Key Points
Zebrafish IL-4–like cytokines promote type 2 and suppress type 1 immunity.
Zebrafish IL-10 is potently anti-inflammatory and essential for gill homeostasis.
Abstract
Mucosal surfaces such as fish gills interface between the organism and the external environment and as such are major sites of foreign Ag encounter. In the gills, the balance between inflammatory responses to waterborne pathogens and regulatory responses toward commensal microbes is critical for effective barrier function and overall fish health. In mammals, IL-4 and IL-13 in concert with IL-10 are essential for balancing immune responses to pathogens and suppressing inflammation. Although considerable progress has been made in the field of fish immunology in recent years, whether the fish counterparts of these key mammalian cytokines perform similar roles is still an open question. In this study, we have generated IL-4/13A and IL-4/13B mutant zebrafish (Danio rerio) and, together with an existing IL-10 mutant line, characterized the consequences of loss of function of these cytokines. We demonstrate that IL-4/13A and IL-4/13B are required for the maintenance of a Th2-like phenotype in the gills and the suppression of type 1 immune responses. As in mammals, IL-10 appears to have a more striking anti-inflammatory function than IL-4–like cytokines and is essential for gill homeostasis. Thus, both IL-4/13 and IL-10 paralogs in zebrafish exhibit aspects of conserved function with their mammalian counterparts.
Introduction
In mammals, IL-4 and IL-13 are structurally and functionally related cytokines that participate in several physiological processes but are mainly known for stimulating type 2 immune responses (1), characterized by mucus and IgE production, eosinophilia, and differentiation of alternatively activated (M2) macrophages (2). Many aspects of type 2 immunity reinforce and protect barrier surfaces, including enhanced mucus production, rapid tissue repair, and IgE blockade of toxins (3–5). These responses confer protection against parasitic infection and some venoms but, when inappropriately activated, contribute to the development of asthma, allergic inflammation, and fibrosis (1, 6–8).
IL-4 or IL-13 or both are secreted by CD4+ Th2 cells, basophils, eosinophils, mast cells, and group 2 innate lymphoid cells (ILCs) but also by granulocytes associated with allergic responses, including basophils, eosinophils, and mast cells (9). IL-4 and IL-13 exert their functions by binding to two types of receptor complexes: a type I receptor, constituted by the IL-4Rα-chain and the IL-2R common γ-chain and a type II receptor, which comprises IL-4Rα-chain and IL-13Rα1 subunits. Type I receptor is mainly expressed in hematopoietic cells, particularly in lymphocytes, whereas the type II receptor is expressed on nonhematopoietic cells, such as epithelial cells. Interestingly, myeloid cells express both type I and type II receptors (1). Both type I and type II receptors signal through STAT6 transcription factor binding to promoter elements within IL-4/IL-13 responsive genes (10). IL-4 and IL-13 suppress inflammatory responses by antagonizing the production of TNF-α, IL-1β, and other proinflammatory mediators (11) and act in opposition to IFN-γ, the canonical Th1 (type 1) cytokine (12). Indeed, coordinately with inducing type 2 immune response, IL-4 and IL-13 suppress type 1 responses that are characterized by inflammation caused by immune cell–mediated destruction of cells infected with intracellular pathogens (13).
Although IL-4 and IL-13 can negatively regulate inflammatory responses, IL-10 has the more central anti-inflammatory role in mammals, potently suppressing IFN-γ responses. IL-10 can suppress a range of aberrant immune responses, including both type 1 and type 2 responses (14). IL-10 is produced by many cell types (15). It regulates CD4+ regulatory T cell differentiation and function, and it is important in maintaining homeostasis at mucosal surfaces (16). In the gastrointestinal tract, IL-10 regulates the immune response to the gut microbiota (17). A decrease of IL-10 production from intestinal epithelial cells leads to the development of gut inflammatory disorders (18). Moreover, lack of IL-10 induces the development of spontaneous inflammation at mucosal surfaces such as the lungs and intestine (19).
Evidence is emerging that fish immune responses can also be classified as type 1 or type 2 and that Th cells similar to mammalian counterparts likely do exist (20). Relevant cytokine receptors and downstream signaling pathway molecules are conserved between fish and mammals (21). However, there is still the need to understand how specific immune cells and cytokines function, in particular in mucosal tissues such as the gills. Fish gills represent an important mucosal surface that is constantly exposed to both waterborne pathogens and commensals and is able to mount immune responses (22, 23). Teleost gills are composed of four arches on each side of the head supported by cartilage or bone tissue. Each arch displays two files of filaments (primary lamellae). In turn, each filament is made of plate-like structures projecting on both sides (secondary lamellae) where gas exchange occurs (24). Lamellae are covered with a microridged respiratory epithelium, comprising pavement cells, mitochondria-rich chloride cells, and mucus-producing goblet cells (25). Several immune cells, including macrophages, neutrophils, and lymphocytes, are found dispersed within the lamellar epithelium, forming the so-called gill-associated lymphoid tissue (26). Moreover, an organized lymphoid area at the origin of the filaments, namely the interbranchial lymphoid tissue (ILT), has been described in salmonids (27, 28). ILT is rich in T cells that are embedded in a meshwork of epithelial cells (28), and it has been shown to play a role in maintaining immune tolerance and homeostasis in the gills (29), suggesting that the ILT represents a vital part of the gill-associated lymphoid tissue. A commensal microbial community has also been discovered in fish gills (30). As with all mucosal surfaces, the balance between inflammatory and regulatory responses is critical for the maintenance of tissue homeostasis and barrier integrity.
Two type 2 cytokines that are related to mammalian IL-4 and IL-13 have been described in teleost fish (31). It is likely that a single il4/13 gene existed in ancestral jawed vertebrates, which has been duplicated in subsequent vertebrate lineages by whole genome duplication and/or tandem duplication events (32). Because of an additional round of whole genome duplication that occurred in fish, teleosts acquired two il4/13 loci (ohnologues), namely il4/13a and il4/13b (33). Analysis of these loci shows that teleost il4/13 genes share the following important characteristics with genes encoding tetrapod IL-4 and IL-13: their positioning relative to rad50 and kif3a gene neighbors (34), the typical short-chain type 1 cytokine organization, and conserved structural motifs (33). The identification of putative binding motifs for GATA3, a transcription factor that directs mammalian Th2 and ILC2 development (35), in the promoter regions of teleost il4/13 genes further supports that these genes encode Th2 cytokines (33).
Enriched levels of mRNA encoding IL-4/13A together with the transcription factor GATA3 have been detected in salmonid fish mucosal tissues, such as the gills and the skin, indicating these to represent Th2-skewed environments that protect fish from parasites and inflammatory responses (36). We have also identified a population of CD4+ Th2-like lymphocytes and their signature cytokines in zebrafish gills (37). To date, a comprehensive characterization of fish IL-4/13 paralogs is lacking, and whether il4/13a and il4/13b represent authentic IL-4 and IL-13 orthologs is still a matter of debate. IL-10 has been described in several fish species (38–41), and its constitutive expression was reported in zebrafish kidney, gut, and gills (41), suggesting a role in maintaining homeostasis in these tissues. Exploring the conservation and divergence of IL-4/13 and IL-10 cytokines would further our understanding of fish immune responses and may also provide an alternative model for dissecting aspects of mammalian immunity.
In this study, we used zebrafish to study the functions of fish IL-4/13A, IL-4/13B, and IL-10 cytokines. We generated zebrafish knockouts for il4/13a and il4/13b genes and addressed the effects of their downregulation in both larvae and adult fish immunity. We showed the importance of IL-4/13A and IL-4/13B in suppressing inflammation as well as maintaining a Th2 phenotype in the gills. To gain further insight into the regulation of inflammation, the gills of il10-deficient zebrafish were examined, revealing the requirement for IL-10 in maintaining homeostasis in this mucosal tissue.
Materials and Methods
Zebrafish care
Zebrafish (Danio rerio) were maintained under standard conditions (∼28°C under a 14-h light/10-h dark cycle) within the Biological Services Unit (The University of Manchester) and the Central Biomedical Services (Imperial College London). Embryos were collected and raised in egg water (Instant Ocean Salt 60 μg/ml) up to 5 d postfertilization (dpf) and then transferred to the main aquarium system. Younger fry were fed powder food and rotifers, whereas older fry and adults were fed powder food and brine shrimp. All food was supplied by Zebrafish Management fish food. il10e46/e46 mutant zebrafish were a kind gift from S. Johnston (University of Sheffield) and were generated by the Sanger Institute through the Zebrafish Mutation Project (42). All regulated procedures received ethical approval from the institutions’ ethical review boards and were performed under Home Office License (project licenses PF74F0848 and P5D71E9B0), according to the United Kingdom’s Animal Act.
CRISPR/Cas9-mediated generation of mutant lines
Creation of single chimeric guide RNA targeting il4/13a and il4/13b.
Guide RNAs (gRNAs) were designed to target the first exon of either il4/13a or il4/13b zebrafish genes using the Harvard chopchop program (https://chopchop.rc.fas.harvard.edu). The il4/13a target site was 5′-GGGTTTTACGTTGAAAGGCA-3′, and the il4/13b target site was 5′-GAAATCATCCAGAGTGTGAA-3′ and were incorporated into a gRNA template for transcription using PCR. The forward primer contained the target gene specific sequence followed by a constant sequence that overlaps with the remaining sequence of Streptococcus pyogenes chimeric gRNA DNA template (plasmid no. 51132; Addgene), whereas the reverse primer was complementary to the gRNA DNA template (see Table I for primer sequences). PCR amplification of the gRNA DNA template using a high fidelity Phusion Taq polymerase was performed, and PCR products were gel purified and used for the synthesis of gRNAs using an Ambion MEGAshortscript T7 kit, according to the manufacturer’s instructions.
Table I. List of primers used to generate the single gRNA templates.
| Name | Forward | Reverse |
|---|---|---|
| il4/13a | 5′-TAATACGACTCACTATAGGGTTTTACGTTGAAAGGCAGTTTTAGAGC-3′ | 5′-AAAAGCACCGACTCGGTGCCACTTTTTCAAG-3′ |
| il4/13b | 5′-TAATACGACTCACTATAGAAATCATCCAGAGTGTGAAGTTTTAGAGC-3′ | 5′-AAAAGCACCGACTCGGTGCCACTTTTTCAAG-3′ |
Injecting CRISPR reagents.
Injection solutions included 30 pg/nl of either gRNA, 250 pg/nl of codon-optimized S. pyogenes nls-zCas9-nls mRNA synthesized using a mMESSAGE mMACHINE SP6 Kit (Life Technologies) from a pCS2 construct (plasmid no. 47929; Addgene), 100 pg/nl H2B-mCerulean3 tracer mRNA similarly generated from a pCS2 construct, and 0.05% (w/v) phenol red to allow visualization of injections. Embryos were injected at the one-cell stage and screened for fluorescence at 24 h postfertilization to identify positively injected fish, which were then raised to adulthood.
Identifying and raising mutant lines.
To identify modified alleles in the F1 progeny for the establishment of il4/13a and il4/13b zebrafish mutant lines, genomic DNA was amplified by PCR using gene-specific primers flanking the targeted site (see Table II for primer sequences), and abnormal sized products were gel purified. One microliter of sample was cloned into a TOPO cloning vector (Invitrogen). Cloning reactions were performed according to the manufacturer’s instructions. Inserts were sequenced by Sanger sequencing using Eurofins Genomics services. Fish carrying selected mutations were out crossed and their F2 progeny genotyped for the presence of mutations (see Table II for primer sequences). Il4/13a+/− and il4/13b+/− heterozygous fish were in crossed to generate homozygous mutants for both il4/13a (il4/13a−/−) and il4/13b (il4/13b−/−) in the F3 generation. To generate il4/13a;b−/− double mutants, il4/13a−/− and il4/13b−/− homozygous fish were intercrossed and their progeny in crossed. il4/13a;b−/− double mutants were in crossed to maintain the line.
Table II. List of primers used for genotyping PCR.
| Name | Forward | Reverse |
|---|---|---|
| il4/13a | 5′-GCACTGTATTCGTCTCGGGTTTTA-3′ | 5′-TTTTCCCCAGATCTACAAGGAAGA-3′ |
| il4/13b | 5′-CTGTTGGTACTTACATTGGTCCCC-3′ | 5′-AGTGTCCTGTCTCATATATGTCAGGT-3′ |
Whole mount in situ hybridization
Three-day-old embryos (n = 9–10) were fixed in 4% (w/v) paraformaldehyde in PBS at 4°C overnight. Fish were then washed in 0.1% (v/v) Tween 20 PBS solution and dehydrated through an ascending series of methanol. Digoxigenin-labeled RNA antisense probe for zebrafish mpx was kindly supplied by S. Renshaw (University of Sheffield), and whole mount in situ hybridization was performed as described in (43). Embryos fixed in glycerol were mounted on a glass slide, and images were collected on a Leica MZ16FA stereomicroscope equipped with a DC490 camera. Quantification of mpx+ cells was performed using ImageJ software.
Real-time quantitative PCR
RNA was isolated from homogenized larvae and/or adult gills (n > 3) using the RNeasy Mini Kit (QIAGEN) and reverse transcribed using the ProtoScript II First Strand cDNA Synthesis Kit (New England BioLabs) with Oligo (dT) primers, according to the manufacturer’s instructions. Quantitative PCR (qPCR) was performed using SYBR Green JumpStart Taq ReadyMix (Sigma), ROX Reference Dye, and cDNA (diluted 1:2). All samples were run in triplicate on 96-well PCR plates (BioLabs). Negative controls were run without cDNA. Data were analyzed by the ΔCt method using bactin for normalization. The primers used for qPCR are listed in Table III.
Table III. List of primers used for qPCR.
| Name | Forward | Reverse |
|---|---|---|
| β actin | 5′-CGAGCTGTCTTCCCATCCA-3′ | 5′-TCACCAACGTAGCTGTCTTTCTG-3′ |
| tnfa | 5′-ACAAGATGGAAGTGTGCTGAGA-3′ | 5′-ATTTCAAGCCACCTGAAGAAAA-3′ |
| il6 | 5′-CCTCTCCTCAAACCTTCAGACC-3′ | 5′-TGCTGTGTTTGATGTCGTTCAC-3′ |
| il1b | 5′-GGACTTCGCAGCACAAAATGAA-3′ | 5′-TTCACTTCACGCTCTTGGATGA-3′ |
| il4/13b | 5′-GCAGGAATGGCTTTGAAGGGTAAA-3′ | 5′-AAACTCCTTCATTGTGCATTCCCC-3′ |
| il4/13a | 5′-GCACTGTATTCGTCTCGGGTTTTA-3′ | 5′-TTTTCCCCAGATCTACAAGGAAGA-3′ |
| il10 | 5′-CTTTAAAGCACTCCACAACCCCAA-3′ | 5′-CTTGCATTTCACCATATCCCGCTT-3′ |
| mpx | 5′-GAGCTGCAGACTACATGGCACA-3′ | 5′-CTAGCCACTGCAGATGCTGACATAG-3′ |
| ifng1.2 | 5′-CCTGGGGAGTATGTTTGCTGTTTT-3′ | 5′-GGGTGTGCATTATGTAGCTGAGAA-3′ |
Histology
Whole heads were collected from wildtype (n = 3), il4/13a−/− (n = 4), il4/13b−/− (n = 4), il4/13a;b−/− (n = 4), and il10e46/e46 (n = 6) as well as wildtype and il10e46/e46 animals following 8-h resiquimod (R848) challenge (n = 3) and fixed in 4% (w/v) paraformaldehyde overnight at 4°C. The next day, the samples were washed in PBS and decalcified in 0.25 M EDTA solution for 3 d at room temperature. After decalcification, heads were washed with tap water and placed in 70% (v/v) EtOH. All specimens were dehydrated, and paraffin wax embedded. Sections (5 μm) were cut on a Leica rotary microtome RM2235 and mounted on slides (SuperFrost Plus, Menzel-Glaser; Thermo Fisher Scientific). Sections (n = 3) were deparaffinized, rehydrated, and stained with H&E. Staining and coverslipping was performed on a Leica workstation operated by the Histology Facility at The University of Manchester. Slides were scanned on a slide scanner (Pannoramic P250 Flash III), and images were processed using 3DHISTECH software. Gill tissue damage was quantified using a score adapted from Mitchell et al. (44) ranging from 0 to 3, where 0 indicates the absence of lamellar fusion, epithelial hyperplasia, or altered tissue architecture, 1 indicates <10% of gill tissue affected, 2 indicates 10–50% of gill tissue affected, and 3 indicates >50% of gill tissue affected.
RNA sequencing
Gills were dissected from 6-mo-old fish; RNA was isolated and subjected to sequencing analysis. Prior to performing RNA sequencing (RNA-seq), the integrity of RNA samples was assessed using a 2200 TapeStation (Agilent Technologies), according to the manufacturer’s instructions. Total RNA was submitted to the Genomic Technologies Core Facility of the University of Manchester, and libraries were generated using the TruSeq Stranded mRNA assay (Illumina), according to the manufacturer’s protocol. Adapter indices were used to multiplex libraries, which were pooled prior to cluster generation using a cBot instrument. The loaded flow cell was then paired-end sequenced (76 + 76 cycles, plus indices) on an Illumina HiSeq 4000 instrument. Finally, the output data were demultiplexed (allowing one mismatch) and BCL-to-Fastq conversion performed using Illumina’s bcl2fastq software (version 2.17.1.14). Unmapped paired-end sequences were tested by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Sequence adapters were removed, and reads were quality trimmed using Trimmomatic_0.36 (45). The reads were mapped against the reference zebrafish genome (danRer10), and counts per gene were calculated using the annotation from Ensembl (http://ftp.ensembl.org/pub/current_gtf/danio_rerio/Danio_rerio.GRCz10.84.gtf.gz) using STAR_2.5.3 (46). The RNA-seq datasets reported in this paper have been deposited in the ArrayExpress database (https://www.ebi.ac.uk/arrayexpress/; accession number E-MTAB-8958). Normalization, principal component analysis (PCA), and differential expression were identified using DESeq2_1.16.1 (47) as genes having a corrected p value <0.05 and log2 fold change >|1|. Venn diagrams were generated using Venny 2.1.0 (48) to identify the presence of common upregulated (log2 fold change > 1) and downregulated (log2 fold change < −1) genes in il4/13a−/−, il4/13b−/−, and il4/13a;b−/− gills.
Functional analysis of differentially expressed genes
Cluster analysis was performed using k-means clustering (Manhattan distance) using maxdView software. Each cluster was then hierarchically clustered. Clustering was performed on the means of each sample group (log 2) that had been z-transformed (for each gene, the mean set to zero, SD to one). Gene ontology analysis and pathway analysis were performed with Enrich (49) using human orthologs of zebrafish genes obtained from Ensembl genome browser version 99. Gene set enrichment analysis (GSEA) was performed using GSEA software (50). Several gene sets consisting of different immune cell markers, spanning innate and adaptive immune cell types, were generated using the human immunology panel from NanoString Technologies. Normalized enrichment scores (NES) were used to generate heatmaps with Morpheus software (https://software.broadinstitute.org/morpheus). xCell analysis was used to perform cell type enrichment analysis from gene expression data for 64 immune cell types (https://xcell.ucsf.edu).
R848 gill challenge
R848 gill challenge was performed as previously described (51) using 5 μl of R848 (0.5 mg/ml; InvivoGen) applied to the gills for 5 min. Following completion of the procedure, fish were returned to fresh system water and monitored for their recovery. Fish were then sacrificed with an overdose of MS222 (800 mg/l) at the indicated timepoint to harvest the gills for RNA extraction.
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 8 (GraphPad Software). The p values <0.05 (where necessary, corrected for multiple testing) were considered statistically significant. Throughout, error bars represent SEM.
Results
Generation of il4/13a and il4/13b zebrafish mutants
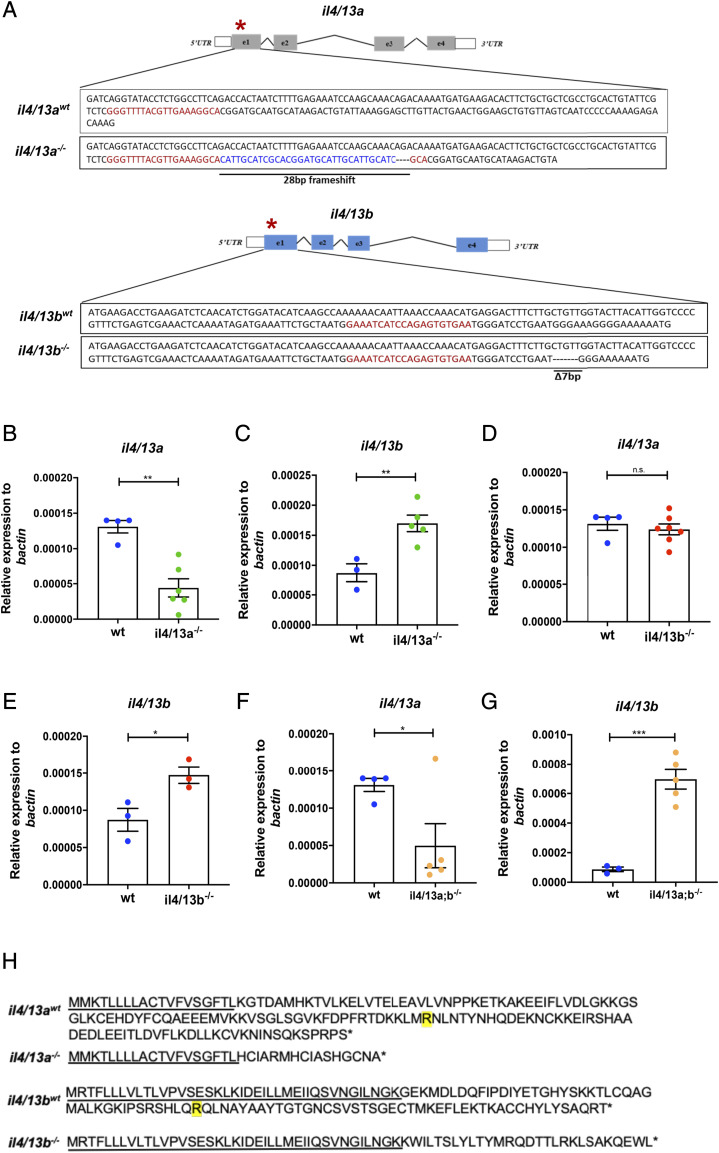
In zebrafish, two similar il4-like genes, namely il4/13a and il4/13b, have been described (33). To generate insight into the function of these two genes, we used CRISPR technology to create mutant lines lacking functional alleles. An il4/13a allele carrying a 28-bp frameshift mutation (32-bp insertion and 4-bp deletion) and an il4/13b allele carrying a 7-bp deletion were isolated (Fig. 1A). Homozygous mutants for both il4/13a (il4/13a−/−) and il4/13b (il4/13b−/−) were found to be viable, and they were intercrossed to generate il4/13a;b−/− double mutants. These were also found to be viable and fertile.
FIGURE 1.
Generation of il4/13a−/− and il4/13b−/− zebrafish mutants by CRISPR/Cas9. (A) Zebrafish il4/13a and il4/13b genes. (B–G) qPCR analysis showing the levels of il4/13a and il4/13b transcripts in wildtype, il4/13a−/−, il4/13b−/−, and il4/13a;b-/- 5 dpf larvae. Gene expression was normalized to the expression of β−actin. Error bars represent SEM; n > 3, *p < 0.05, **p < 0.01, ***p < 0.001. (H) Sequence of wildtype and mutated IL-4/13A and IL-4/13B proteins. Underlined is the unchanged portion of the protein sequence in mutant animals. Yellow-shaded “R” indicates the conserved arginine residue required in the human homologs for receptor binding. The asterisks indicate a stop codon.
To evaluate whether the mutations affected cognate mRNA levels, qPCR analysis was performed. A significant reduction of il4/13a mRNA in il4/13a−/− embryos was observed (Fig. 1B), suggesting mRNA degradation by the nonsense-mediated decay pathway (52). Perhaps as a consequence of nonsense-mediated decay, an increase of il4/13b mRNA (1.6-fold) was detected in il4/13a−/− embryos (Fig. 1C), suggesting a potential mechanism of genetic compensation, as recently described (53). No changes in the levels of il4/13a mRNA were observed in il4/13b−/− larvae (Fig. 1D). The expression of il4/13b mRNA was found increased in il4/13b−/− larvae (Fig. 1E), suggesting that the effects of the mutation might only be evident at the translational level and that increased mRNA might be an attempt to compensate for loss of function of the IL-4/13B protein. qPCR analysis also revealed that the expression of il4/13a was significantly reduced in il4/13a;b−/− embryos (Fig. 1F), whereas the expression of il4/13b was significantly increased (Fig. 1G), in agreement with what was observed in the single mutants.
The selected mutations were predicted to introduce a premature termination codon in both il4/13a and il4/13b genes, leading to the formation of severely truncated proteins. Indeed, only 16 and 28 aa remained in the truncated IL-4/13A and IL-4/13B proteins, respectively (Fig. 1H). Previous analysis of zebrafish protein sequences revealed conservation of an arginine in the αC helix (54), which is critical for the binding of human IL-4 to the IL-4Rα receptor. The mutations identified in IL-4/13A and IL-4/13B proteins abolished this residue, implying that the proteins should not be able to bind to their receptors and thus lack any functional activity.
Disruption of both il4/13a and il4/13b genes leads to a proinflammatory phenotype in larvae
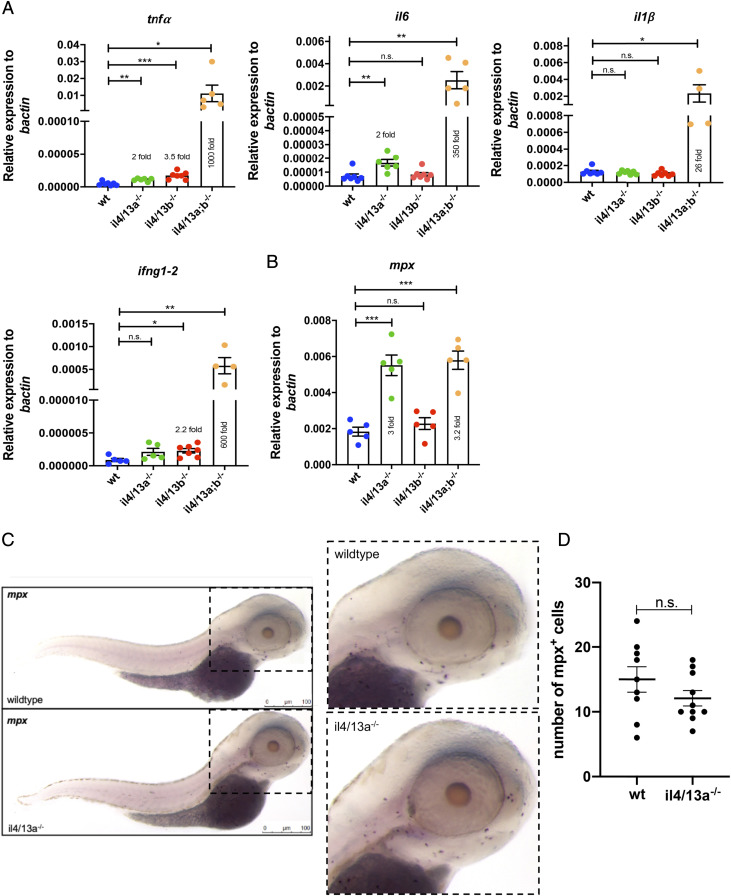
The functions of il4/13a and il4/13b genes were first investigated in larvae, which possess an innate immune system only (55). Whereas a modest increase of tnfα, il6, and ifng1–2 mRNA encoding proinflammatory cytokines was observed in il4/13a−/− and il4/13b−/− single mutants, more pronounced upregulation was observed in il4/13a;b−/− double mutants (Fig. 2A), indicating that il4/13a and il4/13b genes have a fundamental albeit redundant role in suppressing inflammation.
FIGURE 2.
Increased levels of proinflammatory cytokines in il4/13a;b−/− larvae. (A and B) qPCR analysis showing levels of il1β, il6, tnfα, ifng1–2, and mpx mRNA in wildtype, il4/13a−/−, il4/13b−/−, and il4/13a;b−/− 5 dpf larvae. Dots indicate individual fish. Gene expression was normalized to the expression of β−actin. Error bars represent SEM; n > 3, *p < 0.05, **p < 0.01, ***p < 0.001. (C) Representative image of mpx in situ hybridization in wildtype and il4/13a−/− 3 dpf larvae (n = 9–10). Purple dots indicate mpx+ cells. Scale bar, 100 μm. (D) Quantification of mpx+ cells in wildtype and il4/13a−/− larvae head region. Dots indicate individual fish.
Next, we investigated whether the inflammatory phenotype was accompanied by an increase of neutrophils by measuring the levels of myeloperoxidase mpx transcript in larvae. A significant upregulation of mpx was detected in il4/13a;b−/− double mutants (3.2-fold), supporting the inflammatory phenotype. A similar increase of mpx was observed in il4/13a−/− larvae (3-fold) but not in il4/13b−/− larvae (Fig. 2B), suggesting the increase in mpx transcript might be due to loss of functional IL-4/13A. Thus, in situ hybridization for mpx mRNA was performed on wildtype and il4/13a−/− larvae to further investigate changes in neutrophils. However, no difference in neutrophil numbers or distribution were detected in il4/13a−/− embryos (Fig. 2C, 2D).
Transcriptome analysis of the gills reveals a shift toward type 1 immunity in il4/13a;b−/− mutants
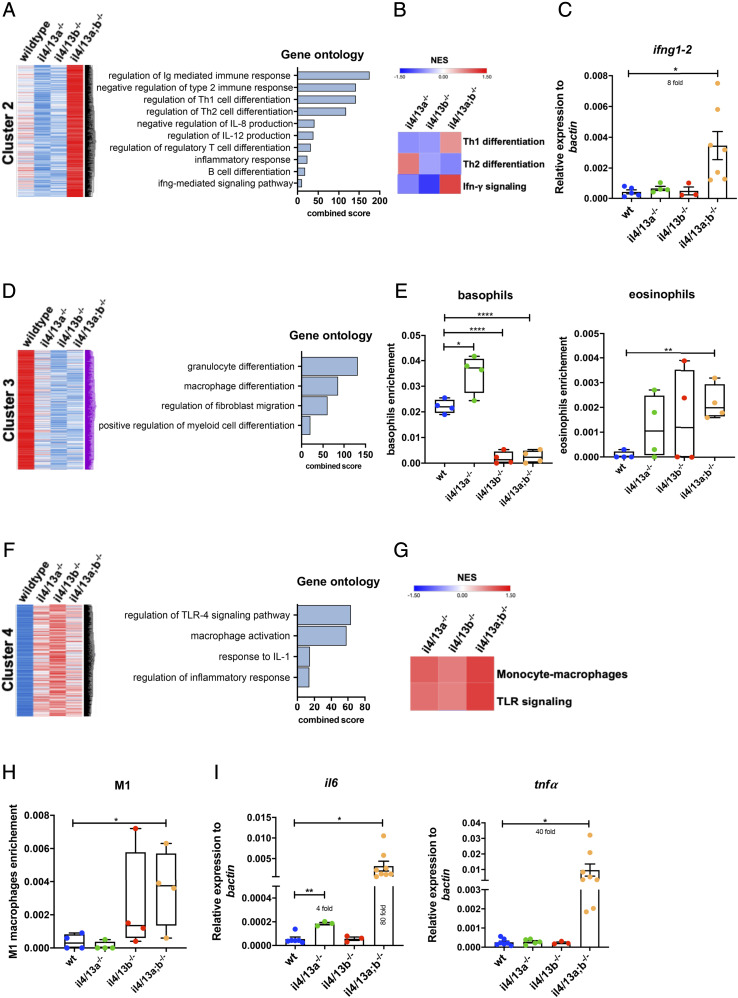
Our previous work demonstrated the expression of il4/13a in the gill tissue of adult zebrafish as well as il4/13b expression in gill-resident CD4+ T cells, suggesting the presence of a Th2-skewed environment in the gills (37). To investigate the gene expression profile of gills from il4/13a−/−, il4/13b−/−, and il4/13a;b−/− mutant animals, RNA-seq was performed on RNA isolated from the gills of adult fish. An overview of the RNA-seq analysis pipeline is shown in Supplemental Fig. 1A. PCA revealed clear segregation and clustering of mRNA profiles according to genotype (Supplemental Fig. 1B). Significantly differentially expressed genes were identified in the gills of mutant animals compared with wildtype (Supplemental Table I). Venn diagram analysis revealed numerous genes uniquely affected by IL-4/13a or IL-4/13b loss, fewer common targets affected by loss of either, and a significant number of redundant targets only affected upon loss of both cytokines (Supplemental Fig. 1C). To identify coordinated changes in gene expression within our samples, hierarchical clustering was performed. Differentially expressed genes were grouped in 10 clusters that were subjected to gene ontology analysis using Enrichr. Enriched gene ontologies were mainly associated with immune-related biological processes (Supplemental Fig. 2).
To characterize the transcriptional profile of il4/13a;b−/− double mutants, we focused on the genes belonging to cluster 2 that were more highly expressed in il4/13a;b−/− double mutants than any other genotype. Gene ontology analysis revealed that these genes were mainly associated with the negative regulation of type 2 immune responses, Th1 differentiation, and IFN-γ signaling (Fig. 3A). GSEA corroborated the inference that enhanced Th1 differentiation, diminished Th2 differentiation, and increased IFN-γ signaling was occurring in the gills of il4/13a;b−/− double mutants (Fig. 3B). Moreover, qPCR confirmed increased levels of ifng1–2 mRNA in the gills of il4/13a;b−/− double mutants (Fig. 3C). All together, these data suggest that in the absence of both il4/13a and il4/13b, there might be a shift toward type 1 immunity, comparable to the role of IL-4 in mammals.
FIGURE 3.
Transcriptome analysis of il4/13a−/−, il4/13b−/−, and il4/13a;b−/− gills. (A, D, and F) Clustering of differentially expressed genes identified in gills harvested from 6-mo-old fish. Blue indicates low expression, red indicates high expression, and white indicates unchanged expression. Bar graphs show significantly enriched ontologies for each cluster of genes. (B and G) Heatmap of NES for the indicated gene sets generated by GSEA in mutant gills compared with wildtype gills. (C and I) qPCR analysis showing transcript levels of ifng1–2, il6, and tnfa in wildtype and mutant gills. Gene expression was normalized to the expression of β−actin. Error bars represent SEM; n > 3, *p < 0.05, **p < 0.01, ****p < 0.0001. (E and H) Enrichment score of basophils, eosinophils, and M1 macrophages obtained by xCell analysis.
Next, we sought to investigate the gene signature of cluster 3 as these genes were downregulated in gills from il4/13a−/−, il4/13b−/−, and il4/13a;b−/− animals. Gene ontology analysis revealed that genes from cluster 3 were mainly associated with granulocyte differentiation, differentiation of myeloid cells, and regulation of fibroblast migration (Fig. 3D). To characterize potential enrichment of immune cell populations, xCell analysis was performed. Among granulocytes, basophils were predicted to be significantly reduced in gills from il4/13b−/− single mutant and il4/13a;b−/− double mutants, whereas an increase of eosinophils was predicted for il4/13a;b−/− double mutants at least (Fig. 3E).
Genes belonging to cluster 4 were found highly upregulated in gills from il4/13a−/−, il4/13b−/−, and il4/13a;b−/− animals, and they were found associated with processes such as TLR4 signaling, macrophage activation, response to IL-1, and regulation of the inflammatory response (Fig. 3F). Accordingly, an enrichment of monocytes–macrophage expression profile as well as TLR signaling signature was detected by GSEA (Fig. 3G). Moreover, xCell analysis predicted an increase of M1 macrophages in the gills of il4/13a;b−/− double mutants (Fig. 3H). To further validate these data, we measured the expression of proinflammatory cytokines in the gills by qPCR and found an increased expression of il6 (80-fold) and tnfα (40-fold) mRNA in the gills of il4/13a;b−/− double mutants (Fig. 3I), suggesting overall heightened inflammatory activity.
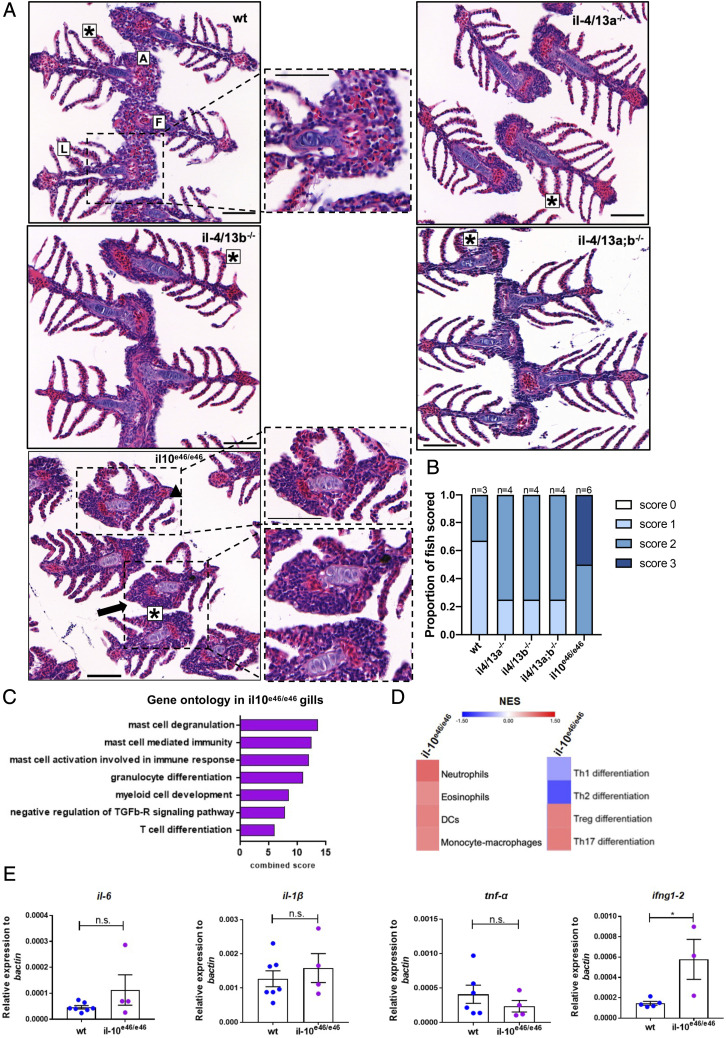
Loss of il4/13a and il4/13b does not affect gill morphology, whereas lack of functional il10 does
To further evaluate the effects of il4/13a and il4/13b on zebrafish gill homeostasis, histology was performed. No significant morphological changes were observed in the gills of il4/13a−/−, il4/13b−/−, or il4/13a;b−/− fish compared with wildtype (Fig. 4A), suggesting that loss of functional IL-4/13A and IL-4/13B does not affect tissue integrity despite a shift toward type 1 immunity. For comparison, we examined the gills of il10e46/e46 mutants lacking functional IL-10, which have also been reported to shift toward type 1 immune responses (56). A thickening of the gill arch and filaments, as well as lamellar epithelial changes, were detected in the absence of functional IL-10 (Fig. 4A, 4B), indicating a role for this cytokine in maintaining gill homeostasis in adult zebrafish.
FIGURE 4.
Mutation of il10 induces morphological changes in the gills of adult zebrafish. (A) Representative images of sections (n = 3) of gills from wildtype, il4/13a−/−, il4/13b−/−, il4/13a;b−/−, and il10e46/e46 animals stained with H&E. Gill arch (A), filaments (F), and lamellae (L) are indicated. Lamellar epithelial cell alteration (asterisks), filament thickening (triangle), and arch thickening (arrow) are indicated. (B) Quantification of gill tissue damage in wildtype, il4/13a−/−, il4/13b−/−, il4/13a;b−/−, and il10e46/e46. Widespread alteration is observed in the gills of il10e46/e46 animals. (C) Bar graph showing significantly enriched ontologies in gills of il10e46/e46 animals. (D) Heatmap of NES for indicated gene sets generated by GSEA in gills of il10e46/e46 compared with wildtype animals. (E) qPCR analysis showing transcript levels of il1β, il6, tnfα, and ifng1–2 in gills of il10e46/e46 compared with wildtype animals. Gene expression was normalized to the expression of β−actin. Error bars represent SEM; n > 3, *p < 0.05.
To further investigate the role of il10 in gill homeostasis, RNA-seq was performed on RNA extracted from the gills of il10e46/e46 mutant animals. Again, PCA confirmed a distinct expression profile for il10e46/e46 mutant animals (Supplemental Fig. 1B). Gene ontology analysis was performed using Enrichr, and an enrichment of immune-related ontologies, particularly related to mast cell and granulocyte differentiation, was observed (Fig. 4C). We also performed pathway analysis using Enrichr and found further evidence for an ongoing inflammatory response, highlighting the action of the complement system, chemokine and cytokine signaling, and other mediators of inflammation (Table IV). To investigate potential enrichment by immune cell populations, GSEA was performed. Enrichment of neutrophils, eosinophils, dendritic cells, and monocyte–macrophages was predicted in gills from il10e46/e46 mutant animals. Moreover, a depletion of Th1 and Th2 cells but an enrichment of regulatory T cells and Th17 was also predicted (Fig. 4D). To validate these data, we measured gene expression changes by qPCR analysis. No significant changes were observed in the levels of il1β, tnfα, and il6 mRNA encoding inflammatory cytokines but an increase of ifng1–2 mRNA was detected in il10e46/e46 gills compared with wildtype (Fig. 4E).
Table IV. Significantly enriched pathways in the gills of il10e46/e46 animals.
| il-10e46/e46 Gills | p Value | Combined Score |
|---|---|---|
| Hematopoietic cell lineage (Kyoto Encyclopedia of Genes and Genomes) | 0.009404725 | 21.44861305 |
| Cells and molecules involved in local acute inflammatory response (WP4493) | 0.038843362 | 11.08262125 |
| Human complement system (WP2806) | 0.002197324 | 5.694253785 |
| Inflammation mediated by chemokine and cytokine signaling pathway (Panther) | 0.024404585 | 5.291463582 |
| IL-23–mediated signaling events (National Cancer Institute) | 0.030898252 | 4.721254283 |
| Cell to cell adhesion signaling (BioCarta) | 0.020030828 | 3.088771356 |
Combined scores and p values are indicated.
Loss of IL-4/13a and IL-4/13b leads to an enhanced type 1 response to R848 in the gills of adult zebrafish
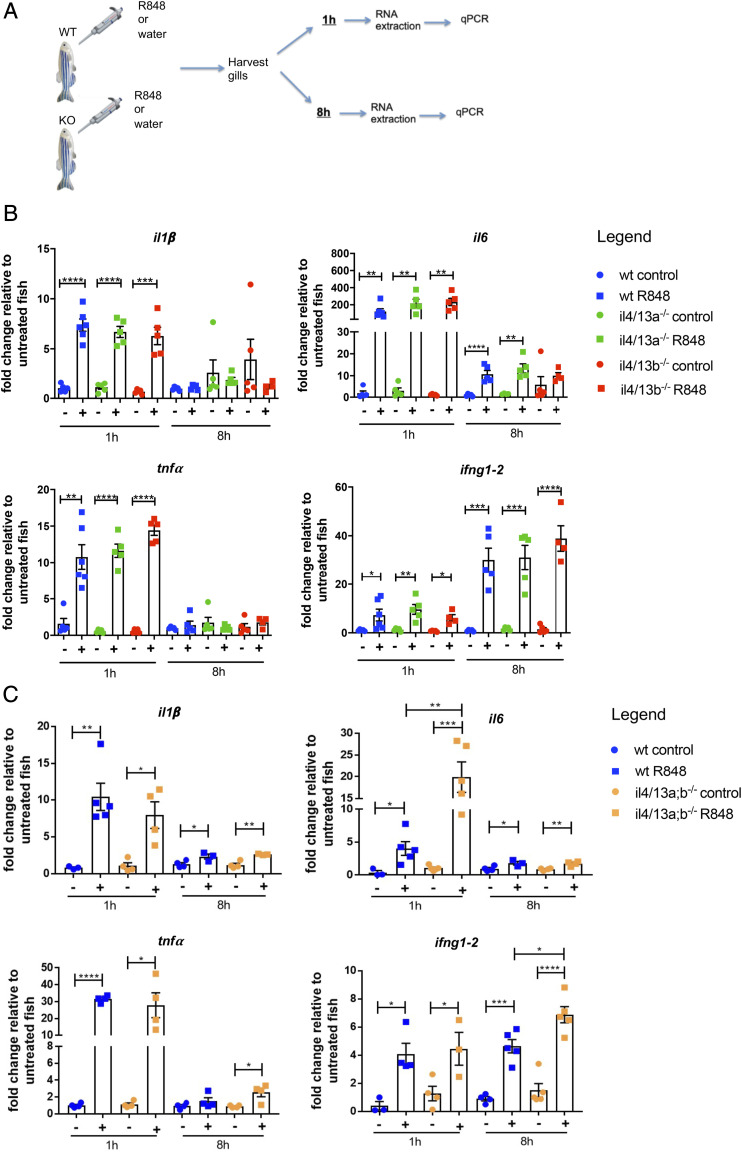
We next sought to investigate the importance of IL-4/13a and IL-4/13b in modulating an inflammatory response in the gills to an exogenous source of irritant. To induce gill inflammation, we used an intervention we have recently established, whereby a solution of R848, a synthetic compound that mimics viral ssRNA and interacts with TLR7/8 in mammals (57), is applied directly to the gills (51). Gene expression was analyzed by qPCR at 1 and 8 h to evaluate the kinetics of the response (Fig. 5A). An inflammatory response was observed in the gills of wildtype, il4/13a−/−, and il4/13b−/− animals after 1 h as shown by significantly enhanced levels of il1β, il6, tnfα, and ifng1–2 mRNA encoding inflammatory cytokines in R848-treated fish compared with untreated controls (Fig. 5B). However, no significant difference was observed between mutants and wildtype. Next, we investigated the later inflammatory response (8 h posttreatment). We observed a comparable downregulation of mRNA for the inflammatory cytokines il1β, il6, and tnfα in both wildtype and mutant fish compared with the 1-h timepoint. ifng1–2 displayed a different kinetics, being significantly upregulated in both wildtype and mutant fish still at 8 h (Fig. 5B). Again, this was comparable for wildtype and mutant fish. All together, these data suggest that neither loss of IL-4/13a nor loss of IL-4/13b impairs the response to R848-induced inflammation in the gills.
FIGURE 5.
R848 induced an enhanced type 1 response in the gills of il4/13a;b−/− double mutants. (A) Schematic overview of R848 gill challenge. (B and C) qPCR analysis showing transcript levels of il1β, il6, tnfα, and ifng1–2 in gills from wildtype, il4/13a−/−, il4/13b−/−, and il4/13a;b−/− animals following R848 challenge for 1 and 8 h. Gene expression was normalized to the expression of β−actin. Fold change is shown. Error bars represent SEM; n > 3, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Next, we evaluated the response to R848 in the absence of both IL-4/13a and IL-4/13b. R848 challenge was performed on the gills of il4/13a;b−/− double mutants for 1 and 8 h. Significantly enhanced levels of il1β, il6, tnfα, and ifng1–2 mRNA were observed in the gills of wildtype and il4/13a;b−/− animals after 1 h. Moreover, levels of il6 were significantly higher in il4/13a;b−/− compared with wildtype (Fig. 5C). At 8 h posttreatment, the levels of il1β, il6, and tnfα were reduced almost to baseline in both wildtype and double mutants. However, ifng1–2 levels were still elevated and again significantly higher in the gills of il4/13a;b−/− double mutants compared with wildtype (Fig. 5C), suggesting an enhanced type 1 response in agreement with the phenotype observed in the steady state.
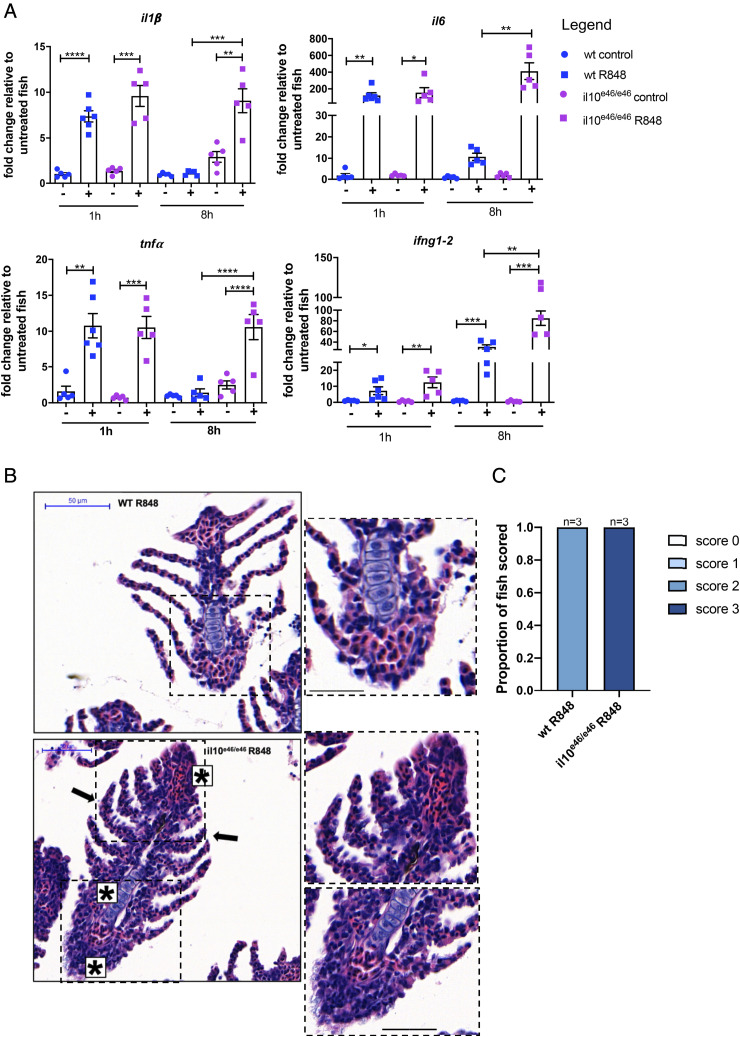
Loss of IL-10 leads to an enhanced inflammatory response to R848 in the gills of il10e46/e46 adult zebrafish
Loss of IL-10 resulted in marked alteration of gill morphology in the steady state as shown by the thickening of the gill arch and filaments as well as lamellar epithelial changes (Fig. 4A, 4B) and changes in gene expression indicative of smoldering inflammation (Fig. 4C–E, Table IV). We predicted that inflammatory responses to exogenous irritants would be exaggerated in the absence of IL-10. Therefore, the gills of il10e46/e46 fish were challenged with R848 for 1 and 8 h, as described before. Significant upregulation of il1β, il6, tnfα, and ifng1–2 mRNA were observed in the gills of stimulated wildtype and il10e46/e46 fish compared with untreated controls after 1 h. However, no difference was observed in the response between wildtype and mutant fish (Fig. 6A), suggesting that loss of IL-10 does not affect the early phase of an inflammatory response. Next, we evaluated the later response (8 h posttreatment), which revealed that compared with wildtype, il1β, il6, and tnfα transcripts were still significantly upregulated in gills from il10e46/e46 animals, and ifng1–2 transcript levels were even more pronounced (Fig. 6A), indicating a prolonged inflammatory response in the absence of il10. In keeping with the exaggerated inflammatory response at 8 h posttreatment, thickening of gill filaments and lamellae were observed in gills from R848-treated il10e46/e46 animals but not wildtype animals (Fig. 6B, 6C). All together, these data suggest that loss of the il10 gene induced an exaggerated inflammatory response in the gills following R848 stimulation.
FIGURE 6.
Prolonged inflammatory response in il10e46/e46 gills following R848 stimulation. (A) qPCR analysis showing transcript levels of il1β, il6, tnfα, and ifng1–2 in gills from wildtype and il10e46/e46 animals following R848 challenge for 1 and 8 h. Gene expression was normalized to the expression of β−actin. Fold change is shown. Error bars represent SEM; n > 3, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (B) Representative images of sections (n = 3) of gills from wildtype and il10e46/e46 animals (n = 3) stained with H&E following 8-h R848 challenge. Epithelial cell alteration in the lamellae (black arrows) and gill filament (stars) are indicated in gills from il10e46/e46 animals. Scale bar, 50 μm. (C) Quantification of gill tissue damage in wildtype and il10e46/e46 animals following R848 challenge.
Discussion
In recent years, the field of fish immunology has progressed considerably with many molecules and cell components of both innate and adaptive immune systems having been identified (20, 22, 58). However, our understanding of immune responses, especially in the gills that constitute important mucosal surfaces in fish, is still patchy. In particular, the identity and characteristics of the cells and signals that maintain homeostasis at this mucosal surface have not been fully defined.
In this study, we have used the zebrafish as a model organism to shed light on fish il4/13a and il4/13b and characterized the consequences of loss of function of these genes under both steady state and inflammatory conditions. Under controlled laboratory conditions, zebrafish lacking IL-4/13A and IL-4/13B were viable, fertile, and did not show any overt phenotype, indicating that such cytokines are not essential for survival and development in zebrafish. Similar findings are observed in IL-4−/− and IL-13−/− mice that are healthy and display no phenotypic abnormalities (59).
We found that disruption of both il4/13a and il4/13b induced a proinflammatory state in zebrafish larvae, indicating the role for IL-4/13A and IL-4/13B in suppressing inflammation. An anti-inflammatory role for fish type 2 cytokines has been previously demonstrated in trout in which the treatment of head kidney cells with rIL-4/13A and rIL-4/13B2 lead to the downregulation of proinflammatory cytokines (31), resembling the situation in mammals (11). Neutrophils are one of the main components of the zebrafish innate immune system and the first cells to respond to inflammation (60). We found significantly elevated levels of the neutrophil marker mpx in il4/13a;b−/− double mutants, supporting the inflammatory phenotype. The increase in mpx transcripts in the il4/13a−/− single mutants indicates that il4/13a may have a potential neutrophil-specific effect albeit one that does not drive a corresponding change in neutrophil number. Effects on mpx transcript were not observed in the il4/13b−/− mutant, suggesting a divergent function for these paralogs in neutrophils in zebrafish. In mammals, neutrophils express IL-4 type I receptor (61, 62), and recent evidence suggests that IL-4– and IL-13–mediated signaling in both mouse and human neutrophils inhibits their migration and effector functions (63, 64) to reduce inflammation and tissue damage during type 2 responses (65). To date, there is no definitive evidence for a role for fish IL-4/13 paralogs in controlling neutrophils. The generation of an il4/13a;b−/−;mpx:EGFP zebrafish line would be valuable to address neutrophil behavior in the absence of functional IL-4/13A and IL-4/13B.
Previously, we demonstrated that CD4-1+ Th2-like lymphocytes resident in the zebrafish gill mucosa were a source of il4/13b expression but not il4/13a that was detected in gill tissue (37). Recent work has described the presence of ILC-like cells in zebrafish gut, which express high levels of il4/13a following helminth Ag exposure, resembling mammalian ILC2 cells (66). In mammals, ILCs contribute to lung homeostasis as well as to lung diseases (67). ILCs might also be present in zebrafish gills, therefore, and be responsible for the expression of il4/13a. To shed light on the function of IL-4/13A and IL-4/13B in zebrafish gills, transcriptome analysis was performed in mutants for these cytokines. We found an enrichment of genes implicated in Th1 differentiation and IFN-γ signaling in the gills of il4/13a;b−/− double mutants, further supported by qPCR analysis. These data suggest that IL-4/13A and IL-4/13B are jointly required for the maintenance of a Th2-like phenotype in zebrafish gills, and their loss leads to an imbalance toward type 1 immunity. Evidence for IL-4/13 cytokines regulating type 2 immunity was previously reported in fish. In zebrafish, the interaction of IL-4/13A with IL-4Rα on B cell surfaces promoted B cell proliferation and Ab production (68); in trout, treatment of head kidney cells with rIL-4/13 proteins downregulated IFN-γ (31), and upregulation of il4/13 paralogs was observed in the gills of Atlantic salmon following parasite infection (69).
The transcriptome data predicted a reduction in basophils but an increase in eosinophils in the gills of il4/13a;b−/− double mutants. In mammals, basophils and eosinophils represent innate sources of IL-4 and IL-13, and their infiltration into tissues is one of the hallmarks of type 2 immune responses (70). Basophils have been described in teleosts (71), and a conserved role for eosinophils was found in response to helminth Ags and infection (72). It might be speculated that decreased basophils and increased eosinophils in the gills of il4/13a;b−/− double mutants is somehow connected, perhaps reflecting a divergent effect of IL-4/13 on the recruitment of these cell populations to gills. This is unanticipated from mammals in which basophils and eosinophils infiltrate tissues coordinately (73).
The increase in the fraction of M1 macrophages as well as enhanced levels of tnfα and il6 transcripts in the gills of il4/13a;b−/− double mutants indicate an inflammatory phenotype. Recent studies have identified subsets of polarized macrophages in zebrafish, describing a proinflammatory M1-like phenotype and anti-inflammatory M2-like phenotype, resembling the situation found in mammals (74). Our data suggest that IL-4/13A and IL-4/13B have a conserved function with mammalian IL-4 and IL-13 in driving macrophage polarization as found in other teleost fish (75–78). The increase of IFN-γ in the gills of il4/13a;b−/− double mutants might also contribute to the induction of M1 macrophages, as previously demonstrated (79), and to the inflammatory phenotype. All together, these findings indicate that loss of IL-4/13A and IL-4/13B leads to an inflammatory phenotype in the gills. This agrees with the phenotype observed in larvae, further supporting an anti-inflammatory function for zebrafish IL-4/13 paralogs.
Despite the induction of proinflammatory signals, loss of IL-4/13A and IL-4/13B did not affect tissue morphology, suggesting that there might be further regulatory mechanisms in place to avoid the disruption of homeostasis in the gills. IL-10 is known to control mammalian immune responses to avoid inflammation and maintain tissue homeostasis. We examined the gills of il10e46/e46 fish and found that IL-10 might be fundamental in maintaining homeostasis in the gills. Transcriptome analysis revealed evidence of smoldering inflammation in the gills of il10e46/e46 fish. In mammals, a lack of IL-10 leads to the development of spontaneous inflammation in the skin, intestine, and lungs (19). In this context, zebrafish IL-10 might have a similar function to its mammalian counterpart.
Recent work has described the use of R848 in inducing inflammation and cytokine response in zebrafish gills (51), highlighting similarities with mouse and human nasal mucosa. In mammals, R848 targets the TLR7 and TLR8 receptors (57), which in fish are mainly expressed in primary lymphoid organs and the gills (80, 81). R848 injection in Atlantic salmon induced IFN expression, whereas stimulation of salmon cell lines failed to induce IFN expression, consistent with the low expression of TLR8 and the lack of expression of TLR7 in these cells (81). These findings suggest that the R848 response in fish might also be mediated by the TLR7/TLR8 pathway, although definitive evidence is lacking. R848 induces a strong inflammatory and IFN response (51, 80). An IFN response is also observed following viral infections in several fish species (82–84), and upregulation of both TLR7 and TLR8 transcripts was detected in infected Atlantic salmon (85). In addition to the IFN response, R848 also induced changes in neutrophil and lymphocyte distribution in zebrafish gills (51). No changes in the distribution but a depletion of large neutrophil-like cells was observed following viral infection in Atlantic salmon (86). Similar to R848 stimulation, an upregulation of proinflammatory cytokines is also observed in response to parasitic infection in salmon and trout (87, 88). In contrast to R848 stimulation, parasitic and bacterial gill infections often lead to major histopathological changes in the tissue (89, 90). This difference might be explained by the acute nature of R848-induced inflammation, which causes only transient effects.
We used R848 to induce inflammation in the gills and found that loss of both IL-4/13A and IL-4/13B led to an enhanced response in the gills of double mutants, which appeared to be type 1 driven, in agreement with transcriptome data in the steady state. In mammals, IL-4 and IL-13 act together to ensure a successful Th2 inflammatory response, and a Th1 phenotype with increased expression of IFN-γ is observed in IL-4/IL-13–deficient mice (59). We infer from our data that in the absence of functional IL-4/13A and IL-4/13B, a Th1-like response dominates in zebrafish, indicating a similar function of these cytokines to their mammalian counterparts.
IL-10 appeared to be fundamental in the maintenance of gill homeostasis, and its loss enhanced the inflammatory response to R848 in the gills, supporting a potent anti-inflammatory function for this cytokine. An anti-inflammatory role for IL-10 has been previously shown in carp (91) and in zebrafish gut (92). Moreover, IL-10–mediated suppression of Th1 cell response and cytokine production was reported in a zebrafish Mycobacterium marinum model (56). For the first time, to our knowledge, we addressed the function of zebrafish IL-10 in the gills and demonstrated its importance in preventing inflammation in this mucosal tissue.
Previous studies indicate that fish IL-4/13 paralogs might have divergent functions, with IL-4/13A providing a basal level of type 2 immunity and IL-4/13B required for specific T cell–mediated immunity (31). Our findings indicate that zebrafish IL-4/13 paralogs have a redundant function in suppressing inflammation and maintaining a Th2 phenotype in the gills. Further investigation would be needed to discover unique functions for IL-4/13 cytokines in zebrafish immunity, and our RNA-seq data will be a useful resource in this respect. We propose that the il4/13a−/−, il4/13b−/−, and il4/13a;b−/− zebrafish mutants would be valuable models to investigate the function of il4/13 paralogs in the context of infection and disease. This would further our knowledge of fish immunology and provide comparative models to answer outstanding questions in mammals.
Supplementary Material
Acknowledgments
We thank our colleagues Peter Walker and Grace Bako (Histology Facility), Dr. Andy Hayes (Genomic Technologies), Peter March (Bioimaging), and the staff from the Biological Service Unit (all Faculty of Biology, Medicine and Health, The University of Manchester) for technical assistance. We also thank the Central Biomedical Services within Imperial College London for assistance. We thank Simon Johnston and Stephen Renshaw (University of Sheffield) for providing il10e46/e46 mutant zebrafish and mpx RNA probe, respectively.
M.J.D. and J.E.A. are cosenior authors.
This work was supported by Biotechnology and Biological Sciences Research Council Grant BB/L007401/1 (to A.F.L.H.). This work was funded by Wellcome Trust Ph.D. studentships 102171/Z/13/Z (to F.B.) and 109057/Z/15/Z (to M.W.) and Grant 106898/A/15/Z (work in the Allen laboratory) and Medical Research Council Grant MR/K01207X/2 (work in the Allen laboratory).
The online version of this article contains supplemental material.
- dpf
- day postfertilization
- gRNA
- guide RNA
- GSEA
- gene set enrichment analysis
- ILC
- innate lymphoid cell
- ILT
- interbranchial lymphoid tissue
- NES
- normalized enrichment score
- PCA
- principal component analysis
- qPCR
- quantitative PCR
- R848
- resiquimod
- RNA-seq
- RNA sequencing.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Junttila I. S. 2018. Tuning the cytokine responses: an update on interleukin (IL)-4 and IL-13 receptor complexes. Front. Immunol. 9: 888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen J. E., Maizels R. M. 2011. Diversity and dialogue in immunity to helminths. Nat. Rev. Immunol. 11: 375–388. [DOI] [PubMed] [Google Scholar]
- 3.Sharpe C., Thornton D. J., Grencis R. K. 2018. A sticky end for gastrointestinal helminths; the role of the mucus barrier. Parasite Immunol. 40: e12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen J. E., Sutherland T. E. 2014. Host protective roles of type 2 immunity: parasite killing and tissue repair, flip sides of the same coin. Semin. Immunol. 26: 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marichal T., Starkl P., Reber L. L., Kalesnikoff J., Oettgen H. C., Tsai M., Metz M., Galli S. J. 2013. A beneficial role for immunoglobulin E in host defense against honeybee venom. Immunity 39: 963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palm N. W., Rosenstein R. K., Medzhitov R. 2012. Allergic host defences. Nature 484: 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gieseck R. L., III, Wilson M. S., Wynn T. A. 2018. Type 2 immunity in tissue repair and fibrosis. Nat. Rev. Immunol. 18: 62–76. [DOI] [PubMed] [Google Scholar]
- 8.Lambrecht B. N., Hammad H., Fahy J. V. 2019. The cytokines of asthma. Immunity 50: 975–991. [DOI] [PubMed] [Google Scholar]
- 9.Neill D. R., Wong S. H., Bellosi A., Flynn R. J., Daly M., Langford T. K. A., Bucks C., Kane C. M., Fallon P. G., Pannell R., et al. 2010. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 464: 1367–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCormick S. M., Heller N. M. 2015. Commentary: IL-4 and IL-13 receptors and signaling. Cytokine 75: 38–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon S. 2003. Alternative activation of macrophages. Nat. Rev. Immunol. 3: 23–35. [DOI] [PubMed] [Google Scholar]
- 12.Kara E. E., Comerford I., Fenix K. A., Bastow C. R., Gregor C. E., McKenzie D. R., McColl S. R. 2014. Tailored immune responses: novel effector helper T cell subsets in protective immunity. PLoS Pathog. 10: e1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Prete G. 1992. Human Th1 and Th2 lymphocytes: their role in the pathophysiology of atopy. Allergy 47: 450–455. [DOI] [PubMed] [Google Scholar]
- 14.Cope A., Le Friec G., Cardone J., Kemper C. 2011. The Th1 life cycle: molecular control of IFN-γ to IL-10 switching. Trends Immunol. 32: 278–286. [DOI] [PubMed] [Google Scholar]
- 15.Saraiva M., O’Garra A. 2010. The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 10: 170–181. [DOI] [PubMed] [Google Scholar]
- 16.Barnes M. J., Powrie F. 2009. Regulatory T cells reinforce intestinal homeostasis. Immunity 31: 401–411. [DOI] [PubMed] [Google Scholar]
- 17.Kühn R., Löhler J., Rennick D., Rajewsky K., Müller W. 1993. Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75: 263–274. [DOI] [PubMed] [Google Scholar]
- 18.Shah N., Kammermeier J., Elawad M., Glocker E. O. 2012. Interleukin-10 and interleukin-10-receptor defects in inflammatory bowel disease. Curr. Allergy Asthma Rep. 12: 373–379. [DOI] [PubMed] [Google Scholar]
- 19.Rubtsov Y. P., Rasmussen J. P., Chi E. Y., Fontenot J., Castelli L., Ye X., Treuting P., Siewe L., Roers A., Henderson W. R., Jr, et al. 2008. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28: 546–558. [DOI] [PubMed] [Google Scholar]
- 20.Yamaguchi T., Takizawa F., Fischer U., Dijkstra J. M. 2015. Along the axis between type 1 and type 2 immunity; principles conserved in evolution from fish to mammals. Biology (Basel) 4: 814–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venkatesh B., Lee A. P., Ravi V., Maurya A. K., Lian M. M., Swann J. B., Ohta Y., Flajnik M. F., Sutoh Y., Kasahara M., et al. 2014. Elephant shark genome provides unique insights into gnathostome evolution. [Published erratum appears in 2014 Nature 513: 574.] Nature 505: 174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomez D., Sunyer J. O., Salinas I. 2013. The mucosal immune system of fish: the evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol. 35: 1729–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campos-Perez J. J., Ward M., Grabowski P. S., Ellis A. E., Secombes C. J. 2000. The gills are an important site of iNOS expression in rainbow trout Oncorhynchus mykiss after challenge with the gram-positive pathogen Renibacterium salmoninarum. Immunology 99: 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson J. M., Laurent P. 2002. Fish gill morphology: inside out. J. Exp. Zool. 293: 192–213. [DOI] [PubMed] [Google Scholar]
- 25.Koppang E. O., Kvellestad A., Fischer U. 2015. Fish mucosal immunity: gill. In Mucosal Health in Aquaculture. Beck B. H., Peatman E., eds. Elsevier Inc., Amsterdam, p. 93–133. [Google Scholar]
- 26.Salinas I., Zhang Y. A., Sunyer J. O. 2011. Mucosal immunoglobulins and B cells of teleost fish. Dev. Comp. Immunol. 35: 1346–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haugarvoll E., Bjerkås I., Nowak B. F., Hordvik I., Koppang E. O. 2008. Identification and characterization of a novel intraepithelial lymphoid tissue in the gills of Atlantic salmon. J. Anat. 213: 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koppang E. O., Fischer U., Moore L., Tranulis M. A., Dijkstra J. M., Köllner B., Aune L., Jirillo E., Hordvik I. 2010. Salmonid T cells assemble in the thymus, spleen and in novel interbranchial lymphoid tissue. J. Anat. 217: 728–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aas I. B., Austbø L., Falk K., Hordvik I., Koppang E. O. 2017. The interbranchial lymphoid tissue likely contributes to immune tolerance and defense in the gills of Atlantic salmon. Dev. Comp. Immunol. 76: 247–254. [DOI] [PubMed] [Google Scholar]
- 30.Ringø E., Holzapfel W. 2000. Identification and characterization of carnobacteria associated with the gills of Atlantic salmon (Salmo salar L.). Syst. Appl. Microbiol. 23: 523–527. [DOI] [PubMed] [Google Scholar]
- 31.Wang T., Johansson P., Abós B., Holt A., Tafalla C., Jiang Y., Wang A., Xu Q., Qi Z., Huang W., et al. 2016. First in-depth analysis of the novel Th2-type cytokines in salmonid fish reveals distinct patterns of expression and modulation but overlapping bioactivities. Oncotarget 7: 10917–10946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitra S., Alnabulsi A., Secombes C. J., Bird S. 2010. Identification and characterization of the transcription factors involved in T-cell development, t-bet, stat6 and foxp3, within the zebrafish, Danio rerio. FEBS J. 277: 128–147. [DOI] [PubMed] [Google Scholar]
- 33.Ohtani M., Hayashi N., Hashimoto K., Nakanishi T., Dijkstra J. M. 2008. Comprehensive clarification of two paralogous interleukin 4/13 loci in teleost fish. Immunogenetics 60: 383–397. [DOI] [PubMed] [Google Scholar]
- 34.Avery S., Rothwell L., Degen W. D. J., Schijns V. E. J. C., Young J., Kaufman J., Kaiser P. 2004. Characterization of the first nonmammalian T2 cytokine gene cluster: the cluster contains functional single-copy genes for IL-3, IL-4, IL-13, and GM-CSF, a gene for IL-5 that appears to be a pseudogene, and a gene encoding another cytokinelike transcript, KK34. J. Interferon Cytokine Res. 24: 600–610. [DOI] [PubMed] [Google Scholar]
- 35.Tindemans I., Serafini N., Di Santo J. P., Hendriks R. W. 2014. GATA-3 function in innate and adaptive immunity. Immunity 41: 191–206. [DOI] [PubMed] [Google Scholar]
- 36.Takizawa F., Koppang E. O., Ohtani M., Nakanishi T., Hashimoto K., Fischer U., Dijkstra J. M. 2011. Constitutive high expression of interleukin-4/13A and GATA-3 in gill and skin of salmonid fishes suggests that these tissues form Th2-skewed immune environments. Mol. Immunol. 48: 1360–1368. [DOI] [PubMed] [Google Scholar]
- 37.Dee C. T., Nagaraju R. T., Athanasiadis E. I., Gray C., Fernandez Del Ama L., Johnston S. A., Secombes C. J., Cvejic A., Hurlstone A. F. L. 2016. CD4-transgenic zebrafish reveal tissue-resident Th2- and regulatory T cell-like populations and diverse mononuclear phagocytes. J. Immunol. 197: 3520–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zou J., Clark M. S., Secombes C. J. 2003. Characterisation, expression and promoter analysis of an interleukin 10 homologue in the puffer fish, Fugu rubripes. Immunogenetics 55: 325–335. [DOI] [PubMed] [Google Scholar]
- 39.Savan R., Igawa D., Sakai M. 2003. Cloning, characterization and expression analysis of interleukin-10 from the common carp, Cyprinus carpio L. Eur. J. Biochem. 270: 4647–4654. [DOI] [PubMed] [Google Scholar]
- 40.Inoue Y., Kamota S., Ito K., Yoshiura Y., Ototake M., Moritomo T., Nakanishi T. 2005. Molecular cloning and expression analysis of rainbow trout (Oncorhynchus mykiss) interleukin-10 cDNAs. Fish Shellfish Immunol. 18: 335–344. [DOI] [PubMed] [Google Scholar]
- 41.Zhang D. C., Shao Y. Q., Huang Y. Q., Jiang S. G. 2005. Cloning, characterization and expression analysis of interleukin-10 from the zebrafish (Danio rerion). J. Biochem. Mol. Biol. 38: 571–576. [DOI] [PubMed] [Google Scholar]
- 42.Kettleborough R. N. W., Busch-Nentwich E. M., Harvey S. A., Dooley C. M., de Bruijn E., van Eeden F., Sealy I., White R. J., Herd C., Nijman I. J., et al. 2013. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature 496: 494–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thisse C., Thisse B. 2008. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3: 59–69. [DOI] [PubMed] [Google Scholar]
- 44.Mitchell S. O., Baxter E. J., Holland C., Rodger H. D. 2012. Development of a novel histopathological gill scoring protocol for assessment of gill health during a longitudinal study in marine-farmed Atlantic salmon (Salmo salar). Aquacult. Int. 20: 813–825. [Google Scholar]
- 45.Bolger A. M., Lohse M., Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Love M. I., Huber W., Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oliveros J. C. 2007. VENNY. An interactive tool for comparing lists with Venn diagrams. Available at: https://bioinfogp.cnb.csic.es/tools/venny/index.html. Accessed: April 21, 2020.
- 49.Kuleshov M. V., Jones M. R., Rouillard A. D., Fernandez N. F., Duan Q., Wang Z., Koplev S., Jenkins S. L., Jagodnik K. M., Lachmann A., et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44: W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Progatzky F., Jha A., Wane M., Thwaites R. S., Makris S., Shattock R. J., Johansson C., Openshaw P. J., Bugeon L., Hansel T. T., Dallman M. J. 2019. Induction of innate cytokine responses by respiratory mucosal challenge with R848 in zebrafish, mice, and humans. J. Allergy Clin. Immunol. 144: 342–345.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagy E., Maquat L. E. 1998. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 23: 198–199. [DOI] [PubMed] [Google Scholar]
- 53.El-Brolosy M. A., Kontarakis Z., Rossi A., Kuenne C., Günther S., Fukuda N., Kikhi K., Boezio G. L. M., Takacs C. M., Lai S. L., et al. 2019. Genetic compensation triggered by mutant mRNA degradation. Nature 568: 193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stocchi V., Wang T., Randelli E., Mazzini M., Gerdol M., Pallavicini A., Secombes C. J., Scapigliati G., Buonocore F. 2017. Evolution of Th2 responses: characterization of IL-4/13 in sea bass (Dicentrarchus labrax L.) and studies of expression and biological activity. Sci. Rep. 7: 2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herbomel P., Thisse B., Thisse C. 2001. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev. Biol. 238: 274–288. [DOI] [PubMed] [Google Scholar]
- 56.Harjula S.-K. E., Ojanen M. J. T., Taavitsainen S., Nykter M., Rämet M. 2018. Interleukin 10 mutant zebrafish have an enhanced interferon gamma response and improved survival against a Mycobacterium marinum infection. Sci. Rep. 8: 10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hemmi H., Kaisho T., Takeuchi O., Sato S., Sanjo H., Hoshino K., Horiuchi T., Tomizawa H., Takeda K., Akira S. 2002. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat. Immunol. 3: 196–200. [DOI] [PubMed] [Google Scholar]
- 58.Wang T., Secombes C. J. 2013. The cytokine networks of adaptive immunity in fish. Fish Shellfish Immunol. 35: 1703–1718. [DOI] [PubMed] [Google Scholar]
- 59.McKenzie G. J., Fallon P. G., Emson C. L., Grencis R. K., McKenzie A. N. J. 1999. Simultaneous disruption of interleukin (IL)-4 and IL-13 defines individual roles in T helper cell type 2-mediated responses. J. Exp. Med. 189: 1565–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Novoa B., Figueras A. 2012. Zebrafish: model for the study of inflammation and the innate immune response to infectious diseases. Adv. Exp. Med. Biol. 946: 253–275. [DOI] [PubMed] [Google Scholar]
- 61.Girard D., Paquin R., Beaulieu A. D. 1997. Responsiveness of human neutrophils to interleukin-4: induction of cytoskeletal rearrangements, de novo protein synthesis and delay of apoptosis. Biochem. J. 325: 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ratthé C., Ennaciri J., Garcês Gonçalves D. M., Chiasson S., Girard D. 2009. Interleukin (IL)-4 induces leukocyte infiltration in vivo by an indirect mechanism. Mediators Inflamm. 2009: 193970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Egholm C., Heeb L. E. M., Impellizzieri D., Boyman O. 2019. The regulatory effects of interleukin-4 receptor signaling on neutrophils in type 2 immune responses. Front. Immunol. 10: 2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heeb L. E. M., Egholm C., Impellizzieri D., Ridder F., Boyman O. 2018. Regulation of neutrophils in type 2 immune responses. Curr. Opin. Immunol. 54: 115–122. [DOI] [PubMed] [Google Scholar]
- 65.Heeb L. E. M., Egholm C., Boyman O. 2020. Evolution and function of interleukin-4 receptor signaling in adaptive immunity and neutrophils. Genes Immun. 21: 143–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hernández P. P., Strzelecka P. M., Athanasiadis E. I., Hall D., Robalo A. F., Collins C. M., Boudinot P., Levraud J. P., Cvejic A. 2018. Single-cell transcriptional analysis reveals ILC-like cells in zebrafish. Sci. Immunol. 3: eaau5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Borger J. G., Lau M., Hibbs M. L. 2019. The influence of innate lymphoid cells and unconventional T cells in chronic inflammatory lung disease. Front. Immunol. 10: 1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu L. Y., Pan P. P., Fang W., Shao J. Z., Xiang L. X. 2012. Essential role of IL-4 and IL-4Rα interaction in adaptive immunity of zebrafish: insight into the origin of Th2-like regulatory mechanism in ancient vertebrates. J. Immunol. 188: 5571–5584. [DOI] [PubMed] [Google Scholar]
- 69.Marcos-López M., Calduch-Giner J. A., Mirimin L., MacCarthy E., Rodger H. D., O’Connor I., Sitjà-Bobadilla A., Pérez-Sánchez J., Piazzon M. C. 2018. Gene expression analysis of Atlantic salmon gills reveals mucin 5 and interleukin 4/13 as key molecules during amoebic gill disease. Sci. Rep. 8: 13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Voehringer D., Reese T. A., Huang X., Shinkai K., Locksley R. M. 2006. Type 2 immunity is controlled by IL-4/IL-13 expression in hematopoietic non-eosinophil cells of the innate immune system. [Published erratum appears in 2006 J. Exp. Med. 203: 1617.] J. Exp. Med. 203: 1435–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Odaka T., Suetake H., Maeda T., Miyadai T. 2018. Teleost basophils have IgM-dependent and dual Ig-independent degranulation systems. J. Immunol. 200: 2767–2776. [DOI] [PubMed] [Google Scholar]
- 72.Balla K. M., Lugo-Villarino G., Spitsbergen J. M., Stachura D. L., Hu Y., Bañuelos K., Romo-Fewell O., Aroian R. V., Traver D. 2010. Eosinophils in the zebrafish: prospective isolation, characterization, and eosinophilia induction by helminth determinants. Blood 116: 3944–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Voehringer D., Shinkai K., Locksley R. M. 2004. Type 2 immunity reflects orchestrated recruitment of cells committed to IL-4 production. Immunity 20: 267–277. [DOI] [PubMed] [Google Scholar]
- 74.Nguyen-Chi M., Laplace-Builhe B., Travnickova J., Luz-Crawford P., Tejedor G., Phan Q. T., Duroux-Richard I., Levraud J.-P., Kissa K., Lutfalla G., et al. 2015. Identification of polarized macrophage subsets in zebrafish. eLife 4: e07288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jiang X., Wang J., Wan S., Xue Y., Sun Z., Cheng X., Gao Q., Zou J. 2020. Distinct expression profiles and overlapping functions of IL-4/13A and IL-4/13B in grass carp (Ctenopharyngodon idella). Aquacult. Fish. 5: 72–79. [Google Scholar]
- 76.Wiegertjes G. F., Wentzel A. S., Spaink H. P., Elks P. M., Fink I. R. 2016. Polarization of immune responses in fish: the ‘macrophages first’ point of view. Mol. Immunol. 69: 146–156. [DOI] [PubMed] [Google Scholar]
- 77.Hodgkinson J. W., Fibke C., Belosevic M. 2017. Recombinant IL-4/13A and IL-4/13B induce arginase activity and down-regulate nitric oxide response of primary goldfish (Carassius auratus L.) macrophages. Dev. Comp. Immunol. 67: 377–384. [DOI] [PubMed] [Google Scholar]
- 78.Joerink M., Ribeiro C. M. S., Stet R. J. M., Hermsen T., Savelkoul H. F. J., Wiegertjes G. F. 2006. Head kidney-derived macrophages of common carp (Cyprinus carpio L.) show plasticity and functional polarization upon differential stimulation. J. Immunol. 177: 61–69. [DOI] [PubMed] [Google Scholar]
- 79.Zou J., Carrington A., Collet B., Dijkstra J. M., Yoshiura Y., Bols N., Secombes C. 2005. Identification and bioactivities of IFN-γ in rainbow trout Oncorhynchus mykiss: the first Th1-type cytokine characterized functionally in fish. J. Immunol. 175: 2484–2494. [DOI] [PubMed] [Google Scholar]
- 80.Purcell M. K., Smith K. D., Hood L., Winton J. R., Roach J. C. 2006. Conservation of toll-like receptor signaling pathways in teleost fish. Comp. Biochem. Physiol. Part D Genomics Proteomics 1: 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Svingerud T., Solstad T., Sun B., Nyrud M. L. J., Kileng Ø., Greiner-Tollersrud L., Robertsen B. 2012. Atlantic salmon type I IFN subtypes show differences in antiviral activity and cell-dependent expression: evidence for high IFNb/IFNc-producing cells in fish lymphoid tissues. J. Immunol. 189: 5912–5923. [DOI] [PubMed] [Google Scholar]
- 82.Landis E. D., Purcell M. K., Thorgaard G. H., Wheeler P. A., Hansen J. D. 2008. Transcriptional profiling of MHC class I genes in rainbow trout infected with infectious hematopoietic necrosis virus. Mol. Immunol. 45: 1646–1657. [DOI] [PubMed] [Google Scholar]
- 83.Grove S., Austbø L., Hodneland K., Frost P., Løvoll M., McLoughlin M., Thim H. L., Braaen S., König M., Syed M., et al. 2013. Immune parameters correlating with reduced susceptibility to pancreas disease in experimentally challenged Atlantic salmon (Salmo salar). Fish Shellfish Immunol. 34: 789–798. [DOI] [PubMed] [Google Scholar]
- 84.McBeath A. J. A., Ho Y. M., Aamelfot M., Hall M., Christiansen D. H., Markussen T., Falk K., Matejusova I. 2014. Low virulent infectious salmon anaemia virus (ISAV) replicates and initiates the immune response earlier than a highly virulent virus in Atlantic salmon gills. Vet. Res. (Faisalabad) 45: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moore L. J., Jarungsriapisit J., Nilsen T. O., Stefansson S., Taranger G. L., Secombes C. J., Morton H. C., Patel S. 2017. Immune gene profiles in Atlantic salmon (salmo salar L.) post-smolts infected with SAV3 by bath-challenge show a delayed response and lower levels of gene transcription compared to injected fish. Fish Shellfish Immunol. 62: 320–331. [DOI] [PubMed] [Google Scholar]
- 86.Rønneseth A., Pettersen E. F., Wergeland H. I. 2006. Neutrophils and B-cells in blood and head kidney of Atlantic salmon (Salmo salar L.) challenged with infectious pancreatic necrosis virus (IPNV). Fish Shellfish Immunol. 20: 610–620. [DOI] [PubMed] [Google Scholar]
- 87.Olsen M. M., Kania P. W., Heinecke R. D., Skjoedt K., Rasmussen K. J., Buchmann K. 2011. Cellular and humoral factors involved in the response of rainbow trout gills to Ichthyophthirius multifiliis infections: molecular and immunohistochemical studies. Fish Shellfish Immunol. 30: 859–869. [DOI] [PubMed] [Google Scholar]
- 88.Bridle A. R., Morrison R. N., Cupit Cunningham P. M., Nowak B. F. 2006. Quantitation of immune response gene expression and cellular localisation of interleukin-1β mRNA in Atlantic salmon, Salmo salar L., affected by amoebic gill disease (AGD). Vet. Immunol. Immunopathol. 114: 121–134. [DOI] [PubMed] [Google Scholar]
- 89.Speare D. J., Markham R. J. F., Despres B., Whitman K., MacNair N. 1995. Examination of gills from salmonids with bacterial gill disease using monoclonal antibody probes for Flavobacterium branchiophilum and Cytophaga columnaris. J. Vet. Diagn. Invest. 7: 500–505. [DOI] [PubMed] [Google Scholar]
- 90.Speare D. J., Ferguson H. W. 1989. Clinical and pathological features of common gill diseases of cultured salmonids in Ontario. Can. Vet. J. 30: 882–887. [PMC free article] [PubMed] [Google Scholar]
- 91.Piazzon M. C., Savelkoul H. S. J., Pietretti D., Wiegertjes G. F., Forlenza M. 2015. Carp Il10 has anti-inflammatory activities on phagocytes, promotes proliferation of memory T cells, and regulates B cell differentiation and antibody secretion. J. Immunol. 194: 187–199. [DOI] [PubMed] [Google Scholar]
- 92.Coronado M., Solis C. J., Hernandez P. P., Feijóo C. G. 2019. Soybean meal-induced intestinal inflammation in zebrafish is T cell-dependent and has a Th17 cytokine profile. Front. Immunol. 10: 610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.