Abstract
The entire spectrum of diagnostic testing, from reagent supply to test performance, has been a major focus during the coronavirus disease 2019 (COVID-19) pandemic. The hope for serologic testing is that it will provide both epidemiologic information about seroprevalence as well as individual information about previous infection. This information is particularly helpful for high-risk individuals who may be outside of the viral shedding window, such as children with suspected multisystem inflammatory syndrome. It is not yet understood whether serologic testing can be interpreted in terms of protective immunity. These concerns must be addressed using highly sensitive and specific tests.
Keywords: SARS-CoV-2, COVID-19, COVID-19 serology
Key points
-
•
Early in the coronavirus disease 2019 (COVID-19) pandemic, the release of poorly characterized antibody tests caused concern about the quality of serologic results and a national discussion about test performance.
-
•
The positive predictive value of COVID-19 serologic tests varies with seroprevalence and is a major concern.
-
•
The kinetics of the antibody response to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are characterized by the appearance of immunoglobulin (Ig) G in most individuals by at least 2 weeks after symptom onset, slightly after IgM and IgA.
-
•
There is continued uncertainty about the significance of antibody tests in terms of the degree and durability of immunity.
-
•
The differential and quantitative detection of viral antigens may prove to be important and will require the development of test platforms to answer these nuanced questions.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic began in December 2019 with several cases of a pneumonia of unknown cause in Wuhan, China.1 The causative agent was quickly identified using molecular techniques as the Betacoronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).2 This discovery was followed by a global effort to develop accurate molecular diagnostics at tremendous scale and pace in order to diagnose acute infection and contain spread of the virus. Questions about serologic testing followed soon after.
Early in the pandemic, lack of regulatory restrictions on serologic testing led to a wave of more than 40 antibody tests to detect immunoglobulin (Ig) G and/or IgM binding to SARS-CoV-2 spike (S) and nucleocapsid (NP) proteins.3, 4, 5, 6, 7, 8, 9 Many of these tests were poorly characterized, which caused concern about the quality of serologic data. Several discussions about so-called immunity passports caused further concern about the use of serologic data.10 In response, the US Food and Drug Administration (FDA) moved to tighten regulation of new commercial serologic tests; more than 30 were subsequently withdrawn from distribution.11 These events have highlighted the importance of rigorous testing evaluation and scope of use. At the same time, tremendous effort has been focused on understanding the immune response to SARS-CoV-2, both from data coming out of the hardest-hit regions and from previous knowledge gleaned from the study of SARS-CoV-1, Middle East respiratory syndrome (MERS), and circulating human coronaviruses (HCoVs). This knowledge has helped to refine the questions of who to test for antibodies to SARS-CoV-2 and for what purpose they should be tested.12
This article reviews the basis of antibody-mediated immunity to SARS-CoV-2 and other coronaviruses (CoVs), with a focus on kinetics and correlates of protection. It then discusses currently available testing options for SARS-CoV-2 antibodies in the context of the rapidly evolving knowledge of disease immunopathogenesis, and how testing may be used to inform a diverse and complicated set of questions.
Viral antigens
SARS-CoV-2 is a member of the family Coronaviridae, which consists of 2 genera that infect mammals: Alphacoronavirus and Betacoronavirus.13 Strains that are relevant to human infections include the 4 circulating seasonal coronaviruses: 229E and NL63 (both Alphacoronavirus), and OC43, HKU1, MERS-CoV, and SARS-CoV-1 (all Betacoronavirus).13 All of the CoVs are enveloped RNA viruses with genomes in the range of 30 kilobases (Fig. 1 ). The first portion of the genome consists of open reading frames (ORFs) 1a and 1b, encoding the replicase-transcriptase polyprotein (pp1ab), followed by 4 structural proteins: S, NP, envelope (E), and membrane (M). Studies on SARS-CoV-1 indicated that the structural NP and S proteins are the dominant antigens for host immune responses to SARS-CoV-2.14 , 15
Fig. 1.
The SARS-CoV-2 genome: SARS-CoV-2 isolate Wuhan-Hu-1, complete genome (NC_045512). Genes encoding nonstructural proteins are shown in gray. Genes encoding structural proteins S, E, M, and NP are shown in blue. ORF, open reading frame.
The mature S protein is a ∼180 kDa glycosylated homotrimer that protrudes from the viral surface, giving the characteristic halo appearance for which CoVs are named.16 The extracellular region is organized into the S1 and S2 domains. S1 comprises the outermost region, contains the receptor-binding domain (RBD) for the target human ACE2 receptor, and initiates host cell entry.17, 18, 19, 20 Studies of SARS-CoV-1 show that receptor binding and proteolytic cleavage at the S1/S2 junction triggers a conformational change in S2 that mediates entry via the membrane fusion peptide sequence.21 The S protein is moderately conserved among members of the Betacoronaviridae, particularly the S2 region proximal to the viral surface (Fig. 2 ).22 The S1 region, including the RBD, is less conserved and of great interest as a target for immunoassays because of its prime role in interaction with the human host.22 Several studies have shown that anti-S antibodies can neutralize virus in cell culture, and efforts toward vaccine development are heavily focused on this protein.16 , 17 , 23, 24, 25
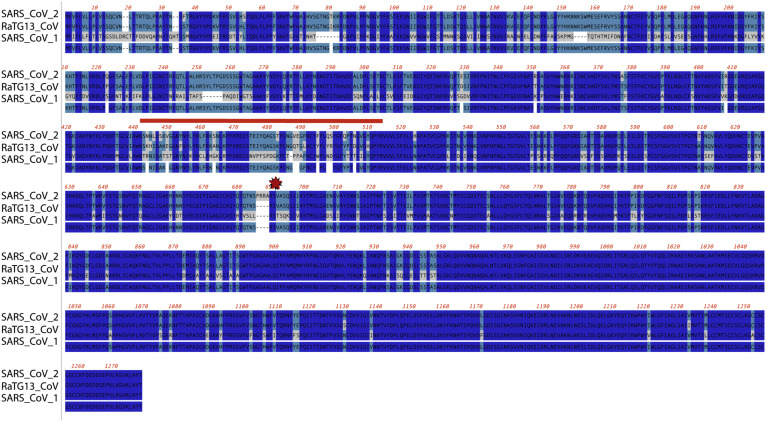
Fig. 2.
Alignment of the S protein among closely related betacoronaviruses: peptide sequences from SARS-CoV-2 (NCBI [National Center for Biotechnology Information] YP_009724390), RaTG13-CoV (NCBI QHR63300), and SARS-CoV-1 (NCBI BAE93401) were aligned using ClustalW. Conserved residues (3/3) are shown in dark blue, (2/3) in teal, (1/3) in gray. The S1/S2 cleavage site is indicated by a red star. The receptor-binding motif is designated by the red line.
Functional and biochemical studies on the ∼50-kDa SARS-CoV-1 and SARS-CoV-2 NP proteins show roles in replication, transcription, and packaging of the genome26 , 27 (Fig. 3 ). The high abundance and antigenicity of NP have made it a focus of both diagnostic and vaccine work for SARS-CoV-1 as well as SARS-CoV-2.28, 29, 30 NP sequences show 99% identity with related bat CoVs (RaTG13) and ∼90% identity with SARS-CoV-1.31
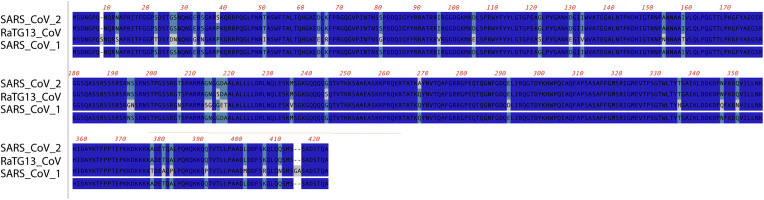
Fig. 3.
Alignment of the NP protein among closely related betacoronaviruses: peptide sequences from SARS-CoV-2 (NCBI YP_009724390), RaTG13-CoV (NCBI QHR63300), and SARS-CoV-1 (NCBI BAE93401) were aligned using ClustalW. Conserved residues (3/3) are shown in dark blue, (2/3) in teal, (1/3) in gray.
These sequence homologies have implications for antibody testing. By sequence analysis, the normal circulating CoV strain OC43 has moderate homology with SARS-CoV-2 in both the NP and S proteins (Fig. 4 ). Sequence similarities can translate into cross-strain antibody binding, decreasing test specificity when used to determine infection prevalence. In contrast, increased specificity may occur from strain differences on surface sequences that are likely targets of antibody-mediated responses.
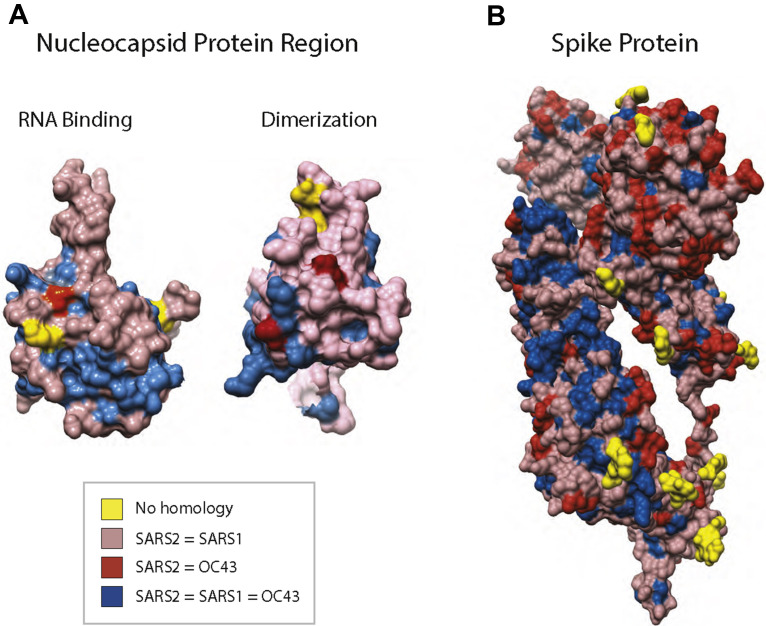
Fig. 4.
Conservation of surface amino acids between SARS-CoV-2, SARS-CoV-1, and CoV-OC43: (A) space-filling model of the NP protein RNA-binding region (PDB [Protein Data Bank] 6M3M) and dimerization region (PDB 2GIB) from SARS-CoV-1 with sequence conservation mapped on the surface projection from ClustalW alignments between SARS-CoV-1 (NCBI BAE93401), SARS-CoV-2 (NCBI YP_009724390), and CoV-OC43 (NCBI YP_009555245). (B) Space-filling model of the full-length SARS-CoV-2 S protein monomer (PDB CVYB) with sequence conservation mapped on the surface projection from ClustalW alignments from SARS-CoV-2 (NCBI YP_009724390), SARS-CoV-1 (NCBI BAE93401), and CoV-OC43 (NCBI YP_009555241).
Antibody kinetics
Using a variety of technologies and antigens, the kinetics of the antibody response to SARS-CoV-2 are being explored. The earliest information came from several groups in China who characterized the serologic responses of patients at the beginning of the pandemic using enzyme-linked immunosorbent assays (ELISAs) based either exclusively on the detection of NP alone or on NP in conjunction with the RBD domain of the S protein. These 2 antigens performed equivalently as assay targets, with antibodies becoming detectable in some individuals within the first week of symptom onset.32, 33, 34, 35, 36
There are conflicting reports regarding the disease course kinetics of anti-NP and anti-S antibody detection, and thus the relative sensitivity of serologic tests. Some reports have shown detection of anti-NP slightly earlier than anti-S antibodies, whereas others have shown the contrary, possibly because of differences in assay format.36 , 37 Similarly, human SARS-CoV-1 anti-NP antibodies were detectable by serologic assays slightly earlier than anti-S Ig.38 , 39 Additional studies have suggested that assays using more restricted epitopes, namely S1 or RBD, may be more specific than those using the full S protein.32, 33, 34 , 36 , 40 Importantly, although antibody responses may be detectable in the first week after symptom onset, a full 2 to 3 weeks is required for a robust response.9 , 36 , 41 Over the course of infection, IgM and IgA appeared earliest, often within 5 days of symptom onset, although IgG appeared in close succession.32, 33, 34 , 36 , 40
With respect to the kinetics of viral RNA (the viral shedding window), antibodies seem to become detectable as the viral load diminishes.33, 34, 35 However, patients with both mild and severe clinical presentations may generate a detectable antibody response before viral clearance.34 , 35 , 42 In contrast, there have been reports that asymptomatic (reverse transcriptase polymerase chain reaction confirmed) individuals may show late onset or even no seroconversion.35
Test validation
Much of the controversy surrounding SARS-CoV-2 serologic testing has centered on fundamental aspects of test validation, and this was also true for SARS-CoV-1.43 , 44 SARS-CoV-2 antibody detection tests have used several platforms, including ELISAs, chemiluminescent, lateral flow, and multiplex methods. All have previously been used to detect antibodies to other viral pathogens.45 In general, these methods involve solid phase coupling of recombinant S or NP as fully trimerized (S protein), monomeric (NP), or peptide fragments (S1-RBD, linear peptide fragments). The solid phase coupled proteins are incubated with serum to allow immunoglobulin binding, which is detected by a secondary anti-IgG/IgA/IgM reagent. Assay readouts include serum dilution titers, colorimetric absorbance, changes in surface reflectivity, or fluorescence intensity.
Assay validation requires a set of known positive and negative serum samples. Archived samples from the pre-COVID-19 era (eg, pre-2020) can be used for a negative gold standard. There is particular interest in determining the false-positive rate because the presence of antibodies will be interpreted as proof of prior infection. Across multiple studies, specificity of ELISA-format assays has been reported to be between 95% and 100%.9 , 32, 33, 34 , 36 , 41 For studies that include samples from individuals with known HCoV infections (HKU1, NL63, OC43, or 229E), SARS-CoV-1, or MERS-CoV, cross-reactivity was generally found to be low, with the exception of sera taken from individuals infected with SARS-CoV-1. Similarly, several clinical conditions are associated with broadly cross-reactive antibodies, including acute respiratory infections, autoimmune diseases (eg, lupus, rheumatoid arthritis), and other infections (eg, syphilis, Lyme disease). Including such serologic samples from the pre-COVID-19 era is critical for test specificity validation. Defining a known positive gold standard poses some challenges. Given the postinfection kinetics of anti-S and anti-NP antibody, it seems prudent to choose positive validation sera from patients taken at least 2 weeks after SARS-CoV-2 infection confirmed by a nucleic acid test.
For any clinical assay, the trade-off between specificity and sensitivity is a key factor in how it will be ultimately used. For example, if a positive value is to be used as a surrogate marker for infection, with potential translation for immunity, minimizing the false-positive rate is critical. In contrast, other considerations are a focus for a test that is used to monitor antibody kinetics after infection, in vaccine clinical trials, or to monitor convalescent plasma or monoclonal antibody therapy. Here, binary (positive or negative) or titer (ranked categorical) test results are less useful than a continuous readout (ie, absolute antibody concentration; eg, nanograms per milliliter), especially when performing statistical comparisons of vaccine efficacy in clinical trials46 or determining who should donate convalescent plasma. Identifying antibody subsets poses another challenge for test validation. The acceptable threshold for anti-S1/RBD antibodies may need to be different for convalescent plasma donor screening than for identifying postinfectious immunity.
Current clinical testing options
Serologic testing for SARS-CoV-2 infection has become available in the United States over the first few months of the pandemic in 2 main formats: point of care and clinical laboratory. Several companies produced rapid, lateral flow–type devices with readouts of total antibody or separate IgM and IgG. Whitman and colleagues47 performed an assessment of many of these and found widely varying performance: between 81.8% and 100% sensitivity at more than 20 days after symptom onset and 84.3% to 100% specificity. Gradually, established manufacturers have begun releasing product for testing in high-complexity clinical laboratories. At the time of writing, 13 assays by 11 different manufacturers have been granted Emergency Use Authorization (EUA) by the FDA, and these represent a variety of targets and technologies (Table 1 ).48
Table 1.
Performance of severe acute respiratory syndrome coronavirus 2 immunoassays with Food and Drug Administration Emergency Use Authorizationa
| Manufacturer | Antigen | Ab Class | Format | Sensitivity (%) | Specificity (%) | Platform |
|---|---|---|---|---|---|---|
| Abbott | NP | IgG | CLIA | 100.0 | 99–99.6 | Architect/Alinity |
| Roche | NP | IgG | ECLIA | 100.0 | 99.8 | Elecsys |
| Ortho | S | IgG, Tot Ab | CLIA | 90–100 | 100.0 | Vitros |
| Diasorin | S1/S2 | IgG | CMIA | 97.6 | 99.3 | Liaison XL |
| Euroimmun | S1 | IgG | ELISA | 90.0 | 97.8–100 | None |
| Wadsworth | NP | Tot Ab | MIA | 88.0 | 98.8 | FlexMap |
| Mt Sinai | RBD and S | IgG | 2-step ELISA | 92.5 | 100.0 | None |
| Cellex | NP and S | IgG and IgM | LFA | 93.8b | 96.0b | None |
| Bio-Rad | NP | Tot Ab | ELISA | 92.2 | 99.6 | None |
| Autobio | S | IgG and IgM | LFA | 99b | 99.0b | None |
Abbreviations: Ab, antibody; CLIA, chemiluminescent immunoassay; CMIA, chemiluminescent microparticle immunoassay; ECLIA, electrochemiluminescence immunoassay; LFA, lateral flow assay; MIA, microsphere immunofluorescence assay; Tot, total.
Performance was assessed using EUA data and FDA assessment as described (https://www.fda.gov/medical-devices/emergency-situations-medical-devices/eua-authorized-serology-test-performance).
Combined IgM/IgG performance.
NP-based assays include chemiluminescent immunoassay (CLIA) and electrochemiluminescence immunoassay technologies offered by Abbott and Roche, respectively. Both of these assays measure IgG and seem to be highly specific and sensitive. The Abbott platform has been evaluated independently in a large cohort in Idaho with confirmation of its performance: 99.90% specificity and 100% sensitivity 17 days after symptom onset.3 Additional technologies with EUA based on detection of NP include a microsphere immunoassay developed by the New York State Department of Health at the Wadsworth. S and S derivative (S1 and RBD)–based assays include a CLIA assay developed by Ortho-Clinical Diagnostics and ELISAs from Euroimmun and Mount Sinai Hospital.9 False-positive results have been noted for sera from patients infected with HCoV-OC43 in the Euroimmun assay.41
Implications for immunity
Some questions where antibody testing can provide clarity include seroprevalence and, on an individual level, whether a person has previously been infected with SARS-CoV-2.49 This assessment can be straightforward, such as when an individual had clinically suggestive symptoms weeks prior but could not, or did not, get a molecular test. Beyond simply assessing previous infection, there is considerable interest in interpreting serologic test results as correlates of protection against future infection (Box 1 ). When using serologic testing to answer such questions, it is important to consider (1) the durability of the immune response, (2) the neutralization potential of antibodies, and (3) the translation of these findings into an assessment of functional in vivo immunity. Critically, there currently are no data regarding the association between the presence and titers of anti–SARS-CoV-2-S or anti-–SARS-CoV-2-NP antibodies and protection from reinfection.
Box 1. Questions for severe acute respiratory syndrome coronavirus 2 antibody testing.
Has the individual been infected (if the molecular result is negative and the patient is likely outside of the window of viral shedding)?
Does the pediatric patient have multisystem inflammatory syndrome?
Are the infection prevention policies and personal protective equipment guidelines adequate for protecting health care workers in the institution?
Can a recovered COVID-19 patient donate plasma for therapeutic use? What is the half-life of anti–SARS-CoV-2 antibodies in a recipient of convalescent plasma?
Does a given serologic response indicate a successful trial vaccine?
Has the individual mounted an adequate response to a vaccine (once one becomes available)?
The COVID-19 pandemic is still in its early stages, so there has not been the opportunity to assess the longevity of the antibody response, although small cohort studies show that IgG antibodies are detectable for at least 6 weeks after symptom onset,35 , 40 whereas IgM is diminished within the first month postinfection.36 , 40 A recent study of IgG levels in asymptomatic versus symptomatic individuals found that neutralizing titers were more likely to diminish in the early convalescent phase in those without a history of symptoms.50 Furthermore, the same study showed that 40.0% of asymptomatic individuals became seronegative in the convalescent phase versus only 12.9% of those who were symptomatic.50 Previous reports indicate that individuals infected with SARS-CoV-1 have detectable IgG antibodies for at least 8 to 24 months after symptom onset, whereas IgM and IgA were markedly decreased.51, 52, 53 In contrast, patients infected with HCoV 229E have markedly diminished antibody levels 1 year postinfection.54 Such results may complicate future interpretation of serologic test results, especially if SARS-CoV-2 infection becomes endemic and seasonal.
Plaque reduction neutralization tests can assess whether infected patients mount a neutralizing antibody response to the virus. Serum or plasma from individuals infected with SARS-CoV-1 or SARS-CoV-2 has been shown in several studies to neutralize viral infectivity in vitro.24 , 41 , 55, 56, 57 Several purified neutralizing antibodies have been characterized against epitopes of both SARS-CoV-1 and SARS-CoV-2, most of which are directed against the RBD of the S protein.14 , 16 , 23, 24, 25 , 58 It remains to be seen how these findings can be translated in vivo to natural immunity or therapeutics.
Neutralizing antibodies are the basis for convalescent plasma therapy, which showed promise in treating patients with SARS-CoV-1 and is currently being tested for those with SARS-CoV-2. Monoclonal antibodies directed at the S1-RBD and other antigenic sites are also in development. Both classes of therapies pose opportunities and challenges for clinical serologic testing. Convalescent plasma donors will need to be screened for high anti–SARS-CoV-2 protective titers, although it is not known which antibodies confer protection. Similarly, such tests may be used to monitor postinfusion antibody levels during convalescent plasma treatment, monoclonal antibody therapies, or after vaccination. Further complicating this issue is evidence from SARS-CoV-1 that antibody-dependent enhancement (ADE) of infection may occur, where antibodies facilitate, rather than block, viral infection.59 Current serologic tests do not distinguish between protective and ADE-inducing antibodies. Such uses of a clinical test will need to be carefully monitored to assure that anti–SARS-CoV-2 antibody tests are suited to these goals.
Summary
Serologic testing is an evolving and complex area of testing for COVID-19. Challenges include test validation and performance, usage, and interpretation across multiple contexts. As knowledge accumulates about the significance of differential and quantitative detection of viral antigens, it will be beneficial to have test platforms that can address questions going beyond a qualitative assessment of previous infection. Careful test validation and appropriate matching to the questions to be answered will help ensure clinical and scientific research rigor and reproducibility. It is likely that the testing landscape for measuring the serologic response to SARS-CoV-2 will evolve rapidly over the next several years. Furthermore, if it is determined that neutralizing antibodies do not confer protection (which may be known within 6–12 months), serologic testing for SARS-CoV-2 may be of purely epidemiologic value and thus less likely to occur in clinical laboratories.
Acknowledgments
Disclosure
Nicole Pecora has received research support from Luminex.
Footnotes
Funding: This work was supported by the National Institutes of Health Institute of Allergy, Immunology and Infectious Diseases grants R01 AI129518 and R21 AI138500 (M.S. Zand), and the University of Rochester Clinical and Translational Science Award UL1 TR002001 from the National Center for Advancing Translational Sciences of the National Institutes of Health (M.S. Zand). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. None of the mentioned funders had any role in study design, data collection and analysis, decision to publish, or preparation of the article.
References
- 1.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryan A., Pepper G., Wener M.H., et al. Performance characteristics of the Abbott Architect SARS-CoV-2 IgG assay and seroprevalence in Boise, Idaho. J Clin Microbiol. 2020;58(8):e00941-20. doi: 10.1128/JCM.00941-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffman T., Nissen K., Krambrich J., et al. Evaluation of a COVID-19 IgM and IgG rapid test; an efficient tool for assessment of past exposure to SARS-CoV-2. Infect Ecol Epidemiol. 2020;10(1):1754538. doi: 10.1080/20008686.2020.1754538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zainol Rashid Z., Othman S.N., Abdul Samat M.N., et al. Diagnostic performance of COVID-19 serology assays. Malays J Pathol. 2020;42(1):13–21. [PubMed] [Google Scholar]
- 6.Infantino M., Grossi V., Lari B., et al. Diagnostic accuracy of an automated chemiluminescent immunoassay for anti-SARS-CoV-2 IgM and IgG antibodies: an Italian experience. J Med Virol. 2020 doi: 10.1002/jmv.25932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takita M., Matsumura T., Yamamoto K., et al. Challenges of community point-of-care antibody testing for COVID-19 herd-immunity in Japan. QJM. 2020 doi: 10.1093/qjmed/hcaa182. hcaa182. PMCID: 7313848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan Y., Chang L., Wang L. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): Current status, challenges, and countermeasures. Rev Med Virol. 2020;30(3):e2106. doi: 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amanat F., Stadlbauer D., Strohmeier S., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26(7):1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phelan A.L. COVID-19 immunity passports and vaccination certificates: scientific, equitable, and legal challenges. Lancet. 2020;395(10237):1595–1598. doi: 10.1016/S0140-6736(20)31034-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Administration USFaD What tests should no longer be distributed for COVID-19? 2020. https://www.fda.gov/medical-devices/emergency-situations-medical-devices/faqs-testing-sars-cov-2 - nolonger Available at: Accessed May 28, 2020.
- 12.Abbasi J. The promise and Peril of antibody testing for COVID-19. JAMA. 2020;323(19):1881–1883. doi: 10.1001/jama.2020.6170. [DOI] [PubMed] [Google Scholar]
- 13.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu M., Shi Y., Guo Z., et al. Antibody responses to individual proteins of SARS coronavirus and their neutralization activities. Microbes Infect. 2005;7(5–6):882–889. doi: 10.1016/j.micinf.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tay M.Z., Poh C.M., Renia L., et al. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du L., He Y., Zhou Y., et al. The spike protein of SARS-CoV--a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7(3):226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walls A.C., Park Y.J., Tortorici M.A., et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shang J., Ye G., Shi K., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wrapp D., Wang N., Corbett K.S., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q., Zhang Y., Wu L., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Millet J.K., Whittaker G.R. Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology. 2018;517:3–8. doi: 10.1016/j.virol.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaimes J.A., Andre N.M., Chappie J.S., et al. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J Mol Biol. 2020;432(10):3309–3325. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y., Wang F., Shen C., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368(6496):1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tai W., Zhang X., He Y., et al. Identification of SARS-CoV RBD-targeting monoclonal antibodies with cross-reactive or neutralizing activity against SARS-CoV-2. Antiviral Res. 2020;179:104820. doi: 10.1016/j.antiviral.2020.104820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wrapp D., De Vlieger D., Corbett K.S., et al. Structural basis for potent neutralization of Betacoronaviruses by single-domain Camelid antibodies. Cell. 2020;181(5):1004–1015.e15. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Surjit M., Lal S.K. The SARS-CoV nucleocapsid protein: a protein with multifarious activities. Infect Genet Evol. 2008;8(4):397–405. doi: 10.1016/j.meegid.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng W., Liu G., Ma H., et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem Biophys Res Commun. 2020;527(3):618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Che X.Y., Hao W., Wang Y., et al. Nucleocapsid protein as early diagnostic marker for SARS. Emerg Infect Dis. 2004;10(11):1947–1949. doi: 10.3201/eid1011.040516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12(3) doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S.J., Leng C.H., Lien S.P., et al. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine. 2006;24(16):3100–3108. doi: 10.1016/j.vaccine.2006.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tilocca B., Soggiu A., Sanguinetti M., et al. Comparative computational analysis of SARS-CoV-2 nucleocapsid protein epitopes in taxonomically related coronaviruses. Microbes Infect. 2020;22(4–5):188–194. doi: 10.1016/j.micinf.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiang F., Wang X., He X., et al. Antibody Detection and Dynamic Characteristics in Patients with COVID-19 [published online ahead of print, 2020 Apr 19] Clin Infect Dis. 2020:ciaa461. doi: 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo L., Ren L., Yang S., et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin Infect Dis. 2020;71(15):778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J., Yuan Q., Wang H., et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019 [published online ahead of print, 2020 Mar 28] Clin Infect Dis. 2020:ciaa344. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yongchen Z., Shen H., Wang X., et al. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID-19 patients. Emerg Microbes Infect. 2020;9(1):833–836. doi: 10.1080/22221751.2020.1756699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu W., Liu L., Kou G., et al. Evaluation of nucleocapsid and spike protein-based ELISAs for detecting antibodies against SARS-CoV-2. J Clin Microbiol. 2020;58(6):e00461-20. doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burbelo P.D., Riedo F.X., Morishima C., et al. Sensitivity in Detection of Antibodies to Nucleocapsid and Spike Proteins of Severe Acute Respiratory Syndrome Coronavirus 2 in Patients With Coronavirus Disease 2019. The Journal of infectious diseases. 2019;222(2):206–213. doi: 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan Y.J., Goh P.Y., Fielding B.C., et al. Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin Diagn Lab Immunol. 2004;11(2):362–371. doi: 10.1128/CDLI.11.2.362-371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woo P.C., Lau S.K., Wong B.H., et al. Differential sensitivities of severe acute respiratory syndrome (SARS) coronavirus spike polypeptide enzyme-linked immunosorbent assay (ELISA) and SARS coronavirus nucleocapsid protein ELISA for serodiagnosis of SARS coronavirus pneumonia. J Clin Microbiol. 2005;43(7):3054–3058. doi: 10.1128/JCM.43.7.3054-3058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang G., Nie S., Zhang Z., et al. Longitudinal Change of Severe Acute Respiratory Syndrome Coronavirus 2 Antibodies in Patients with Coronavirus Disease 2019. The Journal of infectious diseases. 2020;222(2):183–188. doi: 10.1093/infdis/jiaa229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okba N.M.A., Muller M.A., Li W., et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease 2019 patients. Emerg Infect Dis. 2020;26(7):1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wajnberg A., Mansour M., Leven E., et al. Humoral immune response and prolonged PCR positivity in a cohort of 1343 SARS-CoV 2 patients in the New York City region. medRxiv. 2020 2020.04.30.20085613. [Google Scholar]
- 43.Niedrig M., Leitmeyer K., Lim W., et al. First external quality assurance of antibody diagnostic for SARS-new coronavirus. J Clin Virol. 2005;34(1):22–25. doi: 10.1016/j.jcv.2005.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meyer B., Drosten C., Muller M.A. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J., Wiltse A., Zand M.S. A complex dance: measuring the multidimensional worlds of influenza virus evolution and anti-influenza immune responses. Pathogens. 2019;8(4):238. doi: 10.3390/pathogens8040238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li D., Wang J., Garigen J., et al. Continuous readout versus titer-based assays of influenza vaccine trials: sensitivity, specificity, and false discovery rates. Comput Math Methods Med. 2019;2019:9287120. doi: 10.1155/2019/9287120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitman J.D., Hiatt J., Mowery C.T., et al. Test performance evaluation of SARS-CoV-2 serological assays. medRxiv. 2020 2020.2004.2025.20074856. [Google Scholar]
- 48.FDA. https://www.fda.gov/medical-devices/emergency-situations-medical-devices/eua-authorized-serology-test-performance Available at: Accessed May 30, 2020.
- 49.Huang A.T., Garcia-Carreras B., Hitchings M.D.T., et al. A systematic review of antibody mediated immunity to coronaviruses: antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. medRxiv. 2020 doi: 10.1038/s41467-020-18450-4. 2020.2004.2014.20065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Long Q.X., Tang X.J., Shi Q.L., et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26(8):1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 51.Woo P.C., Lau S.K., Wong B.H., et al. Longitudinal profile of immunoglobulin G (IgG), IgM, and IgA antibodies against the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in patients with pneumonia due to the SARS coronavirus. Clin Diagn Lab Immunol. 2004;11(4):665–668. doi: 10.1128/CDLI.11.4.665-668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu W., Fontanet A., Zhang P.H., et al. Two-year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. J Infect Dis. 2006;193(6):792–795. doi: 10.1086/500469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu L.P., Wang N.C., Chang Y.H., et al. Duration of antibody responses after severe acute respiratory syndrome. Emerg Infect Dis. 2007;13(10):1562–1564. doi: 10.3201/eid1310.070576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Callow K.A., Parry H.F., Sergeant M., et al. The time course of the immune response to experimental coronavirus infection of man. Epidemiol Infect. 1990;105(2):435–446. doi: 10.1017/s0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu F., Wang A., Liu M., et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv. 2020 2020.2003.2030.20047365. [Google Scholar]
- 56.Perera R.A., Mok C.K., Tsang O.T., et al. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Euro Surveill. 2020;25(16):2000421. doi: 10.2807/1560-7917.ES.2020.25.16.2000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuchun N., Guangwen W., Xuanling S., et al. Neutralizing antibodies in patients with severe acute respiratory syndrome-associated coronavirus infection. J Infect Dis. 2004;190(6):1119–1126. doi: 10.1086/423286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang C., Li W., Drabek D., et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun. 2020;11(1):2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang J, Zand MS. The potential for antibody-dependent enhancement of SARS-CoV-2 infection: Translational implications for vaccine development. Journal of Clinical and Translational Science, 1–4. 10.1017/cts.2020.39. [DOI]