Introduction
A hallmark of biology is the cyclical nature of organismal physiology, which is driven by networks of biological rhythms. These rhythms are remarkable for their breadth throughout biology as showcased by their observation across the phylogenetic tree from bacteria, fungi, plants, and animals that include fruit flies and mammals. (Pittendrigh, 1954; Cui et al., 2013; Kaczmarek et al., 2017; Lam et al., 2018) Evolutionary theory would suggest that organisms must benefit from the ability to align physiological function to external and internal cues across highly variable environments. (Ouyang et al., 1998; Panda et al., 2002; Dvornyk et al., 2003; Dodd et al., 2005; Spoelstra et al., 2016) Further supporting the significance, the molecular apparatus that drives biological rhythms in mammals including humans has been shown to directly or indirectly lead to the rhythmic oscillation of up to ≈43% of protein-coding genes. (Zhang et al., 2014; Patel et al., 2015)
Further investigation into these oscillations has revealed that: 1) the molecular apparatus is a hierarchical set of transcription factors whose expression changes in reciprocal fashion across the 24-hour period and has thus been termed the circadian clock, and 2) regulation of individual elements of the molecular apparatus enables fine-tuning of physiological function to meet the multitude of constantly changing conditions and demands. (Czeisler et al., 1999) A central circadian clock is located in the suprachiasmatic nucleus (SCN) that is primarily entrained by light cues across the 24-hour period which are received by specialized retinal cells. (Berson et al., 2002; Radziuk, 2013) Yet, peripheral tissues also have the intrinsic molecular machinery, which collectively have been denoted as peripheral clocks. Entrainment of the peripheral clocks occurs both top-down from the central clock via neuronal regulation and laterally by more localized cues such as endocrine hormones and nutrients across the day and night. Communally, internal and external stimuli that have been shown to have significant ability to alter the biological rhythms have been termed zeitgebers. In the context of physiology, special awareness has been paid to disruptions in sleep duration, light/dark cycles, and meal timing. (Knutson et al., 2007; Van Cauter et al., 2008; Hatori et al., 2012; Garaulet et al., 2013; Longo and Panda, 2016)
Studies have been conducted investigating the extent and conditions under which zeitgebers exert influence on circadian clock function in physiology and what changes occur in pathophysiological disorders. The latter aspect has been bolstered by the increasingly clear connections between misalignment of biological rhythms and development of high-impact human diseases. For example, societal pressures such as sleep curtailment or shift work, in which sleep and eating occur at misaligned circadian times of day, have been associated with the development of obesity, insulin resistance and type 2 diabetes mellitus. (Bass and Takahashi, 2010; Arble et al., 2015) Further, genetic alternations observed in C57BL/6J mice have shown similar phenotypes through two integral circadian clock genes: 1) an adipocyte-specific Bmal1 knockout was shown to induce increased adiposity and disordered eating while ultimately affecting systemic appetite control, and 2) homozygous Clock mutant mice displayed hyperphagia, developed obesity, and developed metabolic syndrome characteristics as compared to WT mice. (Turek et al., 2005; Paschos et al., 2012) Adipose tissue, in particular, is integral to the onset of pathophysiology through two closely-related roles: 1) modulation of systemic energy balance, and 2) indirect modulation of systemic inflammatory state. Adipokine and cytokine production from adipose tissue plays a major role in feeding behavior (e.g. leptin), insulin sensitivity (e.g. adiponectin, resistin, and tumor necrosis factor alpha), and energy balance during fasting through lipolysis. (Kershaw and Flier, 2004; Ouchi et al., 2011; McGown et al., 2014; Ryden et al., 2014) Lastly, adipose tissue insulin sensitivity plays an indirect role in systemic inflammation via free-fatty-acid-mediated lipotoxicity. (Grill and Zhou, 1995; Shulman, 2000; Drosatos and Schulze, 2013) Predictably, the peripheral clock in adipose tissue from humans has been reported to exhibit robust patterns of circadian gene expression; also, these endogenous circadian rhythms persist following removal and short-term culture in vitro. (Fain et al., 2003; Phillips et al., 2008; Rogers et al., 2008; Gómez-Santos et al., 2009; Garaulet et al., 2011; Gómez-Abellán et al., 2012; Aubin et al., 2015; Wang et al., 2015) Thus, circadian patterns of gene expression in adipose tissue are highly relevant in physiological and pathophysiological states.
A large portion of the molecular basis of central and peripheral circadian rhythms has been examined via quantitative reverse transcriptase polymerase chain reaction (qRT-PCR). Since its introduction roughly thirty years ago, qRT-PCR remains the gold-standard method to measure individual mRNA species. In this period, reproducibility and comparability of qRT-PCR experiments has been bolstered by establishment of the MIQE guidelines, which are the minimum information for publication of quantitative RT-PCR experiments. (Bustin et al., 2009) Raw quantification cycle (Cq) of a qRT-PCR run remains reflective of linear-modelling of background-corrected fluorescent units, which informs the relative amount of the target sequence in the reaction well. (Tichopad et al., 2003) In order to make the amount comparable it must be adjusted to a corrected cycle called ΔCq, which is gene expression on a logarithmic scale. This adjustment step is called normalization, and is most commonly performed using a reference gene. Although a straightforward correction of the data, normalization has remained a difficult experimental condition to solve. The reference gene is operationally defined as having consistent expression relative to total cDNA concentration, the adjustment is simply conducted as a linear subtraction of the reference gene raw Cq. (Vandesompele et al., 2002; Tichopad et al., 2003) However, the difficulty has been that potential reference genes deviate significantly from the operational definition across experimental conditions, especially those generated by patterns of oscillating gene expression. The consequence of the deviation is that the adjustment step may either accentuate, or attenuate, circadian gene expression differences across a 24-hour period thus skewing the actual results.
There is growing understanding of the need for a library of genes that under precise experimental conditions have been determined to be adequately consistent as a single normalization reference. (Bustin et al., 2013) And, for more heterogeneous data sets, reports are needed for sets of reference genes that can create a composite reference that more closely resembles invariant gene expression. The primary goal of this study was to characterize the consistency of candidate reference genes for repeated measurements of circadian-clock-related gene expression across a 36-hour period with primary human subcutaneous adipose tissue obtained by needle biopsy or surgical excision and maintained by sterile culture. (Carswell et al., 2012) The outcome of the study was the optimal set of candidate reference genes to best fulfill invariant gene expression for these samples. The methods described that were used to identify candidate reference genes and to minimize variability across samples have broad applicability to other studies by: 1) describing the framework by which the suitability of reference genes can be determined, and 2) for experimental design, by addressing the potential significance of time of day on reference gene variation.
Materials and Methods:
Subject characteristics:
Four pre-menopausal morbidly obese women were voluntarily enrolled in the clinical research study (University of Chicago IRB 09–337-B) following approval and scheduling for laparoscopic sleeve gastrectomy surgery. A summary of subject characteristics was reported in Supplemental Table 1. Subjects did not exhibit irregular lifestyles or irregular menstrual cycles, or have diabetes managed with medication (HbA1c: 5.6± 0.6). No malignant diseases were identified by complete blood count nor was pregnancy detected via urine. Additionally, patients were monitored by a registered polysomnographic technologist to ensure the absence of sleep-related disorders. The one subject with obstructive sleep apnea met inclusion criteria with documented compliance using a continuous positive airway pressure machine.
Laboratory Sessions:
In-laboratory sessions were conducted one-to-two weeks prior to the scheduled surgery date, and twelve-to-thirteen weeks following surgery date. Each session lasted two evenings and one day. During which light exposure and meal timing was tightly controlled. To limit evening light exposure, ambient room light was reduced three hours before scheduled bedtimes. Four mini meals were administered per day at: 10 a.m., 1 p.m., 4 p.m., and 7 p.m. In the morning of the second day, there was an aspiratory subcutaneous peri-umbilical fat biopsy, conducted as previously described. (Broussard et al., 2012) The post-surgery session was identical to the pre-surgery session. The in-surgery biopsy yielded mesenteric adipose tissue collected via scalpel-assisted excision, which occurred after the patient had been placed under general anesthesia but prior to the start of the laparoscopic vertical sleeve gastrectomy procedure.
Biopsy Processing:
The processing of the tissue was a modification of the Fried Protocol. (Carswell et al., 2012) A laminar flow hood was used to wash tissue with sterile M199 media (Gibco cat# 12350–039, +1% low endotoxin BSA, +200 nM N6-Rphenylisopropyladenosine) and mince it into approximately five milligram segments. These steps removed blood clots and balanced cellular hypoxia while maintaining tissue architecture that facilitated conventional paracrine signaling. Following initial processing, the tissue was washed once in BSA-free media and rested for four hours in a cell incubator (37 °C, 5% CO2) in ten volumes of sterile culture media (Gibco cat#12350–059, +1 nM insulin, +40 nM dexamethasone). Then, one hundred microliters of packed tissue (± 5%) was aliquoted and measured volumetrically from a 40 μm cell strainer (CellTreat cat# 229481) into one milliliter of culture media (1:10 by volume) per well into a twenty-four well tissue culture plate (Falcon cat# 353047). The first collection was approximately twelve hours following the biopsy in an effort to allow for the majority of cytokine secretion and acute inflammation to abate. Subsequently, a well of tissue was collected periodically (20:00, 24:00, 08:00, 12:00, 16:00, 20:00, 24:00, 08:00), weighed, and snap frozen in liquid nitrogen before storage at −80°C. Similar protocols have shown that human adipose tissue circadian clock gene and clock-controlled gene rhythms can be maintained without direct SCN signal. (Fain et al., 2003; Phillips et al., 2008; Rogers et al., 2008; Gómez-Santos et al., 2009; Garaulet et al., 2011; Gómez-Abellán et al., 2012; Aubin et al., 2015; Wang et al., 2015) Yet, to preserve the SCN-mediated imprint of endogenous entrainment of the peripheral circadian clock, the tissue was not serum shocked.
Sample Preparation:
Tissue was homogenized by two ten-second-pulses using a dispersion-based homogenizer (VWR VDI 12). RNA was isolated from the tissue homogenate using a cold phenol-chloroform-isoamyl-alcohol liquid-liquid separation followed by acid guanidinium-ethanol minicolumn isolation and final elution with 1 μM sodium citrate (Omega cat#R68300). RNA purity was verified and concentration determined by Nanodrop UV-spectroscopy. For reverse transcription step of cDNA synthesis, 500 ng sample RNA per 20 μL was converted using Quanta Qscript Master Mix (cat# 95048) chemistry in a standard thermocycler (Applied Biosystems GeneAmp PCR System 9700). RNA integrity was conducted using a 5’−3’ primer assay on the reference gene: YWHAZ; all samples were assessed to have high integrity (1.00 ± 0.72).
Primer Validation
Efficiency and dynamic range were collected via the gold-standard method of a dilution curve. Maximal cDNA template concentration was a ten-fold dilution from the completed cDNA synthesis thermocycler reaction volume; successive serial dilutions were eight-fold. The slope of the line (m) was calculated by a trendline from three or more consecutive dilutions. Efficiency was calculated using Equation 1 below with m as the slope and d as the dilution:
| (Equation 1) |
Perfect efficiency was 100%. According to MIQE guidelines, primer efficiency estimated above 0.9 can presumed to be perfect efficiency for ΔΔCq relative quantification. Primer sets with lesser efficiency are suggested to use the Pfaffl correction to adjust raw Cq values to be as if the primer set had optimal efficiency. RPII was the only gene that required a Pfaffl correction (E = 0.89), the formula for which is listed below (Equation 2):
| (Equation 2) |
Dynamic range was experimentally defined by the template concentrations used to calculate efficiency. Within this concentration range, no extrapolation is needed to confirm high efficiency amplification. No raw Cq values obtained fell outside the dynamic ranges calculated from the dilution curves.
RT-qPCR:
The selected screen for effective reference genes was limited to the bolded genes in Figure 1, which illustrated reference gene suitability from previously published experimental manipulations. (Fink et al., 2008; Foldager et al., 2009; Curtis et al., 2010; del Pozo et al., 2010; Mehta et al., 2010; Amable et al., 2013; Jacob et al., 2013) The primer sequences are listed in Table 1. (Radonić et al., 2004; Curtis et al., 2010; Valadan et al., 2015) Dbp and Bmal1 primers (Qiagen cat# PPH19697A & PPH06229F) were purchased with a documented guarantee for high efficiency and broad dynamic range but without a precise sequence. To prepare the qRT-PCR experiment, all equipment was thoroughly cleaned with 70% ethanol. Only autoclaved tubes and unopened pipette tip boxes were utilized. Per well, the total reaction volume was 10 μL which consisted of: 4 μL cDNA template, 5 μL SYBR green master mix (Quantabio cat# 95071), and 0.5 μL of each forward and reverse primers. Template concentration of cDNA was 1:100 relative to the reverse-transcription-reaction-derived cDNA. One plate was used per gene per subject with three technical replicates, three positive controls to serve as inter-plate calibration, and three no-template negative controls. Loading was completed over ice before plates were sealed (BioRad cat# MSB1001) and centrifuged (1000 xg, 1 min) to eliminate any bubbles. Running conditions on the CFX Connect Real-time PCR Detection system were: (40 cycles, 55°C annealing temperature). A melt curve was collected for each plate; no contamination or primer dimers were found.
Figure 1: Experimental conditions that informed selection of candidate reference genes.
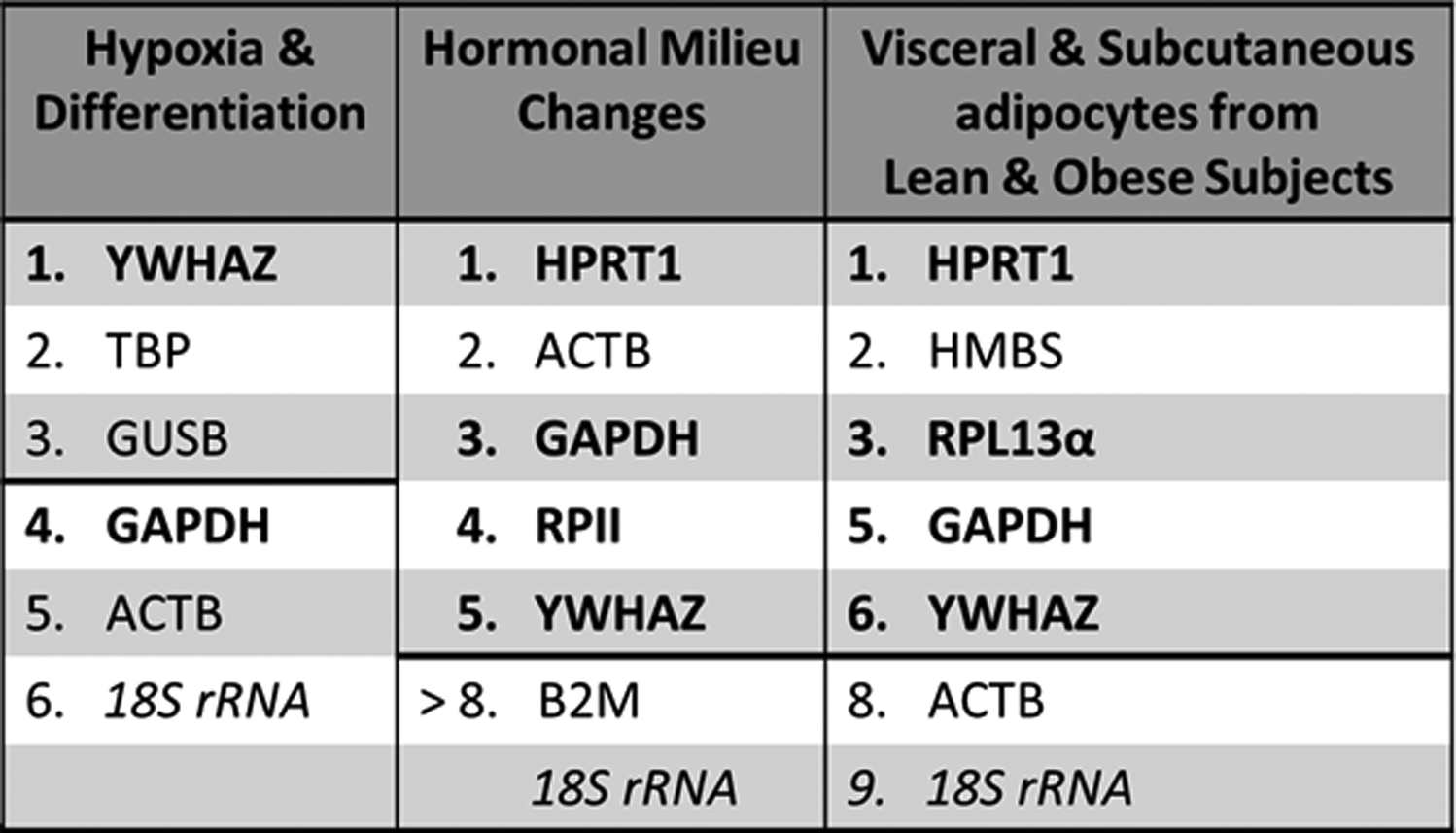
Columns represent ranked stability of reference genes under each different experimental manipulation from previous studies. The thick black dividing line indicated genes that were considered unsuitable for use as reference genes. Bolded genes indicated the reference genes chosen as candidate reference genes.
Table 1:
Summary table of candidate reference gene primer nucleotide sequences with efficiency (E) and source of prior publication of the primer sequence.
| Gene List | Nucleotide Sequence | E | Source |
|---|---|---|---|
| 18S | F: 5’- CGG CTA CCA CAT CCAA AGG A −3’ R: 5’- GCT GGA ATT ACC GCG GCT −3’ |
1.00 | |
| YWHAZ | F: 5’- TGC TTG CATCCC ACA GAC TA-3’ R: 5’- AGGCAGACAATGACAGACCA-3’ |
1.00 | Curtis et al. (2010) |
| RPL13α | F: 5’- AAG GTC GTG CGT CTG AAG-3’ R: 5’- GAG TCC GTG GGT CTT GAG-3’ |
0.94 | Radonić et al. (2004) |
| RPII | F: 5’- GCA CCA CGT CCA ATG ACA T-3’ R: 5’- GTG CGG CTG CTT CCA TAA-3’ |
0.89 | Radonić et al. (2004) |
| GAPDH | F: 5’- TGC ACC ACC AAC TGC TTA GC-3’ R: 5’- GGC ATG GAC TGT GGT CAT GAG-3’ |
0.96 | Curtis et al. (2010) |
| HPRT1 | F: 5’- GGA CTA ATT ATG GAC AGG ACT G −3’ R: 5’- GCT CTT CAG TCT GAT AAA ATC TAC −3’ |
1.00 | Valadan et al. (2015) |
Curtis KM, Gomez LA, Rios C, Garbayo E, Raval AP, Perez-Pinzon MA, and Schiller PC (2010) EF1 alpha and RPL13a represent normalization genes suitable for RT-qPCR analysis of bone marrow derived mesenchymal stem cells. Bmc Molecular Biology 11:15.
Radonić A, Thulke S, Mackay IM, Landt O, Siegert W, and Nitsche A (2004) Guideline to reference gene selection for quantitative real-time PCR. Biochemical and Biophysical Research Communications 313:856–862.
Valadan R, Amjadi O, Tehrani M, Rafiei A, Hedayatizadeh-Omran A, and Alizadeh-Navaei R (2015) Pseudogene-free amplification of HPRT1 in quantitative reverse transcriptase polymerase chain reaction. Analytical Biochemistry 485:46–48.
Results
Clinical Study and Analysis Overview
The novel role of this study was a direct assessment of the circa-24-hour stability of candidate reference genes in cultured primary subcutaneous adipose samples collected before, in-surgery, and after metabolic surgery from women. These conditions were utilized to allow for a complex example of adipose tissue health and endocrine regulation on circadian rhythmicity.
This study tested expression stability across two biological conditions: time-of-day and time-relation to weight-loss surgery. A biopsy was collected in the morning from each subject up to two weeks prior to surgery, at surgery, and twelve weeks following laparoscopic sleeve gastrectomy. Tissue explants were cultured ex vivo under sterile conditions in a time-series experiment. Fewer total subjects were utilized than prior experiments by others; however, due to the multiple collections within the time series protocol, the total samples generated and analyzed were at-minimum comparable to prior published experiments. (Fink et al., 2008; Foldager et al., 2009; Curtis et al., 2010; del Pozo et al., 2010; Mehta et al., 2010; Amable et al., 2013; Jacob et al., 2013)
Suitability of each reference gene was evaluated using raw quantification cycle (Cq) for each sample. Under this paradigm, the goal was to identify genes that: 1) maintained most consistent expression relative to total cDNA concentration, and 2) whose variation captured target gene variation tangential to the experimental manipulation. Three tools were used to evaluate these study goals. First, the statistical tool called BestKeeper provided thorough simple summary statistics in Microsoft Excel. (Pfaffl et al., 2004) Further analysis was also conducted through multiple tools including Normfinder and geNorm. For each, a stability value was returned for each gene that was, under optimal stability, minimized to zero. Normfinder is a plug-in for Microsoft Excel. geNorm is a statistical software module within the qRT-PCR experiment management software, qBase+. For each tool, an analysis was conducted to determine optimal stability of the individual genes, and the optimal reference gene set. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Candidate Reference Gene Panel Selection
Candidate reference genes were chosen by a review of relevant previous literature regarding the axes of variation mentioned above. As recommended, all genes studied had standard verification of primer efficiency, specificity, and dynamic range. (Bustin et al., 2009) Time-relation to weight-loss-surgery was simulated by two factors: 1) modification of endocrine hormone milieu, and 2) ultrastructural tissue variation during substantial weight-loss. These two properties were partially addressed by interrogation of mesenchymal stem cells, the precursor cells of preadipocytes and adipocytes. The experimental manipulations tested were differentiation, hypoxia, and overt hormone administration. Genetic heterogeneity and ultrastructural tissue differences were further examined by incorporation of literature that utilized primary adipose tissue. In these papers, the comparisons were between obese and lean subjects and between visceral and subcutaneous fat. (Fink et al., 2008; Foldager et al., 2009; Curtis et al., 2010; del Pozo et al., 2010; Mehta et al., 2010; Amable et al., 2013; Jacob et al., 2013) The genes that showed the greatest consistency under these conditions were: YWHAZ, RPL13α, GAPDH, HPRT1, and RPII (Figure 1). Each of these genes were evaluated in this study. Genes that were excluded by the screen were ACTB, B2M, GUSB, HMBS, and TBP. Despite poor performance in the cited literature, the widespread usage of 18S warrants its inclusion as a candidate reference gene.
BestKeeper
A test of normality was first needed to justify further statistical analysis of collected data. In the current data set there were approximately one hundred data points, which is enough to visualize the sample distribution. To prepare the data, raw Cq values were converted to the relative quantities. Secondly, the mean and standard error was calculated using the relative quantities. Next, estimated data points were produced from a normal distribution with a mean and standard deviation equivalent to the calculated sample mean and sample standard error. The values of the estimated data points were calculated from the normal distribution at z-scores that reflected the percentile of the rank-ordered data set. The rank-ordered experimental data set was then plotted against the estimated data set in a quantile-quantile plot (Supplemental Figure 1). If the experimental data set was perfectly normal, this plot would reflect a line with slope of one. The plots were approximately linear for each of the candidate reference genes with each gene suggesting a slightly higher dispersion (slope > 1) than standard for a normal distribution.
Given acceptable data distributions, general measures of each distribution were calculated using the Microsoft Excel Add-in BestKeeper. Statistics that provided a general idea of the suitability of the candidate reference genes were: mean Cq, and standard error. Mean raw Cq is probative, or qualitative, evidence of stability. Extremely high or low mean Cq (above/below the range of 20–30) suggested potential for instability in future experiments. Two explanations inform this general property: 1) the technical limitations of a qRT-PCR assay, and 2) the dynamic range of each primer set. High Cq, which reflected low transcript copy number, did warrant greater concern for failed wells and randomness-induced Cq variability. Mean Cq values for each gene were calculated arithmetically, geometrically, or logarithmically by relative quantities (Figure 2). The mean Cq value for 18S was higher than the optimal range (geo μ= 16.77). Additionally, RPII and HPRT1 had mean Cq values near the upper boundary (respective geo μ = 29.41; 29.65). The three that were in the middle of the range were YWHAZ, RPL13α, and GAPDH. The respective geometric mean Cq for these genes were 25.19, 22.5, and 24.35. These results suggested that using three of the six genes (RPII, HPRT1, or 18S) may add experimental difficulty to potential future experiments.
Figure 2: General characteristics of candidate reference gene sample Cq distributions.
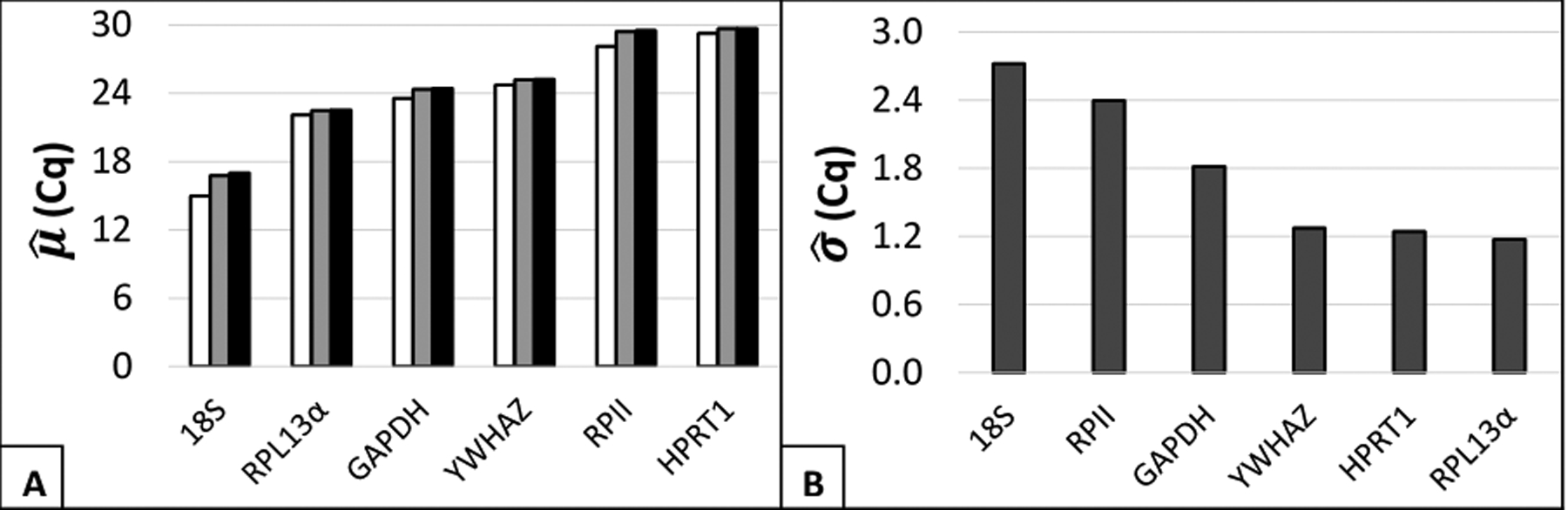
(A) Respective estimates of mean Cq () were calculated for sample Cq values using [white] relative transcript quantities, [grey] geometric averaging, and [black] arithmetic averaging. (B) Respective estimates of standard deviation () were calculated from sample Cq values.
The second statistic used within the software was standard error of the raw Cq values (Figure 2). Given the normality of the sample distributions, a minimized standard error strongly suggested superior reference gene stability. The suggested cut-off points for individual reference gene suitability (± Cq) have been recommended to be 0.5 for genetically homogenous data sets and 1.0 for heterogeneous data sets. For three genes (18S, RPII, and GAPDH), respective standard error was markedly elevated (2.37, 1.94, and 1.46) from the recommended standard. F-test comparison of variances indicated that 18S rRNA and RPII had comparable variances (0.22), while GAPDH had different variance from either (1.3e-4 and 8.3e-3, respectively) (Supplemental Figure 2). For the remaining three genes tested (YWHAZ, HPRT1, and RPL13α), comparable standard error was found (1.08, 0.96, 1.01). Yet, it is notable that none of the reference genes exhibited standard error that was clearly below the heterogeneous data set standard.
GeNorm
The module geNorm within the subscription software qBase+ was next used to assess stability by a proprietarily determined variable (M). Under optimal suitability M is minimized to zero. Similar to the prior analysis, the cut-off for suitable genes was 0.5 for genetically homogenous data sets and 1.0 for heterogeneous data sets. Due to the software adjusting for input sample size, this software was used to look at overall stability of the genes and also stability of the genes when segregated by biopsy collection. Given that there were three biopsies collected with identical collection times, the size of the data sets for the individual biopsy experiments are one-third the size of the cumulative data set.
For each gene, it was clear that one-to-two weeks prior to surgery exhibited the highest M, and therefore the worst stability (Supplemental Figure 3). However, twelve-to-thirteen weeks following surgery was more stable for three genes (GAPDH, YWHAZ, and RPL13α) and less stable for three genes (18S, RPII, and HPRT1) as compared to immediately prior to surgery. As a result of these findings, the cumulative data set closely paralleled the stability value of the biopsy obtained one-to-two weeks prior to surgery.
As an overall ranking, three genes exhibited comparable stability values (GAPDH, YWHAZ, and RPL13α) at respective M: 1.09, 1.051, and 1.03 (Figure 3). HPRT1, despite indicating higher stability in the previous analysis, was indicated to have notably lower stability (1.307). RPII and 18S rRNA exhibited M values of 1.60 and 2.11, respectively. As individual reference genes, none of the genes exhibited overall stability values below one. Yet, the three highest rated genes were, again, only slightly offset from the soft cut-off point.
Figure 3: A) Stability values for each candidate reference gene as obtained by qBase+ software.
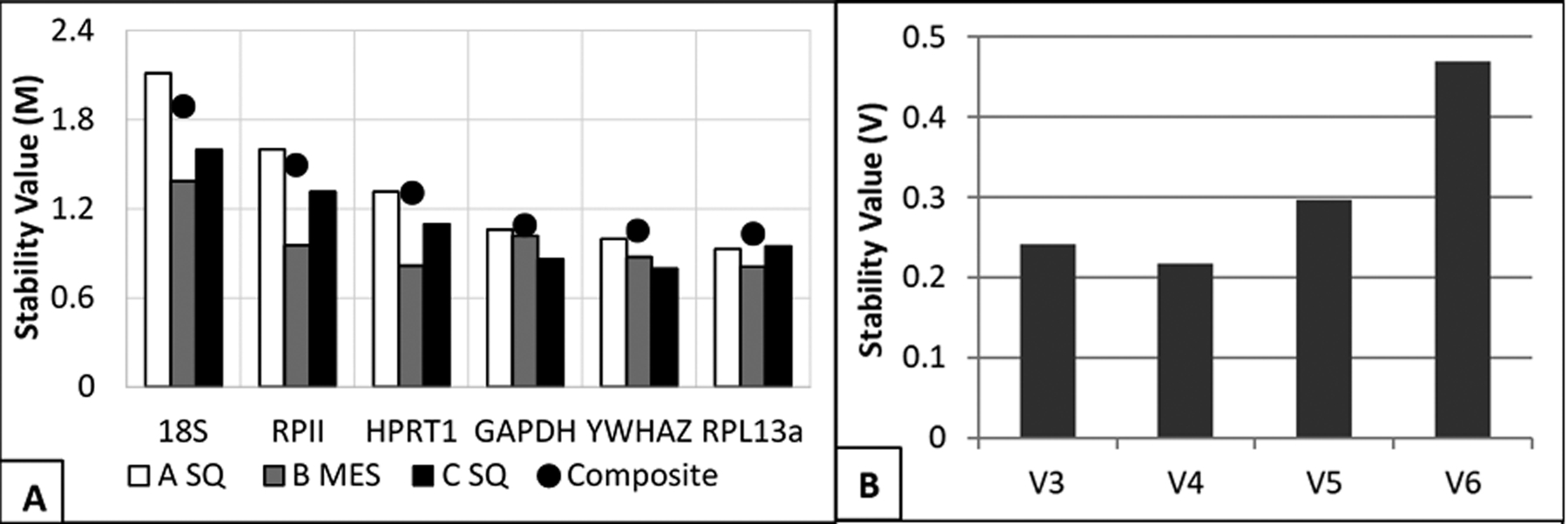
Smaller values indicated better suitability (suitable for homogenous data < 0.5; suitable for heterogeneous data < 1.0). Data set was partitioned and analyzed separately by biopsy: (white) subcutaneous adipose tissue, one-to-two weeks prior to bariatric surgery [A SQ]; (grey) mesenteric adipose tissue, immediately prior to surgery [B MES]; (black) subcutaneous adipose tissue, twelve- to-thirteen weeks following surgery [C SQ]. The plotted points represented the data analyzed as one set. B) Output from geNorm V panel in qBase+ software. Panel suggested the optimal number of genes to include in the reference gene set. Columns represented stability value associated with geometrically-averaged top three gene Cq values (V3 = YWHAZ, RPL13a, GAPDH), and then the successive stability values that included HPRT1, then RPII, and lastly 18S.
Normfinder
In the Normfinder algorithm, stability values were not independently determined for each candidate gene. Rather, each candidate reference gene was given a stability value estimating the instability that it provided to the ordered set of geometrically-averaged relative quantities (2-Cq). As designed, the dependency was to facilitate construction of candidate reference gene sets, which will be discussed later. As a result of the program design, suitability in this program was qualitative and was relative to the other genes in the set. For clarity, reported stability values were calculated including each of the six candidate genes (Figure 4). As the last parameter, the program allowed explicit grouping of samples to explain a portion of the variation. In Figure 4, three different sample groupings were used: 1) collection time, 2) biopsy, or 3) both collection time and biopsy.
Figure 4: Stability values as calculated by Normfinder Excel Add-in with explanatory variable grouping.
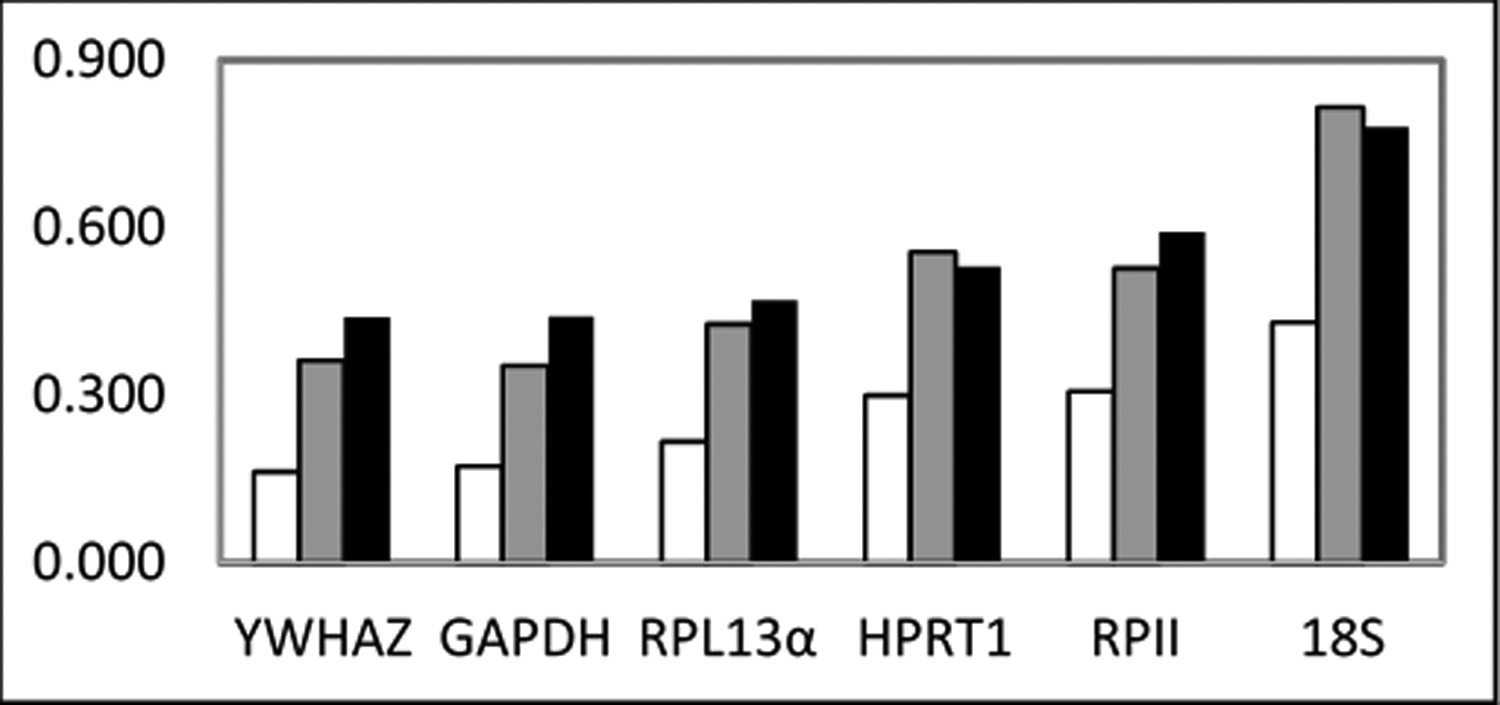
Increased suitability of respective reference genes was qualitatively indicated by lower stability values. Measurements were grouped to define an explanatory variable for relative quantity variation. Group were defined as: (white) collection times, (grey) biopsies, or (black) both biopsies and collection times.
For each of the ways to group samples, the genes measured to be the most stable were: YWHAZ, GAPDH, and RPL13α. 18S, again, was assessed to be the least stable candidate reference gene irrespective of sample grouping. HPRT1 was ranked to be more stable than RPII under two groupings, but worse when grouping samples by biopsy. Grouping by collection time did result in lower stability values compared to the other groupings, which suggested that more variation could be explained by collection time.
BestKeeper, Composite
Since no individual reference gene was definitively sufficient to serve as the internal reference, the analysis shifted to identifying the suitable composite reference gene set. (Vandesompele et al., 2002) The procedural definition was that, upon geometric-averaging of multiple reference gene Cq values, the composite Cq for the derived standard will exhibit minimal standard error across samples. Improved stability has been accomplished through deconstructed stochastic variation resulting from opposing biological regulation which should be tangential to the experimental manipulation. However, this effect was balanced by retention of more individually unsuitable genes in the reference gene set.
Three statistics were used to conduct the analysis: 1) individual gene standard error, 2) correlation between the genes, and 3) the composite standard error. The process of finding the optimal set was to, until the composite standard error increased, to iteratively remove the: 1) the gene with highest remaining standard error, or 2) given similar standard errors, the gene with the highest correlation to remaining genes. This iterative progression enabled the most stable genes to be retained and to maximize the deconstructive variation. The two worst genes were removed from the model leading to the overall variance to decrease markedly. The complete model had a total standard error across samples of 1.480. By the removal of 18S the standard error decreased to 1.256; the subsequent removal of RPII decreased the standard error to 1.074. Following the three individual analyses, it was not clear whether HPRT1 or GAPDH was less suitable.
To gauge the potential consequences of including GAPDH or HPRT1 in the reference gene set, estimated of Pearson correlation were used to estimate covariation between candidate reference genes. Contextually, low covariation suggested that geometric averaging would have an effect to lower experimental variance in future experiments. In this data set, all remaining genes had significant positive correlation with the other genes except GAPDH and HPRT1 (ρ = 0.166, p = 0.111) (Table 2). HPRT1, while significantly related to both YWHAZ (ρ = 0.266, p = 0.009) and RPL13α (ρ = 0.275, p = 0.007), the correlation is notably less than GAPDH for the same. Therefore, it was expected that inclusion of HPRT1 would improve future stability of a composite reference set that included YWHAZ, RPL13α, and/or GAPDH.
Table 2:
Experimental estimation of Pearson correlation between candidate reference genes calculated from respective Cq values from the complete set of samples
| HPRT1 | ||||||
|---|---|---|---|---|---|---|
| 18S | -- | 0.4376 | 0.3531 | 0.5646 | 0.5627 | 0.6119 |
| -- | 1.0 E-05 | 4.8 E-04 | 3.1 E-09 | 3.6 E-09 | 5.69E-11 | |
| YWHAZ | 0.4376 | -- | 0.5630 | 0.7670 | 0.7148 | 0.2663 |
| 1.0 E-05 | -- | 3.5 E-09 | 2.0 E-19 | 5.9 E-16 | 0.00947 | |
| RPL13α | 0.3531 | 0.5630 | -- | 0.8328 | 0.6134 | 0.275 |
| 4.8 E-04 | 3.5 E-09 | -- | 2.3 E-25 | 4.9 E-11 | 0.00726 | |
| RPII | 0.5646 | 0.7670 | 0.8328 | -- | 0.8605 | 0.1917 |
| 3.1 E-09 | 2.0 E-19 | 2.3 E-25 | -- | 1.1 E-28 | 0.0642 | |
| GAPDH | 0.5627 | 0.7148 | 0.6134 | 0.8605 | -- | 0.1659 |
| 3.6 E-09 | 5.9 E-16 | 4.9 E-11 | 1.1 E-28 | -- | 0.1101 | |
| HPRT1 | 0.6119 | 0.2663 | 0.2752 | 0.1917 | 0.1659 | -- |
| 5.7 E-11 | 0.0095 | 0.0073 | 0.0642 | 0.1101 | -- |
The p-values represented the probability that the correlation coefficient resulted from random chance.
To test the prescription, a table was constructed with all permutations of the four most suitable genes (Supplemental Figure 4). When looking at the standard error of gene sets (2 or 3 total) including these genes, the only sets of genes that were overall more stable included HPRT1. YWHAZ and RPL13α had almost no difference in effect on overall stability. And, due to the higher estimate of standard deviation and high covariation with YWHAZ and RPL13α the overall effect of GAPDH was to increase the reference gene set estimates of standard deviation. The suggestion from this analysis was HPRT1 inclusion would improve future composite stability.
Normfinder, Composite
RPII and 18S were excluded from this analysis due to two variables: 1) previously established poor performance of each in all three analyses, and 2) dependent calculation of each reference gene stability in the analysis module. Similar to the prior test, a selective exclusion was conducted (Table 3). Yet, since three genes are needed for the macro to function, the analysis only included the differential effect of a single gene exclusion. Similar to the preliminary analysis, YWHAZ and RPL13a did not notably change the overall data set. However, the effect of GAPDH and HPRT1 on the stability of the reference gene set was reversed in effect compared to the preliminary analysis. Exclusion of GAPDH led to a reduction in average stability of 10–15%, which was commensurate with the effect of excluding YWHAZ and RPL13a. And, exclusion of HPRT1 led to a 30% improvement in reference gene stability. These results suggested that the HPRT1 was the least suited remaining gene for the model. It is notable that this is the opposite recommendation from the BestKeeper analysis.
Table 3:
Single exclusion analysis conducted through Normfinder Add-in.
| Best 4 | (−) GAPDH | (−) HPRT1 | (−) RPL13a | (−) YWHAZ | , gene | |
|---|---|---|---|---|---|---|
| GAPDH | 0.535 | -- | 0.369 | 0.532 | 0.608 | 0.511 |
| HPRT1 | 0.515 | 0.549 | -- | 0.555 | 0.523 | 0.535 |
| RPL13α | 0.377 | 0.468 | 0.365 | -- | 0.398 | 0.402 |
| YWHAZ | 0.364 | 0.507 | 0.230 | 0.387 | -- | 0.372 |
| , set | 0.448 | 0.508 | 0.321 | 0.492 | 0.509 |
To identify the best three-gene-set, each gene was iteratively excluded from the reference set, and the stability values were estimated from the remaining genes. Higher stability was evidenced by lower stability values. Additionally, the stability values of the set of the four best suited genes were reported. Lastly, estimated mean stability values () were calculated for each gene and reference gene set.
GeNorm V
In addition to stability value estimates, qBase+ contains a panel, geNorm V, that conducted a selective inclusion analysis. Genes with higher calculated M were iteratively included into the reference gene set. Similar to the earlier stability value, the lower the resulting output the more stable the reference gene set. The goal output for homogeneous data sets is 0.10, and for heterogeneous data sets the output is optimally 0.15. The result of the analysis was to indicate that at least three genes ought to be included in the optimal reference set and that there was evidence to support the inclusion of four genes (Figure 3B). The relative improvement from three genes to four genes was a decrease from 0.24 to 0.21. Addition of RPII and 18S into the gene set decreased the relative stability, which agrees with the other analyses that were conducted.
Circadian Gene Normalization
At the completion of the above analysis, it was concluded that YWHAZ and RPL13α were optimally included in the reference gene set with GAPDH. There is an argument for HPRT1 to be included instead of GAPDH. Yet, it presents increased technical challenges due to high raw Cq that could overshadow the benefit of greater coverage of stochastic variation due to lower correlation to the other genes in the panel. The disputed suggestion was addressed thusly: 1) illustrating the geometrically averaged relative quantities with four different combinations of reference gene sets (Figure 5), and 2) one subject had mRNA expression measured for Dbp, a circadian clock gene, from samples derived from the biopsies prior to and following surgery normalized by the same four reference gene sets (Figure 6). For the latter, reported fold changes are relative to the biopsy from which the samples were derived. Lastly, the relative quantities for each candidate reference gene were plotted across time after calibration to the experimental average Cq for the data collected from the biopsies: 1) one-to-two weeks prior to surgery, and 2) twelve-to-thirteen weeks following surgery (Supplemental Figure 5).
Figure 5: Profiles of relative quantities for geometrically-averaged reference gene sets.
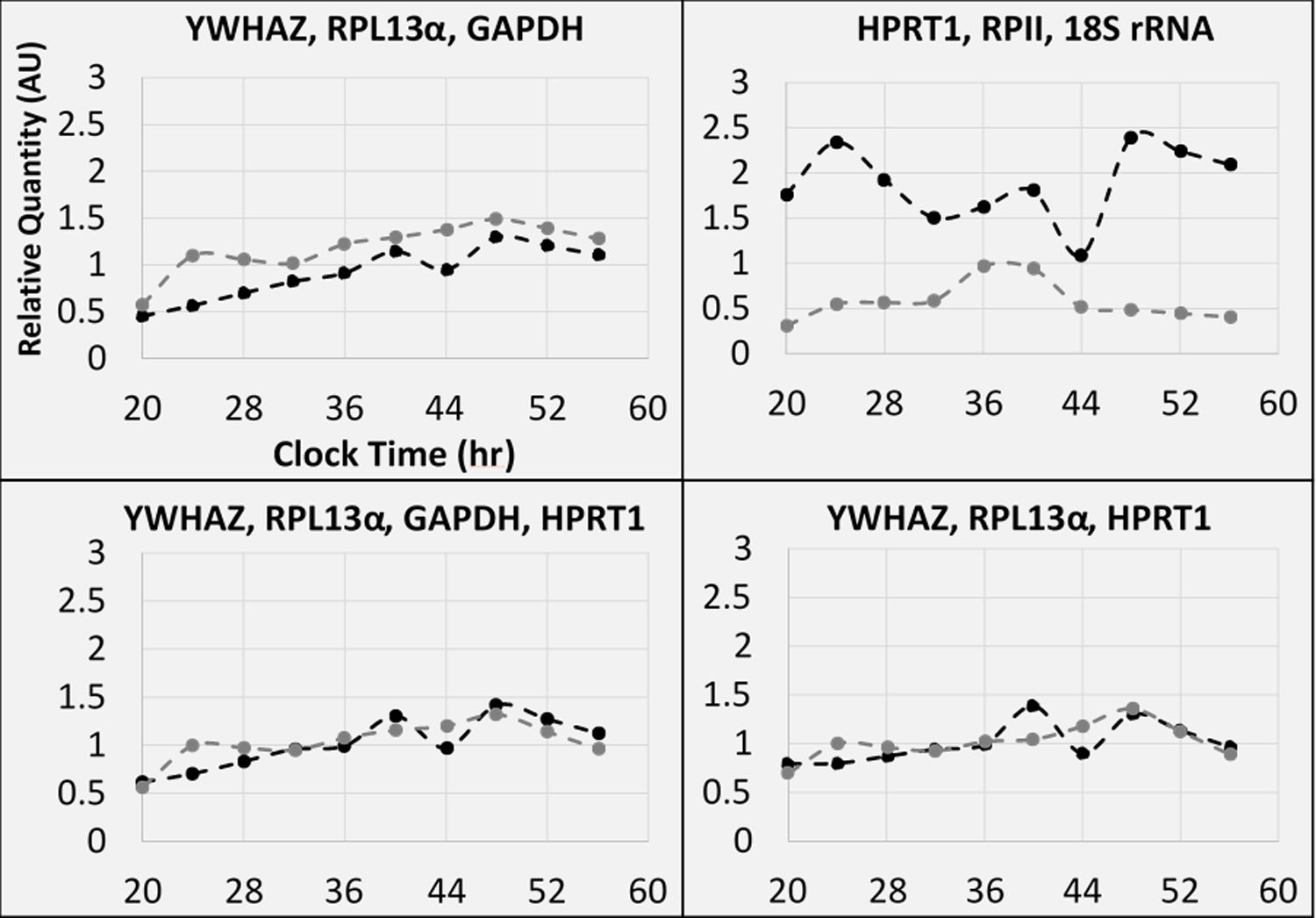
Sample Cq values for each included gene were geometrically averaged together then were calibrated to the mean Cq values of both included biopsy data sets, which were: (black) one-to-two weeks prior to surgery and (grey) twelve-to-thirteen weeks following surgery. Clock time is counting up from day 0, which included the initial biopsy collection in the morning (20 CT = Day 0, 8 PM).
Figure 6: Rhythmic gene expression of clock genes, Dbp and Bmall, as output from least squares cosinor analysis following normalization by four different reference gene sets.
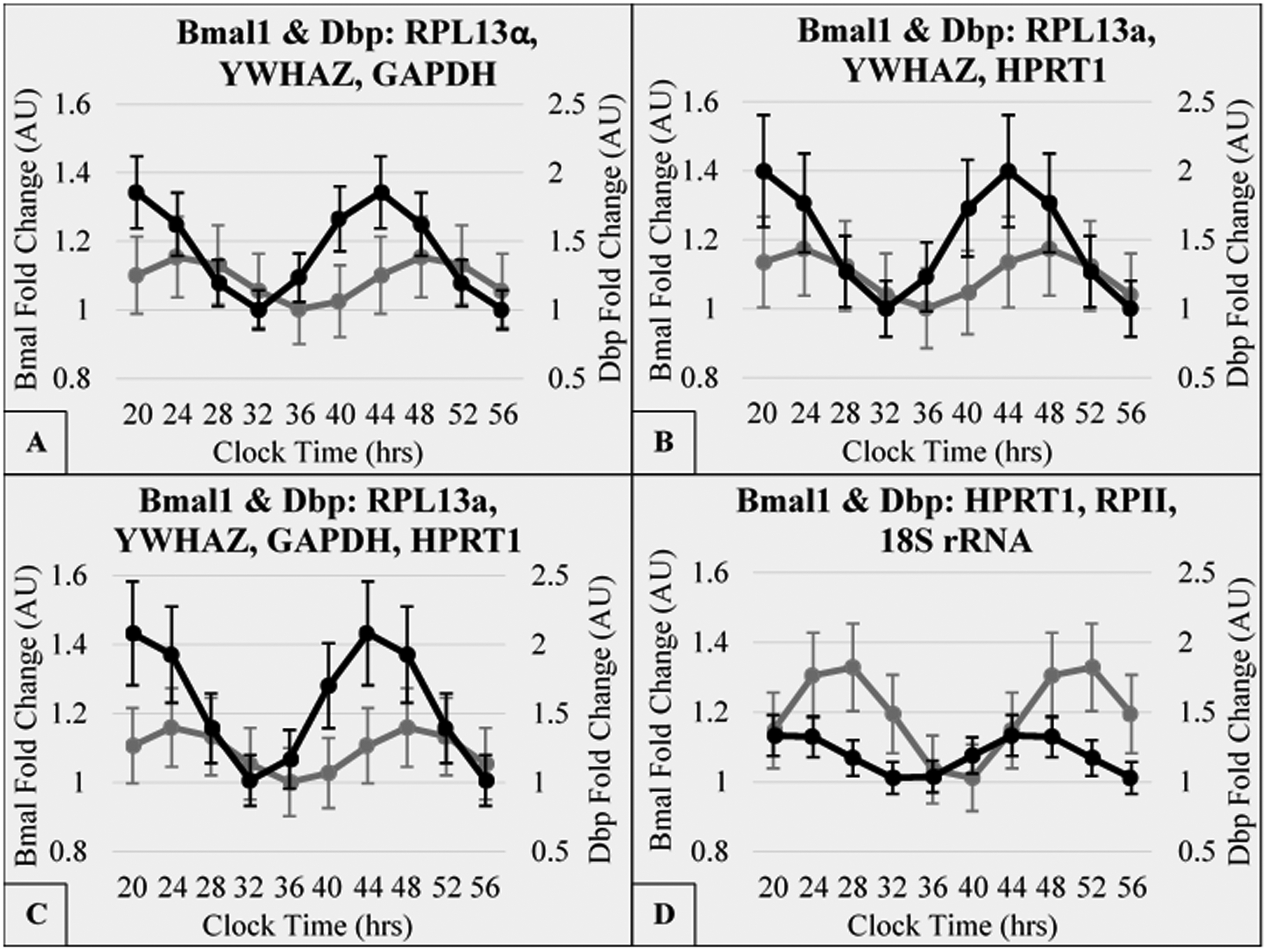
Each gene was measured (n = 3) across the time series experiment for Bmall (grey) and Dbp (black) at: twelve-to-thirteen weeks following surgery. Gene expression was normalized to one of four geometrically-averaged reference gene sets. Fold change was calibrated to maximal ΔCq of each gene’s normalized data set. Time is clock time relative to day 0, which had the biopsy during the morning (20 CT = Day 0, 8 PM). Cosinor analysis was conducted using R project package: cosinor2.
For Figure 5, the four reference gene sets were: 1) YWHAZ, RPL13α, and GAPDH; 2) YWHAZ, RPL13α, and HPRT1; 3) YWHAZ, RPL13α, GAPDH, and HPRT1; and 4) HPRT1, RPII, and 18S rRNA. The first three reference gene sets corresponded to the suggested options remaining after the conclusion of the rhythm-blind analysis. The fourth reference gene set corresponded to the geometric average of the three worst individually suitable genes (Figure 5B). For three potential reference gene sets (Figure 5A, 5C, and 5D), there was fluctuation across the time series (between 0.5 and 1.5 relative quantity). For the post-surgery biopsy, there were minor peaks that occurred at midnight (24 and 48 clock time). For the pre-surgery biopsy, there were minor peaks at 4 PM of day 1 and 12 AM of day 2. In Figure 5B, the pre-surgery biopsy additionally had a peak at midnight of day 1 and the post-surgery biopsy only had a peak between noon and 4 PM of day 1. Lastly, the difference in relative quantity was of a greater magnitude than other panels (Figure 5A, C, D) and in a reverse direction. The individual reference gene relative quantity plots were represented in Supplemental Figure 5.
The circadian clock genes, Bmal1 and Dbp, were measured for the post-surgery biopsy of three subjects. The normalizations were to each reference gene set previously shown in Figure 5. These data sets were used to conduct a least-squares cosinor analysis using the method cosinor.lm from R-project package cosinor2. For each gene, cycle length was set to twenty-four hours and time was the only explanatory variable used for the output variable: fold change. The object output from cosinor.lm had defined acrophase and amplitude, which were used to calculate the modeled data set for each normalization condition (Figure 6). The curves were calibrated such that the minimal data point was set to one. The calculated amplitudes for Dbp were, respectively: 0.43, 0.50, 0.55, and 0.18. The calculated amplitudes for Bmal1 were, respectively: 0.08, 0.09, 0.08, and 0.17. In each panel, the acrophase of Dbp was calculated to be prior to Bmal1 in agreement with previous work in mice. (Takahashi, 2017) The range for acrophase differences was between 3:47 and 5:19 hours (Figure 6A, 6B, and Supplemental Figure 6). For Bmal1, the calculated acrophases were, respectively: 0:54, 23:46, 0:43, and 2:39. In contrast, prior reports of Bmal1 acrophase preceded our report. (Gómez-Santos et al., 2009; Gómez-Abellán et al., 2012) However, in our current study we: 1) utilized no FBS serum shock, which allowed us to maintain the endogenous rhythmicity from each subject; 2) utilized base media of M199 instead of DMEM; and 3) utilized, in media, low insulin (1 nM) and very low dexamethasone treatment (40 nM) to support cell viability. For Dbp, the calculated acrophases were, respectively: 19:36, 19:58, 20:46, and 21:42 (Supplemental Figure 6). Thus the optimization of reference genes used to normalize the data affect both the amplitude of gene rhythms of circadian genes as well as the phase relationship between the two clock genes.
Discussion
Circadian rhythms are indispensable for understanding of normal physiology, and for research addressing the transition into pathophysiology. This is made clear in studies that have examined the disruption of circadian rhythms by sleep curtailment or shift work, and the resulting positive association with obesity, insulin resistance, and diabetes. Improving precision of the assays that observe circadian rhythms is still on-going. This work is meant to add to this knowledge, particularly, in this context, qRT-PCR assays for measuring target gene expression.
To accomplish this goal, primary subcutaneous human adipose tissue was ex vivo cultured and periodically harvested. Previous literature documented that these culture conditions allowed for the preservation of circadian patterns of gene expression in primary adipose tissue, and the results shown in Figure 6 support this conclusion. Four morbidly-obese female subjects were included in the data set, and for each subject biopsies were collected at three times around bariatric surgery. These conditions were utilized to allow for a complex example of adipose tissue health and endocrine regulation on circadian rhythmicity. With this data, we were able to analyze the proof-of-concept differences in circadian rhythmicity, and assess how suitable previously verified reference genes were under the tested experimental conditions.
The aim of this study was to assess whether any of the candidate genes were sufficient to function individually as reference genes, a practice widely employed in research. Despite vetting by other papers assessing that the genes used were stable under hormonal treatment, BMI-variation, and in adipose tissue, in our hands under the experimental conditions and human subjects used, none of the genes resulted in sufficient stability to function across these conditions. Yet, even under these extremely heterogeneous conditions it was possible to create a reference gene set (YWHAZ, RPL13α, and GAPDH) that was most advantageous to interpret experimental results. At the least, these results strongly suggest that more attention is owed to verifying that the reference standard is sufficiently stable for any given experiment. It is plausible to create a system with an unwieldy oversight burden for disseminating qRT-PCR results. Yet, the process is not sufficiently transparent regarding the effect of normalization on experimental results. This critique either overtly or implicitly undermines the confidence in a technique that is still widely relied upon.
Each method to determine the stability of the reference genes relied on different assumptions of what was required for an acceptable reference gene standard. Use of more than one method is encouraged to verify the provisional stability assessment offered by a single method. The overlap for genes that performed well or poorly did agree between the methods utilized in this study. However, the suitability became more muddled between the approaches if under the experimental conditions there was no gene that clearly performed well. In this case, using the multiple analytic tools offers two options: 1) finer distinctions between suitability of the individual genes, or 2) incorporation of multiple genes into a reference gene set. Since the genes that are being considered are likely to be nearer to the border of acceptability, use of more than a single method fosters a more balanced understanding of the ability of each gene to serve within the reference gene set.
There were two genes that deserved to be discussed in particular. Despite still being widely used, 18S rRNA performed the worst of the six genes tested in the study. This provides increasing evidence that 18S rRNA has certain experimental conditions that lead to notably high variation. Since this is the case, this study shows evidence that corroborates that 18S rRNA should be rigorously tested before acceptance as a reference gene, particularly in regards to circadian clock genes assayed over a period of time. The other gene of note was HPRT1. As would be expected with the previously noted high average Cq, there were a high rate of failed qRT-PCR runs. Yet, also supporting previous literature is that re-running the failed wells did result in an average Cq across conditions that was near to the stability of the best performing genes. It remains uncertain whether the stability is due to actual gene expression or manipulation of stability by the rejection of any results that fall out of the narrow acceptable range.
This current study does rely on the fundamental assumption that random enrollment yields representative subject selection. More subjects are always advantageous for suitable representative of subject populations, and there were a comparatively small number of subjects included in this analysis. However, the paired aspect of the study design enabled a more thorough interrogation of the stability of the genes in these individuals. It is true that individual time course measurements were not entirely independent of the other measurements, but there is not consensus of how dependency should be effectively controlled given that biological rhythm research already involves one layer of linear modeling. This study instead made more measurements to account for the stated dependency.
In the tool BestKeeper, there is explicit regard for the concern of co-regulation of reference genes and target genes. The conceptual risk is that since neither reference genes nor target genes operate outside the cellular regulatory mechanisms, there exists the possibility that the experimental manipulation will be reflected in both the reference gene and target gene expression. The downside of this result is that the ΔCq normalization step will eliminate, or dampen, the actual change in target gene expression. This is an example of a false negative error. Therefore, care should be taken in the selection of candidate reference genes. Yet, there is no firm guideline as to what constitutes unacceptable co-regulation. So, instead, it is suggested that this error is conceptually and experimentally addressed by considering and measuring covariance of a number of potential reference genes for each set of experimental protocol used to obtain the greatest fidelity in these critical tools for data normalization.
Supplementary Material
Acknowledgments and Funding
This work was supported by R01 DK103014 (to MJB) and the University of Chicago DRTC P30DK020595, while JMW was supported by a fellowship from T32HL007909. Each author was involved in the design of the experiments, was involved in the writing, and approved the final version for submission. We thank Dr. Eve Van Cauter for guidance and productive conversations. We thank the bariatric surgeons at the University of Chicago for performing the biopsies. And, we thank Kyle Kunze for assistance in carrying out the experiments.
Footnotes
Conflict of Interest
The authors declare that there is no conflict of interest regarding the publication of this article.
References:
- Amable PR, Teixeira MVT, Carias RBV, Granjeiro JM, and Borojevic R (2013) Identification of Appropriate Reference Genes for Human Mesenchymal Cells during Expansion and Differentiation. Plos One 8:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arble DM, Bass J, Behn CD, Butler MP, Challet E, Czeisler C, Depner CM, Elmquist J, Franken P, Grandner MA, Hanlon EC, Keene AC, Joyner MJ, Karatsoreos I, Kern PA, Klein S, Morris CJ, Pack AI, Panda S, Ptacek LJ, Punjabi NM, Sassone-Corsi P, Scheer FA, Saxena R, Seaquest ER, Thimgan MS, Van Cauter E, and Wright KP (2015) Impact of Sleep and Circadian Disruption on Energy Balance and Diabetes: A Summary of Workshop Discussions. Sleep 38:1849–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin K, Safoine M, Proulx M, Audet-Casgrain M-A, Côté J-F, Têtu F-A, Roy A, and Fradette J (2015) Characterization of In Vitro Engineered Human Adipose Tissues: Relevant Adipokine Secretion and Impact of TNF-α. PloS one 10:e0137612–e0137612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass J, and Takahashi JS (2010) Circadian Integration of Metabolism and Energetics. Science 330:1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson DM, Dunn FA, and Takao M (2002) Phototransduction by Retinal Ganglion Cells That Set the Circadian Clock. Science 295:1070. [DOI] [PubMed] [Google Scholar]
- Broussard JL, Ehrmann DA, Van Cauter E, Tasali E, and Brady MJ (2012) Impaired insulin signaling in human adipocytes after experimental sleep restriction: a randomized, crossover study. Annals of internal medicine 157:549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson J, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley G, Wittwer CT, Schjerling P, Day PJ, Abreu M, Aguado B, Beaulieu J-F, Beckers A, Bogaert S, Browne JA, Carrasco-Ramiro F, Ceelen L, Ciborowski K, Cornillie P, Coulon S, Cuypers A, De Brouwer S, De Ceuninck L, De Craene J, De Naeyer H, De Spiegelaere W, Deckers K, Dheedene A, Durinck K, Ferreira-Teixeira M, Fieuw A, Gallup JM, Gonzalo-Flores S, Goossens K, Heindryckx F, Herring E, Hoenicka H, Icardi L, Jaggi R, Javad F, Karampelias M, Kibenge F, Kibenge M, Kumps C, Lambertz I, Lammens T, Markey A, Messiaen P, Mets E, Morais S, Mudarra-Rubio A, Nakiwala J, Nelis H, Olsvik PA, Pérez-Novo C, Plusquin M, Remans T, Rihani A, Rodrigues-Santos P, Rondou P, Sanders R, Schmidt-Bleek K, Skovgaard K, Smeets K, Tabera L, Toegel S, Van Acker T, Van den Broeck W, Van der Meulen J, Van Gele M, Van Peer G, Van Poucke M, Van Roy N, Vergult S, Wauman J, Tshuikina-Wiklander M, Willems E, Zaccara S, Zeka F, and Vandesompele J (2013) The need for transparency and good practices in the qPCR literature. Nature Methods 10:1063. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, and Wittwer CT (2009) The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clinical Chemistry 55:611–622. [DOI] [PubMed] [Google Scholar]
- Carswell KA, Lee MJ, and Fried SK (2012) Culture of Isolated Human Adipocytes and Isolated Adipose Tissue In Human Cell Culture Protocols, Third Edition, Mitry RR, and Hughes RD, eds, pp 203–214, Humana Press Inc, Totowa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Lu F, Li Y, Xue Y, Kang Y, Zhang S, Qiu Q, Cui X, Zheng S, Liu B, Xu X, and Cao X (2013) Ubiquitin-specific proteases UBP12 and UBP13 act in circadian clock and photoperiodic flowering regulation in Arabidopsis. Plant physiology 162:897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis KM, Gomez LA, Rios C, Garbayo E, Raval AP, Perez-Pinzon MA, and Schiller PC (2010) EF1 alpha and RPL13a represent normalization genes suitable for RT-qPCR analysis of bone marrow derived mesenchymal stem cells. Bmc Molecular Biology 11:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czeisler CA, Duffy JF, Shanahan TL, Brown EN, Mitchell JF, Rimmer DW, Ronda JM, Silva EJ, Allan JS, Emens JS, Dijk D-J, and Kronauer RE (1999) Stability, Precision, and Near-24-Hour Period of the Human Circadian Pacemaker. Science 284:2177. [DOI] [PubMed] [Google Scholar]
- del Pozo CH, Calvo RM, Vesperinas-Garcia G, Gomez-Ambrosi J, Fruhbeck G, Corripio-Sanchez R, Rubio MA, and Obregon MJ (2010) IPO8 and FBXL10: New Reference Genes for Gene Expression Studies in Human Adipose Tissue. Obesity 18:897–903. [DOI] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kévei E, Tóth R, Nagy F, Hibberd JM, Millar AJ, and Webb AAR (2005) Plant Circadian Clocks Increase Photosynthesis, Growth, Survival, and Competitive Advantage. Science 309:630. [DOI] [PubMed] [Google Scholar]
- Drosatos K, and Schulze PC (2013) Cardiac lipotoxicity: molecular pathways and therapeutic implications. Current heart failure reports 10:109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvornyk V, Vinogradova O, and Nevo E (2003) Origin and evolution of circadian clock genes in prokaryotes. Proceedings of the National Academy of Sciences 100:2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fain JN, Cheema PS, Bahouth SW, and Lloyd Hiler M (2003) Resistin release by human adipose tissue explants in primary culture. Biochemical and Biophysical Research Communications 300:674–678. [DOI] [PubMed] [Google Scholar]
- Fink T, Lund P, Pilgaard L, Rasmussen JG, Duroux M, and Zachar V (2008) Instability of standard PCR reference genes in adipose-derived stem cells during propagation, differentiation and hypoxic exposure. Bmc Molecular Biology 9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foldager CB, Munir S, Ulrik-Vinther M, Soballe K, Bunger C, and Lind M (2009) Validation of suitable house keeping genes for hypoxia-cultured human chondrocytes. Bmc Molecular Biology 10:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaulet M, Gomez-Abellan P, Alburquerque-Bejar JJ, Lee YC, Ordovas JM, and Scheer F (2013) Timing of food intake predicts weight loss effectiveness. International Journal of Obesity 37:604–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaulet M, Ordovás JM, Gómez-Abellán P, Martínez JA, and Madrid JA (2011) An approximation to the temporal order in endogenous circadian rhythms of genes implicated in human adipose tissue metabolism. Journal of cellular physiology 226:2075–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill V, and Zhou YP (1995) Long term exposure to fatty acids and ketones inhibits B-cell functions in human pancreatic islets of Langerhans. The Journal of Clinical Endocrinology & Metabolism 80:1584–1590. [DOI] [PubMed] [Google Scholar]
- Gómez-Abellán P, Díez-Noguera A, Madrid JA, Luján JA, Ordovás JM, and Garaulet M (2012) Glucocorticoids Affect 24 h Clock Genes Expression in Human Adipose Tissue Explant Cultures. PLOS ONE 7:e50435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Santos C, Gómez-Abellán P, Madrid JA, Hernández-Morante JJ, Lujan JA, Ordovas JM, and Garaulet M (2009) Circadian Rhythm of Clock Genes in Human Adipose Explants. Obesity 17:1481–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JAJ, Ellisman MH, and Panda S (2012) Time-Restricted Feeding without Reducing Caloric Intake Prevents Metabolic Diseases in Mice Fed a High-Fat Diet. Cell Metabolism 15:848–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Guertler R, Naim S, Nixdorf S, Fedier A, Hacker NF, and Heinzelmann-Schwarz V (2013) Careful Selection of Reference Genes Is Required for Reliable Performance of RT-qPCR in Human Normal and Cancer Cell Lines. Plos One 8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarek JL, Thompson SV, and Holscher HD (2017) Complex interactions of circadian rhythms, eating behaviors, and the gastrointestinal microbiota and their potential impact on health. Nutrition reviews 75:673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw EE, and Flier JS (2004) Adipose Tissue as an Endocrine Organ. The Journal of Clinical Endocrinology & Metabolism 89:2548–2556. [DOI] [PubMed] [Google Scholar]
- Knutson KL, Spiegel K, Penev P, and Van Cauter E (2007) The metabolic consequences of sleep deprivation. Sleep medicine reviews 11:163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam VH, Li YH, Liu X, Murphy KA, Diehl JS, Kwok RS, and Chiu JC (2018) CK1α collaborates with DOUBLETIME to regulate PERIOD function in the Drosophila circadian clock. The Journal of Neuroscience. [DOI] [PMC free article] [PubMed]
- Longo VD, and Panda S (2016) Fasting, Circadian Rhythms, and Time-Restricted Feeding in Healthy Lifespan. Cell metabolism 23:1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGown C, Birerdinc A, and Younossi ZM (2014) Adipose Tissue as an Endocrine Organ. Clinics in Liver Disease 18:41–58. [DOI] [PubMed] [Google Scholar]
- Mehta R, Birerdinc A, Hossain N, Afendy A, Chandhoke V, Younossi Z, and Baranova A (2010) Validation of endogenous reference genes for qRT-PCR analysis of human visceral adipose samples. BMC Molecular Biology 11:39–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouchi N, Parker JL, Lugus JJ, and Walsh K (2011) Adipokines in inflammation and metabolic disease. Nature Reviews Immunology 11:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y, Andersson CR, Kondo T, Golden SS, and Johnson CH (1998) Resonating circadian clocks enhance fitness in cyanobacteria. Proceedings of the National Academy of Sciences of the United States of America 95:8660–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Hogenesch JB, and Kay SA (2002) Circadian rhythms from flies to human. Nature 417:329. [DOI] [PubMed] [Google Scholar]
- Paschos GK, Ibrahim S, Song W-L, Kunieda T, Grant G, Reyes TM, Bradfield CA, Vaughan CH, Eiden M, Masoodi M, Griffin JL, Wang F, Lawson JA, and Fitzgerald GA (2012) Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nature medicine 18:1768–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VR, Ceglia N, Zeller M, Eckel-Mahan K, Sassone-Corsi P, and Baldi P (2015) The pervasiveness and plasticity of circadian oscillations: the coupled circadian-oscillators framework. Bioinformatics (Oxford, England) 31:3181–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, and Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – Excel-based tool using pair-wise correlations. Biotechnology Letters 26:509–515. [DOI] [PubMed] [Google Scholar]
- Phillips SA, Ciaraldi TP, Oh DK, Savu MK, and Henry RR (2008) Adiponectin secretion and response to pioglitazone is depot dependent in cultured human adipose tissue. American journal of physiology Endocrinology and metabolism 295:E842–E850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh CS (1954) ON TEMPERATURE INDEPENDENCE IN THE CLOCK SYSTEM CONTROLLING EMERGENCE TIME IN DROSOPHILA. Proceedings of the National Academy of Sciences of the United States of America 40:1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radonić A, Thulke S, Mackay IM, Landt O, Siegert W, and Nitsche A (2004) Guideline to reference gene selection for quantitative real-time PCR. Biochemical and Biophysical Research Communications 313:856–862. [DOI] [PubMed] [Google Scholar]
- Radziuk JM (2013) The suprachiasmatic nucleus, circadian clocks, and the liver. Diabetes 62:1017–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers PM, Mashtalir N, Rathod MA, Dubuisson O, Wang Z, Dasuri K, Babin S, Gupta A, Markward N, Cefalu WT, and Dhurandhar NV (2008) Metabolically favorable remodeling of human adipose tissue by human adenovirus type 36. Diabetes 57:2321–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryden M, Andersson DP, Bergstrom IB, and Arner P (2014) Adipose Tissue and Metabolic Alterations: Regional Differences in Fat Cell Size and Number Matter, But Differently: A Cross-Sectional Study. Journal of Clinical Endocrinology & Metabolism 99:E1870–E1876. [DOI] [PubMed] [Google Scholar]
- Shulman GI (2000) Cellular mechanisms of insulin resistance. The Journal of clinical investigation 106:171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoelstra K, Wikelski M, Daan S, Loudon ASI, and Hau M (2016) Natural selection against a circadian clock gene mutation in mice. Proceedings of the National Academy of Sciences 113:686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS (2017) Transcriptional architecture of the mammalian circadian clock. Nature reviews Genetics 18:164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichopad A, Dilger M, Schwarz G, and Pfaffl MW (2003) Standardized determination of real-time PCR efficiency from a single reaction set-up. Nucleic Acids Research 31:e122–e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, and Bass J (2005) Obesity and metabolic syndrome in circadian Clock mutant mice. Science (New York, NY) 308:1043–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadan R, Amjadi O, Tehrani M, Rafiei A, Hedayatizadeh-Omran A, and Alizadeh-Navaei R (2015) Pseudogene-free amplification of HPRT1 in quantitative reverse transcriptase polymerase chain reaction. Analytical Biochemistry 485:46–48. [DOI] [PubMed] [Google Scholar]
- Van Cauter E, Spiegel K, Tasali E, and Leproult R (2008) Metabolic consequences of sleep and sleep loss. Sleep medicine 9 Suppl 1:S23–S28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, and Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology 3:research0034.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, You T, Murphy K, Lyles MF, and Nicklas BJ (2015) Addition of Exercise Increases Plasma Adiponectin and Release from Adipose Tissue. Medicine and science in sports and exercise 47:2450–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Lahens NF, Ballance HI, Hughes ME, and Hogenesch JB (2014) A circadian gene expression atlas in mammals: Implications for biology and medicine. Proceedings of the National Academy of Sciences 111:16219–16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


