Abstract
Antibiotic resistance is a global public health issue and it is even more daunting in developing countries. The main objective of present study was to investigate molecular responses of antibiotic-resistant bacteria. The 48 bacterial strains, which were previously isolated and identified were subjected to disc diffusion and MIC (minimum inhibitory concentration) determination, followed by investigating the production of the three beta-lactamases (ESBLs (Extended-spectrum Beta-lactamases), MBLs (Metallo Beta-lactamases), AmpCs) and exploring prevalence of the two antibiotic-resistant genes (ARGs); blaTEM and qnrS. Higher MIC values were observed for penicillin(s) than that for fluoroquinolones (ampicillin > amoxicillin > ofloxacin > ciprofloxacin > levofloxacin). Resistance rates were high (58–89%) for all of the tested beta-lactams. Among the tested strains, 5 were ESBL producers (4 Aeromonas spp. and 1 Escherichia sp.), 2 were MBL producers (1 Stenotrophomonas sp. and 1 Citrobacter sp.) and 3 were AmpC producers (2 Pseudomonas spp. and 1 Morganella sp.). The ARGs qnrS2 and blaTEM were detected in Aeromonas spp. and Escherichia sp. The results highlighted the role of Aeromonas as a vector. The study reports bacteria of multidrug resistance nature in the wastewater environment of Pakistan, which harbor ARGs of clinical relevance and could present a public health concern.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-02366-3) contains supplementary material, which is available to authorized users.
Keywords: Antibiotic resistance, Antibiotic resistance genes, Beta-lactamases, Resistant bacteria, Wastewater
Introduction
Overuse and misuse of antibiotics by humans, veterinary field, aquaculture and agriculture are causing development and spread of antibiotic resistance in environment throughout the world (French 2005; Saima et al. 2020). Pakistan stands third among the lower middle-income countries for antibiotic consumption. Over a period of 15 years (2000–2015), there has been 68% rise in antibiotic consumption in Pakistan (Klein et al. 2015). Apart from chemical pollution (Mukhtar et al. 2020), the presence of antibiotics in the environment also causes development of ARGs (Arshad and Zafar 2020), which shade health impacts and can transfer antibiotic resistance to humans and animals through direct or indirect contact (Osińska et al. 2016). The ARGs and associated genes, which function in acquisition and dissemination of antibiotic resistance have been reported in clinical and environmental bacteria (Pormohammad et al. 2019). There is growing evidence that clinical antibiotic resistance is intimately associated with environmental bacteria and ARGs. In fact, it is believed that ARGs now present in the pathogenic bacteria have evolved from microbes, which naturally produce antibiotics or their natural competitors (Aminov et al. 2007; Arshad and Zafar 2020). They may evolve due to recombinatorial events under the influence of anthropogenic impact in the aquatic environment (Baquero et al. 2008), which may result in development of novel resistance mechanisms, thus making it hard to deal with while antibiotics continue to lose their effectiveness.
Even though antibiotic resistance is a global health issue, it has been majorly overlooked in the aquatic environment (Khan et al. 2013), especially in developing countries. A study reported that typhoid-causing Salmonella are gaining resistance against second line antibiotics, which mostly include beta-lactams and fluoroquinolones. This has resulted in development of multidrug-resistant and extensively drug-resistant Salmonella typhi in Pakistan (Azhar et al. 2019). Only a few studies have been conducted, which report about the prevalence of ARGs in aquatic environment of Pakistan. Shah et al. (2012) reported that the ESBL-encoding gene blaTEM was the most prevalent ARG in bacterial DNA directly isolated from fish, fish pond water and sediments of Pakistan and Tanzania. Similarly, DNA isolated from sediments of the aquatic environment of six main rivers of Pakistan had high levels of antibiotics and ARGs (sulI, dfrAI) and other associated genes (intl1), which indicates that the rivers may act as reservoirs of ARGs and disseminate them (Khan et al. 2013). Despite the obvious importance of water and wastewater in emergence and spread of antibiotic resistance and its determinants (Djenadi et al. 2018; Khan et al. 2013; Haller et al. 2018; Oliveira et al. 2017), it remains poorly investigated in Pakistan.
The objectives of this study were to (1) investigate antibiotic-resistant bacterial (ARBs) strains previously isolated from various wastewater streams for MICs determinations against five broad-spectrum antibiotics, beta-lactamase (A-C) characterization, and (2) PCR detection of the ARGs blaTEM and qnrS to observe their possible co-existence. The plasmids that carry ESBL genes (blaTEM, blaCTX-M) often lodge resistance determinants for fluoroquinolones and other antibiotics (Carattoli 2011) and, thus, present a risk of transfer of resistance to more than one antibiotic at the same time. This work has contributed significant information on rarely studied antibiotic-resistant bacteria isolated from wastewater streams for MIC determination, beta-lactamase characterization and ARGs detection in Pakistan.
Materials and methods
Bacterial strains
The bacterial strains (48) used in the study were previously isolated from wastewater streams of Islamabad (33.67 N, 73.06 E) (33.70 N, 73.04 E) in Faisalabad (31.38 N, 73.06 E) and Rawalpindi (33.63 N, 73.04 E). All of the wastewater streams were near hospitals and small health clinics. The samples were diluted by tenfold and dominant colonies were selected on nutrient agar (data not shown here) and stored at – 70 °C in 70% glycerol stocks. The strains were identified by sequencing of 16 s rRNA gene. All of the identified strains were Gram-negative except Bacillus sp., which was Gram-positive. The bacterial population consisted of Pseudomonas spp., Aeromonas spp., Acinetobacter spp., Escherichia spp., Proteus sp., Morganella sp., Bacillus sp., and Citrobacter sp., Shigella sp., Comamonas spp. and Stenotrophomonas sp. All of the following experiments were repeated three times. The details of the strains are summarized in Table 1.
Table 1.
Bacterial genera with their strain IDs along with their respective locations form where they were isolated and antibiogram of the isolated strains
| Strain ID | Location | Genus | Antibiogram | |||||
|---|---|---|---|---|---|---|---|---|
| CTX | CAZ | CPD | IPM | FOX | ||||
| NCCP-1704 | 33 38 09.03 N | 73 02 03.06 E | Aeromonas spp. | R | I | R | S | R |
| NCCP-1714 | – | I | I | R | I | R | ||
| NCCP-1724 | – | I | R | R | I | R | ||
| NCCP-1757 | 31 22 48.39 N | 73 04 20.19 E | R | R | R | R | R | |
| NCCP-1751 | – | R | R | R | R | R | ||
| NCCP-1748 | – | R | R | R | I | R | ||
| NCCP-1749 | – | R | R | R | I | R | ||
| NCCP-1769 | – | I | R | R | I | R | ||
| NCCP-1728 | 33 38 09.03 N | 73 02 03.06 E | Escherichia spp. | R | R | R | R | R |
| NCCP-1731 | – | R | R | R | R | R | ||
| NCCP-1753 | 31 22 48.39 N | 73 04 20.19 E | R | R | R | R | R | |
| NCCP-1754 | – | R | R | R | R | R | ||
| NCCP-1802 | 33 42 10.82 N | 73 02 45.34 E | R | I | R | R | R | |
| NCCP-1717 | 33 38 09.03 N | 73 02 03.06 E | Acinetobacter spp. | R | R | N.D | R | N.D |
| NCCP-1718 | – | R | R | N.D | R | N.D | ||
| NCCP-1720 | – | R | R | N.D | I | N.D | ||
| NCCP-1721 | – | R | R | N.D | I | N.D | ||
| NCCP-1723 | – | R | R | N.D | R | N.D | ||
| NCCP-1813 | 33 42 10.82 N | 73 02 45.34 E | R | R | N.D | S | N.D | |
| NCCP-1758 | 31 22 48.39 N | 73 04 20.19 E | R | R | N.D | R | N.D | |
| NCCP-1760 | – | R | R | N.D | S | N.D | ||
| NCCP-1761 | – | R | R | N.D | I | N.D | ||
| NCCP-1762 | – | R | R | N.D | I | N.D | ||
| NCCP-1763 | – | R | R | N.D | I | N.D | ||
| NCCP-1744 | – | Citrobacter spp. | R | R | R | R | R | |
| NCCP-1750 | – | R | R | R | R | R | ||
| NCCP-1742 | 33 38 09.03 N | 73 02 03.06 E | Comamonas spp. | R | R | N.D | R | N.D |
| NCCP-1786 | 33 40 30.75 N | 73 04 05.31 E | R | R | N.D | S | N.D | |
| NCCP-1771 | – | Proteus sp. | R | R | R | R | R | |
| NCCP-1830 | – | Shigella sp. | R | R | R | R | R | |
| NCCP-1756 | 31 22 48.39 N | 73 04 20.19 E | Bacillus sp. | S | S | S | S | S |
| NCCP-1829 | 33 40 30.75 N | 73 04 05.31 E | Stenotrophomonas sp. | R | R | R | R | R |
| NCCP-1791 | – | Morganella sp. | R | R | R | R | R | |
| NCCP-1709 | 33 38 09.03 N | 73 02 03.06 E | Pseudomonas spp. | R | R | N.D | R | R |
| NCCP-1712 | – | R | R | N.D | R | R | ||
| NCCP-1713 | – | R | R | N.D | R | R | ||
| NCCP-1773 | 33 40 30.75 N | 73 04 05.31 E | R | R | N.D | R | R | |
| NCCP-1780 | – | R | R | N.D | R | R | ||
| NCCP-1781 | – | R | R | N.D | R | R | ||
| NCCP-1782 | – | R | R | N.D | R | R | ||
| NCCP-1783 | – | R | R | N.D | I | R | ||
| NCCP-1784 | – | R | R | N.D | I | R | ||
| NCCP-1787 | – | R | R | N.D | I | R | ||
| NCCP-1788 | – | R | R | N.D | R | R | ||
| NCCP-1801 | 33 42 10.82 N | 73 02 45.34 E | R | R | N.D | R | R | |
| NCCP-1803 | – | R | R | N.D | I | I | ||
| NCCP-1805 | – | R | R | N.D | R | R | ||
| NCCP-1808 | – | I | I | N.D | S | I | ||
| Total strains (48) | Resistant strains | 42 | 43 | 19 | 28 | 32 | ||
| Resistant strains (percentage) | 87.5 | 89.5 | * | 58.3 | * | |||
The dash (–) here indicates that the location is the same as above, while the boxes with N.D. mean that the susceptibility test wasn’t conducted for these strains due to lack of inhibition zone interpretation criteria, that is why the total resistant strains in percentage for those antibiotics were not calculated, as indicated by the asterisk (*)
CPD cefpodoxime, CTX cefotaxime, CAZ ceftazidime, IPM imipenem, FOX cefoxitin, R resistant, I intermediate, S susceptible, ND not determined
MICs (minimum inhibitory concentrations)
The antibiotics chosen for testing in this study are broad-spectrum and are commonly used drugs not only in Pakistan but also the whole world (Wilke et al. 2005; CDDEP 2017) as they are considered a cure all by consumers, which is one of the reasons for their increased consumption. MICs of AMP (ampicillin), AMX (amoxicillin), CIP (ciprofloxacin), OFX (ofloxacin) and LEV (levofloxacin) (Sigma Aldrich) of the strains were determined with agar-dilution method. A range of concentrations (256–0.125 μg mL−1) recommended by CLSI (Clinical and Laboratory Standards Institute) (2012) were tested.
Disc diffusion test
Strains were further screened by disc diffusion test for resistance against CTX (cefotaxime, 30 μg), CAZ (ceftazidime, 30 μg), CPD (cefpodoxime, 10 μg), IPM (imipenem, 10 μg) and FOX (cefoxitin, 30 μg) (CLSI 2018). The strains were incubated overnight in tryptic soy agar (Oxoid) and adjusted to a turbidity of 0.5 McFarland standard in 0.85% saline (NaCl) and streaked on Mueller Hinton Agar (MHA). The MHA plates were then incubated for 18–20 h at 35 °C. The quality control strain Escherichia coli (ATCC 25922) was used. All the discs were purchased from Bioanalyse. The inhibition zones were interpreted according to the Table 2.
Table 2.
The inhibition zone interpretation criteria used for each genus
| Genus | CTX | CAZ | CPD | IMP | FOX |
|---|---|---|---|---|---|
|
Aeromonas spp. Escherichia spp. Citrobacter spp. Proteus sp. Shigella sp. Morganella sp. Stenotrophomonas sp. |
Enterobacteriaceae (CLSI 2018) | Enterobacteriaceae (CLSI 2018) | Enterobacteriaceae (CLSI 2018) | Enterobacteriaceae (CLSI 2018) | Enterobacteriaceae (CLSI 2018) |
| Acinetobacter spp. | CLSI (2018) | CLSI (2018) | N.D | CLSI (2018) | N.D |
| Pseudomonas spp. | Andrews and Howe (2011) | Andrews and Howe (2011) | N.D | Andrews and Howe (2011) | Upadhyay et al. (2010) |
| Comamonas spp. | Pseudomonas spp. | Pseudomonas spp. | N.D | Pseudomonas spp. | N.D |
| Bacillus sp. | Predicted from FOX result | Predicted from FOX result | Predicted from FOX result | Predicted from FOX result | Staphylococcus spp. (CLSI, 2018) |
Although Stenotrophomonas and Aeromonas spp. are not Enterobacteriaceae, the same inhibition zone interpretation criterion has been used for it because, it has been used previously in published literature (Harmon et al. 2018; Rameshkumar et al. 2016). The empty boxes indicate that susceptibility tests have not been conducted for those genera. As CLSI doesn’t provide inhibition zone breakpoints for Pseudomonas spp., they have been interpreted according to published literature (Andrews and Howe 2011; Upadhyay et al. 2010)
Beta-lactamases
ESBL detection was done only in Aeromonas spp. and Enterobacteriaceae with double-disc synergy (DDST) and combination disc test (CDT), which had been previously done by Rameshkumar et al. (2016). In CDT, ESBL production was confirmed using CAZ and CTX alone and in combination with clavulanic acid (10 + 30 μg) in combination disc assay (CLSI 2018). Inhibition zone difference of ≥ 5 mm between CTX and CAZ alone and in combination with clavulanic acid was considered to be positive for ESBL production. In DDST, CAZ and CTX were placed on both sides of amoxicillin + clavulanate (AMC, 20 + 10 μg) disc (Jarlier et al. 1988). Appearance of synergistic zone between CAZ or CTX and AMC was considered to be positive for ESBL production.
The method described by Yong et al. (2002) was used for detection of MBLs only in Stenotrophomonas sp. and Citrobacter sp. which had also been used by Daef et al. (2017) and Rizvi et al. (2009). In this confirmation test, imipenem was used alone and in combination with EDTA (10 + 750 μg). Zone difference of ≥ 6 mm was considered to be positive for MBL production.
AmpC production confirmation was done for Pseudomonas spp. and Morganella sp. only with the disc-approximation method (Mathur et al. 2014) as had also been previously done by Oliveira et al. (2017). In confirmation test, IPM, CAZ, AMC and FOX were used. The Zone flattening of CAZ towards any of the substrate disc was considered to be positive for AmpC production.
PCR for detection of blaTEM and qnrS
The ARG blaTEM, is a TEM enzyme-encoding gene (hydrolyzed beta-lactams). They are of high significance with regard to prevalence, distribution, and diversity. Wherein, qnrS is qnr protein-encoding gene, which is commonly associated with ESBLs (TEM, CTX-M). Rapid preparation of template DNA was done by heating (95 °C for 10 min) a single pure colony in PCR-grade water followed by a centrifugation step (Dallenne et al. 2010). The supernatant of 2 μL was subjected to PCR in a 50 μL mixture containing 25 μL PCR mix (Thermo Fisher), 2 μL (0.4 pmol μL−1) of both forward and reverse primers (Table 3). The cycling conditions were: Initial denaturation at 94 °C for 7 min; 35 cycles of denaturation, annealing and amplification at 94 °C for 40 s, 64 °C (qnrS), 56 °C (blaTEM) for 40 s and 72 °C for 1 min, respectively, and final extension for 7 min at 72 °C. Finally, the PCR product was run on agarose gel (1%) stained with ethidium bromide at 100 V for 1 h and visualized with UV transilluminator, 100 bps ladder (Thermo Fisher) was used as size marker. After gel confirmation, the PCR product was sequenced by Macrogen (Korea). The obtained sequences were run on BLAST algorithm for checking homology. The sequences have been submitted into Genbank. ExPASy was used for translation of the nucleotides to amino acid sequences.
Table 3.
Primers used in this study with their base length and product size
Results
MICs (minimum inhibitory concentrations)
The detailed results related to MICs are presented in Table 4. MICs values were of the following order: ampicillin > amoxicillin > ofloxacin > ciprofloxacin > levofloxacin (Table 4). About 31% of the strains had the highest tested MIC of ampicillin, which in this case was 256 μg mL−1. Among them, 47% were Aeromonas spp. followed by 13% Escherichia spp., 13% Citrobacter spp., and 13% Pseudomonas spp. Similar results were observed for amoxicillin with 28.8% of the strains having the highest tested MIC, among which 37% of the strains were Aeromonas spp., 19% Pseudomonas spp., followed by 12% Escherichia spp. and 12% Citrobacter spp.
Table 4.
Minimal inhibitory concentrations (MICs, μg mL−1) of the bacterial strains for five antibiotics
| Genus (total isolates) | No. of strains (MIC μg mL−1) | ||||
|---|---|---|---|---|---|
| AMP | AMX | OFX | CIP | LEV | |
| Acinetobacter spp. (11) | 1 (0.125) | 1 (0.25) | 2 (0.125) | 1 (0.125) | 7 (0.125) |
| 1 (0.5) | 2 (2) | 1 (0.25) | 4 (0.25) | 2 (0.25) | |
| 2 (2) | 2 (4) | 7 (2) | 3 (1) | 1 (0.5) | |
| 1 (8) | 2 (8) | 1 (8) | 2 (2) | 1 (8) | |
| 4 (32) | 1 (16) | 1 (8) | |||
| 1 (64) | 1 (32) | ||||
| 1 (256) | 1 (64) | ||||
| 2 (128) | |||||
| Aeromonas spp. (8) | 1 (64) | 1 (8) | 1 (0.125) | 1 (0.25) | 5 (0.125) |
| 7 (256) | 1 (64) | 1 (1) | 1 (0.5) | 1 (0.25) | |
| 6 (256) | 1 (4) | 3 (2) | 2 (4) | ||
| 1 (8) | 1 (64) | ||||
| 2 (16) | 2 (128) | ||||
| 1 (128) | |||||
| 1 (256) | |||||
| Citrobacter spp. (2) | 2 (256) | 2 (256) | 1 (256) | 2 (256) | 2 (4) |
| 1 (16) | |||||
| Comamonas spp. (2) | 2 (32) | 1 (32) | 1 (0.25) | 1 (2) | 2 (0.125) |
| 1 (8) | 1 (4) | 1 (128) | |||
| Escherichia spp. (5) | 2 (32) | 1 (0.5) | 2 (0.25) | 2 (0.25) | 2 (0.125) |
| 1 (64) | 1 (32) | 3 (256) | 1 (2) | 2 (4) | |
| 2 (256) | 1 (64) | 1 (128) | 1 (8) | ||
| 2 (256) | 1 (256) | ||||
| Morganella sp. (1) | 16 | 4 | 0.125 | 16 | 0.125 |
| Proteus sp. (1) | 32 | 4 | 1 | 0.25 | 0.125 |
| Pseudomonas spp. (15) | 3 (0.25) | 2 (0.25) | 8 (0.125) | 3 (0.125) | 13 (0.125) |
| 1 (0.5) | 2 (0.5) | 2 (0.25) | 1 (0.25) | 1 (0.25) | |
| 8 (32) | 4 (4) | 1 (0.5) | 3 (0.5) | 1 (4) | |
| 1 (64) | 1 (8) | 1 (4) | 5 (1) | ||
| 2 (256) | 1 (16) | 1 (16) | 2 (2) | ||
| 2 (32) | 1 (64) | 1 (32) | |||
| 3 (256) | 1 (256) | ||||
| Shigella sp. (1) | 32 | 2 | 0.25 | 1 | 2 |
| Stenotrophomonas sp. (1) | 256 | 256 | 0.5 | 64 | 0.125 |
| Bacillus sp. (1) | 64 | 32 | 8 | 16 | 2 |
Outside of bracket, the digits represent the no. of strains, while inside the bracket is minimal inhibitory concentration (MIC) of that strain calculated in μg mLˉ1
AMP ampicillin, AMX amoxicillin, OFX ofloxacin, CIP ciprofloxacin, LEV levofloxacin
Among the fluoroquinolones, 29% of the strains had high MICs (256 and 128 μg mL−1) of ciprofloxacin and ofloxacin together. Among them, 28% were Aeromonas spp., 28% were Escherichia spp., followed by 21% Citrobacter spp. and 7% Pseudomonas spp. MICs for levofloxacin were generally low with the only one Escherichia sp. and one Acinetobacter sp. showing the highest MIC, which was 8 μg mL−1.
Disc diffusion
The strains were highly resistant to third-generation cephalosporins. The bacterial resistance towards CAZ, CTX and IPM was 89% > 87% > 58%, respectively. Percentage resistance to CPD and FOX was not calculated because their susceptibility tests were not conducted for Pseudomonas spp., Acinetobacter spp. and Comamonas spp. The genus Aeromonas exhibited high-resistance rates to all of the tested antibiotics. Whereas, the most effective antibiotic among the tested ones was observed to be imipenem. The detailed data can be observed in Table 1.
Beta-lactamases
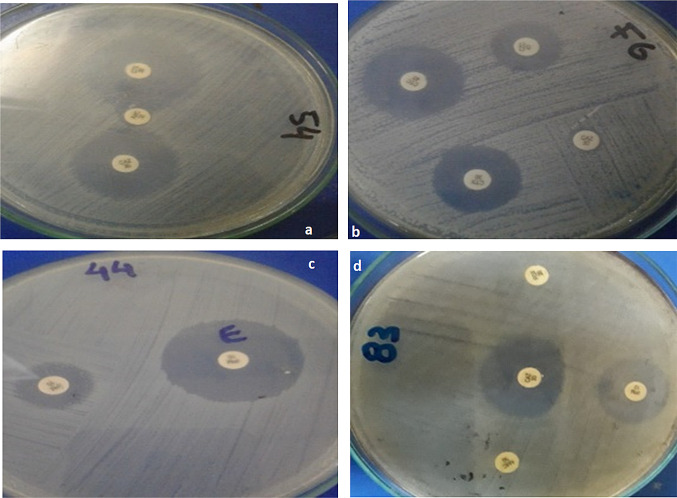
In total, there were 21% (10 out of 48) beta-lactamase producers, among which 5 strains were ESBL producers, 2 strains were MBL producers and only 3 strains were AmpC producers. The AmpC production by the two strains could be inducible. All of the ESBL producers were from the genus Aeromonas except for one strain, which was Escherichia sp. The MBL producers consisted of one Stenotrophomnas sp. and one Citrobacter sp. Two of the AmpC producers were from the genus Pseudomonas, while only one was from the genus Morganella. The pictorial evidence is shown in Fig. 1 and further details are given in Table 5.
Fig. 1.
Beta-lactamase production by resistant bacteria; a Escherichia sp. (ESBL production confirmed by double disc synergy test) b Aeromonas sp. (ESBL production confirmed by combination disc test) c Citrobacter sp. (MBL production confirmed by combination disc test using EDTA) d Pseudomonas sp. (AmpC production confirmed by disc approximation test)
Table 5.
Antibiotic resistance profile of the ARG carrier and beta-lactamase producer bacterial strains
| Strain ID | Bacterial strain | Antibiotics resistance Profile | Beta-lactamases | ARGs | ||
|---|---|---|---|---|---|---|
| ESBL | MBL | AmpC | ||||
| NCCP-1754 | Escherichia sp. | CTX + CAZ + CPD + FOX + IPM | + | blaTEM | ||
| NCCP-1751 | Aeromonas sp. | CTX + CAZ + CPD + IPM + FOX | + | |||
| NCCP-1749 | Aeromonas sp. | CTX + CAZ + CPD + FOX | + | qnrS2 | ||
| NCCP-1757 | Aeromonas sp. | CTX + CAZ + CPD + IPM + FOX | + | |||
| NCCP-1769 | Aeromonas sp. | CAZ + CPD + FOX | + | blaTEM | ||
| NCCP-1744 | Citrobacter sp. | CTX + CAZ + CPD + IPM + FOX | + | |||
| NCCP-1829 | Stenetrophomonas sp. | CTX + CAZ + CPD + IPM + FOX | + | |||
| NCCP-1782 | Pseudomonas sp. | CTX + CAZ + IPM + FOX | + | |||
| NCCP-1787 | Pseudomonas sp. | CTX + CAZ + FOX | + | |||
| NCCP-1791 | Morganella sp. | CTX + CPD + IPM + FOX | + | |||
The symbol (+) indicates that the strain is a producer of that beta-lactamase
CPD cefpodoxime, CTX cefotaxime, CAZ ceftazidime, IPM imipenem, FOX cefoxitin
ARGs
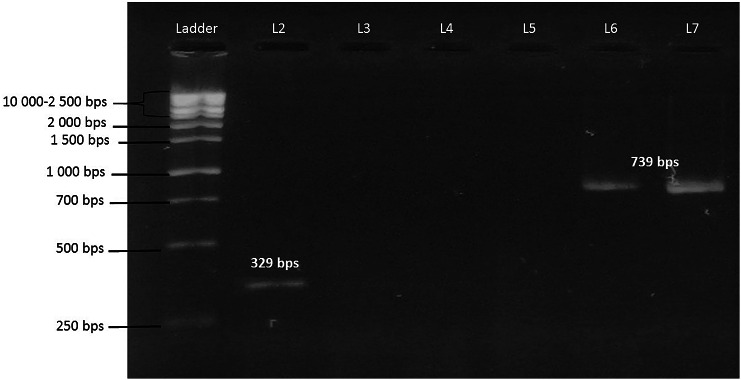
The genes qnrS2 and blaTEM were up to 100% match to sequences detected in Escherichia coli and Klebsiella pneumoniae (Accession no. CP053728, CP054237, MH684532) isolated from clinical samples. The sequences of qnrS2 and blaTEM were deposited in Genbank under the accession numbers MK359204, MN218676 and MN218677. The gel image of bands is shown in Fig. 2. Blast search and alignment of all the three sequences are presented in supplementary material (Figures S1, S2 and S3).
Fig. 2.
PCR amplification; gel image of blaTEM and qnrS2 in comparison with the molecular ladder. Ladder; 100 bps molecular ladder, L2; qnrS2 identified in Aeromonas sp., L6 and L7; blaTEM identified in Escherichia sp. and Aeromonas sp.
Discussion
Antibiotic susceptibility/resistance of bacteria isolated from wastewater streams in Pakistan is rarely studied. There are various reports of typhoid-causing extensively beta-lactam-resistant Salmonella (Azhar et al. 2019). Third-generation cephalosporins and fluoroquinolones are drugs now more commonly prescribed in Pakistan (CDDEP 2017) and carbapenems (imipenem) are our last resort antibiotics for some bacterial infections (Djenadi et al. 2018). High resistance to these antibiotics among environmental bacteria is concerning since there are not many other treatment options and their resistance determinants may be plasmid-borne and transferrable. We report that the environmental bacteria isolated, showed high resistance to the third-generation cephalosporins, cephamycin and carbapenem. Environmental bacteria resistant to third-generation cephalosporins, imipenem and cefoxitin have been reported from Brazil (Oliveira et al. 2017), Algeria (Djenadi et al. 2018), Singapore (Haller et al. 2018) and Thailand (Tansawai et al. 2018). Resistance to third-generation cephalosporins was conferred to production of ESBLs in 5 (Aeromonas spp. and Escherichia sp.) of the resistant strains. Despite high-resistance rates of Pseudomonas spp. and Acinetobacter spp. to the third-generation cephalosporins, no ESBL producers were detected among the genera. ESBL production has been reported in Aeromonas spp. and Escherichia sp. from environmental settings (Tacão et al. 2014). It is well established that beta-lactamases make up one of the major defence mechanisms against beta-lactams (Weldhagen 2004).
High resistance rates (58%) were observed for imipenem. Comparable rates of resistance were also reported by Haller et al. (2018) for bacterial isolates from hospital effluents. Two of the strains (Stenotrophomnas sp. and Citrobacter sp.) were confirmed for MBL production. MBL production has been previously reported in Stenotrophomonas sp. isolated from faeces of a migratory bird (Kenzaka and Tani 2018) and Citrobacter sp. isolated from clinical samples (Rizvi et al. 2009).
Among the resistant strains, only 3 were AmpC producers, which were from genus Pseudomonas and Morganella. The AmpC producing clinical and environmental strains of Pseudomonas spp. were reported from Brazil (Oliveira et al. 2017). Pseudomonas spp. especially Pseudomonas aeruginosa are adept at acquiring antibiotic resistance to various antibiotics by having selective porins, over expressed efflux pump and inducible chromosomal beta-lactamase gene (Hauser 2018). Similarly, the AmpC production has been reported in Morganella morganii isolated from food samples in China (Ye et al. 2017). Besides production of beta-lactamases, resistance to beta-lactams is also largely attributed to other resistance mechanisms like loss of porins (impermeability) and over active efflux systems (Marangon et al. 2004).
High MICs and resistance rates of ampicillin and amoxicillin in Gram-negative environmental bacterial isolates have been reported (Li et al. 2009; Pontes et al. 2009; Sidrach-Cardona et al. 2014; Goni-urriza 2000). In another study, Acinetobacter spp. isolated from pharmaceutical plant and hospital wastewater with comparable MICs were also reported (Paiva et al. 2017). Beta-lactams are antibiotics of frequent use; hence, resistance to these is more commonly observed in environmental bacteria (Pontes et al. 2009).
High MICs of ciprofloxacin were reported (Osińska et al. 2016) for some environmental bacteria isolated from discharge of WWTP and river water. The same was observed in our study, where ciprofloxacin MICs as high as 256 μg mL−1 were noted for Aeromonas. Other studies report results different than ours where no environmental Aeromonas strain with MIC higher than 32 μg mL−1 was observed (Cattoir et al. 2008; Goni-Urriza 2000). Levofloxacin was noted to be the most effective antibiotic in this study. It inhibited most of the strains at low concentrations (0.0125–8 μg mL−1) compared to the other antibiotics. Similar results for environmental Enterobacteriaceae have been reported (Paiva et al. 2017). Most of the strains had low MICs of levofloxacin, even though some of the same isolates had the highest tested MICs (256 μg mL−1) of other fluoroquinolones (ciprofloxacin and ofloxacin). Environmental Aeromonas spp. and Pseudomonas spp. with low-resistance rates to levofloxacin have been reported (Blasco et al. 2008). Levofloxacin is a relatively newer drug from its class and has not been used extensively in prophylaxis, which might explain the low rates of resistance to this antibiotic (Marangon et al. 2004).
The present study showed that not many strains were beta-lactamase producers (only 10), which makes us think that the high-resistance rates to such multitude of antibiotics (mostly beta-lactams) could be caused by the presence of other multiple resistance mechanisms as was also reported by Li et al. (2009).
Keeping in view the results of the preliminary tests (MICs, disc diffusion) and importance of the tested antibiotics, it was imperative to investigate the ARGs blaTEM and qnrS, the latter is considered to be of environmental origin although it confers only low level of resistance to fluoroquinolones. The blast search for qnrS sequences showed them to be highly identical to extracted sequences from E. coli and E. roggenkampii (Acession no. CP053728, LC545449) found in clinical samples and hospital sewage in China and Japan, respectively. We were able to detect qnrS2 in only one Aeromonas sp., which allows us to think that the genetic machinery responsible for fluoroquinolone resistance in the environmental isolates in this study is much more diverse. Other resistance mechanisms could include, overactive efflux system, alterations in genes encoding enzymes, such as DNA gyrase and topoisomerase IV, and further plasmid-mediated resistance mechanisms (Paiva et al. 2017; Tacão et al. 2014). The PMQR gene, qnrS2 was detected in ESBL phenotype Aeromonas sp. There are several studies reporting qnrS2 in Aeromonas spp. from environmental settings (Cattoir et al. 2008). It is not uncommon for ESBL producer to carry resistance genes for other antibiotics like fluoroquinolone and plasmid carrying ESBL-encoding genes often ensnare ARGs encoding resistance to other non-beta-lactam drugs (Cattoir et al. 2008).
The ARG blaTEM highly matched extracted sequences from Escherichia coli, Klebsiella pneumoniae and Salmonella enterica (Accession no. CP054237, MH684532, CP053703), which were isolated from clinical samples in Texas, India. In our study, only two strains from genera Aeromonas sp. and Escherichia sp. carried the ARG blaTEM, which was unexpected since high MICs (32–256 μg mL−1) of ampicillin and amoxicillin and high-resistance rates to the third-generation cephalosporin were observed for many strains. It is possible that the ESBL producers in our study may have harbored other ESBL-encoding genes (such as, CTX-M, SHV) but as they were not investigated, we were not able to deduce anything. A similar case was presented in another study, where highly beta-lactam-resistant bacterial strains were isolated from penicillin production wastewater but they did not carry any beta-lactamase-encoding genes (Li et al. 2009). Another study reported that the prevalence of blaTEM(1) is greater in natural waters where anthropogenic influence is high and attributed most of penicillin resistance to the aforementioned gene (Pontes et al. 2009). From the results of PCR and phenotypic tests for beta-lactamases, it can be assumed that beta-lactamase production may not be primarily causing beta-lactam resistance in the bacterial species in this study, other resistance mechanisms, such as multidrug efflux systems, should be investigated. Although both of the ARGs were harbored by Aeromonas, none of the blaTEM carrier strain carried the ARG qnrS2, hence their co-existence could not be proved. The role of Aeromonas as a potential reservoir and vehicle for dissemination of ARGs has been discussed and this study further confirms the environmental concerns associated with it.
Conclusion
Out of the total 48 strains tested, 29% and 31% strains exhibited the highest MIC (256 μg mL−1) for fluoroquinolones and ampicillin, respectively. There were 21% strains beta-lactamase producers. Two ARGs qnrS2 and blaTEM were detected in Aeromonas spp. and Escherichia sp. Although the targeted ARGs were not found to be as prevalent in the antibiotic-resistant bacteria as was expected but the role of Aeromonas as a vector of ARGs of clinical relevance is strengthened by the study findings. Attention should be paid, and further investigations are needed to do in aquatic environment of Pakistan to better understand the intensity of the situation. Also, the use of nonculture techniques is necessary to understand the dynamics of transfer of ARGs in the local aquatic environment.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The work was part of project funded by HEC, Pakistan under project “Ensuring food security: development of an integrated approach for safe usage of antibiotics containing wastewater for irrigation of crop plants” (NRPU project # 20-4155/R&D/HEC/14).
Author contributions
SS did the experimental work, MF collected the strains from wastewater, MM and RZ assisted in experimental work and preparation of the manuscript. IA, UN and MA reviewed the manuscript and improved the discussion. MA (PI) obtained research funding, supported by IA (Co-PI).
Compliance with ethical standards
Conflict of interest
Authors have no conflict of interest regarding findings and publication of this research. There are no financial competing interests relevant to the work presented here.
Accession numbers
The sequences of the ARGs (qnrS2 and blaTEM) were deposited in Genbank under the accession numbers MK359204, MN218676 and MN218677.
Contributor Information
Saima Saima, Email: saima123amin@gmail.com.
Marium Fiaz, Email: mfiaz.phdiese@student.nust.edu.pk.
Maria Manzoor, Email: marea.manzoor@gmail.com.
Rabeea Zafar, Email: rzafar.phd2017iese@student.nust.edu.pk.
Iftikhar Ahmed, Email: iftikharnarc@hotmail.com.
Uzma Nawaz, Email: uzm_69@yahoo.com.
Muhammad Arshad, Email: marshad@iese.nust.edu.pk.
References
- Aminov RI, Mackie RI. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol Lett. 2007;271(2):147–161. doi: 10.1111/j.1574-6968.2007.00757.x. [DOI] [PubMed] [Google Scholar]
- Andrews JM, Howe RA. BSAC standardized disc susceptibility testing method (version 10) J Antimicrob Chemother. 2011;66(12):2726–2757. doi: 10.1093/jac/dkr359. [DOI] [PubMed] [Google Scholar]
- Arshad M, Zafar R (2020) Antibiotics, AMRs, and ARGs: fate in the environment. In: Hashmi MZ (eds) Antibiotics and antimicrobial resistance genes in the environment. Elsevier, Inc. Netherlands. https://doi.org/10.1016/B978-0-12-818882-8.00009-7, pp 138–154
- Azhar AB, Khalid A, Shah S. The implications of extensive drug resistant typhoid fever: a case report. Cereus. 2019;11(6):5032. doi: 10.7759/cureus.5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero F, Martínez JL, Cantón R. Antibiotics and antibiotic resistance in water environments. Curr Opin Biotechnol. 2008;19(3):260–265. doi: 10.1016/j.copbio.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Blasco MD, Esteve C, Alcaide E. Multiresistant waterborne pathogens isolated from water reservoirs and cooling systems. J Appl Microbiol. 2008;105(2):469–475. doi: 10.1111/j.1365-2672.2008.03765.x. [DOI] [PubMed] [Google Scholar]
- Carattoli A. Plasmids in Gram negatives: molecular typing of resistance plasmids. Int J Med Microbiol. 2011;301(8):654–658. doi: 10.1016/j.ijmm.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Cattoir V, Poirel L, Aubert C, Soussy CJ, Nordmann P. Unexpected occurrence of plasmid-mediated quinolone resistance determinants in environmental Aeromonas spp. Emerg Infect Dis. 2008;14(2):231. doi: 10.3201/eid1402.070677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2012) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically ; approved standard (9th edition). M07-A09. Retrieved from 10.4103/0976-237X.91790. Accessed 23 Oct 2019.
- CLSI (2018) Performance standards for antimicrobial susceptibility testing. Clinical and laboratory standards institute. M100. Retrieved from https://www.clsi.orgmance standards for antimicrobial susceptibility testing. Accessed 23 Oct 2019.
- Daef EAM, Elsherbiny NM, Thabit AG, Wageah EM. Multidrug resistant Stenotrophomonas maltophilia: an emerging cause of hospital acquired infections in Assiut University Hospitals. Egypt Int J Inf Control. 2017;13(1):1–13. [Google Scholar]
- Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- Djenadi K, Zhang L, Murray AK, Gaze WH. Carbapenem resistance in bacteria isolated from soil and water environments in Algeria. J Glob Antimicrob. 2018;15:262–267. doi: 10.1016/j.jgar.2018.07.013. [DOI] [PubMed] [Google Scholar]
- French GL. Clinical impact and relevance of antibiotic resistance. Adv Drug Deliv Rev. 2005;57(10):1514–1527. doi: 10.1016/j.addr.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Goni-Urriza M. Antimicrobial resistance of mesophilic Aeromonas spp. isolated from two European rivers. J Antimicrob Chemother. 2000;46(2):297–301. doi: 10.1093/jac/46.2.297. [DOI] [PubMed] [Google Scholar]
- Haller L, Chen H, Ng C, Le TH, Koh TH, Barkham T. Occurrence and characteristics of extended-spectrum β-lactamase- and carbapenemase- producing bacteria from hospital effluents in Singapore. Sci Total Environ. 2018;80:47–56. doi: 10.1016/j.scitotenv.2017.09.217. [DOI] [PubMed] [Google Scholar]
- Harmon DE, Miranda OA, McCarley A, Eshaghian M, Carlson N, Ruiz C. Prevalence and characterization of carbapenem‐resistant bacteria in water bodies in the Los Angeles–Southern California area. MicrobiologyOpen. 2018;8(4):e00692. doi: 10.1002/mbo3.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser AR. Antibiotics basics for clinicians the ABCs of choosing the right antibacterial agent. 3. USA: Lippincott Williams & Wilkins; 2018. [Google Scholar]
- Jarlier V, Nicolas M, Fournier G, Philippon A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae : hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10(4):867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- Kenzaka T, Tani K. Draft genome sequence of multidrug-resistant Stenotrophomonas pavanii BWK1, Isolated from Mareca penelope Feces. Genome Announc. 2018;6(12):1. doi: 10.1128/genomeA.00187-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan GA, Berglund B, Khan KM, Lindgren PE, Fick J. Occurrence and abundance of antibiotics and resistance genes in rivers, canal and near drug formulation facilities: a study in Pakistan. PLoS ONE. 2013;8(6):e62712. doi: 10.1371/journal.pone.0062712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein EY, Van Boeckel TP, Martinez EM, Pant S, Gandra S, Levin SA. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc Natl Acad Sci USA. 2015;115(15):E3463–3470. doi: 10.1073/pnas.1717295115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Yang M, Hu J, Zhang J, Liu R, Gu X. Antibiotic-resistance profile in environmental bacteria isolated from penicillin production wastewater treatment plant and the receiving river. Environ Microbiol. 2009;11(6):1506–1517. doi: 10.1111/j.1462-2920.2009.01878.x. [DOI] [PubMed] [Google Scholar]
- Marangon BF, Miller D, Muallem MS, Romano AC, Alfonso EC. Ciprofloxacin and levofloxacin resistance among methicillin-sensitive Staphylococcus aureus isolates from Keratitis and conjunctivitis. Am J Opthalmol. 2004;137(3):453–458. doi: 10.1016/j.ajo.2003.10.026. [DOI] [PubMed] [Google Scholar]
- Mathur P, Tak V, Gupta G. Detection of Amp C β lactamases in gram-negative bacteria. J Lab Physicians. 2014;6(1):1. doi: 10.4103/0974-2727.129082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar A, Manzoor M, Gul I, Zafar R, Jamil HI, Niazi AK, Ali MA, Park TJ, Arshad M. Phytotoxicity of different antibiotics to rice and stress alleviation upon application of organic amendments. Chemosphere. 2020;258:127353. doi: 10.1016/j.chemosphere.2020.127353. [DOI] [PubMed] [Google Scholar]
- Oliveira LG, Ferreira LGR, Nascimento AMA, Reis MDP, Dias MF. Lima WG (2018) Antibiotic resistance profile and occurrence of Amp C between Pseudomonas aeruginosa isolated from a domestic full-scale WWTP in southeast Brazil. Water Sci Technol. 2017;1:108–114. doi: 10.2166/wst.2018.091. [DOI] [PubMed] [Google Scholar]
- Osińska A, Harnisz M, Korzeniewska E. Prevalence of plasmid-mediated multidrug resistance determinants in fluoroquinolone-resistant bacteria isolated from sewage and surface water. Environ Sci Pollut Res. 2016;23(11):10818–10831. doi: 10.1007/s11356-016-6221-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva MC, Reis MP, Costa PS, Dias MF, Bleicher L, Scholte LLS. Identification of new bacteria harboring qnrS and aac(6′)-Ib/cr and mutations possibly involved in fluoroquinolone resistance in raw sewage and activated sludge samples from a full-scale WWTP. Water Res. 2017;110:27–37. doi: 10.1016/j.watres.2016.11.056. [DOI] [PubMed] [Google Scholar]
- Pontes DS, Pinheiro FA, Lima-Bittencourt CI, Guedes RLM, Cursino L, Barbosa F. Multiple antimicrobial resistance of gram-negative bacteria from natural oligotrophic lakes under distinct anthropogenic influence in a tropical region. Microb Ecol. 2009;58(4):762–772. doi: 10.1007/s00248-009-9539-3. [DOI] [PubMed] [Google Scholar]
- Pormohammad A, Pouriran R, Azimi H, Goudarzi M. Prevalence of integron classes in gram-negative clinical isolated bacteria in Iran: a systematic review and meta-analysis. Iran J Basic Med Sci. 2019;22(2):118. doi: 10.22038/ijbms.2018.32052.7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameshkumar G, Ramakrishnan R, Shivkumar C, Meenakshi R, Anitha V, Venugopal Reddy Y, Maneksha V. Prevalence and antibacterial resistance patterns of extended-spectrum beta-lactamase producing Gram-negative bacteria isolated from ocular infections. Indian J Ophthalmol. 2016;64(4):303–311. doi: 10.4103/0301-4738.182943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi M, Fatima N, Rashid M, Shukla I, Malik A, Usman A. Extended spectrum AmpC and metallo-beta-lactamases in Serratia and Citrobacter spp. in a disc approximation assay. J Infect Dev Ctries. 2009;3(4):285–294. doi: 10.3855/jidc.126. [DOI] [PubMed] [Google Scholar]
- Saima S, Fiaz M, Zafar R, Ahmed I, Arshad M (2020) Dissemination of antibiotic resistance in the environment. In: Hashmi MZ (ed) Antibiotics and antimicrobial resistance genes in the environment. Elsevier, Inc., The Netherlands. https://doi.org/10.1016/B978-0-12-818882-8.00006-1, pp 99–116
- Shah SQA, Colquhoun DJ, Nikuli HL, Sørum H. Prevalence of antibiotic resistance genes in the bacterial flora of integrated fish farming environments of Pakistan and Tanzania. Environ Sci Technol. 2012;46(16):8672–8679. doi: 10.1021/es3018607. [DOI] [PubMed] [Google Scholar]
- Sidrach-Cardona R, Hijosa-Valsero M, Marti E, Balcázar JL, Becares E. Prevalence of antibiotic-resistant fecal bacteria in a river impacted by both an antibiotic production plant and urban treated discharges. Sci Total Environ. 2014;488–489:220–227. doi: 10.1016/j.scitotenv.2014.04.100. [DOI] [PubMed] [Google Scholar]
- Situation analysis report on antimicrobial resistance in Pakistan (2017) CDDEP.org. https://cddep.org/wp-content/uploads/2018/03/Situational-Analysis-Report-on-Antimicrobial-Resistance-in-Pakistan.pdf
- Tacão M, Moura A, Correia A, Henriques I. Co-resistance to different classes of antibiotics among ESBL-producers from aquatic systems. Water Res. 2014;48:100–107. doi: 10.1016/j.watres.2013.09.021. [DOI] [PubMed] [Google Scholar]
- Tansawai U, Sanguansermsri D, Na-udom A, Walsh TR, Niumsup PR. Occurrence of extended spectrum β-lactamase and AmpC genes among multidrug-resistant Escherichia coli and emergence of ST131 from poultry meat in Thailand. Food Control. 2018;84:159–164. doi: 10.1016/j.foodcont.2017.07.028. [DOI] [Google Scholar]
- Upadhyay S, Sen MR, Bhattacharjee A. Presence of different beta-lactamase classes among clinical isolates of Pseudomonas aeruginosa expressing AmpC beta-lactamase enzyme. J Infect Dev Ctries. 2010;4(04):239–242. doi: 10.3855/jidc.497. [DOI] [PubMed] [Google Scholar]
- Weldhagen GF. Integrons and β-lactamases - A novel perspective on resistance. Int J Antimicrob Agents. 2004;23(6):556–562. doi: 10.1016/j.ijantimicag.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Wilke MS, Lovering AL, Strynadka NC. β-Lactam antibiotic resistance: a current structural perspective. Curr Opin Microbiol. 2005;8(5):525–533. doi: 10.1016/j.mib.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Ye Q, Wu Q, Zhang S, Zhang J, Yang G, Wang H. Antibiotic-resistant extended spectrum β-lactamase- and plasmid-mediated AmpC-producing enterobacteriaceae isolated from retail food products and the pearl river in Guangzhou. China Front Microbiol. 2017;8:96. doi: 10.3389/fmicb.2017.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong D, Lee K, Yum JH, Shin HB, Rossolini GM, Chong Y. Imipenem-EDTA disk method for differentiation of metallo-beta-lactamase-producing clinical isolates of Pseudmonas spp. and Acinetobacter spp. J Clin Microbiol. 2002;40(10):3798–3801. doi: 10.1128/JCM.40.10.3798-3801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.