Abstract
SARS-CoV-2 is the etiologic agent of COVID-19. There is currently no effective means of preventing infections by SARS-CoV-2, except through restriction of population movement and contact. An understanding of the origin, evolution and biochemistry (molecular biology) of SARS-CoV-2 is a prerequisite to its control. Mutations in the phosphorylation sites of SARS-CoV-2 encoded nucleocapsid protein isolated from various populations and locations, are described. Mutations occurred in the phosphorylation sites, all located within a stretch which forms a phosphorylation dependent interaction site, including C-TAK1 phosphorylation sites for 14-3-3. The consequences of these mutations are discussed and a structure-based model for the role of protein 14-3-3 in the sequestration and inhibition of SARS-CoV-2 nucleocapsid protein’s function is presented. It is proposed that the phosphorylation of SARS-CoV-2 nucleocapsid protein and its sequestration by Protein 14-3-3 is a cellular response mechanism for the control and inhibition of the replication, transcription and packaging of the SARS-CoV-2 genome.
Keywords: SARS-CoV-2, Nucleocapsid protein, Protein 14-3-3, Phosphorylation dependent binding and sequestration, Protein kinases, C-TAK1
Highlights
-
•
Mutations in the phosphorylation sites of SARS-CoV-2 encoded nucleocapsid protein.
-
•
Phosphorylation dependent binding and sequestration sites for protein 14-3-3.
-
•
Structure-model of the phosphorylation dependent binding and sequestration.
-
•
Proposal that sequestration by Protein 14-3-3 is a cellular response mechanism.
-
•
Sequestration inhibits replication, transcription and packaging of NCp genome.
1. Introduction
SARS-CoV-2 is the etiologic agent of COVID-19 which is now considered a pandemic [[1], [2], [3], [4]]. Currently, the only effective means of avoiding morbidity due to infections by SARS-CoV-2 is through population control and contact restriction which appear to be quite effective in some parts of the world and not others. However, the number of deaths in various populations and regions of the world are not uniform [5,6]. Although the medical infrastructure and response strategy and protocol play an important role in mitigating illness and death as a result of SARS-CoV-2 infection, the possibility that variability in the number of deaths following SARS-CoV-2 infections may be due to SARS-CoV-2 mutations cannot be ruled out.
Several reports have documented mutations in several SARS-CoV-2 encoded proteins, in particular the Spike protein [7] which determines the infectivity of SARS-CoV-2 by binding to ACE2 [8,9]. Other reports have described mutations in other proteins encoded by SARS-CoV-2, including the nucleocapsid protein [[10], [11], [12], [13], [14]]. SARS-CoV-2 nucleocapsid protein has an essential role in the replication, transcription and assembly of SARS-CoV-2 genome [15,16]. There is also interest in developing drugs and vaccines that target the SARS-CoV-2 nucleocapsid protein [17,18] which has also been shown to control the cell cycle of the host cell [19,20]. The cell cycle control is regulated in large part by protein phosphorylation and dephosphorylation mechanisms involving a large number of protein kinases and protein phosphatases [[21], [22], [23], [24], [25]]. Development of drugs and vaccines that target SARS-CoV-2 nucleocapsid protein must take into account mutations in SARS-CoV-2 nucleocapsid protein that occur strains/sub-strains isolated in various populations and locations, and their functional significance. Here, we describe mutations of the phosphorylation sites of SARS-CoV-2 nucleocapsid protein that were isolated from several populations and locations. We identify the binding of the phosphorylation rich region of SARS-CoV-2 nucleocapsid protein to protein 14-3-3, an important signaling molecule that controls the cell cycle, cell survival and cell death [[26], [27], [28], [29], [30]]. We present a structure-based model of phosphorylation dependent binding and sequestration of SARS-CoV-2 nucleocapsid protein by protein 14-3-3. The consequences of these mutations are also discussed.
2. Materials and methods
The sequences of SARS-CoV-2 nucleocapsid protein from SARS-CoV-2 strains/sub-strains isolated from various countries were curated from the National Center for Biotechnology Information (NCBI) [31] and analyzed for alignments and identities by using the blastp program [32].
Rendering of the structure of SARS-CoV-2 Nucleocapsid phosphoprotein encompassing amino acids 123 to 310 which contains the phosphorylation rich stretch (amino acids 180–210) was performed by QUARK2 Protein Analysis Program pursuant to Xu and Zhang [33,34, https://zhanglab.ccmb.med.umich.edu/QUARK2/]. The structures of SARS-COV-2 Nucleocapsid protein rendered in this work and Protein 14-3-3 (1YZ5) based on the structure determination of Benzinger et al. [35] were visualized and analyzed by the CCP4 Molecular Graphics Program Version 2.10.11 as described by Mc Nicolas et al. [36] and the ZMM Molecular Modeling Program as described by Garden and Zhorov [37].
Phosphorylation of SARS-CoV-2 nucleocapsid protein on serines 197 and 206 were performed using FOLD-X pursuant to Ref. [38]. SARS-CoV-2 nucleocapsid protein phosphorylated on serines 197 and 206 was then analyzed for binding with protein 14-3-3 by protein-protein docking experiments pursuant to Pierce et al. [39].
3. Results
Fig. 1 shows the protein sequences of SARS-CoV-2 encoded nucleocapsid protein of SARS-CoV-2 strains/sub-strains isolated from various countries including, China (NC_045,512), Iran (MT459928; MT447177) Spain (MT655134; MT292671), India (MT451879), Israel (MT276598), Russia (MT510643), Italy (MT527178; MT531537) Poland (MT511066), Bangladesh (MT509958), Greece (MT459920) and Czech Republic (MT517420). Using the sequence of one SARS-CoV-2 nucleocapsid protein of one strain isolated from China as reference, mutations were identified in SARS-COV-2 Nucleocapsid protein of strains/sub-strains isolated from several countries, including Iran, Spain, India, Israel. Russia, Italy, Poland, Bangladesh and Czech Republic. SARS-CoV-2 nucleocapsid proteins of several SARS-CoV-2 strains/sub-strains have undergone mutations on serines 186, 197, and 202 to Phenylalanine, Leucine and Asparagine respectively. Serine 186 is a phosphorylation site for CKI which is involved in the control a wide variety of cellular processes, including the control of the cell cycle [[40], [41], [42]]. Serine 197 is a phosphorylation site for Aurora A and B kinases which are involved in the control of the cell cycle and proper chromosome segregation [[43], [44], [45]]. Next to serine 197, is threonine 198 which is a phosphorylation site for proline directed protein kinases, including Cdk1 and GSK-3 [[46], [47], [48], [49], [50], [51], [52], [53]]. Serine 202 is a phosphorylation site for GSK-3 which is involved in a wide variety of cellular processes including, the control of the cell cycle [[48], [49], [50], [51]]. In addition, arginine 203 and glycine 204 are frequently mutated to lysine and arginine. Arginine 203 and glycine 204 (RG 203–204) lie sandwiched between two phosphorylation sites, including upstream phospho-serine 202 which is phosphorylated by GSK-3 and downstream phospho-threonine 205 which is phosphorylated by PKA [52,53]. Of particular interest, further downstream of RG 203–204 lies serine 206 which is a phosphorylation site for C-TAK1 [54,55]. Serine 197 is also a phosphorylation site for C-TAK1 [54,55].
Fig. 1.
Protein Sequence Alignment of SARS-COV-2 encoded Nucleocapsid protein of SARS-COV-2 isolated from different countries including China, Iran, Spain, India, Israel, Russia, Italy, Poland, Bangladesh, Greece and Czech Republic (between amino acids 180–210). Amino acids in bold letters are mutations observed in this work.
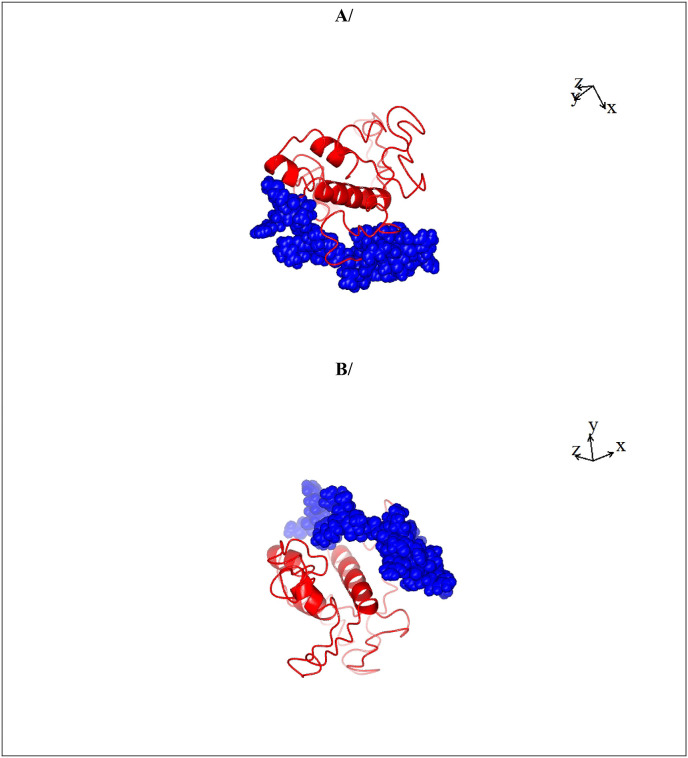
Phosphorylation sites 186, 197, 202, 204 and 206 are located in a stretch that is sandwiched between the RNA binding domain and dimerization domain of SARS-CoV- 2 nucleocapsid protein [56]. Together with phospho-serines 188 and 198 which are nearby, and in particular, phosphoserines 197 and 206 [54,55], the phosphorylation rich stretch forms a phosphorylation dependent binding domain for Protein 14-3-3 which is an important signaling molecule that is involved in the control of a wide variety of cellular processes, including the control of the cell cycle, cell survival and cell death [[26], [27], [28], [29], [30],[57], [58], [59], [60], [61], [62], [63]]. There is no structure described for the full-length SARS-CoV-2 nucleocapsid protein. Only the structures of the N terminal and C terminal portions lacking the phosphorylation rich region have been described [[64], [65], [66], [67], [68], [69]]. A model structure encompassing amino acid residues 123 to 310 was therefore rendered pursuant to Xu and Zhang’s method of protein structure prediction [33,34] (Fig. 2 ). The phosphorylation rich domain of SARS-CoV-2 nucleocapsid protein is localized at the surface of the protein (Fig. 2).
Fig. 2.
A rendering of the structure of SARS-COV-2 Nucleocapsid protein (amino acids 123–309). The structure of SARS-COV-2 Nucleocapsid protein (amino acids 123–309) depicted here was rendered pursuant to Xu and Zhang [33,34]. The phosphorylation rich stretch encompassing amino acids 180 to 210 is shown as spheres in blue and is localized at the surface of SARS-COV-2 Nucleocapsid protein. The sequence of the phosphorylation rich domain of SARS-COV-2 Nucleocapsid protein is: SQASSRSSSRSRNSSRNSTPGSSRGTSPARM.
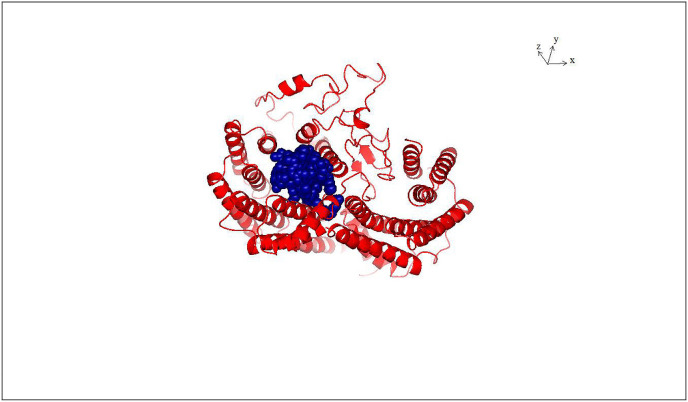
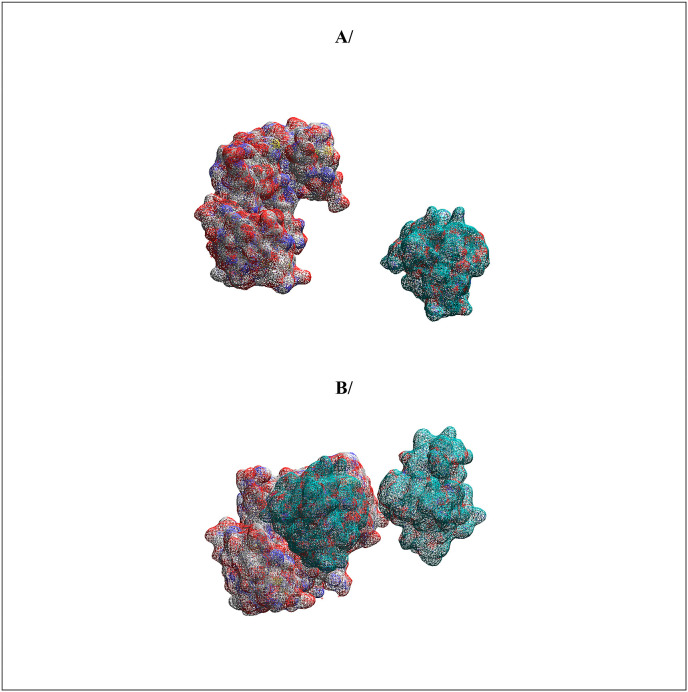
To show that SARS-CoV-2 nucleocapsid protein can bind protein 14-3-3 in a phosphorylation dependent manner, SARS-CoV-2 nucleocapsid protein phosphorylated on serines 197 and 206 which together with their surrounding amino acid motifs form recognition sites for protein 14-4-4, protein-protein docking experiments were performed using protein 14-3-3 as the receptor protein. Fig. 3 shows that the phosphorylation rich stretch encompassing amino acid residues 180 to 210 forms a binding site for protein 14-3-3 as it can be fitted nicely in the protein binding groove of protein 14-3-3 [70,71]. The binding free energy of the complex of protein 14-3-3 to SARS-CoV-2 nucleocapsid protein phosphorylated on serines 197 and 206 was estimated to be −21.72 kcal/mol. Protein-protein docking experiments between protein 14-3-3 and unphosphorylated SARS-CoV-2 nucleocapsid protein shows only unstable complexes with high binding free energies, high binding affinities and high dissociation constants (Data not shown). The molecular mass of Protein 14-3-3 dimer is ∼56 kDa while the molecular mass of SARS-CoV-2 Nucleocapsid protein is∼46 kDa indicating that only monomeric form of the latter can bind 1 mol of dimeric Protein 14-3-3 (Fig. 3, Fig. 4 ). Interestingly, Fig. 3, Fig. 4 shows that only the monomeric SARS-CoV-2 nucleocapsid protein can form a complex with protein 14-3-3, indicating that protein 14-3-3 acts to prevent the dimerization of SARS-COV-2 Nucleocapsid protein.
Fig. 3.
Protein-protein docking of Protein 14-3-3 (as the receptor) and SARS-COV-2 Nucleocapsid protein. The phosphorylation rich domain encompassing amino acids 180 to 210 is shown as spheres in blue. The sequence of the phosphorylation rich domain of SARS-COV-2 Nucleocapsid protein is: SQASSRSSSRSRNSSRNSTPGSSRGTSPARM. The phosphorylation rich domain of SARS-COV-2 Nucleocapsid protein is in contact with the binding groove of Protein 14-3-3. The phosphorylation rich stretch encompassing amino acids 180 to 210 fits into the binding groove of Protein 14-3-3 dimer. Only monomeric of SARS-COV-2 Nucleocapsid protein can bind Protein 14-3-3 dimer.
Fig. 4.
Realistic representations of the structures of SARS-COV-2 Nucleocapsid protein (amino acids 123–309) (blue and red on the right) and Protein 14-3-3 (red, grey and blue of the left). The phosphorylation rich stretch encompassing amino acids 180 to 210 fits into the binding groove of Protein 14-3-3 dimer. Only monomeric SARS-COV-2 Nucleocapsid protein can bind Protein 14-3-3 dimer.
4. Discussion
The sequences of several SARS-CoV-2 strains have been determined [[1], [2], [3], [4],31]. Lu et al. [2] reported that they sequenced SARS-COV-2 genomes from nine individuals from the same location and found that they had 99.98 sequence identity. However, analysis of the sequences of SARS-CoV-2 isolated in different populations and environments must be made to determine differences in infectivity and virulence that are populations and environments specific. Analysis of mutations in SARS-CoV-2 encoded proteins from SARS-CoV-2 strains/sub-strains isolated from various populations and regions have been reported [[10], [11], [12], [13], [14]]. It is anticipated that the consequences of identified mutations will be studied as they may explain how SARS-CoV-2 encoded proteins interact with cellular proteins in order to maximize its viability and replication. A number of reports have documented the role of phosphorylation in the control of SARS-CoV and SARS-CoV-2 nucleocapsid protein’s activity in the cell. However, the functions of the phosphorylation reactions have not been delineated [19, 20, [72], [73], [74]].
The present work shows that SARS-COV-2 encoded Nucleocapsid protein have undergone certain mutations in strains/sub-strains of SARS-CoV-2 isolated from various populations and locations. The mutations of serine 186, serine 197 and serine 202 to phenylalanine, leucine and asparagine respectively of the SARS-CoV-2 nucleocapsid protein are of major consequences because (i) serine 186 is a phosphorylation site for CKI which is involved in the control a wide variety of cellular processes, including the control of the cell cycle [[40], [41], [42]], (ii) serine 197 is a phosphorylation site for Aurora A and B kinases which are involved in the control of the cell cycle and proper chromosome segregation and C-TAK1 which is also involved in the control of the cell cycle [[43], [44], [45],54,55], and (iii) serine 202 is a phosphorylation site for GSK-3 which has been shown to be involved in a wide variety of cellular processes including, the control of the cell cycle [[47], [48], [49], [50], [51], [52], [53]]. The mutation of serine 197 would also have major ramification for the adjacent threonine 198 which is a phosphorylation site for the proline-directed protein kinases, including Cdk-1 and GSK-3 [[46], [47], [48], [49], [50], [51], [52], [53]]. The destruction of phospho-serine 197 would prevent the regulation of phosphorylation of threonine 198 by GSK-3 [Tung, H.Y.L., Manuscript in preparation]. The mutations of arginine 203 and glycine 204 (AG 203–204) of the SARS-CoV-2 nucleocapsid protein will also affect the regulation of the phosphorylations of serines 202 and 206 which are phosphorylation sites for GSK-3 and C-TAK1 respectively [[46], [47], [48], [49], [50], [51], [52], [53], [54], [55]]. It is significant that mutations take place within a stretch that is phosphorylation rich and forms a phosphorylation dependent binding site for protein 14-3-3 [54,55], an important signaling molecule that is involved in the control of the cell cycle, cell survival and cell death [[26], [27], [28], [29], [30]]. Of particular interest is phospho-serines 197 and 206 which are C-TAK1 phosphorylation sites recognized by protein 14-3-3. Mutations of the phosphorylation sites described in this paper would result in the inability of protein 14-3-3 to bind to the SARS-CoV-2 nucleocapsid protein, the consequence of which would be its unfettered actions, including its ability to dimerize, bind to the SARS-COV-2 genome, control and up-regulate the replication, transcription and packaging of the SARS-CoV-2 genome [15,16]. Surijit et al. [74] have described the role of protein 14-3-3 in the translocation of SARS-CoV-2 nucleocapsid protein from the nucleus to the cytoplasm. In this work, structural analysis of SARS-CoV-2 nucleocapsid protein and protein 14-3-3 shows that only monomeric SARS-CoV-2 nucleocapsid protein can form a complex with protein 14-3-3. Here, we propose that in response to SARS-CoV-2 infection, cellular protein 14-3-3 acts to prevent the dimerization of SARS-CoV-2 nucleocapsid protein, replication, transcription and packaging of the SARS-CoV-2 genome. Protein 14-3-3 achieves this by binding to SARS-CoV-2 Nucleocapsid protein and sequestrating it in a phosphorylation dependent manner following phosphorylation of SARS-CoV-2 nucleocapsid protein by several protein kinases that control the cell cycle, including C-TAK1 and possibly CKI, Cdk1, Cdk-5, GSK-3 (Manuscript in preparation). It is also proposed that the phosphorylation of SARS-CoV-2 nucleocapsid protein and its sequestration by protein 14-3-3 is a cellular response mechanism for the control and inhibition of the replication, transcription and packaging of the SARS-CoV-2 genome.
Acknowledgments
This work was supported by the Nacbraht Biomedical Research Institute Fund. The authors would like to thank the Zhang Lab for the rendering of the structure of SARSCOV-2 Nucleocapsid protein (amino acids 123–310).
References
- 1.Zhu N., Zhang D., Wang W. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2019;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.R. Lu, X. Zhao, J. Li, et al. Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395 (2020), 565-574. [DOI] [PMC free article] [PubMed]
- 3.Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu F., Zhao S., Yu B. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tung H.Y.L., Limtung P. SARS-COV-2 infection and virulence: reverse correlations with temperature, humidity and far infrared radiation. Elevated temperature, humidity and far infrared irradiation will significantly lower the viability, infectivity and virulence of SARS-COV-2. ResearchGate. 2020 doi: 10.13140/RG.2.2.14741.55522. [DOI] [Google Scholar]
- 6.Tung H.Y.L., Limtung P. SARS-COV-2 infection and virulence: comparison between United Kingdom and Germany, and other European countries. Researchgate. 2020 doi: 10.13140/RG.2.2.25358.38723. [DOI] [Google Scholar]
- 7.Ou J., Zhou Z., Zhang J. RBD mutations from circulating SARS-CoV-2 strains enhance the structure stability and infectivity of the spike proteins. bioRxiv. 2020 doi: 10.1101/2020.03.15.991844. [DOI] [Google Scholar]
- 8.Li W., Moore M.J., Vasilieva N. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuba K., Imai Y., Rao S. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nature. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panchetti M., Marini B., Benedetti F. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020 doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020 doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawood A.A. Mutated COVID-19 may foretell a great risk for mankind in the future. New Microbes New Infect. 2020 doi: 10.1016/j.nmni.2020.1000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin C. Genotyping coronavirus SARS-CoV-2: methods and implications. Genomics. 2020 doi: 10.1016/j-ygeno.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benvenuoto D., Demir A.B., Giovanetti M. 2020. Evidence for Mutations in SARS-CoV-2 Italian Isolates Potentially Affecting Virus Transmission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cong Y., Ulasli M., Schepers H. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J. Virol. 2020 doi: 10.1128/JVI.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang C.K., Hou M.H., Chang C.F. The SARS coronavirus nucleocapsid protein--forms and functions. Antivir. Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H.-J., Leng C.-H., Lien H.-P. Immunological characterization of the nucleocapsid protein based SARS vaccine. Vaccine. 2006;24:3100–3108. doi: 10.1016/j.vaccine.2006.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Q., Peng W., Ou Y. Prediction and analysis of key protein structures of 2029-nCoV. Future Virol. 2020 doi: 10.2217/fvl-2020-0020. [DOI] [Google Scholar]
- 19.Surjit M., Liu B., Chow V.T.K. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclindependent kinase complex and blocks S phase progression in mammalian cells. J. Biol. Chem. 2006;281:10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizutani T., Fukushi S., Lizuka D. Inhibition of cell proliferation by SARS-CoV infection in Vero E6 cells. FEMS Immunol. 2006;46:236–243. doi: 10.1111/j.1574-695X.2005.00028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgan D. Principles of CDK regulations. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 22.Meijer L., Guidet S., Tung H.Y.L. vol. 1. Plenum Press; New York. U.S.A.,: 1995. (Progress in Cell Cycle Research). [Google Scholar]
- 23.Doree M., Hunt T. From cdc 2 to Cdk1: when did the cell cycle kinase join its cyclin partner. J. Cell Sci. 2002;115:2461–2464. doi: 10.1242/jcs.115.12.2461. 2002. [DOI] [PubMed] [Google Scholar]
- 24.Nurse P. Cyclin dependent kinases and cell cycle control. Chembiochem. 2002;3:596–603. doi: 10.1002/1439-7633(20020703)3:7<596::AID-CBIC596>3.0.CO;2-U. 2002. [DOI] [PubMed] [Google Scholar]
- 25.Tung H.Y.L. Role of protein phosphatase-2A (PP-2A) in the control of mitosis and meiosis New Biomed. Sci. Asia. 2020;1:97–154. [Google Scholar]
- 26.Chan T.A., Hermeking H., Lengauer C. 14-3-3Sigma is required to prevent mitotic catastrophe after DNA damage. Nature. 1999;401:616–620. doi: 10.1038/44188. 10. [DOI] [PubMed] [Google Scholar]
- 27.Peng C.Y., Graves P.R., Ogg S. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1998;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 28.Dalal S.N., Yaffe M.B., DeCaprio J.A. 14-3-3 family members act coordinately to regulate mitotic progression. Cell Cycle. 2004;3(2004):672–677. [PubMed] [Google Scholar]
- 29.Masters S.C., Fu H. 14-3-3 proteins mediate an essential anti-apoptotic signal. J. Biol. Chem. 2001;276:45193–45200. doi: 10.1074/jbc.M105971200. [DOI] [PubMed] [Google Scholar]
- 30.Won J., Kim D.Y., La M. Cleavage of 14-3-3 protein by caspase-3 facilitates bad interaction with bcl-x(L) during apoptosis. J. Biol. Chem. 2003;278:19347–19351. doi: 10.1074/jbc.M213098200. [DOI] [PubMed] [Google Scholar]
- 31.https://www.ncbi.nlm.nih.gov/genbank/sars-cov2-seqs
- 32.https://blast.ncbi.nlm.nih.gov.Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&BLAST_SPEC=blast2seqLINK_LOC=blasttab
- 33.Xu D., Zhang Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins. 2012;80:1715–1735. doi: 10.1002/prot.24065. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu D., Zhang Y. Toward optimal fragment generations for ab initio protein structure assembly. Proteins. 2013;81:229–239. doi: 10.1002/prot.24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benzinger A., Popowicz G.M., Joy J.K. The crystal structure of the nonliganded 14-3-3sigma protein: insights into determinants of isoform specific ligand binding and dimerization. Cell Res. 2005;15:219–227. doi: 10.1038/sj.cr.7290290. [DOI] [PubMed] [Google Scholar]
- 36.McNicholas S., Potterson E., Wilson K.S. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr. 2011;D67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garden D.P., Zhorov B.S. Docking flexible ligands in proteins with a solvent exposure- and distance-dependent dielectric function. J. Comput. Aided Mol. Des. 2010;25:91–105. doi: 10.1007/s10822-009-9317-9. [DOI] [PubMed] [Google Scholar]
- 38.Schymkowitz J., Borg J., Stricher F. The FoldX web server: an online force field. Nucleic Acids Res. 2005 doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pierce B.G., Wiehe K., Huang H. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brockman J.L., Gross S.D., Sussman M.R. Cell cycle-dependent localization of casein kinase I to mitotic spindles. Proc. Natl. Acad. Sci. Unit. States Am. 1992;89:9454–9458. doi: 10.1073/pnas.89.20.9454. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L., Li C., Pan Y. Regulation of p53-MDMX interaction by casein kinase 1 alpha. Mol. Cell Biol. 2005;25:6509–6520. doi: 10.1128/MCB.25.15.6509-6520.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bradley C., Bryan J.D., Jerome A. The Yck2 yeast casein kinase 1 isoform shows cell cycle-specific localization to sites of polarized growth and is required for proper septin organization. Mol. Biol. Cell. 2017 doi: 10.1091/mbc.10.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Francisco L., Wang W., Chan C.S., S C. Type 1 protein phosphatase acts in opposition to IpL1 protein kinase in regulating yeast chromosome segregation. Mol. Cell Biol. 1994;14:4731–4740. doi: 10.1128/mcb.14.7.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tung H.Y.L., Wang W., Chan C.S. Regulation of chromosome segregation by Glc8p, a structural homolog of mammalian inhibitor 2 that functions as both an activator and an inhibitor of yeast protein phosphatase 1. Mol. Cell Biol. 1995;15:6064–6074. doi: 10.1128/mcb.15.11.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu J., Bian M., Zhang Q.C. Roles of Aurora kinases in mitosis and tumorigenesis. Mol. Canc. Res. 2007;5:1–10. doi: 10.1158/1541-7786.MCR-06-0208. [DOI] [PubMed] [Google Scholar]
- 46.Holt L.J., Tuch B.B., Villén J. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:pp1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Songyang Z., Blechner S., Hoagland N. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr. Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 48.Fiol C.J., Mahrenholz A.M., Wang Y. Formation of protein kinase recognition sites by covalent modification of the substrate. Molecular mechanism for the synergistic action of casein kinase II and glycogen synthase kinase 3. J. Biol. Chem. 1987;262:14042–14048. [PubMed] [Google Scholar]
- 49.Frame S., Cohen P. GSK-3 takes centre stage more than 20 years after its discovery. Biochem. J. 2001;359:1–16. doi: 10.1042/0264-6021:3590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaidanovich-Beilin O., Woodgett J.R., Gsk-3: Functional Insights from Cell Biology and Animal Models Frontiers in molecular neuroscience. 2011. 10.3389/fnmol.2011.00040 (2011) [DOI] [PMC free article] [PubMed]
- 51.Rashid M.S., Mazur T., Ji W. Analysis of the role of GSK3 in the mitotic checkpoint. Sci. Rep. 2018 doi: 10.1038/s41598-018-32435-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kemp B.E., Graves D.J., Benjamini E., Krebs E.G. Role of multiple basic residues in determining the substrate specificity of cyclic AMP-dependent protein kinase. J. Biol. Chem. 1977;252:4888–4894. [PubMed] [Google Scholar]
- 53.Kennelly P.J., Krebs E.G. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J. Biol. Chem. 1991;266:1555–15556. 1991. [PubMed] [Google Scholar]
- 54.Ogg S., Gabrielli B., Piwnica-Worms H. Purification of a serine kinase that associates with and phosphorylates human Cdc25C on serine 216. J. Biol. Chem. 1994;269:30461–30469. [PubMed] [Google Scholar]
- 55.Muller J., Ritt D.A., Copeland T.D., Morrison D.K. Functional analysis of CTAK1 substrate binding and identification of PKP2 as a new C-TAK1 substrate. EMBO J. 2003;22:4431–4442. doi: 10.1093/emboj/cdg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang C.K., Hou M.H., Chang C.F. The SARS coronavirus nucleocapsid protein--forms and functions. Antivir. Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moore B.W., McGregor D. Chromatographic and electrophoretic fractionation of soluble proteins of brain and liver. J. Biol. Chem. 1965;240:1647–1653. [PubMed] [Google Scholar]
- 58.Yamauchi T., Nakata H., Fujisawa H. A new activator protein that activates tryptophan 5-monooxygenase and tyrosine 3-monooxygenase in the presence of Ca2+-, calmodulin-dependent protein kinase. Purification and characterization. J. Biol. Chem. 1981;256:5404–5409. [PubMed] [Google Scholar]
- 59.Ichimura T., Isobe T., Okuyama T. Brain 14-3-3 protein is an activator protein that activates tryptophan 5-monooxygenase and tyrosine 3 monooxygenase in the presence of Ca2+, calmodulin-dependent protein kinase II. FEBS Lett. 1987;219:79–82. doi: 10.1016/0014-5793(87)81194-8. [DOI] [PubMed] [Google Scholar]
- 60.Aitken A. 14-3-3 proteins: a historic overview. Semin. Canc. Biol. 2006;16:162–172. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 61.Furukawa Y., Ikuta N., Omata S. Demonstration of the phosphorylation dependent interaction of tryptophan hydroxylase with the 14-3-3 protein. Biochem. Biophys. Res. Commun. 1993;194:144–149. doi: 10.1006/bbrc.1993.1796. [DOI] [PubMed] [Google Scholar]
- 62.Muslin A.J., Tanner J.W., Allen P.M. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 63.Yaffe M.B., Rittinger K., Volinia S. The structural basis for 14-3- 3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 64.Jayaram H., Fan H., Bowman B.R. X-ray structures of the N- and CTerminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J. Virol. 2006;80:6612–6620. doi: 10.1128/JVI.00157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kang S., Yang M., Hong Z. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. bioRxiv. 2020 doi: 10.1101/2020.03.06.977876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou R.J., Zeng R., Lai J. 2020. Crystal Structure of the C-Terminal Domain of SARSCoV- 2 Nucleocapsid Protein.www.rcsb.org/structure/7c22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ye Q., Corbett K.D. 2020. Structure of SARS-CoV-2 Nucleocapsid Dimerization Domain.www.rcsb.org/structure/6WZO#entity-1 [Google Scholar]
- 68.Minasov G., Shuvalova L., Wiersum G., Satchell K.J.F. 2.05 angstrom resolution crystal structure of C-terminal dimerization domain of nucleocapsid phosphoprotein from SARS-CoV-2. www.rcsb.org/structure/6WJI#entity-1
- 69.Zinzula L., Basquin J., Nagy J.I., Bracher A. 2020. 1.45 Angstrom Resolution Crystal Structure of C-Terminal Dimerization Domain of Nucleocapsid Phosphoprotein from SARS-CoV-2.https://www.rcsb.org/structure/6YUN#entity-1 [Google Scholar]
- 70.Xiao B., Smerdon S.J., Jones D.H. Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature. 1995;376:188-–191. doi: 10.1038/376188a0. [DOI] [PubMed] [Google Scholar]
- 71.Liu D., Blenkowska J., Petosa C. Crystal structure of the zeta isoform of the 14-3-3 protein. Nature. 1995;376:191–194. doi: 10.1038/376191a0. [DOI] [PubMed] [Google Scholar]
- 72.Wu C.H., Chen P.J., Yeh S.-H. Nucleocapsid phosphorylation and RNA helicase DDX1 recruitment enables Coronavirus transition from discontinuous transcription. Cell Host Microbe. 2014;16:462–472. doi: 10.1016/j.chom.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu C.H., Yeh S.-H., Tsay Y.G. Glycogen synthase kinase-3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. J. Biol. Chem. 2009;284:5229–52239. doi: 10.1074/jbc.M805747200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Surijit M., Kumar R., Mishra R.N. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J. Virol. 2005;79:11476–11486. doi: 10.1128/JVI.79.17.11476-11486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]