Abstract
Colitis is associated with the development of colorectal cancer (CRC) by largely undefined mechanisms that are critical for understanding the link between inflammation and cancer. Intestinal stem cells (ISCs) marked by leucine-rich repeat–containing G protein–coupled receptor 5 (LGR5) expression are of importance in both the inflammatory response to colitis and progression to colitis-associated colon cancer (CACC). Here, we report in human mucin 1–transgenic (MUC1-transgenic) mouse models of CACC, targeting the MUC1-C oncogenic protein suppresses the (a) Lgr5+ ISC population, (b) induction of Myc and core pluripotency stem cell factors, and (c) severity and progression of colitis to dysplasia and cancer. By extension to human colon cancer cells, we demonstrate that MUC1-C drives MYC, forms a complex with MYC on the LGR5 promoter, and activates LGR5 expression. We also show in CRC cells that MUC1-C induces cancer stem cell (CSC) markers (BMI1, ALDH1, FOXA1, LIN28B) and the OCT4, SOX2, and NANOG pluripotency factors. Consistent with conferring the CSC state, targeting MUC1-C suppresses the capacity of CRC cells to promote wound healing, invasion, self-renewal, and tumorigenicity. In analysis of human tissues, MUC1 expression associates with activation of inflammatory pathways, development of colitis, and aggressiveness of CRCs. These results collectively indicate that MUC1-C is of importance for integrating stemness and pluripotency in colitis and CRC. Of clinical relevance, the findings further indicate that MUC1-C represents a potentially previously unrecognized target that is druggable for treating progression of colitis and CRC.
Keywords: Oncology, Therapeutics
Keywords: Adult stem cells, Colorectal cancer, Inflammatory bowel disease
The MUC1-C oncoprotein drives stemness and pluripotency in the progression of colitis to colorectal cancer, establishing a link between inflammation and carcinogenesis.
Introduction
Chronic inflammation is associated with cancer initiation and progression (1). MUC1 is a heterodimeric protein that is upregulated in the response to inflammation and is overexpressed in diverse carcinomas (2, 3). However, a functional role for MUC1 in linking inflammation and cancer is not known.
The oncogenic human MUC1 C-terminal (MUC1-C) contributes to multiple hallmarks of the cancer cell, including induction of epithelial-mesenchymal transition (EMT) and stemness (4–6). MUC1-C also induces (a) epigenetic reprogramming by activating expression of DNA methyltransferase 1 (DNMT1) and DNMT3b and thereby alterations in DNA methylation patterns (5, 7, 8), (b) BMI1 and other components of the polycomb repressive complex 1 (PRC1) with ubiquitylation of H2A (9), and (c) EZH2, a histone methyltransferase and subunit of PRC2, with changes in H3K27 trimethylation (10). Other work has shown that MUC1-C activates the nucleosome remodeling and deacetylase (NuRD) chromatin remodeling and acetylation complex in association with dedifferentiation and induction of the Yamanaka pluripotency factors (11). These findings have supported the involvement of MUC1-C in driving EMT, stemness, epigenetic reprogramming, and pluripotency, which are associated with cancer stem cell (CSC) populations.
The importance of chronic inflammation in promoting tumorigenesis is supported by the association between colitis and the development of colorectal cancer (CRC) (12). Prolonged inflammation and repetitive cycles of injury/repair in the intestinal mucosa have been linked to the expansion of intestinal stem cells (ISCs) in colitis and CSCs in CRC (13, 14). ISCs marked by expression of leucine-rich repeat–containing G protein–coupled receptor 5 (mouse Lgr5/human LGR5) are cycling populations located at the crypt base (15). Bmi1+ ISCs detectable above the crypt base represent a distinct quiescent pool with the capacity to regenerate Lgr5+ cells in response to damage (16, 17). Populations of Lgr5+ or Bmi1+ ISCs can give rise to colon cancer in mice (15, 16, 18). Human LGR5+ and BMI1+ ISCs have also been linked to CSCs in CRC (19–21). However, the regulation of LGR5 and BMI1 has not been fully investigated in CRC CSCs.
Involvement of the oncogenic MUC1-C protein in the link between inflammation and cancer is unknown. The present work identifies a functional role for MUC1-C in inflammation-associated intestinal carcinogenesis by demonstrating the importance of this oncoprotein in driving (a) Lgr5+ ISCs in mouse models of colitis and (b) LGR5+ CSCs in colon cancer. Our results show that MUC1-C links the expansion of ISCs in colitis to the function of CSCs in CRC by promoting LGR5, BMI1, and core pluripotency factor expression. Of potential clinical significance, these findings indicate that MUC1-C is a druggable target for suppressing progression of colitis and CRC.
Results
MUC1-C promotes inflammation in a MUC1+/– IL-10–/– mouse model of colitis.
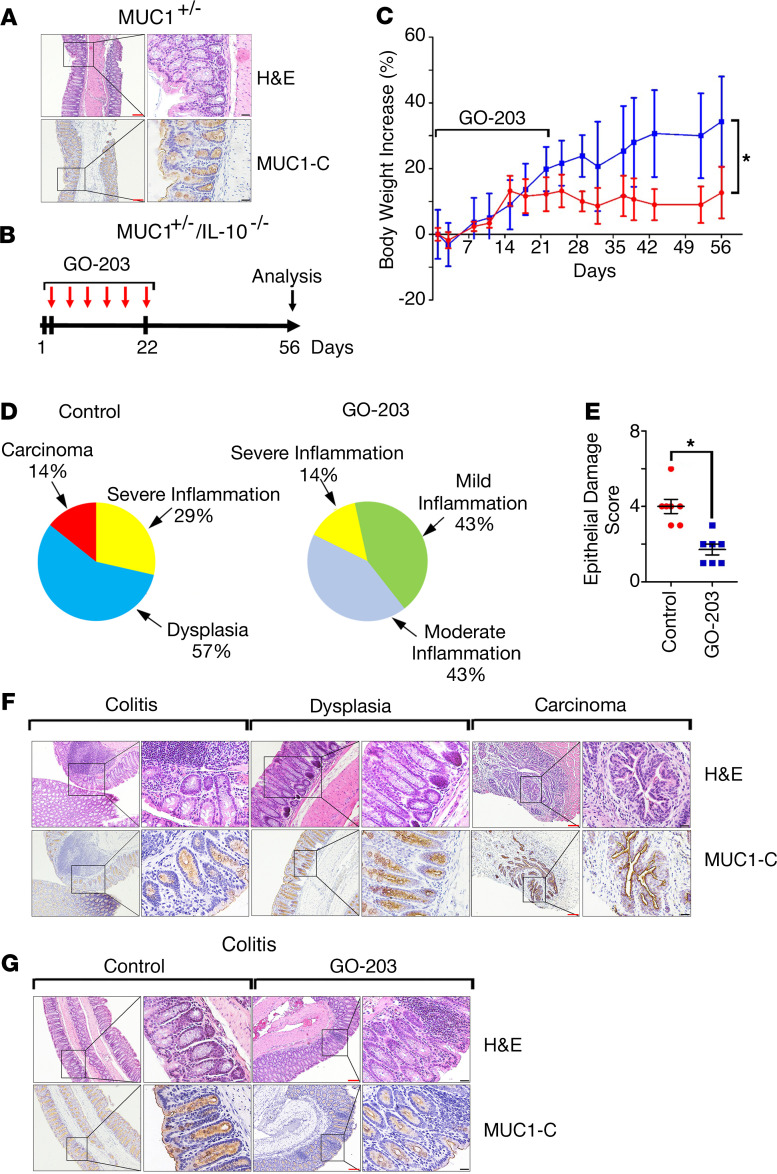
Mice transgenic for human MUC1 (MUC1+/–) express MUC1 in a pattern similar to that in humans with localization to the apical borders of intestinal epithelial cells (Figure 1A) (22, 23). Human MUC1 differs from mouse Muc1; therefore, MUC1+/– mice represent a model for studying the role of the MUC1 protein in vivo. MUC1+/– mice do not develop characteristics of colitis, as evidenced by a normal colonic mucosa (Figure 1A). Crossing MUC1+/– mice with IL-10–/– mice, which develop inflammatory epithelial hyperplasia (24), results in upregulation of MUC1 expression in association with exacerbation of colitis, diarrhea, and rectal prolapse (23, 25). Here, we found that treatment of MUC1+/– IL-10–/– mice with the GO-203 inhibitor (Figure 1B), which blocks MUC1-C homodimerization and oncogenic function (26–28), showed a trend in attenuating the development of rectal prolapse (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.137112DS1). GO-203 treatment of MUC1+/– IL-10–/– mice was also associated with significantly greater increases in body weight compared with controls (Figure 1C). As reported previously (23, 25), analysis of colon tissues from the MUC1+/– IL-10–/– mice demonstrated the presence of moderate to severe inflammation, dysplasia, and progression to carcinoma (Figure 1D, left; Supplemental Table 1). By contrast, GO-203 treatment (a) decreased the degree of inflammation and dysplasia (Figure 1D, right; Supplemental Table 1) and (b) resulted in significant suppression of the epithelial damage score (Figure 1E and Supplemental Table 2). In further support of MUC1-C involvement, we found by IHC that MUC1-C is increased in progression of colitis to dysplasia and carcinoma in control MUC1+/– IL-10–/– mice (Figure 1F) and that GO-203 treatment is associated with decreases in MUC1-C expression (Figure 1G).
Figure 1. Targeting MUC1-C attenuates inflammation in MUC1+/– IL-10–/– mice.
(A) Images of descending colonic mucosa from a MUC1+/– mouse stained with H&E (upper) and for MUC1-C by IHC (lower). Red scale bars: 200 μm. Black scale bars: 50 μm. (B) Schema for GO-203 treatment of MUC1+/– IL-10–/– mice. GO-203 nanoparticles (GO-203/NPs) were administered i.p. twice a week for 3 weeks. (C) Body weight increase for untreated (shown in red) and GO-203–treated (shown in blue) MUC1+/– IL-10–/– mice. The results are expressed as the percentage increase (mean ± SEM) of baseline weight on day 1. Body weights on day 56 were compared using Student’s t test. The asterisk denotes a significant difference (P < 0.05). (D) Pie charts representing the percentage of control untreated (left) and GO-203–treated (right) MUC1+/– IL-10–/– mice with inflammation, dysplasia, and adenocarcinoma as determined by microscopic analysis and scoring of H&E staining (Supplemental Table 1). (E) Epithelial damage score of H&E-stained colons from control and GO-203–treated MUC1+/– IL-10–/– mice as determined by microscopic analysis (Supplemental Table 2). (F) Images of colons with colitis, dysplasia, and adenocarcinoma from control MUC1+/– IL-10–/– mice stained with H&E (upper panels) and for MUC1-C by IHC (lower panels). Red scale bars: 200 μm. Black scale bars: 50 μm. (G) Images of colons with colitis from control and GO-203–treated MUC1+/– IL-10–/– mice stained with H&E (upper panels) and for MUC1-C (lower panels). Red scale bars: 200 μm. Black scale bars: 50 μm.
MUC1-C potentiates carcinogen-induced colitis-associated colon cancer.
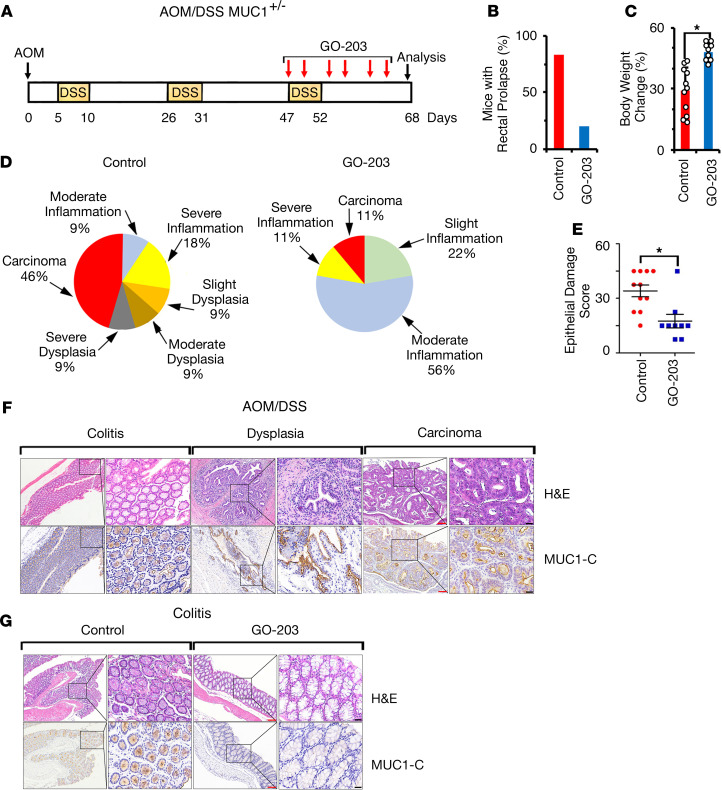
Administration of the carcinogen azoxymethane (AOM) with cycles of dextran sulfate sodium (DSS) is another model of colitis-associated colon cancer (CACC) (29). In the DSS/AOM model (Figure 2A), GO-203 administration was delayed to assess the effects of targeting MUC1-C in a setting of more established colitis than that with early treatment in the MUC1+/– IL-10–/– mice. AOM/DSS treatment of MUC1+/– mice was associated with development of rectal prolapse and effects on body weight gain, which were attenuated by GO-203 administration (Figure 2, B and C; and Supplemental Figure 1C). In the AOM/DSS model, inflammation and dysplasia were more pronounced than in MUC1+/– IL-10–/– mice (Figure 2D and Supplemental Table 3). Still, we found that GO-203 treatment associated with (a) decreases in inflammation, dysplasia, and carcinogenesis (Figure 2D and Supplemental Table 3) and (b) a significant reduction in epithelial damage score (Figure 2E and Supplemental Table 4). Progression from colitis to dysplasia and adenocarcinoma was associated with increases in MUC1-C expression as determined by IHC (Figure 2F). Moreover, as found in the MUC1+/– IL-10–/– model, GO-203–induced downregulation of inflammation and progression was associated with decreased MUC1-C expression (Figure 2G).
Figure 2. MUC1-C promotes AOM/DSS-induced inflammation and tumorigenesis in MUC1+/– mice.
(A) Schema of GO-203 administration to AOM/DSS-induced MUC1+/– mice. Mice were treated with GO-203/NPs i.p. twice a week for 3 weeks. (B) Percentage of AOM/DSS MUC1+/– mice left untreated (n = 11; red bar) or treated with GO-203 (n = 9; blue bar) with rectal prolapse at day 68. (C) Body weight increase for control (red bar) and GO-203–treated (blue bar) AOM/DSS-induced MUC1+/– mice. The results are expressed as the percentage increase (mean ± SEM) of baseline weight on day 1. (D) Pie charts representing the percentage of untreated control (left) and GO-203–treated (right) AOM/DSS-induced MUC1+/– mice with inflammation, dysplasia, and adenocarcinoma as determined by microscopic analysis and scoring of H&E-stained colons (Supplemental Table 3). (E) Epithelial damage score of H&E-stained colons from control and GO-203–treated AOM/DSS-induced MUC1+/– mice as determined by microscopic analysis (Supplemental Table 4). (F) Images of colons with colitis, dysplasia, and adenocarcinoma from control AOM/DSS-induced MUC1+/– mice stained with H&E (upper) and for MUC1-C (lower). Red scale bars: 200 μm. Black scale bars: 50 μm. (G) Images of colons with colitis from control and GO-203–treated AOM/DSS-induced MUC1+/– mice stained with H&E (upper) and for MUC1-C (lower). Red scale bars: 200 μm. Black scale bars: 50 μm.
Targeting MUC1-C suppresses Lgr5 and Bmi1 expression.
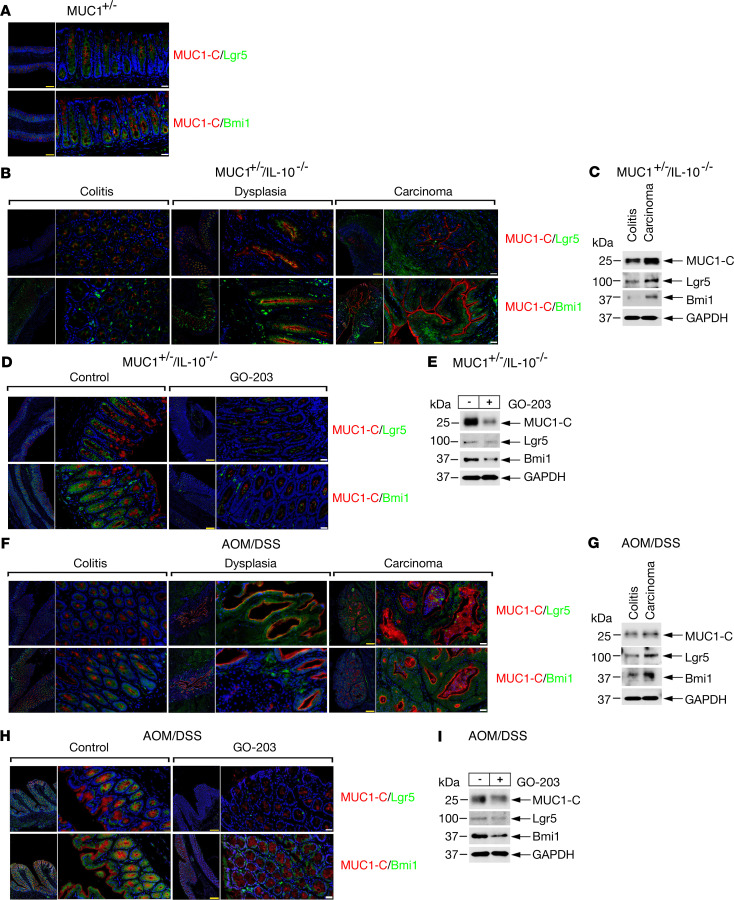
Immunofluorescence studies of the MUC1+/– mouse colon demonstrated Lgr5+ cells largely localized to the crypt base with some spreading to the luminal surface as has been observed in WT mice (Figure 3A, upper and lower) (15, 30). Similar patterns were observed for Bmi1+ ISCs (Figure 3A, upper and lower panels). In contrast, MUC1-C+ cells were found predominantly in more patchy distribution patterns toward the luminal surface (Figure 3A, upper and lower). In MUC1+/– IL-10–/– mice, we found increased (a) MUC1-C, Lgr5, and Bmi1 expression and (b) colocalization of MUC1-C with Lgr5 and Bmi1 with progression from colitis to dysplasia and carcinoma (Figure 3, B and C; see complete unedited blots in supplemental material). Analysis of colitis tissues further showed that GO-203 treatment decreased MUC1-C, Lgr5, and Bmi1 expression (Figure 3, D and E). Similar results obtained in the AOM/DSS-induced MUC1+/– model demonstrated increases in MUC1-C, Lgr5, and Bmi1 expression in progression of colitis to dysplasia and carcinoma (Figure 3, F and G), which were attenuated by GO-203 treatment (Figure 3, H and I), suggesting that MUC1-C contributes to expansion of ISCs in CACC.
Figure 3. MUC1-C drives Lgr5 and Bmi1 in CACC models.
(A) Images of colon tissue from MUC1+/– mice stained for MUC1-C (red) and Lgr5 (green) (upper) or MUC1-C (red) and Bmi1 (green) (lower). Nuclei were stained with DAPI (blue). Scale bars: 200 μm (yellow), 50 μm (white). (B) Images of colitis, dysplasia, and carcinoma tissues from MUC1+/– IL-10–/– mice stained for MUC1-C (red) and Lgr5 (green) (upper) or MUC1-C (red) and Bmi1 (green) (lower). Nuclei were stained with DAPI (blue). Scale bars: 200 μm (yellow), 50 μm (white). (C) Lysates of colitis and carcinoma tissues from MUC1+/– IL-10–/– mice were immunoblotted with antibodies against the indicated proteins. (D) Images of colitis tissues from control and GO-203–treated MUC1+/– IL-10–/– mice stained for MUC1-C (red) and Lgr5 (green) (upper) or MUC1-C (red) and Bmi1 (green) (lower). Nuclei were stained with DAPI (blue). Scale bars: 200 μm (yellow), 50 μm (white). (E) Lysates of colitis tissues from control and GO-203–treated MUC1+/– IL-10–/– mice were immunoblotted with antibodies against the indicated proteins. (F) Images of colitis, dysplasia, and carcinoma tissues from AOM/DSS-induced MUC1+/– mice stained for MUC1-C (red) and Lgr5 (green) (upper) or MUC1-C (red) and Bmi1 (green) (lower). Nuclei were stained with DAPI (blue). Scale bars: 200 μm (yellow), 50 μm (white). (G) Lysates of colitis and carcinoma tissues from AOM/DSS-induced MUC1+/– mice were immunoblotted with antibodies against the indicated proteins. (H) Images of colitis tissues from control and GO-203–treated AOM/DSS-induced MUC1+/– mice stained for MUC1-C (red) and Lgr5 (green) (upper) or MUC1-C (red) and Bmi1 (green) (lower). Nuclei were stained with DAPI (blue). Scale bars: 200 μm (yellow), 50 μm (white). (I) Lysates of colitis tissues from control and GO-203–treated AOM/DSS-induced MUC1+/– mice were immunoblotted with antibodies against the indicated proteins.
MUC1-C induces pluripotency factors in CACC.
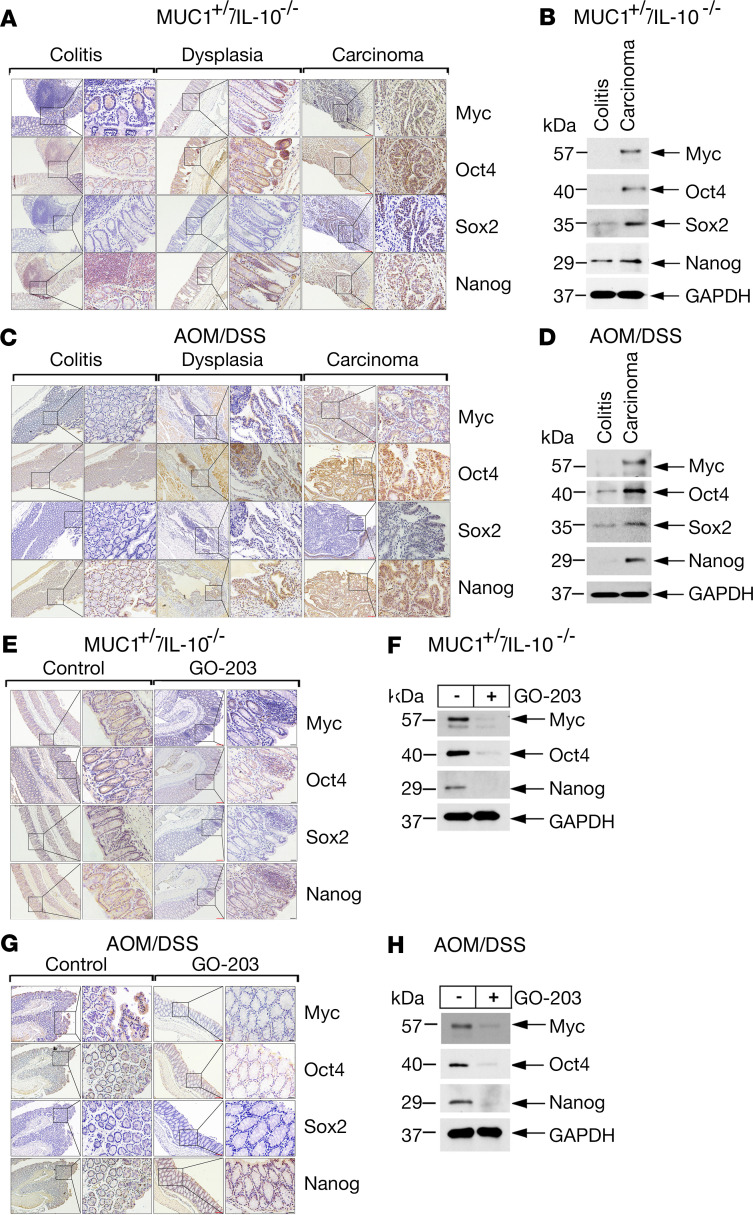
The seminal findings that somatic cells can be reprogrammed to induced pluripotent stem cells by introduction of Oct4, Sox2, Klf4, and Myc (Yamanaka OSKM factors) has had a substantial impact on our understanding of stem cells in normal and cancerous tissues (31, 32). Expression of the core Oct4, Sox2, and Nanog (OSN) pluripotency factors is restricted in somatic cells to maintain lineage specificity and is upregulated in damage and wound healing (32, 33). Relatively little is known about pluripotency factor expression in CACC models. Accordingly, we studied colons from MUC1+/– IL-10–/– mice and found upregulation of Myc and OSN expression (Figure 4, A and B). These observations were extended in studies of AOM/DSS-induced MUC1+/– mice (Figure 4, C and D). We also found that GO-203 treatment of the MUC1+/– IL-10–/– (Figure 4, E and F) and AOM/DSS-induced MUC1+/– (Figure 4, G and H) mice decreased Myc and OSN expression as determined by IHC staining (Figure 4, E and G), IHC scores (Supplemental Figure 2, A and B; Supplemental Tables 5 and 6), and immunoblot analysis (Figure 4, F and H), indicating that, as found for Lgr5 and Bmi1, MUC1-C promotes expression of pluripotency factors in progression of CACC.
Figure 4. Targeting MUC1-C decreases pluripotency factor expression in CACC models.
(A) Images of colitis, dysplasia, and carcinoma tissues from MUC1+/– IL-10–/– mice stained for Myc, Oct4, Sox2, and Nanog. Red scale bars: 200 μm. Black scale bars: 50 μm. (B) Lysates of colitis and carcinoma tissues from MUC1+/– IL-10–/– mice run in the same gel as shown for data in Figure 3C were immunoblotted with antibodies against the indicated proteins and include the same GAPDH blots. (C) Images of colitis, dysplasia, and carcinoma tissues from AOM/DSS-induced MUC1+/– mice stained for Myc, Oct4, Sox2, and Nanog. Red scale bars: 200 μm. Black scale bars: 50 μm. (D) Lysates of colitis and carcinoma tissues from AOM/DSS-induced MUC1+/– mice run in the same gel as shown for data in Figure 3G were immunoblotted with antibodies against the indicated proteins and include the same GAPDH blots. (E) Images of colitis tissues from control and GO-203–treated MUC1+/– IL-10–/– mice stained for Myc, Oct4, Sox2, and Nanog. (F) Lysates of colitis tissues from control and GO-203–treated MUC1+/– IL-10–/– mice run in the same gel as shown for the data in Figure 3E were immunoblotted with antibodies against the indicated proteins and include the same GAPDH blots. (G) Images of colitis tissues from control and GO-203–treated AOM/DSS-induced MUC1+/– mice stained for Myc, Oct4, Sox2, and Nanog. (H) Lysates of colitis tissues from control and GO-203–treated AOM/DSS-induced MUC1+/– mice run in the same gel as shown for the data in Figure 3I were immunoblotted with antibodies against the indicated proteins and include the same GAPDH blots.
MUC1-C drives inflammation and stemness in human colon cancer cells.
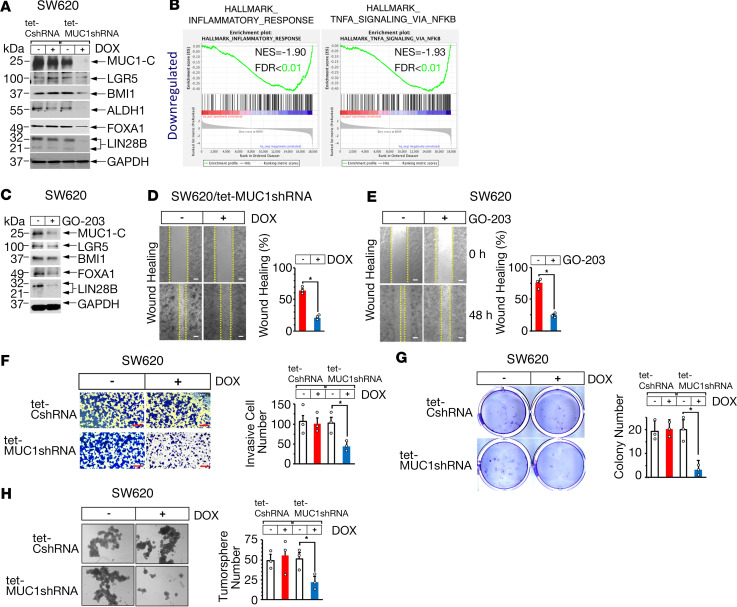
To extend these studies, we established human SW620 colon cancer cells expressing a tet-inducible control shRNA (tet-CshRNA) or a tet-inducible MUC1-C shRNA (tet-MUC1shRNA). Doxycycline (DOX) treatment was associated with downregulation of MUC1-C mRNA and protein in SW620 tet-MUC1shRNA, and not SW620 tet-CshRNA, cells (Figure 5A and Supplemental Figure 3). Of interest, we found that silencing MUC1-C associated with suppression of LGR5 and BMI1 expression (Figure 5A and Supplemental Figure 3). In addition, aldehyde dehydrogenase 1 (ALDH1), forkhead box A1 (FOXA1), and lin-28 homolog B (LIN28B), which have been linked to intestinal CSCs (34–36), were downregulated by MUC1-C silencing (Figure 5A). RNA-Seq analysis further demonstrated that MUC1-C drives these CSC markers in association with the HALLMARK_INFLAMMATORY_RESPONSE and HALLMARK_TNFA_SIGNALING_VIA_NFKB (Figure 5B) pathways, consistent with evidence that inflammatory signaling in cancer cells can drive their stemness (37–39). The association of MUC1-C with CRC cell stemness was extended by targeting MUC1-C with GO-203 treatment, which resulted in downregulation of LGR5 and BMI1, as well as ALDH1, FOXA1, and LIN28B (Figure 5C). In a second model, similar results were obtained with human SK-CO-1 colon cancer cells (Supplemental Figure 4, A and B), further supporting a role for MUC1-C in driving the CSC phenotype. In this respect, we found that targeting MUC1-C significantly attenuated CSC characteristics of wound healing (Figure 5D, left and right; Figure 5E, left and right; Supplemental Figure 4C) and invasion (Figure 5F, left and right; Supplemental Figure 4D). Targeting MUC1-C also significantly decreased the capacity to generate colonies (Figure 5G, left and right; Supplemental Figure 4E) and tumorspheres (Figure 5H, left and right; Supplemental Figure 4F). These results collectively supported the involvement of MUC1-C in driving CRC cell stemness and self-renewal capacity.
Figure 5. Targeting MUC1-C in human colon cancer cells downregulates LGR5, BMI1, and stemness.
(A) Human SW620 colon cancer cells stably expressing a tet-CshRNA or tet-MUC1shRNA were treated with vehicle or 500 ng/mL DOX for 7 days. Lysates were immunoblotted with antibodies against the indicated proteins. (B) RNA-Seq was performed in triplicate on SW620 MUC1shRNA cells treated with vehicle or 500 ng/mL DOX for 7 days. Silencing MUC1-C expression was significantly associated with suppression of the indicated HALLMARK gene sets. (C) SW620 cells were left untreated or treated with 5 μM GO-203 for 48 hours. Lysates were immunoblotted with antibodies against the indicated proteins. (D) SW620 tet-MUC1shRNA cells treated with vehicle or 500 ng/mL DOX for 48 hours were monitored for wound healing in the scratch assay (left). The results are expressed as a percentage (mean ± SD of 3 biologic replicates) of the control at 0 hours (right). Scale bars: 100 μm. (E) SW620 cells left untreated or treated with 5 μM GO-203 for 48 hours were monitored for wound healing in the scratch assay (left). The results (mean ± SD of 3 biologic replicates) are expressed as a percentage of the control at 0 hours (right). Scale bars: 100 μm. (F) SW620 tet-CshRNA and SW620 tet-MUC1shRNA cells treated with vehicle or 500 ng/mL DOX for 24 hours were assayed for invasion (left). The results (mean ± SD of 3 biologic replicates) are expressed as the invasive cell number (right). Scale bar: 200 μm. (G) SW620 tet-CshRNA and SW620 tet-MUC1shRNA cells treated with vehicle or 500 ng/mL DOX for 7 days were assayed for colony formation (left). The results (mean ± SD of 3 biologic replicates) are expressed as colony number per field (right). (H) SW620 tet-CshRNA and SW620 tet-MUC1shRNA cells treated with vehicle or 500 ng/mL DOX for 5 days were assayed for tumorsphere formation (left). The results (mean ± SD of 3 biologic replicates) are expressed as tumorsphere number per field (right). Scale bar: 200 μm.
MUC1-C integrates stemness and pluripotency by a MYC-mediated mechanism.
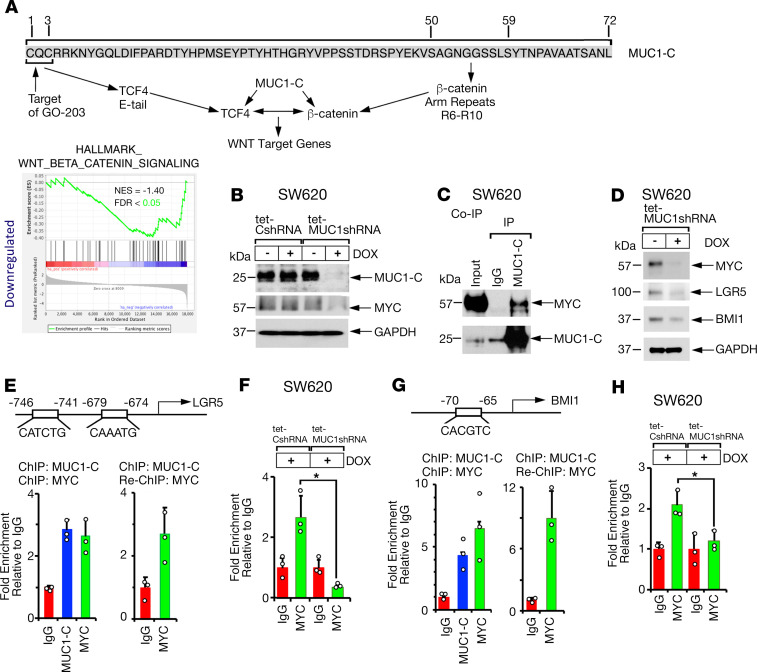
MYC is dysregulated in colitis and is a critical effector in CACC and CRC CSCs (40–42). The MUC1-C cytoplasmic domain binds directly to β-catenin (43, 44) and to TCF4 (45) to promote activation of WNT target genes, such as CCND1 and MYC (Figure 6A) (45, 46). Along these lines, targeting MUC1-C suppressed the HALLMARK_WNT_BETA_CATENIN_SIGNALING pathway (Figure 6A) and MYC expression (Figure 6B and Supplemental Figure 5A). In addition to inducing MYC, MUC1-C binds to the MYC HLH-LZ domain, which is of importance for the MYC transactivation function (11). Consistent with this direct interaction, we detected the presence of nuclear MUC1-C/MYC complexes (Figure 6C). To assess the significance of MUC1-C/MYC signaling, we silenced MYC and, interestingly, found downregulation of LGR5 and BMI1 (Figure 6D and Supplemental Figure 5, B–D). LGR5 has been identified as a WNT pathway target gene in ISCs (15, 30); however, to our knowledge, LGR5 has not been directly linked to MYC-induced activation. This aside, motif analysis of the LGR5 promoter identified putative consensus E-box MYC binding sites (Figure 6E). ChIP-PCR studies further demonstrated that MUC1-C and MYC form a complex on the LGR5 promoter (Figure 6E, left and right). In association with downregulation of LGR5 expression, silencing MUC1-C also decreased MYC occupancy on the LGR5 promoter (Figure 6F). Consistent with studies in other carcinomas (9), we also found that (a) MUC1-C/MYC complexes were detectable on the BMI1 promoter (Figure 6G, left and right) and (b) silencing MUC1-C decreased MYC occupancy (Figure 6H), supporting involvement of a MUC1-C/MYC pathway in driving LGR5 and BMI1 expression in CRC cells.
Figure 6. MUC1-C/MYC signaling induces LGR5 and BMI1 expression.
(A) Amino acid sequence of the MUC1-C cytoplasmic domain with the SAGNGGSSLS motif that binds directly to β-catenin (44). The MUC1-C CQC motif binds directly to the TCF4 E-tail C-clamp region (45). MUC1-C thereby forms a ternary complex with β-catenin and TCF4 on WNT target genes, such as CCND1 and MYC, to activate their transcription (45, 46). The MUC1-C CQC motif is targeted by the GO-203 inhibitor, which disrupts the interaction with TCF4 and thereby activation of the WNT pathway (45, 46). RNA-Seq was performed in triplicate on SW620 MUC1shRNA cells treated with vehicle or 500 ng/mL DOX for 7 days. Silencing MUC1-C expression was significantly associated with suppression of the indicated HALLMARK gene set. (B) SW620 tet-CshRNA and SW620 tet-MUC1shRNA cells were treated with vehicle or 500 ng/mL DOX for 7 days. (C) Nuclear lysates from SW620 cells were precipitated with anti–MUC1-C and an IgG control antibody. Input proteins and the precipitates were immunoblotted with antibodies against MUC1-C and MYC. (D) SW620 cells expressing a tet-MYCshRNA were treated with vehicle or 500 ng/mL DOX for 7 days. (E) Schema of the LGR5 promoter region. Soluble chromatin from SW620 cells was precipitated with a control IgG, anti–MUC1-C, or anti-MYC (left). Soluble chromatin was precipitated with anti–MUC1-C (ChIP) and then reprecipitated with anti-MYC or a control IgG (re-ChIP) (right). (F) Soluble chromatin from SW620 tet-CshRNA and SW620 tet-MUC1shRNA cells treated with 500 ng/mL DOX for 7 days was precipitated with anti-MYC or a control IgG. The DNA samples were amplified by quantitative PCR (qPCR) with primers for the LGR5 promoter. (G) Schema of the BMI1 promoter region (9). Soluble chromatin from SW620 cells was precipitated with a control IgG, anti-MUC1-C or anti-MYC (left). Soluble chromatin was precipitated with anti-MUC1-C (ChIP) and then reprecipitated with anti-MYC or a control IgG (re-ChIP) (right). (H) Soluble chromatin from SW620 tet-CshRNA and SW620 tet-MUC1shRNA cells treated with 500 ng/mL DOX for 7 days was precipitated with anti-MYC or a control IgG. The DNA samples were amplified by qPCR with primers for the BMI1 promoter. The results (mean ± SD of 3 determinations) are expressed as the relative fold enrichment compared with that obtained with the IgG control (assigned a value of 1).
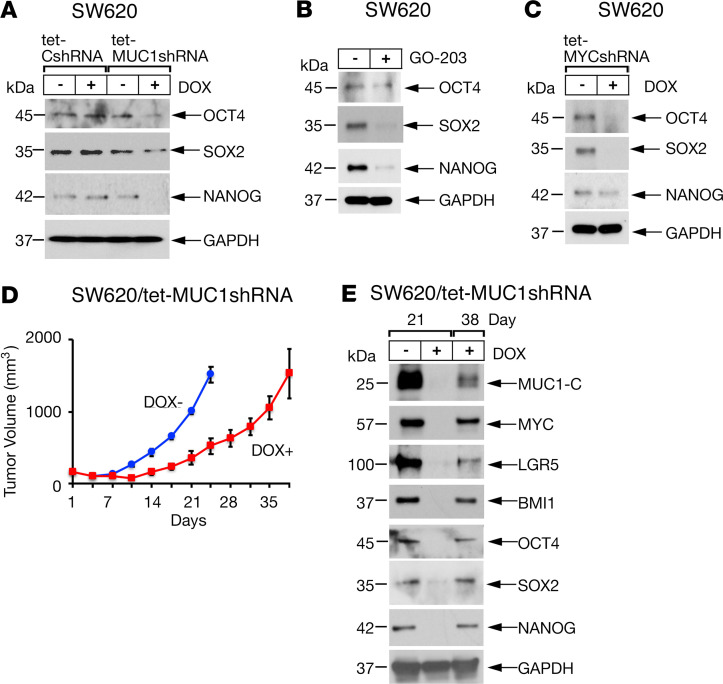
Stemness of CSCs is associated with expression of pioneer pluripotency factors (47–50). As reported for MYC (40–42), OCT4, SOX2, and NANOG are upregulated in CRC as adverse prognostic factors (51, 52). In addition to MYC, we found that targeting MUC1-C with silencing and GO-203 treatment resulted in suppression of OCT4, SOX2, and NANOG expression (Figure 7, A and B; Supplemental Figure 6A). Silencing MYC also suppressed OCT4, SOX2, and NANOG (Figure 7C and Supplemental Figure 6B), indicating that, in addition to inducing stemness as evidenced by LGR5 and BMI1, MUC1-C/MYC signaling drives pluripotency factor expression. Accordingly, we investigated the effects of DOX-induced silencing of MUC1-C in SW620 tet-MUC1shRNA tumor xenografts growing in mice. Tumorigenicity was inhibited by suppressing MUC1-C expression and thereby downregulation of LGR5, BMI1, and the core pluripotency factors (Figure 7, D and E). Similar results were obtained in SK-CO-1 tet-MUC1shRNA xenografts (Supplemental Figure 7, A and B). In both models, increases in MUC1-C expression attributed to overriding of the MUC1shRNA were associated with progression of tumor growth. These findings provided support for a model in which MUC1-C integrates stemness, pluripotency, and tumorigenicity in CRC.
Figure 7. Targeting MUC1-C decreases pluripotency factor expression and tumorigenicity.
(A) Lysates from SW620 tet-CshRNA and SW620 tet-MUC1shRNA cells treated with vehicle or 500 ng/mL DOX for 7 days run in the same gel as shown for the data in Figure 6B were immunoblotted with antibodies against the indicated proteins and include the same GAPDH blots. (B) Lysates from SW620 cells left untreated or treated with 5 μM GO-203 for 48 hours were immunoblotted with antibodies against the indicated proteins. (C) Lysates from SW620 tet-MYCshRNA treated with vehicle or 500 ng/mL DOX for 7 days were immunoblotted with antibodies against the indicated proteins. (D and E) Nude 6-week-old male mice were injected subcutaneously in the flank with 3 × 106 SW620 tet-MUC1shRNA cells. Mice were pair matched into 2 groups when tumors reached 100–150 mm3 and were fed without and with DOX. Tumor volumes are expressed as the mean ± SD for 6 mice (D). Lysates from tumors obtained on the indicated days were immunoblotted with antibodies against the indicated proteins (E).
MUC1 expression associates with inflammation and progression in tissues from patients with colitis and CRC.
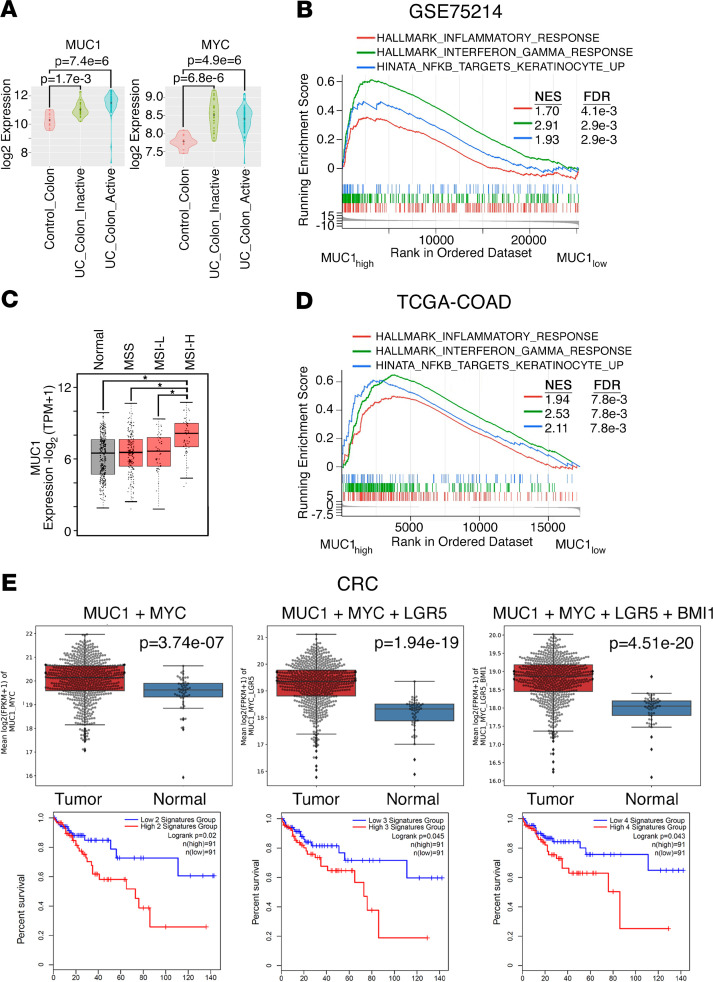
In analyses of gene expression data sets from human cohorts, we found upregulation of MUC1 and MYC expression in involved tissues from patients with inactive and active ulcerative colitis (UC) as compared with normal colon (Figure 8A). Moreover, consistent with involvement of MUC1-C in the mouse CACC models, we found that UC tissues with elevated MUC1 expression (MUC1hi, top quartile) were enriched for inflammatory (e.g., HALLMARK_INFLAMMATORY_RESPONSE), interferon (e.g., HALLMARK_INTERFERON_GAMMA_RESPONSE), and NF-κB signaling (e.g., HINATA_NFKB_TARGETS_KERATINOCYTE_UP) pathways relative to UC tissues with low expression (MUC1lo, bottom quartile) (Figure 8B). These studies were extended to CRCs by analysis of the The Cancer Genome Atlas colon adenocarcinoma (TCGA-COAD) data set, which showed that MUC1 mRNA was significantly upregulated in microsatellite instability-high (MSI-H) cancers (Figure 8C). Of particular interest in this regard, MSI is prevalent in UC and is linked to inflammation (53). Importantly, and similar to observations in UC, CRC with elevated MUC1 expression associated with inflammatory, interferon, and NF-κB signaling pathways (Figure 8D). Further analysis demonstrated that (a) expression of MYC, LGR5, and BMI1 was significantly increased in CRCs compared with normal colonic mucosa (Supplemental Figure 8A), and (b) MYC significantly correlated with LGR5 (Supplemental Figure 8B, left) and BMI1 (Supplemental Figure 8B, right) expression, supporting the central premise that MUC1-C/MYC signaling drives LGR5 and BMI1. In assessing the combined expression of these genes, we found that (a) MUC1 + MYC, (b) MUC1 + MYC + LGR5, and (c) MUC1 + MYC + LGR5 + BMI1 expression were significantly elevated in CRC relative to adjacent normal colonic mucosa (Figure 8E, upper) and that tumors with elevated gene set expression significantly associated with decreases in relapse-free survival (Figure 8E, lower). Of note, analysis of gene expression data sets generated from bulk CRCs is not necessarily informative for CSCs, which are likely to represent a small component of the entire CRC cell population. Regarding expression of MUC1 at the protein level, meta-analyses have shown that the shed MUC1-N subunit is significantly associated with metastatic disease and decreased overall survival (54, 55). However, to our knowledge, previous work has not evaluated expression of the oncogenic MUC1-C subunit. To this end, we performed IHC analysis of CRCs from a cohort of 70 patients and found that MUC1-C was (a) detectable in 90% of these tumors (Supplemental Table 7), (b) selectively expressed in CRC tissues as compared with normal mucosa (Supplemental Figure 9A), and (c) expressed at higher levels in poorly and moderately differentiated, as compared with well-differentiated, CRCs (Supplemental Figure 9B and Supplemental Table 7). We also found that MUC1-C expression associated with increases in invasion (Supplemental Figure 9B and Supplemental Table 7) and tumor stage (Supplemental Table 7), indicating that the MUC1-C protein is upregulated in CRC progression. Additional studies will now be needed in larger cohorts to determine whether MUC1-C expression associates with metastatic disease and poor patient outcomes.
Figure 8. Upregulation of MUC1 expression in patients with UC and CRC.
(A) The GSE75214 data set containing expression profiles from control normal colon (n = 11), UC colon inactive (n = 23), and UC colon active (n = 74) samples was analyzed for MUC1 (left) and MYC (right) expression. The results are presented as violin plots. The dot in the center is the mean. Differences between groups were determined by the Wilcoxon rank sum test. (B) MUC1hi versus MUC1lo differential expression from UC samples (GSE75214) was assessed for functional enrichment with gene set enrichment analysis (GSEA) against the hallmark and canonical pathways. Enrichment plots for select gene signatures are shown. (C) The TCGA-COAD data set was analyzed for MUC1 expression in microsatellite stable (MSS) tumors (n = 175), microsatellite instability-low (MSI-L) tumors (n = 48), and microsatellite instability-high (MSI-H) tumors (n = 48) as compared with that in normal colon tissue (n = 41). (D) MUC1hi versus MUC1lo differential expression from CRC samples was assessed for functional enrichment with GSEA against hallmark and canonical pathways. Enrichment plots for select gene signatures are shown. (E) CRC samples (n = 638) (UCSC Xena data hub) were compared with normal colonic mucosa (n = 51) (upper). Multiple probe set IDs for the indicated genes were averaged for each patient sample after normalization to obtain a representative expression value. The center line indicates the median value, bounds of the box denote 25th (lower) and 75th (upper) percentiles, and whiskers indicate minimum (lower) and maximum (upper) values. Student’s t test was used to compare groups. Kaplan-Meier curves for relapse-free survival of patient groups with CRCs expressing the indicated genes (lower). Blue lines show low signatures groups; red lines, high signatures groups. P = 0.020, 0.045, and 0.043 for MUC1 + MYC, MUC1 + MYC + LGR5, and MUC1 + MYC + LGR5 + BMI1, respectively (log-rank test). For both high and low for MUC1 + MYC, MUC1 + MYC + LGR5, and MUC1 + MYC + LGR5 + BMI1, n = 91. TPM, transcripts per kilobase million; FPKM, fragments per kilobase million.
Discussion
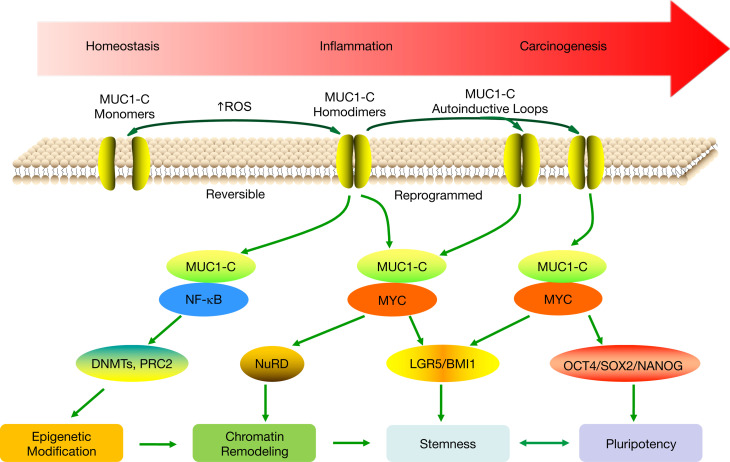
A role for the MUC1-C oncoprotein in the progression of chronic inflammation to cancer is unknown. MUC1-C induces EMT, stemness, and epigenetic reprogramming in cancer cells by activating in part the inflammatory NF-κB p65 pathway (5). In this way, MUC1-C (a) activates transforming growth factor–β activated kinase 1 (TAK1) and the I-κ B kinase complex (IKK) and (b) binds directly to NF-κB p65 to drive NF-κB target genes, which among others include MUC1 itself in an autoinductive circuit (Figure 9) (23, 56, 57). The involvement of MUC1-C in integrating NF-κB inflammatory signaling with hallmark traits of the cancer cell has underscored the notion that MUC1-C may functionally contribute to the association between chronic inflammation and cancer (Figure 9). Our studies here in 2 MUC1-transgenic mouse models of CACC demonstrate that MUC1-C is upregulated in the progression of colitis to dysplasia and carcinoma. We found that treatment with the MUC1-C inhibitor GO-203 attenuated the upregulation of MUC1-C expression and the inflammatory response as evidenced by decreases in (a) rectal prolapse, (b) severity of colitis, and (c) the epithelial damage score. These results could be explained in part by GO-203–induced disruption of the nuclear MUC1-C/NF-κB circuit and thereby suppression of inflammatory NF-κB target genes (Figure 9) (23, 56–58). In concert with such a model, we found that, in addition to suppressing inflammation, GO-203 treatment attenuated the progression of colitis to dysplasia. Moreover, targeting MUC1-C with GO-203 attenuated progression to carcinoma in the AOM/DSS model; however, further studies will now be needed to functionally define the basis for this potentially important observation. Indeed, MUC1-C may integrate 2 pathways in driving intestinal carcinogenesis; that is, in addition to activating NF-κB and inflammation (23, 56–58), the present findings indicate that MUC1-C/MYC signaling contributes to expression of (a) LGR5 and BMI1, which are linked to ISCs, and (b) pluripotency factors that drive dedifferentiation, therapeutic resistance, and tumorigenicity (Figure 9).
Figure 9. Proposed model for involvement of MUC1-C in progression of intestinal inflammation and carcinogenesis.
MUC1-C activates the TAK1/IKK/NF-κB inflammatory pathway in colitis and colon cancers (23). In turn, MUC1-C/NF-κB signaling induces changes in gene expression that include upregulation of (a) MUC1 itself in an autoinductive circuit (57) and (b) epigenetic modifiers, such as DNMTs and EZH2 (5). MUC1-C also activates the WNT/β-catenin/TCF4 pathway, induces MYC expression, and binds directly to MYC (11, 43–46). In this way, MUC1-C contributes to the regulation of MYC target genes involved in chromatin remodeling and dedifferentiation (11, 63). The present results link MUC1-C/MYC signaling to induction of LGR5 and BMI1 in support of a role in driving stemness in colitis and CRC. Stemness is associated with pluripotency, and, in this context, we show that MUC1-C/MYC signaling also drives OCT4, SOX2, and NANOG expression. Stemness and pluripotency are essential for wound healing and in the response to damage associated with colitis. The prolonged autoinduction of MUC1-C in settings of chronic inflammation with repetitive cycles of damage and repair could therefore drive NF-κB, MYC, and epigenetic modifications associated with stemness and pluripotency that become reprogrammed and promote carcinogenesis.
Damage to the intestinal epithelium in mice is repaired by expansion of (a) Lgr5+ ISCs and (b) Bmi1+ ISCs that repopulate the Lgr5+ pool (15–17, 59). Lgr5+ and Bmi1+ ISCs have been linked to the development of colon cancer in mice (15, 16, 18). In mouse CACC models, we found that targeting MUC1-C results in downregulation of Lgr5 and Bmi1 expression, suggesting that MUC1-C may contribute to driving these ISC pools. An alternative explanation was that targeting MUC1-C with suppression of the inflammatory response and thereby extent of intestinal damage indirectly decreases the requirement for expansion of Lgr5+ and Bmi1+ ISC pools. To further address the association of MUC1-C with these ISC markers, we turned to studies in colon cancer cells and found that targeting MUC1-C decreases LGR5 and BMI1 expression. An expectation was that MUC1-C–induced activation of the NF-κB pathway and inflammation would be responsible for driving LGR5 and BMI1; however, targeting NF-κB had little if any effect on their expression. MUC1-C induces MYC by activation of the WNT/β-catenin/TCF4 pathway (46). MUC1-C also interacts directly with the MYC HLH-LZ domain, which regulates the MYC transactivation function (11). Along these lines, we found that, like MUC1-C, silencing MYC suppresses LGR5 expression. We also found that MUC1-C/MYC complexes occupy the LGR5 promoter and that targeting MUC1-C decreases MYC occupancy in association with suppression of LGR5 expression. In addition, and in concert with the present demonstration that MUC1-C and MYC drive BMI1 in CRC cells, other work has shown that MUC1-C activates the BMI1 promoter by a MYC-mediated mechanism (9). These findings uncover MUC1-C/MYC signaling as a previously unrecognized pathway that contributes to LGR5 and BMI1 expression in CACC and CRC (Figure 9), the potential importance of which is supported by the link between LGR5+ and BMI1+ ISCs to CSCs in CRC and the functional role for LGR5 in driving CRC tumorigenicity (19–21).
Studies in a genetic mouse model of intestinal tumorigenesis have demonstrated that inflammatory NF-κB signaling initiates dedifferentiation of nonstem cells and in turn the generation of CSCs (37). Inflammatory signaling mediated by NF-κB in cancer cells drives their stemness, activates β-catenin, and increases the proportion of CSCs in tumors (37–39). There are marked similarities between normal stem cells and CSCs in terms of stemness, gene signatures, and expression of pioneer pluripotency factors (47–50). Pluripotency endows cells with the capacity for oncogenic dedifferentiation and stemness, transdifferentiation with direct cell fate switching, and interconversion between CSCs and non-CSCs, which collectively represents a major challenge responsible for drug resistance, immune evasion, and poor clinical outcomes (50, 60–62). MUC1-C has been linked to cancer cell dedifferentiation and stemness in association with driving tumorigenicity (6, 11, 63). Here, we found that targeting MUC1-C in CACC models and in human colon cancer cells resulted in suppression of core pluripotency factor expression. These apparent parallels between MUC1-C–induced pluripotency factors in settings of inflammation and cancer could hold potentially important implications. In this respect, the appropriation of pluripotency associated with epithelial damage and wound healing could endow cancer cells with the capacity for dedifferentiation and resistance to treatment. Indeed, the present findings that targeting MUC1-C in colon cancer cells suppresses LGR5, BMI1, stemness, pluripotency factors, and tumorigenicity lends credence to the notion that MUC1-C functions in integrating inflammation and wound healing with cancer progression (Figure 9). Our results thus support a model in which intestinal inflammation activates MUC1-C and downstream MYC pathways that induce chromatin remodeling and stemness (Figure 9). Such MUC1-C/MYC signaling would be reversible upon resolution of the inflammatory response and wound healing. However, in the setting of chronic inflammation, prolonged activation of these MUC1-C/MYC pathways establishes irreversible reprogramming of stemness and pluripotency that promotes carcinogenesis, which is intrinsic to the CSC and independent of an inflammatory tumor immune microenvironment (Figure 9).
Of potential clinical relevance, a phase I trial of the GO-203 inhibitor administered intravenously to patients with advanced cancer demonstrated an acceptable safety profile. A short circulating half-life of GO-203 necessitated daily dosing to maintain plasma levels, which was a challenge in the clinic. Accordingly, GO-203 was formulated in NPs for less frequent delivery and further evaluation for cancer treatment (64). GO-203, which is stable at low pH, is also being formulated for oral administration. Biologic therapies, such as anti-TNF antibodies, have had a profound impact on suppressing the inflammatory process; however, certain patients with IBD fail to respond to these agents, eventually become resistant, or experience significant adverse effects, emphasizing the need for other therapeutic options (65). Based on the present findings, MUC1-C represents a potential target for suppressing colitis, preventing the development of CACC, and treating CRC.
Methods
Mouse models of CACC.
Human MUC1-transgenic (MUC1+/−) C57BL/6 mice were purchased from S. Gendler (Mayo Clinic, Scottsdale, Arizona, USA). IL-10−/− C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, Maine, USA). The MUC1+/− mice were crossed with IL-10−/− mice, and then the F1 MUC1+/− IL-10+/− mice were bred with IL-10−/− mice. In a second model, 8-week-old MUC1+/− mice were injected i.p. with 12 mg/kg AOM (A5486; MilliporeSigma, Burlington, Massachusetts, USA). DSS (31395, 2.0%; MilliporeSigma) was fed in the drinking water over 5 days, followed by 16 days of regular water (1 cycle). The DSS cycle was repeated 3 times. Mice were treated i.p. with GO-203/NPs (15 mg/kg each week for 3 weeks).
IHC staining.
IHC staining was performed on paraffin-embedded mouse colon tissues. After antigen retrieval and blocking, sections were stained with anti–MUC1-C (ab15481, 1:100 dilution, and ab109185, 1:250 dilution; Abcam, Cambridge, Massachusetts, USA), anti-MYC (ab32072, 5 μg/mL; Abcam), anti-OCT4 (ab18976, 1:100 dilution; Abcam), anti-NANOG (ab80892, 1:400 dilution; Abcam), and anti-SOX2 (14962, 1:300 dilution; Cell Signaling Technology [CST], Danvers, Massachusetts, USA) at 4°C overnight. Slides were then probed with secondary antibodies (21234, 1:2000 dilution; Thermo Fisher Scientific) and stained with 3,5-diaminobenzidine peroxidase substrate (34002, 1:10 dilution; Thermo Fisher Scientific, Waltham, Massachusetts, USA). The IHC slides were scanned and the resolution was adjusted to 600 dpi.
Immunofluorescence staining.
Mouse colon tissues were dewaxed and hydrated before antigen retrieval and blocking. Sections were coincubated with anti–MUC1-C (ab15481, 1 μg/mL; Abcam) and anti-LGR5 (MA5-25644, 1:100 dilution; Thermo Fisher Scientific) or anti–MUC1-C and anti-BMI1 (ab269678, 2 μg/mL; Abcam). The sections were then stained with secondary antibodies (anti–MUC1-C: a10520, 1:200 dilution; Thermo Fisher Scientific; and anti-LGR5 and anti-BMI1: F-2761, 10 μg/mL; Thermo Fisher Scientific). Nuclei were stained with DAPI (0100-20, SouthernBiotech, Birmingham, Alabama, USA).
Epithelial damage score.
Histologic examination was performed in a blinded manner by a pathologist. Epithelial damage was scored according to the degree of lesions. No mucosal damage was scored as 0; colitis was scored as 1–3 (1, mild; 2, moderate; 3, severe); dysplasia was scored as 4; adenoma was scored as 5; and carcinoma was scored as 6 (66).
IHC score.
IHC staining was graded with a semiquantitative score according to the percentage of positive-staining cells (0: ≤10%, 1: <25%, 2: <50%, 3: <75%, and 4: ≥75%) and intensity of staining (0: negative, 1: weak, 2: intermediate, and 3: strong). The final score was obtained by multiplying the percentage of positive cells by the intensity.
Cell culture.
Human SW620 cells (ATCC, Manassas, Virginia, USA) were grown in RPMI1640 medium (Corning Life Sciences, Corning, New York, USA) containing 10% FBS, 100 μg/mL streptomycin, and 100 U/mL penicillin. SK-CO-1 cells (ATCC) were cultured in MEM (Corning, Manassas, Virginia, USA) containing 10% FBS, 100 μg/mL streptomycin, and 100 U/mL penicillin. Authentication of the cells was performed by short tandem repeat analysis. Cells were monitored for mycoplasma contamination using the MycoAlert Mycoplasma Detection Kit (Lonza, Rockland, Maine, USA).
Inducible gene silencing.
MUC1shRNA (MISSION shRNA TRCN0000122938), MYCshRNA (MISSION shRNA), or a control scrambled shRNA (CshRNA) (MilliporeSigma) was inserted into the pLKO-tet-puro vector (plasmid 21915; Addgene, Cambridge, Massachusetts, USA). The viral vectors were produced in HEK293T cells as described (63). Cells transduced with the vectors were selected for growth in 1–3 μg/mL puromycin. Cells were treated with 0.1% DMSO as the vehicle control or 500 ng/mL DOX (MilliporeSigma).
Quantitative reverse transcription PCR.
Total cellular RNA was isolated using TRIzol reagent (Thermo Fisher Scientific). cDNAs were synthesized using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Thermo Fisher Scientific, Grand Island, New York, USA). The cDNA samples were amplified using the Power SYBR Green PCR Master Mix (Applied Biosystems, Thermo Fisher Scientific) and the CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, California, USA) as described (63). Primers used for quantitative reverse transcription PCR are listed in Supplemental Table 8.
Immunoblotting.
Total lysates prepared from subconfluent cells as described (63) were immunoblotted with anti–MUC1-C (HM-1630-P1ABX, 1:1000 dilution; Thermo Fisher Scientific), anti-MYC (ab32072, 1:1000 dilution; Abcam), anti-LGR5 (PA5-49691, 1:1000 dilution; Thermo Fisher Scientific), anti-BMI1 (6964S, 1:1000 dilution; CST), anti-Bmi1 (ab269678, 2 μg/mL; Abcam), anti-SOX2 (3579, 1:1000 dilution; CST), anti-Sox2 (ab92494, 1:1000 dilution; Abcam), anti-OCT4 (2750S, 1:1000 dilution; CST), anti-Oct4 (NB100-2379SS, 1:500 dilution; Novus Biologicals, Centennial, Colorado, USA) anti-NANOG (4903S, 1:2000 dilution; CST), anti-Nanog (8822T, 1:1000 dilution; CST), anti-ALDH1 (ab134188, 1:1000 dilution; Abcam), anti-FOXA1 (58613S, 1:1000 dilution; CST), anti-LIN28B (11965S, 1:1000 dilution; CST), and anti-GAPDH (5174S, 1:10,000 dilution; CST).
Wound healing assays.
Cells (5 × 105) were cultured in 6-well plates until confluence and then scratched with a 200-μL pipette tip. The cells were treated with vehicle or 500 ng/mL DOX. Migration photos were captured at 0 and 48 hours after scratching. The results were quantitated in 3 independent experiments.
Invasion assays.
Cell invasion assays were performed using Corning Transwell plates (lot MKCC1255, MilliporeSigma). Briefly, cells were seeded into 24-well upper chambers precoated with Matrigel (MilliporeSigma). The lower chamber was filled with cell culture medium containing 10% FBS. After culturing for 24 hours, the invaded cells were stained with 0.5% Crystal Violet (MilliporeSigma) for 15 minutes and counted under a light microscope (Leica ICC50 E). The results were quantitated in 3 independent experiments.
Colony formation assays.
Cells seeded at 500/well in 6-well plates were treated with vehicle or 500 ng/mL DOX for 7 days and then stained with 0.5% crystal violet in 25% methanol. Colonies greater than 25 cells were counted in 3 independent experiments.
Tumorsphere assays.
Single-cell suspensions cultured in MammoCult Human Medium Kit (catalog 05620; STEMCELL Technologies, Cambridge, Massachusetts, USA) at a density of 3000 cells per well of a 6-well ultralow attachment culture plate (Corning) were treated with vehicle or 500 ng/mL DOX for 7 days. Tumorspheres with a diameter at least 60 μm were counted under a microscope (Nikon ECLIPSE TE2000-S).
ChIP assays.
Soluble chromatin was precipitated with anti–MUC1-C, anti-MYC (ab56; Abcam), or a control nonimmune IgG (clone SC2027; Santa Cruz Biotechnology, Santa Cruz, California, USA). The precipitates were analyzed by quantitative PCR using the Power SYBR Green PCR Master Mix and the ABI Prism 7300 sequence detector (Applied Biosystems, Thermo Fisher Scientific). Data are reported as relative fold enrichment. Primers used for ChIP quantitative PCR are listed in Supplemental Table 9.
Human tumor xenograft models.
Male 6-week-old nude mice (Taconic Farms, Hudson, New York, USA) were injected subcutaneously in the flank with 1 × 107 SW620 tet-MUC1shRNA or SK-CO-1 tet-MUC1shRNA cells in 50% Matrigel. When tumor size reached approximately 125 mm3, the mice were pair matched and fed without or with DOX (625 parts per million, daily). Tumor measurements and body weights were recorded twice each week. Mice were sacrificed when tumors reached greater than 1000 mm3 as calculated by the formula width2 × length/2.
RNA-Seq analysis.
Total RNA from cells cultured in triplicates was isolated using TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Carlsbad, California, USA). TruSeq Stranded mRNA (Illumina, San Diego, California, USA) was used for library preparation. Raw sequencing reads were aligned to the human genome (GRCh38.74) using STAR. Raw feature counts were normalized and differential expression analysis was performed using DESeq2. Differential expression rank order was used for subsequent GSEA performed using software developed by the Broad Institute (colon cancer cell RNA-Seq data set) or fgsea (v1.8.0) package in R (TCGA-COAD expression by RNA-Seq data set).
Analyses of publicly available expression data sets from UC and CRC human cohorts.
Two human cohorts, one representing UC (GSE75214) and another representing CRC (TCGA-COAD), were used to assess relationships between MUC1 expression and target gene expression, disease stage, and relapse-free survival. Normalized data were obtained from the Gene Expression Omnibus database (GSE75214) using the GEOquery pipeline or from the NIH Genomic Data Commons (TCGA-COAD) using the TCGAbiolinks pipeline in R. Normalized expressions were ranked by MUC1 expression and MUC1hi (top quartile) samples compared with MUC1lo (bottom quartile) using limma (GSE75214) or edgeR (TCGA-COAD). GSEA was performed using the clusterProfiler package in R. Assessment of gene expression correlation to disease stage and relapse-free survival in the TCGA-COAD cohort was performed using Gene Expression Profiling Interactive Analysis software (http://gepia.cancer-pku.cn/index.html).
Data and materials availability.
There are no restrictions on any of the materials used. The RNA-Seq data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus database under accession code GSE144319 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144319). The gene expression cohorts referenced during the study are available from GSE75214, TCGA-COAD, TCGA rectum adenocarcinoma, and UCSC Xena data sets. All data associated with this study are available in the main text or the supplemental materials.
Statistics.
Each experiment was performed at least 3 times. Student’s 2-tailed t test was used to determine differences between means of groups. A P value of less than 0.05, denoted by an asterisk in all figures, was considered statistically significant.
Study approval.
The mouse colitis and tumor xenograft studies were performed under animal protocol numbers 12-029 and 03-029, respectively, approved by the Dana-Farber Institutional Animal Care and Use Committee.
Author contributions
WL, NZ, MDL, SL, and DK conceived the study. WL, CJ, HR, YY, and MH designed the methodology. WL, NZ, CJ, HR, YY, AF, NY, and MH investigated. MDL, RZ, JW, LK, QH, and SL performed bioinformatics analysis. DK wrote the original draft. WL, NZ, MDL, SL, and DK reviewed and edited the draft. SL and DK acquired funding. HS and SK provided resources. SL and DK supervised the study.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Cancer Institute of the NIH under grant numbers CA97098, CA166480, CA229716, and CA233084 awarded to DK and CA232979 awarded to SL.
Version 1. 05/19/2020
In-Press Preview
Version 2. 06/18/2020
Electronic publication
Funding Statement
D Kufe
D Kufe
D Kufe
D Kufe
S Liu
Footnotes
Conflict of interest: DK has equity interests in Genus Oncology, Reata Pharmaceuticals, Hillstream BioPharma, and Victa Biotherapeutics; served as a member of the board of directors of Genus Oncology and Victa; and is a paid consultant for Reata, CanBas, and Victa.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(12):e137112.https://doi.org/10.1172/jci.insight.137112.
Contributor Information
Wei Li, Email: weili06@tongji.edu.cn.
Ning Zhang, Email: ning_zhang@dfci.harvard.edu.
Caining Jin, Email: cainingjin@gmail.com.
Mark D. Long, Email: Mark.Long@RoswellPark.org.
Hasan Rajabi, Email: hasan_rajabi@dfci.harvard.edu.
Yota Yasumizu, Email: y.yasumizu0707@gmail.com.
Atsushi Fushimi, Email: atsushi_fushimi@dfci.harvard.edu.
Nami Yamashita, Email: nami_yamashita@dfci.harvard.edu.
Masayuki Hagiwara, Email: masayuki_hagiwara@dfci.harvard.edu.
Rongbin Zheng, Email: zhengrongbin1992@gmail.com.
Jin Wang, Email: jinwang0611@gmail.com.
Ling Kui, Email: kuiling2008@163.com.
Harpal Singh, Email: harpal2000@yahoo.com.
Surender Kharbanda, Email: Surender_Kharbanda1@dfci.harvard.edu.
Qiang Hu, Email: Qiang.Hu@RoswellPark.org.
Song Liu, Email: Song.Liu@RoswellPark.org.
References
- 1.Greten FR, Grivennikov SI. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity. 2019;51(1):27–41. doi: 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer. 2009;9(12):874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kufe DW. MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene. 2013;32(9):1073–1081. doi: 10.1038/onc.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajabi H, et al. MUC1-C oncoprotein activates the ZEB1/miR-200c regulatory loop and epithelial-mesenchymal transition. Oncogene. 2014;33(13):1680–1689. doi: 10.1038/onc.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajabi H, Hiraki M, Kufe D. MUC1-C activates polycomb repressive complexes and downregulates tumor suppressor genes in human cancer cells. Oncogene. 2018;37(16):2079–2088. doi: 10.1038/s41388-017-0096-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hata T, et al. Targeting MUC1-C inhibits TWIST1 signaling in triple-negative breast cancer. Mol Cancer Ther. 2019;18(10):1744–1754. doi: 10.1158/1535-7163.MCT-19-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajabi H, et al. DNA methylation by DNMT1 and DNMT3b methyltransferases is driven by the MUC1-C oncoprotein in human carcinoma cells. Oncogene. 2016;35(50):6439–6445. doi: 10.1038/onc.2016.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajabi H, et al. MUC1-C represses the RASSF1A tumor suppressor in human carcinoma cells. Oncogene. 2019;38(47):7266–7277. doi: 10.1038/s41388-019-0940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiraki M, et al. MUC1-C activates BMI1 in human cancer cells. Oncogene. 2017;36(20):2791–2801. doi: 10.1038/onc.2016.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajabi H, et al. MUC1-C activates EZH2 expression and function in human cancer cells. Sci Rep. 2017;7(1):7481. doi: 10.1038/s41598-017-07850-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hata T, et al. MUC1-C Activates the NuRD complex to drive dedifferentiation of triple-negative breast cancer cells. Cancer Res. 2019;79(22):5711–5722. doi: 10.1158/0008-5472.CAN-19-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16(12):713–732. doi: 10.1038/s41575-019-0189-8. [DOI] [PubMed] [Google Scholar]
- 13.Katoh M. Multi‑layered prevention and treatment of chronic inflammation, organ fibrosis and cancer associated with canonical WNT/β‑catenin signaling activation (review) Int J Mol Med. 2018;42(2):713–725. doi: 10.3892/ijmm.2018.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, et al. Long-term culture captures injury-repair cycles of colonic stem cells. Cell. 2019;179(5):1144–1159.e15. doi: 10.1016/j.cell.2019.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457(7229):608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 16.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40(7):915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan KS, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A. 2012;109(2):466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Sousa e Melo F, et al. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature. 2017;543(7647):676–680. doi: 10.1038/nature21713. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi H, et al. Significance of Lgr5(+ve) cancer stem cells in the colon and rectum. Ann Surg Oncol. 2011;18(4):1166–1174. doi: 10.1245/s10434-010-1373-9. [DOI] [PubMed] [Google Scholar]
- 20.Shimokawa M, et al. Visualization and targeting of LGR5+ human colon cancer stem cells. Nature. 2017;545(7653):187–192. doi: 10.1038/nature22081. [DOI] [PubMed] [Google Scholar]
- 21.Yamada M, et al. Clinical significance of Bmi1 expression in inflammatory bowel disease. Oncology. 2017;93(suppl 1):20–26. doi: 10.1159/000481225. [DOI] [PubMed] [Google Scholar]
- 22.Rowse GJ, Tempero RM, VanLith ML, Hollingsworth MA, Gendler SJ. Tolerance and immunity to MUC1 in a human MUC1 transgenic murine model. Cancer Res. 1998;58(2):315–321. [PubMed] [Google Scholar]
- 23.Takahashi H, et al. MUC1-C activates the TAK1 inflammatory pathway in colon cancer. Oncogene. 2015;34(40):5187–5197. doi: 10.1038/onc.2014.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berg DJ, et al. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4(+) TH1-like responses. J Clin Invest. 1996;98(4):1010–1020. doi: 10.1172/JCI118861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beatty PL, Plevy SE, Sepulveda AR, Finn OJ. Cutting edge: transgenic expression of human MUC1 in IL-10–/– mice accelerates inflammatory bowel disease and progression to colon cancer. J Immunol. 2007;179(2):735–739. doi: 10.4049/jimmunol.179.2.735. [DOI] [PubMed] [Google Scholar]
- 26.Raina D, et al. Dependence on the MUC1-C oncoprotein in non-small cell lung cancer cells. Mol Cancer Ther. 2011;10(5):806–816. doi: 10.1158/1535-7163.MCT-10-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raina D, Ahmad R, Rajabi H, Panchamoorthy G, Kharbanda S, Kufe D. Targeting cysteine-mediated dimerization of the MUC1-C oncoprotein in human cancer cells. Int J Oncol. 2012;40(5):1643–1649. doi: 10.3892/ijo.2011.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raina D, et al. Characterization of the MUC1-C cytoplasmic domain as a cancer target. PLoS One. 2015;10(8):e0135156. doi: 10.1371/journal.pone.0135156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okayasu I, Ohkusa T, Kajiura K, Kanno J, Sakamoto S. Promotion of colorectal neoplasia in experimental murine ulcerative colitis. Gut. 1996;39(1):87–92. doi: 10.1136/gut.39.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seishima R, Barker N. A contemporary snapshot of intestinal stem cells and their regulation. Differentiation. 2019;108:3–7. doi: 10.1016/j.diff.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi K, Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol. 2016;17(3):183–193. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- 33.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taniguchi H, et al. Cancer stem cells in human gastrointestinal cancer. Cancer Sci. 2016;107(11):1556–1562. doi: 10.1111/cas.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma W, et al. The clinical significance of forkhead box protein A1 and its role in colorectal cancer. Mol Med Rep. 2016;14(3):2625–2631. doi: 10.3892/mmr.2016.5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tu HC, et al. LIN28 cooperates with WNT signaling to drive invasive intestinal and colorectal adenocarcinoma in mice and humans. Genes Dev. 2015;29(10):1074–1086. doi: 10.1101/gad.256693.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwitalla S, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152(1-2):25–38. doi: 10.1016/j.cell.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 38.Schwitalla S, et al. Loss of p53 in enterocytes generates an inflammatory microenvironment enabling invasion and lymph node metastasis of carcinogen-induced colorectal tumors. Cancer Cell. 2013;23(1):93–106. doi: 10.1016/j.ccr.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 39.Kryczek I, et al. IL-22(+)CD4(+) T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L. Immunity. 2014;40(5):772–784. doi: 10.1016/j.immuni.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yaeger R, et al. Genomic alterations observed in colitis-associated cancers are distinct from those found in sporadic colorectal cancers and vary by type of inflammatory bowel disease. Gastroenterology. 2016;151(2):278–287.e6. doi: 10.1053/j.gastro.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sipos F, Firneisz G, Műzes G. Therapeutic aspects of c-MYC signaling in inflammatory and cancerous colonic diseases. World J Gastroenterol. 2016;22(35):7938–7950. doi: 10.3748/wjg.v22.i35.7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elbadawy M, Usui T, Yamawaki H, Sasaki K. Emerging roles of C-Myc in cancer stem cell-related signaling and resistance to cancer chemotherapy: a potential therapeutic target against colorectal cancer. Int J Mol Sci. 2019;20(9):E2340. doi: 10.3390/ijms20092340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamamoto M, Bharti A, Li Y, Kufe D. Interaction of the DF3/MUC1 breast carcinoma-associated antigen and beta-catenin in cell adhesion. J Biol Chem. 1997;272(19):12492–12494. doi: 10.1074/jbc.272.19.12492. [DOI] [PubMed] [Google Scholar]
- 44.Huang L, Chen D, Liu D, Yin L, Kharbanda S, Kufe D. MUC1 oncoprotein blocks glycogen synthase kinase 3beta-mediated phosphorylation and degradation of beta-catenin. Cancer Res. 2005;65(22):10413–10422. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 45.Rajabi H, et al. MUC1-C oncoprotein induces TCF7L2 transcription factor activation and promotes cyclin D1 expression in human breast cancer cells. J Biol Chem. 2012;287(13):10703–10713. doi: 10.1074/jbc.M111.323311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouillez A, et al. Inhibition of MUC1-C suppresses MYC expression and attenuates malignant growth in KRAS mutant lung adenocarcinomas. Cancer Res. 2016;76(6):1538–1548. doi: 10.1158/0008-5472.CAN-15-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40(5):499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riggs JW, Barrilleaux BL, Varlakhanova N, Bush KM, Chan V, Knoepfler PS. Induced pluripotency and oncogenic transformation are related processes. Stem Cells Dev. 2013;22(1):37–50. doi: 10.1089/scd.2012.0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iglesias JM, Gumuzio J, Martin AG. Linking pluripotency reprogramming and cancer. Stem Cells Transl Med. 2017;6(2):335–339. doi: 10.5966/sctm.2015-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hepburn AC, et al. The induction of core pluripotency master regulators in cancers defines poor clinical outcomes and treatment resistance. Oncogene. 2019;38(22):4412–4424. doi: 10.1038/s41388-019-0712-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Müller M, et al. The role of pluripotency factors to drive stemness in gastrointestinal cancer. Stem Cell Res. 2016;16(2):349–357. doi: 10.1016/j.scr.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 52.Voutsadakis IA. The pluripotency network in colorectal cancer pathogenesis and prognosis: an update. Biomark Med. 2018;12(6):653–665. doi: 10.2217/bmm-2017-0369. [DOI] [PubMed] [Google Scholar]
- 53.Munakata K, et al. Inflammation-associated microsatellite alterations caused by MSH3 dysfunction are prevalent in ulcerative colitis and increase with neoplastic advancement. Clin Transl Gastroenterol. 2019;10(12):e00105. doi: 10.14309/ctg.0000000000000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeng Y, et al. MUC1 predicts colorectal cancer metastasis: a systematic review and meta-analysis of case controlled studies. PLoS One. 2015;10(9):e0138049. doi: 10.1371/journal.pone.0138049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu F, Liu F, Zhao H, An G, Feng G. Prognostic significance of mucin antigen MUC1 in various human epithelial cancers: a meta-analysis. Medicine (Baltimore) 2015;94(50):e2286. doi: 10.1097/MD.0000000000002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahmad R, et al. MUC1 oncoprotein activates the IkappaB kinase beta complex and constitutive NF-kappaB signalling. Nat Cell Biol. 2007;9(12):1419–1427. doi: 10.1038/ncb1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahmad R, et al. MUC1-C oncoprotein functions as a direct activator of the nuclear factor-kappaB p65 transcription factor. Cancer Res. 2009;69(17):7013–7021. doi: 10.1158/0008-5472.CAN-09-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taniguchi K, Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol. 2018;18(5):309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- 59.Metcalfe C, Kljavin NM, Ybarra R, de Sauvage FJ. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell. 2014;14(2):149–159. doi: 10.1016/j.stem.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 60.Smith BA, et al. A human adult stem cell signature marks aggressive variants across epithelial cancers. Cell Rep. 2018;24(12):3353–3366.e5. doi: 10.1016/j.celrep.2018.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Angelis ML, Francescangeli F, La Torre F, Zeuner A. Stem cell plasticity and dormancy in the development of cancer therapy resistance. Front Oncol. 2019;9:626. doi: 10.3389/fonc.2019.00626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miranda A, et al. Cancer stemness, intratumoral heterogeneity, and immune response across cancers. Proc Natl Acad Sci U S A. 2019;116(18):9020–9029. doi: 10.1073/pnas.1818210116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yasumizu Y, et al. MUC1-C regulates lineage plasticity driving progression to neuroendocrine prostate cancer. Nat Commun. 2020;11(1):338. doi: 10.1038/s41467-019-14219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hasegawa M, et al. Intracellular targeting of the oncogenic MUC1-C protein with a novel GO-203 nanoparticle formulation. Clin Cancer Res. 2015;21(10):2338–2347. doi: 10.1158/1078-0432.CCR-14-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chapman TP, Gomes CF, Louis E, Colombel JF, Satsangi J. De-escalation of immunomodulator and biological therapy in inflammatory bowel disease. Lancet Gastroenterol Hepatol. 2020;5(1):63–79. doi: 10.1016/S2468-1253(19)30186-4. [DOI] [PubMed] [Google Scholar]
- 66.Arthur JC, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338(6103):120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.