We used a relatively new method, HyperTRIBE, to identify in vivo RNA targets of the translation inhibitor 4E-BP.
Abstract
4E-BP (eIF4E-BP) represses translation initiation by binding to the 5′ cap–binding protein eIF4E and inhibiting its activity. Although 4E-BP has been shown to be important in growth control, stress response, cancer, neuronal activity, and mammalian circadian rhythms, it is not understood how it preferentially represses a subset of mRNAs. We successfully used HyperTRIBE (targets of RNA binding proteins identified by editing) to identify in vivo 4E-BP mRNA targets in both Drosophila and mammals under conditions known to activate 4E-BP. The protein associates with specific mRNAs, and ribosome profiling data show that mTOR inhibition changes the translational efficiency of 4E-BP TRIBE targets more substantially compared to nontargets. In both systems, these targets have specific motifs and are enriched in translation-related pathways, which correlate well with the known activity of 4E-BP and suggest that it modulates the binding specificity of eIF4E and contributes to mTOR translational specificity.
INTRODUCTION
The signaling pathway downstream of the insulin receptor (InR) and the mammalian (or mechanistic) target of rapamycin kinase (mTOR) is well conserved in all eukaryotes and integrates different environmental signals to govern organismal growth. Under favorable circumstances such as nutrient-rich conditions, InR-mTOR signaling accelerates growth by increasing processes involved in anabolic metabolism and promoting growth in cell size and number (1). The same processes are inhibited in nutrient-restricted or stress conditions. A major process regulated by InR-mTOR signaling is translation. This is so that this high energy–consuming process can be increased under progrowth conditions or decreased under low-growth or no-growth conditions, e.g., under stress conditions.
4E-BP [eukaryotic initiation factor 4E (eIF4E)–binding protein] is a crucial regulator of this translational control pathway. There are three homologs (4E-BP1, 4E-BP2, and 4E-BP3) in mammals but only one in Drosophila melanogaster (Thor/d4E-BP), the product of the Thor gene. d4E-BP is crucial for life-span extension under oxidative stress and nutrient-restricted conditions [Zid et al. (2)], consistent with a prominent role in restricting growth. The impact of 4E-BP extends far beyond cellular stress management, e.g., it affects cancer, proper neuronal activity, and mammalian circadian entrainment (2–6).
4E-BP inhibits translation initiation by blocking the formation of the translation initiation complex eIF4F (1, 7, 8). eIF4F consists of the 5′ cap–binding protein eIF4E, the RNA helicase eIF4A, and the scaffold protein eIF4G. Under favorable growth conditions, the mTOR signaling pathway phosphorylates 4E-BP, which prevents it from binding to eIF4E. Under less favorable or stress conditions, inactivation of mTOR signaling results in dephosphorylation and activation of 4E-BP. Because eIF4G and hypophosphorylated 4E-BP share the same binding site on eIF4E, active 4E-BP blocks the eIF4G-binding site on eIF4E and thereby inhibits eIF4F formation and translation (9).
To identify mRNAs regulated by 4E-BP, others have examined global mRNA translation using ribosome profiling under mTOR inhibition conditions (10, 11). The results showed that active 4E-BP preferentially represses translation of a subset of mRNAs. Although inhibition of eIF4F formation can explain how the general rate of cap-dependent translation is decreased, it does not explain how the translation of specific mRNAs is selectively inhibited, i.e., whether this is a direct or indirect effect of active 4E-BP.
In vitro assays have suggested that 4E-BP may indirectly associate with mRNA through the cap-binding protein eIF4E. There are several pieces of evidence supporting this hypothesis, including that 4E-BP can be copurified with eIF4E from cell extracts with an mRNA-cap analog pull-down assay and that 4E-BP enhances the in vitro interaction of eIF4E with a capped short RNA oligonucleotide (12, 13). However, these interactions do not address the specificity issue, and it is uncertain whether they are physiologically relevant. This is because there are few tools to detect the association of proteins like 4E-BP with specific mRNA within cells.
To address this deficiency, i.e., to identify the in vivo target transcripts of RNA binding proteins (RBPs) in Drosophila, we recently developed TRIBE (targets of RBPs identified by editing) (14). This technique characterizes fusion proteins between the catalytic domain of the Drosophila RNA editing enzyme ADAR (ADARcd) and RBPs of interest. An RBP-ADARcd fusion protein can bind to target transcripts of the RBP and edit adenosine nucleotides to inosines near the fusion protein binding site; these are read as guanosines by high-throughput sequencing and identified as A-to-G editing sites by computational analysis. Transcripts hosting these A-to-G editing sites are considered binding targets of the RBP. The upgraded version of TRIBE, HyperTRIBE, contains the one amino acid substitution E488Q in the ADARcd region (hyper-ADARcd), which improves target identification efficiency by reducing bias originating from the intrinsic editing preference of the ADARcd (15–17).
Here, we applied HyperTRIBE to both flies and mammals and characterized 4E-BP direct target transcripts within cells. These transcripts were enriched for mRNAs encoding translation factors, including the translation initiation factor eIF3. Translation factors fit with previous results describing 4E-BP–regulated transcripts, suggesting that much of this regulation is due to a direct interaction of 4E-BP–containing protein complexes with target transcripts.
RESULTS
mTOR-dependent translational repression affects specific genes, suggesting that 4E-BP may associate with a subset of mRNAs and/or that some mRNAs selectively escape from 4E-BP–mediated repression (11, 18). The TRIBE method was developed in the Drosophila system and is ideal to address the possibility that d4E-BP associates with specific mRNAs. To perform Thor-TRIBE experiments in cultured Drosophila S2 cells, we constructed a plasmid in which the hyperactive Drosophila ADARcd (hyper-dADARcd) was fused to Drosophila 4E-BP (d4E-BP; fig. S1A). If d4E-BP associates with specific eIF4E–5′ capped mRNAs in cells, the hyper-dADARcd should deaminate nearby adenosines, leaving A-to-I editing marks on the associated mRNAs. A plasmid that only expresses the hyper-dADARcd was used as a negative control (16). We used Western blotting to confirm their similar expression (fig. S1B) and also that Thor–hyper-dADARcd could be efficiently dephosphorylated after mTOR inhibition (fig. S1C).
To address whether TRIBE can identify Thor-associated mRNA targets, mRNA libraries were generated from positively transfected cells and sequenced with the Illumina NextSeq 500 System. To generate lists of editing sites and target genes, the sequencing data were then analyzed with our published TRIBE computational analysis pipeline [Rahman et al. (15)]. In wild-type S2 cells or S2 cells expressing hyper-dADARcd alone, very few editing events were detected. However, thousands of editing sites were detected after induction of Thor-TRIBE expression (Fig. 1A, Thor-Hyper). Because mTOR inhibition dephosphorylates 4E-BP and enhances its activity and its interaction with eIF4E (11), we reasoned that mTOR inhibition should increase the number of target mRNAs identified by Thor-TRIBE. More editing sites and target genes were detected after inhibiting mTOR activity with serum depletion and rapamycin. There were also more editing sites and genes after addition of the mTOR inhibitor Torin-1, indicating that editing may reflect Thor activity (Fig. 1A). Moreover, the biological repeats were reproducible (fig. S1D). The data taken together suggest that Thor-TRIBE reflects d4E-BP activity and that the fusion protein associates with a subset of specific mRNAs in cells (Fig. 1A).
Fig. 1. Thor-HyperTRIBE identifies specific RNAs as 4E-BP targets in Drosophila S2 cells.
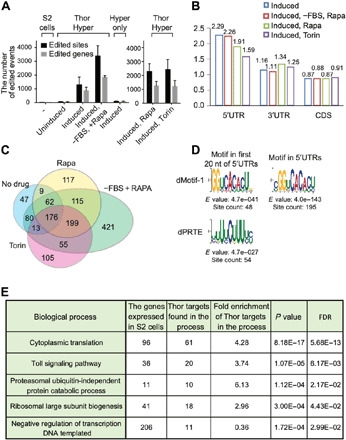
(A) Thor-HyperTRIBE but not hyper-ADARcd alone (Hyper only) edits transcripts after copper induction. The editing sites (black bars) and editing genes (gray bars) in Thor-HyperTRIBE significantly increased in rapamycin- or Torin-1–treated cells and more than doubled in rapamycin-treated plus serum-deprived cells. The number of editing events in cells expressing hyper-ADARcd alone is comparable to that of control S2 cells (N = 2, +SEM). (B) Editing sites identified by Thor-HyperTRIBE are enriched in 5′UTR of mRNAs. (C) Venn diagram of Thor-HyperTRIBE target RNAs reproducibly identified under different conditions shows that the targets are consistent, although significantly more were identified with serum deprivation and rapamycin treatment combined. The transcripts edited in S2 or Hyper only control were removed from the target list. One hundred seventy-six target genes were identified in all conditions. (D) Consensus motifs from the 5′UTRs of 968 Thor-HyperTRIBE targets (listed in table S1), which were reproducibly detected in rapamycin or Torin-1 treatment condition. The GGUCACACU motif is identified in both cases with 195 counts (~20%) in the entire 5′UTR of the targets. (E) Table of enriched GO term biological processes in 968 Thor-HyperTRIBE targets reproducibly detected in rapamycin or Torin-1 treatment condition. FDR, false discovery rate.
We next performed metagene analysis to examine the distribution of editing sites. The distribution of sequencing reads from all exonic regions was used as background. The editing sites of Thor-TRIBE were relatively enriched in 5′ untranslated regions (5′UTRs) compared to this background, consistent with a preferred association of d4E-BP with the eIF4E–5′ cap complex (Fig. 1B). The target transcripts identified under these different conditions overlapped well, although many more were detected after mTOR inhibition (Fig. 1C). We carried out de novo motif searches from the 5′UTRs or 3′UTRs of the 968 Thor targets (table S1), which were reproducibly detected in Thor-HyperTRIBE after rapamycin or Torin-1 treatment. As a background control, we randomly selected 968 highly expressed genes, which were not identified as Thor targets.
The search for consensus sequences from the first 20 nucleotides (nt) of target 5′UTRs identified a GGUCACACUG motif (dMotif-1) and a pyrimidine-rich motif (dPRTE; Fig. 1D). A further motif search within the full 5′UTRs of targets identified a similar CGGUCACACU motif in 195 mRNAs (>20%) with an even more significant E value and site count than that from the first 20 nt; no significant motifs were found within the 3′UTRs (Fig. 1D).
The dPRTE resembles the 5′ terminal oligopyrimidine tract (5′TOP) and the pyrimidine-rich translational element (PRTE) identified as translational control elements in mammals (fig. S1E) (10, 19–21). These motifs are also enriched within the 5′UTRs of mTOR-responsive mammalian mRNAs (10, 11), consistent with the results shown here. Both the 5′TOP and the PRTE are stretches of uninterrupted pyrimidine nucleotides, but the 5′TOP starts with a cytosine residing from position +1 of the 5′UTR, and the PRTE is defined as a pyrimidine-rich element within the 5′UTR including an invariant uridine at position 6 of the motif. The activity of mammalian La-related protein (LARP) is also under mTOR regulation and is reported to bind to a similar 5′TOP or PRTE motif (see Discussion) (22, 23).
Furthermore, gene ontology (GO) term analysis indicates that these Thor targets are enriched in protein synthesis pathways, consistent with the notion that translation is a key process regulated directly by Thor (Fig. 1E). Toll signaling and ubiquitin-independent proteasomal proteins are also enriched, whereas negative regulators of transcription are depleted in the Thor targets. As the mTOR pathway has been shown to be an important mediator of immune responses (24, 25), these results implicate Thor in regulating the innate immune response via Toll signaling.
To verify that d4E-BP inhibits the translation of these mRNAs, protein synthesis was assayed in a number of different ways. We first used metabolic labeling by SUnSET (surface sensing of translation) to assay the effect of mTOR inhibition on general translation. This method uses puromycin incorporation into newly synthesized peptides to monitor the rate of general protein synthesis (26). Puromycin-attached peptides are detected by Western blotting using an anti-puromycin antibody. Cells were pulsed with puromycin, and we compared general protein synthesis after treating cells with several mTOR inhibitors: rapamycin, Torin-1, or Ink128. All three inhibitors reduced protein synthesis, with rapamycin being the most effective (Fig. 2A). We suspect that the effect of the mTOR inhibitors on general protein synthesis may be different in different cells. Translational inhibition by rapamycin is weaker than that by Torin-1 or Ink128 in mouse embryo fibroblasts (MEFs), but rapamycin and Ink128 inhibition are similar in mouse lymphocytes (11, 27).
Fig. 2. Ribosome profiling in S2 cells identifies mRNAs regulated by mTOR pathway.
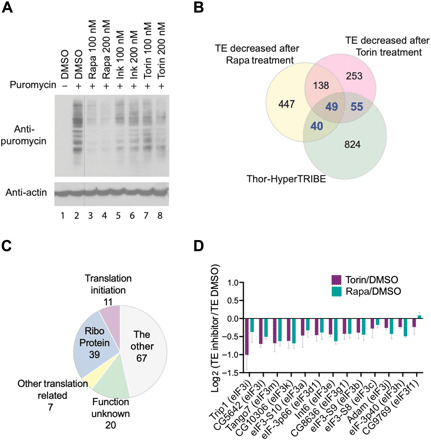
(A) Metabolic labeling by SUnSET shows global protein synthesis reduced after treating S2 cells with mTOR inhibitor, rapamycin, Ink128, or Torin-1 for 2 hours compared to vehicle DMSO. Puromycin was incorporated into newly synthesized peptides, being measured by Western blotting using antibody against puromycin. (B) Ribosome profiling identifies 144 Thor-TRIBE targets (40 + 49 + 55, gene numbers are marked by blue color), TE of which decreased after treating cells with 100 nM rapamycin or Torin-1. Venn diagram overlap of genes between 968 Thor-TRIBE targets, genes with decreased TE after adding rapamycin, and genes with decreased TE after adding Torin-1 (n = 3, TE fold change of mTOR inhibitor versus DMSO < 0.8, P < 0.1). (C) Functional classification of 144 Thor-TRIBE targets, which are from the overlapped portion in (B). The main functions of these genes include ribosome components (Ribo Protein), translational initiation, and other translation-related roles. (D) The translation of most eIF3 mRNAs is repressed by mTOR inhibitor. The bar chart showed log2 TE fold-change value of all expressed eIF3 transcripts (rapamycin/DMSO, blue bar; Torin-1/DMSO, purple bar; n = 3, +SEM).
To examine translational activity at the whole-transcriptome level, we carried out ribosome profiling with and without mTOR inhibition, i.e., 100 nM rapamycin or Torin-1 for 2 hours. The strategy of ribosome profiling in mammals and yeast is to obtain ribosome-protected fragments (RPFs) by digesting cell lysates with ribonuclease I (RNase I) (28, 29). However, Drosophila ribosomes are reported to be too sensitive to RNase I, so micrococcal nuclease (MNase) is commonly used for ribosome profiling in this species. Although this makes P-site mapping difficult because of the strong 3′ A/T bias of MNase, it is still useful for measuring translation rates (30).
Treating cell lysates with MNase confirmed that RPFs are enriched between 28 and 34 nt in size (fig. S2A). We then purified these RPFs, made sequencing libraries using the adaptor ligation–based method, and sequenced them. Input mRNAs were purified from cell lysates without MNase treatment and fragmented to make sequencing libraries following similar protocols to those for RPFs. The length of sequencing reads peaked at ~32 nt from RPFs and ~42 nt from input mRNAs (fig. S2B). The measurements of input and RPF from both replicates correlated well (fig. S2C). Consistent with the results of the metabolic labeling (Fig. 2A), more translationally repressed genes were detected after treating with rapamycin than that with Torin-1, i.e., 674 mRNAs decreased in translational efficiency (TE) after rapamycin treatment and 495 mRNAs decreased in TE after Torin-1 treatment (Fig. 2B and table S2).
To orthogonally validate the Thor-TRIBE target data, we compared these genes with those that manifest decreased TEs in response to rapamycin or Torin-1 treatment. One hundred forty-four of these transcripts were also identified by Thor-TRIBE, suggesting that they are direct targets of d4E-BP (40 + 49 + 55; Fig. 2B). They encode ribosomal proteins (RPs), translational initiation factors, and other translation-related proteins (Fig. 2C). These transcripts are highly enriched for subunits of the eIF3 complex. Of the 13 expressed subunits of eIF3, 10 fall in this overlapping category, namely, eIF3b, eIF3d1, eIF3e, eIF3g1, eIF3h, eIF3i, eIF3j, eIF3k, eIF3l, and eIF3m (table S3). The TE of these 10 genes decreased after mTOR inhibition (Fig. 2D).
We next asked whether the motifs enriched in Thor-TRIBE targets appear in the translational repression assays. We compared the mean TE change after mTOR inhibitor treatment among different groups of transcripts expressed in S2 cells, including non–Thor-TRIBE targets, all Thor-TRIBE targets, Thor-TRIBE targets with dMotif-1, or Thor-TRIBE targets with dPRTE (Fig. 3A).
Fig. 3. Ribosome profiling data show that the TE change of Thor-TRIBE target after mTOR inhibition has a down-regulation compared with non–Thor-TRIBE target.
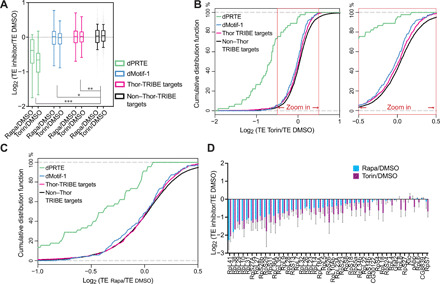
(A) The mean TE change of Thor-TRIBE targets is down-regulated compared with nontargets when mTOR pathway is inhibited by mTOR inhibitor, rapamycin, or Torin-1. The degree of decrease varies with different groups of Thor-TRIBE targets. Box plot of mean TE change (log2 value) after a 2-hour treatment with mTOR inhibitor versus vehicle DMSO from different groups of expressed transcripts, including all non–Thor-TRIBE targets (black, n = 5604), Thor-TRIBE targets (purple, n = 919), Thor-TRIBE targets with dMotif-1 (blue, n = 142), and Thor-TRIBE with dPRTE (green, n = 45). Ordinary one-way analysis of variance (ANOVA): *P < 0.05, **P < 0.01, and ***P < 0.001. (B and C) Cumulative frequency distribution of log2 TE fold-change values from non–Thor-TRIBE targets (black, n = 5604) and different groups of Thor-TRIBE targets showed that Thor targets have significantly different TE change from nontargets. Torin-1 versus DMSO shown in (B); rapamycin versus DMSO shown in (C). (D) TE of most Thor-TRIBE targets with dPRTE decreased after treatment of mTOR inhibitor. The bar chart showed log2 TE fold-change value of Thor-TRIBE targets with dPRTE (rapamycin/DMSO, blue bar; Torin-1/DMSO, purple bar; n = 3, +SEM).
Thor-TRIBE targets containing the dPRTE/TOP motif showed marked translational repression in response to mTOR inhibition. This is consistent with previous reports that regulation of TOP mRNA translation by mTORC1 is 4E-BP dependent (10, 11, 31). Although much less marked, the mean TE changes of all Thor-TRIBE targets and of targets with dMotif-1 were also significantly different from that of nontargets. Cumulative frequency distributions of TE changes of targets with a dPRTE motif also had a marked distribution difference compared to nontargets, and all Thor-TRIBE targets or targets with dMotif-1 showed a much weaker but still significant difference from nontargets, especially when Torin-1 was used as the mTOR inhibitor (Fig. 3, B and C). These ribosome profiling data are notable because most dPRTE-containing Thor-TRIBE targets had substantially decreased TE after treatment with the mTOR inhibitors compared to the dimethyl sulfoxide (DMSO) control (Fig. 3D). Although the 5′TOP and the PRTE cis-regulatory motifs are well known in mammals and predicted to be translation elements in Drosophila (fig. S1E) (21), our results confirm that they play a conserved role in flies (Fig. 3D).
We then used CLIP (crosslinking immunoprecipitation) to address RNA recognition and determine whether d4E-BP is in physical contact with RNA. The RBP and its ultraviolet (UV)–cross-linked RNAs in the cell lysate were partially digested with RNase A. After immunoprecipitating d4E-BP with a V5 antibody, cross-linked RNAs in the RBP were labeled with [γ-32P]adenosine triphosphate ([γ-32P]ATP) at their 5′ ends and detected on denaturing polyacrylamide gels (Fig. 4A). A parallel control experiment was carried out with untransfected S2 cells and an antibody directed against the Drosophila heterogeneous nuclear RNP (HnRNP) protein Hrp48. Although the Thor-TRIBE signal was not as strong as a typical RBP like Hrp48, radioactive signals were reproducibly detected by phosphoimager at 5 to 10 kDa above the expected protein size, indicating that d4E-BP is in close proximity to RNA and may contribute to RNA binding activity like Hrp48 (Fig. 4B). Transcript specificity of the CLIP tags was poor compared to the TRIBE data, suggesting that Thor interacts broadly and transiently with mRNA and that TRIBE may be more successful at pointing to a biologically meaningful subset of transcripts (see Discussion).
Fig. 4. CLIP of Thor-V5 identifies a direct RNA binding activity.
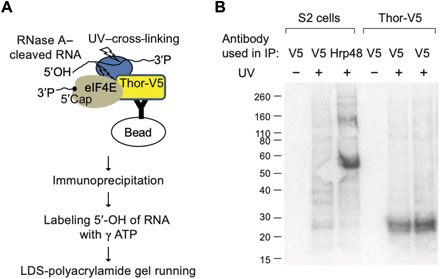
(A) Schematic illustration of CLIP. UV–cross-linked and lysed Thor-V5 cells were treated with RNase A, and then immunoprecipitation was carried out from cell lysate using an antibody against V5 tag. The partially digested RNAs, which had been cross-linked to RBP, were labeled at their 5′ end by [γ-32P]ATP. Radioisotope-labeled RNA-RBP could be separated and visualized in denaturing LDS polyacrylamide gel. LDS, lithium dodecyl sulfate. (B) CLIP of Thor-V5 shows that Thor has an RNA binding activity. A well-known RBP, Hrp48, was used as a positive control. S2 or Thor-V5 cells were UV–cross-linked (UV+) or not (UV−). Immunoprecipitation was carried out using antibody against V5 tag or Hrp48.
Last, we examined whether 4E-BP also associates with target mRNAs in mammalian cells. As mentioned above, there are three 4E-BP proteins in mammals, namely, 4E-BP1, 4E-BP2, and 4E-BP3. They have somewhat different expression patterns in adult mouse tissues, and we chose 4E-BP1 (human or h4E-BP1) for HyperTRIBE experiments because it is the most broadly expressed (32). To carry out h4E-BP1-HyperTRIBE in human cells, we first tried the hyperactive Drosophila ADARcd previously used for HyperTRIBE editing (16). However, few editing events were detected when Drosophila hyper-ADARcd was fused to h4E-BP1 and HyperTRIBE carried out in human prostate cancer PC3 cells. We therefore substituted the hyper-dADARcd with the catalytic domain of human ADAR2 containing the E488Q point mutation at the corresponding position (hyper-hADAR2cd).
The results showed that h4E-BP1-HyperTRIBE but not the hyper-hADAR2cd alone successfully identified h4E-BP1 targets in PC3 cells. mTOR inhibition with Ink128/PP242 increased h4E-BP1-HyperTRIBE editing compared to the DMSO control (Fig. 5A), similar to the Drosophila results shown above (Fig. 1A). Also similar to those results, these human cell data indicate that 4E-BP inhibits translation by associating with its target mRNAs.
Fig. 5. h4E-BP1 HyperTRIBE identifies h4E-BP1 targets in PC3 cells.
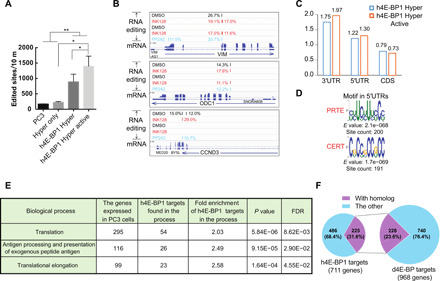
(A) The number of edited sites is significantly higher in h4E-BP1 HyperTRIBE than in wild-type PC3 cells or in Hyper only. The number of edited sites was increased after Ink128 or PP242 treatment (h4E-BP1 Hyper Active). Expressing hyperADARcd alone (Hyper only) did not result in more editing sites than in control PC3 cells. N = 2 to 3, +SEM; *P < 0.05, paired one-tailed Student’s t test; **P < 0.005, unpaired one-tailed Student’s t test. (B) IGV view of three examples of the editing sites and editing percentages of mTOR-sensitive genes VIM, ODC1, and CCND3. Background editing sites detected in control PC3 cells or Hyper only were removed. Samples were incubated with DMSO (black), Ink128 (red), or PP242 (blue). The alignment of mRNA-seq reads is also shown. (C) The editing sites of h4E-BP1 HyperTRIBE are enriched in 5′UTR and 3′UTR of mRNA. (D) Consensus motifs from the 5′UTRs of 711 h4E-BP HyperTRIBE targets (listed in table S4). These targets were detected at least twice after mTOR inhibition. (E) Table of enriched GO term biological processes in 711 h4E-BP1 HyperTRIBE targets. (F) Ortholog mapping between 711 h4E-BP1 HyperTRIBE targets and 968 Thor-HyperTRIBE targets shows that 32% of h4E-BP1 targets are conserved in Drosophila.
Transcripts of VIM (vimentin), ODC1 (ornithine decarboxylase), and CCND3 (cyclin D3) were previously identified as 4E-BP targets by polysome analysis or ribosome profiling using 4E-BP1/2 double knockout MEFs (10, 33). These mRNAs were specifically edited by h4E-BP1-HyperTRIBE (Fig. 5B). Metagene analysis showed that these edited sites in h4E-BP1-HyperTRIBE were enriched in 5′UTR and 3′UTR sequences (Fig. 5C).
De novo motif searches from the UTRs of TRIBE targets identified the PRTE as well as cytosine-enriched regulator of translation (CERT) within the entire 5′UTR. The CERT motif was previously reported to be enriched in eIF4E-sensitive mRNAs, i.e., the translation of these transcripts was sensitive to eIF4E expression levels (34).
These PC3 cell experiments identified 711 h4E-BP1 target genes (table S4), and their functions were enriched in translation processes and the immune response like in fly cells (Fig. 5E). Comparing these 711 h4E-BP1 targets in human cells with the 968 d4E-BP targets in fly cells identified 180 sets of targeted homologs. As 225 human genes have 228 orthologs in flies, at least 32% of the 4E-BP human targets are conserved in flies (Fig. 5F and table S5). These data point to conserved 4E-BP functions and mechanisms, and they also indicate that TRIBE works well in mammalian systems as well as in Drosophila.
DISCUSSION
TRIBE was first developed in Drosophila and worked well both in S2 tissue culture cells and in fly neurons (14, 16, 35). To adapt this method to mammalian systems, we first attempted to use the Drosophila ADAR catalytic domain as the editing component. For unknown reasons, however, its editing efficiency was weak in PC3 cells despite normal expression of the fusion protein. So, we generated a hyper-ADARcd from human ADAR2, which worked well in this human prostate cancer cell line. We suspect that the inefficiency of dADARcd in mammalian cells is due to the different cellular environment compared to Drosophila, e.g., a lack or presence of cofactors/helper proteins and a substantial temperature difference.
The results indicate that 4E-BP associates in both systems with overlapping sets of target mRNAs, which are highly enriched in translation and immune response transcripts. TRIBE therefore captures the two major roles of mTOR, growth control and the immune response. Specific targets include translation apparatus mRNAs like those encoding RPs; ribosomes are major growth effectors. In addition, 4E-BP represses the translation of most subunits of the translation initiation factor eIF3, although to a lesser extent than RPs. Several eIF3 subunits have been shown to exhibit features of TOP mRNAs (36). The function of these eIF3 targets is somewhat less straightforward to interpret, but one possibility is that these interactions also serve to inhibit growth: Reducing eIF3 protein levels would inhibit the translation of eIF3-dependent transcripts, which not only affects the canonical eIF4E-cap–dependent translation but also inhibits the eIF4E-independent cap-dependent translation (37).
There is only modest overlap between the many Thor-TRIBE targets and mRNAs with decreased TE after rapamycin or Torin-1 treatment (Fig. 2B). It is possible that a significant fraction of mRNAs with decreased TE is due to secondary effects of mTOR inhibition, e.g., direct inhibition of a positive translation factor, which would account for the poor representation of many Thor-TRIBE targets. There is also only a modest overlap between the effects of rapamycin and Torin-1 on TE (Fig. 2B). This might reflect different quantitative as well as qualitative effects of the two drugs and be related to the fact that rapamycin had a stronger overall translational inhibitory effect in S2 cells, for unknown reasons (Fig. 2A). This is despite the fact that Torin-1 is considered the more comprehensive mTOR inhibitor and had a stronger inhibitory effect on Thor-TRIBE targets (Figs. 2 and 3).
There are, to our knowledge, no previous data describing in vivo 4E-BP target transcripts. Otherwise put and despite previous results indicating that a subset of mRNAs is preferentially affected by mTOR and suggesting that 4E-BP is its principle downstream translational effector (10, 11), it was unclear how mRNA specificity is determined. It had been proposed that expression of eIF4E is rate-limiting under some conditions such as tumorigenesis and that the sequestration of eIF4E by activated 4E-BP reduces the level of the translation initiation complex eIF4F (34, 38). This results in the translational repression of mRNAs that are more sensitive to eIF4F levels (fig. S3, model 1). This makes sense in light of the fact that 4E-BP and eIF4G compete for the same region of eIF4E. However, this sequestration model is paradoxical as it suggests that the weakest eIF4E-binding mRNAs should be the most strongly affected by 4E-BP activity. Yet, it is known that translation component–encoding transcripts like RP mRNAs are efficiently translated, suggesting that the targets of 4E-BP are normally efficient binders of the canonical translational initiation complex.
This problem has been somewhat ameliorated by the recent identification of LARP1 as a second translational effector, which has been shown to bind directly to the 5′TOP and/or PRTE sequence elements as well as to the 5′ cap (22, 23). It is possible that Thor specificity for 5′TOP and/or PRTE elements comes, in part, from the specificity of dephosphorylated LARP1 for these elements (fig. S3, model 2). In this context, the much stronger repression of Thor targets with dPRTE than with dMotif-1 (Fig. 3) is consistent with a functional collaboration of LARP1 and Thor to repress the expression of dPRTE-containing transcripts. We note that the impact of 4E-BP on translation is likely to be more complicated as it may depend on eIF4E expression levels relative to those of 4E-BP as well as the status of the multiple phosphorylation sites of 4E-BP. There are also three orthologs of mammalian 4E-BP (1/2/3) with tissue-specific expression.
Moreover, LARP1 and 4E-BP have distinct mRNA targets except for PRTE mRNAs (22). It is also notable that Thor-TRIBE targets containing the dPRTE motif and those containing the CGGUCACACU motif (dMotif-1) rarely overlap; they therefore probably reflect different groups of regulated targets. This also suggests that Thor collaborates with different RBPs that affect and may even determine mRNA recognition (fig. S3, model 2). It is even possible in this context that Thor has no RNA recognition activity, which appears to conflict with the positive CLIP result (Fig. 4).
A positive CLIP signal may occur despite a very low intrinsic affinity of a protein for RNA. For example, 95% of the binding energy between eIF4E-BP and RNA could originate from a protein-protein interaction, perhaps an interaction with eIF4E and/or with another RBP, and only 5% from a direct eIF4E-BP-RNA interaction. A similar logic may apply to many of the large number of RBPs revealed by in vivo cross-linking procedures, i.e., perhaps only a minority of them are RBPs capable of binding to RNA in vitro as single recombinant proteins.
It may be relevant in this context that a comparable role of Drosophila LARP1 to that of mammalian LARP1 in growth control has not been established. There is considerable divergence between the fly and the mammalian protein, and Drosophila LARP1 is known to have other functions (39). These considerations indicate that even recognition of the dPRTE motif by Drosophila LARP1 is currently uncertain.
Although 4E-BP–mediated editing is enriched in the 5′UTR, editing occurs elsewhere within the transcript, in fly cells as well as in mammalian cells. This could reflect 4E-BP overexpression, which might also help explain the large number of Thor-TRIBE targets without a strong TE effect from rapamycin or Torin-1 addition (Fig. 2B). However, it is likely that editing outside of the 5′UTR is also because the ADARcd can access and edit a susceptible sequence element a considerable distance away from the binding site of its protein complex (14, 16). A slight enrichment in editing sites is also observed in the 3′UTR with no enrichment in the coding region. This 3′UTR enrichment may reflect associations between the 5′ and 3′ ends of translating mRNAs during 4E-BP–dependent translational inhibition, based, for example, on known protein-protein interactions between 5′ end and 3′ end translation components. This “reach” interpretation is supported by the fact that the Thor-TRIBE target genes have no significantly enriched 3′UTR motif. Moreover, no 5′UTR enrichment was observed in previous TRIBE editing experiments with other RBPs (14, 16). These considerations indicate that the editing data reflect the true binding specificity of Thor rather than any technical bias.
In summary, TRIBE works well for eIF4E-BP, in mammalian cells and in Drosophila cells. Recent results from elsewhere on different RBPs show that TRIBE also works well in other mammalian systems (40–42). Moreover, our current results suggest that TRIBE demonstrates more target transcript specificity than CLIP. Although work in this paper does not constitute a proper head-to-head comparison of TRIBE and CLIP, this suggestion is similar to conclusions from other TRIBE versus CLIP comparisons in mammalian cells (41). Because ADAR deamination is slow (17), TRIBE editing may reflect targets with longer RBP dwell time and therefore tighter binding, whereas cross-linking may be better able to capture ephemeral associations between a RBP and RNA. At a minimum, this progress indicates that TRIBE will add to the arsenal of techniques available to identify and study the mRNA targets of diverse RBPs in many different cells and systems.
MATERIALS AND METHODS
TRIBE in S2 cells
The region including the 5′UTR and CDS (protein coding sequence) of Thor was cloned into pMT-Linker-ADARcd-E488Q-V5 plasmid (100–amino acid flexible linker) to make pMT-Thor-Linker-HyperTRIBE construct using Gibson Assembly (New England Biolabs) (16). pMT HyperTRIBE plasmid (400 ng) and pAct5.1-eGFP (400 ng) (enhanced green fluorescent protein) were cotransfected into Drosophila S2 cells using Cellfectin II from Thermo Fisher Scientific. Twenty-four hours after transfection, TRIBE protein expression was induced with copper sulfate. Another 24 hours later, cells were treated with 50 nM rapamycin (Fisher Scientific) plus serum depletion, 50 nM rapamycin, 50 nM Torin-1 (Fisher Scientific), or vehicle DMSO for 5 hours. More than 10,000 GFP-positive cells were sorted and collected with a BD FACSAria II machine. Total RNA was extracted from the sorted cells with TRIzol LS reagent. Expression of proteins was assayed by Western blot using antibodies against V5 tag (Abcam, ab9116), phospho–4E-BP1 (Thr37/46; Cell Signaling Technology, # 2855), and β-actin (Santa Cruz Biotechnology, sc-47778).
Standard Illumina TruSeq RNA Library Kit was used to construct an RNA-sequencing (RNA-seq) library for TRIBE experiments. Fluorescence-activated cell sorting (FACS)–sorted cells were subjected to RNA-seq library protocol as previously described [McMahon et al. (14)]. All libraries were sequenced by Illumina NextSeq 500 sequencing system using NextSeq High Output Kit v2 (single end, 75 cycles). Each sample was covered by ∼10 million raw reads. The TRIBE analyses were performed according to the process of (15). Briefly, mRNA sequencing reads were mapped to dm3 using Tophat2 (-m 1 -p 5 -g 2 -I 50000 --microexon-search --no-coverage-search), allowing one mismatch. Polymerase chain reaction (PCR) duplicates were removed for editing analysis. Only events with minimum 20 reads and 10% editing were considered to be an editing event; genomic DNA (gDNA) coverage and uniformity of nucleotide identity were also required to avoid inclusion of single-nucleotide polymorphisms.
Background distribution was calculated by read distribution tool in RSeQC package (43). Editing site distribution was calculated by Bedtools intersect with dm3 Ref-seq annotation. Enrichment score was calculated by dividing the percentage of editing sites localized in each region by the percentage of sequencing coverage in the same region.
TRIBE in PC3 cells
The pCMV-hADAR2cd-E488Q plasmid was previously created by making a point mutation on the corresponding position of hADAR2 catalytic domain (42). The region of hADARcd-E488Q was cloned into pcDNA3-3HA-h4E-BP1 (a gift from N. Sonenberg) by Gibson Assembly to create pcDNA3-3HA-h4E-BP1-hADARcd-E488Q vector. Correct insertion was verified by Sanger sequencing.
PC3 cells were obtained from the American Type Culture Collection (ATCC) and maintained in F-12K medium (ATCC) with 10% of fetal bovine serum (FBS) as suggested by ATCC. pAAV-CMV-eGFP3 was cotransfected into PC3 cells with TRIBE construct using Lipofectamine 2000 (Thermo Fisher Scientific). One or 2 days later, PC3 cells were treated with 200 nM Ink128 (MedChemExpress), 2.5 μM PP242 (Selleckchem), or DMSO for 5 hours. GFP-positive cell sorting, RNA-seq library generation, and high-throughput sequencing were carried out as those in S2 cells. The TRIBE analyses were performed according to the processes of (15). Briefly, mRNA-seq reads were mapped to hg38 genome using Star3 (--runThreadN 8 --outFilterMismatchNoverLmax 0.07 --outFilterMatchNmin 16 --outFilterMultimapNmax 1). PCR duplicates were removed for editing analysis. The gDNA-sequencing data of PC3 were obtained from published data (44) to avoid inclusion of single-nucleotide polymorphisms. Only events with minimum 20 reads and 10% editing were considered to be editing events.
Ribosome profiling in S2 cells
Lysate preparation
The preparation of lysate and RPFs for ribosome profiling was performed according to the protocol described in (30). Eighteen million S2 cells were plated to 10-cm dishes in 12 ml of cell culture medium [HyClone SFX-Insect cell culture medium with 10% heat-inactivated FBS (Thermo Fisher Scientific)] 1 day before the experiment. Cells were treated with 100 nM rapamycin, Torin-1, or vehicle DMSO for 2 hours (six dishes of cells for each drug). After treating cells with emetine (20 μg/ml) (Sigma-Aldrich, St. Louis, MO) for 2 min, cells were washed twice with ice-cold phosphate-buffered saline supplemented with emetine [20 μg/ml] and four to six cell volumes (200 μl/10-cm dish) of cold polysome lysis buffer [50 mM tris buffer (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 0.5% Triton X-100, 1 mM dithiothreitol (DTT), SUPERase•In RNase Inhibitor (20 U/ml; Ambion), and emetine (20 μg/ml)] were collected. Cells were then homogenized on ice in a prechilled Dounce homogenizer (seven times with A pestle and seven times with B pestle). The lysate was clarified by spinning 10 min at 20,000g at 4°C. The supernatant was separated to three sets for generating RNA (input) and RPF sequencing libraries. Then, 750 μl of TRIzol LS (Thermo Fisher Scientific) was added to 250 μl of lysate, and total RNA (input RNA) purification was performed according to the manufacturer’s instructions. Other sets were flash-frozen in liquid nitrogen and stored at −80°C for RPF preparation. When ready to proceed, frozen samples were thawed on ice in the cold room.
RPF preparation
For each sample, lysate was diluted 2:1 in digestion mixture [50 mM tris (pH 7.5), 5 mM MgCl2, 0.5% Triton X-100, 1 mM DTT, SUPERase• In (20 U/ml), emetine (20 μg/ml), 15 mM CaCl2, and MNase (150 U per dish; Roche Applied Science)]. Samples were digested for 40 min at 25°C in a thermomixer (Eppendorf, Hamburg, Germany). Digestions were stopped by adding EGTA to a final concentration of 6.25 mM and placing the reactions on ice. For purification of monosomes, Sephacryl S-400 columns (GE Healthcare) were used. Columns were vortexed to mix resin well (avoid bubbles), and the storage buffer on resin was removed by gravity flow. Columns were equilibrated by repeating five times the step of loading 600 μl of polysome buffer and removing it by gravity flow. The columns were spun down at 600g for 4 min. Sample (250 μl) was loaded to the column and spun down at 600g for 2 min to collect the solution flown through into a new tube. The RNA was then purified from the solution using TRIzol LS.
rRNA depletion
Ten micrograms of input RNAs and 2.5 μg of RPF RNAs were treated with Ribo-Zero reaction (MRZH11124C, Epicentre) kit by following the manufacturer’s instructions. For ribosomal RNA (rRNA) depletion from RPF RNAs, the 50°C incubation in the manufacturer’s instructions was omitted.
The collected 100 μl of supernatant from Ribo-Zero reaction is precipitated by adding 500 μl of 100% ethanol, 10 μl of 3 M NaOAc, and 1 μl of glycoBlue (Invitrogen). The RNA was resuspended in 10 μl of nuclease-free water.
Sequencing library preparation
Input RNA was fragmented by partial alkaline hydrolysis. Input RNA (10 μl) was mixed with 10 μl of 2× fragmentation buffer [2 mM EDTA, 12 mM Na2CO3, and 88 mM NaHCO3, (pH ~9.3)] and incubated at 95°C for 18 to 20 min. Then, input RNA and RPF were resolved on a 15% tris-borate EDTA (TBE)–urea gel (Invitrogen). A gel slab corresponding to 34 to 55 nt for input and 28 to 34 nt for RPF was excised from the gel, eluted, and precipitated. 3′ phosphoryl groups were removed from RNA using T4 polynucleotide kinase (New England Biolabs) in the buffer [70 mM tris-HCl, 10 mM MgCl2, and 0.8 mM DTT (pH 6.5)] at 37°C for 20 min. After phenol extraction, library preparation was performed according to the manufacturer’s instructions of TruSeq small RNA kit (Illumina) with modifications. 3′ Adaptor (TruSeq small RNA kit, RA3, preadenylated) was ligated to the RNA using T4 RNA ligase 2 truncated (New England Biolabs) by incubating at 25°C for 6 hours and 22°C for 6 hours. 5′ End of RNA was phosphorylated and labeled by [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs) at 37°C for 45 min. 3′ Adaptor–ligated RNA was resolved on a 15% TBE–urea gel (Invitrogen), and the RNA in the size of 54 to 75 nt for input and 48 to 54 nt for RPF was purified. 5′ Adaptor (TruSeq small RNA kit, RA5, a degenerate bar code, four random nucleotides, was added at 3′ end of RA5) was ligated to RNA using T4 RNA ligase (New England Biolabs) at 25°C for 6 hours and 22°C for 6 hours. Reverse transcription was carried out using RT primer (TruSeq small RNA kit, RTP) and SuperScript III RT enzyme (Invitrogen). Library was amplified by 10 to 14 cycles of PCR using indexing primers (TruSeq small RNA kit) and Phusion polymerase (Thermo Fisher Scientific). Amplification product was size-selected on 6% TBE gels (Invitrogen). Samples were then quantitated using the Bioanalyzer High-Sensitivity DNA Assay (Agilent Technologies) and single-end–sequenced in Illumina NextSeq 500 sequencing system using NextSeq High Output Kit v2 (75 cycles).
Sequence processing and alignment of ribosome profiling
The alignment of sequencing reads was performed according to steps 90 to 96 of (45). Briefly, the raw reads were filtered and preprocessed according to quality scores (fastq_filter), the exact sequence duplicates were collapsed (fasta2collapse), 3′ adaptor sequence was trimmed (fastx_clipper), and degenerate bar-code sequences were removed (stripBarcode). The reads were aligned to the reference genome dm3 (novoalign -t 85 -l 23 -s 1 -r None) with at least 23 high-quality matches and with alignment cost score “−t 85.” Then, the potential PCR duplicates by coordinates were collapsed and unique reads were identified.
mRNA abundance (input RNA) and ribosome density (RPF) for each genomic feature were measured in FPKM (fragments per kilobase of feature length per million mapped reads) aligning to genomes. mRNA abundance reflects the total number of RNA fragments aligning to all countable exonic positions for a given gene. Ribosome density refers to all RPF fragments that align to countable positions of a coding region (CDS) for a given gene. We calculate TE as the ratio of footprint (RPF) FPKM in the CDS to the RNA fragment (input RNA) FPKM across the exons. FPKM more than 2 was used for further analysis.
Motif analysis and GO
We performed motif analysis using MEME version 4.11.2 (parameters: -minw 6 -maxw 10 -maxsize 10000000 -dna -nmotifs 5 -maxsites 200) (46). As the background control of the motif analysis, a similar number of transcripts were randomly selected from highly expressed genes that are not TRIBE targets. GO analyses were performed using PANTHER (47). TRIBE target genes were analyzed against a background of all genes expressed in the S2 cells or PC3 cells (>2 FPKM). Gene expression levels were quantified using Cufflinks2.
Supplementary Material
Acknowledgments
We thank M. Marr, A. Lee, and K. C. Abruzzi for comments on the manuscript as well as N. Sonenberg for the pcDNA3-3HA-4E-BP1 plasmid. Funding: The work was supported by the Howard Hughes Medical Institute and the NIH (grants DA037721 and R01AG052465). The work in BIT was supported by the General Program of National Natural Science Foundation of China (31970622). Author contributions: H.J. and M.R. are responsible for conceptualization. H.J., D.N., A.F., and W.X. performed the experiments. H.J., D.N., W.S., S.L., R.R., and C.L. analyzed the data. H.J., M.R., and W.X. wrote the manuscript. Competing interests: H.J., W.X., and M.R. declare that a PCT patent application (PCT patent application no.: PCT/US2016/054525) has been filed on the TRIBE technique. The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Raw and processed next-generation sequencing data from this study were deposited at National Center for Biotechnology Information Gene Expression Omnibus database with accession number GSE153346. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/33/eabb8771/DC1
REFERENCES AND NOTES
- 1.Saxton R. A., Sabatini D. M., mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zid B. M., Rogers A. N., Katewa S. D., Vargas M. A., Kolipinski M. C., Lu T. A., Benzer S., Kapahi P., 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell 139, 149–160 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teleman A. A., Chen Y.-W., Cohen S. M., 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes Dev. 19, 1844–1848 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tettweiler G., Miron M., Jenkins M., Sonenberg N., Lasko P. F., Starvation and oxidative stress resistance in Drosophila are mediated through the eIF4E-binding protein, d4E-BP. Genes Dev. 19, 1840–1843 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mamane Y., Petroulakis E., LeBacquer O., Sonenberg N., mTOR, translation initiation and cancer. Oncogene 25, 6416–6422 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Cao R., Robinson B., Xu H., Gkogkas C., Khoutorsky A., Alain T., Yanagiya A., Nevarko T., Liu A. C., Amir S., Sonenberg N., Translational control of entrainment and synchrony of the suprachiasmatic circadian clock by mTOR/4E-BP1 signaling. Neuron 79, 712–724 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma X. M., Blenis J., Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10, 307–318 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Kong J., Lasko P., Translational control in cellular and developmental processes. Nat. Rev. Genet. 13, 383–394 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Marcotrigiano J., Gingras A. C., Sonenberg N., Burley S. K., Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 3, 707–716 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Hsieh A. C., Liu Y., Edlind M. P., Ingolia N. T., Janes M. R., Sher A., Shi E. Y., Stumpf C. R., Christensen C., Bonham M. J., Wang S., Ren P., Martin M., Jessen K., Feldman M. E., Weissman J. S., Shokat K. M., Rommel C., Ruggero D., The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thoreen C. C., Chantranupong L., Keys H. R., Wang T., Gray N. S., Sabatini D. M., A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ptushkina M., von der Haar T., Karim M. M., Hughes J. M., McCarthy J. E., Repressor binding to a dorsal regulatory site traps human eIF4E in a high cap-affinity state. EMBO J. 18, 4068–4075 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pause A., Belsham G. J., Gingras A. C., Donzé O., Lin T. A., Lawrence J. C. Jr., Sonenberg N., Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5'-cap function. Nature 371, 762–767 (1994). [DOI] [PubMed] [Google Scholar]
- 14.McMahon A. C., Rahman R., Jin H., Shen J. L., Fieldsend A., Luo W., Rosbash M., TRIBE: Hijacking an RNA-editing enzyme to identify cell-specific targets of RNA-binding proteins. Cell 165, 742–753 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rahman R., Xu W., Jin H., Rosbash M., Identification of RNA-binding protein targets with HyperTRIBE. Nat. Protoc. 13, 1829–1849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu W., Rahman R., Rosbash M., Mechanistic implications of enhanced editing by a HyperTRIBE RNA-binding protein. RNA 24, 173–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuttan A., Bass B. L., Mechanistic insights into editing-site specificity of ADARs. Proc. Natl. Acad. Sci. U.S.A. 109, E3295–E3304 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marr M. T. II, D’Alessio J. A., Puig O., Tjian R., IRES-mediated functional coupling of transcription and translation amplifies insulin receptor feedback. Genes Dev. 21, 175–183 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang H., Hornstein E., Stolovich M., Levy G., Livingstone M., Templeton D., Avruch J., Meyuhas O., Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol. Cell. Biol. 21, 8671–8683 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyuhas O., Synthesis of the translational apparatus is regulated at the translational level. Eur. J. Biochem. 267, 6321–6330 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Meyuhas O., Dreazen A., Ribosomal protein S6 kinase from TOP mRNAs to cell size. Prog. Mol. Biol. Transl. Sci. 90, 109–153 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Hong S., Freeberg M. A., Han T., Kamath A., Yao Y., Fukuda T., Suzuki T., Kim J. K., Inoki K., LARP1 functions as a molecular switch for mTORC1-mediated translation of an essential class of mRNAs. eLife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lahr R. M., Fonseca B. D., Ciotti G. E., Al-Ashtal H. A., Jia J.-J., Niklaus M. R., Blagden S. P., Alain T., Berman A. J., La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs. eLife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Powell J. D., Pollizzi K. N., Heikamp E. B., Horton M. R., Regulation of immune responses by mTOR. Annu. Rev. Immunol. 30, 39–68 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weichhart T., Hengstschlager M., Linke M., Regulation of innate immune cell function by mTOR. Nat. Rev. Immunol. 15, 599–614 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt E. K., Clavarino G., Ceppi M., Pierre P., SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277 (2009). [DOI] [PubMed] [Google Scholar]
- 27.So L., Lee J., Palafox M., Mallya S., Woxland C. G., Arguello M., Truitt M. L., Sonenberg N., Ruggero D., Fruman D. A., The 4E-BP-eIF4E axis promotes rapamycin-sensitive growth and proliferation in lymphocytes. Sci. Signal. 9, ra57 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ingolia N. T., Ghaemmaghami S., Newman J. R., Weissman J. S., Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ingolia N. T., Lareau L. F., Weissman J. S., Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunn J. G., Foo C. K., Belletier N. G., Gavis E. R., Weissman J. S., Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife 2, e01179 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nandagopal N., Roux P. P., Regulation of global and specific mRNA translation by the mTOR signaling pathway. Translation 3, e983402 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsukiyama-Kohara K., Poulin F., Kohara M., DeMaria C. T., Cheng A., Wu Z., Gingras A.-C., Katsume A., Elchebly M., Spiegelman B. M., Harper M.-E., Tremblay M. L., Sonenberg N., Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat. Med. 7, 1128–1132 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Dowling R. J. O., Topisirovic I., Alain T., Bidinosti M., Fonseca B. D., Petroulakis E., Wang X., Larsson O., Selvaraj A., Liu Y., Kozma S. C., Thomas G., Sonenberg N., mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 328, 1172–1176 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Truitt M. L., Conn C. S., Shi Z., Pang X., Tokuyasu T., Coady A. M., Seo Y., Barna M., Ruggero D., Differential requirements for eIF4E dose in normal development and cancer. Cell 162, 59–71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo W., Guo F., Mahon A. M., Couvertier S., Jin H., Diaz M., Fieldsend A., Weerapana E., Rosbash M., NonA and CPX link the circadian clockwork to locomotor activity in Drosophila. Neuron 99, 768–780.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iadevaia V., Caldarola S., Tino E., Amaldi F., Loreni F., All translation elongation factors and the E, F, and H subunits of translation initiation factor 3 are encoded by 5′-terminal oligopyrimidine (TOP) mRNAs. RNA 14, 1730–1736 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee A. S., Kranzusch P. J., Doudna J. A., Cate J. H. D., eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 536, 96–99 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alain T., Sonenberg N., Topisirovic I., mTOR inhibitor efficacy is determined by the eIF4E/4E-BP ratio. Oncotarget 3, 1491–1492 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y., Wang Z.-H., Liu Y., Chen Y., Sun N., Gucek M., Zhang F., Xu H., PINK1 inhibits local protein synthesis to limit transmission of deleterious mitochondrial DNA mutations. Mol. Cell 73, 1127–1137.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen D. T. T., Lu Y., Chu K. L., Yang X., Park S. M., Choo Z. N., Chin C. R., Prieto C., Schurer A., Barin E., Savino A. M., Gourkanti S., Patel P., Vu L. P., Leslie C. S., Kharas M. G., HyperTRIBE uncovers increased MUSASHI-2 RNA binding activity and differential regulation in leukemic stem cells. Nat. Commun. 11, 2026 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.J. Biswas, R. Rahman, V. Gupta, M. Rosbash, R. H. Singer, MS2-TRIBE evaluates protein-RNA interactions and nuclear organization of transcription by RNA editing. bioRxiv 829606 [Preprint]. 4 November 2019. https://doi.org/10.1101/829606. [DOI] [PMC free article] [PubMed]
- 42.Herzog J. J., Xu W., Deshpande M., Rahman R., Suib H., Rodal A. A., Rosbash M., Paradis S., TDP-43 dysfunction restricts dendritic complexity by inhibiting CREB activation and altering gene expression. Proc. Natl. Acad. Sci. U.S.A. 11, 11760–11769 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L., Wang S., Li W., RSeQC: Quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Seim I., Jeffery P. L., Thomas P. B., Nelson C. C., Chopin L. K., Whole-genome sequence of the metastatic PC3 and LNCaP human prostate cancer cell lines. G3 7, 1731–1741 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore M. J., Zhang C., Gantman E. C., Mele A., Darnell J. C., Darnell R. B., Mapping argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat. Protoc. 9, 263–293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., Ren J., Li W. W., Noble W. S., MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P. D., PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/33/eabb8771/DC1


