Abstract
Mycoplasma hyopneumoniae is the most costly pathogen for swine production. Although several studies have focused on the host-bacterium association, little is known about the changes in gene expression of swine cells upon infection. To improve our understanding of this interaction, we infected swine epithelial NPTr cells with M. hyopneumoniae strain J to identify differentially expressed mRNAs and miRNAs. The levels of 1,268 genes and 170 miRNAs were significantly modified post-infection. Up-regulated mRNAs were enriched in genes related to redox homeostasis and antioxidant defense, known to be regulated by the transcription factor NRF2 in related species. Down-regulated mRNAs were enriched in genes associated with cytoskeleton and ciliary functions. Bioinformatic analyses suggested a correlation between changes in miRNA and mRNA levels, since we detected down-regulation of miRNAs predicted to target antioxidant genes and up-regulation of miRNAs targeting ciliary and cytoskeleton genes. Interestingly, most down-regulated miRNAs were detected in exosome-like vesicles suggesting that M. hyopneumoniae infection induced a modification of the composition of NPTr-released vesicles. Taken together, our data indicate that M. hyopneumoniae elicits an antioxidant response induced by NRF2 in infected cells. In addition, we propose that ciliostasis caused by this pathogen is partially explained by the down-regulation of ciliary genes.
Subject terms: Gene expression, Bacteriology, Pathogens, miRNAs, Transcriptomics
Introduction
Respiratory diseases are among the major health problems in the pig farming industry. Mycoplasma hyopneumoniae is the causative agent of swine enzootic pneumonia, a chronic respiratory disease that affects herds worldwide. Although M. hyopneumoniae does not cause high mortality, it is considered the most expensive pathogen for swine production1. This is mainly due to the costs of treatment and vaccination and to losses related to decreased animal performance. In addition, M. hyopneumoniae is essential for the establishment of secondary pathogens in the host, which leads to a significant increase in mortality2. M. hyopneumoniae attaches to the cilia of the tracheal epithelial cells with participation of adhesins3, resulting in ciliostasis and cell death4. Besides adhesins, virulence factors are not well understood in this bacterium. Nevertheless, a recent study from our group indicated hydrogen peroxide production from glycerol and myo-inositol metabolism as important traits that might be related with pathogenesis and with the predominance of M. hyopneumoniae in the swine respiratory tract5.
MicroRNAs (miRNAs) belong to a class of small non-coding RNAs (ncRNAs) of 18–24 nucleotides (nt) in part responsible for post-transcriptional gene regulation in eukaryotes. These evolutionarily conserved molecules influence fundamental biological processes, including cell proliferation, differentiation, apoptosis, immune response, and metabolism6, 7. The binding of miRNAs to target mRNAs changes the mRNA stability and translation efficiency8, leading to degradation, suppression or up-regulation of the target mRNAs6, 9. Interactions between miRNA and mRNA are complex; one single miRNA can target a large number of genes belonging to diverse functional groups. Alternatively, the 3’-UTR of a single mRNA can be targeted by multiple miRNAs10, 11. By modulating miRNA abundance, it is thus possible to fine-tune the expression of proteins within the cell in a very precise manner6, 11.
Recently, it was widely reported that miRNAs can be packed into exosomes and transferred to neighboring or distant cells to regulate cell function7, 12–14. Exosomes are small membrane vesicles (50–150 nm) released from eukaryotic cells both constitutively and upon induction, under normal and pathological conditions13, 15. These vesicles are involved in several cellular functions and have the potential to selectively interact with specific target cells16, 17. In addition to miRNAs, exosomes can transmit information among cells by transferring proteins, lipids and nucleic acids that seem to be selected non-randomly, with some specific populations of molecules being preferentially packaged into the vesicles13, 15. As an efficient cellular signaling and communication system, the release of exosomes by infected host cells has been recognized as a common phenomenon, in some cases beneficial to the host and in others beneficial to the pathogen18.
Host-pathogen interactions result in signaling and physiological modifications in the host cells that induce differential miRNA expression and miRNA-mediated post-transcriptional regulation of genes involved in immune response and several other cellular pathways19, 20. Therefore, simultaneous identification of differentially expressed miRNAs and mRNAs provides a comprehensive view on host-pathogen interactions during the infection and the disease establishment process. In recent years, efforts have been made to identify miRNAs regulated by infection in several mammalian hosts8, 20–22. However, the identification of miRNAs during infection of swine cells with M. hyopneumoniae has not been investigated so far. To improve our understanding on the M. hyopneumoniae-host interaction, we sequenced and analyzed both the mRNAs and miRNAs of a swine tracheal epithelial cell line infected with M. hyopneumoniae strain J. In addition, we identified miRNAs differentially expressed (DE) in the extracellular milieu and in exosome-like vesicles released by the infected cells, which play an important role in cell-cell communication and in the dissemination of host and pathogen-derived molecules during infection15. The simultaneous identification of miRNAs and mRNAs will help us draw a full picture of the changes in gene expression and the possible regulatory mechanisms of host cells during the disease establishment.
Results and discussion
M. hyopneumoniae strain J adhered to NPTr cells
To analyze the differential expression of New-born Pig Trachea (NPTr) cells during the infection with M. hyopneumoniae, we first observed the infection by immunofluorescence microscopy. These analyses were performed to detect the adherence of M. hyopneumoniae strain J to NPTr cells. Supplementary Figure S1 shows the co-localization of M. hyopneumoniae with NPTr cells, corroborating the success of the infection. As few bacteria adhered to the cells within a short period of time (1 vs. 24 h), we chose to analyze the transcriptional alterations of NPTr cells at 24 h post-infection. M. hyopneumoniae strain J was chosen because previous infection assays showed that highly virulent strains, such as 7448 or 7422, damaged the host cells and we were not able to recover RNA with good sequencing quality. Although this strain is considered attenuated and incapable of causing disease in vivo23, 24, our results show that M. hyopneumoniae is capable of adhering to the swine epithelial cells, as previously reported by Burnett et al.25.
mRNA expression profiles
A total of 6 mRNA libraries were generated with the Illumina HiSeq2500 platform. A summary of the samples is provided in Table 1 and a diagram to explain the experimental design is given in Supplementary Fig. S2. The raw reads were submitted to the NCBI Sequence Read Archive under accession number PRJNA545822. After removing adapters and filtering low quality reads, mRNA-seq yielded around 40 million paired-end reads in all 6 samples (approx. 97% of raw reads). Trimming and mapping information for each mRNA sample is available in Supplementary Table S1. Around 85% of the filtered reads were mapped against the porcine genome (Sscrofa10.2—Ensembl release 89) and 40% against annotated genes (Supplementary Table S1).
Table 1.
Samples detailed information. The experimental design with the description of the samples is also explained in Fig. S2.
| Sample ID | Sample name | RNA type | Compartment | Condition | Replicates | |
|---|---|---|---|---|---|---|
| mRNA 1-3 | mRNA CTRL | mRNA | Intracellular | Control | 3 | |
| mRNA 4-6 | mRNA INF | Infected with MHP | ||||
| sRNA 1-3 | sRNA INTRA CTRL | sRNA | Intracellular | Control | ||
| sRNA 4-6 | sRNA INTRA INF | Infected with MHP | ||||
| sRNA 7-8 | sRNA EXTRA CTRL | Extracellular | Total extracellular sRNA | Control | 2 | |
| sRNA 9-10 | sRNA EXTRA INF | Infected with MHP | ||||
| sRNA 11 | sRNA EXO CTRL | sRNA from exosome-like vesicles | Control | Pool (50 biological replicates) | ||
| sRNA 12 | sRNA EXO INF | Infected with MHP | ||||
| sRNA 13 | sRNA SN CTRL | sRNA from vesicle-free supernatant | Control | |||
| sRNA 14 | sRNA SN INF | Infected with MHP | ||||
In the present study, we detected a total of 20,274 (out of 23,215) genes expressed with at least 10 counts across the 6 mRNA libraries from NPTr cells (Supplementary Table S2). Since we used an attenuated strain of M. hyopneumoniae to infect the swine cells, we did not expect extreme changes in gene expression. Indeed, most of the significant log2 of Fold Changes (LogFC) of the DE genes analyzed in our results were below 1 (for up-regulated genes) or above -1 (for down-regulated genes) (Supplementary Table S3). Nevertheless, even if the strain used (J) is considered attenuated, it generated a considerable immune response. A similar situation occurs in vaccine development, in which attenuated strains are used to generate immune response without causing lethal effects. Thus, we believe that infections with highly virulent strains of M. hyopneumoniae would be able to generate a more pronounced effect on gene expression than what was observed here. Regardless, our results provide a general overview of M. hyopneumoniae infection in the host cells.
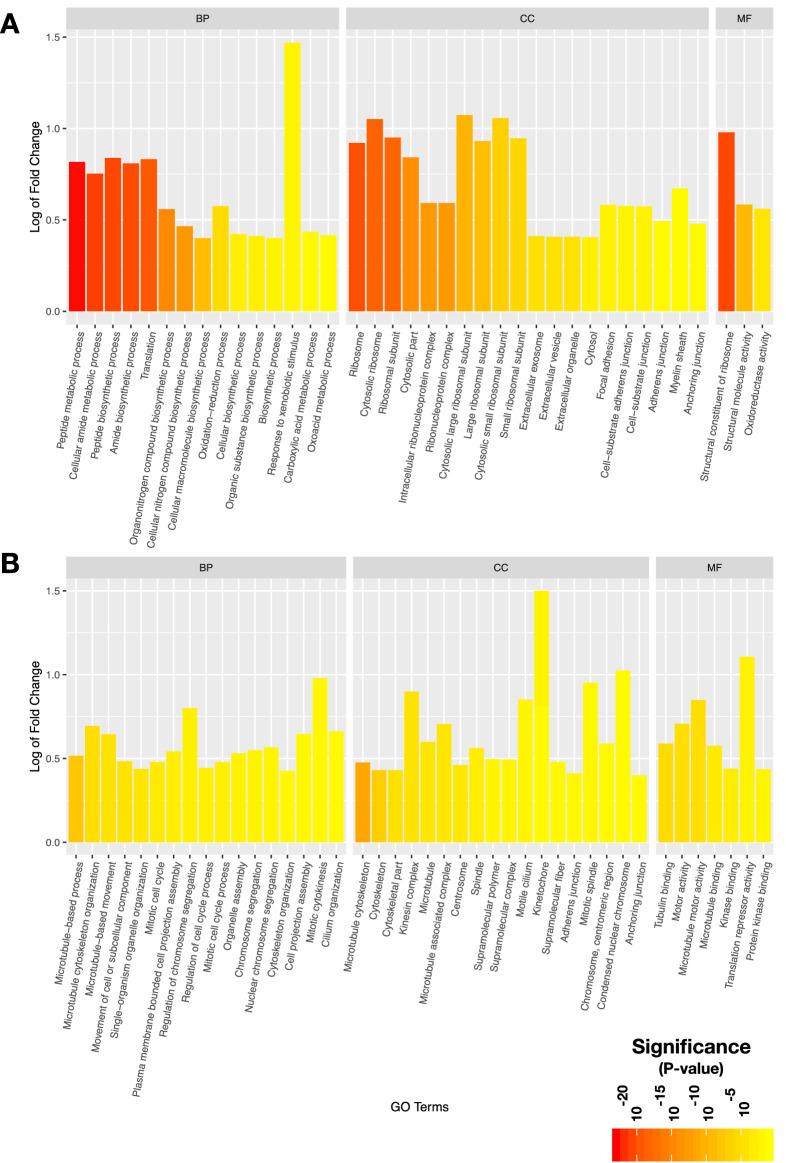
In total, we detected 1,268 DE genes (p-adj , 517 up-regulated and 751 down-regulated), from which 502 were common to two well known methods for the detections of DE genes, 721 were exclusive to DESeq2 and 45 were exclusive to EdgeR. Information from the top 15 up-regulated and down-regulated DE genes is provided in Table 2. The complete DE gene results are found in Supplementary Table S3. The results of gene ontology (GO) enrichment analysis for up-regulated and down-regulated genes in DESeq2 are shown separately in Fig. 1 (p-adj and absolute fold enrichment 0.1) and the complete results are found in Supplementary Table S4. For the up-regulated genes, the enriched terms either in biological process (BP), molecular function (MF) or cellular component (CC) were related to protein synthesis (translation/ribosome), oxidation reduction activity and cell-cell communication (such as exosomes, anchoring and focal adhesion functions) (Fig. 1A). For the down-regulated genes, the majority of the overrepresented terms in all three GO categories were related to cell cycle, cell division, cilia and cytoskeleton (Fig. 1B).
Table 2.
Selected up- and down-regulated genes. Information about the top 15 up- and 15 down-regulated DE genes from both the DeSeq2 and EdgeR methods. Ordered by LogFC calculated by EdgeR.
| DE genes | DESeq2 | EdgeR | UP/DOWN | ||||
|---|---|---|---|---|---|---|---|
| Gene ID | Associated gene name | Description | LogFC | P-adj | LogFC | P-adj | |
| ENSSSCG00000028871 | LOC396866 | Cystatin-A1 | 0.691 | 4,35E24 * | 5.375 | 1.02E14 | Upregulated |
| ENSSSCG00000030385 | C3 | Complement C3 | 1.427 | 4.69E63 | 2.698 | 2.60E43 | Upregulated |
| ENSSSCG00000021728 | LGALS2 | Galectin 2 | 0.646 | 5,38E15 * | 1.507 | 3.71E05 | Upregulated |
| ENSSSCG00000028525 | SAA3 | Serum Amyloid A3 | 0.841 | 4.67E25 | 1.211 | 2.88E21 | Upregulated |
| ENSSSCG00000001101 | SCGN | Secretagogin, EF-Hand Calcium Binding Protein | 0.554 | 6.74E09 | 1.018 | 9.06E05 | Upregulated |
| ENSSSCG00000028982 | AKR1C4 | Aldo-Keto Reductase Family 1 Member C4 | 0.869 | 2.61E76 | 0.953 | 4.61E54 | Upregulated |
| ENSSSCG00000025277 | AKR1CL1 | Aldo-Keto Reductase Family 1, Member C-Like 1 | 0.866 | 3.96E76 | 0.949 | 5.46E54 | Upregulated |
| ENSSSCG00000004678 | DUOX2 | Dual Oxidase 2 | 0.515 | 1.99E09 | 0.721 | 6.93E09 | Upregulated |
| ENSSSCG00000010146 | LGALS8 | Galectin 8 | 0.453 | 1.41E06 | 0.691 | 2.39E15 | Upregulated |
| ENSSSCG00000025273 | CYP11A1 | Cytochrome P450 Family 11 Subfamily A Member 1 | 0.51 | 1.39E11 | 0.648 | 3.80E11 | Upregulated |
| ENSSSCG00000003402 | PGD | Phosphogluconate Dehydrogenase | 0.611 | 3.62E61 | 0.647 | 2.71E36 | Upregulated |
| ENSSSCG00000008959 | CXCL2 | C-X-C Motif Chemokine Ligand 2 | 0.491 | 5.15E13 | 0.59 | 1.10E11 | Upregulated |
| ENSSSCG00000007435 | PLTP | Phospholipid Transfer Protein | 0.436 | 7.49E09 | 0.544 | 3.54E08 | Upregulated |
| ENSSSCG00000010853 | EPHX1 | Epoxide Hydrolase 1 | 0.444 | 5.20E15 | 0.503 | 1.76E06 | Upregulated |
| ENSSSCG00000000843 | TXNRD1 | Thioredoxin Reductase 1 | 0.426 | 7.45E14 | 0.482 | 7.33E13 | Upregulated |
| ENSSSCG00000017135 | ZNF750 | Zinc Finger Protein 750 | 0.741 | 7,84E26 * | 4.899 | 3.43E15 | Downregulated |
| ENSSSCG00000009399 | CYSLTR2 | Cysteinyl Leukotriene Receptor 2 | 0.439 | 1,01E07 * | 1.034 | 2.57E02 | Downregulated |
| ENSSSCG00000013401 | DKK3 | Dickkopf WNT Signaling Pathway Inhibitor 3 | 0.581 | 5.15E13 | 0.774 | 1.35E12 | Downregulated |
| ENSSSCG00000016273 | HTR2B | 5-Hydroxytryptamine Receptor 2B | 0.434 | 1.37E05 | 0.735 | 2.12E03 | Downregulated |
| ENSSSCG00000014232 | LOX | Lysyl Oxidase | 0.464 | 1.06E06 | 0.718 | 1.39E04 | Downregulated |
| ENSSSCG00000011217 | NEK10 | NIMA Related Kinase 10 | 0.418 | 2.80E05 | 0.689 | 4.20E03 | Downregulated |
| ENSSSCG00000003810 | UBE2U | Ubiquitin Conjugating Enzyme E2 U (Putative) | 0.392 | 1.38E04 | 0.676 | 8.30E03 | Downregulated |
| ENSSSCG00000011441 | TNNC1 | Troponin C1, Slow Skeletal And Cardiac Type | 0.399 | 5.92E05 | 0.633 | 3.27E03 | Downregulated |
| ENSSSCG00000015413 | FGL2 | Fibrinogen Like 2 | 0.487 | 1.81E15 | 0.554 | 1.64E14 | Downregulated |
| ENSSSCG00000008334 | MXD1 | MAX Dimerization Protein 1 | 0.385 | 3.21E06 | 0.486 | 3.02E05 | Downregulated |
| ENSSSCG00000027157 | SLC40A1 | Solute Carrier Family 40 Member 1 | 0.365 | 1.23E05 | 0.462 | 3.19E05 | Downregulated |
| ENSSSCG00000006072 | VPS13B | Vacuolar Protein Sorting 13 Homolog B | 0.359 | 4.30E05 | 0.469 | 5.52E04 | Downregulated |
| ENSSSCG00000028322 | BTG2 | BTG Anti-Proliferation Factor 2 | 0.394 | 2.61E09 | 0.452 | 2.29E04 | Downregulated |
| ENSSSCG00000016059 | STAT4 | Signal Transducer And Activator Of Transcription 4 | 0.354 | 6.36E06 | 0.43 | 6.75E05 | Downregulated |
| ENSSSCG00000028196 | MFF | Mitochondrial Fission Factor | 0.352 | 6.40E06 | 0.427 | 3.85E05 | Downregulated |
* In the cases where EdgeR detected a DE gene with high significance and the padj was not calculated in DESeq2 due to the presence of one outlier, we checked case by case and validated as DE, whenever suitable. The adjusted P-value showed in these cases is the value calculated prior to multitest for demonstration purposes only.
Figure 1.
Significantly enriched Gene Ontology functions of DE mRNAs. A. Up-regulated genes were enriched in terms related to protein synthesis, oxidation-reduction activity and cell-cell communication (such as exosomes, anchoring and focal adhesion), either in biological process (BP), molecular function (MF) or cellular component (CC). B. Down-regulated genes showed an enrichment of terms related to cytoskeleton, ciliary function, cell cycle and cell division in all the three categories of GO.
M. hyopneumoniae elicited an antioxidant response and induced the accumulation of NRF2 in the nuclei of NPTr cells
Among the up-regulated genes, we found several ones related to immune response and inflammation, such as C3 complement, SAA3, chemokines (CXCL2 and CCL20) and galectins (LGALS2 and LGALS8) (Supplementary Table S3). Interestingly, we also detected 64 up-regulated genes related to redox homeostasis and antioxidant defense (Table 3). We observed that 46 out of 64 have already been described as targets of the nuclear factor erythroid 2-related factor 2 (NRF2) in closely related species (Table 3). This transcription factor is involved in the protection of the cell against oxidative damage through transcription activation of cytoprotective genes26, 27. More specifically, several studies have shown the protective role of NRF2 in bacterial lung infections in rodents, being a critical factor for assembling the innate immune response in the host28–31.
Table 3.
Up-regulated genes involved in redox homeostasis. Detailed information for differential expression of genes (from both the DeSeq2 and EdgeR methods) related to antioxidant and redox homeostasis functions. The great majority of such genes has been demonstrated to be activated by the NRF2 transcription factor in related species. Duplicated entries related to different transcripts of a given gene were deleted in this table, however calculation of the statistics provided in Supplementary Table S5 was made taking into account the total amount of 64 genes (including duplications).
| DE genes | DESeq2 | EdgeR | Reference | ||||
|---|---|---|---|---|---|---|---|
| Gene ID | Associated gene name | Description | LogFC | padj | LogFC | FDR | |
| ENSSSCG00000028982 | AKR1C4 | Aldo-Keto Reductase Family 1 Member C4 | 0.869 | 2.61E76 | 0.953 | 4.61E54 | Hayes and McMahon117 |
| ENSSSCG00000025277 | AKR1CL1 | Aldo-Keto Reductase Family 1, Member C-Like 1 | 0.866 | 3.96E76 | 0.949 | 5.46E54 | Hayes and McMahon117 |
| ENSSSCG00000003402 | PGD | Phosphogluconate Dehydrogenase | 0.611 | 3.62E61 | 0.647 | 2,71E36 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000025273 | CYP11A1 | Cytochrome P450 Family 11 Subfamily A Member 1 | 0.510 | 1.39E11 | 0.648 | 3.80E11 | |
| ENSSSCG00000010853 | EPHX1 | Epoxide Hydrolase 1 | 0.444 | 5.20E15 | 0.503 | 1.76E06 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000014540 | FTH1 | Ferritin Heavy Chain 1 | 0.436 | 1.81E24 | 0.470 | 1.85E14 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000003153 | FTL | Ferritin Light Chain | 0.427 | 1.25E24 | 0.459 | 5.31E14 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000000843 | TXNRD1 | Thioredoxin Reductase 1 | 0.426 | 7.45E14 | 0.482 | 7.33E13 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000002754 | NQO1 | NAD(P)H Quinone Dehydrogenase 1 | 0.415 | 7.89E18 | 0.456 | 1.64E12 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000002626 | GSTA1 | Glutathione S-Transferase A1 | 0.407 | 1.02E12 | 0.461 | 4.18E09 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000016312 | UGT1A6 | UDP Glucuronosyltransferase Family 1 Member A6 | 0.367 | 1.88E18 | 0.394 | 7.49E12 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000011147 | AKR1C1 | Aldo-Keto Reductase Family 1 Member C1 | 0.360 | 1.28E04 | 0.511 | 1.20E03 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000021386 | PTGR1 | Prostaglandin Reductase 1 | 0.358 | 6.02E12 | 0.397 | 1.55E04 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000001488 | GCLC | Glutamate-Cysteine Ligase Catalytic Subunit | 0.350 | 2.78E10 | 0.392 | 2.89E09 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000002825 | CES1 | Carboxylesterase 1 | 0.348 | 5.58E12 | 0.384 | 2.93E08 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000012136 | PIR | Pirin | 0.334 | 9.34E07 | 0.392 | 7.21E06 | Brzóska et al.119 |
| ENSSSCG00000025762 | GSR | Glutathione SReductase | 0.314 | 2.46E03 | 0.489 | 2.57E02 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000029144 | SRXN1 | Sulfiredoxin 1 | 0.309 | 3.31E06 | 0.359 | 1.12E05 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000010055 | GGT5 | Gamma-Glutamyltransferase 5 | 0.305 | 1.38E03 | 0.421 | 3.33E03 | |
| ENSSSCG00000014833 | UCP2 | Uncopling Protein 2 | 0.303 | 5.10E03 | 0.509 | 4.52E02 | |
| ENSSSCG00000028099 | SLC6A6 | Solute Carrier Family 6 Member 6 | 0.295 | 2.36E03 | 0.412 | 5.50E03 | Hayes and McMahon117 |
| ENSSSCG00000026759 | HMOX1 | Heme Oxygenase 1 | 0.272 | 9.35E03 | 0.402 | 4.36E02 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000008311 | CYP26B1 | Cytochrome P450 Family 26 Subfamily B Member 1 | 0.267 | 4.82E03 | 0.359 | 8.22E03 | |
| ENSSSCG00000028996 | ALDH1A1 | Aldehyde Dehydrogenase 1 Family Member A1 | 0.261 | 1.47E18 | 0.274 | 2.92E07 | Hayes and Dinkova-Kostova18 |
| ENSSSCG00000006717 | PHGDH | Phosphoglycerate Dehydrogenase | 0.260 | 1.20E07 | 0.285 | 4.62E05 | |
| ENSSSCG00000021067 | BLVRB | Biliverdin Reductase B | 0.259 | 2.00E04 | 0.302 | 1.22E03 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000013366 | LDHA | Lactate Dehydrogenase A | 0.255 | 9.07E07 | 0.281 | 8.19E05 | |
| ENSSSCG00000029781 | SELENOM | Selenoprotein M | 0.254 | 2.25E02 | 0.406 | 5.78E02 | |
| ENSSSCG00000001723 | PLA2G7 | Phospholipase A2 Group VII | 0.250 | 4.58E04 | 0.294 | 2.13E03 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000004093 | IYD | Iodotyrosine Deiodinase | 0.250 | 1.09E02 | 0.339 | 1.83E02 | |
| ENSSSCG00000001963 | EGLN3 | Egl-9 Family Hypoxia Inducible Factor 3 | 0.248 | 7.36E03 | 0.324 | 9.69E03 | |
| ENSSSCG00000003914 | PRDX1 | Peroxiredoxin 1 | 0.231 | 1.02E07 | 0.249 | 1.27E04 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000017092 | GPX3 | Glutathione Peroxidase 3 | 0.224 | 7.69E05 | 0.250 | 1.89E03 | Kensler et al.32 |
| ENSSSCG00000018048 | SLC5A10 | Solute Carrier Family 5 Member 10 | 0.215 | 3.21E03 | 0.251 | 9.13E03 | Hayes and McMahon117 |
| ENSSSCG00000003491 | AKR7A2 | Aldo-Keto Reductase Family 7 Member A2 | 0.205 | 1.69E02 | 0.252 | 3.10E02 | Li et al.120 |
| ENSSSCG00000021408 | TKT | Transketolase | 0.190 | 2.39E06 | 0.204 | 1.49E03 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000010340 | FAM213A | Family With Sequence Similarity 213 Member A | 0.184 | 1.62E05 | 0.198 | 4.20E03 | |
| ENSSSCG00000010056 | GGT1 | Gamma-Glutamyltransferase 1 | 0.178 | 1.95E02 | 0.209 | 2.50E02 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000013030 | PRDX5 | Peroxiredoxin 5 | 0.177 | 1.04E02 | 0.204 | 2.22E02 | Graham et al.121 |
| ENSSSCG00000012847 | TALDO1 | Transaldolase 1 | 0.177 | 3.29E03 | 0.199 | 2.33E01 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000030461 | HEPHL1 | Hephaestatin Like 1 | 0.170 | 8.19E05* | 3.003 | 3.60E04 | |
| ENSSSCG00000018084 | ND3 | Mitochondrially Encoded NADH:Ubiquinone Oxidoreductase Core Subunit 3 | 0.166 | 2.19E03 | 0.183 | 2.77E02 | |
| ENSSSCG00000012327 | HSD17B10 | Hydroxysteroid 17-Beta Dehydrogenase 10 | 0.164 | 1.66E02 | 0.188 | 1.94E02 | |
| ENSSSCG00000003007 | BCKDHA | Branched Chain Keto Acid Dehydrogenase E1, Alpha Polypeptide | 0.163 | 3.52E02 | 0.191 | 5.54E02 | |
| ENSSSCG00000010928 | KDM5B | Lysine Demethylase 5B | 0.161 | 2.36E04 | 0.174 | 1.58E02 | |
| ENSSSCG00000014336 | EGR1 | Early Growth Response 1 | 0.159 | 2.52E04 | 0.172 | 1.42E02 | Gomez et al.31 |
| ENSSSCG00000008550 | SLC5A6 | Solute Carrier Family 5 Member 6 | 0.155 | 2.36E03 | 0.170 | 2.44E02 | Hayes and McMahon117 |
| ENSSSCG00000006324 | ALDH9A1 | Aldehyde Dehydrogenase 9 Family Member A1 | 0.155 | 6.71E04 | 0.168 | 2.07E02 | |
| ENSSSCG00000022742 | PRDX6 | Peroxiredoxin 6 | 0.151 | 1.49E03 | 0.164 | 2.28E02 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000001701 | HSP90AB1 | Heat Shock Protein 90 Alpha Family Class B Member 1 | 0.149 | 2.11E04 | 0.160 | 2.15E02 | Hayes and McMahon117 |
| ENSSSCG00000028739 | CEBPB | CCAAT/Enhancer Binding Protein Beta | 0.148 | 1.32E02 | 0.165 | 3.70E01 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000004454 | ME1 | Malic Enzyme 1 | 0.145 | 5.30E03 | 0.160 | 2.96E01 | Hayes and Dinkova-Kostova118 |
| ENSSSCG00000010682 | PRDX3 | Peroxiredoxin 3 | 0.139 | 1.35E02 | 0.154 | 7.06E02 | Gomez et al.31 |
| ENSSSCG00000027525 | DHCR24 | 24-Dehydrocholesterol Reductase | 0.123 | 3.68E03 | 0.133 | 5.52E02 | |
| ENSSSCG00000029876 | SOD2 | Superoxide Dismutase 2 | 0.112 | 1.66E02 | 0.123 | 1.39E01 | Reszka et al.122 |
References related to genes regulated by NRF2.
Genes with more than one ID identified as DE.
* The P-value showed in these cases is the value calculated prior to multitesting for demonstration purposes only (see Table 2).
Indeed, M. hyopneumoniae infection induced the expression of genes related to the two biggest redox systems in NPTr cells - glutathione (GGT1, GGT5, GGLC, GSR) and thioredoxin (TXNRD1, PRDX5, PRDX6) -, and also genes coding for NADPH-regenerating enzymes (used by the aforementioned redox systems), as analogously reported for activation of NRF2 targets in mice32. Moreover, a number of antioxidant genes and genes coding for detoxification enzymes (such as the AKR gene family, NQO1, HMOX and GST) which were up-regulated during M. hyopneumoniae infection, were also reported to be activated by NRF232 (Table 3). We performed a Fisher’s exact test to compare the proportion of genes within the genome expected to be targets of NRF2 with the proportion of up-regulated genes putatively regulated by this transcription factor. The results indicate an extremely significant correlation of an NRF2 target being an upregulated gene (p-value , Supplementary Table S5). The up-regulation of several targets of NRF2 was further validated by reverse-transcription quantitative PCR (RT-qPCR) and the results are available in Supplementary Fig. S3.
One of the many compounds known to activate the NRF2 pathway is hydrogen peroxide33. The production of hydrogen peroxide was previously described to have an important cytotoxic effect in several Mycoplasma species, such as M. pneumoniae34 and M. mycoides subsp. mycoides35. More specifically, the cytotoxity of M. mycoides subsp. mycoides was correlated to the bacterium’s ability to translocate hydrogen peroxide directly into the host cell36. Previously, we have identified that M. hyopneumoniae was capable of producing this toxic product from glycerol metabolism5. Although the production of hydrogen peroxide by the M. hyopneumoniae strain J was not detected in that work, novel analyses indicate that in conditions where glucose is scarce, the attenuated strain J is able to produce this toxic metabolite (Supplementary Fig. S4). In addition, we were able to detect hydrogen peroxide in the medium of the NPTr cells infected with M. hyopneumoniae in the presence of glycerol (Supplementary Fig. S4C). Although the production was higher in the cells infected with the pathogenic strain 7448, M. hyopneumoniae strain J was also able to produce the toxic product, indicating that both strains could potentially cause oxidative damage to the host cells. These results are in accordance with the gene expression of the putative enzyme responsible for hydrogen peroxide production (GlpO) in M. hyopneumoniae strain J, which did not differ from the pathogenic strains5. During infection in general, bacteria need to compete with host cells (and other organisms in the environment) for glucose and other energy sources. For instance, in M. pneumoniae, Halbedel et al. (2004)37 showed that even though glucose was the most efficient carbon source for biomass yield (as is the case for M. hyopneumoniae), the authors propose that glycerol is not only a carbon source, but it could be used by this species as an indicator that they reached their preferred ecological niche, a lipid-rich mucosal surface. Thus, it is plausible to say that M. hyopneumoniae also uses glycerol as a carbon source in vivo, however whether this is a result of competition against other fast-growing bacteria or if this species is targeting a glycerol-rich niche, we cannot affirm at this point. What we can affirm is that glycerol is not the most efficient carbon source for both M. pneumoniae and M. hyopneumoniae, and yet these species make use of it for energy uptake and also for other advantages, such as hydrogen peroxide production. Thus, it is possible that the hydrogen peroxide produced by M. hyopneumoniae strain J as a result of glycerol metabolism might be one of the triggers that activated the transcription factor NRF2.
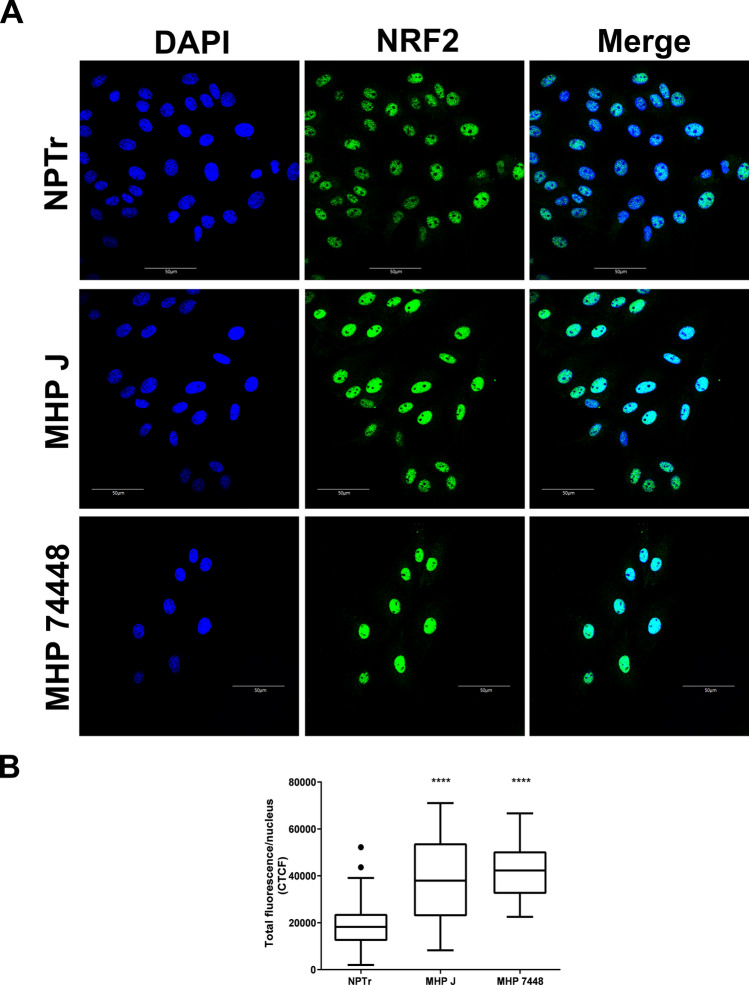
It has been demonstrated in other species that NRF2 is largely regulated at the post-transcriptional level by its sub-cellular distribution, which is controlled by the Kelch-like ECH-associated protein (KEAP1). Under normal conditions, a portion of NRF2 is retained in the cytoplasm via its interaction to KEAP1 and it is subsequently ubiquitinated and degraded by the proteasome. In response to oxidative stress, reactive cysteines in KEAP1 are modified, generating conformational changes in the complex and releasing NRF2, which is translocated and accumulates into the nucleus27, 32, 38. In our analysis, we did not detect a difference in the expression of the mRNAs of NRF2 (base mean expression around 8000 reads, a logFC between infected and non-infected conditions close to zero and a non-significant adjusted p-value of 0.99) and KEAP1 (base mean expression around 4500 reads, logFC between infected and non-infected conditions close to zero and non-significant adjusted p-value of 0.94). However, we were able to demonstrate by confocal immunofluorescence microscopy (Fig. 2) that both the attenuated and the virulent strains of M. hyopneumoniae induced a statistically significant accumulation of this transcription factor in the nuclei of NPTr cells (Fig. 2B). In the nucleus, NRF2 is able to recognize and bind to antioxidant response element (ARE) motifs in the promoter region of target genes, activating their transcription39. In this work, we detected at least one conserved ARE sequence upstream the start codon in the promoter regions of 44 out of the 46 NRF2 predicted targets (with stringent search to TG/TAnnnnGC) with the use of fuzznuc software from EMBOSSv6.6.0 package40 (Supplementary Table S6).
Figure 2.
Localization and expression pattern of the NRF2 protein in cells infected with M. hyopneumoniae.A. Results of the immunofluorescence microscopy analysis indicating the accumulation of NRF2 in the nuclei of infected cells after 1 h of incubation. Both attenuated (J) and virulent (7448) strains of M. hyopneumoniae induced the accumulation of the transcription factor in the nuclei of the epithelial cells. Eukaryotic cell NRF2 was labeled with anti-NRF2 antibody (green) and nuclei were stained with DAPI (blue). Scale bars: . B. Total NRF2 green fluorescence per nucleus of control and infected cells. Both M. hyopneumoniae strains significantly increased the concentration of NRF2 in the nuclei of infected cells. Boxplots represent the measures of at least 25 nuclei in each condition. Outliers are represented as black dots. CTCF—corrected total cell fluorescence. NPTr - uninfected cells. MHP J - NPTr cells infected with M. hyopneumoniae strain J. MHP 7448 - NPTr cells infected with M. hyopneumoniae strain 7448. ****
In this way, it seems that M. hyopneumoniae infection has the potential to cause oxidative stress to the host cells, which in turn activate antioxidant response genes induced by NRF2 to fight the infection and maintain cellular homeostasis. We believe that this oxidative stress was in part related to the hydrogen peroxide produced by this bacterium, although more experiments are needed to prove the association between this mechanism and the up-regulation of antioxidant response genes.
M. hyopneumoniae induced down-regulation of cytoskeleton and ciliary genes as well as a decrease of actin stress fibers in NPTr cells
We identified several down-regulated genes related to ciliary function, cytoskeleton and cell cycle/cell division. The impairment of the ciliary motility is a well-known effect caused by several Mycoplasma respiratory species41, such as M. pneumoniae and M. gallisepticum42, 43. It is also well established that M. hyopneumoniae attaches to cilia of epithelial cells and promotes ciliostasis and loss of cilia, causing damage to the mucociliary apparatus4, 24. Therefore, our hypothesis is that one of the reasons for epithelial damage, besides physical adhesion, could be associated with modulations in gene expression induced by the infection, which is a running hypothesis for at least two epithelial pathogens from the genus Mycobacterium44. In agreement with this hypothesis, we were able to identify the down-regulation of genes coding for axonemal dyneins (DNAH11, DNAH12, DNAI2, DNAL1), which are essential for the ciliary motility45. In addition, genes necessary for axonemal dynein assembly (DYX1C1)46, genes related to ciliogenesis (CEP162, DCDC2, MACF1, IFT57)47–50, ciliary polarization (INTU)51, ciliary beating (MYO1D)52 and several others in which the mutation or knockout is associated with ciliopathies (LRRC6, MNS1, AK7)53–55 were down-regulated in the infected cells (Table 4). Interestingly, in line with our results, a recent study compared the transcriptional response of unvaccinated and vaccinated chicken infected with M. gallisepticum and the authors identified enrichment of GO terms in down-regulated genes related to cilia and cytoskeleton in unvaccinated animals56. Protein functions encoded by the top down-regulated genes were involved in microtubule assembly and stability, axonemal dynein complex assembly, and formation and motor movement of cilia, indicating that at least in one Mycoplasma species the ciliary damage caused by infection could be also explained by the down-regulation of genes involved in the ciliary function.
Table 4.
Down-regulated genes involved in cytoskeleton and cilia.
| DE genes | DESeq2 | EdgeR | ||||
|---|---|---|---|---|---|---|
| Gene ID | Associated gene name | Description | LogFC | P-adj | LogFC | P-adj |
| ENSSSCG00000011441 | TNNC1 | Troponin C1, Slow Skeletal And Cardiac Type | 0.399 | 5.92E05 | 0.633 | 3.27E03 |
| ENSSSCG00000001791 | SAXO2 | Stabilizer Of Axonemal Microtubules 2 | 0.328 | 2.08E03 | 0.547 | 4.38E02 |
| ENSSSCG00000014191 | FER | FER Tyrosine Kinase | 0.331 | 1.75E04 | 0.426 | 4.07E04 |
| ENSSSCG00000015036 | DIXDC1 | DIX Domain Containing 1 | 0.352 | 3.48E07 | 0.405 | 2.57E06 |
| ENSSSCG00000009079 | INTU | Inturned Planar Cell Polarity Protein | 0.273 | 9.94E03 | 0.403 | 4.79E02 |
| ENSSSCG00000015531 | CEP350 | Centrosomal Protein 350 | 0.314 | 3.28E04 | 0.400 | 2.58E03 |
| ENSSSCG00000024357 | DNAI2 | Dynein Axonemal Intermediate Chain 2 | 0.276 | 7.42E03 | 0.395 | 4.04E02 |
| ENSSSCG00000002347 | DNAL1 | Dynein Axonemal Light Chain 1 | 0.285 | 2.87E03 | 0.378 | 8.99E03 |
| ENSSSCG00000004705 | MAP1A | Microtubule Associated Protein 1A | 0.271 | 7.62E03 | 0.376 | 2.25E02 |
| ENSSSCG00000001728 | CD2AP | CD2 Associated Protein | 0.330 | 5.69E08 | 0.366 | 8.97E05 |
| ENSSSCG00000000874 | GAS2L3 | Growth Arrest Specific 2 Like 3 | 0.270 | 5.65E03 | 0.361 | 1.70E02 |
| ENSSSCG00000010896 | ASPM | Abnormal Spindle Microtubule Assembly | 0.291 | 3.90E05 | 0.332 | 3.79E02 |
| ENSSSCG00000015329 | PPP1R9A | Protein Phosphatase 1 Regulatory Subunit 9A | 0.245 | 1.54E02 | 0.328 | 3.17E02 |
| ENSSSCG00000016542 | LRGUK | Leucine Rich Repeats And Guanylate Kinase Domain Containing | 0.230 | 2.63E02 | 0.309 | 5.49E02 |
| ENSSSCG00000016725 | TNS3 | Tensin 3 | 0.239 | 1.28E02 | 0.306 | 2.94E02 |
| ENSSSCG00000016608 | IQUB | IQ Motif And Ubiquitin Domain Containing | 0.248 | 2.68E03 | 0.295 | 1.01E02 |
| ENSSSCG00000002780 | TPPP3 | Tubulin Polymerization Promoting Protein Family Member 3 | 0.251 | 1.52E03 | 0.294 | 8.99E03 |
| ENSSSCG00000006394 | CFAP45 | Cilia And Flagella Associated Protein 45 | 0.251 | 1.56E03 | 0.294 | 6.49E03 |
| ENSSSCG00000013332 | KIF18A | Kinesin Family Member 18A | 0.227 | 2.00E02 | 0.290 | 3.88E02 |
| ENSSSCG00000021571 | KIF27 | Kinesin Family Member 27 | 0.246 | 1.78E03 | 0.287 | 9.15E03 |
| ENSSSCG00000020990 | DNAH12 | Dynein Axonemal Heavy Chain 12 | 0.235 | 6.82E03 | 0.283 | 1.75E02 |
| ENSSSCG00000000530 | FGD4 | FYVE, RhoGEF and PH domain containing 4 | 0.231 | 7.32E03 | 0.277 | 3.30E02 |
| ENSSSCG00000003931 | KIF2C | Kinesin Family Member 2C | 0.253 | 4.30E05 | 0.276 | 3.27E02 |
| ENSSSCG00000015523 | RALGPS2 | Ral GEF With PH Domain And SH3 Binding Motif 2 | 0.255 | 1.06E06 | 0.270 | 3.30E02 |
| ENSSSCG00000008768 | ARAP2 | ArfGAP With RhoGAP Domain, Ankyrin Repeat And PH Domain2 | 0.220 | 7.54E03 | 0.256 | 3.11E02 |
| ENSSSCG00000006936 | ODF2L | Outer Dense Fiber Of Sperm Tails 2 Like | 0.213 | 1.57E02 | 0.255 | 4.25E02 |
| ENSSSCG00000001869 | PEAK1 | Pseudopodium Enriched Atypical Kinase 1 | 0.210 | 1.58E02 | 0.250 | 3.89E02 |
| ENSSSCG00000011941 | IFT57 | Intraflagellar Transport 57 | 0.223 | 3.97E04 | 0.242 | 2.12E03 |
| ENSSSCG00000011745 | PRKCI | Protein Kinase C Iota | 0.225 | 5.70E05 | 0.239 | 8.27E02 |
| ENSSSCG00000002504 | AK7 | Adenylate Kinase 7 | 0.209 | 2.77E03 | 0.231 | 4.20E03 |
| ENSSSCG00000017884 | TEKT1 | Tektin 1 | 0.199 | 1.40E02 | 0.229 | 5.40E02 |
| ENSSSCG00000005952 | LRRC6 | Leucine Rich Repeat Containing 6 | 0.199 | 1.28E02 | 0.227 | 3.64E02 |
| ENSSSCG00000005078 | DAAM1 | Dishevelled Associated Activator Of Morphogenesis 1 | 0.206 | 1.97E03 | 0.225 | 8.79E03 |
| ENSSSCG00000009629 | BIN3 | Bridging Integrator 3 | 0.195 | 1.39E02 | 0.221 | 3.88E02 |
| ENSSSCG00000015379 | DNAH11 | Dynein Axonemal Heavy Chain 11 | 0.204 | 1.68E03 | 0.221 | 3.21E03 |
| ENSSSCG00000016658 | ANLN | Anillin Actin Binding Protein | 0.212 | 1.05E05 | 0.220 | 1.89E03 |
| ENSSSCG00000016794 | MYO10 | Myosin X | 0.210 | 6.68E05 | 0.220 | 3.93E03 |
| ENSSSCG00000004969 | KIF23 | Kinesin Family Member 23 | 0.205 | 6.08E05 | 0.214 | 4.68E02 |
| ENSSSCG00000003654 | MACF1 | Microtubule-Actin Crosslinking Factor 1 | 0.201 | 4.70E04 | 0.214 | 1.08E02 |
| ENSSSCG00000015570 | IVNS1ABP | Influenza Virus NS1A Binding Protein | 0.205 | 1.05E05 | 0.212 | 1.91E02 |
| ENSSSCG00000004289 | CEP162 | Centrosomal Protein 162 | 0.191 | 1.01E02 | 0.212 | 3.17E02 |
| ENSSSCG00000010457 | KIF20B | Kinesin Family Member 20B | 0.192 | 1.92E03 | 0.206 | 4.54E02 |
| ENSSSCG00000010351 | CCSER2 | Coiled-Coil Serine Rich Protein 2 | 0.201 | 2.27E06 | 0.206 | 2.70E03 |
| ENSSSCG00000007235 | TPX2 | TPX2, Microtubule Nucleation Factor | 0.189 | 3.76E04 | 0.198 | 4.85E02 |
| ENSSSCG00000017728 | MYO1D | Myosin ID | 0.184 | 5.05E05 | 0.189 | 2.22E02 |
| ENSSSCG00000010471 | KIF11 | Kinesin Family Member 11 | 0.179 | 5.69E04 | 0.186 | 2.09E02 |
| ENSSSCG00000016781 | TRIO | Trio Rho Guanine Nucleotide Exchange Factor | 0.174 | 1.51E03 | 0.182 | 2.87E02 |
| ENSSSCG00000009348 | STARD13 | StAR Related Lipid Transfer Domain Containing 13 | 0.174 | 4.69E04 | 0.180 | 2.09E02 |
| ENSSSCG00000009793 | CLIP1 | CAP-Gly Domain Containing Linker Protein 1 | 0.174 | 1.19E04 | 0.178 | 9.64E03 |
| ENSSSCG00000001085 | DCDC2 | Doublecortin Domain Containing 2 | 0.162 | 2.19E03 | 0.168 | 3.95E02 |
| ENSSSCG00000005235 | KANK1 | KN Motif And Ankyrin Repeat Domains 1 | 0.163 | 2.54E04 | 0.166 | 5.13E02 |
| ENSSSCG00000009125 | ANK2 | Ankyrin 2 | 0.159 | 4.57E03 | 0.166 | 4.54E02 |
| ENSSSCG00000010313 | VCL | Vinculin | 0.159 | 1.43E05 | 0.159 | 1.20E02 |
| ENSSSCG00000016061 | MYO1B | Myosin IB | 0.150 | 1.13E03 | 0.152 | 5.52E02 |
Detailed information for differential expression of genes (from both the DeSeq2 and EdgeR methods) related to cytoskeleton and ciliary functions.
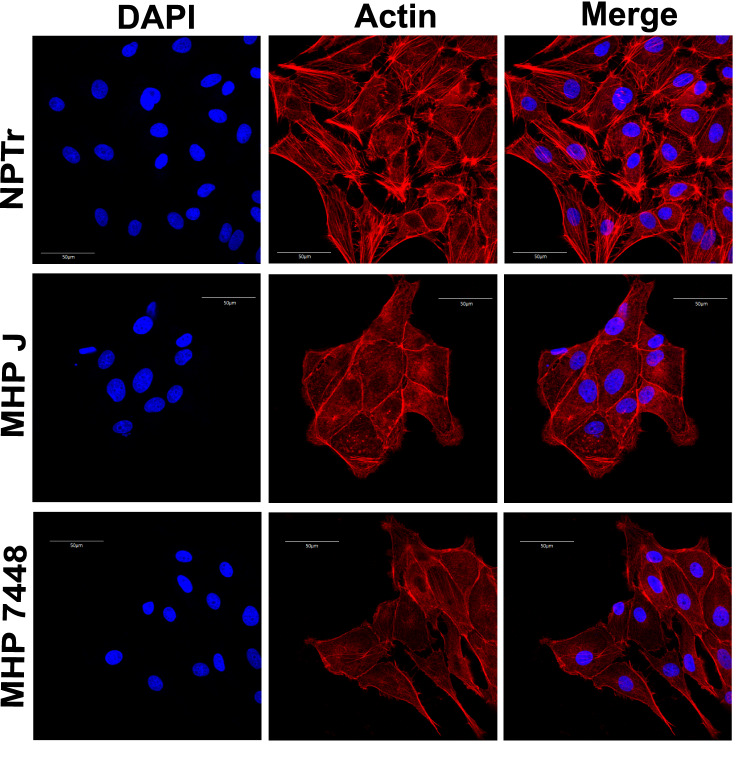
Besides ciliary genes, we also detected the down-regulation of cytoskeleton-related genes, both from microtubules and actin filaments, involved in the organization, rearrangement and stability of these structures (Table 4). It was previously described that the intracellular species M. penetrans is able to trigger reorganization of the host cell cystoskeleton, promoting aggregation of tubulin and -actinin and condensation of phosphorylated proteins57. To investigate if M. hyopneumoniae indeed affected the host cell cytoskeleton, we verified the organization of actin fibers in infected cells by confocal immunofluorescence microscopy. The actin stress fibers were notably less evident in the infected cells, as opposed to the control condition, in which they were abundant and more evenly distributed (Fig. 3). However, whenever present in the infected conditions, these stress fibers were either disorganized and/or at the periphery of the cells. These results corroborate studies from Raymond et al. (2018)58, which suggested that this species induces cytoskeletal rearrangements in the porcine respiratory tract. Although actin is not a major component of the ciliary axoneme, actin cytoskeleton has been implicated in every stage of ciliogenesis and many aspects of ciliary function59, directly associating these two down-regulated functions. In addition, it has recently been shown that M. hyopneumoniae expresses surface-accessible actin-binding proteins and that the host’s extracellular actin may act as a receptor for this bacterium in PK-15 epithelial cells60, indicating its importance for successful infection. Furthermore, the reduction of visible actin stress fibers caused by M. hyopneumoniae may also be related to the activation of NRF2, since the actin cytoskeleton is a scaffold necessary to maintain the transcription factor in the cytoplasm61.
Figure 3.
Organization of actin fibers in cells infected with M. hyopneumoniae. Results of the immunofluorescence microscopy analysis indicating the reduction and change in the pattern of actin stress fibers in infected cells. Eukaryotic cell actin was labeled with phalloidin (red) and nuclei were stained with DAPI (blue). Both attenuated (J) and virulent (7448) strains of M. hyopneumoniae altered the organization and abundance of actin fibers after 24 h of infection, however this effect can already be seen after 1 h of incubation (not shown). NPTr - uninfected cells. MHP J - NPTr cells infected with M. hyopneumoniae strain J. MHP 7448 - NPTr cells infected with M. hyopneumoniae strain 7448. Scale bars:
We also identified the down-regulation of cytoskeleton-related genes that play a role during cell division, such as BUB1B, CENP-I, NEK2 and SPAG5 (Supplementary Table S3). Many of these genes are involved with the mitotic spindle and chromosome segregation62–69. Genes encoding microtubule dependent motor proteins that physically affect chromosome segregation, such as kinesins (usually up-regulated during mitosis)70, as well as genes related to cell cycle progression were down-regulated during infection (Supplementary Table S3). These results suggest a repression of cell division of infected cells. The manipulation of cell cycle by pathogens has been extensively reported, with different pathogens being able to arrest different points of the cell cycle71–74. Therefore, it is possible that M. hyopneumoniae infection also interferes with the host cell cycle.
miRNA expression profiles
A total of 14 small RNA (sRNA) libraries were generated with the Illumina HiSeq2500 platform (Supplementary Table S7). A complete description of the samples is provided in Table 1 and Fig. S2. The raw reads were submitted to the NCBI Sequence Read Archive under accession number PRJNA545822. After removing adapters and filtering low quality reads, sRNA-seq yielded from 1 to 21 million single-end clean reads. Trimming and mapping information for each sRNA sample is given in Supplementary Table S7.
For intracellular sRNAs (INTRA: samples S1 to S6), we were able to map around 93% of the reads against the porcine genome. We also mapped all sRNA reads against the M. hyopneumoniae strain 7448 genome and samples had no more than 0.05% of unique mycoplasmal reads (Supplementary Table S7). All intracellular sRNA samples had similar sequencing depth and showed a typical miRNA distribution length curve ranging from 18 to 26 nt, with a peak at 22 nt (Supplementary Fig. S5A). We also performed a homology analysis of raw reads and we provide the percentage of intracellular clean reads clustered by classical types of sRNAs (Supplementary Fig. S5B), showing an enrichment of miRNA-homolog reads (around 40%).
As expected, total extracellular sRNA samples (EXTRA: S7 to S10) had more RNA degradation due to the presence of RNAses and degraded mRNAs in the extracellular environment. Extracellular sRNA samples of cells infected with the bacterium (S9 and S10) had up to 20% of reads mapped to M. hyopneumoniae, as they were presumed to have more remnants of mycoplasmal cells (Supplementary Table S7). However, the great majority of reads mapping to the bacteria were mainly product of mRNA degradation, as they did not have any specific signature for sRNAs when compared to previous results from Siqueira et al. (2016)75. In this way, we filtered out (i) sequences nt and (ii) sequences mapped to the M. hyopneumoniae genome. Sample S8 (EXTRA) had a sequencing depth smaller than its replicate (S7); however, the distribution of counts was overall similar between replicates and we included it in the further analyses.
Extracellular exosomal sRNA samples (EXO: S11 and S12) had some RNA degradation ( nt), but maintained a pronounced peak at 22 nt. Extracellular sRNAs from vesicle-free supernatant (SN: samples S13 and S14) were more problematic, probably due to (i) too many pre-processing steps and (ii) the presence of RNAses in the extracellular environment. Sample S14 did not yield a minimum amount of reads necessary for the subsequent steps and this condition (SN) was not used for further analyses.
Annotation of known and novel miRNAs
In order to perform miRNA prediction with mirDeep276, we only took into account reads from intracellular samples. We predicted a total of 1,041 miRNAs, which were further clustered into 773 groups. From these, 478 were completely novel (Supplementary Table S8). We created a Sus scrofa miRNA database (ssc-miRNA-DB) with 1,906 different entries from three different sources: 411 known miRNAs in Sus scrofa from miRBase (release 21)77, 722 annotated by Martini et al., (2014)78 and the 773 clusters of mature miRNAs predicted by our analysis. More than 50% of intracellular sRNA clean reads were aligned against the ssc-miRNA-DB (Supplementary Table S7). All other samples were also mapped against this database. The complete pipeline used in this study for miRNA prediction is described in Supplementary Fig. S6.
miRNA differential expression
In all sRNA samples, we detected (with at least 10 counts across all libraries) 290 from the 411 miRNAs from miRBase and 214 of the miRNAs described by Martini et al., (2014)78. Also, 263 predicted miRNAs had more than 10 counts across all libraries. We only took into account the 491 miRNAs that had more than 50 counts across all libraries for differential expression analysis (Supplementary Table S9).
In total, we identified 170 DE miRNAs (121 up-regulated, 48 down-regulated and 1 ambiguous). Table 5 shows 10 selected up-regulated and down-regulated miRNAs detected in this study, and the complete list of DE miRNAs is summarized in Supplementary Table S10. Several homologs of these miRNAs have already been linked to bacterial infection response or immune system response in the literature and these references are listed in Table 5. With the exception of one ambiguous case (ssc-miR-9842-5p), whenever a miRNA was detected as DE in more than one condition (INTRA, EXTRA or EXO), the change in expression (either up- or down-regulated) was in accordance between them (Supplementary Table S10).
Table 5.
Selected DE miRNAs. Information about selected up- and down-regulated miRNAs in intracellular, extracellular and exosome samples.
| miRNA ID | DB Type | Intracellular | Extracellular | Exosomes | Play a role in other bacterial infection | |||
|---|---|---|---|---|---|---|---|---|
| DE | LogFC | DE | LogFC | DE | GFOLD | |||
| Novel-Chr13-miR-10 | novel | Upregulated | 4.121 | Upregulated | 9.420 | Upregulated | 3.659 | |
| Novel-Chr4-miR-57 | novel | Upregulated | 1.218 | Upregulated | 12.164 | Upregulated | 11.582 | |
| Novel-Chr9-miR-16 | novel | Upregulated | 0.768 | Upregulated | 13.079 | Upregulated | 7.947 | |
| Novel-Chr9-dna-26 | novel | Upregulated | 1.448 | Upregulated | 11.270 | Upregulated | 3.592 | Pseudomonas aeruginosa123 |
| mmu-mir-2143-2_6_251367_251451_-_3p_-5-424-HC | Martini et al., 2014 | Upregulated | 2.053 | Upregulated | 5.899 | Upregulated | 2.187 | |
| ssc-miR-1285 | mirBase | NS | Upregulated | 4.965 | NS | Chlamydia trachomatis124, 125 | ||
| ssc-miR-196b-5p | mirBase | NS | Upregulated | 3.971 | NS | Mycobacterium avium126 | ||
| ssc-miR-212 | mirBase | NS | Upregulated | 8.723 | NS | Mycobacterium tuberculosis127 | ||
| ssc-miR-24-1-5p | mirBase | NS | Upregulated | 9.742 | NS | Escherichia coli and Staphylococcus aureus128 | ||
| ssc-miR-146a-5p | mirBase | Upregulated | 1.015 | ND | ND |
LPS stimulation132 |
||
| ssc-miR-9842-5p | mirBase | NS | Upregulated | 4.484 | Downregulated | 2.497 | ||
| ssc-miR-184 | mirBase | Downregulated | 0.594 | ND | NS | Chlamydia trachomatis133 | ||
| ssc-miR-140-3p | mirBase | NS | NS | Downregulated | 3.536 | Mycobacterium tuberculosis134 | ||
| ssc-miR-769-3p | mirBase | NS | Downregulated | 3.924 | NS | |||
| ssc-miR-101 | mirBase | NS | Downregulated | 4.154 | Downregulated | 2.986 | Helicobacter pylori135 | |
| ssc-miR-107 | mirBase | NS | Downregulated | 2.515 | Downregulated | 2.052 | Gut microbiota136 | |
| ssc-miR-31 | mirBase | NS | Downregulated | 1.167 | Downregulated | 2.004 |
Helicobacter pylori135 T-cell activation137 |
|
| ssc-miR-532-5p | mirBase | NS | Downregulated | 3.343 | Downregulated | 3.916 | LPS stimulation132 | |
| antisense-pn8/bta-mir-2320_6_43886629_43886722_-_NA-424-HC | Martini et al., 2014 | NS | Downregulated | 3.150 | Downregulated | 4.440 | Actinobacillus pleuropneumoniae138 | |
|
antisense-ssc-mir-320a*_14_6473893_6473973_-_NA;antisense- sscmir-320a*_14_6473894_6473960_-_NA-424-HC |
Martini et al., 2014 | NS | Downregulated | 1.907 | Downregulated | 2.933 | Helicobacter pylori139 | |
| ssc-mir-107-shorter/ssc-isomir-107_14_106321702_106321788_-_ NA-424-HC | Martini et al., 2014 | NS | Downregulated | 2.582 | Downregulated | 2.206 | Gut microbiota136 | |
Several homologues of these miRNAs were already described to be involved with bacterial infection in other species, and references are provided whenever we found a correlation in the same direction of expression as in this work. The only miRNA that showed ambiguous expression among conditions was ssc-miR-9842-5p, which had inverse expression between the extracellular (up-regulated) and exosome (down-regulated) samples. NS: Not significant; ND: Not detected.
: Whenever LogFC from DESeq2 was not available, we provide LogFC from EdgeR;
: Reference related to cel-miR-233-5p, a possible homolog of novel miRNA 9-dna-26;
: A. pleuropneumoniae was also related to a swine cell infection.
Targets of DE miRNAs were enriched in genes related to redox homeostasis, translation and cytoskeleton
In order to better understand the biological functions that could be involved with the 170 DE miRNAs, we performed different analyses to predict their potential targets. In this sense, all DE miRNAs had at least one DE gene as a predicted target. The complete pipeline used for target prediction in this study is provided in Supplementary Fig. S7.
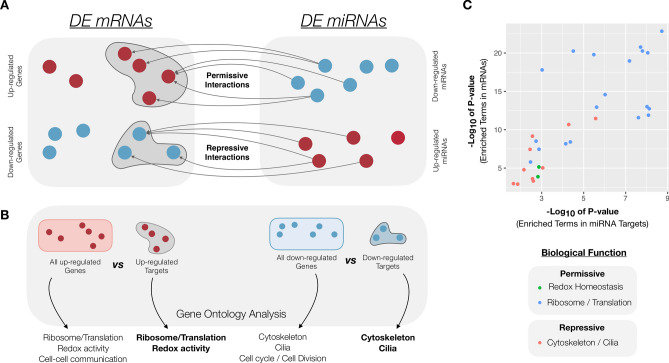
We predicted a total of 79,276 interaction pairs between miRNAs and mRNAs. In this way, based only on the predictions of the software used here, a miRNA could potentially target, on average, 465 genes in the entire porcine genome. However, we only considered as targets the mRNAs that were detected as DE in this study, which significantly decreased our list to a total of 4,287 interaction pairs. We chose to focus on anticorrelations between miRNA and mRNA expression (and kept 1,939 interaction pairs), as the main mode of action of miRNAs is a destabilization of mRNAs79 and the fact that most of experimental validations in the literature are related to interaction pairs with inversed regulation. In this context, a permissive interaction is generally described as one that occurs between a down-regulated miRNA and an up-regulated mRNA, while a repressive interaction is one in which the miRNA is up-regulated with consequent down-regulation of the target mRNA78 (Fig. 4A). In our results, permissive interactions represented 267 genes and 50 miRNAs, while repressive interactions occurred between 425 genes and 121 miRNAs, accounting for 598 permissive and 1341 repressive interactions between DE mRNAs and miRNAs. Supplementary Table S11 summarizes the permissive and repressive target pairs predicted in this study.
Figure 4.
Correlation of GO terms between DE mRNAs and targets of DE miRNAs. A A permissive interaction occurs between a down-regulated miRNA (depicted in blue) and an up-regulated mRNA (depicted in red), whereas a repressive interaction is one in which the miRNA is up-regulated (red) and its target mRNA is down-regulated (blue). B. We performed GO enrichment analyses with the complete up- and down-regulated lists of mRNAs and also with the subset of miRNA targets among each of these lists. C. A correlation of some of the GO terms from DE mRNAs and targets of DE miRNAs was detected, indicating that these functions might also be regulated by miRNAs.
We performed GO analyses to compare the DE genes and targets of DE miRNAs in order to investigate whether their functions could be correlated (Fig. 4B and Supplementary Fig. S8). Interestingly, we were able to identify a correlation between the enriched terms in miRNA targets with some of the GO terms detected in the mRNA up-regulated or down-regulated GO results (Fig. 4C). Target genes from permissive interactions were associated with ribosome/translation and oxidation-reduction activity, whereas target genes from the repressive interactions had enriched terms related to cytoskeleton and ciliary function (Fig. 4C). It is important to highlight the relevance of the miRNAs found in exosome-like vesicles and in extracellular samples in the identification of GO enriched terms of the miRNA targets, since only a small part of the permissive and repressive interactions involved intracellular miRNAs. Complete GO enrichemnt of the miRNA targets is found in Supplementary Table S12.
Regulatory network reconstruction and analysis
In order to gain a broader view of the host’s response to the presence of M. hyopneumoniae, we built a general regulatory network by integrating mRNA gene expression with predicted miRNA-mRNA interactions collected and analyzed in this work (Supplementary Fig. S9). We also included information about physical and genetic validated interactions from the BioGRID v3.4 database80, 81. As previously mentioned, at this point we only took into account interaction pairs that were either repressive or permissive. The global interaction network was composed by 774 nodes and 1965 arcs and our objective was to identify miRNA-mRNA expression patterns. We performed a functional analysis with the use of CLueGO82 and we also detected within the permissive interactions the enrichment of several processes related to immune response and inflammation (Supplementary Fig. S10).
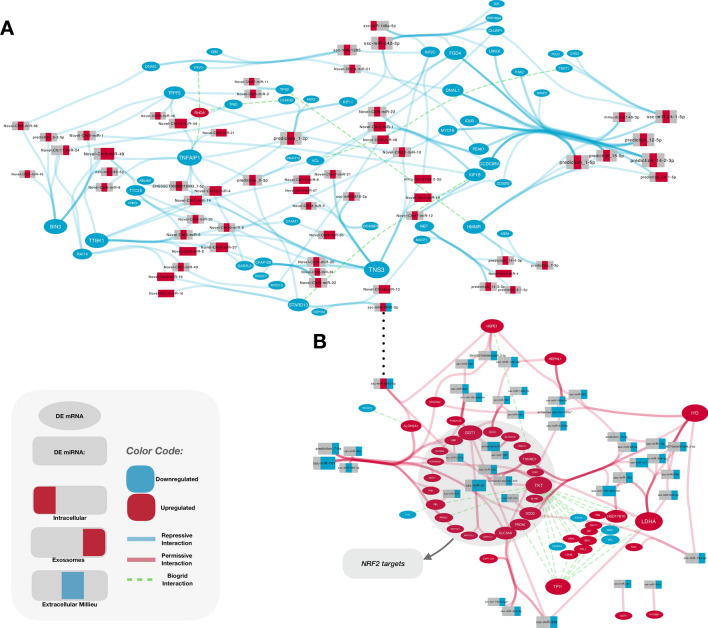
Furthermore, we created two separate regulatory networks, one related to cytoskeleton and cilia (repressive interactions, Fig. 5A) and one related to redox homeostasis (permissive interactions, Fig. 5B) to better refine and understand the possible co-regulation of several of these targets. The networks show a high level of connectivity and we were able to detect miRNAs that interacted only with NRF2 activated targets (such as ssc-miR-31) (Table 6). Furthermore, we were able to detect miRNAs that seemed to regulate more generally redox homeostasis genes and also miRNAs whose targets came from sets of genes with distinct functions (such as glycolysis, immune system defense, ribosomes, among others), indicating that this response might be related to several other important functions within the cell. We believe that this network can be a powerful tool for analyzing the influence of M. hyopneumoniae on the host gene expression in future studies.
Figure 5.
Interaction networks of DE miRNAs and genes involved with cytoskeleton/cilia and redox homeostasis. Rectangles depict miRNAs and ellipses represent genes. Blue ellipses/rectangles indicate that the gene/miRNA was down-regulated and red indicate gene/miRNAs detected as up-regulated. BioGrid interactions are seen as green dashed arcs. These represent both physical and genetic interactions between either genes or proteins from such genes. A. Cytoskeleton/cilia interaction network. We detected many repressive interactions involving genes related to cytoskeleton and ciliary function (blue arcs). B. Redox homeostasis interaction network. Permissive interactions of genes related to redox homeostasis are shown as red arcs. The great majority of the miRNAs targeting these genes are DE only in exosomes (rectangles partially colored in blue, far right). NRF2 targets: genes described in other species to be activated by NRF2 are highlighted in grey.
Table 6.
DE genes putatively activated by NRF2 predicted to be targets of DE miRNAs. Expression of miRNAs is reported for intracellular, extracellular and exosome-like vesicles. NS: Not significant; ND: Not detected.
| Upregulated gene ID | Downregulated miRNA | miRNA expression | ||
|---|---|---|---|---|
| Intracellular | Extracellular | Exosome | ||
| AKR1C1 | ssc-miR-101 | NS | Downregulated | Downregulated |
| AKR1C4 | ssc-miR-31 | NS | Downregulated | Downregulated |
| AKR1CL1 | ssc-miR-31 | NS | Downregulated | Downregulated |
| BLVRB | prediction_13_40037950_40038026_-_3p-353-MC | NS | ND | Downregulated |
| EGR1 | antisense-ssc-mir-423_12_44150500_44150579_-_NA-353-MC | NS | NS | Downregulated |
| GCLC | ssc-miR-148b-3p | NS | NS | Downregulated |
| ssc-miR-19a | NS | NS | Downregulated | |
| ssc-miR-19b | NS | NS | Downregulated | |
| GGT1 | ssc-miR-338 | NS | ND | Downregulated |
| ssc-miR-140-3p | NS | NS | Downregulated | |
|
ssc-mir-92-shorter/ssc-isomir-92_X_108178486_108178565_-_NA;ssc-mir- 92-shorter/ssc-isomir-92_X_108212408_108212487_-_NA-424-HC |
NS | NS | Downregulated | |
| ssc-miR-92a | NS | NS | Downregulated | |
| prediction_7_91732076_91732132_+_5p-353-MC | NS | NS | Downregulated | |
| ssc-miR-31 | NS | Downregulated | Downregulated | |
| antisense-pn8/bta-mir-2320_6_43886629_43886722_-_NA-424-HC | NS | Downregulated | Downregulated | |
| GSR | ssc-miR-101 | NS | Downregulated | Downregulated |
| ssc-miR-22-3p | NS | NS | Downregulated | |
| HMOX1 | ssc-miR-182 | NS | NS | Downregulated |
| HSP90AB1 | ssc-miR-101 | NS | Downregulated | Downregulated |
| NQO1 | ssc-miR-34a | NS | NS | Downregulated |
| PGD | ssc-miR-769-3p | NS | Downregulated | NS |
| PIR | ssc-miR-378 | NS | NS | Downregulated |
| PRDX1 | ssc-miR-101 | NS | Downregulated | Downregulated |
| PRDX6 | ssc-miR-10a-5p | NS | NS | Downregulated |
| ssc-miR-10b | NS | NS | Downregulated | |
| ssc-miR-338 | NS | ND | Downregulated | |
| SLC5A10 | ssc-miR-1307 | NS | ND | Downregulated |
| antisense-pn8/bta-mir-2320_6_43886629_43886722_-_NA-424-HC | NS | Downregulated | Downregulated | |
| SLC5A6 | ssc-miR-10b | NS | NS | Downregulated |
| ssc-miR-338 | NS | ND | Downregulated | |
| ssc-miR-423-3p | NS | ND | Downregulated | |
| ssc-miR-27b-3p | NS | NS | Downregulated | |
| prediction_7_91732076_91732132_+_5p-353-MC | NS | NS | Downregulated | |
| ssc-miR-340 | NS | ND | Downregulated | |
| SOD2 | prediction_7_91732076_91732132_+_5p-353-MC | NS | NS | Downregulated |
| ssc-miR-184 | Downregulated | ND | ND | |
| ssc-miR-28-3p | NS | NS | Downregulated | |
| TKT | antisense-ssc-mir-423_12_44150500_44150579_-_NA-353-MC | NS | NS | Downregulated |
| TXNRD1 | ssc-miR-103 | NS | NS | Downregulated |
| ssc-miR-182 | NS | NS | Downregulated | |
| ssc-mir-107-shorter/ssc-isomir-107_14_106321702_106321788_-_NA-424-HC | NS | Downregulated | Downregulated | |
| ssc-miR-107 | NS | Downregulated | Downregulated | |
| ssc-miR-31 | NS | Downregulated | Downregulated | |
| UGT1A6 | ssc-miR-28-3p | NS | NS | Downregulated |
| antisense-ssc-mir-103_16_52667657_52667738_+_NA-424-HC | NS | NS | Downregulated | |
| ssc-miR-769-3p | NS | Downregulated | NS | |
Permissive interactions were enriched in genes regulated by NRF2
Analyzing more in detail the permissive interactions between mRNAs and miRNAs, we identified several miRNAs targeting NRF2 regulated genes (Table 6). GGT1, for instance, is predicted to be targeted by 7 different miRNAs, while ssc-mir-31 is predicted to target 4 different genes induced by NRF2 (AKR1C4, AKR1CL1, GGT1, and TXNRD1).
It was previously reported that NRF2 can be regulated independently of KEAP1 by miRNAs in human breast cancer cells83. Our miRNA target analysis predicted NRF2 as target of three miRNAs down-regulated in exosome-like vesicles: ssc-mir-340, ssc-mir-19a, and ssc-mir-19b. The first was also predicted to target the transporter gene SLC6A6, while ssc-mir-19a and ssc-mir-19b were predicted to target GCLC, necessary for glutathione synthesis. In addition, miR-340 has been previously identified negatively regulating NRF2 expression in human84. Conversely, miR-19a and miR-19b have been reported to be down-regulated in the presence of hydrogen peroxide in rats85, 86, linking these miRNAs to a response to oxidative stress.
In the literature, other miRNAs were also shown to negatively regulate the expression of NRF2: miR-2883, miR-10187, miR-92a88, miR-27b89, and miR-34a90. All of them were down-regulated in our data and were predicted by this study to regulate NRF2 activated genes (Table 6). In this way, it seems that during M. hyopneumoniae infection, there is a global change in gene expression in an attempt to activate antioxidant genes, in association with the down-regulation of miRNAs that negatively regulate these genes.
Repressive interactions were related to cytoskeleton and cilia
As concerns the repressive interactions of miRNAs and mRNAs, we identified within the targets of up-regulated miRNAs an enrichment of genes related to cytoskeleton and cilia (Supplementary Table S11). For instance, the gene TNS3, related to cytoskeleton, is predicted as target of 16 up-regulated miRNAs (Supplementary Table S11). In addition, several genes encoding for dyneins were predicted as targets of up-regulated miRNAs, with DNAL1 being target of 7 different miRNAs. Mutations and down-regulation of genes coding ciliary proteins, especially dyneins, were related to primary ciliary dyskinesia46, 48, 53, 55, 91, but had not yet been reported in M. hyopneumoniae infection. More investigations on this matter should be carried out, since these results seem to be related to one of the main adverse effects caused by this bacterium: ciliostasis.
Most DE miRNAs were detected in exosome-like vesicles
Analyzing the up-regulated genes, we identified that besides antioxidant response and protein synthesis, there is an enrichment in GO terms related to cell-cell communication (Fig. 1). The up-regulation of cell-cell communication is noteworthy considering that most DE miRNAs were detected only in the extracellular and exosome-like samples (Supplementary Table S10). Moreover, most of the down-regulated miRNAs of exosome-like vesicles were DE only in these vesicles, indicating that the cells might be selecting specific populations of miRNAs to be packaged into these structures, which could be seen as a specific message they are trying to communicate to other cells. In the same direction, the increase in the number of proteins secreted in vesicles was reported in NPTr cells infected with M. hyopneumoniae92, indicating that the communication via exosome-like vesicles is important for swine cells during infection. This corroborates our data, suggesting that M. hyopneumoniae induces modification on the protein and RNA composition of NPTr-released vesicles and is likely important in the cross-talks between epithelial cells.
The relationship between cell–cell communication and DE miRNAs released by infected cells becomes especially interesting when considering genes related to the NRF2 pathway: we identified DE miRNAs predicted to regulate such genes only in extracellular and exosome-like vesicle samples, with no difference of expression in intracellular samples (Table 6). This might suggest the existence of a mechanism in which infected cells send signals to neighboring cells in order to prevent the repression of these genes or degradation of their RNA products. This becomes even more important due to the fact that exosomal miRNAs were already shown to regulate the inflammatory response in receptor cells from mice93. While miRNAs identified in the extracellular total samples might contain sRNAs that reflect degradation remnants due to the presence of RNAses and mRNAs in the extracellular environment, we believe that this is less likely to happen in exosomes, since these vesicles have a membrane that protects their content from extracellular degradation. As exosomes have an important role in cell communication, we should consider the difference in miRNA expression in these vesicles as relevant and speculate on how they might interfere in the gene regulation of neighboring cells. In addition, as exosomes may also affect other cell types, they may contain miRNAs that target genes that were not detected as DE in our samples.
Concluding remarks
M. hyopneumoniae is considered a pathogen with huge negative impact on swine production. However, apart from studies related to adherence to the host cells, little is known about the relationship between this pathogen and the swine host. In this work, we analyzed the changes that M. hyopneumoniae induced in gene and miRNA expression in tracheal epithelial cells. As far as we know, this is the first study to establish a link between gene expression of the swine cells and the most deleterious pathogenic effects of M. hyopneumoniae, namely its cytotoxic epithelial damage (possibly via hydrogen peroxide production) and induced ciliostasis. However, we are aware that infection with this species involves a large component of the immune system, which is even said to be responsible for the major tissue damage related to the infection. Thus, the results found here reflect the effects of M. hyopneumoniae on epithelial cells, while the general picture of the respiratory tract might present different responses to the infection. In this way, it is important to observe that besides the hydrogen peroxide production by this bacterium, other factors can trigger the activation of the cell antioxidant defense during infection. M. hyopneumoniae infection is characterized by the infiltration of a large number of leukocytes in the lung tissue, which produce reactive oxygen and nitrogen species, causing tissue damage38, 94. Accordingly, systemic infections with M. hyopneumoniae have a great potential of causing oxidative stress, so that the transcription of genes activated by NRF2 seems to be important to fight the infection and maintain cellular homeostasis.
In conclusion, we conducted the first study that analyzes mRNA and miRNA differential expression by NGS in epithelial tracheal cells infected with M. hyopneumoniae. Our results bring new insights into the interaction between this bacterium and swine epithelial cells, notably the host cellular response through the activation of genes related to antioxidant response and the repression of cytoskeleton and ciliary genes (possibly related to ciliostasis), and open several perspectives related to the understanding of the pathogenicity of this bacterial species.
Methods
Cultivation of NPTr cells
NPTr cells95 negatively tested for mycoplasma, purchased from Instituto Zooprofilattico Sperimentale della Lombardia e dell’Emilia Romagna, Brescia, Italy, were grown in Minimum Essential Medium with Earle’s Balanced Salts (MEM/EBSS) and 2 mM L-Glutamine (GE Healthcare) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) (Gibco) and 1% penicillin streptomycin (GE Healthcare). Cells were cultured at with 5% CO2 in a humid atmosphere in T75 flasks and 6-well tissue culture plates. Sub-passages were made when cells reached 70-80% confluence. To isolate exosome-like vesicles, the culture medium was removed, confluent cells were washed with phosphate-buffered saline (PBS) and kept in medium without FBS and antibiotics (exosome depleted) for 24 h.
Infection of NPTr cells with M. hyopneumoniae
Mycoplasma hyopneumoniae strain J (ATCC25934) was cultivated in Friis medium96 at for 48 h with gentle agitation in a roller drum. Prior to NPTr infection, Mycoplasma cells were pelleted, washed with PBS and resuspended in MEM/EBSS medium. Based on the methodology described by Assunção et al. (2005)97, we calculated an MOI of 5 M. hyopneumoniae for the immunofluorescence experiments and an MOI of 2 for the RNA extraction experiments. NPTr cells were challenged with M. hyopneumoniae for 24 h.
Immunofluorescence microscopy
NPTr cells were grown in cover slips coated with poly-L-lysine (Sigma-Aldrich) for 48 h and infected (or not, in the control conditions) with M. hyopneumoniae for 24 h or 1 h in the case of the NFR2 localization experiment. The cells were washed with 1x PBS three times and fixed with 4% paraformaldehyde for 15 min. After permeabilization with 0.2% Triton X-100 for 10 min and blocking with 4% BSA in PBS for 20 min, the cells were incubated with the primary antibodies (rabbit polyclonal anti-SPAse I of M. hyopneumoniae98, 1:10,000, and mouse monoclonal anti-Sodium/Potassium ATPase alpha (Invitrogen), 1:20, or rabbit polyclonal anti-NRF2 (Thermo Fisher Scientific), 1:200,) overnight at followed by secondary antibodies (Alexa Fluor488-conjugated anti-rabbit (Invitrogen), 1:1,000, and Alexa Fluor555-conjugated anti-mouse (Invitrogen), 1:1,000) incubation for 1 h in the dark. The cells were washed with PBS and the DNA was stained with 100 nM DAPI and actin was marked with 50 nM phalloidin for 20 min. The slides were mounted with Fluoromount (Sigma-Aldrich). Samples were imaged using an Olympus FluoView 1000 confocal microscope. NRF2 fluorescence was quantified by drawing margin to each individual nucleus and measuring the green fluorescence against the cell background. At least 25 nuclei were quantified in each condition with the ImageJ software99. The Corrected Total Fluorescence (CTCF) of each nucleus was calculated with the following formula: CTCF = Integrated density – (Area of the selected nucleus x fluorescence of the background reading). Statistical significance was analyzed by One-way ANOVA followed by Dunnett’s multiple comparison test () in GraphPad Prism 7.0 software.
Exosome extraction
Exosome-like vesicles were purified from 250 mL (25 F75 flasks) of NPTr conditioned medium (as described by Forterre et al.17). Briefly, after 24 hours of incubation, the medium was recovered and cell debris and organelles were eliminated by centrifugation at 2,000 x g for 20 min and at 10,000 x g for 30 min. The resulting supernatant was filtered through a filter in order to remove large particles. Exosome-like vesicles were pelleted by ultracentrifugation at 100,000 x g for 90 min (Beckman-Coulter, Optima MAX-XP ultracentrifuge, MLA-55 rotor). The supernatant was recovered and saved for posterior use (SN) and the exosome sediment was washed with 25 mL of cold PBS, centrifuged at 100,000 x g for 70 min and resuspended in PBS. Vesicle extractions were performed in duplicates in both conditions of NPTr cells non-infected and infected with M. hyopneumoniae. SN portion was submitted to nucleic acid precipitation with 1/10 volume of 3 M sodium acetate and 3 volumes of 100% cold ethanol at overnight. After centrifugation, the nucleic acid precipitated was allowed to dry at for 1 hour and was ressuspended in of ultrapure RNAse free water.
Sample preparation, RNA extraction and sequencing
For mRNA sequencing, a total of 6 samples were prepared: 3 in a control group (CTL) and 3 in the infected group (INF). For miRNA sequencing, we prepared the samples as follows: total intracellular miRNA (INTRA: 3 CTL vs 3 INF), total extracellular small RNA (EXTRA: 2 CTL vs 2 INF), extracellular exosomal miRNA (EXO: 1 pool CTL vs 1 pool INF) and extracellular miRNA from vesicle-free supernatant (SN: 1 pool CTL vs 1 pool INF: a single library was constructed from a pool of 50 biological replicates). Total RNA for mRNA sequencing and sRNA enriched (bp) for sRNA sequencing and miRNA analyses were extracted with mirVana kit (Ambion), according to the manufacturer’s instructions. Total extracellular RNA for sRNA sequencing was directly extracted from the culture NPTr cell supernatant after centrifugation of cell debris. Exosome sRNA was extracted after vesicle purification. RNA quality was assessed with Bioanalyzer 2100 (Agilent Genomics). A total of 6 mRNA libraries were prepared (one for each sample) using TruSeq Stranded Total RNA Sample Preparation kit (Illumina) and a total of 14 small RNA libraries were prepared using TruSeq Small RNA Sample Prep kit (Illumina). After quality control, the sequencing of all libraries was performed by HiSeq2500 platform (Illumina). RNA library preparation and sequencing was performed by the Duke University GCB Sequencing Platform (Durham, USA).
RT-qPCR of selected differentially expressed mRNAs
For RT-qPCR, total RNA of NPTr cells infected and non-infected with M. hyopneumoniae was isolated with TRizol (Invitrogen) according to the manufacturer’s instructions. A first-strand cDNA synthesis reaction was conducted by adding of total RNA to 500 ng of oligo(dT)15 primer (Promega) and 10 mM deoxynucleotide triphosphates. The mixture was heated for for 5 min and then incubated on ice for 5 min. First-strand buffer (Invitrogen), 0.1 M dithiothreitol and 200 U M-MLV RT (Moloney Murine Leukemia Virus Reverse Transcriptase – Invitrogen) were then added to a total volume of . The reaction was incubated at for 10 min and at for 50 min followed by 15 min at for enzyme inactivation. Quantitative PCR (qPCR) assay was performed using 1:5 cDNA as template and Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen) with specific primers (Supplementary Table S13) on 7500 Real-Time PCR Systems (Applied Biosystems). The qPCR reactions were carried out at for 2 min and for 10 min followed by 40 cycles of for 15 s and for 1 min. A melting curve analysis was done to verify the specificity of the synthesized products and the amplification efficiency for each primer was calculated with the LinRegPCR software application100. The mRNA levels were normalized against porcine DNA topoisomerase II beta (TOP2B) and the relative expression of mRNA was calculated by method. The threshold cycle (CT) of each target test represents the average of three reactions. Three independent biological replicates were done for each target gene. Statistical analyses were performed using the GraphPad Prism 7.0 software with a two-tailed unpaired t-test to compare the relative expression between infected and non-infected NPTr cells ().
Computational analysis of sequencing data
Preprocessing of raw reads
The reads from sRNA-seq and total RNA-seq were processed separately. The raw reads were filtered for low quality, and adapter sequences were trimmed with CUTADAPT101. Total mRNA-seq clean reads were mapped against the porcine reference genome from Ensembl (Sscrofa10.2) with STAR102 and the raw counts of genes were obtained with HTSeq103. Only uniquely mapped reads were used for further analyses.
For miRNA prediction, clean sRNA-seq reads were mapped against the porcine genome with Bowtie104. We used intracellular samples as input for miRDeep276 and kept predictions that had a score of at least 5. After this, we collapsed similar predictions and obtained a total of 773 clusters (773 miRNAs), of which 478 were novel miRNAs. We created a porcine miRNA DB (ssc-miRNA-DB), by including the 411 annotated porcine miRNAs in version 21 of miRBase77, 105 along with the 722 miRNAs characterized by Martini et al. (2014)78 and our novel predictions. Reads from all samples were mapped against this database and a matrix of counts was generated in order to identify DE miRNAs.
Differential expression of mRNA data
Raw counts were used as input for DE gene analysis. We detected possible DE genes in both EdgeR106 and DESeq2107 packages in R. DESeq2 was run for genes that had a total count of at least 10 in all libraries, with the method’s default normalization. EdgeR was used with TMM normalization and general linear model fit, only for genes with cpm greater than 1 in at least 2 libraries. After testing, the adjusted p-values (p-adj) for both methods were adjusted with the Benjamini-Hochberg108 correction for multitesting. Genes were considered DE when p-adj and genes with the most pronounced logFC were selected individually for further investigation. Overall, DESeq2 detected more significant p-adj with less accentuated LogFC, while EdgeR detected less significant false discovery rates (FDR) with more extreme LogFCs. Both techniques have been widely used separately and together in several publications, and in order to select good candidates for testing, we took into account the results of both methods. For GO functional analyses, we also used the complete list of DE genes whenever LogFC was greater than 0.1 (up-regulated) or smaller than (down-regulated). The complete lists from each method were used separately, and we compared the overall outcomes to check the robustness of our results.
Differential expression of miRNA data
The same pipeline used for DE mRNA was performed for total intracellular and total extracellular miRNAs, since we had biological replicates in both cases. In the case where we had no replicates (instead, a single library was constructed from a pool of 50 biological replicates), we used GFOLD109 which provides a generalized fold change for ranking DE genes. GFOLD is said to overcome the shortcomings of p-value and fold change of the existing methods and can provide a more stable and biological meaningful gene ranking when a single biological replicate is available. In this case, we selected miRNAs with GFOLD or for functional analyses.
miRNA target prediction
DE miRNAs were used as input to detect putative interactions with the UTRs of Ensembl transcripts in the porcine genome. We used three methods to detect target pairs: miRanda110, TargetScan111 and PITA112, and one method to validate the hybridization of a target pair, RNAhybrid113. We kept only targets that were predicted by at least two distinct tools and used the following thresholds: score in miRanda , DDG from PITA , score in RNAhybrid . Since TargetScan does not provide a continuous scoring system, we only validated whenever there was a prediction of at least 6mers. Based on these, we chose from the target list only genes that were detected as DE in this study, and subsequentially we only considered target pairs of miRNA-mRNA that had inversed LogFC expression.
Functional analysis of DE mRNAs and targets of DE miRNAs
Functional analysis took into account as input either the list of DE mRNAs itself or the list of targets predicted for the DE miRNAs. We performed a GO enrichment analysis114 to find out which functions were over or underrepresented in each gene list. P-values for enriched GO terms were adjusted with the Benjamini-Hochberg108 correction for multitesting. GO terms and pathways with p-adj were defined as significantly enriched. The GO terms were reduced to representative non redundant terms with the use of the REVIGO tool115.
Regulatory network reconstruction and analysis
We created a general regulatory network of the host response to the bacterial infection with the DE miRNAs and target mRNAs detected in this study (Supplementary Fig. S9 and Supplementary File S1). In this network, we also included information about interactions from the BioGRID v3.4 database80, 81, a general repository that includes experimentally validated physical and genetic interactions. We used human-based official gene symbols to include information from BioGRID, Cytoscape116 to draw the networks, and the ClueGO plugin82 to perform functional enrichment analysis. We further manually curated the networks for genes and miRNAs related with redox homeostasis (permissive regulatory network), as well as cytoskeleton and cilia (repressive regulatory network).
Supplementary Information
Acknowledgements
We thank Henrique B. Ferreira, Lucas Moitinho-Silva and Jéssica A. Paes for the anti-SPAse I antibody and the Centro de Microscopia e Microanálise (CMM-UFRGS) for technical support with confocal microscopy. This work was performed using the computing facilities of NLHPC (ECM-02) in Chile, the CC/PRABI/LBBE in France and the France Génomique e-infrastructure (ANR-10-INBS-09-08). This work was supported by grants from CAPES-COFECUB 782/13 and the European Research Council under the European Community’s Seventh Framework Programme (FP7 /2007-2013)/ERC grant agreement no. [247073]10. SGM was the recipient of a CAPES and a CAPES-Cofecub doctoral fellowship and a CAPES post-doctoral fellowship (CAPES-Biologia Computacional - Process Number: 611 23038.010043/2013-02). CM was the recipient of a doctoral fellowship CONICYT BECAS CHILE DOCTORADO 2016/FOLIO 72170320.
Author contributions
S.G.M., M.G.F., A.Z., and M.F.S. conceived the experiments. S.G.M. conducted the experiments with help from M.G.F., S.R. and F.T. ADG designed and performed the processing of total RNA-seq data. C.M. designed and performed the computational prediction of miRNAs and its targets. M.G.F. performed the statistical analysis of mRNA and miRNA sequencing data. S.G.M., M.G.F. and C.M. reconstructed and refined the regulatory networks. S.G.M., M.G.F., C.M., A.D.G., and L.G. analyzed the results. S.G.M., M.G.F., A.Z., and M.F.S. wrote the manuscript with inputs from all other authors. All authors reviewed the manuscript.
Accession codes
All sequences were submitted to the Sequence Read Archive from NCBI: SRA accession: PRJNA545822.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Scheila G. Mucha and Mariana G. Ferrarini.
Contributor Information
Marie-France Sagot, Email: marie-france.sagot@inria.fr.
Arnaldo Zaha, Email: zaha@cbiot.ufrgs.br.
Supplementary information
is available for this paper at 10.1038/s41598-020-70040-y.
References
- 1.Maes D, Verdonck M, Deluyker H, de Kruif A. Enzootic pneumonia in pigs. Vet. Q. 1996;18:104–109. doi: 10.1080/01652176.1996.9694628. [DOI] [PubMed] [Google Scholar]
- 2.Maes D, et al. Update on Mycoplasma hyopneumoniae infections in pigs: knowledge gaps for improved disease control. Transbound. Emerg. Dis. 2018;65:110–124. doi: 10.1111/tbed.12677. [DOI] [PubMed] [Google Scholar]
- 3.Djordjevic SP, Cordwell SJ, Djordjevic MA, Wilton J, Minion FC. Proteolytic processing of the Mycoplasma hyopneumoniae cilium adhesin. Infect. Immun. 2004;72:2791–2802. doi: 10.1128/IAI.72.5.2791-2802.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeBey MC, Ross RF. Ciliostasis and loss of cilia induced by Mycoplasma hyopneumoniae in porcine tracheal organ cultures. Infect. Immun. 1994;62:5312–5318. doi: 10.1128/iai.62.12.5312-5318.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrarini MG, et al. Hydrogen peroxide production and myo-inositol metabolism as important traits for virulence of Mycoplasma hyopneumoniae. Mol. Microbiol. 2018;108:683–696. doi: 10.1111/mmi.13957. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. Micrornas: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Luo Y, et al. Detection of dietetically absorbed maize-derived micrornas in pigs. Sci. Rep. 2017;7:645. doi: 10.1038/s41598-017-00488-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawless, N., Foroushani, A. B., McCabe, M. S., O’Farrelly, C. & Lynn, D. J. Next generation sequencing reveals the expression of a unique mirna profile in response to a gram-positive bacterial infection. PLoS ONE8, e57543 (2013). [DOI] [PMC free article] [PubMed]
- 9.Duval, M., Cossart, P. & Lebreton, A. Mammalian micrornas and long noncoding rnas in the host-bacterial pathogen crosstalk. In Seminars in cell & developmental biology, vol. 65, 11–19 (Elsevier, 2017). [DOI] [PMC free article] [PubMed]
- 10.Lim LP, et al. Microarray analysis shows that some micrornas downregulate large numbers of target mrnas. Nature. 2005;433:769. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 11.Kim VN, Nam J-W. Genomics of microrna. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Valadi H, et al. Exosome-mediated transfer of mrnas and micrornas is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 13.Rome S. Are extracellular micrornas involved in type 2 diabetes and related pathologies? Clin. Biochem. 2013;46:937–945. doi: 10.1016/j.clinbiochem.2013.02.018. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, et al. Epigenetic regulation of connective tissue growth factor by microrna-214 delivery in exosomes from mouse or human hepatic stellate cells. Hepatology. 2014;59:1118–1129. doi: 10.1002/hep.26768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schorey JS, Cheng Y, Singh PP, Smith VL. Exosomes and other extracellular vesicles in host–pathogen interactions. EMBO Rep. 2015;16:24–43. doi: 10.15252/embr.201439363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rana S, Yue S, Stadel D, Zöller M. Toward tailored exosomes: the exosomal tetraspanin web contributes to target cell selection. Int. J. Biochem. Cell Biol. 2012;44:1574–1584. doi: 10.1016/j.biocel.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 17.Forterre A, et al. Myotube-derived exosomal mirnas downregulate sirtuin1 in myoblasts during muscle cell differentiation. Cell Cycle. 2014;13:78–89. doi: 10.4161/cc.26808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silverman JM, Reiner NE. Exosomes and other microvesicles in infection biology: organelles with unanticipated phenotypes. Cell. Microbiol. 2011;13:1–9. doi: 10.1111/j.1462-5822.2010.01537.x. [DOI] [PubMed] [Google Scholar]
- 19.Muxel SM, Laranjeira-Silva MF, Zampieri RA, Floeter-Winter LM. Leishmania (Leishmania) amazonensis induces macrophage mir-294 and mir-721 expression and modulates infection by targeting nos2 and l-arginine metabolism. Sci. Rep. 2017;7:44141. doi: 10.1038/srep44141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bao H, et al. Genome-wide whole blood micrornaome and transcriptome analyses reveal mirna-mrna regulated host response to foodborne pathogen Salmonella infection in swine. Sci. Rep. 2015;5:12620. doi: 10.1038/srep12620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staedel C, Darfeuille F. Micro rna s and bacterial infection. Cell. Microbiol. 2013;15:1496–1507. doi: 10.1111/cmi.12159. [DOI] [PubMed] [Google Scholar]
- 22.Maudet C, Mano M, Eulalio A. Micrornas in the interaction between host and bacterial pathogens. FEBS Lett. 2014;588:4140–4147. doi: 10.1016/j.febslet.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Zielinski G, Ross R. Effect of growth in cell cultures and strain on virulence of Mycoplasma hyopneumoniae for swine. Am. J. Vet. Res. 1990;51:344–348. [PubMed] [Google Scholar]
- 24.Young TF, Thacker EL, Erickson BZ, Ross RF. A tissue culture system to study respiratory ciliary epithelial adherence of selected swine mycoplasmas. Vet. Microbiol. 2000;71:269–279. doi: 10.1016/s0378-1135(99)00176-5. [DOI] [PubMed] [Google Scholar]
- 25.Burnett TA, et al. P159 is a proteolytically processed, surface adhesin of Mycoplasma hyopneumoniae: defined domains of p159 bind heparin and promote adherence to eukaryote cells. Mol. Microbiol. 2006;60:669–686. doi: 10.1111/j.1365-2958.2006.05139.x. [DOI] [PubMed] [Google Scholar]
- 26.Cho H-Y, Reddy SP, Kleeberger SR. Nrf2 defends the lung from oxidative stress. Antioxid. Redox Signal. 2006;8:76–87. doi: 10.1089/ars.2006.8.76. [DOI] [PubMed] [Google Scholar]
- 27.Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thimmulappa RK, et al. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J. Clin. Investig. 2016;116:984–995. doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Athale J, et al. Nrf2 promotes alveolar mitochondrial biogenesis and resolution of lung injury in staphylococcus aureus pneumonia in mice. Free Radical Biol. Med. 2012;53:1584–1594. doi: 10.1016/j.freeradbiomed.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reddy NM, et al. Innate immunity against bacterial infection following hyperoxia exposure is impaired in nrf2-deficient mice. J. Immunol. 2009;183:4601–4608. doi: 10.4049/jimmunol.0901754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomez JC, Dang H, Martin JR, Doerschuk CM. Nrf2 modulates host defense during Streptococcus pneumoniae pneumonia in mice. J. Immunol. 2016;197:2864–2879. doi: 10.4049/jimmunol.1600043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the keap1-nrf2-are pathway. Annu. Rev. Pharmacol. Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 33.Fourquet S, Guerois R, Biard D, Toledano MB. Activation of nrf2 by nitrosative agents and h2o2 involves keap1 disulfide formation. J. Biol. Chem. 2010;285:8463–8471. doi: 10.1074/jbc.M109.051714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hames C, Halbedel S, Hoppert M, Frey J, Stülke J. Glycerol metabolism is important for cytotoxicity of Mycoplasma pneumoniae. J. Bacteriol. 2009;191:747–753. doi: 10.1128/JB.01103-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vilei EM, Frey J. Genetic and biochemical characterization of glycerol uptake in Mycoplasma mycoides subsp. mycoides sc: Its impact on h2o2production and virulence. Clin. Diagn. Lab. Immunol. 2001;8:85–92. doi: 10.1128/CDLI.8.1.85-92.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bischof DF, Janis C, Vilei EM, Bertoni G, Frey J. Cytotoxicity of mycoplasma mycoides subsp. mycoides small colony type to bovine epithelial cells. Infect. Immun. 2008;76:263–269. doi: 10.1128/IAI.00938-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halbedel S, Hames C, Stülke J. In vivo activity of enzymatic and regulatory components of the phosphoenolpyruvate: sugar phosphotransferase system in mycoplasma pneumoniae. J. Bacteriol. 2004;186:7936–7943. doi: 10.1128/JB.186.23.7936-7943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biswal S, Thimmulappa RK, Harvey CJ. Experimental therapeutics of nrf2 as a target for prevention of bacterial exacerbations in copd. Proc. Am. Thorac. Soc. 2012;9:47–51. doi: 10.1513/pats.201201-009MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Identification of a novel nrf2-regulated antioxidant response element (are) in the mouse nad (p) h: quinone oxidoreductase 1 gene: reassessment of the are consensus sequence. Biochem. J. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rice, P., Longden, I. & Bleasby, A. Emboss: the European molecular biology open software suite (2000). [DOI] [PubMed]
- 41.Araake M. Comparison of ciliostasis by mycoplasmas in mouse and chicken tracheal organ cultures. Microbiol. Immunol. 1982;26:1–14. doi: 10.1111/j.1348-0421.1982.tb00148.x. [DOI] [PubMed] [Google Scholar]
- 42.Abu-Zahr M, Butler M. Growth, cytopathogenicity and morphology of Mycoplasma gallisepticum and M. gallinarum in tracheal explants. J. Comp. Pathol. 1976;86:455–463. doi: 10.1016/0021-9975(76)90014-1. [DOI] [PubMed] [Google Scholar]
- 43.Chaudhry R, Ghosh A, Chandolia A, et al. Pathogenesis of Mycoplasma pneumoniae: an update. Indian J. Med. Microbiol. 2016;34:7. doi: 10.4103/0255-0857.174112. [DOI] [PubMed] [Google Scholar]
- 44.Matsuyama M, et al. Transcriptional response of respiratory epithelium to nontuberculous mycobacteria. Am. J. Respir. Cell Mol. Biol. 2018;58:241–252. doi: 10.1165/rcmb.2017-0218OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kobayashi D, Takeda H. Ciliary motility: the components and cytoplasmic preassembly mechanisms of the axonemal dyneins. Differentiation. 2012;83:S23–S29. doi: 10.1016/j.diff.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 46.Tarkar A, et al. Dyx1c1 is required for axonemal dynein assembly and ciliary motility. Nat. Genet. 2013;45:995. doi: 10.1038/ng.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang W-J, et al. Cep162 is an axoneme-recognition protein promoting ciliary transition zone assembly at the cilia base. Nat. Cell Biol. 2013;15:591. doi: 10.1038/ncb2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schueler M, et al. Dcdc2 mutations cause a renal-hepatic ciliopathy by disrupting wnt signaling. Am. J. Human Genet. 2015;96:81–92. doi: 10.1016/j.ajhg.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.May-Simera HL, et al. Loss of macf1 abolishes ciliogenesis and disrupts apicobasal polarity establishment in the retina. Cell Rep. 2016;17:1399–1413. doi: 10.1016/j.celrep.2016.09.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi L, Koll F, Arnaiz O, Cohen J. The ciliary protein ift 57 in the macronucleus of paramecium. J. Eukaryot. Microbiol. 2018;65:12–27. doi: 10.1111/jeu.12423. [DOI] [PubMed] [Google Scholar]
- 51.Yasunaga T, et al. The polarity protein inturned links nphp4 to daam1 to control the subapical actin network in multiciliated cells. J. Cell Biol. 2015;211:963–973. doi: 10.1083/jcb.201502043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hegan PS, Ostertag E, Geurts AM, Mooseker MS. Myosin id is required for planar cell polarity in ciliated tracheal and ependymal epithelial cells. Cytoskeleton. 2015;72:503–516. doi: 10.1002/cm.21259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernandez-Gonzalez A, Kourembanas S, Wyatt TA, Mitsialis SA. Mutation of murine adenylate kinase 7 underlies a primary ciliary dyskinesia phenotype. Am. J. Respir. Cell Mol. Biol. 2009;40:305–313. doi: 10.1165/rcmb.2008-0102OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou J, Yang F, Leu NA, Wang PJ. Mns1 is essential for spermiogenesis and motile ciliary functions in mice. PLoS Genet. 2012;8:e1002516. doi: 10.1371/journal.pgen.1002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Inaba Y, et al. Transport of the outer dynein arm complex to cilia requires a cytoplasmic protein lrrc6. Genes Cells. 2016;21:728–739. doi: 10.1111/gtc.12380. [DOI] [PubMed] [Google Scholar]
- 56.Arachchige, S. N. K. et al. Differential response of the chicken trachea to chronic infection with virulent Mycoplasma gallisepticum strain ap3as and vaxsafe mg (strain ts-304): a transcriptional profile. Infect. Immun.88, (2020). [DOI] [PMC free article] [PubMed]
- 57.Girón JA, Lange M, Baseman JB. Adherence, fibronectin binding, and induction of cytoskeleton reorganization in cultured human cells by Mycoplasma penetrans. Infect. Immun. 1996;64:197–208. doi: 10.1128/iai.64.1.197-208.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raymond B, et al. Mycoplasma hyopneumoniae resides intracellularly within porcine epithelial cells. Sci. Rep. 2018;8:17697. doi: 10.1038/s41598-018-36054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mirvis M, Stearns T, Nelson WJ. Cilium structure, assembly, and disassembly regulated by the cytoskeleton. Biochem. J. 2018;475:2329–2353. doi: 10.1042/BCJ20170453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raymond B, et al. Extracellular actin is a receptor for Mycoplasma hyopneumoniae. Front. Cell. Infect. Microbiol. 2018;8:54. doi: 10.3389/fcimb.2018.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kang M-I, Kobayashi A, Wakabayashi N, Kim S-G, Yamamoto M. Scaffolding of keap1 to the actin cytoskeleton controls the function of nrf2 as key regulator of cytoprotective phase 2 genes. Proc. Nat. Acad. Sci. 2004;101:2046–2051. doi: 10.1073/pnas.0308347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mack GJ, Compton DA. Analysis of mitotic microtubule-associated proteins using mass spectrometry identifies astrin, a spindle-associated protein. Proc. Nat. Acad. Sci. 2001;98:14434–14439. doi: 10.1073/pnas.261371298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim YH, Choi JY, Jeong Y, Wolgemuth DJ, Rhee K. Nek2 localizes to multiple sites in mitotic cells, suggesting its involvement in multiple cellular functions during the cell cycle. Biochem. Biophys. Res. Commun. 2002;290:730–736. doi: 10.1006/bbrc.2001.6212. [DOI] [PubMed] [Google Scholar]
- 64.Woolner, S., O’Brien, L. L., Wiese, C. & Bement, W. M. Myosin-10 and actin filaments are essential for mitotic spindle function. J. Cell Biol.182, 77–88 (2008). [DOI] [PMC free article] [PubMed]
- 65.Bolanos-Garcia VM, Blundell TL. Bub1 and bubr1: multifaceted kinases of the cell cycle. Trends Biochem. Sci. 2011;36:141–150. doi: 10.1016/j.tibs.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wolter P, et al. Gas2l3, a target gene of the dream complex, is required for proper cytokinesis and genomic stability. J. Cell Sci. 2012;125:2393–2406. doi: 10.1242/jcs.097253. [DOI] [PubMed] [Google Scholar]
- 67.Xu X-L, et al. The microtubule-associated protein aspm regulates spindle assembly and meiotic progression in mouse oocytes. PLoS ONE. 2012;7:e49303. doi: 10.1371/journal.pone.0049303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matson DR, Stukenberg PT. Cenp-i and aurora b act as a molecular switch that ties rzz/mad1 recruitment to kinetochore attachment status. J. Cell Biol. 2014;205:541–554. doi: 10.1083/jcb.201307137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wadsworth P. Tpx2. Curr. Biol. 2015;25:R1156–R1158. doi: 10.1016/j.cub.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cho RJ, et al. Transcriptional regulation and function during the human cell cycle. Nat. Genet. 2001;27:48. doi: 10.1038/83751. [DOI] [PubMed] [Google Scholar]
- 71.Shirin H, et al. Helicobacter pylori inhibits the g1 to s transition in ags gastric epithelial cells. Cancer Res. 1999;59:2277–2281. [PubMed] [Google Scholar]
- 72.Marchès O, et al. Enteropathogenic and enterohaemorrhagic Escherichia coli deliver a novel effector called cif, which blocks cell cycle g2/m transition. Mol. Microbiol. 2003;50:1553–1567. doi: 10.1046/j.1365-2958.2003.03821.x. [DOI] [PubMed] [Google Scholar]
- 73.Jones A, Jonsson A-B, Aro H. Neisseria gonorrhoeae infection causes a g1 arrest in human epithelial cells. FASEB J. 2007;21:345–355. doi: 10.1096/fj.06-6675com. [DOI] [PubMed] [Google Scholar]
- 74.Alekseeva L, et al. Staphylococcus aureus-induced g2/m phase transition delay in host epithelial cells increases bacterial infective efficiency. PLoS ONE. 2013;8:e63279. doi: 10.1371/journal.pone.0063279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Siqueira FM, et al. Mycoplasma non-coding rna: identification of small rnas and targets. BMC Genom. 2016;17:743. doi: 10.1186/s12864-016-3061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. mirdeep2 accurately identifies known and hundreds of novel microrna genes in seven animal clades. Nucl. Acids Res. 2011;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kozomara A, Griffiths-Jones S. mirbase: annotating high confidence micrornas using deep sequencing data. Nucl. Acids Res. 2013;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martini P, et al. Tissue-specific expression and regulatory networks of pig micrornaome. PLoS ONE. 2014;9:e89755. doi: 10.1371/journal.pone.0089755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bartel DP. Metazoan micrornas. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stark C, et al. Biogrid: a general repository for interaction datasets. Nucl. Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chatr-Aryamontri A, et al. The biogrid interaction database: 2017 update. Nucl. Acids Res. 2017;45:D369–D379. doi: 10.1093/nar/gkw1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bindea G, et al. Cluego: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang M, Yao Y, Eades G, Zhang Y, Zhou Q. Mir-28 regulates nrf2 expression through a keap1-independent mechanism. Breast Cancer Res. Treat. 2011;129:983–991. doi: 10.1007/s10549-011-1604-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shi L, et al. mir-340 reverses cisplatin resistance of hepatocellular carcinoma cell lines by targeting nrf2-dependent antioxidant pathway. Asian Pac. J. Cancer Prev. 2015;15:10439–10444. doi: 10.7314/apjcp.2014.15.23.10439. [DOI] [PubMed] [Google Scholar]
- 85.Xu J, et al. mir-19b attenuates h2o2-induced apoptosis in rat h9c2 cardiomyocytes via targeting pten. Oncotarget. 2016;7:10870. doi: 10.18632/oncotarget.7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hong J, et al. Transcriptional downregulation of microrna-19a by ros production and nf-b deactivation governs resistance to oxidative stress-initiated apoptosis. Oncotarget. 2017;8:70967. doi: 10.18632/oncotarget.20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gao A-M, Zhang X-Y, Ke Z-P. Apigenin sensitizes bel-7402/adm cells to doxorubicin through inhibiting mir-101/nrf2 pathway. Oncotarget. 2017;8:82085. doi: 10.18632/oncotarget.18294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu H, et al. Regulation of mir-92a on vascular endothelial aging via mediating nrf2-keap1-are signal pathway. Eur. Rev. Med. Pharmacol. Sci. 2017;21:2734–2742. [PubMed] [Google Scholar]
- 89.Yang H, et al. Activation of a novel c-myc-mir27-prohibitin 1 circuitry in cholestatic liver injury inhibits glutathione synthesis in mice. Antioxid. Redox Signal. 2015;22:259–274. doi: 10.1089/ars.2014.6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alural B, Ozerdem A, Allmer J, Genc K, Genc S. Lithium protects against paraquat neurotoxicity by nrf2 activation and mir-34a inhibition in sh-sy5y cells. Front. Cell. Neurosci. 2015;9:209. doi: 10.3389/fncel.2015.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Milara J, Armengot M, Mata M, Morcillo EJ, Cortijo J. Role of adenylate kinase type 7 expression on cilia motility: possible link in primary ciliary dyskinesia. Am. J. Rhinol. Allergy. 2010;24:181–185. doi: 10.2500/ajra.2010.24.3468. [DOI] [PubMed] [Google Scholar]
- 92.Zimmer FMdAL, Paludo GP, Moura H, Barr JR, Ferreira HB. Differential secretome profiling of a swine tracheal cell line infected with mycoplasmas of the swine respiratory tract. J. Proteom. 2019;192:147–159. doi: 10.1016/j.jprot.2018.08.018. [DOI] [PubMed] [Google Scholar]
- 93.Alexander M, et al. Exosome-delivered micrornas modulate the inflammatory response to endotoxin. Nat. Commun. 2015;6:7321. doi: 10.1038/ncomms8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Muneta Y, et al. Immune response of gnotobiotic piglets against Mycoplasma hyopneumoniae. J. Vet. Med. Sci. 2008;70:1065–1070. doi: 10.1292/jvms.70.1065. [DOI] [PubMed] [Google Scholar]
- 95.Ferrari M, et al. Establishment and characterization of two new pig cell lines for use in virological diagnostic laboratories. J. Virol. Methods. 2003;107:205–212. doi: 10.1016/s0166-0934(02)00236-7. [DOI] [PubMed] [Google Scholar]
- 96.Friis N. Some recommendations concerning primary isolation of Mycoplasma suipneumoniae and Mycoplasma flocculare a survey. Nordisk veterinaermedicin. 1975;27:337–339. [PubMed] [Google Scholar]
- 97.Assunção P, et al. Evaluation of Mycoplasma hyopneumoniae growth by flow cytometry. J. Appl. Microbiol. 2005;98:1047–1054. doi: 10.1111/j.1365-2672.2005.02536.x. [DOI] [PubMed] [Google Scholar]
- 98.Moitinho-Silva L, et al. Mycoplasma hyopneumoniae type i signal peptidase: expression and evaluation of its diagnostic potential. Vet. Microbiol. 2012;154:282–291. doi: 10.1016/j.vetmic.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 99.Schneider CA, Rasband WS, Eliceiri KW. Nih image to imagej: 25 years of image analysis. Nat. Methods. 2012;9:671. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ruijter J, et al. Amplification efficiency: linking baseline and bias in the analysis of quantitative pcr data. Nucl. Acids Res. 2009;37:e45–e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. [Google Scholar]
- 102.Dobin A, et al. Star: ultrafast universal rna-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Anders S, Pyl PT, Huber W. Htseq—a python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short dna sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Griffiths-Jones S. The microrna registry. Nucl. Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Robinson MD, McCarthy DJ, Smyth GK. edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for rna-seq data with deseq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc.: Ser. B (Methodol.) 1995;57:289–300. [Google Scholar]
- 109.Feng J, et al. Gfold: a generalized fold change for ranking differentially expressed genes from rna-seq data. Bioinformatics. 2012;28:2782–2788. doi: 10.1093/bioinformatics/bts515. [DOI] [PubMed] [Google Scholar]
- 110.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microrna targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lewis BP, Shih I-H, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microrna targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 112.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microrna target recognition. Nat. Genet. 2007;39:1278. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 113.Krüger J, Rehmsmeier M. Rnahybrid: microrna target prediction easy, fast and flexible. Nucl. Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ashburner M, et al. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Supek F, Bošnjak M, Škunca N, Šmuc T. Revigo summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hayes JD, McMahon M. Nrf2 and keap1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem. Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 118.Hayes JD, Dinkova-Kostova AT. The nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 119.Brzóska K, Stępkowski TM, Kruszewski M. Basal pir expression in hela cells is driven by nrf2 via evolutionary conserved antioxidant response element. Mol. Cell. Biochem. 2014;389:99–111. doi: 10.1007/s11010-013-1931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li D, Ma S, Ellis EM. Nrf2-mediated adaptive response to methyl glyoxal in hepg2 cells involves the induction of akr7a2. Chem. Biol. Interact. 2015;234:366–371. doi: 10.1016/j.cbi.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 121.Graham DB, et al. Nitric oxide engages an anti-inflammatory feedback loop mediated by peroxiredoxin 5 in phagocytes. Cell Rep. 2018;24:838–850. doi: 10.1016/j.celrep.2018.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Reszka E, et al. Expression of nrf2 and nrf2-modulated genes in peripheral blood leukocytes of bladder cancer males. Neoplasma. 2013;60:123–128. doi: 10.4149/neo_2013_016. [DOI] [PubMed] [Google Scholar]
- 123.Dai L-L, Gao J-X, Zou C-G, Ma Y-C, Zhang K-Q. mir-233 modulates the unfolded protein response in C. elegans during Pseudomonas aeruginosa infection. PLoS Pathog. 2015;11:e1004606. doi: 10.1371/journal.ppat.1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Derrick T, et al. Conjunctival microrna expression in inflammatory trachomatous scarring. PLoS Negl. Trop. Dis. 2013;7:e2117. doi: 10.1371/journal.pntd.0002117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Derrick T, et al. Inverse relationship between microrna-155 and-184 expression with increasing conjunctival inflammation during ocular Chlamydia trachomatis infection. BMC Infect. Dis. 2015;16:60. doi: 10.1186/s12879-016-1367-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liang G, et al. Altered microrna expression and pre-mrna splicing events reveal new mechanisms associated with early stage Mycobacterium avium subspecies paratuberculosis infection. Sci. Rep. 2016;6:24964. doi: 10.1038/srep24964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Siddle KJ, et al. Bacterial infection drives the expression dynamics of micrornas and their isomirs. PLoS Genet. 2015;11:e1005064. doi: 10.1371/journal.pgen.1005064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Naqvi AR, Fordham JB, Nares S. mir-24, mir-30b, and mir-142-3p regulate phagocytosis in myeloid inflammatory cells. J. Immunol. 2015;194:1916–1927. doi: 10.4049/jimmunol.1401893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu Z, et al. Up-regulated microrna-146a negatively modulate Helicobacter pylori-induced inflammatory response in human gastric epithelial cells. Microb. Infect. 2010;12:854–863. doi: 10.1016/j.micinf.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 130.Li N, et al. H. pylori related proinflammatory cytokines contribute to the induction of mir-146a in human gastric epithelial cells. Mol. Biol. Rep. 2012;39:4655–4661. doi: 10.1007/s11033-011-1257-5. [DOI] [PubMed] [Google Scholar]
- 131.Liu Z, et al. Microrna-146a negatively regulates ptgs2 expression induced by helicobacter pylori in human gastric epithelial cells. J. Gastroenterol. 2013;48:86–92. doi: 10.1007/s00535-012-0609-9. [DOI] [PubMed] [Google Scholar]
- 132.Cheng Y, et al. Downregulation of mir-27a* and mir-532-5p and upregulation of mir-146a and mir-155 in lps-induced raw264. 7 macrophage cells. Inflammation. 2012;35:1308–1313. doi: 10.1007/s10753-012-9443-8. [DOI] [PubMed] [Google Scholar]
- 133.Derrick, T. et al. mirnas that associate with conjunctival inflammation and ocular Chlamydia trachomatis infection do not predict progressive disease. Pathog. Dis.75, (2017). [DOI] [PMC free article] [PubMed]
- 134.Zheng L, et al. Differential microrna expression in human macrophages with Mycobacterium tuberculosis infection of beijing/w and non-beijing/w strain types. PLoS ONE. 2015;10:e0126018. doi: 10.1371/journal.pone.0126018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Matsushima K, et al. Microrna signatures in Helicobacter pylori-infected gastric mucosa. Int. J. Cancer. 2011;128:361–370. doi: 10.1002/ijc.25348. [DOI] [PubMed] [Google Scholar]
- 136.Xue X, et al. Downregulation of micro rna-107 in intestinal cd 11c+ myeloid cells in response to microbiota and proinflammatory cytokines increases il-23p19 expression. Eur. J. Immunol. 2014;44:673–682. doi: 10.1002/eji.201343717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xue F, et al. mir-31 regulates interleukin 2 and kinase suppressor of ras 2 during t cell activation. Genes Immun. 2013;14:127. doi: 10.1038/gene.2012.58. [DOI] [PubMed] [Google Scholar]
- 138.Podolska A, et al. Profiling micrornas in lung tissue from pigs infected with Actinobacillus pleuropneumoniae. BMC Genom. 2012;13:459. doi: 10.1186/1471-2164-13-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Noto JM, et al. Strain-specific suppression of microrna-320 by carcinogenic Helicobacter pylori promotes expression of the antiapoptotic protein mcl-1. Am. J. Physiol.-Gastrointest. Liver Physiol. 2013;305:G786–G796. doi: 10.1152/ajpgi.00279.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences were submitted to the Sequence Read Archive from NCBI: SRA accession: PRJNA545822.