Abstract
Dead leaves of Musa sp. (banana) were collected in northern Thailand during an investigation of saprobic fungi. Preliminary morphological observations revealed that three specimens belong to Dictyoarthrinium. Phylogenetic analyses of combined SSU, LSU, ITS and tef1-α sequence data revealed that Dictyoarthrinium forms a clade in Didymosphaeriaceae (Massarineae, Pleosporales, Dothideomycetes) sister to Spegazzinia. Based on contrasting morphological features with the extant taxa of Dictyoarthrinium, coupled with the multigene analyses, Dictyoarthrinium musae sp. nov. is introduced herein. Our study provides the first detailed molecular investigation for Dictyoarthrinium and supports its placement in Didymosphaeriaceae (Massarineae, Pleosporales, Dothideomycetes). Previously, Dictyoarthrinium was classified in Apiosporaceae (Xylariales, Sordariomycetes).
Keywords: Banana, Dictyoarthrinium sacchari, DNA sequences, Musaceae , one new species, saprobes, taxonomy
Introduction
Hughes (1953) documented seven hyphomycete genera (Arthrinium, Catenospegazzinia, Cordella, Dictyoarthrinium, Endocalyx, Pteroconium and Spegazzinia) that had unique basauxic conidiogenous cell development. Hyde et al. (1998) accommodated Dictyoarthrinium, Endocalyx, Scyphospora (= Arthrinium) and Spegazzinia in Apiosporaceae (Xylariales, Sordariomycetes), based on morphological characteristics. Based on molecular phylogenetic data (LSU and ITS), Cordella and Pteroconium were synonymised under Arthrinium by Crous and Groenewald (2013) and Arthrinium was confirmed as the asexual morph of Apiospora. With the availability of molecular data (SSU, LSU, ITS and tef1-α), Tanaka et al. (2015) transferred Spegazzinia to Didymosphaeriaceae. Wijayawardene et al. (2018) and Hyde et al. (2020) accommodated Arthrinium, Dictyoarthrinium and Endocalyx, all with basauxic conidiogenous cell development, in Apiosporaceae.
Dictyoarthrinium was introduced by Hughes (1952) with D. quadratum as the type species. Dictyoarthrinium africanum was simultaneously introduced. Damon (1953) re-examined the type material, descriptions and illustrations of Tetracoccosporium sacchari (Johnston and Stevenson 1917) and mentioned that T. sacchari was congeneric with Dictyoarthrinium quadratum. Therefore, Damon (1953) combined T. sacchari as Dictyoarthrinium sacchari. Damon (1953) also named D. quadratum as the heterotypic synonym of D. sacchari. Rao and Rao (1964) introduced D. lilliputeum and D. microsporum, while Kobayasi et al. (1971) introduced D. rabaulense as novel taxa to the genus. Somrithipol (2007) introduced D. synnematicum and currently seven epithets of Dictyoarthrinium are listed in Index Fungorum (2020). All Dictyoarthrinium species were introduced, based only on morphological data. Vu et al. (2019) sequenced D. sacchari (CBS 529.73) and submitted LSU data to GenBank as the only valid molecular record for the genus.
Dictyoarthrinium is characterised by basauxic conidiogenous cell development (Hughes 1952; Damon 1953; Matsushima 1971). Basauxic development is demonstrated by conidiogenous cells in which elongation occurs at a basal growing point after formation of a single, terminal blastic conidium at its apex (Cole 1976). Conidiophores of Dictyoarthrinium are minutely verruculose, subhyaline and transversely septate (Ellis 1971). Usually, the septa are dark brown and appear as thick stripes on the conidiophore. Conidiophore mother cells are often hyaline or pale brown and cup-shaped (Hughes 1952) or subspherical (Ellis 1971). The length of conidiophores varies within the genus, but in some species, the dimensions are more or less similar. Conidia of Dictyoarthrinium arise from the conidiophore at terminal or lateral parts. Conidiogenesis is monoblastic or polyblastic and integrated (Ellis 1971). Conidia are simple, solitary, dematiaceous and often four-celled. Some taxa (e.g. D. africanum) have 16-celled conidia (Hughes 1952). The surface of conidia is verruculose and most species have warts on the surface. However, the conidia of D. rabaulense are densely echinulate with long spines (Kobayasi 1971). The conidia vary in shape from square to spherical, subspherical or oblong. Most conidia appear flattened on one side. As a specific feature, only D. synnematicum possesses synnemata with filaments (Somrithipol 2007). Stroma, setae and hyphopodia have not been observed in Dictyoarthrinium.
Many Dictyoarthrinium species are saprobes that colonise dead plant materials, although D. rabaulense was recorded even from soil and air (Kobayasi et al. 1971; Ellis 1976). Most Dictyoarthrinium species occur on monocotyledonous plants. The genus is widely distributed across the tropics, mainly in terrestrial environments (Ellis 1971; 1976). The sexual morph of Dictyoarthrinium is unknown. Hosts, substrates and geographical distributions of extant Dictyoarthrinium species are listed in Table 1.
Table 1.
Hosts, substrates and geographical distribution of Dictyoarthrinium species.
| Species | Hosts/substrates | Geographical distribution | References |
|---|---|---|---|
| Dictyoarthrinium africanum S. Hughes | Miscanthus , Panicum, Paspalum virgatum, Saccharum, leaf litter of Typha latifolia | Argentina, Ghana, Solomon Islands, Venezuela | Hughes (1952); Ellis (1971); McKenzie and Jackson (1986); Urtiaga (1986); Tarda et al. (2019) |
| D. lilliputeum P. Rag. Rao and D. Rao | Leaf litter of Bambusa | India | Rao and Rao (1964); Sushma et al. (2020) |
| D. microsporum P. Rag. Rao and D. Rao | Dead leaves of Borassus flabellifer | India | Rao and Rao (1964) |
| D. rabaulense Matsush. | Brassica campestris, Dendrocalamus strictus, Gossypium, Xylia xylocarpa, air and soil | Bismarck Archipelago, Britain, Congo, India, New Caledonia, Nigeria, Tanzania. | Kobayasi et al. (1971); Ellis (1976); Bhat (2010) |
| D. sacchari (J.A. Stev.) Damon = D. quadratum S. Hughes | Dead stems and leaves of Ananas, Bambusa, Borassus, Cassia, Cosmos bipinnatus, Cymbopogon, Delonix elata, Dracaena, Erythrina, Lithachne pauciflora, Musa acuminata, M. paradisiaca, Neolitsea scrobiculata, Pandanus, Persea mechrantha, Phragmites, Prunus amygdalus, Saccharum sp., S. officinarum, S. spontanium, Zinnia, leaf litter of Typha latifolia, decaying plant materials of dicots | Brazil, Cuba, Federated Ghana, India, Malaysia, Pakistan, Puerto Rico, Solomon Islands, Spain, States of Micronesia, Thailand, Venezuela, Zambia | Hughes (1952); Subramanian (1952); Nair and Tyagi (1961); Srivastava et al. (1964); Dennis (1970); Ellis (1971); Matsushima (1971); Stevenson (1975); Srivastava and Gupta (1981); Arnold (1986); McKenzie and Jackson (1986); Paul and Singh (1986); Gene et al. (1990); McKenzie and Jackson (1990); Ahmad et al. (1997); Pande and Rao (1998); Lumyong et al. (2003); Saravanan and Vittal (2007); Leão-Ferreira et al. (2010); Tarda et al. (2019) |
| D. synnematicum Somrith. | Decaying leaves of Musa sp. | India, Thailand | Somrithipol (2007) |
A study was undertaken to determine the saprobic fungi associated with Musa sp. (banana) in Thailand, during the dry season. Three hyphomycetous taxa that morphologically resembled Dictyoarthrinium were examined. According to our phylogenetic analyses of combined SSU, LSU, ITS and tef1-α sequence data, Dictyoarthrinium clustered in Didymosphaeriaceae (Pleosporales, Dothideomycetes) with strong statistical support, sister to Spegazzinia. Hence, we propose to transfer Dictyoarthrinium from Apiosporaceae (Xylariales, Sordariomycetes) to Didymosphaeriaceae (Pleosporales, Dothideomycetes) and introduce Dictyoarthrinium musae sp. nov. as a saprobe recorded from Musa sp. We also provide detailed morphological illustrations, descriptions and DNA sequence data for D. sacchari, recorded on Musa sp. from Thailand, which further validates the novel taxonomic placement of Dictyoarthrinium in Didymosphaeriaceae.
Materials and methods
Sample collection, morphological studies and isolation
Dead leaves of Musa sp. were collected from Thailand during the dry season (December to August) of 2018 and 2019. Specimens were transferred to the laboratory in cardboard boxes. Samples were examined with a Motic SMZ 168 Series microscope. Powder-like masses of fungal conidia were mounted in water for microscopic studies and photomicrography. The specimens were examined using a Nikon ECLIPSE 80i compound microscope and photographed with a Canon 550D digital camera fitted to the microscope. Measurements were made with the Tarosoft (R) Image Frame Work programme and images used for figures were processed with Adobe Photoshop CS3 Extended v. 10.0 software (Adobe Systems, USA).
Single spore isolation was carried out following the method described in Chomnunti et al. (2014). Germinated spores were individually transferred to potato dextrose agar (PDA) plates and incubated at 25 °C in daylight. Colony characteristics were observed and measured after 3 weeks at 25 °C. Herbarium specimens were deposited in the Mae Fah Luang University (MFLU) Herbarium, Chiang Rai, Thailand. Living cultures were deposited in the Culture Collection of Mae Fah Luang University (MFLUCC). Faces of fungi numbers (Jayasiri et al. 2015) and MycoBank numbers (http://www.MycoBank.org) were obtained for the respective taxa.
DNA extraction, PCR amplification and sequencing
Fungal isolates grown on potato dextrose agar (PDA) for 4 weeks at 25 °C were used to extract total genomic DNA. DNA was extracted from 50 to 100 mg of axenic mycelium of the 4-weeks-old growing cultures. The mycelium was ground to a fine powder in liquid nitrogen and fungal DNA was extracted using the Biospin Fungus Genomic DNA Extraction Kit-BSC14S1 (BioFlux, P.R. China) according to the manufacturer’s instructions. Four gene regions, the internal transcribed spacer (ITS), partial 18S small sub unit (SSU), partial 28S large sub unit (LSU) and partial translation elongation factor 1-alpha gene (tef1-α) were amplified using ITS5/ITS4 (White et al. 1990), NS1/NS4 (White et al. 1990), LR0R/LR5 (Vilgalys and Hester 1990) and EF1-983F /EF1-2218R (Rehner 2001) primers, respectively.
Polymerase chain reactions (PCR) were conducted according to the following protocol. The total volume of the PCR reaction was 25 μl and consisted of 12.5 μl of 2 × Power Taq PCR MasterMix (a premix and ready to use solution, including 0.1 Units/μlTaq DNA Polymerase, 500 μm dNTP Mixture each (dATP, dCTP, dGTP, dTTP), 20 mM Tris-HCl pH 8.3, 100 mMKCl, 3 mM MgCl2, stabiliser and enhancer), 1 μl of each primer (10 pM), 2 μl genomic DNA extract and 8.5 μl double distilled water (ddH2O). The reaction was conducted by running for 40 cycles. The annealing temperature was 56 °C for ITS and LSU, 57.2 °C for tef1-α and 55 °C for SSU and initially 95 °C for 3 min, denaturation at 95 °C for 30 seconds, annealing for 1 min, elongation at 72 °C for 30 seconds and final extension at 72 °C for 10 min for all gene regions. PCR amplification was confirmed on 1% agarose electrophoresis gels stained with ethidium bromide. The amplified PCR fragments were sent to a commercial sequencing provider (TsingKe Biological Technology Co., Beijing, China). The nucleotide sequence data acquired were deposited in GenBank.
Sequence alignment
Sequences obtained in this study were subjected to BLAST search in GenBank (https://blast.ncbi.nlm.nih.gov/Blast.cgi). BLAST search results and initial morphological studies supported that our isolates belong to Didymosphaeriaceae. Other sequences used in the analyses were obtained from GenBank based on recently published papers (Tanaka et al. 2015; Jayasiri et al. 2019) (Table 2) and BLAST search results. The single gene alignments were done by MAFFT v. 7.036 (http://mafft.cbrc.jp/alignment/server/large.html; Katoh et al. 2019) using the default settings and later refined, where necessary, using BioEdit v. 7.0.5.2 (Hall 1999).
Table 2.
Selected taxa with their corresponding GenBank accession numbers in the family Didymosphaeriaceae that are used in the phylogenetic analyses. Type strains are indicated as superscript T and newly-generated strains are indicated in bold.
| Taxa | Culture collection | ITS | LSU | SSU | tef1-α |
|---|---|---|---|---|---|
| Alloconiothyrium aptrootii | CBS 980.95T | JX496121 | JX496234 | NA | NA |
| A. aptrootii | CBS 981.95T | JX496122 | JX496235 | NA | NA |
| Austropleospora archidendri | CBS 168.77T | JX496049 | JX496162 | NA | NA |
| A. keteleeriae | MFLUCC 18-1551T | NR_163349 | MK348021 | MK347910 | MK360045 |
| Bambusistroma didymosporum | MFLU 15-0057T | KP761733 | KP761730 | KP761737 | KP761727 |
| B. didymosporum | MFLU 15-0058 | KP761734 | KP761731 | KP761738 | KP761728 |
| Bimuria novae zelandiae | CBS 107.79T | MH861181 | AY016356 | AY016338 | DQ471087 |
| Chromolaenicola lampangensis | MFLUCC 17-1462T | MN325016 | MN325004 | MN325010 | MN335649 |
| C. thailandensis | MFLUCC 17-1510T | MN325018 | MN325006 | MN325012 | MN335651 |
| Cylindroaseptospora leucaenae | MFLUCC 17-2424T | NR_163333 | NG_066310 | MK347856 | MK360047 |
| Deniquelata barringtoniae | MFLUCC 11-0422T | NR_111779 | NG_042696 | JX254656 | NA |
| D. vittalii | NFCCI4249T | MF406218 | MF182395 | MF622059 | MF182398 |
| Dictyoarthrinium musae | MFLUCC 20-0105T | MT482323 | MT482320 | MT482326 | MT495602 |
| D. musae | MFLUCC 20-0106T | MT482324 | MT482321 | MT482327 | MT495603 |
| D. sacchari | MFLUCC 20-0107 | MT482325 | MT482322 | MT482328 | NA |
| D. sacchari | CBS 529.73 | NA | MH872479 | NA | NA |
| Didymocrea sadasivanii | CBS 438.65T | MH858658 | DQ384103 | NA | NA |
| Didymosphaeria rubi-ulmifolii | MFLUCC 14-0023T | NA | KJ436586 | NG_063557 | NA |
| D. rubi-ulmifolii | MFLUCC 14-0024 | NA | KJ436585 | KJ436587 | NA |
| Kalmusia italica | MFLUCC 14-0560T | KP325440 | KP325441 | KP325442 | NA |
| K. variisporum | CBS 121.517T | NR_145165 | JX496143 | NA | NA |
| Kalmusibambusa triseptata | MFLUCC 13-0232T | KY682697 | KY682695 | KY682696 | NA |
| Karstenula rhodostoma | CBS 690.94 | NA | GU301821 | GU296154 | GU349067 |
| K. rhodostoma | CBS 691.94 | LC014559 | AB807531 | AB797241 | AB808506 |
| Laburnicola hawksworthii | MFLUCC 13-0602T | KU743194 | KU743195 | KU743196 | NA |
| L. muriformis | MFLUCC 14-0921T | KU743200 | KU743201 | KU743202 | NA |
| Letendraea cordylinicola | MFLUCC 11-0150 | KM213996 | KM213999 | KM214002 | NA |
| L. cordylinicola | MFLUCC 11-0148T | NR_154118 | NG_059530 | KM214001 | NA |
| Montagnula bellevaliae | MFLUCC 14-0924T | KT443906 | KT443902 | KT443904 | KX949743 |
| M. cirsii | MFLUCC 13-0680 | KX274242 | KX274249 | KX274255 | KX284707 |
| M. scabiosae | MFLUCC 14-0954T | KT443907 | KT443903 | KT443905 | NA |
| Neokalmusia brevispora | KT 1466T | LC014573 | AB524600 | AB524459 | AB539112 |
| N. scabrispora | KT 1023 | LC014575 | AB524593 | AB524452 | AB539106 |
| Neptunomyces aureus | CMG12T | MK912121 | NA | NA | MK948000 |
| N. aureus | CMG13 | MK912122 | NA | NA | MK948001 |
| Paracamarosporium fagi | CPC 24890 | KR611886 | KR611904 | NA | NA |
| P. fagi | CPC 24892T | KR611887 | KR611905 | NA | NA |
| Paraconiothyrium cyclothyrioides | CBS 972.95T | JX496119 | JX496232 | AY642524 | NA |
| Paramassariosphaeria anthostomoides | CBS 615.86 | MH862005 | GU205223 | GU205246 | NA |
| P. anthostomoides | MFLU 16-0172T | KU743206 | KU743207 | KU743208 | NA |
| Paraphaeosphaeria rosae | MFLUCC 17-2549T | MG828937 | MG829046 | MG829152 | MG829223 |
| P. rosicola | MFLUCC 15-0042T | NR_157528 | MG829047 | MG829153 | NA |
| Phaeodothis winteri | CBS 182.58 | NA | GU301857 | GU296183 | NA |
| Pseudocamarosporium propinquum | MFLUCC 13-0544 | KJ747049 | KJ813280 | KJ819949 | NA |
| P. pteleae | MFLUCC 17-0724T | NR_157536 | MG829061 | MG829166 | MG829233 |
| Pseudopithomyces entadae | MFLUCC 17-0917T | NA | NG_066305 | MK347835 | MK360083 |
| P. rosae | MFLUCC 15-0035T | MG828953 | MG829064 | MG829168 | NA |
| Spegazzinia bromeliacearum | URM 8084T | MK804501 | MK809513 | NA | NA |
| S. deightonii | MFLUCC 20-0002 | MN956768 | MN956772 | MN956770 | NA |
| S. intermedia | CBS 249.89T | MH862171 | MH873861 | NA | NA |
| S. lobulata | CBS 361.58T | MH857812 | MH869344 | NA | NA |
| S. musae | MFLUCC 20-0001T | MN930512 | MN930514 | MN930513 | NA |
| S. neosundara | MFLUCC 15-0456T | KX965728 | KX954397 | KX986341 | NA |
| S. radermacherae | MFLUCC 17-2285T | MK347740 | MK347957 | MK347848 | MK360088 |
| S. tessarthra | SH 287 | JQ673429 | AB807584 | AB797294 | AB808560 |
| Tremateia arundicola | MFLU 16-1275T | KX274241 | KX274248 | KX274254 | KX284706 |
| T. guiyangensis | GZAAS01T | KX274240 | KX274247 | KX274253 | KX284705 |
| T. murispora | GZCC 18-2787T | NR_165916 | MK972751 | MK972750 | MK986482 |
| Verrucoconiothyrium nitidae | CBS:119209 | EU552112 | NA | NA | NA |
| Xenocamarosporium acaciae | CBS:139895 | NR_137982 | NG_058163 | NA | NA |
| X. acaciae | MFLUCC 17-2432 | MK347766 | MK347983 | MK347873 | MK360093 |
*Abbreviations of culture collections: CBS: Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands, CPC: Working collection of Pedro Crous housed at CBS, GZAAS: Guizhou Academy of Agricultural Sciences Herbarium, China, KT: K. Tanaka, MFLU: Mae Fah Luang University, Chiang Rai, Thailand, MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand, SH: Academia Sinica People’s Republic of China. Shanghai, URM: Universidade Federal de Pernambuco.
Phylogenetic analyses
Maximum Likelihood (ML) trees were generated using the RAxML-HPC2 on XSEDE (8.2.8) (Stamatakis et al. 2008; Stamatakis 2014) in the CIPRES Science Gateway platform (Miller et al. 2010) using GTR+I+G model of evolution. Bootstrap supports were obtained by running 1000 pseudo-replicates. Maximum Likelihood bootstrap values (ML) ≥ 60% are given above each node of the phylogenetic tree in blue (Fig. 1).
Figure 1.
Maximum Likelihood tree revealed by RAxML from an analysis of SSU, LSU and ITS and tef1-α sequence data of the genera of Didymosphaeriaceae, showing the phylogenetic position of Dictyoarthrinium musae (MFLUCC 20-0105, MFLUCC 20-0106) and D. sacchari (MFLUCC 20-0107). ML bootstrap supports (≥ 60%) and Bayesian posterior probabilities (≥ 0.95 BYPP) are given above the branches, respectively. The tree is rooted with Bambusistroma didymosporum (MFLU 15-0057 and MFLU 15-0058). Strains generated in this study are indicated in brown bold type. Ex-type strains are indicated in black bold. The scale bar represents the expected number of nucleotide substitutions per site.
Bayesian analysis was conducted with MrBayes v. 3.1.2 (Huelsenbeck and Ronquist 2001) to evaluate posterior probabilities (PP) (Rannala and Yang 1996; Zhaxybayeva and Gogarten 2002) by Markov Chain Monte Carlo sampling (BMCMC). Two parallel runs were conducted, using the default settings, but with the following adjustments: four simultaneous Markov chains were run for 2,000,000 generations, trees were sampled every 100th generation and 20,001 trees were obtained. The first 4,000 trees, representing the burn-in phase of the analyses, were discarded. The remaining 16,001 trees were used for calculating PP in the majority rule consensus tree. Branches with Bayesian posterior probabilities (BYPP) ≥ 0.95 are indicated above each node of the phylogenetic tree (Fig. 1). Phylogenetic trees were visualised with the FigTree v1.4.0 programme (Rambaut 2011).
Results
Phylogenetic analyses
The combined SSU, LSU, ITS and tef1-α matrix comprised 61 sequences that represents the genera in Didymosphaeriaceae. The best scoring RAxML tree is shown (Fig. 1) with a final ML optimisation likelihood value of -19278.64. The matrix had 1091 distinct alignment patterns, with 39.08% of undetermined characters or gaps. Estimated base frequencies were: A = 0.234095, C = 0.252628, G = 0.278053, T = 0.235224; substitution rates AC = 1.252730, AG = 2.198875, AT = 1.318760, CG = 0.953798, CT = 5.276095, GT = 1.000000; proportion of invariable sites I = 0.491333; gamma distribution shape parameter α = 0.446418. All trees (ML and BYPP) were similar in topology and did not differ at the generic relationships, which are in agreement with multi-gene phylogeny of Tanaka et al. (2015) and Jayasiri et al. (2019). All Dictyoarthrinium strains analysed herein clustered as a highly-supported monophyletic clade (ML = 100%, BYPP = 1.00) in Didymosphaeriaceae (Fig. 1) sister to Spegazzinia (ML = 75%, BYPP = 0.98). We have included LSU sequence data of D. sacchari (CBS 529.73) of Vu et al. (2019) in our phylogenetic analyses. According to GenBank, CBS 529.73 was classified in Apiosporaceae (Sordariomycetes). In our analyses, D. sacchari (CBS 529.73) clustered with MFLUCC 20-0105, MFLUCC 20-0106 and MFLUCC 20-0107 strains in Didymosphaeriaceae with a strong statistical support (ML = 100%, BYPP = 1.00). Our strain MFLUCC 20-0107 grouped with D. sacchari (CBS 529.73). The novel isolates of D. musae (MFLUCC 20-0105 and MFLUCC 20-0106) were sister to D. sacchari (CBS 529.73 and MFLUCC 20-0107) with strong statistical support (ML = 100%, BYPP = 1.00).
Taxonomy
Dictyoarthrinium musae
Samarakoon, Chomnunti & K.D. Hyde sp. nov.
7BDFC66F-9636-52E8-8C6F-AC016DFBC029
MycoBank No: 835764
Facesoffungi Number: FoF08467
Figure 2.
Dictyoarthrinium musae (MFLU 20-0437, holotype) a conidia on the host b conidiophore and conidia with conidiophore mother cell c–f conidia with conidiophores on stalk g developmental stage of an immature lateral conidium h four-celled terminal conidium i conidiophore j conidiophores and conidia with terminal conidium k, l conidiophores without terminal conidium m attachment of a mature lateral conidium n–q warted four-celled mature conidia r, s mature conidia that split at septa t colony on PDA after 21 days. Scale bars: 500 μm (a); 50 μm (b, c); 20 μm (d–g, i); 10 μm (h); 5 μm (j–s).
Etymology.
Name reflects the host genus, Musa (Musaceae).
Holotype.
MFLU 20-0437
Description.
Saprobic on dead leaves of Musa sp. Sexual morph: Undetermined. Asexual morph: Colonies compact or effuse, black, often pulvinate. Mycelium superficial, a close network of branched and anastomosing hyphae. Stromata none. Setae and hyphopodia absent. Conidiophores 30–140 × 1–2 μm (x¯ 81.5 × 1.6 μm, n = 25), basauxic, arising usually singly from subspherical, subhyaline to light brown conidiophore mother cells, 4.5–4.8 × 4.3–4.5 μm (x̄ = 4.6 × 4.4 μm, n = 10), macronematous, mononematous, straight or flexuous, narrow, cylindrical, rough, subhyaline to pale brown, with thick brown or dark brown transverse septa that appear as stripes with distances of 6.3–5.8 μm at apex and 2.3–3 μm at base of the conidiophore. Conidiogenous cells 4.1–4.5 × 4.3–4.7 μm (x̄ = 4.4 × 4.5 μm, n = 10), blastic, integrated, terminal and intercalary, cylindrical, smooth, denticles absent, hyaline. Conidia 7–11.5 × 6.5–9 μm (x̄ = 8.7 × 7.9 μm, n = 40), solitary, dry, acropleurogenous, simple, square, rounded at the corners, 4-celled, spherical or subspherical, often flattened in one plane, pale to dark brown at maturity, verrucose, with light brown to dark brown warts, immature conidia often 1-celled and subhyaline. Terminal conidium with four cells, sometimes absent or fallen before lateral conidia, mature conidia split along one line of the septa, most conidia arranged obliquely downwards on the conidiophore, conidial formation observed as a bunch starting after conidiophore 1–3 septate.
Culture characteristics.
Conidia germinating on PDA within 18 hrs. Colonies on PDA reaching a diameter of 50 mm after 14 days at 25 °C, slightly raised, hairy, filamentous, moderately dense, middle light grey, periphery white; reverse white to greyish-white.
Material examined.
THAILAND. Chiang Rai. On dead leaves of Musa sp. (Musaceae), 7 December 2018, M. C. Samarakoon, BNS265 (MFLU 20-0437, holotype), ex-type living culture (MFLUCC 20-0105); ibid. 20 February 2019, B. C. Samarakoon BNS2239 (MFLU 20-0438, paratype), ex-paratype living culture (MFLUCC 20-0106).
Notes.
Based on BLAST search results of SSU, LSU, ITS and tef1-α sequence data, Dictyoarthrinium musae (MFLUCC 20-0105 and MFLUCC 20-0106) showed high similarity as follows: SSU = 99.15% to Paraconiothyrium hawaiiense (CBS 120025), LSU = 95.57% to Cylindroaseptospora siamensis (MFLUCC 17-2527), ITS = 98.24% to Kalmusia italica (isolate 5), tef1-α = 97.75% to Spegazzinia neosundara (MFLUCC 13-0211) with 100%, 100%, 87% and 99% query covers, respectively. In the multigene phylogeny, the Dictyoarthrinium clade was sister to Spegazzinia (ML = 75%, BYPP = 0.98). Within the Dictyoarthrinium clade, D. musae (MFLUCC 20-0105 and MFLUCC 20-0106) separated from the sister taxon, D. sacchari with strong statistical support (ML = 100%, BYPP = 1.00). ITS sequence comparison revealed 7.84% base pair differences between D. musae and D. sacchari (MFLUCC 20-0107), which is in agreement with the new species concept outlined by Jeewon and Hyde (2016). Dictyoarthrinium musae differs from D. sacchari by its unique conidial development in the apex. The terminal conidia of D. musae are always 4-celled and similar in colour to mature lateral conidia. In addition, the terminal conidia of D. musae are sometimes absent or fallen before the lateral conidia. In contrast, the terminal conidia of D. sacchari can be 2-celled or 4-celled, pale brown with respect to lateral mature conidia and always persist on the conidiophore. In addition, the mature conidia of D. musae split along one line of the septa and this specific feature is absent in D. sacchari. Dictyoarthrinium musae has a subhyaline, spherical conidiophore mother cell while D. sacchari has a distinct cup-shaped, brown conidiophore mother cell. Therefore, based on contrasting morphological differences to D. sacchari and strong statistical support from our molecular phylogeny, D. musae is herein introduced as a new species.
Dictyoarthrinium sacchari
(J.A. Stev.) Damon, Bull. Torrey bot. Club 80: 164 (1953)
639055B7-FFFC-5BE2-B06C-341C7E53270D
Facesoffungi Number: FoF08468
Figure 3.
Dictyoarthrinium sacchari (MFLU 20-0439) a conidia on the host b developmental stage of terminal conidium attached to the conidiophore c–f Conidiophores and conidia (e, with distinct mother cell) g, h mature conidiophores with four-celled terminal conidium i conidiophore with two celled terminal conidium j developmental stages of conidia on conidiophore k colony on PDA after 21 days l–q conidia. Scale bars: a = 1000 μm (a); 20 μm (b, j); 50 μm (c–i); 5 μm (l–q).
Description.
Saprobic on dead leaves of Musa sp. Sexual morph: Undetermined. Asexual morph: Colonies compact or effuse, black, often pulvinate. Mycelium superficial, a close network of branched and anastomosing hyphae. Stromata none. Setae and hyphopodia absent. Conidiophores 50–110 × 1–2 μm (x̄ = 72.0 × 1.6 μm, n = 15), basauxic, arising from cup-shaped, brown, distinct conidiophore mother cells, 3.4–4.4 × 2.9–4.7 μm (x̄ = 4 × 3.7 μm, n = 10), macronematous, mononematous, usually straight or flexuous, narrow, cylindrical, rough-walled, subhyaline to pale brown, with dark brown transverse septa as stripes with distances of 6.3–5.8 μm at apex and 2.3–3 μm at base of the conidiophore. Conidiogenous cells 4–4.5 × 4.3–4.7 μm (x̄ = 4.4 × 4.5 μm, n = 10), blastic, integrated, terminal and intercalary, cylindrical, smooth, hyaline. Conidia at maturity 8.5–11.5 × 8.5–10 μm (x̄ = 9.9 × 9.3 μm, n = 40), solitary, dry, acropleurogenous, simple, square, rounded at the corners, 4-celled, but difficult to distinguish the cells due to their blackish-brown nature, spherical or subspherical, often flattened in one plane, blackish-brown at maturity, with brown warts on surface of the cells, terminal conidium always 4-celled or 2-celled, light brown when compared with lateral conidia, most conidia arranged perpendicular to the conidiophore, some directed obliquely upwards.
Culture characteristics.
Conidia germinating on PDA within 18 hrs. Colonies on PDA reaching a diameter of 55 mm after 14 days at 25 °C, raised, moderately dense, entire margined, brownish-grey at maturity; reverse white to greyish-white.
Material examined.
Thailand, Chiang Mai. On mid-rib of a dead leaf of Musa sp. (Musaceae), S. Phongeun, 18 July 2018, BNS2287, (MFLU 20-0439), living culture MFLUCC 20-0107.
Notes.
Based on BLAST search results of SSU, LSU, ITS and tef1-α sequence data, our strain (MFLUCC 20-0107) showed high similarity to the taxa in GenBank as follows (SSU = 99.26% to Paraconiothyrium brasiliense (isolate GF1), LSU = 96.14% to Alloconiothyrium aptrooti (CBS 981.95), ITS = 93.00% to Kalmusia italica (MFLUCC 13-0066). In the multigene phylogeny, MFLUCC 20-0107 groups with Dictyoarthrinium sacchari, sister to D. musae with strong statistical support (ML = 100%, BYPP = 1.00). Our strain shares similar morphological features with D. sacchari (Subramanium 1952; Ellis 1971) and did not differ significantly. There are slight differences in conidial dimensions and the length of conidiophores of our collection and other D. sacchari collections by previous studies. Conidial dimensions and the length of conidiophores may differ due to diverse environmental effects and host associations. LSU sequence data of D. sacchari (CBS 529.73) are identical with our strain (MFLUCC 20-0107). Unfortunately, ITS, SSU and tef1-α sequence data of CBS 529.73 are not available in GenBank to compare with our strain. LSU data of Dictyoarthrinium musae have 2.24% of base pair difference with D. sacchari (CBS 529.73 and MFLUCC 20-0107). Dictyoarthrinium sacchari was reported on Musa sp. from Thailand in Lumyong et al. (2003) without morpho-molecular justifications. In this study, we document D. sacchari with detailed morphological illustrations, description, herbarium material and a living culture coupled with DNA sequence data (SSU, LSU, ITS) for a better taxonomic resolution.
Discussion
Both Dictyoarthrinium and Spegazzinia are characterised by basauxic conidiophores (Hughes 1952; Ellis 1971; Tanaka et al. 2015). Spegazzinia often has stellate (α) and disc-shaped (β) conidia (Ellis 1971; Tanaka et al. 2015). The conidia of Dictyoarthrinium (except D. africanum) share some similar characteristics with disc-shaped, β conidia of Spegazzinia. Both conidia are brown, 4-celled and constricted at the septa. Conidia of Dictyoarthrinium have characteristic hyaline or brown warts. Rarely, some taxa of Spegazzinia, for example, S. deightonii, also bear blunt ended spines. Most disc-shaped conidia of Spegazzinia are not warted. In addition, stellate conidia of Spegazzinia are always 4–5-celled and spinulose (Ellis 1971; Tanaka et al. 2015). There are contrasting morphological features of the basauxic conidiophores of both genera. The conidiophores of Dictyoarthrinium are hyaline to subhyaline with septa that appear as dark brown or light brown stripes throughout the conidiophore. The conidiophores (in stellate conidia) of Spegazzinia are more elongated, narrow, aseptate and dematiaceous.
Dictyoarthrinium quadratum (type of Dictyoarthrinium) is the heterotypic synonym of D. sacchari. Dictyoarthrinium quadratum has a terminal mature conidium with one to two cells. As described in Hughes (1952), these 2-celled conidia remain on the conidiophore, even when other conidia fall off. This feature is absent in D. musae. The terminal conidium of D. musae always ends up with four cells. The conidia of D. quadratum are obliquely upwardly directed, whereas the conidia of D. musae are obliquely downwardly directed (Fig. 2). The conidiophores of D. quadratum are erect and straight while D. musae has more curved conidiophores.
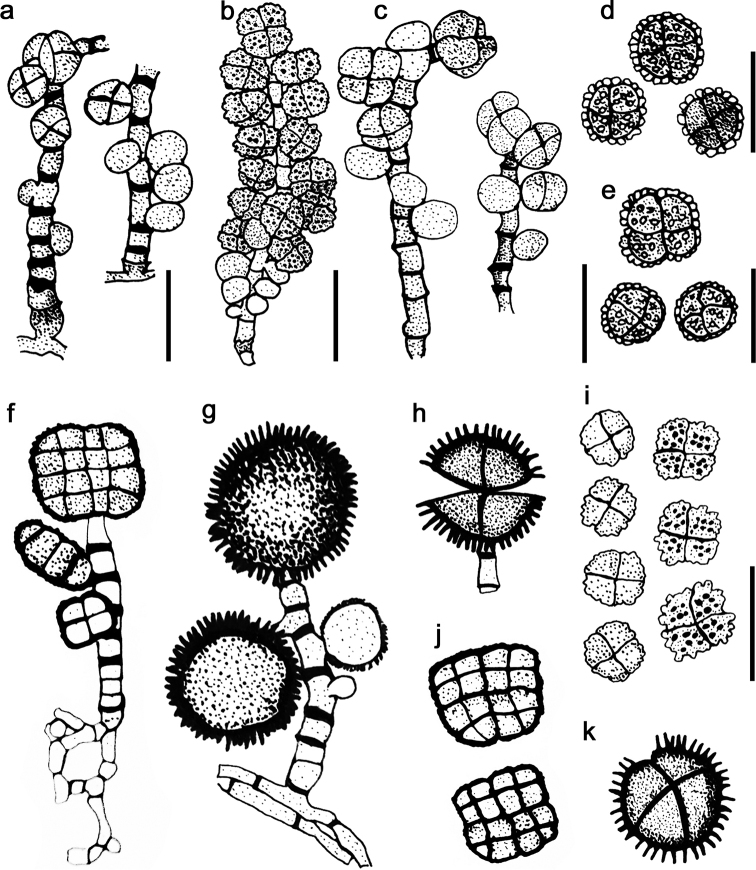
Dictyoarthrinium africanum differs significantly from D. musae by having 16-celled conidia. The conidia of D. rabaulense are completely black and densely echinulate with spines sometimes up to 4 μm long (Ellis 1976). However, D. musae has brown warts on the surface of conidia, while D. lilliputeum has hyaline warts. Dictyoarthrinium microsporum has longer conidiophores (250 μm) than D. musae. Morphological features of Dictyoarthrinium species are illustrated in Fig. 4. A key to the species of Dictyoarthrinium is provided below.
Figure 4.
Morphology of conidia and conidiophores of previously described Dictyoarthrinium species a, dD. microsporumb, iD. synnematicumc, eD. lilliputeumf, jD. africanumg, h, kD. rabaulense. Scale bars: 20 μm (a, c, d, e); 10 μm (b, i). Magnification × 650 (f, g, h, j, k). Redrawn from Rao and Rao (1964), Ellis (1971), Kobayasi et al. (1971) and Somrithipol (2007).
Key to the species of Dictyoarthrinium
| 1 | Synnemata present | D. synnematicum |
| – | Synnemata absent | 2 |
| 2 | Conidia 2- or 4-celled | 3 |
| – | Conidia 16-celled | D. africanum |
| 3 | Conidia with brown warts | 4 |
| – | Conidia with hyaline warts | D. lilliputeum |
| 4 | Conidiophores up to 130 μm long | 5 |
| – | Conidiophores up to 250 μm long | D. microsporum |
| 5 | Terminal conidium always 4-celled, mature conidia split along one line of the septa D. musae | |
| – | Terminal conidium 2- or 4-celled, mature conidia do not split along septa | D. sacchari |
To date, the taxonomy and phylogeny of most genera that have basauxic conidiogenesis (Hughes 1952) have been resolved with their correct taxonomic placements. Dictyoarthrinium and Endocalyx represented the sole unresolved genera. We transferred Dictyoarthrinium to Didymosphaeriaceae based on morphological and molecular evidence. This study uses multigene sequence data of SSU, LSU, ITS and tef1-α for the first time to confirm the taxonomic placement of Dictyoarthrinium in Didymosphaeriaceae.
Supplementary Material
Acknowledgements
Samantha C. Karunarathna would like to thank the CAS President’s International Fellowship Initiative (PIFI) young staff under the grant number: 2020FYC0002 for funding his postdoctoral research and the National Science Foundation of China (NSFC, project code 31851110759) for partially funding this work. Rungtiwa Phookamsak thanks CAS President’s International Fellowship Initiative (PIFI) for young staff (grant no. Y9215811Q1), the National Science Foundation of China (NSFC) project code 31850410489 (grant no. Y81I982211) and Chiang Mai University for their partial support of this research work. Dhanushka Wanasinghe thanks CAS President’s International Fellowship Initiative (PIFI) for funding his postdoctoral research (number 2019PC0008), the National Science Foundation of China and Chinese Academy of Sciences for financial support under the grant 41761144055. K.D Hyde thanks Thailand research grants entitled “The future of specialist fungi in a changing climate: baseline data for generalist and specialist fungi associated with ants, Rhododendron species and Dracaena species (Grant No: DBG6080013) and “Impact of climate change on fungal diversity and biogeography in the Greater Mekong Sub region (Grant No: RDG6130001). Binu C. Samarakoon offers her sincere gratitude to S. Phongeun, G. Samarakoon, Seetha Malani, Thiue Samarakoon and A.J Gajanayake for the valuable support they have given.
Citation
Samarakoon BC, Wanasinghe DN, Samarakoon MC, Phookamsak R, McKenzie EHC, Chomnunti P, Hyde KD, Lumyong S, Karunarathna SC (2020) Multi-gene phylogenetic evidence suggests Dictyoarthrinium belongs in Didymosphaeriaceae (Pleosporales, Dothideomycetes) and Dictyoarthrinium musae sp. nov. on Musa from Thailand. MycoKeys 71: 101–118. https://doi.org/10.3897/mycokeys.71.55493
Contributor Information
Saisamorn Lumyong, Email: scboi009@gmail.com.
Samantha C. Karunarathna, Email: samanthakarunarathna@gmail.com.
References
- Ahmad S, Iqbal SH, Khalid AN. (1997) Fungi of Pakistan. Sultan Ahmad Mycological Society of Pakistan, 248 pp.
- Arnold GRW. (1986) Lista de Hongos Fitopatogenos de Cuba. Ministerio de Cultura Editorial Cientifico-Tecnica, 207 pp.
- Bhat DJ. (2010) Fascinating Microfungi (Hyphomycetes) of Western Ghats – India. Broadway Publishing House, 190–221.
- Chomnunti P, Hongsanan S, Hudson BA, Tian Q, Peršoh D, Dhami MK, Alias AS, Xu J, Liu X, Stadler M, Hyde KD. (2014) The sooty moulds. Fungal Diversity 66: 1–36. 10.1007/s13225-014-0278-5 [DOI] [Google Scholar]
- Cole GT. (1986) Models of cell differentiation in conidial fungi. Microbiological Reviews 50: 95–132. 10.1128/MMBR.50.2.95-132.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ. (2013) A phylogenetic re-evaluation of Arthrinium. IMA Fungus 4: 133–154. 10.5598/imafungus.2013.04.01.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damon SC. (1953) Notes on the hyphomycetous genera, Spegazzinia Sacc. and Isthmospora Bulletin of the Torrey Botanical Club: 155–165. 10.2307/2482189 [DOI]
- Dennis RWG. (1970) Fungus Flora of Venezuela and Adjacent Countries. Kew Bulletin Additional Series III. Verlag von J. Cramer, 531 pp.
- Ellis MB. (1971) Dematiaceous Hyphomycetes. Commonwealth Mycological Institute, Kew.
- Ellis MB. (1976) More Dematiaceous Hyphomycetes. Commonwealth Mycological Institute, Kew.
- Gene J, Cano J, Guarro J. (1990) [Contribution to the study of the Spanish hyphomycetes XI]. Revista Iberoamericana de Micología 7: 31–33. [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleaic Acids Symposium Series 41: 95–98. [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Hughes SJ. (1952) Fungi from the Gold Coast. 11. Mycological Papers 50: 1–104. [Google Scholar]
- Hughes SJ. (1953) Conidiophores, conidia, and classification. Canadian Journal of Botany 39: 577–679. 10.1139/b53-046 [DOI] [Google Scholar]
- Hyde KD, Fröhlich J, Taylor JE. (1998) Fungi from palms XXXVI Reflections on unitunicate ascomycetes with apiospores. Sydowia 50: 21–80. [Google Scholar]
- Hyde KD, Norphanphoun C, Maharachchikumbura SSN, Bhat DJ, Jones EBG, Bundhun D, Chen YJ, Bao DF, Boonmee S, Calabon MS, Chaiwan N, Chethana KWT, Dai DQ, Dayarathne MC, Devadatha B, Dissanayake AJ, Dissanayake LS, Doilom M, Dong W, Fan XL, Goonasekara ID, Hongsanan S, Huang SK, Jayawardena RS, Jeewon R, Karunarathna A, Konta S, Kumar V, Lin CG, Liu JK, Liu NG, Luangsa-ard J, Lumyong S, Luo ZL, Marasinghe DS, McKenzie EHC, Niego AGT, Niranjan M, Perera RH, Phukhamsakda C, Rathnayaka AR, Samarakoon MC, Samarakoon SMBC, Sarma VV, Senanayake IC, Shang QJ, Stadler M, Tibpromma S, Wanasinghe DN, Wei DP, Wijayawardene NN, Xiao YP, Yang J, Zeng XY, Zhang SN, Xiang MM. (2020) Refined families of Sordariomycetes. Mycosphere 11: 305–1059. 10.5943/mycosphere/11/1/7 [DOI] [Google Scholar]
- Index Fungorum (2020) Index Fungorum. http://www.indexfungorum.org/Names/Names.asp [Retrieved 5 May 2020]
- Jayasiri SC, Hyde KD, Ariyawansa HA, Bhat J, Buyck B, Cai L, Dai YC, Abd-Elsalam KA, Ertz D, Hidayat I, Jeewon R. (2015) The Faces of fungi database: fungal names linked with morphology, molecular and human attributes. Fungal Diversity 74(1): 3–18. 10.1007/s13225-015-0351-8 [DOI] [Google Scholar]
- Jayasiri SC, Hyde KD, Jones EBG, McKenzie EHC, Jeewon R, Phillips AJL, Bhat DJ, Wanasinghe DN, Liu JK, Lu YZ, Kang JC, Xu J, Karunarathna SC. (2019) Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 10: 1–186. 10.5943/mycosphere/10/1/1 [DOI] [Google Scholar]
- Jeewon R, Hyde KD. (2016) Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7: 1669–1677. 10.5943/mycosphere/7/11/4 [DOI] [Google Scholar]
- Johnston JR, Stevenson JA. (1917) Sugar cane fungi and diseases of Porto Rico. Journal of the Department of Agriculture, Porto Rico 1: 177–264. [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20: 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayasi Y. (1971) Mycological reports from New Guinea and the Solomon Islands. Bulletin of the National Museum of Nature and Science, 1–11.
- Leão-Ferreira SM, Gusmão LFP. (2010) Conidial fungi from the semi-arid Caatinga biome of Brazil. New species of Endophragmiella and Spegazzinia with new records for Brazil, South America, and Neotropica. Mycotaxon 111: 1–10. 10.5248/111.1 [DOI] [Google Scholar]
- Lumyong P, Photita W, McKenzie EHC, Hyde KD, Lumyong S. (2003) Saprobic fungi on dead wild banana. Mycotaxon 85: 345–346. [Google Scholar]
- Matsushima T. (1971) Microfungi of the Solomon Islands and Papua-New Guinea. Nippon Printing Publishing Company, Osaka, 78 pp. [Google Scholar]
- McKenzie EHC, Jackson GVH. (1986) The fungi, bacteria and pathogenic algae of Solomon Islands. Strengthening Plant Protection and Root Crops Development in the South Pacific. RAS/83/001, Field Document 11. Suva, Fiji, 282 pp https://trove.nla.gov.au/version/42503674 [Google Scholar]
- McKenzie EHC, Jackson GVH. (1990) The fungi, bacteria and pathogenic algae of the Federated States of Micronesia. SPC Technical Paper 199. 67 pp.
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), November 14, 2010, New Orleans, Louisiana 1–8. 10.1109/GCE.2010.5676129 [DOI]
- Nair MC, Tyagi PD. (1961) Notes on some hyphomycetes-I. Proceedings of the Indian Academy of Sciences 54: 269–275. [Google Scholar]
- Pande A, Rao VG. (1998) A Compendium of Fungi on Legumes from India. Scientific Publishers (India), Jodhpur, 188 pp. [Google Scholar]
- Paul YS, Singh BM. (1986) Addition to fungi of India. Indian Phytopathology 39: 748–751. [Google Scholar]
- Rambaut A. (2011) FigTree Tree figure drawing tool version 131, Institute of Evolutionary Biology, University of Edinburgh. Available from: http://treebioedacuk/software/figtree/ [accessed 24 May 2020]
- Rannala B, Yang Z. (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of Molecular Evolution 43: 304–311. 10.1007/BF02338839 [DOI] [PubMed] [Google Scholar]
- Rao PR, Rao D. (1964) Some allied dematiaceae-dictyosporae from India. Mycopathologia 23: 23–28. 10.1007/BF02049180 [DOI] [PubMed] [Google Scholar]
- Rehner S. (2001) Primers for Elongation Factor 1-alpha (EF1-alpha). Insect Biocontrol Laboratory: USDA, ARS, PSI.
- Saravanan T, Vittal BPR. (2007) Some rare and interesting hyphomycetes from eastern Ghats in Tamil Nadu, India. Kavaka 35: 21–44. [Google Scholar]
- Somrithipol S. (2007) A synnematous species of Dictyoarthrinium from Thailand. Mycologia 99: 792–796. 10.1080/15572536.2007.11832542 [DOI] [PubMed] [Google Scholar]
- Srivastava MP, Tandon RN, Bilgrami KS, Ghosh AK. (1964) Study on fungal diseases of some tropical fruits–I. A list of fungi isolated from fruits and fruit trees. Phytopathology 50: 250–251. 10.1111/j.1439-0434.1964.tb02923.x [DOI] [Google Scholar]
- Srivastava RN, Gupta JS. (1981) Seed mycoflora from Indian seed lots of Cosmos bipinnatus and their control. Indian Phytopathology 34: 383–385. [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. (2008) A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology 57: 758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- Stevenson JA. (1975) Fungi of Puerto Rico and the American Virgin Islands. Contribution of Reed Herbarium 23: 743 pp.
- Subramanian CV. (1952) Fungi imperfecti from Madras–Ii. Proceedings of the Indian Academy of Sciences Section B 34: 160–168. [Google Scholar]
- Sushma, Prasher IB, Verma RK. (2020) Some interesting hyphomycetous fungi from India. Vegetos 33: 74–82. 10.1007/s42535-019-00083-8 [DOI] [Google Scholar]
- Tanaka K, Hirayama K, Yonezawa H, Sato G, Toriyabe A, Kudo H, Hashimoto A, Matsumura M, Harada Y, Kurihara Y, Shirouzu T. (2015) Revision of the Massarineae (Pleosporales, Dothideomycetes). Studies in Mycology 82: 75–136. 10.1016/j.simyco.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarda AS, Saparrat MCN, Gómez N. (2019) Assemblage of dematiaceous and Ingoldian fungi associated with leaf litter of decomposing Typha latifolia L. (Typhaceae) in riverine wetlands of the Pampean plain (Argentina) exposed to different water quality. Journal of Environmental Management 250: 109–409. 10.1016/j.jenvman.2019.109409 [DOI] [PubMed] [Google Scholar]
- Urtiaga R. (1986) Indice de enfermedades en plantas de Venezuela y Cuba. Impresos en Impresos Nuevo Siglo. S.R.L. , Barquisimeto, Venezuela, 202 pp. [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. 10.1128/JB.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu D, Groenewald M, De Vries M, Gehrmann T, Stielow B, Eberhardt U, Al-Hatmi A, Groenewald JZ, Cardinali G, Houbraken J, Boekhout T. (2019) Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Studies in Mycology 92: 135–154. 10.1016/j.simyco.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayawardene NN, Hyde KD, Lumbsch T, Liu JK, Maharachchikumbura SSN, Ekanayaka AH, Tian Q, Phookamsak R. (2018) Outline of Ascomycota – 2017. Fungal Diversity 88: 167–263. 10.1007/s13225-018-0394-8 [DOI] [Google Scholar]
- White TJ, Bruns T, Lee SJWT, Taylor JL. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18: 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Zhaxybayeva O, Gogarten JP. (2002) Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. BMC genomics 3: 1–4. 10.1186/1471-2164-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.