Abstract
Electric field–induced release and measurement (EFIRM) is a novel, plate-based, liquid biopsy platform capable of detecting circulating tumor DNA containing EGFR mutations directly from saliva and plasma in both early- and late-stage patients with non–small-cell lung cancer. We investigated the properties of the target molecule for EFIRM and determined that the platform preferentially detects single-stranded DNA molecules. We then investigated the properties of the EFIRM assay and determined the linearity, linear range, precision, and limit of detection for six different EGFR variants (the four most common g.Exon19del variants), p.T790M, and p.L858R). The limit of detection was in single-digit copy number for the latter two mutations, and the limit of detection for Exon19del was 5000 copies. Following these investigations, technical validations were performed for four separate EFIRM liquid biopsy assays, qualitative and quantitative assays for both saliva and plasma. We conclude that EFIRM liquid biopsy is an assay platform that interrogates a biomarker not targeted by any other extant platform (namely, circulating single-stranded DNA molecules). The assay has acceptable performance characteristics in both quantitative and qualitative assays on both saliva and plasma.
The detection and analysis of cell-free circulating tumor DNA (ctDNA) is becoming a useful tool in the care of patients with cancer.1, 2, 3, 4, 5 Commonly referred to as liquid biopsy (LB), this process has both advantages and disadvantages when compared with the gold standard of tissue biopsy.1 Because LB provides no histologic staining or spatial analysis, staging is currently possible only by conventional tissue biopsy. However, when tissue is unavailable, LB may be the only possible source of information regarding the genetic makeup of a solid tumor.1,3 In addition, LB samples can be obtained by minimally invasive (venipuncture) or noninvasive (saliva collection) techniques and can therefore be used for serial monitoring. LBs are also less affected by tumor heterogeneity than tissue biopsy.1
Liquid biopsy has been applied to many solid tumors,4 including lung,3,5,6 breast,7 pancreas,8 melanoma,9 and prostate cancer.10 Heretofore, tissue samples obtained by biopsy or at the time of surgery were the only available specimen source. As such, the amount of tissue available was often limiting. More recently, technology has allowed us to detect and measure cell-free DNA in the blood11 and saliva,12,13 providing ready access to a limitless supply of specimens.
Although liquid biopsy is still in its infancy as a clinical tool, it has already demonstrated value in detecting EGFR mutations in patients with non–small-cell lung cancer (NSCLC) at the time of presentation and relapse,14, 15, 16, 17, 18, 19, 20 prompting Roche Molecular Systems (Pleasanton, CA) to commercially release the Cobas EGFR Mutation Test in 2017. This assay was specifically validated for plasma testing.
There is, as yet, no consensus regarding the clinical utility of LB in NSCLC or other solid tumors. A joint review by the American Society of Clinical Oncology and the College of Molecular Pathologists, published in 2018, asserted that there was insufficient evidence of clinical validity to recommend ctDNA analysis in the routine clinical setting.21 In contrast, a statement article from the International Association for the Study of Lung Cancer that same year concluded that immediate implementation of LB in the clinic is justified in several therapeutic settings relevant to NSCLC.22
As more data accumulate, LB is rapidly gaining acceptance by clinicians and insurers in certain clinical situations. Commercial laboratories now offer single or panel laboratory-developed mutation tests using cell-free DNA obtained by liquid biopsy for patients with NSCLC.3,16 In NSCLC, there are some data to suggest that liquid biopsy is preferable to tissue-based diagnosis for EGFR mutations in some situations, and indications for the use of liquid biopsy in NSCLC are increasing.3,19,20
Our earlier work describes a new method for cell-free DNA analysis that demonstrated both superior sensitivity and specificity to existing PCR-based or next-generation sequencing (NGS) methods in patients with both early-stage (stages I and II) and late-stage NSCLC.11, 12, 13 The electric field–induced release and measurement (EFIRM) LB (eLB) method uses untreated plasma or saliva as input. As no special specimen preparation is required, pre-analytic variables are few. In addition, the eLB platform is in a 96-well microtiter plate format, increasing the opportunities for automation and greatly increasing specimen throughput and reducing turnaround time to as little as 3 hours. Furthermore, the 96-well format allows a dramatic reduction in assay cost to as low as ≤$100/mutation.
In this article, we describe the eLB process and experiments to determine what eLB is measuring, followed by the technical development and validation of four separate eLB assays for six clinically actionable variants in the EGFR gene and four variants in g.exon19del, p.L858R, and p.T790M. There are qualitative assays for both plasma and saliva for potential screening purposes or recurrence detection and quantitative assays for plasma and saliva for the purpose of monitoring disease progression or drug response.
We will present data, including reference range determinations, for the quantitative assay for both plasma and serum and the precision, sensitivity, specificity, level of detection, and intra-assay and interassay reproducibility. We also examine the nature of the nucleic acid sequences that are targeted by eLB.
Materials and Methods
EFIRM Work Flow and Method
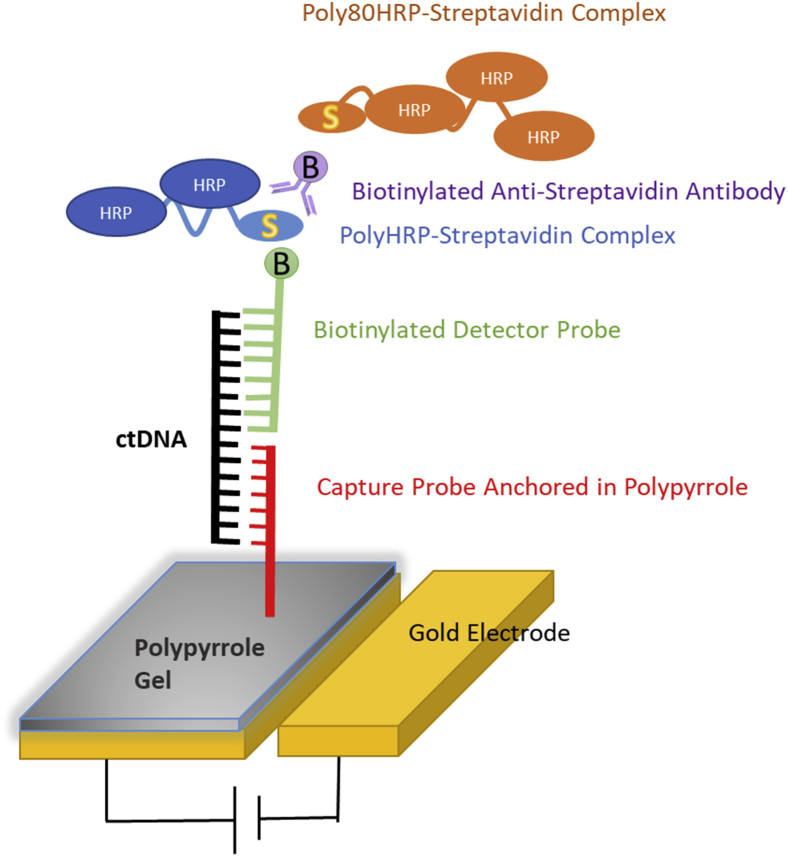
Figure 1 represents a schematic of the EFIRM method. Initially, a single allele-specific capture probe is added to a polypyrrole solution and added to a microtiter plate containing a gold electrode at the bottom of each well. After an electric field has been applied, the liquid becomes polymerized into a conducting gel and the capture oligonucleotides are anchored in the gel. Subsequently, the biological sample is applied and electrically facilitated allele-specific hybridization is performed. In this step, the kinetic energy imparted to the system increases the specificity of the hybridization. A biotinylated detector probe, whose sequence continues immediately after the capture probe sequence ends, is added and another electrically aided hybridization is performed. Subsequently, signal amplification and signal production are accomplished using standard methods. The final reaction generates an nA current that is measured by the EFIRM reader.
Figure 1.
Schematic of electric field–induced release and measurement liquid biopsy design. ctDNA, circulating tumor DNA.
A step-by-step workflow is as follows. Initially, a monomer solution is generated, consisting of 0.3 mol/L KCl (number 60137; Sigma Aldrich, St. Louis, MO) and 144.1 mmol/L pyrrole (W336805; Sigma Aldrich). An appropriate capture probe (Integrated DNA Technologies, Coralville, IA) is added to a final concentration of 2.5 μmol/L. The solution is transferred into a microcentrifuge tube and vortexed to thoroughly mix the contents. Subsequently, 60 μL of monomeric solution is transferred into each of 96 wells of an EFIRM E-Plate (EZLife Bio Inc., Los Angeles, CA). The E-Plate is then mounted into the EFIRM E-Reader (EZLife Bio Inc.) and a cyclic-square wave consisting of 350 mV and 1100 mV for 1 second each is applied for a total of 8 seconds. This step causes the monomer to polymerize into a conducting gel, coating the surface of the gold electrode while anchoring the capture probe to the gel. The plate is then removed from the E-Reader and placed into a 96-well plate washer (model 405LS; BioTek, Winooski, VT). A single wash is then performed using a 2× standard saline citrate 0.5% SDS solution. The plate is now ready for the addition of the clinical sample. Table 1 lists the capture probe/signal probe combinations used is this assay.
Table 1.
Capture and Detection Oligonucleotides for eLB EGFR Assay
| Variant | Probes |
|---|---|
| Capture Probes | |
| Exon 19del | 5′-TGTTGCTTCCTTG-3′ |
| p.L858R | 5′-GTTTGACCCGCCCA-3′ |
| p.T790M | 5′-GAGCGGCATGATGA-3′ |
| Detector Probes∗ | |
| Exon 19del | 5′-ATAGCGACGGGAATTTTAACTTTCTCACCT-3′ |
| p.L858R | 5′-AAAATCTGTGATCTTGACATGCTGCGGTGTTTTGTGCAG-3′ |
| p.T790M | 5′-GCTGCACGGTGGAGGTGAGGCAGATGCCCAGC-3′ |
eLB, electric field–induced release and measurement liquid biopsy.
Detector probes are biotin labeled at the terminal 3′ nucleotide.
To prepare clinical samples for testing, 20 to 30 μL of either plasma or saliva is diluted 1:2 in Ultrahyb Oligo Hybridization buffer (Thermo Fisher Scientific, Waltham, MA). Subsequently, 25 μL of the mixture is transferred to the bottom of each well. Once all samples have been added, the E-Plate is placed back into the E-Reader and a cyclic-square wave consisting of 300 mV and 500 mV for 1 second each is applied for a total of 300 seconds, followed by a 30-minute room temperature incubation in the E-Reader. The plate is then removed from the instrument and washed with 2× standard saline citrate 0.5% SDS in the plate washer. After this wash, 25 μL of a 100 nmol/L solution of detector probe (Integrated DNA Technologies) in casein/phosphate-buffered saline (PBS; 37528; Thermo Fisher Scientific) is added to each, the plate is returned to the E-Reader, and hybridization is performed using a cyclic-square wave consisting of −300 mV and 500 mV for 1 second each for a total of 300 seconds, followed by a 30-minute incubation at room temperature in the E-Reader. The plate is then returned to the plate washer and washed with 2× standard saline citrate 0.5% SDS.
The plate is then removed from the plate washer, and a streptavidin poly–horseradish peroxidase (Thermo Fisher Scientific) solution is prepared by diluting the reagent 1:1000 in casein/PBS and then 60 μL is added to each well and incubated at room temperature for 30 minutes. The plate is then washed again using 1× PBS with 0.05% Tween-20. Biotinylated Anti-Streptavidin Antibody (BA-0500; Vector Laboratories, Burlingame, CA) is diluted 1:10,000 in casein/PBS and 60 μL is pipetted into each well, followed by a 30-minute incubation at room temperature and another wash step with 1× PBS with 0.05% Tween-20. A 1:1000 dilution of Poly80HRP-Streptavidin (Fitzgerald Industries, Acton, MA) casein/PBS is made, and 60 μL is pipetted into each electrode well and incubated at room temperature for 30 minutes, followed by a final wash using 1× PBS with 0.05% Tween-20. Finally, 60 μL of 1-Step Ultra TMB substrate (34028; Thermo Fisher Scientific) solution is added to each well, the plate is placed into the E-Reader, and electrochemical current in nA is measured at a potential of −200 mV for 1 minute.
The technologist is guided in all steps by the EZL-Reader Software version 1.0 (EZLife Bio, Woodland Hills, CA), which provides sequential directions using a graphical user interface. There are five 30-minute incubations, and combined with the pipetting steps, the assay can be easily completed by a single technologist in 3 hours. Once the clinical samples have been added to the E-Plate, walk-away automation would be possible because all steps are performed with the plate washer, the E-Reader, or routine liquid handling.
RNase Treatment Method
RNase treatment was performed by adding RNase cocktail reagent, consisting of 0.025 U/μL of RNase A and 1 U/μL of T1 RNase in casein/PBS RNase (AM2286; Thermo Fisher Scientific), and incubating for 30 minutes at room temperature. Washing was performed using 2× standard saline citrate 0.5% SDS wash buffer. This treatment was performed following the sample capture step of the EFIRM protocol.
DNase Treatment Method
DNase treatment was performed to assess the strandedness of the DNA. First, Proteinase K (P8107S; New England Biolabs, Boston, MA) was added to achieve a concentration of 2 μg/μL in plasma samples. The samples were then incubated for 30 minutes at 50°C. Following this digest, proteinase K was then heated to 65°C for 10 minutes to deactivate the enzyme and then the solution was cooled to 4°C for 1 hour (using a cooling rate of 0.1°C/second) in a thermocycler. Following this initial proteinase K digest to remove interfering proteins, Exonuclease VII (M0379S; New England Biolabs) was introduced at a concentration of 0.33 units/μL and incubated at 37°C for 30 minutes. Exonuclease was subsequently deactivated by heat treatment at 95°C for 10 minutes, followed by cooling to 4°C for 1 hour (with a ramp rate of 0.1°C/second) to reanneal the double-stranded DNA present in the samples. The final solution was then assayed using the EFIRM protocol described in RNase Treatment Method.
Cell Lines
To generate genomic DNA reference standards for the point mutations p.L858R and p.T790M, genomic DNA was isolated using a QuickgDNA miniprep (Zymo Research, Irvine, CA) from the NCI-H1975 cell line (ATCC, Manassas, VA). For g.Exon19del testing, genomic DNA from four different cell lines harboring the top four variants of g.Exon19del mutations (COSM6623/6225/12370/12382) was acquired (Applied Stem Cell, Milpitas, CA).
Limit of Detection Measurements
DNA extracted from the cell lines was diluted in water to a 50 ng/mL concentration. Shearing was accomplished by heat treatment in a thermocycler at 4°C for 3 minutes, followed by heating to 95°C for 7 minutes at a ramp rate of 4°C/second. Following heat treatment, the temperature was lowered to 4°C for 10 minutes at a ramp rate of 4°C/second. The sheared samples were then serially diluted in water and Ultrahyb Oligo Hybridization buffer, maintaining the same ratio of water/Ultrahyb Oligo buffer in each sample. Finally, 30 μL of each dilution was assayed using the EFIRM platform.
Software
The multichannel EFIRM Reader is controlled via a USB 2.0 connection, and parameters are set by a custom software suite called EZL-Reader version 1.0. Data from each EFIRM experiment were exported to the comma-separated value file format and analyzed using the R-Language for statistical analysis.
Biological Samples
Saliva was collected from healthy individual volunteers at meetings of the American Dental Association between 2006 and 2011. Consent was obtained under institutional review board approval (University of California, Los Angeles Institutional Review Board number 06-05-042). There was a mixture of males/females, mostly non-smokers, between 18 and 80 years of age, and a mixture of ethnicities. All subjects consented before collection. Each subject would expectorate approximately 5 mL of whole saliva in a 50 μL conical tube set on ice. The saliva was processed within 0.5 hours of collection. Samples were centrifuged in a refrigerated centrifuge at 2600 × g for 15 minutes at 4°C. The supernatant (cell-free saliva) was then pipetted into two 2-mL cryotubes and the following reagents were added to preserve the RNA and DNA: 1.1 μL Superase-In/1 mL supernatant (Ambion, Austin, TX). After the additional reagents were added, each tube was inverted to mix. The samples were then frozen on dry ice and later stored in −80°C.
One 5-mL tube of blood was collected from consented subjects using a BD Vacutainer Safety-LokBlood Collection Set (367283; BD Biosciences, Franklin Lakes, NJ) and Lavender-top K2 EDTAtubes (367525; BD Biosciences). The tubes were filled and inverted and then centrifuged at 2500 × g at 4°C for 10 minutes within 2 hours of collection. The buffy coat free supernatant (plasma) was then removed, frozen, and stored at −70°C until assayed.
Results
DNA or RNA?
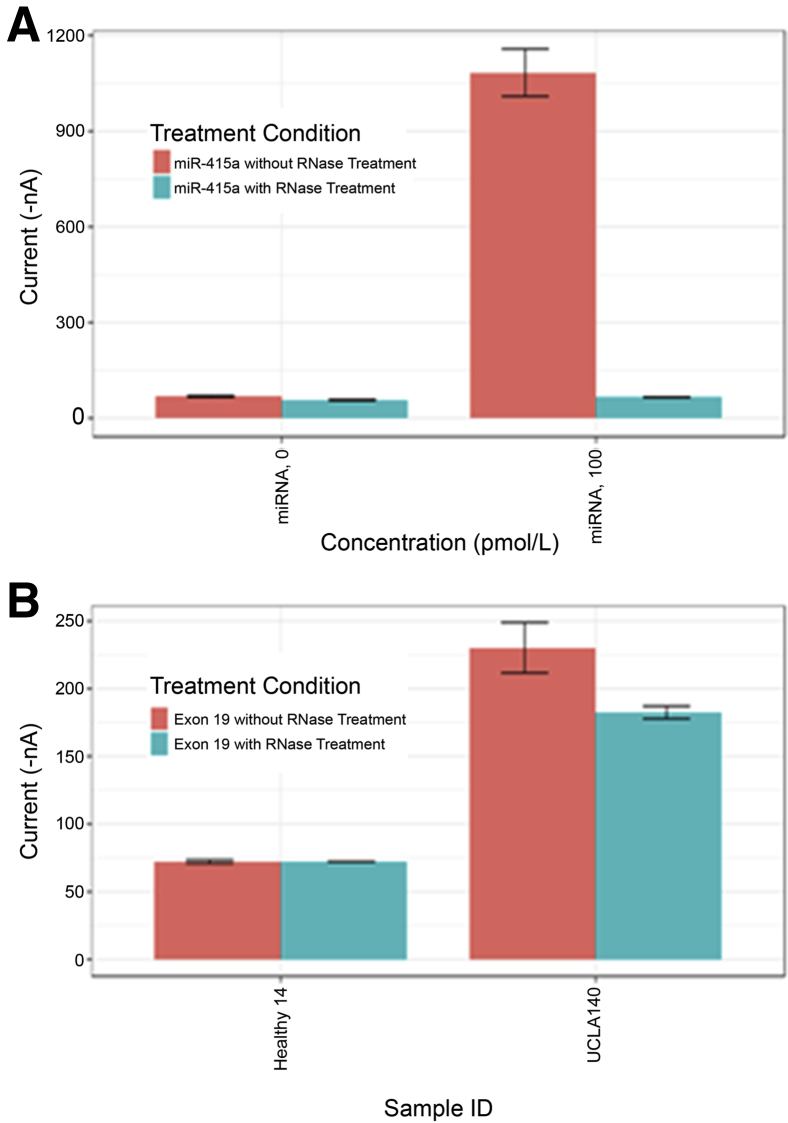
The EFIRM technology is capable of detecting either DNA or RNA molecules. It was investigated whether the EGFR variants detected in clinical samples from patients with NSCLC and reported previously8, 9, 10 were derived from circulating DNA or RNA. An RNase treatment method (Materials and Methods) was developed. As a control for completeness of digestion, a previously developed eLB assay was used for the miRNA species miR-415a. In the control experiment, synthetic miR-415a was added to normal plasma and subjected to eLB analysis both before and after RNase treatment. This result is shown in Figure 2A. The RNase treatment completely eliminated the signal from the eLB reaction for the miRNA, confirming that the RNase digestion had gone to completion under our conditions (Figure 2A).
Figure 2.
Electric field–induced release and measurement liquid biopsy (eLB) assay experiment with RNase treatment to evaluate whether the target detected by eLB is DNA or RNA. A: RNase treatment experiment applied to an RNA-based assay. B: RNase treatment applied to a healthy control and a subject (UCLA140) bearing exon 19 deletion. ID, identifier.
Figure 2B represents pretreated and post-treated normal plasma samples analyzed for the Exon19del variant in EGFR. Plasma from a patient with advanced NSCLC whose tumor was positive for g.Exon19del and whose eLB results were also positive for the same variant is shown.3 There was only a 10% reduction in signal following RNase treatment, indicating that the circulating nucleic acid measured in the eLB assay is primarily DNA and not RNA.
Single- versus Double-Stranded DNA
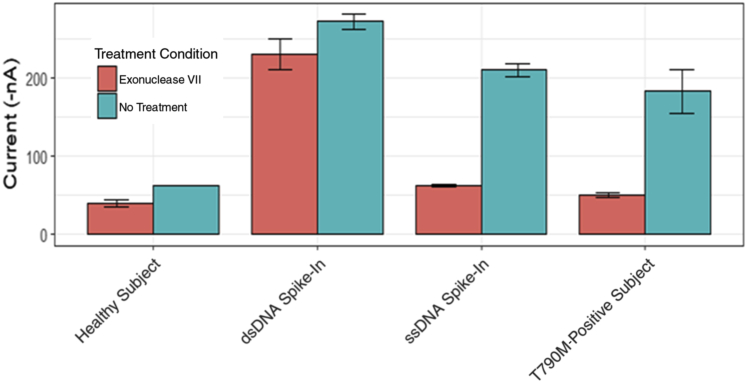
It was then investigated whether the molecule measured by EFIRM is single- or double-stranded DNA. For these experiments, enzymatic treatment with the single-strand specific nuclease Exonuclease VII (Materials and Methods) was used. Figure 3 shows the results of EFIRM analysis before and after Exonuclease VII treatment. The healthy control yielded only background signal in both the treated and the untreated experiments. The double-stranded DNA spike-in experiment demonstrates that when double-stranded DNA containing the variant is spiked into plasma, that there is no reduction in signal following Exonuclease VII digestion. This confirms the specificity of the enzyme. As expected, when synthetic single-stranded DNA is added, Exonuclease VII digestion results in a significant reduction in eLB signal. The plasma sample from the late-stage NSCLC patient, whose prior tissue and eLB results demonstrated the p.T790M EGFR variant, demonstrated that the Exonuclease VII treatment resulted in a reduction in eLB signal to background levels. These data, taken together, demonstrate that the eLB is primarily measuring single-stranded DNA targets.
Figure 3.
Electric field–induced release and measurement liquid biopsy (eLB) assay experiment with Exonuclease VII treatment to evaluate whether the target detected by eLB is DNA or RNA. Exonuclease VII treatment applied to a healthy subject, Exonuclease VII treatment applied to a healthy plasma sample with a synthetic double-stranded T790M oligonucleotide spike in, Exonuclease VII treatment applied to a healthy plasma with a synthetic single-stranded T790M spike in, and Exonuclease VII treatment applied to a p.T790M-bearing subject are all shown. dsDNA, double-stranded DNA; ssDNA, single-stranded DNA.
Technical Validation of Qualitative eLB Assay for Six EGFR Variants
The variant g.Exon19del is not a single variant, but a family of closely related variants of slightly varying locations. Because of the nature of eLB, it was possible to design a capture probe–signal probe combination that detects most, if not all, of the g.Exon19del variants. Data are presented in EGFR Exon 19 Deletion that demonstrate that the single capture probe–detector probe pair is capable of detecting the four most common Exon19del variants. A further discussion of this issue is found below in the quantitative assay description.
Plasma Qualitative Assay
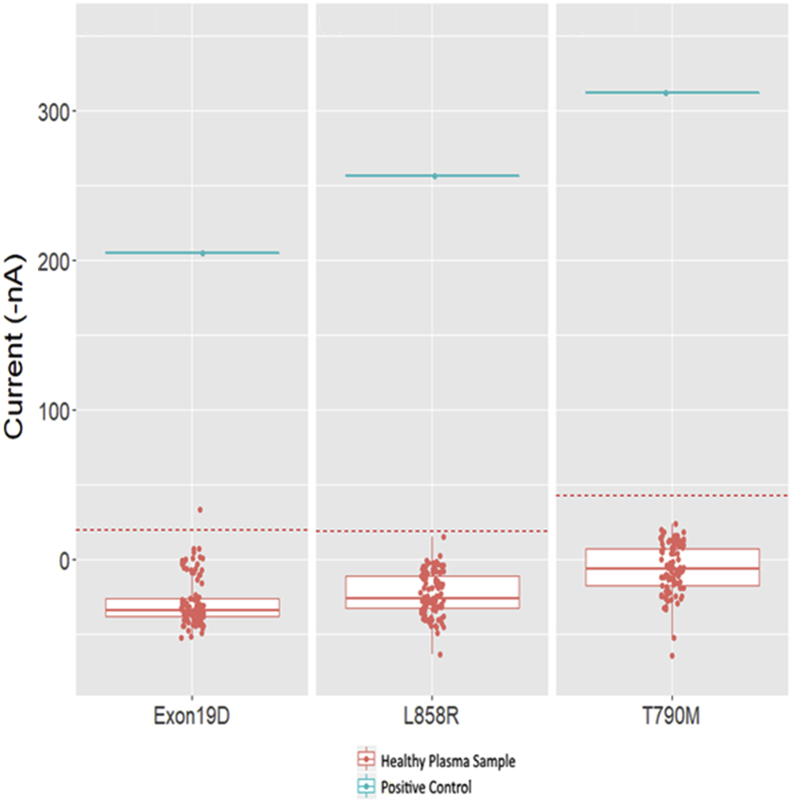
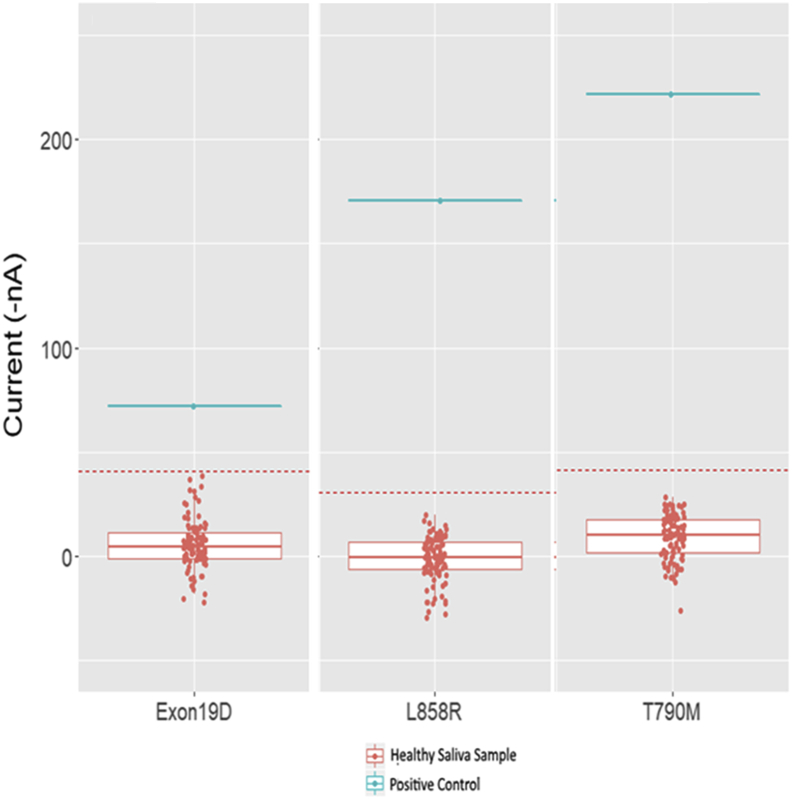
For validation of the qualitative assay, plasma was purchased from 100 healthy individuals (Materials and Methods). These samples were run in duplicate using the eLB technology for the six EGFR variants. The results are shown in Figure 4. Current, in nA, is plotted on the y axis. There are two no template controls on each plate, and the average nA value of the two no template controls is subtracted for all samples during analysis. A standard 3-SD reference range is plotted as the red dotted line. The blue line is a control oligonucleotide added to each plate to confirm assay performance. The established reference range varies slightly with each variant. For the plasma qualitative assay, the cutoff values are shown in Table 2 for g.Exon19del, p.L858R, and p.T790M. The reference range was set to be below 3 SDs of the unaffected controls. The internal standard is well above the cutoff values for each variant (Figure 4).
Figure 4.
Reference range study for 100 healthy saliva samples tested using electric field–induced release and measurement liquid biopsy (eLB) assay. The 2-SD line is indicated by a red dotted lines. eLB assay for exon 19 deletion mutation (left), eLB assay results for p.L858R mutation (middle), and eLB assay for p.T790M mutation (right).
Table 2.
Result of Reference Range Studies for Qualitative EGFR Assay
| Matrix | Variant | Median | Mean current, nA | SD | 2 SDs | 3 SDs | Reference range (3 SDs), nA |
|---|---|---|---|---|---|---|---|
| Plasma | T790M | −6.43771 | −5.96 | 16.18 | 32.37 | 48.55 | <43 |
| Plasma | L858R | −25.84727 | −22.79 | 14.07 | 28.14 | 42.21 | <20 |
| Plasma | Exon 19 | −34.24165 | −28.05 | 15.90 | 31.79 | 47.69 | <20 |
| Saliva | T790M | 10.49715 | 8.82 | 11.58 | 23.16 | 34.75 | <44 |
| Saliva | L858R | −0.5126045 | −1.13 | 10.56 | 21.13 | 31.69 | <31 |
| Saliva | Exon 19 | −3.927202 | −4.46 | 12.08 | 24.16 | 36.24 | <32 |
Interassay and intra-assay variability experiments were performed for the qualitative assay using the derived reference range demonstrated above (Figure 4). For intra-assay variability, a negative sample and a positive sample were run multiple times on a single plate. Each sample was assayed at least eight times (8 replicates of mutation-negative controls and 12 replicates of mutation-positive controls were tested for each of the three mutations). There were no discrepancies, with all negative samples being less than the cutoff and all positive samples being above the cutoff.
For interassay variability, the mutation-negative controls and mutation-positive controls (12 pmol/L oligonucleotide spiked in biofluid) were run on three different plates by two different operators (each plate having 8 replicates of mutation-negative normal and 12 replicates of mutation-positive normal). Once again, there were no discrepant results, yielding both intra-assay and interassay variability of zero.
Validation of Qualitative eLB Assay on Saliva
Figure 5 represents the same experiments as in the previous two sections using saliva samples obtained from consented healthy dentists at annual meetings of the American Dental Association. Plate design and validation design were identical to that of the plasma validation. The assay performance is similar to the plasma assay. Intra-assay and interassay variability were also zero, with all replicates concordant for positive or negative results. As with plasma, the reference ranges vary slightly with each variant. Table 2 is a summary of the reference range determinations for the six assays (namely, plasma and saliva for the six EGFR mutations). Although there are small variations in the reference range for each assay, as can be seen, the positive clinical samples are well above these cutoff values for all variants. The internal positive controls are well above the 3-SD cutoff for all variants.
Figure 5.
Reference range study for 100 healthy plasma samples tested using electric field–induced release and measurement liquid biopsy (eLB) assay. The 2-SD line is indicated by a red dotted lines. eLB assay for exon 19 deletion mutation (left), eLB assay results for p.L858R mutation (middle), and eLB assay for p.T790M mutation (right).
As with any potential screening test, the reference range may need to be adjusted according to clinical performance. One advantage of eLB over other techniques is the ability to analyze both saliva and plasma from the same patient. It is possible that both tests could be performed simultaneously on each patient and only those with positive results for both plasma and saliva considered to be screen positive. For those individuals with one positive and one negative test result, repeated samples could be obtained and analyzed to resolve the discrepancy. The dual specimen analysis would greatly benefit a screening test where high specificity is essential.
eLB Quantitative Assays
The evaluation of the quantitative eLB was done in two parts. Initially, the limit of detection, linearity, and linear range were established using genomic DNA controls (Materials and Methods). However, biological materials are not optimal for use as internal calibrators for assays. Therefore, following the experiments to determine limits of detection, linearity, and linear range using genomic DNA, synthetic oligonucleotides were used as internal standards for the clinical assay.
EGFR Exon 19 Deletion
The g.Exon19del variant of EGFR is actually a family of closely related variants. Table 3 lists the five most prevalent variants in this family. Because the break points of these deletions are proximate to each other, it was possible to design a capture/detector probe set theoretically capable of detecting all reported g.Exon19del family of variants. To demonstrate that the eLB assay could detect the four most common variants, DNA was obtained from four cell lines, each containing one of these common variants (Materials and Methods). The genomic DNA was treated as described above (Materials and Methods) and placed into the eLB reaction at varying concentrations. There are no data to determine whether EFIRM can detect the less prevalent exon 19 del variants. However, these other variants are rare.
Table 3.
List of Common Exon19del Variants
| Amino acid | Nucleotide | COSMIC no.∗ | Prevalence, %22 | Prevalence, % |
|---|---|---|---|---|
| p.E746_A750delELREA (1) | c.2235_2249del15 | 6623 | 70 | 45 |
| p.E746_A750delELREA (2) | c.2236_2250del15 | 6225 | 29.6 | |
| p.L747_P753>S | c.2240_2257del15 | 12,370 | 5.9 | 8.4 |
| p.L747_A750>P | c.2239_2248TTAAGAGAAG>C | 12,382 | 2.0 | 3.4 |
| p.L747_E749delLRE | c.2239_27delTTAAGAGAA | 6218 | 0.3 | na |
COSMIC, Catalogue of Somatic Mutations in Cancer; na, not applicable.
COSMIC Database (https://cancer.sanger.ac.uk/cosmic, last accessed December 12, 2019).
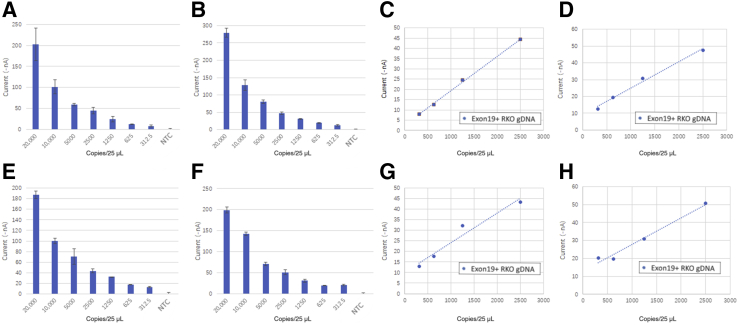
Figure 6 shows the results obtained using serial dilutions of genomic DNA containing the four most common EGFR g.exon19del variants; however, as can be seen, the EGFR assay is capable of detecting all four of these variants with similar performance.
Figure 6.
Electric field–induced release and measurement liquid biopsy assay results of serially diluted genomic DNA (gDNA) for the top four EGFR exon 19 variants. A–D: Plot of measurement results for variants c.2235_2249 del18, c2236_2250 del18, c2240_2257 del18, and c.2239_2248 del 11, respectively. E–H: Linear range of c.2235_2249 del18, c2236_2250 del18, c2240_2257 del18, and c.2239_2248 del 11, respectively. The blue dashed lines represent the best linear least squares fit for the data. NTC, no template control; RKO, reference cell line genomic DNA.
The assays are linear for all four variants, with R2 values of 0.99, 0.99, 0.95, and 0.94 for variants c.2235_2249 del 15, c2236_2250 del 15, c2240_2257 del 15, and c.2239_2248 del 15, respectively. The linear range extends from 20,000 copies/25 μL to 500 copies/25 μL. The sheared genomic DNA fragments are not the optimal template for the eLB reaction. Therefore, the lower limit of detection may be grossly underestimated. These data should, therefore, be considered as a minimal sensitivity for the eLB reaction.
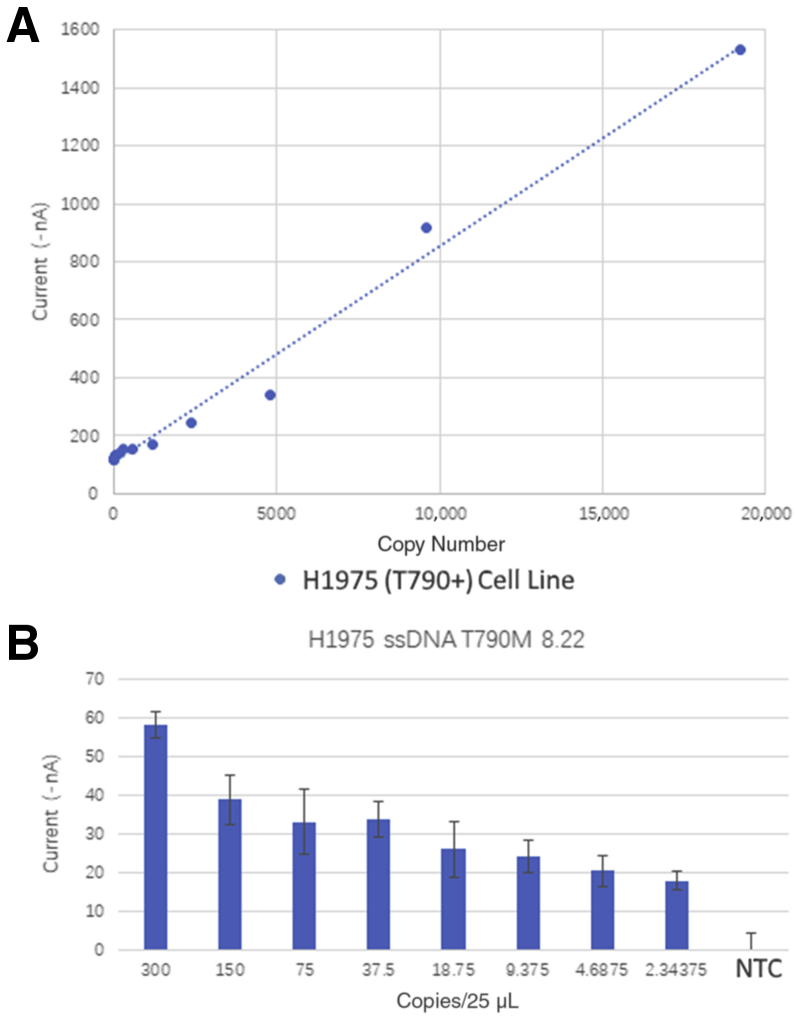
Linearity and Limit of Detection of eLB for p.T790M
Figure 7 displays the data from various dilutions of genomic DNA containing the variant p.T790M. The assay is linear, with an R2 of 0.99 over a range of 300 to single-digit copy number per 25 μL. Once again, the template in these samples is denatured, sheared, genomic DNA, which is not the optimum template for the EFIRM assay, and we would expect the true sensitivity to be better.
Figure 7.
Electric field–induced release and measurement (EFIRM) liquid biopsy assay results of serially diluted genomic DNA for p.T790M mutations. A: Scatterplot of diluted genomic DNA and EFIRM measurements. B: Bar plot of the lower ranges (300 to 2 copies) of the serially diluted genomic DNA. The blue dashed line represents the best linear least squares fit for the data. NTC, no template control; ssDNA, single-stranded DNA.
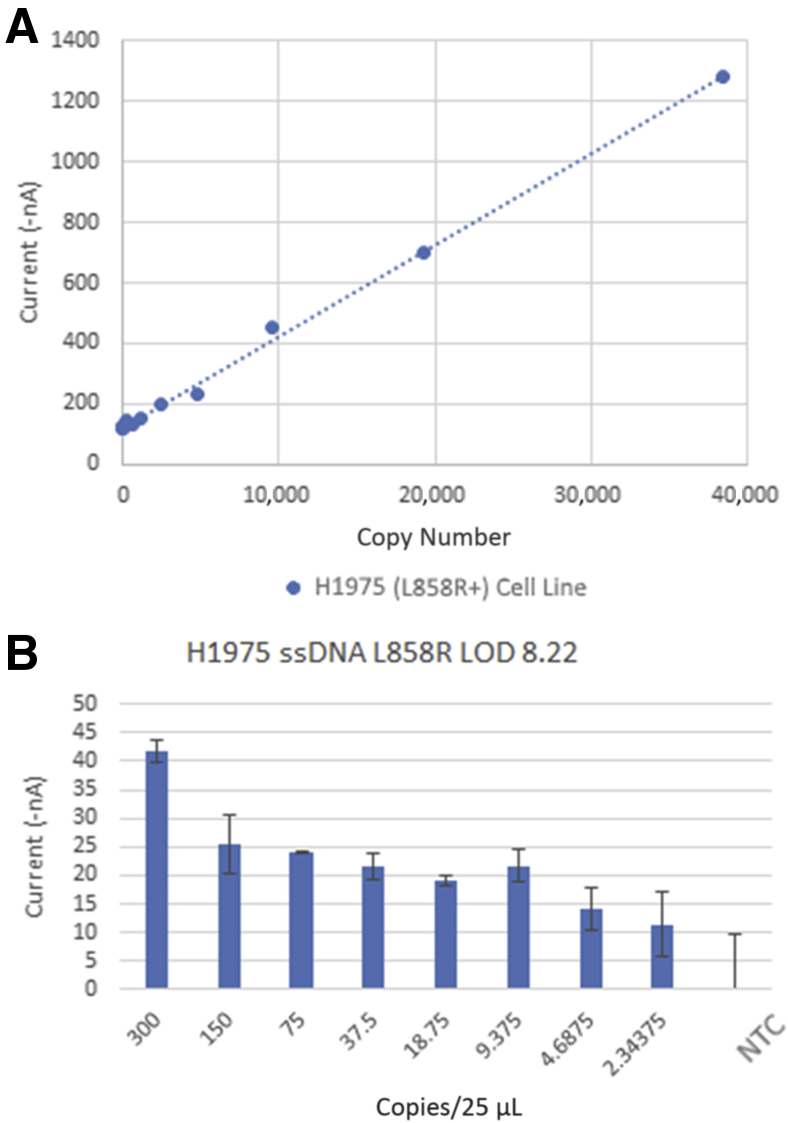
Linearity at Limit of Detection of eLB for p.L858R
Figure 8 displays the linearity and range for p.L858R. Linearity is excellent, with an R2 of 0.998 through a range of 300 copies to single-digit copy number per 25 μL. Taken together, these data show that the EFIRM assay has excellent linearity and sensitivity for the variants detected by the assay.
Figure 8.
Electric field–induced release and measurement (EFIRM) liquid biopsy assay results of serially diluted genomic DNA for p.L858R mutations. A: Scatterplot of diluted genomic DNA and EFIRM measurements. B: Bar plot of the lower ranges (300 to 2 copies) of the serially diluted genomic DNA. The blue dashed line represents the best linear least squares fit for the data. LOD, limit of detection; NTC, no template control; ssDNA, single-stranded DNA.
Analytical Validation of eLB for Six EGFR Variants
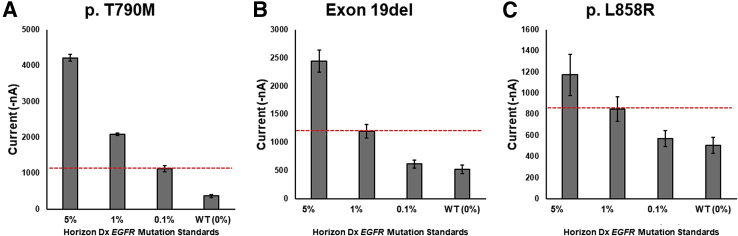
Once the linearity and limit of detection for each variant was determined separately, the performance of the eLB for the variants combined into a single assay was examined. One measure of the sensitivity of a liquid biopsy platform is its ability to detect variant sequences in a background of nonvariant sequences (minor allele detection). Reference materials are available for this purpose, containing artificial mixtures of wild-type and variant sequences. Such standards were purchased from Horizon Diagnostics (Cambridge, UK) and these samples were subjected to eLB analysis. Figure 9 demonstrates the eLB results for the three EGFR variants and reveals that, for p.T790M, eLB is capable of detecting variants at a 0.1% level. The g.exon19del and p.L858R assays are slightly less sensitive, but are able to clearly detect variant sequences at the 1% level. Standards were not available between 1% and 0.1%, so it is not possible to determine how much lower than the 1% minor allele fraction EFRIM was capable of detecting for g.Exon19del and p.L858R.
Figure 9.
Electric field–induced release and measurement liquid biopsy (eLB) assay results of standards from Horizon Diagnostics reference standards. A: eLB for p.T790M. B: eLB for exon 19 deletion. C: eLB for p.L858R. The red dashed lines represent the observed limit of detection for this series. Dx, Diagnostics; WT, wild type.
For the quantitative eLB six variant assays, synthetic oligonucleotide standards were used for calibration and internal standards. Titration was performed in quadruplicate using varying dilutions of oligonucleotides, containing each of the variants at concentrations of 0.02 to 1600 pmol/L. Linearity was excellent, with R2 values of 0.99, 0.98, and 0.99 for p.T790M, p.L858R, and g.Exon19del, respectively (data not shown).
Validation Performance Results (Linearity, Interassay and Intra-Assay CV, and Reference Range)
Quantitative validation of eLB was accomplished by performing a series of experiments with plasma and saliva matrices to evaluate assay linearity, assay CV, and reference range for p.L858R, g.Exon19 Del, and p.T790M. For linearity, twofold dilutions, ranging from 1000 pmol/L to 24 fmol/L, were run with synthetic oligonucleotide targets spiked into plasma or saliva matrices. Each dilution was performed four times. The results demonstrated an R2 ≥ 0.99 for p.L858R (R2 = 0.99), g.Exon19del (R2 = 0.99), and p.T790M (R2 = 0.99) in the 0 to 10 pmol/L range, the 0 to 50 pmol/L range, and the 0 to 12.5 pmol/L range, respectively, in saliva. Similarly, an R2 ≥ 0.99 was achieved for p.L858R (R2 = 0.996), g.Exon19del (R2 = 0.999), and p.T790M (R2 = 0.996) in plasma in the range of 0 to 6.25 pmol/L, 0 to 12.5 pmol/L, and 0 to 12.5 pmol/L, respectively. Limit of detection was calculated using a copy number to oligonucleotide target equivalence method, with the limit of detection of saliva being measured at 27.51 fmol/L (3 copies), 144.86 fmol/L (31 copies), and 60.38 fmol/L (9 copies) for the p.L858R, g.Exon19del, and p.T790M variants, respectively; and for plasma at 41.63 fmol/L (119 copies), 28.26 fmol/L (211 copies), and 27.72 fmol/L (142 copies), respectively.
Intra-assay variation analysis used a microtiter plate containing 88 replicates of each mutation containing 3.25 pmol/L of oligonucleotide target spiked in biofluid and a three-point standard curve (3.25, 1.56, and 0.78 pmol/L). The intra-assay CVs were 8.72%, 18.6%, and 12.5% for p.L858R, Exon19del, and p.T790M, respectively, in plasma and 29.3%, 25.2%, and 23.3% for p.L858R, Exon19del, and p.T790M, respectively, in saliva.
For interassay CV, four runs were made wherein each run assayed 24 replicates of each mutation using 3.25 pmol/L concentrations in biofluid using a three-point standard curve. The interassay CVs were 42.2%, 45.9%, and 30.7% for plasma and 40.6%, 35.9%, and 29.0% for saliva for p.L858R, g.Exon19del, and p.T790M, respectively. The interassay CV high is higher than the intra-assay CVs. This may be because of the stability of short oligonucleotides in the biological fluids. In the initial use of this assay, it would be prudent to perform the tests in duplicate, to identify samples with large variations. We are also initiating experiments to determine the optimal methods for sample collection, processing, and storage. Optimization of these parameters should allow the interassay CVs to be reduced. Alternatively, both plasma and saliva could be analyzed to minimize difficulties with interassay variability. When levels of these variants increase, it is often a greater than fourfold increase, which would allow trends to be seen even with these higher CVs.
To determine reference ranges, plasma and saliva from 40 healthy subjects were run across four or five plates for each of the variants with a three-point standard curve to assess the variability of signal levels within a healthy population. Cutoff values were established on the basis of 3 SDs from the mean at 4.04, 4.34, and 6.40 nA above normalized background current measurement for saliva and 7.10, 9.18, and 12.57 nA above normalized background current measurement for plasma for the p.L858R, g.Exon19del, and p.T790M mutations, respectively.
Discussion
Clinical Sensitivity and Specificity for eLB Qualitative Assay
The utility of any assay is determined by the sensitivity/specificity and positive and negative predictive values for the test. For the eLB platform, there are limited data on these parameters for the detection of EGFR variants in NSCLC patients. In two previously published studies involving late-stage (III and IV) patients,7,8 the data show that qualitative eLB assays detected all 23 patients with p.L858R variants and all 15 patients with g.Exon19del variants in their respective tumors. There were no false positives in 28 NSCLC patients whose tumors did not contain either of these variants. If qualitative eLBs were used for treatment selection, there would be 100% sensitivity and specificity in these late-stage NSCLC patients.
A study of the performance of plasma qualitative eLB in early-stage (I and II) NSCLC patients revealed a sensitivity of 92% for p.L858R and 77% for g.Exon19del.11 The specificities were 95% for the two variants in a control group of patients with benign pulmonary nodules. Of note, the concordance was 100% between tissue variant analysis and eLB results. Whenever the eLB showed a circulating mutant target, the variant was confirmed in the tissue biopsy.11
Currently there are two popular liquid biopsy platforms, NGS and digital droplet PCR (ddPCR).17,23 Both of these platforms require several milliliters of blood (10 to 20 mL for NGS methods and 1 to 10 mL for ddPCR) to perform the analysis. The eLB requires only 30 μL of either plasma or saliva. Previously reported data have demonstrated that eLB is capable of detecting circulating EGFR variant DNA in patients with early-stage lung cancer.11 EFIRM is the only platform capable of this level of discrimination in stage I and II NSCLC patients. The eLB method requires only 30 μL of plasma for each variant; this is >100 times less sample required than for either NGS- or ddPCR-based methods. Neither NGS nor ddPCR platforms have been demonstrated to be able to perform LB on saliva specimens.
One must ask the question, how is it possible for eLB to have increased sensitivity and specificity in early-stage lung cancer patients using <1% of the sample volume of either ddPCR or NGS? There are several factors that might explain this phenomenon.
The eLB is performed on untreated, unpurified plasma or saliva. Both ddPCR and NGS require a DNA isolation step before assay. It is likely that a significant percentage of the ctDNA fragments is lost during the DNA isolation procedures or subsequent manipulations. NGS requires several enzymatic treatments and purifications between steps, and small fragments of DNA are likely lost during these procedural steps. In our experience, each of the enzymatic steps required before library formation are not 100% efficient, so material is lost at each step because of incomplete reactions.
In addition, both the NGS and ddPCR methods assume a target molecular amount of double-stranded DNA of approximately 150 bp in length. This is the reported length of nucleosome protected apoptotic DNA. It is assumed that ctDNA fragments are similar in size and nature to the fetal sequences found in the maternal circulation in noninvasive prenatal screening. It is possible, or even likely, that ctDNA does not arise from orderly apoptosis. In fact, most cancer cells lack the programmed cell death that results in apoptotic DNA fragments. Unlike fetal DNA in the maternal circulation, ctDNA is more likely the result of necrosis from outgrowing the blood supply or by immune lysis. It is possible that this process would result in smaller fragments and potentially single-stranded DNA fragments. Both ddPCR and NGS methods are designed to detect double-stranded DNA fragments of approximately 150 bp, whereas EFIRM's preferred substrate is single-stranded DNA of <100 bp in length (see paragraph below).
In fact, there is evidence that in early-stage cancers, ctDNA containing actionable mutations is smaller in size than both the wild-type DNA and the ctDNA present in more advanced disease. In fact, ctDNA derived from early-stage pancreatic cancer had a fragment size of significantly <100 bp.24 This study used single-stranded DNA sequencing strategies, so it is possible that the observed ultrashort ctDNA fragments were not only <100 bp, but were also single stranded. Other studies using single-stranded sequencing have also demonstrated that ctDNA is smaller than that of circulating DNA from other origins.25,26 Preliminary work from our laboratory indicates that the mutant ctDNA sequences detected by EFIRM in NSCLC patients is actually between 35 and 44 bp in length (data not shown). Other modalities, such as ddPCR, would not be able to detect fragments this short and standard NGS methods would not sequence single-stranded DNA molecules. The unique property of the eLB platform is that its preferred template is ultrasmall single-stranded DNA, allowing eLB to preferentially detect ctDNA while ignoring circulating DNA from other sources.
Another possibility is that the eLB is measuring exosomal DNA. Fernando et al27 report that most exosomal DNA is approximately 76 bp in length. The electric current applied to the biological samples would lyse exosomes. Further work will be necessary to delineate the origin and nature of the fragments analyzed by the eLB.
ddPCR is a technology that is more comparable to EFIRM than NGS-based methods. This method shares the limitation of EFIRM in that only a small number of variants can be assayed simultaneously, but shares its strength in that it is lower in cost and can give rapid turnaround times.2 However, ddPCR currently requires a minimum of 1 mL of blood as opposed to microliters of blood for a single variant and, as currently constituted, will not detect ultrashort single-stranded fragments of DNA. This may be why ddPCR has not been shown to be sensitive in detecting cell-free DNA fragments in early-stage NSCLC patients.
Another advantage of eLB over other technologies is the ability to use non-standard sample types. In this report, we were able to validate both a qualitative and a quantitative eLB assay on saliva. It is probable that other samples types, such as urine, pleural fluid, and cerebrospinal fluid, could be successfully analyzed using the EFIRM technology.
EFIRM is the only liquid biopsy platform reported to be capable of detecting ctDNA in patients with early-stage NSCLC.11 This may provide an opportunity to use liquid biopsy to help evaluate either asymptomatic individuals with a smoking history or individuals with indeterminate nodules discovered by screening radiographic examination. We are currently participating in a prospective trial of using EFIRM to evaluate patients with indeterminate lung nodules. A qualitative assay with positive or negative result could allow a more informed decision for patients found to have an indeterminate nodule on a radiographic examination.
The EFIRM method is plate based, and the entire assay can be performed easily in 3 hours, with little hands-on time. This is similar to the parameters of ddPCRs, as both platforms allow a significantly faster turnaround time than NGS-based methods, which often take several days to complete and 10 to 14 days to report.
A final advantage of eLB is the cost. An eLB can be performed for a few hundred dollars and, therefore, could be used for serial monitoring, for treatment response, early detection of recurrence, or detection of the presence of minimal residual disease. If sensitivities and specificities are sufficient, eLB could also be used to help screen smokers or individuals with indeterminate pulmonary nodules on computed tomographic scan. The sensitivity for eLB as a screening test is currently limited by the prevalence of the six EGFR variants among NSCLC patients. Although the frequency is as high as 40% in China,28 the fraction of NSCLC tumors harboring one of these six variants is lower in the United States. The overall incidence of EGFR mutations in NSCLC is 2.7%, with confidence that the true incidence does not exceed 3.6%, according to Forbes et al.22 For adenocarcinoma, the overall frequency is significantly higher at 23%: for men, it is 19%; and for women, it is 28%. As for smokers with adenocarcinoma, the incidence is 47% overall and 14% for never smokers.29
To increase the sensitivity of detection in the United States, we are in the process of adding nine additional variants to the eLB that should increase the detection rate to 50% of NSCLC patients.
Currently, the EFIRM platform is limited to one variant per well. Multiplexing would be desirable, but given the current hardware configuration, the only potential for multiplexing would be to add two or more capture probes to the same well. Fortunately, the assay requires only 30 μL of biofluid, so it would be possible to perform 50 separate EFIRM assays on 0.5 mL of saliva or plasma. Assay development on the platform is straightforward; capture probes and signal probes are easily designed, and assays can usually be optimized within a few weeks. We are investigating the possibility of multiplexing in single wells, but this will require complete revamping of the plates and EFIRM Reader.
The EFIRM platform is also capable of performing protein-based analysis. Assays can be designed similar to enzyme-linked immunosorbent assay, and preliminary work has shown that autoantibodies can be detected in saliva and serum. The EFIRM reader is about the size of a fax machine, and the entire footprint consists of the reader plus a plate washer, so <4 feet of bench space is required for the entire setup.
In summary, this report shows the technical validation of four eLB assays, which include both qualitative and quantitative assays for plasma and saliva. The assay has a minor allele fraction detection of 0.1% for p.T790M and 1% for Exon19del and p.L858R.
Further investigation will determine the clinical sensitivities and specificities for eLB in the screening setting. However, the quantitative saliva assay is currently being used for treatment monitoring and recurrence detection on an investigational basis. Preliminary data have demonstrated that saliva-based EFIRM reactions can be used for treatment monitoring of NSCLC patients receiving thymidine kinase inhibitor therapy.
The eLB liquid biopsy platform is a promising advance in liquid biopsy technology and may be applicable in several clinical settings. The cost, convenience, turnaround time, and ability to use small samples of both peripheral blood and saliva are significant improvements over current technologies.
Footnotes
Supported by NIH/Public Health Service (PHS) grants UH2/UH3 CA206126 (D.T.W.W.) and U01 CA233370 (D.T.W.W.); the Ronnie Dio Stand Up to Cancer Fund (D.T.W.W.); Center of Applied Nanomedicine, National Cheng Kung University, from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education in Taiwan; and the Canadian Institute of Health Doctoral Foreign Student Award and Tobacco Related Disease Research Program Predoctoral Fellowship (J.C.).
M.T. and J.C. contributed equally to this work.
Disclosures: W.L. is Founder/CEO of EZLife Bio Inc. and holds equity; M.T. is employed by Liquid Diagnostics LLC and holds equity in EZLife Bio Inc.; C.S. is Founder/CEO of Liquid Diagnostics LLC and holds equity.
Contributor Information
David T.W. Wong, Email: dtww@ucla.edu.
Charles M. Strom, Email: strom@liquid-dx.com.
References
- 1.Poulet G., Massias J., Taly V. Liquid biopsy: general concepts. Acta Cytol. 2019;63:449–455. doi: 10.1159/000499337. [DOI] [PubMed] [Google Scholar]
- 2.Xu C., Offin M., Paik P.K., Li B.T. Liquid biopsy guided precision therapy for lung cancers. J Thorac Dis. 2018;10 Suppl 33:S4173–S4175. doi: 10.21037/jtd.2018.11.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rolfo C., Mack P.C., Scagliotti G.V., Baas P., Narlesi F., Bivona T.G., Herbst R.S., Mok T.S., Peled N., Pirker R., Raez L.E., Reck M., Reiss J.W., Sequist L.V., Shepherd F.A., Sholl L.M., Tan D.S.W., Wakelee H.A., Wistuba I.I., Wynes M.W., Carbone D.P., Hirsch F.R., Gandara D.R. Liquid biopsy for advanced non-small cell lung cancer (NSCLC): a statement paper from the IASLC. J Thorac Oncol. 2018;13:1248–1268. doi: 10.1016/j.jtho.2018.05.030. [DOI] [PubMed] [Google Scholar]
- 4.Schwaederle M., Husain H., Fanta P.T., Piccioni D.E., Kesari S., Schwab R.B., Patel S.P., Harismendy O., Ikeda M., Parker B.A., Kurzrock R. Use of liquid biopsies in clinical oncology: pilot experience in 168 patients. Clin Cancer Res. 2016;22:5497–5505. doi: 10.1158/1078-0432.CCR-16-0318. [DOI] [PubMed] [Google Scholar]
- 5.Almodovar K., Iams W.T., Meador C.B., Zhao Z., York S., Horn L., Yan Y., Hernandez J., Chen H., Shyr Y., Lim L.P., Raymond C.K., Lovly C.M. Longitudinal cell-free DNA analysis in patients with small cell lung cancer reveals dynamic insights into treatment efficacy and disease relapse. J Thorac Oncol. 2018;13:112–123. doi: 10.1016/j.jtho.2017.09.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma C., YangX, Xing W., Yu H., Si T., Guo Z. Detection of circulating tumor DNA from non-small cell lung cancer brain metastasis in cerebrospinal fluid samples. Thorac Cancer. 2020;11:588–593. doi: 10.1111/1759-7714.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandez-Garcia D., Hills A., Page K., Gastings R.K., Toghill B., Goddard K.S., Ion C., Ogle C., Boydell A.R., Gleason K., Rutherford M., Lim A., Guttery D.S., Coombes R.C., Shaw J.A. Plasma cell-free DNA (cfDNA) as a predictive and prognostic marker in patients with metastatic breast cancer. Breast Cancer Res. 2019;21:149. doi: 10.1186/s13058-019-1235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Y., Zhang H., Chen N., Hao J., Jin H., Ma X. Diagnostic value of various liquid biopsy methods for pancreatic cancer: a systematic review and meta-analysis. Medicine (Baltimore) 2020;99:e18581. doi: 10.1097/MD.0000000000018581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alrabadi N., Haddad R., Alomari A.K. Detection of gene mutations in liquid biopsy of melanoma patients: overview and future perspectives. Curr Treat Options Oncol. 2020;21:19–26. doi: 10.1007/s11864-020-0708-4. [DOI] [PubMed] [Google Scholar]
- 10.Stuopelyte K., Sabaliauskaite R., Bakavicius A., Hadliadottir B.S., Visakorpi T., Vaananen R.M., Patel C., Danila D.C., Lilja H., Lazutka J.R., Ulys A., Jankevicius J.F., Jarmalaite S. Analysis of AR-FL and AR-V1 in whole blood of patients with castration resistant prostate cancer as a tool for predicting response to abiraterone acetate. J Urol. 2020;204:71–78. doi: 10.1097/JU.0000000000000803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei F., Strom C.M., Cheng J., Lin C.-C., Hsu C.-Y., Soo Hoo G.W., Chia D., Kim Y., Li F., Elashoff D., Grognan T., Tu M., Liao W., Xian R., Grody W.W., Su W.-C., Wong D.T.W. Electric field–induced release and measurement liquid biopsy for noninvasive early lung cancer assessment. J Mol Diagn. 2018;20:738–742. doi: 10.1016/j.jmoldx.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei F., Lin C.-C., Joon A., Feng Z., Troche G., Lira M.E., Chia D., Mao M., Ho C.-L., Su W.-C., Wong D.T.W. Noninvasive saliva-based EGFR gene mutation detection in patients with lung cancer. Am J Respir Crit Care Med. 2014;190:1117–1126. doi: 10.1164/rccm.201406-1003OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pu D., Liang H., Wei F., Akin D., Feng Z., Yan Q., Li Y., Zhen Y., Xu L., Dong G., Wan H., Dong J., Qiu X., Qin C., Zhu D., Wang X., Sun T., Zhang W., Li C., Tang X., Qiao Y., Wong D.T.W., Zhou Q. Evaluation of a novel saliva-based epidermal growth factor receptor mutation detection for lung cancer: a pilot study: saliva-based EGFR mutation detection. Thorac Cancer. 2016;7:428–436. doi: 10.1111/1759-7714.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elazezy M., Joosse S.A. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput Struct Biotechnol J. 2018;16:370–378. doi: 10.1016/j.csbj.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahama T., Azuma K., Shimokawa M., Takeda M., Ishii H., Kato T., Saito H., Daga H., Tsuboguchi Y., Okamato I., Ossubo K., Akamatsu H., Teraoka S., Takahashi T., Ono A., Ohira T., Yokoyama T., Sakai K., Yamamoto N., Nishio K., Nagagawa K. Plasma screening for the T790M mutation of EGFR and phase 2 study of osimertinib efficacy in plasma T790M-positive non-small cell lung cancer: West Japan Oncology Group 8815L/LPS study. Cancer. 2020;126:1940–1948. doi: 10.1002/cncr.32749. [DOI] [PubMed] [Google Scholar]
- 16.Papadopoulou E., Tsoulos N., Tsantikidi K., Metaxa-Mariatou V., Stamou P.E., Kladi-Skandali A., Kapeni E., Tsaousis G., Pentheroudakis G., Petrakis D., Lampropoulou S.I., Aravantinos G., Varthalitis G., Kesesis G., Boukovinas I., Papakotoulas P., Katirtzoglou N., Athanasiadis E., Stavridu F., Christodoulou C., Koumarianou A., Eralp Y., Nasioulas G. Clinical feasibility of NGS liquid biopsy analysis in NSCLC patients. PLoS One. 2019;14:1371–1378. doi: 10.1371/journal.pone.0226853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y.-C., Zhou Q., Wu Y.-L. The emerging roles of NGS-based liquid biopsy in non-small cell lung cancer. J Hematol Oncol. 2017;10:167. doi: 10.1186/s13045-017-0536-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Offin M., Chabon J.J., Razavi P., Isbell J.M., Rudin C.M., Diehn M., Li B.T. Capturing genomic evolution of lung cancers through liquid biopsy for circulating tumor DNA. J Oncol. 2017;2017:4517834. doi: 10.1155/2017/4517834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlovich C., Goldman J.W., Sun J.M., Mann E., Sequist L.V., Konopa K., Wen W., Angendendt P., Horn L., Spiegel D., Soria J.-C., Solomon B., Camidge D.R., Gadgeel S., Paweletz C., Wu L., Chien S., O'Donnell P., Matheny S., Despain D., Rolfe L., Raponi M., Allen A.R., Park K., Wakelee H. Assessment of EGFR mutation status in matched plasma and tumor tissue of NSCLC patients from a phase 1 study of Rociletimib (CO-1686) Clin Cancer Res. 2016;22:2386–2395. doi: 10.1158/1078-0432.CCR-15-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y.L., Sequist L.V., Hu C.P., Feng J., Lu S., Huang Y., Li W., Hou M., Schuler M., Mok T., Yamamoto N., O'Byrne K., Hirsh V., Gibson N., Massey D., Kim M., Yang J.C.-H. EGFR mutation detection in circulating cell-free DNA of lung adenocarcinoma patients: analysis of LUX-lung 3 and 6. Br J Cancer. 2017;116:175–185. doi: 10.1038/bjc.2016.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merker J., Oxnard G., Compton C., Diehn M., Hurley P., Lazar A.J., Lindeman N., Lockwood C.M., Rai A.J., Schilsky R.L., Tsimberidou A.M., Vasalos P., Billman B.L., Oliver T.K., Bruinooge S.S., Hayes D.F., Turner N.C. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J Clin Oncol. 2018;36:1631–1641. doi: 10.1200/JCO.2017.76.8671. [DOI] [PubMed] [Google Scholar]
- 22.Forbes S.A., Bhamra G., Bamford S., Dawson E., Kok C., Clements J., Menzies A., Teague J.W., Futreal P.A., Stratton M.R. The catalogue of somatic mutations in cancer (COSMIC) Curr Protoc Hum Genet. 2008;57:10.11.1–10.11.26. doi: 10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olmedillas-López S., García-Arranz M., García-Olmo D. Current and emerging applications of droplet digital PCR in oncology. Mol Diagn Ther. 2017;21:493–510. doi: 10.1007/s40291-017-0278-8. [DOI] [PubMed] [Google Scholar]
- 24.Liu X., Liu L., Ji Y., Li C., Wei T., Yang X., Zhang Y., Cai X., Gao Y., Xu W., Rao S., Jin D., Lou W., Qiu Z., Wang X. Enrichment of short mutant cell-free DNA fragments enhanced detection of pancreatic cancer. EBioMedicine. 2019;41:345–356. doi: 10.1016/j.ebiom.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burnham P., Kim M.S., Agbor-Enoh S., Luikart H., Valantine H.A., Khush K.K., De Vlaminck I. Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma. Sci Rep. 2016;6:27859. doi: 10.1038/srep27859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Underhill H.R., Kitzman J.O., Hellwig S., Welker N.C., Daza R., Baker D.N., Gligorich K.M., Rostomily R.C., Bronner M.P., Shendure J. Fragment length of circulating tumor DNA. PLoS Genet. 2016;12:e1006162. doi: 10.1371/journal.pgen.1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernando M.R., Jiang C., Krzyzanowski G.D., Ryan W.L. Analysis of human blood plasma cell-free DNA fragment size distribution using EvaGreen chemistry based droplet digital PCR assays. Clin Chim Acta. 2018;483:39–47. doi: 10.1016/j.cca.2018.04.017. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y.-L., Yuan J.-Q., Wang K.-F., Fu X.-H., Han X.-R., Threapleton D., Yang Z.-Y., Mao C., Tang J.-L. The prevalence of EGFR mutation in patients with non-small cell lung cancer: a systematic review and meta-analysis. Oncotarget. 2016;7:78985–78993. doi: 10.18632/oncotarget.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Midha A., Dearden S., McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII) Am J Cancer Res. 2015;5:2892–2911. [PMC free article] [PubMed] [Google Scholar]