Abstract
A multiplex-PCR assay based on mitochondrial cytochrome c oxidase subunit I (COI) sequences was developed for identification of five members of the Barbirostris Complex which occur in Thailand: Anopheles barbirostris s.s., An. dissidens, An. saeungae, An. wejchoochotei and An. barbirostris species A3. Anopheles campestris was not included in the assay due to the lack of unequivocal sequences. Allele-specific primers were designed for specific nucleotide segments of COI sequences of each species. Mismatch method and addition of long GC tail were applied for some primers. The assay provided products of 706 bp for An. barbirostris s.s., 238 bp for An. dissidens, 611 bp for An. saeungae, 502 bp for An. wejchoochotei and 365 bp for An. barbirostris A3. The assay was tested using 111 wild-caught female mosquitoes from Bhutan, Cambodia, Indonesia (Sulawesi) and Thailand. The results of the multiplex PCR were in complete agreement with COI sequencing; however, one of three specimens from Bhutan and all 11 specimens from Indonesia were not amplifiable by the assay due to their distinct COI sequences. This, together with the distinct rDNA sequences of these specimens, suggests the presence of at least two additional new species in the Barbirostris Complex.
Keywords: Anopheles barbirostris, COI, multiplex PCR, species complex, Thailand
1. Introduction
Anopheles barbirostris van der Wulp, a member of the Myzorhynchus Series of the subgenus Anopheles Meigen, was formally described in 1884 from a single female collected in eastern Java, Indonesia [1]. Mosquitoes identified morphologically as An. barbirostris s.l. are common in the Oriental Region, ranging from Timor, the Indonesian Archipelago, Guam westward across mainland Southeast Asia, southern China and southern Asia [2]. However, genetic and molecular studies revealed that the taxon is a complex of sibling species, the Barbirostris Complex [3], consisting of six formally recognized species: An. barbirostris s.s., An. campestris Reid, An. dissidens Taai and Harbach, An. saeungae Taai and Harbach, An. vanderwulpi Townson and Harbach and An. wejchoochotei Taai and Harbach [4]. The complex includes at least one additional species, i.e., An. barbirostris species A3 of Saeung et al. (2008) [5], which is known from Kanchanaburi Province, western Thailand.
The distributions of the members of the complex are not completely known. Anopheles barbirostris s.s. is currently known from Indonesia (Java and Kalimantan) and Thailand [6,7]. The role of An. barbirostris s.s. as a vector of malarial protozoa is questionable. This species is common in Java, but is not important as a malaria vector due to its strong zoophilic behaviour [8]. In contrast, An. barbirostris s.l. in Sulawesi, the Lesser Sunda Island group (Lombok and Flores) and Timor–Leste is more anthropophilic and is an important vector of malarial protozoa and the filarial nematode Brugia malayi. Current evidence has revealed that the internal transcribed spacer 2 (ITS2, rDNA) sequence of An. barbirostris s.l. from Sulawesi [9] differs from An. barbirostris s.s. from Kalimantan [6] by 2.76%, suggesting that it is a distinct species. Laboratory experiments have revealed that An. barbirostris s.s. in Thailand is refractory to Plasmodium falciparum and P. vivax [10]. In Sri Lanka, An. barbirostris s.l. is a malaria vector [11], but molecular identification based on COI and ITS2 sequences revealed a new molecular type which differs from An. barbirostris s.s. by 5.1 and 5.4%, respectively [12]. Anopheles barbirostris s.l. is common in India, but the role of this taxon as a malaria vector is unknown [13].
Anopheles dissidens is currently known from Thailand, China and Myanmar [4,14]. Anopheles saeungae is known from Thailand, Indonesia (Sumatra) and China [4,6,14]. The role of An. dissidens, An. saeungae and An. barbirostris species A3 as natural vectors of malarial parasites is unknown. Anopheles dissidens and An. saeungae are zoophilic and susceptible to P. vivax in the laboratory, with low sporozoite infection rates (<10%) [4,10].
Anopheles campestris, first described from specimens collected in Rantau Panjang, Klang, Selangor, Malaysia [15], is an important vector of malarial protozoa and B. malayi in peninsular Malaysia [16]. Based on morphological identification, An. campestris is also known to occur in Cambodia [17], Thailand [18] and Vietnam [19]. Adults of An. campestris differ principally from those of An. barbirostris s.s. in having darker scales on the wing veins and more white scales on the abdominal terga; larvae and pupae of the former usually have some abdominal setae with more numerous branches [15]. However, Harrison and Scanlon (1975) [18] reported that these characters were so variable in Thai populations that the two species were difficult to identify with certainty. Baimai et al. (1995) [20] reported that the metaphase karyotype of An. campestris from Ayutthaya Province, central Thailand, is distinct from those of An. barbirostris s.l. from elsewhere in Thailand and Indonesia (Java). According to Rattanarithikul et al. (2006) [21], An. campestris in Thailand appears to be mainly confined to flat rice plains in the central and eastern regions and is absent in hilly areas. This species is often confused with An. wejchoochotei (formerly An. campestris-like), which is currently only known from Thailand [4], because it is indistinguishable from An. campestris in the larval, pupal and adult stages. Unfortunately, no molecular data for An. campestris from the type locality is available in GenBank for comparison with An. campestris from elsewhere. Anopheles wejchoochotei is relatively more anthropophilic and probably a vector of malarial parasites in Sa Kaeo Province in eastern Thailand [22,23]. Anopheles vanderwulpi is known from Java and Sumatra of Indonesia and may not be a vector of parasites of human diseases due to its zoophilic behavior [7]. The role of An. barbirostris s.l. as a vector of P. knowlesi, a primate malarial protozoan, is unknown.
Correct identification of members of the Barbirostris Complex is essential for epidemiology studies and the control of malaria because not all of the sibling species are vectors of agents of human disease. However, members of the complex are difficult to distinguish by morphology due to overlapping morphological characters [4]. Consequently, their distributions and the roles they play in the transmission of disease agents can only be known when specimens are identified by a molecular or cytogenetic method. The latter method, however, is less convenient as it requires laboratory colonies [5,24].
Recently, a double multiplex PCR based on ITS2 sequences was developed for the identification of five members of the Barbirostris Complex in Thailand [25]. However, this method requires two steps of PCR reactions and did not detect An. barbirostris species A3, which also occurs in Thailand [5]. Because phylogenetic analyses of the COI and ITS2 sequences produced phylogenetic trees with similar topologies [5,6,24,26], in the present study we developed a multiplex PCR method based on COI sequences for identification of the species of the Barbirostris Complex that occur in Thailand and other countries in the region, except An. campestris due to the lack of available sequences for this nominal taxon in GenBank.
2. Materials and Methods
2.1. Mosquitoes
Adult mosquitoes were collected by aspirator in cattle sheds in Bhutan, Cambodia, Sulawesi and Thailand, during 2016–2019 (Table 1). They were killed with chloroform vapour and kept dried in vials with silica gel. Identification was done under a stereomicroscope following Rattanarithikul et al. (2006) [21]. The morphological terminology and abbreviations found in the Anatomical Glossary of the online Mosquito Taxonomic Inventory (http://mosquito-taxonomic-inventory.info/node/11027) are used herein.
Table 1.
Details of collections of An. barbirostris s.l. made in Bhutan, Cambodia, Indonesia and Thailand, and comparison of results from multiplex PCR and mitochondrial cytochrome c oxidase subunit I (COI) sequencing.
| Country | Location | Location Code | Coordinates | Multiplex PCR /COI Sequences (No. of Specimens) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| An. barbirostris s.s. | An. dissidens | An. saeungae | An. wejchoochotei | An. barbirostris A3 | Unknown | Total | ||||
| Thailand | Mae On, Chiang Mai | MO | 18°44′45″ N 99°13′27″ E | 7/7 | 7/7 | |||||
| Hang Dong, Chiang Mai | HD | 18°43′45″ N 98°54′22″ E | 3/3 | 3/3 | ||||||
| San Sai, Chiang Mai | SS | 18°59′09″ N 99°05′11″ E | 6/6 | 6/6 | ||||||
| Chiang Dao, Chiang Mai | CD | 19°23′27″ N 98°55′51″ E | 2/2 | 2/2 | ||||||
| Mueang Chiang Mai, Chiang Mai | MH | 18°45′21″ N 98°56′22″ E | 2/2 | 2/2 | ||||||
| Ko Kha, Lampang | LP | 18°07′44″ N 99°20′43″ E | 2/2 | 2/2 | ||||||
| Ban Thi, Lamphun | Banti | 18°39′16″ N 99°10′04″ E | 8/8 | 8/8 | ||||||
| Mae Sariang, Mae Hong Son | MS | 18°09′24″ N 97°56′15″ E | 3/3 | 3/3 | ||||||
| Tha Song Yang, Tak | Tak | 17°27′05″ N 98°10′40″ E | 3/3 | 1/1 | 9/9 | 13/13 | ||||
| Makham, Chanthaburi | CB | 12°40′17″ N 102°11′51″ E | 4/4 | 4/4 | ||||||
| Na Chaluai, Ubon Ratchathani | UB | 14°32′54″ N 105°14′33″ E | 38/38 | 3/3 | 41/41 | |||||
| Cambodia | Ratanakiri | Camb | 13°51′26″ N 107°06′04″ E | 5/5 | 1/1 | 6/6 | ||||
| Bhutan | Singye, Sarpang | Bhutan | 26°50′44″ N 90°12′26″ E | 2/2 | 2/2 | |||||
| Gelephu, Sarpang | GE | 26°54′47″ N 90°30′07″ E | 0/1 | 0/1 | ||||||
| Indonesia | Bantaeng, South Sulawesi | Ban | 05°26′58″ S 119°54′15″ E | 0/1 | 0/1 | |||||
| Makassar, South Sulawesi | Lak | 05°07′20″ S 119°28′05″ E | 0/5 | 0/5 | ||||||
| UNHAS | 05°07′54″ S 119°29′02″ E | 0/2 | 0/2 | |||||||
| Maros, South Sulawesi | Pucak | 05°09′29″ S 119°42′01″ E | 0/2 | 0/2 | ||||||
| Wulai, West Sulawesi | Wulai | 01°02′42″ S 119°31′27″ E | 0/1 | 0/1 | ||||||
| Total | 2/2 | 49/49 | 7/7 | 30/30 | 11/11 | 0/12 | 99/111 | |||
2.2. DNA Extraction, Amplification and Sequencing of the COI Gene
Genomic DNA was extracted from the legs of individual mosquitoes using the Pure Link™ Genomic DNA Mini Kit (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s instructions. The DNA was eluted with 20 μL (first elusion) followed by 40 μL (second elusion) buffer. Specimens were retained for morphological examination. The mitochondrial COI gene was amplified by PCR using the LCO-1490 forward primer (5′-GGTCAACAAATCATAAAGATATTGG-3′) and the HCO-2198 reverse primer (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) [27]. The 20 μL PCR reaction consisted of 1X PCR buffer (Invitrogen, Carlsbad, CA, USA), 3.0 mM MgCl2 (Invitrogen, Carlsbad, CA, USA), 0.2 mM dNTPs (Invitrogen, Carlsbad, CA, USA), 0.4 U Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA), 0.2 μM of each primer and 1 μL (5 ng) of extracted DNA. PCR reaction was conducted by using the Dw-T960 Smart Gradient PCR Thermal Cycler (Drawell, Shanghai, China) and the PCR conditions were as follows: an initial denaturation at 95 °C for 2 min, 40 cycles of denaturation at 95 °C for 30 s, annealing at 45 °C for 30 s and extension at 72 °C for 30 s, and a final extension at 72 °C for 5 min. Amplicons were visualized on 2% agarose gels stained with Ethidium bromide (Invitrogen, Carlsbad, CA, USA). PCR products were purified using the Illustra™ ExoProStar™ 1-Step (GE Healthcare Life Sciences, Buckinghamshire, UK) and sequenced using the BigDye® Terminator v3.1 cycle sequencing kit chemistry (First BASE, Selangor, Malaysia).
2.3. Sequencing Alignment and Phylogenetic Analysis
The new COI sequences were compared with those of An. barbirostris s.l. available in GenBank using the Basic Local Alignment Search Tool (BLAST, available at http://blast.ncbi.nlm.nih.gov/Blast.cgi) under default parameters. GenBank included sequences from Thailand: An. barbirostris s.s. (AB373942.1–AB373944.1), An. dissidens (LC333239.1, LC333240.1), An. saeungae (AB971326.1, AB971327.1), An. wejchoochotei (AB971339.1, AB971340.1) and An. barbirostris A3 (AB362238.1–AB362240.1). In addition, COI sequences listed as An. campestris from Chiang Mai (AB436105.1) and Sa Kaeo (AB436124.1) Provinces, Thailand, reported by Thongsahuan et al. (2009) [28], An. barbirostris s.l. from mainland India (AY729982.1, HM773367.1, MN166188.1, MN25480.1, MN264217.1) and Andaman and Nicobar Islands (MK184151.1, MK184158.1, MK184180.1, MK184190.1, MK184200.1), Singapore (KF564681.1–KF564683.1) and Vietnam (MH425426.1–MH425429.1, MH425437.1) were also included in the phylogenetic analysis. One specimen of An. vanderwulpi from the Natural History Museum, London, was sequenced for COI and included in the tree.
Sequences were aligned using Clustal W version 2.0 under default parameters [29]. Ragged ends were trimmed using MEGA version 10.0.5 [30]. Phylogenetic analyses were conducted using maximum likelihood (ML) with MEGA version 10.0.5 [30]. Anopheles pullus Yamada was used as the outgroup, which belongs to the Hyrcanus Group of the Myzorhynchus Series. The best evolutionary model of nucleotide substitution that fits the data was obtained using jModelTest v. 2.1.10 [31]. Robustness of the ML tree was tested with 1000 bootstrapped data sets with bootstrap support values of 70% or more indicated on the tree. This widely used level of bootstrap support [32] was chosen to allow full consideration of how clades inferred in this way correspond to putative species, given that when the latter are closely related they are not expected to have high levels of bootstrap support from a single gene.
2.4. Primer Design and Allele-Specific PCR
Allele-specific primers were designed from published mitochondrial COI sequences available in GenBank: An. barbirostris s.s. (AB373942–AB373944), An. dissidens (AB331574–AB3315780), An. saeungae (AB435997, AB436002, AB436007, AB436013–AB436015, AB331570–AB331572), An. wejchoochotei (AB971336–AB971338, and AB331582–AB331588 as An. campestris) and An. barbirostris species A3 (AB362238–AB362240) (see alignments in Supplementary Figure S1). Sequences were aligned using the Clustal W algorithm [33] implemented in MEGA version 10.0.5 [30]. Primers were designed using the web-based Primer3Plus software [34]. Our initial study revealed difficulties in optimizing conditions for multiplex PCR, in particular high annealing temperatures did not show all diagnostic bands whereas low annealing temperatures showed all diagnostic bands, but there were also nonspecific bands. The mismatch technique [35] was then used to increase specificity and reduce background amplification by positioning the mismatches in some allele-specific primers at the third nucleotide from the 3’ end. To distinguish between the amplification products based on size of the products, the long GC tail was attached to the allele-specific primer [36]. Each allele-specific forward primer and the universal COI reverse primer (HCO-2198) were tested with definitively identified specimens of Saeung et al. (2007, 2008) [5,24] and Suwannamit et al. (2009) [26] in order to check the length of the amplified fragments and assess primer specificity. PCR was conducted using 20 μL volumes containing 1X PCR buffer (Invitrogen, Carlsbad, CA, USA), 3.0 mM MgCl2 (Invitrogen, Carlsbad, CA, USA), 0.2 mM dNTPs (Invitrogen, Carlsbad, CA, USA), 0.4 U Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA), 0.2 μM of each primer and 1 μL (5 ng) of extracted DNA. The PCR reactions were carried out at 95 °C for 2 min, 40 cycles at 95 °C for 30 s, 45 °C for 30 s and 72 °C for 30 s, and a final extension at 72 °C for 5 min. Amplicons were visualised on 2% agarose gels stained with Ethidium bromide (Invitrogen, Carlsbad, CA, USA).
2.5. Multiplex PCR and Validation
The reaction mixtures were prepared from 20 μL volumes that contained 0.2 μM of each primer (six primers). PCR reaction mixture, conditions for amplifications and gel electrophoresis were performed in the same way as the allele-specific PCR method. Results of the novel multiplex-PCR assay were validated by comparing with COI sequences of feral specimens of the Barbirostris Complex collected from different locations in Bhutan, Cambodia, Indonesia (Sulawesi) and Thailand (Table 1).
3. Results
3.1. Phylogenetic Analysis
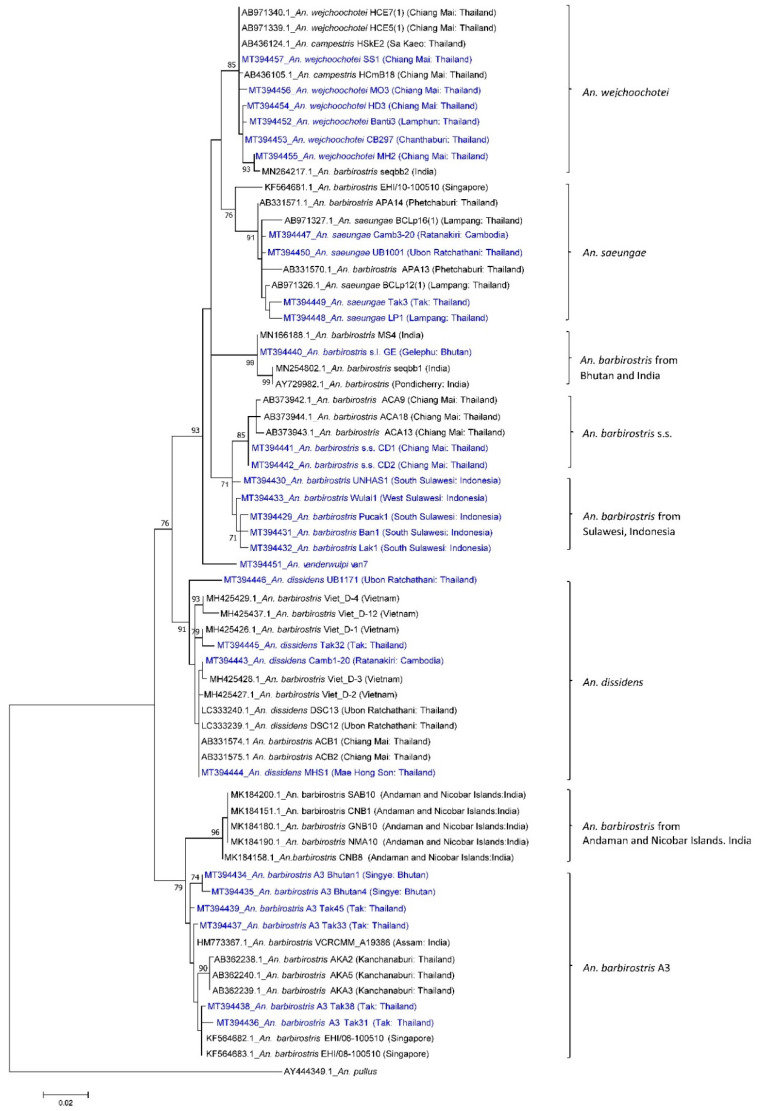
A total of 111 specimens collected from various locations were sequenced for a fragment of the COI gene (Table 1, Supplementary Figure S2). All sequences in this study were deposited in the DDBJ/EMBL/GenBank nucleotide sequence database (MT394429–MT394457, MT450754–MT450836) and Barcode of Life Database (BOLD) (THBAR001-20–THBAR111-20). The best-fit model of nucleotide substitution for ML analysis was GTR+G+I. Twenty-nine representative COI sequences from the current study were included in the phylogenetic tree (highlighted as blue in Figure 1).
Figure 1.
Maximum likelihood tree of COI sequences. Representative specimens of An. barbirostris s.l. in the current study are highlighted in blue. GenBank sequences included specimens from the Andaman and Nicobar Islands, India, Singapore, Thailand and Vietnam, and An. pullus as the outgroup. Bootstrap values of ≥70% are shown.
Phylogenetic analysis of COI sequences (642 bp) revealed that specimens from Thailand collected in the current study were recovered in five clades of the Barbirostris Complex, including An. barbirostris s.s., An. dissidens, An. saeungae, An. wejchoochotei and An. barbirostris A3, and distinguish from the clades consisting of specimens from Gelephu of Bhutan, Sulawesi, and the Andaman and Nicobar Islands, with supportive bootstrap values (71–99%) (Figure 1). The An. dissidens clade also included specimens from Cambodia (MT394443) and GenBank sequences from Vietnam (MH425426.1–MH425429.1, MH425437.1). The sequences of specimens listed as An. campestris (AB436105.1, AB436124.1) by Thongsahuan et al. (2009) [28] fell in the An. wejchoochotei clade, which also included one GenBank sequence of An. barbirostris s.l. from India (MN264217.1). One specimen from Cambodia (MT394447) and one GenBank sequence from Singapore (KF564681.1) fell in the An. saeungae clade. The An. barbirostris A3 clade includes specimens from Tak Province (MT394436–MT394439), Thailand. Interestingly, two specimens from Singye (MT394434, MT394435), Sarpang of Bhutan fell in this clade, which also included one GenBank sequence from India (HM773367.1) and two GenBank sequences from Singapore (KF564682.1, KF564683.1). Five GenBank sequences from the Andaman and Nicobar Islands (MK184151.1, MK184158.1, MK184180.1, MK184190.1, MK184200.1) formed a distinct clade (ML bootstrap value 96%), which is close to the An. barbirostris A3 clade. The sole specimen from Gelephu (MT394440), Sarpang of Bhutan and three GenBank sequences from India (AY729982.1, MN166188.1, MN254802.1) formed a distinct clade (ML bootstrap value 99%). Specimens of An. barbirostris s.s. formed a distinct clade (ML bootstrap value 85%) which is closely related to specimens from southern and western Sulawesi (MT394429–MT394433).
3.2. Primer Design and Multiplex PCR
Five species-specific reverse primers were designed, based on nucleotide alignment of the complete COI region of each species (684 bp) (Figure 2). Primer names, sequences and sizes of the PCR products are shown in Table 2. To increase specificity, mismatches were applied to the third nucleotide from the 3′ end of the allele-specific primers of An. barbirostris s.s., An. dissidens and An. wejchoochotei. Since the allele-specific sites of primers for An. barbirostris s.s. and An. saeungae were very close, the long GC tail (5′-GCGGGCAGGGCGGCGGGGGCGGGGCC-3′) was attached to the allele-specific primer of An. barbirostris s.s. to increase the product size so that the PCR products of both are clearly distinguishable.
Figure 2.
Alignment of COI sequences of five species of the Barbirostris Complex. The primer selection sites are highlighted in colors.
Table 2.
Primers designed for the multiplex-PCR assay. Long GC tail was added to the allele-specific primer for An. barbirostris s.s. Mismatches of nucleotide bases are underlined.
| Species | Primer Name | Primer Sequence (5′–3′) | Product Size (bp) |
|---|---|---|---|
| An. barbirostris s.s. | BARA4_F | [long GC tail] a AATAGTAGGAACTTCTTTATGA | 706 |
| An. dissidens | BARA1_F | ATTACTACTGTTATTAATATAGGA | 238 |
| An. saeungae | BARA2_F | TTAGGTCACCCAGGAGCA | 611 |
| An. wejchoochotei | BARWEJ_F | GATTTGGAAACTGATTACTG | 502 |
| An. barbirostris A3 | BARA3_F | CGGAACTGGATGAACTGTA | 365 |
| Universal reverse primer | HCO 2198_R | TAAACTTCAGGGTGACCAAAAAATCA |
a Long GC tail sequence: 5′-GCGGGCAGGGCGGCGGGGGCGGGGCC-3′.
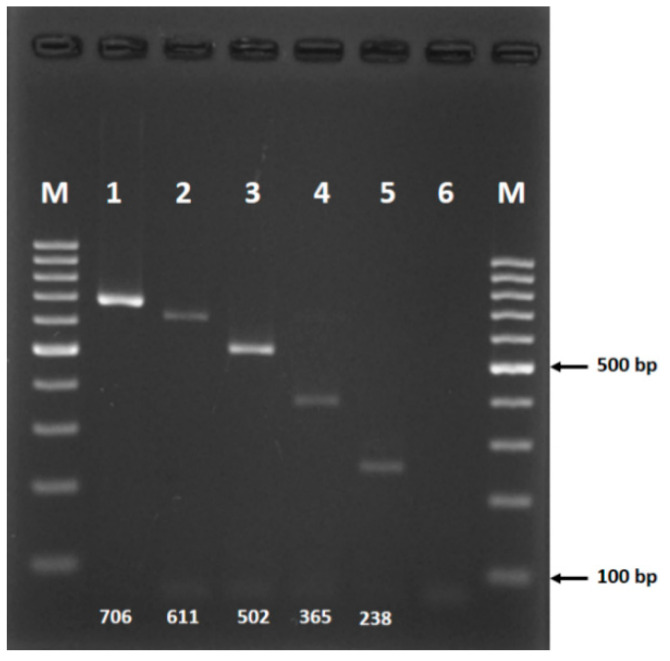
The novel species-specific forward primers that were combined with the universal reverse primer (HCO-2198) successfully simultaneously distinguish all five species of the Barbirostris Complex. This multiplex-PCR assay gave amplified products of 706 bp for An. barbirostris s.s., 238 bp for An. dissidens, 611 bp for An. saeungae, 502 bp for An. wejchoochotei and 365 bp for An. barbirostris A3 (Table 2, Figure 3). The product bands were clearly visible without nonspecific bands.
Figure 3.
PCR products from the multiplex-PCR assay run on 2% agarose gel. Lanes: 1, An. barbirostris s.s.; 2, An. saeungae; 3, An. wejchoochotei; 4, An. barbirostris A3; 5, An. dissidens; 6, negative control. The 100-bp molecular weight ladders are loaded in M lanes.
3.3. Validation
The assay was validated with the 111 sequenced specimens listed in Table 1. The majority of specimens tested were individuals of An. wejchoochotei and An. dissidens, followed by lesser numbers of An. barbirostris species A3 and An. saeungae, and only two specimens of An. barbirostris s.s. Most specimens were successfully amplified without ambiguous bands, as indicated in Figure 3, the exceptions being one specimen from Gelephu, Bhutan, and 11 specimens from Sulawesi, Indonesia, which have sequences different from those of known members of the Barbirostris Complex (Supplementary Figure S2) and form distinct clades (Figure 1). The results from the multiplex PCR and DNA sequencing for amplifiable COI products were in complete agreement.
4. Discussion
Multiplex PCR assays using mtDNA sequences have been used successfully for the identification of members of species complex, e.g., the An. culicifacies complex [37]. However, there may be limitations to its use for identification of closely related species due to incomplete lineage sorting and cross-species hybridization events that can result in shared mitochondria between sibling species, e.g., siblings of the An. lindesayi complex [38]. The multiplex-PCR assay based on COI sequences developed in the present study is relatively simple, fast and cheap compared with DNA sequencing and the multiplex-PCR assay based on ITS2 sequences [25] because it requires only one reaction. The cost of our assay per specimen is about 5 and 10 times cheaper, and 1.2 and 50 times faster, than allele-specific PCR and DNA sequencing, respectively. The designed allele-specific primers and PCR conditions are reliable for identification of specimens of the five species of the Barbirostris Complex that occur in Thailand, including some specimens from Cambodia and Bhutan. The method should facilitate identification of a large number of specimens from Thailand and neighbouring countries, and, thus, should increase knowledge of the distributions of species of the complex and elucidation of the roles they play in the transmission of pathogens. However, there are relatively few sequences for validation of some species, particularly An. barbirostris s.s., An. saeungae and An. barbirostris A3; hence, more specimens from various geographical areas are needed to confirm the reliability of our assay. In addition, the limits of detection depend on the quality of field-caught specimens (such as age and the method of preservation). When freshly captured specimens are used, one or two legs are sufficient to obtain a DNA product and a quantity as low as 1 ng of DNA is amplifiable. In our assay, 5 ng of DNA was generally used with no problem for amplification. However, if there had been no amplified product, the first action to be taken would be to increase the quantity of DNA, up to 10 ng. If there was no product at all, which could be due to poor DNA quality, intraspecific variation or a different species, DNA sequencing would be necessary.
In this study, however, we did not include An. vanderwulpi, which occurs in Indonesia, nor did we include An. campestris due to the lack of unequivocal COI sequences in GenBank. The COI, COII and ITS2 sequences of An. wejchoochotei [4], which were derived from the An. campestris-like specimens collected in Sa Kaeo, Chiang Mai and other provinces in Thailand, were often listed as An. campestris in GenBank accessions (e.g., Saeung et al., 2007 [24]; Suwannamit et al., 2009 [26]; Thongsahuan et al. 2009 [28] and Paredes-Esquivel et al., 2009 [6]), and this name was subsequently used in publications (e.g., Townson et al., 2013 [7]; Gajapathy et al., 2014 [12]; Sum et al., 2014 [39]). In recent years, attempts to collect and identify An. campestris in Thailand based on the metaphase karyotyping reported by Baimai et al. [20], were unsuccessful [5,24,26,28]. The multiplex-PCR assay based on ITS2 sequences developed by Brosseau et al. (2019) [25] included a specific primer designed from a single specimen collected in Chanthaburi Province of eastern Thailand that was identified morphologically. However, its ITS2 sequence has not been deposited in GenBank and whether it corresponds to An. campestris from the type locality in Malaysia is not known. Their assay was tested with mosquitoes collected throughout Thailand, but none were amplified by that primer. We also tested four specimens (kindly provided by T. Chareonviriyaphap) from the same area in Chanthaburi Province (see Table 1), but they were An. wejchoochotei, which appears to be common in Thailand, in agreement with Brosseau et al. (2019) [25]. Based on current evidence, An. campestris may be absent or very rare in Thailand.
Interestingly, the ITS2 sequences of 12 specimens from Selangor, Malaysia included in the phylogenetic analysis of Sum et al. (2014) [39] formed a distinct clade within a larger clade of sequences from sibling species of the Barbirostris Complex collected in Thailand and Indonesia. The larger clade, including the Selangor specimens, comprised five strongly supported clades: three plus the clade of Selangor specimens were labelled as “An. barbirostris” and one clade was labelled as An. campestris. However, as mentioned above, the clade considered to be An. campestris consists of sequences of An. wejchoochotei from Thailand, and the clade of Selangor specimens is an unrecognised species. Since Selangor includes the type locality of An. campestris [15], it seems quite possible that the specimens of Sum et al. were An. campestris, a possibility that needs to be confirmed by further study.
We also did not test our assay with many specimens of An. barbirostris s.s., which appears to be rare in Thailand and mainland Asia. Anopheles barbirostris s.s. has not been detected in China [13], peninsular Malaysia [39] and Sri Lanka [12]. We also did not find evidence that this species occurs in Bhutan, Cambodia, India, Singapore and Vietnam. Anopheles dissidens, An. saeungae and An. wejchoochotei are more common in Thailand and other countries in Asia [4,14,19]. Apart from Thailand, China and Myanmar, our study shows that An. dissidens also occurs in Cambodia and Vietnam. Anopheles saeungae is now known from Singapore in addition to Indonesia (Sumatra), China and Thailand. Besides Thailand, Anopheles wejchoochotei is now known from India. Anopheles barbirostris species A3 was originally found in Kanchanaburi Province of western Thailand, based on metaphase karyotypes, cross-mating experiments and DNA sequencing [5,26]. This species may have a wider distribution in Asia as it has been detected in Tak Province located immediately north of Kanchanaburi Province, and also Bhutan, India and Singapore.
The results of the current study suggest the presence of additional new species in the Barbirostris Complex. The first is the specimen from Gelephu, Bhutan, which has its COI sequence distinct from those of known members of the complex. Also, its ITS2 sequence differs (>10%) from those of other members of the complex (P. Somboon, unpublished). This putative species is also detected in India. The second is the specimens from Sulawesi, which are closely related to An. barbirostris s.s., having COI sequences that are distinct from those of other members of the complex. Moreover, their ITS2 sequences are different from An. barbirostris s.s. (Kimura 2 parameter genetic distances 2.4–3.2%, unpublished data), agreeing with the different ITS2 sequence (MN203102.1) reported by Davidson et al. (2019) [9]. The distinctiveness of both CO1 and ITS2 has previously indicated that An. barbirostris s.l. from Sri Lanka is a distinct species [12]. An. barbirostris s.l. from the Andaman and Nicobar Islands, shown here to be closely related to but genetically distinct from An. barbirostris A3, could be a further species, but further studies are required to distinguish this from the possibility that it is a geographically distinct population of An. barbirostris A3.
5. Conclusions
The multiplex PCR based on COI sequences is a reliable assay for identification of five of the six species of the Barbirostris Complex, i.e., An. barbirostris s.s., An. dissidens, An. saeungae, An. wejchoochotei and An. barbirostris species A3. In places like Thailand where An. campestris may be rare or absent, our multiplex-PCR assay is unlikely to be compromised. Further, as the two putative new species in Bhutan and Sulawesi based on distinct COI sequences were not amplified by our allele-specific primers, their presence would not compromise the validity of the test for identification of the five species for which it is designed. When applying the test to specimens from a new location, it would be advisable to do some initial DNA sequencing to make sure that only these five species are present. In some countries where the five species do not occur, modifications of the assay should be performed to detect those species that are present. However, it is understood that unrecognised species may not produce a PCR product in electrophoresis; hence, further collections, DNA sequencing and phylogenetic analyses are required in these cases.
Acknowledgments
We thank the Faculty of Medicine, Chiang Mai University, for providing a scholarship to Parinya Wilai for her PhD program and Post-Doctoral Fellowship 2020, Chiang Mai University, to Jassada Saingamsook. We also thank Wannapa Suwonkerd and Theeraphap Chareonviriyaphap for providing specimens from Cambodia and Chantaburi Province, Thailand, respectively.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4450/11/7/409/s1: Figure S1. Alignments of COI sequences of An. barbirostris s.l. from GenBank used for designing specific primers for An. barbirostris s.s. (yellow), An. saeungae (green), An. wejchoochotei (pink), An. barbirostris A3 (blue) and An. dissidens (gray). Dots (·) indicate identity of nucleotides within the alignment; Figure S2. Alignments of the COI sequences of the 111 specimens and GenBank sequences included in the study. Dashes (–) indicate alignment gaps and dots (·) indicate identity of nucleotides within the alignment.
Author Contributions
Conceptualization, supervision, project administration, funding acquisition, writing—original draft, P.S.; methodology, formal analysis, investigation, writing—original draft, P.W.; validation, P.W., J.S. and P.S.; resources, P.S., A.S., R.N., A.J. and R.S.M.A.; data curation, P.W.; writing—review and editing, C.W., P.S. and R.E.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Faculty of Medicine (Grant No. PAR-2563-07268) and Office of Research Administration, Chiang Mai University, and the Bio and Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (Grant No. 2015M3A9B6073666).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Van der Wulp F.M. Note XXXVIII. On exotic Diptera. Notes Leyden Mus. 1884;6:248–256. [Google Scholar]
- 2.Sinka M.E., Bangs M.J., Manguin S., Chareonviriyaphap T., Patil A.P., Temperley W.H., Gething P.W., Elyazar I.R., Kabaria C.W., Harbach R.E., et al. The dominant Anopheles vectors of human malaria in the Asia-Pacific region: Occurrence data, distribution maps and bionomic precis. Parasit. Vectors. 2011;4:89. doi: 10.1186/1756-3305-4-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Satoto T.B.T. Ph.D. Thesis. University of Liverpool; Liverpool, UK: 2001. Cryptic Species within Anopheles barbirostris van der Wulp, 1884, Inferred from Nuclear and Mitochondrial Gene Sequence Variation. [Google Scholar]
- 4.Taai K., Harbach R.E. Systematics of the Anopheles barbirostris species complex (Diptera: Culicidae: Anophelinae) in Thailand. Zool. J. Linn. Soc. 2015;174:244–264. doi: 10.1111/zoj.12236. [DOI] [Google Scholar]
- 5.Saeung A., Baimai V., Otsuka Y., Rattanarithikul R., Somboon P., Junkum A., Tuetun B., Takaoka H., Choochote W. Molecular and cytogenetic evidence of three sibling species of the Anopheles barbirostris Form A (Diptera: Culicidae) in Thailand. Parasitol. Res. 2008;102:499–507. doi: 10.1007/s00436-007-0788-0. [DOI] [PubMed] [Google Scholar]
- 6.Paredes-Esquivel C., Donnelly M.J., Harbach R.E., Townson H. A molecular phylogeny of mosquitoes in the Anopheles barbirostris Subgroup reveals cryptic species: Implications for identification of disease vectors. Mol. Phylogenet. Evol. 2009;50:141–151. doi: 10.1016/j.ympev.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Townson H., Dyer N., McAlister E., Satoto T.B.T., Bangs M.J., Harbach R.E. Systematics of Anopheles barbirostris van der Wulp and a sibling species of the Barbirostris Complex (Diptera: Culicidae) in eastern Java, Indonesia. Syst. Entomol. 2013;38:180–191. doi: 10.1111/j.1365-3113.2012.00653.x. [DOI] [Google Scholar]
- 8.Elyazar I.R., Sinka M.E., Gething P.W., Tarmidzi S.N., Surya A., Kusriastuti R., Baird J.K., Hay S.I., Bangs M.J. The distribution and bionomics of Anopheles malaria vector mosquitoes in Indonesia. Adv. Parasitol. 2013;83:173–266. doi: 10.1016/B978-0-12-407705-8.00003-3. [DOI] [PubMed] [Google Scholar]
- 9.Davidson J.R., Wahid I., Sudirman R., Makuru V., Hasan H., Arfah A.M., Nur N., Hidayat M.Y., Hendershot A.L., Xiao H., et al. Comparative field evaluation of kelambu traps, barrier screens and barrier screens with eaves for longitudinal surveillance of adult Anopheles mosquitoes in Sulawesi, Indonesia. Parasit. Vectors. 2019;12:399. doi: 10.1186/s13071-019-3649-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thongsahuan S., Baimai V., Junkum A., Saeung A., Min G.-S., Joshi D., Park M.-H., Somboon P., Suwonkerd W., Tippawangkosol P. Susceptibility of Anopheles campestris-like and Anopheles barbirostris species complexes to Plasmodium falciparum and Plasmodium vivax in Thailand. Mem. Inst. Oswaldo Cruz. 2011;106:105–112. doi: 10.1590/S0074-02762011000100017. [DOI] [PubMed] [Google Scholar]
- 11.Amerasinghe P.H., Amerasinghe F.P., Konradsen F., Fonseka K.T., Wirtz R.A. Malaria vectors in a traditional dry zone village in Sri Lanka. Am. J. Trop. Med. Hyg. 1999;60:421–429. doi: 10.4269/ajtmh.1999.60.421. [DOI] [PubMed] [Google Scholar]
- 12.Gajapathy K., Jude P.J., Goodacre S.L., Peiris L.B., Ramasamy R., Surendran S.N. Molecular characterization of the malaria vector Anopheles barbirostris van der Wulp in Sri Lanka. Parasit. Vectors. 2014;7:348. doi: 10.1186/1756-3305-7-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhattacharyya D.R., Rajavel A.R., Mohapatra P.K., Jambulingam P., Mahanta J., Prakash A. Faunal richness and the checklist of Indian mosquitoes (Diptera: Culicidae) Check List. 2014;10:1342–1358. doi: 10.15560/10.6.1342. [DOI] [Google Scholar]
- 14.Wang Y., Xu J., Ma Y. Molecular characterization of cryptic species of Anopheles barbirostris van der Wulp in China. Parasit. Vectors. 2014;7:592. doi: 10.1186/s13071-014-0592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reid J.A. The Anopheles barbirostris group (Diptera, Culicidae) Bull. Entomol. Res. 1962;53:1–57. doi: 10.1017/S0007485300047945. [DOI] [Google Scholar]
- 16.Reid J.A. Anopheline mosquitoes of Malaya and Bornea. Stud. Inst. Med. Res. Malaya. 1968;31:1–520. [Google Scholar]
- 17.Harrison B.A., Klein J.M. A revised list of the Anopheles of Cambodia. Mosq. Syst. 1975;7:9–12. [Google Scholar]
- 18.Harrison B.A., Scanlon J.E. Medical entomology studies–II. The subgenus Anopheles in Thailand (Diptera: Culicidae) Contri. Am. Entomol. Inst. (Ann. Arbor) 1975;12:iv + 1–307. [Google Scholar]
- 19.Ngo C.T., Dubois G., Sinou V., Parzy D., Le H.Q., Harbach R.E., Manguin S. Diversity of Anopheles mosquitoes in Binh Phuoc and Dak Nong Provinces of Vietnam and their relation to disease. Parasit. Vectors. 2014;7:316. doi: 10.1186/1756-3305-7-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baimai V., Rattanarithikul R., Kijchalao U. Metaphase karyotypes of Anopheles of Thailand and Southeast Asia: IV. The Barbirostris and Umbrosus Species Groups, subgenus Anopheles (Diptera: Culicidae) J. Am. Mosq. Control. Assoc. 1995;11:323–328. [PubMed] [Google Scholar]
- 21.Rattanarithikul R., Harrison B.A., Harbach R.E., Panthusiri P., Coleman R.E. Illustrated keys to the mosquitoes of Thailand. IV. Anopheles. Southeast. Asian J. Trop. Med. Public Health. 2006;37(Suppl. 2):1–128. [PubMed] [Google Scholar]
- 22.Limrat D., Rojruthai B., Apiwathnasorn C., Samung Y., Prommongkol S. Anopheles barbirostris/campestris as a probable vector of malaria in Aranyaprathet, Sa Kaeo Province. Southeast. Asian J. Trop. Med. Public Health. 2001;32:739–744. [PubMed] [Google Scholar]
- 23.Apiwathnasorn C., Prommongkol S., Samung Y., Limrat D., Rojruthai B. Potential for Anopheles campestris (Diptera: Culicidae) to transmit malaria parasites in Pa Rai subdistrict (Aranyaprathet, Sa Kaeo Province), Thailand. J. Med. Entomol. 2002;39:583–586. doi: 10.1603/0022-2585-39.4.583. [DOI] [PubMed] [Google Scholar]
- 24.Saeung A., Otsuka Y., Baimai V., Somboon P., Pitasawat B., Tuetun B., Junkum A., Takaoka H., Choochote W. Cytogenetic and molecular evidence for two species in the Anopheles barbirostris complex (Diptera: Culicidae) in Thailand. Parasitol. Res. 2007;101:1337–1344. doi: 10.1007/s00436-007-0645-1. [DOI] [PubMed] [Google Scholar]
- 25.Brosseau L., Udom C., Sukkanon C., Chareonviriyaphap T., Bangs M.J., Saeung A., Manguin S. A multiplex PCR assay for the identification of five species of the Anopheles barbirostris complex in Thailand. Parasit. Vectors. 2019;12:223. doi: 10.1186/s13071-019-3494-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suwannamit S., Baimai V., Otsuka Y., Saeung A., Thongsahuan S., Tuetun B., Apiwathnasorn C., Jariyapan N., Somboon P., Takaoka H., et al. Cytogenetic and molecular evidence for an additional new species within the taxon Anopheles barbirostris (Diptera: Culicidae) in Thailand. Parasitol. Res. 2009;104:905–918. doi: 10.1007/s00436-008-1272-1. [DOI] [PubMed] [Google Scholar]
- 27.Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 28.Thongsahuan S., Baimai V., Otsuka Y., Saeung A., Tuetun B., Jariyapan N., Suwannamit S., Somboon P., Jitpakdi A., Takaoka H. Karyotypic variation and geographic distribution of Anopheles campestris-like (Diptera: Culicidae) in Thailand. Mem. Inst. Oswaldo Cruz. 2009;104:558–566. doi: 10.1590/S0074-02762009000400004. [DOI] [PubMed] [Google Scholar]
- 29.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 30.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hillis D.M., Bull J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993;42:182–192. doi: 10.1093/sysbio/42.2.182. [DOI] [Google Scholar]
- 33.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okimoto R., Dodgson J.B. Improved PCR amplification of multiple specific alleles (PAMSA) using internally mismatched primers. Biotechniques. 1996;21:20–26. doi: 10.2144/96211bm03. [DOI] [PubMed] [Google Scholar]
- 36.Germer S., Higuchi R. Single-tube genotyping without oligonucleotide probes. Genome Res. 1999;9:72–78. [PMC free article] [PubMed] [Google Scholar]
- 37.Gunathilaka N., Karunaraj P. Identification of sibling species status of Anopheles culicifacies breeding in polluted water bodies in Trincomalee district of Sri Lanka. Malar. J. 2015;14:214. doi: 10.1186/s12936-015-0726-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Namgay R., Pemo D., Wangdi T., Phanitchakun T., Harbach R.E., Somboon P. Molecular and morphological evidence for sibling species within Anopheles (Anopheles) lindesayi Giles (Diptera: Culicidae) in Bhutan. Acta Trop. 2020;207:105455. doi: 10.1016/j.actatropica.2020.105455. [DOI] [PubMed] [Google Scholar]
- 39.Sum J.S., Lee W.C., Amir A., Braima K.A., Jeffery J., Abdul-Aziz N.M., Fong M.Y., Lau Y.L. Phylogenetic study of six species of Anopheles mosquitoes in Peninsular Malaysia based on inter-transcribed spacer region 2 (ITS2) of ribosomal DNA. Parasit. Vectors. 2014;7:309. doi: 10.1186/1756-3305-7-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.