Significance
Sex differences in brain organization are theoretically important for our understanding of sex differences in human cognition and behavior. However, neurobiological sex differences have been easier to characterize in mice than in humans. Recent murine work has revealed a highly reproducible spatial patterning of gray matter volume (GMV) sex differences that is centered on systems for socioreproductive behavior and correlated with regional expression of sex chromosome genes. We integrate neuroimaging and transcriptomic data to establish that these same characteristics also apply to GMV sex differences in humans. These findings establish conserved aspects of sex-biased brain development in humans and mice, and update our understanding of the consistency, candidate causes, and potential functional corollaries of sex-biased brain anatomy in humans.
Keywords: sex differences, gray matter volume, imaging-transcriptomics, sex chromosome
Abstract
Humans display reproducible sex differences in cognition and behavior, which may partly reflect intrinsic sex differences in regional brain organization. However, the consistency, causes and consequences of sex differences in the human brain are poorly characterized and hotly debated. In contrast, recent studies in mice—a major model organism for studying neurobiological sex differences—have established: 1) highly consistent sex biases in regional gray matter volume (GMV) involving the cortex and classical subcortical foci, 2) a preponderance of regional GMV sex differences in brain circuits for social and reproductive behavior, and 3) a spatial coupling between regional GMV sex biases and brain expression of sex chromosome genes in adulthood. Here, we directly test translatability of rodent findings to humans. First, using two independent structural-neuroimaging datasets (n > 2,000), we find that the spatial map of sex-biased GMV in humans is highly reproducible (r > 0.8 within and across cohorts). Relative GMV is female biased in prefrontal and superior parietal cortices, and male biased in ventral occipitotemporal, and distributed subcortical regions. Second, through systematic comparison with functional neuroimaging meta-analyses, we establish a statistically significant concentration of human GMV sex differences within brain regions that subserve face processing. Finally, by imaging-transcriptomic analyses, we show that GMV sex differences in human adulthood are specifically and significantly coupled to regional expression of sex-chromosome (vs. autosomal) genes and enriched for distinct cell-type signatures. These findings establish conserved aspects of sex-biased brain development in humans and mice, and shed light on the consistency, candidate causes, and potential functional corollaries of sex-biased brain anatomy in humans.
Biological sex robustly modifies risk for diverse human brain disorders (1, 2), with some conditions occurring more frequently in males (e.g., autism spectrum disorder), and others more commonly in females (e.g., major depressive disorder). Humans also show consistent sex biases in several cognitive-behavioral domains, including aggression (3), risk taking (4), mental rotation (5), and face processing (both emotional valence and identity) (6–8). These sex biases in disease risk and behavior emerge with stereotyped developmental timing and are highly consistent across different settings (2, 6)—suggesting a potential contribution from patterned sex differences in brain organization. Consequently, characterization of sex differences in human brain organization provides a critical context for understanding how biological sex might contribute to variation in disease risk and behavior.
The bulk of available data regarding sex biases in human brain organization has come from in vivo neuroimaging studies, and the majority of such studies have focused on sex differences in regional brain anatomy (9, 10). Although there is marked variability across reports, some putative foci of sex-biased brain anatomy have now been reported by independent large-scale neuroimaging studies. Specifically, after controlling for the fact that total brain volume is on average ∼10% larger in males than females, these independent reports have found that males show greater mean gray matter volume (GMV) of ventral occipitotemporal cortices, amygdala, putamen, and cerebellum than females, while the opposite sex difference in mean GMV is seen for superior frontal and lateral parietal cortices (10–12). These findings suggest that the human brain may show reproducible sex differences in regional GMV, prompting questions regarding the potential causes and consequences of sex-biased regional GMV in humans. These questions are not only hotly debated (13), but particularly hard to tackle empirically in humans, where access to experimental paradigms is limited.
To date, our most direct information regarding the reproducibility, causes, and consequences of sex-biased regional brain anatomy in mammals has come from work in rodents. Consequently, rodent data—especially from mice—have often served as a critical grounding for theoretical models of sex-biased brain organization in other mammals, including humans. Canonical foci of sex-biased brain volume in mice—as determined by a large corpus of targeted histological studies—include the bed nucleus of the stria terminalis (BNST), the medial amygdala (MeA), and the medial preoptic area (MPOA), all of which show higher volume in males relative to females (14, 15). These sex differences in regional brain volume are colocalized with sex differences in regional cellular composition (16), reside within key circuits for reproductive and social behaviors in rodents, and have been shown to largely arise through the masculinizing effects of gonadal steroids during perinatal life (17). However, this “subcortico-centric” and “gonad-centric” view of sex-biased mammalian brain development has recently been updated by a series of whole-brain neuroimaging studies in transgenic mice (18), posing important questions for our understanding of sex-biased brain development in humans.
First, in addition to replicating the classical histological reports of sex-biased BNST, MeA, and MPOA volume, recent high-resolution structural neuroimaging studies in mice have also identified several reproducible foci of sex-biased volume within the cortical sheet (19–21). The resulting spatially distributed set of regions with sex-biased GMV in mice involves several brain circuits that are known to support domains of sex-biased reproductive and social behavior (20). This observation and related experimental studies in mice suggest that regional GMV sex differences spanning both cortical and subcortical structures may also be highly reproducible in humans, and potentially may serve as a useful marker for capturing regions of sex-biased brain organization that are relevant for behavioral sex differences (22). To date, however, we lack both a formal test for the reproducibility of regional GMV sex differences in humans across independent samples, and a comprehensive comparison of sex-biased regional GMV in humans with functional neuroanatomy of the human brain.
Second, recent murine studies combining in vivo neuroimaging and postmortem gene expression data have indicated that the spatial map of GMV sex differences in mice is preferentially correlated with regional expression of sex-chromosome genes as measured in adult brains postmortem (20). This finding challenges the gonad-centric view of sex differences in mammalian brain anatomy by suggesting an important potential role for direct effects of sex-chromosome genes in shaping or maintaining regional sex differences in brain volume. However, it remains unknown if this close spatial association between volumetric sex differences and brain expression of sex-chromosome genes is also present in humans. More broadly, we lack a systematic transcriptomic annotation of regional sex differences in human brain anatomy.
Here, we seek to translate these recent advances in our understanding of neuroanatomical sex differences from mouse models to humans. We first map and directly quantify the spatial reproducibility of regional GMV sex differences in humans within and across two independent large-scale in vivo neuroimaging datasets totaling >2,000 scans (23, 24). We then functionally annotate the spatial pattern of regional GMV sex differences in humans through a systematic comparison with 50 metaanalytic maps from >11,000 functional neuroimaging studies (25, 26). Finally, we transcriptomically annotate regional sex differences in human brain anatomy by coregistering and comparing our spatial map of cortical GMV sex differences with a public atlas of expression for ∼16,000 genes in the human brain (27) provided by the Allen Institute for Brain Sciences (AIBS). This imaging-transcriptomic comparison 1) allows us to test whether sex differences in humans are indeed preferentially correlated with regional expression of sex-chromosome genes, as has been recently described in mice (20); and 2) provides a systematic ranking of brain-expressed genes in humans by the degree to which their expression is spatially coupled to regional variations of anatomical sex differences. We submit this ranked gene list to a detailed bioinformatic analysis to determine if regional sex biases in human brain anatomy possess a transcriptomic signature that is enriched for specific biological processes, cellular compartments, and cell types.
We find that the adult human brain shows a stereotyped pattern of regional sex differences in GMV (controlling for sex differences in total GMV) that is highly reproducible within and between cohorts (spatial correlation r > 0.8) and exists above and beyond sex differences in overall brain size. Functional annotation of this map identifies a statistically significant conjunction between regions of sex-biased GMV and a distributed set of brain regions that are preferentially engaged during face processing in humans. Finally, our comparison of neuroimaging and transcriptomic maps reveals that the spatial coupling between regional GMV sex differences and brain expression of sex chromosome genes, which was recently discovered in mice (20), is indeed also present in humans. In humans, this spatial coupling is strongest for a number of X- and Y-linked transcriptional regulators with established roles in forebrain development. Finally, we determine that the broader transcriptomic correlates of sex-biased GMV in humans are enriched for signatures of specific cell types and compartments, which differ for regions of male- vs. female-biased GMV.
Collectively, these empirical insights from large-scale integration of structural, functional, and molecular data provide an updated framework for our thinking regarding the potential causes and consequences for sex-biased regional brain development in humans.
Results
Sex Biases in Human Brain Volume Are Regionally Specific and Highly Reproducible.
We first quantified sex differences in regional GMV between age- and education-matched young healthy adults from the Human Connectome Project (HCP) dataset (488 males vs. 488 females, ages 22 to 35 y, mean 28.21 ± 3.24, SI Appendix, Table S1) using a well-established deformation-based morphometry (DBM)-based pipeline, the Diffeomorphic Anatomical Registration Through Exponentiated Lie (DARTEL) algebra (28). This pipeline provides better registration accuracy than other widely used nonlinear deformation algorithms (29). Using a unified model, the brain is precisely classified into distinct tissue classes (e.g., “gray matter,” “white matter”), and all of these tissue segmentations are warped into a standard imaging space of the Montreal Neurological Institute (MNI) (30). We estimated voxelwise sex differences in GMV (represented by the product of Jacobian determinant of the deformation field and gray matter probability at each voxel) while covarying for effects of age and total GMV.
In the primary HCP dataset, after correction for multiple comparisons to ensure familywise error (FWE) <0.05, we observed that mean relative GMV was significantly greater in females than males within the medial and lateral prefrontal, orbitofrontal, superior temporal, and lateral parietal cortices, and the insula. In contrast, GMV was significantly greater in males than females within: 1) ventral temporal and occipital regions including the temporal pole, fusiform gyrus, and primary visual primary cortex; and 2) a distributed set of subcortical regions encompassing the hypothalamus, BNST, amygdala, hippocampus, putamen, and cerebellum (Fig. 1A and SI Appendix, Table S2). The absolute effect sizes (Cohen’s d) associated with these regions of statistically significant GMV sex differences ranged from 0.23 to 0.63, with a mean of 0.33 across clusters (SI Appendix, Table S2). By repeating our analysis in 1,000 independent split-halves of the HCP dataset, we determined that the mean spatial correlation in GMV sex differences between independent halves of the HCP dataset was r = 0.86, and the range of this spatial correspondence statistic was 0.75 to 0.9 across all 1,000 tests (Fig. 1B). Thus, we observed a highly reproducible spatial pattern of GMV sex biases in this large cohort of healthy human adults.
Fig. 1.
Reproducible regional gray matter volume sex differences in the HCP and UK Biobank datasets. (A) Slice maps and surface projections showing statistically significant male-female differences in GMV after correction for multiple comparisons and controlling for covariates (age and total GMV) in the HCP dataset (full regional list in SI Appendix, Table S2). (B) Scatter and box plot showing cross-voxel spatial correlations for the unthresholded sex-difference t-statistic between 1,000 randomly split-halves of the HCP dataset (averaged Pearson’s r = 0.86, range: 0.75 to 0.90). (C) Density plot showing the observed cross-voxel spatial correlation (red line, Pearson’s r = 0.85) between unthresholded sex-difference t-statistic maps in the HCP dataset and UK Biobank sample (SI Appendix, Fig. S1 A and B) and a null distribution for this correlation statistic (from 1,000 reestimations of the spatial correlations after permutation of sex labels in the HCP dataset, Materials and Methods). The observed correlation lies outside the null distribution (i.e., Pperm < 0.001).
Next, we directly quantified the spatial reproducibility of HCP findings in a second fully independent cohort drawn from the UK Biobank (UKB) neuroimaging dataset (560 males vs. 560 females ages 44 to 50 y old, mean age 48.87 ± 1.14 y, SI Appendix, Table S1) (31). Of note, the HCP dataset and UKB sample represent nonoverlapping sets of individuals from different countries, drawn from nonoverlapping yet neighboring segments of the adult lifespan, and scanned using different MRI platforms. Despite these demographic and methodological contrasts between cohorts, the overall spatial pattern of sex differences in GMV was highly consistent between HCP and UKB datasets. Specifically, the observed spatial correlation between regional GMV sex differences in the HCP and UKB cohorts was r = 0.85. This correspondence statistic was significantly elevated relative to an empirical null distribution from comparison of the UKB sex-difference map with those from 1,000 permutations of sex labels in HCP (Pperm < 0.001, Fig. 1C and SI Appendix, Fig. S1). Taken together, these analyses firmly establish that human adults show highly reproducible sex differences in regional GMV after controlling for sex differences in overall brain size. We have posted statistical maps for sex-biased anatomy in the HCP and UKB datasets as reference datasets for wider use at https://neurovault.org/collections/OHYRQZTP/. A summary movie panning coronally through the brain is provided as Movie S1—showing regions of overlapping statistically significant sex differences in GMV across the HCP and UKB samples.
Sex Biases in Regional Human Brain Volume Overlap with Functional Systems That Support Sex-Biased Domains of Cognition.
To refine hypotheses regarding the potential functional consequences of sex differences in regional GMV, we used a well-validated and publicly available platform for “cognitive annotation” of brain maps from metaanalysis of over 11,000 functional neuroimaging studies (“NeuroSynth”) (25). A prior textual analysis of this public database has indicated that the large corpus of existing functional neuroimaging literature can be mapped onto a set of 50 underling “topics” which are each composed of conceptually related cognitive terms (e.g., an “inhibition,” “stroop,” and “suppression” topic vs. a “memory,” “encoding,” and “recall” topic), and each linked to an associated metaanalytic brain activation map (26). By examining the spatial correlation between regional GMV sex differences in the primary HCP cohort (i.e., t-statistic map for male-female contrast in GMV, Fig. 2A) and each of these 50 metaanalytic brain activation maps (called association statistic maps in z scores), we screened for specific topics with associated brain activation maps which were similar to the spatial distribution of sex differences in human GMV. Statistical significance of these spatial correlations between GMV sex differences and topic-specific brain activation maps (“Pperm”) was determined relative to a distribution of 1,000 null correlations from permutation of sex status in the HCP dataset.
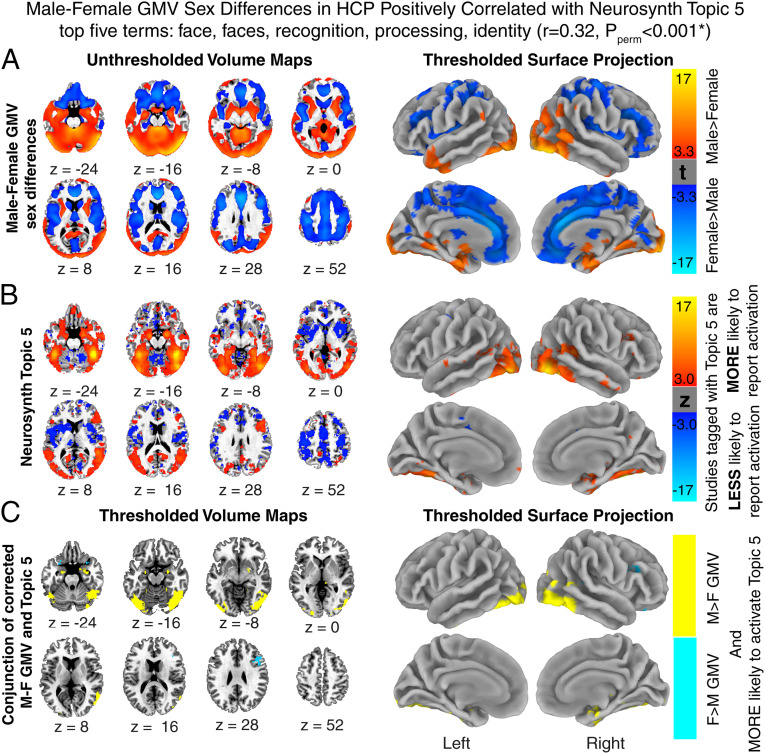
Fig. 2.
Colocalization of gray matter volume sex differences in humans with a distributed brain system that subserves face processing. (A) Reproduction of slice maps and surface projections showing unthresholded (slice maps) and statistically significant (thresholded surface projections) male-female GMV differences in the HCP dataset (compare Fig. 1A). (B) Slice maps and surface projections of the NeuroSynth association statistic in z scores show regions that are more (in warm colors) likely to be activated during tasks related to topic 5, mainly face processing. Pearson’s correlation of the unthresholded maps, with signs, of male-female GMV differences (Left in A) and association statistics of topic 5 (Left in B) is statistically significant (r = 0.32, Pperm < 0.001). (C) A binary conjunction map between cortical regions associated with NeuroSynth topic 5, and cortical regions of significant GMV sex differences. Note that this conjunction is largely composed of regions with male-biased (yellow) as opposed to female-biased (blue) GMV.
At an uncorrected empirical Pperm value of 0.05 (Materials and Methods), we established that the spatial distribution of sex-biased GMV in humans is significantly correlated at |r| ≥ 0.2 with metaanalytic maps for five broad cognitive domains: visual object recognition, face processing, cognitive control, inhibition, and conflict (SI Appendix, Table S3). Specifically, those cortical regions where GMV is greater in males than females tend to be involved in object recognition and face processing, whereas cortical regions showing the opposite sex differences in mean GMV are associated with inhibition, task control, and conflict. After Bonferroni correction for multiple comparisons (i.e., corrected Pperm value threshold across all 50 topics = 0.001), there remained a significant positive correlation (cross-voxel r > 0.3) between the spatial distribution of sex-biased GMV and the metaanalytic functional map for processing of faces and facial identity (Fig. 2 A and B). A further conjunction analysis illustrated that this correlation is reflected by a predominant overlap between cortical regions that are activated during face processing, and those that show significantly male-biased GMV, including the inferior occipital, inferior temporal, hippocampal, and cerebellar areas (Fig. 2C). The emergence of this significant overlap after strict permutation testing with added correction for multiple comparisons is striking, given that there is robust evidence from large datasets that mean performance is greater in females than males for processing of facial identify and emotions (6–8). Thus, there is a nonrandom association between the spatial distribution of sex biases in human brain volume and the spatial distribution of a distributed brain system that subserves a sex-biased domain of social cognition.
Sex Biases in Regional Brain Anatomy Show Preferential Spatial Coupling to Brain Expression of Sex-Chromosome Genes in Humans.
To test if humans also show the spatial association between sex chromosome gene expression and sex differences in GMV that has recently been reported in mice (20), we coregistered an imaging-derived map of regional GMV sex differences (i.e., a t-statistic map for male-female contrast in GMV, Fig. 3A) with publicly available maps of postmortem brain tissue gene expression from the AIBS, which detail expression of 16,906 genes in 1,317 bulk tissue cortical samples from a total of six healthy human donors (27) (Materials and Methods, Fig. 3A, and SI Appendix, Text S2 and Tables S4 and S5). By restricting these imaging-transcriptomic comparisons to the cortical sheet, we were able to interrogate gene-expression correlates for the full map of sex-biased GMV using a large number of postmortem samples from the same transcriptomic class of brain tissue (27).
Fig. 3.
Spatial coupling between human gray matter volume sex differences and expression of sex-chromosome genes. (A) Schematic of method used to rank genes based on the spatial coupling of brain expression with unthresholded regional sex differences (male-female) in cortical GMV. (B) Schematic showing how relative gene ranking relates to the relationship between gene expression and GMV sex biases. Bold borders denote genes within a set of interest (e.g., sex-linked genes), and the asterisk indicates the median rank for this gene set upon which inferences are made. The polarity of gene rankings is set so that more negative ranks indicate more positive correlation between gene expression and the t-statistic of GMV in males vs. females. This ranking positions sex-linked genes first in a “left-right” reading of C. (C) Point-range plot of the median rank (marked by circle with SD as error bar) of genes on the sex chromosomes and each autosome. Sex chromosomes (X- and Y-linked) genes uniquely showed a statistically significant extreme median rank (relative to the middle line/zero rank, Prand = 0.0014, all chromosome ranks given in SI Appendix, Table S7). The polarity of this rank extremity indicates a positive spatial correlation (marked in red) between sex-chromosome gene expression and male-female differences in GMV (i.e., relative high expression where GMV is greater in males than females and relatively low expression where GMV is greater in females than males). (D) Scatter plot of expression for X- and Y-linked genes ranked at the top 5% of SI Appendix, Table S6 (i.e., those with most extremely positive correlations with the t-statistic map of male-female GMV) vs. their aligned t-values of GMV sex differences (see details in Materials and Methods).
By associating each AIBS cortical sample with the test statistic for male-female GMV differences at its nearest voxel in neuroimaging space, we ranked all 16,906 genes in descending order by the cross-sample correlation between their expression and observed male-female differences in cortical GMV (Materials and Methods and Fig. 3 A and B). We first used this ranked gene list (SI Appendix, Table S6) to test if the spatial distribution of regional GMV sex differences in humans is preferentially correlated with the spatial expression of sex-chromosome genes. Supporting this hypothesis, we observed that median gene rank was more extreme for the set of sex-chromosome genes than for any autosomal gene set (Fig. 3C). Moreover, the observed median rank for sex-chromosome genes was statistically significantly extreme relative to null gene rankings based on 10,000 random reorderings of gene ranks (empirical Prand = 0.0014 < Bonferroni-corrected p threshold = 0.0022 [0.05/23]). In contrast, no autosomal gene sets possessed a median rank that was statistically significantly extreme relative to null expectations from rank permutation. These findings indicate that the spatial patterning of sex differences in human GMV shows a specific and statistically significant spatial association with cortical expression of sex-chromosome genes. The polarity of this coupling is such that cortical regions with relatively high expression of sex-chromosome genes tend to show greater GMV in males than females, and vice versa for regions with relatively low expression of sex-chromosome genes.
By next considering each sex chromosome’s gene set separately, we determined that the Y-linked and X-linked gene sets ranked first and second (respectively) relative to autosomal gene sets (Prand = 0.0105, SI Appendix, Fig. S2). Furthermore, an omnibus test of the sum of absolute distances of median ranks of X-linked and Y-linked genes to the center of the ranked gene list was also significantly different from zero (Prand = 0.0183, Materials and Methods)—indicating that in humans, these two chromosomal gene sets are collectively enriched for spatial associations between their expression with normative sex differences in regional GMV. The extreme ranking of sex-chromosome genes was reproduced when analyses were restricted to use of expression data from male AIBS donors (SI Appendix, Fig. S3). Our analytic approach was also able to specify which X- and Y-linked genes possess the strongest spatial correlation between their brain expression and sex differences in regional GMV (Fig. 3D and SI Appendix, Table S6). The top-four ranked X-linked and Y-linked genes (with relatively greater expression in regions of male-biased vs. female-biased GMV) included three from the protocadherin gene family (the paralog pair of PCDH11Y and PCDH11X, and PCDH19) that play a critical role in cell-cell recognition essential for development of the central nervous system (32), and ZNF711, which encodes a zinc finger protein transcription factor implicated in X-linked intellectual disability (33).
We conducted a series of sensitivity analyses to test for robustness of the observed spatial association between sex differences in regional GMV and regional expression of sex-chromosome genes. First, to assess the possibility that preferential correlation between regional GMV sex differences and regional sex chromosome gene expression could be confounded by variation among genes for the degree to which they are expressed in brain, we tested and confirmed that median brain expression was uncorrelated with rank position across genes (r = 0.15, Prand = 0.38). Second, we verified that our ranking of genes by their correlation with the unthresholded continuous test statistic for regional GMV sex differences (SI Appendix, Table S6) was convergent with results from an alternative “categorical” analysis. In this alternative categorical analysis, we identified all genes with statistically significant differential expression between those cortical regions that did show statistically significant sex differences in GMV and those that did not (SI Appendix, Text S3 and Fig. S4A and Materials and Methods). The four gene sets identified by this categorical analysis [overexpressed where GMV is greater in males (SI Appendix, Table S8), underexpressed where GMV is greater in males (SI Appendix, Table S10), and the same two differentially expressed gene groups where GMV is greater in females (SI Appendix, Tables S9 and S11)] were all extremely ranked (SI Appendix, Fig. S4B) in the ranked gene list from our main correlational analysis (SI Appendix, Table S6). This alternative approach to imaging-transcriptomic analysis also specified brain-expressed genes with strikingly reciprocal relationships between their expression and GMV sex differences: 1) 24 genes were both significantly up-regulated where GMV is greater in males and significantly down-regulated where GMV is greater in females (SI Appendix, Table S12), and 2) 37 genes showed the opposite reciprocal expression relationship with regions of statistically significant GMV sex differences (SI Appendix, Table S13). These two sets represent a previously unknown group of high-value genes for further study, given the exceptionally close and reciprocal relationship between their brain expression and the polarity of GMV sex differences.
Third, given that the AIBS postmortem gene expression data are derived from five male donors and one female donor, we took a number of steps to assess if the observed spatial coupling between GMV sex differences and brain expression of sex-chromosome genes was comparable between analyses using the female vs. the male AIBS donors. These analyses were necessarily restricted to X-linked genes, given the absence of Y-linked gene expression in females. As an initial test, we reran the imaging-transcriptomic workflow described in Fig. 3 A and B using male-only and female-only subsets of the AIBS brain gene expression dataset and observed that the ranking of X-linked genes was correlated at 0.33 (Spearman’s rho) between the male-only and female-only analyses, which was significantly different from 0 at an empirical P < 0.001. This result indicates that variation across X-linked genes in their spatial coupling to GMV sex biases is moderately stable across AIBS donor sex. For a further statistical test to examine whether donor sex has a significant effect on this imaging-transcriptomic relationship, we included donor sex as an interaction term in the regression models that were used to interrelate AIBS gene expression and regional GMV sex differences (SI Appendix, Text S2), and identified that donor sex was only a significant modifier of these relationships for two autosomal genes after correction for multiple comparisons, indicating that donor sex has a minimal effect on the imaging-transcriptomic relationship in the majority of 16,000 genes. Finally, to complement these tests, we also examined influences of AIBS donor sex on results from our categorical analyses of imaging-transcriptomic relationships (SI Appendix, Text S3, Fig. S4, and Tables S8–S13). Specifically, we calculated genewise estimates of expression fold change between regions of statistically significant male- vs. female-biased GMV, and compared these between analyses using expression data from male-only vs. female-only AIBS donors. For those X-linked genes that ranked in the top 5% of spatial correlation with the GMV sex differences (SI Appendix, Table S6, i.e., specific ones with higher expression in regions of larger-in-males and lower expression in regions of larger-in-females), the observed fold-change correlation between female and male-only AIBS donor analyses was 0.58 (Pearson’s r), significantly different from 0 at an empirical P < 0.001. This result indicates that the relative differential expression of X-linked genes between regions of male- vs. female-biased GMV is moderately stable across AIBS donor sex. Thus, to the limited extent permitted by the presence of only one female donor in the AIBS dataset, these three analyses collectively suggest that our core findings regarding spatial coupling between cortical sex-chromosome gene expression and cortical GMV sex biases are broadly stable across AIBS donor sex.
Collectively, these imaging-transcriptomic findings 1) establish that regionally specific sex differences in human brain anatomy are indeed preferentially correlated with regional expression of sex-linked genes, 2) specify which sex-linked genes display this spatial correlation with regional GMV sex differences most strongly, and 3) provide a systematic ranking of ∼16,000 genes by the strength of the spatial coupling between their regional expression and male-female differences in GMV.
Further Characterizing the Transcriptomic Correlates of Sex Biases in Human Brain Anatomy.
We next took several steps to more fully characterize the transcriptomic correlates of sex-biased regional GMV in humans and to determine if those genes that are most strongly coupled to sex-biased brain anatomy show enriched annotation for particular biological processes, cellular compartments, and cell types.
First, rank-based gene ontology (GO) enrichment analysis (34) identified annotations for several biological processes (BPs) and cellular compartments (CCs) that were enriched among those genes with expression patterns that strongly correlated with regional GMV sex differences. Of note, bidirectional rank-based analysis of the full-ranked gene list (SI Appendix, Table S6) revealed contrasting annotation enrichments for genes that are positively (SI Appendix, Table S14) vs. negatively (SI Appendix, Table S15) correlated with regional variation in male-female GMV differences (SI Appendix, Fig. S5). Specifically, genes that tended to be more highly expressed in regions of male-biased GMV are localized to the axonal growth cone and neuronal synapses and involved in synapse organization and G protein coupled receptor signaling (SI Appendix, Fig. S5 A and B and Table S14). In contrast, genes that tended to be more highly expressed in regions of female-biased GMV are most strongly enriched for compartment annotations relating to the extracellular matrix, organelles, the major histocompatibility protein complex (MHC), and for biological process annotations including regulation of cell proliferation, axonal guidance, and neuron recognition (SI Appendix, Fig. S5 C and D and Table S15). A notable negative finding was that the ranked gene list (SI Appendix, Table S6) for transcriptomic correlates of sex differences in regional GMV did not show any directional enrichment for Kyoto Encyclopedia of Genes and Genomes (KEGG) (35) or hand-annotated gene sets relating to sex-steroid biosynthesis or sex-steroid receptors (SI Appendix, Table S16).
Second, given that regional differences in cellular composition are thought to be a major driver for regional differences in bulk tissue expression in the cortex (36), we submitted extreme-ranking genes from our imaging-transcriptomic comparison (i.e., top and bottom 10% of the ranked gene list in SI Appendix, Table S6) to a publicly available platform for gene set cell signature enrichment analysis (CSEA) (37). Comparison with cell-specific gene sets indicated that genes with higher expression in regions of male-biased GMV were significantly enriched with signatures for deep-layer (5/6) cortical neurons (Fig. 4A), whereas genes with higher expression in regions of female-biased GMV were significantly enriched for oligodendrocyte and astrocyte gene sets (Fig. 4B). Taken together, these findings propose that cortical regions of sex-biased GMV in the human brain may show a distinctive histological patterning, with different cellular compositions for regions with a male vs. female bias in GMV.
Fig. 4.
Directional enrichment of cell type signatures in transcriptomic correlates of regional gray matter volume sex differences. Using CSEA (38), we identified cell types where genes were significantly overrepresented in the Top Left (A) and Bottom Right (B) 10% of the ranked gene list (SI Appendix, Table S6). CSEA was conducted in the seven cortical cell types (x axis) at specificity index thresholds (pSI) of varying stringency (color coded). Statistical significance is set as −log10. BH FDR-corrected P values in y axis is above the dash line corresponding to BH FDR-corrected P = 0.05.
Discussion
Our findings provide several insights into sex-biased human brain anatomy, which 1) address active controversies regarding the consistency of neuroanatomical sex differences, 2) demonstrate that sex differences in regional GMV are aligned with functional systems for face processing, 3) provide evidence for a close spatial relationship between sex differences in human brain anatomy and regional expression sex-chromosome genes, and 4) establish that genes which are most closely coupled to regional sex differences in cortical GMV anatomy are strongly associated with specific biological processes and cell types. We address each of these insights below.
We quantitatively demonstrate that the spatial patterning of regional GMV sex differences in humans (after control for sex differences in total GMV) is highly reproducible within and between two large cohorts of healthy adults (r > 0.8 correlation across voxels, Fig. 1). Such consistency in the spatial patterning of sex-biased brain anatomy in humans could arise through 1) sex differences in the innate biological programs that shape regional brain volume, and/or 2) systematic sex differences in exposure to, or experience of, environmental factors that can modify regional brain volume. Definitively arbitrating between these two contrasting mechanistic scenarios requires experimental approaches that are hard to implement in humans. However, four lines of evidence argue that sex-biased influences on regional brain volume can operate in a manner that is largely independent from environmental input. First, where experimental data are available from rodent models, stereotyped sex differences in regional brain volume have been linked to intrinsic male-female differences in sex-steroid signaling, which reflect genetically determined sexual differentiation of the gonads rather than sex-biased external environmental exposures (17). Second, in humans, observational studies in rare medical disorders have established that the two fundamental candidate sources for intrinsic, programmed sex differences in human development—sex steroids and sex chromosome dosage—can both influence the volume of those brain regions which show sex-biased anatomy in the general population (39–43). Third, sex biases in regional human brain anatomy have been reported at birth (44), when sources of neuroanatomical variation are limited to genes and the in utero environment. Finally, our observation in this study of a close spatial coupling between regional GMV sex differences and regional expression of sex-chromosome genes (further discussed below) is hard to parsimoniously incorporate into a purely environmental account for the stereotyped patterning of regional GMV sex differences. Thus, while human males and females undoubtedly experience systematically different environments at multiple levels of analysis, and while it is crucial to recognize the complex interplay between sex and gender (45), we propose that environmental factors are unlikely to be a primary driver of the highly reproducible spatial patterning of GMV sex differences observed in our current report.
Although our findings clearly establish that humans show highly reproducible sex differences in regional brain volume from in vivo imaging, it remains to be seen if these differences have any bearing for the many well-established sex biases in cognition, behavior, and mental health (1–8). We identify a statistically significant spatial overlap—albeit of relatively weak effect size—between regions of male-biased GMV, and a set of brain regions that are preferentially activated during face processing. By so highlighting face-processing systems after a broad screen of functional neuroanatomy through metaanalysis of >11,000 neuroimaging studies, our study provides an objective evidence base for targeting studies on the potential behavioral relevance of sex-biased regional GMV in humans. However, because experimentally testing for a causal relationship between neuroanatomical and behavioral sex differences is especially challenging in humans, these future studies would need to leverage emerging datasets (46) that, when fully assembled, will capture interindividual variation in multivariate neuroimaging and face-processing measures within large longitudinal samples of males and females over development.
Our study also newly establishes a conserved relationship between regional anatomical sex biases and regional sex-chromosome gene expression in humans, which has previously been known to exist only in mice (20). This observation is consistent with, but not causal evidence for, the hypothesis that sex-chromosome genes may be involved in establishing or maintaining some neuroanatomical differences between males and females. Such a hypothesis fundamentally challenges the classical gonad-centric view of sex-biased mammalian brain development (18), but has already received some complementary experimental support from neuroimaging studies in transgenic mice which have established that that sex chromosomes and gonadal steroids interact to influence GMV in several regions of sex-biased brain volume in wild-type groups (47). Equivalent transgenic studies are not possible in humans, although neuroimaging studies in patients with sex-chromosome aneuploidies have also identified spatially patterned sex-chromosome dosage effects on GMV that overlap with foci of sex-biased GMV in health (39–41). Furthermore, a similar conjunction between X-chromosome aneuploidy and sex effects on regional GMV has also been reported in mice (19). Taken together with these prior reports, our current findings strengthen the case that gonadal steroids are unlikely to be lone players in patterning regional GMV sex differences in humans. Our analytic approach also helps to guide future refinement of this claim by identifying which sex-chromosome genes are expressed in closest spatial alignment with (and therefore optimally localized to potentially shape) human GMV sex differences. Strikingly, the top four sex-chromosome genes identified by our methodology are all known to play important roles in structural and functional development of the central nervous system. In particular, three of these top four genes (PCDH11Y, PCDH11X, and PCDH19) all encode members of the nonclustered protocadherin protein subfamily that support axonal genesis, outgrowth, and targeting, as well as regulation of dendritic spines and synapses (48). Moreover, mutations in three of these genes have been linked to severe developmental disability syndromes (33, 49–52), providing quasiexperimental evidence for the neurodevelopmental relevance of these genes in humans. An important area for future work would be testing if those genes that show closest spatial alignment with regional variation in GMV sex differences also show significant sex differences in their brain expression. Challenges in pursuing this goal currently include the lack of spatiotemporally comprehensive comparative datasets of brain gene expression in males and females.
Finally, by extending our bioinformatic analysis beyond sex-chromosome genes, we provide a broader molecular and cellular annotation of the gene-expression signatures that track with regional GMV sex differences in the human cortical sheet. Better understanding the molecular and cellular features that distinguish cortical regions of sex-biased GMV from other cortices will help to narrow the search for those aspects of microstructural organization that underlie the macroanatomical sex biases visible to in vivo imaging. Our observation of a spatial coupling between cortical expression of astrocytic gene sets and cortical GMV sex differences is especially notable, given the evidence from animal models that astrocytes are key players in sculpting microstructural sex differences within some canonical regions of sex-biased brain volume such as the MPOA (53). An important next step toward validating these in silico results would be direct histological and cellular characterization in postmortem human tissue from those cortical regions which show the largest and most reproducible mesoscale volumetric sex differences in our current report (Movie S1).
Our findings should be considered in light of certain caveats and study limitations. First, we focus on mapping sex differences in regional GMV during adulthood and we compare these anatomical maps to gene-expression data that are also derived from adults. This approach has the advantage of resolving anatomical sex differences during a relatively stable developmental window which overlaps with the only age range for which we have anatomically comprehensive maps of postmortem gene expression from the brain (27). Furthermore, the use of adult neuroimaging and gene expression data also aligns our study with the mouse study which first reported a spatial association between regional GMV sex differences and regional expression of sex-chromosome genes (20). However, a limitation of this study design is that it cannot directly assess the earlier developmental emergence of sex biases in regional brain volume or their contemporaneous transcriptomic correlates. Thus, for the spatial coupling of sex chromosome gene expression and regional GMV sex differences in adulthood to reflect a causal relationship, one of two different scenarios must apply. First, this spatial coupling may be a passive “echo” of anatomical and expression profiles that were causally linked in earlier development. This hypothesis is supported by the broad stability of cortical expression gradients across postnatal (vs. prenatal) development (54) and the partial congruence between those regional GMV sex differences we report in adulthood, and those that have been reported at birth (44). Alternatively, regional expression of sex-chromosome genes may well play an active role in maintaining regional GMV sex differences in postnatal life. A similar phenomenon has been described for the importance of intact gonads for maintenance of MPOA volumetric sex differences postnatally (47).
A second limitation of our study is that we focus on GMV as our anatomical phenotype of interest, although regional volume is only one aspect of brain morphology (55, 56). However, GMV provides an anatomical phenotype that is equally applicable to cortical and subcortical structures, and more directly translatable between humans and mice, as compared to other potential phenotypes (c.f. cortical gyrification). Nevertheless, an important goal for future studies will be probing transcriptomic correlates of sex-biased brain organization using multivariate methods that can simultaneously incorporate multiple imaging phenotypes and modalities. A key part of this work will be using alternative (e.g., other nonlinear deformation, multiatlas, and surface-based algorithms) methods for image registration that open up consideration of more diverse phenotypes such as cortical thickness, surface area, and folding.
Lastly, although our analyses suggest that the spatial alignment between gene expression and GMV sex differences is broadly similar between the five male AIBS donors and the sole female AIBS donor, it will be important to more closely revisit this issue as suitably sex-balanced and spatially comprehensive maps of human cortical gene expression become available.
Notwithstanding these limitations and caveats, our study provides several key insights into sex-biased brain organization in humans. Specifically, we demonstrate that humans show highly reproducible sex differences in regional brain volume which coincide with systems involved in face processing and are spatially coupled with expression of gene sets tagging contrasting biological features in regions of male-biased vs. female-biased brain volume. Most strikingly, we verify that sex differences in human brain volume are indeed preferentially correlated with the expression gradients for sex-chromosome genes—as has been recently reported in mice. This phenomenon points toward a close and evolutionarily conserved relationship between regional expression of sex-chromosome genes and regional sex biases in brain development. The existence of this conserved relationship combines with newly emerging basic and clinical research on sex chromosome biology (39, 40, 47, 57, 58) to urge a reappraisal of the direct roles that X- and Y-chromosome genes may play in shaping sex-biased brain organization in humans and other mammals.
Materials and Methods
Participants and Neuroimaging Data.
The primary HCP sample included a sex-balanced set of 3T T1-weighted 0.7-mm isotropic anatomical scans for 976 healthy adults aged 22 to 35 y (488 males and females) downloaded from the HCP 1200 release (23). Males and females in this sample were group matched (p of two-sample t tests >0.05) for mean age (28 y) and years of education (15 y). Participant characteristics are detailed in SI Appendix, Table S1. Procedures for recruitment of HCP participants are detailed in ref. 23. A subset of the UKB dataset (31) (www.ukbiobank.ac.uk), defined by the available 5-y age range (44 to 50 y) closest to that of the HCP sample, was used as a replication dataset. This sex-balanced UKB subsample included T1-weighted images acquired on a single 3T scanner (Siemens Skyra, Siemens Healthcare) at an isotropic resolution of 1 mm (24), from 1,120 healthy adults (560 males and females). Males and females were group matched (p of two-sample t tests >0.05) for mean age (49 y) and years of educations (17 y). Participant characteristics are detailed in SI Appendix, Table S1. Procedures for recruitment of HCP participants are detailed in ref. 24. Informed consent was obtained from all participants. Analyses of the HCP and UKB data and the research protocol were approved by the institutional review board at the National Institute of Mental Health. All included participant scans passed quality examination as detailed in SI Appendix, Text S1.
Voxelwise Analysis of Sex Differences in Gray Matter Volume.
Nonlinear deformation-based DARTEL analyses (28) (see details in SI Appendix, Text S1) were applied independently to the HCP and UKB datasets to map sex differences in voxelwise GMV. This image-processing pipeline ensures that 1) individual images are precisely aligned in a standard imaging space (MNI) (59) and 2) segmentation and normalization of gray matter, white matter, and cerebrospinal fluid (CSF), skull, dura, and background are achieved in a unified model (30). All normalized images used in neuroimaging analysis (including tissue segmentation probability maps) passed visual inspection for artifacts or inaccurate segmentation and normalization (see details in SI Appendix, Text S1). Gray matter volume was calculated at each voxel in the cortical and subcortical gray matter areas and compared between male and female groups using a voxelwise ANOVA model with covariates of age and total GMV. Voxelwise t-statistics for the effect of sex on GMV were corrected for whole-brain comparisons using AFNI (60) 3dClustSim, after the bug fix in May 2015, estimating the spatial autocorrelation function from the data to ensure FWE <0.05. Anatomical labeling for the peak voxel from each cluster of statistically significant sex differences was determined using the Automated Anatomical Labeling (AAL) atlas (61). As this parcellation does not include the BNST or hypothalamus—structures of a priori interest from murine studies of sex-biased GMV—we identified voxels that lay within these structures using separate atlases (62, 63). The same pipeline for voxelwise analysis of GMV sex differences was separately applied to both HCP (Fig. 1A and SI Appendix, Fig. S1A and Table S2) and UKB (SI Appendix, Fig. S1B) datasets.
Reproducibility of Sex Differences within the HCP Dataset.
To determine the reproducibility of regional GMV sex differences within the HCP dataset, we randomly split the full HCP sample into half and recalculated the voxelwise GMV sex differences in each split-half sample. This procedure was repeated 1,000 times. The spatial correspondence in GMV sex differences between the two halves of each split was calculated as the Pearson correlation coefficient across voxels for the t-value associated with the beta coefficient for sex. The distribution of this correlation coefficient across all 1,000 split-halves represents the topographical stability of GMV sex differences within the HCP sample (Fig. 1B).
Replication of HCP Sex Differences in UKB Dataset.
We quantified the spatial correspondence in GMV sex differences between the HCP and UKB datasets as the Pearson correlation coefficient across voxels for the t-value associated with the beta coefficient for sex in each (SI Appendix, Fig. S1 A and B). We compared this observed correlation coefficient against an empirical null distribution generated by recomputing the agreement between cohorts for 1,000 GMV sex-difference maps in the HCP cohort with sex permuted across participants (Fig. 1C). This permutation procedure yields a nonparametric P value (Pperm) for the significance of spatial correlations between two maps while accounting for 1) the shared spatial autocorrelation between maps, and 2) the arbitrary variations in power that would otherwise arise from changes in voxel resolution.
Functional Annotation of Regional GMV Sex Differences Using NeuroSynth.
We compared the spatial pattern of GMV sex differences to functional anatomy of the human brain using NeuroSynth (neurosynth.org), an online platform for metaanalysis of functional neuroimaging literature (25). In prior research, a topic modeling technique applied to the text of over 11,406 studies in the NeuroSynth database defined a set of 50 topics capturing conceptually distinct aspects of human cognition. Each topic represents a cluster of weighted text terms with shared relevance for a common overarching cognitive construct (26). NeuroSynth uses these weighted term clusters to calculate topic-specific association test maps, in which each voxel’s association statistic (z score) represents the likelihood that that voxel is preferentially activated by the topic in question over all other topics. We calculated cross-voxel correlations (Pearson’s r) between the unthresholded t-statistic for GMV sex differences in the HCP dataset (Fig. 2A) and the NeuroSynth association statistics for each of these 50 topic maps, and signs of both maps were kept in calculation. The statistical significance of each correlation was established relative to a null distribution of correlation coefficients from 1,000 permutations of sex status in the HCP dataset (i.e., Pperm). We report findings at two thresholds: 1) a liberal joint threshold of |r| > 0.2 and Pperm < 0.05 for display purpose (SI Appendix, Table S3) and 2) a stringent Bonferroni-corrected threshold of Pperm < 0.05/50 (Fig. 2). Following Bonferroni correction, only topic 5 showed a significant correlation with the GMV sex differences. To illustrate this correlation, we created a binary conjunction map (Fig. 2C) between maps of significant GMV sex differences (Right in Fig. 2A) and activated regions associated with NeuroSynth topic 5 (Right in Fig. 2B).
Ranking Genes on the Spatial Correlations Between Their Expression in Brain and Sex Differences in Regional Cortical GMV.
Using our recently published method (64), we ranked genes on the spatial correlations between their regional expression in health [using postmortem data for six donors (SI Appendix, Table S4) from the AIBS (27)], and the regional t-statistic for male-female GMV differences in the HCP data (see schematic of method in Fig. 3A, and details in SI Appendix, Text S2). The AIBS adult human brain atlas offers normalized microarray expression profiles for 58,692 probes at 1,317 spatially distributed bulk tissue samples from the left hemisphere cortex of six donors (SI Appendix, Table S4). We mapped probe-level expression data to 16,906 unique genes as previously described (64, 65). Briefly, each AIBS postmortem sample is ascribed to unique coordinates in MNI stereotaxic space. These coordinates were used to assign each postmortem sample a t-statistic from the map of GMV sex differences (from the voxel closest to the sample). This procedure resulted in a unique pairing of expression values for 16,906 genes with an imaging estimate of male-female GMV differences at each of 1,317 cortical locations (SI Appendix, Table S5). We used linear mixed-effect (LME) models with a fixed effect for GMV variations across samples and a random intercept for donor to model variance dependence between samples from the same brains (Fig. 3A). The outputs from these genewise models enabled us to rank 16,906 genes (by default in descending order) based on their spatial correlations with sex differences (male-female) in regional cortical GMV (Fig. 3 A and B and SI Appendix, Table S6). Of note, our image-transcriptomic comparison was intentionally constrained to the cerebral cortex without the hippocampus and cerebellum and subcortical areas to avoid biases due to gross transcriptional dissimilarities of gene expression levels in these regions (27).
Testing for a Spatial Correspondence Between GMV Sex Differences and Regional Expression of Sex-Chromosome Genes.
The ranking of genes by their spatial correlations with GMV sex differences (demonstrated in Fig. 3B, and see the full list in SI Appendix, Table S6) was used to test for a preferential spatial coupling between GMV sex differences and expression of sex-chromosome genes. We first calculated median ranks for 23 nonoverlapping gene sets representing each autosome (1 through 22) and a combined gene set including all X- and Y-linked genes. For each chromosomal gene set, we then tested if the observed gene set median rank was significantly extreme relative to a null distribution of median ranks identified from 10,000 same-sized fake lists generated by randomly reordering the original ranked list (Fig. 3C). This random-ordering permutation test provided a nonparametric test for statistical significance (Prand), which was also used in subsequent gene enrichment analyses. For the cross-chromosome comparisons, statistical significance was defined using a Bonferroni-corrected threshold of Prand < 0.0022 (0.05/23). To further isolate the spatial correlations at the level of each sex chromosome, we 1) separately tested the enrichment of X- and Y-chromosome genes by comparing their gene set median ranks relative to those for all other chromosomes (SI Appendix, Fig. S2) and 2) also used an omnibus test to examine whether the total absolute displacement of X- and Y-chromosome gene set median ranks from the middle of our ranked gene list was significantly large relative to null expectations random size-matched gene set resampling (i.e., Prand < 0.05). To directly illustrate the spatial correspondence between sex-chromosome gene expression and GMV sex differences, we generated regional estimates of average expression across donors for sex-linked genes that were most closely coupled to GMV sex differences in our main analyses (i.e., genes ranked at the top 5% in SI Appendix, Table S6) and correlated these regional expression estimates with corresponding regional averages of the t-statistic for GMV sex differences (Fig. 3D). In these analyses, regional assignments were given by AIBS anatomical nomenclature, and gene expression values were residuals after regressing out the main effects of AIBS donor.
Testing the Robustness of Sex Chromosome Gene Enrichment.
We used several complementary analyses to test the robustness of sex chromosome gene enrichment. First, to exclude the possibility that this ranked gene list from our primary analysis (Fig. 3 A and B and SI Appendix, Table S6) was determined by the gene expression level rather than by its intrinsic pattern of spatial distribution as designed, we verified that lack of a statistically significant correspondence (i.e., Prand > 0.05) between gene rank (SI Appendix, Table S6) and the mean amplitude of gene expression across all AIBS cortical samples. Second, we confirmed that gene ranking from our core method of imaging-transcriptomic analysis (based on the spatial correlations with the unthresholded t-statistic for GMV sex differences as detailed above and in Fig. 3A) were congruent with results from an alternative imaging-transcriptomic analysis based on defined regions of statistically significant sex differences in GMV (illustrated in SI Appendix, Fig. S3A and detailed in SI Appendix, Text S3). Third, we examined the influence of AIBS donor sex on observed imaging-transcriptomic relationships through three complementary tests as described in Results above.
Gene Ontology and Cell-Type-Specific Expression Analyses.
We used three complementary analyses to place the transcriptomic correlates of GMV sex differences (i.e., SI Appendix, Table S6 ranked gene list) in a richer biological context. First, we conducted rank-based GO enrichment analysis for BP and CC terms using GOrilla (34) (cbl-gorilla.cs.technion.ac.il/). Significant GO terms (Benjamini–Hochberg (BH) false discovery rate [FDR] corrected P < 0.05) were visualized using REViGO (revigo.irb.hr) for semantic space reduction (66) (SI Appendix, Fig. S5 and Tables S14 and S15). Second, to test for potential sex-steroid signaling associations with regional GMV sex differences, we tested two sets of genes (SI Appendix, Table S16) related to sex-steroid signaling for rank enrichment (at Prand < 0.05) in the ranked gene list (Fig. 3 A and B and SI Appendix, Table S6): 1) genes associated with the synthesis of sex-related steroids (testosterone, estrogen, progesterone, adrenal androgens: dehydroepiandrosterone and androstenedione, progesterone-derived neurosteroids: allopregnanolone and pregnenolone) via KEGG (rest.kegg.jp/get/hsa00140) and 2) a hand-curated list of genes encoding steroid sex hormone receptors (SI Appendix, Table S16). Third, using an online platform for CSEA (genetics.wustl.edu/jdlab/csea-tool-2/) (38), we examined whether genes with strong positive or negative spatial correlation with regional GMV sex differences (i.e., top/bottom 10% of ranked list in SI Appendix, Table S6) showed preferential enrichment for expression signatures for any of seven cortical cell types (Fig. 4, Pnoc+, layer 6, layer 5b, Cort+ neurons and layer 5A and immune cells, oligodendrocyte progenitors, astrocytes, and oligodendrocytes).
Supplementary Material
Acknowledgments
This study was supported by the intramural research program of the National Institute of Mental Health (NIMH) (project funding: 1ZIAMH002949-03, clinical trials identifier: NCT00001246, protocol : 89-M-0006). HCP data were provided (in part) by the Washington University - University of Minnesota Consortium of the Human Connectome Project (principal investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University. In addition, this research has been conducted using the UK Biobank Resource under application 22875. We would also like to thank Dr. Peter Schmidt of the NIMH Intramural Research Program for his assistance with curation of steroidal gene lists.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919091117/-/DCSupplemental.
Data Availability.
The HCP data used in this study are available through the database at https://db.humanconnectome.org/. The UKB data used in this study can be obtained at https://www.ukbiobank.ac.uk.
References
- 1.McCarthy M. M., Multifaceted origins of sex differences in the brain. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150106 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rutter M., Caspi A., Moffitt T. E., Using sex differences in psychopathology to study causal mechanisms: Unifying issues and research strategies. J. Child Psychol. Psychiatry 44, 1092–1115 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Archer J., Sex differences in aggression in real-world settings: A meta-analytic review. Rev. Gen. Psychol. 8, 291–322 (2004). [Google Scholar]
- 4.Cross C. P., Copping L. T., Campbell A., Sex differences in impulsivity: A meta-analysis. Psychol. Bull. 137, 97–130 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Lippa R. A., Collaer M. L., Peters M., Sex differences in mental rotation and line angle judgments are positively associated with gender equality and economic development across 53 nations. Arch. Sex. Behav. 39, 990–997 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Gur R. C. et al., Age group and sex differences in performance on a computerized neurocognitive battery in children age 8-21. Neuropsychology 26, 251–265 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herlitz A., Lovén J., Sex differences and the own-gender bias in face recognition: A meta-analytic review. Vis. Cogn. 21, 1306–1336 (2013). [Google Scholar]
- 8.Olderbak S., Wilhelm O., Hildebrandt A., Quoidbach J., Sex differences in facial emotion perception ability across the lifespan. Cogn. Emotion 33, 579–588 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Kaczkurkin A. N., Raznahan A., Satterthwaite T. D., Sex differences in the developing brain: Insights from multimodal neuroimaging. Neuropsychopharmacology 44, 71–85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruigrok A. N. V. et al., A meta-analysis of sex differences in human brain structure. Neurosci. Biobehav. Rev. 39, 34–50 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ritchie S. J. et al., Sex differences in the adult human brain: Evidence from 5216 UK Biobank participants. Cereb. Cortex 28, 2959–2975 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lotze M. et al., Novel findings from 2,838 adult brains on sex differences in gray matter brain volume. Sci. Rep. 9, 1671 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joel D., McCarthy M. M., Incorporating sex as a biological variable in neuropsychiatric research: Where are we now and where should we be? Neuropsychopharmacology 42, 379–385 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorski R. A., Gordon J. H., Shryne J. E., Southam A. M., Evidence for a morphological sex difference within the medial preoptic area of the rat brain. Brain Res. 148, 333–346 (1978). [DOI] [PubMed] [Google Scholar]
- 15.Hines M., Allen L. S., Gorski R. A., Sex differences in subregions of the medial nucleus of the amygdala and the bed nucleus of the stria terminalis of the rat. Brain Res. 579, 321–326 (1992). [DOI] [PubMed] [Google Scholar]
- 16.Kim Y. et al., Brain-wide maps reveal stereotyped cell-type-based cortical architecture and subcortical sexual dimorphism. Cell 171, 456–469.e22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris J. A., Jordan C. L., Breedlove S. M., Sexual differentiation of the vertebrate nervous system. Nat. Neurosci. 7, 1034–1039 (2004). [DOI] [PubMed] [Google Scholar]
- 18.McCarthy M. M., Arnold A. P., Reframing sexual differentiation of the brain. Nat. Neurosci. 14, 677–683 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raznahan A. et al., Triangulating the sexually dimorphic brain through high-resolution neuroimaging of murine sex chromosome aneuploidies. Brain Struct. Funct. 220, 3581–3593 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu L. R. et al., Mouse MRI shows brain areas relatively larger in males emerge before those larger in females. Nat. Commun. 9, 2615 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spring S., Lerch J. P., Henkelman R. M., Sexual dimorphism revealed in the structure of the mouse brain using three-dimensional magnetic resonance imaging. Neuroimage 35, 1424–1433 (2007). [DOI] [PubMed] [Google Scholar]
- 22.McCarthy M. M., Wright C. L., Schwarz J. M., New tricks by an old dogma: Mechanisms of the organizational/activational hypothesis of steroid-mediated sexual differentiation of brain and behavior. Horm. Behav. 55, 655–665 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Essen D. C. et al.; WU-Minn HCP Consortium , The human connectome project: A data acquisition perspective. Neuroimage 62, 2222–2231 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller K. L. et al., Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat. Neurosci. 19, 1523–1536 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yarkoni T., Poldrack R. A., Nichols T. E., Van Essen D. C., Wager T. D., Large-scale automated synthesis of human functional neuroimaging data. Nat. Methods 8, 665–670 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poldrack R. A. et al., Discovering relations between mind, brain, and mental disorders using topic mapping. PLOS Comput. Biol. 8, e1002707 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hawrylycz M. J. et al., An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashburner J., A fast diffeomorphic image registration algorithm. Neuroimage 38, 95–113 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Klein A. et al., Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage 46, 786–802 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashburner J., Friston K. J., Unified segmentation. Neuroimage 26, 839–851 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Collins R., What makes UK Biobank special? Lancet 379, 1173–1174 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Frank M., Kemler R., Protocadherins. Curr. Opin. Cell Biol. 14, 557–562 (2002). [DOI] [PubMed] [Google Scholar]
- 33.van der Werf I. M. et al., Mutations in two large pedigrees highlight the role of ZNF711 in X-linked intellectual disability. Gene 605, 92–98 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z., GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanehisa M., Goto S., KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lein E. S., Belgard T. G., Hawrylycz M., Molnár Z., Transcriptomic perspectives on neocortical structure, development, evolution, and disease. Annu. Rev. Neurosci. 40, 629–652 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Xu X., Wells A. B., O’Brien D. R., Nehorai A., Dougherty J. D., Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J. Neurosci. 34, 1420–1431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dougherty J. D., Schmidt E. F., Nakajima M., Heintz N., Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res. 38, 4218–4230 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raznahan A. et al., Globally divergent but locally convergent X- and Y-chromosome influences on cortical development. Cereb. Cortex 26, 70–79 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mankiw C. et al., Allometric analysis detects brain size-independent effects of sex and sex chromosome complement on human cerebellar organization. J. Neurosci. 37, 5221–5231 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reardon P. K. et al., An allometric analysis of sex and sex chromosome dosage effects on subcortical anatomy in humans. J. Neurosci. 36, 2438–2448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nadig A. et al., Carriage of supernumerary sex chromosomes decreases the volume and alters the shape of limbic structures. eNeuro 5, ENEURO.0265-18.2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giedd J. N. et al., Puberty-related influences on brain development. Mol. Cell. Endocrinol. 254–255, 154–162 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Knickmeyer R. C. et al., Impact of sex and gonadal steroids on neonatal brain structure. Cereb. Cortex 24, 2721–2731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rippon G., Jordan-Young R., Kaiser A., Fine C., Recommendations for sex/gender neuroimaging research: Key principles and implications for research design, analysis, and interpretation. Front. Hum. Neurosci. 8, 650 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garavan H. et al., Recruiting the ABCD sample: Design considerations and procedures. Dev. Cogn. Neurosci. 32, 16–22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vousden D. A. et al., Impact of X/Y genes and sex hormones on mouse neuroanatomy. Neuroimage 173, 551–563 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Peek S. L., Mah K. M., Weiner J. A., Regulation of neural circuit formation by protocadherins. Cell. Mol. Life Sci. 74, 4133–4157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolc K. L. et al., A systematic review and meta-analysis of 271 PCDH19-variant individuals identifies psychiatric comorbidities, and association of seizure onset and disease severity. Mol. Psychiatry 24, 241–251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith L. et al., PCDH19-related epilepsy is associated with a broad neurodevelopmental spectrum. Epilepsia 59, 679–689 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Speevak M. D., Farrell S. A., Non-syndromic language delay in a child with disruption in the Protocadherin11X/Y gene pair. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 156B, 484–489 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Veerappa A. M., Saldanha M., Padakannaya P., Ramachandra N. B., Genome-wide copy number scan identifies disruption of PCDH11X in developmental dyslexia. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 162B, 889–897 (2013). [DOI] [PubMed] [Google Scholar]
- 53.McCarthy M. M., Wright C. L., Convergence of sex differences and the neuroimmune system in autism spectrum disorder. Biol. Psychiatry 81, 402–410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu Y. et al., Spatiotemporal transcriptomic divergence across human and macaque brain development. Science 362, eaat8077 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gennatas E. D. et al., Age-related effects and sex differences in gray matter density, volume, mass, and cortical thickness from childhood to young adulthood. J. Neurosci. 37, 5065–5073 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lerch J. P. et al., Studying neuroanatomy using MRI. Nat. Neurosci. 20, 314–326 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Bellott D. W. et al., Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508, 494–499 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raznahan A. et al., Sex-chromosome dosage effects on gene expression in humans. Proc. Natl. Acad. Sci. U.S.A. 115, 7398–7403 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grabner G. et al., . “Symmetric atlasing and model based segmentation: An application to the Hippocampus in older adults” in Springer Berlin Heidelberg, Larsen R., Nielsen M., Sporring J., Eds. (2006), pp. 58–66. [DOI] [PubMed] [Google Scholar]
- 60.Cox R. W., AFNI: Software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 29, 162–173 (1996). [DOI] [PubMed] [Google Scholar]
- 61.Tzourio-Mazoyer N. et al., Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15, 273–289 (2002). [DOI] [PubMed] [Google Scholar]
- 62.Torrisi S. et al., Resting state connectivity of the bed nucleus of the stria terminalis at ultra-high field. Hum. Brain Mapp. 36, 4076–4088 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pauli W. M., Nili A. N., Tyszka J. M., A high-resolution probabilistic in vivo atlas of human subcortical brain nuclei. Sci. Data 5, 180063 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reardon P. K. et al., Normative brain size variation and brain shape diversity in humans. Science 360, 1222–1227 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Richiardi J. et al.; IMAGEN consortium , BRAIN NETWORKS. Correlated gene expression supports synchronous activity in brain networks. Science 348, 1241–1244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Supek F., Bošnjak M., Škunca N., Šmuc T., REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The HCP data used in this study are available through the database at https://db.humanconnectome.org/. The UKB data used in this study can be obtained at https://www.ukbiobank.ac.uk.