Abstract
Aspergillus fumigatus is an environmental fungus that can cause invasive pulmonary aspergillosis when spores are inhaled into the respiratory tract and invade airway or lung tissue. Influenza is a common respiratory virus that can cause severe respiratory disease and post-influenza invasive pulmonary aspergillosis, which is becoming a well-recognized clinical problem, typically occurs in critically ill patients. Mice challenged with influenza A PR/8/34 H1N1 and subsequently challenged with A. fumigatus had increased fungal burden, viral burden, inflammation and mortality compared to single infected mice. Neutrophil recruitment in the lung of super-infected mice was decreased; however, mice were not neutropenic and there was no difference in absolute blood neutrophils between groups. Additionally, CXCL1 and CXCL2 were decreased in lungs of super-infected mice compared to controls. Interferon levels were increased in mice that received influenza and deletion of STAT1 resulted in decreased fungal burden, increased airway and lung neutrophils, and increased CXCL1 compared to wild-type mice; while deletion of STAT2 did not change fungal burden or airway neutrophilia compared to wild-type mice. These data demonstrate a mechanism by which influenza A-induced STAT1 signaling inhibits neutrophil recruitment and increases susceptibility to post-influenza invasive pulmonary aspergillosis.
Introduction
Influenza is a common respiratory virus and seasonal epidemics cause 3 to 5 million severe influenza cases and an estimated 300,000 to 500,000 deaths worldwide each year from influenza-related pulmonary disease. Although most patients have self-limiting disease that resolves, some develop respiratory failure and require intensive care management. Risk factors for influenza-related complications that increase risk of intensive care unit (ICU) admission include cardiac disease, sleep apnea, obesity, and pulmonary super-infection with bacterial or fungal organisms(1). Bacterial pneumonia is a well-recognized complication of influenza, but additional organisms, including the opportunistic fungal pathogen Aspergillus fumigatus, have been demonstrated to cause severe morbidity and mortality in influenza-infected patients(2).
Aspergillus is a genus of filamentous saprotrophic fungi that are commonly found in the soil and air(3). Aspergillus species reproduce asexually through the generation and dissemination of conidia, which can be inhaled into the human lung. Although Aspergillus species are commonly isolated from the lungs of healthy individuals(4), some species, most often A. fumigatus, can cause significant harm to the host. A. fumigatus has been implicated in allergic bronchopulmonary aspergillosis (ABPA), chronic pulmonary aspergillosis (CPA), and invasive pulmonary aspergillosis (IPA), the latter of which occurs when A. fumigatus hyphae invade lung tissue(5).
IPA primarily affects immunocompromised hosts; however recent reports have demonstrated the threat of IPA in influenza-infected patients who are not otherwise immunocompromised. In a retrospective study of 36 patients, Shah et al. found that 72% of patients with influenza-A. fumigatus super-infection died and of those who died, 81% were receiving antifungal treatment(6). Several other studies have found similar results(7–9), demonstrating the need to improve detection of invasive pulmonary aspergillosis, generate new antifungal drugs, and to further understand the mechanism by which influenza makes the host susceptible to A. fumigatus super-infection. To date, there are no published animal models of influenza related IPA.
IPA is difficult to diagnose and although it is estimated that more than 200,000 cases of invasive aspergillosis occur each year, this likely represents only 50–65% of actual cases(22). Although antifungal therapies have improved, treatment is still limited by lack of early diagnosis, fungal identification, route of administration and spectrum of activity(23, 24). Invasive aspergillosis has a 50% mortality rate if diagnosed and treated but is close to 100% lethal if undiagnosed(22).
In this study, we establish a murine model of post-influenza IPA and demonstrate that influenza-A. fumigatus super-infection is more severe than A. fumigatus infection alone, leading to increased fungal burden, viral burden, inflammation and mortality. Fewer neutrophils were recruited to the lung in response to A. fumigatus in the setting of preceding influenza infection and we observed a decrease in CXCL1 and CXCL2 in super-infected mice. Additionally, we demonstrated that mice lacking STAT1, but not STAT2, clear A. fumigatus more effectively and had more airway and lung neutrophils than wild-type mice during influenza-A. fumigatus super-infection, suggesting that STAT1 is required for impaired fungal clearance and may limit neutrophil recruitment during super-infection. These findings establish a potential mechanism for how influenza makes an otherwise immunocompetent host susceptible to IPA.
Methods
Mice
Six to eight week old male wild-type, C57BL/6 male mice were purchased from Taconic Farms (Germantown, NY). STAT1−/−, STAT1/2−/− and STAT2−/− mice on the C57BL/6 background were obtained from Dr. C. Schindler (Columbia University) and Dr. D. Levy (New York University). IFNγ−/− and C57BL/6 matched controls were purchased from Jackson Laboratories (Bar Harbor, Maine). Mice were maintained under pathogen-free conditions within the animal facilities at the UPMC Children’s Hospital of Pittsburgh. All of the studies were performed on age- and sex-matched mice. Animal studies were conducted with approval from the University of Pittsburgh Institutional Animal Care and Use Committee.
A. fumigatus infection and detection
Aspergillus fumigatus (American Type Culture Collection (ATCC) 42202) was provided as a generous gift from Dr. Jay Kolls (Tulane University). A. fumigatus was maintained on potato dextrose agar for 5–7 d at 37°C. Conidia were harvested by washing the culture flask with 25 mL of sterile PBS supplemented with 0.1% Tween 20. The conidia were passed through a sterile 40-μm nylon membrane to remove hyphal fragments and enumerated on a hemocytometer. Mice were inoculated with 2.5×107 conidia of A. fumigatus (in 50 μl sterile PBS) or control PBS by oropharyngeal aspiration, and lungs were harvested 24 – 120 hours later. For survival experiments, 5×107 conidia of A. fumigatus were delivered. For lung fungal burden analysis, the right upper lobe of each mouse was mechanically homogenized in 1 ml of sterile PBS and plated for fungal CFU counting. Burden was determined by plating 100 μl of a 1:100 dilution of lung homogenate on potato dextrose agar plates. Additionally, the middle and caudal lobes of the right lung were frozen in liquid nitrogen. Samples were then crushed and resuspended in lysis buffer RLT (Qiagen, Hilden, Germany) and homogenized in Lysing Matrix E (MP Biomedicals, Solon, OH) with TissueLyser LT bead mill (Qiagen, Hilden, Germany). Total RNA was extracted from lung homogenate using the Qiagen RNEasy Mini Kit (Qiagen, Hilden, Germany), which includes a DNase treatment step to eliminate genomic DNA. Total RNA was also extracted from serial 1:10 dilutions of live A. fumigatus conidia (103–108) and treated with DNase to form a standard curve. The MasterPure Yeast RNA Purification kit was used on samples containing only conidia with the following modifications due to its ability to isolate RNA from as few as 1000 conidia. Conidia were resuspended in 600μL of Extraction Buffer and homogenized in Lysing Matrix E with TissueLyser LT. RNA was precipitated in isopropanol and 3M sodium acetate with RNA-grade glycogen (ThermoFisher, Waltham, MA). All other reagents were proportionally modified due to the extra lysis buffer used. Lung A. fumigatus burden was analyzed with real-time PCR measurement of A. fumigatus 18S rRNA [GenBank accession number AB008401 (22) http://www.ncbi.nlm.nih.gov/genbank] and quantified using a standard curve of A. fumigatus conidia(10). Forward Primer: 5′-GGCCCTTAAATAGCCCGGT-3′, Reverse Primer: 5′TGAGCCGATAGTCCCCCTAA-3′
Probe: 5′-/56-FAM/AGCCAGCGGCCCGCAAATG-36/TAM/−3′. As a validation of the real-time PCR method, no-amplification controls (i.e., no reverse transcriptase included in the cDNA reaction) yielded a signal, <0.5% by real-time PCR, indicating that the DNase treatment step efficiently eliminated contaminating A. fumigatus DNA. Fungal burden was normalized per μg of total RNA used. For histological analysis, lungs were filled with and stored in 10% buffered formalin phosphate and sections were stained with GMS using standard histological techniques to assess conidia count and fungal germination.
Influenza A/PR/8/34 H1N1 infection
Influenza A/PR/8/34 H1N1 was propagated in chicken eggs as previously described(11). Mice were infected with 100 PFU of influenza A/PR/8/34 H1N1 (in 50μL sterile PBS) from a frozen stock or control PBS by oropharyngeal aspiration. Infected mice were incubated for 6 days, at which time mice received A. fumigatus inoculum or control PBS. After an additional 24–120 hours, lungs were harvested. Viral burden was determined by quantitative real-time RT-PCR on lung RNA for viral matrix protein as described previously (12). Forward Primer: 5′-GGACTGCAGCGTAGACGCTT-3′,ReversePrimer: 5′-CATCCTGTTGTATATGAGGCCCAT-3′, Probe: 5′-/56-FAM/CTCAGTTAT/ZEN/TCTGCTGGTGCACTTGCCA/3IABkFQ/−3′.
Analysis of lung inflammation
At the indicated time points, mouse lungs were lavaged with sterile PBS for inflammatory cell differential counts. The cranial lobe of the right lung was homogenized in sterile PBS by mechanical grinding. The resulting lung homogenate was used for fungal colony counting and cytokine analysis by Lincoplex (Invitrogen, Carlsbad, CA ). The middle and caudal lobes of the right lung were homogenized under liquid nitrogen for RNA extraction using the Absolutely RNA miniprep kit (Agilent Technologies, Santa Clara, CA). RNA was reverse transcribed into cDNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA), which was assayed by real-time PCR for gene expression with Assay on Demand TaqMan primer and probe sets (Life Technologies, Grand Island, NY).(13) The left lung was fixed in 10% neutral buffered formalin for histopathology scoring of H&E stained sections. Scoring was conducted by two sample blinded scorers using a 0–4 scale (4 being the most inflammation, 0 the least) for parenchymal, peribronchial, and perivascular inflammation.
Flow cytometry
Whole mouse lungs were digested with collagenase as previously described. Cells were stained with fluorescent conjugated antibodies for F480, CD11b, CD11c, Ly6G, and Siglec-F (BD Biosciences). Gating strategy can be found in Supplemental Figure 2.
Blood cell quantification
Blood was taken from the heart via cardiac puncture upon tissue collection, placed into K2-EDTA tubes, and assayed within 30 min of recovery using a Hemavet 950FS hematology system (Drew Scientific, Miami Lakes, FL).
Statistical Analysis
All of the data are presented as the mean +/− SEM. Significance was tested by unpaired t test (for two means) or one-way ANOVA (for multiple data groups) followed by Tukey posthoc test. Data was analyzed using the GraphPad Prism software package.
Results
Preceding influenza A infection exacerbates A. fumigatus infection in the lung
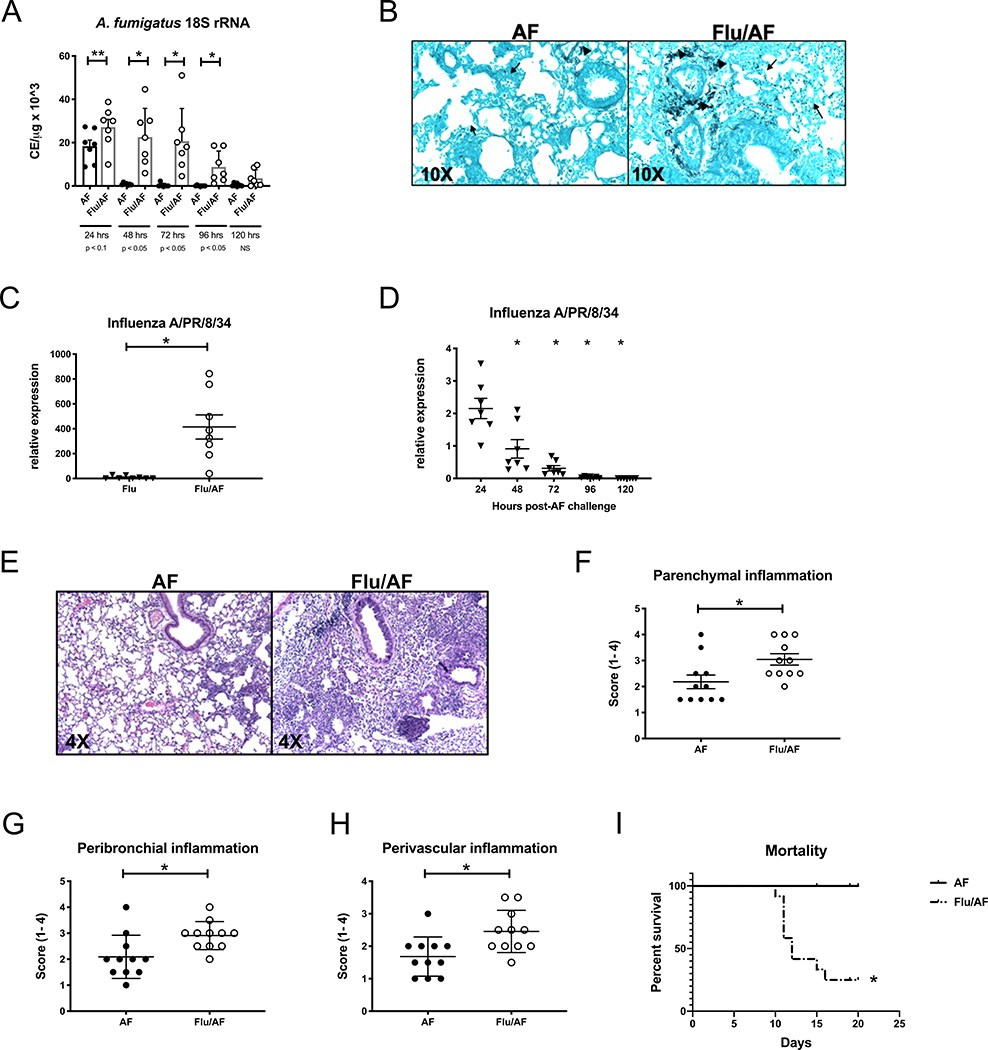
Recent reports have established the clinical link between influenza infection and the development of IPA (7, 9, 14–18). These data support the concept that super-infection with A. fumigatus, leading to invasive pulmonary aspergillosis, may be a severe consequence of influenza. To model this hypothesis in an animal, C57BL/6J male mice were challenged with a sublethal dose of influenza A PR/8/34 H1N1 (100 pfu) for 6 days followed by 2.5 × 107 A. fumigatus (ATCC strain 42202) conidia and after 24 – 120 hours, fungal burden was assessed. Mice were challenged with A. fumigatus on day 6 post-influenza infection to mimic severe influenza in critically ill patients and this timing corresponds to the timing of aspergillus diagnosis after influenza diagnosis (mean 5 days) based on human epidemiologic data(8). Preceding influenza infection resulted in increased fungal burden in the lung at 24 – 96 hours post-A. fumigatus challenge (Figure 1A). We observed the greatest difference in fungal burden between the post-influenza IPA and singularly infected mice at 48 hours post-A. fumigatus challenge and therefore, investigated additional endpoints at this time point. Histologic analysis using Grocott-Gomori methenamine silver (GMS) staining of lung tissue revealed both increased conidia and presence of increased germinating A. fumigatus in super-infected mice compared to singular fungal infection (Figure 1B). We observed similar increases in fungal burden when using both C57BL/6J female mice and influenza A/California/07/2009 H1N1 (Supplemental Figure 1). Viral burden was also increased in super-infected mice compared to singular viral infection (Figure 1C) and viral burden was highest in the super-infected at 24 hours post-A. fumigatus challenge, however was cleared by 120 hours post- A. fumigatus challenge (Figure 1D). Histology of lungs stained with hematoxylin and eosin demonstrated increased inflammation and lung injury in super-infected mice (Figure 1E–H). Finally, super-infected mice demonstrate increased mortality compared to mice singularly infected with A. fumigatus. Notably, the influenza inoculum that mice receive in our model is not lethal(19). These data show that preceding influenza A infection increases susceptibility to A. fumigatus super-infection in the murine lung and allows for the development of invasive pulmonary aspergillosis with increased fungal burden, increased fungal germination, cellular inflammation, lung injury, and mortality.
Figure 1. Preceding influenza A infection exacerbates Aspergillus fumigatus infection in the lung.
C57BL/6 mice were infected with 100 PFU of influenza A PR/8/34 for 6 days, then challenged with 2.5×107 resting conidia of A. fumigatus for 24–120 hours (n=7 mice). A – lung fungal burden was assessed by RT-PCR analysis of A. fumigatus 18S rRNA levels (n=7). C57BL/6 mice were infected with 100 PFU of influenza A PR/8/34 for 6 days, then challenged with 2.5×107 resting conidia of A. fumigatus for 48 hours (n=8 mice). B – Representative histology of GMS stained left lung sections. C – Influenza matrix protein expression in lung RNA (n=8). C57BL/6 mice were infected with 100 PFU of influenza A PR/8/34 for 6 days, then challenged with 2.5×107 resting conidia of A. fumigatus for 24–120 hours (n=7 mice). D – Influenza matrix protein expression in lung RNA (n=7). C57BL/6 mice were infected with 100 PFU of influenza A PR/8/34 for 6 days, then challenged with 2.5×107 resting conidia of A. fumigatus for 48 hours (n=11 mice). (E) Representative histology of H&E-stained left lung sections. Arrows indicate blood vessels, and arrowheads indicate airways. (F-H) Scoring of parenchymal, peribronchial, and perivascular inflammation (n = 11). (I) Survival curves of mice that received 5 3 107 resting conidia of A. fumigatus on day 6 postinfluenza or PBS control (n = 12). Significance was tested by unpaired t test or one-way ANOVA. Each experiment was independently performed two to three times and data are shown from combined experiments. *p, 0.05, **p, 0.1.
Preceding influenza A infection decreases neutrophils in the lungs and airway in response to secondary A. fumigatus infection
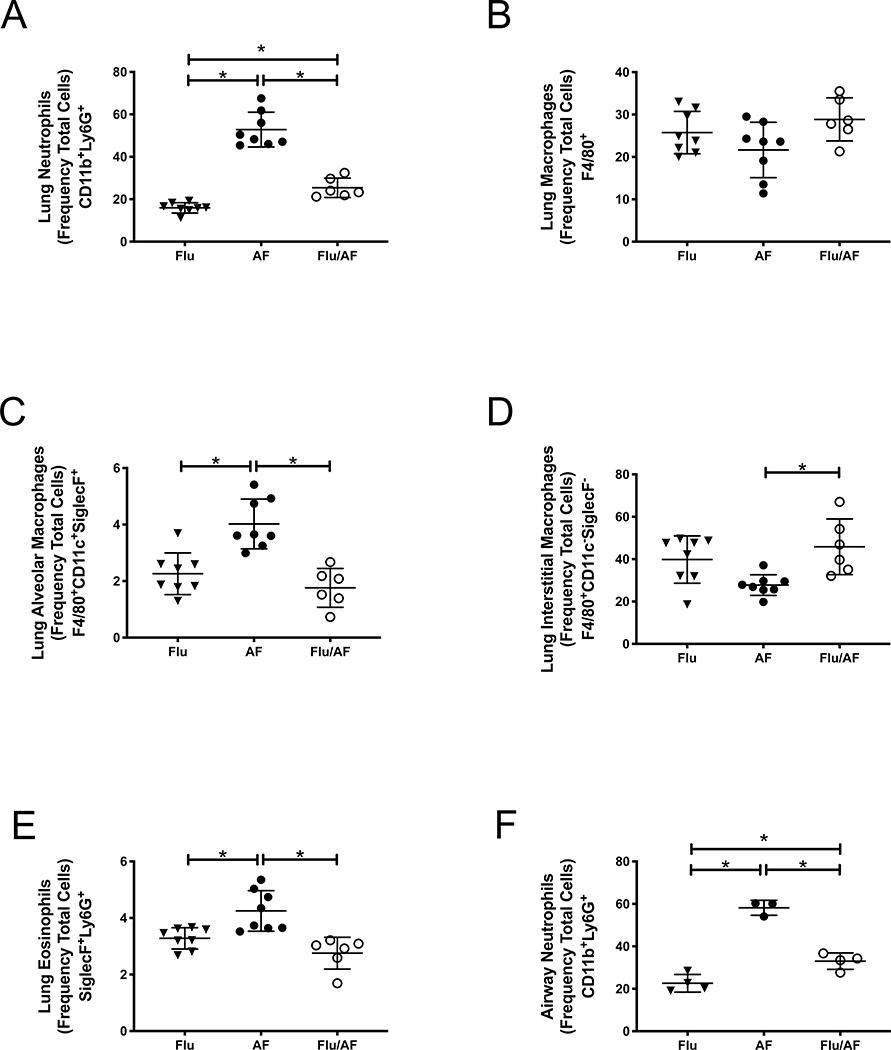
To investigate the host response to A. fumigatus during influenza infection, we performed flow cytometry of lung tissue at 48 hours post- A. fumigatus challenge. We observed decreased neutrophils (CD11b+Ly6G+) in the lungs of super-infected mice compared to singular infection with A. fumigatus (Figure 2A). We did not observe a difference in macrophages (F4/80+) between groups; however, there were decreased numbers of alveolar macrophages (F4/80+CD11c+SiglecF+) and increased numbers of interstitial macrophages (F4/80+CD11c−SiglecF−) in super-infected mice compared to those singularly infected with A. fumigatus (Figure 2B–D). There was also a significant decrease in eosinophils in the super-infected mice (Figure 2E). Next, we performed flow cytometry of bronchoalveolar lavage fluid at 48 hours post- A. fumigatus challenge and also observed decreased neutrophils (CD11b+Ly6G+) in the airway of super-infected mice compared to singular infection with A. fumigatus (Figure 2F). These data suggest that preceding influenza alters the lung inflammatory response to secondary A. fumigatus infection and that fewer neutrophils migrate to the lung and airway during post-influenza IPA, potentially allowing for increased fungal burden.
Figure 2. Preceding influenza A infection decreases neutrophils in the lungs and airway in response to secondary A. fumigatus infection.
C57BL/6 mice were infected with 100 PFU of influenza A PR/8/34 for 6 days, then challenged with 2.5×107 resting conidia of A. fumigatus for 48 hours (n=6–8 mice). A – CD11b+Ly6G+ cells in the lung by flow cytometry (n=6–8). B –F4/80+ cells in the lung by flow cytometry (n=6–8). C- F480+CD11c+SiglecF+cells in the lung by flow cytometry (n=6–8). D - F480+CD11c−SiglecF− cells in the lung by flow cytometry (n=6–8). E - SiglecF+Ly6G+ cells in the lung by flow cytometry (n=6–8). F - CD11b+Ly6G+ cells in the bronchoalveolar lavage fluid by flow cytometry (n=3–4).* p < 0.05. Significance was tested by one-way ANOVA. Each experiment was independently performed 2 times and data are shown from combined experiments with the exception of Panel F which was performed once.
Mice are not neutropenic during post-influenza invasive pulmonary aspergillosis, but CXCL1 and CXCL2 are decreased in the lung
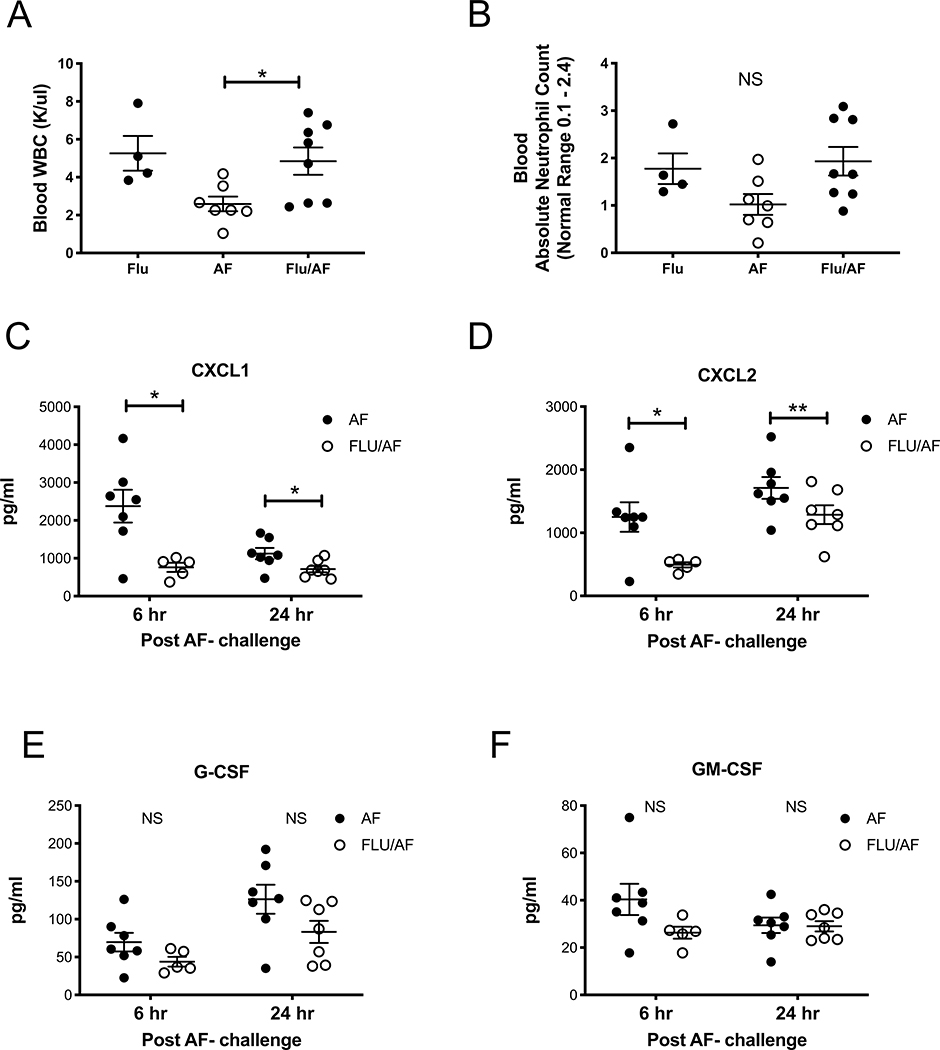
To determine if super-infected mice had fewer neutrophils in the blood, or were potentially neutropenic during post-influenza invasive pulmonary aspergillosis, resulting in fewer neutrophils migrating to the lung, we performed complete blood counts and measured both total white blood cells and absolute neutrophil counts in our mouse groups. We observed higher levels of white blood cells in super-infected mice compared to those singularly infected with A. fumigatus (Figure 3A). Additionally, there was no significant difference in absolute neutrophil counts; however, there was a trend (p=0.07) towards more neutrophils in the super-infected mice (Figure 3B). With a robust neutrophil response to super-infection seen in the blood, we next measured neutrophil chemoattractants in the lung. We observed decreased levels of both CXCL1 and CXCL2 in super-infected mice at early time points, 6 and 24 hours, following fungal challenge. There was no difference in G-CSF or GM-CSF. These data demonstrate that post-influenza IPA does not affect neutrophil numbers in blood, and suggests that bone marrow is adequately responding to infection; however, neutrophil recruitment to the lung is inhibited and the mechanism may be related to decreased neutrophil recruiting chemokines in the lung.
Figure 3. CXCL1 and CXCL2 are decreased in the lung during post-influenza invasive pulmonary aspergillosis.
C57BL/6 mice were infected with 100 PFU of influenza A PR/8/34 for 6 days, then challenged with 2.5×107 resting conidia of A. fumigatus for 48 hours (n=4–8 mice). A – Complete blood count quantification of total white bloods (n=4–8). B - Complete blood count quantification of absolute neutrophil count (n=4–8). C57BL/6 mice were infected with influenza and A. fumigatus for 6–24 hours (n=5–7 mice). C-F - Protein production of chemokines in lung homogenate as measured by Lincoplex (n=5–7). * p < 0.05. Significance was tested by unpaired t test or one-way ANOVA. Each experiment was independently performed 2 times and data are shown from combined experiments.
STAT1 signaling is required for the development of post-influenza invasive pulmonary aspergillosis
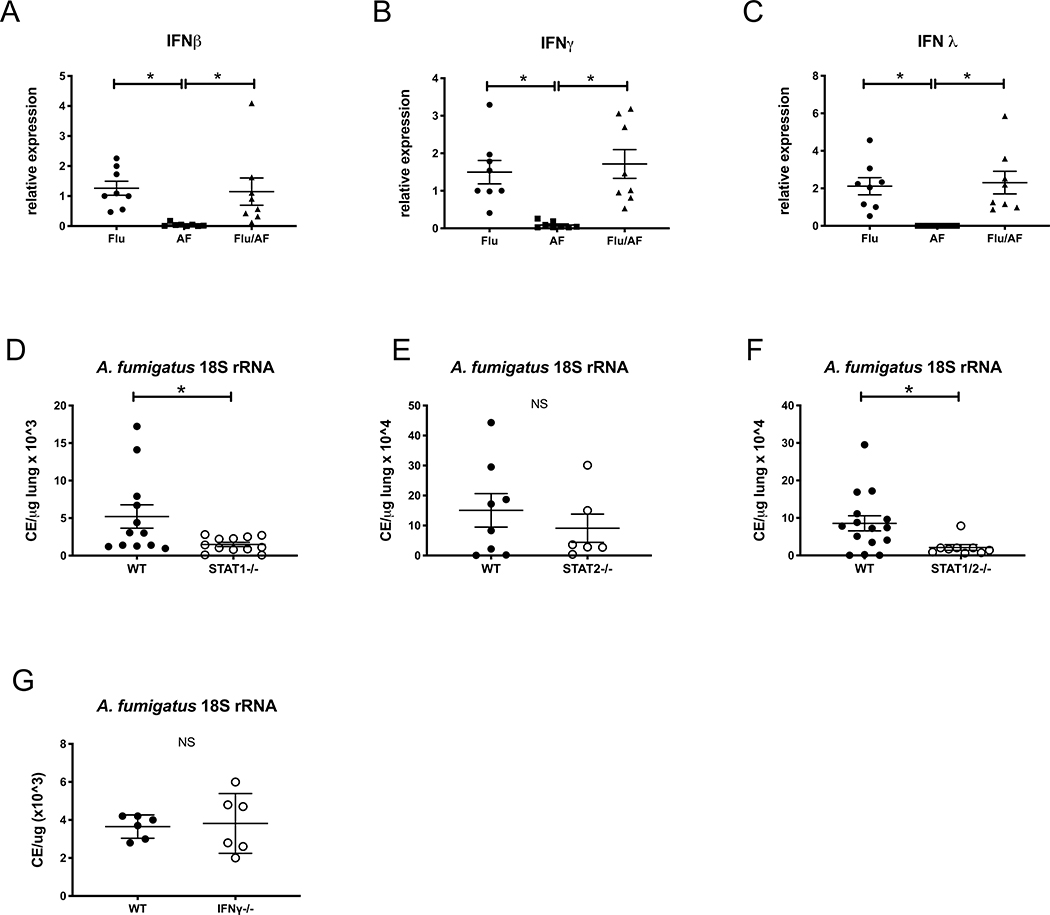
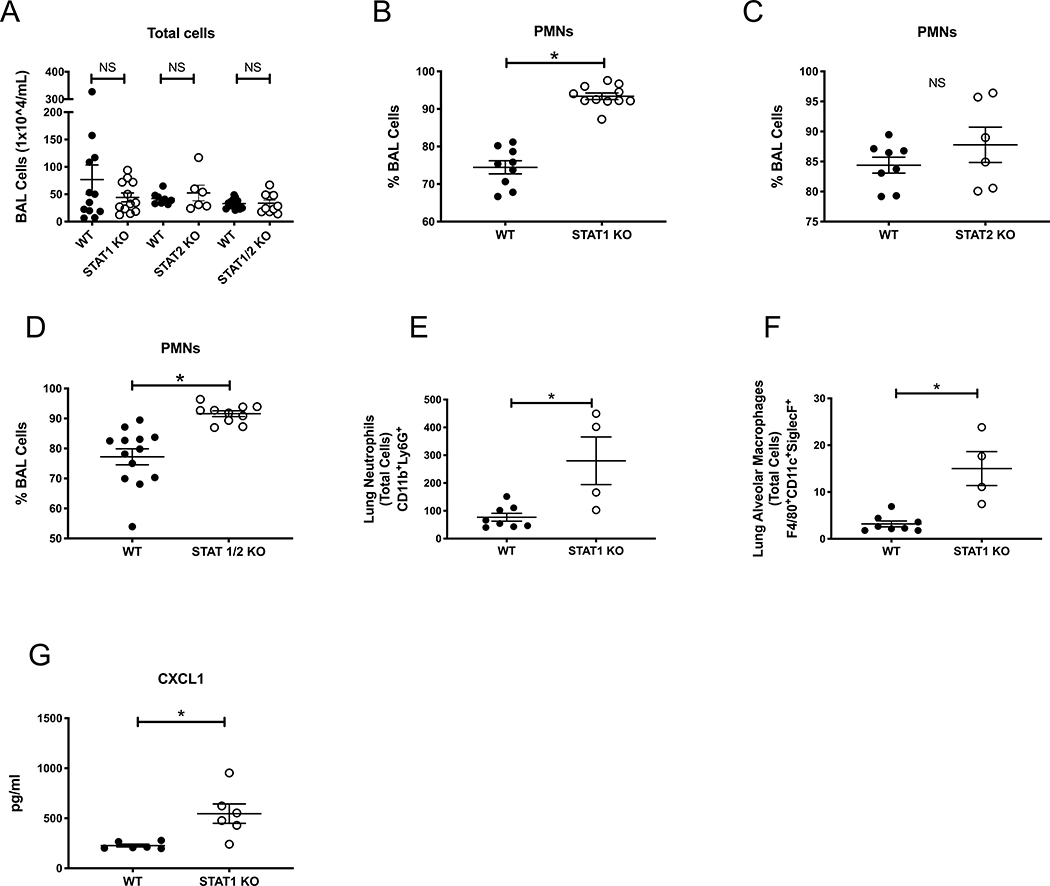
To determine a mechanism by which influenza A inhibits neutrophil recruitment to the lung and airway during post-influenza IPA, we focused on the production of interferons during super-infection. C57BL/6J mice were challenged with a sublethal dose of influenza A PR/8/34 H1N1 (100 pfu) for 6 days followed by 2.5 × 107 A. fumigatus (ATCC strain 42202) conidia and after 48 hours, measured gene expression of interferons. We observed increased levels of IFNβ (type I IFN), IFNγ (type II IFN), and IFNλ (type III IFN) in both post-influenza IPA and singular infection with influenza compared to singular infection with A. fumigatus (Figure 4A–C). These data confirm that influenza induces interferons during post-influenza IPA at similar levels compared to influenza infection alone. Type I and type III IFNs signal through both STAT 1 and STAT 2 while type II IFNs signal through STAT 1 alone. In order to determine if STAT signaling plays a role in exacerbation of A. fumigatus in the lung, we challenged STAT1−/−, STAT2−/− and STAT1/2−/− mice with influenza A follow by A. fumigatus. Deletion of STAT1 or both STAT1 and STAT2 resulted in less fungal burden during super-infection compared to wild-type mice (Figure 4D,F), while deletion of STAT2 alone did not alter fungal burden compared to wild-type mice (Figure 4E). STAT1 deletion blocks type I, II and III interferons while STAT2 deletion blocks only type I and III interferons. STAT2 deletion does not affect type II interferons. Next, we collected bronchoalveolar lavage fluid and measured total cells in the airway of super-infected STAT1−/−, STAT2−/− and STAT1/2−/− mice and found no differences compared to wild-type controls (Figure 5A). We performed cytospin differentials and observed an increased neutrophilic response in both STAT1−/− and STAT1/2−/− mice compared to wild-type (Figure 5B,D) but no difference between STAT2−/− and wild-type mice (Figure 5C), suggesting STAT1 signaling inhibits neutrophil recruitment to the airways during post-influenza IPA. Next, we measured neutrophils and alveolar macrophages by flow cytometry in the lung of wild-type and STAT1−/− mice infected with post-influenza IPA and found an increased number of neutrophils (Figure 5E) and alveolar macrophages (Figure 5F) in the super-infected STAT1−/− mice. Finally, super-infected STAT1−/− mice had increased levels of CXCL1 compared to wild-type controls (Figure 5G).
Figure 4. STAT1 inhibits fungal clearance during post-influenza invasive pulmonary aspergillosis.
C57BL/6 mice were infected with 100 PFU of influenza A PR/8/34 for 6 days, then challenged with 2.5×107 resting conidia of A. fumigatus for 48 hours (n=8 mice). A-C – Interferon gene expression in lung tissue measured by RT-PCR (n=8). STAT1−/− and C57BL/6 mice were infected influenza and A. fumigatus for 48 hours (n=12 mice). D - lung fungal burden was assessed by RT-PCR analysis of A. fumigatus 18S rRNA levels (n=12). STAT2−/− and C57BL/6 mice were infected influenza and A. fumigatus for 48 hours (n=6–8 mice). E - lung fungal burden was assessed by RT-PCR analysis of A. fumigatus 18S rRNA levels (n=6–8). STAT1/2−/− and C57BL/6 mice were infected influenza and A. fumigatus for 48 hours (n=9–15 mice). F - lung fungal burden was assessed by RT-PCR analysis of A. fumigatus 18S rRNA levels (n=9–15). IFNγ−/− and C57BL/6 mice were infected influenza and A. fumigatus for 48 hours (n=6 mice). G - lung fungal burden was assessed by RT-PCR analysis of A. fumigatus 18S rRNA levels (n=6). * p < 0.05. Significance was tested by unpaired t test or one-way ANOVA. Each experiment was independently performed at least 2 times and data are shown from combined experiments with the exception of Panel G which was performed once.
Figure 5. STAT1 deletion increases airway and lung neutrophilia during post-influenza invasive pulmonary aspergillosis.
C57BL/6, STAT1−/−, STAT2−/− and STAT1/2−/− were infected influenza and A. fumigatus for 48 hours (n=6–15 mice). A – Bronchoalveolar lavage cell counts (n=6–15). B-D, Bronchoalveolar lavage differential neutrophils (n=6–15). C57BL/6 and STAT1−/− were infected with influenza and A. fumigatus for 48 hours (n=4–8 mice). E – CD11b+Ly6G+ total cells in the lung by flow cytometry (n=4–8). F - F480+CD11c+SiglecF+cells in the lung by flow cytometry (n=4–8). G - Protein production of chemokines in lung homogenate as measured by Lincoplex (n=6). * p < 0.05. Significance was tested by unpaired t test or one-way ANOVA. Each experiment was independently performed 2 times and data are shown from combined experiments with the exception of Panels E-G data, which were performed once.
Discussion
The findings of this study demonstrate a cellular mechanism by which influenza A infection impairs pulmonary host defense against secondary A. fumigatus challenge. Post-influenza IPA has been described in previously immunocompetent hospitalized patients yet remains relatively understudied. In our current study, we demonstrate for the first time that post-influenza IPA can be effectively modeled in mice.
The immune response to A. fumigatus is complex and includes coordination between macrophages and neutrophils. Although both neutrophils and alveolar macrophages are important to fungal host defense, we chose to focus primarily on neutrophils as prior murine investigation suggests that neutrophils, but not alveolar macrophages provide essential host defense against A. fumigatus conidia(25). Neutrophils have been shown to be essential for clearing aspergillus infection in humans. The duration of neutropenia in patients with acute leukemia is directly related to the risk of developing invasive pulmonary aspergillosis (26). During infection, the murine orthologs of IL-8, CXCL1 and CXCL2, are secreted primarily by resident tissue macrophages within one to two hours of infection (29). CXCL1 and CXCL2 are both strong neutrophil chemoattractants that signal through CXC chemokine Receptor-2 (CXCR2).
Interestingly, we observed a decreased neutrophil response in both the lungs and airway following A. fumigatus challenge during influenza viral infection compared to singular A. fumigatus challenge. Other investigators have shown that neutrophil numbers peak in the murine airway at Day 5 post-influenza PR8 and are decreased back to naïve levels by Day 7 post-influenza(30). In our studies, influenza-infected mice receive secondary challenge with A. fumigatus on Day 6 post-influenza and neutrophil numbers in the airway are likely decreasing by Day 6 of influenza infection. We initially hypothesized that post-influenza IPA mice would have decreased neutrophils in the blood compared to A. fumigatus-infected mice, potentially as a result of being able to mobilize fewer neutrophils from the bone marrow in the context of influenza infection; however, mice in all three infection groups had similar absolute neutrophil counts in the blood. These data suggest that the decreased neutrophils seen in the airway and lung during post-influenza IPA are related to decreased neutrophil recruitment to the lung.
The secretion of chemokines that induce chemotactic migration of neutrophils to the lung is the first critical step for recruitment of neutrophils to the lung(31). Murine ELR+ CXC chemokines include MIP-2 (CXCL2), KC (CXCL1), lipopolysaccharide-induced CXC chemokine (LIX/CXCL5), and Lungkine (CXCL15) and are potent neutrophil chemoattractants. Neutralization of the common ELR+ CXC receptor, CXCR2, results in attenuation of neutrophil recruitment to the lung, increased hyphal germination, and increased mortality as a result of IPA. Both CXCL1 and CXCL2 are increased in immunocompetent and immunosuppressed mice in response to A. fumigatus infection(33). In our current study, CXCL1 and CXCL2 are decreased at both 6 and 24 hours post A. fumigatus challenge, likely contributing to the decreased neutrophil response seen in the lungs and airway at 48 hours post A. fumigatus challenge. CXCL1 and CXCL2 are secreted primarily by resident tissue macrophages within 1–2 hours of infection(29) and the decreased alveolar macrophages seen in post-influenza IPA could be linked to the decreased recruitment of neutrophils to the lung. Interestingly, we measured both increased alveolar macrophages and increased CXCL1 in the STAT1−/− mice compared to wild-type mice. The relationship between alveolar macrophages and neutrophils in post-influenza IPA should be explored in future mechanistic studies. Influenza A virus is known to deplete alveolar macrophages by seven days post-infection in BALB/c mice but not in C57BL/6J mice(34, 35).
Notably, G-CSF and GM-CSF levels were not significantly different between post-influenza IPA and singular infection with A. fumigatus. G-CSF has been used in murine models of systemic aspergillosis and does not affect survival when administered alone but has a synergistic effect when given in combination with antifungals(36). GM-CSF plays a critical role in host defense against A. fumigatus and mice lacking the GM-CSF receptor β chain develop invasive fungal growth and decreased survival after challenge with A. fumigatus; however lung neutrophils are similar in wild-type and GM-CSFRβ−/− mice at 48 hours after infection(37). Neutrophil sequestration in the lung involves multiple steps: activation of transcription factors, production of chemokines, upregulation of cell adhesion molecules and increased cell-to-cell interactions. Production of chemokines CXCL1 and CXCL2 are decreased in post-influenza IPA, but the other components of neutrophil recruitment and sequestration in the lung warrant further investigation in future studies.
Recent studies have been directed at investigating the role of interferons during A. fumigatus infection. Espinosa et. al. identified that CCR2+ monocytes are a source of type I interferons during A. fumigatus infection and prime optimal expression of type III interferons. Type III interferons act on neutrophils to enhance fungicidal activity against A. fumigatus. Mice lacking both type I and III IFN receptors are susceptible to A. fumigatus and have reduced recruitment of neutrophils. Neutrophil specific deletion of STAT1 increase susceptibility to A. fumigatus and suggests that fungal host defense requires expression of STAT1 on neutrophils(38). Type II interferons have also been studied during A. fumigatus infection and interestingly, have been found to have variable effects on invasive aspergillosis. In vitro, IFNγ enhanced phagocytosis and killing by neutrophils. In vivo, systemic IFNγ did not improve phagocytosis or killing by neutrophils, or affect mortality. Local nasal IFNγ enhanced phagocytic recruitment to the lungs but worsened mortality(39). IFNγ has also been used as adjunctive therapy to standard antifungal treatment in humans and improved outcomes(40). IFNγ has also been found to improve outcomes in mice with systemic aspergillosis(41). In our current studies, interferons were highly induced, as expected, in mice that received influenza compared to singular infection with aspergillus. During post-influenza IPA, deletion of STAT1 resulted in decreased fungal burden and increased lung and airway neutrophilia compared to wild-type mice while deletion of STAT2 did not change fungal burden or airway neutrophilia compared to wild-type mice. These data suggest that STAT1 signaling inhibits neutrophil recruitment and increases susceptibility to post-influenza IPA. Additionally, deletion of STAT1 resulted in increased CXCL1 levels, indicating that STAT1 signaling impairs CXCL1 production during post-influenza IPA. STAT2 signaling could be necessary but not sufficient to alter fungal burden during post-influenza IPA and this is another area that we could investigate in the future. Type I, II, and III interferons all signal through STAT1 while only Type I and III interferons signal through STAT2. IFNγ−/− (Type II IFN) mice were not more or less susceptible to post-influenza IPA and future studies will need to be conducted to examine the role of other cytokines that signal through STAT1 such as IL-27, IL-6, and the role of individual interferons.
Although post-influenza IPA remains poorly understood, a significant body of literature currently exists regarding post-influenza bacterial pneumonia. Neutrophils are recruited to the lung during post-influenza bacterial super-infection(42, 43) and are critical to bacterial clearance during influenza at early time points following bacterial challenge(44) but during the period of enhanced susceptibility to bacterial super-infection at days 6–7 post-influenza, neutrophil depletion does not affect bacterial clearance(45, 46). Impaired bacterial killing by neutrophils has been shown to occur through influenza-induced decreased levels of reactive oxygen species(47), suggesting that neutrophil function plays a role during post-influenza bacterial pneumonia. Influenza-induced type I IFN signaling inhibits production of CXCL1 and CXCL2 and impairs neutrophil recruitment to the airway in response to pneumococcal pneumonia(48). Mice lacking STAT1 signaling are protected from post-influenza staphylococcal pneumonia and have increased neutrophils in the airways during both singular influenza infection and influenza, MRSA super-infection(49). Our observations in a post-influenza IPA model suggest that STAT1 signaling plays a detrimental role through the inhibition of CXCL1 and decreased neutrophil recruitment to the lung. Influenza A-induced interferons potentially inhibit CXCL1 production during post-influenza IPA. These initial findings underscore the need for additional investigation to elucidate the mechanisms that result in the development of post-influenza IPA.
Post-influenza IPA is associated with high mortality and understanding how preceding influenza predisposes to secondary fungal infection through examination of the molecular and cellular pathways involved in host defense is critical. Future studies are needed to delineate the specific cell types and immune pathways necessary for fungal host defense. Both neutrophils and interferons play key roles in fungal host defense and additional examination is needed to elucidate the complicated interplay between viral and fungal infections. There may be therapeutic potential for interferons during fungal infections but the exact pathways and degree of interferon induction and interferon route administration requires additional investigation. Identifying severe cases of viral and fungal infection and increased understanding of how the two organisms modulate host defense will be important to patient outcomes.
Supplementary Material
Key Points.
Post-influenza IPA can be effectively modeled in mice
Neutrophil recruitment to the lung is decreased in post-influenza IPA
STAT1 signaling increases susceptibility to post-influenza IPA
Acknowledgments
Funding sources include NHLBI R01HL107380 (JFA), NHLBI K08HL133445 (KMR), and Samuel and Emma Winters Foundation (KMR)
References
- 1.Beumer MC, Koch RM, van Beuningen D, OudeLashof AM, van de Veerdonk FL, Kolwijck E, van der Hoeven JG, Bergmans DC, and Hoedemaekers CWE. 2019. Influenza virus and factors that are associated with ICU admission, pulmonary co-infections and ICU mortality. J Crit Care 50: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verweij PE, Bruggemann RJM, Wauters J, Rijnders BJA, Chiller T, and van de Veerdonk FL. 2020. Influenza Coinfection: Be(a)ware of Invasive Aspergillosis. Clin Infect Dis 70: 349–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Latge JP, and Chamilos G. 2019. Aspergillus fumigatus and Aspergillosis in 2019. Clin Microbiol Rev 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denning DW, Park S, Lass-Florl C, Fraczek MG, Kirwan M, Gore R, Smith J, Bueid A, Moore CB, Bowyer P, and Perlin DS. 2011. High-frequency triazole resistance found In nonculturable Aspergillus fumigatus from lungs of patients with chronic fungal disease. Clin Infect Dis 52: 1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kosmidis C, and Denning DW. 2015. The clinical spectrum of pulmonary aspergillosis. Thorax 70: 270–277. [DOI] [PubMed] [Google Scholar]
- 6.Shah MM, Hsiao EI, Kirsch CM, Gohil A, Narasimhan S, and Stevens DA. 2018. Invasive pulmonary aspergillosis and influenza co-infection in immunocompetent hosts: case reports and review of the literature. Diagn Microbiol Infect Dis 91: 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schauwvlieghe A, Rijnders BJA, Philips N, Verwijs R, Vanderbeke L, Van Tienen C, Lagrou K, Verweij PE, Van de Veerdonk FL, Gommers D, Spronk P, Bergmans D, Hoedemaekers A, Andrinopoulou ER, van den Berg C, Juffermans NP, Hodiamont CJ, Vonk AG, Depuydt P, Boelens J, Wauters J, and g. Dutch-Belgian Mycosis study. 2018. Invasive aspergillosis in patients admitted to the intensive care unit with severe influenza: a retrospective cohort study. Lancet Respir Med 6: 782–792. [DOI] [PubMed] [Google Scholar]
- 8.Vanderbeke L, Spriet I, Breynaert C, Rijnders BJA, Verweij PE, and Wauters J. 2018. Invasive pulmonary aspergillosis complicating severe influenza: epidemiology, diagnosis and treatment. Curr Opin Infect Dis 31: 471–480. [DOI] [PubMed] [Google Scholar]
- 9.Wauters J, Baar I, Meersseman P, Meersseman W, Dams K, De Paep R, Lagrou K, Wilmer A, Jorens P, and Hermans G. 2012. Invasive pulmonary aspergillosis is a frequent complication of critically ill H1N1 patients: a retrospective study. Intensive Care Med 38: 1761–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowman JC, Abruzzo GK, Anderson JW, Flattery AM, Gill CJ, Pikounis VB, Schmatz DM, Liberator PA, and Douglas CM. 2001. Quantitative PCR assay to measure Aspergillus fumigatus burden in a murine model of disseminated aspergillosis: demonstration of efficacy of caspofungin acetate. Antimicrob Agents Chemother 45: 3474–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson KM, Ramanan K, Clay ME, McHugh KJ, Rich HE, and Alcorn JF. 2018. Novel protective mechanism for interleukin-33 at the mucosal barrier during influenza-associated bacterial superinfection. Mucosal Immunol 11: 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van der Velden J, Janssen-Heininger YM, Mandalapu S, Scheller EV, Kolls JK, and Alcorn JF. 2012. Differential requirement for c-Jun N-terminal kinase 1 in lung inflammation and host defense. PLoS One 7: e34638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson KM, Ramanan K, Tobin JM, Nickolich KL, Pilewski MJ, Kallewaard NL, Sellman BR, Cohen TS, and Alcorn JF. 2019. Survival during influenza-associated bacterial superinfection improves following viral- and bacterial-specific monoclonal antibody treatment. JCI Insight 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van de Groep K, Verboom DM, van de Veerdonk FL, Haas PA, van der Poll T, Schultz MJ, Bonten MJM, Cremer OL, and consortium M. 2019. Detection of Invasive Aspergillosis in Critically Ill Patients with Influenza: the Role of Plasma Galactomannan. Am J Respir Crit Care Med. [DOI] [PubMed] [Google Scholar]
- 15.van de Veerdonk FL, Kolwijck E, Lestrade PP, Hodiamont CJ, Rijnders BJ, van Paassen J, Haas PJ, Oliveira Dos Santos C, Kampinga GA, Bergmans DC, van Dijk K, de Haan AF, van Dissel J, van der Hoeven HG, Verweij PE, and Dutch Mycoses Study G. 2017. Influenza-Associated Aspergillosis in Critically Ill Patients. Am J Respir Crit Care Med 196: 524–527. [DOI] [PubMed] [Google Scholar]
- 16.Crum-Cianflone NF 2016. Invasive Aspergillosis Associated With Severe Influenza Infections. Open Forum Infect Dis 3: ofw171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ku YH, Chan KS, Yang CC, Tan CK, Chuang YC, and Yu WL. 2017. Higher mortality of severe influenza patients with probable aspergillosis than those with and without other coinfections. J Formos Med Assoc 116: 660–670. [DOI] [PubMed] [Google Scholar]
- 18.Huang L, Zhang N, Huang X, Xiong S, Feng Y, Zhang Y, Li M, and Zhan Q. 2019. Invasive pulmonary aspergillosis in patients with influenza infection: A retrospective study and review of the literature. Clin Respir J 13: 202–211. [DOI] [PubMed] [Google Scholar]
- 19.Pociask DA, Robinson KM, Chen K, McHugh KJ, Clay ME, Huang GT, Benos PV, Janssen-Heininger YMW, Kolls JK, Anathy V, and Alcorn JF. 2017. Epigenetic and Transcriptomic Regulation of Lung Repair during Recovery from Influenza Infection. Am J Pathol 187: 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.2017. Stop neglecting fungi. Nat Microbiol 2: 17120. [DOI] [PubMed] [Google Scholar]
- 21.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, and White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4: 165rv113. [DOI] [PubMed] [Google Scholar]
- 22.Denning DW, Pleuvry A, and Cole DC. 2013. Global burden of allergic bronchopulmonary aspergillosis with asthma and its complication chronic pulmonary aspergillosis in adults. Med Mycol 51: 361–370. [DOI] [PubMed] [Google Scholar]
- 23.Ostrosky-Zeichner L, Casadevall A, Galgiani JN, Odds FC, and Rex JH. 2010. An insight into the antifungal pipeline: selected new molecules and beyond. Nat Rev Drug Discov 9: 719–727. [DOI] [PubMed] [Google Scholar]
- 24.Denning DW, and Hope WW. 2010. Therapy for fungal diseases: opportunities and priorities. Trends Microbiol 18: 195–204. [DOI] [PubMed] [Google Scholar]
- 25.Mircescu MM, Lipuma L, van Rooijen N, Pamer EG, and Hohl TM. 2009. Essential role for neutrophils but not alveolar macrophages at early time points following Aspergillus fumigatus infection. J Infect Dis 200: 647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerson SL, Talbot GH, Hurwitz S, Strom BL, Lusk EJ, and Cassileth PA. 1984. Prolonged granulocytopenia: the major risk factor for invasive pulmonary aspergillosis in patients with acute leukemia. Ann Intern Med 100: 345–351. [DOI] [PubMed] [Google Scholar]
- 27.Gazendam RP, van Hamme JL, Tool AT, Hoogenboezem M, van den Berg JM, Prins JM, Vitkov L, van de Veerdonk FL, van den Berg TK, Roos D, and Kuijpers TW. 2016. Human Neutrophils Use Different Mechanisms To Kill Aspergillus fumigatus Conidia and Hyphae: Evidence from Phagocyte Defects. J Immunol 196: 1272–1283. [DOI] [PubMed] [Google Scholar]
- 28.Feldmesser M 2006. Role of neutrophils in invasive aspergillosis. Infect Immun 74: 6514–6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Filippo K, Henderson RB, Laschinger M, and Hogg N. 2008. Neutrophil chemokines KC and macrophage-inflammatory protein-2 are newly synthesized by tissue macrophages using distinct TLR signaling pathways. J Immunol 180: 4308–4315. [DOI] [PubMed] [Google Scholar]
- 30.Tate MD, Ioannidis LJ, Croker B, Brown LE, Brooks AG, and Reading PC. 2011. The role of neutrophils during mild and severe influenza virus infections of mice. PLoS One 6: e17618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukacs NW, Hogaboam C, Campbell E, and Kunkel SL. 1999. Chemokines: function, regulation and alteration of inflammatory responses. Chem Immunol 72: 102–120. [DOI] [PubMed] [Google Scholar]
- 32.Phadke AP, and Mehrad B. 2005. Cytokines in host defense against Aspergillus: recent advances. Med Mycol 43 Suppl 1: S173–176. [DOI] [PubMed] [Google Scholar]
- 33.Mehrad B, Strieter RM, Moore TA, Tsai WC, Lira SA, and Standiford TJ. 1999. CXC chemokine receptor-2 ligands are necessary components of neutrophil-mediated host defense in invasive pulmonary aspergillosis. J Immunol 163: 6086–6094. [PubMed] [Google Scholar]
- 34.Ghoneim HE, Thomas PG, and McCullers JA. 2013. Depletion of alveolar macrophages during influenza infection facilitates bacterial superinfections. J Immunol 191: 1250–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Califano D, Furuya Y, and Metzger DW. 2018. Effects of Influenza on Alveolar Macrophage Viability Are Dependent on Mouse Genetic Strain. J Immunol 201: 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sionov E, Mendlovic S, and Segal E. 2005. Experimental systemic murine aspergillosis: treatment with polyene and caspofungin combination and G-CSF. J Antimicrob Chemother 56: 594–597. [DOI] [PubMed] [Google Scholar]
- 37.Kasahara S, Jhingran A, Dhingra S, Salem A, Cramer RA, and Hohl TM. 2016. Role of Granulocyte-Macrophage Colony-Stimulating Factor Signaling in Regulating Neutrophil Antifungal Activity and the Oxidative Burst During Respiratory Fungal Challenge. J Infect Dis 213: 1289–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Espinosa V, Dutta O, McElrath C, Du P, Chang YJ, Cicciarelli B, Pitler A, Whitehead I, Obar JJ, Durbin JE, Kotenko SV, and Rivera A. 2017. Type III interferon is a critical regulator of innate antifungal immunity. Sci Immunol 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson CP, Edmiston CE Jr., Zhu YR, Adams MB, Roza AM, and Kurup V. 2005. A murine model of invasive aspergillosis: variable benefit of interferon-gamma administration under in vitro and in vivo conditions. Surg Infect (Larchmt) 6: 397–407. [DOI] [PubMed] [Google Scholar]
- 40.Delsing CE, Gresnigt MS, Leentjens J, Preijers F, Frager FA, Kox M, Monneret G, Venet F, Bleeker-Rovers CP, van de Veerdonk FL, Pickkers P, Pachot A, Kullberg BJ, and Netea MG. 2014. Interferon-gamma as adjunctive immunotherapy for invasive fungal infections: a case series. BMC Infect Dis 14: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagai H, Guo J, Choi H, and Kurup V. 1995. Interferon-gamma and tumor necrosis factor-alpha protect mice from invasive aspergillosis. J Infect Dis 172: 1554–1560. [DOI] [PubMed] [Google Scholar]
- 42.Kudva A, Scheller EV, Robinson KM, Crowe CR, Choi SM, Slight SR, Khader SA, Dubin PJ, Enelow RI, Kolls JK, and Alcorn JF. 2011. Influenza A inhibits Th17-mediated host defense against bacterial pneumonia in mice. J Immunol 186: 1666–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCullers JA, and Rehg JE. 2002. Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet-activating factor receptor. J Infect Dis 186: 341–350. [DOI] [PubMed] [Google Scholar]
- 44.Rynda-Apple A, Robinson KM, and Alcorn JF. 2015. Influenza and Bacterial Superinfection: Illuminating the Immunologic Mechanisms of Disease. Infect Immun 83: 3764–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson KM, McHugh KJ, Mandalapu S, Clay ME, Lee B, Scheller EV, Enelow RI, Chan YR, Kolls JK, and Alcorn JF. 2014. Influenza A virus exacerbates Staphylococcus aureus pneumonia in mice by attenuating antimicrobial peptide production. J Infect Dis 209: 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McNamee LA, and Harmsen AG. 2006. Both influenza-induced neutrophil dysfunction and neutrophil-independent mechanisms contribute to increased susceptibility to a secondary Streptococcus pneumoniae infection. Infect Immun 74: 6707–6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun K, and Metzger DW. 2014. Influenza infection suppresses NADPH oxidase-dependent phagocytic bacterial clearance and enhances susceptibility to secondary methicillin-resistant Staphylococcus aureus infection. J Immunol 192: 3301–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shahangian A, Chow EK, Tian X, Kang JR, Ghaffari A, Liu SY, Belperio JA, Cheng G, and Deng JC. 2009. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J Clin Invest 119: 1910–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee B, Gopal R, Manni ML, McHugh KJ, Mandalapu S, Robinson KM, and Alcorn JF. 2017. STAT1 Is Required for Suppression of Type 17 Immunity during Influenza and Bacterial Superinfection. Immunohorizons 1: 81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.