Abstract
The importance of long noncoding RNA (lncRNA) in tumorigenesis has been supported by increasing evidence in recent years. However, the mechanism linking lncRNA function with cancer progression remains poorly understood. lncRNA LCPAT1 plays a role in lung cancer. However, how it works in breast cancer (BC) is largely unclear. In this study, we found that LCPAT1 was highly expressed in BC tissues and cell lines. High LCPAT1 expression predicted a low survival rate in BC patients. LCPAT1 promoted BC cell proliferation, migration, and invasion while inhibiting apoptosis in vitro. LCPAT1 knockdown suppressed BC growth in vivo and vice versa. LCPAT1 interacted with RBBP4 and recruited it to the MFAP2 (microfibril-associated protein 2) promoter and then activated MFAP2 transcription. Restoration of MFAP2 rescued the effects of LCPAT1 knockdown in BC cells. In sum, LCPAT1 promotes BC progression through recruiting RBBP4 to initiate MFAP2 transcription.
Keywords: LCPAT1, RBBP4, MFAP2, breast cancer, progression
Graphical Abstract
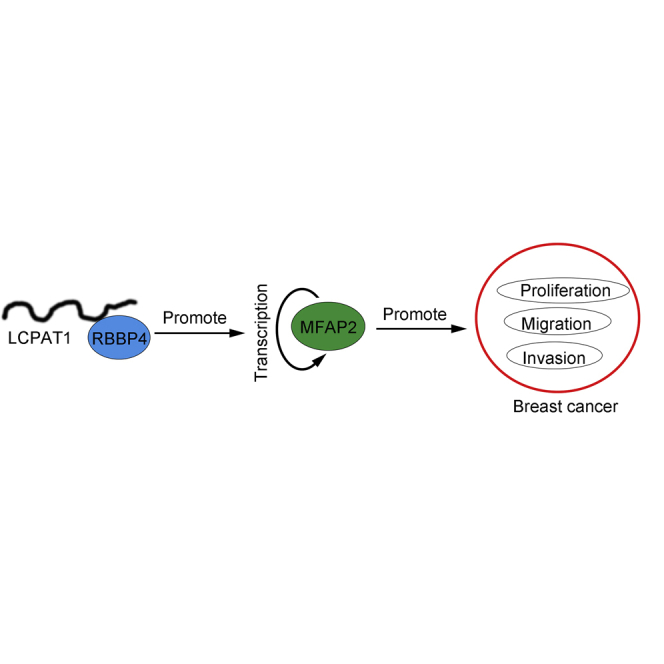
Gong et al. demonstrate that LCPAT1 was highly expressed in breast cancer tissues and promoted breast cancer cell proliferation, migration, and invasion while inhibiting apoptosis through recruiting RBBP4 to initiate MFAP2 transcription. Their work illustrates a new mechanism involved in breast cancer progression and suggests that LCPAT1 may be a new therapeutic target.
Introduction
Breast cancer (BC) is a very common malignancy among women worldwide and causes a large number of cancer-associated deaths.1 The incidence of BC is still increasing gradually. In China, about 270,000 new cases of BC are diagnosed every year.2 Studies show that BC is a very heterogeneous cancer and could be categorized into five major subtypes, such as triple-negative BC (TNBC), according to the gene expression pattern (ER, PR, and HER2).3 Notably, mutation in the BRCA1 or BRCA2 gene increases the risk of BC greatly.4 The clinical outcomes and therapeutic strategies are different among these BC subtypes.5 Although much effort has been made and some advances have been achieved, the pathogenesis of BC is still enigmatic, leading to a poor outcome of patients. The 5-year survival rate of BC patients also requires improvement.6 Therefore, elucidating the molecular mechanism of BC progression will be pivotal in developing specific therapeutic targets and blocking tumor metastasis and recurrence.
About 80% of the genomic transcripts are noncoding RNAs.7 Among them, long noncoding RNAs (lncRNAs) are defined by their lengths of more than 200 nt and little potential for protein coding.8 Emerging work has indicated that lncRNA is involved in expression regulation at the transcriptional or post-transcriptional level.9,10 Many studies have reported the crucial roles of lncRNA in tumorigenesis, including BC.11 Increasing evidence shows that many lncRNAs are aberrantly transcribed in cancer and regulate various biological processes, including growth, differentiation, and invasiveness, among others.12 For example, lncRNA GIHCG promotes colorectal cancer (CRC) growth and enhances tumor cell resistance to chemotherapy.13 LINC01093 is downregulated in liver cancer and inhibits cell proliferation by interacting with IGF2BP1.14 lncRNA TUG1 upregulation in pancreatic cancer contributes to proliferation and reduces apoptosis.15 Additionally, TINCR activation promotes epithelial-mesenchymal transition (EMT) and development of BC.16 Thus, lncRNAs may be potential biomarkers for BC prognosis and therapeutic targets after understanding their functional mechanism well.
LCPAT1 (also named as RCC2 antisense RNA 1) is a functionally unclear lncRNA. A previous study indicated that LCPAT1 regulates lung cancer.17 However, other roles of LCPAT1 have not been explored. We found that LCPAT1 was highly expressed in BC tissues and correlated with poor prognosis. LCPAT1 knockdown reduced proliferation, migration, and invasion of BC in vitro and in vivo. Mechanistically, LCPAT1 interacted with chromatin remodeler RBBP4 and recruited it to MFAP2 (microfibril-associated protein 2) promoter, and then activated MFAP2 transcription. In conclusion, LCPAT1 promotes BC progression through RBBP4-dependent activation of MFAP2 transcription.
Results
Identification of LCPAT1 Upregulation in BC Tissues
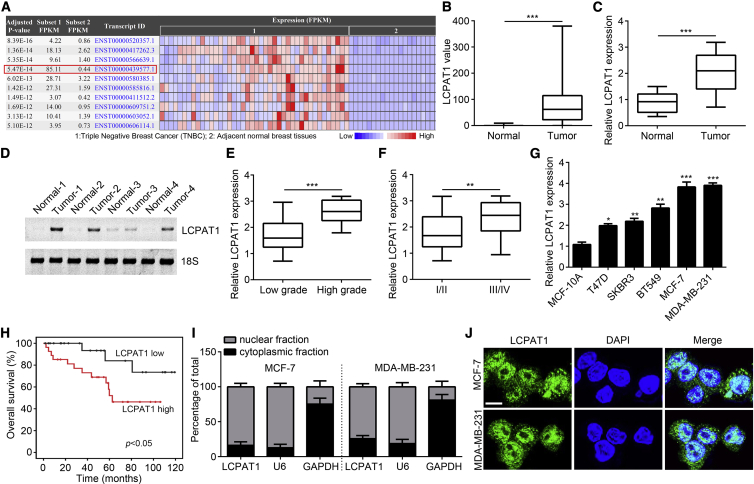
In order to elucidate the correlation between lncRNA dysregulation and BC development, we analyzed differentially expressed lncRNAs in BC tissues according to the bioinformatics approach. Several upregulated lncRNAs were identified by using the Cancer RNA-Seq Nexus online tool (Figure 1A). Among them, LCPAT1 (311 nt in length) was significantly upregulated in tumor tissues (Figure 1B). Then, its overexpression was further confirmed by quantitative real-time polymerase chain reaction (PCR) in 51 BC tissues (Figure 1C). Consistently, northern blotting also achieved a similar result (Figure 1D). Notably, the expression of LCPAT1 was higher in BC tissues with high grade and advanced stages (Figures 1E and 1F), suggesting that LCPAT1 may be involved in BC progression. Then, LCPAT1 levels in BC cell lines were measured. Results indicated that LCPAT1 was highly expressed in BC cell lines compared to MCF-10A cells (Figure 1G). Moreover, we found that higher expression of LCPAT1 was associated with a lower survival rate in BC patients (Figure 1H). Then, we analyzed the subcellular localization of LCPAT1 in BC cells. Quantitative real-time PCR and RNA fluorescence in situ hybridization (FISH) suggested that LCPAT1 was mainly located in the nucleus of MCF-7 and MDA-MB-231 cells (Figures 1I and 1J).
Figure 1.
Identification of LCPAT1 Upregulation in BC Tissues
(A) Upregulated lncRNAs in BC tissues compared to normal controls using the Cancer RNA-Seq Nexus online tool (http://syslab4.nchu.edu.tw/). (B) Expression value of LCPAT1 according to the Cancer RNA-Seq Nexus online tool. (C) Relative expression of LCPAT1 in 51 BC tissues and their matched normal control tissues by quantitative real-time PCR. (D) Northern blotting analysis of LCPAT1 in four pairs of BC tissues and their matched normal controls. 18S was a loading control. (E) LCPAT1 expression was analyzed in BC tissues with low (n = 31) or high (n = 20) grades. (F) LCPAT1 levels were determined in BC tissues with tumor, node, metastasis (TNM) stage I/II (n = 23) or III/IV (n = 28). (G) Relative expression of LCPAT1 in BC cell lines. (H) Overall survival rate analysis according to LCPAT1 median expression value. (I) Nucleocytoplasmic separation assay was performed to analyze LCPAT1 localization in BC cells. (J) LCPAT1 was mainly located in the nucleus of BC cells as analyzed by a FISH assay. Scale bar, 10 μm. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
LCPAT1 Knockdown Inhibits the Malignant Behaviors of BC Cells
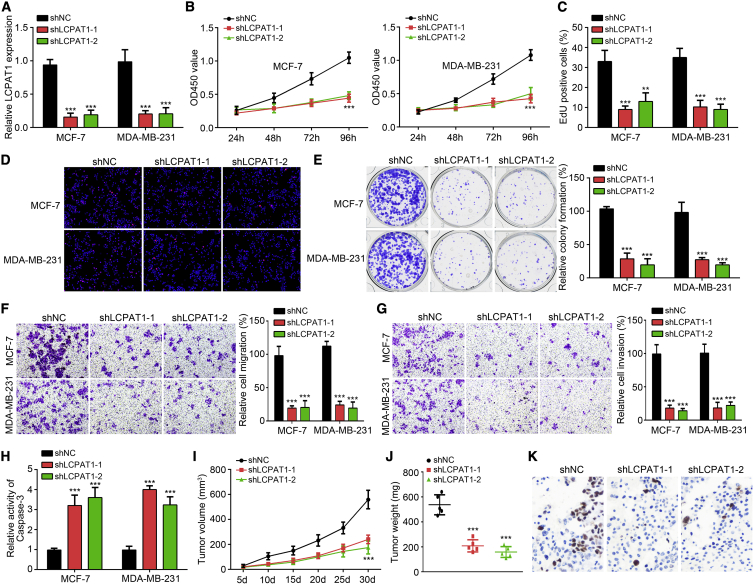
To explore the potential roles of LCPAT1, we constructed LCPAT1-silenced cell lines using two independent short hairpin RNAs (shRNAs). Quantitative real-time PCR validated the silencing of LCPAT1 in these two cell lines (Figure 2A). Then, Cell Counting Kit-8 (CCK8) and 5-ethynyl-2′-deoxyuridine (EdU) incorporation assays were performed and illustrated that knockdown of LCPAT1 markedly impaired the proliferation rate of MCF-7 and MDA-MB-231 cells (Figures 2B and 2C). Clearly, the Ki67-positive cells were also reduced after LCPAT1 knockdown (Figure 2D). The effect of LCPAT1 on proliferation was further demonstrated by decreased colony numbers (Figure 2E). Then, the effects of LCPAT1 on metastasis were evaluated by a Transwell assay. Results suggested that LCPAT1 knockdown gave rise to reduced cell numbers of migration and invasion (Figures 2F and 2G). Additionally, we also found that LCPAT1 silencing increased the activity of caspase-3 (Figure 2H), suggesting that LCPAT1 regulates apoptosis. To further investigate the role of LCPAT1 in vivo, a xenograft experiment was conducted. LCPAT1-silenced or control MDA-MB-231 cells were injected into nude mice and tumor volumes were monitored. Results showed that LCPAT1 knockdown inhibited the tumor volumes (Figure 2I). Also, tumor weights were decreased at the end point of this assay (Figure 2J). Moreover, Ki67 expression in tumor tissues was analyzed by an immunohistochemistry (IHC) assay. As shown, Ki67-positive cells were diminished after LCPAT1 silencing (Figure 2K), indicating that LCPAT1 suppressed BC cell growth in vivo.
Figure 2.
LCPAT1 Knockdown Inhibits the Malignant Behaviors of BC Cells
(A) shRNAs-mediated knockdown of LCPAT1 was validated by quantitative real-time PCR. (B and C) CCK8 (B) and EdU (C) incorporation assays for cell proliferation analysis. (D) Ki67 immunofluorescence (IF) staining was performed to test cell proliferation. Scale bar, 100 μm. (E) LCPAT1 knockdown reduced the colony numbers of MCF-7 and MDA-MB-231 cells. (F) Transwell assay for migration detection. (G) Transwell assay for invasion analysis. (H) The activity of caspase-3 was measured using a caspase-3 activity assay kit. (I) MDA-MB-231 cells were used for tumorigenesis in vivo, and tumor volumes were measured every 5 days. (J) Tumor weights were examined on day 30 after mice were sacrificed. (K) Ki67 expression was analyzed by IHC in tumor tissues to analyze cell proliferation. ∗∗p < 0.01, ∗∗∗p < 0.001.
Overexpression of LCPAT1 Enhances BC Cell Growth and Invasion
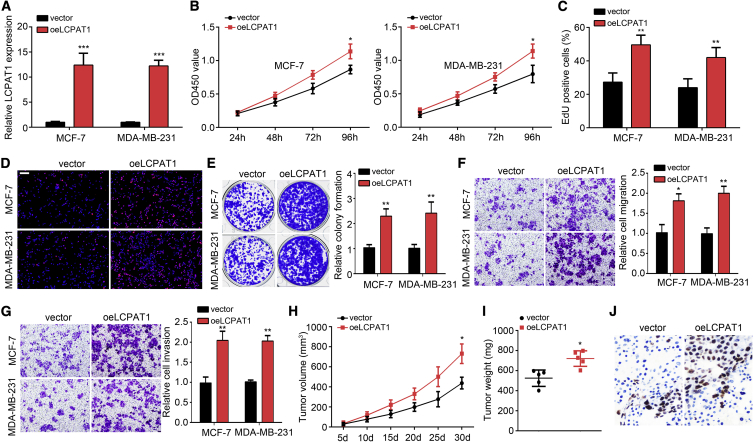
To further confirm the role of LCPAT1, we overexpressed LCPAT1 and performed functional experiments again. PCR confirmed the upregulation of LCPAT1 (Figure 3A). Similarly, CCK8, EdU incorporation, Ki67 staining, and colony formation assays were carried out. All results showed that LCPAT1 overexpression promoted proliferation and EdU incorporation, and led to an increase of Ki67-positive cells and colony numbers (Figures 3B–3E). Furthermore, a Transwell assay proved that LCPAT1 overexpression facilitated the migration and invasion of MCF-7 and MDA-MB-231 cells (Figures 3F and 3G). We also performed a xenograft assay using LCPAT1-overexpressing cell lines. As shown, the tumor volumes and weights were increased in the LCPAT1-overexpressing group (Figures 3H and 3I). Consistently, the Ki67-positive cells were increased in LCPAT1-overexpressing tumor tissues (Figure 3J). Taken together, LCPAT1 promotes BC progression in vitro and in vivo.
Figure 3.
Overexpression of LCPAT1 Enhances BC Cell Growth and Invasion
(A) Stable overexpression of LCPAT1 was validated by quantitative real-time PCR in MCF-7 and MDA-MB-231 cells. (B) CCK8 assay for proliferation evaluation. (C) EdU assay for analysis of proliferation. (D) LCPAT1 overexpression increased Ki67 expression as shown by IF staining. Scale bar, 100 μm. (E) Colony formation ability was enhanced after LCPAT1 upregulation. (F) Migration analysis by Transwell assay. (G) Invasion detection by Transwell assay. (H) Tumor volumes were measured every 5 days. LCPAT1 overexpression accelerated tumor growth. (I) Tumor weights were determined on day 30. (J) Ki67 expression levels in tumor tissues were analyzed by an IHC assay. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
LCPAT1 Promotes MFAP2 Expression
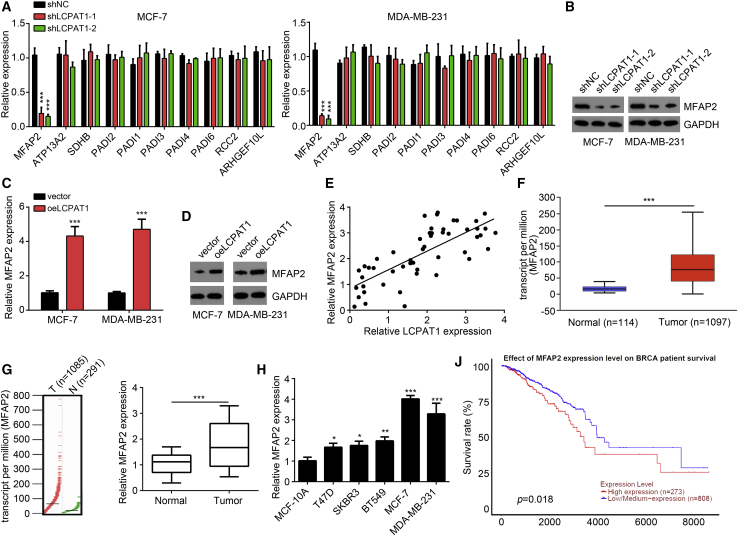
We have noticed that LCPAT1 was mainly located in the nucleus of BC cells. Also, increasing evidence suggests that lncRNAs could regulate the transcription of their neighboring genes in the nucleus.21 Hence, we wondered whether LCPAT1 regulates the expression of its neighboring genes. We selected LCPAT1 neighboring genes using the UCSC online tool (http://genome.ucsc.edu/) (Table S1). We performed quantitative real-time PCR analysis. Interestingly, the expression of MFAP2 was significantly downregulated after LCPAT1 knockdown in MCF-7 and MDA-MB-231 cells (Figure 4A), which was further confirmed by western blotting analysis (Figure 4B). Similarly, upregulation of LCPAT1 promoted expression of MFAP2 (Figures 4C and 4D). Additionally, LCPAT1 expression was positively correlated with MFAP2 in BC tissues (Figure 4E). Then, we performed bioinformatics analysis using online tools (UALCAN and GEPIA) and found that MFAP2 was upregulated in BC tissues compared to normal tissues (Figures 4F and 4G). We further confirmed its upregulation in 51 BC tissues and BC cell lines by quantitative real-time PCR (Figures 4H and 4I). Moreover, online data also indicated that MFAP2 upregulation predicted a low survival rate in BC patients (Figure 4J), suggesting that MFAP2 may act in oncogenic roles.
Figure 4.
LCPAT1 Promotes MFAP2 Expression
(A) Relative expression of the neighboring genes of LCPAT1 was analyzed by quantitative real-time PCR in MCF-7 and MDA-MB-231 cells. (B) Western blotting analysis for MFAP2 expression after LCPAT1 knockdown. (C and D) Quantitative real-time PCR (C) and western blotting (D) analyses for MFAP2 expression after LCPAT1 overexpression. (E) Expression correlation between LCPAT1 and MFAP2 in 51 BC tissues was analyzed. (F) Expression levels of MFAP2 in BC tissues and normal tissues were analyzed by using online tool UALCAN. (G) Expression levels of MFAP2 in BC tissues and normal tissues were analyzed by using online tool GEPIA. (H) Relative expression of MFAP2 in 51 BC tissues and normal controls. (I) Relative expression of MFAP2 in BC cell lines was determined by quantitative real-time PCR. (J) Overall survival rate was analyzed by using an online bioinformatics tool (UALCAN). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
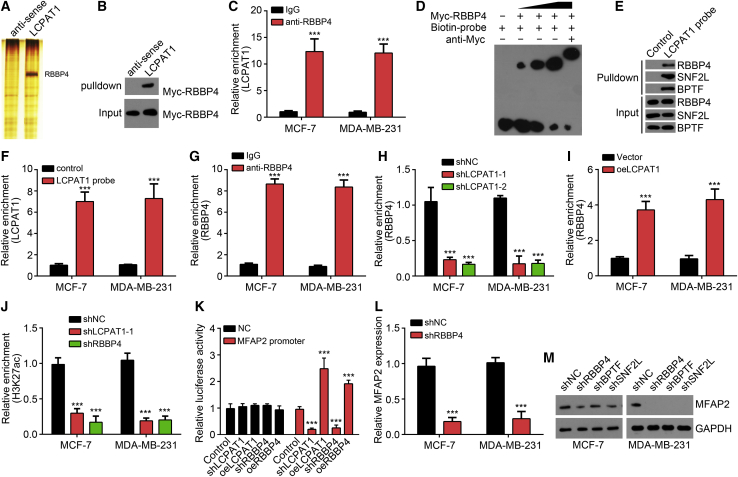
LCPAT1 Interacts with RBBP4 to Initiate MFAP2 Transcription
Afterward, we sought to determine the molecular mechanism in regulating MFAP2 expression. We utilized biotin-labeled LCPAT1 to perform a pull-down assay. The precipitants by biotin-labeled LCPAT1 in MCF-7 cell lysates were separated by SDS-PAGE gel, followed by silver staining. The differential band in the biotin-labeled LCPAT1 lane was identified by mass spectrometry (MS). RBBP4 is an important subunit of the chromatin remodeling complex NURF (Figure 5A). The interaction between LCPAT1 and RBBP4 was validated by a pull-down assay and RNA immunoprecipitation (RIP) assay (Figures 5B and 5C). An electrophoretic mobility shift assay (EMSA) was further conducted to examine their direct interaction. Results showed that RBBP4 could directly interact with LCPAT1 (Figure 5D). Then, the LCPAT1 probes were incubated with MCF-7 cell lysates for a chromatin isolation by RNA purification-western blotting (CHIRP-WB) assay. Results showed that LCPAT1 precipitated RBBP4, SNF2L, and BPTF (three subunits of the NURF complex) (Figure 5E), suggesting that LCPAT1 associated with the NURF complex in BC cells. Then, we analyzed whether LCPAT1 directly bound to the MFAP2 promoter. We performed a CHIRP assay and found that LCPAT1 directly interacted with the promoter of MFAP2 (Figure 5F). Moreover, LCPAT1 interacted with the MFAP2 promoter and the MFAP2 promoter relying on a different region (Figures S1A and S1B). Interestingly, a chromatin immunoprecipitation (ChIP) assay showed that RBBP4 also bound to the same region of the MFAP2 promoter as LCPAT1 (Figure 5G), implying that LCPAT1 and RBBP4 may regulate MFAP2 transcription. Previous studies report that lncRNAs could recruit chromatin remodeler to regulate the accessibility of the gene promoter.21 Also, the NURF complex is a classical activator of gene transcription.10 Thus, we speculated that LCPAT1 might recruit RBBP4 to the MFAP2 promoter. We performed a ChIP assay and found that LCPAT1 knockdown led to reduced association between RBBP4 and the MFAP2 promoter and vice versa (Figures 5H and 5J). Moreover, the enrichment of active histone modification H3K27ac was attenuated on the MFAP2 promoter after knockdown of either LCPAT1 or RBBP4 (Figure 5J). A luciferase reporter assay also showed that knockdown of LCPAT1 or RBBP4 reduced the luciferase activity of the MFAP2 promoter reporter and vice versa, in MCF-7 cells (Figure 5K), suggesting that LCPAT1 promotes MFAP2 transcription through the NURF complex. In effect, the expression of MFAP2 was significantly inhibited by silencing of the NURF complex (Figures 5L and 5M).
Figure 5.
LCPAT1 Interacts with RBBP4 to Initiate MFAP2 Transcription
(A) Biotin-labeled LCPAT1 was incubated with MCF-7 cell lysates for pull-down, followed by SDS-PAGE separation, silver staining, and MS identification. RBBP4 was identified as a LCPAT1 interacting protein. (B) RNA pull-down assay indicated that LCPAT1 interacted with Myc-RBBP4. (C) RIP assay using anti-RBBP4 showed that LCPAT1 was precipitated by anti-RBBP4. (D) An EMSA showed that LCPAT1 directly interacted with RBBP4 in vitro. (E) RNA pulldown assay using LCPAT1 probes indicated that LCPAT1 interacted with the NURF complex subunits, including RBBP4, SNF2L, and BPTF. (F) CHIRP assay showed that LCPAT1 interacted with the promoter of MFAP2. (G) ChIP assay showed that RBBP4 bound to the same region of MFAP2 promoter as LCPAT1. (H) ChIP assay indicated that LCPAT1 knockdown attenuated the interaction between RBBP4 and MFAP2 promoter. (I) ChIP assay indicated that LCPAT1 overexpression promoted the interaction between RBBP4 and MFAP2 promoter. (J) ChIP assay showed that knockdown of either LCPAT1 or RBBP4 reduced the modification of H3K27ac on the MFAP2 promoter. (K) Luciferase reporter assay showed that knockdown of LCPAT1 or RBBP4 suppressed the luciferase activity of the MFAP2 promoter and vice versa in MCF-7 cells. (L and M) Knockdown of RBBP4 suppressed the expression of MFAP2 by quantitative real-time PCR (L) and western blotting (M) analyses. ∗∗∗p < 0.001.
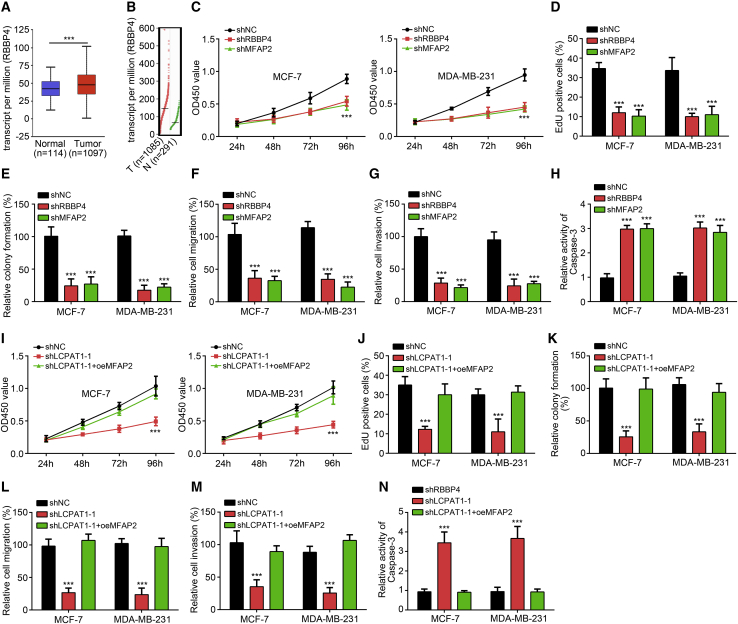
LCPAT1 Regulates BC Progression through Activation of MFAP2
To our knowledge, the roles of RBBP4 and MFAP2 in BC are poorly researched. Bioinformatics analysis showed that RBBP4 was upregulated in BC tissues (Figures 6A and 6B). To investigate the roles of RBBP4 and MFAP2, we performed various experiments. CCK8, EdU incorporation, and colony formation assays showed that knockdown of RBBP4 or MFAP2 dramatically suppressed the proliferation of BC cells (Figures 6C–6E). Also, the migration and invasion were also impaired after silencing of RBBP4 or MFAP2 (Figures 6F and 6G) while cellular apoptosis was increased (Figure 6H). Thus, RBBP4 and MFAP2 acted as oncogenes in BC. To further confirm whether LCPAT1 exerts functions via MFAP2, we restored its expression in LCPAT1-silenced cell lines. CCK8, EdU incorporation, and colony formation assays showed that restoration of MFAP2 increased cellular proliferation rate (Figures 6I–6K). Also, the ability of migration and invasion was also enhanced after MFAP2 restoration (Figures 6L and 6M) while apoptosis was decreased (Figure 6N). Taken together, LCPAT1 associates with RBBP4 to initiate MFAP2 transcription and BC progression.
Figure 6.
LCPAT1 Regulates BC Progression through Activation of MFAP2
(A) Expression value of RBBP4 in BC tissues and normal tissues according to online tool UALCAN. (B) Expression value of RBBP4 in BC tissues and normal tissues according to online tool GEPIA. (C–E) The effects of RBBP4 and MFAP2 on cell proliferation were tested by CCK8 (C), EdU incorporation (D), and colony formation (E) assays. (F and G) Transwell assay was performed to analyze cell migration (F) and invasion (G). (H) Apoptosis was evaluated by measuring the activity of caspase-3. (I–K) Cell proliferation was measured by CCK8 (I), EdU incorporation (J), and colony formation (K) assays after MFAP2 restoration. (L and M) MFAP2 restoration rescued the ability of migration (L) and invasion (M). (N) MFAP2 overexpression decreased the activity of caspase-3. ∗∗∗p < 0.001.
Discussion
In this work, we explored the functions and mechanisms of lncRNA LCPAT1 in BC development. We found that LCPAT1 was highly expressed in BC tissues and correlated with clinical grade, stage, and prognosis. Our data indicated that LCPAT1 knockdown suppressed proliferation, migration, and invasion whereas it promoted apoptosis of BC cells in vitro and vice versa. Moreover, LCPAT1 contributed to BC growth in vivo by xenograft assay. Specifically, we found that LCPAT1 directly interacted with RBBP4 and associated with the MFAP2 promoter. By cooperating with RBBP4, LCPAT1 facilitated MFAP2 transcription and promoted BC progression.
Recently, many studies have indicated the critical roles of lncRNAs in tumor initiation and development.9,13 Some works also show that lncRNAs are potential indicators for tumor diagnosis and prognosis.22 The importance of lncRNAs in BC progression has been emphasized. For instance, NLIPMT upregulation suppresses BC cell proliferation, migration, and invasion.23 lncRNA MEG3 is downregulated in BC tissues and its ectopic expression represses growth of BC cells via phosphatidylinositol 3-kinase (PI3K)/AKT signaling.24 lncRNA-CDC6 promotes BC development via regulating the miR-215/CDC6 axis.25 Additionally, a recent study also showed that LINC01857 transcriptionally activates CREB1 expression through associating with CREBBP protein in BC and promotes tumor progression.26 LCPAT1 has been reported to promote lung cancer cell proliferation, and it participates in DNA damage.17,27 To our knowledge, whether LCPAT1 is involved in BC regulation remains totally unclear. Our study reveals that LCPAT1 is upregulated in BC cells. Moreover, LCPAT1 regulates proliferation, migration, invasion, and apoptosis of BC cells in vitro and in vivo. Its oncogenic roles in BC were first demonstrated by our research.
Previous studies have shown that lncRNAs may regulate the expression of their neighboring genes.21 To explore the downstream signaling, we analyzed the effects of LCPAT1 on the expression of its neighboring genes. Intriguingly, knockdown of LCPAT1 significantly suppressed the expression of MFAP2 in MCF-7 and MDA-MB-231 cells and vice versa. MFAP2 is a component of microfibrils.28 An in vivo assay shows that MFAP2 regulates hemostasis and thrombosis.29 Few studies were conducted to investigate MFAP2 function in cancer. Only one work indicated that MFAP2 promotes gastric cancer progression in vitro.28 In our study, we showed that MFAP2 expression was increased in BC tissues, and its overexpression predicted poor prognosis. Knockdown of MFAP2 caused inhibition of proliferation, migration, and invasion in BC cells, suggesting that LCPAT1/MFAP2 signaling may be involved in BC progression. To further confirm it, rescue assays were performed by overexpression of MFAP2. In fact, restoration of MFAP2 markedly reversed the effects of LCPAT1 knockdown. Thus, our work supports that LCPAT1 regulates BC progression by promoting MFAP2 expression.
Recent studies have suggested that lncRNAs may utilize several mechanisms to play functions, such as sponging microRNAs (miRNAs),25 regulating the stability of specific protein,30 or modifying chromatin structure.10 LCPAT1 was mainly distributed in the nucleus of BC cells, implying it may interact with some proteins to exert effects. In order to elucidate the molecular mechanism of LCPAT1 in regulating MFAP2 expression, a pull-down assay was performed. RBBP4, a chromatin remodeler and an important subunit of the NURF complex, was identified by MS. The NURF complex is a classical activator of gene transcription.10 Several reports indicated that the NURF complex is a tumor initiator.31, 32, 33 Unexpectedly, no study about the role of RBBP4 or the NURF complex in BC has been conducted to date. In this study, we first demonstrated that LCPAT1 and RBBP4 were enriched on the promoter of MFAP2. Moreover, LCPAT1 is indispensable for RBBP4 enrichment on the MFAP2 promoter. Importantly, we did not find any appropriate base paring between the MEAP2 promoter and LCPAT1. Thus, how LCPAT1 interacts with the MEAP2 promoter needs more investigation. Next, ChIP and a luciferase assay illustrated that LCPAT1 recruited RBBP4 to initiate MFAP2 transcription. In effect, silencing of the subunits of the NURF complex led to downregulation of MFAP2 expression. Finally, RBBP4 was found to be upregulated in BC cells. Also, knockdown of RBBP4 also reduced proliferation, migration, and invasion of BC cells. Collectively, LCPAT1 promotes MFAP2 transcription relying on RBBP4 recruitment in BC.
Conclusions
In this study, we identified LCPAT1 as a novel onco-lncRNA in BC and revealed that LCPAT1 promoted BC progression through activation of MFAP2 transcription via recruiting the chromatin remodeler RBBP4. Our work illustrates a new mechanism involved in BC progression and suggests that LCPAT1 may be a new therapeutic target.
Materials and Methods
Human Samples
51 BC samples and corresponding normal control tissues were collected from our hospital. Patients were not treated with chemotherapy or radiotherapy before tissue collection. Tissues were stored in liquid nitrogen. This study was approved by the Ethics Committee of our hospital. Also, the participants provided written informed consent.
Cell Culture and Transfection
Human BC cell lines and the normal breast epithelial cell line MCF-10A were from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences, Shanghai Institute of Biochemistry and Cell Biology. Cells were cultured according to the manufacturer’s instructions. Transfection was completed using Lipofectamine 3000 (Thermo Fisher Scientific). For construction of stably LCPAT1-silenced cell lines, shRNAs targeting LCPAT1 (#1: 5′-CAATGTTGTTGTTTATTTA-3′, and #2: 5′-AGACAATTACAGCACTAAA-3′) were cloned into the pLKO.1 vector. Then, shRNA-expressing plasmids and packaging plasmids were transfected into 293T cells for lentivirus production. After lentivirus infection, the stable cell lines were screened. LCPAT1 knockdown efficiency was validated by quantitative real-time PCR. For LCPAT1 overexpression, the sequence of LCPAT1 was inserted into pCDNA3-puro vector. After transfection, stable cell lines were screened.
Quantitative Real-Time PCR
RNA isolation and quantitative real-time PCR analysis were performed according to a previous study.18
Cell Proliferation Detection
For CCK8 assay, 2,000 cells per well were seeded into the 96-well plates and cultured for the indicated times. Then, 10 μL of CCK8 solution (Dojindo, Japan) was added into each plate and incubated for 2 h. The absorbance at 450 nm was measured by a microplate reader (BioTek Instruments, Winooski, VT, USA).
An EdU incorporation assay was performed as described before.19
For colony formation assays, 500 cells per well were seeded into six-well plates and cultured for 14 days. Then, clones were fixed and stained using crystal violet. Colony numbers were counted.
Transwell Assay
Cell migration and invasion were examined by a Transwell assay using 24-well Transwell chambers with an 8-μm pore size and a track-etched membrane (Corning, New York, NY, USA) as previously described.20
FISH
A FISH assay was performed as described before.16 In brief, the probes targeting LCPAT1 were purchased from Sangon Biotech. Then, cells were fixed using 4% formaldehyde for 10 min and washed with PBS buffer followed by permeabilization using 70% EtOH. Then, the probes were added and hybridized overnight. The imaging was determined using a fluorescence microscope.
RIP and ChIP
The RIP and ChIP assay were carried out according to a previous work.16
In Vivo Animal Experiment
The female BALB/c nude mice (4 weeks old) were randomly divided into two or three subgroups (n = 5 for each group). MDA-MB-231 cells (shRNA negative control [shNC] or shLCPAT1) were subcutaneously injected into the flanks. Then, the tumor volumes were measured at indicated time points and tumor weights were determined on day 30. Animal experiments were approved by the Ethics Committee of our hospital.
Statistical Analysis
The statistical analysis was carried out with GraphPad Prism version 6.0 software. Results were displayed as the mean ± standard deviation (SD). The t test and ANOVA were used for calculating statistical difference. The Kaplan-Meier method and the log-rank test were used to analyze survival rate. p values <0.05 were considered as significant. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author Contributions
X.G. performed experiments; T.D., M.N., X.L., S.S., and Y.Z. analyzed the data; Y.L. and D.L. wrote this manuscript. All authors have read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the Haiyan Fund, Harbin Medical University Cancer Hospital (JJZD2020-11).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.07.015.
Contributor Information
Yue Li, Email: liyue_0617@163.com.
Dalin Li, Email: dalinli2@sina.com.
Supplemental Information
References
- 1.Forouzanfar M.H., Foreman K.J., Delossantos A.M., Lozano R., Lopez A.D., Murray C.J.L., Naghavi M. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet. 2011;378:1461–1484. doi: 10.1016/S0140-6736(11)61351-2. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Jia X., Shi L., Wang X., Luo L., Ling L., Yin J., Song Y., Zhang Z., Qiu N., Liu H. KLF5 regulated lncRNA RP1 promotes the growth and metastasis of breast cancer via repressing p27kip1 translation. Cell Death Dis. 2019;10:373. doi: 10.1038/s41419-019-1566-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Chin K., DeVries S., Fridlyand J., Spellman P.T., Roydasgupta R., Kuo W.L., Lapuk A., Neve R.M., Qian Z., Ryder T. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Sørlie T., Perou C.M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M.B., van de Rijn M., Jeffrey S.S. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adem C., Reynolds C., Ingle J.N., Nascimento A.G. Primary breast sarcoma: clinicopathologic series from the Mayo Clinic and review of the literature. Br. J. Cancer. 2004;91:237–241. doi: 10.1038/sj.bjc.6601920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 8.Bartonicek N., Maag J.L., Dinger M.E. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol. Cancer. 2016;15:43. doi: 10.1186/s12943-016-0530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mondal T., Subhash S., Vaid R., Enroth S., Uday S., Reinius B., Mitra S., Mohammed A., James A.R., Hoberg E. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015;6:7743. doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu B., Ye B., Yang L., Zhu X., Huang G., Zhu P., Du Y., Wu J., Qin X., Chen R. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat. Immunol. 2017;18:499–508. doi: 10.1038/ni.3712. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Z.L., Zhang J., Chen M.L., Li K. Efficacy and safety of trastuzumab added to standard treatments for HER2-positive metastatic breast cancer patients. Asian Pac. J. Cancer Prev. 2013;14:7111–7116. doi: 10.7314/apjcp.2013.14.12.7111. [DOI] [PubMed] [Google Scholar]
- 12.Schmitt A.M., Chang H.Y. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang X., Li Q., Zhang S., Song C., Zheng P. Long noncoding RNA GIHCG induces cancer progression and chemoresistance and indicates poor prognosis in colorectal cancer. OncoTargets Ther. 2019;12:1059–1070. doi: 10.2147/OTT.S192290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He J., Zuo Q., Hu B., Jin H., Wang C., Cheng Z., Deng X., Yang C., Ruan H., Yu C. A novel, liver-specific long noncoding RNA LINC01093 suppresses HCC progression by interaction with IGF2BP1 to facilitate decay of GLI1 mRNA. Cancer Lett. 2019;450:98–109. doi: 10.1016/j.canlet.2019.02.033. [DOI] [PubMed] [Google Scholar]
- 15.Hui B., Xu Y., Zhao B., Ji H., Ma Z., Xu S., He Z., Wang K., Lu J. Overexpressed long noncoding RNA TUG1 affects the cell cycle, proliferation, and apoptosis of pancreatic cancer partly through suppressing RND3 and MT2A. OncoTargets Ther. 2019;12:1043–1057. doi: 10.2147/OTT.S188396. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Dong H., Hu J., Zou K., Ye M., Chen Y., Wu C., Chen X., Han M. Activation of lncRNA TINCR by H3K27 acetylation promotes trastuzumab resistance and epithelial-mesenchymal transition by targeting microRNA-125b in breast cancer. Mol. Cancer. 2019;18:3. doi: 10.1186/s12943-018-0931-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Yu X., Ye X., Lin H., Feng N., Gao S., Zhang X., Wang Y., Yu H., Deng X., Qian B. Knockdown of long non-coding RNA LCPAT1 inhibits autophagy in lung cancer. Cancer Biol. Med. 2018;15:228–237. doi: 10.20892/j.issn.2095-3941.2017.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y., Cheng Q., Liu J., Dong M. leukemia stem cell-released microvesicles promote the survival and migration of myeloid leukemia cells and these effects can be inhibited by microRNA34a overexpression. Stem Cells Int. 2016;2016:9313425. doi: 10.1155/2016/9313425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan L., Mai D., Zhang B., Jiang X., Zhang J., Bai R., Ye Y., Li M., Pan L., Su J. PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Mol. Cancer. 2019;18:9. doi: 10.1186/s12943-019-0940-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li R.H., Chen M., Liu J., Shao C.C., Guo C.P., Wei X.L., Li Y.C., Huang W.H., Zhang G.J. Long noncoding RNA ATB promotes the epithelial-mesenchymal transition by upregulating the miR-200c/Twist1 axe and predicts poor prognosis in breast cancer. Cell Death Dis. 2018;9:1171. doi: 10.1038/s41419-018-1210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu T., Han Z., Li H., Zhu Y., Sun Z., Zhu A. lncRNA DLEU1 contributes to colorectal cancer progression via activation of KPNA3. Mol. Cancer. 2018;17:118. doi: 10.1186/s12943-018-0873-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quan J., Pan X., Zhao L., Li Z., Dai K., Yan F., Liu S., Ma H., Lai Y. lncRNA as a diagnostic and prognostic biomarker in bladder cancer: a systematic review and meta-analysis. OncoTargets Ther. 2018;11:6415–6424. doi: 10.2147/OTT.S167853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y., Lin L., Zhong S., Cai Y., Zhang F., Wang X., Miao R., Zhang B., Gao S., Hu X. Overexpression of novel lncRNA NLIPMT inhibits metastasis by reducing phosphorylated glycogen synthase kinase 3β in breast cancer. J. Cell. Physiol. 2019;234:10698–10708. doi: 10.1002/jcp.27738. [DOI] [PubMed] [Google Scholar]
- 24.Zhu M., Wang X., Gu Y., Wang F., Li L., Qiu X. MEG3 overexpression inhibits the tumorigenesis of breast cancer by downregulating miR-21 through the PI3K/Akt pathway. Arch. Biochem. Biophys. 2019;661:22–30. doi: 10.1016/j.abb.2018.10.021. [DOI] [PubMed] [Google Scholar]
- 25.Kong X., Duan Y., Sang Y., Li Y., Zhang H., Liang Y., Liu Y., Zhang N., Yang Q. lncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J. Cell. Physiol. 2019;234:9105–9117. doi: 10.1002/jcp.27587. [DOI] [PubMed] [Google Scholar]
- 26.Xiong Y., Gu Y., Wang F., Li L., Zhu M., Wang N., Mi H., Qiu X. LINC01857 as an oncogene regulates CREB1 activation by interacting with CREBBP in breast cancer. J. Cell. Physiol. 2019;234:14031–14039. doi: 10.1002/jcp.28090. [DOI] [PubMed] [Google Scholar]
- 27.Gao S., Lin H., Yu W., Zhang F., Wang R., Yu H., Qian B. lncRNA LCPAT1 is involved in DNA damage induced by CSE. Biochem. Biophys. Res. Commun. 2019;508:512–515. doi: 10.1016/j.bbrc.2018.11.171. [DOI] [PubMed] [Google Scholar]
- 28.Wang J.K., Wang W.J., Cai H.Y., Du B.B., Mai P., Zhang L.J., Ma W., Hu Y.G., Feng S.F., Miao G.Y. MFAP2 promotes epithelial-mesenchymal transition in gastric cancer cells by activating TGF-β/SMAD2/3 signaling pathway. OncoTargets Ther. 2018;11:4001–4017. doi: 10.2147/OTT.S160831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Werneck C.C., Vicente C.P., Weinberg J.S., Shifren A., Pierce R.A., Broekelmann T.J., Tollefsen D.M., Mecham R.P. Mice lacking the extracellular matrix protein MAGP1 display delayed thrombotic occlusion following vessel injury. Blood. 2008;111:4137–4144. doi: 10.1182/blood-2007-07-101733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu P., Wang Y., Huang G., Ye B., Liu B., Wu J., Du Y., He L., Fan Z. lnc-β-Catm elicits EZH2-dependent β-catenin stabilization and sustains liver CSC self-renewal. Nat. Struct. Mol. Biol. 2016;23:631–639. doi: 10.1038/nsmb.3235. [DOI] [PubMed] [Google Scholar]
- 31.Ding L., Zhao Y., Dang S., Wang Y., Li X., Yu X., Li Z., Wei J., Liu M., Li G. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol. Cancer. 2019;18:45. doi: 10.1186/s12943-019-1006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao X., Zheng F., Li Y., Hao J., Tang Z., Tian C., Yang Q., Zhu T., Diao C., Zhang C. BPTF promotes hepatocellular carcinoma growth by modulating hTERT signaling and cancer stem cell traits. Redox Biol. 2019;20:427–441. doi: 10.1016/j.redox.2018.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Zhu P., Wu J., Wang Y., Zhu X., Lu T., Liu B., He L., Ye B., Wang S., Meng S. LncGata6 maintains stemness of intestinal stem cells and promotes intestinal tumorigenesis. Nat. Cell Biol. 2018;20:1134–1144. doi: 10.1038/s41556-018-0194-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.