. coli) than for smaller autochthonous organisms; higher decay rates for Gram-negative bacteria than for Gram-positive bacteria; decay rates also affected by motility and virulence factors. | 39, 122, 154, 158, 163, 166 | |
| Predator/prey densities | Culturable E. coli, E. faecalis | Decay rates were positively correlated with bacterial densities. | 157, 158 | |
| Indigenous microbiota (competition + predation) | Location (water/sediment) | Culturable E. coli, E. faecalis, E. coli O157:H7, S. enterica | Higher decay rate of FIB and pathogens in the water column than in sediment. | 45, 122, 156 |
| Water type (fresh/marine) | Culturable, E. coli, enterococci | Greater decay of FIB in marine water than in freshwater, but the effect of indigenous microbiota was greater in freshwater than in marine water. | 27, 28 | |
| Nutrients | Culturable E. coli | Competition and predation increased E. coli decay rates, but higher nutrient levels mitigated these effects. | 83, 90 | |
| Source | E. coli, enterococci (by culture and qPCR), general and human-associated MST markers | FIB (from cattle manure and septage) and FIB/MST markers from septage decayed at a lower rate than FIB and MST markers from sewage and human feces. | 4, 27 |
Several studies investigated the effect of the indigenous aquatic microbiota on microorganisms other than bacteria (e.g., bacteriophages and C. parvum oocysts) in marine water and freshwater (159, 160), albeit with mixed results. While the decay rates of C. parvum were greater in the presence of the indigenous microbiota, suggesting that biotic interactions play a role in its survival (160), the presence of indigenous microbiota increased the decay rates of enterococcal bacteriophages (159), but not somatic and F+ coliphages or GB-124, a bacteriophage that infects Bacteroides spp. (4). While laboratory feeding experiments suggest that protozoan predators do feed on both of these groups under controlled conditions (171, 172), additional work is needed to verify their role under ambient conditions.
In marine and freshwater sediments and sands, removal of all indigenous microbiota generally extended the survival of various culturable FIB (fecal coliforms, E. coli, and enterococci) (28, 152, 156), but studies investigating isolated effects of either predation or competition generally reported that while predation typically increases decay rates, competition appears to be the main driver of decay (28, 131, 156). It is noteworthy that the detrimental effects of competition can also vary by FIB type, since the presence of indigenous bacteria led to greater decay of E. coli than of E. faecalis in water and sediment microcosms (156) and in beach sands (131).
No data exist to date regarding the effect of the indigenous microbiota (and associated predation/competition interactions) on the decay rates of genetic MST markers, bacteriophages, and viral or protozoan pathogens in sands and sediments, but several studies have addressed the effect of predation on the decay rates of bacterial pathogens. In one study, neither removal of all indigenous microbiota nor addition of the predatory protozoan Tetrahymena pyriformis had any effect on the survival of E. coli O157:H7 (156). On the other hand, addition of the same protozoan predator increased the decay rates of S. enterica (122).
Intrinsic Factors
Above, we explored unifying trends from research findings that allow some generalizations of the effects of environmental factors on microbial decay in aquatic environments. For example, in general, exposure to sunlight increases decay rates, particularly for culturable bacteria (Fig. 2). However, for each environmental factor considered thus far, exceptions have been noted. While some of these exceptions may be attributable to differences in experimental design or environmental conditions, others are doubtless influenced by the intrinsic characteristics of the bacterial population(s) in question. The gut microbiota is shaped by the selective pressures exerted by the varied physiologies, diets, and life histories of their animal hosts, which result in different subpopulations of indicators and pathogens in the gastrointestinal tract of each host. In addition, a myriad of waste collection systems (e.g., wastewater treatment plants, septic tanks, manure pits, and lagoons) present different sets of environmental stressors to microbial populations and therefore may select for different subpopulations used in decay experiments.
The diversity and variability of fecal microbial populations, even within a species, further complicate comparison of the effects of a given environmental stressor on pathogens, indicators, and MST markers. Because they are used in a regulatory context worldwide, the persistence of E. coli is frequently compared to that of enterococci. While E. coli is a (nominal) species with great genetic diversity (173), the enterococci include the entire genus Enterococcus, which contains some 36 species that are differentially distributed among various hosts and environmental sources (1). Furthermore, culturable enterococci are defined by the growth of characteristic colonies on selective differential media, providing even greater potential for phylogenetic and phenotypic diversity within the group. E. coli also contains great genetic diversity; while all the strains share a suite of core genes, nearly 80% of the genome may differ among strains (173). Thus, any comparison of decay rates between E. coli and Enterococcus spp. is fraught with potential pitfalls depending on the groups, species, strains, and/or sources used in the study.
Here, we focus on studies that compared the decay rates of different microbes under the same conditions in order to ensure that they experienced the same stressors and that factors intrinsic to the microorganism were driving any observed differences. In the first case, where we considered the source of microorganisms, we included only those studies that directly compared the decay rates of microorganisms from at least two discrete sources. Only general fecal indicators were included, and several studies that mixed fecal sources prior to inoculation were excluded (23, 24, 35, 71, 104, 174). Lastly, whenever possible, we included comparisons with organisms isolated from water column, soil, and sediments (9, 87, 101). Depending on the study design, decay was assessed in both the water column and sediments (9, 87, 121, 175) or in only one of these matrices (4, 27, 29, 36, 101, 129, 176).
In these studies, decay of E. coli and enterococci from cattle, bovine, deer, goose, and ovine feces was considerably slower than that of organisms originating from sewage (27, 175) or human feces (36, 176). In contrast, FIB from dog (9) and seagull (121, 129) feces decayed more rapidly than those from sewage and human feces. FIB isolated from environmental water, soil, and sediments typically decayed more slowly than FIB from sewage (9) or organisms originating from dog, bovine, deer, or goose feces (9, 87). Two studies comparing the decay rates of FIB from primary (human feces) and postprimary (raw and treated wastewater and septage) sources found that organisms from septage decayed more slowly than organisms from feces and raw wastewater (4) while there was no difference in decay for FIB derived from raw versus treated wastewater (29). The source of the inoculum also affected the bacterial response to the environmental stressors, as the decay rate of E. coli from cattle feces, but not human feces, was significantly higher under light than under dark conditions (36).
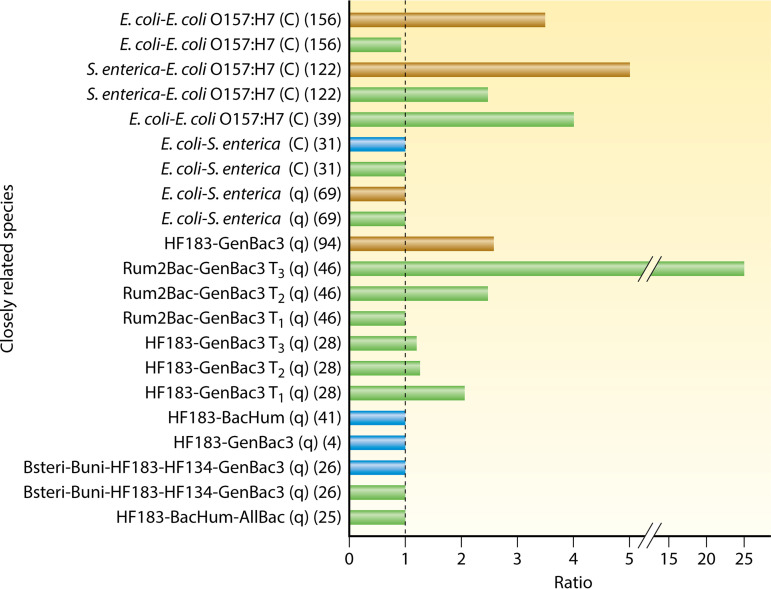
To further test this assertion, we compared the reported decay rates of closely related species (e.g., E. coli to E. coli O157:H7 or Salmonella spp. and various MST markers targeting Bacteroides spp. or Bacteroidales). We focused only on studies or specific treatments within the study that attempted to simulate environmental conditions (4, 25, 26, 28, 31, 39, 41, 46, 69, 94, 122, 156) (Fig. 6). These conditions included exposure to ambient or artificial sunlight and the presence of a full complement of indigenous aquatic microbiota, while it excluded any data where the water and sediment compositions were artificially altered (including addition of nutrients and other chemicals [e.g., cycloheximide or antibiotics], autoclaving and/or baking of water and sediments, and filter sterilization of water). Furthermore, comparisons were made only between analogous measurement techniques (e.g., culture based to culture based or qPCR to qPCR). Since viruses may have seven different types of genomes (DNA/RNA, single stranded [ss]/double stranded [ds], positive sense/negative sense, and reverse-transcribing ssRNA/dsDNA) and the presence of a membrane, as well as a myriad of different capsid proteins that affect their decay outside the host (177), they were not included in this comparison.
FIG 6.
Decay ratio of closely related species in marine water (blue bars), freshwater (green bars), and sands/sediments (brown bars). The dashed line represents a ratio of 1 (no difference in decay rates); values of >1 indicate that the first target decayed more rapidly than the second. Citations for individual studies are provided in parentheses (including references 4, 25, 26, 28, 31, 39, 41, 46, 69, 94, 122, and 156). C, culture-based measurements; q, molecular (e.g., qPCR) measurements; Tn, measurements at multiple time points within a study.
The results of this analysis revealed that the decay rates of even closely related species or strains were frequently not comparable (Fig. 6). For example, in one study, nonpathogenic E. coli decayed faster than E. coli O157:H7 in freshwater during winter (39), while another freshwater study conducted across three seasons in a subtropical climate found a similar trend of E. coli decay being faster than that of E. coli O157:H7 in sediments but no difference in the water column (156) (Fig. 6). Another study comparing the decay rates of S. enterica and E. coli O157:H7 in freshwater and sediments noted more rapid decay of S. enterica in both matrices (122). Other studies comparing the decay rates of nonpathogenic E. coli and S. enterica in marine water and freshwater found no difference between the two, regardless of whether they were measured by culture (31) or qPCR (69).
Observations on the decay of various MST markers measured by qPCR are more straightforward. Most of the studies reported no difference in decay rates (4, 25, 26, 41), and those that did noted that general MST markers (e.g., GenBac3) decayed more slowly than the host-associated subset (e.g., HF183 and Rum2Bac) (28, 46, 94) (Fig. 6). This finding is not surprising, considering that general MST markers also target some environmental Bacteroidales (178, 179), a subset of the order known to persist longer than fecal-associated members (101). Lastly, the decay ratio of MST markers (and likely other, related species) is influenced by the time elapsed since the “pollution event” or, within the studies, since the start of the experiment. For example, the decay ratio between Rum2Bac and GenBac3 (46) increased with time, but the ratio between HF183 and GenBac3 decreased with time (28). As evidenced by the examples above, the differential decay of closely related taxa is one reason that generalizations about microbial persistence in aquatic environments are problematic.
CONCLUSIONS
After more than 30 years of research investigating the decay rates of various fecal indicators and enteric pathogens in aquatic habitats, there has been a progression from simplified laboratory-based experimental designs to more complex in situ systems that better mimic environmental conditions, including experimental designs that include predator and competitor populations. Similarly, we have seen movement toward more realistic sources of microbial inocula (e.g., various primary and postprimary sources or environmental isolates) as opposed to relying on a laboratory-derived strain(s). Gaining a better understanding of the behavior of these organisms in their secondary habitats is essential for accurate evaluation of predictive relationships between indicators and pathogens, which could have profound implications for public health and water management strategies.
The literature on decay rates of FIB, enteric pathogens, and MST genetic markers in environmental waters indicates that the effects of environmental factors on the decay of the microorganisms varies according to the microbial species or group and how it is measured. Sunlight is a good example, as it consistently increased the decay rate of culturable indicators and pathogens but much less frequently affected the decay of microbes measured by qPCR. The effect of water type (saline versus fresh water) is similarly influenced by the measurement method, as decay rates of fecal microorganisms measured by culture tend to be greater in salt water, while those measured by qPCR tend to be greater in freshwater. Another consistent observation is that protozoan grazing and competition with indigenous bacteria lead to increased decay rates of fecal bacteria, although most studies have been conducted on FIB. However, additional research is needed to further elucidate the interplay between the effects of nutrients and intrinsic and extrinsic biotic interactions and to better understand the effects of biotic factors on MST markers and pathogens. Thus, what might seem like a simple question of teasing out the generalizable effects of the environment on microbial decay can rapidly become a complex problem. Lastly, currently accepted generalizations regarding the effects of temperature and physical location (soil/sediments/water column) on the decay rates of indicators and enteric pathogens may be defensible, since there was a general consensus in the literature concerning extended survival at lower temperatures and in sediments/soils (compared to the water column), irrespective of the organism or measurement strategy.
Future decay studies are needed to broaden our general knowledge about the decay rates of pathogens and their relationship to various indicator decay rates. Furthermore, future studies should evaluate and report the statistical significance (or lack thereof) of these relationships and provide the concentration data on a per sample basis to enable meta-analyses across multiple studies, which may yield a more robust assessment of relationships between indicator and pathogen decay rates.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kate Field and Jill Stewart for valuable discussion of this topic, John Griffith and Steve Weisberg for their support of the decay studies, and Orin C. Shanks and Brian R. McMinn for their help with editing the manuscript.
The U.S. Environmental Protection Agency through its Office of Research and Development funded and managed the research described here. It has been subjected to the Agency’s administrative review and approved for publication.
The views expressed in this article are those of the authors and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Biographies

Asja Korajkic, Ph.D., received her doctoral degree in Dr. Harwood’s laboratory at the University of South Florida. She was a postdoctoral fellow at the U.S. Environmental Protection Agency from 2011 to 2015 prior to becoming a research microbiologist with the same agency. She has published in a number of peer-reviewed journals and has presented at numerous national and international conferences. Her earlier research focused on studying microbial water quality at Florida beaches by detecting and quantifying FIB, MST markers, and viral pathogens. More recently, she expanded her research focus to include development and evaluation of concentration methods for pathogenic viruses and bacteriophages, as well as studies of the ecology of these two viral groups in the aquatic habitats. Her special interest is investigating the fate and transport of FIB, MST markers, and pathogens in aquatic habitats and elucidating the effects various biotic and abiotic factors have on their survival.

Pauline Wanjugi, Ph.D., is a Research Scientist II in the Environmental Biology Laboratory at the Wadsworth Center, New York State Department of Health. She has an extensive research background in the assessment of biotic and abiotic factors influencing the survival of fecal indicator organisms in surface waters. At the Wadsworth Center, she is involved in the detection and enumeration of fecal-associated contamination sources in surface waters, as well as assessment of Legionella in premise plumbing and cooling towers. She is also doing research to develop rapid methods for detection of Legionella in environmental samples to decrease response times to potential risks and thereby better protect public health.

Lauren Brooks, Ph.D., is an Assistant Professor in the Biology Department at Utah Valley University. Her dissertation focused on exploring the ways in which experimental design and mathematical modeling could impact and ultimately improve decay rate estimates for fecal indicators. Since finishing her Ph.D., she has broadened her research interests to include investigating environmental sources and sinks of antibiotic resistance genes and how they are transmitted to pathogens. She is currently continuing this research with undergraduate student researchers while teaching courses in biology and microbiology.

Yiping Cao, Ph.D., trained in Molecular Microbiology (Ph.D.) and Applied Statistics (M.A.) at the University of California, Santa Barbara; in Environmental Engineering (M.S.) at California State University, Fullerton; and in Environmental Chemistry (B.S.) at Nanjing University. She was a senior scientist at the Southern California Coastal Water Research Project Authority and is currently the Chief Technology Officer at Source Molecular. She has published over 40 peer-reviewed articles on subjects ranging from public health to statistical modeling, bioremediation, microbial community responses to environmental stress, microbial source tracking, and advanced molecular technology. In the past decade, she conducted extensive research on fecal contamination in urban coastal environments, where the fate and transport of fecal microbes are critical to accurate interpretation of microbial water quality, to the design and maintenance of natural treatment systems, and to the assessment of public health risk. This led to the desire for a synthesis of the literature on the topic.

Valerie J. Harwood, Ph.D., completed the Ph.D. program at Old Dominion University and Eastern Virginia Medical School. She was an Assistant Professor at the University of North Florida from 1995 to 1998 and has been a faculty member at the University of South Florida (USF) since 1998. She is a Professor in and Chair of the Department of Integrative Biology at USF. She has published over 110 peer-reviewed articles on subjects including hyperthermophile biochemistry, the ecology of Vibrio spp., antibiotic-resistant bacteria in environmental waters, microbial source tracking, and the persistence of fecal indicator bacteria in aquatic habitats and their relationship to pathogens. The fate of fecal bacteria in environmental waters has multifaceted implications for the connections between indicator organisms, pathogens, and human health. Discussion with colleagues led to the idea of pulling the exhaustive but disconnected literature on the subject into a coherent narrative, from which general principles and knowledge gaps could be identified.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MMBR.00005-19.
REFERENCES
- 1.Byappanahalli MN, Nevers MB, Korajkic A, Staley ZR, Harwood VJ. 2012. Enterococci in the environment. Microbiol Mol Biol Rev 76:685–706. doi: 10.1128/MMBR.00023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harwood VJ, Staley C, Badgley BD, Borges K, Korajkic A. 2014. Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol Rev 38:1–40. doi: 10.1111/1574-6976.12031. [DOI] [PubMed] [Google Scholar]
- 3.Geeraerd AH, Valdramidis VP, Van Impe JF. 2005. GInaFiT, a freeware tool to assess non-log-linear microbial survivor curves. Int J Food Microbiol 102:95–105. doi: 10.1016/j.ijfoodmicro.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 4.Wanjugi P, Sivaganesan M, Korajkic A, Kelty CA, McMinn B, Ulrich R, Harwood VJ, Shanks OC. 2016. Differential decomposition of bacterial and viral fecal indicators in common human pollution types. Water Res 105:591–601. doi: 10.1016/j.watres.2016.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tymensen L, Booker CW, Hannon SJ, Cook SR, Zaheer R, Read R, McAllister TA. 2017. Environmental growth of enterococci and Escherichia coli in feedlot catch basins and a constructed wetland in the absence of fecal input. Environ Sci Technol 51:5386–5395. doi: 10.1021/acs.est.6b06274. [DOI] [PubMed] [Google Scholar]
- 6.Jang J, Hur HG, Sadowsky MJ, Byappanahalli MN, Yan T, Ishii S. 2017. Environmental Escherichia coli: ecology and public health implications: a review. J Appl Microbiol 123:570–581. doi: 10.1111/jam.13468. [DOI] [PubMed] [Google Scholar]
- 7.Weidhaas J, Mantha S, Hair E, Nayak B, Harwood VJ. 2015. Evidence for extraintestinal growth of bacteroidales originating from poultry litter. Appl Environ Microbiol 81:196–202. doi: 10.1128/AEM.02354-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staley C, Dunny GM, Sadowsky MJ. 2014. Environmental and animal-associated enterococci. Adv Appl Microbiol 87:147–186. doi: 10.1016/B978-0-12-800261-2.00004-9. [DOI] [PubMed] [Google Scholar]
- 9.Anderson KL, Whitlock JE, Harwood VJ. 2005. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl Environ Microbiol 71:3041–3048. doi: 10.1128/AEM.71.6.3041-3048.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goosen N, Moolenaar GF. 2008. Repair of UV damage in bacteria. DNA Repair 7:353–379. doi: 10.1016/j.dnarep.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Nelson KL, Boehm AB, Davies-Colley RJ, Dodd MC, Kohn T, Linden KG, Liu Y, Maraccini PA, McNeill K, Mitch WA, Nguyen TH, Parker KM, Rodriguez RA, Sassoubre LM, Silverman AI, Wigginton KR, Zepp RG. 2018. Sunlight-mediated inactivation of health-relevant microorganisms in water: a review of mechanisms and modeling approaches. Environ Sci Process Impacts 20:1089–1122. doi: 10.1039/c8em00047f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imlay JA. 2013. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Storz G, Hengge-Aronis R. 2000. Oxidative stress, p 47–59. In Storz G, Hengge-Aronis R (ed), Bacterial stress responses. American Society for Microbiology, Washington, DC. [Google Scholar]
- 14.Blanc-Mathieu R, Ogata H. 2016. DNA repair genes in the Megavirales pangenome. Curr Opin Microbiol 31:94–100. doi: 10.1016/j.mib.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Srinivasan V, Schnitzlein WM, Tripathy DN. 2001. Fowlpox virus encodes a novel DNA repair enzyme, CPD-photolyase, that restores infectivity of UV light-damaged virus. J Virol 75:1681–1688. doi: 10.1128/JVI.75.4.1681-1688.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srinivasan V, Tripathy DN. 2005. The DNA repair enzyme, CPD-photolyase restores the infectivity of UV-damaged fowlpox virus isolated from infected scabs of chickens. Vet Microbiol 108:215–223. doi: 10.1016/j.vetmic.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 17.Rastogi RP, Richa Kumar A, Tyagi MB, Sinha RP. 2010. Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J Nucleic Acids 2010:592980. doi: 10.4061/2010/592980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hijnen WA, Beerendonk EF, Medema GJ. 2006. Inactivation credit of UV radiation for viruses, bacteria and protozoan (oo)cysts in water: a review. Water Res 40:3–22. doi: 10.1016/j.watres.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 19.Belosevic M, Craik SA, Stafford JL, Neumann NF, Kruithof J, Smith DW. 2001. Studies on the resistance/reactivation of Giardia muris cysts and Cryptosporidium parvum oocysts exposed to medium-pressure ultraviolet radiation. FEMS Microbiol Lett 204:197–203. doi: 10.1111/j.1574-6968.2001.tb10885.x. [DOI] [PubMed] [Google Scholar]
- 20.Morita S, Namikoshi A, Hirata T, Oguma K, Katayama H, Ohgaki S, Motoyama N, Fujiwara M. 2002. Efficacy of UV irradiation in inactivating Cryptosporidium parvum oocysts. Appl Environ Microbiol 68:5387–5393. doi: 10.1128/aem.68.11.5387-5393.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rochelle PA, Fallar D, Marshall MM, Montelone BA, Upton SJ, Woods K. 2004. Irreversible UV inactivation of Cryptosporidium spp. despite the presence of UV repair genes. J Eukaryot Microbiol 51:553–562. doi: 10.1111/j.1550-7408.2004.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 22.Rochelle PA, Upton SJ, Montelone BA, Woods K. 2005. The response of Cryptosporidium parvum to UV light. Trends Parasitol 21:81–87. doi: 10.1016/j.pt.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 23.Bae S, Wuertz S. 2009. Rapid decay of host-specific fecal Bacteroidales cells in seawater as measured by quantitative PCR with propidium monoazide. Water Res 43:4850–4859. doi: 10.1016/j.watres.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 24.Bae S, Wuertz S. 2012. Survival of host-associated bacteroidales cells and their relationship with Enterococcus spp., Campylobacter jejuni, Salmonella enterica serovar Typhimurium, and adenovirus in freshwater microcosms as measured by propidium monoazide-quantitative PCR. Appl Environ Microbiol 78:922–932. doi: 10.1128/AEM.05157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dick LK, Stelzer EA, Bertke EE, Fong DL, Stoeckel DM. 2010. Relative decay of Bacteroidales microbial source tracking markers and cultivated Escherichia coli in freshwater microcosms. Appl Environ Microbiol 76:3255–3262. doi: 10.1128/AEM.02636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Green HC, Shanks OC, Sivaganesan M, Haugland RA, Field KG. 2011. Differential decay of human faecal Bacteroides in marine and freshwater. Environ Microbiol 13:3235–3249. doi: 10.1111/j.1462-2920.2011.02549.x. [DOI] [PubMed] [Google Scholar]
- 27.Korajkic A, McMinn BR, Harwood VJ, Shanks OC, Fout GS, Ashbolt NJ. 2013. Differential decay of enterococci and Escherichia coli originating from two fecal pollution sources. Appl Environ Microbiol 79:2488–2492. doi: 10.1128/AEM.03781-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korajkic A, McMinn BR, Shanks OC, Sivaganesan M, Fout GS, Ashbolt NJ. 2014. Biotic interactions and sunlight affect persistence of fecal indicator bacteria and microbial source tracking genetic markers in the upper Mississippi river. Appl Environ Microbiol 80:3952–3961. doi: 10.1128/AEM.00388-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noble RT, Lee IM, Schiff KC. 2004. Inactivation of indicator micro-organisms from various sources of faecal contamination in seawater and freshwater. J Appl Microbiol 96:464–472. doi: 10.1111/j.1365-2672.2004.02155.x. [DOI] [PubMed] [Google Scholar]
- 30.Sassoubre LM, Nelson KL, Boehm AB. 2012. Mechanisms for photoinactivation of Enterococcus faecalis in seawater. Appl Environ Microbiol 78:7776–7785. doi: 10.1128/AEM.02375-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinton L, Hall C, Braithwaite R. 2007. Sunlight inactivation of Campylobacter jejuni and Salmonella enterica, compared with Escherichia coli, in seawater and river water. J Water Health 5:357–365. doi: 10.2166/wh.2007.031. [DOI] [PubMed] [Google Scholar]
- 32.Sinton LW, Davies-Colley RJ, Bell RG. 1994. Inactivation of enterococci and fecal coliforms from sewage and meatworks effluents in seawater chambers. Appl Environ Microbiol 60:2040–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sinton LW, Finlay RK, Lynch PA. 1999. Sunlight inactivation of fecal bacteriophages and bacteria in sewage-polluted seawater. Appl Environ Microbiol 65:3605–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinton LW, Hall CH, Lynch PA, Davies-Colley RJ. 2002. Sunlight inactivation of fecal indicator bacteria and bacteriophages from waste stabilization pond effluent in fresh and saline waters. Appl Environ Microbiol 68:1122–1131. doi: 10.1128/aem.68.3.1122-1131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sokolova E, Astrom J, Pettersson TJ, Bergstedt O, Hermansson M. 2012. Decay of Bacteroidales genetic markers in relation to traditional fecal indicators for water quality modeling of drinking water sources. Environ Sci Technol 46:892–900. doi: 10.1021/es2024498. [DOI] [PubMed] [Google Scholar]
- 36.Walters SP, Field KG. 2009. Survival and persistence of human and ruminant-specific faecal Bacteroidales in freshwater microcosms. Environ Microbiol 11:1410–1421. doi: 10.1111/j.1462-2920.2009.01868.x. [DOI] [PubMed] [Google Scholar]
- 37.King BJ, Hoefel D, Daminato DP, Fanok S, Monis PT. 2008. Solar UV reduces Cryptosporidium parvum oocyst infectivity in environmental waters. J Appl Microbiol 104:1311–1323. doi: 10.1111/j.1365-2672.2007.03658.x. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez S, Araujo R. 2012. Effect of environmental parameters on the inactivation of the waterborne pathogen Campylobacter in a Mediterranean river. J Water Health 10:100–107. doi: 10.2166/wh.2011.044. [DOI] [PubMed] [Google Scholar]
- 39.Jenkins MB, Fisher DS, Endale DM, Adams P. 2011. Comparative die-off of Escherichia coli O157:H7 and fecal indicator bacteria in pond water. Environ Sci Technol 45:1853–1858. doi: 10.1021/es1032019. [DOI] [PubMed] [Google Scholar]
- 40.Maraccini PA, Mattioli MC, Sassoubre LM, Cao Y, Griffith JF, Ervin JS, Van De Werfhorst LC, Boehm AB. 2016. Solar inactivation of enterococci and Escherichia coli in natural waters: effects of water absorbance and depth. Environ Sci Technol 50:5068–5076. doi: 10.1021/acs.est.6b00505. [DOI] [PubMed] [Google Scholar]
- 41.Mattioli MC, Sassoubre LM, Russell TL, Boehm AB. 2017. Decay of sewage-sourced microbial source tracking markers and fecal indicator bacteria in marine waters. Water Res 108:106–114. doi: 10.1016/j.watres.2016.10.066. [DOI] [PubMed] [Google Scholar]
- 42.Love DC, Silverman A, Nelson KL. 2010. Human virus and bacteriophage inactivation in clear water by simulated sunlight compared to bacteriophage inactivation at a southern California beach. Environ Sci Technol 44:6965–6970. doi: 10.1021/es1001924. [DOI] [PubMed] [Google Scholar]
- 43.Silverman AI, Peterson BM, Boehm AB, McNeill K, Nelson KL. 2013. Sunlight inactivation of human viruses and bacteriophages in coastal waters containing natural photosensitizers. Environ Sci Technol 47:1870–1878. doi: 10.1021/es3036913. [DOI] [PubMed] [Google Scholar]
- 44.Silverman AI, Nguyen MT, Schilling IE, Wenk J, Nelson KL. 2015. Sunlight inactivation of viruses in open-water unit process treatment wetlands: modeling endogenous and exogenous inactivation rates. Environ Sci Technol 49:2757–2766. doi: 10.1021/es5049754. [DOI] [PubMed] [Google Scholar]
- 45.Korajkic A, Wanjugi P, Harwood VJ. 2013. Indigenous microbiota and habitat influence Escherichia coli survival more than sunlight in simulated aquatic environments. Appl Environ Microbiol 79:5329–5337. doi: 10.1128/AEM.01362-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Korajkic A, McMinn BR, Ashbolt NJ, Sivaganesan M, Harwood VJ, Shanks OC. 2019. Extended persistence of general and cattle-associated fecal indicators in marine and freshwater environment. Sci Total Environ 650:1292–1302. doi: 10.1016/j.scitotenv.2018.09.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Craggs RJ, Sukias JP, Tanner CT, Davies-Colley RJ. 2004. Advanced pond system for dairy-farm effluent treatment. New Zealand J Agricultural Res 47:449–460. doi: 10.1080/00288233.2004.9513613. [DOI] [Google Scholar]
- 48.Curtis TP, Mara DD, Silva SA. 1992. Influence of pH, oxygen, and humic substances on ability of sunlight to damage fecal coliforms in waste stabilization pond water. Appl Environ Microbiol 58:1335–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davies-Colley RJ, Craggs RJ, Nagels JW. 2003. Disinfection in a pilot-scale “advanced” pond system (APS) for domestic sewage treatment in New Zealand. Water Sci Technol 48:81–87. doi: 10.2166/wst.2003.0091. [DOI] [PubMed] [Google Scholar]
- 50.Davies-Colley RJ, Donnison AM, Speed DJ. 1997. Sunlight wavelengths inactivating faecal indicator microorganisms in waste stabilisation ponds. Water Sci Technol 35:219–225. doi: 10.2166/wst.1997.0737. [DOI] [Google Scholar]
- 51.Davies-Colley RJ, Donnison AM, Speed DJ, Ross CM, Nagels JW. 1999. Inactivation of faecal indicator microorganisms in waste stabilisation ponds: Interactions of environmental factors with sunlight. Water Res 33:1220–1230. doi: 10.1016/S0043-1354(98)00321-2. [DOI] [Google Scholar]
- 52.Davies CM, Evison LM. 1991. Sunlight and the survival of enteric bacteria in natural waters. J Appl Bacteriol 70:265–274. doi: 10.1111/j.1365-2672.1991.tb02935.x. [DOI] [PubMed] [Google Scholar]
- 53.Rijal GK, Fujioka RS. 2001. Synergistic effect of solar radiation and solar heating to disinfect drinking water sources. Water Sci Technol 43:155–162. [PubMed] [Google Scholar]
- 54.Solic M, Krstulovic N. 1992. Separate and combined effects of solar radiation, temperature, salinity, and pH on the survival of fecal coliforms in seawater. Mar Pollut Bull 24:411–416. doi: 10.1016/0025-326X(92)90503-X. [DOI] [Google Scholar]
- 55.Christensen J, Linden KG. 2003. How particles affect UV light in the UV disinfection of unfiltered drinking water. J Am Water Works Assoc 95:179–189. doi: 10.1002/j.1551-8833.2003.tb10344.x. [DOI] [Google Scholar]
- 56.Davies-Colley RJ, Craggs RJ, Park J, Nagels JW. 2005. Optical characteristics of waste stabilization ponds: recommendations for monitoring. Water Sci Technol 51:153–161. doi: 10.2166/wst.2005.0452. [DOI] [PubMed] [Google Scholar]
- 57.Fujioka RS, Narikawa OT. 1982. Effect of sunlight on enumeration of indicator bacteria under field conditions. Appl Environ Microbiol 44:395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vreeland RH. 1987. Mechanisms of halotolerance in microorganisms. Crit Rev Microbiol 14:311–356. doi: 10.3109/10408418709104443. [DOI] [PubMed] [Google Scholar]
- 59.Kunin CM, Rudy J. 1991. Effect of NaCl-induced osmotic stress on intracellular concentrations of glycine betaine and potassium in Escherichia coli, Enterococcus faecalis, and staphylococci. J Lab Clin Med 118:217–224. [PubMed] [Google Scholar]
- 60.Empadinhas N, da Costa MS. 2008. Osmoadaptation mechanisms in prokaryotes: distribution of compatible solutes. Int Microbiol 11:151–161. [PubMed] [Google Scholar]
- 61.Solheim M, La Rosa SL, Mathisen T, Snipen LG, Nes IF, Brede DA. 2014. Transcriptomic and functional analysis of NaCl-induced stress in Enterococcus faecalis. PLoS One 9:e94571. doi: 10.1371/journal.pone.0094571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anderson IC, Rhodes M, Kator H. 1979. Sublethal stress in Escherichia coli: a function of salinity. Appl Environ Microbiol 38:1147–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rozen Y, Belkin S. 2001. Survival of enteric bacteria in seawater. FEMS Microbiol Rev 25:513–529. doi: 10.1111/j.1574-6976.2001.tb00589.x. [DOI] [PubMed] [Google Scholar]
- 64.Cordova A, Deserno M, Gelbart WM, Ben-Shaul A. 2003. Osmotic shock and the strength of viral capsids. Biophys J 85:70–74. doi: 10.1016/S0006-3495(03)74455-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ahmed W, Gyawali P, Sidhu JP, Toze S. 2014. Relative inactivation of faecal indicator bacteria and sewage markers in freshwater and seawater microcosms. Lett Appl Microbiol 59:348–354. doi: 10.1111/lam.12285. [DOI] [PubMed] [Google Scholar]
- 66.Bae S, Wuertz S. 2015. Decay of host-associated Bacteroidales cells and DNA in continuous-flow freshwater and seawater microcosms of identical experimental design and temperature as measured by PMA-qPCR and qPCR. Water Res 70:205–213. doi: 10.1016/j.watres.2014.10.032. [DOI] [PubMed] [Google Scholar]
- 67.Gonzalez JM. 1995. Modelling enteric bacteria survival in aquatic systems. Hydrobiologia 316:109–116. doi: 10.1007/BF00016892. [DOI] [Google Scholar]
- 68.Jeanneau L, Solecki O, Wery N, Jarde E, Gourmelon M, Communal PY, Jadas-Hecart A, Caprais MP, Gruau G, Pourcher AM. 2012. Relative decay of fecal indicator bacteria and human-associated markers: a microcosm study simulating wastewater input into seawater and freshwater. Environ Sci Technol 46:2375–2382. doi: 10.1021/es203019y. [DOI] [PubMed] [Google Scholar]
- 69.Nayak B, Weidhaas J, Harwood VJ. 2015. LA35 poultry fecal marker persistence is correlated with that of indicators and pathogens in environmental waters. Appl Environ Microbiol 81:4616–4625. doi: 10.1128/AEM.00444-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Menon P, Billen G, Servais P. 2003. Mortality rates of autochthonous and fecal bacteria in natural aquatic ecosystems. Water Res 37:4151–4158. doi: 10.1016/S0043-1354(03)00349-X. [DOI] [PubMed] [Google Scholar]
- 71.Okabe S, Shimazu Y. 2007. Persistence of host-specific Bacteroides-Prevotella 16S rRNA genetic markers in environmental waters: effects of temperature and salinity. Appl Microbiol Biotechnol 76:935–944. doi: 10.1007/s00253-007-1048-z. [DOI] [PubMed] [Google Scholar]
- 72.Solecki O, Jeanneau L, Jarde E, Gourmelon M, Marin C, Pourcher AM. 2011. Persistence of microbial and chemical pig manure markers as compared to faecal indicator bacteria survival in freshwater and seawater microcosms. Water Res 45:4623–4633. doi: 10.1016/j.watres.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 73.Hurst CJ, Gerba CP. 1980. Stability of simian rotavirus in fresh and estuarine water. Appl Environ Microbiol 39:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schulz CJ, Childers GW. 2011. Fecal bacteroidales diversity and decay in response to variations in temperature and salinity. Appl Environ Microbiol 77:2563–2572. doi: 10.1128/AEM.01473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lo S, Gilbert J, Hetrick F. 1976. Stability of human enteroviruses in estuarine and marine waters. Appl Environ Microbiol 32:245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fayer R, Graczyk TK, Lewis EJ, Trout JM, Farley CA. 1998. Survival of infectious Cryptosporidium parvum oocysts in seawater and eastern oysters (Crassostrea virginica) in the Chesapeake Bay. Appl Environ Microbiol 64:1070–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rodriguez-Zaragoza S. 1994. Ecology of free-living amoebae. Crit Rev Microbiol 20:225–241. doi: 10.3109/10408419409114556. [DOI] [PubMed] [Google Scholar]
- 78.Berney C, Romac S, Mahe F, Santini S, Siano R, Bass D. 2013. Vampires in the oceans: predatory cercozoan amoebae in marine habitats. ISME J 7:2387–2399. doi: 10.1038/ismej.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wieltschnig C, Fischer UR, Velimirov B, Kirschner AK. 2008. Effects of deposit-feeding macrofauna on benthic bacteria, viruses, and protozoa in a silty freshwater sediment. Microb Ecol 56:1–12. doi: 10.1007/s00248-007-9318-y. [DOI] [PubMed] [Google Scholar]
- 80.Thelaus J, Forsman M, Andersson A. 2008. Role of productivity and protozoan abundance for the occurrence of predation-resistant bacteria in aquatic systems. Microb Ecol 56:18–28. doi: 10.1007/s00248-007-9320-4. [DOI] [PubMed] [Google Scholar]
- 81.Craig DL, Fallowfield HJ, Cromar NJ. 2004. Use of microcosms to determine persistence of Escherichia coli in recreational coastal water and sediment and validation with in situ measurements. J Appl Microbiol 96:922–930. doi: 10.1111/j.1365-2672.2004.02243.x. [DOI] [PubMed] [Google Scholar]
- 82.McCrary KJ, Case CLH, Gentry TJ, Aitkenhead-Peterson JA. 2013. Escherichia coli regrowth in disinfected sewage effluent: effect of DOC and nutrients on regrowth in laboratory incubations and urban streams. Water Air Soil Pollut 224:1412. doi: 10.1007/s11270-012-1412-1. [DOI] [Google Scholar]
- 83.Surbeck CQ, Jiang SC, Grant SB. 2010. Ecological control of fecal indicator bacteria in an urban stream. Environ Sci Technol 44:631–637. doi: 10.1021/es903496m. [DOI] [PubMed] [Google Scholar]
- 84.Staley ZR, Senkbeil JK, Rohr JR, Harwood VJ. 2012. Lack of direct effects of agrochemicals on zoonotic pathogens and fecal indicator bacteria. Appl Environ Microbiol 78:8146–8150. doi: 10.1128/AEM.01815-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Staley ZR, Rohr JR, Harwood VJ. 2011. Test of direct and indirect effects of agrochemicals on the survival of fecal indicator bacteria. Appl Environ Microbiol 77:8765–8774. doi: 10.1128/AEM.06044-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Staley ZR, Rohr JR, Harwood VJ. 2010. The effect of agrochemicals on indicator bacteria densities in outdoor mesocosms. Environ Microbiol 12:3150–3158. doi: 10.1111/j.1462-2920.2010.02287.x. [DOI] [PubMed] [Google Scholar]
- 87.Kiefer LA, Shelton DR, Pachepsky Y, Blaustein R, Santin-Duran M. 2012. Persistence of Escherichia coli introduced into streambed sediments with goose, deer and bovine animal waste. Lett Appl Microbiol 55:345–353. doi: 10.1111/j.1472-765X.2012.03296.x. [DOI] [PubMed] [Google Scholar]
- 88.Garzio-Hadzick A, Shelton DR, Hill RL, Pachepsky YA, Guber AK, Rowland R. 2010. Survival of manure-borne E. coli in streambed sediment: effects of temperature and sediment properties. Water Res 44:2753–2762. doi: 10.1016/j.watres.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 89.Shelton DR, Pachepsky YA, Kiefer LA, Blaustein RA, McCarty GW, Dao TH. 2014. Response of coliform populations in streambed sediment and water column to changes in nutrient concentrations in water. Water Res 59:316–324. doi: 10.1016/j.watres.2014.04.019. [DOI] [PubMed] [Google Scholar]
- 90.Wanjugi P, Fox GA, Harwood VJ. 2016. The interplay between predation, competition, and nutrient levels influences the survival of Escherichia coli in aquatic environments. Microb Ecol 72:526–537. doi: 10.1007/s00248-016-0825-6. [DOI] [PubMed] [Google Scholar]
- 91.Gregory LF, Karthikeyan R, Aitkenhead-Peterson JA, Gentry TJ, Wagner KL, Harmel RD. 2017. Nutrient loading impacts on culturable E. coli and other heterotrophic bacteria fate in simulated stream mesocosms. Water Res 126:442–449. doi: 10.1016/j.watres.2017.09.043. [DOI] [PubMed] [Google Scholar]
- 92.Chudoba EA, Mallin MA, Cahoon LB, Skrabal SA. 2013. Stimulation of fecal bacteria in ambient waters by experimental inputs of organic and inorganic phosphorus. Water Res 47:3455–3466. doi: 10.1016/j.watres.2013.03.047. [DOI] [PubMed] [Google Scholar]
- 93.Atlas RM, Bartha R. 1998. Physiological ecology of microorganisms: adaptations to environmental conditions, p 281–331. In Atlas RM, Bartha R (ed), Microbial ecology fundamentals and applications. Benjamin/Cummings Science Publishing, Menlo Park, CA. [Google Scholar]
- 94.Zimmer-Faust AG, Thulsiraj V, Marambio-Jones C, Cao Y, Griffith JF, Holden PA, Jay JA. 2017. Effect of freshwater sediment characteristics on the persistence of fecal indicator bacteria and genetic markers within a Southern California watershed. Water Res 119:1–11. doi: 10.1016/j.watres.2017.04.028. [DOI] [PubMed] [Google Scholar]
- 95.Panoff JM, Corroler D, Thammavongs B, Boutibonnes P. 1997. Differentiation between cold shock proteins and cold acclimation proteins in a mesophilic gram-positive bacterium, Enterococcus faecalis JH2-2. J Bacteriol 179:4451–4454. doi: 10.1128/jb.179.13.4451-4454.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Phadtare S, Yamanaka K, Inouye M. 2000. The cold shock response, p 33–45. In Storz G, Hengge-Aronis R (ed), Bacterial stress responses. ASM Press, Washington, DC. [Google Scholar]
- 97.Giladi H, Goldenberg D, Koby S, Oppenheim AB. 1995. Enhanced activity of the bacteriophage lambda PL promoter at low temperature. FEMS Microbiol Rev 17:135–140. doi: 10.1111/j.1574-6976.1995.tb00195.x. [DOI] [PubMed] [Google Scholar]
- 98.Gabig M, Obuchowski M, Srutkowska S, Wegrzyn G. 1998. Regulation of replication of lambda phage and lambda plasmid DNAs at low temperature. Mol Gen Genet 258:494–502. doi: 10.1007/s004380050760. [DOI] [PubMed] [Google Scholar]
- 99.Ptashne M. 2004. A genetic switch: phage lambda revisited. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 100.Obuchowski M, Shotland Y, Koby S, Giladi H, Gabig M, Wegrzyn G, Oppenheim AB. 1997. Stability of CII is a key element in the cold stress response of bacteriophage lambda infection. J Bacteriol 179:5987–5991. doi: 10.1128/jb.179.19.5987-5991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Balleste E, Blanch AR. 2010. Persistence of Bacteroides species populations in a river as measured by molecular and culture techniques. Appl Environ Microbiol 76:7608–7616. doi: 10.1128/AEM.00883-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bell A, Layton AC, McKay L, Williams D, Gentry R, Sayler GS. 2009. Factors influencing the persistence of fecal Bacteroides in stream water. J Environ Qual 38:1224–1232. doi: 10.2134/jeq2008.0258. [DOI] [PubMed] [Google Scholar]
- 103.Beversdorf LJ, Bornstein-Forst SM, McLellan SL. 2007. The potential for beach sand to serve as a reservoir for Escherichia coli and the physical influences on cell die-off. J Appl Microbiol 102:1372–1381. doi: 10.1111/j.1365-2672.2006.03177.x. [DOI] [PubMed] [Google Scholar]
- 104.Kim M, Wuertz S. 2015. Survival and persistence of host-associated Bacteroidales cells and DNA in comparison with Escherichia coli and Enterococcus in freshwater sediments as quantified by PMA-qPCR and qPCR. Water Res 87:182–192. doi: 10.1016/j.watres.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 105.Brooks Y, Aslan A, Tamrakar S, Murali B, Mitchell J, Rose JB. 2015. Analysis of the persistence of enteric markers in sewage polluted water on a solid matrix and in liquid suspension. Water Res 76:201–212. doi: 10.1016/j.watres.2015.02.039. [DOI] [PubMed] [Google Scholar]
- 106.Sampson RW, Swiatnicki SA, Osinga VL, Supita JL, McDermott CM, Kleinheinz GT. 2006. Effects of temperature and sand on E. coli survival in a northern lake water microcosm. J Water Health 4:389–393. doi: 10.2166/wh.2006.024b. [DOI] [PubMed] [Google Scholar]
- 107.Lee HS, Sobsey MD. 2011. Survival of prototype strains of somatic coliphage families in environmental waters and when exposed to UV low-pressure monochromatic radiation or heat. Water Res 45:3723–3734. doi: 10.1016/j.watres.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 108.Ngazoa ES, Fliss I, Jean J. 2008. Quantitative study of persistence of human norovirus genome in water using TaqMan real-time RT-PCR. J Appl Microbiol 104:707–715. doi: 10.1111/j.1365-2672.2007.03597.x. [DOI] [PubMed] [Google Scholar]
- 109.Ravva SV, Sarreal CZ. 2016. Persistence of F-specific RNA coliphages in surface waters from a produce production region along the central coast of California. PLoS One 11:e0146623. doi: 10.1371/journal.pone.0146623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Staley ZR, Robinson C, Edge TA. 2016. Comparison of the occurrence and survival of fecal indicator bacteria in recreational sand between urban beach, playground and sandbox settings in Toronto, Ontario. Sci Total Environ 541:520–527. doi: 10.1016/j.scitotenv.2015.09.088. [DOI] [PubMed] [Google Scholar]
- 111.Wait DA, Sobsey MD. 2001. Comparative survival of enteric viruses and bacteria in Atlantic Ocean seawater. Water Sci Technol 43:139–142. [PubMed] [Google Scholar]
- 112.Skraber S, Gassilloud B, Schwartzbrod L, Gantzer C. 2004. Survival of infectious Poliovirus-1 in river water compared to the persistence of somatic coliphages, thermotolerant coliforms and Poliovirus-1 genome. Water Res 38:2927–2933. doi: 10.1016/j.watres.2004.03.041. [DOI] [PubMed] [Google Scholar]
- 113.Yang Y, Griffiths MW. 2013. Comparative persistence of subgroups of F-specific RNA phages in river water. Appl Environ Microbiol 79:4564–4567. doi: 10.1128/AEM.00612-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Burkhardt W, Calci KR, Watkins WD, Rippey SR, Chirtel SJ. 2000. Inactivation of indicator microorganisms in estuarine waters. Water Res 34:2207–2214. doi: 10.1016/S0043-1354(99)00399-1. [DOI] [Google Scholar]
- 115.Ives RL, Kamarainen AM, John DE, Rose JB. 2007. Use of cell culture to assess Cryptosporidium parvum survival rates in natural groundwaters and surface waters. Appl Environ Microbiol 73:5968–5970. doi: 10.1128/AEM.00347-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hassard F, Gwyther CL, Farkas K, Andrews A, Jones V, Cox B, Brett H, Jones DL, McDonald JE, Malham SK. 2016. Abundance and distribution of enteric bacteria and viruses in coastal and estuarine sediments—a review. Front Microbiol 7:1692. doi: 10.3389/fmicb.2016.01692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Verhougstraete MP, Byappanahalli MN, Rose JB, Whitman RL. 2010. Cladophora in the Great Lakes: impacts on beach water quality and human health. Water Sci Technol 62:68–76. doi: 10.2166/wst.2010.230. [DOI] [PubMed] [Google Scholar]
- 118.Whitman R, Harwood VJ, Edge TA, Nevers M, Byappanahalli M, Vijayavel K, Brandao J, Sadowsky MJ, Alm EW, Crowe A, Ferguson D, Ge Z, Halliday E, Kinzelman J, Kleinheinz G, Przybyla-Kelly K, Staley C, Staley Z, Solo-Gabriele HM. 2014. Microbes in beach sands: integrating environment, ecology and public health. Rev Environ Sci Biotechnol 13:329–368. doi: 10.1007/s11157-014-9340-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Badgley BD, Nayak BS, Harwood VJ. 2010. The importance of sediment and submerged aquatic vegetation as potential habitats for persistent strains of enterococci in a subtropical watershed. Water Res 44:5857–5866. doi: 10.1016/j.watres.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 120.Chandran A, Varghese S, Kandeler E, Thomas A, Hatha M, Mazumder A. 2011. An assessment of potential public health risk associated with the extended survival of indicator and pathogenic bacteria in freshwater lake sediments. Int J Hyg Environ Health 214:258–264. doi: 10.1016/j.ijheh.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 121.Curtis K, Trapp JM. 2016. Examining the colonization and survival of E. coli from varying host sources in drainage basin sediments and stormwater. Arch Environ Contam Toxicol 71:183–197. doi: 10.1007/s00244-016-0289-1. [DOI] [PubMed] [Google Scholar]
- 122.Wanjugi P, Harwood VJ. 2014. Protozoan predation is differentially affected by motility of enteric pathogens in water vs. sediments. Microb Ecol 68:751–760. doi: 10.1007/s00248-014-0444-z. [DOI] [PubMed] [Google Scholar]
- 123.Haller L, Amedegnato E, Pote J, Wildi W. 2009. Influence of freshwater sediment characteristics on persistence of fecal indicator bacteria. Water Air Soil Pollut 203:217–227. doi: 10.1007/s11270-009-0005-0. [DOI] [Google Scholar]
- 124.Abia AL, Ubomba-Jaswa E, Momba MN. 2016. Competitive survival of Escherichia coli, Vibrio cholerae, Salmonella typhimurium and Shigella dysenteriae in riverbed sediments. Microb Ecol 72:881–889. doi: 10.1007/s00248-016-0784-y. [DOI] [PubMed] [Google Scholar]
- 125.Badgley BD, Thomas FI, Harwood VJ. 2010. The effects of submerged aquatic vegetation on the persistence of environmental populations of Enterococcus spp. Environ Microbiol 12:1271–1281. doi: 10.1111/j.1462-2920.2010.02169.x. [DOI] [PubMed] [Google Scholar]
- 126.Imamura GJ, Thompson RS, Boehm AB, Jay JA. 2011. Wrack promotes the persistence of fecal indicator bacteria in marine sands and seawater. FEMS Microbiol Ecol 77:40–49. doi: 10.1111/j.1574-6941.2011.01082.x. [DOI] [PubMed] [Google Scholar]
- 127.Quilliam RS, Jamieson J, Oliver DM. 2014. Seaweeds and plastic debris can influence the survival of faecal indicator organisms in beach environments. Mar Pollut Bull 84:201–207. doi: 10.1016/j.marpolbul.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 128.Brown KI, Boehm AB. 2015. Comparative decay of Catellicoccus marimmalium and enterococci in beach sand and seawater. Water Res 83:377–384. doi: 10.1016/j.watres.2015.06.055. [DOI] [PubMed] [Google Scholar]
- 129.Cloutier DD, McLellan SL. 2017. Distribution and differential survival of traditional and alternative indicators of fecal pollution at freshwater beaches. Appl Environ Microbiol 83:e02881-16. doi: 10.1128/AEM.02881-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Eichmiller JJ, Borchert AJ, Sadowsky MJ, Hicks RE. 2014. Decay of genetic markers for fecal bacterial indicators and pathogens in sand from Lake Superior. Water Res 59:99–111. doi: 10.1016/j.watres.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 131.Feng F, Goto D, Yan T. 2010. Effects of autochthonous microbial community on the die-off of fecal indicators in tropical beach sand. FEMS Microbiol Ecol 74:214–225. doi: 10.1111/j.1574-6941.2010.00921.x. [DOI] [PubMed] [Google Scholar]
- 132.Hartz A, Cuvelier M, Nowosielski K, Bonilla TD, Green M, Esiobu N, McCorquodale DS, Rogerson A. 2008. Survival potential of Escherichia coli and enterococci in subtropical beach sand: implications for water quality managers. J Environ Qual 37:898–905. doi: 10.2134/jeq2007.0312. [DOI] [PubMed] [Google Scholar]
- 133.Mika KB, Imamura G, Chang C, Conway V, Fernandez G, Griffith JF, Kampalath RA, Lee CM, Lin CC, Moreno R, Thompson S, Whitman RL, Jay JA. 2009. Pilot- and bench-scale testing of faecal indicator bacteria survival in marine beach sand near point sources. J Appl Microbiol 107:72–84. doi: 10.1111/j.1365-2672.2009.04197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yamahara KM, Sassoubre LM, Goodwin KD, Boehm AB. 2012. Occurrence and persistence of bacterial pathogens and indicator organisms in beach sand along the California coast. Appl Environ Microbiol 78:1733–1745. doi: 10.1128/AEM.06185-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang Q, He X, Yan T. 2015. Differential decay of wastewater bacteria and change of microbial communities in beach sand and seawater microcosms. Environ Sci Technol 49:8531–8540. doi: 10.1021/acs.est.5b01879. [DOI] [PubMed] [Google Scholar]
- 136.Mohammed RL, Echeverry A, Stinson CM, Green M, Bonilla TD, Hartz A, McCorquodale DS, Rogerson A, Esiobu N. 2012. Survival trends of Staphylococcus aureus, Pseudomonas aeruginosa, and Clostridium perfringens in a sandy south Florida beach. Mar Pollut Bull 64:1201–1209. doi: 10.1016/j.marpolbul.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 137.Williams AP, Avery LM, Killham K, Jones DL. 2007. Persistence, dissipation, and activity of Escherichia coli O157:H7 within sand and seawater environments. FEMS Microbiol Ecol 60:24–32. doi: 10.1111/j.1574-6941.2006.00273.x. [DOI] [PubMed] [Google Scholar]
- 138.Goodwin KD, Schriewer A, Jirik A, Curtis K, Crumpacker A. 2017. Consideration of natural sources in a bacteria TMDL—lines of evidence, including beach microbial source tracking. Environ Sci Technol 51:7775–7784. doi: 10.1021/acs.est.6b05886. [DOI] [PubMed] [Google Scholar]
- 139.Pace ML, Cole JJ. 1994. Comparative and experimental approaches to top-down and bottom-up regulation of bacteria. Microb Ecol 28:181–193. doi: 10.1007/BF00166807. [DOI] [PubMed] [Google Scholar]
- 140.Thingstad TF. 2000. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol Oceanogr 45:1320–1328. doi: 10.4319/lo.2000.45.6.1320. [DOI] [Google Scholar]
- 141.Feichtmayer J, Deng L, Griebler C. 2017. Antagonistic microbial interactions: contributions and potential applications for controlling pathogens in the aquatic systems. Front Microbiol 8:2192. doi: 10.3389/fmicb.2017.02192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gasol JM, Pedrós-Alió C, Vaqué D. 2002. Regulation of bacterial assemblages in oligotrophic plankton systems: results from experimental and empirical approaches. Antonie Van Leeuwenhoek 81:435–452. doi: 10.1023/A:1020578418898. [DOI] [PubMed] [Google Scholar]
- 143.Baltar F, Aristegui J, Gasol JM, Herndl GJ. 2012. Microbial functioning and community structure variability in the mesopelagic and epipelagic waters of the subtropical northeast Atlantic ocean. Appl Environ Microbiol 78:3309–3316. doi: 10.1128/AEM.07962-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Stubbendieck RM, Straight PD. 2016. Multifaceted interfaces of bacterial competition. J Bacteriol 198:2145–2155. doi: 10.1128/JB.00275-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pernthaler J. 2005. Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol 3:537–546. doi: 10.1038/nrmicro1180. [DOI] [PubMed] [Google Scholar]
- 146.Lee WJ, Patterson DJ. 2002. Abundance and biomass of heterotrophic flagellates, and factors controlling their abundance and distribution in sediments of Botany Bay. Microb Ecol 43:467–481. doi: 10.1007/s00248-002-2000-5. [DOI] [PubMed] [Google Scholar]
- 147.Simek K, Jurgens K, Nedoma J, Comerma M, Armengol J. 2000. Ecological role and bacterial grazing of Halteria spp.: small freshwater oligotrichs as dominant pelagic ciliate bacterivores. Aquat Microb Ecol 22:43–56. doi: 10.3354/ame022043. [DOI] [Google Scholar]
- 148.Sherr BF, Sherr EB, Rassoulzadegan F. 1988. Rates of digestion of bacteria by marine phagotrophic protozoa: temperature dependence. Appl Environ Microbiol 54:1091–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nakano S, Ishii N, Manage PM, Kawabata Z. 1998. Trophic roles of heterotrophic nanoflagellates and ciliates among planktonic organisms in a hypereutrophic pond. Aquat Microb Ecol 16:153–161. doi: 10.3354/ame016153. [DOI] [Google Scholar]
- 150.Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C, Keller H, Lambert C, Evans KJ, Goesmann A, Meyer F, Sockett RE, Schuster SC. 2004. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- 151.Muniesa M, Jofre J. 2004. Factors influencing the replication of somatic coliphages in the water environment. Antonie Van Leeuwenhoek 86:65–76. doi: 10.1023/B:ANTO.0000024909.75523.be. [DOI] [PubMed] [Google Scholar]
- 152.Davies CM, Long JA, Donald M, Ashbolt NJ. 1995. Survival of fecal microorganisms in marine and freshwater sediments. Appl Environ Microbiol 61:1888–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Enzinger RM, Cooper RC. 1976. Role of bacteria and protozoa in the removal of Escherichia coli from estuarine waters. Appl Environ Microbiol 31:758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gonzalez JM, Sherr EB, Sherr BF. 1990. Size-selective grazing on bacteria by natural assemblages of estuarine flagellates and ciliates. Appl Environ Microbiol 56:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kreader CA. 1998. Persistence of PCR-detectable Bacteroides distasonis from human feces in river water. Appl Environ Microbiol 64:4103–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wanjugi P, Harwood VJ. 2013. The influence of predation and competition on the survival of commensal and pathogenic fecal bacteria in aquatic habitats. Environ Microbiol 15:517–526. doi: 10.1111/j.1462-2920.2012.02877.x. [DOI] [PubMed] [Google Scholar]
- 157.Anderson IC, Rhodes MW, Kator HI. 1983. Seasonal variation in survival of Escherichia coli exposed in situ in membrane diffusion chambers containing filtered and nonfiltered estuarine water. Appl Environ Microbiol 45:1877–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Iriberri J, Azua I, Labirua-Iturburu A, Artolozaga I, Barcina I. 1994. Differential elimination of enteric bacteria by protists in a freshwater system. J Appl Bacteriol 77:476–483. doi: 10.1111/j.1365-2672.1994.tb04390.x. [DOI] [PubMed] [Google Scholar]
- 159.Booncharoen N, Mongkolsuk S, Sirikanchana K. 2018. Comparative persistence of human sewage-specific enterococcal bacteriophages in freshwater and seawater. Appl Microbiol Biotechnol 102:6235–6246. doi: 10.1007/s00253-018-9079-1. [DOI] [PubMed] [Google Scholar]
- 160.Chauret C, Nolan K, Chen P, Springthorpe S, Sattar S. 1998. Aging of Cryptosporidium parvum oocysts in river water and their susceptibility to disinfection by chlorine and monochloramine. Can J Microbiol 44:1154–1160. doi: 10.1139/cjm-44-12-1154. [DOI] [PubMed] [Google Scholar]
- 161.Thorn CE, Quilliam RS, Williams AP, Malham SK, Cooper D, Reynolds B, Jones DL. 2011. Grazing intensity is a poor indicator of waterborne Escherichia coli O157 activity. Anaerobe 17:330–333. doi: 10.1016/j.anaerobe.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 162.Korhonen LK, Martikainen PJ. 1991. Survival of Escherichia coli and Campylobacter jejuni in untreated and filtered lake water. J Appl Bacteriol 71:379–382. doi: 10.1111/j.1365-2672.1991.tb03804.x. [DOI] [PubMed] [Google Scholar]
- 163.Dominguez MS, Escalante AH, Folabella AM, Zamora AS. 2012. Selective grazing by protists upon enteric bacteria in an aquatic system. Rev Argent Microbiol 44:43–48. doi: 10.1590/S0325-75412012000100009. [DOI] [PubMed] [Google Scholar]
- 164.McCambridge J, Mcmeekin TA. 1980. Relative effects of bacterial and protozoan predators on survival of Escherichia coli in estuarine water samples. Appl Environ Microbiol 40:907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ravva SV, Sarreal CZ, Mandrell RE. 2010. Identification of protozoa in dairy lagoon wastewater that consume Escherichia coli O157:H7 preferentially. PLoS One 5:e15671. doi: 10.1371/journal.pone.0015671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ravva SV, Sarreal CZ, Mandrell RE. 2014. Strain differences in fitness of Escherichia coli O157:H7 to resist protozoan predation and survival in soil. PLoS One 9:e102412. doi: 10.1371/journal.pone.0102412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pirlot S, Unrein F, Descy JP, Servais P. 2007. Fate of heterotrophic bacteria in Lake Tanganyika (East Africa). FEMS Microbiol Ecol 62:354–364. doi: 10.1111/j.1574-6941.2007.00396.x. [DOI] [PubMed] [Google Scholar]
- 168.Servais P, Garcia-Armisen T, George I, Billen G. 2007. Fecal bacteria in the rivers of the Seine drainage network (France): sources, fate and modelling. Sci Total Environ 375:152–167. doi: 10.1016/j.scitotenv.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 169.Zingel P, Agasild H, Noges T, Kisand V. 2007. Ciliates are the dominant grazers on pico- and nanoplankton in a shallow, naturally highly eutrophic lake. Microb Ecol 53:134–142. doi: 10.1007/s00248-006-9155-4. [DOI] [PubMed] [Google Scholar]
- 170.Korajkic A, Parfrey LW, McMinn BR, Baeza YV, VanTeuren W, Knight R, Shanks OC. 2015. Changes in bacterial and eukaryotic communities during sewage decomposition in Mississippi River water. Water Res 69:30–39. doi: 10.1016/j.watres.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 171.Siqueira-Castro IC, Greinert-Goulart JA, Bonatti TR, Yamashiro S, Franco RM. 2016. First report of predation of Giardia sp. cysts by ciliated protozoa and confirmation of predation of Cryptosporidium spp. oocysts by ciliate species. Environ Sci Pollut Res Int 23:11357–11362. doi: 10.1007/s11356-016-6689-y. [DOI] [PubMed] [Google Scholar]
- 172.Pinheiro MD, Power ME, Butler BJ, Dayeh VR, Slawson R, Lee LE, Lynn DH, Bols NC. 2007. Use of Tetrahymena thermophila to study the role of protozoa in inactivation of viruses in water. Appl Environ Microbiol 73:643–649. doi: 10.1128/AEM.02363-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lukjancenko O, Wassenaar TM, Ussery DW. 2010. Comparison of 61 sequenced Escherichia coli genomes. Microb Ecol 60:708–720. doi: 10.1007/s00248-010-9717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tambalo DD, Fremaux B, Boa T, Yost CK. 2012. Persistence of host-associated Bacteroidales gene markers and their quantitative detection in an urban and agricultural mixed prairie watershed. Water Res 46:2891–2904. doi: 10.1016/j.watres.2012.02.048. [DOI] [PubMed] [Google Scholar]
- 175.Perkins TL, Perrow K, Rajko-Nenow P, Jago CF, Jones DL, Malham SK, McDonald JE. 2016. Decay rates of faecal indicator bacteria from sewage and ovine faeces in brackish and freshwater microcosms with contrasting suspended particulate matter concentrations. Sci Total Environ 572:1645–1652. doi: 10.1016/j.scitotenv.2016.03.076. [DOI] [PubMed] [Google Scholar]
- 176.Liang Z, He Z, Zhou X, Powell CA, Yang Y, Roberts MG, Stoffella PJ. 2012. High diversity and differential persistence of fecal Bacteroidales population spiked into freshwater microcosm. Water Res 46:247–257. doi: 10.1016/j.watres.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 177.Cann AJ. 2001. Principles of molecular virology, 3rd ed Academic Press, London, United Kingdom. [Google Scholar]
- 178.Vierheilig J, Farnleitner AH, Kollanur D, Bloschl G, Reischer GH. 2012. High abundance of genetic Bacteroidetes markers for total fecal pollution in pristine alpine soils suggests lack in specificity for feces. J Microbiol Methods 88:433–435. doi: 10.1016/j.mimet.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lamendella R, Santo Domingo JW, Yannarell AC, Ghosh S, Di Giovanni G, Mackie RI, Oerther DB. 2009. Evaluation of swine-specific PCR assays used for fecal source tracking and analysis of molecular diversity of swine-specific “bacteroidales” populations. Appl Environ Microbiol 75:5787–5796. doi: 10.1128/AEM.00448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.