Abstract
Oncogenesis is driven by germline, environmental and stochastic factors. It is unknown how these interact to produce the molecular phenotypes of tumours. We therefore quantified the influence of germline polymorphisms on the somatic epigenome of 589 localized prostate tumours. Predisposition risk loci influence a tumour’s epigenome, uncovering a mechanism for cancer susceptibility. We identify and validate 1,178 loci associated with altered methylation in tumour but not non-malignant tissue. These tumour methylation quantitative trait loci (tumour meQTLs) influence chromatin structure and RNA and protein abundance. One prominent tumour meQTL is associated with AKT1 expression and is predictive of relapse after definitive local therapy in both discovery and validation cohorts. These data reveal intricate crosstalk between the germline and the epigenome of primary tumours, which may help identify germline biomarkers of aggressive disease to aid patient triage and optimize use of more invasive or expensive diagnostic assays.
Cancer is defined by a set of deregulated cellular processes, termed hallmarks1, which ultimately arise from genomic and epigenomic aberrations2,3. There are three sources for these aberrations: environmental (e.g. DNA-damaging xenobiotics), stochastic (e.g. replication-associated mutations) and genetic (e.g. inherited predisposition polymorphisms)4. Genome-wide association studies (GWAS) have revealed hundreds of germline variants associated with elevated risk of cancer diagnosis5-7. Further, some highly-penetrant polymorphisms in tumour suppressor genes including RB1, APC, BRCA1 and BRCA2 induce unique mutational phenotypes, including epigenomic dysregulation8-10.
Epigenetic aberrations associated with chemical modification of DNA provide additional modes of tumour-specific regulation11. Tumours can hijack epigenetic regulatory systems to silence tumour suppressors12,13 and large-scale rewiring of DNA methylation is common in many cancer types14. Susceptibility loci are enriched at regulatory regions15,16 and these loci can modulate the tumour epigenome17. For example, prostate tumours arising in men with deleterious germline BRCA2 mutations show a genome-wide reduction in methylation relative to sporadic tumours, which may account for their increased aggressivity18.
These data suggest that common germline polymorphisms may influence development of aggressive prostate tumours. GWAS have failed to identify loci robustly associated with prostate cancer survival19, perhaps due to insufficiently large cohort sizes. Because single nucleotide polymorphisms (SNPs) can confer susceptibility by modulating DNA methylation17, we reasoned that interrogating the more direct link between germline and methylation would yield associations with larger effect sizes than germline-survival analyses. As already observed in neuroscience20, germline-methylation analyses might overcome the limitations of small cohorts and identify loci otherwise not selected at genome-wide significance levels. Prognostic germline loci would be attractive minimally-invasive biomarkers to aid early clinical stratification of indolent vs. aggressive disease, and provide prior probabilities to maximize utility of more expensive fluid, tissue or radiologic assays.
We focus on prostate cancer, the second most common malignancy in men21, with few known risk factors22,23 and large molecular and clinical heterogeneity24,25. We compare germline whole-genomes and tumour methylomes of 589 patients with localized prostate cancer (ndiscovery=241 and nvalidation=348) and identify 7,590 validated cis-methylation quantitative trait loci (meQTLs), i.e. germline loci associated with altered methylation levels. Germline variants are not unique to the tumour, therefore we introduce a novel class of functional variants: tumour meQTLs. These are loci associated with altered methylation in tumour but not in non-malignant tissue (i.e. larger effect in tumour vs. reference tissue). We identify and validate 1,178 tumour meQTLs, and show that 17 of these demonstrate tumour-specific RNA or protein abundance changes (termed tumour meQTL-eQTLs). Tumour meQTLs are enriched at tumour-specific regulatory regions in prostate cell lines and primary tumours, and preferentially target sites of chromatin looping. Some tumour meQTLs target known prognostic cancer driver genes, including TCERG1L and AKT1. Indeed, the tumour meQTL targeting AKT1 is predictive of aggressive disease in both our discovery cohort (HR=2.85; P=5.8x10−3) and a validation cohort of 101 clinically matched samples (HR=2.2; P=1.7x10−2). Taken together, these data highlight how germline genotypes can modulate the tumour epigenome to contribute to the tumourigenesis of aggressive prostate cancers. This phenomenon may apply to other tumour types, providing a strategy to create robust, minimally-invasive biomarkers for early-detection of aggressive disease.
Results
Prostate cancer susceptibility loci associated with tumour methylation dysregulation
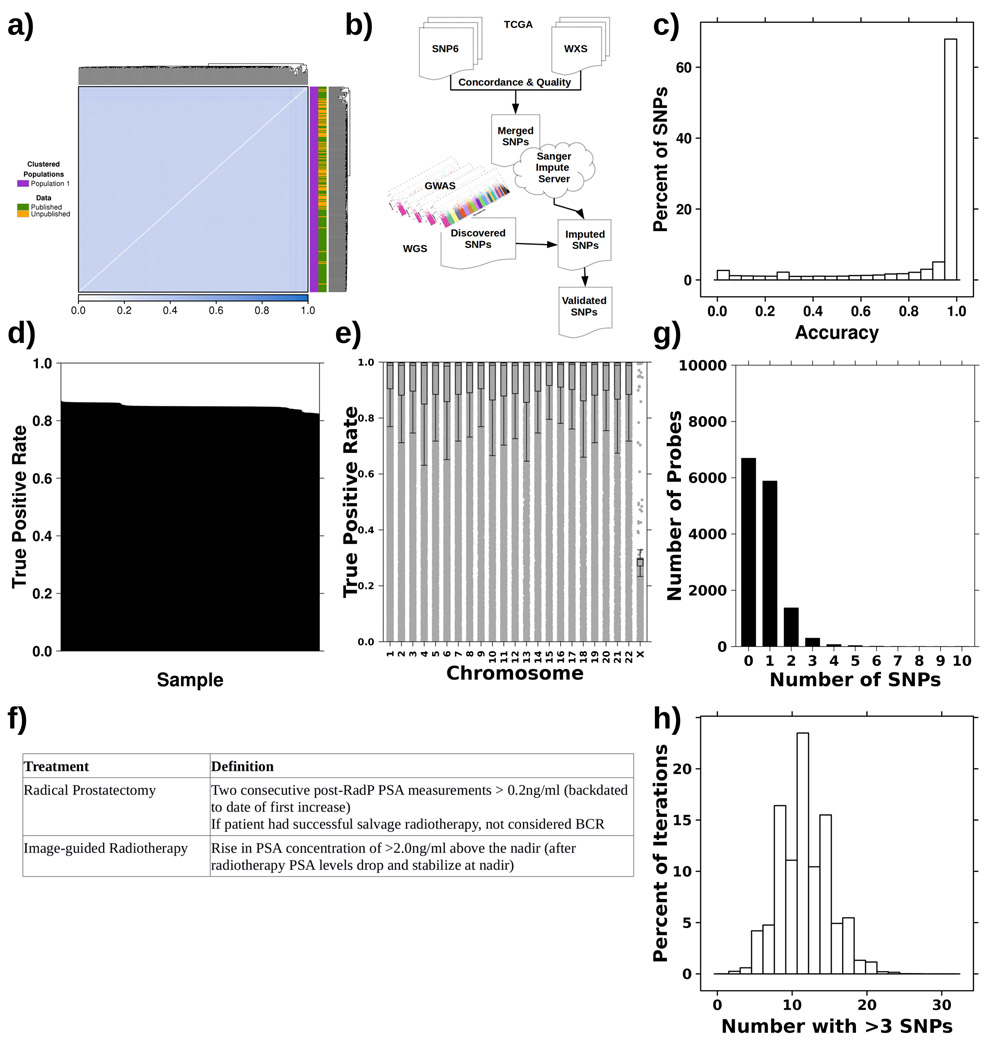
We assembled 241 patients with treatment-naïve prostate cancer that had germline whole-genome sequencing and methylation profiling by array, including 80 new genomes and 161 from the literature25,26. All patients had organ-confined clinically intermediate-risk disease and were treated by image-guided radiotherapy or surgery. Median clinical follow-up was 8.76 years. Identity-by-state clustering did not show population stratification (Extended Data 1a). Supplementary Table 1 summarizes this discovery cohort.
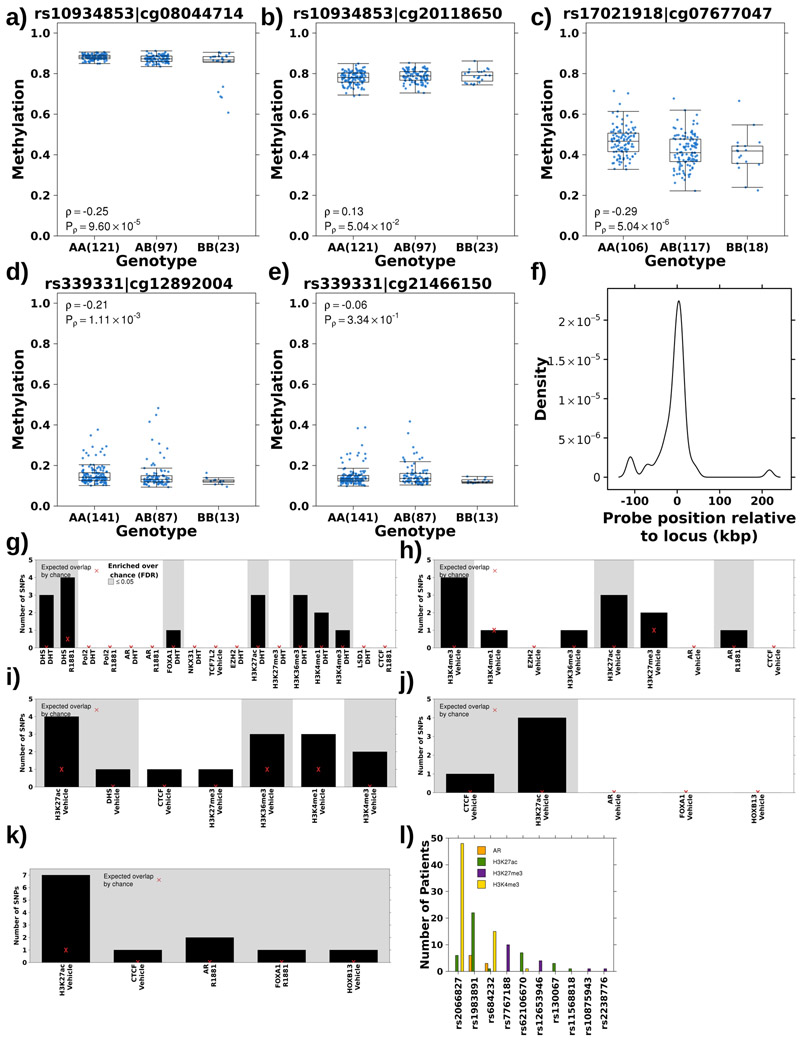
We sought to quantify polymorphisms that module specific epigenetic features of tumour evolution, termed tumour meQTLs to distinguish them from meQTLs which exert effects in normal epithelial tissue. First, we validated previous work17 showing evidence for the association of germline risk loci with tumour methylome alterations. We validated 3/5 of these meQTLs (P<0.01; see Methods; rs10934853:cg08044714, rs17021918:cg07677047, rs339331:cg12892004; Extended Data 2a-e; Supplementary Table 2).
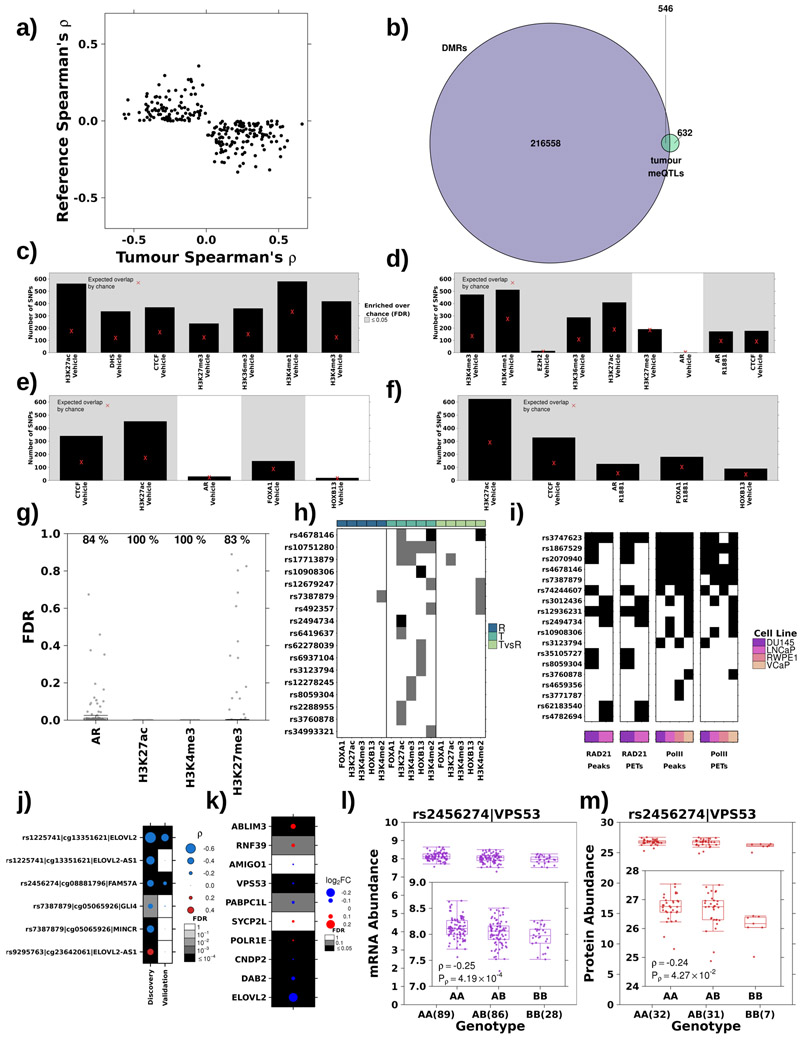
Next, we comprehensively analysed 160 validated germline susceptibility loci associated with prostate cancer incidence that account for 34.4% of familial risk15,25,27-32 (analysis 1a; Figure 1a; Supplementary Table 2). Each risk locus was tested for methylation associations methylome-wide, identifying 79 meQTLs covering 30 loci and 77 probes (P<7x10−10 Bonferroni-adjusted threshold; Spearman’s correlation; Figure 1b; Supplementary Table 2). Of these, 75/79 associations were in cis: the risk locus was located proximal to the methylated site (median distance 11.5±37.7 kbp; Extended Data 2f). There were four trans associations: rs2238776 (chromosome 22) associated with cg11491381 (AVP; chromosome 20), rs4976790 (chromosome 5) associated with cg05952543 and cg20792895 (MKRN3; chromosome 15) and rs7295014 (chromosome 12) associated with cg26860994 (SND1; chromosome 7). None of the risk variants within chromosome 8q24, a well characterized locus proven to be a major contributor to prostate cancer risk, were identified as meQTLs33.
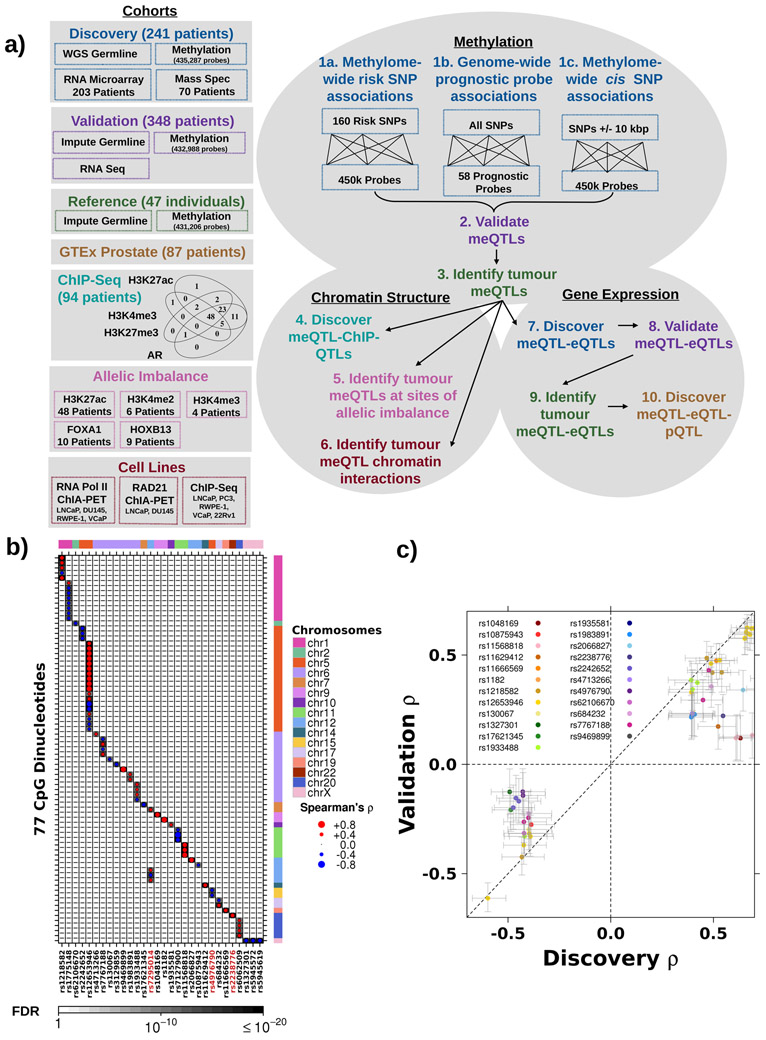
Figure 1 – Prostate cancer susceptibility loci associated with tumour methylation dysregulation.
a) Schematic of datasets and workflow. The boxes along the left and their corresponding colours indicate the dataset used at each step. b) Thirty risk loci (x-axis) were associated with 77 methylation probes (y-axis). Dot size represents Spearman’s ρ magnitude while colour indicates directionality. Background shading represents FDR. Covariate along the top represents the chromosome of each SNP, while the covariate along the right indicates the chromosome of each methylation site. Red ID indicates SNP is involved in a trans meQTL. c) Comparison of Spearman’s ρ in discovery and validation cohorts for 23 risk loci significantly associated (FDR < 0.05) with 55 methylation probes in the validation cohort. Diagonal dotted line represents the y=x line.
To validate these candidate meQTLs, we evaluated 348 cases from The Cancer Genome Atlas (TCGA) with tumour methylation data along with germline SNP array and whole exome sequencing of blood samples (WXS; analysis 2; Figure 1a)34. Following a stringent QC and imputation process (Extended Data 1b), we estimated our SNP detection accuracy in this validation cohort to be 98.8% (Extended Data 1c-e). After imputation, 69/79 risk loci meQTLs were genotyped in the validation cohort and 55/69 validated (23 loci and 55 methylation probes; FDR<0.05; Spearman’s correlation; Figure 1c; Supplementary Table 2). Three trans associations, rs2238776-cg11491381, rs4976790-cg05952543 and rs4976790-cg20792895, replicated in this independent validation dataset (FDR<2.22x10−2; Spearman’s ∣ρ∣>0.13; Supplementary Table 2). Thus, 14% of known prostate cancer risk loci may influence risk by modulating tumour methylation.
We quantified the enrichment of validated meQTLs in transcription factor binding sites or chromatin marks defined in four prostate cancer cell lines, LNCaP, PC3, 22Rv1 and VCaP, and one prostate epithelial cell line, RWPE-1 using ChIP-Seq data (see Methods; Supplementary Table 3). Risk meQTLs were enriched at active regulatory regions, including H3K27ac and H3K4me3 modifications indicative of active promoters, in all cell lines (FDR<0.05; permutation test n=105; Extended Data 2g-k). We confirmed the enrichment of risk meQTLs at regulatory regions in 94 primary prostate samples35. Ten out of 23 loci overlapped at least one of AR, H3K27ac, H3K4me3 or H3K27me3 sites in at least one patient (P=1.51x10−3; permutation test n=105; Extended Data 2l). There was allele specific H3K27ac modification at rs1983891 (β=0.72, P=0.05; logistic regression; Supplementary Table 4). Validated meQTL methylation targets were enriched in CpG islands on chromosomes 5 and 6 (OR>2.3; FDR≤8.43x10−3; Fisher’s Exact Test; Supplementary Table 4). To distinguish tumour meQTLs from meQTLs (loci that affect methylation in prostate epithelial tissue), we considered 47 tumour-adjacent prostate samples from TCGA (analysis 3; Figure 1a). Each tumour-adjacent sample, henceforth referred to as reference, was confirmed to be morphologically normal by pathology review, and had no detectable prostate cancer mutations34. Of these 55 validated meQTLs, 52 were evaluated in reference tissue and 14 were tumour-specific (defined as FDR>0.05 in reference tissue and FDR<0.05 in matched tumour tissue (n=47); Spearman’s correlation; Supplementary Table 2). Only 3/14 tumour meQTLs were proximal to a gene (within 1,500bp) and none were significantly associated with mRNA changes (FDR<0.05; Spearman’s correlation; Supplementary Table 2). Finally, we identified meQTLs missed in the discovery cohort by conducting meQTL discovery in the TCGA cohort. We discovered 165 meQTLs (32 loci and 144 probes) in TCGA of which 32 novel meQTLs validated in the discovery cohort (18 loci and 30 probes; FDR<0.05; Spearman’s correlation; Supplementary Table 2). These results expand our understanding of the role of risk loci in modulating tumour methylation and suggest that we are likely underestimating the extent of this modulation.
Germline variants associate with prognostic methylation levels
Upon validating risk loci tumour meQTLs, we discovered novel loci candidates by identifying tumour meQTLs genome-wide associated with tumour aggressivity. Germline loci that could delineate indolent from aggressive disease would provide a minimally invasive, early-detection biomarker filling a important clinical gap. We selected 58 methylation probes based on their association with biochemical relapse defined by increasing PSA levels following primary treatment; a trigger of salvage therapy and, when occurring within 18 months of primary treatment, a surrogate for prostate-cancer specific mortality36 (Extended Data 1f; Extended Data 3a; Supplementary Table 5). We identified candidate loci genome-wide for each of the 58 prognostic methylation probes (analysis 1b; Figure 1a) and discovered 292 meQTLs targeting 28% of these probes (16/58), covering 223 distinct loci (P<5x10−8; Spearman’s correlation; Figure 2a). For each of these loci, the presence of one or more alternative alleles was associated with significant changes in methylation.
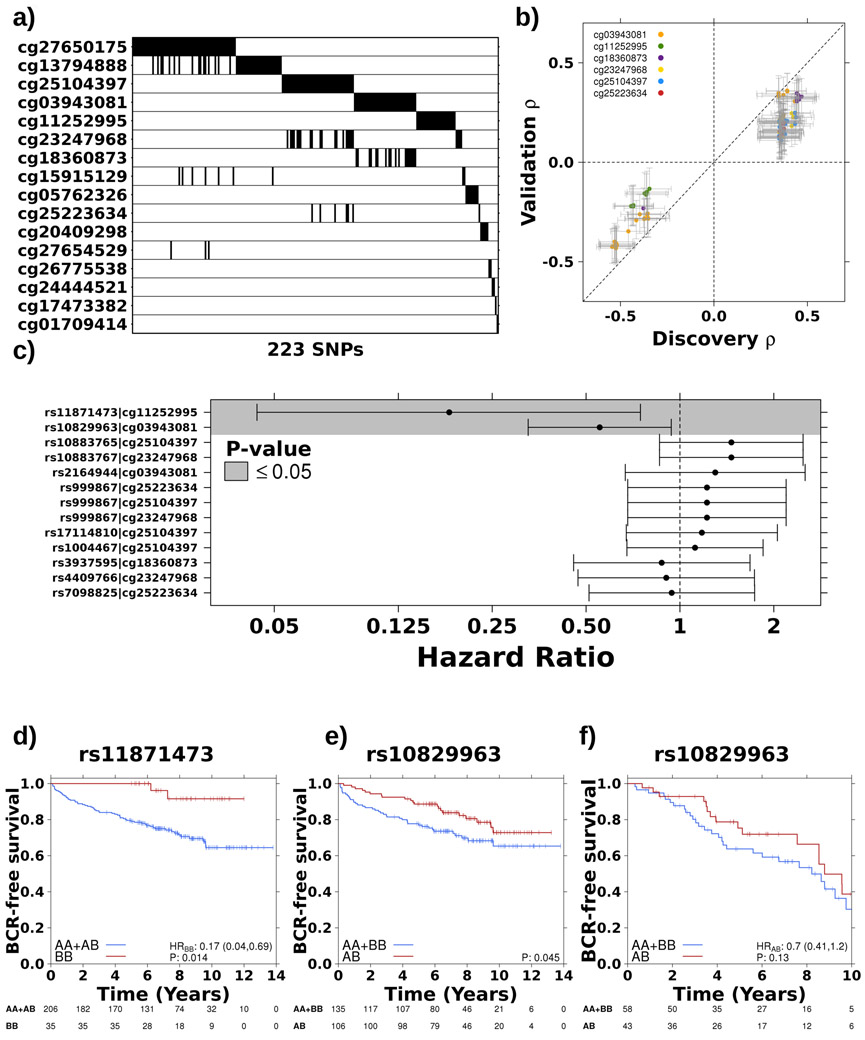
Figure 2 – Germline variants associate with prognostic methylation levels.
a) Summary of 223 significant SNPs (x-axis) for each prognostic methylation probe (y-axis). Black indicates that a SNP is significantly associated (p-value < 5x10−8) with methylation levels at that probe. b) Comparison of Spearman’s ρ in discovery and validation cohorts for 75 SNPs that were significantly associated (FDR < 0.05) with six methylation probes in the validation cohort. Diagonal dotted line represents the y=x line. c) Two cis-meQTLs were prognostic. Dots and error bars represent hazard ratios and 95% confidence intervals, respectively, for the tag SNP from each of the seven haplotypes. Dotted line indicates HR = 1 and grey background shading indicates P ≤ 0.05. d) The homozygous alternative genotype of a haplotype on chromosome 17, associated with methylation of ATP2A3, gives a survival advantage. Hazard ratio and p-value from CoxPH model. e) A haplotype on chromosome 10, associated with methylation of TCERG1L, is co-dominantly associated with BCR. f) Co-dominant association with BCR replicated at rs10829963 in independent cohort (n=101).
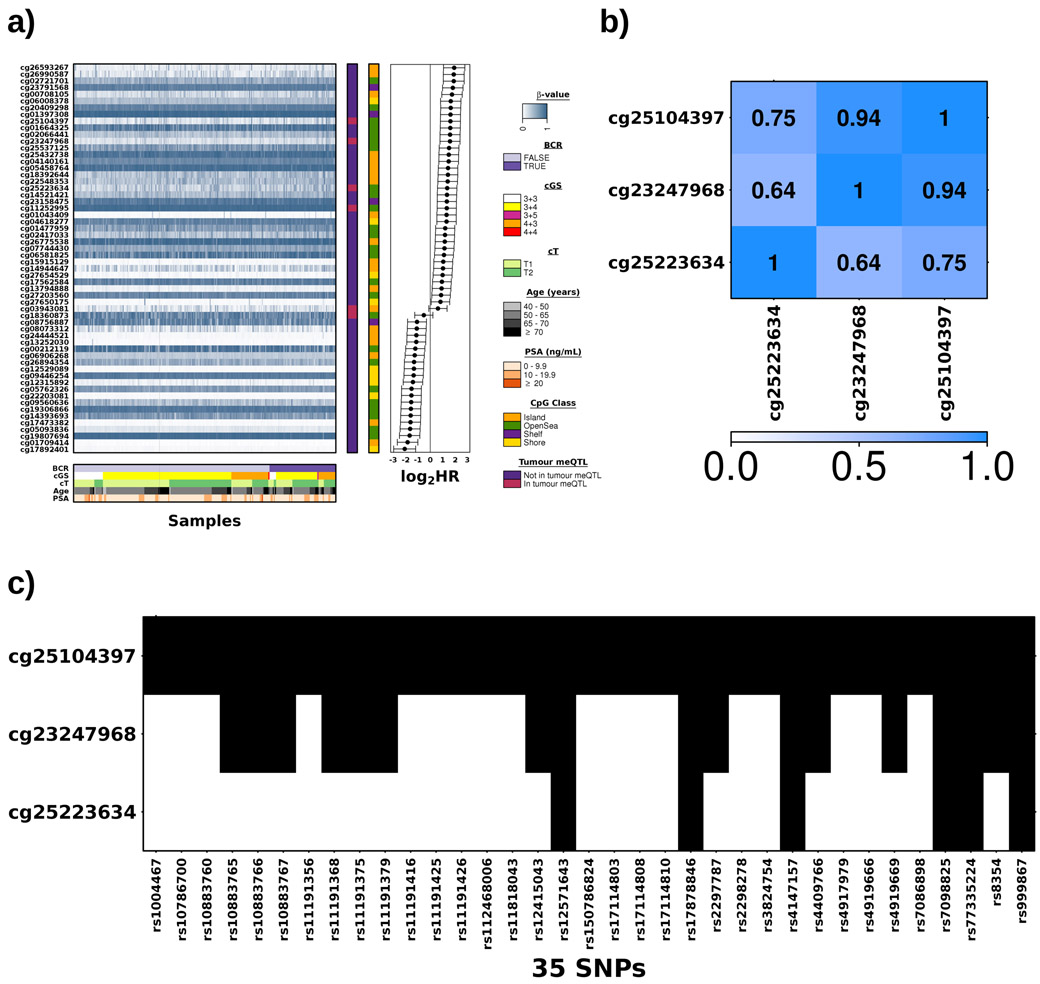
The TCGA dataset34 was used to validate these prognostic meQTLs, providing genotype information for 151/292 loci (analysis 2; Figure 1a). Of these, 113/151 meQTLs validated, representing seven cis-haplotypes covering six methylation probes (FDR<0.05; Figure 2b; Supplementary Table 6). These included 35 loci associated with methylation of cg25104397 (FDR≤4.92x10−2; Spearman’s ∣ρ∣=0.11-0.25), 17 with cg23247968 (FDR≤1.24x10−3; ∣ρ∣=0.18-0.25) and six with cg25223634 (FDR≤2.02x10−2; ∣ρ∣=0.13-0.20; Supplementary Table 6). These three probes are located within 41 bp on chromosome 10, within an open sea region of C10orf26. Their methylation was highly correlated and six loci were associated with all three sites (Extended Data 3b-c). We used paired tumour/reference samples to determine if these meQTLs were tumour-specific (i.e. FDRtumour<0.05 & FDRreference>0.05; Spearman’s correlation; analysis 3; Figure 1a) and identified 38 tumour meQTLs (FDRreference>0.25), all of which target methylation at two sites, cg18360873 and cg03943081 (Supplementary Table 6). These methylation probes are located 5’ and 3’, respectively, of Transcription Elongation Regulator 1 Like (TCERG1L), an epigenetic driver event in prostate cancer26.
Given the prognostic value of the methylation sites targeted by these meQTLs, we evaluated the prognostic value of the loci themselves. As seen earlier, not all risk meQTLs were tumour specific, suggesting meQTLs that have a role in reference tissue are also biologically important. Therefore, we considered tag SNPs for all seven haplotypes involved in validated meQTLs. Two cis-meQTLs were predictive of biochemical recurrence (HR=0.554 & 0.180; P=2.92x10−2 & 1.73x10−2; CoxPH model; Figure 2c-e) and one, rs10829963, showed the same survival trend in an independent cohort26,27,37,38 of 101 clinically-matched patients (HR=0.70; P=0.13; Figure 2f). The validation cohort was insufficiently powered to test rs11871473 (nBB=11; 1–β=0.44). Taken together, these results suggest that the germline may shape tumour aggressivity via tumour methylation dysregulation.
The landscape of cis-tumour meQTLs
All validated tumour meQTLs were in cis associations (i.e. the loci were within 59,151 bp of the methylation site). To quantify cis-tumour meQTL frequency in prostate cancer, we systematically evaluated loci within a 10 kbp window around each of the 434,504 methylation probes (analysis 1c; Figure 1a). We identified 169,562 loci associated with the methylation status of 3.3% of all CpGs quantified (14,287 distinct probes; P<3x10−9 representing 1.5x107 independent tests; Spearman’s correlation; Figure 3a). These associations are not driven by variants affecting the hybridization of probes on the methylation array (Extended Data 1g-h).
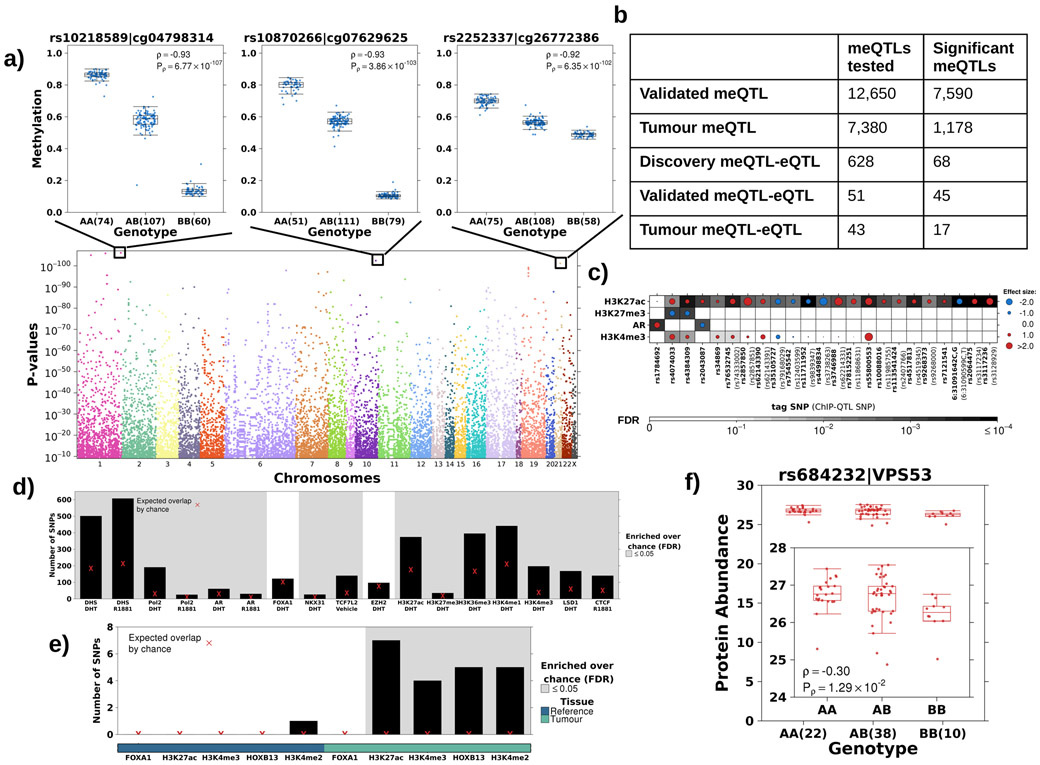
Figure 3 – The landscape of cis tumour meQTLs.
a) Identification of cis tumour meQTLs methylome-wide (P<3x10−9; Bonferroni adjustment). Each point represents a SNP, ordered by chromosome along x-axis. Y-axis gives p-value from Spearman’s correlation. Representative boxplots, showing methylation (y-axis) discretized by genotype (x-axis), for three germline-methylation associations. Boxplot represents median, 0.25 and 0.75 quantiles with whiskers at 1.5x interquartile range. Blue points refer to methylation values and number of samples with each genotype is given in brackets. b) Table shows number of meQTLs tested and statistically significant (FDR<0.05 based on Spearman’s correlation) at each stage. c) Tumour meQTLs demonstrated allele specific AR binding or histone modification (n=30; 23 unique SNPs). Circle size and colour represents magnitude and sign of coefficient from logistic regression model (i.e. red indicates alternate allele associated with increased binding and blue decreased binding). Background shading represents FDR. X-axis labels show tag tumour meQTL-ChIP-QTL and SNP ID in brackets indicate the ChIP-QTL SNP in the case that the ChIP-QTL SNP is not the tag SNP. d) Tumour meQTLs were enriched at transcription factor binding sites and active regulatory elements in LNCaP cells. Y-axis shows number of tumour meQTLs that overlap each target/treatment pair. Background shading indicates FDR<0.05 from permutation analysis (n=105 permutations). Red X represents expected number of overlapping loci by chance. e) Tumour meQTLs were interrogated for overlap with sites of allelic imbalance at FOXA1, H3K27ac, H3K4me3, HOXB13 and H3K4me2 peaks in tumour and reference tissue. Y-axis shows number of tumour meQTLs that overlap each target. Background shading indicates FDR<0.05 from permutation analysis (n=105 permutations). The bottom covariate indicates sites of allelic imbalance in tumour or reference tissue. Red X represents expected number of overlapping loci by chance. f) The tumour meQTL-eQTL-pQTL identified in this analysis is in LD with the risk locus, rs684232, which is also a pQTL for VPS53. Boxplot represents median, 0.25 and 0.75 quantiles with whiskers at 1.5x interquartile range and red points refer to protein abundance values. The number of samples with each genotype is given in brackets.
We validated the locus with the lowest p-value for each probe, provided it was genotyped on the TCGA platform (12,650 loci; analysis 2; Figure 1a; Supplementary Table 7) and 7,590/12,650 (60%) cis-meQTLs validated in this independent cohort (FDR<0.05; Spearman’s correlation; Figure 3b). Of the 7,590 validated meQTLs, 7,380 had genotype and methylation data for the 47 reference prostate samples34 (analysis 3; Figure 1a). A third (1,178/7,380) were consistent with tumour meQTLs, meaning they had associations in tumour tissue (FDRtumour<0.05 in matched tumour samples) but not reference prostate epithelial (FDRreference≥0.05) or had opposite effects in tumour and reference tissue (234 meQTLs; sign(ρtumour)≠sign(ρreference); Figure 3b; Extended Data 4a). Almost half (546/1,178) of these tumour meQTLs were differentially methylated between tumour and reference tissue, suggesting at least a subset of tumour meQTLs target dysregulated methylation sites (Extended Data 4b). These probes were enriched for open seas in intergenic regions on chromosome 6 (OR>1.07; FDR≤3.10x10−2; Fisher’s Exact Test; Supplementary Table 8). By contrast, CpG islands in promoter regions on chromosome X were significantly depleted of validated associations (OR<0.86, FDR≤2.19x10−3, Fisher’s Exact Test; Supplementary Table 8). The depletion on chromosome X may result from less accurate imputation on this chromosome (Extended Data 1e).
Tumour meQTLs target active regulatory regions and sites of chromatin looping
To determine whether tumour meQTLs influence transcription regulation, we quantified the enrichment of tumour meQTLs at regulatory regions in four prostate cancer cell lines (LNCaP, PC3, VCaP and 22Rv1) and one prostate epithelial cell line (RWPE-1) (Supplementary Table 3). Tumour meQTLs were enriched at AR and CTCF bindings sites in LNCaP as well as active enhancers and promoters, as seen by enrichment at H3K27ac, H3K4me1 and H3K4me3 marks (FDR<1x10−26; permutation test n=105; Figure 3d). Enrichment at regulatory regions was replicated in PC-3, VCaP, 22Rv1 and RWPE-1 cell lines (Extended Data 4c-f). Tumour meQTLs were more strongly enriched at the repressive chromatin mark H3K27me3 in the epithelial cell line, RWPE-1 (FDR<1x10−26; Extended Data 4c), than the cancer cell lines PC3 (FDR=0.36; Extended Data 4d) or LNCaP (FDR=0.006; Figure 4d), suggesting a subset of these sites may activate during tumourigenesis. To confirm this enrichment at active regulatory regions, we considered H3K27ac, H3K27me3, H3K4me3 and AR ChIP-Seq data from 94 primary prostate cancer samples35 (analysis 4; Figure 1a). Tumour meQTLs were significantly enriched at H3K27ac and H3K4me3 sites in all primary samples and at AR binding sites in 84% of samples (FDR<0.05; permutation test n=105; Extended Data 4g). To identify specific tumour meQTLs modulating chromatin structure, we tested for allele-specific AR binding and H3K27ac, H3K27me3 and H3K4me3 histone modifications (i.e. ChIP-QTLs) in primary prostate cancer samples35 (analysis 4; Figure 1a). We discovered 30 tumour meQTL-ChIP-QTLs, 23 unique loci, targeting one of the four marks (FDR<0.05; H3K27ac=22, H3K27me3=2, AR=2 & H3K4me3=4; logistic regression; Figure 3c). The variant rs2043087 is located within ALDH1A2, a prostate cancer tumour suppressor39, and is associated with increased H3K27ac (β=1.27; FDR=6.04x10−3) but decreased AR binding (β=−1.49; FDR=9.00x10−3).
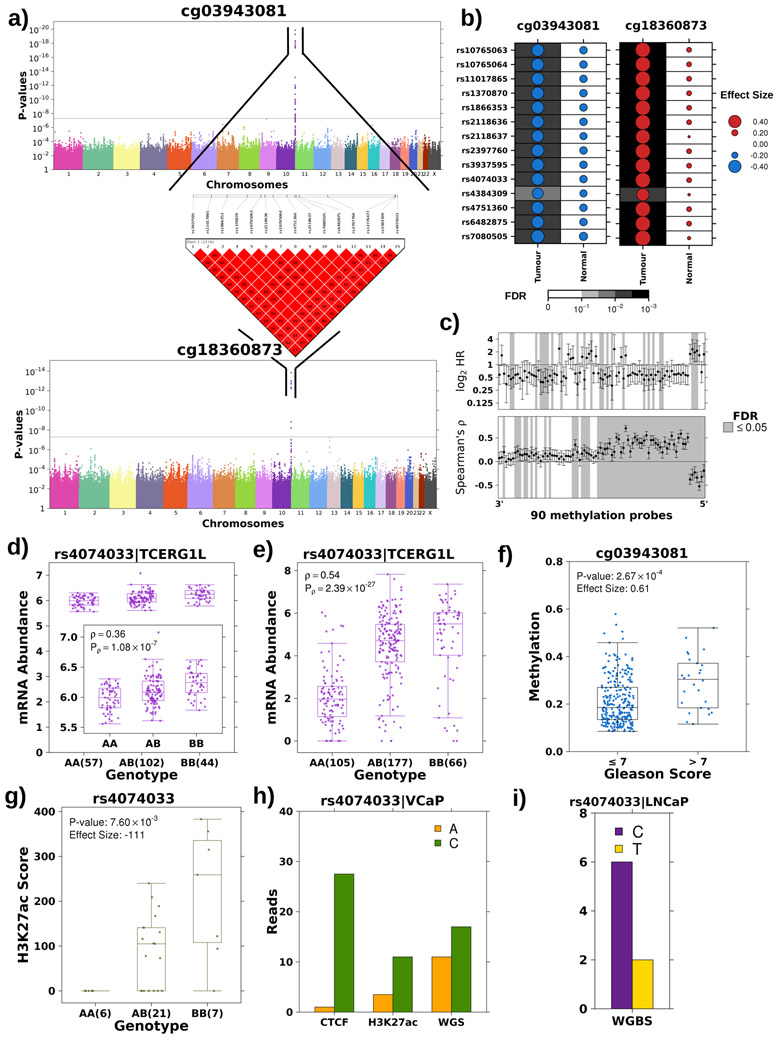
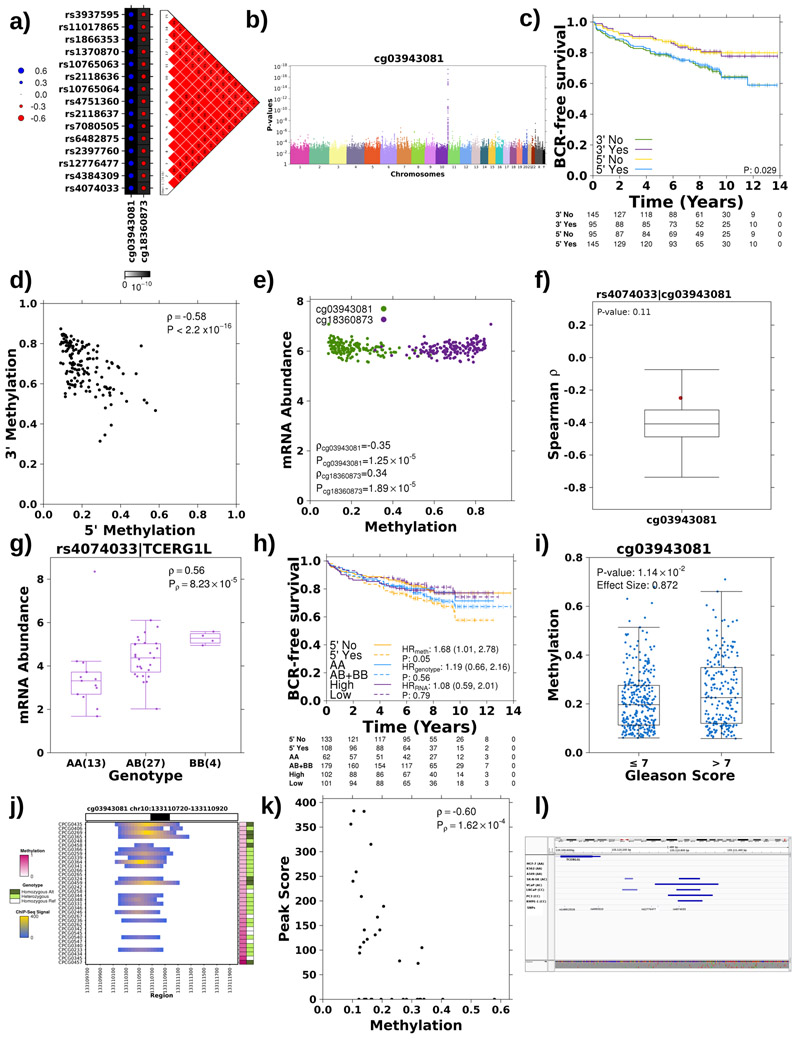
Figure 4 – Tumour meQTL associated with TCERG1L regulation.
a) Haplotype on chromosome 10 strongly associated with methylation probes at both 5’ and 3’ ends of TCERG1L. Manhattan plot presents p-values (y-axis; Spearman’s correlation) for association of each SNP, x-axis ordered by chromosome, with methylation at both 5’ (cg03943081) and 3’ (cg18360873) ends of TCERG1L. The grey line represents the Bonferroni adjustment. All associated SNPs are in strong LD. LD plot shows pairwise D’ values between all associated SNPs where a solid red square indicates a D’ value of 1. b) TCERG1L meQTLs were tumour-specific. Dot size reflects the magnitude and dot colour reflects the directionality of Spearman’s ρ between genotype and methylation at probes 5’ and 3’ of TCERG1L. The background shading indicates the FDR. c) Genotype at rs4074033 was associated with methylation levels of 64/90 probes spanning TCERG1L. Bottom forest plot shows Spearman’s correlation and 95% CI for association of each methylation probe (x-axis) and genotype at rs4074033. Horizontal line represents ρ=0. Top forest plot shows HR and 95% CI for association of methylation probes with BCR using a CoxPH model. Horizontal line represents HR = 1. Grey shading in both plots indicates significant association (FDR<0.05). d-e) The alternative (B) allele exhibits a dominant effect resulting in increased mRNA abundance of TCERG1L in both the discovery (d) and the validation cohort (e). Boxplot represents median, 0.25 and 0.75 quantiles with whiskers at 1.5x interquartile range and purple points refer to mRNA abundance values. The number of samples with each genotype is given in brackets. Difference in abundance levels was quantified using Spearman’s correlation. f) Methylation 5’ of TCERG1L (cg03943081) is significantly associated with Gleason Score in the discovery cohort. Effect quantified by Mann-Whitney and effect size is fold change. Blue points refer to methylation levels. g) The alternative allele showed increased H3K27ac modification in the discovery cohort. Effect quantified using Mann-Whitney test (AA vs. AB+BB) and effect size represents differences in medians. Green points refer to H3K27ac peak signal. h) The alternative allele at rs4074033 preferentially shows H3K27ac modification and is preferentially bound by CTCF. The VCaP cell line is heterozygous at rs4074033 (i.e. genotype: AC). The y-axis shows the number of reads with each allele at rs4074033 from CTCF and H3K27c ChIP-Seq and WGS data. i) The alternative allele at rs4074033 is methylated in LNCaP cell lines (genotype: CC). Y-axis shows number of reads with the methylated C allele vs. the unmethylated T allele (from WGBS).
We further characterized a high-confidence subset of 59 tumour meQTLs associated with biochemical recurrence (P<0.05; CoxPH model; Supplementary Table 7). To support the tumour-specific role of tumour meQTLs in modulating protein-DNA interactions, we identified sites of allelic imbalance in transcription factor binding and histone modification genome-wide in paired tumour and reference samples. Sites of allelic imbalance reflect loci with high regulatory potential. Specifically, we discovered sites of allelic imbalance in tumour and reference samples for FOXA1, HOXB13, H3K27ac, H3K4me3 and H3K4me2. We observed a strong enrichment of tumour meQTLs at H3K27ac, H3K4me3, HOXB13 and H3K4me2 sites in tumour (FDR<0.01; permutation test n=105) but not reference samples (FDR>0.19), supporting their tumour-specific role (Figure 3e; Extended Data 4h; Supplementary Table 7). Next, we explored the impact of tumour meQTLs on chromatin structure, specifically RAD21 and RNA polymerase II (RNA Pol-II) chromatin loops in LNCaP, DU145, VCaP and RWEP-1 cell lines (analysis 6; Figure 3a). Fourteen tumour meQTLs overlapped with RNA Pol-II peaks in at least one cell line, most (12/14) were involved in chromatin looping (Extended Data 4i; Supplementary Table 7). Eleven overlapped with RAD21 binding sites and 9/11 were involved in chromatin loops. Seven overlapped with RNA Pol-II and RAD21 sites suggesting these tumour meQTLs are targeting active enhancer-promoter interactions. These results show tumour meQTLs preferentially target cis-regulatory elements in a tumour-specific manner. Tumour meQTL mechanisms are likely myriad, including disrupting AR binding (rs1784692 or rs2043087), deregulating RNA Pol-II looping (rs3747623 or rs1867529) and others not recognized in this first study.
Tumour meQTLs drive aggressive gene expression program
DNA methylation can directly dysregulate transcription, thus we quantified tumour meQTLs modulation of the transcriptome (microarray profiling) of 203 patients in the discovery cohort. We focused on validated tumour meQTLs with methylation sites proximal to a gene (within 1,500 bp; 628 associations; analysis 7; Figure 1a). We identified 68 tumour meQTLs associated with mRNA abundance in the discovery cohort (termed tumour meQTL-eQTLs; FDR<0.05, Spearman’s ∣ρ∣=0.20-0.55), of which 45 also associated with mRNA abundances in the TCGA validation cohort (analysis 8; FDR<0.05, ∣ρ∣=0.11-0.75; Figure 3b; Supplementary Table 7). Utilizing RAD21 and RNA Pol-II ChIA-PET profiling of prostate cancer cell lines, we identified additional targets for 17 tumour meQTLs (distance between locus and target: 0-148.5 Mbp; median=13.9 Mbp) and four were significantly associated with mRNA abundance of five transcripts (Extended Data 4j). We discovered a significant association between a tumour meQTL-eQTL targeting MINCR, a MYC-induced long non-coding RNA that has been implicated in Burkitt Lymphoma and Gallbladder cancer40,41. Only three of these eQTLs could be tested in TCGA and 2/3 validated, one was previously reported15 (rs2456274:FAM57A and rs1225741:ELOVL2; FDR<0.05; Extended Data 4j). We confirmed 17/43 tumour meQTL-eQTLs were tumour-specific at the RNA level using prostate epithelial eQTL statistics from Genotype-Tissue Expression (GTEx) project42 (FDR>0.05; Figure 3b; Table 1; Supplementary Table 7). These 17 were not enriched in any specific pathway, however 6/10 genes involved in these tumour meQTL-eQTLs were differentially abundant in tumour vs. reference tissue (FDR<0.05; Extended Data 4k).
Table 1:
Significant tumour meQTL-eQTLs
| SNP | Methylation Probe | Gene |
|---|---|---|
| rs1225741 | cg13351621 | SYCP2L |
| rs16934152 | cg13558087 | POLR1E |
| rs2456274 | cg08881796 | VPS53 |
| rs2570972 | cg08367326 | AMIGO1 |
| rs3761188 | cg09328228 | PABPC1L |
| rs3761188 | cg15588266 | PABPC1L |
| rs3764509 | cg14963724 | CNDP2 |
| rs3807032 | cg24330456 | RNF39 |
| rs3807033 | cg05563515 | RNF39 |
| rs3807033 | cg17322683 | RNF39 |
| rs3807033 | cg23793213 | RNF39 |
| rs3849767 | cg18264728 | DAB2 |
| rs4147470 | cg03997398 | ABLIM3 |
| rs4147470 | cg04669407 | ABLIM3 |
| rs9261309 | cg13918754 | RNF39 |
| rs9261309 | cg20249327 | RNF39 |
| rs9295763 | cg20249327 | ELOVL2 |
As an exploratory analysis, we tested if the ten genes in these 17 tumour meQTL-eQTLs were dysregulated at the protein level (analysis 10; Figure 1a). We exploited a dataset of 70 tumours with mass spectrometric quantitation of protein abundances38. Only 3/10 transcripts had their protein abundances quantified, and the small sample-size led to very low statistical power (1–β<0.39). Nevertheless, Vacuolar Protein Sorting-Associated Protein 53 Homolog (VPS53) was a strong tumour meQTL (P=6.95×10−12; ρ=−0.42) that associated with both RNA (FDR=8.22x10−3; ρ=−0.25) and more modestly protein abundances (P=4.27x10−2, ρ=−0.24; Extended Data 4l-m). This tumour meQTL-eQTL-pQTL is of particular interest because rs2456274 is in linkage disequilibrium (LD) with the risk locus rs684232 (D’=1; P=1.29x10−2; ρ=−0.30; Figure 3f) which has been reported as an eQTL for VPS5315. Thus, tumour meQTLs discovery recapitulated a known risk loci, confirming the value of this approach in identifying novel susceptibility loci.
Tumour meQTL associated with TCERG1L regulation
To further characterize novel loci of interest, we focused on tumour meQTLs targeting prognostic methylation sites within and 5’ to TCERG1L (i.e. identified in analysis 1b; Figure 1a). TCERG1L was previously identified as a strong epigenetic driver of aggressive prostate cancer (HR=2.90; 95% CI: 1.30-6.30; P=0.007; n=130)26 and its paralog, TCERG1, is recurrently mutated in prostate cancer43. Further, TCERG1L promoter hyper-methylation has been reported in colorectal cancer44,45. In the discovery cohort, methylation of TCERG1L was strongly associated with a 15-locus region on chromosome 10q26.3 adjacent to and inside of its gene body (P<4.35x10−9; Spearman’s ∣ρ∣=0.42-0.58; Figure 4a). These loci were in strong LD and were associated with both the 5’ and 3’ probes even when correcting for tumour cellularity (Extended Data 5a-b). The haplotype had opposite effects on the 5’ and 3’ probe – i.e. the alternative allele was associated with decreased methylation of the 5’ probe but increased methylation at the 3’ probe (Extended Data 5a). Concordantly, methylation at these two probes was anti-correlated and had opposing effects on patient outcome (Extended Data 5c-e). The TCERG1L meQTL was confirmed to be tumour specific at the 3’ and 5’ probes (FDRreference>0.14; Spearman’s ∣ρ∣=0.08-0.27; permutation P=0.11, see Methods; Figure 4b; Extended Data 5f).
To further interrogate the TCERG1L tumour meQTL, we assessed the methylation profile of 90 probes spanning TCERG1L. Methylation of 64/90 probes was significantly associated with the tag SNP, rs4074033 (Figure 4c; Supplementary Table 9), and 25/90 were associated with biochemical relapse (FDR<0.05; Cox PH model), expanding TCERG1L-methylation from an epigenetic driver26 to a tumour meQTL driver.
Additionally, tumour meQTLs in TCERG1L correlated with mRNA abundance: the non-reference allele was dominantly associated with increased TCERG1L mRNA in our discovery cohort and the TCGA validation cohort (P=2.67x10−8 & 4.53x10−26, respectively, Mann-Whitney; effect size=−0.38 & −2.87; Figures 4d-e). While rs4074033 was identified as a tumour-specific meQTL, it was significantly associated with TCERG1L mRNA abundance in reference tissue, an association also observed in GTEx42 (Extended Data 5g). Out of genotype, tumour methylation and tumour mRNA abundance, tumour methylation was the strongest prognostic measure (HR=1.68; 95% CI=1.01-2.78; P=0.05; Extended Data 5h), concordant with the literature26, suggesting tumourigenic dysregulation is targeted at methylation. Methylation is also significantly associated with Gleason Score in the discovery and validation cohorts (FCdiscovery=0.61; Pdiscovery=2.67x10−4; FCvalidation=0.87; Pvalidation=1.14x10−2; Mann-Whitney; Figure 4f; Extended Data 5i).
Next, we evaluated the effect of TCERG1L germline-dependent tumour methylation on chromatin organization, specifically H3K27ac modifications46. In agreement with the mRNA abundance data, three SNPs within the haplotype (rs12776477, rs4384309, rs4074033) were located within 100 bp of an H3K27ac peak, and the alternative allele dominantly increased the peak score (medianAA-medianAB+BB=−111, Mann-Whitney; P=7.60 x 10−3; Figure 4g; Extended Data 5j). As further confirmation, H3K27ac modification was negatively correlated with 5’ methylation of the gene (Spearman’s ρ=−0.60, P=2.65x10−4; Extended Data 5k) and was replicated in an independent cohort35 (β=1.82; P=7.65x10−3; logistic regression). The alternative allele was significantly associated with decreased H3K27me3 (β=−1.72; P=3.40x10−4) and increased H3K4me3 (β=1.66; P=7.51x10−4) modifications (Figure 3c). Finally, across eight cell lines, only cell lines with at least one alternative allele showed CTCF binding (Extended Data 5l). In VCaP prostate cancer cells, which are heterozygous at rs4074033, the alternative allele was preferentially bound by CTCF and preferentially subject to H3K27ac modification (Figure 4h). The two alleles of rs4074033 differ by an A-C transversion, with the C allele harbouring a CpG not present in the A allele. This CpG is methylated in LNCaP cell lines (Figure 4i). This methylation is consistent with differential CTCF binding, which is associated with altered poly(ADP-ribose) polymerase 1 (PARP1) activity and subsequently DNA (cystosine-5)-methyltransferase 1 (DNMT1) activity47. Taken together, these data show that germline loci in TCERG1L may influence the methylation and chromatin organization of the gene via CTCF binding in a tumour-specific manner supporting reports of TCERG1L as an epigenetic driver of aggressive prostate cancer26.
Tumour meQTL associated with AKT1 regulation
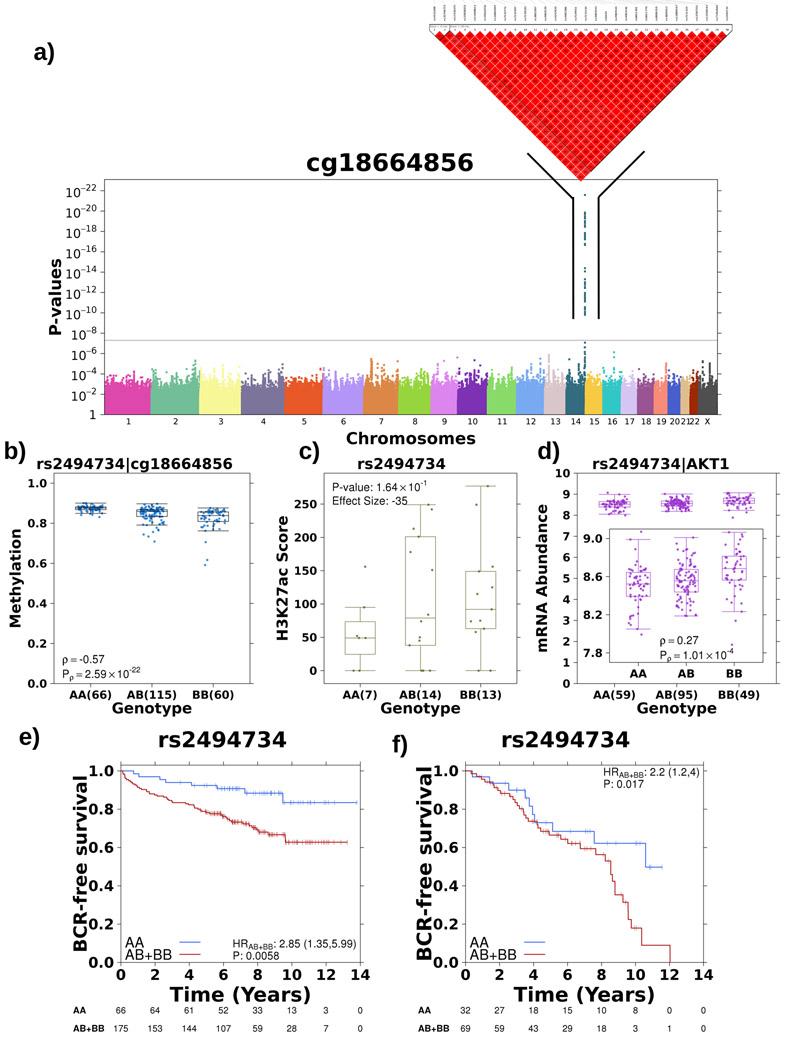
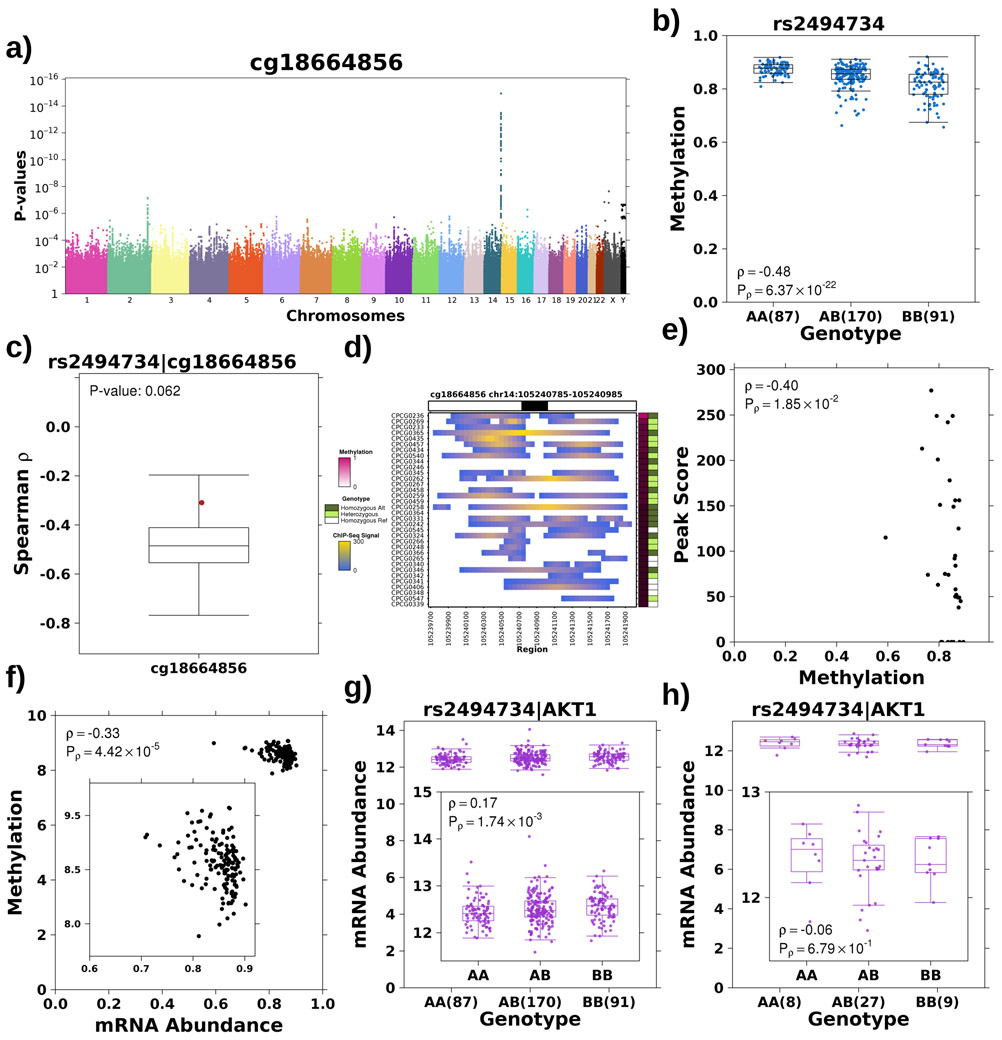
Next, we screened other driver genes that account for prostate cancer aggression and observed a similar link between germline loci, tumour methylation and histone organization for AKT1, which with MYCN is sufficient to transform prostate epithelial cells into adenocarcinomas48 and is associated with elevated risk of prostate cancer incidence49-51. We discovered an association between a 30-loci haplotype both 5’ and spanning into the oncogene AKT1 on chromosome 14 and a methylation probe within a CpG island in the gene body (cg18664856; Figure 5a). The alternative allele additively decreased the methylation of this probe, quantified using the tag SNP rs2494734 (Spearman’s ρ=−0.57, P=2.59x10−22; Figure 5b). The meQTL was robust to correction for tumour cellularity, validated in the TCGA cohort and was tumour specific (Spearman’s ρtumour=−0.39, FDRtumour=0.015, ρreference=−0.31, FDRreference=0.054; permutation P=0.06; see Methods; Extended Data 6a-c; Supplementary Table 7). Furthermore, the alternative allele dominantly associated with increased H3K27ac modification46 (effect size=−35, P=0.164, Mann-Whitney; Figure 5c) and H3K27ac modification was negatively correlated with cg18664856 methylation (Spearman’s ρ=−0.39, P=2.76x10−2; Extended Data 6d-e). Because methylation of cg18664856 was also negatively correlated with AKT1 mRNA abundance (Spearman’s ρ=−0.38, P=1.54x10−5; Extended Data 6f), we checked the effect of rs2494734 genotype on ATK1 mRNA abundance. Consistently, the alternative allele was additively associated with increased AKT1 mRNA abundance (Spearman’s ρ=0.27; P=1.01x10−4; Mann-Whitney; Figure 5d). This effect was validated in the TCGA cohort (Spearman’s ρtumour=0.17, Ptumour=1.74x10−3; Extended Data 6g). While no association was seen in the TCGA reference tissue (ρreference=−0.06, Preference=0.68; Extended Data 6h), this eQTL was reported in GTEx42 with a p-value above the genome-wide significance level (P>1.62x10−10; Supplementary Table 7).
Figure 5 – Tumour meQTL associated with AKT1 regulation.
a) Haplotype on chromosome 14 strongly associated with methylation of a probe within the gene body of AKT1. Manhattan plot represents p-values from Spearman’s correlation as outlined previously. b) The alternative allele is associated with decreased methylation of cg18774856, effect quantified by Spearman’s correlation. Boxplot represents median, 0.25 and 0.75 quantiles with whiskers at 1.5x interquartile range and blue points refer to methylation values. The number of samples with each genotype is given in brackets. c) The alternative allele showed increased H3K27ac modification in this region, effect quantified by Mann-Whitney test (AA vs. AB+BB) and effect size gives difference in medians. Green points refer to H3K27ac ChIP-Seq signal. d) The alternative allele was associated with increased mRNA abundance of AKT1, effect quantified by Spearman’s correlation. Purple points refer to mRNA abundance. e) The presence of the alternative allele confers a survival disadvantage as presented in Kaplan-Meier plot with time along the x-axis in years and estimated proportion of individuals without biochemical recurrence event on y-axis. The hazard ratio from a CoxPH model is also presented along with the number of individuals without an event in each group at each time point along the bottom. f) Alternative allele at rs2456274 is dominantly associated with rapid biochemical recurrence in an independent cohort (n=101).
Finally, given the robust literature on AKT1’s oncogenic functions and therapeutic value52, we tested the effect of rs2494734 genotype on survival. The alternative allele was dominantly associated with increased risk of relapse (HR=2.85; 1.35-5.99 95% confidence intervals; P=5.80x10−3; CoxPH model; Figure 5e) and was validated in an independent cohort of 101 patients26,27,37,38 (HR=2.2; 1.2-4.0 95% confidence intervals; P=0.017; Figure 5f). These findings highlight another example of the interplay between germline haplotypes and tumour methylation, regulating downstream gene expression and impacting the clinical behaviour of prostate cancer.
Discussion
Tumour meQTLs occur when a germline locus influences the epigenetic profile of a tumour, but not its predecessor non-malignant cells. The resulting regulatory effects can ripple through the central dogma, facilitating interactions between the germline and the somatic tissue leading to tumourigenesis decades after birth. While specific driver mutations can be driven by environmental or replicative factors, they arise in the context of the germline genome that biases towards or against them4. Understanding this interaction can help identify determinants of disease susceptibility and aggressivity; particularly important in prostate cancer where current clinical factors do not fully predict the interpatient heterogeneity in tumour behaviour and treatment response. As first presented by Heyn et al.17 and confirmed here, some GWAS loci modulate risk via dysregulation of DNA methylation. Measuring this direct effect of germline on methylation generates large effect sizes, overcoming power limitations of small cohorts. We validate this aspect of germline modulators of tumour methylation by re-identifying the rs684232 haplotype, a previously reported risk loci17. Further, we identify novel loci predictive of aggressive disease, including loci targeting prostate cancer driver events like TCERG1L26 and AKT148-51. Interestingly, not all risk meQTLs were tumour-specific. MeQTLs detected in reference tissue may facilitate tumour initiation, i.e. modulating pre-neoplastic methylation, while, tumour meQTLs may facilitate tumour progression, i.e. modulating oncogenic methylation.
The mechanisms by which germline loci affect tumour methylation are largely unknown, and are likely many. First, the most direct would be a SNP breaking a methylated CpG dinucleotide; in our data this accounted for only 0.1% of tumour meQTLs. Second, SNPs can influence CTCF binding, supported by their enrichment and allele specific-binding at these sites (Figure 3d & 4h). Changes in CTCF binding can impact local methylation by modulating PARP1 activity, and subsequently DNMT1 activity47. Third, SNPs can create or destroy DNA motifs that alter protein binding affinities, thereby promoting or antagonizing methylation53. Finally, tumour meQTLs may represent a secondary effect of the germline modulating processes that co-occur with methylation changes, e.g. chromatin modifications.
The cohort analyzed here was modest in size relative to contemporary GWAS studies (n=589 patients), yet we identified and validated 7,590 meQTLs and 1,178 tumour-specific meQTLs, suggesting they are very widespread in prostate cancer. The tumour meQTLs reported represent tag loci and require fine mapping to determine the casual loci. Additionally, cell type composition can play a role in meQTL identification as different cell types can have different methylation profiles54. For example, loci modulating the tumour microenvironment might alter measured methylation unrelated to methylation in cancer cells. Our approach focused on cis associations – loci proximal to the methylation site – due to their strong signal. We also detected trans tumour meQTLs, despite being under-powered to explore these. Larger cohorts are needed to quantify the trans influences of the germline on the tumour epigenome, and suggest an even larger landscape of germline aberrations influence the tumour epigenome and gene-expression.
These data reveal a novel mechanism through which the germline genome influences the somatic landscape of a tumour. These germline-somatic interactions can be exploited to identify prognostic germline loci that might be minimally-invasive biomarkers to aid triage of patients to more expensive tissue- or radiology-based assays. These data support further exhaustive study of germline-somatic interactions in prostate and other tumour types.
Methods
Discovery patient cohort
All patients had pathologically confirmed prostate cancer and were hormone naive at the time of therapy. All patients were treated with either image-guided radiotherapy (IGRT) or radical prostatectomy (surgery). Single ultrasound-guided needle biopsies were obtained for the IGRT cohort prior to the start of therapy, as previously described26. Fresh-frozen radical prostatectomy specimens were obtained from the University Health Network (UHN) Pathology BioBank or from the Genito-Urinary BioBank of the Centre Hospitalier Universitaire de Quebec – Université Laval (CHUQ). In accordance with local Research Ethics Board (REB) and International Cancer Genome Consortium (ICGC) guidelines, whole blood and informed consent was collected at the time of clinical follow-up. Previously collected tumour tissue was utilized based on UHN REB approved study protocols (UHN 06-0822-CE, UHN 11-0024-CE, CHUQ 2012-913:H12-03-192). Two genitourinary (GU) pathologists (TvdK, BT) independently evaluated scanned H&E-stained slides to confirm Gleason score and tumour cellularity for all tumour specimens. Clinical T category was reported using standard National Comprehensive Cancer Network (NCCN) criteria (https://www.tri-kobe.org/nccn/guideline/urological/english/prostate.pdf). Serum prostate specific antigen (PSA) was reported based on the reading at the time of diagnosis, measured in ng/mL. The discovery cohort consisted of samples from 161 cases previously characterized26,27 along with 80 new cases collected and processed in the same manner. These additional 80 cases were chosen to match the clinical features of the original 130– i.e. similar age, Gleason score, tumour stage, proportion of biochemical recurrences (BCR) and time to BCR. For IGRT patients, BCR was defined as the rise in PSA concentration of at least 2.0 ng/mL above the nadir. The nadir refers to the stable PSA level that follows a slight rise directly after radiotherapy. For surgery patients, BCR was defined as two consecutive post-surgery PSA measurements over 0.2 ng/mL or triggered salvage therapy.
Sample-processing
At UHN, selected prostate samples were cut into 60 x 10 μm sections, with an H&E-stained 4 μm section every 10 cuts. H&E-stained sections were marked by a GU pathologist (TvdK, BT) to indicate areas suitable for macro-dissection (i.e. >70% tumour cellularity). Manual macro-dissection was performed using sterile scalpel blades, and DNA was obtained by phenol:chloroform extraction, as previously reported26. DNA was extracted from whole blood using an ArchivePure DNA Blood Kit (5 PRIME, Inc., Gaithersburg, MD) at the Applied Molecular Profiling Laboratory at the Princess Margaret Cancer Centre. At CHU de Québec, the size of the prostate tissues from the biobank has allowed an easier, yet very efficient procedure for sampling prior to DNA extraction. After histology, quality control was processed the same way as described above and surface of tumoural glands considered large enough, two cores of 1 mm diameter were taken from the tumoural zone using a sterile biopsy punch (Miltex). Tissues were immediately disrupted in ATL buffer using Minilys homogeneizer (Bertin Technologies, Montigny, France). DNA was finally extracted from the lysate using QIAmp DNA mini kit (Qiagen, Hilden, Germany). The same kit was used to generate DNA extractions on blood samples from this site. All DNA samples were quantified using a Qubit 2.0 Fluorometer (Life Technologies, Burlington, ON) and assessed for purity using a Nanodrop ND-1000 spectrophotometer.
Methylation array data generation
Methylation microarray data generation were carried out as previously described26. Briefly, Illumina Infinium HumanMethylation 450k BeadChip kits were used to assess global methylation, using 500 ng of input genomic DNA at the McGill University and Genome Quebec Innovation Centre (Montreal, QC). All samples were processed from fresh frozen prostate cancer tissue.
Methylation array data analysis
Methylation microarray data was processed in the R statistical environment (v.3.2.3) as outlined elsewhere55. Briefly, raw methylation intensity levels were pre-processed using Dasen56 and filtered according to detectability above background noise, non-CpG methylation and cross hybridization using the DMRcate package (v1.6.53). Chromosome location, probe position and gene symbol were annotated using the IlluminaHumanMethylation450kanno.ilmn12.hg19 package (v0.6.0).
Whole-genome sequencing
WGS was conducted as previously reported26. Briefly, sequencing libraries were prepared using 50 ng gDNA and enzymatic reagents from KAPA Library Preparation Kits (KAPA Biosystems, Woburn, MA USA Cat#KK8201) according to protocols as described for end repair, A-tailing, and adapter ligation57. Sequencing was carried out using HiSeq 2000 platform (Illumina Inc.) and samples were sequenced to a minimum coverage depth of 30x and a median coverage of 44.2x ± 4.7x (standard deviation).
mRNA Microarray Generation
Total RNA was extracted from alternating adjacent sections, using the mirVana miRNA Isolation Kit (Life Technologies), according to the manufacturer’s instructions, as described previously26. In total, three batches were profiled at two locations. For batch 1 samples, 150 ng total RNA was assayed on the Affymetrix Human Gene 2.0 ST array (HuGene 2.0 ST) at The Centre for Applied Genomics (The Hospital for Sick Children, Ontario, Canada). For samples in batches 2 and 3, 100 ng total RNA was assayed on the Affymetrix Human Transcriptome Array 2.0 (HTA 2.0) and HuGene 2.0 ST, respectively, at the London Regional Genomics Centre (Robarts Research Institute, London, Ontario, Canada).
Whole-genome sequencing data analysis
Raw sequencing reads were aligned to the human reference genome, GRCh37, using BWA-mem (v0.7.12+)58 at the lane level (Supplementary Table 1). Picard (v1.92) merged these lane-level BAMs from the same library and marked duplicates. Picard was also used to merge library level BAMs from the same sample without marking duplicates. Local realignment and base quality recalibration was completed on tumour/normal pairs together with the Genome Analysis Toolkit (GATK v3.4.0+) (Supplementary Table 1)59. Normal samples were extracted, headers corrected (Samtools v0.1.9)60, and files indexed (Picard v1.92) into individual sample-level BAMs.
mRNA abundance analysis
Raw mRNA data was downloaded from GSE107299 and pre-processed under R (v3.2.5). Background correction, normalization algorithms and annotation were implemented in the oligo (v1.34.2) package from the BioConductor (v3.2) open-source project. The Robust multichip average algorithm was applied to the raw intensity data61. Probes were mapped to Entrez gene ID using custom CDF files (v20) for HTA 2.0 and HuGene 2.0 ST array from http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/CDF_download.asp. The sva package (v3.18.0) was used to correct for batch effects between different arrays. mRNA abundance levels from HuGene 2.0 ST and HTA 2.0 were combined into one dataset based on Entrez Gene IDs. The mRNA abundance levels were averaged amongst duplicated Entrez Gene IDs. Entrez gene IDs were then converted into gene symbols and chromosome locations based on the human reference genome GRCh37 from UCSC table browser (download date: 02/08/2016).
Identification of germline SNPs
GATK (v3.4.0+) (Supplementary Table 1) was used to call germline SNPs by first running HaplotypeCaller on the realigned and recalibrated tumour/normal pair (Supplementary Table 1). Next, VariantRecalibrator and ApplyRecalibration were applied to ensure high quality calls. GATK best-practices filters were applied to the resulting VCFs. We only considered biallelic SNPs in this analysis and 98.54% of autosomal SNPs (4,894,225/4,966,931) had all three genotypes.
Candidate risk meQTL replication
We conducted a candidate meQTL analysis to replicate the 8 prostate meQTLs reported in Hyen et al.17. Associations were tested using Spearman’s correlation. Spearman’s correlation tested the additivity of the alternative allele – i.e. the correlation between methylation and the genotype coded 0 (homozygous reference), 2 (heterozygous) and 3 (homozygous alternative). We considered a significant threshold of P<0.01 (Bonferroni-adjustment). Three of the 8 meQTLs could not be tested in this cohort as the probes were filtered out during methylation processing (cg20129853, cg13762704, cg02340056; Supplementary Table 2).
Risk loci associations
A list of 160 germline polymorphisms associated with prostate cancer risk was cultivated from the literature15,25,27-32. SNPs from these studies were chosen if they were associated with the risk of prostate cancer or prognosis of prostate cancer patients. Associations were tested using Spearman’s correlation. Spearman’s correlation tested the additivity of the alternative allele – i.e. the correlation between the event and the genotype coded 0 (homozygous reference), 2 (heterozygous) and 3 (homozygous alternative). Significant associations were defined as false discovery rates less than 0.05. We chose Spearman’s correlation to avoid violating distributional assumptions made in linear models given that methylation data does not follow traditional distributions. Additionally, we selected Spearman’s correlation over the previously reported multivariate random forest selection frequency method17 given the subsequent genome-wide and methylome-wide approaches in this work (see “Discovery genome wide association studies” and “Discovery cis germline-methylation associations” methods sections). This approach was too computationally intensive to apply to the 1x107 independent tests conducted in the following sections so for consistency we applied Spearman’s correlation for all associations. However, we did implement the multivariate random forest selection frequency method17 to confirm a subset of our high confidence hits (see Validation of germline-methylation associations).
Survival analysis
Survival analysis was conducted in the R statistical environment (v3.3.1). Where the assumption of proportional hazards applied, a Cox proportional hazards model was implemented testing the association between methylation – median dichotomized m-value – with biochemical recurrence, as defined previously26. Probes with p-value < 1x10−4 were carried forward to the analysis. For survival analysis of TCERG1L methylation levels, cutp from the survMisc (v.0.4.5) package was used to determine a dichotomization threshold to replicate thresholds used in previous work28. Survival associations were validated in an independent cohort of 101 clinically-matched primary samples26,27,37,38.
Discovery prognostic germline-methylation associations
Genome wide associations were tested for all 58 prognostic methylation probes. Germline SNPs were filtered based on a minimum allele frequency (MAF > 0.1) and Hardy-Weinberg equilibrium violation (P > 1x10−8). Associations between the remaining SNPs and the 58 prognostic methylation probes were evaluated using the R-plugin feature of the plink software (v1.07) to implement a Spearman’s correlation test62. Spearman’s correlation tested the additivity of the alternative allele (i.e. the correlation between the event and the genotype coded 0 (homozygous reference), 2 (heterozygous) and 3 (homozygous alternative)). Manhattan plots were generated to visualize the results for each SNP. QQ plots were generated to assess bias in the model fit. A stringent Bonferroni adjustment was applied to correct for multiple hypothesis testing, therefore, SNPs with P < 5x10−8 were considered significantly associated. LD was calculated and visualized using Haploview (v4.2)63. Pairwise LD was quantified using D’ and haplotypes defined according to Gabriel et al64.
Discovery cis germline-methylation associations
All methylation probes were tested for cis germline-methylation associations by looking at SNPs that were in a +/− 10 kbp window around the probe. Associations were tested using Spearman’s correlation, as outline earlier, and power was tested using a one-way ANOVA, as outlined above. Associations were deemed significant for p-values < 3x10−9 as this represented the Bonferroni threshold (1.5x107 independent tests).
TCGA validation cohort
The TCGA PRAD data was used as a validation cohort34. Concordance between SNP6 microarray (SNP6) genotypes and whole exome sequencing (WXS) of blood sample calls was evaluated and only samples with >80% concordance were retained (348 samples; excluded 3 samples for concordance < 80%: TCGA-HC-7738, TCGA-EJ-7312, TCGA-EJ-5505). Genotypes were imputed using the Sanger Imputation Service – pre-phasing using Shapeit265, imputation using PBWT66 and the Haplotype Reference Consortium (release 1.1) panel67. The accuracy of the imputed genotypes was evaluated against WXS blood sample calls. A median accuracy of 0.988 was estimated. Genotypes were imputed a second time using combined SNP6 and WXS calls and the same imputation pipeline as described above. In the event that SNP6 and WXS disagreed on the genotype at a particular position, the WXS call was used. A final list of 40,405,505 SNPs were then available for validation studies.
Validation of germline-methylation associations
Associated SNPs from the discovery cohort (P < 5x10−8 from genome-wide analysis and p-value < 4x10−9 from the cis germline-methylation analysis) were tested in the imputed TCGA cohort using the same Spearman’s correlation method outlined above. False discovery adjustment was applied to the remaining SNPs and associations were considered to validate if FDR < 0.05 and the directionality of the Spearman’s ρ was consistent in the discovery and validation cohorts. We implemented the multivariate random forest selection frequency method from Hyen et al.17 for 59 high confidence tumour meQTLs and found all 59 had q-value = 0, calculated as proportion of null models with random forest selection frequency (RFSF) > fit model as described in Hyen et al17, supporting the validity of our approach (Supplementary Table 7).
Tumour-specific germline-methylation associations
Tumour-specific germline methylation associations were determined using the TCGA tumour and reference methylomes34. Similar to the discovery phase, associations were tested using a Spearman’s correlation test. Associations were considered tumour specific if the FDR < 0.05 in the tumour while FDR ≥ 0.05 in the reference in a subset of samples with both tumour and reference methylation profiling (n=47). Tumour specificity was further confirmed for the two stated examples, TCERG1L and AKT1, via a permutation test. The normal Spearman’s ρ was compared to a distribution of tumour Spearman’s ρ based on 1,000,000 random subsets of 47 tumour samples. P-values were calculated based on the number of iterations where the normal ∣ρ∣ was larger than the tumour ∣ρ∣. To identify differentially methylated regions (DMRs) between tumour and normal tissue, raw intensity values were re-normalized together using Dasen56 and DMRs were identified using the R package DMRcate (v1.12.1) with default parameters.
ChIP-Seq data analysis
A subset of 34 samples in the discovery cohort had H3K27ac ChIP-Seq profiling as previously described46. Peak bed files and raw FASTQs for H3K27ac (n = 92), H3K27me3 (n=76), AR (n=88) and H3K4me3 (n=56) were downloaded for an independent cohort from the Gene Expression Omnibus (GSE120738)35. Tumour meQTLs overlapping each target were identified using the downloaded bed files. Here we considered all SNPs within the same haplotype as the tag tumour meQTL. The raw FASTQ files were aligned using bwa (v.0.7.15) and the aligned BAM files from each target were merged for each patient (i.e. H3K27ac, H3K27me3, H3K4me3 and AR BAMs from the same patient were merged). Using the merged BAM files, patients were genotyped at overlapping sites of interest using GATK (v3.4.0+) HaplotypeCaller. Differential binding analysis was conducted using logistic regression to quantify the contribution of genotype on binding variation. We considered the loci significant if FDR < 0.05. For each tumour meQTL we tested all SNPs within the tumour meQTL haplotype reporting the SNP with the minimum p-value. ChIP-Seq data for LNCaP, PC3, 22Rv1, VCaP and RWPE-1 cell lines was downloaded from the sources outlined in Supplementary Table 346,68-80.
Regulatory region enrichment analysis
To detect whether these probes are enriched in certain chromosomes, genomic locations and CpG classes, Fisher’s Exact test followed by multiple test correction (FDR) were applied. Methylation promoter region (transcription start site (TSS) 200, TSS1500 and 5’UTR), gene body (1st Exon and gene body, 3’UTR) and intergenic region were defined as previously described81. Enrichment at transcription factor binding sites and regulatory elements was conducted with previously published ChIP-Seq data from primary tumours35 and LNCaP, PC3, 22Rv1, VCaP and RWPE-1 cell lines46,68-80 (Supplementary Table 3). If multiple target:treatment pairs existed the median number of overlapping SNPs was used. Enrichment was quantified using a permutation test that randomly sampled 23 SNPs when interrogating risk loci meQTLs and 1,031 SNPs when interrogating cis tumour meQTLs genome-wide from a list of observed SNPs in our cohort. P-values were calculated as the number of null iterations with equal to or more SNPs overlapping ChIP-Seq peaks than tumour meQTLs divided by the total number of iterations (105). P-values were FDR-adjusted to account for multiple hypothesis testing. For novel cis tumour meQTLs, we considered the full haplotype of the tag SNPs, i.e. the tumour meQTLs or randomly sampled SNPs, and considered the haplotype overlapping if at least one SNP within the haplotype overlapped with the ChIP-Seq peaks.
Allele-imbalance ChIP-Seq analysis
Prostate tissue was collected from 48 patients with localized primary prostate adenocarcinoma. Each patient yielded a sample of the adenocarcinoma and a sample from surrounding non-malignant prostate tissue. We performed ChIP-Seq for H3k27ac (N=48), H3k4me2 (N=6), H3k4me3 (N=4), FOXA1 (N=10), and HOXB13 (N=9) on these samples, as well as germline SNP genotyping from blood. Germline variants were phased and imputed to the Haplotype Reference Consortium panel67. Mapping and aligning was performed using bwa58; allele-specific reads were processed according to the WASP pipeline82 to remove mapping bias; peaks were identified using the MACS2 software83. Allele-specific read counts were generated by the GATK ASEReadCounter59. We tested for allele-specific signal using a haplotype beta-binomial test that accounts for read over-dispersion. Beta-binomial over-dispersion parameters were estimated for each individual/experiment from the aligned allele-specific counts and were found to be consistently low (<0.01). For each peak and individual, haplotype-specific read counts were merged across all heterozygous read-carrying sites in the peak for a single measure of allele specificity. Every SNP within 100 kbp of the peak center and containing at least one heterozygous individual was then tested for allelic imbalance. All heterozygous individuals were tested together under the expectation of a consistent allele-specific effect. Each test was performed once for samples from normal, tumour, or both, as well as a test for difference in imbalance between tumour and normal. Finally, peaks were considered “imbalanced” in each of these four test categories if any of the variants tested for that peak exhibited allele-specific signal at a 5% FDR.
Overlap between SNP and peak anchor regions
The accession numbers for RAD21 ChIA-PET data from LNCaP and DU145 cells, from ENCODE, are ENCLB189DLP and ENCLB678KEV, respectively. ChIA-PET2 was utilized to process the raw data and obtain the intra-chromosomal interactions84. Peaks with interactions represent a subgroup of the total peaks identified from the ChIA-PET data. We employed intersectBed (bedtools) to overlap the coordinates of SNP sites and peak regions. Overlap analysis of SNPs with total peaks or interaction peaks are summarized in Supplementary Table 7.
Prediction of potential target of risk loci
Peak anchors that overlapped with loci regions were acquired. Genes located in the paired peak anchors were predicted as potential targets of these risk loci.
Pathway enrichment
Genes harbouring tumour meQTL-eQTLs were processed using g:Profiler85 (v. r1741_e90_eg37; significance set at FDR; output set to generic enrichment map; GO, KEGG and REACTOME databases; background set to all annotated genes; minimum number of genes per pathway was set to 2).
Germline-RNA (eQTL) and germline-protein (pQTL) associations
Germline-RNA associations were tested for tumour meQTLs. These associations were first interrogated in the discovery cohort using a Spearman’s correlation test (N = 203) and then validated in the TCGA PRAD-RNA-Seq cohort34. For stringency, only tumour-specific eQTLs that were not observed as GTEx prostate epithelial eQTLs42 were retained. Tumour-specific associations were defined as FDR > 0.05 from published GTEx results where FDR was applied over the candidate list of eQTLs (n=87). Germline-protein associations were identified in an exploratory analysis of 70 primary prostate cancers38 using Spearman’s correlation test.
CTCF mechanism
VCaP CTCF and H3K27ac ChIP-Seq and WGS BAM files were downloaded from ENCODE. Whole genome bisulfite sequencing FASTQ files were downloaded from GEO (accession GSE86832) for three replicates. FASTQ files were aligned using Bismark86 (v0.15.0) with one mismatch allowed in a seed alignment.
Data visualization
Visualizations were generated in the R statistical environment (v3.3.1) with the lattice (v0.24-30), latticeExtra (v0.6-28) and BPG (v5.6.23) packages87. Haplotypes were visualized using Haploview (v4.2)63.
Data Availability
Methylation data are available in the Gene Expression Omnibus under accession GSE84043. Raw sequencing data are available in the European Genome-phenome Archive under accession EGAS00001000900 (https://www.ebi.ac.uk/ega/studies/EGAS00001000900). Processed variant calls are available through the ICGC Data Portal under the project PRAD-CA (https://dcc.icgc.org/projects/PRAD-CA). TCGA WGS/WXS data are available at Genomic Data Commons Data Portal (https://gdc-portal.nci.nih.gov/projects/TCGA-PRAD). Primary samples ChIP-Seq data was retrieved from Gene Expression Omnibus under accession GSE120738. Cell line data sources are outlined in Supplementary Table 3. Detailed information on experimental design can be found in the included Life Sciences Reporting Summary.
Extended Data
Extended Data Figure 1.
Data analysis and quality controls.
Extended Data Figure 2.
Characterizing risk meQTLs.
Extended Data Figure 3.
Characterizing meQTLs targeting prognostic methylation sites.
Extended Data Figure 4.
Characterizing tumor meQTLs.
Extended Data Figure 5.
Characterizing TCERG1L tumor meQTLs.
Extended Data Figure 6.
Characterizing AKT1 tumor meQTLs.
Supplementary Material
Acknowledgements
The authors thank all members of the Boutros lab and Ken Kron and Alice Meng for helpful suggestions and support. The results described here are based in part upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. This study was conducted with the support of Movember through Prostate Cancer Canada and with the additional support of the Ontario Institute for Cancer Research, funded by the Government of Ontario. We thank the Princess Margaret Cancer Centre Foundation and Radiation Medicine Program Academic Enrichment Fund for support (to R.G.B.). R.G.B. is a recipient of a Canadian Cancer Society Research Scientist Award. This work was supported by Prostate Cancer Canada and is proudly funded by the Movember Foundation (grant #RS2014-01 to P.C.B.; grant #RS2014-02 to M.L.; grant #RS-2016-01 to H.H.H.). P.C.B. was supported by a Terry Fox Research Institute New Investigator Award and a CIHR New Investigator Award. H.H.H. was supported by CIHR operating grant 142246 and CCSRI grant 703800. This project was supported by Genome Canada through a Large-Scale Applied Project contract to P.C.B., R. Morin and S. P. Shah. K.E.H was supported by a CIHR Vanier Fellowship. R.S.M. acknowledges funding from the Prostate Cancer Research Program (PCRP) Impact Award-US Department of Defense (W81XWH-17-1-0675), and the Individual Investigator Research Award from CPRIT (RP190454). M.L.F. acknowledges funding from NIH (5R01CA193910), the Challenge Award from the Prostate Cancer Foundation and the H.L. Snyder Medical Foundation. B.G. acknowledges funding from National Human Genome Research Institute (R01HG009120). This work was supported by the NIH/NCI under award number P30CA016042 and by an operating grant from the National Cancer Institute Early Detection Research Network (1U01CA214194-01) to PCB and TK.
Footnotes
Conflict of Interest Statement
All authors declare that they have no conflicts of interest.
References
- 1.Hanahan D & Weinberg RA Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B et al. Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garraway LA & Lander ES Lessons from the cancer genome. Cell 153, 17–37 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Tomasetti C, Li L & Vogelstein B Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 355, 1330–1334 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomlinson IP et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat. Genet 40, 623–630 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Peterson GM et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet 42, 224–228 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michailidou K et al. Association analysis identifies 65 new breast cancer risk loci. Nature 551, 92–94 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knudson AG Two genetic hits (more or less) to cancer. Nat. Rev. Cancer 1, 157–62 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Fearon ER & Vogelstein B A genetic model for colorectal tumourigenesis. Cell 61, 759–767 (1990). [DOI] [PubMed] [Google Scholar]
- 10.Nik-Zainal S Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones PA & Baylin SB The epigenomics of cancer. Cell 128, 683–692 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reynolds PA et al. Tumour suppressor p16INK4A regulates polycomb-mediated DNA hypermethylation in human mammary epithelial cells. J Biol Chem 281, 24790–24802 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Suzuki H et al. Epigenetic inaction of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet 36, 417–422 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Saghafinia S et al. Pan-cancer landscape of aberrant DNA methylation across human tumors. Cell Reports 25, 1066–1080 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Whitington T et al. Gene regulatory mechanisms underpinning prostate cancer susceptibility. Nat. Genet 48, 387–397 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Cowper-Sal-lari R, et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat. Genet 44, 1191–1198 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heyn H et al. Linkage of DNA methylation quantitative trait loci to human cancer risk. Cell Reports 24, 331–338 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Taylor RA et al. Germline BRCA2 mutations drive prostate cancers with distinct evolutionary trajectories. Nat. Commun 8, 13671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szulkin R et al. Genome-wide association study of prostate cancer-specific survival. Cancer Epidemiol Biomarkers Prev 24, 1796–1800 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng B et al. An xQTL map integrates the genetic architecture of the human brain's transcriptome and epigenome. Nat. Neurosci 20, 1418–1426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferlay J et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359–E386 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Klotz L et al. Long-Term Follow-Up of a Large Active Surveillance Cohort of Patients With Prostate Cancer. J. Clin. Oncol 33, 272–277 (2015). [DOI] [PubMed] [Google Scholar]
- 23.D’Amico AV et al. Cancer-specific mortality after surgery or radiation for patients with clinically localized prostate cancer managed during the prostate-specific antigen era. J. Clin. Oncol 21, 2163–2172 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Boutros PC et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat. Genet 47, 736–745 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Cooper CS et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat. Genet 47, 367–372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraser M et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature 541, 359–364 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Espiritu SG et al. The evolutionary landscape of localized prostate cancers drives clinical aggression. Cell 173, 1003–1013 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Lin DW et al. Genetic variants in the LEPR, CRY1, RNASEL, IL4, and ARVCF genes are prognostic markers of prostate cancer-specific mortality. Cancer Epidemiol. Biomark. Prev 20, 1928–1936 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eeles RA et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat. Genet 41, 1116–1121 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eeles RA et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat. Genet 45, 385–391, 391e1–2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lévesque E et al. Steroidogenic germline polymorphism predictors of prostate cancer progression in the estradiol pathway. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res 20, 2971–2983 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Schumacher FR et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet 50, 928–936 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matejcic M et al. , Germline variation at 8q24 and prostate cancer risk in men of European ancestry. Nat. Commun 9, 4616 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas Research Network. The Molecular Taxonomy of Primary Prostate Cancer. Cell 163, 1011–1025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stelloo S et al. Integrative epigenetic taxonomy of primary prostate cancer. Nat. Commun 9, 4900 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson WC et al. Intermediate endpoints after postprostatectomy radiotherapy: 5-year distant metastasis to predict overall survival. Eur Urol 74, 413–419 (2018) [DOI] [PubMed] [Google Scholar]
- 37.Bhandari V et al. Molecular landmarks of tumor hypoxia across cancer types. Nat Genet. 51, 308–318 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Sinha A et al. The proteogenomic landscape of curable prostate cancer. Cancer Cell 35, 414–427 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim H The retinoic acid synthesis gene ALDH1a2 is a candidate tumor supporessor in prostate cancer. Cancer Res. 65, 8118–8124 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Doose G et al. MINCR is a MYC-induced lncRNA able to modulate MYC’s transcriptional network in Burkitt lymphoma. PNAS 112, E5261–E5280 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S et al. Long non-coding RNA MINCR promotes gallbladder cancer progression through stimulating EZH2 expression. Cancer Lett 380, 122–133 (2016). [DOI] [PubMed] [Google Scholar]
- 42.GTex Consortium. Genetic effects on gene expression across human tissues. Nature 550, 204–2013 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Armenia J et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet. 50, 645–651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi JM et al. Genomic and epigenomic integration identifies a prognostic signature in colon cancer. Clin. Cancer. Res 17, 1535–1545 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yi JM et al. DNA methylation biomarker candidates for early detection of colon cancer. Tumour Biol. 33, 363–372 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kron KJ et al. TMPRSS2-ERG fusion co-opts master transcription factors and activates NOTCH signaling in primary prostate cancer. Nat. Genet 49, 1336–1345 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Zampieri M et al. ADP-ribose polymers localized on Ctcf–Parp1–Dnmt1 complex prevent methylation of Ctcf target sites. Biochem. J 441, 645–652 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee JK et al. N-Myc drives neuroendocrine prostate cancer initiated from human prostate epithelial cells. Cancer Cell 29, 536–547 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwon EM et al. Genetic polymorphisms in inflammation pathway genes and prostate cancer risk. Cancer Epidemiol Biomarkers Prev 20, 923–933 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karyadi DM et al. Confirmation of genetic variants associated with lethal prostate cancer in a cohort of men from hereditary prostate cancer families. Int J Cancer 136, 2166–2171 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu JM et al. Association between single nucleotide polymorphisms in AKT1 and the risk of prostate cancer in the Chinese Han population. Genet Mol Res 16 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Song M et al. AKT as a therapeutic target for cancer. Cancer Res, 79, (2019). [DOI] [PubMed] [Google Scholar]
- 53.Stadler MB et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480, 490–495 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Bernstein BE et al. The NIH roadmap epigenomics mapping consortium. Nat Biotechnol. 28, 1045–1048 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 55.Shiah Y-J, Fraser M, Bristow RG & Boutros PC Comparison of Pre-processing Methods for Infinium HumanMethylation450 BeadChip Array. Bioinformatics 33, 3151–3157 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Pidsley R et al. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics 14, 293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fisher S et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 12, R1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma. Oxf. Engl 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McKenna A et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinforma. Oxf. Engl 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Irizarry RA et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31, e15 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barrett JC, Fry B, Maller J & Daly MJ Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005). [DOI] [PubMed] [Google Scholar]
- 64.Gabriel SB et al. The Structure of Haplotype Blocks in the Human Genome. Science 296, 2225–2229 (2002). [DOI] [PubMed] [Google Scholar]
- 65.Delaneau O, Marchini J & Zagury J-F A linear complexity phasing method for thousands of genomes. Nat. Methods 9, 179–181 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Durbin R Efficient haplotype matching and storage using the positional Burrows-Wheeler transform (PBWT). Bioinforma. Oxf. Engl 30, 1266–1272 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Consortium, the H. R. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet 48, 1279–1283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu J et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell 17, 443–54 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang D et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474, 390–4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tan PY et al. Integration of regulatory networks by NKX3–1 promotes androgen-dependent prostate cancer survival. Mol Cell Biol 32, 399–414 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hazelett DJ et al. Comprehensive functional annotation of 77 prostate cancer risk loci. PLoS Genet 10, e1004102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jin HJ et al. Cooperativity and equilibrium with FOXA1 define the androgen receptor transcriptional program. Nat Commun 5, 3972 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu K et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 338, 1465–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang X et al. Integrative functional genomics identifies an enhancer looping to the SOX9 gene disrupted by the 17q24.3 prostate cancer risk locus. Genome Res 22, 1437–46 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen Y et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med 19, 1023–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liang Y et al. LSDI-mediated epigenetic reprogramming drives CENPE expression and prostate cancer progression. Cancer Res 77, 5479–5490 (2017). [DOI] [PubMed] [Google Scholar]
- 78.Sutinen P et al. SUMOylation modulates the transcriptional activity of androgen receptor in a target gene and pathway selective manner. Nucleic Acids Res 42, 8310–8319 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taberlay PC et al. Reconfiguration of nucleosome-depleted regions at distal regulatory elements accompanies DNA methylation of enhancers and insulators in cancer. Genome Res 24, 1421–1432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rickman DS et al. Oncogene-mediated alterations in chromatin conformation. Proc Natl Acad Sci U S A 109, 9083–9088 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mehrmohamadi M et al. Integrative modelling of tumour DNA methylation quantifies the contribution of metabolism. Nat. Commun 7, 13666 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van de Geijn B, et al. WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nat Methods 12, 1061–1063 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li G, et al. ChIA-PET2: a versatile and flexible pipeline for ChIA-PET data analysis. Nucleic Acids Res 45, e4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reimand J, et al. G:Profiler – a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krueger F and Andrews SR Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinforma. 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.P’ng C, et al. BPG: seamless, automated and interactive visualization of scientific data. BMC Bioinformatics 20, 42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Methylation data are available in the Gene Expression Omnibus under accession GSE84043. Raw sequencing data are available in the European Genome-phenome Archive under accession EGAS00001000900 (https://www.ebi.ac.uk/ega/studies/EGAS00001000900). Processed variant calls are available through the ICGC Data Portal under the project PRAD-CA (https://dcc.icgc.org/projects/PRAD-CA). TCGA WGS/WXS data are available at Genomic Data Commons Data Portal (https://gdc-portal.nci.nih.gov/projects/TCGA-PRAD). Primary samples ChIP-Seq data was retrieved from Gene Expression Omnibus under accession GSE120738. Cell line data sources are outlined in Supplementary Table 3. Detailed information on experimental design can be found in the included Life Sciences Reporting Summary.