Abstract
Background
Community-acquired urinary tract infections are associated with significant morbidity, and uropathogenic Escherichia coli (UPEC) alone causes 90% of urinary tract infections. This bacterium retains a diverse armament of virulence factors including fimbria, hemolysins, and siderophores production. In a post invasion scenario, formation of intracellular communities mimic biofilm-like characteristics and are linked to recurrent urinary tract infections. We investigated the effects of different frontline antibiotics on the formation, inhibition, and eradication of biofilms of virulent UPEC strains.
Materials and Methods
A total of 155 UPEC strains were scrutinized for various virulence factors including gelatinase, cell surface hydrophobicity, hemagglutination, and serum bactericidal activity. Biofilm formation was confirmed by three different methods: Congo red agar, test tube, and tissue culture plate method. Biofilm inhibition and eradication assays were performed according to the standard protocols. Topographical analysis of biofilms was done by scanning electronic microscopy (SEM).
Results
Out of 155 strains, 113 (73%) were strong biofilm formesr, while 37 (24%) produced biofilms at moderate level. Significant differences were observed between MICs of planktonic cells (MIC-p) and MICs of UPEC biofilms (MIC-b). Among tested frontline antibiotics, levofloxacin successfully inhibited biofilms at a concentration of 32 µg/mL, while trimethoprim eradicated biofilms at higher concentrations (512–1024 µg/mL). Ciprofloxacin treatment at sub-MIC level significantly enhanced biofilm formation (P<0.05).
Conclusion
The majority of UPEC strains are strong biofilm formers and show higher tolerance towards frontline antibiotics in biofilm form. We observed significant inhibitory effects of levofloxacin (32 µg/mL) on UPEC biofilms, while treatment with sub-minimal concentrations of ciprofloxacin significantly enhanced biofilm formation. Out of all tested antibiotics, trimethoprim (512–1024 µg/mL) eradicated UPEC biofilms.
Keywords: biofilms, MIC, MBEC, sub-MIC, ciprofloxacin, uropathogenic Escherichia coli
Introduction
Urinary tract infections (UTIs) are becoming increasingly challenging, mainly because of higher recurrence rates and resistance to frontline antibiotics.1 Each year 150–250 million cases of UTIs are reported across the globe.2,3 Uropathogenic E. coli (UPEC) alone accounts for 90% of all UTIs, including nosocomial and community-acquired infections.4 As an opportunistic intracellular pathogen, UPEC colonizes the urinary bladder producing variable clinical manifestations ranging from cystitis to severe pyelonephritis.5,6 Bacterial strains possess various virulence factors contributing to effective colonization, higher persistence, and pathogenesis of the disease.7 Moreover, UPEC has the ability to form micro-colonies in the mucosal lining of the bladder, known as intracellular bacterial communities (IBCs) which mimic biofilm-like properties and facilitate bacterial persistence inside the host.8 Their ability to form a biofilm is linked to hospital-acquired infections, particularly catheter associated urinary tract infections (CAUTI). Since biofilms possess a higher concentration of polysaccharides, they render immune components and antibiotics ineffective. In contrast to planktonic state, bacterial cells in consortia possess greater abilities to tolerate antibiotics and remain challenging in clinical settings.9 MDR (multi-drug resistant) biofilm former bacteria have been particularly difficult to treat with β-lactams and fluoroquinolones.10 Although frontline antibiotics such as co-trimoxazole (trimethoprim/sulfamethoxazole), fluoroquinolones (eg, levofloxacin, ciprofloxacin), aminoglycosides (gentamicin), and third generation cephalosporins (eg, ceftazidime, ceftriaxone) are widely used therapeutic options, empirical treatment of both uncomplicated and complicated UTIs at different geographical locations rely on locally assessed susceptibility profiles. Because of their excellent tissue penetration, trimethoprim and second generation fluoroquinolones are important therapeutic choices for the treatment of male prostatitis. Similarly, for many cases of acute cystitis in women usually a 3-day course of trimethoprim is recommended. Unfortunately resistance against frontline antibiotics has been mounting significantly over the last few decades. For example, in Austria 15%, Greece 18%, Portugal 16%, Sweden 16%, UK 14%, Korea 35%, and Europe 25% strains were resistant to trimethoprim.11,14 Alternatively, fluoroquinolones have been widely and frequently used against different types of infections including UTIs.15 Likewise, third and fourth generation cephalosporins such as ceftriaxone and ceftazidime remained reliable therapeutic options for the eradication of co-trimoxazole resistant uropathogens. In addition, aminoglycosides are used in combination with β-lactam or glycopeptides.16,17
The current studyevaluated the effect of these frontline antibiotics on various aspects of UPEC biofilms including inhibition, eradication, and enhancement. Since not much is known about the effect of sub-minimal concentration of ciprofloxacin on biofilms of extraintestinal E. coli, we further investigated the role of sub-MIC of ciprofloxacin on biofilms of UPEC.
Materials and Methods
Bacterial Sampling and Phenotypic Characterization
A total of 155 E. coli isolates were scrutinized from UTI patients. Samples were collected from a tertiary care hospital located in Islamabad during August 2012 to August 2014. Ethical Review Board of Pakistan Institute of Medical Sciences approved this study. Verbal consent of the patients was taken and based on important personal data such as name, age, gender, location a unique identification number was assigned for each participant. Samples under scrutiny were from community-acquired urinary tract infections. Both biochemical and molecular methods were used for the bacterial identification.18 Isolates were tested for phenotypic virulence factors including gelatinase, hemolysin, cell surface hydrophobicity, serum bactericidal activity and hemagglutination. Isolates were further tested for mannose sensitivity (MSHA) or mannose resistance hemagglutination (MRHA).19 Gelatinase production was confirmed on a plate containing mercuric chloride.20 Hemolysin production was assessed on blood agar plates.7 Cell surface hydrophobicity and serum bactericidal activity was determined as described elsewhere.21 ESBL detection was done by double disk synergy testing (DDST).7,22
Minimum Inhibitory Concentrations (MICs) and Biofilm Formation
Antibacterial activity of trimethoprim, levofloxacin, ceftazidime, and gentamicin were measured by micro-dilution broth method (CLSI, 2017).22 E. coli strain ATCC 25,922 was used as an experimental control throughout the experiments. All 155 strains were tested for biofilm formation. Qualitative assessment of biofilms was performed by Congo red agar (CRA) and tube method (TM).23,24 For quantitative assessments, TCP method (tissue culture plate method) was used and is considered gold standard.25 Briefly 200 µL of standardized bacterial suspension (corresponding to 0.5 McFarland 1×106cfu/mL) was inoculated on each well of a flat-bottom 96 well clear polystyrene plate. The plate was incubated at 37°C for 24 hours. After incubation, the content of each well was removed and washed three times with normal saline (PBS 0.9%). Attached bacteria were stained with crystal violet for 15 minutes, subsequently excessive stain was removed, and each well was washed with PBS. The optical density (OD) of adherent bacteria was measured at 540 nm using an ELISA plate reader (BiotekTM). All samples were tested in triplicate. Based on acquired readings, standard deviations were calculated, a cut-off value was adjusted to 3 standard deviations above the mean ODc* (cut off OD). Biofilm formation was assessed at different time points; 2, 4, 10, 18, and 24 hours. Each isolate was categorized according to the following criteria:
Weak biofilm producer: OD=2×ODc
Moderate biofilm producer: 2×ODc≤4×ODc
Strong biofilm producer: OD≥4×ODc
* The cut off OD for the crystal violet was 0.055.
Measurement of MIC-b, MBIC, and MBEC
For the four frontline antibiotics, levofloxacin, ceftazidime, gentamicin, and trimethoprim, minimum inhibitory concentrations of biofilm (MIC-b), minimum biofilm inhibition concentration (MBIC),and minimum biofilm eradication concentration (MBEC) were determined as described elsewhere.25,26 A total of 21 strong biofilm producing strains simultaneously sensitive to all four antibiotics were selected for biofilm assays. Because of the higher frequency of resistance against trimethoprim only seven strong biofilm former strains simultaneously sensitive to trimethoprim and other three classes of frontline antibiotics could be available (included in total 21 strains). These selected sensitive strains were used for the measurements of MIC-b, MBIC, and MBEC.25 For the measurement of MIC-b, 75 µL of standardized bacterial suspension was inoculated in a 96-well microtiter plate. The plate was incubated at 37°C for 24 hours. After incubation plates were washed with phosphate buffer saline (PBS). Two-fold serial dilutions of all four antibiotics (levofloxacin, ceftazidime, gentamicin, and trimethoprim), ranging from 0.5–2048 µg/mL were prepared and 100 µL of appropriate concentration of each antibiotic was added to each well of a micro-titer plate. The plate was incubated for 18–24 hours at 37°C. MIC-b for each tested sample was visually estimated as the lowest concentration of antibiotic capable of inhibiting biofilm formation of planktonic bacteria. Subsequently, each non-turbid well used to estimate MIC-b was culture plated to assess cell viability.
Likewise, MBIC was determined by inoculating a 75 µL standardized bacterial suspension in a 96 well microtiter plate that was incubated for 24 hours at 37°C. Following the incubation step, medium was removed gently; wells were washed thrice with PBS. A two-fold serial dilution of all four antibiotics (levofloxacin, ceftazidime, gentamicin, and trimethoprim) ranging from 0.5–2048 µg/mL was prepared and a 100 µL of appropriate concentration of each antibiotic was added to each well. Wells were washed and 100 μL of recovery media (MH broth) was added to the wells and incubated for 24 hours at 37°C. The lowest concentration of antibiotic that inhibited re-growth of bacteria was considered MBIC.26 For measurement of MBECs, the treatment procedure was essentially the same as described for MBIC, except after the incubation step, wells without visible growth were scraped thoroughly and particular attention was given to the edges of wells. Scraped material was transferred to 1 mL PBS. Each sample was briefly vortexed to disrupt biofilm and a 100 μL sample was subsequently plated on a fresh tryptic soy agar (TSA) plate. Antibiotic concentration on which no bacterial growth was observed on the TSA plate was considered MBEC.26
Effect of Ciprofloxacin Sub-MIC on Biofilm Formation of UPEC
Ciprofloxacin was diluted at sub-MIC level in the following concentrations: 0.5 µg/mL, 0.25 µg/mL, 0.125 µg/mL, and 0.0625 µg/mL. Antibiotic concentrations were prepared by using Mueller Hinton broth (MH broth). To determine the effects of sub-MIC level treatments, 100 µL of each ciprofloxacin concentration (0.5 µg/mL, 0.25 µg/mL, 0.125 µg/mL, and 0.0625 µg/mL) was added to each treatment well, containing 100 µL of standardized bacterial inoculums.27 Microtiter plates were incubated for 24 hrs at 37°C, biofilm quantification was done by adding crystal violet dye and measuring OD at 540 nm. The antibiotic-free standardized bacterial suspension was used as a positive control and bacteria free ciprofloxacin solution was used as a negative control. The readings taken for positive and negative controls were compared to analyze test samples for the biofilm formation.
Scanning Electron Microscopy of Biofilms
Biofilms were visualized by scanning electron microscopy. Antibiotic treated and untreated biofilms were analyzed at different time points. Biofilms were allowed to grow in a 96 well polystyrene plate (with or without levofloxacin treatment) as described elsewhere.26 For SEM analysis each sample was fixed with 200 µL (2.5%) glutaraldehyde for at least 4 hours at 4°C. Samples were washed with PBS. After washing samples were dehydrated by treating with absolute ethanol and isoamyl acetate. Samples were then air dried and coated with gold-palladium using an ion sputter coater (JEOL, Japan). Finally, each sample was observed under the SEM equipment (Model: JSM5910, Manufacturer: JEOL, Japan). Biofilms collected at time points 4, 10, and 18 hours were treated with different concentrations of levofloxacin. Fresh media without antibiotic was used as a negative control.
Statistical Analysis
Each assay was performed in triplicate and repeated at least twice to ensure reproducibility. Statistical correlation between phenotypic markers, antibiotic resistance, and biofilm formation was done by chi-square test. Test was performed by using GraphPad Prism v 7.04. A P-value<0.05 was considered statistically significant.
Results
Phenotypic Identification of Virulence Factors
Out of 155 scrutinized UPEC strains, 67 (43%) showed alpha-hemolysin activity, while 61 (39%) were gelatinase producers. β-hemolysin activity and cell surface hydrophobicity was confirmed in three (2%) and 48 (31%) of the strains, respectively. Serum bactericidal resistance was observed in 132 (85%) of the strains. Likewise, MSHA (Type 1 fimbriae) and MRHA (P fimbriae) was confirmed in three (2%) and 37 (24%) of the strains, respectively.
Qualitative and Quantitative Analysis of Biofilms
Scrutiny of 155 UPEC strains for biofilm production was carried out by three different methods. Overall, 145 (94%) of the strains were confirmed as biofilm producers by CRA (Congo red agar) method. By tube method 154 (99%) and by TCP (tissue culture plate) method all 155 UPEC strains were confirmed as biofilm producers. However, only 113 (73%) of UPEC strains were confirmed as strong biofilm producers, while another 37 (24%) produced biofilms at moderate level. Out of these isolates a total of 21 strong biofilm producing strains simultaneously sensitive to (levofloxacin, ceftazidime, gentamicin, and trimethoprim) were used for further assays to determine MIC-b, MBIC, and MBEC.25 Initially, biofilms of 21 selected strains were analyzed by measuring optical densities (ODs) values at different time points (4, 8, 10, and 24 hours). The mean OD values confirmed colonization of the polystyrene matrix within the initial 4 hours of incubation (shown in Figure 1). Within 8 hours of incubation, an exponential growth phase was attained that reached maximum level within 18 hours of incubation (plateau phase). Biofilms of different time intervals were then analyzed by scanning electron microscopy (Figure 2).
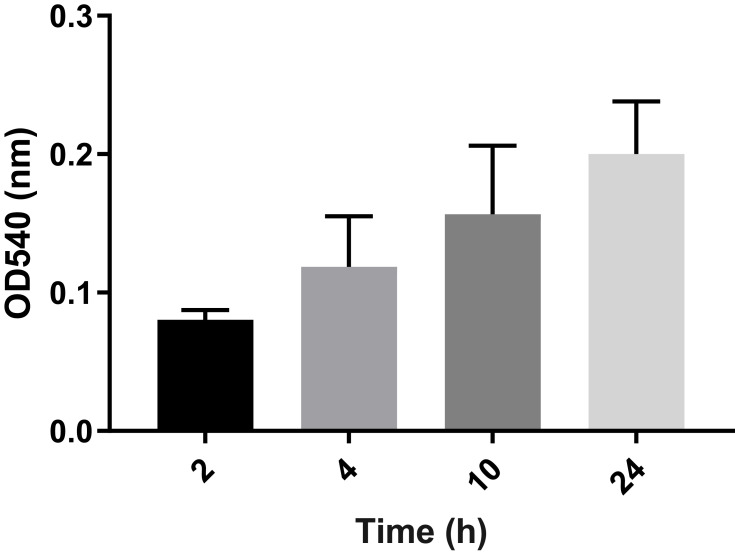
Figure 1.
Average ODs of E. coli biofilm after different time intervals (2, 4, 10, and 24 hours). Bar graph shows weak biofilm formation at 2 and 4 hours (OD~0.08–0.11), moderate biofilm formation at 10 hours (OD~0.157), and mature biofilm at 24 hours (OD~0.20). Error bars show SD from the mean value of all 21 strains.
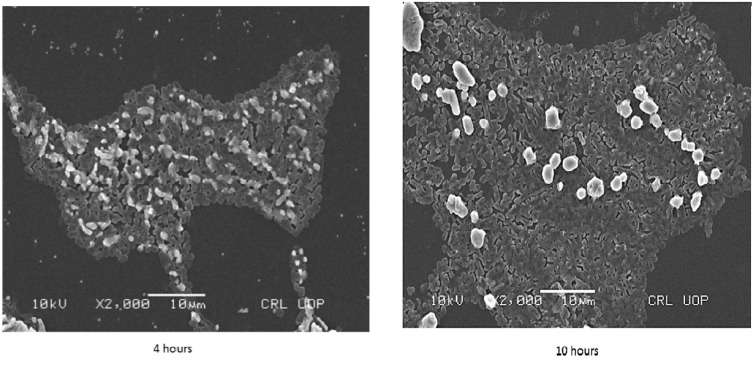
Figure 2.
Scanning electron microscopy (SEM) images of Uropathogenic E. coli strain biofilms after 4 hours (left) and 10 hours (right).
MIC-p and MIC-b of UPEC Biofilm
A comparison based on minimum inhibitory concentrations of antibiotic sensitive planktonic bacterial cells (MIC-p) and minimum inhibitory concentrations of biofilm (MIC-b) confirmed strong differences, indicating a higher level of bacterial tolerance towards frontline antibiotics (levofloxacin, ceftazidime, gentamicin, and trimethoprim) in biofilm form (Table 1). For levofloxacin, 86% of strains showed MIC-b of 32 µg/mL, while MIC-b for 14% of the strains was 64 µg/mL. MIC-b values for gentamicin, ceftazidime, and trimethoprim are shown in Table 1.
Table 1.
Comparison of Minimum Inhibitory Concentration of Planktonic and Biofilm Forms Among UPEC Isolates
| Sample ID | MIC-p (µg/mL) | MIC-b (µg/mL) | ||||||
|---|---|---|---|---|---|---|---|---|
| LEV | GEN | CAZ | TMP | LEV | GEN | CAZ | TMP | |
| ATCC 25,922 | ≤0.125 | ≤0.125 | 0.25 | ≤0.125 | – | – | – | – |
| 2 | 1 | 16 | 16 | 0.125 | 32 | >512 | >2048 | 1024 |
| 3 | 0.12 | 8 | 0.12 | 2 | 32 | >512 | >2048 | 256 |
| 9 | 1 | 1 | 0.5 | – | 32 | 512 | >2048 | – |
| 22 | 0.12 | 1 | 0.12 | – | 32 | >512 | 512 | – |
| 33 | 4 | 16 | 16 | – | 32 | >512 | >2048 | – |
| 34 | 4 | 0.25 | 0.12 | – | 32 | 128 | >2048 | – |
| 47 | 0.12 | 0.12 | 0.12 | – | 32 | 512 | 128 | – |
| 49 | 16 | 4 | 16 | 1 | 32 | 256 | >2048 | 1024 |
| 63 | 16 | 8 | 0.12 | 16 | 32 | 256 | >2048 | 1024 |
| 68 | 0.25 | 0.5 | <0.125 | 16 | 32 | 64 | 128 | 1024 |
| 69 | 8 | 1 | 16 | 1 | 32 | 512 | 256 | 1024 |
| 72 | 16 | 4 | 4 | – | 32 | 64 | 128 | – |
| 74 | 0.12 | 16 | <0.12 | – | 32 | 128 | >2048 | – |
| 81 | 2 | 16 | 4 | – | 64 | 256 | 128 | – |
| 105 | 0.12 | 16 | 4 | 1 | 32 | 256 | 512 | 1024 |
| 107 | 0.12 | 16 | 8 | – | 64 | 1024 | >2048 | – |
| 117 | 0.12 | 16 | <0.12 | – | 64 | 128 | 1024 | – |
| 120 | 0.12 | 1 | <0.12 | – | 32 | 128 | 1024 | – |
| 161 | 2 | 8 | 16 | – | 32 | 256 | 2048 | – |
| 168 | 4 | 16 | 4 | – | 32 | 256 | 1024 | – |
| 176 | 0.25 | 0.5 | <0.12 | – | 32 | 256 | 1024 | – |
Abbreviations: MIC-p, minimum inhibitory concentration of floating bacteria; MIC-b, minimum inhibitory concentration of bacteria in biofilm form; LEV, levofloxacin; GEN, gentamicin; CAZ, ceftazidime; TMP, trimethoprim.
Measurement of MBIC and MBEC of UPEC Biofilms
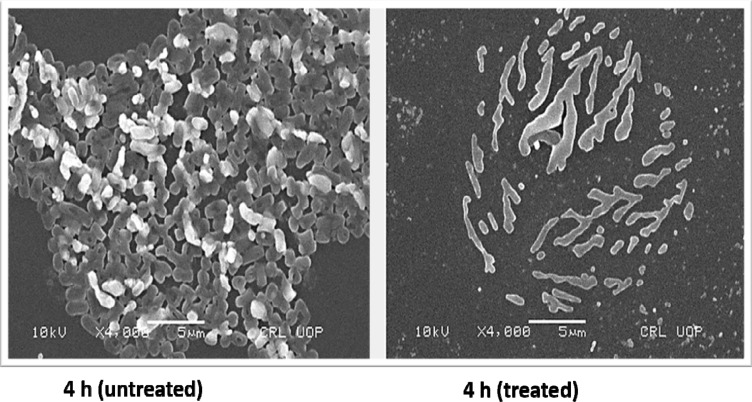
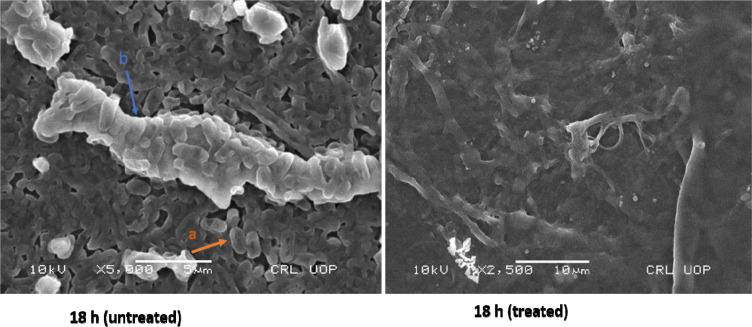
For tested UPEC strains, MBICs for levofloxacin, gentamicin, and ceftazidime were higher than 2048 µg/mL, while trimethoprim inhibited biofilm at 512–1024 µg/mL. Our findings based on MBEC confirm that levofloxacin, gentamicin, and ceftazidime were less effective in eradicating UPEC biofilm even at higher antibiotic concentrations (2048 µg/mL). However, trimethoprim eradicated UPEC biofilm at 512–1024 µg/mL (Table 2). Morphology of a 4 hour biofilm treated with levofloxacin is shown (Figure 3). Likewise, antibiotic treatment of an 18 hour biofilm is also shown (Figure 4).
Table 2.
Comparison of Antimicrobial Susceptibility Between MBIC and MBEC Among UPEC Isolates
| Sample ID | LEV | GEN | CAZ | TMP | ||||
|---|---|---|---|---|---|---|---|---|
| MBIC (µg/mL) | MBEC (µg/mL) | MBIC (µg/mL) | MBEC (µg/mL) | MBIC (µg/mL) | MBEC (µg/mL) | MBIC (µg/mL) | MBEC (µg/mL) | |
| 2 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | 1024 | 1024 |
| 3 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | 512 | 512 |
| 9 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | – | – |
| 22 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | – | – |
| 33 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | – | – |
| 34 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | – | – |
| 47 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | – | – |
| 49 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | 1024 | 1024 |
| 63 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | 1024 | 1024 |
| 68 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | 1024 | 1024 |
| 69 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | 1024 | 1024 |
| 72 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | – | – |
| 74 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | – | – |
| 81 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | – | – |
| 105 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | 1024 | 1024 |
| 107 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | – | – |
| 117 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | – | – |
| 120 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | – | – |
| 161 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | – | – |
| 168 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | – | – |
| 176 | >2048 | >2048 | >2048 | >2048 | >2048 | >2048 | – | – |
Abbreviations: MBIC, minimum biofilm inhibition concentration; MBEC, minimum biofilm eradication concentration; LEV, levofloxacin; GEN, gentamicin; CAZ, ceftazidime; TMP, trimethoprim.
Figure 3.
Scanning electron microscopy (SEM) images showing untreated and levofloxacin treated 4 hours E. coli biofilm cells. Concentrations of levofloxacin was 50 μg/mL. Magnification: ×4000.
Figure 4.
Scanning electron microscopy (SEM) images showing untreated and levofloxacin treated 18 hours E. coli biofilm cells. Concentrations of levofloxacin was 50 μg/mL. Magnification: ×5000.
Notes: a, Individual E. coli cells; b, exopolysaccharide.
Effect of Sub-MIC of Ciprofloxacin on UPEC Biofilm
The effect of sub-minimal concentration of ciprofloxacin on UPEC biofilm was investigated. A variable reduction and induction of biofilm for six selected strains is shown in Table 3. Sub-minimal level of ciprofloxacin was used in the following concentrations, 0.5 µg/mL, 0.25 µg/mL, 0.125 µg/mL, and 0.0625 µg/mL. Biofilm formation for ciprofloxacin treated strains was significantly higher (P<0.05).
Table 3.
Comparison of sub-MIC, MBIC, and MBEC of Ciprofloxacin Against Selected UPEC Strains
| Sample ID | Biofilm Analysis via TCP at 540 nm | Effect of Sub-Minimal and Higher Concentration of CIP on Biofilm Fomation | |||||
|---|---|---|---|---|---|---|---|
| Control | 1/2 MIC | 1/4MIC | 1/8 MIC | 1/16 MIC | MIC-b | MBIC (MRC) | |
| 168 | 0.617±0.22 | 1.550±0.86 | 0.5603±0.43 | 0.885±0.35 | 1.624±1.69 | 128 | >2048 |
| 169 | 0.6233±0.02 | 1.297±0.04 | 2.754±0.14 | 1.33±0.73 | 0.857±0.34 | 2048 | >2048 |
| 176 | 1.109±0.34 | 2.362±0.19 | 1.813±0.35 | 2.013±0.45 | 2.609±0.14 | 128 | >2048 |
| 181 | 0.870±0.08 | 1.023±0.15 | 1.703±0.48 | 1.889±1.13 | 1.779±0.35 | 512 | >2048 |
| 182 | 0.849±0.67 | 0.548±0.18 | 0.519±0.04 | 0.595±0.2 | 1.765±0.39 | 512 | >2048 |
| 183 | 0.333±0.08 | 1.372±0.04 | 0.845±0.04 | 0.855±0.53 | 0.928±0.3 | 512 | >2048 |
Abbreviations: CIP, ciprofloxacin; Sub-MIC, sub-minimal inhibitory concentration; MIC-b, minimum inhibitory concentration of biofilm; MBIC, minimum biofilm inhibition concentration.
Correlation Between Virulence Factors, Antibiotic Resistant, and Biofilms Formation
UPEC strains resistant to ceftazidime (n=60), levofloxacin (n=50), trimethoprim (n=62), and gentamicin (n=20) were screened for phenotypic virulence traits. Although gelatinase production was frequently observed in ceftazidime resistant strains (37, 38%), a statistically significant correlation between gelatinase production and antibiotic resistance was only observed in levofloxacin resistant strains (P<0.05). In the case of MRHA, UPEC strains with P-fimbriae were frequently resistant to antibiotics, however ceftazidime and trimethoprim resistance was significantly (P≤0.01) correlated with this trait. Likewise, trimethoprim resistance was significantly associated with serum resistance (P≤0.001). A statistically significant correlation (P≤0.01) between alpha-hemolysin production and gentamicin sensitivity was concluded. Overall, level of resistance to tested antibiotics remained independent of cell surface hydrophobicity of UPEC strains (P>0.05). In this study no significant correlation between biofilm production and other virulence traits was found except for hemolysins and MSHA (P<0.05) (Table 4).
Table 4.
Correlation Between Virulence Factors, Antibiotic Resistance, and Biofilm
| Virulence Factors | LEV | GEN | CAZ | SXT | Biofilm | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R (n=98) | S (n=57) | R (n=51) | S (n=104) | R (n=97) | S (n=58) | R (n=132) | S (n=23) | s (n=113) | m (n=37) | w (n=5) | |
| ESBL (n=65) | 50** (51%) | 15 (26%) | 20 (39%) | 45 (43%) | 60*** (62%) | 5(9%) | 62** (47%) | 3 (13%) | 46 (41%) | 17 (46%) | 2 (40%) |
| MDR (n=124) | 96*** (98%) | 28 (49%) | 50*** (98%) | 74 (71%) | 94*** (97%) | 30 (52%) | 117*** (89%) | 7 (30%) | 90 (80%) | 30 (81%) | 4 (80%) |
| Gelatinase (n=61) | 32* (33%) | 29 (51%) | 18 (35%) | 43 (39%) | 37 (38%) | 24 (41%) | 49 (37%) | 12 (52%) | 44 (39%) | 15 (41%) | 2 (40%) |
| α-hemolysin (n=67) | 39 (40%) | 28 (49%) | 15 (29%) | 52** (50%) | 40 (41%) | 27 (47%) | 57 (43%) | 10 (43%) | 42 (37%) | 20** (54%) | 5*** (100%) |
| β-hemolysins (n=3) | 3 (3%) | 0 | 2 (4%) | 1 (1%) | 3 (3%) | 0 | 3 (2%) | 0 | 3 (3%) | 0 | 0 |
| CSH (n=48) | 33 (34%) | 15 (25%) | 18 (35%) | 30 (29%) | 34 (35%) | 14 (24%) | 43 (33%) | 5 (22%) | 79 (70%) | 26 (70%) | 4 (80%) |
| SBA (n=132) | 83 (85%) | 49(86%) | 43 (84%) | 89 (85%) | 85 (88%) | 47 (81%) | 124*** (94%) | 8 (35%) | 94 (83%) | 33 89(%) | 5 (100%) |
| Hemagglutination | |||||||||||
| MSHA (n=3) | 2 (2%) | 1 (2%) | 0 | 3 (2%) | 2 (2%) | 1 (2%) | 3 (2%) | 0 | 0 | 3* (8%) | 0 |
| MRHA (n=37) | 26 (27%) | 11 (19%) | 15 (25%) | 22 (21%) | 30** (31%) | 7 (12%) | 36** (27%) | 1(4%) | 31 (27%) | 5 (13%) | 1 (20%) |
Notes: *P<0.05, **P≤0.01, ***P≤0.001.
Abbreviations: LEV, levofloxacin; GEN, gentamicin; CAZ, ceftazidime; SXT, trimethoprim; R, resistant; S, sensitive; s, strong; m, moderate; w, weak.
Discussion
UPEC is frequently associated with both community and hospital acquired UTI. UPEC strains display an interesting mechanism of forming intracellular bacterial communities (IBCs) within mucosal linings of bladder cells. Formations of IBCs have been linked to recurrent UTIs. Development of biofilm-like aggregates in vivo comprise early, middle, and late stages and takes around 6–8 hours to mature.28,29 UPEC biofilms in-vitro are the major cause for catheter associated UTI. In this study, overall, 73% of UPEC isolates were strong biofilm formers. We could categorize UPEC biofilms in three different stages, early, middle, and final stage. SEM analysis revealed that early biofilm formation of UPEC could be observed just after 4 hours of incubation and it took 18 hours before it reached the final stage. Our findings clearly show that MICs for UPEC biofilm (MIC-b) were higher than that of planktonic forms (MIC-p). Furthermore, we observed that, except trimethoprim, other tested frontline antibiotics were less effective in eradicating UPEC in biofilm forms, even at higher concentrations (MBEC>2048 µg/mL) that support the notion that bacterial consortia such as intracellular bacterial communities (IBCs) within mucosal linings of bladder cells may resist a vigorous antibiotic treatment, yet bacterial cells may remain viable, leading to re-occurrence of the UTI. Interestingly, for trimethoprim, similar findings were made when canine uropathogens were tested against TMP-SMX.30 This could be because of differences in antibiotic permeability in biofilm form, quorum sensing mechanisms, and cell growth heterogeneity. Likewise, pharmacokinetic/pharmacodynamic factors of host and bacteria may also play a role in treatment outcome.31
Use of antibiotics asserts a significant selective pressure upon bacterial pathogens resulting in development of defense mechanisms such as changes in protein expression and genetic mutations. Under certain conditions, loss of pathogenicity and induction of SOS under antibiotics pressure has been reported.32 The situation becomes aggravated when antibiotics reach their sub-MIC level before the next dose can be administered, giving bacterial pathogens an opportunity to modulate their pathogenicity. In this study, five out of six UPEC isolates showed enhanced biofilm production at sub-MIC level of ciprofloxacin, confirming that antibiotic treatments at minute concentration can modulate the community structure and heterogeneity of UPEC biofilm. Underlying molecular factors responsible for sensing and contributing to this significant change remain to be investigated. Moreover, a sub-inhibitory concentration of antibiotics can modulate the virulence and adherence of UPEC.33 Other studies showed that sub-lethal concentrations of amoxicillin are involved in increased antibiotic resistance, induction of biofilm formation, and adhesion of uropathogens to the bladder epithelial lining.33,34 Moreover, it has also been shown that patients who have received antibiotics prior to onset of sepsis were less responsive to antibiotic therapy.35 Besides active biofilm formation of UPEC strains encode an array of virulence factors such as fimbriae, surface hydrophobicity, serum resistance, gelatinase, and toxins, which contributes to its effective colonization, increased persistence, and pathogenesis of the disease. Higher occurrence of virulence factors such as gelatinase, hemolysin, and serum resistance indicates the prevalence of virulent UPEC strains. The significant association between biofilm formation and hemolysin may aid UPEC strains in prostate persistence.36 Likewise, another significant factor, curli fimbriae, was attributed to cell-to-cell and cell-to-surface adhesion during biofilm formation. Fimbriae play an important role in initial adhesion and spreading of biofilms on the surface at a later stage.37 It is also worth noting that prevention of a biofilm-like situation in vitro at initial stages could be an interesting strategy to counterfeit the development of infection. Conclusively, the association of virulence factors such as biofilm formation and resistant phenotypes to frontline antibiotics poses a significant challenge to treat UPEC infections.
Conclusion
Therefore, it is imperative that factors associated with biofilm tolerance and recalcitrance against antibiotic would demand new therapeutic strategies which must breach these barriers. Furthermore, while prioritizing patient’s health, it should be a specialist’s first goal to avoid a biofilm-like scenario that could otherwise produce undesirable consequences. Virulence factors have an important role in adherence with epithelial surfaces and for immune evasion. UPEC isolates in this study showed different phenotypes including ESBLs, gelatinase, hemolysins, HA, hydrophobicity, and serum bactericidal activity. Their identification assists in understanding their role in virulence.
Acknowledgment
We thank the Higher Education Commission Pakistan for supporting doctoral work of Zara Rafaque.
Funding Statement
This work was financially supported by Higher Education Commission Pakistan and Quaid-i-Azam University, Islamabad (PIN#213-53961-2BM2-093; 50025385).
Disclosure
The authors report no conflicts of interest for this work.
References
- 1.Ponnusamy P, Natarajan V, Sevanan M. In vitro biofilm formation by uropathogenic Escherichia coli and their antimicrobial susceptibility pattern. Asian Pac J Trop Med. 2012;5:210–213. doi: 10.1016/S1995-7645(12)60026-1 [DOI] [PubMed] [Google Scholar]
- 2.Ponnusamy P, Nagappan R. Extended spectrum beta-lactamase, biofilm-producing uropathogenic pathogens and their antibiotic susceptibility patterns from urinary tract infection-an overview. Int J Microbiol Res. 2013;4:101–118. [Google Scholar]
- 3.Asadi S, Kargar M, Solhjoo K, Najafi A, Ghorbani-Dalini S. The association of virulence determinants of uropathogenic Escherichia coli with antibiotic resistance. Jundishapur J Microbiol. 2014;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foxman B. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am J Med. 2002;113:5–13. doi: 10.1016/S0002-9343(02)01054-9 [DOI] [PubMed] [Google Scholar]
- 5.Mulvey MA, Schilling JD, Martinez JJ, Hultgren SJ. Bad bugs and beleaguered bladders: interplay between uropathogenic Escherichia coli and innate host defenses. Proc Natl Acad Sci. 2000;97:8829–8835. doi: 10.1073/pnas.97.16.8829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bower JM, Eto DS, Mulvey MA. Covert operations of uropathogenic Escherichia coli within the urinary tract. Traffic. 2005;6:18–31. doi: 10.1111/j.1600-0854.2004.00251.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tabasi M, Karam MRA, Habibi M, Yekaninejad MS, Bouzari S. Phenotypic assays to determine virulence factors of uropathogenic Escherichia coli (UPEC) isolates and their correlation with antibiotic resistance pattern. Osong Public Health Res Perspect. 2015;6:261–268. doi: 10.1016/j.phrp.2015.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tajbakhsh E, Ahmadi P, Abedpour-Dehkordi E, Arbab-Soleimani N, Khamesipour F. Biofilm formation, antimicrobial susceptibility, serogroups and virulence genes of uropathogenic E. coli isolated from clinical samples in Iran. Antimicrob Resist Infect Control. 2016;5:11. doi: 10.1186/s13756-016-0109-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shenoy VAARS, Yadav T. The antibiotic susceptibility patterns of uropathogenic Escherichia coli, with special reference to the fluoroquinolones. J Clin Diagn Res. 2013;7:1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mittal S, Sharma M, Chaudhary U. Biofilm and multidrug resistance in uropathogenic Escherichia coli.. Pathog Glob Health. 2015;109:26–29. doi: 10.1179/2047773215Y.0000000001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalhoff A. Global fluoroquinolone resistance epidemiology and implications for clinical use. Interdiscip Perspect Infect Dis. 2012;2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kahlmeter G, Poulsen HO. Antimicrobial susceptibility of Escherichia coli from community-acquired urinary tract infections in Europe: the ECO.SENS study revisited. Int J Antimicrob Agents. 2012;39:45–51. doi: 10.1016/j.ijantimicag.2011.09.013 [DOI] [PubMed] [Google Scholar]
- 13.Linhares I, Raposo T, Rodrigues A, Almeida A. Frequency and antimicrobial resistance patterns of bacteria implicated in community urinary tract infections: a ten-year surveillance study (2000–2009). BMC Infect Dis. 2013;13:19. doi: 10.1186/1471-2334-13-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiedemann B, Heisig A, Heisig P. Uncomplicated urinary tract infections and antibiotic resistance—epidemiological and mechanistic aspects. Antibiotics. 2014;3:341–352. doi: 10.3390/antibiotics3030341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Florindo C, Costa A, Matos C, et al. Novel organic salts based on fluoroquinolone drugs: synthesis, bioavailability and toxicological profiles. Int J Pharm. 2014;469:179–189. doi: 10.1016/j.ijpharm.2014.04.034 [DOI] [PubMed] [Google Scholar]
- 16.Schmitz F-J, Jones ME. Antibiotics for treatment of infections caused by MRSA and elimination of MRSA carriage. What are the choices? Int J Antimicrob Agents. 1997;9:1–19. doi: 10.1016/S0924-8579(97)00027-7 [DOI] [PubMed] [Google Scholar]
- 17.Soleimani N, Aganj M, Ali L, Shokoohizadeh L, Sakinc T. Frequency distribution of genes encoding aminoglycoside modifying enzymes in uropathogenic E. coli isolated from Iranian hospital. BMC Res Notes. 2014;7:842. doi: 10.1186/1756-0500-7-842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ali I, Rafaque Z, Ahmed S, Malik S, Dasti JI. Prevalence of multi-drug resistant uropathogenic Escherichia coli in Potohar region of Pakistan. Asian Pac J Trop Biomed. 2016;6:60–66. doi: 10.1016/j.apjtb.2015.09.022 [DOI] [Google Scholar]
- 19.Snyder JA, Haugen BJ, Lockatell CV, et al. Coordinate expression of fimbriae in uropathogenic Escherichia coli.. Infect Immun. 2005;73:7588–7596. doi: 10.1128/IAI.73.11.7588-7596.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussain A, Ranjan A, Nandanwar N, Babbar A, Jadhav S, Ahmed N. Genotypic and phenotypic profiles of Escherichia coli isolates belonging to clinical sequence type 131 (ST131), clinical non-ST131, and fecal non-ST131 lineages from India. Antimicrob Agents Chemother. 2014;58:7240–7249. doi: 10.1128/AAC.03320-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mittal S, Sharma M, Chaudhary U. Study of virulence factors of uropathogenic Escherichia coli and its antibiotic susceptibility pattern. Indian J Pathol Microbiol. 2014;57:61. doi: 10.4103/0377-4929.130899 [DOI] [PubMed] [Google Scholar]
- 22.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: 24th Informational Supplement. Document M100–S27. PA: Wayne; 2017. [Google Scholar]
- 23.Solati SM, Tajbakhsh E, Khamesipour F, Gugnani HC. Prevalence of virulence genes of biofilm producing strains of Staphylococcus epidermidis isolated from clinical samples in Iran. AMB Express. 2015;5:47. doi: 10.1186/s13568-015-0134-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christensen GD, Simpson W, Younger J, et al. Adherence of coagulase-negative Staphylococci to plastic tissue culture plates: a quantitative model for the adherence of Staphylococci to medical devices. J Clin Microbiol. 1985;22:996–1006. doi: 10.1128/JCM.22.6.996-1006.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Rossi BP, García C, Calenda M, Vay C, Franco M. Activity of levofloxacin and ciprofloxacin on biofilms and planktonic cells of Stenotrophomonas maltophilia isolates from patients with device-associated infections. Int J Antimicrob Agents. 2009;34:260–264. doi: 10.1016/j.ijantimicag.2009.02.022 [DOI] [PubMed] [Google Scholar]
- 26.Wang A, Wang Q, Kudinha T, Xiao S, Zhuo C. Effects of fluoroquinolones and azithromycin on biofilm formation of Stenotrophomonas maltophilia. Sci Rep. 2016;6:29701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majtan J, Majtánová Ľ, Xu M, Majtan V. In vitro effect of subinhibitory concentrations of antibiotics on biofilm formation by clinical strains of Salmonella enterica serovar Typhimurium isolated in Slovakia. J Appl Microbiol. 2008;104:1294–1301. doi: 10.1111/j.1365-2672.2007.03653.x [DOI] [PubMed] [Google Scholar]
- 28.Mulvey MA, Schilling JD, Hultgren SJ, O’Brien AD. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun. 2001;69(7):4572–4579. doi: 10.1128/IAI.69.7.4572-4579.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson GG, Palermo JJ, Schilling JD, Roth R, Heuser J, Hultgren SJ. Intracellular bacterial biofilm-like pods in urinary tract infections. Science. 2003;301(5629):105–107. doi: 10.1126/science.1084550 [DOI] [PubMed] [Google Scholar]
- 30.Shimizu T, Harada K. Determination of minimum biofilm eradication concentrations of orbifloxacin for canine bacterial uropathogens over different treatment periods. Microbiol Immunol. 2017;61(1):17–22. doi: 10.1111/1348-0421.12461 [DOI] [PubMed] [Google Scholar]
- 31.Ambrose PG, Bhavnani SM, Rubino CM, et al. Pharmacokinetics-pharmacodynamics of antimicrobial therapy: it’s not just for mice anymore. Clin Infect Dis. 2007;44:79–86. doi: 10.1086/510079 [DOI] [PubMed] [Google Scholar]
- 32.Baharoglu Z, Krin E, Mazel D, Hughes D. RpoS plays a central role in the SOS induction by sub-lethal aminoglycoside concentrations in Vibrio cholerae. PLoS Genet. 2013;9(4):e1003421. doi: 10.1371/journal.pgen.1003421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goneau LW. Sub-inhibitory antibiotics enhance virulence, persistence, and pathogenesis of uropathogens. MBio. 2007;6:e00356–1. [Google Scholar]
- 34.Adamus-Białek W, Wawszczak M, Arabski M, et al. Ciprofloxacin, amoxicillin, and aminoglycosides stimulate genetic and phenotypic changes in uropathogenic Escherichia coli strains. Virulence. 2019;10(1):260–276. doi: 10.1080/21505594.2019.1596507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garnacho-Montero J, Garcia-Garmendia JL, Barrero-Almodovar A, Jimenez-Jimenez FJ, Perez-Paredes C, Ortiz-Leyba C. Impact of adequate empirical antibiotic therapy on the outcome of patients admitted to the intensive care unit with sepsis. Crit Care Med. 2003;31:2742–2751. doi: 10.1097/01.CCM.0000098031.24329.10 [DOI] [PubMed] [Google Scholar]
- 36.Baqai R, Aziz M, Rasool G. Urinary tract infections in diabetic patients and biofilm formation of uropathogens. Infect Dis J Pak. 2008;17:21–24. [Google Scholar]
- 37.Wojnicz D, Tichaczek-Goska D. Effect of sub-minimum inhibitory concentrations of ciprofloxacin, amikacin and colistin on biofilm formation and virulence factors of Escherichia coli planktonic and biofilm forms isolated from human urine. Braz J Microbiol. 2013;44:259–265. doi: 10.1590/S1517-83822013000100037 [DOI] [PMC free article] [PubMed] [Google Scholar]