Abstract
Purpose
The U.S. response to the SARS-CoV-2 epidemic has been hampered by early and ongoing delays in testing for infection; without data on where infections were occurring and the magnitude of the epidemic, early public health responses were not data-driven. Understanding the prevalence of SARS-CoV-2 infections and immune response is critical to developing and implementing effective public health responses. Most serological surveys have been limited to localities that opted to conduct them and/or were based on convenience samples. Moreover, results of antibody testing might be subject to high false positive rates in the setting of low prevalence of immune response and imperfect test specificity.
Methods
We will conduct a national serosurvey for SARS-CoV-2 PCR positivity and immune experience. A probability sample of U.S. addresses will be mailed invitations and kits for the self-collection of anterior nares swab and finger prick dried blood spot specimens. Within each sampled household, one adult 18 years or older will be randomly selected and asked to complete a questionnaire and to collect and return biological specimens to a central laboratory. Nasal swab specimens will be tested for SARS-CoV-2 RNA by RNA PCR; dried blood spot specimens will be tested for antibodies to SARS-CoV-2 (i.e., immune experience) by enzyme-linked immunoassays. Positive screening tests for antibodies will be confirmed by a second antibody test with different antigenic basis to improve predictive value of positive (PPV) antibody test results. All persons returning specimens in the baseline phase will be enrolled into a follow-up cohort and mailed additional specimen collection kits 3 months after baseline. A subset of 10% of selected households will be invited to participate in full household testing, with tests offered for all household members aged ≥3 years. The main study outcomes will be period prevalence of infection with SARS-CoV-2 and immune experience, and incidence of SARS-CoV-2 infection and antibody responses.
Results
Power calculations indicate that a national sample of 4000 households will facilitate estimation of national SARS-CoV-2 infection and antibody prevalence with acceptably narrow 95% confidence intervals across several possible scenarios of prevalence levels. Oversampling in up to seven populous states will allow for prevalence estimation among subpopulations. Our 2-stage algorithm for antibody testing produces acceptable PPV at prevalence levels ≥1.0%. Including oversamples in states, we expect to receive data from as many as 9156 participants in 7495 U.S. households.
Conclusions
In addition to providing robust estimates of prevalence of SARS-CoV-2 infection and immune experience, we anticipate this study will establish a replicable methodology for home-based SARS-CoV-2 testing surveys, address concerns about selection bias, and improve positive predictive value of serology results. Prevalence estimates of SARS-CoV-2 infection and immune experience produced by this study will greatly improve our understanding of the spectrum of COVID-19 disease, its current penetration in various demographic, geographic, and occupational groups, and inform the range of symptoms associated with infection. These data will inform resource needs for control of the ongoing epidemic and facilitate data-driven decisions for epidemic mitigation strategies.
Keywords: SARS-CoV-2 infection, Probability sampling methods, PCR testing, Serology, SARS-CoV-2 serology
Introduction
The global pandemic of SARS-CoV-2 and its associated illness (coronavirus disease 2019, or COVID-19) have emerged very quickly, challenging traditional systems of clinical and public health response [1,2]. There is broad consensus that the U.S. response to the COVID-19 epidemic has been hampered by lack of adequate testing for SARS-CoV-2 [[3], [4], [5], [6]]. Globally, available statistics representing the scale and growth of the epidemic are based on the numbers of people diagnosed and reported with SARS-CoV-2 infections and the number of people who have died from COVID-19 disease. These measures are informative but biased: diagnoses of COVID-19 disease predominantly count people who were sufficiently sick and symptomatic that they were tested. Moreover, the data are differentially biased by time and jurisdiction. Testing policies have changed over time as test availability increases, and testing policies in heavily impacted areas may be more restrictive for people with mild illness than policies in less impacted areas. Importantly, there are limited population-based data about the proportion of people who become infected with SARS-CoV-2 who remain asymptomatic or about the proportion of people who may already possess antibodies to the virus.
Traditional public health surveillance programs that are linked to disease prevention efforts focus on diagnosing people with an infectious disease and then helping them take steps to minimize the risks of onward transmission. Surveillance data to characterize epidemics are often collected from testing and intervention programs, and surveillance data improve as public health screening and testing programs grow. In the COVID-19 epidemic, the traditional public health model in the United States has been disrupted because of how fast the epidemic emerged and limited testing and contact tracing capacity. There are limited resources for testing in terms of supplies (e.g., shortages of swabs for collection, viral transport media, and personal protective equipment for health care workers collecting invasive specimens) [7,8] and personnel to collect samples. Because of these limitations, in many areas testing resources have been focused on the sickest people, providing testing data that present an underestimate of the true extent of the epidemic and that differentially undercount mildly symptomatic and asymptomatic people. Therefore, it is critical to develop a representative depiction of the distribution of SARS-CoV-2 infection and immune experience to inform public health policies and prevention and control interventions.
The field of antibody testing is rapidly evolving, and our understanding of the clinical significance of seropositivity is limited. Local serosurveys using a variety of sampling methods have reported relatively low prevalence estimates for seropositivity [[9], [10], [11]]. For low prevalence serosurveys, the predictive value of positive antibody tests is a substantial issue. With overall prevalence findings in many surveys in the single digits, even slight performance problems in specificity could result in substantial changes in outcomes. For instance, if a serosurvey in a population with “true” prevalence less than 6% uses an assay with less than or equal to 97% specificity, most positive specimen findings would be false positives. Moreover, serological surveys are a lagging indicator of infection, with one study identifying median time to detectable seroconversion to be 13 days postexposure across antibody types [12] and another finding 15–20 days postexposure across antibody types [13]. An additional concern of some existing methods, unrelated to bias, regards the appropriateness of conducting in-person testing in the face of limited availability of testing resources for persons who are ill and limited personal protective equipment for health care workers. An optimal study design might avert use of such resources.
To provide less biased estimates of prevalence of SARS-CoV-2 virus and immune experience, we propose a study design that differs from previous studies in four critical ways. First, we propose to use address-based sampling, commonly considered to be the reference standard for developing population-representative estimates, with a sample frame that includes nearly all addresses in the United States [14,15]. Second, we will use home specimen collection and remote laboratory testing procedures, which have higher acceptability than in-person specimen collection and can reach otherwise hard-to-reach populations such as workers and persons in rural areas [16]. Third, the use of a serology screening test followed by a high-specificity confirmatory test will allow for improved predictive value of positive specimens using antibody tests that target different antigenic components. Finally, performing both viral detection and antibody testing will provide a simultaneous understanding of the prevalence of viral shedding (and potential infectiousness) and of the prevalence of antibodies (and potentially immunity). In addition to an initial assessment of prevalence, our initial survey will be a baseline for future serial rounds of viral detection and serology testing, allowing for development of population-based, minimally biased estimates of incidence SARS-CoV-2 infection and immune experience, overall and in key subgroups (e.g. racial/ethnic minorities, rural areas, specific age groups).
Methods
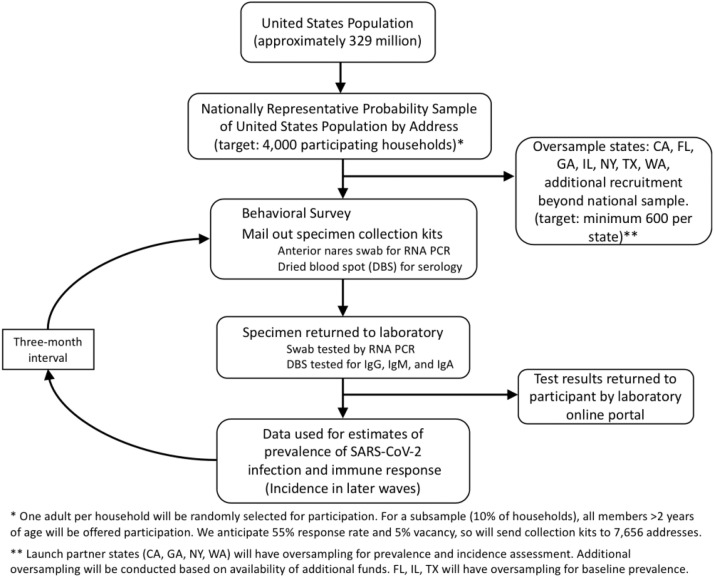
We will use a national address-based household sample to collect survey data on approximately 4000 U.S. participants by collecting survey data and self-collected specimens for SARS-CoV-2 RCA PCR and serology testing. The overall design of the study is illustrated in Figure 1 .
Fig. 1.
Schema for a national household probability sample to estimate prevalence and incidence of SARS-CoV-2 infection and immune experience.
National probability sampling frame
The study will use an address-based sampling (ABS) frame for selection of a probability-based sample, a method commonly considered the reference sampling strategy in the cell phone era, due to its complete coverage of the U.S. households when compared with telephone- and internet-based frames [14,15]. The frame is based on the USPS Computerized Delivery Sequence File that includes roughly 130 million residential addresses, covering all residential delivery points in the United States [17]. Each address is geocoded to a unique latitude and longitude before its related geodemographic data from the Census and commercial databases such as Experian are retrieved. Moreover, approximately 50% of addresses are matched to landline and/or cellular phone numbers that will allow for implementation of a multimodal retention approach and rigorous refusal conversion procedures. This frame, constructed by Marketing Systems Group from the latest Computerized Delivery Sequence File, has been previously used in numerous health research studies [[18], [19], [20], [21]]. For addresses in nonoversampling strata, each address will be selected with an equal probability of selection method to ensure the most efficient estimates at the national level. To increase geographic representation, we will use systematic random sampling, in which the frame is ordered by 9-digit ZIP+4 first. Next, a random starting point is selected and every nth address after the random start is selected. Drop units (multiunit addresses) in the frame each have a drop count, and the frame will be expanded to account for drop units.
National sampling plan
The intial sample for this study will comprise 4000 households selected across the nation using the ABS frame. We will use a two-stage sampling methodology, whereby in the first stage a representative sample of households will be selected, followed by a random selection of one adult in each sample households. We will assume a household-level response rate of 22% and anticipated 5% rate for addresses that may be vacant or otherwise unreachable at the time of survey administration for an overall yield rate of 20.9% (22 % × 95 %). Recent probability samples for COVID testing in Atlanta [22] and Indiana [23] have achieved similar participation rates. We believe this is a feasible yield rate because although contingency valuations can overestimate willingness, a recent national online survey nonetheless found 88% of respondents reported willingness to participate in COVID specimen self-collection research [24]. The total sample will therefore include at least 19,129 addresses (4000/0.209).
Invitations and kits will be sent in waves to allow adjustments for observed response rates and underrepresentation of important subgroups. If there is substantial nonresponse among racial/ethnic minority groups in wave 1, we will oversample geographic areas (e.g., Census blocks) with high representation of African American and/or Hispanic/Latinx populations in subsequent waves. Oversampling strategies will be designed with a goal of attaining sample sizes in racial/ethnic minority subgroups proportional to their size in the underlying U.S. population, facilitating robust estimation for each group. The finite population correction (N-n)/(N-1) will be virtually equal to 1, rendering the difference between sampling with or without replacement in subsequent waves a moot distinction. All waves will be pooled for analysis, and during the weighting process, design weights will be calculated to reflect any oversampling that may be used during the address selection [25,26].
State and locality oversamples
Study methods are amenable to developing locality-specific estimates. We will target a total sample size of at least n = 600 per state although oversampling, additional to the national 4000 household sample, to develop stable state-specific prevalence estimates for up to seven populous states (CA, FL, GA, IL, NY, TX, WA), contingent on availability of resources. These states represent different geographic regions of the United States and include some states that were impacted early. For these oversampled states, we will coordinate with state health departments to maximally inform local public health efforts. Depending on resource availability, we may also include additional oversampling for some states to develop enhanced estimation ability to inform their local public health needs. For instance, in GA, the total sample will include 1200 households with participation offered to all household members aged 3 years or older, allowing for greater precision of local estimation. In total, we anticipate an additional 5156 participants in 3495 households to participate in the state and full household oversamples for a national total of up to 9156 participants in 7495 households.
Household procedures
All households selected will receive a first-class letter introducing the study and its website, an evidence-based practice demonstrated to increase survey response [27]. The study website, hosted on an academic server and featuring multimedia content, will provide basic information regarding the study, address common participant questions, and encourage participant confidence in the study. Each household for which a consent and baseline survey is completed will receive a home participation kit (HPK) in a study-branded box (with branding consistent with the welcome letter) that will detail how they can participate in the research if they are interested. We may assess whether distribution of HPK to households concurrent with the invitation letter increases participation. The HPK will include (1) instructions on accessing electronic or phone procedures for consent, enumeration, and behavioral survey and (2) instructions and materials for self-collection of specimens and return mailing to our central laboratory for testing. Specimen self-collection materials will include a flocked swab for collection of anterior nares specimens and a transport tube containing phosphate-buffered saline; for self-collection of dried blood spot (DBS) cards, the kit will include a finger stick device, alcohol wipe, adhesive bandage, and Whatman 5-spot DBS specimen card.
The HPK will have a unique identifier code that will be used for all baseline procedures, including laboratory testing, incentives, and survey. Instructions will direct participants to an online link or toll-free call for screening consent and the household enumeration procedure (detailed in the following). After the household enumeration is complete, one adult household member will be randomly selected based on an automated algorithm in our survey platform. The selected person will be asked to complete the study consent and baseline questionnaire. Households not completing the enumeration survey will receive two additional postcard reminders to complete the questionnaire and home test kit. Using a multimodal approach, households in the sampling frame with phone numbers (approximately 50%) and those who provided their contact information at enrollment will also receive up to three text messages and/or calls as additional reminders from a call center specializing in research partnerships. Households will have a one-month period to complete their kits for inclusion in the study. We will adjust for differential nonresponse for households lacking phone number information by using information available at the Census block level [28]. Consenting persons who complete baseline participation will be enrolled into an incidence cohort for a follow-up period. We will use similar contact procedures, and HPK, to encourage follow-up participation at 3 months after the baseline survey.
Consent and survey processes
All consents and survey data will be hosted on a secure, HIPAA-compliant electronic survey platform. Participants will be able to self-complete the consent and surveys directly in the online platform or can call a toll-free number to have assistance in completing these processes. Participants who wish to complete surveys by phone will be assisted by staff trained in human subjects protocols and study-specific procedures, with trained staff entering data into the same secure electronic platform as the one used by self-completing survey participants. Call center procedures such as hours of operation, number of contact attempts, and handling of participant questions will be adapted from procedures used in other large surveys [29]. All study materials will be available in English and Spanish. Study procedures have been approved by the Emory University Institutional Review Board (protocol 00000695).
Enumeration and eligibility
The enumeration process will be adapted from procedures used in other national studies [29]. An adult aged 18 years or older in the household will complete enumeration, providing for each member of their household first, middle, and last initials, age, gender. One adult aged 18 years or older be randomly selected from the enumeration list to complete the baseline questionnaire and HPK. There is no exclusion criterion other than age. Persons who have a clotting disorder, are on blood thinners, or are aged 80 years or older will only complete the anterior nares specimen procedure.
Specimen self-collection
The sampled household member will be asked to use the HPK to provide two specimens for laboratory testing. An anterior nares (nasal) swab will allow for detection of SARS-CoV-2 RNA by RNA PCR; The CDC identifies at-home self-collected anterior nares swabs as a suitable specimen type for PCR analysis [30], and self-collected anterior nares swabs have high sensitivity for RNA detection when compared with NP swabs [31]. Swabs will be stored and shipped in phosphate-buffered saline. Participants will also perform a finger prick with an automated lancet and fill in a Whatman 903 protein saver specimen card for the detection of antibodies to SARS-CoV-2 [8]. Specimen collection instructions will represent standard instructions, similar to those we have previously published for self-collected specimens [32,33], customized with branding for this study. Instructions will also include videos that demonstrate each component of the self-collection process [34]. Instructions guide participants to return specimens within 48 hours of collection. Stability testing indicates a longer window, allowing for shipping and laboratory processing time. Laboratorians will determine adequacy of sample based on visual inspection of the specimen and of the date collected and will request retesting for specimens not determined to be adequate. All specimens will be returned through U.S. mail to the central study laboratory in biohazard bags and sturdy outer boxes.
Survey instrument
The 15–20 minute baseline questionnaire will assess domains of demographics, COVID-19 knowledge, SARS-CoV-2 testing history, medical history, symptomatic history, illness in household, social distancing and isolation practices, and life changes due to COVID-19 (Supplement 1). Demographic measures will be adapted from the Census Bureaus’ American Community Survey [35], and include age, race/ethnicity, gender, education, income, and health insurance. COVID-19 knowledge items will be adapted from several sources, focusing on information relevant to protective and proactive health behaviors. SARS-CoV-2 testing history will be based on items previously used for HIV testing history in validated questionnaires [36]. We will assess symptom history at two time points relevant for virus and antibody testing, respectively, using 1-month recall and time since January 2020. Clinical history will be based on symptomology of COVID-19 identified by the U.S. Centers for Disease Control and Prevention, and will use a severity index for experienced symptoms based on an instrument validated for flu [37]. A number of studies have used different considerations to classify persons as “mildly” or “moderately” symptomatic for COVID-19 disease, including assessments based on fever [[38], [39], [40]], respiratory symptoms and their severity [38,[40], [41], [42]], cough [39], nonspecific or other symptoms [38,39], and risk factors [42], yet there is not currently a consensus method to assess symptoms remotely [43]. We are therefore implementing a broad list of symptoms and severity assessments to allow for us to meet emerging consensus definitions for case classification. Social distancing and isolation practices will be based on measures previously used to inform modeling studies, assessing the number of persons with close contact [44,45].
Laboratory testing
RNA PCR
Anterior nares swab specimens will be processed as previously described [34]. Specimens will first be checked for quality. The samples will then undergo total nucleic acid extraction using the Thermo Kingfisher platform (Fisher Scientific, Waltham, MA). Isolated RNA will be reverse-transcribed to DNA using a one-step, one-tube system using reagents from Thermo (Fisher Scientific). The second half of the one-tube system will involve qPCR. The reverse-transcribed DNA will undergo qPCR with primers and probes targeting three gene regions of the SARS-CoV-2 genome (N, S, ORF1), using reagents from Thermo. The results will be analyzed and an interpretation will be made based on cycle threshold values and positive identification of the nucleic acid.
Serology tests
A two-step process will be used: all specimens will be tested with a screening ELISA, and all specimens testing positive on the screening assay will also be tested a second time with a second ELISA. We implement this strategy by screening with a test with relatively high sensitivity to detect total antibodies (BioRad, Hercules, CA: Sens: 92.2%; Spec: 99.6% for IgG, IgM, and IgA) and confirming with a test with high specificity for IgG and IgA (EUROIMMUN IgG, Mountain Lakes, NJ; Sens: 90%, Spec: 100%; EUROIMMUN IgA performance not yet documented by FDA) [46]. This combination of confirmatory isotypes addresses both early and long-term immune responses, and IgA appears at approximately the same time after infection as IgM and at higher concentrations [47]. Based on the FDA-evaluated sensitivity and specificity of the two tests, the predictive value of the algorithm for a positive test (PPV) is 100% in all cases, and the predictive value of a negative test (NPV) ranges from 99.4% to 99.99%, depending on prevalence of antibodies in the population (Table 1 ). Performance data from FDA also contain 95% confidence intervals (CIs) for sensitivity and specificity of tests. If worst-case scenarios (e.g., the lower 95% CI for sensitivity of the screening test and the lower 95% CI for the specificity of the confirmatory test), predictive value of positive tests has more variability, especially for low prevalence scenarios.
Table 1.
Performance of an alogorithm to detect immune response to SARS-CoV-2 infection, by population prevalence and for overall, best-case and worst-case estimates of sensitivity and specificity
| Population Prevalence | FDA estimated |
Worst case∗ |
||
|---|---|---|---|---|
| Algorithm PPV | Algorithm NPV | Algorithm PPV | Algorithm NPV | |
| 0.1% | 100% | 99.99% | 29.6% | 99.9% |
| 1% | 100% | 99.89% | 80.9% | 99.89% |
| 2% | 100% | 99.78% | 89.6% | 99.77% |
| 5% | 100% | 99.4% | 95.7% | 99.41% |
Worst case utilizes the lower 95% CI for sensitivity for the screening test, and the lower 95% CI estimate of specificity for the confirmatory test.
DBS specimens will first be checked for visual quality [34]. A 6 mm punch will be obtained from the DBS and the material will undergo a standard antibody extraction method using TRIS buffer. Once the material is added to the reaction tube, the enzyme immunoassay primary and secondary antibodies (SARS-CoV-2 assay, total immunoglobulins; BioRad) will be added using an automated liquid handler instrument (DSX; Dynex Technologies, Chantilly, VA). All serologic and molecular tests have FDA Emergency Use Authorization (EUA). The protocol will follow the manufacturer's guidelines for reaction conditions, data interpretation, and control checks. For specimens with reactive results on the screening test, a second eluate will be tested with the confirmatory test using the same elution procedures.
Return of results
If use of study assays with self-collected specimens has been approved or cleared under EUA by the FDA at the time of testing, laboratory results will be returned to participants and they will be instructed to seek follow-up care with their usual physician if they have concerns or questions about their results. Results of the overall antibody algorithm will be reported (e.g., if negative on first test, a nonreactive result will be reported; if positive on both tests, a reactive result will be reported). For specimens with discordant results, a third antibody test on a different platform will be run as a tiebreaker and the result of the tiebreaker test will be reported. If the assays used in the study have not received EUA, we will return test results as research results to the extent allowable by the FDA, under the auspices of Emory's IRB approval and an informed consent process. For specimens with insufficient quantity or other extraction failure, participants will be mailed an additional kit to allow for repeat specimen collection.
Primary outcomes
The primary outcomes will be (1) the weighted proportion and 95% CI of SARS-CoV-2 RNA-PCR detected specimens and (2) the weighted proportion and 95% CI of SARS-CoV-2 persons with detectable antibodies (i.e., algorithm-determined positive). These outcomes will be prepared with inference to the United States and to the oversampled states. Table 2 provides more detail regarding study outcomes.
Table 2.
Outcomes for a national serosurvey for COVID-19 infection and immune experience
| Study data source | External data source | Answerable question | Outcome measure | Analysis |
|---|---|---|---|---|
| Baseline PCR tests | U.S. Census for weighting | Prevalence of COVID-19 disease | Period prevalence | Upweighted estimates, confidence intervals (CIs) |
| Baseline antibody (Ab) tests | U.S. Census for weighting | Prevalence of SARS-CoV-2-specific immune response | Period prevalence | Upweighted estimates, CI |
| Baseline + follow-up PCR, Ab tests | U.S. Census for weighting | Incidence of COVID-19 disease | Period incidence | Upweighted estimates, CI |
| Baseline + follow-up PCR, Ab tests | U.S. Census for weighting | Incidence of SARS-CoV-2-specific immune response | Period incidence | Upweighted estimates, CI |
| Baseline + follow-up PCR, Ab tests | U.S. Census for weighting | Prevalent exposure to SARS-Cov-2 (PCR or AB test) | Period prevalence | Upweighted estimates, CI |
| Baseline PCR, Ab tests, reported diagnosis | Public diagnosis data | Estimated proportion of SARS-CoV-2 cases that lead to diagnosed COVID-19 | Proportion | Upweighted estimates, CI sensitivity analyses on diagnosis data |
| Baseline PCR, Ab tests | Public NDI data, excess mortality data | Estimated proportion of SARS-CoV-2 cases that lead to fatality | Proportion | Upweighted estimates, CI sensitivity analyses on fatality data |
| Baseline PCR, Ab tests, reported symptoms | U.S. Census for weighting | Estimated proportion of SARS-CoV-2 cases asymptomatic or mildly symptomatic | Proportion | Upweighted estimates, CI |
| Baseline PCR, Ab tests, reported perception | U.S. Census for weighting | Estimated proportion of SARS-CoV-2 cases that perceived themselves as infected | Proportion | Upweighted estimates, CI |
| Baseline PCR, Ab tests, self-report survey measures | U.S. Census for weighting | Predictors of positive COVID-19 disease such as social distance, occupation, family structure | Odds ratios | Weighted regression estimates |
Data system
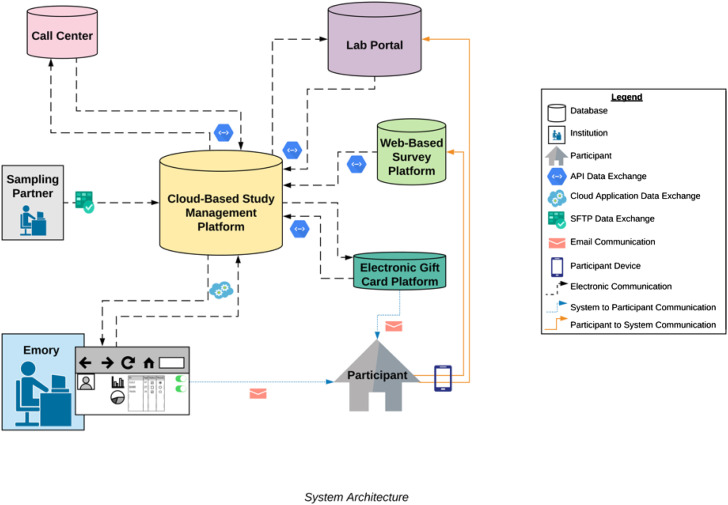
A unified data system that leverages information from research partners will be used for participant management. Figure 2 represents the data sources, including sources of participant-related data (call center, survey data, laboratory data, and incentive), which will be combined with data from Emory and from the sample frame provider into a secure, unified cloud-based participant data management system (DMS). The DMS will allow for real-time tracking of participant progress in the study, and system-automated responses to facilitate scalability and rapid response. This is best illustrated with a description of how participant data will flow through the system. The sample frame provider will enter the list of randomly selected addresses in the sample frame into the DMS, allowing mailing of invitation letters and for the laboratory to mail an HPK to the selected address. The sending of an HPK will be registered in the DMS, for management purposes. When the household respondent completes their questionnaire, the DMS will receive notification of this from the survey automated programming interface (API). After a participant has completed their specimen self-collection and returned the specimen to the laboratory, the laboratory API will place completion data into the DMS, which will trigger the DMS to automatically order a $40 electronic incentive card to be provided to the participant, using the API of the gift card provider. In addition, the DMS will communicate to the call center that further follow-up calls should cease. The system will be built to accommodate different participant pathways, such as for participants who need additional reminders or have opted out of study participation. Study staff will remotely manage the DMS and participant contact process, ensuring participants needs are met and that those interested in participating are on target to complete study tasks in a timely manner. The overarching goal of developing the data system, and engaging with external partners accustomed to handling large volumes of interactions including the study call center, sampling partner, and laboratory, is to facilitate feasibility of conducting the project on an expedited timeline.
Fig. 2.
Unified study data system flowchart.
Primary analysis
Sample weights will be computed and applied to generate estimates of study outcomes that are representative of the United States and of the oversampled states of interest. First, base (inverse probability) weights will be computed to reflect selection probabilities for both households and persons within households. Next, poststratification weights will be computed by ratio-adjusting base weights to characteristics of the survey population, based on the latest population estimates from the Census American Community Survey. For this purpose, we will use a “raking” procedure to ensure alignment with the U.S. population (and oversampled states) with respect to various geodemographic characteristics, including gender, age, race-ethnicity, education level, region, income, home ownership, and metropolitan area. The resulting design effect, which can be approximated by , will be examined to assess the weighting efficiency. We will use Taylor series linearization for variance estimation.
We will develop weighted estimates for study outcomes, including period prevalence and incidence of SARS-CoV-2 infection and SARS-CoV-2-specific immune response, estimates of the proportion of cases that lead to diagnosed COVID-19 or to fatality, and of the numbers and proportions of cases that are mildly symptomatic or asymptomatic. We will create estimates for each of these main outcomes for subgroups including age group, gender, race/ethnicity, symptomology, region, and urbanicity.
We will conduct a series of analyses using regression models appropriate for each type of data, such as logistic regression models for assessment of the predictive utility of various symptoms on prevalence of study outcomes. Several sensitivity analyses will be conducted to assess potential bias, such as full household estimation (more detail in the following) to explore potential deviations from random selection of household members, and a separate analysis to assess the impact of enumerated household members who are unavailable to participate because of hospitalization for a respiratory condition. It will also be important to correct for known imperfection in the diagnostic performance of laboratory tests. Our confirmatory testing procedure will serve to minimize false positives; we will nonetheless account for imperfect performance of the overall test algorithm sensitivity and specificity. We will use the procedures proposed by Diggle, implemented in a Bayesian framework, such that uncertainty from design effects and diagnostic inaccuracy are both quantified in prevalence estimates [48].
Study sample size estimation
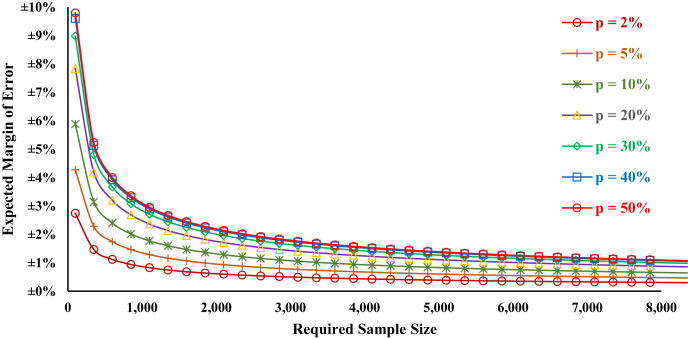
Early serosurvey data have identified a range of antibody prevalence values, 2.8% weighted prevalence in Santa Clara County [10], 4.3% antibody prevalence in Los Angeles County [11], and 12.3% in New York state [9]. Figure 3 displays the overall margin of error as a function of different sample sizes and prevalence levels. In Table 3 , we present margins of error for assessing antibody prevalence for our national sample, given our proposed 4000-person national sample. We find that a scenario with 2% prevalence of infection or immune experience would have a margin of error (MoE) of ±0.43, with 5% prevalence a MoE ±0.68%, and 10% prevalence a MoE ±0.93%. We also anticipate acceptable margins for subgroup analyses, for instance persons 18–39 years old 2% prevalence scenario with MoE ±0.80%, 5% prevalence a MoE ±1.24%, and 10% prevalence a MoE ±1.71%. In Table 4 , we present similar estimates for a statewide sample of 600 persons in New York State. For other oversampled states, these numbers are substantially similar, although differing marginally due to differences in the distributions of age and gender. When sample weights are applied to estimates, MoE may increase. All estimates are based on a two-sided interval, with alpha = 0.05.
Fig. 3.
Margin of error as a function of sample size and period prevalence estimate at 95% confidence.
Table 3.
Margin of error as a function of period prevalence estimate and demographic categories for estimation for a national sample of 4000 persons
| Prevalence | National estimate |
18–39 y |
40–59 y |
60+ y |
Males |
Females |
|---|---|---|---|---|---|---|
| n = 4000 | n = 1181 | n = 1004 | n = 905 | n = 1961 | n = 2039 | |
| 2% | ±0.43% | ±0.80% | ±0.87% | ±0.91% | ±0.62% | ±0.61% |
| 5% | ±0.68% | ±1.24% | ±1.35% | ±1.42% | ±0.96% | ±0.95% |
| 10% | ±0.93% | ±1.71% | ±1.86% | ±1.95% | ±1.33% | ±1.30% |
| 20% | ±1.24% | ±2.28% | ±2.47% | ±2.61% | ±1.77% | ±1.74% |
| 30% | ±1.42% | ±2.61% | ±2.83% | ±2.99% | ±2.03% | ±1.99% |
| 40% | ±1.52% | ±2.79% | ±3.03% | ±3.19% | ±2.17% | ±2.13% |
| 50% | ±1.55% | ±2.85% | ±3.09% | ±3.26% | ±2.21% | ±2.17% |
Table 4.
Margin of error as a function of period prevalence estimate and demographic categories for estimation for a state sample of 600 persons
| Prevalence | New York state sample |
18–39 y |
40–59 y |
60+ y |
Males |
Females |
|---|---|---|---|---|---|---|
| n = 600 | 185 | 152 | 138 | 292 | 308 | |
| 2% | ±1.12% | ±2.02% | ±2.23% | ±2.34% | ±1.61% | ±1.56% |
| 5% | ±1.74% | ±3.14% | ±3.47% | ±3.64% | ±2.50% | ±2.43% |
| 10% | ±2.40% | ±4.33% | ±4.77% | ±5.01% | ±3.44% | ±3.35% |
| 20% | ±3.20% | ±5.77% | ±6.36% | ±6.68% | ±4.59% | ±4.47% |
| 30% | ±3.67% | ±6.61% | ±7.29% | ±7.65% | ±5.26% | ±5.12% |
| 40% | ±3.92% | ±7.07% | ±7.79% | ±8.18% | ±5.62% | ±5.47% |
| 50% | ±4.00% | ±7.21% | ±7.95% | ±8.34% | ±5.74% | ±5.58% |
Full household assessment
Our study design calls for testing a single household member, based on a rationale that full household testing would likely reduce response rates (leading to bias) and that the additional laboratory testing would greatly add to the study cost without substantially increasing study power due to within-household correlation of outcomes. There is potential for bias in selecting a single household respondent: some households will be more likely to select a member who has symptoms, leading to overestimation of primary outcomes. Conversely, other households may have one member who is sick and assumed to have COVID-19, and they may instead test a household member with more mild symptoms, leading to endpoint underestimation. To describe potential bias in selection of a single household member for testing, we will randomly select a group of households to receive additional testing for all household members aged 3 years or older. We will target participation of 400 households, or 10% of households in the national sample, for this purpose. Based on the proportion of single-person households, and average U.S. household size of 2.6 [35], we anticipate this will lead to an additional 396 tests and surveys, allowing for assessment of possible bias in selection of household members for testing. Potential bias will be characterized by comparing test positivity by participant characteristics among fully sampled households to standard households in which one person was sampled. This subset of fully enumerated households will additionally allow for characterizing within-household transmission for households with symptomatic and asymptomatic positive members.
Incidence cohort design
Participants completing baseline procedures will be mailed identical follow-up testing kits at 3 months after the return of their initial test, regardless of the baseline SARS-CoV-2 infection and immune experience at the baseline. We will retest participants using the same laboratory methods as for baseline testing and calculate incidence in the period for each laboratory assessment by calculating the weighted number of participants with newly positive assessment at time T1/the weighted number of susceptible people at time T0. Persons testing PCR- or serology-positive will be retested at the follow-up period to characterize ongoing viral shedding, development of SARS-CoV-2-specific antibodies and behavioral changes at subsequent time points. This will allow for improved understanding of these outcomes for a representative sample of persons who are asymptomatic or mildly symptomatic. In past studies we have observed 80% cohort retention [32,[49], [50], [51]]; we anticipate slightly lower (70%) retention at 3 months for this cohort study. We may also include additional incidence- or resample-assessment time points based on the interests and identified needs of collaborating state and local health departments.
Transmission model analyses
Seroprevalence data serve as a key input into dynamic transmission models of infectious disease [52,53]. For SARS-CoV-2, susceptible, exposed, infectious, and recovered model frameworks are being widely used for a range of applications including forecasting, inferring transmission patterns, and examining the potential impact of interventions such as social distancing [[54], [55], [56], [57]]. We will use our study's estimates of seroprevalence to set the size of the R (recovered) compartment in both simple and age-structured transmission models, thereby increasing the realism of the model scenarios. Our seroprevalence estimates can also be used by the wider modeling community to refine other models for a range of applications. We will specifically use our seroprevalence estimates to model how social distancing can be relaxed in an acceptable safe manner. In doing so, we will investigate how serological testing at the individual and population level may provide data for public health actions.
Results
Participant recruitment and data collection began in July 2020. We will report the main outcomes, of period prevalence of infection with SARS-CoV-2 and SARS-CoV-2-specific immune response, by variables including symptoms, symptom history, underlying conditions, county population concentration (urbanicity), region, gender, age, family size, and isolation practices. Given the urgent nature of the pandemic, we will develop a study website for results dissemination to supplement reporting of study outcomes through academic publication. The study website, COVIDVu.org, will serve as a venue to report preliminary prevalence estimation, as a portal for sharing nonidentifying public use study data sets, and to share infographics to communicate key study findings to a broader audience.
Discussion
The COVID-19 pandemic has had wide-reaching economic and social impact in the United States: most daycares have closed, students are receiving virtual schooling, adults with nonessential positions are teleworking, and many persons have lost their jobs, are working fewer hours, or are being furloughed. For policy makers to be best informed, it is critical to have high-quality, minimally biased data regarding the current status of the epidemic. Several local surveys to establish antibody prevalence have been launched to better characterize local epidemics. In New York State, a sample of over 15,000 persons recruited at grocery stores and community centers during April and early May found overall antibody prevalence of 12.3%, with wide ranges of prevalence from 1.2% in North Country to 19.9% in New York City [9]. In Los Angeles County, a survey of 1952 adults selected from a proprietary database representative of the county (maintained by a market research firm) had 865 participants (50.9%) and found 4.3% weighted antibody prevalence (95% CI: 2.6%–6.2%) [11]. In Santa Clara County, CA, a widely criticized study with convenience sample of 3300 residents had 1.5% (95% CI: 1.1%–2.0%) unweighted antibody prevalence, with weighted estimate of 2.8% (95% CI: 1.3%–4.7%) [10]. The CDC conducted serosurveys with door-to-door outreach using probability samples in two Georgia counties and the State of Indiana in April–May 2020; the results suggested 2.6% (95% CI: 1.1%–6.3%) and 2.8% (95% CI: 2.0%–3.7%), respectively [22,23].
Such assessments are important, but the nonprobability studies are subject to several key areas of potential bias. Because of convenience sampling, data from some of these surveys may not be representative of the underlying population. Persons seeking testing, or willing to receive testing, may be more likely to have experienced illness. Conversely, for household-based testing with health care–worker specimen collection, persons who are working and at higher risk of exposure may not be home for such assessments. Nonrepresentative surveys may also not be able to adequately speak to less advantaged communities, whether in rural areas or in lower-income urban settings.
We propose to leverage a probability sample and home-based testing to produce estimates of the prevalence of SARS-CoV-2 infection and SARS-CoV-2-specific immune response that will include people without symptoms, who might otherwise not seek testing, and who might be unable to travel to a testing location or who might be unwilling to test at a clinical site for testing. This latter concern is important: our preliminary data indicate that nearly half of people may be unwilling to attend a laboratory testing venue for SARS-CoV-2 as part of a research study [24]. Higher participation rates will minimize selection bias. Importantly, willingness to participate in a home test study for COVID-19 was high overall, and not differential across key demographic variables such as age and race/ethnicity [24]. Furthermore, our mail-based testing platform is highly acceptable to participants [32], and initial evaluation of self-specimen collection indicated that the great majority of participants are able to self-collect specimens for SARS-CoV-2 PCR and serology testing [33]. To date, a number of important studies on the prevalence of antibodies have been biased because they likely reflected an over-representation of people seeking care [9,10,58]; because they recruited people in ways that encouraged those concerned about their possible exposures to participate [10]; and because they recruited at convenience locations where attendance in a venue might be selectively associated with lower perceived vulnerability to infection [9]. A probability-based, nationally representative survey will provide a context through which to understand studies that are local and/or based on convenience samples.
We recognize that our strategy, while emphasizing representativeness and minimizing selection bias, has important limitations. Some critical populations will not be reached with our sampling strategy. For example, people who are homeless or who reside in informal lodgings without individual postal addresses will be missed in our sampling; this is an important limitation because there is evidence that homeless persons may be at especially high risk [59]. People who are incarcerated will also be excluded and are also at high risk [59]. It is critical that other types of serosurveillance efforts be developed and implemented to compliment household probability-based serosurveys; triangulation based on an understanding of the weaknesses of a surveillance system is a well-accepted approach to develop an overall understanding of the impact of a health condition [60,61].
Our approach is vulnerable to nonresponse at the baseline, and challenges with retention at the follow-up period. We will attempt to mitigate nonresponse by maximizing opportunities for participants to corroborate the authenticity of the study: using distinctive, professional branding that will be recognizable across interactions; providing a website that will be hosted on an academic web platform with multiple modes of information about the study (e.g., video, FAQ formats); and by mailing a professionally branded and designed home participation kit to all selected households. We will assess for differential nonresponse by characteristics of the census tract of selected participants—for example, median income, racial/ethnic distribution, health insurance coverage, health literacy, etc., and weight to adjust for such nonresponse. We have estimated that we will have 22% response, a level lower than reported willingness to participate in home testing studies for COVID-19 research [24], to account for the possibility that the willingness contingency valuation overestimated willingness. We will seek to enhance retention abilities by collecting a full set of detailed contact information at the baseline, and by using multiple modes of contact strategies at follow-up, strategies we have successfully used in other remote home testing studies [32,62,63]. We will perform analyses at the follow-up period to assess ways in which differential retention across subgroups might impact study findings. We will also seek to consider the impact of nonresponse and loss to follow-up by considering data triangulation, assessing data across different sources including other infection and antibody surveys and local case surveillance data.
Data for public health action are most powerful when they are most local. Our proposal aims to develop national estimates with systematic sampling to ensure adequate geographic diversity. It also represents a framework and a set of tools that will be applied to state-level estimation and could be expanded to smaller jurisdictional levels such as metropolitan areas. As part of the initial study, we will also collect data to develop overall state-level estimates of RNA PCR and antibody positivity for a number of highly populated states.
Understanding the prevalence of SARS-CoV-2 infection and SARS-CoV-2-specific antibodies that might indicate immunity can provide the roadmap to estimating levels of required resources, to improving our understanding of the spectrum of COVID-19 disease, and to understanding the differences in infection in key subpopulations in the U.S. epidemic. By combining traditional methods of recruiting a representative sample with novel methods to allow laboratory assessment of participants without requiring participants to visits a clinical location for specimen collection, we will be able to represent all housed persons in the United States, including otherwise hard to access populations such as people who live in rural areas and people who are hesitant to go into health care settings because of concern of contracting the virus. Public health surveillance is the conscience of an epidemic [64]; our planned national survey will be an important data source that will provide a representative national picture and help to put case reporting and other types of serosurveys in context.
CRediT authorship contribution statement
Aaron J. Siegler: Conceptualization, Writing - original draft, Methodology, Funding acquisition, Writing - review & editing. Patrick S. Sullivan: Conceptualization, Writing - original draft, Methodology, Funding acquisition, Writing - review & editing. Travis Sanchez: Writing - review & editing. Ben Lopman: Writing - original draft, Writing - review & editing. Mansour Fahimi: Writing - review & editing. Charles Sailey: Writing - review & editing. Martin Frankel: Writing - review & editing. Richard Rothenberg: Writing - review & editing. Colleen F. Kelley: Writing - review & editing. Heather Bradley: Writing - original draft, Writing - review & editing.
Acknowledgments
The authors acknowledge the important work of Mariah Valentine-Graves, Laura Gravens, Nicole Luisi, Shelby Mullin, and Ryan Zahn. The authors thank Salesforce for its donation of Salesforce.org licenses and system development for the project, and to acknowledge the contributions of development team members Lee Evans, John Thrasher, Stephen Noe, Anurag Jaiswal, Ryan Williams, David Affentranger, and Todd Siegler. They also thank the Kaiser Family Foundation for their design and thoughtful contributions to this important effort.
Author Patrick Sullivan is the Editor in Chief of Annals of Epidemiology. This article was reviewed and handled by an independent editor. Dr. Sullivan was not involved in the editorial decision of the submission.
This work was supported by the US National Institute of Allergy and Infectious Diseases (3R01AI143875-02S1). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Funding: NIAID 3R01AI143875-02S1, Woodruff Foundation.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.annepidem.2020.07.015.
Supplementary data
References
- 1.World Health Organization Coronavirus disease 2019 (COVID-19): Situation report 51. [online surveillance report] https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports Available at:
- 2.Watkins J. Preventing a covid-19 pandemic. BMJ. 2020;368:m810. doi: 10.1136/bmj.m810. [DOI] [PubMed] [Google Scholar]
- 3.Goldstein N.D., Burstyn I. 2020. On the importance of early testing even when imperfect in a pandemic such as COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adalja A.A., Toner E., Inglesby T.V. Priorities for the US health community responding to COVID-19. JAMA. 2020;323(14):1343–1344. doi: 10.1001/jama.2020.3413. [DOI] [PubMed] [Google Scholar]
- 5.Rimmer A. Covid-19: BMA calls for rapid testing and appropriate protective equipment for doctors. BMJ. 2020;368:m1099. doi: 10.1136/bmj.m1099. [DOI] [PubMed] [Google Scholar]
- 6.Sharfstein J.M., Becker S.J., Mello M.M. Diagnostic testing for the novel coronavirus. JAMA. 2020;323(15):1437–1438. doi: 10.1001/jama.2020.3864. [DOI] [PubMed] [Google Scholar]
- 7.Binnicker M.J. Emergence of a Novel Coronavirus Disease (COVID-19) and the importance of diagnostic testing: why partnership between clinical laboratories, public health agencies, and industry is essential to control the outbreak. Clin Chem. 2020;66(5):664–666. doi: 10.1093/clinchem/hvaa071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacofsky D., Jacofsky E.M., Jacofsky M. Understanding antibody testing for COVID-19. J Arthroplasty. 2020;35:S74–S81. doi: 10.1016/j.arth.2020.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.New York State Amid Ongoing COVID-19 Pandemic, Governor Cuomo Announces Results of Completed Antibody Testing Study of 15,000 People Show 12.3 Percent of Population has Covid-19 Antibodies. Governor’s Office. 2020. https://www.governor.ny.gov/news/video-audio-photos-rush-transcript-amid-ongoing-covid-19-pandemic-governor-cuomo-announces-17
- 10.Bendavid E., Mulaney B., Sood N., Shah S., Ling E., Bromley-Dulfano R. COVID-19 Antibody Seroprevalence in Santa Clara County, California. medRxiv. 2020 doi: 10.1101/2020.04.14.20062463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sood N., Simon P., Ebner P., Eichner D., Reynolds J., Bendavid E. Seroprevalence of SARS-CoV-2–specific antibodies among adults in Los Angeles County, California, on April 10-11, 2020. JAMA. 2020;323(23):2425–2427. doi: 10.1001/jama.2020.8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.-K. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 13.Lou B., Li T.D., Zheng S.F., Su Y.Y., Li Z.Y., Liu W. Serology characteristics of SARS-CoV-2 infection since exposure and post symptom onset. Eur Respir J. 2020;56(2):2000763. doi: 10.1183/13993003.00763-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Couper M.P. New developments in survey data collection. Annu Rev Sociol. 2017;43(1):121–145. [Google Scholar]
- 15.Fahimi M., Kulp D. Quirk's Marketing Research Review: Quirk’s Media. 2009. Address-based sampling – alternatives for surveys that require representative samples of households.https://www.quirks.com/articles/address-based-sampling-may-provide-alternatives-for-surveys-that-require-contacts-with-representative-samples-of-households [Google Scholar]
- 16.Siegler A.J., Hall E., Luisi N., Zolotorzynska M., Wilde G., Sanchez T. Willingness to seek laboratory testing for SARS-CoV-2 with home, drive-through, and clinic-based specimen collection locations. medRxiv. 2020;2020 doi: 10.1101/2020.05.06.20093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valliant R., Hubbard F., Lee S., Chang C. Efficient Use of Commercial Lists in U.S. Household Sampling. J Surv Stat Methodol. 2014;2(2):182–209. doi: 10.1093/jssam/smu006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaVange L.M., Kalsbeek W.D., Sorlie P.D., Aviles-Santa L.M., Kaplan R.C., Barnhart J. Sample design and cohort selection in the Hispanic Community Health Study/Study of Latinos. Ann Epidemiol. 2010;20(8):642–649. doi: 10.1016/j.annepidem.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chido-Amajuoyi O.G., Yu R.K., Agaku I., Shete S. Exposure to court-ordered tobacco industry antismoking advertisements among US adults. JAMA Netw Open. 2019;2(7):e196935. doi: 10.1001/jamanetworkopen.2019.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerel J., Maple M., van de Venne J., Moore M., Flaherty C., Brown M. Exposure to suicide in the community: prevalence and correlates in One U.S. State. Public Health Rep. 2016;131(1):100–107. doi: 10.1177/003335491613100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomson T.L., Nemeth J.M., Peng J., Lu B., Ferketich A.K., Paskett E.D. Address-based sampling for recruiting rural subpopulations: A 2-Phase, multimode approach. J Rural Health. 2018;34(2):193–201. doi: 10.1111/jrh.12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jennifer B Harris, Holly M Biggs, Lucy Breakwell, F Scott Dahlgren. Estimated community seroprevalence of SARS-CoV-2 antibodies — two Georgia Counties, April 28–May 3, 2020. MMWR Morb Mortal Wkly Rep. 2020;69(29):965–970. doi: 10.15585/mmwr.mm6929e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menachemi N., Yiannoutsos C.T., Dixon B.E. Population point prevalence of SARS-CoV-2 infection based on a statewide random sample — Indiana, April 25–29, 2020. MMWR Morb Mortal Wkly Rep. 2020;69(29):960–964. doi: 10.15585/mmwr.mm6929e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall E.W., Luisi N., Zlotorzynska M., Wilde G., Sullivan P.S., Sanchez T., Bradley H., Siegler A.J. SARS-CoV-2/COVID-19 Testing for Research: Willingness to Use Home Specimen Collection Methods. JMIR. 2020;22(9):e19471. doi: 10.2196/19471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen T.C., Clark J., Riddles M.K., Mohadjer L.K., Fakhouri T.H.I. Vital and Health Statistics, National Health and Nutrition Examination Survey, 2015 – 2018: Sample Design and Estimation Procedures. National Center for Health Statistics. Vital Health Stat. 2020;2020(2):184. [PubMed] [Google Scholar]
- 26.Fahimi M. American Statistical Association; 1994. Poststratification of Pooled Survey Data. Proceedings of the Survey Research Methods Section. [Google Scholar]
- 27.Groves R.M., Fowler F.J., Jr., Couper M.P., Lepkowski J.M., Singer E., Tourangeau R. Vol. 561. John Wiley & Sons; 2011. (Survey methodology). [Google Scholar]
- 28.Link M.W., Lai J.W. Cell-phone-only households and problems of differential nonresponse using an address-based sampling design. Public Opin Q. 2011;75(4):613–635. [Google Scholar]
- 29.US Centers for Disease Control and Prevention Behavioral Risk Factor Surveillance System (BRFSS) 2020. https://www.cdc.gov/brfss/index.html
- 30.Centers for Disease Control and Prevention . 2020. Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens from Persons for Coronavirus Disease 2019 (COVID-19) [online testing guidance]https://www.cdc.gov/coronavirus/2019-nCoV/lab/guidelines-clinical-specimens.html Available at: [Google Scholar]
- 31.Tu Y.-P., Jennings R., Cangelosi G.A., Wehber K. Swabs collected by patients or health care workers for SARS-CoV-2 testing. N Engl J Med. 2020;383:494–496. doi: 10.1056/NEJMc2016321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siegler A.J., Mayer K.H., Liu A.Y., Patel R.R., Ahlschlager L.M., Kraft C.S. Developing and assessing the feasibility of a home-based PrEP monitoring and support program. Clin Infect Dis. 2019;68(3):501–504. doi: 10.1093/cid/ciy529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guest J.L., Sullivan P.S., Valentine-Graves M., Valencia R., Adam E., Luisi N. Suitability and sufficiency of telehealth clinician-observed, participant-collected Samples for SARS-CoV-2 testing: The iCollect Cohort Pilot Study. JMIR Public Health Surveill. 2020;6(2):e19731. doi: 10.2196/19731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sullivan P.S., Sailey C., Guest J.L., Guarner J., Kelley C., Siegler A.J. Detection of SARS-CoV-2 RNA and antibodies in diverse samples: protocol to validate the sufficiency of provider-observed, home-collected blood, saliva, and oropharyngeal samples. JMIR Public Health Surveill. 2020;6(2):e19054. doi: 10.2196/19054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.United States Census Bureau . 2020. American Community Survey [online program information]https://www.census.gov/programs-surveys/acs Available at: [Google Scholar]
- 36.Sanchez T.H., Sineath R.C., Kahle E.M., Tregear S.J., Sullivan P.S. The Annual American Men's Internet Survey of behaviors of men who have sex with men in the United States: protocol and key indicators report 2013. JMIR Public Health Surveill. 2015;1(1):e3. doi: 10.2196/publichealth.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powers J.H., Guerrero M.L., Leidy N.K., Fairchok M.P., Rosenberg A., Hernandez A. Development of the Flu-PRO: a patient-reported outcome (PRO) instrument to evaluate symptoms of influenza. BMC Infect Dis. 2016;16:1. doi: 10.1186/s12879-015-1330-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bi Q., Wu Y., Mei S., Ye C., Zou X., Zhang Z. Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: a retrospective cohort study. Lancet Infect Dis. 2020;20:911–919. doi: 10.1016/S1473-3099(20)30287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Böhmer M.M., Buchholz U., Corman V.M., Hoch M., Katz K., Marosevic D.V. Investigation of a COVID-19 outbreak in Germany resulting from a single travel-associated primary case: a case series. Lancet Infect Dis. 2020;29:920–928. doi: 10.1016/S1473-3099(20)30314-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li K., Fang Y., Li W., Pan C., Qin P., Zhong Y. CT image visual quantitative evaluation and clinical classification of coronavirus disease (COVID-19) Eur Radiol. 2020:1–10. doi: 10.1007/s00330-020-06817-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han C., Duan C., Zhang S., Spiegel B., Shi H., Wang W. Digestive symptoms in COVID-19 patients with mild disease severity: clinical presentation, stool viral RNA testing, and outcomes. Am J Gastroenterol. 2020;115(6):916–923. doi: 10.14309/ajg.0000000000000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moein S.T., Hashemian S.M., Mansourafshar B., Khorram-Tousi A., Tabarsi P., Doty R.L. Smell dysfunction: a biomarker for COVID-19. Int Forum Allergy Rhinol. 2020;10(8):944–950. doi: 10.1002/alr.22587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greenhalgh T., Koh G.C.H., Car J. Covid-19: a remote assessment in primary care. BMJ. 2020;368:m1182. doi: 10.1136/bmj.m1182. [DOI] [PubMed] [Google Scholar]
- 44.Mossong J., Hens N., Jit M., Beutels P., Auranen K., Mikolajczyk R. Social contacts and mixing patterns relevant to the spread of infectious diseases. PLoS Med. 2008;5(3):e74. doi: 10.1371/journal.pmed.0050074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiti M.C., Melegaro A., Cattuto C., Nokes D.J. Study design and protocol for investigating social network patterns in rural and urban schools and households in a coastal setting in Kenya using wearable proximity sensors. Wellcome Open Res. 2019;4:84. doi: 10.12688/wellcomeopenres.15268.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.US Food and Drug Administration EUA Authorized Serology Test Performance. [online test performance data]. Updated 5/22/2020. https://www.fda.gov/medical-devices/emergency-situations-medical-devices/eua-authorized-serology-test-performance Available at:
- 47.Yu H.-Q., Sun B.-Q., Fang Z.-F., Zhao J., Liu X.Y., Li Y.M. Distinct features of SARS-CoV-2-specific IgA response in COVID-19 patients. Eur Respir J. 2020:2001526. doi: 10.1183/13993003.01526-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diggle P.J. Estimating prevalence using an imperfect test. Epidemiol Res Int. 2011;2011 [Google Scholar]
- 49.Siegler A.J., Rosenthal E.M., Sullivan P.S., Ahschlager L., Kelley C.F., Mehta C.C. Double-blind, single-center, randomized three-way crossover trial of fitted, thin, and standard condoms for vaginal and anal sex: C-PLEASURE study protocol and baseline data. JMIR Res Protoc. 2019;8(4):e12205. doi: 10.2196/12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siegler A.J., Rosenthal E.M., Sullivan P.S., Mehta C.C., Moore R.H., Ahlschlager L. Levels of clinical condom failure for anal sex: A randomized cross-over trial. EClinicalMedicine. 2019;17:100199. doi: 10.1016/j.eclinm.2019.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sullivan P.S., Rosenberg E.S., Sanchez T.H., Kelley K.F., Luisi N., Cooper H.L. Explaining racial disparities in HIV incidence in black and white men who have sex with men in Atlanta, GA: a prospective observational cohort study. Ann Epidemiol. 2015;25(6):445–454. doi: 10.1016/j.annepidem.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Farrington C.P., Whitaker H.J. Estimation of effective reproduction numbers for infectious diseases using serological survey data. Biostatistics. 2003;4(4):621–632. doi: 10.1093/biostatistics/4.4.621. [DOI] [PubMed] [Google Scholar]
- 53.Glass K., Mercer G.N., Nishiura H., McBryde E.S., Becker N.G. Estimating reproduction numbers for adults and children from case data. J R Soc Interf. 2011;8(62):1248–1259. doi: 10.1098/rsif.2010.0679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weitz J.S., Beckett S.J., Coenen A.R., Demory D., Dominguez-Mirazo M., Dushoff J. Modeling shield immunity to reduce COVID-19 epidemic spread. Nat Med. 2020;26:849–854. doi: 10.1038/s41591-020-0895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sullivan Noah, Okell L.C., Dorigatti I., Winskill P., Wittaker C., Imai N. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis. 2020;20(6):669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kucharski A.J., Russell T.W., Diamond C., Liu Y., Edmunds J., Funk S. Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infect Dis. 2020;20(5):553–558. doi: 10.1016/S1473-3099(20)30144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kissler S.M., Tedijanto C., Goldstein E., Grad Y.H., Lipsitch M. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science. 2020;368(6493):860–868. doi: 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Los Angeles County Department of Public Health USC-LA county study: early results of antibody testing suggest number of COVID-19 infections far exceeds number of confirmed cases in Los Angeles County. 2020. http://publichealth.lacounty.gov/phcommon/public/media/mediapubhpdetail.cfm?prid=2328
- 59.Okonkwo N.E., Aguwa U.T., Jang M., Barre I.A., Page K.R., Sullivan P.S. COVID-19 and the US response: accelerating health inequities. BMJ Evid Based Med. 2020 doi: 10.1136/bmjebm-2020-111426. Epub ahead print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thurmond V.A. The point of triangulation. J Nurs Scholarsh. 2001;33(3):253–258. doi: 10.1111/j.1547-5069.2001.00253.x. [DOI] [PubMed] [Google Scholar]
- 61.German R.R., Lee L.M., Horan J.M., Lee L.M., Milstein B., Pertowski C.A. Updated guidelines for evaluating public health surveillance systems: recommendations from the Guidelines Working Group. MMWR Recomm Rep. 2001;50(RR-13):1–35. quiz CE31-37. [PubMed] [Google Scholar]
- 62.Siegler A.J., Brock J.B., Hurt C.B., Ahlschlager L., Dominguez K., Kelley C.F. An electronic pre-exposure prophylaxis initiation and maintenance home care system for nonurban young men who have sex with men: protocol for a randomized controlled trial. JMIR Res Protoc. 2019;8(6):e13982. doi: 10.2196/13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sullivan P.S., Driggers R., Stekler J.D., Siegler A.J., Goldenberg T., McDougal S.J. Usability and acceptability of a mobile comprehensive HIV prevention App for men who have sex with men: a pilot study. JMIR Mhealth Uhealth. 2017;5(3):e26. doi: 10.2196/mhealth.7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sullivan P.S., Delpech V. HIV surveillance: the conscience of the epidemic. Open AIDS J. 2012;6:65. doi: 10.2174/1874613601206010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.