Abstract
Tumor location and immunity play important roles in the progression of colorectal cancer (CRC). This study aimed to investigate the differences in the immunosurveillance pattern between right‐ and left‐sided CRC and analyze their association with clinicopathologic features, including mismatch repair (MMR) status. We included surgically resected stage II/III CRC cases and evaluated the immunohistochemical findings of HLA class I, HLA class II, programmed cell death‐ligand 1 (PD‐L1), PD‐1, CTLA‐4, CD3, CD4, CD8, TIA‐1, T‐bet, GATA3, RORγT, Foxp3, and CD163. A total of 117 patients were included in the analyses; of these, 30 and 87 had right‐ and left‐sided cancer, respectively. Tumor immunity varied according to the tumor location in the overall cohort. Analysis of the tumors excluding those with DNA mismatch repair (MMR) deficiency also revealed that tumor immunity differed according to the tumor location. In right‐sided colon cancer (CC), high expression of Foxp3 (P = .0055) and TIA‐1 (P = .0396) were associated with significantly better disease‐free survival (DFS). High CD8 (P = .0808) and CD3 (P = .0863) expression tended to have better DFS. Furthermore, in left‐sided CRC, only high PD‐L1 expression in the stroma (P = .0426) was associated with better DFS. In multivariate analysis, high Foxp3 expression in right‐sided CC was an independent prognostic factor for DFS (hazard ratio, 7.6445; 95% confidence interval, 1.2091‐150.35; P = .0284). In conclusion, the immunosurveillance pattern differs between right‐ and left‐sided CRC, even after adjusting for MMR deficiency.
Keywords: colorectal cancer, immune‐checkpoint molecule, mismatch repair, tumor location, tumor‐infiltrating lymphocyte
Comparison of Kaplan‐Meier curves of disease‐free survival (DFS) in each tumor location excluding DNA mismatch repair deficiency. In right‐sided colon cancer, high Foxp3 (P = .0055) and TIA‐1 expression (P = .0396) were associated with significantly better DFS. High CD8 (P = .0808) and CD3 (0.0863) expression showed a tendency towards better DFS. In left‐sided colorectal cancer, only high sPD‐L1 expression (P = .0426) was associated with better DFS.

Abbreviations
- CC
colon cancer
- CI
confidence interval
- CIMP
CpG island methylator phenotype
- CRC
colorectal cancer
- DFS
disease‐free survival
- dMMR
mismatch repair deficiency
- EGFR
epidermal growth factor receptor
- HR
hazard ratio
- IHC
immunohistochemistry
- MMR
mismatch repair
- MSI‐H
microsatellite instability high
- OS
overall survival
- PD‐L1
programmed cell death‐ligand 1
- pMMR
mismatch repair proficiency
- sPD‐L1
PD‐L1 in stromal cells
- TIL
tumor‐infiltrating lymphocyte
- TMA
tissue microarray
- TME
tumor microenvironment
- tPD‐L1
PD‐L1 in tumor cells
- VEGF
vascular endothelial growth factor
1. INTRODUCTION
Colorectal cancer is among the leading causes of cancer mortality worldwide. 1 Despite improvements in prognosis due to treatment advances, including in surgery, chemotherapy, and molecular targeted therapy, the outcomes of CRC remain unsatisfactory. To improve survival, a more comprehensive understanding of the molecular biological mechanism underlying CRC is needed.
Prognosis differs according to tumor sidedness in CRC, with right‐sided CC (ie from the cecum to the splenic flexure) having worse survival than left‐sided CRC (ie from the splenic flexure to the rectum). 2 This could be due to various biological and clinical differences including embryonic origin, vascular supply, and physiological function. Right‐sided CC is derived from the midgut. It is supplied by the superior mesenteric artery and frequently presents with BRAF mutation, MSI‐H, and CIMP. In contrast, left‐sided CRC is derived from the hindgut. It is supplied by the inferior mesenteric artery and frequently includes chromosomal instability. 3 Accordingly, the efficacy of molecular targeted agents differs according to the tumor location. Anti‐VEGF mAbs, which prevent angiogenesis by binding VEGF‐A and/or B, are more effective in right‐sided CC, whereas anti‐EGFR mAb, which inhibits cell proliferation and survival by combining with EGFR, is more effective in left‐sided CRC. 4 , 5 Thus, tumor location should be considered when deciding the treatment plan.
The host immune state also plays a pivotal role in tumor progression. Several recent studies have reported that both the TME, including the TILs, macrophages, and immune‐checkpoint molecules, and clinicopathologic features influence cancer prognosis. 6 , 7 , 8 , 9 , 10 In CRC, a high number of TILs are associated with better prognosis, 6 , 7 and the expression of some immune‐checkpoint molecules can be useful prognostic biomarkers. 8 , 9 , 11 , 12 , 13 However, it remains unclear whether immunity differs according to the tumor location and which cells or molecules are involved in cancer prognosis.
DNA MMR is a system that recognizes and repairs erroneous DNA insertions and deletions. MMR deficiency results in the accumulation of insertion/deletion mutations in short repetitive sequence stretches called microsatellites, leading to the MSI phenotype. Some MSI‐induced mutations create several cancer neoantigens, which can be targeted by the immune cells. Thus, dMMR strongly influences tumor immunity. 14 , 15 , 16 In general, dMMR is more frequent in right‐sided CC than in left‐sided CRC and is associated with better survival than pMMR. 17
Therefore, this study aimed to investigate the clinicopathologic differences according to the immunosurveillance pattern between right‐sided and left‐sided CRC, using IHC staining of MMR proteins to identify biomarkers and prognostic factors.
2. MATERIALS AND METHODS
2.1. Patients
We reviewed formalin‐fixed, paraffin‐embedded tissue specimens from CRC patients who underwent surgical resection in Kurume University between 2007 and 2008. All patients had stage II or III disease as classified based on the 7th edition of the UICC TNM classification of malignant tumors. Clinical data were obtained from the patients’ medical records. All patients were followed up until death or censorship. This study was approved by the Research Ethics Committee of Kurume University and was carried out according to the tenets of the Declaration of Helsinki.
2.2. Immunohistochemistry
The primary Abs used for IHC were as follows: mouse monoclonal anti‐HLA class I ABC Ab (ab70328 [EMR8‐5]; Abcam), mouse monoclonal anti‐HLA DR + DP + DQ Ab (ab7856 [CR3/43]; Abcam), rabbit monoclonal anti‐PD‐L1 Ab (#13684 [E1L3N]; Cell Signaling Technology), mouse monoclonal anti‐PD‐1 Ab (ab52587 [NAT105]; Abcam), mouse monoclonal anti‐CTLA‐4 Ab (UM800141 [UMAB249]; OriGene), mouse monoclonal anti‐CD3 Ab (M7254 [F7.2.38]; Dako), rabbit polyclonal anti‐CD4 Ab (790‐4423 [SP35]; Ventana), mouse monoclonal anti‐CD8 Ab (ab75129 [C8/144B]; Abcam), mouse monoclonal anti‐TIA‐1 Ab (IM2550 [2G9A10F5]; Beckman Coulter), mouse monoclonal anti‐T‐bet Ab (ab91109 [4B10]; Abcam), rabbit monoclonal anti‐GATA3 Ab (#5852 [D13C9]; Cell Signaling Technology), mouse monoclonal anti‐RORγT Ab (MABF81 [6F3.1]; Merck Millipore), rabbit monoclonal anti‐Foxp3 Ab (ab99963 [SP97]; Abcam), and mouse monoclonal anti‐CD163 Ab (CD163‐L‐CE [10D6]; Leica), mouse monoclonal anti‐MSH2 Ab (M3639 [FE11]; Dako), rabbit monoclonal anti‐MSH6 Ab (M3646 [EP49]; Dako), rabbit monoclonal anti‐PMS2 Ab (M3647 [EP51]; Dako), and mouse monoclonal anti‐MLH1 Ab (M3640 [ES05]; Dako). We used the Dako ChemMate EnVision Kit system and a peroxidase/DAB kit for IHC. Some of them were stained in our previous study. 10 Tissue microarray was constructed as reported in our previous study. 10 Briefly, 1 tissue cylinder measuring 3.0 mm in diameter was punched from the center of the tumor using a tissue microarrayer.
2.3. Evaluation of IHC
Immunostaining was evaluated by 2 observers (HK and HM) blinded to the clinical data. The positive expression rate of HLA class I, HLA class II, and PD‐L1 on tumor cells was calculated. For HLA class I, positive cell membrane staining was considered positive expression. Expression of PD‐L1 was measured separately in both tumor and stromal cells. The total number of TILs with positive expression was counted. In every sample, 3 well‐stained hotspots were evaluated at ×400 magnification, which was equivalent to 0.19625 mm2, and the average of the 3 measurements was used for analyses. In evaluation of each protein expression, the comparison with internal positive or negative control was carried out in each TMA core. Some were counted in our previous study using ImageJ software. 10 The numbers generated using automated counting by ImageJ were almost equal to the numbers obtained by manual visual counting. 10 The median values were used as the cut‐off point in every analysis. Representative IHC images are shown in Figure S1.
2.4. Definition of dMMR
The MMR protein content was evaluated for the absence or presence of the following 4 MMR proteins: MSH2, MSH6, PMS2, and MLH1. Tumors showing total absence of nuclear staining in at least 1 of the 4 MMR proteins were defined as dMMR. The expression of MMR proteins in the normal epithelium and lymphocytes was used as the positive internal control for all cases. Four MMR deficiency patterns were assessed: (i) dMLH1/dPMS2; (ii) dMSH2/dMSH6; (iii) dMSH6; and (iv) dPMS2.
2.5. Statistical analysis
The clinicopathologic characteristics were compared between right‐ and left‐sided CRC using the χ2 test for categorical variables and the Wilcoxon rank‐sum test for continuous variables. Survival curves were created using the Kaplan‐Meier method and compared using log‐rank test. Disease‐free survival was defined as the time from surgery to recurrence or death; OS was defined as the time from surgery to death. The Cox proportional hazards model was used for uni‐ and multivariate analyses. All statistical analyses were undertaken using JMP version 13 software (SAS Institute, Cary, NC, USA), and P < .05 was considered to indicate statistical significance.
3. RESULTS
3.1. Comparison of clinicopathologic features according to tumor location
A total of 117 cases were included in the analyses; of these, 30 were right‐sided CC, and 87 were left‐sided CRC. The clinicopathologic features between the 2 groups are summarized in Table S1. There were more elderly patients in the right‐sided CC group (P = .0360). The number of patients with positive lymph node involvement was significantly lower in the right‐sided group than in the left‐sided group (7 [23%] vs. 44 [51%], P = .0078). Moreover, dMMR was significantly more frequent in the right‐sided group (20% vs. 5%, P = .0157). Other patient characteristics were not significantly different between the 2 groups.
3.2. Relationship between tumor location and IHC staining results
A comparison of each IHC staining result between the 2 groups is shown in Figure S2. There were more biomarkers with positive expression in left‐sided CRC. The median HLA class I expression rate was significantly different between right‐ and left‐sided CRC (32% vs 77%, P = .0011). The CD4, RORγT, and CD163 infiltration was significantly more pronounced in left‐sided CRC (P = .0075, P = .0448, and P = .0050, respectively).
3.3. Comparison of DFS and OS curves
The comparison of DFS according to tumor sidedness is shown in Figure 1. In right‐sided CC, low HLA class I (P = .0060), high PD‐1 (P = .0190), high CD3 (P = .0099), high CD8 (P = .0151), high TIA‐1 (P = .0140), and high Foxp3 expression (P = .0190) were associated with significantly better DFS. In left‐sided CRC, only high sPD‐L1 expression (P = .0335) was associated with better DFS.
FIGURE 1.
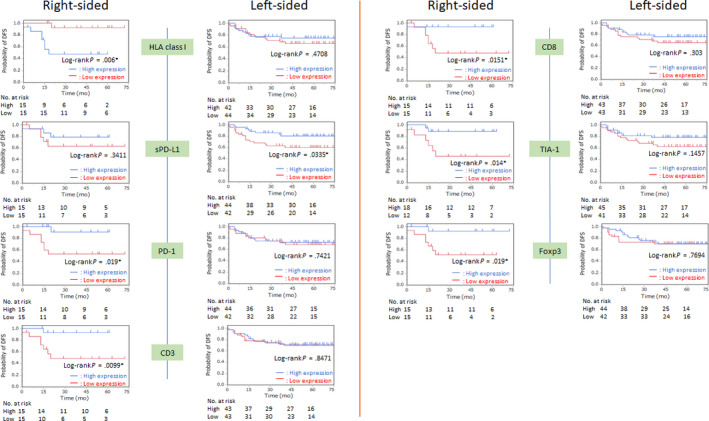
Kaplan‐Meier curves of disease‐free survival (DFS) according to the location of colorectal tumor based on the expression of HLA class I, programmed cell death‐ligand 1 on stromal cells (sPD‐L1), programmed cell death‐1 (PD‐1), CD3, CD8, TIA‐1, and Foxp3. Median values were used as the cut‐off point
A comparison of OS according to tumor sidedness is shown in Figure 2. Low HLA class I expression was associated with better OS in the right‐sided CC group, but there was no significant difference in OS between those with high and with low HLA class I expression (P = .0689). High sPD‐L1 (P = .0053) and high CD3 expression (P = .0305) were correlated with better OS in the left‐sided CRC group.
FIGURE 2.
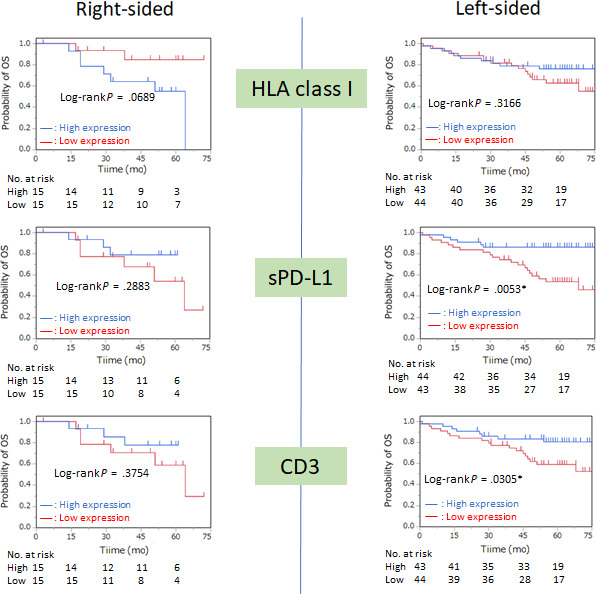
Kaplan‐Meier curves of overall survival (OS) according to location of colorectal tumor based on the expression of HLA class I, programmed cell death‐ligand 1 on stromal cells (sPD‐L1), and CD3. Median values were used as the cut‐off point
Kaplan‐Meier curves of DFS and OS in other biomarkers are shown in Figures [Link], [Link], [Link].
3.4. Comparison of clinicopathologic features and survival between tumor location in MMR proficiency
The clinicopathologic features between the 2 groups in MMR proficiency are summarized in Table 1. There were more elderly patients in the right‐sided CC group (P = .0145). The number of patients with positive lymph node involvement was significantly lower in the right‐sided group than in the left‐sided group (7 [29%] vs. 43 [52%], P = .0470). There were no other significant differences in patient characteristics between the 2 groups. Neither DFS nor OS was significantly different between the 2 groups (Figure S6).
TABLE 1.
Relationship between tumor location and clinicopathologic characteristics of DNA mismatch repair proficient (pMMR) colorectal cancer patients
| pMMR | ||||||
|---|---|---|---|---|---|---|
| Characteristic (n = 107) | Right‐sided (n = 24) | % | Left‐sided (n = 83) | % | Total | P value |
| Sex | ||||||
| Male | 15 | 63 | 56 | 67 | 71 | .0652 |
| Female | 9 | 38 | 27 | 33 | 36 | |
| Age (y), median (range) | ||||||
| ≤70 | 8 | 33 | 51 | 61 | 59 | .0145* |
| >70 | 16 | 67 | 32 | 39 | 48 | |
| Tumor depth | ||||||
| T1‐2 | 2 | 8 | 7 | 8 | 9 | .9875 |
| T3‐4 | 22 | 92 | 76 | 92 | 98 | |
| Lymph node metastasis | ||||||
| Negative | 17 | 71 | 40 | 48 | 57 | .0470* |
| Positive | 7 | 29 | 43 | 52 | 50 | |
| Tumor differentiation | ||||||
| Well/moderate | 21 | 88 | 73 | 88 | 94 | .9526 |
| Others | 3 | 12 | 10 | 12 | 13 | |
| Lymphatic invasion | ||||||
| Negative | 12 | 50 | 35 | 42 | 47 | .4971 |
| Positive | 12 | 50 | 48 | 58 | 60 | |
| Venous invasion | ||||||
| Negative | 5 | 21 | 20 | 24 | 29 | .7370 |
| Positive | 19 | 79 | 63 | 76 | 88 | |
| Perineural invasion | ||||||
| Negative | 20 | 83 | 64 | 77 | 84 | .5039 |
| Positive | 4 | 17 | 19 | 23 | 23 | |
| Adjuvant chemotherapy | ||||||
| No | 17 | 71 | 49 | 59 | 66 | .2885 |
| Yes | 7 | 29 | 34 | 41 | 41 | |
| Recurrence | ||||||
| No | 8 | 33 | 23 | 28 | 33 | .5963 |
| Yes | 16 | 67 | 60 | 72 | 84 | |
P < .05
3.5. Relationship between tumor location and IHC staining results in pMMR
A comparison of each IHC staining result between the 2 groups excluding dMMR is shown in Figure 3. There were more biomarkers with positive expression in left‐sided CRC, even in pMMR. The median expression of HLA class I and HLA class II was significantly different between right‐ and left‐sided CRC (39% vs. 77%, P = .0208 and 0.5% vs. 5%, P = .0498, respectively). The PD‐1, CD4, CD8, and CD163 infiltration was significantly more pronounced in left‐sided CRC (P = .0296, P = .0017, P = .0129, and P = .0025, respectively).
FIGURE 3.
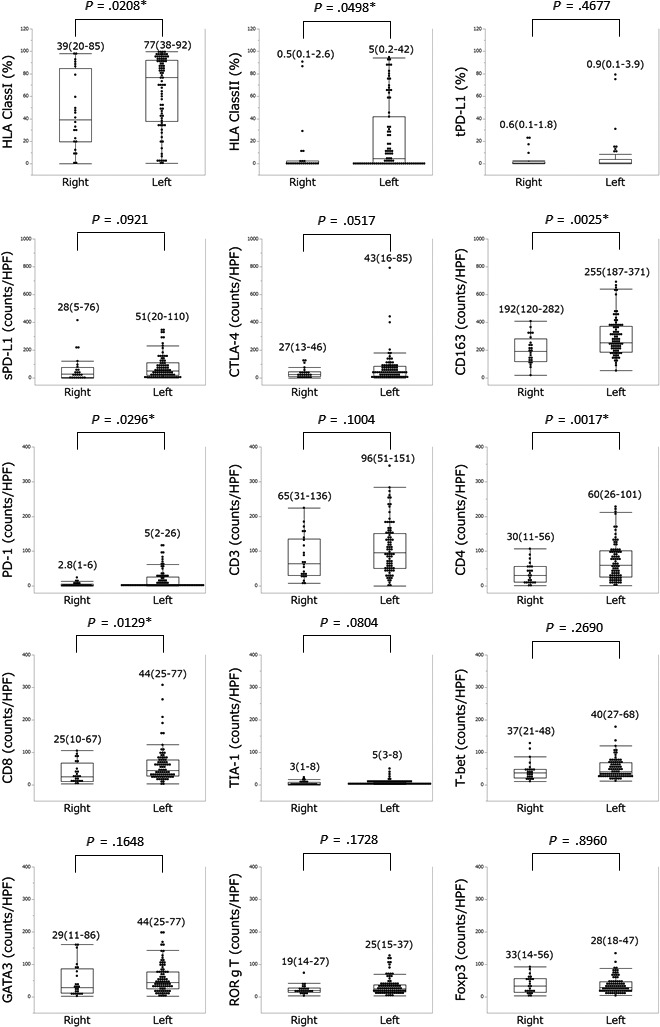
Molecular expression rate/number in right‐ and left‐sided colorectal tumors in DNA mismatch repair proficiency. Numbers above each plot represent the median and interquartile range. HPF, high power field; PD‐1, programmed cell death‐1; sPD‐L1, programmed cell death‐ligand 1 in stromal cells; tPD‐L1, programmed cell death‐ligand 1 in tumor cells
3.6. Comparison of DFS and OS curves between 2 groups in pMMR
A comparison of DFS between the 2 groups excluding dMMR is shown in Figure 4. In right‐sided CC, high expression of Foxp3 (P = .0055) and TIA‐1 (P = .0396) was associated with significantly better DFS. Patients with high CD8 (P = .0808) and CD3 (.0863) expression tended to have better DFS. In left‐sided CRC, only high sPD‐L1 expression (P = .0426) was associated with better DFS.
FIGURE 4.
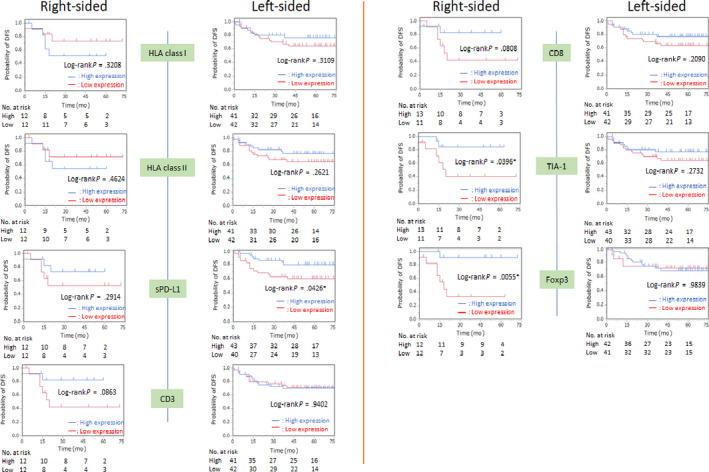
Kaplan‐Meier curves of disease‐free survival (DFS) according to location of colorectal tumor excluding DNA mismatch repair deficiency based on the expression of HLA class I, HLA class II, programmed cell death‐ligand 1 on stromal cells (sPD‐L1), CD3, CD8, TIA‐1, and Foxp3. Median values were used as the cut‐off point
A comparison of OS between the 2 groups excluding dMMR is shown in Figure 5. In right‐sided CC, high expression of CTLA‐4 (P = .0496) and Foxp3 (P = .0479) was associated with better OS. In left‐sided CRC, high expression of sPD‐L1 (P = .0204) and CD3 (P = .0486) correlated with better OS.
FIGURE 5.
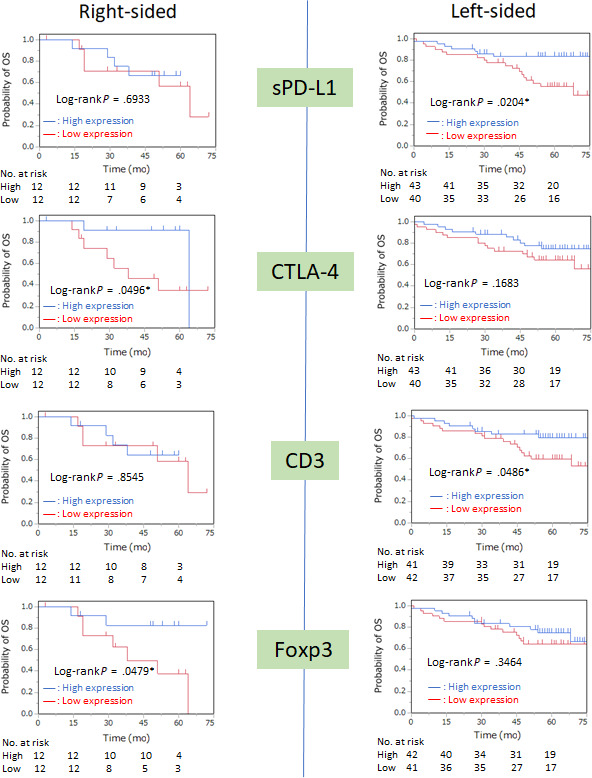
Kaplan‐Meier curves of overall survival (OS) according to location of colorectal tumor excluding DNA mismatch repair deficiency based on the expression of programmed cell death‐ligand 1 on stromal cells (sPD‐L1), CTLA‐4, CD3, and Foxp3. Median values were adopted as the cut‐off point
Kaplan‐Meier curves of DFS and OS according to the other biomarkers are shown in Figures [Link], [Link], [Link].
3.7. Univariate and multivariate analyses for DFS and OS in pMMR
The results of univariate and multivariate analyses of DFS and OS in pMMR are shown in Tables 2 and 3. Univariate analysis of DFS in right‐sided CC excluding dMMR showed that the prognostic factors were TIA‐1 (HR, 4.5348; 95% CI, 1.0431‐30.969; P = .0435) and Foxp3 (HR, 10.642; 95% CI, 1.8744‐199.65; P = .0052) expression. However, only Foxp3 expression (HR, 7.6445; 95% CI, 1.2091‐150.35; P = .0284) remained significant on multivariate analysis. For OS, CTLA‐4 expression (HR, 4.2365; 95% CI, 1.0184‐28.519; P = .0470) and Foxp3 expression (HR, 4.2911; 95% CI, 1.0283‐28.943; P = .0454) were significant factors on univariate analysis.
TABLE 2.
Uni‐ and multivariate analyses of disease‐free survival (DFS) in each tumor location in DNA mismatch repair proficiency
| DFS | ||||
|---|---|---|---|---|
| Univariate | Multivariate | |||
| HR(95% CI) | P‐value | HR (95% CI) | P‐value | |
| Right‐sided | ||||
| Sex (M vs F) | 0.6220 (0.1462‐2.6455) | .5058 | — | — |
| Age (≤70 vs 70<) | 2.5263 (0.5955‐10.718) | .1987 | — | — |
| Tumor depth (T1‐2 vs T3‐4) | 1.4230 (0.0760‐8.0387) | .7525 | — | — |
| Lymph node metastasis (− vs +) | 0.5232 (0.1279‐2.5580) | .3924 | — | — |
| Tumor differentiation (well/mod vs others) | 0.3386 (0.0772‐2.3240) | .2318 | — | — |
| Lymphatic invasion (− vs +) | 0.5531 (0.1132‐2.2516) | .4080 | — | — |
| Venous invasion (− vs +) | 1.9345 (0.3957‐7.9113) | .3839 | — | — |
| Perineural invasion (− vs +) | not estimated | .1240 | — | — |
| HLA classI (low vs high) | 0.4950 (0.1012‐2.0265) | .3279 | — | — |
| HLA classII (low vs high) | 0.5922 (0.1214‐2.4163) | .4663 | — | — |
| tPD‐L1 (low vs high) | 1.0683 (0.2509‐4.5490) | .9260 | — | — |
| sPD‐L1 (low vs high) | 2.1146 (0.5135‐10.407) | .2993 | — | — |
| PD‐1 (low vs high) | 2.9767 (0.6845‐20.334) | .1516 | — | — |
| CTLA‐4 (low vs high) | 0.9789 (0.2312‐4.1445) | .9760 | — | — |
| CD3 (low vs high) | 3.6488 (0.8367‐24.968) | .0868 | — | — |
| CD4 (low vs high) | 2.1619 (0.5256‐10.629) | .2848 | — | — |
| CD8 (low vs high) | 3.7213 (0.8541‐25.451) | .0817 | — | — |
| TIA‐1 (low vs high) | 4.5348 (1.0431‐30.969) | .0435* | 2.3141 (0.4948‐16.784) | 0.3010 |
| T‐bet (low vs high) | 0.6873 (0.1409‐2.8030) | .6032 | — | — |
| GATA3 (low vs high) | 3.4926 (0.7988‐23.938) | .0992 | — | — |
| Foxp3 (low vs high) | 10.642 (1.8744‐199.65) | .0052* | 7.6445 (1.2091‐150.35) | 0.0284* |
| RorγT (low vs high) | 1.1726 (0.2772‐4.9607) | .8220 | — | — |
| CD163 (low vs high) | 1.7702 (0.4341‐8.6335) | .4264 | — | — |
| Left‐sided | ||||
| Sex (M vs F) | 1.3592 (0.5645‐3.7677) | .5085 | — | — |
| Age (≤70 vs 70<) | 1.3729 (0.5858‐3.5732) | .4763 | — | — |
| Tumor depth (T1‐2 vs T3‐4) | 0.3869 (0.0216‐1.8445) | .2809 | — | — |
| Lymph node metastasis (N− vs N+) | 0.4422 (0.1778‐1.0194) | .0556 | — | — |
| Tumor differentiation (well/mod vs others) | 0.1198 (0.0498‐0.3190) | .0001* | 0.0975 (0.0289‐0.3185) | 0.0002* |
| Lymphatic invasion (− vs +) | 0.3804 (0.1370‐0.9178) | .0308* | 0.5718 (0.1832‐1.5692) | 0.2860 |
| Venous invasion (− vs +) | 0.2495 (0.0399‐0.851) | .0237* | 0.1751 (0.0234‐0.7904) | 0.0213* |
| Perineural invasion (− vs +) | 0.3462 (0.1510‐0.8357) | .0197* | 0.6546 (0.2561‐1.7916) | 0.3959 |
| HLA classI (low vs high) | 1.5318 (0.6718‐3.6781) | .3127 | — | — |
| HLA classII (low vs high) | 1.6024 (0.7023‐3.8497) | .2642 | — | — |
| tPD‐L1 (low vs high) | 1.2984 (0.5694‐3.1178) | .5379 | — | — |
| sPD‐L1 (low vs high) | 2.3519 (1.0210‐5.8450) | .0445* | 1.9256 (0.7099‐5.2939) | 0.1952 |
| PD‐1 (low vs high) | 1.1986 (0.5253‐2.7622) | .6642 | — | — |
| CTLA‐4 (low vs high) | 1.6485 (0.7246‐3.8642) | .2325 | — | — |
| CD3 (low vs high) | 0.9694 (0.4205‐2.2126) | .9406 | — | — |
| CD4 (low vs high) | 1.3970 (0.6141‐3.2749) | .4248 | — | — |
| CD8 (low vs high) | 1.6932 (0.7425‐4.0657) | .2117 | — | — |
| TIA‐1 (low vs high) | 1.5849 (0.6950‐3.8055) | .2753 | — | — |
| T‐bet (low vs high) | 0.7746 (0.3306‐1.7611) | .5421 | — | — |
| GATA3 (low vs high) | 1.9569 (0.8498‐4.8618) | .1159 | — | — |
| Foxp3 (low vs high) | 1.0084 (0.4373‐2.3027) | .9841 | — | — |
| RorγT (low vs high) | 1.6612 (0.7281‐3.9908) | .2292 | — | — |
| CD163 (low vs high) | 1.3319 (0.5856‐3.1216) | .4940 | — | — |
Abbreviations: CI, confidence interval; DFS, disease ‐ free survival; F, female; HR, hazard ratio; M, male; mod, moderate; PD‐1, programmed cell death‐1; sPD‐L1, programmed cell death‐ligand 1 in stromal cells; tPD‐L1, programmed cell death‐ligand 1 in tumor cells.
P < .05.
TABLE 3.
Uni‐ and multivariate analyses of overall survival (OS) in each tumor location in DNA mismatch repair proficiency
| OS | ||||
|---|---|---|---|---|
| Univariate | Multivariate | |||
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Right‐sided | ||||
| Sex (M vs F) | 1.4708 (0.3868‐6.9864) | .5792 | — | — |
| Age, y (≤70 vs 70<) | 0.5468 (0.0792‐2.4145) | .4446 | — | — |
| Tumor depth (T1‐2 vs T3‐4) | Not estimated | .1556 | — | — |
| Lymph node metastasis (− vs +) | 0.5803 (0.1414‐2.8439) | .4705 | — | — |
| Tumor differentiation (well/mod vs others) | 0.9602 (0.1691‐18.012) | .9700 | — | — |
| Lymphatic invasion (− vs +) | 1.8034 (0.4408‐8.8175) | .4125 | — | — |
| Venous invasion (− vs +) | 0.4018 (0.0215‐2.2668) | .3418 | — | — |
| Perineural invasion (− vs +) | 0.3743 (0.0858‐2.5630) | .2726 | — | — |
| HLA class I (low vs high) | 0.7919 (0.1958‐2.9964) | .7275 | — | — |
| HLA class II (low vs high) | 1.3584 (0.3589‐5.4955) | .6471 | — | — |
| tPD‐L1 (low vs high) | 0.8374 (0.2068‐3.1734) | .7913 | — | — |
| sPD‐L1 (low vs high) | 1.3198 (0.3102‐5.6158) | .6966 | — | — |
| PD‐1 (low vs high) | 1.4554 (0.3550‐7.1293) | .6043 | — | — |
| CTLA‐4 (low vs high) | 4.2365 (1.0184‐28.519) | .0470* | 3.4053 (0.7683‐23.727) | .1105 |
| CD3 (low vs high) | 1.1376 (0.2685‐4.8206) | .8556 | — | — |
| CD4 (low vs high) | 1.3014 (0.3060‐5.5363) | .7112 | — | — |
| CD8 (low vs high) | 1.0485 (0.2473‐4.4458) | .9467 | — | — |
| TIA‐1 (low vs high) | 1.5232 (0.4013‐6.1790) | .5303 | — | — |
| T‐bet (low vs high) | 1.7270 (0.4546‐7.0084) | .4155 | — | — |
| GATA3 (low vs high) | 3.6282 (0.8704‐24.463) | .0787 | — | — |
| Foxp3 (low vs high) | 4.2911 (1.0283‐28.943) | .0454* | 3.4420 (0.7787‐23.947) | .1066 |
| RorγT (low vs high) | 1.5860 (0.4180‐6.4309) | .4914 | — | — |
| CD163 (low vs high) | 1.4638 (0.3854‐5.9416) | .5701 | — | — |
| Left‐sided | ||||
| Sex (M vs F) | 0.8082 (0.3679‐1.9001) | .6104 | — | — |
| Age, y (≤70 vs 70<) | 0.5784 (0.2648‐1.2825) | .1739 | — | — |
| Tumor depth (T1‐2 vs T3‐4) | 0.3058 (0.0171‐1.4537) | .1614 | — | — |
| Lymph node metastasis (N− vs N+) | 0.3339 (0.1303‐0.7605) | .0082* | 0.6190 (0.2189‐1.6265) | .3344 |
| Tumor differentiation (well/mod vs others) | 0.1891 (0.0800‐0.4964) | .0014* | 0.2895 (0.1018‐0.8644) | .0274* |
| Lymphatic invasion (− vs +) | 0.2622 (0.0873‐0.6459) | .0027* | 0.3336 (0.1037‐0.9127) | .0318* |
| Venous invasion (− vs +) | 0.3366 (0.0796‐0.9727) | .0437* | 0.3776 (0.0843‐1.2222) | .1090 |
| Perineural invasion (− vs +) | 0.3405 (0.1538‐0.7864) | .0129* | 0.7228 (0.2996‐1.7867) | .4742 |
| HLA class I (low vs high) | 1.6634 (0.7644‐3.8001) | .2011 | — | — |
| HLA class II (low vs high) | 1.1777 (0.5421‐2.5977) | .6782 | — | — |
| tPD‐L1 (low vs high) | 0.5936 (0.2626‐1.3081) | .1949 | — | — |
| sPD‐L1 (low vs high) | 2.5843 (1.1600‐6.3016) | .0196* | 1.7512 (0.6893‐4.6548) | .2393 |
| PD‐1 (low vs high) | 2.1281 (0.9656‐5.0206) | .0612 | — | — |
| CTLA‐4 (low vs high) | 1.7347 (0.7948‐3.9707) | .1676 | — | — |
| CD3 (low vs high) | 2.2688 (1.0144‐5.5475) | .0460* | 2.3560 (0.9905‐6.0541) | .0526 |
| CD4 (low vs high) | 1.5236 (0.6964‐3.4939) | .2941 | — | — |
| CD8 (low vs high) | 1.6338 (0.7483‐3.7475) | .2197 | — | — |
| TIA‐1 (low vs high) | 1.1064 (0.5106‐2.4350) | .7970 | — | — |
| T‐bet (low vs high) | 1.0622 (0.4836‐2.3383) | .8792 | — | — |
| GATA3 (low vs high) | 1.0641 (0.4911‐2.3413) | .8745 | — | — |
| Foxp3 (low vs high) | 1.4531 (0.6679‐3.2590) | .3464 | — | — |
| RorγT (low vs high) | 1.5666 (0.7171‐3.5898) | .2623 | — | — |
| CD163 (low vs high) | 1.2735 (0.5856‐2.8553) | .5428 | — | — |
Abbreviations: CI, confidence interval; F, female; HR, hazard ratio; M, male; mod, moderate; PD‐1, programmed cell death‐1; sPD‐L1, programmed cell death‐ligand 1 in stromal cells; tPD‐L1, programmed cell death‐ligand 1 in tumor cells.
P < .05
Univariate analysis of DFS in left‐sided CRC showed that tumor differentiation (HR, 0.1198; 95% CI, 0.0498‐0.3190; P = .0001), lymphatic invasion (HR, 0.3804; 95% CI, 0.1370‐0.9178; P = .0308), venous invasion (HR, 0.2495; 95% CI, 0.0399‐0.8510; P = .0237), perineural invasion (HR, 0.3462; 95% CI, 0.1510‐0.8357; P = .0197), and sPD‐L1 expression (HR, 2.3519; 95% CI, 1.0210‐5.8450; P = .0445) were prognostic factors. However, only tumor differentiation (HR, 0.0975; 95% CI, 0.0289‐0.3185; P = .0002) and venous invasion (HR, 0.1751; 95% CI, 0.0234‐0.7904; P = .0213) remained significant on multivariate analysis. For OS, lymph node metastasis (HR, 0.3339; 95% CI, 0.1303‐0.7605; P = .0082), tumor differentiation (HR, 0.1891; 95% CI, 0.0800‐0.4964; P = .0014), lymphatic invasion (HR, 0.2622; 95% CI, 0.0873‐0.6459; P = .0027), venous invasion (HR, 0.3366; 95% CI, 0.0796‐0.9727; P = .0437), perineural invasion (HR, 0.3405; 95% CI, 0.1538‐0.7864; P = .0129), sPD‐L1 expression (HR, 2.5843; 95% CI, 1.1600‐6.3016; P = .0196), and CD3 expression (HR, 2.2688; 95% CI, 1.0144‐5.5475; P = .0460) were the significant factors of prognosis on univariate analysis. On multivariate analysis, only tumor differentiation (HR, 0.2895; 95% CI, 0.1018‐0.8644; P = .0274) and lymphatic invasion (HR, 0.3336; 95% CI, 0.1037‐0.9127; P = .0318) remained significant.
4. DISCUSSION
The present study investigated the differences in immunosurveillance pattern between right‐sided and left‐sided CRC and analyzed their association with clinicopathologic features, including clinical outcomes. The results showed that the immunosurveillance pattern differed between right‐sided and left‐sided CRC. We found lower HLA class I expression in right‐sided CC and higher CD4, RORγT, and CD163 expression in left‐sided CRC (Figure 3). Log‐rank test showed that low HLA class I, high PD‐1, high CD3, high CD8, high TIA‐1, and high Foxp3 expression were associated with better DFS in right‐sided CC, whereas only high sPD‐L1 and CD3 expression were associated with better DFS in left‐sided CRC. Additionally, analyses excluding dMMR showed a significant difference in the immunosurveillance pattern according to tumor sidedness even in pMMR. Right‐sided CC presented lower HLA class I, HLA class II, PD‐1, CD4, CD8, and CD163 expression than left‐sided CRC. Log‐rank test showed that high expression of CD3, CD8, TIA‐1, and Foxp3 was associated with better DFS in right‐sided CC, whereas high sPD‐L1 and CD3 expression was associated with better DFS in left‐sided CRC. Multivariate analysis of DFS in right‐sided CC excluding dMMR showed that high Foxp3 expression was the only prognostic factor. Despite the more profound immune cell infiltration in left‐sided CRC, immunity had a stronger prognostic influence in right‐sided CC.
MMR deficiency markedly affects tumor immunity and some dMMR‐induced mutations create cancer neoantigens that can be targeted by the immune cells. MMR deficiency is more common in right‐sided than in left‐sided CRC. 17 We analyzed the 2 groups excluding dMMR and found differences even in pMMR. This suggests that tumor immunity is not only affected by the genetic background but also by the tumor location itself. Other methylator phenotypes, such as CIMP, might influence these differences. However, Takahashi et al reported no difference in the frequency of DNA methylation status between right‐ and left‐sided CRC in the microsatellite stable group. 18 Additionally, CIMP‐H frequently occurs simultaneously with MSI‐H. 19
We also found that high expression of the TILs (CD3, CD8, TIA‐1, and Foxp3) indicates better prognosis in right‐sided CC, which is partly in line with the results of some studies. 20 , 21 , 22 Berntsson et al and Zhang et al reported that a high number of PD‐1, CD3, and/or CD8 is related to better prognosis in right‐sided CC. High Foxp3 expression is generally associated with poor prognosis in various carcinomas. 23 , 24 However, some studies reported that high number of Foxp3 expression is associated with favorable prognosis in CRC. 20 , 25 , 26 The reason for the association of high Foxp3 expression with good prognosis only in right‐sided remains unknown.
High sPD‐L1 expression indicates good prognosis in left‐sided CRC, but the influence of PD‐1/PD‐L1 expression in CRC prognosis remains controversial. 21 , 27 , 28 , 29 , 30 , 31 Keir et al suggested that PD‐L1 might be upregulated in activated macrophages and dendritic cells. 32 , 33 Spranger et al also reported that interferon‐γ secreted by the activated lymphocytes and macrophages induces PD‐L1 upregulation on the cell surface. 34 Thus, high sPD‐L1 expression could reflect immune activation.
Snyder et al 35 reported that the efficacy of immune checkpoint inhibitors in melanoma differs according to the number of infiltrative CD8 in the tumor adjacent tissue. In the present study, the immune environment varied according to the tumor location, even when excluding dMMR. This shows that the impact of immune checkpoint inhibitors or any other immune therapies for CRC could vary with the tumor’s anatomical site. Therefore, the tumor location should be considered during immune therapy.
The IHC analyses in this study were undertaken using TMA. Although the validity of IHC analysis of TMA might be controversial, there have been several IHC studies using TMA since it was first introduced. 36 , 37 In the present study, we evaluated some biomarkers through whole‐section staining in 10 cases and confirmed the significant correlation between TMA and whole‐section staining (Figure S10). The results of the present study were concordant with the other previous reports that high number of TILs showed better survival, especially in right‐sided tumors. 20 , 21 , 22 The use of IHC analyses through TMA in the present study was considered to be validated.
This study has some limitations. First, there is a potential risk of selection bias due to the single‐center, retrospective study design. Our findings should be validated in a larger cohort study. Second, other immune‐related proteins should have been considered for a more comprehensive analyses of TME. Finally, only proteins were evaluated in the present study. mRNA and gene analyses, including CIMP and BRAF, are needed to validate our results.
In conclusion, the immunosurveillance pattern and its influence on the clinical outcome differs between right‐sided and left‐sided CRC, even in pMMR. Therefore, planning the treatment according to the tumor location could improve the prognosis of CRC patients.
CONFLICT OF INTEREST
None declared.
ETHICAL APPROVAL
This study was approved by the Research Ethics Committee of Kurume University (Approval number: 399) and was conducted in accordance with the Declaration of Helsinki. The need for informed consent was waived owing to the retrospective nature of the study.
Supporting information
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Fig S7
Fig S8
Fig S9
Fig S10
Table S1
ACKNOWLEDGMENTS
The authors thank Mayumi Miura, Kanako Miyazaki, and Chie Kuroki for their technical assistance. We also thank Editage for editing the manuscript.
Kanno H, Miyoshi H, Yoshida N, et al. Differences in the immunosurveillance pattern associated with DNA mismatch repair status between right-sided and left-sided colorectal cancer. Cancer Sci. 2020;111:3032–3044. 10.1111/cas.14495
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7‐30. [DOI] [PubMed] [Google Scholar]
- 2. Gao XH, Yu GY, Gong HF, et al. Differences of protein expression profiles, KRAS and BRAF mutation, and prognosis in right‐sided colon, left‐sided colon and rectal cancer. Sci Rep. 2017;7(1):7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee GH, Malietzis G, Askari A, Bernardo D, Al‐Hassi HO, Clark SK. Is right‐sided colon cancer different to left‐sided colorectal cancer? ‐ a systematic review. Eur J Surg Oncol. 2015;41(3):300‐308. [DOI] [PubMed] [Google Scholar]
- 4. Tejpar S, Stintzing S, Ciardiello F, et al. Prognostic and predictive relevance of primary tumor location in patients with RAS wild‐type metastatic colorectal cancer: retrospective analyses of the CRYSTAL and FIRE‐3 trials. JAMA Oncol. 2017;3(2):194‐201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arnold D, Lueza B, Douillard JY, et al. Prognostic and predictive value of primary tumour side in patients with RAS wild‐type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Ann Oncol. 2017;28(8):1713‐1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pagès F, Berger A, Camus M, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;353(25):2654‐2666. [DOI] [PubMed] [Google Scholar]
- 7. Galon J, Costes A, Sanchez‐Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313(5795):1960‐1964. [DOI] [PubMed] [Google Scholar]
- 8. Powell AG, Horgan PG, Edwards J. The bodies fight against cancer: is human leucocyte antigen (HLA) class 1 the key? J Cancer Res Clin Oncol. 2012;138(5):723‐728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sconocchia G, Eppenberger‐Castori S, Zlobec I, et al. HLA class II antigen expression in colorectal carcinoma tumors as a favorable prognostic marker. Neoplasia. 2014;16(1):31‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yoshida N, Kinugasa T, Miyoshi H, et al. A high RORgammaT/CD3 ratio is a strong prognostic factor for postoperative survival in advanced colorectal cancer: analysis of helper T cell lymphocytes (Th1, Th2, Th17 and Regulatory T Cells). Ann Surg Oncol. 2016;23(3):919‐927. [DOI] [PubMed] [Google Scholar]
- 11. Watson NF, Ramage JM, Madjd Z, et al. Immunosurveillance is active in colorectal cancer as downregulation but not complete loss of MHC class I expression correlates with a poor prognosis. Int J Cancer. 2006;118(1):6‐10. [DOI] [PubMed] [Google Scholar]
- 12. Zeestraten EC, Reimers MS, Saadatmand S, et al. Combined analysis of HLA class I, HLA‐E and HLA‐G predicts prognosis in colon cancer patients. Br J Cancer. 2014;110(2):459‐468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reimers MS, Engels CC, Putter H, et al. Prognostic value of HLA class I, HLA‐E, HLA‐G, and Tregs in rectal cancer: a retrospective cohort study. BMC Cancer. 2014;14:486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoon YS, Yu CS, Kim TW, et al. Mismatch repair status in sporadic colorectal cancer: immunohistochemistry and microsatellite instability analyses. J Gastroenterol Hepatol. 2011;26(12):1733‐1739. [DOI] [PubMed] [Google Scholar]
- 15. Green AR, Aleskandarany MA, Ali R, et al. Clinical impact of tumor DNA repair expression and T‐cell infiltration in breast cancers. Cancer Immunol Res. 2017;5(4):292‐299. [DOI] [PubMed] [Google Scholar]
- 16. Williams DS, Mouradov D, Jorissen RN, et al. Lymphocytic response to tumour and deficient DNA mismatch repair identify subtypes of stage II/III colorectal cancer associated with patient outcomes. Gut. 2019;68:465‐474. [DOI] [PubMed] [Google Scholar]
- 17. Li P, Xiao Z, Braciak TA, Ou Q, Chen G, Oduncu FS. A relationship to survival is seen by combining the factors of mismatch repair status, tumor location and age of onset in colorectal cancer patients. PLoS One. 2017;12(3):e0172799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takahashi Y, Sugai T, Habano W, et al. Molecular differences in the microsatellite stable phenotype between left‐sided and right‐sided colorectal cancer. Int J Cancer. 2016;139(11):2493‐2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jia M, Jansen L, Walter V, et al. No association of CpG island methylator phenotype and colorectal cancer survival: population‐based study. Br J Cancer. 2016;115(11):1359‐1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berntsson J, Svensson MC, Leandersson K, et al. The clinical impact of tumour‐infiltrating lymphocytes in colorectal cancer differs by anatomical subsite: A cohort study. Int J Cancer. 2017;141(8):1654‐1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Berntsson J, Eberhard J, Nodin B, Leandersson K, Larsson AH, Jirström K. Expression of programmed cell death protein 1 (PD‐1) and its ligand PD‐L1 in colorectal cancer: Relationship with sidedness and prognosis. Oncoimmunology. 2018;7(8):e1465165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang L, Zhao Y, Dai Y, et al. Immune landscape of colorectal cancer tumor microenvironment from different primary tumor location. Front Immunol. 2018;9:1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Qian F, Qinging Y, Linguan W, et al. High tumor‐infiltrating FoxP3+ T cells predict poor survival in estrogen receptor‐positive breast cancer: A meta‐analysis. Eur J Surg Oncol. 2017;43(7):1258‐1264. [DOI] [PubMed] [Google Scholar]
- 24. Song JJ, Zhao SJ, Fang J, et al. Foxp3 overexpression in tumor cells predicts poor survival in oral squamous cell carcinoma. BMC Cancer. 2016;16:530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Salama P, Phillips M, Grieu F, et al. Tumor‐infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer. J Clin Oncol. 2009;27(2):186‐192. [DOI] [PubMed] [Google Scholar]
- 26. Ladoire S, Martin F, Ghiringhelli F. Prognostic role of FOXP3+ regulatory T cells infiltrating human carcinomas: the paradox of colorectal cancer. Cancer Immunol Immunother. 2011;60(7):909‐918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang L, Ren F, Wang Q, et al. Significance of Programmed Death Ligand 1 (PD‐L1) Immunohistochemical Expression in Colorectal Cancer. Mol Diagn Ther. 2016;20(2):175‐181. [DOI] [PubMed] [Google Scholar]
- 28. Enkhbat T, Nishi M, Takasu C, et al. Programmed cell death ligand 1 expression is an independent prognostic factor in colorectal cancer. Anticancer Res. 2018;38(6):3367‐3373. [DOI] [PubMed] [Google Scholar]
- 29. Koganemaru S, Inoshita N, Miura Y, et al. Prognostic value of programmed death‐ligand 1 expression in patients with stage III colorectal cancer. Cancer Sci. 2017;108(5):853‐858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee KS, Kim BH, Oh HK, et al. Programmed cell death ligand‐1 protein expression and CD274/PD‐L1 gene amplification in colorectal cancer: Implications for prognosis. Cancer Sci. 2018;109(9):2957‐2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wyss J, Dislich B, Koelzer VH, et al. Stromal PD‐1/PD‐L1 expression predicts outcome in colon cancer patients. Clin Colorectal Cancer. 2019;18(1):e20‐e38. [DOI] [PubMed] [Google Scholar]
- 32. Keir ME, Liang SC, Guleria I, et al. Tissue expression of PD‐L1 mediates peripheral T cell tolerance. J Exp Med. 2006;203(4):883‐895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Keir ME, Francisco LM, Sharpe AH. PD‐1 and its ligands in T‐cell immunity. Curr Opin Immunol. 2007;19(3):309‐314. [DOI] [PubMed] [Google Scholar]
- 34. Spranger S, Spaapen RM, Zha Y, et al. Up‐regulation of PD‐L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med. 2013;5(200):200ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA‐4 blockade in melanoma. N Engl J Med. 2014;371(23):2189‐2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high‐throughput molecular profiling of tumor specimens. Nat Med. 1998;4(7):844‐847. [DOI] [PubMed] [Google Scholar]
- 37. Nocito A, Kononen J, Kallioniemi OP, Sauter G. Tissue microarrays (TMAs) for high‐throughput molecular pathology research. Int J Cancer. 2001;94(1):1‐5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Fig S7
Fig S8
Fig S9
Fig S10
Table S1


