Abstract
The National Center for Biotechnology Information (NCBI) Taxonomy includes organism names and classifications for every sequence in the nucleotide and protein sequence databases of the International Nucleotide Sequence Database Collaboration. Since the last review of this resource in 2012, it has undergone several improvements. Most notable is the shift from a single SQL database to a series of linked databases tied to a framework of data called NameBank. This means that relations among data elements can be adjusted in more detail, resulting in expanded annotation of synonyms, the ability to flag names with specific nomenclatural properties, enhanced tracking of publications tied to names and improved annotation of scientific authorities and types. Additionally, practices utilized by NCBI Taxonomy curators specific to major taxonomic groups are described, terms peculiar to NCBI Taxonomy are explained, external resources are acknowledged and updates to tools and other resources are documented.
Database URL: https://www.ncbi.nlm.nih.gov/taxonomy
Introduction
As a central resource utilized by all major public sequence databases in the International Nucleotide Sequence Database Collaboration (INSDC; 1; http://www.insdc.org), the National Center for Biotechnology Information (NCBI) Taxonomy plays a vital role in structuring communication concerning all forms of life on Earth. Association of the correct organismal names with genetic and genomic data is foundational to nearly every aspect of biomedical, agricultural and ecological research. Accurate taxonomy is a crucial link between natural history and experimental science (2) and essential to investigation of phenomena related to human welfare such as emergence of pathogens, dispersal of invasive species, loss of biological diversity and climate change.
The NCBI Taxonomy consists of a single, hierarchically arranged list of organismal names across all domains of life. These names are correct, current and valid according to the best authorities within the separate taxonomic disciplines and codes of nomenclature. The NCBI Taxonomy also contains numerous informal names existing outside of the codes of nomenclature. The classification used is phylogenetic, to the degree feasible, reflecting our current understanding of organismal relationships and is regularly updated to reflect new information.
Communication about the identity of research organisms is complicated by the fact that organismal names do not remain static. Taxonomists commonly change names to reflect revised species concepts, following rules laid out in the several codes of nomenclature. Two names may be merged, and one made a synonym of the other if data indicate two described species are in fact one. Name combinations change when taxonomists move species from one genus to another. Under certain conditions, spellings of names are emended. Hence, over time the scientific literature may refer to a single species by different names and authors may fail to use the most up-to-date nomenclature when they publish.
A desire to better capture these complexities and to make NCBI Taxonomy data more findable, accessible, interoperable and reusable (the FAIR data principles; 3) prompted several recent enhancements to the way we capture, curate and display information on organismal names. These are described below, within an overview of the NCBI Taxonomy.
From One Database to Several
The NCBI Taxonomy was initiated in 1991 with the implementation of Entrez, the search and retrieval system for NCBI’s databases. Entrez provided the first system to link nucleotide and protein sequences from numerous disparate sources with different taxonomic classification systems (4). As described in more detail in an earlier paper (5), the need for a centralized classification quickly became apparent as multiple resources had to exchange and link their records. A draft classification was initially provided to a taxonomy database management tool developed by Scott Federhen of NCBI. A subsequent series of taxonomy workshops, involving a broad range of biological specialists, produced a universal classification. In 1995, NCBI Taxonomy was reimplemented in a Sybase SQL Server (subsequently migrated to a Microsoft SQL Server). A year later the first version of NCBI Taxonomy Web Browser (NCBI TaxBrowser) was presented to the public. In the same year, the INSDC decided to use the NCBI Taxonomy as the sole source for taxonomic classification in order to maintain consistency among databases. This included the decision that all issues regarding nomenclature and classification would be resolved prior to the public release of any sequence data.
INSDC partners now send requests for any new organism name (taxonomy consultations) to NCBI Taxonomy curators before final data release. Consequently, the NCBI Taxonomy pages display only those names that are linked to public sequence entries. This involves several processes initiated to maintain the organism names in GenBank. One example, the Taxon3 data service allows for comprehensive use of taxonomic information in various NCBI processes since 2004. Other improvements include the initiation of the NCBI BioCollections database (to verify data on biorepositories linked to sequence data) in 2006 with a public release in 2018 (6) and the implementation of NameBank in 2007. NameBank forms the core of NCBI’s improved array of taxonomic databases, providing comprehensive taxonomic data only some of which is tied to public records. A transition from the previous database to a more comprehensive NameBank-centered data system was completed by 2018 (Supplementary Figure S1). A major enhancement was the addition of type strain and type specimen information to the database (7). This advanced the development of enhanced taxonomic information attached to public sequence records (8).
Figure 1 summarizes the flow of taxonomic information and its usage by various resources. An example of a taxonomic data entry is partially shown with various unique identifiers, tracking synonyms, authorities, publications and type information. The full entry is shown in Supplementary Figure S2. The information flow from the NCBI submission process (exemplified by the NCBI Submissions Portal), INSDC partners and other resources are curated via the TaxEdit submission tool. Taxonomic information from the NCBI Taxonomy servers is then propagated to various external and internal resources via public and non-public file transfer protocol (FTP) files and NCBI Taxonomy services and displayed in the NCBI TaxBrowser. It also serves to inform the submissions tools, modulating automated queries to submitters. Only a few NCBI applications and tools utilizing taxonomic information are highlighted, such as the pathogen pipeline and the Genome Workbench that enable users to prepare genome data for NCBI submission, but many others also rely on taxonomic input. Solid lines indicate direct database interactions, while broken lines indicate indirect ways of sharing information, e.g. emailed taxonomy consultation requests and files downloaded by taxonomic curators. NCBI Taxonomy resources are indicated in orange, general NCBI resources are in blue and the external INSDC resources in grey. Some additional data flows such as taxonomy corrections generated by RefSeq and other NCBI resources are not shown. Several aspects highlighted in Figure 1 will be discussed in more detail throughout this document, but additional information on the structure of NCBI and INSDC resources can be found in previous publications and references therein (1, 9, 10).
Figure 1.
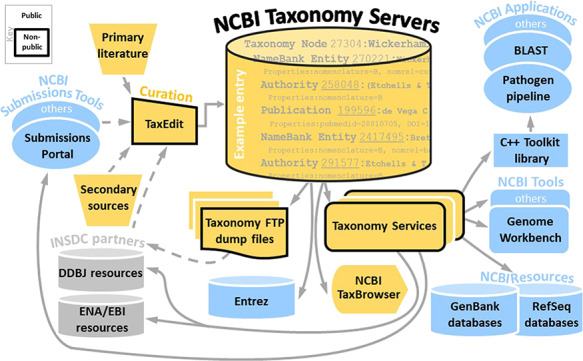
Summarized flow of NCBI Taxonomy information.
Tracking Multiple Entities for Each Taxonomy Node in NameBank
In the NCBI Taxonomy, the term ‘name’ is used in a broad sense, applying to any string of text used to indicate the organism or lineage values in a record. There is a distinction drawn between formal and informal names. Formal names are governed by the rules of the codes of nomenclature and can be associated with type material or defined ranks (see later sections). Informal names follow internal curation rules that are modified and changed by practical considerations and driven by common usage and needs of submitters. For example, informal names lacking species epithets are commonly applied to GenBank records. These terms do not follow the exact application in all the codes of nomenclature, and strategies for dealing with such instances are discussed further on.
Each entry in the INSDC databases maps onto an entry in the NCBI Taxonomy at the rank of species or below (an exception is made for patent entries). Since October 2018, the NCBI Taxonomy has been migrated from a single database to a system that incorporates several databases, focused around the central resource NameBank. This resource provides a framework for data that are not included in NCBI Taxonomy and contextualizes them. A streamlined schema of the database and the relationship between various resources and NameBank is shown in Supplementary Figure S1.
Each TaxNode (equating to a node in a taxonomic tree) has the following:
Taxonomy identifier and primary name
This is shared by all names for a specific TaxNode. Each TaxNode has a stable, unique numerical identifier, the taxonomy identifier (TaxId). Each TaxId has a labelled primary name (a formal or informal name) that appears on the NCBI records.
In publications, this can be standardized as a primary name with its TaxId displayed as:
‘NCBI:txid’ followed by a number, e.g.
Homo sapiens NCBI:txid9606
This information is also displayed in the NCBI TaxBrowser.
NameBank entity identifiers and secondary names
The biggest change in the new system is the addition of separate, stable and unique name entity identifiers for secondary names and their properties managed in NameBank. Formal secondary names under the NCBI system will consist of a Latin binomial or trinomial (consisting of the genus name and species epithet and infraspecies if present) as well as its authority (the person(s) who described the species) and the year of its valid publication. In the NCBI Taxonomy, this is the current name for the TaxNode labelled as Homo sapiens:
Homo sapiens Linneaus, 1758 (with NameBank Entity Id N3004444, which is not displayed publicly).
Informal secondary names are also tracked. A taxonomic example with various identifiers and properties as displayed in the non-public TaxEdit curation tool is shown in Supplementary Figure S2. Additionally, the relational terms, specifying relationships among the different names are shown in Table 1. This makes it possible to accurately label the original name attached to a species description (basionym, also referred to as basonym) as well as the currently accepted name in NCBI Taxonomy (current name). The labels can also distinguish between different synonyms: heterotypic synonyms and homotypic synonyms (see discussion under Codes of Nomenclature). Homotypic (or objective) synonyms are names based on the same type (see discussion of type material). Heterotypic (or subjective) synonyms are based on different types that were considered distinct taxa when first proposed, but subsequently considered to belong to the same taxon as documented in a publication or other authoritative source. This relationship often relies on taxonomic opinion and, consequently, is treated with careful verification by NCBI curators. Heterotypic synonyms can also include their own separate basionyms. The other relational terms used by NCBI Taxonomy are listed in Table 1.
Table 1.
Relational terms in NCBI Taxonomy
| Name category | Description | Number per TaxNode/TaxId |
|---|---|---|
| Primary name | This is the designated label for the TaxNode and its TaxId. | One per TaxNode. |
| Formal relational terms for secondary names | ||
| Current name | The currently accepted name chosen out of all synonyms for the TaxNode. Will often overlap with primary name (except in a few cases). | Up to one per TaxNode (where indicated). |
| Basionym | The originally described name, attached to the type material and species description. | Up to one per name (where indicated), one to more per TaxNode. |
| Homotypic synonym | Names generated after the basionym (e.g. by moving it to a different genus), but sharing the same type. | None to several per TaxNode. |
| Heterotypic synonym | Names with a different basionym and type from those mentioned above. | None to several per TaxNode. |
| Informal relational terms for secondary names | ||
| Acronym | Mainly used for viruses. | None to several per TaxNode. |
| Equivalent | Used for informal names which are related but not synonyms. | None to several per TaxNode. |
| Includes | Used for informal names which forms a subset of a name. | None to several per TaxNode. |
| In-part | Used for formal names which forms a subset of a name. | None to several per TaxNode—can be duplicate across TaxNodes. |
| Blast name | Informal name for groups of organisms. | Up to one per TaxNode (where indicated). |
| Common name | Informal names in common usage—these are not comprehensively added. | None to several per TaxNode. |
| Genbank acronym | Ensures an acronym name type is displayed prominently in flat files. | Up to one per TaxNode but excluding other genbank name types (where indicated). |
| Genbank synonym | Ensures a second synonym is displayed prominently in flat files. | Up to one per TaxNode but excluding other genbank name types (where indicated). |
| Genbank common name | Ensures a vernacular name is displayed prominently in flat files. | Up to one per TaxNode but excluding other genbank name types (where indicated). |
| Nonpublic terms for secondary names | ||
| Misspelling | Used for searches only. | None to several per TaxNode. |
| Unpublished name | Used for searches only. | None to several per TaxNode. |
Another set of name-related terms comprises those pertaining to nomenclatural status and is listed in Supplementary Table S2. These rely mainly on definitions from the codes of nomenclature, addressed in more detail in the section on the four codes of nomenclature below.
Processing Taxonomic Information
Taxonomy services are provided to several databases internal to NCBI, e.g. those in GenBank (9) and RefSeq (11; Figure 1). One such service is the Taxon3 service, which is not public, and provides several taxonomy-related attributes when queried with a name. NCBI has developed a specialized system to keep the content of sequence records synchronized with changes made in NCBI Taxonomy. The process runs weekly, refreshing organism name, type material, lineage and other values on anywhere from tens of thousands to tens of millions of sequence records each cycle. This ensures that taxonomic-related queries in Entrez will return accurate subsets of sequences for the organisms that a user is interested in and that the organism names will reflect the latest classification. Importantly, the definition lines at the top of GenBank records, and shown in Basic Local Alignment Search Tool (BLAST) results, updates on a different schedule, so the most up to date taxonomy information will always be found in the organism source modifier on a record. The external databases in the INSDC (1) consumes a set of non-public FTP dump files and also relies on several taxonomy services. When an organism name on a new record is not present, a taxonomic consultation request is generated and is handled by one of the specialists in the NCBI Taxonomy group. These curators maintain the database with TaxEdit, a customized software tool. A new javascript version that interacts with the updated taxonomy data system was released in 2018. Curators make changes to the NCBI classification as they become aware of updates in the taxonomic literature with an emphasis on changes resulting from molecular phylogenies. There is daily interaction between taxonomy curators and indexers processing new data entries for GenBank, as well as with the other INSDC partners.
Four codes of nomenclature
Independent codes of nomenclature have been drawn up by disparate scientific communities to provide rules for naming. The NCBI Taxonomy deals with names validated in four principal codes. These are the International Code of Nomenclature for algae, fungi and plants, (12; also abbreviated as ICN but referred to here as ICNafp), the International Code of Nomenclature of Prokaryotes (13; abbreviated as ICNP) and the International Code of Zoological Nomenclature (14; abbreviated as ICZN). The viruses are governed by the International Code of Virus Classification and Nomenclature (15; also referred to as the ICTV Code and abbreviated here as ICVCN).
The independent codes are focused on names within their purview. Only rarely are names of a group of organisms governed by more than one code. This is the case for Cyanobacteria (or Cyanophyta). Both the ICNafp and ICNP apply to this group and this makes tracking these names more complicated. Additionally, the codes do not treat names similarly. For example, the ICNP explicitly states: ‘the nomenclature of prokaryotes is not independent of botanical and zoological nomenclature’. However, this is not the case for the other codes and as a result, multiple genus names and a few species names are duplicated within the NCBI classification and can present a challenge to curators and database managers. NCBI Taxonomy deals with three types of duplicated names:
Independent use within separate codes of nomenclature, e.g. genus Morganella (including species of enterobacteria, mushrooms and scale insects covered by three codes of nomenclature; a total of 89 unique names across 194 TaxNodes).
Valid duplication at different ranks within a single code, e.g. the fly genus and subgenus Drosophila (which also has a duplicate genus of mushrooms mentioned in point 1; a total of 23 unique names across 47 TaxNodes).
Unresolved lineage placement, e.g. the yeast genus Candida that consists of many unrelated species; a total of 211 unique names across 430 TaxNodes).
In all cases, these names should be identified by a note ‘duplicate name’ in the NCBI TaxBrowser and have unique TaxIds and other NameBank Entity Ids. For more details on historical treatment of these names and dealing with duplicate binomials see (5).
Resources online and in print
Nomenclatural and taxonomic databases have long been available to verify the validity of taxonomic names and have greatly expanded in number and sophistication. A list of the top sources of information for taxonomic curators is presented in Table 2 with 23 principal sites that are used weekly (16–37) and a running list of additional sites that are used occasionally (Supplementary Table S1). There is no direct association between these sources, and NCBI Taxonomy and curators rely on the information provided on their web interfaces and other data services. When required, there is email communication with external database managers to clarify specific queries. No database is error-free or complete, including NCBI Taxonomy, so consulting the primary literature remains essential. Fortunately, descriptions of new taxa as well as older papers, through organizations such as the Biodiversity Heritage Library, are becoming increasingly accessible online. The expanded in-house taxonomy curation tool, TaxEdit, allows for links using document identifiers (e.g. PMC, PMID or DOI) and citations to be attached to TaxNodes and names in the database, which are then displayed in the NCBI TaxBrowser (Figure 5).
Table 2.
A selection of external resources relied on by NCBI Taxonomy curators
| Database name | URL | Note |
|---|---|---|
| Principal sites (used weekly) | ||
| AlgaeBase (16) | https://www.algaebase.org | Covers algae in the broad sense as photosynthetic eukaryotes excluding embryophytes. |
| Amphibian Species of the World (17) | http://research.amnh.org/vz/herpetology/amphibia/index.php | Regularly updated published by the American Museum of Natural History. |
| American Society of Mammalogists (ASM) Mammal Diversity Database (18) | https://mammaldiversity.org/ | Regularly updated database of mammal taxonomic and biodiversity information. |
| Avibase (19) | https://avibase.bsc-eoc.org/ | Complete data on birds. |
| Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) (20) | https://www.dsmz.de/services/online-tools/prokaryotic-nomenclature-up-to-date | Compilation of all names of bacteria and archaea that have been validly published according to the ICNP. |
| Eschmeyer’s Catalog of Fishes (21) | https://www.calacademy.org/scientists/projects/eschmeyers-catalog-of-fishes | Authoritative reference for taxonomic fish names. |
| Global Lepidoptera Names Index (22) | https://www.nhm.ac.uk/our-science/data/lepindex/ | Searchable database for world’s Lepidoptera names |
| International Committee on Taxonomy of Viruses (ICTV) (23) | https://talk.ictvonline.org/taxonomy/ | Provides list of species, classification and exemplar GenBank accessions. |
| Index Fungorum (24) | http://www.indexfungorum.org | Comprehensive data on fungi. |
| Index Herbariorum (25) | http://sweetgum.nybg.org/science/ih | Database of the world’s herbaria. |
| Integrated Taxonomic Information System (ITIS) (26) | https://www.itis.gov | A partnership of federal and international agencies to provide an authoritative taxonomic information on plants, animals, fungi and microbes. |
| International Plant Names Index (IPNI) (27) | https://www.ipni.org | Most complete tracheophyte database. |
| List of Prokaryotic names with Standing in Nomenclature (LPSN) (28) | http://lpsn.dsmz.de | List of prokaryotic names with standing in nomenclature. |
| MycoBank (29) | http://www.mycobank.org | Comprehensive data on fungi. |
| Nomenclator Zoologicus (30) | http://ubio.org/NomenclatorZoologicus/ | List of the names of genera and subgenera in zoology from the tenth edition of Linnaeus, 1758, to the end of 2004. |
| Pan-European Species directories Infrastructure (31) | http://www.eu-nomen.eu/portal/ | Annotated checklist of species occurring in Europe, aiming to cover the Western Palearctic biogeographic region. |
| Reptile Database (32) | http://www.reptile-database.org | Comprehensive data on reptiles |
| Tropicos (33) | https://www.tropicos.org | Especially good for bryophytes and New World tracheophytes |
| Wilson & Reeder’s Mammal Species of the World (34) | https://www.departments.bucknell.edu/biology/resources/msw3/ | Online version of 3rd edition (2005) without subsequent updates. |
| World Checklist of Selected Plant Families (35) | https://wcsp.science.kew.org/home.do | Especially good for monocots. |
| World Flora Online (36) | http://www.worldfloraonline.org | Supersedes The Plant List (http://www.theplantlist.org). |
| World Register of Marine Species (WoRMS) (37) | http://www.marinespecies.org | A comprehensive and updated list of names and synonymies for marine organisms and for some terrestrial invertebrate groups. |
Figure 5.
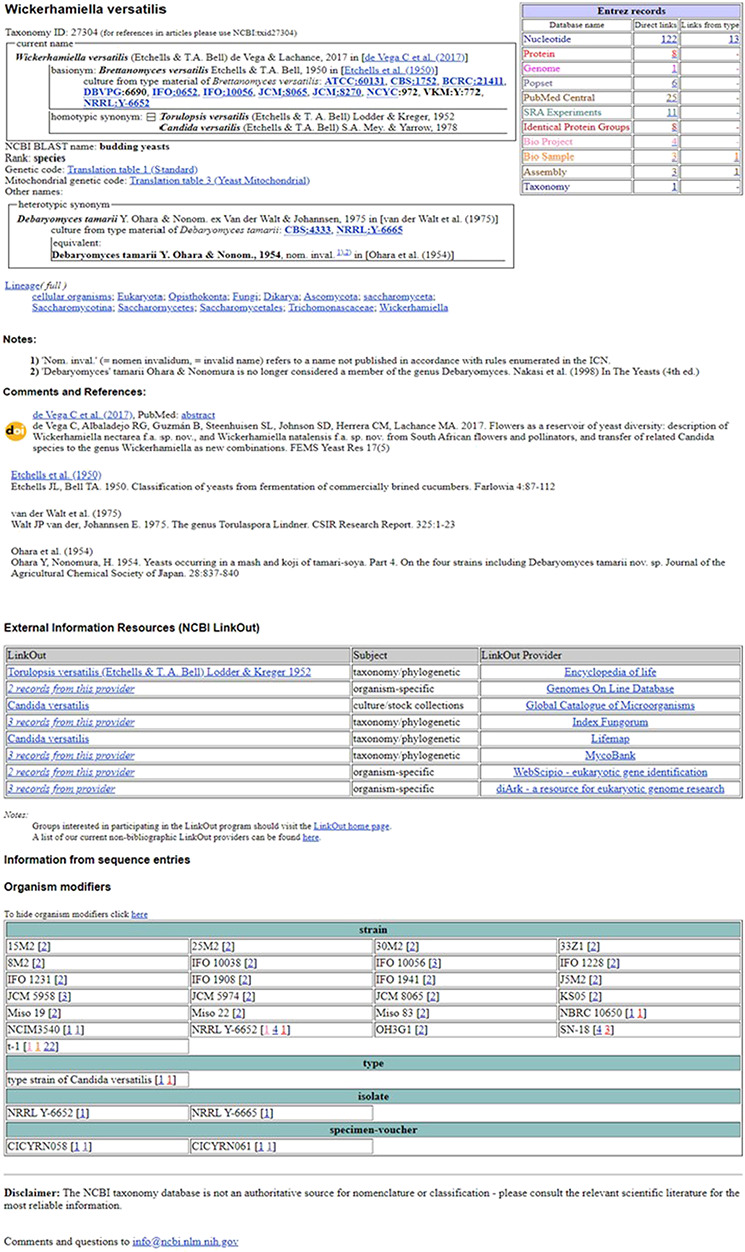
NCBI TaxBrowser example page.
Labelling Hierarchical Information and the Limitations of Ranks
The NCBI Taxonomy is grounded in phylogenetic systematics but also uses traditional hierarchical ranks first proposed by Linneaus in the 18th century. These rank names remain in use even though they cannot fully reflect phylogenetic relationships. The NCBI Taxonomy uses most Linnaean ranks defined in the four codes of nomenclature, but also uses group names that are in common use and cannot be assigned a traditional rank. We assume that any taxonomic group should be monophyletic, i.e. contain all descendants of a single ancestor (although in practice this is not always fulfilled). Where possible, the following seven traditional ranks are universally applied throughout the classification. The highest level in NCBI Taxonomy is superkingdom (viruses, eukaryota, archaea, bacteria), followed by phylum (245 public entries), class (~380 public entries), order (~1500 public entries), family (~9200 public entries), genus (~92 000 public entries) and species (~1.8 million public entries). In the NCBI classification kingdom (Metazoa or animals, Viridiplantae or green plants and Fungi) is only applied to eukaryotes. Besides these, several additional expanded ranks, e.g. subphylum and superfamily are also used. These are defined as formal ranks in NCBI Taxonomy. It should be noted that some of the higher ranks, e.g. superkingdom, are treated differently in various sources. Such instances will have to be reevaluated continually by curators. Complete statistics on the numbers of different groups are on the NCBI Taxonomy statistics page: https://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/index.cgi?chapter=statistics.
A second group of ranks comprises those that are included in NCBI classification out of practical necessity. This includes incertae sedis, unclassified and environmental names. They are non-hierarchical names that reflect either uncertainty of placement in a specific rank or represent informal and poorly defined names in the NCBI classification. Any other designated rank that matches no existing formally defined rank is assigned a ‘no rank’ value in NCBI Taxonomy. ‘No rank’ names may appear in between any ranked TaxNodes in the lineage without breaking the ranking order and can be found above and below species rank. For example, a number of these names include strains that were originally assigned for genome data and placed below species, but as the number of entries grew this practice was abandoned out of practical necessity. The legacy strain level names are being kept as part of the classification but new names are generally not being created (38). Another group with a mix of formal and informal names and names in common use that cannot be assigned to formal rank is made up of ‘clades’, monophyletic groups recognized in phylogenetic studies and which have not been assigned a formal rank. These include PACMAD and BOP clades in grasses, the numerous groups and subgroups in the fly genus Drosophila and several important TaxNodes near the root of Metazoa, e.g. Eumetazoa, Bilateria, Amoebozoa or the Sar clade. PhyloCode is a nomenclatural code separate from the four ones mentioned earlier. It is specifically intended to address the problems stemming from the treatment of ranks when applied to phylogenetic trees and is dedicated to the naming of clades. A recent update has been published and it remains to be seen how broad its usage will be (39). Recent NCBI Taxonomy updates have resulted in some previous rank names being made public and not lumped together as ‘no rank’ anymore. A list of these and other most common rank and group names is shown in Supplementary Table S3.
Progress on Documenting Sequences for All Known Species
The addition of new species to science and thus also to the INSDC databases continues apace. How many more species are still awaiting discovery? This question was posed in a review of the taxonomy resources at NCBI in 1996 (40). At the time, the number of TaxNodes with formal species names in the NCBI Taxonomy was roughly 20 000. When the NCBI Taxonomy was last described in 2012, the number had grown to about 235 000 (5) and today it surpasses 460 000. These account for nearly a quarter of total described species of all organisms, which are estimated as more than 1.85 million based on several sources. In addition, over 1.34 million species-ranked TaxNodes in NCBI Taxonomy are not identified with formal names, with a majority that can be referred to as ‘dark taxa’ (41). Estimates of total species on earth vary widely (42–45) but in any scenario, the ones that are registered in our databases represent only a small fraction of this total.
The accumulation of biodiversity in NCBI databases is visualized in Figure 2. This analysis was made of the addition of new species names to NCBI Taxonomy for each year from the inception of the database to the present. Scientific binomials for most major groups display a steady increase, with metazoans demonstrating the fastest rate of accumulation. This is largely due to intensified research activities on invertebrates, especially insects, which account for about 75% of all known metazoan species, based on numbers from Species2000 (46). An increased slope for their number can also be observed between 2008 and 2009, a result of submissions from a series of Barcode of Life projects (47, 48).
Figure 2.
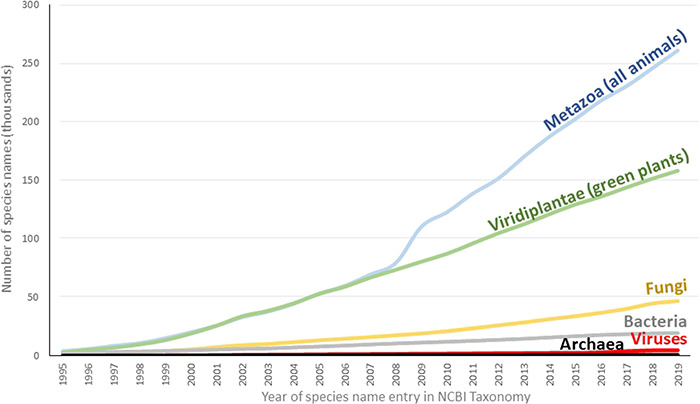
Species names added over time to NCBI Taxonomy. The first occurrence of each species in the NCBI Taxonomy was determined by the created date of its associated TaxNode. This date represents the first addition of the species into the database irrespective of subsequent name changes.
In order to further reveal the progress towards documenting known species, an estimate was done of valid species still absent from NCBI Taxonomy (Figure 3). For each principal group of organisms, the proportion of validated, described species in NCBI Taxonomy to total described species recorded in related source databases was calculated (17–19, 21, 27, 32) for every year since 1995. Bacterial names were recorded over time from several publications and validation lists in the International Journal of Systematic and Evolutionary Microbiology (IJSEM). All groups show a near linear accumulation with no hint of reaching a plateau, with bacteria being the only group that approaches a complete coverage of known species. Metazoa were divided into vertebrates and invertebrates in this analysis to highlight the stark difference in their relative representation. Invertebrates, together with fungi, are the most ‘incomplete’ groups, with only 17% of total known species present in the sequence databases. However, difficulties in tracking relationships of synonyms among databases undoubtedly decreased the accuracy of these estimates.
Figure 3.
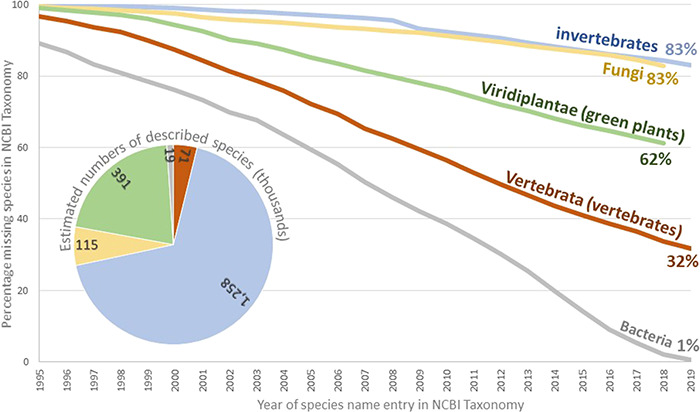
Estimate of the percentage of formal species names missing from the public NCBI databases. Curves were generated by plotting the number of formal species in the NCBI Taxonomy against the running total of described species in the corresponding group by the end of the year. The IJSEM was used as the source for bacteria. The International Plant Names Index (IPNI; 27) was used as the source for the green plants. The Species 2000 Annual Checklist (46) was used as the source for invertebrates and Fungi. Vertebrate data were collected from the Catalogue of Fishes (21), Amphibian Species of the World (17), the Reptile Database (32), Avibase (19) and the American Society of Mammalogists (18). Archaea and viruses were omitted for having a small number of species and a specialized process for reporting new species, respectively.
Curation: Structuring Information from Varied Sources
Differences in dealing with names and sequences
It is important to make a distinction between the curation of organismal names on records and curating their underlying data. Taxonomy curators are focused on the names themselves and publications associated with them. In the past names attached to submissions were wholly reliant on submitters’ input. More recently, however, NCBI has started to verify grossly misidentified submissions by comparing them against references. Validated marker gene libraries have proven to be effective tools in identifying contaminants and misidentified organisms during sequence submission. RNA sequences are routinely compared to reference sets derived from type material. Sequences that diverge significantly from established type material are returned to the submitter for comment or correction. More comprehensive changes have also been made by comparing prokaryotic genome data. These specific areas of curation and the focus on challenges presented by specific taxonomic groups are discussed in more detail below.
Defining and annotating type material ties sequences to names
The type concept is fundamental in the codes of nomenclature. One or more types are designated as an objective standard to fix the scientific name of the species or infraspecies (subspecies, variety or forma). The type of a species or infraspecies is designated when it is newly described and illustrates the trait(s) that distinguish it. Viruses rely on a list of names approved by the International Committee on Taxonomy of Viruses (ICTV) and a regular updated file that contains the name of the exemplar isolate and the corresponding GenBank accession number(s).
NCBI Taxonomy curation over the past decade has greatly improved documentation and data tracking of type strains and specimens (7). This spurred the development of additional tools to improve and enhance the taxonomic information attached to public sequence records. Under the ICNafp and ICZN, types usually consist of dried or pickled specimens or physiologically inactivated cultures. Prokaryotic types under the ICNP (bacteria and archaea) are nearly always living cultures (type strains). Types may be indicated by collector name and number plus institution code where a specimen is deposited and/or institution accession or bar code number. A summary of the type terminology used by NCBI Taxonomy and formally accepted by the INSDC is shown in Table 3. The complete set of terms is listed online (http://www.insdc.org/controlled-vocabulary-typematerial-qualifer). The terms are divided into name-bearing types (also known as nomenclatural types) and non-name-bearing types. The latter are considered of lower nomenclatural importance because they are derived from specimens that serve to inform and expand the concept of the preceding set of types and will generally be different genetically, although still potentially closely related.
Table 3.
Commonly used type terminology (for complete list of accepted terms see www.insdc.org)
| Kind of type | Code of nomenclature | Definition |
|---|---|---|
| Name-bearing types | ||
| Holotype | ICNafp, ICZN | There is only one holotype, usually a single specimen, and the ‘name-bearer’ of its described taxon. It serves as the standard to which all subsequent examples of the described taxon are compared. |
| Type strain | ICNP | Equivalent term to holotype used for prokaryotes. There can be multiple co-identical type strains, cultured from a single source. |
| Neotype | ICNafp, ICZN | If the holotype is lost or destroyed, a neotype specimen is designated from a collection considered to be representative of the original holotype. There is only one neotype. |
| Neotype strain | ICNP | The equivalent term to neotype used for prokaryotes. |
| Isotype | ICNafp | One or more duplicate specimens from the holotype collection can be deposited in other institutions. Usually the collection number is the same as the holotype, but the institution code has to be different. Iso- can be appended to other kinds of types to indicate duplicates, e.g. isosyntype, etc. Isotype is not a formally accepted term in the Zoological Code. |
| Non name-bearing types and additional terms | ||
| Paratype | ICNafp, ICZN | One or more additional specimens chosen to further illustrate traits in the described taxon. |
| Epitype | ICNafp | In botanical nomenclature only, a type designated to expand on the original holotype concept. There should be only one epitype. |
| Culture from –type | ICNafp | Also, sometimes designated ex-type, e.g. ex-holotype, etc. There can be multiple of these. The types of cultivable, microbial eukaryotes must be inactivated and one or preferably more living cultures is extracted from the type and maintained in living culture collections. |
| Reference material/Reference strain | not designated in any code | The reference material and reference strain qualifiers are not types, but internal INSDC terms used to capture any reference strain or material exclusively of types. |
Certain terms not used in nomenclature are used as internal GenBank terms. For example, the term ‘type material’ refers to specimens or cultures where the kind of type is unknown. An additional term included is ‘reference material’ or ‘reference strain’. These terms can include specimens or cultures that do not have nomenclatural standing but nevertheless could have value for taxonomic identification. These include ‘Candidatus’ prokaryotic names, which are names proposed for new species that have not been formally described by the ICNP. ‘Candidatus’ names can be searched in Entrez Taxonomy (discussed further in section) using a term as a filter in Entrez: candidatus current name[filter].
Facilitating access to specimen and strain information with NCBI BioCollections
An increasingly important part of the curation of type material is the use of standardized terms to indicate in which biorepository they reside. This applies to any record with a physical sample. To address this, the NCBI BioCollections database has been created. This is a curated data set of metadata for culture collections, museums, herbaria and other natural history collections connected to sequence records in GenBank (6). The NCBI Biocollection database is used to support ‘structured voucher’ annotation in the sequence entries submitted to INSDC. Darwin Core data standards developed by the Biodiversity Information Standards (TDWG, formerly the Taxonomic Database Working Group) is used for the structured annotation. The Darwin Core standard triplet format for specimen data consists of three parts: the universally recognized acronym for the institution that holds the voucher specimen, the institution’s code for the collection in which the voucher specimen is kept and the unique specimen identifier, all separated by colons, for example:
/specimen_voucher = “USNM:FISH:425122”.
Unlike museum specimens and culture collections, the standard format for herbarium and fungarium vouchers includes the collector’s name and number followed by the herbarium code. This differs from the Darwin Core format adopted by the NCBI Taxonomy and is discussed in more detail elsewhere (6). While most specimen voucher identifiers submitted with GenBank records are listed in the correct /specimen_voucher field, there are a large number that are not and will therefore escape annotation. There are limitations to the Darwin Core format, as it is not universally unique, and future endeavors will include assessing changes and adaptations to utilize other commonly applied standards (49).
Curation: unique challenges for each taxonomic group
Curation of prokaryotes (NCBI:txid2, NCBI:txid2157)
The term ‘prokaryotes’ designates two main groups of superficially similar, but evolutionarily divergent organisms, which have traditionally been studied using similar methods—bacteria and archaea. New names and new combinations published in the IJSEM and other sources are monitored. Priority is given to taxonomic names validly published under the ICNP (13). New names published in other journals besides IJSEM are added to the taxonomy with an ‘effective name’ flag. Effectively published names have no standing in the nomenclature and can be submitted to IJSEM for validation (50). Effectively published names can now be searched in Entrez Taxonomy (see section) using a filter as an additional search term in Entrez (effective current name[filter]). For instance, this search currently yields more than 2000 names: root[organism] and effective current name[filter]. Unpublished prokaryotic names receive placeholder names usually of the form <Genus> sp. <strain_identifier> or < higher_rank> bacterium/archaeon <strain identifier> until the proposed new name is either effectively or validly published.
Obligately endosymbiotic bacteria that are not identified at species level are added with their host name (e.g. Wolbachia endosymbiont of Drosophila simulans; Rickettsia endosymbiont of Achalcus cinereus). Informal Phytoplasma names are added with host name in single quotes and disease type (if available, e.g. ‘Echinacea purpurea’ witches’-broom phytoplasma; ‘Phoenix canariensis’ lethal yellowing phytoplasma). Single quotes are placed around the scientific name of the host organism to clarify that the data are from the phytoplasma and not the host organism. There are many legacy names for which common names are used for host species instead of the scientific name. In such cases, single quotes are not used.
NCBI offers a BLAST data base of validated 16S ribosomal RNA sequences from bacterial and archaeal type strains. A comparison of new 16S ribosomal RNA sequences to this data base is an effective way to check sequence quality and taxonomic identity of the source organism. As part of the NCBI prokaryotic genome submission process, GenBank now performs an average nucleotide identity analysis to identify and correct misidentified genomes during submissions (51). This method is also applied as a routine consistency check to support identification and classification of existing public genome assemblies in GenBank. Data on type strains are collected mostly from original publications and from external sources, such as the German Collection of Microorganisms and Cell Cultures at the Leibniz Institute (DSMZ; 20) and the National Collection of Type Cultures (NCTC), one of four Culture Collections of Public Health England (52). The collected data are analyzed and curated and then used to find and correct misidentified and contaminated genome assemblies. A full list of available type strains can be obtained via FTP (see link under FTP Resources). The process provides taxonomic validation of genomes at the time of submission to GenBank, corrects many misassigned genomes already in GenBank and aids in flagging contamination. Using the methods described in a recent paper (51), over 2000 previously misidentified prokaryotic genomes were identified and corrected. Approximately 70 new prokaryotic genome submissions per month are found to be misidentified and submitters are contacted before the data are released.
Until 2014, strain-level TaxNodes were assigned for all genome samples, primarily for those of prokaryotes (38). This practice has now been halted for prokaryotes and eukaryotes, but legacy strain-level names remain with unique TaxIds in the NCBI Taxonomy. Another change, since August 2017, was to discontinue the practice of assigning specific TaxNodes for each metagenome-assembled genome. It is anticipated that the number of such submissions will continue to grow and will begin to include more organismal data from taxa outside of the prokaryotes.
Curation of green plants (NCBI:txid33090)
Green plants or Viridiplantae are a clade covered by the ICNafp (12) and comprise 18 classes of green algae, plus embryophytes. As of January 2020, over 167 000 species and infraspecies of Viridiplantae were linked to public records in the NCBI Taxonomy, comprising ~40% of all described species. Embryophytes have been the focus of intensive phylogenetic research for decades, culminating in comprehensive, consensus-based classifications by collaborations such as the Angiosperm Phylogeny Group (APG; 53) and the Pteridophyte Phylogeny Group (54). This has had the effect of largely stabilizing higher classification down to the family level. However, classification at this level, and especially below it, remains in flux. For example, relationships within the pea family (Fabaceae) remains uncertain, and teasing out phylogenetic structure at the subfamily, tribal and infrageneric levels is an ongoing focus of study.
Relationships among green algal groups (Viridiplantae minus embryophytes) remain poorly understood, presenting challenges to correct classification. Consensus-based phylogenies such as the APG classifications are still a distant goal for green algae. Further complicating matters is the relative lack of morphological traits in many green algal taxa, such as unicellular coccoid species of the genus Chlorella. Formal species may be poorly circumscribed and molecular analyses often show that strains attributed to the same species may in fact be widely separated. For this reason, taxa submitted as a simple ‘Genus sp.’ (e.g. ‘Chlorella sp.’) are usually assigned unique TaxIds because individual strains may later be recognized as new species.
Curation of green plant names poses some unique challenges. As one example, hybrids among species, and even among multiple genera, are common in plants. Hybrids between two species are treated as a species like any other, either as the hybrid formula (e.g. Populus alba x Populus glandulosa) or as a named hybrid (e.g. Populus x canadensis Moench, 1785). Although hybrids are typically indicated with a multiplication sign (‘×’), the letter ‘x’ is used in the NCBI Taxonomy because many external databases encounter difficulties in translating non-ASCII characters. Because the hybrid sign (× or x) is used very inconsistently in the literature, the same name without the hybrid sign may be added to a hybrid for search purposes, e.g. Populus canadensis is added as a synonym of Populus x canadensis. Intergeneric hybrids are effectively treated in the same way as a named genus (e.g. x Triticosecale) or as a hybrid formula (Thinopyrum x Triticum) in cases where no formal name has been described. Complex hybrids of uncertain parentage, which are particularly common among cultivated plants, are given the non-unique name of ‘Genus hybrid cultivar’, e.g. ‘Rosa hybrid cultivar’, which is unsatisfactory in some ways, but avoids the problem of creating cultivar level TaxNodes for many cultivated plants.
Only the ICNafp recognizes variety (varietas) and form (forma) as infraspecies below subspecies. In practice, most cases apply to green plant names. Although the ICNafp accepts a name in the format ‘Genus species subsp. X var. Y f. Z’, the shorter version ‘Genus species f. Z’ is adopted in NCBI Taxonomy with the longer format added as a synonym. Infraspecies are often treated in databases and monographs as synonyms of the parent species because morphological traits used to distinguish such infraspecies may be variable or unreliable. However, the NCBI Taxonomy curators have adopted a more lenient approach on the grounds that infraspecies may be distinct at the genetic level if not at the morphological level. Autonyms can pose a problem, especially for cultivated plants, where two distinct TaxNodes exist for the same taxonomic entity, such as Zea mays and Zea mays subsp. mays. Autonyms, especially for economically important plants, are generally avoided where possible. Some of these issues may concern fungi as well, which is governed by the same code and are discussed in more detail below.
Curation of fungi (NCBI:txid4751, NCBI:txid4762)
Fungal names are governed under the latest ICNafp (12) with specific sections only applying to this group (55). Currently, the NCBI classification contains nine phyla based on genome comparisons (56) but up to 16 are accepted in the literature (57, 58). This will have to be reassessed as more data emerge. NCBI’s classification includes phyla that have disputed classifications outside of the fungi, such as the Cryptomycota (also known as Rozellomycota or Rozellida) and Microsporidia but follows the majority opinion of the mycological community (59). Placing these lineages and several other unicellular eukaryotic groups within fungi remains under scrutiny and it is possible that this will have to be readjusted in accordance with new data. It should be noted that the plant pathogenic group, Oomycota (NCBI:taxid4762), traditionally studied by mycologists, are curated similarly to fungi, although these are an unrelated group of protists.
A major challenge in fungal taxonomy is dealing with adjustments to the classification from the dual name system after changes to the nomenclature adoptions of the Melbourne Code (60). Historically, sexual forms (teleomorphs) and asexual forms (anamorphs) could not always be linked with certainty in fungi. This resulted in the practice of using different genus and species names for a single species, depending on whether an investigator observed a sexual or asexual stage. Where connections were known, the teleomorph name was recommended to be treated preferentially although in practice, this rule was applied variably. With the increasing use of DNA sequence data for classification, this system has become untenable as exemplified by the declaration of a universal DNA barcode (61). With adoption of the Melbourne Code, all fungal names are treated equally, resulting in synonymy of teleomorph and anamorph genera where data supported it. Curators at NCBI have focused on one such example (species in Hypocrea synonymized with their Trichoderma anamorphs) to introduce a large scale update to the NCBI Taxonomy based on recommendations by the scientific community (62, 63). The work of updating these names will continue.
The annotation of type material in NCBI Taxonomy (7) has greatly enhanced the NCBI curated database Refseq (11), specifically focusing on certain targeted loci (set up as a separate BioProject; 64). The interaction between NCBI Refseq curators, selecting and verifying high quality sequence markers and NCBI taxonomists investigating and correcting parts of the classification has resulted in the current release of a high-quality set of marker sequences for ribosomal genes from type material covering the major lineages of fungi. The RefSeq group uses the nomenclature, classification and type material curation provided by NCBI Taxonomy. However, during fungal RefSeq curation of targeted loci such as ITS, 28S and 18S data as well as genomes, discrepancies of the phylogenetic placement are sometimes observed. After ruling out the possibility of bookkeeping errors, these taxonomic disagreements are reported for review by a taxonomist, resulting in several improvements to the NCBI Taxonomy.
Curation of unicellular eukaryotes other than green algae (NCBI:txid2759 excluding NCBI:txid33090, NCBI:txid33208)
The higher classification of the eukaryotes largely follows the Adl et al. consensus classification proposed by a large group of experts published in 2012 and revisited in 2019 (59, 65). However, in the interest of nomenclatural and taxonomic stability, NCBI has been conservative in adopting close to 10 eukaryotic supergroups that have been suggested in the past 20 years (66). Instead, NCBI has opted to present a simpler hierarchy with 20 basal eukaryotic taxa such as the Viridiplantae, Rhodophyta, Opisthokonta, Alveolata or Stramenopiles. The monophyly of these taxa is well established and often based both on molecular phylogenies and morphological or cell-biological characteristics.
In contrast to the traditional kingdoms/supergroups such as the green plants, fungi or multicellular animals, the eukaryotic supergroups are often based entirely on molecular phylogenetic studies. Of the original six eukaryotic supergroups (66), NCBI has adopted three (Opisthokonta, Amoebozoa and Rhizaria) all of which are still considered to be monophyletic. Among the three original supergroups that NCBI did not recognize, Excavata and Chromalveolata are now superseded by taxa with different compositions whereas the monophyly of the Archaeplastida is currently not strongly supported (66). In 2019, NCBI adopted additional high-level eukaryotic groups (59): Haptista (Centroplasthelida and Haptophyta), Sar (Telonemia, Stramenopiles, Alveolata and Rhizaria), Discoba (Euglenozoa, Heterolobosea, Jakobida) and the Metamonada.
Curation of Metazoa (NCBI:txid33208)
The monophyletic kingdom Metazoa comprises all multicellular animals. Their formal names are regulated by the ICZN. With few exceptions, all metazoan organism names in the database associated with sequence records are treated as either species or subspecies. The NCBI Taxonomy contains ~220 000 formal animal species names, nearly 15% of the total described living animal species, estimated at 1.5 million in a 2013 study (67). These also represent approximately half of all names in the NCBI Taxonomy. Vertebrates are relatively well represented, but a large portion of invertebrate taxa remain unsampled (Figure 3). For example, although roughly one-fourth of all formal species in the database belong to insects (~119 000), they account for barely over 10% of all published insect species that are estimated to number more than 1 million (67).
The classification of major metazoan lineages generally follows the broad consensus of recent phylogenomic studies (68–71) while taking a conservative approach to areas that remain contentious or unresolved (e.g. whether Xenacoelomorpha is the sister group to all other Bilateria (72) or is a clade inside Deuterostomia (73)). Names and concepts of major metazoan lineages have remained largely stable among the 35 phyla and superphyla recognized in the NCBI Taxonomy: 27 were registered at the inception of NCBI Taxonomy in 1995, with only 5 added since 2000. However, evolutionary relationships among these groups have been adjusted several times. For example, Mesozoa (Dicyemida, Orthonectida) that had been placed outside of Eumetazoa are now moved next to Platyhelminthes and Gastrotricha within Lophotrochozoa (74), and Placozoa also has been moved from outside to inside Eumetazoa (75). Cheatognatha, formerly considered a deuterostome group, is now sibling to Rotifera, Gnathostomulida and Micrognathozoa within the protostome clade Gnathifera (68). At class and ordinal levels, the classification has been revised in recent years for several groups, e.g. mammals, birds, fishes and various invertebrates. Significant taxonomic changes at the family rank and below have been the norm during this time in Metazoa (e.g. 76)
Like other groups in the NCBI Taxonomy, curation of metazoan taxa typically includes adding new names, updating synonyms and other actions based on peer-reviewed publications. For competing taxonomic opinions and treatments, newer research, incorporating phylogenetic studies and using advanced methods, is often given more weight. Prevalence of opinions in the field is considered but is not always a deciding factor in making decisions. In specific cases, tracing long and complicated histories of taxonomic and nomenclatural revisions and locating old or rare literature, especially in languages other than English, is required. Other taxonomy databases that are actively maintained by experts are frequently consulted when metazoan names and classifications need to be verified. Some examples of these databases are listed in Table 2. However, it is very common for zoological taxonomy databases to be specialized to a limited scope (for invertebrates, often at the level of order or lower-ranked groups; Supplementary Table S1).
The increase in zoological names has accelerated in recent years (Figure 3). Along with larger phylogenetic studies and taxonomic revisions, research projects on DNA barcoding have contributed significantly to the volume of species names entered in the NCBI Taxonomy. Typically, users submit data sets to GenBank from their accounts in the BOLD system (47, 77) before the associated articles are published. Also, NCBI periodically receives direct update requests from data managers at BOLD to revise organism names for larger quantities of records.
Curation of viruses (NCBI:txid10239)
NCBI’s virus taxonomic treatment is largely based on the classification and nomenclature provided by the ICTV. The ICTV (https://talk.ictvonline.org) provides two regularly updated key files, the Master Species List and the Virus Metadata Resource (VMR; 15, 23). The VMR contains the name of the exemplar virus (isolate) for each ICTV species as well as the corresponding GenBank accession number(s).
The nomenclature of viruses is different from that of cellular organisms in that species names can include numbers and hyphens and can consist of a varying number of elements. Some names resemble the binary species names of animals and plants, e.g. Giessen reptarenavirus is a species in the genus Reptarenavirus. However, a single word name like ‘Astarnavirus’ is currently a valid ICTV viral species name, as is a multi-word name like ‘Tomato mild yellow leaf curl Aragua virus’. According to the ICTVN, names approved by the ICTV are called ‘accepted names’ while names that are not accepted but conform to the ICTVN are called ‘valid names’. Most of the viral names submitted at NCBI are not from ICTV accepted names. For example, although the ICTVN states that species names shall not consist only of a host name and the word virus, names like ‘Rat astrovirus’, ‘Mouse cyclovirus’ etc. are still submitted in great numbers to the sequence databases. The current release (01 May 2020) of the VMR recognizes 7917 exemplars and additional isolates organized into 6590 viral species. Of these, 240 virus species names are without a GenBank accession number (labeled ‘No entry in GenBank’) and they are therefore not present in the NCBI Taxonomy either. In addition to the ICTV names, the NCBI Taxonomy comprises another 30 000 viral species currently not accepted by the ICTV so that the ICTV species currently represent only about 20% of the total. However, it should be noted that ICTV names have a higher average nucleotide count because many non-ICTV virus names are associated with very few sequence records.
The NCBI taxonomy group makes great efforts to stay current and update viral taxon names and classification following the release of each ICTV update. One of the major recent changes was the introduction of several taxa at the highest level of the virus classification, including among others, the Riboviria, for most RNA viruses and the Duplodnaviria, for double-stranded DNA viruses. In addition, the information stored in NCBI Taxonomy can now be used to retrieve organism names and sequences directly by their type of genome. For example, to get the names of ssRNA(+) viruses, one could query Entrez Taxonomy using this query: ‘positive sense single stranded dna virus’[filter]. All these search terms are available as filters in the online dictionaries for Entrez search terms.
Historically, GenBank has largely relied on submitter information for the classification of viruses that are not (yet) recognized by the ICTV. That caused a large number of viruses to be classified only into the ‘unclassified viruses’ or ‘unclassified phages’ because no further information was available at the time of sequence submission. Now, however, complete or nearly complete phage genomes are classified according to phylogenies provided by the NCBI RefSeq virus group that is responsible for resources dedicated to virus information (78, 79). The validations of taxonomic data by the NCBI virus group use several approaches including BLAST, PASC HMM models and nucleotide and protein phylogenies. The group frequently interacts with outside stakeholders to direct subspecific classification for human pathogens as, e.g. flu, dengue and Ebola virus. Regular interactions with the ICTV also occur to clarify problematic placements and drive conversation. For the NCBI taxonomy group, the results of the NCBI virus group taxonomy validation are now the most important tool to allow placement of the non-ICTV viruses within the framework of the ICTV classification.
Curation of artificial and non-organismal sequences (NCBI:txid81077)
Vectors are treated with specific naming conventions, e.g. Cloning vector <identifier>, Expression vector <identifier>, Transposon vector <identifier>, Shuttle vector <identifier>.
Plasmids are annotated with their host organism:
/organism=“Escherichia coli”
/plasmid=“<plasmid name>”
Plasmids that are isolated from the environment are annotated as:
/organism=“uncultured bacterium”
/plasmid=“<plasmid name>“
/lab_host=“Escherichia coli”
Anything with “Plasmid” in the /organism name and “Plasmid” in the lineage is a legacy entry, not corresponding to current rules. Artificial and other sequences are dealt with on a case-by-case basis, but single names such as “synthetic organism” can cover a range of possible submissions.
Formatting informal names
Unpublished or provisional names
Unpublished taxonomic names are not allowed in NCBI records (with a few exceptions). Consequently, these names are internally flagged as ‘unpublished’ in the NCBI Taxonomy. The public records in INSDC databases and the NCBI TaxBrowser are labelled with the relevant strain identifier to facilitate the updates when the new species name is published (for prokaryotes) or with a temporary name consisting of submitter initial and the submission year (all other organisms). Unpublished names are not visible to the public but are searchable in Entrez. It is made clear to submitters that it is their responsibility to notify NCBI when a new name associated with their sequence submission(s) has been published. Unfortunately, most submitters do not do so.
NCBI Taxonomy staff attempt to find such names, but this is not a primary responsibility and there is no specific time cycle associated with such updates. When potential updates are found, a taxonomist specializing in the group in question reviews the paper, updates the NCBI Taxonomy record if appropriate and adds additional information such as authority, citation and type specimens. Since NCBI Taxonomy staff started documenting updates 2 years ago (80), ~7000 additional names that were initially submitted as unpublished are now accurately released (Figure 4). When a provisional name is substantially changed during publication, it is unlikely to be discovered in this process. In order to address this problem fully, other ways of encouraging submitters and journals to communicate updates should be explored in the future. One possibility is to improve the standardization of keywords in PubMed and other abstract aggregators in order to communicate clearly the presence of novel taxonomic names (80).
Figure 4.
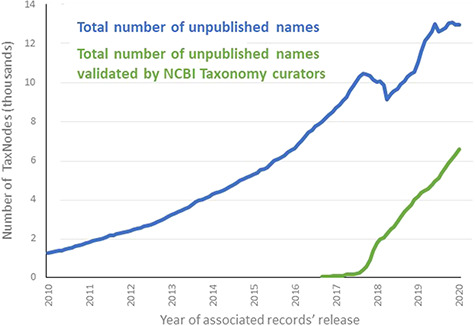
Total number of names labeled as unpublished in NCBI Taxonomy, over time.
Names lacking species epithets require several formats
NCBI Taxonomy curators and GenBank indexers are often required to deal with unique situations. In many instances the codes of nomenclature do not apply, and the resultant names reflect practical solutions that may have to be changed over time. These names include so called ‘open nomenclature terms’ that can express varying degrees of uncertainty in labelling. Examples for these terms and abbreviations include: sp. (species), aff. (affinis = ‘related, but not identical to’) nr. (‘near’), cf. (confer = ‘compare to’), etc.
Historically, the NCBI Taxonomy used a unique name when a record was submitted without a species name. To cut down on the number of taxonomic updates required, NCBI Taxonomy has been adding names without requiring the addition of a strain or another unique identifier since 2017. Currently, this is restricted to viruses and most microbes, including prokaryotes (bacteria and archaea) and eukaryotes (fungi, stramenopiles and unicellular eukaryotes), whereas the remaining names in Metazoa and Viridiplantae will continue to be treated as before. If a text string provided by a submitter is not sufficiently different to separate it from other similar names generated from separate submissions, an epithet is appended to it, which comprises submitter’s initial and year of submission. For prokaryotes informal names for genome sequences are added with strain identifier except for genome assembled from metagenomes. If a sequence is from a potentially new species, it is treated differently (see section on unpublished names).
Quotation marks are used to demarcate certain informal names
These (single or double) are used in several different circumstances. For example, effectively published prokaryotic names are indicated with double quotes. Elsewhere in the NCBI Taxonomy, single quote names have been used to indicate manuscript names (nomina inedita) where the name has either made its way into the literature or become public in the NCBI Taxonomy without formal nomenclatural description.
Square brackets communicate known misclassification
In bacteriology, the standard and accepted practice is to use square brackets to indicate names that are validly published but misclassified and have not yet undergone a formal nomenclatural revision. Outside of prokaryotes this usage, although not the convention, is applied to communicate:
Valid species names known to be misclassified but for which the correct classification is uncertain.
Valid species names not formally transferred to the generally accepted genus through a nomenclatural act.
The following citation is added to such names in the NCBI TaxBrowser: ‘Square brackets ([ ]) around a genus indicates that the name awaits appropriate action by the research community to be transferred to another genus.’
Multiple Ways to Access NCBI Taxonomy Information
The NCBI Taxonomy links to numerous internal and external resources (Figure 1). Under the NCBI TaxBrowser, two different kinds of web pages are supported. Hierarchy pages present the taxonomic classification, while taxon-specific pages summarize all the information associated with a taxonomic entry. The hierarchy pages are also customized to display a table of linked counts of entries in other Entrez databases. Taxon-specific pages will display the names associated with that entry (except for misspellings and unpublished names). The lineage can display a full or abbreviated classification. Manually curated information is displayed as well. This includes type material and comments annotated by the taxonomic curators as well as relevant literature and type material information with hotlinks as appropriate (Figure 5). This is explained in detail in a previous publication (5).
In 2019, the NCBI TaxBrowser was updated in several ways. The Entrez table has been expanded to include a column with links to records from type material for relevant resources and links were introduced directly to the biorepositories as well as specimen and strain pages from the type material listed. Additionally, the page layout has been updated to display homotypic and heterotypic synonyms, current names, authorities and type strains. An example species page is illustrated in Figure 5. This is a yeast species first described from brined cucumbers in 1950 as Brettanomyces versatilis (https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=27304). Taxonomic changes are documented, indicated by homotypic synonyms. In several cases, publications for the name changes are indicated and linked where possible as in the most currently accepted name change to Wickerhamiella versatilis. Type material is indicated with the original species name and links to associated NCBI records are shown in the Entrez table. A name with a separate type, Debaryomyces tamarii, is also indicated as a heterotypic synonym. This was first described as an invalid name in 1954 and only validated in 1975. In both cases, publications are indicated and notes attached by the curators.
At the bottom of the taxon-specific pages, additional layered information sets are found as shown in Figure 5. For relevant organisms, this can include genome information with links to resource information in other databases. Another set of links are External Information Resources or NCBI LinkOut (see below). Another block below this displays the modifiers such as strain, isolate or culture collection associated with the organism in GenBank sequence entries. Finally, each page includes a disclaimer emphasizing that primary sources should be used to confirm taxonomic information.
In contrast to the hierarchical view in the NCBI TaxBrowser, Entrez Taxonomy provides a uniform indexing, search and retrieval engine. This supports Boolean queries and includes search fields common across all Entrez databases. A number of common Entrez queries are presented in (5). Two recently added search options were added as filters, candidatus current name[filter] and effective current name[filter], as mentioned previously.
LinkOut is a service that allows for direct links from NCBI databases to external, validated resources that are provided by third parties. Taxonomically informative links can be made from sequence records or from TaxBrowser pages by request (Figure 5). LinkOut can also be built into catalog database software (e.g. Arctos). More details are in the NCBI help resources: https://www.ncbi.nlm.nih.gov/books/NBK3805/.
The full text FTP files of the complete database are updated and deposited every hour at the Taxonomy FTP site as taxdump files. There are now two versions of the FTP taxdump files—the previous unchanged version and a new version with additional options that include the taxonomic lineage of taxa, information on type strains and material and host information. The most important files in both sets of FTP files are nodes.dmp (which maps TaxIds to their parent TaxIds) and names.dmp (which maps names to TaxIds). Other files in both sets are delnodes.dmp that lists TaxNodes that have been deleted from the database, as well as TaxNodes that were once public but are no longer linked to any public sequence entries. Also, merged.dmp maps secondary TaxIds onto primary TaxIds for taxa that have been synonymized in the database. The new FTP option includes important information on type material, changed lineages and hosts. We recommend that the new version be used, but both options will be supported for the foreseeable future.
ftp://ftp.ncbi.nih.gov/pub/taxonomy (legacy version)
ftp://ftp.ncbi.nlm.nih.gov/pub/taxonomy/new_taxdump (updated, expanded version)
ftp://ftp.ncbi.nih.gov/pub/taxonomy/taxdump_archive/ (archive of taxdump updates for each month since 2014)
ftp://ftp.ncbi.nlm.nih.gov/genomes/ASSEMBLY_REPORTS/prokaryote_type_strain_report.txt (complete set of type stains, including co-identical strains)
Yet another powerful option to retrieve data from NCBI Taxonomy is the public NCBI taxonomy services. Those services provide an application programming interface to NCBI Taxonomy that allows search, browsing and data retrieval in real time. C++ programmers have the option of accessing taxonomy lookup services using NCBI C++ toolkit (https://ncbi.github.io/cxx-toolkit/). In other programming languages, the eutils can be used to access NCBI Taxonomy. Eutils also has a server-side mechanism that allows users to perform complex searches and download results in a user-defined format. A detailed guide on eutils can be found here: https://www.ncbi.nlm.nih.gov/books/NBK1058/.
The Batch Entrez tool provides a more efficient method to download bulk numbers of records from a variety of databases. More information can be found here: https://www.ncbi.nlm.nih.gov/sites/batchentrez?db=taxonomy.
SourceCheck (srcchk) reads a set of GenBank accessions and returns associated metadata, such as taxonomy information, strain identifiers, specimen vouchers, etc. It is now available as a standalone tool: ftp://ftp.ncbi.nih.gov/toolbox/ncbi_tools/cmdline/.
The Taxonomy Common Tree generates a taxonomic tree rooted at the last common ancestor of user-defined nodes (species or other ranks) https://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi.
A live summary of the numbers of Taxonomy Statistics, all public entries in the NCBI Taxonomy, broken down by user-defined taxonomic group, date or rank is available: https://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/index.cgi?chapter=statistics.
Taxonomy Status Report. This allows users to enter names, individually or in bulk and retrieve reports on whether the names are in the NCBI Taxonomy, their TaxIds and their status (for example: primary name, synonym, etc.) https://www.ncbi.nlm.nih.gov/Taxonomy/TaxIdentifier/tax_identifier.cgi.
The NCBI Tree Viewer (TV) is a graphic display tool for phylogenetic trees. It can read tree data in ASN, Newick and Nexus formats and supports functions such as zooming and navigation, displaying in different formats and layouts, collapsing and expanding branches and rooting at midpoint or user-selected nodes. https://www.ncbi.nlm.nih.gov/tools/treeviewer/. More capabilities are available in the NCBI Genome Workbench: https://www.ncbi.nlm.nih.gov/tools/gbench/.
A complete set of NCBI tools can be found here: https://www.ncbi.nlm.nih.gov/guide/all/.
Future Challenges: Expanding Taxonomic Information and Improving Accuracy
The NCBI Taxonomy is the product of a core team of eight curators and four developers with contributions from other NCBI staff. This team relies on a larger environment of resources and scientific publications to produce a working taxonomy. We have highlighted some of our work and sources in this paper, with a focus on newly released ones.
Dealing with taxonomic accuracy is a longstanding challenge (81), affected by misidentified, incomplete and out of date records. We urge submitters to update the taxonomic names on their data, particularly for records with informal names (82). An additional, persistent problem remains: dealing with splitting one species into two or more. GenBank records attached to the name will not adjust automatically and will consist of a mixture of the species before and after the split. These can be manually updated, but this is too time consuming to be practical and it rarely happens. Records obtained from type vouchers and other reference material present a potent link between old and new names, emphasizing the importance of attaching type annotations to records. In organisms where genomes and type material are readily available, improvements are most likely. NCBI has made a major shift by setting up a pipeline to verify the taxonomic names assigned to prokaryotic genomes and has already extended it to fungi, but these changes are purposefully quite conservative and limited by the number of trustworthy genomes obtained from type material. Large-scale sequencing projects, focused on extending the data from type strains will have a major impact on this work (83, 84).
There is also a broader focus on annotating all specimens, strains and other samples acting as sources of biological data (85). Processes relying on voucher information to update taxonomy will necessitate the careful and precise treatment of any voucher material during submission. Where it is feasible, NCBI Taxonomy intends to follow the work of external groups setting up data standards (e.g. 49, 86–88) and highlight the contribution of various biorepositories (89) by making explicit links possible. In addition to processing large data sets from barcoding projects and keeping the NCBI Taxonomy updated (90), a large increase in taxonomic information attached to genome data from several large biodiversity projects, such as the Darwin Tree of Life project (https://www.darwintreeoflife.org/) and UniEuk (91) through our partners at European Bioinformatics Institute (EMBL-EBI), is expected. There are several other efforts underway.
Many other challenges remain. In the next few years, NCBI Taxonomy curators will have to extend the known taxonomic information for each TaxNode and its constituent entries, the vast majority of which remain incomplete. At a minimum, if the original name (basionym) information is associated with every relevant name, it will eliminate unintentional duplications in the NCBI Taxonomy. Other efforts will include extending links to primary literature, improving the annotation of type material and adapting to changes in classification driven by in-depth genome sampling. We will continue to be dependent on essential work done by those producing the external sources referenced in this paper and the input of diverse experts extending sampling across the tree of life. Collaborative projects such as Catalogue of Life Plus (92) could be especially valuable. Members of the research community are encouraged to communicate errors, updates and inconsistencies via the NCBI helpdesk (info@ncbi.nlm.nih.gov) or to send these directly to gb-admin@ncbi.nlm.nih.gov.
Despite these and other challenges, public sequence data can serve as a reliable source for biodiversity research in the future (93) but it will require continued commitment, development and input from reliable, external sources.
Supplementary Material
Acknowledgements
The authors were supported by the Intramural Research Program of the National Library of Medicine, National Institutes of Health. In addition to acknowledging the collaborative efforts of our colleagues at NCBI, INSDC and beyond, we remember the contributions of Scott Federhen, who initiated and guided many improvements to the NCBI Taxonomy over the past two decades. Patrik Inderbitzin, Pam Soltis and Peter Uetz are acknowledged for their helpful comments on earlier versions of this manuscript.
References
- 1. Karsch-Mizrachi I., Takagi T. and Cochrane G. (2018) The international nucleotide sequence database collaboration. Nucleic Acids Res., 46, D48–D51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Strasser B.J. (2008) GenBank—natural history in the 21st century? Science, 322, 537–538. [DOI] [PubMed] [Google Scholar]
- 3. Wilkinson M.D., Dumontier M., Aalbersberg I.J. et al. (2016) The FAIR guiding principles for scientific data management and stewardship. Sci. Data, 3, 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schuler G.D., Epstein J.A., Ohkawa H. et al. (1996) Entrez: molecular biology database and retrieval system. Methods Enzymol., 266, 141–162. [DOI] [PubMed] [Google Scholar]
- 5. Federhen S. (2012) The NCBI taxonomy database. Nucleic Acids Res., 40, D136–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sharma S., Ciufo S., Starchenko E. et al. (2018) The NCBI BioCollections database. Database, 2018, bay006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Federhen S. (2015) Type material in the NCBI taxonomy database. Nucleic Acids Res., 43, D1086–D1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Federhen S., Rossello-Mora R., Klenk H.-P. et al. (2016) Meeting report: GenBank microbial genomic taxonomy workshop (12–13 May, 2015). Stand Genomic Sci., 11, 15. [Google Scholar]
- 9. Sayers E.W., Cavanaugh M., Clark K. et al. (2019) GenBank. Nucleic Acids Res., 47, D94–D99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. O’Sullivan C., Busby B. and Mizrachi I.K. (2017) Managing sequence data In: Keith JM. (ed). Bioinformatics: Volume I: Data, Sequence Analysis, and Evolution. Springer New York, New York, NY, 10.1007/978-1-4939-6622-6_4, pp. 79–106. [Google Scholar]
- 11. O’Leary N.A., Wright M.W., Brister J.R. et al. (2015) Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res., 44, D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Turland NJ, Wiersema JH, Barrie FR et al. (eds) (2018) International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017. Regnum Vegetabile 159. Koeltz Botanical Books, Glashütten, p. 254. [Google Scholar]
- 13. Parker C.T., Tindall B.J. and Garrity G.M. (2019) International Code of Nomenclature of Prokaryotes Prokaryotic Code (2008 revision). Int. J. Syst. Evol. Microbiol., 69, S7–S111. [Google Scholar]
- 14. ICZN (1999) International Code of Zoological Nomenclature 4th ed. http://www.nhm.ac.uk/hosted-sites/iczn/code/ In: International Commission on Zoological Nomenclature. International Trust for Zoological Nomenclature, London, continuously accessed. [Google Scholar]
- 15. Walker P.J., Siddell S.G., Lefkowitz E.J. et al. (2019) Changes to virus taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2019). Arch. Virol., 164, 2417–2429. [DOI] [PubMed] [Google Scholar]
- 16. Guiry M.D. and Guiry G.M. (2020) AlgaeBase. World-wide Electronic Publication, National University of Ireland, Galway. https://www.algaebase.org, continuously accessed.
- 17. Frost D.R. (2020) Amphibian Species of the World: An Online Reference, http://research.amnh.org/herpetology/amphibia/index.htmlAmerican Museum of Natural History, continuously accessed.
- 18. ASM (2020) Mammal Diversity Database, https://www.mammaldiversity.org, American Society of Mammalogists, continuously accessed.
- 19. Lepage D. (2020) Avibase—The World Bird Database, https://avibase.bsc-eoc.org/avibase.jsp, continuously accessed.
- 20. DSMZ (2020) DSMZ-German Collection of Microorganisms and Cell Cultures, Leibniz Institute continuously accessed.
- 21. Fricke R., Eschmeyer W.N. and van der, Laan R. (2020) Eschmeyer’s Catalog of Fishes: Genera, Species, References, http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp, continuously accessed.
- 22. Beccaloni G., Scoble M., Kitching I. et al. (2020) LepIndex: The Global Lepidoptera Names Index, https://www.nhm.ac.uk/our-science/data/lepindex/, Natural History Museum, London, continuously accessed.
- 23. Lefkowitz E.J., Dempsey D.M., Hendrickson R.C. et al. (2018) Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res., 46, D708–D717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kirk P. (2020) Index Fungorum, www.indexfungorum.org, Royal Botanic Gardens Kew, Landcare Research-NZ, Chinese Academy of Science, continuously accessed.
- 25. Thiers B. (2020) Index Herbariorum: A global directory of public herbaria and associated staff In: New York Botanical Garden’s Virtual Herbarium, http://sweetgum.nybg.org/science/ih/, continuously accessed.
- 26. ITIS (2020) The Integrated Taxonomic Information System (ITIS), http://www.itis.gov, continuously accessed.
- 27. Croft J., Cross N., Hinchcliffe S. et al. (1999) Plant names for the 21st century: the International Plant Names Index, a distributed data source of general accessibility. Taxon, 48, 317–324. [Google Scholar]
- 28. Parte A.C. (2018) LPSN—list of prokaryotic names with standing in nomenclature (bacterio.net), 20 years on. Int. J. Syst. Evol. Microbiol., 68, 1825–1829. [DOI] [PubMed] [Google Scholar]
- 29. Robert V., Vu D., Amor A.B.H. et al. (2013) MycoBank gearing up for new horizons. IMA Fungus, 4, 371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Neave S.A. (1939) Foreword to Nomenclator Zoologicus, Volume 1 (A–C). Zoological Society of London, London. [Google Scholar]
- 31. PESI (2020) Pan-European Species Directories Infrastructure, www.eu-nomen.eu/portal, continuously accessed.
- 32. Uetz P., Freed P. and Hošek J. (2020) The Reptile Database, http://www.reptile-database.org, continuously accessed.
- 33. Tropicos (2020) The Tropicos Database https://www.tropicos.org/, Missouri Botanical Garden, Saint Louis, Missouri, continuously accessed.
- 34. Wilson D.E. and Reeder D.M. (2005) Mammal Species of the World. A Taxonomic and Geographic Reference (3rd ed). Johns Hopkins University Press, Baltimore, MD, p. 2142. [Google Scholar]
- 35. WCSP (2020) World Checklist of Selected Plant Families, http://wcsp.science.kew.org, Royal Botanic Gardens, Kew, continuously accessed.
- 36. WFO (2020) World Flora Onlinehttp://www.worldfloraonline.org, World Flora Online Consortium, continuously accessed.
- 37. Horton T., Kroh A., Ahyong S. et al. (2020) World Register of Marine Species, http://www.marinespecies.org, continuously accessed.
- 38. Federhen S., Clark K., Barrett T. et al. (2014) Toward richer metadata for microbial sequences: replacing strain-level NCBI taxonomy taxids with BioProject, BioSample and Assembly records. Stand. Genomic Sci., 9, 1275–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. De, Queiroz K. and Cantino P.D. (2020) International Code of Phylogenetic Nomenclature (PhyloCode). CRC Press, Boca Raton, FL, p. 149. [Google Scholar]
- 40. Leipe D.D. (1996) Biodiversity, genomes, and DNA sequence databases. Curr. Opin. Genet. Dev., 6, 686–691. [DOI] [PubMed] [Google Scholar]
- 41. Page R.D. (2016) DNA barcoding and taxonomy: dark taxa and dark texts. Philos. Trans. R. Soc. B, 371, 20150334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mora C., Tittensor D.P., Adl S. et al. (2011) How many species are there on earth and in the ocean? PLoS Biol., 9, e1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stork N.E. (2018) How many species of insects and other terrestrial arthropods are there on earth? Annu. Rev. Entomol., 63, 31–45. [DOI] [PubMed] [Google Scholar]
- 44. Larsen B.B., Miller E.C., Rhodes M.K. et al. (2017) Inordinate fondness multiplied and redistributed: the number of species on earth and the new pie of life. Q. Rev. Biol., 92, 229–265. [Google Scholar]
- 45. Locey K.J. and Lennon J.T. (2016) Scaling laws predict global microbial diversity. Proc. Natl. Acad. Sci. U. S. A., 113, 5970–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roskov Y., Ower G., Orrell T. et al. (2020) Species 2000 & ITIS Catalogue of Life www.catalogueoflife.org/col, Species 2000: Naturalis. Leiden, the Netherlands, continuously accessed.
- 47. Ratnasingham S. and Hebert P.D.N. (2007) BOLD: the barcode of life data system (www.barcodinglife.org). Mol. Ecol. Notes, 7, 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hebert P.D.N., Cywinska A., Ball S.L. et al. (2003) Biological identifications through DNA barcodes. Proc. R. Soc. B, 270, 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kissling W.D., Ahumada J.A., Bowser A. et al. (2018) Building essential biodiversity variables (EBVs) of species distribution and abundance at a global scale. Biol. Rev., 93, 600–625. [DOI] [PubMed] [Google Scholar]
- 50. Oren A., Garrity G.M. and Parte A.C. (2018) Why are so many effectively published names of prokaryotic taxa never validated? Int. J. Syst. Evol. Microbiol., 68, 2125–2129. [DOI] [PubMed] [Google Scholar]
- 51. Ciufo S., Kannan S., Sharma S. et al. (2018) Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int. J. Syst. Evol. Microbiol., 68, 2386–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. NCTC (2020) The National Collection of Type Cultures (NCTC) for bacteria. Public Health England, continuously accessed. [Google Scholar]
- 53. Group, T.A.P (2016) An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc., 181, 1–20. [Google Scholar]
- 54. The Pteridophyte Phylogeny Group (2016) A community-derived classification for extant lycophytes and ferns. J. Syst. Evol., 6, 563–603. [Google Scholar]
- 55. May T.W., Redhead S.A., Bensch K. et al. (2019) Chapter F of the International Code of Nomenclature for algae, fungi, and plants as approved by the 11th International Mycological Congress, San Juan, Puerto Rico, July 2018. IMA Fungus, 10, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Spatafora J.W., Chang Y., Benny G.L. et al. (2016) A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia, 108, 1028–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tedersoo L., Sanchez-Ramirez S., Koljalg U. et al. (2018) High-level classification of the fungi and a tool for evolutionary ecological analyses. Fungal Divers., 90, 135–159. [Google Scholar]
- 58. Wijayawardene N.N., Pawlowska J., Letcher P.M. et al. (2018) Notes for genera: basal clades of fungi (including Aphelidiomycota, Basidiobolomycota, Blastocladiomycota, Calcarisporiellomycota, Caulochytriomycota, Chytridiomycota, Entomophthoromycota, Glomeromycota, Kickxellomycota, Monoblepharomycota, Mortierellomycota, Mucoromycota, Neocallimastigomycota, Olpidiomycota, Rozellomycota and Zoopagomycota). Fungal Divers., 92, 43–129. [Google Scholar]
- 59. Adl S.M., Bass D., Lane C.E. et al. (2019) Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol., 66, 4–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hawksworth D.L. (2011) A new dawn for the naming of fungi: impacts of decisions made in Melbourne in July 2011 on the future publication and regulation of fungal names. Mycokeys, 1, 7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schoch C.L., Seifert K.A., Huhndorf S. et al. (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci. U. S. A., 109, 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bissett J., Gams W., Jaklitsch W.M. et al. (2015) Accepted Trichoderma names in the year 2015. IMA Fungus, 6, 263–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Robbertse B., Strope P.K., Chaverri P. et al. (2017) Improving taxonomic accuracy for fungi in public sequence databases: applying ‘one name one species’ in well-defined genera with Trichoderma/Hypocrea as a test case. Database, 2017, bax072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schoch C.L., Robbertse B., Robert V. et al. (2014) Finding needles in haystacks: linking scientific names, reference specimens and molecular data for fungi. Database, 2014, bau061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Adl S.M., Simpson A.G.B., Lane C.E. et al. (2012) The revised classification of eukaryotes. J. Eukaryot. Microbiol., 59, 429–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Burki F., Roger A.J., Brown M.W. et al. (2020) The new tree of eukaryotes. Trends Ecol. Evol., 35, 43–55. [DOI] [PubMed] [Google Scholar]
- 67. Zhang Z.Q. (2013) Animal biodiversity: an update of classification and diversity in 2013. Zootaxa, 3703, 5–11. [DOI] [PubMed] [Google Scholar]
- 68. Marletaz F., Peijnenburg K.T.C.A., Goto T. et al. (2019) A new spiralian phylogeny places the enigmatic arrow worms among gnathiferans. Curr. Biol., 29, 312–318. [DOI] [PubMed] [Google Scholar]
- 69. Philippe H., Poustka A.J., Chiodin M. et al. (2019) Mitigating anticipated effects of systematic errors supports sister-group relationship between Xenacoelomorpha and Ambulacraria. Curr. Biol., 29, 1818–1826. [DOI] [PubMed] [Google Scholar]
- 70. Edgecombe G.D., Giribet G., Dunn C.W. et al. (2011) Higher-level metazoan relationships: recent progress and remaining questions. Org. Divers. Evol., 11, 151–172. [Google Scholar]
- 71. Hejnol A., Obst M., Stamatakis A. et al. (2009) Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc. R. Soc. B, 276, 4261–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cannon J.T., Vellutini B.C., Smith J. et al. (2016) Xenacoelomorpha is the sister group to Nephrozoa. Nature, 530, 89–93. [DOI] [PubMed] [Google Scholar]
- 73. Philippe H., Brinkmann H., Copley R.R. et al. (2011) Acoelomorph flatworms are deuterostomes related to Xenoturbella. Nature, 470, 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lu T.M., Kanda M., Satoh N. et al. (2017) The phylogenetic position of dicyemid mesozoans offers insights into spiralian evolution. Zool. Lett., 3, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Laumer C.E., Fernandez R., Lemer S. et al. (2019) Revisiting metazoan phylogeny with genomic sampling of all phyla. Proc. R. Soc. B, 286, 20190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Betancur-R R., Wiley E.O., Arratia G. et al. (2017) Phylogenetic classification of bony fishes. BMC Evol. Biol., 17, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ratnasingham S. and Hebert P.D.N. (2013) A DNA-based registry for all animal species: the barcode index number (BIN) system. Plos One, 8, e66213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sayers E.W., Beck J., Brister J.R. et al. (2020) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., D1, D9–D16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Brister J.R., Ako-Adjei D., Bao Y. et al. (2015) NCBI viral genomes resource. Nucleic Acids Res., 43, D571–D577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Schoch C.L., Aime M.C., de, Beer W. et al. (2017) Using standard keywords in publications to facilitate updates of new fungal taxonomic names. IMA Fungus, 8, 70–73. [Google Scholar]
- 81. Bidartondo M. (2008) Preserving accuracy in GenBank. Science, 319, 1616–1616. [DOI] [PubMed] [Google Scholar]
- 82. Garg A., Leipe D. and Uetz P. (2019) The disconnect between DNA and species names: lessons from reptile species in the NCBI taxonomy database. Zootaxa, 4706, 401–407. [DOI] [PubMed] [Google Scholar]
- 83. Wu L. and Ma J. (2019) The global catalogue of microorganisms (GCM) 10K type strain sequencing project: providing services to taxonomists for standard genome sequencing and annotation. Int. J. Syst. Evol. Microbiol., 69, 895–898. [DOI] [PubMed] [Google Scholar]
- 84. Whitman W.B., Klenk H.P., Arahal D.R. et al. (2019) Genomic Encyclopedia of Bacteria and Archaea (GEBA) VI: learning from type strains. Microbiol. Aust., 40, 125–129. [Google Scholar]
- 85. Becker P., Bosschaerts M., Chaerle P. et al. (2019) Public microbial resource centers: key hubs for findable, accessible, interoperable, and reusable (FAIR) microorganisms and genetic materials. Appl. Environ. Microbiol., 85, e01444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Godden G.T. and Soltis P.S. (2014) A new iDigBio web feature links DNA banks and genetic resources repositories in the United States In: Applequist WA, Campbell LM (eds). DNA Banking for 21st Century. Missouri Botanical Garden, St. Louis, MO, pp. 173–181. [Google Scholar]
- 87. Droege G., Barker K., Seberg O. et al. (2016) The global genome biodiversity network (GGBN) data standard specification. Database, 2016, baw125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Güntsch A., Hyam R., Hagedorn G. et al. (2017) Actionable, long-term stable and semantic web compatible identifiers for access to biological collection objects. Database, 2017, bax003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Boundy-Mills K., McCluskey K., Elia P. et al. (2020) Preserving US microbe collections sparks future discoveries. J. Appl. Microbiol., 129, 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Meiklejohn K.A., Damaso N. and Robertson J.M. (2019) Assessment of BOLD and GenBank—their accuracy and reliability for the identification of biological materials. Plos One, 14, e0217084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Berney C., Ciuprina A., Bender S. et al. (2017) UniEuk: time to speak a common language in protistology! J. Eukaryot. Microbiol., 64, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bánki O., Döring M., Holleman A. et al. (2018) Catalogue of life plus: innovating the CoL systems as a foundation for a clearinghouse for names and taxonomy. Biodivers. Inf. Sci. Stand., 2, e26922. [Google Scholar]
- 93. Leray M., Knowlton N., Ho S.L. et al. (2019) GenBank is a reliable resource for 21st century biodiversity research. Proc. Natl. Acad. Sci. U. S. A., 116, 22651–22656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


