Abstract
Hepatocellular carcinoma (HCC) is difficult to detect, carries a poor prognosis, and is one of few cancers with an increasing yearly incidence. Molecular defects in complement factor H (CFH), a critical regulatory protein of the complement alternative pathway (AP), are typically associated with inflammatory diseases of the eye and kidney. Little is known regarding the role of CFH in controlling complement activation within the liver. While studying aging CFH-deficient (fH–/–) mice, we observed spontaneous hepatic tumor formation in more than 50% of aged fH–/– males. Examination of fH–/– livers (3–24 months) for evidence of complement-mediated inflammation revealed widespread deposition of complement-activation fragments throughout the sinusoids, elevated transaminase levels, increased hepatic CD8+ and F4/80+ cells, overexpression of hepatic mRNA associated with inflammatory signaling pathways, steatosis, and increased collagen deposition. Immunostaining of human HCC biopsies revealed extensive deposition of complement fragments within the tumors. Investigating the Cancer Genome Atlas also revealed that increased CFH mRNA expression is associated with improved survival in patients with HCC, whereas mutations are associated with worse survival. These results indicate that CFH is critical for controlling complement activation in the liver, and in its absence, AP activation leads to chronic inflammation and promotes hepatic carcinogenesis.
Keywords: Immunology, Oncology
Keywords: Complement, Liver cancer
Introduction
Key functions of the immune system are the ability to discriminate self from nonself, and to rapidly eliminate invasive pathogens while causing minimal injury to the host. Rapid induction of inflammation is critical for an efficient immune response, and its rapid resolution prevents unintended damage. If left unresolved, however, inflammation can damage surrounding tissues, eventually leading to fibrosis, organ failure, and carcinogenesis (1, 2). The complement cascade is an important arm of the immune system and is controlled through a balance of activating and regulatory proteins (3). The classical and mannose-binding lectin pathways are activated by antibodies and other pattern-recognition molecules (4). The alternative pathway (AP), however, is distinct in that it is continually activated in plasma through a process called tick-over, indiscriminately depositing activated C3b on nearby surfaces. The deposited C3b catalyzes further AP activation, self-amplifying unless adequately controlled on the target surface by complement regulatory proteins (CRPs). CRPs selectively protect host cells but not invasive pathogens, thereby creating a system of rapid and continuous immune surveillance.
CRPs prevent autologous complement-mediated injury by cleaving and inactivating C3b (a component of the C3 and C5 convertases) and/or accelerating the decay of existing C3 and C5 convertases (3). Because the AP is constitutively activated, inadequate regulation can lead to complement-mediated injury. Congenital and acquired defects in complement regulation are associated with severe diseases, including atypical hemolytic uremic syndrome (aHUS), C3 glomerulopathy (C3G), and protein-losing enteropathy (5–7). Genetic variants of AP proteins are also associated with accelerated progression of some chronic diseases, including age-related macular degeneration (AMD) and IgA nephropathy (8, 9). Complement activation has been linked with the development and spread of several cancers (10–12), raising the possibility that impaired complement regulation could be a risk factor for some types of cancer.
Complement factor H (CFH) is a soluble CRP that regulates AP activation in plasma and on host surfaces through its ability to bind to self-surface ligands. Regions within the CFH protein bind to anionic molecules, including glycosaminoglycans (GAGs), which are highly expressed on basement membranes and extracellular matrix (ECM), and sialic acid, a common component of cellular membranes (13, 14). Engagement of these molecules tethers CFH to host tissues, controlling AP activation at those sites (15). Although sulfated GAGs and sialic acids are ubiquitously expressed, CFH binding and regulatory activity are particularly important for specific tissues (16). For example, CFH is critical for controlling AP activation on ECM and basement membranes, as these surfaces do not intrinsically express other CRPs. Mutations in CFH-binding regions are primarily associated with diseases of the kidneys and eyes (8, 17), possibly because these organs have specialized basement membranes that are highly exposed to plasma proteins.
While studying complement-mediated kidney disease, we observed spontaneous liver tumor development in fH–/– male mice at a significantly higher rate than in control mice. We hypothesized that CFH deficiency causes complement-mediated liver inflammation. Chronic inflammation can promote development of HCC (18), and complement activation can also impair the elimination of tumors by the immune system (19). Consequently, inadequate complement regulation within the liver could contribute to carcinogenesis by several mechanisms. CFH is primarily generated in the liver, as are factor B (CFB) and C3 (activating components of the AP) (20, 21). Proteins produced by hepatocytes enter circulation by passage through the ECM-rich space of Disse and fenestrated endothelial cells lining the sinusoids, continually exposing these regions to high concentrations of complement proteins. The mechanisms of complement activation and regulation within the liver are incompletely understood and a direct connection between CFH dysfunction and liver disease, to our knowledge, has not been previously described.
In the current study, we examined the role of CFH in regulating complement activation in the liver, and the extent to which complement activation is associated with hepatocellular injury. We examined the link between local complement activation in fH–/– mice with changes in the hepatic immune milieu. Finally, we analyzed biopsies and clinical data from patients with HCC to look for evidence of complement dysregulation in human HCC.
Results
Mice lacking CFH spontaneously develop hepatocellular carcinoma.
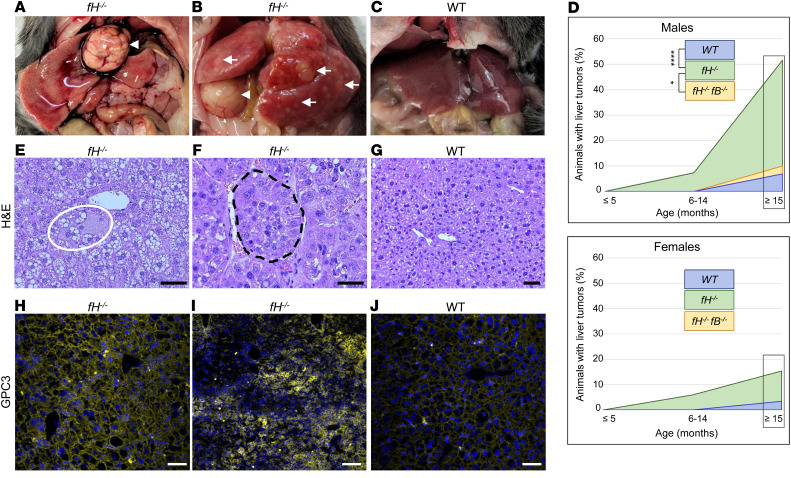
While studying aged fH–/– mice, we noticed a high incidence of spontaneous liver tumor development, which was especially pronounced in males (Figure 1, A and B) compared with WT mice (Figure 1C). In male mice 15 months and older, we observed grossly visible liver tumors in 54% of fH–/– (n = 35) compared with 10% of fH–/–fB–/– mice (which are unable to activate the AP; n = 10) and 7% of WT mice (n = 29). The rate of liver tumor incidence was reduced in female mice 15 months or older, with 15% incidence in fH–/– (n = 26), 3% in WT (n = 29), and 0% in fH–/–fB–/– (n = 10) mice (Figure 1D). Histological evaluation of livers from WT and fH–/– mice from 3 different age groups (≤5 months, 6–14 months, and ≥15 months) revealed notable differences between the 2 strains. Hepatocyte ballooning (an indicator of hepatocyte stress or injury) was present in fH–/– males at all ages (Figure 1E), with multiple instances per field of view observed in over half of the 15-month-old and older males, compared with 14% of WT males of the same age. In male mice 15 months of age or older, multifocal tumor rosettes or nests were seen in all fH–/– mice with visible liver tumors (Figure 1F), and in 50% of those without. Generally, normal liver parenchyma was observed in WT males of the same age (Figure 1G).
Figure 1. fH–/– mice spontaneously develop hepatic tumors.
(A–C) Necropsy images of large focal tumors (A and B, arrowheads) or multilobular tumors (B, arrows) in fH–/– livers. WT livers (C) are generally normal in appearance. (D) Of mice aged 15 months or older, 54% of fH–/– males had visible liver tumors compared with only 10% of fH–/–fB–/– and 7% of WT males. *P = 0.0271 and ****P < 0.0001, respectively; 2-sided Fisher’s exact test; fH–/– n = 35, WT n = 29, fH–/–fB–/– n = 10. Tumor incidence in fH–/– females aged 15 months or older was 15% compared with 0% in fH–/–fB–/– and 3% in WT mice. P = 0.5586 and P = 0.1777, respectively; 2-sided Fisher’s exact test; fH–/– n = 26, WT n = 29, and fH–/–fB–/– n = 10. (E–G) Hepatocyte ballooning (E) and tumor nests in 18-month-old fH–/– livers (F), and generally normal tissue in WT livers (G). Scale bars: 20 μm (E and F), 100 μm (G); n = 20 males (≥15 months) per group. (H–J) Membranous (H) and cytoplasmic (I) GPC3 deposition in fH–/– livers; punctate GPC3 in WT livers (J). Nuclei stained with DAPI (blue). Scale bars: 200 μm (H and J), 100 μm (I); n = 20 males (≥15 months) per group.
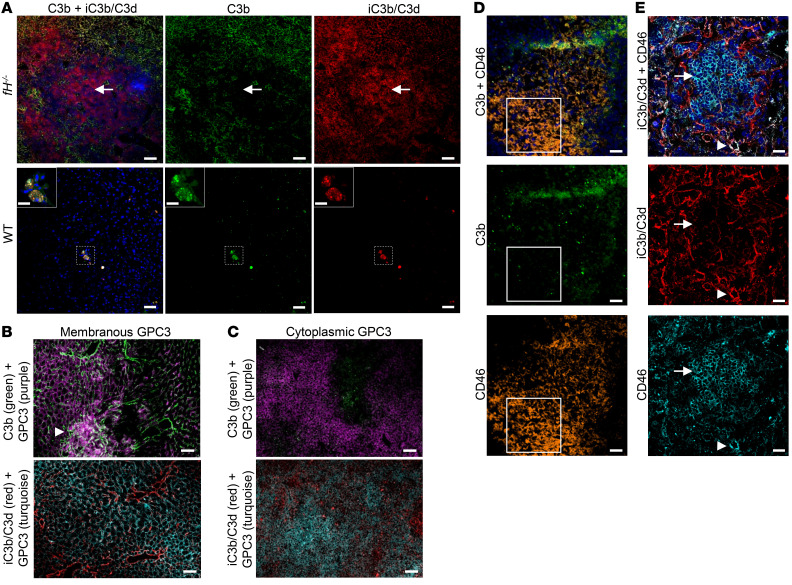
Glypican-3 (GPC3) is a heparan sulfate proteoglycan that is overexpressed in HCC and has been used as a tumor biomarker (22, 23). Variation in the location of GPC3 expression has been noted in human HCC, with localization occurring on cell membranes in some and in the cell cytoplasm of other tumors (24). As further support that tumors in fH–/– mice are HCC, we stained livers collected from animals with and without visible tumors for GPC3 expression. We observed 2 distinct GPC3 expression patterns in the fH–/– mice, which correlated with the macroscopic findings. Livers from fH–/– mice with large focal tumors surrounded by otherwise normal-appearing parenchyma (Figure 1A) expressed GPC3 in a continuous, membranous pattern (Figure 1H). In contrast, GPC3 was predominantly cytoplasmic (Figure 1I) in fH–/– livers with multifocal tumors extending throughout the parenchyma of each lobe (Figure 1B, yellow arrows). Only membranous GPC3 deposition was seen in WT livers with tumors (Figure 1J).
C3 fragments are deposited throughout the livers of fH–/– mice.
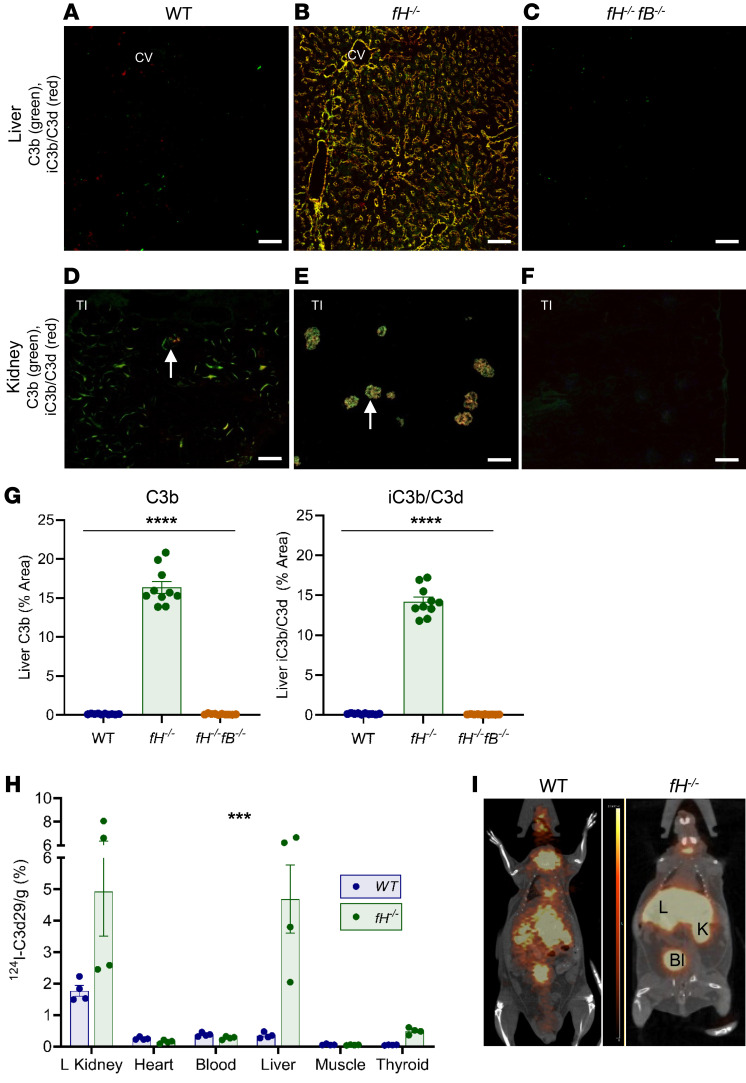
During AP activation, intact fluid-phase C3 is cleaved to form C3b, which covalently attaches to nearby tissues. C3b is subsequently cleaved, generating iC3b and C3d. We immunostained liver and kidney sections from 3- to 5-month-old WT mice, fH–/– mice, CFH heterozygotes (fH+/–), and fH–/–fB–/– mice to evaluate hepatic C3 fragment deposition. In WT and fH+/– mice, we found only trace amounts of punctate C3b and iC3b/C3d in the liver sinusoids (Figure 2A and Supplemental Figure 1A, respectively; supplemental material available online with this article; https://doi.org/10.1172/JCI135105DS1). In fH–/– mice, however, C3b and iC3b/C3d were deposited extensively throughout the sinusoids (Figure 2B). C3b-iC3b/C3d deposition was linear and continuous within the sinusoidal spaces, and appeared to deposit in a gradient, with concentrated C3 in the perivenous regions adjacent to the central vein, becoming more diffuse in the periportal regions. C3b deposits colocalized with collagen IV (COL4) (a component of the basement membrane [ref. 25]) and adjacent to MECA-32 (an endothelial cell marker), further supporting that complement was activated within the sinusoidal wall (Supplemental Figure 2, A and B). Deficiency of both CFH and CFB completely abrogated C3 deposition in livers of fH–/–fB–/– mice (Figure 2C). fH–/– mice are most frequently used to study C3G, which is characterized by C3 deposition localized in the glomeruli (Figure 2E), a location known to be particularly vulnerable to AP activation in the absence of CFH. C3b-iC3b/C3d deposition for each strain has been previously described in detail (26), and representative staining is shown (Figure 2, D and F, and Supplemental Figure 1A). These data demonstrate that, as in the glomeruli, hepatic C3 deposition is significantly greater in fH–/– mice (Figure 2G) and is largely due to AP activation.
Figure 2. C3 fragments preferentially deposit in the livers and kidneys of fH–/– mice.
(A–C) Abundant C3b and iC3b/C3d deposition in fH–/– livers (B), but little to no deposition in WT (A) or fH–/–fB–/– (C) livers. Scale bars: 50 μm; n = 10 males (3–5 months) per group. (D–F) Kidney C3b and iC3b/C3d staining in each strain (glomeruli indicated by arrows). Scale bars: 100 μm; n = 5 males (3–5 months old) per group. (G) Quantification of hepatic C3b and iC3b/C3d deposition. ****P < 0.0001; C3b: F2, 27 = 461.1; iC3b/C3d: F2, 27 = 584.7; 1-way ANOVA with post hoc Tukey’s t test; mean ± SEM; n = 10 per group. (H and I) PET/CT showing focal 124I-C3d29 signal in the left kidney and the liver of the fH–/– mouse (I) and quantification of localized 124I-C3d29 in organs (H). ***P = 0.0001; 124I-C3d29: F5, 36 = 7.067; 2-way ANOVA interaction effect; n = 4 males (3 months old) per group. CV, central vein; TI, tubulointerstitium; L, liver; K, kidney; Bl, bladder.
CFH is a systemic regulator; therefore, fH–/– mice could also be susceptible to AP activation in other organs. To determine other possible locations of AP activation in fH–/– mice, we performed whole-animal PET imaging to detect localized iodinated C3d29 (124I-C3d29) (C3d29 is the monoclonal antibody specific for the complement fragments iC3b and C3d) (27). 124I-C3d29 localization was seen only in the livers and kidneys of fH–/– mice (nonspecific signal in thyroid and bladder is likely due to uptake or clearance of free iodine) (Figure 2I). Bound 124I-C3d29 was quantified (28) 96 hours after imaging (Figure 2H), which confirmed that the liver and kidneys are the predominant sites of C3 deposition in fH–/– mice.
AP activation leads to hepatocellular injury in fH–/– mice.
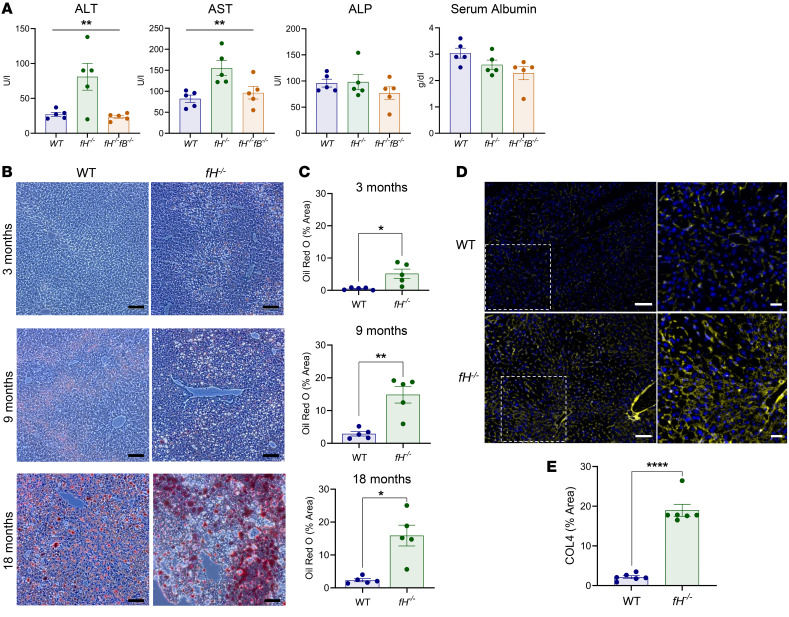
To assess the pathological effects of unregulated AP activation in the liver, we measured levels of alanine transaminase (ALT), aspartate transaminase (AST), alkaline phosphatase (ALP), and albumin in 3-month-old WT, fH–/–, and fH–/–fB–/– mice. ALT and AST were significantly higher in fH–/– mice than in WT or fH–/–fB–/– mice (Figure 3A), confirming hepatocellular injury in fH–/– mice. ALP and albumin were not different among the strains, indicating that the injury had not caused biliary obstruction or impaired synthetic function of the liver at this time point (29).
Figure 3. Hepatocellular injury in fH–/– mice.
(A) Elevated ALT and AST in fH–/– mice. **P < 0.001. ALT, F2, 12 = 8.199; AST, F2, 12 = 7.281; ALP, P = 0.3976; albumin, P = 0.0694; 1-way ANOVA with post hoc Tukey’s t test; mean ± SEM; n = 5 males (3 months) per group. (B and C) Zonal lipid accumulation shown by Oil Red O staining in fH–/– and WT livers at 3, 9, and 18 months. Scale bars: 100 μm. (C) Oil Red O quantification: ** P < 0.01, * P < 0.05; 3 months, t = 3.225, df = 4.106; 9 months, t = 4.556, df = 4.563; and 18 months, t = 4.255, df = 4.176; unpaired, 2-tailed t test with Welch’s correction; mean ± SEM; n = 5 males (each at 3, 9, and 18 months) per group. (D and E) COL4 deposition in WT (D, top) and fH–/– sinusoids (D, bottom). Nuclei stained with DAPI (blue). Images are representative of 4 independent experiments. Scale bars: 100 μm, inset 50 μm; n = 6 males (14 months) per group (E) COL4 quantification. ****P < 0.0001, t = 10.87, df = 5.563; unpaired, 2-tailed t test with Welch’s correction; mean ± SEM; n = 6 males (14 months) per group. ALT, alanine aminotransferase; AST, aspartate aminotransferase; ALP, alkaline phosphatase; COL4, collagen IV.
We stained liver sections from fH–/– and WT males at 3, 9, and 18 months of age with Oil Red O, a lysochrome used to detect lipid. Lipid accumulation was mild and limited to the perivenous hepatocytes of 3-month-old fH–/– mice, but became progressively more widespread with age (Figure 3B, right panels). Positive staining was seen in the 9- and 18-month-old WT mice (Figure 3B, left panels), but when quantified, it was found to be significantly less than in age-matched fH–/– mice (Figure 3C).
We next immunostained livers from 14-month-old fH–/– and WT mice for COL4, a structural component of basement membranes, which, when deposited in excess, leads to the development of fibrosis (30). There was greater COL4 deposition in fH–/– mice compared with WT mice (Figure 3E), occurring in a continuous pattern in the sinusoids of fH–/– mice (Figure 3D, bottom). Only punctate deposits were seen in age-matched WT livers (Figure 3D, top). This suggests that the chronic inflammatory environment triggered by uncontrolled AP activation in fH–/– livers contributes to the development of steatosis and fibrosis as the mice age.
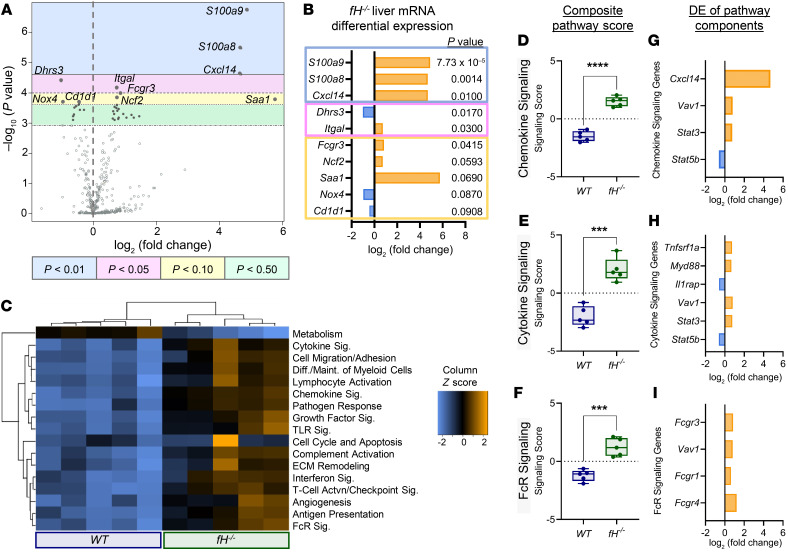
Hepatic mRNA expression reveals a strong inflammatory signature in fH–/– mice.
We examined the mRNA expression profile of myeloid innate immune markers in the livers of 3-month-old fH–/– mice compared with WT mice. When comparing the most differentially expressed (DE) mRNA to WT controls, fH–/– livers had greater than 4.65-fold higher mRNA expression of S100a9 and S100a8, the proinflammatory chemokine Cxcl14, and serum amyloid A 1 (Saa1) (Figure 4, A and B). Of note, S100A9 is expressed on monocytes and neutrophils (31) and has been implicated in the development of cancer (32, 33) and tumor metastasis (34). fH–/– mice also had increased expression of mRNA corresponding to proteins involved in the migration and activation of leukocytes, including Itgal (35), Fcgr3 (36), and neutrophil cytosolic factor 2 (Ncf2) (Figure 4B).
Figure 4. Analysis of liver mRNA reveals upregulation of multiple inflammatory signaling pathways in fH–/– mice.
(A) Volcano plot of fH–/– liver mRNA DE compared with WT covariate and (B) the fold change of the 10 most DE mRNA. Bonferroni’s P values are shown below. (C) Heatmap of myeloid innate immune pathways (data are displayed on the same scale due to Z-transformation; increasing, neutral, or reduced expression indicated by orange, black, or blue, respectively). (D–I) DE of mRNA in fH–/– livers compared with WT livers. Box-and-whisker plots (D–F) showing the composite pathway scores for 3 of the most differentially expressed pathways. ****P < 0.0001, t = 11.01, df = 7.954 (D); ***P = 0.0002, t = 6.777, df = 7.816 (E); ***P = 0.0006, t = 6.033, df = 6.752 (F); unpaired, 2-tailed t test with Welch’s correction. For box-and-whisker plots: box = 25th–75th percentile, line = median, and whiskers = maximum and minimum values. (G–I) The most DE genes (Bonferroni-adjusted P ≤ 0.25) within each of the 3 corresponding composite pathways (D–F). For all experiments, n = 5 males (3 months old) per group. DE, differential expression; Sig, signaling.
Pathway analysis revealed an increase in fH–/– liver mRNA expression in 16 of the 17 gene sets relevant to innate myeloid immune function (Figure 4C). Directed global significance (GS) scores were used to identify the most DE gene sets in fH–/– mice as a group. Three of the gene sets with high directed GS scores (indicating high expression compared with WT covariates), were the chemokine, cytokine, and Fc receptor signaling pathways. Composite scores for these 3 pathways (Figure 4, C and D–F), and the most DE genes corresponding to each of the 3 pathways with relative expression frequencies (Figure 4, G–I), are shown. These data provide transcript level evidence of a proinflammatory environment in the livers of young fH–/– mice.
T cell infiltration and reduced neutrophils in fH–/– livers.
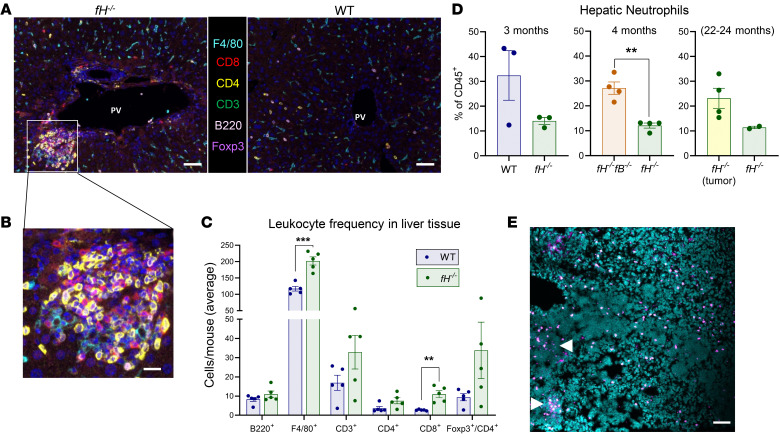
To evaluate the immune milieu within the livers of fH–/– mice, we stained liver sections for 6 general leukocyte markers (F4/80, CD8, CD4, CD3, B220, and Foxp3) and collected images with a Vectra quantitative pathology imaging system. Dense clusters of immune cells were observed in all fH–/– livers near portal veins (Figure 5A, left, and Figure 5B). Similar immune clusters were not seen in WT livers (Figure 5A, right). Significantly more F4/80+ and CD8+ T cells were seen in fH–/– livers (Figure 5C), as well as a trend toward greater numbers of CD4+, CD3+, and CD4+Foxp3+ cells.
Figure 5. Inflammation and leukocyte infiltration in fH–/– livers.
(A and B) Large pockets of inflammatory infiltrate captured by multiplex imaging in fH–/– (A, left, and B) but not in WT livers (A, right). Nuclei stained with DAPI (blue). Original magnification ×200. Scale bars: 50 μm (A), 10 μm (B). (C) Increased populations of each cell type in fH–/– livers compared with WT. F4/80+, ***P = 0.0002, t = 6.802, df = 7.559; CD8+, **P = 0.0069, t = 4.949, df = 4.178; CD4+, P = 0.1228; CD3+, P = 0.1868; Foxp3+/CD4+, P = 0.1409; B220+, P = 0.2894. For A–C, n = 5 males (3 months) per group; 2-tailed t test with Welch’s correction; mean ± SEM. (D) By flow cytometry, young fH–/– males have fewer hepatic neutrophils compared with WT (P = 0.2067) and fH–/–fB–/– (**P = 0.0051, t = 5.596, df = 3.963) mice. Neutrophils trend higher in fH–/– males with liver tumors compared with those without (P = 0.061). Data were analyzed by unpaired, 2-tailed t test with Welch’s correction; mean ± SEM; n = 3 per group (fH–/– and WT, 3 months); n = 4 per group (fH–/– and fH–/–fB–/–, 4 months); n = 4 and 2 (fH–/– with and without liver tumors, respectively; 22–24 months). (E) Clusters of Ly6G+ cells (pink) are observed in livers with multiple tumor foci (arrows). Scale bars: 50 μm; n = 10 independent fH–/– liver tumors. Nuclei stained with DAPI (turquoise). All images are representative of 2 independent experiments.
We assessed hepatic neutrophil populations in young fH–/–, WT, and fH–/–fB–/– males by flow cytometry. There was greater than a 50% reduction in neutrophils (CD45+CD11b+Ly6G+) in fH–/– livers compared with those in WT and fH–/–fB–/– livers (Figure 5D, left and middle). Because neutrophil infiltration within tumors may influence tumor growth (37), we assessed livers with and without tumors from 22- to 24-month-old fH–/– mice. There was a trend toward a greater number of neutrophils in livers with tumors than in those without (Figure 5D, right). We stained fH–/– liver sections containing tumor foci for neutrophils (Ly6G+ cells). Neutrophils were seen within and around tumors (Figure 5E, arrows), providing evidence of neutrophil infiltration in fH–/– liver tumors.
Skewed myeloid cell populations in fH–/– livers.
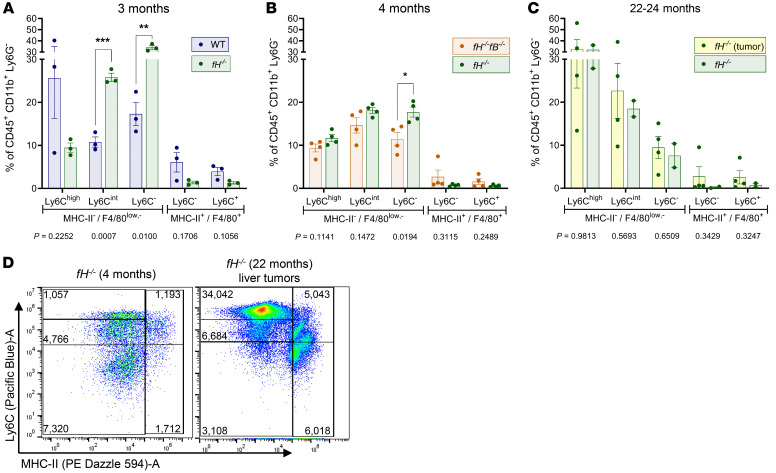
Complement fragments are chemoattractants for infiltrating monocytes, which have been shown to play an important role in shaping the tumor immune microenvironment (38). Previous studies have shown that MHC-II+ monocytes can inhibit tumor growth (39), whereas MHC-II– monocytes can be proangiogenic, suppress T cell function, and promote tumor growth (39, 40). To examine whether the monocyte population is altered in livers of fH–/– mice, we isolated and analyzed hepatic myeloid cells from young fH–/–, WT, and fH–/–fB–/– males, as well as from aged fH–/– males with and without liver tumors. Compared with WT mice, fH–/– mice had fewer Ly6Chi MHC-II– F4/80lo/– proinflammatory infiltrating monocytes (41) and significantly more Ly6C– MHC-II– F4/80lo/– antiinflammatory patrolling monocytes and Ly6Cint MHC-II– F4/80– monocytes (Figure 6A). Only patrolling monocytes (Ly6C– MHC-II– F4/80lo/–) were increased in fH–/– livers when compared with livers from fH–/–fB–/– mice (Figure 6B). There were no significant differences in the monocyte populations between aged fH–/– mice with liver tumors and those without. It is notable, however, that the Ly6Chi MHC-II– F4/80lo/– infiltrating monocytes are the most numerous of these cell types in the aged fH–/– livers, regardless of tumor status (Figure 6C), in contrast to what was seen in young fH–/– mice.
Figure 6. Altered myeloid cell populations in the livers of fH–/– male mice.
(A–C) The frequency of monocyte (Ly6Chi/int/– MHC-II– F4/80lo/–) and macrophage Ly6C+/– MHC-II+ F4/80+) populations in young fH–/– livers compared with WT (A, 3 months) and fH–/–fB–/– (B, 4 months), and livers from aged fH–/– mice with and without visible liver tumors (C, 22–24 months). Data were collected by multispectral flow cytometry and analyzed by unpaired, 2-tailed t test with Welch’s correction; mean ± SEM; n = 3 males per group (fH–/– and WT, 3 months); n = 4 males per group (fH–/– and fH–/–fB–/–, 4 months); n = 4 (fH–/– males with liver tumors) and 2 (fH–/– males without liver tumors). P values for each cell population are shown below each graph; (A) Ly6Cint MHC-II– F4/80lo/– (t = 9.896, df = 3.809) and Ly6C– MHC-II– F4/80lo/– (t = 5.121, df = 3.474); (B) Ly6C– MHC-II– F4/80lo/– (t = 3.250, df = 5.568). (D) Pseudo color dot plot of representative hepatic Ly6C/MHC-II myeloid populations within CD45+CD11b+Ly6G– cells from a young fH–/– mouse (4 months) compared with an aged fH–/– with liver tumors (22 months). Corresponding cell counts for each gate are shown. Detailed gating strategy shown in Supplemental Figure 3.
There were no significant differences in the Ly6C– MHC-II+ F4/80+ and Ly6C+ MHC-II+ F4/80+ macrophage populations between young fH–/– mice and either WT or fH–/–fB–/– mice (Figure 6, A and B) or between aged fH–/– mice with and without liver tumors (Figure 6C). Young and aged fH–/– mice without tumors tended to have fewer macrophages than young WT or fH–/–fB–/– mice or aged fH–/– mice with tumors, respectively.
These findings suggest that the hepatic monocyte population in young fH–/– mice is primarily comprised of antiinflammatory patrolling monocytes. As fH–/– mice age and develop liver tumors, fewer proinflammatory Ly6Chi MHC-II– F4/80lo/– monocytes transition to antiinflammatory Ly6C–MHC-II– F4/80lo/– patrolling monocytes (Figure 6D).
Recombinant murine factor H binds within the liver sinusoids and inactivates C3b.
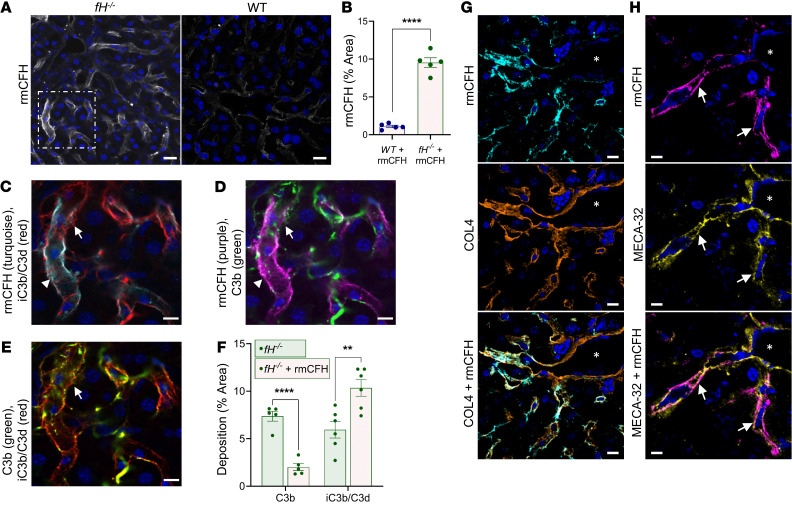
Given the pattern of C3 fragment deposition in fH–/– livers, we examined whether CFH controls AP activation on basement membrane within the sinusoids and hepatic vasculature. We produced and functionally tested a recombinant murine CFH (rmCFH) (42), which was then conjugated with Alexa 647 and injected into male fH–/– and WT mice. The mice were euthanized 24 hours later, and livers were examined for bound rmCFH. The rmCFH deposited along the sinusoids of fH–/– livers (Figure 7, A, left, C, and D) in a pattern much like that of C3b deposition in unmanipulated fH–/– mice (Figure 2B). We observed a similar pattern of rmCFH deposition in WT sinusoids (Figure 7A, right), although in significantly lower amounts (Figure 7B).
Figure 7. Recombinant factor H binds in the sinusoids of fH–/– mice.
(A and B) rmCFH binds within the sinusoids of fH–/– mice in a continuous pattern (A, left), but is reduced and punctate in WT mice (A, right). Scale bars: 20 μm. (B) Quantification of bound rmCFH. ****P < 0.0001, t = 12.98, df = 4.544; 2-tailed t test with Welch’s correction; mean ± SEM. For A and B, n = 5 males per group (WT, 9 months; fH–/–, 7 months). (C–E) Enlarged view of fH–/– image identified by dashed white box in A. Abundant rmCFH deposition (arrowhead) near iC3b/C3d (C, turquoise) and C3b (D, red). Intermittent C3b (C and E; arrows) colocalized with linear iC3b/C3d. Scale bars: 10 μm. (F) Compared with unmanipulated fH–/–, rmCFH-reconstituted fH–/– mice have reduced sinusoidal C3b (****P < 0.0001, t = 8.424, df = 7.179) and increased iC3b/C3d, demonstrating rmCFH-mediated conversion of C3b to iC3b/C3d (**P = 0.0015, t = 4.729, df = 7.931; 2-tailed t test with Welch’s correction; mean ± SEM; n = 5 males [7 months] per group). (G and H) rmCFH colocalized with COL4 (G) and MECA-32 (H, arrows) in fH–/– sinusoids, but not on the large vasculature (vascular lumen denoted by asterisks). Nuclei stained with DAPI (blue). Scale bars: 10 μm; n = 5. Representative images shown from 4 independent experiments.
To assess the functionality of the rmCFH bound within the sinusoids, we stained liver tissues from the rmCFH-injected fH–/– mice for C3b and iC3b/C3d and evaluated the sinusoids for differences in complement deposition (Figure 7, C–E). In rmCFH-reconstituted fH–/– mice, we observed continuous deposition of iC3b/C3d in the sinusoids (Figure 7, C and E) and a shift in C3b deposition from linear to punctate (Figure 7, D and E). There was colocalization of rmCFH with iC3b/C3d (Figure 7C), and only sparse colocalization with C3b (Figure 7D). We quantified C3b and iC3b/C3d deposition in the reconstituted fH–/– mice with that of unmanipulated fH–/– males of the same age. In reconstituted mice, there was a significant decrease in mean percentage of area C3b deposition (2% compared with 7% in unmanipulated mice) and a 2-fold increase in iC3b/C3d (11% compared with 5% in unmanipulated mice) (Figure 7F). These data demonstrate that the injected rmCFH mediated cleavage of bound C3b in the sinusoids, which generated local iC3b/C3d.
Finally, to further characterize the specific location of rmCFH deposition in the sinusoids, we stained liver tissues from rmCFH-injected mice for COL4 and MECA-32. Similar to localization of C3b in the sinusoids, we observed regions of colocalization of rmCFH with both COL4 (Figure 7G) and MECA-32 (Figure 7H), indicating that rmCFH binds ECM and endothelial cells within the sinusoids and is critical for controlling alternative pathway activation at these sites.
Evidence of C3 inactivation within fH–/– liver tumors.
To examine complement regulation within liver tumors, we dual-stained livers from tumor-bearing mice for C3b and iC3b/C3d. In WT livers, we observed small tumor foci with heavy deposits of both C3b and iC3b/C3d, surrounded by normal-appearing parenchyma with very little C3 deposition (Figure 8A, left panels). In contrast, fH–/– tumors and surrounding parenchymal tissue displayed extensive C3b-iC3b/C3d deposition (Figure 8A, right panels). There were distinct regions within some of the fH–/– tumors with minimal C3b and abundant iC3b/C3d deposition, suggesting that there was cofactor activity and inactivation of C3b within the tumor environment. To examine this possibility further, we dual-stained tumor tissues with GPC3 (to indicate tumor boundary) and either C3b or iC3b/C3d (Figure 8, B and C). In tumors with membranous GPC3, we observed both C3b and iC3b/C3d within the tumor lesion (Figure 8B). However, in tissues with cytoplasmic GPC3, C3b deposits were less apparent, suggesting better complement regulation within these tumors (Figure 8C).
Figure 8. Complement deposition in fH–/– liver tumors.
(A) C3b-iC3b/C3d deposition in WT liver tumors (A, bottom) is sparse and focal. Generally dense C3b-iC3b/C3d deposition is seen throughout fH–/– tumors and the surrounding parenchyma (A, top), with distinct regions lacking C3b (white arrow). Scale bars: 100 μm (A, WT), 20 μm (A, WT inset), and 50 μm (A, fH–/–); n = 10 (fH–/–) and 2 (WT) males with liver tumors (≥15 months). Images shown are from 20-month-old mice with visible liver tumors. (B and C) Variable patterns of C3b-iC3b/C3d and GPC3 within and surrounding individual fH–/– tumors. Sinusoidal C3b and iC3b/C3d deposition surrounding fH–/– tumors with membranous GPC3 deposition (B), and dense C3b within the tumor (B, arrowhead). Nearly absent C3b deposition (C, top) and granular iC3b/C3d deposition (C, bottom) in a fH–/– tumor with cytoplasmic GPC3 deposition. Scale bars: 50 μm; n = 10 fH–/– males with liver tumors (≥15 months). (D and E) CD46 expression with C3b (D) and iC3b/C3d (E) deposition in tumor-bearing fH–/– livers. C3b is absent from (D, white box) and iC3b/C3d (E, arrow) is reduced in regions with dense CD46. Additionally, iC3b/C3d deposition is seen colocalizing with CD46 (E, arrowhead). Scale bars: 50 μm (D), 20 μm (E); shown n = 5 tumor-bearing fH–/– livers. Representative images from n > 5 (A) and 4 (B–E) independent experiments.
The presence of iC3b/C3d within fH–/– liver tumors indicates that another protein serves as a cofactor for cleavage of C3b in the absence of CFH. Studies of human HCC have shown that tumors can upregulate membrane-bound complement regulators such as CD46 (membrane cofactor protein or MCP) (43, 44). Similar to CFH, CD46 is a cofactor for the cleavage of C3b, although it is reported to have limited expression in healthy mice (45). We stained fH–/– liver tumors for CD46 and C3b and found prominent CD46 expression in some regions where C3b was absent (Figure 8D, white box). We also dual-stained fH–/– liver tumors for CD46 and iC3b/C3d. There were regions of high CD46 expression in which iC3b/C3d was not seen, as well as regions in which CD46 and iC3b/C3d colocalized (Figure 8E). These results suggest that expression of CD46 within the HCC lesions may inhibit alternative pathway activation and it may also serve as a cofactor for the cleavage of C3b.
C3 fragments are deposited in human HCC biopsies.
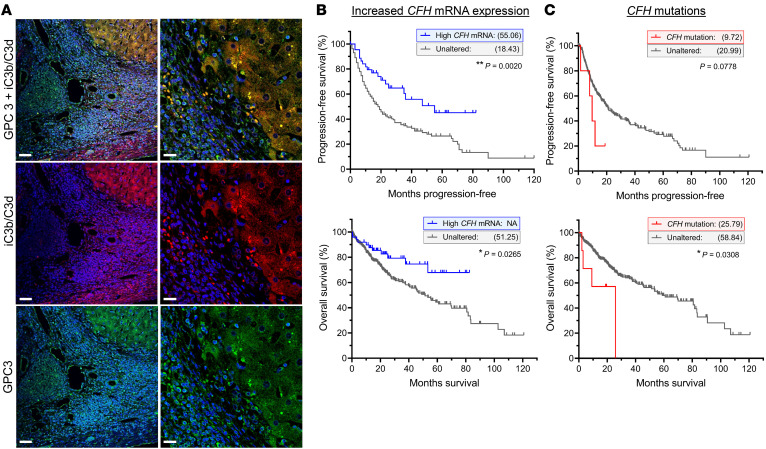
We immunostained liver biopsy samples from 6 confirmed HCC patients for C3d/iC3b and GPC3 (Figure 9A), 5 of which were strongly positive for C3d/iC3b. Among these 5 samples, there was colocalization of GPC3 and C3d/iC3b, indicating that there is complement activation within the tumors.
Figure 9. Complement activation and factor H in human HCC.
(A) iC3b/C3d and GPC3 deposition in human HCC biopsies. Scale bars: 50 μm (left panels), 20 μm (right panels); n = 6. (B and C) Kaplan-Meier plots and statistical analysis based on the work of the TCGA Firehose Legacy study, showing the effect of increased CFH mRNA expression (B) and CFH mutations (C) within the tumors of HCC patients, on survival and disease progression. Increased tumor-associated CFH mRNA improved both progression-free survival (B, top, **P = 0.0020; n = 18 relapse out of 45 [high CFH mRNA], 161 out of 280 [unaltered]) and overall survival (B, bottom: *P = 0.0265; n = 11 deceased out of 50 [high CFH mRNA] and 121 out of 326 [unaltered]). Tumor-associated CFH mutations reduced the number of disease/progression-free months (C, top: P = 0.0778; n = 4 relapsed/progressed out of 5 [CFH mutation] and 175 out of 320 [unaltered]) and overall survival (C, bottom: *P = 0.0308; n = 4 deceased out of 7 [CFH mutation] and 128 out of 369 [unaltered]). Median months survival and progression-free survival shown in parentheses for each group. Data were analyzed by log rank Mantel-Cox test.
We next queried The Cancer Genome Atlas (TCGA) through cBioPortal for Cancer Genomics (46, 47) to determine if CFH mutations or mRNA expression correlate with outcomes for patients with HCC. We found that CFH is altered in 23% (86 of 337) of the patients comprising this data set. Of those 86 patients, 50 had increased tumor CFH mRNA expression, 7 had missense or truncating mutations, and 29 had an amplification in CFH copy number (Supplemental Figure 4A). Increased expression of CFH is expected to help control complement activation, and we found that high CFH mRNA correlated with improved disease/progression-free survival as well as overall survival when compared with HCC patients with unaltered CFH mRNA (Figure 9B). The median disease/progression-free time for patients with high CFH mRNA was 55.06 months compared with 18.43 months for those with unaltered CFH mRNA expression. Conversely, CFH mutations are expected to increase complement activation in the liver and HCC tumors. Patients with CFH mutations had significantly worse overall survival, with a median disease/progression-free time of only 9.72 months compared with 20.99 months for those without CFH mutations (Figure 9C). These findings demonstrate that the complement cascade is activated in the livers of patients with HCC, and that expression levels of CFH mRNA as well as mutations in the CFH gene significantly affect the prognosis for HCC patients.
To determine whether these survival and prognostic outcomes relating to CFH mutations and increased CFH mRNA are unique to HCC, we queried CFH for all cancers in the TCGA Firehose Legacy study (Supplemental Table 1). We found that most cancer types could be grouped according to 3 categories: (a) worse outcome with CFH mutations, improved outcome with increased CFH mRNA; (b) worse outcomes for both CFH mutations and/or increased CFH mRNA; and (c) improved outcomes for both. We found 3 cancers for which the correlations were similar to HCC (worse outcomes with CFH mutations, improved outcomes with increased CFH mRNA): lymphoid neoplasm diffuse large B cell lymphoma, lung adenocarcinoma, and esophageal carcinoma.
Discussion
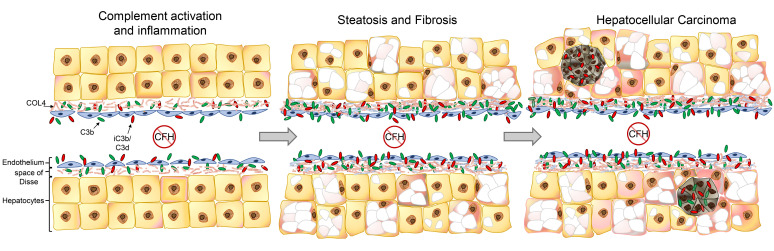
Specific locations such as the eyes and kidneys are known to be particularly dependent on CFH to control the AP. These sites are rich in ECM, have specialized basement membranes, and are susceptible to AP-mediated inflammation in patients with CFH mutations. In the current study, we report that CFH is also critical for controlling AP activation in the liver. There is extensive complement activation throughout the sinusoids of fH–/– mice, which is ameliorated through combined deficiency of CFH and CFB, confirming a principal role of the AP. AP activation in male fH–/– mice was associated with hepatocellular inflammation and injury at 3 months of age, chronic liver damage and steatosis at 9 months, and the development of liver tumors for more than 50% of those aged to 15 months or more. This suggests that the hepatic inflammation resulting from chronic AP activation promotes a favorable environment for tumorigenesis.
C3b and iC3b/C3d were deposited in a continuous pattern along the sinusoids within the livers of fH–/– mice. In mice reconstituted with rmCFH, the injected protein colocalized with COL4 (Figure 7G) and MECA-32 (Figure 7H) within the sinusoids and mediated cleavage of the deposited C3b to inactive iC3b/C3d. We developed a whole-body PET method to identify other sites of AP activation in fH–/– mice and confirmed that the liver and kidneys are the primary sites of complement activation in the absence of CFH.
There is substantial experimental and clinical research linking genetic defects with CFH in kidney disease. Much less is known, however, regarding the role of CFH in liver disease. Although aHUS is primarily regarded as a kidney disease, it is noteworthy that abnormal liver function tests may be seen in up to 46% of patients (48). It has also been reported that complement-mediated injury can lead to hepatic failure in aHUS patients undergoing liver transplantation (49). The presumed mechanism for this outcome is inadequately controlled AP activation in the ischemic liver allograft, resulting from dysfunctional endogenous CFH. It is possible that the liver is a target organ in patients with aHUS-associated CFH mutations but is overlooked due to the severe renal involvement. It is also possible that the binding mechanism of CFH to liver sinusoids and glomerular capillaries involves slightly different interactions. This is the case for AMD and aHUS, in which distinct genetic variants of CFH impair binding of the protein to either the eyes or the kidneys, thereby predisposing patients to one of these diseases but not the other (50).
CFH mutations have not previously been linked with the risk for HCC, although it has been reported that complement activation fragments are elevated in affected patients (51–53). In one study, complement fragments were the only independent plasma biomarker predictive of HCC by multivariate analysis (53). In the current study, we found evidence of complement activation in biopsy samples from 6 patients with HCC. This demonstrates that complement activation can occur within the tumors, similarly to observations in mice. Furthermore, examination of the TCGA reveals that expression of CFH mRNA significantly correlates with survival in patients with HCC (Figure 9B). Although fH–/– mice have complete deficiency of CFH, the TCGA data suggest that variations in CFH function and expression levels may also influence the prognosis of HCC. Future studies can examine whether CFH influences the growth of tumors by inhibiting complement within the tumor itself or through interactions in the tumor microenvironment.
It has been estimated that chronic liver inflammation may account for as much as 90% of HCCs, although the mechanisms linking inflammation with carcinogenesis are incompletely understood (54). Multiple studies have also reported that the complement cascade is activated in the livers of patients with chronic hepatitis B (55, 56), hepatitis C (51, 52, 56), and alcoholic hepatitis (57), inflammatory diseases strongly associated with an increased risk of HCC. Based on the results of the current study, it is possible that inflammation resulting from uncontrolled complement activation within the liver is a common downstream pathway that links these chronic liver diseases with carcinogenesis. The heterogeneous pattern of complement activation within the tumors may be due to upregulation of CD46. It is also possible that additional factors promote complement activation once a cancer has formed. Antibodies may recognize tumor neoantigens, for example. Some tumor cells also produce enzymes that are capable of directly activating C3, such as cathepsins B and L (58).
Complement activation within the liver may contribute to the development of HCC by several mechanisms. Complement activation has been shown to trigger NF-κB activation in Kupffer cells and STAT3 activation in hepatocytes (59), which may facilitate recovery of the liver after acute injury, but chronically promote hepatocyte proliferation and the development of HCC (18, 60, 61). Complement-mediated inflammation in the livers of fH–/– mice could also promote DNA damage, increasing the rate of cancer formation. Complement activation has also been shown to inhibit anti–tumor immunity in multiple types of cancer (10–12, 62, 63). For example, work has shown that C3a and C5a can induce suppressive myeloid cells or directly inhibit T cell proliferation and function (11, 63, 64). We found that the myeloid cell population is altered in the livers of fH–/– mice and that a greater number of Ly6Cint MHC-II– monocytes were present in the livers of these mice (Figure 6, A and B). These cells can be immunosuppressive and may reduce anti–tumor immunity in fH–/– livers. Thus, complement activation within the liver may both directly accelerate the development of HCC as well as impair the ability of the immune system to eliminate these tumors once they have formed.
A limitation of our study is that it uses mice that are genetically deficient in CFH from birth. Many different signaling pathways are activated in these mice, and the development of chronic liver damage and HCC takes a relatively long time. An advantage of this model, however, is that it is characterized by spontaneous tumor formation. Most cancer models in immunocompetent mice involve genetic manipulation of tumor suppressor genes, chemical carcinogenesis, or transplant of cancer cells into recipient mice. Furthermore, in tumor transplant models, the cell lines are often derived from advanced-stage tumors (65). These tumors therefore start with a high mutational burden and have already undergone immune selection. The fH–/– mice, in contrast, provide the opportunity to study spontaneous HCCs as they develop.
In conclusion, we have found that CFH is necessary for controlling spontaneous AP activation in the liver. In the absence of CFH, dysregulated AP activation is associated with hepatocellular injury, steatosis, and development of HCC. We found evidence of complement activation in human HCC biopsies, and that CFH mutations and levels of CFH mRNA correlate with patient survival. There is clinical evidence that the complement system is activated in multiple chronic inflammatory diseases associated with increased risk of HCC. Complement activation may, therefore, be a common pathway linking viral, toxic, or autoimmune injury of the liver with carcinogenesis. Several therapeutic complement inhibitors are currently in development (66). Given the potential role of complement activation in liver carcinogenesis identified in this study, complement inhibition may represent a novel approach for treating HCC.
Methods
Antibodies, equipment, and software.
Manufacturer/supplier, catalog number, clone, and working concentration for all antibodies are provided in Supplemental Table 2. The mouse IgG2 isotype antibody was generated and validated using previously described methods (27). Specific details regarding equipment and software are provided in Supplemental Tables 3 and 4, respectively.
Study design.
Spontaneous hepatic tumor development was observational; therefore, group sizes were dependent on the number of animals reaching advanced age (≥15 months). Ethical and humane endpoints were rigorously followed. All other group sizes were determined based on previous experience (minimal phenotypic variation) and the minimum number of animals required to provide reliable statistical results. Endpoints, when applicable, were determined in advance and were based on age or time (hours following administration of recombinant protein or imaging reagent). No data were excluded. A minimum of 3 independent experiments with successful replication were performed for all studies when possible (except in cases of limited resources). All reported microscopy studies were successfully replicated in at least 5 independent experiments with varying conditions and equipment. Additionally, microscopy data were collected, background adjusted, converted to binary format, and evaluated in a blinded manner. All data are reported as biological replicates.
Human HCC liver tissues were provided as deidentified, presectioned slides by the University of Colorado Biorepository Core facility and the University of Colorado Cancer Center Tissue Biobanking and Histology Shared Resource. The following known BRISQ Tier 1 criteria were provided: solid tissue was removed from the livers of living patients with confirmed HCC diagnosis by surgical/clinical procedure; both clinical and pathology diagnosis of HCC were confirmed for each sample; resected tissues were stabilized and fixed in 10% neutral buffered formalin before long-term preservation in paraffin-embedded blocks; preserved tissues were shipped and stored at ambient temperature (storage duration ranges from 10 to 20 years). HCC liver samples provided for this study were selected based on more than 20% tumor composition by the University of Colorado Biorepository Core.
Mice.
Male and female C57BL/6J WT mice were purchased from the Jackson Laboratory. The fH–/– mice were created and initially provided by MCP (26). Generation of fB–/– mice was previously described (67). Generation of fH–/–fB–/– mice was achieved through interbreeding of fH–/– and fB–/– mice. All mice were bred for at least 7 generations before experimentation.
Liver chemistries.
Fibrin-free serum samples were analyzed using the Heska DriChem 7000 analyzer.
Light microscopy and evaluation of histopathological injury.
Portions of the left lateral liver lobes were collected from each animal at the time of euthanasia and fixed in 10% neutral buffered formalin at 4°C. Fixed tissues were transferred to 70% ethanol and embedded in paraffin. Slides with 3-μm-thick FFPE sections were processed and stained with hematoxylin and eosin. Stained tissues were imaged at original magnifications of ×100 or ×200 and evaluated in a blinded fashion in groups of age- and sex-matched mice.
Oil Red O staining.
Slides with 8-μm-thick frozen sections were warmed to room temperature and fixed in cold 10% neutral buffered formalin for 30 minutes at 4°C. Fixed sections were washed in deionized water 5 times and rinsed with 60% isopropanol before staining in freshly prepared Oil Red O working solution (0.5% stock diluted 3:2 with deionized water) for 15 minutes. Stained tissues were rinsed once more in 60% isopropanol followed by deionized water and mounted with a solution containing 1:1 glycerol and PBS. Slides were imaged in a blinded fashion with a Zeiss Axio Observer D1 microscope in positive phase contrast at ×100 original magnification. Images were quantified with Image J (NIH) by thresholding (for tissue segmentation) and then measuring the thresholded area. Ten separate fields of view were examined for each animal. Numerical values for percentage of area deposition obtained in Image J were analyzed with Prism by a 2-tailed t test with Welch’s correction.
Immunofluorescence staining.
Six-micrometer-thick sagittal sections from frozen, OCT-embedded tissues were warmed to room temperature, fixed in absolute acetone, and washed twice with PBS. Nonspecific binding and endogenous Fc receptors were blocked with 1% BSA and 5% heat-inactivated goat/rat/rabbit serum diluted in PBS for 1 hour at room temperature. Antibodies were diluted in PBS containing 2% heat-inactivated fetal bovine serum (HI-FBS) and 1% BSA, applied to blocked tissue sections, and incubated overnight at 4°C in a humidified chamber. Nuclei were stained with DAPI, washed in cold PBS, and mounted with a 1:1 solution of PBS and glycerol. Kidney autofluorescence was blocked with 0.05% Sudan Black B in 70% ethanol for 15 minutes at room temperature, washed twice in deionized water (10 minutes each), and mounted as described above. Slides were sealed and imaged in a blinded fashion with either an Olympus FV1000 FCS confocal (at ×100, ×200, or ×600 original magnification) or a Zeiss Axio Observer D1 epifluorescent microscope (×100, ×200, or ×400 magnification). A minimum of 10 fields of view (FOV) were captured per sample. Images were converted from binary data format with either Olympus FV-10ASW (version 04.02.02.09) or Zeiss Zen Blue (version 2.6) software. Representative isotype images are shown in Supplemental Figure 1, B–H. Image quantification was performed according to methods described in Oil Red O staining.
PET imaging.
A quantity of 0.5 mCi 124I (Nordion) was added to 100 μg mAb C3d29 (suspended in 200 μL 0.1 M PBS, pH 7.5) in a glass tube coated with 50 μg Iodogen (1,3,4,6-tetrachloro-3α,6α-diphenyl glycoluril) (Pierce) to prevent antibody oxidation. After 5 minutes, the reaction mixture was loaded onto a P6 size exclusion column (Bio-Rad) preequilibrated with 1% BSA/PBS, from which 124I-C3d29 was eluted. Purity of 124I-C3d29 was assessed by thin-layer chromatography using ITLC-SG strips (Varian) and 10% trichloroacetic acid running solvent, and immunoreactivity was verified by extrapolating the binding to recombinant C3d at an infinite excess of antigen.
Male C57BL/6 and fH–/– mice (n = 4 per group) were injected intravenously with 124I-C3d29 (120 μCi = 20 μg protein in 200 μL) and underwent whole-body PET/CT imaging using a Siemens Inveon microPET/CT scanner at 4, 24, 48, 72, and 96 hours after injection. Upon PET scan completion, each animal was moved automatically into the adjunct microCT scanner and low-resolution CT scans covering the entire mouse body in the axial, coronal, and sagittal directions were obtained. Images were reconstructed and analyzed with AsiProVM software (version 6.7.1.1). Animals were sacrificed after the 96-hour scan, and all major organs were collected. Organs were weighed and counted in a gamma counter along with a known amount of the radioactive injectate to determine the biodistribution of the antibody (28). Tissue radioactivity is reported as the percentage of injected radioactive dose per gram of tissue (percentage ID/g) corrected for background and decay.
Liver mRNA analysis.
A portion of the right medial lobe was collected, snap-frozen in liquid nitrogen, and stored at –80°C until RNA extraction. Liver mRNA was purified using the Qiagen RNeasy Mini Kit (Qiagen Sciences) according to the manufacturer’s instructions. Purified RNA was qualitatively assessed by RNA TapeStation (Agilent) and then analyzed using the nCounter Mouse Myeloid Innate Immunity V2 panel (NanoString Technologies). Data were collected for 754 genes of interest and 22 housekeeping genes, and evaluated for quality issues, outliers, or confounders using nSolver (version 4.0) software. Genes failing to register counts above background threshold were not included. A final data analysis was performed using nCounter Advanced Analysis (version 2.0.115) and R 3.3.2 software. Briefly, probe counts were background subtracted and then normalized to the best fit housekeeping genes (Supplemental Table 5) using the GeNorm algorithm (68) within the Advanced Analysis software. The resulting data set used for analysis consisted of 414 genes. DE and pathway analyses were determined by specifying WT as the baseline covariate. The pathways or gene sets with the greatest differential expression were identified by directed global significance scoring, followed by analysis with composite pathway scoring. The composite pathway scoring algorithm used in NanoString advanced analysis is calculated as the first principal component of the pathway genes’ normalized expression, and it was previously described in detail (69).
Multiplex immunofluorescence of hepatic immune populations.
Three-micrometer-thick FFPE liver sections from 3-month-old fH–/– and WT male mice were processed and stained using the Leica Bond-III Fully Automated IHC Autostainer. Leica ER2-pH 9 antigen retrieval buffer was used for CD3ε, FoxP3, CD4, and CD8 antibodies; Dako-pH 6 was used for the F4/80 antibody; and Leica ER1-pH 6 was used for the B220 (CD45R) antibody. Primary antibodies were visualized using Opal dyes. Stained slides were scanned at low magnification with the Vectra 3.0 Quantitative Pathology Imaging System. PhenoChart (version 1.0.8) was used to review the initial scans and to specify the unbiased selection of 10 – 0.75 × 0.5 mm regions of interest (ROI) per animal by choosing the auto select feature. Each selected ROI was scanned again at higher magnification and resulting multispectral images (MSIs) were used for final analysis in the inForm Cell Analysis software (version 2.4.1). Briefly, each MSI was spectrally unmixed, background subtracted, and segmented by tissue, membrane, and nuclear markers. Cells were phenotyped with the inForm cellular phenotyping algorithms. Quantification of data generated based on the aforementioned criteria was performed (approximately 7000–7500 cells per animal). For each animal, total cell count for each antigen was determined by taking the mean of the sum of all MSIs for each specific marker.
Liver myeloid cell isolation.
Liver myeloid cell isolation and flow cytometry methods were adapted from published methods (70). Male fH–/– (with and without liver tumors), WT, and fH–/–fB–/– mice (3, 4, and/or 22 months of age) were euthanized individually by CO2 asphyxiation. Circulating blood was removed by cardiac puncture. Liver lobes were excised, briefly washed in sterile PBS, and placed on ice in 5 mL cold RPMI cell culture medium. Intact lobes were cut into approximately 1.5-mm pieces and placed in 120 U/mL collagenase 3 (Worthington) diluted in 5–10 mL ice-cold HBSS. Samples were transferred to a 37°C water bath, placed on an orbital shaker, and homogenized for 45 minutes (200 rpm) at 37°C (samples were briefly vortexed twice during digestion). Digestion was stopped with ice-cold 2% (vol/vol) heat-inactivated fetal bovine serum (HI-FBS) and 5 mM EDTA in cold HBSS, briefly vortexed, filtered through a 70-μm cell strainer, and centrifuged (8 minutes at 450g at 4°C). The cell pellet was subject to erythrocyte lysis, followed by neutralization, filtration, and centrifugation. The cell pellet was resuspended in cold RPMI with 5% (vol/vol) HI-FBS and fractionated with a Lymphoprep gradient. The interphase was collected and stored in 2% (vol/vol) HI-FBS and 3 mM EDTA in HBSS at 4°C. The remaining lower fraction and pellet were washed and centrifuged twice, resuspended in RPMI with 5% (vol/vol) HI-FBS, and fractionated with a 33% Percoll (MilliporeSigma) gradient followed by centrifugation and 2 additional washes. The pellet was resuspended a final time in RPMI with 5% (vol/vol) HI-FBS and combined with fraction 1, and cell number and viability were calculated.
Flow cytometry analysis of isolated hepatic myeloid cells.
Single cell preparations were adjusted to a concentration of 3 × 106 cells in 150 μL. Cells were incubated on ice for 20 minutes with anti-mouse CD16/CD32 (1 μg/106 cells) to block Fc receptors and for another 20 minutes with the conjugated primary antibody cocktail (each diluted to 0.2 μg/106 cells). Cells were washed, centrifuged, and resuspended in 2 mL cold HBSS with 2% (vol/vol) HI-FBS and 3 mM EDTA. Unstained cells were used to determine cellular autofluorescence. Single-stain cells and positive/negative compensation beads were used to determine true signal, while Fluorescence Minus One (FMO) controls were used to minimize the effects of spectral overlap. Approximately 1 million to 2 million events per sample were collected on the Cytek Aurora Multi-Spectral Flow Cytometer. SpectroFlo (version 2.1.0) was used for event acquisition and spectral unmixing. FlowJo (version 10.6.0) was used for final gating and determination of population composition. GraphPad Prism was used for statistical analysis.
Generation of recombinant murine factor H and reconstitution of mice.
rmCFH was generated using the codon-optimized (Homo sapiens) DNA sequence for CFH (comprising residues 19-1234; Uniprot identifier: P06909) and has been previously described in detail (71). The resulting expression construct was stably transfected into adherent CHO cells and grown to confluence under humified conditions at 37°C at 5% CO2. Exhausted medium from confluent cells was collected; filtered through a 0.22 μm polyethersulfone membrane; equilibrated for purification with the addition of stock purification buffer to a final concentration of 0.02 M sodium phosphate, 0.5 M sodium chloride, and 0.02 M imidazole; and applied to a HisTrap Excel nickel sepharose column (GE Healthcare Bioscience) using a peristaltic pump. Column-bound protein was washed and eluted from the column using a 0.02 M sodium phosphate/0.5 M sodium chloride/0.2 M imidazole buffer. All purification buffers were pH-adjusted to 7.8 ± 0.1 and purification steps were done at 4°C. Eluted protein was concentrated at 4°C using 30K centrifuge filters and then buffer exchanged into Dulbecco’s PBS, pH 7.8. Purified rmCFH was sterile-filtered and conjugated with Alexa Fluor 647 using the SAIVI protein/antibody-labeling kit (Invitrogen) under sterile conditions. The final concentration of conjugated rmCFH was determined by NanoDrop and checked for contamination by Western blot analysis. Functionality was confirmed with a guinea pig erythrocyte hemolysis assay (42).
Male mice (fH–/– [7 months] and WT [9 months]; n = 5 per group) selected for rmCFH reconstitution were similar in size (35 ± 1.5 g) and healthy in appearance. Mice were injected with 400 μg Alexa Fluor 647–conjugated rmCFH by tail vein injection and observed for 20 minutes for adverse reactions. All animals were euthanized 24 hours later, and their organs were harvested for analysis. Liver sections were stained as previously discussed and imaged by confocal microscopy. Quantification of C3b and iC3b/C3d deposition in unmanipulated and rmCFH-reconstituted fH–/– mice was done in Image J (NIH) by thresholding (for tissue segmentation) and then measuring the thresholded area. Five separate fields of view at ×600 magnification were thresholded for each animal. Numerical values for percentage of area deposition obtained in Image J were analyzed with Prism by a 2-tailed t test with Welch’s correction.
Immunofluorescence staining of human HCC tissues.
Three-micrometer-thick, deidentified FFPE sections from 6 HCC patients were subject to standard deparaffinization and rehydration methods (xylene followed by ethanol gradient). Following 2 washes in deionized water, slides were incubated in antigen retrieval citrate buffer, pH 6, at 96°C for 30 minutes, allowed to cool to room temperature for 20 minutes, and washed with running deionized water. Tissues were blocked with 5% HI-FBS and 1% BSA in PBS for 1 hour at room temperature and washed once with cold PBS. Primary antibodies were diluted in 2% HI-FBS and 1% BSA in PBS, applied to blocked tissues, and left to incubate overnight at 4°C in a humidified chamber. Tissues were washed 3 times with cold PBS before incubation with secondary antibody for 1 hour at room temperature, followed by a short incubation with DAPI diluted in PBS and 2 PBS washes. Slides were dried, mounted with 1:1 PBS and glycerol, sealed, and viewed with an Olympus FV1000 FCS confocal microscope. Images were collected at ×100, ×200, and ×600 original magnification. Isotype images are shown (Supplemental Figure 1, B–D).
Human cBioPortal query.
TCGA data were accessed at cBioPortal (46, 47). CFH mutations, copy number alterations, and mRNA expression in patients with hepatocellular carcinoma were queried in TCGA Firehose Legacy study. Overall survival and disease/progression-free survival curves were analyzed in Prism using the logrank (Mantel-Cox) test.
Statistics.
No statistical methods were used to predetermine sample size and independent biological replicates were used for all quantitative analyses. GraphPad Prism was used for analysis, unless otherwise indicated. Quantitative data are represented as dot plots, where the center line indicates the mean and the error bars indicate the standard error of the mean (mean ± SEM). All data points are shown. P values of 0.05 or less were considered statistically significant. Equal variances were not assumed. Comparisons between 2 groups were analyzed by unpaired, 2-tailed t test (95% confidence) with Welch’s correction and null hypothesis testing (Welch’s corrected t and df [degrees of freedom] values are provided for P values reaching significance). Analysis of PET imaging (Figure 2, H and I) was done by ordinary 2-way ANOVA (alpha = 0.05). Comparisons among 3 groups were analyzed by 1-way ANOVA with post hoc Tukey’s t test. The GeNorm normalization (68) and composite pathway scoring (69) algorithms used in the NanoString mRNA analysis are described in detail. See Supplemental Table 6 for a complete listing of experimental group sizes (delineated according to figure) categorized by sex, age, strain, and tumor status.
Study approval.
Animals were maintained under standard housing conditions and all procedures were performed in an AAALAC-accredited facility in accordance with the NIH Guide for the Use and Care of Laboratory Animals. Housing conditions include a 12-hour/12-hour dark-light cycle, temperature and humidity control, and ad libitum access to standard rodent chow. All experiments and protocols were approved by the University of Colorado IACUC, and guidelines establishing humane endpoints were strictly followed.
Clinical remnant archival human specimens were provided as deidentified sectioned slides in compliance with the policies and approval of the Colorado Multiple Institutional Review Board (COMIRB) under IRB-approved protocol 15-1461. Studies were conducted in accordance with the Declaration of Helsinki, the Belmont Report, and US Common Rule. The University of Colorado Biorepository Core Facility provided samples from patients with confirmed HCC status (both clinical and pathology diagnoses) and specimen tumor content of 20% or more.
Author contributions
JL designed the project, developed and performed the experiments (except for PET imaging), collected and analyzed data, adapted the flow cytometry experiment and analyzed flow cytometry data, prepared the figures, wrote and revised the manuscript, and bred, maintained, and harvested the mice. BR performed liver myeloid cell isolation and myeloid cell flow cytometry as well as critical review of the manuscript. MCP generated fH–/– mice, provided technical advice, and contributed to study design. NJS and PMSJ performed PET imaging and associated analyses. ETC and RAN provided conceptual and technical guidance and critical review of the manuscript. JMT contributed to experimental design and wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01DK076690 and R01DK113586 to JMT and R01CA236222 to JMT and RN. MCP is a Wellcome Trust Senior Fellow in Clinical Sciences (212252/Z/18/Z).
We would like to thank Linda K. Johnson for thoughtful discussions and histological evaluation of H&E-stained tissues. The data shown in Figure 9, B and C, Supplemental Figure 4, and Supplemental Table 1 are based on the work of the Cancer Genome Atlas and cBioPortal for Cancer Genomics (46, 47). Imaging experiments were performed in the University of Colorado Anschutz Medical Campus Advance Light Microscopy Core, supported in part by NIH/National Center for Advancing Translational Sciences, Colorado Clinical and Translational Sciences Institute grant UL1TR001082. We would like to acknowledge the University of Colorado Biorepository Core Facility and University of Colorado Cancer Center Tissue Biobanking and Histology Shared Resource (P30CA046934) and thank the following University of Colorado facilities: the Comparative Pathology Shared Resource, the Genomics and Microarray core, the Barbara Davis Center Flow Cytometry core, and the Human Immune Monitoring Shared Resource within the University of Colorado Human Immunology and Immunotherapy Initiative.
Version 1. 05/05/2020
In-Press Preview
Version 2. 06/22/2020
Electronic publication
Version 3. 08/03/2020
Print issue publication
Footnotes
Conflict of interest: JMT receives royalties from Alexion Pharmaceuticals Inc. He is a consultant for AdMIRx Inc., a company developing complement inhibitors. He holds stocks and will receive royalty income from AdMIRx. MCP is principal investigator for a natural history study of C3 glomerulopathy funded by an unrestricted grant from Achillion Pharma. He has research funding from Alexion Pharma for preclinical studies of complement inhibitors, and consultancy agreements with Gemini, Achillion, and Appelis Pharma.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(8):4039–4054.https://doi.org/10.1172/JCI135105.
Contributor Information
Jennifer Laskowski, Email: jennifer.laskowski@cuanschutz.edu.
Brandon Renner, Email: brandon.renner@cuanschutz.edu.
Matthew C. Pickering, Email: matthew.pickering@imperial.ac.uk.
Natalie J. Serkova, Email: natalie.serkova@cuanschutz.edu.
Peter M. Smith-Jones, Email: cs137@hotmail.com.
Eric T. Clambey, Email: eric.clambey@cuanschutz.edu.
Raphael A. Nemenoff, Email: raphael.nemenoff@cuanschutz.edu.
Joshua M. Thurman, Email: joshua.thurman@ucdenver.edu.
References
- 1.Koyama Y, Brenner DA. Liver inflammation and fibrosis. J Clin Invest. 2017;127(1):55–64. doi: 10.1172/JCI88881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shalapour S, Karin M. Immunity, inflammation, and cancer: an eternal fight between good and evil. J Clin Invest. 2015;125(9):3347–3355. doi: 10.1172/JCI80007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zipfel PF, Skerka C. Complement regulators and inhibitory proteins. Nat Rev Immunol. 2009;9(10):729–740. doi: 10.1038/nri2620. [DOI] [PubMed] [Google Scholar]
- 4.Walport MJ. Complement. First of two parts. N Engl J Med. 2001;344(14):1058–1066. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- 5.Bresin E, et al. Combined complement gene mutations in atypical hemolytic uremic syndrome influence clinical phenotype. J Am Soc Nephrol. 2013;24(3):475–486. doi: 10.1681/ASN.2012090884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao X, Pickering MC, Smith RJ. C3 glomerulopathy: the genetic and clinical findings in dense deposit disease and C3 glomerulonephritis. Semin Thromb Hemost. 2014;40(4):465–471. doi: 10.1055/s-0034-1376334. [DOI] [PubMed] [Google Scholar]
- 7.Kurolap A, Eshach-Adiv O, Baris HN. CD55 deficiency and protein-losing enteropathy. N Engl J Med. 2017;377(15):1500. doi: 10.1056/NEJMc1709131. [DOI] [PubMed] [Google Scholar]
- 8.Edwards AO, Ritter R, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308(5720):421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 9.Xie J, et al. Fine mapping implicates a deletion of CFHR1 and CFHR3 in protection from IgA nephropathy in Han Chinese. J Am Soc Nephrol. 2016;27(10):3187–3194. doi: 10.1681/ASN.2015111210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwak JW, et al. Complement activation via a C3a receptor pathway alters CD4+ T lymphocytes and mediates lung cancer progression. Cancer Res. 2018;78(1):143–156. doi: 10.1158/0008-5472.CAN-17-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Markiewski MM, et al. Modulation of the antitumor immune response by complement. Nat Immunol. 2008;9(11):1225–1235. doi: 10.1038/ni.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonavita E, et al. PTX3 is an extrinsic oncosuppressor regulating complement-dependent inflammation in cancer. Cell. 2015;160(4):700–714. doi: 10.1016/j.cell.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt CQ, Hipgrave Ederveen AL, Harder MJ, Wuhrer M, Stehle T, Blaum BS. Biophysical analysis of sialic acid recognition by the complement regulator Factor H. Glycobiology. 2018;28(10):765–773. doi: 10.1093/glycob/cwy061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kajander T, et al. Dual interaction of factor H with C3d and glycosaminoglycans in host-nonhost discrimination by complement. Proc Natl Acad Sci USA. 2011;108(7):2897–2902. doi: 10.1073/pnas.1017087108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt CQ, Lambris JD, Ricklin D. Protection of host cells by complement regulators. Immunol Rev. 2016;274(1):152–171. doi: 10.1111/imr.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langford-Smith A, Keenan TD, Clark SJ, Bishop PN, Day AJ. The role of complement in age-related macular degeneration: heparan sulphate, a ZIP code for complement factor H? J Innate Immun. 2014;6(4):407–416. doi: 10.1159/000356513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noris M, et al. Relative role of genetic complement abnormalities in sporadic and familial aHUS and their impact on clinical phenotype. Clin J Am Soc Nephrol. 2010;5(10):1844–1859. doi: 10.2215/CJN.02210310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He G, Karin M. NF-κB and STAT3 - key players in liver inflammation and cancer. Cell Res. 2011;21(1):159–168. doi: 10.1038/cr.2010.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reis ES, Mastellos DC, Ricklin D, Mantovani A, Lambris JD. Complement in cancer: untangling an intricate relationship. Nat Rev Immunol. 2018;18(1):5–18. doi: 10.1038/nri.2017.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alper CA, Johnson AM, Birtch AG, Moore FD. Human C’3: evidence for the liver as the primary site of synthesis. Science. 1969;163(3864):286–288. doi: 10.1126/science.163.3864.286. [DOI] [PubMed] [Google Scholar]
- 21.Alper CA, Raum D, Awdeh ZL, Petersen BH, Taylor PD, Starzl TE. Studies of hepatic synthesis in vivo of plasma proteins, including orosomucoid, transferrin, alpha 1-antitrypsin, C8, and factor B. Clin Immunol Immunopathol. 1980;16(1):84–89. doi: 10.1016/0090-1229(80)90169-5. [DOI] [PubMed] [Google Scholar]
- 22.Yao S, et al. Diagnostic value of immunohistochemical staining of GP73, GPC3, DCP, CD34, CD31, and reticulin staining in hepatocellular carcinoma. J Histochem Cytochem. 2013;61(9):639–648. doi: 10.1369/0022155413492771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capurro M, et al. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology. 2003;125(1):89–97. doi: 10.1016/S0016-5085(03)00689-9. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki M, et al. Up-regulation of glypican-3 in human hepatocellular carcinoma. Anticancer Res. 2010;30(12):5055–5061. [PubMed] [Google Scholar]
- 25.Martinez-Hernandez A, Amenta PS. The hepatic extracellular matrix. I. Components and distribution in normal liver. Virchows Arch A Pathol Anat Histopathol. 1993;423(1):1–11. doi: 10.1007/BF01606425. [DOI] [PubMed] [Google Scholar]
- 26.Pickering MC, et al. Uncontrolled C3 activation causes membranoproliferative glomerulonephritis in mice deficient in complement factor H. Nat Genet. 2002;31(4):424–428. doi: 10.1038/ng912. [DOI] [PubMed] [Google Scholar]
- 27.Thurman JM, et al. Detection of complement activation using monoclonal antibodies against C3d. J Clin Invest. 2013;123(5):2218–2230. doi: 10.1172/JCI65861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith-Jones PM, Vallabhajosula S, Navarro V, Bastidas D, Goldsmith SJ, Bander NH. Radiolabeled monoclonal antibodies specific to the extracellular domain of prostate-specific membrane antigen: preclinical studies in nude mice bearing LNCaP human prostate tumor. J Nucl Med. 2003;44(4):610–617. [PubMed] [Google Scholar]
- 29.Kwo PY, Cohen SM, Lim JK. ACG clinical guideline: evaluation of abnormal liver chemistries. Am J Gastroenterol. 2017;112(1):18–35. doi: 10.1038/ajg.2016.517. [DOI] [PubMed] [Google Scholar]
- 30.Younossi ZM, et al. Diagnostic modalities for nonalcoholic fatty liver disease, nonalcoholic steatohepatitis, and associated fibrosis. Hepatology. 2018;68(1):349–360. doi: 10.1002/hep.29721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgeworth J, Gorman M, Bennett R, Freemont P, Hogg N. Identification of p8,14 as a highly abundant heterodimeric calcium binding protein complex of myeloid cells. J Biol Chem. 1991;266(12):7706–7713. [PubMed] [Google Scholar]
- 32.Markowitz J, Carson WE. Review of S100A9 biology and its role in cancer. Biochim Biophys Acta. 2013;1835(1):100–109. doi: 10.1016/j.bbcan.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donato R, et al. Functions of S100 proteins. Curr Mol Med. 2013;13(1):24–57. doi: 10.2174/156652413804486214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Källberg E, Stenström M, Liberg D, Ivars F, Leanderson T. CD11b+Ly6C++Ly6G- cells show distinct function in mice with chronic inflammation or tumor burden. BMC Immunol. 2012;13:69. doi: 10.1186/1471-2172-13-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang E, et al. Essential role of CD11a in CD8+ T-cell accumulation and activation in adipose tissue. Arterioscler Thromb Vasc Biol. 2014;34(1):34–43. doi: 10.1161/ATVBAHA.113.302077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jakus Z, Németh T, Verbeek JS, Mócsai A. Critical but overlapping role of FcgammaRIII and FcgammaRIV in activation of murine neutrophils by immobilized immune complexes. J Immunol. 2008;180(1):618–629. doi: 10.4049/jimmunol.180.1.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mantovani A, Cassatella MA, Costantini C, Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol. 2011;11(8):519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- 38.DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol. 2019;19(6):369–382. doi: 10.1038/s41577-019-0127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang B, Li Q, Qin L, Zhao S, Wang J, Chen X. Transition of tumor-associated macrophages from MHC class II(hi) to MHC class II(low) mediates tumor progression in mice. BMC Immunol. 2011;12:43. doi: 10.1186/1471-2172-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Movahedi K, et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 2010;70(14):5728–5739. doi: 10.1158/0008-5472.CAN-09-4672. [DOI] [PubMed] [Google Scholar]
- 41.Ju C, Tacke F. Hepatic macrophages in homeostasis and liver diseases: from pathogenesis to novel therapeutic strategies. Cell Mol Immunol. 2016;13(3):316–327. doi: 10.1038/cmi.2015.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hannan JP, Laskowski J, Thurman JM, Hageman GS, Holers VM. Mapping the complement factor H-related protein 1 (CFHR1):C3b/C3d interactions. PLoS ONE. 2016;11(11):e0166200. doi: 10.1371/journal.pone.0166200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kinugasa N, et al. Expression of membrane cofactor protein (MCP, CD46) in human liver diseases. Br J Cancer. 1999;80(11):1820–1825. doi: 10.1038/sj.bjc.6690604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu Z, et al. Bioinformatic analysis of the membrane cofactor protein CD46 and microRNA expression in hepatocellular carcinoma. Oncol Rep. 2014;31(2):557–564. doi: 10.3892/or.2013.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miwa T, Nonaka M, Okada N, Wakana S, Shiroishi T, Okada H. Molecular cloning of rat and mouse membrane cofactor protein (MCP, CD46): preferential expression in testis and close linkage between the mouse Mcp and Cr2 genes on distal chromosome 1. Immunogenetics. 1998;48(6):363–371. doi: 10.1007/s002510050447. [DOI] [PubMed] [Google Scholar]
- 46.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fakhouri F, et al. Terminal complement inhibitor eculizumab in adult patients with atypical hemolytic uremic syndrome: a single-arm, open-label trial. Am J Kidney Dis. 2016;68(1):84–93. doi: 10.1053/j.ajkd.2015.12.034. [DOI] [PubMed] [Google Scholar]
- 49.Remuzzi G, et al. Hemolytic uremic syndrome: a fatal outcome after kidney and liver transplantation performed to correct factor h gene mutation. Am J Transplant. 2005;5(5):1146–1150. doi: 10.1111/j.1600-6143.2005.00783.x. [DOI] [PubMed] [Google Scholar]
- 50.Zipfel PF, Heinen S, Józsi M, Skerka C. Complement and diseases: defective alternative pathway control results in kidney and eye diseases. Mol Immunol. 2006;43(1-2):97–106. doi: 10.1016/j.molimm.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 51.Kanmura S, et al. The complement component C3a fragment is a potential biomarker for hepatitis C virus-related hepatocellular carcinoma. J Gastroenterol. 2010;45(4):459–467. doi: 10.1007/s00535-009-0160-5. [DOI] [PubMed] [Google Scholar]
- 52.Lee IN, et al. Identification of complement C3a as a candidate biomarker in human chronic hepatitis C and HCV-related hepatocellular carcinoma using a proteomics approach. Proteomics. 2006;6(9):2865–2873. doi: 10.1002/pmic.200500488. [DOI] [PubMed] [Google Scholar]
- 53.Ferrín G, et al. Plasma protein biomarkers of hepatocellular carcinoma in HCV-infected alcoholic patients with cirrhosis. PLoS ONE. 2015;10(3):e0118527. doi: 10.1371/journal.pone.0118527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Llovet JM, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 55.Munoz LE, De Villiers D, Markham D, Whaley K, Thomas HC. Complement activation in chronic liver disease. Clin Exp Immunol. 1982;47(3):548–554. [PMC free article] [PubMed] [Google Scholar]
- 56.Vasei M, et al. Immunoglobulin and complement depositions in the liver of chronic hepatitis patients. Hepatogastroenterology. 2008;55(84):1066–1070. [PubMed] [Google Scholar]
- 57.Shen H, French BA, Liu H, Tillman BC, French SW. Increased activity of the complement system in the liver of patients with alcoholic hepatitis. Exp Mol Pathol. 2014;97(3):338–344. doi: 10.1016/j.yexmp.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Satyam A, et al. Intracellular activation of complement 3 is responsible for intestinal tissue damage during mesenteric ischemia. J Immunol. 2017;198(2):788–797. doi: 10.4049/jimmunol.1502287. [DOI] [PubMed] [Google Scholar]
- 59.Cressman DE, Diamond RH, Taub R. Rapid activation of the Stat3 transcription complex in liver regeneration. Hepatology. 1995;21(5):1443–1449. doi: 10.1002/hep.1840210531. [DOI] [PubMed] [Google Scholar]
- 60.Park EJ, et al. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140(2):197–208. doi: 10.1016/j.cell.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He G, et al. Hepatocyte IKKbeta/NF-kappaB inhibits tumor promotion and progression by preventing oxidative stress-driven STAT3 activation. Cancer Cell. 2010;17(3):286–297. doi: 10.1016/j.ccr.2009.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Medler TR, et al. Complement C5a fosters squamous carcinogenesis and limits T cell response to chemotherapy. Cancer Cell. 2018;34(4):561–578.e6. doi: 10.1016/j.ccell.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nabizadeh JA, et al. The complement C3a receptor contributes to melanoma tumorigenesis by inhibiting neutrophil and CD4+ T cell responses. J Immunol. 2016;196(11):4783–4792. doi: 10.4049/jimmunol.1600210. [DOI] [PubMed] [Google Scholar]
- 64.Wang Y, et al. Autocrine complement inhibits IL10-dependent T-cell-mediated antitumor immunity to promote tumor progression. Cancer Discov. 2016;6(9):1022–1035. doi: 10.1158/2159-8290.CD-15-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pettit SJ, Seymour K, O’Flaherty E, Kirby JA. Immune selection in neoplasia: towards a microevolutionary model of cancer development. Br J Cancer. 2000;82(12):1900–1906. doi: 10.1054/bjoc.2000.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thurman JM, Le Quintrec M. Targeting the complement cascade: novel treatments coming down the pike. Kidney Int. 2016;90(4):746–752. doi: 10.1016/j.kint.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsumoto M, et al. Abrogation of the alternative complement pathway by targeted deletion of murine factor B. Proc Natl Acad Sci USA. 1997;94(16):8720–8725. doi: 10.1073/pnas.94.16.8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tomfohr J, Lu J, Kepler TB. Pathway level analysis of gene expression using singular value decomposition. BMC Bioinformatics. 2005;6:225. doi: 10.1186/1471-2105-6-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stijlemans B, et al. Murine liver myeloid cell isolation protocol. Bio-protocol. 2015;5(10):e1471 [Google Scholar]
- 71.Laskowski J, et al. Distinct roles for the complement regulators factor H and Crry in protection of the kidney from injury. Kidney Int. 2016;90(1):109–122. doi: 10.1016/j.kint.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.