Abstract
Breast Cancer Stem Cells (BCSC) are competent to initiate tumor formation and growth, refractory to conventional therapies, and consequently are implicated in tumor recurrence. Many signaling cascades associated with BCSCs are critical for epithelial-to-mesenchymal transition (EMT). We developed a model system to mechanistically examine BCSCs in basal-like breast cancer using MCF10AT1 FACS sorted for CD24 (negative/low in BCSCs) and CD44 (positive/high in BCSCs). Ingenuity Pathway Analysis comparing RNAseq on the CD24-/low versus CD24+/high MCF10AT1 indicates that the top activated upstream regulators include TWIST1, TGFB1, OCT4, and other factors known to be increased in BCSCs and during EMT. The top inhibited upstream regulators include ESR1, TP63, and FAS. Consistent with our results, many genes previously demonstrated to be regulated by RUNX factors are altered in BCSCs. The RUNX2 interaction network is the top significant pathway altered between CD24-/low and CD24+/high MCF10AT1. RUNX1 is higher in expression at the RNA level than RUNX2. RUNX3 is not expressed. While, human specific qPCR primers demonstrate that RUNX1 and CDH1 decrease in human MCF10CA1a cells that have grown tumors within the murine mammary fat pad microenvironment, RUNX2 and VIM increase. Treatment with an inhibitor of RUNX binding to CBFβ for five days followed by a seven-day recovery period results in EMT suggesting that loss of RUNX1, rather than increase in RUNX2, is a driver of EMT in early stage breast cancer. Increased understanding of RUNX regulation on BCSCs and EMT will provide novel insight into therapeutic strategies to prevent recurrence.
Introduction
Among the heterogeneous population of cells within a tumor, Breast Cancer Stem Cells (BCSCs) are posited to be a small fraction (Chaffer, San Juan, Lim, & Weinberg, 2016; Ming, Michael, & Max, 2015) that are capable of self-renewal and reconstituting the original cellular hierarchy within secondary tumors (Visvader & Lindeman, 2008) (Meacham & Morrison, 2013). BCSCs are highly resistant to conventional therapies and have an increased metastatic potential (Zhao, 2016) (Abdullah & Chow, 2013). The signaling cascades (Notch, WNT, TGFβ, etc) and transcription factors (TWIST, OCT4, SNAI1, ZEB, etc) that regulate stem-like properties in BCSCs control epithelial-to-mesenchymal transition (EMT) (Hadjimichael et al., 2015; G. Li et al., 2018; Scheel & Weinberg, 2012; Shibue & Weinberg, 2017; Singh & Settleman, 2010; Venkatesh et al., 2018). It has been suggested that partial activation of the EMT promotes plasticity that allows reprogramming of the epithelial cell to acquire both migratory and stem-like features (Grigore, Jolly, Jia, Farach-Carson, & Levine, 2016). There is a compelling requirement to increase understanding of the regulatory mechanisms contributing to BCSC physiology.
Recently, our lab and others have demonstrated the importance of the RUNX family of transcription factors in pathways that regulate EMT and BCSCs (Fritz et al., 2019; Hong et al., 2019; D. Hong et al., 2017; Kulkarni et al., 2018; Owens et al., 2014; Valenti et al., 2016). The RUNX proteins consist of RUNX1, RUNX2 and RUNX3. Each of these factors function as key lineage determinants in specific tissues(Y. Ito, S.-C. Bae, & L. S. H. Chuang, 2015). These factors control cell differentiation, proliferation, and the cell cycle during normal development (C. Q. Wang, Jacob, Nah, & Osato, 2010). RUNX1 regulates hematopoietic (Jacob et al., 2010; Yokomizo et al., 2001) (Chelsia Q. Wang et al., 2014), hair follicle (Hoi et al., 2010; Osorio, Lilja, & Tumbar, 2011), gastric (Matsuo et al., 2017) and oral epithelial stem cells (Scheitz, Lee, McDermitt, & Tumbar, 2012). RUNX2 regulates epithelial differentiation by promoting CDH1 in adipose-derived stem cells (Q. Li et al., 2018), and is a crucial regulator of osteogenesis of stem cells (Dalle Carbonare et al., 2019; Javed et al., 2009; Zou et al., 2015).
In the mammary gland, while RUNX2 is critical to maintain the mammary stem/progenitor population (Ferrari et al., 2015), RUNX1 is implicated in luminal development (Sokol et al., 2015). During mammary branching morphogenesis, the level of RUNX2 is increased and accompanied by the upregulation of EMT activators such as SNAI2 (Cao et al., 2017; Ferrari, McDonald, Morris, Cameron, & Blyth, 2013). Overexpressing RUNX2 in mammary epithelial cells activates differentiation and induces EMT (N. O. Chimge et al., 2011; Owens et al., 2014). In contrast, RUNX2 depletion in mouse mammary glands disrupted ductal outgrowth at puberty and progenitor cell differentiation during pregnancy (Ferrari et al., 2015; Owens et al., 2014). These findings establish RUNX factors as obligatory components of physiological control for EMT in biological contexts.
Beyond their impact on normal development, dysregulated RUNX functioning is implicated in cancer (Ito) (Yoshiaki Ito et al., 2015). RUNX1 is frequently translocated (e.g., Runx1-ETO (Hatlen, Wang, & Nimer, 2012), TEL-Runx1 (Fischer et al., 2005) and Runx1-EVI (Mitani et al., 1994)) and mutated (Sood, Kamikubo, & Liu, 2017) in hematopoietic malignancies. Recently, mutations in RUNX1 and CBFB, a critical coregulatory component of RUNX transcription factor complexes, have been shown to be breast cancer drivers. In breast cancer, RUNX1 regulates WNT signaling and crucial transcription factors including ESR1 and ELF5 (Barutcu et al., 2016; N.-O. Chimge et al., 2016; Yoshiaki Ito et al., 2015; van Bragt, Hu, Xie, & Li, 2015). Our group has demonstrated that RUNX1 exhibits tumor suppressor activity and is required to maintain the epithelial phenotype and repress EMT (Deli Hong et al., 2017) and breast cancer stemness (Hong et al., 2018). There is a necessity to elucidate mechanisms underlying RUNX1 influence on BCSCs phenotypes including the impact of compromised RUNX1 regulation for tumor growth in vivo.
While RUNX1 demonstrates tumor suppressor activity, RUNX2 has been well documented to function as an oncogene in breast cancer (Chuang, Ito, & Ito, 2017). Similarly, RUNX2 functions as an oncogene in lymphoma, where it is a frequent target for viral insertion (Stewart et al., 1997). In osteosarcoma, increased RUNX2 expression is also associated with tumorigenicity, metastasis, lower survival and poor prognosis by directly activating the PI3K/AKT pathways (Cohen-Solal, Boregowda, & Lasfar, 2015; Martin, Zielenska, Stein, van Wijnen, & Squire, 2011). Upregulation of RUNX2 has been linked to bone metastasis in breast cancer (N. O. Chimge et al., 2011; Pratap et al., 2005) as well as in multiple epithelial cancer types including colon, prostate, and thyroid cancer (Akech et al., 2010; Cohen-Solal et al., 2015; Niu et al., 2012). RUNX2 contributes to metastatic events through regulation of bone metastatic- related genes, that include osteopontin, bone sialoprotein, matrix metalloproteinases, and activation of signaling pathways including WNT and TGFβ (Pratap et al., 2005).
To mechanistically investigate stemness in basal-like breast cancer, we FACS isolated CD24 (negative/low in BCSCs) and CD44 (positive/high in BCSCs) in MCF10AT1 cells. We observed that many genes previously demonstrated to be regulated by RUNX factors are altered in BCSCs. The RUNX2 interaction network is the top significant pathway altered between CD24-/low and CD24+/high MCF10AT1. While, RUNX1 and CDH1 decrease in human MCF10CA1a cells that have grown tumors within the murine mammary fat pad microenvironment, RUNX2 and VIM increase. Treatment with an inhibitor of RUNX binding to CBFβ for five days followed by a seven-day recovery period results in EMT suggesting that loss of RUNX1, rather than increase in RUNX2, is a driver of EMT in early stage breast cancer. Increased understanding of RUNX regulation on BCSCs and EMT will provide novel insight into therapeutic strategies to prevent recurrence.
Materials and methods
Cell culture:
MCF10AT1 were grown in DMEM: F12 (Hyclone: SH30271, Thermo Fisher Scientific, Waltham, MA) with 5% (v/v) horse serum (Gibco: 16050, Thermo Fisher Scientific, Waltham, MA, USA) + 10 μg/ml human insulin (Sigma Aldrich, St. Louis, MO: I-1882) + 20 ng/ml recombinant hEGF (Peprotech, Rocky Hill, NJ, USA: AF-100–15) + 100 ng/ml cholera toxin (Sigma Aldrich: C-8052) + 0.5 μg/ml hydrocortisone (Sigma Aldrich: H-0888) 50 IU/ml penicillin/50 μg/ml streptomycin and 2 mM glutamine (Life Technologies, Carlsbad, CA, USA: 15140–122 and 25030–081, respectively). MCF10CA1a cells were grown in DMEM: F with 12, 5% (v/v) horse serum with 50 IU/ml penicillin/50 μg/ml streptomycin and 2 mM glutamine.
CD24/CD44 flow cytometry
Flow cytometry for CD24 (PE-cy7, Biolegend 311120) and CD44 (APC, BD Pharmigen 559942) was performed using the optimal conditions as described previously (Fillmore & Kuperwasser, 2008; Hong et al., 2018; Quan, 2013). Gating for negative signal was determined using isotype controls.
RNAseq analysis
Total RNA was isolated using Trizol (Thermo Fisher Scientific, Waltham, MA); (Life Technologies) and purified using the Direct-zol RNA kit (Zymo Research, Irvine, CA: R2050). The quality and quantity of RNA was assessed using the RNA 6000 Nano Kit with the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA quantity was further assessed using a Nanodrop2000 (Thermo Scientific, Lafayette, CO) and Qubit HS RNA assay (Thermo Fisher Scientific). Total RNA was depleted of ribosomal RNA, reverse transcribed using a random hexamer strategy, and strand-specific adapters were added following manufacturer’s protocol (TruSeq Stranded Total RNA Library Prep kit with Ribo-Zero Gold, Illumina, San Diego, CA) with the exception that the final cDNA libraries were amplified using the Real-time Library Amplification Kit (Kapa Biosystems, Wilmington, MA) to reduce over-amplification of libraries. Generated cDNA libraries were assayed for quality using the High Sensitivity DNA Kit on the Agilent 2100 Bioanalyzer (Agilent Technologies) then sequenced as single-end 100 bp reads (IlluminaHiSeq1000, UVM Advanced Genome Technologies Core). Sequencing base calls were generated on the HiSeq 1500 instrument in the UVM Advanced Genome Technologies Core Massively Parallel Sequencing Facility. Fastq conversion and demultiplexing were done using bcl2fastq (Illumina, v1.8.4, San Diego, CA) and evaluated using Fastqc. Sequence files (fastq) were mapped to the most recent assemblies of the human genome (hg38) using TopHat2 (Kim et al., 2013). Expression counts were determined by HTSeq (Anders, Pyl, & Huber, 2015) with recent gene annotations (Gencode v23/v24)(Harrow et al., 2012). Differential expression was analyzed by DESeq2 (Love, Huber, & Anders, 2014). Correlation between replicates and differential gene expression between samples was assessed by principal component analysis (PCA). RNA-Seq datasets have been deposited in the GEO under accession code GSE141503.
Western blotting
Cells were lysed in RIPA buffer and 2X SDS sample buffer supplemented with cOmplete, EDTA-free protease inhibitors (Roche Diagnostics) and MG132 (EMD Millipore San Diego, CA, USA). Lysates were fractionated in an 8.5% acrylamide gel and subjected to immunoblotting. The gels are transferred to PVDF membranes (EMD Millipore) using a wet transfer apparatus (Bio-Rad Laboratories, Hercules, CA, USA). Membranes were blocked using 5% Blotting Grade Blocker Non-Fat Dry Milk (Bio-Rad Laboratories) and incubated overnight at 4°C with the following primary antibodies: a rabbit polyclonal Runx1 (Cell Signaling Technology, Danvers, MA, USA: #4334, 1:1000); a mouse monoclonal to E-cadherin (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA: sc21791, 1:1000); a mouse monoclonal Vimentin (Santa-Cruz Biotechnology sc-6260, 1:1000); a mouse monoclonal to β-Actin (Cell Signaling Technology #3700, 1:1000); a rabbit polyclonal Twist1 (Santa Cruz Biotechnology sc-15393, 1:2000); a rabbit polyclonal Zeb1 (Sigma-Aldrich HPA027524–100UL, 1:1000). Secondary antibodies conjugated to HRP (Santa Cruz Biotechnology) were used for immunodetection, along with the Clarity Western ECL Substrate (Bio-Rad Laboratories) on a Chemidoc XRS+ imaging system (Bio-Rad Laboratories).
Animal studies
Female SCID mice 7 weeks of age were used for mammary fat pad injection. The mice were randomly divided into two groups (seven for each group). In all, 1X106 MCF10CA1a cells suspended in 0.1 ml of saline were mixed with 0.1 ml of Matrigel (BD) and were injected under mammary fat pads. Bioluminescence images were acquired by using the IVIS Imaging System (Xenogen) 5 min after injection 150 mg/kg of D-Luciferin (Gold BioTech, St. Louis, MO) in PBS. All animals were housed in a pathogen-free environment and handled according to protocol number 12–051 approved by the Institutional Animal Care and Use Committee at the University of Vermont. In conducting using animals, the investigators adhere to the laws of the United States and regulations of the Department of Agriculture.
Quantitative PCR
RNA was isolated with Trizol (Life Technologies) and cleaned by DNase digestion (Zymo Research, Irvine, CA, USA). RNA was reversed transcribed using SuperScript II and random hexamers (Life Technologies). cDNA was then subjected to quantitative PCR using SYBR Green technology (Applied Biosystems, Foster City, CA, USA).
Runx1 Forward: AACCCTCAGCCTCAGAGTCA,
Runx1 Reverse: CAATGGATCCCAGGTATTGG;
Runx2 Forward: CAGCCCCAACTTCCTGTG;
Runx2 Reverse: CCGGAGCTCAGCAGAATAAT;
FN1 Forward: CATGAAGGGGGTCAGTCCTA;
FN1 Reverse: CTTCTCAGCTATGGGCTTGC;
VEGF Forward: CCTTGCTGCTCTACCTCCAC;
VEGF Reverse: CCATGAACTTCACCACTTCG;
CXCR4 Forward: TACACCGAGGAAATGGGCTCA;
CXCR4 Reverse: TTCTTCACGGAAACAGGGTTC;
CXCL12 Forward: GTGGTCGTGCTGGTCCTC;
CXCL12 Reverse: AGATGCTTGACGTTGGCTCT;
MMP13 Forward: ATGAGCCAGAGTGTCGGTTC;
MMP13 Reverse: GTTAGTAGCGACGAGCAGGAC;
MMP9 Forward: ATAGACTACTACAGGCT;
MMP9 Reverse: TAGCACGGATAGACCA;
GAPDH Forward: TGTGGTCATGAGTCCTTCCA,
GAPDH Reverse: ATGTTCGTCATGGGTGTGAA;
HPRT Forward: TGCTGACCTGCTGGATTACA,
HPRT Reverse: TCCCCTGTTGACTGGTCATT;
α-Actin Forward: AGCACAGAGCCTCGCCTTT,
β-Actin Reverse: CGGCGATATCATCATCCAT.
Results
Characterization of a breast cancer stem cell population within the MCF10AT1 cell line.
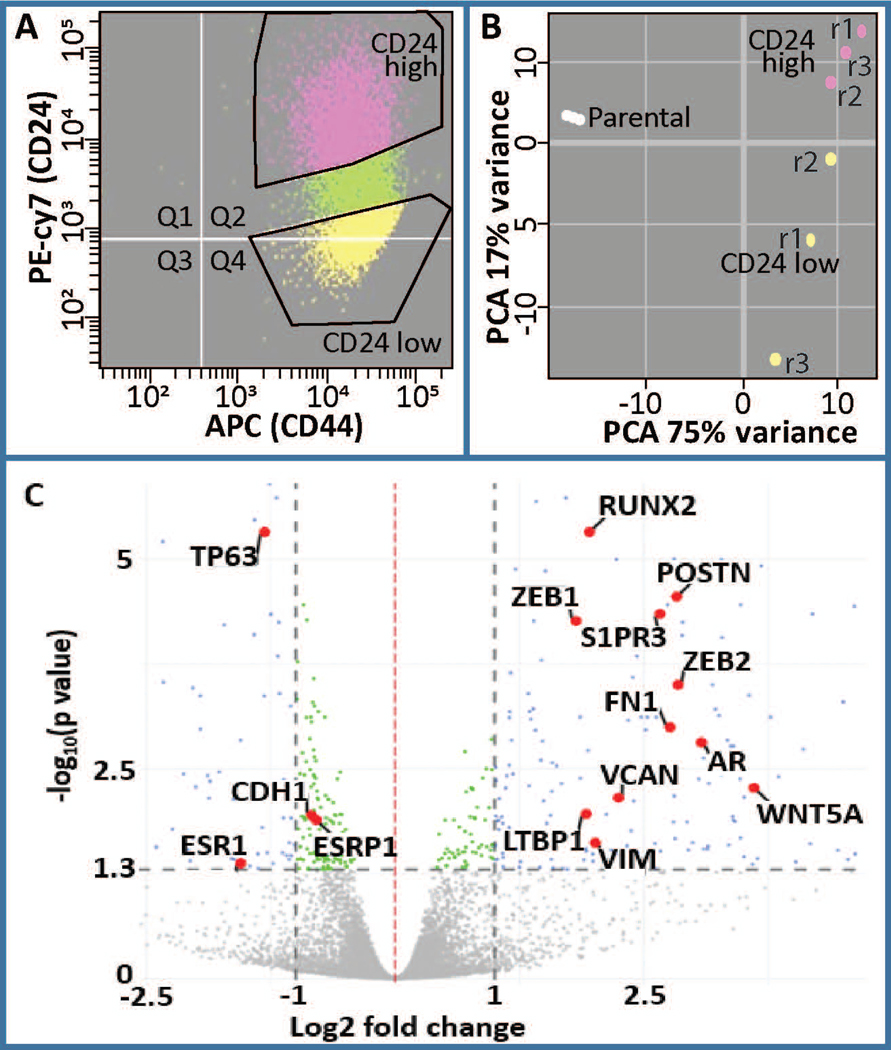
The distribution of CD24 and CD44 was determined by flow cytometry to evaluate the percent of cells that are indicative of breast cancer stemness (CD24-/low/CD44+/high). While all cells are positive for CD44, there is a population of cells that are CD24-/low. We subsequently isolated CD24-/low and CD24+/high subpopulations as demonstrated by the gating in Figure 1A. RNAseq was performed on these two populations and principle component analysis demonstrated that these two populations are distinct in their expression profiles (Fig 1B). Well-known stemness and EMT related genes including CD24, CDH1, VIM, FN1, TGFB1, SNAI1, TWIST1, TWIST2, ZEB1 and ZEB2 are among the differentially expressed (DE) genes and are shown within a volcano plot (Fig 1C, Fig S1). Furthermore, while HER2 and glucorticoid receptor (GR) do not change in expression at the RNA level, ESR1 and THRB are downregulated and AR is upregulated (Fig 1C, Fig S1). Together these results establish a model for investigating cancer stem cells in the basal breast cancer phenotype.
Figure 1.
Analysis of differentially expressed genes in CD24-/low/CD44+/high BCSCs versus CD24+/high/CD44high premalignant basal MCF10AT1 breast cancer cells. (a) FACS was performed on MCF10AT1 using CD24 (PE-cy7) and CD44 (APC) antibodies to isolate BCSCs (CD24−/low/CD44+/high, yellow) and bulk non-BCSC (CD24+/high/CD44+, pink). r1, r2, r3 refer to different replicates of FACS. (b) Principle component analysis demonstrated clustering of these two populations from each other. Parental cells were not subjected to FACS therefore suggesting an affect of FACS on gene expression. (c) A volcano plot demonstrating differentially expressed genes highlighting several genes that are well known to be altered in BCSCs including RUNX2, AR, ESR1, POSTN (Morra & Moch, 2011), CDH1, ESRP1, VIM, ZEB1, ZEB2, LTBP1, VCAN, WNT5A, and S1PR3 (Milara et al., 2012). BCSC, breast cancer stem cell; FACS, fluorescence-activated cell sorting; PCA, principal component analysis
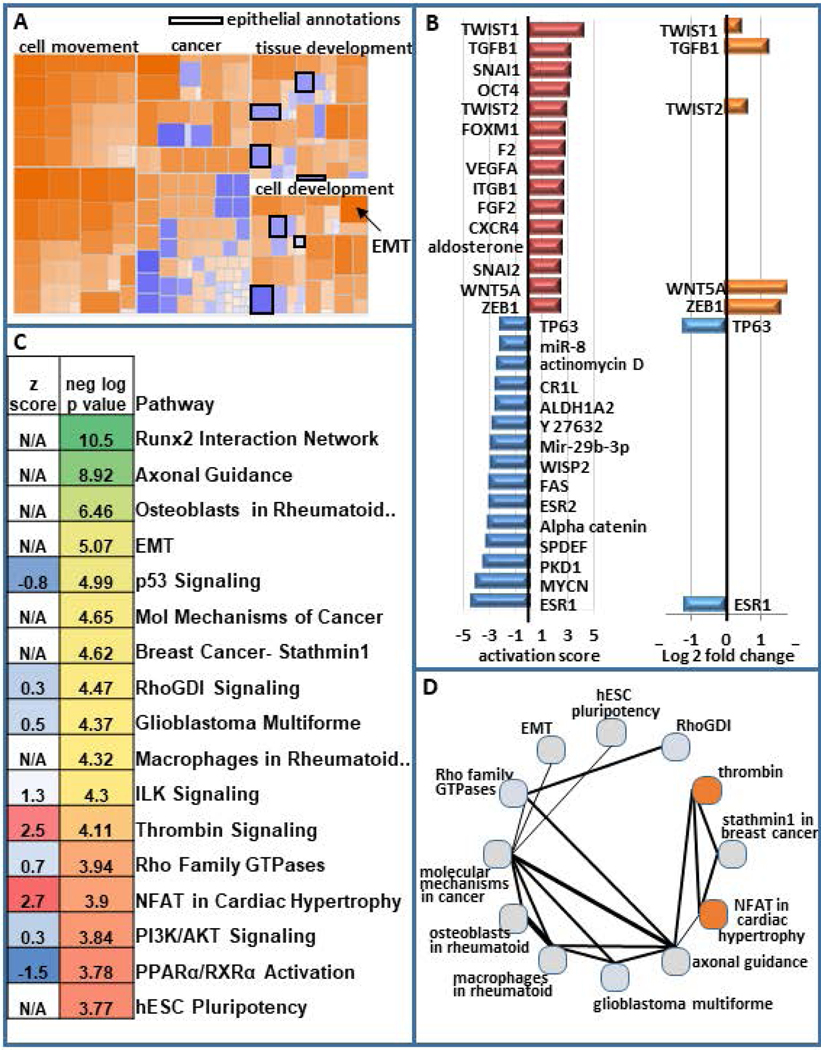
To elucidate key pathways that are integral to stemness within MCF10AT1, we performed Ingenuity Pathway Analysis (IPA). This analysis demonstrated that annotations reflecting pathways involved in cellular movement, invasion and migration are activated within the CD24-/low subpopulation. Pathways related to differentiation are altered, with those associated with epithelial differentiation being inhibited and EMT being activated (Fig 2A). IPA was used to cross-reference our geneset with genesets that are known to be associated with upstream regulators that can be transcription factors, drugs, hormones, etc. Many of the top upstream regulators that are activated among the DE genes in CD24-/low vs CD24+/high are well-known to increase stemness and EMT. For example, the top 5 activated upstream regulators are TWIST1, TGFB1, SNAI1, OCT4, and TWIST2. Several of the top inhibited upstream regulators are known to be decreased in EMT and stemness including ESR1, ESR2, FAS, and TP63. Unexpectedly, a geneset associated with MYC-N is inhibited. Other upstream regulators may mediate loss of cell-cell adhesion and/or increased migration and invasion include PKD1, alpha catenin, VEGFA, and integrin beta-1 (ITGB1). Among these top upstream regulators, seven are altered in their expression at the RNA level (TWIST1, TGFB1, TWIST2, WNT5A, ZEB1 are upregulated, while TP63 and ESR1 are downregulated). Despite the other upstream regulators not being significantly altered in the RNAseq, they may be altered at the protein level (Fig 2B).
Figure 2.
Pathway analysis of differentially expressed genes in BCSCs. (a) Ingenuity pathway analysis on differentially expressed genes in BCSC (CD24−/low/CD44+/high) versus bulk non-BCSC (CD24+/high/CD44+) demonstrates increased cellular movement, invasion, migration, and EMT in BCSCs and decreased epithelial differentiation. (b) The activation score of the top 15 activated and top 15 inhibited upstream regulators are presented. The log2 fold change for these upstream regulators is also shown. (c) Some of the top most significant pathways are displayed (color scale represents the most significant being green and the least being red) as is their activation scores when IPA was able to calculate them (blue being inhibited and red being activated). The top 30 significant pathways that are altered are displayed in Figure S2. (d) The number of genes that are shared between these pathways is depicted with thicker lines indicating more genes in common between them. The circles are colored to indicate the degree of inhibition (blue) and activation (red). BCSC, breast cancer stem cell; EMT, epithelial-to-mesenchymal transition; IPA, Ingenuity Pathway Analysis
Among the top significant pathways that were altered in the RNAseq, were expected including EMT and hESC pluripotency, but additional pathways were also identified as significantly altered in BCSCs and EMT (e.g., p53, ILK, and PI3K/AKT, Fig 2C, Fig S2). The extent to which these pathways share genes is demonstrated in figure 2D, with thicker lines indicating more genes in common. Of potential importance, the most significant pathway that was altered was a RUNX2 associated geneset (Fig 2C).
RUNX is integral to BCSC biology
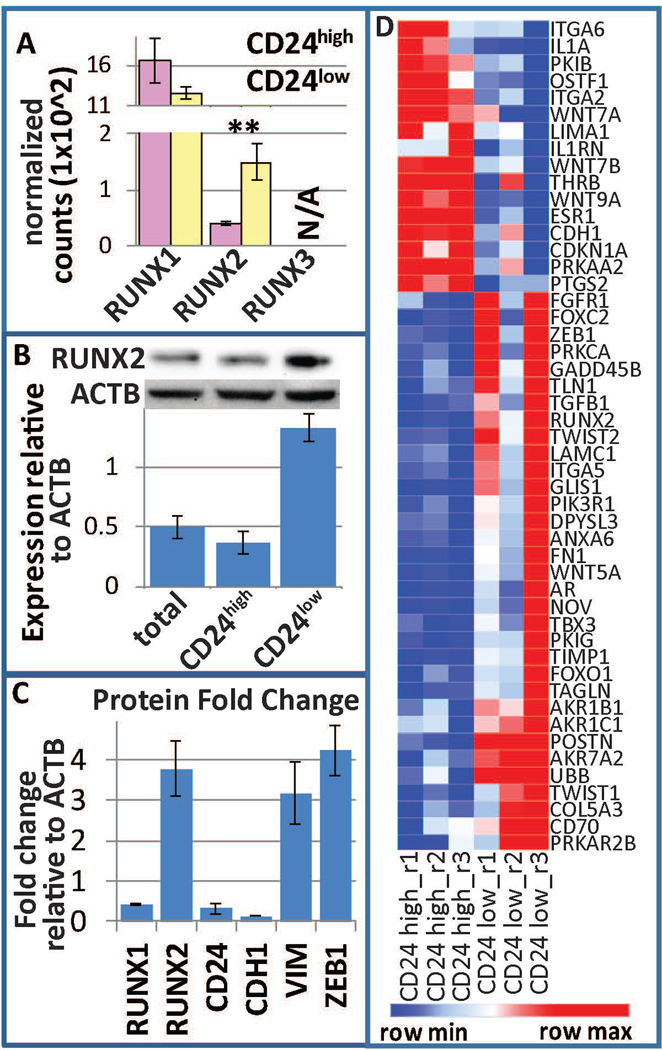
Given our previous results that RUNX1 suppresses EMT and breast cancer stemness (Hong et al., 2018), and the significant association with RUNX2 revealed by gene expression profiling, we investigated the involvement of RUNX factors in breast cancer stemness. We determined that RUNX1 is the predominantly expressed RUNX gene, with RUNX2 being lower in expression, and RUNX3 not expressed in MCF10A or MCF10AT1. While RUNX1 was not significantly altered, RUNX2 was increased at the RNA level in CD24-/low vs CD24+/high MCF10AT1 cells (Fig 3A). Western blot analysis on sorted CD24-/low vs CD24+/high MCF10AT1 demonstrated that RUNX2 is increased within the CD24-/low subpopulation (Fig 3B). As we previously demonstrated, RUNX1 is decreased within this subpopulation, and these cells are more mesenchymal as reflected by their expression of VIM, CDH1, and ZEB1 (Fig 3C). A heatmap of the RUNX2 interaction network is displayed in figure 3D. These CD24-low cells demonstrated greater long-term self-renewal capacity as measured by tumorsphere formation assays (Hong et al., 2018). Collectively, these data provide evidence that cell populations with BCSC properties express higher levels of RUNX2 compared to cells with high levels of CD24.
Figure 3.
RUNX regulation within BCSCs. (a) Normalized counts within the RNAseq demonstrate that RUNX1 is the predominant RUNX factor expressed within MCF10AT1, while RUNX2 is differentially expressed between BCSC (CD24−/low/CD44+/high, yellow) and bulk non-BCSC (CD24+/high/CD44+, pink). RUNX3 is not expressed. (b) FACS for these subpopulations followed by western blot analysis demonstrated an increase of RUNX2 within BCSCs. (c) Densitometry of westerns previously published (Hong et al., 2018) demonstrate a decrease in RUNX1, CD24, and CDH1 and an increase in RUNX2, ZEB1, and VIM. (d) A heatmap of the RUNX2 interaction network is displayed. BCSC, breast cancer stem cell; FACS, fluorescence-activated cell sorting; RUNX, Runt-related transcription factor
RUNX2 increases in human tumor cells within the murine mammary fat pad microenvironment
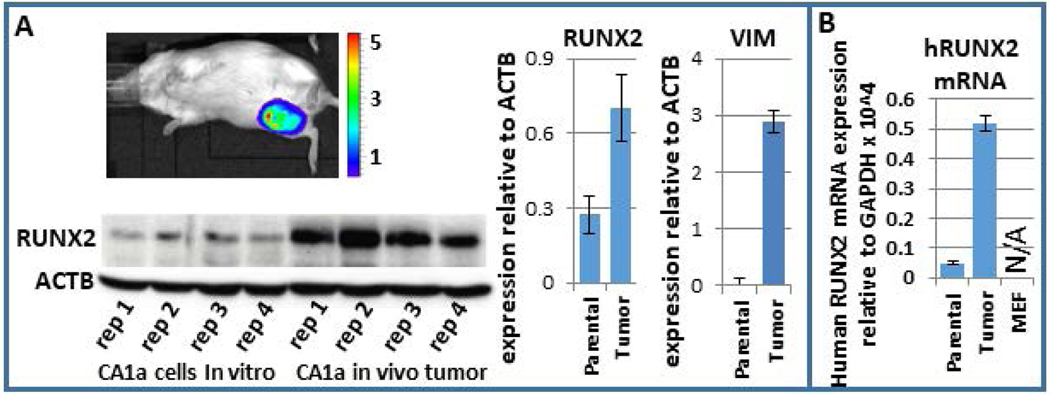
BCSCs are a more tumorigenic subpopulation of cells. Consequently, we investigated whether RUNX2 increases during breast tumor growth in vivo. We utilized a mouse xenograft model to examine RUNX2 expression during tumor progression. MCF10CA1a cells, which are aggressive breast cancer cells, were injected into the mammary fat pad of immunocompromised SCID mice and tumor growth was monitored weekly. One month after injection, mice were euthanized and tumors were removed and analyzed for RUNX2 and other factors at both the protein and mRNA levels. Tumors had a 3.3-fold higher level of RUNX2 protein than the parental MCF10CA1a, as quantified by western blot analysis (Fig 4A). Quantification of western replicates demonstrated that the epithelial marker CDH1 was decreased in tumor samples, while the mesenchymal marker VIM was increased (Fig 4A; (Hong et al., 2018)). Given the potential that infiltrating mouse cells may have increased expression of RUNX2, human-specific qPCR primers were used to determine RUNX2 transcript levels. qPCR analysis in mouse embryonic fibroblasts validated the human specificity of these primers. This finding demonstrated that RUNX2 mRNA is increased specifically within the human MCF10CA1a cancer cells within the tumor environment (Fig. 4B). The human-specific mRNA levels of several cancer-related genes such as VEGF, FN1, MMP13, MMP9, CXCR4, CXCL12 are upregulated in the tumor samples (Hong et al., 2018). These results indicate that the human breast cancer cells that have formed a tumor in the mouse mammary fat pad acquire a more aggressive phenotype.
Figure 4.
RUNX2 increases within MCF10CA1a in the murine mammary fat pad tumor microenvironment. (a) Western blot analysis of four replicates of parental MCF10CA1a and four tumor samples post-injection within the mammary fat pad demonstrate an increase in RUNX2 within the tumor sample. (b) Since RUNX2 may be higher in expression specifically within infiltrating mouse cells within the tumor, human-specific primers for RUNX2 were used to establish that RUNX2 increases specifically within the human MCF10CA1a in the tumor microenvironment. A control qPCR using these primers on mouse embryonic fibroblasts cells resulted in no detectable expression. qPCR, quantitative polymerase chain reaction; RUNX, Runt-related transcription factor
Simultaneous inhibition of RUNX1 and RUNX2 results in EMT
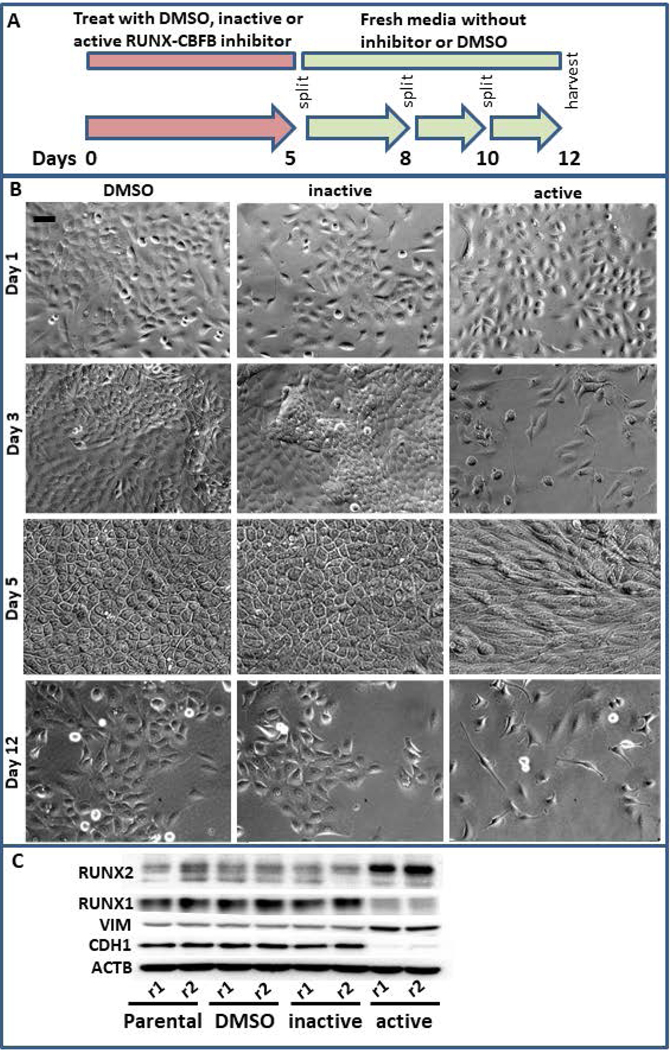
The concurrent decrease in RUNX1 and increase in RUNX2 within the CD24-/low subpopulation suggests an interplay between these factors within BCSCs and EMT. We therefore utilized an inhibitor of the interaction between RUNX factors and CBFβ, to decrease the genomic occupancy of both RUNX1 and RUNX2 (Fig 5A). Treatment of MCF10AT1 cells with this inhibitor for 5 days resulted in cell death, decreased cell growth and a more mesenchymal-like morphology (Fig 5B). The resulting cell population demonstrated decreased RUNX1 and CDH1, and increased RUNX2 and VIM. STR analysis confirmed that this population of cells is not a result of a cross contamination with a cell type that is RUNX-independent. These findings suggest that a general loss of RUNX regulation drives EMT associated with breast cancer and potentially supports the concept that loss of RUNX1, rather than increased RUNX2, is a driver of EMT in early stage breast cancer.
Figure 5.
Simultaneous RUNX1 and RUNX2 inhibition results in epithelial to mesenchymal transition. (a) MCF10AT1 were treated for 5 days with DMSO, an inhibitor that interferes with the interaction between CBFβ and RUNX factors, or an inactive version of this inhibitor that is chemically similar but does not interfere with these interactions. These were then allowed to recover for 7 days in fresh media and passaged as normal. (b) This treatment regimen results in slowed cell growth, cell death, and a more mesenchymal-like morphology. Bar in the upper left panel indicates 20 μm. (c) Western blot analysis demonstrates that the resulting cells are higher in RUNX2 and lower in RUNX1 and are more mesenchymal in their expression of CDH1 and VIM. CBFβ, core binding factor β; DMSO, dimethyl sulfoxide; RUNX, Runt-related transcription factor
Discussion
We provide multiple lines of evidence that RUNX factors play critical roles in BCSCs and during EMT, with RUNX1 suppressing and RUNX2 activating mesenchymal and stemness traits. Basal-like premalignant breast cancer MCF10AT1 cells sorted for the level of CD24, a negative marker for BCSCs, demonstrated altered expression for many genesets associated with upstream regulators that are well-known to be aberrantly expressed during EMT and/or in breast cancer stemness (Hadjimichael et al., 2015; G. Li et al., 2018; Lindsay, McDade, Pickard, McCloskey, & McCance, 2011; Scheel & Weinberg, 2012; Shibue & Weinberg, 2017; Singh & Settleman, 2010; Venkatesh et al., 2018). RUNX factors are critical components of the pathways downstream of these many of these regulators (Y. Ito, S. C. Bae, & L. S. Chuang, 2015; Masse et al., 2012). For example, the genesets associated with TGFB1 and SNAI1 are statistically activated within the MCF10AT1 CD24-/low subpopulation relative to the CD24+/high cells, and ESR1 and TP63 associated genesets are inhibited. Some of these genesets associated with upstream regulators are altered while the expression of the upstream regulator itself is not changed. For example, OCT4 does not change in expression at the RNA level according to our RNAseq. This could be a result of it not being detected at statistical significance or it might be regulated at the post-transcriptional level. Together these findings support the key role for RUNX factor involvement in breast cancer stemness.
Within our RNAseq dataset, the RUNX2 interaction network was the most significantly altered pathway. While our previous results indicate that RUNX1, CD24, and CDH1 are lower and VIM and ZEB1 are higher in the BCSC subpopulation, here we show by FACS followed by western analysis that RUNX2 is increased within these stem-like cells. Consistent with tumor promoter activity, RUNX2 increases within human tumor cells in the murine mammary fat pad microenvironment. Finally, since RUNX1 decreases and RUNX2 increases within the BCSC subpopulation, we addressed the effect that inhibiting both factors simultaneously would have on EMT. These experiments demonstrated that loss of RUNX1, rather than a gain of RUNX2, within early stage basal breast cancer cells is a driver of EMT.
Breast cancer is a leading cause of cancer death in women second only to lung cancer (Siegel, Miller, & Jemal, 2017; Torre et al., 2015). While there have been significant advances in early detection and treatment of breast cancer, further study is required to elucidate the mechanisms of breast cancer progression and metastasis. Accumulating evidence indicates that perturbed RUNX factor regulation is associated with progression of breast cancer (N.-O. Chimge et al., 2016; Deli Hong et al., 2017; van Bragt et al., 2015). Reduced RUNX1 and increased RUNX2 levels in tumors correlate with poor survival for breast cancer patients. Moreover, the expression of RUNX1 is lower in higher histological grade tumors relative to low-to mid-grade tumors (Kadota et al., 2010). RUNX1 and its obligatory cofactor CBFβ loss-of-function somatic mutations have been detected in several breast cancer subtypes (Banerji et al., 2012; Ellis et al., 2012; Network, 2012; Pereira et al., 2016). In ER+ breast cancer, RUNX1 depletion activates WNT signaling and increases ELF5 levels (N.-O. Chimge et al., 2016; van Bragt et al., 2015). In basal-like ER low MCF10AT1 RUNX1 knockdown results in increased ZEB1 (Hong et al., 2018), and in MCF10A its loss results in increased NOTCH3 expression (Malik et al., 2019). Together these findings suggest that a deeper understanding of the role(s) that RUNX factors play within breast cancer will provide novel insight into therapeutic strategies.
Previously, we determined that RUNX1 depletion stimulates EMT in both normal and breast cancer cells (Deli Hong et al., 2017) and that the level of RUNX1 is decreased during tumor growth (Hong et al., 2018). Moreover, forced expression of RUNX1 suppresses tumor growth post injection of MCF10CA1a in the mouse xenografts (Hong et al., 2018). RUNX1 or CBFβ knockout in MCF10A normal-like mammary epithelial cells resulted in tumorigenicity in murine mammary fat pads (Malik et al., 2019). Taken together, these studies strongly support the concept that RUNX1 is a tumor suppressor of breast cancer in vivo. However, the possibility that RUNX1 has other functions in breast cancer, particularly in later stages cannot be excluded. Contrary to the increased tumorigenicity upon RUNX1 knockout in MCF10A and upon its depletion in MCF10CA1a, in the MMTV-PyMT mouse model, RUNX1 expression is positively correlated with tumor progression (Browne et al., 2015). Furthermore, RUNX1 promotes expression of genes involved in tumor growth in late-stage triple negative MDA-MB-231 breast cancer cells (Recouvreux et al., 2016). These contradictory findings suggest a context dependent dual role for RUNX1 in late stage breast cancer.
While we have identified RUNX1 as a suppressor of EMT in normal and early stage breast cancer cells, RUNX2 has been identified to be involved within a regulatory network that controls EMT (N. O. Chimge et al., 2011; Cohen-Solal et al., 2015; Valenti et al., 2016). Ectopic expression of RUNX2 induces EMT by regulating key pathways such as TGFβ and Wnt. Furthermore, RUNX2 promotes invasiveness in luminal ER+ MCF7 breast cancer cells (N. O. Chimge et al., 2011). This increased invasiveness is consistent with the positive regulation of matrix metalloproteinases by RUNX2 in MCF7 and MDA-MB-231 breast cancer cells (Pratap et al., 2005). Consistent with this result, overexpression of RUNX2 mutated in a critical subnuclear matrix targeting signal domain in MDA-MB-231 breast cancer cells resulted in reduced invasiveness (Javed et al., 2005). The impact of RUNX2 on invasiveness is consistent with our finding that RUNX2 enriched cancer stem cells have activated pathways that are associated with increased cellular movement, invasiveness, and migration (Fig 2). Clinically, increased RUNX2 results in an osteomimetic phenotype for cancer cells, an increased metastasis to bone (Akech et al., 2010; Baniwal et al., 2010; Leong et al., 2010), and chemoresistance (Ozaki et al., 2015; Ozaki et al., 2018). The increased level of RUNX2 in BCSCs may be a critical mechanism by which BCSC resist chemotherapies and cause recurrence.
Given the shared binding motif for the RUNX factors, one fundamental question is whether the loss of RUNX1 in breast cancer results in a switch to RUNX2 thereby inducing EMT. In order to address this question, we utilized an inhibitor for the interaction between RUNX factors and CBFβ (Illendula et al., 2016). Upon inhibition, RUNX factors dramatically decrease their affinity with chromatin. We determined that simultaneous inhibition of both RUNX1 and RUNX2 for five days resulted in EMT, with post treatment cells exhibiting an increase in RUNX2 and a decrease in RUNX1. These results suggest that in early stage breast cancer the loss of RUNX1, rather than the gain of RUNX2, is a critical driver of EMT.
Together, our findings support the concept that RUNX factors are critical components within the signaling cascades that regulate EMT and stemness. The resulting BCSCs may be responsible for metastasis, resistance to therapeutics, and tumor recurrence in breast cancer. A deeper understanding of the mechanistic interplay between RUNX1 and RUNX2 in breast cancer will provide novel insight into therapeutic strategies.
Supplementary Material
Acknowledgments
We thank the members of our laboratory, as well as Roxana del Rio-Guerra, PhD of the UVM Flow Cytometry and Cell Sorting Facility for her help with cell sorting. The flow cytometry data we presented were obtained at the Harry Hood Bassett Flow Cytometry and Cell Sorting Facility, University of Vermont College of Medicine. This work was supported by NIH grants U01 CA196383 and P01 CA082834 (G.S.S., J.L.S.), R01 CA139322 (G.S.S.), R37 DE012528 (J.B.L.) and F32 CA220935, as well as the Charlotte Perelman Fund for Cancer Research.
Footnotes
Conflict of Interest
The authors have declared that no conflict of interest exists.
Data Availability
RNA-Seq datasets have been deposited in the GEO under accession code GSE141503. Any other data are readily available upon email request to the corresponding author.
References
- Abdullah LN, & Chow EK-H (2013). Mechanisms of chemoresistance in cancer stem cells. Clinical and Translational Medicine, 2, 3–3. doi: 10.1186/2001-1326-2-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akech J, Wixted JJ, Bedard K, van der Deen M, Hussain S, Guise TA, . . . Lian JB (2010). Runx2 association with progression of prostate cancer in patients: mechanisms mediating bone osteolysis and osteoblastic metastatic lesions. Oncogene, 29(6), 811–821. doi: 10.1038/onc.2009.389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, & Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics, 31(2), 166–169. doi: 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, . . . Meyerson M (2012). Sequence analysis of mutations and translocations across breast cancer subtypes. Nature, 486(7403), 405–409. doi:http://www.nature.com/nature/journal/v486/n7403/abs/nature11154.html#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniwal SK, Khalid O, Gabet Y, Shah RR, Purcell DJ, Mav D, . . . Frenkel B (2010). Runx2 transcriptome of prostate cancer cells: insights into invasiveness and bone metastasis. Mol Cancer, 9, 258. doi: 10.1186/1476-4598-9-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barutcu AR, Hong D, Lajoie BR, McCord RP, van Wijnen AJ, Lian JB, . . . Stein GS (2016). RUNX1 contributes to higher-order chromatin organization and gene regulation in breast cancer cells. Biochim Biophys Acta, 1859(11), 1389–1397. doi: 10.1016/j.bbagrm.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne G, Taipaleenmäki H, Bishop NM, Madasu SC, Shaw LM, van Wijnen AJ, . . . Lian JB (2015). Runx1 is associated with breast cancer progression in MMTV-PyMT transgenic mice and its depletion in vitro inhibits migration and invasion. Journal of Cellular Physiology, 230(10), 2522–2532. doi: 10.1002/jcp.24989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Sun B, Zhao X, Zhang Y, Gu Q, Liang X, . . . Zhao N (2017). The Expression and Functional Significance of Runx2 in Hepatocellular Carcinoma: Its Role in Vasculogenic Mimicry and Epithelial-Mesenchymal Transition. Int J Mol Sci, 18(3). doi: 10.3390/ijms18030500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer CL, San Juan BP, Lim E, & Weinberg RA (2016). EMT, cell plasticity and metastasis. Cancer and Metastasis Reviews, 35(4), 645–654. doi: 10.1007/s10555-016-9648-7 [DOI] [PubMed] [Google Scholar]
- Chimge N-O, Little GH, Baniwal SK, Adisetiyo H, Xie Y, Zhang T, . . . Frenkel B (2016). RUNX1 prevents oestrogen-mediated AXIN1 suppression and β-catenin activation in ER-positive breast cancer. Nature Communications, 7, 10751. doi:10.1038/ncomms1075110.1038/ncomms10751http://www.nature.com/articles/ncomms10751#supplementary-informationhttp://www.nature.com/articles/ncomms10751#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimge NO, Baniwal SK, Little GH, Chen YB, Kahn M, Tripathy D, . . . Frenkel B (2011). Regulation of breast cancer metastasis by Runx2 and estrogen signaling: the role of SNAI2. Breast Cancer Res, 13(6), R127. doi: 10.1186/bcr3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang LS, Ito K, & Ito Y (2017). Roles of RUNX in Solid Tumors. Adv Exp Med Biol, 962, 299–320. doi: 10.1007/978-981-10-3233-2_19 [DOI] [PubMed] [Google Scholar]
- Cohen-Solal KA, Boregowda RK, & Lasfar A (2015). RUNX2 and the PI3K/AKT axis reciprocal activation as a driving force for tumor progression. Mol Cancer, 14, 137. doi: 10.1186/s12943-015-0404-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle Carbonare L, Mottes M, Cheri S, Deiana M, Zamboni F, Gabbiani D, . . . Valenti MT. (2019). Increased Gene Expression of RUNX2 and SOX9 in Mesenchymal Circulating Progenitors Is Associated with Autophagy during Physical Activity. Oxid Med Cell Longev, 2019, 8426259. doi: 10.1155/2019/8426259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, . . . Mardis ER (2012). Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature, 486(7403), 353–360. doi:http://www.nature.com/nature/journal/v486/n7403/abs/nature11143.html#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari N, McDonald L, Morris JS, Cameron ER, & Blyth K (2013). RUNX2 in mammary gland development and breast cancer. J Cell Physiol, 228(6), 1137–1142. doi: 10.1002/jcp.24285 [DOI] [PubMed] [Google Scholar]
- Ferrari N, Riggio AI, Mason S, McDonald L, King A, Higgins T, . . . Blyth K (2015). Runx2 contributes to the regenerative potential of the mammary epithelium. Sci Rep, 5, 15658. doi: 10.1038/srep15658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillmore CM, & Kuperwasser C (2008). Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Research : BCR, 10(2), R25-R25. doi: 10.1186/bcr1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Schwieger M, Horn S, Niebuhr B, Ford A, Roscher S, . . . Stocking C (2005). Defining the oncogenic function of the TEL//AML1 (ETV6//RUNX1) fusion protein in a mouse model. Oncogene, 24(51), 7579–7591. [DOI] [PubMed] [Google Scholar]
- Fritz AJ, Gillis NE, Gerrard DL, Rodriguez PD, Hong D, Rose JT, . . . Stein GS (2019). Higher order genomic organization and epigenetic control maintain cellular identity and prevent breast cancer. Genes Chromosomes Cancer, 58(7), 484–499. doi: 10.1002/gcc.22731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigore AD, Jolly MK, Jia D, Farach-Carson MC, & Levine H (2016). Tumor Budding: The Name is EMT. Partial EMT. Journal of Clinical Medicine, 5(5), 51. doi: 10.3390/jcm5050051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjimichael C, Chanoumidou K, Papadopoulou N, Arampatzi P, Papamatheakis J, & Kretsovali A (2015). Common stemness regulators of embryonic and cancer stem cells. World Journal of Stem Cells, 7(9), 1150–1184. doi: 10.4252/wjsc.v7.i9.1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, . . . Hubbard TJ (2012). GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res, 22(9), 1760–1774. doi: 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatlen MA, Wang L, & Nimer SD (2012). AML1-ETO driven acute leukemia: insights into pathogenesis and potential therapeutic approaches. Frontiers of Medicine, 6(3), 248–262. doi: 10.1007/s11684-012-0206-6 [DOI] [PubMed] [Google Scholar]
- Hoi CSL, Lee SE, Lu S-Y, McDermitt DJ, Osorio KM, Piskun CM, . . . Tumbar T (2010). Runx1 Directly Promotes Proliferation of Hair Follicle Stem Cells and Epithelial Tumor Formation in Mouse Skin. Molecular and Cellular Biology, 30(10), 2518–2536. doi: 10.1128/MCB.01308-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D, Fritz AJ, Finstad KH, Fitzgerald MP, Weinheimer A, Viens AL, . . . Stein GS (2018). Suppression of Breast Cancer Stem Cells and Tumor Growth by the RUNX1 Transcription Factor. Mol Cancer Res, 16(12), 1952–1964. doi: 10.1158/1541-7786.MCR-18-0135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D, Fritz AJ, Gordon JA, Tye CE, Boyd JR, Tracy KM, . . . Stein GS (2019). RUNX1-dependent mechanisms in biological control and dysregulation in cancer. J Cell Physiol, 234(6), 8597–8609. doi: 10.1002/jcp.27841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D, Messier TL, Tye CE, Dobson JR, Fritz AJ, Sikora KR, . . . Stein GS (2017). Runx1 stabilizes the mammary epithelial cell phenotype and prevents epithelial to mesenchymal transition. Oncotarget, 8(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D, Messier TL, Tye CE, Dobson JR, Fritz AJ, Sikora KR, . . . Stein GS (2017). Runx1 stabilizes the mammary epithelial cell phenotype and prevents epithelial to mesenchymal transition. Oncotarget, 8(11), 17610–17627. doi: 10.18632/oncotarget.15381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illendula A, Gilmour J, Grembecka J, Tirumala VSS, Boulton A, Kuntimaddi A, . . . Bushweller JH (2016). Small Molecule Inhibitor of CBFbeta-RUNX Binding for RUNX Transcription Factor Driven Cancers. EBioMedicine, 8, 117–131. doi: 10.1016/j.ebiom.2016.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y (2004). Oncogenic potential of the RUNX gene family: ‘overview’. . Oncogene, 23(24), 4198–4208. [DOI] [PubMed] [Google Scholar]
- Ito Y, Bae S-C, & Chuang LSH (2015). The RUNX family: developmental regulators in cancer. Nat Rev Cancer, 15(2), 81–95. doi:10.1038/nrc387710.1038/nrc3877http://www.nature.com/nrc/journal/v15/n2/abs/nrc3877.html#supplementary-informationhttp://www.nature.com/nrc/journal/v15/n2/abs/nrc3877.html#supplementary-information [DOI] [PubMed] [Google Scholar]
- Ito Y, Bae SC, & Chuang LS (2015). The RUNX family: developmental regulators in cancer. Nat Rev Cancer, 15(2), 81–95. doi: 10.1038/nrc3877 [DOI] [PubMed] [Google Scholar]
- Jacob B, Osato M, Yamashita N, Wang CQ, Taniuchi I, Littman DR, . . . Ito Y (2010). Stem cell exhaustion due to Runx1 deficiency is prevented by Evi5 activation in leukemogenesis. Blood, 115(8), 1610–1620. doi: 10.1182/blood-2009-07-232249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed A, Afzal F, Bae JS, Gutierrez S, Zaidi K, Pratap J, . . . Lian JB (2009). Specific residues of RUNX2 are obligatory for formation of BMP2-induced RUNX2-SMAD complex to promote osteoblast differentiation. Cells Tissues Organs, 189(1–4), 133–137. doi: 10.1159/000151719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed A, Barnes GL, Pratap J, Antkowiak T, Gerstenfeld LC, van Wijnen AJ, . . . Stein GS (2005). Impaired intranuclear trafficking of Runx2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo. Proc Natl Acad Sci U S A, 102(5), 1454–1459. doi: 10.1073/pnas.0409121102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota M, Yang HH, Gomez B, Sato M, Clifford RJ, Meerzaman D, . . . Lee MP (2010). Delineating Genetic Alterations for Tumor Progression in the MCF10A Series of Breast Cancer Cell Lines. PLOS ONE, 5(2), e9201. doi: 10.1371/journal.pone.0009201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, & Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol, 14(4), R36. doi: 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni M, Tan TZ, Syed Sulaiman NB, Lamar JM, Bansal P, Cui J, . . . Ito Y (2018). RUNX1 and RUNX3 protect against YAP-mediated EMT, stem-ness and shorter survival outcomes in breast cancer. Oncotarget, 9(18), 14175–14192. doi: 10.18632/oncotarget.24419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong DT, Lim J, Goh X, Pratap J, Pereira BP, Kwok HS, . . . van Wijnen AJ (2010). Cancer-related ectopic expression of the bone-related transcription factor RUNX2 in non-osseous metastatic tumor cells is linked to cell proliferation and motility. Breast Cancer Res, 12(5), R89. doi: 10.1186/bcr2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Su Q, Liu H, Wang D, Zhang W, Lu Z, . . . Bi J (2018). Frizzled7 Promotes Epithelial-to-mesenchymal Transition and Stemness Via Activating Canonical Wnt/beta-catenin Pathway in Gastric Cancer. Int J Biol Sci, 14(3), 280–293. doi: 10.7150/ijbs.23756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhao H, Xia S, Wei H, Chen F, & Jin P (2018). RUNX2 promotes epithelial differentiation of ADSCs and burn wound healing via targeting E-cadherin. Oncotarget, 9(2), 2646–2659. doi: 10.18632/oncotarget.23522 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lindsay J, McDade SS, Pickard A, McCloskey KD, & McCance DJ (2011). Role of DeltaNp63gamma in epithelial to mesenchymal transition. J Biol Chem, 286(5), 3915–3924. doi: 10.1074/jbc.M110.162511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, & Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 15(12), 550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik N, Yan H, Moshkovich N, Palangat M, Yang H, Sanchez V, . . . Huang J (2019). The transcription factor CBFB suppresses breast cancer through orchestrating translation and transcription. Nat Commun, 10(1), 2071. doi: 10.1038/s41467-019-10102-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JW, Zielenska M, Stein GS, van Wijnen AJ, & Squire JA (2011). The Role of RUNX2 in Osteosarcoma Oncogenesis. Sarcoma, 2011, 282745. doi: 10.1155/2011/282745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse I, Barbollat-Boutrand L, Molina M, Berthier-Vergnes O, Joly-Tonetti N, Martin MT, . . . Lamartine J (2012). Functional interplay between p63 and p53 controls RUNX1 function in the transition from proliferation to differentiation in human keratinocytes. Cell Death Dis, 3, e318. doi: 10.1038/cddis.2012.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo J, Kimura S, Yamamura A, Koh CP, Hossain MZ, Heng DL, . . . Ito Y (2017). Identification of Stem Cells in the Epithelium of the Stomach Corpus and Antrum of Mice. Gastroenterology, 152(1), 218–231.e214. doi: 10.1053/j.gastro.2016.09.018 [DOI] [PubMed] [Google Scholar]
- Meacham CE, & Morrison SJ (2013). Tumour heterogeneity and cancer cell plasticity. Nature, 501(7467), 328–337. doi: 10.1038/nature12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming L, Michael B, & Max SW (2015). Epithelial-Mesenchymal Plasticity of Breast Cancer Stem Cells: Implications for Metastasis and Therapeutic Resistance. Current Pharmaceutical Design, 21(10), 1301–1310. doi: 10.2174/1381612821666141211120604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani K, Ogawa S, Tanaka T, Miyoshi H, Kurokawa M, Mano H, . . . Hirai H (1994). Generation of the AML1-EVI-1 fusion gene in the t(3;21)(q26;q22) causes blastic crisis in chronic myelocytic leukemia. The EMBO Journal, 13(3), 504–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network TCGA (2012). Comprehensive molecular portraits of human breast tumours. Nature, 490(7418), 61–70. doi:http://www.nature.com/nature/journal/v490/n7418/abs/nature11412.html#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu DF, Kondo T, Nakazawa T, Oishi N, Kawasaki T, Mochizuki K, . . . Katoh R (2012). Transcription factor Runx2 is a regulator of epithelial-mesenchymal transition and invasion in thyroid carcinomas. Lab Invest, 92(8), 1181–1190. doi: 10.1038/labinvest.2012.84 [DOI] [PubMed] [Google Scholar]
- Osorio KM, Lilja KC, & Tumbar T (2011). Runx1 modulates adult hair follicle stem cell emergence and maintenance from distinct embryonic skin compartments. J Cell Biol, 193(1), 235–250. doi: 10.1083/jcb.201006068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens TW, Rogers RL, Best S, Ledger A, Mooney AM, Ferguson A, . . . Naylor MJ (2014). Runx2 is a novel regulator of mammary epithelial cell fate in development and breast cancer. Cancer Res, 74(18), 5277–5286. doi: 10.1158/0008-5472.CAN-14-0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki T, Sugimoto H, Nakamura M, Hiraoka K, Yoda H, Sang M, . . . Nagase H (2015). Runt-related transcription factor 2 attenuates the transcriptional activity as well as DNA damage-mediated induction of pro-apoptotic TAp73 to regulate chemosensitivity. FEBS J, 282(1), 114–128. doi: 10.1111/febs.13108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki T, Yu M, Yin D, Sun D, Zhu Y, Bu Y, & Sang M (2018). Impact of RUNX2 on drug-resistant human pancreatic cancer cells with p53 mutations. BMC Cancer, 18(1), 309. doi: 10.1186/s12885-018-4217-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira B, Chin S-F, Rueda OM, Vollan H-KM, Provenzano E, Bardwell HA, . . . Caldas C (2016). The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nature Communications, 7, 11479. doi:10.1038/ncomms1147910.1038/ncomms11479https://www.nature.com/articles/ncomms11479#supplementary-informationhttps://www.nature.com/articles/ncomms11479#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap J, Javed A, Languino LR, van Wijnen AJ, Stein JL, Stein GS, & Lian JB (2005). The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion. Mol Cell Biol, 25(19), 8581–8591. doi: 10.1128/MCB.25.19.8581-8591.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan YY, Yingb; Wang, Xiaolib; Fu, Qibina Wang, Weikanga Wu, Jingwena Yang, Gena; Ren, Junb; Wang, Yuganga. (2013). Impact of cell dissociation on identification of breast cancer stem cells. Cancer Biomarkers, 12(3), 123–133. [DOI] [PubMed] [Google Scholar]
- Recouvreux MS, Grasso EN, Echeverria PC, Rocha-Viegas L, Castilla LH, Schere-Levy C, . . . Rubinstein N (2016). RUNX1 and FOXP3 interplay regulates expression of breast cancer related genes. Oncotarget, 7(6), 6552–6565. doi: 10.18632/oncotarget.6771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel C, & Weinberg RA (2012). Cancer stem cells and epithelial–mesenchymal transition: Concepts and molecular links. Seminars in Cancer Biology, 22(5–6), 396–403. doi: 10.1016/j.semcancer.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheitz CJF, Lee TS, McDermitt DJ, & Tumbar T (2012). Defining a tissue stem cell-driven Runx1/Stat3 signalling axis in epithelial cancer. The EMBO Journal, 31(21), 4124–4139. doi: 10.1038/emboj.2012.270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibue T, & Weinberg RA (2017). EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol, 14(10), 611–629. doi: 10.1038/nrclinonc.2017.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, & Jemal A (2017). Cancer statistics, 2017. CA: A Cancer Journal for Clinicians, 67(1), 7–30. doi: 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- Singh A, & Settleman J (2010). EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene, 29(34), 4741–4751. doi: 10.1038/onc.2010.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol ES, Sanduja S, Jin DX, Miller DH, Mathis RA, & Gupta PB (2015). Perturbation-Expression Analysis Identifies RUNX1 as a Regulator of Human Mammary Stem Cell Differentiation. PLoS Comput Biol, 11(4), e1004161. doi: 10.1371/journal.pcbi.1004161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood R, Kamikubo Y, & Liu P (2017). Role of RUNX1 in hematological malignancies. Blood, 129(15), 2070–2082. doi: 10.1182/blood-2016-10-687830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M, Terry A, Hu M, O’Hara M, Blyth K, Baxter E, . . . Neil JC (1997). Proviral insertions induce the expression of bone-specific isoforms of PEBP2alphaA (CBFA1): evidence for a new myc collaborating oncogene. Proc Natl Acad Sci U S A, 94(16), 8646–8651. doi: 10.1073/pnas.94.16.8646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, & Jemal A (2015). Global cancer statistics, 2012. CA: A Cancer Journal for Clinicians, 65(2), 87–108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- Valenti MT, Serafini P, Innamorati G, Gili A, Cheri S, Bassi C, & Dalle Carbonare L (2016). Runx2 expression: A mesenchymal stem marker for cancer. Oncol Lett, 12(5), 4167–4172. doi: 10.3892/ol.2016.5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bragt MPA, Hu X, Xie Y, & Li Z (2015). RUNX1, a transcription factor mutated in breast cancer, controls the fate of ER-positive mammary luminal cells. eLife, 3. doi: 10.7554/eLife.03881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh V, Nataraj R, Thangaraj GS, Karthikeyan M, Gnanasekaran A, Kaginelli SB, . . . Basalingappa KM (2018). Targeting Notch signalling pathway of cancer stem cells. Stem Cell Investig, 5, 5. doi: 10.21037/sci.2018.02.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, & Lindeman GJ (2008). Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer, 8(10), 755–768. [DOI] [PubMed] [Google Scholar]
- Wang CQ, Jacob B, Nah GSS, & Osato M (2010). Runx family genes, niche, and stem cell quiescence. Blood Cells, Molecules, and Diseases, 44(4), 275–286. doi: 10.1016/j.bcmd.2010.01.006 [DOI] [PubMed] [Google Scholar]
- Wang, Chelsia Q, Krishnan V, Tay, Lavina S, Chin, Desmond Wai L, Koh, Cai P, Chooi, Jing Y, . . . Osato M (2014). Disruption of Runx1 and Runx3 Leads to Bone Marrow Failure and Leukemia Predisposition due to Transcriptional and DNA Repair Defects. Cell Reports, 8(3), 767–782. doi: 10.1016/j.celrep.2014.06.046 [DOI] [PubMed] [Google Scholar]
- Yokomizo T, Ogawa M, Osato M, Kanno T, Yoshida H, Fujimoto T, . . . Ito Y (2001). Requirement of Runx1/AML1/PEBP2αB for the generation of haematopoietic cells from endothelial cells. Genes to Cells, 6(1), 13–23. doi: 10.1046/j.1365-2443.2001.00393.x [DOI] [PubMed] [Google Scholar]
- Zhao J (2016). Cancer stem cells and chemoresistance: The smartest survives the raid. Pharmacology & Therapeutics, 160, 145–158. doi: 10.1016/j.pharmthera.2016.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Kidwai FK, Kopher RA, Motl J, Kellum CA, Westendorf JJ, & Kaufman DS (2015). Use of RUNX2 expression to identify osteogenic progenitor cells derived from human embryonic stem cells. Stem Cell Reports, 4(2), 190–198. doi: 10.1016/j.stemcr.2015.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.