Abstract
Emerging viral diseases pose a major threat to public health worldwide. Nearly all emerging viruses, including Ebola, Dengue, Nipah, West Nile, Zika, and coronaviruses (including SARS-Cov2, the causative agent of the current COVID-19 pandemic), have zoonotic origins, indicating that animal-to-human transmission constitutes a primary mode of acquisition of novel infectious diseases. Why these viruses can cause profound pathologies in humans, while natural reservoir hosts often show little evidence of disease is not completely understood. Differences in the host immune response, especially within the innate compartment, have been suggested to be involved in this divergence. Natural killer (NK) cells are innate lymphocytes that play a critical role in the early antiviral response, secreting effector cytokines and clearing infected cells. In this review, we will discuss the mechanisms through which NK cells interact with viruses, their contribution towards maintaining equilibrium between the virus and its natural host, and their role in disease progression in humans and other non-natural hosts.
Current Opinion in Virology 2020, 40:97–111
This review comes from a themed issue on Viral immunology
Edited by Dirk Dittmer and Blossom Damania
For a complete overview see the Issue and the Editorial
Available online 9th August 2020
https://doi.org/10.1016/j.coviro.2020.07.003
1879-6257/© 2020 Elsevier B.V. All rights reserved.
Preface
Emerging viral diseases pose an ongoing threat to mankind, especially in a globalized and highly interconnected world. A prime example of such a threat is the current COVID-19 pandemic, with more than eighteen million confirmed cases and over 680 000 deaths worldwide at the time of this publication [1]. All of the diseases with the greatest potential to cause a public health emergency, as identified by the World Health Organization, are driven by viruses of zoonotic origin [2]. These include viruses that cause haemorrhagic fever (e.g. Ebola, Marburg, Dengue, and Lassa viruses), highly pathogenic respiratory coronaviruses (e.g. those causing MERS and SARS), and other viruses (e.g. Nipah, Zika, and Chikungunya).
These zoonotic viruses are extremely diverse in nature, as some are transmitted through vectors such as mosquitoes or ticks (e.g. Dengue, Tick-borne encephalitis), whereas human-to-human is the main mode of transmission for others (e.g. Ebola, SARS-Cov2). Some zoonotic viruses have a wide range of natural hosts (e.g. Huaiyangshan, West Nile), whereas others are restricted to specific species such as bats (e.g. Marburg, Nipah) (Table 1 ). However, most zoonotic viruses have shared characteristics, such as being single-stranded RNA (ssRNA) viruses, and causing mild or asymptomatic infections in its natural host animal, while provoking profound pathologies in humans [3••]. Understanding the immune mechanisms that allow animal reservoirs to tolerate these viruses will shed light into how viral zoonotic infections progress to severe illness in humans. This review describes the role of Natural killer (NK) cells, a critical component of early antiviral immunity, in the establishment of tolerance to viral infections in natural hosts, as well as their role in the development of disease in non-natural hosts.
Table 1.
Overview of NK cell response to emerging viruses of zoonotic origin, as identified by the WHO [2,177]. General epidemiological data sourced from the World Health Organization and Center for Disease Control and Prevention [178,179]
| Virus | Family | Natural Host(s)/reservoir | Main mode of transmission | Human to human transmission? | Pathology in humans | Avoidance of NK cell recognition/activation | Protective role of NK cells | Detrimental effects of NK cells |
|---|---|---|---|---|---|---|---|---|
| Lassa | Arenaviridae | Multimammate rat | Rodent excrements and body fluids | Llimited, requires contact with body fluids | 80% asymptomatic. General malaise. In severe cases (1%), hemorrhage, shock and death. Specially severe in late pregnancy. | Upregulation of MHC class I, downregulation of NKG2D ligands [103,104] | Likely. Associations of inhibitory KIRs with case fatality [105]. Association of NK cell expansion with recovery [106]. | Unknown. Unlikely |
| MERS-Cov | Coronaviridae | Dromedaries, (bats?) | Droplets | Limited, requires close contact | High Fever, dry cough, shortness of breath. In severe cases, pneumonia and lung failure. | Inhibition of type I IFN responses [66,67] | Probable. Reduced NK cell numbers in mouse models [73] | Unknown. |
| SARS-Cov | Coronaviridae | Bats, (civets?) | Droplets, (airborne?) | Moderate | High Fever, dry cough, shortness of breath. In severe cases, pneumonia and lung failure. | Inhibition of type I IFN responses [69,117] | Unclear. Not required in mouse models [74], but likely protective in humans [71] | Unknown. |
| SARS-Cov2 | Coronaviridae | Bats, (pangolins?) | Droplets, (stool?, body fluids?) | Extensive | High Fever, dry cough, shortness of breath. In severe cases, pneumonia and lung failure. | likely inhibition of type I IFN responses | Probable. Profound lymphopenia and NK cell exhaustion in severe cases [72,118] | Unknown. Possible? |
| Ebola | Filoviridae | African fruit bats | Human direct contact, body fluids, sexual | Moderate | Fever, muscle pain, rash, diarrhea / vomiting. In severe cases, extensive Hemorrhage. ∼50% fatality rate | Inhibition type I IFN responses [78,81], concealment of activating ligands [84] | Very Likely. VLP-primed NK cells protect against Ebola [79••]. Recognition through activating NKp30 and NKG2D [83,84] | Likely. NK cells may kill antiviral T cells [85]. Higher IFN-γ in deceased patients [86] |
| Marburg | Filoviridae | African fruit bats | Human direct contact, body fluids, sexual | Moderate | Fever, muscle pain, rash diarrhea / vomiting. In severe cases, extensive Hemorrhage. ∼50% fatality rate | Similar to Ebola, but lower inhibition of type I IFN signaling [119] | Unknown. Likely similar to Ebola responses [120] | Unknown. Likely similar to Ebola responses[120] |
| Dengue | Flaviviridae | Primates, (humans) | Mosquitoes | Rare cases of mother to child and sexual transmission | 75% asymptomatic. Fever, muscle / joint pain, nausea. In severe cases (∼1%) facial bleeding and frequent vomiting, blood in vomit and stool. | Inhibition of type I IFN responses, upregulation MHC class I [96] | Very Likely. NK cell IFN-γ required for early control [89••]. Recognition through activating KIR2DS2 and NKp44 [90]. Associations of inhibitory KIRs with case incidence [98]. | Possible. Higher IFN-γ and NK cell activation in critical phase [99,102] |
| Tick Borne encephalitis | Flaviviridae | Small rodents | Ticks | None documented | ∼30% asymptomatic. General malaise. In severe cases (∼20%), meningoencephalitis (seizures, confusion, paralysis). ∼2–20% fatality rate | Unclear. Elevated IFN-α levels but reduced NK cell killing [121]. Dampened responsiveness to type I IFN? [122] | Likely. NK cells strongly activated and proliferative during acute phase. High levels of IFN-γ in human patients [121] | Possible. NK cells increased in CSF of TBE patients [123,124] |
| West Nile | Flaviviridae | Crows, other birds | Mosquitoes | Rare cases of mother to child, transfusions | 80% asymptomatic. Fever, muscle / joint pain, vomit, rash. In severe cases (<1%), meningoencephalitis. (<0.1% fatality rate) | Inhibition of type I IFN responses [125]. Upregulation of MHC class I [126] | Very Likely. Strong NK cell activation and killing to WN-infected cells [127]. Increased IFN-γ production and mature phenotype of NK cells from patients with previous WN infection [128,129]. | Unclear. NK cells increase in CNS but depletion in mice does not change disease outcome [130] |
| Yellow Fever | Flaviviridae | Monkeys (lemurs?) | Mosquitoes | Extremely rare | ∼60% asymptomatic. Fever, malaise, vomit. In severe cases (∼10%), jaundice, hemorrhage, shock, organ failure. ∼5% fatality rate. | Inhibition of type I IFN responses [131] | Very Likely. Strong NK cell expansion, Increased IFN-γ production and cytokine responsiveness [132, 133, 134]. Correlates with better protection in humanized mouse model [135]. | Possible. Increased NK cell numbers in liver compared to healthy livers or those from bacterial liver infection [136] |
| Zika | Flaviviridae | Monkeys (livestock?) | Mosquitoes, sexual transmission | Mother to child, transfusions, sexual transmission | 80% asymptomatic. Mild fever, rash, muscle / joint pain, conjunctivitis. May cause birth defects (microcephaly, brain damage, joint problems) | Dampened responsiveness to type I IFN [137,138]. Upregulation of MHC class I? [139] | Conflicting reports. NK cells are activated early [140], proliferate [141], and activation correlates with viral clearance [142]. Others report NK cells do not proliferate nor respond during Zika infection [143,144] | Possible. Increased NK cell infiltration in CNS in fatal Zika cases and mouse models [145,146] |
| Crimean-Congo Hemorrhagic fever | Nairoviridae | Livestock and cattle | Ticks | Limited, requires contact with body fluids | High fever, muscle and back pain, light sensitivity. At late stages, severe internal hemorrhage, liver failure. ∼30% fatality rate | Unknown. | Possible. High IFN-γ levels early during infection [147,148], Accumulation of NK cells in sites of viral replication[147]. | Unclear. Higher NK cell number in severe cases in humans [149], but others report no correlation [150]. |
| Avian influenza | Orthomyxoviridae | Poultry, (other birds) | Birds secretions | Extremely rare | Fever, cough, shortness of breath. Fatality rate ranges from ∼2% (H1N1 strain) to 60% (H5N1 strain). | Unclear. Induction of Type I IFN responses [151,152] and downregulation of MHC class I [153]. | Strain specific? Lower NK cell activation and killing in infection with highly pathogenic H5N1strain, compared to low pathogenic strains [154,155]. H3N2 strain induces hyper-responsiveness in human lung NK cells [156]. | Strain specific? NK cell depletion improves disease outcome in mice infected with H1N1 strain [157,158]. |
| Nipah | Paramyxoviridae | Asian fruit bats, pigs | Excrements and body fluids | Limited, requires close contact | Initially fever, muscle pain, vomiting. At Later stages, encephalitis (mental confusion, seizures, coma) Fatality rate ∼40%. | Inhibition of Type I IFN responses [159]. However, IFN-β produced in vitro [160]. | Unclear. Increased cytotoxic cell markers and NK cell markers in survivors monkeys [161], and increase in NK cell numbers and proliferation in one survivor [162] | Possible. NK cell signature predicted to be contribute to fatal outcomes [161]. |
| Huaiyangshan (SFTS) | Phenuiviridae | Rodents, (other mammals) | Ticks | Extremely rare | Severe fever, vomiting diarrhea. In severe cases, thrombocytopenia, multiple organ failure. Fatality rate 10∼30%. | Dampening of Type I IFN responses [163,164]. Induction of anti-inflammatory cytokines (Il-10) [165] | Possible. Increase in NK cell percentage during early disease stages in recovery patients [166]. | Unlikely. No difference in NK cell percentage between fatal and recovered cases [166] |
| Rift valley fever | Phenuiviridae | Livestock and cattle | Livestock meat and body fluids, mosquitoes | None documented | Fever, muscle / joint pain. In sever cases (<1%), meningo-encephalitis and hemorrhagic fever. | Inhibition of type I IFN responses [167, 168, 169] | Unclear. Reduced NK cell numbers in hyper-susceptible mice [170], but depletion does not increase susceptibility in resistant mice [171] | Unlikely. NK cell depletion does not improve disease outcome in mice [171] |
| Rabies | Rhabdoviridae | Bats, dogs, raccoons | Bites | Extremely rare | Fever, nausea, vomiting. Aggressiveness, hallucinations, spasm, seizures. ∼100% fatality rate if unvaccinated. | Dampening of Type I IFN responses? Infection of immuno-privileged sites [172] | Very Likely. NK cells are required for protection following vaccination [109••] | Very unlikely |
| Chikungunya | Togaviridae | Primates | Mosquitoes | Rare cases of mother to child | Most infections are symptomatic. Fever, nausea and severe joint pain. Low fatality rate (<0.1%) | Unclear. Induction of strong I IFN production and downregulation of MHC class I [173]. Dissemination? | Likely. Associations of inhibitory KIRs with case incidence [174]. Strong NK cell activation and expansion during acute phase [175] | Very Likely. NK cell depletion ameliorates joint pathology [176] |
Natural killer cells in innate immunity
NK cells are lymphocytes of the innate immune system with the unique ability to rapidly destroy virally infected cells and tumors without previous sensitization [4,5]. They play a critical role in the control of viral infections, as deficiencies in the NK cell compartment are associated with increased susceptibility to certain viral infections in humans [6, 7, 8]. Unlike T and B cells, NK cells do not express a rearranged antigen receptor, but instead express a wide array of germline-encoded receptors, which can be activating or inhibitory [4,9]. NK cells are ‘educated’ through their inhibitory receptors to recognize ‘self’ (e.g. ligands normally present on healthy cells, such as MHC class I molecules) [9,10]. In contrast, activating receptors can recognize ‘non-self’ pathogen components (e.g. viral ligands, proteins, or peptides expressed during infection) or stress signals (e.g. NKG2D ligands expressed during injury, infection, or tumorigenesis) [9,10]. NK cells also express activating Fc receptors (e.g. CD16), which helps them recognize antibody-coated target cells and clear them via antibody-dependent cell-mediated cytotoxicity (ADCC) [11]. Moreover, during viral infection, pro-inflammatory cytokines (e.g. IFN-α, IL-12, and IL-2/15) can directly activate NK cells [12, 13, 14]. When engaging a potential target, all these signals are integrated to determine whether the NK cell will kill, make cytokines, and proliferate. Upon activation, NK cells can release granzymes and perforin to lyse infected target cells, produce effector cytokines such as IFN-γ to alert the surrounding tissue of infection, and in some cases undergo robust proliferation [4,8,15,16••,17].
NK cells throughout evolution: keeping an eye on the viral niche
NK cells, along with other lymphocytes such as T and B cells, likely arose in a common vertebrate progenitor around 500 million years ago [18]. The antiviral and antitumoral activities of NK cells (or NK-like cells) have been described across many non-mammalian species, including lampreys [18,19], fish [20,21], amphibians [22,23], reptiles [24] and birds [25,26]. The transcription factors that define the NK cell lineage (e.g. Id2, Nfil3, Eomes), as well as molecules required for their cytotoxic potential (e.g. Rab27a, Prf1), are evolutionarily conserved [18]. However, unlike with T and B cell receptors derived from gene rearrangement, where antigen recognition is largely shared from fishes to mammals [27], antigen recognition through NK cell receptors is evolutionarily diverse. Fish, birds, reptiles, and amphibians are predicted to express different classes of NK cell receptors, such as the novel immune-type receptors (NITRs) in fish, chicken Ig-like receptors (ChIRs) in birds, and Xenopus Ig-like receptors (xILRs) in amphibians [28]. Mammalian NK cells on the other hand typically recognize their targets through killer cell Ig-like receptors (KIRs), killer cell lectin-like receptors (KLRs) and leukocyte Ig-like receptors (LILRs) [28].
NK cells are diverse on a population level, as each cell stochastically expresses a unique combination of activating and inhibitory receptors [29]. Furthermore, although the genes encoding NK cell receptors (NKR) can be present as a single copy and exhibit low polymorphism in some mammals, they can be highly polymorphic and expanded into superfamilies (likely by gene duplication) in closely related species [28,30,31]. Such interspecies variability (with the Klrk gene encoding the NKG2D receptor being an exception) makes NKRs some of the most rapidly evolving gene families in eukaryotes [28]. These features may be responsible for the unique responsiveness of NK cells to variations in genetic and environmental factors that impact the host immune response to viral infections. Interestingly, other forms of innate immune sensing such as Toll-like receptors (TLRs), RIG-I-like receptors (RLRs) and NOD-like receptors (NLRs), which recognize pathogen-associated molecular patterns, are remarkably conserved down to invertebrates [29,32].
The striking diversity of NKRs suggests an intense selective pressure from the pathogens they encounter, especially viruses. This co-evolution of the NKR repertoire with the viral interface is arguably best studied in the mouse and human NK cell response to cytomegalovirus (CMV) [33, 34, 35]. CMV is a species-specific herpesvirus that persists through latency in healthy hosts, and thought to intimately interact with the host immune system. To avoid CD8+ T cell recognition, human and mouse CMV (HCMV and MCMV, respectively) possess multiple genes encoding immunoevasins that interfere with antigen presentation by degrading, retaining, or preventing the assembly of MHC class I on the cell surface of infected cells [36]. However, because downregulation of MHC class I causes CMV-infected cells to be susceptible to NK cell recognition of “missing self”, CMV has evolved MHC class I-like proteins as a decoy to engage inhibitory NKRs and tune down NK cell reactivity [37]. As perhaps an evolutionary countermeasure, activating NKRs recognizing the same decoy ligands have appeared in mouse and human genomes that allow for NK cell recognition of CMV-infected cells [33]. Examples of this host-pathogen evolutionary adaptation include the inhibitory Ly49I and activating Ly49H receptor pair in mice, which both recognize the MCMV m157 glycoprotein [38], and the inhibitory NKG2A and activating NKG2C receptors in humans, which both recognize the viral UL40 peptides presented on the nonclassical MHC class I molecule HLA-E [39,40]. In further evidence of this evolutionary ‘tug-of-war’ between host and virus, MCMV strains isolated from wild mice have been shown to exhibit high polymorphism in its m157 gene, and HCMV isolates that promote strong NKG2C engagement are far rarer than those inducing weak activation in humans [41,42••]. Thus, both mouse and human immune systems have evolved remarkably similar molecular mechanisms of viral control of CMV using different families of NKRs, representing a strong example of convergent evolution.
Innate immunity in reservoirs of zoonotic viruses
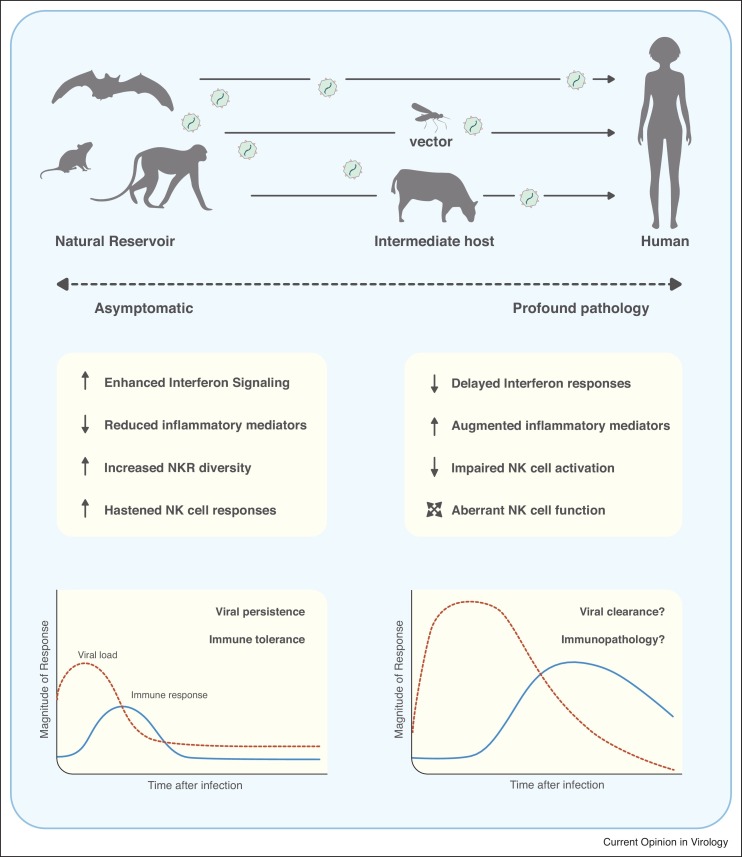
Animals with the greatest potential to transmit zoonotic diseases to humans include mammals such as bats, primates, and rodents [3••]. While ecological and biological factors certainly play a role in the transmission of zoonotic diseases, a growing body of evidence suggests that transmission across species may occur because these animals possess a more permissive innate immune response allowing them to carry a high viral burden (Figure 1 ).
Figure 1.
The immune response against zoonotic viruses in natural reservoirs compared to humans. Top: Zoonotic viruses can directly pass from natural hosts (e.g. bats, mice, monkeys) to humans, or be transmitted through intermediate hosts or vectors (e.g. mosquitoes, cattle). Middle and bottom: Some natural reservoirs have evolved enhanced interferon responses while reducing pro-inflammatory mediators. Increased NKR complexity and diversity, and improved NK cell responsiveness may also contribute to viral persistence while keeping the host asymptomatic. When zoonotic viruses jump to novel hosts such as humans, a slower interferon response and impaired early NK cell activation may lead to poor virus clearance, aberrant immune responses, heighten inflammation, and profound pathology.
The example of bats
Bats are reservoirs to Ebola and Marburg viruses, Hendra and Nipah viruses, and SARS coronaviruses, among others [43,44]. Their roosting behaviour, ability to fly, widespread distribution, and consumption as bushmeat makes them especially well-positioned to transmit zoonotic viruses to humans [45]. Despite having a high metabolic rate associated with the energetic cost of flight, bats have unusually long lifespans [46]. Consistent with this energetic demand, genes involved in oxidative phosphorylation and DNA damage response are under positive selection in bats [47]. In turn, bats may have fine-tuned their response to high cellular stress in order to avoid overt inflammatory responses, which has been hypothesized to provide a unique niche for certain viruses. Indeed, bat cells show dampened activation of the NLRP3 inflammasome during viral infection, and the production of interferons (IFNs) following viral entry is rapid but more transient and lower in magnitude compared to other mammals [45,48]. Moreover, interferon-stimulated genes (ISG) such as ISG15 may have been positively selected for in certain bat species, and some IFN gene families are greatly expanded [47,49,50••]. Altogether, it is believed that quick control of viral infections and reduced induction of pro-inflammatory cytokines have allowed bat viruses to rapidly co-evolve with their host without provoking major immune-mediated pathologies [51].
Type I IFNs provide a strong activating signal for NK cells, and a dysregulation in the IFN-NK cell axis in bats could have detrimental consequences. Recently, ISG15 has been shown to boost the cytolytic activity of NK cells [52], which may contribute to control of viral replication in bats. Interestingly, the bat genome does not encode for KIR genes, but shows an amplification of LILR and KLR genes, especially NKG2A/B [30,50••]. Six of these NKG2A/B genes putatively encode activating and inhibitory interaction motifs simultaneously, adding an extra layer of complexity to the regulation of NKRs in bats. Although NK cells have been characterized in bat peripheral blood using cross-reacting antibodies [53], their functionality and responsiveness against bat viruses have not been tested. Altogether, the current evidence suggests that the NK cell activation threshold in bats may be tightly controlled to avoid detrimental immune responses.
Lessons from monkeys
Primates are a reservoir for Chikungunya and Yellow Fever virus (YFV), among others [3••]. Furthermore, although Human Immunodeficiency Virus (HIV) is now restricted to humans, it is thought to have originated from the Simian Immunodeficiency Virus (SIV) [54]. Whereas some monkey species are tolerant to SIV (e.g. sooty mangabeys (SM)), other related species are susceptible (e.g. rhesus macaques (RM)), which allows for comparative analyses of the respective immune response in natural and non-natural hosts. SM pDCs have been shown to produce less IFN-α in response to SIV and YFV infection, in part due to dampened TLR7 signaling [55••,56]. Lower levels of such pro-inflammatory cytokines in serum may be partly responsible for the diminished NK cell activation and proliferation observed in SM compared to RM and humans [55••,57]. Interestingly, it has been reported that SM NK cells respond more rapidly to SIV infection than RM NK cells, suggesting that, as with bats, early control of viral infection by the innate immune system prevents aberrant immune responses later on [58••].
Turning to rodents for answers
Rodents are reservoirs to Lassa fever virus and a wide range of hantaviruses. Although the rodent immune system is arguably as extensively studied as the human, relatively little is known about the immune mechanisms promoting viral tolerance in wild natural hosts. Syrian hamsters are tolerant to certain hantaviruses, such as Sin Nombre Virus (SNV), but succumb to the closely related Andes Virus (ANDV), with symptoms remarkably similar to the human manifestation of the disease [59, 60, 61]. Although comparative analyses are scarce, ANDV appears to avoid early innate immune recognition and elicit a stronger immune response later on [59,62]. Priming with poly I:C or SNV protects hamsters from ANDV-driven pulmonary disease, suggesting that rapid IFN responses may allow for viral control while avoiding later immunopathology [63], similar to bats. ANDV produces severe lymphopenia, and immunosuppressants make hamsters susceptible to SNV [60,64], highlighting the critical role of immune responses for keeping the virus in check. Interestingly, T cell depletion does not alter the course of disease in ADNV and SNV-infected hamsters, suggesting that perhaps innate lymphocytes, such as NK cells, may play a role in the pathology (or avoidance of pathology) of hantavirus diseases [61,65].
The careful study of the immune system in reservoirs of zoonotic diseases will certainly offer insights into how these animals carry high viral loads while remaining asymptomatic. A growing body of evidence suggests that differences in the innate immune system, particularly in viral recognition and induction of pro-inflammatory cytokines, may explain such phenomenon (Figure 1). Since NK cells are key players in the integration and amplification of these signals, some animal reservoirs may have adapted their NKR repertoire and functionality to avoid excessive immune activation leading to immunopathology. As such, a deeper understanding of the NK cell response to zoonotic viruses in its natural host may shed light into their role in disease pathology in humans.
NK cells and viral diseases of zoonotic origin
NK cells have been proposed to contribute to immunopathology in some zoonotic diseases, while having protective effects against other viral infections. Here, we will discuss their role in the progression of diseases caused by SARS coronaviruses and some Haemorrhagic fever viruses (HFVs). This data is summarized in Table 1 and Figure 2 , alongside with other relevant viral diseases of zoonotic origin, including Zika, West Nile, and Nipah. Finally, we will examine their contribution to the protection conferred by vaccines against zoonotic viruses.
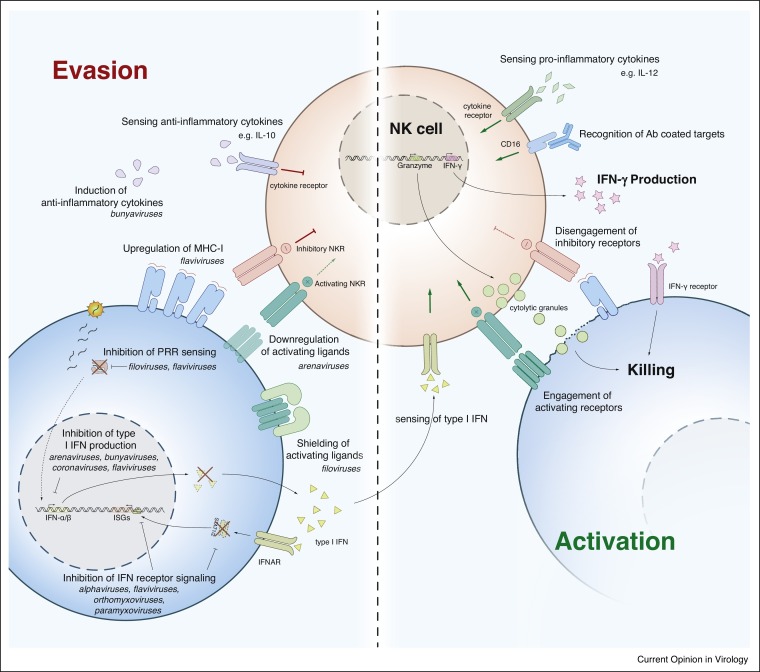
Figure 2.
Mechanisms of NK cell evasion or activation by zoonotic viruses. Left: Zoonotic viruses can avoid interferon responses by blocking PRR sensing, inhibiting interferon production, and dampening interferon receptor signalling. These viruses also evade NK cell recognition by upregulating ligands for inhibitory NKR, downregulating or shielding activating NKR ligands, and inducing anti-inflammatory cytokines. Right: When activating signals (green arrows) outweigh inhibitory signals (red flat-end arrows), NK cells become activated and secrete pro-inflammatory cytokines (e.g. IFN-γ), release cytotoxic granules to kill target cells, and undergo proliferation. See also Table 1 for a complete list of references. PRR, pattern recognition receptors. NKR, NK cell recognition receptors. ISG, interferon stimulated genes.
SARS coronaviruses
The coronaviruses MERS-CoV, SARS-Cov, and SARS-Cov2 have caused three global outbreaks in the past two decades, with the latter being the causative agent behind the current COVID-19 pandemic. Negative outcomes have been associated with aberrant immune responses and subsequent lung pathology. MERS-CoV induced high levels of inflammatory cytokines associated with strong myeloid responses (e.g. IL-β, IL-6, and IL-8), but failed to induce early type I IFNs [66, 67, 68]. Common human coronaviruses induce strong type I IFN responses compared to SARS and MERS, which may account for their lower fatality rate [67,69]. Interestingly, IFN-α production anecdotally correlated with survival of MERS patients, as did serum levels of IFN-γ [70]. Although definitive data is lacking, these studies suggest that early IFN production (e.g. IFN-γ production by NK cells) may contribute to viral control in these patients. Conversely, delayed recognition and responses may result in virus-mediated cytopathy and aberrant production of highly inflammatory cytokines leading to lung pathology.
All three coronaviruses mentioned above have been documented to provoke severe lymphopenia and lower circulating NK cell numbers in some patients, although the exact role of NK cells in disease pathology is not clear [71, 72, 73]. Although mouse models of SARS infection suggest that lymphocytes do not play a major role in pathology, these models do not accurately replicate disease progression in humans [74,75]. Given our understanding of viral infections in hamsters and macaques, we would argue that developing more relevant animal models is key to studying virus-immune interactions, and immune-mediated pathobiology. Currently, one US-based company has obtained approval from the FDA to use NK cell-based immunotherapy as a potential treatment for COVID-19, with others following suit [76,77]. Although the available data indicates that NK cell-targeted therapy may have beneficial effects on virus control and disease progression early on, these data also suggest that NK cells can exacerbate the severity of the disease at later time points. Thus, extreme caution should be exercised when designing such treatment strategies, and there should not be a rush to haphazardly implement such cellular treatments without careful design.
Haemorrhagic fever viruses
HFVs are a diverse group of viruses with the common denominator of causing fever and severe bleeding, although disease severity is highly variable. HFVs include arenaviruses (e.g. Lassa virus), filoviruses (e.g. Ebola virus), flaviviruses (e.g. Dengue virus) and hantaviruses. Here we attempt to summarize the NK cell response to representative examples of these viruses.
Ebola
Ebola virus-infected cells have been shown to be resistant to NK cell-mediated killing [78,79••]. Ebola is known to potently inhibit type I IFN induction in infected myeloid cells, and IFN-α and IL-12 are difficult to detect in infected individuals [78,80, 81, 82]. In contrast, viral-like particles (VLPs) of Ebola, which contain no RNA or inhibitory proteins, are able to readily activate DCs, and promote NK cell cytotoxicity and IFN-γ production [79••,83]. These findings suggest that NK cells can more efficiently target Ebola-infected cells when they receive the proper cytokine signals. Indeed, transfer of VLP-primed NK cells provided almost complete protection against lethal Ebola challenge in mice, where protection was dependent on cytotoxicity and not on IFN-γ production by NK cells [79••].
Ebola can evade NK cell recognition through additional mechanisms. VLP-treated NK cells recognize Ebola-infected cells through NKp30 and possibly NKG2D; however, Ebola uses its glycoprotein to conceal NKp30 and NKG2D ligands expressed on the surface of infected cells, thereby shielding them from NK cell killing [84]. Interestingly, while the glycoprotein also impairs MHC class I presentation and thus recognition by the T cells, engagement of inhibitory NKRs can still occur. Altogether, these VLP studies suggest that NK cells can recognize viral proteins present in the VLP, but additional evasion proteins in the Ebola genome can impair NK cell activation. The direct mechanisms and relevance of these Ebola immunoevasins in human infections remain to be determined.
Ebola infections are also characterized by profound lymphopenia, lymphocyte apoptosis, and extensive cellular damage. Do NK cells directly (via cytotoxicity) or indirectly (via IFN-γ secretion) contribute to these phenotypes? One study suggested that NK cells can contribute to pathology by migrating to infected organs and killing infiltrating T cells [85]. Furthermore, IFN-γ was markedly elevated in serum samples from patients that succumbed to haemorrhagic shock, but not in those who survived [86,87]. IFN-γ mRNA levels in PBMCs were surprisingly similar in both patient groups, suggesting that translation into IFN-γ protein may be exacerbated in those that led to fatality, a step-wise mechanism for IFN-γ production that has been recently characterized in NK cells [88].
Dengue
NK cells and IFN-γ production appear to be important for Dengue virus control, as NK cell depletion and IFN-γ blockade led to high viremia and virus-mediated pathology in humanized mouse models [89••]•. Furthermore, NK cells have been suggested to recognize and lyse Dengue-infected cells in vitro through activating receptors such KIR2DS2 (proposed to recognize NS3 peptides in the context of HLA-C0102) and NKp44 (proposed to recognize Dengue envelope protein) [90,91]. However, despite a strong IFN response and potent NK cell activation, viral replication can reach high levels in patients and in vitro [92, 93, 94, 95]. Rather than directly impeding the engagement of NK cell activating receptors, like Ebola, Dengue instead can evade NK cell targeting by upregulating HLA molecules on infected cells [96]. Indeed, blocking HLA in vitro restores NK cell degranulation against Dengue-infected cells, and NK cell cytotoxicity is diminished at early disease stages when viral titers are highest [95,97]. The upregulation of HLA molecules by Dengue suggests that inhibitory NKRs are engaged during NK cell surveillance of infected cells, which is supported by the observation that expression of inhibitory receptors KIR2DL1, KIR3DL1, and KIR2DL5 are more prevalent in Dengue-infected patients than healthy controls [98]. Furthermore, KIR3DL1 has been suggested to directly recognize Dengue NS1-derived peptides presented on HLA-B57 [99]. However, given that KIR3DL1 can already directly recognize endogenous peptides presented on HLA-Bw4, whether NS1-derived peptides can directly alter NK cell function in vivo is unknown.
After an initial febrile phase, Dengue viral titers dwindle and a latter critical phase ensues which can progress into serious complications, including haemorrhage and fatal shock [100,101]. This latter critical phase is likely immune-mediated. NK cell activation and IFN-α titers were higher in haemorrhagic patients compared to milder symptomatic patients, suggesting that these cellular and soluble immune components may directly contribute to pathology at these later stages [93,99,102]. By comparison, infection with Chikungunya virus elicits similar initial symptoms, but very rarely results in fatal haemorrhage and shock. Furthermore, whereas NK cells only increased in number late during Dengue infection, their numbers increased early during Chikungunya infection [95]. Lastly, NK cells from Dengue-infected patients produced more IFN-γ but were less cytotoxic than those from Chikungunya patients [95]. Many other factors beyond the specific immune components herein presented distinguish the pathologies driven by these viral infections. Nonetheless, the available data suggest that NK cells may play a role in the onset of haemorrhagic complications.
Lassa fever
Lassa virus has previously been described to evade NK cell killing by maintaining high levels of MHC class I on infected cells while inhibiting expression of NKG2D ligands [103,104]. Indeed, expression of the inhibitory NKR KIR2DL2 has been associated with increased fatality rate [105]. Although NK cells can be activated by Lassa virus-infected cells, they do not produce IFN-γ [104,106]. The reduced inflammatory profile may partly explain the lower occurrence of haemorrhage and mortality in comparison with other HFVs. Interestingly, NK cells will proliferate to a greater degree in Lassa-infected monkeys that recover from the infection, but are reduced in number in those that succumb to the virus [106]. Whether NK cell proliferation provides protection against Lassa virus has not been formally addressed.
Altogether, it is tempting to speculate that although NK cells provide critical protection in the early phases of HFV infection, the exacerbated NK cell response observed in the later stages contributes to the onset of haemorrhagic complications.
Vaccinating through the NK cell compartment
Among the biggest challenges we face with zoonotic viruses is the lack of effective vaccines. To date, only vaccines against rabies, tick-borne encephalitis, Japanese encephalitis, yellow fever, and recently Ebola, are available. Rabies infection leads to delirium and certain death if untreated, but vaccination after suspected exposure effectively controls the infection [107,108]. Interestingly, in prime-boost studies (where three doses are recommended for full protection), NK cells are among the first and strongest responders during each re-vaccination dose [109••]. IFN-γ production and degranulation predominantly occurred through an ADCC mechanism, suggesting that protective antiviral NK cell responses may depend on rabies-specific antibodies generated following vaccination [109••].
An effective vaccine against Ebola developed in Canada was recently approved for use in the US and Europe [110, 111, 112]. A systems vaccinology approach revealed a correlation between a strong NK cell activation signature and development of an antibody response to this Ebola vaccine [113]. Furthermore, and similar to the rabies vaccine, NK cells from Ebola-vaccinated individuals mounted an antibody-dependent response against Ebola glycoprotein, where IFN-γ production and degranulation correlated in magnitude with neutralizing antibody titers [114].
Currently, two recombinant rabies-based vaccines are being developed against Marburg and Lassa viruses [115••,116]. Although these vaccines provided almost complete protection from viral challenge, the antibodies generated were non-neutralizing. Instead, these virus-specific antibodies elicited strong NK cell-mediated ADCC against infected cells expressing the viral glycoprotein used for vaccination. Blocking Fc receptors on NK cells or directly depleting NK cells decreased killing of target cells, and immunized mice lacking the Fc receptor became susceptible to viral challenge [115••,116]. Altogether, these studies highlight the critical role of NK cells in conferring protection through vaccination against zoonotic diseases.
Concluding remarks
NK cells are critical for the control of viral infections. The receptors they use to recognize virally infected cells have rapidly evolved to adapt to the viral niche in each species. Indeed, some animal reservoirs of emerging viruses (e.g. bats) have exquisitely complex and expanded NK cell receptor repertoires. Emerging viruses may persist in their natural hosts by avoiding the induction of strong pro-inflammatory responses. In turn, natural hosts may have evolved enhanced interferon responses to prevent pathology. When this equilibrium is disturbed, or transmission occurs, the virus can cause severe disease in closely related or unrelated species (Figure 1).
Although some emerging viruses can evade NK cell recognition by dampening type I IFN signalling, others increase the expression of inhibitory ligands while downregulating activating ligands for NK cell receptors in the infected cells. Furthermore, while NK cells contribute to viral clearance early during infection, exacerbated or prolonged NK cell activation at later stages of infection may contribute to severe immunopathology. The complex interaction of viral ligands with NK cells is the result of a frenetic arms-race between host and pathogen (Figure 2). While this delicate equilibrium has developed over millennia and will likely continue for many more, we hope that new insights into the biology of antiviral NK cells will tilt the balance in our favour.
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
•• of outstanding interest
Acknowledgements
We thank members of the Sun lab, Dr. Lewis Lanier, Dr. Daniel Calabrese, and Regina Bou Puerto for the useful discussion and comments on this review. The Sun lab was supported by grants from the Burroughs Wellcome Fund, the American Cancer Society, and the National Institutes of Health (AI100874, AI130043, and P30CA008748).
References
- 1.Johns Hopkins University: Coronavirus Resource Center. https://coronavirus.jhu.edu/map.html.
- 2.Blueprint for R&D preparedness and response to public health emergencies due to highly infectious pathogens. http://www.who.int/csr/research-and-development/meeting-report-prioritization.pdf.
- 3••.Mandl J.N., Ahmed R., Barreiro L.B., Daszak P., Epstein J.H., Virgin H.W., Feinberg M.B. Reservoir host immune responses to emerging zoonotic viruses. Cell. 2015;160:20–35. doi: 10.1016/j.cell.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this review, the authors provide a detailed analysis of immune mechanisms in natural hosts of emerging zoonotic viruses that allows for viral tolerance and persistance.
- 4.Yokoyama W.M., Kim S., French A.R. The dynamic life of natural killer cells. Annu Rev Immunol. 2004;22:405–429. doi: 10.1146/annurev.immunol.22.012703.104711. [DOI] [PubMed] [Google Scholar]
- 5.Kiessling R., Klein E., Pross H., Wigzell H. “Natural” killer cells in the mouse. II. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Characteristics of the killer cell. Eur J Immunol. 1975;5:117–121. doi: 10.1002/eji.1830050209. [DOI] [PubMed] [Google Scholar]
- 6.Orange J.S. Natural killer cell deficiency. J Allergy Clin Immunol. 2013;132:515–525. doi: 10.1016/j.jaci.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee S.H., Miyagi T., Biron C.A. Keeping NK cells in highly regulated antiviral warfare. Trends Immunol. 2007;28:252–259. doi: 10.1016/j.it.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Jost S., Altfeld M. Control of human viral infections by natural killer cells. Annu Rev Immunol. 2013;31:163–194. doi: 10.1146/annurev-immunol-032712-100001. [DOI] [PubMed] [Google Scholar]
- 9.Orr M.T., Lanier L.L. Natural killer cell education and tolerance. Cell. 2010;142:847–856. doi: 10.1016/j.cell.2010.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lanier L.L. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W., Erbe A.K., Hank J.A., Morris Z.S., Sondel P.M. NK cell-mediated antibody-dependent cellular cytotoxicity in cancer immunotherapy. Front Immunol. 2015;6:368. doi: 10.3389/fimmu.2015.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madera S., Rapp M., Firth M.A., Beilke J.N., Lanier L.L., Sun J.C. Type I IFN promotes NK cell expansion during viral infection by protecting NK cells against fratricide. J Exp Med. 2016;213:225–233. doi: 10.1084/jem.20150712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun J.C., Madera S., Bezman N.A., Beilke J.N., Kaplan M.H., Lanier L.L. Proinflammatory cytokine signaling required for the generation of natural killer cell memory. J Exp Med. 2012;209:947–954. doi: 10.1084/jem.20111760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orange J.S., Biron C.A. Characterization of early IL-12, IFN-alphabeta, and TNF effects on antiviral state and NK cell responses during murine cytomegalovirus infection. J Immunol. 1996;156:4746–4756. [PubMed] [Google Scholar]
- 15.Sun J.C., Beilke J.N., Lanier L.L. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Adams N.M., Geary C.D., Santosa E.K., Lumaquin D., Le Luduec J.B., Sottile R., van der Ploeg K., Hsu J., Whitlock B.M., Jackson B.T., et al. Cytomegalovirus infection drives avidity selection of natural killer cells. Immunity. 2019;50:1381–1390. doi: 10.1016/j.immuni.2019.04.009. e1385. [DOI] [PMC free article] [PubMed] [Google Scholar]; In these studies, the authors show that single NK cell clones can expand ∼104 fold, and expansion is controlled by the avidity of NK cell receptors to its activating ligand.
- 17.Grassmann S., Pachmayr L.O., Leube J., Mihatsch L., Andrae I., Flommersfeld S., Oduro J., Cicin-Sain L., Schiemann M., Flossdorf M., et al. Distinct surface expression of activating receptor Ly49H drives differential expansion of NK cell clones upon murine cytomegalovirus infection. Immunity. 2019;50:1391–1400.e1394. doi: 10.1016/j.immuni.2019.04.015. [DOI] [PubMed] [Google Scholar]; In these studies, the authors show that single NK cell clones can expand ∼104 fold, and expansion is controlled by the avidity of NK cell receptors to its activating ligand.
- 18.Vivier E., van de Pavert S.A., Cooper M.D., Belz G.T. The evolution of innate lymphoid cells. Nat Immunol. 2016;17:790–794. doi: 10.1038/ni.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boehm T., Hirano M., Holland S.J., Das S., Schorpp M., Cooper M.D. Evolution of alternative adaptive immune systems in vertebrates. Annu Rev Immunol. 2018;36:19–42. doi: 10.1146/annurev-immunol-042617-053028. [DOI] [PubMed] [Google Scholar]
- 20.Utke K., Bergmann S., Lorenzen N., Kollner B., Ototake M., Fischer U. Cell-mediated cytotoxicity in rainbow trout, Oncorhynchus mykiss, infected with viral haemorrhagic septicaemia virus. Fish Shellfish Immunol. 2007;22:182–196. doi: 10.1016/j.fsi.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Purcell M.K., Laing K.J., Winton J.R. Immunity to fish rhabdoviruses. Viruses. 2012;4:140–166. doi: 10.3390/v4010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Jesus Andino F., Chen G., Li Z., Grayfer L., Robert J. Susceptibility of Xenopus laevis tadpoles to infection by the ranavirus Frog-Virus 3 correlates with a reduced and delayed innate immune response in comparison with adult frogs. Virology. 2012;432:435–443. doi: 10.1016/j.virol.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen G., Robert J. Antiviral immunity in amphibians. Viruses. 2011;3:2065–2086. doi: 10.3390/v3112065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munoz F.J., De la Fuente M. The immune response of thymic cells from the turtle Mauremys caspica. J Comp Physiol B. 2001;171:195–200. doi: 10.1007/s003600000159. [DOI] [PubMed] [Google Scholar]
- 25.Gobel T.W., Kaspers B., Stangassinger M. NK and T cells constitute two major, functionally distinct intestinal epithelial lymphocyte subsets in the chicken. Int Immunol. 2001;13:757–762. doi: 10.1093/intimm/13.6.757. [DOI] [PubMed] [Google Scholar]
- 26.Gobel T.W., Chen C.L., Shrimpf J., Grossi C.E., Bernot A., Bucy R.P., Auffray C., Cooper M.D. Characterization of avian natural killer cells and their intracellular CD3 protein complex. Eur J Immunol. 1994;24:1685–1691. doi: 10.1002/eji.1830240734. [DOI] [PubMed] [Google Scholar]
- 27.Scott-Browne J.P., Crawford F., Young M.H., Kappler J.W., Marrack P., Gapin L. Evolutionarily conserved features contribute to alphabeta T cell receptor specificity. Immunity. 2011;35:526–535. doi: 10.1016/j.immuni.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoder J.A., Litman G.W. The phylogenetic origins of natural killer receptors and recognition: relationships, possibilities, and realities. Immunogenetics. 2011;63:123–141. doi: 10.1007/s00251-010-0506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strauss-Albee D.M., Blish C.A. Human NK cell diversity in viral infection: ramifications of ramification. Front Immunol. 2016;7:66. doi: 10.3389/fimmu.2016.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hilton H.G., Rubinstein N.D., Janki P., Ireland A.T., Bernstein N., Fong N.L., Wright K.M., Smith M., Finkle D., Martin-McNulty B., et al. Single-cell transcriptomics of the naked mole-rat reveals unexpected features of mammalian immunity. PLoS Biol. 2019;17 doi: 10.1371/journal.pbio.3000528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Averdam A., Petersen B., Rosner C., Neff J., Roos C., Eberle M., Aujard F., Munch C., Schempp W., Carrington M., et al. A novel system of polymorphic and diverse NK cell receptors in primates. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwasaki A. A virological view of innate immune recognition. Annu Rev Microbiol. 2012;66:177–196. doi: 10.1146/annurev-micro-092611-150203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun J.C., Lanier L.L. The natural selection of herpesviruses and virus-specific NK cell receptors. Viruses. 2009;1:362. doi: 10.3390/v1030362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammer Q., Rückert T., Romagnani C. Natural killer cell specificity for viral infections. Nat Immunol. 2018;19:800–808. doi: 10.1038/s41590-018-0163-6. [DOI] [PubMed] [Google Scholar]
- 35.Lanier L.L. Evolutionary struggles between NK cells and viruses. Nat Rev Immunol. 2008;8:259–268. doi: 10.1038/nri2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orange J.S., Fassett M.S., Koopman L.A., Boyson J.E., Strominger J.L. Viral evasion of natural killer cells. Nat Immunol. 2002;3:1006–1012. doi: 10.1038/ni1102-1006. [DOI] [PubMed] [Google Scholar]
- 37.Beck S., Barrell B.G. Human cytomegalovirus encodes a glycoprotein homologous to MHC class-I antigens. Nature. 1988;331:269–272. doi: 10.1038/331269a0. [DOI] [PubMed] [Google Scholar]
- 38.Adams E.J., Juo Z.S., Venook R.T., Boulanger M.J., Arase H., Lanier L.L., Garcia K.C. Structural elucidation of the m157 mouse cytomegalovirus ligand for Ly49 natural killer cell receptors. Proc Natl Acad Sci U S A. 2007;104:10128–10133. doi: 10.1073/pnas.0703735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braud V.M., Allan D.S., O’Callaghan C.A., Soderstrom K., D’Andrea A., Ogg G.S., Lazetic S., Young N.T., Bell J.I., Phillips J.H., et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 40.Tomasec P., Braud V.M., Rickards C., Powell M.B., McSharry B.P., Gadola S., Cerundolo V., Borysiewicz L.K., McMichael A.J., Wilkinson G.W. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science. 2000;287:1031. doi: 10.1126/science.287.5455.1031. [DOI] [PubMed] [Google Scholar]
- 41.Voigt V., Forbes C.A., Tonkin J.N., Degli-Esposti M.A., Smith H.R., Yokoyama W.M., Scalzo A.A. Murine cytomegalovirus m157 mutation and variation leads to immune evasion of natural killer cells. Proc Natl Acad Sci U S A. 2003;100:13483–13488. doi: 10.1073/pnas.2233572100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42••.Hammer Q., Rückert T., Borst E.M., Dunst J., Haubner A., Durek P., Heinrich F., Gasparoni G., Babic M., Tomic A., et al. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat Immunol. 2018;19:453–463. doi: 10.1038/s41590-018-0082-6. [DOI] [PubMed] [Google Scholar]; The authors show that adaptive NKG2C+ NK cell responsiveness is dictated by the affinity of NKG2C to the variable HCMV UL40 peptides from clinical isolates.
- 43.Brook C.E., Dobson A.P. Bats as’ special’ reservoirs for emerging zoonotic pathogens. Trends Microbiol. 2015;23:172–180. doi: 10.1016/j.tim.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mandl J.N., Schneider C., Schneider D.S., Baker M.L. Going to Bat(s) for studies of disease tolerance. Front Immunol. 2018;9:2112. doi: 10.3389/fimmu.2018.02112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banerjee A., Baker M.L., Kulcsar K., Misra V., Plowright R., Mossman K. Novel insights into immune systems of bats. Front Immunol. 2020;11:26. doi: 10.3389/fimmu.2020.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hayman D.T.S. Bat tolerance to viral infections. Nat Microbiol. 2019;4:728–729. doi: 10.1038/s41564-019-0430-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang G., Cowled C., Shi Z., Huang Z., Bishop-Lilly K.A., Fang X., Wynne J.W., Xiong Z., Baker M.L., Zhao W., et al. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339:456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahn M., Anderson D.E., Zhang Q., Tan C.W., Lim B.L., Luko K., Wen M., Chia W.N., Mani S., Wang L.C., et al. Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host. Nat Microbiol. 2019;4:789–799. doi: 10.1038/s41564-019-0371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papenfuss A.T., Baker M.L., Feng Z.P., Tachedjian M., Crameri G., Cowled C., Ng J., Janardhana V., Field H.E., Wang L.F. The immune gene repertoire of an important viral reservoir, the Australian black flying fox. BMC Genomics. 2012;13:261. doi: 10.1186/1471-2164-13-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50••.Pavlovich S.S., Lovett S.P., Koroleva G., Guito J.C., Arnold C.E., Nagle E.R., Kulcsar K., Lee A., Thibaud-Nissen F., Hume A.J., et al. The Egyptian rousette genome reveals unexpected features of bat antiviral immunity. Cell. 2018;173:1098–1110. doi: 10.1016/j.cell.2018.03.070. e1018. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, the authors suggest that bats, reservoir for many zoonotic diseases, possess expanded NK cell receptor repertoires with unusual inhibitory motifs.
- 51.Brook C.E., Boots M., Chandran K., Dobson A.P., Drosten C., Graham A.L., Grenfell B.T., Muller M.A., Ng M., Wang L.F., et al. Accelerated viral dynamics in bat cell lines, with implications for zoonotic emergence. eLife. 2020;9 doi: 10.7554/eLife.48401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iglesias-Guimarais V., Ahrends T., de Vries E., Knobeloch K.-P., Volkov A., Borst J. IFN-stimulated gene 15 is an alarmin that boosts the CTL response via an innate, NK cell–dependent route. J Immunol. 2020 doi: 10.4049/jimmunol.1901410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martinez Gomez J.M., Periasamy P., Dutertre C.A., Irving A.T., Ng J.H., Crameri G., Baker M.L., Ginhoux F., Wang L.F., Alonso S. Phenotypic and functional characterization of the major lymphocyte populations in the fruit-eating bat Pteropus alecto. Sci Rep. 2016;6 doi: 10.1038/srep37796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharp P.M., Hahn B.H. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. 2011;1 doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55••.Mandl J.N., Barry A.P., Vanderford T.H., Kozyr N., Chavan R., Klucking S., Barrat F.J., Coffman R.L., Staprans S.I., Feinberg M.B. Divergent TLR7 and TLR9 signaling and type I interferon production distinguish pathogenic and nonpathogenic AIDS virus infections. Nat Med. 2008;14:1077–1087. doi: 10.1038/nm.1871. [DOI] [PubMed] [Google Scholar]; Mandl et al. show that Sooty Mangabeys, natural host for SIV, may allow virus persistence through dampened TLR7 signaling.
- 56.Mandl J.N., Akondy R., Lawson B., Kozyr N., Staprans S.I., Ahmed R., Feinberg M.B. Distinctive TLR7 signaling, type I IFN production, and attenuated innate and adaptive immune responses to yellow fever virus in a primate reservoir host. J Immunol. 2011;186:6406–6416. doi: 10.4049/jimmunol.1001191. [DOI] [PubMed] [Google Scholar]
- 57.Giavedoni L.D., Velasquillo M.C., Parodi L.M., Hubbard G.B., Hodara V.L. Cytokine expression, natural killer cell activation, and phenotypic changes in lymphoid cells from rhesus macaques during acute infection with pathogenic simian immunodeficiency virus. J Virol. 2000;74:1648–1657. doi: 10.1128/jvi.74.4.1648-1657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58••.Pereira L.E., Johnson R.P., Ansari A.A. Sooty mangabeys and rhesus macaques exhibit significant divergent natural killer cell responses during both acute and chronic phases of SIV infection. Cell Immunol. 2008;254:10–19. doi: 10.1016/j.cellimm.2008.06.006. [DOI] [PubMed] [Google Scholar]; In this article, the authors show that monkeys that resist SIV infection have enhanced, early NK cell responses, compared to those that succumb.
- 59.Safronetz D., Zivcec M., Lacasse R., Feldmann F., Rosenke R., Long D., Haddock E., Brining D., Gardner D., Feldmann H., et al. Pathogenesis and host response in Syrian hamsters following intranasal infection with Andes virus. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wahl-Jensen V., Chapman J., Asher L., Fisher R., Zimmerman M., Larsen T., Hooper J.W. Temporal analysis of Andes virus and Sin Nombre virus infections of Syrian hamsters. J Virol. 2007;81:7449–7462. doi: 10.1128/JVI.00238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prescott J., Safronetz D., Haddock E., Robertson S., Scott D., Feldmann H. The adaptive immune response does not influence hantavirus disease or persistence in the Syrian hamster. Immunology. 2013;140:168–178. doi: 10.1111/imm.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simons M.J., Gorbunova E.E., Mackow E.R. Unique interferon pathway regulation by the Andes virus nucleocapsid protein is conferred by phosphorylation of serine 386. J Virol. 2019;93 doi: 10.1128/JVI.00338-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brocato R.L., Wahl V., Hammerbeck C.D., Josleyn M.D., McElroy A.K., Smith J.M., Hooper J.W. Innate immune responses elicited by Sin Nombre virus or type I IFN agonists protect hamsters from lethal Andes virus infections. J Gen Virol. 2018;99:1359–1366. doi: 10.1099/jgv.0.001131. [DOI] [PubMed] [Google Scholar]
- 64.Brocato R.L., Hammerbeck C.D., Bell T.M., Wells J.B., Queen L.A., Hooper J.W. A lethal disease model for hantavirus pulmonary syndrome in immunosuppressed Syrian hamsters infected with Sin Nombre virus. J Virol. 2014;88:811–819. doi: 10.1128/JVI.02906-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hammerbeck C.D., Hooper J.W. T cells are not required for pathogenesis in the Syrian hamster model of hantavirus pulmonary syndrome. J Virol. 2011;85:9929–9944. doi: 10.1128/JVI.05356-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou J., Chu H., Li C., Wong B.H., Cheng Z.S., Poon V.K., Sun T., Lau C.C., Wong K.K., Chan J.Y., et al. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J Infect Dis. 2014;209:1331–1342. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lau S.K.P., Lau C.C.Y., Chan K.H., Li C.P.Y., Chen H., Jin D.Y., Chan J.F.W., Woo P.C.Y., Yuen K.Y. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: implications for pathogenesis and treatment. J Gen Virol. 2013;94:2679–2690. doi: 10.1099/vir.0.055533-0. [DOI] [PubMed] [Google Scholar]
- 68.Alosaimi B., Hamed M.E., Naeem A., Alsharef A.A., AlQahtani S.Y., AlDosari K.M., Alamri A.A., Al-Eisa K., Khojah T., Assiri A.M., et al. MERS-CoV infection is associated with downregulation of genes encoding Th1 and Th2 cytokines/chemokines and elevated inflammatory innate immune response in the lower respiratory tract. Cytokine. 2020;126 doi: 10.1016/j.cyto.2019.154895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheung C.Y., Poon L.L., Ng I.H., Luk W., Sia S.F., Wu M.H., Chan K.H., Yuen K.Y., Gordon S., Guan Y., et al. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J Virol. 2005;79:7819–7826. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Faure E., Poissy J., Goffard A., Fournier C., Kipnis E., Titecat M., Bortolotti P., Martinez L., Dubucquoi S., Dessein R., et al. Distinct immune response in two MERS-CoV-infected patients: can we go from bench to bedside? PLoS One. 2014;9 doi: 10.1371/journal.pone.0088716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.National Research Project for SARS Beijing Group The involvement of natural killer cells in the pathogenesis of severe acute respiratory syndrome. Am J Clin Pathol. 2004;121:507–511. doi: 10.1309/WPK7Y2XKNF4CBF3R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leist S.R., Jensen K.L., Baric R.S., Sheahan T.P. Increasing the translation of mouse models of MERS coronavirus pathogenesis through kinetic hematological analysis. PLoS One. 2019;14 doi: 10.1371/journal.pone.0220126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yasui F., Kohara M., Kitabatake M., Nishiwaki T., Fujii H., Tateno C., Yoneda M., Morita K., Matsushima K., Koyasu S., et al. Phagocytic cells contribute to the antibody-mediated elimination of pulmonary-infected SARS coronavirus. Virology. 2014;454-455:157–168. doi: 10.1016/j.virol.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Glass W.G., Subbarao K., Murphy B., Murphy P.M. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J Immunol. 2004;173:4030–4039. doi: 10.4049/jimmunol.173.6.4030. [DOI] [PubMed] [Google Scholar]
- 76.Slater H: FDA Accepts IND for NK Cell Therapy CYNK-001 to Treat Patients with COVID-19. https://http://www.cancernetwork.com/immuno-oncology/fda-accepts-ind-nk-cell-therapy-cynk-001-treat-patients-covid-19.
- 77.CYTOVIA Therapeutics and MACROMOLTEK to Develop Dual-Acting Natural Killer Immunotherapy Against SARS CoV2 (COVID-19). https://http://www.globenewswire.com/news-release/2020/04/07/2012885/0/en/CYTOVIA-Therapeutics-and-MACROMOLTEK-to-Develop-Dual-Acting-Natural-Killer-Immunotherapy-Against-SARS-CoV2-COVID-19.html 2020.
- 78.Lubaki N.M., Younan P., Santos R.I., Meyer M., Iampietro M., Koup R.A., Bukreyev A. The Ebola interferon inhibiting domains attenuate and dysregulate cell-mediated immune responses. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79••.Warfield K.L., Perkins J.G., Swenson D.L., Deal E.M., Bosio C.M., Aman M.J., Yokoyama W.M., Young H.A., Bavari S. Role of natural killer cells in innate protection against lethal ebola virus infection. J Exp Med. 2004;200:169–179. doi: 10.1084/jem.20032141. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this article, the authors show that NK cells primed with Ebola virus like particles completely protect mice from lethal Ebola infection.
- 80.Leroy E.M., Baize S., Volchkov V.E., Fisher-Hoch S.P., Georges-Courbot M.C., Lansoud-Soukate J., Capron M., Debre P., McCormick J.B., Georges A.J. Human asymptomatic Ebola infection and strong inflammatory response. Lancet. 2000;355:2210–2215. doi: 10.1016/s0140-6736(00)02405-3. [DOI] [PubMed] [Google Scholar]
- 81.Baize S., Leroy E.M., Georges A.J., Georges-Courbot M.C., Capron M., Bedjabaga I., Lansoud-Soukate J., Mavoungou E. Inflammatory responses in Ebola virus-infected patients. Clin Exp Immunol. 2002;128:163–168. doi: 10.1046/j.1365-2249.2002.01800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wauquier N., Becquart P., Padilla C., Baize S., Leroy E.M. Human fatal Zaire Ebola virus infection is associated with an aberrant innate immunity and with massive lymphocyte apoptosis. PLoS Negl Trop Dis. 2010;4:e837. doi: 10.1371/journal.pntd.0000837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fuller C.L., Ruthel G., Warfield K.L., Swenson D.L., Bosio C.M., Aman M.J., Bavari S. NKp30-dependent cytolysis of filovirus-infected human dendritic cells. Cell Microbiol. 2007;9:962–976. doi: 10.1111/j.1462-5822.2006.00844.x. [DOI] [PubMed] [Google Scholar]
- 84.Edri A., Shemesh A., Iraqi M., Matalon O., Brusilovsky M., Hadad U., Radinsky O., Gershoni-Yahalom O., Dye J.M., Mandelboim O., et al. The Ebola-glycoprotein modulates the function of natural killer cells. Front Immunol. 2018;9:1428. doi: 10.3389/fimmu.2018.01428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fausther-Bovendo H., Qiu X., He S., Bello A., Audet J., Ippolito G., Wong G., Kobinger G. NK cells accumulate in infected tissues and contribute to pathogenicity of Ebola virus in mice. J Virol. 2019;93:e01703–01718. doi: 10.1128/JVI.01703-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Villinger F., Rollin P.E., Brar S.S., Chikkala N.F., Winter J., Sundstrom J.B., Zaki S.R., Swanepoel R., Ansari A.A., Peters C.J. Markedly elevated levels of interferon (IFN)-gamma, IFN-alpha, interleukin (IL)-2, IL-10, and tumor necrosis factor-alpha associated with fatal Ebola virus infection. J Infect Dis. 1999;179(Suppl. 1):S188–191. doi: 10.1086/514283. [DOI] [PubMed] [Google Scholar]
- 87.Baize S., Leroy E.M., Georges-Courbot M.C., Capron M., Lansoud-Soukate J., Debré P., Fisher-Hoch S.P., McCormick J.B., Georges A.J. Defective humoral responses and extensive intravascular apoptosis are associated with fatal outcome in Ebola virus-infected patients. Nat Med. 1999;5:423–426. doi: 10.1038/7422. [DOI] [PubMed] [Google Scholar]
- 88.Piersma S.J., Pak-Wittel M.A., Lin A., Plougastel-Douglas B., Yokoyama W.M. Activation receptor–dependent IFN-γ production by NK cells is controlled by transcription, translation, and the proteasome. J Immunol. 2019;203:1981–1988. doi: 10.4049/jimmunol.1900718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89••.Costa V.V., Ye W., Chen Q., Teixeira M.M., Preiser P., Ooi E.E., Chen J. Dengue virus-infected dendritic cells, but not monocytes, activate natural killer cells through a contact-dependent mechanism involving adhesion molecules. mBio. 2017;8 doi: 10.1128/mBio.00741-17. [DOI] [PMC free article] [PubMed] [Google Scholar]; Costa et al. show that NK cells provide early control against Dengue infection, and that NK cell activation by Dengue virus-infected DCs is contact dependent.
- 90.Naiyer M.M., Cassidy S.A., Magri A., Cowton V., Chen K., Mansour S., Kranidioti H., Mbiribindi B., Rettman P., Harris S., et al. KIR2DS2 recognizes conserved peptides derived from viral helicases in the context of HLA-C. Sci Immunol. 2017;2 doi: 10.1126/sciimmunol.aal5296. eaal5296. [DOI] [PubMed] [Google Scholar]
- 91.Hershkovitz O., Rosental B., Rosenberg L.A., Navarro-Sanchez M.E., Jivov S., Zilka A., Gershoni-Yahalom O., Brient-Litzler E., Bedouelle H., Ho J.W., et al. NKp44 receptor mediates interaction of the envelope glycoproteins from the West Nile and dengue viruses with NK cells. J Immunol. 2009;183:2610–2621. doi: 10.4049/jimmunol.0802806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kurane I., Ennis F.A. Production of interferon alpha by dengue virus-infected human monocytes. J Gen Virol. 1988;69:445–449. doi: 10.1099/0022-1317-69-2-445. [DOI] [PubMed] [Google Scholar]
- 93.Kurane I., Innis B.L., Nimmannitya S., Nisalak A., Meager A., Ennis F.A. High levels of interferon alpha in the sera of children with dengue virus infection. Am J Trop Med Hyg. 1993;48:222–229. doi: 10.4269/ajtmh.1993.48.222. [DOI] [PubMed] [Google Scholar]
- 94.Sudiro T.M., Zivny J., Ishiko H., Green S., Vaughn D.W., Kalayanarooj S., Nisalak A., Norman J.E., Ennis F.A., Rothman A.L. Analysis of plasma viral RNA levels during acute dengue virus infection using quantitative competitor reverse transcription-polymerase chain reaction. J Med Virol. 2001;63:29–34. [PubMed] [Google Scholar]
- 95.Petitdemange C., Wauquier N., Devilliers H., Yssel H., Mombo I., Caron M., Nkoghe D., Debre P., Leroy E., Vieillard V. Longitudinal analysis of natural killer cells in dengue virus-infected patients in comparison to chikungunya and chikungunya/dengue virus-infected patients. PLoS Negl Trop Dis. 2016;10 doi: 10.1371/journal.pntd.0004499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mathew A. Defining the role of NK cells during dengue virus infection. Immunology. 2018;154:557–562. doi: 10.1111/imm.12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McKechnie J.L., Beltrán D., Pitti A., Saenz L., Araúz A.B., Vergara R., Harris E., Lanier L.L., Blish C.A., López-Vergès S. HLA upregulation during dengue virus infection suppresses the natural killer cell response. Front Cell Infect Microbiol. 2019;9:268. doi: 10.3389/fcimb.2019.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Beltrame L.M., Sell A.M., Moliterno R.A., Clementino S.L., Cardozo D.M., Dalalio M.M., Fonzar U.J., Visentainer J.E. Influence of KIR genes and their HLA ligands in susceptibility to dengue in a population from southern Brazil. Tissue Antigens. 2013;82:397–404. doi: 10.1111/tan.12256. [DOI] [PubMed] [Google Scholar]
- 99.Townsley E., O’Connor G., Cosgrove C., Woda M., Co M., Thomas S.J., Kalayanarooj S., Yoon I.K., Nisalak A., Srikiatkhachorn A., et al. Interaction of a dengue virus NS1-derived peptide with the inhibitory receptor KIR3DL1 on natural killer cells. Clin Exp Immunol. 2016;183:419–430. doi: 10.1111/cei.12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Simmons C.P., Farrar J.J., van Vinh Chau N., Wills B. Dengue. New Engl J Med. 2012;366:1423–1432. doi: 10.1056/NEJMra1110265. [DOI] [PubMed] [Google Scholar]
- 101.Rothman A.L. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat Rev Immunol. 2011;11:532–543. doi: 10.1038/nri3014. [DOI] [PubMed] [Google Scholar]
- 102.Green S., Pichyangkul S., Vaughn D.W., Kalayanarooj S., Nimmannitya S., Nisalak A., Kurane I., Rothman A.L., Ennis F.A. Early CD69 expression on peripheral blood lymphocytes from children with dengue hemorrhagic fever. J Infect Dis. 1999;180:1429–1435. doi: 10.1086/315072. [DOI] [PubMed] [Google Scholar]
- 103.Baize S., Kaplon J., Faure C., Pannetier D., Georges-Courbot M.C., Deubel V. Lassa virus infection of human dendritic cells and macrophages is productive but fails to activate cells. J Immunol. 2004;172:2861–2869. doi: 10.4049/jimmunol.172.5.2861. [DOI] [PubMed] [Google Scholar]
- 104.Russier M., Reynard S., Tordo N., Baize S. NK cells are strongly activated by Lassa and Mopeia virus-infected human macrophages in vitro but do not mediate virus suppression. Eur J Immunol. 2012;42:1822–1832. doi: 10.1002/eji.201142099. [DOI] [PubMed] [Google Scholar]
- 105.Wauquier N., Petitdemange C., Tarantino N., Maucourant C., Coomber M., Lungay V., Bangura J., Debre P., Vieillard V. HLA-C-restricted viral epitopes are associated with an escape mechanism from KIR2DL2(+) NK cells in Lassa virus infection. EBioMedicine. 2019;40:605–613. doi: 10.1016/j.ebiom.2019.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Baize S., Marianneau P., Loth P., Reynard S., Journeaux A., Chevallier M., Tordo N., Deubel V., Contamin H. Early and strong immune responses are associated with control of viral replication and recovery in lassa virus-infected cynomolgus monkeys. J Virol. 2009;83:5890–5903. doi: 10.1128/JVI.01948-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stokstad E. Taming rabies. Science. 2017;355:238–242. doi: 10.1126/science.355.6322.238. [DOI] [PubMed] [Google Scholar]
- 108.Overduin L.A., van Dongen J.J.M., Visser L.G. The cellular immune response to rabies vaccination: a systematic review. Vaccines (Basel) 2019;7 doi: 10.3390/vaccines7030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109••.Horowitz A., Behrens R.H., Okell L., Fooks A.R., Riley E.M. NK cells as effectors of acquired immune responses: effector CD4+ T cell-dependent activation of NK cells following vaccination. J Immunol. 2010;185:2808–2818. doi: 10.4049/jimmunol.1000844. [DOI] [PubMed] [Google Scholar]; Horowitz et al. demonstrate that NK cell-mediated ADCC is crucial for protection against rabies virus challenge in rabies-vaccinated mice.
- 110.Plummer F.A., Jones S.M. The story of Canada’s Ebola vaccine. CMAJ. 2017;189:E1326–e1327. doi: 10.1503/cmaj.170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.U.S. Food and Drug Administration . 2019. Ervebo (Ebola Zaire Vaccine, Live) STN: 125690. [Google Scholar]
- 112.European Medicines Agency . 2019. Ervebo (Ebola Zaire Vaccine [rVSVΔG-ZEBOV-GP, live]), EMA/615676/2019. [Google Scholar]
- 113.Rechtien A., Richert L., Lorenzo H., Martrus G., Hejblum B., Dahlke C., Kasonta R., Zinser M., Stubbe H., Matschl U., et al. Systems vaccinology identifies an early innate immune signature as a correlate of antibody responses to the Ebola vaccine rVSV-ZEBOV. Cell Rep. 2017;20:2251–2261. doi: 10.1016/j.celrep.2017.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wagstaffe H.R., Clutterbuck E.A., Bockstal V., Stoop J.N., Luhn K., Douoguih M., Shukarev G., Snape M.D., Pollard A.J., Riley E.M., et al. Antibody-dependent natural killer cell activation after Ebola vaccination. J Infect Dis. 2019 doi: 10.1093/infdis/jiz657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115••.Abreu-Mota T., Hagen K.R., Cooper K., Jahrling P.B., Tan G., Wirblich C., Johnson R.F., Schnell M.J. Non-neutralizing antibodies elicited by recombinant Lassa-Rabies vaccine are critical for protection against Lassa fever. Nat Commun. 2018;9 doi: 10.1038/s41467-018-06741-w. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, the authors show that non-neutralizing antibodies generated via vaccination confer protection to Lassa virus via NK cell-mediated ADCC.
- 116.Keshwara R., Hagen K.R., Abreu-Mota T., Papaneri A.B., Liu D., Wirblich C., Johnson R.F., Schnell M.J. A recombinant rabies virus expressing the Marburg virus glycoprotein is dependent upon antibody-mediated cellular cytotoxicity for protection against Marburg virus disease in a murine model. J Virol. 2019;93 doi: 10.1128/JVI.01865-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Spiegel M., Pichlmair A., Martinez-Sobrido L., Cros J., Garcia-Sastre A., Haller O., Weber F. Inhibition of Beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J Virol. 2005;79:2079–2086. doi: 10.1128/JVI.79.4.2079-2086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Edwards M.R., Liu G., Mire C.E., Sureshchandra S., Luthra P., Yen B., Shabman R.S., Leung D.W., Messaoudi I., Geisbert T.W., et al. Differential regulation of interferon responses by Ebola and Marburg virus VP35 proteins. Cell Rep. 2016;14:1632–1640. doi: 10.1016/j.celrep.2016.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fernando L., Qiu X., Melito P.L., Williams K.J.N., Feldmann F., Feldmann H., Jones S.M., Alimonti J.B. Immune response to Marburg virus Angola infection in nonhuman primates. J Infect Dis. 2015;212:S234–S241. doi: 10.1093/infdis/jiv095. [DOI] [PubMed] [Google Scholar]
- 121.Blom K., Braun M., Pakalniene J., Lunemann S., Enqvist M., Dailidyte L., Schaffer M., Lindquist L., Mickiene A., Michaëlsson J., et al. NK cell responses to human tick-borne encephalitis virus infection. J Immunol. 2016;197:2762–2771. doi: 10.4049/jimmunol.1600950. [DOI] [PubMed] [Google Scholar]
- 122.Robertson S.J., Mitzel D.N., Taylor R.T., Best S.M., Bloom M.E. Tick-borne flaviviruses: dissecting host immune responses and virus countermeasures. Immunol Res. 2009;43:172–186. doi: 10.1007/s12026-008-8065-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tomazic J., Ihan A. Flow cytometric analysis of lymphocytes in cerebrospinal fluid in patients with tick-borne encephalitis. Acta Neurol Scand. 1997;95:29–33. doi: 10.1111/j.1600-0404.1997.tb00064.x. [DOI] [PubMed] [Google Scholar]
- 124.Toczylowski K., Grygorczuk S., Osada J., Wojtkowska M., Bojkiewicz E., Wozinska-Klepadlo M., Potocka P., Sulik A. Evaluation of cerebrospinal fluid CXCL13 concentrations and lymphocyte subsets in tick-borne encephalitis. Int J Infect Dis. 2020;93:40–47. doi: 10.1016/j.ijid.2020.01.023. [DOI] [PubMed] [Google Scholar]
- 125.Fredericksen B.L., Gale M., Jr West Nile virus evades activation of interferon regulatory factor 3 through RIG-I-dependent and -independent pathways without antagonizing host defense signaling. J Virol. 2006;80:2913–2923. doi: 10.1128/JVI.80.6.2913-2923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cheng Y., King N.J.C., Kesson A.M. Major histocompatibility complex Class I (MHC-I) induction by West Nile Virus: involvement of 2 signaling pathways in MHC-I up-regulation. J Infect Dis. 2004;189:658–668. doi: 10.1086/381501. [DOI] [PubMed] [Google Scholar]
- 127.Zhang M., Daniel S., Huang Y., Chancey C., Huang Q., Lei Y.F., Grinev A., Mostowski H., Rios M., Dayton A. Anti-West Nile virus activity of in vitro expanded human primary natural killer cells. BMC Immunol. 2010;11:3. doi: 10.1186/1471-2172-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yao Y., Strauss-Albee D.M., Zhou J.Q., Malawista A., Garcia M.N., Murray K.O., Blish C.A., Montgomery R.R. The natural killer cell response to West Nile virus in young and old individuals with or without a prior history of infection. PLoS One. 2017;12 doi: 10.1371/journal.pone.0172625. e0172625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Strauss-Albee D.M., Fukuyama J., Liang E.C., Yao Y., Jarrell J.A., Drake A.L., Kinuthia J., Montgomery R.R., John-Stewart G., Holmes S., et al. Human NK cell repertoire diversity reflects immune experience and correlates with viral susceptibility. Sci Transl Med. 2015;7 doi: 10.1126/scitranslmed.aac5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bai F., Thompson E.A., Vig P.J.S., Leis A.A. Current understanding of west Nile virus clinical manifestations, immune responses, neuroinvasion, and immunotherapeutic implications. Pathogens. 2019;8 doi: 10.3390/pathogens8040193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Laurent-Rolle M., Morrison J., Rajsbaum R., Macleod J.M.L., Pisanelli G., Pham A., Ayllon J., Miorin L., Martinez C., tenOever B.R., et al. The interferon signaling antagonist function of yellow fever virus NS5 protein is activated by type I interferon. Cell Host Microbe. 2014;16:314–327. doi: 10.1016/j.chom.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Neves P.C., Matos D.C., Marcovistz R., Galler R. TLR expression and NK cell activation after human yellow fever vaccination. Vaccine. 2009;27:5543–5549. doi: 10.1016/j.vaccine.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 133.Marquardt N., Ivarsson M.A., Blom K., Gonzalez V.D., Braun M., Falconer K., Gustafsson R., Fogdell-Hahn A., Sandberg J.K., Michaelsson J. The human NK cell response to Yellow Fever Virus 17D is primarily governed by NK cell differentiation independently of NK cell education. J Immunol. 2015;195:3262–3272. doi: 10.4049/jimmunol.1401811. [DOI] [PubMed] [Google Scholar]
- 134.Lam L.K.M., Watson A.M., Ryman K.D., Klimstra W.B. Gamma-interferon exerts a critical early restriction on replication and dissemination of yellow fever virus vaccine strain 17D-204. NPJ Vaccines. 2018;3:5. doi: 10.1038/s41541-017-0039-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Douam F., Ziegler C.G.K., Hrebikova G., Fant B., Leach R., Parsons L., Wang W., Gaska J.M., Winer B.Y., Heller B., et al. Selective expansion of myeloid and NK cells in humanized mice yields human-like vaccine responses. Nat Commun. 2018;9 doi: 10.1038/s41467-018-07478-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Quaresma J.A., Barros V.L., Pagliari C., Fernandes E.R., Guedes F., Takakura C.F., Andrade H.F., Jr, Vasconcelos P.F., Duarte M.I. Revisiting the liver in human yellow fever: virus-induced apoptosis in hepatocytes associated with TGF-beta, TNF-alpha and NK cells activity. Virology. 2006;345:22–30. doi: 10.1016/j.virol.2005.09.058. [DOI] [PubMed] [Google Scholar]
- 137.Grant A., Ponia S.S., Tripathi S., Balasubramaniam V., Miorin L., Sourisseau M., Schwarz M.C., Sanchez-Seco M.P., Evans M.J., Best S.M., et al. Zika Virus targets human STAT2 to inhibit Type I interferon signaling. Cell Host Microbe. 2016;19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Matusali G., Houzet L., Satie A.P., Mahe D., Aubry F., Couderc T., Frouard J., Bourgeau S., Bensalah K., Lavoue S., et al. Zika virus infects human testicular tissue and germ cells. J Clin Invest. 2018;128:4697–4710. doi: 10.1172/JCI121735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Glasner A., Oiknine-Djian E., Weisblum Y., Diab M., Panet A., Wolf D.G., Mandelboim O. Zika virus escapes NK cell detection by upregulating major histocompatibility complex class I molecules. J Virol. 2017;91 doi: 10.1128/JVI.00785-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Aid M., Abbink P., Larocca R.A., Boyd M., Nityanandam R., Nanayakkara O., Martinot A.J., Moseley E.T., Blass E., Borducchi E.N., et al. Zika virus persistence in the central nervous system and lymph nodes of rhesus monkeys. Cell. 2017;169:610–620. doi: 10.1016/j.cell.2017.04.008. e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dudley D.M., Aliota M.T., Mohr E.L., Weiler A.M., Lehrer-Brey G., Weisgrau K.L., Mohns M.S., Breitbach M.E., Rasheed M.N., Newman C.M., et al. A rhesus macaque model of Asian-lineage Zika virus infection. Nat Commun. 2016;7 doi: 10.1038/ncomms12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Osuna C.E., Lim S.Y., Deleage C., Griffin B.D., Stein D., Schroeder L.T., Omange R.W. Zika viral dynamics and shedding in rhesus and cynomolgus macaques. Nat Med. 2016;22:1448–1455. doi: 10.1038/nm.4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.O’Connor M.A., Tisoncik-Go J., Lewis T.B., Miller C.J., Bratt D., Moats C.R., Edlefsen P.T., Smedley J., Klatt N.R., Gale M., et al. Early cellular innate immune responses drive Zika viral persistence and tissue tropism in pigtail macaques. Nat Commun. 2018;9 doi: 10.1038/s41467-018-05826-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Silveira E.L.V., Rogers K.A., Gumber S., Amancha P., Xiao P., Woollard S.M., Byrareddy S.N., Teixeira M.M., Villinger F. Immune Cell dynamics in rhesus macaques infected with a Brazilian strain of Zika virus. J Immunol. 2017;199:1003–1011. doi: 10.4049/jimmunol.1700256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hayashida E., Ling Z.L., Ashhurst T.M., Viengkhou B., Jung S.R., Songkhunawej P., West P.K., King N.J.C., Hofer M.J. Zika virus encephalitis in immunocompetent mice is dominated by innate immune cells and does not require T or B cells. J Neuroinflammation. 2019;16:177. doi: 10.1186/s12974-019-1566-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Azevedo R.S.S., de Sousa J.R., Araujo M.T.F., Martins Filho A.J., de Alcantara B.N., Araujo F.M.C., Queiroz M.G.L., Cruz A.C.R., Vasconcelos B.H.B., Chiang J.O., et al. In situ immune response and mechanisms of cell damage in central nervous system of fatal cases microcephaly by Zika virus. Sci Rep. 2018;8:1. doi: 10.1038/s41598-017-17765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Welch S.R., Ritter J.M., McElroy A.K., Harmon J.R., Coleman-McCray J.D., Scholte F.E.M., Kobinger G.P., Bergeron É, Zaki S.R., Nichol S.T., et al. Fluorescent Crimean-Congo hemorrhagic fever virus illuminates tissue tropism patterns and identifies early mononuclear phagocytic cell targets in Ifnar-/- mice. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1008183. e1008183-e1008183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bente D.A., Alimonti J.B., Shieh W.-J., Camus G., Ströher U., Zaki S., Jones S.M. Pathogenesis and immune response of crimean-Congo hemorrhagic fever virus in a STAT-1 knockout mouse model. J Virol. 2010;84:11089–11100. doi: 10.1128/JVI.01383-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yilmaz M., Aydin K., Akdogan E., Sucu N., Sonmez M., Omay S.B., Koksal I. Peripheral blood natural killer cells in Crimean-Congo hemorrhagic fever. J Clin Virol. 2008;42:415–417. doi: 10.1016/j.jcv.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 150.Akinci E., Yilmaz M., Bodur H., Onguru P., Bayazit F.N., Erbay A., Ozet G. Analysis of lymphocyte subgroups in Crimean-Congo hemorrhagic fever. Int J Infect Dis. 2009;13:560–563. doi: 10.1016/j.ijid.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 151.Penski N., Härtle S., Rubbenstroth D., Krohmann C., Ruggli N., Schusser B., Pfann M., Reuter A., Gohrbandt S., Hundt J., et al. Highly pathogenic avian influenza viruses do not inhibit interferon synthesis in infected chickens but can override the interferon-induced antiviral state. J Virol. 2011;85:7730–7741. doi: 10.1128/JVI.00063-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.To K.K., Hung I.F., Li I.W., Lee K.L., Koo C.K., Yan W.W., Liu R., Ho K.Y., Chu K.H., Watt C.L., et al. Delayed clearance of viral load and marked cytokine activation in severe cases of pandemic H1N1 2009 influenza virus infection. Clin Infect Dis. 2010;50:850–859. doi: 10.1086/650581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Koutsakos M., McWilliam H.E.G., Aktepe T.E., Fritzlar S., Illing P.T., Mifsud N.A., Purcell A.W., Rockman S., Reading P.C., Vivian J.P., et al. Downregulation of MHC class I expression by influenza A and B viruses. Front Immunol. 2019;10:1158. doi: 10.3389/fimmu.2019.01158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jansen C.A., de Geus E.D., van Haarlem D.A., van de Haar P.M., Löndt B.Z., Graham S.P., Göbel T.W., van Eden W., Brookes S.M., Vervelde L. Differential lung NK cell responses in avian influenza virus infected chickens correlate with pathogenicity. Sci Rep. 2013;3 doi: 10.1038/srep02478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Achdout H., Meningher T., Hirsh S., Glasner A., Bar-On Y., Gur C., Porgador A., Mendelson M., Mandelboim M., Mandelboim O. Killing of avian and swine influenza virus by natural killer cells. J Virol. 2010;84:3993–4001. doi: 10.1128/JVI.02289-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Scharenberg M., Vangeti S., Kekäläinen E., Bergman P., Al-Ameri M., Johansson N., Sondén K., Falck-Jones S., Färnert A., Ljunggren H.-G., et al. Influenza A virus infection induces hyperresponsiveness in human lung tissue-resident and peripheral blood NK cells. Front Immunol. 2019;10 doi: 10.3389/fimmu.2019.01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Abdul-Careem M.F., Mian M.F., Yue G., Gillgrass A., Chenoweth M.J., Barra N.G., Chew M.V., Chan T., Al-Garawi A.A., Jordana M., et al. Critical role of natural killer cells in lung immunopathology during influenza infection in mice. J Infect Dis. 2012;206:167–177. doi: 10.1093/infdis/jis340. [DOI] [PubMed] [Google Scholar]
- 158.Zhou G., Juang S.W., Kane K.P. NK cells exacerbate the pathology of influenza virus infection in mice. Eur J Immunol. 2013;43:929–938. doi: 10.1002/eji.201242620. [DOI] [PubMed] [Google Scholar]
- 159.Basler C.F. Nipah and hendra virus interactions with the innate immune system. Curr Top Microbiol Immunol. 2012;359:123–152. doi: 10.1007/82_2012_209. [DOI] [PubMed] [Google Scholar]
- 160.Lo M.K., Miller D., Aljofan M., Mungall B.A., Rollin P.E., Bellini W.J., Rota P.A. Characterization of the antiviral and inflammatory responses against Nipah virus in endothelial cells and neurons. Virology. 2010;404:78–88. doi: 10.1016/j.virol.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 161.Prasad A.N., Woolsey C., Geisbert J.B., Agans K.N., Borisevich V., Deer D.J., Mire C.E., Cross R.W., Fenton K.A., Broder C.C., et al. Resistance of Cynomolgus monkeys to Nipah and Hendra virus disease is associated with cell-mediated and humoral immunity. J Infect Dis. 2020;221:S436–S447. doi: 10.1093/infdis/jiz613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lara A., Cong Y., Jahrling P.B., Mednikov M., Postnikova E., Yu S., Munster V., Holbrook M.R. Peripheral immune response in the African green monkey model following Nipah-Malaysia virus exposure by intermediate-size particle aerosol. PLoS Negl Trop Dis. 2019;13 doi: 10.1371/journal.pntd.0007454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chen X., Ye H., Li S., Jiao B., Wu J., Zeng P., Chen L. Severe fever with thrombocytopenia syndrome virus inhibits exogenous Type I IFN signaling pathway through its NSs invitro. PLoS One. 2017;12 doi: 10.1371/journal.pone.0172744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Moriyama M., Igarashi M., Koshiba T., Irie T., Takada A., Ichinohe T. Two conserved amino acids within the NSs of severe fever with thrombocytopenia syndrome phlebovirus are essential for anti-interferon activity. J Virol. 2018;92:e00706–00718. doi: 10.1128/JVI.00706-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Choi Y., Park S.-J., Sun Y., Yoo J.-S., Pudupakam R.S., Foo S.-S., Shin W.-J., Chen S.B., Tsichlis P.N., Lee W.-J., et al. Severe fever with thrombocytopenia syndrome phlebovirus non-structural protein activates TPL2 signalling pathway for viral immunopathogenesis. Nat Microbiol. 2019;4:429–437. doi: 10.1038/s41564-018-0329-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liu J., Wang L., Feng Z., Geng D., Sun Y., Yuan G. Dynamic changes of laboratory parameters and peripheral blood lymphocyte subsets in severe fever with thrombocytopenia syndrome patients. Int J Infect Dis. 2017;58:45–51. doi: 10.1016/j.ijid.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 167.Bouloy M., Janzen C., Vialat P., Khun H., Pavlovic J., Huerre M., Haller O. Genetic evidence for an interferon-antagonistic function of Rift Valley Fever Virus nonstructural protein NSs. J Virol. 2001;75:1371–1377. doi: 10.1128/JVI.75.3.1371-1377.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.McElroy A.K., Nichol S.T. Rift Valley fever virus inhibits a pro-inflammatory response in experimentally infected human monocyte derived macrophages and a pro-inflammatory cytokine response may be associated with patient survival during natural infection. Virology. 2012;422:6–12. doi: 10.1016/j.virol.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Le May N., Mansuroglu Z., Leger P., Josse T., Blot G., Billecocq A., Flick R., Jacob Y., Bonnefoy E., Bouloy M. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS Pathog. 2008;4:e13. doi: 10.1371/journal.ppat.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lathan R., Simon-Chazottes D., Jouvion G., Godon O., Malissen M., Flamand M., Bruhns P., Panthier J.J. Innate immune basis for Rift Valley fever susceptibility in mouse models. Sci Rep. 2017;7 doi: 10.1038/s41598-017-07543-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Harmon J.R., Spengler J.R., Coleman-McCray J.D., Nichol S.T., Spiropoulou C.F., McElroy A.K. CD4 T cells, CD8 T cells, and monocytes coordinate to prevent Rift valley fever virus encephalitis. Journal of Virology. 2018;92:e01270–01218. doi: 10.1128/JVI.01270-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Scott T.P., Nel L.H. Subversion of the immune response by rabies virus. Viruses. 2016;8 doi: 10.3390/v8080231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Maucourant C., Petitdemange C., Yssel H., Vieillard V. Control of acute Arboviral infection by natural killer cells. Viruses. 2019;11 doi: 10.3390/v11020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Petitdemange C., Wauquier N., Jacquet J.M., Theodorou I., Leroy E., Vieillard V. Association of HLA class-I and inhibitory KIR genotypes in Gabonese patients infected by chikungunya or dengue type-2 viruses. PLoS One. 2014;9 doi: 10.1371/journal.pone.0108798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Thanapati S., Das R., Tripathy A.S. Phenotypic and functional analyses of NK and NKT-like populations during the early stages of chikungunya infection. Front Microbiol. 2015;6:895. doi: 10.3389/fmicb.2015.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Teo T.H., Her Z., Tan J.J., Lum F.M., Lee W.W., Chan Y.H., Ong R.Y., Kam Y.W., Leparc-Goffart I., Gallian P., et al. Caribbean and la reunion chikungunya virus isolates differ in their capacity to induce proinflammatory Th1 and NK cell responses and acute joint pathology. J Virol. 2015;89:7955–7969. doi: 10.1128/JVI.00909-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mehand M.S., Millett P., Al-Shorbaji F., Roth C., Kieny M.P., Murgue B. World Health Organization methodology to prioritize emerging infectious diseases in need of research and development. Emerg Infect Dis. 2018;24 doi: 10.3201/eid2409.171427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Centers for Disease Control and Prevention: Zoonotic Diseases. http://www.cdc.gov/onehealth/basics/zoonotic-diseases.html.
- 179.World Health Organization: Zoonoses. http://www.who.int/topics/zoonoses/en/.