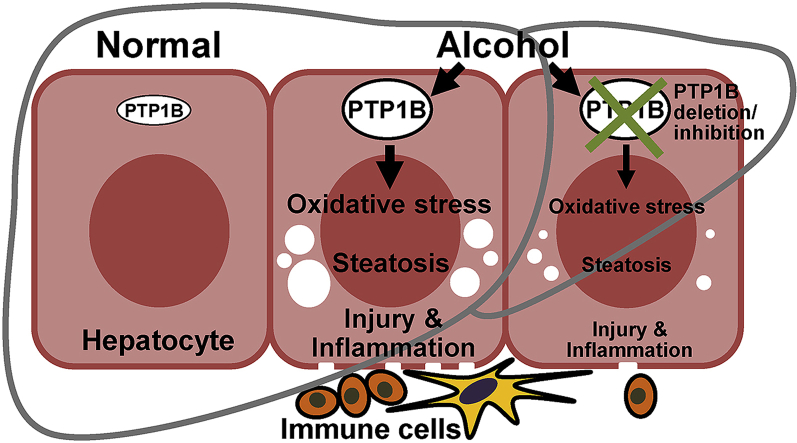
Abstract
Alcoholic liver disease (ALD) is a major health problem and a significant cause of liver-related death. Currently, the mainstay for ALD therapy is alcohol abstinence highlighting the need to develop pharmacotherapeutic approaches. Protein-tyrosine phosphatase 1B (PTP1B) is an established regulator of hepatic functions, but its role in ALD is mostly unexplored. In this study, we used mice with liver-specific PTP1B disruption as well as pharmacological inhibition to investigate the in vivo function of this phosphatase in ALD. We report upregulation of hepatic PTP1B in the chronic plus binge mouse model and, importantly, in liver biopsies of alcoholic hepatitis patients. Also, mice with hepatic PTP1B disruption attenuated ethanol-induced injury, inflammation, and steatosis compared with ethanol-fed control animals. Moreover, PTP1B deficiency was associated with decreased ethanol-induced oxidative stress in vivo and ex vivo. Further, pharmacological modulation of oxidative balance in hepatocytes identified diminished oxidative stress as a contributor to the salutary effects of PTP1B deficiency. Notably, PTP1B pharmacological inhibition elicited beneficial effects and mitigated hepatic injury, inflammation, and steatosis caused by ethanol feeding. In summary, these findings causally link hepatic PTP1B and ALD and define a potential therapeutic target for the management of this disease.
Keywords: Alcoholic liver disease, Hepatocyte, Protein-tyrosine phosphatase 1B, Oxidative stress, Inflammation, Steatosis
Graphical abstract
Highlights
-
•
Hepatic PTP1B expression is elevated in a mouse model of ALD and AH patients.
-
•
Liver-specific PTP1B disruption ameliorated ALD in mice.
-
•
PTP1B deficiency was associated with decreased ethanol-induced oxidative stress.
-
•
PTP1B pharmacological inhibition attenuated alcohol-induced hepatic injury in mice.
1. Introduction
Chronic excessive alcohol consumption is a significant contributor to the development of alcoholic liver disease (ALD), which is a global healthcare problem [1,2]. The disease encompasses a broad spectrum of pathologies that range from steatosis to alcoholic hepatitis (AH) and may progress to hepatocellular carcinoma. While the majority of heavy drinkers develop fatty liver, only minority progress to AH, and 10–15% develop cirrhosis [3,4]. Also, up to 40% of severe AH patients die within six months [5]. The current foundation for ALD therapy is alcohol cessation underscoring the need to develop novel prevention and treatment strategies [6,7]. Deciphering the intricate mechanisms underlying ALD pathogenesis is vital for identifying new targets and developing effective mechanism-based interventions.
Protein tyrosine phosphorylation is a crucial post-translational mechanism that maintains homeostasis and is dynamically regulated by protein-tyrosine kinases and protein-tyrosine phosphatases (PTPs) [8]. Protein-tyrosine phosphatase 1B (PTP1B; encoded by PTPN1) is a widely expressed non-receptor phosphatase that modulates many facets of signaling (reviewed in Ref. [[9], [10], [11]]). Also, PTP1B is a long-standing therapeutic target for obesity, type 2 diabetes (T2D), and some cancers [12,13]. Notably, PTP1B antisense oligonucleotides enhance insulin sensitivity in overweight, T2D patients [14]. Compelling evidence implicate PTP1B in the liver metabolic and non-metabolic functions. Mice with liver-specific PTP1B disruption exhibit increased insulin sensitivity and decreased accumulation of hepatic triglyceride and cholesterol when fed a high-fat diet (HFD) [15,16]. Moreover, PTP1B deficiency confers protection in rodent models of liver injury, including FAS-induced hepatic failure, liver regeneration following partial hepatectomy, hepatotoxicity caused by acetaminophen overdose, and bile duct ligation-induced fibrosis [[17], [18], [19], [20], [21]].
A growing body of evidence suggests a role for PTP1B in ALD. Ethanol leads to the upregulation of PTP1B in the muscle and liver of rats and muscle of macaque [[22], [23], [24]]. Additionally, binge drinking increases hypothalamic PTP1B and induces systemic insulin resistance in rats, which is prevented by the administration of a PTPB inhibitor [25]. Moreover, PTP1B modulates NF-κB signaling and enhances macrophage activation under the ethanol challenge [26]. However, knowledge gaps persist, including the role of hepatic PTP1B in ALD, the underlying molecular mechanism, and the suitability of this phosphatase as a therapeutic target for this disease. In this study, we evaluated the expression of hepatic PTP1B under ethanol challenge, determined the effects of PTP1B disruption and pharmacological inhibition in the chronic plus binge preclinical mouse model of ALD, and investigated the molecular mechanism.
2. Materials and methods
2.1. Reagents and human samples
Antibodies for GAPDH (3683), F4/80 (70076), phospho–NF–κB p65 Ser536 (3033), NF-κB p65 (8242), phospho-AKT Ser473 (4060), phospho-ERK1/2 Thr202/Tyr204 (4370) and ERK1/2 (4695) were from Cell Signaling Technology; AKT (sc-81434), phospho-eNOS Ser1177 (sc-21871-R), eNOS (sc-376751), iNOS (sc-7271), SOD1 (sc-271014) and GPx1/2 (sc-133160) were from Santa Cruz Biotechnology; CYP2E1 (ab28146), NOX2 (ab129068), NOX4 (ab216654) and 4-hydroxynonenal (4-HNE, ab46545) were from Abcam. Antibodies for murine (AF3954) and human (MABS197) PTP1B were from R&D Systems and Millipore, respectively. Information on the used antibodies is detailed in Supplementary Table 1. N-acetylcysteine (NAC; A9165) and hydrogen peroxide (H2O2; 386790) were from Sigma. PTP1B pharmacological inhibitor (DPM-1001) was from Glixx Laboratories (GLXC-11401). Dr. Zhaoli Sun (Johns Hopkins University) kindly provided de-identified human liver biopsies from healthy donors and alcoholic hepatitis patients.
2.2. Mouse studies
Mice with liver-specific PTP1B disruption were generated by crossing PTP1B floxed (Ptpn1fl/fl) mice [27] to Alb-Cre mice, with both strains on the C57BL/6J background. We housed mice in a temperature- and humidity-controlled animal facility under a 12-h light/dark cycle with free access to food and water. For the ethanol challenge, we used the chronic plus single binge mouse model [28]. Briefly, age-matched (8–12 weeks old) female control (Ptpn1fl/fl) mice and those with hepatic PTP1B disruption (Ptpn1fl/fl, Alb-Cre) were adapted to Lieber-DeCarli control liquid diet (F1259SP, Bio-Serv) for five days. After the acclimation period, the mice were fed 5% (v/v) ethanol liquid diet (cat #F1258SP, Bio-Serv) ad libitum or pair-fed an isocaloric control liquid diet (cat #F1259SP, Bio-Serv) for ten days. On day eleven, ethanol- and pair-fed mice were dosed by oral gavage with a single bolus of ethanol (5 g/kg body weight) or isocaloric maltose dextrin solution for 9 h then sacrificed and tissues collected and stored for further analysis. For PTP1B pharmacological inhibition, wild-type female mice (C57BL/6J background, 12–16 weeks old) were treated daily with 5 mg/kg of DPM-1001/DMSO in the ethanol liquid diet at the initiation of ethanol feeding. An equal amount of DMSO was applied to the control group. All mouse studies were approved by the Institutional Animal Care and Use Committee guidelines at the University of California Davis.
2.3. Histology
4% paraformaldehyde-fixed liver samples were paraffin-embedded, sectioned, and hematoxylin/eosin (H&E)-stained by the Anatomic Pathology Service (UC Davis). Images were acquired by the Olympus BX51 microscope. For immunofluorescence, liver sections were deparaffinized in xylene, and heat-mediated antigen retrieval was performed with citrate buffer (10 mM sodium citrate, pH 6.0) for F4/80 antibodies and Tris-EDTA buffer (10 mM Tris Base, 1 mM EDTA, pH 9.0) for 4-HNE and human PTP1B antibodies. Samples were blocked by 3% BSA at room temperature for 1 h then incubated with primary antibodies at 4 °C overnight. Images were visualized with appropriate Alexa Fluor-conjugated secondary antibodies (Thermo Fisher Scientific) and detected by an Olympus FV1000 laser scanning confocal microscope.
2.4. Biochemical analyses
Frozen liver samples were ground by mortar and pestle in the presence of liquid nitrogen. Protein was extracted by radioimmunoprecipitation assay buffer containing 10 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate, 5 mM EDTA, 20 mM NaF, 2 mM sodium orthovanadate and protease inhibitors. Whole lysates were clarified by centrifugation at 12,000 rcf for 10 min at 4 °C, and protein concentrations quantified using a BCA protein assay kit (Pierce). For immunoblotting, tissue lysates were resolved by SDS-PAGE and transferred to PVDF membranes (Bio-Rad). Target proteins were recognized with the relevant primary and secondary antibodies incubated at 4 °C overnight and at room temperature for 1 h, respectively. Blots were incubated with the HyGLO Chemiluminescent HRP antibody detection kit (Denville Scientific) then exposed to HyBlot autoradiography films (Denville Scientific). Band intensities were quantitated using the FluorChem 9900 program (Alpha Innotech). Protein phosphorylation was normalized to the corresponding protein expression. Blood plasma samples were collected by centrifugation at 2,000 rcf for 15 min at 4 °C, and alanine aminotransferase (ALT) determined using ALT/SGPT color endpoint kit (A526-120, Teco Diagnosis). For hepatic triglycerides, liver (~25 mg) was homogenized in equal amounts (1:1 v/v) of PBS and chloroform/methanol (2:1 v/v) solutions. After vortexing for 3 min, the mixture was centrifuged at 3,000 rcf for 10 min at room temperature, and the lower layer was collected to air-dry overnight. The pellet was re-suspended in isopropanol and measured using Infinity Triglycerides Liquid Stable Reagent kit (TR22421, Thermo Fisher Scientific). All assays were conducted following the manufacturer's instructions.
2.5. Quantitative real-time PCR
Frozen livers were homogenized, and RNA extracted using TRIzol reagent (Invitrogen) with the quantity and quality determined using NanoDrop One (Thermo Fisher Scientific). After that, cDNA was generated using a high-capacity cDNA reverse transcription kit (Applied Biosystems). Samples were mixed with SsoAdvanced Universal SYBR Green Supermix (Thermo Fisher Scientific) and relevant primer pairs to determine the threshold cycle (Ct) by CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Gene expression was normalized with TATA box-binding protein (Tbp) and beta-2-microglobulin (B2M) as the housekeeping genes for mouse and human samples, respectively. Relative fold change of mRNA level was calculated using the ΔCT method. The sequences of primers are listed in Supplementary Table 2.
2.6. Isolation and culture of primary hepatocytes
Hepatocyte isolation and culture were performed as previously described with few modifications [29]. Briefly, stock solutions were prepared (1L, pH 7.3) as detailed and filtered using a 0.22 μm PVDF membrane (SCGVU10RE, Millipore). MEM buffer: Minimum essential medium (M0268, Sigma), 2.2 g NaHCO3. Solution I: Hanks' balance salts without Ca2+ (H2387, Sigma), 2.38 g HEPES, 0.35 g NaHCO3, 0.19 g EDTA. Solution II: MEM buffer plus 0.1 mg/ml Collagenase P (11249002001, Sigma), freshly prepared before use. Solution III: Hanks' balanced salts (H6136, Sigma), 2.38 g HEPES, 0.35 g NaHCO3. WE medium: Williams’ E medium (W4125, Sigma), 2.6 g HEPES, 2.2 g NaHCO3, 100 ml 10× penicillin/streptomycin (15140122, Gibco). Mice were sacrificed, and the liver was perfused slowly through a portal vein by 10 ml warm Solution I then 10 ml warm Solution II with periodically applying pressure on inferior vena cava to swell the tissue. After that, the liver was collected and incubated with 10 ml Solution II in a 50 ml polypropylene tube for 8 min at 37 °C. Next, the liver lobes were torn apart gently for hepatocyte release with 10 ml Solution III plus 10% FBS. The cell suspension was filtered through a 100 μm strainer and washed again by 10 ml Solution III (10% FBS). Samples were centrifuged at 50 rcf for 2 min to pellet hepatocytes, and the pellet was resuspended with 20 ml Solution III. The wash step was repeated once, and then the pellet was resuspended in WE medium for cell counting by hemocytometer. Isolated hepatocytes were seeded on a collagen-coated culture surface at 1.5 × 105/well for 24-well plate (354408, Corning) and 6 × 105/well for a 35 mm dish (356456, Corning). After 6h incubation with WE medium containing 10% FBS at 37 °C, primary hepatocytes were maintained with WE medium plus 1% FBS at 37 °C until further experiments.
2.7. Measurement of reactive oxygen species (ROS)
The primary hepatocytes were pre-loaded with the ROS probe (CM-H2DCFDA, Thermo Fisher Scientific) at 50 μg/24-well plates for 30 min at 37 °C. Then hepatocytes were treated with ethanol (100 mM) and NAC (2 mM) or H2O2 (10 μM) for an additional 60 min at 37 °C. Data were collected using a plate reader (BioTek Synergy H1) with excitation at 492 nm and emission at 522 nm.
2.8. Statistics
Data were expressed as means + standard error of the mean (SEM) and statistical analyses performed using Excel with Real Statistics plug-in (www.real-statistics.com). Statistical significance was determined by one-way ANOVA with post-hoc Tukey's test or two-tailed t-test as appropriate. The details of each experiment, including the number of samples, are included in the figures legends. Differences were considered significant at p < 0.05.
3. Results
3.1. Elevated hepatic PTP1B in ethanol-fed mice and AH patients
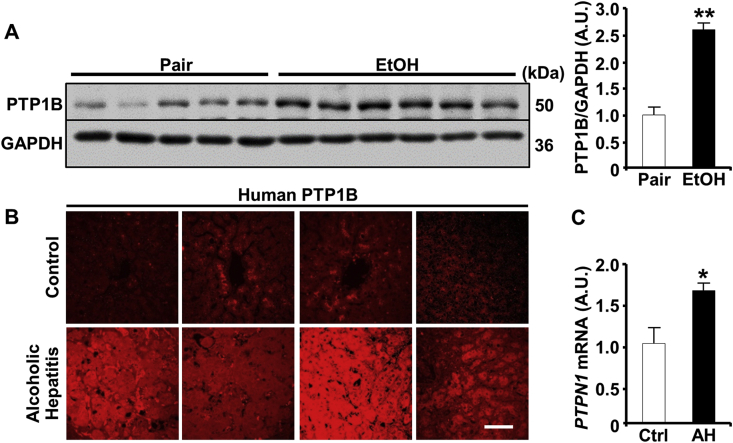
Ethanol elicits the upregulation of PTP1B in rat and macaque models [[22], [23], [24]]. Herein, we determined the expression of hepatic PTP1B in the chronic plus binge mouse model of ALD and AH patients. Immunoblotting of hepatic lysates demonstrated a significant increase in PTP1B protein expression in ethanol-fed compared with pair-fed wild-type mice (Fig. 1A). Additionally, we evaluated PTP1B expression in de-identified human liver biopsies from healthy subjects and AH patients. Notably, the biopsies from AH patients displayed significantly increased expression of PTP1B mRNA and more intense immunostaining compared to healthy subjects (Fig. 1B and C). These findings ascertained elevated hepatic PTP1B upon ethanol challenge and suggested that dysregulation of PTP1B signaling might be relevant to ethanol-induced hepatic dysfunction.
Fig. 1.
Elevated hepatic PTP1B expression in the chronic plus binge mouse model and alcoholic hepatitis patients. A) Immunoblots of PTP1B and GAPDH expression in liver lysates from wild-type female mice used in the chronic plus binge model (left panel). Pair (n = 5): control diet + maltose gavage, EtOH (n = 6): ethanol diet + ethanol gavage. Each lane represents an independent animal. PTP1B protein level was quantitated and normalized with GAPDH, then expressed as means + SEM (right panel. **p < 0.01 Pair vs. EtOH. (B) Immunostaining of PTP1B in liver sections (n = 4 per group), and (C) hepatic PTPN1 mRNA from healthy subjects (control; Ctrl) and alcoholic hepatitis (AH) patients. Gene expression was determined by qPCR, normalized to B2M mRNA, then expressed as means + SEM (n = 5 for Ctrl and n = 6 for AH). *p < 0.05 Ctrl vs. AH by a two-tailed t-test. A.U.: arbitrary unit. Scale bar: 50 μm.
3.2. Hepatic PTP1B disruption mitigated ethanol-induced injury, inflammation, and steatosis
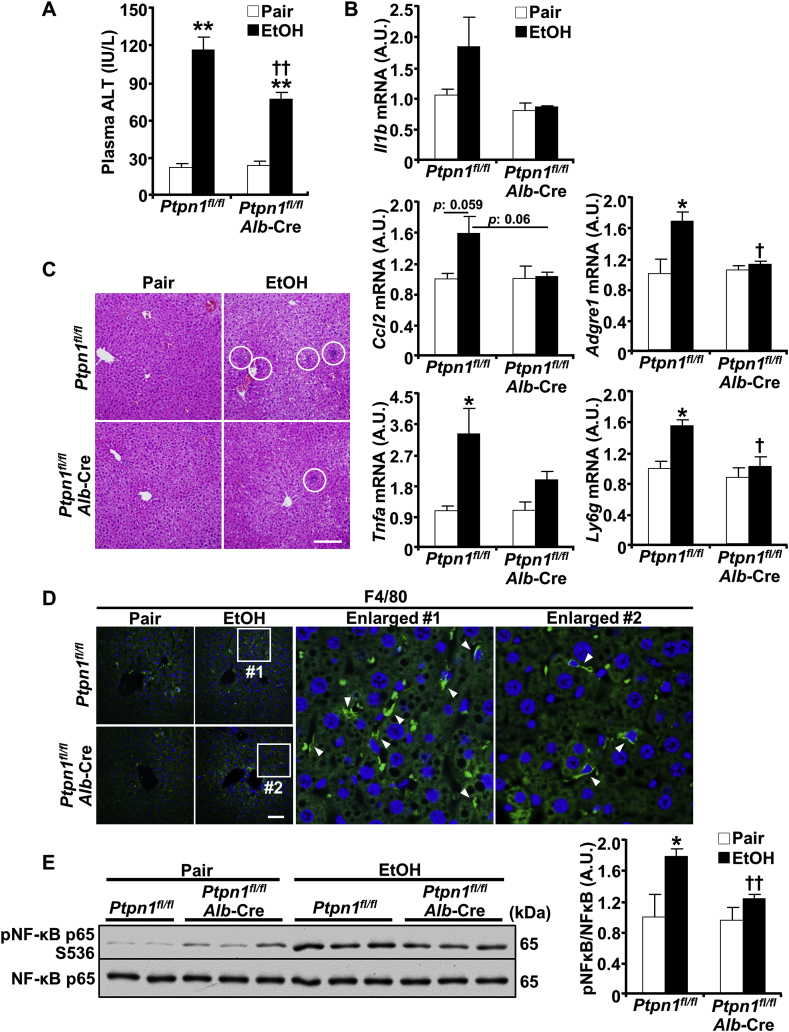
To determine the extent to which the altered PTP1B expression might contribute to ALD, we used mice with liver-specific PTP1B deficiency (Ptpn1fl/fl, Alb-Cre; termed LPTP1B KO) [15,27]. We subjected age-matched control (Ptpn1fl/fl) and LPTP1B KO female mice to an ethanol challenge using the chronic plus binge model and assessed ensuing alterations. This experimental model recapitulates several aspects of human disease, including hepatic injury, inflammation, and steatosis [28]. The body weights of pair- and ethanol-fed control and LPTP1B KO mice were comparable (data not sown). To evaluate the hepatic injury, we determined blood alanine aminotransferase (ALT) in control and LPTP1B KO mice under pair- and ethanol-fed states. Ethanol increased ALT in both genotypes, but that was significantly lower in the LPTP1B KO mice (Fig. 2A). Also, we assessed the expression of inflammatory cytokines and marker genes of immune cells. Hepatic mRNA expression of interleukin‐1β (Il1b), monocyte chemotactic protein 1 (MCP1; Ccl2), and tumor necrosis factor-alpha (Tnfa) under ethanol feeding exhibited a trend of lower induction in LPTP1B KO mice (Fig. 2B). Similarly, we observed significantly lower ethanol-induced expression of macrophage F4/80 (Adgre1) and lymphocyte antigen 6 complex locus G6D (Ly6g) mRNA in LPTP1B KO mice compared to control animals (Fig. 2B). In keeping with this, hematoxylin and eosin and F4/80 immunostaining showed fewer ethanol-induced immune cells and macrophages in LPTP1B KO mice compared with controls (Fig. 2C and D). Consistent with these findings, LPTP1B KO mice exhibited significantly lower ethanol-induced hepatic NF-κB phosphorylation compared with control animals (Fig. 2E).
Fig. 2.
Hepatic PTP1B disruption mitigates ethanol-induced injury and inflammation. Ctrl (Ptpn1fl/fl) and KO (Ptpn1fl/fl, Alb-Cre) female mice were used in the chronic plus binge model. Pair: control diet + maltose gavage, EtOH: ethanol diet + ethanol gavage. A) Plasma ALT presented in a bar chart as means + SEM (n = 5 pair-fed Ctrl mice and n = 6 for each of the remaining groups). **p < 0.01 Pair vs. EtOH and ††p < 0.01 Ctrl vs. KO by one-way ANOVA with post-hoc Tukey's test. B) Hepatic mRNA of Il1b, Ccl2, Tnfa, Adgre1, and Ly6g were determined by qPCR normalized to Tbp then expressed as means + SEM (n = 3 per group). *p < 0.05 Pair vs. EtOH and †p < 0.05 Ctrl vs. KO by a two-tailed t-test. C) H&E-stained liver sections of Ctrl and KO mice with ethanol-induced inflammatory cell infiltration indicated by white circles. Scale bar: 100 μm. D) Confocal images of liver sections immunostained with the macrophage marker F4/80 (green) and DAPI counterstain (blue). Boxed areas are enlarged, and arrowheads indicate macrophages. Scale bar: 50 μm. E) Immunoblots of pNF-κB and NF-κB in hepatic lysates and each lane represents an independent animal. The phosphorylation level was normalized to protein expression and represented as means + SEM (right panel; n = 5 pair-fed Ctrl mice and n = 6 for each of the remaining groups). *p < 0.05 Pair vs. EtOH and ††p < 0.01 Ctrl vs. KO by a two-tailed t-test. A.U.: arbitrary unit.
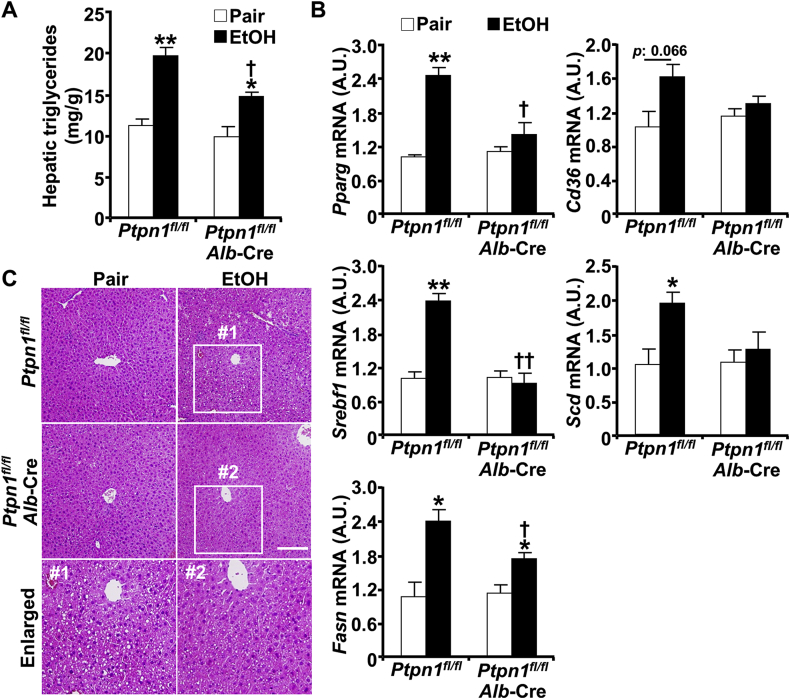
Having established the effects of hepatic PTP1B disruption on ethanol-induced injury and inflammation, next, we sought to assess its impact on hepatic steatosis. Ethanol oxidative metabolites, including acetaldehyde and reactive oxygen species (ROS), exert a myriad of effects that alter lipid homeostasis and promote steatosis [6]. Ethanol increased hepatic triglycerides, the predominant lipids implicated in steatosis, in control (Ptpn1fl/fl) and LPTP1B KO mice, but to a significantly lower level in the latter (Fig. 3A). This phenotype was accompanied by alterations in the expression of genes involved in hepatic de novo lipogenesis and fatty acid uptake. Indeed, LPTP1B KO mice presented diminished ethanol-induced expression of mRNA of lipogenesis-related genes namely peroxisome proliferator-activated receptor gamma (Pparg), as well as sterol regulatory element-binding protein 1c (Srebf1) and its targets fatty acid synthase (Fasn) and sterol CoA desaturase (Scd) (Fig. 3B). Additionally, the cluster of differentiation 36 (Cd36) that is implicated in FA uptake exhibited a trend for lower induction in LPTP1B KO mice compared with controls. In keeping with this, histological analysis and gross morphology revealed decreased ethanol-induced accumulation of lipid vacuoles in the livers of LPTP1B KO mice compared with controls (Fig. 3C). Altogether, these findings demonstrate that hepatic PTP1B disruption mitigates ethanol-induced injury and inflammation, as well as hepatic steatosis.
Fig. 3.
Hepatic PTP1B disruption attenuates ethanol-induced steatosis. Ctrl (Ptpn1fl/fl) and KO (Ptpn1fl/fl, Alb-Cre) female mice were used in the chronic plus binge model. Pair: control diet + maltose gavage, EtOH: ethanol diet + ethanol gavage. A) Hepatic triglycerides concentrations presented as means + SEM (n = 5 pair-fed Ctrl mice and n = 6 for each of the remaining groups). *p < 0.05, **p < 0.01 Pair vs. EtOH and †p < 0.05 Ctrl vs. KO by one-way ANOVA with post-hoc Tukey's test. B) Hepatic mRNA of Pparg, Srebf1, Fasn, Cd36, and Scd were determined by qPCR normalized to Tbp then expressed as means + SEM (n = 3 per group). *p < 0.05, **p < 0.01 Pair vs. EtOH and †p < 0.05, ††p < 0.01 Ctrl vs. KO by a two-tailed t-test. A.U.: arbitrary unit. C) H&E-stained liver sections of Ctrl and KO mice demonstrating lipid accumulation and boxed areas are enlarged. Scale bar: 100 μm.
3.3. Hepatic PTP1B disruption ameliorated ethanol-induced oxidative stress in vivo and ex vivo
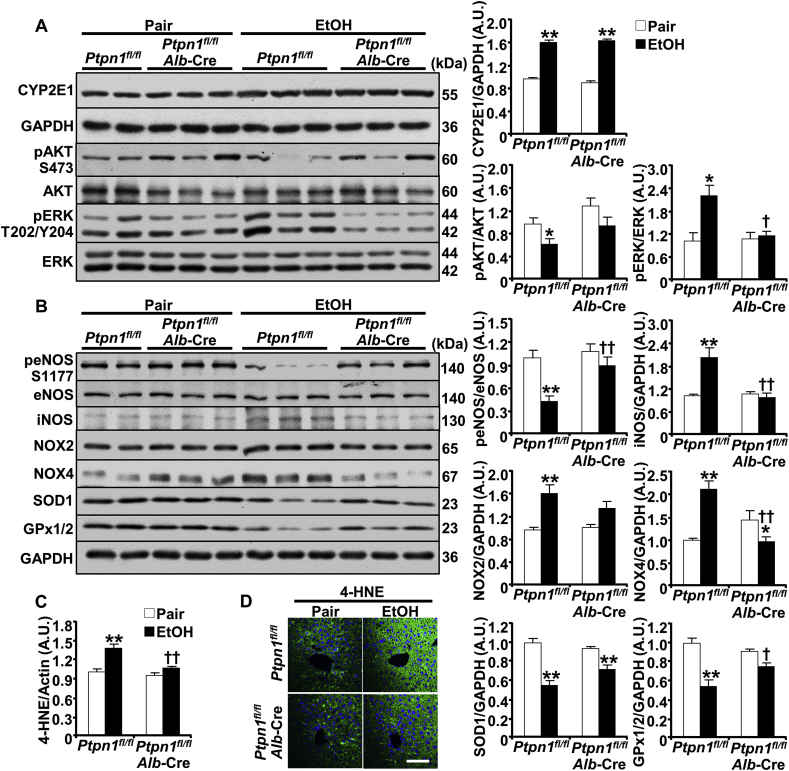
We sought to delineate the pathways that might mediate hepatic PTP1B actions under ethanol challenge. Ethanol oxidative metabolism is a significant contributor to ALD [30,31]. In hepatocytes, alcohol is degraded by inducible (CYP2E1) and non-inducible (alcohol dehydrogenase) enzymes that generate acetaldehyde and ROS, thereby contributing to cellular injury [6,[31], [32], [33]]. We evaluated the effects of hepatic PTP1B disruption on ethanol-induced oxidative stress and lipid peroxidation in pair- and ethanol-fed control (Ptpn1fl/fl) and LPTP1B KO mice. Ethanol increased hepatic CYP2E1 expression as expected [32] and to a comparable level in control and LPTP1B KO mice (Fig. 4A). Notably, PTP1B deficiency mitigated ethanol-induced attenuation of AKT phosphorylation and elevation of ERK phosphorylation (Fig. 4A). Consistently, ethanol-induced attenuation of endothelial nitric oxide synthase (eNOS) phosphorylation was significantly lower in LPTP1B KO mice compared with controls (Fig. 4B). Moreover, ethanol increased the expression of inducible nitric oxide synthase (iNOS), NADPH oxidase 2 (NOX2), and NADPH oxidase 4 (NOX4) in both genotypes but to a significantly lower level in the LPTP1B KO mice. Additionally, hepatic PTP1B deficiency diminished the ethanol-induced reduction of the antioxidants glutathione peroxidase 1/2 (GPx1/2) and superoxide dismutase 1 (SOD1) (Fig. 4B). In keeping with this, the oxidative stress-induced lipid peroxidation evaluated by 4-HNE immunoblotting (Supplementary Fig. S1 and Fig. 4C) and immunostaining (Fig. 4D), was significantly lower in ethanol-fed LPTP1B KO mice compared with controls. Thus, hepatic PTP1B disruption in vivo was associated with diminished ethanol-induced activation of critical components in oxidative stress signaling and lipid peroxidation.
Fig. 4.
Hepatic PTP1B disruption is associated with decreased ethanol-induced oxidative stress. Ctrl (Ptpn1fl/fl) and KO (Ptpn1fl/fl, Alb-Cre) female mice were used in the chronic plus binge model. Pair: control diet + maltose gavage, EtOH: ethanol diet + ethanol gavage. Immunoblots of (A) CYP2E1, pAKT, AKT, pERK, ERK and GAPDH, and (B) peNOS, eNOS, iNOS, NOX2, NOX4, SOD1, GPx1/2, and GAPDH in mouse liver lysates (left panels). Each lane represents an independent animal. Protein expression of CYP2E1, iNOS, NOX2, NOX4, SOD1, and GPx1/2 was normalized with GAPDH. Phosphorylation of AKT, ERK, and eNOS was normalized with their respective protein. C) Lipid peroxidation was determined by immunoblotting liver lysates with 4-hydroxynonenal (4-HNE) then quantitated (full lane intensity of 4-HNE immunoblot was normalized with Actin), as well as immunostaining of liver sections (D). All quantitative results were plotted as means + SEM (n = 5 pair-fed Ctrl mice and n = 6 for each of the remaining groups). *p < 0.05, **p < 0.01 Pair vs. EtOH and †p < 0.05, ††p < 0.01 Ctrl vs. KO by a two-tailed t-test (A), and One-way ANOVA with post-hoc Tukey's test (B, C). A.U.: arbitrary unit. Scale bar: 50 μm.
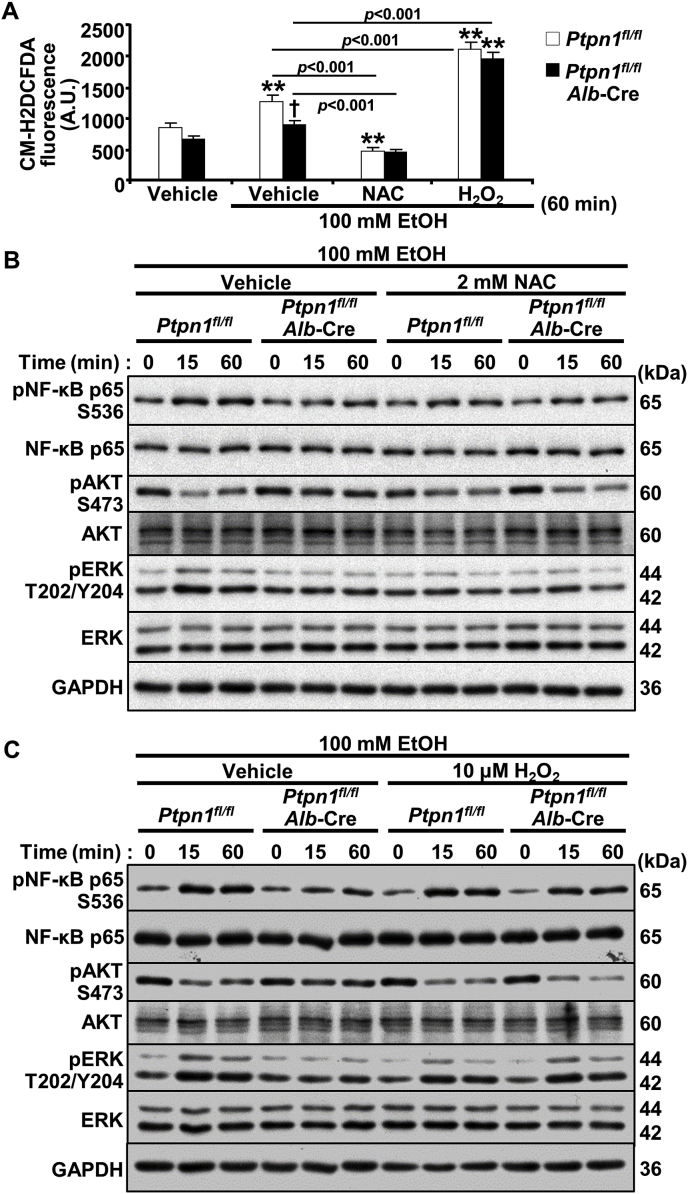
Having established that hepatic PTP1B disruption can attenuate ethanol-induced oxidative stress in vivo, we next evaluated whether this might occur in ex vivo. To this end, we isolated primary hepatocytes from control and LPTP1B KO mice and determined the impact of ethanol on oxidative stress. Ethanol increased ROS in primary hepatocytes but to a significantly lower level in LPTP1B KO compared to control hepatocytes (Fig. 5A). To ascertain that alterations in intracellular ROS contribute to the effects of PTP1B deficiency, we sought to refashion the ROS levels using pharmacological approaches. To this end, we treated primary hepatocytes from control and LPTP1B KO mice with NAC and H2O2 to diminish and increase intracellular ROS, respectively. We found that these interventions altered ROS levels as expected and rendered them comparable between genotypes (Fig. 5A). Subsequently, we evaluated inflammation, and oxidative stress signaling in the hepatocytes from control and LPTP1B KO mice. The ethanol-induced suppression of AKT and elevation of NF-κB and ERK phosphorylation were lower in LPTP1B KO compared to control hepatocytes (Fig. 5B and C). Remarkably, NAC treatment of control hepatocytes rendered them comparable to LPTP1B KO cells with lower ethanol-induced AKT suppression and NF-κB and ERK activation (Fig. 5B). On the other hand, treating LPTP1B KO hepatocytes with H2O2 abolished the protective effects of PTP1B deficiency, and the cells exhibited elevated ethanol-induced AKT suppression and NF-κB and ERK activation (Fig. 5C). Collectively, these findings established that the beneficial effects of hepatic PTP1B disruption are mediated, at least in part, by diminished ethanol-induced oxidative stress, and are consistent with being cell-autonomous.
Fig. 5.
PTP1B disruption modulates oxidative stress signaling in ethanol-treated hepatocytes. Primary hepatocytes isolated from Ctrl (Ptpn1fl/fl) and KO (Ptpn1fl/fl, Alb-Cre) male mice were incubated without and with 100 mM ethanol (EtOH) plus 2 mM N-acetylcysteine (NAC) and 10 μM hydrogen peroxide (H2O2) as indicated. A) Intracellular ROS levels as measured by CM-H2DCFDA upon EtOH stimulation and with interventions of NAC and H2O2 for 60 min. The CM-H2DCFDA fluorescence was plotted in the bar chart as means + SEM. **p < 0.01 without vs. with EtOH and †p < 0.05 Ctrl vs. KO. Statistical significance was assessed using one-way ANOVA with post-hoc Tukey's test. A.U.: arbitrary unit. Immunoblots of pNF-κB, NF-κB, pAKT, AKT, pERK, ERK, and GAPDH in lysates from hepatocytes co-treated with EtOH and NAC (B) or H2O2 (C) for the indicated periods.
3.4. PTP1B pharmacological inhibition alleviated the harmful effects of ethanol feeding
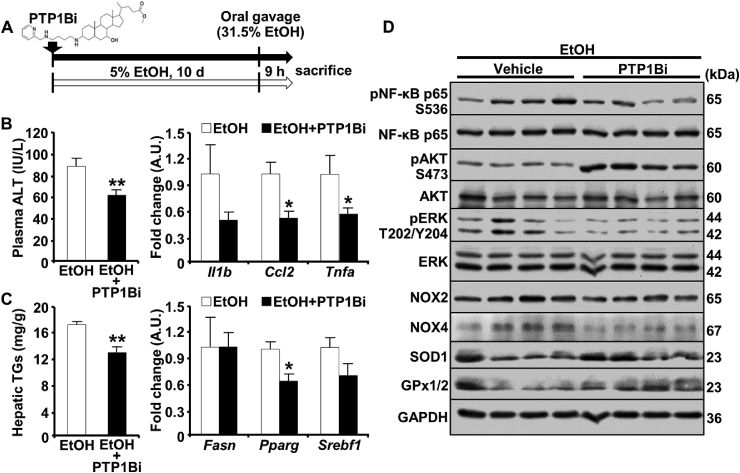
Given the salutary effects of PTP1B disruption, we determined the effects of its pharmacological targeting in the chronic plus binge model as outlined (Fig. 6A). To this end, we used the potent, non-competitive and orally bioavailable PTP1B inhibitor methyl (3β,7α)-3-((4-((pyridin-2-ylmethyl)amino)butyl)amino)-7-hydroxycholan-24-oate (PTP1Bi; DPM-1001) [34]. Briefly, wild-type female mice were fed the ethanol diet without and with the PTP1B inhibitor (5 mg/kg orally). Notably, PTP1B inhibition alleviated ethanol-induced hepatic injury and inflammation, as evidenced by decreased plasma ALT and hepatic expression of Il1b, Ccl2, and Tnfa mRNA (Fig. 6B). Moreover, ethanol-induced triglycerides accumulation and expression of key lipogenesis-related genes (Pparg, Srebf1) were lower in inhibitor-treated mice compared with non-treated control animals (Fig. 6C). Further, PTP1B pharmacological inhibition attenuated ethanol-evoked hepatic NF-κB and ERK activation, reduced NOX2 and NOX4 expression, and increased AKT activation as well as SOD1 and GPx1/2 expression (Fig. 6D). Altogether, these findings are consistent with PTP1B inhibition alleviating ALD in a preclinical mouse model, at least partly, by attenuating ethanol-induced inflammation and oxidative stress.
Fig. 6.
PTP1B pharmacological inhibition attenuates ethanol-induced hepatic injury, inflammation, steatosis, and oxidative stress. A) Schematic depicting the administration of PTP1B inhibitor (PTP1Bi; DPM-1001) in the chronic plus binge model. Wild-type female mice were provided 5% EtOH liquid diet without (white arrow) and with PTP1B inhibitor (black arrow) for ten days. Mice were sacrificed 9 h post oral gavage of 31.5% EtOH (0.02 ml/g body weight) on day 11. (B) Plasma ALT level (left panel; n = 6 EtOH and n = 7 EtOH + PTP1Bi) and hepatic mRNA of Il1b, Ccl2 and Tnfa (right panel; n = 4 per group), and (C) hepatic triglycerides (TGs; n = 7 EtOH and n = 8 EtOH + PTP1Bi) and mRNA of Fasn, Pparg and Srebf1 (right panel; n = 4 each group) are presented as means + SEM. *p < 0.05, **p < 0.01 EtOH vs. EtOH + PTP1Bi by a two-tailed t-test. D) Immunoblots of pNF-κB, NF-κB, pAKT, AKT, pERK, ERK, NOX2, NOX4, SOD1, GPx1/2, and GAPDH in mouse liver lysates. Each lane represents an independent animal.
4. Discussion
ALD is a significant cause of liver-related death, and alcohol cessation is the mainstay of therapy for this deadly malady. Given the challenges of sustaining changes in lifestyle and heavy alcohol consumption, there is an unmet need for pharmacotherapeutic interventions. The current findings implicated PTP1B, a long-standing therapeutic target for obesity and T2D, in ALD. Ethanol increased the expression of hepatic PTP1B, suggesting that dysregulation of its signaling might be relevant in ALD. Indeed, using a tissue-specific loss-of-function approach, we reported beneficial effects of hepatic PTP1B deficiency in a preclinical mouse model of the disease. Importantly, we explored a pharmacological intervention with translational potential by targeting PTP1B in vivo and identified this phosphatase as a likely target for ALD.
Several interrelated factors, including the regulation of expression, fine-tune PTP1B signaling. The observed elevation of hepatic PTP1B could be ascribed, in large part, to changes in hepatocytes as they constitute about 80% of the liver volume [35]. The functional significance of PTP1B upregulation remains to be elucidated but may exacerbate the disease. Indeed, the converse, PTP1B deficiency and pharmacological inhibition, elicited beneficial effects, and ameliorated ALD. Additionally, the ethanol-induced increase of PTP1B in mice is in keeping with the elevated expression of this phosphatase in animal models of disease [[36], [37], [38]]. In particular, chronic ethanol consumption increases PTP1B protein expression and activity in rat skeletal muscle [22] and liver [24], as well as macaque [23]. Notably, the elevated hepatic PTP1B in AH patients endowed potential translational relevance to the current observations. The expression of the closely-related T cell phosphatase (TCPTP) [39] that is also implicated in hepatic function [40] was not altered in biopsies of AH patients compared with healthy subjects indicating differential expression of these PTPs (Hsu and Haj, unpublished data). The molecular mechanism by which PTP1B is upregulated in ALD is currently uncharted. Also, the level of regulation is undetermined and may include alterations in translation and protein expression and degradation. Of note, HFD-induced inflammation in mice increases PTP1B mRNA and protein expression in vivo (liver, adipose tissue, and skeletal muscle) with TNFα inducing PTP1B transcription through NF-κB [38]. So it is reasonable to speculate that ethanol-triggered inflammation may contribute, at least in part, to the marked upregulation of hepatic PTP1B in ALD. Also, it is not clear if the elevated expression translates to a concomitant increase in enzymatic activity. Numerous post-translational modifications including, but not limited to, oxidation regulate PTP1B activity. Indeed, PTP1B contains an essential catalytic cysteine with a low pKa that enhances the catalytic function but also renders the phosphatase susceptible to oxidation leading to reversible inactivation [[41], [42], [43]]. Of note, hepatic PTP1B is oxidized in mice fed a HFD [44] and those with hepatic GPx-1 deficiency [45]. The ROS that is generated by ethanol metabolism may amplify PTP1B oxidation and contribute to irreversible oxidation, but the magnitude and ensuing impact on phosphatase activity and expression remain to be determined.
The increase of hepatic PTP1B suggested that the upregulation of this enzyme might be benign or a contributor to ALD pathogenesis. To differentiate between the two scenarios and investigate the role of hepatic PTP1B in ALD, we generated mice with liver-specific PTP1B disruption using PTP1B floxed mice [27] that have been used to ablate PTP1B in various tissues [[15], [46], [47]]. The Alb-Cre transgenics yield efficient disruption in hepatocytes [[48], [49], [50]] but do not significantly impact nonparenchymal cells that also are implicated in ALD. Several rodent models of ALD are available, each with limitations and translational relevance [51]. While no animal model faithfully recapitulates all the human disease features, the model herein reproduces several aspects [28] but is not amenable for assessing fibrosis as with the hybrid model (high cholesterol/fat diet with intragastric ethanol) [52]. Of note, the present studies were performed using female mice, given their higher sensitivity to ethanol than males, as observed in patients [53]. Also, the current studies addressed the effects of PTP1B disruption, mostly in the liver. It is plausible that hepatic PTP1B disruption, via a direct and/or indirect mechanism, might grant protection against the deleterious effects of alcohol in other tissue(s). In keeping with this notion, hepatic PTP1B deficiency protects against HFD-induced endothelial dysfunction [54].
We demonstrated beneficial effects of hepatic PTP1B disruption in a mouse model of ALD and established mitigation of ethanol-induced injury, inflammation, and steatosis (Fig. 2, Fig. 3, Fig. 4). The attenuation of ethanol-induced injury is in line with the protective effects of PTP1B deficiency in hepatic injury. Indeed, PTP1B deficiency decreases hepatocyte apoptosis in a FAS-induced injury model [17], improves liver regeneration and decreases liver injury during partial hepatectomy [18,19], and mitigates APAP-induced cell death and ROS [20]. Also, hepatic PTP1B disruption attenuated ethanol-induced hepatic and systemic inflammation, as evidenced by reduced ethanol-induced inflammatory cytokines, inflammatory cell infiltration, and NF-κB activation (Fig. 2). Consistent with this observation, it was recently reported that PTP1B silencing attenuates alcohol and LPS-induced inflammatory response and NF-κB signaling in macrophages, whereas overexpression induces inflammation [26]. PTP1B is an established modulator of the inflammatory response but is likely to be a tissue- and stimulus-dependent [55]. Further, hepatic PTP1B disruption attenuated hepatic steatosis caused by ethanol, as evidenced by decreased hepatic triglycerides, expression of genes involved in lipogenesis, and histology (Fig. 3). These findings are consistent with the impact of PTP1B deficiency on steatosis in response to xenobiotic and metabolic challenges. In particular, chronic ethanol consumption induces hepatic steatosis in rats and is associated with increased PTP1B and fatty acid translocase [24]. Additionally, in response to a HFD challenge, hepatic PTP1B disruption decreases hepatic and serum triglycerides and reduces the expression of lipogenic genes [15].
Amelioration of ethanol-induced oxidative stress is a substantial mediator of the salutary effects of hepatic PTP1B deficiency. Ethanol oxidative metabolism generates a significant amount of ROS, including superoxide anion (O2•‾), hydrogen peroxide (H2O2), and the hydroxyl radical (•OH) [56], and is a contributor to ALD pathogenesis [30,31]. The augmented production of reactive oxygen/nitrogen species (ROS/RNS) and the depletion of antioxidants result in an imbalance of redox homeostasis [6]. Importantly, oxidative stress is reported in patients with ALD, and the severity of disease correlates with elevated lipid peroxidation and reduced antioxidants [57]. Biochemical evaluation of oxidative stress provided compelling evidence supporting the attenuation of this pathway in LPTP1B KO mice. Chronic alcohol abuse increases CYP2E1 [32], which leads to elevated ROS [58]. We observed comparable ethanol-induced upregulation of CYP2E1 in control and LPTP1B KO mice suggesting similar ethanol metabolism. The association of ethanol-impaired AKT activation [59,60] and elevated ERK activation [61,62] and hepatic injury is documented. In line with these reports, the ethanol-altered AKT and ERK phosphorylation was significantly mitigated in LPTP1B KO mice. Moreover, eNOS activation is attenuated under chronic ethanol feeding [63], whereas AKT-induced eNOS phosphorylation (Ser1177) enhances its activity [64]. In keeping with this, the ethanol-induced attenuation of eNOS phosphorylation was mitigated in LPTP1B KO mice compared with controls. Furthermore, alcohol-induced activation of iNOS and NOX is observed in multiple hepatic cell types, including hepatocytes [65]. Consistently, the ethanol-induced upregulation of oxidative stress proteins and downregulation of antioxidants in control animals were diminished in LPTP1B KO mice. The highly reactive hydroxyl radicals contribute to various reactions, including lipid peroxidation that leads to hepatic injury by the formation of toxic aldehydes such as 4-HNE [66,67]. In keeping with the decreased oxidative stress in LPTP1B KO mice, the lipid peroxidation was diminished in these mice. Notably, primary hepatocytes recapitulated the effects of hepatic PTP1B disruption in vivo consistent with those being cell-autonomous. Furthermore, pharmacological modulations that refashioned ROS in hepatocytes established diminished oxidative stress as a mediator of the beneficial effects of PTP1B deficiency under ethanol challenge. Noteworthy is that PTP1B disruption may modulate additional signaling pathways including, but not limited to, endoplasmic reticulum (ER) stress to impact ALD. Indeed, ethanol and its metabolites impair proper protein folding within the ER lumen leading to stress that is countered by the unfolded protein response [68,69]. Hepatic PTP1B disruption attenuates HFD-induced ER stress [15,16,70], and if its disruption comparably impacts ethanol-induced stress remains to be established.
Using the orally bioavailable PTP1B inhibitor (DPM-1001) [34], we demonstrated that targeting PTP1B ameliorated ethanol-induced hepatic injury, inflammation, steatosis, and oxidative stress. Given the systemic delivery of DPM-1001, the effects of PTP1B inhibition are likely ascribed to hepatic and non-hepatic tissue(s). In keeping with this notion, preliminary studies suggested diminished ethanol-induced anxiety in inhibitor-treated mice compared with untreated animals as evaluated by the open field test (Hsu and Haj, data not shown). Systemic delivery of PTP1B inhibitor may be of value as alcohol can permeate a plethora of tissues with adverse consequences [71]. Indeed, PTP1B inhibition using antisense oligonucleotides enhances insulin sensitivity and decreases weight in T2D patients [14]. However, it is worth noting that myeloid-specific PTP1B deficiency in mice leads to the development of acute leukemia [72], suggesting the need for a targeted inhibitor delivery. Indeed, intracerebroventricular administration of a small-molecule PTPB inhibitor prevents binge drinking-induced systemic insulin resistance and glucose intolerance [25]. Given the resurging interest in targeting PTP1B [12], newly developed inhibitors may be repurposed for ALD. Of note, DPM-1001 is a therapeutic agent for Wilson's disease [73]. In summary, the current findings identified hepatic PTP1B as a novel target for ALD. They suggested that PTP1B pharmacological inhibition, likely in combination therapy, might afford a viable therapeutic approach for this disease.
Declaration of competing interest
The authors declare no conflicts of interest.
Acknowledgments
The authors thank Drs. Zhaoli Sun and and Ali Reza Ahmadi (Johns Hopkins University) for providing liver biopsies of healthy and AH patients (NIAAA grant R24AA025017 to Dr. Sun). This research was supported in part by NIAAA grant R21AA027633 to Dr. Ming-Fo Hsu. Research in the Haj laboratory is funded by NIDDK grants R01DK090492 and R01DK095359 and NIEHS grant P42ES04699. Dr. Haj is a co-leader of the Endocrinology and Metabolism Core of UC Davis Mouse Metabolic Phenotyping Center, which is funded by U24DK092993. Dr. Nagy is the leader of an NIAAA grant P50AA024333. NIH grant 1S10RR019266 supports the MCB Light Microscopy Imaging Facility.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101658.
Contributor Information
Ming-Fo Hsu, Email: mfhsu@ucdavis.edu.
Fawaz G. Haj, Email: fghaj@ucdavis.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Global Status Report on Alcohol and Health 2018. World Health Organization; Geneva: 2018. [Google Scholar]
- 2.Seitz H.K., Bataller R., Cortez-Pinto H., Gao B., Gual A., Lackner C., Mathurin P., Mueller S., Szabo G., Tsukamoto H. Alcoholic liver disease. Nat Rev Dis Primers. 2018;4(1):16. doi: 10.1038/s41572-018-0014-7. [DOI] [PubMed] [Google Scholar]
- 3.Mann R.E., Smart R.G., Govoni R. The epidemiology of alcoholic liver disease. The Dionysos Study Group, Gut. 2003;27(3):209–219. [PMC free article] [PubMed] [Google Scholar]
- 4.Philips C.A., Augustine P., Yerol P.K., Rajesh S., Mahadevan P. Severe alcoholic hepatitis: current perspectives. Hepat. Med. 2019;11:97–108. doi: 10.2147/HMER.S197933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lucey M.R., Mathurin P., Morgan T.R. Alcoholic hepatitis. N. Engl. J. Med. 2009;360(26):2758–2769. doi: 10.1056/NEJMra0805786. [DOI] [PubMed] [Google Scholar]
- 6.Ceni E., Mello T., Galli A. Pathogenesis of alcoholic liver disease: role of oxidative metabolism. World J. Gastroenterol. 2014;20(47):17756–17772. doi: 10.3748/wjg.v20.i47.17756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh S., Osna N.A., Kharbanda K.K. Treatment options for alcoholic and non-alcoholic fatty liver disease: A review. World J. Gastroenterol. 2017;23(36):6549–6570. doi: 10.3748/wjg.v23.i36.6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tonks N.K. Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 2006;7(11):833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 9.Tonks N.K. PTP1B: from the sidelines to the front lines! FEBS Lett. 2003;546(1):140–148. doi: 10.1016/s0014-5793(03)00603-3. [DOI] [PubMed] [Google Scholar]
- 10.Bakke J., Haj F.G. Protein-tyrosine phosphatase 1B substrates and metabolic regulation. Semin. Cell Dev. Biol. 2015;37C:58–65. doi: 10.1016/j.semcdb.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z.Y., Dodd G.T., Tiganis T. Protein tyrosine phosphatases in hypothalamic insulin and leptin signaling. Trends Pharmacol. Sci. 2015;36(10):661–674. doi: 10.1016/j.tips.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Maheshwari N., Karthikeyan C., Trivedi P., Moorthy N. Recent advances in protein tyrosine phosphatase 1B targeted drug discovery for type II diabetes and obesity. Curr. Drug Targets. 2018;19(5):551–575. doi: 10.2174/1389450118666170222143739. [DOI] [PubMed] [Google Scholar]
- 13.Kostrzewa T., Styszko J., Gorska-Ponikowska M., Sledzinski T., Kuban-Jankowska A. Inhibitors of protein tyrosine phosphatase PTP1B with anticancer potential. Anticancer Res. 2019;39(7):3379–3384. doi: 10.21873/anticanres.13481. [DOI] [PubMed] [Google Scholar]
- 14.Digenio A., Pham N.C., Watts L.M., Morgan E.S., Jung S.W., Baker B.F., Geary R.S., Bhanot S. Antisense inhibition of protein tyrosine phosphatase 1B with IONIS-PTP-1BRx improves insulin sensitivity and reduces weight in overweight patients with type 2 diabetes. Diabetes Care. 2018;41(4):807–814. doi: 10.2337/dc17-2132. [DOI] [PubMed] [Google Scholar]
- 15.Delibegovic M., Zimmer D., Kauffman C., Rak K., Hong E.G., Cho Y.R., Kim J.K., Kahn B.B., Neel B.G., Bence K.K. Liver-specific deletion of protein-tyrosine phosphatase 1B (PTP1B) improves metabolic syndrome and attenuates diet-induced endoplasmic reticulum stress. Diabetes. 2009;58(3):590–599. doi: 10.2337/db08-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Owen C., Lees E.K., Grant L., Zimmer D.J., Mody N., Bence K.K., Delibegovic M. Inducible liver-specific knockdown of protein tyrosine phosphatase 1B improves glucose and lipid homeostasis in adult mice. Diabetologia. 2013;56(10):2286–2296. doi: 10.1007/s00125-013-2992-z. [DOI] [PubMed] [Google Scholar]
- 17.Sangwan V., Paliouras G.N., Cheng A., Dube N., Tremblay M.L., Park M. Protein-tyrosine phosphatase 1B deficiency protects against Fas-induced hepatic failure. J. Biol. Chem. 2006;281(1):221–228. doi: 10.1074/jbc.M507858200. [DOI] [PubMed] [Google Scholar]
- 18.Revuelta-Cervantes J., Mayoral R., Miranda S., Gonzalez-Rodriguez A., Fernandez M., Martin-Sanz P., Valverde A.M. Protein Tyrosine Phosphatase 1B (PTP1B) deficiency accelerates hepatic regeneration in mice. Am. J. Pathol. 2011;178(4):1591–1604. doi: 10.1016/j.ajpath.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samino S., Revuelta-Cervantes J., Vinaixa M., Rodriguez M.A., Valverde A.M., Correig X. A (1)H NMR metabolic profiling to the assessment of protein tyrosine phosphatase 1B role in liver regeneration after partial hepatectomy. Biochimie. 2013;95(4):808–816. doi: 10.1016/j.biochi.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 20.Mobasher M.A., Gonzalez-Rodriguez A., Santamaria B., Ramos S., Martin M.A., Goya L., Rada P., Letzig L., James L.P., Cuadrado A., Martin-Perez J., Simpson K.J., Muntane J., Valverde A.M. Protein tyrosine phosphatase 1B modulates GSK3beta/Nrf2 and IGFIR signaling pathways in acetaminophen-induced hepatotoxicity. Cell Death Dis. 2013;4:e626. doi: 10.1038/cddis.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Ruiz I., Blanes Ruiz N., Rada P., Pardo V., Ruiz L., Blas-Garcia A., Valdecantos M.P., Grau Sanz M., Solis Herruzo J.A., Valverde A.M. Protein tyrosine phosphatase 1b deficiency protects against hepatic fibrosis by modulating nadph oxidases. Redox Biol. 2019;26 doi: 10.1016/j.redox.2019.101263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao L., Zhang X., Wang F.R., Cao M.F., Zhang X.J., Sun N.N., Zhang J., Gao L., Zhao J.J. Chronic ethanol consumption up-regulates protein-tyrosine phosphatase-1B (PTP1B) expression in rat skeletal muscle. Acta Pharmacol. Sin. 2010;31(12):1576–1582. doi: 10.1038/aps.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LeCapitaine N.J., Wang Z.Q., Dufour J.P., Potter B.J., Bagby G.J., Nelson S., Cefalu W.T., Molina P.E. Disrupted anabolic and catabolic processes may contribute to alcohol-accentuated SAIDS-associated wasting. J. Infect. Dis. 2011;204(8):1246–1255. doi: 10.1093/infdis/jir508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shirpoor A., Heshmati E., Kheradmand F., Gharalari F.H., Chodari L., Naderi R., Majd F.N., Samadi M. Increased hepatic FAT/CD36, PTP1B and decreased HNF4A expression contributes to dyslipidemia associated with ethanol-induced liver dysfunction: rescue effect of ginger extract. Biomed. Pharmacother. 2018;105:144–150. doi: 10.1016/j.biopha.2018.05.121. [DOI] [PubMed] [Google Scholar]
- 25.Lindtner C., Scherer T., Zielinski E., Filatova N., Fasshauer M., Tonks N.K., Puchowicz M., Buettner C. Binge drinking induces whole-body insulin resistance by impairing hypothalamic insulin action. Sci. Transl. Med. 2013;5(170) doi: 10.1126/scitranslmed.3005123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang L., Sun Y.Y., Liu Y.R., Yin N.N., Bu F.T., Yu H.X., Du X.S., Li J., Huang C. PTP1B promotes macrophage activation by regulating the NF-kappaB pathway in alcoholic liver injury. Toxicol. Lett. 2020;319:11–21. doi: 10.1016/j.toxlet.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Bence K.K., Delibegovic M., Xue B., Gorgun C.Z., Hotamisligil G.S., Neel B.G., Kahn B.B. Neuronal PTP1B regulates body weight, adiposity and leptin action. Nat. Med. 2006;12(8):917–924. doi: 10.1038/nm1435. [DOI] [PubMed] [Google Scholar]
- 28.Bertola A., Mathews S., Ki S.H., Wang H., Gao B. Mouse model of chronic and binge ethanol feeding (the NIAAA model) Nat. Protoc. 2013;8(3):627–637. doi: 10.1038/nprot.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W.C., Ralphs K.L., Tosh D. Isolation and culture of adult mouse hepatocytes. Methods Mol. Biol. 2010;633:185–196. doi: 10.1007/978-1-59745-019-5_13. [DOI] [PubMed] [Google Scholar]
- 30.Cichoz-Lach H., Michalak A. Oxidative stress as a crucial factor in liver diseases. World J. Gastroenterol. 2014;20(25):8082–8091. doi: 10.3748/wjg.v20.i25.8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li S., Tan H.Y., Wang N., Zhang Z.J., Lao L., Wong C.W., Feng Y. The role of oxidative stress and antioxidants in liver diseases. Int. J. Mol. Sci. 2015;16(11):26087–26124. doi: 10.3390/ijms161125942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lieber C.S. Cytochrome P-4502E1: its physiological and pathological role. Physiol. Rev. 1997;77(2):517–544. doi: 10.1152/physrev.1997.77.2.517. [DOI] [PubMed] [Google Scholar]
- 33.Ambade A., Mandrekar P. Oxidative stress and inflammation: essential partners in alcoholic liver disease. Int J Hepatol. 2012;2012:853175. doi: 10.1155/2012/853175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krishnan N., Konidaris K.F., Gasser G., Tonks N.K. A potent, selective, and orally bioavailable inhibitor of the protein-tyrosine phosphatase PTP1B improves insulin and leptin signaling in animal models. J. Biol. Chem. 2018;293(5):1517–1525. doi: 10.1074/jbc.C117.819110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao B., Jeong W.I., Tian Z. Liver: an organ with predominant innate immunity. Hepatology. 2008;47(2):729–736. doi: 10.1002/hep.22034. [DOI] [PubMed] [Google Scholar]
- 36.Krishnan N., Krishnan K., Connors C.R., Choy M.S., Page R., Peti W., Aelst L.V., Shea S.D., Tonks N.K. PTP1B inhibition suggests a therapeutic strategy for Rett syndrome. J. Clin. Invest. 2015;125(8):3163–3177. doi: 10.1172/JCI80323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang S., Song G.J., Jung M., Kim J.H., Park H., Rahman M.H., Zhang Z., Park D.H., Kook H., Lee I., Suk K. A novel role for protein tyrosine phosphatase 1B as a positive regulator of neuroinflammation. J. Neuroinflammation. 2016;13(1):86. doi: 10.1186/s12974-016-0545-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zabolotny J.M., Kim Y.B., Welsh L.A., Kershaw E.E., Neel B.G., Kahn B.B. Protein-tyrosine phosphatase 1B expression is induced by inflammation in vivo. J. Biol. Chem. 2008;283(21):14230–14241. doi: 10.1074/jbc.M800061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cool D.E., Tonks N.K., Charbonneau H., Walsh K.A., Fischer E.H., Krebs E.G. cDNA isolated from a human T-cell library encodes a member of the protein-tyrosine-phosphatase family. Proc. Natl. Acad. Sci. U. S. A. 1989;86(14):5257–5261. doi: 10.1073/pnas.86.14.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukushima A., Loh K., Galic S., Fam B., Shields B., Wiede F., Tremblay M.L., Watt M.J., Andrikopoulos S., Tiganis T. T-cell protein tyrosine phosphatase attenuates STAT3 and insulin signaling in the liver to regulate gluconeogenesis. Diabetes. 2010;59(8):1906–1914. doi: 10.2337/db09-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee S.R., Kwon K.S., Kim S.R., Rhee S.G. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J. Biol. Chem. 1998;273(25):15366–15372. doi: 10.1074/jbc.273.25.15366. [DOI] [PubMed] [Google Scholar]
- 42.Peters G.H., Frimurer T.M., Olsen O.H. Electrostatic evaluation of the signature motif (H/V)CX5R(S/T) in protein-tyrosine phosphatases. Biochemistry. 1998;37(16):5383–5393. doi: 10.1021/bi971187i. [DOI] [PubMed] [Google Scholar]
- 43.Tiganis T. Reactive oxygen species and insulin resistance: the good, the bad and the ugly. Trends Pharmacol. Sci. 2011;32(2):82–89. doi: 10.1016/j.tips.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 44.Gurzov E.N., Tran M., Fernandez-Rojo M.A., Merry T.L., Zhang X., Xu Y., Fukushima A., Waters M.J., Watt M.J., Andrikopoulos S., Neel B.G., Tiganis T. Hepatic oxidative stress promotes insulin-STAT-5 signaling and obesity by inactivating protein tyrosine phosphatase N2. Cell Metabol. 2014;20(1):85–102. doi: 10.1016/j.cmet.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merry T.L., Tran M., Dodd G.T., Mangiafico S.P., Wiede F., Kaur S., McLean C.L., Andrikopoulos S., Tiganis T. Hepatocyte glutathione peroxidase-1 deficiency improves hepatic glucose metabolism and decreases steatohepatitis in mice. Diabetologia. 2016;59(12):2632–2644. doi: 10.1007/s00125-016-4084-3. [DOI] [PubMed] [Google Scholar]
- 46.Delibegovic M., Bence K.K., Mody N., Hong E.G., Ko H.J., Kim J.K., Kahn B.B., Neel B.G. Improved glucose homeostasis in mice with muscle-specific deletion of protein-tyrosine phosphatase 1B. Mol. Cell Biol. 2007;27(21):7727–7734. doi: 10.1128/MCB.00959-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu S., Xi Y., Bettaieb A., Matsuo K., Matsuo I., Kulkarni R.N., Haj F.G. Disruption of protein-tyrosine phosphatase 1B expression in the pancreas affects beta-cell function. Endocrinology. 2014:3329–3338. doi: 10.1210/en.2013-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Postic C., Magnuson M.A. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26(2):149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 49.Weisend C.M., Kundert J.A., Suvorova E.S., Prigge J.R., Schmidt E.E. Cre activity in fetal albCre mouse hepatocytes: utility for developmental studies. Genesis. 2009;47(12):789–792. doi: 10.1002/dvg.20568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greenhalgh S.N., Conroy K.P., Henderson N.C. Cre-ativity in the liver: transgenic approaches to targeting hepatic nonparenchymal cells. Hepatology. 2015;61(6):2091–2099. doi: 10.1002/hep.27606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghosh Dastidar S., Warner J.B., Warner D.R., McClain C.J., Kirpich I.A. Rodent models of alcoholic liver disease: role of binge ethanol administration. Biomolecules. 2018;8(1):3. doi: 10.3390/biom8010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lazaro R., Wu R., Lee S., Zhu N.L., Chen C.L., French S.W., Xu J., Machida K., Tsukamoto H. Osteopontin deficiency does not prevent but promotes alcoholic neutrophilic hepatitis in mice. Hepatology. 2015;61(1):129–140. doi: 10.1002/hep.27383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teschke R. Alcoholic liver disease: current mechanistic aspects with focus on their clinical relevance. Biomedicines. 2019;7(3):68. doi: 10.3390/biomedicines7030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Agouni A., Tual-Chalot S., Chalopin M., Duluc L., Mody N., Martinez M.C., Andriantsitohaina R., Delibegovic M. Hepatic protein tyrosine phosphatase 1B (PTP1B) deficiency protects against obesity-induced endothelial dysfunction. Biochem. Pharmacol. 2014;92(4):607–617. doi: 10.1016/j.bcp.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 55.Feldhammer M., Uetani N., Miranda-Saavedra D., Tremblay M.L. PTP1B: a simple enzyme for a complex world. Crit. Rev. Biochem. Mol. Biol. 2013;48(5):430–445. doi: 10.3109/10409238.2013.819830. [DOI] [PubMed] [Google Scholar]
- 56.Bondy S.C. Ethanol toxicity and oxidative stress. Toxicol. Lett. 1992;63(3):231–241. doi: 10.1016/0378-4274(92)90086-y. [DOI] [PubMed] [Google Scholar]
- 57.Masalkar P.D., Abhang S.A. Oxidative stress and antioxidant status in patients with alcoholic liver disease. Clin. Chim. Acta. 2005;355(1–2):61–65. doi: 10.1016/j.cccn.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 58.Lu Y., Cederbaum A.I. CYP2E1 and oxidative liver injury by alcohol. Free Radic. Biol. Med. 2008;44(5):723–738. doi: 10.1016/j.freeradbiomed.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abdelmegeed M.A., Banerjee A., Jang S., Yoo S.H., Yun J.W., Gonzalez F.J., Keshavarzian A., Song B.J. CYP2E1 potentiates binge alcohol-induced gut leakiness, steatohepatitis, and apoptosis. Free Radic. Biol. Med. 2013;65:1238–1245. doi: 10.1016/j.freeradbiomed.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun Y.Y., Zhao Y.X., Li X.F., Huang C., Meng X.M., Li J. Beta-arrestin 2 promotes hepatocyte apoptosis by inhibiting akt pathway in alcoholic liver disease. Front. Pharmacol. 2018;9:1031. doi: 10.3389/fphar.2018.01031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aroor A.R., Jackson D.E., Shukla S.D. Elevated activation of ERK1 and ERK2 accompany enhanced liver injury following alcohol binge in chronically ethanol-fed rats. Alcohol Clin. Exp. Res. 2011;35(12):2128–2138. doi: 10.1111/j.1530-0277.2011.01577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bukong T.N., Iracheta-Vellve A., Gyongyosi B., Ambade A., Catalano D., Kodys K., Szabo G. Therapeutic benefits of spleen tyrosine kinase inhibitor administration on binge drinking-induced alcoholic liver injury, steatosis, and inflammation in mice. Alcohol Clin. Exp. Res. 2016;40(7):1524–1530. doi: 10.1111/acer.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karaa A., Kamoun W.S., Clemens M.G. Chronic ethanol sensitizes the liver to endotoxin via effects on endothelial nitric oxide synthase regulation. Shock. 2005;24(5):447–454. doi: 10.1097/01.shk.0000180616.13941.7d. [DOI] [PubMed] [Google Scholar]
- 64.Schleicher M., Yu J., Murata T., Derakhshan B., Atochin D., Qian L., Kashiwagi S., Di Lorenzo A., Harrison K.D., Huang P.L., Sessa W.C. The Akt1-eNOS axis illustrates the specificity of kinase-substrate relationships in vivo. Sci. Signal. 2009;2(82):ra41. doi: 10.1126/scisignal.2000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu H., Jia Z., Misra H., Li Y.R. Oxidative stress and redox signaling mechanisms of alcoholic liver disease: updated experimental and clinical evidence. J Dig Dis. 2012;13(3):133–142. doi: 10.1111/j.1751-2980.2011.00569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brooks P.J. DNA damage, DNA repair, and alcohol toxicity--a review. Alcohol Clin. Exp. Res. 1997;21(6):1073–1082. [PubMed] [Google Scholar]
- 67.Albano E. Free radical mechanisms in immune reactions associated with alcoholic liver disease. Free Radic. Biol. Med. 2002;32(2):110–114. doi: 10.1016/s0891-5849(01)00773-0. [DOI] [PubMed] [Google Scholar]
- 68.Longato L., Ripp K., Setshedi M., Dostalek M., Akhlaghi F., Branda M., Wands J.R., de la Monte S.M. Insulin resistance, ceramide accumulation, and endoplasmic reticulum stress in human chronic alcohol-related liver disease. Oxid Med Cell Longev. 2012;2012 doi: 10.1155/2012/479348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ji C. New insights into the pathogenesis of alcohol-induced ER stress and liver diseases. Int J Hepatol. 2014;2014:513787. doi: 10.1155/2014/513787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Agouni A., Mody N., Owen C., Czopek A., Zimmer D., Bentires-Alj M., Bence K.K., Delibegovic M. Liver-specific deletion of protein tyrosine phosphatase (PTP) 1B improves obesity- and pharmacologically induced endoplasmic reticulum stress. Biochem. J. 2011;438(2):369–378. doi: 10.1042/BJ20110373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Souza-Smith F.M., Lang C.H., Nagy L.E., Bailey S.M., Parsons L.H., Murray G.J. Physiological processes underlying organ injury in alcohol abuse. Am. J. Physiol. Endocrinol. Metab. 2016;311(3):E605–E619. doi: 10.1152/ajpendo.00270.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Le Sommer S., Morrice N., Pesaresi M., Thompson D., Vickers M.A., Murray G.I., Mody N., Neel B.G., Bence K.K., Wilson H.M., Delibegovic M. Deficiency in protein tyrosine phosphatase PTP1B shortens lifespan and leads to development of acute leukemia. Canc. Res. 2018;78(1):75–87. doi: 10.1158/0008-5472.CAN-17-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krishnan N., Felice C., Rivera K., Pappin D.J., Tonks N.K. DPM-1001 decreased copper levels and ameliorated deficits in a mouse model of Wilson's disease. Genes Dev. 2018;32(13–14):944–952. doi: 10.1101/gad.314658.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.