Abstract
The intracellular innate antiviral response in human cells is an essential component of immunity against virus infection. As obligate intracellular parasites, all viruses must evade the actions of the host cell’s innate immune response in order to replicate and persist. Innate immunity is induced when pathogen recognition receptors of the host cell sense viral products including nucleic acid as “non-self”. This process induces downstream signaling through adaptor proteins to activate latent transcription factors that drive the expression of genes encoding antiviral and immune modulatory effector proteins that restrict virus replication and regulate adaptive immunity. The interferon regulatory factors (IRFs) are transcription factors that play major roles in innate immunity. In particular, IRF3 is activated in response to infection by a range of viruses including RNA viruses, DNA viruses and retroviruses. Among these viruses, human immunodeficiency virus type 1 (HIV-1) remains a major global health problem mediating chronic infection in millions of people wherein recent studies show that viral persistence is linked with the ability of the virus to dysregulate and evade the innate immune response. In this review, we discuss viral pathogen sensing, innate immune signaling pathways and effectors that respond to viral infection, the role of IRF3 in these processes and how it is regulated by pathogenic viruses. We present a contemporary overview of the interplay between HIV-1 and innate immunity, with a focus on understanding how innate immune control impacts infection outcome and disease.
Keywords: interferon, ISG, HIV, virus, innate immunity
Introduction
All nucleated cells have the capacity to respond to pathogens by sensing certain pathogen-associated molecular patterns (PAMPs) through germline-encoded cellular pattern recognition receptors (PRRs). This process leads to the production of type I (α and β) interferons (IFN). The distribution of PRRs varies; for example, toll-like receptors (TLRs) are distributed within various cell types and expressed on the cell surface or within endosomes. On the other hand, the RIG-I-like receptors (RLRs) are cytosolic PRRs that are widely expressed across most cell types [1]. One common feature of PRR signaling is the induction of IFN production. IFN binds to the type 1 IFN receptor to signal the induction of hundreds of interferon-stimulated genes (ISGs) in the local uninfected tissue and infected cells, thus creating an antiviral state that suppresses virus infection and serves to promote the onset of adaptive immunity [2,3]. A major feature of this cellular antiviral state is the actions of ISG products that may interact directly with viruses or may operate to shut down host protein translation machinery to prevent appropriation by viruses. Concomitantly, expressed ISGs may impart dendritic cells maturation for ramping up antigen presentation to adaptive immune cells [4,5], including CD8+ and CD4+ T cells [6,7]. In addition, B and T cells themselves express a varying complement of PRRs that are important for adaptive immune function and cell survival [8,9]. In total, innate immune cells and innate immune signaling provide the initial response to virus infection. Viral clearance results from coordinated actions of innate and adaptive immunity with complete pathogen clearance arising from innate immunity-stimulated gene products and appropriate training of the adaptive immune response through the actions of innate immune effector genes.
Among pathogenic human viruses, human immunodeficiency virus type 1 (HIV-1) remains a major global health problem and is the leading cause of adult death in sub-Saharan Africa [10]. HIV-1 is a sexually transmitted virus that initiates infection typically through exposure via mucosal surfaces. Despite the advent of highly effective therapy, treatment is not curative, and there is no preventative vaccine. Of note is that heterosexual transmission of HIV-1 is inefficient compared to parenteral transmission, in which mucosal innate immune defenses are bypassed. These differences in HIV-1 transmission efficiency may be due to virus restriction through innate immune defenses at mucosal barriers triggered by virus exposure [11]. Studies that have evaluated the actual sequence of incoming viral genomes during acute infection in patients after HIV-1 exposure reveal that a single founder viral genome is typically linked with the virus’s transit through the mucosal barrier to infect CD4+ T lymphocytes [12], indicating that the initial target cell of the acute infection is a mucosal CD4+ T cell. At this point in acute HIV-1 infection, the ability of the infected T cell to mount its own innate intracellular antiviral response and counteractions by HIV-1 to evade this response will play a major role in defining the outcome of infection. While a successful innate immune response by the infected T cell will suppress or contain the infection and can facilitate spontaneous clearance, viral strategies to successfully evade this response will support proviral integration and virus replication to establish a focus of infection in the mucosal epithelium, recruit additional target cells of infection and facilitate spread to lymphoid tissues for systemic HIV-1 dissemination [11,13–15].
There prevail major questions regarding the interactions between HIV-1 and the processes within the infected CD4+ cell that recognize HIV-1 as non-self to trigger cell-intrinsic innate antiviral immunity and what effector pathways, molecules and their activities, as well as what viral evasion strategies, serve to determine the outcome of HIV-1 infection and immunity. Understanding these processes of the HIV-1/host interactions is essential for informing design of new therapies and an effective vaccine to control infection. Here, we provide an overview of the major PRR families, signaling pathways and transcription factor effector actions in programming cell-intrinsic innate immunity. In the second half of this presentation, we include a contemporary focus on HIV-1 to review recent advances in understanding host defense and viral evasion strategies of innate immunity that impart the outcome of HIV-1 infection.
PRR Signaling
PRR sensing of virus infection plays a critical role in discriminating self from non-self to initiate the immune response. PRR signaling and downstream effector actions must be activated quickly during virus infection but are required to be held in check in the absence of infection to prevent constant inflammation and host responses leading to autoimmune disease. Thus, PRRs have evolved to sense biochemical structures or PAMPs unique to the invading pathogen. For example, during virus infection, the presence and accumulation of double-stranded RNA (dsRNA) or single-stranded RNA (ssRNA) species in the cytoplasm of the host cell is detected by RLRs to induce immune signaling [16], while DNA present in the cytosol is detected as non-self by cyclic GMP/AMP synthase (cGAS), a recently identified PRR of cytosolic DNA [17]. Moreover, TLRs serve to identify PAMPs presented in endosomes and at the cell surface, enabling phagocytic cells to constantly sample the extracellular milieu [18]. Typically such PRR/PAMP interactions do not occur in the absence of viral infection. As a result, PRRs demonstrate location and substrate specificity in PAMP recognition. The PRR families include the TLRs, RLRs, Nod-like receptors (NLRs) and the novel DNA sensor cGAS, with each family displaying roles in viral pathogen sensing innate immune triggering. Below we summarize the role of each in viral sensing and response, and we finish with a focus on HIV-1. For further information specific to viral PAMP structure and host PRR function, we refer the reader to recent reviews on this topic [16,19,20].
TLR/PAMP interaction
The TLRs comprise nine related members in humans. All TLRs share some basic structural features, including transmembrane domains, extracellular (or endosomal) leucine-rich regions that interact with PAMPs and cytoplasmic signaling domains that initiate cascades through adaptor molecules [18]. All of the TLRs except TLR3 signal through the adaptor proteins TIRAP/MyD88, while TLR3 and TLR4 signal through TRIF/TRAM [21]. MyD88 signaling leads to the activation of nuclear factor κ-light-chain enhancer of activated B cells (NFκB), while TRIF signaling leads to activation of the transcription factors NFκB via TRAF6 and interferon regulatory factors (IRFs) 3 and 7 (IRF3 and IRF7) via TBK1 [18]. The MyD88 and TRIF signaling adaptors have toll-interleukin 1 receptor (TIR) domains that interact with cytoplasmic TLR TIR domains, and these TIR/TIR interactions are essential for TLR function [21].
TLRs are mainly located within membranes of the cell surface and endosomes. They are highly expressed in conventional dendritic cells (cDCs) and other immune cells, with TLR7 and TLR9 being especially highly expressed on plasmacytoid dendritic cells (pDCs), the major interferon-producing cells of the body [22,23]. TLRs are also expressed by various mucosal and epithelial cells, thus allowing them to mediate pathogen sensing and signaling along key virus entry points. In terms of microbial infections, TLR1, TLR2, TLR4, TLR5 and TLR6 respond primarily to bacterial PAMPs [18]. TLR7 and TLR9 recognize ssRNA and single-stranded DNA (ssDNA), respectively, and signal through MyD88 to NFκB and IRF7, while TLR3 binds dsRNA and signals through TRIF to NFκB and IRF3. TLR8 appears to function and be expressed similarly to TLR7 but is less essential for immunity [24]. TLR3, TLR7, TLR8 and TLR9 are all expressed in endosomes and bind nucleic acid PAMPs derived from phagocytosed cell debris, viruses and bacteria [18]. There is also evidence that infected pDCs can bring cytosolic PAMPs to the endolysosomal TLRs through autophagy, a process by which endosomes form around cytosolic contents [25]. Moreover, pDCs can detect viral RNA PAMPs from neighboring infected cells through exosomal transfer via cell-to-cell contacts [26,27].
RLRs recognize cytosolic RNA PAMPs
RLRs are expressed in the cytosol of all nucleated cells. The RLR family consists of three members: retinoic acid-inducible gene-I (RIG-I), melanoma differentiation-associated gene 5 (MDA5) and laboratory of genetics and physiology 2 (LGP2). All are DexD/H box RNA helicases that bind RNA, hydrolyze ATP and scan RNA substrates for recognition motifs [28–30]. RIG-I exhibits a preference for shorter dsRNA structures and ssRNA containing 5′ triphosphate and specific sequence motifs (described below) while MDA5 binds longer dsRNA structures [30]. This property of MDA5 likely reflects its ability to form long filaments on dsRNA [31]. Structurally, RIG-I and MDA5 contain N-terminal caspase activation and recruitment domains (CARDs) and a helicase domain, and RIG-I but not MDA5 has a C-terminal repressor domain (RD) [32]. LGP2 also contains a helicase domain and RD but lacks the CARD domains, and it functions as a regulator of RIG-I and MDA5 signaling [33]. The RD of RIG-I prevents unwanted activation of signaling in the absence of appropriate RNA ligand by auto-regulating CARD exposure to other signaling partners [33,34].
RIG-I binds to RNA ligands that contain a 5′ triphosphate and, in the case of hepatitis C virus, has sequence specificity for poly-U/UC RNA [33–35]. Upon binding to PAMPs, the RIG-I RD is displaced from the CARDs allowing for interaction with TRIM25 and 14–3-3ε, proteins that respectively modify RIG-I and serve as a chaperone to promote the relocation of RIG-I from the cytosol to the mitochondrial-associated endoplasmic reticulum membrane [36,37]. At the mitochondrial-associated endoplasmic reticulum membrane, RIG-I CARD interaction with the CARD located on the adaptor protein mitochondrial antiviral signaling protein (MAVS) leads to the assembly of a signaling complex that may include the adaptor protein stimulator of interferon genes (STING) [38]. Complex assembly results in the activation of NFκB and IRF3/IRF7 [39]. The ligand specificity for MDA5 is less clear, though MDA5 signaling also requires MAVS [40,41]. MDA5 is the RLR that predominantly binds and signals in response to the long forms of the synthetic RNA mimetic poly(I:C) [42]. MAVS itself is essential for signaling interferon production and antiviral immunity induced by the RLRs [40,41,43,44].
Overall, signaling in response to foreign RNA is complex with multiple sensors feeding into or augmenting the RLR pathway. For example, binding of viral dsRNA to 2′–5′ oligoadenylate synthase (OAS1) leads to the activation of RNase L, which generates RLR ligands from self and viral RNA [45]. Other DexD/H box RNA helicases, HMGB proteins, NOD2 and LRRFIP1 are reviewed elsewhere [46] for their roles in RNA sensing, and additional RNA sensors or proteins that interact with RLR pathways remain to be discovered. Interestingly, RIG-I is also activated by RNA generated as a result of the bacterial infection-induced unfolded protein response [47] and by cellular RNA polymerase III products of cytosolic DNA transcription (further described below).
Nucleotide-binding domain and leucine-rich repeat-containing proteins
The NLR proteins are expressed in the cytosol of various cell types including epithelia, antigen-presenting cells and adaptive immune cells [8]. NLRs represent a diverse family of PRRs, but they all share an ATPase domain and a C-terminal leucine-rich repeat domain [48]. The N-terminal domains vary among the different NLR family members. NOD1 and NOD2 have N-terminal CARD domains, respond to antimicrobial PAMPs in the cytosol (such as bacterial peptidoglycan derivatives or viral ssRNA) and signal to the activation of NFκB and IRF3 [49]. The NLRP proteins contain N-terminal pyrin domains and (for those that have known functions) interact with caspase-1 upon activation and thus drive interleukin-1β maturation and secretion for induction of the inflammatory response. Signals for NLRP activation include damage or danger-associated molecules, like uric acid, ATP (representing dying cells), reactive oxygen species (suggesting bacterial infection and inflammation) or ion flux (suggesting loss of membrane integrity as could occur during bacterial or viral infection) [48]. The NLRs and related NLRPs serve essential roles to detect PAMPs and signal the initiation of the inflammatory response, thus driving inflammation coincident with innate immune signaling by RLRs and TLRs during acute infection.
DNA sensors
Among TLRs, PAMP DNA sensing is accomplished by TLR9. TLR9 is located within endosomes, allowing cells to sense DNA from a wide range of microbes. Additionally, eukaryotic cells express PRRs that detect cytosolic DNA to trigger innate immune responses, as revealed by autoinflammatory diseases caused by accumulation of cytosolic DNA or defects of dNTP clearance [50–53]. The cytosolic DNA sensors discovered to date include IFI16, DNA-dependent activator of IRFs (DAI), RIG-I (RIG-I indirectly senses foreign DNA via recognition of RNA polymerase III products of cytosolic pathogen DNA transcription), DDX41, LRRFIP1, Ku70 and cGAS. DAI binds B-form DNA and activates IRF3, but surprisingly, it is not essential for innate immune responses to viral DNA in mice [54]. RNA polymerase III transcribes AT-rich DNA that can accumulate in the cytosol upon DNA virus infection. This processes was realized upon study of the cytosolic DNA analog poly(dA:dT) and its pol-III-mediated transcription into poly(rA:rU) possessing 5′ triphosphate, which is recognized by RIG-I for signaling through MAVS [55]. However, this pathway is not essential for sensing of DNA virus infection [55]. DDX41 is a DExD/H helicase family member, like the RLRs, that binds nucleic acids and signals through STING for the activation of IRF3 and NFκB [56]. IFI16 is a pyrin and HIN-200 domain-containing (PYHIN) family member that binds DNA through its HIN domain and can oligomerize with other pyrin domain-containing proteins. Similar to DDX41, IFI16 signals the activation of IRF3 in a STING-dependent manner [57]. LRRFIP1 binds DNA through an N-terminal domain and activates β-catenin, which then binds to the C-terminus of IRF3 in the nucleus and enhances transcriptional cofactor interaction [58]. Ku70 is a DNA double strand break repair protein that binds exposed DNA termini in the cytoplasm and signals to the promoter of IFN-λ1, an antiviral type III interferon [59]. Together, these factors serve to recognize cytosolic DNA as a PAMP to induce innate antiviral immunity.
However, despite the many DNA sensing mechanisms described above, and while the adaptor STING is essential for the cytoplasmic DNA response [60], no individual DNA-sensing PRR appears to be essential for DNA-dependent type I IFN production in all cell types, and this notion is still true after the recent discovery of cGAS [17]. Structurally similar to OAS1, cGAS binds DNA in the cytosol and generates the second messenger cyclic hetero-dinucleotide cGAMP, which then binds and activates STING, leading to the activation of IRF3 and production of type I IFN [17,61,62]. It should be noted that STING itself can bind cyclic di-GMP while DDX41, a binding partner of STING, can bind cyclic di-GMP and cyclic di-AMP directly to drive IFN expression [63,64]. The structure of cGAMP that is produced by mammalian cGAS appears to have an asymmetric phosphodiester linkage > Gp(2′–5′)Ap(3′–5′)> [65], unlike the symmetric > Gp(3′–5′)Ap(3′–5′) > structure originally proposed by Wu et al. [61]. The symmetric cyclic dinucleotide > Gp(3′–5′)Ap(3′–5′) > is a bacterial second messenger molecule, while the asymmetric > Gp(2′–5′)Ap(3′–5′) > appears to be unique to metazoans and thus far exclusively produced by cGAS in response to cytoplasmic DNA binding wherein it serves as a second messenger to activate STING [65–67]. Thus, further investigation of the sensing of cytosolic DNA may focus on the generation of asymmetric cGAMP by cGAS during infection by DNA viruses or retroviruses that expose viral DNA in the cytoplasm.
Sensing of HIV-1
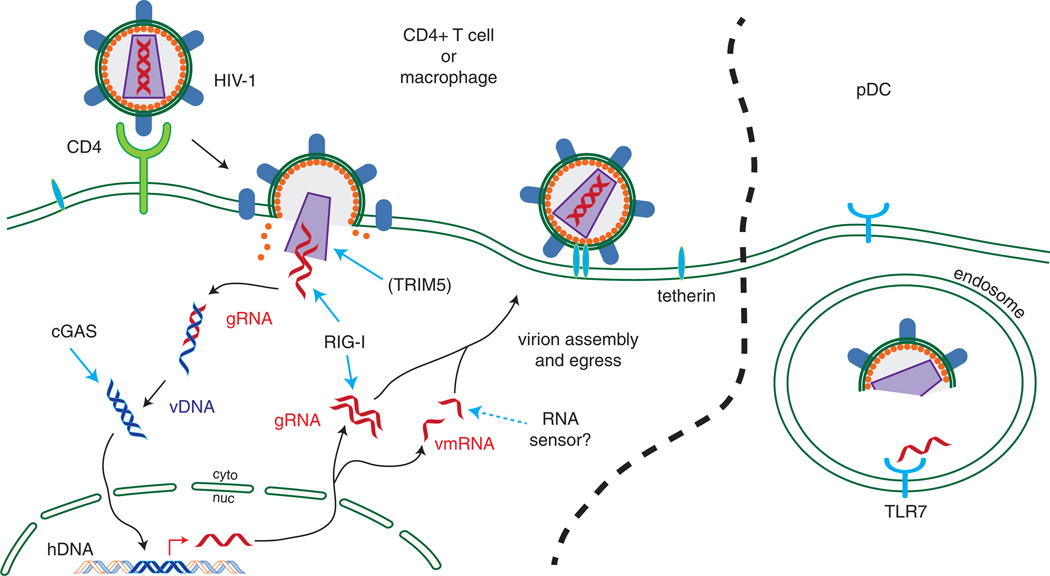
HIV-1 utilizes the cell surface molecule CD4 as its primary receptor along with CCR5 or CXCR4 as coreceptors [68]. CD4 is expressed on a subset of T cells, macrophages and dendritic cells. Whereas CCR5 is expressed on macrophages and activated CD4+ T cells, CXCR4 is expressed on naïve CD4+ T cells [68]. The life cycle of HIV-1 is depicted in Fig. 1, during which virions deliver viral capsid to the cytoplasm, which then dissociates to expose viral RNA that is then quickly reverse transcribed to DNA using cellular dNTPs. During this cycle, viral nucleic acid in the form of ssRNA (appearing perhaps briefly), RNA:DNA hybrids, ssDNA and double-stranded DNA could all theoretically be exposed as PAMPs for varying amounts of time to putative cytosolic sensors (Fig. 1). The final double-stranded DNA proviral genome translocates to the nucleus and is integrated into the host cell genome. Activation of HIV-1-infected T cells leads to viral mRNA expression, including genomic RNA, and protein production. These products assemble in new virions to mediate further rounds of infection, while viral nucleic acids can theoretically serve as PAMPs to trigger innate immune programs within the infected cell.
Fig. 1.
The HIV-1 life cycle and points of PRR sensing. Upon binding to CD4/coreceptor and viral entry into target cells (left), the HIV-1 capsid is exposed, which is sensed albeit inefficiently by TRIM5. The related SIV is sensed robustly by TRIM5 while HIV-1 is sensed effectively by TRIM5 in nonhuman primate cells. Once the capsid uncoats, genomic RNA (gRNA) accesses the cytoplasm for reverse transcription, which generates single-stranded and double-stranded viral DNA (vDNA). This DNA then integrates into the host genome (hDNA). Transcription of the integrated vDNA into viral messenger RNA (vmRNA) is necessary for HIV-1 protein translation, while new gRNA is transcribed for nascent virions. RIG-I may sense gRNA, while sensing of vDNA and vmRNA occurs through DNA and RNA sensing pathways, including cGAS/STING and other pathways that signal IRF3 activation. Tetherin senses accumulating virions at the cell surface. In pDCs (right), HIV-1 RNA is sensed by TLR7 in the endosome and may arrive there by endocytosis or autophagy.
It has been reported that HIV-1 presents PAMPs to the cell in the form of capsid structure [69] and nucleic acid [70,71]. Sensing of incoming capsid structure by the host cell TRIM5 protein signals to the transcription factors AP-1 and NFκB [69], though human TRIM5 has lost this ability in comparison to TRIM5 from closely related primates infected with the related simian virus, SIV. RIG-I has been shown to bind HIV-1 genomic RNA to trigger IRF3-dependent signaling [71], though the validation of these observations in vivo has not yet been reported. In CD4+ T cells ex vivo, it has been shown that cGAS can bind to HIV-1 reverse transcription intermediates to produce cGAMP that binds to STING to signal IRF3 activation, IFN-β expression and cell-intrinsic innate immune defenses [72]. Additional cytosolic PRRs appear to identify HIV-1 products to drive sensing of reverse transcribed DNA products to induce IFN production and innate immune defenses [73], as well as caspase activation and cell death [70]. In particular, constitutively expressed Trex1, a cellular nuclease, was found to digest ssDNA derived from endogenous retroelements and prevent their triggering of cytosolic DNA sensors [50]. In this respect, Trex1 was expected mask the presence of HIV-1 infection by digesting any potential PAMPs that are not subject to normal viral metabolism. Notably, the absence of Trex1 then provides an intracellular environment wherein HIV-1 infection results in accumulation of viral PAMP (ssDNA) ligand to trigger IRF3 activation through pathogen sensing and signaling via STING [73,74]. Tetherin likely represents another sensor of HIV-1 and is an ISG that functions to restrict the release of certain viruses (including HIV-1). In this case, tetherin may also serve to sense HIV-1 virion accumulation at the cell surface and activate downstream signaling of NFκB, thus driving innate immune gene expression and response against accumulated HIV-1 virions [75–77]. Moreover, a recent genetic screen of Trex1−/− cells infected with HIV-1-based retroviruses identified a number of host factors implicated in PRR function or signaling of HIV-1 reverse transcription DNA products. These include TRIM56, STING, TBK1 and IRF3, in addition to at least 10 other factors that likely operate independently or in conjunction with cGAS in HIV-1 recognition [73].
In addition to virus sensing mechanisms in CD4+ T cells and macrophages (the main target cells of HIV-1 infection), there are also sensing mechanisms in bystander cells not known to be productively infected with HIV-1. As an example, pDCs, via a TLR7-dependent pathway, sense endocytosed HIV-1 virions [78] or infected CD4+ T cell debris [79] and produce IFN. Interestingly, HIV-1 virion membrane fusion with the pDC endosomal membrane appears to enhance TLR7 signaling, suggesting that endocytosed virions deliver ssRNA to the cytoplasm that is then delivered from the cytoplasm back to the endosome, as through a process akin to autophagy [79,80]. Further, co-culture of HIV-1-infected CD4+ T cells with 293T cells (which lack TLR expression) led to activation of IRF3 in the 293T cells, indicating the presence of cytosolic sensing of HIV-associated PAMPs, such as DNA or RNA viral products [79]. Conversely to pDCs, HIV-1-exposed CD4+ cDCs are not efficiently infected due to the expression of the restriction factor SAMHD1 (further discussed below) and demonstrate an absence of innate immune signaling in response to HIV-1 [81].
IRF Family and IRF3
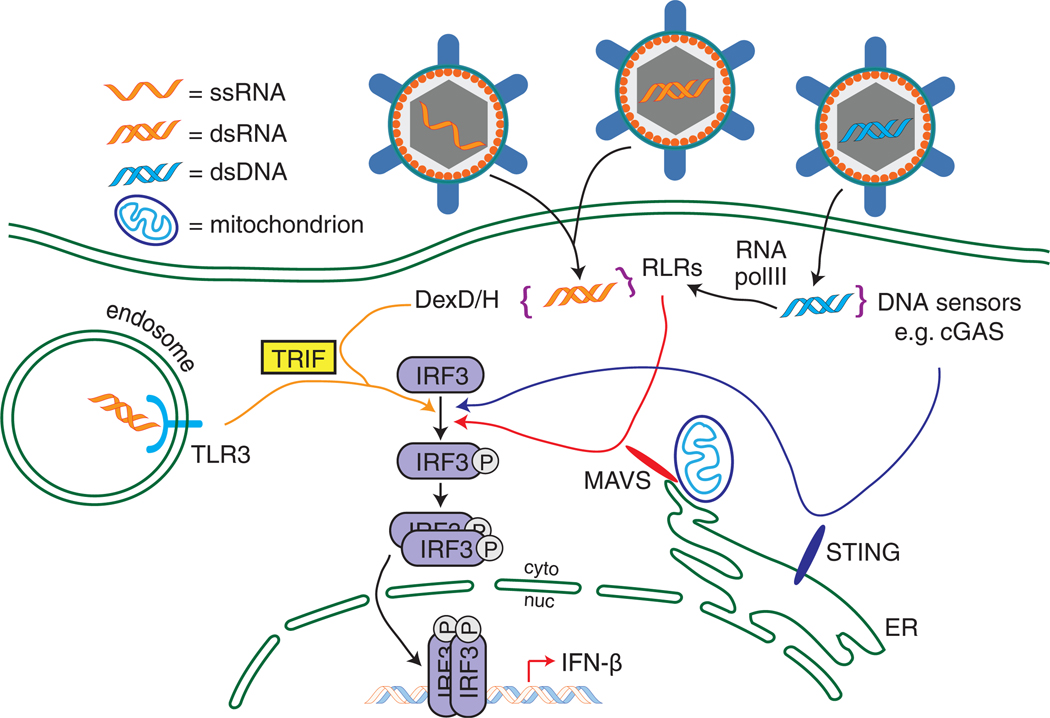
Several viral PAMP/PRR signaling pathways converge on IRF3 to induce type I IFN expression (Fig. 2). IRF3 is one of the nine members of the IRF family of transcription factors. IRFs are constitutively expressed to various levels with IRF members being differentially induced to increased levels by IFN, other proinflammatory cytokines, or virus infection. An example is IRF7, which is induced by IFN in most cell types but is constitutively expressed in pDCs. On the other hand, IRF3 is constitutively expressed to a high level in most cells [82,83]. The IRFs share an N-terminal DNA-binding domain (DBD) that contains a helix–turn–helix domain and is conserved across the family. While the different IRF DBDs have slightly different sequence binding preferences [84], the consensus DNA sequence recognized by these DBDs is called the IRF-element or IRF-E, also referred to as the IFN-stimulated response element (ISRE) by various groups. This motif is of the general sequence 5′-GAAANNGAAAG/CT/C-3′, where “N” can be any nucleotide [85]. Binding by one IRF may enhance the accessibility of nearby transcription factor binding sites [86]. All of the IRFs except IRF1 and IRF2 have an IRF association domain (IAD) near the C-terminus, which allows for homodimer formation (e.g., IRF3/IRF3), heterodimer formation (e.g., IRF3/IRF7) or other cofactor interaction [e.g., CREB-binding protein (CBP); CBP/IRF3] [82]. Some IRFs, like IRF3 and IRF7, also have C-terminal serine-rich regions containing bioactive phosphorylation sites that serve to activate the IRF and promote its nuclear accumulation and dimerization activities [85].
Fig. 2.
Convergence of antiviral sensing pathways on IRF3. Viruses containing RNA genomes, when endocytosed, present RNA ligands for TLR3 in endosomes, which then signals through TRIF to activate IRF3. RNA PAMPs generated during viral replication can be detected in the cytosol by the RLRs or other DexD/H helicases that signal through MAVS or TRIF, respectively. Viral DNA in the cytoplasm either is sensed directly by molecules such as cGAS, which signals through STING, or is transcribed by RNA pol III to generate RIG-I ligands. For simplicity, MAVS is shown on the mitochondrion, and STING on the endoplasmic reticulum (ER), though both molecules are localized to multiple intracellular membranes. Activation of these various pathways leads to IRF3 phosphorylation, dimerization, nuclear translocation and the transcription of IFN-β.
IRFs play varying roles in development and innate immunity. Among other functions, IRF1 serves to amplify TLR signaling through an interaction with MyD88 in cDCs [87]. Along with other transcription factor binding sites, IRF1 binding sites are located in the HIV-1 LTR, and IRF1 binding is required to drive HIV-1 genome expression prior to expression of the viral Tat protein [88,89]. IRF3 is activated by several PRRs, including the cytoplasmic nucleic acid sensors. Activated IRF3 drives the transcription of IRF3-dependent genes, including IFN-β, which then drives signaling through the type I IFN receptor (IFNAR). Signaling through IFNAR activates a Jak-STAT pathway leading to the formation of a transcriptional complex containing activated STAT1 and STAT2 with IRF9, which like IRF3 is constitutively expressed. This transcriptional complex, called interferon-stimulated gene factor 3 (ISGF3), drives the transcription of IRF7, which then cooperates with IRF3 to drive the expression of a large set of ISGs.
In addition to its central role in driving the IFN response to PRR signaling, IRF3 activation promotes a pro-apoptotic response [90]. This pathway involves the binding of IRF3 to Bax, translocation of IRF3/Bax to the mitochondria and the initiation of the intrinsic apoptosis pathway leading to caspase-3 activation [90]. The IRF3 activation program is important for suppression of viral infection and, if abrogated via IRF3 depletion, leads to chronic viral infection due to a failure of the host tissues to contain the spread of viral infection [91]. Further, the IRF3-mediated process of apoptotic viral restriction is further amplified by the IRF3-dependent gene, IFIT2, which itself drives a similar Bax- or Bak-mediated apoptotic response [92]. In summary, innate immune signaling in response to virus infection culminates in the actions of members of the IRF transcription factor family. IRF3 plays a central role in the induction of the innate antiviral response and apoptotic response for restriction of virus infection.
IRF3 Activation
IRF3 is a 427-amino-acid protein with the domain structure indicated in Fig. 3. The C-terminal serine-rich region of IRF3 contains several phosphorylation sites. While IRF3 is basally phosphorylated within its amino-terminus in the resting state in non-infected cells, activation of IRF3 is driven by virus-induced phosphorylation of residues S385, S386, T390 and S396 [93]. Phosphorylation at S385 or S386 is required for IRF3 dimerization, while phosphorylation at S396 is required for IRF3 dimerization, translocation to the nucleus and association with CBP/p300, histone acetyltransferases necessary for IRF3 transcriptional activity [94,95]. Activation-specific phosphorylation is governed by the IκB kinase-related protein kinases TBK1 and IKKε [96,97], which are activated downstream of TRIF-, MAVS- or STING-dependent PRR signaling [97–99]. After phosphorylation, IRF3 dimerizes, which is thought to occur as a result of charged repulsion of the C-terminal phosphorylation domain from the IAD, exposing the IAD for IAD/IAD interaction between two IRF3 molecules [100]. IRF3 then translocates to the nucleus where it interacts with the cofactor CBP/p300. This complex then binds cooperatively to promoters containing the IRF-E/ISRE sequence leading to IRF3-dependent gene transcription [84]. Dimeric IRF3 actually engages two IRF-E/ISRE sequences often in close proximity to each other. Indeed, the tight packing of the IRF3 homodimer at these DNA binding sites may be important for alteration of DNA structure to facilitate assembly of functional transcription initiation complexes to drive gene expression [101].
Fig. 3.
Schematic of IRF3 functional domains. NLS, nuclear localization sequence; NES, nuclear export sequence; PRO, proline-rich region; SRR, serine-rich region.
IRF3 activity is tightly regulated, such that its activity is shut down once it has performed its duty in transcription of innate immune genes. Thus, if the innate immune response is successful in clearing the cell of virus and associated PAMP, the cell then returns to a normal metabolic state lacking IFN and proinflammatory cytokine expression, bringing resolution to the innate immune and inflammatory responses. This process of response resolution is accompanied by a variety of processes designed to dampen the host response to infection. For example, the transcription factor MafB blocks recruitment of cofactors to IRF3 in the nucleus [102], while the activation of caspase-8 by RLR signaling leads to caspase-8-mediated cleavage of IRF3, preparing it for ubiquitination [103]. Importantly, the RBCK1 is a key virus-inducible E3 ubiquitin ligase that ubiquitinates phosphorylated/active IRF3 in a reaction requiring the cellular Pin1 protein, thus leading to proteasome-dependent degradation of IRF3 [104,105]. SUMOylation of IRF3 has also been implicated in IRF3 turnover and regulation of IRF3 function [106]. As IRF3 mRNA is highly stable and constitutively expressed, any decrease in total IRF3 protein abundance due to activation and depletion can be rectified rapidly, thus providing a pool of IRF3 for subsequent virus stimulation and metabolism [107].
IRF3-Responsive Genes and ISGs
The archetype IRF3-dependent gene is the antiviral cytokine IFN-β, which employs a well-defined virus-inducible enhancer element located − 102 to − 47 bp from the transcription start site of the IFN-β gene [108]. Strong activation of this enhancer, which is only basally activated at best in resting cells [109], involves the cooperation of several transcription factors: NFκB (composed of subunits p50 and p65/RelA), an IRF3 dimer, an IRF7 dimer and AP-1 (composed of subunits ATF-2 and c-Jun) [110]. IRF7, like IRF3, binds to the IRF-E/ISRE sequence but its DBD stringency is less than that for IRF3 so it engages a wider variety of promoters with ISRE elements (including IFN-α promoters) to induce and amplify a broad ISG response [84]. On the IFN-β promoter, these factors assemble in very close proximity to bind most nucleotides in a 51-base stretch of DNA. This complex, termed the enhanceosome, is governed by the post-translational modification of its constituent factors [110]. Each factor is stabilized on the DNA by interactions with CBP/p300 within the enhanceosome structure [108]. While IRF7 is expressed at low levels prior to IFN signaling to allow IFN-β transcription at early time points during acute virus infection, IRF7 expression is highly increased upon IFN-β signaling, thus amplifying the IFN response [111]. IRF3 and IRF7 are sufficient to drive a level of virus-dependent IFN-β production in the absence of NFκB and AP-1. While NFκB and AP-1 may be responsible for the weak basal IFN-β promoter activity to support tonic signaling by IFN, it is the full assembly of the enhanceosome that mediates the robust production of IFN-β during initial PAMP signaling in acute virus infection [112].
In addition to IFN, IRF3 directly drives the expression of many other genes by binding to an IRF-E/ISRE in their promoters. These genes include several with known antiviral function, such as viperin, IFIT1, IFIT2, IFIT3, ISG15 and IFITM3, as well as IFN-α subtypes, and also other ISGs [113,114]. In addition, IRF3 directly increases expression of immunomodulatory molecules such as chemokines CCL4/MIP-1β and CCL5/RANTES, as well as the DNA sensor IFI16 [114,115]. For some genes, like IFIT2, IRF3 activation alone is sufficient to drive expression, while CCL5 expression involves cooperation of IRF3 with NFκB [114]. The functions of these and other select genes directly stimulated by IRF3 are listed in Table 1.
Table 1.
IRF3 target genes in virus infection
| Gene name | Function | Reference |
|---|---|---|
| CCL4 (MIP-1β) | Proinflammatory cytokine, immune cell activation and chemotaxis | [114] |
| CCL5 (RANTES) | Proinflammatory cytokine, immune cell activation and chemotaxis | [114,115] |
| CXCL10 | Proinflammatory cytokine, immune cell activation and chemotaxis | [114] |
| IFI16 | Sensor of intracellular DNA | [57,114] |
| IFIT1 (ISG56) | Inhibition of protein translation by phosphorylation of eIF3, viral triphosphate RNA binding | [113,114,116–118] |
| IFIT2 (ISG54) | Viral, AU-rich RNA binding, association with IFIT1 | [114,118,119] |
| IFIT3 (ISG60) | Association with IFIT1/IFIT2 other functions unknown | [114,118,119] |
| IFITM3 | Disrupts cellular cholesterol homeostasis and inhibits viral entry by blocking viral membrane fusion with plasma membrane | [114,120,121] |
| IFN-α4 | Type I interferon, ligates IFNAR | [114] |
| IFN-α5 | Type I interferon, ligates IFNAR | [114] |
| IFN-β | Type I interferon, ligates IFNAR | [114,122] |
| ISG15 | Attached to proteins in a process similar to ubiquitination; activates PKR | [113,114,118,123] |
| ISG20 | 3′–5′ RNA exonuclease | [114,124,125] |
| Major histocompatibility complex class I | Viral antigen presentation | [113,114] |
| OAS1 | RNA binding, RNase L activation, RIG-I activation | [45,118] |
| Viperin/RSAD2 | Inhibition of lipid raft synthesis, regulation of cellular metabolism | [114,118,126,127] |
| ZAP | Viral mRNA degradation | [128,129] |
Once IFN is transcribed, translated and secreted, it binds to IFNAR on the same or nearby cells. Ligation of IFNAR by extracellular IFN results in the phosphorylation of associated kinases Jak1 and Tyk2 at the IFNAR cytoplasmic domain, resulting in recruitment and phosphorylation of STAT1 and STAT2 [130,131]. STAT1/STAT2 heterodimers dissociate from the membrane signaling complex and associate with cytosolic IRF9 to form a trimeric complex called ISGF3 [132]. This complex binds ISRE sites in gene promoters in the nucleus and drives transcription of ISGs, most importantly IRF7. IRF7 is then expressed in the cytosol and, when activated, functions as a homodimer or in a heterodimeric complex with IRF3 to drive expression of a large set of ISGs, including IFN-β and all IFN-α subtypes, broadening the antimicrobial response [133]. As noted above, the differential abilities of IRF3 and IRF7 to drive transcription of genes of host defense is highlighted by the respective promoter sequences they recognize, reflecting that IRF7 DNA binding sequence is less restricted than that for IRF3 [84].
To date, of the hundreds of ISGs present in human cells, only a relative few have been functionally characterized. Notably, many PRRs are ISGs themselves, and many enzymes of cell metabolism are also ISGs directly involved in antiviral activity [3]. ISGs may also modify existing cellular proteins; for example, the TRIM family contains many ISGs, some of which ubiquitinate, SUMOylate or ISGylate host proteins with varying effects on the antiviral response [134,135]. A summary of select human ISGs with known functions particularly enriched for those with anti-HIV-1 activity (and not listed in Table 1) is displayed in Table 2.
Table 2.
ISG examples and their functions in virus infection
| Gene name | Function | Reference |
|---|---|---|
| APOBEC3G | Deaminates retroviral reverse transcription intermediate DNA | [136–138] |
| cGAS | Senses intracellular DNA | [3,17] |
| Herc5 | Inhibits HIV-1 assembly | [139] |
| IFITM1 and IFITM2 | Inhibits viral entry by blocking viral membrane fusion with plasma membrane | [3,120] |
| IRF1 | ISRE binding, signal amplification | [3,87] |
| IRF2 | Attenuates IRF1 activity | [3,140] |
| MDA5 | PRR for cytoplasmic dsRNA | [3,40] |
| PKR | Phosphorylates eIF2α, inhibiting translation | [141] |
| RIG-I | PRR for cytoplasmic dsRNA | [3,40] |
| Schlafen 11 | Depletes tRNA pool, disadvantaging HIV-1 protein translation due to codon bias | [142] |
| Tetherin/BST2 | Restricts enveloped virus release by tethering virions to the cell membrane, activates NFκB | [75–77] |
| TRIM21 | Inhibits normal IRF3 degradation | [143] |
| TRIM28/KAP1 | Deacetylates HIV-1 integrase, inhibiting proviral integration | [144] |
| TRIM5α | Destabilizes incoming retroviral capsid, activates NFκB | [69,135,145] |
Viral Antagonism of IRF3
While IRF3-dependent gene induction is attenuated by ubiquitination and proteasome-dependent degradation of IRF3 protein in the course of normal cell metabolism following resolution of acute of virus infection, IRF3 function is itself targeted by many different viruses in order to evade innate immunity. Viral control of IRF3 allows for the linked inhibition of innate immunity, and virus-mediated blockades that impose indirectly or directly on IRF3 or that disrupt ISG expression are observed at the level of PAMP sensing, PRR signaling, IRF3 activation, IRF3 stability, IFN signaling and the function of specific ISGs [146,147]. We refer the reader to several excellent reviews on viral evasion of IRF3 function [46,148,149]. Table 3 provides an overview of RNA and DNA virus antagonism strategies for targeting of IRF3, while HIV-1 strategies for IRF3 and innate immune regulation are described below.
Table 3.
Examples of virus-encoded antagonists of innate immunity
| Virus | Antagonist encoded | Effect on host cell | Reference |
|---|---|---|---|
| Bocavirus | NP1 | Binds to IRF3 DBD, blocking binding to IFN-β promoter | [150] |
| Borna disease virus | Phosphoprotein P | Competes with IRF3 for TBK1 binding, preventing IRF3 phosphorylation | [151] |
| Bovine viral diarrhea Virus | Npro | Ubiquitinates IRF3, targeting it for proteasomal degradation | [152] |
| Dengue virus | NS2B3 | Cleaves STING, blocking IFN induction | [153] |
| Ebola | VP35 | Binds dsRNA, masking PAMPs from RLRs; inhibits IRF3 activation downstream of MAVS | [154,155] |
| Epstein-Barr virus (EBV) | BRLF1 | Inhibits IRF3 and IRF7 mRNA transcription and protein expression | [156] |
| EBV | BGLF4 | Phosphorylates IRF3 in a proline-rich region, inhibiting IRF3 activation | [157] |
| Hepatitis C virus | NS3/4A | Cleaves MAVS, preventing RLR signaling; cleaves TRIF, preventing TLR3 signaling | [98,158–161] |
| Herpes simplex virus 1 | ICP0 | Degrades IFI16, preventing sensing of nuclear viral DNA; sequesters IRF3 and CBP away from chromatin | [162,163] |
| HIV-1 | Vpu | Targets tetherin and CD4 for sequestration and degradation and displaces tetherin from virions, enhancing virus release; targets IRF3 and prevents IRF3-driven gene induction | [164–169] |
| HIV-1 | Vif | Targets APOBEC3G for degradation, preventing hypermutation of reverse transcribed DNA | [136,170] |
| HIV-1 | Protease | Targets RIG-I for lysosomal degradation | [171] |
| Human herpesvirus 6 | IE1 | Prevents IRF3 activation | [172] |
| Kaposi’s sarcoma herpes virus (KSHV) | RTA | Ubiquitinates IRF7, targeting it for proteasomal degradation | [173] |
| KSHV | vIRF1 | Sequesters CBP/p300 away from IRF3, preventing IRF3-driven gene transcription | [174] |
| KSHV | vIRF2 | Prevents induction of IRF3-driven genes as well as ISGF3-dependent ISGs | [175] |
| KSHV | vIRF3 | Binds to DBD or IAD on IRF7, preventing DNA binding | [176] |
| Respiratory syncytial virus | NS1 | Binds to IRF3, preventing CBP interaction | [177] |
| Rhesus cytomegalovirus | Virion component | Prevents IRF3 activation after virus entry | [178] |
| Rotavirus | NSP1 | Targets IRF3 for proteasomal degradation | [179] |
| Varicella-zoster virus | ORF61 | Binds and targets activated IRF3 for proteasomal degradation | [180] |
| West Nile virus | NS5 | Prevents STAT1 phosphorylation, preventing IFNAR signaling | [147] |
Modulation of Innate Immunity by HIV-1
HIV-1 infection is currently incurable without allogeneic bone marrow transplant in chronically infected adults or without aggressive and rapid combination antiretroviral therapy in neonates and acutely infected adults [181–183]. HIV-1 success at establishing chronic infection is linked with a variety of immune evasion strategies beyond the basic integration of the HIV-1 genome into target cells. These include an overall ability to maintain a latent infection and to avoid antibody responses by varying or shielding immunodominant epitopes within the viral envelope glycoprotein [184,185], variation of CD8+ T cell epitopes among viral proteins [186], regulation of innate immune effector gene function [187] and control of innate immune signaling after PRR recognition (described below).
Of particular importance to macrophages and cDCs in HIV-1 infection was the discovery of the cellular SAMHD1 protein as a restriction factor for HIV-1 infection in these cells [188]. As a result of SAMHD1 actions, HIV-1 infection is highly restricted in these antigen-presenting cells such that the cells process and present a paucity of viral antigen at best, thus reducing the onset and breadth of T-cell-mediated immunity. The mechanism for HIV-1 restriction is attributed to SAMHD1-mediated decrease of the cytosolic free intracellular dNTP pool, resulting in inhibition of viral DNA synthesis during the HIV-1 reverse transcription phase [189]. In contrast, HIV-2, which can productively infect cDCs and macrophages, encodes the protein Vpx, which effectively targets SAMHD1 for degradation and enables HIV-2 to infect these cell types [190]. The activity of SAMHD1 to deplete the dNTP pool is inhibited by phosphorylation, while this inhibition is relieved or “derepressed” by IFN treatment of cells [191]. In resting CD4+ T cells, which are non-permissive to HIV-1 infection, ectopic Vpx expression imparts permissiveness for HIV-1 infection, implicating SAMHD1 as an HIV-1 restriction factor [192]. SAMHD1 is phosphorylated and inactivated in dividing cells, including activated CD4+ T cells, which then become permissive to HIV-1 [191,193]. The connection with IFN as a “derepressor” of SAMHD1 thereby confers regulation by IFN and PRR signaling programs to SAMHD1 function.
Upon HIV-1 recognition by specific PRRs (described above), the activation of IRF3 and induction of IFN and innate intracellular defenses drives the expression of genes that impart suppression of HIV-1 infection. These HIV-1 restriction factors are also ISGs and include TRIM5α, which recognizes and disassembles incoming capsid [145,194]; IFITM proteins, which prevent viral membrane fusion [120,195]; tetherin, which prevents viral escape from infected cells [75–77]; APOBEC3G, a member of a family of deaminases that mutates reverse transcribed DNA [136–138,170]; and schlafen 11, which depletes the cytosolic tRNA pool to preferentially inhibit viral codon-biased translation [142]. Mechanisms employed by HIV-1 to avoid the effects of the IFITM proteins or schlafen 11 have not yet been defined, while HIV-1 is known to antagonize or evade other molecules of innate antiviral defense. In particular, human TRIM5α does not effectively recognize HIV-1 capsid but does recognize SIV. TRIM5α is thought to have evolved in humans to more specifically recognize a now extinct retrovirus that shared homology to SIV but not to current HIV strains, which might have developed evasion mechanisms against TRIM5α actions [196]. On the other hand, tetherin is actively antagonized by HIV-1 through the actions of the Vpu protein to promote virus release [75], while APOBEC3G is targeted for inhibition by the HIV-1 Vif protein to prevent its hypermutation of the viral genome [170]. In addition, RIG-I functions as a putative PRR of HIV-1 RNA products and is antagonized by the viral protease [171]. Thus HIV-1 has evolved both passive and active mechanisms to regulate the innate immune response to avoid restriction and support viral replication and spread.
IRF3 regulation by HIV-1
IRF3 is a central transcriptional regulator of innate immune gene induction, but until recently, its role in the cell-intrinsic innate immune defenses of CD4+ T cells had not been described. IRF3 was found to play an essential role in protecting T cells from virus infection, such that IRF3 activation is rapidly induced in T cells infected with a variety of RNA virus pathogens. Importantly, active IRF3 was shown to robustly suppress HIV-1 infection when ectopically expressed in T cell lines [107]. Remarkably, however, acute HIV-1 infection failed to trigger IFN expression or innate immune gene induction in these cells, suggesting that HIV-1 may regulate or suppress IRF3 activation during acute infection. Thus, two independent groups have now reported that HIV-1 causes the degradation of IRF3 and blocks IRF3 activation of innate immune gene induction [107,197]. This ability of HIV-1 to block innate immune signaling may be important in the early replication and spread of the virus. In the first report of IRF3 antagonism by HIV-1, Okumura et al. found that both Vif and Vpr contributed to IRF3 depletion in infected cells [197]. In subsequent work, Doehle et al. found that Vpu-deficient strains of HIV-1 allowed for the induction of IRF3-dependent genes whereas Vpu-expressing strains blocked IRF3-induced gene expression concomitant with driving the reduction of IRF3 protein levels in the infected cell, suggesting that Vpu plays a role in antagonism of IRF3 function [164]. In particular, Vpu was shown to block virus-induced IFN-β promoter signaling but not NFκB signaling, implying that it mediates a specific block to IRF3 activation rather than PRR signaling in general. IRF3-directed regulation by HIV-1 therefore disrupts innate immune induction while sparing NFκB activity, the latter of which is essential for viral RNA expression. During HIV-1 infection, Vpu was found to be co-localized with IRF3 and lysosomal markers such that lysosome inhibitors rescued IRF3 expression during HIV-1 infection [165]. These findings suggest that IRF3 is degraded during HIV-1 infection and that Vpu mediates a block in IRF3-dependent signaling by directing the lysosome-mediated destruction of IRF3. The regulation of IRF3 by HIV-1 has been called into question when it was reported that HIV-1 targets NFκB signaling but not IRF3 [198], in contrast to previous reports [164,197]. Of relevance is that Vpu has also been proposed to inhibit the activation of the IFN-β promoter by blocking tetherin-driven NFκB signaling [198]. This Vpu regulation of NFκB through tetherin interaction could in part explain the discrepancies in these studies. Vpu has been additionally implicated in the suppression of ligands displayed on infected cells and recognized by NK cells, thus allowing HIV to evade NK-cell mediated killing [199–201]. It is intriguing to speculate the expression of these NK cell ligands could be regulated by IRF3 and/or NFκB, thus connecting NK cell killing activity of targets with these HIV-1 innate immune evasion strategies.
Despite evidence that HIV-1 counteracts the induction of interferon through innate immune regulation strategies, the level of IFN in patient sera is strongly elevated during the early days and weeks of infection [11] and is reported to be elevated overall during chronic infection compared to healthy controls [202]. The source of the acute IFN is attributed to pDCs recruited to infectious foci in the mucosa [203], especially since IFN production by cDCs is inefficient in HIV-1 infection [204], though seeding of infection to gut-associated tissue or other lymphoid tissue could also be a source of IFN in vivo [70,202]. Importantly, the chronic activation of pDCs and IFN production is associated with an immune exhaustion and dysfunction phenotype that precipitates opportunistic infections during advanced HIV disease [205]. The sensing by pDCs of HIV-1 virions or infected cell debris through TLR7 or cytosolic sensors likely contributes to chronic pDC activation in vivo, though IRF7 rather than IRF3 is thought to be the central transcription factor for pDC IFN production [111]. Of note is that IRF7 is not suppressed by HIV-1 [107]. As such, while antagonism of innate immune signaling by HIV-1 through IRF3 or other mechanisms in acute infection may be effective enough to evade innate immunity and promote viral spread, these processes do not suppress IRF7-driven systemic IFN production by pDCs, thus contributing to the chronic immune activation that can accompany HIV-1 infection.
Concluding Remarks
Investigation of innate immune induction and antagonism by viruses should drive the identification of factors as therapeutic targets to intervene with acute and chronic infection. Moreover, defining the processes of PRR signaling will facilitate PRR targeting with artificial or natural ligands in strategies for immune enhancement and vaccine adjuvant design. Defining the HIV-1/host interactions that regulate innate antiviral immunity directly within CD4+ cells is paramount to rendering novel approaches that would intervene with infection before replication and spread of the virus to therapeutically inaccessible tissue reservoirs. Such strategies will likely need to be targeted to mucosal sites of virus exposure where innate antiviral defenses would have the greatest impact during early HIV-1 infection. We note that, in Dr. Charles Janeway’s 1989 discussion on the nature of innate immunity, he predicted that: “A virus that did not induce costimulator activity and also did not infect dendritic cells might be particularly dangerous” [5]. Indeed, this discussion today defines HIV-1. Interventions to boost the innate immune response against HIV-1 are therefore warranted and sorely needed.
Acknowledgments
We thank Dr. Stacy M. Horner for critical reading of the manuscript. A.R. is funded by University of Washington Medical Scientist Training Program (5T32GM007266-34) and University of Washington STD/AIDS Training Grant Program (T32 AI07140-32). The Gale laboratory is supported by National Institutes of Health grants.
Abbreviations used:
- IRF
interferon regulatory factor
- HIV-1
human immunodeficiency virus type 1
- PAMP
pathogen-associated molecular pattern
- PRR
pattern recognition receptor
- dsRNA
double-stranded RNA
- ssRNA
single-stranded RNA
- cGAS
cyclic GMP/AMP synthase
- NLR
Nod-like receptor
- cDC
conventional dendritic cell
- pDC
plasmacytoid dendritic cell
- ssDNA
single-stranded DNA
- CARD
caspase activation and recruitment domain
- RD
repressor domain
- DBD
DNA-binding domain
- ISRE
IFN-stimulated response element
- IAD
IRF association domain
- CBP
CREB-binding protein
References
- [1].Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science 2010;327:291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity 2006;25:373–81. [DOI] [PubMed] [Google Scholar]
- [3].Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 2011;472:481–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Medzhitov R. Approaching the asymptote: 20 years later. Immunity 2009;30:766–75. [DOI] [PubMed] [Google Scholar]
- [5].Janeway CA. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harbor Symp Quant Biol 1989;54:1–13. [DOI] [PubMed] [Google Scholar]
- [6].Welsh RM, Bahl K, Marshall HD, Urban SL. Type 1 interferons and antiviral CD8 T-cell responses. PLoS Pathog 2012;8:e1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tough DF. Modulation of T-cell function by type I interferon. Immunol Cell Biol 2012;90:492–7. [DOI] [PubMed] [Google Scholar]
- [8].Michallet MC, Rota G, Maslowski K, Guarda G. Innate receptors for adaptive immunity. Curr Opin Microbiol 2013;16:296–302. [DOI] [PubMed] [Google Scholar]
- [9].Suthar MS, Ramos HJ, Brassil MM, Netland J, Chappell CP, Blahnik G, et al. The RIG-I-like receptor LGP2 controls CD8(+) T cell survival and fitness. Immunity 2012;37:235–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012;380:2095–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cohen MS, Shaw GM, McMichael AJ, Haynes BF. Acute HIV-1 infection. N Engl J Med 2011;364:1943–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med 2009;206:1273–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Haase AT. Targeting early infection to prevent HIV-1 mucosal transmission. Nature 2010;464:217–23. [DOI] [PubMed] [Google Scholar]
- [14].Haase AT. Early events in sexual transmission of HIV and SIV and opportunities for interventions. Annu Rev Med 2011;62:127–39. [DOI] [PubMed] [Google Scholar]
- [15].McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. The immune response during acute HIV-1 infection: clues for vaccine development. Nat Rev Immunol 2010;10:11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Loo YM, Gale M. Immune signaling by RIG-I-like receptors. Immunity 2011;34:680–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013;339:786–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 2010;11:373–84. [DOI] [PubMed] [Google Scholar]
- [19].Wilkins C, Gale M. Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol 2010;22:41–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ramos HJ, Gale M. RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr Opin Virol 2011;1:167–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Botos I, Segal DM, Davies DR. The structural biology of Toll-like receptors. Structure 2011;19:447–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell 2006;124:783–801. [DOI] [PubMed] [Google Scholar]
- [23].Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol 2006;7:131–7. [DOI] [PubMed] [Google Scholar]
- [24].Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 2004;303:1526–9. [DOI] [PubMed] [Google Scholar]
- [25].Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 2007;315:1398–401. [DOI] [PubMed] [Google Scholar]
- [26].Dreux M, Garaigorta U, Boyd B, Decembre E, Chung J, Whitten-Bauer C, et al. Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host Microbe 2012;12:558–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Takahashi K, Asabe S, Wieland S, Garaigorta U, Gastaminza P, Isogawa M, et al. Plasmacytoid dendritic cells sense hepatitis C virus-infected cells, produce interferon, and inhibit infection. Proc Natl Acad Sci U S A 2010;107:7431–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Patel SS, et al. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature 2011;479:423–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, et al. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science 2009;323:1070–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 2008;205:1601–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Peisley A, Jo MH, Lin C, Wu B, Orme-Johnson M, Walz T, et al. Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments. Proc Natl Acad Sci U S A 2012;109: E3340–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol 2005;175:2851–8. [DOI] [PubMed] [Google Scholar]
- [33].Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, et al. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci U S A 2007;104:582–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 2008;454:523–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Schnell G, Loo YM, Marcotrigiano J, Gale M. Uridine composition of the poly-U/UC tract of HCV RNA defines non-self recognition by RIG-I. PLoS Pathog 2012;8:e1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liu HM, Loo YM, Horner SM, Zornetzer GA, Katze MG, Gale M. The mitochondrial targeting chaperone 14–3-3epsilon regulates a RIG-I translocon that mediates membrane association and innate antiviral immunity. Cell Host Microbe 2012;11:528–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Horner SM, Liu HM, Park HS, Briley J, Gale M. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci U S A 2011;108:14590–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008;455:674–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Paz S, Sun Q, Nakhaei P, Romieu-Mourez R, Goubau D, Julkunen I, et al. Induction of IRF-3 and IRF-7 phosphorylation following activation of the RIG-I pathway. Cell Mol Biol (Noisy-le-grand) 2006;52:17–28. [PubMed] [Google Scholar]
- [40].Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol 2008;82:335–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 2005;6:981–8. [DOI] [PubMed] [Google Scholar]
- [42].Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A 2006;103:8459–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, et al. The specific and essential role of MAVS in antiviral innate immune responses. Immunity 2006;24:633–42. [DOI] [PubMed] [Google Scholar]
- [44].Suthar MS, Ma DY, Thomas S, Lund JM, Zhang N, Daffis S, et al. IPS-1 is essential for the control of West Nile virus infection and immunity. PLoS Pathog 2010;6:e1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Malathi K, Dong B, Gale M, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 2007;448:816–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Goubau D, Deddouche S, Reis ESC. Cytosolic sensing of viruses. Immunity 2013;38:855–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cho JA, Lee AH, Platzer B, Cross BC, Gardner BM, De Luca H, et al. The unfolded protein response element IRE1alpha senses bacterial proteins invading the ER to activate RIG-I and innate immune signaling. Cell Host Microbe 2013;13:558–69. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [48].Kufer TA, Sansonetti PJ. NLR functions beyond pathogen recognition. Nat Immunol 2011;12:121–8. [DOI] [PubMed] [Google Scholar]
- [49].Ting JP, Duncan JA, Lei Y. How the noninflammasome NLRs function in the innate immune system. Science 2010;327:286–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 2008;134:587–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, et al. Mutations involved in Aicardi–Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet 2009;41:829–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Goncalves A, Karayel E, Rice GI, Bennett KL, Crow YJ, Superti-Furga G, et al. SAMHD1 is a nucleic-acid binding protein that is mislocalized due to aicardi–goutieres syndr ome-associ ated mutati ons. H um M u tat 2012;33:1116–22. [DOI] [PubMed] [Google Scholar]
- [53].Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, et al. Mutations in the gene encoding the 3′–5′ DNA exonuclease TREX1 cause Aicardi–Goutieres syndrome at the AGS1 locus. Nat Genet 2006;38:917–20. [DOI] [PubMed] [Google Scholar]
- [54].Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature 2008;451:725–9. [DOI] [PubMed] [Google Scholar]
- [55].Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 2009;138:576–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol 2011;12:959–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 2010;11:997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, et al. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat Immunol 2010;11:487–94. [DOI] [PubMed] [Google Scholar]
- [59].Zhang X, Brann TW, Zhou M, Yang J, Oguariri RM, Lidie KB, et al. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J Immunol 2011;186:4541–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 2009;461:788–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wu J, Sun L, Chen X, Du F, Shi H, Chen C, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 2013;339:826–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature 2013;498:332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 2011;478:515–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol 2012;13:1155–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, et al. cGAS produces a 2′−5′-linked cyclic dinucleotide second messenger that activates STING. Nature 2013;498:380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 2013;153:1094–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, et al. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep 2013;3:1355–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Arrildt KT, Joseph SB, Swanstrom R. The HIV-1 env protein: a coat of many colors. Curr HIV/AIDS Rep 2012;9:52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 2011;472:361–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Doitsh G, Cavrois M, Lassen KG, Zepeda O, Yang Z, Santiago ML, et al. Abortive HIV infection mediates CD4 T cell depletion and inflammation in human lymphoid tissue. Cell 2010;143:789–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Berg RK, Melchjorsen J, Rintahaka J, Diget E, Soby S, Horan KA, et al. Genomic HIV RNA induces innate immune responses through RIG-I-dependent sensing of secondary-structured RNA. PLoS One 2012;7:e29291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 2013;341:903–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Lee MN, Roy M, Ong SE, Mertins P, Villani AC, Li W, et al. Identification of regulators of the innate immune response to cytosolic DNA and retroviral infection by an integrative approach. Nat Immunol 2013;14:179–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol 2010;11:1005–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 2008;451:425–30. [DOI] [PubMed] [Google Scholar]
- [76].Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, Johnson MC, et al. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 2009;139:499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Galao RP, Le Tortorec A, Pickering S, Kueck T, Neil SJ. Innate sensing of HIV-1 assembly by Tetherin induces NFkappaB-dependent proinflammatory responses. Cell Host Microbe 2012;12:633–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Beignon AS, McKenna K, Skoberne M, Manches O, DaSilva I, Kavanagh DG, et al. Endocytosis of HIV-1 activates plasmacytoid dendritic cells via Toll-like receptorviral RNA interactions. J Clin Invest 2005;115:3265–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Lepelley A, Louis S, Sourisseau M, Law HK, Pothlichet J, Schilte C, et al. Innate sensing of HIV-infected cells. PLoS Pathog 2011;7:e1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Iwasaki A. Innate immune recognition of HIV-1. Immunity 2012;37:389–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Goujon C, Arfi V, Pertel T, Luban J, Lienard J, Rigal D, et al. Characterization of simian immunodeficiency virus SIVSM/human immunodeficiency virus type 2 Vpx function in human myeloid cells. J Virol 2008;82:12335–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Honda K, Takaoka A, Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 2006;25:349–60. [DOI] [PubMed] [Google Scholar]
- [83].Au WC, Moore PA, Lowther W, Juang YT, Pitha PM. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci U S A 1995;92:11657–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lin R, Genin P, Mamane Y, Hiscott J. Selective DNA binding and association with the CREB binding protein coactivator contribute to differential activation of alpha/beta interferon genes by interferon regulatory factors 3 and 7. Mol Cell Biol 2000;20:6342–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol 2006;6:644–58. [DOI] [PubMed] [Google Scholar]
- [86].Fujii Y, Shimizu T, Kusumoto M, Kyogoku Y, Taniguchi T, Hakoshima T. Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. EMBO J 1999;18:5028–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Negishi H, Fujita Y, Yanai H, Sakaguchi S, Ouyang X, Shinohara M, et al. Evidence for licensing of IFN-gamma-induced IFN regulatory factor 1 transcription factor by MyD88 in Toll-like receptor-dependent gene induction program. Proc Natl Acad Sci U S A 2006;103:15136–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Sgarbanti M, Borsetti A, Moscufo N, Bellocchi MC, Ridolfi B, Nappi F, et al. Modulation of human immunodeficiency virus 1 replication by interferon regulatory factors. J Exp Med 2002;195:1359–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Sgarbanti M, Remoli AL, Marsili G, Ridolfi B, Borsetti A, Perrotti E, et al. IRF-1 is required for full NF-kappaB transcriptional activity at the human immunodeficiency virus type 1 long terminal repeat enhancer. J Virol 2008;82:3632–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Chattopadhyay S, Yamashita M, Zhang Y, Sen GC. The IRF-3/Bax-mediated apoptotic pathway, activated by viral cytoplasmic RNA and DNA, inhibits virus replication. J Virol 2011;85:3708–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Chattopadhyay S, Fensterl V, Zhang Y, Veleeparambil M, Yamashita M, Sen GC. Role of interferon regulatory factor 3-mediated apoptosis in the establishment and maintenance of persistent infection by Sendai virus. J Virol 2013;87:16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Stawowczyk M, Van Scoy S, Kumar KP, Reich NC. The interferon stimulated gene 54 promotes apoptosis. J Biol Chem 2011;286:7257–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Bergstroem B, Johnsen IB, Nguyen TT, Hagen L, Slupphaug G, Thommesen L, et al. Identification of a novel in vivo virus-targeted phosphorylation site in interferon regulatory factor-3 (IRF3). J Biol Chem 2010;285:24904–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Panne D, McWhirter SM, Maniatis T, Harrison SC. Interferon regulatory factor 3 is regulated by a dual phosphorylation-dependent switch. J Biol Chem 2007;282:22816–22. [DOI] [PubMed] [Google Scholar]
- [95].Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J 1998;17:1087–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science 2003;300:1148–51. [DOI] [PubMed] [Google Scholar]
- [97].Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 2003;4:491–6. [DOI] [PubMed] [Google Scholar]
- [98].Lin R, Lacoste J, Nakhaei P, Sun Q, Yang L, Paz S, et al. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKepsilon molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3–4A proteolytic cleavage. J Virol 2006;80:6072–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signaling 2012;5:ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Qin BY, Liu C, Lam SS, Srinath H, Delston R, Correia JJ, et al. Crystal structure of IRF-3 reveals mechanism of autoinhibition and virus-induced phosphoactivation. Nat Struct Biol 2003;10:913–21. [DOI] [PubMed] [Google Scholar]
- [101].Shukla H, Vaitiekunas P, Majumdar AK, Dragan AI, Dimitriadis EK, Kotova S, et al. The linker of the interferon response factor 3 transcription factor is not unfolded. Biochemistry 2012;51:6320–7. [DOI] [PubMed] [Google Scholar]
- [102].Kim H, Seed B. The transcription factor MafB antagonizes antiviral responses by blocking recruitment of coactivators to the transcription factor IRF3. Nat Immunol 2010;11:743–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Sears N, Sen GC, Stark GR, Chattopadhyay S. Caspase-8-mediated cleavage inhibits IRF-3 protein by facilitating its proteasome-mediated degradation. J Biol Chem 2011;286:33037–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Saitoh T, Tun-Kyi A, Ryo A, Yamamoto M, Finn G, Fujita T, et al. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat Immunol 2006;7:598–605. [DOI] [PubMed] [Google Scholar]
- [105].Zhang M, Tian Y, Wang RP, Gao D, Zhang Y, Diao FC, et al. Negative feedback regulation of cellular antiviral signaling by RBCK1-mediated degradation of IRF3. Cell Res 2008;18:1096–104. [DOI] [PubMed] [Google Scholar]
- [106].Kubota T, Matsuoka M, Chang TH, Tailor P, Sasaki T, Tashiro M, et al. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J Biol Chem 2008;283:25660–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Doehle BP, Hladik F, McNevin JP, McElrath MJ, Gale M. Human immunodeficiency virus type 1 mediates global disruption of innate antiviral signaling and immune defenses within infected cells. J Virol 2009;83:10395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Panne D. The enhanceosome. Curr Opin Struct Biol 2008;18:236–42. [DOI] [PubMed] [Google Scholar]
- [109].Taniguchi T, Takaoka A. A weak signal for strong responses: interferon-alpha/beta revisited. Nat Rev Mol Cell Biol 2001;2:378–86. [DOI] [PubMed] [Google Scholar]
- [110].Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell 2007;129:1111–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 2005;434:772–7. [DOI] [PubMed] [Google Scholar]
- [112].Balachandran S, Beg AA. Defining emerging roles for NF-kappaB in antivirus responses: revisiting the interferon-beta enhanceosome paradigm. PLoS Pathog 2011;7:e1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A 1998;95:15623–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Andersen J, VanScoy S, Cheng TF, Gomez D, Reich NC. IRF-3-dependent and augmented target genes during viral infection. Genes Immun 2008;9:168–75. [DOI] [PubMed] [Google Scholar]
- [115].Lin R, Heylbroeck C, Genin P, Pitha PM, Hiscott J. Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol Cell Biol 1999;19:959–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Guo J, Hui DJ, Merrick WC, Sen GC. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J 2000;19:6891–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Pichlmair A, Lassnig C, Eberle CA, Gorna MW, Baumann CL, Burkard TR, et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat Immunol 2011;12:624–30. [DOI] [PubMed] [Google Scholar]
- [118].Grandvaux N, Servant MJ, tenOever B, Sen GC, Balachandran S, Barber GN, et al. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol 2002;76:5532–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Yang Z, Liang H, Zhou Q, Li Y, Chen H, Ye W, et al. Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms. Cell Res 2012;22:1328–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Li K, Markosyan RM, Zheng YM, Golfetto O, Bungart B, Li M, et al. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog 2013;9:e1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Amini-Bavil-Olyaee S, Choi YJ, Lee JH, Shi M, Huang IC, Farzan M, et al. The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe 2013;13:452–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Schafer SL, Lin R, Moore PA, Hiscott J, Pitha PM. Regulation of type I interferon gene expression by interferon regulatory factor-3. J Biol Chem 1998;273:2714–20. [DOI] [PubMed] [Google Scholar]
- [123].Okumura F, Okumura AJ, Uematsu K, Hatakeyama S, Zhang DE, Kamura T. Activation of double-stranded RNA-activated protein kinase (PKR) by interferon-stimulated gene 15 (ISG15) modification down-regulates protein translation. J Biol Chem 2013;288:2839–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Espert L, Degols G, Gongora C, Blondel D, Williams BR, Silverman RH, et al. ISG20, a new interferon-induced RNase specific for single-stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. J Biol Chem 2003;278:16151–8. [DOI] [PubMed] [Google Scholar]
- [125].Espert L, Degols G, Lin YL, Vincent T, Benkirane M, Mechti N. Interferon-induced exonuclease ISG20 exhibits an antiviral activity against human immunodeficiency virus type 1. J Gen Virol 2005;86:2221–9. [DOI] [PubMed] [Google Scholar]
- [126].Seo JY, Yaneva R, Cresswell P. Viperin: a multifunctional, interferon-inducible protein that regulates virus replication. Cell Host Microbe 2011;10:534–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Nasr N, Maddocks S, Turville SG, Harman AN, Woolger N, Helbig KJ, et al. HIV-1 infection of human macrophages directly induces viperin which inhibits viral production. Blood 2012;120:778–88. [DOI] [PubMed] [Google Scholar]
- [128].Wang N, Dong Q, Li J, Jangra RK, Fan M, Brasier AR, et al. Viral induction of the zinc finger antiviral protein is IRF3-dependent but NF-kappaB-independent. J Biol Chem 2010;285:6080–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Gao G, Guo X, Goff SP. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 2002;297:1703–6. [DOI] [PubMed] [Google Scholar]
- [130].Yan H, Krishnan K, Lim JT, Contillo LG, Krolewski JJ. Molecular characterization of an alpha interferon receptor 1 subunit (IFNaR1) domain required for TYK2 binding and signal transduction. Mol Cell Biol 1996;16:2074–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Li X, Leung S, Kerr IM, Stark GR. Functional subdomains of STAT2 required for preassociation with the alpha interferon receptor and for signaling. Mol Cell Biol 1997;17:2048–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Fu XY, Schindler C, Improta T, Aebersold R, Darnell JE. The proteins of ISGF-3, the interferon alpha-induced transcriptional activator, define a gene family involved in signal transduction. Proc Natl Acad Sci U S A 1992;89:7840–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Lin R, Mamane Y, Hiscott J. Multiple regulatory domains control IRF-7 activity in response to virus infection. J Biol Chem 2000;275:34320–7. [DOI] [PubMed] [Google Scholar]
- [134].Ozato K, Shin DM, Chang TH, Morse HC. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol 2008;8:849–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Rajsbaum R, Stoye JP, O’Garra A. Type I interferon-dependent and -independent expression of tripartite motif proteins in immune cells. Eur J Immunol 2008;38:619–30. [DOI] [PubMed] [Google Scholar]
- [136].Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 2002;418:646–50. [DOI] [PubMed] [Google Scholar]
- [137].Peng G, Lei KJ, Jin W, Greenwell-Wild T, Wahl SM. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J Exp Med 2006;203:41–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Mohanram V, Skold AE, Bachle SM, Pathak SK, Spetz AL. IFN-alpha induces APOBEC3G, F, and A in immature dendritic cells and limits HIV-1 spread to CD4+ T cells. J Immunol 2013;190:3346–53. [DOI] [PubMed] [Google Scholar]
- [139].Woods MW, Kelly JN, Hattlmann CJ, Tong JG, Xu LS, Coleman MD, et al. Human HERC5 restricts an early stage of HIV-1 assembly by a mechanism correlating with the ISGylation of Gag. Retrovirology 2011;8:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Harada H, Willison K, Sakakibara J, Miyamoto M, Fujita T, Taniguchi T. Absence of the type I IFN system in EC cells: transcriptional activator (IRF-1) and repressor (IRF-2) genes are developmentally regulated. Cell 1990;63:303–12. [DOI] [PubMed] [Google Scholar]
- [141].Meurs EF, Watanabe Y, Kadereit S, Barber GN, Katze MG, Chong K, et al. Constitutive expression of human double-stranded RNA-activated p68 kinase in murine cells mediates phosphorylation of eukaryotic initiation factor 2 and partial resistance to encephalomyocarditis virus growth. J Virol 1992;66:5805–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Li M, Kao E, Gao X, Sandig H, Limmer K, Pavon-Eternod M, et al. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature 2012;491:125–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Yang K, Shi HX, Liu XY, Shan YF, Wei B, Chen S, et al. TRIM21 is essential to sustain IFN regulatory factor 3 activation during antiviral response. J Immunol 2009;182:3782–92. [DOI] [PubMed] [Google Scholar]
- [144].Allouch A, Di Primio C, Alpi E, Lusic M, Arosio D, Giacca M, et al. The TRIM family protein KAP1 inhibits HIV-1 integration. Cell Host Microbe 2011;9:484–95. [DOI] [PubMed] [Google Scholar]
- [145].Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 2004;427:848–53. [DOI] [PubMed] [Google Scholar]
- [146].Lee HR, Kim MH, Lee JS, Liang C, Jung JU. Viral interferon regulatory factors. J Interferon Cytokine Res 2009;29:621–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Laurent-Rolle M, Boer EF, Lubick KJ, Wolfinbarger JB, Carmody AB, Rockx B, et al. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J Virol 2010;84:3503–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Taylor KE, Mossman KL. Recent advances in understanding viral evasion of type I interferon. Immunology 2013;138:190–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Loo YM, Gale M. Viral regulation and evasion of the host response. Curr Top Microbiol Immunol 2007;316:295–313. [DOI] [PubMed] [Google Scholar]
- [150].Zhang Z, Zheng Z, Luo H, Meng J, Li H, Li Q, et al. Human bocavirus NP1 inhibits IFN-beta production by blocking association of IFN regulatory factor 3 with IFNB promoter. J Immunol 2012;189:1144–53. [DOI] [PubMed] [Google Scholar]
- [151].Unterstab G, Ludwig S, Anton A, Planz O, Dauber B, Krappmann D, et al. Viral targeting of the interferon-{beta}-inducing Traf family member-associated NF-{kappa}B activator (TANK)-binding kinase-1. Proc Natl Acad Sci U S A 2005;102:13640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Hilton L, Moganeradj K, Zhang G, Chen YH, Randall RE, McCauley JW, et al. The NPro product of bovine viral diarrhea virus inhibits DNA binding by interferon regulatory factor 3 and targets it for proteasomal degradation. J Virol 2006;80:11723–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, et al. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog 2012;8:e1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Basler CF, Mikulasova A, Martinez-Sobrido L, Paragas J, Muhlberger E, Bray M, et al. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J Virol 2003;77:7945–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Cardenas WB, Loo YM, Gale M, Hartman AL, Kimberlin CR, Martinez-Sobrido L, et al. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J Virol 2006;80:5168–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Bentz GL, Liu R, Hahn AM, Shackelford J, Pagano JS. Epstein-Barr virus BRLF1 inhibits transcription of IRF3 and IRF7 and suppresses induction of interferon-beta. Virology 2010;402:121–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Wang JT, Doong SL, Teng SC, Lee CP, Tsai CH, Chen MR. Epstein-Barr virus BGLF4 kinase suppresses the interferon regulatory factor 3 signaling pathway. J Virol 2009;83:1856–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Foy E, Li K, Wang C, Sumpter R, Ikeda M, Lemon SM, et al. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 2003;300:1145–8. [DOI] [PubMed] [Google Scholar]
- [159].Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, et al. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci U S A 2006;103:6001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 2005;437:1167–72. [DOI] [PubMed] [Google Scholar]
- [161].Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, et al. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A 2005;102:2992–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Orzalli MH, DeLuca NA, Knipe DM. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A 2012;109:E3008–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Melroe GT, Silva L, Schaffer PA, Knipe DM. Recruitment of activated IRF-3 and CBP/p300 to herpes simplex virus ICP0 nuclear foci: potential role in blocking IFN-beta induction. Virology 2007;360:305–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Doehle BP, Chang K, Fleming L, McNevin J, Hladik F, McElrath MJ, et al. Vpu-deficient HIV strains stimulate innate immune signaling responses in target cells. J Virol 2012;86:8499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Doehle BP, Chang K, Rustagi A, McNevin J, McElrath MJ, Gale M. Vpu mediates depletion of interferon regulatory factor 3 during HIV infection by a lysosome-dependent mechanism. J Virol 2012;86:8367–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].McNatt MW, Zang T, Bieniasz PD. Vpu binds directly to tetherin and displaces it from nascent virions. PLoS Pathog 2013;9:e1003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 2008;3:245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Douglas JL, Viswanathan K, McCarroll MN, Gustin JK, Fruh K, Moses AV. Vpu directs the degradation of the human immunodeficiency virus restriction factor BST-2/Tetherin via a {beta}TrCP-dependent mechanism. J Virol 2009;83:7931–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Magadan JG, Perez-Victoria FJ, Sougrat R, Ye Y, Strebel K, Bonifacino JS. Multilayered mechanism of CD4 downregulation by HIV-1 Vpu involving distinct ER retention and ERAD targeting steps. PLoS Pathog 2010;6:e1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med 2003;9:1404–7. [DOI] [PubMed] [Google Scholar]
- [171].Solis M, Nakhaei P, Jalalirad M, Lacoste J, Douville R, Arguello M, et al. RIG-I-mediated antiviral signaling is inhibited in HIV-1 infection by a protease-mediated sequestration of RIG-I. J Virol 2011;85:1224–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Jaworska J, Gravel A, Fink K, Grandvaux N, Flamand L. Inhibition of transcription of the beta interferon gene by the human herpesvirus 6 immediate-early 1 protein. J Virol 2007;81:5737–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Yu Y, Wang SE, Hayward GS. The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity 2005;22:59–70. [DOI] [PubMed] [Google Scholar]
- [174].Lin R, Genin P, Mamane Y, Sgarbanti M, Battistini A, Harrington WJ, et al. HHV-8 encoded vIRF-1 represses the interferon antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene 2001;20:800–11. [DOI] [PubMed] [Google Scholar]
- [175].Fuld S, Cunningham C, Klucher K, Davison AJ, Blackbourn DJ. Inhibition of interferon signaling by the Kaposi’s sarcoma-associated herpesvirus full-length viral interferon regulatory factor 2 protein. J Virol 2006;80:3092–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Joo CH, Shin YC, Gack M, Wu L, Levy D, Jung JU. Inhibition of interferon regulatory factor 7 (IRF7)-mediated interferon signal transduction by the Kaposi’s sarcoma-associated herpesvirus viral IRF homolog vIRF3. J Virol 2007;81:8282–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Ren J, Liu T, Pang L, Li K, Garofalo RP, Casola A, et al. A novel mechanism for the inhibition of interferon regulatory factor-3-dependent gene expression by human respiratory syncytial virus NS1 protein. J Gen Virol 2011;92:2153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].DeFilippis V, Fruh K. Rhesus cytomegalovirus particles prevent activation of interferon regulatory factor 3. J Virol 2005;79:6419–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Barro M, Patton JT. Rotavirus nonstructural protein 1 subverts innate immune response by inducing degradation of IFN regulatory factor 3. Proc Natl Acad Sci U S A 2005;102:4114–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Zhu H, Zheng C, Xing J, Wang S, Li S, Lin R, et al. Varicella-zoster virus immediate-early protein ORF61 abrogates the IRF3-mediated innate immune response through degradation of activated IRF3. J Virol 2011;85:11079–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Allers K, Hutter G, Hofmann J, Loddenkemper C, Rieger K, Thiel E, et al. Evidence for the cure of HIV infection by CCR5Delta32/Delta32 stem cell transplantation. Blood 2011;117:2791–9. [DOI] [PubMed] [Google Scholar]
- [182].New Dolgin E., intensive trials planned on heels of Mississippi HIV “cure”. Nat Med 2013;19:380–1. [DOI] [PubMed] [Google Scholar]
- [183].Saez-Cirion A, Bacchus C, Hocqueloux L, Avettand-Fenoel V, Girault I, Lecuroux C, et al. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog 2013;9:e1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 1998;280:1884–8. [DOI] [PubMed] [Google Scholar]
- [185].Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, et al. Antibody neutralization and escape by HIV-1. Nature 2003;422:307–12. [DOI] [PubMed] [Google Scholar]
- [186].Allen TM, Altfeld M, Geer SC, Kalife ET, Moore C, O’Sullivan KM, et al. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J Virol 2005;79:13239–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Kirchhoff F. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe 2010;8:55–67. [DOI] [PubMed] [Google Scholar]
- [188].Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 2011;474:654–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol 2012;13:223–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Manel N, Hogstad B, Wang Y, Levy DE, Unutmaz D, Littman DR. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature 2010;467:214–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Cribier A, Descours B, Valadao AL, Laguette N, Benkirane M. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep 2013;3:1036–43. [DOI] [PubMed] [Google Scholar]
- [192].Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, et al. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med 2012;18:1682–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen LA, Kim B, et al. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 2013;13:441–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A 2006;103:5514–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Lu J, Pan Q, Rong L, He W, Liu SL, Liang C. The IFITM proteins inhibit HIV-1 infection. J Virol 2011;85:2126–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Kaiser SM, Malik HS, Emerman M. Restriction of an extinct retrovirus by the human TRIM5alpha antiviral protein. Science 2007;316:1756–8. [DOI] [PubMed] [Google Scholar]
- [197].Okumura A, Alce T, Lubyova B, Ezelle H, Strebel K, Pitha PM. HIV-1 accessory proteins VPR and Vif modulate antiviral response by targeting IRF-3 for degradation. Virology 2008;373:85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Hotter D, Kirchhoff F, Sauter D. HIV-1 Vpu does not degrade interferon regulatory factor 3. J Virol 2013;87:7160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Matusali G, Potesta M, Santoni A, Cerboni C, Doria M. The human immunodeficiency virus type 1 Nef and Vpu proteins downregulate the natural killer cell-activating ligand PVR. J Virol 2012;86:4496–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Shah AH, Sowrirajan B, Davis ZB, Ward JP, Campbell EM, Planelles V, et al. Degranulation of natural killer cells following interaction with HIV-1-infected cells is hindered by downmodulation of NTB-A by Vpu. Cell Host Microbe 2010;8:397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Bolduan S, Hubel P, Reif T, Lodermeyer V, Hohne K, Fritz JV, et al. HIV-1 Vpu affects the anterograde transport and the glycosylation pattern of NTB-A. Virology 2013;440:190–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Hardy GA, Sieg S, Rodriguez B, Anthony D, Asaad R, Jiang W, et al. Interferon-alpha is the primary plasma type-I IFN in HIV-1 infection and correlates with immune activation and disease markers. PLoS One 2013;8:e56527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Li Q, Estes JD, Schlievert PM, Duan L, Brosnahan AJ, Southern PJ, et al. Glycerol monolaurate prevents mucosal SIV transmission. Nature 2009;458:1034–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Harman AN, Lai J, Turville S, Samarajiwa S, Gray L, Marsden V, et al. HIV infection of dendritic cells subverts the IFN induction pathway via IRF-1 and inhibits type 1 IFN production. Blood 2011;118:298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Miller E, Bhardwaj N. Dendritic cell dysregulation during HIV-1 infection. Immunol Rev 2013;254:170–89. [DOI] [PMC free article] [PubMed] [Google Scholar]