Abstract
NSUN5, a gene encodes a cytosine-5 RNA methyltransferase, is rarely mentioned in cancers. Our study is the first one to evaluate the role of NSUN5 in the progression of colorectal cancer. Data from TCGA was used to show the different expression of NSUN5 between CRC tumor tissues and adjacent normal ones. The NSUN5 expression in the tissue microarray was detected by immunohistochemistry (IHC). qRT-PCR was conducted for NSUN5 expression examination in CRC cell lines. Cell proliferation was analyzed by the Celigo machine. GESA and correlation analysis were performed to reveal the possible underlying mechanism. The effects of NSUN5 expression on CRC cell behavior in vitro were analyzed by flow cytometry and β-galactosidase staining. The expression of cell-cycle related proteins were evaluated by western blot. Subcutaneously implanted tumor model was carried out for animal experiment. NSUN5 expression was up-regulated in CRC tumor tissues and cells, and associated with advanced tumor stages (III, IV). NSUN5 could promote cell proliferation, trigger cell cycle arrest in vitro and boost tumor growth in vivo. In addition, knockdown of NSUN5 could lead to a higher expression of Rb and a lower expression of CDK4, CDK6, p-Rb and CCNE1, but made no difference on P21, Bcl-2, caspase3 and C-Caspase3 of CRC cells. Taken together, we identify NSUN5 as a promoter in CRC development via cell cycle regulation.
Keywords: NUSN5, new targets, colorectal cancer, cell cycle, cell proliferation, tumor growth
Introduction
The clinical therapeutic of colorectal cancer (CRC) has been improving over the years mainly due to molecular-target therapies. However, the incidence and mortality rate of CRC are still major concerns in the prevention and control of malignant tumors in China and worldwide. In addition, a younger trend in the affected population, drug resistance and limited therapeutic targets are urgent to be solved [1,2]. Although few patients, such as advanced patients with microsatellite instability-high (MSI-h), can benefit from immunotherapies [3,4], finding more driven genes is still an irreplaceable strategy. Compare to study discovered therapeutic targets in-depth or apply targets in other tumors to CRC, identifying novel genes is also significant.
NSUN5 is a member of the NOL1/Nop2/sun protein family. The family is highly conserved from archaea to eukaryotes and has close contact with RNA methylation because of their S-adenosyl methionine binding-domain and their relationship to Escherichia coli Sun/Fmu protein that methylates C967 in 16S ribosomal RNA (rRNA) [5,6]. There are six members in NSUN family: Nsun2 (Misu), Nsun3, Nsun4, Nsun5 (Wbscr20, Wbscr20a), Nsun6 (NOPD1) and Nsun7. These members are currently reported to function as an RNA methyltransferase. Among them, NSUN2, as the most studied member, was vital for mediating m5C installation on message RNA (mRNA) and involved in transfer RNA (tRNA) modification, neurocognitive condition and stem cell differentiation [7,8]. What’s more, NSUN2 was associated with kinds of cancer progression by promoting cell proliferation and could be a potential marker for prognosis [7,9]. And NSUN6, apart from a regulator of tRNA [10], recently was also reported to be involved in promoting bone metastasis [11]. NSUN4 played an essential role in translation and assembly of mitochondria by MTERF4-NSUN4 protein complex [12,13] and NSUN3 also focused on mitochondrial tRNA modifications and regulated embryonic stem cell differentiation [14-16]. While NSUN7 mainly functioned in germ cell motility and NSUN7 mutation might result in male sterility or subfertility [17,18]. As for NSUN5, most current researches are still based on its function as an RNA methyltransferase. However, its role in other biological processes, especially the tumor progression, is rarely mentioned.
Therefore, we conducted this study to elucidate the involvement of NSUN5 in CRC, showing that its role in CRC progression. We found that NSUN5 was highly expressed in CRC tissue compared to normal ones, both in TCGA data and our tissue microarray. Then, cell proliferation was hugely inhibited after the knockdown of NSUN5. Accompany with this, cell cycle arrest was enhanced, but not apoptosis and senescence. By GESA, NSUN5 was involved in cell cycle and had relevance with cell-cycle related genes and the relevance was verified by in RKO and HT29 cells transfected with shNC and shNSUN5. Finally, knockdown of NSUN5 also inhibited tumor growth in vivo. In all, this research suggested that NSUN5 promoted CRC proliferation and progression mainly via cell cycle regulation.
Materials and methods
Integrative analysis of TCGA and GESA
mRNA-seq data of CRC patients was downloaded from The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) [19]. The NSUN5 expression of tumor tissues in all CRC patients were compared with that of paired normal tissues. According to the tumor stage, CRC patients were divided into four subgroups (stage I, II, III, IV), each of which was compared with the expression of NSUN5. Gene set enrichment analysis (GSEA) is performed by GSEA 3.0 software to determine whether a predefined set of genes is significantly different between the two groups: high and low NSUN5.
Tissue microarray and immunohistochemistry
Paraffin-embedded tissues were deparaffinized at 60°C about 2 hours, hydrated in ethylalcohol, and blocked by 3% H2O2 for 30 minutes. After put into citric acid retrievals at 100°C for 15 minutes and blockage with 5% bovine serum albumin for 1 hour, the slides were incubated with primary antibody at 4°C overnight, then incubated with HRP labeled secondary antibody (YESEN, Shanghai, China) at room temperature for 1 h. Finally, the slides were stained with diaminobenzidine (DAB, Gene Tech, Shanghai, China) and the results were captured using Nikon Eclipse 80i microscope.
NSUN5 staining was evaluated based on intensity scores (0, negative; 1, weak; 2, moderate; and 3, strong staining) and the percentage scores (1, ≤ 25%; 2, 25%-50%; 3, 50%-75%; 4, > 75%). The positive cell percentage and the staining intensity of the target cells were scored at each tissue point on the tissue microarray. The products of the two were viewed as the final scores: 0, negative (-); 1 to 4, weak (+), 5 to 8, moderate (++), and 9 to 12, strong (+++).
Cell culture and reagents
CRC cell lines including RKO, HT29 and HEK293 cells were purchased from ATCC company (Manassas, VA, USA). RKO, HT29, and HEK293 were cultured in RPMI1640, Macoy 5A and DMEM medium (SIGMA, New York, USA) respectively supplemented with 10% fetal bovine serum (Life Technologies, Shanghai, China), and antibiotics (100 μg/mL streptomycin and 100 U/mL penicillin) in a 5% CO2 incubator at 37°C.
Cell transfection
The lentivirus contain NSUN5 shRNA (shNSUN5) and corresponding controls (shNC) were synthesis from Genechem (Shanghai, China), the sequence (5’-CCGGCCCCAATGAGGTCCTGTTGGACTCGAGTCCAACAGGTCCTCATTCCGGTTTTT-3’) led to the largest decrease was selected. The lentivirus were transfected into RKO and HT-29 by a Lipofectamine 3000 (Invitrogen, New York, USA) according to the manufacturer’s specification. After transfection for 48 hours, the knockdown efficiency was evaluated by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and Western blot (WB).
Cell proliferation
CRC cells were plated at a density of 2000/well. From the second day after the boarding, Celigo Image Cytometer checked the plate once a day in the same view and continuously for 3-5 days; The numbers of fluorescent cells were accurately calculated and photoed in each scanning.
Cell cycle, apoptosis and senescence detection
To analyze the cell cycle, cells transfected with shNSUN5 and shNC were digested and washed twice with precooled D-Hanks and fixed with 75% precooled ethanol. Preparation of cell staining solution: 40× PI (2 mg/mL):100× RNase (10 mg/mL):1× D-Hanks = 25:10:1000. Then, the cells were washed with D-Hanks and resuspended in 600 μl of staining solution in the dark at room temperature for 20 min. Subsequently, the cell cycle analysis was performed by flow cytometry using a Beckmen LX instrument.
For the apoptosis analysis, an APC-Annexin V Apoptosis Detection Kit (Biolegend, New York, USA) was used according to the manufacturer’s instructions. Cells transfected with shNSUN5 and shNC were digested and washed twice with precooled PBS and resuspended in 1× binding buffer. Then, the cells were stained with 5 μl Annexin V-APC and 5 μl propidium iodide (PI) in the dark for 15 min at room temperature. The apoptosis rate was measured by flow cytometry using a Beckmen LX instrument.
The cell senescence was detected by the Cellular senescence β-galactosidase staining kit (Beyotime, Shanghai, China) and performed according to the protocol.
Subcutaneously implanted tumor model
The animal experiments were approved by the Animal Ethics Committee of Second Hospital, School of Medicine, Zhejiang University. HT29-shNC and HT29-shNSUN5 cells in logarithmic growth phase were digested and suspended in a mixture of PBS and Matrigel (ratio 3:1) to form a single cell suspension. The cell concentration was adjusted to 8×106/200 μl. Eight BALB/C female nude mice (4-6 weeks old; 18-20 g) were divided into two groups with 4 mice in each group. The 200 ul of homogeneous suspension of HT29-shNC and HT29-shNSUN5 cells (containing 8×106 cells) was injected subcutaneously into the right armpit skin of nude mice. The size of subcutaneous tumors was measured with vernier caliper every 3 days. The length (a) and shortdiameter (b) of the transplanted tumors were recorded. The volume of the transplanted tumors was calculated and the growth curve was drawn according to the volume formula: V (mm3) = (a×b2)/2. On the 28st day after inoculation, nude mice were killed under anesthesia, and the tumors were removed and weighed.
Western blot (WB)
The protein of tissue or cells were extracted using radio immunoprecipitation assay (RIPA) buffer (Beyotime, Shanghai, China) and the protein concentration was measured by bicinchoninic (BCA) kit (Thermofisher, New York, USA). Protein samples extracted from CRC cells were separated via 10% SDS-polyacrylamide gel electrophoresis, then transferred to polyvinylidene difluoride (PVDF) membrane and blocked with 5% non-fat milk. The membranes were incubated at 4°C overnight with primary antibody, followed by secondary HRP-linked antibody for 2 hours at room temperature. At last, the signal was detected by enhanced chemiluminescence (ECL) detection kit (Thermofisher, New York, USA). The primary antibodies included in this study were in Table S1.
Quantitative RT-PCR
Total RNAs from cells and tissues were obtained by using a Total RNA extraction kit (Solarbio). The Takara PrimeScript RT Master Mix kit (Takara Biotechnology, China) was used to conduct reverse transcription. Real-time PCR analysis was performed by the SYBR Premix Ex Taq II kit (Takara Biotechnology) and Applied Biosystems 7500 Fast Real-Time PCR System. GAPDH were used for endogenous reference. Experiments were repeated three times and the ΔΔCt method was applied to calculate the relative expression level of mRNA. The primers (Genechem) used are as following: NSUN5: Forward, TGTACTCCAGCAACTTCCAGA; Reverse, CTTCCAACAGGTCCTCATTCC. GAPDH: Forward, TGACTTCAACAGCGACACCCA; Reverse, CACCCTGTTGCTGTAGCCAAA.
Statistical analysis
The results were exhibited as mean ± SD. Student’s t-test or one-way ANOVA was used to analyze data from two or multiple groups, respectively. Such as the expression of NSUN5 between normal and tumor tissues, the expression of NSUN5 between cell lines transfected with lentivirus of sh-NSUN5 or sh-NC. P<0.05 was considered statistically significant. All the experiments were repeated at least three times.
Results
NSUN5 was up-regulated in CRC
Preliminary data from the TCGA database showed that the expression of NSUN5 was significantly increased in CRC tissues as compared with adjacent normal tissues (P<0.001, Figure 1A). Moreover, CRC patients in stage III-IV exhibited a higher NSUN5 expression than the patients in stage I-II (P<0.05, Figure 1B) and seemly a shorter disease-free survival (DFS) (the data were not shown). Furthermore, we validated the expression difference in our tissue microarray, which contained 30 pairs of tumor and adjacent normal tissues. The expression of NSUN5 was evaluated by IHC (Figure 1C) and the results showed that 19 tumor tissues were strong and 11 were moderate while 9 normal tissues were strong, 16 were moderate and 5 were weak (Table 1). The results identified that NSUN5 was markedly up-regulated in CRC (P<0.001, Figure 1D).
Figure 1.
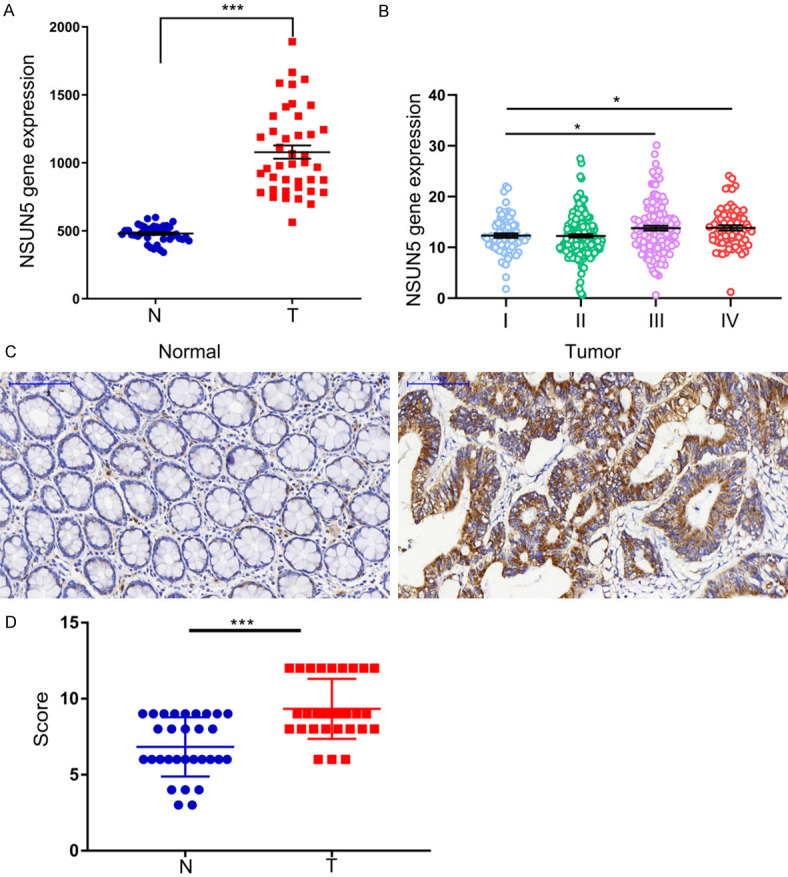
The expression of NSUN5 was increased in CRC. A. From TCGA database, the expression of NSUN5 was increased in CRC tissues. B. From TCGA database, the expression of NSUN5 in different stages of CRC patients. C. From the CRC tissue microarray, the protein expression of NSUN5 in 30 pairs of CRC tissues (T) and adjacent normal tissues (AT) were measured by immunohistochemical analysis. The representative image was shown. D. The intensity of IHC was scored to reflect the expression of NSUN5 and the scores of the tumor tissues were significantly higher than the normal ones. The Data are shown as mean ± SD. *P<0.05, ***P<0.001.
Table 1.
The expression of NSUN5 of tumor (T) and normal (N) tissues in the tissue microarray
| Numbers | IHC intensity scores | |
|---|---|---|
|
| ||
| Tumor | Normal | |
| 1 | 12 | 4 |
| 2 | 8 | 6 |
| 3 | 8 | 6 |
| 4 | 9 | 6 |
| 5 | 8 | 6 |
| 6 | 8 | 6 |
| 7 | 8 | 9 |
| 8 | 9 | 9 |
| 9 | 12 | 8 |
| 10 | 9 | 6 |
| 11 | 9 | 6 |
| 12 | 12 | 9 |
| 13 | 9 | 4 |
| 14 | 9 | 9 |
| 15 | 12 | 9 |
| 16 | 8 | 9 |
| 17 | 9 | 6 |
| 18 | 12 | 4 |
| 19 | 12 | 8 |
| 20 | 12 | 6 |
| 21 | 6 | 8 |
| 22 | 9 | 9 |
| 23 | 9 | 8 |
| 24 | 9 | 8 |
| 25 | 6 | 3 |
| 26 | 9 | 6 |
| 27 | 9 | 6 |
| 28 | 12 | 9 |
| 29 | 12 | 9 |
| 30 | 6 | 3 |
| T value | 5.876 | |
| P value | <0.0001 | |
Knockdown of NSUN5 inhibited cells growth in vitro
By qRT-PCR, NSUN5 was identified as a highly expressed gene in four CRC cell lines, including RKO, HCT 116, LoVo and HT-29 (Figure 2A). To evaluate the function of NSUN5 in CRC, HT-29 and RKO were chosen to be transfected with shNSUN5 and shNC. The efficiency of knockdown was examed by qRT-PCR (P<0.01, Figure 2B) and WB (Figure 2C). Celigo Image Cytometer was applied to count the cell and take photo of fluorescence images. The results showed that the cell proliferation of the RKO-shNSUN5 was obviously slower than that of RKO-shNC and fluorescence images of cells in same views from day1 to day5 were taken photo (P<0.001, Figure 2D).
Figure 2.
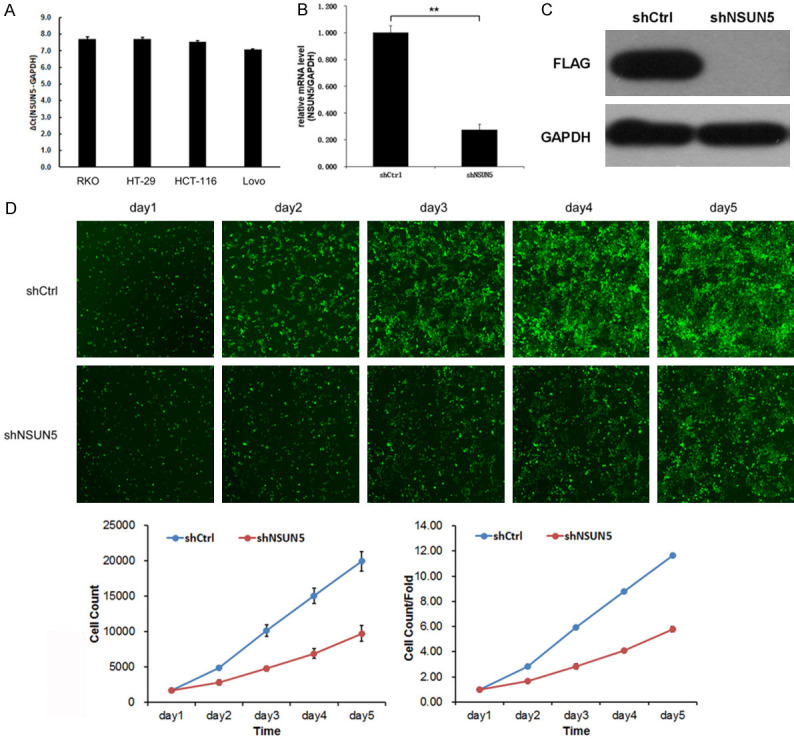
NSUN5 knockdown inhibited cell proliferation in vitro. A. The expression of NSUN5 in CRC cell lines (RKO, HT-29, HCT-116, Lovo) was detected by qRT-PCR assay. B, C. HT-29 cells were transfected with lentivirus of sh-NSUN5 or sh-NC, and NSUN5 expression was identified by qRT-PCR and WB. D. Cell proliferation was examined by cell-counting machine Celigo, and the fluorescent image of RKO-shNSUN5 and RKO-shNC was taken photo by the machine from day1 to day5. The two curves showed the cell number and cell number/fold change over time of HT-29-shNSUN5 and HT-29-shNC. The Data are shown as mean ± SD. *P<0.05, ***P<0.001.
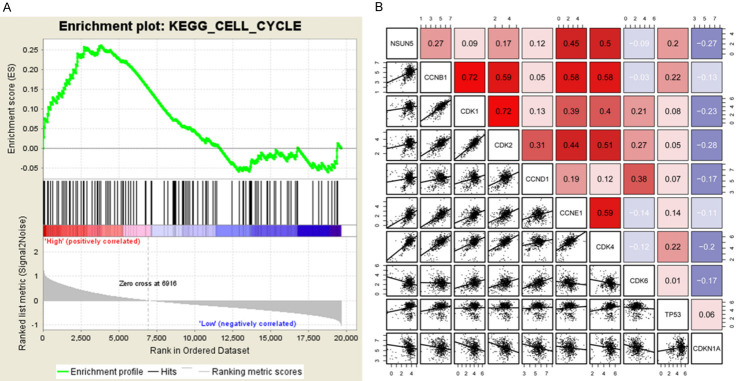
NSUN5 was associated with cell cycle according to bioinformatic analysis
NUSN5 was rarely mentioned in cancers, so there were few researches about the mechanism that NSUN5 promoted cancer progression. To elucidate the mechanism of NSUN5 to promote cell proliferation, we searched for the pathways NSUN5 might involved in via KEGG enrichment analysis and the result showed NSUN5 was markedly cell cycle-related (Figure 3A). Further, we performed the correlation analysis between NSUN5 and cell cycle genes and the result showed NSUN5 had a positive relation with CDK1/2/4, CCNB1, CCNE1, CCND1 and TP53, especially CDK4 and CCNE1 while a negative relation with CDK6 and CDKN1A (Figure 3B).
Figure 3.
The correlation between NSUN5 and cell cycle pathways. A. GSEA analysis showed NSUN5 was enriched in cell cycle-related genes in CRC. B. Correlation analysis showed NSUN5 was correlated with CDK4, CCNE1, CCND1, CDK2, CDK1, CCNB1, TP53. GSEA, Gene set enrichment analysis.
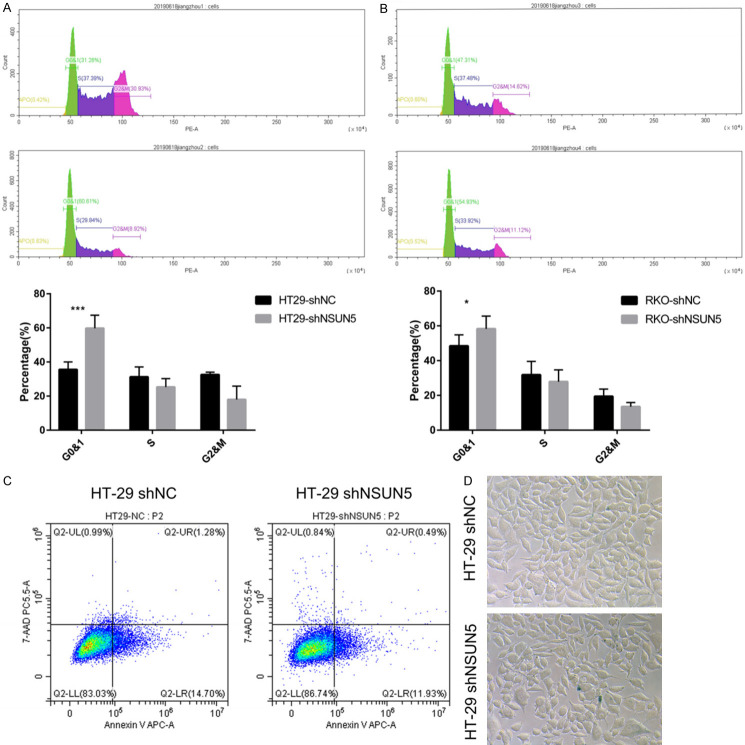
Knockdown of NSUN5 triggered CRC cell cycle arrest
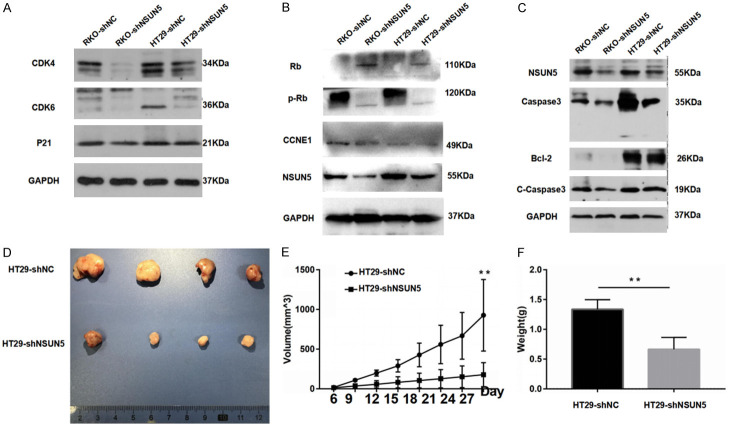
Next, we explored the mechanism behind the promotion. According to the clue from the GSEA, we first supposed that the reason was related with the cell cycle, while the apoptosis, necrosis and senescence were also examed. After knockdown of NSUN5, both HT-29 and RKO cells showed significant cell cycle arrest, especially HT-29. The flow cytometry analysis showed that compared to the HT-29-NC, the percentage of HT-29-shNSUN5 in G0/G1 phase was markedly increased (P<0.01, Figure 4A), but the percentage in S and G2/M phase was decreased (P<0.05, Figure 4A). The consistent result was also observed in the RKO-shNSUN5 and RKO-shNC (P<0.05, Figure 4B). In addition, we measured the effect of NSUN5 knockdown on HT-29 and RKO cell apoptosis and senescence. There was no difference in apoptosis and necrosis after NSUN5 knockdown, as determined by flow cytometry analysis (Figure 4C). Downregulation of NSUN5 also made no difference in HT-29 cells senescence, as suggested by β-galactosidase staining (Figure 4D). Then, we examined the expression of cell cycle associated proteins in RKO and HT29 cells transfected with shNC and shNSUN5. First, a significantly higher expression of Rb while a lower expression of p-Rb were observed. Further, a decline expression of CCNE1, CDK4 and CDK6 was acquired after knockdown, but the expression of P21 had no difference (Figures 5A, 5B, S1, S3, S4). As for apoptosis associated proteins that caspase3, cleaved-caspase3 and Bcl-2 did not show a difference (Figures 5C, S1, S2). The results proved that the cell cycle arrest was a considerable reason why NSUN5 promoted CRC cell growth.
Figure 4.
NSUN5 knockdown triggered cell cycle arrest. A, B. Representative flow cytometry analysis of the cell cycle distribution of RKO and HT29 cells transfected with shNC and shNSUN5. C. Apoptosis rates of HT29 cells transfected with shNC and shNSUN5, as determined by flow cytometry. D. The cell senescence status of HT29 cells transfected with shNC and shNSUN5, as detected by β-Galactosidase staining. The Data are shown as mean ± SD. *P<0.05, ***P<0.001.
Figure 5.
Knockdown of NSUN5 inhibited cell cycle-related signaling activity and cell proliferation in vivo. A. Western blotting detection of CDK4, CDK6 and P21 expression in RKO and HT29 cells transfected with shNC and shNSUN5. B. Western blotting detection of Rb, p-Rb and CCNE1 expression in RKO and HT29 cells transfected with shNC and shNSUN5. C. The expression of apoptosis proteins that caspase3, cleaved-caspase3 and Bcl-2 was tested by western blot assay in RKO and HT29 cells transfected with shNC and shNSUN5. D-F. Subcutaneously implanted tumor model in nude mouse was used for evaluating the effect of NSUN5 on tumor growth. D. The image of subcutaneous xenograft tumors of HT29 cells transfected with shNC and shNSUN5. E. The curve of xenograft volume over time. F. The comparison of tumor weight between the two groups. The Data are shown as mean ± SD. **P<0.01.
Knockdown of NSUN5 showed inhibiting effect on cell growth in vivo
To evaluate the oncogenic role of NSUN5 in vivo, we observed the effect of NSUN5 on tumor growth based on a subcutaneously implanted tumor model in nude mouse. Tumor volume was recorded every 3 days, and the mice were finally sacrificed after 28 days. We verified that NSUN5 knockdown effectively slowed the tumor growth (Figure 5D) and reduced tumor volume (P<0.01, Figure 5E) and weight (P<0.01, Figure 5F).
Discussion
No matter the incidence or mortality, CRC is a major issue to be addressed. Apart from traditional chemo or radiotherapies, and considering emerging immunotherapy (such as immune checkpoint blockade) is currently just for minority patients (such as microsatellite instability-high, MSI) [20], molecular target therapies remain the current irreplaceable area for the breakthrough of CRC treatment. However, current targets are limited to oncogenes or antiangiogenesis, such as EGFR or VEGF [21,22]. Identifying new targets may be a wise strategy to breakthrough the bottleneck. Here, we found a rarely-reported target named NSUN5, which is an RNA methyltransferase. All the time, NSUN5 was considered to be linked to the nerve system, but not tumor, and its function was about cerebral cortex developing, agenesis and hypomyelination of the corpus callosum and cognitive deficits [23,24]. Until recently, epigenetic loss of NSUN5 was proved to drive a stress adaptive translational program in glioma by targeting ribosomes [25].
Here, we are the first one who confirmed NSUN5 was an oncogene in CRC. Data from TCGA showed that NSUN5 was highly expressed in CRC tumor tissues and associated with advanced tumor stages, which revealed its role in promoting CRC development. These differences have not been reported before. Then we verified the results by the tissue microarray. Also we found that several CRC cell lines were highly expressed NSUN5. So we knocked down NSUN5 in cell lines, which resulted in an apparent reduction in cells proliferation but an enhancement in cell cycle arrest, but not apoptosis or senescence. Therefore, we inferred that NSUN5 promoted CRC promotion via a cell cycle-related pathway. The analysis of GSEA and correlation analysis were consistent, which suggested NSUN5 was linked to cell cycle-related genes such as CDK4 and CCNE1. Such correlations were also verified by WB in cell lines. Futhermore, a strong correlation between NSUN5 and Rb was identified while P21 was little influenced. The results suggested that NSUN5 promoted CRC cells proliferation mainly through Rb-CDKs signal transduction. Finally, in vivo, CRC cell lines knocked down of NSUN5 showed a rather slower proliferation than control groups.
As we all know in recent years, the progression of tumors may not lay at only genes, but also epigenetic modification, such as mRNA, even long-non-coding RNA (lncRNA) [26]. Growing evidences led us to pay attention to the dysfunction of RNA modification [27,28]. NSUN5 is just such a member of RNA methyltransferases. In Janin’s model, NSUN5 showed tumor-suppressor function in vivo glioma models, and an unmethylated status of NSUN5 led to long-term survival in glioma patients [25]. While in our study, NSUN5 promoted CRC progression. The difference may due to mechanisms in two kinds of tumors. In glioma, the silencing of NSUN5 by DNA methylation affected never system, which was consistent with other researches [27,29]. While in the CRC microenvironment, NSUN5 promoted progression through the cell cycle. NSUN5 was also reported to be associated with cell growth before. The study investigated how NSUN5 deficiency influenced the development of the cerebral cortex revealed that the cortical of NSUN5-KO mice was thinner than wild type ones due to an abnormal laminar organization and a shorter processes of pyramidal cells [29]. Methylation of NSUN5 could also modulate organismal lifespan and enhance stress resistance in a conserved mechanism [5]. In our study, we also detected the senescence, but not a significant difference.
In conclusion, we proved firstly that NSUN5 was a promoter in the progression of CRC mainly through cell cycle regulation. The decrease of NSUN5 expression resulted in a significant cell cycle arrest. Therefore, we supposed that NSUN5 could be a potential target in CRC.
Acknowledgements
This study was funded by the National Natural Science Foundation of China (Grant no. 81902629, PC) and the National Natural Science Foundation of China (Grant no. 81702803, GMH).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.DeSantis CE, Miller KD, Goding Sauer A, Jemal A, Siegel RL. Cancer statistics for African Americans, 2019. CA Cancer J Clin. 2019;69:211–233. doi: 10.3322/caac.21555. [DOI] [PubMed] [Google Scholar]
- 2.Punt CJ, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol. 2017;14:235–246. doi: 10.1038/nrclinonc.2016.171. [DOI] [PubMed] [Google Scholar]
- 3.Llosa NJ, Luber B, Siegel N, Awan AH, Oke T, Zhu Q, Bartlett BR, Aulakh LK, Thompson ED, Jaffee EM, Durham JN, Sears CL, Le DT, Diaz LA Jr, Pardoll DM, Wang H, Housseau F, Anders RA. Immunopathologic stratification of colorectal cancer for checkpoint blockade immunotherapy. Cancer Immunol Res. 2019;7:1574–1579. doi: 10.1158/2326-6066.CIR-18-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alves CA, Vicente ED, Vicente AMP, Rienda IC, Tomé M, Querol X, Amato F. Inactivation of proprotein convertases in t cells inhibits PD-1 expression and creates a favorable immune microenvironment in colorectal cancer. Sci Total Environ. 2019;79:5008–5021. doi: 10.1158/0008-5472.CAN-19-0086. [DOI] [PubMed] [Google Scholar]
- 5.Dominissini D, Rechavi G. 5-methylcytosine mediates nuclear export of mRNA. Cell Res. 2017;27:717–719. doi: 10.1038/cr.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma S, Yang J, Watzinger P, Kotter P, Entian KD. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res. 2013;41:9062–9076. doi: 10.1093/nar/gkt679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Li A, Sun BF, Yang Y, Han YN, Yuan X, Chen RX, Wei WS, Liu Y, Gao CC, Chen YS, Zhang M, Ma XD, Liu ZW, Luo JH, Lyu C, Wang HL, Ma J, Zhao YL, Zhou FJ, Huang Y, Xie D, Yang YG. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol. 2019;21:978–990. doi: 10.1038/s41556-019-0361-y. [DOI] [PubMed] [Google Scholar]
- 8.Okabe M, Taga K, Yoshino A, Yamamoto Y, Taneda A, Shinoda S, Kanamasa S, Shervani Z. Mammalian NSUN2 introduces 5-methylcytidines into mitochondrial tRNAs. Eur Biophys J. 2019;47:8734–8745. doi: 10.1093/nar/gkz575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alshaker H, Wang Q, Brewer D, Pchejetski D. Transcriptome-wide effects of sphingosine kinases knockdown in metastatic prostate and breast cancer cells: implications for therapeutic targeting. Front Pharmacol. 2019;10:303. doi: 10.3389/fphar.2019.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alshaker H, Wang Q, Brewer D, Pchejetski D. Transcriptome-wide effects of sphingosine kinases knockdown in metastatic prostate and breast cancer cells: implications for therapeutic targeting. Front Pharmacol. 2019;10:303. doi: 10.3389/fphar.2019.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Li H, Long T, Dong H, Wang ED, Liu RJ. Archaeal NSUN6 catalyzes m5C72 modification on a wide-range of specific tRNAs. Nucleic Acids Res. 2019;47:2041–2055. doi: 10.1093/nar/gky1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C, Wang S, Xing Z, Lin A, Liang K, Song J, Hu Q, Yao J, Chen Z, Park PK, Hawke DH, Zhou J, Zhou Y, Zhang S, Liang H, Hung MC, Gallick GE, Han L, Lin C, Yang L. A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat Cell Biol. 2017;19:106–119. doi: 10.1038/ncb3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camara Y, Asin-Cayuela J, Park CB, Metodiev MD, Shi Y, Ruzzenente B, Kukat C, Habermann B, Wibom R, Hultenby K, Franz T, Erdjument-Bromage H, Tempst P, Hallberg BM, Gustafsson CM, Larsson NG. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011;13:527–539. doi: 10.1016/j.cmet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Spahr H, Habermann B, Gustafsson CM, Larsson NG, Hallberg BM. Structure of the human MTERF4-NSUN4 protein complex that regulates mitochondrial ribosome biogenesis. Proc Natl Acad Sci U S A. 2012;109:15253–15258. doi: 10.1073/pnas.1210688109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haag S, Sloan KE, Ranjan N, Warda AS, Kretschmer J, Blessing C, Hubner B, Seikowski J, Dennerlein S, Rehling P, Rodnina MV, Hobartner C, Bohnsack MT. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 2016;35:2104–2119. doi: 10.15252/embj.201694885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakano S, Suzuki T, Kawarada L, Iwata H, Asano K, Suzuki T. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNA(Met) Nat Chem Biol. 2016;12:546–551. doi: 10.1038/nchembio.2099. [DOI] [PubMed] [Google Scholar]
- 17.Van Haute L, Dietmann S, Kremer L, Hussain S, Pearce SF, Powell CA, Rorbach J, Lantaff R, Blanco S, Sauer S, Kotzaeridou U, Hoffmann GF, Memari Y, Kolb-Kokocinski A, Durbin R, Mayr JA, Frye M, Prokisch H, Minczuk M. Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat Commun. 2016;7:12039. doi: 10.1038/ncomms12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khosronezhad N, Hosseinzadeh Colagar A, Mortazavi SM. The Nsun7 (A11337)-deletion mutation, causes reduction of its protein rate and associated with sperm motility defect in infertile men. J Assist Reprod Genet. 2015;32:807–815. doi: 10.1007/s10815-015-0443-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khosronezhad N, Colagar AH, Jorsarayi SG. T26248G-transversion mutation in exon7 of the putative methyltransferase Nsun7 gene causes a change in protein folding associated with reduced sperm motility in asthenospermic men. Reprod Fertil Dev. 2015;27:471–480. doi: 10.1071/RD13371. [DOI] [PubMed] [Google Scholar]
- 20.Fakih M, Ouyang C, Wang C, Tu TY, Gozo MC, Cho M, Sy M, Longmate JA, Lee PP. Immune overdrive signature in colorectal tumor subset predicts poor clinical outcome. J Clin Invest. 2019;129:4464–4476. doi: 10.1172/JCI127046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cremolini C, Loupakis F, Antoniotti C, Lupi C, Sensi E, Lonardi S, Mezi S, Tomasello G, Ronzoni M, Zaniboni A, Tonini G, Carlomagno C, Allegrini G, Chiara S, D’Amico M, Granetto C, Cazzaniga M, Boni L, Fontanini G, Falcone A. FOLFOXIRI plus bevacizumab versus FOLFIRI plus bevacizumab as first-line treatment of patients with metastatic colorectal cancer: updated overall survival and molecular subgroup analyses of the open-label, phase 3 TRIBE study. Lancet Oncol. 2015;16:1306–1315. doi: 10.1016/S1470-2045(15)00122-9. [DOI] [PubMed] [Google Scholar]
- 22.Carli F, Bousquet-Dion G, Awasthi R, Elsherbini N, Liberman S, Boutros M, Stein B, Charlebois P, Ghitulescu G, Morin N, Jagoe T, Scheede-Bergdahl C, Minnella EM, Fiore JF Jr. Effect of multimodal prehabilitation vs postoperative rehabilitation on 30-day postoperative complications for frail patients undergoing resection of colorectal cancer: a randomized clinical trial. JAMA Surg. 2020;155:233–42. doi: 10.1001/jamasurg.2019.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen P, Zhang T, Yuan Z, Shen B, Chen L. Expression of the RNA methyltransferase Nsun5 is essential for developing cerebral cortex. Mol Brain. 2019;12:74. doi: 10.1186/s13041-019-0496-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venook AP, Niedzwiecki D, Lenz HJ, Innocenti F, Fruth B, Meyerhardt JA, Schrag D, Greene C, O’Neil BH, Atkins JN, Berry S, Polite BN, O’Reilly EM, Goldberg RM, Hochster HS, Schilsky RL, Bertagnolli MM, El-Khoueiry AB, Watson P, Benson AB 3rd, Mulkerin DL, Mayer RJ, Blanke C. Effect of first-line chemotherapy combined with cetuximab or bevacizumab on overall survival in patients with KRAS wild-type advanced or metastatic colorectal cancer: a randomized clinical trial. JAMA. 2017;317:2392–2401. doi: 10.1001/jama.2017.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janin M, Ortiz-Barahona V, de Moura MC, Martínez-Cardús A, Llinàs-Arias P, Soler M, Nachmani D, Pelletier J, Schumann U, Calleja-Cervantes ME, Moran S, Guil S, Bueno-Costa A, Piñeyro D, Perez-Salvia M, Rosselló-Tortella M, Piqué L, Bech-Serra JJ, De La Torre C, Vidal A, Martínez-Iniesta M, Martín-Tejera JF, Villanueva A, Arias A, Cuartas I, Aransay AM, La Madrid AM, Carcaboso AM, Santa-Maria V, Mora J, Fernandez AF, Fraga MF, Aldecoa I, Pedrosa L, Graus F, Vidal N, Martínez-Soler F, Tortosa A, Carrato C, Balañá C, Boudreau MW, Hergenrother PJ, Kötter P, Entian KD, Hench J, Frank S, Mansouri S, Zadeh G, Dans PD, Orozco M, Thomas G, Blanco S, Seoane J, Preiss T, Pandolfi PP, Esteller M. Epigenetic loss of RNA-methyltransferase NSUN5 in glioma targets ribosomes to drive a stress adaptive translational program. Acta Neuropathol. 2019;138:1053–1074. doi: 10.1007/s00401-019-02062-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang T, Chen P, Li W, Sha S, Wang Y, Yuan Z, Shen B, Chen L. Cognitive deficits in mice lacking Nsun5, a cytosine-5 RNA methyltransferase, with impairment of oligodendrocyte precursor cells. Glia. 2019;67:688–702. doi: 10.1002/glia.23565. [DOI] [PubMed] [Google Scholar]
- 27.Kelly AD, Issa JJ. The promise of epigenetic therapy: reprogramming the cancer epigenome. Curr Opin Genet Dev. 2017;42:68–77. doi: 10.1016/j.gde.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 28.Janin M, Ortiz-Barahona V, de Moura MC, Martinez-Cardus A, Llinas-Arias P, Soler M, Nachmani D, Pelletier J, Schumann U, Calleja-Cervantes ME, Moran S, Guil S, Bueno-Costa A, Pineyro D, Perez-Salvia M, Rossello-Tortella M, Pique L, Bech-Serra JJ, De La Torre C, Vidal A, Martinez-Iniesta M, Martin-Tejera JF, Villanueva A, Arias A, Cuartas I, Aransay AM, La Madrid AM, Carcaboso AM, Santa-Maria V, Mora J, Fernandez AF, Fraga MF, Aldecoa I, Pedrosa L, Graus F, Vidal N, Martinez-Soler F, Tortosa A, Carrato C, Balana C, Boudreau MW, Hergenrother PJ, Kotter P, Entian KD, Hench J, Frank S, Mansouri S, Zadeh G, Dans PD, Orozco M, Thomas G, Blanco S, Seoane J, Preiss T, Pandolfi PP, Esteller M. Epigenetic loss of RNA-methyltransferase NSUN5 in glioma targets ribosomes to drive a stress adaptive translational program. Acta Neuropathol. 2019;138:1053–1074. doi: 10.1007/s00401-019-02062-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan Z, Chen P, Zhang T, Shen B, Chen L. Agenesis and hypomyelination of corpus callosum in mice lacking Nsun5, an RNA methyltransferase. Cells. 2019;8:552. doi: 10.3390/cells8060552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.