Abstract
Background
Most gut microbiome studies have been performed using stool samples. However, the small intestine is of central importance to digestion, nutrient absorption, and immune function, and characterizing its microbial populations is essential for elucidating their roles in human health and disease.
Aims
To characterize the microbial populations of different small intestinal segments and contrast these to the stool microbiome.
Methods
Male and female subjects undergoing esophagogastroduodenoscopy without colon preparation were prospectively recruited. Luminal aspirates were obtained from the duodenum, jejunum, and farthest distance reached. A subset also provided stool samples. 16S rRNA sequencing was performed and analyses were carried out using CLC Genomics Workbench.
Results
16S rRNA sequencing identified differences in more than 2000 operational taxonomic units between the small intestinal and stool microbiomes. Firmicutes and Proteobacteria were the most abundant phyla in the small intestine, and Bacteroidetes were less abundant. In the small intestine, phylum Firmicutes was primarily represented by lactic acid bacteria, including families Streptococcaceae, Lactobacillaceae, and Carnobacteriaceae, and Proteobacteria was represented by families Neisseriaceae, Pasteurellaceae, and Enterobacteriaceae. The duodenal and FD microbial signatures were markedly different from each other, but there were overlaps between duodenal and jejunal and between jejunal and FD microbial signatures. In stool, Firmicutes were represented by families Ruminococcaceae, Lachnospiraceae, Christensenellaceae, and Proteobacteria by class Deltaproteobacteria.
Conclusions
The small bowel microbiome is markedly different from that in stool and also varies between segments. These findings may be important in determining how compositional changes in small intestinal microbiota contribute to human disease states.
Electronic supplementary material
The online version of this article (10.1007/s10620-020-06173-x) contains supplementary material, which is available to authorized users.
Keywords: Microbiome, Duodenum, Jejunum, Stool, 16S metagenomic analysis
Introduction
The Human Microbiome Project [1] was a groundbreaking effort to understand the microbiome of the human body. The microbial populations of several important body sites were analyzed, including the mouth, nose, skin, vaginal tract, and gut [2]. The gut microbiome was specifically characterized using stool as a surrogate, due to the ease of obtaining stool samples. However, while stool is useful in analyzing the microbial populations of the distal colon, it does not adequately represent the entire gastrointestinal tract [3]. Conditions such as acidity, transit time, and microbial biomass vary tremendously along the length of the intestinal tract, and these differences have significant effects on the microbial populations present [3].
A number of parameters in the evaluation of the human gut microbiome have been framed by this initial work using stool. First, Bacteroidetes and Firmicutes are the two dominant phyla in the stool microbiome [4, 5], and the proportion and composition of these two phyla may be important in obesity and other conditions [6, 7]. A second important parameter is microbial diversity, which has been found to be an important determinant of human health. Many gut conditions, such as inflammatory bowel disease, are associated with reduced microbial diversity in stool samples [8, 9].
In contrast to the colon, the small intestine, which is divided into the duodenum, jejunum, and ileum, is of central importance to digestion and nutrient absorption as well as immune function [10]. Of these, the duodenum is perhaps the most important, as it is the principal site of convergence for chyme from the stomach, enzymes from the pancreas and bile salts from the hepatobiliary system. Recognizing the importance of characterizing the microbial populations of the small intestine and their roles in human health and disease, we devised the REIMAGINE (Revealing the Entire Intestinal Microbiota and its Associations with the Genetic, Immunologic, and Neuroendocrine Ecosystem) study [11]. A critical part of studying the small bowel microbiome is the development of techniques to acquire and analyze small intestinal samples. The small bowel is a low microbial biomass environment, in contrast to the high microbial biomass of stool [3, 12], and the open aspiration catheters traditionally used for aspirating fluid are associated with a high risk of oral contamination, which is extremely problematic for microbiome analysis [13, 14]. The amount of fluid in the small bowel is also limited, particularly when patients are in a fasted state, and it is often viscous, making microbes in this matrix poorly accessible. Therefore, as part of the REIMAGINE study, we optimized techniques to reduce cross-contamination from other sites such as the mouth and stomach, developed methods for releasing bacteria from viscous mucous, and improved techniques for low biomass fluids [15]. In this study, we used these validated techniques to characterize the microbial populations in different parts of the small intestine and to contrast these to the microbiome in stool.
Methods
Study Subjects
The REIMAGINE study is a large-scale study designed to examine the relationship between small bowel microbiome in human health and disease [15]. Consecutive male and female patients aged 18–85 years and undergoing upper gastrointestinal endoscopy [esophagogastroduodenoscopy (EGD)] or antegrade double-balloon enteroscopy (DBE) without colonoscopy for the standard of care purposes are eligible to participate. After completing a comprehensive health information questionnaire, small bowel aspirates are collected [15]. Optional stool samples are also obtained from subjects who are willing to provide them. The study protocol was approved by the Cedars-Sinai Medical Center Institutional Review Board, and all subjects provided informed written consent prior to participation in the study.
Study Procedures
Questionnaires
Prior to EGD/DBE, all subjects completed a study questionnaire which documented their demographics and medical and family history, including medication use, use of alcohol and recreational drugs, travel history, and dietary habits and changes. Medical information provided by participants was verified using medical records audits for all subjects. All patient data were de-identified prior to analysis. Although subjects taking antibiotics are included in the parent REIMAGINE study, samples from subjects who had taken antibiotics in the 6 months prior to endoscopy were excluded from this study.
Small Intestinal Sample Collection
During EGD, samples of luminal fluid (up to 2 mL) were obtained from the second portion of the duodenum using a custom sterile aspiration catheter (Hobbs Medical, Inc.) as described previously [15]. During DBE, samples of luminal fluid (up to 2 mL) were obtained from the duodenum, jejunum, and the farthest distance reached during the procedure (FD). Again, a custom sterile catheter was used to collect fluid from each segment, to mitigate the risk of oral and gastric acid cross-contamination.
Stool Sample Collection
Subjects who agreed to provide stool samples for comparative analysis of the small intestinal and stool microbiomes were given collection kits after the EGD or DBE procedure. Stool samples were collected using the OMNIgene-GUT Microbiome kit (DNA Genotek, Ottawa, ON, Canada) according to the manufacturer’s instructions. Subjects collected the stool samples at home 1–2 days after the procedure and mailed the samples to the laboratory. Samples were stored at 25 °C, then frozen and transferred to − 80 °C until analyzed.
Aspirate Processing
Immediately after aspiration, samples were processed using a novel validated protocol that optimizes microbial isolation specifically from small bowel aspirates [15]. In brief, samples were treated with sterile 1 × Dithiothreitol (EMD Millipore Corp., Darmstadt, Germany) in a 1:1 ratio and vortexed for 30 s. Samples were then centrifuged at high speed (~ 14,000 RPM) for 10 min, and the supernatant was carefully removed and stored at − 80 °C for untargeted metabolomic analysis. Five hundred microliters of sterile Allprotect reagent (Qiagen, Hilden, Germany) was added to each pellet for stabilization of DNA, RNA, and proteins, and the pellets were stored at − 80 °C prior to DNA isolation and analysis of the small bowel microbiome.
DNA Extraction and Quantification
For small bowel aspirate samples, microbial pellets stored under Allprotect reagent were thawed on ice and sterile 1 × DTT was added in a 1:1 ratio to liquefy the Allprotect as described previously. [15] DNA extraction was then performed as described previously, [15] using the MagAttract PowerSoil DNA KF Kit (Qiagen, cat. No. 27000-4-KF). For stool samples, the OMNIgene-GUT tubes were vortexed vigorously for 10 s. 250 µL of the samples were transferred to a sterile 2-mL tube where the lysis step was carried out. DNA extraction was then performed using the MagAttract PowerSoil DNA KF Kit. DNAs were quantified using Qubit dsDNA BR Assay kits or Quant-iT™ PicoGreen® dsDNA assay (Invitrogen by Thermo Fisher Scientific, Waltham, MA, USA) on a Qubit 4 Fluorometer (Invitrogen).
Library Preparation and 16S rRNA Sequencing
16S library preparation for DNAs from small bowel aspirates and stool was performed according to the Illumina (Illumina, San Diego, CA, USA) protocol https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf as described previously [15], using the gene-specific primers S-D-Bact-0341-b-S-17 and S-D-Bact-0785-a-A-21 published and validated by Klindworth et al. [16] to amplify the V3 and V4 regions. The final libraries were quantified using Qubit 1X dsDNA HS Assay kits on a Qubit 4 Fluorometer and analyzed using Agilent DNA 1000 chips (Agilent Technologies, Santa Clara, CA) on an Agilent 2100 Bioanalyzer System (Agilent Technologies, Santa Clara, CA).
16S Metagenomic Sequencing and Analysis
As described previously, 15 to 20 pM of the pooled libraries were paired-end sequenced on a MiSeq System (Illumina, San Diego, California) [15]. Operational Taxonomic Unit (OTU) clustering and taxonomic analyses were performed using CLC Genomics Workbench v. 10.1.1 and CLC Microbial Genomics Module v. 2.5 (Qiagen). Sequences were first trimmed and merged and then were clustered into OTUs at 97% sequence similarity level using the Amplicon-Based OTU clustering tool. The most abundant sequences were selected as representative of each cluster and then assigned to a taxonomy level using CLC Microbial Genomics default values and the Greengenes Database 2013 release. Low-depth samples (less than 9000 sequences per sample) were removed from the analysis. Alpha diversity indexes were calculated using the Abundance Analysis tool. The weighted Unifrac metric was used to calculate inter-sample diversity (beta-diversity).
Statistical Analysis
Predictions for significant differentially abundant OTUs within each small bowel segment and stool were performed following recommendations from McMurdie and Holmes [17], and from Weiss et al. [18], used when the average library size for each group is approximately equal and/or the fold difference between groups is not high (> 2–3x on average).
Multiple comparisons and statistical analyses were performed using CLC Genomics Workbench v. 10.1.1 and CLC Microbial Genomics Module v. 2.5 (Qiagen) as described previously [15]. A negative binomial GLM model was used to obtain maximum likelihood estimates for an OTU’s log-fold change between two conditions, and the Wald test was used to determine significance. False discovery rate (FDR) was performed to correct P values. Graph construction and PLS-DA analysis were performed using GraphPad Prism 7.02 (GraphPad Software, La Jolla, CA, USA) and MetaboAnalyst (Xia Lab @ McGill University), respectively.
Results
Subjects and Samples
At the time of this analysis, the REIMAGINE study had prospectively collected duodenal aspirates from 232 consecutive subjects undergoing any form of upper endoscopic procedure [esophagogastroduodenoscopy (EGD) or oral double-balloon endoscopy (DBE)] and who had not taken any antibiotics in the 6 months prior to endoscopy. Of these, 53 subjects who underwent EGD provided duodenal aspirates and matching stool samples (Table 1). This group served as the basis to compare the proximal small bowel microbiome to stool. A second group of 23 subjects had undergone DBE to traverse the full length of the small intestine and provided samples of luminal fluid from the duodenum and jejunum and the farthest distance (FD) reached during the procedure. Of these 23 DBE subjects, eight provided matching stool samples, forming a third group for analysis (see Table 1).
Table 1.
Subject demographics
| Duodenal aspirates with matching stool samples (Group 1, n = 53) |
Small intestinal segmental aspirates (duodenum, jejunum, and FD) (Group 2, n = 23) |
Small intestinal segmental aspirates with matching stool samples (Group 3, n = 8) |
|
|---|---|---|---|
| Gender (Male/Female) | 20/33 | 8/15 | 4/4 |
|
Age (years) (Mean ± SD) |
61.86 ± 12.32 | 57.69 ± 11.64 | 60.25 ± 11.51 |
| Reasons for endoscopy | Screening, abdominal pain, dyspepsia, GERD, bloating, diarrhea, IBS, workup of celiac disease, Crohn’s disease, anemia, GI bleed, etc | ||
Microbial Profiles in Duodenum and Stool (Group 1)
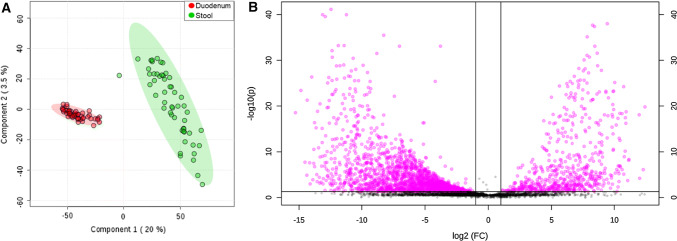
For the 53 subjects who provided both duodenal samples during EGD and stool samples, beta-diversity analysis of the corresponding microbiomes revealed two completely distinct clusters on a PCoA plot (Fig. 1a), and differences in more than 2000 OTUs were identified between the duodenal and stool microbiomes (Fig. 1b).
Fig. 1.
a PCoA plot of the duodenal and stool microbial profiles in the same subjects (Group 1, n = 53). Duodenum—red; stool—green. Transparent circles indicate the 95% confidence region. b Volcano plot of OTUs that were statistically different in the duodenal microbiome and stool microbiome in the same subjects (Group 1, n = 53) (FDR P value < 0.05 and fold change (FC) threshold of 2.0 shown in pink)
The duodenal microbiome was characterized by higher relative abundances of the phyla Firmicutes (~ 55%) and Proteobacteria (~ 21%), whereas the stool microbiome exhibited higher relative abundances of Bacteroidetes (~ 50%) and Firmicutes (~ 40%) (Supplemental Table 1). When compared to the microbial profile in stool, the duodenum exhibited markedly increased relative abundance of phyla Proteobacteria, Fusobacteria, Actinobacteria, and TM7, the last of which was present only in the duodenal microbiome (Supplemental Figure 1). The relative abundance of phylum Firmicutes was also increased in the duodenal microbiome compared to stool (Fold Change, FC = 1.40, FDR P value = 0.004) (Table 2). In contrast, phylum Bacteroidetes was significantly decreased in the duodenal microbiome when compared to stool, and the phylum Verrucomicrobia was nearly undetectable in the duodenum (Supplemental Figure 1) (Table 2). At the genus level, the relative abundances of a total of 156 genera were statistically different in the duodenal microbiome compared to the stool microbiome, and Euclidean Ward clustering analysis revealed two distinct microbial signatures (Supplemental Figure 2).
Table 2.
Phylum-level differences between the duodenal and stool microbiomes in the same subjects (Group 1, n = 53)
| Duodenal microbiome versus stool microbiome (n = 53) | |||
|---|---|---|---|
| Phylum | Fold Change (FC) | log2(FC) | FDR P value |
| Bacteroidetes | 0.10753 | − 3.2172 | 2.61E−20 |
| Verrucomicrobia | 0.002806 | − 8.4771 | 1.19E−16 |
| TM7 | 129.17 | 7.0131 | 1.19E−16 |
| Actinobacteria | 8.837 | 3.1436 | 1.91E−15 |
| Fusobacteria | 7.2287 | 2.8537 | 3.29E−13 |
| Cyanobacteria | 0.040278 | − 4.6339 | 1.47E−08 |
| Spirochaetes | 98.719 | 6.6253 | 5.18E−06 |
| SR1 | 422.43 | 8.7226 | 0.000274 |
| Proteobacteria | 3.6101 | 1.852 | 0.000505 |
| Euryarchaeota | 0.005182 | − 7.5924 | 0.000702 |
| Chloroflexi | 32.155 | 5.007 | 0.003565 |
| Lentisphaerae | 0.009193 | − 6.7653 | 0.003959 |
| Firmicutes | 1.4011 | 0.4866 | 0.004179 |
| Thermi | 13.653 | 3.7712 | 0.06949 |
Microbial Profiles in Different Small Intestinal Segments (Group 2)
A total of 23 subjects had the microbiome mapped in three different three segments of the small intestine (duodenum, jejunum, and farthest distance [FD] reached during the DBE procedure) (Table 1). Microbial alpha diversity was similar in all three segments, as determined by Simpson’s index, Shannon entropy, and Total OTU number (ANOVA P value > 0.05). Beta-diversity in all three small intestinal segments was also similar, as determined by the unweighted UniFrac distance, and no distinct clusters were observed on the PCoA plots (Supplemental Figure 3). The major phyla observed in these small intestinal segments were Firmicutes, Proteobacteria, and Actinobacteria, which together represented almost 90% of the total relative abundance (Fig. 2, Supplemental Table 2). The remaining phyla including Fusobacteria, Bacteroidetes, and TM7 together had a relative abundance of < 10% in the small bowel (Supplemental Table 2). While this pattern was consistent in all segments analyzed, there was some variation within this. Specifically, the relative abundances of the phyla Bacteroidetes, Proteobacteria, and Fusobacteria varied in the different small intestinal segments (likelihood ratio test (LRT) FDR P value < 0.05), and the relative abundance of phylum Proteobacteria was higher in the jejunum than in the duodenum (FC = 9.99, FDR P value = 5.39E−4) and FD (FC = 8.16, FDR P value = 2.64E−3). In these 23 subjects, the top five most abundant classes regardless of small intestinal segment were Bacilli and Clostridia (phylum Firmicutes), Gammaproteobacteria (phylum Proteobacteria), Actinobacteria (phylum Actinobacteria), and Fusobacteriia (phylum Fusobacteria). However, the relative abundances of classes Gammaproteobacteria and Fusobacteriia were different between segments (LRT FDR P value < 0.001, FDR P value = 2.72E−7, respectively), and Gammaproteobacteria was increased in the jejunum compared to duodenum and FD (FDR P value = 2.86E−4, FDR P value = 0.02, respectively).
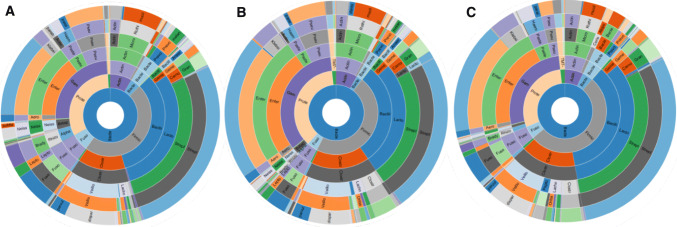
Fig. 2.
The 16S rRNA microbiome profiles of aspirates collected from a—duodenum, b—jejunum, and c—farthest distance (FD), in the same subjects (Group 2, n = 23)
At the family level, 111 families were identified in the duodenal microbiome, 99 in the jejunal microbiome, and 101 in the FD microbiome (Supplemental Figure 4). The most abundant family in all small intestinal segments tested was Streptococcaceae (p_Firmicutes,c_Bacilli,o_Lactobacillales), which represented more than 26% of the entire microbiome composition in each segment, followed by Enterobacteriaceae (p_Proteobacteria,c_Gammaproteobacteria,o_Enterobacteriales), which represented 21% in each segment, and Veillonellaceae (p_Firmicutes,c_Clostridia,o_Clostridiales), which represented 9% in each segment. The relative abundances of 11 microbial families varied depending on location in the small intestine (Supplemental Table 3). The duodenal microbiome exhibited decreased relative abundance of family Clostridiaceae and increased relative abundance of family Bradyrhizobiaceae compared to the jejunum and FD (P < 0.05, Supplemental Table 3). The FD microbiome exhibited increased relative abundance of family Peptostreptococcaceae when compared to duodenum and jejunum (P < 0.05, Supplemental Table 3). In addition, the relative abundances of many other families were different in FD when compared to duodenum, including Bacillaceae, Bacteroidaceae, Enterococcaceae, and many families from phylum Proteobacteria, such as Enterobacteriaceae, Moraxellaceae, and Neisseriaceae (Supplemental Table 3).
PLS discriminant analysis (PLS-DA) at the family level, performed in order to sharpen the separation between segments by rotating PCoA components such that a maximum separation among intestinal segments was obtained, revealed that the microbial signature within the jejunum overlapped with the duodenal and FD microbial signatures (Supplemental Figure 5A). In contrast, the family-level microbial signature in duodenal samples was markedly different from the signature identified in FD (Supplemental Figure 5B), and the top 3 variable importance in projection (VIP) scores associated with the separation observed on the PLS-DA plot were for families Prevotellaceae, Neisseriaceae, and Moraxellaceae (Supplemental Figure 6).
Microbial Profiles in Different Small Intestinal Segments Versus Stool (Group 3)
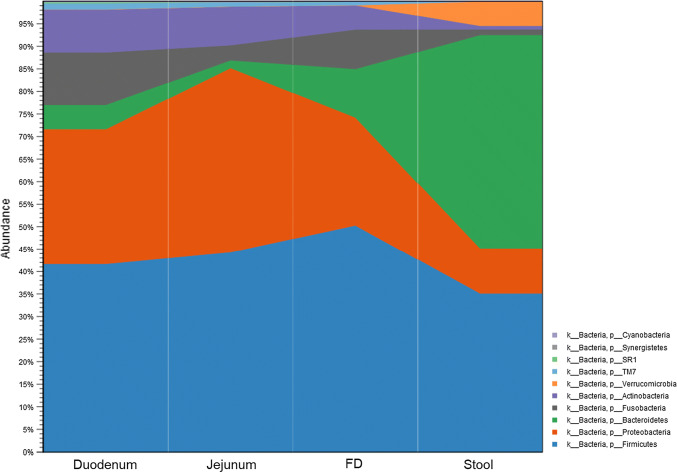
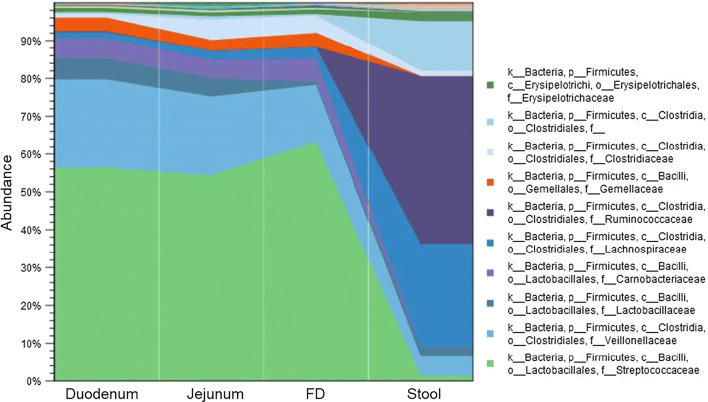
Lastly, we compared the microbiome profiles in the different segments of the small bowel, as represented by duodenal, jejunal and FD samples, to the microbiome in stool samples from the same individuals (n = 8). The results revealed a clear and marked difference in the microbiome from duodenum to stool (Fig. 3, Supplemental Figure 7). Phylum Firmicutes was highly abundant in both the small bowel and stool. It was the major phylum detected in the small intestine, representing more than 50% of the total microbial relative abundance in all segments, and the second major phylum detected in stool. Overall, the relative abundance of Firmicutes in the duodenum was slightly higher than in stool (Table 2). However, the microbial composition within this phylum was markedly different in the small intestinal segments when compared to stool (Fig. 4). In the small intestinal segments, phylum Firmicutes was primarily represented by the order Lactobacillales (68% of all Firmicutes), which includes lactic acid bacteria (which are facultative anaerobes) from the families Streptococcaceae, Lactobacillaceae, and Carnobacteriaceae, the last of which was absent in the stool (Fig. 4). In contrast, phylum Firmicutes in stool was primarily represented by the order Clostridiales (93% of all Firmicutes), which includes obligate anaerobes from the families Ruminococcaceae, Lachnospiraceae, and Christensenellaceae, the last of which was absent in the small intestine. The relative abundances of these three families were increased in the stool microbiome when compared to all segments of the small intestine (Supplemental Table 4). The relative abundance of phylum Proteobacteria was increased in the duodenal microbiome when compared to stool (Table 2). In the small intestine, this phylum was characterized by higher abundance of family Neisseriaceae (class Betaproteobacteria) (FC = 5.57, FDR P value = 2.13E−14), and families Pasteurellaceae and Enterobacteriaceae (class Gammaproteobacteria) (FC = 4.93, FDR P value = 2.30E−12 and FC = 3.35, FDR P value = 1.03E−4, respectively) (Supplemental Figure 8A). In contrast, phylum Proteobacteria in stool was characterized by the Alcaligenaceae family and H2S producers from class Deltaproteobacteria (FC = 3.32, FDR P value = 2.10E−03, and FC = 1.82 FDR, P value = 0.03, respectively) (Supplemental Figure 8B).
Fig. 3.
Relative abundance of microbial populations at the phylum level in the duodenum, jejunum, and farthest distance (FD) and stool, in the same subjects (Group 3, n = 8)
Fig. 4.
Relative abundance of the major Firmicutes families in the duodenum, jejunum, FD, and stool, in the same subjects (Group 3, n = 8)
Discussion
Over the last decade, there has been a tremendous interest in the role of the human microbiome in human health and disease. The human microbiome project began the tremendous volume of work that has since dominated the literature [1]. The original description of the human microbiome included analyses of the mouth, nose, skin, vagina, and gut. Stool was used as a surrogate for the entire “gut,” and a large number of studies identified differences in the proportions of Firmicutes and Bacteroidetes, the two major phyla of the stool microbiome. Two key findings from these studies using stool are that imbalances in the relative abundances of different bacterial populations are important, and the association between reductions in microbial diversity and various diseases and conditions. For example, initial data suggested that obesity could be associated with a higher Bacteroidetes/Firmicutes ratio [6, 7]. In the case of diversity, data consistently show that serious intestinal diseases such as inflammatory bowel disease are associated with reductions in the diversity of the stool microbiome [8, 9].
While findings related to the microbiome continue to evolve, there are limitations to using stool a surrogate for the entire gut. Stool is easy to access and has a high bacterial biomass, making DNA extraction and amplification easier. However, the colon only represents 4–5 feet of the entire intestine. The small intestine, which is 20 feet long, is an important part of the gastrointestinal tract. It is the site of convergence of the gastric contents (food), bile, and pancreatic secretions, and is the major absorptive surface of the intestinal tract. In addition, given the villi and microvilli, the small intestine has a large surface area. Therefore, the influence of small bowel microbes is potentially enormous, and yet there has been no systematic study of the microbiome across the full length of the small and large intestines, although previous studies have assessed small intestinal microbiome composition based on biopsy and luminal contents analysis in healthy volunteers and in disease states [19–25]. This study is part of the REIMAGINE study, which is a large-scale effort to examine and understand the microbial composition of the small intestine in human health and disease, and provides a detailed analysis of the tremendous differences between the small bowel and stool microbiomes. Our findings demonstrate that the small bowel microbiome is unique and that stool is not a surrogate for the entire gut microbiome. Numerous differences were identified between the small bowel and stool microbiomes. The stool microbiome is characterized by a high relative abundance of two major phyla, Bacteroidetes and Firmicutes. The small bowel microbiome is also dominated by two major phyla, but these are Firmicutes and Proteobacteria, and there is a low relative abundance of Bacteroidetes, possibly due to high levels of bile acids in the small intestine [26]. While it appears that the common link between the stool and small bowel microbiomes is the phylum Firmicutes, closer examination reveals that the composition of Firmicutes in the small bowel is also very different from that in stool and is mainly characterized by the families Streptococcaceae, Veillonellaceae, and Lactobacillaceae. Families Streptococcaceae and Lactobacillaceae produce lactic acid as the final product of glucose fermentation [27, 28]. The role of lactic acid bacteria in the gut is not fully understood, but many studies have associated these bacteria with gut and immune system improvements, and with mediating the effects of the gut microbiota on the development of obesity [29–31].
The composition of phylum Proteobacteria in the small bowel is also different from that in stool. Species from class Deltaproteobacteria are more abundant in stool, including hydrogen sulfite producers such as Desulfovibrio sp. and Bilophila sp. [32, 33]. In contrast, in the small intestine phylum Proteobacteria is strongly represented by class Gammaproteobacteria, which comprises several known pathogens with medical and scientific importance [34]. In addition, Proteobacteria species commonly isolated in saliva, such as Haemophilus sp. and Neisseria subflava, are also abundant in the small bowel [35]. The small intestinal microbiome also contains distinct taxa, such as TM7. The candidate phylum TM7 is mainly found in the small intestine, where it represents almost 2% of the total bacterial abundance. TM7 has been found in trace amounts in stool. This unique phylum is associated with human diseases, such as inflammatory mucosal disease, and despite its global prevalence, still cannot be cultivated, making it one of the most enigmatic phyla known [36, 37]. Similarly, the stool microbiome exhibits a distinct abundance of taxa from the phylum Verrucomicrobia.
Until recently, the deep small bowel was inaccessible. The endoscopic technique known as double-balloon endoscopy has allowed access to nearly all levels of the small intestine and in some cases can reach the cecum. This technique enables the evaluation of the microbiome along almost the entire length of bowel. In this study, it initially appeared that the small bowel had a consistent bacterial profile. However, further analysis revealed that there is a gradual shift in the microbiome in samples from more distal locations within the small bowel. These are highlighted by subtle shifts in specific taxa, such as a gradual increase in Bacteroidetes and reduction in Proteobacteria, but these still do not reach the levels found in the colon. Many of our findings are consistent with the results of previous studies which examined and compared the small intestinal microbiome in conditions such as irritable bowel syndrome (IBS) and celiac disease and in healthy controls, using techniques that ranged from fluorescent in situ hybridization (FISH) to pyrosequencing and 16S rRNA gene sequencing [22–25]. For example, Dlugosz et al. [23] examined the mucosa-associated microbiome in the jejunum of IBS subjects and healthy controls, and found a similar order of abundance of the major phyla (Firmicutes 43/ %, Proteobacteria 23%, Bacteriodetes 15%, and Actinobacteria 9.3%) in both IBS subjects and controls, and identified high abundances of the genera Streptococcus and Veillonella, but lower Escherichia. However, Chung et al. [24] found higher abundance of Proteobacteria (47.7%), similar abundance of Bacteroidetes (15.3%) and lower abundance of Firmicutes (11%) in mucosa-associated jejunal microbiome of healthy controls. Interestingly, that study found that family Prevotellaceae was more abundant, and Neisseriaceae less abundant, in the mucosa-associated jejunal microbiome of IBS subjects versus controls, and found an association between Neisseriaceae and severity of IBS symptoms [24]. Li et al. [19] examined duodenal luminal and mucosa-associated microbiome, as well as fecal and rectal mucosa-associated microbiomes, in a small study of 9 healthy volunteers, and also found that phyla Firmicutes and Proteobacteria dominated in the duodenal samples, with the genera Prevotella, Stenotrophomonas, and Streptococcus dominating in the duodenal luminal samples. Lastly, in a study of duodenal biopsies from patients with celiac disease, patients with celiac disease on a gluten-free diet, and controls, D’Argenio et al. [38] found highest relative abundances of genera Streptococcus, Prevotella, Propioibacterium, and Acinetobacter in healthy controls and found that genus Neisseria was significantly more abundant in duodenal samples from patients with celiac disease versus controls.
As this study is forward-looking, it is also important to look at historical methods for sampling the small bowel as a contrast. While there are new emerging devices to sample the small bowel microbiome (electronic capsules), the Watson capsule has been around for decades and was predominantly used to biopsy the small bowel but has been used to also assess the small bowel microbiome [39]. The challenge with this capsule is its lack of sterility as it must traverse the upper passages and stomach. The other challenge is localization, which requires radiology. Another method to assess the small bowel microbiome dates even earlier. Ileal effluents have been studied as early as 1969 for microbiology, but these are predominantly done in patients with ileostomies [40, 41]. The challenge here is that the colon has an enormous volume of bacteria that can influence the ileum. In the intact bowel, ileal effluents would require colonoscopy and colonoscopy preparation which is known to impact the small bowel and colonic microbiomes.
Although the REIMAGINE study is intended to be a definitive large-scale study of the microbial composition of the small intestine, there are limitations to the current work. One of the pitfalls of the study is that these subjects were undergoing upper endoscopy for the assessment of intestinal complaints, including dyspepsia, GERD, anemia, and others. It is possible that these conditions influence the results of the study. However, one argument against this is the concept of regression to the mean assuming adequate sample size. Another possible limitation is the consideration of biomass. Relative abundance is one consideration in microbiome analysis, but another is the absolute number of bacteria in a particular taxonomic group. For example, there may be a larger relative abundance of Proteobacteria in the small intestine compared to stool, but the absolute number of Proteobacteria/mL may be greater in stool. However, this does not take into account the biomass in relation to the very large surface area of the small intestine. As such, this impact and the true biomass are difficult to calculate.
In conclusion, small bowel microbiome is unique in many ways. Considering the surface area and length of the small bowel and its importance to human nutrition and immune function, analysis of the small bowel microbiome may have a greater impact on our understanding of human disease than analysis of stool. As this study demonstrates, the human small intestinal microbiome is dominated by the phyla Firmicutes and Proteobacteria. Deeper analysis reveals that within the major phyla, the distributions of particular taxa vary in different parts of the small bowel. Understanding these compositional changes in the microbiome in all parts of the gut, including the small intestine, will be important to finding future links to human health and disease.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Data availability
The datasets generated during the current study are available in the National Center for Biotechnology Information (NCBI) BioProject Repository https://www.ncbi.nlm.nih.gov/bioproject under BioProject ID PRJNA590519.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hillman ET, Lu H, Yao T, Nakatsu CH. Microbial ecology along the gastrointestinal tract. Microbes Environ. 2017;32:300–313. doi: 10.1264/jsme2.ME17017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sears CL. A dynamic partnership: celebrating our gut flora. Anaerobe. 2005;11:247–251. doi: 10.1016/j.anaerobe.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Consortium HMP. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 7.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durban A, Abellan JJ, Jimenez-Hernandez N, et al. Structural alterations of faecal and mucosa-associated bacterial communities in irritable bowel syndrome. Environ Microbiol Rep. 2012;4:242–247. doi: 10.1111/j.1758-2229.2012.00327.x. [DOI] [PubMed] [Google Scholar]
- 10.Leser TD, Molbak L. Better living through microbial action: the benefits of the mammalian gastrointestinal microbiota on the host. Environ Microbiol. 2009;11:2194–2206. doi: 10.1111/j.1462-2920.2009.01941.x. [DOI] [PubMed] [Google Scholar]
- 11.Sedighi R, Mathur R, Celly S, et al. Sa1912—revealing the entire intestinal microbiota and its associations to the genetic, immunologic, and neuroendocrine ecosystems: methodology for the REIMAGINE study. Gastroenterology. 2019;156:S-450. doi: 10.1016/S0016-5085(19)37978-8. [DOI] [Google Scholar]
- 12.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erdogan A, Rao SS, Gulley D, Jacobs C, Lee YY, Badger C. Small intestinal bacterial overgrowth: duodenal aspiration vs glucose breath test. Neurogastroenterol Motil. 2015;27:481–489. doi: 10.1111/nmo.12516. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs C, Coss Adame E, Attaluri A, Valestin J, Rao SS. Dysmotility and proton pump inhibitor use are independent risk factors for small intestinal bacterial and/or fungal overgrowth. Aliment Pharmacol Ther. 2013;37:1103–1111. doi: 10.1111/apt.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leite G, Morales W, Weitsman S, et al. Optimizing microbiome sequencing for small intestinal aspirates: validation of novel techniques through the REIMAGINE study. BMC Microbiol. 2019 doi: 10.1186/s12866-12019-11617-12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klindworth A, Pruesse E, Schweer T, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiss S, Xu ZZ, Peddada S, et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5:27. doi: 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li G, Yang M, Zhou K, et al. Diversity of duodenal and rectal microbiota in biopsy tissues and luminal contents in healthy volunteers. J Microbiol Biotechnol. 2015;25:1136–1145. doi: 10.4014/jmb.1412.12047. [DOI] [PubMed] [Google Scholar]
- 20.Saffouri GB, Shields-Cutler RR, Chen J, et al. Small intestinal microbial dysbiosis underlies symptoms associated with functional gastrointestinal disorders. Nat Commun. 2019;10:2012. doi: 10.1038/s41467-019-09964-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seekatz AM, Schnizlein MK, Koenigsknecht MJ, et al. Spatial and temporal analysis of the stomach and small-intestinal microbiota in fasted healthy humans. mSphere. 2019;4:e00126-19. doi: 10.1128/mSphere.00126-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerckhoffs AP, Ben-Amor K, Samsom M, et al. Molecular analysis of faecal and duodenal samples reveals significantly higher prevalence and numbers of Pseudomonas aeruginosa in irritable bowel syndrome. J Med Microbiol. 2011;60:236–245. doi: 10.1099/jmm.0.022848-0. [DOI] [PubMed] [Google Scholar]
- 23.Dlugosz A, Winckler B, Lundin E, et al. No difference in small bowel microbiota between patients with irritable bowel syndrome and healthy controls. Sci Rep. 2015;5:8508. doi: 10.1038/srep08508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung CS, Chang PF, Liao CH, et al. Differences of microbiota in small bowel and faeces between irritable bowel syndrome patients and healthy subjects. Scand J Gastroenterol. 2016;51:410–419. doi: 10.3109/00365521.2015.1116107. [DOI] [PubMed] [Google Scholar]
- 25.Li G, Yang M, Jin Y, et al. Involvement of shared mucosal-associated microbiota in the duodenum and rectum in diarrhea-predominant irritable bowel syndrome. J Gastroenterol Hepatol. 2018;33:1220–1226. doi: 10.1111/jgh.14059. [DOI] [PubMed] [Google Scholar]
- 26.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr Opin Gastroenterol. 2014;30:332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felis GE, Pot B. The family Lactobacillaceae. In: Holzapfel WH, Wood BJB, editors. Lactic Acid Bacteria: Biodiversity and Taxonomy. New York: Wiley; 2014. [Google Scholar]
- 28.du Toit M, Huch M, Cho GS, Franz CMAP. The genus Streptococcus. In: Holzapfel WH, Wood BJB, editors. Lactic Acid Bacteria: Biodiversity and Taxonomy. New York: Wiley; 2014. [Google Scholar]
- 29.Pessione E. Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Front Cell Infect Microbiol. 2012;2:86. doi: 10.3389/fcimb.2012.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perdigon G, Maldonado Galdeano C, Valdez JC, Medici M. Interaction of lactic acid bacteria with the gut immune system. Eur J Clin Nutr. 2002;56:S21–S26. doi: 10.1038/sj.ejcn.1601658. [DOI] [PubMed] [Google Scholar]
- 31.Tsai YT, Cheng PC, Pan TM. Anti-obesity effects of gut microbiota are associated with lactic acid bacteria. Appl Microbiol Biotechnol. 2014;98:1–10. doi: 10.1007/s00253-013-5346-3. [DOI] [PubMed] [Google Scholar]
- 32.Warren YA, Citron DM, Merriam CV, Goldstein EJ. Biochemical differentiation and comparison of Desulfovibrio species and other phenotypically similar genera. J Clin Microbiol. 2005;43:4041–4045. doi: 10.1128/JCM.43.8.4041-4045.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peck SC, Denger K, Burrichter A, Irwin SM, Balskus EP, Schleheck D. A glycyl radical enzyme enables hydrogen sulfide production by the human intestinal bacterium Bilophila wadsworthia. Proc Natl Acad Sci USA. 2019;116:3171–3176. doi: 10.1073/pnas.1815661116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rizzatti G, Lopetuso LR, Gibiino G, Binda C, Gasbarrini A. Proteobacteria: a common factor in human diseases. Biomed Res Int. 2017;2017:9351507. doi: 10.1155/2017/9351507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keijser BJ, Zaura E, Huse SM, et al. Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res. 2008;87:1016–1020. doi: 10.1177/154405910808701104. [DOI] [PubMed] [Google Scholar]
- 36.He X, McLean JS, Edlund A, et al. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci USA. 2015;112:244–249. doi: 10.1073/pnas.1419038112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferrari B, Winsley T, Ji M, Neilan B. Insights into the distribution and abundance of the ubiquitous candidatus Saccharibacteria phylum following tag pyrosequencing. Sci Rep. 2014;4:3957. doi: 10.1038/srep03957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D’Argenio V, Casaburi G, Precone V, et al. Metagenomics reveals dysbiosis and a potentially pathogenic N. flavescens strain in duodenum of adult celiac patients. Am J Gastroenterol. 2016;111:879–890. doi: 10.1038/ajg.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullivan A, Tornblom H, Lindberg G, et al. The micro-flora of the small bowel in health and disease. Anaerobe. 2003;9:11–14. doi: 10.1016/S1075-9964(03)00060-X. [DOI] [PubMed] [Google Scholar]
- 40.Percy-Robb IW, Brunton WA, Jalan KN, McManus JP, Gould JC, Sircus W. The relationship between the bacterial content of ileal effluent and the metabolism of bile salts in patients with ileostomies. Gut. 1969;10:1049–1050. [PubMed] [Google Scholar]
- 41.Booijink CC, El-Aidy S, Rajilic-Stojanovic M, et al. High temporal and inter-individual variation detected in the human ileal microbiota. Environ Microbiol. 2010;12:3213–3227. doi: 10.1111/j.1462-2920.2010.02294.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available in the National Center for Biotechnology Information (NCBI) BioProject Repository https://www.ncbi.nlm.nih.gov/bioproject under BioProject ID PRJNA590519.