While enteric parasitic infections are among the most important infections in lower- and middle-income countries, their impact on gut microbiota is poorly understood. We reasoned that clinical symptoms associated with these infections may be influenced by alterations of the microbiome that occur during infection. To explore this notion, we took a two-pronged approach. First, we studied a cohort of dogs naturally infected with various enteric parasites and found a strong association between parasite infection and altered gut microbiota composition. Giardia, one of the most prevalent parasite infections globally, had a particularly large impact on the microbiome. Second, we took a database-driven strategy to integrate microbiome data with clinical data from large human field studies and found that Giardia infection is also associated with marked alteration of the gut microbiome of children, suggesting a possible explanation for why Giardia has been reported to be associated with protection from moderate to severe diarrhea.
KEYWORDS: GEMS, Giardia, MAL-ED, database, diarrhea, microbiome, parasite
ABSTRACT
Enteric parasitic infections are among the most prevalent infections in lower- and middle-income countries (LMICs) and have a profound impact on global public health. While the microbiome is increasingly recognized as a key determinant of gut health and human development, the impact of naturally acquired parasite infections on microbial community structure in the gut, and the extent to which parasite-induced changes in the microbiome may contribute to gastrointestinal symptoms, is poorly understood. Enteric parasites are routinely identified in companion animals in the United States, presenting a unique opportunity to leverage this animal model to investigate the impact of naturally acquired parasite infections on the microbiome. Clinical, parasitological, and microbiome profiling of a cohort of 258 dogs revealed a significant correlation between parasite infection and composition of the bacterial community in the gut. Relative to other enteric parasites, Giardia was associated with a more pronounced perturbation of the microbiome. To compare our findings to large-scale epidemiological studies of enteric diseases in humans, a database mining approach was employed to integrate clinical and microbiome data. Substantial and consistent alterations to microbiome structure were observed in Giardia-infected children. Importantly, infection was associated with a reduction in the relative abundance of potential pathobionts, including Gammaproteobacteria, and an increase in Prevotella—a profile often associated with gut health. Taken together, these data show that widespread Giardia infection in young animals and humans is associated with significant remodeling of the gut microbiome and provide a possible explanation for the high prevalence of asymptomatic Giardia infections observed across host species.
IMPORTANCE While enteric parasitic infections are among the most important infections in lower- and middle-income countries, their impact on gut microbiota is poorly understood. We reasoned that clinical symptoms associated with these infections may be influenced by alterations of the microbiome that occur during infection. To explore this notion, we took a two-pronged approach. First, we studied a cohort of dogs naturally infected with various enteric parasites and found a strong association between parasite infection and altered gut microbiota composition. Giardia, one of the most prevalent parasite infections globally, had a particularly large impact on the microbiome. Second, we took a database-driven strategy to integrate microbiome data with clinical data from large human field studies and found that Giardia infection is also associated with marked alteration of the gut microbiome of children, suggesting a possible explanation for why Giardia has been reported to be associated with protection from moderate to severe diarrhea.
INTRODUCTION
Enteric parasites, including helminths and protozoa, are among the most prevalent infections in lower- and middle-income countries (LMICs) with an estimated 3.5 billion people affected worldwide (1, 2). Infection with eukaryotic pathogens often results in acute, moderate to severe diarrheal disease and/or chronic malnutrition and stunting, which has significant consequences for morbidity and mortality (3–5). Conversely, some intestinal parasites are frequently associated with asymptomatic infections (6, 7). Giardia, for example, was found in 18 of 1,093 (1.6%) of healthy volunteers in Melbourne, Australia (8) and in 286 of 1,359 (21%) of healthy schoolchildren in Madrid, Spain (9). It is important to understand whether and how these abundant and pervasive parasites impact gut health.
While the microbiome is increasingly recognized as a key determinant of gut health and human development, the impact of naturally acquired parasite infections on the microbial community in the gut is poorly understood. Many studies of parasites and their impact on the microbiome involve experimental infections of laboratory animals (10–15). While such studies can be powerful for elucidating mechanisms, they often involve laboratory-adapted parasite strains, specialized animal husbandry practices, or high infectious doses, all of which can impact host immunity and the composition of the microbiome. Conversely, studies of parasite infections in human populations are challenging due to the relatively low prevalence of these infections in developed countries and the presence of confounding variables in LMICs, such as malnourishment and coinfections (16–19). These issues are largely overcome by studying enteric parasite infections in companion animals. Various enteric parasites are frequently found in screenings of domestic dogs and cats in the United States (20). For example, a study of over one million dogs throughout the United States in 2006 found that 12.5% were infected with at least one enteric parasite, with the most prevalent being Giardia which infected 4% of dogs (21). As companion animals, dogs are increasingly recognized as an ideal model system for translational gut microbiome research. In addition to harboring similar gut microbiota as humans, dogs often share their environment with humans, consume a similar omnivorous diet, and can spontaneously develop gastrointestinal (GI) disease that shares many features in common with inflammatory bowel disease in humans (22–29). In addition, like humans, dogs frequently become infected with enteric parasites in early life. Here, we performed 16S rRNA sequencing of fecal samples from 258 dogs naturally infected with one or more eukaryotic parasites to evaluate the impact of parasite infection on gut microbiota composition. We found that parasite infections are associated with significant perturbations to the microbiome and that Giardia is associated with the largest changes in canine gut microbiota.
We also investigated whether Giardia—a frequent infection among humans residing in LMICs—causes similar perturbations in human gut microbiota composition. The Global Enteric Multicenter Study (GEMS) investigated the causes of pediatric moderate to severe diarrhea (MSD) in LMICs (30). In addition to reporting a strong association between infection with rotavirus or Cryptosporidium and the development of MSD, this study also reported the surprising observation that Giardia was found more often among asymptomatic participants than those with MSD in this cohort (30), despite the association between Giardia and serious chronic health conditions, including growth stunting (31), irritable bowel syndrome (IBS), and fatigue (32). A follow-up to the GEMS study performed 16S sequencing of fecal samples from approximately 1,000 GEMS participants (33), but this study only considered the relationship between the microbiome and MSD and did not examine a role for parasite infections in influencing this relationship. Similarly, The Etiology, Risk Factors, and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health (MAL-ED) study investigated the hypothesis that infection with enteric pathogens contributes to undernutrition in children (34). A follow-up to the MAL-ED study performed 16S sequencing on nearly 1,000 fecal samples from participants in the Peruvian cohort to assess the taxa associated with the burden of Campylobacter and other enteric pathogens (35). We used a database mining approach to determine whether Giardia infection perturbs the human gut microbiome, in both the GEMS and MAL-ED cohorts, similarly to how it perturbs the canine gut microbiome, and to gain insight into possible mechanisms by which Giardia infection may be linked to protection against diarrhea in some individuals.
RESULTS
Enteric parasite infections perturb the canine microbiome.
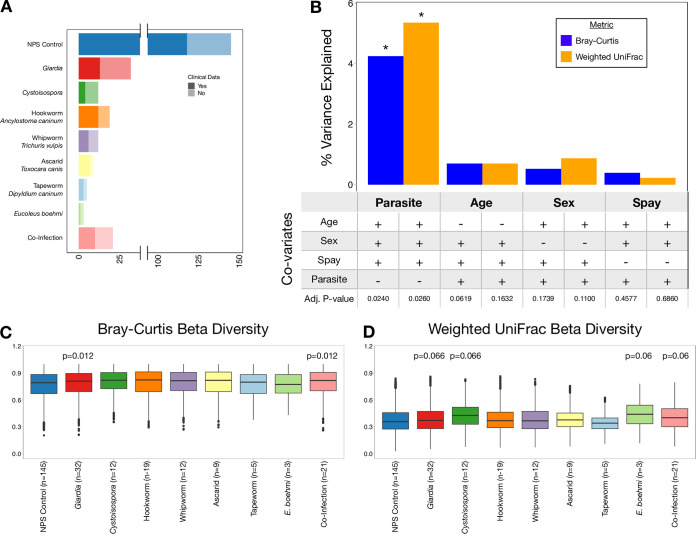
A stool bank was generated from samples screened at a veterinary clinical parasitology service as part of our Companion Animal Microbiome during Parasitism (CAMP) study (see Materials and Methods). A total of 258 canine fecal samples were split into 9 groups based on parasite infection status (Fig. 1A): (i) no parasite seen (NPS) controls; (ii) Giardia, the causative agent of giardiasis; (iii) Cystoisospora, the causative agent of coccidiosis, an intestinal tract infection; (iv) hookworm, which causes intestinal distress and anemia; (v) whipworm, which causes severe irritation to the large intestine; (vi) ascarid, which causes weakness, diarrhea, and vomiting; (vii) tapeworm, which is generally asymptomatic but indicative of flea infestation and may cause perianal pruritis; (viii) Eucoleus boehmi, a parasite whose eggs are shed in the stool and that can cause chronic rhinitis; and (ix) dogs with coinfections of two or more of these parasites. Since certain enteric parasites, such as Giardia, are more prevalent in young animals, age and other potential confounding variables were controlled for in our statistical analyses. Parasite infection status was associated with significant changes in beta diversity, as determined by both Bray-Curtis and weighted UniFrac metrics, even when covariates such as age, sex, and spay/neuter status were controlled for as confounding variables (adjusted P value [Adj P] < 0.05 by permutational multivariate analysis of variance [PERMANOVA]) (Fig. 1B). The significance of parasite infection remains unchanged when infections represented by fewer than 9 samples were removed from the analysis (tapeworm and Eucoleus boehmi). Approximately 5% of the variation in microbiome composition was explained by parasite infection status compared to <1% explained by age, sex, or spay/neuter status alone (Adj P > 0.05 by PERMANOVA) (Fig. 1B). Specifically, Giardia- and coinfected animals displayed the most significant differences in beta diversity compared to NPS controls by both Bray-Curtis (Fig. 1C) and weighted UniFrac metrics (Fig. 1D).
FIG 1.
Parasite infection perturbs canine gut microbiota. 16S sequencing of fecal samples from 258 dogs infected with none, one, or multiple enteric parasites was performed. (A) The number of no parasite seen (NPS) controls, dogs infected with each parasite, and dogs infected with more than one parasite are shown. The number of dogs for which clinical data are unavailable are represented by lighter shading. (B) The percent variance in Bray-Curtis and weighted UniFrac beta diversity explained by each variable is represented by blue and orange bars, respectively. Whether or not age, sex, spay/neuter status, or parasite infection were controlled and the significance of each variable are shown below each bar. Asterisks highlight variables with adjusted P values of <0.05. (C and D) Boxplots showing the difference in Bray-Curtis beta diversity (C) and weighted UniFrac beta diversity (D) between samples in each infection and NPS controls. Adjusted P values of <0.1 are shown above each box.
Canine Giardia infection is associated with significant alterations in gut microbiota composition.
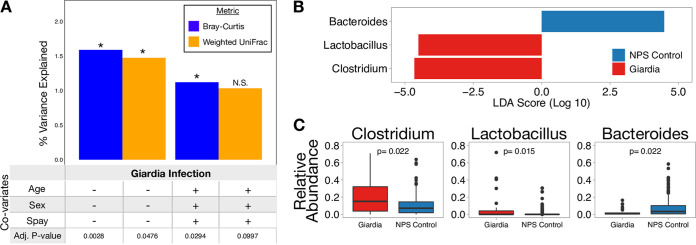
Given the diverse range of parasites detected in our animals, we set out to determine whether specific types of parasites were associated with more pronounced microbiome alterations. Giardia infection is associated with a change in Bray-Curtis (Adj P < 0.01; 1.6% of total variation) and weighted UniFrac (Adj P < 0.05; 1.5% of total variation) beta diversity compared to NPS controls, without controlling for age, sex, and spay/neuter status (Fig. 2A). When controlling for age, sex, and spay/neuter status, beta diversity is still significantly altered during Giardia infection as measured by Bray-Curtis (Adj P < 0.05; 1.1% of total variation), but no longer meets the 0.05 cutoff for significance for weighted UniFrac (Adj P = 0.0997; 1.0% of total variation) (Fig. 2A). The differences in beta diversity between Giardia infection and NPS controls were driven by several bacterial taxa as determined by linear discriminant analysis (LDA) effect size (LEfSe) analysis (Fig. 2B and C). Giardia is associated with enrichment of Clostridium, a genus that contains several commensal taxa, as well as an enrichment of Lactobacillus. However, Giardia was also associated with a reduction in Bacteroides, a genus that includes important commensal bacteria. In order to verify the taxa associated with Giardia infection, point-biserial correlation coefficients were calculated for each taxon with average relative abundance of >1%. Consistent with our LEfSe results, point-biserial correlation coefficients also showed enrichment of Clostridium and Lactobacillus, and a reduction in Bacteroides in addition to a reduction in Megamonas (see Table S1 in the supplemental material). The high relative abundance of Clostridium and Lactobacillus and the low relative abundance of Bacteroides in Giardia-infected dogs compared with NPS controls show that Giardia infection in animals is associated with an altered gut microbiota composition.
FIG 2.
Giardia infection is associated with enrichment of several key bacterial taxa in the canine gut. (A) Histogram showing that Giardia infection is associated with a significant difference in beta diversity compared to NPS controls. Bar height reflects the percentage of total beta diversity variance that is explained by Giardia infection. Plus symbols below the bars show when age, sex, and spay/neuter status are controlled for. Asterisks denote bars with adjusted P values of <0.05. (B) LEfSe graph shows the magnitude of enrichment with LDA score of > comparing Giardia-infected dogs to NPS control dogs. (C) Boxplots showing the relative abundance of differentially enriched taxa. Clostridium is among the most highly enriched bacterial taxa associated with Giardia infection compared to controls.
Differentially abundant taxa associated with Giardia infection compared to NPS controls, as identified by point-biserial correlation coefficient. Point-biserial correlation coefficients showing that Clostridium, Lactobacillus, and Bacteroides are significantly associated with Giardia infection, as seen in the LEfSe analysis (Fig. 2). Download Table S1, PDF file, 0.04 MB (39.8KB, pdf) .
Copyright © 2020 Berry et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
It is possible that the microbiota changes observed in Giardia-infected dogs could be driven, in part, by clinical variables such as diarrhea or antibiotic use. To discriminate between changes linked to diarrhea or antibiotics versus those linked to infection, we evaluated medical records, when available (n = 174), to identify animals with a recent history of diarrhea or antibiotic use (Fig. 1A). Interestingly, Giardia infection was not associated with diarrhea or antibiotic use (P > 0.25 by chi-squared test): among dogs with clinical data, 4 of 13 (30.1%) Giardia-infected dogs had diarrhea compared to 21 of 118 (18%) NPS control dogs with diarrhea (see Fig. S1A in the supplemental material), while 4 of 13 (15.4%) Giardia-infected dogs received antibiotics compared to 15 of 118 (12.7%) NPS control dogs (Fig. S2A). In contrast, antibiotic use was strongly correlated with diarrhea (P < 0.01; chi-squared test), with most dogs on antibiotics having diarrhea (11/15) and over half of dogs with diarrhea being on antibiotics (11/21). Among NPS control animals with clinical data available (n = 118 dogs) (Fig. S1A), those with diarrhea had significantly different Bray-Curtis beta diversity (Adj P < 0.001;, 2.9% of total variation) and weighted UniFrac beta diversity (Adj P < 0.01; 3.7% of total variation) compared to asymptomatic animals; and those receiving antibiotics had significantly different Bray-Curtis beta diversity (Adj P < 0.001, 3.5% of total variation) and weighted UniFrac beta diversity (Adj P < 0.01; 3.9% of total variation) compared to those not receiving antibiotics, when controlling for age, sex, and spay/neuter status (Fig. S1B). Next, we used our NPS control group (n = 118) to define a microbiome signature associated with diarrhea in the absence of observable parasites, allowing us to compare this signature with Giardia-infected animals. LEfSe analysis identified Escherichia as enriched in animals with diarrhea and in those receiving antibiotics, while Bacteroides and Fusobacterium were enriched in asymptomatic dogs (Fig. S1C and S1D) and those not receiving antibiotics (Fig. S2B). Taken together, these data define a microbiome profile associated with diarrhea and antibiotic use in NPS animals that is marked by enrichment of Escherichia and Fusobacterium and show that this signature is distinct from that observed during Giardia infection (Fig. 2B and C).
Diarrhea is associated with enrichment of a different set of taxa compared to Giardia. (A) Pie charts showing the relative proportion of dogs with diarrhea and asymptomatic animals among NPS control and Giardia-infected dogs. The proportion of symptomatic dogs is not significantly different between groups (P > 0.05 by chi-squared test). (B) Histogram showing that diarrhea and antibiotic use are associated with a significant difference in beta diversity compared to asymptomatic and no antibiotic use. Bar height reflects the percentage of total beta diversity variance that is explained by each variable. Age, sex, and spay/neuter status were controlled for all calculations. Asterisks denote bars with adjusted P < 0.05. (C) LEfSe graph showing the magnitude of enrichment for each taxon with a LDA score of >2 comparing animals with and without diarrhea. (D) Boxplots showing the relative abundance of taxa that are differentially abundant between animals with and without diarrhea. Download FIG S1, PDF file, 0.1 MB (126.1KB, pdf) .
Copyright © 2020 Berry et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Antibiotic use in dogs is associated with a similar gut microbiota profile as dogs with diarrhea due to strong correlation between antibiotic use and diarrhea. (A) Pie chart showing the relative proportion of dogs receiving antibiotics and those not among all samples that have associated clinical data (n = 174), NPS controls with clinical data (n = 118), and Giardia-positive dogs with clinical data (n = 15). (B) LEfSe graph showing the magnitude of enrichment for each taxon with LDA score > 2 comparing NPS control dogs receiving and not receiving antibiotics. Escherichia is highly enriched in dogs receiving antibiotics and in dogs with diarrhea, while Bacteroides and Fusobacterium are reduced in dogs receiving antibiotics and in dogs with diarrhea, likely because most dogs receiving antibiotics have diarrhea. Megamonas and Faecalibacterium are reduced in dogs receiving antibiotics, but not in dogs with diarrhea. Download FIG S2, PDF file, 0.07 MB (70.6KB, pdf) .
Copyright © 2020 Berry et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The effect of Giardia on the microbiome persists during coinfection.
We reasoned that if Giardia—compared to other parasites observed in our samples—is driving changes in the microbiome, then we should observe a similar profile in animals harboring coinfections with Giardia and at least one other parasite. Ten out of 21 dogs harboring multiple parasites (“coinfection”) were infected with Giardia and one or more other parasites. These 10 Giardia coinfected samples were indistinguishable from Giardia singly infected animals by Bray-Curtis (Adj P > 0.1) and weighted UniFrac (Adj P > 0.1) beta diversity (Fig. S3). In contrast, Giardia singly infected samples were significantly different from the remaining 11 coinfected samples not involving Giardia by Bray-Curtis (Adj P < 0.05) and weighted UniFrac (Adj P < 0.05) beta diversity; however, false discovery rate correction raises these P values slightly above the 0.05 significance threshold (Fig. S3). Taken together, these results show that Giardia infection in dogs is associated with a unique and significant change in gut microbiota composition compared to NPS controls that persist even in the context of coinfection with other parasites.
The effects of Giardia in canines persist even when one or more other parasites are present. Fecal samples from dogs infected with multiple parasites, one of which is Giardia, are not different from those singly infected with Giardia in terms of Bray-Curtis or weighted UniFrac beta diversity (Adj P > 0.1). In contrast, fecal samples from dogs infected with multiple, non-Giardia parasites are different from those singly infected with Giardia (P < 0.05; adjusted P < 0.1), and infection status here represents a larger percentage of the variance in beta diversity. Age, sex, and spay/neuter status were controlled for all calculations. Download FIG S3, PDF file, 0.05 MB (48.1KB, pdf) .
Copyright © 2020 Berry et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Giardia infection is among the largest predictors of the pediatric gut microbiota structure in the GEMS case-control study.
After finding that parasites, in particular Giardia, perturb the canine gut microbiome, we asked whether Giardia similarly affected the human gut microbiome. To this end, we employed a database mining approach to integrate and query data from the Global Enteric Multicenter Study (GEMS) and the Etiology, Risk Factors, and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health (MAL-ED) study. Clinical and epidemiological data from GEMS were made available on ClinEpiDB.org (Fig. 3A and B) from over 22,000 participants. Previously published microbiome data from a subset of the same participants (n = 1,004) were loaded on MicrobiomeDB.org (36, 37). Clinical and microbiome data were manually integrated, leading to the identification of 215 participants who were positive for Giardia and for which microbiome data were available. Not surprisingly, age and moderate to severe diarrhea (MSD) were strongly correlated with Bray-Curtis beta diversity (Adj P < 0.001), explaining 11% and 5.5% of the total variation in microbiome structure, respectively (Fig. 3C). Giardia infection was associated with a similarly large perturbation of the gut microbiota (Adj P < 0.001; 1.9% of the total variation), while Cryptosporidium and rotavirus infection were each associated with <0.5% of the variation in microbiota composition in this cohort (Adj P < 0.01). Only 14 of the 215 Giardia-infected children were also coinfected with either Cryptosporidum or rotavirus. Infection with Giardia was also significantly associated with gut microbiota composition in each of the four countries individually (Fig. S4A).
FIG 3.
Enteric parasites are associated with gut microbiota perturbations in children. (A) The number of participants with moderate to severe diarrhea (MSD) (green) and without MSD (blue) in each of five age cohorts is shown. (B) Giardia is more frequently found in children without MSD compared to children with MSD. (C) The percent variation in Bray-Curtis beta diversity explained by several variables is shown by bars. Whether the analysis was stratified by age, country, and/or MSD status is shown below each bar. Giardia is significantly associated with a change in gut microbiota and explains more microbiota variation than any other enteric pathogen detected here.
Giardia infection is associated with significant changes in gut microbiome composition in each of the four countries sampled. (A) Bars depict the percent variation in Bray-Curtis beta diversity explained by Giardia infection in each of the four countries. Each analysis was stratified by age and MSD status. Giardia is significantly associated with a change in gut microbiota in all four countries; however, the amount of variation explained by Giardia infection varies across countries. (B) LEfSe graph showing the magnitude of enrichment for all taxa meeting the adjusted P value threshold of 0.1, comparing children with and without Giardia infection. Only taxa with relative abundance of >1% across all samples were used in the analysis. Asterisks denote the significance threshold for each taxon in each country. Download FIG S4, PDF file, 0.09 MB (97.8KB, pdf) .
Copyright © 2020 Berry et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
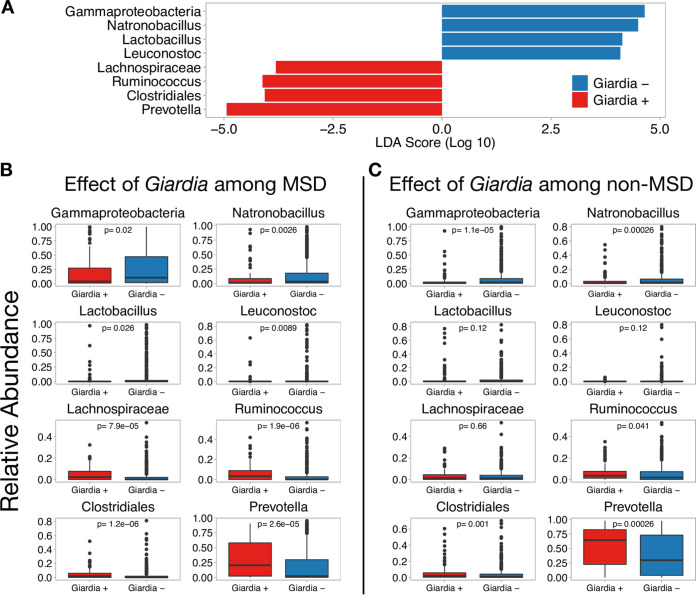
We observed that Giardia infection among GEMS participants was associated with enrichment of Prevotella and a reduction in Gammaproteobacteria (Fig. 4A)—an effect that was evident in children with MSD (Fig. 4B) and without MSD (Fig. 4C). LEfSe analyses performed on data partitioned by country showed that Giardia infection is associated with enrichment of Prevotella and a reduction in Gammaproteobacteria in all four countries (Fig. S4B). Diarrhea is commonly associated with a reduction in Prevotella and an increased abundance of Gammaproteobacteria. Moreover, age strongly influences the relative abundance of Prevotella and Gammaproteobacteria (Fig. S5A and S5B, respectively), as well as Giardia prevalence (Fig. S5C and S5D). To control for these factors, the impact of Giardia was assessed among 12- to 17-month-old GEMS participants, a cohort with high relative abundance of both Prevotella and Gammaproteobacteria and high prevalence of Giardia infections (29.1%; n = 51) and for which Giardia prevalence is not correlated with age (P = 0.99 by chi-squared test). Among 12- to 17-month-old children, the association between Giardia infection and reduction in Gammaproteobacteria and enrichment of Prevotella remained (Fig. S6).
FIG 4.
Giardia infection in children is associated with a reduction in Gammaproteobacteria regardless of disease status. (A) LEfSe graph showing the magnitude of enrichment for each taxon with LDA score of >2 comparing children with and without Giardia infection. (B and C) Boxplots showing the relative abundance of differentially enriched taxa among children with MSD (B) and those without MSD (C). A very similar set of taxa are differentially expressed during Giardia infection regardless of clinical disease. Although the relative abundance of Gammaproteobacteria and Prevotella are different between MSD and non-MSD, Giardia infection is significantly associated with a reduction of Gammaproteobacteria and enrichment of Prevotella regardless of MSD status. Taxa were collapsed to the genus level when possible; however, Gammaproteobacteria, Clostridiales, and Lachnospiraceae were only able to be collapsed to the class, order, and family, respectively.
Prevotella abundance, Gammaproteobacteria abundance, and Giardia prevalence are correlated with age in young children. (A and B) Boxplots showing that, over the first 5 years of life, the relative abundance of the Prevotella genus increases with age (A) and the relative abundance of the Gammaproteobacteria class decreases with age (B). (C and D) Bar graphs showing that the proportion of children infected with Giardia is low for children in the first year of life compared to 1- to 5-year-old children. Download FIG S5, PDF file, 0.1 MB (106.1KB, pdf) .
Copyright © 2020 Berry et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Giardia is associated with a reduction in Gammaproteobacteria and enrichment of Prevotella among 12- to 17-month-old children. (A) LEfSe graph showing the magnitude of enrichment for each taxon with LDA score > 2 comparing 12- to 17-month-old children with (n = 51) and without (n = 124) Giardia infection. (B) Boxplots showing the differences in relative abundance in taxa associated with Giardia infection among all 12- to 17-month-old participants. (C and D) Boxplots showing that Giardia is associated with a reduction in the Gammaproteobacteria class among 12- to 17-month-old children with MSD (C) but that Giardia is not significantly associated with Gammaproteobacteria among 12 to 17-month-old children without MSD (D), when the relative abundance of Gammaproteobacteria is low. Download FIG S6, PDF file, 0.1 MB (114.1KB, pdf) .
Copyright © 2020 Berry et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Longitudinal tracking of Giardia status reveals infection-dependent alterations in microbiome composition.
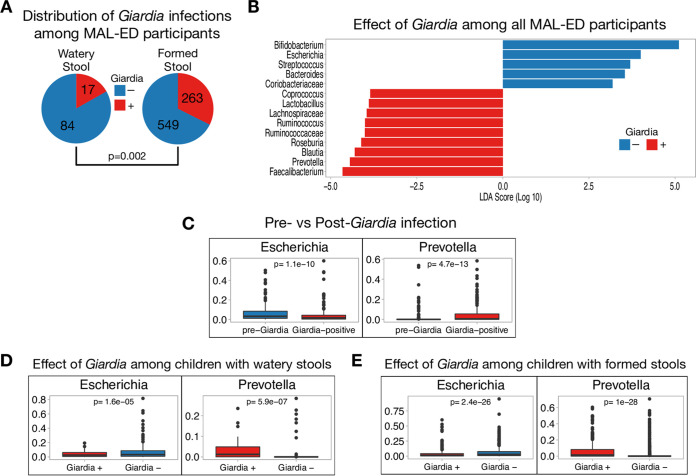
We employed the same database mining strategy to combine clinical and microbiome data from the MAL-ED study. Clinical and epidemiological data for 2,145 participants sampled longitudinally (>1.8 million observations) were stored on ClinEpiDB.org and were manually integrated with fecal microbiome data (available on microbiomeDB.org) from 271 participants from the Peru cohort, each sampled up to four times at 6, 12, 18, and 24 months of life (n = 913 samples) (34, 38). Of the 913 fecal samples, 280 were positive for Giardia by enzyme-linked immunosorbent assay (ELISA). Giardia was observed more frequently in formed or soft stool samples compared to liquid or watery stools (P = 0.002 by chi-squared test) (Fig. 5A), consistent with the GEMS study, where Giardia infection was more prevalent among participants without MSD. Giardia infection among MAL-ED participants was associated with enrichment of Prevotella and a reduction in Escherichia, a member of the Gammaproteobacteria class (Fig. 5B). MAL-ED sequenced the V4 region of the 16S gene and was classified against the SILVA database, while GEMS sequenced the V1-V2 region and was classified against the Greengenes database, which likely led to some differences in taxonomic classification, including the identification of the Escherichia genus specifically in MAL-ED compared to the broader Gammaproteobacteria class in GEMS.
FIG 5.
Giardia is associated with a reduction in Escherichia and enrichment of Prevotella among Peruvian children in the MAL-ED study. (A) Among all Peruvian participants in the MAL-ED study, Giardia is more frequently found in children with formed or soft stools than in children with liquid or watery stools (P = 0.002 by chi-squared test). (B) LEfSe graph showing the magnitude of enrichment for all taxa meeting the adjusted P value threshold of 0.05, comparing children with (red) and without (blue) Giardia infection. Only taxa with relative abundance of >1% across all samples were used in the analysis. (C) Boxplots showing the differences in relative abundance of Escherichia and Prevotella immediately before and after acquiring Giardia infection. (D) Boxplots showing that Giardia is associated with a reduction in Escherichia and enrichment of Prevotella among children with liquid or watery stools and (E) among children with formed or soft stools. All P values were adjusted using Benjamini-Hochberg multiple testing correction.
We reasoned that if Giardia infection was driving the change in microbiome composition observed in the GEMS data, then the longitudinal aspect of MAL-ED should allow identification of individuals that convert from Giardia negative to positive during the study period, providing a unique context for examining parasite-microbiome interactions. A total of 170 pairs of MAL-ED observations were identified where a Giardia-positive stool sample could be compared to the Giardia-negative sample obtained from the same individual 6 months prior. Analysis of the microbiome in these discordant pairs showed that becoming infected with Giardia was marked by decreased relative abundance of Escherichia and increased Prevotella (Adj P < 1e−9) (Fig. 5C). Since samples were collected 6 months apart, an effect of age cannot be ruled out. To address this, we also evaluated 53 pairs of observations where individuals converted from Giardia positive to negative over a 6-month period. In these cases, clearing Giardia was not associated with a change in relative abundance of Escherichia or Prevotella (Adj P > 0.1), suggesting that the differences observed upon infection cannot be explained by age alone. These data also suggest that clearing Giardia does not have an inversely proportional effect on the gut microbiota composition as acquiring Giardia. Finally, the enrichment of Prevotella and reduction in Escherichia were evident in children with liquid or watery stools (Fig. 5D) as well as those with formed or soft stools (Fig. 5E). Taken together, these results demonstrate that Giardia infection leads to an altered gut microbiome structure in humans, marked by changes in the relative abundance of taxa linked to gut health (39–41).
DISCUSSION
Enteric parasite infections are among the most common causes of diarrhea in humans in the developing world. While bacterial infections and the gut microbiome have been well-studied, the impact of enteric eukaryotic parasites on the microbiome is not well understood, with some reports showing altered microbiome composition (17, 42–46, 105) while others showed either modest or no impact (47–50). Because these studies often rely on experimental infection with one or few parasite species or observations using a small number of participants, they provide limited insight into the broader impact of enteric parasites on the gut microbiome. By combining clinical parasitology and microbiome profiling from humans and canines infected with a phylogenetically diverse range of enteric parasites, we show that naturally acquired enteric parasite infections are a major factor associated with microbiome composition, that this effect is observed across host species, and that Giardia is associated with the largest impact among all parasites surveyed in dogs and humans.
Giardia is one of the most common enteric parasites in the world and is remarkable in its ability to cause an array of clinical phenotypes, ranging from asymptomatic infection to severe acute diarrheal disease to chronic gastrointestinal disease. Giardia is the causative agent of giardiasis, a diarrheal illness, and is clearly implicated in serious growth stunting and long-term health consequences (51), cementing its role as a pathogen. However, our observations (Fig. 3), as well as other reports in humans and animals suggest that Giardia infection is frequently asymptomatic (8, 51–58). Intriguingly, several large epidemiological case-control studies recently showed higher Giardia prevalence in asymptomatic participants compared to those with moderate to severe diarrheal disease, revealing a possible protective role (30, 34, 51, 56, 59, 60). The negative association between Giardia and MSD may be due, in part, to asymptomatic participants shedding hardier cysts that preserve DNA better than the trophozoites typically associated with severe Giardia infection; however, data from this study and others suggest biological explanations for the phenomenon. Giardia infection may modulate symptoms in some individuals by modulating the immune response to other pathogens (56, 61), as seen in a recent study showing that Giardia coinfection attenuates the severity of disease caused by other enteric pathogens (62). We reasoned that parasite-induced perturbations in the microbiome could also be an important factor influencing gastrointestinal symptoms. Our results, in combination with similar results from recent smaller-scale mouse and human experiments (62, 63), raise the possibility that the shift in microbiome composition during Giardia infection—marked by a reduction in Gammaproteobacteria and an increase in Prevotella—may explain, at least in part, the apparent protective effect of Giardia against diarrhea in some age/site cohorts (30, 34, 56).
One intriguing extension of our findings is the notion that Giardia may benefit directly from manipulation of the microbiome as seen during infection with the intestinal parasite Blastocystis which engulfs highly abundant bacterial taxa to meet its nutritional demands, causing drastic changes to gut microbiota composition (64). Infection by another protozoan parasite, Entamoeba histolytica, results in enrichment of Escherichia coli that protects the parasite from oxidative damage by producing malate dehydrogenase (65). Similarly, during infection with the helminth Trichuris muris, Proteobacteria directly interact with parasite eggs to induce hatching, thereby enhancing worm reproduction (66). Taken together, these studies highlight that eukaryotic parasites impact the microbiome in ways that can influence host health, immunity, and parasite biology.
Our data show an association between Giardia infection and microbiome composition but do not resolve whether infection is the primary driver of these changes. It is possible that certain microbiome compositions confer susceptibility or resistance to colonization by the parasite, as suggested by previous studies (67). Although it may seem surprising that a pathogen of the upper small intestine could have the potential to impact microbiome composition in the stool, recent studies of mice experimentally infected with Giardia revealed alterations of the gut microbiome throughout the small and large intestine, indicating both a causal role for infection in inducing these changes and the ability of the parasite to profoundly alter bacterial community structure far from the site of infection (12). Moreover, our analysis of longitudinal data from the MAL-ED study, which show a clear shift in composition commensurate with infection, argue in favor of microbiome changes being a consequence, rather than a cause, of infection.
Taken together, these data make a strong case for pursuing mechanistic studies that address interactions between Giardia, the microbiome, and the host. Interestingly, Giardia infection is associated with malabsorption of fats, leading to intestinal steatosis and increased transit of lipids into the distal small intestine and colon (68), malabsorption of sugars and proteins, and diffuse shortening of the intestinal brush border microvilli (69, 70). These pathological changes could alter substrate availability for commensal bacteria, providing a possible explanation for compositional changes in the microbiome during this infection. Additionally, gut microbiota composition changes could be caused by Giardia-induced disturbances to biofilm composition and structure which have been linked to dysbiosis (71, 72).
The age at which humans or animals are exposed to Giardia is thought to impact clinical manifestations. For example, there appears to be a window of time early in childhood development when Giardia infection is negatively associated with diarrhea. Studies of several pediatric cohorts show either no correlation or a negative correlation between Giardia infection and diarrhea (30, 51, 53–55, 73), although definitions of diarrhea vary from “loose stool” to clinically severe diarrhea. Notably, those studies that specify moderate to severe diarrhea show a significant association between Giardia infection and not having diarrhea (30, 54), suggesting that Giardia infection may be negatively associated with severe diarrhea but not loose stools. Indeed, clinical manifestations associated with Giardia infection are variable, likely due in part to variation among Giardia genotypes, and may explain why children in countries where Giardia is not endemic are more likely than children in countries where Giardia is endemic to have symptomatic Giardia infection (74–77). In contrast, adults—especially those in areas where Giardia is not endemic—show a positive correlation between Giardia infection and diarrhea (51, 78). Previous studies suggest that the association between growth stunting and Giardia infection is dependent on age (79), with some studies showing that asymptomatic Giardia infection is associated with growth stunting among children older than 18 months, but not infants or in children during their first 18 months (80), while others show an association between Giardia and growth stunting at 2 years of age (31). The effects of Giardia on gut microbiota may also be age dependent. The MAL-ED study of Peruvian children found that gut microbiota associated with Giardia burden varied by age (35). For example, high Giardia burden was associated with enrichment of Prevotella only in fecal samples of 24-month-old children. Here, we show an association between Giardia infection and altered gut microbiota composition in specific age cohorts as well, raising the possibility that parasite-microbiome interactions may partially explain the age-dependent disease presentation during Giardia infection. Collectively, these data point to the gut microbiome, host immunity, parasite genotype, and age (81–84) as variables that may interact or operate independently to augment the balance between protection and pathogenesis during Giardia infection.
One major obstacle to investigating relationships between clinical variables and microbiome composition in large-scale studies like GEMS and MAL-ED is that these data are not always collected at the same time, by the same researchers, with the goal of being analyzed together. For example, although extensive clinical and epidemiological data were collected from over 22,000 participants in GEMS (30) and over 1.8 million observations in MAL-ED (34), microbiome profiling data were collected from a subset of approximately 1,000 samples from each study, and were published separately and with relatively sparse metadata (33, 38). Our study highlights that a database-driven approach that integrates microbiome data with extensive clinical and epidemiological data allows for the identification of novel associations and an opportunity to compare microbiome phenotypes across host species and across studies.
MATERIALS AND METHODS
A dockerized environment containing code and software is available on Code Ocean (https://codeocean.com/capsule/2815529/tree) and fully reproduces all analyses and figures in the article.
Canine sample collection.
Fecal samples for our Companion Animal Microbiome during Parasitism (CAMP) study were acquired from patients seen at the Ryan Hospital at the University of Pennsylvania’s School of Veterinary Medicine (PennVet) as part of both sick and wellness visits, as well as from healthy dogs in animal shelters that were brought to the Ryan Hospital to be spayed or neutered. Fecal samples were examined for parasites by fecal flotation (using a zinc sulfate solution at a specific gravity of 1.18 g/ml) at the Clinical Parasitology Laboratory of PennVet. Dogs either had no observable parasites (n = 145), one (n = 92), or multiple (n = 21) protozoan parasites, including Giardia (n = 32) and Cystoisospora (n = 12), and helminths, including hookworm (Ancylostoma caninum) (n = 19), whipworm (Trichuris vulpis) (n = 12), ascarid (Toxocara canis) (n = 9), tapeworm (Dipylidium caninum) (n = 5), and Eucoleus boehmi (n = 3) (Fig. 1A). Although zinc flotation is less sensitive for detecting Giardia infection compared to antigen immunoassays, it has the advantage of higher specificity for detecting active Giardia infections rather than detecting antigen that can persist even after the infection is cleared. Samples containing yeast were excluded from the study. Age, sex, and spay and neuter status were recorded at the time of fecal sample collection for all samples. Fecal samples from 113 infected dogs and 145 dogs without detectable parasites were stored at −80°C until DNA extraction. Clinical data from patient visits were obtained for 174 PennVet patients to determine whether gastrointestinal symptoms or antibiotic use occurred within 1 week of fecal sample collection.
16S rRNA gene sequencing and analysis.
DNA was extracted from fecal samples using Qiagen PowerSoil DNA extraction kit. 16S rRNA gene sequencing was performed as described previously (85). Briefly, the V4 region of the 16S rRNA gene was amplified using PCR, which was performed using Accuprime Pfx supermix and custom primers for 30 cycles (85). PicoGreen quantification was used to normalize post-PCR products, and AMPureXP beads were used to clean the combined pools. Libraries were quantified and sized using a Qubit 2.0 and Tapestation 4200, respectively. Then, 250-bp paired-end sequencing was performed using an Illumina MiSeq. The QIIME2 pipeline (86) was used to process and analyze 16S sequencing data. Samples were demultiplexed using q2-demux and denoised using Dada2 (87). Sequences were aligned using maaft (88), and phylogenetic trees were reconstructed using fasttree (89). Weighted UniFrac (90) and Bray-Curtis (91) beta diversity metrics were estimated using q2-core-metrics-diversity after samples were rarefied to 4,100 reads per sample, and P values were adjusted for multiple hypothesis testing using Benjamini-Hochberg (B-H) false discovery rate (FDR) corrections (92). Taxonomy was assigned to sequences using q2-feature-classifier classify-sklearn (93) against the Greengenes 13-8 99% operational taxonomic unit (OTU) reference sequences (94). Taxa were collapsed to the genus level, when possible. OTUs with less than 1% average relative abundance across all samples were removed.
Correlation analysis and differential feature selection.
The correlation between variables such as parasite infection and microbiota composition was determined using permutational multivariate analysis of variance (PERMANOVA) as implemented in the vegan package (95) in R (96). Differentially abundant taxa were determined using linear discriminant analysis (LDA) effect size (LEfSe) (97), and P values were adjusted for multiple hypothesis testing using B-H FDR corrections in R. Boxplots and LEfSe plots were visualized using ggplot2 (98), patchwork (99), and ggthemes (100). Point-biserial correlation coefficients were calculated to identify differentially abundant taxa between Giardia-infected and no parasite seen (NPS) controls with 10,000 permutations using the indicspecies package in R (101), adjusting for multiple hypothesis testing using B-H FDR corrections (see Table S1 in the supplemental material).
Integration and analysis of GEMS data.
The Global Enteric Multicenter Study (GEMS) investigated the causes, incidence, and impact of moderate to severe diarrhea in 23,567 0- to 59-month-old children in Asia and Africa (30). Clinical and epidemiological data and anthropometric measurements for each participant were downloaded from ClinEpiDB.org (36, 37). The presence of Giardia, Cryptosporidium, and rotavirus were determined using ELISA on participant fecal samples. Additionally, sequencing of the V1-V2 region of the 16S rRNA gene was performed on stool samples from 1,007 participants (33). Taxonomy was determined by classifying sequences against the Greengenes 99% OTU reference sequences. Here, clinical data from 1,004 GEMS participants was downloaded from ClinEpiDB.org, and the relative abundances of bacterial taxa for the same 1,004 participants was downloaded from MicrobiomeDB.org (102). The data sets were manually combined so that clinical and epidemiological data were matched to gut bacterial taxon abundance data.
Correlations between clinical variables (e.g., Giardia infection) and Bray-Curtis beta diversity were calculated using the vegan package in R. Patients were divided among five age groups (0 to 6, 6 to 12, 12 to 18, 18 to 24, and 24 to 59 months) to control for the effects associated with age. Here, associations with age were stratified by country, associations with country were stratified by age, and all other associations were stratified by age group and country (Fig. 5C), as done by Kotloff et al. (30). Taxonomy was collapsed to the genus level, when possible, and taxa with mean relative abundance across all samples of <1% were removed. Differentially abundant taxa between Giardia-positive versus Giardia-negative and MSD cases versus controls were determined using LEfSe, adjusting P values for multiple hypothesis testing using B-H FDR corrections. LEfSe plots and boxplots were visualized using ggplot2 (98), patchwork (99), and ggthemes (100).
Integration and analysis of MAL-ED data.
The Etiology, Risk Factors, and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health (MAL-ED) study investigated the burden of enteropathogens and malnutrition by monitoring 2,145 children across eight sites in Africa, Asia, and South America starting at 17 days of life and followed longitudinally for up to 60 months (34). Clinical and epidemiological data and anthropometric measurements for each participant were downloaded from ClinEpiDB.org (36, 37). Fecal samples were collected from participants, and the presence of Giardia was determined by ELISA. Additionally, sequencing of the V4 region of the 16S rRNA gene was performed on stool samples from 913 participants from the Peruvian cohort (38). Taxonomy was determined by classifying sequences against the SILVA reference database (103, 104). Here, clinical data from 913 MAL-ED observations were downloaded from ClinEpiDB.org, and the relative abundances of bacterial taxa for the same observations were downloaded from MicrobiomeDB.org (102). The data sets were manually combined so that clinical and epidemiological data were matched to gut bacterial taxa abundance data. Taxonomy was collapsed to the genus level, when possible, and taxa with mean relative abundance across all samples of <1% were removed. Differentially abundant taxa between Giardia-positive versus Giardia-negative children were determined using LEfSe, adjusting P values for multiple hypothesis testing using B-H FDR corrections. The effects of Giardia on gut microbiota composition were also stratified by stool consistency: the effects among watery or liquid stools and the effects among formed or soft stools. LEfSe plots and boxplots were visualized using ggplot2 (98), patchwork (99), and ggthemes (100).
Data availability.
All sequencing data analyzed here are publicly available on the Sequence Read Archive (SRA) under study accession number PRJNA594732. All sequencing data used for the canine analyses is also publicly available on MIcrobiomeDB.org as part of the CAMP study. All code is available and reproducible on CodeOcean (https://codeocean.com/capsule/2815529/tree).
ACKNOWLEDGMENTS
We thank Lisa Mattei and Huanjia Zhang at the Children’s Hospital of Philadelphia for assistance in sequencing.
A Tobacco Formula grant provided partial support for the project and for A.S.F.B. and R.N.B. This Tobacco Formula grant is under the Commonwealth Universal Research Enhancement (CURE) program with the grant number SAP 4100068710. The funders had no role in data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Haque R. 2007. Human intestinal parasites. J Health Popul Nutr 25:387–391. [PMC free article] [PubMed] [Google Scholar]
- 2.Menu E, Mary C, Toga I, Raoult D, Ranque S, Bittar F. 2019. A hospital qPCR-based survey of 10 gastrointestinal parasites in routine diagnostic screening, Marseille, France. Epidemiol Infect 147:e100. doi: 10.1017/S0950268819000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fletcher SM, Stark D, Harkness J, Ellis J. 2012. Enteric protozoa in the developed world: a public health perspective. Clin Microbiol Rev 25:420–449. doi: 10.1128/CMR.05038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine MM, Kotloff KL, Nataro JP, Muhsen K. 2012. The Global Enteric Multicenter Study (GEMS): impetus, rationale, and genesis. Clin Infect Dis 55(Suppl 4):S215–S224. doi: 10.1093/cid/cis761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moosavi A, Haghighi A, Mojarad EN, Zayeri F, Alebouyeh M, Khazan H, Kazemi B, Zali MR. 2012. Genetic variability of Blastocystis sp. isolated from symptomatic and asymptomatic individuals in Iran. Parasitol Res 111:2311–2315. doi: 10.1007/s00436-012-3085-5. [DOI] [PubMed] [Google Scholar]
- 6.Holtman GA, Kranenberg JJ, Blanker MH, Ott A, Lisman-van Leeuwen Y, Berger MY. 2017. Dientamoeba fragilis colonization is not associated with gastrointestinal symptoms in children at primary care level. Fam Pract 34:25–29. doi: 10.1093/fampra/cmw111. [DOI] [PubMed] [Google Scholar]
- 7.Jokelainen P, Hebbelstrup Jensen B, Andreassen BU, Petersen AM, Röser D, Krogfelt KA, Nielsen HV, Stensvold CR. 2017. Dientamoeba fragilis, a commensal in children in Danish day care centers. J Clin Microbiol 55:1707–1713. doi: 10.1128/JCM.00037-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hellard ME, Sinclair MI, Hogg GG, Fairley CK. 2000. Prevalence of enteric pathogens among community based asymptomatic individuals. J Gastroenterol Hepatol 15:290–293. doi: 10.1046/j.1440-1746.2000.02089.x. [DOI] [PubMed] [Google Scholar]
- 9.Reh L, Muadica AS, Köster PC, Balasegaram S, Verlander NQ, Chércoles ER, Carmena D. 2019. Substantial prevalence of enteroparasites Cryptosporidium spp., Giardia duodenalis and Blastocystis sp. in asymptomatic schoolchildren in Madrid, Spain, November 2017 to June 2018. Euro Surveill 24(43):1900241. doi: 10.2807/1560-7917.ES.2019.24.43.1900241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang S, El-Fahmawi A, Christian DA, Fang Q, Radaelli E, Chen L, Sullivan MC, Misic AM, Ellringer JA, Zhu X-Q, Winter SE, Hunter CA, Beiting DP. 2019. Infection-induced intestinal dysbiosis is mediated by macrophage activation and nitrate production. mBio 10:e00935-19. [CrossRef] doi: 10.1128/mBio.00935-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gimblet C, Meisel JS, Loesche MA, Cole SD, Horwinski J, Novais FO, Misic AM, Bradley CW, Beiting DP, Rankin SC, Carvalho LP, Carvalho EM, Scott P, Grice EA. 2017. Cutaneous Leishmaniasis induces a transmissible dysbiotic skin microbiota that promotes skin inflammation. Cell Host Microbe 22:13–24.e4. doi: 10.1016/j.chom.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barash NR, Maloney JG, Singer SM, Dawson SC. 2017. Giardia alters commensal microbial diversity throughout the murine gut. Infect Immun 85:e00948-16. [CrossRef] doi: 10.1128/IAI.00948-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerbaba TK, Gupta P, Rioux K, Hansen D, Buret AG. 2015. Giardia duodenalis-induced alterations of commensal bacteria kill Caenorhabditis elegans: a new model to study microbial-microbial interactions in the gut. Am J Physiol Gastrointest Liver Physiol 308:G550–G561. doi: 10.1152/ajpgi.00335.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerbaba TK, Green-Harrison L, Buret AG. 2017. Modeling host-microbiome interactions in Caenorhabditis elegans. J Nematol 49:348–356. [PMC free article] [PubMed] [Google Scholar]
- 15.Bartelt LA, Bolick DT, Mayneris-Perxachs J, Kolling GL, Medlock GL, Zaenker EI, Donowitz J, Thomas-Beckett RV, Rogala A, Carroll IM, Singer SM, Papin J, Swann JR, Guerrant RL. 2017. Cross-modulation of pathogen-specific pathways enhances malnutrition during enteric co-infection with Giardia lamblia and enteroaggregative Escherichia coli. PLoS Pathog 13:e1006471. doi: 10.1371/journal.ppat.1006471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freeman CD, Klutman NE, Lamp KC. 1997. Metronidazole. A therapeutic review and update. Drugs 54:679–708. doi: 10.2165/00003495-199754050-00003. [DOI] [PubMed] [Google Scholar]
- 17.Jenkins TP, Peachey LE, Ajami NJ, MacDonald AS, Hsieh MH, Brindley PJ, Cantacessi C, Rinaldi G. 2018. Schistosoma mansoni infection is associated with quantitative and qualitative modifications of the mammalian intestinal microbiota. Sci Rep 8:12072. doi: 10.1038/s41598-018-30412-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosa BA, Supali T, Gankpala L, Djuardi Y, Sartono E, Zhou Y, Fischer K, Martin J, Tyagi R, Bolay FK, Fischer PU, Yazdanbakhsh M, Mitreva M. 2018. Differential human gut microbiome assemblages during soil-transmitted helminth infections in Indonesia and Liberia. Microbiome 6:33. doi: 10.1186/s40168-018-0416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renelies-Hamilton J, Noguera-Julian M, Parera M, Paredes R, Pacheco L, Dacal E, Saugar JM, Rubio JM, Poulsen M, Köster PC, Carmena D. 2019. Exploring interactions between Blastocystis sp., Strongyloides spp. and the gut microbiomes of wild chimpanzees in Senegal. Infect Genet Evol 74:104010. doi: 10.1016/j.meegid.2019.104010. [DOI] [PubMed] [Google Scholar]
- 20.Gates MC, Nolan TJ. 2009. Endoparasite prevalence and recurrence across different age groups of dogs and cats. Vet Parasitol 166:153–158. doi: 10.1016/j.vetpar.2009.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Little SE, Johnson EM, Lewis D, Jaklitsch RP, Payton ME, Blagburn BL, Bowman DD, Moroff S, Tams T, Rich L, Aucoin D. 2009. Prevalence of intestinal parasites in pet dogs in the United States. Vet Parasitol 166:144–152. doi: 10.1016/j.vetpar.2009.07.044. [DOI] [PubMed] [Google Scholar]
- 22.Wang S, Martins R, Sullivan MC, Friedman ES, Misic AM, El-Fahmawi A, De Martinis ECP, O’Brien K, Chen Y, Bradley C, Zhang G, Berry ASF, Hunter CA, Baldassano RN, Rondeau MP, Beiting DP. 2019. Diet-induced remission in chronic enteropathy is associated with altered microbial community structure and synthesis of secondary bile acids. Microbiome 7:126. doi: 10.1186/s40168-019-0740-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerquetella M, Spaterna A, Laus F, Tesei B, Rossi G, Antonelli E, Villanacci V, Bassotti G. 2010. Inflammatory bowel disease in the dog: differences and similarities with humans. World J Gastroenterol 16:1050–1056. doi: 10.3748/wjg.v16.i9.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jergens AE, Simpson KW. 2012. Inflammatory bowel disease in veterinary medicine. Front Biosci (Elite Ed) 4:1404–1419. doi: 10.2741/470. [DOI] [PubMed] [Google Scholar]
- 25.Peiravan A, Bertolini F, Rothschild MF, Simpson KW, Jergens AE, Allenspach K, Werling D. 2018. Genome-wide association studies of inflammatory bowel disease in German shepherd dogs. PLoS One 13:e0200685. doi: 10.1371/journal.pone.0200685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vázquez-Baeza Y, Hyde ER, Suchodolski JS, Knight R. 2016. Dog and human inflammatory bowel disease rely on overlapping yet distinct dysbiosis networks. Nat Microbiol 1:16177. doi: 10.1038/nmicrobiol.2016.177. [DOI] [PubMed] [Google Scholar]
- 27.Suchodolski JS, Markel ME, Garcia-Mazcorro JF, Unterer S, Heilmann RM, Dowd SE, Kachroo P, Ivanov I, Minamoto Y, Dillman EM, Steiner JM, Cook AK, Toresson L. 2012. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS One 7:e51907. doi: 10.1371/journal.pone.0051907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simpson KW, Dogan B, Rishniw M, Goldstein RE, Klaessig S, McDonough PL, German AJ, Yates RM, Russell DG, Johnson SE, Berg DE, Harel J, Bruant G, McDonough SP, Schukken YH. 2006. Adherent and invasive Escherichia coli is associated with granulomatous colitis in boxer dogs. Infect Immun 74:4778–4792. doi: 10.1128/IAI.00067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coelho LP, Kultima JR, Costea PI, Fournier C, Pan Y, Czarnecki-Maulden G, Hayward MR, Forslund SK, Schmidt TSB, Descombes P, Jackson JR, Li Q, Bork P. 2018. Similarity of the dog and human gut microbiomes in gene content and response to diet. Microbiome 6:72. doi: 10.1186/s40168-018-0450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acácio S, Biswas K, O’Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 31.Rogawski ET, Liu J, Platts-Mills JA, Kabir F, Lertsethtakarn P, Siguas M, Khan SS, Praharaj I, Murei A, Nshama R, Mujaga B, Havt A, Maciel IA, Operario DJ, Taniuchi M, Gratz J, Stroup SE, Roberts JH, Kalam A, Aziz F, Qureshi S, Islam MO, Sakpaisal P, Silapong S, Yori PP, Rajendiran R, Benny B, McGrath M, Seidman JC, Lang D, Gottlieb M, Guerrant RL, Lima AAM, Leite JP, Samie A, Bessong PO, Page N, Bodhidatta L, Mason C, Shrestha S, Kiwelu I, Mduma ER, Iqbal NT, Bhutta ZA, Ahmed T, Haque R, Kang G, Kosek MN, Houpt ER, MAL-ED Network Investigators. 2018. Use of quantitative molecular diagnostic methods to investigate the effect of enteropathogen infections on linear growth in children in low-resource settings: longitudinal analysis of results from the MAL-ED cohort study. Lancet Glob Health 6:e1319–e1328. doi: 10.1016/S2214-109X(18)30351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Litleskare S, Rortveit G, Eide GE, Emberland KE, Hanevik K, Langeland N, Wensaas K-A. 2019. Quality of life and its association with irritable bowel syndrome and fatigue ten years after giardiasis. Neurogastroenterol Motil 31:e13559. doi: 10.1111/nmo.13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pop M, Walker AW, Paulson J, Lindsay B, Antonio M, Hossain MA, Oundo J, Tamboura B, Mai V, Astrovskaya I, Corrada Bravo H, Rance R, Stares M, Levine MM, Panchalingam S, Kotloff K, Ikumapayi UN, Ebruke C, Adeyemi M, Ahmed D, Ahmed F, Alam MT, Amin R, Siddiqui S, Ochieng JB, Ouma E, Juma J, Mailu E, Omore R, Morris JG, Breiman RF, Saha D, Parkhill J, Nataro JP, Stine OC. 2014. Diarrhea in young children from low-income countries leads to large-scale alterations in intestinal microbiota composition. Genome Biol 15:R76. doi: 10.1186/gb-2014-15-6-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MAL-ED Network Investigators. 2014. The MAL-ED study: a multinational and multidisciplinary approach to understand the relationship between enteric pathogens, malnutrition, gut physiology, physical growth, cognitive development, and immune responses in infants and children up to 2 years of age in resource-poor environments. Clin Infect Dis 59(Suppl 4):S193–S206. doi: 10.1093/cid/ciu653. [DOI] [PubMed] [Google Scholar]
- 35.Rouhani S, Griffin NW, Yori PP, Olortegui MP, Siguas Salas M, Rengifo Trigoso D, Moulton LH, Houpt ER, Barratt MJ, Kosek MN, Gordon JI. 2019. Gut microbiota features associated with campylobacter burden and postnatal linear growth deficits in a Peruvian birth cohort. Clin Infect Dis doi: 10.1093/cid/ciz906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aurrecoechea C, Barreto A, Basenko EY, Brestelli J, Brunk BP, Cade S, Crouch K, Doherty R, Falke D, Fischer S, Gajria B, Harb OS, Heiges M, Hertz-Fowler C, Hu S, Iodice J, Kissinger JC, Lawrence C, Li W, Pinney DF, Pulman JA, Roos DS, Shanmugasundram A, Silva-Franco F, Steinbiss S, Stoeckert CJ, Spruill D, Wang H, Warrenfeltz S, Zheng J. 2017. EuPathDB: the eukaryotic pathogen genomics database resource. Nucleic Acids Res 45:D581–D591. doi: 10.1093/nar/gkw1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruhamyankaka E, Brunk BP, Dorsey G, Harb OS, Helb DA, Judkins J, Kissinger JC, Lindsay BR, Roos DS, Stoeckert CJ, Zheng J, Shah Tomko S. 2019. ClinEpiDB: an open-access clinical epidemiology database resource encouraging online exploration of complex studies. Zenodo doi: 10.5281/zenodo.3522209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouhani S, Griffin NW, Yori PP, Gehrig JL, Olortegui MP, Salas MS, Trigoso DR, Moulton LH, Houpt ER, Barratt MJ, Kosek MN, Gordon JI. 2019. Diarrhea as a potential cause and consequence of reduced gut microbial diversity among undernourished children in Peru. Clin Infect Dis doi: 10.1093/cid/ciz905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kostic AD, Xavier RJ, Gevers D. 2014. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146:1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torres J, Hu J, Seki A, Eisele C, Nair N, Huang R, Tarassishin L, Jharap B, Cote-Daigneault J, Mao Q, Mogno I, Britton GJ, Uzzan M, Chen C-L, Kornbluth A, George J, Legnani P, Maser E, Loudon H, Stone J, Dubinsky M, Faith JJ, Clemente JC, Mehandru S, Colombel J-F, Peter I. 2020. Infants born to mothers with IBD present with altered gut microbiome that transfers abnormalities of the adaptive immune system to germ-free mice. Gut 69:42–51. doi: 10.1136/gutjnl-2018-317855. [DOI] [PubMed] [Google Scholar]
- 41.Heidarian F, Alebouyeh M, Shahrokh S, Balaii H, Zali MR. 2019. Altered fecal bacterial composition correlates with disease activity in inflammatory bowel disease and the extent of IL8 induction. Curr Res Transl Med 67:41–50. doi: 10.1016/j.retram.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 42.Holm JB, Sorobetea D, Kiilerich P, Ramayo-Caldas Y, Estellé J, Ma T, Madsen L, Kristiansen K, Svensson-Frej M. 2015. Chronic Trichuris muris infection decreases diversity of the intestinal microbiota and concomitantly increases the abundance of lactobacilli. PLoS One 10:e0125495. doi: 10.1371/journal.pone.0125495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Houlden A, Hayes KS, Bancroft AJ, Worthington JJ, Wang P, Grencis RK, Roberts IS. 2015. Chronic Trichuris muris infection in C57BL/6 mice causes significant changes in host microbiota and metabolome: effects reversed by pathogen clearance. PLoS One 10:e0125945. doi: 10.1371/journal.pone.0125945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cattadori IM, Sebastian A, Hao H, Katani R, Albert I, Eilertson KE, Kapur V, Pathak A, Mitchell S. 2016. Impact of helminth infections and nutritional constraints on the small intestine microbiota. PLoS One 11:e0159770. doi: 10.1371/journal.pone.0159770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee SC, Tang MS, Easton AV, Devlin JC, Chua LL, Cho I, Moy FM, Khang TF, Lim YAL, Loke P. 2019. Linking the effects of helminth infection, diet and the gut microbiota with human whole-blood signatures. PLoS Pathog 15:e1008066. doi: 10.1371/journal.ppat.1008066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toro-Londono MA, Bedoya-Urrego K, Garcia-Montoya GM, Galvan-Diaz AL, Alzate JF. 2019. Intestinal parasitic infection alters bacterial gut microbiota in children. PeerJ 7:e6200. doi: 10.7717/peerj.6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cooper P, Walker AW, Reyes J, Chico M, Salter SJ, Vaca M, Parkhill J. 2013. Patent human infections with the whipworm, Trichuris trichiura, are not associated with alterations in the faecal microbiota. PLoS One 8:e76573. doi: 10.1371/journal.pone.0076573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneeberger PHH, Coulibaly JT, Panic G, Daubenberger C, Gueuning M, Frey JE, Keiser J. 2018. Investigations on the interplays between Schistosoma mansoni, praziquantel and the gut microbiome. Parasit Vectors 11:168. doi: 10.1186/s13071-018-2739-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee NN, Bidot WA, Ericsson AC, Franklin CL. 2020. Effects of Giardia lamblia colonization and fenbendazole treatment on canine fecal microbiota. J Am Assoc Lab Anim Sci 59:423–429. doi: 10.30802/AALAS-JAALAS-19-000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujishiro MA, Lidbury JA, Pilla R, Steiner JM, Lappin MR, Suchodolski JS. 2020. Evaluation of the effects of anthelmintic administration on the fecal microbiome of healthy dogs with and without subclinical Giardia spp. and Cryptosporidium canis infections. PLoS One 15:e0228145. doi: 10.1371/journal.pone.0228145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muhsen K, Levine MM. 2012. A systematic review and meta-analysis of the association between Giardia lamblia and endemic pediatric diarrhea in developing countries. Clin Infect Dis 55(Suppl 4):S271–S293. doi: 10.1093/cid/cis762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robertson ID, Thompson RC. 2002. Enteric parasitic zoonoses of domesticated dogs and cats. Microbes Infect 4:867–873. doi: 10.1016/s1286-4579(02)01607-6. [DOI] [PubMed] [Google Scholar]
- 53.Liu J, Kabir F, Manneh J, Lertsethtakarn P, Begum S, Gratz J, Becker SM, Operario DJ, Taniuchi M, Janaki L, Platts-Mills JA, Haverstick DM, Kabir M, Sobuz SU, Nakjarung K, Sakpaisal P, Silapong S, Bodhidatta L, Qureshi S, Kalam A, Saidi Q, Swai N, Mujaga B, Maro A, Kwambana B, Dione M, Antonio M, Kibiki G, Mason CJ, Haque R, Iqbal N, Zaidi AKM, Houpt ER. 2014. Development and assessment of molecular diagnostic tests for 15 enteropathogens causing childhood diarrhoea: a multicentre study. Lancet Infect Dis 14:716–724. doi: 10.1016/S1473-3099(14)70808-4. [DOI] [PubMed] [Google Scholar]
- 54.Breurec S, Vanel N, Bata P, Chartier L, Farra A, Favennec L, Franck T, Giles-Vernick T, Gody J-C, Luong Nguyen LB, Onambélé M, Rafaï C, Razakandrainibe R, Tondeur L, Tricou V, Sansonetti P, Vray M. 2016. Etiology and epidemiology of diarrhea in hospitalized children from low income country: a matched case-control study in Central African Republic. PLoS Negl Trop Dis 10:e0004283. doi: 10.1371/journal.pntd.0004283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Platts-Mills JA, Babji S, Bodhidatta L, Gratz J, Haque R, Havt A, McCormick BJ, McGrath M, Olortegui MP, Samie A, Shakoor S, Mondal D, Lima IF, Hariraju D, Rayamajhi BB, Qureshi S, Kabir F, Yori PP, Mufamadi B, Amour C, Carreon JD, Richard SA, Lang D, Bessong P, Mduma E, Ahmed T, Lima AA, Mason CJ, Zaidi AK, Bhutta ZA, Kosek M, Guerrant RL, Gottlieb M, Miller M, Kang G, Houpt ER, MAL-ED Network Investigators. 2015. Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob Health 3:e564–e575. doi: 10.1016/S2214-109X(15)00151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bartelt LA, Platts-Mills JA. 2016. Giardia: a pathogen or commensal for children in high-prevalence settings? Curr Opin Infect Dis 29:502–507. doi: 10.1097/QCO.0000000000000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tysnes KR, Skancke E, Robertson LJ. 2014. Subclinical Giardia in dogs: a veterinary conundrum relevant to human infection. Trends Parasitol 30:520–527. doi: 10.1016/j.pt.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 58.Covacin C, Aucoin DP, Elliot A, Thompson RCA. 2011. Genotypic characterisation of Giardia from domestic dogs in the USA. Vet Parasitol 177:28–32. doi: 10.1016/j.vetpar.2010.11.029. [DOI] [PubMed] [Google Scholar]
- 59.Muhsen K, Cohen D, Levine MM. 2014. Can Giardia lamblia infection lower the risk of acute diarrhea among preschool children? J Trop Pediatr 60:99–103. doi: 10.1093/tropej/fmt085. [DOI] [PubMed] [Google Scholar]
- 60.Veenemans J, Mank T, Ottenhof M, Baidjoe A, Mbugi EV, Demir AY, Wielders JPM, Savelkoul HFJ, Verhoef H. 2011. Protection against diarrhea associated with Giardia intestinalis Is lost with multi-nutrient supplementation: a study in Tanzanian children. PLoS Negl Trop Dis 5:e1158. doi: 10.1371/journal.pntd.0001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roxström-Lindquist K, Palm D, Reiner D, Ringqvist E, Svärd SG. 2006. Giardia immunity–an update. Trends Parasitol 22:26–31. doi: 10.1016/j.pt.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 62.Manko-Prykhoda A, Allain T, Motta J-P, Cotton JA, Feener T, Oyeyemi A, Bindra S, Vallance BA, Wallace JL, Beck P, Buret AG. 2020. Giardia spp. promote the production of antimicrobial peptides and attenuate disease severity induced by attaching and effacing enteropathogens via the induction of the NLRP3 inflammasome. Int J Parasitol 50:263–275. doi: 10.1016/j.ijpara.2019.12.011. [DOI] [PubMed] [Google Scholar]
- 63.Mejia R, Damania A, Jeun R, Bryan PE, Vargas P, Juarez M, Cajal PS, Nasser J, Krolewiecki A, Lefoulon E, Long C, Drake E, Cimino RO, Slatko B. 2020. Impact of intestinal parasites on microbiota and cobalamin gene sequences: a pilot study. Parasit Vectors 13:200. doi: 10.1186/s13071-020-04073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nieves-Ramírez ME, Partida-Rodríguez O, Laforest-Lapointe I, Reynolds LA, Brown EM, Valdez-Salazar A, Morán-Silva P, Rojas-Velázquez L, Morien E, Parfrey LW, Jin M, Walter J, Torres J, Arrieta MC, Ximénez-García C, Finlay BB. 2018. Asymptomatic intestinal colonization with protist blastocystis is strongly associated with distinct microbiome ecological patterns. mSystems 3:e00007-18. [CrossRef] doi: 10.1128/mSystems.00007-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shaulov Y, Shimokawa C, Trebicz-Geffen M, Nagaraja S, Methling K, Lalk M, Weiss-Cerem L, Lamm AT, Hisaeda H, Ankri S. 2018. Escherichia coli mediated resistance of Entamoeba histolytica to oxidative stress is triggered by oxaloacetate. PLoS Pathog 14:e1007295. doi: 10.1371/journal.ppat.1007295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hayes KS, Bancroft AJ, Goldrick M, Portsmouth C, Roberts IS, Grencis RK. 2010. Exploitation of the intestinal microflora by the parasitic nematode Trichuris muris. Science 328:1391–1394. doi: 10.1126/science.1187703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singer SM, Nash TE. 2000. The role of normal flora in Giardia lamblia infections in mice. J Infect Dis 181:1510–1512. doi: 10.1086/315409. [DOI] [PubMed] [Google Scholar]
- 68.Farthing MJG. 1993. Pathogenesis of giardiasis. Trans R Soc Trop Med Hyg 87:17–21. doi: 10.1016/0035-9203(93)90531-T. [DOI] [PubMed] [Google Scholar]
- 69.Hartong WA, Gourley WK, Arvanitakis C. 1979. Giardiasis: clinical spectrum and functional-structural abnormalities of the small intestinal mucosa. Gastroenterology 77:61–69. doi: 10.1016/S0016-5085(79)80011-6. [DOI] [PubMed] [Google Scholar]
- 70.Khanna R, Vinayak VK, Mehta S, Kumkum, Nain CK. 1988. Giardia lamblia infection in immunosuppressed animals causes severe alterations to brush border membrane enzymes. Dig Dis Sci 33:1147–1152. doi: 10.1007/BF01535792. [DOI] [PubMed] [Google Scholar]
- 71.Beatty JK, Akierman SV, Motta J-P, Muise S, Workentine ML, Harrison JJ, Bhargava A, Beck PL, Rioux KP, McKnight GW, Wallace JL, Buret AG. 2017. Giardia duodenalis induces pathogenic dysbiosis of human intestinal microbiota biofilms. Int J Parasitol 47:311–326. doi: 10.1016/j.ijpara.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 72.Buret AG, Motta J-P, Allain T, Ferraz J, Wallace JL. 2019. Pathobiont release from dysbiotic gut microbiota biofilms in intestinal inflammatory diseases: a role for iron? J Biomed Sci 26:1. doi: 10.1186/s12929-018-0495-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Soriano-Arandes A, García-Carrasco E, Serre-Delcor N, Treviño-Maruri B, Sulleiro E, Ruiz-Giardín JM, Sanmartín JV, Torrús D, Rojo-Marcos G, Cuadros J, Martín-Echevarría E, López-Vélez R, Molina I, Pérez-Molina JA, Redivi Study Group. 2016. Travelers’ diarrhea in children at risk: an observational study from a Spanish database. Pediatr Infect Dis J 35:392–395. doi: 10.1097/INF.0000000000001049. [DOI] [PubMed] [Google Scholar]
- 74.Cacciò SM, Ryan U. 2008. Molecular epidemiology of giardiasis. Mol Biochem Parasitol 160:75–80. doi: 10.1016/j.molbiopara.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 75.Haque R, Roy S, Kabir M, Stroup SE, Mondal D, Houpt ER. 2005. Giardia assemblage A infection and diarrhea in Bangladesh. J Infect Dis 192:2171–2173. doi: 10.1086/498169. [DOI] [PubMed] [Google Scholar]
- 76.Eckmann L. 2003. Mucosal defences against Giardia. Parasite Immunol 25:259–270. doi: 10.1046/j.1365-3024.2003.00634.x. [DOI] [PubMed] [Google Scholar]
- 77.Homan WL, Mank TG. 2001. Human giardiasis: genotype linked differences in clinical symptomatology. Int J Parasitol 31:822–826. doi: 10.1016/s0020-7519(01)00183-7. [DOI] [PubMed] [Google Scholar]
- 78.Andersson T, Forssell J, Sterner G. 1972. Outbreak of giardiasis: effect of a new antiflagellate drug, tinidazole. Br Med J 2:449–451. doi: 10.1136/bmj.2.5811.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Donowitz JR, Alam M, Kabir M, Ma JZ, Nazib F, Platts-Mills JA, Bartelt LA, Haque R, Petri WA. 2016. A prospective longitudinal cohort to investigate the effects of early life giardiasis on growth and all cause diarrhea. Clin Infect Dis 63:792–797. doi: 10.1093/cid/ciw391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lehto K-M, Fan Y-M, Oikarinen S, Nurminen N, Hallamaa L, Juuti R, Mangani C, Maleta K, Hyöty H, Ashorn P. 2019. Presence of Giardia lamblia in stools of six- to 18-month old asymptomatic Malawians is associated with children’s growth failure. Acta Paediatr 108:1833–1840. doi: 10.1111/apa.14832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, Nikkïla J, Monti D, Satokari R, Franceschi C, Brigidi P, De Vos W. 2010. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One 5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shulzhenko N, Morgun A, Hsiao W, Battle M, Yao M, Gavrilova O, Orandle M, Mayer L, Macpherson AJ, McCoy KD, Fraser-Liggett C, Matzinger P. 2011. Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat Med 17:1585–1593. doi: 10.1038/nm.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kolling G, Wu M, Guerrant RL. 2012. Enteric pathogens through life stages. Front Cell Infect Microbiol 2:114. doi: 10.3389/fcimb.2012.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sahagún J, Clavel A, Goñi P, Seral C, Llorente MT, Castillo FJ, Capilla S, Arias A, Gómez-Lus R. 2008. Correlation between the presence of symptoms and the Giardia duodenalis genotype. Eur J Clin Microbiol Infect Dis 27:81–83. doi: 10.1007/s10096-007-0404-3. [DOI] [PubMed] [Google Scholar]
- 85.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2 — approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lozupone CA, Hamady M, Kelley ST, Knight R. 2007. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sorensen T. 1948. A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of vegetation on Danish Commons. Kongelige Danske Videnskabernes Selskab, Biologiske Skrifter Bind 5, Nr. 4. [Google Scholar]
- 92.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B (Methodological) 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 93.Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Caporaso JG. 2018. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2019. vegan: community ecology package. Computer software, R. [Google Scholar]
- 96.R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 97.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wickham H. 2016. ggplot2 − elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 99.Pedersen TL. 2017. patchwork: the composer of ggplots. Computer software, R. [Google Scholar]
- 100.Arnold JB. 2019. ggthemes: extra themes, scales and geoms for “ggplot2.” Computer software, R. [Google Scholar]
- 101.De Cáceres M, Legendre P. 2009. Associations between species and groups of sites: indices and statistical inference. Ecology 90:3566–3574. doi: 10.1890/08-1823.1. [DOI] [PubMed] [Google Scholar]
- 102.Oliveira FS, Brestelli J, Cade S, Zheng J, Iodice J, Fischer S, Aurrecoechea C, Kissinger JC, Brunk BP, Stoeckert CJ, Fernandes GR, Roos DS, Beiting DP. 2018. MicrobiomeDB: a systems biology platform for integrating, mining and analyzing microbiome experiments. Nucleic Acids Res 46:D684–D691. doi: 10.1093/nar/gkx1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glöckner FO. 2014. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res 42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hansen MEB, Rubel MA, Bailey AG, Ranciaro A, Thompson SR, Campbell MC, Beggs W, Dave JR, Mokone GG, Mpoloka SW, Nyambo T, Abnet C, Chanock SJ, Bushman FD, Tishkoff SA. 2019. Population structure of human gut bacteria in a diverse cohort from rural Tanzania and Botswana. Genome Biol 20:16. doi: 10.1186/s13059-018-1616-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differentially abundant taxa associated with Giardia infection compared to NPS controls, as identified by point-biserial correlation coefficient. Point-biserial correlation coefficients showing that Clostridium, Lactobacillus, and Bacteroides are significantly associated with Giardia infection, as seen in the LEfSe analysis (Fig. 2). Download Table S1, PDF file, 0.04 MB (39.8KB, pdf) .
Copyright © 2020 Berry et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Diarrhea is associated with enrichment of a different set of taxa compared to Giardia. (A) Pie charts showing the relative proportion of dogs with diarrhea and asymptomatic animals among NPS control and Giardia-infected dogs. The proportion of symptomatic dogs is not significantly different between groups (P > 0.05 by chi-squared test). (B) Histogram showing that diarrhea and antibiotic use are associated with a significant difference in beta diversity compared to asymptomatic and no antibiotic use. Bar height reflects the percentage of total beta diversity variance that is explained by each variable. Age, sex, and spay/neuter status were controlled for all calculations. Asterisks denote bars with adjusted P < 0.05. (C) LEfSe graph showing the magnitude of enrichment for each taxon with a LDA score of >2 comparing animals with and without diarrhea. (D) Boxplots showing the relative abundance of taxa that are differentially abundant between animals with and without diarrhea. Download FIG S1, PDF file, 0.1 MB (126.1KB, pdf) .
Copyright © 2020 Berry et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Antibiotic use in dogs is associated with a similar gut microbiota profile as dogs with diarrhea due to strong correlation between antibiotic use and diarrhea. (A) Pie chart showing the relative proportion of dogs receiving antibiotics and those not among all samples that have associated clinical data (n = 174), NPS controls with clinical data (n = 118), and Giardia-positive dogs with clinical data (n = 15). (B) LEfSe graph showing the magnitude of enrichment for each taxon with LDA score > 2 comparing NPS control dogs receiving and not receiving antibiotics. Escherichia is highly enriched in dogs receiving antibiotics and in dogs with diarrhea, while Bacteroides and Fusobacterium are reduced in dogs receiving antibiotics and in dogs with diarrhea, likely because most dogs receiving antibiotics have diarrhea. Megamonas and Faecalibacterium are reduced in dogs receiving antibiotics, but not in dogs with diarrhea. Download FIG S2, PDF file, 0.07 MB (70.6KB, pdf) .
Copyright © 2020 Berry et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The effects of Giardia in canines persist even when one or more other parasites are present. Fecal samples from dogs infected with multiple parasites, one of which is Giardia, are not different from those singly infected with Giardia in terms of Bray-Curtis or weighted UniFrac beta diversity (Adj P > 0.1). In contrast, fecal samples from dogs infected with multiple, non-Giardia parasites are different from those singly infected with Giardia (P < 0.05; adjusted P < 0.1), and infection status here represents a larger percentage of the variance in beta diversity. Age, sex, and spay/neuter status were controlled for all calculations. Download FIG S3, PDF file, 0.05 MB (48.1KB, pdf) .
Copyright © 2020 Berry et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Giardia infection is associated with significant changes in gut microbiome composition in each of the four countries sampled. (A) Bars depict the percent variation in Bray-Curtis beta diversity explained by Giardia infection in each of the four countries. Each analysis was stratified by age and MSD status. Giardia is significantly associated with a change in gut microbiota in all four countries; however, the amount of variation explained by Giardia infection varies across countries. (B) LEfSe graph showing the magnitude of enrichment for all taxa meeting the adjusted P value threshold of 0.1, comparing children with and without Giardia infection. Only taxa with relative abundance of >1% across all samples were used in the analysis. Asterisks denote the significance threshold for each taxon in each country. Download FIG S4, PDF file, 0.09 MB (97.8KB, pdf) .
Copyright © 2020 Berry et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Prevotella abundance, Gammaproteobacteria abundance, and Giardia prevalence are correlated with age in young children. (A and B) Boxplots showing that, over the first 5 years of life, the relative abundance of the Prevotella genus increases with age (A) and the relative abundance of the Gammaproteobacteria class decreases with age (B). (C and D) Bar graphs showing that the proportion of children infected with Giardia is low for children in the first year of life compared to 1- to 5-year-old children. Download FIG S5, PDF file, 0.1 MB (106.1KB, pdf) .
Copyright © 2020 Berry et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Giardia is associated with a reduction in Gammaproteobacteria and enrichment of Prevotella among 12- to 17-month-old children. (A) LEfSe graph showing the magnitude of enrichment for each taxon with LDA score > 2 comparing 12- to 17-month-old children with (n = 51) and without (n = 124) Giardia infection. (B) Boxplots showing the differences in relative abundance in taxa associated with Giardia infection among all 12- to 17-month-old participants. (C and D) Boxplots showing that Giardia is associated with a reduction in the Gammaproteobacteria class among 12- to 17-month-old children with MSD (C) but that Giardia is not significantly associated with Gammaproteobacteria among 12 to 17-month-old children without MSD (D), when the relative abundance of Gammaproteobacteria is low. Download FIG S6, PDF file, 0.1 MB (114.1KB, pdf) .
Copyright © 2020 Berry et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All sequencing data analyzed here are publicly available on the Sequence Read Archive (SRA) under study accession number PRJNA594732. All sequencing data used for the canine analyses is also publicly available on MIcrobiomeDB.org as part of the CAMP study. All code is available and reproducible on CodeOcean (https://codeocean.com/capsule/2815529/tree).