Summary
Methods for one-photon fluorescent imaging of calcium dynamics can capture the activity of hundreds of neurons across large fields of view, at a low equipment complexity and cost. In contrast to two-photon methods, however, one-photon methods suffer from higher levels of crosstalk from neuropil, resulting in decreased signal-to-noise ratio and artifactual correlations of neural activity. We address this problem by engineering cell body-targeted variants of the fluorescent calcium indicators GCaMP6f and GCaMP7f. We screened fusions of GCaMP to natural as well as artificial peptides, and identified fusions that localized GCaMP to within 50 microns of the cell body of neurons in mice and larval zebrafish. One-photon imaging of soma-targeted GCaMP in dense neural circuits reported fewer artifactual spikes from neuropil, increased signal-to-noise ratio, and decreased artifactual correlation across neurons. Thus, soma targeting of fluorescent calcium indicators facilitates usage of simple, powerful, one-photon methods for imaging neural calcium dynamics.
eTOC Blurb
One-photon fluorescent imaging of calcium signals can capture the activity of hundreds of neurons across large fields of view, but suffers from crosstalk from neuropil. Shemesh et al. engineer cell body-targeted variants of fluorescent calcium indicators, and show in mice and zebrafish that artifactual spikes and correlations are greatly reduced.
Introduction
Methods for one-photon fluorescent imaging of calcium dynamics are popular for neural activity mapping in the living brain. These techniques capture, at high speeds (e.g., >20 Hz), the dynamics of hundreds of neurons across large fields of view, at a low equipment complexity and cost (Alivisatos et al. 2013; Grienberger & Konnerth 2012; Keller et al. 2015). Neuroscientists often focus on analyzing data from cell bodies of neurons being imaged, but these signals are contaminated by those from closely passing axons and dendrites (Fig. 1A, B) (Harris et al. 2016; Peron et al. 2015; Chen et al. 2013). Computational methods attempt to algorithmically correct somatic signals for the neuropil contribution (Pinto & Dan 2015; Andilla & Hamprecht 2014; Mukamel et al. 2009; Pnevmatikakis et al. 2016; Pnevmatikakis et al. 2014). Although such algorithms are widely used in two-photon calcium imaging, one-photon calcium imaging is subject to higher neuropil contamination levels, which remains an open problem for ongoing computational research (Zhou et al. 2016). Alternatively, genetically encoded calcium indicators can be expressed in the nucleus, which eliminates the neuropil signal (Kim et al. 2014; Nguyen et al. 2016; Schrödel et al. 2013; Bengtson et al. 2010; Vladimirov et al. 2014); however, the requirement for calcium to enter the nucleus slows the temporal precision of such imaging, compared to classical cytosolic calcium imaging.
Figure 1. Somatic GCaMP variants.
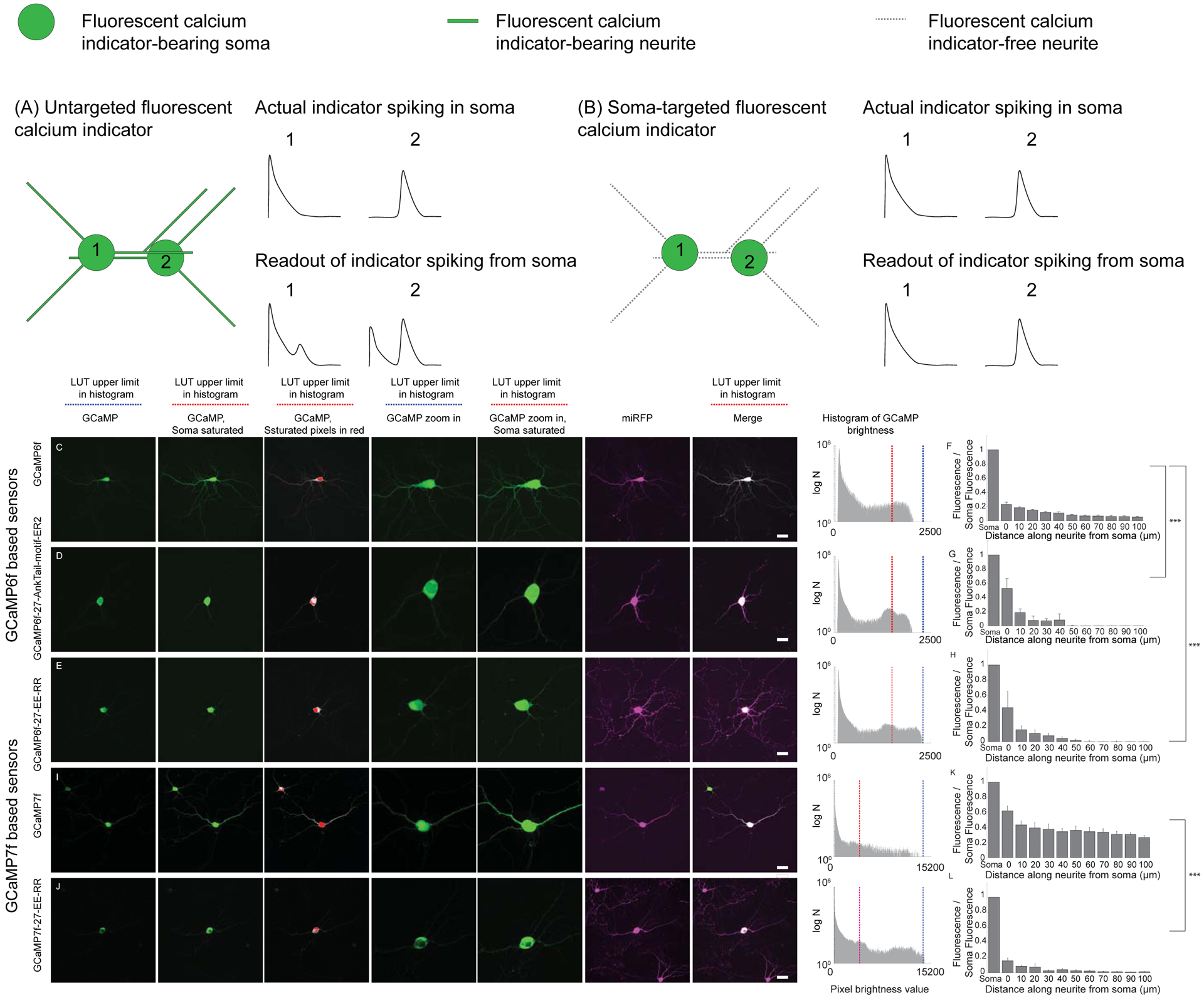
Untargeted GCaMP expresses throughout the neural cytosol, so that GCaMP-bearing neurites from nearby cells (A) can bleed into the signals attributed to a given cell body (compare “actual” to “readout”). Restricting GCaMP expression to the cell body would improve imaging (B) by eliminating these neurite signals. (C, D, E, I, J) Representative confocal max projection images of cultured hippocampal neurons expressing wild-type vs. selectively soma-targeted GCaMP6f or GCaMP7f variants, as well as the countermarker miRFP. (C, left panel) A hippocampal neuron in culture expressing GCaMP6f and miRFP, seen in the GFP channel. Look up table (LUT) was adjusted for minimal saturation (blue bar over image refers to the LUT limit in the histogram at far right of C). (C, Second to left panel) The same neuron seen in the GFP channel, LUT adjusted to saturate the cell body (red bar over image refers to the LUT limit in the histogram at far right of C), to help neurites be more visible. (C, third to left panel) The same soma-saturated image, wherein non saturated pixels are presented in grey and saturated pixels in red. (C, middle panel) A zoom-in on the image presented in the left panel. (C, third to right panel) A zoom-in on the image presented in the second to left panel. (C, second to right panel). The neuron seen in the miRFP channel (magenta). (Right panel) Merge of the second to left (soma-saturated GCaMP channel) and the second to right (miRFP channel) panels. A histogram of pixel values is given in the far right. The upper limit for the LUT of the left GCaMP image is given in blue, and the upper limit for the LUT of the second-to-left image is given in red. (D) As in C, for a neuron expressing GCaMP6f-27-AnkTail-motif-ER2 (SomaGCaMP6f1). (E) As in C, for a neuron expressing GCaMP6f-27-EE-RR (SomaGCaMP6f2). (I) As in C, for a neuron expressing GCaMP7f. (J) As in C, for a neuron expressing GCaMP7f-27-EE-RR (SomaGCaMP7f). Scalebar: 20 μm. (F) Bar plot (mean + standard error) of GCaMP6f brightness versus position along a neurite, normalized to somatic brightness, of neurons as in C (n = 8 neurites from 8 cells from 3 cultures). (G) As in F, for neurons expressing SomaGCaMP6f1 (n = 5 neurites from 5 cells from 2 cultures). ***P < 0.001, Kruskal-Wallis analysis of variance of neurite brightness followed by post-hoc test via Steel’s test with GCaMP6f as control; Supplemental Table 2, statistics for Figure 1). (H) As in F, for neurons expressing GCaMP6f-27-EE-RR (SomaGCaMP6f2; n = 5 neurites from 5 cells from 3 cultures). ***P < 0.001, Kruskal-Wallis analysis of variance of neurite brightness followed by post-hoc test via Steel’s test with GCaMP6f as control). (K) As in F, for neurons expressing GCaMP7f (n = 6 neurites from 6 cells from 2 cultures). (L) As in F, for neurons expressing SomaGCaMP7f (n = 6 neurites from 6 cells from 2 cultures). ***P < 0.001, Wilcoxon rank sum test of neurite brightness followed by post-hoc test via Steel’s test with GCaMP6f as control). Supplemental Table 8, percentage of saturated pixels in GCaMP images.
We hypothesized that if we could localize a genetically encoded calcium indicator such as GCaMP6f (Chen et al. 2013) or GCaMP7f (Dana et al. 2019) to the cytosol near the cell body, we could greatly reduce neuropil fluorescence, similar to the effect of nuclear-localized GCaMP6f, while not sacrificing speed. While soma-targeting of membrane proteins such as optogenetic actuators has been done before (Greenberg et al. 2011; Shemesh et al. 2017; Pégard et al. 2017; Baker et al. 2016; Wu et al. 2013a; Forli et al. 2018), this strategy has not been adapted for genetically encoded calcium indicators. We screened through natural and artificial peptides and discovered two motifs that, when fused to GCaMP, enabled it to express within 50 microns of the cell body. Kinetics were similar to those mediated by conventional GCaMP. In intact brain circuits of larval zebrafish and mice, such soma-targeted GCaMPs greatly reduced the number of neuropil contamination spikes mistakenly attributed to a given neural cell body, and reduced artifactual correlations between nearby neurons.
Results
Designing and screening cell-body targeted GCaMP variants
We fused various peptides to GCaMP6f and GCaMP7f and assessed their ability to target GCaMP to the cell body (Supplemental Table 1A, list of the proteins; Supplemental Table 7, sequences of the fragments used). These included the kainate receptor subunit KA2 (Valluru et al. 2005; Shemesh et al. 2017), the potassium channel KV2.1 (Lim et al. 2000), the sodium channels NaV1.2 and NaV1.6 (Garrido et al. 2003), the adaptor protein AnkyrinG (Zhang & Bennett 1998), and the rat small conductance calcium-activated potassium channel rSK1 (Bowden et al. 2001). In addition, we explored de novo designed coiled-coil proteins that self-assemble into complexes, hypothesizing that their mutual binding could potentially slow their diffusion from the cell body (Moll et al. 2001; Selgrade et al. 2013). For some proteins, earlier work analyzed cell body expression by fusing the full-length proteins to reporters – specifically, NaV1.2, NaV1.6, AnkyrinG, and rSK1 were fused to fluorescent proteins (FPs) (Garrido et al. 2003; Schäfer et al. 2010; Zhang & Bennett 1998; Moruno Manchon et al. 2015), KA2 to a Myc-tag (Valluru et al. 2005), and KV2.1 to an HA-tag (Lim et al. 2000). In some cases, key fragments were found to cause soma targeting of a reporter: for NaV1.2 and NaV1.6, 326- and 27-amino acid segments within intracellular loops between transmembrane domains, termed NaV1.2(I-II) and NaV1.6(II-III) respectively, were sufficient for somatic localization (Garrido et al. 2001; Garrido et al. 2003). For KV2.1, a 65–amino acid segment within the intracellular loop between transmembrane domains IV and V (KV2.1-motif) sufficed (Wu et al. 2013b; Lim et al. 2000). For rSK1, the tail region (rSK1-tail) sufficed (Fletcher et al. 2003). For AnkyrinG it was found that the spectrin-binding domain (AnkSB-motif), the tail domain (AnkTail-motif), the membrane-binding domain (AnkMB-motif), the COOH-terminal domain (AnkCT-motif) and the serine-rich domain (AnkSR-motif) were all targeted to the axon and the cell body of neurons (Zhang & Bennett 1998).
We made over 30 fusions between GCaMP6f and the protein fragments reported above (Supplemental Table 1B, fusions screened; Supplemental Table 7, sequences). For NaV1.2, NaV1.6, KV2.1, and rSK1 we performed fusions in which the previously characterized localization fragment was attached to the C-terminus of GCaMP6f. In a recent study (Shemesh et al. 2017), we fused the channelrhodopsin CoChR (Klapoetke et al. 2014) to the first 150 amino acids of the KA2 receptor subunit (KA2(1–150)) thereby creating a somatic CoChR. Since both N and C terminal fusions of KA2(1–150) with CoChR caused somatic localization, we made similar upstream and downstream fusions of this fragment with GCaMP6f. In the present study, we also found that the first 100 amino acids of KA2 (KA2(1–100)) were sufficient to introduce somatic localization of GCaMP6f, therefore we made additional upstream and downstream fusions of KA2(1–100) with GCaMP6f. In some cases, we inserted into the construct a superfolder GFP (sfGFP (Pédelacq et al. 2006), which contains three mutations to EGFP in order to enhance folding), with a mutation to abolish its fluorescence (here called nullsfGFP). For AnkyrinG fragments, we made fusions both upstream and downstream of GCaMP6f. For de-novo coiled-coil proteins, we made downstream fusions only.
We expressed each of these GCaMP6f fusion proteins in cultured mouse hippocampal neurons. Using wide-field fluorescence microscopy we screened for expression level (fluorescence under baseline conditions), somatic localization, toxicity (assessed as the percentage of dead cells), and whether there were fluorescence changes (df/f0) in response to spontaneous neural activity. Five constructs did not result in obvious toxicity, exhibited somatic localization, and displayed df/f0 similar to that of GCaMP6f. These were GCaMP6f fused to the fragments mentioned below: NaV1.2(I-II) (GCaMP6f-27-NaV1.2(I-II)-ER2); nullsfGFP and KA2(1–100) (GCaMP6f-24-nullsfGFP-24-KA2(1–100)-ER2); a zero-photocurrent CoChR mutant called nullCoChR followed by the KV2.1-motif (nullCoChR-12-GCaMP6f-KV2.1-motif); AnkTail-motif (GCaMP6f-27-AnkTail-motif-ER2, Figure 1D); and the coiled-coil peptide set EE-RR (GCaMP6f-27-EE-RR, Figure 1E).
We next screened these somatic GCaMP6f candidates in mouse brain slices exposed to 1 mM 4-aminopyridine (4-AP, which resulted in ~5–20 GCaMP fluorescent transients per minute, aka GCaMP spikes). We assessed df/f0, the ratio of df/f0 between the cell body and the neuropil, and the brightness. We found that GCaMP6f-24-nullsfGFP-24-KA2(1–100)-ER2 expressed in the neurites of pyramidal neurons in the cortex, in contrast to the culture data, and did not pursue this construct further. The remaining constructs had a similar df/f0 compared to GCaMP6f and a soma to neuropil df/f0 ratio higher than that of GCaMP6f. GCaMP6f-27-AnkTail-motif-ER2 and GCaMP6f-27-EE-RR had the highest baseline brightness (Figure 2; Supplemental Table 2, statistics for Figure 2), and thus we pursued these two constructs for more detailed characterization, naming them SomaGCaMP6f1 and SomaGCaMP6f2 respectively.
Figure 2. Brain slice screening of soma-targeted GCaMP6f candidates.
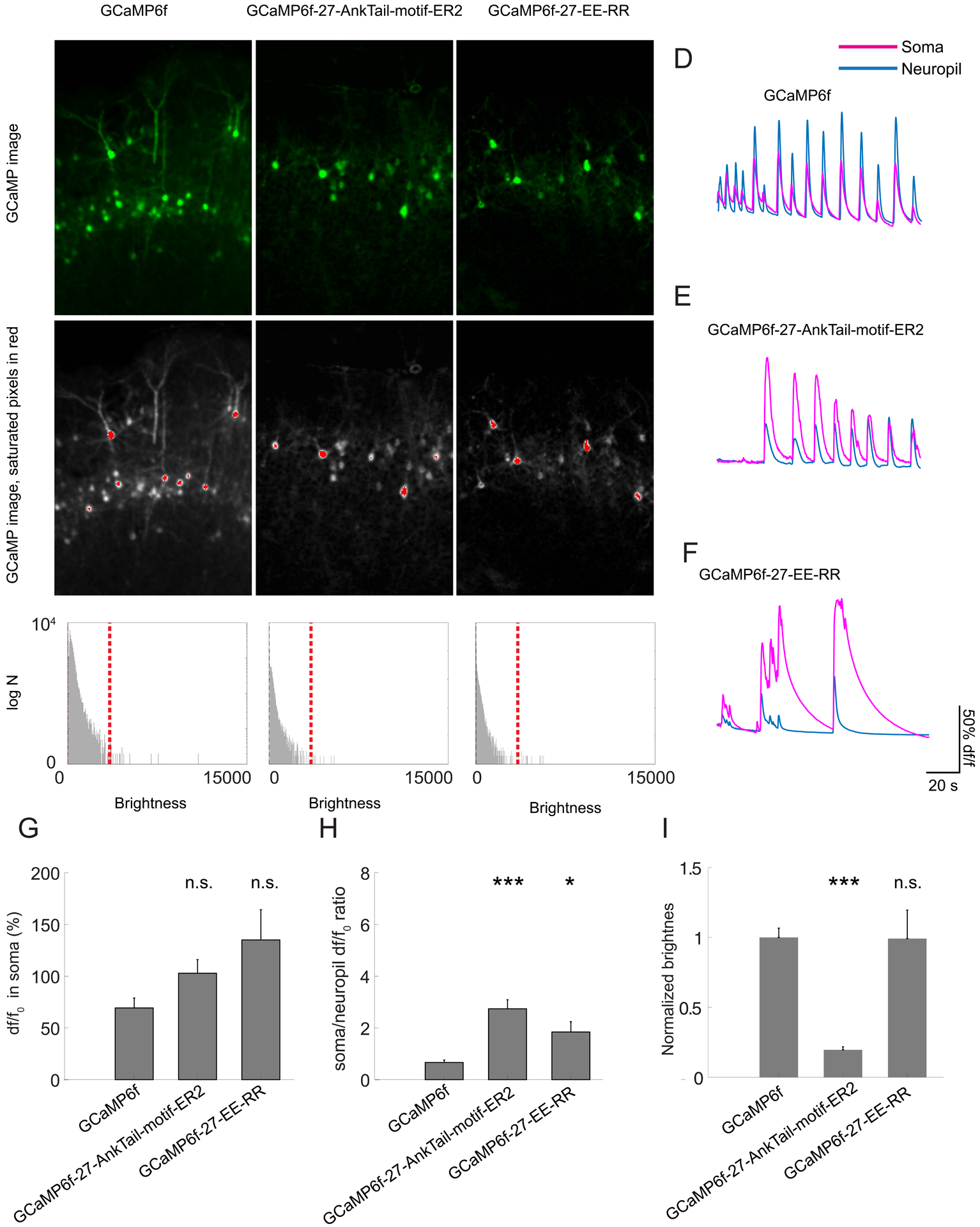
(A-C) Representative max projections of confocal images of mouse cortical slices expressing GCaMP6f variants. Scale bar: 200μm. (A) GCaMP6f. (B) GCaMP6f-27-AnkTail-motif-ER2 (SomaGCaMPf1). (C) GCaMP6f-27-EE-RR (SomaGCaMPf2). Top, GCaMP channel shown in green; bottom, GCaMP nonsaturated pixels shown in gray, with saturated pixels shown in red. Brightness histograms of the images presented in A, B, and C are shown below the respective images. Red line denotes the upper LUT limit. Supplemental Table 8, percentage of saturated pixels in GCaMP images. (D-F) Representative traces of the GCaMP signals from the soma (magenta) and the neuropil (blue). (G) Bar chart showing df/f0 in the somata of neurons expressing different GCaMP6f targeting variants (n = 20 cells from 2 slices from 2 mice for each variant). n.s., not significant, Kruskal-Wallis analysis of variance followed by post-hoc test via Steel’s test with GCaMP6f as control group; see Supplemental Table 2, statistics for Figure 2. Plotted is mean plus or minus standard error throughout. (H) Bar chart showing the ratio between df/f0 at the cell body vs. the neuropil for different GCaMP6f targeting variants (n = 20 cells from 2 slices from 2 mice for each variant). *P < 0.05, ***P < 0.001, Kruskal-Wallis analysis of variance followed by post-hoc test via Steel’s test with GCaMP6f as control group. (I) Bar chart showing the baseline brightness of the cell body for different GCaMP6f targeting variants (n = 20 cells from 2 slices from 2 mice for each variant). ***P < 0.001, n.s., not significant, Kruskal-Wallis analysis of variance followed by post-hoc test via Steel’s test with GCaMP6f as control group.
Characterization of SomaGCaMP variants in mouse hippocampal cultures
We co-expressed GCaMP6f, GCaMP6f-27-AnkTail-motif-ER2 (SomaGCaMP6f1) or GCaMP6f-27-EE-RR (SomaGCaMP6f2) with the red fluorescent protein miRFP to serve as a cellular tracer, using cultured mouse hippocampal neurons (Figure 1C, D, E). We found that fluorescence decreased at a higher rate along the neurites in SomaGCaMP6f1 (Figure 1F, G) and SomaGCaMP6f2 (Figure 1F, H) expressing cells compared to GCaMP6f expressing cells (Supplemental Table 2, statistics for Figure 1). We also fused GCaMP7 (Dana et al. 2019) to the EE-RR sequence, which we selected due to its bioorthogonality and high level of expression in vivo (Figure 2I), to yield GCaMP7f-27-EE-RR, termed SomaGCaMP7f (Figure 1I, J), which was also soma-localized (Figure 1K, L). The baseline fluorescences of GCaMP6f, SomaGCaMP6f1, SomaGCaMP6f2 and GCaMP6f-NLS (nuclear localization sequence) expressing cells in culture were similar to each other (Figure 3A; Supplemental Table 2, statistics for Figure 3), as were the baseline fluorescences of GCaMP7f and SomaGCaMP7f (Figure 3A). The fluorescent response of each molecule to a single action potential (AP; Figure 3B) were similar between targeted and untargeted GCaMPs (Figure 3C). SomaGCaMPs had SNRs (signal-to-noise ratios; defined as the magnitude of fluorescence change caused by a single AP divided by the standard deviation of the baseline fluorescence) similar to untargeted GCaMPs, whereas GCaMP6f-NLS had an SNR lower than that of GCaMP6f (Figure 3D). We found that SomaGCaMPs had rise (τon) and decay (τoff) times, for a single AP, similar to those of untargeted GCaMP, but that, as expected from previous work, GCaMP6f-NLS had rise and decay times significantly slower than those of GCaMP6f (Figure 3E, F). The resting potential, membrane capacitance, holding current, and membrane resistance of cultured hippocampal neurons did not differ for cells expressing conventional vs. soma-targeted GCaMPs, nor did AP width, amplitude, or threshold (Figure S1; Supplemental Table 5, statistics for Figure S1). In addition, we quantified the distribution of native proteins along axons and found no differences in the locations of any of the endogenous proteins assessed (the potassium channel KV2.1, the sodium channel NaV1.2, and the calcium channel CaV2.1, as well as the scaffolding protein AnkG) between conventional vs. soma-targeted GCaMP-expressing neurons (Figure S2; Supplemental Table 6, statistics for Figure S2). Both SomaGCaMPs and conventional GCaMPs appeared to express in the cytosol, as opposed to on the membrane (Figure S3).
Figure 3. Kinetics and sensitivity of SomaGCaMPs.
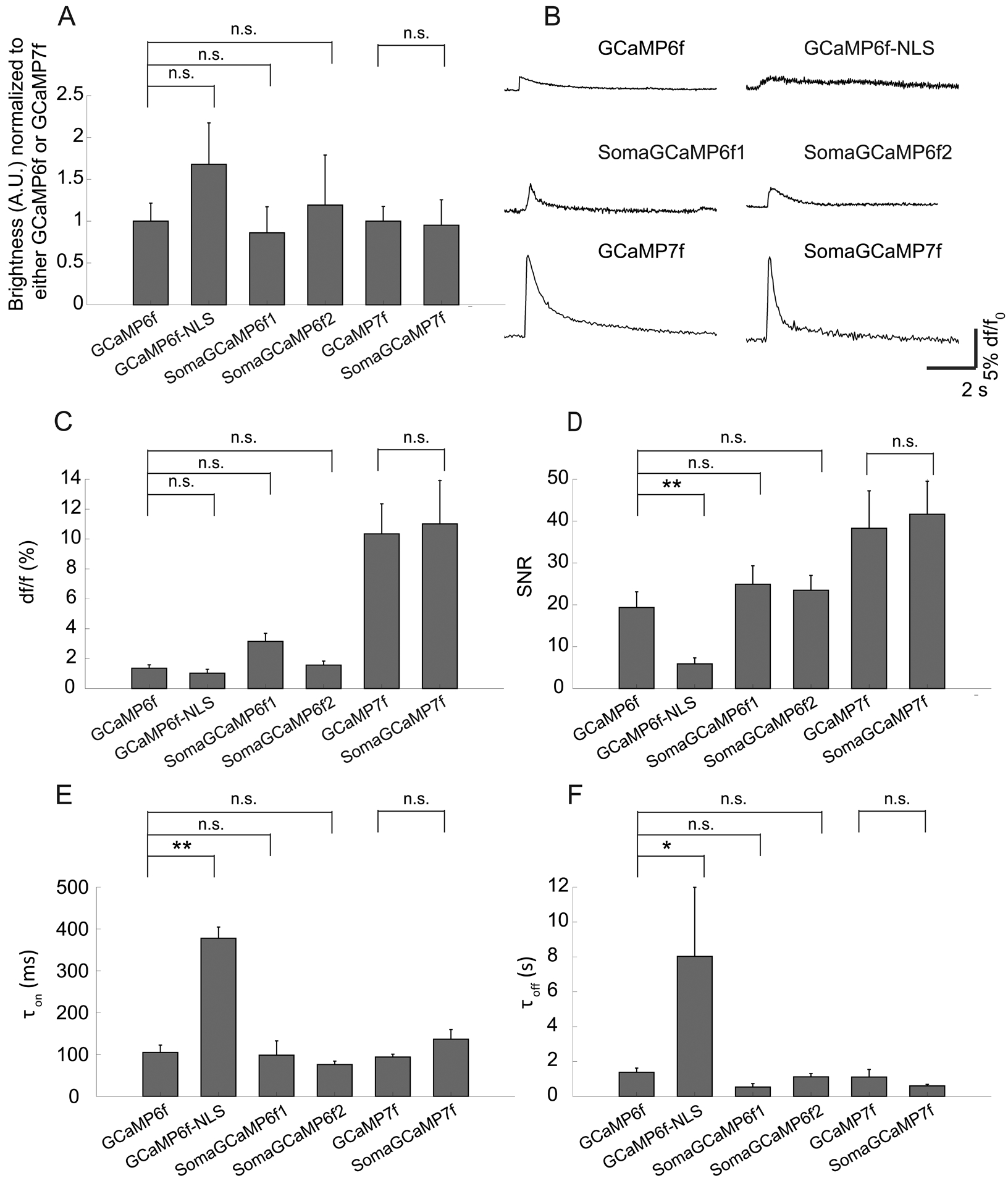
GCaMP6f, GCaMP6f-NLS (nuclear localization sequence), SomaGCaMP6f1, SomaGCaMP6f2, GCaMP7f and SomaGCaMP7f were expressed in hippocampal neurons. (A) Baseline brightness values for GCaMP variants (n = 8 cells from 2 cultures for GCaMP6f; n = 7 cells from 2 cultures for SomaGCaMP6f1; n = 5 cells from 2 cultures for SomaGCaMP6f2; n = 7 cells from 2 cultures for GCaMP6f-NLS; n = 6 cells from 2 cultures for GCaMP7f; n = 7 cells from 3 cultures for SomaGCaMP7f). Brightness was normalized to GCaMP brightness. n.s., not significant; for GCaMP6f, GCaMP6f-NLS, SomaGCaMP6f1, and SomaGCaMP6f2, Kruskal-Wallis analysis of variance followed by post-hoc test via Steel’s test with GCaMP6f as control group; for GCaMP7f and SomaGCaMP7f, Wilcoxon rank sum test; Supplemental Table 2, statistics for Figure 3. Plotted is mean plus or minus standard error throughout. (B) Representative fluorescence response for one action potential (AP) in the cell body for cultured neurons expressing GCaMPs and SomaGCaMPs. (C) df/f0 for GCaMPs and SomaGCaMPs (n = 8 cells from 2 cultures for GCaMP6f; n = 5 cells from 2 cultures for SomaGCaMP6f1; n = 7 cells from 2 cultures for SomaGCaMP6f2; n = 8 cells from 2 cultures for GCaMP6f-NLS; n = 6 cells from 2 cultures for GCaMP7f; n = 7 cells from 3 cultures for SomaGCaMP7f). n.s., not significant; statistical tests as in A. (D) Signal-to-noise ratio (SNR), defined as the magnitude of the fluorescence change caused by a single AP divided by the standard deviation of the baseline fluorescence, for GCaMPs and SomaGCaMPs (n’s as in panel C). **, P < 0.01; n.s., not significant; statistical tests as in A. (E) Time constant for signal rise (Ton) during a single AP for GCaMPs and SomaGCaMPs (n = 8 cells from 2 cultures for GCaMP6f; n = 5 cells from 2 cultures for SomaGCaMP6f1; n = 6 cells from 2 cultures for SomaGCaMP6f2; n = 8 cells from 2 cultures for GCaMP6f-NLS; n = 6 cells from 2 cultures for GCaMP7f; n = 7 cells from 3 cultures for SomaGCaMP7f). **, P < 0.01; n.s., not significant; statistical tests as in A. (F) Time constant for signal decay (Toff) after a single AP for GCaMPs and SomaGCaMPs (n = 7 cells from 2 cultures for GCaMP6f; n = 5 cells from 2 cultures for SomaGCaMP6f1; n = 7 cells from 2 cultures for SomaGCaMP6f2; n = 8 cells from 2 cultures for GCaMP6f-NLS; n = 6 cells from 2 cultures for GCaMP7f; n = 7 cells from 3 cultures for SomaGCaMP7f). *, P < 0.05; n.s., not significant; statistical tests as in A.
Characterization of SomaGCaMP variants in brain slices
We repeated some of the localization experiments of Figure 1, in neurons in mouse brain slices, focusing on GCaMP6f variants for concreteness. We co-expressed GCaMP variants with a red fluorescent protein (mScarlet) in layer 2/3 neurons of the mouse cortex. We used mScarlet to manually trace cells and quantified fluorescence brightness for various GCaMP6f variants (Figure 4A). We normalized the GCaMP6f fluorescence in the green channel by the mScarlet fluorescence to control for the varying size and shape of neural processes, and found that for SomaGCaMPs this ratio decreased to a few percent of the ratio for GCaMP6f-expressing cells over the first 150 microns of neurite (Figure 4B; Supplemental Table 3, statistics for Figure 4). Similar patterns held when we looked at GCaMP6f brightness without mScarlet normalization (Figure 4D).
Figure 4. Decreased neuropil crosstalk in mouse brain slices expressing SomaGCaMP.
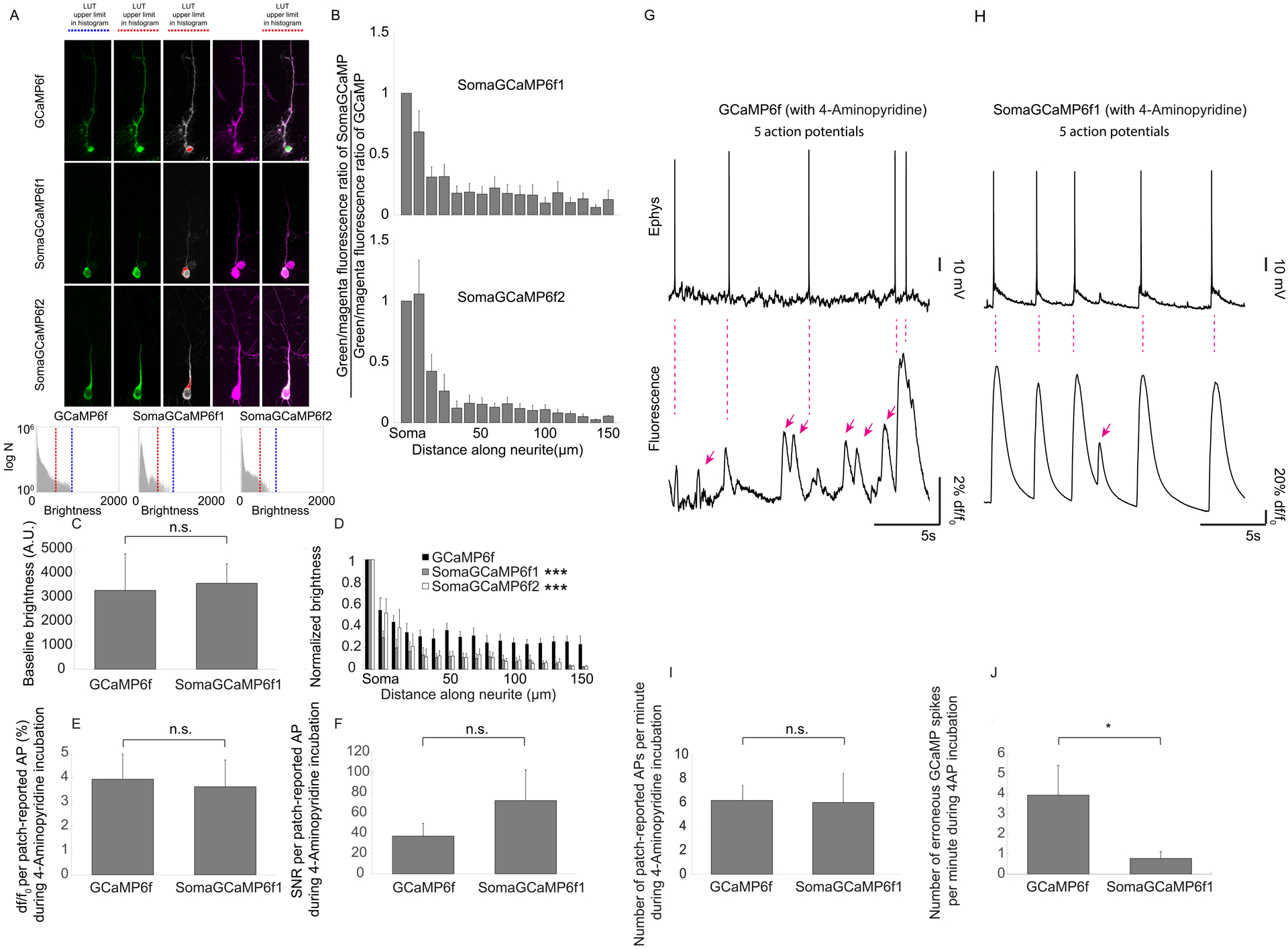
(A) Representative maximum intensity projections of confocal stacks of neurons expressing (from top to bottom) GCaMP6f, SomaGCaMP6f1, and SomaGCaMP6f2 in mouse cortical brain slices. Non-saturated images of the GCaMP channel are presented in the leftmost panels (LUT histogram shown below, with blue bar the upper end of the range), soma-saturated images that highlight GCaMP fluorescence in neurites are given in the next column (LUT histogram below, with red bar the upper end of the range), soma-saturated images with nonsaturated pixels in grey and saturated pixels in red are shown in the third column, mScarlet is shown in magenta in the fourth column, and finally the fifth column shows merged images (between the soma-saturated GCaMP and mScarlet images). Scale bar: 20μm. Supplemental Table 8, percentages of saturated pixels in GCaMP images. (B, top) Bar plots of (SomaGCaMP6f1 brightness / mScarlet brightness) divided by (GCaMP6f brightness / mScarlet brightness) versus position along a neurite. (GCaMP6f, n = 5 neurons from 4 slices from 2 mice; SomaGCaMP6f1, n = 9 neurons from 4 slices from 2 mice). Plotted is mean plus or minus standard error throughout this figure. (B, bottom) As in B, top, but for SomaGCaMP6f2. (GCaMP6f, n = 5 neurons from 4 slices from 2 mice; SomaGCaMP6f2, n = 6 neurons from 3 slices from 2 mice). Supplemental Table 3, statistics for Figure 4. (C) Bar chart showing baseline brightness for GCaMP6f or SomaGCaMP6f1 in brain slice, following light power tuning to make them equal (n = 7 neurons from 2 slices from 2 mice for GCaMP6f; n = 22 neurons from 6 slices from 3 mice for SomaGCaMP6f1). n.s., not significant, Wilcoxon rank sum test. (D) Bar plot of brightness versus position along a neurite, normalized to brightness at the soma, from neurons expressing GCaMP6f variants (GCaMP6f, n = 5 neurons from 4 slices from 2 mice; SomaGCaMP6f1, n = 9 neurons from 4 slices from 2 mice; SomaGCaMP6f2, n = 6 neurons from 3 slices from 2 mice.). ***P < 0.001, Kruskal-Wallis analysis of variance followed by post-hoc test via Steel’s test, comparing to GCaMP6f. (E) Bar chart showing df/f0 at somata of neurons expressing GCaMP6f variants (n = 14 APs from 3 neurons from 3 slices from 2 mice for GCaMP6f; n = 6 APs from 3 neurons from 3 slices from 3 mice for SomaGCaMP6f1). Statistics as in C. (F) Bar chart showing SNR at somata of neurons expressing GCaMP6f variants (n = 14 APs from 3 neurons from 3 slices from 2 mice for GCaMP6f; n = 6 APs from 3 neurons from 3 slices from 3 mice for SomaGCaMP6f1). Statistics as in C. (G, top) Representative patch recording of a cell expressing GCaMP6f under 4-AP stimulation. (G, bottom) GCaMP6f signal from the cell recorded in G, top. Magenta arrows, GCaMP spikes that lack patch spikes. (H) As in G, but for a cell expressing SomaGCaMP6f1. (I) Bar chart showing patch-reported APs per minute in neurons expressing GCaMP6f variants (n = 8 neurons from 8 slices for GCaMP6f from 4 mice; n = 6 neurons from 6 slices for SomaGCaMP6f1 from 3 mice). Statistics as in C. (J) Bar chart showing erroneous GCaMP spikes per minute in neurons expressing GCaMP6f variants (n = 8 neurons from 8 slices from 4 mice for GCaMP6f; n = 6 neurons from 6 slices from 3 mice for SomaGCaMP6f1). *P < 0.05, Wilcoxon rank sum test.
We next characterized whether soma targeting of GCaMP6f could reduce neuropil contamination in brain slices, choosing SomaGCaMP6f1 for this experiment. We found that the baseline brightness of the cell body of SomaGCaMP6f1-expressing neurons was about 5-fold lower than that of GCaMP6f-expressing neurons in live brain slices (Figure 2I, S4; Supplemental Table 6, statistics for Figure S4). Slices expressing GCaMP6f vs. SomaGCaMP6f1 at comparable densities (18 ± 7 cells and 21 ± 5 cells per 106 μm3, respectively; mean ± standard error of the mean is reported unless otherwise indicated; n = 3 slices from 3 mice each). We increased the excitation light power in SomaGCaMP6f1 experiments to match the baseline brightness of GCaMP6f (Figure 4C), for the remaining experiments of Figure 4; in such a condition, the df/f0 of and SNR of GCaMP spikes per single patch-reported spikes observed during 4-AP evoked activity were similar between GCaMP6f and SomaGCaMP6f1 expressing cells (Figure 4E, 4F). The df/f0 of the GCaMP transient driven by a burst (5–20 spikes) was significantly higher in SomaGCaMP6f1 vs. GCaMP6f expressing cells (Figure S5; Supplemental Table 6, statistics for Figure S5).
We then measured the number of fluorescent GCaMP-reported spikes that lack an associated patch-reported spike in brain slices expressing GCaMP6f vs. SomaGCaMP6f1, when slices were exposed to 0.1 mM 4-AP to cause asynchronous spiking (Figure 4G vs. Figure 4H, respectively). Patch-reported spike rates were similar between GCaMP6f- and SomaGCaMP6f1-expressing cells (Figure 4I). GCaMP6f neurons exhibited a roughly 2:3 ratio of erroneous spikes to actual spikes, but in SomaGCaMP6f1 slices, the ratio was reduced to 1:6 (Figure 4J). GCaMP6f-expressing neurons exhibited 10.4±2.2 GCaMP spikes per minute (Figure S5D), similar to the number of electrophysiology-derived APs (Figure 4I, 6.2+/−1.3), plus the number of erroneous spikes (Figure 4J, 3.9+/−1.4). SomaGCaMP6f1 expressing neurons exhibited 6.7±3.0 GCaMP spikes per minute (Figure S5D), similar to the number of electrophysiology-derived APs (Figure 4I, 6.0+/−2.4), plus the number of erroneous spikes (Figure 4J, 0.65+/−0.3).
Simulating the benefits of SomaGCaMP reduction of neuropil contamination vs. post-hoc computational reduction of neuropil contamination
Algorithms for neuropil contamination reduction for one-photon calcium imaging have been developed for neuroscience use. A popular algorithm is constrained nonnegative matrix factorization (CNMF) (Pnevmatikakis et al. 2016), which enables identification of GCaMP-expressing neurons with subsequent demixing and deconvolution of their fluorescence spikes. We simulated calcium transients in mouse (Figure 5A–C) and larval zebrafish (Figure 5D–F) brain, to help us understand the impact of SomaGCaMP vs. CNMF on live brain imaging. We simulated the “ground truth” spikes in the cell bodies (Figure 5A, D) as well as how the data would look in isolated volumes, imaged through a lightsheet microscope (chosen due to its high spatial resolution), reported by GCaMP6f (Figure 5B, E) vs. SomaGCaAMP6f variants (Figure 5C, F). We simulated in-plane and out-of-plane artifacts of neuropil driven by the point-spread-function of the microscope, and then calculated the correlation between the simulated ground truth spiking and the microscope-observed spiking that would be observed, when expressing GCaMP6f vs. SomaGCaMP6f variants. We found that for both mice (Figure 5G) and fish (Figure 5H), the correlation between the simulated ground truth spiking and the microscope-observed spiking reported by SomaGCaMP variants were significantly higher than when the microscope-observed spiking was reported by GCaMP6f (Supplemental Table 4, statistics for Figure 5). CNMF, in contrast, did not increase the correlation between simulated ground truth spiking and the microscope-observed spiking reported by either GCaMP6f or SomaGCaMP variants. Thus CNMF may reduce correlations in calcium imaging data, but at least in simulation, some of that reduction in correlation may be reduction in actual signal.
Figure 5. Simulation of soma-targeting of GCaMP vs. post-hoc computational demixing using CNMF.
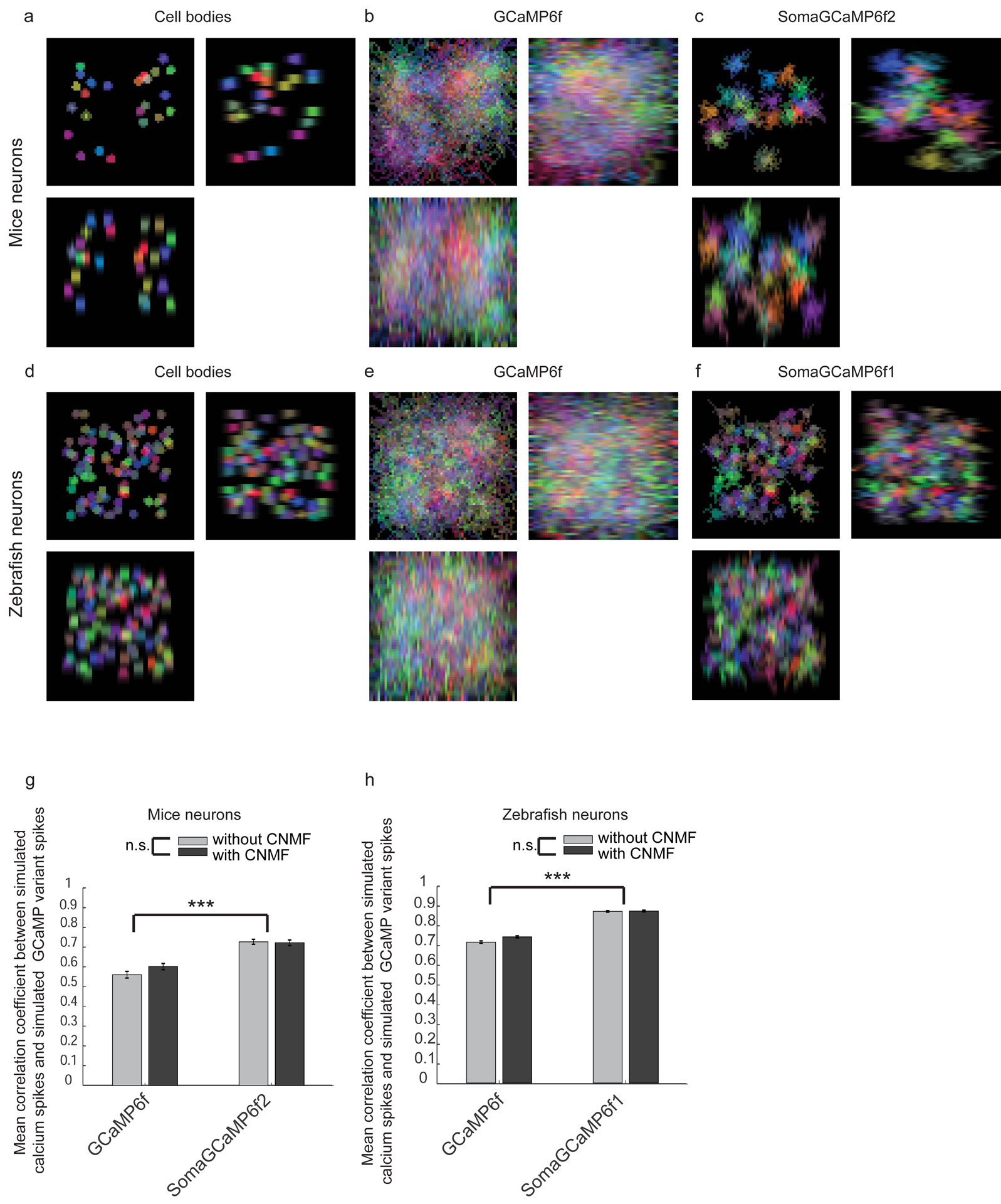
(A) Simulated images of cell bodies from mouse cortical in vivo imaging. Scale bars: 10 μm for XY images, 5 μm for XZ and YZ images, throughout the figure. (B) Simulated images of GCaMP from mouse in vivo imaging. (C) Simulated images of SomaGCaMP from mouse in vivo imaging. (D-F) As in A-C, but for the zebrafish midbrain. (G) Mean correlation coefficient between simulated ground-truth calcium dynamics and simulated recorded calcium dynamics for the mouse brain as in A-C, before (light gray) and after (dark gray) CNMF (n = 300 neurons from 10 simulations for each GCaMP variant). ***P < 0.001, two-way analysis of variance (ANOVA), followed by post-hoc Tukey’s HSD test; Supplemental Table 4, statistics for Figure 5. Plotted is mean plus or minus standard error in G, H. (H) As in G but for the zebrafish (n = 1200 neurons from 10 simulations for each GCaMP variant).
SomaGCaMP reduces crosstalk between neurons in larval zebrafish brain
We transiently and sparsely expressed GCaMP6f, SomaGCaMP6f1, SomaGCaMP6f2, GCaMP7f or SomaGCaMP7f, along with mCherry as a cell morphology marker, in the brains of larval zebrafish (Figure 6A, B). All of these molecules expressed successfully, but we focused on comparing GCaMP6f vs. SomaGCaMP6f1 since the SomaGCaMP6f1 fish transgenic line was the first to be ready for experimentation. We evaluated the green-to-red ratio for SomaGCaMP6f1 and SomaGCaMP7f and found it to decrease to a few percent over the first 150 microns of neurite coming out of the cell body (Figure 6C).
Figure 6. Decreased neuropil crosstalk in SomaGCaMP-expressing larval zebrafish.
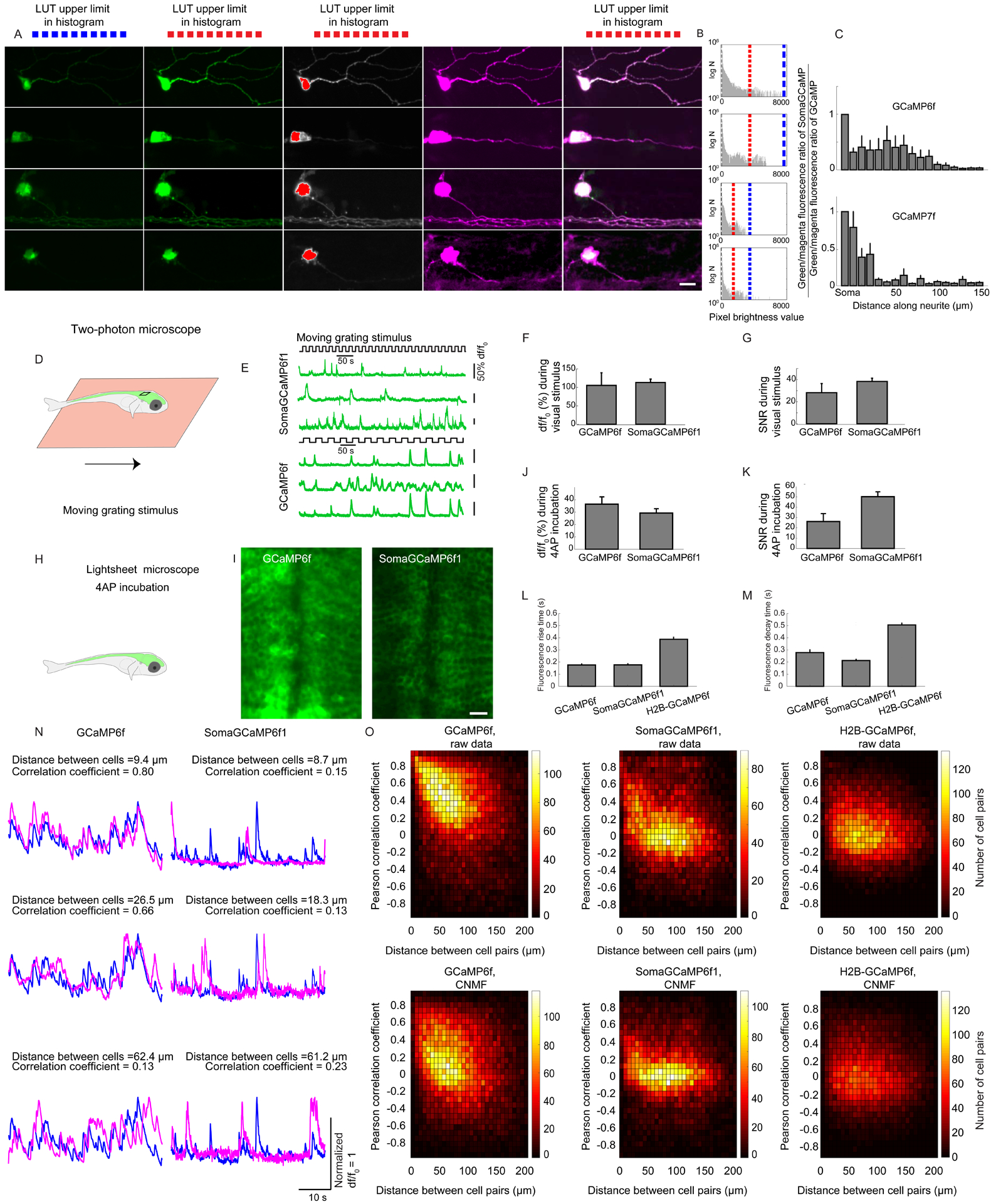
(A) Representative images of neurons expressing GCaMP6f, SomaGCaMP6f1, GCaMP7f, or SomaGCaMP7f in zebrafish larvae at 5 dpf. Images and histograms are formatted as in Figure 4A. Scale bar: 5μm. Supplemental Table 8, percentages of saturated pixels in GCaMP images. (B) Histograms of pixel brightnesses for the images of A, formatted as in Figure 4A. (C, top) Bar plots of (SomaGCaMP brightness/mCherry brightness) divided by (GCaMP brightness/mCherry brightness) versus position along a neurite (GCaMP6, n = 8 neurons from 4 fishes; SomaGCaMP6f1, n = 7 neurons from 6 fishes). Supplemental Table 3, statistics for Figure 6. (C, bottom) As in C, top, but for GCaMP7f (n = 5 neurons from 3 fishes) and SomaGCaMP7f (n = 5 neurons from 3 fishes). (D) Fish were imaged under the 2-photon microscope, and exposed to a moving grating as a visual stimulus. (E) Representative calcium traces for the experiment of D. (F) Bar chart showing df/f0 at somata of neurons in the optic tectum for the experiment of D (n = 6 neurons from 3 fishes for GCaMP6f; n = 5 neurons from 3 fishes for SomaGCaMP6f1). n.s., not significant, Wilcoxon rank sum test. (G) As in F but for signal-to-noise ratio (SNR). (H) Fish were imaged using a lightsheet microscope and 4-AP pharmacological stimulation. (I) Images of neurons expressing GCaMP6f variants taken at a depth of 70 μm from the top of the brain, in the zebrafish midbrain. Scale bar: 10μm. (J) Bar chart showing df/f0 at the somata of zebrafish neurons in the forebrain for the experiment of H (n = 5 neurons from 2 fishes for GCaMP6f; n = 5 neurons from 2 fishes for SomaGCaMP6f1). n.s., not significant, Wilcoxon rank sum test. (K) As in J but for signal-to-noise ratio (SNR). *P < 0.05, Wilcoxon rank sum test. (L) Bar chart of fluorescence rise time at somata of neurons for the experiment of H (n = 101 neurons from 5 fishes for GCaMP6f; n = 146 neurons from 4 fishes for SomaGCaMP6f1; n = 513 neurons from 6 fishes for H2B-GCaMP6f). ***P < 0.001, Kruskal-Wallis analysis of variance followed by post-hoc test via Steel’s test. (M) As in L but for fluorescence decay time. (N) Traces, normalized to maximum, of representative cell pairs in the forebrain expressing GCaMP6f variants that are ~10 μm (top row), ~20 μm (middle row) and ~50 μm (bottom row) apart, during 4-AP stimulation. (O) Density plot showing Pearson correlation coefficients of cell pairs in the forebrain as a function of distance for GCaMP6f (n = 426 cells from 5 fishes), SomaGCaMP6f1 (n = 340 cells from 4 fishes) or H2B-GCaMP6f (n = 676 cells from 6 fishes), during 4-AP stimulation. Top, without CNMF; bottom, with CNMF. ***P < 0.001, two-dimensional Kolmogorov-Smirnov test with GCaMP6f as control condition.
We imaged the tectum of the fish brain with a two-photon microscope while presenting a visual stimulus consisting of a moving grating (Figure 6D) and found that cells expressing GCaMP6f or SomaGCaMP6f1 exhibited fluorescence transients (Figure 6E) with similar df/f0 and SNR (Figure 6F,G; Supplemental Table 3, statistics for Figure 6), when measured with similar light powers. For the following experiments, we generated stably pan-neuronally expressing fish lines. We imaged these fish with a one-photon lightsheet microscope (Figure 6H), and found that in SomaGCaMP6f1 fish cell bodies could be more easily seen because of less neurite fluorescence (Figure 6I). We increased the light power so that SomaGCaMP6f1 would have similar brightness as GCaMP6f, for the remaining panels of Figure 6. For neural activity induced by 1 mM 4-AP, the df/f0 for GCaMP6f and SomaGCaMP6f1 cells in the forebrain were similar, and the SNR for SomaGCaMP6f1 was twice that of GCaMP6f (Figure 6J, K). We detected approximately 3 times more GCaMP spikes in SomaGCaMP6f1 fish or in H2B-GCaMP6f (nuclear GCaMP6f) fish compared to non-targeted GCaMP6f fish (Figure S6; Supplemental Table 6, statistics for Figure S6), suggesting a general utility in avoiding crosstalk for increasing spike count accuracy. GCaMP6f and SomaGCaMP6f1 fish had GCaMP-spikes with similar τon and τoff speeds, but those of H2B-GCaMP6f fish were approximately twice as slow (Figure 6L, M), highlighting the improved kinetics associated with somatic vs. nuclear targeting.
We calculated Pearson correlation coefficients between all the possible neuron pairs in the field of view (Figure 6N) and plotted them vs. distance between neurons (Figure 6O, top panels). We found that in GCaMP6f, SomaGCaMP6f1 and H2B-GCaMP6f expressing brains, the shorter the distance between neuron pairs, the higher the correlation between their GCaMP-spikes (Figures 6O top panels and S6C). However, the mean correlation coefficient in the GCaMP6f case was approximately twice that of the SomaGCaMP6f1 case and approximately three times higher than that of the H2B-GCaMP6f case (Figure 6O top panels, Figure S6B; Supplemental Tables 3 and 6 for full statistics). This suggested that the contamination of cell body signals by neuropil signals could manifest as an artifactual increase in correlation between neural activity patterns.
We applied post hoc cleanup with CNMF (Figure 6O, bottom panels), and found that CNMF decreased the mean correlation coefficient between nearby neurons for all three GCaMP6f variants (Figure S6B), but the mean correlation coefficient was still twice and thrice higher in the GCaMP6f case compared to the SomaGCaMP6f1 and H2B-GCaMP6f cases respectively. Thus, even with CNMF usage (with all the caveats mentioned above), soma-targeting still offered reduced neuron-neuron correlations in the dense larval zebrafish brain.
SomaGCaMP reduces crosstalk, increases SNR and enables detecting more GCaMP spikes in brains of behaving mice
For in vivo mouse experiments, we expressed SomaGCaMP variants in the dorsal striatum of mice, where it has been suggested that medium spiny neurons form populations of clustered cells with highly correlated neural activity (Barbera et al., 2016): measuring such correlations would ideally be done without neuropil contamination (Klaus et al., 2017). We expressed SomaGCaMP6f1 and SomaGCaMP6f2 in the dorsal striatum of the living mouse brain, and imaged both using a conventional wide-field one-photon imaging system (Ali I. Mohammed et al. 2016). Consistent with the zebrafish and mouse cortical slice experiments, we found that the SomaGCaMP6f1 was approximately 5 times dimmer than GCaMP6f, but that SomaGCaMP6f2 was not (Figure S7; Supplemental Table 6, statistics for Figure S7). Therefore we focused on SomaGCaMP6f2 and SomaGCaMP7f (which has a similar design to SomaGCaMP6f2, and is thus also bright) in the live mouse brain. We measured calcium signals in the dorsal striatum between GCaMP- vs. SomaGCaMP-expressing mice while they ran on a spherical treadmill. SomaGCaMP mice exhibited a substantial reduction in neuropil fluorescence as compared to GCaMP mice (Figure 7A–D). We detected GCaMP spikes in both GCaMP- and SomaGCaMP-expressing mouse brains (Figure 7E–H). SomaGCaMP6f2 decay times were faster than GCaMP6f decay times (Figure S7B). SomaGCaMP6f2 reported approximately slightly more calcium events than GCaMP6f, and SomaGCaMP7 reported a similar number of calcium events compared to GCaMP7f (Figure 7I; Supplemental Table 3, statistics for Figure 7).
Figure 7. SomaGCaMP reduces neuropil contamination in the striatum of behaving mice.
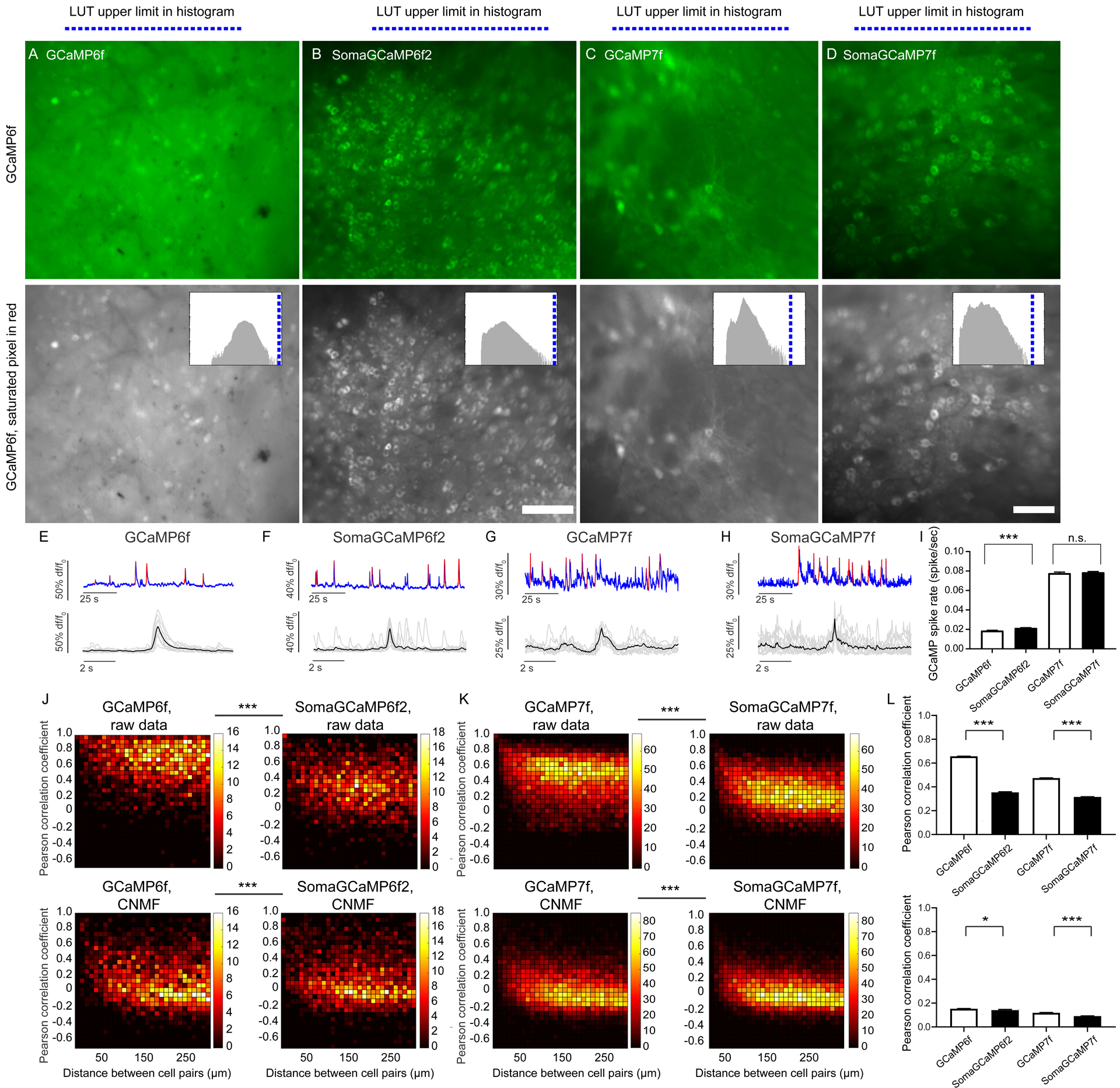
(A, B, C, D, top row) Representative projection images showing the fluorescence summed across the frames of an epifluorescent imaging session, from the dorsal striatum in GCaMP vs. SomaGCaMP mice. Scale bar: 100 μm. (A, B, C, D, bottom row) The images in A, B, C, D presented in grayscale, with saturated pixels in red (note: none are red). Histograms of pixel values in upper right corners; blue line, upper limit of LUT. Supplemental Table 8, percentages of saturated pixels in GCaMP images. (E-H) Top, representative calcium traces from the experiments of A-D. Blue, calcium traces; red, calcium events identified based on thresholding. Bottom, calcium events from the top traces, aligned, with individual events in gray and averages in black. (I) Bar chart of GCaMP-spike rates (n = 930 neurons from 6 GCaMP6f mice, n = 594 neurons from 4 mice expressing SomaGCaMP6f2, n = 634 neurons from 4 GCaMP7f mice, n = 1098 neurons from 5 mice expressing SomaGCaMP7f). ***P <0.001, Kruskal-Wallis analysis of variance followed by post-hoc test via Dunn’s test; Supplemental Table 3, statistics for Figure 7. Shown throughout this figure is mean plus or minus standard error. (J) Correlated fluorescence vs. distance for cell pairs from mice expressing GCaMP6f (left; n = 860 cells from 6 mice) or SomaGCaMP6f2 (right; n = 149 cells from 4 mice), without (top) and with (bottom) CNMF. ***P < 0.001, two-dimensional Kolmogorov-Smirnov test. (K) As in J but for GCaMP7f (left; n = 634 cells from 4 mice) and SomaGCaMP7f (right; n = 1098 cells from 5 mice). (L) Bar plot showing Pearson correlation coefficients (n = 67795 cell-pairs from 6 GCaMP6f mice, n = 44890 cell-pairs from 4 SomaGCaMP6f2 mice, n = 12582 cell-pairs from 4 GCaMP7f mice, n = 10420 cell-pairs from 5 SomaGCaMP7f mice), without (top) and with (bottom) CNMF. *P < 0.05, ***P < 0.001, Kruskal-Wallis analysis of variance followed by post-hoc test via Dunn’s test.
We calculated Pearson correlation coefficients between all the possible neuron pairs within the imaging field for mice expressing either GCaMP vs. SomaGCaMP (Figures 7J–K, top left panels). In GCaMP6f or GCaMP67f-expressing mice, we identified high correlations for nearby cells that fell off with increasing distance. In contrast, SomaGCaMP6f2 or SomaGCaMP7f expressing mice had far lower correlations across the board (Figure 7J–K, top right panels), approximately 1/3 to 1/2 less than for GCaMP (Figure 7L); we even found instances of strong negatively correlated pairs that were not apparent in non-targeted GCaMP mice. We analyzed the mean correlation coefficient for three distance ranges (0–50 μm, 50–150 μm, 100–300 μm) and found that correlation fell off with distance (Figure S7), but this correlation was lower when expressing SomaGCaMP6f2 or SomaGCaMP7f then with GCaMP6f or GCaMP7f respectively (Figure S7C–D). Following CNMF application, the pairwise correlations in GCaMP6f brains decreased, almost to the level mediated by SomaGCaMP6f2 (Figure 7J–K, bottom panels), with the caveats mentioned above.
In addition to wide-field imaging, the advent of gradient-refractive index (GRIN) lenses and miniature head-mounted microscopes has allowed for cellular-resolution one-photon calcium imaging in deep regions of the rodent brain during naturalistic behaviors (Flusberg et al. 2008). This has rapidly seen widespread use in many subfields of behavioral neuroscience such as memory (Cai et al., 2016; Grewe et al., 2017), spatial navigation (Sun et al., 2015; Ziv et al., 2013) motivation and learning (Jennings et al., 2015; Pinto and Dan, 2015), and addiction (Xia et al., 2017). Miniaturized microscopes, however, suffer from poor axial resolution which can result in exacerbation of neuropil contamination and crosstalk. Therefore, extracting activity from microendoscopic videos with confidence is an ongoing challenge in the field, and the optimal methods for addressing this issue is a highly debated topic (Resendez et al., 2016; Siciliano and Tye, 2019). We expressed GCaMP6f or SomaGCaMP6f2 in the medial prefrontal cortex of mice (Figure 8A–D) and gained optical access via a chronically implanted GRIN lens. We imaged both GCaMP6f and SomaGCaMP6f2 using the same imaging parameters, recorded GCaMP spikes (Figure 8E–F), and found that the SNR of the GCaMP spikes was 1.4 times higher in SomaGCaMP6f2 expressing mice compared to mice expressing GCaMP6f (Figure 8G; Supplemental Table 3, statistics for Figure 8). In addition, SomaGCaMP6f2 reported approximately 92% more calcium events than GCaMP6f (Figure 8H). SomaGCaMP6f2 rise and decay times were faster than GCaMP6f decay times reminiscent of that observed in zebrafish larvae (Figure 8I, J).
Figure 8. SomaGCaMP imaging improvements in medial prefrontal cortex of awake mice imaged with endoscopic microscopes.
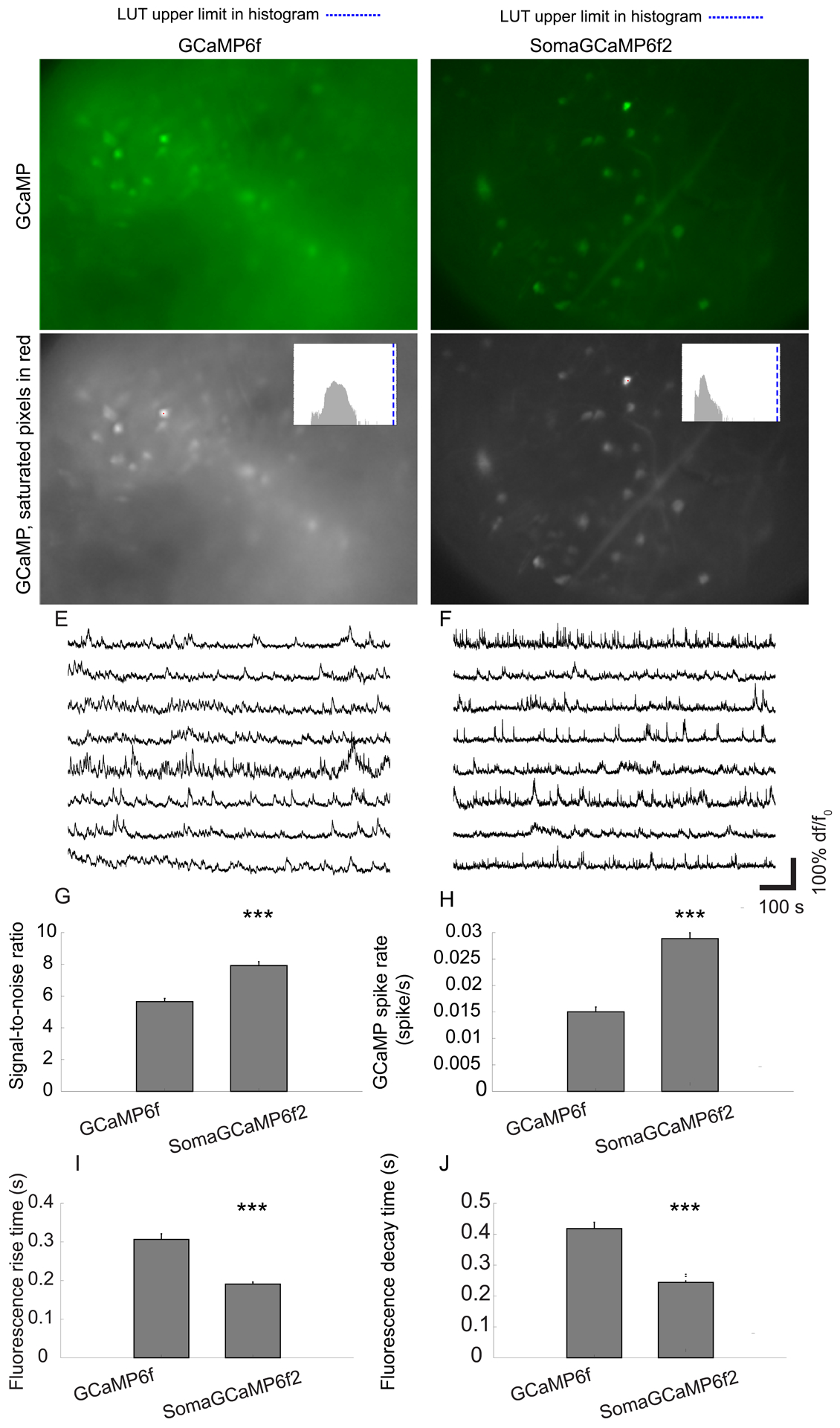
(A, B) Representative standard deviation images showing fluorescence fluctuation across the frames of an epifluorescent imaging session, from the medial prefrontal in GCaMP6f- (A) or SomaGCaMP6f2- (B) expressing mice. (C, D) The images in A, B presented in grayscale, with saturated pixels in red (one pixel in each image). Histogram of pixel values, in upper right corner. Blue line, upper value of histogram LUT. Supplemental Table 8, percentages of saturated pixels. (E, F) Representative calcium traces from neurons in the experiments of A-D. (G) Bar chart of SNR (n = 222 neurons from 4 mice expressing SomaGCaMP6f2, n = 107 neurons from 2 GCaMP6f mice). ***P <0.001, Wilcoxon rank sum test; Supplemental Table 3, statistics for Figure 8. Plotted is mean plus or minus standard error throughout. (H) As in G, but for GCaMP spike rates. (I) As in G, but for fluorescence rise times. (J) As in G but for fluorescence decay times.
We plotted the pairwise correlograms for the microendoscopic data (Figure S8; Supplemental Table 6, statistics for Figure S8), and found that overall pairwise correlations were quite high, and similar between GCaMP6f and SomaGCaMP6f2 (Figure S8A–C), and similar across different distances taken from the small field of view (Figure S8D); CNMF decreased all these correlations by a large factor (Figure S8C,E) compared to the other CNMF analyses described above, possibly because of the low density of expression in this experiment and the high magnitude of background signal present in microendoscope data (see Supplemental Table 3 for statistics on cell density vs. CNMF effects, for Figures 6–8).
Discussion
We here report that it is possible to target genetically encoded calcium sensors to cell bodies in multiple species in vivo. The variants we focused on for further characterization and validation, SomaGCaMP6f1, SomaGCaMP6f2 and SomaGCaMP7f, demonstrated satisfactory brightness (with that of SomaGCaMP6f2 comparable to that of conventional GCaMP6f; and SomaGCaMP7f comparable to that of conventional GCaMP7f; SomaGCaMP6f1 was dimmer than conventional GCaMP6f and may not be preferred), sensitivity, and kinetics in mouse and zebrafish brains. We observed decreased crosstalk, as reflected by lower numbers of artifactual (e.g., not detectable via patch pipette) spikes, and reduced artifactual correlation between neurons that are nearby, in both zebrafish and mouse brain. Although nuclear-localized GCaMP can also achieve isolation between neurons, soma-targeting results in several-fold higher SNR and several-fold faster kinetics, compared to nuclear GCaMP. In addition, computational cleanup of non-targeted GCaMP via application of CNMF decreased correlations between nearby neurons, but in a fashion that may not actually improve the accurate reconstruction of spikes (at least when probed by simulations), a problem avoided by somatic GCaMP that improves the direct reporting of neural activity from individual neurons.
The general principles by which proteins are localized to the soma of neurons are not yet fully elucidated. However, earlier studies of protein trafficking in neurons provide some clues as to how SomaGCaMPs are targeted to the cell body of neurons. The AnkTail-motif used in SomaGCaMP6f1 was shown to be sufficient for GFP targeting to the proximal axon hillock of the cell body in cultured DRG neurons (Zhang & Bennett 1998). It was proposed that AnkTail-motif was capable of binding directly to docking sites in the axon proximal segments and neural cell bodies, although the exact nature of these docking sites is not known. Curiously, the EE-RR-motif employed in SomaGCaMP6f2 is orthogonal to native proteins in neurons and therefore most likely utilizes a different soma-targeting mechanism. The EE-RR peptides may bind each other due to hydrophobic interactions that result in the formation of large intermolecular complexes. These molecular complexes of SomaGCaMP6f2 or SomaGCaMP7f may be sufficient to slow down trafficking away from the cell body (Ramírez et al. 2011). In both cases, expression of these proteins did not alter active nor passive membrane properties, nor the distribution of all endogenous channels that we examined.
Having fewer artifactual spikes will increase the accuracy of the assessment of neural codes in the living brain. Reducing artifactual correlation may also help with studies of functional connectivity. Single-photon calcium imaging has a speed advantage compared to two-photon imaging, and wide-field calcium imaging is simple, feasible and robust. The advantage of SomaGCaMP in performing single-photon imaging in these model systems is that they may enable separation of bona-fide physiological correlation from non-physiological correlation, something that post hoc computational methods cannot guarantee.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ed Boyden, edboyden@mit.edu.
Materials Availability
All SomaGCaMP sequences are posted on Genbank (MK695671, MK695670 and MT364350 for SomaGCaMP6f1, SomaGCaMP6f2 and SomaGCaMP7f respectively), and DNA reagents posted to Addgene for distribution upon publication. Further information and requests for reagents may be directed to, and will be fulfilled by, the Lead Contact and corresponding author, Dr. Ed Boyden (edboyden@mit.edu).
Data and Code Availability
The code generated during this study is available at: https://github.com/dgoodwin208/SomaGCaMP-Processing. The sequences of SomaGCaMP variants can be found through GenBank and Addgene.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Procedures
Procedures at MIT involving animals were in accordance with the National Institutes of Health Guide for the care and use of laboratory animals and approved by the Massachusetts Institute of Technology Animal Care and Use Committee. Procedures at BU were approved by the Boston University Institutional Animal Care and Use Committee. Zebrafish experiments at Janelia were conducted according to protocols approved by the Institutional Animal Care and Use Committee of the Howard Hughes Medical Institute, Janelia Research Campus. Hippocampal neuron culture was prepared from postnatal day 0 or day 1 Swiss Webster (Taconic) mice as previously described (Klapoetke et al. 2014). In-utero electroporation was performed on female Swiss Webster mice (Taconic).
Zebrafish animals and transgenesis
For Figure 6, we used previously published transgenic zebrafish lines expressing either GCaMP6f in the cytosol Tg(elavl3:GCaMP6f)jf1 (Freeman et al. 2014) or GCaMP6f in the nucleus Tg(elavl3:H2B-GCaMP6f) (Dunn et al. 2016). Transgenic zebrafish line Tg(elavl3:SomaGCaMP6f1) and zebrafishes transiently expressing GCaMP6f, SomaGCaMP6f1, SomaGCaMP6f2, GCaMP7f or SomaGCaMP7f, along with mCherry as a cell morphology marker, were generated using previous protocols (Freeman et al. 2014) using the Tol2 transposon system, in which indicators (and mCherry for zebrafishes with transient expression) were subcloned into a Tol2 vector that contained the zebrafish elavl3 promoter. The transgene construct and transposase RNA were injected into 1–2-cell-stage embryos and the transgenic lines were isolated by the high expression of bright green fluorescence in the central nervous system in the next generation. The larvae were reared in 14:10 light-dark cycles according to a standard protocol at 28.5°C, in a solution containing Instant Ocean salt from Carolina Biological Supply Company (65mg/L Instant Ocean, 30 mg/L Sodium bicarbonate). Experiments were performed on animals 5–7 days post fertilization (dpf) at room temperature. DNA constructs for elavl3:SomaGCaMP6f1 and elavl3:SomaGCaMP7f and zebrafish line Tg(elavl3:SomaGCaMP6f1) are available upon request.
METHOD DETAILS
Neuronal culture, transfection, and AAV transduction
For neuronal expression of GCaMP6f fusions with trafficking sequences during the screen for soma targeting sequences (e.g., Figures 1C, D, E, I, J), and for neuronal expression of miRFP (e.g., Figures 1C, D, E, I, J), we transfected primary mouse hippocampal neurons at 4 days in vitro (DIV) with a commercial calcium phosphate kit (Invitrogen). We added an additional washing step with acidic MEM buffer (pH 6.8 – 6.9) after calcium phosphate precipitate incubation to completely re-suspend residual precipitates (Jiang & Chen 2006). We used 1μg of DNA. Neurons were imaged 14–18 DIV (days in vitro; 10–14 days post-transfection). For neuronal expression of GCaMP6f, GCaMP6f-NLS, SomaGCaMP6f1, SomaGCaMP6f2, and/or mCherry for electrophysiology, antibody staining, and membrane staining in Figures 3A–F, S1A–H, S2A–D, and S3A–C, we transduced primary mouse hippocampal neurons at DIV 7–9 with the corresponding AAV(s) with a DJ serotype under CAG or Syn promoter (UNC vector core).
Gene synthesis
All genes were synthesized (by Epoch Life Science) with mammalian codon optimization and subcloned into pAAV backbone under CAG or Syn promoters, see Supplemental Tables 1, 2 and 7 for descriptions and amino acid sequences. Briefly, for the final selected variants, 1200 bp from the tail region of the human AnkyrinG protein (Zhang & Bennett 1998) (AnkTail-motif) were cloned followed by the ER2 (Hofherr et al. 2005) trafficking sequence from the potassium channel Kir2.1, with the resulting molecule being GCaMP6f-27-AnkTail-motif-ER2, named SomaGCaMP6f1, and 264 bp of a de novo designed coiled-coil peptide EE-RR fused to the C-terminus of GCaMP6f via a 27 amino acid flexible linker, named SomaGCaMP6f2. A nuclear localization sequence (NLS) was synthesized based on a sequence found in the literature (Kosugi et al. 2009). SomaGCaMP7f was generated by replacing GCaMP6f with GCaMP7f in SomaGCaMP6f2.
Image analysis
Analysis of GCaMP variant brightness and red fluorescent protein brightness along neurites, and calculation of green-to-red ratio as a function of position in brain slices and zebrafish brains.
Images for this analysis were taken for fixed brain slices prepared as described below using mice at P12 – P24 (Figure 4A, B, D), and for fixed zebrafish larvae at 5–7 dpf (Figure 6A, B, C). Images in the red channel (representing mScarlet in mouse brain slices or mCherry in zebrafish) and in the green channel (representing GCaMP variants) were collected using a spinning disk CSUW1 confocal unit (Yokogawa, Tokyo, Japan). We imaged the neurons using the same parameters for all GCaMP variants (GCaMP6f, SomaGCaMP6f1, SomaGCaMP6f2, GCaMP7f, and SomaGCaMP7f). We chose to image the native GCaMP fluorescence of the GCaMP variants after fixation, as it represented the natural state of the protein, and presented any brightness differences among the GCaMP variants, while not fluctuating in time due to neural activity since the samples were fixed. The image analysis was performed in ImageJ as follows: we used the images in the red channel in order to trace neurons. This was because the SomaGCaMP variant signal decreased from the cell body as one entered the neurites, while mCherry or mScarlet was not somatically targeted. For each neuron we first defined the boundaries of the soma. To that end, we drew a 20 μm diameter circle 5–20 μm away from the soma, inside which there was no apparent fluorescence from the soma or from neurites. Then, we defined the average fluorescence in the circle as the background fluorescence for this neuron, focusing on the red channel. We considered pixels with fluorescence intensity of at least 10% above background levels as part of the soma or processes, and we defined the boundary between soma and its processes manually by examining the cell morphology. Then, we drew a polygon with 5–15 sides along the soma boundary and measured the average fluorescence inside of it, and subtracted the previously calculated background value. The resulting value was considered to be the soma fluorescence in the red channel. We repeated this analysis in the green channel, using the boundaries previously defined by the analysis of the red channel. The resulting value was considered the soma fluorescence in the green channel. To measure fluorescence intensities along neurites, we defined 1μm2 rectangles along the neurite that were up to 150 μm away from the soma at increments of 10 μm in the red channel. (For some neurons we could only measure up to 140 μm because after that the neurites became very dim and/or not traceable with the distal neurites from multiple neurons packed together. As a result, the majority of neurons were measured up to 150 μm, while some cells were only measured up to 140 μm.) The distance between each rectangle and the soma was measured along the respective neurites (not the minimal linear distance from the soma, since neurites were curved). We made sure that the pixel intensity values at the boundaries of each rectangle were at least 10% above the background fluorescence defined above, to be considered inside the neurite. We averaged the fluorescence intensity in each rectangle, then subtracted the background fluorescence, then divided it by the average soma fluorescence and plotted the resulting ratio with respect to distance along the neurite. The ratios for each distance were averaged across neurites and data was plotted (using Matlab) as average and standard error of the mean. We repeated this analysis in the green channel, using the boundaries defined by the red channel. In summary, these analyses yielded the dendrite/soma ratios out to 140–150 μm in increments of 10 μm, in both the green and the red channels. We divided the values in the green channel by the corresponding values in the red channels. We then normalized the green/red ratio values to the green/red value at the soma, to achieve our final measure of green to red fluorescence ratio. Importantly, we chose dendrites by tracablity: if a dendrite was crossing other dendrites and thereby we could not determine its continuity, we excluded it from the analysis.
Analysis of GCaMP brightness along neurites, in cultured neurons.
Images for this analysis were taken for cultured neurons (Figure 1F, G, H, K, L) at 14–18 DIV (10–14 days post-transfection). The image analysis was performed in ImageJ. For each neuron we first defined the boundaries of the soma. To that end, we drew a 20 μm diameter circle near the soma, inside which there was no apparent fluorescence from the soma or from neurites. We defined the average fluorescence in the circle as background fluorescence. We considered pixels with fluorescence intensity of at least 10% above background levels as part of the soma and processes, and we defined the boundary between soma and its processes by the apparent cell morphology. Then, we drew a polygon along the defined soma boundary and measured the average fluorescence inside of it, and subtracted the previously calculated background value. The resulting value was considered soma fluorescence. To measure fluorescence intensities along neurites, we defined 1μm2 rectangles along the neurite that were up to 100 μm away from soma at increments of 10 μm. The distance between each rectangle and the soma was measured along the neurites (not the minimal linear distance from the soma, since neurites were curved). We then defined the background value exactly as described above for the soma. We made sure that the pixel intensity values at the boundaries of the rectangle were at least 10% above background levels, to be considered inside the neurite. We averaged the fluorescence intensity in each rectangle, then subtracted the background, then divided it by the average soma fluorescence and plotted the resulting ratio with respect to distance along the neurite. The ratios for each distance were averaged across neurites and data was plotted (using Matlab) as average and standard error of the mean.
Analysis of ion channel and scaffold protein distribution in cultured neurons.
Primary mouse hippocampal neurons (Figure S2A–D) were transduced with either GCaMP6f + mCherry, SomaGCaMP6f1 + mCherry or SomaGCaMP6f2 + mCherry. Images for this analysis were taken from neurons fixed at 14–18 DIV (10–14 days post-transduction). Following fixation, we stained the proteins as described below. Images in the red channel (representing mCherry), in the green channel (representing GCaMP6f variants), and in the near-infrared channel (representing protein staining) were collected using a spinning disk CSUW1 confocal (Yokogawa, Tokyo, Japan). We imaged the neurons using the same parameters for GCaMP6f, SomaGCaMP6f1 and SomaGCaMP6f2. The image analysis was performed in ImageJ as follows: we used the images in the red channel in order to trace neurons. This was because the SomaGCaMP variant signal decreased from the cell body and into the neurites, while mCherry was non-targeted. For each neuron we first defined the boundaries of the soma using the red channel as described in the previous sections. Then, we switched to the near-infrared channel, and calculated the background value for this neuron as described above. We next drew a polygon along the defined soma boundary and measured the average fluorescence inside of it, and subtracted the previously calculated background value. The resulting value was considered soma fluorescence in the near-infrared channel. To measure fluorescence intensities along neurites, we defined 1μm2 rectangles along the neurite that were up to 100 μm away from soma at increments of 10 μm in the red channel. The distance between each rectangle and the soma was measured along the neurites (not the minimal linear distance from the soma, since neurites were curved). We chose to trace the neurite which had the highest intensity for each neuron, meaning it was the axon. This is based on past reports showing that the highest labeling for KV2.1 (Jensen et al. 2017), Nav1.2 (Tian et al. 2014), AnkyrinG (Zhang & Bennett 1998) and CaV2.1 (Yu et al. 2010) is along the axon. We averaged the fluorescence intensity in the near-infrared channel for each rectangle, then subtracted the background value of this neuron, then divided it by the average soma fluorescence and plotted the resulting ratio with respect to distance along the neurite. The ratios for each distance were averaged across neurites and data was plotted (using Matlab) as average and standard error of the mean.
Analysis of membrane distribution in cultured neurons.
Primary mouse hippocampal neurons (Figure S3A–C) were transduced with either GCaMP + mCherry, SomaGCaMP6f1 + mCherry or SomaGCaMP6f2 + mCherry. Images for this analysis were taken from neurons fixed at 14–18 DIV (10–14 days post-transduction). Following fixation, we stained the membrane as described below. Images in the red channel (representing mCherry) and in the green channel (representing GCaMP6f variants) and a near infrared channel (representing the membrane staining) were collected using a spinning disk CSUW1 confocal (Yokogawa, Tokyo, Japan). We imaged the neurons using the same parameters for GCaMP6f, SomaGCaMP6f1 and SomaGCaMP6f2. The image analysis was performed on single confocal z slices in ImageJ as follows: we first defined the boundaries of the soma using the red channel as described above. Then, we switched to the near-infrared channel (membrane staining) and the green channel (GCaMP6f variant), and calculated the background values for each channel as described above. We next drew a rectangle with a width of 32 microns and a height of 1 micron that went through the cell body from side to side. We chose this width since the diameter of cell bodies is smaller than 32 μm, and therefore we were able to analyze the fluorescence from one side of the cell body to the other. We then measured the fluorescence along the wide dimension of the rectangle, at increments of 0.3 μm, in both the green (GCaMP6f variant) and the near infrared channel (membrane staining), subtracted the background for each channel and plotted them along the same x-axis. We noticed that in several cases the nucleus was included in the rectangle and in some not, however we were focused on the plasma membrane and its relation to the GCaMP6f variants, and found, as discussed in the results that the membrane staining fluorescent signal was starting to rise further away from the center of the cell body compared to the GCaMP fluorescent signal.
Analyzing brightness, df/f0, SNR, fluorescent rise-time and fluorescence decay time following 1 action potential in-vitro
For Figure 3, hippocampal cells expressing the GCaMP6f trafficking variants were bathed with synaptic blockers (0.01 mM NBQX and 0.01 mM GABAzine) and patched (in current clamp), and at the same time images were acquired with a Hamamatsu Orca Flash 4.0 with an exposure of 20 ms. An action potential was elicited in the neuron using a 10 ms, 50–200 pA current injection, and the resulting fluorescence change was recorded for a period of 20 seconds, to allow the GCaMP6f fluorescence to return to baseline. To avoid sampling bias, we imaged and patched the first 2–3 cells detected according to the GCaMP fluorescence brightness in each plate. To calculate the GCaMP6f brightness at the soma of each cell, we defined the boundary of the soma by the apparent cell morphology in the image and subtracted the background fluorescence (as defined above) from the average fluorescence inside the soma boundary. To calculate df/f0 we first calculated baseline fluorescence. Baseline fluorescence was defined as the average fluorescence during the 1-second period right before the beginning of the fluorescence response. df/f0 was calculated by dividing the maximum fluorescence change by the baseline fluorescence. To calculate the SNR we divided the maximum fluorescence change by the standard deviation of baseline fluorescence during the 1-second period right before the onset of a GCaMP-spike. We calculated τon by extracting the time constant from the exponential fit of the rising segment of the fluorescence response. We calculated τoff by extracting the time constant from the exponential fit of the falling segment of the fluorescence.
Measuring df/f0 and soma-to neuropil ratio in acute brain slices for SomaGCaMP variant screening
For Figure 2A–I, regions of interest (ROIs) denoting cell bodies and neuropil were determined manually on a projection of the standard deviation of the fluorescence per pixel in the movies using ImageJ: twenty cells and one neuropil section were traced by hand using ImageJ’s freehand selection and ROI manager tools, from which 21 time histories of average fluorescence values F were extracted of length 2000 frames (40 seconds at 50Hz). The baseline fluorescence was defined as a 4-second time window with no apparent action potentials, from which we define B as the mean value in the baseline. For each neuron, we defined the df/f0 as . We next calculated the soma to neuropil df/f0 ratio by dividing the soma df/f0 by the neuropil df/f0.
Measuring the fluorescent signals from cell bodies in slice patching and imaging crosstalk experiments in mouse brain slices
When choosing a region of interest (Figure 4E–J, Figure S5A–D), we chose an area that was inside the cell body. We avoided choosing the ROI as the entire cell body, since that ROI may contain GCaMP6f filled processes originating from neighboring cells. We defined a cell body by the apparent cell morphology as was done as in the in vitro current clamp experiments. We then chose an ROI inside the cell body, approximately 1μm from the cell body’s apparent boundaries.
Analyzing brightness, df/f0 and SNR in acute slice patching experiments of GCaMP6f or SomaGCaMPf1
For Figure 4E, F and Figure S5A, we defined the boundary of the soma by the apparent cell morphology from the movies recorded in slice patching experiments, and measured the average fluorescence inside the soma boundary in each frame. To calculate df/f0 we first calculated baseline fluorescence. Baseline fluorescence was defined as the average fluorescence during the 100 to 500 ms period right before the beginning of fluorescence response. df/f0 was calculated by dividing the maximum fluorescence changes over baseline fluorescence in each cell body. To calculate the SNR we divided the maximum fluorescence change by the standard deviation of baseline fluorescence during the 100 to 500 ms period right before the onset of GCaMP-spikes.
Analyzing brightness, df/f0, SNR and correlations in zebrafish larvae with either transient expression or stable pan-neuronal expression of GCaMP6f or SomaGCaMPf1
The movies recorded from zebrafish larvae with stable pan-neuronal expression using a lightsheet microscope (Figure 6H–O) were first motion corrected using NormCorre (Pnevmatikakis and Giovanucci 2017). The movies recorded from zebrafish larvae with transient expression using a 2-photon microscope (Figure 6D–G) were not motion corrected because little motion was observed. We defined the boundary of the soma by the apparent cell morphology from the movies, and measured the average fluorescence inside the soma boundary in each frame. To calculate df/f0 we first calculated baseline fluorescence. Baseline fluorescence was defined as the average fluorescence during the 1-second period right before the beginning of a fluorescence transient. df/f0 was calculated by dividing the maximum fluorescence change by baseline fluorescence in each cell body. To calculate the SNR we divided the maximum fluorescence change by the standard deviation of baseline fluorescence during the 1-second period right before the onset of a GCaMP-spike. To calculate correlation-coefficients between neuronal pairs in zebrafish larvae with stable pan-neuronal expression of GCaMP6f or SomaGCaMPf1, we processed the motion corrected movies with CaImAn (Pnevmatikakis et al 2016) to segment all putative neurons in the field of view, then denoised and deconvolved the fluorescence traces. An additional manual review was done for each candidate neuron from CaImAn to examine the spatial footprint and temporal characteristics to confirm it was a neuron. These filtered sets of neurons were then used for pairwise correlations (Figure 6 N,O) of the denoised time signal and pairwise distance measurements using the centroid of the spatial footprints.
Analysis of in vivo calcium imaging data in live mice (for Figure 7A–G)
a). Motion correction
Sessions varied between 5 and 12 minutes in length and imaging sessions were analyzed from four SomaGCaMP6f2 mice and six GCaMP6f expressing mice. Motion correction was performed with a custom python script. For each imaging session, a reference image was generated by projecting the mean values of every pixel in the first 2047 frames of the recording session. The reference image and each frame of the video underwent a series of image processing steps to enhance the contrast and the character of the image. We first high-pass filtered the image with a Gaussian filter (python SciPy package, ndimage.gaussian_filter, sigma=50) to remove any potential non-uniform background. We then enhanced the edges of the high intensity areas by sharpening the image as described in http://www.scipy-lectures.org/advanced/image_processing/. In brief, we consecutively low-pass filtered the image with Gaussian filters at two levels (sigma = 2 and 1). The differences in the two images, which represent the edges of high intensity areas, were multiplied by 100 and added back to the first low-pass filtered image, resulting in a sharpened image. Finally, to compensate for potential bleaching that may affect the overall intensity of the whole image, we normalized the intensity of each image by shifting the mean intensity to zero and divided by the standard deviation of the intensity.
We then calculated the cross-correlations between the enhanced reference image and each frame to obtain the displacement between the location of max correlation coefficient and the center of the image. The shift that countered the displacement was then applied to the original, unenhanced image to complete the motion correction.
b). Identification of regions of interest from mouse in-vivo experiments:
To identify the regions of interest (ROIs) that represent neurons, we first generated time-collapsed images by subtracting the average intensity value of each pixel over all videos from its maximum intensity. We then applied ACSAT (Shen et al. 2018) to generate ROIs with the following parameters: iteration=2, minimum size=50 pixels, and maximum size=300 pixels. In brief, ACSAT is a threshold-based ROI segmentation algorithm that adaptively adjusts the threshold at both global and local levels to capture ROIs with various intensities. Due to the shifting process during motion correction, the time-collapsed image often contains high intensity strips at the edge, which cause false-positive ROIs in ACSAT. Therefore, we excluded any ROIs within 10 pixels of the edge. Also, ROIs that were identified which were exceedingly large or small in size (less than 50 pixels or greater than 500 pixels) were excluded. Centroids were then identified for each ROI using the MATLAB command “regionprops” with the “centroid” argument.
c). Trace interpolation for mouse in-vivo experiments:
While SomaGCaMP6f2 sessions were recorded at a constant rate of 20Hz by the camera, the sampling frequency for GCaMP6f sessions was triggered by a MATLAB script which accidentally introduced an unintentional slight variability within the sampling rate (21.31 +/−0.02 Hz (+/− s.d)). Therefore, traces for GCaMP6 were interpolated between the first and last time point in each 4-video sequence given by the time stamps of the corresponding Tiff files. Interpolation was performed with a constant sampling interval of 50ms (20 Hz) using linear interpolation (“interp1” in MATLAB).
d). Computation of df/f0 and linear detrending for mouse in-vivo experiments:
After interpolating the traces from GCaMP6f sessions, df/f0 values were computed for each trace by subtracting its mean and dividing by its initial fluorescence. Each trace was then subject to a linear detrending using the MATLAB command “detrend”. Following this step, traces were each manually inspected to ensure that they had a dynamic nature and represented actual neurons. Traces that didn’t meet these qualifications were excluded from further analysis (n=12 SomaGCaMP6f2 and n=15 GCaMP6f cells).
e). Identification of homologous subregions from GCaMP6f session for mouse in-vivo experiments:
To equalize the number of neurons recorded from each session and to keep the range of distances between cells consistent from different imaging sessions, only a portion of the full field was analyzed from each recording session. To do so, we highlighted subregions from each GCaMP6f session for further analysis. First, we characterized the visible brain region in each GCaMP6f session by computing a bounding box around the area of cell labeling, and computed the total number of neurons in each bounding box. These computations were performed as follows:
First, an ROI mask was constructed for each session. Each mask was then morphologically closed using the MATLAB function imclose(*,strel), with “strel” a structuring element, in this case set to the shape of a disk with a radius of 30 pixels (strel(‘disk’,30)). Second, this image was morphologically eroded using the MATLAB command “imerode”, again using a “disk”-type structuring element but in this case with a radius of 10 pixels. Finally, the image was morphologically dilated using the MATLAB command “imdilate”, and a structuring element of a disk with radius 20 pixels. This produced an image with an opaque region encompassing the region of the image most densely laden with ROIs. Following these procedures, we computed a bounding box around this region using the command “regionprops” with a second argument of “boundingbox”. Finally, the number of ROIs with centroids in this bounding box was computed for each session. Limits of the bounding box used for calculating relative positions of the centroids were computed by rounding the coordinates of the x and y starting points of the bounding box, and taking those points between these values through the values (extent of x = round(x+width-1), extent of y = round(y+height-1)), where height and width are the properties of the bounding box returned by MATLAB. Centroids were rounded to their nearest whole pixel values for this analysis.
To compute the factors necessary to identify a bounding box across all other sessions, we computed summary statistics of these bounding boxes for each GCaMP6f session. To identify the height of our bounding box, we divided the height of each bounding box by the bounding box’s area, averaged these quantities, and then multiplied them by the average area across all bounding boxes. An analogous procedure was performed to find a suitable bounding box width. Lastly, the number of ROIs identified in each bounding box were averaged to find a target number of neurons. In summary, our target region had a height of approximately 396 μm, a width of approximately 804 μm, yielding an area of 3.1856e+05 μm2, with approximately 177 neurons in this region. Our SomaGCaMP6f2 data had an average bounding box height of approximately 373 μm, a width of approximately 715 μm, and an average area of 2.64e05 μm2. To locate an area that fulfilled these requirements, the height and width estimated were first both rounded to whole numbers. Then, first by vertical pixels and then by horizontal pixels, areas constituting the required widths and heights were searched and the number of neurons with (rounded) centroids within these areas were counted. After all rectangles with these characteristics were searched, the region identified that had a number of neurons closest to the average number of neurons in bounding boxes from all other sessions was used as the region for analysis. If multiple regions had the same number of ROIs or were equally close in number, the first region that was identified was used. For the remainder of these analyses (peak characteristic comparison and pairwise-correlation analysis), only the identified ROIs within this region were used.
f). Event identification for mouse in-vivo experiments:
Spectral frequency analysis has been shown to be a reliable tool for estimating calcium fluorescence events as it is less influenced by drifts in baseline activity (Patel et al. 2015; Ruffinatti et al. 2013; Deneux et al. 2016). Within our data we noticed that the onsets of Ca events could be detected using Fourier analysis where event onset coincided with increasing low frequency power (powerevent). To take advantage of this observation, we first calculated the spectrogram from traces (Matlab chronux, mtspecgramc with tapers=[2 3] and window=[1 0.05]), and averaged the power below 2 Hz. To detect any significant increase in power, we calculated the change in the power at each time point (powerdiff), and identified the outliers (3 median absolute deviations away from the median power) in powerdiff (Matlab function isoutlier). For outliers that occurred at consecutive time points, we only kept the first outliner, which represented the start of the change. We further selected the outliers with positive powerdiff as they were indicators for the increase in the power. After identifying the time points of the significant increase, we then determined the end of powerevent by identifying the first time point where the power decreased.
To obtain the peaks and start points of Ca events, we first extended the end point of powerevent to the second time point with decreased Ca signal. After extension, the peak was defined as the time point within powerevent where the maximum Ca signal occurred, and the start point was defined as the time point with minimum Ca signal between the peak and the start of powerevent. To ensure the quality of Ca events, we excluded any Ca event with amplitude (the signal difference between the peak and the onset) less than 4 standard deviations of the trace in the 20 second time window prior to Ca event onset. At the end of this process, some Ca events were found to overlap. To address this issue, the final set of Ca events was set to be the union of all of the identified Ca events, and the peak amplitude of each new event was defined as the maximum of the event minus the minimum of the event.
g). Computation of peak characteristics for mouse in-vivo experiments:
Once peaks were identified, we then determined their waveforms. Waveforms were defined as 10 seconds flanking (5 seconds before and 5 seconds following) an event peak. Once identified, we subtracted the minimum value off the waveform. Then, event rate, rise time and decay times were computed as follows. To compute the event rate for a particular session, the number of waveforms identified over the course of the session were totaled for each region of interest, and this number was then divided by the total length of the session. Next, rise times were computed using the mean post-minimum subtracted peak waveform taken across all waveforms for a given ROI. These waveforms are aligned naturally because each is centered around its peak. To obtain the rise and decay time for each ROI, we first calculated a threshold as following: all events were averaged together, centered around their peak maxima, and the following equation was used to determine a threshold value:
For rise time, the number of data points between the maximum of each identified event and the first point prior to the event where the trace fell to less than or equal to a significance threshold were computed. Falling times were computed by determining the number of data points between the maximum of an event and the first point following this maximum whose value dropped to a value less than or equal to the significance threshold. Any trace that lacked either an identified rise time or decay time, or both, was excluded from statistical analyses, and were also excluded from the computation of pairwise correlations. Event rates, fall times, and rise times computed ROI-wise from SomaGCaMP6f2 mice were compared with the respective values from ROIs in GCaMP6f mice via a Wilcoxon rank-sum test.
h). Pairwise-correlation analysis for mouse in-vivo experiments:
Following application of the CNMF algorithm, traces for each region of interest were truncated into 50 time point (2.5 second) segments in order to reduce the risk of non-stationarity of the df/f0 time traces, and correlation coefficients were computed pairwise over the course of each session. Pairwise correlation coefficients were then averaged over all of the segments of each session for each pair of ROIs. For statistical analysis, the average pairwise correlation coefficient across all ROI pairs for each recording session was computed, and results from GCaMP6f and SomaGCaMP6f2 animals were compared using a Wilcoxon rank-sum test.
Image Processing and Analysis for mice in vivo endoscopic recordings
Image preprocessing of endoscope data (Figure 8 and Figure S8) was accomplished using Mosaic software (v.1.1.2., Inscopix). Raw videos were pre-processed by applying ×4 spatial downsampling to reduce file size and processing time. Lateral movement was corrected for by using a portion of a single reference frame (typically a window surrounding a constellation of neurons) as previously described (Vander Weele et al. 2018) . Images were cropped to remove post-registration borders and sections in which cells were not observed.
After motion correction and cropping, we used extracted fluorescence activity traces from single cells either as they were (Figure S8A,B, upper panels) or by using a modified version of the constrained non-negative matrix factorization algorithm optimized for micro-endoscopic imaging (CNMF-E) developed by Zhou and colleagues (2016) (Figure S8A,B, bottom panels). As described previously (Vander Weele et al. 2018), our analysis differed from CNMF-E (Zhou et al. 2016) in that cells were identified manually by hand-selecting seeding pixels, based on visual inspection of the video and an image generated by plotting the peak-to-noise ratio for each pixel over the length of the video.
Simulation of calcium imaging in densely labeled tissue in mouse and zebrafish with GCaMP6f and SomaGCaMP variants
To simulate calcium imaging in densely labeled tissue in mouse and zebrafish with GCaMP6f and SomaGCaMP variants (Figure 5), we utilized the simulation software from Yoon et al 2019 (provided to the editors at Neuron as requested by the editors via the online submission forms). Briefly, a given number of neuron cell bodies are generated randomly in space of a specified volume. Neuronal processes are created as a random walk of specified length starting from the cell bodies, and the trafficking of calcium indicator is modeled by parameters observed experimentally (see table below). The optics of a given microscope are then modeled; in this case we modeled the population of neurons as it would be viewed through a 1-photon microscope, under the following resolution: 1 micron × 1 micron × 2 microns. For a 25 s duration, ground truth activity was randomly generated per neuron and a video of the optical calcium dynamics was simulated, as if it were taken from a microscope. We then applied CNMF to the data and calculated the correlation in time between the ground truth neuron activity and the observed simulated neuron activity. To simulate densely labeled tissue in mouse, we used 30 neurons in a 64×64×64um volume as the neuronal density and 20 neuronal processes per neuron. To simulate densely labeled tissue in zebrafish, we used 120 neurons in a 64×64×64um volume as the neuronal density and 5 neuronal processes per neuron.
Parameter table:
| Mouse | Zebrafish | |
|---|---|---|
| Number of neurons per 64×64×64um volume | 30 | 120 |
| Number of neural processes per neuron | 20 | 5 |
| Length of GCaMP6f fluorescent signal process (in microns) | 50 | 50 |
| Length of SomaGCaMP6f variant fluorescent signal in process (in microns) | 5 | 5 |
Antibody and membrane staining of fixed neuron culture
Primary mouse hippocampal neurons were fixed at 14–21 days in vitro with 1x phosphate-buffered saline (PBS) + 4% paraformaldehyde for 10 min at room temperature (RT), quenched with 1x PBS + 100 mM glycine for 5 min at RT, and washed twice in 1x PBS for 5 min at RT. Fixed neurons were permeabilized in 1x PBS + 0.1% Triton X-100 for 15 min at RT, and then blocked in the BLOTTO-T solution, which was made by adding 0.1% Triton X-100 into the commercially available BLOTTO solution (#37530, Thermo Fisher Scientific), for 45 min at RT with gentle shaking. Fixed neurons were incubated in primary antibodies in BLOTTO-T for 45 min at RT with gentle shaking, and then washed in BLOTTO-T for 3 times for 5 min each at RT with gentle shaking. Fixed neurons were incubated in secondary antibodies in BLOTTO-T for 45 min at RT with gentle shaking, and then washed in 1x PBS for 3 times for 5 min each at RT with gentle shaking. For samples stained with mouse monoclonal primary antibodies (including anti-Nav1.2 and anti-Kv2.1), anti-mouse-IgG-subclass specific secondary antibodies were used (anti-mouse-IgG2a and anti-mouse-IgG1, respectively).
Primary and secondary antibodies and concentrations used:
Anti-AnkyrinG, Santa Cruz Biotechnology (sc-12719) at 1:50; anti-mouse IgG (H+L) Alexa 647 (A-21235) at 1:200. Anti-Nav1.2, NeuroMab (75–024) at 1:1000; anti-mouse IgG2a Alexa 647 (A-21241) at 1:1500. Anti-Kv2.1, NeuroMab (75–014) at 1:1000; anti-mouse IgG1 Alexa 647 (A-21240) at 1:1500. Anti-Cav2.1, Alomone (ACC-001) at 1:250; anti-rabbit IgG (H+L) Alexa 647 (A-21245) at 1:1000. Anti-mCherry, Kerafast (EMU106) at 1:1000; anti-rabbit IgG (H+L) Alexa 546 (A-11035) at 1:300. Anti-mCherry, Thermo Fisher Scientific (M11217) at 1:1000; anti-rat IgG (H+L) Alexa 546 (A-11081) at 1:300.
Membrane staining was performed with fluorophore-conjugated wheat germ agglutinin (#W32466, Thermo Fisher Scientific) following the manufacturer’s protocol. Briefly, neurons were fixed in the same way as described above, but without the permeabilization and blocking procedures. Fixed neurons were washed 3 times in Hank’s Balanced Salt Solution (HBSS) for 5 min each at RT, and then incubated with Alexa Fluor 647-conjugated wheat germ agglutinin in HBSS at 5 μg/mL for 10 min at RT. Neurons were then washed twice in HBSS and once in 1x PBS, for 5 min each at RT.
Electrophysiology
Current and voltage clamp recordings of cultured neurons
Whole cell patch clamp recordings in culture (for Figure 3 and Figure S1) were made using Axopatch 200B or Multiclamp 700B amplifiers, a Digidata 1440 digitizer, and a PC running pClamp (Molecular Devices). For in vitro current-clamp recordings, neurons were patched 14–18 DIV (7–11 days after AAV transduction) to allow for sodium channel maturation. Neurons were bathed in room temperature Tyrode containing 125 mM NaCl, 2 mM KCl, 3 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 30 mM glucose and the synaptic blockers 0.01 mM NBQX and 0.01 mM GABAzine. The Tyrode pH was adjusted to 7.3 with NaOH and the osmolarity was adjusted to 300 mOsm with sucrose. For in vitro voltage-clamp recordings, neurons were patched 19–21 DIV (17–20 days post-transfection) and were done under similar conditions as current-clamp recordings, except the Tyrode also contained 1 μM tetrodotoxin (TTX, Tocris Bioscience). For recordings, borosilicate glass pipettes (Warner Instruments) with an outer diameter of 1.2 mm and a wall thickness of 0.255 mm were pulled to a resistance of 5–10 MΩ with a P-97 Flaming/Brown micropipette puller (Sutter Instruments) and filled with a solution containing 155 mM K-gluconate, 8 mM NaCl, 0.1 mM CaCl2, 0.6 mM MgCl2, 10 mM HEPES, 4 mM Mg-ATP, and 0.4 mM Na-GTP. The pipette solution pH was adjusted to 7.3 with KOH and the osmolarity was adjusted to 298 mOsm with sucrose.
Electrophysiology and calcium imaging in acute brain slice for cross talk analysis and assessment of sensitivity for spike number
Individual living slices (Figure 4 and Figure S5) were transferred to a recording chamber mounted on an upright microscope (Olympus BX51WI) and continuously superfused (2–3 ml/min) with artificial cerebrospinal fluid (ACSF) (124 mM NaCl, 2.5 mM KCl, 1.2 mM NaH2PO4, 24 mM NaHCO3, 5 mM HEPES, 12.5 mM glucose, 2 mM MgSO4, 2 mM CaCl2 with the pH adjusted to 7.3–7.4 with NaOH or HCl and the osmolarity adjusted to 300–310 mOsm with glucose) at room temperature. Cells were visualized through a 40x NA0.8 water-immersion objective to identify GCaMP6f-positive cells. Whole-cell current-clamp recordings were obtained from GCaMP6f-positive pyramidal neurons in layer 2/3 of motor cortex, using an Axopatch 700B amplifier (Molecular Devices) and Digidata 1440 digitizer (Molecular Devices). For recordings, borosilicate glass pipettes (Warner Instruments) with an outer diameter of 1.2 mm and a wall thickness of 0.255 mm were pulled to a resistance of 3–5 MΩ with a P-97 Flaming/Brown micropipette puller (Sutter Instruments) and filled with a solution containing 155 mM K-gluconate, 8 mM NaCl, 0.1 mM CaCl2, 0.6 mM MgCl2, 10 mM HEPES, 4 mM Mg-ATP, and 0.4 mM Na-GTP. The pipette solution pH was adjusted to 7.3 with KOH and the osmolarity was adjusted to 298 mOsm with sucrose. GCaMP fluorescence was excited by a SPECTRA X light engine (Lumencor) with 470/24 nm excitation filter (Semrock). To perform fair comparison of GCaMP6f1 and SomaGCaMP6f1 for Figure 4 and S5 excitation light power was adjusted on a cell-to-cell basis, in the range of 0.5 to 20 mW/mm2, to achieve similar intensity of fluorescence baseline between the two constructs. Fluorescence was collected through the same objective through a 525/50 nm emission filter and imaged onto an sCMOS camera (Andor Zyla5.5 or Hamamatsu Orca-Flash4.0 V2) at 50Hz acquisition frequency. For assessing the sensitivity of the GCaMP6f variants to action potential number using whole-cell patch clamp (Figure S5A) we performed 500 pA current injections (50 Hz current injections, 5 ms, in trains of 5, 10, or 20 pulses). For assessing crosstalk we performed the imaging as described above while stimulating cells in the slice with 0.1 mM 4-aminopyridine, aimed at producing low spike rates (as seen in Figure 4I).
Imaging
Imaging GCaMP targeting variants in culture
GCaMP6f trafficking variants that were found to localize predominantly in the soma of cultured neurons (Figure 1, Figure 3 and Supplemental Table 1) were imaged with an LED (X-Cite XLED1, Excelitas Tecnologies) mounted on a microscope for wide-field illumination (Leica 3000B), through a Leica HCX APO L 40x objective (air, NA=0.6). Imaging was performed with a Hamamatsu Orca Flash 4.0 camera using a 480 nm LED and GFP-3035D filter cube (Semrock) for GFP fluorescence (power, 34.84 mW/mm2).
Calcium imaging in acute brain slices for screening of somatic GCaMP6f variants
For Figure 2, individual slices were transferred to a recording chamber mounted on an inverted epifluorescnce microscope (Nikon Eclipse Ti inverted microscope equipped with 10x NA 0.3 objective lens, a SPECTRA X light engine (Lumencor) with 475/28 nm exciter (Semrock), and a 5.5 Zyla camera (Andor), controlled by NIS-Elements AR software) and continuously superfused (2–3 ml/min) with ACSF at room temperature. Cells were visualized through a 10x objective to identify GCaMP6f-positive cells under excitation light power in the range from 0.5 to 4 mW/mm2 adjusted to achieve comparable levels of baseline fluorescence for all screened constructs. 4-aminopyridine at a final concentration of 1 mM was added to induce neuronal activity.
Imaging GCaMP and SomaGCaMP6f1 in zebrafish
For Figure 6D–G, individual zebrafish larvae at 4–5 dpf expressing either GCaMP6f or SomaGCaMP6f1 were exposed to the paralytic agent alpha-bungarotoxin (Sigma Aldrich) for 30–45 seconds, at a concentration of 1 mg/ml. Then, the paralyzed fish were embedded in 1.5% ultralow-melting agarose (Sigma Aldrich) prepared in E3 medium, and imaged using a custom built 2-photon microscope. A forward moving grating was used as a stimulus as GCaMP6f or SomaGCaMP6f1 expressing cells were imaged at 15 Hz: for GCaMP6f experiments, 20s on / 20s off stimulus periods were used; for SomaGCaMP6f1, 10s on / 10s off (the difference in frequencies between GCaMP6f and SomaGCaMP6f1 was inadvertent).
For Figure 6H–O and Figure S6, individual zebrafish larvae at 4–5 dpf expressing GCaMP6f, SomaGCaMP6f1, or H2B-GCaMP6f were exposed to the paralytic agent pancuronium bromide (Sigma Aldrich) for 30–45 seconds, at a concentration of 0.20 mg/ml. The fish were under visual inspection until they stopped swimming. Then, the paralyzed fish were embedded in 1.5% ultralow-melting agarose (Sigma Aldrich) prepared in E3 medium. The embedded larvae were mounted in an imaging chamber flooded with E3 medium, in a Lightsheet Z.1 microscope (Zeiss). For imaging, the fish were illuminated with an excitation laser line at 488 nm with maximum power of 50mW, through 10x/0.2NA illumination optics, and imaged through a 20x/1.0NA water dipping detection objective. Since the baseline fluorescence of SomaGCaMP6f1 was approximately 4.7 fold lower compared to GCaMP6f, the percentage of light power for GCaMP6f imaging was 5% while the light power for SomaGCaMP6f1 imaging was 22.5–25%. The fish were imaged at 25 Hz, downsampled to 1Hz, for periods of 10–20 minutes, while incubated with 1 mM 4-aminopyridine to induce spiking.
In vivo mouse imaging in the striatum:
For Figure 7 and Figure S7, animals were positioned underneath a microscope, and imaged while freely locomoting on a spherical treadmill. For each animal, full session recordings (5–12 min) were performed while monitoring GCaMP fluorescence using the specifications noted below. Image acquisition occurred via a custom microscope equipped with a scientific CMOS (sCMOS) camera (ORCA-Flash4.0 LT Digital CMOS camera C11440–42U; Hamamatsu, Boston, MA). GCaMP was excited using a 5W LED (LZ1–00B200, 460 nm; LedEngin, San Jose CA). The custom microscope included a Leica N Plan 10X 0.25 PH1 microscope objective lens, a dual band excitation filter (FF01–468/553–25), a dichroic mirror (FF493/574-Di01–25×36), and a dual band emission filter (FF01–512/630–25; Semrock, Rochester, NY). Image acquisition was performed using HC Image Live (HC Image Live; Hamamatsu; Boston, MA). The exact sampling intervals varied based on demands of the Windows 7 operating system but was approximately 20Hz. For each image frame, exposure time was fixed at 20ms. Image data were stored as multi-page tagged image file format (mpTIFF’s).
For Figure 8, animals were gently restrained and connected with the miniaturized microscope (single channel epifluorescence, 475-nm blue LED, Inscopix) via the baseplate and secured with a small screw on the baseplate. After adjustments were made to optimize the focus, animals were placed into an operant conditioning chamber (Med Associates). Grayscale images were collected at 20 frames per second on an Inscopix miniaturized microscope (nVista HD V2).
Animal surgery, training and behavior
Mouse surgery and virus injection in the striatum (Figure 7 and Figure S7):
All animal procedures were approved by the Boston University Institutional Animal Care and Use Committee. Breeding pairs were obtained from Jackson Laboratory (Maine). A total of 11 mice (PV-cre mice; B6;129P2-Pvalbtm1(cre)Arbr/J; we did not use the Cre function in these experiments), 8–12 weeks old at the start of the experiments, were used in these experiments. Both male and female mice were used in this study. Animals first underwent viral injection surgery targeting the left striatum under stereotaxic conditions (AP: +0.5, ML:-1.8 mm, DV: -1.6). Mice were injected with 500 nL of either (AAV8-Syn-GCaMP6f; n=7; titer: 6.6 e12 GC/ml) or 500nL AAVDJ-Syn-SomaGCaMP6f2; n=1; titer:5.6e12 GC/ml or 500 nL AAVDJ-CAG-SomaGCaMP6f2; n=3; titer: 2.4e12 GC/ml. We used AAV8 GCaMP6f due to its availability. We moved to DJ for all new constructs including the somatic GCaMP6f. AAV8-Syn-GCaMP6f was obtained from the University of Pennsylvania Vector Core and AAVDJ-CAG-SomaGCaMP6f2 and AAVDJ-Syn-SomaGCaMP6f2 were obtained from the University of North Carolina Vector Core. All injections were made via pulled glass pipettes (diameter: 1.2 mm) pulled to a sharp point and then broken at the tip to a final inner diameter of ~20 μm. Virus was delivered via slow pressure ejection (10–15 psi, 15–20 ms pulses delivered at 0.5 Hz). The pipette was lowered over 3 min and allowed to remain in place for 3 min before infusion began. The rate of the infusion was 100 nL/min. At the conclusion of the infusion, the pipette remained in place for 10 min before slowly being withdrawn over 2–3 minutes. Upon complete recovery (7+ days after virus injection, mice underwent a second procedure for the implantation of a sterilized custom imaging cannula (OD: 0.317 cm, ID: 0.236 cm, height, 2 mm diameter), fitted with a circular coverslip (size 0; OD: 3mm) adhered using a UV-curable optical adhesive (Norland Products). To access the dorsal striatum, the cortical tissue overlying the striatum was carefully aspirated away to expose the corpus callosum. The white matter was then thinned until the underlying striatal tissue could be visualized through the surgical microscope. The window was then placed and centered above the striatum. During the same surgery, a custom aluminum head-plate was attached to the skull, anterior to the imaging cannula.
Mouse Training (Figure 7 and Figure S7):
Following surgery for virus infusion and window implantation (typically about 21–28 days), mice were handled for several days before being headfixed to the treadmill/imaging apparatus. Mice then were habituated to running on the spherical treadmill while headfixed, 3–4 days per week, over the next two weeks at the same time of day as subsequent recordings. Each animal received at least 6 habituation sessions prior to the first recording day. Habituation was performed in the dark with the imaging LED illuminated to the same intensity as it would be for recording sessions.
Movement data acquisition (Figure 7 and Figure S7):
The spherical treadmill was constructed similar to that previously described by Dombeck et al.6. Briefly, the treadmill consisted of a 3D printed plastic housing and a Styrofoam ball supported with air. Movement was monitored using two computer USB mouse sensors affixed to the plastic housing at the midline of the Styrofoam ball. Each mouse sensor was mounted 3–4mm away from the surface of the ball to prevent interference with ball movement. The LED sensors projected on the ball surface 78 degrees apart. The x- and y- surface displacement measured by each mouse was acquired using a separate computer running a Linux OS (minimal CentOS 6), and a simple multi-threaded python script that asynchronously read and accumulated mouse motion events, and sent packaged <dx,dy> data at 100Hz to the image acquisition computer via a RS232 serial link. Packaged motion data were received on the imaging computer using a Matlab script that stored the accumulated motion between frame triggers synchronized to each acquired frame.
Subjects and Surgery in the mPFC (Figure 8)
Male wild-type C57BL/6J mice were group-housed (2–4 subjects per cage) on a 12:12 h reverse light:dark cycle (lights off at 09.00) with ad libitum access to food and water. Subjects were prepared for in vivo epifluorescent calcium imaging as previously described (Vander Weele et al. 2018). Briefly, viral vectors carrying either AAVDJ-CAG-SomaGCaMP6f2 or AAVDJ-CAG-GCaMP6f (UNC Vector Core, titers were matched at 2.4×1012) were injected into the medial prefrontal cortex (mPFC) (from bregma in mm: AP: +1.8, ML: +0.3, DV: −2.75 and −2.4) (300 nL each, at 100 nL/minute) using a beveled microinjection needle (33 gauge for mice) with a 10 μl microsyringe (Nanofil; WPI) and pump (UMP3 and Micro4; WPI). The most ventral injection (DV: -2.75) was completed first and the injection needle was immediately raised to the more dorsal location (DV: -2.4) for the next injection. After completion of the second injection, 10 min were allowed to pass before the needle was raised another 0.1 mm and allowed to rest another 5 minutes before being slowly withdrawn.
After virus infusions, the craniotomy was enlarged to >1 mm in diameter, dura removed, and surface of the tissue was perforated with a 30 gauge beveled needle, but no tissue was aspirated. A 1 mm diameter, ~4 mm length gradient refractive index lens (GRIN lens; GLP-1040, Inscopix) was held by vacuum on the tip of a blunted needle surrounded by plastic tubing for stability and was lowered stereotaxically through the craniotomy under constant saline perfusion to minimize tissue/blood desiccation. Lenses were implanted slightly posterior and lateral of the needle track for virus infusions to avoid tissue damage in the imaging plane, and were lowered to the mPFC (AP: −1.77, ML: −0.4, DV: −2.32, mm from bregma). Lens implants were secured to the skull with a thin layer of adhesive cement (C&B Metabond; Parkell), followed by black cranioplastic cement (Ortho-Jet; Lang). Lenses were covered with the top of an eppendorf tube and cemented in place with cranioplastic cement for protection during the virus incubation period (at least 3 weeks). Following virus incubation, mice were again anaesthetized with isoflurane, stereotaxically secured, and baseplates (Inscopix) were cemented around the lens to support the connection of the miniaturized microscope for freely moving imaging.
In utero electroporation (Figure 4 and Figure 2)
Embryonic day (E) 15.5 timed-pregnant female Swiss Webster mice (Taconic) were deeply anesthetized with 2% isoflurane. Uterine horns were exposed and periodically rinsed with warm sterile phosphate buffered saline (PBS). A plasmid encoding GCaMP6f or SomaGCaMP6f variants under control of CAG promoter at final concentration 1–2 μg/μl diluted with PBS was injected into the lateral ventricle of the right cerebral hemisphere. Five voltage pulses (40 V, 50 ms duration, 1 Hz) were delivered using 5 mm round plate electrodes (ECM™ 830 Electroporation Generator, Harvard Apparatus). Injected embryos were placed back into the dam, and allowed to mature to delivery. All experimental manipulations were performed in accordance with protocols approved by the Massachusetts Institute of Technology Committee on Animal Care and were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Acute brain slice preparation
Acute brain sections for cross talk analysis, and spike number sensitivity assessment (Figure 4 and Figure S5 respectively) were prepared using in utero electroporated mice at P12 – P24, as described above. Mice were used without regard for sex. Mice were anaesthetized by isoflurane inhalation, euthanized, and cerebral hemispheres were removed, placed in ice cold choline-based cutting solution consisting of (in mM): 110 choline chloride, 25 NaHCO3, 2.5 KCl, 7 MgCl2, 0.5 CaCl2, 1.25 NaH2PO4, 25 glucose, 11.6 ascorbic acid, and 3.1 pyruvic acid (339–341 mOsm/kg; pH 7.75 adjusted with NaOH), blocked and transferred into a slicing chamber containing ice-cold choline-based cutting solution. Coronal slices (300 μm thick) were cut with a Compresstome VF-300 slicing machine, transferred to a holding chamber with ACSF, and recovered for 10 min at 34 °C, followed by another 50 min at room temperature. Slices were subsequently maintained at room temperature until use. Both cutting solution and ACSF were constantly bubbled with 95% O2/5% CO2.
Histological analysis of mouse brains expressing GCaMP variants
For Figure 4A,B,D, deeply anesthetized mice were perfused transcardially with 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.3) and brains were postfixed for 4–16 h at 4°C. 50–100 μm sections were cut with a Leica VT1000s vibratome and imaged using an inverted Nikon Eclipse Ti microscope equipped with a spinning disk sCSUW1 confocal scanner unit (Yokogawa, Tokyo, Japan), a 40x, NA 1.15 objective (Nikon), and a 4.2 PLUS Zyla camera (Andor), controlled by NIS-Elements AR software.
Supplementary Material
Supplemental Table 3: Statistical analysis for mouse brain slice, and for fish and mouse in vivo experiments (which include Figures 4, 6, 7, 8). Related to Figures 4, 6, 7, and 8.
Supplemental Table 5: Statistical analysis for membrane and action potential properties for Figure S1. Related to Figure 3.
Supplemental Table 6: Statistical analysis for Figures S2, S4, S5, S6, S7, S8. Related to Figures 1–8.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| 4-Aminopyridine | Sigma-Aldrich | Cat# 275875 |
| Agarose, low gelling temperature | Sigma-Aldrich | Cat# A9414 |
| Pancuronium bromide | Sigma-Aldrich | Cat# P1918 |
| E3 medium | This paper | http://cshprotocols.cshlp.org/content/2011/10/pdb.rec66449 |
| Antibodies | ||
| Anti-AnkyrinG | Santa Cruz Biotechnology | sc-12719, RRID:AB_626674 |
| anti-mouse IgG (H+L) Alexa 647 | Thermo Fisher Scientific | A-21235, RRID:AB_2535804 |
| Anti-Nav1.2 | NeuroMab | 75–024, RRID:AB_2184030 |
| anti-mouse IgG2a Alexa 647 | Thermo Fisher Scientific | A-21241, RRID:AB_2535810 |
| anti-mouse IgG1 Alexa 647 | Thermo Fisher Scientific | A-21240, RRID:AB_2535809 |
| Anti-Cav2.1 | Alomone | ACC-001, RRID:AB_2039764 |
| anti-rabbit IgG (H+L) Alexa 647 | Thermo Fisher Scientific | A-21245, RRID:AB_2535814 |
| Anti-mCherry | Kerafast | EMU106 |
| anti-rabbit IgG (H+L) Alexa 546 | Thermo Fisher Scientific | A-11035, RRID:AB_2534093 |
| anti-rat IgG (H+L) Alexa 546 | Thermo Fisher Scientific | A-11081, RRID:AB_2534125 |
| Anti-Kv2.1 | NeuroMab | 75–014, RRID:AB_10673392 |
| Critical Commercial Assays | ||
| Calcium Phosphate Transfection Kit | Invitrogen | Cat# K278001 |
| QIAprep Spin Miniprep Kit | QIAGEN | Cat# 27106 |
| Plasmid Plus Midi Kit | QIAGEN | Cat# 12945 |
| In-Fusion HD Cloning Kit | Clontech | Cat# 638910 |
| Experimental Models: Organisms/Strains | ||
| GcaMP6f fish | Dr. Misha Ahrens | N/A |
| SomaGCaMP6f1 fish | Dr. Misha Ahrens | N/A |
| Mouse: C57BL/6J | Jackson | Cat# 000664, RRID:IMSR_JAX:000664 |
| Mouse: Swiss Webster | Taconics | Timed Pregnant E10 |
| Sequence-Based Reagents | ||
| AnkTail-motif: REGRIDDEEPFKIVEKVKEDLVKVSEILKKDVCVESKGPPKSPKSDKGHSPEDDWTEFSSEEIREARQAAASHAPSLPERVHGKANLTRVIDYLTNDIGSSSLTNLKYKFEEAKKDGEERQKRILKPAMALQEHKLKMPPASMRPSTSEKELCKMADSFFGADAILESPDDFSQHDQDKSPLSDSGFETRSEKTPSAPQSAESTGPKPLFHEVPIPPVITETRTEVVHVIRSYEPSSGEIPQSQPEDPVSPKPSPTFMELEPKPTTSSIKEKVKAFQMKASSEEEDHSRVLSKGMRVKEETHITTTTRMVYHSPPGGECASERIEETMSVHDIMKAFQSGRDPSKELAGLFEHKSAMSPDVAKSAAETSAQHAEKDSQMKPKLERIIEVHIEKGPQSPCE | This paper | N/A |
| EE-RR: LEIEAAFLEQENTALETEVAELEQEVQRLENIVSQYETRYGPLGSLEIRAAFLRRRNTALRTRVAELRQRVQRLRNIVSQYETRYGPL | This paper | N/A |
| Recombinant DNA | ||
| AAV8-Syn-GCaMP6f | UNC vector core | N/A |
| AAV8-Syn-SomaGCaMP6f1 | UNC vector core | N/A |
| AAV-DJ-Syn-SomaGCaMP6f1 | UNC vector core | N/A |
| AAV-DJ-Syn-SomaGCaMP6f2 | UNC vector core | N/A |
| AAV-DJ-CAG-GCaMP6f | UNC vector core | N/A |
| AAV-DJ-CAG-SomaGCaMP6f1 | UNC vector core | N/A |
| AAV-DJ-CAG-SomaGCaMP6f2 | UNC vector core | N/A |
| AAV8-Syn-mCherry | UNC vector core | N/A |
| AAV-DJ-Syn-GCaMP7f | Janelia vector core | N/A |
| AAV-DJ-Syn-SomaGCaMP7f | Janelia vector core | N/A |
| Software and Algorithms | ||
| pClamp10 | Molecular devices | N/A |
| Clampfit 10.3 | Molecular devices | N/A |
| MatLab | Mathworks | N/A |
| Photoshop CS6 | Adobe | N/A |
| Illustrator CS6 | Adobe | N/A |
| Excel 2010 | Microsoft | N/A |
| Nikon Elements | Nikon | NIS-Elements AR |
| Calman | N/A | N/A |
| NormCorre | N/A | https://www.ncbi.nlm.nih.gov/pubmed/28782629 |
Highlights.
One-photon calcium imaging of brain activity can suffer from neuropil crosstalk.
Targeting GCaMPs to the cell body reduces neuropil crosstalk.
One-photon imaging of somatic GCaMP reduces artifactual spikes and correlations.
Somatic GCaMPs can be used in multiple species, such as mice and zebrafish.
Acknowledgements
We thank Eftychios Pnevmatikakis and Andrea Giovannucci from the Flatiron Institute at the Simons Foundation for useful discussions about computational methods for neuropil contamination reduction, and for their active development of NormCorre and CaImAn. K.M.T. was a New York Stem Cell Foundation - Robertson Investigator and McKnight Scholar and this work was supported by funding from R01-MH102441 (NIMH) and Pioneer Award DP1-AT009925 (NCCIH). C.A.S. was supported by NIH grants F32 MH111216 (NIMH) and K99 DA045103 (NIDA), and a NARSAD Young Investigator Award (Brain and Behavior Research Foundation). J.S.T and E.S.B. were supported by NIH U24 NS109113. E.S.B. was supported by Lisa Yang, John Doerr, NIH 1R01DA045549, NIH 1R01MH114031, NIH RF1NS113287, the HHMI-Simons Faculty Scholars Program, Human Frontier Science Program RGP0015/2016, U. S. Army Research Laboratory and the U. S. Army Research Office under contract/grant number W911NF1510548, US-Israel Binational Science Foundation Grant 2014509, NSF 1848029 and 1734870, NIH 2R01DA029639, NSF CBET 1344219, the MIT Media Lab, the Open Philanthropy Project, NIH 1R24MH106075, NIH R44EB021054, NIH 1R01GM104948, and NIH Director’s Pioneer Award 1DP1NS087724. J.T. acknowledges NIH grant U01 NS099714. Joyce Wang is supported by the NSF Graduate Research Fellowship grant 174530.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The MIT Media Laboratory, Massachusetts Institute of Technology (MIT), Cambridge, Massachusetts, USA.
Or A. Shemesh, Changyang Linghu, Kiryl D. Piatkevich, Daniel Goodwin, Limor Freifeld, Ishan Gupta, Nikita Pak, Young-Gyu Yoon, Burcu Guner-Ataman, Chih-Chieh (Jay) Yu, Ruixuan Gao, Edward S. Boyden
Department of Biological Engineering, MIT, Cambridge, Massachusetts, USA.
Or A. Shemesh, Kiryl D. Piatkevich, Chih-Chieh (Jay) Yu, Ishan Gupta, Amy E. Keating, Edward S. Boyden
MIT Center for Neurobiological Engineering, MIT, Cambridge, Massachusetts, USA.
Or A. Shemesh, Changyang Linghu, Kiryl D. Piatkevich, Daniel Goodwin, Limor Freifeld, Ishan Gupta, Nikita Pak, Young-Gyu Yoon, Edward S. Boyden
Department of Brain and Cognitive Sciences, MIT, Cambridge, Massachusetts, USA.
Or A. Shemesh, Kiryl D. Piatkevich, Edward S. Boyden
MIT McGovern Institute for Brain Research, MIT, Cambridge, Massachusetts, USA.
Or A. Shemesh, Changyang Linghu, Kiryl D. Piatkevich, Daniel Goodwin, Limor Freifeld, Ishan Gupta, Nikita Pak, Young-Gyu Yoon, Orhan Tunc Celiker, Chih-Chieh (Jay) Yu, Ruixuan Gao, Edward S. Boyden
Department of Electrical Engineering and Computer Science, MIT, Cambridge, Massachusetts, USA.
Changyang Linghu, Young-Gyu Yoon, Orhan Tunc Celiker
Boston University, Department of Biomedical Engineering, Boston, MA 02215.
Howard Gritton, Seth Bensussen, Hua-An Tseng, Michael F. Romano, Xue Han
The Picower Institute for Learning and Memory, Department of Brain and Cognitive Sciences, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Cody Siciliano, Kay M. Tye, Joyce Wang, Mark F. Bear, Habiba Noamany
Department of Mechanical Engineering, MIT, Cambridge, Massachusetts, USA.
Nikita Pak.
Janelia Research Campus, Howard Hughes Medical Institute, Ashburn, Virginia, USA.
Sujatha Narayan, Chao-Tsung Yang, Misha B. Ahrens
Department of Biology, MIT, Cambridge, Massachusetts, USA.
Won Min Park, Amy E. Keating
Epilepsy Genetics Program, Department of Neurology, Boston Children’s Hospital & Harvard Medical School, Massachusetts, USA.
Jeremy F.P. Ullmann
Koch Institute, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Edward S. Boyden
Department of Physiology and Membrane Biology; University of California, Davis School of Medicine; Davis, California, USA.
James S. Trimmer
School of Electrical Engineering, KAIST, Daejeon, Republic of Korea.
Young-Gyu Yoon
Internal Medicine Research Unit, Pfizer Inc., Cambridge, MA, USA.
Shoh Asano
Department of Neuroscience, Baylor College of Medicine, Houston, TX USA.
Jacob Reimer, Andreas S. Tolias
Center for Neuroscience and AI, Baylor College of Medicine, Houston, TX USA.
Jacob Reimer, Andreas S. Tolias
Department of Electrical and Computer Engineering, Rice University, Houston TX USA.
Andreas S. Tolias
Salk Institute for Biological Studies, La Jolla, CA 92037.
Kay M. Tye
Faculty of Biomedical Engineering; Technion; Haifa; Israel.
Limor Freifeld
Declaration of Interests
The authors OAS, CL, KDP, WMP, and ESB declare they applied for a US patent based on the work presented in this paper, Application No.: PCT/US2019/065773.
References
- Ahrens MB et al. , 2013. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nature methods, 10(5), pp.413–20. Available at: 10.1038/nmeth.2434 [Accessed December 19, 2015]. [DOI] [PubMed] [Google Scholar]
- Alivisatos AP et al. , 2013. Nanotools for Neuroscience and Brain Activity Mapping. ACS Nano, 7(3), pp.1850–1866. Available at: http://pubs.acs.org/doi/10.1021/nn4012847 [Accessed April 16, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J-M & Martinez LM, 1998. Functional connectivity between simple cells and complex cells in cat striate cortex. Nature Neuroscience, 1(5), pp.395–403. Available at: http://www.nature.com/articles/nn0998_395 [Accessed March 23, 2018]. [DOI] [PubMed] [Google Scholar]
- Andilla FD & Hamprecht FA, 2014. Sparse Space-Time Deconvolution for Calcium Image Analysis. , pp.64–72. Available at: https://papers.nips.cc/paper/5342-sparse-space-time-deconvolution-for-calcium-image-analysis [Accessed April 12, 2018].
- Baker CA et al. , 2016. Cellular resolution circuit mapping with temporal-focused excitation of soma-targeted channelrhodopsin. eLife, 5 Available at: http://elifesciences.org/lookup/doi/10.7554/eLife.14193 [Accessed June 12, 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbera G et al. , 2016. Spatially Compact Neural Clusters in the Dorsal Striatum Encode Locomotion Relevant Information. Neuron, 92(1), pp.202–213. Available at: http://www.ncbi.nlm.nih.gov/pubmed/27667003 [Accessed February 15, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtson CP et al. , 2010. Nuclear Calcium Sensors Reveal that Repetition of Trains of Synaptic Stimuli Boosts Nuclear Calcium Signaling in CA1 Pyramidal Neurons. Biophysical Journal, 99(12), pp.4066–4077. Available at: https://www.sciencedirect.com/science/article/pii/S0006349510013329 [Accessed April 10, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdyyeva T et al. , 2014. Zolpidem Reduces Hippocampal Neuronal Activity in Freely Behaving Mice: A Large Scale Calcium Imaging Study with Miniaturized Fluorescence Microscope T. Amédée, ed. PLoS ONE, 9(11), p.e112068 Available at: http://www.ncbi.nlm.nih.gov/pubmed/25372144 [Accessed April 19, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdyyeva TK et al. , 2016. Direct Imaging of Hippocampal Epileptiform Calcium Motifs Following Kainic Acid Administration in Freely Behaving Mice. Frontiers in Neuroscience, 10, p.53 Available at: http://journal.frontiersin.org/Article/10.3389/fnins.2016.00053/abstract [Accessed April 19, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden SEH et al. , 2001. Somatic Colocalization of Rat SK1 and D class (Cav 1.2) L-type Calcium Channels in Rat CA1 Hippocampal Pyramidal Neurons. Journal of Neuroscience, 21(20). Available at: http://www.jneurosci.org/content/21/20/RC175.long [Accessed June 18, 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T-W et al. , 2013. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature, 499(7458), pp.295–300. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3777791&tool=pmcentrez&rendertype=abstract [Accessed July 9, 2014]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H et al. , 2019. High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nature Methods, 16(7), pp.649–657. Available at: http://www.nature.com/articles/s41592-019-0435-6 [Accessed December 5, 2019]. [DOI] [PubMed] [Google Scholar]
- Deneux T et al. , 2016. Accurate spike estimation from noisy calcium signals for ultrafast three-dimensional imaging of large neuronal populations in vivo. Nature Communications, 7, p.12190 Available at: http://www.nature.com/doifinder/10.1038/ncomms12190 [Accessed June 14, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn TW et al. , 2016. Brain-wide mapping of neural activity controlling zebrafish exploratory locomotion. eLife, 5, p.e12741 Available at: http://www.ncbi.nlm.nih.gov/pubmed/27003593 [Accessed March 13, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher S, Bowden SEH & Marrion NV, 2003. False interaction of syntaxin 1A with a Ca2+-activated K+ channel revealed by co-immunoprecipitation and pull-down assays: implications for identification of protein–protein interactions. Neuropharmacology, 44(6), pp.817–827. Available at: https://www.sciencedirect.com/science/article/pii/S0028390803000492?via%3Dihub [Accessed March 25, 2018]. [DOI] [PubMed] [Google Scholar]
- Flusberg BA et al. , 2008. High-speed, miniaturized fluorescence microscopy in freely moving mice. Nature Methods, 5(11), pp.935–938. Available at: http://www.nature.com/articles/nmeth.1256 [Accessed February 13, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forli A et al. , 2018. Two-Photon Bidirectional Control and Imaging of Neuronal Excitability with High Spatial Resolution In Vivo. Cell Reports, 22(11), pp.3087–3098. Available at: https://www.sciencedirect.com/science/article/pii/S2211124718302572 [Accessed April 16, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman J et al. , 2014. Mapping brain activity at scale with cluster computing. Nature Methods, 11(9), pp.941–950. Available at: http://www.nature.com/articles/nmeth.3041 [Accessed June 1, 2018]. [DOI] [PubMed] [Google Scholar]
- Garrido JJ et al. , 2003. A targeting motif involved in sodium channel clustering at the axonal initial segment. Science (New York, N.Y.), 300(5628), pp.2091–4. Available at: http://www.ncbi.nlm.nih.gov/pubmed/12829783 [Accessed February 11, 2015]. [DOI] [PubMed] [Google Scholar]
- Garrido JJ et al. , 2001. Identification of an axonal determinant in the C-terminus of the sodium channel Na(v)1.2. The EMBO journal, 20(21), pp.5950–61. Available at: http://www.ncbi.nlm.nih.gov/pubmed/11689435 [Accessed May 2, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg KP, Pham A & Werblin FS, 2011. Differential targeting of optical neuromodulators to ganglion cell soma and dendrites allows dynamic control of center-surround antagonism. Neuron, 69(4), pp.713–20. Available at: http://www.cell.com/article/S0896627311000729/fulltext [Accessed January 27, 2015]. [DOI] [PubMed] [Google Scholar]
- Greschner M et al. , 2011. Correlated firing among major ganglion cell types in primate retina. The Journal of physiology, 589(Pt 1), pp.75–86. Available at: http://www.ncbi.nlm.nih.gov/pubmed/20921200 [Accessed March 23, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienberger C & Konnerth A, 2012. Imaging Calcium in Neurons. Neuron, 73(5), pp.862–885. Available at: https://www.sciencedirect.com/science/article/pii/S0896627312001729 [Accessed April 26, 2018]. [DOI] [PubMed] [Google Scholar]
- Grubb MS & Burrone J, 2010. Channelrhodopsin-2 localised to the axon initial segment. PloS one, 5(10), p.e13761 Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2966437&tool=pmcentrez&rendertype=abstract [Accessed December 17, 2014]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KD et al. , 2016. Improving data quality in neuronal population recordings. Nature Neuroscience, 19(9), pp.1165–1174. Available at: http://www.nature.com/articles/nn.4365 [Accessed February 15, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofherr A, Fakler B & Klöcker N, 2005. Selective Golgi export of Kir2.1 controls the stoichiometry of functional Kir2.x channel heteromers. Journal of cell science, 118(Pt 9), pp.1935–43. Available at: http://www.ncbi.nlm.nih.gov/pubmed/15827083 [Accessed November 19, 2015]. [DOI] [PubMed] [Google Scholar]
- Jennings JH et al. , 2015. Visualizing Hypothalamic Network Dynamics for Appetitive and Consummatory Behaviors. Cell, 160(3), pp.516–527. Available at: http://www.ncbi.nlm.nih.gov/pubmed/25635459 [Accessed February 13, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen CS et al. , 2017. Trafficking of Kv2.1 Channels to the Axon Initial Segment by a Novel Nonconventional Secretory Pathway. The Journal of Neuroscience, 37(48), pp.11523–11536. Available at: http://www.ncbi.nlm.nih.gov/pubmed/29042434 [Accessed April 2, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M & Chen G, 2006. High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nature protocols, 1(2), pp.695–700. Available at: http://www.ncbi.nlm.nih.gov/pubmed/17406298 [Accessed November 19, 2015]. [DOI] [PubMed] [Google Scholar]
- Keller PJ, Ahrens MB & Freeman J, 2015. Light-sheet imaging for systems neuroscience. Nature Methods, 12(1), pp.27–29. Available at: http://www.nature.com/articles/nmeth.3214 [Accessed April 26, 2018]. [DOI] [PubMed] [Google Scholar]
- Kim CK et al. , 2014. Prolonged, brain-wide expression of nuclear-localized GCaMP3 for functional circuit mapping. Frontiers in neural circuits, 8, p.138 Available at: http://www.ncbi.nlm.nih.gov/pubmed/25505384 [Accessed December 21, 2015]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH et al. , 2016. Long-Term Optical Access to an Estimated One Million Neurons in the Live Mouse Cortex. Cell reports, 17(12), pp.3385–3394. Available at: http://www.ncbi.nlm.nih.gov/pubmed/28009304 [Accessed February 15, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T et al. , 2015. Entorhinal Cortical Ocean Cells Encode Specific Contexts and Drive Context-Specific Fear Memory. Neuron, 87(6), pp.1317–1331. Available at: https://www.sciencedirect.com/science/article/pii/S0896627315007308#fig1 [Accessed April 19, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapoetke NC et al. , 2014. Independent optical excitation of distinct neural populations. Nature methods, 11(3), pp.338–46. Available at: 10.1038/nmeth.2836 [Accessed July 9, 2014]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus A et al. , 2017. The Spatiotemporal Organization of the Striatum Encodes Action Space. Neuron, 96(4), p.949 Available at: http://www.ncbi.nlm.nih.gov/pubmed/29144975 [Accessed April 11, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S et al. , 2009. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. The Journal of biological chemistry, 284(1), pp.478–85. Available at: http://www.ncbi.nlm.nih.gov/pubmed/19001369 [Accessed November 26, 2015]. [DOI] [PubMed] [Google Scholar]
- Kumar J, Schuck P & Mayer ML, 2011. Structure and assembly mechanism for heteromeric kainate receptors. Neuron, 71(2), pp.319–31. Available at: http://www.ncbi.nlm.nih.gov/pubmed/21791290 [Accessed April 24, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim ST et al. , 2000. A Novel Targeting Signal for Proximal Clustering of the Kv2.1 K+ Channel in Hippocampal Neurons. Neuron, 25(2), pp.385–397. Available at: http://www.sciencedirect.com/science/article/pii/S0896627300809022 [Accessed April 25, 2016]. [DOI] [PubMed] [Google Scholar]
- Ma D et al. , 2001. Role of ER export signals in controlling surface potassium channel numbers. Science (New York, N.Y.), 291(5502), pp.316–9. Available at: http://www.ncbi.nlm.nih.gov/pubmed/11209084 [Accessed November 19, 2015]. [DOI] [PubMed] [Google Scholar]
- Mohammed AI et al. , 2016. An integrative approach for analyzing hundreds of neurons in task performing mice using wide-field calcium imaging. Scientific Reports, 6(1), p.20986 Available at: http://www.nature.com/articles/srep20986 [Accessed February 13, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed AI et al. , 2016. An integrative approach for analyzing hundreds of neurons in task performing mice using wide-field calcium imaging. Scientific reports, 6, p.20986 Available at: http://www.ncbi.nlm.nih.gov/pubmed/26854041 [Accessed March 23, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll JR et al. , 2001. Designed heterodimerizing leucine zippers with a ranger of pIs and stabilities up to 10–15 M. Protein Science, 10(3), pp.649–655. Available at: http://www.ncbi.nlm.nih.gov/pubmed/11344333 [Accessed April 3, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moruno Manchon JF et al. , 2015. Cytoplasmic sphingosine-1-phosphate pathway modulates neuronal autophagy. Scientific Reports, 5(1), p.15213 Available at: http://www.nature.com/articles/srep15213 [Accessed March 25, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamel EA, Nimmerjahn A & Schnitzer MJ, 2009. Automated analysis of cellular signals from large-scale calcium imaging data. Neuron, 63(6), pp.747–60. Available at: http://www.ncbi.nlm.nih.gov/pubmed/19778505 [Accessed April 12, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen JP et al. , 2016. Whole-brain calcium imaging with cellular resolution in freely behaving Caenorhabditis elegans. Proceedings of the National Academy of Sciences, 113(8), pp.E1074–E1081. Available at: http://www.ncbi.nlm.nih.gov/pubmed/26712014 [Accessed February 15, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley MG & Kim PS, 1998. A buried polar interaction can direct the relative orientation of helices in a coiled coil. Biochemistry, 37(36), pp.12603–10. Available at: http://pubs.acs.org/doi/abs/10.1021/bi981269m [Accessed April 3, 2018]. [DOI] [PubMed] [Google Scholar]
- Pédelacq J-D et al. , 2006. Engineering and characterization of a superfolder green fluorescent protein. Nature Biotechnology, 24(1), pp.79–88. Available at: http://www.ncbi.nlm.nih.gov/pubmed/16369541 [Accessed June 17, 2017]. [DOI] [PubMed] [Google Scholar]
- Patel TP et al. , 2015. Automated quantification of neuronal networks and single-cell calcium dynamics using calcium imaging. Journal of Neuroscience Methods, 243, pp.26–38. Available at: http://www.ncbi.nlm.nih.gov/pubmed/25629800 [Accessed June 14, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pégard NC et al. , 2017. Three-dimensional scanless holographic optogenetics with temporal focusing (3D-SHOT). Nature Communications, 8(1), p.1228 Available at: http://www.nature.com/articles/s41467-017-01031-3 [Accessed April 16, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peron SP et al. , 2015. A Cellular Resolution Map of Barrel Cortex Activity during Tactile Behavior. Neuron, 86(3), pp.783–799. Available at: https://www.sciencedirect.com/science/article/pii/S0896627315002512?via%3Dihub [Accessed February 15, 2018]. [DOI] [PubMed] [Google Scholar]
- Pinto L & Dan Y, 2015. Cell-Type-Specific Activity in Prefrontal Cortex during Goal-Directed Behavior. Neuron, 87(2), pp.437–50. Available at: http://www.ncbi.nlm.nih.gov/pubmed/26143660 [Accessed April 19, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnevmatikakis EA et al. , 2014. A structured matrix factorization framework for large scale calcium imaging data analysis. Available at: http://arxiv.org/abs/1409.2903 [Accessed April 12, 2018]. [Google Scholar]
- Pnevmatikakis EA et al. , 2016. Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron, 89(2), pp.285–99. Available at: http://www.ncbi.nlm.nih.gov/pubmed/26774160 [Accessed February 15, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez OA, Härtel S & Couve A, 2011. Location matters: the endoplasmic reticulum and protein trafficking in dendrites. Biological Research, 44(1), pp.17–23. Available at: http://www.scielo.cl/scielo.php?script=sci_arttext&pid=S0716-97602011000100003&lng=en&nrm=iso&tlng=en [Accessed February 12, 2019]. [DOI] [PubMed] [Google Scholar]
- Romano SA et al. , 2017. An integrated calcium imaging processing toolbox for the analysis of neuronal population dynamics. PLoS computational biology, 13(6), p.e1005526 Available at: http://www.ncbi.nlm.nih.gov/pubmed/28591182 [Accessed April 19, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffinatti FA et al. , 2013. Spatial Wavelet Analysis of Calcium Oscillations in Developing Neurons M. Perc, ed. PLoS ONE, 8(10), p.e75986 Available at: http://dx.plos.org/10.1371/journal.pone.0075986 [Accessed June 14, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer MKE et al. , 2010. L1 syndrome mutations impair neuronal L1 function at different levels by divergent mechanisms. Neurobiology of disease, 40(1), pp.222–37. Available at: http://www.sciencedirect.com/science/article/pii/S0969996110001919 [Accessed February 11, 2015]. [DOI] [PubMed] [Google Scholar]
- Schrödel T et al. , 2013. Brain-wide 3D imaging of neuronal activity in Caenorhabditis elegans with sculpted light. Nature Methods, 10(10), pp.1013–1020. Available at: http://www.nature.com/nmeth/journal/v10/n10/full/nmeth.2637.html?message-global=remove [Accessed October 1, 2015]. [DOI] [PubMed] [Google Scholar]
- Sekiguchi KJ et al. , 2016. Imaging large-scale cellular activity in spinal cord of freely behaving mice. Nature communications, 7, p.11450 Available at: http://www.ncbi.nlm.nih.gov/pubmed/27121084 [Accessed March 24, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selgrade DF et al. , 2013. Protein scaffold-activated protein trans-splicing in mammalian cells. Journal of the American Chemical Society, 135(20), pp.7713–9. Available at: http://www.ncbi.nlm.nih.gov/pubmed/23621664 [Accessed February 15, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh OA et al. , 2017. Temporally precise single-cell-resolution optogenetics. Nature Neuroscience, 20(12), pp.1796–1806. Available at: http://www.nature.com/articles/s41593-017-0018-8 [Accessed February 13, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen SP et al. , 2018. Automatic Cell Segmentation by Adaptive Thresholding (ACSAT) for large scale calcium imaging datasets. bioRxiv, p.260075 Available at: https://www.biorxiv.org/content/early/2018/02/05/260075 [Accessed April 30, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit AK et al. , 2016. Calcium Imaging of Neuronal Activity in Drosophila Can Identify Anticonvulsive Compounds B. McCabe D, ed. PLOS ONE, 11(2), p.e0148461 Available at: http://dx.plos.org/10.1371/journal.pone.0148461 [Accessed March 23, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C et al. , 2014. Molecular identity of axonal sodium channels in human cortical pyramidal cells. Frontiers in Cellular Neuroscience, 8, p.297 Available at: http://www.ncbi.nlm.nih.gov/pubmed/25294986 [Accessed April 2, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valluru L et al. , 2005. Ligand binding is a critical requirement for plasma membrane expression of heteromeric kainate receptors. The Journal of biological chemistry, 280(7), pp.6085–93. Available at: http://www.jbc.org/content/280/7/6085.full [Accessed February 2, 2015]. [DOI] [PubMed] [Google Scholar]
- Vladimirov N et al. , 2014. Light-sheet functional imaging in fictively behaving zebrafish. Nature Methods, 11(9), pp.883–884. Available at: http://www.ncbi.nlm.nih.gov/pubmed/25068735 [Accessed June 10, 2018]. [DOI] [PubMed] [Google Scholar]
- Vander Weele CM et al. , 2018. Dopamine enhances signal-to-noise ratio in cortical-brainstem encoding of aversive stimuli. Nature, 563(7731), pp.397–401. Available at: http://www.ncbi.nlm.nih.gov/pubmed/30405240 [Accessed February 21, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C et al. , 2013a. rAAV-mediated subcellular targeting of optogenetic tools in retinal ganglion cells in vivo. Taylor WR, ed. PloS one, 8(6), p.e66332 Available at: http://dx.plos.org/10.1371/journal.pone.0066332 [Accessed March 29, 2016]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C et al. , 2013b. rAAV-mediated subcellular targeting of optogenetic tools in retinal ganglion cells in vivo. PloS one, 8(6), p.e66332 Available at: http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0066332 [Accessed March 29, 2016]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y et al. , 2010. P/Q and N channels control baseline and spike-triggered calcium levels in neocortical axons and synaptic boutons. The Journal of neuroscience : the official journal of the Society for Neuroscience, 30(35), pp.11858–69. Available at: http://www.ncbi.nlm.nih.gov/pubmed/20810905 [Accessed April 2, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X & Bennett V, 1998. Restriction of 480/270-kD ankyrin G to axon proximal segments requires multiple ankyrin G-specific domains. The Journal of cell biology, 142(6), pp.1571–81. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2141775&tool=pmcentrez&rendertype=abstract [Accessed April 29, 2016]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P et al. , 2016. Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. Available at: http://arxiv.org/abs/1605.07266 [Accessed April 18, 2018]. [DOI] [PMC free article] [PubMed]
- Ziv Y et al. , 2013. Long-term dynamics of CA1 hippocampal place codes. Nature neuroscience, 16(3), pp.264–6. Available at: http://www.ncbi.nlm.nih.gov/pubmed/23396101 [Accessed February 13, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 3: Statistical analysis for mouse brain slice, and for fish and mouse in vivo experiments (which include Figures 4, 6, 7, 8). Related to Figures 4, 6, 7, and 8.
Supplemental Table 5: Statistical analysis for membrane and action potential properties for Figure S1. Related to Figure 3.
Supplemental Table 6: Statistical analysis for Figures S2, S4, S5, S6, S7, S8. Related to Figures 1–8.
Data Availability Statement
The code generated during this study is available at: https://github.com/dgoodwin208/SomaGCaMP-Processing. The sequences of SomaGCaMP variants can be found through GenBank and Addgene.


