Abstract
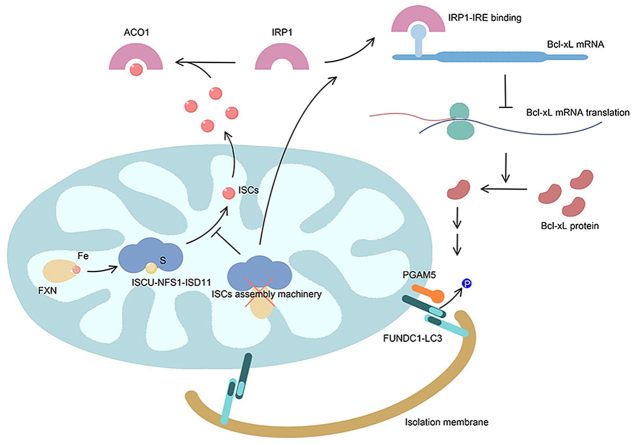
Both iron metabolism and mitophagy, a selective mitochondrial degradation process via autolysosomal pathway, are fundamental for the cellular well-being. Mitochondria are the major site for iron metabolism, especially the biogenesis of iron-sulfur clusters (ISCs) via the mitochondria-localized ISCs assembly machinery. Here we report that mitochondrial ISCs biogenesis is coupled with receptor-mediated mitophagy in mammalian cells. Perturbation of mitochondrial ISCs biogenesis, either by depleting iron with the iron chelator or by knocking down the core components of the mitochondrial ISCs assembly machinery, triggers FUNDC1-dependent mitophagy. IRP1, one of the cellular iron sensors to maintain iron homeostasis, is crucial for iron stresses induced mitophagy. Knockdown of IRP1 disturbed iron stresses induced mitophagy. Furthermore, IRP1 could bind to a newly characterized IRE in the 5’ untranslated region of the Bcl-xL mRNA and suppress its translation. Bcl-xL is an intrinsic inhibitory protein of the mitochondrial phosphatase PGAM5, which catalyzes the dephosphorylation of FUNDC1 for mitophagy activation. Alterations of the IRP1/Bcl-xL axis navigate iron stresses induced mitophagy. We conclude that ISCs serve as physiological signals for mitophagy activation, thus coupling mitophagy with iron metabolism.
Keywords: Mitophagy, Bcl-xL, ISCs, FUNDC1
Graphical abstract
Highlights
-
•
Perturbation of ISCs biogenesis triggers FUNDC1 dependent mitophagy.
-
•
IRP1 targets a newly characterized IRE in Bcl-xL mRNA to suppress its translation.
-
•
IRP1/Bcl-xL axis navigates iron stresses induced mitophagy and dominates mitochondrial redox response.
1. Introduction
Mitochondria are essential organelles for a myriad of fundamental cellular processes, including oxidative phosphorylation associated energy supply and programmed cell death. Mitochondria also generate reactive oxygen species (ROS) as the inevitable by-product of electron transport chain. To fulfill such diverse functions, graded mechanisms have been evolved to maintain mitochondrial homeostasis in response to dynamic metabolic and environmental cues. Mitochondria themselves are highly dynamic, undergoing cycles of fission and fusion, as well as constantly turning over. Mitochondrial homeostasis is largely determined by the balanced mitochondrial biogenesis and mitophagy, a process that selectively removes damaged or superfluous mitochondria through autophagic and lysosomal degradation [1]. To date, Pink1/Parkin pathway and the receptor-mediated mitophagy pathway are characterized in mammalian cells. Upon the loss of mitochondrial membrane potential, Pink1 becomes stabilized on the mitochondrial outer membrane [2], where it is able to activate the E3 ubiquitin ligase Parkin to ubiquitinate numerous mitochondrial outer membrane proteins [3], and to recruit selective autophagy receptors (such as NDP52 and OPTN) to mediate mitophagy via their interaction with LC3 [4,5]. Additionally, several mitochondrially localized mitophagy receptors were identified to mediate selective recognition and removal of damaged mitochondria in mammalian cells. These include Nix/Bnip3 [6], FUNDC1 [7,8], Prohibitin 2 [9], FKBP8 [10] and Bcl2l13 [11]. These receptor proteins contain a LIR (LC3 interacting region) motif and directly interact with LC3 or its homologs to mediate the selective recognition of mitochondria by autophagosomal membranes in response to distinct mitochondrial stresses. Significant progress has been achieved in understanding the mechanism of mitophagy execution; however, the molecular details of mitophagy regulation in response to metabolic cues remain elusive. This is important because defective mitophagy leads to the accumulation of dysfunctional mitochondria, excessive production of ROS, the disruption of cellular metabolism, and even cell death. Moreover, dysfunctional mitochondria accompanied with increased oxidative stress are widely observed in diseased tissues of human patients and animal models, which is strongly associated with the pathogenesis of human diseases [12].
Mitochondria are centers for cellular iron metabolism. Cytosolic iron is transported into mitochondria where it is utilized to synthesize heme and iron-sulfur clusters (ISCs), two kinds of iron-containing molecules structurally and functionally important for numerous proteins involved in fundamental cellular processes, such as the tricarboxylic acid (TCA) cycle, the electron transport chain (ETC), DNA damage/repair and oxygen metabolism [13,14]. ISCs are mainly assembled in the mitochondrial matrix by the highly conserved ISCs assembly machinery [15]. The cysteine desulfurase NFS1 is required to abstract the sulfur atom from cysteine and donate it to a scaffold protein, ISCU, on which nascent clusters are assembled. The assembled [2Fe–2S] or [4Fe–4S] clusters are transferred to recipients, including mitochondrial proteins, cytosolic proteins and even nuclear proteins [16]. In mammalian systems, ISCs serve as signal molecules to dominate cellular iron homeostasis through the iron regulatory proteins (IRP1/2) and iron-responsive element (IRE) systems. In iron-replete conditions, IRP1 binds to a [4Fe–4S] and displays cytosolic aconitase (ACO1) activity, which catalyzes the interconversion of citrate and isocitrate in TCA cycle, while IRP2 is ubiquitinated for proteasomal degradation. In iron depleted conditions, the apo-IRP1 escapes from binding to ISCs and IRP2 is stabilized. IRP1/2 can then target to the conserved stem-loop structures in the untranslated regions (UTRs) of target mRNAs, which are known as IREs. This binding suppresses the translation of the target mRNAs if the IREs are located in the 5′UTR, as is the case for the mRNAs encoding iron storage associated L- and H-Ferritin, heme synthesis associated ALAS2 (5-Aminolevulinate Synthase 2), and iron export associated FPN1 (Ferroportin 1). Binding of IRP1/2 to IREs stabilizes the target mRNAs if the IRE is located in the 3′UTR, as is the case for iron uptake-associated TfR1 (Transferrin Receptor Protein 1). The IRPs/IREs system thus leads to increased iron absorption and decreased iron export, iron utilization and iron storage, thereby maintaining appropriate intracellular iron concentrations (reviewed in Ref. [17]). Moreover, because of the importance of ISCs in the mitochondrial behaviors, defective ISCs assembly or dysregulated IRPs-IREs system would lead to abnormal mitochondrial integrity and result in excessive ROS production. Previous work has shown that iron depletion by DFP (deferiprone), a widely used iron chelator, initiates mitophagy program in a Pink1/Parkin-independent fashion [18]. However, the molecular mechanism is not clear.
Both iron metabolism and mitophagy are fundamental processes for the cellular well-being. Dysregulation of these processes leads to the redox imbalance and contributes to a number of hematological, metabolic, and neurodegenerative diseases [19]. In particular, the accumulation of dysfunctional mitochondria and dysregulated iron homeostasis are commonly observed in ageing-related diseases [20,21]. It is still not clear whether these two events are causally linked or not. We have previously identified that the mitochondrial outer membrane protein FUNDC1 acts as a novel mitophagy receptor through its interaction with LC3 [7]. Biochemical and structural analyses have shown that dephosphorylation of FUNDC1 at Ser13 by mitochondrial phosphatase PGAM5 enhances the FUNDC1-LC3 interaction to initiate mitophagy [8]. We further demonstrated that Bcl-xL is the intrinsic inhibitor of PGAM5 [22], and suppresses the dephosphorylation of Ser13 to inhibit FUNDC1-dependent mitophagy [22]. We now provide evidence to show that mitochondrial ISCs biogenesis is coupled with FUNDC1-mediated mitophagy through fine tuning the translational efficiency of Bcl-xL mRNA by IRP1, which defines a novel mechanism of mitophagy regulation in response to altered iron metabolism.
2. Materials and methods
2.1. Plasmids and regents
The full-length IRP1 CDS was cloned from a human cDNA library and then inserted into the bacterial expression vector pGEX-4T-1 (Amersham, VYA0221) and the mammalian expression vector pcDNA4/TO-Myc (Invitrogen, V102020), respectively. The other constructs were maintained in the lab.
For site-directed mutagenesis, upstream and downstream primers were synthesized containing the desired mutations. Briefly, 15 ng wildtype plasmid as the template, the corresponding primers and Pfu DNA Polymerase (Agilent, 600135) were employed for site-directed mutagenesis PCR according to the manufacturer's protocol. The PCR product was processed with the endonuclease DpnI (New England Biolabs, R0176L) to degrade the template wildtype plasmid. The product was then transformed into competent E. coli cells, and positive clones were sequenced to confirm the mutagenesis.
The regents used in this study are following: deferoxamine (DFO, D9533), deferiprone (DFP, 379409), Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP, C2920), chloroquine (CQ, C6628), and actinomycin D (A1410) were purchased from Sigma-Aldrich. EBSS (SH30014.08) was purchased from HyClone.
2.2. Antibodies
The following primary antibodies were used in this study: α-TIM23 (BD Biosciences, 611223), α-TOM20 (BD Biosciences, 612278), α-COX4 (ProteinTech, 11242-1-AP), α-VDAC1 (Abcam, ab14734), α-LC3 (MBL, PM036), α-Bcl-xL (Cell Signaling Technology, 2762), α-ISCU (Santa Cruz, sc-373694), α-FXN (Santa Cruz, ab113691), α-NFS1 (Abcam, ab204355), α-ISD11 (Sigma-Aldrich, HPA030362), α-CIAO1 (GeneTex, GTX53698), α-NARFL (GeneTex, GTX116764), α-IRP1 (Cell Signaling Technology, 20272), α-HSP60 (Cell Signaling Technology, 12165), α-GFP (Santa Cruz, sc-9996), α-PGAM5 (Abcam, ab126534), α-ATG5 (Cell Signaling Technology, 2630), α-NRF1 (ProteinTech, 12482-1-AP), α-PGC1α (ProteinTech, 20658-1-AP), α-TFAM (ProteinTech, 19998-1-AP), α-Flag (Sigma-Aldrich, F1804), α-Myc (Sigma-Aldrich, M4439), α-Actin (Sigma-Aldrich, A8481). The rabbit α-FUNDC1 and rabbit α-F1-P13 phosphorylation polyclonal antibodies were generated by immunizing rabbits with a purified FUNDC1 peptide or a purified FUNDC1 phosphorylated peptide, respectively. The polyclonal antibodies were affinity purified before use. HRP-conjugated secondary antibodies were purchased from Jackson Immuno Research Laboratories. The secondary antibodies used for immunofluorescence staining were: donkey anti-mouse IgG Alexa Fluor-555 (Molecular Probes, A31570), donkey anti-rabbit IgG Alexa Fluor-555 (Molecular Probes, A31572) and donkey anti-rabbit IgG Alexa Fluor-488 (Molecular Probes, A21206).
2.3. Cell culture and transfection
HeLa, HepG2, and U2OS cells were maintained in Dulbecco's modified Eagle medium (DMEM, Gibco, 12100–061) supplemented with 10% fetal bovine serum (Hyclone, SV30087.02) and 1% penicillin-streptomycin at 37 °C with 5% CO2.
For DNA transfection, cells were plated in 6-well plates for 24 h and the medium was then changed to serum-free OPTI-MEM (Gibco, 31985) medium before transfection. The cells were transfected with corresponding plasmids by using MegaTran 1.0 transfection reagent (OriGene, TT200003). After 6 h of transfection, the medium was changed to fresh DMEM supplemented with 10% fetal bovine serum.
For siRNA transfection, 3–5 × 104 cells (per well) were plated in 24-well plates for 24 h and then transfected with siRNA using Lipofectamine RNAiMAX (Invitrogen, 13778) according to the manufacturer's protocol. The siRNA sequences targeting each gene are listed in Supplementary Table 1. All of the siRNAs were synthesized by Ribo Bio.
2.4. Construction of stable cell lines
The FUNDC1 knockdown HeLa cells (sh-F1) and rescued HeLa cells (sh-F1/WT, sh-F1/△LIR), PGAM5 knockout HeLa cells, Tet-inducible Bcl-xL expression HeLa cells, GFP-Parkin HeLa cell, and GFP-LC3 HeLa cells were maintained in our lab. The HeLa cell stably expressed mt-Keima was kindly gifted by Dr. Xin Pan from the Academy of Military Medical Sciences, Beijing.
The pLKO.1 vector was obtained from Sigma-Aldrich (SHC001). Plasmids expressing hairpin siRNA were constructed by inserting pairs of annealed DNA oligonucleotides into pLKO.1 according to the manufacturer's instructions. The targeting sequence to IRP1 was 5′-CCATAAGACCTTTATCTAT-3′. A nontargeting sequence, 5′-ACTACCGTTGTTATAGGTG-3′, was used as a scramble control (sc). To generate the stable knockdown cells, HeLa cells were transfected with pLKO.1-IRP1 or pLKO.1-sc. 24 h after transfection, 0.5 μg/ml puromycin (Sigma-Aldrich, P9620) was added to the medium for selection. Stable clones were analyzed by western blotting to confirm IRP1 knockdown.
2.5. SDS-PAGE and western blotting
Cells were harvested by trypsin and collected by centrifugation. After washing with ice-cold PBS (phosphate buffered saline) twice, cell pellets were lysed with appropriate volume of lysis buffer (20 mM Tris, 1% NP40, 10% glycerol, 2 mM EDTA, 137 mM NaCl, pH 7.2, 1 mM PMSF, 1 mM Na3VO4, 10 mM NaF) on ice for 30 min. The mixture was then centrifuged to collect the supernatant as the whole cell extract. An equivalent quantity (20 μg) of protein from each whole cell extract was then subjected to SDS-PAGE and transferred to nitrocellulose membranes. Membrane-bound proteins were blocked with 5% fat-free milk, and subsequently incubated with the indicated primary antibodies and HRP-conjugated secondary antibodies. Finally, immunoreactive protein bands were visualized with a chemiluminescence kit (TianGen, PA110). The relative protein levels to Actin were quantitatively analyzed via the band density by the Image J software.
2.6. Immunofluorescence
Cells were plated in 6-well plate with sterilized glass coverslips. After the indicated treatments or transfections, cells were fixed with 3.7% formaldehyde in DMEM at 37 °C for 15 min, and then washed three times with ice-cold PBS. Subsequently, cells on coverslips were permeabilized by 0.2% Triton X-100 at RT for 15 min, blocked by 5% fetal bovine serum, then incubated with indicated primary and corresponding secondary conjugated antibodies at room temperature for 120 min and 60 min, respectively. Images were captured with a LSM 510 Zeiss confocal microscope.
2.7. Immunoprecipitation
After the indicated treatments or transfections, cells plated in a 10 cm dish were lysed in 0.8 ml lysis buffer (20 mM Tris, 1% NP40, 10% glycerol, 2 mM EDTA, 137 mM NaCl, pH 7.2) plus protease and phosphatase inhibitors (1 mM PMSF, 1 mM Na3VO4, 10 mM NaF) for 30 min on ice. After centrifugation at 12,000 g for 15 min, the cell lysates were immunoprecipitated with specific antibodies and protein-A/G plus-Sepharose (Santa Cruz, sc-2003) overnight at 4 °C. Thereafter, the precipitants were washed 4 times with ice-cold lysis buffer, and the immune complexes were eluted with loading buffer for 3 min at 100 °C and analyzed via SDS-PAGE and western blotting.
2.8. GST pulldown assay
GST and GST-LC3 fusion proteins were incubated with glutathione-Sepharose 4 Fast Flow beads (Amersham Biosciences, 20182003–2) at 4 °C for 2 h and then washed 3 times with 1 ml ice-cold PBS. After the indicated treatments or transfections, cell lysates were prepared and subsequently added to the conjugated beads, followed by incubation at 4 °C overnight. The precipitated complexes were washed five times with 1 ml ice-cold PBS and boiled with loading buffer at 100 °C for 3 min and analyzed via SDS-PAGE and western blotting using specific antibodies. The GST and GST-LC3 bands on the membrane were visualized through Ponceau S staining.
2.9. Purification of GST-IRP1 fusion protein
The coding sequence of IRP1 was sub-cloned and inserted into the prokaryotic expression vector pGEX-4T-1. pGEX-4T-1-IRP1 plasmid was then transformed into competent Rosetta cells. The transformants were maintained in LB medium and GST-IRP1 expression was induced by addition of 0.1 mM IPTG for 16 h at 16 °C. The bacteria were pelleted and resuspended in lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.05% NP-40) supplemented with protease inhibitor PMSF and 0.5 mg/ml lysozyme at 4 °C for 30 min. The cells were ruptured by ultrasonication, and the lysate supernatant was harvested following centrifugation. Washed glutathione beads were then added to the supernatant to bind the GST-IRP1 fusion protein at 4 °C for 4 h. After washing in ice-cold lysis buffer for three times, elution buffer (100 mM Tris, pH 8.0) supplemented with 10 mg/ml glutathione was added to the beads to harvest the fusion protein. The protein concentration was determined by Bradford assay, and the protein purity was checked by SDS-PAGE and Coomassie blue staining.
2.10. REMSA
REMSAs were done as described previously [23]. Briefly, biotin-labeled human FTH IRE probe, unlabeled FTH IRE probe, mutant FTH IRE probe, Bcl-xL IRE1 probe, mutant Bcl-xL IRE1 probe, Bcl-xL IRE2 probe, and mutant Bcl-xL IRE2 probe were synthesized in GenePharma, Suzhou, China. The biotin-labeled FTH IRE probe was incubated with 20 ng of purified GST-IRP1 protein for 25 min at room temperature with or without excessive unlabeled competitor RNA probe as indicated. RNA-protein complexes were resolved by nondenaturing PAGE, transferred to a nylon membrane. The biotin-labeled RNA is detected by using the Streptavidin Horseradish Peroxidase Conjugate and a highly sensitive chemiluminescent substrate.
2.11. Polysome profiling assay
The sh-IRP1 and sc cells plated in 15 cm dishes were grown to 80% confluency, then the cells were washed twice with ice-cold PBS and lysed with 1 ml polysome lysis buffer (20 mM Tris-HCl, pH 7.4, 10 mM NaCl, 10 mM MgCl2, 1% Triton X-100, 2% Tween 20, and 1% sodium deoxycholate) for 10 min on ice. After centrifugation at 20,000 g for 15 min at 4 °C, the supernatant was collected and loaded onto 10%–50% (wt/vol) sucrose density gradients (buffered in 20 mM Tris-HCl, pH 7.4, 10 mM NaCl, 10 mM MgCl2) and centrifuged at 36,000 rpm (SW 40 Ti rotor, Beckman Coulter) for 3 h at 4 °C. Gradients were fractionated and the optical density at 260 nm was continuously recorded using a fractionator. RNA from each fraction and input was extracted by using TRNzol (TianGen, DP405) according to the manufacturer's instructions, and the corresponding cDNA was synthesized by using a cDNA synthesis kit (Takara, 6130). Semi-quantitative PCR reactions were carried out on a PCR machine (Bio-Rad).
2.12. mt-Keima assay
The mt-Keima assay to measure mitophagy activity was performed as reported previously [24]. Briefly, cell plated in a 6-well plate (for flow cytometry analysis) or a confocal dish (for confocal imaging) was transfected with mt-Keima plasmid. After 24 h of expression, cells were then subjected to the indicated treatments or transfections. For flow cytometry analysis, the cells were collected by trypsin digestion and excited with a 405 nm laser with emission detected at 610 ± 10 nm and with a 561 nm laser with emission detected at 610 ± 10 nm simultaneously. For confocal scanning, the living cells were observed and the images were captured with a LSM 510 Zeiss confocal microscope. Fluorescence of mt-Keima was imaged in two channels via two sequential excitations (458 nm, green; 543 nm, red) and using a 620-nm emission range.
2.13. Flow cytometric assay to measure mitochondrial ROS and mitochondrial membrane potential
After indicated treatment, cells were digested by trypsin without EDTA. After centrifugation at 1000 rpm for 5 min, cells were stained with 10 μM mitochondrial ROS indicator MitoSOX (Molecular Probes, M36008) or 100 nM mitochondrial membrane potential indicator TMRE (Molecular Probes, T669) for 30 min at 37 °C away from light. After staining, flow cytometric analysis was performed to monitor the relative intensity of the red fluorescence. The flow cytometric data were analyzed using FCS Express software.
2.14. Real-time PCR
Cells were harvested by direct addition of 1 ml TRNzol (TianGen, DP405) after the indicated treatments or transfections. Total cellular RNA was extracted according to the manufacturer's protocol. The corresponding cDNA was synthesized by using a cDNA synthesis kit (Takara, 6130). Quantitative RT-PCR was performed on diluted cDNA with SYBR green Master Mix (NEWBIO INDUSTRY, 308102) on a real-time PCR machine (Bio-Rad). The primers used in this study are listed in Supplementary Table 2.
2.15. Isolation of mitochondria
After the indicated treatments or transfections, cells plated in a 10 cm dish were harvested by trypsin digestion. After washing with ice-cold PBS twice, the cells were resuspended in 150 μl isolation buffer (10 mM Tris-MOPS, 1 mM EGTA-Tris, 200 mM sucrose, pH 7.4) with protease and phosphatase inhibitors (1 mM PMSF, 1 mM Na3VO4, 10 mM NaF) for 30 min on ice, then lysed by passing 15 times through a 27.5 g needle attached to 1 ml syringe. Nuclei and intact cells were pelleted by centrifugation for 10 min at 800 g, 4 °C. The supernatant was transferred to a fresh tube and centrifuged for 15 min at 10,000 g, 4 °C. The pellet was collected as crude mitochondria.
2.16. Oxyblot assay
Mitochondria were isolated from cells challenged with the indicated treatments or transfections as described above. The mitochondria were lysed in lysis buffer (137 mM NaCl, 20 mM Tris, pH 7.4, 2 mM EGTA, 1% NP-40) supplemented with protease inhibitors. The total protein concentration of each mitochondrial lysate was analyzed with a Bradford Protein Assay kit (Beyotime, P0006), mitochondrial lysates with equivalent protein quantities were reacted using the OxyBlot kit (Millipore, S7150) according to the manufacturer's protocol. The levels of oxidized protein were analyzed by western blotting.
2.17. Actinomycin D assay to measure Bcl-xL mRNA stability
After the indicated treatments or transfections, 6.5 μg/ml actinomycin D was added into the cell medium for the indicated time. The total RNA was extracted by using TRNzol (TianGen, DP405), and cDNA was synthesized by using a cDNA synthesis kit (Takara, 6130). Quantitative RT-PCR was performed on diluted cDNA with SYBR green Master Mix (NEWBIO INDUSTRY, 308102) on a real-time PCR machine (Bio-Rad). The primers for Bcl-xL and reference Actin used in this study are listed in Supplementary Table 2.
2.18. Statistical analysis
For quantitative analyses presented as histograms, values were obtained from at least three independent experiments and are expressed as the mean ± SEM. Statistical significances were determined by a Student's t-test. p < 0.05 was considered as statistically significant. All statistical analyses were performed with GraphPad Prism software.
3. Results
3.1. Iron deficiency induces FUNDC1-dependent mitophagy
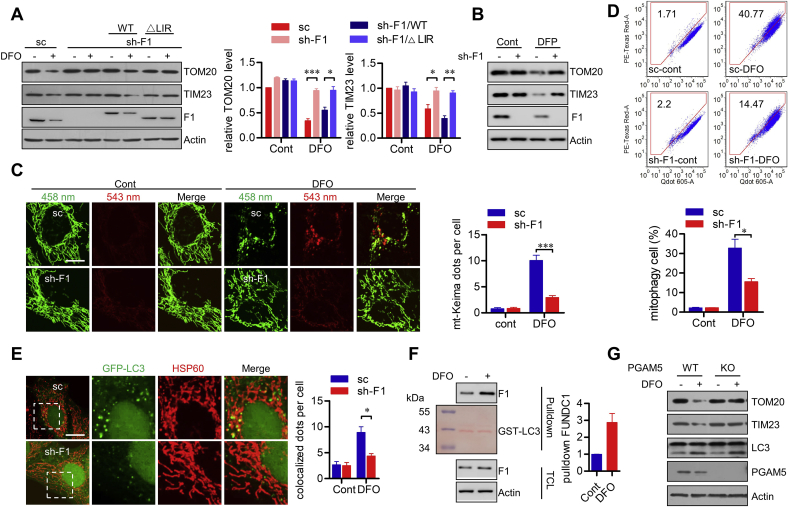
Previous research showed that iron depletion induced by iron chelator initiates mitophagy, which is independent on Pink1/Parkin pathway [18]. As iron depletion mimics chemical hypoxia [25], and reversely, hypoxia could regulate iron homeostasis [26]. We thus addressed whether iron deficiency induced mitophagy is dependent on mitophagy receptor FUNDC1, which is crucial for hypoxia induced mitophagy. Exposure of cells to DFO, a widely used iron chelator, induced the degradation of mitochondrial marker proteins (TOM20 and TIM23) (Fig. 1A), similar to previous report [18]. DFO-induced mitochondrial protein degradation was sharply suppressed in stable FUNDC1 knockdown (sh-F1) cells. Re-introduction of WT FUNDC1, but not the LIR deleted mutant (△LIR, which loses the interaction with LC3 and thus is a mitophagy defective mutant), could restore the degradation of TOM20 and TIM23 upon DFO exposure (Fig. 1A). Another widely used iron chelator, DFP, also induced degradation of mitochondrial proteins. Similarly, FUNDC1 knockdown suppressed DFP induced degradation of these mitochondrial proteins (Fig. 1B). We further measured the mitophagy activity by employing mt-Keima assay. Keima is a pH-sensitive fluorescent protein and mt-Keima is targeted into the mitochondria by fusing a tandem repeats of the COX VIII presequence to Keima. A low ratio of mt-Keima-derived fluorescence (543 nm/458 nm) indicates a neutral environment, while a high ratio indicates an acidic pH, which shows the mitochondria engulfed by lysosomes [24]. In scramble (sc) cells that stably express mt-Keima, increase of multiple round structures with a high 543 nm signal formed following DFO challenge, while FUNDC1 knockdown significantly decreased the formation of mt-Keima puncta under the identical condition (Fig. 1C). Additionally, the mt-Keima activities were also detected by flow cytometry analysis. Following DFO treatment, the percentage of mitophagic cells with higher red/green fluorescence increased, while FUNDC1 knockdown leads to an obvious decrease in this percentage (Fig. 1D). Furthermore, knockdown of FUNDC1 suppressed the co-localization of GFP-LC3 puncta with mitochondria during iron chelation (Fig. 1E). These data thus collaboratively suggest an indispensable role of FUNDC1 in DFO-induced mitophagy. FUNDC1 dephosphorylation by mitochondrial phosphatase PGAM5 is crucial for FUNDC1 activation and the enhanced FUNDC1-LC3 interaction. GST pulldown assays showed that DFO treatment increased the interaction between FUNDC1 and LC3 (Fig. 1F), further demonstrating FUNDC1 activation upon DFO treatment. Also, knockout of PGAM5 suppressed DFO-induced mitochondrial protein degradation (Fig. 1G). Taken together, these data show that iron deprivation results in the activation of FUNDC1 by PGAM5, leading to the enhanced FUNDC1-LC3 interaction and facilitating FUNDC1-dependent mitophagy.
Fig. 1.
Iron deficiency induces FUNDC1-dependent mitophagy. (A) HeLa cells stably expressed scramble control (sc) shRNA, FUNDC1 shRNA (sh-F1), or sh-F1 together with shRNA-resistant WT or △LIR mutated FUNDC1, were treated with DFO for 24 h. Cells were then harvested for immunoblotting analysis. Relative levels of TOM20 and TIM23 are shown in the right histograms. Data represent the mean ± SEM of three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001. (B) sc or sh-F1 stably expressed HeLa cells were treated with DFP for 24 h. Cells were then harvested for immunoblotting analysis. (C) sc or sh-F1 stably expressed HeLa cells expressed mt-Keima were treated with DFO for 24 h, and then imaged with 458 nm or 543 nm light excitation using a LSM 510 Zeiss confocal microscope. Representative images are presented. Scale bar, 10 μm. The mt-Keima dots per cell were counted and shown in the right histogram. Data represent the mean ± SEM of three independent experiments, each with >200 cells counted per condition. ***p < 0.001. (D) Cells as in (C) were treated with DFO for 24 h, signals from green and red channels were detected by flow cytometry. The percentage of mitophagic cells with higher red/green fluorescence was showed in the below histograms as mean ± SEM of three experiments. *p < 0.05. (E) Cells as in (B) were transiently transfected with GFP-LC3 and then treated with DFO. Cells were then fixed, stained by α-HSP60 antibody, and imaged with a LSM 510 Zeiss confocal microscope. Representative images are presented. Scale bar, 10 μm. The HSP60-LC3 colocalized dots per cell were counted and shown in the right histograms. Data represent the mean ± SEM of three independent experiments, each with >200 cells counted per condition. *p < 0.05. (F) HeLa cells were challenged with DFO for 12 h, and cell lysates were prepared and incubated with purified GST-LC3 fusion protein immobilized on sepharose beads. The total cell lysates (TCL) and the bead-associated fractions (Pulldown) were subjected to immunoblotting analysis. Ponceau S staining was used to visualize GST-LC3 bands. Relative levels of FUNDC1 in pulldown samples are shown in the right histograms. Data represent the mean ± SEM of three independent experiments. (G) WT and PGAM5 KO HeLa cells were treated with DFO for 24 h. Cells were then harvested and subjected for immunoblotting analysis.
3.2. Defective ISCs assembly activates mitophagy
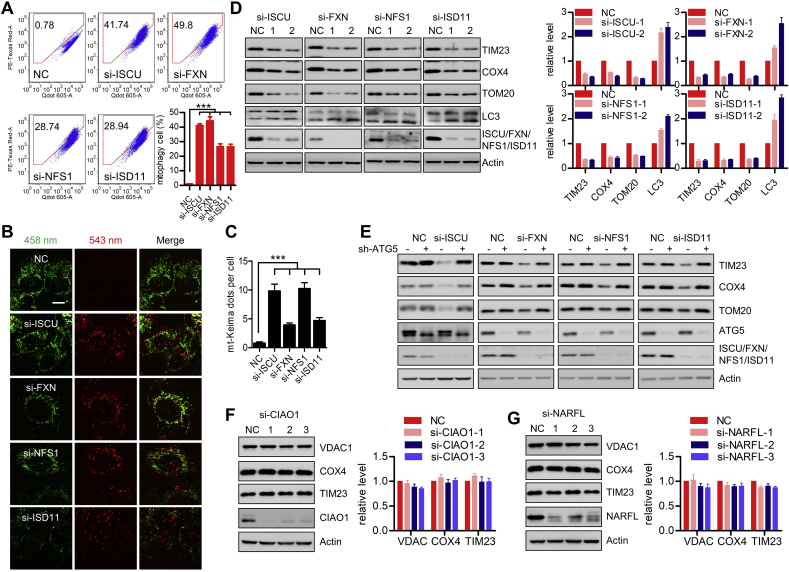
Cytosolic iron is imported into mitochondria for the biosynthesis of ISCs. The core components of the mitochondrial ISCs machinery are essential for ISCs assembly and thus essential for the maturation of mitochondrial, cytosolic and nuclear ISCs-containing proteins [27]. Given the significance of ISCs biogenesis in mitochondrial functions and iron homeostasis, we hypothesized that perturbing the mitochondrial ISCs assembly machinery may impair mitochondrial integrity. To test this, we knocked down the core components of mitochondrial ISCs assembly (including the dedicated scaffold protein ISCU, the iron donor molecule Frataxin (FXN), the cysteine desulfurase NFS1, and the small partner protein ISD11), and evaluated the mitophagy activity by mt-Keima assay. In cells that stably express mt-Keima, the percentage of mitophagic cells with higher red/green fluorescence increased following the transfection of these siRNA targeting the four genes involved in mitochondrial ISCs assembly (ISCU, FXN, NFS1, ISD11), which is consistent to the previous report that FXN silencing triggers mitophagy in worms and in mammalian cells (Fig. 2A). Additionally, increased formation of multiple round structures with a higher 543 nm signal was observed by using confocal microscope following transfection of these siRNAs (Fig. 2B and C). Furthermore, we found that transfection of siRNAs targeting each of these crucial components induced decrease of mitochondrial marker proteins (including TIM23, COX4 and TOM20), accompanied with upregulation of LC3-II conversion (Fig. 2D). The similar decrease of mitochondrial proteins was also observed in HepG2 and U2OS cells transfected of these siRNAs (Figs. S1A–B). Knockdown of ATG5, an essential component involved in autophagosome formation, diminished the degradation of mitochondrial proteins upon these siRNAs transfection, further indicating an involvement of autophagy/mitophagy in this degradation of mitochondrial proteins (Fig. 2E).
Fig. 2.
Defective mitochondrial ISCs assembly activates mitophagy. (A) mt-Keima stable HeLa cells were transfected with siRNAs targeting ISCU, FXN, NFS1 or ISD11, or the NC siRNA. After 72 h of transfection, the cells were collected and signals from green and red channels were detected by flow cytometry. The percentage of mitophagic cells with higher red/green fluorescence was shown in the right below histograms as mean ± SEM of three experiments. ***p < 0.001. (B) Cells as in (A) were imaged with 458 nm (shown in green) or 543 nm (shown in red) light excitation using a LSM 510 Zeiss confocal microscope. Representative images are presented. Scale bar, 10 μm. (C) Graph showing the number of mt-Keima dots per cell in (B). Data represent the mean ± SEM of three independent experiments, each with >200 cells counted per condition. ***p < 0.001. (D) HeLa cells were transfected with siRNAs targeting ISCU, FXN, NFS1 or ISD11, or the NC siRNA. After 72 h of transfection, the cells were harvested for immunoblotting analysis. The relative levels of indicated bands are shown in the right histograms. Data represent the mean ± SEM of three independent experiments. (E) ATG5 stable knockdown HeLa cells and the control cells were transfected with siRNAs targeting ISCU, FXN, NFS1 or ISD11, or the NC siRNA. After 72 h of transfection, the cells were harvested for immunoblotting analysis. (F–G) HeLa cells were transfected with siRNAs targeting CIAO1 (F) or NARFL (G). After 72 h of transfection, the cells were harvested for immunoblotting. The relative levels of indicated bands are shown in the right histograms. Data represent the mean ± SEM of three independent experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Transfection of these siRNAs failed to decrease the mRNA level of mitochondrial genes including Cox4, Tim23 and Tom20 (Fig. S1C). Furthermore, these siRNAs failed to affect the protein level of mitochondrial biogenesis regulators (NRF1, PGC1α and TFAM) (Fig. S1D), excluding the possibilities of transcription repression and impaired mitochondrial biogenesis upon these siRNAs transfection. ISCs could also be synthesized in cytosol by the so-called cytosolic ISCs assembly (CIA) mechanism [28], we checked whether CIA is also coupled with mitophagy and found that silencing two key CIA components, CIAO1 and NARFL [28] has no obvious effect on the levels of mitochondrial marker proteins (Fig. 2F and G). Taken together, these data suggest that defective mitochondrial ISCs assembly, but not CIA mechanism, activates mitophagy.
3.3. FUNDC1 is indispensable for mitophagy induced by defective ISCs assembly
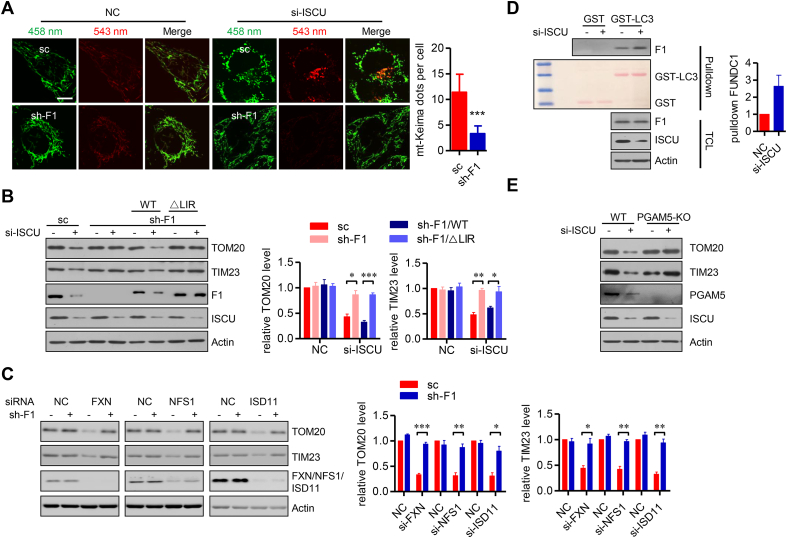
ISCs are initially assembled on the highly conserved scaffold protein ISCU, which coordinates the iron donor FXN and the sulfur donor NFS1-ISD11 complex inside the mitochondrial matrix, and then transfers newly synthesized ISCs to the recipient apo-proteins. We first addressed the mechanism underlying the induction of mitophagy by ISCU knockdown. Unlike the mitochondrial uncoupler FCCP, transfection of siRNA targeting ISCU did not induce translocation of GFP-Parkin onto mitochondria, as revealed by immunofluorescence assays and mitochondrial fractionation assays (Figs. S1E–F). It is known that Pink1/Parkin-dependent mitophagy is triggered by the loss of mitochondrial membrane potential [29]. However, ISCU knockdown could not decrease the mitochondrial membrane potential (Fig. S1G). This excludes an involvement of the Pink1/Parkin axis in ISCU knockdown-induced mitophagy. In contrast, FUNDC1 knockdown decreased the ISCU knockdown-induced delivery of mitochondria to lysosomes, as shown by the mt-Keima assay (Fig. 3A). Furthermore, FUNDC1 knockdown prevented the degradation of mitochondrial proteins upon ISCU siRNA transfection, which could be restored by re-introduction of WT FUNDC1, but not the △LIR mutant, further suggesting a critical role of FUNDC1 in this si-ISCU induced mitophagy (Fig. 3B). Similarity, FUNDC1 knockdown also diminished the degradation of mitochondrial proteins upon transfection of siRNAs targeting FXN, NFS1 or ISD11, as revealed by the western blotting analysis (Fig. 3C).
Fig. 3.
FUNDC1 is indispensable for mitophagy induced by defective mitochondrial ISCs assembly. (A) HeLa cells stably expressed sc shRNA and mt-Keima, or sh-F1 and mt-Keima were transfected with si-ISCU or the NC siRNA. After 72 h of transfection, the cells were imaged with 458 nm (shown in green) or 543 nm (shown in red) light excitation using a LSM 510 Zeiss confocal microscope. Representative images are presented. Scale bar, 10 μm. The mt-Keima dots per cell were counted and shown in the right histogram. Data represent the mean ± SEM of three independent experiments, each with >200 cells counted per condition. ***p < 0.001. (B) HeLa cells stably expressed sc, sh-F1, or sh-F1 with shRNA-resistant WT or △LIR mutated FUNDC1, were transfected with si-ISCU or the NC siRNA. After 72 h of transfection, the cells were harvested and analyzed by immunoblotting. The relative levels of indicated bands are shown in the right histograms. Data represent the mean ± SEM of three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001. (C) HeLa cells stably expressed sc shRNA or sh-F1 were transfected with the indicated siRNAs or the NC siRNA. After 72 h of transfection, the cells were harvested and analyzed by immunoblotting. The relative levels of indicated bands are shown in the right histograms. Data represent the mean ± SEM of three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001. (D) HeLa cells were transfected with si-ISCU or the NC siRNA for 48 h, and the cell lysates were prepared and incubated with purified GST-LC3 fusion protein or GST protein immobilized on sepharose beads. The total cell lysates (TCL) and the bead-associated fractions (Pulldown) were subjected to SDS-PAGE for immunoblotting analysis. Ponceau S staining was used to visualize GST and GST-LC3 bands. The relative levels of FUNDC1 bands in pulldown samples are shown in the right histograms. Data represent the mean ± SEM of three independent experiments. (E) WT and PGAM5 KO HeLa cells were transfected with si-ISCU or the NC siRNA. After 72 h of transfection, the cells were harvested and analyzed by immunoblotting. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The mitochondrial phosphatase PGAM5, which is known to dephosphorylate FUNDC1 at Ser13, and thus enhance the interaction of FUNDC1 and LC3, is indispensable for FUNDC1-dependent mitophagy [8]. GST pulldown assays showed that ISCU knockdown enhanced the interaction between FUNDC1 and LC3 (Fig. 3D). Moreover, PGAM5 knockout decreased the degradation of mitochondrial proteins induced by ISCU knockdown (Fig. 3E). Altogether, these data demonstrate that FUNDC1 is indispensable for mitophagy execution induced by defective mitochondrial ISCs assembly.
3.4. IRP1 senses iron stress and cellular ISCs status to induce mitophagy
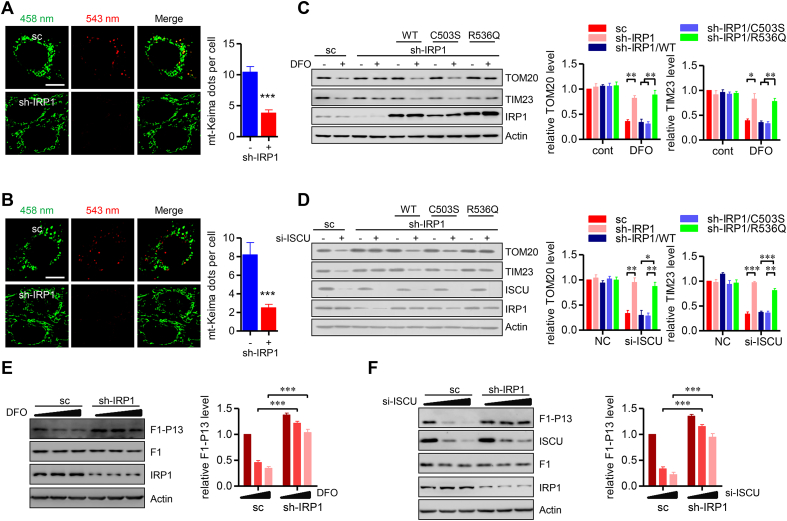
Cellular iron and ISCs levels are sensed by iron regulatory protein 1/2 (IRP1/2) [30]. We next explored the potential role of IRP1/2 in sensing the cellular iron level to modulate FUNDC1-dependent mitophagy. We found that knockdown of IRP1 disturbed mitophagy, as evidenced by the mt-Keima assay, when cells were challenged with DFO (Fig. 4A) or ISCU knockdown (Fig. 4B). Western blotting assays revealed that knockdown of IRP1 suppressed DFO and ISCU knockdown induced degradation of mitochondrial marker proteins (Fig. 4C and D). Previous studies have showed that the C503S IRP1 mutant possesses constitutive IRE-binding activity with limited aconitase activity regardless of the cellular iron level, while the R536Q mutant has decreased IRE-binding activity. We thus addressed the question of whether the iron regulatory activity/IRE-binding activity or aconitase activity of IRP1/ACO1 is responsible for mitophagy regulation during iron stresses. Reintroduction of WT IRP1 and the C503S mutant into IRP1 stable knockdown cells restored the degradation of mitochondrial proteins induced by DFO challenge and ISCU knockdown (Fig. 4C and D). In striking contrast, reintroduction of the R536Q mutant, which has minimal IRE binding activity, fails to do so, suggesting that the iron regulatory activity/IRE-binding activity of IRP1/ACO1 is responsible for mitophagy regulation (Fig. 4C and D). Short time exposure of DFO (12 h) or si-ISCU transfection (36 h) led to FUNDC1 dephosphorylation at Ser13 (Fig. 4E and F). IRP1 knockdown could maintain the FUNDC1 phosphorylated level under the identical conditions (Fig. 4E and F). In addition, IRP2 is another well-characterized iron sensor for maintaining iron homeostasis. Depletion of IRP2 showed a defective mitophagy induced by iron chelation, as evidenced by the mt-Keima assay and western blotting assays (Figs. S2A–B). Altogether, these data suggest that IRPs are functionally important for FUNDC1 activation and mitophagy during iron stresses.
Fig. 4.
IRP1 is indispensable for iron stresses induced mitophagy. (A and B) HeLa cells stably expressed sc shRNA or sh-IRP1 were transiently transfected with mt-Keima. Following 24 h of expression, cells were treated with DFO for another 24 h (A), or transfected with si-ISCU for another 72 h (B), and then cells were imaged with 458 nm (shown in green) or 543 nm (shown in red) light excitation using a LSM 510 Zeiss confocal microscope. Representative images are presented. Scale bar, 10 μm. Graph at right showing the number of mt-Keima puncta per cell. Data represent the mean ± SEM of three independent experiments, each with >200 cells counted per condition. ***p < 0.001. (C and D) HeLa cells stably expressed sc shRNA or sh-IRP1 were transfected with WT IRP1, or C503S, R536Q mutant, then treated with DFO for another 24 h (C), or transfected with si-ISCU for another 72 h (D). The cells were then harvested and analyzed by immunoblotting. The relative levels of indicated bands are shown in the right histograms. Data represent the mean ± SEM of three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001. (E and F) HeLa cells stably expressed sc shRNA or sh-IRP1 were exposed to 100 μM or 250 μM DFO for 12 h (E), or transfected with 10 pmol or 30 pmol si-ISCU or the NC siRNA for 48 h (F). Cells were harvested for blotting phosphorylated FUNDC1 level at Ser13 (F1–P13). The relative levels of F1–P13 to F1 are shown in the right histograms. Data represent the mean ± SEM of three independent experiments. Data represent the mean ± SEM of three independent experiments. ***p < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We next aimed to understand whether IRP1 specifically acts on mitophagy, or just as a modulator of general autophagy. Ectopic expression of IRP1 failed to induce the formation of LC3 puncta in GFP-LC3 stable HeLa cells (Fig. S3A). Additionally, IRP1 knockdown failed to influence autophagic flux induced by EBSS (starvation medium) or DFO, as revealed by western blotting (Figs. S3B–C). Furthermore, IRP1 knockdown had limited effect on Parkin mediated mitophagy, as shown by the equivalent translocation of Parkin onto mitochondria upon FCCP exposure (Fig. S3D), while IRP1 knockdown obviously decreased hypoxia induced mitophagy, as shown by the mt-Keima assay (Fig. S3E). These data thus suggested that IRP1 specifically acts on FUNDC1 mediated mitophagy (iron stresses induced and hypoxia induced mitophagy) without any apparent effect on bulk autophagy regulation.
3.5. IRP1 targets Bcl-xL mRNA to modulate mitophagy
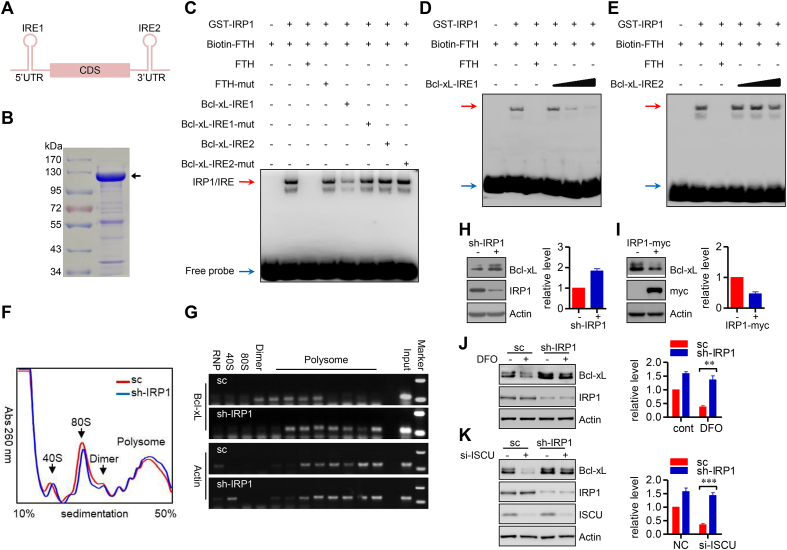
IRP1 modulates expression of iron metabolism associated genes mainly by binding to IREs in the UTRs of target mRNAs [31]. Based on this information, we hypothesized that some of the mitophagy associated genes may have such IRE sequences. Using SIREs software (http://ccbg.imppc.org/sires/index.html), we screened autophagy genes (Atg5, Atg7 and ULK1) and mitophagy-specific genes (FUNDC1, PGAM5, Bcl-xL, Nix, Pink1 and Parkin). We found that Bcl-xL mRNA contains two putative IREs, one in the 5′UTR (putative IRE1) and the other in the 3′UTR (putative IRE2) (Fig. 5A). To investigate whether these are functional IREs, we purified GST-IRP1 fusion protein (Fig. 5B) and performed competitive RNA electrophoretic mobility shift assays (REMSAs). A biotin-labeled FTH IRE probe was incubated with purified GST-IRP1 protein with or without an unlabeled IRE competitor. As a positive control, the unlabeled wild-type (WT) FTH IRE (10 folds to the biotin-labeled FTH IRE probe) efficiently competed with the biotin-labeled FTH IRE for binding to IRP1, whereas a mutated competitor as a negative control did not. The high dose of unlabeled Bcl-xL IRE1 (250 folds to the biotin-labeled FTH IRE probe) partially competed with the biotin-labeled FTH IRE for binding to IRP1, while the unlabeled Bcl-xL IRE2 (250 folds to the biotin-labeled FTH IRE probe) failed to compete. In the meantime, neither of the mutants could compete (Fig. 5C). Moreover, the competition of Bcl-xL IRE1 with the biotin-labeled FTH IRE is dose dependent (Fig. 5D), while Bcl-xL IRE2 failed to compete, even at higher dose (500 folds to the biotin-labeled FTH IRE probe) (Fig. 5E). This suggests that the IRE in the 5′UTR is functional and responsible for the IRP1 binding to Bcl-xL mRNA, although not so strong as the FTH IRE.
Fig. 5.
IRP1 targets to Bcl-xL mRNA. (A) A schematic showing two putative IREs in Bcl-xL mRNA, one in the 5′UTR and the other in the 3′UTR. (B) Coomassie blue staining of the purified GST-IRP1 recombinant protein. The black arrowhead indicates the GST-IRP1 fusion protein, while other bands are non-specific or degraded bands. (C) Biotin-labeled FTH IRE probe was incubated with purified GST-IRP1 protein, in presence or absence of 250-fold unlabeled competitor probe as indicated. The RNA-protein was subjected into nondenaturing PAGE, and visualized with an ECL kit. The red arrow indicates IRP1/IRE complex, and the blue arrow shows the free probe. (D–E) Biotin-labeled FTH IRE probe was incubated with purified GST-IRP1 protein, in presence of unlabeled FTH IRE, or increasing amount (100-fold, 250-fold and 500-fold) of Bcl-xL IRE1 (D) or IRE2 (E) as indicated. The RNA-protein complex was analyzed by REMSA. The red arrow indicates IRP1/IRE complex, and the blue arrow shows the free probe. (F) HeLa cells stably expressed sc shRNA or sh-IRP1 were lysed in polysome lysis buffer, and the cell lysates were separated by sucrose density gradient centrifugation. Gradients were fractionated and the optical density at 260 nm was continuously recorded using a fractionator. (G) Distribution of Bcl-xL mRNAs across the density gradients from (F) was determined by semi-quantitative RT-PCR. Actin mRNA was used as the control. (H) Cell lysates from HeLa cells transfected of NC or si-IRP1 were subjected to SDS-PAGE and analyzed by immunoblotting. The relative levels of Bcl-xL are shown in the right histograms. Data represent the mean ± SEM of three independent experiments. (I) HeLa cells were transfected with IRP1. After 24 h of transfection, cells were harvested and analyzed by immunoblotting. The relative levels of Bcl-xL are shown in the right histograms. Data represent the mean ± SEM of three independent experiments. (J–K) HeLa cells stably expressed sc shRNA or sh-IRP1 treated by DFO (J), or transfected with siRNA targeting to ISCU (K), were immunoblotted for Bcl-xL level. The relative levels of Bcl-xL are shown in the right histograms. Data represent the mean ± SEM of three independent experiments. **p < 0.01; ***p < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Next, we wished to understand the biological significance of the IRP1 binding to Bcl-xL mRNA. As the IRE responsible for the IRP1-Bcl-xL mRNA interaction is in the 5′UTR, we hypothesized that IRP1 may modulate the translational efficiency of Bcl-xL. To directly verify this, we sedimented polysomes from IRP1 stable knockdown cells and the control cells in sucrose density gradients to separate mRNAs that are efficiently translated (associated with heavy polysomes) from those that are poorly translated (associated with light polysomes). IRP1 knockdown did not induce an overall change of polysome pattern (Fig. 5F). Indeed, IRP1 knockdown accelerated translation of Bcl-xL mRNAs, as illustrated by a shift of Bcl-xL mRNA toward heavy polysomes, whereas the control Actin mRNA was unaffected (Fig. 5G), suggesting an enhanced translation of Bcl-xL mRNA in IRP1 knockdown cells. We also examined the possibility that the IRP1-Bcl-xL mRNA interaction modulates the mRNA stability. The abundance of Bcl-xL mRNA was unaffected by overexpression of IRP1 or knockdown of IRP1 (Figs. S4A–B). Utilizing the actinomycin D chase assay, we found that overexpression of IRP1, as well as knockdown of IRP1, failed to change the Bcl-xL mRNA stability (Figs. S4C–D). In contrast, knockdown of IRP1 increased the level of Bcl-xL protein (Fig. 5H). Ectopic expression of IRP1 decreased the level of Bcl-xL protein (Fig. 5I). Because IRP1 knockdown impairs mitophagy, we also addressed whether impaired mitophagy is account for the elevated Bcl-xL in IRP1 knockdown cells or not. Actually, the decrease of Bcl-xL in ATG5 knockdown cells was observed under iron stresses, including DFO challenge and ISCU knockdown (Figs. S4E–F), excluding the possibility of autophagy or mitophagy involved in modulating Bcl-xL level by IRP1. Next, we wanted to address the potential role of IRP1 in regulating Bcl-xL under iron stressed condition. DFO treatment and ISCU knockdown decreased Bcl-xL level. However, IRP1 knockdown could stabilize Bcl-xL, even under iron stressed condition (Fig. 5J and K). Altogether, these data suggest that IRP1 binds to the IRE located in the 5′UTR of the Bcl-xL mRNA, suppresses the translational efficiency and thus downregulates expression of the Bcl-xL protein.
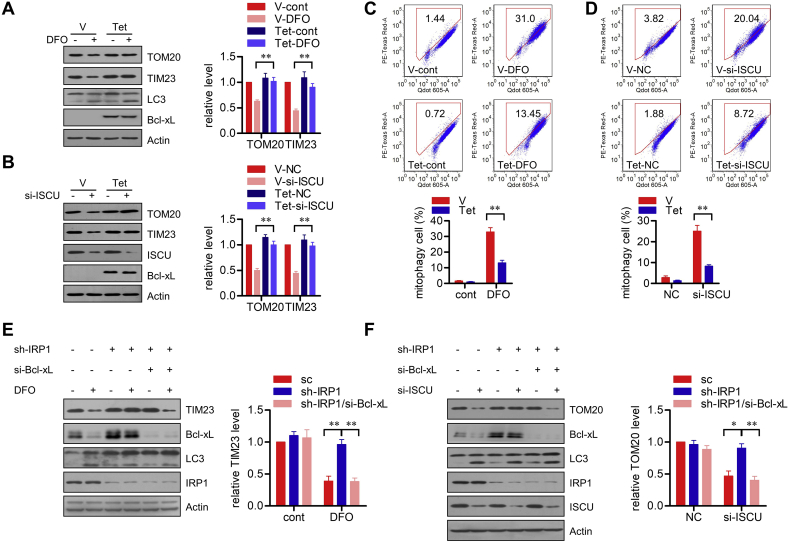
We previously identified that Bcl-xL acts as an intrinsic inhibitory molecule for FUNDC1-dependent mitophagy by inhibiting the phosphatase activity of PGAM5, and thus suppressing the dephosphorylation and activation of FUNDC1. Knockdown of Bcl-xL can't directly initiate, but can sensitize hypoxia induced mitophagy [22]. We thus prompted to address the potential role of Bcl-xL in iron stresses-induced mitophagy. Tetracycline-inducible Bcl-xL expression suppressed the degradation of mitochondrial proteins induced by DFO exposure or ISCU knockdown (Fig. 6A and B). Furthermore, the mt-Keima assay showed that tetracycline-inducible Bcl-xL expression reduces the percentage of mitophagic cells during iron depletion or ISCU knockdown (Fig. 6C and D). This suggests that iron stresses-induced mitophagy, like hypoxia-induced mitophagy, can also be navigated by Bcl-xL. Moreover, knockdown of Bcl-xL in IRP1 stable knockdown cells reversed the degradation of mitochondrial proteins induced by DFO exposure or ISCU knockdown (Fig. 6E and F), suggesting that Bcl-xL acts downstream of IRP1 in mitophagy regulation. In conclusion, our results thus establish that IRP1 senses iron stresses to navigate mitophagy by modulating Bcl-xL translation.
Fig. 6.
IRP1 targets Bcl-xL to modulate mitophagy. (A and B) HeLa cells with stable tetracycline (Tet)-induced Bcl-xL expression were pre-treated with Tet or the vehicle (V) for 6 h, and then incubated with DFO for 24 h (A) or transfected with si-ISCU or NC siRNA for 72 h (B). The cells were harvested and analyzed by immunoblotting. The relative levels of indicated bands are shown in the right histograms. Data represent the mean ± SEM of three independent experiments. **p < 0.01. (C and D) HeLa cells stably co-expressing mt-Keima and Tet-induced Bcl-xL were pre-treated with Tet or the vehicle (V) for 6 h, then incubated with DFO for 24 h (C) or transfected with si-ISCU or NC siRNA for 72 h (D). The cells were collected and signals from green and red channels were detected by flow cytometry. The percentage of mitophagic cells with higher red/green fluorescence was showed in the below histograms as mean ± SEM from three experiments; **p < 0.01. (E) HeLa cells stably expressed sc shRNA or sh-IRP1 were transiently transfected with si-Bcl-xL or NC siRNA. After 48 h of transfection, the cells were challenged with DFO for 24 h. The cells were harvested and analyzed by immunoblotting. The relative levels of TIM23 are shown in the right histograms. Data represent the mean ± SEM of three independent experiments. **p < 0.01. (F) HeLa cells stably expressed sc shRNA or sh-IRP1 were transiently transfected with si-Bcl-xL accompanied with si-ISCU or the NC siRNA. After 72 h of transfection, the cells were harvested for immunoblotting analysis. The relative levels of TOM20 are shown in the right histograms. Data represent the mean ± SEM of three independent experiments. *p < 0.05; **p < 0.01.
Mitophagy is now considered as one of the major mechanisms for mitochondrial quality control. Indeed, DFO treatment had limited effect on the production of mitochondrial ROS in the control cells, whereas IRP1 knockdown led to an increase in mitochondrial ROS production in response to DFO exposure (Fig. S5A). Excessive production of ROS could lead to protein oxidation and result in the formation of protein carbonyl groups on protein side chains, which could be detected by a commercial Oxyblot kit. DFO treatment resulted in a marginal increase of carbonyl groups of mitochondrial proteins, whereas IRP1 knockdown gave rise to a significant increase in this carbonylation of mitochondrial proteins (Fig. S5B). This is similar to FUNDC1, as FUNDC1 knockdown also resulted in the increased production of mitochondrial ROS (Fig. S5C), as well as the elevated carbonylation of mitochondrial proteins (Fig. S5D). These findings suggest that both FUNDC1 and IRP1, which are important modulators for mitophagy execution in response to the altered iron metabolism, are crucial for the mitochondrial redox response.
4. Discussion
Previous work has shown that iron depletion triggers mitophagy via a Pink1/Parkin-independent pathway in mammalian cells [18] and in C. elegans [32]. In the current study, we provide conclusive evidence to show that defective mitochondrial ISCs biogenesis, either by knockdown of mitochondrial ISCs assembly machinery associated components or by chelator mediated iron depletion, initiates FUNDC1-mediated mitophagy. We further suggest that ISCs serve as intrinsic and physiological signaling molecules to support Bcl-xL translation, and disturbed mitochondrial ISCs biogenesis leads to the suppression of Bcl-xL translation via the iron sensor IRP1, and the sensitization of mitophagy activation. Our results thus disclose a novel mechanism for coupling mitophagy with iron homeostasis, and a retrograde signaling pathway for mitophagy activation in response to ISCs status.
Mitochondria are one of the major sites for iron metabolism. Cellular iron is transported into mitochondria via the mitochondrial transporter mitoferrin1/2 [33], and is then utilized for ISCs biogenesis by the ISCs assembly machinery [15,16]. Because of the chemical reactivities of iron and sulfur, ISCs are versatile cofactors that accept or donate electrons for mitochondrial respiration as well as catalyzing other cellular enzymatic reactions [31]. ISCs can also be transported outside of mitochondria via the transporter ABCB7 [34,35] where they then bind to IRP1 and other enzymes for iron metabolism and other cellular processes. It is thus not surprising that knockdown of mitochondrial ISCs assembly machinery associated components leads to defective ISCs biogenesis and mitophagy. This is in agreement with previous observations suggested that FXN knockdown could induce mitophagy in C. elegans and in mammalian cells [36]. Furthermore, previous siRNA studies showing that depletion of mammalian ISCs assembly factors inactivates various metabolic enzymes including aconitase and xanthine oxidase, thereby leading to reprogramming of cellular metabolism [37,38]. Our results argue that ISCs act as novel signaling molecules for mitophagy regulation, and extend the physiological functions of ISCs beyond iron metabolism, energy metabolism and other cellular processes. Clinical studies have revealed that defects in mitochondrial ISCs assembly give rise to severe and even fatal disorders, and mutations of associated genes cause human diseases that manifest distinctive combinations of tissue-specific impairments [16]. Metabolic reprogramming and accumulation of abnormal mitochondria are commonly observed in ISCs-related disorders in animal models and diseased tissues isolated from human patients [[39], [40], [41], [42], [43], [44]]. For example, mutations of the ISCU gene, such as splice mutations which are predicted to decrease protein expression, or the missense mutation G50E, lead to ISCU myopathy, with main clinical features of severe exercise intolerance and mitochondrial dysfunction [[45], [46], [47]]. Moreover, it has been widely understood the tight association between iron metabolism and cancers. Excessive dietary iron intake statistically elevates risks of several malignancies, including esophageal cancer, colorectal cancer, liver cancer and lung cancer [48]. Cancer cells addict high level of intracellular iron and thus shift iron metabolism toward enhanced iron uptake and decelerated iron export [49]. Iron chelators perform highly potent antitumor activity [50]. Specifically, ISCU was reported to be p53-reponsiable and suppress liver tumor by supporting ISCs assembly and maintaining cellular iron homeostasis [51]. It is reasonable that defective ISCs assembly leading to disturbed mitochondrial bioenergetics results in Warburg effect in cancer cells [52]. Although several studies reported that mitophagy limits tumorigenesis and suppresses tumor growth by maintaining mitochondrial integrity [53], further work will be directed towards understanding if targeting induction of FUNDC1-dependent mitophagy or its combinations with iron chelators or other chemotherapy could be used for therapeutic design of cancers.
IRP1 senses and maintains cellular iron levels via reversible binding to ISCs and interaction with IREs of target mRNAs, which mostly encode proteins involved in iron metabolism [54]. IRP1 activation was reported to preserve mitochondrial function and to dominate mitochondrial iron levels, as well as to maintain ISCs and heme biosynthesis [55]. We now show that IRP1 has an expanded function in mitophagy modulation by fine tuning Bcl-xL level and the FUNDC1 phosphorylation at Ser13. Knockdown of IRP1 disturbs iron stresses induced mitophagy, which could be restored by knockdown of Bcl-xL. Moreover, IRP1 associated mitophagy is involved in mitochondrial redox response. Knockdown of IRP1 leads to an increase in mitochondrial ROS production and oxidized mitochondrial proteins in response to iron depletion. This is parallel to FUNDC1, as knockdown of FUNDC1 results in a similar phenotype. These findings suggest that both FUNDC1 and IRP1 are important for mitophagy associated mitochondrial redox response in response to the altered cellular iron metabolism.
One of our major findings is that IRP1 modulates mitophagy activity by fine tuning the translational efficiency of Bcl-xL mRNA. In particular, IRP1 senses the cellular iron and ISCs levels, binds to the newly characterized IRE located in the 5′UTR of Bcl-xL mRNA, and thus suppresses the translational efficiency. The IRE in the 5′UTR of Bcl-xL mRNA is functional as evidenced by the competitive binding of IRP1 to FTH IRE, although not so strong as the FTH IRE, which further explains why previous genome-wide screening study didn't find any functional IRE in Bcl-xL mRNA [56]. IRP1 binding to the newly characterized IRE in the 5′UTR leads to a suppression of translation, but not changing in the mRNA stability, as evidenced by the polysome profiling assay, as well as by the negative correlation of protein levels between IRP1 and Bcl-xL. Knockdown of Bcl-xL in IRP1 defective cells restores mitophagy induced by DFO exposure and ISCU knockdown. This suggests that Bcl-xL functions downstream of IRP1 in mitophagy regulation in response to iron stresses. Bcl-xL was originally identified to inhibit apoptosis through its interaction with proapoptotic proteins such as Bax or other Bcl-2-family members [57,58]. Several other molecules such as the mitochondrial fission factor Drp1 [59] and the autophagy regulator Beclin1 [60] have been reported as Bcl-xL binding factors, linking Bcl-xL to other cellular processes including mitochondrial dynamics and autophagy. Bcl-xL was also found to regulate mitochondrial mass and mitochondrial energetics through interactions with the ATPase beta subunit [61]. These studies thus highlighted that Bcl-xL is a master regulator of mitochondrial integrity in response to distinct stresses including iron stresses, which largely overlap with the hypoxic signaling pathway as previously documented. Our results extend the function of Bcl-xL in iron metabolism and imply that the Bcl-xL regulates a wide array of mitochondrial behaviors in a coordinated fashion in response to iron stresses. Our delineation of the signaling mechanism between mitochondrial ISCs assembly and FUNDC1-dependent mitophagy may offer a new strategy for therapeutic intervention in diseases with aberrant mitochondrial ISCs biogenesis and abnormal iron homeostasis.
Author contributions
Hao Wu, Huifang Wei, Yan Qin, Lei Liu and Quan Chen designed the overall study; Hao Wu, Huifang Wei, Sheikh Arslan Sehgal, Di Zhang, Dejiu Zhang, and Xiaohui Wang performed research; Hao Wu, Huifang Wei, Lei Liu and Quan Chen analyzed data and wrote the paper.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
The authors are grateful to Tong Zhao from Institute of Microbiology, Chinese Academy of Sciences, for her technical support of flow cytometry assay. This research was supported by the Natural Science Foundation of China (31790404, 31520103904), the Ministry of Sciences and Technology (2016YFA0500201) to Quan Chen; the Natural Science Foundation of China (91854105, 31671446) to Lei Liu; and the Fundamental Research Funds for the Central Universities (Project NO. 2662020DKPY009) to Hao Wu.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101661.
Contributor Information
Hao Wu, Email: whao.1988@mail.hzau.edu.cn.
Lei Liu, Email: liulei@ioz.ac.cn.
Quan Chen, Email: chenq@ioz.ac.cn.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Lemasters J.J. Perspective - selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 2.Jin S.M., Lazarou M., Wang C., Kane L.A., Narendra D.P., Youle R.J. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.S., Saiki S., Kawajiri S., Sato F., Kimura M., Komatsu M., Hattori N., Tanaka K. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazarou M., Sliter D.A., Kane L.A., Sarraf S.A., Wang C., Burman J.L., Sideris D.P., Fogel A.I., Youle R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heo J.M., Ordureau A., Paulo J.A., Rinehart J., Harper J.W. The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol. Cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J., Ney P.A. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu L., Feng D., Chen G., Chen M., Zheng Q., Song P., Ma Q., Zhu C., Wang R., Qi W., Huang L., Xue P., Li B., Wang X., Jin H., Wang J., Yang F., Liu P., Zhu Y., Sui S., Chen Q. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 8.Chen G., Han Z., Feng D., Chen Y., Chen L., Wu H., Huang L., Zhou C., Cai X., Fu C., Duan L., Wang X., Liu L., Liu X., Shen Y., Zhu Y., Chen Q. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol. Cell. 2014;54:362–377. doi: 10.1016/j.molcel.2014.02.034. [DOI] [PubMed] [Google Scholar]
- 9.Wei Y., Chiang W.C., Sumpter R., Jr., Mishra P., Levine B. Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell. 2017;168:224–238 e210. doi: 10.1016/j.cell.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhujabal Z., Birgisdottir A.B., Sjottem E., Brenne H.B., Overvatn A., Habisov S., Kirkin V., Lamark T., Johansen T. FKBP8 recruits LC3A to mediate Parkin-independent mitophagy. EMBO Rep. 2017;18:947–961. doi: 10.15252/embr.201643147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murakawa T., Yamaguchi O., Hashimoto A., Hikoso S., Takeda T., Oka T., Yasui H., Ueda H., Akazawa Y., Nakayama H., Taneike M., Misaka T., Omiya S., Shah A.M., Yamamoto A., Nishida K., Ohsumi Y., Okamoto K., Sakata Y., Otsu K. Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat. Commun. 2015;6 doi: 10.1038/ncomms8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pieczenik S.R., Neustadt J. Mitochondrial dysfunction and molecular pathways of disease. Exp. Mol. Pathol. 2007;83:84–92. doi: 10.1016/j.yexmp.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Richardson D.R., Lane D.J.R., Becker E.M., Huang M.L.H., Whitnall M., Rahmanto Y.S., Sheftel A.D., Ponka P. Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. Proc. Natl. Acad. Sci. U.S.A. 2010;107:10775–10782. doi: 10.1073/pnas.0912925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braymer J.J., Lill R. Iron-sulfur cluster biogenesis and trafficking in mitochondria. J. Biol. Chem. 2017;292:12754–12763. doi: 10.1074/jbc.R117.787101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rouault T.A. Biogenesis of iron-sulfur clusters in mammalian cells: new insights and relevance to human disease. Dis. Model. Mech. 2012;5:155–164. doi: 10.1242/dmm.009019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rouault T.A., Tong W.H. Iron-sulfur cluster biogenesis and human disease. Trends Genet. 2008;24:398–407. doi: 10.1016/j.tig.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang D.L., Ghosh M.C., Rouault T.A. The physiological functions of iron regulatory proteins in iron homeostasis - an update. Front. Pharmacol. 2014;5:124. doi: 10.3389/fphar.2014.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen G.F., Toth R., James J., Ganley I.G. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 2013;14:1127–1135. doi: 10.1038/embor.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hentze M.W., Muckenthaler M.U., Andrews N.C. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 20.Bratic A., Larsson N.G. The role of mitochondria in aging. J. Clin. Invest. 2013;123:951–957. doi: 10.1172/JCI64125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mallikarjun V., Sriram A., Scialo F., Sanz A. The interplay between mitochondrial protein and iron homeostasis and its possible role in ageing. Exp. Gerontol. 2014;56:123–134. doi: 10.1016/j.exger.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 22.Wu H., Xue D.F., Chen G., Han Z., Huang L., Zhu C.Z., Wang X.H., Jin H.J., Wang J., Zhu Y.S., Liu L., Chen Q. The BCL2L1 and PGAM5 axis defines hypoxia-induced receptor-mediated mitophagy. Autophagy. 2014;10:1712–1725. doi: 10.4161/auto.29568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez M., Galy B., Muckenthaler M.U., Hentze M.W. Iron-regulatory proteins limit hypoxia-inducible factor-2 alpha expression in iron deficiency. Nat. Struct. Mol. Biol. 2007;14:420–426. doi: 10.1038/nsmb1222. [DOI] [PubMed] [Google Scholar]
- 24.Sun N., Yun J., Liu J., Malide D., Liu C., Rovira, Holmstrom K.M., Fergusson M.M., Yoo Y.H., Combs C.A., Finkel T. Measuring in vivo mitophagy. Mol. Cell. 2015;60:685–696. doi: 10.1016/j.molcel.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez M., Galy B., Muckenthaler M.U., Hentze M.W. Iron-regulatory proteins limit hypoxia-inducible factor-2alpha expression in iron deficiency. Nat. Struct. Mol. Biol. 2007;14:420–426. doi: 10.1038/nsmb1222. [DOI] [PubMed] [Google Scholar]
- 26.Peyssonnaux C., Zinkernagel A.S., Schuepbach R.A., Rankin E., Vaulont S., Haase V.H., Nizet V., Johnson R.S. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs) J. Clin. Invest. 2007;117:1926–1932. doi: 10.1172/JCI31370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rouault T.A., Tong W.H. Iron-sulphur cluster biogenesis and mitochondrial iron homeostasis. Nat. Rev. Mol. Cell Biol. 2005;6:345–351. doi: 10.1038/nrm1620. [DOI] [PubMed] [Google Scholar]
- 28.Sharma A.K., Pallesen L.J., Spang R.J., Walden W.E. Cytosolic iron-sulfur cluster assembly (CIA) system: factors, mechanism, and relevance to cellular iron regulation. J. Biol. Chem. 2010;285:26745–26751. doi: 10.1074/jbc.R110.122218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pickrell A.M., Youle R.J. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muckenthaler M.U., Galy B., Hentze M.W. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu. Rev. Nutr. 2008;28:197–213. doi: 10.1146/annurev.nutr.28.061807.155521. [DOI] [PubMed] [Google Scholar]
- 31.Lill R., Muhlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 32.Kirienko N.V., Ausubel F.M., Ruvkun G. Mitophagy confers resistance to siderophore-mediated killing by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 2015;112:1821–1826. doi: 10.1073/pnas.1424954112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw G.C., Cope J.J., Li L.T., Corson K., Hersey C., Ackermann G.E., Gwynn B., Lambert A.J., Wingert R.A., Traver D., Trede N.S., Barut B.A., Zhou Y., Minet E., Donovan A., Brownlie A., Balzan R., Weiss M.J., Peters L.L., Kaplan J., Zon L.I., Paw B.H. Mitoferrin is essential for erythroid iron assimilation. Nature. 2006;440:96–100. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- 34.Kispal G., Csere P., Prohl C., Lill R. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 1999;18:3981–3989. doi: 10.1093/emboj/18.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Csere P., Lill R., Kispal G. Identification of a human mitochondrial ABC transporter, the functional orthologue of yeast Atm1p. FEBS Lett. 1998;441:266–270. doi: 10.1016/s0014-5793(98)01560-9. [DOI] [PubMed] [Google Scholar]
- 36.Schiavi A., Maglioni S., Palikaras K., Shaik A., Strappazzon F., Brinkmann V., Torgovnick A., Castelein N., De Henau S., Braeckman B.P., Cecconi F., Tavernarakis N., Ventura N. Iron-starvation-induced mitophagy mediates lifespan extension upon mitochondrial stress in C. elegans. Curr. Biol. 2015;25:1810–1822. doi: 10.1016/j.cub.2015.05.059. [DOI] [PubMed] [Google Scholar]
- 37.Fosset C., Chauveau M.J., Guillon B., Canal F., Drapier J.C., Bouton C. RNA silencing of mitochondrial m-Nfs1 reduces Fe-S enzyme activity both in mitochondria and cytosol of mammalian cells. J. Biol. Chem. 2006;281:25398–25406. doi: 10.1074/jbc.M602979200. [DOI] [PubMed] [Google Scholar]
- 38.Tong W.H., Rouault T.A. Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metabol. 2006;3:199–210. doi: 10.1016/j.cmet.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 39.Saha P.P., Kumar S.K.P., Srivastava S., Sinha D., Pareek G., D'Silva P. The presence of multiple cellular defects associated with a novel G50E iron-sulfur cluster scaffold protein (ISCU) mutation leads to development of mitochondrial myopathy. J. Biol. Chem. 2014;289:10359–10377. doi: 10.1074/jbc.M113.526665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kollberg G., Tulinius M., Melberg A., Darin N., Andersen O., Holmgren D., Oldfors A., Holme E. Clinical manifestation and a new ISCU mutation in iron-sulphur cluster deficiency myopathy. Brain. 2009;132:2170–2179. doi: 10.1093/brain/awp152. [DOI] [PubMed] [Google Scholar]
- 41.Puccio H., Simon D., Cossee M., Criqui-Filipe P., Tiziano F., Melki J., Hindelang C., Matyas R., Rustin P., Koenig M. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat. Genet. 2001;27:181–186. doi: 10.1038/84818. [DOI] [PubMed] [Google Scholar]
- 42.Martelli A., Friedman L.S., Reutenauer L., Messaddeq N., Perlman S.L., Lynch D.R., Fedosov K., Schulz J.B., Pandolfo M., Puccio H. Clinical data and characterization of the liver conditional mouse model exclude neoplasia as a non-neurological manifestation associated with Friedreich's ataxia. Dis. Model. Mech. 2012;5:860–869. doi: 10.1242/dmm.009829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gonzalez-Cabo P., Palau F. Mitochondrial pathophysiology in Friedreich's ataxia. J. Neurochem. 2013;126:53–64. doi: 10.1111/jnc.12303. [DOI] [PubMed] [Google Scholar]
- 44.Farhan S.M., Wang J., Robinson J.F., Lahiry P., Siu V.M., Prasad C., Kronick J.B., Ramsay D.A., Rupar C.A., Hegele R.A. Exome sequencing identifies NFS1 deficiency in a novel Fe-S cluster disease, infantile mitochondrial complex II/III deficiency. Mol. Genet. Genom. Med. 2014;2:73–80. doi: 10.1002/mgg3.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mochel F., Knight M.A., Tong W.H., Hernandez D., Ayyad K., Taivassalo T., Andersen P.M., Singleton A., Rouault T.A., Fischbeck K.H., Haller R.G. Splice mutation in the iron-sulfur cluster scaffold protein ISCU causes myopathy with exercise intolerance. Am. J. Hum. Genet. 2008;82:652–660. doi: 10.1016/j.ajhg.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olsson A., Lind L., Thornell L.E., Holmberg M. Myopathy with lactic acidosis is linked to chromosome 12q23.3-24.11 and caused by an intron mutation in the ISCU gene resulting in a splicing defect. Hum. Mol. Genet. 2008;17:1666–1672. doi: 10.1093/hmg/ddn057. [DOI] [PubMed] [Google Scholar]
- 47.Saha P.P., Kumar S.K., Srivastava S., Sinha D., Pareek G., D'Silva P. The presence of multiple cellular defects associated with a novel G50E iron-sulfur cluster scaffold protein (ISCU) mutation leads to development of mitochondrial myopathy. J. Biol. Chem. 2014;289:10359–10377. doi: 10.1074/jbc.M113.526665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cross A.J., Leitzmann M.F., Gail M.H., Hollenbeck A.R., Schatzkin A., Sinha R. A prospective study of red and processed meat intake in relation to cancer risk. PLoS Med. 2007;4:e325. doi: 10.1371/journal.pmed.0040325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torti S.V., Manz D.H., Paul B.T., Blanchette-Farra N., Torti F.M. Iron and cancer. Annu. Rev. Nutr. 2018;38:97–125. doi: 10.1146/annurev-nutr-082117-051732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitnall M., Howard J., Ponka P., Richardson D.R. A class of iron chelators with a wide spectrum of potent antitumor activity that overcomes resistance to chemotherapeutics. Proc. Natl. Acad. Sci. U. S. A. 2006;103:14901–14906. doi: 10.1073/pnas.0604979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Funauchi Y., Tanikawa C., Yi Lo P.H., Mori J., Daigo Y., Takano A., Miyagi Y., Okawa A., Nakamura Y., Matsuda K. Regulation of iron homeostasis by the p53-ISCU pathway. Sci. Rep. 2015;5:16497. doi: 10.1038/srep16497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan S.Y., Zhang Y.Y., Hemann C., Mahoney C.E., Zweier J.L., Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metabol. 2009;10:273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panigrahi D.P., Praharaj P.P., Bhol C.S., Mahapatra K.K., Patra S., Behera B.P., Mishra S.R., Bhutia S.K. The emerging, multifaceted role of mitophagy in cancer and cancer therapeutics. Semin. Canc. Biol. 2019;579(19) doi: 10.1016/j.semcancer.2019.07.015. 30115–4. [DOI] [PubMed] [Google Scholar]
- 54.Hentze M.W., Muckenthaler M.U., Galy B., Camaschella C. Two to tango: regulation of mammalian iron metabolism. Cell. 2010;142:24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 55.Martelli A., Schmucker S., Reutenauer L., Mathieu J.R.R., Peyssonnaux C., Karim Z., Puy H., Galy B., Hentze M.W., Puccio H. Iron regulatory protein 1 sustains mitochondrial iron loading and function in Frataxin deficiency. Cell Metabol. 2015;21:311–322. doi: 10.1016/j.cmet.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 56.Sanchez M., Galy B., Schwanhaeusser B., Blake J., Bahr-Ivacevic T., Benes V., Selbach M., Muckenthaler M.U., Hentze M.W. Iron regulatory protein-1 and -2: transcriptome-wide definition of binding mRNAs and shaping of the cellular proteome by iron regulatory proteins. Blood. 2011;118:e168–179. doi: 10.1182/blood-2011-04-343541. [DOI] [PubMed] [Google Scholar]
- 57.Chipuk J.E., Moldoveanu T., Llambi F., Parsons M.J., Green D.R. The BCL-2 family reunion. Mol. Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chipuk J.E., Green D.R. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol. 2008;18:157–164. doi: 10.1016/j.tcb.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Delivani P., Adrain C., Taylor R.C., Duriez P.J., Martin S.J. Role for CED-9 and Egl-1 as regulators of mitochondrial fission and fusion dynamics. Mol. Cell. 2006;21:761–773. doi: 10.1016/j.molcel.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 60.Pattingre S., Tassa A., Qu X.P., Garuti R., Liang X.H., Mizushima N., Packer M., Schneider M.D., Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 61.Berman S.B., Chen Y.B., Qi B., McCaffery J.M., Rucker E.B., Goebbels S., Nave K.A., Arnold B.A., Jonas E.A., Pineda F.J., Hardwick J.M. Bcl-x(L) increases mitochondrial fission, fusion, and biomass in neurons. JCB (J. Cell Biol.) 2009;184:707–719. doi: 10.1083/jcb.200809060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.