Abstract
Background:
Chronic exposure to inorganic arsenic (iAs) is a significant public health problem. Methylation of iAs by arsenic methyltransferase (AS3MT) controls iAs detoxification and modifies risks of iAs-induced diseases. Mechanisms underlying these diseases have been extensively studied using animal models. However, substantive differences between humans and laboratory animals in efficiency of iAs methylation have hindered the translational potential of the laboratory studies.
Objectives:
The goal of this study was to determine whether humanization of the As3mt gene confers a human-like pattern of iAs metabolism in mice.
Methods:
We generated a mouse strain in which the As3mt gene along with the adjacent Borcs7 gene was humanized by syntenic replacement. We compared expression of the mouse As3mt and the human AS3MT and the rate and pattern of iAs metabolism in the wild-type and humanized mice.
Results:
AS3MT expression in mouse tissues closely modeled that of human and differed substantially from expression of As3mt. Detoxification of iAs was much less efficient in the humanized mice than in wild-type mice. Profiles for iAs and its methylated metabolites in tissues and excreta of the humanized mice were consistent with those reported in humans. Notably, the humanized mice expressed both the full-length AS3MT that catalyzes iAs methylation and the human-specific splicing variant that has been linked to schizophrenia.
Conclusions:
These results suggest that AS3MT is the primary genetic locus responsible for the unique pattern of iAs metabolism in humans. Thus, the humanized mouse strain can be used to study the role of iAs methylation in the pathogenesis of iAs-induced diseases, as well as to evaluate the role of in schizophrenia. https://doi.org/10.1289/EHP6943
Introduction
Inorganic arsenic (iAs) is the chemical form of As commonly found in drinking water (ATSDR 2007) and in some foods (Cubadda et al. 2017). Chronic exposure to iAs has been associated with risk of skin, bladder, lung, and liver cancers (IARC 2004), as well as with other diseases (Naujokas et al. 2013), including diabetes (Maull et al. 2012), cardiovascular (Abhyankar et al. 2012; Moon et al. 2012; Saquib et al. 2012; States et al. 2009), respiratory (Sanchez et al. 2016), and neurological diseases (Caito and Aschner 2015; Parvez et al. 2011). Humans and most other mammalian species have developed a mechanism for detoxifying iAs that involves the sequential conversion of iAs to monomethyl-As (MAs) and MAs to dimethyl-As (DMAs) (Thomas et al. 2007). Both methylation steps are catalyzed by orthologs of a single enzyme, arsenic () methyltransferase (AS3MT) (Lin et al. 2002). The methylation of iAs by AS3MT not only promotes whole-body clearance of As (Drobná et al. 2009; Hughes et al. 2010) but also produces methylated intermediates that contain highly reactive and toxic trivalent As () (Watanabe and Hirano 2013). and , unlike their pentavalent counterparts (), exceed iAs in potency as cytotoxins, genotoxins, and enzyme inhibitors (Thomas et al. 2001). Hence, and may be critical determinants of toxic and carcinogenic effects associated with chronic exposure to iAs. Altered capacity to methylate iAs has been linked to an increased risk of diseases associated with iAs exposure (Ahsan et al. 2007; Pierce et al. 2013; Vahter 2000).
In most mammals the gene encoding AS3MT is present as a single copy from the gene encoding BLOC-1 Related Complex Subunit 7 (BORCS7). To date the methylation of iAs is the only known function of AS3MT; however, recent studies have identified the AS3MT/BORCS7 locus as conferring risk for schizophrenia (Duarte et al. 2016). Compared with healthy individuals, expression of BORCS7 and of the human-specific splicing variant of AS3MT () has been found to be consistently higher in the brains of patients with schizophrenia (Li et al. 2016). Notably, expression levels of AS3MT and BORCS7 were found to be weakly correlated (Li et al. 2016), suggesting that the two genes may share transcriptional regulatory elements. Although published data showed that iAs exposure can affect As3mt expression in mice (Stýblo et al. 2019), the role of iAs exposure in the expression of BORCS7 or , which is thought to lack iAs methylation activity, has never been studied.
Laboratory studies of the mechanistic basis of iAs-associated diseases have been hindered by substantial differences between laboratory animals and humans in their capacity to metabolize and detoxify iAs. Rats, unlike humans, sequester significant portions of ingested iAs in erythrocytes in the form of (Lu et al. 2007). Mice, the species most commonly used in mechanistic studies, differ from humans, displaying very high rates of As methylation. This species difference is associated with faster rates of urinary clearance of methylated metabolites and the predominance of DMAs as the main urinary metabolite of iAs in mice (Vahter 1999). Interspecies differences in As metabolism may contribute to difficulties encountered replicating some of the effects of iAs exposure reported in humans, including the carcinogenic effects, in laboratory mice. Thus, producing an iAs methylation phenotype in mice that resembles the phenotype found in humans could create a better animal model to study the role of iAs metabolism in adverse health effects of chronic iAs exposure. To generate such a model, we have humanized the entire Borcs7/As3mt locus in 129S6 mice by syntenic replacement. In this process, the segment of mouse DNA carrying these two genes was removed and replaced with the syntenic region of human DNA in mouse embryonic stem (ES) cells using homologous recombination. This ensured that the junctions between the human and mouse DNA are known to the base pair level. Mice were generated from the ES cells, with the expectation that they would express the human AS3MT and BORCS7 proteins.
The present study describes in detail the creation of the humanized mouse strain, expression of human AS3MT and BORCS7 in tissues of the humanized mice, and metabolism of iAs in these mice after a single dose and during a subchronic exposure to iAs in drinking water.
Methods
Rationale for Humanizing the Borcs7/As3mt Locus
We chose to excise the segment of mouse DNA carrying the As3mt and Borc7 genes and replaced it with the corresponding segment of human DNA. The promoter of AS3MT abuts the untranslated region of BORC7, and the weak correlation in the expression pattern of the genes suggests possible shared transcriptional regulatory elements (Li et al. 2016). In addition, the current human transcript databases, for example, the University of California Santa Cruz Genome Browser (UCSC 2013) lists a putative read-through BORCS7/AS3MT transcript lacking the final exon of BORCS7 and the initial exon of AS3MT. This suggests the possibility that at least some AS3MT activity may be linked to transcription driven by the BORCS7 promoter. Thus, the inclusion of Borcs7 in the humanized region increases the likelihood that the elements directing the tissue species differences in the expression of AS3MT are included in the humanized region.
Assembly of Borcs7/As3mt Displacer
The Borcs7/As3mt displacer construct was assembled using a standard recombineering approach (Figure 1). The mouse arms of homology were derived from the 129s7/AB2.2 bMQ BAC library (Source BioScience plc, Nottingham, UK). The short homology arm was derived from BAC bMQ455L14 and the long arm of homology from BAC bMQ345L20. The segment of human genomic DNA containing the BORCS7 and AS3MT loci was derived from the human tile path BAC RP11-753C18 (BACPAC Resources Center, Children’s Hospital Oakland Research Institute, Oakland, CA). The single nucleotide polymorphisms (SNPs) previously associated with interindividual differences in iAs metabolism (Apata et al. 2017) or schizophrenia risk (Li et al. 2016), which were included in the this BORCS7/AS3MT segment, are described in Table S1. A DNA segment () extending from chr19:46,683,032 to 46,744,011 were deleted from the mouse genome and replaced with a segment of human DNA extending from chr10:102,845,563 to 102,904,823. The haplotype of the AS3MT gene carried in the displacer construct is *1A (Wood et al. 2006). The resistance marker gene used for selection of ES cells in which the displacer construct underwent genomic integration consisted of a phosphoglycerate kinase (PGK)-neo cassette flanked by mutant loxP sites. The marker gene can be excised, leaving only a nonfunctional lox site in its place.
Figure 1.
Scheme for humanization of the mouse Borcs7/As3mt locus: (A) Human BORCS7/AS3MT locus showing the relative positions of the BORCS7 and AS3MT genes, as well as the closest flanking genes. (B) Mouse Borcs7/As3mt locus with flanking genes. (C) Schematic of the Borcs7/As3mt displacer construct. (D) Humanized locus prior to Cre-mediated marker excision. (The names of human genes are capitalized.)
All polymerase chain reactions (PCRs) carried out during the displacer assembly used Taq polymerase (Bio Basic), with an initial denaturation temperature of 98°C for 20 s followed by 20 cycles of denaturation for 15 s at 96°C, annealing for 30 s at a 55°C, and extension at 72°C with an extension time of 1 min per of product length. All Red/ET recombination reactions were carried out using a commercially available kit (Gene Bridges; Cat. no. K001). All predesigned primers used during the construction of the displacer vector, were from Sigma-Aldrich and are listed in Table S2. In descriptions of specific assays, each of these primers is listed by the corresponding number (#).
As stated above, the short arm of the displacer construct was derived from the bMQ455L15 BAC library. This was accomplished by first replacing the Borcs7/As3mt region of the BAC with an ampicillin resistance marker by Red/ET recombination. The ampicillin resistance marker was amplified from the pBlueScript II SK () cloning vector (Stratagene; primers #215334 and #215335). The homology arms, referred to as the Red arm (primers #215319 and #215320) and the D/S arm (primers #215331 and #215333), respectively, were generated by PCR amplification of segments of the bMQ455L15 BAC. The homology arms were fused to the ampicillin resistance gene by overlap PCR.
The displacer short arm, together with the inserted ampicillin gene, was subcloned from the modified bMQ455L15 BAC into a plasmid vector by Red/ET recombination. The plasmid vector was prepared by ligation of four PCR products. The vector backbone carrying a ColE1 origin of replication and a spectinomycin gene was derived by amplification of the pFloxxerX plasmid previously assembled in Koller’s laboratory (see Table S3) (primers #215323 and #215324). The bMQ455L15 BAC was used as a template for the other three PCRs: U/S arm (primers #215325 and #215326), short arm (primers #21327 and #21328), and D/S arm (primers #215330 and #215332). All PCR products were gel purified on a 1.2% agarose gel using a commercially available kit (Qiagen; Cat. no. 28115), digested with BbsI (NEB; Cat. no. R3539S) and MluI (NEB; Cat. no. R0198S), and column purified (Qiagen; Cat. no. 28104). Fifty nanograms of each PCR product was combined and ligated (NEB; Cat. no. M0202S) in a reaction for 1 h at 15°C. The ligation reaction was heat inactivated at 65°C for 15 min, and then of the reaction was transformed into chemically competent E. coli cells (Zymo Research; Cat. no. T3002) according to the manufacturer’s directions and plated on Luria Broth agar plates supplemented with spectinomycin at . The resulting plasmid vector was digested with MluI and used for Red/ET recombination with the modified bMQ455L15 BAC. The resulting plasmid was in turn digested with BbsI to release the displacer short arm and ampicillin resistance gene flanked by the U/S and D/S homology arms. This BbsI restriction fragment was introduced into the bMQ345L20 BAC by Red/Recombination to create a BAC vector in which the ampicillin resistance marker was flanked by the displacer arms of homology.
The segment of the human BORCS7/AS3MT locus included in the displacer vector was prepared for excision from the RP11-753C18 BAC in two steps. In the first step, a neomycin resistance gene was inserted into the BAC upstream of the BORCS7 gene. To accomplish this, the neo gene, flanked by mutant loxP sites was ligated to homology arms, referred to as Black-Red and Purple, respectively. The neomycin resistance gene was excised from the vector pSfiI-JT15-neoJTZ17-SfiI previously assembled in Koller’s laboratory (see Table S4) by digestion with SfiI (NEB; Cat. no. R0123S). The Black-Red homology arm was amplified (primers #215317 and #215320) using the bMQ455L15 BAC as a template, and the Purple homology arm was amplified (primers #215336 and #215337) using the RP11-753C18 BAC as a template. All fragments were gel purified as described above, and the Black-Red and Purple homology arm PCRs were digested with BglI (NEB; Cat. no. R0143S) and then column purified as described above. The loxP-flanked neomycin resistance gene was ligated to the homology arms as described above in a reaction containing a total of of DNA, with the three fragments at an equimolar ratio. Two microliters of the ligation reaction was used for a Red/ET reaction with the RP11-753C18 BAC. In the second step, an ampicillin gene was inserted downstream of the AS3MT gene in the RP11-753C18 BAC carrying the neomycin resistance marker. To accomplish this, homology arms, referred to as Blue-Green and Yellow, respectively, were added to the ampicillin resistance gene. The bMQ455L14 BAC was used as the template for the Blue-Green PCR (primers #215318, #215338) and pBluescript II SK ()was used as the template for the ampicillin-Yellow PCR (primers #215339 and #56719). The two PCR products were gel purified and then combined in an overlap PCR (primers #215318 and #56719). The resulting PCR product was gel purified and was used for a Red/ET reaction, with the RP11-735C18 BAC carrying the neomycin resistance cassette.
Approximately of the modified RP11-753C18 BAC was digested with MluI in a digest, and of the digest was used to insert the human BORCS7/AS3MT segment and neomycin resistance cassette by Red/ET recombination into the bMQ354L20 BAC carrying the short arm and ampicillin gene, simultaneously displacing the ampicillin gene. This reaction yielded the final BORCS7/AS3MT displacer vector, which was digested with NotI to release the BAC backbone before transfection into ES cells.
Generation of Mouse ES Cells
The parental ES cell line (phnx43) used in these studies was generated as follows: Male and female 129S6/SvEvTac mice (Taconic Bioscience) were mated. Females were checked every morning for copulatory plugs, indicative of successful mating. Three days later, females were euthanized by lethal carbon dioxide () exposure followed by physical euthanasia, and the 3.5-d embryos (blastocysts) were collected by flushing the uterus with -independent medium (Gibco; Cat. no. 18045-088) supplemented with 1% bovine serum albumin (Sigma-Aldrich; Cat. no. A3675). Individual embryos were collected from the lavage fluid with a pipette and placed in wells that had been seeded 24 h earlier with primary mouse embryo fibroblasts (MEFs; Gibco; Cat. no. GSC-6005M). Blastocysts and MEFs were cultured in Gibco KnockOut™ (KO) medium (Cat. no. 10829-018) supplemented with ES-qualified 15% fetal bovine serum (FBS; Gibco; Cat. no. 10439024) and l-glutamine (Gibco; Cat. no. 25030-081). After 5–10 d in culture, when the inner cell mass of the embryos reached in diameter, the embryos were removed and disrupted with a pipette and placed in a new well seeded with MEFs. This process of serial expansion of a single inner cell mass was continued until a single inner cell mass was expanded to at least 10 colonies of cells. Cells were then further expanded by disruption and exposure to 0.25% trypsin for 5 min at 37°C. After expansion and cryopreservation in 10% dimethyl sulfoxide (Sigma-Aldrich; Cat. no. D2660), a sample of the cell line was subjected to karyotype analysis in KaryoLogic Inc. (RTP, NC). The cell line used in this study had a 40, XY (normal male mouse) karyotype.
Expression of Human BORCS7/AS3MT in Mouse ES Cells
Single-cell suspensions of the parental ES cell were prepared by treatment with 0.25% trypsin as described above. In a volume of , cells were electroporated in the presence of of DNA. The apparatus used was a BTX Electro cell manipulator 600 with cuvette BTX 45-0125 (Cheshire Analytical). The settings used were 270 V, , R8 (). After electroporation, the cells were distributed onto seven tissue culture plates (Corning; Cat. no. 430167) in Gibco KO medium supplemented with 15% ES-qualified FBS (Gibco; Cat. no. 10439024) and l-glutamine (Gibco; Cat. no. 25030-081). After 24 h, selection was initiated with the addition of Geneticin™ (final concentration ; Gibco; Cat. no. 10131-35). After 7 d, individual neomycin resistant colonies became visible. Cells from colonies were pooled and used for RNA isolation to confirm that the human genes were expressed (see details below). Once this was established, individual colonies from the remaining plates were transferred individually by pipette to a 96-cell plate. Each colony was trypsinized, and a portion used for PCR analysis to identify those in which the DNA had integrated by homologous recombination, as described below. The remaining cells were allowed to grow, and cultures carrying a targeted allele, as determined by the assay detailed below, were transferred sequentially to 2-, 10, and tissue culture plates. At this point, some cells were cryopreserved while a portion of each was used for karyotype analysis and to confirm the expression of human AS3MT by Probe-based droplet digital PCR (ddPCR), using commercially available primers (Applied Biosystems; Hs00960526). Here, a reaction mixture was pipetted into a DG8™ cartridge along with of Droplet Generation Oil for Probes (Bio Rad) and loaded into the QX200 Droplet Generator (Bio Rad) according to the manufacturer’s instructions. After droplet generation, the droplets were transferred to an Eppendorf twin.tec® 96-well plate and placed in a C100 Touch Thermal Cycler (Bio Rad), using the manufacturer’s recommended cycling conditions. After cycling, the plate was read in the QX200 Droplet Reader (Bio Rad) QuantaSoft™ software (version 1864011).
A portion of each of the ES colonies () was disrupted by incubation for 45 min at 55°C in lysis buffer consisting of Tris-EDTA [ Trisma Base (Thermo Fisher Scientific; Cat. no. BP152), ethylenediaminetetraacetic acid (EDTA; Thermo Fisher Scientific; Cat. no. 02-002-786), 10% vol/vol Triton™ X-100 (Millipore Sigma; Cat. no. 108643) and Proteinase K (Roche; Cat. no. 03115879001)]. Lysates from all colonies were screened for homologous recombination by PCR using primers Screen F and #176268 (see Table S4) with GXL polymerase (Takara Bio) according to the manufacturer’s suggested protocol. The PCR program used was 98°C initial denaturation for 10 s, followed by 35 cycles with 10 s denaturation at 98°C, 15 s annealing at 57°C, 4 min at 68°C, and a final 10-min extension at 68°C. PCR products were analyzed by agarose gel electrophoresis (0.8% agarose; Bio Basic; Cat. no. D0012), in Tris-acetate-EDTA running buffer. The size of the PCR product was determined by comparison with Ladder (NEB; Cat. no. N3200L).
Twenty-seven clones with homologous recombination were identified and were subjected to further analysis. Because of this high recombination frequency, it was not necessary to use clustered regularly interspaced short palindromic repeats and CRISPR-associated protein (9CRISPR/cas9) to stimulate recombination events. RNA was prepared from pools of the cells and examined for the expression of the human genes. RNA was also prepared from a number of the individual clones after expansion. Briefly, cells were lysed directly in the culture dish and homogenized by repeated pipetting. RNA was isolated using RNA Bee or STAT60 (Tel-Test) according to the manufacturer’s instructions. RNA yield and quality were assessed using a NanoDrop 2000 (Thermo Fisher). One to two micrograms total RNA was reverse transcribed with the High Capacity Reverse Transcription Kit (Applied Biosystems). Expression of the human AS3MT was examined by qualitative PCR (qPCR) on a C100 Touch Thermal Cycler (Bio Rad) using commercially available primers (Applied Biosystems; Hs00960526).
Generation of Mice Expressing Human BORCS7/AS3MT
For injection into blastocysts, the ES cells carrying the human BORCS7/AS3MT locus were again trypsinized to generate single-cell suspensions. Collection of recipient blastocysts, blastocyst injection, and transfer to pseudopregnant dams was carried out by the UNC Chapel Hill Animal Models Core following procedures described by Longenecker and Kulkarni (2009) and by Koller et al. (1989). To maintain the mutation on the 129S6/SvEvTac background, the chimeric mice were bred to 129S6/SvEvTac mice (Taconic Bioscience). The resulting F1 mice were identified by Taq as wild-type (WT) homozygotes for the mouse As3mt/Borcs7 locus (WT/WT), or heterozygous (WT/Hs) or homozygous (Hs/Hs) for the human AS3MT/BORCS7 locus. The primers used were common and endo for the endogenous locus and common and displaced for the displaced locus (see Table S2). The PCR assays were carried out using Taq polymerase (Bio Basic) with an initial denaturation temperature of 98°C for 20 s followed by 35 cycles of denaturation for 15 s at 96°C, annealing for 30 s at a 54°C, and extension at 72°C for 30 s with a final extension of 1 min. The F2 generation—consisting of Hs/Hs homozygotes, Hs/WT heterozygotes, and WT/WT homozygotes—was produced by mating of F1 Hs/WT heterozygotes. The Hs/Hs, Hs/WT, and WT/WT offspring from the F2 generation were housed together, one litter per cage under controlled conditions with 12-h light/dark cycle at and relative humidity. The parental mice and mice in the F1 and F2 generations were fed Purina LabDiet 5V5R and drank deionized water ad libitum.
All procedures involving mice were approved by the University of North Carolina Institutional Animal Care and Use Committee.
Collection of Tissues for mRNA Expression Analysis
Tissues for mRNA expression analysis were collected from F2 Hs/WT mice, both males and females. Measurement of RNA expression in Hs/WT mice, which were heterozygous for the human and mouse gene, allowed direct comparison of expression in the same tissue/RNA sample, minimizing the effects of physiological variation between animals or quality of sample, which could result in subtle differences in RNA quality and cDNA formation.
Mice were euthanized by exposure to followed by physical euthanasia. The following tissues were collected: whole brain, parts of brain (cortices, pons, medulla, hippocampus, cerebellum, pituitary), spinal cord, adrenals, liver, spleen, kidney, duodenum, jejunum, colon, stomach, uterus, ovaries, testes, heart, and skin (from back of neck). Neuronal and glial cells were also dissected. Brain parts and spinal cord were collected from male mice at 24 d of age. Neuronal cells were isolated from embryonic day 17 (E17) embryos and glial cells from postnatal day 1 (P1) pups. All other tissues were collected from 8- to 12-wk-old mice. Three mice were used for each tissue. Blood was collected from lethally anesthetized mice via cardiac puncture with an EDTA-coated syringe just prior to thoracotomy. To isolate mononuclear cells from blood, of whole blood were layered onto of Histopaque® 1083 (Sigma) and centrifuged at room temperature for 30 min with no brakes at . The upper layer was discarded and the interface containing the mononuclear cells was transferred to a conical tube. The cells were then washed twice with phosphate-buffered saline and then lysed with RNA Bee.
Neuronal cell isolation and culture.
Brains were harvested from eight E17 embryos and placed in a petri dish containing Hanks’s balanced salt solution (HBSS; Gibco). Meninges were removed, cortices dissected, and placed in a new petri dish containing HBSS. Cortices were transferred to a conical tube containing HBSS and centrifuged at for 1 min at 4°C. The tissue was digested with TrypLE Express (Gibco; Cat. no. 12605-010) and DNAse I (Roche; Cat. no. 10104159001) for 30 min at 37°C. The digested cortices were centrifuged at for 1 min at 4°C. The TrypLE + DNAse I was aspirated and the cortices were washed for 2 min with FBS to inactivate the trypsin. The tissue was centrifuged again and resuspended in complete Dulbecco’s Modified Eagle Medium (DMEM; Gibco) with 10% FBS (VWR), GlutaMax (Gibco) with penicillin/streptomycin (Gibco), and 2-mercaptoethanol (Sigma). The cortices were titurated 15 times with a pipette and then 10 times with a flamed-tip Pasteur pipette. The cells were then passed through a cell strainer into a clean conical tube. Cells were then pelleted by centrifugation at for 5 min. The cell pellet was resuspended in complete DMEM and plated in culture dishes coated with poly d-lysine (Sigma); cells per dish. After 24 h, cytosine B-d-arabinofuranoside (Ara-C; Sigma; Cat. no. C6645) was added using a 50% media change without exposing the cells to air. The cells were harvested for RNA isolation on Day 7.
Mixed glial cell culture.
Brains were harvested from six P1 pups and placed in a dish containing HBSS. Meninges were removed, cortices dissected, and placed in a conical tube containing cold DMEM. Cortices were centrifuged at for 1 min at 4°C and media replaced with cold DMEM. The cortices were titurated 10 times with a flamed-tip Pasteur pipette coated in serum. The cells were then passed through a cell strainer into a fresh conical tube. Cells were pelleted by centrifugation at for 5 min. The cell pellet was resuspended in complete DMEM, and of the suspension was plated on three culture plates. The medium was changed 24 h later and then daily. The cells were harvested for RNA on Day 7.
Isolation of adrenal cortex and medulla.
To obtain samples enriched for each of these two distinct functional regions of the gland, the adrenal cortex was removed from medulla of male and female adrenal gland by microdissection. Enrichment was quantitated by qPCR analysis of genes known to be expressed in only one of these two regions: Chgb in the medulla and Cyp11b1 in the cortex (The Human Protein Atlas 2020), using Applied Biosystem Taqman probes Mm00483287 and Mm01204952, respectively. RNA was isolated from cells and tissues using the method described for ES cells; however, the tissues were first homogenized in STAT60 in a desktop homogenizer (Fast-Prep 24; MP Bio) using ceramic beads.
Analysis of AS3MT, , BORCS7, As3mt, and Borcs7 Expression in Collected Tissues and Cells
Both ddPCR and qPCR were used to detect and quantify AS3MT, , BORCS7, As3mt, and Borcs7 mRNA. The specific method is indicated in each figure legend. ddPCR is minimally influenced by differences in the efficiency of the mouse and human primer sets (Quan et al. 2018) and therefore provides a means of direct comparison of expression between the endogenous mouse locus and the humanized locus. The distribution of the AS3MT transcript in the tissues was compared with AS3MT protein distribution described by The Human Protein Atlas for the human tissues. SYBR green qPCR was used to compare RNA expression in whole brain, cortex, hippocampus, pituitary, cerebellum, pons, medulla, spinal cord, glial cells, neurons, adrenals, ovaries, testes, heart, kidney, liver, colon, jejunum, spleen, and blood. ddPCR was used to assess RNA expression in ES cells, whole brain, liver, adrenals, spinal cord, ovaries, testes, spleen, and heart. For all assays, total RNA was reverse transcribed with the High Capacity Reverse Transcription Kit (Applied Biosystems).
For SYBR green qPCR, the relative expression of full-length AS3MT, the d2d3 variant, and the BORCS7/AS3MT fusion transcript were assessed on a QuantStudio6 Flex™ Real-Time PCR System (Applied Biosystems). SYBR green reactions were run using iTaq Universal SYBR Green Supermix (Bio Rad), of each primer, and of cDNA. Primers used for quantification of AS3MT isoforms were d2d3-F and d2d3-R for the d2d3 isoform and full-F and full-R for the full-length isoform (see Table S2). For analysis of BORCS7/AS3MT read-through transcripts expression in mouse tissues, RNA was reverse transcribed and SYBR green quantitative PCR was run using primers -GAACAGTCATCGGATCTACAGGA- and -GAACAGTCATCGGATCTACAGGA- and iTaq SYBR green Universal Supermix (BioRad).
For ddPCR, absolute concentration of As3mt, AS3MT, Borcs7, and BORCS7 was acquired using the ddPCR Supermix for Probes (no dUTP) (Bio Rad) and TaqMan Gene Expression Assays, As3mt (Mm00491075_m1), AS3MT (Hs00960526_g1), Borcs7 (Mm01205060_m1), and BORCS7 (Hs00376014_m1), all from Applied Biosystems. The ddPCR analysis was carried out as described above.
Evaluation of the Metabolism of a Single Dose of iAs
The metabolism of iAs was examined in 18- to 22-wk-old Hs/Hs male () and female () F2 offspring and their WT/WT male () and female () littermates. The mice were given a single dose of iAs (sodium arsenite, pure; Sigma-Aldrich) in deionized water (DIW) by gavage: of body weight. Immediately after dosing, each mouse was placed in a metabolic cage (1 mouse/cage) and urine and feces were collected in 24-h intervals for 3 d. During these 3 d, all mice drank DIW ad libitum. The mice were fasted during the first 24 h but had free access to purified laboratory diet (Envigo Teklad; Cat. no. AIN-93G) during the second and the third days. Our published data showed that iAs content in this type of diet ranges from 10 to (Douillet et al. 2017; Huang et al. 2018; Murko et al. 2018). The 24-h urine and feces samples were frozen in dry ice and stored at .
Evaluation of the Metabolism of iAs during Subchronic Exposure
After collection of urine and feces following the single-dose administration, the Hs/Hs and WT/WT mice were again caged together (one litter per cage) and maintained on the purified diet and DIW for 5 wk to allow for clearance of As from the body, which was confirmed by measuring total As (tAs) levels in urine (see the “Results” section for details). Both WT and Hs/Hs mice (now 23- to 27-wk-old) were then exposed to iAs (sodium arsenite, 99% pure; Sigma-Aldrich) in drinking water () for 4 wk. Spot urine samples () were collected weekly. Body weights were recorded before and after the exposure. After 4 wk, all mice were sacrificed by cervical dislocation without anesthesia and tissues were collected, including the liver, kidneys, pancreas, spleen, heart, lung, adrenals, kidneys, bladder, visceral fat, calf muscle, testes or ovaries, and cecum and colon. All tissues were flash-frozen in dry ice and stored at .
Analysis of As Species in Urine, Feces, and Tissues
Speciation analysis of As was carried out in 24-h urines and feces collected after the single dose of iAs and in spot urine samples collected during subchronic iAs exposure. The speciation analysis was also performed in livers and kidneys collected after subchronic exposure at sacrifice. Ten percent homogenates of liver and kidney were prepared in ice-cold DIW (10% wt/vol) using Wheaton Potter-Elvehjem–style tissue grinders with a PTFE pestle and Wheaton overhead stirrer apparatus (DWK Life Sciences). Feces were snap frozen in liquid nitrogen and pulverized to powder using a steel mortar and pestle placed in dry ice. The powder was mixed with 2N ultrapure phosphoric acid 1:5 (Thermo Fisher) and digested in a MARS5 microwave (CEM Corp.) for 10 h at 90°C. This method eliminates the biological matrix but does not alter As speciation (Currier et al. 2011). The digestates were neutralized by sodium hydroxide (Sigma-Aldrich) to pH 6. The urines, tissue homogenates, and neutralized digestates were treated with 2% l-cysteine (Sigma-Aldrich) at room temperature for 1 h to reduce pentavalent As species to their trivalent counterparts prior to the analysis (Currier et al. 2011). The cysteine-treated samples were analyzed by hydride-generation atomic absorption spectrometry coupled with a cryotrap (HG-CT-AAS), as previously described (Currier et al. 2011; Hernández-Zavala et al. 2008). This analysis determined the concentrations of iAs, MAs, and DMAs. tAs concentration in urine, feces, and tissues was calculated as the sum of iAs, MAs, and DMAs. For assessment of the efficiency of iAs metabolism after a single oral dose, the amount of tAs was expressed as the percentage of the dose by comparing the amount of elemental As administered as iAs to each mouse with the amount of tAs in 24-h urine and feces samples collected from that mouse.
The instrumental limits of detection (LOD) for iAs, MAs, and DMAs using this method are 14, 8, and As, respectively (Hernández-Zavala et al. 2008). An imputed value of 0 was used for measurements below the LOD. HG-CT-AAS is also capable of detecting and quantifying trimethylarsine oxide (TMAsO) (Hernández-Zavala et al. 2008), a product of iAs metabolism by rat As3mt (Waters et al. 2004). However, because TMAsO is rarely detected in human urine and because TMAsO was not a product of iAs methylation by recombinant human AS3MT in our published study (Ding et al. 2012), we did not include TMAsO analysis in this study.
Statistical Analysis
One-way analysis of variance with Student-Newman-Keuls or Tukey’s multiple comparison post-test was used to assess differences in gene expression and in the concentrations and proportions of As species among WT/WT and Hs/Hs mice. Student’s t-test was applied for comparison of mouse and human gene expression in a single tissue of Hs/WT mice and for comparison of As species concentrations or proportions between male and female mice and between Hs/Hs and WT/WT mice of the same sex. The statistical analyses were performed using Prism 8 software (GraphPad Software). Differences with were considered statistically significant.
Results
Expression of BORCS7 and AS3MT in Hs/WT Mice
Expression of AS3MT and BORCS7 were directly compared with those of endogenous mouse genes in tissues collected from F2 male Hs/WT mice that carried one copy of the mouse locus and one copy of the human locus (Figure 2A) using both conventional qPCR and, when appropriate, ddPCR. Expression of human AS3MT and BORCS7 was easily detected in all tissues expressing the corresponding mouse genes, although the level of expression of the human and mouse genes differed (Figure 2B,C). Notable in this analysis was the species difference in expression of AS3MT/As3mt in the liver and adrenal glands. This difference was examined in more detail in samples obtained from both female and male animals (Figure 2D). The expression of AS3MT in the mouse liver was significantly lower than that of the mouse ortholog in both males and females. In comparison, expression of human AS3MT was significantly higher than mouse in the adrenal gland. Robust expression of the mouse transcript was, however, noted, with levels similar to that of the liver.
Figure 2.

Expression of the mouse and human BORCS7/AS3MT locus in tissues of heterozygotes carrying one human (Hs) and one wild-type (WT) allele: (A) Breeding scheme used to ensure that all animals carry a single WT and Hs locus. Levels of (B) Borcs7/BORCS7 and (C) As3mt/AS3MT transcripts in tissues of male and female mice determined by droplet digital PCR (ddPCR) (, ; *, **, and *** by Student’s t-test for differences between expression of the human and mouse genes). (D) Expression of As3mt/AS3MT in the adrenal gland and liver of male and female mice determined by ddPCR (, for males and for females; *, **, and *** by ANOVA with Tukey’s post-test for differences between expression of the human and mouse genes). (E) Expression of AS3MT in cortex and medulla enriched adrenal tissues by quantitative PCR; the enrichment was quantitated by expression of the medulla- and cortex- specific genes, Chgb and Cyp11b1, respectively (medulla and cortex were pooled samples from four mice); (F) Pearson’s correlation between expression of AS3MT and BORCS7 (, ), and As3mt and Borcs7 (, ) in ES cells and tissues shown in (B) and (C). All tissues were harvested from 8- to 12-wk-old mice. Note: ANOVA, analysis of variance; ddPCR, droplet digital polymerase chain reaction; ES, embryonic stem; PCR, polymerase chain reaction; SE, standard error.
To further characterize the distribution of AS3MT/As3mt in the adrenal gland, we examined expression of both genes separately in medulla, which is a neuroendocrine tissue responsible for catecholamine production, and cortex, which produces corticosterone, the major glucocorticoid in rodents, and aldosterone, the major mineralocorticoid. Tissue specificity of RNA isolated from each section of the adrenals was assessed by quantitative analysis of mRNA species previously reported to be expressed specifically in either the medulla (Chgb) or cortex (Cyp11b1) (The Human Protein Atlas 2020). We found that mouse As3mt and human AS3MT were expressed in both sections of the adrenal gland (Figure 2E).
We also used Pearson correlation analysis to evaluate the association between expression of the mouse As3mt and Borcs7, as well as the human AS3MT and BORCS7, in ES cells and the tissues of Hs/WT mice, as shown in Figure 2B,C. In spite of the tight linkage, we did not find statistically significant correlation between either the mouse genes (, ) or human genes (, ) (Figure 2F).
To verify that the use of heterozygous animals for expression analysis was appropriate, and specifically to exclude the possibility that expression of the human locus was influenced by the presence of the mouse locus or vice versa, we compared AS3MT and As3mt expression in the adrenals and liver of mice homozygous and heterozygous for each gene (see Figure S1). As expected, expression levels in the heterozygotes were lower by as compared with homozygotes.
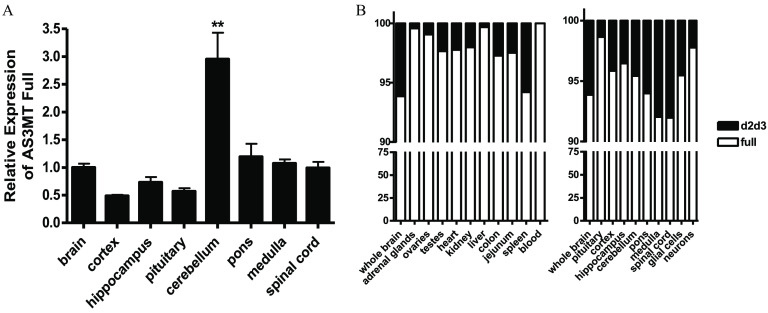
Because the levels of AS3MT expression in the brain approached those observed in the liver, we used qPCR analysis to determine whether this reflected a generalized expression throughout the brain or whether expression was localized to a particular region. Figure 3 shows that the highest levels of expression were observed in the brain stem.
Figure 3.
Expression of AS3MT and the variant in tissues of heterozygotes carrying one human (Hs) and one wild-type (WT) allele: (A) Relative expression of the full-length AS3MT in regions of brain as compared with the whole brain (, ; ** by ANOVA with Tukey’s post-test for comparison of cerebellum with other brain regions). (B) Relative expression of the full-length AS3MT and in tissues with focus on brain and brain regions and cells. The brain tissues were harvested from 8- to 12-wk-old mice, neurons were obtained from E17 embryos, and glial cells from P1 pups. Note: ANOVA, analysis of variance; E, embryonic day; P, postnatal day; SE, standard error.
Expression of the Isoform in the Humanized Mice
We next determined whether the information and enzymatic machinery for direct expression of the schizophrenia associated AS3MT splice variant () was present in this humanized model and, if so, whether the tissue distribution of the variant transcript would mimic that observed in humans. To accomplish this, PCR primers specific for this variant were designed (see Table S2), and expression of the common and variant transcripts was determined by qPCR analysis. The percentage of total transcript contributed by the variant was determined. Similar to reports for human tissue, the variant represented over 5% of the total AS3MT transcript of the brain, with the contribution varying slightly between regions. In contrast, the variant represented a very small percentage of the transcript present in non–central nervous tissue, such as the adrenal gland and liver. (Figure 3A,B). To examine this further, RNA was isolated from neuronal cultures prepared from E17 embryos and from glial cell culture prepared from P1 newborn brains. AS3MT expression was observed in both (Figure 3B).
Expression of the BORCS7/AS3MT Read-through Transcripts
We also analyzed the liver, adrenals, and testes from one male Hs/Hs mouse for expression of the previously reported human-specific read-through transcript composed of exons from both the BORCS7 and AS3MT genes (Lu et al. 2015; Pierce et al. 2012; Prakash et al. 2010; Ripke et al. 2013; Schizophrenia Working Group of the Psychiatric Genomics Consortium 2014). Testes were included in this analysis because they express relatively high levels of BORCS7 (Figure 2B), and expression of the BORCS7/AS3MT read-through variant is driven by the BORCS7 promoter given that BORCS7 is located of AS3MT (Li et al. 2016). We found this variant to be expressed in all three tissues (see Figure S2A). To verify the identity and structure of the transcript, of cDNA from testes of WT/WT, Hs/WT, and Hs/Hs mice (1 mouse per genotype) were PCR amplified and resolved on a 1.6% agarose gel. As expected, a fragment was observed, consistent with the previously described read-through transcript (see Figure S2B). However, an additional product was found of about . To establish the identity of this product, the DNA band was excised from the gel and subjected to Sanger sequence analysis. We found that in this transcript, the exon shared with the penultimate exon of the lower two BORCS7 isoforms spliced to a cryptic splice acceptor site located downstream of the position corresponding to the transcription start site of the AS3MT gene (see Figure S2C). The BORCS7 stop codon was present in both the previously reported and this novel read-through transcript, thus preventing the AS3MT coding sequence in these transcripts from being translated and, presumably, rendering them susceptible to nonsense-mediated decay.
Metabolism of a Single Oral Dose of iAs in Hs/Hs and WT/WT Mice
The concentrations of iAs and its metabolites were measured in urine and feces of F2 male and female Hs/Hs mice and their WT/WT littermates in three 24-h intervals during 72 h after a single oral dose of iAs ( of body weight). The concentrations of iAs and DMAs were above the LOD in all urine samples collected from the Hs/Hs and WT/WT mice; MAs concentrations were above the LOD in urines of Hs/Hs mice but below the LOD in most urines (52 of 54 samples) collected from WT/WT mice regardless of sex. In feces, DMAs concentrations were below the LOD in 48 of 54 samples collected from male and female Hs/Hs mice.
Urinary excretion of As.
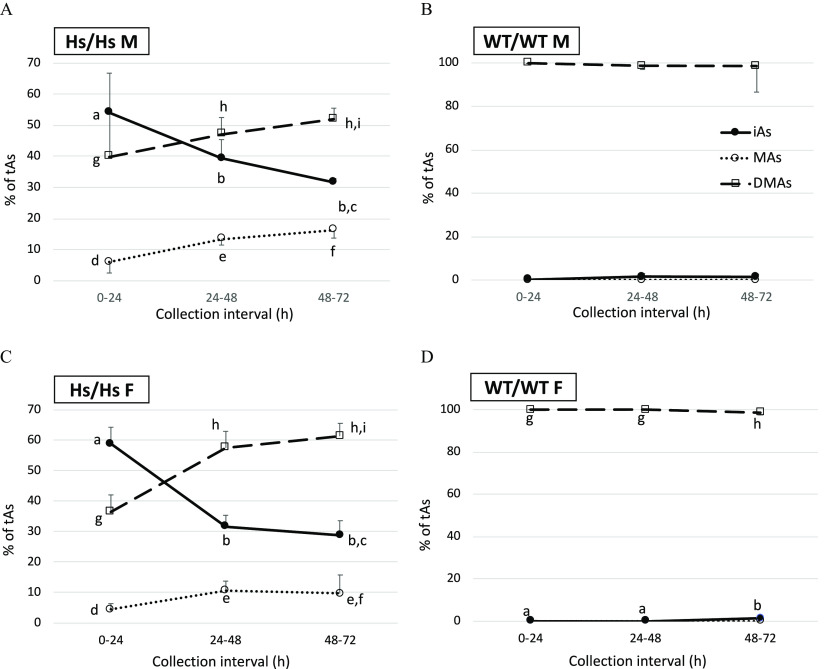
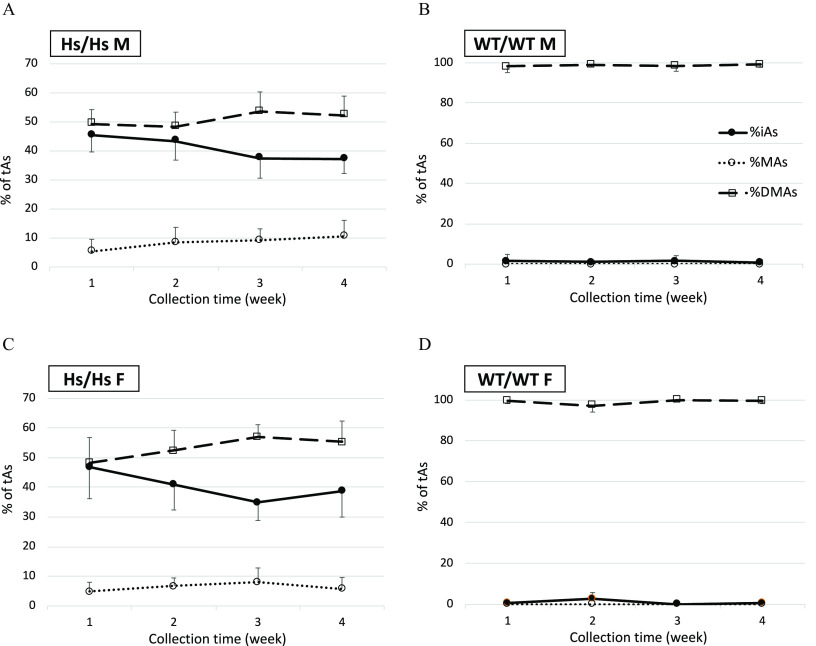
During the first 24 h, WT/WT mice excreted significantly greater amounts of total As () in urine than their Hs/Hs counterparts (Figure 4). On average, WT/WT males and females excreted 34.9% and 60.7% of the administered dose, respectively, as compared with 8.2% excreted by Hs/Hs males and 16.5% by Hs/Hs females. For Hs/Hs and WT/WT mice, tAs amounts excreted between 24 and 48 h or between 48 and 72 h did not differ significantly. Overall, WT/WT males and females respectively excreted 47.7% and 75.9% of the dose in urine during 72 h postdosing. Hs/Hs males and females excreted only 21.4% and 34.8%, respectively, in urine during this interval. Differences between sexes for each genotype were not statistically significant when the test accounted for multiple comparisons. However, sex differences within genotype were statistically significant when the t-test was used to compare WT/WT males and females () and Hs/Hs males and females (). Percentages of urinary tAs represented by iAs (%iAs), MAs (%MAs), and DMAs (%DMAs) also differed by genotype (Figure 5). DMAs was the major urinary metabolite in WT/WT mice () but represented only 40–52% and 37–61% of urinary tAs for Hs/Hs males and females, respectively. During the 72-h collection, urinary %DMAs increased significantly and %iAs decreased for Hs/Hs mice. By contrast, urinary %DMAs and %iAs were little changed for WT/WT mice. Notably, MAs accounted for 4–16% of urinary tAs for Hs/Hs mice but was not detected in the urine of WT/WT mice.
Figure 4.
Total arsenic (tAs) in urine of humanized (Hs/Hs) and wild-type (WT/WT) male (M) and female (F) mice during 72 h after oral administration of a single dose of inorganic arsenic (). Urine was collected in 24-h intervals. tAs was calculated as the sum of inorganic arsenic, methyl-arsenic, and dimethyl-arsenic and was expressed as a percentage of the dose (; for Hs/Hs males, for Hs/Hs females, for WT/WT males, and for WT/WT females). MAs concentrations were below the LOD in 52 of 54 urine samples collected from WT/WT mice during the three collection intervals; a value of was imputed for MAs concentrations in these samples. In each collection interval, values marked with different letters are significantly different () by ANOVA with Student-Newman-Keuls post-test. Note: ANOVA, analysis of variance; LOD, limit of detection; MAs, methyl-arsenic; SE, standard error.
Figure 5.
Proportions of total arsenic (%tAs) represented by inorganic arsenic (iAs), methyl-arsenic (MAs), and dimethyl-arsenic (DMAs) in the urine of humanized (Hs/Hs) and wild-type (WT/WT) male (M) and female (F) mice collected during 24-h intervals after oral administration of a single dose of iAs (). (A) male Hs/Hs; (B) male WT/WT; (C) female Hs/Hs; and (D) female WT/WT. ; for Hs/Hs males, for Hs/Hs females, for WT/WT males, and for WT/WT females. MAs concentrations were below the LOD in 52 of 54 urine samples collected from WT/WT mice during the three collection intervals; a value of was imputed for MAs concentrations in these samples. For each of the metabolites, statistically significant differences in %tAs values between the collection intervals are marked with different letters: a, b, c for differences in %iAs; d, e, f for differences in %MAs; and g, h, i for differences in %DMAs. (ANOVA with Student-Newman-Keuls post-test.) The absence of marking indicates the lack of significant differences between the collection intervals. Note: ANOVA, analysis of variance; LOD, limit of detection; SD, standard deviation; %DMAs, percentage DMAs; %MAs, percentage MAs.
Urinary concentrations and proportions of As metabolites differed significantly between HS/HS males and females. Urinary concentrations of iAs and MAs were higher in males than in females in the 48-72 h collection interval (see Figure S3). Similarly, urinary %iAs and %MAs were higher in males between 24 and 48 h and between 48 and 72 h, respectively (see Figure S4). No sex-related differences were found among WT/WT mice.
Fecal excretion of As.
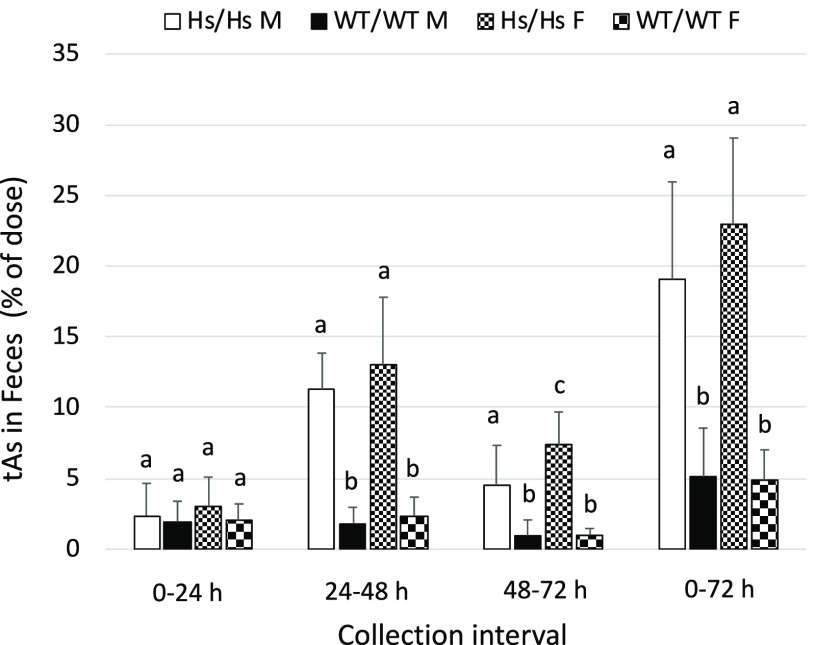
Compared with urine, very little tAs (2–3% of the dose) was excreted in feces of either WT/WT or Hs/Hs mice during the first 24 h after gavage (Figure 6). Between 24 and 48 h postdosing, fecal tAs increased to 11–13% of the dose for Hs/Hs mice but did not increase for WT/WT mice. During the 0- to 72-h collection period, fecal excretion accounted for of the dose in WT/WT mice and 19–21% in Hs/Hs mice. For both genotypes, iAs was the major metabolite, accounting for up to 46% of fecal tAs for Hs/Hs mice and up to 99% of fecal tAs for WT/WT mice (Figure 7). However, although fecal %iAs decreased over time for Hs/Hs mice, fecal %iAs increased or remained unchanged for WT/WT mice. MAs was a major metabolite in feces of Hs/Hs mice, reaching 54–56% of fecal tAs between 48 and 72 h after gavage. In contrast, DMAs was a minor metabolite (0–3% of tAs). Only traces of MAs were detected in feces of WT/WT mice, and DMAs represented up to 24% of fecal tAs. There were no statistically significant sex-related differences in concentrations of iAs, MAs, or DMAs either among Hs/Hs or WT/WT mice (see Figure S5). However, significant sex-related differences were found in the proportions of As metabolites. Feces of WT/WT males collected during the 48- to 72-h interval had lower %iAs and higher %DMAs than did feces of WT females. During the 24- to 48-h collection, feces of Hs/Hs males had lower %MAs than did feces of Hs/Hs females (see Figure S6).
Figure 6.
Total arsenic (tAs) in feces of humanized (Hs/Hs) and wild-type (WT/WT) male (M) and female (F) mice during 72 h after oral administration of a single dose of inorganic arsenic ( body weight). Feces were collected in 24-h intervals. tAs was calculated as the sum of inorganic arsenic, methyl-arsenic, and dimethyl-arsenic, and was expressed as the percentage of the dose (; for Hs/Hs males, for Hs/Hs females, for WT/WT males, and for WT/WT females). DMAs concentrations were below the LOD in 48 of 54 fecal samples collected during the three collection intervals from male and female Hs/Hs mice; a value of was imputed for DMAs concentrations in these samples. In each collection interval, values marked with different letters are significantly different () by ANOVA with Student-Newman-Keuls post-test. Note: ANOVA, analysis of variance; DMAs, dimethyl-arsenic; LOD, limit of detection; SD, standard deviation.
Figure 7.
Proportions of total As (%tAs) represented by inorganic arsenic (iAs), methyl-arsenic (MAs) and dimethyl-arsenic (DMAs) in feces of humanized (Hs/Hs) and wild-type (WT/WT) male (M) and female (F) mice collected during 24-h intervals after oral administration of a single dose of iAs ( body weight): (A) male Hs/Hs; (B) male WT/WT; (C) female Hs/Hs; and (D) female WT/WT. , for Hs/Hs males, for Hs/Hs females, for WT/WT males, and for WT/WT females. DMAs concentrations were below the LOD in 48 of 54 fecal samples collected during the three collection intervals from male and female Hs/Hs mice; a value of was imputed for DMAs concentrations in these samples. For each of the metabolites, statistically significant changes in %tAs values between the collection intervals are marked with different letters: a, b, c for differences in %iAs; d, e, f for differences in %MAs; and g, h for differences in %DMAs. (ANOVA with Student-Newman-Keuls post-test.) The absence of marking indicates the lack of significant differences between the collection intervals. Note: ANOVA, analysis of variance; LOD, limit of detection; %DMAs, percentage DMAs; %MAs, percentage MAs.
Disposition of As in Urine and Tissues of Hs/Hs and WT/WT Mice During Subchronic Exposure to iAs
After completion of the experiments examining metabolism of the single oral dose of iAs, the F2 Hs/Hs and WT/WT mice were caged together (one litter per cage) and maintained on the purified diet and DIW for 5 wk to allow for clearance of iAs from the body. After this time, the concentrations of total As in urine of Hs/Hs and WT/WT mice ranged from 4 to , that is, the levels that are typical for urine of unexposed mice (Douillet et al. 2017; Drobná et al. 2009). These mice were then exposed to iAs in drinking water ( as arsenite) for 4 wk. The numbers of mice per genotype and treatment groups were the same as those specified for the single-dose experiment. Spot urine samples were collected weekly. Body weights were assessed before and after the exposure, before sacrifice. Tissues were harvested during sacrifice at the end of Week 4.
There were no statistically significant differences in body weights of WT/WT and H/Hs males, vs. (), or females, vs. , prior to subchronic exposure to iAs. Similarly, no significant differences in body weights of WT/WT and Hs/Hs mice were found after the exposure: vs. for males, and vs. for females. No differences in weight gains were found between the WT/WT and Hs/Hs mice of the same sex.
The concentrations of iAs and its metabolites were measured in the spot urine samples and in liver and kidneys. iAs and DMAs were detected in all urine samples. MAs levels were below the LOD in 70 of 72 urine samples collected from WT/WT mice. DMAs levels were below the LOD in 16 of 18 liver samples collected from Hs/Hs mice. In the kidneys, all As species were above the LOD.
Arsenic metabolites in urine.
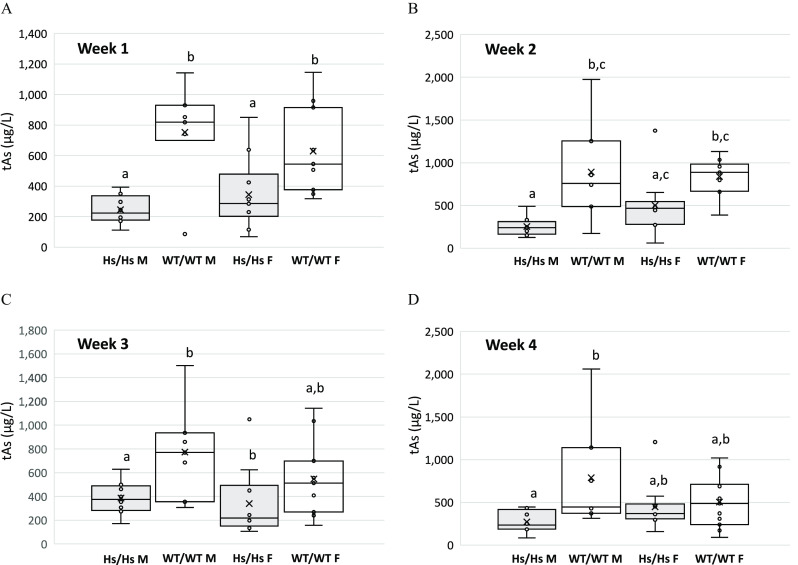
After the first week of the exposure, the concentrations of tAs in urine of Hs/Hs male and female mice were significantly lower than in urine of their WT/WT counterparts (Figure 8). Although this trend persisted during the next 3 wk, only differences between WT/WT and Hs/Hs males remained statistically significant. Urinary tAs concentrations in male and female mice of either genotype were not significantly different. Striking differences were found between Hs/Hs and WT/WT mice in proportions of As metabolites. These differences were consistent throughout the 4-wk exposure interval (Figure 9, Figure S7). In the urine of WT/WT mice, DMAs accounted for of tAs; only traces of iAs were found and MAs was not detected at any time point (Figure 9B,D). The Hs/Hs genotype had striking effects on proportions of As species in urine. Here, DMAs accounted for only 48–55% of urinary tAs and iAs and MAs accounted for 35–47% and for 5–11% of urinary tAs, respectively. In the urine of Hs/Hs males and females, %iAs decreased and %DMAs increased between Week 1 and Week 4 (Figure 9A,C); however, these changes were not statistically significant. For either genotype, percentages of As metabolites in the urine did not differ significantly between males and females except for Week 4, when urinary %MAs of Hs/Hs males (11%) was significantly greater than in the urine of Hs/Hs females (6%) (see Figure S7).
Figure 8.
Total As (tAs) in urine of humanized (Hs/Hs) and wild-type (WT/WT) male (M) and female (F) mice during 4 wk of chronic exposure to inorganic arsenic in drinking water (). (A) Week 1; (B) Week 2; (C) Week 3; and (D) Week 4. Spot urine samples were collected at the end of each week. tAs was calculated as the sum of inorganic arsenic, methyl-arsenic, and dimethyl-arsenic. Mean (x), median (―), 25th and 75th percentiles (box), maximum and minimum (whiskers), and individual values including outliers are shown ( for Hs/Hs males, for Hs/Hs females, for WT/WT males, and for WT/WT females). MAs concentrations were below the LOD in 70 of 72 urine samples collected from WT/WT mice during the four collection intervals; a value of was imputed for MAs concentrations in these samples. Within each panel, values marked with different letters are significantly different () by ANOVA with Student-Newman-Keuls post-test. Note: ANOVA, analysis of variance; LOD, limit of detection; MAs, methyl-arsenic.
Figure 9.
Proportions of total As (%tAs) represented by inorganic arsenic (%iAs), methyl-arsenic (%MAs), and dimethyl-arsenic (%DMAs) in urine of humanized (Hs/Hs) and wild-type (WT/WT) male (M) and female (F) mice collected during 4 wk of exposure to iAs in drinking water (). (A) male Hs/Hs; (B) male WT/WT; (C) female Hs/Hs; and (D) female WT/WT. , for Hs/Hs males, for Hs/Hs females, for WT/WT males, and for WT/WT females. MAs concentrations were below the LOD in 70 of 72 urine samples collected from WT/WT mice during the four collection intervals; a value of was imputed for MAs concentrations in these samples. The absence of marking indicates the lack of significant differences between the collection intervals. (ANOVA with Student-Newman-Keuls post-test.) Note: ANOVA, analysis of variance; SD, standard deviation.
Arsenic metabolites in liver and kidneys.
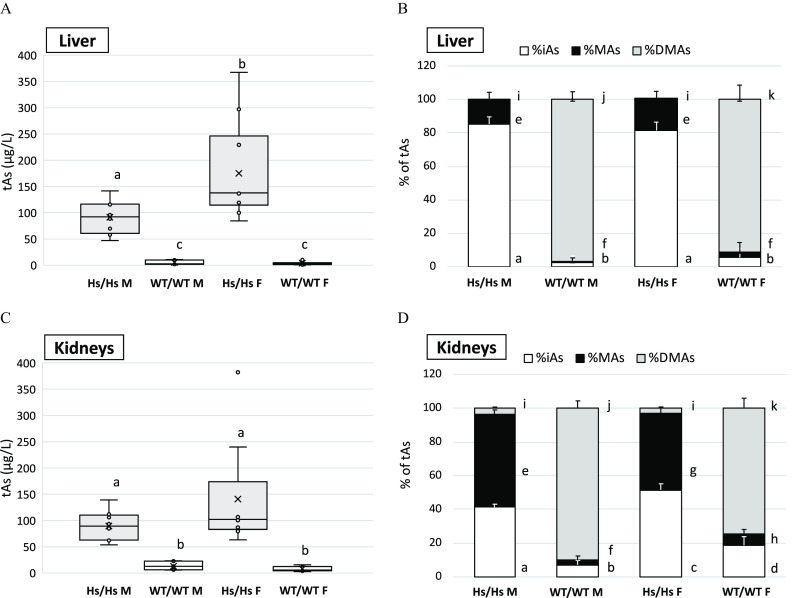
Differences between Hs/Hs and WT/WT mice in concentrations and proportions of iAs and its metabolites in the liver and kidneys (Figure 10) reflected those found in the urine. tAs concentrations were significantly higher in livers and kidneys of Hs/Hs mice than in livers and kidneys of WT/WT mice (Figure 10A,B). In addition, tAs, iAs, and MAs concentrations tended to be higher in the organs of Hs/Hs females as compared with Hs/Hs males (see Figure S8). Proportions of As metabolites also differed between genotypes and were influenced by sex. In WT/WT mice, DMAs was the major metabolite, accounting for of tAs in the livers and in kidneys (Figure 10B,D). %DMAs was significantly higher in the livers and kidneys of WT/WT males than females and %iAs and %MAs tended to be lower. In contrast, only traces of DMAs were found in the livers of male and female Hs/Hs mice; iAs accounted for and MAs for of tAs, respectively, in this organ. In kidneys of Hs/Hs mice, %iAs and %MAs accounted for similar proportions of tAs. Average %iAs was significantly higher and %MAs was lower in the kidneys of females as compared with males: 52% vs. 41% iAs and 45% vs. 55% MAs.
Figure 10.
Total As (tAs) and arsenic metabolites in liver and kidneys of humanized (Hs/Hs) and wild-type (WT/WT) male (M) and female (F) mice after 4-wk exposure to inorganic arsenic (iAs) in drinking water (). tAs was calculated as the sum of iAs, methyl-arsenic (MAs), and dimethyl-arsenic (DMAs). tAs in (A) liver and (C) kidneys; mean (x), median (―), 25th and 75th percentiles (box), maximum and minimum (whiskers), and individual values including outliers are shown; values marked with different letters are significantly different () by ANOVA with Student-Newman-Keuls post-test. Proportions of tAs (%tAs) represented by iAs (%iAs), methyl-arsenic (%MAs), and dimethyl-arsenic (%DMAs) in (B) liver and (D) kidneys; . DMAs concentrations were below the LOD in 16 of 18 liver samples collected from Hs/Hs mice; a value of was imputed for DMAs concentrations in these samples. Different letters indicate significant differences between strains and sexes in proportions of each of the individual metabolites (): a, b, c, d for differences in %iAs; e, f, g, h for differences in %MAs; and i, j, k for differences in %DMAs. (ANOVA with Student-Newman-Keuls post-test.) for Hs/Hs males, for Hs/Hs females, for WT/WT males, and for WT/WT females. Note: ANOVA, analysis of variance; LOD, limit of detection; SE, standard error.
Comparison of as Disposition in Hs/Hs and WT/WT Mice and in Humans
Concentrations of tAs in livers and kidneys of mice exposed for 4 wk to in this study were compared with tAs concentrations predicted for human tissues by a physiologically based pharmacokinetic (PBPK) model (El-Masri and Kenyon 2008). This model consisted of interconnected individual PBPK submodels for iAs, MAs, and DMAs. Each submodel was constructed using flow-limited compartments describing the mass balance of the chemicals in the gastrointestinal tract (lumen and tissue), lung, liver, kidney, muscle, skin, heart, and brain. Levels of tAs were obtained by adding the overall model-predicted concentrations of As, MMA, and DMA in tissues, assuming chronic exposure to and water consumption rate of . This model predicted tAs concentrations to reach 118.5 and in human livers and kidneys, respectively. These values are similar to tAs concentrations found in the livers and kidneys of Hs/Hs mice but are an order of magnitude higher than tAs concentrations in the livers and kidneys of WT/WT mice (Table 1).
Table 1.
Total arsenic levels () in livers and kidneys of humans and of the humanized (Hs/Hs) and wild-type (WT/WT) male and female mice exposed to in drinking water.
Steady-state total arsenic levels calculated using the human physiologically based pharmacokinetic model for inorganic arsenic, assuming consumption of 2 L of water/d (El-Masri and Kenyon 2008).
Total arsenic levels determined in the present study ( for Hs/Hs males, for Hs/Hs females, for WT/WT males, and for WT/WT females).
Discussion
Earlier studies reported significant differences in iAs metabolism between mice and humans. In humans chronically exposed to iAs, DMAs is the major urinary metabolite, but both MAs and iAs account for substantial fractions of urinary tAs. For example, in a U.S. population chronically exposed to iAs in drinking water, iAs and MAs accounted for 4.4–37.9% and 5.9–29.3% of urinary tAs, respectively, and DMAs accounted for 47.4–86.0% of tAs (Hudgens et al. 2016). Similar proportions of urinary As metabolites have been reported in other iAs-exposed populations (Vahter 1999, 2000). In contrast, in the urine of WT C57BL/6 mice (Douillet et al. 2017; Huang et al. 2018) and mice from 12 Collaborative Cross strains (Stýblo et al. 2019) exposed to iAs in drinking water (0.1, 1, or ), 95–99% of tAs was present as DMAs, whereas MAs accounted for a very small fraction of tAs at or near the limits of detection and quantification. Thus, the conversion of iAs to DMAs is more efficient in mice as compared with humans (Vahter 1999, 2000). Consequently, mice given a single dose of iAs excrete of the dose in 48 h, whereas the biological half-time of iAs in humans is (Vahter 1999). Similarly, chronic exposures to iAs would result in a lower body burden in mice as compared with humans. We have previously shown that tAs concentrations in the livers of mice exposed chronically to of drinking water were in the range of tAs concentrations reported in the livers of humans chronically exposed to only in drinking water (Mazumder 2005; Paul et al. 2007). Because of the higher efficiency of iAs methylation and lower body burden, mice are less susceptible than humans to the adverse effects of iAs exposure. The goal of the present study was to generate a mouse strain in which iAs metabolism would resemble that in humans and in which the effects of iAs could be studied at the environmentally relevant exposure levels.
The mouse and human AS3MT share only of primary structure (Drobná et al. 2009), which alone could explain the differences between mice and humans in the efficiency of iAs metabolism. Because AS3MT is the key enzyme in the pathway for iAs methylation, we hypothesized that replacing the mouse As3mt gene with its human ortholog would confer a human-like pattern of iAs metabolism in the mouse. The mice we have generated have a single copy of human DNA replacing the syntenic segment of mouse DNA, with the transition between the DNA of the two species known to the base pair. Homozygous humanized mice can be bred without the need to frequently track the humanized locus. The neighboring genes are defined and not altered by the recombination event, and, because there is a single copy of the human DNA, intrageneic recombination events do not occur. The vector sequences used in the genetic manipulations are removed, and the only evidence of the engineering is a mutant loxP site. This approach circumvents problems that are typically associated with the introduction of a human gene as a transgene, such as variable copy number, instability of transgene copy number, impact of transgene expression on neighboring endogenous genes, inclusion of genes in the transgenic BAC, and complex breeding schemes, when a study requires the absence of an endogenous mouse gene (Chandler et al. 2007; Cheng et al. 2011; Mukai et al. 2006; Yueh et al. 2011).
We included Borcs7 in our humanization scheme because the promoter of AS3MT abuts the untranslated region of BORC7, and the weak correlation in the expression pattern of these two genes (Li et al. 2016) suggests possible shared transcriptional regulatory elements. Although we were unable to observe this weak correlation in expression between the two genes, we were able to detect the read-through BORCS7/AS3MT transcript lacking the final exon of BORCS7 and the initial exon of AS3MT listed on the UCSC Genome Browser (UCSC 2013). The inclusion of BORCS7 in the humanized region increases the likelihood that the elements directing the species differences in the expression of AS3MT are included in the humanized region. The mouse Borcs7 is a subunit of an eight-protein complex known as BORC, which has been shown to be essential for centrifugal transport of lysosomes given that loss of its function results in the centripetal collapse of lysosomes as well as reduction in cell spreading and migration (Pu et al. 2015). Because of the high degree of conservation between Borcs7 and its human ortholog, the human protein is likely to compensate for the loss of the endogenous mouse protein in the Hs/Hs mice.
We chose to humanize mice from the 129S6 strain because of the genetic stability of the ES cells from this strain, the lower cost of growth of 129 ES lines compared with lines from other strains, and because of a high rate of homologous recombination. The 129S6 subline breeds well, with an average litter size of seven pups. In addition, should it be required for future studies, ES lines can be generated from the blastocysts of the humanized mice, quickly and at low cost, compared with other strains. Such a line would be an ideal starting point for modification of additional loci that might be identified as relevant to iAs metabolism or diseases related to iAs exposure.
The results of the present study are consistent with our working hypothesis. Humanizing the As3mt/Borcs7 locus changed profoundly both the patterns of AS3MT expression and iAs metabolism. The mouse As3mt was highly expressed in multiple tissues, including the liver, heart, adrenals, testes, and ovaries. In contrast, the human AS3MT was expressed primarily in the adrenals. AS3MT expression in the adrenals exceeded by an order of magnitude the expression in any of the other tissues examined. In the liver, the expression of AS3MT was significantly lower than the expression of As3mt, suggesting lower capacity of the humanized mice to metabolize iAs. Indeed, our studies demonstrated that the metabolism of iAs in Hs/Hs mice was slower and tissue retention higher than in WT/WT mice. The distribution of As metabolites in urine of Hs/Hs mice, with major fractions of tAs found in the forms of iAs and MAs, was also consistent with a lower capacity for methylation of iAs and resembled patterns of iAs metabolism reported in human studies. Overall, the expression of human AS3MT in Hs/Hs mice promoted tissue retention of iAs and MAs. Because the trivalent form of MAs () is the most toxic metabolite formed in the pathway for iAs methylation (Vahter and Concha 2001), we expect Hs/Hs mice to be more susceptible to iAs toxicity than WT/WT mice. However, no obvious signs of toxicity (e.g., severe weight loss or gross pathology in tissues) were observed in either male of female Hs/Hs mice exposed to As for 4 wk in the present study.
The urine is thought to be the main route for excretion of the metabolites of iAs (ATSDR 2007). Little is known about As excretion and speciation in feces. In humanized Hs/Hs mice, fecal excretion appeared to partially compensate for the inefficient urinary excretion. Hs/Hs mice excreted a larger fraction of the dose of iAs in feces than did WT mice. iAs and MAs accounted for most of tAs in feces of Hs/Hs mice, whereas iAs was the predominant As species in feces of WT/WT mice. For both strains, DMAs was a minor contributor to fecal As. The source of fecal As is iAs that was not absorbed in the gastrointestinal tract and arsenicals produced from iAs by intestinal microflora, as well as absorbed As that is excreted in bile. Given the time frame of sample collection in this study and the short () gastrointestinal transit time in mice (Padmanabhan et al. 2013), most fecal As was likely derived from biliary clearance. In mice treated with iAs, MAs (as ) has been shown to be the predominant form of As in bile (Csanaky and Gregus 2002). Our results suggest that the humanized AS3MT genotype that favored MAs production is associated with increased biliary excretion of this metabolite.
Laboratory studies using mice showed that gut bacteria can metabolize iAs to its methylated metabolites and thus affect the speciation of As in feces (Lu et al. 2014b) as well as iAs toxicity (Coryell et al. 2018). Exposure to iAs has also been shown to modify the gut microbiome (Lu et al. 2014a). We cannot at this time rule out that humanization of the Borcs7/As3mt locus or the changes in iAs metabolism associated with the humanization of this locus resulted in modification of gut microbiome in Hs/Hs mice and thus contributed to the differences between WT/WT and Hs/Hs mice in the distribution of As species in feces.
Results of population studies have suggested that women are more efficient than men in metabolizing iAs (Lindberg et al. 2007, 2008a, 2008b). This suggestion has been based entirely on the fact that %DMAs in the urine of women exposed to iAs is somewhat higher than in the urine of men. Our results show that sex played a significant role in the metabolism of iAs in Hs/Hs mice. Female mice excreted a greater fraction of the single iAs dose in urine than did male mice. After subchronic exposure to iAs in drinking water, livers of female mice contained higher concentrations of tAs than livers of male mice, although no differences were found in tAs concentration in urine collected before sacrifice. In kidneys, %MAs was significantly higher in males as compared with females. No statistically significant differences between Hs/Hs males and females were found in %DMAs in urine after the single dose of iAs or after iAs exposure in drinking water. However, %MAs was higher in the urine of males as compared with females. Although these results suggest a role of sex in iAs metabolism in the Hs/Hs mice, the implication for the overall capacity of males vs. females to metabolize iAs remains unclear. The use of a single dose of iAs in the acute and single iAs concentration in the subchronic studies represents a major limitation. To properly characterize the role of sex in iAs metabolism by Hs/Hs mice, multiple doses and concentrations of iAs will have to be used in future studies.
Genome-wide association studies have linked SNPs in the genomic region spanning the AS3MT/BORCS7 locus to risk of schizophrenia. One study has shown that the schizophrenia risk alleles are associated with an increased expression of BORCS7 and of a previously uncharacterized AS3MT isoform (), which is more abundant in individuals with schizophrenia than in controls (Duarte et al. 2016; Li et al. 2016). Expression of this isoform, which reportedly lacks iAs methyltransferase activity (Li et al. 2016), was found to be associated with a polymorphism in variable number tandem repeat in the first exon of AS3MT (Li et al. 2016). Expression of was also reported to be about 2-fold higher in the brain than in somatic tissues (Li et al. 2016). Here we show that Hs/Hs mice express in multiple tissues, with the highest ratio of to the full-length AS3MT in neural tissues, including brain and spinal cord. Thus, Hs/Hs mice may represent a novel model to study schizophrenia in laboratory settings.
Conclusions
We have created a humanized mouse strain in which AS3MT expression and iAs metabolism resemble those described in humans. The rate and patterns of iAs metabolism indicate that this strain will be more sensitive to adverse effects of iAs exposure than are existing mouse strains. Notably, the human-specific isoform, which has been linked to risk of schizophrenia in clinical studies, is highly expressed in the brain of Hs/Hs mice. The human AS3MT and BORCS7 genes incorporated into the Hs/Hs genome by syntenic replacement can be modified in future studies using CRISPR/cas9 to introduce SNPs associated with altered iAs metabolism or expression. Thus, the Hs/Hs mouse strain represents a novel and promising model for laboratory studies in the field of iAs toxicology as well as for studies exploring mechanisms underlying the development of schizophrenia.
Supplementary Material
Acknowledgments
This work was funded by National Institutes of Health (NIH) grants R21ES029050 to M.S. and B.H.K. and R01ES022697 to M.S. and by the UNC Superfund Program grant P42ES031007. Additional support was provided by NIH grant DK 056350 to the UNC NORC. Blastocyst injections were performed by the UNC Chapel Hill Animal Models Core, which is supported in part by Cancer Center Core Support grant P30 CA016086 to the UNC Lineberger Comprehensive Cancer Center.
References
- Abhyankar LN, Jones MR, Guallar E, Navas-Acien A. 2012. Arsenic exposure and hypertension: a systematic review. Environ Health Perspect 120(4):494–500, PMID: 22138666, 10.1289/ehp.1103988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahsan H, Chen Y, Kibriya MG, Slavkovich V, Parvez F, Jasmine F, et al. . 2007. Arsenic metabolism, genetic susceptibility, and risk of premalignant skin lesions in Bangladesh. Cancer Epidemiol Biomarkers Prev 16(6):1270–1278, PMID: 17548696, 10.1158/1055-9965.EPI-06-0676. [DOI] [PubMed] [Google Scholar]
- Apata M, Arriaza B, Llop E, Moraga M. 2017. Human adaptation to arsenic in Andean populations of the Atacama Desert. Am J Phys Anthropol 163(1):192–199, PMID: 28206677, 10.1002/ajpa.23193. [DOI] [PubMed] [Google Scholar]
- ATSDR (Agency for Toxic Substances and Disease Registry). 2007. Toxicological profile for arsenic. Atlanta, GA: U.S. DHHS, Public Health Service; https://www.atsdr.cdc.gov/toxprofiles/tp.asp?id=22&tid=3 [accessed 21 July 2020]. [Google Scholar]
- Caito S, Aschner M. 2015. Neurotoxicity of metals. Handb Clin Neurol 131:169–189, PMID: 26563789, 10.1016/B978-0-444-62627-1.00011-1. [DOI] [PubMed] [Google Scholar]
- Chandler KJ, Chandler RL, Broeckelmann EM, Hou Y, Southard-Smith EM, Mortlock DP. 2007. Relevance of BAC transgene copy number in mice: transgene copy number variation across multiple transgenic lines and correlations with transgene integrity and expression. Mamm Genome 18(10):693–708, PMID: 17882484, 10.1007/s00335-007-9056-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Ma X, Gonzalez FJ. 2011. Pregnane X receptor- and CYP3A4-humanized mouse models and their applications. Br J Pharmacol 163(3):461–468, PMID: 21091656, 10.1111/j.1476-5381.2010.01129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coryell M, McAlpine M, Pinkham NV, McDermott TR, Walk ST. 2018. The gut microbiome is required for full protection against acute arsenic toxicity in mouse models. Nat Commun 9(1):5424, PMID: 30575732, 10.1038/s41467-018-07803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csanaky I, Gregus Z. 2002. Species variations in the biliary and urinary excretion of arsenate, arsenite and their metabolites. Comp Biochem Physiol C Toxicol Pharmacol 131(3):355–365, PMID: 11912060, 10.1016/s1532-0456(02)00018-2. [DOI] [PubMed] [Google Scholar]
- Cubadda F, Jackson BP, Cottingham KL, Van Horne YO, Kurzius-Spencer M. 2017. Human exposure to dietary inorganic arsenic and other arsenic species: state of knowledge, gaps and uncertainties. Sci Total Environ 579:1228–1239, PMID: 27914647, 10.1016/j.scitotenv.2016.11.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currier JM, Svoboda M, de Moraes DP, Matousek T, Dĕdina J, Stýblo M. 2011. Direct analysis of methylated trivalent arsenicals in mouse liver by hydride generation-cryotrapping-atomic absorption spectrometry. Chem Res Toxicol 24(4):478–480, PMID: 21361335, 10.1021/tx200060c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Saunders RJ, Drobná Z, Walton FS, Xun P, Thomas DJ, et al. . 2012. Methylation of arsenic by recombinant human wild-type arsenic (+3 oxidation state) methyltransferase and its methionine 287 threonine (M287T) polymorph: role of glutathione. Toxicol Appl Pharmacol 264(1):121–130, PMID: 22868225, 10.1016/j.taap.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douillet C, Huang MC, Saunders RJ, Dover EN, Zhang C, Stýblo M. 2017. Knockout of arsenic (+3 oxidation state) methyltransferase is associated with adverse metabolic phenotype in mice: the role of sex and arsenic exposure. Arch Toxicol 91(7):2617–2627, PMID: 27847981, 10.1007/s00204-016-1890-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobná Z, Naranmandura H, Kubachka KM, Edwards BC, Herbin-Davis K, Styblo M, et al. . 2009. Disruption of the arsenic (+3 oxidation state) methyltransferase gene in the mouse alters the phenotype for methylation of arsenic and affects distribution and retention of orally administered arsenate. Chem Res Toxicol 22(10):1713–1720, PMID: 19691357, 10.1021/tx900179r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte RRR, Troakes C, Nolan M, Srivastava DP, Murray RM, Bray NJ. 2016. Genome-wide significant schizophrenia risk variation on chromosome 10q24 is associated with altered cis-regulation of BORCS7, AS3MT, and NT5C2 in the human brain. Am J Med Genet B Neuropsychiatr Genet 171(6):806–814, PMID: 27004590, 10.1002/ajmg.b.32445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Masri HA, Kenyon EM. 2008. Development of a human physiologically based pharmacokinetic (PBPK) model for inorganic arsenic and its mono- and di-methylated metabolites. J Pharmacokinet Pharmacodyn 35(1):31–68, PMID: 17943421, 10.1007/s10928-007-9075-z. [DOI] [PubMed] [Google Scholar]
- Hernández-Zavala A, Matoušek T, Drobná Z, Paul DS, Walton F, Adair BM, et al. . 2008. Speciation of arsenic in biological matrices by automated hydride generation-cryotrapping-atomic absorption spectrometry with multiple microflame quartz tube atomizer (multiatomizer). J Anal At Spectrom 23:342–351, PMID: 18677417, 10.1039/b706144g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MC, Douillet C, Dover EN, Stýblo M. 2018. Prenatal arsenic exposure and dietary folate and methylcobalamin supplementation alter the metabolic phenotype of C57BL/6J mice in a sex-specific manner. Arch Toxicol 92(6):1925–1937, PMID: 29721587, 10.1007/s00204-018-2206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudgens EE, Drobna Z, He B, Le XC, Styblo M, Rogers J, et al. . 2016. Biological and behavioral factors modify urinary arsenic metabolic profiles in a U.S. population. Environ Health 15(1):62, PMID: 27230915, 10.1186/s12940-016-0144-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MF, Edwards BC, Herbin-Davis KM, Saunders J, Styblo M, Thomas DJ. 2010. Arsenic (+3 oxidation state) methyltransferase genotype affects steady-state distribution and clearance of arsenic in arsenate-treated mice. Toxicol Appl Pharmacol 249(3):217–223, PMID: 20887743, 10.1016/j.taap.2010.09.017. [DOI] [PubMed] [Google Scholar]
- IARC (International Agency for Research on Cancer). 2004. Some Drinking-Water Disinfectants and Contaminants, Including Arsenic. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, vol 84 Lyon, France: IARC, 269–477. [PMC free article] [PubMed] [Google Scholar]
- Koller BH, Hagemann LJ, Doetschman T, Hagaman JR, Huang S, Williams PJ, et al. . 1989. Germ-line transmission of a planned alteration made in a hypoxanthine phosphoribosyltransferase gene by homologous recombination in embryonic stem cells. Proc Natl Acad Sci USA 86(22):8927–8931, PMID: 2573070, 10.1073/pnas.86.22.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Jaffe AE, Straub RE, Tao R, Shin JH, Wang Y, et al. . 2016. A human-specific AS3MT isoform and BORCS7 are molecular risk factors in the 10q24.32 schizophrenia-associated locus. Nat Med 22(6):649–656, PMID: 27158905, 10.1038/nm.4096. [DOI] [PubMed] [Google Scholar]
- Lin S, Shi Q, Nix FB, Styblo M, Beck MA, Herbin-Davis KM, et al. . 2002. A novel S-adenosyl-l-methionine:arsenic(III) methyltransferase from rat liver cytosol. J Biol Chem 277(13):10795–10803, PMID: 11790780, 10.1074/jbc.M110246200. [DOI] [PubMed] [Google Scholar]
- Lindberg A-L, Ekström E-C, Nermell B, Rahman M, Lönnerdal B, Persson L-A, et al. . 2008a. Gender and age differences in the metabolism of inorganic arsenic in a highly exposed population in Bangladesh. Environ Res 106(1):110–120, PMID: 17900557, 10.1016/j.envres.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Lindberg A-L, Kumar R, Goessler W, Thirumaran R, Gurzau E, Koppova K, et al. . 2007. Metabolism of low-dose inorganic arsenic in a central European population: influence of sex and genetic polymorphisms. Environ Health Perspect 115(7):1081–1086, PMID: 17637926, 10.1289/ehp.10026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg A-L, Rahman M, Persson L-A, Vahter M. 2008b. The risk of arsenic induced skin lesions in Bangladeshi men and women is affected by arsenic metabolism and the age at first exposure. Toxicol Appl Pharmacol 230(1):9–16, PMID: 18336856, 10.1016/j.taap.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Longenecker G, Kulkarni AB. 2009. Generation of gene knockout mice by ES cell microinjection. Curr Protoc Cell Biol 44(1):19.14.1–19.14.36, PMID: 19731226, 10.1002/0471143030.cb1914s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Abo RP, Schlieper KA, Graffam ME, Levine S, Wishnok JS, et al. . 2014a. Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: an integrated metagenomics and metabolomics analysis. Environ Health Perspect 122(3):284–291, PMID: 24413286, 10.1289/ehp.1307429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Mahbub R, Cable PH, Ru H, Parry NMA, Bodnar WM, et al. . 2014b. Gut microbiome phenotypes driven by host genetics affect arsenic metabolism. Chem Res Toxicol 27(2):172–174, PMID: 24490651, 10.1021/tx400454z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Wang H, Li X-F, Arnold LL, Cohen SM, Le XC. 2007. Binding of dimethylarsinous acid to cys-13α of rat hemoglobin is responsible for the retention of arsenic in rat blood. Chem Res Toxicol 20(1):27–37, PMID: 17226924, 10.1021/tx060195+. [DOI] [PubMed] [Google Scholar]
- Lu X, Wang L, Lin X, Huang J, Gu CC, He M, et al. . 2015. Genome-wide association study in Chinese identifies novel loci for blood pressure and hypertension. Hum Mol Genet 24(3):865–874, PMID: 25249183, 10.1093/hmg/ddu478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maull EA, Ahsan H, Edwards J, Longnecker MP, Navas-Acien A, Pi J, et al. . 2012. Evaluation of the association between arsenic and diabetes: a National Toxicology Program workshop report. Environ Health Perspect 120(12):1658–1670, PMID: 22889723, 10.1289/ehp.1104579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder DNG. 2005. Effect of chronic intake of arsenic-contaminated water on liver. Toxicol Appl Pharmacol 206(2):169–175, PMID: 15967205, 10.1016/j.taap.2004.08.025. [DOI] [PubMed] [Google Scholar]
- Moon K, Guallar E, Navas-Acien A. 2012. Arsenic exposure and cardiovascular disease: an updated systematic review. Curr Atheroscler Rep 14(6):542–555, PMID: 22968315, 10.1007/s11883-012-0280-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai HY, Motohashi H, Ohneda O, Suzuki N, Nagano M, Yamamoto M. 2006. Transgene insertion in proximity to the c-myb gene disrupts erythroid-megakaryocytic lineage bifurcation. Mol Cell Biol 26(21):7953–7965, PMID: 16940183, 10.1128/MCB.00718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murko M, Elek B, Styblo M, Thomas DJ, Francesconi KA. 2018. Dose and diet—sources of arsenic intake in mouse in utero exposure scenarios. Chem Res Toxicol 31(2):156–164, PMID: 29244955, 10.1021/acs.chemrestox.7b00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naujokas MF, Anderson B, Ahsan H, Aposhian HV, Graziano JH, Thompson C, et al. . 2013. The broad scope of health effects from chronic arsenic exposure: update on a worldwide public health problem. Environ Health Perspect 121(3):295–302, PMID: 23458756, 10.1289/ehp.1205875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan P, Grosse J, Asad ABMA, Radda GK, Golay X. 2013. Gastrointestinal transit measurements in mice with 99mTc-DTPA-labeled activated charcoal using NanoSPECT-CT. EJNMMI Res 3(1):60, PMID: 23915679, 10.1186/2191-219X-3-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez F, Wasserman GA, Factor-Litvak P, Liu X, Slavkovich V, Siddique AB, et al. . 2011. Arsenic exposure and motor function among children in Bangladesh. Environ Health Perspect 119(11):1665–1670, PMID: 21742576, 10.1289/ehp.1103548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul DS, Hernández-Zavala A, Walton FS, Adair BM, Dedina J, Matousek T, et al. . 2007. Examination of the effects of arsenic on glucose homeostasis in cell culture and animal studies: development of a mouse model for arsenic-induced diabetes. Toxicol Appl Pharmacol 222(3):305–314, PMID: 17336358, 10.1016/j.taap.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce BL, Kibriya MG, Tong L, Jasmine F, Argos M, Roy S, et al. . 2012. Genome-wide association study identifies chromosome 10q24.32 variants associated with arsenic metabolism and toxicity phenotypes in Bangladesh. PLoS Genet 8(2):e1002522, PMID: 22383894, 10.1371/journal.pgen.1002522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce BL, Tong L, Argos M, Gao J, Farzana J, Roy S, et al. . 2013. Arsenic metabolism efficiency has a causal role in arsenic toxicity: Mendelian randomization and gene-environment interaction. Int J Epidemiol 42(6):1862–1871, PMID: 24536095, 10.1093/ije/dyt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash T, Sharma VK, Adati N, Ozawa R, Kumar N, Nishida Y, et al. . 2010. Expression of conjoined genes: another mechanism for gene regulation in eukaryotes. PLoS One 5(10):e13284, PMID: 20967262, 10.1371/journal.pone.0013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J, Schindler C, Jia R, Jarnik M, Backlund P, Bonifacino JS. 2015. BORC, a multisubunit complex that regulates lysosome positioning. Dev Cell 33(2):176–188, PMID: 25898167, 10.1016/j.devcel.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan P-L, Sauzade M, Brouzes E. 2018. dPCR: a technology review. Sensors (Basel) 18(4):1271, PMID: 29677144, 10.3390/s18041271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, O’Dushlaine C, Chambert K, Moran JL, Kähler AK, Akterin S, et al. . 2013. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet 45(10):1150–1159, PMID: 23974872, 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez TR, Perzanowski M, Graziano JH. 2016. Inorganic arsenic and respiratory health, from early life exposure to sex-specific effects: a systematic review. Environ Res 147:537–555, PMID: 26891939, 10.1016/j.envres.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saquib N, Saquib J, Ahmed T, Khanam MA, Cullen MR. 2012. Cardiovascular diseases and type 2 diabetes in Bangladesh: a systematic review and meta-analysis of studies between 1995 and 2010. BMC Public Health 12:434, PMID: 22694854, 10.1186/1471-2458-12-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. 2014. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511(7510):421–427, PMID: 25056061, 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- States JC, Srivastava S, Chen Y, Barchowsky A. 2009. Arsenic and cardiovascular disease. Toxicol Sci 107(2):312–323, PMID: 19015167, 10.1093/toxsci/kfn236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stýblo M, Douillet C, Bangma J, Eaves LA, de Villena FP-M, Fry R. 2019. Differential metabolism of inorganic arsenic in mice from genetically diverse Collaborative Cross strains. Arch Toxicol 93(10):2811–2822, PMID: 31493028, 10.1007/s00204-019-02559-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Human Protein Atlas. 2020. The adrenal gland-specific proteome. https://www.proteinatlas.org/humanproteome/tissue/adrenal+gland [accessed 21 July 2020].
- Thomas DJ, Li J, Waters SB, Xing W, Adair BM, Drobna Z, et al. . 2007. Arsenic (+3 oxidation state) methyltransferase and the methylation of arsenicals. Exp Biol Med (Maywood) 232(1):3–13, PMID: 17202581. [PMC free article] [PubMed] [Google Scholar]
- Thomas DJ, Styblo M, Lin S. 2001. The cellular metabolism and systemic toxicity of arsenic. Toxicol Appl Pharmacol 176(2):127–144, PMID: 11601889, 10.1006/taap.2001.9258. [DOI] [PubMed] [Google Scholar]
- UCSC (University of California Santa Cruz). 2013. Genome Browser Gateway. Santa Cruz, CA: University of California Santa Cruz, Genomics Institute; https://genome.ucsc.edu/cgi-bin/hgGateway?hgsid=834004679_0TKSQmfCdDYnn1sBDBgvD88GKaDR [accessed 21 July 2020]. [Google Scholar]
- Vahter M. 1999. Methylation of inorganic arsenic in different mammalian species and population groups. Sci Prog 82(1):69–88, PMID: 10445007, 10.1177/003685049908200104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahter M. 2000. Genetic polymorphism in the biotransformation of inorganic arsenic and its role in toxicity. Toxicol Lett 112–113:209–217, PMID: 10720733, 10.1016/S0378-4274(99)00271-4. [DOI] [PubMed] [Google Scholar]
- Vahter M, Concha G. 2001. Role of metabolism in arsenic toxicity. Pharmacol Toxicol 89(1):1–5, PMID: 11484904, 10.1111/j.1600-0773.2001.890101.x. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Hirano S. 2013. Metabolism of arsenic and its toxicological relevance. Arch Toxicol 87(6):969–979, PMID: 22811022, 10.1007/s00204-012-0904-5. [DOI] [PubMed] [Google Scholar]
- Waters SB, Devesa V, Fricke MW, Creed JT, Stýblo M, Thomas DJ. 2004. Glutathione modulates recombinant rat arsenic (+3 oxidation state) methyltransferase-catalyzed formation of trimethylarsine oxide and trimethylarsine. Chem Res Toxicol 17(12):1621–1629, PMID: 15606138, 10.1021/tx0497853. [DOI] [PubMed] [Google Scholar]
- Wood TC, Salavagionne OE, Mukherjee B, Wang L, Klumpp AF, Thomae BA, et al. . 2006. Human arsenic methyltransferase (AS3MT) pharmacogenetics: gene resequencing and functional genomics studies. J Biol Chem 281(11):7364–7373, PMID: 16407288, 10.1074/jbc.M512227200. [DOI] [PubMed] [Google Scholar]
- Yueh MF, Mellon PL, Tukey RH. 2011. Inhibition of human UGT2B7 gene expression in transgenic mice by the constitutive androstane receptor. Mol Pharmacol 79(6):1053–1060, PMID: 21415305, 10.1124/mol.110.070649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.