Pediatric supratentorial ependymomas with RELA fusions (RELA-EP) have been identified as a unique novel tumor entity [9, 10]. Fusions between C11orf95 and RELA pathologically activate the NFκB signaling pathway indicated by nuclear accumulation of p65-RelA. Deletions of CDKN2A encoding the negative cell-cycle regulator p16 have been described in a subset of supratentorial ependymomas, associated with worse outcome [2, 5, 7]. We assessed the frequency and prognostic impact of CDKN2A deletions in a cohort of 54 RELA-EP in children treated according to HIT2000-E protocols (for detailed demographic information, see supplementary materials and methods and supplementary table 1).
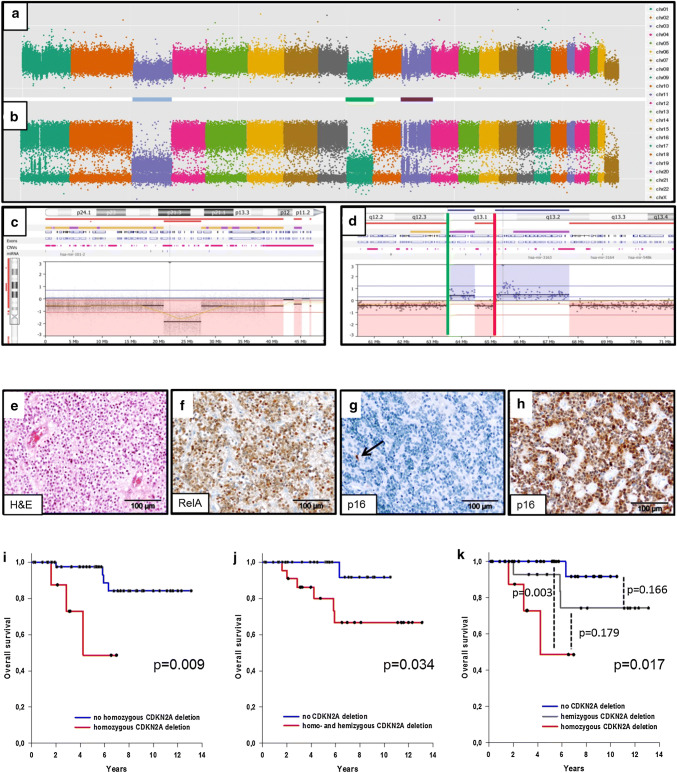
High-resolution, genome-wide copy number profiles were generated by molecular inversion probe (MIP) assay. Chromosomal breaks were identified within the C11orf95 and RELA genes corresponding to fusion transcripts (Fig. 1a, d). All cases showed pathological nuclear accumulation of p65-RelA as a hallmark of RELA-EP (Fig. 1f). Homozygous deletion (complete loss) of CDKN2A was detected in 9 of 54 (16.7%) cases (Fig. 1c); and 8 of these (88.9%) showed a concordant complete loss of p16 protein (Fig. 1g). In one case, few tumor cells expressed p16 protein indicating retained CDKN2A alleles in single cells. Fourteen cases (25.9%) harbored a hemizygous deletion of CDKN2A. In these, p16 protein was retained in 92.9% of cases tested—one case lacked p16 protein expression indicating the inactivation of the second allele by alternative mechanisms. Thirty-one of 54 cases (57.4%) had no deletion of CDKN2A; all showed p16 protein expression (Fig. 1h). Immunohistochemistry for p16, therefore, may serve as a surrogate for complete CDKN2A loss, but cannot differentiate between hemizygous and wild-type status. There was no statistical association between CDKN2A deletions and mitotic activity as previously described in IDH-mutant glioma [1]. The presence of CDKN2A deletions (homo- or hemizygous) correlated with higher age at diagnosis in line with the literature [3, 5, 8]. CDKN2A deletion may also occur as secondary event in tumor progression [7].
Fig. 1.
a Genomic copy number profile and b allele distribution (MIP) of a RELA-EP showing chromothripsis of chromosome 11; c case with homozygous CDKN2A deletion; d case showing breaks in C11orf95 (green bar) and RELA (red bar); e clear cell morphology; f nuclear p65-RelA; g case with homozygous CDKN2A deletion/loss of p16 protein (arrow, endothelial cell as internal positive control); h case without CDKN2A deletion/retained p16; i–k Kaplan–Meier analysis, impact of CDKN2A deletions on OS
To identify possible differences between RELA-EP with versus without CDKN2A deletion on the transcript level, 12 RELA-EP were analyzed by RNA sequencing for differentially expressed genes. After correction for multiple testing, five genes were found significantly downregulated including CDKN2A and CDKN2B and their neighboring gene MTAP (S-methyl-5′-thioadenosine phosphorylase) located in the deleted region. MTAP is a key enzyme in the methionine salvage pathway. Its deletion leads to dependence on the activity of the methyltransferase PRMT5 [6] which can be blocked by PRMT5 inhibitors as interesting novel therapeutics in MTAP deleted tumors. In addition, KIF7 (15q26) encoding a cilia-associated protein and ZNF536 (19q12) encoding a neuronal marker were found downregulated. GABRA2 (4p12) encoding the gamma-aminobutyric acid receptor subunit alpha-2 was found highly upregulated in CDKN2A deleted tumors (supplementary figure 3).
Kaplan–Meier analysis revealed a significant correlation between CDKN2A deletions and overall survival status (OS). Different groups were compared: (1) homozygous CDKN2A deletion vs. hemizygous CDKN2A deletion and tumors with two retained alleles (p = 0.009); (2) homo- or hemizygous CDKN2A deletions vs. tumors with two retained alleles (p = 0.034) and (3) all three strata separately (p = 0.017) (Fig. 1i–k). In contrast to Korshunov et al. [5], neither homozygous nor hemizygous deletion showed prognostic relevance regarding EFS (supplementary figure 2). Predominant clear cell morphology as a histological feature was a favorable prognosticator for OS (p = 0.039), and high mitotic activity (> 17 mitoses/10HPF) was a predictor for tumor relapse (p = 0.004) as well as OS (p = 0.007) (supplementary figure 1). Multivariate analysis confirmed mitotic activity as independent prognostic indicator for EFS (supplementary table 2).
Our data show that deletions of CDKN2A represent an objective parameter for risk stratification in RELA-EP. Molecular inversion probe methodology turned out to represent a sensitive and quantitative tool for CDKN2A assessment in FFPE material. Apart from ependymoma, homozygous deletions of CDKN2A have recently been described as adverse prognostic marker for other CNS tumors, including anaplastic IDH-mutant gliomas and BRAF-mutant low-grade gliomas [1, 4, 11]. The deletion/inactivation of CDKN2A may result in a pathological activation of cyclin-dependent kinases 4/6 targetable by specific inhibitors such as palbociclib. Therefore, CDKN2A inactivation in RELA-ependymomas may represent a potential therapeutical target.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary table 1, demographics, clinical, neuropathological and genetic information of the patient cohort (DOCX 17 kb)
Supplementary table 2, multivariate analysis (DOCX 20 kb)
Supplementary figure 1, Kaplan-Meier analysis of age at diagnosis, clear cell morphology and mitotic activity (PPTX 132 kb)
Supplementary figure 2, KM analysis of CDKN2A deletion in RELA ependymomas (EFS) (PPTX 116 kb)
Supplementary figure 3, differential expression (RNA sequencing) (PPTX 79 kb)
supplementary materialsand methods (DOCX 20 kb)
Acknowledgements
Open Access funding provided by Projekt DEAL. Funding was provided by Deutsche Kinderkrebsstiftung (Grant nos. DKS 2006.03, 2009.19, 2011.01 and 2014.17). We thank Dr. Steffen Albrecht, Montreal, for critical reading of the manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Appay R, Dehais C, Maurage C-A, et al. CDKN2A homozygous deletion is a strong adverse prognosis factor in diffuse malignant IDH-mutant gliomas. Neuro-oncology. 2019;21:1519–1528. doi: 10.1093/neuonc/noz126.000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bortolotto S, Chiadò-Piat L, Cavalla P, et al. CDKN2A/p16 in ependymomas. J Neurooncol. 2001;54:9–13. doi: 10.1023/A:1012537105775. [DOI] [PubMed] [Google Scholar]
- 3.Hirose Y, Aldape K, Bollen A, et al. Chromosomal abnormalities subdivide ependymal tumors into clinically relevant groups. Am J Pathol. 2001;158:1137–1143. doi: 10.1016/S0002-9440(10)64061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korshunov A, Casalini B, Chavez L, et al. Integrated molecular characterization of IDH-mutant glioblastomas. Neuropathol Appl Neurobiol. 2019;45:108–118. doi: 10.1111/nan.12523. [DOI] [PubMed] [Google Scholar]
- 5.Korshunov A, Witt H, Hielscher T, et al. Molecular staging of intracranial ependymoma in children and adults. J Clin Oncol. 2010;28:3182–3190. doi: 10.1200/JCO.2009.27.3359. [DOI] [PubMed] [Google Scholar]
- 6.Mavrakis KJ, McDonald ER, 3rd, Schlabach MR, et al. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science. 2016;351:1208–1213. doi: 10.1126/science.aad5944. [DOI] [PubMed] [Google Scholar]
- 7.Milde T, Pfister S, Korshunov A, et al. Stepwise accumulation of distinct genomic aberrations in a patient with progressively metastasizing ependymoma. Genes Chrom Cancer. 2009;48:229–238. doi: 10.1002/gcc.20635. [DOI] [PubMed] [Google Scholar]
- 8.Nowak J, Jünger ST, Huflage H, et al. MRI phenotype of RELA-fused pediatric supratentorial ependymoma. Clin Neuroradiol. 2019;29:595–604. doi: 10.1007/s00062-018-0704-2. [DOI] [PubMed] [Google Scholar]
- 9.Parker M, Mohankumar KM, Punchihewa C, et al. C11orf95–RELA fusions drive oncogenic NF-κB signalling in ependymoma. Nature. 2014;506:451–455. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pietsch T, Wohlers I, Goschzik T, et al. Supratentorial ependymomas of childhood carry C11orf95–RELA fusions leading to pathological activation of the NF-κB signaling pathway. Acta Neuropathol. 2014;127:609–611. doi: 10.1007/s00401-014-1264-4. [DOI] [PubMed] [Google Scholar]
- 11.Ryall S, Zapotocky M, Fukuoka K, et al. Integrated molecular and clinical analysis of 1,000 pediatric low-grade gliomas. Cancer Cell. 2020;37:569–583. doi: 10.1016/j.ccell.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table 1, demographics, clinical, neuropathological and genetic information of the patient cohort (DOCX 17 kb)
Supplementary table 2, multivariate analysis (DOCX 20 kb)
Supplementary figure 1, Kaplan-Meier analysis of age at diagnosis, clear cell morphology and mitotic activity (PPTX 132 kb)
Supplementary figure 2, KM analysis of CDKN2A deletion in RELA ependymomas (EFS) (PPTX 116 kb)
Supplementary figure 3, differential expression (RNA sequencing) (PPTX 79 kb)
supplementary materialsand methods (DOCX 20 kb)