Abstract
The term metagenomics refers to the use of sequencing methods to simultaneously identify genomic material from all organisms present in a sample, with the advantage of greater taxonomic resolution than culture or other methods. Applications include pathogen detection and discovery, species characterisation, antimicrobial resistance detection, virulence profiling, and study of the microbiome and microecological factors affecting health. However, metagenomics involves complex and multistep processes and there are important technical and methodological challenges that require careful consideration to support valid inference. We co-ordinated a multidisciplinary, international expert group to establish reporting guidelines that address specimen processing, nucleic acid extraction, sequencing platforms, bioinformatics considerations, quality assurance, limits of detection, power and sample size, confirmatory testing, causality criteria, cost, and ethical issues. The guidance recognises that metagenomics research requires pragmatism and caution in interpretation, and that this field is rapidly evolving.
Background
The term metagenome was coined in 1998 to describe the collection of genomes from microbes present in environmental soil samples by using approaches previously used to study single genomes.1 The sequencing of genetic material from clinical samples has become common practice in research on clinical microorganisms. In this context, metagenomics refers to the application of sequencing methods that can identify coexistent genomic material from any organism present in patient samples (ie, microorganism and host nucleic acid), usually with the aim of pathogen identification for clinical diagnosis or research.2, 3, 4 Examples of practical applications include pathogen detection and discovery, species characterisation or subtyping, antimicrobial resistance detection, virulence profiling, and studies of the microbiome and microecological drivers of health and disease.5, 6, 7, 8, 9, 10, 11, 12 Metagenomics is also being introduced as a diagnostic tool for causal studies of clinical syndromes (such as encephalitis),13, 14 for exploring the microbiome,15, 16 and for tracking disease outbreaks.17, 18 A current example of the transformational effect of direct sequencing of clinical samples has been the application for rapid investigation and dissemination of information on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes COVID-19.11, 12
Metagenomics data are generated using high-throughput sequencing methods, also referred to as deep, next-generation, massively parallel, or shotgun sequencing. In this Review, for simplicity, we refer to all these approaches as sequencing. We also include capture probe enrichment-based sequencing methods that use nucleotide probes to increase sensitivity4 and targeted amplicon sequencing—eg, sequencing the 16S ribosomal ribonucleic acid (rRNA) gene to identify bacteria.19 Capture probe enrichment-based sequencing and targeted amplicon sequencing might not be considered true examples of metagenomics and are not the focus of our Review; however, some similar considerations about reporting of results apply.
Metagenomic sequencing has advantages for pathogen identification over conventional methods, such as culture or targeted PCR, because many or most microbial species present within a sample can be detected simultaneously with high taxonomic resolution. Detailed characterisation of microbial communities and population dynamics also enables the study of ecological interactions. Furthermore, this method does not require culture techniques, and therefore can be used for microbial species that are difficult or time consuming to grow. This is particularly relevant for diagnostic applications, where routine culture is in decline.20, 21
Key messages.
-
•
The term metagenomics refers to the use of sequencing methods to simultaneously identify genomic material from all organisms present in a sample, with the advantage of greater taxonomic resolution than culture or other methods.
-
•
Applications include pathogen detection and discovery, species characterisation, antimicrobial resistance detection, virulence profiling, and study of the microbiome and microecological factors affecting health.
-
•
Metagenomics involves complex and multistep processes and there are important technical and methodological challenges that require careful consideration to support valid inference.
-
•
We co-ordinated a multidisciplinary, international expert group to establish reporting guidelines that address specimen processing, nucleic acid extraction, sequencing platforms, bioinformatics considerations, quality assurance, limits of detection, power and sample size, confirmatory testing, causality criteria, cost, and ethical issues.
-
•
The guidance recognises that metagenomics research requires pragmatism and caution in interpretation, and that this field is rapidly evolving. Reporting standards should support clarity, consistency, and robustness of research.
However, appropriate study design for metagenomics research is not well defined and metagenomic technologies pose important technical challenges. These challenges include methodological artefacts introduced by wet laboratory methods and the effect that different computational approaches have on the analysis of multivariate and complex data. Furthermore, the ethical implications of sequencing are substantial and data privacy considerations are increasingly recognised. The multiple steps and different expertise required to generate and analyse metagenomic sequence data involves numerous decision points, which could introduce bias and affect downstream inference about the presence and abundance of microbial species in the sample.
A metagenome result should therefore be interpreted as one of many possible representations of the true sample composition of a given microbiome. Understanding and reporting sources of bias and limitations to valid inference should improve protocol performance and enable metagenomic research to proceed with transparent recognition of the limitations. However, existing reporting statements for epidemiology studies, including STROBE (STrengthening the Reporting of OBservational studies in Epidemiology)22 and its infectious disease molecular epidemiology extension, STROME-ID (Strengthening the Reporting of Molecular Epidemiology for Infectious Diseases),23 do not fully address issues specific to metagenomics. For this reason, scientific journals, and their readers, might not be adequately equipped with a standardised set of guidelines to evaluate and critically appraise clinical and epidemiological studies applying metagenomics. We aimed to improve the clarity and consistency of metagenomics research reporting, ranging from clinical diagnostics to microbiome studies, with suggestions for optimal practice and recommendations for robust and accurate reporting.
Titles and abstracts
The term metagenomics should be included in the title or abstract, and the keywords of the study when these methods contribute substaintially to the results reported
Clear and concise language incorporating standardised terminology, with references if appropriate, enables the accurate indexing of published studies in recognised databases. This is crucial for easy information retrieval and knowledge dissemination. For example, a systematic literature review of studies applying metagenomics in encephalitis using medical subject headings and keyword searches for the terms sequencing or metagenomics in four databases (PubMed, Embase, Web of Science, and Cochrane)13 failed to identify two relevant studies that did not report the terms.25, 26 These studies were identified by experts in the field who were directly involved with the studies.
Describing methods and study design
Describe specimen collection, handling and storage processes, and nucleic acid extraction methods
Steps involved in sample collection, handling, and processing are frequently poorly reported in publications and yet they will have considerable effect on the results and reproducibility of a study and could introduce variability artefacts.27, 28, 29, 30 In particular, many studies use material banked and collected originally for other purposes. In this Review, we describe important potential sources of error and their contribution to bias.
Nucleic acids, particularly RNA, are labile. Consequently, the collection methods, addition of nucleic acid stabilisers, and time to processing can affect the results obtained.31 To address these issues, reporting should include durations, volumes, temperatures, and methods used before, during, and after the storage of samples.32, 33 Extraction methods contribute to another major source of method-induced variation—eg, by being DNA or RNA specific, or tailored to specific organism types—so should be described.34 Other details of sample preparation methods should also be reported including filtration, centrifugation, DNA digestion, rRNA depletion, separation in RNA or DNA, and random amplification. Standardised protocols of sample preparation methods should also be followed, if available and appropriate, and documented clearly in the publication methods. Authors should also consider submitting to standardised protocol repositories to provide transparency in the study design and methodology.
Describe sequencing methods, including sequencing depth
Different metagenomic sequencing platforms might produce different types of reads—eg, single versus paired-end, and short (100–300 bp) versus long (>1000 bp). Sequencing platforms have different error rates, with the probability of a nucleic base being read incorrectly ranging from less than 0·01% for Illumina sequencers to 5–10% for Oxford Nanopore Technologies sequencers (current figures as of February, 2020).35 Additionally, sequencers often read a base incorrectly when processing samples with large homopolymer repeats, GC-rich, structurally repetitive, and other complex regions of the genome. Consequent false-positive and false-negative errors need consideration when reporting species composition.36
Sequencing depth refers to the number of times a particular nucleic base is represented within reads or the redundancy of coverage,37 and has implications for identification of low abundant transcripts and confidence in sequencing data. However, sequencing depth must be balanced according to the research question and the available resources. There are several factors that affect sequencing depth, including the sequencing platform and the sequence that is being read (eg, species diversity of the sample).37, 38, 39
Describe methods used for bioinformatics analysis
For the purposes of this statement, the term bioinformatics applies to all analysis steps involving raw sequencing data, including base calling, de-multiplexing, trimming and removal of reads (eg, reads of low quality, low complexity, adapters and indexes, or of human origin), read normalisation, alignment of sequence reads to reference databases, de-novo assembling of genomes, and taxonomic assignment of reads, assembled contigs, or both. There are multiple viable options for many of these tasks, with ongoing debate in the community about optimal methods, which can depend on the scientific question at hand. The field of metagenomics is developing rapidly and methods once considered best practice can be superseded following new analytical advances.
There should be clear descriptions of the bioinformatics methods used, including, at a minimum, the software name, version, and the main commands run with values for the essential parameters or flags. It is also advisable to make data and programming code open access, whether as supplementary files or shared online—eg, via Github or Figshare. Where possible, a version-controlled container, package, or easily installable version of the complete analytical pipeline (including all dependencies and required databases) could be made available for download and review. The open source release of bioinformatics workflows should be encouraged wherever possible to improve transparency and reproducibility, and should include adequate validation datasets, meaningful documentation, and examples of expected outputs and reports (appendix pp 1–2).
Describe quality assurance methods, including internal and external quality controls
An important strength of metagenomics analyses is their ability to detect any genomic material present within one sample. However, detection applies equally to true sample material and to any contaminating nucleic acids present in a sample, which can be introduced at any stage from sample collection to processing. For example, contamination could come from the extraction kit, the so-called kitome,40 or at the point of specimen collection. Sampling is rarely done under completely sterile conditions, and tissues obtained from tissue banks are therefore often contaminated. Low biomass and low abundance sites (for example tumours, the brain, and fetal tissues such as the placenta) are particularly prone to the risk of misclassifying contaminants.
To show attempts to ensure internal validity and reproducibility and identify potential contamination, internal controls for all extraction and sequencing processes should be reported as part of standard operating procedures.4, 27 Positive controls are usually spiked with DNA or RNA—eg, synthetic nucleic acid standards such as sequins47—and negative controls are usually a blank (eg, water) sample or ideally a similar or identical matrix (tissue, body fluids, etc) that are expected to contain no microorganism genomic material based on patient factors and test results. For clinical metagenomics, formal laboratory implementation involves a system of external controls. Arranging this system of external controls is difficult; however, publicly and commercially available controls and mock community samples are now available and we recommend that their use should be reported.48, 49
Describe use of orthogonal methods to confirm pathogen identity, function, and viability
The conventional methods in microbiology for confirming the presence of a pathogen are culture or growth of the pathogen from a clinical sample and immunohistochemistry, the histological localisation of candidate species in tissue biopsies. However, traditional culture can be difficult when antibiotics have been administered before sampling or for pathogens that are slow growing, fastidious, present in low-concentration, or currently undescribed. Sequencing has high discriminative power and could have higher sensitivity than culture-based methods. For example, in a polymicrobial sample, growth can be affected by presence of other competing bacteria or by inadequate growth conditions. Metagenomics methods have consistently shown higher classification accuracy when comparing taxonomic profiles of synthetic polymicrobial samples obtained from extended quantitative culture with non-selective media.50
Confirmatory assays appropriate to the study setting, justification for the methods used, and a description of their limitations should be reported. For cases in which confirmatory assays are not possible (eg, because of high cost or low volume of samples) an explanation should be provided. Rigorous validation of the method used, particularly for pathogens and proficiency testing, especially in clinical laboratories should be described (appendix pp 2–3).
Describe the criteria used to assess the role of pathogens in disease aetiology
Confirming the presence of microbial DNA or RNA in association with disease is an important step in establishing a causal relationship between a microorganism and disease.51, 52 A major challenge for metagenomics research and diagnostics is distinguishing pathogens from commensals or contaminants.53, 54 Interpretation of microbiome investigations can be further complicated if a misbalance in variation and abundance of different bacteria—sometimes referred to as dysbiosis—is suspected to be the cause of the condition.55 It is also worth considering that the cause of some diseases might involve multiple sequential or interacting species, which can be collectively important.56, 57 Furthermore, sequencing investigations can identify novel organisms, for which the clinical significance will be unknown. These issues are particularly relevant in the investigation of the cause of CNS infections.
Several criteria to establish causality have been proposed over the past century, including the incorporation of metagenomic technologies (appendix 7–9).58, 59
State the time from collection to results and cost consideration
The time from sample collection to processing (transport time), including cold-chain transportation and transit, can affect the compositional profile of microorganisms inferred from metagenomics. Overgrowth or degradation can occur during the period between collection and (cryo)storage with the result that the sequencing profile may not accurately reflect the composition of the sample at the time of collection. An extended duration of storage can result in a shift in the relative representation of bacterial taxa and substantial variability in metagenomics data. For example, faecal samples stored for longer than 3 months at −80°C experience selective loss of Bacteroides spp.6, 60, 61
If the sample is obtained post mortem, it is essential to report the time from death to sample acquisition given extravasation of gut bacteria into the bloodstream that can complicate interpretation of metagenomic data. For some applications, it might be relevant to report the overall turnaround time of the bioinformatic analyses—ie, including computational time for bioinformatics analysis. For example, Oxford Nanopore technology may be deployed in the field or at point of need, allowing sequencing to be done rapidly in near real-time; still, actionable results are also dependent on the time required for computational analysis.62, 62 The turnaround time of bioinformatic analyses is crucial in the context of clinical applications, when metagenomics is used to help to guide or tailor patient treatment. Variables such as sequencing run time and total computational analysis time (with system specifications—eg, number of cores and amount of memory used) should be stated clearly, as should the sequencing depth.64
Setting
State whether sample collection was retrospective or prospective
As described in the STAndards for Reporting of Diagnostic accuracy (STARD) guidelines, clarity is needed regarding the sequence of events in diagnostic testing to ensure that sources of bias are addressed.65 The analyte can degrade if there is a long time in between sample collection and the metagenomics assay. Retrospective sampling might also lead to bias in the samples tested. For instance, when comparing studies of unidentified encephalitis, samples retrospectively selected for metagenomics might be those that are difficult to diagnose (eg, with a low titre) or taken at later timepoints in the course of infection, and therefore more likely to be non-infectious.66
Participants
Consider factors influencing microbiota compositions when selecting participants
Most diagnostic and public health laboratories do not yet use metagenomic technologies routinely. As such, patients included in metagenomics studies are often from tertiary referral or specialist centres, which are unlikely to be representative of the wider population, as discussed in STROBE and STROME-ID.22, 23 This limitation can introduce challenges for appropriate selection of controls for case-control studies and for studies assessing the strength of disease associations.
Species composition of human microbiomes are affected by various host factors, including age, sex, behaviour (eg, diet and lifestyle), and environment.67, 68 Exposure to pharmacological substances can also profoundly influence microbiome composition. For example, a single standard course of antibiotics has been shown to alter species composition of the gut and oral microbiomes for over a year.69, 70 Matching of cases and controls is particularly challenging for metagenomics studies given the broad range of microbes considered.71 Metagenomics studies should aim to minimise and statistically control for host confounders or, at a minimum, list those confounders that might affect interpretation of results.
Bias
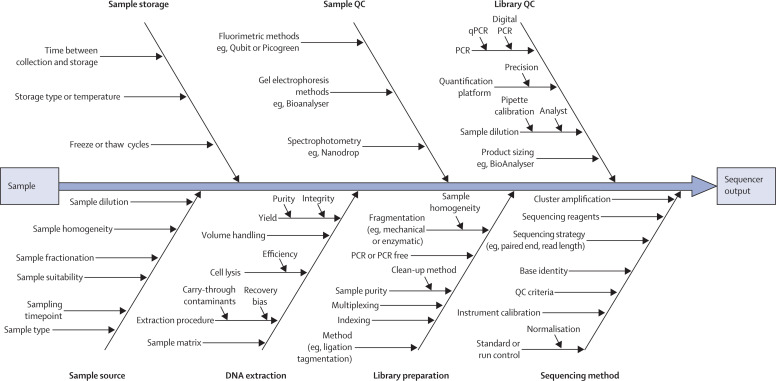
Bias is a source of error that remains constant with replication affecting trueness;72 it is separate to random error, which affects the precision of an experiment. Together, these sources of error contribute to measurement uncertainty that, when conducting metagenomics sequencing, has many potential sources (figure 1 ). Replication, including replication of the whole process, provides a means to estimate random error, which can vary when using different sequencing strategies.72 Adherence to strictly described laboratory protocols can improve random error and reproducibility,21 but it cannot be used alone to remove bias.
Figure 1.
Sources of uncertainty diagram highlighting potential contributing sources
For simplicity, this figure considers the sequencing of DNA from an environment and does not consider the process beyond the data output from the sequencer. The arrows pointing towards the central black arrow show the experimental process from left to right and the sources of variability that could contribute uncertainty. Conceptually it is clear how some of these factors contribute to systematic effects (bias). However, in addition these factors also contribute to the random error (variance) that will influence the precision of a potential finding. QC=quality control.
Address potential sources of bias (sampling, transport, storage, library preparation, and sequencing)
Bias can occur at each step of a diagnostic sequencing pipeline (panel 1 ) and is more difficult to evaluate than random error. For metagenomics studies, microbiological contamination of samples can introduce bias. Experimental bias that is caused at different stages of a metagenomics experiment is more challenging to control for than selection bias or contamination. The fact that the microbiome is composed of many different microorganisms means that a given protocol could lead to certain groups being over-represented in the processed samples. For example, enrichment protocols can introduce bias for pathogen detection.73 Capture probe-targeted sequencing will limit detection to targeted sequences, and 16S rRNA gene sequencing has limitations with regard to the level of taxonomic classification. This precise form of bias does not exist in untargeted metagenomics; however, other experimental bias can occur at different protocol stages, including during sampling, nucleic acid extraction,74 or post-extraction steps.75 Studies using 16S should consider that different primers amplify different bacterial families with varying degrees of success because of mismatches, resulting in potential bias in abundance and diversity metrics,76 which cannot be completely corrected bioinformatically.77
Panel 1. Examples of potential sources of bias in metagenomics studies and implications for result interpretation**This list is not comprehensive, but illustrates how results can be affected by collection, processing, and analysis methods.
Specimen collection methods
Collection without a cold chain, or nucleic acid stabilising agents, can cause nucleic acid degradation and potential false-negative results or overgrowth of selected organisms, which leads to misinterpretation of abundance. Multiple freeze-thaw cycles can also cause nucleic acid degradation.
Nucleic acid extraction method
The absence of a bead-beating step could make the detection of some bacteria difficult (ie, bacteria do not lyse properly so their DNA is not released and will not be sequenced). Small specimen volumes can reduce the ability to detect low-level organisms.
Sequencing library preparation
Poly-A tail enrichment of RNA will not include fragmented pathogen genomes; DNA sequencing alone will not detect RNA viruses.
Targeting of sequences
Capture probe-targeted sequencing will limit detection to targeted, known sequences. 16S targeted sequencing, as opposed to whole genome sequencing, will have limitations for the level of taxonomic classification.
Sequencing methods
High-level sample multiplexing can lead to insufficient read depth to detect organisms present at low levels. Computational contamination can occur between samples pooled on the same sequencing run due to a sample barcode for a sequence being misread and misassigned to another sample on the same run.82 This is termed barcode bleed-through; dual barcodes drop the rate of bleed through dramatically compared with single barcodes. Unique molecular identifiers are an even more powerful way to identify this phenomenon when compared with dual barcodes.
Processing controls
Negative controls allow some contaminating organisms to be identified. Internal positive controls, reference standards such as sequins, reduce bias introduced by experimental variability and can improve recognition of low-level organisms.
Analysis methods
A small curated database, or highly stringent criteria might not include novel or unexpected organisms, leading to false negative results. An uncurated database or lenient criteria might also identify organisms incorrectly.
By reporting the potential sources of bias for a given study (figure 1) their potential influence can be considered with mitigation or compensation strategies or caveats made to improve interpretation. The complexity and multistep nature of microbiome measurement means that any metagenomics experiment should be considered and reported as a representative result, rather than assuming that it perfectly reflects the microbes present and their abundance. It is also why the term unbiased, which is often used when describing metagenomic experiments that do not use enrichment, should be used with caution (or not at all). The term untargeted metagenomics could be used instead (appendix pp 3–4).
Address potential bias introduced by bioinformatics analysis
Classification algorithms rely on alignment of sequencing reads and contigs obtained from overlapping reads against reference genomes. In the case of the alignment of assembled contigs, reads that cannot be built into contigs (unassigned reads) are discarded, which can lead to a potential loss of information.78 Classification of reads might be slow and a smaller database could be built with unique sequences representing certain taxa.79 However, this can lead to bias in the assignment of homologous sequences and should be clearly reported.
Samples containing low abundance pathogens might produce false-negative results by not classifying sequencing reads as relevant or produce false-positive results if reads are non-specific.80 Subsequent alignment of sequence reads against a reference genome of the candidate pathogen(s) identified by the metagenomics analysis can provide necessary validation—wide and distributed coverage of the reference genome and high mapping identity is unlikely to result in a false positive. The level of coverage might be limited in samples with low pathogen load but still can be a true-positive result. Sufficient read depth is not always available for metagenomics data from clinical samples, which often contain a large proportion of reads derived from the host. Additionally, high read depth can generally be achieved only for microbes present at high-copy number. Authors should report where these considerations are relevant.
Assessing the quality of reads before downstream classification is crucial for ensuring accuracy of taxonomic assignment. This quality control usually includes removal of adapters, background sequences (human, host, or known), low-complexity sequence reads, trimming of low-quality bases at the ends of reads, and removal of primer sequences. The total number of reads in each sample can be affected by factors including DNA extraction methods, sample handling, library preparation, differences in sequencing depth. As such, it is generally advisable to normalise read abundance between samples before any analysis and report where this is done.81 Sophisticated statistical modelling approaches can deal with variation in read numbers between samples without loss of data (eg, DESeq2).82
Describe or address limitations of reference databases
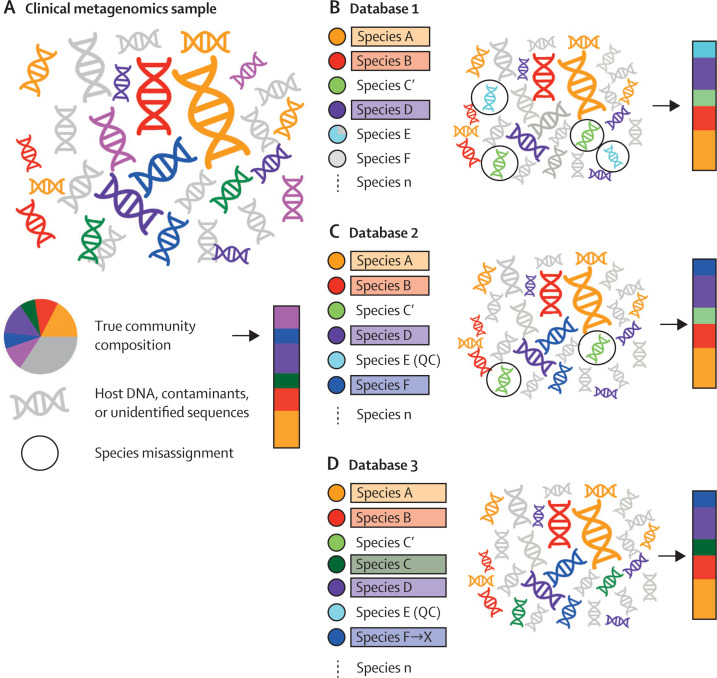
The use of reference databases should be clearly described. It is crucial that the reference database, genomic data download date, and a description of the procedures behind the inclusion and indexing of reference sequences are clearly presented. Limitations of reference databases can interfere with correct assignment of sequences (figure 2 ). Curated reference databases might not include all the relevant microbial diversity. Conversely, non-curated databases can comprise incorrectly named, incomplete, low sequencing quality, or artefactual sequences.83 Studies have shown that sequences arising from sample contamination or incompleteness (eg, an incomplete region of a genome that contains an important mutation) are frequent features of reference databases, particularly when draft genomes are included. For example, over 1000 published microbial genome sequences have been identified as contaminated with phiX174, a bacteriophage used as a control in Illumina sequencing,24 and 2250 NCBI GenBank draft bacterial and archaeal genomes contain spurious human sequences.84 Additionally, false-negative results might be due to a focal species missing taxonomic representation in the databases, which have an inherent curatorial bias to known human associated pathogens (appendix pp 4–5).85
Figure 2.
The importance of reference database choice, design, and versioning in taxonomic profiling of clinical metagenomics samples
(A) Schematic representation of a typical clinical metagenomics sample with species assigned as coloured DNA and grey denoting DNA deriving from the host, contaminants, unidentified taxa, or taxa sequenced at low depth. The pie chart provides the full metagenomic composition with the bar providing the species composition excluding host DNA and contaminants. (B) Taxonomic profiling based on database 1. Species confidently assigned are highlighted by colours with unassigned species shown in grey. Using database 1, species A, B, and D are correctly assigned. Species that are misassigned are outlined with a circle. In this instance, sequences from species C are assigned to the closely related species C' because of the lack of a representative of species C in the reference database. Additionally, the reference database contains a partially contaminated sequence from species E, which is misassigned to contaminant sequences in the test clinical metagenomics sample. This affects the inference of species composition shown in the bar. (C) The addition of species F to database 2 allows assignment of a greater proportion of the species present in the original clinical metagenomics sample. Quality control and improvement of reference species E, now species E (QC), removes the spurious assignment of contaminant species. Species C is still misassigned to species C', its closest representative in the database. (D) Updating the reference database to include species C results in the correct assignment of sequences to species C rather than species C'. Species F is taxonomically reassigned to species X, leading to a change in the assigned species name despite no change in the data in the reference or query datasets. In all cases the pink sequences present in the original clinical metagenomics sample are not assigned as this species is not present in any of the three reference databases.
Study size
Describe clearly how power calculations were made
Whenever comparisons in metagenomic species composition between two or more groups are made, authors should report relevant parameters such as significance level, power threshold, sequencing depth, effect size, number of comparisons, methods used to correct for multiple comparisons, and details of the statistical methods used for power calculations. It should be clearly stated how an effect size was derived and a rationale for the clinical relevance of the specific effect size should be given. If no power calculation was made, an explanation should be given about why this was not considered feasible or useful (appendix pp 5–6).
Statistical methods
State the limit of detection, including analytical sensitivity and specificity
The limit of detection (LOD) refers to the minimum quantity of genomic material from an organism required for its detection and should be stated in metagenomics studies. Determination of the LOD for a metagenomics study is dependent on the sequencing technology, sequencing depth, read length, representation of genomes related to the taxa of interest in the reference database, and the complexity of the community and amount of host nucleic acid in the sample. Simple calculations give estimates for the LOD (eg, for 106 reads per sample, the LOD is one read per sample), which corresponds to a relative abundance of the order of magnitude of 10−6 (ie, ∼0·0001%). Formal calculations of LOD that are needed for clinical validation should be done using probit analysis.86 In practice, the LOD will be considerably higher than that derived from these calculations because a single read from a taxon is very likely to be due to contamination or misclassification. Rather than trusting such calculations, the use of positive (spiked) controls and negative controls in the sequencing run allows assessment of sensitivity and specificity. With a single infection, the number of on-target reads will be correlated with the signal in the sample but mixed infections and coinfections will influence sensitivity.87 Experimentally validating these for model organisms that represent the specific pathogens of interest (eg, a DNA virus, an RNA virus, Gram-negative and Gram-positive bacteria, etc) is recommended, particularly for diagnostic tests.
Discussion
Attempt or acknowledge the need for functional or phenotypic validation
Genotypic data do not always correlate with clinical phenotype; for example, mechanisms that involve inducible resistance, gene expression and regulation, or post-translational modifications. In studies investigating mixed microbial communities it may not always be possible to determine which taxon a particular gene belongs to.88, 89 This is also relevant in the establishment of causality.
Efforts should be made to undertake phenotypic and functional validation to assess the inferred results. If this is not possible, or beyond the scope of the study, the limitations of inferring results solely from genotypic data should be acknowledged and discussed, including known caveats and restrictions on making key assumptions.
Consider the need for species or strain resolution
Different strains or lineages within a species can differ widely in their phenotypic characteristics. For example, sequencing with strain-level resolution enabled identification of specific strains of Escherichia coli associated with necrotising enterocolitis in preterm newborns90 and lineages of Salmonella enterica associated with varying clinical phenotypes.91 Therefore, profiling microbial communities with sub-species resolution can be useful, although de novo assembly of metagenomic reads remains a methodological challenge.
The strain and species resolution capacity of the assay used should be clearly stated with consideration for how the resolution applies to the study in question. In particular, microbial community profiling using 16S rRNA gene sequencing cannot identify individual species within some genera and should never be used to identify to the strain level. As recommended in STROME-ID, a definition or reference to published definitions of a strain should be provided.23
Other information
Report any ethical considerations with specific implications for metagenomics
Metagenomics produces a vast amount of host and pathogen data, which are untargeted and sometimes not of immediate interest.92 Molecular methods to deplete human genomic material exist; however, they remain imperfect. It might be sufficient to detail in a protocol that the host data will be removed, and not analysed, although this approach could lead to bias in microbial reads caused by the in silico host-depletion method—host genomes can contain viable viral genomes and non-viable genetic material derived from or shared with microorganisms. In these cases, the method used to identify and exclude host reads—eg, through mapping of all reads to the host reference genome—should be reported. including the choice of mapping algorithm and programme parameters.
Even if data analysis is restricted to non-human reads, it could still unveil potentially sensitive information,93 such as a new diagnosis of HIV. It has also been shown that more than 80% of individuals can be identified from populations of hundreds using their gut microbiome profile.94 These issues pose real concerns, particularly with the increasing requirement for data to be made publicly available. For all these reasons, specific ethical implications relating to metagenomics data and corresponding approvals should be stated, and appropriate ethical approval should be obtained.
Conclusions
Metagenomics has already made a significant impact on pathogen detection and characterisation, and we probably still underestimate its full potential. Increasing use of metagenomics has been accompanied by recognition of complex issues at every stage in the pipeline—ie, sample collection, sequencing, and analysis. Standards for reporting are therefore needed to ensure clarity, consistency, and robustness of research. The guidance given in this paper constitutes a set of recommendations and we recognise that research studies need to be pragmatic and use available resources. Nonetheless, reporting known and potential limitations should minimise misrepresentation. It is inevitable that the field of metagenomics will continue to advance steadily and these guidelines will need to be updated.
Search strategy and selection criteria
In 2018, a STROBE-metagenomics working group was established, identified through notable researchers in the field, including a geographically diverse group of epidemiologists, statisticians, bioinformaticians, neurologists, virologists, microbiologists, and specialists in public health and infectious diseases. Participants met to agree the structure and content of the statement, and the proposal was registered with the Equator Network.24 Specific issues to be covered were identified (panel 2 ). A systematic approach was taken to gather evidence to support the recommendations, with literature searches performed in PubMed, searching references of articles, and supplemented by expert opinion. Literature searches were done in PubMed using medical subject headings terms and keywords “(?sequenc* OR metagenom* OR Illumina OR RNA-seq OR RNASeq OR (Roche 454) OR (Ion torrent) OR (Proton / PGM) OR MiSeq OR HiSeq OR NextSeq OR MinION OR Nanopore OR PacBio) AND (infectio* OR microorganism OR microorganisms OR pathogen OR pathogens OR bacteria* OR virus OR viral OR fungus OR fungi OR parasite OR parasites OR parasitic)”, searching references of articles, and supplemented by expert opinion from within the group. Articles were limited to those in English language published between January, 2000, and June, 2019. Areas that were adequately addressed in existing STROBE22 and STROME-ID23 statements were not covered. Iterative versions of the guidelines and manuscript were circulated to develop a consensus. The STROBE-metagenomics extension has been developed to complement the STROBE and STROME-ID statements, with the new recommendations organised alongside the existing table. The guidelines discussed therefore cover only the new proposals for reporting.
Panel 2. Key issues to be addressed in publications applying metagenomics.
-
•
Specimen collection, handling, preservation, and storage
-
•
Nucleic acid extraction
-
•
Sequencing instrumentation and processing, including library preparation
-
•
Bioinformatic analysis method, including workflow, database composition, and parameterisation
-
•
Quality assurance measures, including internal quality control, such as the use of adequate internal and external controls
-
•
Limits of detection, including analytical sensitivity, and specificity for clinical testing
-
•
Power and sample size calculations
-
•
Use of orthogonal methods to confirm sequencing results
-
•
Criteria to confirm the role of pathogen(s) in disease aetiology
-
•
Turnaround time
-
•
Cost
-
•
Ethical considerations
-
•
Specific issues related to applications, such as in the diagnosis of CNS infections, and investigation of antimicrobial resistance
For more on protocol sharing see http://www.protocols.io/
This online publication has been corrected. The corrected version first appeared at thelancet.com/infection on October 23, 2020
Acknowledgments
Acknowledgments
AN and LBR are National Institute of Health (NIH) employees and are in receipt on an NIH grant (NS003130). TB is supported by the University of Oxford and the Medical Research Council (grant number MR/N013468/1). MW is funded by a National Institute of Neurological Disorders and Stroke (grant number K08NS096117). LVT is a Wellcome Research Fellow (grant number 204904/Z/16/Z).
Contributors
TB and NF conceived the idea and, together with CO, co-ordinated the Review. DOS and JH designed figure 1 and LvD and FB designed figure 2. All authors were involved in the study design, literature review, writing the manuscript, and editing successive drafts.
Declaration of interests
ME reports personal fees and other financials from PATHOQUEST, none received during the conduct of the study. MRW has a patent issued for Depletion of Abundant Sequences by Hybridization. All other authors declare no competing interests.
Footnotes
This list is not comprehensive, but illustrates how results can be affected by collection, processing, and analysis methods.
Supplementary Material
References
- 1.Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol. 1998;5:R245–R249. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- 2.Quince C, Walker AW, Simpson JT, Loman NJ, Segata N. Shotgun metagenomics, from sampling to analysis. Nature Biotechnol. 2017;35:833–844. doi: 10.1038/nbt.3935. [DOI] [PubMed] [Google Scholar]
- 3.Forbes JD, Knox NC, Peterson C-L, Reimer AR. Highlighting clinical metagenomics for enhanced diagnostic decision-making: a step towards wider implementation. computational and structural biotechnology journal. Comput Struct Biotechnol J. 2018;16:108–120. doi: 10.1016/j.csbj.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiu CY, Miller SA. Clinical metagenomics. Nat Rev Genet. 2019;20:341–355. doi: 10.1038/s41576-019-0113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forbes JD, Knox NC, Ronholm J, Pagotto F, Reimer A. Metagenomics: the next culture-independent game changer. Front Microbiol. 2017;8 doi: 10.3389/fmicb.2017.01069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorzelak MA, Gill SK, Tasnim N, Ahmadi-Vand Z, Jay M, Gibson DL. Methods for improving human gut microbiome data by reducing variability through sample processing and storage of stool. PLoS One. 2015;10 doi: 10.1371/journal.pone.0134802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simner PJ, Miller S, Carroll KC. Understanding the promises and hurdles of metagenomic next-generation sequencing as a diagnostic tool for infectious diseases. Clinical Infect Dis. 2018;66:778–788. doi: 10.1093/cid/cix881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamura S, Yang C-S, Sakon N, et al. Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach. PLoS One. 2009;4 doi: 10.1371/journal.pone.0004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Helm E, Imamovic L, Hashim Ellabaan MM, van Schaik W, Koza A, Sommer MOA. Rapid resistome mapping using nanopore sequencing. Nucleic Acids Res. 2017;45:e61. doi: 10.1093/nar/gkw1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim D, Hofstaedter CE, Zhao C, et al. Optimising methods and dodging pitfalls in microbiome research. Microbiome. 2017;5:52. doi: 10.1186/s40168-017-0267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu N, Zhang D, Wang W, et al. a novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown JR, Bharucha T, Breuer J. Encephalitis diagnosis using metagenomics: application of next generation sequencing for undiagnosed cases. J Infect. 2018;76:225–240. doi: 10.1016/j.jinf.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson MR, Sample HA, Zorn KC, et al. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med. 2019;380:2327–2340. doi: 10.1056/NEJMoa1803396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhernakova A, Kurilshikov A, Bonder MJ, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wirbel J, Pyl PT, Kartal E, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. 2019;25:679–689. doi: 10.1038/s41591-019-0406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greninger AL, Zerr DM, Qin X, et al. Rapid metagenomic next-generation sequencing during an investigation of hospital-acquired human parainfluenza virus 3 infections. J Clin Microbiol. 2017;55:177–182. doi: 10.1128/JCM.01881-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loman NJ, Constantinidou C, Christner M, et al. A culture-independent sequence-based metagenomics approach to the investigation of an outbreak of Shiga-toxigenic Escherichia coli O104:H4. JAMA. 2013;309:1502–1510. doi: 10.1001/jama.2013.3231. [DOI] [PubMed] [Google Scholar]
- 19.Brooks JP, Edwards DJ, Harwich MD, et al. The truth about metagenomics: quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol. 2015;15:66. doi: 10.1186/s12866-015-0351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruppé E, Lazarevic V, Girard M, et al. Clinical metagenomics of bone and joint infections: a proof of concept study. Sci Rep. 2017;7 doi: 10.1038/s41598-017-07546-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt K, Mwaigwisya S, Crossman LC, et al. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J Antimicrob Chemother. 2017;72:104–114. doi: 10.1093/jac/dkw397. [DOI] [PubMed] [Google Scholar]
- 22.von Elm E, Altman DG, Egger M, et al. The strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. Int J Surg. 2008;61:344–349. doi: 10.1016/j.jclinepi.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 23.Field N, Cohen T, Struelens MJ, et al. Strengthening the reporting of molecular epidemiology for infectious diseases (STROME-ID): an extension of the STROBE statement. Lancet Infect Dis. 2014;14:341–352. doi: 10.1016/S1473-3099(13)70324-4. [DOI] [PubMed] [Google Scholar]
- 24.Equator Network EQUATOR (Enhancing the QUAlity and Transparency of health Research) Network home page. 2009. https://www.equator-network.org/
- 25.Duncan CJ, Mohamad SM, Young DF, et al. Human IFNAR2 deficiency: lessons for antiviral immunity. Sci Transl Med. 2015;7 doi: 10.1126/scitranslmed.aac4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morfopoulou S, Brown JR, Davies EG, et al. Human Coronavirus OC43 associated with fatal encephalitis. N Engl J Med. 2016;375:497–498. doi: 10.1056/NEJMc1509458. [DOI] [PubMed] [Google Scholar]
- 27.Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 28.Yohe S, Hauge A, Bunjer K, et al. Clinical validation of targeted next-generation sequencing for inherited disorders. Arch Pathol Lab Med. 2015;139:204–210. doi: 10.5858/arpa.2013-0625-OA. [DOI] [PubMed] [Google Scholar]
- 29.Jennings LJ, Arcila ME, Corless C, et al. Guidelines for validation of next-generation sequencing-based oncology panels: a joint consensus recommendation of the association for molecular pathology and college of american pathologists. J Mol Diagn. 2017;19:341–365. doi: 10.1016/j.jmoldx.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlaberg R, Chiu CY, Miller S, Procop GW, Weinstock G. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med. 2017;141:776–786. doi: 10.5858/arpa.2016-0539-RA. [DOI] [PubMed] [Google Scholar]
- 31.Seelenfreund E, Robinson WA, Amato CM, Tan A-C, Kim J, Robinson SE. Long term storage of dry versus frozen RNA for next generation molecular studies. PLoS One. 2014;9 doi: 10.1371/journal.pone.0111827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panek M, Čipčić Paljetak H, Barešić A, et al. Methodology challenges in studying human gut microbiota—effects of collection, storage, DNA extraction and next generation sequencing technologies. Sci Rep. 2018;8 doi: 10.1038/s41598-018-23296-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu WK, Chen CC, Panyod S, et al. Optimization of fecal sample processing for microbiome study—the journey from bathroom to bench. J Formos Med Assoc. 2019;118:545–555. doi: 10.1016/j.jfma.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Ali N, Rampazzo RCP, Costa ADT, Krieger MA. Current nucleic acid extraction methods and their implications to point-of-care diagnostics. Biomed Res Int. 2017;2017 doi: 10.1155/2017/9306564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minervini CF, Cumbo C, Orsini P, et al. Nanopore sequencing in blood diseases: a wide range of opportunities. Front Genet. 2020;11:76. doi: 10.3389/fgene.2020.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schirmer M, Ijaz UZ, D'Amore R, Hall N, Sloan WT, Quince C. Insight into biases and sequencing errors for amplicon sequencing with the Illumina MiSeq platform. Nucleic Acids Res. 2015;43:e37. doi: 10.1093/nar/gku1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP. Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet. 2014;15:121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- 38.Clark MJ, Chen R, Lam HYK, et al. Performance comparison of exome DNA sequencing technologies. Nat Biotechnol. 2011;29:908–914. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jennings LJ, Arcila ME, Corless C, et al. Guidelines for validation of next-generation sequencing-based oncology panels: a joint consensus recommendation of the association for molecular pathology and college of american pathologists. J Mol Diagn. 2017;19:341–365. doi: 10.1016/j.jmoldx.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hardwick SA, Chen WY, Wong T, et al. Synthetic microbe communities provide internal reference standards for metagenome sequencing and analysis. Nat Commun. 2018;9 doi: 10.1038/s41467-018-05555-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.The Integrative HMP (iHMP) Research Network Consortium The integrative human microbiome project. Nature. 2019;569:641–648. doi: 10.1038/s41586-019-1238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hornung BVH, Zwittink RD, Kuijper EJ. Issues and current standards of controls in microbiome research. FEMS Microbiol Ecol. 2019;95 doi: 10.1093/femsec/fiz045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cummings LA, Kurosawa K, Hoogestraat DR, et al. Clinical next generation sequencing outperforms standard microbiological culture for characterizing polymicrobial samples. Clin Chem. 2016;62:1465–1473. doi: 10.1373/clinchem.2016.258806. [DOI] [PubMed] [Google Scholar]
- 51.Lipkin WI. Microbe hunting. Microbiol Mol Biol Rev. 2010;74:363–377. doi: 10.1128/MMBR.00007-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Granerod J, Cunningham R, Zuckerman M, et al. Causality in acute encephalitis: defining aetiologies. Epidemiol Infect. 2010;138:783–800. doi: 10.1017/S0950268810000725. [DOI] [PubMed] [Google Scholar]
- 53.Fischbach MA. Microbiome: focus on causation and mechanism. Cell. 2018;174:785–790. doi: 10.1016/j.cell.2018.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langelier C, Kalantar KL, Moazed F, et al. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc Natl Acad Sci USA. 2018;115:e12353–e12362. doi: 10.1073/pnas.1809700115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh VP, Proctor SD, Willing BP. Koch's postulates, microbial dysbiosis and inflammatory bowel disease. Clin Microbiol Infect. 2016;22:594–599. doi: 10.1016/j.cmi.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 56.Gyarmati P, Kjellander C, Aust C, Song Y, Öhrmalm L, Giske CG. Metagenomic analysis of bloodstream infections in patients with acute leukemia and therapy-induced neutropenia. Sci Rep. 2016;6 doi: 10.1038/srep23532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grumaz S, Stevens P, Grumaz C, et al. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 2016;8:73. doi: 10.1186/s13073-016-0326-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fredricks DN, Relman DA. Sequence-based identification of microbial pathogens: a reconsideration of Koch's postulates. Clin Microbiol Rev. 1996;9:18–33. doi: 10.1128/cmr.9.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lipkin WI. The changing face of pathogen discovery and surveillance. Nature Rev Microbiol. 2013;11:133–141. doi: 10.1038/nrmicro2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cuthbertson L, Rogers GB, Walker AW, et al. Time between collection and storage significantly influences bacterial sequence composition in sputum samples from cystic fibrosis respiratory infections. J Clin Microbiol. 2014;52:3011–3016. doi: 10.1128/JCM.00764-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cardona S, Eck A, Cassellas M, et al. Storage conditions of intestinal microbiota matter in metagenomic analysis. BMC Microbiol. 2012;12:158. doi: 10.1186/1471-2180-12-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Senol Cali D, Kim JS, Ghose S, Alkan C, Mutlu O. Nanopore sequencing technology and tools for genome assembly: computational analysis of the current state, bottlenecks and future directions. Brief Bioinform. 2019;20:1542–1559. doi: 10.1093/bib/bby017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller RR, Montoya V, Gardy JL, Patrick DM, Tang P. Metagenomics for pathogen detection in public health. Genome Med. 2013;5:81. doi: 10.1186/gm485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bossuyt PM, Reitsma JB, Bruns DE, et al. The STARD statement for reporting studies of diagnostic accuracy: explanation and elaboration. Ann Intern Med. 2003;49:7–18. doi: 10.7326/0003-4819-138-1-200301070-00012-w1. [DOI] [PubMed] [Google Scholar]
- 66.Ambrose HE, Granerod J, Clewley JP, et al. Diagnostic strategy used to establish etiologies of encephalitis in a prospective cohort of patients in England. J Clin Microbiol. 2011;49:3576–3583. doi: 10.1128/JCM.00862-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shaw L, Ribeiro ALR, Levine AP, et al. The human salivary microbiome is shaped by shared environment rather than genetics: evidence from a large family of closely related individuals. mBio. 2017;8:e01237–e01317. doi: 10.1128/mBio.01237-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lassalle F, Spagnoletti M, Fumagalli M, et al. Oral microbiomes from hunter-gatherers and traditional farmers reveal shifts in commensal balance and pathogen load linked to diet. Mol Ecol. 2018;27:182–195. doi: 10.1111/mec.14435. [DOI] [PubMed] [Google Scholar]
- 69.Shaw LP, Bassam H, Barnes CP, Walker AS, Klein N, Balloux F. Modelling microbiome recovery after antibiotics using a stability landscape framework. ISME J. 2019;13:1845–1856. doi: 10.1038/s41396-019-0392-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zaura E, Brandt BW, Teixeira de Mattos MJ, et al. Same exposure but two radically different responses to antibiotics: resilience of the salivary microbiome versus long-term microbial shifts in feces. mBio. 2015;6:e01693–e01715. doi: 10.1128/mBio.01693-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schenk T, Enders M, Pollak S, Hahn R, Huzly D. High prevalence of human parvovirus B19 DNA in myocardial autopsy samples from subjects without myocarditis or dilative cardiomyopathy. J Clin Microbiol. 2009;47:106–110. doi: 10.1128/JCM.01672-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sullivan DM, Laver T, Temisak S, et al. Assessing the accuracy of quantitative molecular microbial profiling. Int J Mol Sci. 2014;15:21476–21491. doi: 10.3390/ijms151121476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pettengill JB, McAvoy E, White JR, Allard M, Brown E, Ottesen A. Using metagenomic analyses to estimate the consequences of enrichment bias for pathogen detection. BMC Res Notes. 2012;5:378. doi: 10.1186/1756-0500-5-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Velásquez-Mejía EP, de la Cuesta-Zuluaga J, Escobar JS. Impact of DNA extraction, sample dilution, and reagent contamination on 16S rRNA gene sequencing of human feces. Appl Microbiol Biotechnol. 2018;102:403–411. doi: 10.1007/s00253-017-8583-z. [DOI] [PubMed] [Google Scholar]
- 75.Huggett JF, Laver T, Tamisak S, et al. Considerations for the development and application of control materials to improve metagenomic microbial community profiling. Accred Qual Assur. 2013;18:77–83. [Google Scholar]
- 76.Cai L, Ye L, Tong AHY, Lok S, Zhang T. Biased diversity metrics revealed by bacterial 16S pyrotags derived from different primer sets. PLoS One. 2013;8 doi: 10.1371/journal.pone.0053649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Edgar RC. UNBIAS: an attempt to correct abundance bias in 16S sequencing, with limited success. bioRxiv. 2017 https://www.biorxiv.org/content/10.1101/124149v1.full.pdf published online April 4. (preprint). [Google Scholar]
- 78.Paez-Espino D, Pavlopoulos GA, Ivanova NN, Kyrpides NC. Nontargeted virus sequence discovery pipeline and virus clustering for metagenomic data. Nat Protoc. 2017;12:1673–1682. doi: 10.1038/nprot.2017.063. [DOI] [PubMed] [Google Scholar]
- 79.Kim D, Song L, Breitwieser FP, Salzberg SL. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 2016;26:1721–1729. doi: 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Breitwieser FP, Baker DN, Salzberg SL. KrakenUniq: confident and fast metagenomics classification using unique k-mer counts. Genome Biol. 2018;19:198. doi: 10.1186/s13059-018-1568-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pereira MB, Wallroth M, Jonsson V, Kristiansson E. Comparison of normalization methods for the analysis of metagenomic gene abundance data. BMC Genomics. 2018;19:274. doi: 10.1186/s12864-018-4637-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mukherjee S, Huntemann M, Ivanova N, Kyrpides NC, Pati A. Large-scale contamination of microbial isolate genomes by Illumina PhiX control. Stand Genomic Sci. 2015;10:18. doi: 10.1186/1944-3277-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Breitwieser FP, Pertea M, Zimin AV, Salzberg SL. Human contamination in bacterial genomes has created thousands of spurious proteins. Genome Res. 2019;29:954–960. doi: 10.1101/gr.245373.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burd EM. Validation of laboratory-developed molecular assays for infectious diseases. Clin Microbiol Rev. 2010;23:550–576. doi: 10.1128/CMR.00074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Greninger AL. The challenge of diagnostic metagenomics. Expert Rev Mol Diagn. 2018;18:605–615. doi: 10.1080/14737159.2018.1487292. [DOI] [PubMed] [Google Scholar]
- 88.Miller S, Naccache S, Samayoa E, et al. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res. 2019;29:831–842. doi: 10.1101/gr.238170.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Relman DA. Actionable sequence data on infectious diseases in the clinical workplace. Clin Chem. 2015;61:38–40. doi: 10.1373/clinchem.2014.229211. [DOI] [PubMed] [Google Scholar]
- 90.Ward DV, Scholz M, Zolfo M, et al. Metagenomic sequencing with strain-level resolution implicates uropathogenic E coli in necrotizing enterocolitis and mortality in preterm infants. Cell Rep. 2016;14:2912–2924. doi: 10.1016/j.celrep.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Khajanchi BK, Yoskowitz NC, Han J, Wang X, Foley SL. Draft genome sequences of 27 Salmonella enterica serovar schwarzengrund isolates from clinical sources. Microbiol Resour Announc. 2019;8:e01687–e01718. doi: 10.1128/MRA.01687-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Charalampous T, Kay GL, Richardson H, et al. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat Biotechnol. 2019;37:783–792. doi: 10.1038/s41587-019-0156-5. [DOI] [PubMed] [Google Scholar]
- 93.Ruppé E, Schrenzel J. Messages from the third international conference on clinical metagenomics (ICCMg3) Microbes Infect. 2019;21:273–277. doi: 10.1016/j.micinf.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 94.Franzosa EA, Huang K, Meadow JF, et al. Identifying personal microbiomes using metagenomic codes. Proc Natl Acad Sci USA. 2015;112:e2930–e3008. doi: 10.1073/pnas.1423854112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Uncited References
- 41.Branton WG, Ellestad KK, Maingat F, et al. Brain microbial populations in HIV/AIDS: alpha-proteobacteria predominate independent of host immune status. PLoS One. 2013;8 doi: 10.1371/journal.pone.0054673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singer E, Andreopoulos B, Bowers RM, et al. Next generation sequencing data of a defined microbial mock community. Sci Data. 2016;3 doi: 10.1038/sdata.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rinke C, Low S, Woodcroft BJ, et al. Validation of picogram- and femtogram-input DNA libraries for microscale metagenomics. PeerJ. 2016;4 doi: 10.7717/peerj.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bowers RM, Clum A, Tice H, et al. Impact of library preparation protocols and template quantity on the metagenomic reconstruction of a mock microbial community. BMC Genomics. 2015;16:856. doi: 10.1186/s12864-015-2063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Branton WG, Lu JQ, Surette MG, et al. Brain microbiota disruption within inflammatory demyelinating lesions in multiple sclerosis. Sci Rep. 2016;6 doi: 10.1038/srep37344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Balloux F, Brønstad Brynildsrud O, van Dorp L, et al. From theory to practice: translating whole-genome sequencing (WGS) into the clinic. Trends Microbiol. 2018;26:1035–1048. doi: 10.1016/j.tim.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.