Abstract
Growing evidence has shown that exercise can affect the gut microbiota. The effects of exercise frequency on the gut microbiota in elderly individuals are still largely unknown. In the present study, samples from 897 elderly and 1,589 adult individuals (18–60 years old) from the American Gut Project were screened. Microbial diversity and composition were analyzed by QIIME2, and microbial function was predicted by PICRUSt2. The outcomes were further analyzed by STAMP. The analysis showed that the α‐diversity of gut microbiota increased with increasing age, and regular exercise reshaped the alterations in microbial composition and function induced by aging. Moreover, the α‐diversity of gut microbiota was higher in overweight elderly individuals than in normoweight elderly individuals, and regular exercise significantly affected the microbial composition and function in overweight elderly individuals. In conclusion, we revealed that regular exercise benefits elderly individuals, especially overweight elderly individuals, by modulating the gut microbiota.
Keywords: elderly, exercise, gut microbiota, overweight
This study investigated whether microbial diversity increases with increasing age. The gut microbiota composition of daily exercising elderly individuals approached that of adults (18–60 years old), and regular exercise resulted in an increased relative abundance of bacterial functional pathways. We also showed that overweight elderly individuals had increased microbial diversity and significant changes in the microbial composition.

1. INTRODUCTION
In the intestine, hundreds of millions of bacteria play essential roles in host health (Bäckhed et al., 2004). The gut microbiota is composed of various microorganisms that form a complex ecological balance. The complexity and diversification of the gut microbiota benefit the host in many ways, such as by providing resistance against potential pathogens and increasing immunity (Round & Mazmanian, 2009). Metabolites produced by bacteria mediate host nutrition absorption and health. Thus, an imbalance in the gut microbiota could result in various disease pathogenicities, such as obesity and diabetes mellitus (Tremaroli & Bäckhed, 2012). The gut microbiota can be regulated by a range of factors, such as diet, antibiotic use, diseases, and exercise (Nicholson et al., 2012). Therefore, the gut microbiota is becoming a novel target for disease therapies.
Exercise is an important means to delay aging and prevent and manage diseases (Biddle & Batterham, 2015). Exercise can effectively reduce the risks of heart disease, stroke, hypertension, diabetes, cancer, and osteoporosis and improve depression (Biddle & Batterham, 2015). Exercise is beneficial to health by reducing body mass, the body mass index (BMI), the fat mass percentage, the fasting glucose level, and the fasting insulin level (Biddle & Batterham, 2015). Accumulating evidence shows that exercise affects the gut microbial composition, which might play a positive role in energy regulation (Mach & Fuster‐Botella, 2017; Monda et al., 2017). Animal studies have shown that exercise improves the diversity of the gut microbiome and changes the composition of bacteria in the gut (Brandt et al., 2018; Feng et al., 2017). Studies show that exercise could change the microbial composition at the phylum level, affecting the abundance of phyla such as Firmicutes, Proteobacteria, Prevotella, and Cyanobacteria (Lambert et al., 2015; Liu et al., 2015). The families Christensenellaceae and Coriobacteriaceae have shown increased abundance, suggesting a link between exercise and health improvement (Liu et al., 2015; Zhao et al., 2018). Furthermore, exercise training increases the production of short‐chain fatty acids (Feng et al., 2017). In humans, following an exercise challenge, the gut microbiota diversity and microbial composition are altered (Taniguchi et al., 2018; Whisner, Maldonado, Dente, Krajmalnik‐Brown, & Bruening, 2018). Similar results have been shown in athletes. Athletes have increased microbial diversity, which might be correlated with metabolic improvement and an inflammation reduction (Barton et al., 2018; Clarke et al., 2014). Thus, the exercise–gut microbiota axis might play an important role in maintaining health.
Overweight (BMI > 25) is becoming a global health problem. Exercise helps regulate body weight, as it is inversely associated with weight gain and contributes to weight loss (Jakicic, Rogers, Davis, & Collins, 2018). In high‐fat diet (HFD)‐induced obese mice, exercise counteracted the microbial imbalance, which was distinct from dietary effects, protected the intestinal barrier, and improved bile acid homeostasis (Carbajo‐Pescador et al., 2019; Evans et al., 2014). Moreover, genetic capacity studies predict that exercise increases tricarboxylic acid (TCA) cycle metabolism (Denou, Marcinko, Surette, Steinberg, & Schertzer, 2016). In humans, exercise‐induced alterations in microbial beta diversity depend on obesity status (Allen et al., 2018).
The gut microbiota undergoes aging‐related changes that may affect health. An early study showed that γ‐proteobacteria were enriched, while Ruminococcus obeum and its closely related phylotypes were hardly detected in elderly individuals (Hayashi, Sakamoto, Kitahara, & Benno, 2003). Studies have shown that the microbial community structure of elderly individuals is significantly different from that of young individuals (Claesson et al., 2011). Shen et al. (2018) found that the microbial changes caused by aging were characterized by reduced Bacteroidetes abundance. Ogawa, T., et al. suggested that adequate exercise was important for maintaining health because it manipulates the gut microbiota in elderly individuals (Shimizu, 2018). However, the effects of exercise frequency on the gut microbiota of elderly individuals remain largely unknown. The present study was carried out to characterize regular exercise‐induced changes in the properties of the gut microbiota in elderly individuals and to further analyze regular exercise‐induced changes that benefit overweight elderly (OE) individuals.
2. METHODS
2.1. Data sources
The data for this study were obtained from the American Gut Project (AGP) (McDonald et al., 2018). This project was the largest crowd‐funded research project created by Rob Knight and Jeff Leach. Samples taken from participants were kept at the Rob Knight Laboratory at the University of California, San Diego. DNA was extracted from the samples and then sequenced using an Illumina MiSeq sequencer.
The sample data were obtained from the Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra), in which 25,376 samples from the American Gut Project were deposited, including 19,988 samples collected from fecal specimens. Upon excluding the samples without basic information (sex, age, BMI, or self‐reported exercise frequency), from patients who received antibiotic treatment within 6 months, from patients with tumors, and from patients who went on a trip within the previous three months, the data of 3,795 samples remained. Of these, samples from 897 elderly individuals and 1,589 18‐ to 60‐year‐old adults (adults18‐60) with normoweight were included in the study.
2.2. Experimental design
To determine the alterations in gut microbiota with age, 1,589 samples from adults18‐60 and 462 samples from elderly individuals with a normal BMI were divided into 6 groups according to age (approximately ten years per age group): the 18‐ to 30‐year‐old group (329 individuals), 31‐ to 40‐year‐old group (433 individuals), 41‐ to 50‐year‐old group (428 individuals), 51‐ to 60‐year‐old group (399 individuals), 61‐ to 70‐year‐old group (371 individuals), and over 70‐year‐old group (91 individuals; Table 1).
TABLE 1.
Number of samples used in different groups
| Adults18−60 (n = 1,589) | Normoweight | Overweight | Underweight |
|---|---|---|---|
| 18‐ to 30‐year‐old | 329 | ||
| 31‐ to 40‐year‐old | 433 | ||
| 41‐ to 50‐year‐old | 428 | ||
| 51‐ to 60‐year‐old | 399 |
| Elderly (n = 897) | Normoweight | Overweight | Underweight |
|---|---|---|---|
| 462 | 413 | 22 | |
| 61‐ to 70‐year‐old | 371 | ||
| over 70‐year‐old | 91 |
| Exercise frequency in elderly (n = 897) | Exercise in overweight elderly (n = 413) | ||
|---|---|---|---|
| Daily exercise | 194 | Daily or regular exercise | 74 |
| Regular exercise | 360 | Never or rare exercise | 222 |
| Occasional exercise | 209 | ||
| Rare exercise | 102 | ||
| Never exercise | 32 | ||
To define the benefit of exercise in elderly individuals, the 897 elderly individual samples were divided into the daily exercise group (194 individuals), regular exercise group (360 individuals), occasional exercise group (209 individuals), rare exercise group (102 individuals), and never exercise group (32 individuals; Table 1).
To determine the alterations in gut microbiota with BMI in elderly individuals, elderly individual samples were further divided into 3 groups according to BMI: the underweight elderly group (BMI < 18.5) (22 elderly individuals), normoweight elderly (NE) group (18.5 < BMI<25) (462 elderly individuals), and overweight elderly (OE) group (BMI > 25) (413 elderly individuals; Table 1).
To define the benefit of exercise in OE individuals, the OE group was further divided into the daily or regular exercise in OE individuals (DROE) group (74 elderly individuals) and the never or rare exercise in OE individuals (NROE) group (222 elderly individuals). OE individuals who occasionally exercised (117) were excluded from the analysis (Table 1).
2.3. FASTQ format conversion
The SRA files were downloaded to a computer, and the sratoolkit tools were used to convert the SRA data into the fastq format. The fastq‐dump.exe command was used to convert the format.
2.4. Data processing
QIIME version 2 was utilized to process the data (Brandt et al., 2018). First, the Deblur plugin was used for sequence quality control. The operational taxonomic units (OTUs) resulting from Deblur in QIIME2 were created by grouping unique sequences that had the equivalent of 100% similarity to OTUs in QIIME1 (Brandt et al., 2018). The feature‐table‐table seqs command mapped feature IDs to sequences. The QIIME diversity alpha‐rarefaction visualizer was utilized to explore alpha diversity. Next, the taxonomic composition of the samples was explored. The naive Bayes classifier and the q2‐feature‐classifier plugin with the Greengenes 13.8 database were used to assign taxonomy to the sequences and map the sequences. The relative abundances at the phylum and family levels were determined based on OTU tables.
PICRUSt2 was used to predict metagenomic functions based on the normalized OTU tables (Douglas et al., 2020).
2.5. Statistical analysis
STAMP was used to calculate the level of significance (Parks, Tyson, Hugenholtz, & Beiko, 2014). Welch's t tests (two‐sided) were used for two‐group comparisons. Welch's inverted confidence interval (CI) method was used for CI calculation. The Benjamini–Hochberg false discovery rate method was used to calculate adjusted p‐values.
2.6. Patient and public involvement statement
Patients were not included in the sampling for this study.
3. RESULTS
3.1. Microbial composition was altered by age
Of the 25,376 samples screened by the American Gut Project, samples from 897 elderly individual samples and 1,589 adults18‐60 (18–60 years old) with a normal BMI (18.5 to 25) were analyzed in this study. The α‐diversity analysis showed that the OTU numbers were 164.3, 168.6, 181.6, 186.3, 181.0, and 191.7 (p < .0001), while the Shannon indices were 4.989, 5.083, 5.222, 5.309, 5.203, and 5.391 (p < .0001) in the 18‐ to 30‐year‐old group, 31‐ to 40‐year‐old group, 41‐ to 50‐year‐old group, 51‐ to 60‐year‐old group, 61‐ to 70‐year‐old group, and over the 70‐year‐old group, respectively (Figure 1a,b). Thus, the microbial α‐diversity significantly increased with age.
FIGURE 1.

Alteration of the microbial composition in elderly individuals. Both (a) OTU number and (b) Shannon index showed that the microbial α‐diversity increased with age in 2,011 people with a normal BMI. Comparing the microbial composition in elderly individuals to that in adults18‐60, (c) three phyla were significantly changed. (d) Twenty‐seven families were significantly changed in the microbial composition in elderly individuals with different exercise frequencies compared with that in adults18‐60
Since α‐diversity significantly changed with age, we detected the gut microbiota composition in elderly individuals and compared it with that of adults18‐60 (Figure 1c,d). At the phylum level, the relative abundances of Proteobacteria and Verrucomicrobia were significantly increased by 20.3% (p < .05) and 71.1% (p < .05), respectively, while the relative abundance of Actinobacteria was significantly decreased by 33.3% (p < .05) in the elderly individuals compared with the adults18‐60. At the family level, the relative abundances of Aerococcaceae, Bifidobacteriaceae, Lactobacillaceae, Pasteurellaceae, Prevotellaceae, Tissierellaceae, Turicibacteraceae, and Veillonellaceae were significantly increased by 70.1%, 64.6%, 60.6%, 46.8%, 27.7%, 55.8%, 45.8%, and 29.6%, respectively (p < .05), while the relative abundances of Barnesiellaceae, Christensenellaceae, Desulfovibrionaceae, Enterobacteriaceae, Odoribacteraceae, Oxalobacteraceae, Rikenellaceae, and Verrucomicrobiaceae were significantly decreased by 31.3%, 101.5%, 20.4%, 46.8%, 58.5%, 70.2%, 38.7%, and 75.8%, respectively (p < .05), in the elderly individuals compared with the adults18‐60.
3.2. Microbial composition and function were altered by exercise frequency
To determine the effect of exercise frequency on microbial α‐diversity in elderly individuals, elderly individual samples were divided into 5 groups (never, rare (a few times/month), occasional (1–2 times/week), regular (3–5 times/week), and daily). The analysis showed that the OTU numbers were 194.9, 195.5, 196.3, 186.7, and 191.9 (p > .05), while the Shannon indices were 5.478, 5.466, 5.489, 5.351, and 5.332, respectively (p > .05; Figure 2a,b). Thus, these results suggested that the microbial α‐diversity was almost unaffected by exercise frequency in elderly individuals.
FIGURE 2.
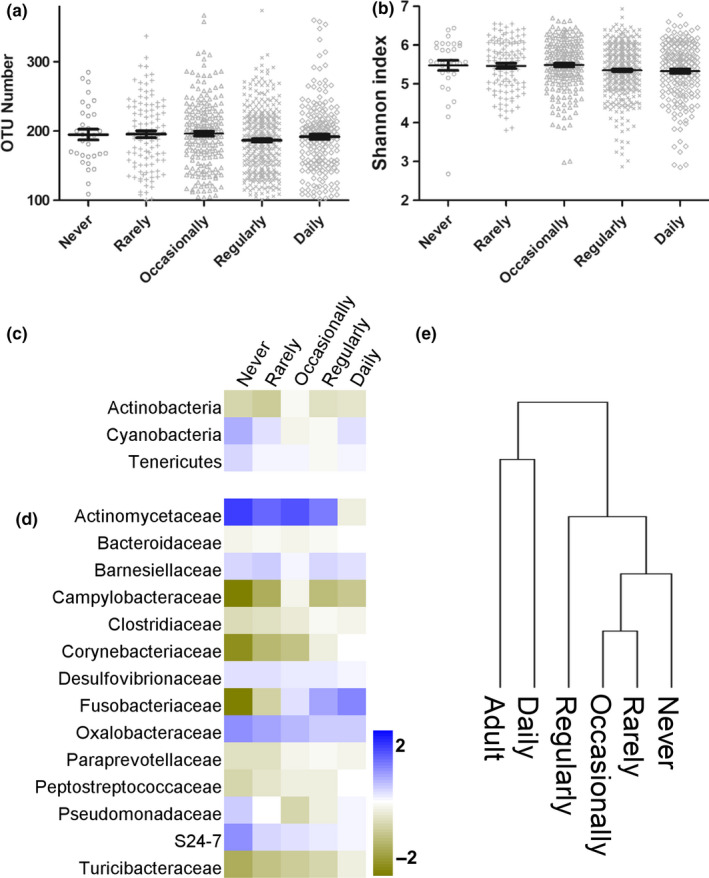
Alteration of microbial function in the DRE group. Both (a) OTU number and (b) Shannon index showed that exercise frequency affected α‐diversity in elderly individuals. Exercise‐induced 3 phyla (c) and 14 families (d) to gradually approach the levels of those in adults18‐60. (e) Evolutionary analysis of the microbial composition at the family level
To define the benefit of exercise in elderly individuals, the microbial abundances of the never, rare, occasional, regular, and daily exercise groups were compared with that of adults18‐60 (Figure 2c,d). At the phylum level, the relative abundances of Actinobacteria, Cyanobacteria, and Tenericutes in elderly individuals gradually approached those in adults18‐60. At the family level, the relative abundances of Actinomycetaceae, Desulfovibrionaceae, S24‐7, Pseudomonadaceae, Barnesiellaceae, and Oxalobacteraceae gradually decreased with exercise frequency. The relative abundances of Campylobacteraceae, Fusobacteriaceae, Turicibacteraceae, Paraprevotellaceae, Clostridiaceae, Peptostreptococcaceae, Corynebacteriaceae, and Bacteroidaceae gradually increased with exercise frequency. As shown in Figure 2e, the evolutionary analysis revealed that the abundances in the daily exercise group were the closest to those in the adults18‐60 at both the phylum and family levels.
For analysis of microbial function, elderly individual samples were recombined into the daily or regular exercise (DRE) group and never or rare exercise (NRE) group (Figure 3). Twenty‐four pathways were identified in the comparison of the DRE and NRE groups (p < .05), including 1 vitamin‐related pathway (8‐amino‐7‐oxononanoate biosynthesis I), 12 nucleotide metabolism‐related pathways (purine nucleotides de novo biosynthesis I, guanosine nucleotides de novo biosynthesis I, pyrimidine deoxyribonucleoside salvage, pyrimidine deoxyribonucleotide phosphorylation, pyrimidine ribonucleosides salvage, pyrimidine deoxyribonucleotides de novo biosynthesis I, purine nucleotides degradation II (aerobic), guanosine nucleotides de novo biosynthesis II, pyrimidine deoxyribonucleotides de novo biosynthesis (E. coli), pyrimidine ribonucleotides de novo biosynthesis, purine ribonucleosides degradation, and purine nucleotides de novo biosynthesis II), 2 cell wall biosynthesis‐related pathways (UDP‐N‐acetylglucosamine‐derived O‐antigen building blocks biosynthesis and UDP‐2,3‐diacetamido‐2,3‐dideoxy‐α‐D‐mannuronate biosynthesis), 7 glucose metabolism‐related pathways (glycolysis II (from fructose 6‐phosphate), pentose phosphate pathway, (R,R)‐butanediol biosynthesis, pyruvate fermentation to propanoate I, glycolysis, β‐D‐glucuronosides degradation, and hexuronide and hexuronate degradation), and 2 amino acid metabolism‐related pathways (l‐glutamate and l‐glutamine biosynthesis and urea cycle). The relative abundances of 18 of those pathways were significantly higher, while the abundances of 5 of those pathways were significantly lower in the DRE group than in the NRE group. Therefore, these results suggest that regular exercise significantly modulated microbial function in elderly individuals.
FIGURE 3.

Alteration of microbial function in the DRE group. Twenty‐nine pathways were significantly changed in the DROE group compared with the NROE group. The 29 pathways were associated with vitamin, nucleotide, glucose, and amino acid metabolism
3.3. Microbial α‐diversity was altered by regular exercise in OE individuals
The analysis showed that the OTU numbers were 165.5, 184.3, and 200.6 (p < .001), while the Shannon indices were 5.054, 5.244, and 5.508 (p < .001) in the underweight elderly, NE, and OE groups, respectively (Figure 4a,b). Thus, microbial α‐diversity significantly increased with BMI.
FIGURE 4.
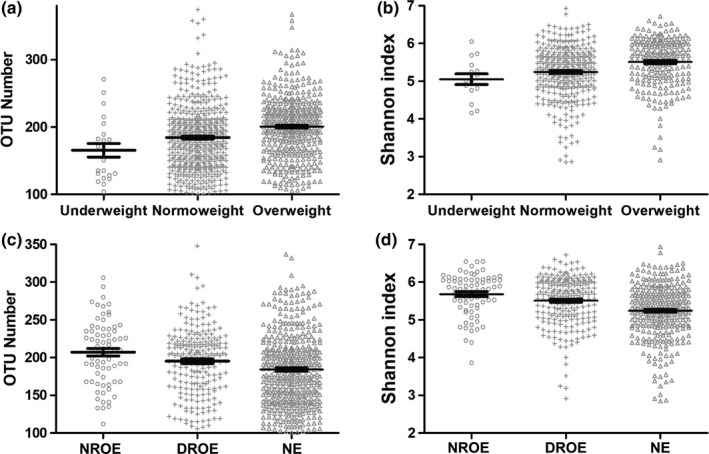
Alteration of the microbial α‐diversity in OE individuals. Both (a) OTU number and (b) Shannon index showed that the microbial α‐diversity increased with BMI. Both (c) OTU number and (d) Shannon index showed that the DROE group had altered microbial α‐diversity
The α‐diversity analysis showed that the OTU numbers were 207.2 and 195.2 (p < .001), while the Shannon indices were 5.681 and 5.508 (p < .001) in the DROE and NROE groups, respectively (Figure 4c,d). Thus, the microbial α‐diversity was significantly affected by exercise in OE individuals.
3.4. Microbial abundance was partially restored by regular exercise in OE individuals
Next, the abundances of gut microbiota constituents were detected at the phylum and family levels in OE individuals (Figure 5). At the phylum level, the relative abundances of Cyanobacteria, Firmicutes, and Verrucomicrobia were increased by 165.7%, 6.1%, and 36.3% (p < .05), respectively, while the Bacteroidetes abundance was decreased by 6.7% in OE individual samples (p < .05) compared with (NE) individual samples. At the family level, the relative abundances of Verrucomicrobiaceae, S24‐7, Prevotellaceae, Oxalobacteraceae, and Lachnospiraceae were increased by 34.2%, 52.7%, 40.4%, 102.4%, and 15.1%, respectively, while the relative abundances of Enterobacteriaceae, Christensenellaceae, Barnesiellaceae, Bacteroidaceae, Porphyromonadaceae, and Peptococcaceae were decreased by 18.1%, 24.3%, 21.4%, 17.2%, 20.3%, and 31.9%, respectively, in the OE individuals (p < .05) compared with the NE individuals.
FIGURE 5.
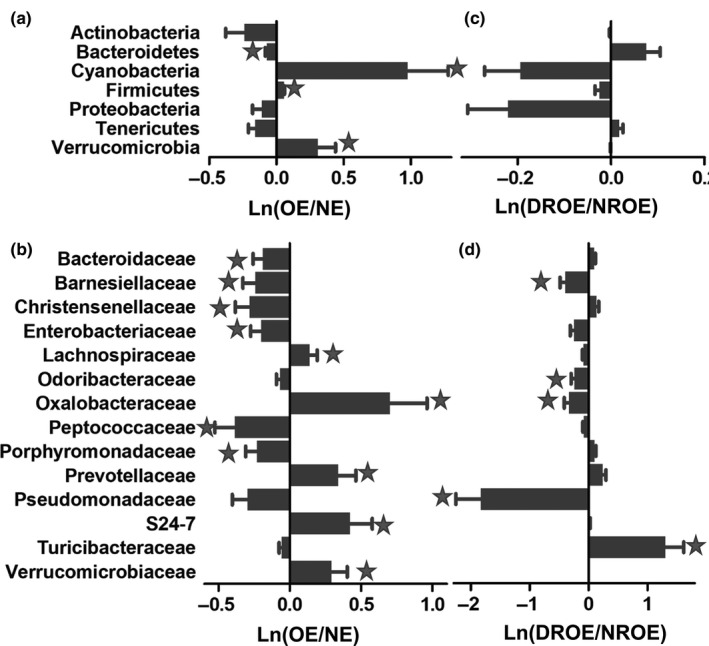
Alteration of the microbial composition in OE individuals. Comparison of the microbial compositions in OE and NE individuals at the (a) phylum and (b) family levels. Comparison of the microbial composition in the DROE and NROE groups at the (c) phylum and (d) family levels. The star represents p < .05
As shown in Figure 5, exercise affected the gut microbial composition in OE individuals. At the phylum level, the relative abundances of Bacteroidetes, Cyanobacteria, Firmicutes, Tenericutes, and Verrucomicrobia were partially restored in exercising (p > .05) compared with nonexercising OE individual samples. At the family level, the relative abundance of Turicibacteraceae was significantly increased, while the relative abundances of Pseudomonadaceae, Oxalobacteraceae, Odoribacteraceae, and Barnesiellaceae were significantly decreased in the frequently exercising OE individuals compared with nonexercising OE individuals.
3.5. Microbial functions were partially restored by regular exercise in OE individuals
Microbial functions were detected in OE individuals, and it was found that the relative abundances of 129 pathways were significantly changed (Appendix 1: Table A1). The relative abundances of 79 pathways were significantly increased, while those of 50 pathways were significantly decreased in the overweight group compared with the normoweight groups. Moreover, 25 pathways were identified by comparing the DROE and NROE groups (Appendix 2: Table A2). Among them, 19 pathways were significantly higher and 6 pathways were significantly lower in the DROE group than in the NROE group.
Notably, 13 common pathways were identified (Figure 6). Among them, 12 pathways that were changed by overweight were restored by frequent exercise. The abundances of purine nucleotides de novo biosynthesis II, pyrimidine deoxyribonucleotides de novo biosynthesis, pyrimidine ribonucleotides de novo biosynthesis purine nucleotides de novo biosynthesis I, guanosine nucleotides de novo biosynthesis I, pyrimidine deoxyribonucleoside salvage, pyrimidine deoxyribonucleosides salvage, pyrimidine deoxyribonucleotide phosphorylation, pyrimidine ribonucleosides salvage, guanosine nucleotides de novo biosynthesis II, and pyruvate fermentation to propanoate I were significantly increased in OE individuals and significantly decreased in those in the frequent exercise group. The abundance of l‐methionine biosynthesis was significantly decreased in OE individuals and significantly increased in the frequent exercise group.
FIGURE 6.
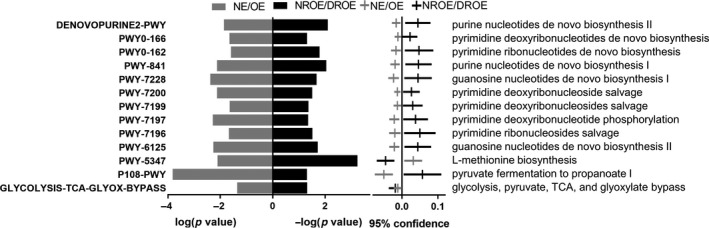
Alteration of microbial function in OE individuals. Thirteen pathways showed opposite trends in the NE/OE and DROE/NROE groups
4. DISCUSSION
The gut microbiota plays a major role in age‐related diseases. Moreover, changes in gut microbiota diversity have profound impacts on host metabolism. In the present study, we detected the effect of exercise on the gut microbiota in elderly individuals by using data obtained from the American Gut Project. The microbial α‐diversity increased with age in the normal BMI population, and the gut microbiota was restored by exercise in elderly individuals.
A recent study showed that microbial α‐diversity is positively associated with age in populations such as the United States, the United Kingdom, and Colombia (de la Cuesta‐Zuluaga et al., 2019). Kasai et al. (2015) showed that bacterial diversity was significantly higher in obese individuals than in nonobese individuals in a Japanese population. The Shannon index was increased in those with an obese BMI in a randomly selected Alabama resident study (Davis, Yadav, Barrow, & Robertson, 2017). Infants who were obese at 6 months of age had higher levels of alpha diversity than nonobese infants (Ville, Levine, Zhi, Lararia, & Wojcicki, 2020). However, some studies showed that obesity/overweight was associated with decreased α‐diversity in animals or humans (Chen et al., 2020; Da Silva, Monteil, & Davis, 2020; van der Merwe et al., 2020). Moreover, a recent study showed no differences in the α‐diversity in obese individuals in an Asian population (Koo et al., 2019). In this study, we analyzed the association of gut microbial α‐diversity with age and BMI. The analysis showed that the microbial α‐diversity increased with increasing age. In addition, the microbial α‐diversity increased with increasing BMI in elderly individuals. We also showed that the microbial α‐diversity was decreased in the overweight group of whole AGP samples (data not shown). Therefore, the analysis results based on AGP are in accordance with the finding that microbial α‐diversity is positively associated with increased age and obesity in elderly individuals.
In humans, the effect of exercise on gut microbial diversity is somewhat controversial. Studies have shown that exercise can increase microbial α‐diversity (Houghton et al., 2018; Keohane et al., 2019). However, other studies have shown that exercise dOEs not affect the α‐diversity of the gut microbiota (Cronin et al., 2018; Kern et al., 2020; Kim & Kang, 2019). Besides, studies have shown that exercise might be negatively associated with microbial α‐diversity (Allen et al., 2015; Mika et al., 2015). The reason for the discrepancies may be that the previous studies were small in scale. In the present study, we found that the microbial α‐diversity decreased with increasing exercise frequency in elderly individuals, and the microbial α‐diversity decreased in the OE group. Therefore, these results seem to be consistent with other research that found a negative association with exercise frequency in elderly individuals, especially in OE individuals.
Comparing the microbial composition of elderly individuals with that of adults18‐60, we found that 3 phyla and 27 families had significantly different abundances. At the phylum level, the abundance of Proteobacteria was significantly increased, while that of Actinobacteria was significantly decreased. The Bacteroidetes abundance was lower in the elderly individual group (37.7%) than in the adults18‐60 (39.0%; p > .05), while the Bacteroidetes/Firmicutes ratio was not significantly different between the two groups. At the family level, the abundances of Desulfovibrionaceae and Enterobacteriaceae were significantly increased, while that of Bifidobacteriaceae was significantly decreased in elderly individuals. Actinobacteria species are key in maintaining gut homeostasis and play an important role in the treatment of gastrointestinal diseases and systemic diseases (Jami, Ghanbari, Kneifel, & Domig, 2015). Bifidobacteria species (phylum: Actinobacteria) are widely used as probiotics and have demonstrated beneficial effects under a variety of pathological conditions (Shen et al., 2018). Studies have indicated that Proteobacteria may be a characteristic microorganism of diseases, including metabolic disorders and inflammatory bowel disease. Proteobacteria can cause inflammation and lead to disease development (Rizzatti, Lopetuso, Gibiino, Binda, & Gasbarrini, 2017). The increase in intestinal Desulfovibrionaceae abundance is an important feature of colitis (Leonardi et al., 2017). Enterobacteriaceae are important pathogens because of their capacity to produce endotoxins (Xie et al., 2017). Therefore, our results suggested that aging induced an increase in harmful bacteria and a decrease in beneficial microbes.
The microbial function analysis predicted that 25 pathways were significantly different between elderly individuals and adults18‐60 (Appendix 2: Table A2). The amino acid metabolism‐related pathways were altered, which might result in decreased biosynthesis of essential amino acids (threonine, phenylalanine, lysine, and tryptophan) and aromatic amino acids (tyrosine, phenylalanine, and tryptophan). Vitamin biosynthesis‐related pathway abundances were decreased. A previous study showed that the gut microbiota was involved in essential amino acid homeostasis (Lin, Liu, Piao, & Zhu, 2017). The gut microbiota can synthesize K and B vitamins, including biotin, riboflavin, cobalamin, folic acid, nicotinic acid, pantothenic acid, pyridoxine, and thiamine (Rowland et al., 2018). It is estimated that vitamins synthesized by gut microbes can provide more than a quarter of the intake (Rowland et al., 2018). Therefore, our results suggest that age induced a decrease in the production of essential amino acids and vitamins by the gut microbiota, thereby reducing these substances in elderly individuals.
Studies have shown that exercise is linked to microbial composition (Murtaza et al., 2019; Petersen et al., 2017; Scheiman et al., 2019). Murtaza et al. (2019) identified two dominant enterotypes (Bacteroides and Prevotella) in elite endurance athletes. Scheiman et al. (2019) showed that Veillonella was linked to exercise performance. Petersen et al suggested that Prevotella was correlated with exercise (Petersen et al., 2017). Barton et al found enrichment of Akkermansia in athletes (Barton et al., 2018). We compared the microbial composition of each exercise frequency group with that of adults18‐60 and found that the abundances of 3 phyla and 14 families gradually approached those in adults18‐60 with an increase in exercise frequency. The abundance of Actinobacteria gradually increased and was similar to that in adults18‐60. Additionally, the abundance of Cyanobacteria gradually decreased and approached that in adults18‐60. Cyanobacteria are associated with diseases and, in some cases, with human and animal death (Lange et al., 2018). Cyanobacteria release their toxins, such as lipopolysaccharides, in B cells in the gut (Swanson‐Mungerson et al., 2017). Furthermore, the evolutionary analysis showed that the microbial composition in the daily exercise group was closest to that in adults18‐60. We also revealed that daily exercise shifts the gut microbiota to a younger phenotype.
In OE individuals, the abundances of Actinobacteria and Bacteroidetes were decreased, while the abundances of Cyanobacteria and Firmicutes were increased at the phylum level. The Bacteroidetes/Firmicutes ratio was significantly decreased. At the family level, the abundances of S24‐7 and Lachnospiraceae were increased, while the abundances of Christensenellaceae, Barnesiellaceae, and Bacteroidaceae were decreased in OE individuals. It was reported that S24‐7 enrichment was associated with a high‐fat diet in diabetes‐sensitive mice (Serino et al., 2012). Kameyama et al showed that intestinal colonization by a Lachnospiraceae bacterium promoted the development of diabetes in obese mice (Kameyama & Itoh, 2014). It was reported that Christensenellaceae bacteria were beneficial for human health and had an increased abundance in lean people (Requena, Martínez‐Cuesta, & Peláez, 2018). Barnesiellaceae could serve as a marker to discriminate lean and obese individuals (Rodriguez, Benninghoff, Aardema, Phatak, & Hintze, 2019). Hakkak, Korourian, Foley, & Erickson (2017) showed that lean rats exhibited much lower Firmicutes to Bacteroidetes ratios than obese rats. A recent report showed that Bacteroidaceae might play a role in bamboo shoot fiber‐mediated suppression of high‐fat diet‐induced obesity (Li, Guo, Ji, & Zhang, 2016). Therefore, our results suggest an increase in harmful bacteria and a decrease in beneficial microbes in OE individuals.
Next, we revealed that the microbial α‐diversity was significantly decreased, while the microbial abundance approached the mean level in elderly individuals in the DROE group. At the phylum level, the abundance of Bacteroidetes was increased, while the abundances of Proteobacteria, Cyanobacteria, and Firmicutes were decreased in the DROE group compared with the NROE group. At the family level, the abundance of Lachnospiraceae was decreased, while the abundances of Christensenellaceae and Bacteroidaceae were increased in OE individuals. Exercise effectively counteracted obesity‐induced microbial imbalances, increased the abundance of the Bacteroides genera, and decreased the abundance of the Porphyromonas genera in male Wistar rats (Carbajo‐Pescador et al., 2019). A 6‐week endurance exercise regimen in overweight women revealed that training slightly increased genus‐level β‐diversity and decreased Proteobacteria abundance (Munukka et al., 2018). Therefore, our results suggest that regular exercise might play a role in decreasing harmful bacteria and increasing beneficial microbes in OE individuals.
Thirteen common pathways were isolated and showed opposite trends in the OE/NE and DROE/NROE groups. Ten pathways related to nucleotide biosynthesis were significantly inhibited in OE individuals and enhanced after regular exercise. Thus, these results suggest that regular exercise might modulate nucleotide biosynthesis, which is important for microorganism growth.
This study relied on the participants' accurate self‐reporting of the frequency of exercise without tracking the intensity of exercise. Studies have shown that insufficient or excessive exercise intensity has a significant impact on the structure and function of the intestinal flora. Moreover, the main research participants in this study were Caucasian, and the amount of data from other ethnic groups is small. Future research should specifically study other ethnic groups for comparative analysis.
5. CONCLUSION
In conclusion, our results revealed that microbial diversity increased with increasing age. The gut microbiota composition of daily exercising elderly individuals approached that of adults18‐60, and regular exercise resulted in an increased relative abundance of bacterial functional pathways related to nucleotide metabolism, glucose metabolism, and lipid metabolism. We also showed that OE individuals had increased microbial diversity and significant changes in the microbial composition, which responded with alterations in bacterial functional pathways related to vitamin, nucleotide, and glucose metabolism. Furthermore, regular exercise partially reshaped the microbial composition. Altogether, our findings support the role of regular exercise in maintaining the stability of the gut microbiota in elderly individuals and reveal that regular exercise benefits OE individuals.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
Qiwei Zhu: Data curation (lead). Shangfei Jiang: Software (lead). Guankui Du: Conceptualization (lead); Formal analysis (lead); Funding acquisition (lead); Investigation (lead); Methodology (lead); Project administration (lead); Resources (lead); Writing – original draft (lead); Writing – review and editing (lead).
ETHICS STATEMENT
None required.
ACKNOWLEDGMENTS
This study was funded by the National Natural Science Foundation of China (No. 81960672) (Guankui Du).
APPENDIX 1.
TABLE A1.
The relative abundances of 129 pathways were significantly changed in OE individuals compared with the NW groups
| Pathway |
NW relative frequency (%) mean ± SD |
OW relative frequency (%) mean ± SD |
p‐Values |
|---|---|---|---|
| PWY‐5971 | 0.1931 ± 0.1349 | 0.2708 ± 0.1734 | 5.45E−11 |
| PENTOSE‐P‐PWY | 0.4021 ± 0.1285 | 0.3532 ± 0.1140 | 4.87E−08 |
| FASYN‐INITIAL‐PWY | 0.1628 ± 0.1234 | 0.2150 ± 0.1393 | 1.23E−07 |
| PWY‐7664 | 0.2106 ± 0.1528 | 0.2746 ± 0.1695 | 1.24E−07 |
| GLCMANNANAUT‐PWY | 0.2290 ± 0.0710 | 0.2037 ± 0.0604 | 1.62E−07 |
| PWYG‐321 | 0.2146 ± 0.1528 | 0.2771 ± 0.1663 | 1.64E−07 |
| PWY‐6282 | 0.1902 ± 0.1449 | 0.2501 ± 0.1612 | 1.85E−07 |
| PWY‐5989 | 0.1918 ± 0.1481 | 0.2520 ± 0.1639 | 2.57E−07 |
| POLYAMINSYN3‐PWY | 0.0616 ± 0.0379 | 0.0759 ± 0.0372 | 3.11E−07 |
| PWY0‐862 | 0.1926 ± 0.1447 | 0.2505 ± 0.1594 | 3.67E−07 |
| PWY‐5505 | 0.5451 ± 0.1688 | 0.6013 ± 0.1448 | 1.11E−06 |
| PWY‐7007 | 0.0008 ± 0.0063 | 0.0035 ± 0.0079 | 1.13E−06 |
| PWY‐5484 | 0.6491 ± 0.1140 | 0.6094 ± 0.1114 | 1.98E−06 |
| GLUCARGALACTSUPER‐PWY | 0.0222 ± 0.0255 | 0.0149 ± 0.0179 | 4.96E−06 |
| GALACTARDEG‐PWY | 0.0222 ± 0.0255 | 0.0149 ± 0.0179 | 4.96E−06 |
| PWY‐6071 | 0.0035 ± 0.0105 | 0.0077 ± 0.0138 | 5.75E−06 |
| REDCITCYC | 0.1191 ± 0.1076 | 0.0889 ± 0.0776 | 9.24E−06 |
| PWY‐7332 | 0.0137 ± 0.0213 | 0.0083 ± 0.0128 | 1.67E−05 |
| GLYCOLYSIS | 0.7464 ± 0.0847 | 0.7200 ± 0.0824 | 1.90E−05 |
| PWY‐6901 | 0.4311 ± 0.1174 | 0.3968 ± 0.1061 | 3.05E−05 |
| PWY‐7446 | 0.0082 ± 0.0151 | 0.0045 ± 0.0098 | 3.95E−05 |
| PWY‐7220 | 0.6884 ± 0.1044 | 0.6587 ± 0.0970 | 6.21E−05 |
| PWY‐7222 | 0.6884 ± 0.1044 | 0.6587 ± 0.0970 | 6.21E−05 |
| METHGLYUT‐PWY | 0.0120 ± 0.0211 | 0.0069 ± 0.0141 | 6.84E−05 |
| PWY‐6519 | 0.2150 ± 0.1206 | 0.2500 ± 0.1223 | 9.87E−05 |
| P108‐PWY | 0.4574 ± 0.1624 | 0.4143 ± 0.1473 | 0.0002 |
| PWY‐5676 | 0.1208 ± 0.0744 | 0.1039 ± 0.0573 | 0.0005 |
| PWY‐5180 | 0.0253 ± 0.0326 | 0.0178 ± 0.0260 | 0.0005 |
| PWY‐5182 | 0.0253 ± 0.0326 | 0.0178 ± 0.0260 | 0.0005 |
| GLUCUROCAT‐PWY | 0.2137 ± 0.0745 | 0.2324 ± 0.0707 | 0.0005 |
| PWY‐6471 | 0.1483 ± 0.1003 | 0.1240 ± 0.0903 | 0.0005 |
| PWY0‐41 | 0.0124 ± 0.0189 | 0.0085 ± 0.0122 | 0.0006 |
| GLUCARDEG‐PWY | 0.0246 ± 0.0256 | 0.0189 ± 0.0192 | 0.0006 |
| BIOTIN‐BIOSYNTHESIS‐PWY | 0.2195 ± 0.1112 | 0.2471 ± 0.1076 | 0.0006 |
| TCA | 0.5675 ± 0.1223 | 0.5385 ± 0.1119 | 0.0007 |
| PWY‐6478 | 0.0414 ± 0.0411 | 0.0517 ± 0.0422 | 0.0008 |
| PWY‐7090 | 0.0022 ± 0.0032 | 0.0016 ± 0.0018 | 0.0008 |
| GALACT‐GLUCUROCAT‐PWY | 0.2319 ± 0.0727 | 0.2490 ± 0.0679 | 0.0009 |
| PWY‐5022 | 0.0369 ± 0.0366 | 0.0292 ± 0.0276 | 0.0010 |
| PWY‐6690 | 0.0101 ± 0.0171 | 0.0067 ± 0.0118 | 0.0013 |
| HCAMHPDEG‐PWY | 0.0101 ± 0.0171 | 0.0067 ± 0.0118 | 0.0013 |
| PWY0‐321 | 0.0051 ± 0.0157 | 0.0090 ± 0.0175 | 0.0014 |
| PYRIDNUCSAL‐PWY | 0.6468 ± 0.0809 | 0.6636 ± 0.0631 | 0.0014 |
| PWY‐7254 | 0.0876 ± 0.0899 | 0.0694 ± 0.0668 | 0.0015 |
| GLYOXYLATE‐BYPASS | 0.0492 ± 0.0623 | 0.0369 ± 0.0434 | 0.0015 |
| PWY‐5677 | 0.0271 ± 0.0244 | 0.0225 ± 0.0166 | 0.0024 |
| PWY‐7663 | 0.9422 ± 0.0817 | 0.9574 ± 0.0553 | 0.0025 |
| PWY‐6588 | 0.1974 ± 0.1010 | 0.1774 ± 0.0814 | 0.0027 |
| RIBOSYN2‐PWY | 0.6539 ± 0.0671 | 0.6670 ± 0.0524 | 0.0027 |
| KETOGLUCONMET‐PWY | 0.0134 ± 0.0231 | 0.0090 ± 0.0166 | 0.0028 |
| P42‐PWY | 0.7444 ± 0.1884 | 0.7062 ± 0.1643 | 0.0031 |
| PWY0‐1533 | 0.0230 ± 0.0272 | 0.0178 ± 0.0211 | 0.0032 |
| PWY0‐1296 | 0.4445 ± 0.1115 | 0.4673 ± 0.0999 | 0.0032 |
| ANAEROFRUCAT‐PWY | 0.6227 ± 0.0880 | 0.6047 ± 0.0804 | 0.0034 |
| PWY‐6969 | 0.6513 ± 0.1314 | 0.6249 ± 0.1149 | 0.0035 |
| PWY‐6126 | 0.7834 ± 0.0636 | 0.7700 ± 0.0612 | 0.0036 |
| PWY‐5913 | 0.4997 ± 0.1519 | 0.4689 ± 0.1391 | 0.0040 |
| PWY‐7228 | 0.4718 ± 0.1020 | 0.4510 ± 0.0956 | 0.0042 |
| P341‐PWY | 0.0014 ± 0.0052 | 0.0006 ± 0.0025 | 0.0051 |
| PWY‐7197 | 0.3681 ± 0.0967 | 0.3488 ± 0.0916 | 0.0052 |
| PWY‐6125 | 0.5017 ± 0.0964 | 0.4826 ± 0.0911 | 0.0054 |
| PWY‐5747 | 0.0136 ± 0.0187 | 0.0103 ± 0.0137 | 0.0060 |
| PWY‐5667 | 0.8401 ± 0.0899 | 0.8569 ± 0.0787 | 0.0065 |
| PWY0‐1319 | 0.8401 ± 0.0899 | 0.8569 ± 0.0787 | 0.0065 |
| P122‐PWY | 0.1046 ± 0.0615 | 0.0939 ± 0.0459 | 0.0065 |
| PWY‐5705 | 0.0176 ± 0.0170 | 0.0146 ± 0.0129 | 0.0069 |
| PWY‐5154 | 0.3929 ± 0.1133 | 0.3715 ± 0.1030 | 0.0070 |
| GLYCOCAT‐PWY | 0.8738 ± 0.1187 | 0.8961 ± 0.1077 | 0.0074 |
| PWY‐841 | 0.5890 ± 0.0899 | 0.5716 ± 0.0869 | 0.0074 |
| PWY‐7200 | 0.3806 ± 0.0613 | 0.3691 ± 0.0566 | 0.0075 |
| PWY‐6163 | 0.7938 ± 0.0703 | 0.8064 ± 0.0595 | 0.0077 |
| PWY‐5347 | 0.3726 ± 0.1086 | 0.3926 ± 0.0957 | 0.0078 |
| PWY‐621 | 0.5584 ± 0.1486 | 0.5841 ± 0.1179 | 0.0083 |
| PWY0‐42 | 0.0110 ± 0.0177 | 0.0082 ± 0.0117 | 0.0083 |
| PWY0‐1277 | 0.0164 ± 0.0261 | 0.0119 ± 0.0199 | 0.0083 |
| PWY‐7184 | 0.3518 ± 0.0778 | 0.3376 ± 0.0701 | 0.0088 |
| PWY0‐845 | 0.2685 ± 0.1161 | 0.2478 ± 0.1005 | 0.0092 |
| PWY‐5973 | 0.8943 ± 0.0760 | 0.9063 ± 0.0516 | 0.0099 |
| GLYCOGENSYNTH‐PWY | 0.8051 ± 0.1419 | 0.8306 ± 0.1305 | 0.0109 |
| TCA‐GLYOX‐BYPASS | 0.0564 ± 0.0703 | 0.0451 ± 0.0517 | 0.0112 |
| PWY‐5532 | 0.0010 ± 0.0020 | 0.0007 ± 0.0012 | 0.0114 |
| GOLPDLCAT‐PWY | 0.0068 ± 0.0127 | 0.0094 ± 0.0152 | 0.0125 |
| PYRIDOXSYN‐PWY | 0.2254 ± 0.1127 | 0.2057 ± 0.1032 | 0.0129 |
| DENOVOPURINE2‐PWY | 0.5938 ± 0.0826 | 0.5789 ± 0.0821 | 0.0137 |
| PWY‐7237 | 0.0543 ± 0.0561 | 0.0630 ± 0.0411 | 0.0144 |
| PWY‐4984 | 0.1049 ± 0.0787 | 0.0935 ± 0.0530 | 0.0178 |
| PWY‐6165 | 0.0016 ± 0.0035 | 0.0025 ± 0.0058 | 0.0204 |
| PWY‐7196 | 0.5429 ± 0.1101 | 0.5243 ± 0.1087 | 0.0214 |
| ARO‐PWY | 0.7795 ± 0.0856 | 0.7929 ± 0.0745 | 0.0214 |
| FAO‐PWY | 0.0545 ± 0.0568 | 0.0460 ± 0.0451 | 0.0223 |
| PHOSLIPSYN‐PWY | 0.7803 ± 0.0894 | 0.7941 ± 0.0763 | 0.0223 |
| PWY0‐166 | 0.4147 ± 0.0681 | 0.4042 ± 0.0579 | 0.0225 |
| P164‐PWY | 0.1413 ± 0.0669 | 0.1315 ± 0.0506 | 0.0225 |
| PWY‐7199 | 0.5239 ± 0.0738 | 0.5119 ± 0.0700 | 0.0227 |
| PWY0‐781 | 0.2635 ± 0.0963 | 0.2488 ± 0.0801 | 0.0228 |
| PWY‐5659 | 0.5976 ± 0.0871 | 0.5835 ± 0.0816 | 0.0232 |
| CODH‐PWY | 0.0179 ± 0.0216 | 0.0213 ± 0.0190 | 0.0234 |
| P124‐PWY | 0.1339 ± 0.0888 | 0.1210 ± 0.0676 | 0.0238 |
| GLYCOL‐GLYOXDEG‐PWY | 0.0291 ± 0.0321 | 0.0245 ± 0.0239 | 0.0245 |
| PWY‐5415 | 0.0065 ± 0.0148 | 0.0045 ± 0.0104 | 0.0254 |
| PWY0‐162 | 0.5985 ± 0.0980 | 0.5828 ± 0.0940 | 0.0255 |
| RUMP‐PWY | 0.0667 ± 0.0562 | 0.0588 ± 0.0413 | 0.0267 |
| PWY‐5304 | 0.1095 ± 0.0954 | 0.1243 ± 0.0881 | 0.0276 |
| COMPLETE‐ARO‐PWY | 0.8166 ± 0.0871 | 0.8297 ± 0.0758 | 0.0278 |
| HISTSYN‐PWY | 0.7138 ± 0.0779 | 0.7017 ± 0.0732 | 0.0290 |
| PWY‐7187 | 0.4234 ± 0.0635 | 0.4139 ± 0.0558 | 0.0299 |
| P163‐PWY | 0.0352 ± 0.0357 | 0.0303 ± 0.0257 | 0.0301 |
| PWY‐6185 | 0.0024 ± 0.0074 | 0.0038 ± 0.0095 | 0.0301 |
| PWY‐5910 | 0.0185 ± 0.0400 | 0.0131 ± 0.0289 | 0.0303 |
| PWY‐6151 | 0.4874 ± 0.1112 | 0.5037 ± 0.0971 | 0.0319 |
| SER‐GLYSYN‐PWY | 0.7444 ± 0.0713 | 0.7552 ± 0.0664 | 0.0328 |
| PWY‐922 | 0.0135 ± 0.0309 | 0.0095 ± 0.0217 | 0.0334 |
| PWY‐5695 | 0.6651 ± 0.1110 | 0.6481 ± 0.1065 | 0.0338 |
| PWY‐7013 | 0.0434 ± 0.0496 | 0.0369 ± 0.0356 | 0.0354 |
| PWY‐6608 | 0.2527 ± 0.1048 | 0.2378 ± 0.0902 | 0.0365 |
| PWY‐6470 | 0.0213 ± 0.0356 | 0.0165 ± 0.0271 | 0.0380 |
| PWY‐6182 | 0.0024 ± 0.0074 | 0.0037 ± 0.0093 | 0.0381 |
| PWY‐5198 | 0.0005 ± 0.0018 | 0.0013 ± 0.0068 | 0.0387 |
| GLUCOSE1PMETAB‐PWY | 0.0470 ± 0.0659 | 0.0393 ± 0.0342 | 0.0389 |
| ARGSYNBSUB‐PWY | 0.6160 ± 0.1288 | 0.6344 ± 0.1157 | 0.0399 |
| TEICHOICACID‐PWY | 0.0879 ± 0.0496 | 0.0950 ± 0.0452 | 0.0412 |
| GLYCOLYSIS‐TCA‐GLYOX‐BYPASS | 0.0819 ± 0.0870 | 0.0706 ± 0.0667 | 0.0441 |
| CATECHOL‐ORTHO‐CLEAVAGE‐PWY | 0.0025 ± 0.0069 | 0.0038 ± 0.0105 | 0.0449 |
| PWY‐3781 | 0.1989 ± 0.1952 | 0.1734 ± 0.1545 | 0.0452 |
| THREOCAT‐PWY | 0.0054 ± 0.0166 | 0.0080 ± 0.0182 | 0.0458 |
| PWY‐5417 | 0.0027 ± 0.0076 | 0.0041 ± 0.0111 | 0.0468 |
| PWY‐5431 | 0.0027 ± 0.0076 | 0.0041 ± 0.0111 | 0.0468 |
| PWY‐7560 | 0.7715 ± 0.0707 | 0.7811 ± 0.0632 | 0.0497 |
| NONMEVIPP‐PWY | 0.7715 ± 0.0707 | 0.7811 ± 0.0632 | 0.0497 |
APPENDIX 2.
TABLE A2.
The relative abundances of 24 pathways were significantly changed in DROE compared with NROE
| Pathway |
NROE relative frequency (%) mean ± SD |
DROE relative frequency (%) mean ± SD |
p‐values |
|---|---|---|---|
| PWY‐7090 | 0.0015 ± 0.0016 | 0.0020 ± 0.0029 | 0.0052 |
| PWY‐7332 | 0.0084 ± 0.0138 | 0.0120 ± 0.0192 | 0.0077 |
| PWY‐5484 | 0.6120 ± 0.1064 | 0.6382 ± 0.1169 | 0.0077 |
| GLYCOLYSIS | 0.7214 ± 0.0761 | 0.7392 ± 0.0873 | 0.0121 |
| PWY‐841 | 0.5670 ± 0.0867 | 0.5853 ± 0.0904 | 0.0201 |
| PWY‐7228 | 0.4474 ± 0.0969 | 0.4669 ± 0.1017 | 0.0274 |
| DENOVOPURINE2‐PWY | 0.5743 ± 0.0830 | 0.5907 ± 0.0835 | 0.0289 |
| P108‐PWY | 0.4183 ± 0.1360 | 0.4460 ± 0.1627 | 0.0305 |
| PWY‐6125 | 0.4791 ± 0.0936 | 0.4972 ± 0.0959 | 0.0335 |
| PWY‐7184 | 0.3343 ± 0.0715 | 0.3481 ± 0.0773 | 0.0351 |
| GLUCUROCAT‐PWY | 0.2332 ± 0.0724 | 0.2194 ± 0.0764 | 0.0367 |
| PWY‐6519 | 0.2480 ± 0.1223 | 0.2251 ± 0.1235 | 0.0379 |
| PWY‐6353 | 0.2724 ± 0.0803 | 0.2575 ± 0.0765 | 0.0380 |
| PWY‐4984 | 0.0908 ± 0.0592 | 0.1024 ± 0.0713 | 0.0381 |
| PENTOSE‐P‐PWY | 0.3659 ± 0.1145 | 0.3875 ± 0.1281 | 0.0415 |
| PWY‐7197 | 0.3461 ± 0.0930 | 0.3633 ± 0.0969 | 0.0423 |
| PWY‐7200 | 0.3666 ± 0.0601 | 0.3776 ± 0.0599 | 0.0433 |
| PWY‐5505 | 0.5909 ± 0.1517 | 0.5628 ± 0.1648 | 0.0437 |
| GALACT‐GLUCUROCAT‐PWY | 0.2503 ± 0.0707 | 0.2374 ± 0.0741 | 0.0441 |
| PWY0‐162 | 0.5776 ± 0.0972 | 0.5953 ± 0.0975 | 0.0448 |
| PWY0‐166 | 0.4008 ± 0.0591 | 0.4118 ± 0.0667 | 0.0456 |
| P125‐PWY | 0.0074 ± 0.0160 | 0.0112 ± 0.0365 | 0.0471 |
| PWY0‐1296 | 0.4674 ± 0.1002 | 0.4492 ± 0.1098 | 0.0483 |
| PWY‐7196 | 0.5194 ± 0.1089 | 0.5388 ± 0.1110 | 0.0491 |
Zhu Q, Jiang S, Du G. Effects of exercise frequency on the gut microbiota in elderly individuals. MicrobiologyOpen. 2020;9:e1053 10.1002/mbo3.1053
Qiwei Zhu and Shangfei Jiang contributed equally.
DATA AVAILABILITY STATEMENT
All data used for this study are available at https://www.ebi.ac.uk/ena/browser/view/PRJEB11419
REFERENCES
- Allen, J. M. , Berg Miller, M. E. , Pence, B. D. , Whitlock, K. , Nehra, V. , Gaskins, H. R. , … Woods, J. A. (2015). Voluntary and forced exercise differentially alters the gut microbiome in C57BL/6J mice. Journal of Applied Physiology, 118(8), 1059–1066. 10.1152/japplphysiol.01077.2014 [DOI] [PubMed] [Google Scholar]
- Allen, J. M. , Mailing, L. J. , Niemiro, G. M. , Moore, R. , Cook, M. D. , White, B. A. , … Woods, J. A. (2018). Exercise alters gut microbiota composition and function in lean and obese humans. Medicine and Science in Sports and Exercise, 50(4), 747–757. 10.1249/MSS.0000000000001495 [DOI] [PubMed] [Google Scholar]
- Backhed, F. , Ding, H. , Wang, T. , Hooper, L. V. , Koh, G. Y. , Nagy, A. , … Gordon, J. I. (2004). The gut microbiota as an environmental factor that regulates fat storage. Proceedings of the National Academy of Sciences of the United States of America, 101(44), 15718–15723. 10.1073/pnas.0407076101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, W. , Penney, N. C. , Cronin, O. , Garcia‐Perez, I. , Molloy, M. G. , Holmes, E. , … O'Sullivan, O. (2018). The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut, 67(4), 625–633. [DOI] [PubMed] [Google Scholar]
- Biddle, S. J. , & Batterham, A. M. (2015). High‐intensity interval exercise training for public health: A big HIT or shall we HIT it on the head? International Journal of Behavioral Nutrition and Physical Activity, 12(1), 95 10.1186/s12966-015-0254-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt, N. , Kotowska, D. , Kristensen, C. M. , Olesen, J. , Lützhøft, D. O. , Halling, J. F. , … Pilegaard, H. (2018). The impact of exercise training and resveratrol supplementation on gut microbiota composition in high‐fat diet fed mice. Physiological Reports, 6(20), e13881 10.14814/phy2.13881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbajo‐Pescador, S. , Porras, D. , García‐Mediavilla, M. V. , Martínez‐Flórez, S. , Juarez‐Fernández, M. , Cuevas, M. J. , … Sánchez‐Campos, S. (2019). Beneficial effects of exercise on gut microbiota functionality and barrier integrity, and gut‐liver crosstalk in an in vivo model of early obesity and non‐alcoholic fatty liver disease. Disease Models & Mechanisms, 12(5), dmm039206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Sun, H. , Jiang, F. , Shen, Y. , Li, X. , Hu, X. , … Wei, P. (2020). Alteration of the gut microbiota associated with childhood obesity by 16S rRNA gene sequencing. PeerJ, 8, e8317 10.7717/peerj.8317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson, M. J. , Cusack, S. , O'Sullivan, O. , Greene‐Diniz, R. , De Weerd, H. , Flannery, E. , … O'Toole, P. W. (2011). Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proceedings of the National Academy of Sciences of the United States of America, 108(Suppl 1), 4586–4591. 10.1073/pnas.1000097107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, S. F. , Murphy, E. F. , O'Sullivan, O. , Lucey, A. J. , Humphreys, M. , Hogan, A. , … Cotter, P. D. (2014). Exercise and associated dietary extremes impact on gut microbial diversity. Gut, 63(12), 1913–1920. 10.1136/gutjnl-2013-306541 [DOI] [PubMed] [Google Scholar]
- Cronin, O. , Barton, W. , Skuse, P. , Penney, N. C. , Garcia‐Perez, I. , Murphy, E. F. , … Shanahan, F. (2018). A prospective metagenomic and metabolomic analysis of the impact of exercise and/or whey protein supplementation on the gut microbiome of sedentary adults. mSystems, 3(3), e00044‐18 10.1128/mSystems.00044-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva, C. C. , Monteil, M. A. , & Davis, E. M. (2020). Overweight and obesity in children are associated with an abundance of Firmicutes and reduction of Bifidobacterium in their gastrointestinal microbiota. Childhood Obesity, 16(3), 204–210. [DOI] [PubMed] [Google Scholar]
- Davis, S. C. , Yadav, J. S. , Barrow, S. D. , & Robertson, B. K. (2017). Gut microbiome diversity influenced more by the Westernized dietary regime than the body mass index as assessed using effect size statistic. MicrobiologyOpen, (4), e00476 10.1002/mbo3.476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cuesta‐Zuluaga, J. , Kelley, S. T. , Chen, Y. , Escobar, J. S. , Mueller, N. T. , Ley, R. E. , … Thackray, V. G. (2019). Age‐ and sex‐dependent patterns of gut microbial diversity in human adults. mSystems, 4(4), e00261‐19 10.1128/mSystems.00261-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denou, E. , Marcinko, K. , Surette, M. G. , Steinberg, G. R. , & Schertzer, J. D. (2016). High‐intensity exercise training increases the diversity and metabolic capacity of the mouse distal gut microbiota during diet‐induced obesity. American Journal of Physiology‐Endocrinology and Metabolism, 310(11), E982–E993. 10.1152/ajpendo.00537.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, G. M. , Maffei, V. J. , Zaneveld, J. , Yurgel, S. N. , Brown, J. R. , Taylor, C. M. , … Langille, M. G. I. (2020). PICRUSt2: An Improved and Customizable Approach for Metagenome Inference., bioRxiv, 672295. [Google Scholar]
- Evans, C. C. , LePard, K. J. , Kwak, J. W. , Stancukas, M. C. , Laskowski, S. , Dougherty, J. , … Ciancio, M. J. (2014). Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet‐induced obesity. PLoS ONE, 9(3), e92193 10.1371/journal.pone.0092193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, X. , Uchida, Y. , Koch, L. , Britton, S. , Hu, J. , Lutrin, D. , & Maze, M. (2017). Exercise prevents enhanced postoperative neuroinflammation and cognitive decline and rectifies the gut microbiome in a rat model of metabolic syndrome. Frontiers in Immunology, 8, 1768 10.3389/fimmu.2017.01768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkak, R. , Korourian, S. , Foley, S. L. , & Erickson, B. D. (2017). Assessment of gut microbiota populations in lean and obese Zucker rats. PLoS ONE, 12(7), e0181451 10.1371/journal.pone.0181451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, H. , Sakamoto, M. , Kitahara, M. , & Benno, Y. (2003). Molecular analysis of fecal microbiota in elderly individuals using 16S rDNA library and T‐RFLP. Microbiology and Immunology, 47(8), 557–570. 10.1111/j.1348-0421.2003.tb03418.x [DOI] [PubMed] [Google Scholar]
- Houghton, D. , Stewart, C. J. , Stamp, C. , Nelson, A. , Aj ami, N. J. , Petrosino, J. F. , … Greaves, L. C. (2018). Impact of age‐related mitochondrial dysfunction and exercise on intestinal microbiota composition. Journals of Gerontology. Series A, Biological Sciences and Medical Sciences, 73(5), 571–578. 10.1093/gerona/glx197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakicic, J. M. , Rogers, R. J. , Davis, K. K. , & Collins, K. A. (2018). Role of physical activity and exercise in treating patients with overweight and obesity. Clinical Chemistry, 64(1), 99–107. 10.1373/clinchem.2017.272443 [DOI] [PubMed] [Google Scholar]
- Jami, M. , Ghanbari, M. , Kneifel, W. , & Domig, K. J. (2015). Phylogenetic diversity and biological activity of culturable Actinobacteria isolated from freshwater fish gut microbiota. Microbiological Research, 175, 6–15. 10.1016/j.micres.2015.01.009 [DOI] [PubMed] [Google Scholar]
- Kameyama, K. , & Itoh, K. (2014). Intestinal colonization by a Lachnospiraceae bacterium contributes to the development of diabetes in obese mice. Microbes and Environments, 29(4), 427–430. 10.1264/jsme2.ME14054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai, C. , Sugimoto, K. , Moritani, I. , Tanaka, J. , Oya, Y. , Inoue, H. , … Takase, K. (2015). Comparison of the gut microbiota composition between obese and non‐obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next‐generation sequencing. BMC Gastroenterology, 15(1), 100 10.1186/s12876-015-0330-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keohane, D. M. , Woods, T. , O'Connor, P. , Underwood, S. , Cronin, O. , Whiston, R. , … Molloy, M. G. M. (2019). Four men in a boat: Ultra‐endurance exercise alters the gut microbiome. Journal of Science and Medicine in Sport, 22(9), 1059–1064. 10.1016/j.jsams.2019.04.004 [DOI] [PubMed] [Google Scholar]
- Kern, T. , Blond, M. B. , Hansen, T. H. , Rosenkilde, M. , Quist, J. S. , Gram, A. S. , … Stallknecht, B. (2020). Structured exercise alters the gut microbiota in humans with overweight and obesity‐A randomized controlled trial. International Journal of Obesity, 44(1), 125–135. 10.1038/s41366-019-0440-y [DOI] [PubMed] [Google Scholar]
- Kim, D. , & Kang, H. (2019). Exercise training modifies gut microbiota with attenuated host responses to sepsis in wild‐type mice. The FASEB Journal, 33(4), 5772–5781. 10.1096/fj.201802481R [DOI] [PubMed] [Google Scholar]
- Koo, S. H. , Chu, C. W. , Khoo, J. J. C. , Cheong, M. , Soon, G. H. , Ho, E. X. P. , … Hsiang, J. C. (2019). A pilot study to examine the association between human gut microbiota and the host's central obesity. JGH Open, 3(6), 480–487. 10.1002/jgh3.12184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, J. E. , Myslicki, J. P. , Bomhof, M. R. , Belke, D. D. , Shearer, J. , & Reimer, R. A. (2015). Exercise training modifies gut microbiota in normal and diabetic mice. Applied Physiology, Nutrition, and Metabolism, 40(7), 749–752. 10.1139/apnm-2014-0452 [DOI] [PubMed] [Google Scholar]
- Lange, J. , Demir, F. , Huesgen, P. F. , Baumann, U. , Von Elert, E. , & Pichlo, C. (2018). Heterologous expression and characterization of a novel serine protease from Daphnia magna: A possible role in susceptibility to toxic cyanobacteria. Aquatic Toxicology, 205, 140–147. 10.1016/j.aquatox.2018.09.013 [DOI] [PubMed] [Google Scholar]
- Leonardi, I. , Gerstgrasser, A. , Schmidt, T. S. B. , Nicholls, F. , Tewes, B. , Greinwald, R. , … Frey‐Wagner, I. (2017). Preventive Trichuris suis ova (TSO) treatment protects immunocompetent rabbits from DSS colitis but may be detrimental under conditions of immunosuppression. Scientific Reports, 7(1), 16500 10.1038/s41598-017-16287-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Guo, J. , Ji, K. , & Zhang, P. (2016). Bamboo shoot fiber prevents obesity in mice by modulating the gut microbiota. Scientific Reports, 6, 32953 10.1038/srep32953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, R. , Liu, W. , Piao, M. , & Zhu, H. (2017). A review of the relationship between the gut microbiota and amino acid metabolism. Amino Acids, 49(12), 2083–2090. 10.1007/s00726-017-2493-3 [DOI] [PubMed] [Google Scholar]
- Liu, T.‐W. , Park, Y.‐M. , Holscher, H. D. , Padilla, J. , Scroggins, R. J. , Welly, R. , … Swanson, K. S. (2015). Physical activity differentially affects the cecal microbiota of ovariectomized female rats selectively bred for high and low aerobic capacity. PLoS ONE, 10(8), e0136150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach, N. , & Fuster‐Botella, D. (2017). Endurance exercise and gut microbiota: A review. Journal of Sport and Health Science, 6(2), 179–197. 10.1016/j.jshs.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, D. , Hyde, E. , Debelius, J. W. , Morton, J. T. , Gonzalez, A. , Ackermann, G. , … Gunderson, B. (2018). American Gut: An Open Platform for Citizen Science Microbiome Research. mSystems, 3(3), e00031‐18 10.1128/mSystems.00031-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika, A. , Van Treuren, W. , Gonzalez, A. , Herrera, J. J. , Knight, R. , & Fleshner, M. (2015). Exercise is more effective at altering gut microbial composition and producing stable changes in lean mass in juvenile versus adult male F344 rats. PLoS ONE, 10(5), e0125889 10.1371/journal.pone.0125889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monda, V. , Villano, I. , Messina, A. , Valenzano, A. , Esposito, T. , Moscatelli, F. , & Monda, M. (2017). Exercise modifies the gut microbiota with positive health effects. Oxidative Medicine and Cellular Longevity, 2017 10.1155/2017/3831972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munukka, E. , Ahtiainen, J. P. , Puigbó, P. , Jalkanen, S. , Pahkala, K. , Keskitalo, A. , … Pekkala, S. (2018). Six‐week endurance exercise alters gut metagenome that is not reflected in systemic metabolism in over‐weight women. Frontiers in Microbiology, 9, 2323 10.3389/fmicb.2018.02323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaza, N. , Burke, L. M. , Vlahovich, N. , Charlesson, B. , O'Neill, H. , Ross, M. L. , … Morrison, M. C. (2019). The effects of dietary pattern during intensified training on stool microbiota of elite race walkers. Nutrients, 11(2). 10.3390/nu11020261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson, J. K. , Holmes, E. , Kinross, J. , Burcelin, R. , Gibson, G. , Jia, W. , & Pettersson, S. (2012). Host‐gut microbiota metabolic interactions. Science, 336(6086), 1262–1267. [DOI] [PubMed] [Google Scholar]
- Parks, D. H. , Tyson, G. W. , Hugenholtz, P. , & Beiko, R. G. (2014). STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics, 30(21), 3123–3124. 10.1093/bioinformatics/btu494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, L. M. , Bautista, E. J. , Nguyen, H. , Hanson, B. M. , Chen, L. , Lek, S. H. , … Weinstock, G. M. (2017). Community characteristics of the gut microbiomes of competitive cyclists. Microbiome, 5(1), 98 10.1186/s40168-017-0320-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Requena, T. , Martínez‐Cuesta, M. C. , & Peláez, C. (2018). Diet and microbiota linked in health and disease. Food & Function, 9(2), 688–704. 10.1039/C7FO01820G [DOI] [PubMed] [Google Scholar]
- Rizzatti, G. , Lopetuso, L. , Gibiino, G. , Binda, C. , & Gasbarrini, A. (2017). Proteobacteria: A common factor in human diseases. BioMed Research International, 2017 10.1155/2017/9351507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, D. M. , Benninghoff, A. D. , Aardema, N. D. , Phatak, S. , & Hintze, K. J. (2019). Basal diet determined long‐term composition of the gut microbiome and mouse phenotype to a greater extent than fecal microbiome transfer from lean or obese human donors. Nutrients, 11(7), 1630 10.3390/nu11071630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round, J. L. , & Mazmanian, S. K. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nature Reviews Immunology, 9(5), 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland, I. , Gibson, G. , Heinken, A. , Scott, K. , Swann, J. , Thiele, I. , & Tuohy, K. (2018). Gut microbiota functions: Metabolism of nutrients and other food components. European Journal of Nutrition, 57(1), 1–24. 10.1007/s00394-017-1445-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiman, J. , Luber, J. M. , Chavkin, T. A. , MacDonald, T. , Tung, A. , Pham, L.‐D. , … Kostic, A. D. (2019). Meta‐omics analysis of elite athletes identifies a performance‐enhancing microbe that functions via lactate metabolism. Nature Medicine, 25(7), 1104–1109. 10.1038/s41591-019-0485-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serino, M. , Luche, E. , Gres, S. , Baylac, A. , Bergé, M. , Cenac, C. , … Burcelin, R. (2012). Metabolic adaptation to a high‐fat diet is associated with a change in the gut microbiota. Gut, 61(4), 543–553. 10.1136/gutjnl-2011-301012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, X. I. , Miao, J. , Wan, Q. , Wang, S. , Li, M. , Pu, F. , … He, F. (2018). Possible correlation between gut microbiota and immunity among healthy middle‐aged and elderly people in southwest China. Gut Pathogens, 10(1), 4 10.1186/s13099-018-0231-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, Y. (2018). Gut microbiota in common elderly diseases affecting activities of daily living. World Journal of Gastroenterology, 24(42), 4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson‐Mungerson, M. , Incrocci, R. , Subramaniam, V. , Williams, P. , Hall, M. L. , & Mayer, A. M. (2017). Effects of cyanobacteria Oscillatoria sp. Lipopolysaccharide on B cell activation and toll‐like receptor 4 signaling. Toxicology Letters, 275, 101–107. 10.1016/j.toxlet.2017.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi, H. , Tanisawa, K. , Sun, X. , Kubo, T. , Hoshino, Y. , Hosokawa, M. , … Higuchi, M. (2018). Effects of short‐term endurance exercise on gut microbiota in elderly men. Physiological Reports, 6(23), e13935 10.14814/phy2.13935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaroli, V. , & Bäckhed, F. (2012). Functional interactions between the gut microbiota and host metabolism. Nature, 489(7415), 242 10.1038/nature11552 [DOI] [PubMed] [Google Scholar]
- Van Der Merwe, M. , Sharma, S. , Caldwell, J. L. , Smith, N. J. , Gomes, C. K. , Bloomer, R. J. , … Pierre, J. F. (2020). Time of Feeding Alters Obesity‐Associated Parameters and Gut Bacterial Communities, but Not Fungal Populations, in C57BL/6 Male Mice. Curr Dev Nutr, 4(2), nzz145 10.1093/cdn/nzz145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ville, A. , Levine, E. , Zhi, D. , Lararia, B. , & Wojcicki, J. M. (2020). Alterations in the gut microbiome at 6 months of age in obese latino infants. Journal of the American College of Nutrition, 39(1), 47–53. 10.1080/07315724.2019.1606744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisner, C. M. , Maldonado, J. , Dente, B. , Krajmalnik‐Brown, R. , & Bruening, M. (2018). Diet, physical activity and screen time but not body mass index are associated with the gut microbiome of a diverse cohort of college students living in university housing: A cross‐sectional study. BMC Microbiology, 18(1), 210 10.1186/s12866-018-1362-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, J. , Peters, B. , Li, B. , Li, L. , Yu, G. , Xu, Z. , & Shirtliff, M. (2017). Clinical features and antimicrobial resistance profiles of important Enterobacteriaceae pathogens in Guangzhou representative of Southern China, 2001–2015. Microbial Pathogenesis, 107, 206 10.1016/j.micpath.2017.03.038 [DOI] [PubMed] [Google Scholar]
- Zhao, X. , Zhang, Z. , Hu, B. , Huang, W. , Yuan, C. , & Zou, L. (2018). Response of gut microbiota to metabolite changes induced by endurance exercise. Frontiers in Microbiology, 9, 765 10.3389/fmicb.2018.00765 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used for this study are available at https://www.ebi.ac.uk/ena/browser/view/PRJEB11419


