Significance
Many potential drug targets have been identified, but development of chemical probes to validate these targets has lagged behind. Computational screening holds promise for providing chemical tools to do so but has long been plagued by high false-positive rates: Many compounds ranked highly against a given target protein do not actually show activity. Machine learning approaches have not solved this problem, which we hypothesize is because models were not trained on sufficiently compelling “decoys.” By addressing this through a unique training strategy, we show that more effective virtual screening is attainable. We expect this insight to enable improved performance across diverse virtual screening pipelines, thus helping to provide chemical probes for new potential drug targets as they are discovered.
Keywords: virtual screening, machine learning classifier, structure-based drug design, protein–ligand complex
Abstract
With the recent explosion in the size of libraries available for screening, virtual screening is positioned to assume a more prominent role in early drug discovery’s search for active chemical matter. In typical virtual screens, however, only about 12% of the top-scoring compounds actually show activity when tested in biochemical assays. We argue that most scoring functions used for this task have been developed with insufficient thoughtfulness into the datasets on which they are trained and tested, leading to overly simplistic models and/or overtraining. These problems are compounded in the literature because studies reporting new scoring methods have not validated their models prospectively within the same study. Here, we report a strategy for building a training dataset (D-COID) that aims to generate highly compelling decoy complexes that are individually matched to available active complexes. Using this dataset, we train a general-purpose classifier for virtual screening (vScreenML) that is built on the XGBoost framework. In retrospective benchmarks, our classifier shows outstanding performance relative to other scoring functions. In a prospective context, nearly all candidate inhibitors from a screen against acetylcholinesterase show detectable activity; beyond this, 10 of 23 compounds have IC50 better than 50 μM. Without any medicinal chemistry optimization, the most potent hit has IC50 280 nM, corresponding to Ki of 173 nM. These results support using the D-COID strategy for training classifiers in other computational biology tasks, and for vScreenML in virtual screening campaigns against other protein targets. Both D-COID and vScreenML are freely distributed to facilitate such efforts.
Advances in biomedical sciences, driven especially by the advent of next-generation genome sequencing technologies, have enabled discovery of many new potential drug targets (1, 2). Ultimately, however, validating a new candidate target for therapeutic intervention requires development of a chemical probe to explore the consequences of pharmacological manipulation of this target (3). In recent years, this step has typically been carried out by using high-throughput screening (HTS) (4) as a starting point for subsequent medicinal chemistry optimization; with improvements in automation, it has become feasible to screen libraries that exceed a million compounds (5).
More recently, however, sets of robust chemical transformations from available building blocks have been used to enumerate huge libraries of compounds that are readily accessible but never before synthesized (6–9). These libraries can comprise billions of compounds, and thus remain far beyond the scale accessible to even the most ambitious HTS campaign. This expansion of chemical space in which to search, along with the high cost of setting up and implementing an HTS screen, has increasingly driven the use of complementary computational approaches.
In broad terms, virtual screening approaches can be categorized into two classes: ligand-based screens and structure-based screens (10–12). Ligand-based screening starts from the (two-dimensional [2D] or three-dimensional [3D]) structure of one or more already-known ligands, and then searches a chemical library for examples that are similar (in either a 2D or a 3D sense). In contrast, structure-based screening does not require a priori knowledge of any ligands that bind to the target protein: Instead, it involves sequentially docking each member of the chemical library against the 3D structure of the target protein (receptor) and using a scoring function to evaluate the “quality” of each modeled protein–ligand complex. The scoring function is intuitively meant to serve as a proxy for the expected strength of a given protein–ligand complex (i.e., its binding affinity) (13), and is typically built upon either a physics-based force field (13–17), an empirical function (18–22), or a set of knowledge-based terms (23–28).
After docking, the scoring function is used to select the most promising compounds for experimental characterization; at this stage, the accuracy of the scoring function is of paramount importance and represents the primary determinant of success or failure in structure-based screening (29). A snapshot of the field was captured by a review summarizing successful outcomes from 54 virtual screening campaigns against diverse protein targets (12); for the most part, all groups screened the same 3 to 4 million compounds from ZINC (8, 30). Excluding G protein-coupled receptors (GPCRs) and artificial cavities designed into protein cores, the median values across the set reveal that an expert in the field—using their own preferred methods of choice, which can include various postdocking filters and human visual inspection (“expert hit-picking”)—can expect about 12% of their predicted compounds to show activity. That said, the hit rate can also be higher in cases where the composition of the screening library is restricted to compounds containing a functional group with natural affinity for the target site (certain well-explored enzyme active sites). Conversely, the hit rate is typically lower when the scoring function is applied without additional filters or human intervention (12). The median value of the most potent hit from each of the collected campaigns had Kd or Ki value of ∼3 μM, although this latter result is strongly impacted by the fact that some of these Kd or Ki values are from custom compounds subsequently optimized via medicinal chemistry, rather than from the initial screening hit.
Despite extensive efforts, the reasons for which active compounds are only identified at a relatively low rate are not quite clear. In addition to factors not evident from the structure of the modeled complex (compound solubility, incorrectly modeled protonation/tautomerization states of the ligand, etc.), we and others have hypothesized that the current bounds of performance may be attributable to limitations in traditional scoring functions (31, 32): These may include inadequate parametrization of individual energy terms, exclusion of potentially important terms, and also failure to consider potential nonlinear interactions between terms. For these reasons, machine learning techniques may be especially well suited for developing scoring functions that will provide a dramatic improvement in the ability to identify active compounds without human expert intervention. However, while machine learning may offer the potential to improve on the high false-positive rate of current scoring function, further analysis has revealed that many methods to date reporting promising results in artificial benchmark experiments may have inadvertently overfit models to the training data (33): This can be a subtle effect of information leakage, occurring when the validation/testing data are not truly nonredundant from the training data. Other studies have shown that apparently impressive performance from deep learning methods can result from detecting systematic differences in the chemical properties of active versus decoy compounds (34). Either of these artifacts inflates expectations based on benchmark performance, but ultimately leads to nontransferrable and disappointing outcomes when the methods are tested in subsequent prospective evaluations (35–38).
Here, we report the development of a dataset aimed to promote training of a machine learning model designed to be maximally useful in real-world (prospective) virtual screening applications. To build this dataset, we compile a set of “compelling” decoy complexes: a set that mimics representative compounds that might otherwise move forward to experimental testing if generated in the course of a typical virtual screening pipeline. We then use this dataset to train a machine learning classifier to distinguish active complexes from these compelling decoys, with the rationale that this is precisely the step at which standard scoring functions must be augmented. Finally, we apply this model in a prospective experiment, by screening against a typical enzyme target (acetylcholinesterase [AChE]) and testing the top-scoring compounds in a biochemical (wet laboratory) assay for inhibition of protein activity.
Results
Developing a Challenging Training Set.
Machine learning methods at varying levels of sophistication have long been considered in the context of structure-based virtual screening (29, 31, 32, 39–54). The vast majority of such studies sought to train a regression model that would recapitulate the binding affinities of known complexes, and thus provide a natural and intuitive replacement for traditional scoring functions (29, 31, 32, 39–47, 50, 51, 53). The downside of such a strategy, however, is that the resulting models are not ever exposed to any inactive complexes in the course of training: This is especially important in the context of docked complexes arising from virtual screening, where most compounds in the library are presumably inactive. We instead anticipated that a binary classifier would prove more appropriate for distinguishing active versus inactive compounds, and that training would prove most effective if decoy complexes closely reflected types of complexes that would be encountered during real applications.
Building first our set of active complexes, we drew examples from available crystal structures in the Protein Data Bank (PDB). Others have used collections of active compounds for which the structure of the complex is not known, and docked these to obtain a considerably larger set of active complexes (41, 49). The downside of this approach, however, is that misdocked examples (which may be numerous) are labeled as active during training; this is problematic because misdocked models do not have appropriate interactions with the protein target that would lead to engagement, and thus should be marked as inactive by the classifier. While restricting examples of active complexes to those available in the PDB drastically limits the number available for training, this strategy ensures that the resulting model will evaluate complexes on the basis of the protein–ligand interactions provided.
Our primary consideration in compiling active compounds for the training set was that the scope of examples should match as closely as possible those anticipated to be encountered when the model is deployed. Training the model on an overly restrictive set of examples would limit its utility (since many cases will be “out of distribution”), whereas training too broadly might limit the resulting model’s performance. Accordingly, we sought to train the model on precisely the type of scenarios that match its intended application. We therefore further filtered the set of active compounds from the PDB to include only ligands that adhere to the same physicochemical properties required for inclusion in our compound library for real screening applications (Methods). This led to a collection of 1,383 active complexes, which were then subjected to energy minimization: This prevented us from inadvertently training a model that simply distinguished between crystal structures and models produced by virtual screening.
Turning next to the set of decoy complexes, our primary consideration in compiling the training set was that the decoy complexes should be as “compelling” as possible. If the decoy complexes can be distinguished from the active complexes in some trivial way—if they frequently have steric clashes, for example, or they are systematically underpacked, or they do not contain intermolecular hydrogen bonds—then the classifier can simply use these obvious differences to readily distinguish active versus inactive compounds. In addition to making compelling decoys, the proportion of decoys-to-actives also has a significant effect on the performance of machine learning trained model (55). In order to achieve a nearly balanced training set, we aimed to include only small number of (very challenging) decoy complexes.
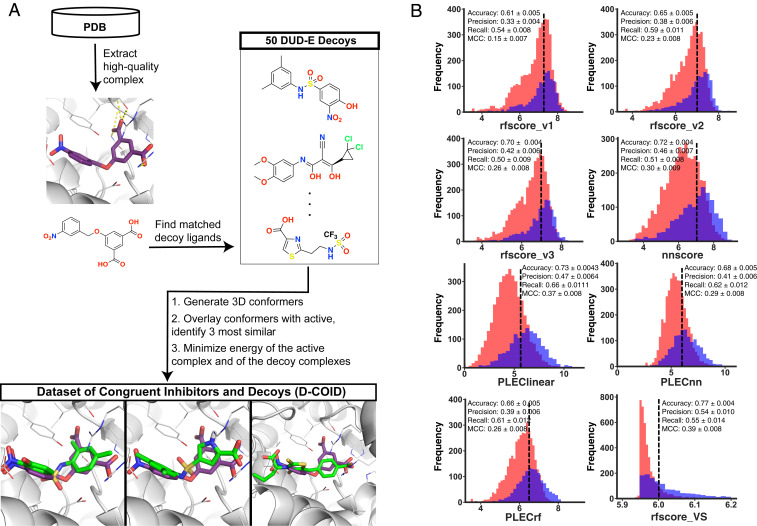
For each active complex, we first used the DUD-E server (56) to identify 50 compounds with physicochemical properties matched to the active compound but completely unrelated chemical structure: This provided a set of compounds compatible in very broad terms for the corresponding protein’s active site, and also ensured that the decoy compounds would not have systematic differences from the active compounds. We then built low-energy conformations of each candidate decoy compound, and screened these against the 3D structure of the active compound using ROCS (57). From among the 50 candidates, we selected those that best matched the overall shape and charge distribution of the active ligand. Using the structural alignment of the decoy compound onto the active compound, we placed the decoy into the protein’s active site and carried out the same energy minimization that was applied to the active complexes (Fig. 1A).
Fig. 1.
Developing a challenging training set (D-COID). (A) Active complexes were assembled from the PDB by filtering for ligands that match those reflected in a screening library. For each active complex, 50 physicochemically matched compounds were selected and overlaid onto the active compounds; the three most similar compounds on the basis of overall shape and electrostatic similarity were aligned into the protein active site, and used as decoy complexes. This strategy mimics the selection of candidate (active) compounds in a realistic pharmacophore-based screening pipeline, and thus generates highly compelling decoy complexes for training/testing. (B) Modern scoring functions cannot distinguish active complexes from decoys in this set. Overlaid histograms are presented for scores obtained using various scoring functions when applied to active complexes (blue) and decoy complexes (red) in D-COID. For all eight methods tested, the distribution of scores assigned to active complexes strongly overlaps with the distribution of scores assigned to decoy complexes. From each model’s continuous scores, 10-fold cross-validation was used to obtain the classification cutoff that maximizes Matthews correlation coefficient (MCC) on each subset of the data, and these cutoffs were used in calculating precision/recall/MCC. Performance measures are presented as the average of 100 bootstrapped models, and uncertainty is presented as 95% confidence intervals.
We note that the protocol used here to build the decoy complexes doubles as an entirely reasonable approach for ligand-based (pharmacophoric) virtual screening: Indeed, ROCS is typically applied to identify compounds with matched 3D properties to a given template, with the expectation that the hits will themselves be active (58–60). Thus, the unique strategy motivating construction of our training set is in essence a form of adversarial machine learning: We intentionally seek to build decoys that we anticipate would be misclassified by most models. We named this dataset D-COID (dataset of congruent inhibitors and decoys) and have made it publicly available for others to use freely (Methods).
To confirm that this decoy-generation strategy indeed led to a challenging classification problem, we applied some of the top reported scoring functions in the literature to distinguish between active and decoy complexes in the D-COID set. For all eight methods tested [nnscore (32), RF-Score v1 (31), RF-Score v2 (40), RF-Score v3 (29), PLEClinear (42), PLECnn (42), PLECrf (42), and RF-Score-VS (41)], we found that the distribution of scores assigned to active complexes was strongly overlapping with those of the decoy complexes (Fig. 1B), indicating that these models showed very little discriminatory power when applied to this set.
Typical scoring functions report a continuous value, because they intend to capture the strength of the protein–ligand interaction. In order to use the scoring function for classification, one must define a threshold value at which complexes are predicted to be either active or inactive. To avoid overestimating performance by selecting the threshold with knowledge of the test set, we carried out 10-fold cross-validation to determine appropriate threshold. In particular, we used 90% of the dataset to define the threshold that maximized the Matthews correlation coefficient (MCC), and then applied this threshold to assign each complex in the unseen 10% as active/inactive. Using this unbiased thresholding measure to assign each complex in the D-COID set, we found MCC for the best-performing scoring function in this experiment to be only 0.39.
A Classifier for Identifying Active Complexes: vScreenML.
Having developed a relevant and challenging training set, we next sought to develop a machine learning model that could discriminate between active and decoy complexes in this set. It has been pointed out in the past that machine learning models built exclusively upon protein–ligand element–element distance counts can yield apparently impressive performance in certain benchmarks without proving useful beyond these (37). To avoid this pitfall, we used as our starting point the Rosetta energy function (61): a classical linear combination of traditional (physics-based) molecular mechanics energy terms, alongside empirical terms added so that distributions of atomic arrangements would quantitatively mimic those observed in the PDB (62). While we acknowledge that the Rosetta energy function is not commonly used for virtual screening, this is primarily because it is too slow to be applied for docking large compound libraries: In one recent benchmark for classification of active versus decoy complexes (63), the Rosetta energy function showed equivalent performance as the popular FRED Chemgauss4 scoring function (64).
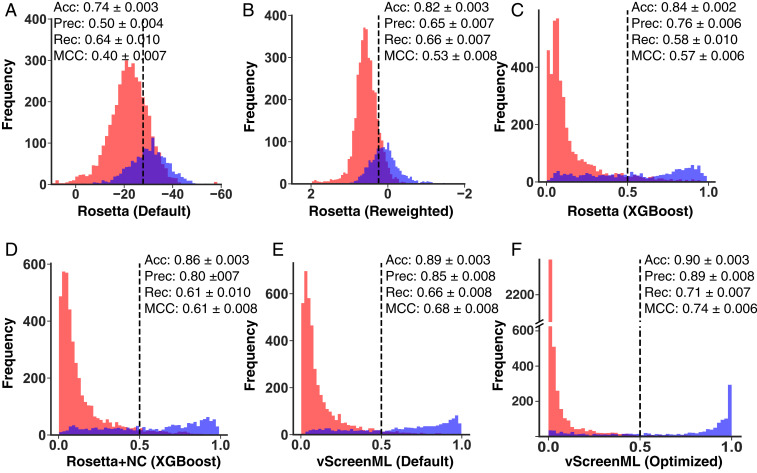
At the outset, we found that applying Rosetta to the D-COID set did not yield results notably different from in our previous experiment (Fig. 2A), and indeed this was confirmed quantitatively through the MCC (0.40). Next, we used 10-fold cross-validation to reweight the terms in this scoring function for improved performance in this D-COID classification task using a perceptron (65, 66) to maintain the linear functional form of the Rosetta energy function: This resulted in a modest improvement in the apparent separation of scores (Fig. 2B), but a notable improvement in MCC (0.53). This observation is unsurprising, because the Rosetta energy function is primarily optimized for proteins rather than protein–ligand complexes, and retraining its component energies for a specific task will naturally lead to improved performance for that task. For precisely this reason, historically a separate linearly reweighted version of the default Rosetta energy function has been used when modeling protein–ligand complexes (67) or when reranking complexes from virtual screening (63).
Fig. 2.
Development of vScreenML. Overlaid histograms are presented for scores obtained when scoring active complexes (blue) and decoy complexes (red) from D-COID. Scoring functions used were: (A) default Rosetta energy function, (B) linearly reweighted Rosetta energy terms, (C) Rosetta energy terms combined via XGBoost, (D) Rosetta energy terms plus structural assessments, (E) Rosetta terms plus additional diverse descriptors (nonoptimized vScreenML), and (F) vScreenML after hyperparameter tuning. Over the course of this sequence, the overlap between the active and decoy complexes is progressively reduced and MCC systematically increases. For the first two panels, 10-fold cross-validation was used to obtain the classification cutoff that maximizes Matthews correlation coefficient (MCC) on each subset of the data, and these cutoffs were used in calculating precision/recall/MCC. Because the remaining panels each report results from classification models, their thresholds are fixed at 0.5. Performance measures are presented as the average of 100 trained models, each of which derived from 10-fold cross-validation (Methods). Uncertainty is presented as 95% confidence intervals. In all cases, performance measures were calculated for a subset of the data that was held out from the training step.
Next, we explored the performance of models that move beyond linear combinations of these energy terms, and instead use these component energies as the basis for building decision trees. Using the XGBoost framework (an implementation of gradient-boosted decision trees), we observed notable separation of the scores assigned to active/decoy complexes (Fig. 2C), along with a slight increase in MCC (0.57). Importantly, here and in the extensions below, the model is evaluated using a held-out subset of the data that was not included in training.
To complement the existing terms in the Rosetta energy function, we next added a series of structural quality assessments calculated by Rosetta that are not included in the energy function (SI Appendix, Fig. S1); inclusion of these terms yielded a model with further improved discriminatory power (Fig. 2D). Inspired by this improvement, we then incorporated additional structural features aiming to capture more sophisticated chemistry than that encoded in Rosetta’s simple energy function, specifically from RF-Score (31) (features that count the occurrence of specific pairwise intermolecular contacts), from BINANA (68) (analysis of intermolecular contacts), from ChemAxon (ligand-specific molecular descriptors), and from Szybki (a term intended to capture ligand conformational entropy lost upon binding). We proceeded to train a model using this collection of features, which we denote “vScreenML,” and were pleased to discover that these again increased the separation between scores assigned to active and decoy complexes (Fig. 2E). Finally, we used hyperparameter tuning to optimize development of the model (SI Appendix, Tables S1 and S2), and accordingly developed a model that provided nearly complete separation of active and decoy complexes (Fig. 2F) and unprecedented MCC for this challenging task (0.74). We have made this model publicly available for others to use freely (Methods).
Through the course of developing of this model, we transitioned from a linear combination of six Rosetta features with clear physical basis, to a collection of 68 diverse and likely nonorthogonal features connected through a more complex underlying model (SI Appendix, Fig. S1). Using the complete set of features that comprise vScreenML, we tested alternate machine learning frameworks, leading us to discover that a different implementation of gradient-boosted decision trees yielded essentially identical performance, and other models built upon decision trees were only slightly worse. By contrast, other models that are not built on decision trees did not provide comparable performance (SI Appendix, Table S3). Importantly, we note that this model has been trained to distinguish actives from decoy complexes in a context where both have been subjected to energy minimization using the Rosetta energy function: The same optimized model is not necessarily expected to recognize actives successfully if they have not been prepared this way (e.g., crystal structures).
To evaluate the contributions of each part of our feature set, we next removed one at a time all features from a given origin, and explored how the lack of these features would affect performance (SI Appendix, Table S4). This experiment showed that only a very small deterioration in performance was observed when either the RF-Score or BINANA features were removed, but removing both had a larger impact; this is unsurprising, given the fact that many of the features in these sets are correlated. Interestingly, removing the Rosetta features had comparable impact as together removing both the RF-Score and the BINANA features, implying that the Rosetta features provide nonoverlapping information relative to these counting-based features. Finally, we find that removal of SZYBKI’s conformational entropy term had no impact on the model’s performance, suggesting either that the change in ligand conformational entropy as described by SZYBKI does not help distinguish active versus decoy complexes in this dataset, or that this effect is already captured through some combination of other features. In principle, features that are unnecessary (either because they are correlated with other features or because they do not help in classification) should be removed to better avoid the risk of overtraining. Because XGBoost is not particularly susceptible to overtraining and our feature set remains relatively small in comparison to our training set, however, in this case we elected instead to simply test our model immediately in orthogonal benchmarks to evaluate potential overtraining.
Benchmarking vScreenML Using Independent Test Sets.
The DEKOIS project (currently at version 2.0) (69, 70) is intended to provide a “demanding” evaluation set for testing virtual screening methods. Acknowledging that a wide variety of factors make some protein targets easier to model than others, this set includes 81 different proteins with available crystal structures. For each protein, a custom library is provided that contains 40 active compounds and 1,200 decoys: Thus, about 3.2% of each library is active. The crystal structures of active complexes are not provided (and indeed, most have not yet been experimentally determined). To evaluate performance of a new scoring function, one typically ranks all 1,240 compounds for a given protein and selects the top-scoring 12; the enrichment factor for this subset of the library (EF-1%) corresponds to the ratio of the percent of active compounds among the selected 12 to the ratio of active compounds in the original library. Scoring perfectly for a given protein in this set would mean ranking 12 active compounds before all 1,200 of the decoys: This would correspond to EF-1% = 1.00/0.032 = 31. Conversely, a method that randomly selects compounds from the library would (on average) select active compounds 3.2% of the time, and thus yield an EF-1% of 1.
Among the 81 proteins in the DEKOIS set, we noted that some were included in our training set as well. To avoid any potential information leakage that might overestimate the performance we could expect in future applications (33), we completely removed these test cases. This left a set of 23 protein targets, each of which vScreenML had never seen before. For each protein, we docked each compound in the corresponding library to the active site (Methods); we note that this unavoidable step could artificially deflate the apparent performance of vScreenML or other models tested, since a misdocked active compound should have no basis for being identified as active. Some of the compounds in the DEKOIS set could not be suitably modeled in all parts of our pipeline, and were therefore removed (this arose primarily due to atom types for which Rosetta lacks parameters, such as boron); each of the 23 proteins considered ultimately was used to generate 30 to 40 active complexes and 800 to 1,200 decoy complexes. Each of these complexes (both actives and decoys) were then subjected to energy minimization using the Rosetta: As noted earlier, vScreenML should only be applied in the context of Rosetta-minimized structures. Along with vScreenML, eight other machine learning scoring functions were then used to rank the docked-and-minimized models: nnscore (32), PLECnn (42), PLECrf (42), PLEClinear (42), RF-Score v1 (31), RF-Score v2 (40), RF-Score v3 (29), and RF-Score-VS (41). We additionally included the (default) Rosetta energy function in this benchmark (61). vScreenML was used exactly as trained on the D-COID set, with no adjustments for this DEKOIS benchmark.
While vScreenML was not trained on precisely the same protein as any of the 23 included in this benchmark, some of these had close homologs in the training set. Among these 23 test cases, the median sequence identity for the closest homolog in the training set was 42%; however, performance of vScreenML was not better in the cases for which a closer homolog was present in the training set (SI Appendix, Table S5), implying that this similarly had not inadvertently allowed vScreenML to recognize certain complexes based on similarity of their binding sites.
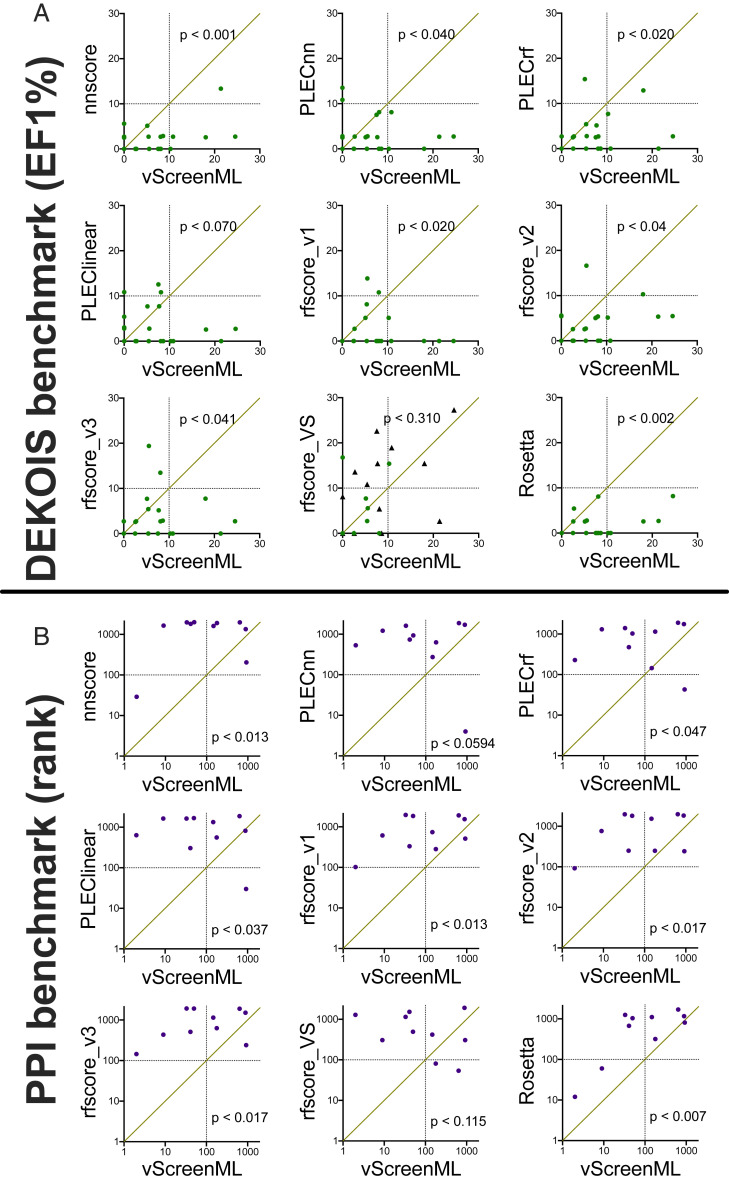
To compare performance between methods, we plot EF-1% using one method (for each of the 23 protein targets) as a function of EF-1% using the other method (Fig. 3A). As plotted here, points below the diagonal are specific protein targets for which vScreenML outperformed the alternate method (higher EF-1% for this protein target). The importance of training on both actives and decoys for this task is immediately apparent in these comparisons, by comparing for example vScreenML against PLECnn (a neural network representing the current state-of-the-art among models trained exclusively on active complexes). For the 23 targets in this experiment, PLECnn out-performs vScreenML in 3 cases (points above the diagonal), whereas vScreenML proves superior in 12 cases (the other 8 cases were ties).
Fig. 3.
Comparing vScreenML to other scoring functions using two independent virtual screening benchmarks. Each benchmark is composed of multiple protein targets, corresponding to points on these plots. (A) DEKOIS benchmark, composed of 23 protein targets. For each target (individual dots), 30 to 40 active complexes and 800 to 1,200 decoy complexes are provided. For a given target, each scoring is used to rank the set of complexes. For a given scoring function, the number of active complexes in the top 1% of all complexes is used to calculate the enrichment of actives relative to randomly selecting complexes; thus, higher numbers indicate better performance. When comparing vScreenML against another method, a point below the diagonal indicates superior performance by vScreenML for this particular target. Targets seen by rfscore_VS during training of this method are marked with black triangles. (B) PPI benchmark, composed of 10 protein targets. For each target, a single active complex is hidden among 2,000 decoy complexes. Instead of using enrichment, the rank of the active compound (relative to the decoys) is calculated: thus, lower numbers indicate better performance. When comparing vScreenML against another method, a point above the diagonal indicates superior performance by vScreenML for this particular target. P values in both cases were computed using the two-tailed Wilcoxon signed-rank test.
To evaluate in a statistically rigorous way which method was superior, we applied the (nonparametric) Wilcoxon signed-rank test: This paired difference test uses the rank values in the data, and thus it takes into account not just which method has higher EF-1%, but also the magnitude of the difference (63). We used a two-tailed test, in order to assume no a priori expectation about what method would outperform the other. At a threshold of P < 0.05, this analysis shows that vScreenML outperformed eight of the nine alternate scoring functions to a statistically significant degree. Only RF-Score-VS was not outperformed by vScreenML at a statistically significant threshold; however, we note that about half of the 23 targets in this benchmark were included in training RF-Score-VS (black points in this figure), which may have provided it with a slight advantage relative to vScreenML (since the latter had not seen any of these targets before).
To test these methods on a second independent virtual screening benchmark, we drew from our own prior studies of inhibitors of protein–protein interactions (63). In the course of evaluating existing scoring functions, we had several years ago assembled a set of small molecules that engage protein interaction sites; 10 of these protein targets had not been included in training vScreenML. For each of these, we had previously compiled 2,000 decoys with dissimilar chemical structure matched to the active compound’s lipophilicity. The decoy compounds were already docked and energy minimized from our studies, making this “PPI set” a natural testbed for the newer methods that were not available at the time this benchmark was developed (63). In contrast to the DEKOIS benchmark, the structures of the active complexes are drawn from (energy-minimized) crystal structures, removing a potential source of variability (since misdocked active compounds should not be labeled “correct” by a scoring function). Here again, vScreenML was used exactly as trained on the D-COID set, with no adjustments for this particular benchmark.
Because each protein target is only associated with a single active compound in this test set, we cannot meaningfully calculate enrichment factor; instead, after scoring each of the complexes, we simply report the rank of the active compound. As there are 2,001 complexes for each protein target, a method that performs as random would be expected to rank the active compound at position 1001, on average. After applying each of the same scoring functions used in our DEKOIS experiment, we find that for 5 of the 10 protein targets vScreenML ranks the active compound among the top 100 (i.e., top 5% of the compounds for a given target) (Fig. 3B). The other scoring functions tested each ranked the active compound in the top 100 for at most one target, except for RF-Score-VS, which met this criterion twice. Once again applying the Wilcoxon signed-rank test to these rankings, we once again conclude that vScreenML outperforms at a statistically significance degree all of these alternate scoring functions except for RF-Score-VS.
To determine whether vScreenML’s impressive performance derived from its training on the D-COID set or from the broad collection of features it includes, we used D-COID to train a model using the features from RF-Score v1; our retrained model preserves the same random forest framework and hyperparameters from the original model (31). As noted earlier (Fig. 1B), RF-Score v1 initially yields very little discriminative power when applied to the D-COID set; after retraining on this set, we find much improved separation of the scores assigned to active versus decoy complexes (SI Appendix, Fig. S2 A and B), although not close to the performance of vScreenML (Fig. 2F). This retrained variant of RF-Score v1 also outperforms the original RF-Score v1 on both the DEKOIS and the PPI benchmarks, albeit not to a level of statistical significance, and for the PPI benchmark it even ranks two actives in the top 100 for their corresponding protein targets (SI Appendix, Fig. S2 C and D). That said, the level of improvement is insufficient for the retrained RF-Score v1 to outperform vScreenML in either benchmark (SI Appendix, Fig. S2 E and F), consistent with their relative performance on D-COID set. Overall, these observations show that training using the D-COID approach can certainly improve performance of existing scoring functions for other unrelated tasks; however, it also suggests that some part of vScreenML’s power derives from the broad and diverse set of features that it uses.
Evaluating vScreenML in a Prospective Experiment.
As noted earlier, it is absolutely essential to test new scoring functions in prospective experiments: This can readily determine whether performance in a given benchmark experiment is likely to extend into real future applications, and rule out any possibility that inadvertent information leakage allowed an overfit model to “cheat” in benchmark experiments. We selected as our representative target human AChE because of its biomedical relevance and the availability of a straightforward functional assay (using commercially available enzyme and substrate).
To ensure that our search for new candidate AChE inhibitors would not be limited by the chemical space present in a small screening library, we turned to a newly available virtual library of “readily accessible” but never-before-synthesized compounds (9). At the time of our screen, this library was comprised of 732 million chemical entities that conform to historic criteria for drug-likeness (71, 72). Because building conformers and docking each entry in this library would be extremely computationally demanding, we instead took a two-step approach to finding candidate inhibitors. First, we explicitly docked a chemically diverse set of 15 million representatives from the library, and applied energy minimization to the top 20,000 models from the crude docking step. We ranked each of these using vScreenML and identified the top 100 candidates. For each of these 100 initial candidates, we returned to the complete compound library and identified 209 analogs on the basis of chemical similarity: After merging these with the parent compounds from each search, this led to a new focused library of 20,213 unique compounds. We structurally aligned each of these compounds back onto the parent docked model that led to their selection, reminimized, and then used vScreenML to rank these second-stage candidates. We collected into a single list the 20 top-scoring compounds from the first round together with the 20 top-scoring compounds from the second round, noting that 4 compounds were included on both lists. We eliminated compounds that were extremely close analogs of one another, and sought to purchase the remainder. Based on a standard filter (73), none of these structures was predicted to be PAINS (pan-assay interference) compounds. Ultimately 23 compounds were successfully synthesized, as selected by vScreenML without any human intervention (SI Appendix, Table S6). While some compounds use a shared scaffold, overall there are multiple diverse chemotypes represented in this collection (SI Appendix, Fig. S3). Interestingly, none of these compounds would have been prioritized by the other scoring functions evaluated in the context of our study (SI Appendix, Table S7). Looking back at the complexes present in D-COID, we also found that the closest ligand to each of these compounds was bound to a protein completely unrelated to AChE (SI Appendix, Table S8); thus, vScreenML had not simply recognized specific binding sites in the training set that resembled that of AChE.
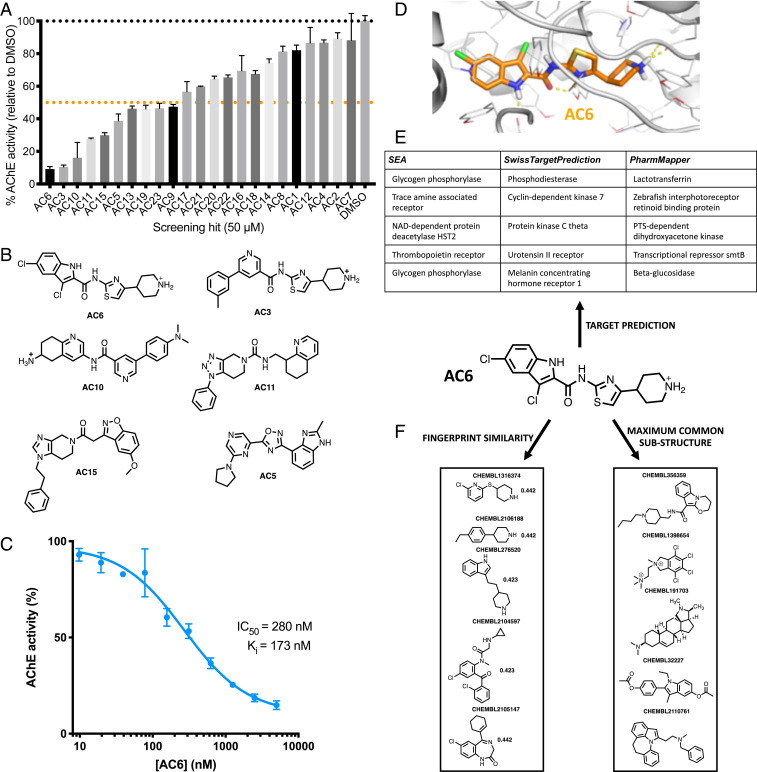
We initially tested these 23 compounds at a concentration of 50 μM for inhibition of AChE, using a colorimetric enzyme assay (Fig. 4A). To our amazement, we found that nearly all of the 23 compounds selected by vScreenML showed detectable enzyme inhibition: All except AC12 and AC7 showed a statistically significant difference in AChE activity relative to dimethyl sulfoxide (DMSO) alone (P < 0.05, one-tailed t test). Of these 23 compounds, 10 of them provided more than 50% inhibition, indicating that these compounds’ IC50 was better than 50 μM. Moreover, the most potent of these used a variety of diverse chemical scaffolds, although the most potent pair (AC6 and AC3) do share an extensive common substructure (Fig. 4B). We then evaluated the activity of the most potent inhibitor, AC6: In the absence of any medicinal chemistry optimization, we found this compound to have an IC50 of 280 nM, corresponding to a Ki value of 173 nM (Fig. 4C). Thus, applying vScreenML led to a much higher hit rate than observed in typical screening campaigns, and also yielded a much more potent starting point than is typically observed.
Fig. 4.
Prospective evaluation of vScreenML in a virtual screen against human acetylcholinesterase (AChE). (A) Of the 23 compounds prioritized by vScreenML for testing, at 50 μM nearly all of these inhibit AChE. Data are presented as mean ± SEM; n = 3. (B) Chemical structures of the most potent hit compounds. (C) Dose–response curve for the most potent hit compound, AC6. Data are presented as mean ± SEM; n = 3. (D) Model of AC6 (orange sticks) in the active site of the AChE (light gray). (E) Predicted activity of AC6 from three target identification tools: None of these identifies AChE as a potential target of this compound, suggesting that this is a new scaffold for AChE inhibition. (F) Similarity searching against all compounds in ChEMBL designated as AChE inhibitors (either by fingerprint similarity of by shared substructure) finds no hits with discernible similarity, confirming that this is a new scaffold for AChE inhibition.
Unsurprisingly, the underlying model of the complex that was used by vScreenML to identify this compound shows extensive and nearly optimal protein–ligand interactions (Fig. 4D). In principle, it should be the quality of these interactions that guided vScreenML to prioritize this compound for experimental validation. To rule out the possibility that vScreenML had instead somehow “recognized” AC6 as an AChE inhibitor from its training, we asked whether chemoinformatic approaches could have been used to find AC6.
We first provided the chemical structure of AC6 to three different “reverse screening” methods: Similarity Ensemble Approach (SEA) (74), SwissTargetPrediction (75, 76), and PharmMapper (77, 78). Each of these tools look for similarity of the query compound against all compounds with known bioactivity, and then they rely on the fact that similar compounds have similar bioactivity to predict the likely target(s) of the query compound. SEA and SwissTargetPrediction carry out this search on the basis of 2D similarity (i.e., similar chemical structures), whereas PharmMapper evaluates 3D similarity (i.e., shared pharmacophores). We took for each method the top five predicted activities for AC6, but found that none of these methods included AChE among their predictions (Fig. 4E); the same also held true of predicted activities for the other compounds tested (SI Appendix, Table S9). All of these methods do include AChE among their list of potential targets, however, as confirmed by ensuring that this prediction emerges when these servers are provided with the structure of previously described AChE inhibitor donepezil (SI Appendix, Fig. S4).
To directly determine the AChE inhibitor described to date that is most similar to AC6, we compiled from ChEMBL all 2,742 compounds reported to have this activity. We then screened this collection to determine their similarity to AC6, as defined by either chemical fingerprints or by shared substructure, and found that the 5 most similar compounds as gauged by either approach bear no obvious similarity to AC6 (Fig. 4F). Analogous analysis revealed no similar chemical scaffolds for the other AC compounds either (SI Appendix, Table S10). Collectively then, these experiments confirm that AC6 and the other AC-series compound are indeed novel chemical scaffolds with respect to their inhibition of AChE and could not possibly have been identified by vScreenML through inadvertent leakage during the model’s training.
Discussion
At the outset of this work, we noted that typical virtual screening studies report hit rates of about 12%, with the most potent reported compound having Kd or Ki value of ∼3 μM (with the caveat that some of these relied on additional optimization beyond the initial screen) (12). Obviously, the results of our screen against AChE using vScreenML far surpass these mileposts; in light of this, it important to carefully consider the potential contributions to vScreenML’s performance in this experiment.
First, we reemphasize the dissimilarity between AC6 and any known AChE inhibitor: This makes it exceedingly unlikely that vScreenML found AC6 simply on the basis of having been trained on some close analog.
Second, we carried out a nonstandard two-step screening strategy to efficiently explore the complete Enamine collection, hoping to essentially carry out an internal round of medicinal chemistry optimization before testing any compounds explicitly. Tracking the provenance of our most potent compounds, however, we discovered that all four of our most potent compounds had already been identified in the first of the two screening steps (SI Appendix, Table S11). A previous virtual screen of the Enamine library (9) explicitly docked all compounds from the library, at a time that the library comprised “only” 138 million compounds, and found through retrospective analysis that picking a single representative compounds from a cluster of analogs would typically not yield sufficient docking score for the cluster to be advanced for further exploration. In essence, both our results and the observations from this previous screen suggest that the SAR landscape may not be sufficiently smooth to allow potentially promising scaffolds to be identified from a single arbitrary representative: Rather, finding the best hits (on the basis of docking scores) does unfortunately require explicitly screening each member of the library individually. In this context, then, it is unlikely that the observed performance of vScreenML can be attributed to having used a two-step strategy for screening the Enamine library.
In this vein, we also note that our screening strategy was allowed to explore an unusually large chemical space comprising 732 million synthetically accessible compounds. However, 7 of our top 10 compounds (those with IC50 values better than 50 μM) had already been identified in the first screening step (SI Appendix, Table S11), owing to the ineffectiveness of identifying useful scaffolds from a single representative compound. The bulk of the success in this screen was essentially achieved by screening a library of 15 million diverse compounds, which is by no means unprecedented and has not led to such dramatic success in the past.
Importantly, we cannot rule out the prospect that the performance we observe here is a result of AChE being an unexpectedly easy target. It is certainly the case that virtual screening hit rates against GPCRs are often much higher those obtained for other target classes (12). Indeed, careful examination of the literature showed that some of the studies reporting virtual screens against AChE (79–83) do indeed find considerably higher hit rates and more potent compounds than the median values we quote across all target classes. In light of these other results, then, a degree of caution must be exercised before extrapolating the performance of vScreenML in this prospective AChE benchmark to other target classes; further evaluation will be needed to explicitly determine whether vScreenML affords similarly outstanding results in future screening experiments.
At the same time, however, results of retrospective benchmarks comparing vScreenML to other scoring functions are unambiguous. As described, vScreenML dramatically outperforms eight other modern machine learning scoring functions on both the DEKOIS and the PPI benchmark sets. Both benchmarks were carried out with careful vigilance to ensure that information from training could not contaminate the test data. In the past, we strongly suspect inadvertent overtraining of this type has limited the utility of other models and at the same time provided artificially inflated performance on initial (retrospective) benchmarks. Indeed, a recurrent disappointment from many past machine learning scoring functions has been their inability to translate performance from retrospective benchmarks into equivalent results in future prospective applications (38). For example, 3 y after publication of nnscore (32), this program was used in a screen against farnesyl diphosphate synthase, and only provided one hit with IC50 of 109 μM (from 10 compounds tested) (84). Where possible, then, we strongly urge incorporation of careful prospective evaluations alongside retrospective benchmarks, as a safeguard against potentially misleading performance from the latter. Already such prospective experiments have been included in other recent studies (39, 85), strongly supporting transferability of the underlying methods. The ability to readily compare vScreenML against other machine learning scoring functions was also greatly facilitated by the Open Drug Discovery Toolkit (ODDT) (86), which provides implementations of multiple methods. Direct head-to-head evaluations of this manner are indeed critical to explore the relative strengths of different approaches, ideally across diverse types of benchmarks.
While vScreenML does incorporate a broad and distinct set of features, these have been largely collected from other approaches: There is nothing particularly unique or special about the features it includes. There are also numerous potential contributions to protein–ligand interactions that are not captured in this collection of features, ranging from inclusion of tightly bound interfacial waters (16, 87, 88) to explicit polarizability and quantum effects (89, 90). In this vein, ongoing research in ligand-based screening has led to new approaches that learn optimal molecular descriptors (and thus the representation that directly leads to the features themselves) at the same time as the model itself is trained (91, 92): These might similarly be used as a means to improve the descriptors used in structure-based screening as well. Thus, there is likely to be considerable future improvement to vScreenML that is possible, by further optimization of the features that it captures.
Rather than the specific features incorporated in this first incarnation of vScreenML, we believe that the impressive performance we observed in our retrospective benchmarks is instead primarily attributable to the strategy used in training the model. Whereas scoring functions have historically focused on recapitulating binding affinities of complexes, vScreenML is unique in having been trained to distinguish active complexes from extremely challenging decoys in the D-COID set. Indeed, the overarching hypothesis of our study was that building truly compelling decoys to better represent the (inactive) compounds selected from actual virtual screens, we would lead to a model capable of distinguishing precisely these cases. The performance of vScreenML in both the retrospective and prospective benchmark strongly supports this hypothesis.
Thus, the D-COID set represents an important resource for driving development of improved scoring functions beyond vScreenML, and accordingly we have made this dataset freely available for this purpose (Methods).
Methods
Accessing These Tools.
The D-COID dataset is available at https://data.mendeley.com/datasets/8czn4rxz68/ (93). vScreenML is available at https://github.com/karanicolaslab/vScreenML. The use of vScreenML requires that features be calculated precisely the same way that the model was trained, and it should be applied only to Rosetta-minimized complexes (given that these were used for training the model).
Software Versions.
Rosetta calculations were carried out using the developer trunk source code, git revision 0831787c75bba750254e86f55acf8b6fe314a7b9. The following versions of other software were used in this study: OMEGA (v2.5.1.4), ROCS (v3.2.1.4), FRED (v3.2.0.2), SZYBKI (v1.9.0.3), ODDT (oddt_cli 0.7), RFSCORE-VS (v1.0), and CHEMAXON Marvin (v1.5.6).
Building the D-COID Set.
The overarching goal of our study was to train a model for real virtual screening applications. We therefore included in D-COID only active complexes that included representative drug-like ligands, and excluded chemical matter that did not reflect the composition of the screening libraries we prefer to use.
We downloaded from the PDB (94) all protein–ligand complexes (56,195 entries as of May 2018), and then restricted this set to crystal structures with resolution better than 2.5 Å (43,148 complexes). We then drew from Ligand Expo (95) to define a set of 2,937 specific ligands found in the PDB that we deemed ineligible for our study: These include nucleotide-like molecules (e.g., ATP), cofactors (e.g., NAD), metal-containing ligands (e.g., heme), crystallographic additives (e.g., PEG), and covalent ligands. Our rationale was that the interactions in these types of complexes may not be representative of those found in the types we wanted to identify using our model, because complexes involving drug-like compounds with proteins may be fundamentally different from each of these examples (e.g., because the protein is extensively evolved to fit the ligand, or because the metal dominates the energetics of the interaction, or because the ligand interacts only weakly with the protein). Instead, our goal was to train a model using a more restricted set of complexes in which the interactions best represent those expected in successful docked models from virtual screening. We filtered to remove any complexes in which the only ligand was ineligible based on the criteria above. Our set included several structures that included a separate (eligible) ligand in addition to one of these ineligible ligands; we retained these eligible ligands for consideration, if they were at least 12 Å away from the ineligible ligand. Filtering out ineligible ligands in this manner reduced our set to 26,271 complexes.
To focus training on precisely the type of chemical matter used in our virtual screens, we then applied to this collection the same stringent filter we use when building our screening libraries: molecular weight between 300 and 400 Da and clogP 1 to 4. This filter drastically cut down the size of our collection (to 2,075 complexes). Finally, complexes with double occupancy or ambiguous density were manually excluded, leaving a high-quality collection of 1,383 active complexes.
For each of these active complexes, we extracted the ligand and used the Database of Useful Decoys Enhanced (DUD-E) (56) server to generate 50 property-matched decoys: compounds with similar physicochemical properties but dissimilar chemical topology. For each of these decoy compounds, we used OpenEye’s OMEGA (96) to generate 300 low-energy conformers, and then used ROCS (57) to align each of these to the structure of the active conformer from the PDB. The three decoys that best matched the 3D shape and pharmacophoric features of the active conformer were identified on the basis of their Tanimoto-Combo score; this led to a total of 4,149 decoy compounds. By virtue of having aligned the conformers of the decoys to the active conformation to evaluate their similarity, already the alignment was available for placing the decoy compound in the corresponding protein’s active site. We later discovered that 39 of these decoy compounds included chemical features that could not be processed by the programs we used to extract structural features for vScreenML; these decoys were removed, leading to a total of 4,110 decoy complexes.
Finally, to present both the active and decoy complexes in a context mimicking that of a virtual screening output, we subjected all complexes to standard energy minimization in Rosetta (61).
Extracting Structural Features.
For each of the minimized active and decoy complexes, structural features were extracted first using the Rosetta (“REF15”) energy function (61). Ligand properties were calculated using ChemAxon’s cxcalc, and the ligand’s conformational entropy was estimated using OpenEye’s SZYBKI tool. The open source implementations of RF-Score (31) and BINANA (68) were used to calculate structural features from these two programs. The complete list of vScreenML’s features is presented in SI Appendix, Fig. S1.
Machine Learning.
We considered a total of eight classification algorithms in this study, using the Python implementations of each: Gradient Boosting (GB) (97), Extreme Gradient Boosting (XGB) (98), Random Forest (RF) (99), Extremely Randomized Trees (ET) (100), Gaussian Naïve Bayes (GNB) (101), k-Nearest Neighbor (kNN) (101), Linear Discriminant Analysis (LDA) (101), and Quadratic Discriminant Analysis (QDA) (101).
To retrain RF-Score v1 under D-COID, we used a standard random forest model with hyperparameters n_estimators = 500 and max_features = 5 [drawing these values from the original study describing RF-Score v1 (31)].
For XGBoost hyperparameter optimization, we carried out a grid search to find the set of parameters that gave the best accuracy upon 10-fold cross-validation. Optimization was carried out by iteratively retraining the model using a fixed partition of the data, and at each step evaluating performance using a separate held-out validation set. This led to a single set of parameters (SI Appendix, Table S2) that were used when evaluating performance.
To evaluate performance of various models, in each case we used 10-fold cross-validation; the dataset was split into 10 subsets in a stratified manner to ensure that the overall ratio of actives to decoys was preserved in each split. This process was repeated 10 times with different seeds, to yield a total of 100 distinct trained models, with a corresponding held-out test set for each model (hyperparameters were held at the same fixed values for all models). We then calculated accuracy, precision, recall, area under the ROC curve (AUC), and MCC for each test set using its corresponding model, and then evaluated the mean and 95% confidence intervals for each parameter over these 100 instances.
In all cases, performance metrics are reported only for a held-out subset of the data, and never for the same data on which the model was trained.
Virtual Screening Benchmarks.
Comparisons between scoring functions was enabled by the Open Drug Discovery Toolkit (ODDT) (86), which provides implementations of nnscore (version 2), RF-Score v1, RF-Score v2, RF-Score v3, PLEClinear, PLECnn, and PLECrf at https://github.com/oddt/oddt. The implementation of RF-Score-VS was obtained from https://github.com/oddt/rfscorevs.
In both the DEKOIS and the PPI benchmark experiments, we carefully sought to minimize any potential information leakage from vScreenML’s training (on D-COID) and the targets present in these benchmark sets. Excluding a specific complex present in both sets is insufficient, because the structure of a close chemical analog bound to the same target protein could still provide an unfair advantage. For this reason, we excluded from these benchmarks sets any protein targets present in D-COID (on the basis of shared Uniprot IDs). This reduced the number of DEKOIS targets from 81 to 23, and the number of PPI targets from 18 to 10.
For the DEKOIS set, we docked both the actives and the decoys to their respective target protein using OpenEye’s FRED (64), and then applied energy minimization in Rosetta. For the PPI set, active complexes were minimized starting from their crystal structures; decoy complexes were generated by docking with FRED, then energy minimized.
Statistical analysis was carried out using the (two-tailed) Wilcoxon signed-rank test as implemented in Python. Comparisons were applied directly to the EF-1% values for the DEKOIS experiment, and to the log10 of the ranks in the PPI experiment.
Virtual Screen Against Acetylcholinesterase.
We began by downloading from the chemical vendor Enamine the “diverse set” of 15 million compounds representative of their REAL database (732 million compounds). For each compound, we used OMEGA (96) to generate 300 low-energy conformers, and used FRED (64) to dock these against the crystal structure of human AChE solved in complex with potent inhibitor donepezil (PDB ID 4ey7) (102). We carried forward the top 20,000 complexes (on the basis of FRED score) for Rosetta minimization, and used each of these minimized models as input for vScreenML.
For each of the top 100 complexes (as ranked by vScreenML), we extracted the ligand and used this to query the Enamine database for analogs. Each query returned 210 analogs; because 787 of these were redundant, this led to a new collection of 20,213 unique compounds for the second stage of screening. Each of the compounds in this new library was used to build 300 conformers, and ROCS was used to select the conformer that allowed for optimal alignment onto the ligand in the complex from the first round of screening. The resulting models were energy minimized in Rosetta, then used as input for vScreenML.
Models from both the first and second rounds of screening were collected together, and the top-ranked models from vScreenML were identified, and the top-scoring 32 compounds were requested for synthesis. Of the requested compounds, 23 were successfully synthesized and delivered for testing.
AChE Inhibition Assay.
Compounds were tested for inhibition of human AChE using a colorimetric assay (103). Acetylthiocholine is provided as substrate, which is hydrolyzed by AChE to thiocholine; the free sulfhydryl then reacts with Ellman’s reagent [5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB)] to yield a yellow product that we detected spectrophotometrically at 410 nm. AChE, acetylthiocholine, and DTNB were acquired together as the Amplite colorimetric assay kit (AAT Bioquest). Assays were carried out in 0.1 M sodium phosphate buffer (pH 7.4), 1% DMSO, with 0.01% Triton. Assays were carried out in 96-well plates in reaction volumes of 100 μL, and absorbance was monitored for 30 min. The rate of product formation was determined by taking the slope of the absorbance as a function of time, and normalized to that of DMSO alone to yield percent inhibition for each well.
IC50 values were obtained from dose–response curves spanning inhibitor concentrations from 10 nM to 50 μM. To determine Ki, we first determined the Km value for substrate acetylthiocholine under our assay conditions. This allowed the Cheng–Prusoff equation (104) to be used for obtaining Ki from IC50, assuming classic competitive inhibition.
Novelty of AC-Series Compounds as AChE Inhibitors.
For each of the target identification methods [SEA (74), SwissTargetPrediction (75, 76), and PharmMapper (77, 78)], we used the corresponding web servers to generate predictions.
To find the most similar known AChE ligands, we searched ChEMBL (105) for AChE and downloaded all 2,742 hits in SDF format. We then used ChemAxon’s Standardizer tool to remove counterions from compounds in salt form. Using RDKit, we generated Morgan fingerprints with radius of 2 for each of the ChEMBL ligands, then evaluated the Dice similarity of these fingerprints relative to each AC-series compound. We also used RDKit to evaluate the maximum common substructure between AC6 and each of the ChEMBL ligands, setting ringMatchesRingOnly = True and completeRingsOnly = True. We ranked the resulting matches based on the number of atoms and bonds in the common substructure.
Supplementary Material
Acknowledgments
We thank Joanna Slusky for a useful suggestion regarding presentation of the figures, and Juan Manuel Perez Bertoldi for his initial application of vScreenML to the PPI benchmark set. We thank ChemAxon for providing an academic research license. This work used the Extreme Science and Engineering Discovery Environment Allocation MCB130049, which is supported by National Science Foundation Grant ACI-1548562. This work was supported by grants from the National Institute of General Medical Sciences (R01GM099959, R01GM112736, and R01GM123336) and the National Science Foundation (CHE-1836950). This research was funded in part through the NIH/NCI Cancer Center Support Grant P30CA006927.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The D-COID dataset has been deposited on Mendeley, https://data.mendeley.com/datasets/8czn4rxz68/. vScreenML has been deposited on GitHub, https://github.com/karanicolaslab/vScreenML.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000585117/-/DCSupplemental.
References
- 1.Vogelstein B. et al., Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang M. T. et al., Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat. Biotechnol. 34, 155–163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bunnage M. E., Chekler E. L., Jones L. H., Target validation using chemical probes. Nat. Chem. Biol. 9, 195–199 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Macarron R. et al., Impact of high-throughput screening in biomedical research. Nat. Rev. Drug Discov. 10, 188–195 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Clare R. H. et al., Industrial scale high-throughput screening delivers multiple fast acting macrofilaricides. Nat. Commun. 10, 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu Q., Peng Z., Kostrowicki J., Kuki A., LEAP into the Pfizer global virtual library (PGVL) space: Creation of readily synthesizable design ideas automatically. Methods Mol. Biol. 685, 253–276 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Hu Q. et al., Pfizer global virtual library (PGVL): A chemistry design tool powered by experimentally validated parallel synthesis information. ACS Comb. Sci. 14, 579–589 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Sterling T., Irwin J. J., ZINC 15—ligand discovery for everyone. J. Chem. Inf. Model. 55, 2324–2337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lyu J. et al., Ultra-large library docking for discovering new chemotypes. Nature 566, 224–229 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McInnes C., Virtual screening strategies in drug discovery. Curr. Opin. Chem. Biol. 11, 494–502 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Lavecchia A., Di Giovanni C., Virtual screening strategies in drug discovery: A critical review. Curr. Med. Chem. 20, 2839–2860 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Irwin J. J., Shoichet B. K., Docking screens for novel ligands conferring new biology. J. Med. Chem. 59, 4103–4120 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrara P., Gohlke H., Price D. J., Klebe G., Brooks C. L. 3rd, Assessing scoring functions for protein-ligand interactions. J. Med. Chem. 47, 3032–3047 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Genheden S., Ryde U., The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 10, 449–461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Athanasiou C., Vasilakaki S., Dellis D., Cournia Z., Using physics-based pose predictions and free energy perturbation calculations to predict binding poses and relative binding affinities for FXR ligands in the D3R Grand Challenge 2. J. Comput. Aided Mol. Des. 32, 21–44 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Labbé C. M. et al., AMMOS2: A web server for protein-ligand-water complexes refinement via molecular mechanics. Nucleic Acids Res. 45, W350–W355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bryce R. A., Physics-based scoring of protein-ligand interactions: Explicit polarizability, quantum mechanics and free energies. Future Med. Chem. 3, 683–698 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Böhm H. J., The development of a simple empirical scoring function to estimate the binding constant for a protein-ligand complex of known three-dimensional structure. J. Comput. Aided Mol. Des. 8, 243–256 (1994). [DOI] [PubMed] [Google Scholar]
- 19.Eldridge M. D., Murray C. W., Auton T. R., Paolini G. V., Mee R. P., Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J. Comput. Aided Mol. Des. 11, 425–445 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Friesner R. A. et al., Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Krammer A., Kirchhoff P. D., Jiang X., Venkatachalam C. M., Waldman M., LigScore: A novel scoring function for predicting binding affinities. J. Mol. Graph. Model. 23, 395–407 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Trott O., Olson A. J., AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mooij W. T., Verdonk M. L., General and targeted statistical potentials for protein-ligand interactions. Proteins 61, 272–287 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Muegge I., Martin Y. C., A general and fast scoring function for protein-ligand interactions: A simplified potential approach. J. Med. Chem. 42, 791–804 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Gohlke H., Hendlich M., Klebe G., Knowledge-based scoring function to predict protein-ligand interactions. J. Mol. Biol. 295, 337–356 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Verdonk M. L., Ludlow R. F., Giangreco I., Rathi P. C., Protein-ligand informatics force field (PLIff): Toward a fully knowledge driven “force field” for biomolecular interactions. J. Med. Chem. 59, 6891–6902 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Zhou H., Skolnick J., GOAP: A generalized orientation-dependent, all-atom statistical potential for protein structure prediction. Biophys. J. 101, 2043–2052 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang C., Liu S., Zhu Q., Zhou Y., A knowledge-based energy function for protein-ligand, protein-protein, and protein-DNA complexes. J. Med. Chem. 48, 2325–2335 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Li H., Leung K. S., Wong M. H., Ballester P. J., Improving AutoDock Vina using random forest: The growing accuracy of binding affinity prediction by the effective exploitation of larger data sets. Mol. Inform. 34, 115–126 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Irwin J. J., Shoichet B. K., ZINC—a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 45, 177–182 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ballester P. J., Mitchell J. B., A machine learning approach to predicting protein-ligand binding affinity with applications to molecular docking. Bioinformatics 26, 1169–1175 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durrant J. D., McCammon J. A., NNScore 2.0: A neural-network receptor-ligand scoring function. J. Chem. Inf. Model. 51, 2897–2903 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wallach I., Heifets A., Most ligand-based classification benchmarks reward memorization rather than generalization. J. Chem. Inf. Model. 58, 916–932 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Chen L. et al., Hidden bias in the DUD-E dataset leads to misleading performance of deep learning in structure-based virtual screening. PLoS One 14, e0220113 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu S. et al., Practical model selection for prospective virtual screening. J. Chem. Inf. Model. 59, 282–293 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaput L., Martinez-Sanz J., Saettel N., Mouawad L., Benchmark of four popular virtual screening programs: Construction of the active/decoy dataset remains a major determinant of measured performance. J. Cheminform. 8, 56 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gabel J., Desaphy J., Rognan D., Beware of machine learning-based scoring functions-on the danger of developing black boxes. J. Chem. Inf. Model. 54, 2807–2815 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Colwell L. J., Statistical and machine learning approaches to predicting protein-ligand interactions. Curr. Opin. Struct. Biol. 49, 123–128 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Ballester P. J. et al., Hierarchical virtual screening for the discovery of new molecular scaffolds in antibacterial hit identification. J. R. Soc. Interface 9, 3196–3207 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ballester P. J., Schreyer A., Blundell T. L., Does a more precise chemical description of protein-ligand complexes lead to more accurate prediction of binding affinity? J. Chem. Inf. Model. 54, 944–955 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wójcikowski M., Ballester P. J., Siedlecki P., Performance of machine-learning scoring functions in structure-based virtual screening. Sci. Rep. 7, 46710 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wójcikowski M., Kukiełka M., Stepniewska-Dziubinska M. M., Siedlecki P., Development of a protein-ligand extended connectivity (PLEC) fingerprint and its application for binding affinity predictions. Bioinformatics 35, 1334–1341 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H., Leung K. S., Wong M. H., Ballester P. J., Substituting random forest for multiple linear regression improves binding affinity prediction of scoring functions: Cyscore as a case study. BMC Bioinformatics 15, 291 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heck G. S. et al., Supervised machine learning methods applied to predict ligand- binding affinity. Curr. Med. Chem. 24, 2459–2470 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Öztürk H., Özgür A., Ozkirimli E., DeepDTA: Deep drug-target binding affinity prediction. Bioinformatics 34, i821–i829 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stepniewska-Dziubinska M. M., Zielenkiewicz P., Siedlecki P., Development and evaluation of a deep learning model for protein-ligand binding affinity prediction. Bioinformatics 34, 3666–3674 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wójcikowski M., Siedlecki P., Ballester P. J., Building machine-learning scoring functions for structure-based prediction of intermolecular binding affinity. Methods Mol. Biol. 2053, 1–12 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Abbasi W. A., Asif A., Ben-Hur A., Minhas F. U. A. A., Learning protein binding affinity using privileged information. BMC Bioinformatics 19, 425 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ragoza M., Hochuli J., Idrobo E., Sunseri J., Koes D. R., Protein-ligand scoring with convolutional neural networks. J. Chem. Inf. Model. 57, 942–957 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brylinski M., Nonlinear scoring functions for similarity-based ligand docking and binding affinity prediction. J. Chem. Inf. Model. 53, 3097–3112 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Ashtawy H. M., Mahapatra N. R., BgN-score and BsN-score: Bagging and boosting based ensemble neural networks scoring functions for accurate binding affinity prediction of protein-ligand complexes. BMC Bioinformatics 16 (suppl. 4), S8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Svetnik V. et al., Random forest: A classification and regression tool for compound classification and QSAR modeling. J. Chem. Inf. Comput. Sci. 43, 1947–1958 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Zilian D., Sotriffer C. A., SFCscore(RF): A random forest-based scoring function for improved affinity prediction of protein-ligand complexes. J. Chem. Inf. Model. 53, 1923–1933 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Li L., Wang B., Meroueh S. O., Support vector regression scoring of receptor-ligand complexes for rank-ordering and virtual screening of chemical libraries. J. Chem. Inf. Model. 51, 2132–2138 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boughorbel S., Jarray F., El-Anbari M., Optimal classifier for imbalanced data using Matthews correlation coefficient metric. PLoS One 12, e0177678 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mysinger M. M., Carchia M., Irwin J. J., Shoichet B. K., Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. J. Med. Chem. 55, 6582–6594 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hawkins P. C., Skillman A. G., Nicholls A., Comparison of shape-matching and docking as virtual screening tools. J. Med. Chem. 50, 74–82 (2007). [DOI] [PubMed] [Google Scholar]
- 58.McGaughey G. B. et al., Comparison of topological, shape, and docking methods in virtual screening. J. Chem. Inf. Model. 47, 1504–1519 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Rush T. S. 3rd, Grant J. A., Mosyak L., Nicholls A., A shape-based 3-D scaffold hopping method and its application to a bacterial protein-protein interaction. J. Med. Chem. 48, 1489–1495 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Muchmore S. W., Souers A. J., Akritopoulou-Zanze I., The use of three-dimensional shape and electrostatic similarity searching in the identification of a melanin-concentrating hormone receptor 1 antagonist. Chem. Biol. Drug Des. 67, 174–176 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Alford R. F. et al., The Rosetta all-atom energy function for macromolecular modeling and design. J. Chem. Theory Comput. 13, 3031–3048 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leaver-Fay A. et al., Scientific benchmarks for guiding macromolecular energy function improvement. Methods Enzymol. 523, 109–143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bazzoli A., Kelow S. P., Karanicolas J., Enhancements to the Rosetta energy function enable improved identification of small molecules that inhibit protein-protein interactions. PLoS One 10, e0140359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McGann M., FRED pose prediction and virtual screening accuracy. J. Chem. Inf. Model. 51, 578–596 (2011). [DOI] [PubMed] [Google Scholar]
- 65.Gallant S. I., Perceptron-based learning algorithms. IEEE Trans. Neural Netw. 1, 179–191 (1990). [DOI] [PubMed] [Google Scholar]
- 66.Pedregosa F. et al., Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011). [Google Scholar]
- 67.Meiler J., Baker D., ROSETTALIGAND: Protein-small molecule docking with full side-chain flexibility. Proteins 65, 538–548 (2006). [DOI] [PubMed] [Google Scholar]
- 68.Durrant J. D., McCammon J. A., BINANA: A novel algorithm for ligand-binding characterization. J. Mol. Graph. Model. 29, 888–893 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vogel S. M., Bauer M. R., Boeckler F. M., DEKOIS: Demanding evaluation kits for objective in silico screening–a versatile tool for benchmarking docking programs and scoring functions. J. Chem. Inf. Model. 51, 2650–2665 (2011). [DOI] [PubMed] [Google Scholar]
- 70.Bauer M. R., Ibrahim T. M., Vogel S. M., Boeckler F. M., Evaluation and optimization of virtual screening workflows with DEKOIS 2.0—a public library of challenging docking benchmark sets. J. Chem. Inf. Model. 53, 1447–1462 (2013). [DOI] [PubMed] [Google Scholar]
- 71.Lipinski C. A., Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods 44, 235–249 (2000). [DOI] [PubMed] [Google Scholar]
- 72.Veber D. F. et al., Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 45, 2615–2623 (2002). [DOI] [PubMed] [Google Scholar]
- 73.Sud M., MayaChemTools: An open source package for computational drug discovery. J. Chem. Inf. Model. 56, 2292–2297 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Keiser M. J. et al., Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 25, 197–206 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Gfeller D. et al., SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Res. 42, W32–W38 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Daina A., Michielin O., Zoete V., SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 47, W357–W364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu X. et al., PharmMapper server: A web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 38, W609–W614 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang X. et al., PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 45, W356–W360 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mizutani M. Y., Itai A., Efficient method for high-throughput virtual screening based on flexible docking: Discovery of novel acetylcholinesterase inhibitors. J. Med. Chem. 47, 4818–4828 (2004). [DOI] [PubMed] [Google Scholar]
- 80.Sopkova-de Oliveira Santos J. et al., Virtual screening discovery of new acetylcholinesterase inhibitors issued from CERMN chemical library. J. Chem. Inf. Model. 50, 422–428 (2010). [DOI] [PubMed] [Google Scholar]
- 81.Chen Y. et al., Discovery of a novel acetylcholinesterase inhibitor by structure-based virtual screening techniques. Bioorg. Med. Chem. Lett. 22, 3181–3187 (2012). [DOI] [PubMed] [Google Scholar]
- 82.Doytchinova I. et al., Novel hits for acetylcholinesterase inhibition derived by docking-based screening on ZINC database. J. Enzyme Inhib. Med. Chem. 33, 768–776 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mokrani E. H. et al., Identification of new potent acetylcholinesterase inhibitors using virtual screening and in vitro approaches. Mol. Inform. 38, e1800118 (2019). [DOI] [PubMed] [Google Scholar]
- 84.Lindert S. et al., Farnesyl diphosphate synthase inhibitors from in silico screening. Chem. Biol. Drug Des. 81, 742–748 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Durrant J. D. et al., Neural-network scoring functions identify structurally novel estrogen-receptor ligands. J. Chem. Inf. Model. 55, 1953–1961 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wójcikowski M., Zielenkiewicz P., Siedlecki P., Open drug discovery toolkit (ODDT): A new open-source player in the drug discovery field. J. Cheminform. 7, 26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang Y., Abdallah A. H. A., Lill M. A., Calculation of thermodynamic properties of bound water molecules. Methods Mol. Biol. 1762, 389–402 (2018). [DOI] [PubMed] [Google Scholar]
- 88.Cuzzolin A., Deganutti G., Salmaso V., Sturlese M., Moro S., AquaMMapS: An alternative tool to monitor the role of water molecules during protein-ligand association. ChemMedChem 13, 522–531 (2018). [DOI] [PubMed] [Google Scholar]
- 89.Ehrlich S., Göller A. H., Grimme S., Towards full quantum-mechanics-based protein-ligand binding affinities. ChemPhysChem 18, 898–905 (2017). [DOI] [PubMed] [Google Scholar]
- 90.Yilmazer N. D., Korth M., Recent progress in treating protein-ligand interactions with quantum-mechanical methods. Int. J. Mol. Sci. 17, 742 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kearnes S., McCloskey K., Berndl M., Pande V., Riley P., Molecular graph convolutions: Moving beyond fingerprints. J. Comput. Aided Mol. Des. 30, 595–608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Altae-Tran H., Ramsundar B., Pappu A. S., Pande V., Low data drug discovery with one-shot learning. ACS Cent. Sci. 3, 283–293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Adeshina Y., Karanicolas J., Dataset of congruent inhibitors and decoys (D-COID), Mendeley Data (Version 1, 2019). dx.doi.org/10.17632/8czn4rxz68.1. Deposited 09 December 2019.
- 94.Berman H. M. et al., The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Feng Z. et al., Ligand Depot: A data warehouse for ligands bound to macromolecules. Bioinformatics 20, 2153–2155 (2004). [DOI] [PubMed] [Google Scholar]
- 96.Hawkins P. C., Skillman A. G., Warren G. L., Ellingson B. A., Stahl M. T., Conformer generation with OMEGA: Algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model. 50, 572–584 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Natekin A., Knoll A., Gradient boosting machines, a tutorial. Front. Neurorobot. 7, 21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen T., Guestrin C., “XGBoost: A Scalable Tree Boosting System ” in Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (ACM, New York, 2016), pp. 785–794. [Google Scholar]
- 99.Breiman L., Random forests. Mach. Learn. 45, 5–32 (2001). [Google Scholar]
- 100.Geurts P., Ernst D., Wehenkel L., Extremely randomized trees. Mach. Learn. 63, 3–42 (2006). [Google Scholar]
- 101.Vapnik V., Statistical Learning Theory, (Wiley, New York, 1998). [Google Scholar]
- 102.Cheung J. et al., Structures of human acetylcholinesterase in complex with pharmacologically important ligands. J. Med. Chem. 55, 10282–10286 (2012). [DOI] [PubMed] [Google Scholar]
- 103.Ellman G. L., Courtney K. D., Andres V. Jr., Feather-Stone R. M., A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 7, 88–95 (1961). [DOI] [PubMed] [Google Scholar]
- 104.Cheng Y., Prusoff W. H., Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22, 3099–3108 (1973). [DOI] [PubMed] [Google Scholar]
- 105.Gaulton A. et al., The ChEMBL database in 2017. Nucleic Acids Res. 45, D945–D954 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.