Abstract
The relationship between kidney cancer, specifically clear cell renal cell carcinoma (ccRCC), and the hypoxia signaling program has been extensively characterized. Its underlying role as the primary driver of the disease has led to the development of the most effective targeted therapies to date. Cellular responses to hypoxia or mutations affecting the von Hippel-Lindau (VHL) tumor suppressor gene stabilize the hypoxia inducible factor (HIF) transcription factors which then orchestrate elaborate downstream signaling events resulting in adaptations to key biological processes, such as reprogramming metabolism. The direct link of hypoxia signaling to glucose uptake and glycolysis has long been appreciated; however, the HIF family of proteins directly regulate many downstream targets, including other transcription factors with their own extensive networks. In this review, we will summarize our current understanding of how hypoxia signaling regulates other metabolic pathways and how this contributes to the development and progression of clear cell renal cell carcinomas.
Keywords: ccRCC, Pseudohypoxia, HIF, Metabolism, Mitochondria
Introduction
Clear cell renal carcinoma (ccRCC) is the most frequent form of RCC, accounting for approximately 75% of diagnosed renal carcinomas [1]. The most up-to-date databases estimate that with incidence rates more than doubling in the past twenty years, ccRCC will be the 8th most commonly diagnosed tumor in the United States with approximately 74,000 new cases expected in 2020 in the United States alone and over 300,000 worldwide. According to the National Cancer Institute, it is also estimated that in this year alone there will be nearly 15,000 deaths due to ccRCC in the United States and approximately 100,000 deaths globally. It is worth noting that the 5-year survival rates plummet from 93% for localized disease to 69% and 12% for regional and distant metastatic disease, respectively [2]. Although 5-year survival rates have seen improvement with the implementation of tyrosine kinase inhibitors (TKI) [3], and further gains are expected with the advent of immunotherapies, survival rates with advanced disease remain unacceptably low.
Clear cell renal carcinomas are distinguishable from other RCC subtypes due to a characteristic clear cell morphology that results from an abundance of lipids and glycogen present in the cytoplasm causing the cells to appear translucent [4, 5], providing the first clue that this cancer would be inextricably linked to metabolic alterations. Further examination into this phenotype ultimately did reveal an overhaul in the metabolic wiring of these tumors, where classical Warburg metabolism is accompanied by a reversal in TCA cycle directionality, causing an upregulation in glutamine and lipid metabolism used to fuel the anabolic demands required for rapid growth [6]. Driving these metabolic adaptations and initiating tumorigenesis, clear cell renal carcinomas harbor genetic alterations that cause major signaling pathways to become dysregulated.
Several of the most common inactivating mutations in ccRCC result from the loss of chromosome 3p, where several chromatin remodeling genes such as BAP1, PBRM1, and SETD2 reside [5, 6], suggesting that ccRCCs favor certain chromatin states. Later considerations in this review will attempt to link chromatin state and metabolism, such that the constellation of genetic driver events in ccRCC are related. However, to set the stage for exploring hypoxia signaling in ccRCC it is critical to first examine the most prevalent mutation observed in ccRCC. Approximately 90% of cases demonstrate inactivation of von Hippel-Lindau (VHL) gene, which encodes the E3 ubiquitin ligase that recognizes hypoxia-inducible factor (HIF) proteins. The HIF family of transcription factors consists of three alpha- subunit isoforms: HIF-1α, HIF-2α, and HIF-3α which then heterodimerize with HIF-1β, bind to hypoxia response elements (HRE) in the promoters of target genes, and orchestrate a complex transcriptional program [7, 8]. The HIF signaling axis (or HIF signaling) is a term used broadly to include all of the HIF dimer transcriptional outputs. Throughout this review, when a specific subunit is under consideration, it will be named as such, and in other cases the broader definition of the HIF family as mediators of the HIF response will be applied. HIF signaling influences many foundational cellular processes such as cell cycle, transcription, and translation resulting in a broad spectrum of responses to pathways involved in angiogenesis, metabolism, proliferation, and cell survival [9]. In the presence of oxygen, prolyl hydroxylases (PHD) add a hydroxyl group to HIFs and VHL specifically recognizes those HIF proteins that have been hydroxylated [10]. VHL binding leads to polyubiquitylation, which targets the HIF transcription factors for proteasomal degradation [11, 12] (Figure 1A). In addition to this canonical HIF degradation pathway, the presence of oxygen also reduces HIF signaling by promoting the activity of the factor inhibiting HIF (FIH), an asparaginyl hydroxylase which hydroxylates an asparagine residue in the C-terminal activation domain and prevents the recruitment of HIF co-activators p300 and CBP [13, 14]. On the contrary, when oxygen concentrations are low the HIF-α subunits are stabilized due to suspended PHD and FIH activity. Under these hypoxic conditions, HIF-α subunits translocate into the nucleus and form a transcriptionally active complex with HIF-1β, as well as the co-activators p300 and CBP [15-17]. Loss of VHL activity, and the subsequent stabilization of the HIF-α subunits results in constitutive HIF signaling and has been established as an essential trigger to the initiation of ccRCC development [11]. In ccRCCs where VHL is lost, a state of pseudohypoxia is acquired by the accumulation of the HIFs in the presence of oxygen (Figure 1B) [18]. While in many VHL−/− ccRCC tumors HIF-1α and HIF-2α functionally coexist to coordinate other major signal transduction cascades, such as the MAPK and Akt-mTOR pathways [19], it has been established that there can be a preferential upregulation of constitutive HIF-2α signaling that can promote the upregulation and activity of the oncogene c-Myc, rather than elevated HIF-1α signaling which is canonically observed in many other cancers, where the driving factor of HIF stabilization is regional hypoxia and c-Myc activity is suppressed [5, 11, 20]. Further supporting this concept, an absence of HIF-1α protein expression and truncated mRNA transcripts was observed in a panel of RCC cell lines [21]. In light of these findings, more recent studies have even suggested that HIF-1α functions as a tumor suppressor when expressed in ccRCC [22-24], which is supported by the finding that a loss of chromosome 14q, or regions of 14q that encompass the HIF1A locus, is observed in 45% of patients [1, 23, 25]. To date, these studies along with functional analyses studying the promoter binding differences between these two HIF-α isoforms [26-28], point to an intricately regulated overlapping HIF network that segues into a dichotomous transcriptional program which can be influenced by the ratio of HIF-α isoforms (Figure 2A) [28-30].
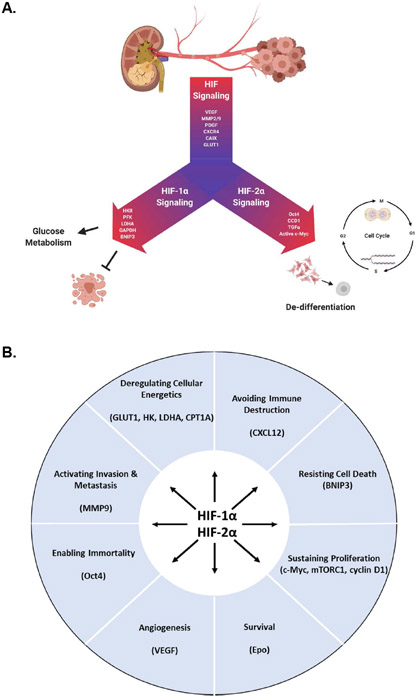
Figure 1. The dysregulation of HIF signaling is the primary driver of ccRCC.
(A) Under normoxic conditions, sufficient oxygen supply fuels the activity of prolyl hydroxylases (PHD) which hydroxylate proline residues of the HIF-α proteins. These hydroxylated HIF-α proteins are specifically recognized by E3 ubiquitin ligase pVHL, which directs the HIF-α proteins for ubiquitin-dependent proteasomal degradation. (B) During hypoxia, PHDs are left without the necessary substrates to perform their catalytic activity and are further inhibited by metabolic by-products generated from unchecked HIF-driven signaling. In approximately 90% of ccRCC cases, VHL is lost resulting in a state of pseudohypoxia and rampant, constitutive HIF signaling.
Figure 2. The HIF signaling axis.
(A) The hypoxia-inducible factor family of transcription factors exhibit unity in their regulation of genes involved in angiogenesis, invasion, and glucose transport. In ccRCC, HIF-1α and HIF-2α facilitate tumorigenesis via unique transcriptional programs. (B) HIF-dependent signaling is a central regulator of the “Hallmarks of Cancer” in ccRCC.
In VHL-deficient ccRCC, the HIF signaling network is an essential mediator of growth and progression independent of oxygen saturation. As transcription factors, the HIFs serve as a central regulator to many genes with functions residing in each of the “Hallmarks of Cancer” and are required to initiate, support, and progress tumor growth (Figure 2B) [11, 25, 31, 32]. While HIF signaling is largely responsible for many of the unique metabolic features observed in ccRCC, it also plays an integral role in supporting the metabolic reprogramming that is common to many cancers which results in the dysregulation of key glycolytic genes such as glucose transporter 1 (GLUT1), hexokinase II (HK2), and lactate dehydrogenase A (LDHA). Additionally, HIF signaling also selectively activates pyruvate dehydrogenase kinase (PDK) to keep glucose-derived carbons from entering the TCA cycle, enhancing glycolytic activity and biomass accumulation [25, 26, 33]. A recent study infused ccRCC patients with [U-13C]glucose to analyze the metabolic differences between the tumors and adjacent tissue, where it was observed that ccRCC tumors displayed enhanced glycolysis coupled with reduced glucose oxidation into the TCA cycle [34]. Furthermore, when the spectra obtained from ccRCC tumors were compared to those generated from tumors of the brain or lung, which are known to rely heavily on glycolysis, ccRCC tumors still displayed enhanced production of glycolytic intermediates with reduced entry into the TCA cycle [34]. This study corroborates findings made previously using in vitro cell systems and mouse models which extensively characterize the role of hypoxia signaling in fueling and regulating glycolysis in ccRCC [23, 26, 35]. Notably, a TCGA study analyzing over 400 RCC tumors made similar conclusions regarding aggressive RCC tumors with poor outcome associating with aerobic glycolysis [1]. These studies and others focusing on glycolysis have been reviewed in detail elsewhere [33, 36, 37].
Despite the prevalence of glycolytic activity and lack of glucose oxidation into the TCA cycle being functionally validated, a comprehensive, integrated metabolomic and transcriptional profiling of ccRCC tumors revealed that a disparity exists between the expression of metabolic genes in transcriptional datasets and functional metabolite readouts [38], which suggests transcriptional data of metabolic genes in ccRCC may be subject to overinterpretation and warrant further investigation with functional metabolomics. Moreover, other studies have gone on to show that ccRCC have a dependence for other bioenergetic substrates, such as glutamine, in a HIF-dependent manner and fluxing into other biosynthetic pathways [39-41]. Additionally, there are other reports of hypoxia signaling in ccRCC regulating other key metabolic pathways such as lipid and cholesterol metabolism [42-44], acetate metabolism [41], pathways shunting from glycolysis [45], and mitochondrial metabolism [46, 47]. For the remainder of this review, we will focus on how HIF signaling serves as a master regulator of global metabolism in ccRCC beyond glycolysis with an emphasis on alternative metabolic pathways.
Pentose Phosphate Pathway
NADPH Production and Redox Balance
In the aforementioned 2013 TCGA analysis that identified an association between high grade RCCs and aerobic glycolysis, it was also discovered that these tumors exhibit a dependence on the pentose phosphate pathway (PPP) [1]. Since ccRCC tumor cells rapidly utilize glycolysis and avoid contribution to the TCA cycle, the PPP provides an avenue to shuttle glucose for the synthesis and accumulation of other biosynthetic precursors and cofactors such as ribose-5-phosphate and NADPH, which are used to fuel nucleotide synthesis, lipid synthesis, and combat reactive oxygen species (ROS). Intriguingly, it has been demonstrated in several different cancer cell lines that glucose-6-phosphate dehydrogenase (G6PD), the rate-limiting enzyme of the PPP, is dynamically O-GlcNAcylated in response to hypoxia and that this post-translational modification activates the enzyme and fuels glucose flux through the PPP, generating precursors for biosynthesis and reducing equivalents for equilibrating ROS [48] (Figure 3). As it relates to ccRCC, recent work from the Zhu Lab observed G6PD expression to be elevated at the mRNA and protein levels across 149 matched ccRCC tumor and normal tissue samples [49]. In a parallel study, the same group performed experiments to characterize the effects of G6PD upregulation in ccRCC and found that G6PD activity supports proliferation by facilitating the cell cycle, resulting in enhanced tumorigenesis in vitro and in vivo [50]. Interestingly, the authors then connect G6PD activity to increased levels of NADPH and NOX4 expression, which resulted in a corresponding increase in ROS production. The connection between increased NADPH, NOX4, and ROS in ccRCC was first described in 2014 where it was shown that NOX4 expression supported HIF-2α signaling [51]. Silencing G6PD was successful at reducing NADPH, NOX4, and ROS implicating G6PD as a key mediator for regulating redox balance in ccRCC. Additionally, they identified p-STAT3 to be elevated under conditions of G6PD upregulation mediating proliferation through downstream signaling such as Cyclin D1 regulation. Lastly, they provide evidence that p-STAT3 enters a positive feedback loop where it can directly bind to the promoter and drive transcription of G6PD [50].
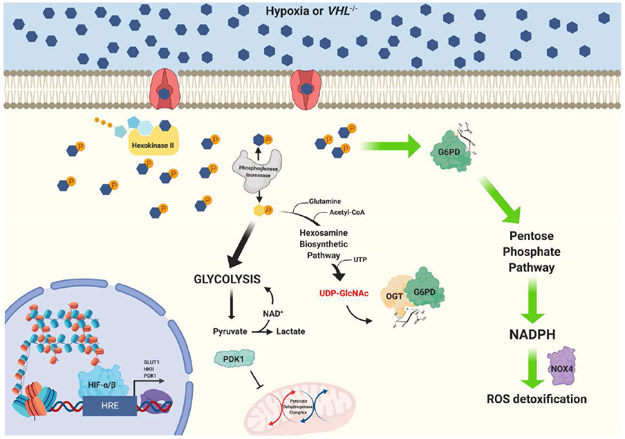
Figure 3. Hypoxia-mediated glucose metabolism promotes dynamic O-GlcNAcylation of G6PD facilitating the utilization of PPP to produce NADPH for ROS detoxification.
Hypoxia signaling facilitates glucose metabolism by upregulating glucose transporters and other glycolytic machinery. As glucose enters the cell it is phosphorylated into glucose-6-phosphate (G6P) by hexokinase II and then enzymatically converted into fructose-6-phosphate (F6P) by phosphoglucose isomerase. While most F6P continues on into glycolysis, a small fraction is shunted into the hexosamine biosynthetic pathway where it is joined by other metabolic inputs to synthesize the charged sugar UDP-GlcNAc. This sugar moiety serves as the substrate for O-linked glycosylation of Ser/Thr residues on target proteins, a post-translational modification catalyzed the enzyme O-GlcNAc transferase (OGT). Under hypoxic conditions, glucose-6-phosphate dehydrogenase (G6PD) is O-GlcNAc modified resulting in enhanced activity and shuttling of G6P through the PPP, producing ample NADPH for biosynthetic and detoxification processes. In ccRCC, G6PD activity has been shown to be elevated and enhance NADPH production and NOX4 expression supporting tumorigenesis.
Future experiments studying G6PD activity in ccRCC should be aimed at regulatory post-translational modifications with a specific focus on O-GlcNAcylation given its relationship to HIF signaling [52]. Intriguingly, the cycling of this modification uses only the enzymes O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA), simplifying experimental design [53]. Furthermore, these enzymes have been suggested as biomarkers in other cancers and have established small molecule inhibitors targeting their activity looking to gain clinical traction [54-56].
Nucleotide Metabolism
Studies such as those previously described magnify a role for PPP activity in ccRCC to drive NADPH accumulation for maintaining redox homeostasis [49-51]; however, it is important to remember that NADPH is also an essential cofactor for biosynthetic processes such as nucleotide and lipid synthesis. As with any rapidly proliferating cell, nucleotides need to be synthesized efficiently; thus, the precursors for nucleotide synthesis accumulate before a cell enters S phase. Ribose-5-phosphate (R5P) is the final product of the oxidative PPP reactions where it can then be acted on by ribose-5-phosphate isomerase (RPI) and enter the non-oxidative reactions, or by ribose-phosphate diphosphokinase which converts R5P into phosphoribosyl pyrophosphate (PRPP), the first step for purine synthesis. One of the enzymes responsible for R5P production, transketolase (TKT), which converts glyceraldehyde-3-phosphate into R5P, has recently been shown to be tightly regulated by transketolase like protein 1 (TKTL1), whose expression is elevated in ccRCC patient samples when compared to adjacent tissue, subsequently resulting in increased R5P concentrations [57]. Mechanistically, the study found TKTL1 expression to prevent TKT homodimer formation by interacting with TKT in a heterodimer complex, which results in the accumulation of R5P by preventing its conversion and removal from the pathway [57].
Despite not linking the mechanisms to hypoxia, the studies described above do highlight an essential role for the pentose phosphate pathway in ccRCC. Through the work of others, it has been demonstrated that hypoxia influences the post-translational modification and regulation of G6PD activity [48], and upregulates TKTL1 expression to support tumor growth in colorectal cancer [58]. Taken together, it would seem like a logical progression to test whether these models translate as contributing features in the metabolic profile of ccRCC via drivers of HIF signaling.
Mitochondria
TCA cycle
HIF signaling has been extensively documented in its ability to upregulate glycolysis, bypass the TCA cycle, and oxidize pyruvate into lactate, a process known as anaerobic glycolysis [59, 60]. Cancer cells undergo metabolic reprogramming in favor of aerobic glycolysis, or the Warburg Effect, enabling cells to continually utilize this pathway even under normoxic conditions as an effort to accumulate biomass [61-63]. The loss of the tumor suppressor gene VHL is a common mechanism employed by cancer cells to hyperactivate Warburg metabolism and fuel tumorigenesis via constitutive HIF signaling [64-67]. Essential to maintaining continuous glycolytic metabolism, HIFs have been shown to upregulate and enhance the activity of the PDK enzymes. Consequently, the activity of the pyruvate dehydrogenase complex (PDH) that functions to convert pyruvate into acetyl-CoA and flux into the TCA cycle is inhibited [35, 68-70]. In a recent study, it was shown that PDK1 protein expression is elevated in ccRCC patient tumors and in the 786-O cell line, an expected result based on the link between activated hypoxia signaling and PDK1 [71]. In an effort to reverse the HIF-PDK1 axis-mediated suppression of mitochondrial metabolism and function, the researchers treated 786-O cells with dichloroacetate (DCA), which inhibits PDK1-dependent phosphorylation of PDH resulting in a reactivation of PDH [72, 73]; in response to DCA treatment, they observed mitochondrial membrane repolarization and an increase in mitochondrial ROS production in ccRCC cells but not in normal samples [71]. Utilizing HIF-luciferase reporter assays, the authors demonstrate that DCA treatment does not alter HIF-2α protein expression, rather, it inhibits HIF transcriptional activity and glucose metabolism, while enhancing p53 signaling; overall, these effects resulted in reduced angiogenesis and tumor growth [71].
Oxidative Phosphorylation
ccRCC tumor cells exhibit an incredible propensity to utilize glycolysis to fuel their expansion and often oxidative phosphorylation (OXPHOS) is viewed as being disconnected from cancer progression and even associated with better outcome. Many of these conclusions have been extrapolated from transcriptomic-based approaches, rather than functional analysis, and a cross-sectional study comparing transcriptomic datasets to metabolomic datasets identified discrepancies in mRNA expression and metabolite output [38]. Findings such as those just described highlight a gap in our ability to integrate gene expression datasets to downstream cellular processes, as these data are often misinterpreted and not the best representation of protein expression and/or function. A recent single-cell sequencing study examined head and neck squamous cell carcinomas (HNSCC) and melanoma patient samples and discovered expression of OXPHOS genes to be generally elevated in single malignant cells and that the activity of these genes positively correlated with glycolysis and hypoxia in all cell types, indicative of a role supporting heterogeneity [74]. This comprehensive analysis concluded that OXPHOS activity is negatively regulated by hypoxia signaling initially; however, as ccRCC tumors progress they reprogram their metabolism to more heavily utilize OXPHOS, suggesting that OXPHOS is overall dysregulated in ccRCC and significantly contributes to disease progression [38]. A comparable integrated proteogenomic approach analyzed the transcriptome and proteome in ccRCC patient samples and came to similar conclusions, finding OXPHOS gene transcripts and protein levels did not correlate; in fact, mRNA appeared to be reduced in tumor samples while the protein was elevated [75].
Interestingly, in more advanced tumors both mRNA and protein were elevated and seemed to correlate with hypoxia signaling [75]. The results described by both groups suggest that OXPHOS is dynamically regulated in cancers and that hypoxia signaling lies at the crux of glycolysis and oxidative phosphorylation [38, 74]. Other reports of OXPHOS significantly contributing to tumor development have recently been published; in one instance, a study demonstrated intratumoral cancer cell heterogeneity in high grade serous ovarian cancer (HGSOC) where molecular subgroups were identified based on OXPHOS expression and activity and these high-OXPHOS tumors were shown to be dependent on oxidative metabolism in a manner supported by glutamine metabolism and beta-oxidation [76].
Recent studies have begun to investigate the functional importance of OXPHOS in ccRCC.In one such study, researchers examined metabolic responses to the loss of a key epigenetic modifier SETD2, a frequently occurring secondary mutation. Using the VHL−/− 786-O cell line, the authorsfound that SETD2 deletion upregulated oxidative phosphorylation genes and enhanced their activity via upregulating PGC1α, resulting in increased mitochondrial mass and respiration [77]. Interestingly, an earlier study found that HIF signaling suppresses PGC1α [78]. By performing VHL reconstitution or shHIF1A and shHIF2A experiments in VHL−/− cell lines, they observed PGC1α expression increase with VHL expression [78]. Conversely, cells or patient samples with a VHL deletion displayed reduced PGC1α expression, decreased TFAM expression and mitochondrial mass [78]. Furthermore, overexpression of PGC1α was shown to enhance oxidative stress and mitochondrial ROS, as well as sensitizing cells to cytotoxic therapies [78]. The results reported in these independent studies appear contradictory, but it could suggest that a secondary mutation to SETD2, commonly found in advanced disease, is a key driver in rewiring metabolism to more heavily utilize oxidative phosphorylation, as suggested in the integrated metabolomic studies [38, 75, 77, 78] (Figure 4). Though OXPHOS may not be the metabolic driver behind ccRCC, a deeper evaluation into its function and role in supporting tumorigenesis is needed and would benefit from further investigation into the contribution of the genetic drivers of ccRCC.
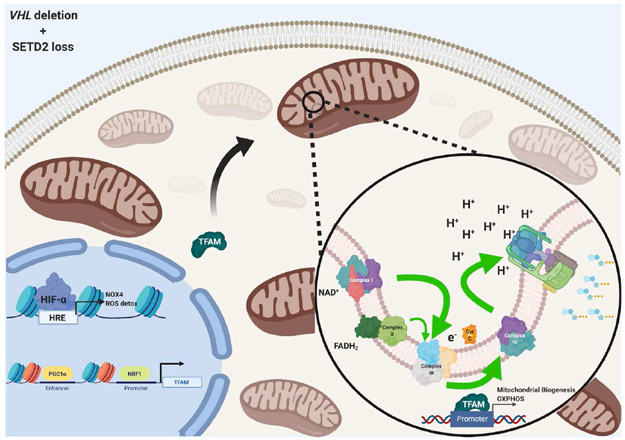
Figure 4. Hypothetical mechanism of cooperation between hypoxia signaling and mitochondrial respiration in VHL−/−/SETD2−/− ccRCC.
Loss of SETD2 enhances chromatin accessibility, potentially alleviating suppression of PGC1α and even promoting a symbiotic balance between HIF-mediated signaling, mitochondrial biogenesis, and oxidative metabolism.
Mitochondrial Dynamics
Another key feature of mitochondrial health, function, and signaling is the cyclical dynamics of these organelles via fission and fusion processes, as well as mitochondrial biogenesis and autophagy [79-81]. Mitochondrial dynamics are essential to maintaining high quality mitochondria and the dysfunction of these processes has been intimately linked to aging and metabolic dysregulation, resulting in pathologies such as diabetes and obesity, as well as the neurodegenerative diseases Charcot-Marie-Tooth subtype 2A and autosomal dominant optic atrophy [80, 82-85]. Since cancer is commonly associated with aging, especially for RCCs where the median age of diagnosis in the United States is 64 years old [18, 86], researchers have been investigating the role of mitochondrial dynamics in the development and progression of cancer. In the context of general biology, hypoxia signaling has been shown to support mitochondrial fragmentation and fission through a mechanism requiring the phosphorylation of dynamin-related protein 1 (Drp1) at serine 616 in pancreatic beta cells [87] or serine 637 in endothelial progenitor cells [88]. In another study utilizing breast, lung, and pancreatic cancer cell line models it was found that hypoxia mediates mitophagy during dissemination, enhancing removal of ROS-producing mitochondria in response to a reduced requirement for oxidative metabolism resulting from increased glycolytic activity and reductive carboxylation [89]. While there is a deficiency in literature on hypoxia-mediated effects on mitochondrial dynamics in ccRCC, it was recently found that defective autophagy supports ccRCC tumorigenesis by downregulating Mul1, an E3 ubiquitin ligase that associates with mitochondria and plays a critical role in the regulation of mitochondrial dynamics [90]. Specifically, it was identified that Mul1 expression promotes autophagic flux and the degradation of p62-associated protein complexes [90]. Previously, it had been shown that p62-mediated autophagy was required for HIF-2α degradation as part of a constitutive process to compensate for ubiquitin-mediated proteasomal degradation [91]; however, ccRCC has been demonstrated to have deletion of or loss-of-function mutations to autophagy-related gene ATG7, and when this couples with VHL deletion, the two major mechanisms responsible for degrading HIF-2α become futile, further supportingunregulated signaling through this pathway [91]. Moving forward, it would be interesting to see if HIF signaling has a direct effect on the fission and fusion mechanisms and/or machinery and how they are altered in response to changes in HIF expression or oxygen levels. Future studies should also investigate how altering proteins that are essential to the structure and function of mitochondria, such as the mitofusins and OPA1 [92], impacts HIF signaling and whether such proteins could be potential therapeutic targets in ccRCC. However, in ccRCC there is an overall lack of literature regarding mitochondrial dynamics and the influence on tumorigenesis. Thus, it is imperative to begin building a fundamental understanding of the requirement for these proteins by applying genetic or pharmacologic manipulations and performing functional assays such as Seahorse metabolic assays, flow cytometry to analyze mitochondrial potential and mitochondrial turnover, and electron microscopy to capture any structural changes to the mitochondria.
Amino Acid Metabolism
Glutaminolysis, Aspartate, and Arginine
In the earlier referenced 2019 study by Courtney et al. where ccRCC patients received intraoperative infusions of heavy isotope glucose or acetate, one of the major conclusions that the authors made was that there was reduced TCA cycle turnover of carbons sourced from glucose or acetate [34]. This finding could suggest that another carbon source is being utilized to fuel the TCA cycle and in ccRCC it is well documented that glutaminolysis and reductive carboxylation significantly contribute to the TCA cycle [39, 40]. In support of this, several studies have confirmed elevated expression utilizing sequencing and histological approaches for the glutamine uptake transporter solute carrier family 1 member 5 (SLC1A5) and found that high expression correlates with reduced survival in ccRCC [1, 93]. Given the metabolic importance of glutamine metabolism in ccRCC, it would be reasonable to speculate that genetic deletion of SLC1A5 would be catastrophic to these cells; however, the authors show that while SLC1A5 deletion starves ccRCCs of amino acids, the cells compensate by upregulating the neutral amino acid transporters SLC38A1 and SLC38A2 which can sustain the import of glutamine, as well as mTOR signaling via exchanging substrates with LAT1 [94]. The link between glutamine transporters and hypoxia has been elegantly documented in other cancer models, where it has been shown that the lactate produced from hypoxia-fueled glycolysis is eventually metabolized back into pyruvate via glycolytic oxidation and this lactate-derived pyruvate functions to stabilize HIF-1α and HIF-2α by inhibiting PHDs [95]. Once stabilized, HIF-2α can then promote c-Myc transcriptional activity, which includes SLC1A5 and glutaminase (GLS), the first enzyme in glutamine oxidation [95]. Curiously, the expression of glutaminase enzymes in ccRCC appear relatively unchanged when compared to normal cells or adjacent tissue [39, 40, 96, 97]. As seen previously, mRNA and protein expression levels can be deceiving and do not always translate into proportionate changes in activity [38, 75]. In 2017, Aboud et al. performed a functional analysis of glutamine uptake and metabolism [98]. Here, the authors first treated 786-O and SN-12 cells with the GLS inhibitors CB-839 or BPTES and found that these cells experienced reduced survival accompanied by elevated apoptotic activity associated with a decrease in the GSH/GSSG rato in response to increased levels of 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG), a marker for DNA damage [98]. Utilizing PET imaging in an orthotopic ccRCC tumor model, the researchers further demonstrate that tumors show enhanced uptake of 18F-(2S,4R)4- fluoroglutamine (18F-FGln) compared to normal kidney [98]. The authors of this study suggest these findings highlight a potential role for implementing glutamine metabolism in diagnostic applications and could even provide insight into patient populations that might respond to glutaminase inhibitors that are currently under testing in clinical trials [98-102].
A recent study in a breast cancer model evaluated the impact of hypoxia on the glutamine oxidation and incorporation of glutamine-derived carbon and nitrogen into downstream metabolites and found that hypoxia stimulated glutamine-derived nitrogen was used to synthesize dihydroorotate and incorporated into nucleotides to fuel tumor growth; on the other hand, glutamine-derived carbon followed a more well-understood path, entering reductive carboxylation and being incorporated into acetyl-CoA [103]. Interestingly, the authors also found that glutamine-sourced nitrogen was not used for the synthesis of arginine, asparagine, or aspartate under hypoxia [103]. Recent work from the lab of Julian Aragones was able to recapitulate these findings in VHL-deficient ccRCC cell line models, where they performed isotopic glutamine flux tracing and observed reduced aspartate biosynthesis which resulted from HIF-1α dependent downregulation of glutamic-oxaloacetic transaminase enzymes (GOT1 and GOT2) [104]. In this study, the control of aspartate biosynthesis is emphasized as a key mechanism orchestrating the tumor suppressor role of HIF-1α that has been described in ccRCC. By utilizing the VHL-deficient 786-O cell line, which only expresses truncated mRNA transcripts for HIF-1α that do not translate into functional protein, the authors ectopically expressed a constitutively active HIF-1α and observed robust downregulation of the GOT enzymes and subsequent reduction in aspartate synthesis, a phenotype that was reversed via aspartate supplementation [104]. Of clinical relevance, when compared to VHL-deficient ccRCC patient tumor samples, non-clear cell renal cell carcinoma (NCRCC) patient samples with functional VHL displayed enhanced expression of GOT1 and GOT2 [104]. A recent report from the lab of Celeste Simon identified robust suppression of the urea cycle enzymes arginase 2 (ARG2), which function together to hydrolyze arginine into ornithine and urea that eventually gets converted into citrulline, and argininosuccinate synthase (ASS1) which utilizes ARG2-derived citrulline and aspartate from various sources to synthesize argininosuccinate [105]. Mechanistically, the authors demonstrate with isotopic arginine flux tracing that ARG2 function inhibits ccRCC growth by depleting pyridoxal phosphate, the active form of vitamin B6 which acts as an essential cofactor for several enzymatic reactions [105].
Together, these studies highlight an importance of amino acid metabolism in ccRCC and suggest a critical role in the development and progression of these tumors. Considering evidence provided so far, it appears glutaminolysis is more critical for supporting reductive carboxylation rather than synthesis of aspartate and other amino acids [104, 105]. Taken together with what has been found in other cancer models [103], it would be intriguing to evaluate the incorporation of glutamine-derived nitrogen into nucleotides and test the impact that this has on ccRCC growth. Moving forward, it would also be interesting to test the effects of manipulating GOT1 and/or GOT2 activity in a variety of RCC models with pharmacological inhibition to test if this can sensitize these tumors to other treatments, as has been shown in preclinical pancreatic ductal adenocarcinoma (PDAC) models [106].
Branched Chain Amino Acids
Leucine, isoleucine, and valine comprise a group of essential amino acids known as the branched-chain amino acids (BCAAs), which are characterized as having branching aliphatic side chains that are nonpolar and hydrophobic [107]. BCAAs primarily function in anabolic metabolism, serving as the building blocks for proteins within cells [107]. In the clinic, cancer patients undergoing aggressive therapies and suffering from advanced disease are often supplied dietary BCAAs in an effort to combat cachexia; however, at the molecular level branched-chain amino acids fuel mTOR signaling and anabolic protein and lipid synthesis pathways in cancer cells, supporting tumor growth and survival [108-110]. Recently, a bioinformatic analysis was applied to data extracted from the TCGA focusing on metabolism-associated genes in ccRCC [111]. Algorithmic models determined differentially expressed genes across various stages of ccRCC; these data were then subjected to enrichment analysis which narrowed the focus on 22 metabolic pathways that were dysregulated dependent on tumor stage [111]. From these analyses, it was determined that amino acid metabolism, particularly BCAA metabolism, was significantly altered in stage IV disease [111]. Expanding on these findings, another group performed metabolomic analysis on the conditioned media from HEK293, 786-O (VHL−/−), and Caki-1 (VHL+/+), as well as serum samples from healthy individuals and ccRCC patients hypothesizing that they would find distinct metabolites associated with cancer, as well as with specific genetic drivers [112]. In the analysis, the authors performed four types of comparisons: HEK293 vs 786-O, HEK293 vs Caki-1, Caki-1 vs 786-O, and healthy vs ccRCC patient serum and observed significant differences in BCAAs and their metabolic byproducts in all of these comparisons [112]. While in the cell line comparisons these metabolites were mostly elevated, ccRCC patient serum had significantly less leucine and isoleucine than in the healthy donor samples; however, the researchers did observe a significant increase in N-lactoyl-leucine, a pseudopeptide formed by combining lactate with leucine [113]. This finding was shown to associate with HIF signaling, where the conditioned media from the 786-O cells accumulated more of this metabolite, extrapolating a novel role for HIF-mediated lactate production in the regulation of BCAA metabolism and peptide synthesis [112]. Studies like these will be important to expand upon, with the intention of better understanding the functional implications of the BCAAs in ccRCC.
Conclusion
VHL deletion is a defining feature of ccRCC, and the downstream signaling ramifications and unique metabolic framework are heavily influenced by the resulting pseudohypoxic state. The contribution of HIF-fueled glucose uptake and glycolysis in ccRCC cannot be overstated; however, it is often overlooked how hypoxia signaling can coordinate glucose flux into other key pathways such as the pentose phosphate pathway or utilize alternative bioenergetic substrates altogether. In this review, we have focused on less frequently discussed metabolic pathways and summarized recent discoveries that are directly linked to hypoxia signaling and/or that highlight the importance of these pathways in supporting ccRCC tumorigenesis.
To date, much of the molecular and mechanistic work has focused narrowly on the cancer cells of ccRCC tumors; however, Ricketts et al. concluded in a recent analysis of the TCGA that ccRCC tumors are abundantly heterogeneous and show a significant infiltration of immune cells [114]. Considering the complexity of these tumors, it would progress our understanding of the disease to take a more holistic approach, expanding on the infusion studies described by Courtney et al. and analyzing the metabolic contribution from various cell types [34], in addition to considering the complexity of involved metabolic pathways. In the examples that we highlight above, it is demonstrated that HIF-driven ccRCCs are reliant to the point of addiction on glutamine to feed reductive carboxylation for the production of biosynthetic precursors such as acetyl-CoA. As it relates to cell type, it has been shown that glutaminase inhibition with the small molecule CB-839, which was recently incorporated into clinical trials treating ccRCC patients, has differential effects resulting in reduced viability in cancer cells and enhanced differentiation and effector function in T cells [115].
In addition to tumor heterogeneity, the genetic drivers of these tumors contribute globally and locally to the make-up and behavior of a tumor. Findings such as those where enhanced oxidative phosphorylation was observed in 786-O cells with a secondary deletion of SETD2 emphasize a need for a deeper understanding of the metabolic environment driven by VHL deletion and how the pseudohypoxic state orchestrates subsequent metabolic adaptations to acquired secondary mutations [77]. Metabolite analysis will undoubtedly provide the field with clarity regarding the pathways that are present as metabolic biomarkers, and those that provide opportunities for synthetic engagement, with the ultimate goal to interfere with key vulnerabilities that are created as a result of metabolic shifts. Given the lipid-rich nature of ccRCC and supporting literature that suggests hypoxia regulates the saturation status of lipids [44, 116], a future frontier for kidney tumor metabolism exists in extracting lipid-omics from tumors of various genetic backgrounds, providing an in-depth view of lipid content, characteristics, and the role of pseudohypoxia in comprehensively mediating the “clear cell” phenotype. Lastly, implementing isotopic tracing with various bioenergetic substrates in tumors with genetic diversity and analyzing the contribution of these substrates into lipid stores could pinpoint the metabolic preferences that specific genetic drivers utilize to support the overall phenotype.
Highlights.
Hypoxia orchestrates complex and dynamic metabolic programs to support ccRCC.
Hypoxia influences the dynamics of post-translational modifications to alter metabolism.
HIF signaling coordinates the utilization of oxidative phosphorylation in ccRCC with secondary mutations.
Defective autophagy supports ccRCC and HIF signaling by altering mitochondrial dynamics and HIF degradation.
HIF-driven ccRCC are dependent on glutamine and amino acid metabolism.
Acknowledgments
The authors give special thanks to their funding sources. WKR has received financial support from the AACR and institutional research funding from Bristol-Myers Squibb, Merck, Pfizer, Calithera Biosciences, Peloton, and Incyte. ZAB is supported by the Integrated Biological Systems Training in Oncology Ruth L. Kirschstein NRSA training grant (5T32CA119925-12).
Abbreviations
- 18F-FGln
18F-(2S,4R)4- fluoroglutamine
- 8-oxodG
8-oxo-7,8-dihydro-2′-deoxyguanosine
- ARG2
arginase 2
- ASS1
arginosuccinate synthase 1
- BCAA
branched chain amino acids
- ccRCC
clear cell renal cell carcinoma
- DCA
dichloroacetate
- Drp-1
dynamin-related protein 1
- FIH
factor inhibiting HIF
- G6PD
glucose-6-phosphate dehydrogenase
- GLS
glutaminase
- GLUT1
glucose transporter 1
- GOT
glutamic oxaloacetic transferase
- HGSOC
high grade serous ovarian cancer
- HIF
hypoxia inducible factor
- HK2
hexokinase 2
- HNSCC
head and neck squamous cell carcinoma
- HRE
hypoxia response element
- LDHA
lactate dehydrogenase A
- NCRCC
non-clear cell renal cell carcinoma
- OGT
O-GlcNAc transferase
- OXPHOS
oxidative phosphorylation
- PDAC
pancreatic ductal adenocarcinoma
- PDK
pyruvate dehydrogenase kinase
- PDH
pyruvate dehydrogenase
- PHD
prolyl hydroxylase
- PPP
pentose phosphate pathway
- PRPP
phosphoribosyl pyrophosphate
- R5P
ribose-5-phosphate
- ROS
reactive oxygen species
- RPI
ribose-5-phosphate isomerase
- SLC1A5
solute carrier family 1 member 5
- TKI
tyrosine kinase inhibitors
- TKT
transketolase
- TKTL1
transketolase-like protein 1
- VHL
von Hippel-Lindau
Footnotes
Conflicts of Interest
ZAB has no conflicts of interest. WKR reports the following conflicts: institutional research funding from Calithera, Peloton, Merck, Pfizer, BMS, Incyte to support clinical trials, and research funding in support of a laboratory project from Incyte.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.The Cancer Genome Atlas Research, N., et al. , Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature, 2013. 499: p. 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noone AM, et al. , Cancer Incidence and Survival Trends by Subtype Using Data from the Surveillance Epidemiology and End Results Program, 1992-2013. Cancer Epidemiol Biomarkers Prev, 2017. 26(4): p. 632–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkins MB and Tannir NM, Current and emerging therapies for first-line treatment of metastatic clear cell renal cell carcinoma. Cancer Treat Rev, 2018. 70: p. 127–137. [DOI] [PubMed] [Google Scholar]
- 4.Muglia VF and Prando A, Renal cell carcinoma: histological classification and correlation with imaging findings. Radiologia brasileira, 2015. 48(3): p. 166–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jonasch E, Gao J, and Rathmell WK, Renal cell carcinoma. The BMJ, 2014. 349: p. g4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wettersten HI, et al. , Metabolic reprogramming in clear cell renal cell carcinoma. Nature Reviews Nephrology, 2017. 13: p. 410. [DOI] [PubMed] [Google Scholar]
- 7.Semenza GL and Wang GL, A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Molecular and Cellular Biology, 1992. 12(12): p. 5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang GL, et al. , Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proceedings of the National Academy of Sciences, 1995. 92(12): p. 5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schofield CJ and Ratcliffe PJ, Oxygen sensing by HIF hydroxylases. Nature Reviews Molecular Cell Biology, 2004. 5(5): p. 343–354. [DOI] [PubMed] [Google Scholar]
- 10.Appelhoff RJ, et al. , Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. Journal of Biological Chemistry, 2004. 279(37): p. 38458–38465. [DOI] [PubMed] [Google Scholar]
- 11.Rathmell WK and Chen S, VHL Inactivation in Renal Cell Carcinoma: Implications for Diagnosis, Prognosis, and Treatment. Expert review of anticancer therapy, 2008. 8(1): p. 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore LE, et al. , Von Hippel-Lindau (VHL) inactivation in sporadic clear cell renal cancer: associations with germline VHL polymorphisms and etiologic risk factors. PLoS genetics, 2011. 7(10): p. e1002312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hewitson KS, et al. , Hypoxia-inducible Factor (HIF) Asparagine Hydroxylase Is Identical to Factor Inhibiting HIF (FIH) and Is Related to the Cupin Structural Family. Journal of Biological Chemistry, 2002. 277(29): p. 26351–26355. [DOI] [PubMed] [Google Scholar]
- 14.Lando D, et al. , FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes & development, 2002. 16(12): p. 1466–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Semenza GL, et al. , Structural and functional analysis of hypoxia-inducible factor 1. Kidney International, 1997. 51(2): p. 553–555. [DOI] [PubMed] [Google Scholar]
- 16.Luo JC and Shibuya M, A variant of nuclear localization signal of bipartite-type is required for the nuclear translocation of hypoxia inducible factors (1α, 2α and 3α). Oncogene, 2001. 20(12): p. 1435–1444. [DOI] [PubMed] [Google Scholar]
- 17.Freedman SJ, et al. , Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1α. Proceedings of the National Academy of Sciences, 2002. 99(8): p. 5367–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh JJ, et al. , Renal cell carcinoma. Nature reviews Disease primers, 2017. 3: p. 17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordan JD, et al. , HIF-α Effects on c-Myc Distinguish Two Subtypes of Sporadic VHL-Deficient Clear Cell Renal Carcinoma. Cancer Cell, 2008. 14(6): p. 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordan JD, Thompson CB, and Simon MC, HIF and c-Myc: Sibling Rivals for Control of Cancer Cell Metabolism and Proliferation. Cancer Cell, 2007. 12(2): p. 108–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shinojima T, et al. , Renal cancer cells lacking hypoxia inducible factor (HIF)-1α expression maintain vascular endothelial growth factor expression through HIF-2α. Carcinogenesis, 2007. 28(3): p. 529–536. [DOI] [PubMed] [Google Scholar]
- 22.Khan MN, et al. , Factor inhibiting HIF (FIH-1) promotes renal cancer cell survival by protecting cells from HIF-1α-mediated apoptosis. British Journal of Cancer, 2011. 104(7): p. 1151–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen C, et al. , Genetic and functional studies implicate HIF1α as a 14q kidney cancer suppressor gene. Cancer discovery, 2011. 1(3): p. 222–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raval RR, et al. , Contrasting Properties of Hypoxia-Inducible Factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-Associated Renal Cell Carcinoma. Molecular and Cellular Biology, 2005. 25(13): p. 5675–5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meléndez-Rodríguez F, et al. , Hypoxia-Inducible Factor 2-Dependent Pathways Driving Von Hippel–Lindau-Deficient Renal Cancer. Frontiers in Oncology, 2018. 8(214). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu C-J, et al. , Differential roles of hypoxia-inducible factor 1α (HIF-1α) and HIF-2α in hypoxic gene regulation. Molecular and cellular biology, 2003. 23(24): p. 9361–9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu C-J, et al. , Differential regulation of the transcriptional activities of hypoxia-inducible factor 1 alpha (HIF-1alpha) and HIF-2alpha in stem cells. Molecular and cellular biology, 2006. 26(9): p. 3514–3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Covello KL, Simon MC, and Keith B, Targeted Replacement of Hypoxia-Inducible Factor-1α by a Hypoxia-Inducible Factor-2α Knock-in Allele Promotes Tumor Growth. Cancer Research, 2005. 65(6): p. 2277–2286. [DOI] [PubMed] [Google Scholar]
- 29.Gordan JD and Simon MC, Hypoxia-inducible factors: central regulators of the tumor phenotype. Current opinion in genetics & development, 2007. 17(1): p. 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee CM, et al. , VHL Type 2B gene mutation moderates HIF dosage in vitro and in vivo. Oncogene, 2009. 28(14): p. 1694–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanahan D and Weinberg RA, The hallmarks of cancer. cell, 2000. 100(1): p. 57–70. [DOI] [PubMed] [Google Scholar]
- 32.Hanahan D and Weinberg RA, Hallmarks of cancer: the next generation. cell, 2011. 144(5): p. 646–674. [DOI] [PubMed] [Google Scholar]
- 33.Rathmell WK, Rathmell JC, and Linehan WM, Metabolic pathways in kidney cancer: Current therapies and future directions. Journal of Clinical Oncology, 2018. 36(36): p. 3540–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Courtney KD, et al. , Isotope tracing of human clear cell renal cell carcinomas demonstrates suppressed glucose oxidation in vivo. Cell metabolism, 2018. 28(5): p. 793–800. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J.-w., et al. , HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell metabolism, 2006. 3(3): p. 177–185. [DOI] [PubMed] [Google Scholar]
- 36.Phillips R, Kidney cancer: FBP1 depletion feeds ccRCC. Nature Reviews Urology, 2014. 11(9): p. 482. [DOI] [PubMed] [Google Scholar]
- 37.Shuch B, Linehan WM, and Srinivasan R, Aerobic glycolysis: a novel target in kidney cancer. Expert review of anticancer therapy, 2013. 13(6): p. 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hakimi AA, et al. , An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer cell, 2016. 29(1): p. 104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metallo CM, et al. , Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature, 2012. 481(7381): p. 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gameiro PA, et al. , In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell metabolism, 2013. 17(3): p. 372–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie H and Simon MC, Oxygen availability and metabolic reprogramming in cancer. Journal of Biological Chemistry, 2017. 292(41): p. 16825–16832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du W, et al. , HIF drives lipid deposition and cancer in ccRCC via repression of fatty acid metabolism. Nature communications, 2017. 8(1): p. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J, et al. , Uptake of HDL-cholesterol contributes to lipid accumulation in clear cell renal cell carcinoma. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids, 2019. 1864(12): p. 158525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ackerman D, et al. , Triglycerides promote lipid homeostasis during hypoxic stress by balancing fatty acid saturation. Cell reports, 2018. 24(10): p. 2596–2605. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li B, et al. , Fructose-1, 6-bisphosphatase opposes renal carcinoma progression. Nature, 2014. 513(7517): p. 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miess H, et al. , The glutathione redox system is essential to prevent ferroptosis caused by impaired lipid metabolism in clear cell renal cell carcinoma. Oncogene, 2018. 37(40): p. 5435–5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siska PJ, et al. , Mitochondrial dysregulation and glycolytic insufficiency functionally impair CD8 T cells infiltrating human renal cell carcinoma. JCI Insight, 2017. 2(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rao X, et al. , O-GlcNAcylation of G6PD promotes the pentose phosphate pathway and tumor growth. Nature Communications, 2015. 6(1): p. 8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Q, et al. , Overexpression of G6PD represents a potential prognostic factor in clear cell renal cell carcinoma. Journal of Cancer, 2017. 8(4): p. 665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Q, et al. , G6PD promotes renal cell carcinoma proliferation through positive feedback regulation of p-STAT3. Oncotarget, 2017. 8(65): p. 109043–109060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gregg JL, et al. , NADPH oxidase NOX4 supports renal tumorigenesis by promoting the expression and nuclear accumulation of HIF2α. Cancer research, 2014. 74(13): p. 3501–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferrer CM, et al. , O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Molecular cell, 2014. 54(5): p. 820–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bacigalupa ZA, Bhadiadra CH, and Reginato MJ, O-GlcNAcylation: key regulator of glycolytic pathways. Journal of Bioenergetics and Biomembranes, 2018. 50(3): p. 189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Starska K, et al. , Gene and protein expression of O-GlcNAc-cycling enzymes in human laryngeal cancer. Clinical and experimental medicine, 2015. 15(4): p. 455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu D, et al. , Increased expression of O-GlcNAc transferase (OGT) is a biomarker for poor prognosis and allows tumorigenesis and invasion in colon cancer. International Journal of Clinical and Experimental Pathology, 2019. 12(4): p. 1305. [PMC free article] [PubMed] [Google Scholar]
- 56.Gloster TM, et al. , Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nature chemical biology, 2011. 7(3): p. 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y, et al. , APC/CCDH1 synchronizes ribose-5-phosphate levels and DNA synthesis to cell cycle progression. Nature Communications, 2019. 10(1): p. 2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bentz S, et al. , Hypoxia Induces the Expression of Transketolase-Like 1 in Human Colorectal Cancer. Digestion, 2013. 88(3): p. 182–192. [DOI] [PubMed] [Google Scholar]
- 59.Lum JJ, et al. , The transcription factor HIF-1α plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes & development, 2007. 21(9): p. 1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Macbeth RA and Bekesi JG, Oxygen consumption and anaerobic glycolysis of human malignant and normal tissue. Cancer research, 1962. 22(2): p. 244–248. [PubMed] [Google Scholar]
- 61.Koppenol WH, Bounds PL, and Dang CV, Otto Warburg's contributions to current concepts of cancer metabolism. Nature Reviews Cancer, 2011. 11(5): p. 325–337. [DOI] [PubMed] [Google Scholar]
- 62.Vander Heiden MG, Cantley LC, and Thompson CB, Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science, 2009. 324(5930): p. 1029–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Warburg O, On the origin of cancer cells. Science, 1956. 123(3191): p. 309–314. [DOI] [PubMed] [Google Scholar]
- 64.Linehan WM, Rubin JS, and Bottaro DP, VHL loss of function and its impact on oncogenic signaling networks in clear cell renal cell carcinoma. The international journal of biochemistry & cell biology, 2009. 41(4): p. 753–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mandriota SJ, et al. , HIF activation identifies early lesions in VHL kidneys: evidence for site-specific tumor suppressor function in the nephron. Cancer cell, 2002. 1(5): p. 459–468. [DOI] [PubMed] [Google Scholar]
- 66.Kondo K and Kaelin WG Jr, The von Hippel–Lindau tumor suppressor gene. Experimental cell research, 2001. 264(1): p. 117–125. [DOI] [PubMed] [Google Scholar]
- 67.Kondo K, et al. , Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer cell, 2002. 1(3): p. 237–246. [DOI] [PubMed] [Google Scholar]
- 68.Samanta D and Semenza GL, Metabolic adaptation of cancer and immune cells mediated by hypoxia-inducible factors. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer, 2018. 1870(1): p. 15–22. [DOI] [PubMed] [Google Scholar]
- 69.Scott BA, Avidan MS, and Crowder CM, Regulation of hypoxic death in C. elegans by the insulin/IGF receptor homolog DAF-2. Science, 2002. 296(5577): p. 2388–2391. [DOI] [PubMed] [Google Scholar]
- 70.Simon MC, Coming up for air: HIF-1 and mitochondrial oxygen consumption. Cell metabolism, 2006. 3(3): p. 150–151. [DOI] [PubMed] [Google Scholar]
- 71.Kinnaird A, et al. , Metabolic modulation of clear-cell renal cell carcinoma with dichloroacetate, an inhibitor of pyruvate dehydrogenase kinase. European urology, 2016. 69(4): p. 734–744. [DOI] [PubMed] [Google Scholar]
- 72.Kato M, et al. , Distinct structural mechanisms for inhibition of pyruvate dehydrogenase kinase isoforms by AZD7545, dichloroacetate, and radicicol. Structure, 2007. 15(8): p. 992–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whitehouse S, Cooper RH, and Randle PJ, Mechanism of activation of pyruvate dehydrogenase by dichloroacetate and other halogenated carboxylic acids. Biochemical Journal, 1974. 141(3): p. 761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiao Z, Dai Z, and Locasale JW, Metabolic landscape of the tumor microenvironment at single cell resolution. Nature Communications, 2019. 10(1): p. 3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Clark DJ, et al. , Integrated Proteogenomic Characterization of Clear Cell Renal Cell Carcinoma. Cell, 2019. 179(4): p. 964–983.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gentric G, et al. , PML-Regulated Mitochondrial Metabolism Enhances Chemosensitivity in Human Ovarian Cancers. Cell metabolism, 2019. 29(1): p. 156–173.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu J, et al. , Loss of SETD2 Induces a Metabolic Switch in Renal Cell Carcinoma Cell Lines toward Enhanced Oxidative Phosphorylation. Journal of Proteome Research, 2019. 18(1): p. 331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.LaGory EL, et al. , Suppression of PGC-1a Is Critical for Reprogramming Oxidative Metabolism in Renal Cell Carcinoma. Cell Reports, 2015. 12(4): p. 717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tilokani L, et al. , Mitochondrial dynamics: overview of molecular mechanisms. Essays in Biochemistry, 2018. 62(3): p. 341–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sebastián D, Palacín M, and Zorzano A, Mitochondrial Dynamics: Coupling Mitochondrial Fitness with Healthy Aging. Trends in Molecular Medicine, 2017. 23(3): p. 201–215. [DOI] [PubMed] [Google Scholar]
- 81.Eisner V, Picard M, and Hajnóczky G, Mitochondrial dynamics in adaptive and maladaptive cellular stress responses. Nature Cell Biology, 2018. 20(7): p. 755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chan DC, Mitochondria: dynamic organelles in disease, aging, and development. Cell, 2006. 125(7): p. 1241–1252. [DOI] [PubMed] [Google Scholar]
- 83.Dai W and Jiang L, Dysregulated Mitochondrial Dynamics and Metabolism in Obesity, Diabetes, and Cancer. Frontiers in endocrinology, 2019. 10: p. 570–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pereira R, et al. , Resistance to Diet-Induced Obesity in Mice Lacking OPA1 in Adipose Tissue Occurs Independently of Fat-Derived FGF-21 and BAT Function. Diabetes, 2018. 67(Supplement 1): p. 276–LB. [Google Scholar]
- 85.Westermeier F, et al. , Defective insulin signaling and mitochondrial dynamics in diabetic cardiomyopathy. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, 2015. 1853(5): p. 1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shuch B, et al. , Defining early-onset kidney cancer: implications for germline and somatic mutation testing and clinical management. Journal of Clinical Oncology, 2014. 32(5): p. 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang D, et al. , Increased mitochondrial fission is critical for hypoxia-induced pancreatic beta cell death. PLOS ONE, 2018. 13(5): p. e0197266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Da Yeon K, et al. , Hypoxia-dependent mitochondrial fission regulates endothelial progenitor cell migration, invasion, and tube formation. The Korean Journal of Physiology & Pharmacology, 2018. 22(2): p. 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Labuschagne CF, et al. , Cell clustering promotes a metabolic switch that supports metastatic colonization. Cell metabolism, 2019. 30(4): p. 720–734. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yuan Y, et al. , Mitochondrial E3 ubiquitin ligase 1 promotes autophagy flux to suppress the development of clear cell renal cell carcinomas. Cancer science, 2019. 110(11): p. 3533–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu XD, et al. , Autophagy mediates HIF2α degradation and suppresses renal tumorigenesis. Oncogene, 2015. 34(19): p. 2450–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Detmer SA and Chan DC, Functions and dysfunctions of mitochondrial dynamics. Nature Reviews Molecular Cell Biology, 2007. 8(11): p. 870–879. [DOI] [PubMed] [Google Scholar]
- 93.Liu Y, et al. , High expression of Solute Carrier Family 1, member 5 (SLC1A5) is associated with poor prognosis in clear-cell renal cell carcinoma. Scientific Reports, 2015. 5(1): p. 16954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bröer A, Rahimi F, and Bröer S, Deletion of Amino Acid Transporter ASCT2 (SLC1A5) Reveals an Essential Role for Transporters SNAT1 (SLC38A1) and SNAT2 (SLC38A2) to Sustain Glutaminolysis in Cancer Cells. Journal of Biological Chemistry, 2016. 291(25): p. 13194–13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pérez-Escuredo J, et al. , Lactate promotes glutamine uptake and metabolism in oxidative cancer cells. Cell cycle, 2016. 15(1): p. 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hoerner CR, Chen VJ, and Fan AC, The ‘Achilles Heel’ of metabolism in renal cell carcinoma: glutaminase inhibition as a rational treatment strategy. Kidney Cancer, 2019. 3(1): p. 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Okazaki A, et al. , Glutaminase and poly (ADP-ribose) polymerase inhibitors suppress pyrimidine synthesis and VHL-deficient renal cancers. The Journal of clinical investigation, 2017. 127(5): p. 1631–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Abu Aboud O, et al. , Glutamine Addiction in Kidney Cancer Suppresses Oxidative Stress and Can Be Exploited for Real-Time Imaging. Cancer research, 2017. 77(23): p. 6746–6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Emberley E, et al. CB-839, a selective glutaminase inhibitor, has anti-tumor activity in renal cell carcinoma and synergizes with cabozantinib and everolimus. in Keystone Symposia, Tumor Metabolism: Mechanisms and Targets, Whistler Canada 2017. [Google Scholar]
- 100.Meric-Bernstam F, et al. , Phase 1 study of CB-839, a small molecule inhibitor of glutaminase (GLS), alone and in combination with everolimus (E) in patients (pts) with renal cell cancer (RCC). 2016, American Society of Clinical Oncology. [Google Scholar]
- 101.Tannir NM, et al. , CANTATA: A randomized phase 2 study of CB-839 in combination with cabozantinib vs. placebo with cabozantinib in patients with advanced/metastatic renal cell carcinoma. 2018, American Society of Clinical Oncology. [Google Scholar]
- 102.Emberley E, et al. , CB-839, a selective glutaminase inhibitor, has anti-tumor activity in renal cell carcinoma and synergizes with everolimus and receptor tyrosine kinase inhibitors. Eur. J. Cancer, 2016. 69: p. S124. [Google Scholar]
- 103.Wang Y, et al. , Coordinative metabolism of glutamine carbon and nitrogen in proliferating cancer cells under hypoxia. Nature Communications, 2019. 10(1): p. 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Meléndez-Rodríguez F, et al. , HIF1α Suppresses Tumor Cell Proliferation through Inhibition of Aspartate Biosynthesis. Cell Reports, 2019. 26(9): p. 2257–2265.e4. [DOI] [PubMed] [Google Scholar]
- 105.Ochocki JD, et al. , Arginase 2 suppresses renal carcinoma progression via biosynthetic cofactor pyridoxal phosphate depletion and increased polyamine toxicity. Cell metabolism, 2018. 27(6): p. 1263–1280. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nelson BS, et al. , Tissue of origin dictates GOT1 dependence and confers synthetic lethality to radiotherapy. Cancer & Metabolism, 2020. 8(1): p. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brosnan JT and Brosnan ME, Branched-Chain Amino Acids: Enzyme and Substrate Regulation. The Journal of Nutrition, 2006. 136(1): p. 207S–211S. [DOI] [PubMed] [Google Scholar]
- 108.O'Connell TM, The complex role of branched chain amino acids in diabetes and cancer. Metabolites, 2013. 3(4): p. 931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ericksen RE, et al. , Loss of BCAA Catabolism during Carcinogenesis Enhances mTORC1 Activity and Promotes Tumor Development and Progression. Cell Metabolism, 2019. 29(5): p. 1151–1165.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee JH, et al. , Branched-chain amino acids sustain pancreatic cancer growth by regulating lipid metabolism. Experimental & Molecular Medicine, 2019. 51(11): p. 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li H-J, et al. , Identification of metabolism-associated genes and pathways involved in different stages of clear cell renal cell carcinoma. Oncology letters, 2018. 15(2): p. 2316–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Knott ME, et al. , Metabolic Footprinting of a Clear Cell Renal Cell Carcinoma in Vitro Model for Human Kidney Cancer Detection. Journal of Proteome Research, 2018. 17(11): p. 3877–3888. [DOI] [PubMed] [Google Scholar]
- 113.Jansen RS, et al. , N-lactoyl-amino acids are ubiquitous metabolites that originate from CNDP2-mediated reverse proteolysis of lactate and amino acids. Proceedings of the National Academy of Sciences of the United States of America, 2015. 112(21): p. 6601–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ricketts CJ, et al. , The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma. Cell Rep, 2018. 23(1): p. 313–326 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Johnson MO, et al. , Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell, 2018. 175(7): p. 1780–1795. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yang H, et al. , SREBP1-driven lipid desaturation supports clear cell renal cell carcinoma growth through regulation of NF-κB signaling. Biochemical and Biophysical Research Communications, 2018. 495(1): p. 1383–1388. [DOI] [PubMed] [Google Scholar]