Abstract
Objective:
22q11.2 deletion syndrome (22q11DS) is among the strongest known genetic risk factors for schizophrenia. Prior studies report variable alterations in subcortical brain structures in 22q11DS. To better characterize subcortical alterations in 22q11DS, including modulating effects of clinical and genetic heterogeneity, we studied a large multicenter neuroimaging cohort from the ENIGMA 22q11.2 Deletion Syndrome Working Group.
Method:
Subcortical structures were measured using harmonized protocols for gross volume and subcortical shape morphometry in 533 individuals with 22q11DS and 330 matched healthy controls (HC) (age: 6–56 years, 49% female).
Results:
Subjects with 22q11DS showed lower intracranial volume (ICV), thalamus, putamen, hippocampus, and amygdala volumes, but greater lateral ventricle, caudate, and accumbens volumes, compared to HC (Cohen’s d = −0.90 – 0.93). Shape analysis revealed complex differences in 22q11DS across all structures. The larger A-D deletion was associated with more extensive shape alterations compared to the smaller A-B deletion. 22q11DS subjects with psychosis (22q+Psy) showed lower ICV, hippocampus, amygdala, and thalamic volumes (Cohen’s d = −0.91 – 0.53) compared to 22q11DS subjects without psychosis. Shape analysis revealed lower thickness and surface area across subregions of these structures. Compared to subcortical findings from other neuropsychiatric disorders studied by the ENIGMA Consortium, there was significant convergence between 22q+Psy and schizophrenia, bipolar, major depression, and obsessive-compulsive disorders.
Conclusions:
Here, in the largest neuroimaging study of 22q11DS, we found widespread alterations to subcortical brain structures, which were impacted by deletion size and psychotic illness. Findings indicate significant overlap between 22q11DS-associated psychosis, idiopathic schizophrenia, and other severe neuropsychiatric illnesses.
Introduction
22q11.2 deletion syndrome (22q11DS), also known as DiGeorge or Velocardiofacial syndrome, is a multisystem disorder resulting from a hemizygous microdeletion on the long arm of chromosome 22, affecting multiple genes involved in neurodevelopment. 22q11DS results in craniofacial, cardiac, and immune system abnormalities, as well as neurocognitive deficits (1). Up to 1 in 4 develop psychotic illness in adolescence or early adulthood, making it one of the strongest known genetic risk factors for schizophrenia (2). There is also considerable psychiatric comorbidity in 22q11DS, as elevated rates of attention deficit hyperactivity disorder (ADHD), anxiety or mood disorders, and/or autism spectrum disorders (ASD) are also observed (2; 3; 4). Taken together, 22q11DS offers a genetically homogeneous framework to study how highly penetrant genetic variants disrupt neurobiological pathways contributing to developmental neuropsychiatric disorders. This genetics-first approach may result in greater power to detect biomarkers of psychiatric illness by providing larger effect sizes than those associated with common genetic variation.
22q11DS-associated psychotic disorder has a similar clinical presentation to idiopathic schizophrenia (5). In the largest coordinated analysis of subcortical brain structure in schizophrenia to date, smaller hippocampal volume was the strongest effect (6). However, the extent to which variations in underlying subcortical structures overlap between 22q11DS and idiopathic schizophrenia is not well understood, largely due to the lack of large, well-characterized cohorts. Elucidating concordant or divergent aspects of subcortical morphometry between 22q11DS-associated psychosis and idiopathic schizophrenia can shed light on brain mechanisms underlying expression of psychotic illness. Further, it will inform whether such subcortical alterations reflect a specific neuroanatomic signature of psychosis, or are characteristic of other neuropsychiatric disorders.
Mouse models of the 22q11.2 deletion have shown disrupted neurogenesis (7) and altered brain development along the anterior-posterior axis (8). Consistent with this, subcortical volume reductions are reported in human 22q11.2 deletion carriers (9; 10; 11), with greater volumetric reductions in more posterior brain regions (12) and thinning in midline structures (13). Even so, most studies have examined small samples, typically ascertained from a single site, limiting power to detect subtle brain abnormalities and determine brain signature consistency across cohorts. Answering these questions definitively requires larger samples employing similar neuroimaging processing and analysis techniques.
Moreover, while most neuroimaging studies examine regional volumes, haploinsufficiency for 22q11.2 genes may differentially impact subregions or subfields of subcortical structures (11). High-resolution shape analysis has been used to map fine-grained subcortical alterations in Alzheimer’s disease (14), and multiple neuropsychiatric disorders (15; 16), offering insights into differential impact on subcompartments or subfields with known structural and functional connectivity patterns. Our recent study of 22q11DS cortical structure (17) revealed distinct disruptions of cortical thickness and surface area (SA), measures linked to known and dissociable developmental determinants (18; 19).
The size of the 22q11.2 microdeletion may also be a source of heterogeneity. Recently, we found smaller deletions were associated with higher IQ and increased cortical SA in 22q11DS (17; 20), but whether these effects extend to subcortical brain morphometry is unknown.
To address the limitations of smaller, single site studies of 22q11DS, we performed a coordinated analysis of the largest MRI dataset to date, ascertained by the ENIGMA 22q11.2 Deletion Syndrome Working Group. To map abnormalities at a finer scale than is possible with regional volumetry, we used a surface-based mapping approach, which is sensitive to subtle variations in subcortical morphometry, thus offering insight into the disruption of known subcompartments or subfields (21; 22).
We assessed overall subcortical brain volumes and pointwise shape differences across the entire surface of each structure, to answer three main questions:
What is the spatial distribution of subcortical differences between 22q11DS and healthy controls (HC)?
Do differences in subcortical structure depend on deletion size?
Do subcortical differences exist between 22q11DS subjects with a history of psychosis (22q+Psy) versus those without (22q-Psy)? And do those 22q+Psy-associated subcortical patterns overlap with those found in idiopathic schizophrenia and other neuropsychiatric disorders?
Methods
Data Sample
A total of 863 unrelated subjects (22q11DS = 533, HC = 330) from 11 study sites were included. Participant demographics are listed in Table 1 (by site in Supplemental Table S1). All individual participating research studies had obtained approval from their local ethics committees and/or institutional review boards, and written informed consent (and/or assent for minors) was obtained for all participants. Comparison of the two most common deletion subtypes (A-D vs. A-B) was conducted on matched samples: 106 22q11DS subjects with A-D deletions, 23 22q11DS subjects with A-B deletions, and 86 HC (Supplemental Methods; Supplemental Table S2). Psychosis diagnosis was assessed by structured clinical interview at each study site, with diagnoses validated across sites using a consensus procedure (22). Sixty-four subjects with 22q11DS with a psychotic disorder diagnosis (22q+Psy) were compared to 64 subjects with no history of psychosis (22q-Psy) by matching +/−Psy participants within each site by sex and the nearest possible age (Supplemental Table S3). Participant ascertainment and inclusion/exclusion is further described in Supplemental Methods and Table S4.
Table 1:
Combined study demographics, including % with history of psychotic disorder, frequency of deletion subtypes (3Mb A-D and 1.5Mb A-B are most common), and common psychotropic medications (at the time of scan). Other deletions include nested A-C, B-D, C-E, D-F, and D-G breakpoints. Groups did not differ in terms of age or sex distribution, but IQ was significantly lower in 22q11DS subjects vs. HC (p = 1.7 x 10−136).
| Healthy Control (HC) | (sd) / (%) | 22q11DS | (sd) / (%) | |
|---|---|---|---|---|
| N | 330 | 533 | ||
| Age (mean (sd)) | 18.14 | −9.24 | 17.85 | −8.6 |
| IQ (mean (sd)) | 110.64 | −15.35 | 74.95 | −12.53 |
| Sex = Female (%) | 148 | −44.88 | 275 | −51.6 |
| Psychotic_Disorder (%) | 0 | 0 | 73 | −13.8 |
| Deletion_Type (%) | ||||
| A-B | 0 | 0 | 28 | −8 |
| A-D | 0 | 0 | 311 | −88.6 |
| Other | 0 | 0 | 12 | −3.5 |
| Current Medication (%) | ||||
| Typical Antipsychotic | 0 | 0 | 15 | −3 |
| Atypical Antipsychotic | 0 | 0 | 72 | −14.5 |
| Lithium | 0 | 0 | 4 | −0.8 |
| Anticonvulsant | 1 | −0.4 | 31 | −6.3 |
| Antidepressant | 5 | −1.8 | 95 | −19.2 |
| Psychostimulant | 5 | −1.8 | 65 | −13.1 |
Image Acquisition and Processing
T1-weighted brain MRI data were collected from 11 sites (see Supplemental Table S5 for acquisition parameters). Sites with more than one scanner or acquisition protocol were treated as separate sites in the analysis. UCLA, UC Davis, and Toronto each acquired data on two scanners, leading to a total of 14 scanning sites. MRI images were centralized on a secure server at the University of Southern California Imaging Genetics Center (IGC) for processing and analysis.
Subcortical Segmentation
All T1-weighted scans were segmented using the FreeSurfer software, version 5.3.0 (23) to derive volumes for 8 bilateral regions of interest (ROIs): lateral ventricle, nucleus accumbens, amygdala, caudate, hippocampus, putamen, pallidum, and thalamus (16 total structures per scan) along with intracranial volume (ICV).
Subcortical Shape Analysis
As subtle and complex variations in local volume may be undetectable by gross volume measures, we applied a novel surface-based parametric mapping technique, the ENIGMA Subcortical Shape Analysis Pipeline (22), to investigate high-definition shape variation within the bilateral subcortical structures described above (14 ROIs excluding the lateral ventricles). We recently applied this technique in a single-site study of reciprocal 22q11.2 CNVs (25).
Briefly, using the subcortical FreeSurfer segmentations as inputs, two measures of shape morphometry were derived across the surface of each ROI. The first, ‘radial distance’ (subsequently referred to as ‘thickness’) is the distance from each surface vertex to a medial curve, and represents a measure of local thickness. The second measure — the logarithm of the Jacobian determinant (Jacobian, or SA dilation/contraction from now on) — is the surface dilation ratio between the template and the individual subject’s structure. The Jacobian can be interpreted as areal dilation or contraction of the ROIs’ surface, where higher Jacobian measures suggest larger local SA.
Both thickness and Jacobian measures were calculated in native space for up to 2,502 homologous points across each of the 14 subcortical shape models to index detailed regional shape differences across subjects (see Supplemental Methods).
Quality Control
Visual quality inspection was performed by a rater trained in neuroanatomy for all volumes and shape models using ENIGMA standardized quality control protocols (see Supplemental Methods).
Statistical Analysis
Primary analyses were conducted using multiple linear regression via the lm function in the R statistical environment, version 3.1.3 (26). The dependent variable was ROI volume for gross volumetric analysis and either thickness or Jacobian for vertex-wise shape analysis. Primary analyses were run on left and right structures separately. The independent variable was the grouping variable of interest (e.g., diagnosis, deletion subtype, or history of psychosis) while adjusting for appropriate covariates.
Basic covariate adjustments included those for age, age2, sex and ICV. Age effects were modeled with both a linear and quadratic term based on model fit (Supplemental Table S6, S7). Sex was included as a covariate, as it was associated with ROI volume (Supplemental Table S8; Supplemental Figure F1), as was ICV (Supplemental Table S9; Supplemental Figure F2). No age-by-sex interactions on ROI volume were detected (Supplemental Table S10). Handedness was largely not associated with ROI volumes and therefore not used as a covariate in follow-up models, in line with our prior large-scale studies of handedness and brain laterality (27) (Supplemental Table S11). As IQ and related measures are consistently found to be associated with brain volume (Supplemental Table S12), IQ was included in secondary analyses.
Medications found to have significant associations with subcortical volume were added as covariates in secondary analyses, and included typical and atypical antipsychotics, anticonvulsants and antidepressants (Supplemental Methods; Supplemental Table S13).
For gross volumetric analyses, Cohen’s d effect size estimates were computed from the t-statistic of the group variable from the regression models (28). To correct for multiple comparisons, a standard false discovery rate (FDR) correction was applied across all ROIs of the 5 main comparisons (22q11DS vs. HC, A-D vs. HC, A-B vs. HC, A-B vs. A-D, and 22q+Psy vs. 22q-Psy) at the conventionally accepted level of 5% (q=0.05) (29). FDR-corrected p-values<0.05 were considered significant. Gross volume results surviving Bonferroni correction (0.05/85 total tests across all 5 main analysis contrasts, p<0.00058) are reported in Supplemental Table S14.
For vertex-wise Jacobian and thickness analyses, the multiple linear regression model was fit at each point across the surface. As these values were calculated in native space (i.e., without scaling the image), ICV was used to adjust for effects of head size. A modified searchlight FDR procedure was applied globally across all structures for each statistical model with FDR-corrected p-values<0.05 considered significant (see Supplemental Methods).
Additional details regarding the main analyses (22q11DS vs. HC, effects of deletion size, 22q+Psy vs. 22q-Psy) can be found in the Supplemental Methods.
Cross-Disorder Comparison: 22q11DS-Psychosis, Idiopathic Schizophrenia, and Other Neuropsychiatric Disorders
To compare the pattern of 22q+Psy to that of other neuropsychiatric disorders, Spearman rank correlations were used to correlate Cohen’s d effect size estimates from the 22q+Psy versus 22q-Psy analyses with comparable case-control analyses from the ENIGMA schizophrenia (30), major depressive disorder (MDD) (31), bipolar disorder (BD) (32), obsessive-compulsive disorder (OCD) (33), autism spectrum disorder (ASD) (34), and attention deficit hyperactivity disorder (ADHD) (35) working group studies. Each of these studies constitutes the largest investigation of subcortical structure to date, and used harmonized processing and quality control protocols (see Supplemental Methods for details).
Results
22q11.2 Deletion vs. Healthy Controls
Gross volumetric analysis revealed significant group differences across the majority of ROIs (14/17), with moderate to large effects (Cohen’s d = −0.90 – 0.93) (Figure 1; Supplemental Table S14). The pattern of effects included significantly lower volumes, in 22q11DS relative to HC for total ICV, thalamus, putamen, hippocampus, and amygdala. In contrast, 22q11DS cases had greater ventricular, caudate and accumbens volumes. Effects were greatest for the lateral ventricles (54.05–60.02% weighted mean difference, larger in 22q11DS) and the hippocampus (10.75–11.85% weighted mean difference, smaller in 22q11DS). These results (in terms of both pattern and effect size) remained essentially unchanged when adjusting for medication (Supplement Table S15), IQ (Supplement Table S16), and when treating scanning site as a random effect (Supplement Table S17). In addition, a significant group-by-age interaction was detected for the bilateral caudate, pallidum and left thalamus. Whereas the left thalamus and bilateral caudate volumes tended to be lower in 22q11DS with increasing age, the pattern was flipped for the pallidum (i.e., greater age-associated decrease in pallidum volume in HC, relative to 22q11DS; Supplemental Table 18; Supplemental Figure F3). No sex-by-diagnosis interactions were detected for any ROI (Supplemental Table S19).
Figure 1:
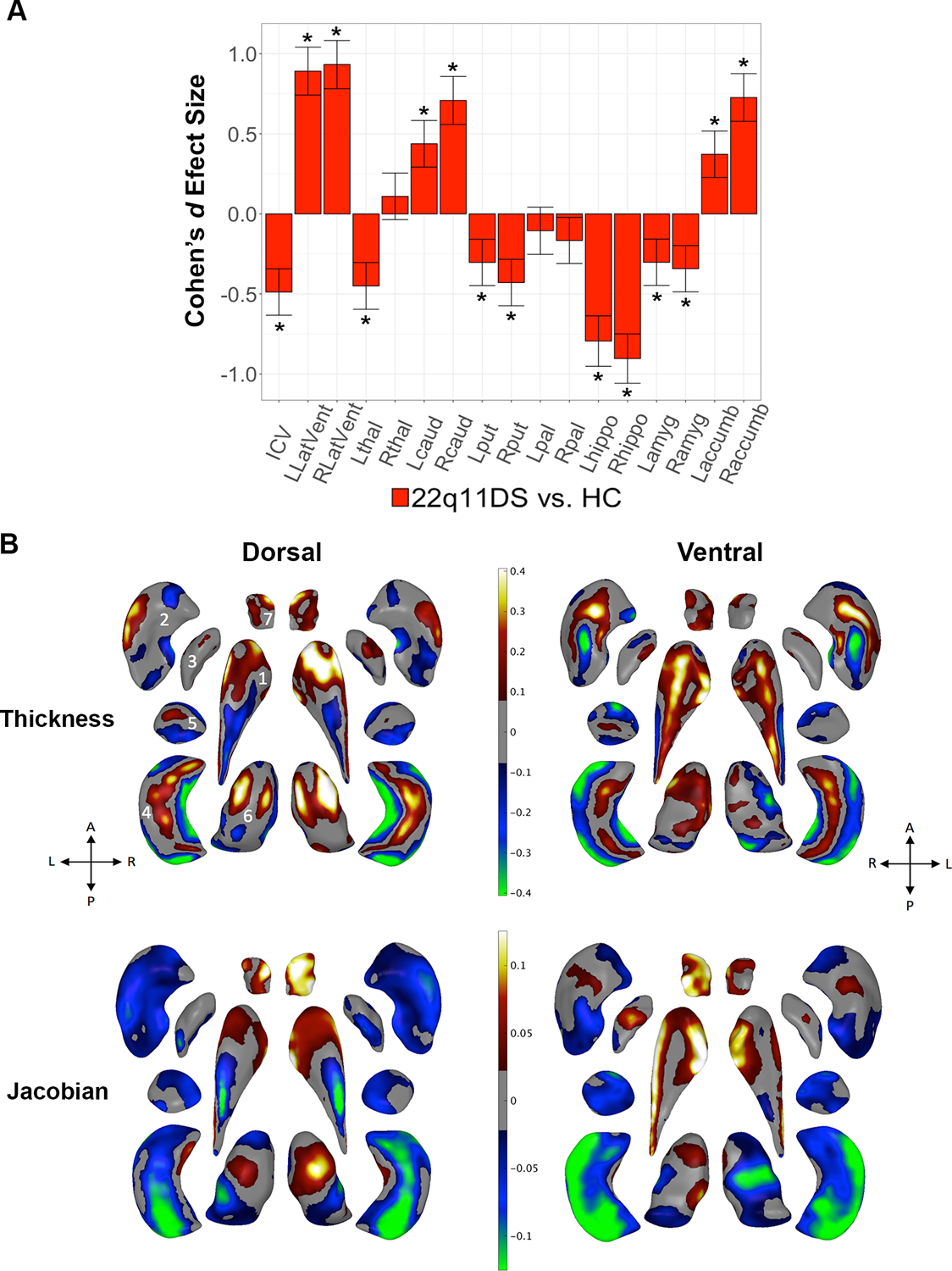
22q11DS vs. HC: Gross volume and shape analysis. A. Cohen’s d effect size (with 95% confidence intervals) plotted for major pairwise gross volumetric comparisons. An asterisk (*) indicates significant group difference after correction for multiple comparisons for 22q11DS versus HC (FDR q<0.05). Positive effect sizes indicate 22q11DS>HC; negative effect sizes indicate HC>22q11DS. Models were adjusted for age, age2, sex, ICV, and scan site. Full statistical model outputs including standard error, coefficient estimates, p-values, and % difference may be found in Supplemental Table S14. Abbreviations: L/R, left/right; LatVent, lateral ventricle; thal, thalamus; caud, caudate; put, putamen; pal, pallidum; hippo, hippocampus; amyg, amygdala; accumb, accumbens; ICV, intracranial volume. B. Shape analysis with regression coefficients plotted in regions passing correction for multiple comparisons (FDR q<0.05). Blue/green colors indicate negative coefficients, or regions of lower thickness (i.e., local radial distance) or Jacobian (i.e., local surface area contraction) measures in 22q11DS versus HC. Red/yellow colors indicate positive coefficients, or regions of greater thickness or Jacobian values in 22q11DS versus HC. The top row includes thickness results; the bottom row includes Jacobian surface area results. Dorsal and ventral views of the structures are provided: A, anterior; P, posterior; L, left; R, right. 1. Caudate; 2. Putamen; 3. Globus Pallidus; 4. Hippocampus; 5. Amygdala; 6. Thalamus; 7. Nucleus Accumbens. Gray regions indicate areas of no significant difference after correction for multiple comparisons.
Subcortical shape analysis revealed complex group differences, involving subregions with both higher and lower thickness and Jacobian values in 22q11DS relative to HC (Figure 1). In particular, greater local thickness was observed in the head of the caudate, thalamus, and dorsal/ventral hippocampal regions, while the caudate body and lateral/medial hippocampal subregions were thinner. The Jacobian analysis revealed SA contraction across large portions of the putamen, amygdala, and hippocampus, and dilation across anterior/lateral regions of the caudate and most of the nucleus accumbens. These effects were robust to adjustments for medication and ROI volume (Supplemental Figure F4).
Effects of Deletion Size
ANCOVA results indicated significant differences between gross volumes across A-D, A-B, and HC matched samples (Supplemental Table S20). While no gross volume differences between 22q11DS subjects with A-B versus A-D deletions surpassed multiple comparison correction (Figure 2; Supplemental Table S14), shape analysis revealed that subjects with A-B deletions had higher local SA (higher Jacobian measures) in the hippocampus, thalamus, and putamen, with lower caudate and accumbens thickness/Jacobian measures, compared to those with A-D deletions (Figure 2). The hippocampus showed complex subregional thickness effects, with thicker medial/lateral aspects and thinner dorsal/ventral regions in A-B versus A-D. These results remained stable when adjusting for ROI and medication (Supplemental Figure F7). Comparisons of both deletion types versus HC are detailed in Figure 2 and the Supplemental Results and Discussion Sections A and B.
Figure 2:
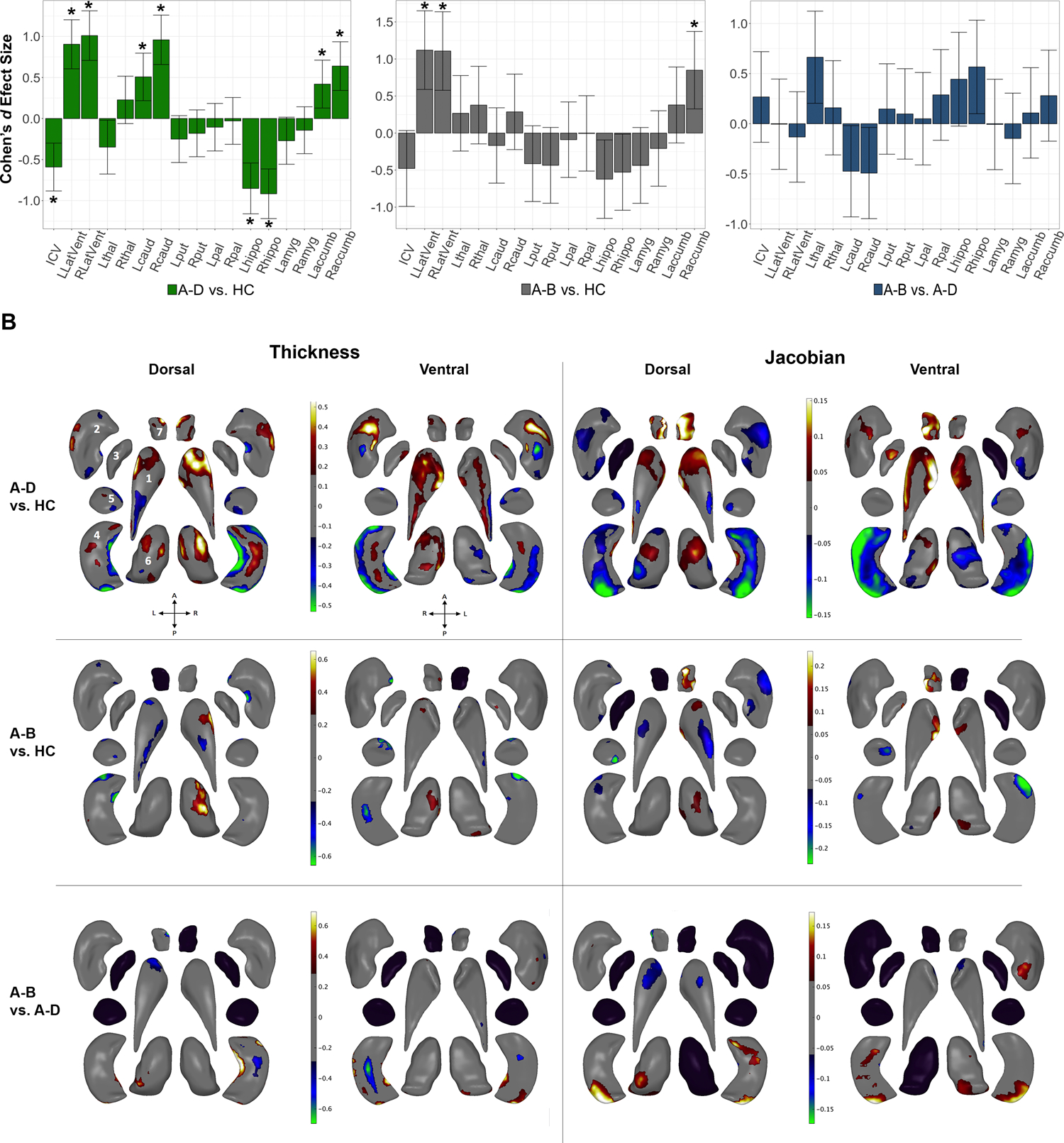
Effects of deletion size: Gross volume and shape analysis. A. Cohen’s d effect size (with 95% confidence intervals) plotted for major pairwise gross volumetric comparisons (Left panel: A-D vs. HC, middle panel: A-B vs. HC, right panel: A-B vs. A-D). An asterisk (*) indicates significant group difference after correction for multiple comparisons (FDR q<0.05). FDR corrected p-values<0.05 were considered significant. All models were adjusted for age, age2, sex, ICV, and scan site. Full statistical model outputs including standard error, coefficient estimates, p-values, and % difference may be found in Supplemental Table S14. Abbreviations: L/R, left/right; LatVent, lateral ventricle; thal, thalamus; caud, caudate; put, putamen; pal, pallidum; hippo, hippocampus; amyg, amygdala; accumb, accumbens; ICV, intracranial volume. B. Shape analysis with regression coefficients plotted in regions passing correction for multiple comparisons (FDR q<0.05). Blue/green colors indicate negative coefficients, or regions of lower thickness or Jacobian measures in cases versus controls (group listed first = case, group listed second = control). Red/yellow colors indicate positive coefficients, or regions of greater thickness or Jacobian values in cases versus controls. The left two columns include thickness results; the right two columns include Jacobian results. Thickness represents local radial distance and Jacobian represents local surface area dilation/contraction. Dorsal and ventral views of the structures are provided: A, anterior; P, posterior; L, left; R, right. 1. Caudate; 2. Putamen; 3. Globus Pallidus; 4. Hippocampus; 5. Amygdala; 6. Thalamus; 7. Nucleus Accumbens. Gray regions indicate areas of no significant difference after correction for multiple comparisons. Black structures are those for which no vertex-wise test was significant after correction for multiple comparisons.
Effects of Psychotic Disorder
The 22q+Psy and 22q-Psy groups were well-matched demographically. However, as expected, 22q+Psy subjects had a higher rate of antipsychotic and anticonvulsant treatment, and lower IQ compared to the 22q-Psy group (Supplemental Table S3). A significant psychosis-by-age interaction was observed for the left and right caudate, in which 22q+Psy had relatively larger caudate volumes with increased age compared to 22q-Psy (Supplemental Table S23).
22q+Psy showed significantly smaller hippocampal, amygdala, right thalamus and ICV volumes compared to the matched 22q-Psy cohort (Figure 3; Supplemental Table S14). These effects were similar when adjusting for medication (Supplemental Table S24) and IQ (Supplemental Table 25). However, when additionally adjusting for ICV, no group differences survived correction for multiple comparisons, likely due to significantly lower overall ICV volumes in the 22q+Psy group (Supplemental Table S26).
Figure 3:
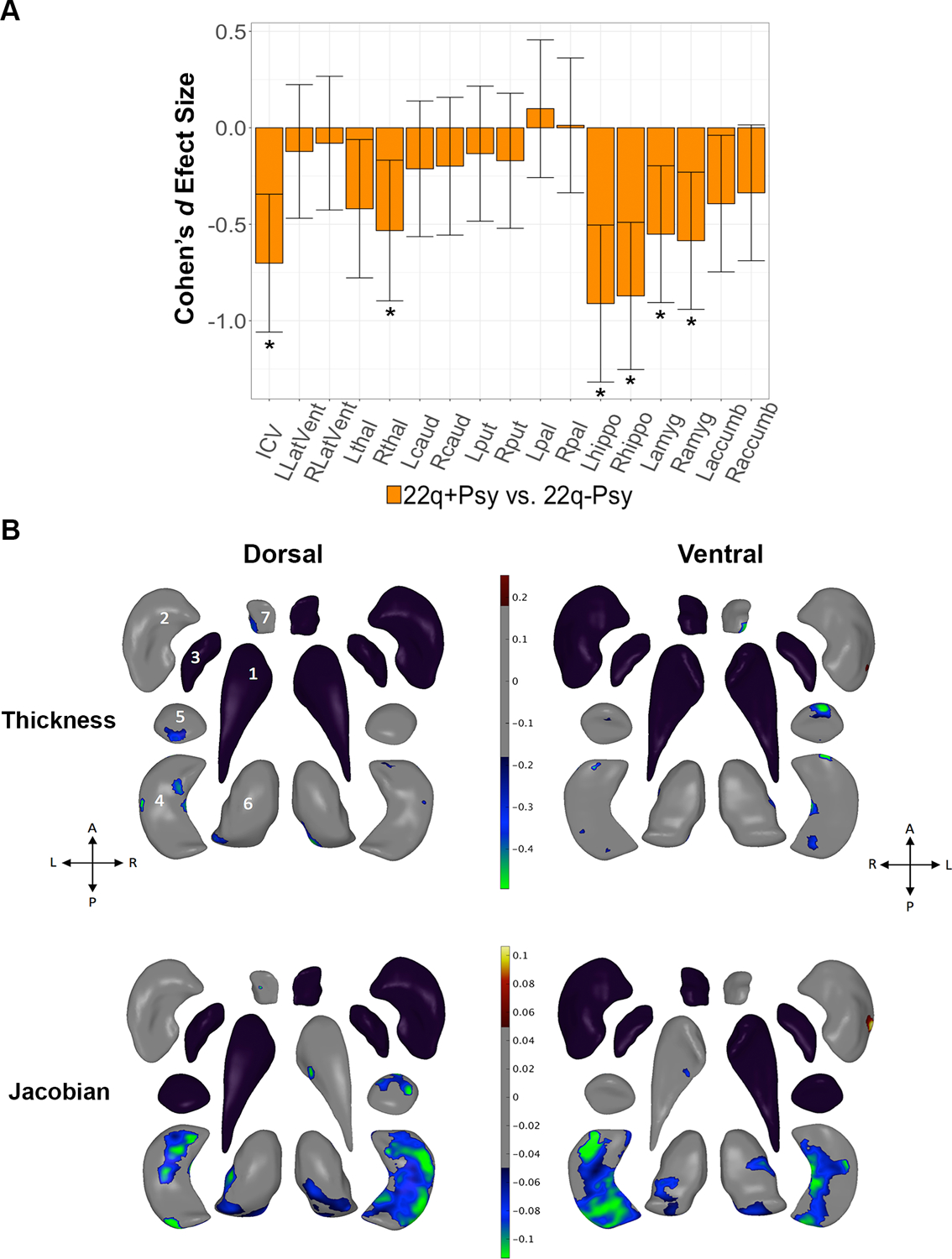
Effects of psychotic illness in 22q11DS: 22q+Psy versus 22q-Psy gross volume and shape analysis. A. Cohen’s d effect size (with 95% confidence intervals), plotted for major pairwise gross volumetric comparisons. An asterisk (*) indicates significant group difference after correction for multiple comparisons (FDR q<0.05) for 22q+Psy versus 22q-Psy. Models were adjusted for age, age2, sex, and scan site. Full statistical model outputs including standard error, coefficient estimates, p-values, and % difference may be found in Supplemental Table S14. Abbreviations: L/R, left/right; LatVent, lateral ventricle; thal, thalamus; caud, caudate; put, putamen; pal, pallidum; hippo, hippocampus; amyg, amygdala; accumb, accumbens; ICV, intracranial volume. B. Shape analysis comparing 22q+Psy versus 22q-Psy with regression coefficient values plotted in regions passing correction for multiple comparisons (q<0.05). Blue/green colors indicate negative coefficients, or regions of lower thickness (i.e., local radial distance) or Jacobian (i.e., local surface area contraction) measures in 22q+Psy versus 22q-Psy. Red/yellow colors indicate positive coefficients, or regions of greater thickness or Jacobian values in 22q+Psy versus 22q-Psy. The top row includes thickness results; the bottom row includes Jacobian results. Dorsal and ventral views of the structures are provided: A, anterior; P, posterior; L, left; R, right. 1. Caudate; 2. Putamen; 3. Globus Pallidus; 4. Hippocampus; 5. Amygdala; 6. Thalamus; 7. Nucleus Accumbens. Gray regions indicate areas of no significant difference after correction for multiple comparisons. Black structures are those for which no vertex-wise test was significant after correction for multiple comparisons.
Subcortical shape analysis revealed lower thalamic, hippocampal, amygdala, and nucleus accumbens thickness and Jacobian measures in 22q+Psy subjects compared to 22q-Psy, with particularly prominent reductions in the hippocampus. There was one region along the left dorsal putamen where the reverse pattern was observed (higher SA in 22q+Psy subjects; Figure 3). When adjusting for medication, effects were diminished but exhibited a similar pattern (Supplemental Figure F8). When also adjusting for ICV, two clusters surpassed correction: a region of higher SA in the left putamen and lower SA in the right hippocampus (Supplemental Figure F9). When adjusting for both ICV and medication, no shape differences survived correction for multiple comparisons.
22q11DS Psychosis Cross-Disorder Comparisons
Effect sizes for 22q+Psy versus 22q-Psy subcortical ROI volumes were significantly correlated with those from the ENIGMA SCZ, MDD, BD, and OCD case-control studies. However, 22q+Psy effect sizes were not significantly correlated with those from the ENIGMA ASD and ADHD case-control studies (Figure 4). In contrast, effect sizes for 22q11DS overall versus HC comparison were not significantly correlated with those observed in any other disorder (Supplemental Tables S27 and S28; Supplemental Figure F10).
Figure 4:
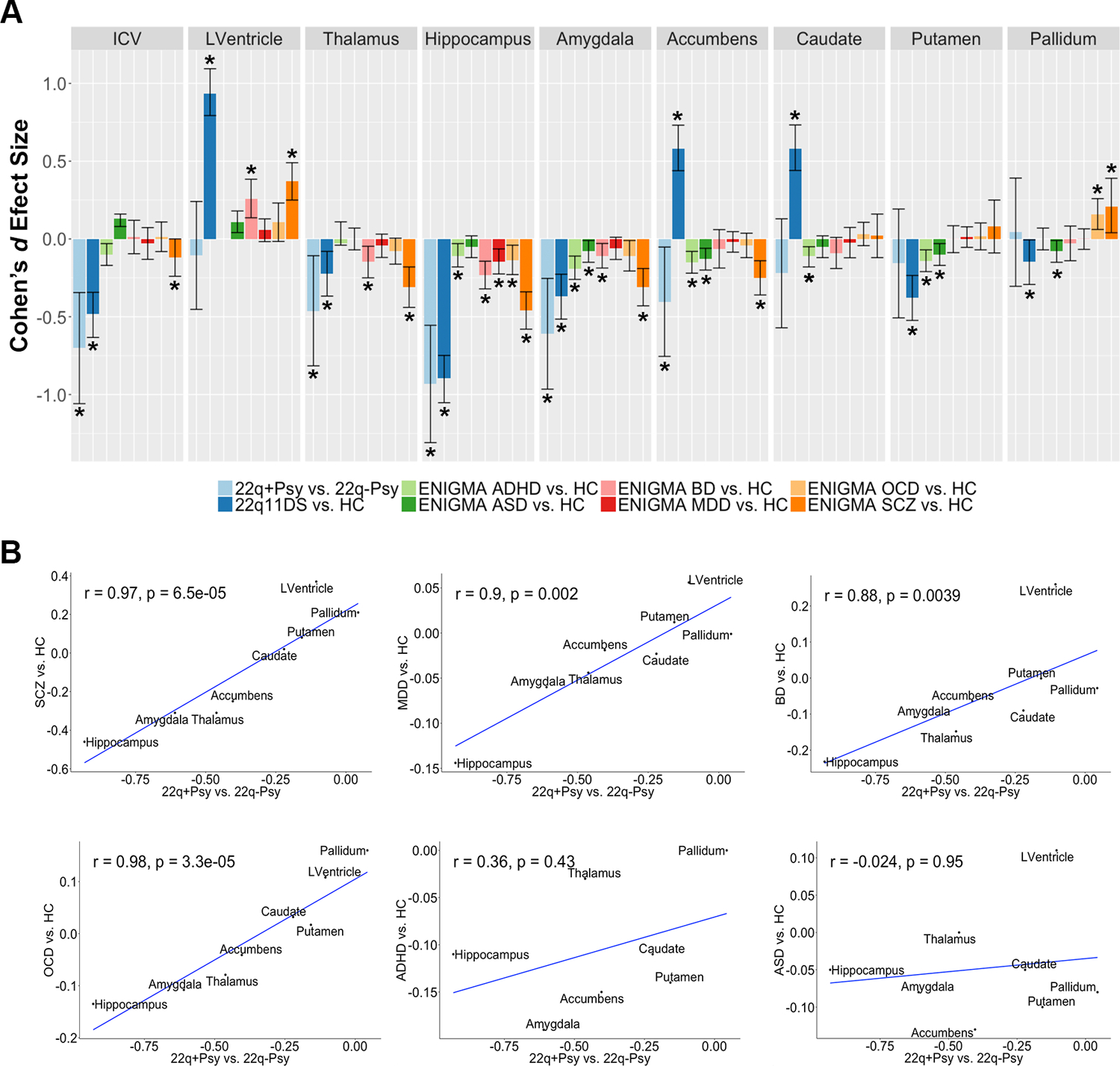
Cross-disorder comparisons from the ENIGMA psychiatric working group subcortical studies. A. Case-control Cohen’s d effect size estimates from the ENIGMA schizophrenia (5), major depression (30), bipolar disorder (31), obsessive-compulsive disorder (32), autism spectrum (33), and attention deficit hyperactivity disorder (34) working group studies. Asterisk (*) indicates significant group difference, including 95% confidence intervals from original study publication. Note that the ENIGMA ADHD group did not assess lateral ventricle volume in their subcortical study. B. Spearman rank correlations between 22q+Psy vs. 22q-Psy effect size estimates and those from the other ENIGMA psychiatric working groups (8 ROIs: lateral ventricle, amygdala, hippocampus, thalamus, caudate, putamen, pallidum, and nucleus accumbens). Significant correlations were found between 22q+Psy and the ENIGMA schizophrenia, major depressive disorder, bipolar disorder, and obsessive-compulsive disorder working group studies.
Discussion
This study represents the largest neuroimaging investigation to date of subcortical brain structure in 22q11DS and provides 5 key findings:
We detected robust group differences between 22q11.2 deletion carriers and HC using conventional measures of gross volume. Even when accounting for overall smaller ICV, we found smaller bilateral hippocampus, amygdala, putamen, and left thalamus volumes, and larger bilateral caudate, accumbens, and lateral ventricle volumes.
Subcortical shape analysis revealed complex local morphometric differences between 22q11DS and HC across most subcortical ROIs, not discernible by conventional gross volumetric analysis.
Shape analysis also revealed, for the first time, significant, localized effects of deletion size on sub-regions of the hippocampus, thalamus, caudate, putamen, and accumbens, with less severe disruptions of subcortical morphometry in those with smaller deletions.
22q11DS subjects with psychotic illness had lower ICV, thalamic, hippocampal, and amygdala volumes compared to 22q11DS subjects without history of psychosis, with effects driven largely by contracted SA across subregions of these structures.
Specifically, subcortical alterations in 22q11DS-psychosis significantly overlapped with effects observed in the largest studies of subcortical structure in schizophrenia, BD, MDD, and OCD, but not with those seen in ASD and ADHD. Effect sizes for 22q11DS overall and 22q+Psy were generally greater than those found in other ENIGMA studies of idiopathic neuropsychiatric disorders.
Our large multisite cohort study revealed overlapping but more extensive group differences than previously detected in our single-site study (25), with significant differences in gross volume observed across 14/17 regions, all with moderate to large effect sizes, and with consistent results across sites. Typically developing controls showed generally expected age effects on subcortical volumes, with age-by-diagnosis interactions in gross volume of the bilateral caudate, pallidum and left thalamus, indicating possible divergent developmental trajectories that are the focus of future longitudinal studies.
Shape analysis revealed subregional patterns of both higher and lower local thickness and SA relative to controls, particularly in larger structures (caudate, putamen, hippocampus and thalamus). Interestingly, these findings parallel our cortical analysis of 22q11DS, in which general patterns of lower cortical SA and greater thickness were reversed in regions with extensive subcortical connections such as the cingulate and parahippocampal gyri (17).
In the hippocampus, 22q11.2 deletion carriers showed thinning along the lateral/medial axis, likely corresponding to CA1 and subiculum subfields (16), with thickening in dorsal/ventral regions, which may correspond to CA2–4 subfields and parts of the subiculum. Jacobian maps indicate a more extensive pattern of contracted SA, consistent with decreased density of dendritic spines and impaired dendritic growth in hippocampal neurons observed in mouse models of 22q11DS (35). Complex alterations to other structures, such as the thalamus and caudate, appear to overlap with underlying nuclei that project to cortical association areas serving higher-order cognitive functions (see Supplement Results and Discussion Section C).
While there were no significant differences in gross volume, shape analysis revealed regions of lower SA in the hippocampus, putamen and thalamus, as well as greater caudate SA and thickness in the large A-D vs. A-B deletion. This pattern again parallels our cortical findings, where the larger A-D deletion was associated with lower cortical SA compared to A-B cases (17). While significant, the observed effects of deletion size on subcortical morphometry warrant replication in larger samples, given the limited number of subjects with smaller (A-B) deletions.
Consistent with findings in the ENIGMA schizophrenia cohort (6), psychosis in 22q11DS was associated with lower ICV, hippocampal, amygdala and right thalamic volumes. Significant correlations between the pattern of subcortical disruptions in 22q+Psy and schizophrenia suggest concordance with idiopathic schizophrenia, also observed at the cortical level (17). These findings further support the genetics-first approach in providing valuable insight into mechanisms underlying the development of psychosis not only in 22q11DS but in the broader population. Here, shape analysis additionally revealed that lower gross volumes in 22q+Psy were driven primarily by contracted SA across these structures, with several regional effects (lower hippocampal and higher putamen SA) that exceeded global brain size effects, after adjusting for ICV. Functionally, altered hippocampal-prefrontal connectivity has been associated with working memory impairments in 22q11DS mice (37); deficits in both spatial working memory and functional connectivity (38) are well-documented in both 22q11DS and idiopathic schizophrenia. Overall smaller hippocampal volumes observed in 22q11DS, and particularly in those with psychosis, may underlie connectivity defects.
Interestingly, 22q+Psy subcortical effect sizes were also correlated with those from ENIGMA studies of BD, MDD, and OCD, suggesting globally similar profiles of subcortical alterations across this set of neuropsychiatric disorders. Though elevated rates of bipolar disorder have not been reported in large studies of 22q11DS (2; 39), the correlation between subcortical patterns in 22q+Psy and other disorders may reflect the underlying genetic overlap between schizophrenia, bipolar disorder, major depression, and other psychiatric illnesses (40). Common subcortical structural abnormalities across disorders further motivates the use of shape analysis techniques to define more localized effects across subcompartments with known structural and functional connectivity patterns.
In contrast, 22q+Psy patterns diverged from those seen in ASD and ADHD, suggesting distinct subcortical disruptions in these earlier-onset disorders. Although these are common comorbidities in 22q11DS, subcortical disruptions in 22q11DS cases overall did not significantly overlap with those seen in the corresponding idiopathic disorders investigated (see Supplemental Figure F10).
The large sample size (the largest ever conducted of 22q11DS) and centralized processing and analysis of raw neuroimaging data were key strengths of our study. However, certain limitations must be noted. First, the relationship between subregional shape measures and underlying cytoarchitecture is not well understood. The correspondence of such shape variations to changes in underlying subfields and gene expression is a focus of ongoing work. Secondly, we cannot rule out that some non-psychotic subjects with 22q11DS may later develop a psychotic disorder, which would likely attenuate the group differences reported here. Further investigation of 22q11DS medical comorbidities (e.g., cardiovascular abnormalities) and comorbid psychiatric diagnoses (e.g., ASD) were outside the scope of the current study, but will be pursued in future work as they may also contribute to variability in brain structure. Future longitudinal studies are underway to investigate the developmental trajectories of psychotic symptom emergence and other psychiatric disorders, which will greatly improve our understanding of both the heterogeneity and developmental effects of 22q11DS.
While our cross-disorder analysis is strengthened by harmonized processing protocols applied across the largest existing neuroimaging studies of their kind, these studies included individuals with different age ranges and demographic profiles that could impact such relationships. Future work directly comparing harmonized measures across demographically matched samples will help address such limitations.
Here, we have shown robust differences in subcortical structure between 22q11DS and demographically comparable HC, with more extreme alterations in those with larger deletions and/or psychosis. Subcortical alterations in 22q11DS-associated psychosis overlapped with those from the largest studies to date of subcortical structure in idiopathic schizophrenia and other serious mental illnesses. This further supports 22q11DS as a biologically applicable framework for understanding brain mechanisms that underlie the development of these disorders, and is the focus of future ENIGMA cross-disorder and genetic analyses.
Supplementary Material
Acknowledgements
Christopher R. K. Ching is supported in part by NIA T32AG058507, NIH/NIMH 5T32MH073526, and NIH grant U54EB020403 from the Big Data to Knowledge (BD2K) Program. Carrie E. Bearden is supported by NIH/NIBIB U54 EB020403 (BD2K), National Institute of Mental Health RO1 MH085953 and R01MH100900, U01MH101719, and U01MH101719-04S1. Paul M. Thompson is supported by NIH grants U54EB020403 from the Big Data to Knowledge (BD2K) Program, as well as R01MH116147, P41 EB015922, R01MH111671 and R56AG058854. Wendy R. Kates is supported by NIH MH064824. Declan G. Murphy is supported by the NIHR Mental Health Biomedical Research Centre (BRC) at South London and Maudsley NHS Foundation Trust and King’s College London and EU-AIMS (European Autism Interventions)/EU AIMS-2-TRIALS, a European Innovative Medicines Initiative Joint Undertaking under Grant Agreements No. 115300 and 777394. David Roalf is supported by NIH K01 MH102609; NARSAD Young Investigators Award; Life Span Brain Institute (LiBI)-a collaboration between the University of Pennsylvania School of Medicine and Children’s Hospital of Philadelphia. Beverly S. Emanuel is supported by NIH PO1-HD070454, UO1-MH191719, R01 MH087636-01A1, R01 GM125757. Donna M. McDonald-McGinn is supported by the US National Institutes of Health grants PO1-HD070454, UO1-MH191719, and R01 MH087636-01A1. T. Blaine Crowley is supported by the US National Institutes of Health grants PO1-HD070454, UO1-MH191719, and R01 MH087636-01A1. Elaine H. Zackai is supported by US National Institutes of Health grants PO1-HD070454, UO1-MH191719, and R01 MH087636-01A1. Anne S. Bassett is supported by grant U01MH101723. Eva W. C. Chow is supported by the Canadian Institutes of Health Research. Fidel Vila-Rodriquez is supported by the Michael Smith Foundation for Health Research and Seedlings Foundation. Joanne Doherty is supported by The Wellcome Trust grant 102003/Z/13/Z. Adam Cunningham and Marianne van den Bree are supported by the MRC Centre Grant No. MR/L010305/1. David E. Linden is supported by the The Wellcome Trust grant 100202/Z/12/Z. Michael J. Owen is supported by MRC Centre Grant No. MR/L010305/1 and The Wellcome Trust grant 100202/Z/12/Z. Haley Moss is supported by MRC Centre Grant No. MR/L010305/1. Gabriela M. Repetto is supported by the FONDECYT Chile Grant 1171014. Nicolas A. Crossley is supported by Grant FONDECYT regular 1160736 from the Corporacin Nacional de Ciencia y Tecnologa, Chile.
We would like to acknowledge the ENIGMA psychiatric disorder working groups for providing published data from their neuroimaging studies of subcortical brain structures in their respective working groups, which allowed for the cross-disorder comparison in this paper. We would especially like to acknowledge Lianne Schmaal and Dick J. Veltman from the ENIGMA Major Depression Working Group, Derrek P. Hibar and Ole A. Andreassen from the ENIGMA Bipolar Disorder Working Group, Theo G. van Erp and Jessica A. Turner from the ENIGMA Schizophrenia Working Group, Premika S. Boedhoe, Dan J. Stein, and Odile A. van den Heuvel from the ENIGMA OCD Working Group, Martine Hoogman and Barbara Franke from the ENIGMA ADHD Working Group, and Daan van Rooij and Jan K. Buitelaar from the ENIGMA Autism Spectrum Disorders Working Group.
We thank Agnes McMahon and Sophia Thomopoulos, former and current ENIGMA program managers, for all their efforts in helping this international consortium effort thrive.
Footnotes
Disclosures
Declan G. Murphy has received honoraria from Roche. Paul M. Thompson and Christopher R. K. Ching have received grant support from Biogen, Inc. (Boston, USA) for work unrelated to the topic of this manuscript. All other authors have no conflicts of interest to disclose.
Contributor Information
Christopher R.K. Ching, Imaging Genetics Center, Mark and Mary Stevens Institute for Neuroimaging and Informatics, Keck School of Medicine, Los Angeles; Department of Psychiatry and Biobehavioral Sciences, Semel Institute for Neuroscience and Human Behavior, Los Angeles; Department of Psychology, UCLA, Los Angeles.
Boris A. Gutman, Department of Biomedical Engineering, Armour College of Engineering, Illinois Institute of Technology, Chicago.
Daqiang Sun, Department of Psychiatry and Biobehavioral Sciences, Semel Institute for Neuroscience and Human Behavior, Los Angeles.
Julio Villalon Reina, Imaging Genetics Center, Mark and Mary Stevens Institute for Neuroimaging and Informatics, Keck School of Medicine, Los Angeles.
Anjanibhargavi Ragothaman, Department of Biomedical Engineering, Oregon Health and Science University, Portland.
Dmitry Isaev, Department of Biomedical Engineering, Duke University, Durham, N.C..
Artemis Zavaliangos-Petropulu, Imaging Genetics Center, Mark and Mary Stevens Institute for Neuroimaging and Informatics, Keck School of Medicine, Los Angeles.
Amy Lin, Department of Psychiatry and Biobehavioral Sciences, Semel Institute for Neuroscience and Human Behavior, Los Angeles; Graduate Interdepartmental Program in Neuroscience, UCLA School of Medicine, Los Angeles.
Rachel K. Jonas, Department of Psychiatry and Biobehavioral Sciences, Semel Institute for Neuroscience and Human Behavior, Los Angeles; Graduate Interdepartmental Program in Neuroscience, UCLA School of Medicine, Los Angeles.
Laura Pacheco-Hansen, Department of Psychiatry and Biobehavioral Sciences, Semel Institute for Neuroscience and Human Behavior, Los Angeles.
Ariana Vajdi, Department of Psychiatry and Biobehavioral Sciences, Semel Institute for Neuroscience and Human Behavior, Los Angeles.
Jennifer K. Forsyth, Department of Psychiatry and Biobehavioral Sciences, Semel Institute for Neuroscience and Human Behavior, Los Angeles; Department of Psychology, UCLA, Los Angeles.
Maria Jalbrzikowski, Department of Psychiatry, University of Pittsburgh, Pittsburgh.
Geor Bakker, Department of Psychiatry and Neuropsychology, Maastricht University, Maastricht, the Netherlands; Department of Radiology and Nuclear Medicine, Amsterdam University Medical Centers, Amsterdam.
Therese van Amelsvoort, Department of Psychiatry and Neuropsychology, Maastricht University, Maastricht, the Netherlands.
Kevin M. Antshel, Department of Psychology, Syracuse University, Syracuse, N.Y..
Wanda Fremont, Department of Psychiatry and Behavioral Sciences, SUNY Upstate Medical University, Syracuse.
Wendy R. Kates, Department of Psychiatry and Behavioral Sciences, SUNY Upstate Medical University, Syracuse.
Linda E. Campbell, School of Psychology, University of Newcastle, Newcastle, Australia.
Kathryn L. McCabe, School of Psychology, University of Newcastle, Newcastle, Australia; MIND Institute and Department of Psychiatry and Behavioral Sciences, University of California Davis, Davis.
Michael C. Craig, Institute of Psychiatry, Psychology, and Neuroscience, Sackler Institute for Translational Neurodevelopment, and Department of Forensic and Neurodevelopmental Sciences, King’s College London; Bethlem Royal Hospital, National Institute for Health Research Maudsley Biomedical Research Centre, and SLaM NHS Foundation Trust, National Autism Unit, London.
Eileen Daly, Institute of Psychiatry, Psychology, and Neuroscience, Sackler Institute for Translational Neurodevelopment, and Department of Forensic and Neurodevelopmental Sciences, King’s College London.
Maria Gudbrandsen, Institute of Psychiatry, Psychology, and Neuroscience, Sackler Institute for Translational Neurodevelopment, and Department of Forensic and Neurodevelopmental Sciences, King’s College London.
Clodagh M. Murphy, Institute of Psychiatry, Psychology, and Neuroscience, Sackler Institute for Translational Neurodevelopment, and Department of Forensic and Neurodevelopmental Sciences, King’s College London; Behavioural Genetics Clinic, Adult Autism Service, Behavioural and Developmental Psychiatry Clinical Academic Group, South London and Maudsley NHS Foundation Trust, London.
Declan G. Murphy, Institute of Psychiatry, Psychology, and Neuroscience, Sackler Institute for Translational Neurodevelopment, and Department of Forensic and Neurodevelopmental Sciences, King’s College London; Behavioural Genetics Clinic, Adult Autism Service, Behavioural and Developmental Psychiatry Clinical Academic Group, South London and Maudsley NHS Foundation Trust, London.
Kieran C. Murphy, Department of Psychiatry, Royal College of Surgeons in Ireland, and Education and Research Centre, Beaumont Hospital, Dublin.
Ania Fiksinski, Department of Psychiatry, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht, the Netherlands; Clinical Genetics Research Program, Centre for Addiction and Mental Health, Toronto; Dalglish Family 22q Clinic; University Health Network, Toronto.
Sanne Koops, Department of Psychiatry, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht, the Netherlands.
Jacob Vorstman, Department of Psychiatry, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht, the Netherlands; Department of Psychiatry, University of Toronto, Toronto; Program in Genetics and Genome Biology, Research Institute, and Department of Psychiatry, Hospital for Sick Children, Toronto.
T. Blaine Crowley, Division of Human Genetics and 22q and You Center, Children’s Hospital of Philadelphia, Philadelphia.
Beverly S. Emanuel, Division of Human Genetics and 22q and You Center, Children’s Hospital of Philadelphia, Philadelphia; Department of Pediatrics, University of Pennsylvania Perelman School of Medicine, Philadelphia.
Raquel E. Gur, Department of Psychiatry, University of Pennsylvania Perelman School of Medicine and Children’s Hospital of Philadelphia, Philadelphia.
Donna M. McDonald-McGinn, Division of Human Genetics and 22q and You Center, Children’s Hospital of Philadelphia, Philadelphia; Department of Pediatrics, University of Pennsylvania Perelman School of Medicine, Philadelphia.
David Roalf, Department of Psychiatry, University of Pennsylvania Perelman School of Medicine, Philadelphia.
Kosha Ruparel, Department of Psychiatry, University of Pennsylvania Perelman School of Medicine, Philadelphia.
J. Eric Schmitt, Departments of Radiology and Psychiatry, Hospital of the University of Pennsylvania, Philadelphia.
Elaine H. Zackai, Division of Human Genetics and 22q and You Center, Children’s Hospital of Philadelphia, Philadelphia; Department of Pediatrics, University of Pennsylvania Perelman School of Medicine, Philadelphia.
Courtney A. Durdle, MIND Institute and Department of Psychiatry and Behavioral Sciences, University of California Davis, Davis; Department of Psychological and Brain Sciences, University of California, Santa Barbara.
Naomi J. Goodrich-Hunsaker, MIND Institute and Department of Psychiatry and Behavioral Sciences, University of California Davis, Davis; Department of Neurology, University of Utah, Salt Lake City.
Tony. J Simon, MIND Institute and Department of Psychiatry and Behavioral Sciences, University of California Davis, Davis.
Anne S. Bassett, Clinical Genetics Research Program, Centre for Addiction and Mental Health, Toronto; Campbell Family Mental Health Research Institute, Centre for Addiction and Mental Health, Toronto; Dalglish Family 22q Clinic; Department of Mental Health, and Toronto General Hospital Research Institute; University Health Network, Toronto; Department of Psychiatry, University of Toronto, Toronto.
Nancy J. Butcher, Child Health Evaluative Sciences, Hospital for Sick Children Research Institute, Toronto.
Eva W. C. Chow, Clinical Genetics Research Program, Centre for Addiction and Mental Health, Toronto; Clinical Genetics Service, Centre for Addiction and Mental Health, Toronto; Department of Psychiatry, University of Toronto, Toronto.
Fidel Vila-Rodriguez, Department Psychiatry, University of British Columbia, Vancouver.
Adam Cunningham, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, U.K..
Joanne Doherty, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, U.K.; Cardiff University Brain Research Imaging Centre, Cardiff, U.K..
David E. Linden, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, U.K.; Cardiff University Brain Research Imaging Centre, Cardiff, U.K..
Haley Moss, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, U.K..
Michael J. Owen, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, U.K..
Marianne van den Bree, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, U.K..
Nicolas A. Crossley, Department of Psychiatry, Pontificia Universidad Católica de Chile, Santiago.
Gabriela M. Repetto, Clinica Alemana, Universidad del Desarrollo, Centro de Genética y Genomica, Facultad de Medicina, Santiago.
Paul M. Thompson, Imaging Genetics Center, Mark and Mary Stevens Institute for Neuroimaging and Informatics, Keck School of Medicine, Los Angeles; Departments of Neurology, Psychiatry, Radiology, Engineering, Pediatrics, and Ophthalmology, University of Southern California, Los Angeles.
Carrie E. Bearden, Department of Psychiatry and Biobehavioral Sciences, Semel Institute for Neuroscience and Human Behavior, Los Angeles; Department of Psychology, UCLA, Los Angeles.
References
- 1.McDonald-McGinn DM, Sullivan KE, Marino B, et al. : 22q11.2 deletion syndrome. Nat Rev Dis Primers 2015; 1:15071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneider M, Debbané M, Bassett AS, et al. : Psychiatric disorders from childhood to adulthood in 22q11.2 deletion syndrome: results from the International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome. Am J Psychiatry 2014; 171:627–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jonas RK, Montojo CA, Bearden CE: The 22q11.2 deletion syndrome as a window into complex neuropsychiatric disorders over the lifespan. Biol Psychiatry 2014; 75:351–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiroi N, Takahashi T, Hishimoto A, et al. : Copy number variation at 22q11.2: from rare variants to common mechanisms of developmental neuropsychiatric disorders. Mol Psychiatry 2013; 18:1153–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassett AS, Chow EW, Abdel Malik P, et al. : The schizophrenia phenotype in 22q11 deletion syndrome. Am J Psychiatry 2003; 160:1580–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Erp TG, Hibar DP, Rasmussen JM, et al. : Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol Psychiatry 2016; 21:585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meechan DW, Tucker ES, Maynard TM, et al. : Diminished dosage of 22q11 genes disrupts neurogenesis and cortical development in a mouse model of 22q11 deletion/DiGeorge syndrome. Proc Natl Acad Sci U S A 2009; 106:16434–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toyoda R, Assimacopoulos S, Wilcoxon J, et al. : FGF8 acts as a classic diffusible morphogen to pattern the neocortex. Development 2010; 137:3439–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kates WR, Burnette CP, Bessette BA, et al. : Frontal and caudate alterations in velocardiofacial syndrome (deletion at chromosome 22q11.2). J Child Neurol 2004; 19:337–42 [DOI] [PubMed] [Google Scholar]
- 10.Bish JP, Nguyen V, Ding L, et al. : Thalamic reductions in children with chromosome 22q11.2 deletion syndrome. Neuroreport 2004; 15:1413–5 [DOI] [PubMed] [Google Scholar]
- 11.Flahault A, Schaer M, Ottet MC, et al. : Hippocampal volume reduction in chromosome 22q11.2 deletion syndrome (22q11.2DS): a longitudinal study of morphometry and symptomatology. Psychiatry Res 2012; 203:1–5 [DOI] [PubMed] [Google Scholar]
- 12.Frank DU, Fotheringham LK, Brewer JA, et al. : An Fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development 2002; 129:4591–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bearden CE, van ETG, Dutton RA, et al. : Alterations in midline cortical thickness and gyrification patterns mapped in children with 22q11.2 deletions. Cereb Cortex 2009; 19:115–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutman BA, Hua X, Rajagopalan P, et al. : Maximizing power to track Alzheimer’s disease and MCI progression by LDA-based weighting of longitudinal ventricular surface features. Neuroimage 2013; 70:386–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Mamah D, Harms MP, et al. : Progressive deformation of deep brain nuclei and hippocampal-amygdala formation in schizophrenia. Biol Psychiatry 2008; 64:1060–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mamah D, Alpert KI, Barch DM, et al. : Subcortical neuromorphometry in schizophrenia spectrum and bipolar disorders. Neuroimage Clin 2016; 11:276–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun D, Ching CRK, Lin A, et al. : Large-scale mapping of cortical alterations in 22q11.2 deletion syndrome: Convergence with idiopathic psychosis and effects of deletion size. Mol Psychiatry 2018; [DOI] [PMC free article] [PubMed]
- 18.Raznahan A, Shaw P, Lalonde F, et al. : How does your cortex grow? J Neurosci 2011; 31:7174–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamnes CK, Herting MM, Goddings AL, et al. : Development of the Cerebral Cortex across Adolescence: A Multisample Study of Inter-Related Longitudinal Changes in Cortical Volume, Surface Area, and Thickness. J Neurosci 2017; 37:3402–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y, Guo T, Fiksinski A, et al. : Variance of IQ is partially dependent on deletion type among 1,427 22q11.2 deletion syndrome subjects. Am J Med Genet A 2018; 176:2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roshchupkin GV, Gutman BA, Vernooij MW, et al. : Heritability of the shape of subcortical brain structures in the general population. Nat Commun 2016; 7:13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutman BA, Fletcher PT, Cardoso MJ, et al. : A Riemannian Framework for Intrinsic Comparison of Closed Genus-Zero Shapes. Inf Process Med Imaging 2015; 24:205–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gur RE, Bassett AS, McDonald-McGinn DM, et al. : A neurogenetic model for the study of schizophrenia spectrum disorders: the International 22q11.2 Deletion Syndrome Brain Behavior Consortium. Molecular Psychiatry 2017; 22:1664–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischl B, Salat DH, Busa E, et al. : Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 2002; 33:341–55 [DOI] [PubMed] [Google Scholar]
- 25.Lin A, Ching CRK, Vajdi A, et al. : Mapping 22q11.2 Gene Dosage Effects on Brain Morphometry. J Neurosci 2017; 37:6183–6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LM Function. Available from: https://stat.ethz.ch/R-manual/R-devel/library/stats/html/lm.html.
- 27.Kong XZ, Mathias SR, Guadalupe T, et al. : Mapping cortical brain asymmetry in 17,141 healthy individuals worldwide via the ENIGMA Consortium. Proc Natl Acad Sci U S A 2018; 115:E5154–E5163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakagawa S, Cuthill IC: Effect size confidence interval and statistical significance: a practical guide for biologists. Biological Reviews 2007; 82:591–605 [DOI] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y: Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society Series B (Methodological) 1995; 289–300
- 30.van Erp TG, Hibar DP, Rasmussen JM, et al. : Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol Psychiatry 2016; 21:585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmaal L, Veltman DJ, van ETG, et al. : Subcortical brain alterations in major depressive disorder: findings from the ENIGMA Major Depressive Disorder working group. Mol Psychiatry 2016; 21:806–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hibar DP, Westlye LT, van ETG, et al. : Subcortical volumetric abnormalities in bipolar disorder. Mol Psychiatry 2016; 21:1710–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boedhoe PS, Schmaal L, Abe Y, et al. : Distinct Subcortical Volume Alterations in Pediatric and Adult OCD: A Worldwide Meta- and Mega-Analysis. Am J Psychiatry 2017; 174:60–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Rooij D, Anagnostou E, Arango C, et al. : Cortical and Subcortical Brain Morphometry Differences Between Patients With Autism Spectrum Disorder and Healthy Individuals Across the Lifespan: Results From the ENIGMA ASD Working Group. Am J Psychiatry 2018; 175:359–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoogman M, Bralten J, Hibar DP, et al. : Subcortical brain volume differences in participants with attention deficit hyperactivity disorder in children and adults: a cross-sectional mega-analysis. Lancet Psychiatry 2017; 4:310–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mukai J, Dhilla A, Drew LJ, et al. : Palmitoylation-dependent neurodevelopmental deficits in a mouse model of 22q11 microdeletion. Nat Neurosci 2008; 11:1302–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sigurdsson T, Stark KL, Karayiorgou M, et al. : Impaired hippocampalprefrontal synchrony in a genetic mouse model of schizophrenia. Nature 2010; 464:763–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukai J, Tamura M, Fénelon K, et al. : Molecular Substrates of Altered Axonal Growth and Brain Connectivity in a Mouse Model of Schizophrenia. Neuron 2015; 86:680–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malhotra D, McCarthy S, Michaelson JJ, et al. : High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron 2011; 72:951–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SH, Ripke S, Neale BM, et al. : Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 2013; 45:984–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


