Abstract
Neuromyelitis Optica (NMO) is an autoimmune disease with a higher prevalence in non-European populations. Because the Mexican population resulted from the admixture between mainly Native American and European populations, we used genome-wide microarray, HLA high-resolution typing and AQP4 gene sequencing data to analyze genetic ancestry and to seek genetic variants conferring NMO susceptibility in admixed Mexican patients. A total of 164 Mexican NMO patients and 1,208 controls were included. On average, NMO patients had a higher proportion of Native American ancestry than controls (68.1% vs 58.6%; p = 5 × 10–6). GWAS identified a HLA region associated with NMO, led by rs9272219 (OR = 2.48, P = 8 × 10–10). Class II HLA alleles HLA-DQB1*03:01, -DRB1*08:02, -DRB1*16:02, -DRB1*14:06 and -DQB1*04:02 showed the most significant associations with NMO risk. Local ancestry estimates suggest that all the NMO-associated alleles within the HLA region are of Native American origin. No novel or missense variants in the AQP4 gene were found in Mexican patients with NMO or multiple sclerosis. To our knowledge, this is the first study supporting the notion that Native American ancestry significantly contributes to NMO susceptibility in an admixed population, and is consistent with differences in NMO epidemiology in Mexico and Latin America.
Subject terms: Genetics, Immunology, Medical research, Risk factors
Introduction
Neuromyelitis optica (NMO) is a chronic autoimmune inflammatory and demyelinating disease of the central nervous system (CNS), which mainly affects the optic nerve and spinal cord. Although NMO was first described in the XIX century, it was considered a clinical variant of multiple sclerosis (MS) for decades1,2. In 2004, the discovery of positive antiaquaporin-4 antibodies (AQP4-IgG) in serum of the majority of NMO patients led to significant progress in the clinical characterization of the disease, now acknowledged as a distinct entity with different immunological, clinical and epidemiological features3,4.
Although it has been difficult to establish the actual prevalence of NMO, mainly because most reports are not comparable due to differences in study design, methodological approaches, and diagnostic criteria, worldwide NMO prevalence has been estimated between 0.51 and 4.4 cases per 100,000 inhabitants6,7. The prevalence and clinical manifestations of NMO vary among different ethnic groups, and several authors have stated that NMO is more frequent in non-European populations8,9. Interestingly, the relative frequency of NMO (estimated as the ratio of NMO/(MS + NMO) cases) has been found to decrease gradually in South America from North (Venezuela) to South (Argentina). Because ethnicity also changes gradually from North to South in this region, with the proportion of European individuals being lower in Venezuela and higher in Argentina, the authors suggested that ethnic origin influences NMO frequency in Latin America10. To date there are no population-based studies of the prevalence of NMO in Mexico, and there is a single study estimating NMO prevalence at 1.3 per 100,000 inhabitants based on the NMO/(MS + NMO) relative frequency at a referral center in Mexico City12.
Like many other autoimmune diseases, NMO is a multifactorial disorder that results from complex interactions between genetic and environmental factors. Recent studies have reported associations of NMO with genetic variation in the Human Leukocyte Antigen (HLA) genome region in chromosome 6, particularly with class II alleles, showing ethnical and geographical differences: The DRB1*03:01 allele has been associated with NMO in European13–16, Brazilian17,18, Afro-Caribbean19 and Mexican patients20; DRB1*16:02 in Southern Han Chinese21, Japanese22 and in Southern Brazilian patients23; DQB1*04:02 in a European-ancestry cohort13,24; and DRB1*04:05 in Southern Brazilians23. Moreover, candidate gene studies have reported associations with variation in non-HLA genes such as AQP4 and others involved in immune function (PD-1, IL-17, IL-7R, CD6 and CD58)24–28. Of the latter, only AQP4 gene variation has been analyzed in various populations by sequencing promoter and/or coding regions of the gene attempting to identify variants involved in the pathogenesis of NMO. However, the association of AQP4 gene variation with NMO remains unclear, with inconsistent findings among populations29–34.
Because the Mexican population of today resulted from a complex and ongoing admixture process involving mainly Native American and European genetic components, we hypothesized that Native American ancestry also contributes to NMO susceptibility in Mexican patients. We thus used genome-wide microarray data, HLA high-resolution typing and AQP4 gene sequencing data to explore genetic ancestry and to seek genetic variants conferring susceptibility to NMO in Mexican patients.
Results
The present study includes 164 patients with Neuromyelitis Optica (NMO) (79% female) and 1,208 controls (61% female).
Population admixture analysis
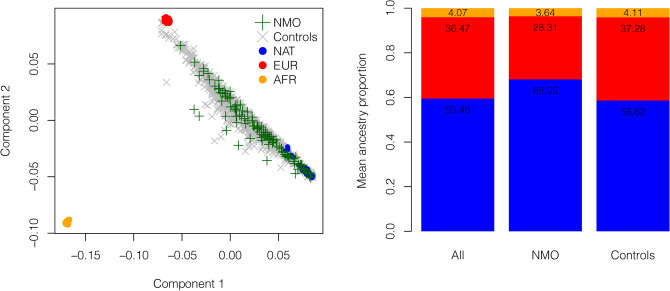
Reference Native American (NAT) and continental populations were used to generate a multidimensional scaling (MDS) plot. Figure 1. The left panel shows that components 1 and 2 distinguish Africans (AFR) and Europeans (EUR) from NAT individuals. The average genome-wide proportion of Native American ancestry was significantly higher in NMO patients than in controls (68.1% vs 58.6%; P = 5 × 10–6). Conversely, the average genome-wide proportion of European ancestry was lower in cases than in controls (28.3% vs 37.3%; P = 3 × 10–6), while African ancestry was similar in both groups (3.6% vs 4.1%; P = 0.143) (Fig. 1, right panel).
Figure 1.
Global ancestry analysis. The left panel shows multidimensional scaling analysis of NMO patients and controls, with Europeans (CEU) and Africans (YRI) from the 1,000 Genomes projects, and unrelated central Native Mexican (NAT) individuals as reference populations. The right panel shows mean ancestry proportion estimated by ADMIXTURE assuming three parental populations (K = 3), in the entire study population, cases and controls.
Genome-wide association analysis
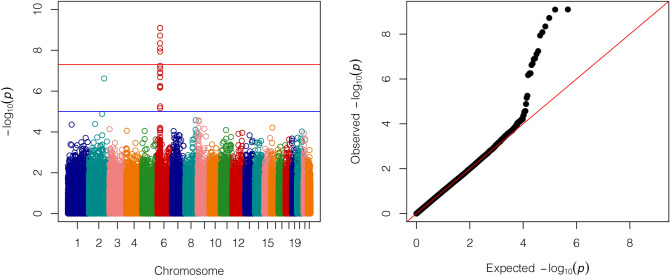
The genome-wide association analysis (GWAS) included 119 NMO cases and 1,208 controls. Figure 2 shows the Manhattan and Q-Q plots after adjusting for sex and two principal components. Six signals surpassed the genome-wide significance threshold (5 × 10–8), spanning a 173.1 kb region within the major histocompatibility complex (MHC) region. Two SNPs in perfect linkage disequilibrium (LD) (r2 = 1) showed the most significant associations under an additive model, rs9272219 (OR = 2.48, P = 8 × 10–10) and rs9273012 (OR = 2.49, P = 8 × 10–10), both within the HLA-DQA1 gene. As described in Table 1, two intergenic (rs1964995 and rs9271588) and two SNPs within the HLA-DRB9 gene (rs9368726 and rs9405108) were also significantly associated with NMO. LD estimations among these 6 SNPs ranged between 0.5 and 1 (0.5 < r2 < 1). In order to assess whether these associations are driven by a single SNP, we conditioned the analysis based on the rs9272219 genotype. No other associations maintained genome-wide significance in the conditioned analysis (Supplementary Fig. 1). Table 1 also shows associations of these SNPs with NMO using the data of Estrada et al. 201816, available at https://github.com/Biogen-Inc/statgen (UTS/ACP data set).
Figure 2.
Genome-wide association analysis. Manhattan plot and Q–Q plot. P-values were obtained by logistic regression adjusting for sex and two principal components. Six signals surpassed the genome-wide significance threshold (5 × 10–8), spanning a 173.1 kb region in the major histocompatibility complex (MHC) region.
Table 1.
Association statistics for genome-wide significant SNPs.
| rsID | Position | Gene | RA | RAF NMO (n = 119) | RAF Ctrl (n = 1,208) | Unconditioned | Conditioned on rs9272219 | UTS/ACP data set | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | OR | P | ||||||
| rs9368726 | 32,438,542 | HLA-DRB9 | A | 0.80 | 0.61 | 2.70 (1.92–3.70) | 1 × 10–8 | 1.75 (1.09–2.86) | 0.022 | 2.22 | 8 × 10–9 |
| rs9405108 | 32,438,648 | HLA-DRB9 | G | 0.80 | 0.61 | 2.70 (1.92–3.85) | 8 × 10–9 | 1.75 (1.08–2.86) | 0.023 | 2.22 | 8 × 10–9 |
| rs1964995 | 32,449,411 | Intergenic | A | 0.78 | 0.56 | 2.63 (1.92–3.70) | 5 × 10–9 | 1.79 (1.09–2.94) | 0.023 | – | – |
| rs9271588 | 32,590,953 | Intergenic | A | 0.75 | 0.53 | 2.63 (1.92–3.57) | 2 × 10–9 | 1.75 (1.05–2.86) | 0.032 | 1.96 | 1 × 10–9 |
| rs9272219 | 32,602,269 | HLA-DQA1 | A | 0.67 | 0.42 | 2.48 (1.86–3.32) | 8 × 10–10 | – | – | – | – |
| rs9273012 | 32,611,641 | HLA-DQA1 | G | 0.67 | 0.42 | 2.49 (1.86–3.32) | 8 × 10–10 | – | – | – | – |
RA, Risk allele. RAF: Risk allele frequency; UTS/ACP data set correspond to data obtained from Estrada et al. 201816.
HLA analysis
Detailed HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 allele and haplotype frequencies in study groups are described in Supplementary Tables 1–9. No HLA variant deviated from Hardy–Weinberg equilibrium. Table 2 shows HLA alleles that differed significantly in frequency between NMO patients and controls. The most significantly NMO-associated HLA class II alleles (P < 0.01) were HLA-DQB1*03:01 (P = 0.00008), -DRB1*08:02 (P = 0.0011), -DRB1*16:02 (P = 0.0014), -DRB1*14:06 (P = 0.00368) and -DQB1*04:02 (P = 0.00552), all associated with increased NMO risk, while -DQB1*03:02 (P = 0.00013) and -DQB1*02:02 (P = 0.00899) were associated with decreased NMO risk. Two-locus HLA haplotypes associated with NMO patients are listed in Table 3. The lead class II haplotype associated with increased NMO risk was HLA-DRB1*16:02-DQB1*03:01 (P = 0.0014), followed by HLA-DRB1*08:02-DQB1*04:02 (P = 0.00177) and -DRB1*14:06-DQB1*03:01 (P = 0.00368), while -DRB1*07:01-DQB1*02:02 (P = 0.00796) was associated with decreased NMO risk. No extended haplotypes differed significantly between patients and controls.
Table 2.
Comparison of HLA class I and class II allele frequencies in NMO patients and controls.
| HLA Allele | NMO (n = 71) | Controls (n = 97) | P | |||
|---|---|---|---|---|---|---|
| AF | n | AF | n | |||
| Protective | B*39:05 | 0.0423 | 6 | 0.0979 | 19 | 0.04088 |
| DRB1*07:01 | 0.0282 | 4 | 0.0876 | 17 | 0.01996 | |
| DRB1*04:07 | 0.0775 | 11 | 0.1598 | 31 | 0.01692 | |
| DQB1*02:02 | 0.0282 | 4 | 0.0979 | 19 | 0.00899 | |
| DQB1*03:02 | 0.1197 | 17 | 0.2887 | 56 | 0.00013 | |
| Risk | B*39:06 | 0.0775 | 11 | 0.0309 | 6 | 0.04818 |
| B*35:14 | 0.0493 | 7 | 0.0103 | 2 | 0.03275 | |
| DQB1*04:02 | 0.2746 | 39 | 0.1546 | 30 | 0.00552 | |
| DRB1*14:06 | 0.0915 | 13 | 0.0206 | 4 | 0.00368 | |
| DRB1*16:02 | 0.1549 | 22 | 0.0515 | 10 | 0.00140 | |
| DRB1*08:02 | 0.2676 | 38 | 0.1289 | 25 | 0.00110 | |
| DQB1*03:01 | 0.3380 | 48 | 0.1546 | 30 | 0.00008 | |
AF, Allele frequency. Only alleles with a frequency ≥ 0.01 in both groups and with significant differences between cases and controls are presented.
Table 3.
Comparison of HLA class I and class II haplotype frequencies in NMO patients and controls.
| HLA Haplotype | NMO (n = 71) | Controls (n = 97) | P | |||||
|---|---|---|---|---|---|---|---|---|
| HF | n | Δ′ | HF | n | Δ′ | |||
| Protective | B*39:05-C*07:02 | 0.0423 | 6 | 1 | 0.0979 | 19 | 1 | 0.04088 |
| DRB1*04:07-DQB1*03:02 | 0.0704 | 10 | 0.8967 | 0.1546 | 30 | 1 | 0.01300 | |
| DRB1*07:01-DQB1*02:02 | 0.0211 | 3 | 0.7428 | 0.0876 | 17 | 1 | 0.00796 | |
| Risk | B*35:14-C*04:01 | 0.0493 | 7 | 1 | 0.0103 | 2 | 1 | 0.03275 |
| B*39:06-C*07:02 | 0.0704 | 10 | 0.8805 | 0.0206 | 4 | 1 | 0.02422 | |
| DRB1*14:06-DQB1*03:01 | 0.0915 | 13 | 1 | 0.0206 | 4 | 1 | 0.00368 | |
| DRB1*08:02-DQB1*04:02 | 0.2606 | 37 | 0.9637 | 0.1289 | 25 | 1 | 0.00177 | |
| DRB1*16:02-DQB1*03:01 | 0.1549 | 22 | 1 | 0.0515 | 10 | 1 | 0.00140 | |
HF, Haplotype frequency. Only haplotypes with a frequency ≥ 0.01 in both groups and with significant differences between cases and controls are presented.
Table 4 shows HLA class II alleles stratified by rs9272219 alleles (C/A). Interestingly, previously reported risk alleles HLA-DQB1*04:02 and -DRB1*16:02, as well as HLA-DQB1*03:01, -DRB1*08:02 and -DRB1*14:06, all associated with NMO risk in the present study, were in strong linkage disequilibrium (LD) with the rs9272219 “A” risk allele. On the other hand, no NMO risk alleles, and two alleles associated with decreased NMO risk (-DQB1*03:02 and -DRB1*04:07) were in strong LD with the rs9272219 “C” allele.
Table 4.
Comparison of HLA-DRB1 and HLA-DQB1 allele frequencies, stratified by rs9272219 genotype, in NMO patients and controls.
| rs9272219 | HLA Allele | NMO (n = 31) | Controls (n = 97) | P | ||||
|---|---|---|---|---|---|---|---|---|
| AF | n | Δ′ | AF | n | Δ′ | |||
| C | DQB1*04:02 | 0.0323 | 2 | 1 | 0.0155 | 3 | 1 | NS |
| DRB1*04:04 | 0.0484 | 3 | 1 | 0.0619 | 12 | 0.8473 | NS | |
| DRB1*04:07 | 0.0484 | 3 | 1 | 0.1598 | 31 | 0.94 | 0.01528 | |
| DQB1*03:02 | 0.0968 | 6 | 1 | 0.2887 | 56 | 1 | 0.00107 | |
| DQB1*03:03 | 0.0161 | 1 | 1 | 0.0103 | 2 | 1 | NS | |
| DQB1*05:01 | 0.0323 | 2 | 1 | 0.0773 | 15 | 1 | NS | |
| DQB1*06:01 | 0.0161 | 1 | 1 | 0.0155 | 3 | 0.6337 | NS | |
| DQB1*06:02 | 0.0161 | 1 | 0.7338 | 0.0567 | 11 | 1 | NS | |
| DQB1*06:04 | 0.0161 | 1 | 1 | 0.0309 | 6 | 1 | NS | |
| DRB1*01:01 | 0.0161 | 1 | 1 | 0.0206 | 4 | 1 | NS | |
| DRB1*01:02 | 0.0161 | 1 | 1 | 0.0258 | 5 | 1 | NS | |
| DRB1*07:01 | 0.0161 | 1 | 1 | 0.0876 | 17 | 1 | 0.04056 | |
| DRB1*13:02 | 0.0161 | 1 | 1 | 0.0361 | 7 | 1 | NS | |
| DRB1*15:01 | 0.0161 | 1 | 0.7338 | 0.0567 | 11 | 1 | NS | |
| DRB1*15:02 | 0.0161 | 1 | 1 | 0.0206 | 4 | 1 | NS | |
| A | DQB1*04:02 | 0.2742 | 17 | 1 | 0.1392 | 27 | 1 | 0.01415 |
| DRB1*03:01 | 0.0806 | 5 | 1 | 0.0412 | 8 | 1 | NS | |
| DRB1*08:02 | 0.2903 | 18 | 1 | 0.1237 | 24 | 1 | 0.00279 | |
| DRB1*14:02 | 0.0161 | 1 | 1 | 0.0309 | 6 | 1 | NS | |
| DRB1*16:02 | 0.1613 | 10 | 1 | 0.0515 | 10 | 1 | 0.00831 | |
| DQB1*03:01 | 0.3065 | 19 | 0.911 | 0.1546 | 30 | 1 | 0.00851 | |
| DQB1*02:01 | 0.0806 | 5 | 0 | 0.0412 | 8 | 1 | NS | |
| DQB1*03:19 | 0.0484 | 3 | 1 | 0.0103 | 2 | 1 | NS | |
| DRB1*11:02 | 0.0323 | 2 | 1 | 0.0155 | 3 | 1 | NS | |
| DRB1*11:04 | 0.0161 | 1 | 1 | 0.0206 | 4 | 1 | NS | |
| DRB1*14:06 | 0.0968 | 6 | 0.7153 | 0.0206 | 4 | 1 | 0.01497 | |
AF, allele frequency. Only Alleles with a frequency ≥ 0.01 in both groups are presented.
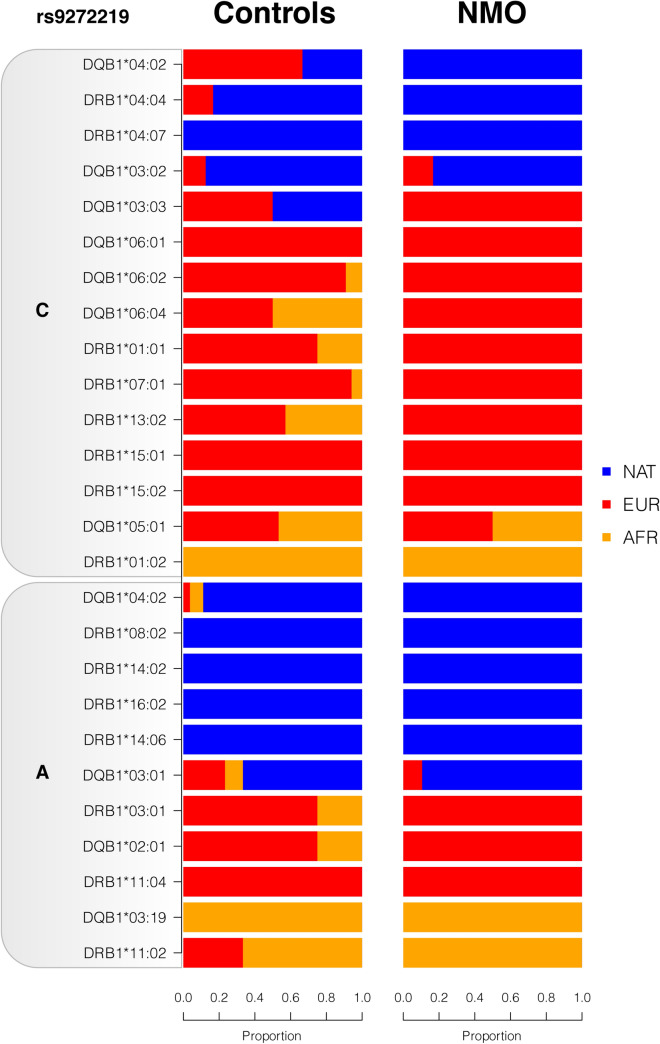
We then used RFMix to infer the local ancestry (NAT, EUR or AFR) of all alleles described in Table 4, in both cases and controls. Notably, all NMO risk alleles HLA-DRB1*08:02, -DRB1*16:02 and -DRB1*14:06 were inferred to be of Native American ancestry, while risk alleles -DQB1*04:02 and -DQB1*03:01 were predominantly of NAT ancestry both in cases and controls. Regarding protective NMO alleles, -DRB1*04:07 and -DQB1*03:02 alleles were also predominantly of NAT ancestry(Fig. 3).
Figure 3.
Local ancestry of HLA alleles. Proportion of HLA alleles described in Table 4 inferred as Native American (NAT), European (EUR) or African (AFR). Local ancestry was estimated using RFMIX using trio-phased populations as reference.
AQP4 sequencing
After sequencing all AQP4 exons including exon–intron boundaries, we identified 35 SNPs in samples from NMO or MS Mexican patients. No novel variants were found. Thirteen of these variants were found in the 3′UTR region, one in the 5′UTR region, and three were synonymous variants. Although some variants were more frequent in NMO than in MS patients, the differences were not statistically significant. Supplementary Table 10, compares the alternative allele frequency of these 35 SNPs in NMO and MS patients, 1,000 Genomes continental populations and in 12 Native Mexican whole genome sequences35.
Discussion
While several studies have stated that the epidemiology of NMO differs from that of MS, being more frequent in non-European populations9–11, reliable comparisons among studies are difficult to establish. Epidemiological data suggest that ethnicity influences NMO prevalence, particularly in Latin America10. In Mexico, there is only one non-population based study estimating NMO prevalence in a referral center located in Mexico City12. To our knowledge, this is the first study in the Mexican admixed population where a genome-wide analysis revealed a higher proportion of Native American ancestry in NMO cases as compared to controls. This contrasts with the higher European genetic component previously observed in the Mexican patients with MS36. The NAT ancestry estimated in our control group recruited in Mexico City (central Mexico) is consistent with previous NAT ancestry estimations in the Mexican Mestizo population (~ 55%), known to gradually decline from South to North throughout the Mexican territory37,38. Unfortunately, NMO epidemiological studies in Mexico are scarce, and there is no information on the geographical distribution or ethnicity of NMO patients in Mexico.
In our GWAS analysis, six SNPs within the MHC region were associated with NMO with genome-wide significance. Both linkage disequilibrium and the conditional association analyses suggest that this association is driven by a single signal, led by two SNPs in perfect linkage disequilibrium (rs9272219 and rs9273012). There is only one previous report of a GWAS for NMO in individuals of European ancestry16, where two independent signals in the same MHC region were significantly associated with NMO: rs28383224, which is 18.6 kb downstream and rs1150757, which is 573.1 kb downstream the lead SNP found in the present study (rs9272219). Although rs28383224 and rs1150757 genotypes were not available in our analysis, both NMO GWAS share data on 3 of the 6 SNPs associated with NMO in the Mexican cohort (rs9368726, rs9405108 and rs9271588). These 3 SNPs were also associated with increased NMO risk in the European cohort, although with slightly lower odds ratio values. It is important to point out that we found no SNPs in proximity of rs1150757 associated with NMO in the Mexican population.
Class II HLA alleles (HLA-DQB1*03:01, -DRB1*08:02, -DRB1*16:02, -DRB1*14:06 and -DQB1*04:02) and class II haplotypes (HLA-DRB1*16:02-DQB1*03:01, -DRB1*08:02-DQB1*04:02 and -DRB1*14:06-DQB1*03:01) showed the most significant associations with increased NMO risk in the present Mexican cohort, while HLA-DQB1*03:02 and -DQB1*02:02 alleles were significantly associated with decreased NMO risk. The HLA-DRB1*16:02 allele has also been associated with NMO in Southern Han Chinese and Japanese populations, and more recently in Southern Brazilians21–23. A very recent meta-analysis showed that the HLA-DRB1*16:02 allele was strongly associated with autoimmune diseases predominantly mediated by autoantibodies5. The frequency of this allele varies across the world but it is highest in Native populations of America (~ 39%), is also frequent in populations from Oceania (~ 28%) and South-East Asia (~ 28%), but is relatively low in Europe (~ 6%) and Africa (~ 4%)39. Furthermore, haplotype HLA-DRB1*16:02-DQB1*03:01 is very frequent in Native American populations from the Southern state of Oaxaca (Mixe, Mixtec and Zapotec) and Xavantes from Central Brazil, but is very rare in other continental populations39–41. Recently, haplotype HLA-DRB1*16-DQB1*03:01 was also associated with Parry-Romberg syndrome, an autoimmune disease affecting the craniofacial nerve in Mexican patients42.
The HLA-DRB1*03:01 allele has been consistently associated with NMO in European populations, and admixed populations with important contribution of the European gene pool (Brazilian mulatto, Afro-Caribbean and a small Mexican mestizo cohort)13–20. The frequency of HLA-DRB1*03:01 is as high as 20% in European, North African, Western Asian populations, but ranges from only 0 to 2% in Native Mexican populations39. Although in the present study the frequency of this allele was two-fold higher in cases as compared to controls (7.04% vs 4.12%, Supplementary Table 4), the difference did not reach statistical significance probably due to low statistical power derived from the small sample and effect sizes. Inconsistencies are not uncommon in genetic association studies and show the complexity of the genetic ancestry contribution in admixed populations. Interestingly, in the European GWAS, the HLA-DRB1*03:01 allele was imputed and found to be associated with AQP4-IgG-seropositive NMO but not with AQP4-IgG-seronegative NMO, and showed a high correlation with rs1150757 (r2 = 0.7) but a poor correlation with rs28383224 (r2 = 0.2)16. In the present study, the HLA-DRB1*03:01 allele was not associated with NMO, nor with any SNP in proximity of rs1150757.
HLA alleles previously associated with NMO in populations with European and/or Native American ancestry (HLA-DRB1*03:0113–20, -DRB1*16:0221–23 and -DQB1*04:0213,24) were in strong LD with the rs9272219 “A” risk allele. In contrast, two alleles significantly associated with decreased NMO risk in the present study (HLA-DRB1*04:07 and -DQB1*03:02) were in strong LD with the rs9272219 “C” allele. Whether the latter are in fact NMO protective alleles needs to be confirmed in independent cohorts. Moreover, no previously reported NMO risk alleles were found in individuals with the rs9272219 “C” allele.
Notably, local ancestry analyses revealed that all HLA alleles most associated with NMO risk and protection in the present study were predominantly inferred as of Native American ancestry. This is consistent with our finding of a higher proportion of NAT ancestry in NMO cases as compared to controls, and with epidemiological data suggesting that NMO is more prevalent in non-European populations9–11. To our knowledge there is only one previous study analyzing local ancestry of demyelinating diseases, where HLA alleles DRB1*16:02 and DRB1*14:02 were inferred as of Native American ancestry in Hispanics43, also in consistency with our local ancestry findings. As expected, the well-known HLA-DRB1*03:01 NMO risk allele was predominantly inferred as of European ancestry. Altogether, our SNP and HLA analyses suggest that a group of HLA alleles predominantly of Native American ancestry are associated with NMO susceptibility in the admixed Mexican population.
A limited number of studies have analyzed the role of AQP4 variants in the pathogenesis of NMO in USA29, Chinese30–32, Japanese33 and Spanish34 populations, with inconclusive results. We sequenced AQP4 coding regions in Mexican patients with NMO and MS, however no novel or missense variants were identified. Interestingly, four 3′UTR variants (rs7240333, rs14393, rs1058424 and rs3763043) were more frequent in NMO as compared to MS patients, although the differences were not significant. Two of these variants (rs1058424 and rs3763043) showed a weak but significant association with NMO in the Han Chinese population31. The highest frequencies of these four 3′UTR polymorphisms have been found in Native Mexicans (29.2%, 79.2%, 50% and 79.2%, respectively)35.
Some limitations of the study must be pointed out. Firstly, because no medical information was obtained from the control group (CANDELA project participants from Mexico), misclassification bias could potentially affect the statistical power of the study. Controls lacking medical information have been previously used in other GWAS studies, as the effect on statistical power is expected to be modest unless the extent of this bias is substantial44. In the present study, it is unlikely albeit possible, that a low number of control participants were affected with NMO or could eventually develop NMO in the future. However, because NMO prevalence is very low, this bias is expected to be small. In addition, because of the possibility of spurious associations, the novel HLA associations here identified should be interpreted with caution and be confirmed in further studies including Mexican and other Latin American populations.
To our knowledge, this is the first study to examine the genetic ancestry of NMO patients supporting the notion that Native American ancestry significantly contributes to Neuromyelitis optica susceptibility in the admixed Mexican population. This finding is consistent with differences in the prevalence of NMO in populations of Mexico and Latin America, and contrasts with the epidemiology and genetics of multiple sclerosis36,45.
Methods
Study population
A total of 164 Mexican NMO patients were included. NMO diagnosis was based on Wingerchuk criteria described in 2006 and revised in 201546,47. All NMO participants had DNA samples stored at the National Institute of Neurology and Neurosurgery “Manuel Velasco Suárez” (INNN) DNA bank. Only DNA samples meeting quality requirements were used to perform microarray genotyping, high-resolution HLA typing and/or AQP4 sequencing. From the 164 NMO patients, it was possible to perform microarray analysis in 119, high-resolution typing of the HLA region in 71 and AQP4 sequencing in 48 cases. Only 25 cases were available for the three platforms and 31 have microarray and high-resolution HLA typing. Moreover, 1,208 individuals from the Consortium for the Analysis of the Diversity and Evolution of Latin America (CANDELA)48 were included as a control group for the genome-wide association analysis. AQP4 sequencing data were compared in 48 NMO cases and 48 MS Mexican patients. All MS patients were recruited at the INNN and fulfilled the McDonald criteria for MS diagnosis49. A sample of 15 trios from central Native Mexican populations (Nahuas and Totonacs) were included as a Native American (NAT) reference panel for ancestry analyses. All experiments were performed in accordance with the relevant guidelines and regulations.
Informed consent was obtained from all participants, and the study was approved by the Ethics Committee of INNN for NMO and MS patients; Universidad Nacional Autónoma de México for CANDELA controls; and Instituto Nacional de Medicina Genómica (INMEGEN) for NAT trios.
Genotyping
Eighty-three NMO samples were genotyped using the Illumina HumanOmniExpress array (~ 700,000 SNPs) and 36 using Illumina expanded Multi-Ethnic Genome Array (~ 1,700,000 SNPs), at the INMEGEN. Controls had been previously genotyped on HumanOmniExpress array as part of the CANDELA Consortium study48. One of the CANDELA-Mexico controls was also genotyped at INMEGEN for quality control purposes, and microarray data concordance was 99.8%.
High-resolution typing of the HLA region
HLA class I (A, B and C) and class II (DRB1 and DQB1) genes were typed by direct sequencing (sequence-based typing, SBT50) in a total of 71 NMO samples and 97 controls. Genotypes were called using Applied Biosystems analysis software (Foster City, CA, USA) and the IMGT/HLA database alignment tool51. Ambiguities were resolved using previously validated group-specific sequencing primers (GSSP)50.
AQP4 sequencing
AQP4 coding and UTR regions were sequenced on a Illumina MiSeq system. Primers were designed manually to span the regions of interest. Quality control of raw sequences was conducted using FastQC52 and the Trimmomatic53 algorithm was used to remove adapter sequences and trim short and low-quality end-read sequences. By using Bowtie254 and SAMTools55, we cleaned the sequence reads and then aligned them to the human reference genome (hg19) and variant calling and annotations.
Statistical analysis
Genome-wide screening
Quality control (QC) of the genotype data was carried out in PLINK56. SNPs and individuals were removed from the analysis based on minor allele frequency < 5%, call rate < 95%, deviation from the Hardy–Weinberg equilibrium (HWE) at P < 1 × 10–5 and genotyping efficiency < 90%. Pairwise identity by descent (IBD) estimates were used to identify related individuals. No discordant sex information was found. After quality control, the data set comprised the genotypes of 119 NMO patients and 1,208 controls for 252,805 SNPs.
Ancestry analyses were carried out using ADMIXTURE57 assuming three parental populations: EUR and AFR from the 1,000 Genomes project, and NAT genotypes of unrelated individuals, i.e. only parents of the NAT trios were included. The total number of autosomal SNPs common to the five populations (NMO patients, CANDELA controls, EUR, AFR and NAT) was 197,323. For each individual, the proportion of European, African and Native American ancestry was estimated at the genomic level. The significance of differences between ancestry proportions in NMO patients as compared to controls was determined using a T-test statistic. Local ancestry was determined through a random forest procedure using RFMix with 5 expectation–maximization (EM) iterations and a minimum of 6 reference haplotypes per tree node58. EUR and AFR from 1,000 Genomes project, along with trio phased NAT genotypes were used as reference populations for local ancestry. Haplotype phasing of NAT, NMO and control subjects was performed with Beagle59.
A genome-wide case–control association study was conducted using logistic regression models, adjusting for sex and two principal components, assuming an additive effect using PLINK56.
HLA region analysis
Allele and haplotype frequencies were obtained by direct counting, and haplotype blocks were built based on previous reports. Allele frequencies for HLA-A, -B, -C, -DRB1 and -DQB1 were compared between NMO patients and the control group. Maximum-likelihood haplotype frequencies for two-point, three-point and four-point associations were estimated in each group using an EM algorithm implemented in Arlequin v3.160. Linkage disequilibrium (LD; Δ and Δ′) and HWE were also calculated using Arlequin. Class II HLA haplotype were stratified by SNP rs9272219 and the significance of the differences between NMO patients and CANDELA controls was determined using a Fisher’s exact test.
Supplementary information
Acknowledgements
We thank Hugo Villamil-Ramírez, Juan Carlos Guevara and Jorge Morales-Morillón for technical support. This work was supported by Consejo Nacional de Ciencia y Tecnología (CONACyT) (Grant No. SALUD-2013-01-201206 to S.R-H).
Author contributions
S.R-H., J.F-R., B.A-P., J.G. and T.C. conceived and designed study. S.R-H., M.V-C., L.R.M–K., R.B., H.P-U., E.Y. and J.G. performed data analysis. J.D.G-R., S.R-M. and C.G-T. participated processing and analyzing AQP4 sequencing. T.C., J.F-R., V.R-A., J.O-M. and I.T-F. were responsible for recruiting NMO patients. J.L.G-C., G.O., M.E-T., M.G-L. and V.H.T-M. were responsible for NMO patients sample handling. N.Y., T.V.L. and S.M.A performed HLA typing. V.A-A., M.C-B., C.G., G.B., F.R., R.G-J., A.R-L. and S.C-Q. are members of steering committee of CANDELA Consortium and responsible for CANDELA samples and data. S.R-H., M.T.V-M., R.B., E.Y., J.G. and T.C wrote the paper, incorporating input from all authors.
Data availability
The dataset for the NMO patients generated and/or analyzed during the current study are available from the corresponding authors on reasonable request. Access to the CANDELA dataset used in this manuscript was obtained through a formal request to the Consortium for the Analysis of the Diversity and Evolution of Latin America steering committee.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sandra Romero-Hidalgo and José Flores-Rivera.
Contributor Information
Sandra Romero-Hidalgo, Email: sromero@inmegen.gob.mx.
Julio Granados, Email: julgrate@yahoo.com.
Teresa Corona, Email: coronav@unam.mx.
Supplementary information
is available for this paper at 10.1038/s41598-020-69224-3.
References
- 1.Jarius S, Wildemann B. The history of neuromyelitis optica. J. Neuroinflamm. 2013;10:8. doi: 10.1186/1742-2094-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trebst C, et al. Update on the diagnosis and treatment of neuromyelitis optica: recommendations of the neuromyelitis optica study group (NEMOS) J. Neurol. 2014;261:1–16. doi: 10.1007/s00415-013-7169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lennon VA, et al. A serum autoantibody marker of neuromyelitis optica: distinction from multiple sclerosis. Lancet. 2004;364:2106–2112. doi: 10.1016/S0140-6736(04)17551-X. [DOI] [PubMed] [Google Scholar]
- 4.Lennon VA, Kryzer TJ, Pittock SJ, Verkman AS, Hinson SR. IgG marker of optic-spinal multiple sclerosis binds to the aquaporin-4 water channel. J. Exp. Med. 2005;202:473–477. doi: 10.1084/jem.20050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, et al. Comprehensive meta-analysis reveals an association of the HLA-DRB1*1602 allele with autoimmune diseases mediated predominantly by autoantibodies. Autoimmun Rev. 2020;19:102532. doi: 10.1016/j.autrev.2020.102532. [DOI] [PubMed] [Google Scholar]
- 6.Asgari N, et al. Epidemiology of neuromyelitis optica spectrum disorder. Acta Neurol. Scand. 2018;137:626–627. doi: 10.1111/ane.12936. [DOI] [PubMed] [Google Scholar]
- 7.Etemadifar M, Nasr Z, Khalili B, Taherioun M, Vosoughi R. Epidemiology of neuromyelitis optica in the world: a systematic review and meta-analysis. Mult. Scler. Int. 2015;2015:174720. doi: 10.1155/2015/174720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim SH, et al. Racial differences in neuromyelitis optica spectrum disorder. Neurology. 2018;91:e2089–e2099. doi: 10.1212/WNL.0000000000006574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flanagan EP, et al. Epidemiology of aquaporin-4 autoimmunity and neuromyelitis optica spectrum. Ann. Neurol. 2016;79:775–783. doi: 10.1002/ana.24617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alvarenga MP, Schimidt S, Alvarenga RP. Epidemiology of neuromyelitis optica in Latin America. Mult. Scler. J. Exp. Transl. Clin. 2017;3:2055217317730098. doi: 10.1177/2055217317730098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyamoto K, et al. Nationwide epidemiological study of neuromyelitis optica in Japan. J. Neurol. Neurosurg. Psychiatry. 2018;89:667–668. doi: 10.1136/jnnp-2017-317321. [DOI] [PubMed] [Google Scholar]
- 12.Rivera JF, Kurtzke JF, Booth VJ, Corona VTT. Characteristics of Devic's disease (neuromyelitis optica) in Mexico. J. Neurol. 2008;255:710–715. doi: 10.1007/s00415-008-0781-2. [DOI] [PubMed] [Google Scholar]
- 13.Zephir H, et al. Is neuromyelitis optica associated with human leukocyte antigen? Mult. Scler. 2009;15:571–579. doi: 10.1177/1352458508102085. [DOI] [PubMed] [Google Scholar]
- 14.Blanco Y, et al. HLA-DRB1 typing in Caucasians patients with neuromyelitis optica. Rev. Neurol. 2011;53:146–152. [PubMed] [Google Scholar]
- 15.Bruijstens AL, et al. HLA association in MOG-IgG- and AQP4-IgG-related disorders of the CNS in the Dutch population. Neurol. Neuroimmunol. Neuroinflamm. 2020 doi: 10.1212/NXI.0000000000000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estrada K, et al. A whole-genome sequence study identifies genetic risk factors for neuromyelitis optica. Nat. Commun. 2018;9:1929. doi: 10.1038/s41467-018-04332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brum DG, et al. HLA-DRB association in neuromyelitis optica is different from that observed in multiple sclerosis. Mult. Scler. 2010;16:21–29. doi: 10.1177/1352458509350741. [DOI] [PubMed] [Google Scholar]
- 18.Alvarenga MP, et al. The HLA DRB1*03:01 allele is associated with NMO regardless of the NMO-IgG status in Brazilian patients from Rio de Janeiro. J. Neuroimmunol. 2017;310:1–7. doi: 10.1016/j.jneuroim.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 19.Deschamps R, et al. Different HLA class II (DRB1 and DQB1) alleles determine either susceptibility or resistance to NMO and multiple sclerosis among the French Afro-Caribbean population. Mult. Scler. 2011;17:24–31. doi: 10.1177/1352458510382810. [DOI] [PubMed] [Google Scholar]
- 20.Alonso VR, et al. Neuromyelitis optica (NMO IgG+) and genetic susceptibility, potential ethnic influences. Cent. Nerv. Syst. Agents Med. Chem. 2018;18:4–7. doi: 10.2174/1871524916666160229115047. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, et al. HLA-DPB1 0501 is associated with susceptibility to anti-aquaporin-4 antibodies positive neuromyelitis optica in southern Han Chinese. J. Neuroimmunol. 2011;233:181–184. doi: 10.1016/j.jneuroim.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Yoshimura S, et al. Distinct genetic and infectious profiles in Japanese neuromyelitis optica patients according to anti-aquaporin 4 antibody status. J. Neurol. Neurosurg. Psychiatry. 2013;84:29–34. doi: 10.1136/jnnp-2012-302925. [DOI] [PubMed] [Google Scholar]
- 23.Kay CSK, Scola RH, Arndt RC, Lorenzoni PJ, Werneck LC. HLA-alleles class I and II associated with genetic susceptibility to neuromyelitis optica in Brazilian patients. Arq. Neuropsiquiatr. 2019;77:239–247. doi: 10.1590/0004-282X20190031. [DOI] [PubMed] [Google Scholar]
- 24.Asgari N, Nielsen C, Stenager E, Kyvik KO, Lillevang ST. HLA, PTPN22 and PD-1 associations as markers of autoimmunity in neuromyelitis optica. Mult. Scler. 2012;18:23–30. doi: 10.1177/1352458511417480. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, et al. Interleukin 17 gene polymorphism is associated with anti-aquaporin 4 antibody-positive neuromyelitis optica in the Southern Han Chinese—a case control study. J. Neurol. Sci. 2012;314:26–28. doi: 10.1016/j.jns.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Zhuang JC, et al. Variants of interleukin-7/interleukin-7 receptor alpha are associated with both neuromyelitis optica and multiple sclerosis among chinese han population in Southeastern China. Chin Med. J. (Engl) 2015;128:3062–3068. doi: 10.4103/0366-6999.169093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park TJ, et al. Associations of CD6, TNFRSF1A and IRF8 polymorphisms with risk of inflammatory demyelinating diseases. Neuropathol Appl. Neurobiol. 2013;39:519–530. doi: 10.1111/j.1365-2990.2012.01304.x. [DOI] [PubMed] [Google Scholar]
- 28.Kim JY, Bae JS, Kim HJ, Shin HD. CD58 polymorphisms associated with the risk of neuromyelitis optica in a Korean population. BMC Neurol. 2014;14:57. doi: 10.1186/1471-2377-14-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matiello M, et al. Genetic analysis of aquaporin-4 in neuromyelitis optica. Neurology. 2011;77:1149–1155. doi: 10.1212/WNL.0b013e31822f045b. [DOI] [PubMed] [Google Scholar]
- 30.Mai W, et al. Preliminary study on the association of AQP4 promoter polymorphism with anti-aquaporin-4 antibody positivity in southern Han Chinese patients with idiopathic demyelinating disorders of central nervous system. J. Neuroimmunol. 2013;255:75–80. doi: 10.1016/j.jneuroim.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Wei Q, et al. Human aquaporin 4 gene polymorphisms in Chinese patients with neuromyelitis optica. J. Neuroimmunol. 2014;274:192–196. doi: 10.1016/j.jneuroim.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 32.Wang QS, Xiao HQ, Chen HX, Liu YP, Ding XD. The single nucleotide polymorphism site of aquaporin-4 gene in patients with neuromyelitis optica. Exp. Ther. Med. 2017;14:6017–6021. doi: 10.3892/etm.2017.5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogasawara M, et al. Genetic analysis of the aquaporin-4 gene for anti-AQP4 antibody-positive neuromyelitis optica in a Japanese population. Jpn. J. Ophthalmol. 2016;60:198–205. doi: 10.1007/s10384-016-0441-5. [DOI] [PubMed] [Google Scholar]
- 34.García-Miranda P, et al. Predictive value of serum antibodies and point mutations of AQP4, AQP1 and MOG in A cohort of spanish patients with neuromyelitis optica spectrum disorders. Int. J. Mol. Sci. 2019 doi: 10.3390/ijms20225810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romero-Hidalgo S, et al. Demographic history and biologically relevant genetic variation of Native Mexicans inferred from whole-genome sequencing. Nat. Commun. 2017;8:1005. doi: 10.1038/s41467-017-01194-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ordonez G, et al. Genomewide admixture study in Mexican Mestizos with multiple sclerosis. Clin. Neurol. Neurosurg. 2015;130:55–60. doi: 10.1016/j.clineuro.2014.11.026. [DOI] [PubMed] [Google Scholar]
- 37.Silva-Zolezzi I, et al. Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico. Proc. Natl. Acad. Sci. USA. 2009;106:8611–8616. doi: 10.1073/pnas.0903045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moreno-Estrada A, et al. Human genetics. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science. 2014;344:1280–1285. doi: 10.1126/science.1251688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalez-Galarza FF, Christmas S, Middleton D, Jones AR. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 2011;39:D913–919. doi: 10.1093/nar/gkq1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cerna M, et al. Differences in HLA class II alleles of isolated South American Indian populations from Brazil and Argentina. Hum. Immunol. 1993;37:213–220. doi: 10.1016/0198-8859(93)90504-t. [DOI] [PubMed] [Google Scholar]
- 41.Hollenbach JA, et al. HLA diversity, differentiation, and haplotype evolution in Mesoamerican Natives. Hum Immunol. 2001;62:378–390. doi: 10.1016/s0198-8859(01)00212-9. [DOI] [PubMed] [Google Scholar]
- 42.Palafox D, et al. Determinación de HLA en pacientes con Síndrome de Parry Romberg atendidos en el Servicio de Cirugía Plástica y Reconstructiva del Hospital General “Dr. Manuel Gea González”. Cirugía Plástica Ibero-Latinoamericana. 2016;42:6. doi: 10.4321/S0376-78922016000200004. [DOI] [Google Scholar]
- 43.Chi C, et al. Admixture mapping reveals evidence of differential multiple sclerosis risk by genetic ancestry. PLoS Genet. 2019;15:e1007808. doi: 10.1371/journal.pgen.1007808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Consortium, W. T. C. C Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson AJ, Baranzini SE, Geurts J, Hemmer B, Ciccarelli O. Multiple sclerosis. Lancet. 2018;391:1622–1636. doi: 10.1016/S0140-6736(18)30481-1. [DOI] [PubMed] [Google Scholar]
- 46.Wingerchuk DM, Lennon VA, Pittock SJ, Lucchinetti CF, Weinshenker BG. Revised diagnostic criteria for neuromyelitis optica. Neurology. 2006;66:1485–1489. doi: 10.1212/01.wnl.0000216139.44259.74. [DOI] [PubMed] [Google Scholar]
- 47.Wingerchuk DM, et al. International consensus diagnostic criteria for neuromyelitis optica spectrum disorders. Neurology. 2015;85:177–189. doi: 10.1212/WNL.0000000000001729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruiz-Linares A, et al. Admixture in Latin America: geographic structure, phenotypic diversity and self-perception of ancestry based on 7,342 individuals. PLoS Genet. 2014;10:e1004572. doi: 10.1371/journal.pgen.1004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Polman CH, et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann. Neurol. 2011;69:292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lebedeva TV, et al. Resolution of HLA class I sequence-based typing ambiguities by group-specific sequencing primers. Tissue Antigens. 2011;77:247–250. doi: 10.1111/j.1399-0039.2010.01616.x. [DOI] [PubMed] [Google Scholar]
- 51.Robinson J, Waller MJ, Parham P, Bodmer JG, Marsh SG. IMGT/HLA Database–a sequence database for the human major histocompatibility complex. Nucleic Acids Res. 2001;29:210–213. doi: 10.1093/nar/29.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andrews, S. FastQC: a quality control tool for high throughput sequence data., <https://www.bioinformatics.babraham.ac.uk/projects/> (2010).
- 53.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maples BK, Gravel S, Kenny EE, Bustamante CD. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 2013;93:278–288. doi: 10.1016/j.ajhg.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online. 2007;1:47–50. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset for the NMO patients generated and/or analyzed during the current study are available from the corresponding authors on reasonable request. Access to the CANDELA dataset used in this manuscript was obtained through a formal request to the Consortium for the Analysis of the Diversity and Evolution of Latin America steering committee.