Abstract
Recent advances in optical mapping have allowed the construction of improved genome assemblies with greater contiguity. Optical mapping also enables genome comparison and identification of large-scale structural variations. Association of these large-scale genomic features with biological functions is an important goal in plant and animal breeding and in medical research. Optical mapping has also been used in microbiology and still plays an important role in strain typing and epidemiological studies. Here, we review the development of optical mapping in recent decades to illustrate its importance in genomic research. We detail its applications and algorithms to show its specific advantages. Finally, we discuss the challenges required to facilitate the optimization of optical mapping and improve its future development and application.
Abbreviations: bp, base pair; DBG, de Bruijn graph; DLS, direct label and strain; DNA, deoxyribonucleic acid; Hi-C, high-throughput chromosome conformation capture; kb, kilobase pair; Mb, million base pair; OLC, overlap-layout-consensus; PacBio, Pacific Biosciences; PCR, polymerase chain reaction; SRS, short-read sequencing; SV, structural variation; 3D, three-dimensional
Keywords: Optical mapping, Genome assembly, Structural variation, Next generation sequencing
1. Introduction
Short-read sequencing (SRS) has made genomic and genetic studies in various species affordable [1], [2], but the limited sequence lengths generated by SRS platforms prevent the reads from spanning most repetitive and complex regions, leading to fragmented and collapsed genome assemblies [2], [3]. The use of assemblies based on SRS can therefore reduce the accuracy of downstream analyses, such as those used for the detection of genomic variations [1], [4]. Although different methods and computational algorithms have been developed to solve this problem [5], [6], [7], [8], [9], it is difficult to fully overcome the deficiencies inherited from the short read length.
Long-read sequencing provided by Pacific Biosciences (PacBio) and Oxford Nanopore technologies now help to cover most repeats and produces more complete genome assemblies [10], [11], [12], [13]. However, the read lengths (~15 kb on average) are still insufficient to cover some large repetitive and complex genomic regions [14], [15], thus hampering biological studies of these regions. In addition, while PacBio or Oxford Nanopore sequencing can produce substantial long-sequence reads for prokaryote genome assembly, for eukaryotic genomes, the reads can generally only be assembled to the scaffold-level, but not to the chromosome-level [16].
Short-read long-insert technologies, linked-read sequencing technologies, high-throughput chromosome conformation capture (Hi-C) technologies and optical mapping are used to solve these assembly problems [10], [17]. Extended from the concept of paired-end reads to contain a fixed length insertion between two sequences at the ends of a DNA fragment, the mate-pair and similar methods construct libraries joining ends of circularized longer fragments (2–5 kb) in single reads to resolve adjacent sequences at longer intervals. Similarly, linked-read sequencing technologies, such as those provided by 10x Genomics, and Hi-C technologies such as those provided by Dovetail Genomics and Phase Genomics, use fluidics and other ligation methods to capture proximity information. The variations in library preparation ultimately rely on short-read sequencing [17]. Although the cost of these technologies is relatively low, sequencing biases, such as those caused by PCR amplification and enzyme selection [18], [19], can make these technologies error-prone [10]. Moreover, Burton et al. [20] and Bickhart et al. [21] found that Hi-C technologies can lead to mis-assembly, such as false inversion and scaffold misplacement, which has to be corrected with data from orthogonal methods such as radiation hybrid maps. Compared to single-molecule technologies, linked reads have coverage drawbacks as they cannot fully cover the source DNA fragments. When using linked reads, they can introduce gaps particularly during eukaryotic genome assembly [22]. Additionally, the current linked-read sequencing methods have a limited ability to process polyploid genomes [23], [24].
In contrast to the above technologies used to capture linkage information or to determine 3D genome architecture, optical mapping uses a light microscope-based technique to physically locate specific enzymes or sequence motifs to produce DNA sequence fingerprints [25]. The resulting optical maps contain only the physical locations of selected enzymes, rather than base-by-base nucleotide information. Specifically, during the generation of optical maps, selected enzymes are used to fluorescently label DNA molecules, and images of the fluorescent signal patterns are then used to produce maps.
The average molecule length of optical maps (~225 kb) is substantially greater than the read length produced by short-read sequencing (typically 150–300 bp) and long-read sequencing (typically ~ 15 kb on average) [26]. Optical maps can easily span genomic regions that are difficult to resolve by DNA sequencing. The physical location and relative separation of the restriction/labelling sites obtained from optical mapping facilitate genome scaffolding, genome-assembly completeness validation, and large-scale structural variation detection [27].
Optical mapping is intensively used in genomic studies of microorganisms, plants, animals, and human diseases [10], [28]. Ongoing efforts to improve optical mapping technology will enable further increases in the accuracy of genomic research. Here we review the development of optical mapping, detail its algorithms and applications, and emphasize future perspectives and applications for optical mapping that could assist genomic research.
2. Development of optical mapping
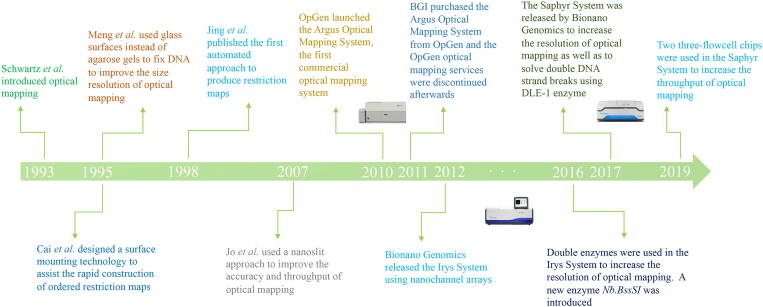
Since being pioneered by Schwartz et al. [25] in 1993, the technology used in optical mapping has been improved substantially (Fig. 1). Initially, optical mapping was used to assist genome assembly of microorganisms, such as yeast and bacteria [29]. However, due to its low throughput, high error rate, and imprecise DNA-length measurement, optical mapping has not been widely adopted for genomic studies in higher species [14]. Recent advances in technologies and algorithms have led to further improvements in optical mapping, making it an important auxiliary tool in genomic research.
Fig 1.
Optical mapping milestone timeline. Major breakthroughs in optical mapping over the past two decades are displayed.
2.1. History of optical mapping
In 1993, Schwartz et al. [25] demonstrated the use of optical mapping by constructing ordered restriction maps of Saccharomyces cerevisiae chromosomes. First, the stained and stretched DNA molecules were fixed in agarose gel and then nicked by a selected restriction enzyme. Next, specialized fluorescence microscopy was used to observe and record the specific patterns of single-molecule DNA restriction fragments. However, it was later found that the thickness of agarose gel can affect image capture by scattering light and attenuating signals.
To solve this problem, in 1995 Cai et al. [30] and Meng et al. [31] fixed DNA molecules on glass surfaces that had been treated with specific chemicals. Consequently, the DNA stretch rate was improved to 60% of its contour length, compared with 30% on agarose [29]. The number of cut sites was also increased. To increase throughput, in 1998 Jing et al. [32] used the “fluid fixation” effect in their design of the first automated approach to generate restriction maps. The improvement of microfluidics and chemistries led to further advances in optical mapping, and in 2007, Jo et al. [33] reported a nanoslit approach that had higher accuracy and throughput in restriction map production.
Throughout this time, companies such as OpGen, Bionano Genomics, and NABsys have invested intensively in single-molecule whole-genome mapping and have extended its application from microorganisms to more complex organisms. OpGen and Bionano Genomics have commercially released different platforms to produce optical maps, such as Argus from the former and Irys and Saphyr from the latter. However, with the acquisition of the Argus platform by BGI in 2011, OpGen’s optical mapping services were terminated. Bionano Genomics currently dominates the optical mapping market and continues to invest in technology development. Yet, given the pending release of the HD-mapping system from NABsys, it is likely that some of the Bionano Genomics market share will be taken by NABsys.
2.1.1. OpGen optical mapping
OpGen was licensed its technology from New York University and the University of Wisconsin and was the first company to commercialize optical mapping technology [34], represented by its release of the Argus platform and its accompanying analysis software, MapSolver [34], [35], [36]. OpGen applied the original restriction-based method for map construction to produce maps of 200 kb in length [10], [29], [36]. At first, Argus targeted simpler organisms, such as bacteria and yeast. Later, OpGen expanded its support to higher organisms across animals and plants, including humans, domestic goat and legume Medicago [34], [36], [37], [38]. A global alignment strategy was applied in the design of MapSolver to help detect complex genome rearrangements [36], [39]. However, with the sale of the Argus system, OpGen discontinued its optical mapping services, and its optical mapping market share was then assumed by its competitors.
2.1.2. Bionano optical mapping
Bionano Genomics is another optical mapping technology provider, originally named BionanoMatrix. The company rose to popularity in 2012 after the release of its massively parallel Irys platform, which applies nanochannel-array fluidics technology to produce optical maps [40]. This technology greatly enhances the throughput and the accuracy of molecule length estimation by use of a more uniform linearization [14], [40], and the use of nicking enzymes to create only single strand breaks preserve molecule contiguity more than OpGen technologies do. The Irys platform came bundled with the IrysView desktop application for data visualization and management. Bionano also provides its proprietary RefAligner for map alignment and Assembler for de novo map assembly, together with an IrysSolve Scripts package to assist data analysis. In 2016, two maps generated from the use of different nicking enzymes were added to the Irys workflow to complement the limited label density and increase map coverage. However, double strand breaks at “fragile sites” where two nicking sites locate closely (~400 bp) on opposite strands still hamper the contiguity of optical maps.
Bionano recently developed Direct Label and Strain (DLS) technology to improve map contiguity to the chromosome-level [41]. The release of the DLS protocol was coupled with the new Saphyr platform, which has improved optics and throughput. DLE-1, the first enzyme in the DLS family, functions akin to a methyltransferase by labelling DNA without damaging it [29], [42]. DLE-1 eliminates the “fragile site” problem [43]. Currently, the Saphyr platform guarantees a production of 1300 Gb of raw data per flow-cell for human samples. The resolution of Bionano optical mapping has been increased up to 500 bp, and there have been attempts to develop algorithms to enable a resolution of 10 bp [40], [44]. Bionano has also updated their data analysis software to a web application supported by Bionano Solve and Bionano Access, to streamlining bioinformatics analysis and data management.
The Bionano Tools are freely accessible by the public. There is constant advancement of instruments and chemistries, and the software is continuously updated with new features by Bionano Genomics, whilst the core components (RefAligner and Assembler) remain proprietary.
2.1.3. Nabsys mapping
In contrast to OpGen and Bionano Genomics, NABsys is not like a traditional optical-mapping technology provider. NABsys eschews image capture and conversion in their technology and instead directly detects the sequence-specific tags using an electronic nanodetector and uses the resulting data to produce maps [45]. NABsys claims that their technology has greater sensitivity and accuracy than existing optical mapping technologies. The resolution of NABsys maps is claimed to be much greater than that of its competitors and is capable of resolving physical distances between tags of fewer than 300 bp [45]. Because the NABsys HD-Mapping platform has not been commercially released, it is difficult to publicly assess its performance. Nevertheless, NABsys mapping is a potential option for use in genome assembly, structural variation (SV) detection, and strain typing.
3. Workflow and algorithms used in optical mapping
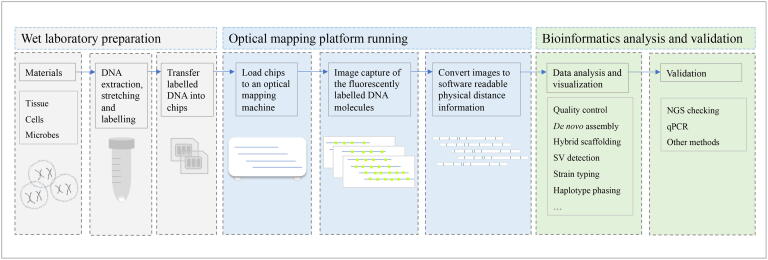
Optical mapping workflows generally commence with experiment design, followed by high-molecular-weight DNA extraction, DNA labelling, DNA molecule loading and image capture, and then data conversion, bioinformatics analyses, and validation (Fig. 2). Standard data processing involves molecule-map quality control, map alignment, de novo assembly, and scaffolding (Fig. 3). Downstream analyses include but are not limited to genome-assembly improvement, SV detection, strain typing, and comparative genomics.
Fig 2.
Workflow used in optical map production and analysis. Generally, the workflow can be divided into three parts: wet laboratory preparation, optical map generation, and bioinformatics analysis and validation. The essential steps are listed in this figure to show how optical mapping works.
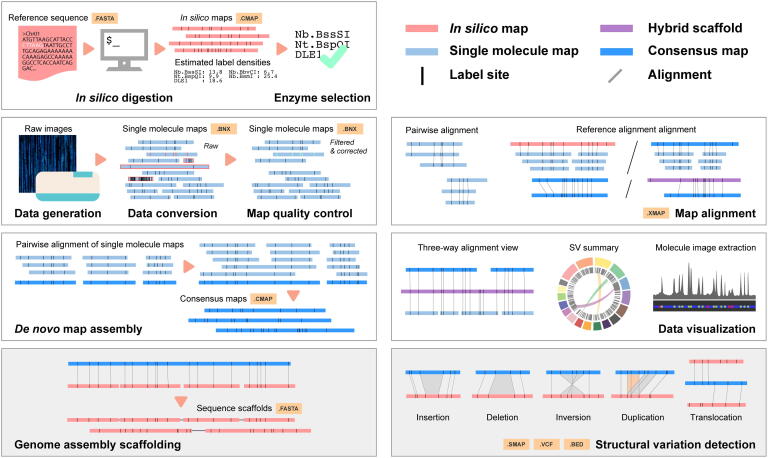
Fig 3.
Optical-mapping data analysis. The major bioinformatics analysis steps are illustrated from in silico genome digestion, data quality control, de novo map assembly, map alignment, scaffolding, and SV detection.
3.1. In silico digestion
It is important to select the correct enzyme and the label density required for an optical mapping experiment, because a low label density may limit the resolution of maps and reduce the coverage of useful molecules, whilst a high label density may confound data grouping [46]. During genome digestion in silico, a draft genome assembly is usually used to check the label density generated by a selected enzyme in a genome (Fig. 3). This process is based on a sequence motif finding and can be performed with several tools. For instance, Bionano Genomics provides Bionano Knickers, Label Density Calculator, and “fa2cmap_multi_color.pl” in the Bionano Solve package to assist with in silico genome digestion. OMTools [47] and Nucleomics (https://github.com/Nucleomics-VIB/bionano-tools) also implement such a function in their packages to assist with enzyme selection.
3.2. Data conversion
Converting images or electronic signals into labels of physical distances is a prerequisite for optical mapping data analysis (Fig. 2). Usually, this process is performed after image capture by Semi-AutoVis in the OpGen Argus platform, AutoDetect in the Bionano Irys platform, or Bionano Access in the Bionano Saphyr platform. The conversion process can be summarized as extracting skeletal segments, tiling restriction maps, grouping segments, and handling sizing errors [48]. The algorithms used in each tool must deal with ambiguities in sizing and end-point location because they can be easily affected by image signal intensities and noise [46]. Moreover, the thermal motion of DNA under imaging may blur fluorescent patterns, which then require correction via kymograph alignment. Weighted Path Align is an algorithm that may enable kymograph alignment of nanochannel images in linear time [49].
3.3. Data correction
The raw restriction maps produced by various platforms may be error prone. This is mitigated by the use of raw-map error-correction to enable efficient use of data. This correction is performed by software such as cOMet, which corrects raw optical-mapping data by detecting maps with significant overlaps using k-mer seeds (Table 1) [50]. However, cOMet is demanding of computing resources, particularly for processing of large genomes. To solve this problem, a spaced-mer method, Elmeri, was developed [51]. On a human data test-set, it was found that Elmeri outperformed cOMet in both running speed and accuracy and used substantially fewer CPU hours than cOMet (15 CPU hours vs. 10 CPU days). Furthermore, the quality of raw maps generated with Elmeri show more than four times improvement of alignment scores over those generated by cOMet.
Table 1.
State-of-the-art tools used in optical mapping analysis.
| Key feature | Tool | Description | URL | Ref |
|---|---|---|---|---|
| Data simulation | OMSim | A tool for producing synthetic Bionano optical maps | https://github.com/biointec/omsim | [144] |
| BMSIM | A tool for producing and mimicking real Bionano molecules | https://github.com/pingchen09990102/BMSIM | [43] | |
| Data correction | cOMet | A tool for raw map error correction using k-mer seeds | https://github.com/kingufl/cOMet | [50] |
| Elmeri | A tool for raw map error correction using spaced-mer seeds | https://github.com/LeenaSalmela/Elmeri | [51] | |
| Map alignment | RefAligner | An aligner for map alignment owned by Bionano Genomics using a dynamic programming framework | https://bionanogenomics.com/support/software-downloads | NA |
| OMBlast | An aligner for map alignment using a modified seed-and-extend method | https://github.com/TF-Chan-Lab/OMBlast | [56] | |
| OMMA | An aligner for multiple map alignment in a population-scale study | https://github.com/TF-Chan-Lab/OMTools | [62] | |
| Maligner | An aligner for raw map alignment based on a dynamic programming framework for large eukaryotic genomes and an indexed method for identifying discordances in the reference | https://github.com/LeeMendelowitz/maligner | [57] | |
| OPTIMA | An aligner for whole genome alignment using a novel seed-and-extend global alignment method | https://github.com/verznet/OPTIMA | [39] | |
| Genome assembly | Assembler | An assembler for de novo map assembly owned by Bionano Genomics | https://bionanogenomics.com/support/software-downloads | NA |
| Novo&Stitch | A tool for assembly reconciliation and scaffolding | https://github.com/ucrbioinfo/Novo_Stitch | [80] | |
| BiSCoT | A tool for scaffold contiguity improvement based on map alignment | https://github.com/institut-de-genomique/biscot | [79] | |
| Assembly evaluation | BionanoAnalyst | A tool for reporting and visualizing potential errors in the genome assemblies based on discordances in the alignment | https://github.com/AppliedBioinformatics/BioNanoAnalyst | [76] |
| misSEQuel | A tool for reporting and resolving mis-assemblies using optical maps and paired-end reads | https://www.cs.colostate.edu/seq/missequel | [85] | |
| SV detection | OMSV | A tool for SV detection based on map alignment from OMBlast and RefAligner using a complete error model | http://yiplab.cse.cuhk.edu.hk/omsv | [117] |
| Structome | A tool for SV detection using Bionano Irys data and map alignment | https://github.com/RyanONeil/structome | NA | |
| Data management and visualization | IrysView | A tool bundled with the Bionano Irys platform for data analysis, data management and visualization on a window system | https://bionanogenomics.com/support/software-downloads | NA |
| Bionano Access | A tool package bundled with the Bionano Saphyr platform for data analysis, data management and visualization on multiple operating systems | https://bionanogenomics.com/support/software-downloads | NA | |
| MapOptics | A tool for viewing comparisons of in silico maps, empirical optical maps, and hybrid scaffolds | https://github.com/FadyMohareb/mapoptics | [75] | |
| OMTools | A toolkit for data processing, map alignment and visualization | https://github.com/TF-Chan-Lab/OMTools | [47] | |
| Pipeline wrapper | runBNG | A wrapper of Bionano IrysSolve Scripts package for data processing and analysis on Linux systems | https://github.com/AppliedBioinformatics/runBNG | [74] |
| Irys-scaffolding | A refined pipeline for genome assembly based on Bionano RefAligner and Assembler | https://github.com/kstatebioinfo/Irys-scaffolding | [26] | |
| OMWare | A refined pipeline for genome assembly based on Bionano RefAligner and Assembler | https://github.com/sharpa/OMWare | [73] |
3.4. Map alignment
Alignment is an essential step in the analysis of optical map data. It can be performed by grouping data to quickly identify maps homologous to target genome regions (Fig. 3). Alignment reveals discordances between query and reference maps, which can provide useful information for SV detection or mis-assembly identification [48]. However, the alignment of optical mapping data is more complicated and challenging than sequence alignment because a greater diversity of errors (e.g. missing and spurious labels) can be introduced during enzyme digestion or labelling. DNA breaks can occur during sample preparation. Image noise can also introduce false-positive and false-negative label signals [46]. Moreover, imprecise scaling can lead to errors in molecule length and label distance estimation. To cope with these problems, algorithms are used in sequence alignment, such as local and global alignment and dynamic programming, accompanied by scoring approaches.
In 2006, Valouev et al. [52] introduced a technique to assist optical map alignment, in which the nature of errors in optical mapping are embedded in a scoring system in a dynamic programming framework. Later, SOMA was developed, based on dynamic programming with a heuristic scoring scheme [53]. During benchmarking, it was found that SOMA performed better than Valouev et al.’s technique [53], [54]. Similarly, the Bionano RefAligner also adopts a dynamic programming framework to perform local alignment between maps. RefAligner then penalizes unaligned labels at alignment ends followed by applying a likelihood to select for global alignment [26].
Another method, TWIN, uses dynamic programming and attempts to accelerate alignment by using an FM-index method [55]. However, the error intolerance of TWIN limits its application [56], [57]. OPTIMA (Table 1), a recently developed application, uses a seed-and-extend global–local alignment method with a technology-agnostic statistical model to evaluate alignment performance [39]. Maligner, on the other hand, combines a fast and stringent dynamic programming framework and an index-based method to align single-molecule maps or in silico maps to a reference. A new alignment-score normalization method has also been implemented in Maligner [57]. OMBlast was recently designed to align maps using a seed-and-extend approach that indexes reference as k-tuples to save memory. After seeding, OMBlast recursively extends the alignment with different scaling factors. Its performance is considerably better than that of other tools [56].
Overlapping unassembled raw maps in the optical map assembly step is challenging but necessary for building consensus optical maps. The index-based Kohdista is a recent solution that formulates the alignment problem as an automaton path-matching problem to index and query restriction maps [58]. Kohdista has been tested on E. coli data to demonstrate its fast alignment performance. It is likely that Kohdista will be used or modified to perform high-quality pairwise restriction-map alignments of large eukaryotic organisms.
Multiple alignment is also useful in comparative analysis to identify similar regions that may have functional, structural, or evolutionary relationships. Multiple alignment of nucleotide sequences has been achieved with a number of algorithms, such as those used in ClustalW [59], Muscle [60], and MAFFT [61]. To solve the multiple alignment problem in optical mapping, Leung et al. [62] published a two-step pipeline based on optical mapping by multiple alignment to separate genomic segments into collinear blocks, thereby enabling population-scale comparison of complex genomic features.
3.5. Map assembly
De novo single-molecule map assembly is useful for identification of novel genomic features, such as large insertions in a genome (Fig. 3). It can also be used to detect genetic features of a species without a reference genome.
The concept of de novo single-molecule map assembly is similar to that of assembly in sequence contexts. The fragment assembly problem was first modelled mathematically by Lander and Waterman, suggesting that two reads should be merged if the overlap exceeds a threshold and that, therefore, the short-read lengths can be compensated by deeper coverage [63], [64]. Contig construction can be well approached using graphs, where fragments are represented as nodes, connected by edges when the overlap justifies a merge. The overlap-layout-consensus (OLC) paradigm [65] and the de Bruijn graph (DBG) [66] are the common methods to construct sequence assemblies, where the former performs pairwise alignment for all combinations and the latter computes the overlap information implicitly between neighbouring artificially chopped fragments of a fixed size k called k-mer [64]. A major difference between the two approaches lies in repeat handling. OLC places repeats as separate nodes and DBG collapses them into single nodes, resulting in higher computation and memory demand in pairwise comparison involving repeats in OLC graphs [66].
While DBG offers more efficient performance in sequence assembly, it is not yet readily adopted in de novo whole genome map assembly due to the error properties of optical mapping data that affect its alignment principles as discussed above [67]. The first optical map assembly algorithm was described by Anantharaman et al. [68] in 1997, modelling the errors using Bayesian probability and determining the best alignment pair by dynamic programming. This algorithm uses a greedy heuristic global search for islands of connected fragments to combine and hence is sensitive to data quality. While the algorithm has been demonstrated in several microorganism genomes, the search space remains unscalable [67]. Valouev et al. [69] implemented the OLC graph by first sorting pairwise alignment with a confidence score to construct a layout graph. Although this solution is scalable, it is time-consuming when performing error correction and could drop potential connectivity during strict graph correction [67]. In 2011, Goldstein et al. [70] briefly proposed an idea of using a DBG algorithm Germinate and Grow to assemble restriction maps tested on the Medicago truncatula genome (~500 Mb). The algorithm first uses geometric k-mer hashing to identify potential error-free nodes in the DBG to assemble subsets of restriction maps as seed maps. Then those assembled restriction maps are iteratively extended and refined to cover more parts of the genome. While the algorithm has been reported to be used in the local map assembly of a domestic cow genome, in regions that are largely discordant with the sequence reference from prior alignment, and whole genome map finishing, the tool has not been officially published [71]. Among the latest development, Li et al. [67] proposed an iterative algorithm (IOMA) for optical map assembly, where the consensus map from one OLC contig construction is taken forward as input for the next iteration, until the alignment coverage stops increasingly generate consensus genome maps. Currently, the iterative OLC-based Bionano assembler is the de facto tool for single-molecule optical map assembly, with a customizable fixed number of extensions (Table 1) [72]. Some pipelines, such as AssembleIrysCluster [26] and OMWare [73] are based on the Bionano assembler.
3.6. Data visualization and other processing
Data visualization is an important way to check the quality of data analysis. IrysView, which is bundled with the Bionano Irys platform, is an application that helps with data visualization by enabling plotting of raw data metrics, repeat content, and map alignment by enabling raw-image viewing. However, its reliance on a Microsoft Windows (Microsoft Inc, USA) operating system limits its application [74].
In contrast, OMView implemented in OMTools is the first application to allow data visualization on multiple platforms [47]. OMView accepts separate input maps, pairwise and multiple alignments in standard data formats, and supports annotation visualization. OMView also marks large-scale SVs, such as insertion or deletion of bases (indels) and inversions, with symbols. Options are also provided in OMView to help visualize segment stretch factors with the use of color. Conversely, whilst OMView provides multiple panes to visualize alignments to the same reference, MapOptics provides three way-alignment view features, thus allowing view comparisons of in silico maps, empirical optical maps, and hybrid scaffolds. This is convenient for assessing the performance of hybrid scaffolding [75].
Another cross-platform software is BioNanoAnalyst, which visualizes mis-assemblies as conflicts between sequence and optical map assemblies [76]. Bionano Genomics recently renewed its data visualization tool and integrated Bionano Solve in a web application that incorporates draggable three-way alignments, conflict viewing, resolution viewing, and a Circos-like whole-genome SV-distribution plot to assist Bionano data analysis. An image displayer is installed on the new Bionano workstation computer to visualize the raw image files in a JXR format. The image displayer provides adjustable contrast and export options to help reduce artefactual problems during data conversion. Irys Extract makes further use of raw images by providing a function to crop raw images for specified molecules, which is useful for showing special label patterns [77].
4. Applications of optical mapping
Optical mapping is increasingly used in various areas of genomic research due to its improved accuracy, decreasing cost, and high throughput. Generally, optical mapping is used to improve genome assembly, to facilitate large-scale SV detection, and for strain typing. Although these applications can also be achieved by other technologies, such as genome scaffolding by using linked reads, mate-pair reads or Hi-C reads, compared to optical mapping, the read length and physical read coverage of these alternatives can be lower, and they generally depend on short-read sequencing to produce reads. Therefore, optical mapping remains one of the best options to facilitate these genomic research tasks mentioned above. With recent advances in technologies, optical mapping is also expected to improve haplotyping on different ploidy levels and to be applicable to other research, such as epigenomic studies.
4.1. Genome assembly improvement
Initially, optical mapping was developed to assist genome assembly, during which two approaches are used to piece together fragmented genome drafts. A scaffolding-based approach is commonly used, in which sequence contigs are aligned, validated, and scaffolded in an a priori assembly fashion. In an alternative approach, the long-linkage information from optical mapping data is incorporated into the de Bruijn graph for assembly building.
An early developed tool, SOMA, applies a dynamic-programming algorithm to scaffold short-read assemblies with assembled maps. SOMA models segment-length distribution and calculates the extent of matches between reference and query maps with scoring functions [53]. OMACC is another tool that further improves the accuracy of genome assembly by rescaling optical maps and applying length constraints in path selection [78]. OMACC considers the gap sizes and copy numbers of repeats in the gap between two contigs to generate better assemblies.
In addition to the hybrid pipeline provided by Bionano Genomics for Bionano scaffolding, the Irys-scaffolding pipeline is the first refined pipeline to help scaffolding [26]. This pipeline applies a threshold minimum percent of total aligned length to filter local alignments. Although the source code of the Irys-scaffolding pipeline is available on GitHub (Table 1), the alignment still relies on RefAligner, which is difficult to optimize due to the many unexplained configuration parameters provided. A pre-released Bionano SCaffolding COrrection Tool (BisCoT) was recently claimed to use pre-existing optical assembly to generate better scaffolds than the Bionano hybrid scaffolding pipeline by closing gaps. However, the accuracy of this new method is yet to be evaluated [79].
Multiple genome assemblies can be merged to improve the contiguity of genome assembly. Novo & Stitch is such a tool to produce the best assembly merged from several assembly versions [80]. The pipeline iteratively selects the optimal non-conflicting alignments and stitches up sequence contigs by aligning two original contigs to the new one. In contrast, OMGS takes a more streamlined scaffolding approach as it incorporates information from multiple optical maps by correcting raw maps with cOMet [81].
There are two algorithms that directly align optical maps to a DBG to help sequence-based genome assembly. Optical maps are used as a guide to connect paths and to search for correct paths instead of grouping repeats into single paths. The first algorithm was AGORA, which uses assembled restriction maps to eliminate inconsistent alternate paths [82]. Then came omGraph, an algorithm designed to use the shorter and error-prone raw restriction maps by first performing map error-correction [83]. Both algorithms were designed to support restriction maps, whilst tests and optimization are needed to apply them to non-restriction-based data, as the latter are known to have different error models [84].
In addition to improving the contiguity of genome assemblies, optical maps can be used to detect mis-assemblies. Several specialized tools are available for this purpose. For instance, BioNanoAnalyst uses a scoring approach to detect mis-assemblies in a draft genome assembly by identifying discordances between consensus optical maps and the reference [76]. MisSEQuel detects breakpoints of errors using paired-end sequence reads and optical mapping data [85]. MisSEQuel can also be used to detect artificial genomic rearrangements in a genome caused by a sequence assembler. Chimericognizer is an alternative that uses one or more sets of Bionano optical maps to detect chimeric contigs or maps by concatenating multiple sequence assemblies [86]. The assemblies can be obtained from different assembly tools or one assembly tool with different parameter settings.
Optical mapping was successfully used in the microbial genome assembly of species such as Deinococcus radiodurans [87] and Plasmodium falciparum [88] during the early development. Later, OpGen enabled optical mapping to the genome assembly of advanced species. In a goat genome assembly, OpGen optical maps helped anchor 2090 fosmid scaffolds into 315 super-scaffolds and demonstrated that optical mapping is a useful technique in genome assembly for species with larger genomes. After that, optical mapping was gradually adopted for analysis of the genomes of animals species such as the ostrich [89] and the yellow croaker [90].
The adoption of optical mapping is further increasing with the development of long-read sequencing. In current genome-assembly pipelines, optical maps are commonly used with a combination of PacBio or Oxford Nanopore reads with Hi-C data to build reference-quality genome assemblies. This technology combination has been applied to genome analysis in various species, such as humans [91], [92], other animals [93], [94], [95], [96], and plants [97], [98], [99], [100], [101], [102], [103], [104], [105], [106], [107], [108], [109], [110]. In particular, the Genome Reference Consortium (GRC), in an effort to improve the quality of reference genomes by error correction, gap closure, and variation representation, has been producing optical mapping data for humans and other model organisms [36]. Another ambitious application of optical mapping is implemented in the Vertebrate Genome Project (VGP) [111], which is affiliated with the Genome 10K Consortium. The data and assemblies for approximately a hundred species among the 260 species targeted in Phase I are currently available at https://vgp.github.io/genomeark.
4.2. Large-scale SV detection
Genetic variations are important resources for the study of evolution and domestication. Nevertheless, the investigation of genetic variations has long been technically limited to small variants, such as single nucleotide polymorphisms (SNPs) and small indels. Large genomic SVs of kilobases or more in length have been challenging to study. In 2008, Kidd et al. [112] mapped eight human genomes from the Human Genome Structural Variation Project and refined 1695 SV locations larger than 6 kb. They used fosmid end-sequence pairs to validate 11 loci of potential SVs that are greater than 40 kb. This study set a foundation for SV study by optical mapping. Optical mapping has since been applied to various comparative genomic studies.
In 2016, Mak et al. [113] demonstrated systematic genome-wide SV detection by comparing assembled optical maps, which were generated with the Nt.BspQI nickase from a father-mother-daughter trio selected from the 1000 Human Genome Project (1KGP) [113]. This study identified 59 insertions and 156 deletions over 5 kb and 16 inversions. The detection of the integration sites of Epstein–Barr Virus (EBV) during cell-line transformation demonstrates the application of optical mapping in large-scale SV identification. In 2019, Levy-Sakin et al. [114] mapped genomes of 154 individuals among the 26 populations in the 1KGP and found that genes located in copy-number variation regions seem to agree with the evolutionary patterns.
In addition to its application in human genomes, large-scale SV detection is also important in other research. For example, the mapping of a highly rearranged liposarcoma cell line generated 72 fusion maps representing 112.3 Mb highly rearranged regions, suggesting that chained fusions and a higher level of complex genomic architecture can occur in cancer [115]. In a soybean study, optical mapping was used to reveal an inversion that causes loss of soybean seed-coat color during domestication [106]. In a primate evolutionary study, SVs were found to be enriched near genes down-regulated in human cerebral organoids compared to those of chimpanzees [116].
There are two tools used for optical mapping SV detection: Bionano SVCaller and OMSV [117]. OMSV is a tool that identifies SVs by combining molecule-to reference alignments generated from RefAligner and OMBlast. During processing, RefAligner is used to generate a homologous alignment, whilst OMBlast handles complex rearrangements by split-alignment. Variations such as extra or missing labels, large segment-size differences and complex SVs are detected separately before being combined and deduplicated.
To facilitate the annotation of identified clinically relevant SVs, an R package called nanotatoR has also been released [118]. It calculates population frequencies of SVs using data from the Database of Genomic Variants (DGV) and, to filter variants, overlaps primary gene lists from the NCBI databases to SVs identified by Bionano SVCaller. Although optical map-based SV detection tools have been developed, their application and performance in higher ploidy contexts have not been demonstrated.
4.3. Microbial strain typing
Pathogen identification is essential in epidemiological surveillance. Precise classification at a sub-species level, known as strain typing, is necessary to provide insight into clonal evolution that helps to trace transmission routes and formulate outbreak control. Strain typing was one of the earliest common applications of optical mapping. In strain typing, the gold standard method is pulse-field gel electrophoresis (PFGE), which enables the analysis of size patterns of DNA restriction fragments, and multilocus sequence typing (MLST), which involves Sanger sequencing of several selected housekeeping gene loci to match the profile of allele combinations [119], [120]. However, MLST relies primarily on the assumption that housekeeping genes are evolutionarily conserved but lacks whole genome information, whilst PFGE does not provide the order of DNA fragments, which makes it difficult to resolve fragments of similar sizes. These drawbacks limit the resolution of these methods. Optical mapping, as a high-throughput profiling alternative, can be easily applied to capture the size and order of fragments from the whole genome.
Bacteria are known to exchange genetic materials via horizontal transfer, including cassette elements that harbor toxin and antibiotic resistance with medical implications [121]. In 2007, Latreille et al. [122] proposed to routinely use optical mapping combined with DNA sequencing for bacterial genome assembly to aid identification, until simultaneous mapping of multiple bacterial genome became possible [35]. Furthermore, optical mapping has proven useful in various cases, such as the tracking of a food-borne outbreak of an enterohemorrhagic Escherichia coli O157:H7 and distinguishing five strains of a toxigenic Vibrio cholerae O1 under the same serotype and biotype from different geographic origin [123].
4.4. Haplotype phasing
Haplotype phasing finds continuous stretches of DNA on the same chromosome and can be viewed as a means of resolving genome assemblies at a higher resolution and detecting more SVs between haploid genomes of the same organism [124]. Capturing accurate allele structures not only refines the genome assembly, but also contributes to better understanding of the population structure and evolutionary process [125]. Various approaches can be used to solve this problem, based on the use of population, family trio, or a single-sample information and different molecular methods [126].
The first comprehensive analysis of the diploid human genome was performed using a hybrid assembly of Illumina short reads, PacBio long reads, and Bionano optical maps [127]. This study achieved 99% consistency with previous trio results. The phased human genome of the Korean individual AK1 is a demonstration of this application [91]. Haplotype phasing in nonhuman species is not yet popular, and haplotype-aware assembly pipelines for nonhuman samples are available but not yet officially supported by Bionano. However, some attempts have been made to use optical mapping to perform haplotype genome phasing in buffalos [128], cattle [129], and pigs [130]. In a recent African cassava study, optical mapping was used to identify allelic variants and allele-specific expression via haplotype comparison [131].
In contrast to DNA sequencing, optical mapping can produce long single molecule maps to cover heterozygous genome regions or loci with different haplotypes that cannot be easily spanned by DNA sequence reads. Using optical mapping, it is possible to resolve different alleles without the requirement of a family-trio or population information. Nevertheless, due to the relatively low resolution of optical mapping, DNA sequencing is needed to retrieve the sequences of different haplotypes in the phased optical map alignment.
4.5. Other genomic feature detection
Although optical mapping produces maps with superior lengths, the molecule mapping rate and hence the accessibility of different genomic regions still depends on the distribution of the enzyme recognition motifs. Moreover, the motifs of standard enzyme choices often have no direct biological implications. For more options and control, scientists are looking to alternative labelling methods to target regions of interest.
Early proposals included labelling with peptide nucleic acids or triplex-forming oligonucleotides [132]. In 2019, Müller et al. [133] published a protocol for enzyme-free optical mapping by competitive binding of YOYO-1 and netropsin. Their method blocks an AT-rich region that highlights a DNA contour, preventing DNA damage by nicking. This method also offers the potential to highlight DNA damage lesions.
In terms of more direct biological relevance, a vibrant research area of optical mapping is epigenetics. Methylation of CpG islands, clusters of cytosine-guanine dinucleotide DNA motifs, is a key mechanism in mammalian imprinting. In 2008, Ananiev et al. [134] tested mapping methylation optically on E. coli and human cell lines by using a methylation-sensitive restriction enzyme. In 2018, Gabrieli et al. [135] looked for 5-hydromethylcytosine, a product of active DNA demethylation, on the Bionano platform. Later, Sharim et al. [136] advanced methylation detection by using a synthetic analog to “trick” methylases into transferring a fluorophore instead of a methyl group. Hence, the protocol is compatible with the high-throughput Bionano platform with the merit of minimizing DNA damage, similar to the DLS method discussed above.
In addition to epigenomics, the application of optical mapping has been extended to other areas. In 2020, Young et al. [137] utilized the CRISPR/Cas9 targeted-labelling technique developed by McCaffrey et al. [138] to extend their previous study on subtelomeric regions that used nick-based genome maps. By specifically tagging fluorophores to the telomeric repeats, the location of population-specific patterns identified in the subtelomeric region using traditional labelling method were verified. The Bionano team has recently announced an improved non-nicking targeted-labelling method called CRISPR-Bind to further improve the application of optical mapping [139].
5. Summary and outlook
While current advances in DNA sequencing have improved genomic research by producing more complete genome assemblies and detecting larger SVs, the use of DNA sequencing alone is often not yet sufficient to produce a genome assembly of desired reference-grade quality, particularly in eukaryotic organisms. The application of other genomic methods such as optical mapping usually help to overcome the shortfalls in sequencing data to achieve a chromosome-level genome assembly for species with large and complex genomes.
After three decades of development, optical mapping has emerged as an important complementary method in genomic studies. Although it cannot replace DNA sequencing due to a lack of base-by-base nucleotide information, it can quickly report genomic structural information that is not easily captured by DNA sequencing. With the ability to map long molecule lengths at low cost, optical mapping has facilitated genome assembly, large-scale SV detection, and strain typing. Nonetheless, the resolution of optical mapping is still low, which makes it unsuitable for analysis of maps shorter than 100 to 150 kb, where alignment reliability is easily compromised by insufficient labelling sites [29].
Although some strategies have been implemented to improve resolution, such as applying multiple enzymes or direct DNA labelling, most designs are based on human genomes and may not be transferable to other species. Direct labelling is a promising solution to increase mapping resolution and avoid double-strand breaks. However, only the enzyme DLE-1 is commercialized for the Bionano Saphyr system. Due to the genome divergence between different species, one sequence motif pattern may not suit all species, and thus other labelling alternatives validated by comprehensive tests and demonstrations are required.
In addition, the key algorithms implemented in Bionano Tools are not open source. Researchers cannot directly study or modify the code used in RefAligner and Assembler to improve their performance. Although other tools such as OMTools [47] and OMSV [117] have been developed to assist optical mapping analysis, no other de novo assembly tools are available to compete with Bionano Tools. The direct use of the Bionano pipeline may also introduce false positives and false negatives, particularly in non-human genomes. Parameter tuning can be useful to customize the pipeline for different studies, yet the limited instructions provided and the lack of clarity on the algorithms behind RefAligner and Assembler make the adjustment difficult. For de novo map assembly, although k-mer hashing of optical maps has been suggested to be impractical in the assembly context [67], with the development of k-mer based alignment tools such as OMTools, this possibility may be worth re-exploring.
Haplotype phasing in different ploidy levels is also important in genomic research. Although Bionano Genomics has released a haplotype-phased de novo assembly pipeline to assist haplotype studies, the pipeline is human-genome oriented and little instruction has been released to assist pipeline tweaking. Different algorithms and methods are needed in optical mapping to help achieve a global phased genome for species with large and complex genomes. With the advantages of optical mapping and an increasing number of polyploid species being studied, optical mapping is expected to overcome the complications arising from polyploidy [140]. Still, a polyploid-specific analysis pipeline remains to be developed, and it is difficult to validate the reported results.
Graphs can be an efficient data structure to represent reference sequence assemblies, where different alleles are separated into different paths, with preserved linkage information and identical sequences grouped into single paths to save space [141]. A condensed graph structure presents advantages in both saving storage and in minimizing extra computational demands during analyses [142], [143]. This might motivate the further development and application of graph-based optical mapping analysis.
In the near future, it is expected that novel methods, such as new DNA stretching, labelling, and analysis approaches will be developed to increase the accuracy of optical mapping from signal conversion, map alignment, and de novo assembly to downstream analyses. Work is still needed to explore the possibility of optical mapping in haplotype phasing and polyploid research. The low efficiency of optical mapping in data visualization and examination impedes its application to large-scale projects. An automated checking and validation method is therefore required to further improve the efficiency of optical mapping.
Overall, optical mapping has proven indispensable for genomic studies. With growing application and development efforts, it is expected that optical mapping will continue to improve genomic studies, enabling the assembly of more complete genomes and the discovery of novel variations. The consequent determination of genotype and phenotype associations from optical mapping is also becoming more practical. Although the current technologies have some pitfalls, it is expected that refined optical-mapping solutions will be developed in the near future to further enrich genomic research.
CRediT authorship contribution statement
Yuxuan Yuan: Visualization, Writing - review & editing. Claire Yik-Lok Chung: Visualization, Writing - review & editing. Ting-Fung Chan: Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Hong Kong Research Grants Council Area of Excellence Scheme (AoE/M-403/16) and Collaborative Research Fund (C4057-18EF), CUHK Group Research Scheme 3110135, a generous donation from Mr. and Mrs. Sunny Yang, and the Innovation and Technology Commission, Hong Kong Special Administrative Region Government to the State Key Laboratory of Agrobiotechnology (CUHK). Any opinions, findings, conclusions or recommendations expressed in this publication do not reflect the views of the Government of the Hong Kong Special Administrative Region or the Innovation and Technology Commission. Armin Scheben copyedited this manuscript.
References
- 1.Alkan C., Sajjadian S., Eichler E.E. Limitations of next-generation genome sequence assembly. Nat Methods. 2011 doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Treangen T.J., Salzberg S.L. Repetitive DNA and next-generation sequencing: Computational challenges and solutions. Nat Rev Genet. 2012 doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salzberg S.L., Yorke J.A. Beware of mis-assembled genomes. Bioinformatics. 2005 doi: 10.1093/bioinformatics/bti769. [DOI] [PubMed] [Google Scholar]
- 4.Cameron D.L., Di Stefano L., Papenfuss A.T. Comprehensive evaluation and characterisation of short read general-purpose structural variant calling software. Nat Commun. 2019 doi: 10.1038/s41467-019-11146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zimin A.V., Marçais G., Puiu D., Roberts M., Salzberg S.L., Yorke J.A. The MaSuRCA genome assembler. Bioinformatics. 2013 doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012 doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012 doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zerbino D.R. Using the Velvet de novo assembler for short-read sequencing technologies. Curr Protoc Bioinforma. 2010 doi: 10.1002/0471250953.bi1105s31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simpson J.T., Wong K., Jackman S.D., Schein J.E., Jones S.J.M., Birol I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009 doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan Y., Bayer P.E., Batley J., Edwards D. Improvements in Genomic Technologies: Application to Crop Genomics. Trends Biotechnol. 2017 doi: 10.1016/j.tibtech.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Goodwin S., McPherson J.D., McCombie W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat Rev Genet. 2016 doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amarasinghe S.L., Su S., Dong X., Zappia L., Ritchie M.E., Gouil Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020;21:30. doi: 10.1186/s13059-020-1935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sedlazeck F.J., Lee H., Darby C.A., Schatz M.C. Piercing the dark matter: Bioinformatics of long-range sequencing and mapping. Nat Rev Genet. 2018 doi: 10.1038/s41576-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 14.Cao H., Hastie A.R., Cao D., Lam E.T., Sun Y., Huang H. Rapid detection of structural variation in a human genome using nanochannel-based genome mapping technology. GigaScience. 2014 doi: 10.1186/2047-217X-3-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Staňková H., Hastie A.R., Chan S., Vrána J., Tulpová Z., Kubaláková M. BioNano genome mapping of individual chromosomes supports physical mapping and sequence assembly in complex plant genomes. Plant Biotechnol J. 2016 doi: 10.1111/pbi.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belser C., Istace B., Denis E., Dubarry M., Baurens F.C., Falentin C. Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps. Nat Plants. 2018 doi: 10.1038/s41477-018-0289-4. [DOI] [PubMed] [Google Scholar]
- 17.Eisenstein M. Startups use short-read data to expand long-read sequencing market. Nat Biotechnol. 2015 doi: 10.1038/nbt0515-433. [DOI] [PubMed] [Google Scholar]
- 18.Ma W., Ay F., Lee C., Gulsoy G., Deng X., Cook S. Fine-scale chromatin interaction maps reveal the cis-regulatory landscape of human lincRNA genes. Nat Methods. 2014 doi: 10.1038/nmeth.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies J.O.J., Oudelaar A.M., Higgs D.R., Hughes J.R. How best to identify chromosomal interactions: A comparison of approaches. Nat Methods. 2017 doi: 10.1038/nmeth.4146. [DOI] [PubMed] [Google Scholar]
- 20.Burton J.N., Adey A., Patwardhan R.P., Qiu R., Kitzman J.O., Shendure J. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat Biotechnol. 2013 doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bickhart D.M., Rosen B.D., Koren S., Sayre B.L., Hastie A.R., Chan S. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat Genet. 2017 doi: 10.1038/ng.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho S.S., Urban A.E., Mills R.E. Structural variation in the sequencing era. Nat Rev Genet. 2019 doi: 10.1038/s41576-019-0180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Everyday de novo assembly n.d. https://www.10xgenomics.com/solutions/assembly/ (accessed February 15, 2020).
- 24.Meleshko D., Marks P., Williams S., Hajirasouliha I. Detection and assembly of novel sequence insertions using Linked-Read technology. BioRxiv. 2019 doi: 10.1101/551028. [DOI] [Google Scholar]
- 25.Schwartz D, Li X, Hernandez L, Ramnarain S, Huff E, Wang Y. Ordered restriction maps of Saccharomyces cerevisiae chromosomes constructed by optical mapping. Science (80) 1993. https://doi.org/10.1126/science.8211116. [DOI] [PubMed]
- 26.Shelton J.M., Coleman M.C., Herndon N., Lu N., Lam E.T., Anantharaman T. Tools and pipelines for BioNano data: Molecule assembly pipeline and FASTA super scaffolding tool. BMC Genomics. 2015 doi: 10.1186/s12864-015-1911-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aston C., Mishra B., Schwartz D.C. Optical mapping and its potential for large-scale sequencing projects. Trends Biotechnol. 1999 doi: 10.1016/S0167-7799(99)01326-8. [DOI] [PubMed] [Google Scholar]
- 28.Tang H., Lyons E., Town C.D. Optical mapping in plant comparative genomics. GigaScience. 2015 doi: 10.1186/s13742-015-0044-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bogas D., Nyberg L., Pacheco R., Azevedo N.F., Beech J.P., Gomila M. Applications of optical DNA mapping in microbiology. Biotechniques. 2017 doi: 10.2144/000114555. [DOI] [PubMed] [Google Scholar]
- 30.Cai W., Aburatani H., Stanton V.P., Housman D.E., Wang Y.K., Schwartz D.C. Ordered restriction endonuclease maps of yeast artificial chromosomes created by optical mapping on surfaces. Proc Natl Acad Sci U S A. 1995 doi: 10.1073/pnas.92.11.5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meng X., Benson K., Chada K., Huff E.J., Schwartz D.C. Optical mapping of lambda bacteriophage clones using restriction endonucleases. Nat Genet. 1995 doi: 10.1038/ng0495-432. [DOI] [PubMed] [Google Scholar]
- 32.Jing J., Reed J., Huang J., Hu X., Clarke V., Edington J. Automated high resolution optical mapping using arrayed, fluid-fixed DNA molecules. Proc Natl Acad Sci U S A. 1998 doi: 10.1073/pnas.95.14.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jo K., Dhingra D.M., Odijk T., De Pablo J.J., Graham M.D., Runnheim R. A single-molecule barcoding system using nanoslits for DNA analysis. Proc Natl Acad Sci U S A. 2007 doi: 10.1073/pnas.0611151104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fong T. OpGen Prepping Optical Mapping System for Analysis of Human, Other Larger Genomes 2010. https://www.genomeweb.com/sequencing/opgen-prepping-optical-mapping-system-analysis-human-other-larger-genomes (accessed February 16, 2020).
- 35.Riley M.C., Lee J.E., Lesho E., Kirkup B.C. Optically mapping multiple bacterial genomes simultaneously in a single run. PLoS ONE. 2011 doi: 10.1371/journal.pone.0027085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Howe K., Wood J.M.D. Using optical mapping data for the improvement of vertebrate genome assemblies. GigaScience. 2015 doi: 10.1186/s13742-015-0052-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang W., Dong Y., Xie M., Jiang Y., Xiao N., Du X. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus) Nat Biotechnol. 2013 doi: 10.1038/nbt.2478. [DOI] [PubMed] [Google Scholar]
- 38.Tang H., Krishnakumar V., Bidwell S., Rosen B., Chan A., Zhou S. An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics. 2014 doi: 10.1186/1471-2164-15-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verzotto D., Teo A.S.M., Hillmer A.M., Nagarajan N. OPTIMA: Sensitive and accurate whole-genome alignment of error-prone genomic maps by combinatorial indexing and technology-agnostic statistical analysis. GigaScience. 2016 doi: 10.1186/s13742-016-0110-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lam E.T., Hastie A., Lin C., Ehrlich D., Das S.K., Austin M.D. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat Biotechnol. 2012 doi: 10.1038/nbt.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Direct Label and Stain Technology n.d. https://bionanogenomics.com/technology/dls-technology/ (accessed February 18, 2020).
- 42.Neely R.K., Dedecker P., Hotta J.I., Urbanaviite G., Klimaauskas S., Hofkens J. DNA fluorocode: A single molecule, optical map of DNA with nanometre resolution. Chem Sci. 2010 doi: 10.1039/c0sc00277a. [DOI] [Google Scholar]
- 43.Chen P., Jing X., Ren J., Cao H., Hao P., Li X. Modelling BioNano optical data and simulation study of genome map assembly. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jain A., Sheats J., Reifenberger J.G., Cao H., Dorfman K.D. Modeling the relaxation of internal DNA segments during genome mapping in nanochannels. Biomicrofluidics. 2016 doi: 10.1063/1.4964927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oliver J.S., Catalano A., Davis J.R., Grinberg B.S., Hutchins T.E., Kaiser M.D. High-Definition Electronic Genome Maps from Single Molecule Data. BioRxiv. 2017 [Google Scholar]
- 46.Ravindran P, Gupta A. Image processing for optical mapping. Gigascience 2015. https://doi.org/10.1186/s13742-015-0096-z. [DOI] [PMC free article] [PubMed]
- 47.Leung A.K.Y., Jin N., Yip K.Y., Chan T.F. OMTools: A software package for visualizing and processing optical mapping data. Bioinformatics. 2017 doi: 10.1093/bioinformatics/btx317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mendelowitz L, Pop M. Computational methods for optical mapping. Gigascience 2014. https://doi.org/10.1186/2047-217X-3-33. [DOI] [PMC free article] [PubMed]
- 49.Noble C., Nilsson A.N., Freitag C., Beech J.P., Tegenfeldt J.O., Ambjörnsson T. A fast and scalable kymograph alignment algorithm for nanochannel-based optical DNA mappings. PLoS ONE. 2015 doi: 10.1371/journal.pone.0121905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mukherjee K, Washimkar D, Muggli MD, Salmela L, Boucher C. Error correcting optical mapping data. Gigascience 2018. https://doi.org/10.1093/gigascience/giy061. [DOI] [PMC free article] [PubMed]
- 51.Salmela L., Mukherjee K., Puglisi S.J., Muggli M.D., Boucher C. Fast and accurate correction of optical mapping data via spaced seeds. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btz663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valouev A., Li L., Liu Y.C., Schwartz D.C., Yang Y., Zhang Y. Alignment of optical maps. J Comput Biol. 2006 doi: 10.1089/cmb.2006.13.442. [DOI] [PubMed] [Google Scholar]
- 53.Nagarajan N., Read T.D., Pop M. Scaffolding and validation of bacterial genome assemblies using optical restriction maps. Bioinformatics. 2008 doi: 10.1093/bioinformatics/btn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarkar D., Goldstein S., Schwartz D.C., Newton M.A. Statistical significance of optical map alignments. J Comput Biol. 2012 doi: 10.1089/cmb.2011.0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muggli M.D., Puglisi S.J., Boucher C. Efficient indexed alignment of contigs to optical maps. Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics) 2014 doi: 10.1007/978-3-662-44753-6_6. [DOI] [Google Scholar]
- 56.Leung A.K.Y., Kwok T.P., Wan R., Xiao M., Kwok P.Y., Yip K.Y. OMBlast: Alignment tool for optical mapping using a seed-and-extend approach. Bioinformatics. 2017 doi: 10.1093/bioinformatics/btw620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mendelowitz L.M., Schwartz D.C., Pop M. Maligner: A fast ordered restriction map aligner. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btv711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muggli M.D., Puglisi S.J., Boucher C. Kohdista: An efficient method to index and query possible Rmap alignments. Algorithms Mol Biol. 2019 doi: 10.1186/s13015-019-0160-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson J.D., Gibson T.J., Higgins D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr Protoc Bioinforma. 2003 doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- 60.Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004 doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013 doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leung A.K.Y., Liu M.C.J., Li L., Lai Y.Y.Y., Chu C., Kwok P.Y. OMMA enables population-scale analysis of complex genomic features and phylogenomic relationships from nanochannel-based optical maps. GigaScience. 2019 doi: 10.1093/gigascience/giz079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lander E.S., Waterman M.S. Genomic mapping by fingerprinting random clones: A mathematical analysis. Genomics. 1988 doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- 64.Li Z., Chen Y., Mu D., Yuan J., Shi Y., Zhang H. Comparison of the two major classes of assembly algorithms: Overlap-layout-consensus and de-bruijn-graph. Brief Funct Genomics. 2012 doi: 10.1093/bfgp/elr035. [DOI] [PubMed] [Google Scholar]
- 65.Myers E.W. Toward Simplifying and Accurately Formulating Fragment Assembly. J Comput Biol. 1995 doi: 10.1089/cmb.1995.2.275. [DOI] [PubMed] [Google Scholar]
- 66.Pevzner P.A., Tang H., Waterman M.S. An Eulerian path approach to DNA fragment assembly. Proc Natl Acad Sci U S A. 2001 doi: 10.1073/pnas.171285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li M, Yiu SM, Chan TF, Lam ET. An iterative algorithm for de novo optical map assembly. Proc. - 2017 IEEE Int. Conf. Bioinforma. Biomed. BIBM 2017, 2017. https://doi.org/10.1109/BIBM.2017.8217958
- 68.Anantharaman T.S. Genomics via optical mapping II: Ordered restriction maps. J Comput Biol. 1997 doi: 10.1089/cmb.1997.4.91. [DOI] [PubMed] [Google Scholar]
- 69.Valouev A., Schwartz D.C., Zhou S., Waterman M.S. An algorithm for assembly of ordered restriction maps from single DNA molecules. Proc Natl Acad Sci U S A. 2006 doi: 10.1073/pnas.0604040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goldstein S, Schwartz DC. Germinate & Grow. Res. Comput. Mol. Biol., 2011.
- 71.Zhou S., Goldstein S., Place M., Bechner M., Patino D., Potamousis K. A clone-free, single molecule map of the domestic cow (Bos taurus) genome. BMC Genomics. 2015 doi: 10.1186/s12864-015-1823-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barseghyan H., Tang W., Wang R.T., Almalvez M., Segura E., Bramble M.S. Next-generation mapping: A novel approach for detection of pathogenic structural variants with a potential utility in clinical diagnosis. Genome Med. 2017 doi: 10.1186/s13073-017-0479-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sharp A.R., Udall J.A. OMWare: A tool for efficient assembly of genome-wide physical maps. BMC Bioinf. 2016 doi: 10.1186/s12859-016-1099-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yuan Y., Bayer P.E., Lee H., Edwards D. runBNG: A software package for BioNano genomic analysis on the command line. Bioinformatics. 2017 doi: 10.1093/bioinformatics/btx366. [DOI] [PubMed] [Google Scholar]
- 75.Burgin J., Molitor C., Mohareb F. MapOptics: A light-weight, cross-platform visualization tool for optical mapping alignment. Bioinformatics. 2019 doi: 10.1093/bioinformatics/bty1013. [DOI] [PubMed] [Google Scholar]
- 76.Yuan Y., Bayer P.E., Scheben A., Chan C.-K.K., Edwards D. BioNanoAnalyst: a visualisation tool to assess genome assembly quality using BioNano data. BMC Bioinf. 2017;18:323. doi: 10.1186/s12859-017-1735-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Arielly R, Ebenstein Y. Irys Extract. Bioinformatics 2018. https://doi.org/10.1093/bioinformatics/btx437. [DOI] [PMC free article] [PubMed]
- 78.Chen Y.M., Yu C.H., Hwang C.C., Liu T. OMACC: an Optical-Map-Assisted Contig Connector for improving de novo genome assembly. BMC Syst Biol. 2013 doi: 10.1186/1752-0509-7-S6-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Istace B., Belser C., Aury J.-M. BiSCoT: Improving large eukaryotic genome assemblies with optical maps. BioRxiv. 2019 doi: 10.1101/674721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pan W., Wanamaker S.I., Ah-Fong A.M.V., Judelson H.S., Lonardi S. Novo&Stitch: Accurate reconciliation of genome assemblies via optical maps. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pan W., Jiang T., Lonardi S. OMGS: Optical Map-Based Genome Scaffolding. J Comput Biol. 2019 doi: 10.1089/cmb.2019.0310. [DOI] [PubMed] [Google Scholar]
- 82.Lin H.C., Goldstein S., Mendelowitz L., Zhou S., Wetzel J., Schwartz D.C. AGORA: Assembly Guided by Optical Restriction Alignment. BMC Bioinf. 2012 doi: 10.1186/1471-2105-13-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mukherjee K., Alipanahi B., Kahveci T., Salmela L., Boucher C. Aligning optical maps to de Bruijn graphs. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz069. [DOI] [PubMed] [Google Scholar]
- 84.Li M, Mak ACY, Lam ET, Kwok PY, Xiao M, Yip KY, et al. Towards a more accurate error model for BioNano optical maps. Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics), 2016. https://doi.org/10.1007/978-3-319-38782-6_6.
- 85.Muggli M.D., Puglisi S.J., Ronen R., Boucher C. Misassembly detection using paired-end sequence reads and optical mapping data. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pan W., Lonardi S. Accurate detection of chimeric contigs via Bionano optical maps. Bioinformatics. 2019 doi: 10.1093/bioinformatics/bty850. [DOI] [PubMed] [Google Scholar]
- 87.Lin J., Qi R., Aston C., Jing J., Anantharaman T.S., Mishra B. Whole-genome shotgun optical mapping of Deinococcus radiodurans. Science. 1999;(80-:). doi: 10.1126/science.285.5433.1558. [DOI] [PubMed] [Google Scholar]
- 88.Lai Z., Jing J., Aston C., Clarke V., Apodaca J., Dimalanta E.T. A shotgun optical map of the entire Plasmodium falciparum genome. Nat Genet. 1999 doi: 10.1038/15484. [DOI] [PubMed] [Google Scholar]
- 89.Zhang J., Li C., Zhou Q., Zhang G. Improving the ostrich genome assembly using optical mapping data. GigaScience. 2015 doi: 10.1186/s13742-015-0062-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiao S., Li J., Ma F., Fang L., Xu S., Chen W. Rapid construction of genome map for large yellow croaker (Larimichthys crocea) by the whole-genome mapping in BioNano Genomics Irys system. BMC Genomics. 2015 doi: 10.1186/s12864-015-1871-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Seo J.S., Rhie A., Kim J., Lee S., Sohn M.H., Kim C.U. De novo assembly and phasing of a Korean human genome. Nature. 2016 doi: 10.1038/nature20098. [DOI] [PubMed] [Google Scholar]
- 92.Jain M., Koren S., Miga K.H., Quick J., Rand A.C., Sasani T.A. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol. 2018 doi: 10.1038/nbt.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ganapathy G., Howard J.T., Ward J.M., Li J., Li B., Li Y. High-coverage sequencing and annotated assemblies of the budgerigar genome. GigaScience. 2014 doi: 10.1186/2047-217X-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roscito J.G., Sameith K., Pippel M., Francoijs K.J., Winkler S., Dahl A. The genome of the tegu lizard Salvator merianae: combining Illumina, PacBio, and optical mapping data to generate a highly contiguous assembly. GigaScience. 2018 doi: 10.1093/gigascience/giy141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang L., Wu J., Liu X., Di D., Liang Y., Feng Y. A high-quality genome assembly for the endangered golden snub-nosed monkey (Rhinopithecus roxellana) GigaScience. 2019 doi: 10.1093/gigascience/giz098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kingan S.B., Urban J., Lambert C.C., Baybayan P., Childers A.K., Coates B. A high-quality genome assembly from a single, field-collected spotted lanternfly (Lycorma delicatula) using the PacBio Sequel II system. GigaScience. 2019 doi: 10.1093/gigascience/giz122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Michael T.P., Jupe F., Bemm F., Motley S.T., Sandoval J.P., Lanz C. High contiguity Arabidopsis thaliana genome assembly with a single nanopore flow cell. Nat Commun. 2018 doi: 10.1038/s41467-018-03016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mascher M., Gundlach H., Himmelbach A., Beier S., Twardziok S.O., Wicker T. A chromosome conformation capture ordered sequence of the barley genome. Nature. 2017 doi: 10.1038/nature22043. [DOI] [PubMed] [Google Scholar]
- 99.Song J.M., Guan Z., Hu J., Guo C., Yang Z., Wang S. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat Plants. 2020 doi: 10.1038/s41477-019-0577-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ning D.-L., Wu T., Xiao L.-J., Ma T., Fang W.-L., Dong R.-Q. Chromosomal-level assembly of Juglans sigillata genome using Nanopore, BioNano, and Hi-C analysis. GigaScience. 2020;9. doi: 10.1093/gigascience/giaa006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhu T., Wang L., You F.M., Rodriguez J.C., Deal K.R., Chen L. Sequencing a Juglans regia × J. microcarpa hybrid yields high-quality genome assemblies of parental species. Hortic Res. 2019 doi: 10.1038/s41438-019-0139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Luo M.C., Deal K.R., Murray A., Zhu T., Hastie A.R., Stedman W. Optical nano-mapping and analysis of plant genomes. Methods Mol Biol. 2016 doi: 10.1007/978-1-4939-3622-9_9. [DOI] [PubMed] [Google Scholar]
- 103.Jiao Y., Peluso P., Shi J., Liang T., Stitzer M.C., Wang B. Improved maize reference genome with single-molecule technologies. Nature. 2017 doi: 10.1038/nature22971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Moll K.M., Zhou P., Ramaraj T., Fajardo D., Devitt N.P., Sadowsky M.J. Strategies for optimizing BioNano and Dovetail explored through a second reference quality assembly for the legume model. Medicago truncatula. BMC Genomics. 2017 doi: 10.1186/s12864-017-3971-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Valliyodan B., Cannon S.B., Bayer P.E., Shu S., Brown A.V., Ren L. Construction and comparison of three reference-quality genome assemblies for soybean. Plant J. 2019 doi: 10.1111/tpj.14500. [DOI] [PubMed] [Google Scholar]
- 106.Xie M., Chung C.Y.L., Li M.W., Wong F.L., Wang X., Liu A. A reference-grade wild soybean genome. Nat Commun. 2019 doi: 10.1038/s41467-019-09142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shen Y., Liu J., Geng H., Zhang J., Liu Y., Zhang H. De novo assembly of a Chinese soybean genome. Sci China Life Sci. 2018 doi: 10.1007/s11427-018-9360-0. [DOI] [PubMed] [Google Scholar]
- 108.Dvorak J., Wang L., Zhu T., Jorgensen C.M., Luo M.C., Deal K.R. Reassessment of the evolution of wheat chromosomes 4A, 5A, and 7B. Theor Appl Genet. 2018 doi: 10.1007/s00122-018-3165-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Udall J.A., Long E., Ramaraj T., Conover J.L., Yuan D., Grover C.E. The Genome Sequence of Gossypioides kirkii Illustrates a Descending Dysploidy in Plants. Front Plant Sci. 2019 doi: 10.3389/fpls.2019.01541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang M., Tu L., Yuan D., Zhu D., Shen C., Li J. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat Genet. 2019 doi: 10.1038/s41588-018-0282-x. [DOI] [PubMed] [Google Scholar]
- 111.A reference standard for genome biology. Nat Biotechnol 2018. https://doi.org/10.1038/nbt.4318 . [DOI] [PubMed]
- 112.Kidd J.M., Cooper G.M., Donahue W.F., Hayden H.S., Sampas N., Graves T. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008 doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mak A.C.Y., Lai Y.Y.Y., Lam E.T., Kwok T.P., Leung A.K.Y., Poon A. Genome-wide structural variation detection by genome mapping on nanochannel arrays. Genetics. 2016 doi: 10.1534/genetics.115.183483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Levy-Sakin M., Pastor S., Mostovoy Y., Li L., Leung A.K.Y., McCaffrey J. Genome maps across 26 human populations reveal population-specific patterns of structural variation. Nat Commun. 2019 doi: 10.1038/s41467-019-08992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chan E.K.F., Cameron D.L., Petersen D.C., Lyons R.J., Baldi B.F., Papenfuss A.T. Optical mapping reveals a higher level of genomic architecture of chained fusions in cancer. Genome Res. 2018 doi: 10.1101/gr.227975.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kronenberg ZN, Fiddes IT, Gordon D, Murali S, Cantsilieris S, Meyerson OS, et al. High-resolution comparative analysis of great ape genomes. Science (80-) 2018. https://doi.org/10.1126/science.aar6343. [DOI] [PMC free article] [PubMed]
- 117.Li L., Leung A.K.Y., Kwok T.P., Lai Y.Y.Y., Pang I.K., Chung G.T.Y. OMSV enables accurate and comprehensive identification of large structural variations from nanochannel-based single-molecule optical maps. Genome Biol. 2017 doi: 10.1186/s13059-017-1356-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Barsheghyan SB, Delot EC VE. nanotatoR: nanotatoR: next generation structural variant annotation and classification 2019. https://doi.org/10.18129/B9.bioc.nanotatoR .
- 119.Maiden M.C.J., Bygraves J.A., Feil E., Morelli G., Russell J.E., Urwin R. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998 doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Urwin R., Maiden M.C.J. Multi-locus sequence typing: A tool for global epidemiology. Trends Microbiol. 2003 doi: 10.1016/j.tim.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 121.Skippington E., Ragan M.A. Lateral genetic transfer and the construction of genetic exchange communities. FEMS Microbiol Rev. 2011 doi: 10.1111/j.1574-6976.2010.00261.x. [DOI] [PubMed] [Google Scholar]
- 122.Latreille P., Norton S., Goldman B.S., Henkhaus J., Miller N., Barbazuk B. Optical mapping as a routine tool for bacterial genome sequence finishing. BMC Genomics. 2007 doi: 10.1186/1471-2164-8-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Miller J.M. Whole-genome mapping: A new paradigm in strain-typing technology. J Clin Microbiol. 2013 doi: 10.1128/JCM.00093-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Browning S.R., Browning B.L. Haplotype phasing: Existing methods and new developments. Nat Rev Genet. 2011 doi: 10.1038/nrg3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Alföldi J., Lindblad-Toh K. Comparative genomics as a tool to understand evolution and disease. Genome Res. 2013 doi: 10.1101/gr.157503.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huang M., Tu J., Lu Z. Recent advances in experimental whole genome haplotyping methods. Int J Mol Sci. 2017 doi: 10.3390/ijms18091944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pendleton M., Sebra R., Pang A.W.C., Ummat A., Franzen O., Rausch T. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat Methods. 2015 doi: 10.1038/nmeth.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ananthasayanam S., Kothandaraman H., Nayee N., Saha S., Baghel D.S., Gopalakrishnan K. First near complete haplotype phased genome assembly of River buffalo (Bubalus bubalis) BioRxiv. 2019 doi: 10.1101/618785. [DOI] [Google Scholar]
- 129.Low W.Y., Tearle R., Liu R., Koren S., Rhie A., Bickhart D.M. Haplotype-Resolved Cattle Genomes Provide Insights Into Structural Variation and Adaptation. BioRxiv. 2019 doi: 10.1101/720797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang Y., Lian J., Xie B., Chen M., Niu Y., Li Q. Chromosome-scale de novo assembly and phasing of a Chinese indigenous pig genome. BioRxiv. 2019 doi: 10.1101/770958. [DOI] [Google Scholar]
- 131.Kuon J.E., Qi W., Schläpfer P., Hirsch-Hoffmann M., Von Bieberstein P.R., Patrignani A. Haplotype-resolved genomes of geminivirus-resistant and geminivirus-susceptible African cassava cultivars. BMC Biol. 2019 doi: 10.1186/s12915-019-0697-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Geron-Landre B. Sequence-specific fluorescent labeling of double-stranded DNA observed at the single molecule level. Nucleic Acids Res. 2003 doi: 10.1093/nar/gng125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Müller V., Dvirnas A., Andersson J., Singh V., Kk S., Johansson P. Enzyme-free optical DNA mapping of the human genome using competitive binding. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ananiev G.E., Goldstein S., Runnheim R., Forrest D.K., Zhou S., Potamousis K. Optical mapping discerns genome wide DNA methylation profiles. BMC Mol Biol. 2008 doi: 10.1186/1471-2199-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gabrieli T., Sharim H., Nifker G., Jeffet J., Shahal T., Arielly R. Epigenetic Optical Mapping of 5-Hydroxymethylcytosine in Nanochannel Arrays. ACS Nano. 2018 doi: 10.1021/acsnano.8b03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sharim H., Grunwald A., Gabrieli T., Michaeli Y., Margalit S., Torchinsky D. Long-read single-molecule maps of the functional methylome. Genome Res. 2019 doi: 10.1101/gr.240739.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Young E., Abid H.Z., Kwok P.Y., Riethman H., Xiao M. Comprehensive Analysis of Human Subtelomeres by Whole Genome Mapping. PLoS Genet. 2020 doi: 10.1371/journal.pgen.1008347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.McCaffrey J., Young E., Lassahn K., Sibert J., Pastor S., Riethman H. High-throughput single-molecule telomere characterization. Genome Res. 2017 doi: 10.1101/gr.222422.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang D., Chan S., Sugerman K., Lee J., Lam E.T., Bocklandt S. CRISPR-bind: a simple, custom CRISPR/dCas9-mediated labeling of genomic DNA for mapping in nanochannel arrays. BioRxiv. 2018 doi: 10.1101/371518. [DOI] [Google Scholar]
- 140.Kyriakidou M., Tai H.H., Anglin N.L., Ellis D., Strömvik M.V. Current strategies of polyploid plant genome sequence assembly. Front Plant Sci. 2018 doi: 10.3389/fpls.2018.01660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Paten B., Novak A.M., Eizenga J.M., Garrison E. Genome graphs and the evolution of genome inference. Genome Res. 2017 doi: 10.1101/gr.214155.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rakocevic G., Semenyuk V., Lee W.P., Spencer J., Browning J., Johnson I.J. Fast and accurate genomic analyses using genome graphs. Nat Genet. 2019 doi: 10.1038/s41588-018-0316-4. [DOI] [PubMed] [Google Scholar]
- 143.Ameur A. Goodbye reference, hello genome graphs. Nat Biotechnol. 2019 doi: 10.1038/s41587-019-0199-7. [DOI] [PubMed] [Google Scholar]
- 144.Miclotte G., Plaisance S., Rombauts S., Van de Peer Y., Audenaert P., Fostier J. OMSim: a simulator for optical map data. Bioinformatics. 2017 doi: 10.1093/bioinformatics/btx293. [DOI] [PMC free article] [PubMed] [Google Scholar]