Significance
Human cytomegalovirus (HCMV) infection is lifelong and persistent. Periodic reactivation of cytomegalovirus poses serious disease risk for immune-compromised patients. A critical driver of reactivation is expression of viral genes from the major immediate early locus. Recent paradigm-shifting evidence shows that reactivation is driven from promoters distinct from those that drive replication in permissive cells. Understanding the contextual control of these promoters and how they specifically respond to cellular cues that drive reactivation is critical for establishing future therapies that prevent reactivation. Our findings mechanistically define a previously enigmatic relationship between differentiation and reactivation and provide potential targets for therapeutic intervention to prevent HCMV reactivation and disease.
Keywords: HCMV, herpesvirus, FOXO, latency, reactivation
Abstract
Human progenitor cells (HPCs) support human cytomegalovirus (HCMV) latency, and their differentiation along the myeloid lineage triggers cellular cues that drive reactivation. A key step during HCMV reactivation in latently infected HPCs is reexpression of viral major immediate early (MIE) genes. We recently determined that the major immediate early promoter (MIEP), which is primarily responsible for MIE gene expression during lytic replication, remains silent during reactivation. Instead, alternative promoters in the MIE locus are induced by reactivation stimuli. Here, we find that forkhead family (FOXO) transcription factors are critical for activation of alternative MIE promoters during HCMV reactivation, as mutating FOXO binding sites in alternative MIE promoters decreased HCMV IE gene expression upon reactivation and significantly decreased the production of infectious virus from latently infected primary CD34+ HPCs. These findings establish a mechanistic link by which infected cells sense environmental cues to regulate latency and reactivation, and emphasize the role of contextual activation of alternative MIE promoters as the primary drivers of reactivation.
Reactivation of latent human cytomegalovirus (HCMV) infection poses a significant threat to patients with compromised immune systems. While primary infections in healthy adults are typically associated with little or mild disease, reactivation in immunocompromised individuals, such as solid organ and stem cell transplant recipients, can lead to significant morbidity and mortality (1). Despite this, little is known regarding the mechanisms controlling HCMV reactivation.
Following an initial burst of viral gene expression upon entering cells that support latency (e.g., CD34+ human progenitor cells [HPCs]), HCMV gene expression is largely silenced, allowing the virus to persist in a quiescent or latent state (2, 3). Proinflammatory cytokines and cellular cues that drive differentiation induce the reexpression of viral lytic cycle genes, culminating in the production of infectious virus that can spread throughout the host and cause disease (4, 5). Critical to HCMV reactivation is the reexpression of the viral immediate early 1 and 2 proteins (IE1 and IE2), which drive the expression of the HCMV lytic cycle (6–8). Defining factors that regulate IE1 and IE2 expression during reactivation is thus critical for understanding the mechanisms controlling reactivation and resulting HCMV disease.
In cells permissive for lytic replication such as fibroblasts, the expression of both UL123 and UL122 transcripts (encoding IE1 and IE2, respectively) is largely driven by the major immediate early promoter (MIEP), which is silenced during latency. Until recently it was presumed that reactivation of the lytic cycle relied on the resumption of MIEP activity. However, we recently discovered two alternative promoters (iP1 and iP2, together referred to as intronic promoters) within the first intron of the canonical major immediate early locus (9). Rather than inducing MIEP activity, HCMV reactivation stimuli instead induce transcription from the iP1 and iP2 promoters, which correlates with the increase in UL123 and UL122 mRNA levels and IE1 and IE2 protein levels (10). Deletion of the intronic promoters significantly attenuates the production of infectious virus after reactivation, revealing that the iP1 and iP2 promoters play critical roles in IE1 and IE2 reexpression and HCMV reactivation.
Here we begin to unravel the mechanisms by which HCMV senses cellular cues to trigger IE1 and IE2 reexpression by defining FOXO transcription factors (TFs) as key players linking cellular differentiation to HCMV reactivation. These data provide mechanistic insights into HCMV reactivation and suggest that manipulating FOXO transcription factor activity may be a future means to limit HCMV disease.
Materials and Methods
Cells and Viruses.
MRC-5 fibroblasts and HeLa cells were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin. HCMV TB40E derived from a bacterial artificial chromosome (BAC) containing an SV40 promoter-driven GFP reporter (11) was used as wild-type (WT) virus, and served as the backbone for the generation of recombinant viruses. Titers of cell-free virus were determined by the 50% tissue culture infective dose (TCID50) method on MRC-5 fibroblasts. Unless otherwise noted, all infections were performed at a multiplicity of infection (MOI) of 1 for 1 h, followed by removal of the inoculum.
Construction of Recombinant Viruses.
BAC-mediated recombineering was used to generate viral mutants on the TB40E genomic background using a two-step recombination approach, as before (9). Briefly, the iP2 locus was replaced with a kanamycin/levansucrase fusion cassette (KanSacB) using homology-mediated recombination in recombination-competent SW105 Escherichia coli (12). The KanSacB cassette was then replaced with the iP2 sequences containing the indicated mutation in a second round of recombination. The entire genomes of the wild-type and recombinant BACs were then sequenced to ensure no additional changes were present other than the intended mutations.
THP-1 Latency Model.
Latency studies were conducted as described previously (10, 13, 14). Briefly, cells were infected with HCMV (TB40/E; refs. 5 and 15) at a multiplicity of 2 plaque-forming units (pfu) per cell. After 5 d, cells were treated with 100 nM 12-o-tetradecanoylphorbol-13-acetate (TPA) to induce reactivation, or with dimethyl sulfoxide (DMSO) solvent control. Whole cell lysates, DNA, and RNA were collected at each of the indicated time points. Detailed experimental methods are found in SI Appendix.
Assay of Infectious Centers for Latency and Reactivation.
The frequency of HCMV reactivation in CD34+ HPCs was quantified as previously described (10, 13, 14). Briefly, pure populations of CD34+ HPCs were infected with TB40E WT or FOXOmut123 recombinant virus (MOI = 2). Twenty-four hours after infection, latently infected cells were purified and cocultured with stromal cells to maintain latent infection. After 10 d, latently infected cells were cocultured with naïve MRC-5 fibroblast monolayers to quantify the number of infectious centers produced. Detailed experimental methods, including isolation and purification of these cells, are found in SI Appendix.
qRT-PCR Analysis.
mRNA abundance was quantified by qRT-PCR as described previously (10, 16). Briefly, RNA was extracted and then treated with DNase. The RNA was reverse transcribed and then quantified by real-time PCR using SYBR green incorporation and transcript-specific primers. For transfected cells, RNA abundance was determined by comparison to a standard curve generated from qPCR analysis of 10-fold serial dilutions of a DNA standard specific for each primer pair. Transcript abundance in latently infected cells was measured as previously described (10). Detailed methods are provided in SI Appendix, Supplementary Materials and Methods.
Plasmid Construction.
Luciferase reporter vectors containing the distal promoter (dP), MIEP, iP1, or iP2 promoters have been previously described (9). Details of additional plasmids used in this study are described in SI Appendix, Supplemental Materials and Methods.
Luciferase Assays.
Luciferase assays were performed as described previously (17). Luciferase activity was measured 24 h after transfection of HeLa cells and normalized to the protein concentration in the sample. Luciferase reactions were performed in duplicate for each sample and averaged, and the graphs show the mean values of at least three biological replicates performed on different days. Detailed methods are provided in SI Appendix, Supplemental Materials and Methods.
Electrophoretic Mobility Shift Assays (EMSA).
Double-stranded DNA probes were incubated with 500 ng purified recombinant FOXO3a protein (Abnova) on ice for 20 min. The reaction was resolved on polyacrylamide gel and then transferred to a nylon membrane. Membranes were probed with streptavidin-horseradish peroxidase (HRP) according to manufacturer’s instructions (Thermo Fisher) and visualized by chemiluminescence (BioExpress). Detailed methods are provided in SI Appendix, Supplementary Materials and Methods.
Western Blotting.
Western blotting was performed as described previously (18). Briefly, cells were lysed in radioimmunoprecipitation assay buffer containing protease inhibitors and equal amounts of protein were resolved on sodium dodecyl sulfate/polyacrylamide gel electrophoresis gels. Proteins were transferred to nitrocellulose membranes (Amersham) and probed with primary antibodies followed by horseradish peroxidase-coupled secondary antibodies. Membranes were visualized by chemiluminescence (ECL) reagent (BioExpress) using a chemiluminescence detection system (Bio-Rad). Detailed methods are provided in SI Appendix, Supplementary Materials and Methods.
In Silico Analysis of FOXO Binding Sites in the MIE Locus.
The program Find Individual Motif Occurrences (FIMO; ref. 19) was used to search intron A of the MIE genomic locus for the two known FOXO binding motifs RWAAAYAA and MMAAAYAA. Only sites with a threshold P value of P <0.01 were considered.
Results
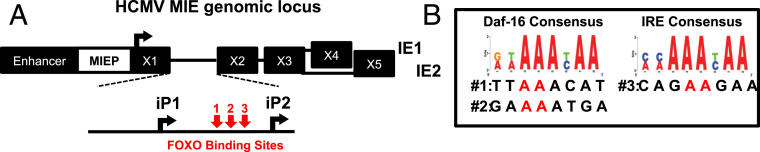
We recently described a mechanism by which the reexpression of the UL123 and UL122 mRNAs during HCMV reactivation is driven by the alternative promoters iP1 and iP2 (Fig. 1A), rather than the MIEP (10). While iP1 and iP2 promoters are necessary for efficient reactivation, the factors regulating their activity are unknown. Reactivation is induced by differentiation of monocytes into macrophages or by treating latently infected cells with phorbol esters (e.g., TPA) (10) or LY294002 (20, 21), a chemical inhibitor of the PI3K/mTOR pathway. Phorbol esters, LY294002, and monocyte differentiation all activate the FOXO family of transcription factors (22). An in silico analysis of the HCMV MIE locus identified three potential FOXO binding sites located between the iP1 and iP2 transcription start sites (TSSs) (Fig. 1B). We thus hypothesized that FOXO transcription factors stimulate iP1, iP2, or both, resulting in increased transcription of the UL123 and UL122 mRNAs and HCMV reactivation.
Fig. 1.
HCMV intronic promoters contain FOXO TF consensus sites. (A) Schematic showing the location of the intronic promoters iP1 and iP2 and potential FOXO TF binding sites in the major immediate early intronic promoter locus. (B) The intronic promoters were searched against the two consensus binding motifs for FOXO transcription factors. The sequences of the three potential FOXO binding sites are shown. The bases highlighted in red were changed to CC in the mutants described in Figs. 2–6.
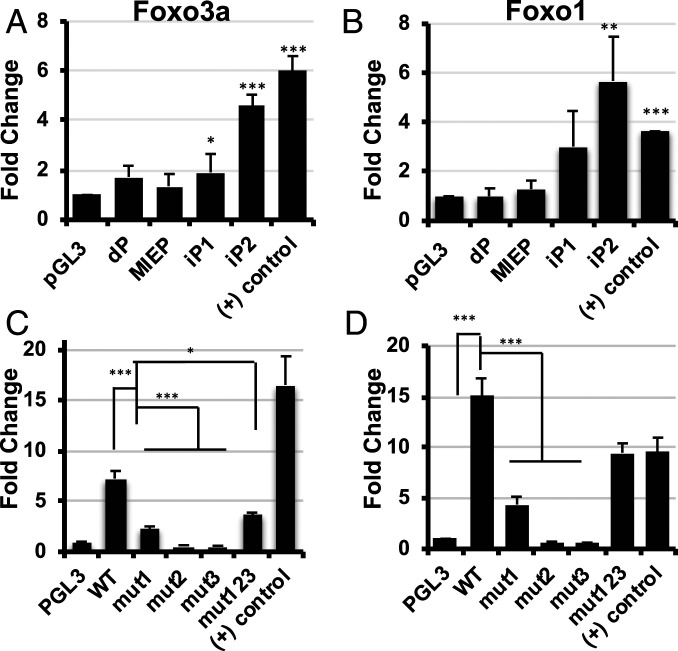
To test this hypothesis, we first determined if FOXO TFs stimulated the activity of the iP1 and iP2 promoters outside the context of infection. Of the four members of the FOXO family of transcription factors, FOXO1 and FOXO3a are ubiquitously expressed in many tissues, including bone marrow, whereas FOXO4 and FOXO6 expression was limited to specific tissues or not detectable (23). Further, FOXO1 and FOXO3a play critical roles in hematopoietic stem cell maintenance and differentiation processes linked to HCMV latency (24, 25). We cotransfected vectors expressing FOXO1 and FOXO3a with reporter constructs containing the dP, MIEP, iP1, or iP2 promoters upstream of luciferase. We found neither FOXO1 nor FOXO3a affected the activity of the MIEP or the dP, but either FOXO1 or FOXO3a significantly increased the activity of both iP1 and iP2 (Fig. 2 A and B). We mutated critical nucleotides in each potential FOXO binding site in the iP2 construct, as iP2 was the most FOXO-responsive promoter (shown in Fig. 1B), and also generated a reporter construct containing mutations in all three binding sites to determine if FOXO binding to multiple sites had a cooperative effect. Mutating any of the three potential FOXO binding sites, either alone or in combination, significantly decreased iP2 induction by FOXO transcription factors (Fig. 2 C and D).
Fig. 2.
(A) MIE intronic promoters are activated by FOXO. (A and B) HeLa cells were cotransfected with FOXO overexpression plasmids and previously described luciferase reporter constructs containing the four promoters in the MIE genomic locus 5′ of the luciferase gene; the distal promoter (dP), the major immediate early promoter (MIEP), intronic promoter 1 (iP1), and intronic promoter 2 (iP2). Luciferase activity was measured at 24 h after transfection and normalized to the amount of protein in the sample. A previously characterized positive control ([+] control) was included which contains three consensus FOXO binding sites 5′ of the luciferase gene. The parental luciferase plasmid pGL3 basic, which lacks a promoter upstream of luciferase, served as a control. The graphs show the fold change in activity of each promoter in the presence of FOXO3a (A) or FOXO1 (B) compared to the pGL3 basic control. (C and D) Each potential FOXO binding site in the iP2 promoter was mutated either alone (mut1, mut2, and mut3) or in combination (mut123).The reporters were cotransfected with either FOXO3a (C) or FOXO1 (D), and luciferase assays were performed as above (n = 3; *P < 0.05, **P < 0.05, ***P < 0.005).
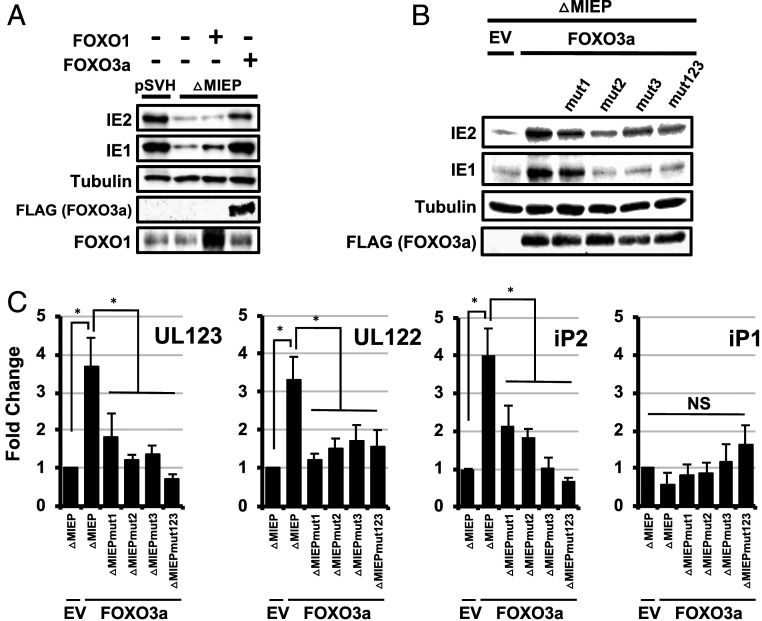
We next determined how FOXO transcription factors affect IE1 and IE2 expression in the context of the MIE locus. To this end, we measured IE1 and IE2 protein levels in cells cotransfected with FOXO expression vectors and the plasmid pSVH, which contains the entire MIE locus, from 840 base pairs upstream of the MIEP TSS through the 3′ UTR of exon 5 (26–28). In this construct, the MIEP is the predominant promoter driving transcription of UL123 and UL122 mRNA, and both iP1 and iP2 are minimally active (9). We found expression of FOXO3a, but not FOXO1, resulted in a slight, but reproducible, increase in IE1 and IE2 protein levels (SI Appendix, Fig. S1).
To confirm that FOXO TFs increase IE1 and IE2 expression independently of the MIEP, we repeated the above experiments using a variant of the pSVH plasmid where the core MIEP is deleted (pSVHΔMIEP). We previously showed that IE1 and IE2 expression from this plasmid is driven exclusively by the iP1 and iP2 promoters (9). As before, deleting the core MIEP promoter greatly decreased IE1 and IE2 protein levels (Fig. 3A). In contrast to the luciferase reporters, FOXO1 expression had minimal effect on IE1 or IE2 protein levels in this setting. However, FOXO3a expression significantly increased IE1 and IE2 protein levels (Fig. 3A). Surprisingly IE1 and IE2 protein levels were comparable to levels in cells transfected with the wild-type MIE locus in the presence of FOXO3a (Fig. 3A).
Fig. 3.
FOXO3a stimulates transcription from iP2 in the context of the MIE genomic locus. (A) A plasmid containing the regulatory and coding regions for the HCMV IE1 and IE2 genes (pSVH) or a variant of pSVH lacking the core MIEP (pSVHΔMIEP) was transfected into HeLa cells together with a FOXO1 or FOXO3a expression plasmid, or an empty expression vector (EV) control. Cells were harvested at 24 h after infection, and cell lysates were analyzed by Western blot using the indicated antibodies. Results are representative of three independent experiments. (B) Each FOXO consensus site in iP2 was mutated in the context of the pSVHΔMIEP plasmid, either alone (mut1, mut2, and mut3) or in combination (mut123). Cells were transfected with the indicated plasmid along with a FOXO3a expression plasmid (FOXO3a) or the EV control and harvested 24 h later. Cell lysates were analyzed by Western blot using the indicated antibodies. The results are representative of three independent experiments. (C) Cells were transfected and harvested as in B. RNA was extracted and analyzed by qRT-PCR using primers specific for UL123 (encoding IE1), UL122 (encoding IE2), iP1, or iP2. The graphs show the fold change in RNA abundance compared to cells transfected with pSVHDMIEP and the empty expression vector control (n = 3; *P < 0.05; NS = not significant).
As FOXO1 had minimal impact on IE1/2 expression in the context of the MIE locus, we focused further studies on FOXO3a. Mutating any of the potential FOXO TF binding sites abrogated the effects of FOXO3a on IE1 and IE2 protein (Fig. 3B) and UL123 and UL122 mRNA levels (Fig. 3C). The increase in UL123 and UL122 mRNA correlated with a matching increase in transcription from the iP1 and iP2 promoters (Fig. 3C), suggesting that FOXO3a increases UL123 and UL122 mRNA by stimulating iP1 and iP2 activity.
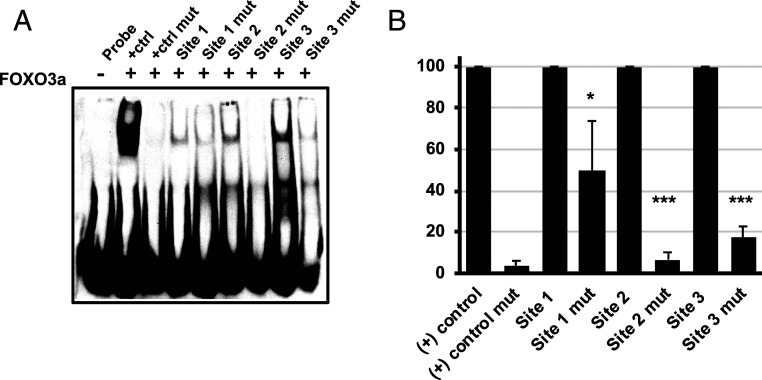
To determine if FOXO TFs directly bind FOXO-responsive sites in the MIE intronic promoters, we used an EMSA to measure binding of recombinant FOXO3a to double-stranded oligonucleotides containing each of the three potential FOXO binding sites. FOXO3a bound to each of the three sites, with FOXO site 3 exhibiting the greatest binding (Figs. 1 and 4A). Mutating each of the potential FOXO sites using the same strategy as above decreased FOXO3a binding to each site (Fig. 4 A and B). Together with the results above, these data show FOXO3a can directly bind specific sequences within the intronic promoter region, suggesting that the effect of FOXO3a on UL123 and UL122 mRNA and IE1 and IE2 protein expression is not due to an indirect effect on other cellular pathways.
Fig. 4.
FOXO3a directly binds to FOXO consensus sites in iP2. (A) Recombinant purified FOXO3a was incubated with double-stranded biotinylated probes containing the indicated FOXO consensus sites in iP2 (site 1, site 2, and site 3), or mutated sequences containing the changes shown in Fig. 1 (site 1 mut, site 2 mut, and site 3 mut). Protein:DNA complexes were resolved on nondenaturing acrylamide gels, and the complexes were visualized by chemiluminescence. The data are representative of three independent experiments. (B) Graph showing the decrease in FOXO3a binding to the mutant probes compared to the wild-type probes, which are set to 100%. The graph shows the mean of three independent experiments (*P < 0.05; ***P < 0.0005).
We next sought to define the role of FOXO TFs in stimulating iPs during HCMV reactivation. We considered depleting FOXO TFs from latently infected cells; however, FOXO TFs play critical roles in cellular differentiation (29). Therefore any reactivation phenotypes found in FOXO-depleted, latently infected cells could be due to defects in differentiation, defects in intronic promoter activation, or both. To circumvent this issue, we generated a recombinant virus containing mutations in the FOXO binding sites between iP1 and iP2 and measured the effects on virus replication and latency. In fibroblasts, the recombinant virus expressed viral immediate early (IE), early (E), and late (L) proteins (SI Appendix, Fig. S2B) similarly to the wild-type virus control, and replicated to equivalent titer and with similar kinetics as wild-type virus (SI Appendix, Fig. S2A). These results are consistent with our previous results showing a minor role for iP1 and iP2 in HCMV lytic replication in fibroblasts (9, 10).
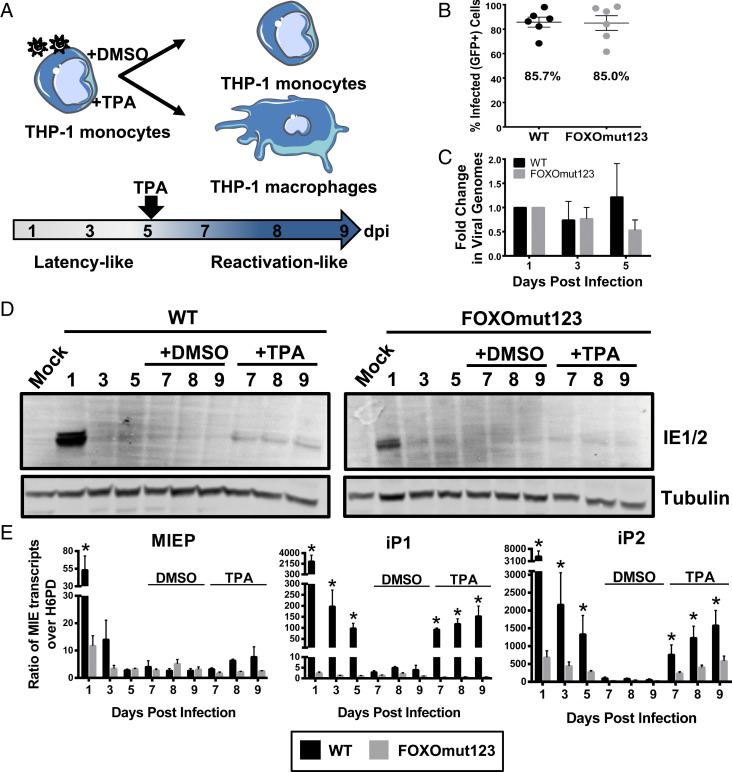
To determine the role of FOXO binding to the iP1 and iP2 promoters in regulating reexpression of major immediate early genes in hematopoietic cells, we infected THP-1 cells, a model system for HCMV latency studies (30–33), with wild-type virus or the recombinant containing mutations in all three FOXO binding sites (Fig. 5A). Although THP-1 cells are limited in their capacity to recapitulate every aspect of HCMV latency and reactivation, our prior work demonstrates that HCMV gene expression is largely silenced in THP-1 cells following a transient burst of gene expression, and that TPA treatment induces a transcriptional reexpression of HCMV MIE mRNAs and proteins, despite their failure to produce high titers of viral progeny (10). THP-1 cells were infected to similar levels with the wild-type and mutant viruses, as determined by the percentage of GFP-positive cells at day 1 after infection (Fig. 5B), and both WT and FOXOmut123 viral genomes were maintained during the latency period (days 1 through 5) prior to TPA treatment (Fig. 5C).
Fig. 5.
FOXO consensus sites are required for reexpression of MIE genes in THP-1 cells. (A) Schematic of THP-1 model for reexpression of MIE genes after stimulation with TPA. THP-1 cells were infected with TB40/E WT or FOXOmut123. At 5 dpi, cells were treated with TPA to stimulate macrophage differentiation and reexpression of viral genes, or with a DMSO vehicle control. (B) Equal infection (GFP+) of THP-1 cells with each virus was determined by flow cytometric analysis at 24 h postinfection (hpi). Biological replicates are represented as single points around the mean. SE is shown. (C) Total DNA was isolated and viral genomes were quantified using a primer to the β2.7 region of the HCMV genome relative to a cellular gene, RNase-P. Ratio of viral-to-cellular DNA at 3 and 5 dpi are normalized to their respective 1 dpi. Data from five independent biological replicates are shown; SEM is depicted. Two-way analysis of variance (ANOVA) showed no statistically significant change in genome copy for either infection over the time course. (D) Protein lysates were harvested at the indicated time points and immunoblotted to detect viral IE1 and IE2 with tubulin as a loading control. (E) MIE transcripts derived from the MIEP, iP1, or iP2 were quantified using reverse transcriptase quantitative PCR relative to the low-abundance housekeeping gene H6PD. Error bars represent the average of three independent experiments amplified in triplicate and SEM is shown. Statistical significance (*P ≤ 0.05) was determined by multiple paired t tests comparing accumulation of each transcript in WT versus mutant virus infection at each time point.
Similar to infection with viruses lacking the entire iP1 and iP2 elements (10), mutating FOXO binding sites in iP2 decreased IE protein accumulation at 1 d postinfection (dpi) (Fig. 5D). A quiescent infection was established after infection with either the WT or FOXOmut123 viruses, as IE proteins were expressed at levels below the limit of detection from 3 dpi through 9 dpi in the DMSO-treated control groups. The mutant virus expressed appreciably less IE1 and IE2 protein relative to the WT infection (Fig. 5D) after the addition of TPA to stimulate reactivation. To determine the promoters driving IE gene expression, we quantified transcripts derived from the MIEP (or distal promoter), iP1, or iP2 using primers specific to sequences unique to transcripts derived from each promoter. Transcripts derived from the MIEP/distal promoter remained very low both before and after TPA treatment of cells infected with wild-type virus (Fig. 5E). MIEP-derived transcript levels were significantly affected by mutation of the FOXO binding sites at 1 dpi, but not following TPA reactivation treatment. In contrast, iP1 and iP2 transcripts accumulated to high levels in cells infected with wild-type virus before being silenced, and were induced following TPA stimulation (Fig. 5E). iP1 and iP2 transcripts were expressed to significantly lower levels in cells infected with the FOXO binding site mutant relative to the wild-type virus over the time course, and were reexpressed to lower levels following TPA treatment. From these data we conclude that FOXO binding sites in the intronic promoters play an important role in the initial early burst of MIE expression, and TPA-induced MIE reexpression in THP-1 cells. Further, we analyzed FOXO TF expression in THP-1 cells pre- and postdifferentiation with TPA. Consistent with FOXO3a playing a critical role in stimulating iP1 and iP2 (Figs. 3 and 4), accumulation of FOXO3a, but not FOXO1, transcripts were induced following TPA treatment (SI Appendix, Fig. S3A), and FOXO3a protein levels were increased (SI Appendix, Fig. S3B. This finding aligns with differentiation-induced changes in host TF abundance as a driver of reactivation (20, 34).
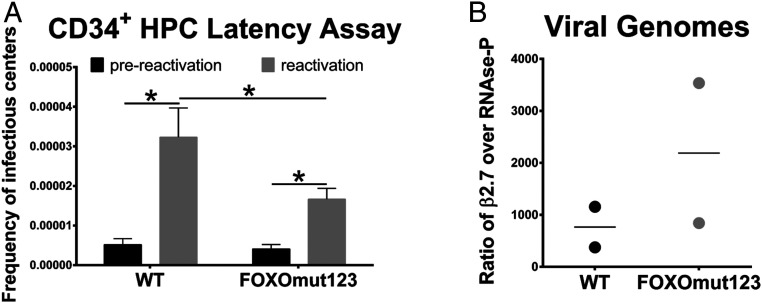
We next measured the impact of FOXO TF binding to the intronic promoters during infection of primary CD34+ bone marrow-derived HPCs, the gold standard experimental latency model. Unlike the THP-1 cell line model, primary cells produce quantifiable numbers of viral progeny during HCMV reactivation (35–37). Pure populations of infected CD34+ HPCs were isolated and cultured for 10 d. The frequency of infectious centers was quantified following reactivation and compared to the frequency present in a lysate prepared at the time of reactivation (prereactivation control). We observed a significant decrease in the amount of infectious virus produced following reactivation of the recombinant virus as compared to wild-type virus (Fig. 6A). The reduced frequency of reactivation for FOXOmut123 virus cannot be attributed to a defect in maintenance of the latent infection, as recombinant genomes are present in similar, or even higher numbers, when compared to wild-type genomes on day 10 of the latency coculture (Fig. 6B). Together these data show that while dispensable for lytic replication, FOXO TF binding sites in the intronic MIE promoters play critical roles in HCMV reactivation.
Fig. 6.
FOXO binding sites are necessary for efficient HCMV reactivation in CD34+ HPCs. CD34+ HPCs were infected with TB40/E WT or FOXOmut123 at a MOI of 2. Pure populations of infected (GFP+) CD34+ HPCs were isolated by FACS and seeded into long-term bone marrow cultures. (A) At 10 dpi, viable CD34+ HPCs (reactivation) or an equivalent cell lysate (prereactivation control) were seeded by limiting dilution onto fibroblast monolayers in a cytokine-rich media to promote myeloid differentiation. Infectious centers (GFP+ fibroblasts) were scored 14 d later and are expressed as frequency of infectious centers. Error bars represent SEM for three independent biological replicates. Statistical significance was determined using two-way analysis of variance (ANOVA) with repeated measures by both factors (wild type vs. mutant and prereactivation vs. reactivation where *P ≤ 0.05). (B) Total DNA was isolated at 10 dpi and viral genomes were quantified relative to cellular DNA by qPCR using primers to the β2.7 region of the HCMV genome and the cellular gene RNase-P. Data from two independent biological replicates are shown; mean is depicted.
Discussion
Contrary to the long-held paradigm that viral MIE gene reexpression during reactivation requires the MIEP, we recently discovered two promoters in the MIE locus, iP1 and iP2 (9), that are necessary for IE gene reexpression and HCMV reactivation (10). Here we begin to unravel the regulatory mechanisms controlling iP1 and iP2 activity by showing that the FOXO transcription factors are critical positive regulators of MIE gene expression from iP1 and iP2 in hematopoietic cells. As FOXO transcription factors also drive cellular differentiation, our results provide a mechanism by which HCMV senses and responds to changes in cellular differentiation to regulate reactivation.
Both FOXO1 and FOXO3a can activate the intronic promoters (Fig. 2); however, our results suggest that FOXO3a is the critical FOXO transcription factor required for reactivation. While FOXO1 increased intronic promoter activity from a luciferase reporter (Fig. 2B), FOXO1 did not significantly impact IE1 and IE2 protein expression in the context of the more complex MIE genomic locus (Fig. 3A). In the context of latent infection, both FOXO1 and FOXO3a are induced above the basal expression levels present in uninfected cells. However, only FOXO3a is induced significantly following TPA-induced reactivation, which mirrors with the reexpression of the HCMV major immediate early genes from the intronic promoters (SI Appendix, Fig. S3). While FOXO1 and FOXO3a are both expressed in hematopoietic cells, FOXO3a more efficiently localizes to the nucleus in myeloid progenitor cells, particularly during cellular stresses such as those that induce reactivation (38, 39). Further, FOXO3a regulates monocyte-to-macrophage differentiation (29, 40), while FOXO1 promotes maintenance of stemness in CD34+ HPCs (29). FOXO3a also directly binds specific sequences in the intronic promoters (Fig. 4) and strongly increases their activity (Fig. 2). While additional studies are needed to more fully elucidate the role of specific FOXO TFs in HCMV biology, these data suggest that FOXO3a is a particularly crucial positive regulator of viral lytic gene expression in the contexts of HCMV latency and reactivation.
Our data also provide new insight into the roles of intronic promoters in the context of latency and reactivation. While deleting either iP1 or iP2 decreases reactivation, transcripts arising from the iP2 promoter were more abundant upon reactivation (10). Our data show that FOXO binding sites in the intronic promoters are critical for efficient reactivation (Fig. 6A), suggesting that activation of iP1 and iP2 by FOXO TFs is critical for MIE gene expression during reactivation. The effect of FOXO binding site mutation on both iP1 and iP2 activity further suggests that transcription from these elements is coordinated.
Our data also show that FOXO binding sites in iP1 and iP2 are critical for the early burst of MIE gene expression immediately after infection of THP-1 cells (Fig. 5), consistent with the requirement for iP1 and iP2 for efficient MIE gene expression early after infection of THP-1 cells (10). Going forward, it will be important to understand how this early burst of MIE gene expression impacts latent infection and reactivation, and how FOXO TFs regulate MIE expression during different stages of infection. It is intriguing that mutation of the FOXO binding sites in the intronic promoter region also leads to a statistically significant reduction in the accumulation of MIEP-derived transcripts at 1 dpi, but not during reactivation induced by TPA treatment (Fig. 5E). These data suggest that the MIEP, iP1, and iP2 share common regulatory elements, and that the activity of these promoters may be coordinated to ensure proper contextual and temporal regulation of MIE gene expression. Further support for this idea comes from recent studies showing that host TF binding sites associated with the MIEP enhancer can also impact the activity of the intronic promoters (34, 41).
While we define a role for FOXO TFs in reactivation, our data also suggest additional factors regulate intronic promoter activity and HCMV reactivation, as mutation of all three FOXO binding sites in the intronic promoter region did not result in a complete loss of reactivation, highlighting the complexity of the MIE locus. Several transcription factors are implicated in reactivation (7, 8, 34, 42, 43) suggesting they may also regulate intronic promoter activity. For example, AP1 transcription factor binding sites in the MIE enhancer are required for efficient IE gene reexpression and intronic promoter activation during reactivation (44). Further studies are needed to identify additional transcription factors that regulate the intronic promoters in conjunction with FOXO TFs to control HCMV reactivation.
Supplementary Material
Acknowledgments
We thank Jim Alwine, John Purdy, Stan Lemon, Mark Heise, Ralph Baric, Blossom Damania, and members of the N.J.M., F.G., and J.P.K. laboratories for helpful conversations and critical feedback. This work was supported by NIH grants AI143191 to N.J.M., J.P.K. and F.G., AI127335 to F.G., AI123811 and AI103311 to N.J.M., and support from the University of North Carolina at Chapel Hill Virology Training Grant (T32 AI07419 to A.E.H.). D.C.-M. is supported by a postdoctoral fellowship (18POST33960140) from the American Heart Association.
Footnotes
Competing interest statement: J.P.K., F.G., and N.J.M. have filed a patent for manipulation of intronic promoters to reduce reactivation of viral vaccines.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002651117/-/DCSupplemental.
Data Availability.
Data and associated protocols for this work are provided herein or in SI Appendix. Unique biochemical reagents are available by contacting the corresponding author.
References
- 1.Nogalski M. T., Collins-McMillen D., Yurochko A. D., Overview of human cytomegalovirus pathogenesis. Methods Mol. Biol. 1119, 15–28 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Shelbourn S. L., Kothari S. K., Sissons J. G., Sinclair J. H., Repression of human cytomegalovirus gene expression associated with a novel immediate early regulatory region binding factor. Nucleic Acids Res. 17, 9165–9171 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinclair J. H., Baillie J., Bryant L. A., Taylor-Wiedeman J. A., Sissons J. G., Repression of human cytomegalovirus major immediate early gene expression in a monocytic cell line. J. Gen. Virol. 73, 433–435 (1992). [DOI] [PubMed] [Google Scholar]
- 4.Söderberg-Nauclér C. et al., Reactivation of latent human cytomegalovirus in CD14(+) monocytes is differentiation dependent. J. Virol. 75, 7543–7554 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Connor C. M., Murphy E. A., A myeloid progenitor cell line capable of supporting human cytomegalovirus latency and reactivation, resulting in infectious progeny. J. Virol. 86, 9854–9865 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keller M. J. et al., Reversal of human cytomegalovirus major immediate-early enhancer/promoter silencing in quiescently infected cells via the cyclic AMP signaling pathway. J. Virol. 81, 6669–6681 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan J. et al., Breaking human cytomegalovirus major immediate-early gene silence by vasoactive intestinal peptide stimulation of the protein kinase A-CREB-TORC2 signaling cascade in human pluripotent embryonal NTera2 cells. J. Virol. 83, 6391–6403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X. et al., Phorbol ester-induced human cytomegalovirus major immediate-early (MIE) enhancer activation through PKC-delta, CREB, and NF-kappaB desilences MIE gene expression in quiescently infected human pluripotent NTera2 cells. J. Virol. 84, 8495–8508 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arend K. C., Ziehr B., Vincent H. A., Moorman N. J., Multiple transcripts encode full-length human cytomegalovirus IE1 and IE2 proteins during lytic infection. J. Virol. 90, 8855–8865 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins-McMillen D. et al., Alternative promoters drive human cytomegalovirus reactivation from latency. Proc. Natl. Acad. Sci. U.S.A. 116, 17492–17497 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Umashankar M. et al., A novel human cytomegalovirus locus modulates cell type-specific outcomes of infection. PLoS Pathog. 7, e1002444 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warming S., Costantino N., Court D. L., Jenkins N. A., Copeland N. G., Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33, e36 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caviness K. et al., Complex interplay of the UL136 isoforms balances cytomegalovirus replication and latency. MBio 7, e01986 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Umashankar M. et al., Antagonistic determinants controlling replicative and latent states of human cytomegalovirus infection. J. Virol. 88, 5987–6002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinzger C. et al., Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J. Gen. Virol. 89, 359–368 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Lenarcic E. M., Ziehr B., De Leon G., Mitchell D., Moorman N. J., Differential role for host translation factors in host and viral protein synthesis during human cytomegalovirus infection. J. Virol. 88, 1473–1483 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziehr B. et al., Human cytomegalovirus TRS1 protein associates with the 7-methylguanosine mRNA cap and facilitates translation. Proteomics 15, 1983–1994 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vincent H. A., Ziehr B., Moorman N. J., Mechanism of protein kinase R inhibition by human cytomegalovirus pTRS1. J. Virol. 91, e01574-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grant C. E., Bailey T. L., Noble W. S., FIMO: Scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buehler J. et al., Host signaling and EGR1 transcriptional control of human cytomegalovirus replication and latency. PLoS Pathog. 15, e1008037 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buehler J. et al., Opposing regulation of the EGF receptor: A molecular switch controlling cytomegalovirus latency and replication. PLoS Pathog. 12, e1005655 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Q. F. et al., Reprogramming of histone methylation controls the differentiation of monocytes into macrophages. FEBS J. 284, 1309–1323 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Uhlén M. et al., Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Miyamoto K. et al., Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell 1, 101–112 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Liang R., Ghaffari S., Stem cells seen through the FOXO lens: An evolving paradigm. Curr. Top. Dev. Biol. 127, 23–47 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Stenberg R. M. et al., Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J. Virol. 64, 1556–1565 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stenberg R. M., Stinski M. F., Autoregulation of the human cytomegalovirus major immediate-early gene. J. Virol. 56, 676–682 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Depto A. S., Stenberg R. M., Regulated expression of the human cytomegalovirus pp65 gene: Octamer sequence in the promoter is required for activation by viral gene products. J. Virol. 63, 1232–1238 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tothova Z., Gilliland D. G., FoxO transcription factors and stem cell homeostasis: Insights from the hematopoietic system. Cell Stem Cell 1, 140–152 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Yee L. F., Lin P. L., Stinski M. F., Ectopic expression of HCMV IE72 and IE86 proteins is sufficient to induce early gene expression but not production of infectious virus in undifferentiated promonocytic THP-1 cells. Virology 363, 174–188 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Weinshenker B. G., Wilton S., Rice G. P., Phorbol ester-induced differentiation permits productive human cytomegalovirus infection in a monocytic cell line. J. Immunol. 140, 1625–1631 (1988). [PubMed] [Google Scholar]
- 32.Lee C. H. et al., Factors affecting human cytomegalovirus gene expression in human monocyte cell lines. Mol. Cells 9, 37–44 (1999). [PubMed] [Google Scholar]
- 33.Arcangeletti M. C. et al., Human cytomegalovirus reactivation from latency: Validation of a “switch” model in vitro. Virol. J. 13, 179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kew V. G., Yuan J., Meier J., Reeves M. B., Mitogen and stress activated kinases act co-operatively with CREB during the induction of human cytomegalovirus immediate-early gene expression from latency. PLoS Pathog. 10, e1004195 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Umashankar M., Goodrum F., Hematopoietic long-term culture (hLTC) for human cytomegalovirus latency and reactivation. Methods Mol. Biol. 1119, 99–112 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Goodrum F., Jordan C. T., Terhune S. S., High K., Shenk T., Differential outcomes of human cytomegalovirus infection in primitive hematopoietic cell subpopulations. Blood 104, 687–695 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Goodrum F. D., Jordan C. T., High K., Shenk T., Human cytomegalovirus gene expression during infection of primary hematopoietic progenitor cells: A model for latency. Proc. Natl. Acad. Sci. U.S.A. 99, 16255–16260 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang R., Rimmelé P., Bigarella C. L., Yalcin S., Ghaffari S., Evidence for AKT-independent regulation of FOXO1 and FOXO3 in haematopoietic stem and progenitor cells. Cell Cycle 15, 861–867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yalcin S. et al., Foxo3 is essential for the regulation of ataxia telangiectasia mutated and oxidative stress-mediated homeostasis of hematopoietic stem cells. J. Biol. Chem. 283, 25692–25705 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Paik J. H. et al., FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 128, 309–323 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collins-McMillen D., Buehler J., Peppenelli M., Goodrum F., Molecular determinants and the regulation of human cytomegalovirus latency and reactivation. Viruses 10, 444 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan J., Li M., Torres Y. R., Galle C. S., Meier J. L., Differentiation-coupled induction of human cytomegalovirus replication by union of the major enhancer retinoic acid, cyclic AMP, and NF-κB response elements. J. Virol. 89, 12284–12298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dupont L. et al., Src family kinase activity drives cytomegalovirus reactivation by recruiting MOZ histone acetyltransferase activity to the viral promoter. J. Biol. Chem. 294, 12901–12910 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krishna B. A., Wass A. B., O’Connor C. M., Activator protein-1 transactivation of the major immediate early locus is a determinant of cytomegalovirus reactivation from latency. bioRxiv:10.1101/2020.02.06.929281 (07 May 2020). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and associated protocols for this work are provided herein or in SI Appendix. Unique biochemical reagents are available by contacting the corresponding author.