Summary
The SARS-CoV-2 pandemic raises many scientific and clinical questions. These include how host genetic factors affect disease susceptibility and pathogenesis. New work is emerging related to SARS-CoV-2; previous work has been conducted on other coronaviruses that affect different species. We reviewed the literature on host genetic factors related to coronaviruses, systematically focusing on human studies. We identified 1,832 articles of potential relevance. Seventy-five involved human host genetic factors, 36 of which involved analysis of specific genes or loci; aside from one meta-analysis, all were candidate-driven studies, typically investigating small numbers of research subjects and loci. Three additional case reports were described. Multiple significant loci were identified, including 16 related to susceptibility (seven of which identified protective alleles) and 16 related to outcomes (three of which identified protective alleles). The types of cases and controls used varied considerably; four studies used traditional replication/validation cohorts. Among other studies, 30 involved both human and non-human host genetic factors related to coronavirus, 178 involved study of non-human (animal) host genetic factors related to coronavirus, and 984 involved study of non-genetic host factors related to coronavirus, including involving immunopathogenesis. Previous human studies have been limited by issues that may be less impactful now, including low numbers of eligible participants and limited availability of advanced genomic methods; however, these may raise additional considerations. We outline key genes and loci from animal and human host genetic studies that may bear investigation in the study of COVID-19. We also discuss how previous studies may direct current lines of inquiry.
Keywords: coronavirus, COVID-19, host genetic factors, SARS-CoV-2
The SARS-CoV-2 pandemic raises many scientific and clinical questions. These include how host genetic factors affect disease susceptibility and pathogenesis. New work is emerging related to SARS-CoV-2; previous work has been conducted on other coronaviruses that affect different species. We reviewed the literature on host genetic factors related to coronaviruses, systematically focusing on human studies. We identified 1,832 articles of potential relevance. Seventy-five involved human host genetic factors, 36 of which involved analysis of specific genes or loci; aside from one meta-analysis, all were candidate-driven studies, typically investigating small numbers of research subjects and loci. Three additional case reports were described. Multiple significant loci were identified, including 16 related to susceptibility (seven of which identified protective alleles) and 16 related to outcomes (three of which identified protective alleles). The types of cases and controls used varied considerably; four studies used traditional replication/validation cohorts. Among other studies, 30 involved both human and non-human host genetic factors related to coronavirus, 178 involved study of non-human (animal) host genetic factors related to coronavirus, and 984 involved study of non-genetic host factors related to coronavirus, including involving immunopathogenesis. Previous human studies have been limited by issues that may be less impactful now, including low numbers of eligible participants and limited availability of advanced genomic methods; however, these may raise additional considerations. We outline key genes and loci from animal and human host genetic studies that may bear investigation in the study of COVID-19. We also discuss how previous studies may direct current lines of inquiry.
Introduction
The ongoing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic raises many scientific and clinical questions. One set of questions involves susceptibility and outcomes related to SARS-CoV-2 infection (COVID-19). Hypotheses suggested to explain observed differences include host sex, age, comorbidities, and genetic factors.1 As with many complex diseases, the reality for most individuals most likely involves a combination of genetic—including viral and host genetics —and non-genetic variables. Large, international studies and collaborations have formed to investigate host genetic factors related to COVID-19. These investigations include analyses of existing public and private datasets, as well as the establishment of new cohorts (e.g., see “The COVID-19 Host Genetics Initiative” and “23andMe/23andMe Research Blog” entries in the Web Resources).
Relative to other coronaviruses, SARS-CoV-2 has unique biological properties and related clinical impact, but data regarding other coronaviruses may be relevant. Previous studies have been disparate in terms of the virus and species studied, as well as the aims and methods. This has resulted in a rich body of literature that is difficult to efficiently leverage for SARS-CoV-2-related work.
To address this, we aimed to perform a review of the literature to outline previous studies of host genetic factors related to coronaviruses. Our first objective is to systematically encapsulate genes and loci interrogated through these efforts. This can help populate lists of genes that—along with data from related biological studies—may bear scrutiny in the developing and important large-scale host genetic studies of SARS-CoV-2. Our second objective is to present an overview of themes from animal and human studies in order to inform current efforts. A systematic analysis may in turn help bolster efforts to identify susceptibility alleles and, eventually, potential avenues for treatment not yet well defined through human studies.
Literature Search and Sources
The methods we used to systematically identify and categorize published articles are described in the Supplemental Materials (see the Supplemental Methods and the PRISMA checklist). Of note, we did not include articles on preprint servers, though a growing number are available.
In summary, our search identified 1,832 unique articles of potential relevance (Figure 1 and Table S1). After initial review, 105 were included in our qualitative synthesis; 75 of these involved study of human host genetic factors related to coronavirus (Table 1). Thirty-six of the 75 human studies involved analysis of specific genes or loci (one was a meta-analysis study of multiple respiratory pathogens), while 39 involved biological, computational, or case report studies of human host genetic factors. Thirty involved both human and non-human host genetic factors (these largely investigated inter-species differences in disease susceptibility and pathogenesis); 178 involved study of non-human (animal) host genetic factors; 984 involved non-genetic host factors, including immunopathogenesis; 17 involved study of other pathogens (not coronavirus); and 528 involved other studies of coronavirus. Twenty studies were assigned to the other categories and removed. We use themes identified in our review of these articles to highlight areas that are particularly relevant to human studies of COVID-19; in addition to the limited references cited here, please refer to Table S1 for the additional literature identified through this search, as well as section-specific references not included in the main manuscript.
Figure 1.
PRISMA Diagram of Systematic Review Process, Including Articles Used for Narrative Review
Table 1.
Summary of Human Studies (Including Those Related to Specific Genes or Loci) on Host Genetic Factors Related to Coronaviruses
| Human Coronavirus Studied (Other Coronaviruses or Pathogens) | Method(s) or Approach(es) | Key Findings | PMID |
|---|---|---|---|
| SARS-CoV-1 | analysis of association of HLA (including MIM: 142800, 142830, 142857) gene polymorphisms with susceptibility to SARS-CoV-1 infection or clinical parameters | association of HLA-B∗4601 with severity of SARS-CoV-1 infection | 12969506 |
| SARS-CoV-1 | analysis of association of HLA gene polymorphisms with susceptibility to SARS-CoV-1 infection | HLA-B∗0703, HLA-DRB1∗0301 and co-inheritance of HLA-B∗0703 and HLA-B60 were associated with susceptibility to SARS-CoV-1 infection | 15243926 |
| SARS-CoV-1 | analysis of association of ACE2 (MIM; 30035) polymorphisms with SARS-CoV-1 clinical parameters | no association of ACE2 polymorphisms with SARS-CoV-1 outcomes | 15331509 |
| SARS-CoV-1 | analysis of association of ACE (MIM; 106180) polymorphism with susceptibility to SARS-CoV-1 or clinical parameters | ACE D allele (rs4646994) was associated with hypoxemia in SARS-CoV-1 infections | 15381116 |
| SARS-CoV-1 | analysis of association of OAS1 (MIM; 164350), PKR (MIM; 176871), and MX1 (MIM; 147150) polymorphisms with susceptibility to SARS-CoV-1 or clinical parameters | OAS1 rs3741981/rs1131454 (NC_000012.12:g.112911065G>A) and rs2660 (NC_000012.12:g.112919637G>A) were associated with SARS-CoV-1 susceptibility; MX1 rs2071430 (NC_000021.9:g.41426138G>T) was associated in hypoxemia in SARS-CoV-1 infections | 15766558 |
| SARS-CoV-1 | analysis of association of ACE insertion/deletion (I/D) polymorphism with susceptibility to SARS-CoV-1 or clinical parameters | no association was found with ACE insertion/deletion (I/D) polymorphism (rs4646994) and susceptibility to SARS-CoV-1 or clinical parameters | 15819995 |
| SARS-CoV-1 | analysis of association of MBL (MIM; 614372) polymorphisms susceptibility to SARS-CoV-1 or clinical parameters and biological study of MBL | serum MBL was lower in patients with SARS-CoV-1 infections than controls, and haplotypes associated with lower serum MBL were more frequent in patients with SARS-CoV-1 infections than in control subjects, but there was not association with mortality | 15838797 |
| SARS-CoV-1 | analysis of association of ACE2 polymorphisms and susceptibility to SARS-CoV-1 infection | no association was found with ACE2 polymorphisms and susceptibility to SARS-CoV-1 infection | 15937940 |
| SARS-CoV-1 | analysis of association of MBL polymorphisms and susceptibility to SARS-CoV-1 infection | MBL rs1800450 (NC_000010.11:g.52771475C>T) was associated with susceptibility to SARS-CoV-1 infection | 16170752 |
| SARS-CoV-1 | analysis of association of FCGR2A (MIM; 146790) and MBL polymorphisms and susceptibility to SARS-CoV-1 infection or clinical parameters | homozygosity for FCGR2A rs1801274 (NC_000001.11:g.161509955A>C), as well as a linear trend of FCGR2A genotypes, was associated with severe SARS-CoV-1 infection | 16185324 |
| SARS-CoV-1 | analysis of association of CLEC4M (MIM; 605872) VNTR polymorphism with susceptibility to SARS-CoV-1 and biological studies of cells with these polymorphisms | homozygosity for the CLEC4M VNTR polymorphism was associated with susceptibility to SARS-CoV-1, and homozygous cells had higher binding capacity for SARS-CoV-1, higher proteasome-dependent viral degradation, and lower capacity for trans infection. | 16369534 |
| SARS-CoV-1 | analysis of association of HLA polymorphisms with SARS-CoV-1 susceptibility | HLA-Cw∗0801 was associated with susceptibility to SARS-CoV-1 infection | 16455884 |
| SARS-CoV-1 | analysis of association of polymorphisms in 65 genes with SARS-CoV-1 viral shedding | SARS-CoV-1 shedding was associated with alleles of IL18, IL1A, RELB, and FLG2 (see Table S2 for alleles) | 16652313 |
| SARS-CoV-1 | analysis of association of OAS1 and MX1 polymorphisms with susceptibility to SARS-CoV-1 | OAS1 rs2660 (NC_000012.12:g.112919637G>A) and MX1 rs2071430 (NC_000021.9:g.41426138G>T) were associated with susceptibility to SARS-CoV-1 | 16824203 |
| SARS-CoV-1 | analysis of association of CLEC4M VNTR polymorphism with susceptibility to SARS-CoV-1 infection | no association was found with homozygosity for the CLEC4M VNTR polymorphism and susceptibility to SARS-CoV-1 | 17534354 |
| SARS-CoV-1 | analysis of association of CLEC4M VNTR polymorphism with susceptibility to SARS-CoV-1 infection | no association was found with homozygosity for the CLEC4M VNTR polymorphism and susceptibility to SARS-CoV-1 | 17534355 |
| SARS-CoV-1 | analysis of association of CCL5 (MIM; 187011), CXCL9 (MIM; 601704), and CXCL10 (MIM; 147310) polymorphisms with susceptibility to SARS-CoV-1 infection or clinical parameters | CCL5 rs2107538 (NC_000017.11:g.35880776C>T) was associated with susceptibility to SARS-CoV-1 in one cohort and severe outcomes of SARS-CoV-1 infection in another cohort | 17540042 |
| SARS-CoV-1 | analysis of association of FCER2 (MIM; 151445) and ICAM3 (MIM; 146631) polymorphisms with susceptibility to SARS-CoV-1 or clinical parameters | homozygosity for ICAM rs2304237 (NC_000019.10:g.10335892T>C) was associated with higher LDH levels and lower total WBC counts | 17570115 |
| SARS-CoV-1 | analysis of association of CD14 (MIM; 158120), TLR2 (MIM; 603028), and TLR4 (MIM; 603030) polymorphisms with susceptibility to SARS-CoV-1 or clinical parameters | CD14 rs2569190 (NC_000005.10:g.140633331A>C) was associated with severe SARS-CoV-1 infection (this data was also combined with previous data, suggesting that this and an FCGR2A allele are risk genotypes for severe SARS-CoV-1 infection) | 17913858 |
| SARS-CoV-1 | analysis of association of TNF (MIM; 191160) polymorphisms with interstitial lung fibrosis and femoral head osteonecrosis in discharged SARS-CoV-1patients | TNF rs1800630 (NC_000006.12:g.31574699C>A) status was associated with susceptibility to SARS-CoV-1 and with femoral head necrosis in discharged SARS-CoV-1patients | 18312678 |
| SARS-CoV-1 | analysis of association of polymorphisms in IL12RB1 (MIM; 601604) with susceptibility to SARS-CoV-1 or clinical outcomes | IL12RB1 rs11575932 (NC_000019.10:g.18063894G>A) was associated with susceptibility to SARS-CoV-1 infection | 18478121 |
| SARS-CoV-1 | analysis of association of polymorphisms in 4 C-type lectin genes with susceptibility to SARS-CoV-1 infection | no association of polymorphisms in C-type lectin genes with SARS-CoV-1 susceptibility | 18697825 |
| SARS-CoV-1 | analysis of association of polymorphisms in 9 inflammatory response genes with susceptibility to SARS-CoV-1 or clinical outcomes | no association of polymorphisms in inflammatory response genes with SARS-CoV-1 susceptibility or clinical outcomes | 18708672 |
| SARS-CoV-1 | analysis of association of polymorphisms in MASP2 (MIM: 605102) with susceptibility to SARS-CoV-1 infection | no association of MASP2 polymorphisms with SARS-CoV-1 susceptibility | 19405982 |
| SARS-CoV-1 | analysis of association of HLA polymorphisms with SARS-CoV-1 susceptibility | HLA-DRB1∗12 was more frequent in SARS-CoV-1 patients versus controls; HLA-DRB1∗1202 showed the strongest association with SARS-CoV-1 infection in a dominant model | 19445991 |
| SARS-CoV-1 | analysis of association of polymorphisms in 64 genes with susceptibility to SARS-CoV-1 infection | CXCL10(−938AA) is protective (but appears jointly with other variants); FGL2(+158T/∗)a is associated with higher susceptibility unless combined with CXCL10/(−938AA), when jointly is associated with lower susceptibility | 19590927 |
| SARS-CoV-1 | analysis of association of CD209 (MIM: 604672) polymorphism with SARS-CoV-1 outcomes | CD209 polymorphism rs4804803 (NC_000019.10:g.7747847A>G) and ICAM3 rs2304237 (NC_000019.10:g.10335892T>C) are associated with lower LDH levels (and therefore, worse prognosis) | 20359516 |
| SARS-CoV-1 | biological study and analysis of MX1 promoter polymorphisms with suppressed interferon beta induction and association of MX1 promoter polymorphisms with susceptibility to SARS-CoV-1 infection | differences were observed in binding affinity to nuclear proteins related to IFN-beta stimulation; MX1 rs2071430 (NC_000021.9:g.41426138G>T) was associated with lower risk of SARS-CoV-1 infection | 20462354 |
| SARS-CoV-1 | analysis of association of HLA gene polymorphisms with SARS-CoV-1 susceptibility | no significant associations (after correction) HLA gene polymorphisms with SARS-CoV-1 susceptibility were identified | 20864745 |
| SARS-CoV-1 | biological study of in vitro functional effects of CD209 polymorphism and analysis of association of CD209 polymorphism with SARS-CoV-1 outcomes | CD209 polymorphism rs4804803 (NC_000019.10:g.7747847A>G) was associated with lower risk of high admission LDH levels, and may contribute to a reduced immune response/reduced lung injury during disease progression | 20864747 |
| SARS-CoV-1 | analysis of association of AHSG (MIM: 138680) and CYP4F3 (MIM: 601270) polymorphisms with SARS-CoV-1 susceptibility | AHSG polymorphism rs2248690 (NC_000003.12:g.186612299T>A) was associated with SARS-CoV-1 susceptibility (as well as higher AHSG serum concentration) | 21904596 |
| SARS-CoV-1 | analysis of association of HLA polymorphisms with SARS-CoV-1 susceptibility | HLA-Cw∗1502 conferred resistance against SARS infection is associated with resistance to SARS-CoV-1 infection | 21958371 |
| SARS-CoV-1 | analysis of association of HLA polymorphisms with SARS-CoV-1 susceptibility and outcome | no association of HLA polymorphisms with SARS-CoV-1 susceptibility and outcome were identified | 24643938 |
| SARS-CoV-1 | analysis of association of CCL2 (MIM: 158105) and MBL polymorphisms with susceptibility to SARS-CoV-1 infection | MBL rs1800450 (NC_000010.11:g.52771475C>T) and CCL2 rs1024611 (NC_000017.11:g.34252769A>G) were cumulatively associated with SARS-CoV-1 susceptibility | 25818534 |
| SARS-CoV-1 (and other respiratory pathogens) | meta-analysis of 386 studies on susceptibility to tuberculosis, influenza, respiratory syncytial virus, SARS-CoV-1, and pneumonia | in a pooled model, IL4 (MIM: 147780) rs2070874 (NC_000005.10:g.132674018C>T) status was positively associated with susceptibility after multiple testing correction | 26524966 |
| SARS-CoV-2 | case report of death due to COVID-19 in three previously healthy adult brothers | suggestion of genetic predisposition due to apparent familial clustering | 32277694 |
| SARS-CoV-2 | case reports of two patients with X-linked agammaglobulinemia (and documented pathogenic variants in BTK [MIM: 300300]) | patients recovered, suggesting that B cell response might not be required to overcome the SARS-CoV-2 infection | 32319118 |
| SARS-CoV-2 | analysis of association of IFITM3 (MIM: 605579) polymorphism with clinical outcomes of SARS-CoV-2 infection | significant association of homozygosity IFITM3 rs12252 (NC_000011.10:g.320772A>G) with disease severity | 32348495 |
| SARS-CoV-2 | case report of a large family cluster with more severe disease compared to other patients presenting at the same time | suggestion of genetic predisposition due to apparent familial clustering of severity | 32492209 |
More details are available in Table S2 (see also Supplemental References). Abbreviations are as follows: CCoV, canine coronavirus; FCoV, feline coronavirus; HCoV-229E, human coronavirus 229E;HCoV NL63, human coronavirus NL63; HCoV OC43, human coronavirus OC43; LDH, lactate-dehydrogenase; MBL, Mannose-binding lectin; MERS-CoV, middle east respiratory syndrome coronavirus; SARS-CoV-1, severe acute respiratory syndrome coronavirus 1; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SL-CoV, SARS-Cov-1-like coronaviruses; TGEV, porcine transmissible gastroenteritis coronavirus; WBC, white blood cell; WT, wild-type.
Data describing variant specifics to enable HGVS nomenclature are not available (i.e., online databases do not appear to contain live data).
Coronaviruses: General Background
Although SARS-CoV-2 has seized recent attention, there are other coronaviruses with a large related body of literature. The Coronavirinae subfamily of the Coronaviridae family consists of four genera.2,3 Among these, the alphacoronaviruses include two major human coronaviruses, HCoV-229E (multiple HCoV-229E-like strains have been identified) and HCoV-NL63.3 Alphacoronaviruses that affect other species include feline coronavirus (FCoV), which includes feline infectious peritonitis virus (FIPV) and feline enteric coronavirus (FECV), canine coronavirus (CCoV), and transmissible gastroenteritis virus (TGEV) and porcine epidemic diarrhea virus (PEDV) in pigs.3 The betacoronaviruses consist of four lineages: lineage A (HCoV-OC43 and HCoV-HKU1, as well as coronaviruses affecting other species, such as mouse hepatitis virus [MHV]), lineage B (SARS-CoV-1 and SARS-CoV-2), lineage C (Middle East respiratory syndrome (MERS) and many bat coronaviruses), and lineage D (coronaviruses only identified in bats to date).4 HCoV-OC43, HCoV-229E, HCoV-HKU1, and HCoV-NL63 can result in a variety of presentations, including “common cold” and severe but rarely fatal disease; they are also frequently detected as co-infections with other viruses.3,5 There are other rare coronaviruses observed in humans as well as in other species2,3 (see for further details as described by the International Committee on Taxonomy of Viruses [ICTV]; see Web Resources).
Animal Studies of Coronavirus: General Background
Coronaviruses affect many species, from Beluga whales to spotted hyenas to turkeys. Sequelae of disease can range from apparently asymptomatic infections to severe or lethal effects on different organ systems, potentially manifesting as diarrheal, encephalitic, nephritic, respiratory, and other findings.6
In addition to ecologic studies of wild animals, there are numerous non-observational animal studies of coronaviruses, such as those involving ferrets,7 hamsters,8 guinea pigs,9 rats,10 and non-human primates.11 Formal host genetic studies have been described for some but not all species. Many studies have simply involved examination of differences in species susceptibility and pathogenesis related to human and non-human coronaviruses without interrogation of specific variants in a particular species.
Among the host genetic work in animals, the objectives and methods used depend on the species studied. For example, in chickens and pigs, the types of published studies predictably differ from those conducted on experimental mice. That is, although MHV represents a problem for mouse colonies, the rationale of the livestock studies may focus more purely on economic repercussions versus attempts to use a model organism to understand immunopathogenesis. The degree to which results may be reported through the scientific literature (versus other routes) is also anticipated to differ between these groups. See Figure 2 for a summary of reported interrogated loci in animal studies.
Figure 2.
Genes Investigated in Animal Studies Related to Coronavirus Disease
See discussion in the text for more details and referenced studies for specific citations; additional citations are given in the Supplemental Materials. Human genes are shown only for those studies that included analysis of multiple species; other human gene details are presented elsewhere.
Species Susceptibility
One type of study of host genetic factors involves trying to understand whether and how different species are susceptible to infections. This has several important implications related to human health. A first implication involves the zoonotic potential of a pathogen.12 Relevant studies have explored host ranges and reservoirs. For example, bats, camels, and humans can be infected by MERS, unlike mice, ferrets, hamsters, and guinea pigs. SARS-CoV-2 replicates better in ferrets and cats than in dogs, pigs, chickens, and ducks. One explanation involves genetic characteristics of the host receptor for the relevant virus (see Receptor Studies below for further discussion).13
As a natural reservoir for many coronaviruses, bats have been investigated more extensively than other species outside of laboratory-based animals and livestock. One interesting aspect involves host/pathogen co-evolution. That is, research has included co-evolutionary studies between coronaviruses and the genomes of bat hosts (e.g., by correlating phylogenetic analyses of bat coronaviruses with CYTB in multiple bat species)14 as well as other genetic/biologic studies related to host genetic factors. These have involved relatively well-characterized genes, such as the ACE2 receptor gene with SARS-CoV-115 and the DPP4 receptor gene with MERS.16 Specific residues in the ANPEP receptor gene influence species susceptibility to multiple different coronaviruses.17 In addition to allowing analyses of host susceptibility, these and similar studies help provide estimates for the time-frame of coronavirus circulation in species and populations.18
As a second example, camels are an important reservoir of coronaviruses that can infect humans; this became especially relevant in the context of MERS. Several host genetic studies have looked at DPP4 receptor characteristics and species tropism, including comparisons between camels, humans, and other species.19 To underscore the importance of considering host factors beyond genetics, many studies have analyzed non-genetic correlations with the spread from camels to humans. Examples in this context include the size of the domesticated camel herds, what the herds were used for (e.g., food or transport), and how active the herds were.20 In the burgeoning studies of COVID-19 host genetic factors, controlling for these types of other variables will be challenging and important. However, it is possible that sheer statistical power may be able to address some of these issues. Similar approaches have achieved significant results for other etiologically and medically complex diseases (such as preterm birth), including using some of the same datasets and approaches being proposed for COVID-19 studies.21
As a final example, palm civets (as well as other species) have been examined in relation to zoonotic implications of coronavirus disease. Specifically, questions about ACE2 have been described in the context of SARS-CoV-1 and the impact of species-specific variants in this and other genes.22 This work has emphasized interactions of viral and host genetics.23 This aspect bears further scrutiny in COVID-19 studies, especially given recent data regarding SARS-CoV-2 genetic changes detected in different areas of the world (e.g., see data from Nextstrain under Web Resources).
In addition to observational studies, experimental approaches have been used to study species susceptibility. Hamsters have been used as model organisms to study coronaviruses, including via standard hamster cell lines as well as other approaches with hamster models.24 For example, hamsters have been used to study species susceptibility to MHV (related to the Ceacam1 receptor),25 how alterations of specific Dpp4 amino acids in hamsters affect susceptibility to MERS,19,26 and the roles of ACE2 and CD209L in SARS-CoV-1 susceptibility.24 Related to the human implications of this type of work, newer gene editing techniques may be an efficient way to provide experimental validation of specific variants that have been implicated in COVID-19.
A second, related implication involves identifying experimental animals that mimic human response to the virus (or that can be used to understand the disease in other species). Among other reasons, this can be important for understanding human infection and developing and testing possible treatments; in addition to the above-mentioned experimental animals, other animals, including non-human primates, have been used to study coronavirus in this way.27,28 As usual, these studies have included host receptors as well as genes and mechanisms involved in downstream viral pathogenesis and have employed a variety of computational and experimental approaches.13,29,30
Beyond receptor studies (see further details below), the site of viral replication appears to vary according to the species and coronavirus. This may be potentially related to tissue-specific receptor expression, such as has been shown in studies of cats and ferrets.31 This line of reasoning may also be relevant to age-specific differences observed with COVID-19 in humans.32 That is, one of several potential factors that may explain why most children are more mildly affected by COVID-19 is age-related differences in ACE2 receptor expression.
Receptor Studies
In various species, efforts have focused on genes encoding the relevant coronavirus receptor, including effects of viral and host genetic changes and how these may impact the disease process. Among other cell surface determinants,33 these receptor genes include ACE2 (MIM: 30035) for HCoV-NL63,34 SARS-CoV-1,35 and SARS-CoV-2,36 ANPEP (MIM: 151530) for HCoV-229,37 FIPV,38 CCoV,39 and TGEV,40 DPP4 (MIM: 102720) for MERS,41 and Ceacam1 for MHV (see Figure 2, which summarizes key genes investigated in animal studies on coronaviruses).42 In animals, significant work has been done related to host genetic factors involving these receptor genes. For example, studies in rats include computational approaches examining receptor characteristics, such as Ace2 in the context of SARS-CoV-1,43 and experimental approaches that suggest that rats are not susceptible to MERS on the basis of Dpp4 characteristics.30
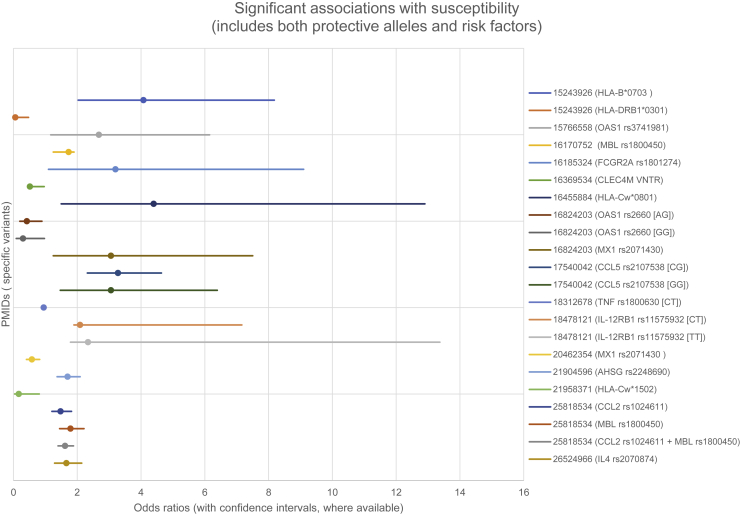
In humans (see Tables 1 and S2 and Figures 3 and 4 for details on human studies of these genes, including specific references), studies of specific ACE2 polymorphisms have not shown significant associations with SARS-CoV-1 susceptibility or outcome. CLEC4M (CD209L) (MIM: 605872) encodes an alternate receptor with lower viral affinity. There is mixed evidence for an association of susceptibility to SARS-CoV-1 with CLEC4M polymorphisms (tandem repeats). Several studies have used (and are using) existing datasets to explore allele frequencies (such as in ACE2) in various geographic/ancestral populations: the hypothesis is that differences in allele frequencies—as well as observed differences in gene expression—may be one reason for differential impacts of COVID-19 in different parts of the world.44,45 In conjunction with population studies, computational functional studies have been performed on identified ACE2 variants.46
Figure 3.
Significant Genetic Associations with Human Susceptibility to Coronavirus Disease
Both protective and permissive genes are shown. Only studies reporting odds ratios (ORs) and confidence intervals (CIs) are shown. See Table S2 and Supplemental References.
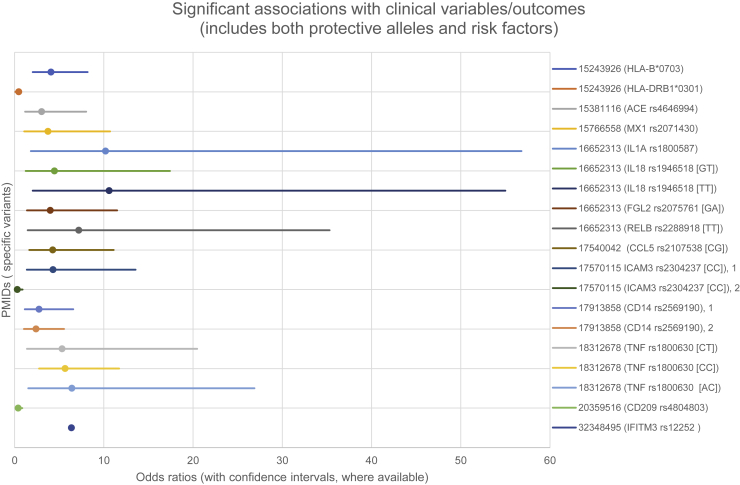
Figure 4.
Significant Genetic Associations with Human Clinical Variables and Outcomes Related to Coronavirus Disease
Both protective and permissive genes are shown. Only studies reporting ORs and CIs are shown (PMID: 32348495 did not include CI). See Table S2 and Supplemental References.
Following up these data through host genetic studies with cases and controls will help further examine variants in these and other genes. Validating findings with biological data will also be helpful—for example, a recent study on COVID-19 showed that certain immune mediators, cytokines, and chemokines correlate with aspects of disease.47 Because of today’s availability of genomic approaches, and in contrast to the previously published studies (described in Tables 1 and S2), emerging studies on COVID-19 will most likely have to consider both rare and common variants in these and other genes, as well as combinatorial models explaining susceptibility and outcomes.
Multiple studies have examined mutant ACE2. Studying the effects of mutant ACE2 on SARS-CoV-1 entry provided evidence that the cytoplasmic tail of ACE2 is not required for SARS-CoV-1 penetration.48 SARS-CoV-2 studies have suggested that truncated ACE2 could act as a COVID-19 therapeutic through inhibition of SARS-CoV-2 spike protein activity.49 Computational models suggest that, although most ACE2 variants result in similar binding affinity for SARS-CoV-2 spike protein, certain variants in the gene (rs73635825 and rs143936283) demonstrate different intermolecular interactions with the spike protein.50
Extensions of Receptor Studies to Interventional Approaches in Animal Studies
Pigs can be infected by TGEV and PEDV, as well as the more recently-identified porcine deltacoronavirus (PDCoV). Similar to coronavirus disease in chickens, these diseases can affect the food industry, and studies have aimed to address ways to ameliorate disease, such as through vaccines and other methods.51 Modern gene editing techniques have been studied in this context; these have also garnered recent interest in COVID-19.
In pig studies, variants (both naturally occurring and experimentally induced) have been shown to have varying effects on different coronaviruses. For example, aminopeptidase N, encoded by ANPEP (also called APN), was reported as a functional receptor for TGEV and PEDV (as well as HCoV-229E), but multiple models, including CRISPR/Cas9-generated knockouts, show differences in cellular susceptibility to TGEV and PEDV.51,52 In another study, infection by PEDV and TGEV correlated positively with ANPEP expression, but PEDV and TGEV could infect ANPEP-positive and ANPEP-negative enterocytes: differences were observed between viral strains. Overall, the results suggested the presence of an additional receptor.53 Similar to work on SARS-CoV-1 in humans, variants in these additional receptor genes may be clinically relevant.24,54
Building on this type of work, site-specific editing of ANPEP has been raised as a potential means to breed resistant animals.55 In a similar vein, knockout of CMAH (hypothesized to affect cellular binding) does not result in immunity to PEDV but appears to improve outcomes.56
This line of thinking can be extended to human studies. In COVID-19, the use of splice-switching antisense oligonucleotides has been proposed to affect ACE2 in order to limit SARS-CoV-2 entry.57 Other modern techniques, such as CRISPR, have emerged as powerful tools for many research and a growing number of potential clinical applications. CRISPR has been described as a potential diagnostic and therapeutic tool to test for and combat SARS-CoV-2 infection.58,59 CRISPR applications to host cells has also been suggested as a therapeutic avenue.60 It is likely that additional ethical and biological questions will arise that may echo previous discussions about these approaches in other clinical areas.61 As with other questions in COVID-19 (e.g., ethical questions pertaining to human challenge studies in vaccine trials62), balancing risks and benefits will be critical.
Major Histocompatibility Complex (MHC)
The major histocompatibility complex (MHC) has been explored in studies of multiple species related to coronavirus, including chickens,63 domestic cats,64 and cheetahs.65 As with the human studies summarized below (see also Tables 1 and S2), the evidence has been mixed and unclear.
Studies of cheetahs present an interesting example related to MHC genes, which may have connections to human COVID-19 studies. Among wild animals, severe population bottlenecks (resulting in reduced genetic diversity) in cheetahs has been used to explain their increased susceptibility to infection by FIPV as well as other infectious diseases. Several such bottlenecks appear to have occurred in cheetahs as a result of a combination of factors.66 Among possible explanations for cheetahs’ coronavirus susceptibility, genetic uniformity of the MHC has been proposed.65
In humans, severe COVID-19 outcomes have already been reported in peer-reviewed literature67 (as well as many lay articles), but specific suggestions of associations with consanguinity have not been identified. However, analyzing such families may be informative, as has been the case for many conditions with genetic underpinnings.
Separate from the above, HLA genes (including MIM: 142800, 142830, 142857) have also been studied in humans in relation to SARS-CoV-1, again with overall mixed evidence (see Tables 1 and S2 and Figures 3 and 4 for details on human HLA studies, including specific references). HLA alleles that appear to be related to susceptibility and/or outcome of disease have been identified. This mixed evidence may reflect issues with study design, such as sample size and ascertainment. The HLA genes remain a logical target of interest in relation to COVID-19.68 As a preface to case-control host genetic studies, a recent report has described peptide-binding affinities between hundreds of HLA class I and class II proteins and the proteomes of seven pandemic viruses, including coronaviruses. Similar to human population work on ACE2 and other genes in humans, the HLA alleles have been examined in relation to peptide-binding affinities.69 An in silico analysis of viral peptide-MHC class I binding affinity in relation to HLA genotypes for SARS-CoV-2 peptides, as well as potential cross-protective immunity related to four common human coronaviruses, provides evidence that HLA-B∗46:01 may be associated with COVID-19 vulnerability, whereas HLA-B∗15:03 may enable cross-protective T-cell-based immunity.70 Correlating these theoretical data with case-control results is a logical next step.
Other Immune Genes
Beyond the HLA genes, other key genes involved in immune processes have been investigated in host genetic studies. We use the extensive mouse studies to illustrate this point.
Differences in the susceptibility of various mouse lines to MHV has been noted for seven decades.71,72 This coronavirus remains a challenge for the health of mouse colonies, although relatively recent improvements in animal care practices have been beneficial.73 Various MHV strains show a range of tissue tropism and host effects on different mouse lines.74 For example, the JHM strain of MHV causes encephalitis in susceptible animal lines.75
Unsurprisingly, the majority of host genetic research in mouse models has centered on pathways known to be implicated in viral infection susceptibility. MHV-based mouse studies have used transgenic models to directly test the role of implicated immunologic and related pathways (summarized in Table 2). Work in humans so far has also concentrated on key immune genes (see Tables 1 and S2); similar work in relation to COVID-19 has been proposed.76
Table 2.
Summary of Relevant Mouse Studies Related to Coronavirus (See Also Figure 2)
| Mouse (Human Gene) | Method(s) or Approach(es) | Pathway: Key Findings | PMID |
|---|---|---|---|
| Ace2 (ACE2 [MIM: 300335]) | humanized mice, SARS-CoV1 | viral receptor: humanized Ace2 mice, increased infection, permissive gene | 18495771 |
| Atg5 (ATG5 [MIM: 604261]) | KO, MHV infection | autophagy: required for MHV replication, permissive gene | 14699140 |
| Atp1a1 (ATP1A1 [MIM: 182310]) | knockdown and chemical inhibition across many coronaviruses | ion channel: chemical inhibition or gene silencing, results in blocking viral entry, permissive gene | 25653449 |
| B2m (B2M [MIM: 109700]) | KO, MHV infection | adaptive immunity: MHC class I/CD8 T cells required for host immune response, protective gene | 8799201; 10023135 |
| Bnip3 (BNIP3 [MIM: 603293]) | cell culture model, MHV infection | apoptosis: pro-apoptotic gene is suppressed upon viral entry, likely protective | 14599795 |
| C3 (C3 [MIM: 120700]) | KO, SARS-CoV1 | complement pathway: decreased complement activation leads to less severe disease, implicated immune driven component of disease, gene is permissive | 30301856 |
| C5ar1(C5AR1 [MIM: 113995]) | KO, MHV infection | complement pathway: complement pathway exacerbates hepatitis, KO decreases manifestations, decreased susceptibility, permissive gene | 24604562 |
| Ccr1 (CCR1 [MIM: 601159]) | KO, MHV infection | cytokine pathways: loss of Ccr1 increased mortality, protective gene | 18158733 |
| Ccr2 (CCR2 [MIM: 601267]) | KO, MHV infection | cytokine pathways: Ccr2 required for clearance of the virus from CNS, KO increased susceptibility, protective gene | 15518805 |
| Ccr5 (CCR5 [MIM: 601373]) | KO, MHV infection | cytokine pathways: KO decreased severity of demyelination disease, permissive gene | 11543653 |
| Cd200r1 (CD200R1 [MIM: 607546]) | KO, MHV infection | immune receptor: Cd200 KO increases clearance of MHV, decreases susceptibility, permissive gene | 22615569 |
| Ceacam1 (CEACAM1 [MIM: 109770]) | isoform specific transgenic and KO, MHV infection | viral receptor: KOs are fully resistant to infection, liver, and CNS manifestations, permissive gene | 11483763; 15331748 |
| Cxcl10 (CXCL10 [MIM: 147310]) | KO, MHV infection | cytokine pathways: interferon related (T2), KO leads to increased mortality, protective gene | 17142734; 17617609 |
| Cxcl9 (CXCL9 [MIM: 601704]) | KO, MHV infection | cytokine pathways: interferon related (T2), KO had increased MHV associated mortality, protective gene | 18973912 |
| Dpp4 (DPP4 [MIM: 102720]) | various transgenic and humanized models, MERS infection | viral receptor: humanized Dpp4 or mutations, deletions in mouse Dpp4 leads to MERS induced ARDS, permissive gene | 24574399; 25653445; 29691378; 30142928; 31883094 |
| Ebi3 (EBI3 [MIM: 605816]) | KO, MHV infection | cytokine pathways: interferon related (T2), KO leads to increased mortality, protective gene | 23102608 |
| Foxn1 (FOXN1 [MIM: 600838]) | KO, MHV infection | adaptive immunity: athymic mice lacking T cells unable to clear infection cause severe disseminated disease, protective gene | 8799201; 15070459 |
| H2-Ab1 (H2AB1 [MIM: 301037]) | KO, MHV infection | adaptive immunity: MHC class I/CD4 T cells required for host immune response, protective gene | 8799201 |
| Ifih1 (IFIH1 [MIM: 606951]) | KO, MHV infection | cytokine pathways: interferon related (T1), KO more severe, disseminated MHV infection, decreased survival, protective gene | 26423942 |
| Ifnar1, (IFNAR1 [MIM: 107450]) | KO, MHV infection | cytokine pathways: interferon related (T1), KO leads to increased mortality and higher viral titers, protective gene | 18667505; 19215224; 19650917 |
| Ifnar1 (IFNAR1 [MIM: 107450]) | KO, SARS-CoV1 | interferon pathway: type I, II, and III interferons do not alter infection for SARS-CoV-1, in contrast to MHV | 20386712 |
| Ifng (IFNG [MIM: 147570]) | KO, MHV infection | cytokine pathways: interferon related (T2), KO has increased mortality, decreased viral clearance, protective gene | 9973424; 11864749 |
| Ifngr1 (IFNGR1 [MIM: 107470]) | KO, MHV infection | cytokine pathways: interferon related (T2), KO has increased mortality, decreased viral clearance, protective gene | 8752933; 15039522; 20042510 |
| Ifngr1 (IFNGR1 [MIM: 107470]) | KO, SARS-CoV1 | interferon pathway: type I, II, and III interferons do not alter infection for SARS-CoV-1, in contrast to MHV | 20386712 |
| Ighm (IGHM [MIM: 147020]) | KO, MHV infection | adaptive immunity: B cell deficient develop subclinical infection and transmit virus for increased time span, protective gene | 15027615 |
| Il1r1 (IL1R1 [MIM: 147810]) | KO, MHV infection | cytokine pathways: KO shows reduced viral replication, mortality, and disease progression, permissive gene | 26367131 |
| Mavs (MAVS [MIM: 609676]) | KO, MHV infection | cytokine pathways: interferon related (T1), viral sensor, studied in the presence of attenuated virus, protective gene | 29717007 |
| Myd88 (MYD88 [MIM: 602170]) | KO, rMA15 infection | cytokine pathways: downstream of multiple pathways, KO increased susceptibility to MHV infection and mortality, protective gene | 19079579 |
| Prkdc (PRKDC [MIM: 600899]) | KO, MHV infection | adaptive immunity: loss of T and B cells causes severe disseminated infection, protective gene | 8799201 |
| Rag1 (RAG1 [MIM: 179615]) | KO, MHV infection | adaptive immunity: loss of mature T and B cells leads to failure to clear infection, protective gene | 17142734; 18973912; 25428866; 27604627 |
| Serpine1 (SERPINE1 [MIM: 173360]) | KO, SARS-CoV1 infection | tissue remodeling: KO mice are more susceptible to infection and inflammation, protective gene | 23919993 |
| Stat1 (STAT1 [MIM: 600555]) | KO/KI, HCoV-229E infection | cytokine pathways: interferon related (T1), KO increased susceptibility HCoV in transgenic APN model, protective | 15919828 |
| Stat1 (STAT1 [MIM: 600555]) | KO, SARS-CoV-1 | cytokine pathways: KO worsens disease, increases susceptibility, protective gene | 20386712; 23142821 |
| Stat6 (STAT6 [MIM: 601512]) | conditional KO, LysM and FoxJ1, Stat1/Stat6 −/− double knockout, SARS-CoV-1 | cytokine pathways: conditional KO of Stat1 in macrophages but not ciliated epithelial cells showed pulmonary disease, double knockout of Stat1 and Stat6 relieves pulmonary disease, implicates alternatively activated macrophages, permissive gene | 23015710 |
| Ticam2 (TICAM2 [MIM: 608321]) | KO, SARS-CoV1 | immune receptor: TLR mediated, KO developed more severe infection, increased viral titer, and increased weight loss, protective gene | 28592648 |
| Tlr2 (TLR2 [MIM: 603028]) | KO, MHV infection | immune receptor: KO decreases inflammatory response, protective gene | 19740307 |
| Tlr3 (TLR3 [MIM: 603029]) | KO, SARS-CoV1 | immune receptor: TLR mediated, KO more susceptible for SARS-CoV-1 infection, although no increased mortality, protective gene | 26015500 |
| Tlr4 (TLR4 [MIM: 603030]) | KO, SARS-CoV1 | immune receptor: TLR mediated, KO more susceptible for SARS-CoV-1 infection, although no increased mortality, protective gene | 26015500 |
| Tlr7 (TLR7 [MIM: 300365]) | KO, MHV infection | immune receptor: viral sensor, KO prolonged infection, protective gene | 29717007 |
| Tram1 (TRAM1 [MIM: 605190]) | KO, SARS-CoV1 | immune receptor: TLR mediated, KO more susceptible for SARS-CoV-1 infection, although no increased mortality, protective gene | 26015500 |
| Trif (TRIF [MIM: 607601]) | KO, SARS-CoV1 | immune receptor: TLR mediated, KO more susceptible to SARS-CoV-1 infection, more severe infection with increased interferon signaling, protective gene | 26015500 |
| Trim55 (TRIM55 [MIM: 606469]) | KO, SARS-CoV1 | uncharacterized pathway: contributed to lung pathology, KO decreased severity, permissive gene | 26452100 |
| Usp18 (USP18 [MIM: 607057]) | KO, MHV infection | cytokine pathways: interferon related (T1), KO leads to increased survival, decreased pathology and viral titer, gene is permissive | 24648452 |
Note that the different studies have disparate objectives, many of which more directly involve aspects of immunopathogenesis versus standard host genetic questions regarding why specific genetic variants may affect disease susceptibility and outcomes. See also Supplemental References. Abbreviations are as follows: ARDS, acute respiratory distress syndrome; CNS, central nervous system; KI, knock-in; KO, knockout; MERS, middle east respiratory syndrome; MHC, major histocompatibility complex; MHV, mouse hepatitis virus; SARS-CoV-1, severe acute respiratory syndrome coronavirus 1; T1, type 1; T2, type 2; TLR, Toll-like receptor.
Mouse host genetic studies include investigations of humoral and cellular adaptive immune responses, specific cytokine and immune receptor pathways, viral receptors, complement pathways, apoptosis, autophagy, and tissue repair. These studies have prominently implicated types I (ɑβ) and II (γ) interferon responses in host response and predominant protection against MHV infection. However, not all pro-inflammatory pathways have been shown to be protective. For example, complement activation promotes tissue damage caused by MHV infection, highlighting the complex interplay between the host and virus. These transgenic models have also returned to questions regarding the susceptibility of different strains.77 In addition to targeted gene disruptions described above, a GWAS using a recombinant inbred mouse panel implicated Trim55, which is involved in vascular cuffing and inflammation in response to SARS-CoV-1.78
Although these studies have provided much better understanding of the disease process, it is not always clear how well the results for one viral strain and mouse line can be extrapolated more broadly. Similar themes emerge in human studies of other conditions. That is, the clinical effects of particular variants may differ from one population to the next, most likely because of other, interacting genetic and non-genetic factors. This may make findings in one population difficult to generalize or may mean that certain genetic variants are most clinically relevant in certain populations. In humans, this issue becomes especially important in clinically-oriented variant analysis.79 Similar concerns might arise in related situations, such as the use of genetic data to help drive therapeutic development. An example of a population-specific consideration has already been mentioned in relation to COVID-19 is that a variant in SCN5A (MIM: 600163) that is common in individuals of recent African descent may increase the risk of cardiovascular morbidity and mortality, including upon exposure to hydroxychloroquine and azithromycin.80
In addition to the human host genetic studies that examined this broad category of genes, biologic investigations have been performed. For example, the TRIM proteins play regulatory roles in innate antiviral responses; TRIM56 had been shown to inhibit replication of the flavivirus bovine viral diarrhea virus. Studies of mutant TRIM56 (MIM: 616996) on antiviral activity against HCoV-OC43 and other viruses showed that anti-HCoV-OC43 activity relies solely upon TRIM56 E3 ligase activity; this appears different from the mechanisms for other viral pathogens.81 Depletion or expression of a catalytically inactive version of PPIA (MIM: 123840), also known as cyclophilin A, results in impaired HCoV-229E replication.82 Cyclophilin A is a peptidyl-prolyl cis/trans isomerase that binds CoV proteins and is required for viral propagation through an unclear mechanism. Specific variants in IFITM genes encoding interferon-induced transmembrane proteins (IFITM1 [MIM: 604456] and IFITM3 [MIM: 605579] were studied) facilitate the entry of multiple human coronaviruses (HCoV-229E, HCoV-NL63, HCoV OC43, MERS-CoV, and SARS-CoV-1 were studied) despite surprisingly inhibiting the entry of other viruses.83 Finally, studies have manipulated various genes/proteins involved in viral pathogenesis to explore functional effects, including GLTSCR2 (MIM: 605691),84 IFITM1, IFITM2 (MIM: 605578), IFITM3,85 and MAVS (MIM: 609676).86
Evidence of Interactions of Viral and Host Genetic Factors
Multiple lines of evidence suggest a complex relationship between viral and host genetics. Again, mouse studies have focused on this area, as well as exploring other questions regarding susceptibility and pathogenesis.87
Examinations of different laboratory mouse strains have suggested that multiple loci are involved in host genetic factors related to MHV.88,89 Early mouse studies yielded various models, including potential monogenic/Mendelian explanations as well as more complex explanations involving interacting loci.74,90,91 Human studies will be more complex than those on inbred mouse lines. Some of the small candidate-driven association studies in humans have tried to use combinatorial models but were most likely hampered by multiple issues, including the numbers of available cases and controls and the ability to query multiple common and rare variants simultaneously (see Tables 1 and S2 for details). In addition to potentially addressing this complexity with large numbers of participants, elegant approaches have been proposed. For example, deep investigation of outliers may yield answers that can be further investigated in the general population.92 These outliers may represent extremes of clinical sequelae, such as those who appear to be unaffected or otherwise young and healthy individuals who are more severely affected than would be anticipated. Specific examples have already been reported in the literature on COVID-19.93 Another area of interest may involve studying individuals with identified pathogenic or severe variants (e.g., “human knockouts”) to determine correlations with COVID-19. Studies of populations that have already been genotyped and extensively studied may be especially powerful.
Sex Effects
As described, work in human and animals has explored various host factors related to coronavirus infection. For example, human94 and animal10,95 studies have implicated age as having significant associations with outcomes in coronavirus infections. Currently, age appears to be strongly correlated with COVID-19 outcomes.96 The overall explanations for this remain unclear but could involve age-related gene expression. Sex also appears to be correlated with outcomes. Animal studies identify sex effects, such as those related to disease severity, in multiple species.97,98 Human studies of SARS-CoV-1 and SARS-CoV-2 suggest a correlation between sex and certain clinical parameters, perhaps rooted in sex-based or related immunologic differences or gene dosage effects.94,99 However, separating biological differences from sex-related cultural practices (e.g., different rates of social distancing) and body habitus (i.e., potential correlations of body mass index with sex separate from strict genetic correlations) may be difficult.
Hypothesis-free versus Candidate Approaches
Human host genetic studies on coronavirus have been largely candidate driven to date (see Tables 1 and S2 and Figures 3 and 4 for details on human studies, including specific references), though many hypothesis-free studies on COVID-19 are in various phases of completion. As shown in Figures 3 and 4, human studies have examined susceptibility to infection as well as questions regarding various outcomes (some studies investigated both areas). Animal studies on coronaviruses have employed hypothesis-free as well as candidate approaches.
In chickens, the infectious bronchitis virus (IBV) coronavirus can cause disease that affects different organ systems and tissues, such as IBV-associated nephritis. As with other species, inbred status and specific chicken lines have been shown to impact host susceptibility, immune response, and outcomes, and virus-host genetic interactions have been described.100, 101, 102 Breeding experiments have suggested different inheritance patterns related to susceptibility and outcomes and have implicated both MHC and non-MHC loci.63,103 Multiple GWASs investigating immune response to IBV have identified significantly associated polymorphisms in the breeds studied;104,105 the implicated or nearest genes include AKT1, AvBD12, CEP170B, CRYL1, CWF19L2, DHRSX, FAM19A2, GABRB3, INTS9, NMNAT3, PINX1, RAB39A, VRK1, YEATS2, and SETBP1 (see Figure 2, including related to genes identified through studies of other animals as described below).104,105
Felines can be infected by FCoV, which includes FIPV and FECV.95 As with other species, cats demonstrate a range of potential effects. In addition to association with traits such as age, sex, and reproductive status, purebred status and loss of heterozygosity has been shown to be associated with the effects of disease. Susceptibility and outcomes also appear to vary between different breeds.95,106, 107, 108 A small study of feline leukocyte antigen (FLA)-DRB alleles did not show a statistically significant association between FLA-DRB alleles and FCoV infection outcome.64 Polymorphisms in IFNG (investigated because FIP can result in decreased interferon-gamma levels) were shown to correlate with plasma interferon-gamma levels and outcomes.109 Polymorphisms in TNFA and CD209 were also shown to be associated with outcomes in one inbred breed.110
In addition to candidate studies, several GWASs have been performed in cats. One small study on outcomes in experimentally induced infections in random-bred cats identified one associated genomic region (which did not harbor any obvious candidate genes).95 Another small study on an inbred breed identified multiple candidate genes (ELMO1, ERAP1, ERAP2, RRAGA, and TNSF10) but none were fully concordant with the FIP disease phenotype.111
The GWAS approach (which has also been used to study mice, resulting in implication of Trim55 as described above78) raises several important issues. A first issue involves immediate clinical applicability. That is, GWAS approaches may reveal findings that were not immediately hypothesized to be involved, such as variants in genes other than those known to be involved in viral pathogenesis and immunity.112 These findings may be statistically significant, but translating results to clinical uses in the near-term may be challenging despite excitement and perhaps incomplete understanding in the lay press. However, these insights may be important for longer-term and equally important purposes, such as in relation to therapeutic development or understanding which populations may be overall more or less vulnerable to disease. In other words, pertinent host genetic findings identified in hypothesis-free ways may unearth unexpected findings (beyond receptor, HLA, and well-characterized immune genes) that may yield important next steps to help combat the disease.
A second issue—which has received more recent attention in many genomic studies—involves important secondary information that may be revealed through host genetic research or through genomic testing and studies done for other purposes. Previously, lists of recommended secondary genes have been compiled in general contexts, and recommendations have been made about informing individuals about these findings (prior to the COVID-19 pandemic). With COVID-19, genomic investigators have newly assembled lists of secondary genetic information that may be relevant to the pandemic. These include genes involved in pharmacogenomics, conditions that involve metabolic or thrombotic crises, and cardiopulmonary conditions.113 Beyond this overarching framework, specific papers have already been published about pharmacogenomic considerations for medications, such as anti-IL-6 agents for the treatment of COVID-19 (as well as hydroxychloroquine and azithromycin).80,114
Human Studies
Details of the human studies are presented in the section on Literature Search and Sources, and in Table 1, S2, and Figures 3, 4, and 5.
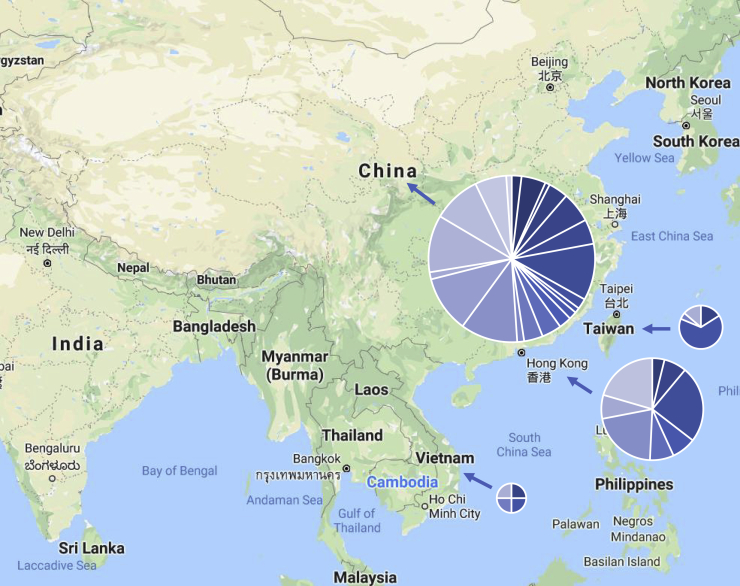
Figure 5.
Previous Cohorts Studied with Relative Numbers of Cases Shown in All the Studies Performed
Controls are not depicted here because relatively large populations from donor banks were used in several studies, skewing the data. Each circle represents the total number of cases from that country (China = 7,429; Hong Kong = 2,333; Taiwan = 406; Vietnam = 176). Each country’s circle is divided into sections, each of which represents an individual study. Studies that recruited in multiple countries are shown in each respective country. Study designs (including those related to both cases and controls) differed markedly. Details for each depicted study are given in Table S2.
Of the 39 human studies on host genetic study factors, 35 (90%) involved SARS-CoV-1, whereas 4 (10%) involved SARS-CoV-2. Thirty-six of the 39 studies examined specific genes and loci; three of the SARS-CoV-2 studies were case reports (two on single families and the other on two patients with a rare immunodeficiency) without specific studies related to host factors. All of the association studies except one were candidate-gene analyses based on genes hypothesized to be important in disease susceptibility or clinical variables/outcome. The exception to date was a meta-analysis of 386 studies on susceptibility to tuberculosis, influenza, respiratory syncytial virus, SARS-CoV-1, and pneumonia.115
As summarized in Figures 3 and 4, candidate studies ranged from studies of single variants to studies of over 50 genes selected because of biological plausibility; seven of these studies focused on HLA alleles. Sixteen significant loci related to susceptibility to coronavirus were reported (seven of which identified protective alleles) (Figure 3). Sixteen significant loci related to outcomes or clinical variables were reported (three which identified protective alleles) (Figure 4). The types of cases and controls used varied considerably. For example, some studies compared healthcare workers with SARS-CoV-1 infection with healthcare workers who tested negative. Others compared data from individuals with documented infection with data from control samples taken from blood donors. Only four studies used separate cohorts for replication/validation.
Four studies conducted laboratory-based biological studies in addition to association analyses. Of note, one study related to allele frequencies and expression in SARS-CoV-2 focused on specific genes but used data generated via exome sequencing and SNP-arrays.44 Large amounts of data generated through these types of genomic assays are currently being analyzed; some results are available on preprint servers and through other data sharing mechanisms.
In addition to the germline variants described in these previous studies, non-germline changes are discussed as possibly pertinent to COVID-19. Correlations between clonal hematopoiesis and COVID-19 mortality have been suggested,116 as have the potential importance of tumor-based ACE2 genetics and epigenetics.117
Limitations to Human Studies to Date
Traditional genome-wide methods have been applied to human viral infections generally,115 but results have not been specific to coronaviruses, and it is unclear to what extent the observations are relevant to the current pandemic. Several dozen studies have investigated human genetic factors related to coronavirus infection. However, these studies have been limited by several factors. Although the previous endemic human coronaviruses are common, the mildness of disease may have deprioritized recruitment into these studies. Similar observations may explain the relative dearth of serologic knowledge related to these pathogens. MERS and SARS-CoV-1 are severe, but the fact that these epidemics were limited more than the COVID-19 pandemic may have fortunately led to a lack of cases to conduct traditional association studies (unlike some other respiratory infections leading to more widespread disease).118 See Figure 5 for a depiction of study locations and the relative numbers of cases included in each study, as well as Table S2 for details of the corresponding studies. Additionally, MERS and SARS-CoV-1 primarily affected humans prior to the technological developments that led to wide availability of much cheaper and faster genomic sequencing.
As shown (Table S2), the small sample sizes of previous studies may have led to the preponderance of candidate gene studies. The sample sizes may also have precluded significant findings because of limitations of statistical power and the ability to replicate or validate findings. As previous research took place in certain countries and regions (Figure 5), it is possible that the results would not extrapolate to other populations. Finally, candidate approaches can be inherently limited because non-hypothesized loci may be significantly involved.
On the basis of announcements about multiple large-scale projects on COVID-19 host genetic factors, as well as the existence of larger genomic datasets that can be mined quickly and new methods that can be used to address biological questions, it is anticipated that considerable efforts—and an unfortunately large pool of research subjects—will yield significant new results quickly.
Implications Related to Genetic Conditions
Although separate from the bulk of the material reviewed here, another area is worthy of brief mention. This is the rapid and sometimes dramatic changes that have been necessary to manage patients with genetic and related conditions. Just as many genetic researchers have pivoted to address the pandemic, clinical genetic experts have modified their practices to support the patients they serve. The literature already reflects specific guidance and lessons learned for many genetic conditions, such as Charcot-Marie-Tooth, G6PD deficiency, Gaucher disease, inherited arrhthymias, and inborn errors of metabolism (see Table S3 for references for COVID-19 guidance related to these conditions). The information takes into account how the known genetic and biologic underpinnings of disease—as well as related considerations such as pharmacogenomics—should be considered to optimize outcomes. In addition to these pragmatic guidelines, understanding gained from studying the impact of COVID-19 on people with these rare diseases may yield insights that can be applied to the population at large, much like how unraveling the causes of primary immunodeficiencies can lead to generalizable knowledge about the immune system.
Limitations to Our Findings
There are multiple limitations to our summaries and analyses. First, it is likely that relevant articles were missed by our search process, and that key findings—including the study of certain genes—were therefore omitted. Along these lines, important findings within identified articles may also have been missed. Due to publication biases, some studies that have been conducted may not have reported relevant data. Second, this analysis focused on DNA-based variants. These DNA-based genetic changes include those studied and identified through association studies as well as genes that were manipulated in experimental approaches, such as via knockout models to understand disease pathogenesis. Related “omic” approaches, such as targeted or broad transcriptomic or proteomic studies, are frequently used to understand important aspects of disease. These approaches can lead to knowledge regarding specific genetic changes. For example, observed transcriptomic changes may enable the identification of important DNA-based variants that explain disease by correlating transcriptomic data with results of DNA sequencing.119 As another example from proteomics, a recent paper describes the human/SARS-CoV-2 protein-protein interactome, which may be highly relevant for understanding host genetic factors.120 However, we categorized non-DNA-based “omic” approaches separately from DNA-based studies and did not attempt to comprehensively recapitulate what is known about host reaction to disease. Finally, as the studies varied in many aspects, such as how cases and controls were defined, and which loci were interrogated, we were careful about comparing or combining data between different studies.
Conclusions
Human studies on other coronaviruses and model organism work has provided us with a guide for potential classes of genomic variants that are relevant to SARS-CoV-2 infection. Although we only addressed one facet of host responses to COVID-19, our analyses may help bolster the investigation of specific candidate loci. Future work involving in-depth phenotypic characterization, extensive patient sequencing (including that of outliers with severe and mild disease), and modeling efforts will allow clinicians and researchers to use this information to directly impact clinical care.
Data and Code Availability
This study did not generate any new data.
Declaration of Interests
B.D.S. previously worked for a subsidiary of Opko Health, a company whose subsidiary companies currently perform genetic testing as well as COVID-19-related testing.
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health. B.D.S. and D.B.B. are National Institutes of Health employees. P.D. was supported by the US National Institutes of Health award 2R01AI148049-21A1. D.A.T.C. was supported by the US National Institutes of Health award R01-AI114703-01.
Published: August 14, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.08.007.
Web Resources
23andMe/23andMe Research Blog, “Could host genetics play a role in the severity of COVID-19,” https://blog.23andme.com/23andme-research/genetics-and-covid-19-severity/
International Committee on Taxonomy of Viruses, https://talk.ictvonline.org/
Nextstrain, http://nextstrain.org/help/general/about-nextstrain/
The COVID-19 Host Genetics Initiative, https://www.covid19hg.org/
Supplemental Data
Articles Included in the Systematic Review, with Article Categorization (Based on Schema Described in Supplemental Methods and Figure 1)
Details of Human Studies Related to Host Genetic Factors
References
- 1.Zhang X., Tan Y., Ling Y., Lu G., Liu F., Yi Z., Jia X., Wu M., Shi B., Xu S. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583:437–440. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- 2.Woo P.C., Lau S.K., Huang Y., Yuen K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. (Maywood) 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 3.Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Groot R.J., Baker S., Baric R., Enjuanes L., Gorbalenya A., Holmes K., Perlman S., Poon L., Rottier P., Talbot P. Family coronaviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy. Elsevier; 2012. pp. 806–828. [Google Scholar]
- 5.Gaunt E.R., Hardie A., Claas E.C., Simmonds P., Templeton K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Poder S. Feline and canine coronaviruses: common genetic and pathobiological features. Adv. Virol. 2011;2011:609465. doi: 10.1155/2011/609465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu Y.K., Ali G.D., Jia F., Li Q., Kelvin D., Couch R.C., Harrod K.S., Hutt J.A., Cameron C., Weiss S.R., Jonsson C.B. The SARS-CoV ferret model in an infection-challenge study. Virology. 2008;374:151–163. doi: 10.1016/j.virol.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan J.F., Zhang A.J., Yuan S., Poon V.K., Chan C.C., Lee A.C., Chan W.M., Fan Z., Tsoi H.W., Wen L. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa325. Published online March 26, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang L., He C., Lei M., Li S., Hao Y., Zhu H., Duan Q. Pathology of guinea pigs experimentally infected with a novel reovirus and coronavirus isolated from SARS patients. DNA Cell Biol. 2005;24:485–490. doi: 10.1089/dna.2005.24.485. [DOI] [PubMed] [Google Scholar]
- 10.Nagata N., Iwata N., Hasegawa H., Fukushi S., Yokoyama M., Harashima A., Sato Y., Saijo M., Morikawa S., Sata T. Participation of both host and virus factors in induction of severe acute respiratory syndrome (SARS) in F344 rats infected with SARS coronavirus. J. Virol. 2007;81:1848–1857. doi: 10.1128/JVI.01967-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McAuliffe J., Vogel L., Roberts A., Fahle G., Fischer S., Shieh W.J., Butler E., Zaki S., St Claire M., Murphy B., Subbarao K. Replication of SARS coronavirus administered into the respiratory tract of African Green, rhesus and cynomolgus monkeys. Virology. 2004;330:8–15. doi: 10.1016/j.virol.2004.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fish I., Boissinot S. Contrasted patterns of variation and evolutionary convergence at the antiviral OAS1 gene in old world primates. Immunogenetics. 2015;67:487–499. doi: 10.1007/s00251-015-0855-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui J., Han N., Streicker D., Li G., Tang X., Shi Z., Hu Z., Zhao G., Fontanet A., Guan Y. Evolutionary relationships between bat coronaviruses and their hosts. Emerg. Infect. Dis. 2007;13:1526–1532. doi: 10.3201/eid1310.070448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou Y., Peng C., Yu M., Li Y., Han Z., Li F., Wang L.F., Shi Z. Angiotensin-converting enzyme 2 (ACE2) proteins of different bat species confer variable susceptibility to SARS-CoV entry. Arch. Virol. 2010;155:1563–1569. doi: 10.1007/s00705-010-0729-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui J., Eden J.S., Holmes E.C., Wang L.F. Adaptive evolution of bat dipeptidyl peptidase 4 (dpp4): implications for the origin and emergence of Middle East respiratory syndrome coronavirus. Virol. J. 2013;10:304. doi: 10.1186/1743-422X-10-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tusell S.M., Schittone S.A., Holmes K.V. Mutational analysis of aminopeptidase N, a receptor for several group 1 coronaviruses, identifies key determinants of viral host range. J. Virol. 2007;81:1261–1273. doi: 10.1128/JVI.01510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leopardi S., Holmes E.C., Gastaldelli M., Tassoni L., Priori P., Scaravelli D., Zamperin G., De Benedictis P. Interplay between co-divergence and cross-species transmission in the evolutionary history of bat coronaviruses. Infect. Genet. Evol. 2018;58:279–289. doi: 10.1016/j.meegid.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Doremalen N., Miazgowicz K.L., Milne-Price S., Bushmaker T., Robertson S., Scott D., Kinne J., McLellan J.S., Zhu J., Munster V.J. Host species restriction of Middle East respiratory syndrome coronavirus through its receptor, dipeptidyl peptidase 4. J. Virol. 2014;88:9220–9232. doi: 10.1128/JVI.00676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miguel E., Chevalier V., Ayelet G., Ben Bencheikh M.N., Boussini H., Chu D.K., El Berbri I., Fassi-Fihri O., Faye B., Fekadu G. Risk factors for MERS coronavirus infection in dromedary camels in Burkina Faso, Ethiopia, and Morocco, 2015. Euro Surveill. 2017;22:30498. doi: 10.2807/1560-7917.ES.2017.22.13.30498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang G., Feenstra B., Bacelis J., Liu X., Muglia L.M., Juodakis J., Miller D.E., Litterman N., Jiang P.P., Russell L. Genetic Associations with Gestational Duration and Spontaneous Preterm Birth. N. Engl. J. Med. 2017;377:1156–1167. doi: 10.1056/NEJMoa1612665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F. Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections. J. Virol. 2008;82:6984–6991. doi: 10.1128/JVI.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C., Chen Q.X., Gao Y.W., Zhou H.Q., Xiang H. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. USA. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeffers S.A., Tusell S.M., Gillim-Ross L., Hemmila E.M., Achenbach J.E., Babcock G.J., Thomas W.D., Jr., Thackray L.B., Young M.D., Mason R.J. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schickli J.H., Thackray L.B., Sawicki S.G., Holmes K.V. The N-terminal region of the murine coronavirus spike glycoprotein is associated with the extended host range of viruses from persistently infected murine cells. J. Virol. 2004;78:9073–9083. doi: 10.1128/JVI.78.17.9073-9083.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Doremalen N., Miazgowicz K.L., Munster V.J. Mapping the Specific Amino Acid Residues That Make Hamster DPP4 Functional as a Receptor for Middle East Respiratory Syndrome Coronavirus. J. Virol. 2016;90:5499–5502. doi: 10.1128/JVI.03267-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martina B.E., Haagmans B.L., Kuiken T., Fouchier R.A., Rimmelzwaan G.F., Van Amerongen G., Peiris J.S., Lim W., Osterhaus A.D. Virology: SARS virus infection of cats and ferrets. Nature. 2003;425:915. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smits S.L., de Lang A., van den Brand J.M., Leijten L.M., van IJcken W.F., Eijkemans M.J., van Amerongen G., Kuiken T., Andeweg A.C., Osterhaus A.D., Haagmans B.L. Exacerbated innate host response to SARS-CoV in aged non-human primates. PLoS Pathog. 2010;6:e1000756. doi: 10.1371/journal.ppat.1000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim Y.I., Kim S.G., Kim S.M., Kim E.H., Park S.J., Yu K.M., Chang J.H., Kim E.J., Lee S., Casel M.A.B. Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe. 2020;27:704–709.e2. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukuma A., Tani H., Taniguchi S., Shimojima M., Saijo M., Fukushi S. Inability of rat DPP4 to allow MERS-CoV infection revealed by using a VSV pseudotype bearing truncated MERS-CoV spike protein. Arch. Virol. 2015;160:2293–2300. doi: 10.1007/s00705-015-2506-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van den Brand J.M., Haagmans B.L., Leijten L., van Riel D., Martina B.E., Osterhaus A.D., Kuiken T. Pathology of experimental SARS coronavirus infection in cats and ferrets. Vet. Pathol. 2008;45:551–562. doi: 10.1354/vp.45-4-551. [DOI] [PubMed] [Google Scholar]
- 32.Bunyavanich S., Do A., Vicencio A. Nasal Gene Expression of Angiotensin-Converting Enzyme 2 in Children and Adults. JAMA. 2020;323:2427–2429. doi: 10.1001/jama.2020.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakkers M.J., Zeng Q., Feitsma L.J., Hulswit R.J., Li Z., Westerbeke A., van Kuppeveld F.J., Boons G.J., Langereis M.A., Huizinga E.G., de Groot R.J. Coronavirus receptor switch explained from the stereochemistry of protein-carbohydrate interactions and a single mutation. Proc. Natl. Acad. Sci. USA. 2016;113:E3111–E3119. doi: 10.1073/pnas.1519881113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pöhlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. USA. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeager C.L., Ashmun R.A., Williams R.K., Cardellichio C.B., Shapiro L.H., Look A.T., Holmes K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tresnan D.B., Levis R., Holmes K.V. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 1996;70:8669–8674. doi: 10.1128/jvi.70.12.8669-8674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benbacer L., Kut E., Besnardeau L., Laude H., Delmas B. Interspecies aminopeptidase-N chimeras reveal species-specific receptor recognition by canine coronavirus, feline infectious peritonitis virus, and transmissible gastroenteritis virus. J. Virol. 1997;71:734–737. doi: 10.1128/jvi.71.1.734-737.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delmas B., Gelfi J., L’Haridon R., Vogel L.K., Sjöström H., Norén O., Laude H. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams R.K., Jiang G.S., Holmes K.V. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. USA. 1991;88:5533–5536. doi: 10.1073/pnas.88.13.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li K.K., Yip C.W., Hon C.C., Lam C.Y., Zeng F., Leung F.C. Characterisation of animal angiotensin-converting enzyme 2 receptors and use of pseudotyped virus to correlate receptor binding with susceptibility of SARS-CoV infection. Hong Kong Med. J. 2012;18(Suppl 3):35–38. [PubMed] [Google Scholar]
- 44.Asselta R., Paraboschi E.M., Mantovani A., Duga S. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. Aging (Albany N.Y.) 2020;12:10087–10098. doi: 10.18632/aging.103415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao Y., Li L., Feng Z., Wan S., Huang P., Sun X., Wen F., Huang X., Ning G., Wang W. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020;6:11. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cruz J.O., Conceição I.M.C.A., Sousa S.M.B., Luizon M.R. Functional prediction and frequency of coding variants in human ACE2 at binding sites with SARS-CoV-2 spike protein on different populations. J. Med. Virol. 2020 doi: 10.1002/jmv.26126. Published online June 3, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao Y., Qin L., Zhang P., Li K., Liang L., Sun J., Xu B., Dai Y., Li X., Zhang C. Longitudinal COVID-19 profiling associates IL-1RA and IL-10 with disease severity and RANTES with mild disease. JCI Insight. 2020;5:139834. doi: 10.1172/jci.insight.139834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inoue Y., Tanaka N., Tanaka Y., Inoue S., Morita K., Zhuang M., Hattori T., Sugamura K. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 2007;81:8722–8729. doi: 10.1128/JVI.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Basit A., Ali T., Rehman S.U. Truncated human angiotensin converting enzyme 2; a potential inhibitor of SARS-CoV-2 spike glycoprotein and potent COVID-19 therapeutic agent. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1768150. Published online May 20, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hussain M., Jabeen N., Raza F., Shabbir S., Baig A.A., Amanullah A., Aziz B. Structural variations in human ACE2 may influence its binding with SARS-CoV-2 spike protein. J. Med. Virol. 2020 doi: 10.1002/jmv.25832. Published online April 6, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li W., Luo R., He Q., van Kuppeveld F.J.M., Rottier P.J.M., Bosch B.J. Aminopeptidase N is not required for porcine epidemic diarrhea virus cell entry. Virus Res. 2017;235:6–13. doi: 10.1016/j.virusres.2017.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitworth K.M., Rowland R.R.R., Petrovan V., Sheahan M., Cino-Ozuna A.G., Fang Y., Hesse R., Mileham A., Samuel M.S., Wells K.D., Prather R.S. Resistance to coronavirus infection in amino peptidase N-deficient pigs. Transgenic Res. 2019;28:21–32. doi: 10.1007/s11248-018-0100-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cui T., Theuns S., Xie J., Van den Broeck W., Nauwynck H.J. Role of Porcine Aminopeptidase N and Sialic Acids in Porcine Coronavirus Infections in Primary Porcine Enterocytes. Viruses. 2020;12:402. doi: 10.3390/v12040402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan V.S., Chan K.Y., Chen Y., Poon L.L., Cheung A.N., Zheng B., Chan K.H., Mak W., Ngan H.Y., Xu X. Homozygous L-SIGN (CLEC4M) plays a protective role in SARS coronavirus infection. Nat. Genet. 2006;38:38–46. doi: 10.1038/ng1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen J., Pan K., Chen Z., Li Y., Ding B., Han C., Cao Z., Bao W., Zhang Y. Production of porcine aminopeptidase N (pAPN) site-specific edited pigs. Anim. Sci. J. 2019;90:366–371. doi: 10.1111/asj.13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tu C.F., Chuang C.K., Hsiao K.H., Chen C.H., Chen C.M., Peng S.H., Su Y.H., Chiou M.T., Yen C.H., Hung S.W. Lessening of porcine epidemic diarrhoea virus susceptibility in piglets after editing of the CMP-N-glycolylneuraminic acid hydroxylase gene with CRISPR/Cas9 to nullify N-glycolylneuraminic acid expression. PLoS ONE. 2019;14:e0217236. doi: 10.1371/journal.pone.0217236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rehman S.U., Tabish M. Alternative splicing of ACE2 possibly generates variants that may limit the entry of SARS-CoV-2: a potential therapeutic approach using SSOs. Clin. Sci. (Lond.) 2020;134:1143–1150. doi: 10.1042/CS20200419. [DOI] [PubMed] [Google Scholar]
- 58.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abbott T.R., Dhamdhere G., Liu Y., Lin X., Goudy L., Zeng L., Chemparathy A., Chmura S., Heaton N.S., Debs R. Development of CRISPR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza. Cell. 2020;181:865–876.e12. doi: 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Faiq M.A. B-cell engineering: A promising approach towards vaccine development for COVID-19. Med. Hypotheses. 2020;144:109948. doi: 10.1016/j.mehy.2020.109948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Daley G.Q., Lovell-Badge R., Steffann J. After the Storm - A Responsible Path for Genome Editing. N. Engl. J. Med. 2019;380:897–899. doi: 10.1056/NEJMp1900504. [DOI] [PubMed] [Google Scholar]
- 62.Schaefer G.O., Tam C.C., Savulescu J., Voo T.C. COVID-19 vaccine development: Time to consider SARS-CoV-2 challenge studies? Vaccine. 2020;38:5085–5088. doi: 10.1016/j.vaccine.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bacon L.D., Hunter D.B., Zhang H.M., Brand K., Etches R. Retrospective evidence that the MHC (B haplotype) of chickens influences genetic resistance to attenuated infectious bronchitis vaccine strains in chickens. Avian Pathol. 2004;33:605–609. doi: 10.1080/03079450400013147. [DOI] [PubMed] [Google Scholar]
- 64.Addie D.D., Kennedy L.J., Ryvar R., Willoughby K., Gaskell R.M., Ollier W.E., Nart P., Radford A.D. Feline leucocyte antigen class II polymorphism and susceptibility to feline infectious peritonitis. J. Feline Med. Surg. 2004;6:59–62. doi: 10.1016/j.jfms.2003.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Brien S.J., Roelke M.E., Marker L., Newman A., Winkler C.A., Meltzer D., Colly L., Evermann J.F., Bush M., Wildt D.E. Genetic basis for species vulnerability in the cheetah. Science. 1985;227:1428–1434. doi: 10.1126/science.2983425. [DOI] [PubMed] [Google Scholar]
- 66.Dobrynin P., Liu S., Tamazian G., Xiong Z., Yurchenko A.A., Krasheninnikova K., Kliver S., Schmidt-Küntzel A., Koepfli K.P., Johnson W. Genomic legacy of the African cheetah, Acinonyx jubatus. Genome Biol. 2015;16:277. doi: 10.1186/s13059-015-0837-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yousefzadegan S., Rezaei N. Case Report: Death Due to Novel Coronavirus Disease (COVID-19) in Three Brothers. Am. J. Trop. Med. Hyg. 2020;102:1203–1204. doi: 10.4269/ajtmh.20-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Debnath M., Banerjee M., Berk M. Genetic gateways to COVID-19 infection: Implications for risk, severity, and outcomes. FASEB J. 2020 doi: 10.1096/fj.202001115R. Published online June 11, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barquera R., Collen E., Di D., Buhler S., Teixeira J., Llamas B., Nunes J.M., Sanchez-Mazas A. Binding affinities of 438 HLA proteins to complete proteomes of seven pandemic viruses and distributions of strongest and weakest HLA peptide binders in populations worldwide. HLA. 2020 doi: 10.1111/tan.13956. Published May 31, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nguyen A., David J.K., Maden S.K., Wood M.A., Weeder B.R., Nellore A., Thompson R.F. Human leukocyte antigen susceptibility map for SARS-CoV-2. J Virol. 2020;94:e00510–e00520. doi: 10.1128/JVI.00510-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gledhill A.W., Andrewes C.H. A hepatitis virus of mice. Br. J. Exp. Pathol. 1951;32:559–568. [PMC free article] [PubMed] [Google Scholar]
- 72.Gallily R., Warwick A., Bang F.B. Effect of Cortisone of Genetic Resistance to Mouse Hepatitis Virus in Vivo and in Vitro. Proc. Natl. Acad. Sci. USA. 1964;51:1158–1164. doi: 10.1073/pnas.51.6.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guénet J.L. Assessing the genetic component of the susceptibility of mice to viral infections. Brief. Funct. Genomics Proteomics. 2005;4:225–240. doi: 10.1093/bfgp/4.3.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kantoch M., Warwick A., Bang F.B. The cellular nature of genetic susceptibility to a virus. J. Exp. Med. 1963;117:781–798. doi: 10.1084/jem.117.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilson G.A., Dales S. In vivo and in vitro models of demyelinating disease: efficiency of virus spread and formation of infectious centers among glial cells is genetically determined by the murine host. J. Virol. 1988;62:3371–3377. doi: 10.1128/jvi.62.9.3371-3377.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mihm S. COVID-19: Possible Impact of the Genetic Background in IFNL Genes on Disease Outcomes. J. Innate Immun. 2020;12:273–274. doi: 10.1159/000508076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kyuwa S., Shibata S., Tagawa Y., Iwakura Y., Machii K., Urano T. Acute hepatic failure in IFN-gamma-deficient BALB/c mice after murine coronavirus infection. Virus Res. 2002;83:169–177. doi: 10.1016/S0168-1702(01)00432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gralinski L.E., Ferris M.T., Aylor D.L., Whitmore A.C., Green R., Frieman M.B., Deming D., Menachery V.D., Miller D.R., Buus R.J. Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross. PLoS Genet. 2015;11:e1005504. doi: 10.1371/journal.pgen.1005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manrai A.K., Funke B.H., Rehm H.L., Olesen M.S., Maron B.A., Szolovits P., Margulies D.M., Loscalzo J., Kohane I.S. Genetic Misdiagnoses and the Potential for Health Disparities. N. Engl. J. Med. 2016;375:655–665. doi: 10.1056/NEJMsa1507092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Giudicessi J.R., Roden D.M., Wilde A.A.M., Ackerman M.J. Genetic susceptibility for COVID-19-associated sudden cardiac death in African Americans. Heart Rhythm. 2020 doi: 10.1016/j.hrthm.2020.04.045. Published online May 5, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu B., Li N.L., Wang J., Shi P.Y., Wang T., Miller M.A., Li K. Overlapping and distinct molecular determinants dictating the antiviral activities of TRIM56 against flaviviruses and coronavirus. J. Virol. 2014;88:13821–13835. doi: 10.1128/JVI.02505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.von Brunn A., Ciesek S., von Brunn B., Carbajo-Lozoya J. Genetic deficiency and polymorphisms of cyclophilin A reveal its essential role for Human Coronavirus 229E replication. Curr. Opin. Virol. 2015;14:56–61. doi: 10.1016/j.coviro.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao X., Sehgal M., Hou Z., Cheng J., Shu S., Wu S., Guo F., Le Marchand S.J., Lin H., Chang J., Guo J.T. Identification of Residues Controlling Restriction versus Enhancing Activities of IFITM Proteins on Entry of Human Coronaviruses. J. Virol. 2018;92:e01535-17. doi: 10.1128/JVI.01535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang P., Meng W., Han S.C., Li C.C., Wang X.J., Wang X.J. The nucleolar protein GLTSCR2 is required for efficient viral replication. Sci. Rep. 2016;6:36226. doi: 10.1038/srep36226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao X., Guo F., Liu F., Cuconati A., Chang J., Block T.M., Guo J.T. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc. Natl. Acad. Sci. USA. 2014;111:6756–6761. doi: 10.1073/pnas.1320856111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lei Y., Moore C.B., Liesman R.M., O’Connor B.P., Bergstralh D.T., Chen Z.J., Pickles R.J., Ting J.P. MAVS-mediated apoptosis and its inhibition by viral proteins. PLoS ONE. 2009;4:e5466. doi: 10.1371/journal.pone.0005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bang F.B. The use of a genetically incompatible combination of host and virus (MHV) for the study of mechanisms of host resistance. Adv. Exp. Med. Biol. 1981;142:359–373. doi: 10.1007/978-1-4757-0456-3_30. [DOI] [PubMed] [Google Scholar]
- 88.Lévy-Leblond E., Oth D., Dupuy J.M. Genetic study of mouse sensitivity to MHV3 infection: influence of the H-2 complex. J. Immunol. 1979;122:1359–1362. [PubMed] [Google Scholar]
- 89.Smith M.S., Click R.E., Plagemann P.G. Control of mouse hepatitis virus replication in macrophages by a recessive gene on chromosome 7. J. Immunol. 1984;133:428–432. [PubMed] [Google Scholar]
- 90.Daya M., Wong F., Cervin M., Evans G., Vennema H., Spaan W.J., Anderson R. Mouse fibroblast mutants selected for survival against mouse hepatitis virus infection show increased resistance to infection and virus-induced cell fusion. Adv. Exp. Med. Biol. 1990;276:59–66. doi: 10.1007/978-1-4684-5823-7_9. [DOI] [PubMed] [Google Scholar]
- 91.Damy S.B., Vassão R.C., Lucchiari M.A., Pereira C.A., Sant’Anna O.A. A comparative study of resistance to MHV3 infection in genetically homogeneous and heterogeneous mouse populations. Braz. J. Med. Biol. Res. 1992;25:1025–1027. [PubMed] [Google Scholar]
- 92.Casanova J.L., Su H.C., COVID Human Genetic Effort A Global Effort to Define the Human Genetics of Protective Immunity to SARS-CoV-2 Infection. Cell. 2020;181:1194–1199. doi: 10.1016/j.cell.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ikitimur H., Borku Uysal B., Cengiz M., Ikitimur B., Uysal H., Ozcan E., Islamoglu M.S., Seyhan S., Yavuzer H., Yavuzer S. “Determining Host Factors Contributing to Disease Severity in a Family Cluster of 29 Hospitalized SARS-CoV-2 Patients: Could Genetic Factors Be Relevant in the Clinical Course of COVID-19?”. J. Med. Virol. 2020 doi: 10.1002/jmv.26106. Published online June 3, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chan K.C., Tang N.L., Hui D.S., Chung G.T., Wu A.K., Chim S.S., Chiu R.W., Lee N., Choi K.W., Sung Y.M. Absence of association between angiotensin converting enzyme polymorphism and development of adult respiratory distress syndrome in patients with severe acute respiratory syndrome: a case control study. BMC Infect. Dis. 2005;5:26. doi: 10.1186/1471-2334-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pedersen N.C., Liu H., Gandolfi B., Lyons L.A. The influence of age and genetics on natural resistance to experimentally induced feline infectious peritonitis. Vet. Immunol. Immunopathol. 2014;162:33–40. doi: 10.1016/j.vetimm.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Verity R., Okell L.C., Dorigatti I., Winskill P., Whittaker C., Imai N., Cuomo-Dannenburg G., Thompson H., Walker P.G.T., Fu H. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect. Dis. 2020;20:669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Norris J.M., Bosward K.L., White J.D., Baral R.M., Catt M.J., Malik R. Clinicopathological findings associated with feline infectious peritonitis in Sydney, Australia: 42 cases (1990-2002) Aust. Vet. J. 2005;83:666–673. doi: 10.1111/j.1751-0813.2005.tb13044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Karnam G., Rygiel T.P., Raaben M., Grinwis G.C., Coenjaerts F.E., Ressing M.E., Rottier P.J., de Haan C.A., Meyaard L. CD200 receptor controls sex-specific TLR7 responses to viral infection. PLoS Pathog. 2012;8:e1002710. doi: 10.1371/journal.ppat.1002710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.de Groot N.G., Bontrop R.E. COVID-19 pandemic: is a gender-defined dosage effect responsible for the high mortality rate among males? Immunogenetics. 2020;72:275–277. [Google Scholar]
- 100.Ignjatovic J., Reece R., Ashton F. Susceptibility of three genetic lines of chicks to infection with a nephropathogenic T strain of avian infectious bronchitis virus. J. Comp. Pathol. 2003;128:92–98. doi: 10.1053/jcpa.2002.0609. [DOI] [PubMed] [Google Scholar]
- 101.Dawes M.E., Griggs L.M., Collisson E.W., Briles W.E., Drechsler Y. Dramatic differences in the response of macrophages from B2 and B19 MHC-defined haplotypes to interferon gamma and polyinosinic:polycytidylic acid stimulation. Poult. Sci. 2014;93:830–838. doi: 10.3382/ps.2013-03511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.da Silva A.P., Hauck R., Zhou H., Gallardo R.A. Understanding Immune Resistance to Infectious Bronchitis Using Major Histocompatibility Complex Chicken Lines. Avian Dis. 2017;61:358–365. doi: 10.1637/11666-050117-RegR. [DOI] [PubMed] [Google Scholar]
- 103.Bumstead N., Huggins M.B., Cook J.K. Genetic differences in susceptibility to a mixture of avian infectious bronchitis virus and Escherichia coli. Br. Poult. Sci. 1989;30:39–48. doi: 10.1080/00071668908417123. [DOI] [PubMed] [Google Scholar]
- 104.Luo C., Qu H., Ma J., Wang J., Hu X., Li N., Shu D. A genome-wide association study identifies major loci affecting the immune response against infectious bronchitis virus in chicken. Infect. Genet. Evol. 2014;21:351–358. doi: 10.1016/j.meegid.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang W., Zhang T., Zhang G., Wang J., Han K., Wang Y., Zhang Y. Genome-wide association study of antibody level response to NDV and IBV in Jinghai yellow chicken based on SLAF-seq technology. J. Appl. Genet. 2015;56:365–373. doi: 10.1007/s13353-014-0269-y. [DOI] [PubMed] [Google Scholar]
- 106.Foley J.E., Poland A., Carlson J., Pedersen N.C. Risk factors for feline infectious peritonitis among cats in multiple-cat environments with endemic feline enteric coronavirus. J. Am. Vet. Med. Assoc. 1997;210:1313–1318. [PubMed] [Google Scholar]
- 107.Pesteanu-Somogyi L.D., Radzai C., Pressler B.M. Prevalence of feline infectious peritonitis in specific cat breeds. J. Feline Med. Surg. 2006;8:1–5. doi: 10.1016/j.jfms.2005.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bell E.T., Malik R., Norris J.M. The relationship between the feline coronavirus antibody titre and the age, breed, gender and health status of Australian cats. Aust. Vet. J. 2006;84:2–7. doi: 10.1111/j.1751-0813.2006.tb13114.x. [DOI] [PubMed] [Google Scholar]
- 109.Hsieh L.E., Chueh L.L. Identification and genotyping of feline infectious peritonitis-associated single nucleotide polymorphisms in the feline interferon-γ gene. Vet. Res. (Faisalabad) 2014;45:57. doi: 10.1186/1297-9716-45-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang Y.T., Hsieh L.E., Dai Y.R., Chueh L.L. Polymorphisms in the feline TNFA and CD209 genes are associated with the outcome of feline coronavirus infection. Vet. Res. (Faisalabad) 2014;45:123. doi: 10.1186/s13567-014-0123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Golovko L., Lyons L.A., Liu H., Sørensen A., Wehnert S., Pedersen N.C. Genetic susceptibility to feline infectious peritonitis in Birman cats. Virus Res. 2013;175:58–63. doi: 10.1016/j.virusres.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Doganay L., Agaoglu N.B., Irvem A., Alkurt G., Yildiz J., Kose B., Demirkol Y.K., Dogan O.A., Doganay G.D. Responding to COVID-19 in Istanbul: Perspective from genomic laboratory. North. Clin. Istanb. 2020;7:311–312. doi: 10.14744/nci.2020.30075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stergachis A.B., Weiss S.T., Green R.C. Biobanks could identify medically actionable findings relevant for COVID-19 clinical care. Nat. Med. 2020;26:991. doi: 10.1038/s41591-020-0953-x. [DOI] [PubMed] [Google Scholar]
- 114.Perricone C., Conigliaro P., Ciccacci C., Marcucci E., Cafaro G., Bartoloni E., Perricone R., Novelli G., Borgiani P., Gerli R. The differential response to anti IL-6 treatment in COVID-19: the genetic counterpart. Clin. Exp. Rheumatol. 2020;38:580. [PubMed] [Google Scholar]
- 115.Patarčić I., Gelemanović A., Kirin M., Kolčić I., Theodoratou E., Baillie K.J., de Jong M.D., Rudan I., Campbell H., Polašek O. The role of host genetic factors in respiratory tract infectious diseases: systematic review, meta-analyses and field synopsis. Sci. Rep. 2015;5:16119. doi: 10.1038/srep16119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shivarov V., Ivanova M. Clonal haematopoiesis and COVID-19: A possible deadly liaison. Int. J. Immunogenet. 2020;47:329–331. doi: 10.1111/iji.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chai P., Yu J., Ge S., Jia R., Fan X. Genetic alteration, RNA expression, and DNA methylation profiling of coronavirus disease 2019 (COVID-19) receptor ACE2 in malignancies: a pan-cancer analysis. J. Hematol. Oncol. 2020;13:43. doi: 10.1186/s13045-020-00883-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Curtis J., Luo Y., Zenner H.L., Cuchet-Lourenço D., Wu C., Lo K., Maes M., Alisaac A., Stebbings E., Liu J.Z. Susceptibility to tuberculosis is associated with variants in the ASAP1 gene encoding a regulator of dendritic cell migration. Nat. Genet. 2015;47:523–527. doi: 10.1038/ng.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Frésard L., Smail C., Ferraro N.M., Teran N.A., Li X., Smith K.S., Bonner D., Kernohan K.D., Marwaha S., Zappala Z., Undiagnosed Diseases Network. Care4Rare Canada Consortium Identification of rare-disease genes using blood transcriptome sequencing and large control cohorts. Nat. Med. 2019;25:911–919. doi: 10.1038/s41591-019-0457-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Articles Included in the Systematic Review, with Article Categorization (Based on Schema Described in Supplemental Methods and Figure 1)
Details of Human Studies Related to Host Genetic Factors
Data Availability Statement
This study did not generate any new data.