Abstract
The MRE11-RAD50-NBS1 (MRN) protein complex is one of the primary vehicles for repairing DNA double strand breaks and maintaining the genomic stability within the cell. The role of the MRN complex to recognize and process DNA double-strand breaks as well as signal other damage response factors is critical for maintaining proper cellular function. Mutations in any one of the components of the MRN complex that effect function or expression of the repair machinery could be detrimental to the cell and may initiate and/or propagate disease. Here, we discuss, in a structural and biochemical context, mutations in each of the three MRN components that have been associated with diseases such as ataxia telangiectasia-like disorder (ATLD), Nijmegen breakage syndrome (NBS), NBS-like disorder (NBSLD) and certain types of cancers. Overall, deepening our understanding of disease-causing mutations of the MRN complex at the structural and biochemical level is foundational to the future aim of treating diseases associated with these aberrations.
Keywords: MRE11-RAD50-NBS1, DNA double-strand break repair, ATLD, NBS, cancer mutations
1. Introduction
Genomic DNA within a cell experiences hundreds of spontaneous damaging events per day. The various forms of DNA damage include DNA base lesions (generated by hydrolytic reactions and non-enzymatic methylations), DNA-DNA crosslinks (both inter- and intra-strand), DNA-protein crosslinks, and single- and double-strand breaks (DSBs). Cells rely on a number of specific, multilayered systems to detect and repair these DNA lesions. These are collectively called the DNA damage response (DDR) and are composed of ~200 proteins that either act directly in repair or have a supporting role. When damage to DNA is not repaired due to a failure of the DDR, cell cycle arrest and/or apoptosis will occur in the best-case scenario; otherwise, mutations and genomic instability can arise. In addition to causing discontinuity in the genetic code, the damaged DNA is subject to further chemical and physical damage that can result in increased susceptibility to cancer, neurological defects, and other diseases. Indeed, mutation of DDR genes resulting in altered protein expression and/or function have been associated with cancer and other diseases [1,2,3,4,5].
The MRE11-RAD50-NBS1/Xrs2 (MRN in humans or MRX in yeast) protein complex is one of the first responders to DNA DSBs and is the primary complex to recognize, signal, and assist in the repair of these lesions [6,7,8]. Knocking out MRE11, RAD50, or NBS1 leads to embryotic lethality in mice [9,10,11]. Although MRN is essential for cell survival, mutations and/or changes in protein levels have been reported in several diseases. For example, hypomorphic germline mutations of the MRN genes give rise to several rare diseases: ataxia telangiectasia-like disorder (ATLD), Nijmegen breakage syndrome (NBS), and NBS-like disorder (NBSLD) [12,13,14]. Moreover, somatic MRN mutations have been found in various cancer types, and the association between mutations of the genes for the MRN complex and cancer susceptibility have been observed in ovarian and breast cancers and glioma [15,16,17,18,19,20,21,22]. However, there appears to be some controversy here as a multigene panel-based clinical study for pathogenic variants in inherited oncogenes demonstrated that the genes of MRN complex do not confer any appreciable risks of breast cancer [23]. Yet, if mutations within the MRN genes are not a driver of oncogenesis, they seem to affect prognosis, outcome, and survival rates [24,25,26,27].
In this review, we will outline many of the currently known disease-related mutations in the MRN genes and describe the physical, biochemical, and structural information reported in the literature for each.
2. DNA DSBs and the Role of MRN in Repair
In DNA DSBs, the two complementary DNA strands are broken simultaneously making this the least frequent but most dangerous form of DNA damage. DNA DSBs can be generated by exogenous agents, such as ionizing or UV radiation, and genotoxic chemicals. The latter includes both base alkylating chemicals, like methyl methane sulfonate (MMS), and crosslinking agents [28]. DNA DSBs are also a regular cellular event resulting from reactive oxygen species produced during normal cellular metabolism as well as from DNA replication, meiotic recombination, and programmed rearrangements of the immunoglobulin and T cell receptor loci during lymphoid cell development. Collapsed replication forks that occur when DNA replication encounters a single-strand DNA (ssDNA) break also resemble DNA DSBs [29,30].
Given the highly cytotoxic nature of DNA DSBs, the cell has evolved several strategies for repairing these lesions with non-homologous end joining (NHEJ) and homologous recombination (HR) serving as the two main repair pathways [31,32]. The MRN/X complex is an essential player in both of these repair pathways. NHEJ is an error-prone repair mechanism that is active throughout the cell cycle and is initiated by heterodimeric Ku70-Ku80 binding to and protecting the free DNA ends [32]. Ku70-Ku80 then activates DNA-PKcs and recruits the DNA ligase IV complex, which ligates the two broken ends. Although the precise role of the MRN/X complex in NHEJ is unknown, in budding yeast MRX is essential for NHEJ where it is thought that the complex tethers the DNA ends and stimulates DNA ligase IV activity [33,34,35]. In mammalian cells, the MRN complex plays a still undefined supporting role [36,37]. Unlike NHEJ, the HR pathway is a relatively error-free solution for repairing DNA DSBs that is only active during the S/G2 phases of the cell cycle, when a sister chromatid is available to act as the repair template. HR is also used to fix stalled replication forks, which occur when replication aborts due to problems with the genomic DNA template (e.g., ssDNA breaks or the presence of protein-DNA covalent adducts). Once MRN/X identifies a DNA DSB, it initiates the process of repair by generating an endonucleolytic cut [38,39], in conjunction with CtIP (Sae2 in yeast) [40], which acts as an entry point for EXO1 and DNA2 nucleases to generate the 3′-overhangs necessary for HR [39]. Additionally, the MRN/X complex activates ATM protein kinase (Tel1 in yeast) [41,42], which phosphorylates NBS1 among other targets, setting off a signaling cascade alerting the cell to the presence of the break. The 3′-overhangs are the substrate for RAD51, and together the RAD51-ssDNA complex performs the homology search, leading to strand invasion and eventual DNA DSB repair.
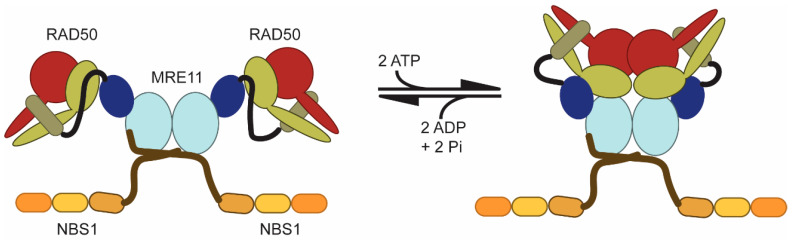
MRE11 and RAD50 constitute the universally conserved catalytic components of the complex, whereas NBS1/Xrs2 is only found in eukaryotes and acts as a flexible scaffold to recruit downstream effector proteins. Briefly, dimeric MRE11 is a nuclease that forms the core of the complex. One RAD50 and one NBS1 associate with each MRE11 protomer to form the complete MRN complex (i.e., M2R2N2; Figure 1) [43]. Generally, the functions of RAD50, an ATPase, alter the structural and functional landscape of the complex: ATP binding induces a “closed” conformation (Figure 1, right) and ATP hydrolysis allows return to the “open” conformation (Figure 1, left) [44,45,46]. When the “open” form is populated, MRE11 is nuclease active. On the other hand, in the ATP-bound “closed” form, MRE11 is inactive, but RAD50 can bind DNA which is important for telomere length maintenance [47]. MRN also activates the ATM kinase from the “closed” form [41,42]. Additionally, cycling between the two conformations is important for processive MRE11 nuclease activity [48]. It is unclear what the effect of ATP binding is on NBS1. A precise understanding of MRN function, and how disease-associated mutations within the complex alter that function, is supported heavily by structural biology studies. Yet, we currently have limited structural information about the eukaryotic MRN complex. The vast majority of structural information we do have for MRN is from X-ray crystallography structures [44,45,46,49,50,51,52,53,54,55] and a recent cryo-electron microscopy structure [56] of archaeal and bacterial MR homologs, although structures do exist of truncated forms of H. sapiens (PDB id: 3T1I) [57], S. pombe (4FCX) [58], and C. thermophilum (4YKE) [59] MRE11. Structures of ATP-bound “closed” RAD50 from C. thermophilum (5DA9) [60] as well as the N-terminal folded region of S. pombe NBS1 (3HUE and 3I0N) [61,62] and the C-terminal MRE11 binding regions of S. pombe NBS1 in complex with MRE11 (4FBW) [58] also exist. Fortunately, the high level of sequence similarity between homologs allows for detailed structure-function analysis of disease-associated mutants. In fact, many of the mutations described below for MRE11 and RAD50 are to highly conserved positions in the sequence and structure between archaea, bacteria, and eukarya, which underscores their importance for function.
Figure 1.
Global conformational changes in the MRN complex induced by ATP binding and hydrolysis. Cartoon representation of MRE11 (blue shades), RAD50 (red and light green), and NBS1 (orange shades). Each monomer in the MRE11 dimer is bound to a RAD50 and NBS1. ATP binding to RAD50 causes RAD50 monomers to associate creating a “closed” complex. Subsequent ATP hydrolysis and ADP + Pi release allows the complex to return to an “open” state.
3. Correlation of MRE11 with Disease
3.1. MRE11 in Brief
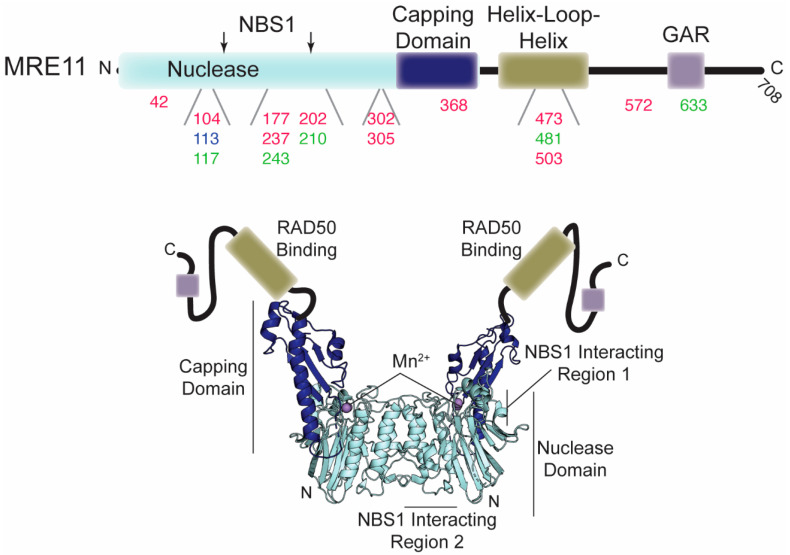
MRE11, a member of the calcineurin-like metallophosphoesterase superfamily of proteins [63], is a dimer. MRE11 has Mn2+-dependent 3′-to-5′ dsDNA exonuclease and ssDNA endonuclease activities that initiate resection of DNA ends [64,65,66]. Sequential endo- and exonuclease activities of MRE11 are important for removing protein adducts on the DNA ends [67,68,69]. Each protomer is comprised of nuclease, capping, helix-loop-helix, and glycine/arginine-rich (GAR) domains (Figure 2). The nuclease domain of MRE11 octahedrally coordinates two catalytically required Mn2+ ions via seven conserved residues in the phosphodiesterase motifs, facilitates MRE11 dimerization via a four-helical bundle, and contains binding sites for NBS1 [50,58]. The capping domain of MRE11 may be responsible for DNA unwinding, as structural studies have shown that a helical wedge inserted into the DNA minor groove causes it to rotate upon substrate DNA binding [52]. The helix-loop-helix motif is responsible for binding to each RAD50 monomeric subunit [53]. Finally, methylation of the arginines in the metazoan specific C-terminal GAR domain has been shown to be important for MRE11 exonuclease activity and activation of the ATR kinase [70,71].
Figure 2.
Disease-associated mutations in MRE11. Top, domain architecture of MRE11. The numbers below indicate the position of mutations outlined in the text and in Table 1. Red, green, and blue numbers correspond to mutations associated with cancer, ATLD, and NBSLD, respectively. The two arrows above indicate the position of the NBS1 interacting regions. Bottom, crystal structure of the MRE11 nuclease and capping domain dimer from C. thermophilum (PDB ID: 4YKE). Since the helix-loop-helix and GAR domains are absent in this structure, they are cartooned in (not to scale).
3.2. Disease-Associated MRE11 Variants
Due to the various roles MRN has in maintaining the integrity of the genomic DNA, mutations affecting the nuclease activity, structure, or expression levels of MRE11 have been correlated with immunodeficiency, sensitivity to ionizing radiation (IR), and/or oncogenesis (Table 1). In fact, ATLD, an autosomal recessive disease characterized by higher sensitivity to ionizing radiation, developmental disorders, immunodeficiency, and neuronal and cerebellar degeneration, is often caused by mutation in MRE11 that leads to lower levels of the enzyme or an inability to interact with NBS1 (Figure 2 and see below). Additionally, sporadic mutations in MRE11 have been observed in different cancers (Figure 2), including colorectal, breast, and ovarian, that contain chromosomal instabilities (e.g., translocations and end-joining).
Table 1.
Disease-associated mutations in MRE11.
| Mutation in MRE11 | Disease/Cancer Type | Location in Gene or Structure | Effect on Structure/Function | Ref. |
|---|---|---|---|---|
| Δ5-7 MRE11 | colon cancer | deletion of exons 5-7; deletes the third and fourth phosphoesterase motifs | Loss of 3′-to-5′ exonuclease activity impairs HR. Weak interactions with RAD50 and NBS1. Suppressed ATM activation. | [76] |
| E42K, E42D, and E42A |
ovarian, uterine, and small cell lung cancers | nuclease domain | Inhibits Tel1/ATM activation. May disrupt an ionic interaction stabilizing the interface of the nuclease and capping domains. | [77] |
| S104C | Breast cancer | nuclease domain, NBS1 interaction region 2 | Reduced NBS1 binding. | [57,78] |
| D113G | NBSLD | nuclease domain, NBS1 interaction region 2 | May affect MRE11 dimer geometry/stability and NBS1 binding. | [58,79] |
| N117S | ATLD3/4 | nuclease domain, NBS1 interaction region 2 | May affect MRE11 dimer stability. Reduced NBS1 binding. | [12,80] |
| A177V | lung cancer | nuclease domain | Impairs DSB repair in yeast. Packing of this buried residue might be disrupted. | [77] |
| R202G | breast cancer | nuclease domain | Could affect the local stability around NBS1 binding region 1. Impaired NBS1 interaction. | [57,82] |
| W210C | ATLD7/8 | nuclease domain | Affects MRN protein expression resulting in dysfunctional MRN complex. | [58,83] |
| W243R | ATLD17/18 | nuclease domain | Impaired interactions with RAD50 and NBS1. Disrupts MRE11 dimerization. Causes defects in ATM activation. | [73,74,81] |
| F237C | breast cancer | nuclease domain | Unknown | [85] |
| H302Y | breast cancer | nuclease domain | Unknown | [85] |
| R305W | ovarian cancer | linker between nuclease and capping domains | Possibly affects structural organization of protein. | [57,86] |
| D368Y | lung and nasopharyn-geal cancers | capping domain | Strongly impairs DSB repair. Possibly disrupts the fold of MRE11. | [77] |
| L473F | colorectal cancers | helix-loop-helix | Impaired interaction with RAD50. | [84] |
| T481 Kcompound heterozyg-ous with R527Stop |
ATLD5/6 | helix-loop-helix | Decreased expression levels of MRN. Increased sensitivity to IR. Attenuated ATM activation. | [87] |
| R503H | breast carcinoma | helix-loop-helix | Unknown | [78] |
| R572Q | lymphoma | GAR motif | Disrupts interactions with RAD50 and NBS1. Defects in ATR activation. | [74,78] |
| R633Stop | ATLD1/2; breast cancer |
truncation near C-terminus | Does not localize to sites of damage. | [12,74,80,82] |
Splice variants—A number of mutations within exons and introns of MRE11 have been reported that lead to different splice variants of the protein. Recently, in a family with ATLD a MRE11 variant was described with a point mutation near the 3′ end of exon 7. Although the mutation did not change the amino acid, in silico analysis suggested that the mutation altered the splicing of exon 7 leading to a frameshift and a subsequent premature stop codon. RT-PCR and immunohistochemical analysis suggested that this frameshift led to degradation of the transcript by nonsense-mediated mRNA decay [72]. In the ATLD17/18 patients, a single nucleotide substitution in an intron near a splice site is compound heterozygous with W243R (see below) [73]. This splice site variant causes skipping of exon 10 and results in a MRE11 9-to-11 variant that maintains the open reading frame. The variant is predicted to encode MRE11 lacking 27 amino acids Δ(340–366) situated in the structurally important capping domain. In mice, even with wild-type expression levels, this deletion causes defects in ATM and ATR activation and the G2/M checkpoint and is unable to interact with NBS1 [74]. Another MRE11 splice variant was described in a screen of mismatch repair-deficient cancer cell lines. Here, heterozygous mutations that shortened a poly(T) tract significantly reduced the correct splicing process and led to the skipping of exon 5, a frameshift, and premature stop codon [75]. Again, these truncated MRE11 transcripts are likely degraded by nonsense-mediated mRNA decay leading to decreased levels of MRE11.
Δ5–7MRE11—This mutation was found in the mismatch repair deficient HCT116 colon cancer cell line that experiences replication fork stress and has an increased sensitivity to the DNA damaging drugs camptothecin (CPT) and thymidine [76]. This deletion in MRE11 results in a loss of exons 5–7, eliminating the third and fourth highly conserved phosphoesterase motifs (amino acids 106–220). Because of the loss of these conserved nuclease motifs, Δ5–7MRE11 lacks 3′-to-5′ exonuclease activity which impairs HR. Interestingly, Δ5–7MRE11 retains its DNA-binding activity and in fact showed increased binding to fork substrates with single-stranded regions. The accumulation of Δ5–7MRE11 at ssDNA of replication forks after thymidine treatment could inhibit HR by preventing DNA processing to proper intermediates for repair or triggering ATM autophosphorylation. In fact, Δ5–7MRE11 cells have suppressed ATM activation. Finally, in co-immunoprecipitation (co-IP) experiments Δ5–7MRE11 showed a weaker interaction with RAD50 and very little affinity for NBS1 as compared to wild type MRE11.
E42K, E42D, and E42A—E42K was documented in ovarian and small cell lung cancer, whereas E42D and E42A were noted in uterine endometrioid carcinoma [77]. In yeast, mutating this glutamate to a lysine resulted in a separation-of-function phenotype that only partially affected DNA DSB repair but inhibited Tel1/ATM activation. Based on structural overlays, glutamate 42 is in a long α-helix within the nuclease domain and points toward the capping domain. Thus, this mutation may disrupt an ionic interaction between the nuclease and capping domains destabilizing their interface.
S104C—This mutation was reported in the tumor tissue of a breast carcinoma patient [78]. The crystal structure of the human MRE11 core shows that serine 104 is in the MRE11-NBS1 interacting loop (NBS1 interaction region 2; Figure 2), and in fact, the S104C mutant has significantly reduced NBS1 binding activity in in vitro pull-down assays [57]. Serine 104 forms a hydrogen bond with the imidazole side chain of histidine 73 of the other MRE11, which could contribute to the stability of loop α2-β3, a part of NBS1 binding site.
D113G—A study of two unrelated patients reported that a compound heterozygous mutation of human MRE11 D113G is linked with NBSLD [79]. In all eukaryotic crystal structures of MRE11, aspartate 113, which is located in an eukaryotic insertion loop within the nuclease domain, forms a salt bridge with an arginine residue across the MRE11 dimer. Additionally, D113 is positioned within the NBS1 interaction region 2; thus, D113 may stabilize the dimer or the conformation of the insertion loop necessary for binding NBS1. It is hypothesized therefore that the observed disease phenotype may be due to perturbations in MRE11 dimer geometry, stability, and/or interaction with NBS1 [58].
N117S—This heterozygous allele of MRE11, with an asparagine in the nuclease domain substituted with a serine, was found in the ATLD3/4 patients and alters the interaction of MRE11 with NBS1 [12,76,80]. Although co-IP assays of MRE11 N117S with NBS1 from mammalian cells showed an impaired interaction, the mutation did not cause DNA DSB repair defects [12,80,81]. Notably, asparagine 117 is located in the same eukaryotic insertion loop as S104 and D113, and a hydrogen bond between the N117 residues of each protomer may bridge the MRE11 dimer [57]. It has been noted that D113G and N117S, which are four residues apart on the same loop, have different phenotypes. Yet, the importance of this region for MRE11 dimer stabilization and NBS1 binding and the effects of the mutations on these features may explain the different disease phenotypes [58].
A177V—This heterozygous mutation in a β-strand of the nuclease domain was found in lung adenocarcinoma. When modeled in yeast, A177V strongly impaired DSB repair [77]. In crystal structures of eukaryotic MRE11, this residue is buried within the core of the protein and is surrounded by two conserved aromatic residues. Therefore, substitution to a bulkier amino acid like valine might disrupt this packing.
R202G—This mutation in the loop β8-β9 of the nuclease domain was found in a breast cancer patient [82]. In the structure of human MRE11 core domain, arginine 202 forms an ion pair with glutamate 207, whereas the aliphatic region stacks between phenylalanine 237 and glutamate 207. The R202G mutation did not show any significant differences in nuclease function or thermal stability but might disrupt the local stability around the NBS1 binding region 1, as NBS1 did not successfully co-IP this mutant [57].
W210C—This mutation of a highly conserved tryptophan was discovered in the ATLD7/8 patients [83]. The initial study of fibroblast cell lines derived from these patients showed that this missense mutation leads to high levels of MRE11 and RAD50 expression but very low levels of NBS1 that ultimately results in a dysfunctional MRN complex. In S. pombe, this mutant was insensitive to DNA damaging agents, but showed reduced interaction with Nbs1 (but not Rad50) in Mre11 co-IPs [81]. This mutant is predicted to alter structural elements of the NBS1 interaction region 1 along the side of the MRE11 nuclease core (Figure 2) [58]. Additionally, a mutation that resulted in a stop codon at tryptophan 210 was found in a large sequencing study of colorectal cancers [84].
W243R—This mutation, which is compound heterozygous with the exon 10 deletion mutation described above, was reported in the ATLD17/18 patients who also developed lung adenocarcinomas with multiple bone metastases [73]. Tryptophan 243 is just upstream of nuclease motif V, but does not affect DNA binding or nuclease activity in vitro [81]. This mutant in S. pombe is acutely sensitive to DNA-damaging agents, on the level of Mre11 deletion, and co-IPs indicated impaired interactions with both Nbs1 and Rad50. Interestingly, telomere maintenance is unaffected giving this mutant a separation-of-function phenotype. Furthermore, in mice, this mutation is unable to interact with NBS1, disrupts MRE11 dimerization, and as a result causes defects in ATM activation [74]. Finally, structural studies of the analogous mutant in archaebacteria P. furiosus (L204R) suggest that allostery between the members of the MRN complex is disrupted [81].
F237C and H302Y—Both of these mutations within the nuclease domain are somatic missense mutations that were identified in patients with breast cancer [85]. Detailed biochemical and structural information about these mutants is still unknown.
R305W—This mutation discovered in a patient with ovarian cancer was reported in a study of 151 people with high familial risk for breast cancer and/or ovarian cancer [86]. An arginine at residue 305, which is in a linker between the nuclease and capping domains, is very highly conserved even in distantly related species. Arginine 305 forms an ionic interaction with an acidic residue in α-helix 1 of the nuclease domain and substitution to the neutral hydrophobic tryptophan would certainly affect the structural organization of that region [57].
D368Y—This heterozygous mutation in the capping domain was found in lung adenocarcinoma and nasopharyngeal carcinoma. When modeled in yeast, D368Y strongly impaired DSB repair [77]. This acidic residue is surface exposed near the end of the capping domain in crystal structures of eukaryotic MRE11. Therefore, substitution to a hydrophobic aromatic amino acid might disrupt the fold of MRE11 in this region.
L473F—This mutation was identified in a sequencing study of 100 genes associated with genomic instability in a panel of 192 colorectal cancers [84]. Leucine 473 is in the helix-loop-helix motif that is responsible in part for interactions with RAD50. Three other mutations (M523K, Q629K, and M675I) in less conserved regions of MRE11 were also found in the same study.
T481K—This compound heterozygous mutation with R527Stop was discovered in the ATLD5/6 siblings [87]. Expression levels of all three MRN proteins were significantly reduced in lymphoblastoid cell lines derived from these patients, which also showed an increased sensitivity to IR. ATM kinase activation was attenuated, as was the downstream signaling of some ATM targets. Threonine 481 is in the helix-loop-helix motif of MRE11 that interacts with RAD50.
R503H and R572Q—These missense mutations of conserved positions of the MRE11 protein were discovered in two patients with breast carcinoma (R503H) and lymphoma (R572Q) [78]. Arginine 503 is at the end of the helix-loop-helix motif. Arginine 572 is in the GAR motif. When the heterozygous R572Q mutation was studied in mice, the mutated MRE11 was able to interact with RAD50 and NBS1, and although the data suggested wild-type ATM activation and G2/M checkpoint signaling, there were defects in ATR activation [74].
R633Stop—This mutation, found in the ATLD1/2 patients (homozygous) and one breast cancer patient (heterozygous), causes a premature truncation of MRE11 76 amino acids from the end [12,82]. This truncated MRE11 can associate with both RAD50 and NBS1, albeit with less affinity as gauged by co-IP, but does not go to the site of DNA damage [12,80]. In mice, however, expression of the R633Stop mutant at wild-type levels does not show any defects in ATM or ATR activation. Furthermore, the mice mutant interacts with both RAD50 and NBS1 in co-IP and yeast two-hybrid assays [74].
4. Correlation of RAD50 with Disease
4.1. RAD50 in Brief
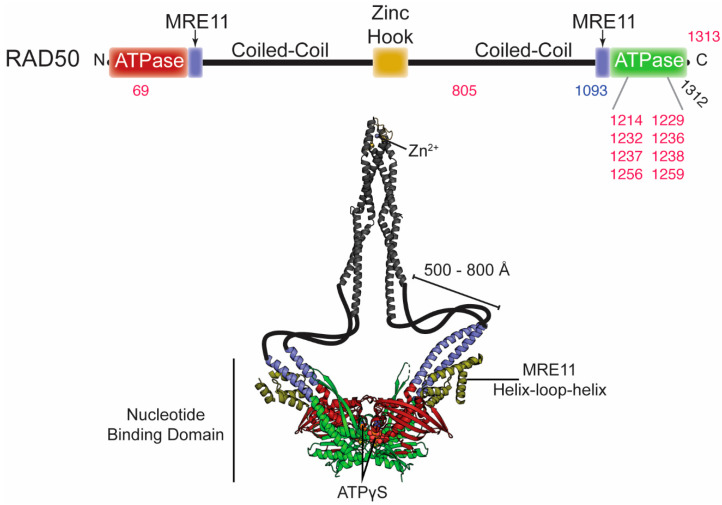
RAD50 is a member of the ATP-binding cassette (ABC) ATPase superfamily of proteins. The C-terminal sub-domain of each RAD50 monomer folds back onto its own N-terminal sub-domain to form the complete ABC ATPase nucleotide binding domain (NBD; Figure 1 and Figure 3) [49]. The two sub-domains are separated by a ~500–800 Å anti-parallel coiled-coil and an apical zinc hook domain that extend away from the NBD, making RAD50 a member of the structural maintenance of chromosomes (SMC) protein family [88]. Two RAD50 protomers dimerize via their zinc hook domains, and although a precise role for RAD50 dimerization by the zinc hook and the coiled-coil domain is not known, both are required for DNA DSB repair [89,90]. Two RAD50 molecules associate with the MRE11 dimer through interactions between the MRE11 helix-loop-helix motif and the base of the RAD50 coiled-coils (Figure 3). The MR head domain, formed by the MRE11 dimer and the NBD of each RAD50, plays a crucial role in DNA damage recognition and resection. ATP binding by RAD50 functions as a molecular switch, causing the global conformational change described above where the two RAD50 NBDs interact to form a “closed” state of MR (Figure 1). The MR complex switches back to the “open” state following RAD50 ATP hydrolysis and subsequent dissociation of the NBDs. According to small angle X-ray scattering (SAXS) and luminescence resonance energy transfer (LRET) data, the “open” and “closed” structures of MR are in a dynamic equilibrium [44,91].
Figure 3.
Disease-associated mutations in RAD50. Top, domain architecture of RAD50. The numbers indicate the position of mutations outlined in the text and Table 2. Red and blue numbers correspond to mutations associated with cancer and NBSLD, respectively. The two arrows above indicate the positions where MRE11 binds. Bottom, crystal structures of the dimer RAD50 nucleotide binding domain in complex with the MRE11 helix-loop-helix domain from C. thermophilum (PDB ID: 5DA9) and the dimer zinc hook domain from H. sapiens (PDB ID: 5GOX). This is the ATPγS-bound “closed” conformation of Rad50. The majority of the coiled-coil domain is absent from these structures, so it is was cartooned in to connect the two structures and is not to scale.
4.2. Disease-Associated RAD50 Variants
Deletions, single point mutations, or altered expression of the RAD50 gene have been shown to cause NBSLD and an increased risk of certain types of cancer (Table 2). One patient has been described with NBSLD, which resulted from inherited compound heterozygous mutations in the RAD50 gene [14]. A correlation between germline mutations of RAD50 and breast cancer risk is still unclear. A RAD50 687delT mutation causes a frameshift to a stop codon, L234Stop, and was associated with increased breast cancer risk in a Finnish population, but this mutation was not seen in other Nordic populations [86,92]. On the other hand, Fan et al. found that although a group of breast cancer patients (n = 7657) with RAD50 germline mutations (and without BRACA1/2 mutations) had unfavorable survival compared to other patients, these mutations were not necessarily associated with an increased risk of breast cancer because the frequency of the mutations was similar to control patients [93]. And Thompson et al. found more RAD50 truncations and missense mutations in control patients than the familial breast cancer panel they screened [94].
Table 2.
Disease-associated RAD50 mutations.
| Mutation in RAD50 | Disease/Cancer Type | Location in the Structure | Effect on Structure/Function | Ref. |
|---|---|---|---|---|
| D69N, D69Y, and D69G | uterine, colorectal, bladder, and lung cancers; myelodysplastic syndrome | Walker A | Impaired Tel1/ATM activation and decreased ATP hydrolysis. Could affect “open” to “closed” global transition dynamics. | [77] |
| R850C | endometrioid, breast/ovarian cancers | coiled coil region | Unknown | [95,96] |
| R1093Stop compound heterozyg-ous with Stop1313Y | NBSLD | truncated at C-terminal sub-domain | RAD50 deficiency. Increased radiosensitivity, chromosomal instability, impaired activation of ATM, impaired G1/S cellcycle-checkpoint activation. | [14] |
| R1214H and R1214C | pancreatic cancer | extended signature helix/basic switch | Impaired ATP hydrolysis activity, NBD association, and allostery within MRN. | [77,97,98,99] |
| A1229D | Burkitt Lymphoma | Walker B | Unknown | [100] |
| E1232K | lung cancer | Walker B | Defective in ATP hydrolysis. | [47,77] |
| N1236D | Burkitt Lymphoma | D-loop | Unknown | [100] |
| L1237F and L1237V | bladder and colorectal cancer; breast cancer | D-loop | Increased ATP hydrolysis. Destabilization of “closed” MRN complex. Loss of ATM signaling and Mre11 exonuclease activity. | [77,91,101] |
| D1238N | breast cancer | D-loop | Severe DSB repair defect. Increased ATP hydrolysis. Destabilization of “closed” MRN complex. Loss of ATM signaling and Mre11 exonuclease activity. | [91,101] |
| R1256C and R1256H | bladder/uterine cancers | between D-loop and His-loop | Impaired Tel1/ATM activation. | [77] |
| Q1259K | endometrial carcinoma | between D-loop and His-loop | Unknown | [101] |
| Q1263H | endometrial carcinoma | between D-loop and His-loop | Unknown | [95,96] |
D69N, D69Y, and D69G—This aspartate is part of the Walker A motif that binds ATP. D69N is associated with uterine endometrioid carcinoma, colorectal adenocarcinoma, bladder and urothelial carcinoma, and myelodysplastic syndrome; D69Y is reported in lung adenocarcinoma and acinic cell carcinoma; and D69G is associated with lung adenocarcinoma [77]. In S. cerevisiae cells, D67N and D67Y did not show sensitivity to clastogens and could repair DNA DSBs but had impaired Tel1/ATM activation. In vitro assays with purified yeast Rad50 D67N/Y showed near wild-type DNA binding, but a decrease in ATP hydrolysis. Molecular dynamics suggest that mutation of aspartate 69 may affect ADP release after hydrolysis, perhaps altering the “open” to “closed” dynamics of the MRN complex [77].
R850C—RAD50 genes were also found to be frequently mutated in endometrioid carcinoma and tended to co-occur with mutations in KMT2D and SETD1B, proteins involved in chromatin remodeling through histone methylation [95]. This particular study identified missense mutations R850C and Q1263H, as well as several splice region variants. R850C was also found in a screen of hereditary breast and ovarian cancer patients [96]. This conserved arginine is located in the coiled coil region of RAD50.
R1093Stop—This mutation, compound heterozygous with Stop1313Y, causes a stop codon at the C-terminus of the coiled-coil region, resulting in a truncated protein where the C-terminal sub-domain of the NBD is not translated. These mutations led to a RAD50 deficiency with cells from the patient showing increased radiosensitivity, chromosomal instability, impaired radiation-induced activation of and downstream signaling through the ATM protein, and impaired G1/S cell cycle-checkpoint activation [14]. This patient was the first identified NBSLD case where NBS-like symptoms were caused by mutation in RAD50 instead of NBS1.
R1214H and R1214C—This arginine residue is in the extended signature helix, also known as the basic switch, and was identified as important for MR function based on a crystal structure of P. furiosus Rad50 [53]. A substitution to a histidine was subsequently identified in a patient with pancreatic cancer [97]. R1214C and R1214H mutations were also identified in cancer genomic data from the clinical sequencing program at Memorial Sloan Kettering Cancer Center, and R1214C conferred defects in DSB repair when modeled in yeast [77]. In in vitro studies, arginine 1214 was observed to be important for Rad50 ATP hydrolysis, NBD association, and allostery between the proteins of the core MR complex [98,99].
E1232K—Mutation of the absolutely conserved glutamate in the Walker B domain has been used in mechanistic studies in yeast and in in vitro structural studies of T. maritima Rad50 as this residue is essential for ATP hydrolysis [44,47]. The E1232K mutation was later found in lung cancer tumors [77]. Yeast harboring this mutation die in response to DNA damaging drugs, and although defective in ATP hydrolysis, the mutant can bind to DNA in vitro [47]. A somatic mutation of the conserved alanine three positions preceding this (A1229D) was found from sequencing endemic Burkitt Lymphoma tumor biopsies [100].
L1237F and D1238N—These two mutations occur to adjacent residues in the universally conserved D-loop motif. L1237F is associated with urothelial bladder and colorectal tumors, whereas D1238N is associated with breast cancer [101]. In yeast, the analogous D1238N mutant could not survive on CPT indicating a severe DSB repair defect, while L1237F had only a negligible effect on repair. In vitro, these mutants showed increased Rad50 ATP hydrolysis activity not from faster association of the ATP-bound NBDs but faster dissociation, making the “closed” complex less stable [91]. Accordingly, both mutations led to the loss of ATM signaling and Mre11 exonuclease activity in mutant MR complexes in yeast [77,101]. A L1237V variant was found in breast invasive lobular carcinoma [77]. Sequencing of endemic Burkitt Lymphoma tumor biopsies also identified a somatic N1236D mutation immediately upstream of the D-loop [100].
R1256C and R1256H—These mutations of a conserved arginine in the helix between the D-loop and the His-loop were also identified from mining the cancer genomic data from the clinical sequencing program at Memorial Sloan Kettering Cancer Center. They were found in bladder/uterine cancers. R1259C yeast have impaired Tel1 activation but only partially affect DSB repair [77].
Q1259K—This mutation of a conserved glutamine on the same helix between the D-loop and His-loop as arginine 1256 was identified in endometrial carcinoma tumors. In yeast, this mutant shows negligible CPT sensitivity similar to that of the L1237F D-loop mutant [101].
5. Correlations of NBS1 with Disease
5.1. NBS1 in Brief
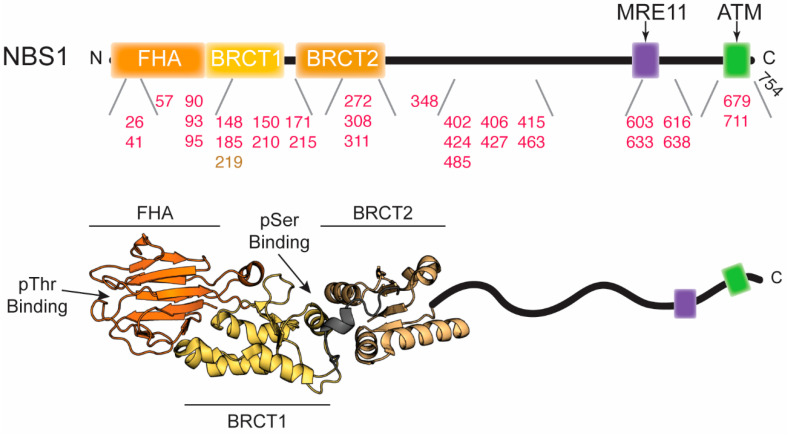
NBS1, also called Nibrin in humans or Xrs2 in budding yeast, is encoded by the NBN gene. It is generally thought of as a scaffolding protein employed in directing and associating with other DNA damage response factors at the site of the DNA DSB. NBS1 interacts with phosphorylated histone variant H2AX (γ-H2AX) near the site of DNA damage and translocates MRE11/RAD50 into the nucleus where the MRN complex senses the breaks and activates ATM [102]. NBS1 is phosphorylated by ATM and this activates downstream proteins like p53, BRCA1, and CHK2 to assist in repair and control cell cycle progression [103]. Structural studies have shown that NBS1 contains an ordered N-terminal region comprised of a fork-head associated (FHA) domain [104,105] followed by two tandem breast cancer associated 1 C-terminus (BRCT) domains [106,107] (Figure 4). This N-terminal region interacts with phosphorylated proteins such as CtlP/Sae2, MDC1, MDM2, and several others. NBS1 also contains an intrinsically disordered C-terminal region spanning amino acids 334 to 754 which has yet to be structurally characterized in its entirety but is known to contain the RAD18, MRE11, RNF20, and ATM interaction motifs, phosphorylation sites, as well as nuclear localization signals (Figure 4).
Figure 4.
Disease-associated mutations in NBS1. Top, domain architecture of NBS1. The numbers indicate the position of mutations outlined in the text and in Table 3. Red and gold numbers correspond to mutations associated with cancer and NBS, respectively. The two arrows above indicate the positions where MRE11 and ATM bind. Bottom, crystal structure of the S. pombe FHA and tandem BRCT domains (PDB ID: 3HUE). The intrinsically disordered C-terminus is cartooned in and is not to scale.
5.2. Disease-Associated NBS1 Variants
Deletions, single point mutations, or altered expression of the NBN gene have been shown to cause Nijmegen Breakage Syndrome (NBS), aplastic anemia, and an increased risk of certain types of cancer (Table 3). NBS is a rare autosomal recessive syndrome that presents with clinical features such as microcephaly, immunodeficiency, radiosensitivity, mental and growth retardation, and an increased risk for infections and cancer, particularly B-cell non-Hodgkin lymphoma [13]. Germline NBS1 mutations may lead to leukemia, malignant melanoma, prostate, breast, or ovarian cancer.
Table 3.
Disease-associated NBS 1 mutations.
| Mutation in NBS1 | Disease/Cancer Type | Location in the Structure | Effect on Structure/Function | Ref. |
|---|---|---|---|---|
| 657del5 | NBS; breast, prostate, and colorectal cancers; medulloblastoma; lymphoblastic leukemia; and non-Hodgkin lymphoma |
transition between BRCT1 and BRCT2 | Truncates NBS1 after BRCT1 and expresses a secondary C-terminal fragment starting near BRCT2. Disrupts tandem BRCT domains and proper phosphoprotein interaction. | [22,110] |
| V26I | medulloblastoma | FHA | Possibly disrupts phosphoprotein binding. | [62,116] |
| I41M | hepatocellular carcinoma | FHA | Possibly disrupts phosphoprotein binding | [61,117] |
| L57M/H711Y double mutation | medulloblastoma | FHA; C-terminus | L57M may disrupt the FHA/BRCT1 interface and destabilize the protein. H711Y may disrupt RNF20, MRE11, and/or ATM binding. |
[61,116] |
| T90S | intrahepatic cholangiocarcinoma | FHA | May disrupt FHA domain structure and/or phosphoprotein binding site. Decreased nuclear localization of MRE11. | [117] |
| S93L | acute lymphoblastic leukemia (ALL) | FHA | May disrupt FHA domain structure and/or phosphoprotein binding site. | [119] |
| D95N | ALL; breast, larynx, and prostate cancers | FHA | May disrupt FHA domain structure and/or phosphoprotein binding site. | [119,120,121,122] |
| T148I/ P427L double mutation | medulloblastoma | BRCT1; Intrinsically disordered region | T148I may disrupt the hydrophobic cluster where it is located and nearby phosphoserine binding cleft. | [116] |
| L150F | breast cancer | BRCT1 | Possible disruption of a hydrophobic cluster and phosphoserine binding cleft. Increases chromosomal instability. | [61,86,92] |
| I171V | ALL; breast, larynx, and colorectal cancers; head and neck tumors; aplastic anemia | BRCT1 | Possible disruption of a hydrophobic cluster and phosphoserine binding cleft. Increased sensitivity to IR and MMS and lower frequency of HR repair. Loss of association with MDC1. | [62,119,123,124,125,126] |
| E185Q | leukemia and lung cancers; urinary system cancer | BRCT1 | Possibly affects the interaction with BRCA1. May cause an increase in tumor aggression. | [120,127,128,129] |
| V210F | ALL and Non-Hodgkin lymphoma | BRCT1/BRCT2 linker | Hydrophobic residue could disrupt phosphoprotein binding and/or protein stability. | [62,119,130] |
| R215W | ALL; Hodgkin and Non-Hodgkin lymphomas; melanoma; prostate, breast, and colorectal cancers | BRCT1/BRCT2 linker | Disruption of salt bridge destabilizes structure. Decreased co-localization with γ-H2AX at sites of DNA damage and decreased repair efficiency. | [61,62,108,124,125,131] |
| D272N | hepatocellular carcinoma | BRCT2 | Could disrupt ATM phosphorylation of serine 278. | [111,117] |
| A308T | medulloblastoma | BRCT2 | May disrupt the structure of BRCT2 or interface between BRCT1/BRCT2. | [62,116] |
| G311R | medulloblastoma | BRCT2 | May disrupt the structure of BRCT2 or interface between BRCT1/BRCT2. | [62,116] |
| V348D | hepatocellular carcinoma | intrinsically disordered region | Could disrupt ATM phosphorylation of serine 343. | [117] |
| T402A | glioblastoma | intrinsically disordered region | Could disrupt ATM phosphorylation of serine 397. | [115] |
| S406F | glioblastoma | intrinsically disordered region | Could disrupt ATM phosphorylation of serine 397. May introduce order to the intrinsically disordered domain. | [115] |
| S415R | hepatocellular carcinoma | intrinsically disordered region | Could interrupt the sequence space of the intrinsically disordered domain, altering the surrounding structure. | [117] |
| M424V | glioblastoma | intrinsically disordered region | Unknown | [115] |
| T463I/ Q616H double mutation | glioblastoma | intrinsically disordered region | T463I could disrupt nuclear localization of MRE11; Q616H may affect ATM phosphorylation of serine 615. | [115] |
| T485M | glioblastoma | intrinsically disordered region | Unknown | [115] |
| F603L | hepatocellular carcinoma | C-terminal region | Could disrupt RAD18 binding. | [117] |
| S633T | hepatocellular carcinoma | C-terminal region | Could disrupt MRE11 binding. Deficient in nuclear localization of MRE11. | [117] |
| S638P | intrahepatic cholangiocarcinoma | C-terminal region | Could disrupt MRE11 binding. Deficient in nuclear localization of MRE11. | [117] |
| Y679H | renal cell carcinoma | C-terminal region | Could disrupt MRE11 binding and nuclear localization of MRE11. | [135] |
657del5—By far the most common mutation seen in patients with NBS is the germline deletion of five nucleotides within exon 6 of the NBN gene. Due to its high prevalence in Slavic populations, it is believed to be a founder mutation. Patients homozygous for this mutation account for approximately 90% of all NBS cases, whereas heterozygotes only show the NBS phenotype of an increased susceptibility to cancer [22,108,109]. This deletion leads to a premature truncation of NBS1 resulting in an N-terminal fragment of the protein that is approximately 26 kDa (p26) as well as the expression of a separate C-terminal fragment of 70 kDa (p70) [110]. p26 encompasses amino acids 1–218, which include the FHA domain and the first BRCT domain, terminating directly before the start of the second BRCT domain. With the addition of 18 amino acids on its N-terminus, p70 includes the remainder of NBS1 from amino acid 221 to the end. p70 therefore contains the second BRCT domain, the nuclear localization signals, both the MRE11 and ATM interaction sites, as well as serines 278, 343, 397, and 615 which are phosphorylated by ATM in response to irradiation [111]. p70 consequently allows interaction with MRE11 necessary for cell viability (e.g., nuclear localization of the MRN/X complex); however, the disruption of the tandem BRCT domains that occurs with this mutation likely disrupts proper phosphoprotein interaction [107,112].
Many other mutations encoding a truncated NBS1 protein have been described in studies looking at mutations in the DNA damage repair genes of cancer patients [113,114,115,116]. These truncations all lack varying degrees of the C-terminus of NBS1, and since the MRE11 and ATM interaction domains lie near the end of the NBS1, they are usually absent in these mutants prompting an increased risk of oncogenesis.
V26I—This polymorphism was found in a study screening for mutations in patients with medulloblastoma that found a total of 15 novel miscoding NBS1 mutations in 7 of 42 (17%) tumors [116]. Valine 26 is found in the FHA domain and lies near the conserved arginine 28 residue that has been observed to be important in phosphoprotein binding and DNA damage sensitivity [62].
I41M—This mutation was found in a patient with hepatocellular carcinoma and lies within the FHA domain of NBS1 [117]. Isoleucine 41 lies directly adjacent to the conserved phosphoprotein binding region of the FHA domain, and the methionine substitution could perturb the orientation of nearby residues such as serine 42 observed to be involved in phospho-threonine interactions [61].
L57M/H711Y—This double mutation was observed in the same screen of patients with medulloblastoma mentioned above [116]. Leucine 57 lies within the FHA domain at the opposite end of the phosphoprotein binding region and along the BRCT1 interface [61]. Disruption of this region could lead to instability of the protein. Histidine 711 lies next to the RNF20 binding domain directly in between the MRE11 and ATM binding domains sequentially. Disruption of either of these domains could impede the DSB repair pathway [118].
T90S, S93L, and D95N—The S93L and D95N point mutations were found in patients with acute lymphoblastic leukemia (ALL), whereas T90S was observed in an intrahepatic cholangiocarcinoma case [117,119]. D95N was identified in breast, larynx, and prostate cancers as well, though this mutation is also often found in control samples [120,121,122]. All three of these mutations are found within the FHA domain of NBS1 although not as close to the phosphoprotein binding region as the previous two mutations listed above. They do however surround several residues that are highly conserved among other FHA domains and could still be disrupting the phosphoprotein binding site from a distance or disrupting the fold of the domain. Interestingly, in the tumor containing the T90S variant, immunohistochemistry showed down regulation of nuclear localization of MRE11 [117].
T148I/P427L—This double mutation was observed in a single medulloblastoma patient [116]. Threonine 148 lies within the first BRCT domain in a hydrophobic cluster, a site where several other mutations found in cancer screens are also located. This hydrophobic cluster, which partially lines the interface between the BRCT1 and BRCT2 linker, lies near a phosphoserine binding cleft. The second residue, proline 427, lies within the unstructured portion of NBS1.
L150F—This mutation, first found in a screen of breast cancer patients, occurs in the first BRCT domain of NBS1 [86]. Structural studies of S. pombe Nbs1 have shown that this highly conserved leucine resides within a hydrophobic core of the BRCT domain adjacent to a phosphoserine interaction cleft [61]. Mutation to a phenylalanine was predicted to disrupt this binding pocket leading to distorted phosphoprotein interaction. In tumor cells carrying this mutation, it has been observed to cause an increase in chromosomal instability [92].
I171V—This mutation has been found in patients with ALL, breast cancer, larynx cancer, head and neck tumors, and colorectal carcinoma; has been implicated as an origin of aplastic anemia; and is one of the most frequently described NBS1 polymorphisms [119,123,124]. Gao et al. determined from meta-analysis based on 60 publications with ~40,000 cancer cases and ~65,000 controls that this variant is associated with a significant increase in overall cancer risk [125]. Leucine 171 lies within the first BRCT domain and resides within the same hydrophobic cluster as leucine 150 described above. It is predicted that variation of this residue could cause a shift in the geometry of this region and impair protein-protein interactions [62]. Indeed, when expressed in human cells homozygous for the 657del5 mutation described above, the I171V NBS1 mutant was more sensitive to IR and MMS as compared to cells expressing wild type NBS1 and showed a 3-fold lower frequency of HR repair. In co-IP assays with lysates from these cells, wild type NBS1 pulled down MDC1 while I171V did not suggesting the I171V variant reduces DSB repair activity through loss of association with MDC1 [126].
E185Q—This mutation is associated with an increased risk of leukemia, lung cancer, and urinary system cancer. It has also been reported in studies screening for mutations in breast, prostate, and bladder cancers and nasopharyngeal carcinomas although its relationship with these remains controversial [5,120,127,128,129]. Glutamate 185 lies within the BRCT1 domain and could possibly affect the interaction of NBS1 with BRCA1. The overexpression of E185Q-mutated NBS1 in nasal pharyngeal cancer cells significantly increased the migration of the cells in cellular migration assays [129]. This mutation therefore may not only increase the risk of developing cancer but could also increase tumor aggression.
V210F—This mutation has been found in patients diagnosed with ALL [119,130]. Valine 210 is found in the linker region connecting BRCT1 and BRCT2 and lies within the same hydrophobic cluster as leucines 171 and 150. Mutation to a much bulkier hydrophobic residue in this region could disrupt phosphoprotein binding and/or protein stability [62].
R215W—Aside from the 657del5 mutation found in the majority of patients with NBS, R215W is probably the most frequently studied NBS1 mutation. It has been found in patients with ALL, Hodgkin lymphoma, Non-Hodgkin lymphoma, melanoma, prostate, breast, and colorectal cancers [124]. As with the I171V variant, Gao et al. concluded that R215W is associated with a significant increase in overall cancer risk [125]. Additionally, R215W was found to be compound heterozygous with the 657del5 mutation in Japanese twins with especially severe NBS. Lymphoblastoid cell extracts from those patients had much lower levels of the full-length NBS1 and the cells were deficient in ATM activation [108]. Like valine 210, arginine 215 also lies within the BRCT1 and BRCT2 linker region near the hydrophobic cluster where leucines 171 and 150 reside and, interestingly, also near where the 657del5 mutation causes truncation. This residue in the S. pombe crystal structure is solvent exposed, and it is likely that a mutation from a polar residue to a bulky hydrophobic residue could strongly disrupt this region [62]. Additionally, arginine 215 was predicted to form a salt bridge with asparagine 205 and glutamate 206, which would be disrupted upon mutation. Combined, these effects could induce instability around this region; in fact, R215W NBS1 shows higher trypsin sensitivity than wild type [61]. In vivo, R215W has been shown to hinder the co-localization of NBS1 with γ-H2AX at the site of DNA damage upon irradiation and accordingly results in a decrease in repair efficiency [131], perhaps because the arginine 215 residue is required for correct orientation of the BRCT domains to recognize γ-H2AX.
D272N—This mutation was found in a patient with hepatocellular carcinoma in conjunction with an alteration in the TP53 pathway [117]. Aspartate 272 lies within the second BRCT domain of NBS1 and begins the serine 278 phosphorylation site loop (amino acids 272–295). Disruption of this loop could cause improper phosphorylation of NBS1 by ATM upon DNA damage, a critical step in the HR repair pathway [111].
A308T and G311R—These mutations were found in patients with medulloblastoma [116]. Alanine 308 and glycine 311 are found in the second BRCT domain on the interface with the BRCT1 domain [62]. In both mutations, small side chains are substituted with much bulkier residues which may disrupt the structure of BRCT2 or perhaps the interface between the two domains.
V348D—This mutation was found in a screen of patients diagnosed with hepatocellular carcinoma [117]. Valine 348 lies directly C-terminal to the end of the second BRCT domain in the beginning of the intrinsically disordered and structurally uncharacterized portion of NBS1. Valine 348 is near serine 343 in primary structure and might disrupt the ability of ATM to phosphorylate at this location in response to DNA damage.
T402A and S406F—These two residues, also located in the intrinsically disordered domain of NBS1 and C-terminal to the second BRCT domain, were both found to be mutated in a screen of glioblastomas [115]. Sequentially, threonine 402 and serine 406 lie near another phosphorylation site of ATM (serine 397) and could play a role in obstructing this signaling step in the DNA damage response pathway. Additionally, serine 406 lies within a region rich in disorder promoting residues. In S406F, the serine is mutated to a phenylalanine which has a much greater propensity for forming ordered structures. This disruption in the sequence space of the intrinsically disordered domain of NBS1 could affect the unique structural properties of this area that are generated by its selective use of disorder promoting amino acids [132].
S415R and M424V—These mutations were found in patients with hepatocellular carcinoma and glioblastoma, respectively [115,117]. Like serine 406, serine 415 also lies within a string of disorder promoting residues and mutation to an arginine, which is less commonly found in intrinsically disordered domains, could promote instability or structural changes at this location.
T463I/Q616H—This double mutation was found in a screen of glioblastoma [115]. The first mutation of threonine 463 to isoleucine lies within the nuclear localization signal shown to interact with the importin KPNA2 [133]. The second mutation lies directly adjacent to serine 615, a serine that is a phosphorylation target of ATM [111].
T485M—Threonine 485 was found to be mutated to methionine in one case of glioblastoma [115]. This mutation does not lie near any post translationally modified residues or interaction domains sequentially; however, it could lie near enough in structure to disrupt NBS1 functionality.
F603L, S633T, and S638P—The F603L and S633T mutations were found in patients with hepatocellular carcinoma and the S638P mutation was found in a patient with intrahepatic cholangiocarcinoma [117]. All three of these mutations occur in the C-terminal region of NBS1. Phenylalanine 603 is near the RAD18 binding domain. Disruption of the RAD18 binding domain leads to impaired localization of RAD18 to sites of DNA damage which could adversely affect both the HR and trans-lesion synthesis DNA repair pathways [134]. Serines 633 and 638 are near the MRE11 binding domain. Disruption of MRE11 binding impairs the nuclear transport of MRN by NBS1 and causes subsequent deficiencies in DNA DSB repair. In fact, immunohistochemistry and immunofluorescent assays demonstrated that tumor cells carrying S633T or S638P mutations were observed to be deficient in the nuclear localization of MRE11 [117].
Y679H—This mutation was observed in a screen of renal cell carcinomas [135]. Tyrosine 679 is also found in the C-terminus near the MRE11 binding domain and, like the three C-terminal NBS1 mutations described just above, could affect interaction with MRE11 and therefore nuclear localization of the MRN complex.
6. Concluding Remarks
In this review, we sought to comprehensively survey the known disease-associated mutations in the MRN complex. When available, we presented the known structural and biochemical effects that each mutation has on MRN function. From our perspective, an understanding of the structure/function relationships of disease-associated mutations offers the opportunity to learn about the function/dysfunction of MRN in DNA DSB repair. More broadly, when structural and biochemical data are coupled with cell-based assays, in vivo studies, and patient data, this complete picture allows for a nano- to macroscale view of the role of MRN in genome stability and the consequences of disrupting that role. However, as is clear above, there are many cases where a molecular mechanism for the effects of a mutation or an understanding for the correlation with disease states are not known. This is especially true for the intrinsically disordered region of NBS1. Thus, there are many avenues for increasing our understanding for how the biological activity of the MRN complex, including how the complex interacts with other repair proteins, plays into the bigger role of the complex in maintaining genome integrity. The study of disease-associated mutations also presents the occasion to explore new anti-cancer therapeutics. Therapeutically controlling the activity of MRN could present an option for the treatment of certain cancer types, where orthologous DNA damage repair pathways are defective, through the synthetic lethality strategy. For example, there is interesting evidence that patients with endometrial cancer and mutations in MRE11 could be treated with PARP inhibitors [136,137]. Overall, understanding these mutations from a structural and biochemical standpoint will enhance our knowledge of the function of the MRN complex and may help in treating or preventing MRN-related diseases in the future.
Acknowledgments
The authors are grateful to the other members of the Latham Laboratory for helpful discussions.
Author Contributions
Conceptualization, S.R. and M.P.L.; validation, M.D.C. and M.P.L.; writing—original draft preparation, S.R., T.A.B., M.D.C. and M.P.L.; writing—review and editing, S.R., T.A.B., M.D.C. and M.P.L.; visualization, M.P.L.; supervision, M.D.C. and M.P.L; funding acquisition, M.P.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Cancer Prevention and Research Institute of Texas (CPRIT), grant number RP180553, and the National Institute of General Medical Sciences (NIGMS), grant number 1R35GM128906.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Romero-Laorden N., Castro E. Inherited mutations in DNA repair genes and cancer risk. Curr. Probl. Cancer. 2017;41:251–264. doi: 10.1016/j.currproblcancer.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Chae Y.K., Anker J.F., Carneiro B.A., Chandra S., Kaplan J., Kalyan A., Santa-Maria C.A., Platanias L.C., Giles F.J. Genomic landscape of DNA repair genes in cancer. Oncotarget. 2016;7:23312–23321. doi: 10.18632/oncotarget.8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo R., DuBoff M., Jayakumaran G., Kris M.G., Ladanyi M., Robson M.E., Mandelker D., Zauderer M.G. Novel Germline Mutations in DNA Damage Repair in Patients with Malignant Pleural Mesotheliomas. J. Thorac. Oncol. 2020;15:655–660. doi: 10.1016/j.jtho.2019.12.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knijnenburg T.A., Wang L., Zimmermann M.T., Chambwe N., Gao G.F., Cherniack A.D., Fan H., Shen H., Way G.P., Greene C.S., et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018;23:239–254.e6. doi: 10.1016/j.celrep.2018.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vineis P., Manuguerra M., Kavvoura F.K., Guarrera S., Allione A., Rosa F., Di Gregorio A., Polidoro S., Saletta F., Ioannidis J.P.A., et al. A field synopsis on low-penetrance variants in DNA repair genes and cancer susceptibility. J. Natl. Cancer Inst. 2009;101:24–36. doi: 10.1093/jnci/djn437. [DOI] [PubMed] [Google Scholar]
- 6.Rupnik A., Lowndes N.F., Grenon M. MRN and the race to the break. Chromosoma. 2010;119:115–135. doi: 10.1007/s00412-009-0242-4. [DOI] [PubMed] [Google Scholar]
- 7.Williams G.J., Lees-Miller S.P., Tainer J.A. Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair (Amst) 2010;9:1299–1306. doi: 10.1016/j.dnarep.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oh J., Symington L.S. Role of the Mre11 complex in preserving genome integrity. Genes (Basel) 2018;9:589. doi: 10.3390/genes9120589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao Y., Weaver D.T. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997;25:2985–2991. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo G., Yao M.S., Bender C.F., Mills M., Bladl A.R., Bradley A., Petrini J.H.J. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl. Acad. Sci. USA. 1999;96:7376–7381. doi: 10.1073/pnas.96.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu J., Petersen S., Tessarollo L., Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol. 2001;11:105–109. doi: 10.1016/S0960-9822(01)00019-7. [DOI] [PubMed] [Google Scholar]
- 12.Stewart G.S., Maser R.S., Stankovic T., Bressan D.A., Kaplan M.I., Jaspers N.G., Raams A., Byrd P.J., Petrini J.H.J., Taylor A.M. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/S0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 13.Chrzanowska K.H., Gregorek H., Dembowska-Bagińska B., Kalina M.A., Digweed M. Nijmegen breakage syndrome (NBS) Orphanet J. Rare Dis. 2012;7:1–19. doi: 10.1186/1750-1172-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waltes R., Kalb R., Gatei M., Kijas A.W., Stumm M., Sobeck A., Wieland B., Varon R., Lerenthal Y., Lavin M.F., et al. Human RAD50 Deficiency in a Nijmegen Breakage Syndrome-like Disorder. Am. J. Hum. Genet. 2009;84:605–616. doi: 10.1016/j.ajhg.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koczkowska M., Krawczynska N., Stukan M., Kuzniacka A., Brozek I., Sniadecki M., Debniak J., Wydra D., Biernat W., Kozlowski P., et al. Spectrum and prevalence of pathogenic variants in ovarian cancer susceptibility genes in a group of 333 patients. Cancers (Basel) 2018;10:442. doi: 10.3390/cancers10110442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brandt S., Samartzis E.P., Zimmermann A.K., Fink D., Moch H., Noske A., Dedes K.J. Lack of MRE11-RAD50-NBS1 (MRN) complex detection occurs frequently in low-grade epithelial ovarian cancer. BMC Cancer. 2017;17:1–9. doi: 10.1186/s12885-016-3026-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H., Liu Y., Zhou K., Zhou C., Zhou R., Cheng C., Wei Q., Lu D., Zhou L. Genetic variations in the homologous recombination repair pathway genes modify risk of glioma. J. Neurooncol. 2016;126:11–17. doi: 10.1007/s11060-015-1892-0. [DOI] [PubMed] [Google Scholar]
- 18.Damiola F., Pertesi M., Oliver J., Le Calvez-Kelm F., Voegele C., Young E.L., Robinot N., Forey N., Durand G., Vallée M.P., et al. Rare key functional domain missense substitutions in MRE11A, RAD50, and NBNcontribute to breast cancer susceptibility: Results from a Breast Cancer Family Registry case-control mutation-screening study. Breast Cancer Res. 2014;16:R58. doi: 10.1186/bcr3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amemiya Y., Bacopulos S., Al-Shawarby M., Al-Tamimi D., Naser W., Ahmed A., Khalifa M., Slodkowska E., Seth A. A comparative analysis of breast and ovarian cancer-related gene mutations in Canadian and Saudi Arabian patients with breast cancer. Anticancer Res. 2015;35:2601–2610. [PubMed] [Google Scholar]
- 20.Uzunoglu H., Korak T., Ergul E., Uren N., Sazci A., Utkan N.Z., Kargi E., Triyaki Ç., Yirmibesoglu O. Association of the nibrin gene (NBN) variants with breast cancer. Biomed. Rep. 2016;4:369–373. doi: 10.3892/br.2016.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan R.T., Siddique A., Shahid N., Khokher S., Fatima W. Breast cancer risk associated with genes encoding DNA repair MRN complex: A study from Punjab, Pakistan. Breast Cancer. 2018;25:350–355. doi: 10.1007/s12282-018-0837-9. [DOI] [PubMed] [Google Scholar]
- 22.Zhang G., Zeng Y., Liu Z., Wei W. Significant association between Nijmegen breakage syndrome 1 657del5 polymorphism and breast cancer risk. Tumor Biol. 2013;34:2753–2757. doi: 10.1007/s13277-013-0830-z. [DOI] [PubMed] [Google Scholar]
- 23.Couch F.J., Shimelis H., Hu C., Hart S.N., Polley E.C., Na J., Hallberg E., Moore R., Thomas A., Lilyquist J., et al. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol. 2017;3:1190–1196. doi: 10.1001/jamaoncol.2017.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Situ Y., Chung L., Lee C.S., Ho V. MRN (MRE11-RAD50-NBS1) Complex in Human Cancer and Prognostic Implications in Colorectal Cancer. Int. J. Mol. Sci. 2019;20:816. doi: 10.3390/ijms20040816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho V., Chung L., Singh A., Lea V., Revoltar M., Lim S., Tut T.-G., Ng W., Lee M., de Souza P., et al. Early Postoperative Low Expression of RAD50 in Rectal Cancer Patients Associates with Disease-Free Survival. Cancers (Basel) 2017;9:163. doi: 10.3390/cancers9120163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho V., Chung L., Singh A., Lea V., Abubakar A., Lim S.H., Ng W., Lee M., de Souza P., Shin J.S., et al. Overexpression of the MRE11-RAD50-NBS1 (MRN) complex in rectal cancer correlates with poor response to neoadjuvant radiotherapy and prognosis. BMC Cancer. 2018;18:1–11. doi: 10.1186/s12885-018-4776-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J., Su T., Yang L., Zhang C., He Y. High expression of MRE11 correlates with poor prognosis in gastric carcinoma. Diagn. Pathol. 2019;14:1–9. doi: 10.1186/s13000-019-0844-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cannan W.J., Pederson D.S. Mechanisms and Consequences of Double-Strand DNA Break Formation in Chromatin. J. Cell. Physiol. 2016;231:3–14. doi: 10.1002/jcp.25048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keeney S., Neale M.J. Initiation of meiotic recombination by formation of DNA double-strand breaks: Mechanism and regulation. Biochem. Soc. Trans. 2006;34:523–525. doi: 10.1042/BST0340523. [DOI] [PubMed] [Google Scholar]
- 30.Dudley D.D., Chaudhuri J., Bassing C.H., Alt F.W. Mechanism and control of V(D)J recombination versus class switch recombination: Similarities and differences. Adv. Immunol. 2005 doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- 31.Han J., Huang J. DNA double-strand break repair pathway choice: The fork in the road. Genome Instab. Dis. 2019 doi: 10.1007/s42764-019-00002-w. [DOI] [Google Scholar]
- 32.Lieber M.R. The Mechanism of Double-Strand DNA Break Repair by the Nonhomologous DNA End-Joining Pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boulton S.J., Jackson S.P. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L., Trujillo K., Ramos W., Sung P., Tomkinson A.E. Promotion of Dnl4-Catalyzed DNA End-Joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 Complexes. Mol. Cell. 2001;8:1105–1115. doi: 10.1016/S1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- 35.Matsuzaki K., Shinohara A., Shinohara M. Forkhead-associated domain of yeast Xrs2, a homolog of human Nbs1, promotes nonhomologous end joining through interaction with a ligase IV partner protein, Lif1. Genetics. 2008;179:213–225. doi: 10.1534/genetics.107.079236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rass E., Grabarz A., Plo I., Gautier J., Bertrand P., Lopez B.S. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 37.Xie A., Kwok A., Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myler L.R., Gallardo I.F., Soniat M.M., Deshpande R.A., Gonzalez X.B., Kim Y., Paull T.T., Finkelstein I.J. Single-Molecule Imaging Reveals How Mre11-Rad50-Nbs1 Initiates DNA Break Repair. Mol. Cell. 2017;67:891–898.e4. doi: 10.1016/j.molcel.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reginato G., Cannavo E., Cejka P. Physiological protein blocks direct the Mre11-Rad50-Xrs2 and Sae2 nuclease complex to initiate DNA end resection. Genes Dev. 2017;31:2325–2330. doi: 10.1101/gad.308254.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langerak P., Mejia-Ramirez E., Limbo O., Russell P. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cassani C., Vertemara J., Bassani M., Marsella A., Tisi R., Zampella G., Longhese M.P. The ATP-bound conformation of the Mre11–Rad50 complex is essential for Tel1/ATM activation. Nucleic Acids Res. 2019;47:3550–3567. doi: 10.1093/nar/gkz038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee J.-H., Mand M.R., Deshpande R.A., Kinoshita E., Yang S.-H., Wyman C., Paull T.T. Ataxia Telangiectasia-Mutated (ATM) Kinase Activity Is Regulated by ATP-driven Conformational Changes in the Mre11/Rad50/Nbs1 (MRN) Complex. J. Biol. Chem. 2013;288:12840–12851. doi: 10.1074/jbc.M113.460378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Syed A., Tainer J.A. The MRE11–RAD50–NBS1 Complex Conducts the Orchestration of Damage Signaling and Outcomes to Stress in DNA Replication and Repair. Annu. Rev. Biochem. 2018;87:263–294. doi: 10.1146/annurev-biochem-062917-012415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lammens K., Bemeleit D.J., Möckel C., Clausing E., Schele A., Hartung S., Schiller C.B., Lucas M., Angermüller C., Söding J., et al. The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell. 2011;145:54–66. doi: 10.1016/j.cell.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Möckel C., Lammens K., Schele A., Hopfner K.-P., Pemberton T. ATP driven structural changes of the bacterial Mre11:Rad50 catalytic head complex. Nucleic Acids Res. 2012;40:914–927. doi: 10.1093/nar/gkr749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim H.S., Kim J.S., Park Y.B., Gwon G.H., Cho Y. Crystal structure of the Mre11-Rad50-ATPγS complex: Understanding the interplay between Mre11 and Rad50. Genes Dev. 2011;25:1091–1104. doi: 10.1101/gad.2037811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rojowska A., Lammens K., Seifert F.U., Direnberger C., Feldmann H., Hopfner K.-P. Structure of the Rad50 DNA double-strand break repair protein in complex with DNA. EMBO J. 2014;33:2847–2859. doi: 10.15252/embj.201488889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herdendorf T.J., Albrecht D.W., Benkovic S.J., Nelson S.W. Biochemical characterization of bacteriophage T4 Mre11-Rad50 complex. J. Biol. Chem. 2011;286:2382–2392. doi: 10.1074/jbc.M110.178871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hopfner K.-P., Karcher A., Shin D.S., Craig L., Arthur L.M., Carney J.P., Tainer J.A. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/S0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 50.Hopfner K.-P., Karcher A., Craig L., Woo T.T., Carney J.P., Tainer J.A. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/S0092-8674(01)00335-X. [DOI] [PubMed] [Google Scholar]
- 51.Hopfner K.-P., Craig L., Moncalian G., Zinkel R.A., Usui T., Owen B.A.L.L., Karcher A., Henderson B., Bodmer J.-L., McMurray C.T., et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 52.Williams R.S., Moncalian G., Williams J.S., Yamada Y., Limbo O., Shin D.S., Groocock L.M., Cahill D., Hitomi C., Guenther G., et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams G.J., Williams R.S., Williams J.S., Moncalian G., Arvai A.S., Limbo O., Guenther G., SilDas S., Hammel M., Russell P., et al. ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat. Struct. Mol. Biol. 2011;18:423–431. doi: 10.1038/nsmb.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sung S., Li F., Park Y.B., Kim J.S., Kim A., Song O.-K., Kim J., Che J., Lee S.E., Cho Y. DNA end recognition by the Mre11 nuclease dimer: Insights into resection and repair of damaged DNA. EMBO J. 2014;33:1–14. doi: 10.15252/embj.201488299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y., Sung S., Kim Y., Li F., Gwon G., Jo A., Kim T., Song O.-K., Lee S.E., Cho Y., et al. ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex. EMBO J. 2016;35:1–16. doi: 10.15252/embj.201592462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Käshammer L., Saathoff J.-H., Lammens K., Gut F., Bartho J., Alt A., Kessler B., Hopfner K.-P. Mechanism of DNA End Sensing and Processing by the Mre11-Rad50 Complex. Mol. Cell. 2019;76:382–394.e6. doi: 10.1016/j.molcel.2019.07.035. [DOI] [PubMed] [Google Scholar]
- 57.Park Y.B., Chae J., Kim Y.C., Cho Y. Crystal structure of human Mre11: Understanding tumorigenic mutations. Structure. 2011;19:1591–1602. doi: 10.1016/j.str.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 58.Schiller C.B., Lammens K., Guerini I., Coordes B., Feldmann H., Schlauderer F., Möckel C., Schele A., Sträßer K., Jackson S.P., et al. Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat. Struct. Mol. Biol. 2012;19:693–700. doi: 10.1038/nsmb.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seifert F.U., Lammens K., Hopfner K.-P. Structure of the catalytic domain of Mre11 from Chaetomium thermophilum. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2015;71:752–757. doi: 10.1107/S2053230X15007566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seifert F.U., Lammens K., Stoehr G., Kessler B., Hopfner K.-P. Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50. EMBO J. 2016;35:759–772. doi: 10.15252/embj.201592934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams R.S., Dodson G.E., Limbo O., Yamada Y., Williams J.S., Guenther G., Classen S., Glover J.N.M., Iwasaki H., Russell P., et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lloyd J., Chapman J.R., Clapperton J.A., Haire L.F., Hartsuiker E., Li J., Carr A.M., Jackson S.P., Smerdon S.J. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell. 2009;139:100–111. doi: 10.1016/j.cell.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matange N., Podobnik M., Visweswariah S.S. Metallophosphoesterases: Structural fidelity with functional promiscuity. Biochem. J. 2015;467:201–216. doi: 10.1042/BJ20150028. [DOI] [PubMed] [Google Scholar]
- 64.Paull T.T. 20 Years of Mre11 Biology: No End in Sight. Mol. Cell. 2018;71:419–427. doi: 10.1016/j.molcel.2018.06.033. [DOI] [PubMed] [Google Scholar]
- 65.Furuse M., Nagase Y., Tsubouchi H., Murakami-Murofushi K., Shibata T., Ohta K. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paull T.T., Gellert M. The 3′ to 5′ Exonuclease Activity of Mre11 Facilitates Repair of DNA Double-Strand Breaks. Mol. Cell. 1998;1:969–979. doi: 10.1016/S1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 67.Hoa N.N., Shimizu T., Zhou Z.W., Wang Z.Q., Deshpande R.A., Paull T.T., Akter S., Tsuda M., Furuta R., Tsusui K., et al. Mre11 Is Essential for the Removal of Lethal Topoisomerase 2 Covalent Cleavage Complexes. Mol. Cell. 2016;64:580–592. doi: 10.1016/j.molcel.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 68.Aparicio T., Baer R., Gottesman M., Gautier J. MRN, CtIP, and BRCA1 mediate repair of topoisomerase II-DNA adducts. J. Cell Biol. 2016;212:399–408. doi: 10.1083/jcb.201504005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Neale M.J., Pan J., Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Déry U., Coulombe Y., Rodrigue A., Stasiak A., Richard S., Masson J.-Y. A Glycine-Arginine Domain in Control of the Human MRE11 DNA Repair Protein. Mol. Cell. Biol. 2008;28:3058–3069. doi: 10.1128/MCB.02025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu Z., Vogel G., Yan C., Dubeau D., Spehalski E., Hébert J., Ferguson D.O., Masson J.Y., Richard S. The MRE11 GAR motif regulates DNA double-strand break processing and ATR activation. Cell Res. 2012;22:305–320. doi: 10.1038/cr.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sedghi M., Salari M., Moslemi A.R., Kariminejad A., Davis M., Goullée H., Olsson B., Laing N., Tajsharghi H. Ataxia-telangiectasia-like disorder in a family deficient for MRE11A, caused by a MRE11 variant. Neurol. Genet. 2018 doi: 10.1212/NXG.0000000000000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uchisaka N., Takahashi N., Sato M., Kikuchi A., Mochizuki S., Imai K., Nonoyama S., Ohara O., Watanabe F., Mizutani S., et al. Two Brothers with Ataxia-Telangiectasia-like Disorder with Lung Adenocarcinoma. J. Pediatr. 2009;155:435–438. doi: 10.1016/j.jpeds.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 74.Regal J.A., Festerling T.A., Buis J.M., Ferguson D.O. Disease-associated MRE11 mutants impact ATM/ATR DNA damage signaling by distinct mechanisms. Hum. Mol. Genet. 2013;22:5146–5159. doi: 10.1093/hmg/ddt368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Giannini G., Ristori E., Cerignoli F., Rinaldi C., Zani M., Viel A., Ottini L., Crescenzi M., Martinotti S., Bignami M., et al. Human MRE11 is inactivated in mismatch repair-deficient cancers. EMBO Rep. 2002;3:248–254. doi: 10.1093/embo-reports/kvf044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wen Q., Scorah J., Phear G., Rodgers G., Rodgers S., Meuth M. A mutant allele of MRE11 found in mismatch repair-deficient tumor cells suppresses the cellular response to DNA replication fork stress in a dominant negative manner. Mol. Biol. Cell. 2008;19:1693–1705. doi: 10.1091/mbc.e07-09-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hohl M., Mojumdar A., Hailemariam S., Kuryavyi V., Ghisays F., Sorenson K., Chang M., Taylor B.S., Patel D.J., Burgers P.M., et al. Modeling cancer genomic data in yeast reveals selection against ATM function during tumorigenesis. PLoS Genet. 2020;16:e1008422. doi: 10.1371/journal.pgen.1008422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fukuda T., Sumiyoshi T., Takahashi M., Kataoka T., Asahara T., Inui H., Watatani M., Yasutomi M., Kamada N., Miyagawa K. Alterations of the double-strand break repair gene MRE11 in cancer. Cancer Res. 2001;61:23–26. [PubMed] [Google Scholar]
- 79.Matsumoto Y., Miyamoto T., Sakamoto H., Izumi H., Nakazawa Y., Ogi T., Tahara H., Oku S., Hiramoto A., Shiiki T., et al. Two unrelated patients with MRE11A mutations and Nijmegen breakage syndrome-like severe microcephaly. DNA Repair (Amst) 2011;10:314–321. doi: 10.1016/j.dnarep.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 80.Lee J.-H., Ghirlando R., Bhaskara V., Hoffmeyer M.R., Gu J., Paull T.T. Regulation of Mre11/Rad50 by Nbs1: Effects on nucleotide-dependent DNA binding and association with ataxiatelangiectasia-like disorder mutant complexes. J. Biol. Chem. 2003;278:45171–45181. doi: 10.1074/jbc.M308705200. [DOI] [PubMed] [Google Scholar]
- 81.Limbo O., Moiani D., Kertokalio A., Wyman C., Tainer J.A., Russell P. Mre11 ATLD17/18 mutation retains Tel1/ATM activity but blocks DNA double-strand break repair. Nucleic Acids Res. 2012;40:11435–11449. doi: 10.1093/nar/gks954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bartkova J., Tommiska J., Oplustilova L., Aaltonen K., Tamminen A., Heikkinen T., Mistrik M., Aittomäki K., Blomqvist C., Heikkilä P., et al. Aberrations of the MRE11-RAD50-NBS1 DNA damage sensor complex in human breast cancer: MRE11 as a candidate familial cancer-predisposing gene. Mol. Oncol. 2008;2:296–316. doi: 10.1016/j.molonc.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fernet M., Gribaa M., Salih M.A.M., Seidahmed M.Z., Hall J., Koenig M. Identification and functional consequences of a novel MRE11 mutation affecting 10 Saudi Arabian patients with the ataxia telangiectasia-like disorder. Hum. Mol. Genet. 2005;14:307–318. doi: 10.1093/hmg/ddi027. [DOI] [PubMed] [Google Scholar]
- 84.Wang Z., Cummins J.M., Shen D., Cahill D.P., Jallepalli P.V., Wang T.-L., Parsons D.W., Traverso G., Awad M., Silliman N., et al. Three classes of genes mutated in colorectal cancers with chromosomal instability. Cancer Res. 2004;64:2998–3001. doi: 10.1158/0008-5472.CAN-04-0587. [DOI] [PubMed] [Google Scholar]
- 85.Sjöblom T., Jones S., Wood L.D., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., et al. The Consensus Coding Sequences of Human Breast and Colorectal Cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 86.Heikkinen K., Karppinen S.-M., Soini Y., Mäkinen M., Winqvist R. Mutation screening of Mre11 complex genes: Indication of RAD50 involvement in breast and ovarian cancer susceptibility. J. Med. Genet. 2003;40:e131. doi: 10.1136/jmg.40.12.e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Delia D., Paine M., Buscemi G., Savio C., Palmeri S., Lulli P., Carlessi L., Fontanella E., Chessa L. MRE11 mutations and impaired ATM-dependent responses in an Italian family with ataxia-telangiectasia-like disorder. Hum. Mol. Genet. 2004;13:2155–2163. doi: 10.1093/hmg/ddh221. [DOI] [PubMed] [Google Scholar]
- 88.Hopfner K.-P., Tainer J.A. Rad50/SMC proteins and ABC transporters: Unifying concepts from high-resolution structures. Curr. Opin. Struct. Biol. 2003;13:249–255. doi: 10.1016/S0959-440X(03)00037-X. [DOI] [PubMed] [Google Scholar]
- 89.Hohl M., Kochańczyk T., Tous C., Aguilera A., Krężel A., Petrini J.H.J. Interdependence of the rad50 hook and globular domain functions. Mol. Cell. 2015;57:479–491. doi: 10.1016/j.molcel.2014.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hohl M., Kwon Y., Galván S.M., Xue X., Tous C., Aguilera A., Sung P., Petrini J.H.J. The Rad50 coiled-coil domain is indispensable for Mre11 complex functions. Nat. Struct. Mol. Biol. 2011;18:1124–1131. doi: 10.1038/nsmb.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Boswell Z.K., Canny M.D., Buschmann T.A., Sang J., Latham M.P. Adjacent mutations in the archaeal Rad50 ABC ATPase D-loop disrupt allosteric regulation of ATP hydrolysis through different mechanisms. Nucleic Acids Res. 2020;48:2457–2472. doi: 10.1093/nar/gkz1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Heikkinen K., Rapakko K., Karppinen S.M., Erkko H., Knuutila S., Lundán T., Mannermaa A., Børresen-Dale A.L., Borg Å., Barkardottir R.B., et al. RAD50 and NBS1 are breast cancer susceptibility genes associated with genomic instability. Carcinogenesis. 2006;27:1593–1599. doi: 10.1093/carcin/bgi360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fan C., Zhang J., Ouyang T., Li J., Wang T., Fan Z., Fan T., Lin B., Xie Y. RAD50 germline mutations are associated with poor survival in BRCA1/2–negative breast cancer patients. Int. J. Cancer. 2018;143:1935–1942. doi: 10.1002/ijc.31579. [DOI] [PubMed] [Google Scholar]
- 94.Thompson E.R., Rowley S.M., Li N., McInerny S., Devereux L., Wong-Brown M.W., Trainer A.H., Mitchell G., Scott R.J., James P.A., et al. Panel testing for familial breast cancer: Calibrating the tension between research and clinical care. J. Clin. Oncol. 2016;34:1455–1459. doi: 10.1200/JCO.2015.63.7454. [DOI] [PubMed] [Google Scholar]
- 95.García-Sanz P., Triviño J.C., Mota A., Pérez López M., Colás E., Rojo-Sebastián A., García Á., Gatius S., Ruiz M., Prat J., et al. Chromatin remodelling and DNA repair genes are frequently mutated in endometrioid endometrial carcinoma. Int. J. Cancer. 2017;140:1551–1563. doi: 10.1002/ijc.30573. [DOI] [PubMed] [Google Scholar]
- 96.Maresca L., Lodovichi S., Lorenzoni A., Cervelli T., Monaco R., Spugnesi L., Tancredi M., Falaschi E., Zavaglia K., Landucci E., et al. Functional Interaction Between BRCA1 and DNA Repair in Yeast May Uncover a Role of RAD50, RAD51, MRE11A, and MSH6 Somatic Variants in Cancer Development. Front. Genet. 2018;9:1–13. doi: 10.3389/fgene.2018.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Young E.L., Thompson B.A., Neklason D.W., Firpo M.A., Werner T., Bell R., Berger J., Fraser A., Gammon A., Koptiuch C., et al. Pancreatic cancer as a sentinel for hereditary cancer predisposition. BMC Cancer. 2018;18:697. doi: 10.1186/s12885-018-4573-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Deshpande R.A., Williams G.J., Limbo O., Williams R.S., Kuhnlein J., Lee J.-H., Classen S., Guenther G., Russell P., Tainer J.A., et al. ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling. EMBO J. 2014;33:482–500. doi: 10.1002/embj.201386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boswell Z.K., Rahman S., Canny M.D., Latham M.P. A dynamic allosteric pathway underlies Rad50 ABC ATPase function in DNA repair. Sci. Rep. 2018;8:1–12. doi: 10.1038/s41598-018-19908-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kaymaz Y., Oduor C.I., Yu H., Otieno J.A., Ong’echa J.M., Moormann A.M., Bailey J.A. Comprehensive transcriptome and mutational profiling of Endemic Burkitt lymphoma reveals EBV type-specific differences. Mol. Cancer Res. 2017;15:563–576. doi: 10.1158/1541-7786.MCR-16-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Al-Ahmadie H., Iyer G., Hohl M., Asthana S., Inagaki A., Schultz N., Hanrahan A.J., Scott S.N., Brannon A.R., McDermott G.C., et al. Synthetic Lethality in ATM-Deficient RAD50-Mutant Tumors Underlies Outlier Response to Cancer Therapy. Cancer Discov. 2014;4:1014–1021. doi: 10.1158/2159-8290.CD-14-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kobayashi J., Tauchi H., Sakamoto S., Nakamura A., Morishima K., Matsuura S., Kobayashi T., Tamai K., Tanimoto K., Komatsu K. NBS1 localizes to γ-H2AX foci through interaction with the FHA/BRCT domain. Curr. Biol. 2002;12:1846–1851. doi: 10.1016/S0960-9822(02)01259-9. [DOI] [PubMed] [Google Scholar]
- 103.Lee J.H., Lim D.S. Dual role of Nbs1 in the ataxia telangiectasia mutated-dependent DNA damage response. FEBS J. 2006;273:1630–1636. doi: 10.1111/j.1742-4658.2006.05191.x. [DOI] [PubMed] [Google Scholar]
- 104.Hofmann K., Bucher P. The FHA domain: A putative nuclear signalling domain found in protein kinases and transcription factors. Trends Biochem. Sci. 1995;20:347–349. doi: 10.1016/S0968-0004(00)89072-6. [DOI] [PubMed] [Google Scholar]
- 105.Durocher D., Henckel J., Fersht A.R., Jackson S.P. The FHA domain is a modular phosphopeptide recognition motif. Mol. Cell. 1999 doi: 10.1016/S1097-2765(00)80340-8. [DOI] [PubMed] [Google Scholar]
- 106.Koonin E.V., Altschul S.F., Bork P. BRCA1 Protein Products ... Functional Motifs. Nat. Genet. 1996;13:266–268. doi: 10.1038/ng0796-266. [DOI] [PubMed] [Google Scholar]
- 107.Becker E., Meyer V., Madaoui H., Guerois R. Detection of a tandem BRCT in Nbs1 and Xrs2 with functional implications in the DNA damage response. Bioinformatics. 2006;22:1289–1292. doi: 10.1093/bioinformatics/btl075. [DOI] [PubMed] [Google Scholar]
- 108.Seemanová E., Sperling K., Neitzel H., Varon R., Hadac J., Butova O., Schröck E., Seeman P., Digweed M. Nijmegen breakage syndrome (NBS) with neurological abnormalities and without chromosomal instability. J. Med. Genet. 2006;43:218–224. doi: 10.1136/jmg.2005.035287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Varon R., Seemanova E., Chrzanowska K., Hnateyko O., Piekutowska-Abramczuk D., Krajewska-Walasek M., Sykut-Cegielska J., Sperling K., Reis A. Clinical ascertainment of Nijmegen breakage syndrome (NBS) and prevalence of the major mutation, 657del5, in three Slav populations. Eur. J. Hum. Genet. 2000;8:900–902. doi: 10.1038/sj.ejhg.5200554. [DOI] [PubMed] [Google Scholar]
- 110.Maser R.S., Zinkel R., Petrini J.H.J. An alternative mode of translation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nat. Genet. 2001;27:417–421. doi: 10.1038/86920. [DOI] [PubMed] [Google Scholar]
- 111.Wu X., Ranganathant V., Weisman D.S., Helne W.F., Ciccone D.N., O’Neill T.B., Crick K.E., Pierce K.A., Lane W.S., Rathbun G., et al. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature. 2000;405:477–482. doi: 10.1038/35013089. [DOI] [PubMed] [Google Scholar]
- 112.Kim J.H., Grosbart M., Anand R., Wyman C., Cejka P., Petrini J.H.J. The Mre11-Nbs1 Interface Is Essential for Viability and Tumor Suppression. Cell Rep. 2017;18:496–507. doi: 10.1016/j.celrep.2016.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ebi H., Matsuo K., Sugito N., Suzuki M., Osada H., Tajima K., Ueda R., Takahashi T. Novel NBS1 heterozygous germ line mutation causing MRE11-binding domain loss predisposes to common types of cancer. Cancer Res. 2007;67:11158–11165. doi: 10.1158/0008-5472.CAN-07-1749. [DOI] [PubMed] [Google Scholar]
- 114.Varon R., Vissinga C., Platzer M., Cerosaletti K.M., Chrzanowska K.H., Saar K., Beckmann G., Seemanová E., Cooper P.R., Nowak N.J., et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/S0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 115.Watanabe T., Nobusawa S., Lu S., Huang J., Mittelbronn M., Ohgaki H. Mutational inactivation of the nijmegen breakage syndrome gene (NBS1) in glioblastomas is associated with multiple TP53 mutations. J. Neuropathol. Exp. Neurol. 2009;68:210–215. doi: 10.1097/NEN.0b013e31819724c2. [DOI] [PubMed] [Google Scholar]
- 116.Huang J., Grotzer M.A., Watanabe T., Hewer E., Pietsch T., Rutkowski S., Ohgaki H. Mutations in the nijmegen breakage syndrome gene in medulloblastomas. Clin. Cancer Res. 2008;14:4053–4058. doi: 10.1158/1078-0432.CCR-08-0098. [DOI] [PubMed] [Google Scholar]
- 117.Wang Y., Hong Y., Li M., Long J., Zhao Y.P., Zhang J.X., Li Q., You H., Tong W.M., Jia J.D., et al. Mutation inactivation of Nijmegen breakage syndrome gene (NBS1) in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. PLoS ONE. 2013;8:e82426. doi: 10.1371/journal.pone.0082426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Saito Y., Komatsu K. Functional role of NBS1 in radiation damage response and translesion DNA synthesis. Biomolecules. 2015;5:1990–2002. doi: 10.3390/biom5031990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Varon R., Reis A., Henze G., Einsiedel H.G.V., Sperling K., Seeger K. Mutations in the Nijmegen Breakage Syndrome gene (NBS1) in childhood acute lymphoblastic leukemia (ALL) Cancer Res. 2001;61:3570–3572. [PubMed] [Google Scholar]
- 120.Desjardins S., Beauparlant J.C., Labrie Y., Ouellette G., Durocher F. Variations in the NBN/NBS1 gene and the risk of breast cancer in non-BRCA1/2French Canadian families with high risk of breast cancer. BMC Cancer. 2009;9:181. doi: 10.1186/1471-2407-9-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ziólkowska I., Mosor M., Wierzbicka M., Rydzanicz M., Pernak-Schwarz M., Nowak J. Increased risk of larynx cancer in heterozygous carriers of the I171V mutation of the NBS1 gene. Cancer Sci. 2007;98:1701–1705. doi: 10.1111/j.1349-7006.2007.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hebbring S.J., Fredriksson H., White K.A., Maier C., Ewing C., McDonnell S.K., Jacobsen S.J., Cerhan J., Schaid D.J., Ikonen T., et al. Role of the Nijmegen breakage syndrome 1 gene in familial and sporadic prostate cancer. Cancer Epidemiol. Biomarkers Prev. 2006;15:935–938. doi: 10.1158/1055-9965.EPI-05-0910. [DOI] [PubMed] [Google Scholar]
- 123.Shimada H., Shimizu K., Mimaki S., Sakiyama T., Mori T., Shimasaki N., Yokota J., Nakachi K., Ohta T., Ohki M. First case of aplastic anemia in a Japanese child with a homozygous missense mutation in the NBS1 gene (I171V) associated with genomic instability. Hum. Genet. 2004;115:372–376. doi: 10.1007/s00439-004-1155-1. [DOI] [PubMed] [Google Scholar]
- 124.Masi A., Antoccia A. NBS1 Heterozygosity and Cancer Risk. Curr. Genom. 2008;9:275–281. doi: 10.2174/138920208784533610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gao P., Ma N., Li M., Tian Q.B., Liu D.W. Functional variants in NBS1 and cancer risk: Evidence from a meta-analysis of 60 publications with 111 individual studies. Mutagenesis. 2013;28:683–697. doi: 10.1093/mutage/get048. [DOI] [PubMed] [Google Scholar]
- 126.Yamamoto Y., Miyamoto M., Tatsuda D., Kubo M., Nakagama H., Nakamura Y., Satoh H., Matsuda K., Watanabe T., Ohta T. A rare polymorphic variant of NBS1 reduces DNA repair activity and elevates chromosomal instability. Cancer Res. 2014;74:3707–3715. doi: 10.1158/0008-5472.CAN-13-3037. [DOI] [PubMed] [Google Scholar]
- 127.Fang W., Qiu F., Zhang L., Deng J., Zhang H., Yang L., Zhou Y., Lu J. The functional polymorphism of NBS1 p.Glu185Gln is associated with an increased risk of lung cancer in Chinese populations: Case-control and a meta-analysis. Mutat. Res. 2014;770:61–68. doi: 10.1016/j.mrfmmm.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 128.He Y.Z., Chi X.S., Zhang Y.C., Deng X.B., Wang J.R., Lv W.Y., Zhou Y.H., Wang Z.Q. NBS1 Glu185Gln polymorphism and cancer risk: Update on current evidence. Tumor Biol. 2014;35:675–687. doi: 10.1007/s13277-013-1093-4. [DOI] [PubMed] [Google Scholar]
- 129.Zheng J., Zhang C., Jiang L., You Y., Liu Y., Lu J., Zhou Y. Functional NBS1 polymorphism is associated with occurrence and advanced disease status of nasopharyngeal carcinoma. Mol. Carcinog. 2011;50:689–696. doi: 10.1002/mc.20803. [DOI] [PubMed] [Google Scholar]
- 130.Mosor M., Ziółkowska I., Pernak-Schwarz M., Januszkiewicz-Lewandowska D., Nowak J. Association of the heterozygous germline I171V mutation of the NBS1 gene with childhood acute lymphoblastic leukemia. Leukemia. 2006;20:1454–1456. doi: 10.1038/sj.leu.2404285. [DOI] [PubMed] [Google Scholar]
- 131.di Masi A., Viganotti M., Polticelli F., Ascenzi P., Tanzarella C., Antoccia A. The R215W mutation in NBS1 impairs γ-H2AX binding and affects DNA repair: Molecular bases for the severe phenotype of 657del5/R215W Nijmegen breakage syndrome patients. Biochem. Biophys. Res. Commun. 2008;369:835–840. doi: 10.1016/j.bbrc.2008.02.129. [DOI] [PubMed] [Google Scholar]
- 132.Uversky V.N. The alphabet of intrinsic disorder. Intrinsically Disord. Proteins. 2013;1:e24684. doi: 10.4161/idp.24684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tseng S.F., Chang C.Y., Wu K.J., Teng S.C. Importin KPNA2 is required for proper nuclear localization and multiple functions of NBS1. J. Biol. Chem. 2005;280:39594–39600. doi: 10.1074/jbc.M508425200. [DOI] [PubMed] [Google Scholar]
- 134.Yanagihara H., Kobayashi J., Tateishi S., Kato A., Matsuura S., Tauchi H., Yamada K., Takezawa J., Sugasawa K., Masutani C., et al. NBS1 Recruits RAD18 via a RAD6-like Domain and Regulates Pol η-Dependent Translesion DNA Synthesis. Mol. Cell. 2011;43:788–797. doi: 10.1016/j.molcel.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 135.Varela I., Tarpey P., Raine K., Huang D., Ong C.K., Stephens P., Davies H., Jones D., Lin M.L., Teague J., et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vilar E., Bartnik C.M., Stenzel S.L., Raskin L., Ahn J., Moreno V., Mukherjee B., Iniesta M.D., Morgan E.A., Rennert G., et al. MRE11 deficiency increases sensitivity to poly(ADP-ribose) polymerase inhibition in microsatellite unstable colorectal cancers. Cancer Res. 2011;71:2632–2642. doi: 10.1158/0008-5472.CAN-10-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bian L., Meng Y., Zhang M., Li D. MRE11-RAD50-NBS1 complex alterations and DNA damage response: Implications for cancer treatment. Mol. Cancer. 2019;18:1–14. doi: 10.1186/s12943-019-1100-5. [DOI] [PMC free article] [PubMed] [Google Scholar]