Abstract
Cryptochromes are blue-light receptors that mediate photoresponses in plants. The genomes of most land plants encode two clades of cryptochromes, CRY1 and CRY2, which mediate distinct and overlapping photoresponses within the same species and between different plant species. Photoresponsive protein–protein interaction is the primary mode of signal transduction of cryptochromes. Cryptochromes exist as physiologically inactive monomers in the dark; the absorption of photons leads to conformational change and cryptochrome homooligomerization, which alters the affinity of cryptochromes interacting with cryptochrome-interacting proteins to form various cryptochrome complexes. These cryptochrome complexes, collectively referred to as the cryptochrome complexome, regulate transcription or stability of photoresponsive proteins to modulate plant growth and development. The activity of cryptochromes is regulated by photooligomerization; dark monomerization; cryptochrome regulatory proteins; and cryptochrome phosphorylation, ubiquitination, and degradation. Most of the more than 30 presently known cryptochrome-interacting proteins are either regulated by other photoreceptors or physically interacting with the protein complexes of other photoreceptors. Some cryptochrome-interacting proteins are also hormonal signaling or regulatory proteins. These two mechanisms enable cryptochromes to integrate blue-light signals with other internal and external signals to optimize plant growth and development.
Keywords: blue light, cryptochrome, CRY1, CRY2, photoreceptor, photomorphogenesis, transcription, proteolysis, protein–protein interactions, Arabidopsis
1. INTRODUCTION
Cryptochromes are found in all major evolutionary lineages, from archaea to bacteria, algae to terrestrial plants, and sponges to humans, and they were among the first photoreceptors to evolve in plants (15, 42, 135). Cryptochromes were first discovered in Arabidopsis (2). Researchers found that the long hypocotyl 4 (HY4) gene responsible for the blue-light inhibition of the hypocotyl elongation of Arabidopsis seedlings encodes a protein homologous to DNA photolyases (2). Photolyases are photoresponsive DNA-repairing enzymes that repair the cyclobutane pyrimidine dimer or 6–4 pyrimidine-pyrimidone photoproducts of ultraviolet (UV)-damaged DNA (134). The HY4 protein, later referred to as cryptochrome 1 or CRY1 (82), contains flavin adenine dinucleotide (FAD), which is the primary chromophore of photolyases and cryptochromes, but generally lacks the DNA-repairing enzymatic activity of the photolyase (83, 101). Soon after the discovery of the first cryptochrome in Arabidopsis, cryptochromes were also found in other lineages, such as insects and mammals, acting as photoreceptors, transcriptional regulators, or integral parts of the circadian oscillator (31, 53, 108, 147, 157). Cryptochromes are defined by their common two-domain structure: the highly conserved FAD-binding photolyase homology region(PHR) domain that is approximately 500 residues in length and the divergent CRY C-terminal extension (CCE) domain of various lengths that often contains intrinsically disordered regions (121, 155). The CCE domains of cryptochromes from moss, fern, and angiosperm all contain an evolutionarily conserved DQXVP-acidic-STAES (DAS) signature, and researchers hypothesized that the ancestral plant cryptochromes arose by fusion of an ancestral photolyase sequence to a DAS-containing sequence, which may or may not have been lost during evolution (84). Although the PHR domain and CCE domain of cryptochromes were previously thought to act as the light-sensing domain and effector domain, respectively, most presently known cryptochrome-interacting proteins, except constitutive photomorphogenic 1 (COP1), physically interact with the PHR domain of cryptochromes, suggesting that both the PHR and CCE of cryptochromes may act as effect domains.
Based on sequence analyses, cryptochromes are grouped into three major classes: plant (and plant-like), animal (and animal-like), and CRY-DASH (cryptochrome-Drosophila, Arabidopsis, Synechocystis, human), which are widely found in microbials and eukaryotic organelles (35, 71, 105, 110). The ancestral cyclobutane pyrimidine dimer-repairing photolyase might have duplicated at least eight times before the divergence of eubacteria and eukaryotes, and cryptochromes in different lineages, such as those of plants and animals, are thought to have evolved independently (15, 63). Different organisms have different numbers of cryptochromes, ranging from two in Arabidopsis and humans to six or seven in soybean and zebrafish (187). Almost all higher plants studied have two phylogenetically distinguishable clades of cryptochromes, CRY1 and CRY2, corresponding to the two Arabidopsis cryptochromes originally discovered (84). Most plant cryptochromes, including those in Chlorophyta (green algae), Bryophyta (mosses), and Angiosperms, act as sensory photoreceptors, whereas animal cryptochromes are either photoresponsive photoreceptors (type I) or nonphotoresponsive transcription coregulators (type II) (14, 136). It appears that regardless of their photoresponsiveness, most cryptochromes in different organisms act as the regulators of gene expression, especially transcription (151, 162). Plant cryptochromes are nucleocytoplasmic proteins. For example, Arabidopsis CRY1 locates and functions in both the nucleus and cytoplasm (172), whereas CRY2 seems to be an exclusively nuclear protein that completes its post-translational life cycle in the nucleus (182). Although cryptochromes were previously defined as photolyase-like proteins without DNA-repairing activity, this earlier definition is modifiable in light of the more recent findings that the CRY-DASH and algal cryptochromes have both DNA-repairing and transcription-regulatory activities (21, 140, 150). The crystal structure of the PHR domain of Arabidopsis CRY1 was the first to be solved, and it exhibits a striking similarity to the structure of Escherichia coli DNA photolyase despite their evolutionary distance (11). However, the crystal structure of the full-length plant cryptochrome has not been reported because of the technical difficulty of crystallizing a protein with the large, intrinsically disordered CCE domain. The crystal structures of the full-length Drosophila and mouse cryptochromes, which have relatively small CCE domains, and their complexes, including the respective partner proteins, have been determined (24, 138, 174, 190). These studies demonstrate the pivotal roles of the FAD-binding pocket and the physical interactions between the PHR and CCE domains in the functions of animal cryptochromes. There seems to be a general consensus that light-dependent changes in the interaction between the PHR and CCE domains of cryptochromes may at least partially explain the photoresponsive conformational changes of plant cryptochromes.
In the last 25 years, the function, photochemistry, and molecular mechanisms of cryptochromes in different evolutionary lineages have been extensively investigated, and the results of these studies have been reviewed periodically. For example, plant cryptochrome has been previously reviewed twice in this journal (18, 84). In this article, we focus on the recent progress of mechanistic studies of cryptochromes in plants, especially the model plant Arabidopsis, but we do not recount all of the discoveries of plant cryptochromes. Readers are encouraged to read the previous reviews and their references for a more comprehensive understanding of plant cryptochromes.
2. CRYPTOCHROME-MEDIATED PHOTORESPONSES
2.1. Cryptochrome-Mediated Photoresponses in Higher Plants
Plant cryptochromes regulate many aspects of the plant life cycle, by mediating blue-light regulation of various steps of the gene expression process (Figure 1). Almost all presently known functions of cryptochromes were originally discovered by phenotypical and physiological analyses of induced mutations or natural variations impairing the cryptochrome genes. Cryptochrome-mediated photoresponses in terrestrial plants include photoresponsive genome transcription (102, 161), entrainment of the circadian clock (146), inhibition of the germination of dormant seeds (7), inhibition of hypocotyl elongation (2), stimulation of cotyledon expansion (86), coordination of temperature sensing (10), modulation of gravitropic responses (73, 118), promotion of root greening (154), stimulation of stomata opening and development (64, 102), regulation of shade avoidance (123), control of programmed cell death (25), enhancement of biotic (173) and abiotic stress responses (102), promotion of floral initiation (30, 40), regulation of fruit development (29), and suppression of leaf senescence (106). Cryptochromes may also be involved in the regulation of phototropism (5, 73) and magnetoreception (3, 43, 126). The above list of cryptochrome-mediated photoresponses is hardly complete: Additional cryptochrome-mediated photoresponses, especially those involved in cross talks between light and other signaling processes, or new functions in plant species other than Arabidopsis are likely to be discovered. Among various cryptochrome-mediated photoresponses in land plants, the blue-light inhibition of hypocotyl elongation is the most widely used readout for the studies of cryptochromes. Although cryptochromes can independently regulate photomorphogenesis in the absence of phytochromes (54, 148), cryptochrome-mediated photoresponses are often redundantly, coordinately, or antagonistically coregulated by other photoreceptors, including phytochromes (109, 128, 130, 176); phototropins (19); the LOV-domain/F-box flavoproteins including ZEITLUPE (ZTL); the flavin-binding, kelch repeat, F-box 1 (FKF1); LOV KELCH PROTEIN 2 (LKP2) (59); and UV-B resistance 8 (UVR8) (45, 61). This is at least partially because cryptochromes mediate blue-light responses by physically interacting with cryptochrome-interacting signaling proteins. Many cryptochrome-signaling proteins can also interact with photoreceptors other than cryptochromes, which may compete with or enhance the signaling process of the respective photoreceptors, and cause change within similar genomic or proteomic expression networks, to modulate similar growth and developmental changes in response to different or similar wavelengths of light.
Figure 1.
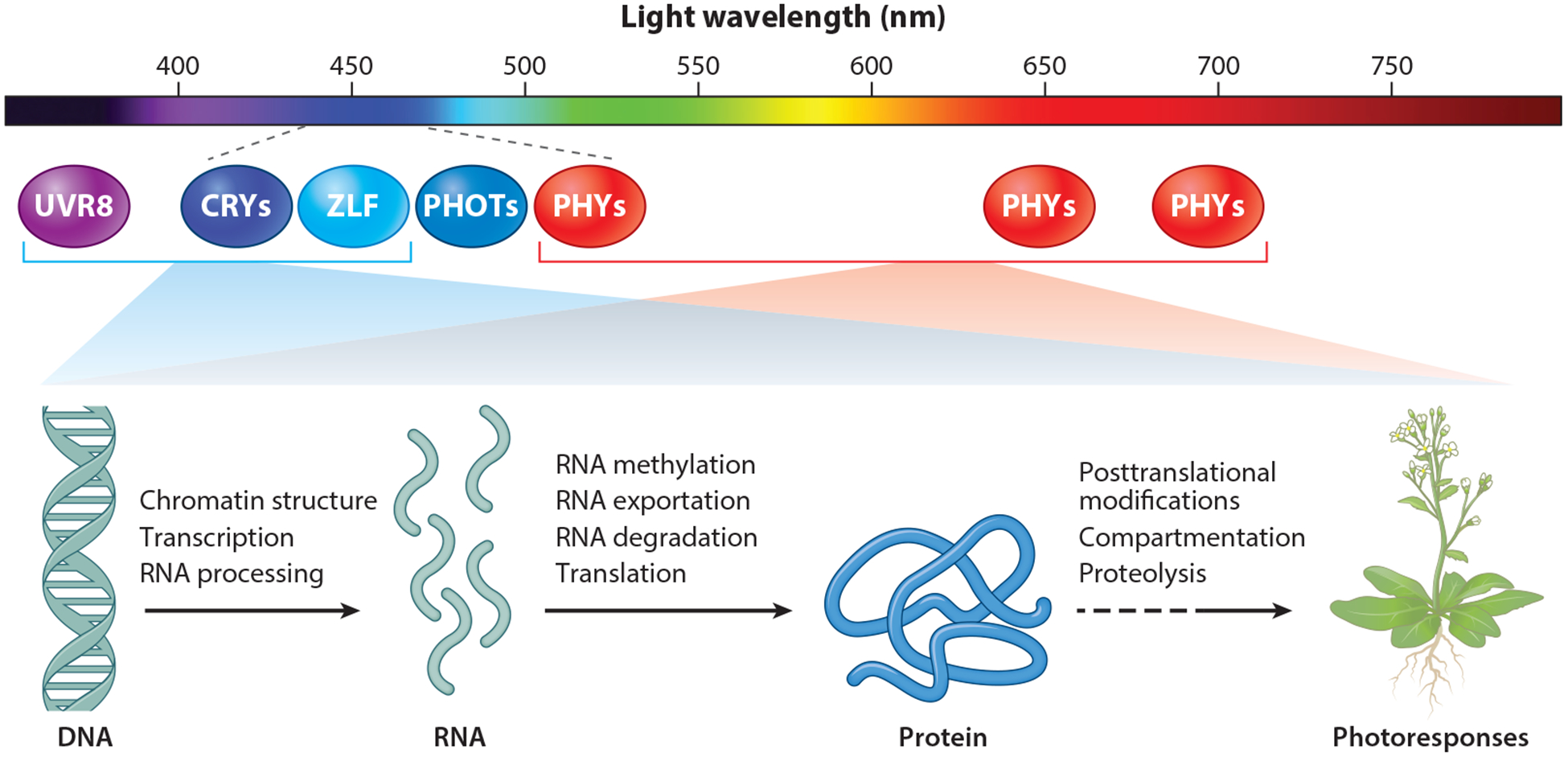
Photoreceptors mediate light regulation of gene expression to modulate plant growth and development. The diagram depicts the general action mechanisms of most plant photoreceptors, except phototropins (PHOTs), which act by modulating gene expression to alter plant growth and development. The ultraviolet (UV)-B receptor UVR8, blue/UV-A light receptors cryptochromes (CRYs), PHOTs, the LOV-domain/F-box proteins ZTL/FKF1/LKP2 represented by ZLF, and red/far-red (FR) light receptors phytochromes (PHYs) have the respective absorption spectra that are indicated by their positions under the light wavelength (nm). Known or potential regulatory mechanisms of gene expression are indicated. PHOTs are found primarily in plasma membrane such that regulation of gene expression is not depicted as the main mechanism of PHOTs. PHYs, especially phyA, are known to absorb blue light, in addition to red/FR light, and to regulate blue light responses.
2.2. Cryptochrome-Mediated Photoresponses in Green Algae
Cryptochromes have also been extensively studied in green algae, which appear to have a more complex set of cryptochromes than higher plants (32, 69, 71). In addition to canonical plant cryptochromes and CRY-DASH, green algae also have plant-like, animal-like, and cryptochrome/photolyase family (CPF) proteins. The algal cryptochromes seem different from that of the higher plants in at least two aspects. First, algal cryptochromes are often bifunctional, acting as both DNA photolyases and transcription regulators. For example, CPF1 of the marine diatom Phaeodactylum tricornutum and green algae Ostreococcus tauri has been shown to exhibit both 6–4 DNA photolyase activity and transcription regulatory activity (21, 26, 46). When algal CPF1 proteins are expressed in heterologous mammalian cells (Chinese hamster ovary cells), they can bind to the mammalian CLOCK/BMAL1 heterodimer and suppress the activity of CLOCK/BMAL1-regulated expression of the E-box-containing reporter promoter (21, 46). Second, algal cryptochromes may mediate photoresponses to a broader spectrum of light than the cryptochromes of the higher plants mediate. For example, Chlamydomonas reinhardtii possesses a plant cryptochrome (pCRY, previously referred to as CPH1) and an animal-like cryptochrome (aCRY), and both aCRY and pCRY respond to red light in addition to blue light (71). pCRY, which is 104 kDa and one of the largest cryptochromes, undergoes proteolysis via the 26S proteasome system in response to either blue light or red light (129), whereas aCRY, which is approximately 70 kDa, regulates the light-dependent expression of genes in response to blue and red light (8, 36). The Chlamydomonas pCRY regulates the circadian clock and life cycle progression; the pcry mutant exhibits many phenotypic defects, including period lengthening and arrhythmicity, phase shift abnormality in response to blue light, defects in the stimulation of mating by light or inhibition of mating by darkness, and light-dependent germination (111). Chlamydomonas aCRY is a bifunctional and broad-spectral photoreceptor that possesses both 6–4 DNA photolyase and photoreceptor activities (36), and it mediates photoresponses to blue, yellow, and red light (but not far-red light) (8, 36). Based on the analyses of absorption spectra of purified aCRY, changes of gene expression in the acry mutant, and the crystal structure of the PHR domain of aCRY, researchers proposed that different redox states of the FAD chromophore of aCRY may explain the broad-spectra photoresponses of aCRY (8, 36). According to the current view, the fully oxidized FAD of aCRY absorbs blue light to convert to the neutral radical state that absorbs blue, yellow, and red light to regulate gene expression (8, 36). Because the cryptochrome-signaling protein has not been reported in algae, exactly how algal cryptochromes regulate gene expression remains unclear.
3. CRYPTOCHROME PHOTOACTIVATION AND INACTIVATION
3.1. Cryptochrome Photoreduction and Photoactivation
Photoactivation of a photoreceptor can be broadly defined as photoresponsive changes that activate the photoreceptor, such as submolecular changes of the energy, orbital, or electronic state of chemical bonds of the chromophore and its interacting amino acids, and subsequent molecular or intermolecular changes, such as conformational changes that affect intermolecular interactions of the photoreceptor. Photoactivation of plant cryptochromes begins with photoexcitation or photon absorption and photoreduction of the FAD chromophore (83, 101, 141), which results in conformational changes, such as disengagement of the PHR and CCE domains (112, 121, 179, 184) and formation of the cryptochrome homooligomers that interact with cryptochrome-interacting proteins to alter gene expression and plant development (91, 131, 137, 161). Partly because the fully oxidized FAD absorbs blue light more effectively than any other redox forms of FAD, the FAD photoreduction has been hypothesized to explain how cryptochromes are photoactivated. The non-redox photolyase-like cyclic electron shuttle mechanism has been proposed as an alternative photochemistry for cryptochrome photoactivation (87). However, this hypothesis has not been experimentally tested nor does it readily explain the long lifetime of the active cryptochromes required for their functions in plant cells (47, 112).
The FAD photoreduction hypothesis argues that the photon-excited FAD is reduced by receiving electrons transferred through three evolutionarily conserved tryptophan residues known as the Trp triad (1, 87). Recently, the fourth residue has been reported to contribute to cryptochrome photoreduction (9, 85, 113, 114, 117), and an Asp residue near the isoalloxazine ring of FAD has been proposed as the proton donor (13, 70). Despite abundant in vitro studies of the cryptochrome photoreduction phenomenon (1), exactly how FAD photoreduction is involved in cryptochrome photoactivation remains controversial (1, 87). At least two questions about cryptochrome photoreduction contribute to the controversy of cryptochrome photochemistry. First, why do mutations of the Trp-triad residues of cryptochromes that abolish cryptochrome photoreduction in vitro fail to abolish their physiological activities in vivo? Most Trp-triad mutants of cryptochromes studied, including Arabidopsis CRY1 (38) and CRY2 (79, 89), fail to undergo photoreduction in vitro but remain physiologically active in vivo (87, 89). Recently, researchers reported that some Trp mutants impaired photoresponses of the Drosophilia cryptochrome (dCRY) in vivo (85), supporting the Trp triad-dependent photoreduction hypothesis for the Drosophilia cryptochrome. However, the Arabidopsis cry2 mutations altered in two or all three Trp residues of the Trp triad have recently been shown to retain the physiological activities in vivo, imposing further challenges to the Trp triad-dependent FAD photoreduction hypothesis (89). Researchers also noticed that free flavins and flavoenzymes, which have no reported light-dependent functions in vivo, commonly undergo photoreduction in vitro (104, 124), making it difficult to establish the causal relationship for the correlations between the cryptochrome photoreduction observed in vitro and the photoactivation mechanism of the respective cryptochrome in vivo. Therefore, new and innovative approaches may be needed to solve this conundrum.
Another aspect of the FAD photoreduction hypothesis is how to maintain FAD in its oxidized state in the dark. Because the oxidized FAD is the predominant redox form that absorbs blue light, it is expected to be the ground state of cryptochromes. However, the midpoint redox potentials of Arabidopsis CRY1 have been estimated by two independent studies to be approximately − 143 mV to −153 mV for FAD/FADH• and −161 mV to −181 mV for FADH•/FADH2 (6, 83). The redox potential of Arabidopsis cytoplasm is measured to be approximately −320 mV (107, 139), which is markedly lower than −143 mV to −153 mV, the potential of the FAD/FADH• couple of CRY1. Under this reduced cellular condition, little CRY1 is expected to spontaneously oxidize to its fully oxidized ground state (87). Researchers have reported recently that the Drosophila dCRY has a redox potential of −316 mV for FAD/FAD•− (85), which is strikingly lower than 125 mV reported previously (122). This new estimation of the midpoint redox potential of dCRY seems to explain how dCRY may be oxidized in the inset cells (85). But it remains to be investigated whether the midpoint redox potential of plant cryptochromes might be lower than what is currently known or whether the subcellular microenvironment of the nucleus that most cryptochromes reside in might provide a redox potential that is more oxidized than the currently known redox potentials of plant cryptochromes. For example, cryptochrome-interacting proteins may create a protein complex to provide a microenvironment so oxidized that it may force cryptochromes to maintain oxidized states despite the reductive cytoplasmic environment.
3.2. Cryptochrome Photooligomerization and Photoactivation
Regardless of the exact photochemical mechanism of cryptochromes, it must lead to a change in protein conformation in order to activate cryptochromes. Light-dependent conformational changes of cryptochromes were first demonstrated by a partial proteolysis experiment, in which light exposure resulted in a five- to tenfold increase in the proteolysis rate of the Arabidopsis CRY1 protein expressed and purified from insect cells (121). Although light-induced conformational change has not been directly observed in a plant cryptochrome due to the lack of a crystal structure, it has been hypothesized that the conformational change may lead to or be associated with a light-induced disengagement of the PHR and CCE domains (112, 121, 179, 184). This domain disengagement model has been demonstrated for the full-length Drosophila dCRY photoreceptor (190). In dCRY, the small C-terminal helix of dCRY docks to the FAD pocket in the groove analogous to the one that binds DNA in the DNA photolyases. The dCRY crystal exposed to X-rays photoreduces FAD to its anionic semiquinone state (FAD•−) to facilitate restructuring of the tail helix of dCRY (190). The light-induced domain disengagement model would predict that overexpression of a truncated cryptochrome fragment devoid of the photon-absorbing PHR domain would interfere with the function of endogenous cryptochromes. Indeed, it has been shown that transgenic expression of the β-glucuronidase (GUS)-CCT1 or GUS-CCT2 fusion proteins, which are GUS fused to the C-terminal CCT (i.e., CCE) domains of Arabidopsis CRY1 or CRY2, resulted in a constitutive photomorphogenic phenotype similar to that of the constitutive photomorphogenesis 1 (cop1) mutant (28, 179). Later studies showed that the constitutive photomorphogenic phenotypes resulting from overexpression of the CCE domain of cryptochromes are caused by physical interaction between the CCE domain of cryptochromes and the WD40 domain of COP1 (52, 74, 75, 125).
Researchers predict that domain disengagement of cryptochromes may lead to changes of intermolecular interactions, resulting in homooligomerization (including homodimer, homotetramer, etc.) of cryptochromes and their interaction with other proteins (131, 137, 161). A number of experimental observations are consistent with this prediction. Blue-light-dependent cryptochrome homooligomerization was first implicated by the finding that the CRY2-RFP (red florescence protein) and CRY2-GFP (green florescence protein) fusion proteins, as well as the endogenous CRY2 protein, rapidly form nuclear speckles (also known as nuclear bodies or photobodies) in Arabidopsis cells exposed to blue light (103, 183). Moreover, Arabidopsis CRY2 protein expressed in heterologous mammalian cells forms morphologically similar photobodies in response to blue light, demonstrating that photoexcited CRY2 is capable of homooligomerizing into photobodies in the absence of other proteins (12, 119). At least three earlier studies showed that cryptochrome homooligomerization is required for the functions of Arabidopsis CRY1 and CRY2 (131, 137, 184). The first study showed that Arabidopsis CRY1 forms homooligomers via its PHR domain [also called Cryptochrome N Terminus (CNT)] (i.e., PHR), that the CNT1 fragment can interact with the endogenous CRY1 to cause dominant-negative inhibition of the activity of CRY1 in transgenic plants, and that the CNT1 fragments of CRY1 mutated in A462V, G347R, or S66N lost their abilities to interact with CRY1 or to inhibit the endogenous CRY1 (137). The second study showed that the chemically induced oligomerization of the C-terminal domain fragments of CRY2 could elicit changes in the expression of the cryptochrome-target genes in the absence of light (131). The third study showed that disengagement of the PHR domain and CCE domain and homooligomerization of Arabidopsis CRY2 may expose a small region spanning the two domains (NC80, residues 486–565) to elicit physiological functions of CRY2 (184). Together, these studies suggest that cryptochrome homooligomerization is necessary for the functions of plant cryptochromes. Although the light response of cryptochrome oligomerization was not detected in these three earlier studies, it was later shown that homooligomerization and heterooligomerization of Arabidopsis cryptochromes were blue-light-dependent photoreactions (92, 161), which are collectively referred to as cryptochrome photooligomerization (92). In these experiments, homooligomerization or heterooligomerization of recombinant Arabidopsis CRY1 or CRY2 proteins fused to different epitope tags were coexpressed in human embryonic kidney 293 (HEK293) cells or Arabidopsis plants, and their interactions were analyzed by co-immunoprecipitation (92, 161). Because HEK293 cells contain no other plant proteins to interfere with the co-immunoprecipitation assay by indirect protein-protein interaction, they can be conveniently used to study the light responses and kinetics of cryptochrome photooligomerization. For example, Arabidopsis CRY2 expressed in HEK293 cells took about 1 min to reach saturation of homooligomerization when the cells were exposed to 100 μmol m−2s−1 blue light (Figure 2a). This method has also been used to demonstrate that Arabidopsis CRY1 and CRY2 can form heterooligomers and that photoresponsive cryptochromes from other organisms, such as rice, soybean, liverwort, Drosophila, monarch butterfly, and zebrafish, could all undergo photooligomerization (92). These observations are consistent with a hypothesis that photooligomerization may be an evolutionarily conserved photoactivation mechanism of cryptochrome photoreceptors from not only higher plants but also other evolutionary lineages.
Figure 2.
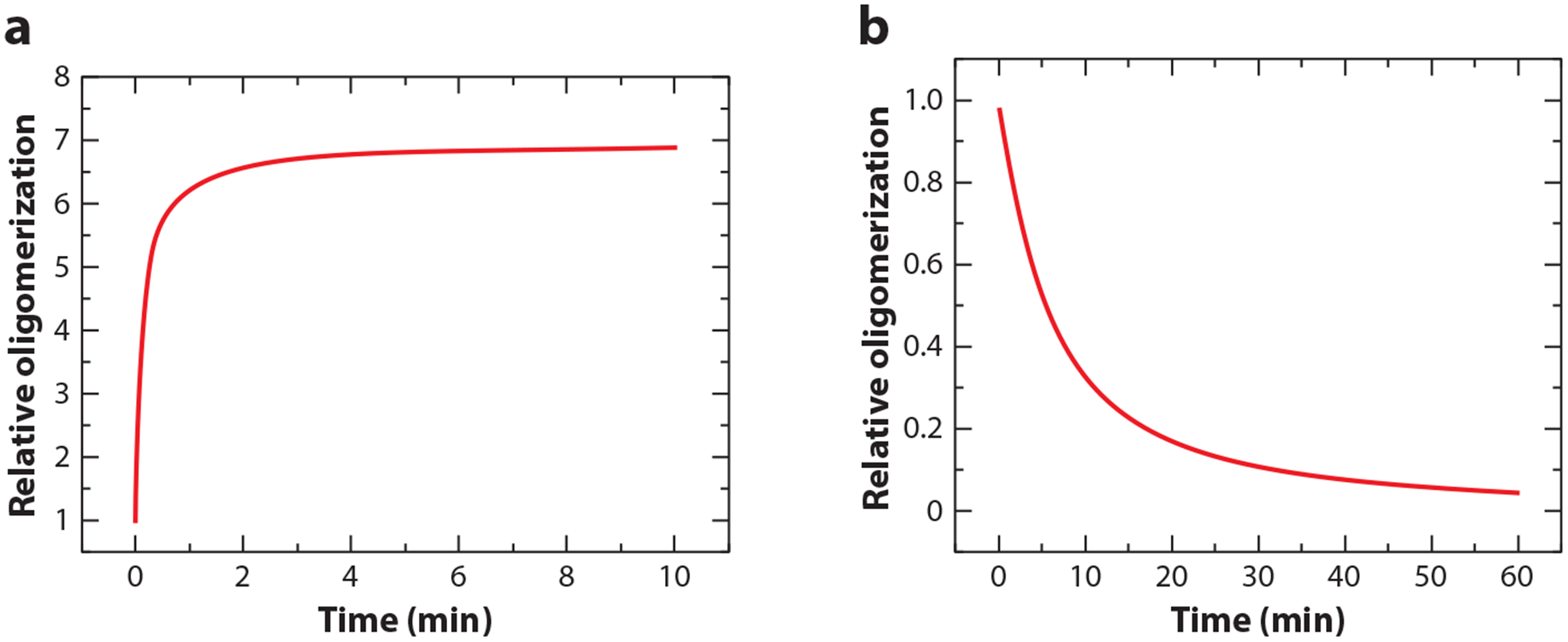
CRY2 photooligomerization and dark-reversion. The Flag- and Myc-tagged Arabidopsis CRY2 recombinant proteins were coexpressed in human embryonic kidney cells (HEK293). (a) HEK293 cells were irradiated with 100 μmol m−2s−1 blue light for the indicated time at 21°C; the kinetics of CRY2 homooligomerization was measured by co-immunoprecipitation at the indicated time after illumination. (b) The kinetics of CRY2 monomerization or dark-reversion was measured by co-immunoprecipitation at the indicated time after light-treated HEK293 cells were transferred to darkness. Results of this experiment indicate that photooligomerization of CRY2 is much faster (~30x) than its dark reversion under the experimental conditions used.
3.3. Cryptochrome Inactivation and the CRY-BIC Complexes
The inactivation of photoreceptors is usually necessary to maintain the sustained photosensitivity and to avoid the excessive photosensitivity of cells. Because photoreduction of the FAD chromophore is likely the first step of cryptochrome photoactivation, FAD oxidation in the absence of light might play a role in the inactivation of cryptochrome oligomers. A direct test of this possibility may help to resolve the controversy of cryptochrome photoreduction. However, the physiologically active cryptochrome homooligomers are expected to be at the higher energy state so they undergo spontaneous reversion to monomers in darkness by thermal relaxation. The rate of cryptochrome monomerization in darkness is much slower than that of the cryptochrome photooligomerization reaction in light. For example, in the heterologous HEK293 cells, the rate of CRY2 monomerization is at least 30 times slower than that of the photooligomerization reaction (Figure 2b). The rates of light-induced oligomerization and dark-dependent monomerization of Arabidopsis CRY2 appear faster in plant cells than those shown in heterologous HEK293 system (92). This observation suggests that the cellular environment and/or cryptochrome-interacting proteins may affect the equilibrium of cryptochrome photooligomerization and monomerization to govern the cryptochrome activity in plants under different light conditions.
In plants, the blue-light-dependent cryptochrome oligomerization can also be suppressed or inactivated by interaction with two closely related cryptochrome inhibitory proteins, known as blue-light inhibitor of cryptochrome 1 (BIC1) and BIC2 (161). These two cryptochrome inhibitors were initially identified in a gain-of-function genetic screen (161). It was found that overexpression of BIC1 or BIC2 in transgenic plants suppressed all known photobiochemical and photophysiological activities of CRY1 and CRY2, including blue-light inhibition of hypocotyl elongation, blue-light-responsive gene expression, photoperiodic promotion of flowering, blue-light-dependent interaction of cryptochromes with cryptochrome-signaling proteins, blue-light-induced formation of photobodies of CRY2, blue-light-dependent phosphorylation of both CRY1 and CRY2, and blue-light-dependent polyubiquitination and degradation of CRY2, whereas the loss-of-function bic1bic2 double mutants are hypersensitive to blue light (161). When BIC proteins and CRY2 are co-expressed in mammalian or plant cells, they physically interact to inhibit CRY2 oligomerization under blue light. However, these experiments did not distinguish whether BIC proteins might bind to the photoexcited cryptochrome monomer or to the photoactivated cryptochrome oligomer. In other words, whether BIC proteins inhibit cryptochrome photoactivation or promote cryptochrome inactivation remains to be elucidated. Photoactivated cryptochromes trigger a negative feedback circuitry, because expression of the messenger RNAs (mRNAs) of the BIC genes increase more than 200–1,200-fold in etiolated seedlings exposed to 100 μmol m−2s−1 blue light for an hour (165). Interestingly, although the function of BIC proteins is blue-light–specific, mRNA expression of BIC genes is induced by not only blue light but also red light, and the photoinduction of BIC expression requires cryptochromes, phytochromes, COP1, and HY5 (165). This result suggests that photoactivation of cryptochromes triggers a negative-feedback reaction that increases the supply of BIC proteins to prevent excessive levels of active cryptochromes in the cells, and this process requires the actions of both cryptochromes and phytochromes.
In addition to cryptochrome monomerization, light-dependent proteolysis may also serve the purpose of actively removing the activated cryptochromes. For example, depending on the plant species, CRY1, CRY2, and other types of cryptochromes have all been shown to undergo light-dependent proteolysis (17, 48, 58, 86, 129, 188). For example, Arabidopsis CRY2 undergoes rapid blue-light-specific ubiquitination and 26S proteasome-dependent degradation in the nucleus (86, 182, 192). Arabidopsis CRY2 has a half-life of longer than 24 h in etiolated seedlings, in contrast to a half-life of 25 min in etiolated seedlings exposed to 16 μmol m−2s−1 blue light (182). The ubiquitination and degradation of Arabidopsis CRY2 are dependent on CRY2 oligomerization and phosphorylation, which suggests that degradation is a mechanism to remove active CRY2 (86, 161, 183). Because both the protein kinases and E3 ubiquitin ligases are required for CRY2 activity and degradation (93, 143, 160, 167), the cryptochrome photoactivation and inactivation processes are expected to be closely coupled in time and space. A model of the photoactivation and inactivation of plant cryptochromes, based on the results of the studies described above, is depicted in Figure 3. According to this model, plant cryptochromes exist as inactive monomers in the absence of light, presumably containing the oxidized FAD; photoexcited FAD undergoes photoreduction or other types of photochemical changes to promote conformational changes and oligomerization of the cryptochrome proteins. Cryptochrome homooligomers actively interact with cryptochrome-signaling proteins to alter gene expression and photomorphogenesis, and photoactivated cryptochrome homooligomers are rapidly inactivated by FAD oxidation, monomerization, and possibly BIC interaction, and then are removed by proteolysis. The cryptochrome-mediated blue-light stimulation of the transcription of the BIC genes constitutes a negative feedback loop to maintain the homeostasis of the active and inactive cryptochrome pools in plant cells (Figure 3; Table 1).
Figure 3.
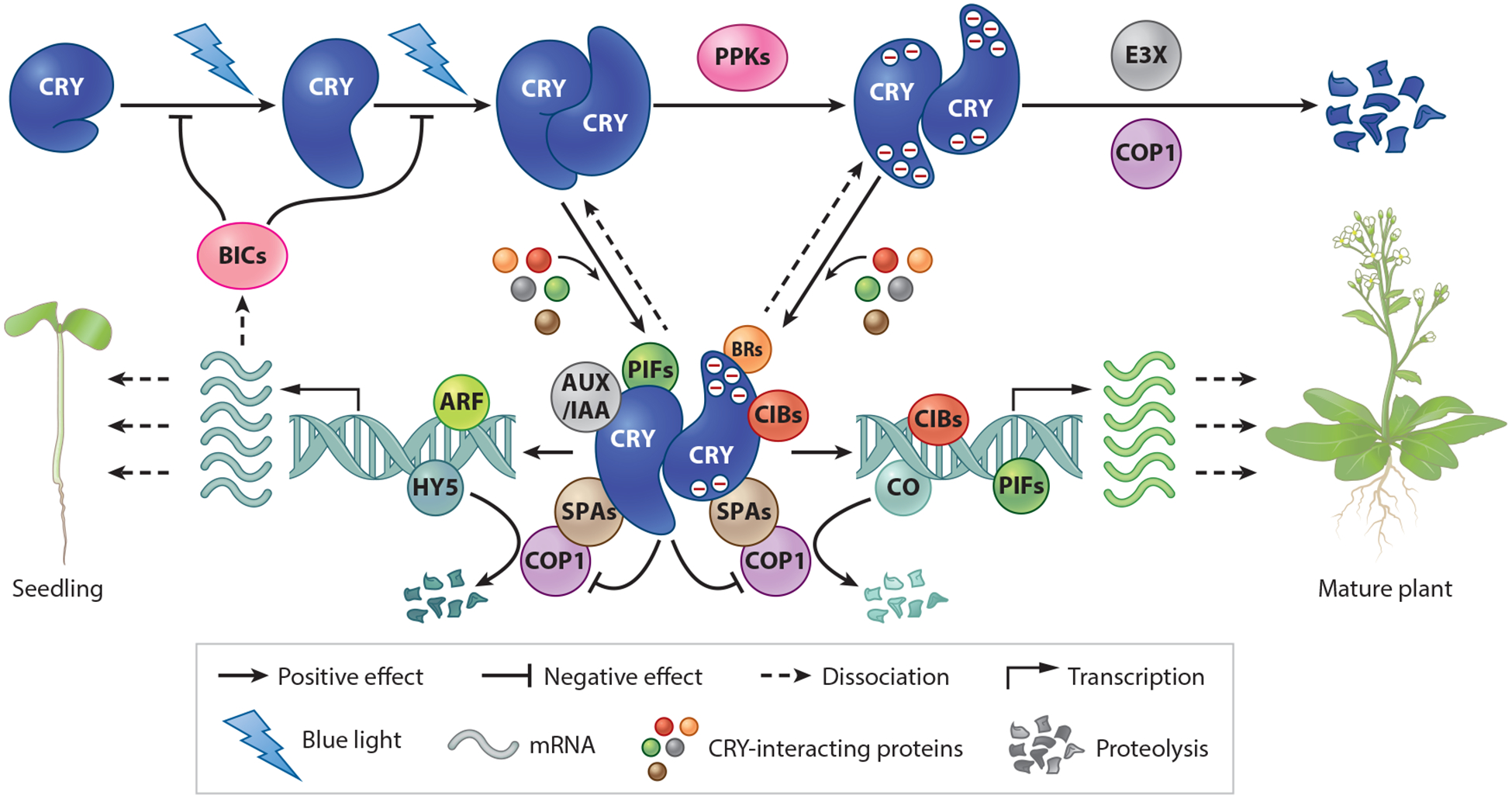
CRY photoactivation, signaling, and inactivation mechanisms. Cryptochromes exist as inactive monomers in darkness. Photoexcited cryptochromes undergo homooligomerization to become biochemically and physiologically active. The cryptochrome homooligomers interact with cryptochrome-interacting proteins. The presently known cryptochrome-interacting proteins, or components of the CRY complexome, include transcription regulators CIB proteins, PIF proteins, AUX/IAA proteins, the cryptochrome regulators BICs and PPKs, the E3 ubiquitin ligase complex COP/SPA, and the BRs (including BZR1, BES1, BIN2, and BIM1). The CRY complexome mediates blue-light regulation of transcription or protein stability. For example, the CRY-COP1-SPA interaction positively regulates the abundance of the HY5 protein, which promotes transcription of the BIC genes. The BIC proteins interact with photoexcited cryptochromes to inhibit cryptochrome homooligomerization and activity. The PPK protein kinases catalyze blue-light-dependent phosphorylation of cryptochromes to positively regulate not only cryptochrome activity but also cryptochrome polyubiquitination. The COP1/SPA proteins and another E3 ubiquitin ligase (E3X) catalyze the polyubiquitination and degradation of phosphorylated cryptochromes. Abbreviations: AUX/IAA, auxin/indole-3-acetic acid; ARFs, auxin response factors; BIC, blue-light inhibitor of cryptochromes; BIM1, bisindolylmaleimide-based protein kinase C inhibitor; BIN2, bridging integrator 2; BR, brassinosteroid regulator; BZR1, brassinosteroid signaling positive regulator; BES1, BRI1-EMS-SUPPRESSOR 1; BIM1, BES1-INTERACTING MYC-LIKE1; BIN2, BR-INSENSITIVE 2; CIB, cryptochrome-interacting basic helix-loop-helix; CO, CONSTANS; COP1, CONSTITUTIVE PHOTOMORPHOGENESIS 1; CRY, cryptochrome; E3X, unknown E3 ubiquitin ligases; HY5, LONG HYPOCOTYL 5; PIF, phytochrome-interacting factor; PPK, photoregulatory protein kinase; SPA, suppressor of phytochrome A.
Table 1.
Components of the Arabidopsis Cryptochrome (CRY) Complexomes
| Accession | Abbreviation | Full name | CRY interaction(s) | CRY effects | Activity | Major function(s) | Reference(s) |
|---|---|---|---|---|---|---|---|
| AT4G34460 | AGB1 | ARABIDOPSIS G-PROTEIN BETA SUBUNIT 1 | CRY1 (blue +), CRY2(blue +) | − | Trimeric G protein | Auxin signaling, HY5-interacting | 80 |
| At3G52740 | BIC1 | Blue-light Inhibitor of Cryptochromes 1 | CRY2 (blue +) | + | Crypto chrome inhibitor | Blue light signaling, hypocotyl growth, flowering-time | 161, 165 |
| AT3G44450 | BIC2 | Blue-light Inhibitor of Cryptochromes 2 | CRY2 (blue +) | + | Crypto chrome inhibitor | Blue light signaling, growth promoter, flowering inhibitor | 161, 165 |
| AT4G18710 | BIN2 | BRASSINOSTEROID-INSENSITIVE2 | CRY1 (blue+) | + | Protein kinase | BR signaling, blue light signaling | 44 |
| AT5G08130 | BIM1 | BES1-INTERACTING MYC-LIKE1 | CRY1 (blue +), CRY2 (blue +) | + | Transcription regulator | BR signaling, blue light signaling | 164 |
| AT1G75080 | BZR1 | BRASSINAZOLE-RESISTANT 1 | CRY1 (blue +), CRY2 (blue +) | − | Transcription factor | BR signaling, blue light signaling | 44, 164 |
| AT1G19350 | BZR2/BES1 | BRASSINAZOLE-RESISTANT 2/BRI1-EMS SUPPRESSOR 1 | CRY1 (blue +), CRY2 (blue +) | − | Transcription factor | BR signaling, blue light signaling | 44, 164 |
| AT4G34530 | CIB1 | CRYPTOCHROME-INTERACTING BASIC-HELIX-LOOP-HELIX 1 | CRY2 (blue +) | − | Transcription factor | Blue light signaling, flowering promotion | 91, 95, 106 |
| AT5G48560 | CIB2 | CRY2-INTERACTING BHLH 2 | CRY2 | + | Transcription factor | Blue light signaling, flowering promotion | 95 |
| AT3G07340 | CIB3 | CRY2-INTERACTING BHLH 3 | CRY2 (in vitro) | + | Transcription factor | Blue light signaling | 95 |
| AT1G10120 | CIB4 | CRY2-INTERACTING BHLH 4 | CRY2 (in vitro) | + | Transcription factor | Blue light signaling, flowering promotion | 95 |
| AT1G26260 | CIB5 | CRYPTOCHROME-INTERACTING BASIC-HELIX-LOOP-HELIX 5 | CRY2 (blue +) | + | Transcription factor | Blue light signaling, flowering promotion | 95 |
| AT2G32950 | COP1 | CONSTITUTIVE PHOTOMORPHOGENIC 1 | CRY1, CRY2 | − | E3 ubiquitin ligase | Light signaling, photo morphogenesis | 159, 178 |
| AT4G08920 | CRY1 | CRYPTOCHROME 1 | CRY1 (blue +), CRY2 (blue +) | − | Blue light receptor | Blue light signaling | 92, 137 |
| AT1G04400 | CRY2 | CRYPTOCHROME 2 | CRY1 (blue +), CRY2 (blue +) | + | Blue light receptor | Blue light signaling | 161, 137 |
| AT2G18300 | HBI1 | HOMOLOG OF BEE2 INTERACTING WITH IBH 1 | CRY1 (blue +), CRY2 | − | Transcription factor | BR signaling, | 163 |
| AT3G23050 | IAA7 | INDOLE-3-ACETIC ACID 7 | CRY1 (blue +), CRY2 (blue +) | + | Transcription regulator | Auxin signaling | 175 |
| AT1G04550 | IAA12 | INDOLE-3-ACETIC ACID 12 | CRY1 (blue +) | + | Transcription regulator | Auxin signaling | 175 |
| AT1G04250 | IAA17 | INDOLE-3-ACETIC ACID 17 | CRY1 (blue +), CRY2 (blue +) | + | Transcription regulator | Auxin signaling | 175 |
| AT1G09570 | phyA | PHYTOCHROME A | CRY1 | + | Red/Far-red light receptor | Red light signaling, blue light signaling | 4 |
| AT2G18790 | phyB | PHYTOCHROME B | CRY2 | + | Red/Far-red light receptor | Red light signaling, blue light signaling | 103 |
| AT3G13670 | PPK1 | photo regulatory protein kinase 1 | CRY2 (blue +) | + | Protein kinase | CRY regulator, PIF3 regulator | 93, 115 |
| AT5G18190 | PPK2 | photo regulatory protein kinase 2 | CRY2 (blue +) | + | Protein kinase | CRY regulator, PIF3 regulator | 93, 115 |
| AT3G03940 | PPK3 | photo regulatory protein kinase 3 | CRY2 (blue +) | + | Protein kinase | CRY regulator, PIF3 regulator | 93, 115 |
| AT2G25760 | PPK4 | photo regulatory protein kinase 4 | CRY2 (blue +) | + | Protein kinase | CRY regulator, PIF3 regulator | 93, 115 |
| AT2G43010 | PIF4 | PHYTOCHROME INTERACTING FACTOR 4 | CRY1 (blue+), CRY2 | − | Transcription factor | Phytochrome signaling, | 98, 123 |
| AT3G59060 | PIF5 | PHYTOCHROME-INTERACTING FACTOR 5 | CRY1, CRY2 | − | Transcription factor | Phytochrome signaling, | 123 |
| AT2G46340 | SPA1 | SUPPRESSOR OF PHYA-105 1 | CRY1 (blue +), CRY2 (blue +) | − | Positive regulator of COP 1 | Light signaling, photomorphogenesis | 81, 88,191 |
| AT4G11110 | SPA2 | SPA 1-RELATED 2 | CRY1 (blue+) | − | Positive regulator of COP 1 | Light signaling, photomorphogenesis | 81 |
| AT3G15354 | SPA3 | SPA1-RELATED 3 | CRY1 (blue+) | − | Positive regulator of COP 1 | Light signaling, photomorphogenesis | 81 |
| AT1G53090 | SPA4 | SPA1-RELATED 4 | CRY1 (blue+) | − | Positive regulator of COP 1 | Light signaling, photomorphogenesis | 81 |
| AT5G57360 | ZTL | ZEITLUPE | CRY1 | + | Blue light receptor, E3 ubiquitin ligase | Blue light signaling, Circadian clock | 60, 90 |
4. THE CRYPTOCHROME COMPLEXOME AND CRYPTOCHROME SIGNAL TRANSDUCTION
More than 30 cryptochrome-interacting proteins have been reported (Table 1). Our understanding of the molecular mechanisms of cryptochrome signal transduction has been advanced primarily by the identification and investigation of these cryptochrome-interacting proteins. Blue-light-responsive protein–protein interactions appear to be the primary mechanisms underlying cryptochrome signal transduction, whereby photoactivated cryptochrome oligomers interact with cryptochrome-interacting proteins to regulate gene expression and photoresponses (Figure 3; Table 1). Plant cryptochromes have been reported to physically interact with CONSTITUTIVE PHOTOMORPHOGENESIS 1 (COP1) (159, 178), CRY2-interacting protein 1–5 (CIB1–CIB5) (91), suppressor of phytochrome A 1–4 (SPA1–SPA4) (81, 88, 191), BIC1 and BIC2 (161), phytochrome interacting factor 1–7 (PIF1–PIF7), photoregulatory protein kinase 1–4 (PPK1–PPK4) (93, 115), auxin/indole-3-ascetic acid (AUX/IAA) (175), AGB1 (a G-protein β subunit) (80), phytochromes (phyA and phyB) (4, 103), and several brassinosteroid signaling proteins (44, 164). Various cryptochrome complexes composed of cryptochromes and individual cryptochrome-interacting proteins are collectively referred to as the cryptochrome complexome. It is conceivable that in plant cells, the cryptochrome complexome exists in dynamic homeostasis by the reversible formation and disintegration of cryptochrome monomers, cryptochrome homooligomers, and cryptochrome heterooligomers composed of different cryptochrome-interacting proteins. One remarkable feature of the cryptochrome complexome is that many cryptochrome-interacting proteins are either regulated by other photoreceptors or physically interacting with other photoreceptors or their signaling proteins (Table 1). COP1/SPA, PIF, PPK, and AGB1 proteins are known to interact with the red/far-red light receptor phytochromes or their signaling proteins. BIC proteins are the only presently known cryptochrome-interacting proteins that exhibit blue-light-specific activity, but their expression is also regulated transcriptionally by phytochromes and post-translationally by the LOV-domain/F-box photoreceptors (90, 165). This feature of the cryptochrome complexome seems to provide a straightforward mechanistic explanation for the longstanding notion of photoreceptor coactions in plants and our naive expectation that plants grown in nature must integrate light signals of different wavelengths into similar genomic, proteomic, or metabolic changes to affect similar cellular activities, such as cell elongation, chloroplast development, metabolism, and cell fate determination.
4.1. The CRY-COP1-SPA Complexes
COP1 is the first cryptochrome-signaling protein identified in plants (159, 178). The loss-of-function cop1 mutant exhibits constitutive photomorphogenic phenotypes, leading to the hypothesis that COP1 is a central repressor of plant photomorphogenesis (28). Indeed, this hypothesis has been supported by studies of many photoreceptors, including cryptochromes. COP1 contains a RING finger motif, a coiled-coil domain, and a WD40-repeat domain (28, 56). COP1 acts by complexing with its related SPA proteins (SPA1 to SPA4), which contain a coiled-coil domain, WD40 repeats, and a kinase-like domain instead of the RING domain of COP1 (50, 51). Like cryptochromes, COP1 is evolutionarily conserved in major evolutionary lineages, including humans, but SPA proteins are specific to the green lineage (42). The COP1-SPA complex is best known for its function as substrate receptors of the cullin 4-based E3 ubiquitin ligase, CUL4COP1/SPAs, which facilitates ubiquitination and degradation of different light-signaling proteins, although they also perform other functions in plant development by interacting with various substrate proteins (50, 56). A conserved VP motif (VPE/D) has been identified in COP1 substrate proteins (52), and it overlaps with the DAS motif of cryptochromes (84). Structural analysis has recently shown that the VP/DAS motif of Arabidopsis CRY2 directly interacts with COP1 (74). Transgenic overexpression of either the oligomerized CCE domain or the DAS-containing 80-residue fragment (NC80) of cryptochromes causes a constitutive photomorphogenic phenotype resembling that of the loss-of-function cop1 mutant or the spaq quadruple mutant (28, 179, 184). These genetic studies support the hypothesis that the VP/DAS motif of the CCE domain of cryptochromes interacts with COP1 to exert cryptochrome functions. Because COP1 interacts with many proteins that contain the VP motif (125), the cryptochrome-mediated blue-light signal is expected to integrate into the COP1 multiple signaling hub to regulate plant development.
Although CRY1 interacts with COP1 in a light-independent manner in heterologous systems (159, 178), the light-dependent formation of the CRY1-COP1 complex was detected in plant cells, which is explained by COP1-SPA interactions (132, 142, 189) and light-dependent CRY-SPA interaction in vivo (81, 88, 191). Arabidopsis CRY1 and CRY2 interact with SPA proteins in a blue-light-dependent manner. The CRY–COP1-SPA interaction and its suppression of COP1/SPA activity can at least partially explain the blue-light-dependent stabilization of transcription factors, such as LONG HYPOCOTYL 5 (HY5) and CONSTANS (CO) (81, 88, 167, 191), which regulate hypocotyl growth and floral initiation, respectively (156, 159, 178). It is interesting that structurally similar CRY1 and CRY2 interact with SPA1 in different ways. The N-terminal PHR domain of CRY2 interacts with the N-terminal kinase-like domain of SPA1 in response to blue light, which enhances the CRY2–COP1 interaction (191). Exactly how the enhanced CRY2–COP1-SPA interaction inhibits the activity of the CUL4COP1-SPAs E3 ubiquitin ligase remains unclear. In contrast, the C-terminal CCE domain of CRY1 interacts with the C-terminal WD40 domain of SPA1 in a blue-light-dependent manner, which results in suppression of the SPA1–COP1 interaction and CUL4COP1-SPAs ligase activity (81, 88). Therefore, CRY1 apparently acts as a light-dependent competitive inhibitor of the COP1-SPA interaction and CUL4COP1-SPAs activity. Regardless of the complex details of the CRY-COP1-SPA tripartite interaction, it seems clear now that cryptochromes can directly interact with COP1 to form the CRY-COP1 complex, probably in a light-independent manner, whereas cryptochromes also interact with SPA1 but in a light-dependent manner. The CRY–SPA interaction conveys the light signal to alter the CRY-SPA-COP1 tripartite complex, suppressing CUL4COP1-SPAs E3 ligase activity to regulate the stability of other light-signaling proteins.
4.2. The CRY-CIB Complexes
The first blue-light-specific cryptochrome-interacting protein identified in plants was a basic helix-loop-helix (bHLH) transcription factor known as cryptochrome-interacting bHLH 1 (CIB1) (91). Because of its relatively high specificity, affinity, and robust photoresponsiveness, the blue-light-dependent CRY2-CIB1 interaction has been widely utilized as an optogenetic tool for biomedical research in mammalian models (66, 185). CIB1 belongs to the BEE/CIB subfamily (family 18) of bHLH transcription factors (37, 91). The BEE/CIB subfamily contains 17 members (152), including genes encoding BR enhanced expression 1–3 (BEE1–BEE3), CESTA, and ILI1 binding bHLH (IBH1), which regulate brassinosteroid signaling (37, 127, 186); BIGPETALp, which interacts with ARF8 to regulate petal growth (158); and activator of cell elongation (ACE) proteins, which also regulate cell elongation (57). Different family members of the BEE/CIB subfamily of bHLH proteins seem to regulate both overlapping and unique aspects of plant development. CRY2 mediates blue-light stimulation of the CIB1 activation of transcription of the FLOWERING LOCUST T (FT) gene, which encodes the central regulator of floral initiation, or florigen, that migrates from leaves to the apical meristem to promote floral meristem development (23, 65, 68, 91, 171, 181). Three other CIB1-like bHLH proteins, CIB2, CIB4, and CIB5, have also been shown to interact with photoactivated CRY2 and bind to the E-box (CANNTG) elements of the promoter of FT (91, 95). CRY2 does not seem to affect the affinity of CIB proteins to DNA or chromatins (91). However, it been recently reported that CIB proteins directly interact with CO to promote floral initiation (96), implying that CRY2 might affect CIB–CO interaction and FT transcription. Recently, more CIB1-related proteins, including BEE2, CIL1, and HBI1, have been shown to interact with photoactivated cryptochromes (163). And at least one of them, HBI1, has been shown to mediate CRY1-dependent blue-light inhibition of hypocotyl elongation (163). Interestingly, in contrast to the lack of effect of CRY2–CIB1 interaction on the affinity of the DNA-binding activity of CIB1, the CRY1–HBI1 interaction inhibits the DNA-binding activity of HBI1 (163). CIB and BIC proteins seem to be the only two groups of cryptochrome-interacting proteins that so far have not been reported to interact with signaling proteins of other photoreceptors. However, the mRNA or protein expression of CIBs and BICs is photoregulated by other photoreceptors. It was found that CIB proteins (CIB1, CIB3, CIB4, CIB5) are degraded in the dark or red light by the 26S proteasome, whereas blue light suppresses degradation of all of them. Surprisingly, neither cryptochromes nor the dark-active CUL4COP1-SPA E3 ubiquitin ligase is involved in the control of CIB degradation (90). It is intriguing that photoreceptors and E3 ligase other than the CIB-interacting photoreceptors cryptochromes or the cryptochrome-interacting CUL4COP1-SPA E3 ubiquitin ligase have emerged in evolution to regulate CIB degradation. Nevertheless, researchers have found that the LOV-domain photoreceptors ZTL and LKP2 mediate blue-light inhibition of CIB degradation (90). The E3 ubiquitin ligase responsible for CIB degradation has not been identified. Cryptochromes are known to interact with ZTL (60) that is a substrate receptor of the SCFZTL E3 ubiquitin ligases (41). However, exactly how cryptochrome- and ZTL-type photoreceptors interact with each other or with CIB proteins to coordinate blue-light suppression of CIB degradation remains unclear.
Blue light-dependent CRY2–CIB interactions are evolutionarily conserved, yet the physiological function of the CRY–CIB interaction may be diverse in different plant species. For example, photoactivated CRY2 interacts with CIB1 to promote CIB1 activation of flowering in Arabidopsis, whereas CRY2 interacts with CIB1 to suppress CIB1 promotion of leaf senescence in soybean (106). Exactly how the CRY2–CIB1 complex mediates the blue-light-responsive alteration of transcription remains unclear. Although few effects of CRY2 or light were observed for the CIB1–DNA or CIB1–chromatin interaction in an earlier study (91), a more recent study argued that CRY2 may enhance the DNA-binding affinity of CIB1 or that CRY2 itself may be a DNA-binding transcription activator (180). In this study, CRY2 and/or CIB1 was coexpressed in mammalian HEK293 cells; the transcription activation activity of CIB1 was analyzed by reporter genes, and the CIB1 DNA-binding affinity was analyzed by the affinity precipitation of a biotin-labeled DNA fragment mixed with cell lysates. In these assays, photoactivated CRY2 appears to enhance the affinity of CIB proteins to its target DNA (the E-box-containing second intron of the FT gene) and the CIB1 transcription activation activity for the reporter gene. However, if CRY2 itself exhibits both DNA-binding and transcription activation activity in the HEK293 system, the above results may be alternatively interpreted by the activity of CRY2. The possibility that CRY2 itself may act as a DNA-binding transcriptional activator seems particularly appealing because the presumed ancestors of cryptochromes, DNA photolyases, are DNA-binding proteins, and animal cryptochromes are transcription repressors, but additional work is needed to further test this possibility.
4.3. The CRY-PIF Complexes
In addition to CIB proteins, plant cryptochromes interact with another group of bHLH transcription factors, PIF (phytochrome interacting factor 1–7) proteins, which are phytochrome-interacting and G-box-binding transcription factors that belong to a phylogenetic clade remotely related to the CIB proteins (77, 120, 152). The pifq quadruple mutants (pif1pif3pif4pif5) exhibit a constitutive photomorphogenic phenotype, including growth arrest of hypocotyls (78, 145). Although PIF proteins are best known for their roles in the phytochrome-mediated photomorphogenesis responses, such as red light inhibition of hypocotyl elongation, these positive regulators of growth appear to act as systems integrators that integrate different signals, including hormones, sugar, circadian timing, and temperature (76, 120). In addition to red light, the pif4pif5 and pifq mutants exhibit a short hypocotyl phenotype in blue light, implying an association of PIF proteins blue-light inhibition of growth (72, 97). Photoactivated CRY1 and CRY2 can physically interact with PIF4 and PIF5 via the PHR domain of cryptochromes and the N-terminal domain of PIF proteins in a region distinct from the phytochrome-binding motif (98, 123). It was shown that under shade conditions, canopies decrease not only the red-to-far-red ratio but also blue light intensity and that both phyB and cryptochromes mediate shade-avoidance responses in low blue light (123). phyB binds PIF3 to promote PIF3 phosphorylation, ubiquitination, and degradation (115, 116), the low blue light in shade conditions may suppress phyB-dependent PIF degradation to promote growth. Indeed, it has been reported that cryptochromes suppress degradation of PIF1 in low blue light (16). The decreased blue-light intensity caused by canopy may decrease cryptochrome oligomerization to weaken the CRY–PIF interaction, resulting in increased accumulation of PIF proteins to promote hypocotyl elongation. Cryptochromes have also been previously shown to play important roles in temperature responses of Arabidopsis (10, 34, 39). A recent study showed that CRY1 interacts with PIF4 and PIF5 in a blue-light-dependent manner to affect the function of CRY1 in hypocotyl growth at high ambient temperatures (98). Similar to the CRY2–CIB1 interaction, the CRY1–PIF4 interaction does not affect the DNA- or chromatin-binding activity of PIF4, but it suppresses the transcription activation activity of PIF4, affecting transcription of the auxin biosynthesis or signaling genes, and hypocotyl elongation of seedlings grown at high ambient temperatures (98).
4.4. The CRY-PPK Complexes
Arabidopsis CRY1 and CRY2 undergo blue-light-dependent phosphorylation in plant cells (143, 144). Phosphorylation of CRY2 not only enhances its activity but also facilitates its ubiquitination and degradation by CUL4COP1/SPA and other E3 ubiquitin ligases (94, 143, 167, 182). Label-free quantitative mass spectrometry analyses of Arabidopsis CRY2 proteins purified from plants identified at least two dozen phosphorylated residues of CRY2, including 18 serine and 6 threonine residues (93, 160). The level of phosphorylation in almost all those phosphorylated residues increased in response to blue light (160). Surprisingly, both the S-to-A and the phosphomimetic S-to-D mutants of CRY2 exhibited a loss of phosphorylation and partial loss of function in physiological activities. Given that an aspartate (−1 charge per residue) carries relatively fewer negative charges than phosphorylated serine (−1.5 to −2 charges per residue) at pH7.2 estimated for the Arabidopsis nuclear compartment, this observation suggests that photoactivation of cryptochromes may change charge distribution to facilitate domain disengagement (184). Although cryptochromes have been shown to undergo autophosphorylation in vitro, their phosphorylation in vivo is primarily catalyzed by protein kinases. Four closely related CRY2-associated protein kinases, referred to as photoregulatory protein kinases (PPK1 to PPK4), have been shown to catalyze blue-light-induced phosphorylation of CRY1 and CRY2 in vivo or ex vivo (93, 115). PPKs are plant-specific protein kinases evolutionarily derived from the ubiquitous casein kinase I, which were previously called MUT9-like kinases (166). All four PPKs preferentially interact with photoexcited but unphosphorylated cryptochromes, and they phosphorylate cryptochromes in a partially redundant manner. The blue-light-dependent phosphorylation of CRY1 or CRY2 appears normal in the monogenic ppk mutants but is largely absent in the ppk1ppk2ppk3 and ppk1ppk2ppk4 triple mutants and the artificial microRNA lines (amiR4k) with reduced expression of all 4 PPK genes. Unphosphorylated CRY2 proteins are not ubiquitinated nor degraded in the ppk triple mutants, confirming the previous prediction that CRY2 phosphorylation is required for its subsequent ubiquitination and degradation (143, 160, 161, 182). These results support a model demonstrating that photoexcited cryptochromes are phosphorylated by four structurally related and functionally redundant PPK kinases; phosphorylation of cryptochromes causes charge-dependent conformational changes to enhance the physiological activity of both CRY1 and CRY2 as well as the polyubiquitination and degradation of CRY2 (Figure 3). PPKs have other substrates in addition to cryptochromes, including the phytochrome-signaling protein PIF3 (115), histone H2A (149), histone H3 (166), and probably circadian clock proteins (55), suggesting that PPKs may act as a hub of multiple signaling interactions.
4.5. The CRY-AUX/IAA and CRY-AGB1 Complexes
Plant hormones are major internal regulatory molecules of plant growth and development, and photoresponses inevitably interact with hormonal regulation networks in plants (27). Cryptochromes mediate genome-wide gene expression changes in response to blue light, including changes in mRNA expression of biosynthesis and signaling genes of various phytohormones, including auxin, gibberellin, and brassinosteroid (33, 67, 99, 100, 118). Regulation of auxin-responsive gene expression has been proposed to explain the cryptochrome-mediated blue-light inhibition of cell elongation (170). Recent studies found that the photoactivated cryptochromes can physically interact with the AUX/IAA proteins to accomplish this. AUX/IAA proteins are a family of small (25 to 35 kDa), auxin-inducible transcription repressors that bind to auxin response factors (ARFs) to suppress ARF activity and the transcription of auxin-responsive genes (133, 168). The Arabidopsis genome encodes 29 AUX/IAA proteins, which, together with 23 ARF proteins, can regulate the diverse auxin responses in plants. The SCFTIR/AFB E3 ubiquitin ligase complex acts as an auxin receptor that binds to auxin to facilitate interaction between the substrate receptor transport inhibitor response 1/auxin signaling F-box (TIR1/AFB) proteins of the SCFTIR/AFB E3 ligase and its substrates, AUX/IAA proteins, leading to the ubiquitination and degradation of the AUX/IAA proteins, release of the inhibition of ARF proteins, and promotion of auxin-responsive gene expression and cell elongation. This general auxin-signaling mechanism allows plants to control growth by regulating the homeostasis of the AUX/IAA and ARF proteins, the TIR/AFB-AUX/IAA interaction, or the AUX/IAA-ARF interaction, in response to different environmental factors, such as light. For example, auxin induces the TIR1–AUX/IAA interaction in the dark to promote cell elongation in etiolated seedlings or plants under shade, whereas light inhibits the auxin-induced TIR1–AUX/IAA interaction and AUX/IAA degradation. In plants exposed to blue light, the PHR domains of photoactivated cryptochromes interact with AUX/IAA proteins, including IAA7, IAA12, and IAA17 (175). The interaction between cryptochromes and those IAA proteins suppresses the TIR1–IAA interaction and IAA degradation (175). Therefore, cryptochromes may act as blue-light-dependent competitive inhibitors of auxin signal transduction and hypocotyl elongation (175). Phytochromes also physically interact with the AUX/IAA proteins in response to red or far-red light (22, 175, 177). And similar to the CRY–AUX/IAA interaction in blue light, the phytochrome–AUX/IAA interaction causes suppression of the TIR1–AUX/IAA interaction, AUX/IAA degradation, and cell elongation under red light or shade conditions.
In addition to interacting with AUX/IAA proteins, Arabidopsis cryptochromes also interact with the G-protein β subunit AGB1 (80). The photoactivated CRY1 interacts with AGB1 to disrupt the AGB1–HY5 interaction, which positively regulates the DNA-binding activity of HY5 to promote photomorphogenesis. Although heterotrimeric G proteins are usually plasma membrane proteins that interact with G protein–coupled receptors in response to light, odors, hormones, and growth factors in animals (169), it is not uncommon that in animal cells the subunits of G proteins are imported to the nucleus to regulate the activity of transcription factors (49). Moreover, both HY5 and G proteins are known to participate in the auxin responses in plants (20, 62, 153), which is consistent with the proposition that CRY1-AGB1 regulation may also be associated with the light-auxin crosstalk.
4.6. The CRY-BR (Brassinosteroid Regulators) Complexes
In addition to auxin, plant cryptochromes also integrate blue-light signals with brassinosteroid signals by interacting with brassinosteroid-signaling proteins or brassinosteroid regulators (BRs). It has been reported recently that photoactivated Arabidopsis cryptochromes interact with several -brassinosteroid-signaling proteins (collectively called brassinosteroid regulators here), including BRI1-EMS-SUPPRESSOR 1 (BES1) and BRASSINAZOLE-RESISTANT 1 (BZR1), related master transcription factors that positively regulate brassinosteroid signaling; BES1-INTERACTING MYC-LIKE1 (BIM1), a bHLH transcription factor interacting with BES1; and BR-INSENSITIVE 2 (BIN2), a GSK3-like protein kinase catalyzing phosphorylation of BZR1 and BES1 to prevent their nuclear importation and function (44, 164). Plant cryptochromes may interact with these BRs to suppress brassinosteroid signaling by multiple mechanisms. For example, CRY1 interacts with BIN2 to enhance BIN2-dependent phosphorylation and cytoplasmic retention of BZR1, whereas CRY1 preferentially interacts with the dephosphorylated forms of BES1 and BZR1 to inhibit their DNA-binding activity and brassinosteroid-responsive transcription (44, 164). Because these brassinosteroid regulators interact with each other, cryptochromes may form a dynamic complex or complexes composed of these proteins to directly regulate the modulation of brassinosteroid-responsive transcriptional regulons in response to both the internal brassinosteroid signal and the external light signal.
5. FUTURE CHALLENGES
We anticipate that new components of the cryptochrome complexome, in addition to those reported so far, will be discovered to further our knowledge of plant cryptochromes. However, there are at least three major challenges to a mechanistic understanding of cryptochrome-mediated photoresponses in plants: What are the exact conformational and structural changes of the photoactivated cryptochrome monomer and oligomers, how do cryptochromes mediate photoresponses in different cells, and what are the mechanisms of cryptochrome regulation of gene expression other than their general roles regulating transcription and protein stability? First, it may be fair to say that cryptochromes are among the structurally least understood plant photoreceptors at present. This holdup is partially explained by the technical difficulty of obtaining suitable protein crystals of a full-length plant cryptochrome. The domain disengagement hypothesis predicts relatively large photoresponsive conformational changes of plant cryptochromes, whereas the crystal-packing effect of protein crystals and lack of stringent dark control due to inevitable X-ray illumination in diffraction crystallography may impose additional challenges to observing the exact photoresponsive conformational changes of cryptochromes in crystallography. Recent advancements in cryo-electron microscopy technology could potentially solve those technical hurdles. Second, plant cryptochromes appear to be expressed ubiquitously, but light is unlikely to have the same effects on different cells and organs of a plant. Therefore, how to distinguish the specific functions of cryptochromes in the specific photoresponses of individual cells is another challenge. Recent advances in single-cell RNA and protein analysis technologies would likely bring new insights about the novel and cell-specific functions of plant cryptochromes. Finally, most mechanistic studies of plant cryptochromes are presently limited to the cryptochrome-mediated regulation of the rate of mRNA transcription and ubiquitination-dependent proteolysis. It remains unclear whether or how cryptochromes affect other steps within the gene expression process or the specific mechanisms of individual steps. For example, it remains unknown whether cryptochromes regulate the initiation, elongation, or termination of transcription, or how cryptochromes alter cotranscriptional or post-transcriptional mRNA modifications, such as RNA methylation, to affect mRNA degradation, nuclear exportation, or translation. Again, newly available methodologies for the analyses of transcriptomes, epitranscriptomes, translatomes, proteomes, or individual cryptochrome-targeting genes and proteins will shed more light on the mechanisms underlying cryptochrome-mediated photoresponses in plants.
Cryptochrome: blue-light receptor that mediates photoresponses in plants
HY4: long hypocotyl 4
FAD: flavin adenine dinucleotide
PHR: photolyase homologous region
CCE: cryptochrome C-terminal extension (also known as CCT, CTT, etc.)
DAS: DQXVP-acidic-STAES
COP1: constitutive photomorphogenic 1
DASH: Drosophila, Arabidopsis, Synechocystis, human
HEK293: human embryonic kidney 293
BIC: blue-light inhibitor of cryptochrome
CIB: cryptochrome-interacting basic helix-loop-helix
SPA: suppressor of phytochrome A and related proteins
AUX/IAA: auxin/indole-3-acetic acid
PIF: phytochrome-interacting factor
PPK: photoregulatory protein kinase
ARF: auxin response factor
TIR1/AFB: Transport Inhibitor Response 1/Auxin Signaling F-Box
BR: brassinosteroid regulator
BES1: BRI1-EMS-SUPPRESSOR 1
BZR1: brassinazole-resistant 1
BIM1: Bes1-interacting Myc-like1
BIN2: BR-insensitive 2
ACKNOWLEDGMENTS
The authors thank Drs. Tiantian Su, Xu Wang, Zecheng Zuo, and Qing Liu for helpful discussions and sharing unpublished experimental results. Work in the authors’ laboratory is supported in part by the National Natural Science Foundation of China (31970265 to Q.W.), Natural Science Foundation of Fujian Province (2019J06014 to Q.W.), and the National Institutes of Health (GM56265 to C.L.).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Ahmad M 2016. Photocycle and signaling mechanisms of plant cryptochromes. Curr. Opin. Plant Biol 33:108–15 [DOI] [PubMed] [Google Scholar]
- 2.Ahmad M, Cashmore AR. 1993. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366:162–66 [DOI] [PubMed] [Google Scholar]; Genetic identification of the first cryptochrome.
- 3.Ahmad M, Galland P, Ritz T, Wiltschko R, Wiltschko W. 2007. Magnetic intensity affects cryptochrome-dependent responses in Arabidopsis thaliana. Planta 225:615–24 [DOI] [PubMed] [Google Scholar]
- 4.Ahmad M, Jarillo JA, Smirnova O, Cashmore AR. 1998. The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol. Cell 1:939–48 [DOI] [PubMed] [Google Scholar]
- 5.Ahmad M, Jarillo JA, Smirnova O, Cashmore AR. 1998. Cryptochrome blue-light photoreceptors of Arabidopsis implicated in phototropism. Nature 392:720–23 [DOI] [PubMed] [Google Scholar]
- 6.Balland V, Byrdin M, Eker AP, Ahmad M, Brettel K. 2009. What makes the difference between a cryptochrome and DNA photolyase? A spectroelectrochemical comparison of the flavin redox transitions. J. Am. Chem. Soc 131:426–27 [DOI] [PubMed] [Google Scholar]
- 7.Barrero JM, Downie AB, Xu Q, Gubler F. 2014. A role for barley CRYPTOCHROME1 in light regulation of grain dormancy and germination. Plant Cell 26:1094–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beel B, Prager K, Spexard M, Sasso S, Weiss D, et al. 2012. A flavin binding cryptochrome photoreceptor responds to both blue and red light in Chlamydomonas reinhardtii. Plant Cell 24:2992–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biskup T, Hitomi K, Getzoff ED, Krapf S, Koslowski T, et al. 2011. Unexpected electron transfer in cryptochrome identified by time-resolved EPR spectroscopy. Angew. Chem. Int. Ed 50:12647–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blazquez MA, Ahn JH, Weigel D. 2003. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat. Genet 33:168–71 [DOI] [PubMed] [Google Scholar]
- 11.Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, et al. 2004. Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. PNAS 101:12142–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV. 2013. Optogenetic protein clustering and signaling activation in mammalian cells. Nat. Methods 10:249–52 [DOI] [PubMed] [Google Scholar]
- 13.Cailliez F, Müller P, Gallois M, de la Lande A. 2014. ATP binding and aspartate protonation enhance photoinduced electron transfer in plant cryptochrome. J. Am. Chem. Soc 136:12974–86 [DOI] [PubMed] [Google Scholar]
- 14.Cashmore AR. 2003. Cryptochromes: enabling plants and animals to determine circadian time. Cell 114:537–43 [PubMed] [Google Scholar]
- 15.Cashmore AR, Jarillo JA, Wu YJ, Liu D. 1999. Cryptochromes: blue light receptors for plants and animals. Science 284:760–65 [DOI] [PubMed] [Google Scholar]
- 16.Castillon A, Shen H, Huq E. 2009. Blue light induces degradation of the negative regulator phytochrome interacting factor 1 to promote photomorphogenic development of Arabidopsis seedlings. Genetics 182:161–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatterjee M, Sharma P, Khurana JP. 2006. Cryptochrome 1 from Brassica napus is up-regulated by blue light and controls hypocotyl/stem growth and anthocyanin accumulation. Plant Physiol. 141:61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, et al. 2011. The cryptochromes: blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol 62:335–64 [DOI] [PubMed] [Google Scholar]
- 19.Christie JM. 2007. Phototropin blue-light receptors. Annu. Rev. Plant Biol 58:21–45 [DOI] [PubMed] [Google Scholar]
- 20.Cluis CP, Mouchel CF, Hardtke CS. 2004. The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J. 38:332–47 [DOI] [PubMed] [Google Scholar]
- 21.Coesel S, Mangogna M, Ishikawa T, Heijde M, Rogato A, et al. 2009. Diatom PtCPF1 is a new cryptochrome/photolyase family member with DNA repair and transcription regulation activity. EMBO Rep. 10:655–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colón-Carmona A, Chen DL, Yeh KC, Abel S. 2000. Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 124:1728–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corchnoy SB, Swartz TE, Lewis JW, Szundi I, Briggs WR, Bogomolni RA. 2003. Intramolecular proton transfers and structural changes during the photocycle of the LOV2 domain of phototropin 1. J. Biol. Chem 278:724–31 [DOI] [PubMed] [Google Scholar]
- 24.Czarna A, Berndt A, Singh HR, Grudziecki A, Ladurner AG, et al. 2013. Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function. Cell 153:1394–405 [DOI] [PubMed] [Google Scholar]
- 25.Danon A, Coll NS, Apel K. 2006. Cryptochrome-1-dependent execution of programmed cell death induced by singlet oxygen in Arabidopsis thaliana. PNAS 103:17036–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Riso V, Raniello R, Maumus F, Rogato A, Bowler C, Falciatore A. 2009. Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res. 37:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Wit M, Galvão VC, Fankhauser C. 2016. Light-mediated hormonal regulation of plant growth and development. Annu. Rev. Plant Biol 67:513–37 [DOI] [PubMed] [Google Scholar]
- 28.Deng X-W, Matsui M, Wei N, Wagner D, Chu AM, et al. 1992. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a Gβ homologous domain. Cell 71:791–801 [DOI] [PubMed] [Google Scholar]
- 29.El-Assal SE-D, Alonso-Blanco C, Hanhart CJ, Koornneef M. 2004. Pleiotropic effects of the Arabidopsis cryptochrome 2 allelic variation underlie fruit trait-related QTL. Plant Biol. 6:370–74 [DOI] [PubMed] [Google Scholar]
- 30.El-Assal SE-D, Alonso-Blanco C, Peeters AJ, Raz V, Koornneef M. 2001. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat. Genet 29:435–40 [DOI] [PubMed] [Google Scholar]
- 31.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. 1998. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95:669–79 [DOI] [PubMed] [Google Scholar]
- 32.Essen L-O, Franz S, Banerjee A. 2017. Structural and evolutionary aspects of algal blue light receptors of the cryptochrome and aureochrome type. J. Plant Physiol 217:27–37 [DOI] [PubMed] [Google Scholar]
- 33.Folta KM, Pontin MA, Karlin-Neumann G, Bottini R, Spalding EP. 2003. Genomic and physiological studies of early cryptochrome 1 action demonstrate roles for auxin and gibberellin in the control of hypocotyl growth by blue light. Plant J. 36:203–14 [DOI] [PubMed] [Google Scholar]
- 34.Foreman J, Johansson H, Hornitschek P, Josse EM, Fankhauser C, Halliday KJ. 2011. Light receptor action is critical for maintaining plant biomass at warm ambient temperatures. Plant J. 65:441–52 [DOI] [PubMed] [Google Scholar]
- 35.Fortunato AE, Annunziata R, Jaubert M, Bouly J-P, Falciatore A. 2015. Dealing with light: the widespread and multitasking cryptochrome/photolyase family in photosynthetic organisms. J. Plant Physiol 172:42–54 [DOI] [PubMed] [Google Scholar]
- 36.Franz S, Ignatz E, Wenzel S, Zielosko H, Putu EPGN, et al. 2018. Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii. Nucleic Acids Res. 46:8010–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, et al. 2002. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162:1445–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao J, Wang X, Zhang M, Bian M, Deng W, et al. 2015. Trp triad-dependent rapid photoreduction is not required for the function of Arabidopsis CRY1. PNAS 112:9135–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gould PD, Ugarte N, Domijan M, Costa M, Foreman J, et al. 2013. Network balance via CRY signalling controls the Arabidopsis circadian clock over ambient temperatures. Mol. Syst. Biol 9:650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo H, Yang H, Mockler TC, Lin C. 1998. Regulation of flowering time by Arabidopsis photoreceptors. Science 279:1360–63 [DOI] [PubMed] [Google Scholar]
- 41.Han L, Mason M, Risseeuw EP, Crosby WL, Somers DE. 2004. Formation of an SCFZTL complex is required for proper regulation of circadian timing. Plant J. 40:291–301 [DOI] [PubMed] [Google Scholar]
- 42.Han X, Chang X, Zhang Z, Chen H, He H, et al. 2019. Origin and evolution of core components responsible for monitoring light environment changes during plant terrestrialization. Mol. Plant 12:847–62 [DOI] [PubMed] [Google Scholar]
- 43.Harris SR, Henbest KB, Maeda K, Pannell JR, Timmel CR, et al. 2009. Effect of magnetic fields on cryptochrome-dependent responses in Arabidopsis thaliana. J. R. Soc. Interface 6:1193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He G, Liu J, Dong H, Sun J. 2019. The blue-light receptor CRY1 interacts with BZR1 and BIN2 to modulate the phosphorylation and nuclear function of BZR1 in repressing BR signaling in Arabidopsis. Mol. Plant 12:689–703 [DOI] [PubMed] [Google Scholar]
- 45.Heijde M, Ulm R. 2012. UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 17:230–37 [DOI] [PubMed] [Google Scholar]
- 46.Heijde M, Zabulon G, Corellou F, Ishikawa T, Brazard J, et al. 2010. Characterization of two members of the cryptochrome/photolyase family from Ostreococcus tauri provides insights into the origin and evolution of cryptochromes. Plant Cell Environ. 33:1614–26 [DOI] [PubMed] [Google Scholar]
- 47.Herbel V, Orth C, Wenzel R, Ahmad M, Bittl R, Batschauer A. 2013. Lifetimes of Arabidopsis cryptochrome signaling states in vivo. Plant J. 74:583–92 [DOI] [PubMed] [Google Scholar]
- 48.Hirose F, Shinomura T, Tanabata T, Shimada H, Takano M. 2006. Involvement of rice cryptochromes in de-etiolation responses and flowering. Plant Cell Physiol. 47:915–25 [DOI] [PubMed] [Google Scholar]
- 49.Ho MKC, Su Y, Yeung WWS, Wong YH. 2009. Regulation of transcription factors by heterotrimeric G proteins. Curr. Mol. Pharmacol 2:19–31 [DOI] [PubMed] [Google Scholar]
- 50.Hoecker U 2017. The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Curr. Opin. Plant Biol 37:63–69 [DOI] [PubMed] [Google Scholar]
- 51.Hoecker U, Tepperman JM, Quail PH. 1999. SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284:496–99 [DOI] [PubMed] [Google Scholar]
- 52.Holm M, Hardtke CS, Gaudet R, Deng XW. 2001. Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 20:118–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsu DS, Zhao X, Zhao S, Kazantsev A, Wang RP, et al. 1996. Putative human blue-light photoreceptors hCRY1 and hCRY2 are flavoproteins. Biochemistry 35:13871–77 [DOI] [PubMed] [Google Scholar]
- 54.Hu W, Franklin KA, Sharrock RA, Jones MA, Harmer SL, Lagarias JC. 2013. Unanticipated regulatory roles for Arabidopsis phytochromes revealed by null mutant analysis. PNAS 110:1542–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang H, Alvarez S, Bindbeutel R, Shen Z, Naldrett MJ, et al. 2016. Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol. Cell. Proteomics 15:201–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang X, Ouyang X, Deng XW. 2014. Beyond repression of photomorphogenesis: role switching of COP/DET/FUS in light signaling. Curr. Opin. Plant Biol 21:96–103 [DOI] [PubMed] [Google Scholar]
- 57.Ikeda M, Fujiwara S, Mitsuda N, Ohme-Takagi M. 2012. A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell 24:4483–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Imaizumi T, Kanegae T, Wada M. 2000. Cryptochrome nucleocytoplasmic distribution and gene expression are regulated by light quality in the fern Adiantum capillus-veneris. Plant Cell 12:81–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ito S, Song YH, Imaizumi T. 2012. LOV domain-containing F-box proteins: light-dependent protein degradation modules in Arabidopsis. Mol. Plant 5:573–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jarillo JA, Capel J, Tang RH, Yang HQ, Alonso JM, et al. 2001. An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature 410:487–90 [DOI] [PubMed] [Google Scholar]
- 61.Jenkins GI. 2014. The UV-B photoreceptor UVR8: from structure to physiology. Plant Cell 26:21–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones AM, Ecker JR, Chen J-G. 2003. A reevaluation of the role of the heterotrimeric G protein in coupling light responses in Arabidopsis. Plant Physiol. 131:1623–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kanai S, Kikuno R, Toh H, Ryo H, Todo T. 1997. Molecular evolution of the photolyase-blue-light photoreceptor family. J. Mol. Evol 45:535–48 [DOI] [PubMed] [Google Scholar]
- 64.Kang CY, Lian HL, Wang FF, Huang JR, Yang HQ. 2009. Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21:2624–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, et al. 1999. Activation tagging of the floral inducer FT. Science 286:1962–65 [DOI] [PubMed] [Google Scholar]
- 66.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. 2010. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 7:973–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kleine T, Kindgren P, Benedict C, Hendrickson L, Strand A. 2007. Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol. 144:1391–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. 1999. A pair of related genes with antagonistic roles in mediating flowering signals. Science 286:1960–62 [DOI] [PubMed] [Google Scholar]
- 69.König S, Juhas M, Jäger S, Kottke T, Büchel C. 2017. The cryptochrome—photolyase protein family in diatoms. J. Plant Physiol 217:15–19 [DOI] [PubMed] [Google Scholar]
- 70.Kottke T, Batschauer A, Ahmad M, Heberle J. 2006. Blue-light-induced changes in Arabidopsis cryptochrome 1 probed by FTIR difference spectroscopy. Biochemistry 45:2472–79 [DOI] [PubMed] [Google Scholar]
- 71.Kottke T, Oldemeyer S, Wenzel S, Zou Y, Mittag M. 2017. Cryptochrome photoreceptors in green algae: unexpected versatility of mechanisms and functions. J. Plant Physiol 217:4–14 [DOI] [PubMed] [Google Scholar]
- 72.Kunihiro A, Yamashino T, Mizuno T. 2010. PHYTOCHROME-INTERACTING FACTORS PIF4 and PIF5 are implicated in the regulation of hypocotyl elongation in response to blue light in Arabidopsis thaliana. Biosci. Biotechnol. Biochem 74:2538–41 [DOI] [PubMed] [Google Scholar]
- 73.Lasceve G, Leymarie J, Olney MA, Liscum E, Christie JM, et al. 1999. Arabidopsis contains at least four independent blue-light-activated signal transduction pathways. Plant Physiol. 120:605–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lau K, Podolec R, Chappuis R, Ulm R, Hothorn M. 2019. Plant photoreceptors and their signaling components compete for COP1 binding via VP peptide motifs. EMBO J. 38:e102140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lau OS, Deng XW. 2012. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17:584–93 [DOI] [PubMed] [Google Scholar]
- 76.Leivar P, Monte E. 2014. PIFs: systems integrators in plant development. Plant Cell 26:56–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leivar P, Quail PH. 2011. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16:19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH. 2009. Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21:3535–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li X, Wang Q, Yu X, Liu H, Yang H, et al. 2011. Arabidopsis cryptochrome 2 (CRY2) functions by the photoactivation mechanism distinct from the tryptophan (trp) triad-dependent photoreduction. PNAS 108:20844–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lian H, Xu P, He S, Wu J, Pan J, et al. 2018. Photoexcited CRYPTOCHROME 1 interacts directly with G-protein β subunit AGB1 to regulate the DNA-binding activity of HY5 and photomorphogenesis in Arabidopsis. Mol. Plant 11:1248–63 [DOI] [PubMed] [Google Scholar]
- 81.Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, et al. 2011. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 25:1023–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin C, Ahmad M, Gordon D, Cashmore AR. 1995. Expression of an Arabidopsis cryptochrome gene in transgenic tobacco results in hypersensitivity to blue, UV-A, and green light. PNAS 92:8423–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin C, Robertson DE, Ahmad M, Raibekas AA, Jorns MS, et al. 1995. Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269:968–70 [DOI] [PubMed] [Google Scholar]; Biochemical characterization of the first cryptochrome.
- 84.Lin C, Shalitin D. 2003. Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol 54:469–96 [DOI] [PubMed] [Google Scholar]
- 85.Lin C, Top D, Manahan CC, Young MW, Crane BR. 2018. Circadian clock activity of cryptochrome relies on tryptophan-mediated photoreduction. PNAS 115:3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR. 1998. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. PNAS 95:2686–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu B, Liu H, Zhong D, Lin C. 2010. Searching for a photocycle of the cryptochrome photoreceptors. Curr. Opin. Plant Biol 13:578–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu B, Zuo Z, Liu H, Liu X, Lin C. 2011. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 25:1029–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu H, Su T, He W, Wang Q, Lin C. 2019. The universally conserved residues are not universally required for stable protein expression or functions of cryptochromes. Mol. Biol. Evol 37:327–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu H, Wang Q, Liu Y, Zhao X, Imaizumi T, et al. 2013. Arabidopsis CRY2 and ZTL mediate blue-light regulation of the transcription factor CIB1 by distinct mechanisms. PNAS 110:17582–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu H, Yu X, Li K, Klejnot J, Yang H, et al. 2008. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322:1535–39 [DOI] [PubMed] [Google Scholar]; Identification of the first blue light-specific cryptochrome-interacting protein.
- 92.Liu Q, Su T, He W, Ren H, Liu S, et al. 2020. Photooligomerization determines photosensitivity and photoreactivity of plant cryptochromes. Mol. Plant In press. 10.1016/j.molp.2020.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu Q, Wang Q, Deng W, Wang X, Piao M, et al. 2017. Molecular basis for blue light-dependent phosphorylation of Arabidopsis cryptochrome 2. Nat. Commun 8:15234. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identification of four protein kinases that phosphorylate cryptochromes.
- 94.Liu Q, Wang Q, Liu B, Wang W, Wang X, et al. 2016. The blue light-dependent polyubiquitination and degradation of Arabidopsis cryptochrome2 requires multiple E3 ubiquitin ligases. Plant Cell Physiol. 57:2175–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu Y, Li X, Li K, Liu H, Lin C. 2013. Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLOS Genet. 9:e1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu Y, Li X, Ma D, Chen Z, Wang J-W, Liu H. 2018. CIB1 and CO interact to mediate CRY2-dependent regulation of flowering. EMBO Rep. 19:e45762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Luo Q, Lian H-L, He S-B, Li L, Jia K-P, Yang H-Q. 2014. COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell 26:2441–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ma D, Li X, Guo Y, Chu J, Fang S, et al. 2016. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. PNAS 113:224–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ma L, Chen C, Liu X, Jiao Y, Su N, et al. 2005. A microarray analysis of the rice transcriptome and its comparison to Arabidopsis. Genome Res. 15:1274–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ma L, Li J, Qu L, Hager J, Chen Z, et al. 2001. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13:2589–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Malhotra K, Kim ST, Batschauer A, Dawut L, Sancar A. 1995. Putative blue-light photoreceptors from Arabidopsis thaliana and Sinapis alba with a high degree of sequence homology to DNA photolyase contain the two photolyase cofactors but lack DNA repair activity. Biochemistry 34:6892–99 [DOI] [PubMed] [Google Scholar]; Biochemical characterization of the first cryptochrome.
- 102.Mao J, Zhang Y-C, Sang Y, Li Q-H, Yang H-Q. 2005. A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. PNAS 102:12270–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mas P, Devlin PF, Panda S, Kay SA. 2000. Functional interaction of phytochrome B and cryptochrome 2. Nature 408:207–11 [DOI] [PubMed] [Google Scholar]; Discovery of the cryptochrome photobody.
- 104.McCormick DB, Koster JF, Veeger C. 1967. On the mechanisms of photochemical reductions of FAD and FAD-dependent flavoproteins. Eur. J. Biochein 2:387–91 [DOI] [PubMed] [Google Scholar]
- 105.Mei Q, Dvornyk V. 2015. Evolutionary history of the photolyase/cryptochrome superfamily in eukaryotes. PLOS ONE 10:e0135940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Meng Y, Li H, Wang Q, Liu B, Lin C. 2013. Blue light-dependent interaction between cryptochrome2 and CIB1 regulates transcription and leaf senescence in soybean. Plant Cell 25:4405–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, et al. 2007. Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J. 52:973–86 [DOI] [PubMed] [Google Scholar]
- 108.Miyamoto Y, Sancar A. 1998. Vitamin B2-based blue-light photoreceptors in the retinohypothalamic tract as the photoactive pigments for setting the circadian clock in man and mouse. PNAS 95:6097–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mockler T, Yang H, Yu X, Parikh D, Cheng Y-C, et al. 2003. Regulation of photoperiodic flowering by Arabidopsis photoreceptors. PNAS 100:2140–45 [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstration of the redundant functions of phyA, CRY1, and CRY2 in flowering.
- 110.Muller M, Carell T. 2009. Structural biology of DNA photolyases and cryptochromes. Curr. Opin. Struct. Biol 19:277–85 [DOI] [PubMed] [Google Scholar]
- 111.Müller N, Wenzel S, Zou Y, Künzel S, Sasso S, et al. 2017. A plant cryptochrome controls key features of the Chlamydomonas circadian clock and its life cycle. Plant Physiol. 174:185–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Müller P, Bouly J-P. 2015. Searching for the mechanism of signalling by plant photoreceptor cryptochrome. FEBS Lett. 589:189–92 [DOI] [PubMed] [Google Scholar]
- 113.Müller P, Bouly J-P, Hitomi K, Balland V, Getzoff ED, et al. 2014. ATP binding turns plant cryptochrome into an efficient natural photoswitch. Sci. Rep 4:5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Müller P, Yamamoto J, Martin R, Iwai S, Brettel K. 2015. Discovery and functional analysis of a 4th electron-transferring tryptophan conserved exclusively in animal cryptochromes and (6–4) photolyases. Chem. Commun 51:15502–5 [DOI] [PubMed] [Google Scholar]
- 115.Ni W, Xu S-L, González-Grandío E, Chalkley RJ, Huhmer AFR, et al. 2017. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun 8:15236. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstration of the common kinases shared by the cryptochrome- and phytochrome-signaling.
- 116.Ni W, Xu S-L, Tepperman JM, Stanley DJ, Maltby DA, et al. 2014. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344:1160–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nohr D, Franz S, Rodriguez R, Paulus B, Essen L-O, et al. 2016. Extended electron-transfer in animal cryptochromes mediated by a tetrad of aromatic amino acids. Biophys. J 111:301–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ohgishi M, Saji K, Okada K, Sakai T. 2004. Functional analysis of each blue light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis. PNAS 101:2223–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ozkan-Dagliyan I, Chiou Y-Y, Ye R, Hassan BH, Ozturk N, Sancar A. 2013. Formation of Arabidopsis cryptochrome 2 photobodies in mammalian nuclei: application as an optogenetic DNA damage checkpoint switch. J. Biol. Chem 288:23244–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Paik I, Kathare PK, Kim J-I, Huq E. 2017. Expanding roles of PIFs in signal integration from multiple processes. Mol. Plant 10:1035–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Partch CL, Clarkson MW, Ozgur S, Lee AL, Sancar A. 2005. Role of structural plasticity in signal transduction by the cryptochrome blue-light photoreceptor. Biochemistry 44:3795–805 [DOI] [PubMed] [Google Scholar]; Demonstration of the light-induced conformational change of cryptochromes.
- 122.Patanjali SR, Parimoo S, Weissman SM. 1991. Construction of a uniform-abundance (normalized) cDNA library. PNAS 88:1943–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pedmale UV, Huang SC, Zander M, Cole BJ, Hetzel J, et al. 2016. Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164:233–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Penzer GR, Radda GK. 1968. The chemistry of flavines and flavoproteins: photoreduction of flavines by amino acids. Biochem. J 109:259–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Podolec R, Ulm R. 2018. Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr. Opin. Plant Biol 45:18–25 [DOI] [PubMed] [Google Scholar]
- 126.Pooam M, Arthaut L-D, Burdick D, Link J, Martino CF, Ahmad M. 2019. Magnetic sensitivity mediated by the Arabidopsis blue-light receptor cryptochrome occurs during flavin reoxidation in the dark. Planta 249:319–32 [DOI] [PubMed] [Google Scholar]
- 127.Poppenberger B, Rozhon W, Khan M, Husar S, Adam G, et al. 2011. CESTA, a positive regulator of brassinosteroid biosynthesis. EMBO J. 30:1149–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Quail PH. 2010. Phytochromes. Curr. Biol 20:R504–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Reisdorph NA, Small GD. 2004. The CPH1 gene of Chlamydomonas reinhardtii encodes two forms of cryptochrome whose levels are controlled by light-induced proteolysis. Plant Physiol. 134:1546–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rockwell NC, Su YS, Lagarias JC. 2006. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol 57:837–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rosenfeldt G, Viana RM, Mootz HD, von Arnim AG, Batschauer A. 2008. Chemically induced and light-independent cryptochrome photoreceptor activation. Mol. Plant 1:4–14 [DOI] [PubMed] [Google Scholar]
- 132.Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, et al. 2003. The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 17:2642–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Salehin M, Bagchi R, Estelle M. 2015. SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27:9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sancar A 1994. Structure and function of DNA photolyase. Biochemistry 33:2–9 [DOI] [PubMed] [Google Scholar]
- 135.Sancar A 2000. Cryptochrome: the second photoactive pigment in the eye and its role in circadian photoreception. Annu. Rev. Biochem 69:31–67 [DOI] [PubMed] [Google Scholar]
- 136.Sancar A 2003. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev 103:2203–37 [DOI] [PubMed] [Google Scholar]
- 137.Sang Y, Li QH, Rubio V, Zhang YC, Mao J, et al. 2005. N-terminal domain–mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell 17:1569–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schmalen I, Reischl S, Wallach T, Klemz R, Grudziecki A, et al. 2014. Interaction of circadian clock proteins CRY1 and PER2 is modulated by zinc binding and disulfide bond formation. Cell 157:1203–15 [DOI] [PubMed] [Google Scholar]
- 139.Schwarzlander M, Fricker MD, Muller C, Marty L, Brach T, et al. 2008. Confocal imaging of glutathione redox potential in living plant cells. J. Microsc 231:299–316 [DOI] [PubMed] [Google Scholar]
- 140.Selby CP, Sancar A. 2006. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. PNAS 103:17696–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Senda T, Senda M, Kimura S, Ishida T. 2009. Redox control of protein conformation in flavoproteins. Antioxid. Redox Signal 11:1741–66 [DOI] [PubMed] [Google Scholar]
- 142.Seo HS, Yang J-Y, Ishikawa M, Bolle C, Ballesteros ML, Chua N-H. 2003. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423:995–99 [DOI] [PubMed] [Google Scholar]
- 143.Shalitin D, Yang H, Mockler TC, Maymon M, Guo H, et al. 2002. Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417:763–67 [DOI] [PubMed] [Google Scholar]
- 144.Shalitin D, Yu X, Maymon M, Mockler T, Lin C. 2003. Blue light-dependent in vivo and in vitro phosphorylation of Arabidopsis cryptochrome 1. Plant Cell 15:2421–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shin J, Kim K, Kang H, Zulfugarov I, Bae G, et al. 2009. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. PNAS 106:7660–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Somers DE, Devlin PF, Kay SA. 1998. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282:1488–90 [DOI] [PubMed] [Google Scholar]; Demonstration of the function of cryptochromes in the circadian clock of plants.
- 147.Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, et al. 1998. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95:681–92 [DOI] [PubMed] [Google Scholar]
- 148.Strasser B, Sanchez-Lamas M, Yanovsky MJ, Casal JJ, Cerdan PD. 2010. Arabidopsis thaliana life without phytochromes. PNAS 107:4776–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Su Y, Wang S, Zhang F, Zheng H, Liu Y, et al. 2017. Phosphorylation of histone H2A at serine 95: a plant-specific mark involved in flowering time regulation and H2A.Z deposition. Plant Cell 29:2197–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tagua VG, Pausch M, Eckel M, Gutiérrez G, Miralles-Durán A, et al. 2015. Fungal cryptochrome with DNA repair activity reveals an early stage in cryptochrome evolution. PNAS 112:15130–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Takahashi JS. 2017. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet 18:164–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Toledo-Ortiz G, Huq E, Quail PH. 2003. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15:1749–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ullah H, Chen J-G, Temple B, Boyes DC, Alonso JM, et al. 2003. The β-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell 15:393–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Usami T, Mochizuki N, Kondo M, Nishimura M, Nagatani A. 2004. Cryptochromes and phytochromes synergistically regulate Arabidopsis root greening under blue light. Plant Cell Physiol. 45:1798–808 [DOI] [PubMed] [Google Scholar]
- 155.Uversky VN. 2019. Intrinsically disordered proteins and their “mysterious” (meta)physics. Front. Phys 7:10 [Google Scholar]
- 156.Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. 2004. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303:1003–6 [DOI] [PubMed] [Google Scholar]
- 157.van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, et al. 1999. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398:627–30 [DOI] [PubMed] [Google Scholar]
- 158.Varaud E, Brioudes F, Szécsi J, Leroux J, Brown S, et al. 2011. AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell 23:973–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang H, Ma LG, Li JM, Zhao HY, Deng XW. 2001. Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294:154–58 [DOI] [PubMed] [Google Scholar]; Demonstration of the first signaling mechanism of plant cryptochromes.
- 160.Wang Q, Barshop WD, Bian M, Vashisht AA, He R, et al. 2015. The blue light-dependent phosphorylation of the CCE domain determines the photosensitivity of Arabidopsis CRY2. Mol. Plant 8:631–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang Q, Zuo Z, Wang X, Gu L, Yoshizumi T, et al. 2016. Photoactivation and inactivation ofArabidopsis cryptochrome 2. Science 354:343–47 [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstration of photoactivation and inactivation mechanisms of a plant cryptochrome.
- 162.Wang Q, Zuo Z, Wang X, Liu Q, Gu L, et al. 2018. Beyond the photocycle—how cryptochromes regulate photoresponses in plants? Curr. Opin. Plant Biol 45:120–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang S, Li L, Xu P, Lian H, Wang W, et al. 2018. CRY1 interacts directly with HBI1 to regulate its transcriptional activity and photomorphogenesis in Arabidopsis. J. Exp. Bot 69:3867–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang W, Lu X, Li L, Lian H, Mao Z, et al. 2018. Photoexcited CRYPTOCHROME1 interacts with dephosphorylated BES1 to regulate brassinosteroid signaling and photomorphogenesis in Arabidopsis. Plant Cell 30:1989–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang X, Wang Q, Han Y-J, Liu Q, Gu L, et al. 2017. A CRY-BIC negative feedback circuitry regulating blue light sensitivity of Arabidopsis. Plant J. 92:426–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang Z, Casas-Mollano JA, Xu J, Riethoven J-JM, Zhang C, Cerutti H. 2015. Osmotic stress induces phosphorylation of histone H3 at threonine 3 in pericentromeric regions of Arabidopsis thaliana. PNAS 112:8487–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Weidler G, zur Oven-Krockhaus S, Heunemann M, Orth C, Schleifenbaum F, et al. 2012. Degradation of Arabidopsis CRY2 is regulated by SPA proteins and phytochrome A. Plant Cell 24:2610–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Weijers D, Wagner D. 2016. Transcriptional responses to the auxin hormone. Annu. Rev. Plant Biol 67:539–74 [DOI] [PubMed] [Google Scholar]
- 169.Weis WI, Kobilka BK. 2018. The molecular basis of G protein–coupled receptor activation. Annu. Rev. Biochem 87:897–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wickland DP, Hanzawa Y. 2015. The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: functional evolution and molecular mechanisms. Mol. Plant 8:P983–97 [DOI] [PubMed] [Google Scholar]
- 171.Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, et al. 2005. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309:1056–59 [DOI] [PubMed] [Google Scholar]
- 172.Wu G, Spalding EP. 2007. Separate functions for nuclear and cytoplasmic cryptochrome 1 during photomorphogenesis of Arabidopsis seedlings. PNAS 104:18813–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wu L, Yang H. 2010. CRYPTOCHROME 1 is implicated in promoting R protein-mediated plant resistance to Pseudomonas syringae in Arabidopsis. Mol. Plant 3:539–48 [DOI] [PubMed] [Google Scholar]
- 174.Xing W, Busino L, Hinds TR, Marionni ST, Saifee NH, et al. 2013. SCFFBXL3 ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature 496:64–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Xu F, He S, Zhang J, Mao Z, Wang W, et al. 2017. Photoactivated CRY1 and phyB interact directly with AUX/IAA proteins to inhibit auxin signaling in Arabidopsis. Mol. Plant 11:P521–41 [DOI] [PubMed] [Google Scholar]
- 176.Xu X, Paik I, Zhu L, Huq E. 2015. Illuminating progress in phytochrome-mediated light signaling pathways. Trends Plant Sci. 20:641–50 [DOI] [PubMed] [Google Scholar]
- 177.Yang C, Xie F, Jiang Y, Li Z, Huang X, Li L. 2018. Phytochrome A negatively regulates the shade avoidance response by increasing auxin/indole acidic acid protein stability. Dev. Cell 44:29–41.e4 [DOI] [PubMed] [Google Scholar]
- 178.Yang H-Q, Tang R-H, Cashmore AR. 2001. The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13:2573–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yang H-Q, Wu Y-J, Tang R-H, Liu D, Liu Y, Cashmore AR. 2000. The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103:815–27 [DOI] [PubMed] [Google Scholar]; Discovery of the function of CCE domains of plant cryptochromes.
- 180.Yang L, Mo W, Yu X, Yao N, Zhou Z, et al. 2018. Reconstituting Arabidopsis CRY2 signaling pathway in mammalian cells reveals regulation of transcription by direct binding of CRY2 to DNA. Cell Rep. 24:585–93.e4 [DOI] [PubMed] [Google Scholar]
- 181.Yanovsky MJ, Kay SA. 2002. Molecular basis of seasonal time measurement in Arabidopsis. Nature 419:308–12 [DOI] [PubMed] [Google Scholar]
- 182.Yu X, Klejnot J, Zhao X, Shalitin D, Maymon M, et al. 2007. Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 19:3146–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yu X, Sayegh R, Maymon M, Warpeha K, Klejnot J, et al. 2009. Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. Plant Cell 21:118–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yu X, Shalitin D, Liu X, Maymon M, Klejnot J, et al. 2007. Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. PNAS 104:7289–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhang K, Cui B. 2015. Optogenetic control of intracellular signaling pathways. Trends Biotechnol. 33:92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhang L-Y, Bai M-Y, Wu J, Zhu J-Y, Wang H, et al. 2009. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21:3767–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zhang Q, Li H, Li R, Hu R, Fan C, et al. 2008. Association of the circadian rhythmic expression of GmCRY1a with a latitudinal cline in photoperiodic flowering of soybean. PNAS 105:21028–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhou T, Zhou L, Ma Y, Gao J, Li W, et al. 2018. Cryptochrome 1b from sweet sorghum regulates photoperiodic flowering, photomorphogenesis, and ABA response in transgenic Arabidopsis thaliana. Plant Mol. Biol. Report 36:13–22 [Google Scholar]
- 189.Zhu D, Maier A, Lee JH, Laubinger S, Saijo Y, et al. 2008. Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20:2307–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zoltowski BD, Vaidya AT, Top D, Widom J, Young MW, Crane BR. 2011. Structure of full-length Drosophila cryptochrome. Nature 480:396–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zuo Z, Liu H, Liu B, Liu X, Lin C. 2011. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol 21:841–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zuo Z-C, Meng Y-Y, Yu X-H, Zhang Z-L, Feng D-S, et al. 2012. A study of the blue-light-dependent phosphorylation, degradation, and photobody formation of Arabidopsis CRY2. Mol. Plant 5:726–33 [DOI] [PMC free article] [PubMed] [Google Scholar]


