Significance
Bacteriophages (phages), viruses that infect bacteria, are the most abundant microbes on Earth. Extreme specificity to host species or even strains of bacteria makes phages amenable to particular biotechnical applications. This specificity is partially governed by the surface-expressed structures of bacteria that phages use as cell-binding receptors. However, these receptors remain less-studied in phage biology, because their discovery relies on laborious methods utilizing well-characterized bacteria. Here we present INSeq screens as a rapid, high-throughput tool to identify candidate phage receptors. This method was successful in identifying receptors for previously well-characterized phages T2, T4, T6, and T7 as well as identifying receptors for newly isolated phages R3, U115, EC14, EC35, and 8S.
Keywords: bacteriophage, Escherichia coli, transposon insertion sequencing, selection, virus
Abstract
As the most abundant microbes on Earth, novel bacteriophages (phages; bacteria-specific viruses) are readily isolated from environmental samples. However, it remains challenging to characterize phage–bacteria interactions, such as the host receptor(s) phages bind to initiate infection. Here, we tested whether transposon insertion sequencing (INSeq) could be used to identify bacterial genes involved in phage binding. As proof of concept, results showed that INSeq screens successfully identified genes encoding known receptors for previously characterized viruses of Escherichia coli (phages T6, T2, T4, and T7). INSeq screens were then used to identify genes involved during infection of six newly isolated coliphages. Results showed that candidate receptors could be successfully identified for the majority (five of six) of the phages; furthermore, genes encoding the phage receptor(s) were the top hit(s) in the analyses of the successful screens. INSeq screens provide a generally useful method for high-throughput discovery of phage receptors. We discuss limitations of our approach when examining uncharacterized phages, as well as usefulness of the method for exploring the evolution of broad versus narrow use of cellular receptors among phages in the biosphere.
At an estimated 1031 particles, the number of bacteriophages (phages; bacteria-specific viruses) in the global biosphere comprises a vast and uninvestigated microbiological diversity (1). This largely untapped “bioprospecting” resource allows for newly discovered, naturally occurring phages to be developed for numerous applications. In stark contrast to this biodiversity are the relatively few phages that have been well-characterized (e.g., lambda, ΦX174, T4, T7, Φ6). These well-studied phages have been valuable for elucidating fundamental aspects of molecular biology and evolutionary genetics (2–4). However, there is increasing interest in utilizing phages for a myriad of purposes, including alleviating current medical challenges posed by antibacterial resistance, exemplified by recent cases where phages were used to treat antibiotic-resistant bacterial infections in humans (5, 6). It is therefore crucial to design approaches whereby newly discovered phages are characterized prior to any intended applications.
Isolating phage strains from environmental sources can be straightforward, though uncovering detailed characteristics of phage biology can be challenging. Sequencing approaches and molecular techniques are increasingly affordable, allowing for easier determination of genome size and nucleic acid content (DNA vs. RNA; single- vs. double-stranded) used to classify newly discovered phages into known or proposed virus families (7). Electron microscopy is typically sufficient to describe basics of phage morphology and capsid structure (8). Classic laboratory methods are useful for estimating host range (infectability across bacterial genotypes or species) and to determine whether a phage has a temperate versus strictly lytic replication cycle (7). However, other details of phage–bacteria interactions can be more difficult to ascertain. In particular, identifying the specific cell-surface receptor(s) that a phage uses in host binding and the bacterial genes that are involved in resistance to phage infection is not as readily determined using the above methods.
To initiate an infection, a phage particle must first recognize and bind one or more receptors on the surface of a susceptible bacterial cell. Many of the phages specific to Gram-positive bacteria use carbohydrate moieties of peptidoglycan or teichoic acid as their receptors (9), whereas others, such as phage γ which infects Bacillus anthracis and phage SPP1 that infects Bacillus subtilis, use protein receptors GamR and YueB, respectively (10, 11). Thus far, receptors used by phages of Gram-negative bacterial hosts appear limited to specific proteins and/or lipopolysaccharide (LPS) moieties present on the outer cell membrane (12–19). Overall, these binding targets are often highly conserved structures of bacteria, indicating that phages have been evolutionarily selected to exploit essential features of their hosts (20). In the simplest cases, a phage may use a single outer-membrane protein as a receptor, such as phage T6 attachment to the Tsx nucleoside channel of Escherichia coli (14). Other phages bind to a specific LPS motif, as seen in several members of the Podoviridae virus family (16). In further examples, phages use a primary receptor to facilitate attachment but require a secondary receptor to irreversibly bind to a host cell (18, 19). Finally, phages such as T2 can use two or more receptors on the surface of a bacterium interchangeably to bind and initiate infection (12, 15).
Elucidating molecular details of phage binding is fundamental to understanding phage biology because the receptors confer host specificity of a virus by allowing phages to differentiate between species or strains of bacteria. Nevertheless, it is challenging to determine phage receptors, especially when host bacteria themselves are not well-characterized. Previously, knockout libraries such as the Keio collection (single-gene deletion mutants of E. coli K-12) (21) have been utilized to identify specific bacterial genes involved in phage T7 infection (22), whereas similar resources do not exist for other nonmodel bacteria. Even in the case of E. coli, however, such methods can be laborious, involving the screening of individual phage samples on very large numbers of mutant bacterial strains if genes for phage receptors are unknown. Transposon mutant libraries have been used to identify bacterial mutants that are resistant to phage infection (23–25). While this method requires relatively less effort, it is limited by the number of transposon mutants chosen for follow-up study, making it potentially challenging to identify the receptor(s) used by a single phage. One other common method to identify possible phage receptors is to characterize spontaneous phage-resistant mutants of bacteria. However, this approach typically necessitates comparing whole-genome sequences of phage-resistant mutants with a well-annotated reference genome to identify candidate mutations, which are not guaranteed to be in genes encoding phage receptors.
Advances in high-throughput sequencing have facilitated the development of new sequencing-based transposon mutant screens, which enable the rapid identification of genes that contribute to bacterial fitness in a particular selective environment (26, 27). This method makes use of a modified transposon to create a diverse transposon mutant library, where each bacterial genome contains a single randomly incorporated insertion mutation. The sites of transposition in the mutant population can be easily identified by sequencing, and the relative abundances of specific insertions can be quantified. Comparison of the relative abundances of transposon mutants pre- and postselection can be used to identify genes contributing to fitness (bacterial growth) under the selective environment of interest.
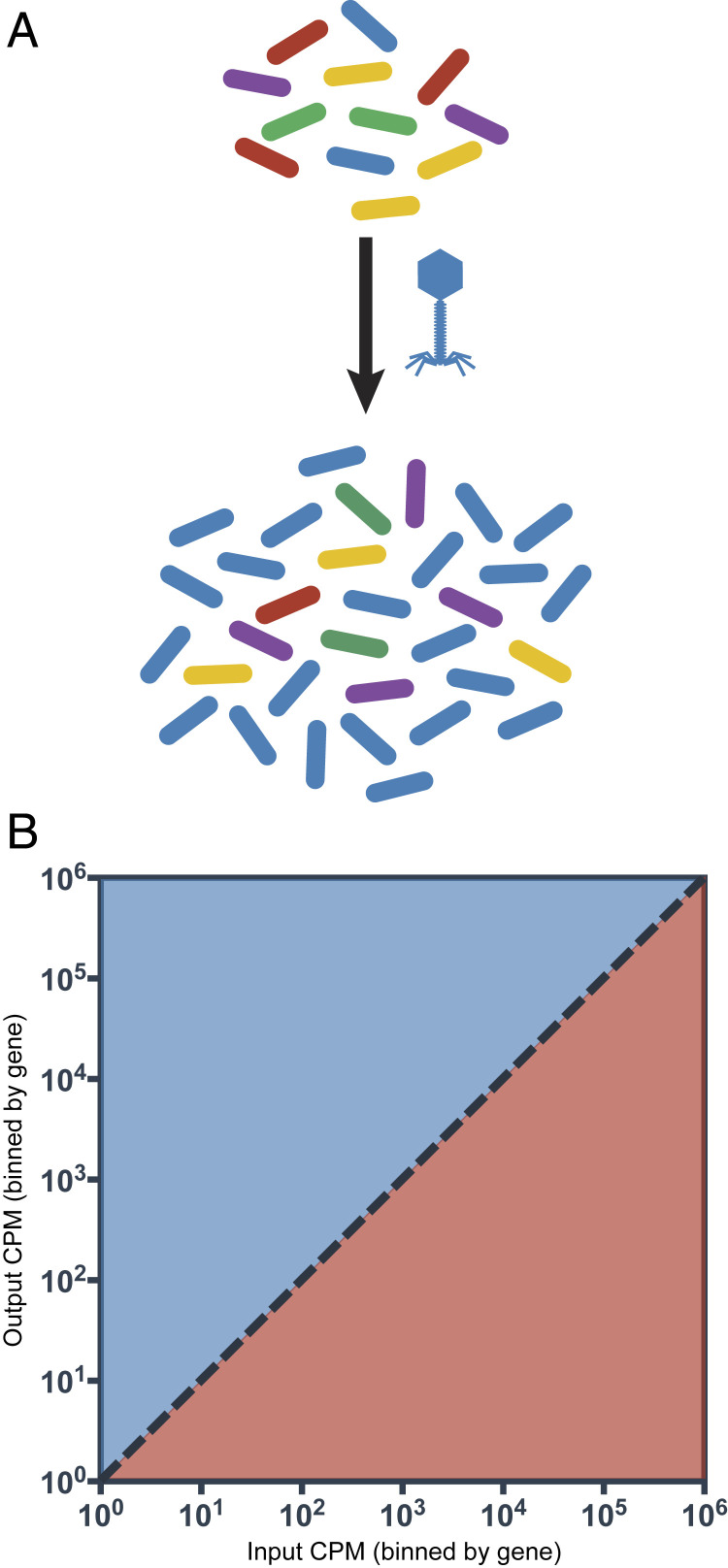
Here, we describe a high-throughput method to identify phage receptors by querying the entire repertoire of bacterial genes that are critical for survival during selection by lytic phages (Fig. 1A). The disruption of a gene involved in phage resistance would result in a fitness disadvantage under the selective pressure of phages; consequently, these mutants would be represented at a lower frequency in the output transposon mutant pool (TMP) (Fig. 1B). Conversely, disrupting a bacterial gene required for phage infection and replication (e.g., genes encoding phage receptors) would provide a fitness advantage under the selective pressure of phages and these mutants should be enriched in the output TMP.
Fig. 1.
Design of the INSeq screen. (A) Experimental setup for selection of the pooled transposon mutant library with phages indicating enrichment for phage-resistant mutants. (B) Mutants in genes involved in resistance to phages will become underrepresented in the output TMP (red) and mutants in genes required for phage infection will become overrepresented in the output TMP (blue).
We hypothesized that this approach could successfully identify bacterial genes responsible for receptor binding in four well-characterized viruses capable of infecting E. coli K-12: phages T2, T4, T6, and T7. Results of our high-throughput method agreed with the published literature in all cases. We further verified these conclusions using classic adsorption (phage attachment) assays, demonstrating that phages fail to attach to bacterial knockout mutants lacking the gene(s) encoding a phage receptor(s). We then tested our second hypothesis that this method could be used to identify receptor-binding genes in each of six newly discovered and previously uncharacterized phages, similarly capable of infecting E. coli K-12. Our approach, along with confirmatory adsorption assays, was successful in discovering the binding targets for five out of six of these viruses, demonstrating both the general utility of the method as well as the enduring challenge of developing an infallible technique for discerning phage receptors in bacteria.
Results and Discussion
INSeq Transposon Mutant Library Covers 80% of Nonessential Genes.
The final pooled transposon mutant library contained 17,100 independent transposon insertions in 3,253 bacterial genes. Visual inspection of results (Fig. 2) showed genome coverage did not appear to be affected by the inherent GC bias of mariner transposons (28). However, there was an increased number of transposition events near the E. coli K-12 origin of replication (Fig. 2). This result likely was attributed to multiple copies of the origin present during DNA replication (28). While the final library was not completely saturated (i.e., not every nonessential gene was represented by a transposon mutant), ∼80% of nonessential E. coli genes had at least one independent transposon insertion event in the final transposon mutant library. We used EcoCyc (29) to compile a list of 137 genes expressed at the cell outer membrane (Table 1); of these genes, 31 were not represented in the TMP used for our screens.
Fig. 2.
Genome plot of E. coli strain BW25113. Track 1 (inside) is a heatmap of GC content, where red represents high GC content and blue represents low GC content. Track 2 plots the abundance of transposon insertions in a coding region (green) or intergenic region (blue). Track 3 shows intergenic regions (red) and track 4 shows coding regions (multicolor).
Table 1.
List of genes encoding membrane proteins
| acrZ | envY | mdtQ | pgaA | yaiW | ynfB |
| amiD | fadL | mepS | pgaB | ybgQ | ypjA |
| appX | fecA | mipA | pgpB | ybhC | ypjB |
| bamA* | fepA | mlaA | phoE | yceK | yqhH |
| bamB | fhuA | mliC | pldA | ychO | yraJ |
| bamC | fhuE | mltA | ppk | yddB | yraP |
| bamD* | fimD | mltB | pqiC | yddL* | yzcX |
| bamE | fiu | mltC | qseG | ydeT | |
| bcsC | flgG | mltD | rcsF | ydiY | |
| bglH | flgH | mltF | rhsB | yehB | |
| bhsA | flu | nanC | rhsD | yfaL | |
| blc | gfcD | nfrA | rlpA | yfaZ | |
| borD | gfcE | nlpD | rsxG | yfcU | |
| btuB | gspD | nlpE | rzoD | yfeN | |
| chiP | hofQ | nlpI | rzoR | yfgH | |
| cirA | htrE | ompA | sfmD | yfiB | |
| csgB | lamB | ompC | skp | yghG | |
| csgE | loiP | ompF | slp | ygiB | |
| csgF | lolB | ompL | slyB | yhcD | |
| csgG | lpoA | ompG | tamA | yiaD* | |
| cusC | lpp | ompN | tamB | yiaT | |
| ecnA | mdtP | ompT | tolC | yjbF | |
| ecnB | lpoB | ompW | tsx | yjbH | |
| ecpC | lptA* | ompX | uidC | yjgL | |
| elfC | lptD* | pagP | wza | yliI | |
| emtA | lptE* | pal | yaiO | yncD |
Bolded genes are not included in the TMP.
Essential genes.
The transposon insertion sequencing (INSeq) screen was initiated with a multiplicity of infection (MOI; ratio of phage to bacteria) roughly equaling 0.01 (Materials and Methods). We noted that a lower MOI could allow for the occurrence of additional spontaneous (non-transposon insertion) mutations that could experience subsequent positive selection in the phage environment. Nevertheless, low MOI was essential to allow the TMP to undergo multiple rounds of cell division under phage selection, which was necessary for transposon mutant enrichment in the designed INSeq screen (Fig. 1). Furthermore, we reasoned that the reduced initial selective pressure at low MOI would allow us to observe transposon-disrupted genes with subtle (i.e., low to moderate) fitness effects (30).
INSeq Identifies Receptors for Phages T6, T2, T4, and T7.
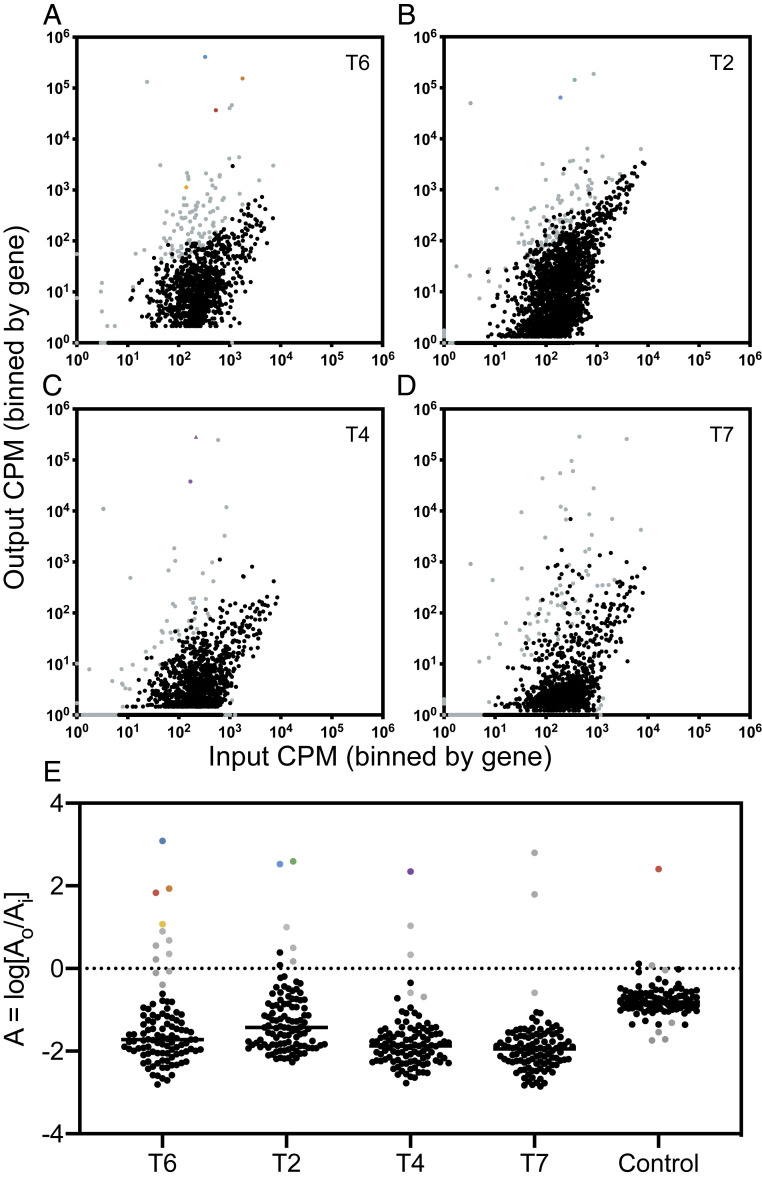
As a proof of concept for our method, we tested whether an INSeq screen could accurately identify the bacterial receptors for four well-characterized phages: T6, T2, T4, and T7. Controls consisted of INSeq screens using “mock infections” that were performed identically but in the absence of test phage. Results (Fig. 3 A–D) are plotted to depict all insertions in bacterial genes on a logarithmic scale as the ratio of input normalized (to counts per million; cpm) reads to output normalized reads in cpm. As compared with the mock infected control (SI Appendix, Fig. S1A), each screen in the presence of a test phage resulted in an observed shift of the TMP with many transposon mutants becoming underrepresented in the output population. This result was consistent with phages imposing strong selection pressure on the TMP. As the entire TMP shifts toward underrepresentation in the output population due to declining abundance of bacteria, it becomes difficult to distinguish truly significant hits that are at a fitness disadvantage in the presence of phage. However, this does not impair our ability to identify genes encoding phage receptors, as mutants in these genes should become overrepresented in the output population. Therefore, candidate receptor hits were considered if they were statistically significant, had a positive log ratio, A, of output (AO) to input (AI) relative abundance (A = log[AO/AI]), and were genes encoding a membrane protein (Fig. 3E).
Fig. 3.
Results of INSeq screens with well-characterized phages. Each point represents the normalized relative abundance of all transposon mutants in a specific gene. All statistically significant transposon mutants are in gray (P < 0.05). Certain hits are colored; other mutants in regulatory genes are triangles in the same color as the gene. (A) Results of a screen with phage T6. Significantly overrepresented hits tsx::Tn, btuB::Tn, lamB::Tn, and ompX::Tn are in blue, orange, red, and yellow, respectively. (B) Results of a screen with phage T2. Significantly overrepresented hits ompF::Tn and fadL::Tn are in green and blue, respectively. (C) Results of a screen with phage T4. Significantly overrepresented hit ompC::Tn is in purple. (D) Results of a screen with phage T7. (E) Log ratio of the relative abundance of 106 transposon mutants in genes encoding membrane proteins for each screen. The outlier in the control screen is lamB::Tn.
The screen in the presence of phage T6 (Fig. 3A) yielded 144 significant hits (SI Appendix, Dataset S1), of which 59 were positively enriched in the presence of phage T6. The log ratio of output to input relative abundance (A) of a subset of the TMP, transposon mutants in genes encoding membrane proteins (Fig. 3E), was queried for candidate receptors. Transposon mutant tsx::Tn (A = 3.09) was the top hit of these screens. This result was consistent with previous observations of phage T6 using the Tsx nucleoside channel as a primary receptor (14). Eight other genes encoding membrane proteins were enriched in the output: lamB::Tn (A = 1.84), btuB::Tn (A = 1.94), ompX::Tn (A = 0.90), rhsD::Tn (A = 0.55), ybgQ::Tn (A = 0.36), ynfB::Tn (A = 0.22), ompW::Tn (A = 1.08), and pgaB::Tn (A = 0.68). However, A values of these hits were far lower than that of tsx::Tn, indicating that in future screens, only the hits with the highest A values should be considered as candidate receptors. Nonetheless, a subset of these other eight hits was assayed in further experiments to demonstrate that these genes were not involved in the binding of phage T6 (see below).
The phage T2 screen (Fig. 3B) resulted in 143 significant hits, of which 86 (SI Appendix, Dataset S1) were positively enriched in the presence of phage T2. Candidate receptors fadL::Tn (A = 2.60) and ompF::Tn (A = 2.53) were identified (Fig. 3E) as the top two hits of these screens. This was a key result, as it was unclear if our approach could successfully identify both receptors used by a phage when the presence of only one was necessary and sufficient for phage adsorption. Thus, these results were consistent with previous observations of OmpF and FadL as interchangeable receptors for phage T2 (12, 15). There were two other enriched hits in genes encoding membrane proteins: rhsD::Tn (A = 1.00) and ynfB::Tn (A = 0.55). These two hits also came up in the screen with phage T6, indicating that they might be commonly required for phage infection. However, as above, their A values were far below those of fadL::Tn and ompF::Tn, and neither were considered for follow-up validation.
The phage T4 screen (Fig. 3C) resulted in 177 significant hits, of which only 20 (SI Appendix, Dataset S1) were positively enriched in the presence of phage T4. The top hit (Fig. 3E) was ompC::Tn (A = 2.35); additionally, envZ::Tn was significantly enriched in the output TMP (Fig. 2C). EnvZ is part of a two-component regulatory system that increases the expression of OmpC (31). Two other identified hits (Fig. 3E) were yjbF::Tn (A = 1.04) and yjbH::Tn (A = 0.34). Another gene in the same operon, yjbE::Tn (A = 1.36), was significantly enriched in the output TMP. The Yjb operon is involved in production of extracellular polysaccharide and biofilm formation (32). Previous literature suggests that phage T4 is not spatially inhibited by extracellular matrixes and is instead able to associate with the surface of biofilms (33). While it is interesting that multiple genes in this operon are significantly enriched in the output, further investigations of the importance of phage T4 with products of the Yjb operon are reserved for future studies, as previous results have demonstrated that phage T4 uses both OmpC and LPS as coreceptors (18). When OmpC is present, phage T4 can infect regardless of the terminal sugar residue of LPS; however, in the absence of OmpC, phage T4 is able to attach to LPS chains with an exposed terminal glucose residue (18). Exposure of this terminal glucose residue in LPS is conferred by a deletion of waaU, a gene that was not included in the TMP. Validation assays, therefore, were conducted on the top hit from this screen, OmpC (see below).
The phage T7 screen (Fig. 3D) resulted in 188 significant hits, of which 44 (SI Appendix, Dataset S1) were enriched in the output TMP. A previous screen of the Keio collection identified 10 genes important for phage T7 infection; however, none of these single-gene deletions were sufficient to abolish infection (22). Phage T7 is predicted to use LPS, specifically lipid A or a keto-deoxyoctulosonate sugar moiety, as a receptor (34); therefore, it is unlikely that deletion of a single bacterial gene would eliminate phage adsorption as many of the genes involved in synthesis of the inner core regions of LPS are essential. Therefore, instead of querying the 44 significantly output-enriched hits from the INSeq screen for a specific gene, these hits were subjected to a Gene Ontology (GO) enrichment analysis (35). The results of GO term analysis indicated that the significant, output-enriched transposon mutants from the phage T7 screen were enriched for 22 genes involved in cellular biosynthetic processes (pyrI, yjbH, mutT, accA, uvrD, mmuP, yaeI, mmuM, hisA, metG, yegS, carB, sufS, opgH, waaF, gmhB, trpS, waaG, yebK, speB, hldD, and dnaQ). Upon closer inspection, approximately one-third of these genes appeared to be involved in LPS metabolism: yaeI, waaF, gmhB, waaG, hldD, and yegS. Phosphodiesterase YaeI is annotated as a protein involved in lipid A biosynthesis and YegS is an annotated lipid kinase, while waaF, gmhB, waaG, and hldD are four out of five genes that make up functional gene cluster 91, which consists of genes involved in synthesis and polymerization of the core heptose region of LPS. This result was encouraging, as it was unclear if our screen would be successful in identifying LPS as a receptor due to the complex nature of LPS biosynthesis and the essentiality of many of the genes involved.
We note that false positives are expected to occur during our screen, especially since previous studies demonstrate that altering expression of one outer-membrane protein affects the expression of other outer-membrane proteins (36). While we observed some false positives (e.g., see phage T6 results above), identification of phage receptors is still expected to succeed with this method in combination with confirmatory assays described below. In each of the screens conducted with phages T2, T4, and T6, the known receptors were all of the top hits from the short list of candidate receptors. The use of LPS as a receptor, as is the case for phage T7, is likely to be successfully identified using the GO term with the overrepresented hits.
Efficiency of Plating and Adsorption Assays Confirm Known Receptors for Characterized Phages.
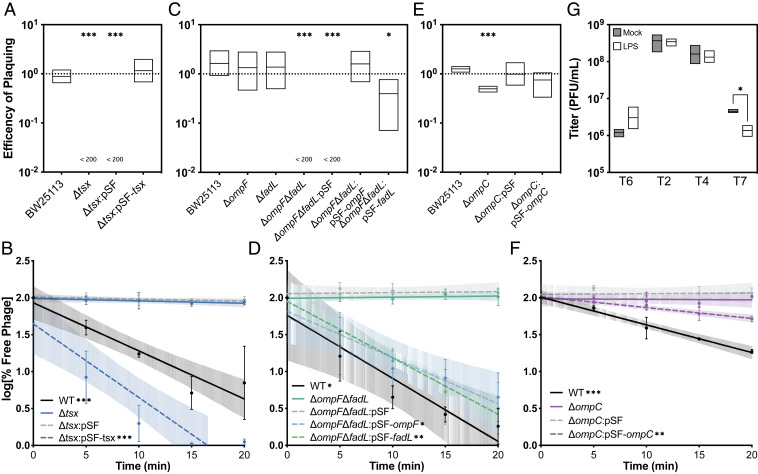
To verify that the top identified hits in the INSeq analyses were genes important for phage infection, we conducted efficiency of plating (EOP) assays to measure phage ability to productively infect bacterial strains lacking expression of the candidate gene (37) relative to productive growth on wild-type (WT) bacteria for a subset of the top hits. Since EOP does not directly measure phage attachment, adsorption assays were used to confirm that the putative genes were phage receptors for a subset of the top hits (Fig. 4). We concluded that EOP and adsorption assays were successful in confirming that the top hit(s) from the screens with phages T6, T2, and T4 was the known receptor(s) (SI Appendix, Results). In addition, results from GO term enrichment for phage T7 were confirmed by reduction in titers upon adsorption with LPS.
Fig. 4.
Assays validating phage receptors of well-characterized phages. (A) Efficiency of plating assays for phage T6 on various E. coli strains. EOP was calculated as a ratio of titer on the test strain to the titer on WT (BW25113ΔicdC). EOPs are plotted as mean and SD of three independent experiments. Significance was calculated using the t test as significantly different from 1; “<200” indicates that the titer on the test strain was below the limit of detection for this assay, which was 200 PFUs per milliliter. (B) Adsorption assays of phage T6 on various E. coli strains plotted as a log of the percent of free phage. Dashed lines indicate strains with an empty vector (pSF) or a complement vector (pSF-tsx) grown in the presence of 100 μg/mL Cb with induction by 1 mM IPTG. Percent free phage was determined as a ratio of free phage at the time point divided by the total phage added at the beginning of the assay and is plotted as a mean and SD of three independent experiments. Linear regression was performed for each strain. Regression lines with a significantly nonzero slope indicate that the particular strain is able to support phage T2 adsorption. (C) EOP assays for phage T2 on various E. coli strains. (D) Adsorption assays of phage T2 on various E. coli strains. (E) EOP assays for phage T4 on various E. coli strains. (F) Adsorption assays of phage T4 on various E. coli strains. (G) LPS phage inactivation assays for phages T2, T4, and T7. Titers for each phage were determined after a 20-min incubation with either 80 ng/mL LPS or water (11). Titers are plotted as means with SD from three independent experiments. A t test was used to determine significant differences between the mock condition and the LPS condition (*P < 0.05, **P < 0.01, ***P < 0.001).
INSeq Identifies Candidate Receptors for Newly Isolated Phages.
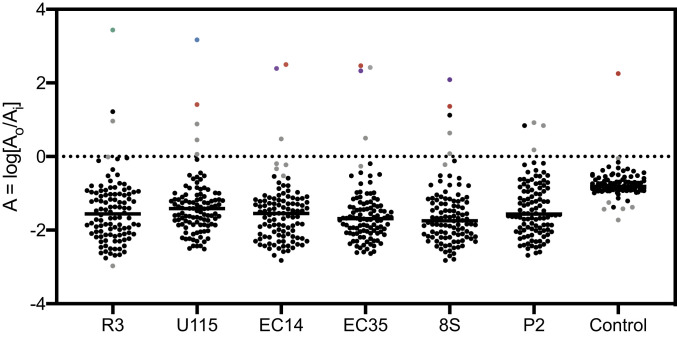
Next, uncharacterized phages were screened for receptors using our validated methods. Six phages (EC14, EC35, R3, P2, U115, and 8S) were chosen randomly from a large library of environmental phage isolates and subjected to an INSeq screen for receptor identification. Prior to the current study, no traits for these phage strains were determined, other than their ability to infect E. coli K-12. Screens with these six coliphages were performed as described above; hits that were statistically significant had a positive log ratio of output to input relative abundance, and were in genes encoding membrane proteins that were considered for further validation.
The phage R3 screen resulted in 161 significant hits (SI Appendix, Fig. S3A), of which 67 (SI Appendix, Dataset S1) were enriched in the presence of phage R3. Only two hits (Fig. 5) were identified, ompA::Tn (A = 3.44) and rhsD::Tn (A = 0.96). As the top hit, OmpA was considered the candidate receptor for phage R3.
Fig. 5.
Log ratio of the relative abundance of 106 transposon mutants in genes encoding membrane proteins for each screen with uncharacterized phages. Gray represents statistically significant hits, and other top hits ompA::Tn, tsx::Tn, ompC::Tn, and lamB::Tn are in green, blue, purple, and red, respectively. The outlier in the control screen is lamB::Tn (P < 0.05).
The phage U115 screen resulted in 150 significant hits (SI Appendix, Fig. S3B), of which 58 (SI Appendix, Dataset S1) were enriched in the presence of phage U115. The top hit (Fig. 5) of these screens was tsx::Tn (A = 3.17). There were four other hits: lamB::Tn (A = 1.41), rhsD::Tn (A = 0.88), ompX::Tn (A = 0.45), and ybgQ::Tn (A = 0.06). The top two hits from these screens, Tsx and LamB, were considered candidate receptors for phage U115.
The phage P2 screen resulted in 143 significant hits (SI Appendix, Fig. S3C), of which 59 (SI Appendix, Dataset S1) were enriched in the presence of phage P2. Three hits (Fig. 5) were slightly enriched: rhsD::Tn (A = 0.92), ynfB::Tn (A = 0.18), and yjbH::Tn (A = 0.84). However, none of these hits were promising, as receptors for the well-characterized phages T6, T2, and T4 were enriched to a log ratio greater than 1. Furthermore, all three of these transposon mutants were background hits in other screens. Therefore, it is likely that either phage P2 uses LPS as a receptor (as in phage T7), or that the receptor for phage P2 is one of the 31 membrane proteins not included in the TMP.
The phage 8S screen resulted in 164 significant hits (SI Appendix, Fig. S3D), of which 49 (SI Appendix, Dataset S1) were enriched in the presence of phage 8S. The top two hits (Fig. 5) were ompC::Tn (A = 2.09) and lamB::Tn (A = 1.36). Other hits included rhsD::Tn (A = 0.64) and ynfB::Tn (A = 0.08). Additionally, OmpC regulators, envZ::Tn (A = 2.50) and ompR::Tn (A = 2.57), were enriched in the output TMP. The top two hits from this screen, OmpC and LamB, were considered candidate receptors for phage 8S infection.
The phage EC35 screen resulted in 160 significant hits (SI Appendix, Fig. S3E), of which 69 (SI Appendix, Dataset S1) were enriched in the presence of phage EC35. The top two hits (Fig. 5) were lamB::Tn (A = 2.47) and ompC::Tn (A = 2.33). Other hits included rhsD::Tn (A = 0.50) and loiP::Tn (A = 2.42). Additionally, OmpC regulators, envZ::Tn (A = 2.38) and ompR::Tn (A = 3.03), as well as LamB regulators, malT::Tn (A = 2.61) and malK::Tn (A = 0.31), were enriched in the output TMP. The top three hits from this screen were LamB, OmpC, and LoiP; however, loiP::Tn was only enriched in one out of three replicate screens. Therefore, LamB and OmpC were considered candidates for phage EC35 receptors.
The phage EC14 screen resulted in 177 significant hits (Fig. 4F), of which 32 (SI Appendix, Dataset S1) were enriched in the presence of phage EC14. Similar to phage EC35, the top two hits from the screen with phage EC14 were lamB::Tn (A = 2.47) and ompC::Tn (A = 2.33). The other hit was rhsD::Tn (A = 0.48). Additionally, OmpC regulators envZ::Tn (A = 2.09) and ompR::Tn (A = 2.77), LamB regulator malT::Tn (A = 2.60), and malK::Tn (A = 0.003) were enriched in the output TMP. LamB and OmpC were considered candidates for phage EC14 infection.
Receptors for Five Uncharacterized Phages Identified with EOP and Adsorption Assays.
EOP and adsorption assays were used to validate the candidate receptors for uncharacterized phages which were identified in each INSeq screen.
Outer-membrane protein OmpA was the candidate with the highest A value in the screen with phage R3 (Fig. 5), and was selected as the likely receptor for further analysis. In an EOP assay, phage R3 had an EOP (Fig. 6A) below the limit of detection on BW25113ΔompA. Plaquing was restored upon plasmid complementation of the ompA gene; however, plaquing was only restored to 46% of the phage growth on the parental strain. This result was likely due to differences in expression levels between chromosomally encoded ompA and exogenous ompA. Furthermore, phage R3 did not adsorb (Fig. 6B) to strain BW25113ΔompA. Similar to plaquing, adsorption was restored upon complementation with OmpA. Results of both the EOP and adsorption assays strongly indicated that OmpA was the primary receptor for the previously uncharacterized phage R3.
Fig. 6.
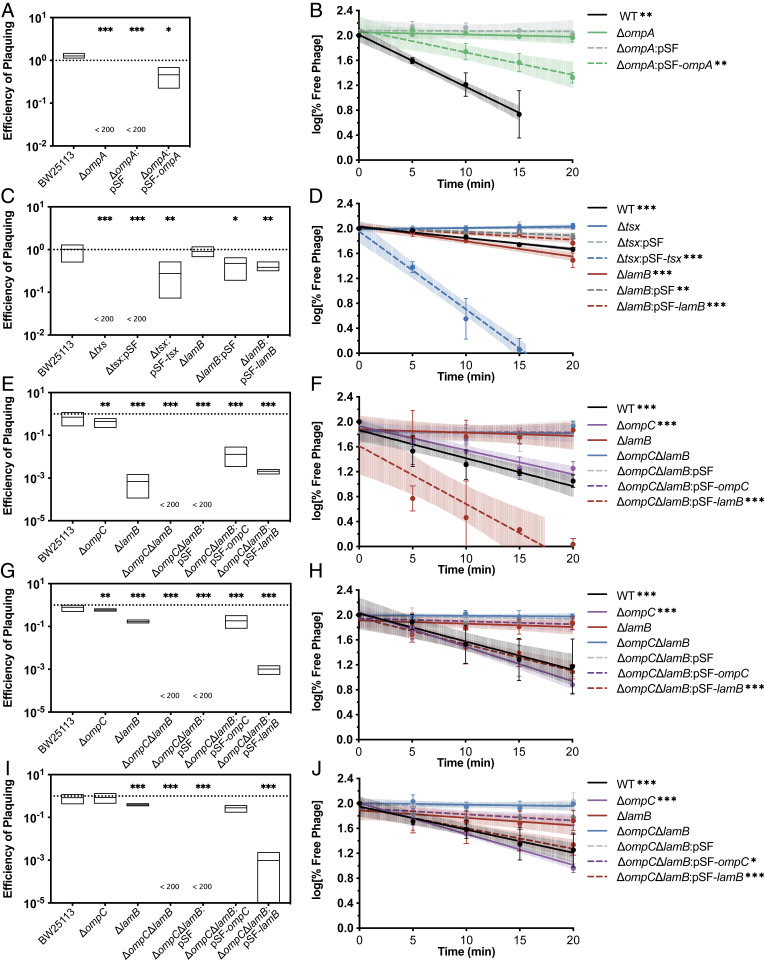
Assays validating phage receptors of newly isolated phages. (A) EOP assays for phage R3 on various E. coli strains. (B) Adsorption assays of phage R3 on various E. coli strains plotted as a log of the percent of free phage. (C) EOP assays for phage U115 on various E. coli strains. (D) Adsorption assays of phage U115 on various E. coli strains. (E) EOP assays for phage 8S on various E. coli strains. (F) Adsorption assays of phage 8S on various E. coli strains plotted as a log of the percent of free phage. (G) EOP assays for phage EC35 on various E. coli strains. (H) Adsorption assays of phage EC35 on various E. coli strains. (I) EOP assays for phage EC145 on various E. coli strains. (J) Adsorption assays of phage EC14 on various E. coli strains (*P < 0.05, **P < 0.01, ***P < 0.001).
The screen of phage U115 identified Tsx, a nucleoside transporter, as well as LamB as potential receptors for phage U115 (Fig. 5). Phage U115 had an EOP (Fig. 6C) below the limit of detection on BW25113Δtsx and plaquing was restored to wild-type levels upon complementation with Tsx. Deletion of lamB had no significant effect on the EOP of phage U115. Phage U115 did not adsorb (Fig. 6D) to BW25113Δtsx or to the empty vector control, BW25113Δtsx::pSF. However, complementation with Tsx not only restored phage U115 adsorption but allowed U115 to adsorb better than wild type; 90% of phage U115 had adsorbed to BW25113Δtsx::pSF-tsx by 15 min while only 46.4% of phage U115 had adsorbed to WT by 20 min. This result was likely due to the overexpression of exogenous tsx resulting in more Tsx on the outer membrane, relative to basal expression in WT bacteria. Phage U115 adsorbed to BW25113ΔlamB at levels comparable to wild type. Results of both the EOP and adsorption assays indicated that phage U115 did not require LamB as a receptor to adsorb or infect E. coli, and that Tsx was the primary receptor for phage U115.
The INSeq screen of phage 8S identified OmpC, an outer-membrane porin, and LamB as potential receptors for the virus (Fig. 5). EOP assays (Fig. 6E) revealed a 2.3-fold decrease in EOP of phage 8S on BW25113ΔompC and a 1,455-fold decrease in EOP on BW25113ΔlamB. Phage 8S was unable to infect the double knockout BW25113ΔompCΔlamB, and infection was restored upon complementation with either ompC or lamB. Phage 8S appeared to adsorb (Fig. 6F) to BW25113ΔompC at levels similar to wild type. However, phage 8S did not significantly adsorb to BW25113ΔlamB or to BW25113ΔompCΔlamB. Adsorption was restored upon complementation of BW25113ΔompCΔlamB with lamB but not ompC. This result indicated that LamB was necessary for adsorption of phage 8S. However, since phage 8S was able to infect BW25113ΔlamB but unable to infect BW25113ΔompCΔlamB, it likely used OmpC as a secondary receptor. Adsorption assays indicated that LamB was the primary receptor for phage 8S as it adsorbed well to BW25113ΔompCΔlamB:pSF-lamB but did not adsorb appreciably to BW25113ΔompCΔlamB complemented with ompC over the course of the assay.
Similar to the screen for phage 8S, the screen for phage EC35 identified OmpC and LamB (Fig. 5). There was a 1.7-fold decrease and a 6-fold decrease in EOP (Fig. 6G) of phage EC35 on BW25113ΔompC and BW25113ΔlamB, respectively. Phage EC35 was unable to infect BW25113ΔompCΔlamB and infection was restored upon complementation with either ompC or lamB. Phage EC35 was unable to adsorb (Fig. 6H) to BW25113ΔlamB or BW25113ΔompCΔlamB, but was able to adsorb to BW25113ΔompC and BW25113ΔompCΔlamB upon complementation with lamB, and not with ompC. Again, these data indicated that phage EC35 used LamB as a primary receptor but likely required OmpC as a secondary receptor, as only deletion of both ompC and lamB abolished infection in an EOP assay.
INSeq screens for a third phage, EC14, also identified OmpC and LamB as potential receptors (Fig. 5). There was a 1.2-fold decrease and a 2.6-fold decrease in EOP (Fig. 6I) of phage EC14 on BW25113ΔompC and BW25113ΔlamB, respectively. Phage EC35 was unable to infect BW25113ΔompCΔlamB. Complementation with either ompC or lamB was sufficient to restore infection. Phage EC14 was unable to adsorb (Fig. 6J) to BW25113ΔlamB and BW25113ΔompCΔlamB but was able to adsorb to BW25113ΔompC and BW25113ΔompCΔlamB upon complementation with lamB or ompC. This result indicated that, unlike phage 8S or EC35, EC14 was able to use either LamB or OmpC as a receptor.
Unlike the screens above, the screen with phage P2 resulted in no statistically significant hits (Fig. 5) with a log ratio of output cpm to input cpm above 1.0 in genes expressed at the outer membrane; the top two hits were rhsD (A = 0.92) and yjbH (A = 0.84). Transposon mutant rhsD::Tn was likely background noise as it came up in every screen and follow-up assays were not done. EOP and adsorption assays were performed with BW25113ΔyjbH. There was no decrease in EOP of phage P2 on BW25113ΔyjbH (SI Appendix, Fig. S4A). Phage P2 was able to adsorb to BW25113ΔyjbH (SI Appendix, Fig. S4B). It is possible that similar to phage T7, phage P2 uses LPS as a receptor. However, incubation with purified LPS did not result in a significant drop in titer (SI Appendix, Fig. S4C). Furthermore, GO term analysis did not uncover any significant enrichment for any biological processes. Another possibility is that similar to phage T2, phage P2 has multiple receptors. However, successful identification of both receptors of phage T2 indicates that our method should allow for the identification of multiple receptors. Therefore, it is likely that the receptor for phage P2 is one of the 31 outer-membrane genes that was not represented in the original TMP; it is possible that a transposon mutant library with higher saturation would allow for the identification of the phage P2 receptor. However, it is also possible that an essential gene encodes the receptor for phage P2, which would not be included in a transposon mutant library.
In all screens performed, whether well-characterized phages or newly isolated ones, lon::Tn was consistently the top hit (Fig. 3 and SI Appendix, Fig. S3). As lon encodes the Lon protease, which functions in the cytoplasm, it was not considered a candidate receptor hit and no follow-up was conducted. However, mutations in lon that disrupt the proteolytic function result in accumulation of rcsA, an activator of capsular polysaccharide biosynthesis (38). Therefore, it is likely that deletion of lon generally results in phage resistance due to overexpression of capsule. Based on these screens, it is possible that mutations in lon result in a smaller fitness cost than mutations in genes that code for phage receptors. This result will be explored further in future studies.
INSeq screens were successful in identifying candidate phage receptors for five out of six previously uncharacterized phages and further analysis of specific hits confirmed that the top hit(s) identified in the screens was the phage receptor(s). We note that our approach is designed to identify the receptor(s) used by a phage when interacting with a single bacterial strain. However, this method could be employed in multiple different strains of bacteria in order to identify receptors for different hosts for phages that encode multiple tailspikes that permit infection across multiple bacterial hosts (39).
Interestingly, each of the phage receptors identified in this study has been identified as a receptor for other phages (40). With 137 annotated membrane proteins and a branching LPS chain consisting of 10 sugar monomers with various phosphate groups in E. coli BW25113, it is surprising that previous reviews on phage receptors reported that only ∼10 membrane proteins and various sugar linkages were exploited as phage receptors (40, 41). Furthermore, three out of five phages chosen randomly from our coliphage library used OmpC as a receptor in some capacity. These observations are interesting because the vast biodiversity of lytic phages suggests that these viruses may have evolved to exploit most, if not all, possible receptors on the surface of bacteria, implicating selection by these “predatory” phages as a possible key driver of genomic diversity in bacteria (42). However, this expectation assumes that all cell-surface structures are equally suitable to serve as phage receptors in the eyes of natural selection. Rather, selection for phages to use a particular structure on the surface of a bacterium is likely influenced by the abundance of that molecule on the surface and whether the protein is well-conserved among bacterial genotypes. Both OmpA and OmpC are reported to be among the top 20 most highly expressed proteins in E. coli, with 207,618 and 163,538 molecules per cell, respectively (43). On one hand, with so many potential binding sites on the surface (approximately one OmpA or OmpC molecule per 30 square nanometers), a bacterium is extremely vulnerable to attack by phages that bind to either OmpA or OmpC, suggesting that phages should be strongly selected to target these structures (44). However, on the other hand, this creates the possibility for intense competition among phages that use common binding sites such as OmpA or OmpC, which likely results in selection for phages to exploit a less common receptor to reduce competition for hosts. Previously, phage receptors have been vastly understudied, likely because of the large effort required to screen phages on individual bacterial mutants. Thus, the higher-throughput INSeq screen presented here demonstrates a method to rapidly identify phage receptors, fostering a broader possibility to efficiently explore the evolution of phage binding sites and interphage competition to use these receptors. This could be a key approach in the ultimate study of whether virus evolution has resulted in some binding targets being over- versus underexploited in the phage world.
Around the time of submission, both a paper came out and a preprint was posted describing similar methods for identifying genes involved in phage infection in Enterococcus faecalis and E. coli, respectively (45, 46). However, there were some differences in outcomes among these studies which may reflect differences in the efficacies of the approaches. It would be interesting to compare these various methods in future work.
Conclusion
Transposon insertion sequencing is a powerful technique that allows for identification of bacterial genes that contribute to fitness in particular selective conditions. Here, we used INSeq screens to identify phage receptors for multiple E. coli phages. Proof-of-concept experiments validated this approach by confirming known phage receptors. In particular, this method was successful in confirming receptors for previously well-characterized phages that use a single protein receptor, dual-protein receptors, a primary LPS receptor and secondary protein receptor, and LPS as receptor. This method was then extended to identify receptors for five out of six previously uncharacterized randomly chosen phages. We expect that a more saturated transposon mutant library would allow for identification of the receptor for newly discovered phage P2. Our approach was particularly promising, because for each screen that resulted in successful protein receptor identification, the genes encoding the receptors were among the top hits for each phage.
In addition to identifying phage receptors, our screens could provide information about the fitness of other bacterial genes in the presence of phages, such as bacterial genes that were involved in phage replication or resistance to phages. Thus, our approach might allow for identification of a subset of bacterial genes that are universally important during phage infection or resistance.
While this study focused on phage receptors for viruses that infect E. coli, transposon insertion sequencing provides information for any transposon mutant in the TMP, and has been employed in many different species of bacteria (37, 47–50). Our method for the high-throughput identification of phage receptors should allow for rapid characterization of newly isolated phages that target various different species of bacteria. If enough phage receptors are identified for sequenced phages, bioinformatic approaches might be extended to allow for receptor identification based on the genomes of phages alone. Furthermore, in addition to using INSeq screens to identify phage receptors, our screens provide information about the fitness of other bacterial genes in the presence of phages, potentially allowing for the identification of a subset of bacterial genes that are universally important during phage infection. Therefore, in addition to allowing for a high-throughput identification of phage receptors, INSeq screens may provide insight into generalized mechanisms of phage resistance.
Materials and Methods
Bacterial Strains, Plasmids, and Phages.
Bacteria used in this study were obtained from the Coli Genetic Stock Center at Yale University, and phages T2, T4, T6, and T7 were kindly provided by J. Wertz, Yale University. The six uncharacterized phages in the study were isolated from sewage or environmental water samples. All bacteria, plasmid, and phage strains are listed in SI Appendix, Table S1. Bacteria were cultured at 37 °C with shaking (200 rpm) in Luria broth (LB) (4) and on LB agar (1.5%) plates, where dilutions in LB followed by plating were used to estimate bacterial densities as colony-forming units (CFUs) per milliliter. Carbenicillin (Cb; 100 μg/mL), gentamicin (Gm; 10 μg/mL), kanamycin (Km; 50 μg/mL), arabinose (0.1%), and isopropyl β-d-1-thiogalactopyranoside (IPTG; 1 mM) were added when appropriate. Phage strains were amplified in shaking liquid culture overnight on the amplification host BW25113ΔicdC (hereafter referred to as WT), and filter sterilized with a 0.22-μm filter to obtain a cell-free lysate. Titers were estimated as plaque-forming units (PFUs) per milliliter and determined by plaques formed via dilution plating in “soft” agar (0.75%) overlays on lawns of WT grown on agar plates, unless otherwise noted.
INSeq Transposon Mutant Library Preparation.
The insertion sequence transposon mutant library was made via conjugation. Donor bacterial strain S17 λ pir (26) containing plasmid pSAM_PA (50) was conjugated with recipient strain WT on plates containing arabinose at a ratio of 1:1 donor to recipient. After a 3-h incubation at 37 °C, conjugation mixtures were plated for single colonies on plates with Gm and Km to isolate transconjugants that were resistant to both markers (Gmr, Kmr). Following a 24-h incubation at 37 °C, colonies were scraped into LB medium using a sterile spatula and stored in 20% glycerol at −80 °C.
INSeq Screen.
Aliquots of the library were thawed on ice, washed once in LB, and resuspended to a concentration of ∼107 CFUs per milliliter in LB and incubated at 37 °C for 1 h, with shaking. A sample of test phage was added, in triplicate, to a final concentration of ∼105 PFUs per milliliter, for MOI ∼ 0.01. Following overnight incubation, cultures were harvested and genomic DNA was extracted and prepared for sequencing (49). DNA libraries were pooled and sequenced at the Yale Center for Genome Analysis via the Illumina HiSeq 2500 System. Sequences were analyzed using scripts modified from Goodman et al. (26). Briefly, using Python scripts adapted from analysis packages previously described (49), sequencing reads were indexed by barcode, and transposon sequence was trimmed leaving 16 bp of adjacent genomic DNA. These 16-bp sequences were aligned to the reference genome (GenBank accession no. CP009273.1) using Bowtie 2 by counting the number of reads for each insertion site, normalizing to counts per million reads, and binning by gene (51). Transposon insertions mapping to the distal 5% ends of any coding region as well as transposon insertions mapping to intergenic regions were filtered out during analysis. A Z test was performed using the log ratio of normalized output count to normalized input count, and Q values from a false discovery rate correction of <0.05 were considered significant for further analysis.
Efficiency of Plating and Adsorption Assays.
Efficiency of plating was measured as the ratio of the test phage titer on the experimental strain to its titer on the WT strain. Phage binding to cells (adsorption) was measured over a 20-min adsorption assay (52), which estimates the rate of “disappearance” of phage particles (i.e., reduction in titer) in liquid LB medium over time when viruses are challenged to attach to bacterial cells.
Supplementary Material
Acknowledgments
We thank members of the P.E.T. laboratory, A. Goodman, B. Kacmierczak, S. Shames, and D. Weinberger for helpful comments and valuable discussions regarding this study and for feedback on early drafts and submitted versions of the manuscript.
Footnotes
Competing interest statement: P.E.T. discloses a financial interest in Felix Biotechnology, Inc., a company that seeks to commercially develop phages for use as therapeutics.
Data deposition: Raw sequencing data reported in this paper have been deposited in Dryad (https://doi.org/10.5061/dryad.t76hdr7z3) and at the National Center for Biotechnology Information Sequence Read Archive (accession no. PRJNA637562).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001888117/-/DCSupplemental.
Data Availability.
Raw sequencing data have been made available at Dryad (https://doi.org/10.5061/dryad.t76hdr7z3), and at the National Center for Biotechnology Information Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/PRJNA637562). Other data can be found in SI Appendix, Dataset S1.
References
- 1.Hendrix R. W., Smith M. C. M., Burns R. N., Ford M. E., Hatfull G. F., Evolutionary relationships among diverse bacteriophages and prophages: All the world’s a phage. Proc. Natl. Acad. Sci. U.S.A. 96, 2192–2197 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crick F. H., Barnett L., Brenner S., Watts-Tobin R. J., General nature of the genetic code for proteins. Nature 192, 1227–1232 (1961). [DOI] [PubMed] [Google Scholar]
- 3.Hershey A. D., Chase M., Independent functions of viral protein and nucleic acid in growth of bacteriophage. J. Gen. Physiol. 36, 39–56 (1952). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luria S. E., Delbrück M., Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28, 491–511 (1943). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan B. K. et al., Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health 2018, 60–66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schooley R. T., et al. , Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother. 61, e00954-17 (2017). Correction in: Antimicrob. Agents Chemother.62, e02221-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clokie M. R. J., Kropinski A. M., Eds., Bacteriophages: Methods and Protocols, Volume 2: Molecular and Applied Aspects, (Humana Press, 2009). [Google Scholar]
- 8.Ackermann H.-W., “Bacteriophage electron microscopy” in Advances in Virus Research, Lobocka M., Szybalski W. T., Eds. (Elsevier, 2012), Vol. 82, pp. 1–32. [DOI] [PubMed] [Google Scholar]
- 9.Dowah A. S. A., Clokie M. R. J., Review of the nature, diversity and structure of bacteriophage receptor binding proteins that target gram-positive bacteria. Biophys. Rev. 10, 535–542 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.São-José C., Baptista C., Santos M. A., Bacillus subtilis operon encoding a membrane receptor for bacteriophage SPP1. J. Bacteriol. 186, 8337–8346 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davison S., Couture-Tosi E., Candela T., Mock M., Fouet A., Identification of the Bacillus anthracis (gamma) phage receptor. J. Bacteriol. 187, 6742–6749 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Black P. N., The fadL gene product of Escherichia coli is an outer membrane protein required for uptake of long-chain fatty acids and involved in sensitivity to bacteriophage T2. J. Bacteriol. 170, 2850–2854 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Click E. M., Webster R. E., The TolQRA proteins are required for membrane insertion of the major capsid protein of the filamentous phage f1 during infection. J. Bacteriol. 180, 1723–1728 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hantke K., Phage T6—Colicin K receptor and nucleoside transport in Escherichia coli. FEBS Lett. 70, 109–112 (1976). [DOI] [PubMed] [Google Scholar]
- 15.Hantke K., Major outer membrane proteins of E. coli K12 serve as receptors for the phages T2 (protein Ia) and 434 (protein Ib). Mol. Gen. Genet. 164, 131–135 (1978). [DOI] [PubMed] [Google Scholar]
- 16.Lindberg A. A., Bacteriophage receptors. Annu. Rev. Microbiol. 27, 205–241 (1973). [DOI] [PubMed] [Google Scholar]
- 17.Morona R., Henning U., New locus (ttr) in Escherichia coli K-12 affecting sensitivity to bacteriophage T2 and growth on oleate as the sole carbon source. J. Bacteriol. 168, 534–540 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Washizaki A., Yonesaki T., Otsuka Y., Characterization of the interactions between Escherichia coli receptors, LPS and OmpC, and bacteriophage T4 long tail fibers. MicrobiologyOpen 5, 1003–1015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu F., Mizushima S., Roles of lipopolysaccharide and outer membrane protein OmpC of Escherichia coli K-12 in the receptor function for bacteriophage T4. J. Bacteriol. 151, 718–722 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith D. L. et al., Short-tailed stx phages exploit the conserved YaeT protein to disseminate Shiga toxin genes among enterobacteria. J. Bacteriol. 189, 7223–7233 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baba T. et al., Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qimron U., Marintcheva B., Tabor S., Richardson C. C., Genomewide screens for Escherichia coli genes affecting growth of T7 bacteriophage. Proc. Natl. Acad. Sci. U.S.A. 103, 19039–19044 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hossain M. J., Rahman K. S., Terhune J. S., Liles M. R., An outer membrane porin protein modulates phage susceptibility in Edwardsiella ictaluri. Microbiology 158, 474–487 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Lee H. S., Choi S., Shin H., Lee J. H., Choi S. H., Vibrio vulnificus bacteriophage SSP002 as a possible biocontrol agent. Appl. Environ. Microbiol. 80, 515–524 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maynard N. D. et al., A forward-genetic screen and dynamic analysis of lambda phage host-dependencies reveals an extensive interaction network and a new anti-viral strategy. PLoS Genet. 6, e1001017 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodman A. L. et al., Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6, 279–289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Opijnen T., Bodi K. L., Camilli A., Tn-seq: High-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 6, 767–772 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chao M. C., Abel S., Davis B. M., Waldor M. K., The design and analysis of transposon insertion sequencing experiments. Nat. Rev. Microbiol. 14, 119–128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keseler I. M. et al., The EcoCyc database: Reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 45, D543–D550 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van den Bergh B., Swings T., Fauvart M., Michiels J., Experimental design, population dynamics, and diversity in microbial experimental evolution. Microbiol. Mol. Biol. Rev. 82, e00008-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aiba H., Mizuno T., Phosphorylation of a bacterial activator protein, OmpR, by a protein kinase, EnvZ, stimulates the transcription of the ompF and ompC genes in Escherichia coli. FEBS Lett. 261, 19–22 (1990). [DOI] [PubMed] [Google Scholar]
- 32.Ferrières L., Aslam S. N., Cooper R. M., Clarke D. J., The yjbEFGH locus in Escherichia coli K-12 is an operon encoding proteins involved in exopolysaccharide production. Microbiology 153, 1070–1080 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Sutherland I. W., Hughes K. A., Skillman L. C., Tait K., The interaction of phage and biofilms. FEMS Microbiol. Lett. 232, 1–6 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Lupo D. et al., The T7 ejection nanomachine components gp15-gp16 form a spiral ring complex that binds DNA and a lipid membrane. Virology 486, 263–271 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Ashburner M. et al.; The Gene Ontology Consortium , Gene Ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schnaitman C. A., McDonald G. A., Regulation of outer membrane protein synthesis in Escherichia coli K-12: Deletion of ompC affects expression of the OmpF protein. J. Bacteriol. 159, 555–563 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shames S. R. et al., Multiple Legionella pneumophila effector virulence phenotypes revealed through high-throughput analysis of targeted mutant libraries. Proc. Natl. Acad. Sci. U.S.A. 114, E10446–E10454 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torres-Cabassa A. S., Gottesman S., Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J. Bacteriol. 169, 981–989 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scholl D., Rogers S., Adhya S., Merril C. R., Bacteriophage K1-5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli. J. Virol. 75, 2509–2515 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bertozzi Silva J., Storms Z., Sauvageau D., Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 363, fnw002 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Chang V., Chen L.-Y., Wang A., Yuan X., The effect of lipopolysaccharide core structure defects on transformation efficiency in isogenic Escherichia coli BW25113 rfaG, rfaP, and rfaC mutants. J. Exp. Microbiol. Immunol. 14, 101–107 (2010). [Google Scholar]
- 42.Rodriguez-Valera F. et al., Explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 7, 828–836 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Li G. W., Burkhardt D., Gross C., Weissman J. S., Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kondev T. J., Phillips R., Garcia H., Physical Biology of the Cell, (Garland Science, 2009). [Google Scholar]
- 45.Chatterjee A. et al., Parallel genomics uncover novel enterococcal-bacteriophage interactions. MBio 11, e03120-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mutalik V. K. A., et al. , High-throughput mapping of the phage resistance landscape in E. coli. bioRxiv:10.1101/2020.02.15.951020 (16 February 2020). [DOI] [PMC free article] [PubMed]
- 47.Bachman M. A. et al., Genome-wide identification of Klebsiella pneumoniae fitness genes during lung infection. MBio 6, e00775 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao B. et al., Metabolic and fitness determinants for in vitro growth and intestinal colonization of the bacterial pathogen Campylobacter jejuni. PLoS Biol. 15, e2001390 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodman A. L., Wu M., Gordon J. I., Identifying microbial fitness determinants by insertion sequencing using genome-wide transposon mutant libraries. Nat. Protoc. 6, 1969–1980 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rundell E. A., Commodore N., Goodman A. L., Kazmierczak B. I., A screen for antibiotic resistance determinants reveals a fitness cost of the flagellum in Pseudomonas aeruginosa. J. Bacteriol. 202, e00682-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kropinski A. M., “Measurement of the rate of attachment of bacteriophage to cells” in Bacteriophages, Clokie M. R., Kropinski A. M., Eds. (Humana Press, 2009), pp. 151–155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data have been made available at Dryad (https://doi.org/10.5061/dryad.t76hdr7z3), and at the National Center for Biotechnology Information Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/PRJNA637562). Other data can be found in SI Appendix, Dataset S1.