Abstract
Pulmonary involvement occurs in up to 95% of sarcoidosis cases. In this pilot study, we examine lung compartment-specific protein expression to identify pathways linked to development and progression of pulmonary sarcoidosis. We characterized bronchoalveolar lavage (BAL) cells and fluid (BALF) proteins in recently diagnosed sarcoidosis cases. We identified 4,306 proteins in BAL cells, of which 272 proteins were differentially expressed in sarcoidosis compared to controls. These proteins map to novel pathways such as integrin-linked kinase and IL-8 signaling and previously implicated pathways in sarcoidosis, including phagosome maturation, clathrin-mediated endocytic signaling and redox balance. In the BALF, the differentially expressed proteins map to several pathways identified in the BAL cells. The differentially expressed BALF proteins also map to aryl hydrocarbon signaling, communication between innate and adaptive immune response, integrin, PTEN and phospholipase C signaling, serotonin and tryptophan metabolism, autophagy, and B cell receptor signaling. Additional pathways that were different between progressive and non-progressive sarcoidosis in the BALF included CD28 signaling and PFKFB4 signaling. Our studies demonstrate the power of contemporary proteomics to reveal novel mechanisms operational in sarcoidosis. Application of our workflows in well-phenotyped large cohorts maybe beneficial to identify biomarkers for diagnosis and prognosis and therapeutically tenable molecular mechanisms.
Subject terms: Proteome informatics, Pathogenesis, Biomarkers, Translational research, Molecular biology, Proteomics
Introduction
Sarcoidosis is a multisystem immune-mediated disease of unknown cause with widely variable disease manifestations, severity, and outcomes1. It affects 45–300/100,000 individuals in the US, all ages, races, and both sexes2,3. Diagnostic delays are frequent as sarcoidosis is a diagnosis of exclusion, with no confirmatory test currently available. Despite a greater understanding of sarcoidosis pathogenesis4,5, the mechanisms contributing to the heterogeneity of disease manifestations and predictors of disease outcomes are poorly defined6. The annual mortality is approximately 2.8/million people1 and rising. Sarcoidosis-related mortality is attributed to four high-risk manifestations that include: treatment-resistant pulmonary sarcoidosis, multi-organ sarcoidosis, cardiac sarcoidosis, and neurosarcoidosis7. Respiratory failure from progressive pulmonary disease is the leading cause of sarcoidosis-related mortality in the US7,8. While remission is common, it is not known if systemic anti-inflammatory therapy decreases the risk of progressive pulmonary disease. Another current knowledge gap is the absence of validated markers to predict which patients with pulmonary sarcoidosis will progress.
The pathologic hallmark of sarcoidosis is the formation of epithelioid granuloma associated with infiltration of CD4 + T cells and scattered macrophages, giant cells, with CD8 + T cells and B cells around the granuloma9. While the exact details are not known, it appears that exposure to a yet unidentified antigen(s) results in an exuberant adaptive immune response with CD4 + T cells10, regulatory T cells (Tregs), and high levels of Th1 cytokines TNF–α, IFNγ, and IL–2. Additionally, an abnormal innate immune response is seen in bronchoalveolar lavage (BAL) cells in sarcoidosis. A less robust immune response is apparent in remitting disease compared to the exuberant response in progressive sarcoidosis, likely due to different T cell populations and abnormal counter-regulatory immune measures. Overall, the immune response is aberrant in sarcoidosis and compartmentalized to the lung with much higher response noted in the lung cells compared to blood cells11,12. The whole blood transcriptional profile of active sarcoidosis overlaps with tuberculosis and chronic beryllium disease, and inactive sarcoidosis overlaps with controls13,14. Genes with differential expression in sarcoidosis map to IFN-signaling, TLR signaling, and Fcɣ receptor-mediated phagocytosis15,16. In chronic progressive sarcoidosis, the gene expression in peripheral blood mononuclear cells demonstrates differential expression of genes participating in CXCL9 and TCR-mediated responses17. Transcriptional studies in BAL cells revealed that pathways linked to adaptive immune response, T-cell signaling, and chemokine signaling such as IFNγ, IL-12, 1L-17, and IL-23 are involved in sarcoidosis18. In lung tissue, gene networks engaged in cell movement, immune function, and in Th1-type responses such as signal transducer and activator of transcription 1 (STAT1), IL-5, IL-7, CXCR5, and CXCR9 were overexpressed in sarcoidosis lung tissues11. However, the approach of examining comprehensive protein changes that result from these differences in transcription is underutilized and has not been well evaluated using contemporary techniques.
Prior studies have used protein microarrays19,20, 2-dimensional electrophoresis (2DE)12,21–23, and top-down24 as well as shotgun proteomics25–27 to examine variable sarcoidosis phenotypes including Lofgren’s syndrome, non-Lofgren’s chest x-ray (CXR) stage I, and stage II/III pulmonary sarcoidosis and compared them to subjects with asthma, IPF, tuberculosis or healthy smoking and non-smoking controls. These studies have identified differences in protein spots on 2DE12,21,22, differentially expressed proteins25,26,28 and also possible mechanisms that could explain the development of sarcoidosis25–27. In a large study that utilized SELDI-TOF MS to compare BAL fluid (BALF) from sarcoidosis subjects with Lofgren’s syndrome and different CXR stages of pulmonary sarcoidosis (n = 65) with healthy controls, 40 differentially expressed peaks were identified compared to healthy controls and included 27 peaks that were specific for a particular CXR stage24. A study using affinity planar antigen microarray proteomics examining BALF and reported that mitochondrial ribosomal protein L43, nuclear receptor coactivator 2, adenosine diphosphate-ribosylation factor GTPase activating protein 1 and zinc finger protein 688 demonstrated higher reactivity in sarcoidosis lungs20. Another study reported several differentially expressed BALF proteins in nine sarcoidosis patients with stage II/III sarcoidosis compared to healthy controls analyzed by 2DE followed by MALDI-TOF MS25. The differentially expressed proteins mapped to canonical PI3K/Akt/mTOR signaling, MAP kinase, hypoxia response, and pluripotency-associated transactional factor pathways. These studies support rigorous evaluation of well-characterized, clinically-meaningful sarcoidosis phenotypes by contemporary techniques to identify novel mechanisms of sarcoidosis which can provide tenable treatment targets and biomarkers for personalized care.
Our goal is to couple contemporary proteomics with data-driven analytics for unbiased discovery of novel disease mechanisms in pulmonary sarcoidosis and progressive pulmonary disease, a known high-risk manifestation of sarcoidosis. As a critical first step in evaluating the proteome in sarcoidosis, we focus on BAL cells as alveolitis is seen in patients with active pulmonary sarcoidosis and immune cells provide an ex vivo model for biological mechanisms in inflammatory lung diseases. The BALF is the most proximate fluid to the site of injury, and thus has a high likelihood to identify disease-specific and potentially pathogenic changes. For this proof-of-concept study, we performed label-based MS for measuring protein abundance to gain insights into the intracellular protein interactions in sarcoidosis. We also employed label-free quantitative proteomics on BALF from controls and untreated sarcoidosis cases who, on follow-up, either were found to have progressive or non-progressive pulmonary disease. We found significant differences in BALF and cellular proteins between cases and controls and progressive versus non-progressive cases suggesting that this approach may find useful application in larger studies.
Results
We characterized the proteins in BAL cells from four controls and four sarcoidosis cases. There was no difference in age, sex, race and smoking status for the two groups. The BAL leucocyte count was not significantly different but the sarcoidosis cases had more lymphocytes and a lower number of macrophages (Table 1). For the studies in BALF, we examined seven controls and ten sarcoidosis subjects (non-progressive = 5, progressive = 5) prior to initiation of any systemic anti-inflammatory therapy. There was no difference in the age, race, smoking status, BAL leucocytes, neutrophils, or lymphocytes and macrophages (Table 2). At enrollment, the forced vital capacity (FVC), forced expiratory volume in 1 second (FEV1) and diffusing capacity for carbon monoxide (DLCO) were also not different in subjects with progressive vs. non-progressive disease.
Table 1.
Clinical and demographic variables for controls and sarcoidosis subjects for BAL cell studies.
| Controls (n = 4) | Sarcoidosis (n = 4) | p-value* | |
|---|---|---|---|
| Age (years) | 46 (39.5, 46.75) | 39.5 (36, 46.75) | 0.88 |
| Sex (M/F) | 2/2 | 2/2 | 1.00 |
| Race (AA/C) | 0/4 | 0/4 | |
| Smoking (smokers/non-smokers)# | 0/4 | 0/4 | |
| BAL WBC count/μL | 162 (122.5, 213.5) | 83.5 (42.75, 166.3) | 0.20 |
| BAL neutrophils (%) | 0.5 (0.27, 0.5) | 1.0 (1.0, 1.75) | 0.02 |
| BAL lymphocytes (%) | 5.1 (1.95, 9.375) | 33.0 (15.0, 54.25) | 0.03 |
| BAL macrophages (%) | 94.4 (90.35, 97.55) | 65 (44.75, 83.0) | 0.02 |
| Percent predicted FVC | 95 (84, 103.75) | ||
| Percent predicted FEV1 | 93 (85.7, 99.2) | ||
| Percent predicted DLCO | 111 (94.5,121) |
Date presented as median (IQR).
*Mann-Whitney test or Chi-square test.
#All subjects were non-smokers (controls: 1 never smoker, 2 former smoker and 1 prior smoking history not known; cases: 3 never smokers and 1 former smoker).
Table 2.
Clinical and demographic variables for controls and sarcoidosis subjects for BALF studies.
| Controls (n = 7) | Non-progressive (n = 5) | Progressive (n = 5) | p-value | |
|---|---|---|---|---|
| Age (years) | 32.0 (23.0, 54.0) | 52.0 (41.0, 54.5) | 53.0 (465, 54) | < 0.05* |
| Sex (M/F) | 4/3 | 2/3 | 3/2 | 0.78 |
| Race (AA/C) | 0/7 | 1/4 | 0/5 | 0.28 |
| Smoking (smokers/non-smokers)# | 0/7 | 0/5 | 1/4 | 0.28 |
| BAL WBC count/μL | 105.5 (81.75, 163) | 175 (120, 542) | 80 (55, 112) | 0.14 |
| BAL neutrophils (%) | 0.50 (0.2, 1.5) | 0.50 (0.35, 1.3) | 0.2 (0.1, 0.85) | 0.56 |
| BAL lymphocytes (%) | 6.0 (4.2, 9.8) | 6 (1.6, 40) | 5 (3.5, 12) | 0.21 |
| BAL macrophages (%) | 93 (90, 95) | 94 (59, 98) | 94 (87, 96) | 0.22 |
| Percent predicted FVC | 91 (75, 100) | 84 (71, 97) | 0.55 | |
| Percent predicted FEV1 | 92.5 (70.25, 107.3) | 68 (53, 86.5) | 0.11 | |
| Percent predicted DLCO | 93 (84.25, 97.25) | 96 (93, 110.0) | 0.40 |
Date presented as median (IQR).
ANOVA with post hoc Tukey test to compare all pairs of columns.
*Significant difference between controls and progressive group.
#All subjects except one were non-smokers (controls: 3 never smokers, 3 former smokers, 1 prior smoking history not known; non-progressive sarcoidosis: 3 never smokers, 2 former smokers; progressive sarcoidosis: 1 current smoker, 2 former smokers, 2 never smokers).
Cellular proteins differ between sarcoidosis BAL cells and controls
The liquid chromatography (LC)-tandem mass spectrometry (MS/MS) identified 23,837 spectra at the given thresholds; 16,890 (71%) were included in quantitation. From these spectra, we identified 4,365 proteins (Supplemental Table S1; ‘Scaffold export’ tab). These included three proteins from the common Repository of Adventitious Proteins (cRAP) (serum albumin precursor, cluster of trypsin precursor and keratin, type 1 cytoskeletal 9) and 56 proteins that matched to the decoy (reverse) sequences, which were removed from further analysis resulting in identification of 4,306 high-confidence proteins (probability of 99%, Supplemental Table S2; ‘Scaffold-cleaned up’ tab).
We used a stringent permutation testing and identified 272 differentially expressed proteins controlling for an FDR of ≤ 5%, Fig. 1 (Supplemental Table S1; ‘DE Proteins’ tab) between cases and controls. Table 3 lists the differentially expressed proteins that showed the most significant changes. Several other proteins that were differentially expressed included myeloperoxidase, T-cell immune regulator, cathepsin G, integrin subunit beta2, integrin subunit alpha M, myosin light chain, matrix metalloproteinase 9, PI3K regulator subunit, APOE, interleukin-13 receptor alpha 1-binding protein (TRAF3-interacting protein 1), and SERPINA1.
Figure 1.
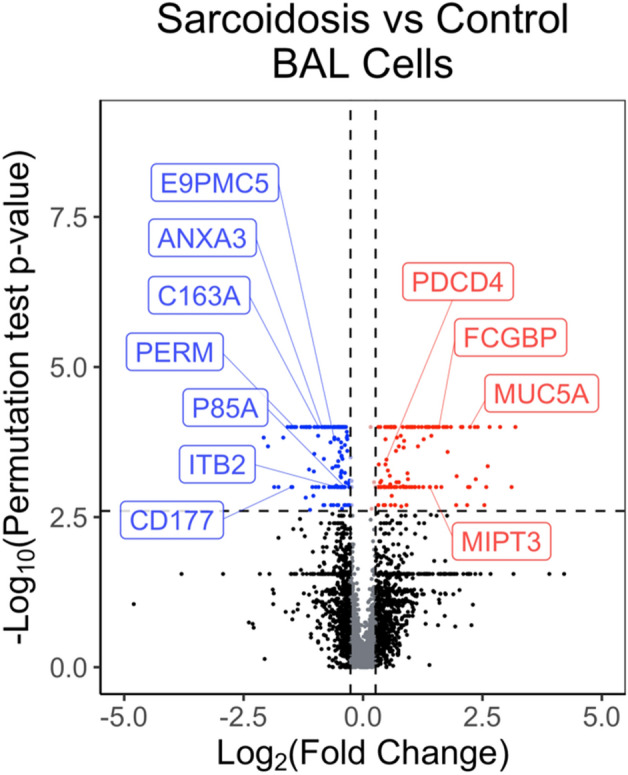
Volcano plot showing the differentially expressed BAL cell proteins. An individual dot represents each protein. The log2 fold change is plotted on the x-axis, and the log2 FDR corrected p-value is plotted on the y-axis. The horizontal dashed line corresponds to statistical significance from the permutation test (B and H corrected p-value = 0.0025) on a numerical scale, and the vertical line corresponds to a 1.2-fold change. The protein depicted by red dots are more abundant in sarcoidosis and the ones in blue dots are more abundant in controls. The black dots indicate the proteins that do not show a statistically significant change. MUC5A Mucin 5A, FCGBP IgG Fc-binding protein, MIPT3 TRAF3-interacting protein (also called Interleukin-13 receptor alpha 1-binding protein), PDCD4 Programmed cell death protein 4, P85A Phosphatidylinositol 3-kinase regulatory subunit alpha, ITB2 Integrin beta-2, E9PMC5 T cell immune regulator 1, ANXA3 Annexin A3, CD163 Scavenger receptor cysteine-rich type 1 protein, CD177 CD177 antigen, PERM Myeloperoxidase.
Table 3.
Top ten differentially expressed cellular proteins comparing controls with sarcoidosis.
| Protein name | B and H corrected p-value | Log2 fold change |
|---|---|---|
| Mucin-5AC | < 0.0001 | 2.24 |
| Glutamate-rich protein 3 | < 0.0001 | 2.65 |
| Long-chain-fatty-acid-CoA ligase 1 | < 0.0001 | − 0.97 |
| Keratin, type I cytoskeletal 18 | < 0.0001 | 1.46 |
| Mucin-5B | < 0.0001 | 1.72 |
| D-3-phosphoglycerate dehydrogenase | < 0.0001 | − 1.48 |
| PDZ and LIM domain protein 1 | < 0.0001 | 1.39 |
| Vinculin | < 0.0001 | − 0.62 |
| Methyl-CpG-binding protein 2 | < 0.0001 | 1.34 |
| Cluster of endoplasmin | < 0.0001 | − 0.55 |
Biological relevance of the differentially expressed proteins in the BAL cells of cases compared to controls
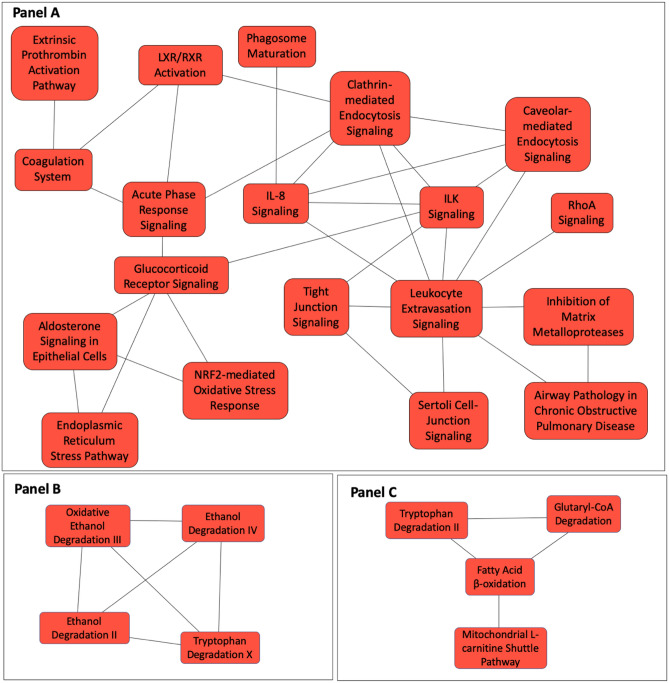
To determine the biological significance of the differentially expressed proteins, we performed IPA core analysis to identify the canonical pathways that map to these proteins. The pathways that met the statistical threshold (− log[p-value] ≥ 1.3) and the proteins assigned to each canonical pathway are listed in Table 4. These include phagosome maturation, leukocyte extravasation signaling, tight junction signaling, ILK signaling, IL-8 signaling, clathrin-mediated endocytosis signaling, caveolin-mediated endocytosis signaling, glucocorticoid receptor signaling, NRF2-mediated oxidative stress response and RhoA signaling (Fig. 2). We also identified pathways linked to matrix turnover and glucocorticoid receptor signaling. Several metabolic pathways such as fatty acid β-oxidation, mitochondrial dysfunction, ethanol degradation, tryptophan metabolism and NRF2-mediated oxidant response also differed between controls and sarcoidosis subjects. The z-score indicating the activation state was available for fatty acid β-oxidation (− 2.5), leukocyte extravasation signaling (− 0.6), coagulation system (− 0.5), inhibition of matrix metalloproteases (1.0), ILK signaling (− 0.4), ethanol degradation (− 1.0), IL-8 signaling (− 1.7) and acute phase response signaling (− 1.9).
Table 4.
Canonical pathways represented by cellular proteins differentially expressed between sarcoidosis and control subjects.
| Ingenuity canonical pathway | log (B-H p-value) | Molecules | Protein names |
|---|---|---|---|
| Fatty acid β-oxidation I | 3.1 | HSD17B10, ACSL3, HSD17B4, HADHA, ACAA2, ACSL1 | Hydroxysteroid 17-beta dehydrogenase 10, Acyl-CoA synthetase long chain family member 3, Hydroxysteroid 17-beta dehydrogenase 4, Hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha, Acetyl-CoA acyltransferase 2, Acyl-CoA synthetase long chain family member 1 |
| Phagosome maturation | 2.58 | DYNC1H1, NSF, MPO, TCIRG1, ATP6V0D1, NCF2, RAB7A, CTSG, RILP, EEA1 | Dynenin cytoplasmic 1 heavy chain 1, N-ethylmaleimide sensitive factor vesicle fusing ATPase, Myeloperoxidase, T cell immune regulator 1, ATPase hydrogen transporting unit v0 subunit d1, Neutrophil cytosolic factor 2, Rab interacting lysosomal protein, Cathepsin G, RAB71 member Ras oncogene family, early endosome antigen 1 |
| Leukocyte extravasation signaling | 2.22 | ITGB2, ITGAM, MYL6, EZR, MMP8, PIK3R1, NCF2, CTNNA1, AFDN, VCL, MMP9 | Integrin subunit beta 2, Integrin subunit alpha M, Myosin light chain 6, Ezrin, Matrix metallopeptidase 8, Phosphoinositide-3-kinase regulatory subunit 1, Neutrophil cytosolic factor 2, Catenin alpha 1, Afadin, Adherens junction formation factor, Vinculin, Matrix metallopeptidase 9 |
| Coagulation system | 2.22 | F5, F13A1, SERPINA1, FGA, A2M | Coagulation factor V, Coagulation factor XIII A chain, Serpin family A member 1, Fibrinogen alpha chain, Alpha-2-macroglobulin |
| Sertoli cell-sertoli cell junction signaling | 2.19 | EPB41, TJP2, CGN, CTNNA1, SPTB, AFDN, SPTA1, VCL, SPTAN1, A2M | Erythrocyte membrane protein band 4.1, Tight junction protein 2, Cingulin, Catenin alpha 1, Spectrin beta, erythrocytic, Afamin, Adherens junction formation factor, Spectrin alpha, Erythrocytic 1, Vinculin, Spectrin alpha, Non-erythrocytic 1, Alpha-2-macroglobulin |
| Inhibition of matrix metalloproteases | 2.19 | HSPG2, ADAM17, MMP8, A2M, MMP9 | Heparan sulfate proteoglycan 2, ADAM metallopeptidase domain 17, Matrix metallopeptidase 8, Alpha-2-macroglobulin, Matrix metallopeptidase 9 |
| Tight junction signaling | 1.92 | EPB41, NSF, TJP2, MYL6, CGN, CTNNA1, AFDN, VCL, SPTAN1 | Erythrocyte membrane protein band 4.1, N-ethylmaleimide sensitive factor, Vesicle fusing ATPase, Tight junction protein 2, Myosin light chain 6, Cingulin, Catenin alpha 1, Afadin, Adherens junction formation factor, Vinculin, Spectrin alpha, Non-erythrocytic 1 |
| Glucocorticoid receptor signaling | 1.74 | HSP90B1, KRT8, PIK3R1, SLPI, KRT18, GTF2E2, KRT5, FKBP5, CD163, HSPA5, A2M, NR3C1, KRT4 | Heat shock protein 90 beta family member 1, Deratin 8, Phosphoinositide-3-kinase regulatory subunit 1, Secretory leukocyte peptidase inhibitor, Keratin 18, General transcription factor IIE subunit 2, Keratin 5, FK506 binding protein 5, CD163 molecule, Heat shock protein family A (Hsp70) member 5, Alpha-2-macroglobulin, nuclear receptor subfamily 3 group C member 1, Keratin 4 |
| ILK signaling | 1.53 | FLNB, ITGB2, MYL6, FLNA, PIK3R1, VIM, KRT18, VCL, MMP9 | Filamin B, Integrin subunit beta 2, Myosin light chain 6, filamin A, Phosphoinositide-3-kinase regulatory subunit 1, Vimentin, Keratin 18, Vinculin, Matrix metallopeptidase 9 |
| Extrinsic prothrombin activation pathway | 1.53 | F5, F13A1, FGA | Coagulation factor V, Coagulation factor XIII A chain, Fibrinogen alpha chain |
| Glutaryl-CoA degradation | 1.53 | HSD17B10, HSD17B4, HADHA | Hydroxysteroid 17-beta dehydrogenase 10, Hydroxysteroid 17-beta dehydrogenase 4, Hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha |
| Ethanol degradation II | 1.53 | ALDH4A1, AKR1A1, ALDH1A1, ACSL1 | Aldehyde dehydrogenase 4 family member A1, Aldo–keto reductase family 1 member A1, Aldehyde dehydrogenase 1 family member A1, Acyl-CoA synthetase long chain family member 1 |
| Mitochondrial L-carnitine shuttle pathway | 1.52 | ACSL3, CPT1A, ACSL1 | Acyl-CoA synthetase long chain family member 3, Carnitine palmitoyltransferase 1A, Acyl-CoA synthetase long chain family member 1 |
| Mitochondrial dysfunction | 1.51 | GSR, HSD17B10, CPT1A, ATP5PO, ACO2, COX5A, VDAC1, UQCRC1 | Glutathione-disulfide reductase, Hydroxysteroid 17-beta dehydrogenase 10, Carnitine palmitoyltransferase 1A, ATP synthase peripheral stalk subunit OSCP, Aconitase 2, Cytochrome c oxidase subunit 5A, Voltage dependent anion channel 1, Ubiquinol-cytochrome c reductase core protein 1 |
| IL-8 signaling | 1.51 | ITGB2, ITGAM, PLD3, MPO, PIK3R1, NCF2, MMP9, LASP1, AZU1 | Integrin β 2, Integrin α M, Phospholipase D member 3, Myeloperoxidase, PI3K regulator subunit, Neutrophil cytosolic factor 2, Matrix metalloproteinase 9, LIM and SH3 protein 1, Azurocidin 1 |
| Aldosterone signaling in epithelial cells | 1.51 | HSP90B1, PIK3R1, DNAJC13, DNAJC3, HSPA5, PI4KA, DNAJB13, AHCY | Heat shock protein 90 beta family member 1, Phosphoinositide-3-kinase regulatory subunit 1, DnaJ heat shock protein family (Hsp40) member C13, DnaJ heat shock protein family (Hsp40) member C3, Heat shock protein family A (Hsp70) member 5, Phosphatidylinositol 4-kinase alpha, DnaJ heat shock protein family (Hsp40) member B13, Adenosylhomocysteinase |
| Folate polyglutamylation | 1.51 | MTHFD1, SHMT2 | Methylenetetrahydrofolate dehydrogenase (cyclohydrolase and formyltetrahydrofolate synthetase 1), Serine hydroxymethyltransferase 2 |
| Acute phase response signaling | 1.48 | ALB, APCS, PIK3R1, SERPINA3, SERPINA1, FGA, NR3C1, A2M | Albumin, Amyloid P component (serum), Phosphoinositide-3-kinase regulatory subunit 1, Serpin family A member 3, Serpin family A member 1, Fibrinogen alpha chain, nuclear receptor subfamily 3 group C member 1, Alpha-2-macroglobulin |
| Caveolar-mediated endocytosis signaling | 1.47 | FLNB, ITGB2, ALB, ITGAM, FLNA | Filamin B, Integrin beta 2, Albumin, Integrin alpha M, Filamin A |
| Oxidative ethanol degradation III | 1.46 | ALDH4A1, ALDH1A1, ACSL1 | Aldehyde dehydrogenase 4 family member A1, Aldehyde dehydrogenase 1 family member A1, Acyl-CoA synthetase long chain family member 1 |
| Endoplasmic reticulum stress pathway | 1.46 | HSP90B1, DNAJC3, HSPA5 | Heat shock protein 90 beta family member 1, DnaJ heat shock protein family (Hsp40) member C3, Heat shock protein family A (Hsp70) member 5 |
| Superpathway of serine and glycine biosynthesis I | 1.31 | PHGDH, SHMT2 | Phosphoglycerate dehydrogenase, Serine hydroxymethyltransferase 2 |
| Tryptophan degradation X (mammalian, via tryptamine) | 1.31 | ALDH4A1, AKR1A1, ALDH1A1 | Aldehyde dehydrogenase 4 family member A1, Aldo–keto reductase family 1 member A1, Aldehyde dehydrogenase 1 family member A1 |
| Tryptophan degradation III (eukaryotic) | 1.31 | HSD17B10, HSD17B4, HADHA | Hydroxysteroid 17-beta dehydrogenase 10, Hydroxysteroid 17-beta dehydrogenase 4, Hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha |
| Ethanol degradation IV | 1.31 | ALDH4A1, ALDH1A1, ACSL1 | Aldehyde dehydrogenase 4 family member A1, Aldehyde dehydrogenase 1 family member A1, Acyl-CoA synthetase long chain family member 1 |
| NRF2-mediated oxidative stress response | 1.3 | GSR, AKR1A1, PIK3R1, DNAJC13, DNAJC3, FKBP5, DNAJB13, FTH1 | Glutathione-disulfide reductase, Aldo–keto reductase family 1 member A1, Phosphoinositide-3-kinase regulatory subunit 1, DnaJ heat shock protein family (Hsp40) member C13, DnaJ heat shock protein family (Hsp40) member C3, FK506 binding protein 5, Ferritin heavy chain 1, DnaJ heat shock protein family (Hsp40) member B13 |
| LXR/RXR activation | 1.3 | APOE, ALB, SERPINA1, FGA, MMP9, CLU | Apolipoprotein E, Albumin, serpin family A member 1, Fibrinogen alpha chain, Matrix metallopeptidase 9, Clusterin |
| Clathrin-mediated endocytosis signaling | 1.3 | APOE, ITGB2, ALB, PIK3R1, RAB7A, SERPINA1, SH3GLB2, CLU | Apoliprotein E, Integrin beta 2, Albumin, PI3K regulator subunit, RAB71 member Ras oncogene family, Serpin family member A1, SH3 domain containing GRB2 like endophilin B2, Clusterin |
| Airway pathology in chronic obstructive pulmonary disease | 1.3 | MMP8, MMP9 | Matrix metallopeptidase 8, Matrix metallopeptidase 9 |
| RhoA signaling | 1.3 | SEPT9, MYL6, EZR, BAIAP2, PI4KA, KTN1 | Septin 9, Myosin light chain 6, Ezrin, BAI1 associated protein 2, Phosphatidylinositol 4-kinase alpha, Kinectin 1 |
Figure 2.
Cellular canonical pathways represented by differentially expressed proteins between sarcoidosis and controls implementing Overlapping Canoncial Pathway functionality in IPA. The 273 differentially expressed proteins map to thirty statistically significant canonical pathways. Each canonical pathway is represented as a node. The edges indicated at least two common proteins between the nodes to indicate shared biological function. Three clusters of overlapping pathways were identified. A larger cluster of overlapping canonical pathways includes diverse biological functions including IL-8, ILK, RhoA signaling, caveolin and clathrin-mediated endocytic signaling, NRF2-mediated oxidant response signaling and glucocorticoid receptor signaling (Panel A). The other two of have limited number of nodes and are involved in metabolic functions (Panels B, C).
Differences in the bronchoalveolar lavage fluid proteins between sarcoidosis and controls and between sarcoidosis phenotypes
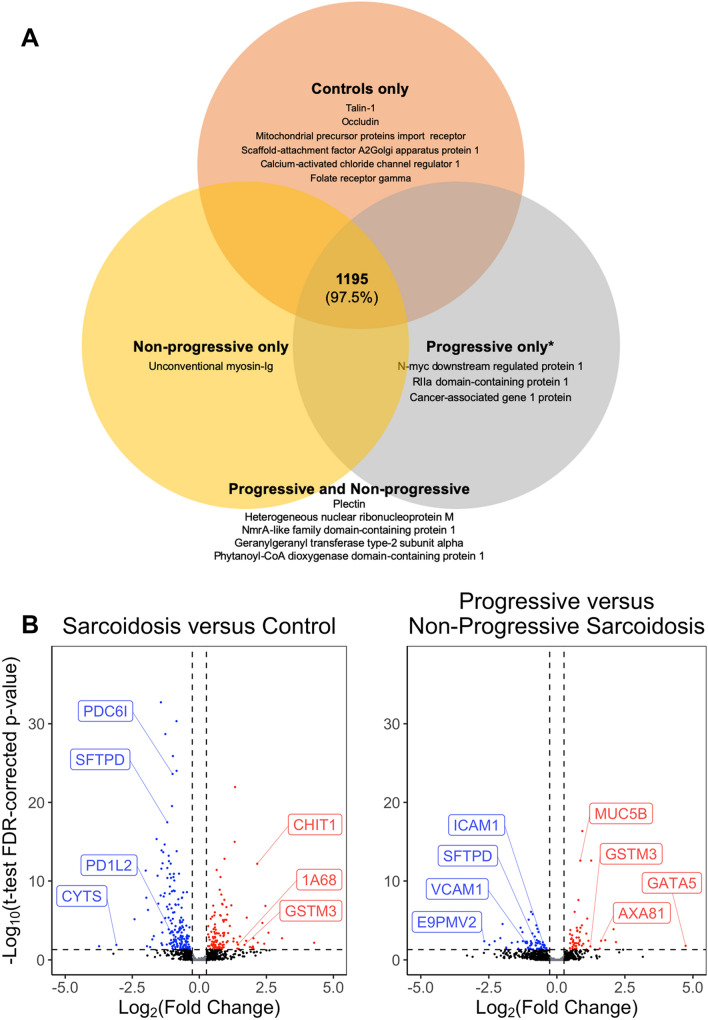
We examined BALF from seven control and ten sarcoidosis subjects. All BALF samples were analyzed by label-free mass spectrometry in triplicates. We identified 1,293 BALF proteins at an FDR of ≤ 1% (Supplemental Table S2; ‘Original File’ tab). These included 62 proteins that matched to the decoy (reverse) sequences or cRAP database such as keratins, filaggrin, cartilage matrix proteins, which were not considered for further analysis. The remaining 1,231 included 1,195 proteins present in all patients and controls. Seven proteins were only detected in controls and not in sarcoidosis cases, while five proteins were present only in sarcoidosis cases but not in control BALF. There were 12 proteins detected in controls and non-progressive cases but not in progressive sarcoidosis, five proteins in control and progressive cases but not in non-progressive sarcoidosis, one protein in only non-progressive but not in controls or progressive sarcoidosis, and four proteins were detected in only progressive but not in non-progressive sarcoidosis or controls (Fig. 3A). Peptides from the 1,231 BALF proteins (Supplemental Table S2; HAP CON REV tab) included proteins that originate from inflammatory cells and epithelial cell such as chitotriosidase-1, macrophage colony stimulating factor, Fc-gamma RIII-alpha, macrophage migration inhibitory factor (macrophage), human neutrophil defensin 3, neutrophil elastase (neutrophils), lymphocyte antigen, lymphocyte cytosolic protein (lymphocytes), aquaporin 1 and 5 (type 1 alveolar epithelial cells), and surfactant protein B (type 2 alveolar epithelial cells). Sixty-nine high abundance and immunoglobulin proteins or immunoglobulin fractions that were not completely removed by the high-abundance protein depletion column were also detected. These proteins were included for functional analysis as these proteins are crucial for many biological functions. Good quality quantitative spectral data was available to compare 1,223 of the 1,231 proteins in sarcoidosis vs. control subjects (Supplemental Table S3; ‘Sarc vs. control’ tab) and 1,206 of 1,231 proteins in progressive vs. non-progressive pulmonary sarcoidosis subjects (Supplemental Table S3; ‘P vs NP’ tab).
Figure 3.
The BALF proteins detected in the controls and sarcoidosis cases. (A) The spectral database search identified 1,231 proteins of which 1,195 were detected in control, progressive and non-progressive subjects. Seven proteins were identified in control subjects but not in sarcoidosis cases. Five protein were present in sarcoidosis cases but not in controls, and four* proteins were detected in progressive sarcoidosis cases. (B) Volcano plot showing the differentially expressed BALF proteins. An individual dot represents each protein. The log2 fold change is plotted on the x-axis, and the log2 FDR corrected p-value is plotted on the y-axis. The horizontal dashed line corresponds to a corrected p-value = 0.05 on a numerical scale, and the vertical line corresponds to a 1.2-fold change. The left panel compares sarcoidosis to controls, and the right panel examines progressive and non-progressive subjects. The proteins depicted by red dots are more abundant in sarcoidosis (left panel), or progressive sarcoidosis (right panel) and have a positive log fold change. The blue dots are more abundant in controls (left panel) or non-progressive sarcoidosis (right panel). The black dots indicate proteins that do not show a statistically significant change. CHIT1 Chitotriosidase, GSTM3 Glutathione-S-transferase, 1A68 HLA class I histocompatibility antigen, SFTPD Pulmonary surfactant-associated protein D, PDC61 Programmed cell death 6-interacting protein, PD1L2 Programmed cell death 1 ligand 2, HMGA1 High mobility group protein HMG-I, CYTS Cystatin-S, VCAM1 Vascular cell adhesion protein, E9PMV2 HLA class II histocompatibility antigen, DQ alpha 1 chain, ICAM1 Intercellular adhesion molecule 1, AXA81 Annexin A8, GATA5 Transcription factor GATA-5, MUC5B Mucin-5B. *One detected protein was an uncharacterized protein.
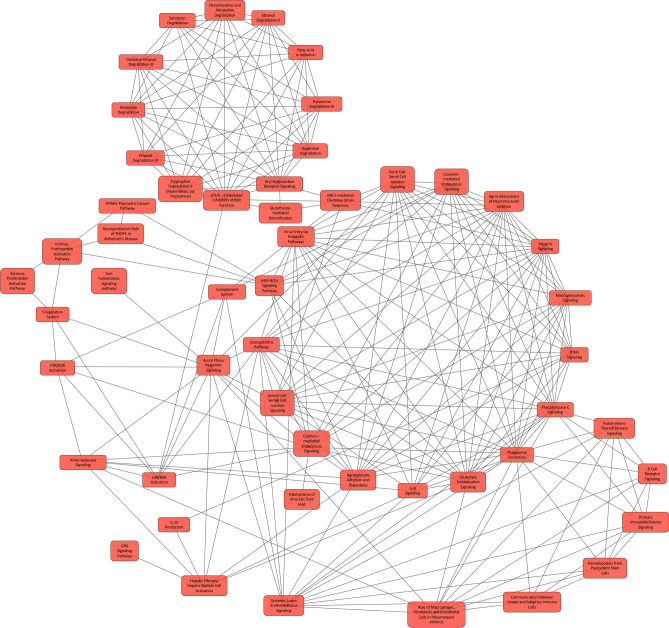
We identified 293 differentially expressed proteins in sarcoidosis (n = 10) compared to the seven control subjects (Supplemental Table S3; Sarcoidosis vs Control tab), Fig. 3B. These proteins included chitotriosidase-1, serum amyloid protein P, surfactant protein D, S100P, inter-alpha-trypsin inhibitor, annexin, glutathione-S-transferase, interleukin-1 receptor accessory protein, cystatin-5, caveolin, choline transport protein, Fc-gamma RII-a, (Fcγ-binding protein), interleukin 6 receptor, programmed cell death 1 ligand 2, and aquaporin-1. The proteins with most significant differences with a higher abundance in sarcoidosis or controls are listed in Table 5. To find the biological relevance of the differentially expressed proteins, we determined the canonical pathways that map to these proteins (Table 6). These pathways include phagosome formation and maturation, IL-8 signaling, IL-12 signaling in macrophages, clathrin and caveolin endocytic signaling, LXR/RXR activation, B cell receptor signaling, communication between innate and adaptive immune cells, aryl hydrocarbon receptor signaling and NRF2-mediated oxidative stress response. Kinases signaling pathways such as PTEN, phospholipase C and GP6 signaling also map to the differentially expressed proteins. Overlapping Canonical Pathway analysis identified highly intricate network of pathways participating in immunological functions, acute phase response and metabolic processes (Fig. 4). The z-score indicating the activation state was available for LXR/RXR activation (2.9), acute phase response signaling (1.39), complement system (− 0.8), coagulation system (− 0.816), agrin interactions at the neuromuscular junction (− 1.633), glutathione-mediate detoxification (1.3), osteoarthritis pathways (− 0.4), SPINK1 pancreatic cancer pathway (1.6), intrinsic prothrombin activation pathway (− 0.5), phospholipase C signaling (− 0.6), serotonin degradation (− 1.3), BAG2 Signaling Pathway (− 1.), neuroprotective role of THOP1 in Alzheimer’s disease (− 2.2), leucocyte extravasation signaling (− 1.4), IL-8 signaling (− 0.4), GP6 signaling Pathway (0.8), PTEN signaling (2.5) and integrin signaling (− 1.9).
Table 5.
Top differentially expressed BALF proteins in sarcoidosis vs controls.
| Uniprot ID | Gene symbol | Protein name | Signal Log-ratio | p-value | p.fdr |
|---|---|---|---|---|---|
| Proteins higher in sarcoidosis cases (compared to control subjects) | |||||
| H3BNG3 | H3BNG3 | 40S ribosomal protein S | 4.28 | 1.13–03 | 6.86–03 |
| M0QZ52 | M0QZ52 | Calmodulin | 3.08 | 2.35–04 | 1.83–03 |
| D6RE83 | D6RE83 | Ubiquitin carboxyl-terminal hydrolase | 2.58 | 3.77–05 | 3.64–04 |
| O60218 | AK1BA | Aldo–Keto Reductase Family 1 | 2.46 | 7.39–09 | 1.46–07 |
| O00764 | PDXK | Pyridoxal Kinase | 2.42 | 1.53–03 | 9.01–03 |
| P02042 | HBD | Hemoglobin Subunit Delta | 2.35 | 1.62–06 | 2.09–05 |
| P07585 | PGS2 | Decorin | 2.18 | 1.04–03 | 6.39–03 |
| Q13231 | CHIT1 | Chitotriosidase-1 | 2.16 | 1.18–14 | 6.59–13 |
| P07451 | CAH3 | Carbonic anhydrase 3 | 2.02 | 7.55–03 | 3.38–02 |
| Q6UWP8 | SBSN | Suprabasin | 2.00 | 4.08–03 | 2.00–02 |
| Proteins higher in controls subjects (compared to sarcoidosis cases) | |||||
| Q53TN4 | CYBR1 | Cytochrome b reductase 1 | − 1.75 | 3.45–04 | 2.53–03 |
| H3BN27 | H3BN27 | Plasmolipin | − 1.83 | 5.08–03 | 2.41–02 |
| C9JKI3 | C9JKI3 | Caveolin | − 1.92 | 2.60–08 | 4.68–07 |
| K7EM38 | K7EM38 | Actin, cytoplasmic 2 | − 1.95 | 3.70–03 | 1.84–02 |
| A0A0C4DGI3 | A0A0C4DGI3 | Citrate synthase | − 1.98 | 5.31–10 | 1.33–08 |
| P20142 | PEPC | Gastricsin | − 2.00 | 1.02–13 | 4.82–12 |
| P06870 | KLK1 | Kallikrein-1 | − 2.42 | 4.84–07 | 7.14–06 |
| P01036 | CYTS | Cystatin 5 | − 3.09 | 2.35–03 | 1.27–02 |
| G3V1K2 | G3V1K2 | Ribitol-5-phosphate xylosyltransferase 1 | − 3.74 | 3.70–03 | 1.84–02 |
| P17096 | HMGA1 | High mobility group protein HMG-I | − 5.10 | 7.45–04 | 4.88–03 |
Fold changes calculated relative to controls resulting in positive log fold change if a protein was more abundant in sarcoidosis subjects and a negative log fold change when it was more abundant in controls.
Signal Log-ratio signal-log ratio (log2 magnitude of change), p-value protein level p-value calculated from beta distribution, p.fdr False discovery rate corrected p-value.
Table 6.
Canonical pathways represented by proteins differentially expressed between sarcoidosis and control subjects in BALF.
| Ingenuity canonical pathways | − log(B-H p-value) | Molecules | Protein name |
|---|---|---|---|
| Acute phase response signaling | 10.1 | A2M, APCS, APOA1, APOH, C3, C9, CP, FGB, FN1, HP, IL1RAP, IL6ST, ITIH2, ITIH3, ITIH4, RRAS, SERPINA1, SERPINA3, SERPINF1, SERPINF2 | Alpha-2-macroglobulin, Serum amyloid P-component, Apolipoprotein A-I, Beta-2-glycoprotein 1, Complement C3, Complement component C9, Fibrinogen beta chain, Fibronectin, Haptoglobin, Interleukin-1 receptor accessory protein, Interleukin-6 receptor subunit beta, Inter-alpha-trypsin inhibitor heavy chain H2, Inter-alpha-trypsin inhibitor heavy chain H3, Inter-alpha-trypsin inhibitor heavy chain H4, Ras-related protein R-Ras, Alpha-1-antitrypsin, Alpha-1-antichymotrypsin, Pigment epithelium-derived factor, Alpha-2-antiplasmin |
| Complement system | 8.26 | C1QB, C1QC, C3, C6, C7, C9, CD55, CFD, CFH, ITGB2 | Complement C1q subcomponent subunit B, Complement C1q subcomponent subunit C, Complement C3, Complement component C6, Complement component C7, Complement component C9, Complement decay-accelerating factor, Complement factor D, Complement factor H, Integrin beta-2 |
| LXR/RXR activation | 6.11 | APOA1, APOA4, APOE, APOH, C3, C9, HADH, IL1RAP, ITIH4, S100A8, SERPINA1, SERPINF1, SERPINF2 | Apolipoprotein A-I, Apolipoprotein A-IV, Apolipoprotein E, Beta-2-glycoprotein 1, Complement C3, Complement component C9, Hydroxyacyl-coenzyme A dehydrogenase (mitochondrial), Interleukin-1 receptor accessory protein, Inter-alpha-trypsin inhibitor heavy chain H4, Protein S100-A8, Alpha-1-antitrypsin, Pigment epithelium-derived factor, Alpha-2-antiplasmin |
| Coagulation system | 3.43 | A2M, FGB, PROS1, SERPINA1,SERPINC1,SERPINF2 | Alpha-2-macroglobulin, Fibrinogen beta chain, Vitamin K-dependent protein S, Alpha-1-antitrypsin, SERPINC1, Alpha-2-antiplasmin |
| FXR/RXR activation | 3.43 | APOA1, APOA4, APOE, APOH, C3, C9, ITIH4, SERPINA1, SERPINF1, SERPINF2 | Apolipoprotein A-I, Apolipoprotein A-IV, Apolipoprotein E, Beta-2-glycoprotein 1, Complement C3, Complement component C9, Inter-alpha-trypsin inhibitor heavy chain H4, Alpha-1-antitrypsin, Pigment epithelium-derived factor, Alpha-2-antiplasmin |
| Phagosome formation | 3.43 | FCGR2A, FN1, IGHG1, IGHG2, IGHG3, ITGA3, ITGB1, ITGB2, MRC1, RHOF | Low affinity immunoglobulin gamma Fc region receptor II-a, Fibronectin, Immunoglobulin heavy constant gamma 1, Immunoglobulin heavy constant gamma 2, Immunoglobulin heavy constant gamma 3, Integrin alpha-3, Integrin beta-1, Integrin beta-2, Macrophage mannose receptor 1, Rho-related GTP-binding protein |
| Agrin interactions at neuromuscular junction | 3.38 | ACTG1, AGRN, DAG1, ITGA3, ITGB1, ITGB2, LAMC1, RRAS | Actin (cytoplasmic 2), Agrin, Dystroglycan, Integrin alpha-3, Integrin beta-1, Integrin beta-2, Laminin subunit gamma-1, Ras-related protein R-Ras2 |
| Caveolar-mediated endocytosis signaling | 2.76 | ACTG1, CAV1, CD55, HLA-A, ITGA3, ITGB1, ITGB2 | Actin (cytoplasmic 2), Caveolin-1, Complement decay-accelerating factor, HLA class I histocompatibility antigen (A alpha chain), Integrin alpha-3, Integrin beta-1, Integrin beta-2 |
| Clathrin-mediated endocytosis signaling | 2.76 | ACTG1, APOA1, APOA4, APOE, ITGB1, ITGB2, MET, S100A8, SERPINA1, TSG101, UBC | Actin (cytoplasmic 2), Apolipoprotein A-I, Apolipoprotein E, Integrin beta-1, Integrin beta-2, Hepatocyte growth factor receptor, Protein S100-A8, Alpha-1-antitrypsin, Tumor susceptibility gene 101 protein, Polyubiquitin-C |
| Primary immunodeficiency signaling | 2.76 | IGHD, IGHG1, IGHG2, IGHG3, IGHM, IGLL1, IGLL5 | Immunoglobulin heavy constant delta, Immunoglobulin heavy constant gamma 1, Immunoglobulin heavy constant gamma 2, Immunoglobulin heavy constant gamma 3, Immunoglobulin heavy constant mu, Immunoglobulin lambda-like polypeptide 1, Immunoglobulin lambda-like polypeptide 1, Immunoglobulin lambda-like polypeptide 5 |
| Glutathione-mediated detoxification | 2.73 | ANPEP, GSTA2, GSTM1, GSTM2, GSTM3 | Aminopeptidase N, Glutathione-S-transferase A2, Glutathione-S-transferase Mu 1, Glutathione-S-transferase Mu 2, Glutathione-S-transferase Mu 3 |
| Virus entry via endocytic pathways | 2.64 | ACTG1, CAV1, CD55, HLA-A, ITGA3, ITGB1, ITGB2, RRAS | Actin (cytoplasmic 2), Caveolin-1, Complement decay-accelerating factor, HLA class I histocompatibility antigen (A alpha chain), Integrin alpha-3, Integrin beta-1, Integrin beta-2, Ras-related protein R-Ras2 |
| Iron homeostasis signaling pathway | 2.64 | ACO2, CD163, CP, CYBRD1, FTH1, HBD, HBG1, HP, LRP1 | Aconitate hydratase (mitochondrial), Scavenger receptor cysteine-rich type 1 protein M130, Fibrinogen beta chain, Cytochrome b reductase 1, Ferritin heavy chain, Hemoglobin subunit delta, Hemoglobin subunit gamma-1, Haptoglobin, Prolow-density lipoprotein receptor-related protein 1 |
| Osteoarthritis pathway | 2.52 | ALPG, ANXA2, DCN, FGFR3, FN1, IL1RAP, ITGA3, ITGB1, LRP1, S100A8, S100A9 | Alkaline phosphatase (germ cell type), Annexin A2, Decorin, Fibroblast growth factor receptor 3, Fibronectin, Interleukin-1 receptor accessory protein, Integrin alpha-3, Integrin beta-1, Prolow-density lipoprotein receptor-related protein 1, Protein S100-A8, Protein S100-A9 |
| SPINK1 pancreatic cancer pathway | 2.49 | CPM, CPN1, CPQ, CTSB, KLK1, KLK11 | Carboxypeptidase M, Carboxypeptidase N catalytic chain, Carboxypeptidase Q, Cathepsin B, Kallikrein-1, Kallikrein-11 |
| Autophagy | 2.48 | CTSB, CTSC, CTSD, CTSH, CTSS, CTSZ | Cathepsin B, Dipeptidyl peptidase 1, Cathepsin D, Pro-cathepsin H, Cathepsin S, Cathepsin Z |
| Hepatic fibrosis/hepatic stellate cell activation | 2.44 | A2M, COL5A1, COL6A1, COL6A3, FGFR2, FN1, ICAM1, IL1RAP, MET, VCAM1 | Alpha-2-macroglobulin, Collagen alpha-1(V) chain, Collagen alpha-1(VI) chain, Collagen alpha-3(VI) chain, Fibroblast growth factor receptor 2, Fibronectin, Intercellular adhesion molecule 1, Interleukin-1 receptor accessory protein, Hepatocyte growth factor receptor, Vascular cell adhesion protein 1 |
| Adenine and adenosine salvage | 2.44 | APRT, PNP | Adenine phosphoribosyltransferase, Purine nucleoside phosphorylase |
| Mechanisms of viral exit from host cells | 2.44 | ACTG1, CHMP2A, PDCD6IP, TSG101, VPS4A | Actin (cytoplasmic 2), Charged multivesicular body protein 2a, Programmed cell death 6-interacting protein, Tumor susceptibility gene 101 protein, Vacuolar protein sorting-associated protein 4A |
| Systemic lupus erythematosus signaling | 2.42 | C6, C7, C9, FCGR2A, HLA-A, IGHG1, IGHG2, IGHG3, IGHM, KLK1, RRAS | Complement component C6, Complement component C7, Complement component C9, Low affinity immunoglobulin gamma Fc region receptor II-a, HLA class I histocompatibility antigen (A alpha chain), Immunoglobulin heavy constant gamma 1, Immunoglobulin heavy constant gamma 2, Immunoglobulin heavy constant gamma 3, Immunoglobulin heavy constant mu, Kallikrein-1, Ras-related protein R-Ras2 |
| Atherosclerosis signaling | 2.42 | APOA1, APOA4, APOE, ICAM1, ITGB2, S100A8, SERPINA1, VCAM1 | Apolipoprotein A-I, Apolipoprotein A-IV, Apolipoprotein E, Intercellular adhesion molecule 1, Integrin beta-2, Protein S100-A8, Alpha-1-antitrypsin, Vascular cell adhesion protein 1 |
| Intrinsic prothrombin activation pathway | 2.42 | FGB, KLK1, KLK11, PROS1, SERPINC1 | Fibrinogen beta chain, Kallikrein-1, Kallikrein-11, Vitamin K-dependent protein S, Antithrombin-III |
| Tryptophan degradation × (Mammalian, via Tryptamine) | 2.32 | AKR1B10, ALDH2, ALDH3B1, ALDH7A1 | Aldo–keto reductase family 1 member B10, Aldehyde dehydrogenase (mitochondrial), Aldehyde dehydrogenase family 3 member B1, Alpha-aminoadipic semialdehyde dehydrogenase |
| Hematopoiesis from pluripotent stem cells | 2.17 | IGHD, IGHG1, IGHG2, IGHG3, IGHM | Immunoglobulin heavy constant delta, Immunoglobulin heavy constant gamma 1, Immunoglobulin heavy constant gamma 2, Immunoglobulin heavy constant gamma 2, Immunoglobulin heavy constant gamma 3, Immunoglobulin heavy constant mu |
| S-adenosyl-l-methionine biosynthesis | 2.11 | MAT2A, MAT2B | S-adenosylmethionine synthase isoform type-2, Methionine adenosyltransferase 2 subunit beta |
| Aryl hydrocarbon receptor signaling | 2.1 | ALDH2, ALDH3B1, ALDH7A1, CTSD, GSTA2, GSTM1, GSTM2, GSTM3 | Aldehyde dehydrogenase (mitochondrial), Aldehyde dehydrogenase family 3 member B1, Alpha-aminoadipic semialdehyde dehydrogenase, Cathepsin D, Glutathione-S-transferase A2, Glutathione-S-transferase Mu 1, Glutathione-S-transferase Mu 2, Glutathione-S-transferase Mu 3 |
| Phospholipase C Signaling | 2.1 | CALM1 (includes others), FCGR2A, IGHG1, IGHG2, IGHG3, ITGA3, ITGB1, PLD3, PPP1CB, RHOF, RRAS | Calmodulin-1, Low affinity immunoglobulin gamma Fc region receptor II-a, Immunoglobulin heavy constant gamma 1, Immunoglobulin heavy constant gamma 2, Immunoglobulin heavy constant gamma 3, Integrin alpha-3, Integrin beta-1, Phospholipase D3, Serine/threonine-protein phosphatase PP1-beta catalytic subunit, Rho-related GTP-binding protein RhoF, Ras-related protein R-Ras2 |
| LPS/IL-1 mediated inhibition of RXR function | 2.01 | ALDH2, ALDH3B1, ALDH7A1, APOE, FABP4, GSTA2, GSTM1, GSTM2, GSTM3, IL1RAP | Aldehyde dehydrogenase (mitochondrial), Aldehyde dehydrogenase family 3 member B1, Alpha-aminoadipic semialdehyde dehydrogenase, Apolipoprotein E, Fatty acid-binding protein, adipocyte, Glutathione-S-transferase A2, Glutathione- S-transferase Mu 1, Glutathione-S-transferase Mu 2, Glutathione-S-transferase Mu 3, Interleukin-1 receptor accessory protein |
| Ethanol degradation II | 2 | ADH1C, ALDH2, ALDH3B1, ALDH7A1 | Alcohol dehydrogenase 1C, Aldehyde dehydrogenase (mitochondrial), Aldehyde dehydrogenase family 3 member B1, Alpha-aminoadipic semialdehyde dehydrogenase |
| Phagosome maturation | 2 | CTSB, CTSC, CTSD, CTSH,CTSS, CTSZ, HLA-A, TSG101 | Cathepsin B, Dipeptidyl peptidase 1, Cathepsin D, Pro-cathepsin H, Cathepsin S, Cathepsin Z, HLA class I histocompatibility antigen (A alpha chain), Tumor susceptibility gene 101 protein |
| Extrinsic prothrombin activation pathway | 1.89 | FGB, PROS1, SERPINC1 | Fibrinogen beta chain, Vitamin K-dependent protein S, Antithrombin-III |
| Noradrenaline and adrenaline degradation | 1.89 | ADH1C, ALDH2, ALDH3B1, ALDH7A1 | Alcohol dehydrogenase 1C, Aldehyde dehydrogenase (mitochondrial), Aldehyde dehydrogenase family 3 member B1, Alpha-aminoadipic semialdehyde dehydrogenase |
| Histamine degradation | 1.82 | ALDH2, ALDH3B1, ALDH7A1 | Aldehyde dehydrogenase (mitochondrial), Aldehyde dehydrogenase family 3 member B1, Alpha-aminoadipic semialdehyde dehydrogenase |
| Communication between innate and adaptive immune cells | 1.75 | HLA-A, IGHD, IGHG1, IGHG2, IGHG3, IGHM | HLA class I histocompatibility antigen (A alpha chain), Immunoglobulin heavy constant delta, Immunoglobulin heavy constant gamma 1, Immunoglobulin heavy constant gamma 2, Immunoglobulin heavy constant gamma 3, Immunoglobulin heavy constant mu |
| Germ cell-sertoli cell junction signaling | 1.71 | A2M, ACTG1, CDH1, ITGA3, ITGB1, NECTIN2, RHOF, RRAS | Alpha-2-macroglobulin, Actin (cytoplasmic 2), Cadherin-1, Integrin alpha-3, Integrin beta-1, Nectin-2, Rho-related GTP-binding protein RhoF, Ras-related protein R-Ras2 |
| serotonin degradation | 1.71 | ADH1C, ALDH2, ALDH3B1, ALDH7A1, B4GAT1 | Alcohol dehydrogenase 1C, Aldehyde dehydrogenase (mitochondrial), Aldehyde dehydrogenase family 3 member B1, Alpha-aminoadipic semialdehyde dehydrogenase, Beta-1,4-glucuronyltransferase 1 |
| Oxidative ethanol degradation III | 1.71 | ALDH2, ALDH3B1, ALDH7A1 | Aldehyde dehydrogenase (mitochondrial), Aldehyde dehydrogenase family 3 member B1, Alpha-aminoadipic semialdehyde dehydrogenase |
| Fatty acid α-oxidation | 1.67 | ALDH2, ALDH3B1, ALDH7A1 | Aldehyde dehydrogenase (mitochondrial), Aldehyde dehydrogenase family 3 member B1, Alpha-aminoadipic semialdehyde dehydrogenase |
| Pyruvate fermentation to lactate | 1.64 | LDHA, LDHB | L-lactate dehydrogenase A chain, L-lactate dehydrogenase B chain |
| Glycogen biosynthesis II (from UDP-d-glucose) | 1.64 | GBE1, UGP2 | 1,4-alpha-glucan-branching enzyme, UTP–glucose-1-phosphate uridylyltransferase |
| Putrescine degradation iii | 1.64 | ALDH2, ALDH3B1, ALDH7A1 | Aldehyde dehydrogenase (mitochondrial), Aldehyde dehydrogenase family 3 member B1, Alpha-aminoadipic semialdehyde dehydrogenase |
| BAG2 signaling pathway | 1.64 | ANXA2, CTSB,HSPA4, HSPA5 | Annexin A2, Cathepsin B, Protein SPA1-RELATED 4, Endoplasmic reticulum chaperone BiP |
| Role of macrophages, fibroblasts and endothelial cells in rheumatoid arthritis | 1.63 | CALM1 (includes others), FN1, ICAM1, IGHG1, IGHG2, IGHG3, IL1RAP, IL6ST, LRP1, RRAS, VCAM1 | Calmodulin-1, Low affinity immunoglobulin gamma Fc region receptor II-a, Fibronectin, Intercellular adhesion molecule 1, Immunoglobulin heavy constant gamma 1, Immunoglobulin heavy constant gamma 3, Interleukin-1 receptor accessory protein, Interleukin-6 receptor subunit beta, Prolow-density lipoprotein receptor-related protein 1, Ras-related protein R-Ras2, Vascular cell adhesion protein 1 |
| B cell receptor signaling | 1.59 | CALM1 (includes others), FCGR2A, IGHD, IGHG1, IGHG2, IGHG3, IGHM, RRAS | Calmodulin-1, Low affinity immunoglobulin gamma Fc region receptor II-a, Low affinity immunoglobulin gamma Fc region receptor II-a, Immunoglobulin heavy constant gamma 1, Immunoglobulin heavy constant gamma 2, Immunoglobulin heavy constant gamma 3, Immunoglobulin heavy constant mu, Ras-related protein R-Ras2 |
| Sertoli cell-sertoli cell junction signaling | 1.59 | A2M, ACTG1, CDH1, F11R, ITGA3, ITGB1, NECTIN2, RRAS | Alpha-2-macroglobulin, Actin (cytoplasmic 2), Cadherin-1, Junctional adhesion molecule A, Integrin alpha-3, Integrin beta-1, Nectin-2, Ras-related protein R-Ras2 |
| Macropinocytosis signaling | 1.59 | ITGB1, ITGB2, MET, MRC1, RRAS | Integrin beta-1, Integrin beta-2, Hepatocyte growth factor receptor, Macrophage mannose receptor 1, Ras-related protein R-Ras2 |
| Ethanol degradation IV | 1.58 | ALDH2, ALDH3B1, ALDH7A1 | Aldehyde dehydrogenase (mitochondrial), Aldehyde dehydrogenase family 3 member B1, Alpha-aminoadipic semialdehyde dehydrogenase |
| NRF2-mediated oxidative stress response | 1.57 | ACTG1, FTH1, GSTA2, GSTM1, GSTM2, GSTM3, RRAS, USP14 | Actin (cytoplasmic 2), Ferritin heavy chain, Glutathione-S-transferase A2, Glutathione- S-transferase Mu 1, Glutathione S-transferase Mu 2, Glutathione-S-transferase Mu 3, Ras-related protein R-Ras2, Ubiquitin carboxyl-terminal hydrolase 14 |
| Aspartate degradation II | 1.57 | GOT2, MDH2 | Aspartate aminotransferase (mitochondrial), Malate dehydrogenase (mitochondrial) |
| TCA cycle II (eukaryotic) | 1.56 | ACO2, CS, MDH2 | Aconitate hydratase (mitochondrial), Citrate synthase (mitochondrial), Malate dehydrogenase (mitochondrial) |
| Agranulocyte adhesion and diapedesis | 1.56 | ACTG1, FN1, ICAM1, ITGA3, ITGB1, ITGB2, PECAM1, VCAM1 | Actin (cytoplasmic 2), Fibronectin, Intercellular adhesion molecule 1, Integrin alpha-3, Integrin beta-1, Integrin beta-2, Platelet endothelial cell adhesion molecule, Vascular cell adhesion protein 1 |
| Autoimmune thyroid disease signaling | 1.56 | HLA-A, IGHG1, IGHG2, IGHG3 | HLA class I histocompatibility antigen (A alpha chain), Immunoglobulin heavy constant gamma 1, Immunoglobulin heavy constant gamma 2, Immunoglobulin heavy constant gamma 3 |
| Neuroprotective role of THOP1 in Alzheimer's disease | 1.53 | CFD, HLA-A, KLK1, KLK11, PRSS8, SERPINA3 | Complement factor D, HLA class I histocompatibility antigen (A alpha chain), Kallikrein-1, Kallikrein-11, Prostasin, Alpha-1-antichymotrypsin |
| Leukocyte extravasation signaling | 1.51 | ACTG1, F11R, ICAM1, ITGA3, ITGB1, ITGB2, PECAM1, VCAM1 | Actin (cytoplasmic 2), Junctional adhesion molecule A, Intercellular adhesion molecule 1, Integrin alpha-3, Integrin beta-1, Integrin beta-2, Platelet endothelial cell adhesion molecule, Vascular cell adhesion protein 1 |
| IL-8 signaling | 1.51 | CDH1, ICAM1, ITGB2, LASP1, PLD3, RHOF, RRAS, VCAM1 | Cadherin-1, Intercellular adhesion molecule 1, Integrin beta-2, LIM and SH3 domain protein 1, Ras-related protein R-Ras2, Vascular cell adhesion protein 1 |
| Glycolysis I | 1.51 | GAPDH, GPI, PFKL | Glyceraldehyde-3-phosphate dehydrogenase, Glucose-6-phosphate isomerase, ATP-dependent 6-phosphofructokinase (liver type) |
| Gluconeogenesis I | 1.51 | GAPDH, GPI, MDH2 | Glyceraldehyde-3-phosphate dehydrogenase, Glucose-6-phosphate isomerase, Malate dehydrogenase (mitochondrial) |
| GP6 signaling pathway | 1.51 | CALM1 (includes others), COL5A1, COL6A1, COL6A3, FGB, LAMC1 | Calmodulin-1, Low affinity immunoglobulin gamma Fc region receptor II-a, Collagen alpha-1(V) chain, Collagen alpha-1(VI) chain, Collagen alpha-3(VI) chain, Fibrinogen beta chain, Laminin subunit gamma-1 |
| IL-15 production | 1.49 | DDR1, EPHB4, FGFR2, FGFR3, MET, ROS1 | Epithelial discoidin domain-containing receptor 1, Ephrin type-B receptor 4, Fibroblast growth factor receptor 2, Fibroblast growth factor receptor 3, Hepatocyte growth factor receptor, Proto-oncogene tyrosine-protein kinase ROS |
| UDP-N-acetyl-d-galactosamine biosynthesis II | 1.43 | GPI, PGM3 | Glucose-6-phosphate isomerase, Phosphoacetylglucosamine mutase |
| PTEN signaling | 1.42 | FGFR2, FGFR3, IGF2R, ITGA3, ITGB1, RRAS | Fibroblast growth factor receptor 2, Fibroblast growth factor receptor 3, Cation-independent mannose-6-phosphate receptor, Integrin alpha-3, Integrin beta-1, Ras-related protein R-Ras2 |
| MSP-RON signaling pathway | 1.38 | ACTG1, ITGB2, KLK1, KLK11 | Actin (cytoplasmic 2), Integrin beta-2, Kallikrein-1, Kallikrein-11 |
| Dopamine degradation | 1.38 | ALDH2, ALDH3B1, ALDH7A1 | Aldehyde dehydrogenase (mitochondrial), Aldehyde dehydrogenase family 3 member B1, Alpha-aminoadipic semialdehyde dehydrogenase |
| Integrin signaling | 1.38 | ACTG1, CAV1, ITGA3, ITGB1, ITGB2, PPP1CB, RHOF, RRAS | Actin (cytoplasmic 2), Caveolin-1, Integrin alpha-3, Integrin beta-1, Integrin beta-2, Serine/threonine-protein phosphatase PP1-beta catalytic subunit, Rho-related GTP-binding protein RhoF, Ras-related protein R-Ras2 |
Figure 4.
The canonical pathway represented by differentially expressed proteins in BALF between sarcoidosis and controls implementing Overlapping Canonical Pathway functionality in IPA. The 293 differentially expressed proteins map to 65 statistically significant canonical pathways. Each canonical pathway is represented as a node. The edges indicated at least two common proteins between the nodes to denote shared biological function. Complex network of pathways with diverse functions including immunological processes, signal transduction by kinases, acute phase response signaling, NRF2-mediated antioxidant response and several metabolic pathways were detected in this analysis.
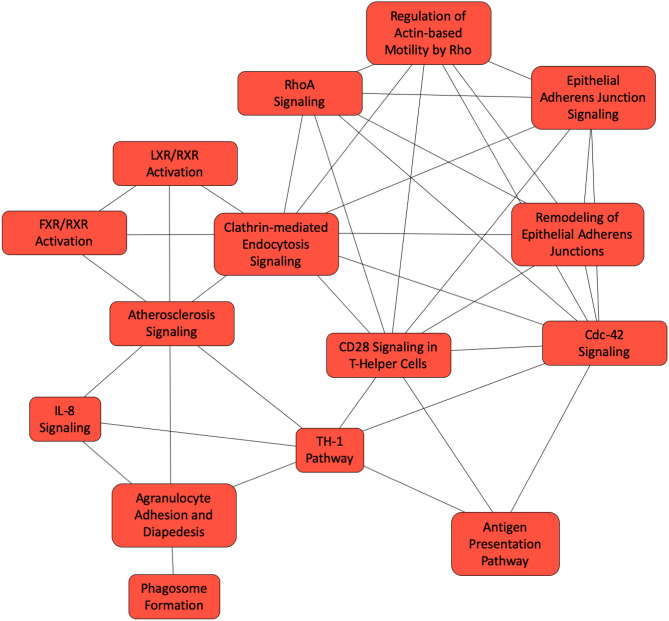
When we compared the BALF proteins between progressive vs. non-progressive sarcoidosis subjects (n = 5 each), there were 121 differentially expressed proteins. The proteins that differed between phenotypes included heat shock protein 90, glutathione-S-transferase, mucin-5B, annexin, CD5 antigen like protein (apoptosis inhibitor expressed by macrophages), chitotriosidase 1, ICAM 1, tropomyosin, integrin beta-2, pulmonary surfactant protein B and D, fatty acid binding protein, and HLA class II histocompatibility antigen DQ-α. The proteins with most significant differences with a higher abundance in cases with progressive disease compared to non-progressive disease are listed in Table 7. To determine the pathways that may contribute to the progression of sarcoidosis, we mapped the differentially expressed proteins between the progressive and non-progressive cases to canonical pathways in IPA (Table 8); these include aryl hydrocarbon receptor signaling, clathrin-mediated endocytic signaling, glutathione redox reaction, glutathione-mediated detoxification, antigen presentation pathway, phagosome formation, CD28 signaling in T-helper cells, CDC-42 signaling, RhoA signaling and PFKFB4 signaling pathway (Fig. 5). The z-score indicating the activation state was available for glycolysis (1.0), LXR/RXR (− 1.6) and IL-8 signaling (1.3).
Table 7.
Top differential expressed BALF proteins comparing progressive to non-progressive cases.
| Uniprot ID | Gene symbol | Protein name | Signal log-ratio | p-value | p.fdr |
|---|---|---|---|---|---|
| Proteins higher in progressive sarcoidosis (compared to non-progressive) | |||||
| Q9BWX5 | GATA5 | Transcription factor GATA-5 | 4.73 | 1.16–03 | 1.64–02 |
| Q96DG6 | CMBL | Carboxymethylenebutenolidase homolog | 2.17 | 2.39–04 | 5.55–03 |
| O00764 | PDXK | Pyridoxal kinase | 2.08 | 1.83–06 | 1.30–04 |
| P68871 | HBB | Hemoglobin subunit beta | 1.78 | 9.67–05 | 3.15–03 |
| Q5VT79 | AXA81 | Annexin A8-like protein 1 | 1.62 | 1.20–04 | 3.79–03 |
| P69905 | HBA | Hemoglobin subunit alpha | 1.58 | 3.21–03 | 3.72–02 |
| P12429 | ANXA3 | Annexin A3 | 1.26 | 6.35–16 | 2.55–13 |
| P21266 | GSTM3 | Glutathione-S-transferase Mu 3 | 1.23 | 1.43–03 | 1.96–02 |
| P08263 | GSTA1 | Glutathione-S-transferase A1 | 1.16 | 1.23–04 | 3.79–03 |
| Q6P5S2 | LEG1H | Protein LEG1 homolog | 1.12 | 3.88–08 | 5.84–06 |
| Proteins higher in non-progressive sarcoidosis (compared to progressive) | |||||
| P11047 | LAMC1 | Laminin subunit gamma-1 | − 1.42 | 2.21–04 | 5.23–03 |
| F8VR50 | F8VR50 | Actin-related protein 2/3complex subunit 3 | − 1.44 | 2.21–04 | 5.23–03 |
| P21695 | GPDA | Glycerol-3-phosphate dehydrogenase | − 1.66 | 3.27–04 | 6.79–03 |
| Q96C19 | EFHD2 | EF-hand domain-containing protein D2 | − 1.86 | 4.01–03 | 4.32–02 |
| P30464 | 1B15 | HLA class I histocompatibility antigen, B alpha chain | − 1.86 | 2.10–03 | 2.72–02 |
| O43866 | CD5L | CD5 antigen-like | − 2.00 | 2.36–07 | 2.84–05 |
| P31025 | LCN1 | Lipocalin-1 | − 2.08 | 4.42–05 | 1.72–03 |
| S4R3A2 | S4R3A2 | Fatty acid-binding protein | − 2.29 | 1.55–04 | 4.30–03 |
| P23246 | SFPQ | Splicing factor, proline- and glutamine-rich | − 2.48 | 7.88–04 | 1.23–02 |
| E9PMV2 | E9PMV2 | HLA class II histocompatibility antigen, DQ alpha 1 chain | − 2.67 | 1.61–04 | 4.31–03 |
Fold changes calculated relative to non-progressive sarcoidosis cases resulting in a positive log fold change for proteins higher in progressive sarcoidosis and a negative log fold change for proteins higher in non-progressive sarcoidosis cases.
Signal Log-ratio signal-log ratio (log2 magnitude of change), p-value protein level p-value calculated from beta distribution, p.fdr false discovery rate corrected p-value.
Table 8.
Canonical pathways mapping to proteins differentially expressed between progressive and non-progressive sarcoidosis.
| Ingenuity canonical pathways | -log (B-H p-value) | Molecules | Protein names |
|---|---|---|---|
| Atherosclerosis signaling | 4.05 | APOA4, CLU, ICAM1, ITGB2, LCAT, LYZ, PON1, VCAM1 | Apolipoprotein A-IV, Clusterin, Intercellular adhesion molecule 1, Integrin beta-2, Phosphatidylcholine-sterol acyltransferase, Lysozyme C, Serum paraoxonase/arylesterase 1, Vascular cell adhesion protein 1 |
| Aryl hydrocarbon receptor signaling | 3.11 | ALDH9A1, BAX, GSTA1, GSTM3, GSTP1, HSP90AB1, HSPB1 | 4-trimethylaminobutyraldehyde dehydrogenase, Apoptosis regulator BAX, Glutathione-S-transferase A1, Glutathione-S-transferase Mu 3, Glutathione-S-transferase P, Heat shock protein HSP 90-beta, Heat shock protein beta-1 |
| Clathrin-mediated endocytosis signaling | 3.11 | APOA4, ARPC3, ARPC4, CLU, ITGB2, LYZ, PON1, TFRC | Apolipoprotein A-IV, Actin-related protein 2/3 complex subunit 3, Actin-related protein 2/3 complex subunit 4, Clusterin, Integrin beta-2, Lysozyme C, Serum paraoxonase/arylesterase 1, Transferrin receptor protein 1 |
| Glycolysis I | 3.11 | ENO2, GPI, PKM, TPI1 | Gamma-enolase, Glucose-6-phosphate isomerase, Pyruvate kinase PKM, Triosephosphate isomerase |
| l-Cysteine degradation III | 2.82 | GOT1, MPST | Aspartate aminotransferase (cytoplasmic), 3-mercaptopyruvate sulfurtransferase |
| LXR/RXR activation | 2.74 | APOA4, CLU, LCAT, LYZ, PON1, VTN | Apolipoprotein A-IV, Clusterin, Phosphatidylcholine-sterol acyltransferase, Lysozyme C, Serum paraoxonase/arylesterase 1, Vitronectin |
| Complement System | 2.74 | C8A, CFH, CFI, ITGB2 | Complement component C8, Complement factor H alpha chain, Integrin beta-2, Complement factor I, Intercellular adhesion molecule 1, Integrin beta-2 |
| Glutathione redox reactions II | 2.34 | GLRX, PDIA3 | Glutaredoxin-1, Protein disulfide-isomerase A3 |
| Phenylalanine degradation I (aerobic) | 2.34 | PCBD1, QDPR | Pterin-4-alpha-carbinolamine dehydratase, Dihydropteridine reductase |
| Epithelial adherens junction signaling | 2.34 | ARPC3, ARPC4, BAIAP2, CDH1, JUP, TUBB4B | Actin-related protein 2/3 complex subunit 3, Actin-related protein 2/3 complex subunit 4, Brain-specific angiogenesis inhibitor 1-associated protein 2, Cadherin-1, Junction plakoglobin, Tubulin beta-4B chain |
| Gluconeogenesis | 2.07 | ENO2, GPI, MDH1 | Gamma-enolase, Glucose-6-phosphate isomerase, Malate dehydrogenase, cytoplasmic |
| FXR/RXR activation | 1.95 | APOA4, CLU, LCAT, PON1, VTN | Apolipoprotein A-IV, Clusterin, Phosphatidylcholine-sterol acyltransferase, Serum paraoxonase/arylesterase 1, Vitronectin |
| Aspartate degradation II | 1.95 | GOT1, MDH1 | Aspartate aminotransferase (cytoplasmic), Malate dehydrogenase, cytoplasmic |
| Remodeling of epithelial adherens junctions | 1.95 | ARPC3, ARPC4, CDH1, TUBB4B | Actin-related protein 2/3 complex subunit 3, Actin-related protein 2/3 complex subunit 4, Cadherin-1, Tubulin beta-4B chain |
| IL-8 signaling | 1.93 | BAX, CDH1, ICAM1, ITGB2, LASP1, VCAM1 | Apoptosis regulator BAX, Cadherin-1, Intercellular adhesion molecule 1, Integrin beta-2, LIM and SH3, Vascular cell adhesion protein 1 |
| Glutathione-mediated Detoxification | 1.93 | GSTA1, GSTM3, GSTP1 | Glutathione-S-transferase A1, Glutathione-S-transferase Mu 3, Glutathione-S-transferase P |
| Antigen presentation pathway | 1.73 | HLA-B, HLA-DQA1, PDIA3 | Protein disulfide-isomerase A3, HLA class II histocompatibility antigen (DQ alpha 1 chain), Protein disulfide-isomerase A3 |
| Regulation of actin-based motility by Rho | 1.62 | ARPC3, ARPC4, BAIAP2, GSN | Actin-related protein 2/3 complex subunit 3, Actin-related protein 2/3 complex subunit 4, Brain-specific angiogenesis inhibitor 1-associated protein 2, Gelsolin |
| Glycogen degradation II | 1.6 | PGM1, TYMP | Phosphoglucomutase-1, Thymidine phosphorylase |
| PFKFB4 signaling pathway | 1.6 | GPI, HK3, TKT | Glucose-6-phosphate isomerase, Hexokinase-3, Transketolase |
| CDC-42 signaling | 1.59 | ARPC3, ARPC4, BAIAP2, HLA-B, HLA-DQA1 | Actin-related protein 2/3 complex subunit 3, Actin-related protein 2/3 complex subunit 4, Brain-specific angiogenesis inhibitor 1-associated protein 2, Protein disulfide-isomerase A3, HLA class II histocompatibility antigen (DQ alpha 1 chain) |
| Glycogen degradation III | 1.52 | PGM1, TYMP | Phosphoglucomutase-1, Thymidine phosphorylase |
| Agranulocyte adhesion and diapedesis | 1.37 | FN1, ICAM1, ITGB2, PECAM1, VCAM1 | Fibronectin, Intercellular adhesion molecule 1, Integrin beta-2, Platelet endothelial cell adhesion molecule, Vascular cell adhesion protein 1 |
| CD28 signaling in T helper cells | 1.36 | ARPC3, ARPC4, HLA-B, HLA-DQA1 | Actin-related protein 2/3 complex subunit 3, Actin-related protein 2/3 complex subunit 4, Protein disulfide-isomerase A3, HLA class II histocompatibility antigen (DQ alpha 1 chain) |
| RhoA signaling | 1.36 | ARPC3, ARPC4, BAIAP2, NRP2 | Actin-related protein 2/3 complex subunit 3, Actin-related protein 2/3 complex subunit 4, Brain-specific angiogenesis inhibitor 1-associated protein 2, Neuropilin-2 |
| Th1 pathway | 1.36 | HLA-B, HLA-DQA1, ICAM1, ITGB2 | Protein disulfide-isomerase A3, HLA class II histocompatibility antigen (DQ alpha 1 chain), Intercellular adhesion molecule 1, Integrin beta-2 |
| Phagosome formation | 1.35 | FN1, ITGB2, PDIA3, VTN | Fibronectin, Integrin beta-2, Protein disulfide-isomerase A3, Vitronectin |
| IL-12 signaling and production in macrophages | 1.28 | APOA4, CLU, LYZ, PON1 | Apolipoprotein A-IV, Clusterin, Lysozyme C, Serum paraoxonase/arylesterase 1 |
Figure 5.
The canonical pathway represented by differentially expressed proteins in the BALF between progressive and non-progressive sarcoidosis implementing Overlapping Canonical Pathway functionality in IPA. The 121 differentially expressed proteins map to twenty-seven canonical pathways. Each canonical pathway is represented as a node. The edges indicated at least two proteins between the nodes to indicate shared biological function. The Th1 pathway, CD28 signaling, CDC-42 signaling and IL-8 signaling are highly-connected nodes detected with this analysis.
Discussion
Use of ‘omics’ tools to improve the understanding of sarcoidosis has been recognized as a high priority area of research in sarcoidosis29. We implemented an approach that coupled state-of-the-art mass spectrometry based proteomics with novel bioinformatics for a comprehensive characterization of the protein changes in the lung compartment in well-phenotyped cases. In the absence of well-characterized animal models, the examination of BAL cells provides an ex vivo model of the immune response in sarcoidosis. While proteomic studies have been conducted previously23,26,28, no prior study has comprehensively characterized mixed BAL cells and BALF. In addition, we established workflows for comprehensive characterization of BALF to obtain unprecedented coverage and detect proteins that originate from diverse cellular and extracellular sources. Ultimately, our approach to characterize mixed BAL cells captured the complex interplay between inflammatory cells in sarcoidosis. Specifically, in BAL cells and fluid we identified several pathways present in macrophages such as clathrin-mediated endocytic signaling and other phagocytic processes as well as redox-related pathways that were previously reported to be upregulated in sarcoidosis23,30. We also identified novel pathways implicated in sarcoidosis such as signaling by integrin-linked kinase, IL-8, and caveolar-mediated endocytic signaling in our studies comparing BAL cells from controls and sarcoidosis cases. The studies in BALF showed higher levels of chitotriosidase, a potential biomarker and an investigational agent for therapy31,32 when comparing cases to controls. Several of the biological pathways identified in the BAL cells were also identified in the BALF, suggesting that BALF is a useful biofluid to investigate mechanistic processes in sarcoidosis. In our comparison of cases with progressive vs. non-progressive sarcoidosis, we identified several novel pathways that may be involved in progression in sarcoidosis. These included CD28 signaling and PFKFB4 signaling. These results suggest that a systematic characterization of BALF may prove fruitful to develop disease models and classifiers with diagnostic and prognostic utility, while BALF and the cellular proteome will provide insight into the mechanisms underlying sarcoidosis as well as the processes that promote progressive disease.
We examined BAL cells as the inflammatory response is aberrant in sarcoidosis with (a) yet unknown antigen(s) triggering an exuberant although dysfunctional immune response with CD4 + T cells, Tregs, high levels of Th1 cytokines TNF-α, IFNγ, and IL-210,33,34, along with inappropriate counter regulatory responses. Previous studies investigating protein changes in alveolar macrophages23,26 and gene expression changes in peripheral blood mononuclear cells35 found phagocytosis-related pathways to be upregulated in sarcoidosis subjects such as Fcγ receptor-mediated phagocytosis and clathrin-mediated endocytic signaling. We identified differences in cellular proteins mapping to phagosome maturation and clathrin-mediated endocytic signaling in sarcoidosis vs. controls BAL cells. Phagocytosis is crucial for innate and adaptive immune response and plays an essential role in antigen presentation, supporting the notion that sarcoidosis results from the response to an unknown external exposure requiring antigen processing and presentation for the development of disease. Similar to previous reports, we observed that the proteins involved in clathrin-mediated endocytic signaling differ in sarcoidosis cases when compared to controls. Additionally, caveolar-mediated endocytic signaling was also different between the two comparison groups. While both these pathways play a role in endocytic internalization of a variety of particles, again implicating exposure in disease ontogeny, these pathways also play a role in signal transduction and the regulation of many plasma membrane activities that have not been studied in sarcoidosis as well as have an influence on the immune response in alveolar macrophages36,37 and peripheral blood mononuclear cells38. In fact, the role of clathrin and caveolar pathways in the development of sarcoidosis has not been systematically studied. Thus, our findings suggest new pathways for investigation of potential disease pathogenesis and or cell regulation in sarcoidosis.
With an unbiased approach, we identified several canonical pathways mapping differentially expressed proteins that have not been previously linked to sarcoidosis, but would be likely to play a role in disease pathogenesis. These include integrin-linked kinase (ILK) signaling, IL-8 signaling, and inhibition of matrix metalloproteinases. ILK is an intracellular protein that primarily functions to connect integrins to the cytoskeletal proteins. The intracellular domain of ILK interacts with different proteins and regulates the phosphorylation of protein kinase B (PI3K)/AKT1 and glycogen synthase kinase 3B39. The downstream signaling cascade of PI3K/AKT activation includes activation of mTOR25, which is implicated in the development and the progression of sarcoidosis and has been proposed as a potential therapeutic target40. Thus, ILK-mediated mTOR activation could be a possible mechanism mediating inflammation in a subset of sarcoidosis cases. ILK signaling also activates c-Jun N-terminal kinase (JNK) via transcription-factor activator protein 1 (AP1) and regulates the gene expression of MMP941 and also IL-8 signaling42. IL-8 is a chemokine in the CXC family and is produced by non-leucocytic and leucocytic cells including macrophages, and binds to CXCR1 and CXCR2 surface receptors43. Several cytokines such as TNF-α induce the production of IL-844. Higher levels of IL-8 have been reported sarcoidosis BALF45 and serum, with the latter correlating with pulmonary46 and chronic disease47. IL-8 signaling has recently been reported to directly regulate adaptive T cell reactivity48 and phagosome function. Thus, our findings are not surprising but suggest that future studies investigating IL-8 signaling could improve the understanding of sarcoidosis pathogenesis and potentially phenotypes. They also highlight the importance of comprehensive characterization of the BAL cell protein changes in providing insight into sarcoidosis development and or progression, an approach that offers promise and is underutilized thus far in sarcoidosis research.
The examination of BALF revealed many proteins that are represented by canonical pathways that were also found in BAL cells. This indicates that biological mechanisms that contribute to the development of sarcoidosis can be identified in the BALF. When we compared BALF from sarcoidosis subjects to controls, similar to the findings from BAL cells, we identified several pathways that are linked to the inflammatory response. These included phagosome formation/maturation, clathrin- and caveolar-mediated endocytic signaling, LXR/RXR activation, IL-8 signaling, fatty acid oxidation, NRF2-mediated oxidative stress response and tryptophan degradation. Several of these pathways are also assigned to the proteins that are differentially expressed between progressive and non-progressive sarcoidosis cases. Some BALF pathways map to proteins that are only differentially expressed between progressive and non-progressive sarcoidosis. Specifically, we identified proteins mapping to CD28 signaling in T-helper cells, PFKFB4 (6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4) signaling and IL-12 signaling and production in macrophages. CD28 is a stimulatory immune checkpoint molecule of B7-CD28 superfamily with diverse roles in naïve and CD4 + T cells. The cytoplasmic tail of CD28 contains signaling motifs that are phosphorylated in response to TCR and CD28 stimulation49. Binding of the adaptor proteins to the activated motif, in turn phosphorylates and activates CDC-4250, culminating in the activation of JNK51. While we did not identify enrichment of canonical JNK pathways, BALF may only reflect some of the processes involved in sarcoidosis pathogenesis with secreted proteins. Regardless, the finding of differentially expressed BALF proteins mapping to CDC-42 and CD28 signaling suggests that they may possibly be involved in disease progression. Additionally, CD28 controls differentiation of Tregs from naïve CD4 T cells, providing novel mechanisms that may explain progression or remission of sarcoidosis. Interestingly, we identified PFKFB4 and IL-12 signaling also mapping to proteins that are differentially expressed in progressive vs. non-progressive cases. PFKFB4 is a bifunctional glycolytic enzyme that synthesizes and degrades fructose 2,6,-biphosphate. PFKFB4 regulates glucose metabolism and cell fate of dendritic cells52 and may provide a link for immunomodulatory effects by 1,25-dihydroxyvitamin D3 (1,25 (OH2) D3). Vanhewegan et al., identified PFKFB4 as a master regulator of 1,25 (OH)2 D3 induced DC tolerogenicity and inhibition of PFKFB4 signaling promotes secretion of proinflammatory cytokines including TNF-α53. The exact role of these pathways in the progression of pulmonary disease remains to be investigated, but our study suggests further investigation should be undertaken.
In pulmonary sarcoidosis, higher oxidant stress is reported in inflammatory cells in the lung54 and BALF55. In our study, the examination of mixed BAL cells indicated altration in redox balance in newly-diagnosed sarcoidosis subjects. Specifically, the mitochondrial l-carnitine shuttle pathway which is involved in fatty acid and lipid degradation, was mapped by proteins with differential abundance in controls compared to sarcoidosis, suggesting that the mitochondrial metabolism is altered. Furthermore, we found differentially expressed proteins in pathways related to β-oxidation of fatty acids and mitochondrial dysfunction. We also identified several cytoprotective enzymes that mapped to NRF2 mediated oxidative stress response were differentially abundant in sarcoidosis compared to controls. NRF2 regulates mitochondrial redox homeostasis by several mechanisms such as detoxification of peroxides, regeneration of GSH, increased synthesis of GSH and NADPH and via the NRF2-Keap 1 response. Mitochondrial dysfunction occurs when the reactive oxygen species (ROS)-mediated stress overpowers the antioxidant defense system56. Bleomycin challenge in NRF2 knockout mice results in increased inflammatory makers, lower level of antioxidant enzymes, a bias towards Th2 response and increased fibrosis57. Taken together, these findings suggest that in sarcoidosis abnormal fatty acid and lipid degradation in the mitochondria cause the production of oxidants, with altered redox balance. It is possible that the detoxification mechanisms are overwhelmed causing mitochondrial dysfunction, production of reactive oxygen species that contribute to the inflammatory response seen in the lungs. NRF2 activators such as curcumin, sulforaphane, resveratrol, and quercetin counteract increased oxidant stress have a potential benefit in acute respiratory distress syndrome58, chronic obstructive pulmonary disease59, asthma60 and idiopathic pulmonary fibrosis61 and could be tested as a possible therapeutic strategy in sarcoidosis. The proteins differentially expressed between controls vs sarcoidosis and progressive vs. non-progressive sarcoidosis cases also mapped to Aryl hydrocarbon receptor signaling. AhR signaling is emerging as an important regulator of immunity in response to endogenous and exogenous ligands62 including tryptophan and serotonin metabolism. The differentially expressed proteins in both of the comparisons mapped to tryptophan /serotonin degradation but only reached statistical significance in the sarcoidosis vs control comparison. AhR signaling controls adaptive immunity by regulating T cell differentiation and by effecting antigen-presenting cells63. AhR regulates T cell response at multiple levels including T cell fate64. AhR is linked to induction of CD4 + Treg or Th1765 and Th22 cell differentiation directing the balance between effector and regulatory T cells. AhR signaling is implicated in other diseases with granulomatous inflammation. In Crohn’s disease, AhR RNA transcripts were markedly downregulated in the inflamed tissue and in the CD4 + T cells66. AhR signaling is also implicated in particulate induced granulomatous inflammation such as silicosis 67.
While we observe differences in the biological processes annotated to the differentially expressed proteins, systematic investigation of the BAL could provide the yet elusive biomarkers with diagnostic and prognostic value in sarcoidosis. In our dataset, we found higher BAL levels of chitotriosidase in sarcoidosis cases compared to controls. Chitotriosidase is a monocyte-macrophage-derived protein that is elevated in plasma and BALF and has been associated with sarcoidosis severity68. Another interesting protein with differential expression higher in the BALF in sarcoidosis compared to controls, programmed cell death 1 ligand 2 (PD-L2), is a ligand for programmed death-1 (PD1) receptor. PD-L2 is a transmembrane protein that is involved in immune checkpoint activity of PD1. In sarcoidosis, PD1 has been linked to the development of T cell exhaustion69 and a blockade of the PD1 pathway restored sarcoidosis CD4 proliferative capacity70. The notion that the PD1 pathway is involved in sarcoidosis is also strengthened by reports of sarcoidosis-like illness in patients receiving PD1 immune checkpoint modulators71. While the presence of individual proteins in our dataset is encouraging, we expect that the systematic examination of global protein changes in the BALF coupled with statistical approaches to construct a parsimonious model consisting of an orthogonal set of proteins will be the best approach for diagnosis and prognostication in sarcoidosis.
A network-based approach is a powerful framework for studying the organizational structure of complex systems72. Networks are represented as a collection of features (nodes) and links (edges) that connect pairs of nodes. The ‘guilt-by-association’ principle73 implies shared biology of pathways. Moreover, biological networks demonstrate scale-free behavior74,75 indicating that they have a relatively large number of low-connectivity nodes and only a few high-connectivity nodes, called ‘hubs,’ that are likely to be essential to network function. In the analysis of cellular proteins, IL-8 signaling, leucocyte extravasation signaling, ILK signaling, glucocorticoid receptor signaling and clathrin-mediated endocytic signaling demonstrated high-connectivity (Fig. 2). The overlapping pathway analysis for the BALF comparison of sarcoidosis and controls identified complex networks with a large number of nodes (Fig. 4). Several immune pathways such as IL-8, leukocyte extravasation signaling, B-cell receptor signaling, phagosome formation, and communication between adaptive and innate immune response signaling demonstrated high-connectivity with each other. Several signal transduction pathways were also highly-connected to immune pathways. Similarly, serval metabolic pathways were highly connected and NRF2 mediated antioxidant response was a ‘node’ that connected the metabolic pathways to immune pathways. Immune pathways were also connected to acute phase response signing mediated by complement and coagulation activation. In the overlapping canonical pathways analysis of BALF proteins in progressive and non-progressive cases, CD28, CDC-42 and IL-8 signaling, and Th1 pathways had high-connectivity suggesting a central role of these pathways in the progression of pulmonary sarcoidosis. Identifying networks of sarcoidosis development and progression in larger samples would allow data partition-based modeling approaches to reveal network topology and may provide valuable insights into disease biology that can not be revealed with conventional reductionist approaches.
Despite the small sample size, we believe our pilot study provides proof of concept for this line of investigation. Our experimental design is robust as we used stringent thresholds for protein identification and a conservative permutation test that decreased the chances of false positives to determine the differentially expressed cellular proteins. Similarly, for the BALF study, we examined each sample in triplicate. We also used a robust PECA procedure that implements algorithms that identifies peptide-level quantitative differences for more robust inferences regarding protein levels76. We expected that this mass spectrometry based bioinformatics workflow would provide a pipeline for application to future large-scale studies in sarcoidosis. A larger sample size would provide more robust inferences regarding the cellular mechanisms of progressive sarcoidosis in a cohort that represents heterogeneity in disease biology and yet allow implementation of resampling methods such as bootstrapping and cross validation for data analysis. We anticipated that workflows developed in this pilot study would identify pathways in peripheral blood mononuclear cells or lymph node tissue, some of which will overlap with the pathways in the lung, as well as some that might differ in direction between BAL and blood or be distinct and apparent in blood only, potentially serving as an easily accessible biomarker. Furthermore, we also hypothesized there would be activation of kinase signal transduction pathways after PBMC recruitment to the lung or other organ and activation of specific canonical and signaling pathways that would govern disease progression or remission.
Conclusions and future directions
The pathophysiologic mechanisms that explain the variability in disease manifestations and course in sarcoidosis are not well understood. A significant challenge is the lack of established disease models that represent the systems contributing to the immune response in sarcoidosis. Single molecule studies are important for understanding the disease biology in sarcoidosis but fail to capture the interactions involved in heterogeneous diseases. Systems levels approaches will be critical to improve our understanding of sarcoidosis. As proteins are the primary effectors of cellular function, characterization of the changes in proteins will be essential to improve our knowledge of sarcoidosis. We established promising proteomics workflows that will be valuable to develop models (classifiers) for diagnosis and prognosis and also identify therapeutically tenable treatment targets in sarcoidosis. Investigating the cellular and BALF protein changes provides an opportunity to examine the complex interplay of protein interactions response for the development and progression of sarcoidosis as well test the validity of protein participating in these biological processes as biomarkers for disease diagnosis and predict progression. The novel mechanisms identified in our pilot study will need to be evaluated with conventional structure function study to determine causal links in sarcoidosis.
Materials and methods
The study was approved by the University of Minnesota (UMN) IRB (protocol number 1501M60321and the National Jewish Health IRB (protocol number HS-2458) and all studies were conducted under the relevant guidelines/regulations. Study participants provided informed consent for the collection of BAL fluid and cells for these studies.
Eligible subjects consisted of individuals with sarcoidosis defined by the criteria outlined in the Joint Statement of American Thoracic Society (ATS), the European Respiratory Society (ERS) and the World Association of Sarcoidosis and Other Granulomatous Disorders (WASOG)3. Subjects without presence of another disease that could significantly affect patient immune response were also enrolled as healthy controls. Bronchoscopy and bronchoalveolar lavage were performed per standard protocol at UMN and NJH77. Four newly diagnosed sarcoidosis subjects were enrolled for examination of BAL cells at UMN (Table 1). Leftover BAL cells from four normal controls were obtained from prior research studies. For the BALF studies, 10 sarcoidosis subjects and 7 healthy controls were enrolled at NJH (Table 2). After collection, the BAL was transported on ice, centrifuged at 500g for 10 min, and the resulting cells and supernatant were stored at − 80 °C using common procedures at the two sites.
For the BALF studies, the sarcoidosis subjects were divided into two distinct phenotypes: those with non-progressive pulmonary disease and those with progressive pulmonary disease using criteria previously established17,78,79. Non-progressive pulmonary disease cases had stable disease and met the following criteria on up to two-years follow-up or more: (1) ≤ 10% decline in FVC or FEV1 and a ≤ 15% decline in DLCO and a stable CXR, and/or (2) ≥ 10% improvement in FVC or a ≥ 15% improvement in DLCO or improved CXR AND (3) no indication for immunosuppressive therapy. Progressive pulmonary disease cases met any of the following criteria from diagnosis/initial evaluation on at least two-year follow-up: (1) ≥ 10% decline in FVC and/or FEV1; or a ≥ 15% decline in DLCO; or (2) worsening CXR as determined by the interpreting radiologist/ investigator; and/or (3) start of immune-suppressive therapy.
Protein isolation and MS spectral-data acquisition
Mixed BAL cells:
BAL cells were resuspended in hypotonic lysis buffer with HALT protease inhibitor cocktail (Thermo Fisher Scientific), and lysed using sequential cell disruption techniques including a freeze–thaw at 98 °C with vortexing and sonication (Sonics) on ice before buffering with 1 M triethylammonium bicarbonate (Sigma). The lysed cells were then centrifuged at 20,000g for 15 minutes and the supernatant was collected for further processing. To increase the protein recovery, the pellet from this step was resuspended in a buffer (containing 7 M urea and 2 M thiourea in 0.4 M triethylammonium bicarbonate at pH 8.5), freeze-thawed, vortexed and centrifuged at 15,000g for 15 min at 20 °C. The supernatants from the two centrifugation steps were combined and concentrated using an Amicon 3-MWCO filter (Millipore). An equal amount of protein was processed for in-gel cleanup and digestion (EMBL Method), reduced with dithiothreitol (Sigma-Aldrich), treated with iodoacetamide (Sigma-Aldrich) to block cysteine residues, digested with trypsin (Promega) and cleaned with an MCX stage tip (3M-Empore 2241). Isobaric labeling of digested peptides was carried out with TMT-10Plex (Thermo Fisher Scientific) reagent followed by MCX and SPE cleanup with appropriate buffer exchanges, and offline fractionation on Shimadzu Prominence with Xbridge 150 × 2.1 mm column (Waters) with two-minute fractions at a flow rate of 200µL/min, and peptide amounts of 15mAU-equivalent aliquots from fractions 7–38 were concatenated. LC–MS data was acquired for each concatenated fraction using an Easy-nLC 1,000 HPLC (Thermo Fisher Scientific, Waltham, MA) in tandem with an Orbitrap Fusion (Thermo Fisher Scientific) MS instrument.
BALF proteins:
The BALF was processed using our previously published protocol80,81. Briefly, BALF was sonicated (Sonics), centrifuged for 15 min at 14,000g at 4 °C and filtered with pre-rinsed (5% methanol and water) syringe (Monoject, Covidien) and 0.22um PES filter to remove remaining particulates. The fluid was then concentrated and desalted using Amicon 3-MWCO filters, and a Bradford assay (Bio-Rad) was used to quantify protein. High-abundance proteins were removed using a Seppro IgY 14 spin-column (Sigma-Aldrich) with appropriate buffer exchanges. Equal amount of enriched medium- and low-abundance protein was processed for in-gel cleanup and digestion similar to the BAL cells above. LC–MS data was acquired for each concatenated fraction using an Easy-nLC 1,000 HPLC in tandem with an Orbitrap Fusion using settings similar to BAL cells analysis with minor differences. The differences were (1) The column was heated to 50 °C and (2) the dynamic exclusion was set to 15 s with a 10-ppm high and low mass tolerance.
Mass spectral dataset analysis by sequence database search for protein identification and quantification
The BAL cell quantification was accomplished using TMT reagent, and the BALF dataset was analyzed using MS1 spectral quantification.
Identification and quantification of TMT-labeled cellular proteins:
The spectral dataset was searched against the target-decoy version of the Human UniProt database (72,886 protein sequences; October 10th 2018) along with the contaminant sequences from the cRAP database (https://www.thegpm.org/crap/). Scaffold Q + (version Scaffold_4.8.9, Proteome Software Inc., Portland, OR) was used to perform TMT-based peptide quantitation and protein identification. The threshold of peptide identifications was set at an FDR < 0.5% by the Scaffold Local FDR algorithm. The protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least one peptide82. Channels were corrected according to the algorithm described in i-Tracker83. Normalization was performed iteratively (across samples and spectra) on intensities, as described in Statistical Analysis of Relative Labeled Mass Spectrometry Data from Complex Samples Using ANOVA84. Medians were used for averaging. Spectra data were log-transformed, pruned off those matched to multiple proteins and those missing a reference value, and weighted by an adaptive intensity-weighting algorithm. Of 23,837 spectra in the experiment at the given thresholds, 16,890 (71%) were included in quantitation. The proteins that matched to the cRAP or the decoy sequence were removed from analysis.
Identification and MS1quant label-free quantification of BALF proteins
Raw files were searched against the target-decoy version of Human UniProt database (73,928 protein sequences, November 21 2019) along with the cRAP database using the MaxQuant 1.6.10.43 algorithm. Default search parameters were used as follows: peptide spectral matching and proteins with 1% FDR modifications include fixed carbamidomethyl of C, variable oxidation of M, and N-terminal acetylation. BALF samples were quantified in label-free quantification (LFQ) mode, and spectra were “matched between runs”.
Statistical analysis
The peptide-level data for the BALF was imported into the GalaxyP (https://galaxyp.org) framework for implementing the Peptide-level Expression Change Averaging (PECA)-procedure76 using the Bioconductor package (https://www.bioconductor.org/packages/release/bioc/html/PECA.html). This method differs from the common approach, where protein expression intensities are precomputed from the peptide data and an expression change between two groups of samples is first calculated for each measured peptide. The corresponding protein-level expression changes are then defined as medians over the peptide-level changes. For this analysis, we determined the modified t-statistic, which is calculated using the linear modeling approach in the Bioconductor limma (linear models for microarray data) package85. To identify differential expression in the BALF dataset, the comparability of relative expression changes between alternative peptides was investigated by considering signal log-ratios by a two-sample t-test with a p-value ≤ 0.05 corrected for multiple hypotheses testing. For the intracellular proteins, given the substantially higher number of proteins detected, we used a conservative permutation test to decrease the possibility of type 1 error rate with an unadjusted significance level p ≤ 0.05 corrected by the Benjamini–Hochberg method for testing multiple hypotheses.
To gain insight into the biological significance of differentially expressed proteins, we performed functional analysis using Ingenuity Pathway Analysis [IPA (IPA QIAGEN, Redwood City https://www.quiagen.com/ingenuty)]. This analysis was performed on proteins with an FDR corrected p-value ≤ 0.05 as the cutoff for differential expression for both BAL cell and fluid datasets. The IPA core analysis was performed using the difference of the weighted log fold change between comparison groups. We focused on canonical pathways that met a Benjamini and Hochberg (B-H)–corrected p-value obtained using the right-tailed Fisher exact test of ≤ 0.05 (equivalent to −log [B-H p-value] ≥ 1.3), as done previously81. We also examined on Overlapping Canonical Pathways functionality in IPA which is designed to visualize the shared biology in pathways through the common features (genes/proteins) in the pathways. The network of overlapping pathways shows each canonical pathway meeting the statistical threshold of −log (B-H p-value) ≥ 1.3 as a single “node”. An edge connects any two pathways when there are at least two common proteins shared between the pathways.
Supplementary information
Acknowledgements
The authors greatly appreciate the patients and the research participants. They also thank the personnel at the Center for Mass Spectrometry and Proteomics, especially LeeAnn Higgins and Todd Markowski for MS analysis. The Minnesota Supercomputing Institute maintains GalaxyP at the University of Minnesota, and the infrastructure is supported by National Cancer Institute Grant U24CA199347 (TJG). Lee S Parsons PhD assisted in implementing Maxquant on Mesabi servers at Minnesota Supercomputing Institute. The project was supported by the University of Minnesota Department of Medicine and the Division of Pulmonary and Critical Care (MB), Sarcoidosis Research and Education Foundation (MB), R01ES023826 (LAM. LL), R01HL140357 (LAM), U01 HL112695 and UL1TR002535 (LAM)
Abbreviations
- BAL
Bronchoalveolar lavage
- BALF
Bronchoalveolar lavage fluid
- TNF
Tumor necrosis factor
- TLR
Toll-like receptor
- IL
Interleukin
- SELDI
Surface-enhanced laser desorption ionization
- TOF
Time-of-flight
- MS
Mass spectrometer
- MALDI
Matrix-assisted laser desorption ionization
- 2 DE
2-Dimension electrophoresis
- CXR
Chest X-ray
- Treg
Regulatory T cells
- FVC
Forced vital capacity
- FEV
Forced expiratory volume
- DLCO
Diffusing capacity for carbon monoxide
- PI3K
Phosphoinositide 3-kinase
- mTOR
Mammalian target of rapamycin complex 1
Author contributions
Study Design (MB, LAM, TEF), statistical analysis and interpretation (MB, LAM, KJV, EJP, RS, LL BPO, TEF), MS data analysis (PDJ, TJG, RS, KJV, MB), enrollment of study subjects and clinical data abstraction (BB, CR, MG, EHD, DMP, MB), manuscript writing and editing (MB, LAM, DMP, TEF, LL, BPO), protein extraction and sample preparation for MS (KJV).
Data availability
The MS proteomics datasets were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD014438 (BAL cells) and PXD016637 (BALF).
Competing interests
CI Restrepo: Dr Restrepo has funding from Mallinckrodt Pharmaceuticals for a randomized double-blind placebo-controlled trial that is not relevant to this manuscript. No other financial conflict of interests to disclose. Maier LA: Dr Maier has funding from Mallinckrodt Pharmaceuticals for a randomized double-blind placebo-controlled trial that is not relevant to this manuscript. No other financial conflict of interests to disclose. Maneesh Bhargava: No conflict of interest. KJ Viken: No conflict of interest. B Barkes: No conflict of interest. TJ Griffin: No conflict of interest. M Gillespie: No conflict of interest. PD Jagtap: No conflict of interest. R Sajulga: No conflict of interest. EJPeterson: No conflict of interest. HE Dincer: No conflict of interest. L Li: No conflict of interest. BP O’Conno: No conflict of interest. TE: Fingerlin: No conflict of interest. DM Perlman: No conflict of interest.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-69281-8.
References
- 1.Gerke AK, Judson MA, Cozier YC, Culver DA, Koth LL. Disease burden and variability in sarcoidosis. Ann. Am. Thorac. Soc. 2017;14:S421–S428. doi: 10.1513/AnnalsATS.201707-564OT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rybicki BA, Major M, Popovich J, Jr, Maliarik MJ, Iannuzzi MC. Racial differences in sarcoidosis incidence: a 5-year study in a health maintenance organization. Am. J. Epidemiol. 1997;145:234–241. doi: 10.1093/oxfordjournals.aje.a009096. [DOI] [PubMed] [Google Scholar]
- 3.Statement on sarcoidosis Joint Statement of the American Thoracic Society (ATS), the European Respiratory Society (ERS) and the World Association of Sarcoidosis and Other Granulomatous Disorders (WASOG) adopted by the ATS board of directors and by the ERS Executive Committee, February 1999. Am. J. Respir. Crit. Care Med. 1999;160:736–755. doi: 10.1164/ajrccm.160.2.ats4-99. [DOI] [PubMed] [Google Scholar]
- 4.Zissel G, Muller-Quernheim J. Cellular players in the immunopathogenesis of sarcoidosis. Clin. Chest Med. 2015;36:549–560. doi: 10.1016/j.ccm.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 5.Schupp JC, Vukmirovic M, Kaminski N, Prasse A. Transcriptome profiles in sarcoidosis and their potential role in disease prediction. Curr. Opin. Pulm. Med. 2017;23:487–492. doi: 10.1097/MCP.0000000000000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maier LA, Crouser ED, Martin WJ, Eu J. Executive summary of the NHLBI workshop report: leveraging current scientific advances to better understand sarcoidosis variability and improve outcomes. Ann. Am. Thorac. Soc. 2017 doi: 10.1513/AnnalsATS.201707-563OT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sauer WH, Stern BJ, Baughman R, Culver DA, Royal W. High risk sarcoidosis: current concepts and research imperatives. Ann. Am. Thorac. Soc. 2017 doi: 10.1513/AnnalsATS.201707-566OT. [DOI] [PubMed] [Google Scholar]
- 8.Swigris JJ, et al. Sarcoidosis-related mortality in the United States from 1988 to 2007. Am. J. Respir. Crit. Care Med. 2011;183:1524–1530. doi: 10.1164/rccm.201010-1679OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gal AA, Koss MN. The pathology of sarcoidosis. Curr. Opin. Pulm. Med. 2002;8:445–451. doi: 10.1097/00063198-200209000-00018. [DOI] [PubMed] [Google Scholar]
- 10.Richmond BW, et al. Sarcoidosis Th17 cells are ESAT-6 antigen specific but demonstrate reduced IFN-gamma expression. J. Clin. Immunol. 2013;33:446–455. doi: 10.1007/s10875-012-9817-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crouser ED, et al. Gene expression profiling identifies MMP-12 and ADAMDEC1 as potential pathogenic mediators of pulmonary sarcoidosis. Am. J. Respir. Crit. Care Med. 2009;179:929–938. doi: 10.1164/rccm.200803-490OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabounchi-Schutt, F., Mikko, M., Eklund, A., Grunewald, J. & J, A. S. Serum protein pattern in sarcoidosis analysed by a proteomics approach. Sarcoidosis Vasc. Diffuse Lung Dis.21, 182–190 (2004). [PubMed]
- 13.Bloom CI, et al. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS ONE. 2013;8:e70630. doi: 10.1371/journal.pone.0070630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, et al. Beryllium-induced lung disease exhibits expression profiles similar to sarcoidosis. Eur. Respir. J. 2016;47:1797–1808. doi: 10.1183/13993003.01469-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su R, et al. Interferon-inducible chemokines reflect severity and progression in sarcoidosis. Respir Res. 2013;14:121. doi: 10.1186/1465-9921-14-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maertzdorf J, et al. Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc. Natl. Acad. Sci. USA. 2012;109:7853–7858. doi: 10.1073/pnas.1121072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su R, et al. Longitudinal analysis of sarcoidosis blood transcriptomic signatures and disease outcomes. Eur. Respir. J. 2014;44:985–993. doi: 10.1183/09031936.00039714. [DOI] [PubMed] [Google Scholar]
- 18.Gharib SA, et al. Sarcoidosis activates diverse transcriptional programs in bronchoalveolar lavage cells. Respir. Res. 2016;17:93. doi: 10.1186/s12931-016-0411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du SS, et al. Screening for differentially expressed proteins relevant to the differential diagnosis of sarcoidosis and tuberculosis. PLoS ONE. 2015;10:e0132466. doi: 10.1371/journal.pone.0132466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haggmark A, et al. Proteomic profiling reveals autoimmune targets in sarcoidosis. Am. J. Respir. Crit. Care Med. 2015;191:574–583. doi: 10.1164/rccm.201407-1341OC. [DOI] [PubMed] [Google Scholar]
- 21.Magi B, et al. Bronchoalveolar lavage fluid protein composition in patients with sarcoidosis and idiopathic pulmonary fibrosis: a two-dimensional electrophoretic study. Electrophoresis. 2002;23:3434–3444. doi: 10.1002/1522-2683(200210)23:19<3434::AID-ELPS3434>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 22.Sabounchi-Schutt F, Astrom J, Hellman U, Eklund A, Grunewald J. Changes in bronchoalveolar lavage fluid proteins in sarcoidosis: a proteomics approach. Eur. Respir. J. 2003;21:414–420. doi: 10.1183/09031936.03.00060902. [DOI] [PubMed] [Google Scholar]
- 23.Silva E, et al. Quantitative intact proteomics investigations of alveolar macrophages in sarcoidosis. Eur. Respir. J. 2013;41:1331–1339. doi: 10.1183/09031936.00178111. [DOI] [PubMed] [Google Scholar]
- 24.Kriegova E, et al. Protein profiles of bronchoalveolar lavage fluid from patients with pulmonary sarcoidosis. Am. J. Respir. Crit. Care Med. 2006;173:1145–1154. doi: 10.1164/rccm.200507-1126OC. [DOI] [PubMed] [Google Scholar]
- 25.Landi C, et al. A functional proteomics approach to the comprehension of sarcoidosis. J. Proteomics. 2015;128:375–387. doi: 10.1016/j.jprot.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 26.Kjellin H, et al. Alterations in the membrane-associated proteome fraction of alveolar macrophages in sarcoidosis. Sarcoidosis Vasc. Diffuse Lung Dis. 2016;33:17–28. [PubMed] [Google Scholar]
- 27.Martinez-Bravo MJ, et al. Pulmonary sarcoidosis is associated with exosomal vitamin D-binding protein and inflammatory molecules. J. Allergy Clin. Immunol. 2017;139:1186–1194. doi: 10.1016/j.jaci.2016.05.051. [DOI] [PubMed] [Google Scholar]
- 28.Silva E, et al. A quantitative proteomic analysis of soluble bronchoalveolar fluid proteins from patients with sarcoidosis and chronic beryllium disease. Sarcoidosis Vasc. Diffuse Lung. Dis. 2007;24:24–32. [PubMed] [Google Scholar]
- 29.Crouser ED, et al. Application of 'Omics' and systems biology to sarcoidosis research. Ann. Am. Thorac. Soc. 2017 doi: 10.1513/AnnalsATS.201707-567OT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koutsokera A, et al. Systemic oxidative stress in patients with pulmonary sarcoidosis. Pulm. Pharmacol. Ther. 2009;22:603–607. doi: 10.1016/j.pupt.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Bargagli E, et al. Chitotriosidase analysis in bronchoalveolar lavage of patients with sarcoidosis. Sarcoidosis. Vasc. Diffuse Lung Dis. 2007;24:59–64. doi: 10.1007/s11083-007-9059-z. [DOI] [PubMed] [Google Scholar]
- 32.Harlander M, Salobir B, Zupancic M, Tercelj M. Bronchoalveolar lavage chitotriosidase activity as a biomarker of sarcoidosis. Sarcoidosis Vasc. Diffuse Lung Dis. 2016;32:313–317. [PubMed] [Google Scholar]
- 33.Chen ES, et al. T cell responses to mycobacterial catalase-peroxidase profile a pathogenic antigen in systemic sarcoidosis. J. Immunol. 2008;181:8784–8796. doi: 10.4049/jimmunol.181.12.8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oswald-Richter KA, et al. Cellular responses to mycobacterial antigens are present in bronchoalveolar lavage fluid used in the diagnosis of sarcoidosis. Infect. Immun. 2009;77:3740–3748. doi: 10.1128/IAI.00142-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Talreja J, et al. RNA-sequencing identifies novel pathways in sarcoidosis monocytes. Sci. Rep. 2017;7:2720. doi: 10.1038/s41598-017-02941-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moulakakis C, et al. Surfactant protein A enhances constitutive immune functions of clathrin heavy chain and clathrin adaptor protein 2. Am. J. Respir. Cell Mol. Biol. 2016;55:92–104. doi: 10.1165/rcmb.2015-0219OC. [DOI] [PubMed] [Google Scholar]
- 37.Harris J, Werling D, Hope JC, Taylor G, Howard CJ. Caveolae and caveolin in immune cells: distribution and functions. Trends Immunol. 2002;23:158–164. doi: 10.1016/S1471-4906(01)02161-5. [DOI] [PubMed] [Google Scholar]
- 38.Jans J, elMoussaoui H, de Groot R, de Jonge MI, Ferwerda G. Actin- and clathrin-dependent mechanisms regulate interferon gamma release after stimulation of human immune cells with respiratory syncytial virus. Virol. J. 2016;13:52. doi: 10.1186/s12985-016-0506-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hannigan GE, McDonald PC, Walsh MP, Dedhar S. Integrin-linked kinase: not so 'pseudo' after all. Oncogene. 2011;30:4375–4385. doi: 10.1038/onc.2011.177. [DOI] [PubMed] [Google Scholar]
- 40.Linke M, et al. Chronic signaling via the metabolic checkpoint kinase mTORC1 induces macrophage granuloma formation and marks sarcoidosis progression. Nat. Immunol. 2017;18:293–302. doi: 10.1038/ni.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu C, Dedhar S. Integrin-linked kinase (ILK) and its interactors: a new paradigm for the coupling of extracellular matrix to actin cytoskeleton and signaling complexes. J. Cell. Biol. 2001;155:505–510. doi: 10.1083/jcb.200108077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oltmanns U, Issa R, Sukkar MB, John M, Chung KF. Role of c-jun N-terminal kinase in the induced release of GM-CSF, RANTES and IL-8 from human airway smooth muscle cells. Br. J. Pharmacol. 2003;139:1228–1234. doi: 10.1038/sj.bjp.0705345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukaida N. Pathophysiological roles of interleukin-8/CXCL8 in pulmonary diseases. Am. J. Physiol. Lung Cell Mol. Physiol. 2003;284:L566–577. doi: 10.1152/ajplung.00233.2002. [DOI] [PubMed] [Google Scholar]
- 44.Matsushima K, et al. Molecular cloning of a human monocyte-derived neutrophil chemotactic factor (MDNCF) and the induction of MDNCF mRNA by interleukin 1 and tumor necrosis factor. J. Exp. Med. 1988;167:1883–1893. doi: 10.1084/jem.167.6.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Girgis RE, Basha MA, Maliarik M, Popovich J, Jr, Iannuzzi MC. Cytokines in the bronchoalveolar lavage fluid of patients with active pulmonary sarcoidosis. Am. J. Respir. Crit. Care Med. 1995;152:71–75. doi: 10.1164/ajrccm.152.1.7599865. [DOI] [PubMed] [Google Scholar]
- 46.Mortaz E, et al. Association of serum TNF-alpha, IL-8 and free light chain with HLA-DR B alleles expression in pulmonary and extra-pulmonary sarcoidosis. J. Inflamm. 2015;12:21. doi: 10.1186/s12950-015-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yokoyama T, Kanda T, Kobayashi I, Suzuki T. Serum levels of interleukin-8 as a marker of disease activity in patients with chronic sarcoidosis. J. Med. 1995;26:209–219. [PubMed] [Google Scholar]
- 48.Meniailo ME, et al. Direct effects of interleukin-8 on growth and functional activity of T lymphocytes. Int. Immunopharmacol. 2017;50:178–185. doi: 10.1016/j.intimp.2017.06.023. [DOI] [PubMed] [Google Scholar]
- 49.Azuma M, et al. B70 antigen is a second ligand for CTLA-4 and CD28. Nature. 1993;366:76–79. doi: 10.1038/366076a0. [DOI] [PubMed] [Google Scholar]
- 50.Collins TL, Deckert M, Altman A. Views on Vav. Immunol Today. 1997;18:221–225. doi: 10.1016/s0167-5699(97)01037-2. [DOI] [PubMed] [Google Scholar]
- 51.Su B, et al. JNK is involved in signal integration during costimulation of T lymphocytes. Cell. 1994;77:727–736. doi: 10.1016/0092-8674(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 52.Vanherwegen AS, et al. Vitamin D controls the capacity of human dendritic cells to induce functional regulatory T cells by regulation of glucose metabolism. J. Steroid. Biochem. Mol. Biol. 2019;187:134–145. doi: 10.1016/j.jsbmb.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 53.Steinman RM. Some interfaces of dendritic cell biology. APMIS. 2003;111:675–697. doi: 10.1034/j.1600-0463.2003.11107802.x. [DOI] [PubMed] [Google Scholar]
- 54.Cassatella MA, et al. Generation of superoxide anion by alveolar macrophages in sarcoidosis: evidence for the activation of the oxygen metabolism in patients with high-intensity alveolitis. Immunology. 1989;66:451–458. [PMC free article] [PubMed] [Google Scholar]
- 55.Rottoli P, et al. Carbonylated proteins in bronchoalveolar lavage of patients with sarcoidosis, pulmonary fibrosis associated with systemic sclerosis and idiopathic pulmonary fibrosis. Proteomics. 2005;5:2612–2618. doi: 10.1002/pmic.200401206. [DOI] [PubMed] [Google Scholar]
- 56.Curtis JM, et al. Downregulation of adipose glutathione S-transferase A4 leads to increased protein carbonylation, oxidative stress, and mitochondrial dysfunction. Diabetes. 2010;59:1132–1142. doi: 10.2337/db09-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kikuchi N, et al. Nrf2 protects against pulmonary fibrosis by regulating the lung oxidant level and Th1/Th2 balance. Respir. Res. 2010;11:31. doi: 10.1186/1465-9921-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei J, et al. Nrf2 activation protects against intratracheal LPS induced mouse/murine acute respiratory distress syndrome by regulating macrophage polarization. Biochem. Biophys. Res. Commun. 2018;500:790–796. doi: 10.1016/j.bbrc.2018.04.161. [DOI] [PubMed] [Google Scholar]
- 59.Malhotra D, et al. Decline in NRF2-regulated antioxidants in chronic obstructive pulmonary disease lungs due to loss of its positive regulator, DJ-1. Am. J. Respir. Crit. Care Med. 2008;178:592–604. doi: 10.1164/rccm.200803-380OC. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Brown RH, Reynolds C, Brooker A, Talalay P, Fahey JW. Sulforaphane improves the bronchoprotective response in asthmatics through Nrf2-mediated gene pathways. Respir. Res. 2015;16:106. doi: 10.1186/s12931-015-0253-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beeh KM, et al. Glutathione deficiency of the lower respiratory tract in patients with idiopathic pulmonary fibrosis. Eur. Respir. J. 2002;19:1119–1123. doi: 10.1183/09031936.02.00262402. [DOI] [PubMed] [Google Scholar]
- 62.Gutierrez-Vazquez C, Quintana FJ. Regulation of the immune response by the aryl hydrocarbon receptor. Immunity. 2018;48:19–33. doi: 10.1016/j.immuni.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banchereau J, et al. Immunobiology of dendritic cells. Annu. Rev. Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 64.Korn T. How T cells take developmental decisions by using the aryl hydrocarbon receptor to sense the environment. Proc. Natl. Acad. Sci. USA. 2010;107:20597–20598. doi: 10.1073/pnas.1015420107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quintana FJ, et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 66.Monteleone I, et al. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology. 2011;141:237–248. doi: 10.1053/j.gastro.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 67.Beamer CA, Shepherd DM. Role of the aryl hydrocarbon receptor (AhR) in lung inflammation. Semin. Immunopathol. 2013;35:693–704. doi: 10.1007/s00281-013-0391-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bargagli E, Maggiorelli C, Rottoli P. Human chitotriosidase: a potential new marker of sarcoidosis severity. Respiration. 2008;76:234–238. doi: 10.1159/000134009. [DOI] [PubMed] [Google Scholar]
- 69.Hawkins C, et al. Local and systemic CD4(+) T cell exhaustion reverses with clinical resolution of pulmonary sarcoidosis. J. Immunol. Res. 2017;2017:3642832. doi: 10.1155/2017/3642832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Braun NA, et al. Blockade of the programmed death-1 pathway restores sarcoidosis CD4(+) T-cell proliferative capacity. Am. J. Respir. Crit. Care Med. 2014;190:560–571. doi: 10.1164/rccm.201401-0188OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lomax AJ, et al. Immunotherapy-induced sarcoidosis in patients with melanoma treated with PD-1 checkpoint inhibitors: case series and immunophenotypic analysis. Int. J. Rheum. Dis. 2017;20:1277–1285. doi: 10.1111/1756-185X.13076. [DOI] [PubMed] [Google Scholar]
- 72.Camacho DM, Collins KM, Powers RK, Costello JC, Collins JJ. Next-generation machine learning for biological networks. Cell. 2018;173:1581–1592. doi: 10.1016/j.cell.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 73.Wolfe CJ, Kohane IS, Butte AJ. Systematic survey reveals general applicability of "guilt-by-association" within gene coexpression networks. BMC Bioinformatics. 2005;6:227. doi: 10.1186/1471-2105-6-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Albert R. Scale-free networks in cell biology. J. Cell. Sci. 2005;118:4947–4957. doi: 10.1242/jcs.02714. [DOI] [PubMed] [Google Scholar]
- 76.Suomi T, Corthals GL, Nevalainen OS, Elo LL. Using peptide-level proteomics data for detecting differentially expressed proteins. J. Proteome. Res. 2015;14:4564–4570. doi: 10.1021/acs.jproteome.5b00363. [DOI] [PubMed] [Google Scholar]
- 77.Moller DR, et al. Rationale and design of the genomic research in alpha-1 antitrypsin deficiency and sarcoidosis (GRADS) study sarcoidosis protocol. Ann. Am. Thorac. Soc. 2015;12:1561–1571. doi: 10.1513/AnnalsATS.201503-172OT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang IV, et al. DNA methylation changes in lung immune cells are associated with granulomatous lung disease. Am. J. Respir. Cell Mol. Biol. 2019;60:96–105. doi: 10.1165/rcmb.2018-0177OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lockstone HE, et al. Gene set analysis of lung samples provides insight into pathogenesis of progressive, fibrotic pulmonary sarcoidosis. Am. J. Respir. Crit. Care Med. 2010;181:1367–1375. doi: 10.1164/rccm.200912-1855OC. [DOI] [PubMed] [Google Scholar]
- 80.Bhargava M, et al. Proteome profiling in lung injury after hematopoietic stem cell transplantation. Biol. Blood Marrow Transpl. 2016;22:1383–1390. doi: 10.1016/j.bbmt.2016.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bhargava M, et al. Bronchoalveolar lavage fluid protein expression in acute respiratory distress syndrome provides insights into pathways activated in subjects with different outcomes. Sci. Rep. 2017;7:7464. doi: 10.1038/s41598-017-07791-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 83.Shadforth IP, Dunkley TP, Lilley KS, Bessant C. i-Tracker: for quantitative proteomics using iTRAQ. BMC Genomics. 2005;6:145. doi: 10.1186/1471-2164-6-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oberg AL, et al. Statistical analysis of relative labeled mass spectrometry data from complex samples using ANOVA. J Proteome Res. 2008;7:225–233. doi: 10.1021/pr700734f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics datasets were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD014438 (BAL cells) and PXD016637 (BALF).