Abstract
Heat shock protein 70 (Hsp70) is an important molecular chaperone that regulates oncoprotein stability and tumorigenesis. However, attempts to develop anti-chaperone drugs targeting molecules such as Hsp70 have been hampered by toxicity issues. Hsp70 is regulated by a suite of co-chaperone molecules that bring “clients” to the primary chaperone for efficient folding. Rather than targeting Hsp70 itself, here we have examined the feasibility of inhibiting the Hsp70 co-chaperone DNAJA1 as a novel anticancer strategy. We found DNAJA1 to be upregulated in a variety of cancers, suggesting a role in malignancy. To confirm this role, we screened the NIH Approved Oncology collection for chemical-genetic interactions with loss of DNAJA1 in cancer. 41 compounds showed strong synergy with DNAJA1 loss, whereas 18 dramatically lost potency. Several hits were validated using a DNAJA1 inhibitor (116-9e) in castration-resistant prostate cancer cell (CRPC) and spheroid models. Taken together, these results confirm that DNAJA1 is a hub for anticancer drug resistance and that DNAJA1 inhibition is a potent strategy to sensitize cancer cells to current and future therapeutics. The large change in drug efficacy linked to DNAJA1 suggests a personalized medicine approach where tumor DNAJA1 status may be used to optimize therapeutic strategy.
Subject terms: Chaperones, Chemical genetics
Introduction
Hsp70 is a molecular chaperone that plays important roles in protein quality control processes such as protein folding, transport, degradation, and the prevention of protein aggregation1. Hsp70 levels are elevated in various cancers and overexpression correlates with poor prognosis for survival and response to cancer therapy2. The elevated levels of Hsp90 and Hsp70 chaperones in cancer and their role in fostering multiple oncogenic pathways has made these proteins attractive drug targets with numerous anti-chaperone compounds having been developed so far3. Problematically, Hsp70 is required for cell survival and protein homeostasis, and thus its inhibition is detrimental to the viability of both normal and cancer cells, with dubious selectivity for tumor cells4.
Hsp70 performs all its functions in association with a large spectrum of helper proteins known as co-chaperones that include J-proteins, tetratricopeptide repeat (TPR) domain-containing proteins and nucleotide exchange factors (NEFs) which fine-tune Hsp70 specificity and activity in the cell. The J-proteins recruit the protein substrates or clients and interact with such clients at the interface of NBD and SBDβ of Hsp70. This interaction leads to increased Hsp70-mediated ATP turnover and activation of protein folding. J-proteins have a highly conserved 70 amino acid motif containing Histidine, Proline and Aspartic acid amino acid residues known as HPD motif which is essential for stimulating ATPase activity of Hsp705. In humans, the J-protein family has about 50 members which are further divided into three groups based on the localization of J-domain within a protein6. The Hsp40 DNAJA1 (more commonly referred to as DNAJA1) associates with unfolded polypeptide chains, preventing their aggregation6. Several Hsp70 inhibitors have failed in clinical trials due to their toxicity. More recently, alternative strategies have focused on sensitizing cells to anticancer agents by either manipulating post-translational modification of chaperones or their interaction with specific co-chaperones4,7–11.
DNAJA1 (mammalian homolog of yeast Ydj1) is an interesting possible anticancer target as a key mediator of Hsp70 function that appears to regulate specific features of tumorigenesis8,12. A recent study demonstrated that CRPCs expressing ARv7 are insensitive to Hsp90 inhibitors but are sensitive to Hsp40 inhibition13. In addition, we have shown that targeting specific oncoprotein complexes (ribonucleotide reductase) with a combination of traditional as well as a DNAJA1 inhibitor produces highly synergistic effects8. We propose that targeting DNAJA1 in cancer may offer an attractive alternative to the toxicity induced by full Hsp90/Hsp70 inhibition.
Anticancer monotherapies using broadly active cytotoxic or molecularly targeted drugs are limited in their ability to demonstrate a reliable clinical response. This is due to redundant signaling pathways, feedback loops and resistance mechanisms in cancer cells14. Thus, combination anticancer therapies have been used clinically for over 50 years to improve the responses achieved by monotherapies alone. Cancer cell line-based models for these combination therapies are easy and inexpensive to perform using high-throughput drug screening protocols (HTS) to identify the most effective drug combination15,16. HTS helps to explore the relationship between the cell line characteristics and drug specific dose responses15. Chemogenomics is one such HTS-based approach where a large collection of anticancer chemical drugs are screened to identify biological targets. These screening sets often contain small molecules that are well annotated and have defined molecular targets. Such an approach is particularly beneficial for cancer research because malignant cells often contain multiple aberrations that require targeted therapy to inactivate cancer driver activities and mitigate deleterious effects of the drugs to normal cells14.
Here, we performed an unbiased screen of the NIH Approved Oncology Drug set containing 131 anti-cancer drugs in combination with HAP1 cancer cell lines depleted of J-protein DNAJA1. We identified 41 compounds showing strong synergy with the loss of DNAJA1, and in contrast 18 molecules that displayed reduced potency in the knockout cell line. We validated three drugs (cabozantinib, clofarabine and vinblastine) in combination with a unique DNAJA1 inhibitor (116-9e) for synergy in the LNCaP cancer cell lines and confirmed omacetaxine mepesuccinate, idarubicin and sorafenib for antagonism (i.e. with reduced potency after DNAJA1 inhibition). This study demonstrates the validity of developing Hsp70 co-chaperone inhibitors to sensitize cells to current anticancer therapies and suggests that determining DNAJA1 status of a tumor may be beneficial in selecting the most appropriate course of treatment.
Results
DNAJA1 is mutated and overexpressed in a variety of cancers
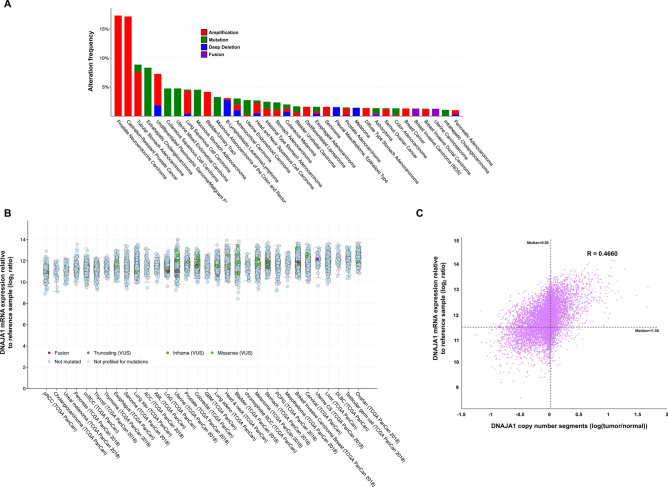
While the roles of Hsp90 and Hsp70 in cancer have been thoroughly studied, much less is known of the role that regulatory co-chaperone proteins such as DNAJA1 play in tumorigenesis. As a first step, we queried the cBioPortal cancer genomic database (cbioportal.org) to determine the incidence of DNAJA1 alterations in cancer. Analysis of data from 176 non-redundant studies representing 44,347 patient samples revealed that DNAJA1 was altered at a frequency of greater than 1% in 35 cancer types (Fig. 1A). Although the majority of alterations in DNAJA1 occur at a relatively low frequency (< 5% of cancers) DNAJA1 is significantly amplified in prostate neuroendocrine cancer (PNC) and castration-resistant prostate cancer at a frequency of 17.31% and 17.14% respectively (Fig. 1A). Hsp70 and Hsp90 are often overexpressed in tumors2. To determine whether the DNAJA1 expression is also overexpressed in cancer, we analyzed DNAJA1 mRNA expression in samples from the TGCA PAN-CAN Atlas. Interestingly, DNAJA1 mRNA was expressed at significantly higher levels in these samples, with a median expression in cancer over 3,000 × relative to WT reference samples (Fig. 1B). To determine if this dramatic overexpression of DNAJA1 was a result of amplification, we plotted DNAJA1 expression vs amplification (Fig. 1C). Interestingly, there was minimal correlation between amount of amplification and DNAJA1 expression (r = 0.45) suggesting that while DNAJA1 may be an important marker in cancer it is not caused by gene amplification.
Figure 1.
DNAJA1 is altered in cancer. (A) Prevalence of DNAJA1 alterations in various cancer genomes analyzed via the cBioPortal. (B) DNAJA1 mRNA expression in cancers (TGCA PanCan) obtained from cBioPortal. mRNA expression value is log2 ratio of expression seen in cancer vs. reference cells (please see www.cbioportal.org/faq for more information). (C) increased DNAJA1 expression is not driven by copy number increase. DNAJA1 copy number vs DNAJA1 expression was plotted and Pearson’s correlation coefficient (R-value) was calculated. Median of both variables is marked by dotted line on the graph.
Characterizing the role of DNAJA1 in anticancer drug resistance
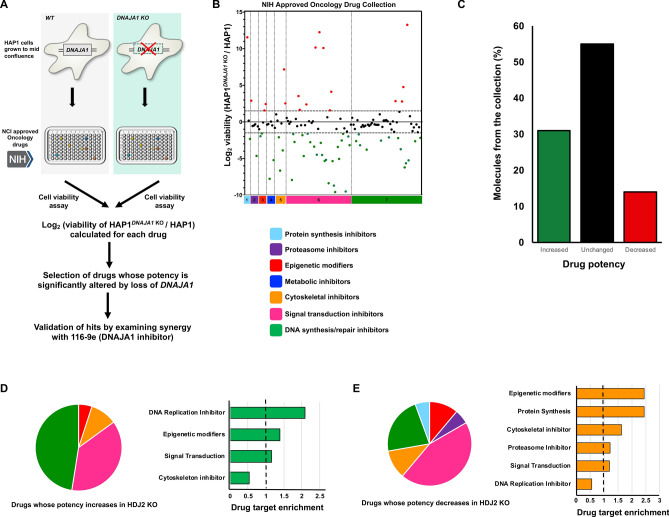
The existing literature is contradictory as to whether DNAJA1 may possess tumor suppressor or driver properties12,17. To clarify whether silencing of DNAJA1 could be beneficial in the treatment of cancer, we screened wildtype HAP1 cells and HAP1 cells lacking DNAJA1 (HAP1DNAJA1 KO) for comparative resistance against the NIH NCI Approved Oncology Collection (Fig. 2A) (https://dtp.cancer.gov/organization/dscb/obtaining/available_plates.html). Prior to screening, we validated the status of the DNAJA1 knockout cell line by Western blotting for DNAJA1 and other major chaperones and co-chaperones (Hsp70, Hsc70, Hsp90, Bag-3 and Hsp110). As expected, we confirmed loss of DNAJA1 and interestingly did not observe any compensatory effects on the levels of the other chaperones/co-chaperones studied (Fig. S1). According to pharmacologic action, the compounds in the library have been divided into seven categories: protein synthesis inhibitors, proteasome inhibitors, epigenetic modifiers, metabolic inhibitors, cytoskeletal inhibitors, signal transduction inhibitors and DNA synthesis/repair inhibitors. Further fold enrichment of each drug category was calculated for the drugs whose potency increased or decreased with loss of DNAJA1. To monitor the screening quality, each screening plate contained control wells treated with vehicle (1% DMSO). The final concentration of the screening compounds was 50 μmol/L. Positive hits (synergistic) or negative hits (antagonistic) were determined by normalizing the log2 ratio of viability of DNAJA1 knockout cells over wildtype cells. A full list of the screening results is shown in Supplementary Table T1 and the sorted data are graphically plotted in Fig. 2B. The effectiveness of a large proportion of anticancer molecules in the collection were impacted, with 41 of (31%) showing increased potency and 18 (14%) showing reduced potency upon loss of DNAJA1 (Fig. 2C). Drug target analysis was carried out by calculating fold enrichment of positive hits (synergistic) or negative hits (antagonistic) over the total number of drugs in that category. Drug target analysis of the synergistic drug hits revealed significant enrichment in DNA synthesis and repair inhibitors, signal transduction inhibitors as well as cytoskeletal inhibitors (Fig. 2D). In contrast, drug target analysis of antagonistic drug hits revealed a higher enrichment in categories such as epigenetic modifiers, protein synthesis inhibitors, cytoskeletal inhibitors and proteasome inhibitors (Fig. 2E). For a full list of drugs in each category and raw data from screen, please see supplemental Table T1.
Figure 2.
Sensitivity of WT and DNAJA1 knockout cells to the NIH Approved Oncology Collection. (A) Workflow of high-throughput cell-based screen. (B) A collection of 132 drugs were screened at 50 μmol/L with Wild-type and DNAJA1 KO cells. Results are the average of at least triplicates and error is SEM. The dotted lines represent a potency change of Log2 > 1.5 or Log2 < − 1.5. The effect of DNAJA1 knockout on drug potency is colored as follows: red (decreased drug potency), green (increased drug potency) or black (no change in drug potency). (C) Summary of effect of DNAJA1 knockout on the potency of the NIH approved oncology collection. (D, E) Drug ontology of synergistic and antagonistic hits based on the pathways affected by the approved oncology drugs in the screen.
Strikingly, a small number of compounds with supposedly related function showed dissimilar alteration of potency upon loss of DNAJA1 function, potentially caused by off-target drug effects (see discussion).
Validation of anticancer drugs significantly altered for potency upon loss of DNAJA1
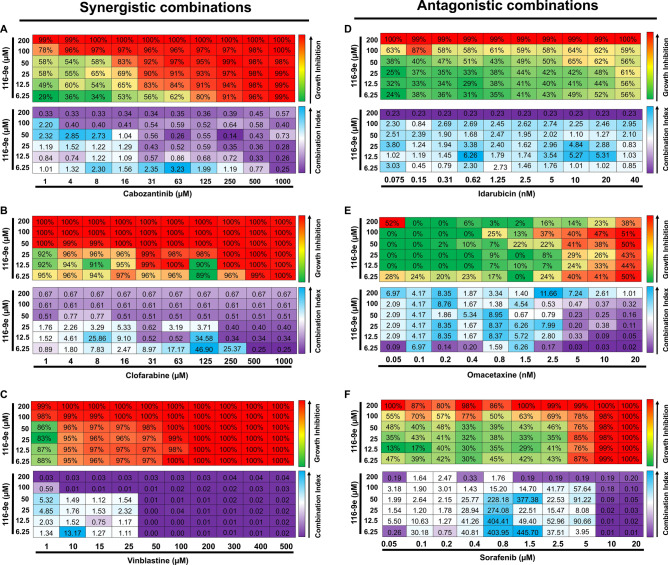
Many anticancer compounds have low potency, poor therapeutic index or suffer from the development of resistance . Monotherapy is rarely efficient and instead drug cocktails are widely used in the clinic16. Establishing these combinations can enhance the scope of preclinical studies and inform the design of future clinical trials. Although knockout of DNAJA1 substantially increased the potency of a number of anticancer molecules, it remained to be determined whether small-molecule inhibition of DNAJA1 could produce a similar result. Our previous bioinformatics analysis indicated that a large proportion of prostate cancer cells contain either amplification or mutation of DNAJA1 (approximately 18%, see Fig. 1). To validate the results of our initial screen, we analyzed the effect of treating prostate cancer cells (LNCaP) with a combination of 116-9e, a small molecule inhibitor of DNAJA118 and selected hits from our screen. We decided to focus on three synergistic drugs discovered in the screen: cabozantinib (receptor tyrosine kinase inhibitor), clofarabine (an RNR inhibitor)19 and vinblastine (microtubule inhibitor/G2 arresting agent)20,21. We also validated three drugs that demonstrated a significant loss of potency in cells lacking DNAJA1: sorafenib (a VEGFR-2 inhibitor)22, omacetaxine mepesuccinate (more commonly known as homoharringtonine, a protein translation inhibitor)23 and idarubicin (topoisomerase II inhibitor)24. To determine synergy in a quantitative manner, we calculated drug synergy (Combination Index values, CI) between 116-9e and either synergistic or antagonistic drugs hits across a broad range of concentrations using the Chou-Talalay method25 (for effects of individual drugs, please see Fig. S2). For three hits identified in our screen (cabozantinib, clofarabine and vinblastine) we confirmed significant synergy (CI < 1) with 116-9e across a range of doses (Fig. 3A–C). In contrast, idarubicin, omacetaxine and sorafenib displayed a significantly antagonistic interaction (CI > 1) across a range of doses (Fig. 3D–F). These data suggest that while DNAJA1 inhibition is a promising strategy to sensitize cells to a number of currently used anticancer drugs, the loss of DNAJA1 can significantly decrease the potency of a small subset of inhibitors.
Figure 3.
Drug interaction between 116-9e (DNAJA1 inhibitor) and selected hits. LNCaP cells were treated with different concentrations of cabozantinib (A), clofarabine (B), vinblastine (C), idarubicin (D), omacetaxine (E) and sorafenib (F) with or without 116-9e for 72 h in RPMI-1640 medium containing 10% FBS. Each point is the mean ± SD for three independent experiments. Growth inhibition was determined using Cell Titer-Glo assay. Combination Index (CI, measure of drug synergy) was determined using Chou-Talalay method via Compusyn software. CI values are as follows: < 0.1 (very strongly synergistic), 0.1–0.3 (strongly synergistic), < 0.9 (synergistic), 0.9–1.1 (additive), 1.1–3.3 (antagonistic), 3.3–10 (strongly antagonistic), > 10 (very strongly antagonistic).
Evaluating the effects of dual targeting of identified drugs with DNAJA1 inhibition on morphology and viability of prostate cancer spheroids
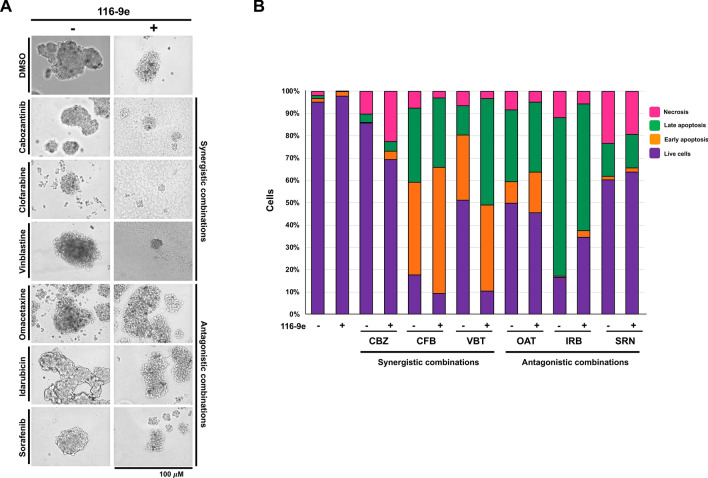
Recent studies have suggested that precision therapy approaches involving the exposure of drugs directly to the primary tumor tissue have the potential to augment the personalized medicine efforts and influence clinical decisions26. Establishing ex vivo three-dimensional (3D) tumor spheroids or organoids derived from primary cancers can be easily established and potentially scaled to screen drug combinations. These 3D cancer models appear to recapitulate features of the tumor of origin in terms of heterogeneity, cell differentiation, histoarchitecture, and clinical drug response and can be used for rapid drug screening27. We therefore next examined the effect of drug combination (three antagonistic and synergistic hits) on LNCaP spheroids. Specifically, changes in spheroid size and shape induced by the 3 antagonistic and synergistic drugs were determined. Visual examination revealed that for the synergistic drugs combination with 116-9e resulted in physical disruption of LNCaP spheroids, resulting in decrease in spheroid size (Fig. 4A). The disruption started on the second day of the treatment. However, when the 3 antagonistic drugs were administered along with 116-9e, there were minimal changes in spheroid morphology indicating that the combination was ineffective.
Figure 4.
Effect of combination treatments on prostate cancer spheroids. (A) Cells were plated on Matrigel-coated 24 well plates. Six drugs (cabozantinib, clofarabine, vinblastine, idarubicin, omacetaxine and sorafenib) were tested on prostate cancer spheroids. These experiments were performed in triplicate and are average of 3 replicates from 3 different wells of a cell culture plate. The pictures are representative images as acquired using an EVOS cell imager. (B) Proliferation of spheroids treated with cabozantinib (CBZ), clofarabine (CFB), vinblastine (VBT), idarubicin (IRB), omacetaxine (OAT) and sorafenib (SRN) measured using AnnexinV/PI staining.
Next, we measured the induction of apoptosis in the spheroids post drug treatments. We determined the kinetics of apoptosis induction using AnnexinV/PI staining. Drug-induced apoptosis was readily detected in the LNCaP spheroids treated with mono and dual drug combinations. In concurrence with the previous results, the combination of the three synergistic drugs with 116-9e displayed enhanced apoptosis as compared to the single drug treatment whereas spheroids treated with the 3 antagonistic drugs showed little or no difference in the rate of apoptosis as compared to the dual drug combination with 116-9e (Fig. 4B).
Discussion
Although inhibitors of Hsp70 and Hsp90 have been developed for research purposes, the conversion of these molecules for use in patient treatment have been hampered by toxicity issues4. We undertook this study to resolve conflicting literature on whether inhibiting DNAJA1, a co-chaperone of Hsp70 may be useful as a novel anticancer strategy. Our bioinformatic analysis of DNAJA1 expression and mutation clearly identify DNAJA1 as being highly altered in a range of cancers, particularly in prostate cancer. Interestingly, DNAJA1 despite being substantially overexpressed in a range of cancers, there was minimal correlation between DNAJA1 copy number and level of expression. While beyond the scope of this study, it is possible that the high levels of DNAJA1 expression observed may be a result of increased transcription brought on hyperactive signaling pathways common in cancer cells. This data in conjunction with a recent finding that Hsp40 is involved in regulation of ARv13 and p5328 makes DNAJA1 inhibition an ideal choice as a novel therapeutic target in Prostate Cancer.
In this study, loss of DNAJA1 increased the potency of a substantial number (31%) of clinically used anticancer drugs. This increased potency may be related to the destabilization of clients that are the target of these small molecules. For example, Hsp70 activates many proteins involved in the DNA damage response and DNA repair pathways (DDR), including ATM, APE1, PARP1, XRCC129. Recently, studies from our group have established roles for both Hsp70 and DNAJA1 in stability of the RNR complex8,29,30. It is unsurprising then that many of the anticancer agents displaying synergy with loss of DNAJA1 are connected to inhibition of the DNA damage response/repair. These include molecules such as 5-fluorouracil (5-FU), premetrexed, clofarabine, olaparib and niraparib etoposide, teniposide and valrubicin. Here we validated synergy with the RNR inhibitor clofarabine. Clofarabine is phosphorylated intracellularly to form cytotoxic active 5′-triphosphate metabolite, which inhibits the enzymatic activities of RNR and DNA polymerase, resulting in inhibition of DNA synthesis and repair31. While most DDR inhibitors displayed increased potency with DNAJA1 depletion, four of them were antagonistic to loss of DNAJA1. These include topoisomerase inhibitors and nucleic acid synthesis inhibitors such as trifluridine, irinotecan, epirubicin (4′-epi-isomer of the antibiotic doxorubicin) and idarubicin (4-demethoxy analogue of daunorubicin)32. While at first these results seem paradoxical, it is worth noting that inrinotecan is a type I topoisomerase inhibitor, whereas Etoposide (synergistic with loss of DNAJA1) is a type II topoisomerase inhibitor. It may be that Hsp70 and DNAJA1 play opposing regulatory roles in the stabilization and activation of these related proteins.
In addition to DDR, DNAJA1 is also involved in signal transduction, with previous reports indicating that the yeast homolog of DNAJA1 (Ydj1) is critical for supporting the integrity of kinase signaling networks33. DNAJA1 is mobilized to specific sites within the nucleus in response to inappropriate targeting or folding of specific mutant receptors. DNAJA1 overexpression ameliorates the defective transactivation and trans-repression activity of mutant Glucocorticoid receptors34. In line with the previous studies, we found that a handful of Receptor Tyrosine kinase inhibitors were synergistic with DNAJA1 depletion. These included Vascular endothelial growth factor receptor (VEGFR) inhibitors such as sunitinib, cabozantinib, lenvatinib and pazopanib. Interestingly, randomized phase III clinical trials are being conducted to validate the efficacy of Cabozantinib in heavily pretreated prostate cancer patients35. One implication from our study is that DNAJA1 inhibition might significantly enhance the effect of cabozantinib monotherapy.
Strikingly, some of the kinase inhibitors were antagonistic to DNAJA1 depletion. These include VEGFR inhibitors such as regorafenib and sorafenib. This disparity can be explained by the different target receptors and mechanisms of action of these drugs. Interestingly, recent studies indicated that these small molecule inhibitors exhibit off-target effects. Some of these drugs are misidentified and mischaracterized for their target specific inhibition, which has contributed to the high failure rate of these drugs in the treatment of cancer patients36.
In addition to its role in signal transduction, DNAJA1 is also important for maintaining the cellular cytoskeleton. Previous studies have suggested that YDJ1 (the yeast homolog of DNAJA1) is important for the proper assembly of microtubules37. Another report showed that DNAJA1 depletion causes relocation of N-cadherin and enhanced activity of metalloproteinases. This leads to changes in the actin cytoskeleton indicating that DNAJA1 is important for prevention of the amoeboid-like transition of tumor cells38. These studies indicated the involvement of DNAJA1 in maintaining cytoskeletal organization. We found 3 anticancer drugs targeting the cytoskeleton to be synergistic with DNAJA1 depletion, including vinblastine sulfate (cytoskeletal inhibitor that disrupts microtubule formation during mitosis and interferes with glutamic acid metabolism), estramustine (binds to microtubule-associated proteins (MAPs) and inhibits microtubule dynamics) and ixabepilone (promotes tubulin polymerization and microtubule stabilization, thereby arresting cells in the G2-M phase39. Strikingly, two of the tubulin inhibitors were found to be antagonistic to DNAJA1 depletion. These include paclitaxel and ixabepilone. Paclitaxel inhibits the disassembly of microtubules resulting in the inhibition of cell division whereas Ixabepilone promotes tubulin polymerization and microtubule stabilization, arresting cells in the G2-M phase of the cell cycle39. This apparent discrepancy may be explained by off-target effects of these molecules (see below).
Epigenetic modifying drugs display substantially modified potency depending on cellular DNAJA1 status. While previous studies have indicated the association between proteomic changes and histone PTMs in response to Hsp90 inhibitor treatment in bladder carcinoma cells, no such association has been shown for DNAJA1 and Histone PTMs40. Interestingly, vorinostat was the only drug that was synergistic to DNAJA1 inhibition. It is a histone deacetylase inhibitor that binds to the catalytic domain of the histone deacetylases (HDACs). However, we also identified two histone deacetylase inhibitor drugs to be antagonistic to DNAJA1 depletion, panobinostat and romidepsin. These inhibit histone deacetylase (HDAC) which may impact cell cycle protein expression, cell cycle arrest in the G2/M phase and apoptosis41. Excitingly, our data suggest a functional link between histones, their modifications and DNAJA1. While these findings require further investigation, it is possible that DNAJA1 may regulate the stability of histones themselves or histone chaperones. Interestingly, bortezomib (a proteasome inhibitor) lost potency when DNAJA1 was either inhibited with 116-9e or knocked out with CRISPR. Interestingly, a similar phenomenon has been observed in B16F10 melanoma cells. While treatment of these cells with 10 nM bortezomib was cytotoxic, this effect was not observed in cells treated with a combination of both quercetin (an Hsp70 inhibitor) and bortezomib42. This apparent antagonism may be explained by their mechanism of action on the heat shock transcription factor, HSF1. While bortezomib acts to trigger the heat shock response in some cancers, the Hsp70/co-chaperone system maintains HSF1 in an less active immature form43–46. It is interesting to note that while there are clear classes of drugs that are made more potent by loss of DNAJA1 function (DNA damage response, cytoskeletal function etc.), there are a small number of drugs in these classes that are not impacted at all or even made less potent. This apparent discrepancy implies that some of these inhibitors might have multiple cellular targets in addition to their proposed primary mechanism of action. This theory has been validated in fascinating studies comparing effects of small molecule therapies, gene knockout and knockdowns that theoretically target the same genes36.
As in the case of any chemogenomic screen, care must be taken to validate screening results with other methods. While CRISPR KO cell lines offer the advantage of complete, specific and permanent gene silencing as compared to transient inhibition seen with small molecules or siRNA, the authors acknowledge that permanent knockout lines may respond by overpexressing/suppressing other genes to compensate, leading to non-specific effects. In this study, we took the approach of following up our intitial screen with small molecule validation in 2D and 3D cell culture models. Going forward, we intend to validate several of these hits in vivo (mouse) model systems. 116-9e is a relatively new and unexplored molecule; while it clearly alters JDP binding, the exact impact on all Hsp70-JDPs has not been characterized8,18. Interestingly, the DNAJA1 knockout cell line grows effectively the same as WT and our examination of key chaperone protein levels do note reveal any major alterations. However, we do acknowledge that it is possible that the DNAJA1 knockout cell line may have adapted by overexpressing other JDPs such as DNAJA2, DNAJB1 or DNAJB6. In future studies, we hope to investigate this further by quantitating the proteome in HAP1 cells (WT and DNAJA1 knockout) in both untreated and 116-9e treated conditions. Given the essential nature of Hsp70/Hsc70 in cancer cells, if 116-9e truly inhibited all JDP interactions it would be highly toxic to cells which we do not observe, suggesting there must be some selectivity in JDP inhibition.
Recent studies from our group and other have described the clear impact of Hsp70/JDP inhibition on individual oncoprotein client stability and prostate cancer cell survival8,13. Overall, this study demonstrates the larger feasibility of inhibiting Hsp70 co-chaperones such as DNAJA1 as a novel anticancer therapy, acting to fine-tune Hsp70 function rather than completely abolishing it. Nearly a third of the anticancer compounds screened demonstrated increased potency in DNAJA1 knockout cells. Rather than attempting to develop co-chaperone inhibitors as a monotherapy, we believe their strength lies as sensitizing agents to existing therapies. Moreover, our data imply that overexpression of DNAJA1 in patient tumors may impact the effectiveness of a number of commonly used anticancer drugs. While further experiments characterizing (1) the specific DNAJA1-mediated effects in cancer proliferation and (2) the specificity of drugs such as 116-9e are required, our studies suggest perhaps a future precision medicine approach that uses tumor DNAJA1 status to guide treatment strategy.
Materials and methods
Cell culture
The HAP1 Chronic Myelogenous Leukemia cancer cell line and DNAJA1 knockout cell line was purchased from Horizon Discovery and were cultured in Iscove’s Modified Eagle Medium (Invitrogen) with 10% fetal bovine serum (Gibco), 100 units/ml penicillin, and 100 μg/ml streptomycin at 5% CO2 and 37° C. The LNCaP cancer cell line was purchased from ATCC and were cultured in RPMI-1640 medium (Invitrogen) with 10% fetal bovine serum (FBS, Clontech), 100 units/ml penicillin, and 100 μg/ml streptomycin at 5% CO2 and 37° C.
Drug screening
Approved Oncology Drug plates consisting of the most current FDA approved anticancer drugs were obtained from the National Cancer Institute (NCI). For experiments delineating the synergy between the loss of DNAJA1 and approved anticancer drug, HAP1 cells and HAP1 (DNAJA1 KO) cells were plated in growth media at 20% confluency 1 day prior to drug treatment. On Day 1 of treatment, cells were treated with DMSO (control), Approved oncology anticancer drugs at 50 µM for 72 h. Following drug treatments, Cell Titer-Glo reagent was added directly to the wells according to the manufacturer’s instructions. The luminescence was measured on Bio-Tek Plate reader. Luminescence reading was normalized to and expressed as a relative percentage of the plate averaged DMSO control. The data shown are the mean and SEM of three independent biological replicates.
Combination index (CI) calculations
For IC50 calculations, LNCaP cells were seeded in triplicates in 96-well white bottom Nunc plates in growth media at 20% confluency 1 day prior to initiation of drug treatment. On Day 1 of treatment, cells were treated with DMSO (control) and ten folds serial dilution of anti-cancer drugs cabozantinib, clofarabine, vinblastine, sorafenib, idarubicin and omacetaxine mepesuccinate and 116-9e. After 72 h, cell viability was measured using Promega Cell Titer-Glo cell viability assay on Bio-Tek plate reader. The combination index was calculated using the Chou-Talalay method using CompuSyn software47.
Spheroid generation
Single-cell suspensions (5,000/well) were plated in one well of 24-well plates in a 1:1 mixture of RPMI medium and Matrigel (BD Bioscience CB-40324). Cells in Matrigel were kept cold at all times and under continuous agitation. Warm PBS was added to all empty wells, if any. Plates were incubated at 37 °C with 5% CO2 for 15 min to solidify the gel before addition of 100 µl of pre-warmed RPMI to each well. Two days after seeding, the media was fully aspirated and replaced with fresh RPMI containing the indicated drugs. The same procedure was repeated daily on two consecutive days. Twenty-four hours after the last treatments, the media was aspirated and the wells were washed with 100 µl of pre-warmed PBS. To prepare for downstream assays, spheroids were released from the Matrigel by incubating at 37 °C for 40 min in 100 µl of 10 mg/mL Dispase (Sigma).
Apoptosis assay
Apoptosis of LNCaP spheroids was detected by the Annexin V–FITC/propidium iodide–binding assay. Cells were treated with either 0.1% DMSO (dimethyl sulfoxide),116-9e, cabozantinib, clofarabine, vinblastine, sorafenib, idarubicin, omacetaxine mepesuccinate and sorafenib alone or in combination with 116-9e for 48 h at the IC50 concentrations, and then stained with Annexin V–FITC and propidium iodide. The rate of apoptosis was determined using a BD LSR Fortessa flow cytometer, and the collected data were analyzed using FlowJo software. Apoptosis was reported as the mean ± SD. The results are representative of three independent experiments.
Bioinformatics
Cancer genome data and Cancer Cell Line Encyclopedia data were accessed from the cBioPortal (www.cbioportal.org) for Cancer Genomics 48. Total patient numbers and detailed information regarding published datasets and associated publications are indicated in Fig. 1A,B.
Statistical analysis
Data were analyzed using GraphPad Prism built-in statistical tests indicated in relevant figure legends. The following asterisk system for P-value was used: P < 0.05; P < 0.01; 0.001; and P < 0.0001.
Western blotting
Protein extracts were made as described8. 30 μg of protein was separated by 4–12% NuPAGE SDS-PAGE (Thermo). Proteins were detected using the following antibodies; anti-DNAJA1/HDJ2 (Thermo # MA5-12748), anti-Actin (CST # 9774), Anti-Hsc70 (Santa Cruz, # sc-7298), anti-Hsp70 (Enzo # C92F3A5), anti-Hsp90 ⍺/ℬ (Santa Cruz # sc-13119), anti-Bag3 (Santa Cruz # sc-136467), anti-Hsp110 (Stress Marq, # SPC-195),) at 1:4,000 dilution in TBST + 1% BSA. The secondary antibody (StarBright Blue 700 Fluorescent Secondary Mouse) was used at 1:3,000 dilution in TBST + 1% BSA. Blots were imaged on a Chemi Doc MP imaging system (Bio-Rad).
Supplementary information
Acknowledgements
The authors thank NIH for providing materials used in this study and J. Gestwicki, D. Dreau and T. Erick for helpful comments. We thank all reviewers for their helpful and constructive comments. This manuscript has been released as a pre-print at BioRxiv49. This project was supported by NCI R15CA208773 and Grant-In-Aid of Research from Sigma Xi (G2018031591887158), The Scientific Research Society.
Author contributions
N. performed all main experiments and wrote manuscript drafts; J.S.B. assisted with the chemogenomic screen; J.E.T. and L.E.K. helped with data analysis, figure drawing and manuscript editing; S.K.C. and A.W.T. conceived the experiments and edited the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-70764-x.
References
- 1.Rosenzweig R, Nillegoda NB, Mayer MP, Bukau B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019;20:665–680. doi: 10.1038/s41580-019-0133-3. [DOI] [PubMed] [Google Scholar]
- 2.Ciocca DR, Calderwood SK. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones. 2005;10:86–103. doi: 10.1379/csc-99r.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gestwicki JE, Shao H. Inhibitors and chemical probes for molecular chaperone networks. J. Biol. Chem. 2019;294:2151–2161. doi: 10.1074/jbc.TM118.002813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans CG, Chang L, Gestwicki JE. Heat shock protein 70 (hsp70) as an emerging drug target. J. Med. Chem. 2010;53:4585–4602. doi: 10.1021/jm100054f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig EA, Marszalek J. How do J-Proteins get Hsp70 to do so many different things? Trends Biochem. Sci. 2017;42:355–368. doi: 10.1016/j.tibs.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nitika & Truman, A. W. Cracking the Chaperone Code: cellular roles for Hsp70 phosphorylation. Trends. Biochem. Sci .42, 932–935. 10.1016/j.tibs.2017.10.002 (2017). [DOI] [PMC free article] [PubMed]
- 8.Sluder, I. T., Nitika, Knighton, L. E. & Truman, A. W. The Hsp70 co-chaperone Ydj1/HDJ2 regulates ribonucleotide reductase activity. PLoS Genet14, e1007462, 10.1371/journal.pgen.1007462 (2018). [DOI] [PMC free article] [PubMed]
- 9.Truman AW, et al. CDK-dependent Hsp70 phosphorylation controls G1 cyclin abundance and cell-cycle progression. Cell. 2012;151:1308–1318. doi: 10.1016/j.cell.2012.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodford MR, et al. Impact of posttranslational modifications on the anticancer activity of Hsp90 inhibitors. Adv. Cancer Res. 2016;129:31–50. doi: 10.1016/bs.acr.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Dushukyan N, et al. Phosphorylation and ubiquitination regulate protein phosphatase 5 activity and its prosurvival role in kidney cancer. Cell Rep. 2017;21:1883–1895. doi: 10.1016/j.celrep.2017.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stark JL, et al. Structure and function of human DnaJ homologue subfamily a member 1 (DNAJA1) and its relationship to pancreatic cancer. Biochemistry. 2014;53:1360–1372. doi: 10.1021/bi401329a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moses MA, et al. Targeting the Hsp40/Hsp70 chaperone axis as a novel strategy to treat castration-resistant prostate cancer. Cancer Res. 2018;78:4022–4035. doi: 10.1158/0008-5472.CAN-17-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmer, A. C. & Sorger, P. K. (2017) Combination cancer therapy can confer benefit via patient-to-patient variability without drug additivity or synergy. Cell 171, 1678–1691. doi: 10.1016/j.cell.2017.11.009 (2017). [DOI] [PMC free article] [PubMed]
- 15.Ding KF, et al. Nonlinear mixed effects dose response modeling in high throughput drug screens: application to melanoma cell line analysis. Oncotarget. 2018;9:5044–5057. doi: 10.18632/oncotarget.23495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koh SB, et al. Mechanistic distinctions between CHK1 and WEE1 inhibition guide the scheduling of triple therapy with gemcitabine. Cancer Res. 2018;78:3054–3066. doi: 10.1158/0008-5472.CAN-17-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang CC, et al. HDJ-2 as a target for radiosensitization of glioblastoma multiforme cells by the farnesyltransferase inhibitor R115777 and the role of the p53/p21 pathway. Cancer Res. 2006;66:6756–6762. doi: 10.1158/0008-5472.CAN-06-0185. [DOI] [PubMed] [Google Scholar]
- 18.Wisen S, et al. Binding of a small molecule at a protein-protein interface regulates the chaperone activity of hsp70-hsp40. ACS Chem. Biol. 2010;5:611–622. doi: 10.1021/cb1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wisitpitthaya S, et al. Cladribine and fludarabine nucleotides induce distinct hexamers defining a common mode of reversible RNR inhibition. ACS Chem. Biol. 2016;11:2021–2032. doi: 10.1021/acschembio.6b00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu CC, et al. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science. 2011;333:459–462. doi: 10.1126/science.1204117. [DOI] [PubMed] [Google Scholar]
- 21.Senra JM, et al. Inhibition of PARP-1 by olaparib (AZD2281) increases the radiosensitivity of a lung tumor xenograft. Mol. Cancer Ther. 2011;10:1949–1958. doi: 10.1158/1535-7163.MCT-11-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dudgeon C, et al. Inhibiting oncogenic signaling by sorafenib activates PUMA via GSK3beta and NF-kappaB to suppress tumor cell growth. Oncogene. 2012;31:4848–4858. doi: 10.1038/onc.2011.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gandhi V, Plunkett W, Cortes JE. Omacetaxine: a protein translation inhibitor for treatment of chronic myelogenous leukemia. Clin. Cancer Res. 2014;20:1735–1740. doi: 10.1158/1078-0432.CCR-13-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hevener K, Verstak TA, Lutat KE, Riggsbee DL, Mooney JW. Recent developments in topoisomerase-targeted cancer chemotherapy. Acta Pharm. Sin. B. 2018;8:844–861. doi: 10.1016/j.apsb.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 26.Vlachogiannis G, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pickl M, Ries CH. Comparison of 3D and 2D tumor models reveals enhanced HER2 activation in 3D associated with an increased response to trastuzumab. Oncogene. 2009;28:461–468. doi: 10.1038/onc.2008.394. [DOI] [PubMed] [Google Scholar]
- 28.Parrales A, et al. DNAJA1 controls the fate of misfolded mutant p53 through the mevalonate pathway. Nat. Cell Biol. 2016;18:1233–1243. doi: 10.1038/ncb3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knighton LE, Delgado LE, Truman AW. Novel insights into molecular chaperone regulation of ribonucleotide reductase. Curr. Genet. 2019;65:477–482. doi: 10.1007/s00294-018-0916-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Truman AW, et al. Quantitative proteomics of the yeast Hsp70/Hsp90 interactomes during DNA damage reveal chaperone-dependent regulation of ribonucleotide reductase. J. Proteom. 2015;112:285–300. doi: 10.1016/j.jprot.2014.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huguet F, et al. Clofarabine for the treatment of adult acute lymphoid leukemia: the Group for Research on Adult Acute Lymphoblastic Leukemia intergroup. Leuk Lymphoma. 2015;56:847–857. doi: 10.3109/10428194.2014.887708. [DOI] [PubMed] [Google Scholar]
- 32.Deng S, et al. Dexrazoxane may prevent doxorubicin-induced DNA damage via depleting both topoisomerase II isoforms. BMC Cancer. 2014;14:842. doi: 10.1186/1471-2407-14-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gillies AT, Taylor R, Gestwicki JE. Synthetic lethal interactions in yeast reveal functional roles of J protein co-chaperones. Mol. Biosyst. 2012;8:2901–2908. doi: 10.1039/c2mb25248a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang Y, Ramakrishnan C, Thomas J, DeFranco DB. A role for HDJ-2/HSDJ in correcting subnuclear trafficking, transactivation, and transrepression defects of a glucocorticoid receptor zinc finger mutant. Mol. Biol. Cell. 1997;8:795–809. doi: 10.1091/mbc.8.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grullich C. Cabozantinib: a MET, RET, and VEGFR2 tyrosine kinase inhibitor. Recent Results Cancer Res. 2014;201:207–214. doi: 10.1007/978-3-642-54490-3_12. [DOI] [PubMed] [Google Scholar]
- 36.Lin, A. et al. Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci. Transl. Med.10.1126/scitranslmed.aaw8412 (2019). [DOI] [PMC free article] [PubMed]
- 37.Oka M, et al. Loss of Hsp70-Hsp40 chaperone activity causes abnormal nuclear distribution and aberrant microtubule formation in M-phase of Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:29727–29737. doi: 10.1074/jbc.273.45.29727. [DOI] [PubMed] [Google Scholar]
- 38.Meshalkina DA, et al. Knock-down of Hdj2/DNAJA1 co-chaperone results in an unexpected burst of tumorigenicity of C6 glioblastoma cells. Oncotarget. 2016;7:22050–22063. doi: 10.18632/oncotarget.7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cobham MV, Donovan D. Ixabepilone: a new treatment option for the management of taxane-resistant metastatic breast cancer. Cancer Manag. Res. 2009;1:69–77. doi: 10.2147/CMAR.S5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li QQ, et al. Proteomic analysis of proteome and histone post-translational modifications in heat shock protein 90 inhibition-mediated bladder cancer therapeutics. Sci. Rep. 2017;7:201. doi: 10.1038/s41598-017-00143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eckschlager, T., Plch, J., Stiborova, M. & Hrabeta, J. Histone Deacetylase Inhibitors as Anticancer Drugs. Int J Mol Sci . doi:10.3390/ijms18071414 (2017). [DOI] [PMC free article] [PubMed]
- 42.Yerlikaya A, Okur E, Eker S, Erin N. Combined effects of the proteasome inhibitor bortezomib and Hsp70 inhibitors on the B16F10 melanoma cell line. Mol. Med. Rep. 2010;3:333–339. doi: 10.3892/mmr_00000262. [DOI] [PubMed] [Google Scholar]
- 43.Mitsiades N, et al. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc. Natl. Acad. Sci. USA. 2002;99:14374–14379. doi: 10.1073/pnas.202445099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shah SP, et al. Bortezomib-induced heat shock response protects multiple myeloma cells and is activated by heat shock factor 1 serine 326 phosphorylation. Oncotarget. 2016;7:59727–59741. doi: 10.18632/oncotarget.10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abravaya K, Myers MP, Murphy SP, Morimoto RI. The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev. 1992;6:1153–1164. doi: 10.1101/gad.6.7.1153. [DOI] [PubMed] [Google Scholar]
- 46.Baler R, Welch WJ, Voellmy R. Heat shock gene regulation by nascent polypeptides and denatured proteins: hsp70 as a potential autoregulatory factor. J. Cell Biol. 1992;117:1151–1159. doi: 10.1083/jcb.117.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 48.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nitika et al. Chemogenomic screening Identifies the Hsp70 Co-chaperone HDJ2 as a Hub for Anticancer Drug Resistance. bioRxiv, 818427, doi:10.1101/818427 (2019). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.