Abstract
Introduction:
Cellular senescence is a rapidly growing field with potential relevance for the treatment of multiple human diseases. In the last decade, cellular senescence and the senescence-associated secretory phenotype (SASP) have emerged as central drivers of aging and many chronic diseases, including cancer, neurodegeneration, heart disease and osteoarthritis. Major efforts are underway to develop drugs that selectively eliminate senescent cells (senolytics) or suppress or alter the SASP (senomorphics) to treat age-related diseases in humans. The translation of senescence-targeting therapies into humans is still in early stages. Nonetheless, it is clear that proteomic approaches will facilitate the discovery of important SASP proteins, development of senescence- and SASP-derived biomarkers, and identification of therapeutic targets for senolytic and senomorphic drugs.
Areas covered:
We review recent proteomic studies of cellular senescence and their translational relevance and, particularly, characterization of the secretory phenotype and preclinical development of biomarkers (from 2008–2020, PubMed). We focus on emerging areas, such as the heterogeneity of senescent cells and the SASP, extracellular vesicles released by senescent cells, and validating biomarkers of aging in vivo.
Expert opinion:
Proteomic and multi-omic approaches will be important for the development of senescence-based biomarkers to facilitate and monitor future therapeutic interventions that target senescent cells.
Keywords: aging, biomarkers, clinical proteomics, data-independent acquisition, plasma, secretome, senescence, senescence-associated secretory phenotype, senolytics, senomorphics
1. Introduction
The development of clinical proteomic biomarkers is an emerging and fast-growing field in human biomedical research. Recently, the focus has been developing senescence-based biomarkers of aging, frailty and age-related diseases [1, 2, 3]. Over the last decade, proteomic studies of human plasma and other biofluids have made significant progress in accurately quantifying proteins and potential biomarkers at increased depth and coverage [4, 5, 6, 7, 8]. Relatively novel mass spectrometric workflows and comprehensive acquisition strategies, such as data-independent acquisition (DIA) [9, 10], are now frequently utilized to discover and develop clinical biomarkers [11, 12]. One of the most promising areas for these emerging technologies is therapies that target a fundamental aging process known as cellular senescence.
Cellular senescence is widely accepted as a basic driver of aging and age-related diseases. In this complex stress response, cells permanently lose the ability to proliferate and alter distal tissues through systemic and local paracrine effects [13, 14, 15]. Cellular senescence can be triggered by stressors, including genotoxic agents, nutrient deprivation, hypoxia, mitochondrial dysfunction and oncogene activation [13, 16, 17, 18]. Although senescent cells irreversibly arrest growth, they remain metabolically active and secrete many biologically active molecules, known as the senescence-associated secretory phenotype (SASP) [19, 20]. The SASP initiates inflammation, wound healing and growth responses in nearby cells [21, 22, 23]. With age, the number of senescent cells or “senescence burden” increases, and this increased senescence burden and chronic SASP drive many pathological hallmarks of aging [24, 25, 26].
Cellular senescence is an example of antagonistic pleiotropy -- a trait that is beneficial early in life but detrimental later in life [27]. In healthy tissues, the SASP is typically transient and contributes to tissue homeostasis, including tissue repair [23] and regeneration [28], fine-tuning embryogenesis [29, 30] and parturition [31]. In contrast, the chronic presence of senescent cells and a SASP is associated with multiple age-related diseases, including Alzheimer’s and Parkinson’s disease [32], atherosclerosis [33], cardiovascular dysfunction [33], cancer metastasis and recurrence [34], cataracts [35], diabetes [36], metabolic syndrome [37], pulmonary fibrosis [38], osteoarthritis [39], osteoporosis [40], and sarcopenia/frailty [41], among others. Importantly, genetic ablation of senescent cells in reduces inflammation in aged or damaged tissues; ultimately, increasing median life span (health span) by postponing or ameliorating age-related pathologies [32, 33, 38, 40, 42, 43, 44]. Eliminating senescent cells and the SASP is considered a highly promising therapeutic strategy for preventing or treating age-associated diseases and extending health span. To selectively kill senescent cells non-genetically, drugs known as ‘senolytics’ are being developed; additionally, drugs termed senomorphics or senostatics are being developed to mitigate the detrimental effects of senescent cells by modifying the SASP [45, 46].
To develop clinical therapeutics that target senescent cells, it is critical to have reliable biomarkers to measure the senescent cell burden in humans both to identify patients with an elevated burden and to track the efficacy of the therapeutics. In this review, we will discuss proteomic strategies to discover senescence-derived biomarkers and their great potential for measuring the senescent cell burden.
Senescent cells secrete many molecules, and the resulting SASP consists of a complex mixture of both proteins, metabolites, and other molecules. However, thorough investigation is required to determine which SASP protein factors or protein panels qualify as biomarkers to quantitatively assess the senescent cell burden, and subsequently which SASP factors can be used efficiently and accurately as a biomarker for aging and age-related diseases. Alternatively, metabolites could be used as potential biomarkers of senescence, as senescent cells feature i) significantly transformed metabolic activity, resulting in dramatic alterations in tricarboxylic acid cycle metabolites, and (ii) impaired mitochondrial function, as exemplified by decreased membrane potential and reduced fission and fusion rates [47, 48]. In fact, the extracellular metabolome of senescent cells is highly altered and dynamic [49], suggesting a ‘metabolomic SASP’ is another promising area of senescence biology and biomarker discovery. Many non-protein molecules, such as DNA and RNA components (secreted in extracellular vesicles) [50], lipids, and other metabolites [49, 51] are potentially secreted by senescent cells and represent a highly promising yet understudied area of research. In this review, however, our discussion will focus on protein biomarkers of senescence and aging, and how to discover and use them in pre-clinical settings. In particular, we will discuss proteomic tools and targeted assays that can aid the development of senolytic and senomorphic therapies in humans.
2. Proteomics of the Senescence-Associated Secretory Phenotype
Translating senescence- and SASP-targeting interventions into humans will require a comprehensive profile of the SASP to identify the deleterious components of the SASP and develop human biomarkers to assess the senescent cell burden in humans. Our review of proteomic studies of the SASP spans the period between the initial characterization of the SASP in 2008 until now. The literature was reviewed for proteomic studies of senescent cells, focusing on mass spectrometry-based and antibody-based studies, using resources such as PubMed as well as data repositories, including MassIVE, ProteomeXchange, and PRIDE.
2.1. Early proteomic characterization of the SASP
The originally identified SASP comprised 40–100 chemokines, growth factors and proteases that were identified by biased or targeted methods (e.g., antibody arrays) and/or transcriptional analyses [19, 20, 52, 53]. Today, more comprehensive and unbiased technologies have identified hundreds of additional SASP factors, including proteins, lipids and other metabolites (Table 1).
Table 1:
Proteomic Studies of Senescent Cells
| Method | Cell type | Cellular compartment | Senescence inducer | Reference |
|---|---|---|---|---|
| Label-free MS | Human bladder cancer cells | PM | p21-induced, p16-induced | [139] |
| Human bone marrow and adipose mesenchymal stromal cells | SASP | GSIS, RS | [66] | |
| Human breast cancer cells | EVs | GSIS | [106] | |
| Human retinal pigment epithelial cells | EVs | GSIS | [108] | |
| Human fetal lung fibroblasts | PM | GSIS | [140] | |
| Label-free quantitative MS | Human fetal lung fibroblasts, proximal tubule epithelial cells | SASP and EVs | GSIS, OIS, atazanavir treatment | [3] |
| Human foreskin fibroblasts | EVs | OIS and GSIS | [64] | |
| SILAC | Human fibroblasts | SASP | GSIS | [65] |
| Human fetal lung fibroblasts | PM | OIS | [141] | |
| Human fetal lung fibroblasts | SASP | OIS | [15] | |
| Antibody-array- based proteomics | Human fibroblasts, renal epithelial cells, epithelial tumor cells | SASP | GSIS, RS | [20] |
| MEF and primary adult mammary gland fibroblasts | SASP | GSIS | [52] | |
| Human fetal lung fibroblasts | SASP | OIS | [19] | |
| Mouse embryonic fibroblasts | SASP | GSIS | [22] |
Abbreviations: EVs = extracellular vesicles; GSIS = genotoxic stress induced senescence; IC = intracellular; MS = mass spectrometry; OIS = oncogene induced senescence; PM = plasma membrane; RS = replicative senescence; SASP = senescence-associated secretory phenotype; SILAC = stable isotope labeling with amino acids in cell culture
Early proteomic studies, such as antibody arrays, quantified about 100 proteins secreted into the conditioned media of cultured senescent cells. For example, an early study applied cytokine antibody arrays to identify a few dozen SASP proteins, many of which were induced by multiple stimuli, such as replicative exhaustion, ionizing radiation (10 Gy) and RAS-oncogene expression [20]. These SASP factors were mostly conserved in multiple human cell types, including fetal lung fibroblasts, neonatal foreskin fibroblasts, adult breast fibroblasts, prostate epithelial cells and prostate tumor cells. Further, two factors, interleukins IL-6 and IL-8, were the main drivers of invasive cancer phenotypes and epithelial-to-mesenchymal transition in cultured non-invasive breast cancer cells [20]. Numerous studies have confirmed the expression or secretion of originally identified SASP factors, including cytokines (IL-6, IL-8) [15, 19, 54], growth factors (GROA, IGFBPs) [23, 55], and proteases or protease inhibitors (MMPs,TIMPs) [56, 57, 58, 59, 60, 61] in cultured cells and in vivo. Importantly, under standard culture conditions of 20% oxygen, the SASP is not conserved between mouse and human cells, but is conserved when cells are cultured at more physiological (3%) oxygen levels [52]. After oncogene-induced senescence (OIS), lung fibroblasts secrete CXCR2-binding proteins, including CXCL1 (GROα), CXCL2 (GROβ), CXCL3 (GROγ), CXCL5 (ENA78), CXCL6 (GCP2), NAP2 and IL-8 (CXCL8) [19]. Further, senescence correlated with the activation of CXCR2, inhibition of CXCR2, CXCL1 or IL-8 alleviated the OIS growth arrest, and CXCR2 expression was elevated in pre-malignant lesions in vivo [19], suggesting that CXCR2 signaling reinforces senescent phenotypes.
While senescent cells and the SASP drive many age-related pathologies and phenotypes, such as cancer invasiveness, paracrine senescence and inflammation [62], senescence also has important beneficial roles in development [30], wound healing [55] and cellular reprogramming [22]. Distinguishing between SASP factors needed for these beneficial effects and those that drive pathology will be important in developing therapies that target senescent cells and the SASP. The distinction between the beneficial and detrimental SASP is not fully understood. Senescent cells and their secreted factors are critical for cellular reprogramming by the Yamanaka factors OCT4, SOX2, KLF4, and cMYC (OSKM), which reprogram differentiated cells into induced pluripotent stem cells (iPSCs) both in culture and in vivo [22]. Among the factors secreted by senescent mouse embryonic fibroblasts is IL-6, which is required for efficient OSKM-driven reprogramming. It will be valuable to apply proteomic approaches to identify SASP components that promote ‘beneficial’ phenotypes, such as wound healing or cellular reprogramming. For example, by performing cell culture screening and testing the ability of fractionated portions of the SASP to induce wound healing, followed by identification of components in the active fractions with unbiased proteomic analysis. Similarly, SASP fractions can be interrogated for detrimental effects in cell culture when incubated with healthy cells.
2.2. Large-scale mass spectrometry-based proteomic studies of the SASP
More advanced proteomic approaches based on unbiased mass spectrometry (MS) have expanded the SASP by about 10-fold (Table 1). The complexity of the SASP, often monitored by the secretion of a few proteins, has been greatly underappreciated, and a small set of factors cannot explain the diverse phenotypes the SASP produces in vivo. This complexity is due, in part, to the dependence of the SASP on senescence-inducing stimuli, cell type, and temporal dynamics of the senescent phenotype [63]. In addition, senescent cells secrete biologically active molecules in extracellular vesicles (EV SASP) [50, 64]. To completely understand the dynamic and context-dependence of the soluble and EV SASP and potential senescence-based biomarkers, MS-based proteomics will be critical to create comprehensive protein profiles. Several studies have begun to apply these approaches to identify new SASP factors and potential biomarkers of senescence and aging [3, 15, 65, 66].
Identifying the factors that mediate the induction and propagation of senescent cells via paracrine senescence is important for developing therapeutics that reduce senescent cells and to identify aging and disease biomarkers. One study of paracrine senescence in healthy fibroblasts utilized forward and reverse ‘Stable Isotope Labeling with Amino acids in Cell culture’ (SILAC) experiments to quantitatively measure the secretomes of OIS (using oncogenic RAS) of human fetal lung fibroblasts compare to non-senescent controls. They identified 127 differentially secreted proteins, with TGF-β family ligands, VEGF family proteins, and chemokines among the top hits [15]. Based on these targets, a drug screen identified chemical compounds that partially mitigated the growth arrest caused by paracrine senescence, induced by the conditioned medium of senescent fibroblasts. Of the compounds that mitigated growth inhibition due to paracrine senescence were several top hits from the proteomic screen, including VEGFR2/FLT3, TGFBR1, ALK4, ALK5, ALK7 and CCR2 receptor targeting compounds. These results suggest that multiple factors secreted by RAS oncogene-induced senescent fibroblasts mediate paracrine senescence. These secreted mediators are potential pro-aging factors that drive pathological phenotypes and may be biomarkers of aging or senescence-burden.
Recently, a SILAC study showed that senescent cells secrete bioactive protein factors that alter hemostasis and drive blood clotting [65]. From this unbiased SILAC MS analysis performed in senescent human lung fibroblasts after genotoxic stress (X-irradiation) in culture, dozens of novel SASP factors were identified. Notably, the SASP contained 44 proteins that participated in hemostasis or blood clotting, including SERPINs (SERPINE1 & 2, SERPINF1 & 2, SERPINB2, SERPINH1, SERPING1) and thrombospondins (THBS1 & 2). This study [65] used a transgenic mouse model that enabled the selective elimination of senescent cells (p16–3MR mice [23]) to test if senescent cells alter hemostasis. When senescent cells were increased in mice with a single intraperitoneal dose (10 mg/kg) of the chemotherapeutic agent doxorubicin [34], blood clotting increased; conversely, when senescent cells were selectively removed, the blood clotting response returned to normal. These results suggest that senescent cells secrete factors into plasma at concentrations sufficient to promote clotting, and, presumably, other phenotypes. Since they enter the circulation, SASP factors hold potential as plasma biomarkers for aging and age-related diseases that are driven by senescent cells. In addition, since senescent cells drive the clotting-related side-effects of anti-cancer therapies, understanding the SASP is important for patients exposed to genotoxic chemotherapies. Furthermore, targeting senescent cells or the SASP may be a clinical strategy to mitigate the side effects of certain chemotherapeutic drugs and, presumably, other drugs that drive cells into senescence, such as HIV protease inhibitors [67, 68].
The SASP is heterogeneous and dynamic, so it will be essential to profile the SASP caused by multiple inducers, cell types, time points, and other variable to ensure robust generalized senescence biomarkers. Several recent studies have begun this process by performing unbiased MS analysis on senescent cells induced by different stimuli, cell types, and, to some extent, at various intervals after senescence induction [3, 66]. Ozcan et al. performed a label-free analysis of senescent secretomes from bone marrow- or adipose-derived mesenchymal stromal cells after inducing senescence by doxorubicin (1 μM), X-irradiation, hydrogen peroxide or replicative exhaustion [66]. By examining proteins that were identified in the majority (9/10) of the senescent secretomes, they found that most SASP proteins fell into several categories: extracellular matrix and cytoskeletal proteins (MMP2, TIMP2, SERPINE1, FLNB, TUBA1C, TUBB), redox factors (PRDX6, PARK7, ERP46), gene expression regulators (MVP, YWHAE, PSMB4, IGFBP3), and miscellaneous (CTSD, CD13).
Our group recently applied data-independent acquisitions (DIA) to comprehensively and quantitatively profile the SASP of lung fibroblasts after X-irradiation, RAS oncogene-overexpression and treatment with the HIV protease inhibitor atazanavir, as well as X-irradiated renal epithelial cells [3]. We identified a “core” fibroblast SASP composed of 150 proteins, regardless of the originating stimulus. Within the core SASP, 39 proteins (24% of total) were also identified as plasma biomarkers of aging in a recent study [69]. Among the proteins we found were most highly secreted by senescent cells, GDF15, STC1, SERPINE1, and MMP1 were also significantly elevated in human plasma with age. The presence of SERPINE1 (PAI-1), MMP1, CXCL1, and GDF15 confirmed several previous proteomic analyses of the SASP, including in other cell types and inducers [3, 15, 19, 20, 65, 66]. In addition to being identified a plasma aging biomarker’, several core soluble SASP biomarkers, have been identified as human disease biomarkers. For example, human cohort studies reported growth/differentiation factor 15 (GDF15) as a biomarker of cardiovascular disease, cardiovascular and cancer mortality and morbidity, renal disease, and all-cause mortality independent of cardiovascular mortality [70, 71, 72, 73, 74, 75, 76], as well as a driver of senescence-associated colon cancer metastasis [77]. In addition, MMP1 is a biomarker for several cancers, pulmonary fibrosis and potentially Alzheimer’s disease [78, 79, 80, 81], whereas STC1 is a diagnostic and prognostic biomarker for cancers, pulmonary fibrosis, renal ischemia/reperfusion injury and Alzheimer’s disease [82, 83, 84, 85, 86, 87].
In parallel with our study of SASP-based biomarkers, we released the “SASP Atlas”, a comprehensive resource for senescence-associated secretory phenotypes [3]. The SASP Atlas (www.SASPAtlas.com) is a curated and freely available database of secretomes of senescent cells, including the soluble and EV SASPs that can be used to identify SASP components or biomarker candidates for senescence burden, aging and related diseases [3]. Although a subset of the SASP is annotated as “core”, the SASP protein profiles are mostly distinct among cell types, senescence-inducing stimuli and intervals after induction. In vivo, it is unknown which “types” of SASPs are primary contributors to aging or primary sources of plasma biomarkers. Thus, biomarkers in human patients will likely vary, depending on the affected tissue, originating cell type, and senescence stimulus. For example, based on multi-omic analysis, we proposed an inducer-specific biomarker signature of senescent lung fibroblasts originating from OIS that comprises the five proteins (CXCL5, MMP9, MMP3, CST4, and CCL3) that were robustly increased in the secretomes and transcriptomes of RAS-induced senescent fibroblasts in multiple omic studies [3, 88, 89]. Additional comprehensive quantitative profiles of the SASP and expansion of the SASP Atlas profiles under a variety of physiological conditions will provide biomarker candidates with a higher degree of selectivity to specific pathologies in humans.
Among both antibody array studies and mass spectrometry studies, a SASP core is beginning to emerge reproducibly across multiple studies, multiple inducers, and multiple cells types. Among the core SASP factors that were originally discovered and reproducibly reported from antibody array studies are inflammatory cytokines (IL-6, IL-8, IL-1α, IL-1β, CXCL1), growth factors (IGFBPs 2, −3, −4, −5, −6, −7), proteases and protease-regulators (MMP1, MMP2, TIMP1, TIMP2). Several of the inflammatory core SASP factors identified in early antibody array studies, such as IL-6, IL-8, CXCL1, have been the focus of numerous SASP-related studies [15, 19, 20, 54]. However, new unbiased mass spectrometry approaches have identified and highlighted either novel or understudied factors that are robustly secreted by senescent cells in multiple contexts. GDF15 is a protein of emerging interest with links to a plethora of age-related diseases and aging [70, 71, 72, 73, 74, 75, 76], which has been identified in SASP from multiple senescence inducers and cell types by both antibody-based and mass spectrometry studies [3, 20, 52]. Few studies have only recently focused on GDF15 in the SASP [77, 90], and its role in senescence biology remains largely unknown. Stanniocalcin-1 (STC1) is a core SASP component reported using multiple inducers, cell types, and across multiple -omic studies [3, 88, 89], however, STC1 that is clearly unstudied in the context of senescence, despite its relevance in multiple age-related diseases. Other ‘core’ SASP proteins identified across multiple inducers, cell types, and large-scale proteomic studies include MMP2, MMP1, TIMP2, THBS1, and SERPINE1 [3, 65, 66].
3. Proteomics of Extracellular Vesicles Released from Senescent Cells
Extracellular vesicles (EVs) are small, lipid-bilayer enclosed, cell-derived particles. They are released from cells and can be carried or transported to target cells where they can fuse with plasma membranes, thereby releasing cargo into target cells and facilitating cell-cell communication [91, 92]. EVs are broadly classified into three major sub-populations, based on their size and biogenesis [93]: i) apoptotic bodies, >1000 nm in diameter, ii) micro vesicles, 100–1000 nm in diameter, and ii) exosomes, 30–150 nm in diameter. EV production and release are regulated processes that can vary with physiological and pathologic conditions, and external stimuli can drastically change the EV-production rate and composition of EV cargo [94]. EVs in bodily fluids could thus be monitoring tools for disease processes [95, 96]. The properties of EVs, including their roles in intracellular communication and as vehicles for molecular exchange, make them the objects of intense interest and research to unravel a novel disease biomarkers with potentially unparalleled precision and specificity for tissues and sites of disease [95]. Evidence is now mounting that senescent cells release increased levels of biologically active factors within EVs that promote aging-related diseases, such as cancer, osteoarthritis and immunosenescence [97]. It will be critical to profile the proteomes of EV SASPs, comprehensively and in multiple contexts (i.e., multiple tissue types and senescence inducing stimuli) to identify the molecules that drive the biological activities of the EV SASP and to develop novel sets of senescence-based disease biomarkers.
Major challenges in EV research are their isolation and analysis of clinical-grade EVs [98, 99]. For example, biological fluids contain complex protein mixtures that can complicate the isolation of EVs. Current isolation methods typically rely on size differences among the different types of EVs and other biological particles, or utilize EV-specific surface markers to isolate pure samples [100, 101]. These methods vary greatly in the purity of the final sample and their compatibility with downstream proteomic applications. Regardless of the methods of exosome isolation used (a predominant workflow seems to be ultracentrifugation), the purity of isolated exosomes should be confirmed by multiple independent quality control assessments, including the presence of exosome-specific markers (CD63, CD9, CD81, ALIX, TSG101, and others) [93], size distribution analysis (by nanoparticle tracking analysis or microscopy), and the absence of contaminating proteins. In the case of plasma exosome isolation, abundant plasma proteins, such as albumin and IGG can be used to assess contamination.
Interestingly, the SASP soluble factors and small extracellular vesicles can transmit paracrine senescence to nearby cells [15, 64, 102]. EVs released by senescent cells show pro-tumorigenic effects [103, 104] and are associated with osteoarthritis [39, 105]. In future studies, it will be important to distinguish between effects that arise from the soluble SASP and the EV SASP.
To date, few studies have performed large-scale or targeted identification or quantification of senescent EV proteins or other cargo [3, 64, 104, 105, 106, 107]. Some studies examined the senescent EV proteomes in cancer cells or after cancer treatment. For example, senescence-associated exosome release was first described in human prostate cancer cells in which radiation induced senescence, and increased release of exosomes containing CD276 (B7H3), a known prostate cancer biomarker, and large amounts of small RNA [104]. Chemotherapy-induced senescent triple negative breast cancer cells (Cal51) secrete exosomes containing increased levels of 142 proteins, including ATPases, annexins, integrins and insoluble SASP factors [106], determined by label-free liquid chromatography (LC)-MS/MS. Interestingly, these drug-resistant breast cancer cells defend against chemotherapy drugs via exosome secretion [106]. As in the tumor-promoting soluble SASP [20], the EV SASP from doxorubicin-induced (200 ng/mL for 4 – 10 days) senescent human retinal pigment epithelial cells promotes cancer cell proliferation via EphA2 [108], and about 100 other proteins enriched in senescent EVs. Interestingly, one study investigated senolytic treatment on the proteome and RNA content of EVs in a mouse model of arthritis [105]. The senolytic drug UB0101 reduced the severity of the disease and the number of senescence-associated EVs in synovial fluid [105]. Additionally, the senescence-associated EVs of diseased synovial fluid are dramatically different in protein and microRNA composition [39].
Two recent studies used unbiased profiling to examine the EV proteomes of senescent cells [3, 64]. One study found that EVs secreted by senescent human foreskin fibroblasts induced paracrine senescence in healthy cells. Hundreds of proteins were enriched in exosomes secreted by OIS and DNA damage–induced senescence in human foreskin fibroblasts [64]. One protein, Interferon Induced Transmembrane Protein 3 (IFITM3), within senescent EVs, partially mediated paracrine senescence. Our group also recently performed quantitative proteomic analysis of EVs secreted by OIS and irradiation-induced senescent lung fibroblasts [3]. Interestingly, we found that the protein signatures were highly specific to each senescence inducer, suggesting that the EV protein signature may distinguish among subtypes of senescent cells. In both proteomic EV studies, the protein composition and changes in the EV SASP were distinct from the proteins and changes in the soluble SASP, suggesting both can offer a novel set of biomarkers.
4. Conclusion
Plasma and other biofluids (e.g., urine, synovial fluid, cerebral spinal fluid) are routinely used to detect biomarkers and offer the possibility to develop proteomic biomarkers to quantify the senescence burden. In general, cell secretomes are ideally suited as sources for disease biomarker candidates [109]. The SASP and released EVs are particularly well suited as a source of biomarker candidates for aging and age-related diseases owing to the senescence burden. Overall, proteins secreted into biofluids by senescent cells are important as mediators of aging-associated diseases and are a promising source of biomarkers.
5. Expert Opinion
The translation of senescence-targeting therapies into clinical applications is promising but still in its infancy, and many opportunities remain to be explored to advance these interventions. The field will greatly benefit from applied proteomics and multi-omic technologies for improved and clinically relevant understanding of the underlying biology of senescent cells, identifying biomarkers of senescent cell burden, and developing senolytic and senomorphic therapies.
Overall, extensive SASP profiles will be critical in three key areas that will drive the translation of senescence-targeted therapies into humans: i) uncovering the SASP factors that cause biological changes, both adaptive and pathological, associated with senescent cells, ii) identifying biomarkers of senescent cell burden, and iii) developing senomorphic therapies targeted at specific SASP factors or modules. Unbiased and comprehensive proteomic profiling of the SASP is an important first step in characterizing the SASP. We now know that the phenotypes of senescent cells, including the SASP, are dynamic and heterogeneous, depending on their originating tissue, senescence-inducing stimulus, and the passage of time [3, 15, 20, 66]. Also, the proteomes of extracellular vesicles released by senescent cells or the EV SASP [64] add another layer of complexity to the SASP. Defining the SASP in all these dimensions is a huge low-hanging fruit opportunity to apply proteomics to senescence research. We anticipate numerous large-scale proteomic studies of the SASP in multiple cell types, originating stimuli, timepoints and disease models. Central databases of the soluble and EV SASPs, such as the SASP Atlas [3], will be paramount for exploring the role of SASPs and their value as biomarkers in multiple disease states.
5.1. Identifying key SASP proteins and developing clinical biomarkers
Establishing comprehensive SASP profiles is a key early goal in our proposed senescence biomarker discovery pipeline (Figure 1). Establishing catalogs of disease-, tissue-, and inducer-specific SASP factors, as well as robust core SASP factors (secreted by senescent cells in multiple contexts/tissues), will be a foundation for developing specific and precise biomarkers. In the second phase, verification will be key to determine the SASP factors in human biofluids by proteomic approaches. We and others identified several SASP factors that are robust aging or disease biomarkers in human plasma, including GDF15, STC1, MMP1 and SERPINE1 [3, 65, 69]. It is important to establish that these factors are detectable in plasma (or other biofluids) and can be measured reproducibly. The dynamic range of proteins in plasma remains an ongoing challenge for the detection and quantification of low-abundance proteins as biomarkers in mass spectrometry. Targeted approaches such as SRM/PRM and unbiased DIA-based approaches are nonetheless ever-improving methods for the detection and quantitative assessment of plasma biomarkers with ever improving sensitivity and coverage of the plasma proteome. Modern unbiased MS approaches, such as DIA, are particularly useful for this type of discovery, verification and qualification because they are unbiased, quantitative and comprehensive when compared with proteomic arrays, while maintaining several advantages of targeted MS approaches when compared with data-dependent acquisition (DDA) approaches. In recent years, DIA methods have significantly improved in standardization and have been tested in multi-lab large-scale biomarker assessments [9]. Additionally, the most sensitive, reproducible, and quantifiable peptides and transitions identified by DIA can be used to develop further high-sensitivity and high-throughput targeted MS assays [8], such as selected reaction monitoring or parallel reaction monitoring assays [12]. Therefore, DIA is excellent for qualification and verification of reproducible and quantitative peptides for the validation phase. For the absolute quantification of plasma biomarkers, synthetic heavy peptide standards corresponding to peptide biomarkers (for which SRM/PRM/DIA assays have been developed) can be spiked into each sample prior to MS analysis. Absolute quantification approaches are more commonly applied to a small number of targets and become challenging and cost-prohibitive to perform on larger panels of biomarkers, but the multiplexing capabilities of absolute quantification approaches are improving with new instrumentation and emerging methods such as triggered by Offset, Multiplexed, Accurate mass, High resolution, and Absolute Quantitation (TOMAHAQ) [110].
Figure 1: Pipeline for the development of senescence-based biomarkers of aging and disease.
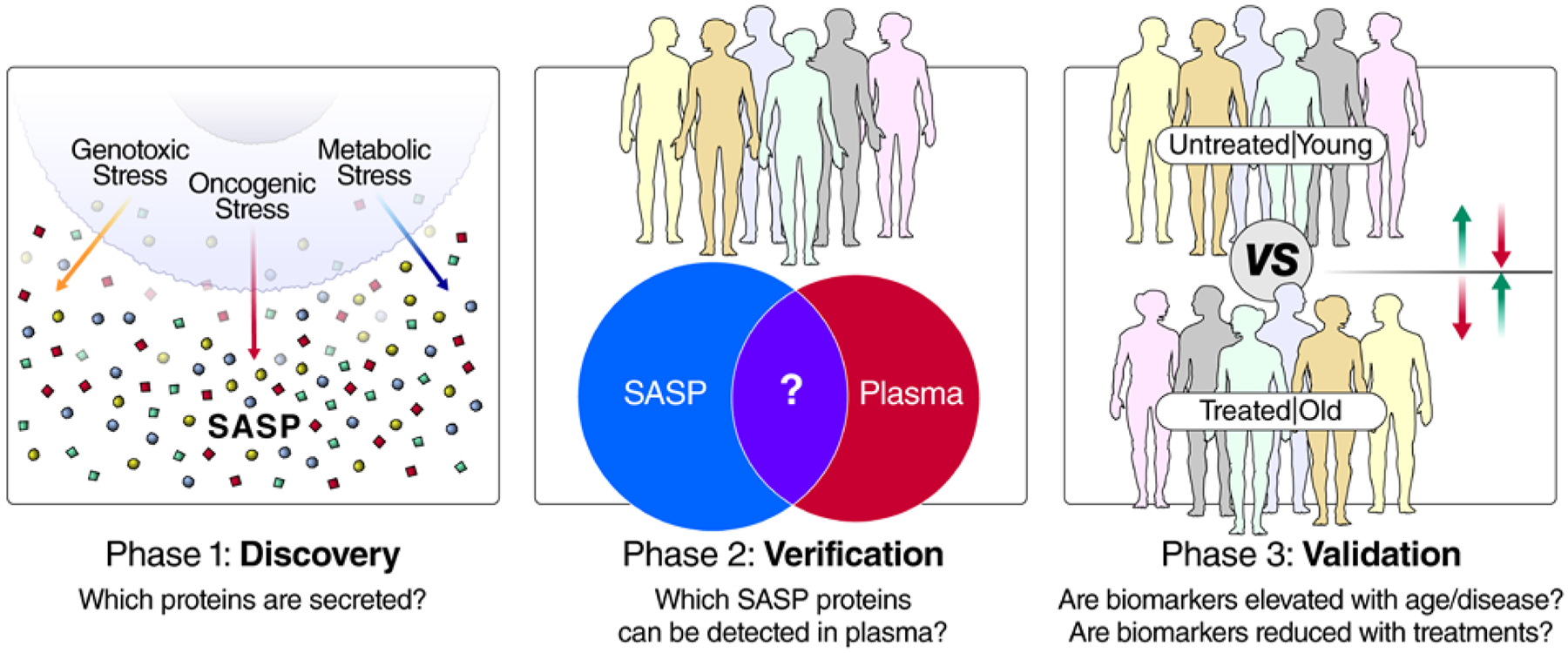
In phase 1, discovery, the secretomes (SASP) of senescent cells are characterized in cell-culture systems to generate biomarker candidates that can be tailored to specific biological contexts, based on the disease and cell type. Multiple cell types and senescence-inducing stimuli (e.g., genotoxic/oncogenic/metabolic stress) can be interrogated, and the most robust biomarkers of all conditions used for later stages of biomarker development. In step 2, verification, biomarker candidates are filtered, based on whether they can be detected in plasma (or other biofluids), and biomarker assays are developed based on the most reproducible and quantitative peptide(s) from each protein biomarker candidate. Finally, these biomarkers are interrogated in human cohorts in step 3, validation. Cohorts for step 3 should be chosen so that they contain subjects with high and low senescent cell burdens, and ideally, an intervention that either increases or reduces senescent cell burden. Examples of cohorts with variable senescence burden are old and young individuals, and doxorubicin-treated patients and untreated patients (or before and after doxorubicin treatment).
During the validation phase, verified biomarker candidates are tested in a bona fide cohort containing patients with high- and low-senescent cell burdens. For human aging studies, there are multiple cohorts, such as the Baltimore Longitudinal Study of Aging [69] (NIA), LonGenity [111] (Albert Einstein College of Medicine), and InCHIANTI (www.inchiantistudy.net), as well as disease contexts with known associations with senescent cells, such as osteoarthritis and neurodegeneration. A more immediately testable clinical use for SASP markers may be in patients taking drugs that induce cellular senescence, such as genotoxic chemotherapies or certain HAART drugs. For example, thrombosis is a major side-effect in patients on genotoxic chemotherapies; the incidence may reach 20–23% in doxorubicin-treated patients [112, 113], but the mechanism is not understood. We recently showed that clearing senescent cells eliminates the thrombotic effects of doxorubicin (10 mg/kg) in mice [65]. This suggests that senolytics or senomorphics could be of immediate clinical value to cancer patients. Most ideal to validate biomarkers would be human trials that include senolytics or senomorphics. Although these drugs are still on the horizon [45, 114], certain lifestyle or dietary interventions with senomorphic effects are being tested in humans, including exercise [115, 116, 117], dietary restriction [118] and intermittent fasting [119]. In human intervention trials, senescence-inducing therapies could be tested for their ability to increase senescence biomarkers or for the ability of senolytics or senomorphics to reduce them.
Experimental design considerations are critical to improve the chance for discovery of sufficiently powered, specific, and generalizable clinical biomarkers. Following a carefully considered set of guidelines by Forshed et al. [120] concerning the research questions at hand, experimental design (randomization, sample size, etc), sample selection, and analytical methods is extremely important to maximize the discovery of significant, effective, and reproducible clinical biomarkers. Of course, extreme care should be taken to follow FDA processes, standards and metrics for development of biomarker-based analyses, including multiplexed targeted proteomic technologies that are refined jointly by the FDA and The National Cancer Institute’s Clinical Proteomic Technologies for Cancer Initiative (NCI-CPTAC) [121, 122, 123, 124, 125].
Additionally, antibody-based proteomic approaches can be complementary and beneficial in combination with MS-based approaches. For example, antibody arrays are valuable and potentially preferable for detecting proteins with few or no unique tryptic peptides, such as interleukins and other small proteins that cannot be unambiguously identified in a traditional MS analysis. Additionally, once biomarker panels and detection antibodies are established, antibody panels are clinically feasible to quantify biomarkers, assuming they meet quality and reproducibility standards required for proteomic biomarkers.
5.2. Defining the key biological activities of SASP proteins
Screening for biologically active SASP factors, particularly those contributing to pathological changes during aging or related diseases, will be vital to identify aging- and disease-driving SASP factors to target with SASP therapeutics. Several approaches can identify important disease-driving or disease-associated SASP factors. One approach, driven by available data, is to compile hypothesized disease-causing SASP candidates by comparing known SASP profiles with known disease biomarkers. We recently compiled several aging- and disease-associated SASP proteins in this manner (e.g., GDF15, STC1, MMP1, and SERPINE1) [3]. These candidates can be screened on an individual basis in cell culture for activities that fully or partially recapitulate the effects of the SASP, discussed more below. Another approach, more systematic and unbiased, is to directly test the activity of fractions or subsets of the SASP. For example, taking conditioned media from senescent cells, fractionating using one of many available methods (e.g., ion exchange chromatography), and testing which fractions recapitulate some or all the activity of the SASP in a phenotypic screen. The biologically active fractions can be fractionated further and eventually profiled by MS-based proteomics.
Groups of candidate proteins or fractions of the SASP identified by either approach can be verified using in vivo or cell culture screens that block or increase the activity of each candidate and then tested for a disease outcome. For example, the SASP drives invasive phenotypes (increased scattering, chemotaxis and EMT markers) in non-invasive breast cancer cells in culture. SASP factors could be tested for those that drive these phenotypes by treating breast cancer cells with conditioned medium from senescent +/− individual SASP factors. Similar screens could be used for other phenotypes associated with senescent cells, such as the induction of paracrine senescence [15, 64], proliferation and activation of pro-inflammatory NAD+ consuming macrophages [126], and cellular reprogramming [22]. Protein fractionation in combination with MS-based proteomics will be important for identifying key biologically active SASP components. Once those SASP proteins are identified, senomorphic drugs can be developed to target one or more for therapeutic use in humans.
5.3. Senescent cell surfaceomics
One of the promising and most translationally relevant proteomic opportunities in senescence research is the application of cell-surface proteome (surfaceome) technologies. State-of-the-art proteomic approaches, such as cell-surface capture (CSC) [127], enable the unbiased detection and quantification of cell surface proteins. A miniaturized automated form of CSC, autoCSC, vastly improves the technology in terms of sensitivity, reproducibility and throughput [128] and makes large-scale characterizations more feasible. Software tools, such as SURFY [129] and SurfaceGenie [130], also exist for in silico prediction of cell-surface proteins based on existing proteomic and transcriptomic data. CSC has been extensively applied in creating cell-type specific surfaceome databases for humans and mice, such as the Cell Surface Protein Atlas [131]. Surfaceomic technology has also been widely utilized in the areas of cancer, stem cells and immunology [128, 132, 133, 134]. The discovery of robust specific senescent cell-surface markers would open the door to novel drug and immunotherapeutic approaches to eliminate senescent cells and new biomarkers for senescent cell burden. Importantly, cell-surface markers may also provide the ability to isolate and study senescent cells from intact tissues, rather than cultured primary cells artificially induced to senesce in culture for the first time. With the ability to isolate and study endogenous senescent cells, in vivo-relevant, or even personalized, phenotypes of senescent cells could be examined and used in drug screens.
5.4. Senescence-associated post-translational modifications (PTMs)
One of the most powerful applications of mass spectrometry-based proteomics is the identification and quantification of post-translational modifications (PTMs), yet these approaches have been rarely applied in the context of senescence and aging biology. However, modern mass spectrometry workflows have enabled comprehensive identification and quantitative assessments of PTMs, including acylomes (acetylation, succinylation, and others), ubiquitinomes, phospho-proteomics, and many other types of PTMs with ever-improving efficiency and coverage [135, 136, 137]. Among the newest approaches to these analyses, emerging data independent acquisition (DIA) workflows have enabled comprehensive identification, quantification, and site-localization of PTMs, in some cases eliminating the need for PTM enrichment prior to analysis [137]. As senescent cells are characterized by the formation of heterochromatin and alterations in histone modifications [138], proteomic profiling of senescence-associated PTMs may become extremely valuable toward understanding of senescence biology. In addition, the existence of PTMs in the SASP has been entirely unexplored and will represent an exciting new area of research, and perhaps uncover novel senescence biomarker candidates.
5.4. A 5-year look into the future
We speculate that new trends will be set in how human clinical specimens will be subjected to proteomic assays to determine senescence burden. Novel proteomic technologies and instrumentation will allow data acquisition with improved dynamic range and scan speed, increased sensitivity and resolution and sophisticated data processing algorithms. Specific senescence biomarker panels have been developed and will continue to be refined, and thus, biofluids will be queried in a high-throughput fashion for markers of senescence. We envision that more robust biomarker panel assays will be applied to large human cohorts for clinical assessments of senescence burden to gain insights into biological aging, and also in clinical settings to tailor specific treatments for patients, based on their proteomic signatures in their own biofluids.
Senescence-derived biomarkers may become key tools for precision medicine therapeutics and clinical care for elderly patients. We anticipate that evaluation of senescence markers could be part of any quantitative proteomics assay targeting human cohorts for investigation of age-related diseases or aging itself. With human therapies involving senolytics and senomorphics, clinicians will be able to measure senescence biomarkers, and targeted proteomics assays will serve to assess treatment success by measuring reductions in SASP factors in the circulation.
Finally, in vivo studies of senescent cells in tissues might take advantage of knowledge gained from senescent cell surfaceomics, enriching senescent cells directly from tissues followed by ultra-sensitive proteomics and/or single-cell RNA sequencing.
Article highlights.
Emerging technologies and standards in clinical proteomics are ideally suited for developing cellular senescence-derived biomarkers of aging and disease.
Promising therapeutics for multiple human age-related diseases include i) senolytics, which selectively eliminate senescent cells, and ii) senomorphics, which inhibit or modify the senescence-associated secretory phenotype (SASP). These drugs improve aging phenotypes in mice and are beginning to be tested in humans.
SASP proteins and other molecules are candidate biomarkers that can monitor senescent cell burden in human plasma and other biofluids and will be critical tools for developing senolytics and senomorphics.
Modern proteomic approaches comprehensively investigate and uncover SASP protein profiles that reveal SASP heterogeneity that depends on cell type, senescence-inducing stimuli, and temporal dynamics.
To develop robust and specific biomarkers of aging, modern proteomic technologies can be used for unbiased screening of SASP factors, profiling the extracellular vesicle proteome, and characterizing the senescent cell surface proteome.
We propose the development of clinically relevant senescence-derived biomarkers in a three-phase pipeline: Discovery, Verification, and Validation.
Funding
This work was supported by the National Institutes of Health under award numbers: U01 G060906-02 and U01 AG060906-02S1 (PI: Schilling), under award numbers P01AG017242 and R01AG051729 (PI: Campisi), and under award number 1S10 OD016281 (PI: Bradford Gibson, Buck Institute). This work was further supported by the Glenn Foundation for Medical Research for a postdoctoral fellowship (NB) and by the SENS Foundation for a postdoctoral fellowship (AK).
Footnotes
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Ferrucci L, Gonzalez-Freire M, Fabbri E, et al. Measuring biological aging in humans: A quest. Aging Cell. 2019. December 12:e13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zampino M, Ferrucci L, Semba RD. Biomarkers in the path from cellular senescence to frailty. Exp Gerontol. 2020. January;129:110750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basisty N, Kale A, Jeon OH, et al. A Proteomic Atlas of Senescence-Associated Secretomes for Aging Biomarker Development. PLOS Biology. 2020;18(1):e3000599. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Quantitative, unbiased, and comprehensive proteomic analysis of the SASP in multiple inducers and cell types and identification of senescence-based biomarkers of aging in human plasma.
- 4.Keshishian H, Burgess MW, Specht H, et al. Quantitative, multiplexed workflow for deep analysis of human blood plasma and biomarker discovery by mass spectrometry. Nat Protoc. 2017. August;12(8):1683–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoofnagle AN, Aebersold R, Anderson NL, et al. Painting a Moving Picture: Large-Scale Proteomics Efforts and Their Potential for Changing Patient Care. Clinical Chemistry. 2011;57(10):1357–1360. [DOI] [PubMed] [Google Scholar]
- 6.Hsu CY, Ballard S, Batlle D, et al. Cross-Disciplinary Biomarkers Research: Lessons Learned by the CKD Biomarkers Consortium. Clin J Am Soc Nephrol. 2015. May 7;10(5):894–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright I, Van Eyk JE. A Roadmap to Successful Clinical Proteomics. Clin Chem. 2017. January;63(1):245–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi T, Song E, Nie S, et al. Advances in targeted proteomics and applications to biomedical research. Proteomics. 2016. August;16(15–16):2160–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins BC, Hunter CL, Liu Y, et al. Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nat Commun. 2017. August 21;8(1):291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillet LC, Navarro P, Tate S, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012. June;11(6):O111 016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sajic T, Liu Y, Aebersold R. Using data-independent, high-resolution mass spectrometry in protein biomarker research: perspectives and clinical applications. Proteomics Clin Appl. 2015. April;9(3–4):307–21. [DOI] [PubMed] [Google Scholar]
- 12.Meyer JG, Schilling B. Clinical applications of quantitative proteomics using targeted and untargeted data-independent acquisition techniques. Expert Review of Proteomics. 2017 2017/May/04;14(5):419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campisi J Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 2001. November;11(11):S27–31. [DOI] [PubMed] [Google Scholar]
- 14.Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007. September;8(9):729–40. [DOI] [PubMed] [Google Scholar]
- 15.Acosta JC, Banito A, Wuestefeld T, et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat Cell Biol. 2013 2013;15(8):978–90. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Proteomic study identifying potential factors and mechanisms important in paracrine senescence.
- 16.d’Adda di Fagagna F Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer. 2008. July;8(7):512–22. [DOI] [PubMed] [Google Scholar]
- 17.Petrova NV, Velichko AK, Razin SV, et al. Small molecule compounds that induce cellular senescence. Aging Cell. 2016. December;15(6):999–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiley CD, Velarde MC, Lecot P, et al. Mitochondrial Dysfunction Induces Senescence with a Distinct Secretory Phenotype. Cell Metab. 2016 2016;23(2):303–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acosta JC, O’Loghlen A, Banito A, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008. June 13;133(6):1006–18. [DOI] [PubMed] [Google Scholar]
- 20.Coppé J-P, Patil CK, Rodier F, et al. Senescence-Associated Secretory Phenotypes Reveal Cell-Nonautonomous Functions of Oncogenic RAS and the p53 Tumor Suppressor. PLOS Biology. 2008 December 2, 2008;6(12):e301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campisi J Cellular senescence: putting the paradoxes in perspective. Curr Opin Genet Dev. 2011. February;21(1):107–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosteiro L, Pantoja C, Alcazar N, et al. Tissue damage and senescence provide critical signals for cellular reprogramming in vivo. Science. 2016. November 25;354(6315). [DOI] [PubMed] [Google Scholar]; • Describes a potential role of the SASP in tissue reprogramming and repair.
- 23.Demaria M, Ohtani N, Youssef SA, et al. An Essential Role for Senescent Cells in Optimal Wound Healing through Secretion of PDGF-AA. Developmental cell. 2014 2014-December-22;31(6):722–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neves J, Demaria M, Campisi J, et al. Of flies, mice, and men: evolutionarily conserved tissue damage responses and aging. Dev Cell. 2015 2015;32(1):9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tchkonia T, Zhu Y, van Deursen J, et al. Cellular senescence and the senescent secretory phenotype: therapeutic opportunities. J Clin Invest. 2013 2013;123(3):966–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tominaga K The emerging role of senescent cells in tissue homeostasis and pathophysiology. Pathobiol Aging Age Relat Dis. 2015;5:27743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giaimo S, d’Adda di Fagagna F. Is cellular senescence an example of antagonistic pleiotropy? Aging Cell. 2012. June;11(3):378–83. [DOI] [PubMed] [Google Scholar]
- 28.Krizhanovsky V, Xue W, Zender L, et al. Implications of cellular senescence in tissue damage response, tumor suppression, and stem cell biology. Cold Spring Harb Symp Quant Biol. 2008;73:513–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storer M, Mas A, Robert-Moreno A, et al. Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell. 2013 2013;155(5):1119–30. [DOI] [PubMed] [Google Scholar]
- 30.Muñoz-Espín D, Cañamero M, Maraver A, et al. Programmed cell senescence during mammalian embryonic development. Cell. 2013 2013;155(5):1104–18. [DOI] [PubMed] [Google Scholar]
- 31.Menon R, Behnia F, Polettini J, et al. Placental membrane aging and HMGB1 signaling associated with human parturition. Aging (Albany NY). 2016. February;8(2):216–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chinta SJ, Woods G, Demaria M, et al. Cellular Senescence Is Induced by the Environmental Neurotoxin Paraquat and Contributes to Neuropathology Linked to Parkinson’s Disease. Cell Rep. 2018. January 23;22(4):930–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Childs BG, Baker DJ, Wijshake T, et al. Senescent intimal foam cells are deleterious at all stages of atherosclerosis. Science. 2016. October 28;354(6311):472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demaria M, O’Leary MN, Chang J, et al. Cellular Senescence Promotes Adverse Effects of Chemotherapy and Cancer Relapse. Cancer Discovery. 2017 February 2017;7(2):165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu Q, Qin Z, Yu J, et al. Effects of senescent lens epithelial cells on the severity of age-related cortical cataract in humans: A case-control study. Medicine (Baltimore). 2016. June;95(25):e3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson PJ, Shah A, Ntranos V, et al. Targeted Elimination of Senescent Beta Cells Prevents Type 1 Diabetes. Cell Metab. 2019. May 7;29(5):1045–1060 e10. [DOI] [PubMed] [Google Scholar]
- 37.Palmer AK, Xu M, Zhu Y, et al. Targeting senescent cells alleviates obesity-induced metabolic dysfunction. Aging Cell. 2019. June;18(3):e12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schafer MJ, White TA, Iijima K, et al. Cellular senescence mediates fibrotic pulmonary disease. Nat Commun. 2017. February 23;8:14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeon OH, Kim C, Laberge R-M, et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nature Medicine. 2017 June 2017;23(6):775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farr JN, Xu M, Weivoda MM, et al. Targeting cellular senescence prevents age-related bone loss in mice. Nat Med. 2017. September;23(9):1072–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baar MP, Perdiguero E, Munoz-Canoves P, et al. Musculoskeletal senescence: a moving target ready to be eliminated. Curr Opin Pharmacol. 2018. June;40:147–155. [DOI] [PubMed] [Google Scholar]
- 42.Baar MP, Brandt RM, Putavet DA, et al. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell. 2017;169:132–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bussian TJ, Aziz A, Meyer CF, et al. Clearance of senescent glial cells prevents tau-dependent pathology and cognitive decline. Nature. 2018. October;562(7728):578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu M, Pirtskhalava T, Farr JN, et al. Senolytics improve physical function and increase lifespan in old age. Nat Med. 2018. August;24(8):1246–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim EC, Kim JR. Senotherapeutics: emerging strategy for healthy aging and age-related disease. BMB Rep. 2019. January;52(1):47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Short S, Fielder E, Miwa S, et al. Senolytics and senostatics as adjuvant tumour therapy. EBioMedicine. 2019. March;41:683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaplon J, Zheng L, Meissl K, et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature. 2013. June 6;498(7452):109–12. [DOI] [PubMed] [Google Scholar]
- 48.Passos JF, Nelson G, Wang C, et al. Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol Syst Biol. 2010;6:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.James ENL, Bennett MH, Parkinson EK. The induction of the fibroblast extracellular senescence metabolome is a dynamic process. Sci Rep. 2018. August 14;8(1):12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takasugi M Emerging roles of extracellular vesicles in cellular senescence and aging. Aging Cell. 2018. April;17(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.James EL, Michalek RD, Pitiyage GN, et al. Senescent human fibroblasts show increased glycolysis and redox homeostasis with extracellular metabolomes that overlap with those of irreparable DNA damage, aging, and disease. J Proteome Res. 2015. April 3;14(4):1854–71. [DOI] [PubMed] [Google Scholar]
- 52.Coppé J-P, Patil CK, Rodier F, et al. A Human-Like Senescence-Associated Secretory Phenotype Is Conserved in Mouse Cells Dependent on Physiological Oxygen. PLoS ONE. 2010 2010-February-12;5(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuilman T, Michaloglou C, Vredeveld LCW, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008 June 13, 2008;133(6):1019–1031. [DOI] [PubMed] [Google Scholar]
- 54.Laberge RM, Sun Y, Orjalo AV, et al. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat Cell Biol. 2015 2015;17(8):1049–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Velarde MC, Demaria M, Melov S, et al. Pleiotropic age-dependent effects of mitochondrial dysfunction on epidermal stem cells. Proc Natl Acad Sci U S A. 2015 2015;112(33):10407–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Comi P, Chiaramonte R, Maier JA. Senescence-dependent regulation of type 1 plasminogen activator inhibitor in human vascular endothelial cells. Exp Cell Res. 1995. July;219(1):304–8. [DOI] [PubMed] [Google Scholar]
- 57.West MD, Shay JW, Wright WE, et al. Altered expression of plasminogen activator and plasminogen activator inhibitor during cellular senescence. Exp Gerontol. 1996. Jan-Apr;31(1–2):175–93. [DOI] [PubMed] [Google Scholar]
- 58.Mu XC, Higgins PJ. Differential growth state-dependent regulation of plasminogen activator inhibitor type-1 expression in senescent IMR-90 human diploid fibroblasts. J Cell Physiol. 1995. December;165(3):647–57. [DOI] [PubMed] [Google Scholar]
- 59.West MD, Pereira-Smith OM, Smith JR. Replicative senescence of human skin fibroblasts correlates with a loss of regulation and overexpression of collagenase activity. Exp Cell Res. 1989. September;184(1):138–47. [DOI] [PubMed] [Google Scholar]
- 60.Millis AJ, Hoyle M, McCue HM, et al. Differential expression of metalloproteinase and tissue inhibitor of metalloproteinase genes in aged human fibroblasts. Exp Cell Res. 1992. August;201(2):373–9. [DOI] [PubMed] [Google Scholar]
- 61.Zeng G, Millis AJ. Differential regulation of collagenase and stromelysin mRNA in late passage cultures of human fibroblasts. Exp Cell Res. 1996. January 10;222(1):150–6. [DOI] [PubMed] [Google Scholar]
- 62.Coppé J-P, Desprez P-Y, Krtolica A, et al. The Senescence-Associated Secretory Phenotype: The Dark Side of Tumor Suppression. Annual review of pathology. 2010 2010;5:99–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malaquin N, Martinez A, Rodier F. Keeping the senescence secretome under control: Molecular reins on the senescence-associated secretory phenotype. Experimental Gerontology. 2016 September 2016;82:39–49. [DOI] [PubMed] [Google Scholar]
- 64.Borghesan M, Fafián-Labora J, Eleftheriadou O, et al. Small Extracellular Vesicles Are Key Regulators of Non-cell Autonomous Intercellular Communication in Senescence via the Interferon Protein IFITM3. Cell Reports. 2019 June 25, 2019;27(13):3956–3971.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]; • First comprehensive proteomic analysis of the exosome SASP.
- 65.Wiley CD, Liu S, Limbad C, et al. SILAC Analysis Reveals Increased Secretion of Hemostasis-Related Factors by Senescent Cells. Cell Reports. 2019 September 24, 2019;28(13):3329–3337.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ozcan S, Alessio N, Acar MB, et al. Unbiased analysis of senescence associated secretory phenotype (SASP) to identify common components following different genotoxic stresses. Aging (Albany NY). 2016. July;8(7):1316–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lefèvre C, Auclair M, Boccara F, et al. Premature senescence of vascular cells is induced by HIV protease inhibitors: implication of prelamin A and reversion by statin. Arteriosclerosis, Thrombosis, and Vascular Biology. 2010 December 2010;30(12):2611–2620. [DOI] [PubMed] [Google Scholar]
- 68.Hernandez-Vallejo SJ, Beaupere C, Larghero J, et al. HIV protease inhibitors induce senescence and alter osteoblastic potential of human bone marrow mesenchymal stem cells: beneficial effect of pravastatin. Aging Cell. 2013 December 2013;12(6):955–965. [DOI] [PubMed] [Google Scholar]
- 69.Tanaka T, Biancotto A, Moaddel R, et al. Plasma proteomic signature of age in healthy humans. Aging Cell. 2018. October;17(5):e12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bidadkosh A, Lambooy SPH, Heerspink HJ, et al. Predictive Properties of Biomarkers GDF-15, NTproBNP, and hs-TnT for Morbidity and Mortality in Patients With Type 2 Diabetes With Nephropathy. Diabetes Care. 2017 June 2017;40(6):784–792. [DOI] [PubMed] [Google Scholar]
- 71.Daniels LB, Clopton P, Laughlin GA, et al. Growth-differentiation factor-15 is a robust, independent predictor of 11-year mortality risk in community-dwelling older adults: the Rancho Bernardo Study. Circulation. 2011 May 17, 2011;123(19):2101–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ho JE, Mahajan A, Chen M-H, et al. Clinical and genetic correlates of growth differentiation factor 15 in the community. Clinical Chemistry. 2012 November 2012;58(11):1582–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ho JE, Hwang S-J, Wollert KC, et al. Biomarkers Of Cardiovascular Stress And Incident Chronic Kidney Disease. Clinical chemistry. 2013 2013-November;59(11):1613–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rohatgi A, Patel P, Das SR, et al. Association of growth differentiation factor-15 with coronary atherosclerosis and mortality in a young, multiethnic population: observations from the Dallas Heart Study. Clinical Chemistry. 2012 January 2012;58(1):172–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wallentin L, Zethelius B, Berglund L, et al. GDF-15 for prognostication of cardiovascular and cancer morbidity and mortality in men. PloS One. 2013 2013;8(12):e78797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wollert KC, Kempf T, Wallentin L. Growth Differentiation Factor 15 as a Biomarker in Cardiovascular Disease. Clinical Chemistry. 2017 2017/January/01;63(1):140–151. [DOI] [PubMed] [Google Scholar]
- 77.Guo Y, Ayers JL, Carter KT, et al. Senescence-associated tissue microenvironment promotes colon cancer formation through the secretory factor GDF15. Aging Cell. 2019. December;18(6):e13013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bhat R, Crowe EP, Bitto A, et al. Astrocyte senescence as a component of Alzheimer’s disease. PloS One. 2012 2012;7(9):e45069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen Y-K, Tung C-W, Lee J-Y, et al. Plasma matrix metalloproteinase 1 improves the detection and survival prediction of esophageal squamous cell carcinoma. Scientific Reports. 2016 2016-July-20;6:30057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rosas IO, Richards TJ, Konishi K, et al. MMP1 and MMP7 as Potential Peripheral Blood Biomarkers in Idiopathic Pulmonary Fibrosis. PLoS Medicine. 2008 2008-April;5(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roy R, Yang J, Moses MA. Matrix Metalloproteinases As Novel Biomarkers and Potential Therapeutic Targets in Human Cancer. Journal of Clinical Oncology. 2009 2009. –November-1;27(31):5287–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chang AC-M, Doherty J, Huschtscha LI, et al. STC1 expression is associated with tumor growth and metastasis in breast cancer. Clinical & Experimental Metastasis. 2015 January 2015;32(1):15–27. [DOI] [PubMed] [Google Scholar]
- 83.Du Y-Z, Gu X-H, Cheng S-F, et al. The oncogenetic role of stanniocalcin 1 in lung adenocarcinoma: a promising serum candidate biomarker for tracking lung adenocarcinoma progression. Tumour Biology: The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2016 April 2016;37(4):5633–5644. [DOI] [PubMed] [Google Scholar]
- 84.Ohkouchi S, Ono M, Kobayashi M, et al. Myriad Functions of Stanniocalcin-1 (STC1) Cover Multiple Therapeutic Targets in the Complicated Pathogenesis of Idiopathic Pulmonary Fibrosis (IPF). Clinical Medicine Insights Circulatory, Respiratory and Pulmonary Medicine. 2015 2015;9(Suppl 1):91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pan JS, Huang L, Belousova T, et al. Stanniocalcin-1 inhibits renal ischemia/reperfusion injury via an AMP-activated protein kinase-dependent pathway. J Am Soc Nephrol. 2015. February;26(2):364–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Su J, Guo B, Zhang T, et al. Stanniocalcin-1, a new biomarker of glioma progression, is associated with prognosis of patients. Tumour Biol. 2015. August;36(8):6333–9. [DOI] [PubMed] [Google Scholar]
- 87.Shahim P, Blennow K, Johansson P, et al. Cerebrospinal Fluid Stanniocalcin-1 as a Biomarker for Alzheimer’s Disease and Other Neurodegenerative Disorders. Neuromolecular Med. 2017. March;19(1):154–160. [DOI] [PubMed] [Google Scholar]
- 88.Hernandez-Segura A, de Jong TV, Melov S, et al. Unmasking Transcriptional Heterogeneity in Senescent Cells. Current biology: CB. 2017 September 11, 2017;27(17):2652–2660.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Casella G, Munk R, Kim KM, et al. Transcriptome signature of cellular senescence. Nucleic Acids Research. 2019 2019/August/22;47(14):7294–7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Park H, Kim CH, Jeong JH, et al. GDF15 contributes to radiation-induced senescence through the ROS-mediated p16 pathway in human endothelial cells. Oncotarget. 2016. March 1;7(9):9634–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Camussi G, Deregibus MC, Bruno S, et al. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010. November;78(9):838–48. [DOI] [PubMed] [Google Scholar]
- 92.Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van der Pol E, Boing AN, Harrison P, et al. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012. July;64(3):676–705. [DOI] [PubMed] [Google Scholar]
- 94.Yanez-Mo M, Siljander PR, Andreu Z, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Boukouris S, Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics Clin Appl. 2015. April;9(3–4):358–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gamez-Valero A, Lozano-Ramos SI, Bancu I, et al. Urinary extracellular vesicles as source of biomarkers in kidney diseases. Front Immunol. 2015;6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang X, Hubal MJ, Kraus VB. Immune cell extracellular vesicles and their mitochondrial content decline with ageing. Immunity & Ageing. 2020 2020/January/04;17(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jablonska J, Pietrowska M, Ludwig S, et al. Challenges in the Isolation and Proteomic Analysis of Cancer Exosomes-Implications for Translational Research. Proteomes. 2019. May 15;7(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ludwig N, Whiteside TL, Reichert TE. Challenges in Exosome Isolation and Analysis in Health and Disease. Int J Mol Sci. 2019. September 21;20(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Greening DW, Xu R, Ji H, et al. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol Biol. 2015;1295:179–209. [DOI] [PubMed] [Google Scholar]
- 101.Yu LL, Zhu J, Liu JX, et al. A Comparison of Traditional and Novel Methods for the Separation of Exosomes from Human Samples. Biomed Res Int. 2018;2018:3634563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nelson G, Wordsworth J, Wang C, et al. A senescent cell bystander effect: senescence-induced senescence. Aging Cell. 2012. April;11(2):345–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jakhar R, Crasta K. Exosomes as Emerging Pro-Tumorigenic Mediators of the Senescence-Associated Secretory Phenotype. Int J Mol Sci. 2019. May 24;20(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lehmann BD, Paine MS, Brooks AM, et al. Senescence-associated exosome release from human prostate cancer cells. Cancer Research. 2008 October 01, 2008;68(19):7864–7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jeon OH, Wilson DR, Clement CC, et al. Senescence cell-associated extracellular vesicles serve as osteoarthritis disease and therapeutic markers. JCI insight. 2019 April 04, 2019;4(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kavanagh EL, Lindsay S, Halasz M, et al. Protein and chemotherapy profiling of extracellular vesicles harvested from therapeutic induced senescent triple negative breast cancer cells. Oncogenesis. 2017. October 9;6(10):e388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Effenberger T, von der Heyde J, Bartsch K, et al. Senescence-associated release of transmembrane proteins involves proteolytic processing by ADAM17 and microvesicle shedding. FASEB J. 2014. November;28(11):4847–56. [DOI] [PubMed] [Google Scholar]
- 108.Takasugi M, Okada R, Takahashi A, et al. Small extracellular vesicles secreted from senescent cells promote cancer cell proliferation through EphA2. Nat Commun. 2017. June 6;8:15729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stastna M, Van Eyk JE. Secreted proteins as a fundamental source for biomarker discovery. Proteomics. 2012. February;12(4–5):722–35. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Review describing why secretomes are valuable sources for plasma biomarkers.
- 110.Erickson BK, Rose CM, Braun CR, et al. A Strategy to Combine Sample Multiplexing with Targeted Proteomics Assays for High-Throughput Protein Signature Characterization. Mol Cell. 2017. January 19;65(2):361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lehallier B, Gate D, Schaum N, et al. Undulating changes in human plasma proteome profiles across the lifespan. Nature Medicine. 2019 2019/December/01;25(12):1843–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zangari M, Siegel E, Barlogie B, et al. Thrombogenic activity of doxorubicin in myeloma patients receiving thalidomide: implications for therapy. Blood. 2002;100(4):1168–1171. [DOI] [PubMed] [Google Scholar]
- 113.Kim S-H, Lim K-M, Noh J-Y, et al. Doxorubicin-Induced Platelet Procoagulant Activities: An Important Clue for Chemotherapy-Associated Thrombosis. Toxicological Sciences. 2011;124(1):215–224. [DOI] [PubMed] [Google Scholar]
- 114.Justice JN, Nambiar AM, Tchkonia T, et al. Senolytics in idiopathic pulmonary fibrosis: Results from a first-in-human, open-label, pilot study. EBioMedicine. 2019. February;40:554–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schafer MJ, White TA, Evans G, et al. Exercise Prevents Diet-Induced Cellular Senescence in Adipose Tissue. Diabetes. 2016. June;65(6):1606–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rossman MJ, Kaplon RE, Hill SD, et al. Endothelial cell senescence with aging in healthy humans: prevention by habitual exercise and relation to vascular endothelial function. Am J Physiol Heart Circ Physiol. 2017. November 1;313(5):H890–H895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Werner C, Furster T, Widmann T, et al. Physical exercise prevents cellular senescence in circulating leukocytes and in the vessel wall. Circulation. 2009. December 15;120(24):2438–47. [DOI] [PubMed] [Google Scholar]
- 118.Fontana L, Nehme J, Demaria M. Caloric restriction and cellular senescence. Mechanisms of Ageing and Development. 2018 2018/December/01/;176:19–23. [DOI] [PubMed] [Google Scholar]
- 119.Brandhorst S, Choi In Y, Wei M, et al. A Periodic Diet that Mimics Fasting Promotes Multi-System Regeneration, Enhanced Cognitive Performance, and Healthspan. Cell Metabolism. 2015 2015/July/07/;22(1):86–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Forshed J Experimental Design in Clinical ‘Omics Biomarker Discovery. J Proteome Res. 2017. November 3;16(11):3954–3960. [DOI] [PubMed] [Google Scholar]
- 121.Boja ES, Fehniger TE, Baker MS, et al. Analytical validation considerations of multiplex mass-spectrometry-based proteomic platforms for measuring protein biomarkers. J Proteome Res. 2014. December 5;13(12):5325–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fehniger TE, Boja ES, Rodriguez H, et al. Four Areas of Engagement Requiring Strengthening in Modern Proteomics Today. Journal of Proteome Research. 2014 2014/December/05;13(12):5310–5318. [DOI] [PubMed] [Google Scholar]
- 123.Carr SA, Abbatiello SE, Ackermann BL, et al. Targeted Peptide Measurements in Biology and Medicine: Best Practices for Mass Spectrometry-based Assay Development Using a Fit-for-Purpose Approach. Molecular & Cellular Proteomics. 2014;13(3):907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rodriguez H, Težak Ž, Mesri M, et al. Analytical Validation of Protein-Based Multiplex Assays: A Workshop Report by the NCI-FDA Interagency Oncology Task Force on Molecular Diagnostics. Clinical Chemistry. 2010;56(2):237–243. [DOI] [PubMed] [Google Scholar]; • Review of proteomic biomarker discovery pipeline and FDA requirements.
- 125.Regnier FE, Skates SJ, Mesri M, et al. Protein-Based Multiplex Assays: Mock Presubmissions to the US Food and Drug Administration. Clinical Chemistry. 2010;56(2):165–171. [DOI] [PubMed] [Google Scholar]
- 126.Covarrubias AJ, Lopez-Dominguez JA, Perrone R, et al. Aging-related inflammation driven by cellular senescence enhances NAD consumption via activation of CD38+ macrophages. bioRxiv. 2019:609438. [Google Scholar]
- 127.Bausch-Fluck D, Hofmann A, Wollscheid B. Cell surface capturing technologies for the surfaceome discovery of hepatocytes. Methods Mol Biol. 2012;909:1–16. [DOI] [PubMed] [Google Scholar]
- 128.van Oostrum M, Muller M, Klein F, et al. Classification of mouse B cell types using surfaceome proteotype maps. Nat Commun. 2019. December 16;10(1):5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bausch-Fluck D, Goldmann U, Muller S, et al. The in silico human surfaceome. Proc Natl Acad Sci U S A. 2018. November 13;115(46):E10988–E10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Waas M, Snarrenberg ST, Littrell J, et al. SurfaceGenie: a web-based application for prioritizing cell-type specific marker candidates. bioRxiv. 2019:575969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bausch-Fluck D, Hofmann A, Bock T, et al. A mass spectrometric-derived cell surface protein atlas. PLoS One. 2015;10(3):e0121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Boheler KR, Gundry RL. Concise Review: Cell Surface N-Linked Glycoproteins as Potential Stem Cell Markers and Drug Targets. STEM CELLS Translational Medicine. 2017;6(1):131–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fujinaka CM, Waas M, Gundry RL. Mass Spectrometry-Based Identification of Extracellular Domains of Cell Surface N-Glycoproteins: Defining the Accessible Surfaceome for Immunophenotyping Stem Cells and Their Derivatives In: Boheler KR, Gundry RL, editors. The Surfaceome: Methods and Protocols. New York, NY: Springer New York; 2018. p. 57–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kuhlmann L, Cummins E, Samudio I, et al. Cell-surface proteomics for the identification of novel therapeutic targets in cancer. Expert Review of Proteomics. 2018 2018/March/04;15(3):259–275. [DOI] [PubMed] [Google Scholar]
- 135.Xie X, Shah S, Holtz A, et al. Simultaneous Affinity Enrichment of Two Post-Translational Modifications for Quantification and Site Localization. J Vis Exp. 2020. February 27(156). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Basisty N, Meyer JG, Wei L, et al. Simultaneous Quantification of the Acetylome and Succinylome by ‘One-Pot’ Affinity Enrichment. Proteomics. 2018. September;18(17):e1800123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nakayasu ES, Burnet MC, Walukiewicz HE, et al. Ancient Regulatory Role of Lysine Acetylation in Central Metabolism. mBio. 2017. November 28;8(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Paluvai H, Di Giorgio E, Brancolini C. The Histone Code of Senescence. Cells. 2020. February 18;9(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Althubiti M, Lezina L, Carrera S, et al. Characterization of novel markers of senescence and their prognostic potential in cancer. Cell Death & Disease. 2014 2014/November/01;5(11):e1528–e1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim KM, Noh JH, Bodogai M, et al. Identification of senescent cell surface targetable protein DPP4. Genes Dev. 2017. August 1;31(15):1529–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hoare M, Ito Y, Kang TW, et al. NOTCH1 mediates a switch between two distinct secretomes during senescence. Nat Cell Biol. 2016. September;18(9):979–92. [DOI] [PMC free article] [PubMed] [Google Scholar]


