Abstract
Applying exome sequencing to populations with unique genetic architecture has the potential to reveal novel genes and variants associated with traits and diseases. We sequenced and analyzed the exomes of 6,716 individuals from a Southwestern American Indian (SWAI) population with well-characterized metabolic traits. We found that the SWAI population has distinct allelic architecture compared to populations of European and East Asian ancestry, and there were many predicted loss-of-function (pLOF) and nonsynonymous variants that were highly enriched or private in the SWAI population. We used pLOF and nonsynonymous variants in the SWAI population to evaluate gene-burden associations of candidate genes from European genome-wide association studies (GWASs) for type 2 diabetes, body mass index, and four major plasma lipids. We found 19 significant gene-burden associations for 11 genes, providing additional evidence for prioritizing candidate effector genes of GWAS signals. Interestingly, these associations were mainly driven by pLOF and nonsynonymous variants that are unique or highly enriched in the SWAI population. Particularly, we found four pLOF or nonsynonymous variants in APOB, APOE, PCSK9, and TM6SF2 that are private or enriched in the SWAI population and associated with low-density lipoprotein (LDL) cholesterol levels. Their large estimated effects on LDL cholesterol levels suggest strong impacts on protein function and potential clinical implications of these variants in cardiovascular health. In summary, our study illustrates the utility and potential of exome sequencing in genetically unique populations, such as the SWAI population, to prioritize candidate effector genes within GWAS loci and to find additional variants in known disease genes with potential clinical impact.
Keywords: exome sequencing, isolated founder population, rare variant, gene-burden association, metabolic traits
Introduction
The genetic architecture of a population is influenced by the specific demographic history that the population has undergone. Founder and bottleneck events and subsequent reproductive isolation can result in a dramatic change in the allele frequency spectrum, potentially increasing the frequency of rare functional variants due to random genetic drift, thus allowing greater statistical power to detect the association of such variants with traits of interest.1, 2, 3, 4, 5, 6, 7 American Indians are predicted to have gone through a series of founder and bottleneck events. One such bottleneck occurred around 15,000 years ago when a small number of Eurasians migrated across the Bering Strait and settled into the American continent.8 In addition, European colonization of the Americas led to other bottleneck events around 500 years ago.9 Consistent with this history, American Indians have a distinct genetic background compared to populations of other ancestries.10,11
The study specifically focuses on a Southwestern American Indian (SWAI) population (i.e., an American Indian population in the Southwestern region of the United States). This population has a very high prevalence of obesity and type 2 diabetes (T2D) and has been deeply characterized for metabolic traits.12, 13, 14 Previously, genetic studies have been conducted in this population with specific focus on metabolic traits, including genome-wide linkage analyses,15 genome-wide association studies (GWASs),16, 17, 18, 19, 20 assessment of genes and/or variants found in GWAS studies in other ancestry groups,21, 22, 23, 24, 25, 26 and targeted sequencing of physiologic candidate genes.27, 28, 29, 30, 31, 32 These approaches have found common and rare variants that are associated with metabolic traits and disease status in this population; however, a systematic examination of coding variation across the genome and its potential impact on metabolic traits has not been fully explored.
In this study, we sequenced the exomes of 6,716 individuals from the SWAI population and found a total of ∼1.2 million variants, including 16,880 predicted loss-of-function (pLOF) variants and 258,306 nonsynonymous variants, many of which are highly enriched or private in this population. The goal of our study was to characterize the exome architecture of the SWAI population in comparison to more cosmopolitan populations, i.e., large populations with lower barriers to migration, and examine the phenotypic impact of rare coding variants that are either private or enriched in this population.
Subjects and Methods
Study Subjects
The study participants were individuals with American Indian ancestry from the Southwestern region of the United States who enrolled in a longitudinal study of metabolic disorders as described previously.14,33 Measurements included height and weight for body mass index (BMI) calculation and fasting lipid levels. Maximum BMI and age at maximum BMI were used for analysis. T2D status was determined on the basis of the criteria of the American Diabetes Association or the review of the medical records. Diabetes in this population has been primarily classified as T2D despite the relatively early onset of the disease because of the absence of key characteristics of type 1 diabetes, including islet autoantibodies, ketoacidosis, and insulin dependence.34, 35, 36, 37 The self-reported number of great grandparents that were American Indian was recorded as a measure of admixture. Individuals with all eight American Indian great grandparents are herein referred to as “full American Indians.” DNA from the blood of the participants was collected to evaluate the genetic etiology of metabolic disorders. The study protocol was approved by the Institutional Review Board (IRB) of the National Institute of Diabetes and Digestive and Kidney Diseases. Informed consent was obtained from all participants.
Individuals from two additional studies were included as references for comparison. The DiscovEHR study is a collaborative project between the Regeneron Genetics Center and the Geisinger Health System based in Pennsylvania with participants who enrolled in Geisinger’s MyCode Community Health Initiative.38 The study was approved by the IRB at the Geisinger. The TAICHI study is a collaborative study with participants recruited at several academic centers in Taiwan.39 The study was approved by the IRBs at all participating centers (Taichung Veteran’s General Hospital, Tri-Service General Hospital, the National Taiwan University Hospital, and the National Health Research Institute of Taiwan) and the IRB of the Los Angeles Biomedical Research Institute. All participants provided written informed consent.
Exome Sequencing, Variant Calling, and Quality Control
DNA samples from 6,809 SWAI individuals were exome sequenced at the Regeneron Genetics Center via sequencing methodology, genome alignment, and genotype calling approaches as previously described.40 Briefly, exonic regions were targeted with an xGEN probe library with additional capture probes. Targeted DNA was sequenced on the Illumina HiSeq 2500 platform with v4 chemistry with 75bp paired-end reads. Sequencing was performed such that >85% of the bases were covered at ≥20× depth. Read alignment to human genome reference GRCh38 and variant calling were performed with BWA-MEM and GATK, respectively. 93 samples were removed on the basis of quality control metrics, including low coverage (<75% of targeted bases with at least 20× depth), low quality, sex mismatch, sample duplicates, and high discordance with array genotypes, resulting in the final count of 6,716 exomes for analysis. Variants were further filtered by missing call rates (<10%) and Hardy-Weinberg equilibrium p values (>1 × 10−15).
DNA samples from 29,575 individuals of European ancestry from the DiscovEHR study were exome sequenced and processed by the same method. DNA samples from 13,947 individuals of East Asian ancestry from the TAICHI study were exome sequenced and processed by an analogous method as previously described.38 Exome sequencing and variant calling of all three studies (SWAI, DiscovEHR, and TAICHI) were conducted at the Regeneron Genetics Center following identical quality control measures, except for the use of two different exome targeting reagents (SWAI and DiscovEHR on xGEN capture versus TAICHI on VCRome capture). To account for the difference in the exome targeting reagents, all analyses that make comparisons across studies were conducted among the subset of variants that map to the intersection of consistently covered regions of each targeting reagent. Consistently covered regions were defined as having ≥20× read depth in ≥90% of a randomly sampled set of 1,000 exomes sequenced with the targeting reagent.
Variant Annotation
Variants were annotated for their predicted effects on all autosomal protein-coding transcripts with annotated start and stop in Ensembl85 (54,214 transcripts corresponding to 19,467 genes) with snpEff.41 Variants were annotated as pLOF when they were predicted to incur a frameshift, premature stop codon, loss of start or stop codon, or disruption of canonical splice dinucleotides. Nonsynonymous variants included missense single nucleotide variants (SNVs) and inframe indels. When a variant had different predicted effects among different transcripts, a more deleterious effect was prioritized. The variants detected in the SWAI exomes were compared to dbSNP (v151)42 and gnomAD exomes (r2.1).43
Principal-Component Analysis
Reference genomes were downloaded from 1000 Genomes Project server.44 The principal-component analysis was performed with independent (r2 measure of linkage disequilibrium [LD] < 0.2) common (minor allele frequency [MAF] ≥ 5%) autosomal bi-allelic variants that were detected in both the reference genomes and the SWAI exomes. To avoid the impact of extended LD and high variability regions, such as the major histocompatibility complex, these regions were omitted from principal-component analysis. We first derived the principal components from the reference genomes and projected individuals from SWAI onto the principal-component space via PLINK2.45
Comparison of Allelic Architecture and Frequency
The allelic architecture of SWAI exomes was compared to European ancestry exomes from the DiscovEHR study and East Asian exomes, predominantly of Han Chinese from Taiwan, from the TAICHI study. For the comparison of proportional site frequency spectra, 6,716 European and 6,716 East Asian exomes were randomly sampled from the DiscovEHR and TAICHI studies, respectively. The number of pLOF and nonsynonymous variants were counted according to the minor allele count (MAC) bins, and the proportion was calculated. For the comparison of allele frequency, we included only self-reported full American Indians from the SWAI study to minimize the impact of admixture. To avoid situations where the minor allele of the same variant differs between studies, all allele frequencies refer to the alternate allele frequencies (AAF) of the variant compared to the human genome reference. For any study, if no alternate alleles were observed within a consistently covered region (as described above), the allele frequency of the variant in that study was inferred to be 0. Allele frequencies in the SWAI population were also compared to the population frequencies from gnomAD exomes r2.1. When a variant was not listed in gnomAD exomes, but the genomic position was called with mean read depth ≥ 20, the allele frequency of the variant in gnomAD was inferred to be 0.
Deriving Candidate Genes from European GWASs
We derived the set of candidate effector genes for BMI,46 T2D,47 and plasma lipid levels48 from previous GWASs consisting entirely or predominantly of European ancestry. Sentinel variants of independent association signals were derived by the conditional and joint (COJO) analysis of GCTA49 using 10,000 randomly selected unrelated individuals of European ancestry from the UK Biobank study50 as the LD reference. Genes that are within the ±250 kb window of the sentinel variants were derived to test their associations for corresponding traits in the SWAI study.
Association Analysis
For gene-burden tests, pLOF and missense variants were grouped into eight masks with two allele frequency cutoffs (AAF < 1% and < 5%) and four functional effect criteria: (1) M1, pLOF variants only, (2) M2, pLOF and all missense variants, (3) M3, pLOF and missense variants predicted to be deleterious by all five prediction algorithms used (SIFT,51 LRT,52 MutationTaster,53 PolyPhen2-HumDiv, and PolyPhen2-HumVar54), and (4) M4, pLOF and missense variants predicted to be deleterious by at least one of the five prediction algorithms.38,55 If different masks of a gene are comprised of the same variants, they were collapsed to one mask with most stringent definition so that only unique masks were tested for association. The Bonferroni corrected p value cutoff was calculated as 0.05/total number of gene-burdens tested for a given trait, i.e., the sum of unique masks across all candidate genes of the trait. The number of gene-burden tests and p value cutoff for each trait are provided in the relevant section dedicated for the trait (see Results). For significant gene-burden associations, the individual variants that were included in the masks were also tested for associations. Only the masks and variants with at least 10 alternate allele counts were tested.
Associations were tested under a linear mixed model using SAIGE56 for T2D status and BOLT57 for quantitative traits to adjust for population structure and cryptic relatedness. For diabetes, age, age2, sex, and five principal components of ancestry were included as covariates. For age of diabetes onset, sex and five principal components were included as covariates. For BMI and lipid traits (triglyceride measures were natural-log transformed), residuals were derived adjusting for age, age2, sex, and five principal components and normalized by rank-based inverse normal transformation.
Results
Characterization of Exome Variants
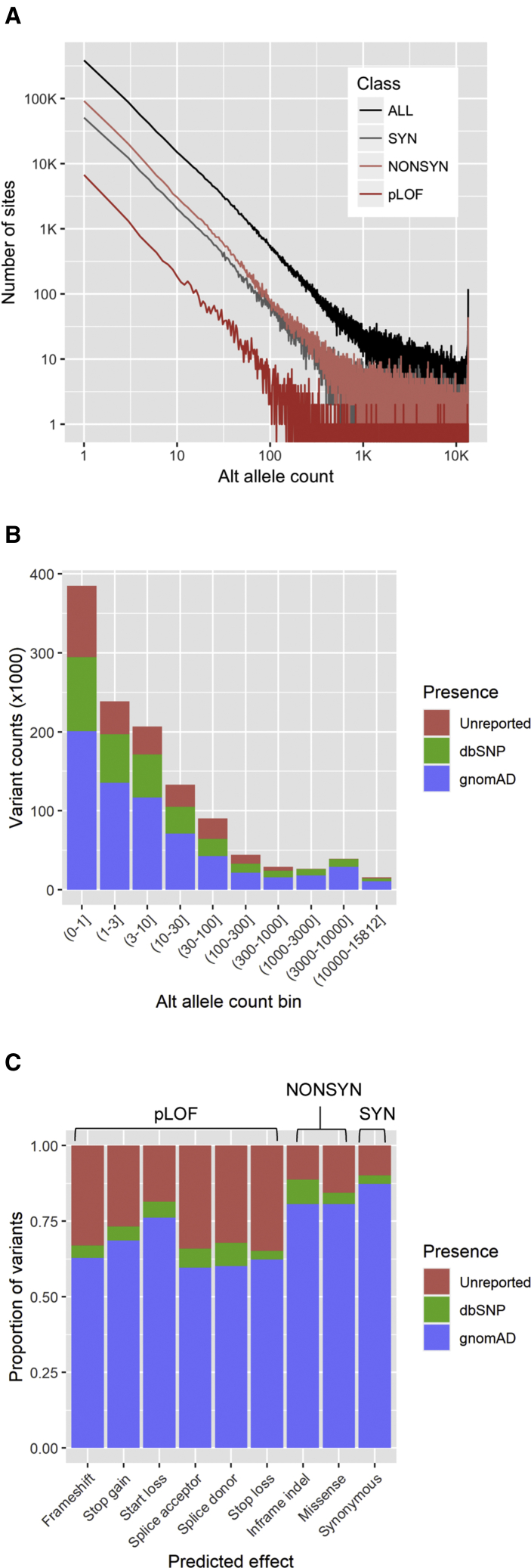
We detected a total of 1,208,812 variants from the exomes of 6,716 SWAI individuals (Table 1 and Figure 1A), of which 1,130,961 (93.6%) were SNVs and 77,851 (6.4%) were indels. When annotated for predicted effects, 16,880 (1.4%) were pLOF variants (frameshift, stop-gain, start-loss, splice acceptor, splice donor, and stop-loss) and 258,306 (21.4%) were nonsynonymous variants (inframe indels and missense). The majority of variants were rare, i.e., less than 10 alternate allele counts (corresponding to the AAF of <0.07%) in SWAI individuals.
Table 1.
Summary Statistics and Annotation of Variants Captured by Exome Sequencing of 6,716 SWAI Individuals
|
All |
Alternate Allele Count ≥10 |
||||||
|---|---|---|---|---|---|---|---|
| Total Number | Number (%) Not in dbSNPa | Number (%) Not in gnomADb | Total Number | Number (%) Not in dbSNPa | Number (%) Not in gnomADb | ||
| Variant Type | |||||||
| All | 1,208,812 | 245,039 (20.3%) | 545,979 (45.2%) | 393,548 | 76,966 (19.6%) | 175,318 (44.5%) | |
| SNVs | 1,130,961 | 228,981 (20.2%) | 505,888 (44.7%) | 366,309 | 72,909 (19.9%) | 162,486 (44.4%) | |
| Indels | 77,851 | 16,058 (20.6%) | 40,091 (51.5%) | 27,239 | 4,057 (14.9%) | 12,832 (47.1%) | |
| Variant Effect | |||||||
| pLOF (n = 16,880) | frameshift | 6,881 | 2,456 (35.7%) | 2,552 (37.1%) | 1,474 | 401 (27.2%) | 418 (28.4%) |
| stop gained | 5,288 | 1,427 (27.0%) | 1,659 (31.4%) | 1,016 | 315 (31.0%) | 354 (34.8%) | |
| start lost | 668 | 125 (18.7%) | 159 (23.8%) | 177 | 33 (18.6%) | 43 (24.3%) | |
| splice acceptor | 1,858 | 675 (36.3%) | 750 (40.4%) | 596 | 185 (31.0%) | 198 (33.2%) | |
| splice donor | 1,858 | 612 (32.9%) | 741 (39.9%) | 465 | 175 (37.6%) | 209 (44.9%) | |
| stop lost | 327 | 117 (35.8%) | 123 (37.6%) | 123 | 59 (48.0%) | 61 (49.6%) | |
| Nonsynonymous (n = 258,306) | in-frame indel | 4,157 | 591 (14.2%) | 801 (19.3%) | 1,323 | 119 (9.0%) | 173 (13.1%) |
| missense | 254,149 | 40,529 (15.9%) | 49,061 (19.3%) | 68,494 | 11,088 (16.2%) | 12,631 (18.4%) | |
| Synonymous | 164,772 | 16,650 (10.1%) | 20,898 (12.7%) | 54,952 | 4,551 (8.3%) | 5,357 (9.7%) | |
Variants detected in SWAI exomes were categorized by their type and predicted functional effect. The number of variants were counted on the basis of whether they have an alternate allele count ≥ 10 in SWAI exomes and whether they have not been reported in dbSNP or gnomAD exomes.
dbSNP v151 was used for comparison.
gnomAD exomes r2.1 was used for comparison.
Figure 1.
Summary Statistics and Annotation of Variants Captured by Exome Sequencing of 6,716 SWAI Individuals
(A) Site frequency distribution of 1,208,812 autosomal variants according to the predicted functional effect class.
(B) The number of variants that are previously reported in gnomAD or dbSNP databases or unreported as a function of alternate allele count.
(C) The proportion of variants that are previously reported in gnomAD or dbSNP databases or unreported stratified by predicted functional effect. pLOF, predicted loss-of-function; NONSYN, nonsynonymous; SYN, synonymous.
When compared to dbSNP and gnomAD exome databases, 241,042 variants (19.9%) were not reported in either database (20.3% not in dbSNP and 45.2% not in gnomAD exome). These previously unreported variants tended to be more rare in frequency (Figure 1B) and more enriched among pLOF variants than among nonsynonymous or synonymous variants (Figure 1C).
Population Structure
The SWAI population has considerable admixture according to the self-reported American Indian ancestry of the study subjects; of the 6,716 sequenced subjects, 4,897 subjects (corresponding to 72.9%) were full American Indians (all eight great grandparents were American Indian), whereas the rest had varying degrees of admixture (Figure S1A). To evaluate the population structure and admixture of the SWAI population on the basis of the genetic data, we constructed principal components from three ancestral super populations (European, East Asian, and African ancestries) from the 1000 Genomes Project and projected SWAI study subjects onto the principal-component space. When only the self-reported full American Indians from the SWAI study were plotted, they clustered about an axis between the European and East Asian clusters (Figure S1B). When all individuals from the SWAI study were plotted, we observed that individuals with greater self-reported admixture tended to deviate further from the full American Indian cluster toward European and African clusters (Figure S1C). These results confirm that the SWAI population is comprised of individuals with complete or partial American Indian ancestry.
Comparison of Allelic Architecture and Frequency
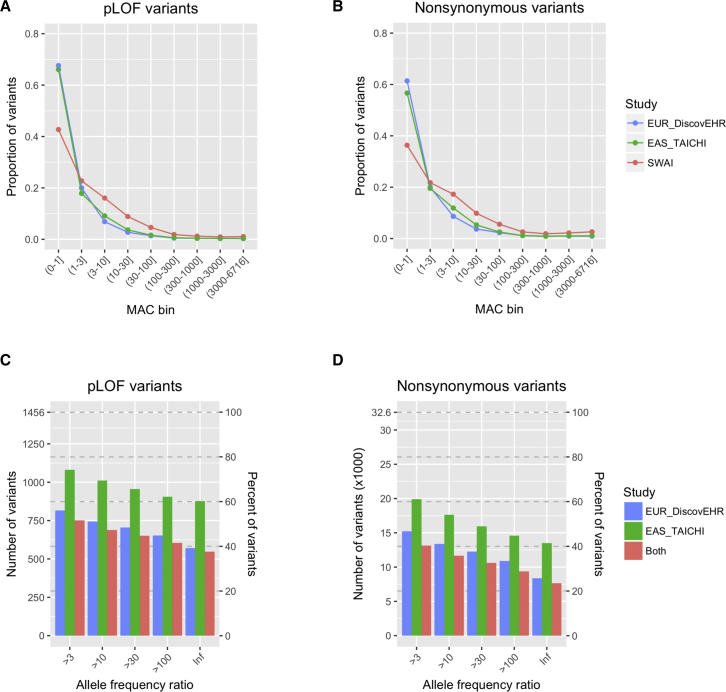
We compared the allelic architecture of SWAI exomes to European ancestry exomes from the DiscovEHR study and East Asian exomes from the TAICHI study that respectively served as the extant proxies for ancestral European and East Asian genomes that influenced the American Indian genome. As described in Subjects and Methods, analyses were restricted to variants in the consistently covered regions of the two exome targeting reagents that were used. We compared the proportional site frequency spectra of SWAI exomes to the same number of European and East Asian ancestry exomes that were randomly sampled. SWAI exomes were relatively depleted of ultra-rare pLOF and nonsynonymous variants (MAC ≤ 3) compared to European ancestry exomes but were enriched for moderately rare pLOF and nonsynonymous variants (3 < MAC ≤ 1,000) compared to both European and East Asian ancestry exomes (Figures 2A and 2B).
Figure 2.
Comparison of the Distribution and Frequency of pLOF and Nonsynonymous Variants among SWAI, European, and East Asian Exomes
(A and B) Comparison of the distribution of pLOF (A) and nonsynonymous (B) variants at different MAC bins among SWAI, European, and East Asian exomes from the SWAI, DiscovEHR, and TAICHI studies, respectively.
(C and D) The number and percentage of pLOF (C) and nonsynonymous (D) variants that are enriched in full American Indian exomes from the SWAI study compared to European exomes, East Asian exomes, or both European and East Asian exomes. The analysis is restricted to variants with alternate allele count ≥10 in full American Indians from the SWAI population.
To examine how many of the variants that were detected in the SWAI exomes are private or enriched in the SWAI population, we compared the allele frequency of pLOF and nonsynonymous variants in full American Indians from the SWAI population to individuals with European and East Asian ancestries. Considering the power for statistical inference, the analysis was restricted to variants with a minimum alternate allele count of 10 in the SWAI exomes. Among the total of 1,456 pLOF variants, 548 (38.4%) were only detected in SWAI exomes and 689 (48.3%) were more than 10 times more enriched in SWAI exomes compared to both European ancestry and East Asian exomes (Figure 2C). Among the total of 32,577 nonsynonymous variants, 7,640 (23.7%) were only detected in SWAI exomes and 11,649 (36.1%) were more than 10 times more enriched in SWAI exomes compared to European and East Asian ancestry exomes (Figure 2D).
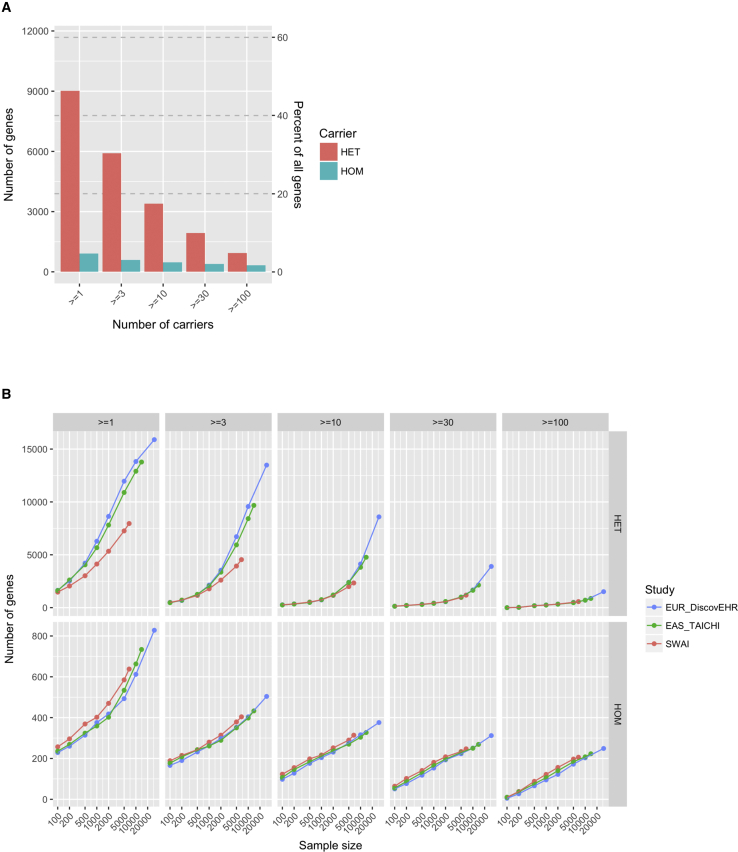
Genes with pLOF Variation
Because pLOF variants can provide a valuable insight on the biological connection between genes and traits, we examined how many genes carried pLOF variation in SWAI exomes. Of the 19,467 autosomal genes annotated, 9,015 genes (46.3%) had at least one heterozygous carrier of pLOF variants and 3,398 genes (17.5%) had at least ten heterozygous carriers (Table 2 and Figure 3A). 907 genes (4.7%) had at least one homozygous carrier of pLOF variants, and 466 genes (2.4%) had at least ten homozygous carriers.
Table 2.
Number and Percentage of Genes with at Least X Number of Carriers of pLOF Variants in 6,716 SWAI Exomes
| Number of Carriers | Number (%) of Genes with Any Carriers | Number (%) of Genes with Heterozygous Carriers | Number (%) of Genes with Homozygous Carriers |
|---|---|---|---|
| ≥1 | 9,016 (46.3%) | 9,015 (46.3%) | 907 (4.7%) |
| ≥3 | 5,910 (30.4%) | 5,907 (30.3%) | 593 (3.0%) |
| ≥10 | 3,407 (17.5%) | 3,398 (17.5%) | 466 (2.4%) |
| ≥30 | 1,948 (10.0%) | 1,936 (9.9%) | 389 (2.0%) |
| ≥100 | 953 (4.9%) | 939 (4.8%) | 327 (1.7%) |
Figure 3.
Comparison of the Number of Genes with pLOF Carriers among SWAI, European, and East Asian Exomes
(A) The number and percentage of genes among 19,467 annotated autosomal genes with at least X number of heterozygous and homozygous pLOF carriers in the SWAI study alone.
(B) The comparison of the number of genes with at least X number of heterozygous (top) and homozygous (bottom) pLOF carriers at fixed sample sizes randomly extracted from the SWAI, European, and East Asian exomes. The analysis was restricted to the consistently covered regions across the three studies.
To see whether population history impacted the number and distribution of pLOF variation, we compared the number of genes with pLOF carriers in the SWAI exomes sampled from the current study to the same number of European and East Asian exomes sampled from DiscovEHR and TAICHI studies, respectively. The analysis was again restricted to variants in the consistently covered regions across the studies for comparison. Consistent with the founder effect, the number of genes with heterozygous pLOF carriers was lower in SWAI exomes than in European and East Asian exomes (Figure 3B, top). On the other hand, the number of genes with homozygous pLOF carriers was greater in SWAI exomes (Figure 3B, bottom), potentially because of the fact that the SWAI population experienced reproductive isolation with small population size.
pLOF variation might accumulate as a result of random genetic drift or specific environmental pressure that populations face that could increase tolerance to loss-of-function of certain genes. We investigated the overlap among the set of genes with ≥10 pLOF carriers in the SWAI (n = 6,716), European (n = 29,575), and East Asian (n = 13,947) exomes. Considering the power for downstream statistical inference, we set the minimum number of carriers at 10. Although the total sample size of SWAI exomes was smaller than the sample sizes of European exomes and East Asian exomes, there were 275 genes with ≥10 heterozygous pLOF carriers and 87 genes with ≥10 homozygous carriers only in SWAI exomes (Figure S2). Of all the genes with ≥10 heterozygous and ≥10 homozygous pLOF carriers in SWAI exomes, ∼11.8% and 27.7% were unique to SWAI exomes, respectively.
Testing Candidate GWAS Genes for Association with Metabolic Traits
Genetic association analysis using SWAI exomes can not only provide additional evidence for the candidate effector genes in GWAS loci but can also find variants with potential clinical impact that are unique or enriched in the SWAI population. We derived the list of candidate effector genes from the latest and largest European GWASs for BMI, T2D, and plasma lipid levels and tested their association with respective traits in the SWAI study. We used a gene-burden approach, aggregating pLOF and missense variants into eight masks with two allele frequency cutoffs (<1% and <5%, indicated as 1 and 5 following the period in the name of the mask) and four functional effect criteria (M1 to M4), as described in detail in Subjects and Methods. The list and frequencies of the individual variants that make up the significant gene-burden associations are shown in Table S1.
Body Mass Index
2,785 genes within the ±250 kb window from independent association signals from the latest European BMI GWAS46 were analyzed for association with maximum BMI measured in the SWAI study (Bonferroni p value cutoff = 0.05 divided by 7,886 gene-burden tests = 6.3 × 10−6). The M3.1 mask of MC4R (MIM: 155541), a gene associated with early-onset obesity (MIM: 618406), was the only gene-burden significantly associated with increased maximum BMI in the SWAI study (Table 3, Beta = 0.56 SD, p = 5.2 × 10−9). The gene-burden association was driven by the aggregate effects of four previously described variants, including a frameshift variant, p.Gly34fs [ss3984446997], and three missense variants, p.Arg165Gly [ss3984446996], p.Ala303Pro [ss3984446994], and p.Arg165Gln [rs747681609], that are either private or enriched in the SWAI population and were associated with maximum BMI individually (Table S2).27 These variants were previously identified by targeted sequencing of MC4R in the SWAI population and were found to impair the activity of MC4R in vitro, suggesting their functional impact.27
Table 3.
Gene-Burden Associations of Candidate Effector Genes from European GWASs for Body Mass Index, Type 2 Diabetes, and Plasma Lipid Levels in the SWAI Study
| Sentinel Variant | Associations from European GWASs |
Gene-Burden Associations in the SWAI Study |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Closest Gene | Variant Effect | AAF | Trait | Effecta | p Value | Gene | Top Maskb | Freq | Effecta | p Value | |
| BMI46 | |||||||||||
| rs6567160 | MC4R | intergenic | 0.23 | BMIc | 0.06 | 1.8E−178 | MC4R | M3.1 | 0.011 | 0.56 | 5.2E−09 |
| Type 2 Diabetes47 | |||||||||||
| rs523288 | MC4R | intergenic | 0.24 | T2D | 1.05 | 7.6E−13 | MC4R | M3.1 | 0.010 | 2.62 | 1.2E−05 |
| rs67254669 | ABCC8 | missense | 0.001 | T2D | 1.89 | 1.1E−08 | ABCC8 | M3.5 | 0.018 | 2.21 | 9.3E−06 |
| Plasma Lipid Levels48 | |||||||||||
| rs541041 | APOB | intergenic | 0.81 | TC | 0.11 | 5.3E−237 | APOB | M4.5 | 0.062 | −0.21 | 1.4E−06 |
| LDLC | 0.12 | 1.3E−287 | −0.29 | 1.6E−09 | |||||||
| rs445925 | APOE | downstream | 0.11 | TC | −0.21 | 0 | APOE | M4.1 | 0.014 | −0.63 | 9.4E−14 |
| LDLC | −0.32 | 0 | −0.86 | 4.7E−20 | |||||||
| rs11591147 | PCSK9 | missense | 0.015 | TC | −0.41 | 0 | PCSK9 | M2.5 | 0.057 | −0.20 | 6.2E−06 |
| LDLC | −0.48 | 0 | M3.5 | 0.028 | −0.44 | 9.1E−11 | |||||
| rs58542926 | TM6SF2 | missense | 0.07 | TC | −0.13 | 7.0E−155 | TM6SF2 | M4.5 | 0.067 | −0.26 | 4.6E−10 |
| LDLC | −0.10 | 6.5E−93 | −0.22 | 7.9E−07 | |||||||
| TG | −0.12 | 3.7E−125 | −0.28 | 2.5E−11 | |||||||
| rs2792751 | GPAM | missense | 0.73 | HDLC | −0.03 | 3.8E−21 | GPAM | M3.5 | 0.026 | 0.58 | 5.1E−15 |
| rs1800588 | LIPC | upstream | 0.24 | HDLC | 0.12 | 0 | LIPC | M4.1 | 0.013 | 0.47 | 9.8E−6 |
| rs622082 | IGHMBP2 | missense | 0.31 | HDLC | −0.02 | 5.90E−10 | CPT1A | M2.1 | 0.014 | −0.50 | 1.30E−06 |
| rs12328675 | COBLL1 | 3′ UTR | 0.12 | HDLC | 0.05 | 3.1E−37 | GRB14 | M2.1 | 0.013 | −0.46 | 1.8E−05 |
| rs964184 | ZPR1 | 3′ UTR | 0.85 | TC | −0.09 | 4.7E−135 | APOC3 | M4.5 | 0.026 | −0.35 | 3.1E−08 |
| HDLC | 0.11 | 2.6E−217 | 0.74 | 6.0E−23 | |||||||
| TG | −0.25 | 0 | −1.16 | 7.5E−72 | |||||||
Abbreviations are as follows: AAF, alternate allele frequency; BMI, body mass index; T2D, type 2 diabetes; TC, total cholesterol; LDLC, low-density lipoprotein cholesterol; HDLC, high-density lipoprotein cholesterol; TG, triglyceride.
The effects are beta coefficients in the standard deviation unit of normalized traits for BMI and lipid traits and odds ratios for T2D.
The mask with strongest trait association is displayed. Refer to Subjects and Methods for a detailed mask definition.
BMI in the SWAI study is maximum BMI.
Type 2 Diabetes
1,251 genes within the ±250 kb window from independent association signals from the latest European T2D GWAS47 were analyzed for association with T2D in the SWAI study (Bonferroni p value cutoff = 0.05 divided by 3,732 gene-burden tests = 1.3 × 10−5). Two gene-burdens were significantly associated with T2D risk: the M3.1 mask of MC4R and M3.5 mask of ABCC8 (MIM: 600509), a gene previously associated with maturity onset diabetes of the young, or MODY (MIM: 606391) (Table 3).
The same M3.1 mask of MC4R, which was associated with maximum BMI, was also associated with T2D (odds ration [OR] = 2.6, p = 1.2 × 10−5). When adjusted for maximum BMI, the association was only partially mitigated (OR = 2.2, p = 5.8 × 10−4), suggesting that MC4R might affect T2D independently of its effect on obesity. The gene-burden association was driven by the aggregate effects of three individual variants that are unique or highly enriched in the SWAI population, p.Gly34fs, p.Arg165Gly, and p.Arg165Gln (Table S3). The mask was also associated with earlier onset of T2D (Beta = −4.3 years, p = 5.5 × 10−3): all three homozygous carriers developed T2D under the age of 30 years (Figure S3A).
The M3.5 mask of ABCC8 was associated with diabetes (OR = 2.2, p = 9.3 × 10−6) mainly driven by a missense variant, p.Arg1420His [rs1272388614] (OR = 2.2, p = 1.5 × 10−5), which was previously reported.30 Notably, this variant was ∼489-fold and ∼115-fold enriched in SWAI individuals compared to individuals with European and East Asian ancestry, respectively (Table S3). Consistent with the known role of ABCC8 in MODY, early-onset forms of diabetes, and what was previously reported for p.Arg1420His alone, the M3.5 mask was associated with earlier age of onset (Beta = −6.9 years, p = 1.8 × 10−7); the one homozygous carrier developed diabetes before the age of 10 years (Figure S3B). ABCC8 encodes sulfonylurea receptor 1 protein (SUR1), which constitutes the ATP-sensitive potassium (KATP) channel, and it was previously shown that the p.Arg1420His mutation in SUR1 leads to impaired activity of the KATP channel in vitro,30 suggesting the functional impact of the variant.
Plasma Lipids
Up to 756 genes within the ±250 kb window from independent association signals from the latest GWAS for plasma lipid traits48 were analyzed for association with fasting total cholesterol, low-density lipoprotein (LDL) cholesterol, high-density lipoprotein (HDL) cholesterol, and triglyceride levels in the SWAI study (Bonferroni p value cutoff = 0.05 divided by up to 2,101 gene-burden tests = 2.4 × 10−5). Nine genes were significantly associated with at least one lipid trait (Table 3), among which six genes, APOB (MIM: 107730), APOE (MIM: 107741), PCSK9 (MIM: 607786), TM6SF2 (MIM: 606563), LIPC (MIM: 151670), and APOC3 (MIM: 107720), have biologically confirmed, either by knockout mice or by pharmacologic intervention, effects on the associated lipid traits.58, 59, 60, 61, 62, 63, 64, 65, 66 On the other hand, although genetic loci near GPAM (MIM: 602395), CPT1A (MIM: 600528), and GRB14 (MIM: 601524) have been associated with HDL cholesterol levels and the genes are involved in lipid metabolism67 or glycemic regulation,68,69 their effects on HDL cholesterol levels have not specifically been demonstrated in experimental models. GPAM gene-burden (M3.5) was associated with increased HDL cholesterol levels (Beta = 0.58 SD, p = 5.1 × 10−15), primarily driven by a missense variant, p.Ser611Arg [ss3984446988] (Beta = 0.57 SD, p = 3.8 × 10−14). This variant has an AAF of 0.025 in the SWAI population, but it was not detected in individuals with European ancestry and was ∼383-fold enriched compared to individuals with East Asian ancestry (Table S4). Notably, although the sentinel variant of GPAM from the European GWAS (rs2792751, encoding p.Ile43Val substitution) was associated with reduced HDL cholesterol levels,48 the p.Ser611Arg variant was associated with increased levels. This suggests that these two missense variants might have opposite effects on GPAM function. CPT1A gene-burden (M2.1) was associated with reduced HDL cholesterol levels in the SWAI study (Beta = −0.50 SD, p = 1.3 × 10−6). The association was mainly driven by two missense variants, p.Asp543Asn [rs1251355160] and p.Ala275Thr [rs2229738], the former of which is private in the SWAI population with an AAF of 0.006 (Table S4). Although CPT1A is not the closest gene to the sentinel GWAS variant, it was the only gene with significant gene-burden association in the SWAI study among the eight genes within the ±250 kb window of the sentinel variant that were tested for association (Table S5). Lastly, GRB14 gene-burden (M2.1) was associated with decreased HDL cholesterol levels (Beta = −0.46 SD, p = 1.8 × 10−5), largely driven by a missense variant, p.Ser220Tyr [rs780131269] (Beta = −0.76 SD, p = 2.2 × 10−7). This variant is present in the SWAI population at an AAF of 0.007 but was not detected in individuals with European or East Asian ancestry (Table S4). Although the GWAS sentinel variant is closest to COBLL1 and is also close to SLC38A11 (within the ±250 kb window), GRB14 was the only gene with significant gene-burden association in the SWAI study (Table S5).
Notably, the gene-burden associations of APOB, APOE, PCSK9, and TM6SF2 with LDL cholesterol levels were mostly driven by variants that are highly enriched or private in the SWAI population and had large estimated effects on LDL cholesterol levels (Table S4 and Figure S4). A frameshift pLOF variant of APOB, p.Ala3175fs [ss3984446986], is private in the SWAI population (AAF = 0.001) and was associated with lower LDL cholesterol levels (Beta = −2.30 SD, p = 1.8 × 10−13). This variant is expected to result in premature truncation at amino acid residue (aa) 3216 and might subject the resulting APOB to intracellular degradation, poor lipidation, and/or impaired secretion, similarly to other known truncating mutations,70,71 leading to lower LDL cholesterol levels, as seen in the carriers (Figure S4A). A missense variant of APOE, p.Ala184Asp [rs981058595], is private in the SWAI population (AAF = 0.007) and was associated with lower LDL cholesterol levels (Beta = −1.18 SD, p = 2.3 × 10−20). This variant was not in LD with the common variants of APOE E2 and E4 haplotypes in the SWAI population (r2 < 0.05). The variant resides in the hinge region between the N-terminal receptor-binding domain and C-terminal lipoprotein-binding domain. Previously, changes in the hinge region were shown to affect the binding of APOE to LDL receptors.72 The association of p.Ala184Asp with reduced LDL cholesterol levels suggests the possibility that the mutation might alter the hinge region in a way that it increases affinity to LDL receptors, lowering LDL cholesterol levels, as observed in the carriers (Figure S4B). A missense variant of PCSK9, p.Gly244Asp [rs370501906], is highly enriched in the SWAI population (AAF = 0.024) and was associated with lower LDL cholesterol levels (Beta = −0.46 SD, p = 4.7 × 10−10). This variant resides in the catalytic domain of PCSK9 and is close to a catalytic triad (aa 226). Previously, another missense variant in this domain, p.Leu253Phe, was shown to inhibit the catalytic activity of the protein,73 suggesting that p.Gly244Asp might affect PCSK9 in a similar manner. Reduced activity of PCSK9 will lead to increased surface expression of LDL receptors, which would be consistent with the lower plasma LDL cholesterol levels seen in p.Gly244Asp carriers (Figure S4C). Lastly, a missense variant of TM6SF2, p.Arg138Trp [rs142056540], highly enriched in the SWAI population (AAF = 0.046), was associated with lower LDL cholesterol levels (Beta = −0.20 SD, p = 1.2 × 10−4), suggesting that the mutation might impair the function of TM6SF2 in hepatic very-low-density lipoprotein (VLDL) processing,64 leading to lower LDL cholesterol levels observed in the carriers (Figure S4D). Further studies are needed to demonstrate the functional impacts of these variants and evaluate their clinical implications for cardiovascular health in the SWAI population.
To address the possibility that the gene-burden associations observed in the SWAI study might simply be tagging the association of previously established European GWAS signals, we examined the frequency and trait association of the sentinel variants from European GWASs in the SWAI study. We found that many of the GWAS sentinel variants were not as common in the SWAI population as they were in the European populations and, as expected on the basis of the small sample size, were not as strongly associated with traits in the SWAI study (Table S6). For five gene-burden associations where the GWAS sentinel variants had comparable p values (difference in p values < 1,000-fold), we performed conditional analysis to re-examine the gene-burden associations upon adjusting for the GWAS sentinel variants. The GWAS sentinel variants did not fully correct for the gene-burden associations, indicating that the gene-burden results in the SWAI study provide additional evidence for the genes beyond the GWAS sentinel variants (Table S6).
Discussion
Our study illustrates that exome sequencing applied to founder populations, such as this SWAI population, can uncover additional genetic variants that are associated with clinical and quantitative traits and expand our understanding of the genetic contribution to these traits. This is enabled by the distinct allelic architecture of the SWAI population: rare functional variants drift to higher frequency, increasing the statistical power to detect their associations with traits. In addition, gene-burden approaches aggregating rare pLOF and nonsynonymous variants affecting the same gene further enhanced the power to evaluate the relationship between genes and traits of interest.
The genetic architecture of the SWAI population is influenced by their unique population history involving bottleneck events followed by isolation. Consistent with the expectation that bottleneck events reduce overall genetic diversity, we observed fewer numbers of pLOF and nonsynonymous variants in SWAI exomes compared to European and East Asian exomes that underwent rapid population growth. Reproductive isolation following bottleneck events can randomly increase the frequency of rare variants. When we compared the proportion of pLOF and nonsynonymous variants across MAC bins, we observed selective enrichment of moderately rare variants in SWAI exomes compared to European and East Asian ancestry exomes, similar to the observation in Finnish populations that also underwent a series of bottleneck events and isolation.5 In addition, reproductive isolation in small populations can increase the homozygosity of genetic variants. As expected, the SWAI population had a greater number of pLOF and nonsynonymous variants in homozygosity compared to equivalent numbers of more cosmopolitan European and East Asian ancestry populations. We found little evidence of positive assortative mating based on kinship in the SWAI population (only 5 out of 648 parent pairs were estimated to be in 3rd degree or closer relationships), therefore the higher pLoF homozygosity in the SWAI population most likely resulted from the reproductive isolation with finite population size.
GWASs have traditionally focused on common variants that are captured by genotyping arrays or imputation, and as a result, many association signals are noncoding, making it challenging to pinpoint the effector genes that mediate the association. In our study, we examined the genes within the GWAS loci associated with BMI, T2D, and plasma lipid traits in European populations for their association in the SWAI study by using a gene-burden approach. We found significant associations for a handful of these genes, primarily driven by variants specific or enriched in the SWAI population, providing additional evidence for prioritizing candidate effector genes in GWAS loci. Of note, gene-burden associations tended to have stronger effects on traits compared to GWAS associations, consistent with the expectation that rare pLOF and nonsynonymous variants have greater impacts than common variants (Table 3). Most of the associated genes, namely MC4R, ABCC8, APOB, APOE, PCSK9, TM6SF2, LIPC, and APOC3, have experimentally validated effects on the traits with which they were associated. On the other hand, the biological effects of GPAM, CPT1A, and GRB14 on HDL cholesterol levels have not been fully determined yet. GPAM encodes mitochondrial glycerol-3-phosphate acyltransferase, which mediates the acylation of glycerol-3-phosphate, the first step in triglyceride synthesis.74 A previous study on Gpam knockout mice observed reduced hepatic triglyceride content and plasma total cholesterol and triglyceride levels,67 indicating its role in hepatic lipid metabolism; however, plasma HDL cholesterol levels were not significantly different between Gpam genotypes, although they trended lower in the knockout mice among both sexes.67 CPT1A encodes carnitine palmitoyltransferase 1A, which plays an essential role in fatty acid oxidation. Earlier studies in Greenlanders and Yup’ik Eskimos reported an association of a missense variant of CTP1A (rs80356779, encoding p.Pro479Leu substitution), highly enriched in these populations, with increased HDL cholesterol levels,75,76 but a later study with a larger number of Greenlanders reported a nominal association in the opposite direction.77 The functional effect of this variant on CPT1A activity is also unclear; skin fibroblasts from a carrier of the variant showed reduced basal activity of CPT1A but elevated activity in the presence of malonyl-CoA, a potent inhibitor of CPT1A.78 GRB14 encodes an adaptor protein that was previously found to inhibit insulin receptors in vitro68 and, when deleted in mice, improve glycemic trait.69 A recent report suggested that Grb14 deletion in mice leads to repression of liver X receptor (LXR) activity.79 Since LXR is a well-known modulator of plasma HDL cholesterol levels,80 the association of GRB14 with HDL cholesterol levels might be meditated through the LXR pathway. Further experimental evidence is needed to confirm the biological effects and direction of effects of these genes on HDL cholesterol levels.
The current study using the exome sequence of the SWAI population complements and extends previous genetic studies that have been conducted in the SWAI population via targeted sequencing or genotyping of candidate genes and variants and high-density genotyping arrays. The exome sequence enabled the systemic examination of all candidate genes for their association with metabolic traits at the gene level, which confirmed significant associations of MC4R and ABCC8 for BMI and T2D that were previously found in the SWAI population by targeted sequencing of these specific genes.27,30 In addition, the exome sequence allowed for the identification of rare coding variants beyond the common variants that have been captured by targeted genotyping or genotyping arrays,22,23,26 leading to a more comprehensive understanding of the impact of genetic variation in the candidate genes on traits. A previous GWAS for T2D performed in the SWAI population via a genotyping array found genome-wide significant associations of two common intronic variants in KCNQ1 and DNER with T2D risk.17,21 We did not find additional associations of pLOF or nonsynonymous variants of KCNQ1 and DNER with T2D risk, suggesting that the previously observed GWAS association signals might be mediated by alteration in transcriptional regulation.
It is worth noting that most gene-burden associations that we found were driven by pLOF and/or nonsynonymous variants that are unique or highly enriched in the SWAI population. Many of these variants were associated with traits with strong effects, warranting further investigation on the clinical implications of these variants in the SWAI population. In addition, further characterization of the functional impact of these protein-sequence-altering variants can broaden our understanding of the structure and regulation of the proteins. Although the current study specifically focused on the exome variants within the GWAS candidate genes, more studies are ongoing to identify genetic associations across the exome and could shed light on additional genetic underpinnings of the high prevalence of metabolic disorders in this population and uncover additional regulators of metabolic traits.
Declaration of Interests
H.K., B.Y., N.G., A.R.S., and C.V.H. are current or former employees and/or stockholders of Regeneron Genetics Center or Regeneron Pharmaceuticals. H.K. is an employee of Pfizer. N.G. is an employee of RTW Investments. The other authors declare no competing interests.
Acknowledgments
We thank the volunteers from the Southwestern American Indian community who participated in the study, the participants of the DiscovEHR study, and the participants of the TAICHI study. We also thank all research staff and teams at the Regeneron Genetics Center, National Institute of Diabetes and Digestive and Kidney Diseases, Geisinger Health System, and TAICHI Consortium who contributed to the current study. We thank the Regeneron postdoctoral program for their support and guidance for H.K. The study is funded by Regeneron Pharmaceuticals and the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases.
Published: July 7, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.06.009.
Contributor Information
Hye In Kim, Email: hyein.kim267@gmail.com.
Cristopher V. Van Hout, Email: cristopher.vanhout@regeneron.com.
Data and Code Availability
The genetic data of SWAI are protected materials and are not publicly available in respect of the IRB regulation. The data that support the reported findings and the codes that are used to generate figures are available from the corresponding authors upon reasonable request. Variants that are specifically described in the manuscript have been submitted to dbSNP (https://www.ncbi.nlm.nih.gov/snp) for public access.
Web Resources
1000 Genomes Projects, https://www.internationalgenome.org
OMIM, https://www.omim.org
PLINK2, www.cog-genomics.org/plink/2.0
Supplemental Data
References
- 1.Lim E.T., Würtz P., Havulinna A.S., Palta P., Tukiainen T., Rehnström K., Esko T., Mägi R., Inouye M., Lappalainen T., Sequencing Initiative Suomi (SISu) Project Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 2014;10:e1004494. doi: 10.1371/journal.pgen.1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Southam L., Gilly A., Süveges D., Farmaki A.E., Schwartzentruber J., Tachmazidou I., Matchan A., Rayner N.W., Tsafantakis E., Karaleftheri M. Whole genome sequencing and imputation in isolated populations identify genetic associations with medically-relevant complex traits. Nat. Commun. 2017;8:15606. doi: 10.1038/ncomms15606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xue Y., Mezzavilla M., Haber M., McCarthy S., Chen Y., Narasimhan V., Gilly A., Ayub Q., Colonna V., Southam L. Enrichment of low-frequency functional variants revealed by whole-genome sequencing of multiple isolated European populations. Nat. Commun. 2017;8:15927. doi: 10.1038/ncomms15927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rivas M.A., Avila B.E., Koskela J., Huang H., Stevens C., Pirinen M., Haritunians T., Neale B.M., Kurki M., Ganna A., International IBD Genetics Consortium. NIDDK IBD Genetics Consortium. T2D-GENES Consortium Insights into the genetic epidemiology of Crohn’s and rare diseases in the Ashkenazi Jewish population. PLoS Genet. 2018;14:e1007329. doi: 10.1371/journal.pgen.1007329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Locke A.E., Steinberg K.M., Chiang C.W.K., Service S.K., Havulinna A.S., Stell L., Pirinen M., Abel H.J., Chiang C.C., Fulton R.S., FinnGen Project Exome sequencing of Finnish isolates enhances rare-variant association power. Nature. 2019;572:323–328. doi: 10.1038/s41586-019-1457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou L., Kember R.L., Roach J.C., O’Connell J.R., Craig D.W., Bucan M., Scott W.K., Pericak-Vance M., Haines J.L., Crawford M.H. A population-specific reference panel empowers genetic studies of Anabaptist populations. Sci. Rep. 2017;7:6079. doi: 10.1038/s41598-017-05445-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sidore C., Busonero F., Maschio A., Porcu E., Naitza S., Zoledziewska M., Mulas A., Pistis G., Steri M., Danjou F. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat. Genet. 2015;47:1272–1281. doi: 10.1038/ng.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitchen A., Miyamoto M.M., Mulligan C.J. A three-stage colonization model for the peopling of the Americas. PLoS ONE. 2008;3:e1596. doi: 10.1371/journal.pone.0001596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Fallon B.D., Fehren-Schmitz L. Native Americans experienced a strong population bottleneck coincident with European contact. Proc. Natl. Acad. Sci. USA. 2011;108:20444–20448. doi: 10.1073/pnas.1112563108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma J., Amos C.I. Principal components analysis of population admixture. PLoS ONE. 2012;7:e40115. doi: 10.1371/journal.pone.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verdu P., Pemberton T.J., Laurent R., Kemp B.M., Gonzalez-Oliver A., Gorodezky C., Hughes C.E., Shattuck M.R., Petzelt B., Mitchell J. Patterns of admixture and population structure in native populations of Northwest North America. PLoS Genet. 2014;10:e1004530. doi: 10.1371/journal.pgen.1004530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennett P.H., Burch T.A., Miller M. Diabetes mellitus in American (Pima) Indians. Lancet. 1971;2:125–128. doi: 10.1016/s0140-6736(71)92303-8. [DOI] [PubMed] [Google Scholar]
- 13.Knowler W.C., Bennett P.H., Hamman R.F., Miller M. Diabetes incidence and prevalence in Pima Indians: a 19-fold greater incidence than in Rochester, Minnesota. Am. J. Epidemiol. 1978;108:497–505. doi: 10.1093/oxfordjournals.aje.a112648. [DOI] [PubMed] [Google Scholar]
- 14.Knowler W.C., Pettitt D.J., Saad M.F., Bennett P.H. Diabetes mellitus in the Pima Indians: incidence, risk factors and pathogenesis. Diabetes Metab. Rev. 1990;6:1–27. doi: 10.1002/dmr.5610060101. [DOI] [PubMed] [Google Scholar]
- 15.Hsueh W.C., Nair A.K., Kobes S., Chen P., Göring H.H.H., Pollin T.I., Malhotra A., Knowler W.C., Baier L.J., Hanson R.L. Identity-by-Descent Mapping Identifies Major Locus for Serum Triglycerides in Amerindians Largely Explained by an APOC3 Founder Mutation. Circ Cardiovasc Genet. 2017;10:e001809. doi: 10.1161/CIRCGENETICS.117.001809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanson R.L., Bogardus C., Duggan D., Kobes S., Knowlton M., Infante A.M., Marovich L., Benitez D., Baier L.J., Knowler W.C. A search for variants associated with young-onset type 2 diabetes in American Indians in a 100K genotyping array. Diabetes. 2007;56:3045–3052. doi: 10.2337/db07-0462. [DOI] [PubMed] [Google Scholar]
- 17.Hanson R.L., Muller Y.L., Kobes S., Guo T., Bian L., Ossowski V., Wiedrich K., Sutherland J., Wiedrich C., Mahkee D. A genome-wide association study in American Indians implicates DNER as a susceptibility locus for type 2 diabetes. Diabetes. 2014;63:369–376. doi: 10.2337/db13-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malhotra A., Kobes S., Knowler W.C., Baier L.J., Bogardus C., Hanson R.L. A genome-wide association study of BMI in American Indians. Obesity (Silver Spring) 2011;19:2102–2106. doi: 10.1038/oby.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bian L., Traurig M., Hanson R.L., Marinelarena A., Kobes S., Muller Y.L., Malhotra A., Huang K., Perez J., Gale A. MAP2K3 is associated with body mass index in American Indians and Caucasians and may mediate hypothalamic inflammation. Hum. Mol. Genet. 2013;22:4438–4449. doi: 10.1093/hmg/ddt291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piaggi P., Masindova I., Muller Y.L., Mercader J., Wiessner G.B., Chen P., Kobes S., Hsueh W.C., Mongalo M., Knowler W.C., SIGMA Type 2 Diabetes Consortium A Genome-Wide Association Study Using a Custom Genotyping Array Identifies Variants in GPR158 Associated With Reduced Energy Expenditure in American Indians. Diabetes. 2017;66:2284–2295. doi: 10.2337/db16-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanson R.L., Guo T., Muller Y.L., Fleming J., Knowler W.C., Kobes S., Bogardus C., Baier L.J. Strong parent-of-origin effects in the association of KCNQ1 variants with type 2 diabetes in American Indians. Diabetes. 2013;62:2984–2991. doi: 10.2337/db12-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanson R.L., Rong R., Kobes S., Muller Y.L., Weil E.J., Curtis J.M., Nelson R.G., Baier L.J. Role of Established Type 2 Diabetes-Susceptibility Genetic Variants in a High Prevalence American Indian Population. Diabetes. 2015;64:2646–2657. doi: 10.2337/db14-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller Y.L., Hanson R.L., Piaggi P., Chen P., Wiessner G., Okani C., Skelton G., Kobes S., Hsueh W.C., Knowler W.C. Assessing the Role of 98 Established Loci for BMI in American Indians. Obesity (Silver Spring) 2019;27:845–854. doi: 10.1002/oby.22433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller Y.L., Piaggi P., Chen P., Wiessner G., Okani C., Kobes S., Knowler W.C., Bogardus C., Hanson R.L., Baier L.J. Assessing variation across 8 established East Asian loci for type 2 diabetes mellitus in American Indians: Suggestive evidence for new sex-specific diabetes signals in GLIS3 and ZFAND3. Diabetes Metab. Res. Rev. 2017;33 doi: 10.1002/dmrr.2869. [DOI] [PubMed] [Google Scholar]
- 25.Nair A.K., Muller Y.L., McLean N.A., Abdussamad M., Piaggi P., Kobes S., Weil E.J., Curtis J.M., Nelson R.G., Knowler W.C. Variants associated with type 2 diabetes identified by the transethnic meta-analysis study: assessment in American Indians and evidence for a new signal in LPP. Diabetologia. 2014;57:2334–2338. doi: 10.1007/s00125-014-3351-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nair A.K., Piaggi P., McLean N.A., Kaur M., Kobes S., Knowler W.C., Bogardus C., Hanson R.L., Baier L.J. Assessment of established HDL-C loci for association with HDL-C levels and type 2 diabetes in Pima Indians. Diabetologia. 2016;59:481–491. doi: 10.1007/s00125-015-3835-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thearle M.S., Muller Y.L., Hanson R.L., Mullins M., Abdussamad M., Tran J., Knowler W.C., Bogardus C., Krakoff J., Baier L.J. Greater impact of melanocortin-4 receptor deficiency on rates of growth and risk of type 2 diabetes during childhood compared with adulthood in Pima Indians. Diabetes. 2012;61:250–257. doi: 10.2337/db11-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Traurig M.T., Perez J.M., Ma L., Bian L., Kobes S., Hanson R.L., Knowler W.C., Krakoff J.A., Bogardus C., Baier L.J. Variants in the LEPR gene are nominally associated with higher BMI and lower 24-h energy expenditure in Pima Indians. Obesity (Silver Spring) 2012;20:2426–2430. doi: 10.1038/oby.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muller Y.L., Piaggi P., Hoffman D., Huang K., Gene B., Kobes S., Thearle M.S., Knowler W.C., Hanson R.L., Baier L.J., Bogardus C. Common genetic variation in the glucokinase gene (GCK) is associated with type 2 diabetes and rates of carbohydrate oxidation and energy expenditure. Diabetologia. 2014;57:1382–1390. doi: 10.1007/s00125-014-3234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baier L.J., Muller Y.L., Remedi M.S., Traurig M., Piaggi P., Wiessner G., Huang K., Stacy A., Kobes S., Krakoff J. ABCC8 R1420H Loss-of-Function Variant in a Southwest American Indian Community: Association With Increased Birth Weight and Doubled Risk of Type 2 Diabetes. Diabetes. 2015;64:4322–4332. doi: 10.2337/db15-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller Y.L., Skelton G., Piaggi P., Chen P., Nair A., Kobes S., Hsueh W.C., Knowler W.C., Hanson R.L., Baier L.J., Bogardus C. Identification and functional analysis of a novel G310D variant in the insulin-like growth factor 1 receptor (IGF1R) gene associated with type 2 diabetes in American Indians. Diabetes Metab. Res. Rev. 2018;34:e2994. doi: 10.1002/dmrr.2994. [DOI] [PubMed] [Google Scholar]
- 32.Muller Y.L., Hanson R.L., Wiessner G., Nieboer L., Kobes S., Piaggi P., Abdussamad M., Okani C., Knowler W.C., Bogardus C., Baier L.J. Assessing FOXO1A as a potential susceptibility locus for type 2 diabetes and obesity in American Indians. Obesity (Silver Spring) 2015;23:1960–1965. doi: 10.1002/oby.21236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olaiya M.T., Hanson R.L., Kavena K.G., Sinha M., Clary D., Horton M.B., Nelson R.G., Knowler W.C. Use of graded Semmes Weinstein monofilament testing for ascertaining peripheral neuropathy in people with and without diabetes. Diabetes Res. Clin. Pract. 2019;151:1–10. doi: 10.1016/j.diabres.2019.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knowler W.C., Bennett P.H., Bottazzo G.F., Doniach D. Islet cell antibodies and diabetes mellitus in Pima Indians. Diabetologia. 1979;17:161–164. doi: 10.1007/BF01219743. [DOI] [PubMed] [Google Scholar]
- 35.Savage P.J., Bennett P.H., Senter R.G., Miller M. High prevalence of diabetes in young Pima Indians: evidence of phenotypic variation in a genetically isolated population. Diabetes. 1979;28:937–942. doi: 10.2337/diab.28.10.937. [DOI] [PubMed] [Google Scholar]
- 36.Dabelea D., Hanson R.L., Bennett P.H., Roumain J., Knowler W.C., Pettitt D.J. Increasing prevalence of Type II diabetes in American Indian children. Diabetologia. 1998;41:904–910. doi: 10.1007/s001250051006. [DOI] [PubMed] [Google Scholar]
- 37.Katzeff H.L., Savage P.J., Barclay-White B., Nagulesparan M., Bennett P.H. C-peptide measurement in the differentiation of type 1 (insulin-dependent) and type 2 (non-insulin-dependent) diabetes mellitus. Diabetologia. 1985;28:264–268. doi: 10.1007/BF00271682. [DOI] [PubMed] [Google Scholar]
- 38.Dewey F.E., Murray M.F., Overton J.D., Habegger L., Leader J.B., Fetterolf S.N., O’Dushlaine C., Van Hout C.V., Staples J., Gonzaga-Jauregui C. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science. 2016;354:aaf6814. doi: 10.1126/science.aaf6814. [DOI] [PubMed] [Google Scholar]
- 39.Assimes T.L., Lee I.T., Juang J.M., Guo X., Wang T.D., Kim E.T., Lee W.J., Absher D., Chiu Y.F., Hsu C.C. Genetics of Coronary Artery Disease in Taiwan: A Cardiometabochip Study by the Taichi Consortium. PLoS ONE. 2016;11:e0138014. doi: 10.1371/journal.pone.0138014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montasser M.E., Mc V.H.C.V., Farland R., Rosenberg A., Callaway M., Shen B., Li N., Daly T.J., Howard A.D., Lin W. Genetic and functional evidence relates a missense variant in B4GALT1 to lower LDL-C and fibrinogen. bioRxiv. 2019 doi: 10.1101/721704. [DOI] [Google Scholar]
- 41.Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sherry S.T., Ward M., Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. [PubMed] [Google Scholar]
- 43.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019 doi: 10.1101/531210. [DOI] [Google Scholar]
- 44.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yengo L., Sidorenko J., Kemper K.E., Zheng Z., Wood A.R., Weedon M.N., Frayling T.M., Hirschhorn J., Yang J., Visscher P.M., GIANT Consortium Meta-analysis of genome-wide association studies for height and body mass index in :700000 individuals of European ancestry. Hum. Mol. Genet. 2018;27:3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahajan A., Taliun D., Thurner M., Robertson N.R., Torres J.M., Rayner N.W., Payne A.J., Steinthorsdottir V., Scott R.A., Grarup N. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 2018;50:1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu D.J., Peloso G.M., Yu H., Butterworth A.S., Wang X., Mahajan A., Saleheen D., Emdin C., Alam D., Alves A.C., Charge Diabetes Working Group. EPIC-InterAct Consortium. EPIC-CVD Consortium. GOLD Consortium. VA Million Veteran Program Exome-wide association study of plasma lipids in >300,000 individuals. Nat. Genet. 2017;49:1758–1766. doi: 10.1038/ng.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 52.Chun S., Fay J.C. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 54.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Purcell S.M., Moran J.L., Fromer M., Ruderfer D., Solovieff N., Roussos P., O’Dushlaine C., Chambert K., Bergen S.E., Kähler A. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou W., Nielsen J.B., Fritsche L.G., Dey R., Gabrielsen M.E., Wolford B.N., LeFaive J., VandeHaar P., Gagliano S.A., Gifford A. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 2018;50:1335–1341. doi: 10.1038/s41588-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loh P.R., Tucker G., Bulik-Sullivan B.K., Vilhjálmsson B.J., Finucane H.K., Salem R.M., Chasman D.I., Ridker P.M., Neale B.M., Berger B. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farese R.V., Jr., Ruland S.L., Flynn L.M., Stokowski R.P., Young S.G. Knockout of the mouse apolipoprotein B gene results in embryonic lethality in homozygotes and protection against diet-induced hypercholesterolemia in heterozygotes. Proc. Natl. Acad. Sci. USA. 1995;92:1774–1778. doi: 10.1073/pnas.92.5.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raal F.J., Santos R.D., Blom D.J., Marais A.D., Charng M.J., Cromwell W.C., Lachmann R.H., Gaudet D., Tan J.L., Chasan-Taber S. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial. Lancet. 2010;375:998–1006. doi: 10.1016/S0140-6736(10)60284-X. [DOI] [PubMed] [Google Scholar]
- 60.Zhang S.H., Reddick R.L., Piedrahita J.A., Maeda N. Spontaneous hypercholesterolemia and arterial lesions in mice lacking apolipoprotein E. Science. 1992;258:468–471. doi: 10.1126/science.1411543. [DOI] [PubMed] [Google Scholar]
- 61.Ghiselli G., Schaefer E.J., Gascon P., Breser H.B., Jr. Type III hyperlipoproteinemia associated with apolipoprotein E deficiency. Science. 1981;214:1239–1241. doi: 10.1126/science.6795720. [DOI] [PubMed] [Google Scholar]
- 62.Rashid S., Curtis D.E., Garuti R., Anderson N.N., Bashmakov Y., Ho Y.K., Hammer R.E., Moon Y.A., Horton J.D. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc. Natl. Acad. Sci. USA. 2005;102:5374–5379. doi: 10.1073/pnas.0501652102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwartz G.G., Steg P.G., Szarek M., Bhatt D.L., Bittner V.A., Diaz R., Edelberg J.M., Goodman S.G., Hanotin C., Harrington R.A., ODYSSEY OUTCOMES Committees and Investigators Alirocumab and Cardiovascular Outcomes after Acute Coronary Syndrome. N. Engl. J. Med. 2018;379:2097–2107. doi: 10.1056/NEJMoa1801174. [DOI] [PubMed] [Google Scholar]
- 64.Smagris E., Gilyard S., BasuRay S., Cohen J.C., Hobbs H.H. Inactivation of Tm6sf2, a Gene Defective in Fatty Liver Disease, Impairs Lipidation but Not Secretion of Very Low Density Lipoproteins. J. Biol. Chem. 2016;291:10659–10676. doi: 10.1074/jbc.M116.719955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Homanics G.E., de Silva H.V., Osada J., Zhang S.H., Wong H., Borensztajn J., Maeda N. Mild dyslipidemia in mice following targeted inactivation of the hepatic lipase gene. J. Biol. Chem. 1995;270:2974–2980. doi: 10.1074/jbc.270.7.2974. [DOI] [PubMed] [Google Scholar]
- 66.Khetarpal S.A., Zeng X., Millar J.S., Vitali C., Somasundara A.V.H., Zanoni P., Landro J.A., Barucci N., Zavadoski W.J., Sun Z. A human APOC3 missense variant and monoclonal antibody accelerate apoC-III clearance and lower triglyceride-rich lipoprotein levels. Nat. Med. 2017;23:1086–1094. doi: 10.1038/nm.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hammond L.E., Gallagher P.A., Wang S., Hiller S., Kluckman K.D., Posey-Marcos E.L., Maeda N., Coleman R.A. Mitochondrial glycerol-3-phosphate acyltransferase-deficient mice have reduced weight and liver triacylglycerol content and altered glycerolipid fatty acid composition. Mol. Cell. Biol. 2002;22:8204–8214. doi: 10.1128/MCB.22.23.8204-8214.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Béréziat V., Kasus-Jacobi A., Perdereau D., Cariou B., Girard J., Burnol A.F. Inhibition of insulin receptor catalytic activity by the molecular adapter Grb14. J. Biol. Chem. 2002;277:4845–4852. doi: 10.1074/jbc.M106574200. [DOI] [PubMed] [Google Scholar]
- 69.Cooney G.J., Lyons R.J., Crew A.J., Jensen T.E., Molero J.C., Mitchell C.J., Biden T.J., Ormandy C.J., James D.E., Daly R.J. Improved glucose homeostasis and enhanced insulin signalling in Grb14-deficient mice. EMBO J. 2004;23:582–593. doi: 10.1038/sj.emboj.7600082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yao Z.M., Blackhart B.D., Linton M.F., Taylor S.M., Young S.G., McCarthy B.J. Expression of carboxyl-terminally truncated forms of human apolipoprotein B in rat hepatoma cells. Evidence that the length of apolipoprotein B has a major effect on the buoyant density of the secreted lipoproteins. J. Biol. Chem. 1991;266:3300–3308. [PubMed] [Google Scholar]
- 71.Peloso G.M., Nomura A., Khera A.V., Chaffin M., Won H.H., Ardissino D., Danesh J., Schunkert H., Wilson J.G., Samani N. Rare Protein-Truncating Variants in APOB, Lower Low-Density Lipoprotein Cholesterol, and Protection Against Coronary Heart Disease. Circ. Genom. Precis Med. 2019;12:e002376. doi: 10.1161/CIRCGEN.118.002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morrow J.A., Arnold K.S., Dong J., Balestra M.E., Innerarity T.L., Weisgraber K.H. Effect of arginine 172 on the binding of apolipoprotein E to the low density lipoprotein receptor. J. Biol. Chem. 2000;275:2576–2580. doi: 10.1074/jbc.275.4.2576. [DOI] [PubMed] [Google Scholar]
- 73.Zhao Z., Tuakli-Wosornu Y., Lagace T.A., Kinch L., Grishin N.V., Horton J.D., Cohen J.C., Hobbs H.H. Molecular characterization of loss-of-function mutations in PCSK9 and identification of a compound heterozygote. Am. J. Hum. Genet. 2006;79:514–523. doi: 10.1086/507488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Igal R.A., Wang S., Gonzalez-Baró M., Coleman R.A. Mitochondrial glycerol phosphate acyltransferase directs the incorporation of exogenous fatty acids into triacylglycerol. J. Biol. Chem. 2001;276:42205–42212. doi: 10.1074/jbc.M103386200. [DOI] [PubMed] [Google Scholar]
- 75.Rajakumar C., Ban M.R., Cao H., Young T.K., Bjerregaard P., Hegele R.A. Carnitine palmitoyltransferase IA polymorphism P479L is common in Greenland Inuit and is associated with elevated plasma apolipoprotein A-I. J. Lipid Res. 2009;50:1223–1228. doi: 10.1194/jlr.P900001-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lemas D.J., Wiener H.W., O’Brien D.M., Hopkins S., Stanhope K.L., Havel P.J., Allison D.B., Fernandez J.R., Tiwari H.K., Boyer B.B. Genetic polymorphisms in carnitine palmitoyltransferase 1A gene are associated with variation in body composition and fasting lipid traits in Yup’ik Eskimos. J. Lipid Res. 2012;53:175–184. doi: 10.1194/jlr.P018952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Skotte L., Koch A., Yakimov V., Zhou S., Søborg B., Andersson M., Michelsen S.W., Navne J.E., Mistry J.M., Dion P.A. CPT1A Missense Mutation Associated With Fatty Acid Metabolism and Reduced Height in Greenlanders. Circ Cardiovasc Genet. 2017;10:e001618. doi: 10.1161/CIRCGENETICS.116.001618. [DOI] [PubMed] [Google Scholar]
- 78.Brown N.F., Mullur R.S., Subramanian I., Esser V., Bennett M.J., Saudubray J.M., Feigenbaum A.S., Kobari J.A., Macleod P.M., McGarry J.D., Cohen J.C. Molecular characterization of L-CPT I deficiency in six patients: insights into function of the native enzyme. J. Lipid Res. 2001;42:1134–1142. [PubMed] [Google Scholar]
- 79.Popineau L., Morzyglod L., Carré N., Caüzac M., Bossard P., Prip-Buus C., Lenoir V., Ragazzon B., Fauveau V., Robert L. Novel Grb14-Mediated Cross Talk between Insulin and p62/Nrf2 Pathways Regulates Liver Lipogenesis and Selective Insulin Resistance. Mol. Cell. Biol. 2016;36:2168–2181. doi: 10.1128/MCB.00170-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tontonoz P., Mangelsdorf D.J. Liver X receptor signaling pathways in cardiovascular disease. Mol. Endocrinol. 2003;17:985–993. doi: 10.1210/me.2003-0061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genetic data of SWAI are protected materials and are not publicly available in respect of the IRB regulation. The data that support the reported findings and the codes that are used to generate figures are available from the corresponding authors upon reasonable request. Variants that are specifically described in the manuscript have been submitted to dbSNP (https://www.ncbi.nlm.nih.gov/snp) for public access.