Abstract
BACKGROUND:
Anthracycline-related cardiomyopathy is a leading cause of late morbidity in childhood cancer survivors. Glutathione S-transferases (GSTs) are a class of phase II detoxification enzymes that facilitate the elimination of anthracyclines. As free-radical scavengers, GSTs could play a role in oxidative damage-induced cardiomyopathy. Associations between the GSTμ1 (GSTM1) null genotype and iron-overload-related cardiomyopathy have been reported in patients with thalassemia.
METHODS:
The authors sought to identify an association between the GSTM1 null genotype and anthracycline-related cardiomyopathy in childhood cancer survivors and to corroborate the association by examining GSTM1 gene expression in peripheral blood and human-induced pluripotent stem cell cardiomyocytes (hiPSC-CMs) from survivors with and without cardiomyopathy. GSTM1 gene deletion was examined by polymerase chain reaction in 75 survivors who had clinically validated cardiomyopathy (cases) and in 92 matched survivors without cardiomyopathy (controls). Conditional logistic regression analysis adjusting for sex, age at cancer diagnosis, chest radiation, and anthracycline dose was used to assess the association between genotype and cardiomyopathy. Proprietary bead array technology and quantitative real-time polymerase chain reaction were used to measure GSTM1 expression levels in samples from 20 cases and 20 matched controls. hiPSC-CMs from childhood cancer survivors (3 with cardiomyopathy, 3 without cardiomyopathy) also were examined for GSTM1 gene expression levels.
RESULTS:
A significant association was observed between the risk of cardiomyopathy and the GSTM1 null genotype (odds ratio, 2.7; 95% CI, 1.3-5.9; P = .007). There was significant downregulation of GSTM1 expression in cases compared with controls (average relative expression, 0.67 ± 0.57 vs 1.33 ±1.33, respectively; P = .049). hiPSC-CMs from patients who had cardiomyopathy revealed reduced GSTM1 expression (P = .007).
CONCLUSIONS:
The current findings could facilitate the identification of childhood cancer survivors who are at risk for anthracycline-related cardiomyopathy.
Keywords: anthracyclines, cardiomyopathy, childhood cancer survivors, gene expression, glutathione S-transferase 1 (GSTM1) null genotype
INTRODUCTION
Over 85% of children who are diagnosed with cancer today are expected to survive for 5 or more years.1 Anthracycline chemotherapy (used to treat >50% of children with cancer) has contributed to the steady improvements in survival observed over the past several decades. However, cardiomyopathy progressing to congestive heart failure is an unfortunate complication of anthracyclines.2,3 Indeed, childhood cancer survivors are at a 5-fold to 15-fold higher risk of anthracycline-related cardiotoxicity compared with age-matched controls without anthracycline exposure.4–9 The outcome is poor, such that 5-year survival rates are <50% after a diagnosis of congestive heart failure.10,11 Cardiomyopathy risk increases with anthracycline dose.9,12,13 However, there is considerable interpatient variability in cardiomyopathy risk at any anthracyclines dose, suggesting that genetic factors possibly modify the association between anthracyclines and cardiomyopathy risk.5,9,14–32
Anthracycline metabolism likely plays a significant role in the individual susceptibility to anthracycline-related cardiomyopathy. Indeed, high-risk genetic variants in the CBR3 gene result in the generation of larger amounts of cardiotoxic alcohol metabolites of anthracyclines33 and have been implicated in increasing the risk of cardiomyopathy after moderate exposure to anthracyclines.9 Glutathione S-transferases (GSTs) are a class of phase II detoxification enzymes that catalyze the conjugation of reduced glutathione to toxic agents (including anthracyclines) and eliminate them from the body.34 GSTs also act as scavengers of free radicals and could play a role in oxidative damage-induced cardiomyopathy—an important pathogenetic mechanism in anthracycline-related cardiotoxicity. Patients with thalassemia major are at risk for cardiomyopathy because of iron overload, and significant associations between the GSTμ1 (GSTM1) null genotype and left ventricular dysfunction have been reported in this population.35,36 Homozygous deletions (null genotype) of the GSTM1 gene (chromosome 1p13.3) result in a complete absence of corresponding enzymatic activity.37–39 However, an evaluation of the role of the GSTM1 null genotype in the development of anthracycline-related cardiomyopathy has not been undertaken. We used peripheral blood from anthracycline-exposed childhood cancer survivors who had cardiomyopathy (cases) matched to those without cardiomyopathy (controls) to genotype the GSTM1 gene and examine GSTM1 gene expression levels in peripheral blood and in human-induced pluripotent stem cell cardiomyocytes (hiPSC-CMs).
MATERIALS AND METHODS
Study Design and Population
Study participants were drawn from a Children’s Oncology Group (COG) study (ALTE03N1; Key Adverse Events After Childhood Cancer; clinicaltrials.gov identifier NCT00082745), NCT00082745; principal investigator, Smita Bhatia). COG member institutions (see Supporting Table 1) enrolled patients after obtaining approval from local institutional review boards. Written informed consent/assent was obtained from patients, parents, or legal guardians. A matched case-control study design was used. Cases consisted of anthracycline-exposed childhood cancer survivors who developed cardiomyopathy. For each case, from 1 to 3 anthracycline-exposed childhood cancer survivors with no signs or symptoms of cardiomyopathy were randomly selected from childhood cancer survivors registered with the COG and matched on primary cancer diagnosis, year of diagnosis (±5 years), and race/ethnicity. The selected controls were required to have had a longer duration of cardiomyopathy-free follow-up compared with the time from cancer diagnosis to cardiomyopathy for the corresponding matched case. Participants provided peripheral blood samples for DNA and RNA.
Phenotype Assessment
Anthracycline-exposed participants had normal cardiac function before anthracycline exposure. Cases fulfilled American Heart Association criteria for cardiac compromise40 by presenting with symptoms (dyspnea, orthopnea, fatigue, etc) and/or signs (edema, hepatomegaly, rales, etc) of cardiac decompensation. None of the participants selected as controls had any symptoms or signs of cardiac compromise. Echocardiographic data were used as source validation for all cases and, when available, for controls.
Therapeutic Exposures
Lifetime anthracycline exposure was calculated by multiplying cumulative dose (mg/m2) of individual anthracyclines received by a factor that reflected the drug’s cardiotoxic potential41 and then summing the results. Chest radiation that included the heart was categorized as a yes/no variable.
DNA and RNA Isolation
Peripheral blood for DNA (n = 167) and RNA (n = 40) was collected directly into K2EDTA tubes and PAXgene Blood RNA tubes, respectively. DNA and RNA were isolated using the Gentra Puregene Blood Kit and the PAXgene Blood RNA kit (both from Qiagen Inc), respectively. The concentration was measured on Nanodrop ND-1000 Spectrophotometers (Thermo Fisher Scientific). DNA integrity was analyzed by using the Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific). RNA quality was checked using a Bioanalyzer Nanochip (Agilent Technologies), and samples with RNA integrity numbers >7 were used for microarray analysis.
GSTM1 Gene Deletion
GSTM1 deletion was examined by multiplex polymerase chain reaction (PCR), as previously reported, using the B2M gene as control (for details, see Supporting Methods and Supporting Fig. 1).42
Gene Expression Analysis
Microarray
Illumina HumanHT-12 v4.0 Expression BeadChips (Illumina, Inc) were used for mRNA transcription profiling (for details, see Supporting Methods).
Quantitative real-time PCR validation of GSTM1 gene expression
Expression of the GSTM1 gene also was measured using quantitative real-time PCR (for details, see Supporting Methods).
RNA sequencing of peripheral blood
Libraries were prepared using the TruSeq RNA Sample Preparation Kit (Illumina, Inc) according to the proto cols recommended by the manufacturer. Each library was paired-end sequenced (2 × 50 bp) by using the TruSeq SBS Kit v4-HS, on a HiSeq2000 platform.
RNA sequencing in cardiomyocytes
Day-30 hiPSC-CMs from 6 anthracycline-exposed childhood cancer survivors (3 with anthracycline-related cardiotoxicity and 3 without cardiotoxicity) were used.43 RNA was extracted using TRI Reagent and the Direct-zol RNA microprep kit (both from Zymo), including on-column DNase digestion to remove genomic DNA. Samples were quantified using an Agilent 2100 Bioanalyzer and passed quality control. Gene expression levels and exon use were estimated using the featureCounts function in the Subread software44. Differential gene expression analysis was performed using the DEseq2 package45 and R (v3.3.3; R Foundation for Statistical Computing). Details are provided in the Supporting Methods.
Statistical Analysis
Analyses were conducted using GenomeStudio (Illumina, Inc.), R (http://www.r-project.org/, accessed July 2, 2019), SAS (SAS Institute Inc), and PLINK (http://zzz.bwh.harvard.edu/plink/, accessed July 2, 2019).. We summarized the characteristics of the survivor population according to case-control status. Continuous and categorical variables were examined using independent-sample t tests or Wilcoxon rank-sum tests and Fisher exact or chi-square tests, respectively.
Statistical analysis of genotype data
Multivariable conditional logistic regression analysis (adjusting for anthracycline dose, chest radiation, age at diagnosis of primary cancer, and sex) was used to determine the association between GSTM1 variants and cardiomyopathy. Odds ratios were calculated if the anthracycline dose was either dichotomized (<250 mg/m2 and ≥250 mg/m2) or treated as a continuous variable. Analyses also were conducted for stratified cumulative anthracycline exposure (<250 mg/m2 or ≥250 mg/m2).
Statistical analysis of gene expression data
To determine differential expression of the GSTM1 gene, we used paired t tests on log2-transformed expression values.
RESULTS
Genotyping of Matched Case-Control Sets for GSTM1 Null Variant and Association With Cardiomyopathy Risk
DNA was available for 167 anthracycline-exposed childhood cancer survivors (75 cases and 92 matched controls). Demographic and clinical characteristics of the study participants are summarized in Table 1. The median age at primary cancer diagnosis for cases and controls was 7.8 and 9.6 years, respectively. The cumulative anthracycline dose was significantly higher among the cases (median, 300 vs 255 mg/m2; P = .02). Although a higher proportion of cases received chest radiation compared with controls (34.7% vs 26.1%), the difference did not reach statistical significance (P = .4). Controls were followed for a significantly longer time (median, 12 years) compared with cases (6 years; P < .0001). All cases were symptomatic; ie, they fulfilled American Heart Association criteria for cardiac compromise. The mean ejection fraction was 39.4%, and the mean fractional shortening was 22.2%. None of the controls had any clinical signs or symptoms of cardiac compromise. Echocardiographic indices available for 61% of the controls showed a mean ejection fraction of 65.9% and a mean fractional shortening of 36.7%. The echocardiographic validation of cases and controls are detailed in Supporting Table 2.
TABLE 1.
Demographic and Clinical Characteristics by Case-Control Status for Glutathione S-Transferase μ1 Genotyping
| Variable | Cases, N = 75 | Controls, N = 92 | Pa |
|---|---|---|---|
| Age at primary cancer diagnosis, y | |||
| Mean ± SD | 8.3 ±5.47 | 9.4 ±6.35 | .2 |
| Median [range] | 7.8 [3.8-11.5] | 9.6 [3.3-14.8] | |
| Sex | |||
| Female | 43 (57.3) | 40 (43.5) | .07 |
| Cumulative anthracycline exposure, mg/m2 | |||
| Mean ± SDb | 306.9 ± 111.6 | 264.4 ± 125.1 | .02c |
| Median [range] | 300 [230.3-375] | 255 [150-368] | |
| Race: No. (%) | |||
| Asian | 4 (5.3) | 6 (6.5) | .9 |
| Black | 2 (2.7) | 4 (4.4) | |
| Hispanic | 4 (5.3) | 6 (6.5) | |
| White | 65 (86.7) | 76 (82.6) | |
| Chest radiation | |||
| Yes: No. (%) | 26 (34.7) | 24 (26.1) | .4 |
| Dose: Mean ± SD, cGy | 1044.1 ± 1640.5 | 811.9 ± 1792.7 | |
| Primary diagnosis: | |||
| No. (%) | |||
| ALL | 15 (20.0) | 18 (19.7) | |
| AML | 5 (6.7) | 6 (6.5) | |
| Bone | 20 (26.7) | 20 (21.7) | .9 |
| HD | 8 (10.7) | 10 (10.9) | |
| Kidney | 4 (5.3) | 4 (4.3) | |
| Neuroblastoma | 6 (8.0) | 9 (9.8) | |
| NHL | 8 (1 0.7) | 14 (15.2) | |
| Sarcoma | 9 (12.0) | 11 (11.9) | |
| Time from diagnosis to cardiac event (cases) or time to enrollment (controls), y | |||
| Mean ± SD | 7.42 ± 6.5 | 12.96 ± 7.1 | <.0001c |
| Median [range] | 6.0 [1.3-11.6] | 12 [7.4-17.2] | |
| Echocardiographic characteristics, %d | |||
| Ejection fraction | 39.4 | 65.9 | |
| Fractional shortening | 22.2 | 36.7 |
Abbreviations: ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; cGy, centigrays; HD, Hodgkin disease; NHL, non-Hodgkin lymphoma.
P values were estimated using either chi-square tests and t tests for categorical variables or the Wilcoxon test for continuous variables.
Doses were not available for 1 case and 1 control.
This P value indicates a significant difference.
Echocardiographic indices were available for 100% of the cases and 61% of the controls; for the remaining controls, physician notes endorsed the absence of clinical signs/symptoms of cardiac compromise according to American Heart Association criteria.
There was overrepresentation of the GSTM1 null genotype among cases versus controls (60.0% vs 38.04%; P = .005) (see Supporting Table 3). Results of the multi variable conditional logistic regression analysis in Table 2 summarizes the association between the GSTM1 null genotype and the risk of cardiomyopathy. After adjusting for cumulative anthracycline exposure dichotomized as low-dose (<250 mg/m2) and high-dose (≥250 mg/m2), chest radiation (yes or no), age at diagnosis of primary cancer, and sex, the odds that patients who had the GSTM1 null genotype would develop cardiomyopathy were 2.7 times greater (95% CI, 1.3-5.9; P = .007) compared with patients who had the GSTM1 positive genotype. We also examined the association after including anthracycline dose as a continuous variable and found that patients with the GSTM1 null genotype had a 2.52-fold higher odds of developing cardiomyopathy (95% CI, 1.2-5.2; P = .01) (see Supporting Table 4) after adjusting for the variables listed above. Finally, conducting separate analyses for those exposed to high-dose anthracyclines and low-dose anthracyclines, the magnitude of GSTM1 cardiomyopathy was generally similar (see Supporting Table 5).
TABLE 2.
Risk Factors Associated With Anthracycline-Related Cardiomyopathy
| Risk Factor | OR (95% CI) | Pa |
|---|---|---|
| Age at primary cancer diagnosis, y | ||
| Per y increase in age | 0.95 (0.88-1.01) | .1 |
| Anthracycline dose, mg/m2b | ||
| <250 | 1.00 | |
| ≥250 | 2.5 (0.99-6.4) | .05 |
| Chest radiation | ||
| No | 1.00 | |
| Yes | 1.5 (0.6-3.6) | .4 |
| Sex | ||
| Male | 1.00 | |
| Female | 1.5 (0.7-2.9) | .3 |
| Genotype | ||
| GSTM1 positive | 1.00 | |
| GSTM1 null | 2.7 (1.3-5.9) | .007c |
Abbreviations: GSTM1, glutathione S-transferase μ1; OR, odds ratio.
P values were estimated from a conditional logistic regression model that included age at cancer diagnosis, sex, chest radiation and anthracycline exposure (categorical).
Doses were not available for 1 case and 1 control.
This P value indicates a significant difference.
Constitutive Gene Expression Analysis of Matched Case-Control Sets for GSTM1 and Association With Cardiomyopathy Risk
Gene expression analysis using peripheral blood
This case-control set included 40 non-Hispanic white anthracycline-exposed childhood cancer survivors (20 cases and 20 matched controls) with high-quality RNA samples. Of these, 14 matched case-control pairs had also participated in the genotyping analysis. As shown in Table 3, the median age at primary cancer diagnosis for the cases and controls was 5.5 and 6.4 years, respectively (P = .8). Cases received a higher cumulative anthracycline exposure (median dose, 295 vs 217.5 mg/m2; P = .002). The median time between cancer diagnosis and cardiomyopathy was 11.4 years; controls were followed for a significantly longer period (median, 15.9 years; P = .02).
TABLE 3.
Demographic and Clinical Characteristics by Case-Control Status for Gene Expression Analysis
| Variable | Cases, N = 20 | Controls, N = 20 | Pa |
|---|---|---|---|
| Age at primary cancer diagnosis, y | |||
| Mean ± SD | 6.6 ± 5.4 | 7.5 ±5.8 | .6 |
| Median [range] | 5.5 [1.9-9.2] | 6.4 [1.4-13.2] | .8 |
| Sex: No. (%) | |||
| Female | 10 (50.0) | 8 (40.0) | .5 |
| Cumulative anthracycline exposure, mg/m2 | |||
| Mean ± SD | 304.6 ± 116.9 | 198.1 ±81.9 | .002c |
| Median [range] | 295 [240.7-350.0] | 217.5 [121.3-266.3] | .002c |
| Chest radiation | |||
| Yes: No. (%) | 7 (35.0) | 7 (35.0) | 1.0 |
| Dose: Mean ± SD, cGy | 797.5 ± 1280.7 | 725.5 ± 1218.3 | 0.9 |
| Primary diagnosis: | |||
| No. (%) | |||
| ALL | 5 (25.0) | 5 (25.0) | 1.0b |
| HL or NHL | 9 (45.0) | 9 (45.0) | |
| Other | 6 (30.0) | 6 (30.0) | |
| Time from cancer diagnosis, yd | |||
| Mean ± SD | 12.5 ± 5.9 | 16.8 ± 5.7 | .02c |
| Median [range] | 11.4 [8.8-18.0] | 15.9 [12.2-21.2] | .02c |
Abbreviations: ALL, acute lymphoblastic leukemia; cGy, centigrays; HL, Hodgkin lymphoma; NHL, non-Hodgkin lymphoma.
P values were estimated using either chi-square tests and t tests for categorical variables or the Wilcoxon test for continuous variables.
This P value was determined with the Fisher exact test.
This P value indicates a significant difference.
The time in years from cancer diagnosis to cardiac event is indicated for cases, and the time in years to enrollment is indicated for controls.
The analysis of all probes from the Illumina HT-12v4 expression array for the GSTM1 gene revealed 1 probe (ILMN_2391861) with signal intensities above those of the housekeeping genes. Figure 1 compares the average intensities for GSTM1 on the array in the 20 matched case-control pairs. GSTM1 showed significant downregulation in 12 of 20 cases compared with controls. Array-based differential expression for GSTM1 was confirmed by real-time quantitative PCR (see Supporting Fig. 2), with significant downregulation in cases versus controls (0.67 ± 0.57 vs 1.33 ± 1.33; P = .049). We also performed RNA sequencing (RNA-Seq) on 7 of the 20 paired samples. RNA-Seq reads ranged from 0 to 4 for null genotypes and from 62 to 180 for positive genotypes; genotypes were derived using the RNA-Seq reads with high confidence.
FIGURE 1.
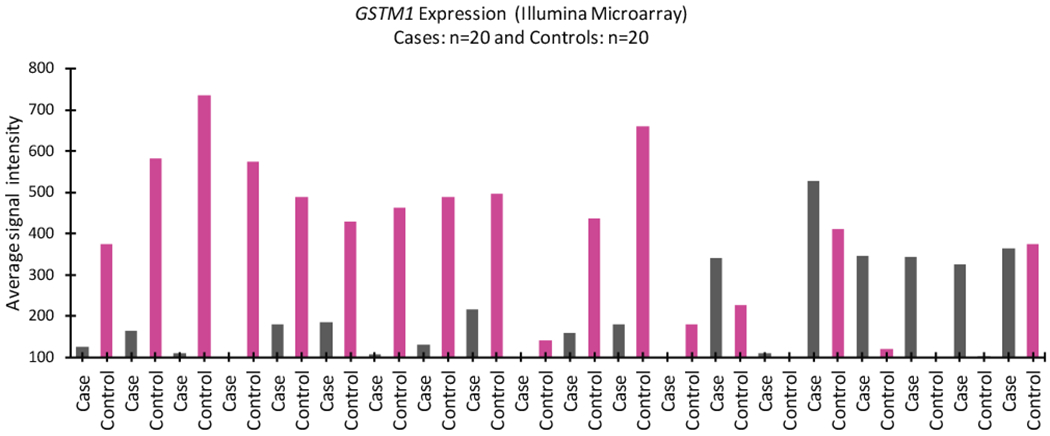
The expression profile of the glutathione S-transferase μ1 (GSTM1) gene is shown on the Illumina Microarray (probe ILMN_2391861) in anthracycline-related cardiomyopathy. Matched cases are indicated in gray, and controls are indicated in pink.
Gene expression analysis using cardiomyocytes
hiPSC-CMs from patients who had anthracycline-related cardiotoxicity showed significantly reduced expression of GSTM1 (P = .007) (Fig. 2).
FIGURE 2.
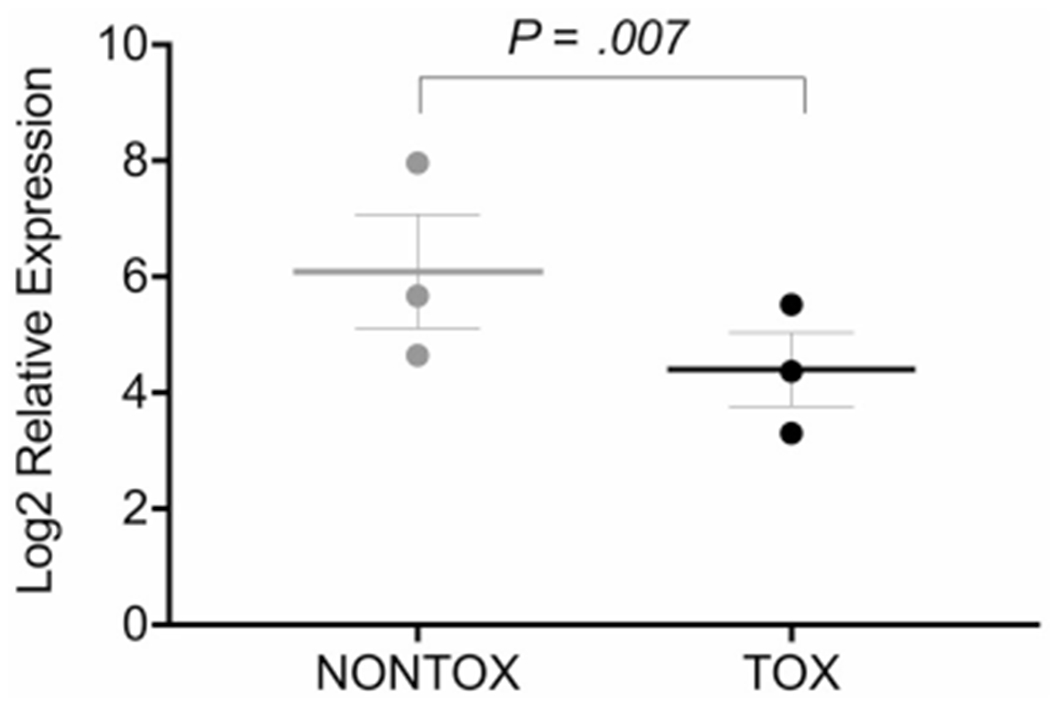
Glutathione S-transferase μ1 (GSTM1) expression was determined by RNA sequencing in human-induced pluripotent stem cell cardiomyocytes derived from 6 childhood cancer survivors, including 3 anthracycline-exposed patients with no cardiomyopathy (NONTOX) and 3 anthracycline-exposed patients with cardiomyopathy (TOX).
DISCUSSION
In this study, we observed that the GSTM1 null variant was associated with anthracycline-related cardiomyopathy in anthracycline-exposed childhood cancer survivors. We extended these findings by demonstrating downregulated GSTM1 gene expression in the peripheral blood as well as reduced GSTM1 expression in hiPSC-CMs generated from childhood cancer survivors who had anthracycline-related cardiomyopathy. Taken together, these findings provide a biologically plausible association between the GSTM1 null variant and anthracycline-related cardiomyopathy.
GSTs are a class of phase II detoxification enzymes that catalyze the conjugation of reduced glutathione to toxic agents, including anthracyclines by making them water soluble and eliminating them from the body.34 GSTs also act as scavengers of free radicals and could play a role in oxidative damage-induced cardiomyopathy. Homozygous deletions and the null genotype of the GSTM1 gene (chromosome 1p13.3) result in the complete absence of corresponding enzymatic activity.37–39 There is significant variability in the GSTM1 null genotype by race/ethnicity and geography; eg, prevalence of the GSTM1 null genotype is approximately 5% in Europeans, 70% to 79% in East Asians, 57% in South Asians, 49% in Africans, and 69% in admixed American populations46–48.
Previous investigations that sought to examine the as sociation with the GSTM1 null genotype were unsuccessful, in part because of the modest sample size of cases with cardiomyopathy27,30–32 or because of challenges associated with analyzing copy number variation using genome-wide arrays.49 However, significant associations between the GSTM1 null genotype and cardiomyopathy have been reported in patients with thalassemia.35,36 Although some studies have examined the functional relevance of genetic variants,9,15,22,26,28,29 the functional relevance often remains unknown, in part because the identified single-nucleotide variations (formerly single-nucleotide polymorphisms) are located in intronic regions where they may play a role in regulating distant loci. Differential constitutive gene expression possibly could help discern the pathogenesis of anthracycline-related cardiomyopathy. Differential gene expression ideally should be measured in the affected tissue. However, procurement of cardiac tissue in anthracycline-exposed individuals is logistically difficult and not without risk, hence the dependence on surrogate tissues, such as peripheral blood. In addition, the establishment of methodologies to convert human-induced pluripotent stem cells (hiPSCs) to cardiomyocytes (hiP-SC-CMs) has enabled human cardiomyocytes to be mass produced in vitro for cardiovascular disease modeling50.51.
This study needs to be considered within the context of its limitations. Members of the GSTμ class of enzymes share 75% to 99% sequence identity, with maximum homology between GSTM1 and GSTM2. Expression of GSTM1 (and GSTM2) was confirmed separately by real-time PCR, and null alleles for only GSTM1 were confirmed by genotyping; a null allele for GSTM2 is not reported. Although a successful replication of this association would have been ideal, the strong biologic plausibility and the demonstration of reduced expression in peripheral blood and in patient-derived cardiomyocytes strengthens the biologic basis of this association.
In summary, to our knowledge, we are the first to report an association between the GSTM1 null genotype and anthracycline-related cardiomyopathy. This finding, along with the previous reports of other genetic variants,6 could be considered in a risk-prediction model to facilitate the identification of childhood cancer survivors who are at increased risk of anthracycline-related cardiomyopathy, such that targeted interventions could be instituted.
Supplementary Material
Acknowledgments
FUNDING SUPPORT
This work was supported in part by an American Cancer Society Institutional Research Grant (IRG-60-001-53-IRG) and the Kaul Pediatric Research Institute Research Grant (to Purnima Singh), Children’s Oncology Group (COG) Chair’s grant U10 CA98543 (Peter Adamson), National Clinical Trials Network (NCTN) Operations Center grant U10CA180886 (Peter Adamson, principal investigator), COG Statistics and Data Center grant U10CA098413 (Peter Adamson, principal investigator), St Baldrick’s Foundation through an unrestricted grant to the COG (National Institutes of Health grants GM 115279 and CA 21765; Peter Adamson, principal investigator), The Leukemia and Lymphoma Society Translational Research Program (6093-08; Smita Bhatia, principal investigator), the NCTN Statistics and Data Center (grant U10CA180899), and the National Cancer Institute Community Oncology Research Program Research Base (UG1CA189955; Peter Adamson, principal investigator).
Footnotes
Additional supporting information may be found in the online version of this article.
CONFLICT OF INTEREST DISCLOSURES
Charles A. Sklar reports personal fees and honoraria from Novo Nordisk outside the submitted work. Paul Burridge reports research funding from Abbott Diagnostics outside the submitted work. Mary V. Relling reports investigator-initiated research funding from Servier outside the submitted work. The remaining authors made no disclosures.
REFERENCES
- 1.Armstrong GT, Chen Y, Yasui Y, et al. Reduction in late mortality among 5-year survivors of childhood cancer. N Engl J Med. 2016;374:833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mulrooney DA, Yeazel MW, Kawashima T, et al. Cardiac outcomes in a cohort of adult survivors of childhood and adolescent cancer: retrospective analysis of the Childhood Cancer Survivor Study cohort. BMJ. 2009;339:b4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oeffinger KC, Mertens AC, Sklar CA, et al. Chronic health conditions in adult survivors of childhood cancer. N Engl J Med. 2006; 355:1572–1582. [DOI] [PubMed] [Google Scholar]
- 4.Bhatia S Role of genetic susceptibility in development of treatment-related adverse outcomes in cancer survivors. Cancer Epidemiol Biomarkers Prev. 2011;20:2048–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grenier MA, Lipshultz SE. Epidemiology of anthracycline cardiotoxicity in children and adults. Semin Oncol. 1998;25:72–85. [PubMed] [Google Scholar]
- 6.Leong SL, Chaiyakunapruk N, Lee SW. Candidate gene association studies of anthracycline-induced cardiotoxicity: a systematic review and meta-analysis. Sci Rep. 2017;7:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneider BP, Shen F, Gardner L, et al. Genome-wide association study for anthracycline-induced congestive heart failure. Clin Cancer Res. 2017;23:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swain SM, Whaley FS, Ewer MS. Congestive heart failure in patients treated with doxorubicin: a retrospective analysis of three trials. Cancer. 2003;97:2869–2879. [DOI] [PubMed] [Google Scholar]
- 9.Blanco JG, Sun CL, Landier W, et al. Anthracycline-related cardiomyopathy after childhood cancer: role of polymorphisms in carbonyl reductase genes—a report from the Children’s Oncology Group. J Clin Oncol. 2012;30:1415–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felker GM, Thompson RE, Hare JM, et al. Underlying causes and long-term survival in patients with initially unexplained cardiomyopathy. N Engl J Med. 2000;342:1077–1084. [DOI] [PubMed] [Google Scholar]
- 11.van der Pal HJ, van Dalen EC, Kremer LC, Bakker PJ, van Leeuwen FE. Risk of morbidity and mortality from cardiovascular disease following radiotherapy for childhood cancer: a systematic review. Cancer Treat Rev. 2005;31:173–185. [DOI] [PubMed] [Google Scholar]
- 12.Volkova M, Russell R 3rd. Anthracycline cardiotoxicity: prevalence, pathogenesis and treatment. Curr Cardiol Rev. 2011;7:214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giantris A, Abdurrahman L, Hinkle A, Asselin B, Lipshultz SE. Anthracycline-induced cardiotoxicity in children and young adults. Crit Rev Oncol Hematol. 1998;27:53–68. [DOI] [PubMed] [Google Scholar]
- 14.Armenian SH, Ding Y, Mills G, et al. Genetic susceptibility to anthracycline-related congestive heart failure in survivors of haematopoietic cell transplantation. Br J Haematol. 2013;163:205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanco JG, Leisenring WM, Gonzalez-Covarrubias VM, et al. Genetic polymorphisms in the carbonyl reductase 3 gene CBR3 and the NAD(P)H:quinone oxidoreductase 1 gene NQO1 in patients who developed anthracycline-related congestive heart failure after childhood cancer. Cancer. 2008;112:2789–2795. [DOI] [PubMed] [Google Scholar]
- 16.Hertz DL, Caram MV, Kidwell KM, et al. Evidence for association of SNPs in ABCB1 and CBR3, but not RAC2, NCF4, SLC28A3 or TOP2B, with chronic cardiotoxicity in a cohort of breast cancer patients treated with anthracyclines. Pharmacogenomics. 2016; 17:231–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krajinovic M, Elbared J, Drouin S, et al. Polymorphisms of ABCC5 and NOS3 genes influence doxorubicin cardiotoxicity in survivors of childhood acute lymphoblastic leukemia. Pharmacogenomics J. 2016;16:530–535. [DOI] [PubMed] [Google Scholar]
- 18.Lipshultz SE, Lipsitz SR, Kutok JL, et al. Impact of hemochromatosis gene mutations on cardiac status in doxorubicin-treated survivors of childhood high-risk leukemia. Cancer. 2013;119:3555–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lubieniecka JM, Graham J, Heffner D, et al. A discovery study of daunorubicin induced cardiotoxicity in a sample of acute myeloid leukemia patients prioritizes P450 oxidoreductase polymorphisms as a potential risk factor. Front Genet. 2013;4:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reichwagen A, Ziepert M, Kreuz M, et al. Association of NADPH oxidase polymorphisms with anthracycline-induced cardiotoxicity in the RICOVER-60 trial of patients with aggressive CD20(+) B-cell lymphoma. Pharmacogenomics. 2015;16:361–372. [DOI] [PubMed] [Google Scholar]
- 21.Semsei AF, Erdelyi DJ, Ungvari I, et al. ABCC1 polymorphisms in anthracycline-induced cardiotoxicity in childhood acute lymphoblastic leukaemia. Cell Biol Int. 2012;36:79–86. [DOI] [PubMed] [Google Scholar]
- 22.Visscher H, Rassekh SR, Sandor GS, et al. Genetic variants in SLC22A17 and SLC22A7 are associated with anthracycline-induced cardiotoxicity in children. Pharmacogenomics. 2015;16:1065–1076. [DOI] [PubMed] [Google Scholar]
- 23.Visscher H, Ross CJ, Rassekh SR, et al. Validation of variants in SLC28A3 and UGT1A6 as genetic markers predictive of anthracycline-induced cardiotoxicity in children. Pediatr Blood Cancer. 2013;60:1375–1381. [DOI] [PubMed] [Google Scholar]
- 24.Wasielewski M, van Spaendonck-Zwarts KY, Westerink ND, et al. Potential genetic predisposition for anthracycline-associated cardiomyopathy in families with dilated cardiomyopathy. Open Heart. 2014;1:e000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wojnowski L, Kulle B, Schirmer M, et al. NAD(P)H oxidase and multidrug resistance protein genetic polymorphisms are associated with doxorubicin-induced cardiotoxicity. Circulation. 2005;112:3754–3762. [DOI] [PubMed] [Google Scholar]
- 26.Aminkeng F, Bhavsar AP, Visscher H, et al. A coding variant in RARG confers susceptibility to anthracycline-induced cardiotoxicity in childhood cancer. Nat Genet. 2015;47:1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vivenza D, Feola M, Garrone O, Monteverde M, Merlano M, Lo Nigro C. Role of the renin-angiotensin-aldosterone system and the glutathione S-transferase mu, pi and theta gene polymorphisms in cardiotoxicity after anthracycline chemotherapy for breast carcinoma. Int J Biol Markers. 2013;28:e336–e347. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Liu W, Sun CL, et al. Hyaluronan synthase 3 variant and anthracycline-related cardiomyopathy: a report from the Children’s Oncology Group. J Clin Oncol. 2014;32:647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Sun CL, Quinones-Lombrana A, et al. CELF4 variant and anthracycline-related cardiomyopathy: a Children’s Oncology Group genome-wide association study. J Clin Oncol. 2016;34:863–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiss JR, Kopecky KJ, Godwin J, et al. Glutathione S-transferase (GSTM1, GSTT1 and GSTA1) polymorphisms and outcomes after treatment for acute myeloid leukemia: pharmacogenetics in Southwest Oncology Group (SWOG) clinical trials. Leukemia. 2006;20:2169–2171. [DOI] [PubMed] [Google Scholar]
- 31.Rajic V, Aplenc R, Debeljak M, et al. Influence of the polymorphism in candidate genes on late cardiac damage in patients treated due to acute leukemia in childhood. Leuk Lymphoma. 2009;50:1693–1698. [DOI] [PubMed] [Google Scholar]
- 32.Rossi D, Rasi S, Franceschetti S, et al. Analysis of the host pharmacogenetic background for prediction of outcome and toxicity in diffuse large B-cell lymphoma treated with R-CHOP21. Leukemia. 2009;23:1118–1126. [DOI] [PubMed] [Google Scholar]
- 33.Lakhman SS, Ghosh D, Blanco JG. Functional significance of a natural allelic variant of human carbonyl reductase 3 (CBR3). Drug Metab Dispos. 2005;33:254–257. [DOI] [PubMed] [Google Scholar]
- 34.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. [DOI] [PubMed] [Google Scholar]
- 35.Mokhtar GM, Sherif EM, Habeeb NM, et al. Glutathione S-transferase gene polymorphism: relation to cardiac iron overload in Egyptian patients with beta thalassemia major. Hematology. 2016;21:46–53. [DOI] [PubMed] [Google Scholar]
- 36.Singh MM, Kumar R, Tewari S, Agarwal S. Association of GSTT1/ GSTM1 and ApoE variants with left ventricular diastolic dysfunction in thalassaemia major patients. Hematology. 2019;24:20–25. [DOI] [PubMed] [Google Scholar]
- 37.Xu S, Wang Y, Roe B, Pearson WR. Characterization of the human class mu glutathione S-transferase gene cluster and the GSTM1 deletion. J Biol Chem. 1998;273:3517–3527. [DOI] [PubMed] [Google Scholar]
- 38.Pemble S, Schroeder KR, Spencer SR, et al. Human glutathione S-transferase theta (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem J. 1994;300(pt 1):271–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Board P, Coggan M, Johnston P, Ross V, Suzuki T, Webb G. Genetic heterogeneity of the human glutathione transferases: a complex of gene families. Pharmacol Ther. 1990;48:357–369. [DOI] [PubMed] [Google Scholar]
- 40.Hunt SA, Abraham WT, Chin MH, et al. 2009 Focused update incorporated into the ACC/AHA 2005 Guidelines for the Diagnosis and Management of Heart Failure in Adults: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines: developed in collaboration with the International Society for Heart and Lung Transplantation. Circulation. 2009;119:e391–e479. [DOI] [PubMed] [Google Scholar]
- 41.Feijen EAM, Leisenring WM, Stratton KL, et al. Derivation of anthracycline and anthraquinone equivalence ratios to doxorubicin for late-onset cardiotoxicity. JAMA Oncol. 2019;5:864–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Covault J, Abreu C, Kranzler H, Oncken C. Quantitative real-time PCR for gene dosage determinations in microdeletion genotypes. Biotechniques. 2003;35:594–596, 598. [DOI] [PubMed] [Google Scholar]
- 43.Burridge PW, Li YF, Matsa E, et al. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat Med. 2016;22:547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;4l:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garte S, Gaspari L, Alexandrie AK, et al. Metabolic gene polymorphism frequencies in control populations. Cancer Epidemiol Biomarkers Prev. 2001;10:1239–1248. [PubMed] [Google Scholar]
- 47.McCarroll SA, Hadnott TN, Perry GH, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. [DOI] [PubMed] [Google Scholar]
- 48.Santos M, Niemi M, Hiratsuka M, et al. Novel copy-number variations in pharmacogenes contribute to interindividual differences in drug pharmacokinetics. Genet Med. 2018;20:622–629. [DOI] [PubMed] [Google Scholar]
- 49.Zollner S, Teslovich TM. Using GWAS data to identify copy number variants contributing to common complex diseases. Stat Sci. 2009;24:530–546. [Google Scholar]
- 50.Magdy T, Burmeister BT, Burridge PW. Validating the pharmacogenomics of chemotherapy-induced cardiotoxicity: what is missing? Pharmacol Ther. 2016;168:113–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burridge PW, Diecke S, Matsa E, Sharma A, Wu H, Wu JC. Modeling cardiovascular diseases with patient-specific human pluripotent stem cell-derived cardiomyocytes. Methods Mol Biol. 2016;1353:119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


